Abstract
Triple negative breast cancers (TNBCs) have a relatively poor prognosis and cannot be effectively treated with current targeted therapies. We searched for genes that have the potential to be therapeutic targets by identifying genes consistently over-expressed when amplified. Fifty-six TNBCs were subjected to high-resolution microarray-based comparative genomic hybridisation (aCGH), of which 24 were subjected to genome-wide gene expression analysis. TNBCs were genetically heterogeneous; no individual focal amplification was present at high frequency, although 78.6% of TNBCs harboured at least one focal amplification. Integration of aCGH and expression data revealed 40 genes significantly overexpressed when amplified, including the known oncogenes and potential therapeutic targets, FGFR2 (10q26.3), BUB3 (10q26.3), RAB20 (13q34), PKN1 (19p13.12), and NOTCH3 (19p13.12). We identified two TNBC cell lines with FGFR2 amplification, which both had constitutive activation of FGFR2. Amplified cell lines were highly sensitive to FGFR inhibitor PD173074, and to RNAi silencing of FGFR2. Treatment with PD173074 induced apoptosis resulting partly from inhibition of PI3K-AKT signalling. Independent validation using publicly available aCGH datasets revealed FGFR2 gene was amplified in 4% (6/165) of TNBC, but not in other subtypes (0/214, p=0.0065). Our analysis demonstrates that TNBCs are heterogeneous tumours with amplifications of FGFR2 in a subgroup of tumours.
Keywords: Triple negative breast cancer, microarrays, gene expression, comparative genomic hybridisation, FGFR2
Introduction
The identification of distinct subgroups of breast cancer has led to the development of therapeutic strategies that exploit the underlying biology of the subtype, with hormonal therapies and HER2 targeting agents for hormone receptor-positive and HER2-positive breast cancers, respectively (Reis-Filho and Tutt, 2008; Schneider et al., 2008). Tumours lacking expression of hormone receptors (oestrogen receptor (ER) and progesterone receptor (PR)) and HER2 (triple negative breast cancers, TNBCs), on the other hand, pose a significant clinical challenge due to a poor understanding of the genetic alternations that underlie the development of TNBC (Reis-Filho and Tutt, 2008; Schneider et al., 2008) and this is reflected in a lack of subtype-specific targeted therapies.
TNBCs comprise a heterogeneous group of breast cancers and account for 10-15% (Agrawal et al., 2007; Carey et al., 2007; Dent et al., 2007; Reis-Filho and Tutt, 2008; Schneider et al., 2008) of all invasive breast cancers. Histologically, the majority of TNBCs are grade III invasive ductal carcinomas of no special type (IDC-NST), although the majority of medullary, metaplastic and adenoid cystic carcinomas also display a triple negative phenotype (Reis-Filho and Tutt, 2008). These tumours are more prevalent in young women (<50 years) and in women of African and Hispanic descent. TNBCs have a poor prognosis characterised by early relapse (Dent et al., 2008; Tischkowitz and Foulkes, 2006), potentially reflecting the high proliferative rate of TNBCs. Similarly, women with TNBCs have a significantly shorter survival following recurrence when compared to those with non-triple negative cancers (Dent et al., 2007; Harris et al., 2006). Therefore, the identification of novel therapeutic targets for TNBCs is important if the outcome of patients with these tumours is to be improved.
Based on the concept of oncogene addiction, we and others have demonstrated that genes that are consistently overexpressed when amplified may be selectively required for the survival of cancer cells harbouring their amplification, and can be exploited as potential therapeutic targets (Bernard-Pierrot et al., 2008; Natrajan et al., 2009a; Reis-Filho et al., 2006). Previous studies have examined TNBCs with expression profiling (Bertucci et al., 2008; Kreike et al., 2007) and microarray-based comparative genomic hybridisation (aCGH) (Andre et al., 2009; Han et al., 2008). These studies have found TNBCs to be heterogeneous, with complex genomic profiles and infrequent amplifications.
To identify amplicon drivers and genes that have the potential to be therapeutic targets in TNBCs, we integrated aCGH and gene expression data from a large series of TNBCs. Our aims were to characterise the genomic and transcriptomic profiles of TNBCs and identify and validate genes that are recurrently amplified and consistently overexpressed when amplified in TNBCs. We found more frequent high level, focal amplifications than previously described (Chin et al., 2006), and identified potential therapeutic targets that are consistently overexpressed when amplified. We validated the fibroblast growth factor receptor 2 (FGFR2) gene as one of these targets and provided functional data to suggest that this tyrosine kinase receptor may be a novel therapeutic target in a subset of TNBCs harbouring FGFR2 gene amplification.
Material and Methods
Tumour samples
Fifty-six fresh-frozen samples of TNBCs were obtained after approval by local Ethic Committees from the authors’ institutions. Triple negative tumours were selected according to their lack of expression to ER, PR and HER2 as defined by immunohistochemistry (Kreike et al., 2007). All tumours were morphologically invasive ductal cancers of no special type. ER, PR and HER2 antibodies, antigen retrieval systems and scoring methods are summarised in Supplementary Table 1. One representative section of each tumour was stained with haematoxiylin-eosin. Samples were either microdissected with a sterile needle under a stereomicroscope (Marchio et al., 2008), or samples from Kreike et al. (Kreike et al., 2007) were only included if there were >70% of neoplastic cells in the section. A complete description of the cohort analysed here is described in Supplementary Table 2. Out of the samples included in this study, the aCGH profiles of 23 cases were reported in Natrajan et al. (Natrajan et al., 2009a) and the expression profiles of 24 cases were reported in Kreike et al. (Kreike et al., 2007). No statistically significant differences were observed in terms of patient age, tumour size, histological grade, prevalence of basal-like phenotype and outcome between the 24 cases included in this study, and the remaining TNBCs from Kreike et al. (2007; Supplementary Table 3).
RNA and DNA extraction
DNA and RNA were extracted as previously described. DNA concentration was measured with Picogreen® (Invitrogen, Paisley UK) according to the manufacturer’s instructions (Marchio et al., 2008).
Microarray-Based Comparative Genomic Hybridisation
The 32K BAC re-array collection (CHORI) tiling path aCGH platform was constructed at the Breakthrough Breast Cancer Research Centre, as described previously (Marchio et al., 2008).This type of BAC array platform has been shown to be as robust as and to have comparable resolution with high density oligonucleotide arrays (Coe et al., 2007; Gunnarsson et al., 2008; Tan et al., 2007). DNA labelling, array hybridisations and image acquisition were performed as previously described (Natrajan et al., 2009a). aCGH data were pre-processed and analysed using an in-house R script (BACE.R) in R version 2.9.0, as previously described (Mackay et al., 2009; Natrajan et al., 2009b). After filtering polymorphic BACs, a final dataset of 31544 clones with unambiguous mapping information according to the August 2009 build (hg19) of the human genome (http://www.ensembl.org) was smoothed using the circular binary segmentation (cbs) algorithm (Mackay et al., 2009; Natrajan et al., 2009b). A categorical analysis was applied to the BACs after classifying them as representing amplification (>0.45), gain (>0.08 and ≤0.045), loss (<−0.08), or no-change according to their cbs-smoothed Log2 ratio values (Natrajan et al., 2009b; Reis-Filho et al., 2008). Threshold values were determined and validated as previously described (Natrajan et al., 2009b).
Tumours were classified according to their pattern of genomic alterations into ‘simplex’, ‘firestorm’ or ‘sawtooth’ as previously described(Hicks et al., 2006; Natrajan et al., 2009a).
Gene expression analysis
RNA labelling, hybridisation, slide scanning and data normalisation were performed as previously described (Kreike et al., 2007; Weigelt et al., 2008). Fluorescent intensities were normalised and corrected for biases (Hannemann et al., 2006), and weighted averages and confidence levels were computed according to the Rosetta Error Model (Hughes et al., 2000). All the expression data can be retrieved from http://www.ebi.ac.uk/microarrayas/ae/browse.html?keywords=E-NCMF-2&species=&array=&exptype=&pagesize=25&sortby=releasedate&sortorder=descending. Of the TNBCs analysed, 94.2% were of basal-like phenotype using the criteria described by Hu et al. (Hu et al., 2006) (Supplementary Table 2).
Identification of genes whose expression correlates with copy number changes
To identify genes whose expression levels correlate with copy number changes, cbs-smoothed Log2 ratios from aCGH data were used to assign the aCGH states for each of the 24,650 genes in the gene expression dataset using the median values for all BACs which overlap with the genomic position of each gene. This resulted in a 1:1 matrix of expression ratios and aCGH cbs values, which were used for downstream statistical analysis. Pearson’s correlations were performed between gene expression array Log2 ratios and cbs-smoothed ratios derived from aCGH analysis for each gene. The p values for each test were adjusted with Benjamini and Hochberg multiple p-value adjustment (Benjamini and Hochberg, 1995). Adjusted p values < 0.05 were considered significant.
To define genes that were upregulated when gained, downregulated when lost or overexpressed when amplified, we performed a multi-Mann–Whitney U test using categorical aCGH states (i.e. gain vs no gain, loss vs no loss or amplification vs no amplification) as the grouping variable and the expression of genes as the dependent variable as previously described (Mackay et al., 2009; Natrajan et al., 2009b). P-values were adjusted using Benjamini and Hochberg multiple comparison p-value adjustment (Benjamini and Hochberg, 1995). Adjusted p values < 0.05 were considered significant.
For regions of recurrent amplification, matched heatmaps were created by retrieving gene expression values and corresponding median-overlay aCGH states for each gene as previously described (Mackay et al., 2009; Natrajan et al., 2009b). Genes were ordered according to chromosomal location and cases were separated into those that harbour amplifications in the region and those which do not. Within these groups the samples were ordered based upon the sum of expression values within the region.
Ingenuity pathway analysis
Ingenuity pathway analysis (IPA) program (http://www.ingenuity.com) was used as previously described (Natrajan et al., 2009b). HUGO gene identifiers were mapped to networks available in the Ingenuity database and ranked by score. The score indicates the likelihood of the genes in a network being found together due to random chance. Using a 99% confidence level, scores of ≥3 are considered significant.
Cell lines, materials and antibodies
Cell lines were obtained from ATCC or DMSZ, and maintained in DMEM or RPMI with 10% FBS (PAA gold) and 2mM L-glutamine (Sigma) (details available on request). S68 was a kind gift of Veronique Catros, Rennes, France. PD173074 was obtained from Sigma, BEZ235 from Axon Medchemicals, and U0126 from Calbiochem. siRNA were purchased from Dharmacon: FGFR2 siGenome SMARTpool (siFGFR2, M-003132-04) and siGENOME Non-Targeting siRNA Pool #1 (siCON, D-001206-13). Antibodies used were FGFR2 (sc-122, Santa-Cruz), phospho-FRS2-Tyr196 (3864, Cell Signaling), phospho-AKT-Ser473 (4058, Cell Signaling), phospho-ERK1/2-Thr202/Tyr204 (4370, Cell Signaling), and β-Actin (sc-1616, Santa-Cruz).
Cell line drug sensitivity, siRNA transfection and FACS analysis
Cell lines were transfected with siCON or siFGFR2 in 96 well plates with Lipofectamine RNAiMax (Invitrogen), Lipofectamine 2000 (Invitrogen), Dharmafect 3 (Thermo Scientific), or Oligofectamine (Invitrogen) according to manufacturers instructions. Transfection of all cell lines was confirmed with positive control PLK1 siRNA (loss of survival < 25% that of siCON transfection in all cell lines, Supplementary Figure 3). Survival was assessed with Cell Titre-Glo® cell viability assay (Promega) after five population doublings or 7 days which ever was shorter. For sensitivity to PD173074, cell lines were plated in 96 well plates, the following day media supplemented with PD173074 at increasing concentrations, and survival assessed with Cell Titre-Glo® after 96 hours exposure. Survival curves and estimated SF50 were calculated with GraphPad prism V5.0. For Combination Index (CI) cells were treated in 96 well plate for 96 hrs with two fold dilutions of PD173074, BEZ235, or combination, and CI calculated using non-mutually exclusive median effect model as described previously (Chou and Talalay, 1984). FACS analysis was performed as described previously (Turner et al., 2008).
Western blotting
Indicated cell lines where grown on 10cm plates, and grown for 24 hrs in serum free medium or normal medium. Where indicated plates were treated with drug for 1 hour prior to lysis. Western blots were carried out with precast TA or Bis-Tris gels (Invitrogen) as described previously (Turner et al., 2008).
Fluorescence in situ hybridisation for FGFR2
FGFR2 gene copy number status was assessed in breast cancer cell lines with in-house generated BAC probes comprising two BACs (RP11-300A10 and RP11-753P11), which map to specifically to the FGFR2 gene locus, as previously described (Lambros et al., 2006).
Results
TNBCs display complex genomic profiles
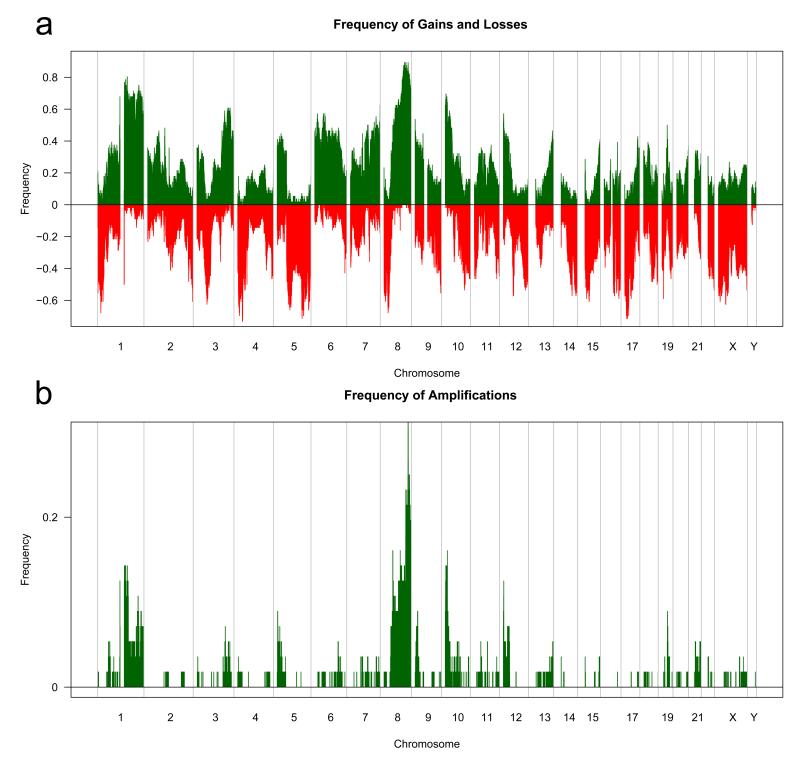
We established the genomic array CGH profiles of 56 TNBCs with an array CGH platform with a previously validated effective resolution of 50Kb. This revealed a high level of genetic instability, with gain or loss affecting a median of 44.4% of the genome (range 9.3%-76.7%). Using the genomic pattern classification proposed by Hicks et al. (2006) 15 of the TNBCs (27%) were classified as ‘simplex’ and 41 (73%) were considered to have ‘complex’ genomic patterns, of which 28 (50%) were ‘sawtooth’ and 13 (23%) were ‘firestorm’ (Hicks et al., 2006). Given that the majority of TNBCs displayed ‘sawtooth’ patterns, it is not surprising that multiple regions of recurrent gains and losses were identified (Figure 1a and Supplementary Table 4). In agreement with previous studies (Andre et al., 2009; Han et al., 2008), loss of 1p36-p34, 5q11-q35, 8p23.3-p12 and 17p13-q21 and gain of 3q22-q26, 6p25-p21, 7q32-q36, 8p11-q24, 10p15.3-p11.21 and 12p13-p11 were found in >30% of TNBCs (Figure 1a). Recurrent high level gains and amplifications were found in TNBCs (Figure 1b and Supplementary Table 5). At least one focal amplification (<10Mb) was found in 78.6% (44/56) of cases; however, the prevalence of each amplification was relatively low, ranging from 4% to 27% (Supplementary Table 5).
Figure 1.
Genomic alterations in triple negative breast cancers. a) Frequency of gains and losses in 56 TNBCs profiled with aCGH. The proportion of tumours in which each clone is gained (green bars) or lost (red bars) is plotted (Y axis) for each BAC clone according to genomic location (X axis). b) The proportion of tumours in which each clone is amplified (green bars) is plotted (Y axis) for each BAC clone according to genomic location (X axis). BAC clones were categorised as amplified if the (log2 ratio) cbs ratios were >0.45.
Integrative aCGH and expression analysis reveal pathways and networks that are enriched for genes whose expression correlates with copy number
To determine the genes whose expression levels correlate with copy number, we overlaid aCGH and expression data in an unbiased, genome-wide fashion (Natrajan et al., 2009b). This analysis revealed that the expression of 4972 out of 24565 genes (20.2%) correlated with copy number changes (Pearson’s correlation adjusted p<0.05, Supplementary Table 5). This analysis suggests that a large proportion of the genes expressed in TNBCs are at least in part regulated by copy number changes.
Within this list we determined the genes whose expression was significantly upregulated or downregulated in the presence of copy number gain or loss respectively (Mann-Whitney U adjusted p<0.05, Supplementary Table 6). We identified 324 genes whose mRNA expression levels were significantly higher in tumours harbouring DNA copy number gains and 39 genes whose mRNA expression levels were significantly lower in tumours displaying DNA copy number loss. The lower number of genes down-regulated when lost may stem from the slightly lower sensitivity of array CGH to detect loss of a single copy of a genomic region in highly aneuploid tumours (Ng et al., 2006), coupled with the limitations of expression arrays to accurately determine the expression of genes expressed at low levels.
Ingenuity pathway analysis (IPA) of genes expressed at higher levels when gained revealed that 5 networks and 27 canonical pathways were significantly enriched (Supplementary Table 8). Importantly, the canonical pathways of growth factors that have been shown to play a role in breast cancer development and progression, or to be potential therapeutic targets, were significantly enriched with genes upregulated when gained (Table 1). Copy number gains of the tyrosine kinase receptors were rare, and the growth factor pathway enrichment reflected frequent copy number gains and over-expression of key adapter molecules and down-stream signal transduction kinases (Table 1). The p53 signalling pathway was enriched for genes that are upregulated when gained, including survivin (BIRC5).
Table 1.
Tyrosine kinase receptor canonical pathways significantly enriched for genes significantly upregulated when gained.
| Ingenuity Canonical Pathways |
p | Ratio | Genes |
|---|---|---|---|
| Angiopoietin Signaling | 0.0030 | 0.0735 | PTK2,PAK1,FOXO1,PIK3C3,BIRC5 |
| HGF Signaling | 0.0047 | 0.0594 | PTK2,RAF1,PAK1,GAB1,PIK3C3,MAPK9 |
| FAK Signaling | 0.0102 | 0.0532 | PTK2,RAF1,PAK1,PIK3C3,CAPN10 |
| FGF Signaling | 0.0380 | 0.0476 | RAF1,GAB1,PIK3C3,MAPKAPK2 |
| VEGF Signaling | 0.0427 | 0.0449 | PTK2,RAF1,FOXO1,PIK3C3 |
| IGF-1 Signaling | 0.0490 | 0.0426 | PTK2,RAF1,FOXO1,PIK3C3 |
Recurrent amplified regions and potential amplicon drivers
We next analysed the data to identify the genes that were significantly overexpressed when amplified in TNBCs, compared to non-amplified cancers. This analysis revealed only 40 genes (Mann-Whitney U test adjusted p<0.05, Table 2) but included multiple genes that have been shown to either have oncogenic properties or to be potential therapeutic targets, such as FGFR2 (10q26), BUB3 (10q26), RAB20 (13q34), NOTCH3 (19p13) and PKN1 (19p13). It should be noted that this analysis, with a non-parametric rank sum test corrected for false discovery, is intentionally conservative and will identify genes that are robustly over-expressed when amplified. Such an analysis is not intended to be exhaustive. Of the 40 genes, 38 also displayed expression levels that correlated with copy number as assessed by Pearson correlation (Supplementary Table 10). Genes that are robustly over-expressed when amplified potential amplicon driver, and therapeutic targets, as has been suggested for NOTCH3 (Yamaguchi et al., 2008), and BUB3 (Yuan et al., 2006) (Table 2).
Table 2.
Genes that are significantly over-expressed when amplified.
| Symbol | Description | Cytoband | Start (Mb) | End (Mb) | MWU Adjusted p value amplification |
|---|---|---|---|---|---|
| SIPA1L2 | Signal-induced proliferation-associated 1-like protein 2 | 1q42.2 | 232533711 | 232697304 | 0.029 |
| IMPA1 | Inositol monophosphatase A1 | 8q21.13 | 82569151 | 82598589 | 0.024 |
| ZFAND1 | AN1-type zinc finger protein 1 | 8q21.13 | 82613958 | 82633530 | 0.024 |
| SNX16 | Sorting nexin-16 | 8q21.13 | 82711822 | 82754521 | 0.024 |
| ATP5C1 | ATP synthase subunit gamma, mitochondrial Precursor | 10p14 | 7830093 | 7849755 | 0.031 |
| TAF3 | Transcription initiation factor TFIID subunit 3 | 10p14 | 7860501 | 8056714 | 0.031 |
| USP6NL | USP6 N-terminal-like protein | 10p14 | 11502509 | 11653753 | 0.047 |
| FGFR2 | Fibroblast growth factor receptor 2 Precursor | 10q26.13 | 123237848 | 123357972 | 0.013 |
| ATE1 | Arginyl-tRNA--protein transferase 1 | 10q26.13 | 123502626 | 123687977 | 0.013 |
| NSMCE4A | Non-SMC element 4 homolog A | 10q26.13 | 123716611 | 123734710 | 0.013 |
| TACC2 | Transforming acidic coiled-coil-containing protein 2 | 10q26.13 | 123748709 | 124014059 | 0.013 |
| PLEKHA1 | Pleckstrin homology domain-containing family A member 1 |
10q26.13 | 124134220 | 124191867 | 0.013 |
| CUZD1 | CUB and zona pellucida-like domain-containing protein 1 Precursor |
10q26.13 | 124591665 | 124610309 | 0.047 |
| PSTK | L-seryl-tRNA(Sec) kinase | 10q26.13 | 124739556 | 124749908 | 0.013 |
| IKZF5 | Zinc finger protein Pegasus | 10q26.13 | 124751965 | 124768321 | 0.013 |
| ACADSB | Short/branched chain specific acyl-CoA dehydrogenase, mitochondrial Precursor |
10q26.13 | 124768429 | 124817806 | 0.013 |
| BUB3 | Budding uninhibited by benzimidazoles 3 homolog | 10q26.13 | 124913760 | 124924886 | 0.013 |
| RAB20 | Ras-related protein Rab-20 | 13q34 | 111175419 | 111214080 | 0.027 |
| CARS2 | Cysteinyl-tRNA synthetase 2, mitochondrial | 13q34 | 111293759 | 111358463 | 0.027 |
| ANKRD10 | Ankyrin repeat domain-containing protein 10 | 13q34 | 111530887 | 111567416 | 0.027 |
| ARHGEF7 | Rho guanine nucleotide exchange factor 7 | 13q34 | 111767624 | 111958078 | 0.021 |
| TUBGCP3 | Gamma-tubulin complex component 3 | 13q34 | 113139326 | 113242481 | 0.021 |
| ATP11A | Probable phospholipid-transporting ATPase IH | 13q34 | 113344643 | 113541482 | 0.027 |
| PCID2 | PCI domain-containing protein 2 | 13q34 | 113831891 | 113863029 | 0.027 |
| CUL4A | Cullin-4A | 13q34 | 113863086 | 113919399 | 0.021 |
| GRTP1 | Growth hormone-regulated TBC protein 1 | 13q34 | 113978506 | 114018463 | 0.021 |
| DCUN1D2 | DCN1-like protein 2 | 13q34 | 114110134 | 114145023 | 0.027 |
| TMCO3 | Transmembrane and coiled-coil domain-containing protein 3 Precursor |
13q34 | 114145308 | 114204542 | 0.021 |
| CDC16 | Cell division cycle protein 16 homolog | 13q34 | 115000362 | 115038198 | 0.021 |
| UPF3A | Regulator of nonsense transcripts 3A | 13q34 | 115047078 | 115071261 | 0.021 |
| C13orf8 | Zinc finger protein 828 | 13q34 | 115079988 | 115092796 | 0.021 |
| DDX39 | ATP-dependent RNA helicase | 19p13.12 | 14519633 | 14530171 | 0.035 |
| PKN1 | Serine/threonine-protein kinase N1 | 19p13.12 | 14544166 | 14582678 | 0.035 |
| GIPC1 | PDZ domain-containing protein | 19p13.12 | 14588572 | 14606944 | 0.042 |
| DNAJB1 | DnaJ homolog subfamily B member 1 | 19p13.12 | 14625582 | 14629201 | 0.035 |
| NDUFB7 | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 7 |
19p13.12 | 14676892 | 14682886 | 0.035 |
| SYDE1 | Rho GTPase-activating protein | 19p13.12 | 15218214 | 15225789 | 0.035 |
| ILVBL | Acetolactate synthase-like protein | 19p13.12 | 15225789 | 15236577 | 0.042 |
| NOTCH3 | Neurogenic locus notch homolog protein 3 Precursor | 19p13.12 | 15270445 | 15311792 | 0.042 |
| BRD4 | Bromodomain-containing protein 4 | 19p13.12 | 15348301 | 15391262 | 0.035 |
MWU: Mann Whitney U test.
FGFR2 is amplified in triple negative breast cancers and breast cancer cell lines
We selected, for further investigation, FGFR2 amplifications from the list amplified and over-expressed, as there are a number of drugs in early phase clinical trials that target the FGFRs. A re-analysis of data from publicly available aCGH datasets (Adelaide et al., 2007; Andre et al., 2009; Chin et al., 2006), including our current data set, revealed FGFR2 amplification in 4% (6/165 95% CI 1.4-7.8%) of TNBCs, with no cases of FGFR2 amplification in other subtypes (0/214, p=0.0065. Fisher’s exact test). Excluding our data set, the frequency was similar (TNBC 4% (4/109) vs other subtypes 0% (0/214), p=0.0125).
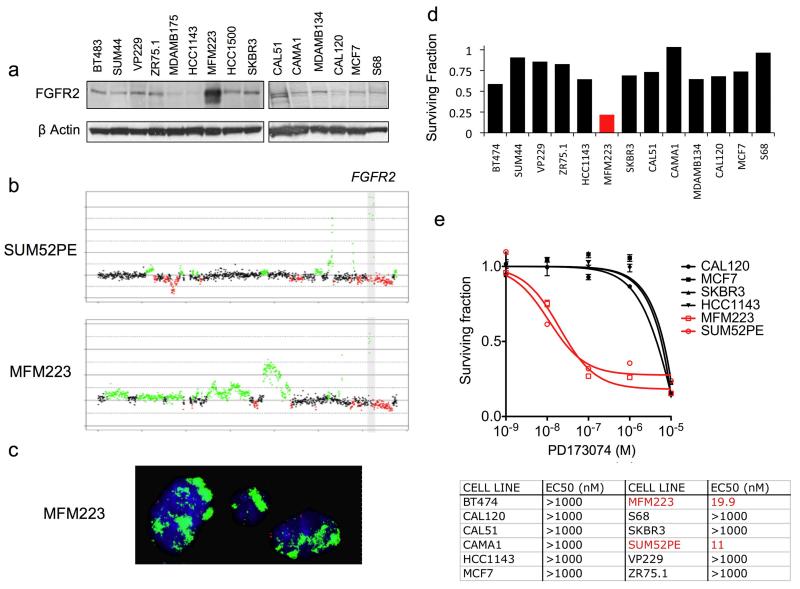
We screened a panel of 51 breast cancer cell lines using a combination of aCGH and western blotting of cell lysates to identify cell lines with FGFR2 gene amplification and protein overexpression. We identified two cell lines with FGFR2 amplification one of which, SUM52PE, had been described previously (Tannheimer et al., 2000). MFM223 was found to express high levels of FGFR2 protein (Figure 3a) and to harbour FGFR2 gene amplification by aCGH (Figure 3b). FGFR2 amplification was confirmed by FGFR2 FISH in MFM223 (Figure 3c), and by copy number PCR (Supplementary Figure 2). Both amplified cell lines expressed substantially higher FGFR2 mRNA than all other cell lines (SUM52PE 67 fold and MFM223 26 fold higher than median expression of non-amplified lines, Supplementary Figure 1). Both MFM223 and SUM52 were found to be triple negative, with neither cell line expressing ER or PR by western blotting nor harbouring HER2 gene amplification (Supplementary Figure 1).
Figure 3.
FGFR2 amplified cell lines are sensitive to FGFR2 silencing and FGFR inhibition. a) Western blot of cell lines of 15 breast cancer cell lines, demonstrating over-expression of FGFR2 protein in MFM223. FGFR2 was frequently observed as a doublet, that reflects different glycosylation states of the extracellular domain. b) Chromosome 10 aCGH profiles of MFM223 and SUM52PE with a gray box indicating the smallest region of amplification of the FGFR2 amplicon. c) FISH for chromosome 10 centromere (red) and FGFR2 (green) on MFM223 cells. MFM223 demonstrate unquantifiable high numbers of FGFR2 signals. The FGFR2 probe was specific for the FGFR2 locus (Supplementary Figure 2). d) Sensitivity of breast cancer cell lines to FGFR2 siRNA, demonstrating sensitivity of MFM223 (red) to FGFR2 silencing. Cell lines were transfected with FGFR2 siRNA, or siCON non-targeting control, and survival assessed at 5-7 days post transfection with Cell Titre-Glo® cell viability assay (Promega). Survival of FGFR2 siRNA transfected cells was expressed relative to that of siCON transfected. e) Graph: Selected cell lines were grown for 96 hrs in media supplemented with a range of concentrations of PD173074 pan FGFR tyrosine kinase inhibitor, and survival expressed relative to that of untreated cells. FGFR2 amplified cell lines in Red. Error bars represent SEM. Table: IC50 of breast cancer cell lines to 96hrs treatment with PD173074.
FGFR2 expression and tyrosine kinase activity are required for the survival of cell lines with FGFR2 gene amplification
We transfected a panel of cell lines with FGFR2 siRNA (Figure 3d). FGFR2 silencing selectively reduced the survival of MFM223 cells, FGFR2 amplified cell line, compared to all transfectable non-amplified cell lines (Figure 3, and Supplementary Figure 3). Silencing of FGFR2 by siRNA has confirmed by western blot (Supplementary Figure 3). We then examined the sensitivity of the cell line panel to PD173074, a highly potent selective pan-FGFR tyrosine kinase inhibitor (Mohammadi et al., 1998). The FGFR2 amplified cell lines were substantially more sensitive to PD173074 than all comparator cell lines (Figure 3e), indicating that FGFR kinase activity was selectively required for the growth/survival of cell lines harbouring FGFR2 gene amplification.
Ligand independent signalling in FGFR2 amplified cell lines
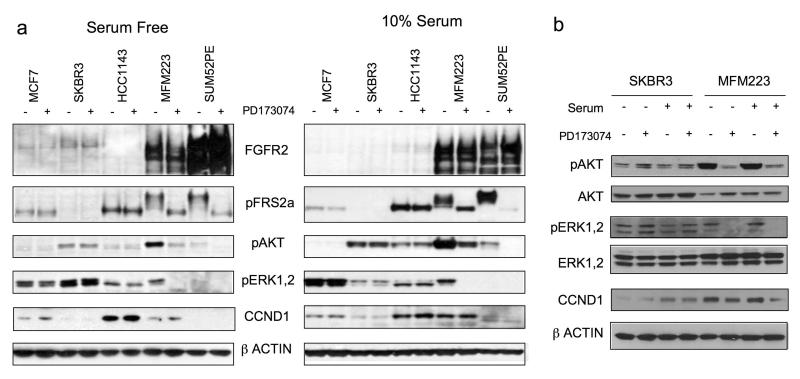
Signalling through the FGFR family of growth receptors is reliant on the adapter protein Fibroblast Receptor Substrate 2 alpha (FRS2), which binds directly to phosphorylated FGFR forming a FRS2-SOS-GRB2 complex to activate MAPK signalling and a FRS2-SOS-GAB1 complex to activate PI3K-AKT signalling. We examined the signal transduction pathways activated downstream of FGFR2 in serum starved conditions with and without PD173074 to inhibit FGFR2 kinase activity. FRS2 exhibits a mobility shift, reflecting phosphorylation, in serum starved amplified cell lines, which is abolished by treatment with PD173074, indicating ligand independent constitutive activation of the receptor (Figure 4a). In both cell lines harbouring FGFR2 gene amplification, AKT Ser473 was phosphorylated in an FGFR kinase dependent manner, with ERK1/2 Thr202/Tyr204 phosphorylated in an FGFR kinase dependent fashion in MFM223. SUM52PE expressed low levels of phosphorylated ERK, which presumably was reflected in the low proliferative rate of this cell line (data not shown). Phosphorylation of AKT and ERK1/2 was independent of FGFR kinase activity in control cell lines MCF7, SKBR3 and HCC1143 (Figure 4).
Figure 4.
a) Signalling downstream of FGFR2 in amplified cell lines. Indicated cell lines were grown either in 10% serum, or serum starved for 24 hrs, and lysates were made after 1hr exposure to 1μM PD173074 (+), or no exposure (−), as indicated. Lysates were subject to SDS-PAGE and western blotting with antibodies against FGFR2, phosphorylated FRS2-Tyr196, phosphorylated AKT1-Ser473, phosphorylated ERK1/2-Thr202/Tyr204, CCND1, and β-Actin. b) Side-by-side comparison of lysates from MFM223 grown in 10% serum or serum starved, with or without 1μM PD173074, with SKBR3 lysates for comparison.
FGFR2 amplified MFM223 die by apoptosis following FGFR inhibition
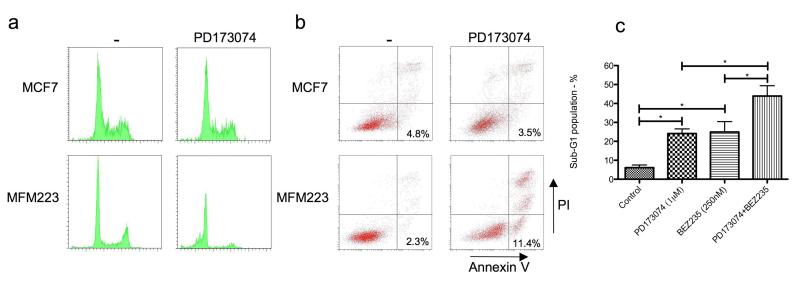
We investigated the mechanism of loss of survival in MFM223 on PD173074 treatment. After 48 hr treatment with PD173074 there was no significant alteration of the cell cycle profile of the MCF7 control cell line, but there was a substantial increase in subG1 cells in MFM223 (Figure 5). To investigate whether the increase in subG1 reflected increased apoptosis, we assessed Annexin V/PI staining in MCF7 and MFM223 cells treated for 48hrs with PD173074 (Figure 5b). There was no difference in the proportion of apoptotic, Annexin V positive/PI negative, cells in MCF7 without and with PD173074 (4.8% vs 3.5%, p=NS), but a substantial increase in apoptotic cells in MFM223 (2.3% vs 11.4% respectively, p<0.001).
Figure 5.
FGFR inhibition induces apoptosis in MFM223 partly through loss of AKT signalling. a) PI cell cycle profiles in MCF7 and MFM223 treated with vehicle or 1μM PD173074 for 48 hrs. b) Annexin V / PI staining in MCF7 and MFM223 treated with vehicle or 1μM PD173074 for 48 hrs. Example plots from one experiment, with proportion of apoptotic Annexin V positive/PI negative cells from three independent experiments: MCF7 without and with PD173074 (4.8% vs 3.5% respectively, p=NS), MFM223 without and with PD173074 (2.3% vs 11.4% respectively, p<0.001 Student’s T Test). c) Fraction of cells in subG1, as assessed by PI FACS, after 24 hours exposure to 1μM PD173074, BEZ235 250nM, or combination of both inhibitors. Displayed mean of three independent experiments. Error bars SEM and * P<0.01 (Student’s T test).
To confirm that these observations represented ligand independent signalling, we grew MFM223 in serum free medium. In the absence of serum MFM223 proliferated at a similar rate compared to cells grown in 10% serum. In the absence of serum MFM223 were dependent on FGFR for proliferation, and underwent apoptosis in the presence of PD173074 (Supplementary Figure 4).
FGFR2 amplified cell lines also have PI3 kinase pathway aberrations
We noted that AKT was highly phosphorylated in MFM223, but that this was substantially decreased in PD173074 treated cells. To investigate whether this reflected an activating mutation in PIK3CA, we sequenced the helical and kinase domains of PIK3CA and identified a classical H1047R kinase mutation in MFM223 (Supplementary Figure 5). The H1047R mutation has been described to active PI3 kinase function and transforms human mammary epithelial cells (Zhao et al., 2005). We therefore examined the effect of dual targeting of PI3 kinase and FGFR. Treatment of MFM223 with the dual PI3 kinase/mTOR inhibitor BEZ235 (Maira et al., 2008) abolished AKT phosphorylation (Supplementary Figure 5) and substantially increased the subG1 fraction (Figure 5c). Treatment with combination of BEZ235 and PD173074 increased the subG1 fraction to a greater extent than either drug given individually (Figure 5c and Supplementary Figure 2). Similarly, dual treatment with both inhibitors had an additive effect as assessed by combination index (CI) assay (Supplementary Figure 5, CI 1.02). SUM52 had wild-type PIK3CA, but did not express PTEN protein (Supplementary Figure 5), and a similar additive CI was observed in SUM52 cells between PD173074 and BEZ235 (CI 0.985). These data suggests that dual targeting of FGFR2 and PIK3CA may be of benefit in cancers with FGFR2 amplification.
Taken together, our data provide strong circumstantial evidence that FGFR2 may be a therapeutic target for a subset of TNBC harbouring FGFR2 gene amplification, and that our method of integrated analysis of transcriptomic and genomic data, focused on genes that are over-expressed when amplified, identifies potential treatment targets.
Discussion
TNBCs are genetically unstable and often harbour complex patterns of genetic aberrations. Using high resolution array CGH and gene expression platforms, tumours with >70% of neoplastic cells, and only invasive ductal cancers of no special type, we have demonstrated that a substantial proportion of genes have expression levels that significantly correlate with copy number in TN cancers. Functional annotation of these genes using IPA revealed that the canonical pathways of several tyrosine kinase receptors involved in tumourigenesis and cancer progression were enriched for the genes upregulated by copy number gain were enriched (Supplementary Table 8). Interestingly, copy number gains of the tyrosine kinase receptors were rare; instead, upregulation of signal transduction kinases downstream of the receptors (e.g. RAF1, PTK2, PIK3C2G, CSNK1D, MAPK9) and adaptor proteins (e.g., GRB2, GAB1) was observed. It is possible that these recurrent copy number gains, of multiple components of the canonical pathways of tyrosine kinase receptors, could create a permissive context for activation of these pathways in TN breast cancers. In addition this observation suggests that in TNBCs it may be possible to identify commonly activated signal transduction pathways that could be targeted effectively for TNBC therapy.
The majority of TNBCs showed losses on 1p, 2q, 3p, 4p, 5q, 8p 9q, 16q, 17p, 19p, and 23p; and gains on 1p, 3q, 6p, 9p, 7q, 8p, 10p, and 12p. Only 11% of the tumours in our cohort showed concurrent 1q gain and 16q loss, the typical changes of low-grade ER positive breast cancers, in agreement with our previous results suggesting that progression from grade I to grade III is an uncommon phenomenon in TNBCs (Natrajan et al., 2009a). It should be noted that many of the regions that were affected by genetic aberrations in TNBCs such as gain of 1q, 3q, 7q, 8q, and 10p and loss of 4p, 5q, 17p, and 8p, have also been found in tumours arising in BRCA1 mutation carriers (Jonsson et al., 2005). These results highlight the similarities between sporadic TNBCs and tumours arising in BRCA1 mutation carriers and provide yet another line of evidence to suggest that, as a group, TNBCs phenocopy familial BRCA1 tumours (Turner et al., 2004).
Similar to the HER2 amplicon, whose smallest regions of amplification encompasses 13 genes, of which only 7 are expressed at significantly higher levels when amplified (Marchio et al., 2008; Orsetti et al., 2004), our analysis demonstrated that only a few genes mapping to regions recurrently amplified in TN breast cancers were consistently overexpressed when amplified. Importantly, genes identified in this study as overexpressed when amplified may constitute potential therapeutic targets, including FGFR2 (10q26), mitotic spindle checkpoint protein BUB3 (10q26), RAS oncogene family member RAB20 (13q34), Notch family member NOTCH3 (19p13) and the protein kinase C super family member PKN1 (19p13).
Our data adds to the body of evidence linking aberrant FGF signaling to breast cancer pathogenesis. Interestingly, a SNP in FGFR2 intron 2 is a common low risk predisposition gene for breast cancer, that predisposes selectively for ER positive breast cancer (Easton et al., 2007; Garcia-Closas et al., 2008). In contrast our data suggest that amplification of FGFR2 is found in ER negative TNBCs. Most TNBCs exhibit high levels of genomic instability, which potentially reflects an underlying defect in the processes that maintain genome stability, and this defect could theoretically provide a mechanism through which some TNBCs acquire FGFR2 amplfication, coupled with the survival advantage conferred by activated FGFR2 signalling. Activating mutations in FGFR2 are found in endometrial cancer (Byron et al., 2008), but are rare in breast cancer (Greenman et al., 2007).
We confirmed that a mechanism underlying oncogene addiction in cell lines with FGFR2 gene amplification is activation of PI3K-AKT signalling, and resulting inhibition of apoptosis. Signalling in cell lines harbouring FGFR2 amplification appears to be ligand independent, with both cell lines activating AKT in an FGFR kinase dependent manner. Interestingly, in MFM223, AKT phosphorylation was predominantly under control of upstream FGFR signalling despite the presence of an activating PI3KCA kinase mutation (Figure 4), suggesting that in this cell line PI3KCA kinase domain mutation predominantly amplified upstream signal. Likewise SUM52 remained sensitive to FGFR inhibition, despite lacking PTEN expression (Supplementary Figure 5), concurring with previous results found in FGFR2 mutant endometrial cancer cell lines (Byron et al., 2008). We note that HCC1143 have been shown to harbour an FGFR2 mutation (Greenman et al., 2007). HCC1143 show no evidence of FGFR dependent signalling (Figure 4), nor dependence on FGFR for proliferation (Figure 3), suggesting the mutation identified (R203C) does not active the receptor.
Here, we have identified a higher number of recurrent amplifications in TN cancers than previously reported (Andre et al., 2009; Han et al., 2008), possibly due to the use of samples with >70% of tumour cells, which reduces the bias introduced by the contamination with diploid non-neoplastic cells (i.e. stromal cells and inflammatory infiltrate). In fact, 76.8% of the tumours analysed in this study harboured at least one focal (<10Mb) amplification. However, the majority of these amplifications were shown to occur at low frequency, confirming that at the genomic level TNBCs are genomically heterogeneous. To develop therapeutic strategies directed at drivers of the recurrent amplicons in TNBC will be challenging, requiring comprehensive molecular pathology analyses of primary tumours to select the appropriate targeted therapy for the individual tumours. A novel approach to clinical trial design would also be required if rare oncogenic targets are to be validated. An alternative approach to the therapy of TNBCs would be to identify shared signal transduction pathways that could be targeted for tumour treatment without targeting the oncogene directly.
Supplementary Material
Figure 2.
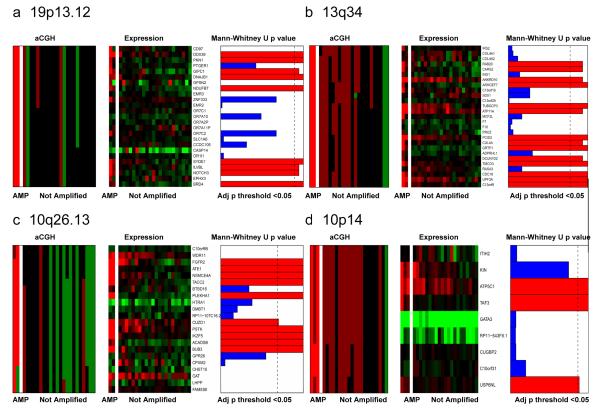
Matched heatmaps of expression and aCGH within regions of recurrent amplification in 24 TNBCs. a) 19p13.12 amplicon; b) 13q34 amplicon; c) 10q26.13 amplicon; d) 10p14 amplicon. For each amplicon, genes within the amplified region are recovered and median aCGH values and states are assigned. Samples are separated into those harbouring an amplification within the region and those that do not. Expression and cbs values are depicted in two matching heatmaps (aCGH states on the left and expression values on the right) in which the genes are ordered according to their chromosomal position and the tumours ordered according to the sum of their aCGH values. In the correlation box, red bars indicate correlated genes whose expression significantly correlates with amplification (Mann-Whitney U test adjusted p<0.05), whereas blue bars indicate non significant genes (Mann-Whitney U test adjusted p<0.05). aCGH: green: copy number loss; black: no copy number change; dark red: copy number gain; bright red: gene amplification; gene expression: green: downregulation, red: upregulation.
Acknowledgements
This work was supported by grants from Cancer Research UK and Breakthrough Breast Cancer. Dr Nicholas Turner is a CRUK clinician scientist. We acknowledge NHS funding to the NIHR Biomedical Research Centre.
Footnotes
Conflict of interest statement We have no conflict of interest.
References
- Adelaide J, Finetti P, Bekhouche I, Repellini L, Geneix J, Sircoulomb F, et al. Integrated profiling of basal and luminal breast cancers. Cancer Res. 2007;67:11565–75. doi: 10.1158/0008-5472.CAN-07-2536. [DOI] [PubMed] [Google Scholar]
- Agrawal G, Chen JH, Baick CH, Chen AE, Mehta RS, Nalcioglu O, et al. Pathological complete response in triple negative poorly differentiated invasive ductal breast carcinoma detected during pregnancy. J Clin Oncol. 2007;25:2618–20. doi: 10.1200/JCO.2007.11.3084. [DOI] [PubMed] [Google Scholar]
- Andre F, Job B, Dessen P, Tordai A, Michiels S, Liedtke C, et al. Molecular characterization of breast cancer with high-resolution oligonucleotide comparative genomic hybridization array. Clin Cancer Res. 2009;15:441–51. doi: 10.1158/1078-0432.CCR-08-1791. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. 1995;57:289–300. [Google Scholar]
- Bernard-Pierrot I, Gruel N, Stransky N, Vincent-Salomon A, Reyal F, Raynal V, et al. Characterization of the recurrent 8p11-12 amplicon identifies PPAPDC1B, a phosphatase protein, as a new therapeutic target in breast cancer. Cancer Res. 2008;68:7165–75. doi: 10.1158/0008-5472.CAN-08-1360. [DOI] [PubMed] [Google Scholar]
- Bertucci F, Finetti P, Cervera N, Esterni B, Hermitte F, Viens P, et al. How basal are triple-negative breast cancers? Int J Cancer. 2008;123:236–40. doi: 10.1002/ijc.23518. [DOI] [PubMed] [Google Scholar]
- Byron SA, Gartside MG, Wellens CL, Mallon MA, Keenan JB, Powell MA, et al. Inhibition of activated fibroblast growth factor receptor 2 in endometrial cancer cells induces cell death despite PTEN abrogation. Cancer Res. 2008;68:6902–7. doi: 10.1158/0008-5472.CAN-08-0770. [DOI] [PubMed] [Google Scholar]
- Carey LA, Dees EC, Sawyer L, Gatti L, Moore DT, Collichio F, et al. The triple negative paradox: primary tumor chemosensitivity of breast cancer subtypes. Clin Cancer Res. 2007;13:2329–34. doi: 10.1158/1078-0432.CCR-06-1109. [DOI] [PubMed] [Google Scholar]
- Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–41. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- Coe BP, Ylstra B, Carvalho B, Meijer GA, Macaulay C, Lam WL. Resolving the resolution of array CGH. Genomics. 2007;89:647–53. doi: 10.1016/j.ygeno.2006.12.012. [DOI] [PubMed] [Google Scholar]
- Dent R, Hanna WM, Trudeau M, Rawlinson E, Sun P, Narod SA. Pattern of metastatic spread in triple-negative breast cancer. Breast Cancer Res Treat. 2008 doi: 10.1007/s10549-008-0086-2. [DOI] [PubMed] [Google Scholar]
- Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007;13:4429–34. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–93. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Closas M, Hall P, Nevanlinna H, Pooley K, Morrison J, Richesson DA, et al. Heterogeneity of breast cancer associations with five susceptibility loci by clinical and pathological characteristics. PLoS Genet. 2008;4:e1000054. doi: 10.1371/journal.pgen.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–8. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunnarsson R, Staaf J, Jansson M, Ottesen AM, Goransson H, Liljedahl U, et al. Screening for copy-number alterations and loss of heterozygosity in chronic lymphocytic leukemia--a comparative study of four differently designed, high resolution microarray platforms. Genes Chromosomes Cancer. 2008;47:697–711. doi: 10.1002/gcc.20575. [DOI] [PubMed] [Google Scholar]
- Han W, Jung EM, Cho J, Lee JW, Hwang KT, Yang SJ, et al. DNA copy number alterations and expression of relevant genes in triple-negative breast cancer. Genes Chromosomes Cancer. 2008;47:490–9. doi: 10.1002/gcc.20550. [DOI] [PubMed] [Google Scholar]
- Hannemann J, Velds A, Halfwerk JB, Kreike B, Peterse JL, van de Vijver MJ. Classification of ductal carcinoma in situ by gene expression profiling. Breast Cancer Res. 2006;8:R61. doi: 10.1186/bcr1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris LN, Broadwater G, Lin NU, Miron A, Schnitt SJ, Cowan D, et al. Molecular subtypes of breast cancer in relation to paclitaxel response and outcomes in women with metastatic disease: results from CALGB 9342. Breast Cancer Res. 2006;8:R66. doi: 10.1186/bcr1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks J, Krasnitz A, Lakshmi B, Navin NE, Riggs M, Leibu E, et al. Novel patterns of genome rearrangement and their association with survival in breast cancer. Genome Res. 2006;16:1465–79. doi: 10.1101/gr.5460106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TR, Roberts CJ, Dai H, Jones AR, Meyer MR, Slade D, et al. Widespread aneuploidy revealed by DNA microarray expression profiling. Nat Genet. 2000;25:333–7. doi: 10.1038/77116. [DOI] [PubMed] [Google Scholar]
- Jonsson G, Naylor TL, Vallon-Christersson J, Staaf J, Huang J, Ward MR, et al. Distinct genomic profiles in hereditary breast tumors identified by array-based comparative genomic hybridization. Cancer Res. 2005;65:7612–21. doi: 10.1158/0008-5472.CAN-05-0570. [DOI] [PubMed] [Google Scholar]
- Kreike B, van Kouwenhove M, Horlings H, Weigelt B, Peterse H, Bartelink H, et al. Gene expression profiling and histopathological characterization of triple-negative/basal-like breast carcinomas. Breast Cancer Res. 2007;9:R65. doi: 10.1186/bcr1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambros MB, Simpson PT, Jones C, Natrajan R, Westbury C, Steele D, et al. Unlocking pathology archives for molecular genetic studies: a reliable method to generate probes for chromogenic and fluorescent in situ hybridization. Lab Invest. 2006;86:398–408. doi: 10.1038/labinvest.3700390. [DOI] [PubMed] [Google Scholar]
- Mackay A, Tamber N, Fenwick K, Iravani M, Grigoriadis A, Dexter T, et al. A high-resolution integrated analysis of genetic and expression profiles of breast cancer cell lines. Breast Cancer Res Treat. 2009 doi: 10.1007/s10549-008-0296-7. [DOI] [PubMed] [Google Scholar]
- Maira SM, Stauffer F, Brueggen J, Furet P, Schnell C, Fritsch C, et al. Identification and characterization of NVP-BEZ235, a new orally available dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor with potent in vivo antitumor activity. Mol Cancer Ther. 2008;7:1851–63. doi: 10.1158/1535-7163.MCT-08-0017. [DOI] [PubMed] [Google Scholar]
- Marchio C, Natrajan R, Shiu KK, Lambros MB, Rodriguez-Pinilla SM, Tan DS, et al. The genomic profile of HER2-amplified breast cancers: the influence of ER status. J Pathol. 2008;216:399–407. doi: 10.1002/path.2423. [DOI] [PubMed] [Google Scholar]
- Mohammadi M, Froum S, Hamby JM, Schroeder MC, Panek RL, Lu GH, et al. Crystal structure of an angiogenesis inhibitor bound to the FGF receptor tyrosine kinase domain. Embo J. 1998;17:5896–904. doi: 10.1093/emboj/17.20.5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natrajan R, Lambros MB, Rodriguez-Pinilla SM, Moreno-Bueno G, Tan DS, Marchio C, et al. Tiling path genomic profiling of grade 3 invasive ductal breast cancers. Clin Cancer Res. 2009a;15:2711–22. doi: 10.1158/1078-0432.CCR-08-1878. [DOI] [PubMed] [Google Scholar]
- Natrajan R, Weigelt B, Mackay A, Geyer FC, Grigoriadis A, Tan DS, et al. An integrative genomic and transcriptomic analysis reveals molecular pathways and networks regulated by copy number aberrations in basal-like, HER2 and luminal cancers. Breast Cancer Res Treat. 2009b doi: 10.1007/s10549-009-0501-3. epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Ng G, Huang J, Roberts I, Coleman N. Defining ploidy-specific thresholds in array comparative genomic hybridization to improve the sensitivity of detection of single copy alterations in cell lines. J Mol Diagn. 2006;8:449–58. doi: 10.2353/jmoldx.2006.060033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsetti B, Nugoli M, Cervera N, Lasorsa L, Chuchana P, Ursule L, et al. Genomic and expression profiling of chromosome 17 in breast cancer reveals complex patterns of alterations and novel candidate genes. Cancer Res. 2004;64:6453–60. doi: 10.1158/0008-5472.CAN-04-0756. [DOI] [PubMed] [Google Scholar]
- Reis-Filho JS, Drury S, Lambros MB, Marchio C, Johnson N, Natrajan R, et al. ESR1 gene amplification in breast cancer: a common phenomenon? Nat Genet. 2008;40:809–10. doi: 10.1038/ng0708-809b. author reply 810-2. [DOI] [PubMed] [Google Scholar]
- Reis-Filho JS, Simpson PT, Turner NC, Lambros MB, Jones C, Mackay A, et al. FGFR1 emerges as a potential therapeutic target for lobular breast carcinomas. Clin Cancer Res. 2006;12:6652–62. doi: 10.1158/1078-0432.CCR-06-1164. [DOI] [PubMed] [Google Scholar]
- Reis-Filho JS, Tutt AN. Triple negative tumours: a critical review. Histopathology. 2008;52:108–18. doi: 10.1111/j.1365-2559.2007.02889.x. [DOI] [PubMed] [Google Scholar]
- Schneider BP, Winer EP, Foulkes WD, Garber J, Perou CM, Richardson A, et al. Triple-negative breast cancer: risk factors to potential targets. Clin Cancer Res. 2008;14:8010–8. doi: 10.1158/1078-0432.CCR-08-1208. [DOI] [PubMed] [Google Scholar]
- Tan DS, Lambros MB, Natrajan R, Reis-Filho JS. Getting it right: designing microarray (and not ‘microawry’) comparative genomic hybridization studies for cancer research. Lab Invest. 2007;87:737–54. doi: 10.1038/labinvest.3700593. [DOI] [PubMed] [Google Scholar]
- Tannheimer SL, Rehemtulla A, Ethier SP. Characterization of fibroblast growth factor receptor 2 overexpression in the human breast cancer cell line SUM-52PE. Breast Cancer Res. 2000;2:311–20. doi: 10.1186/bcr73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischkowitz MD, Foulkes WD. The basal phenotype of BRCA1-related breast cancer: past, present and future. Cell Cycle. 2006;5:963–7. doi: 10.4161/cc.5.9.2713. [DOI] [PubMed] [Google Scholar]
- Turner N, Tutt A, Ashworth A. Hallmarks of ‘BRCAness’ in sporadic cancers. Nat Rev Cancer. 2004;4:814–9. doi: 10.1038/nrc1457. [DOI] [PubMed] [Google Scholar]
- Turner NC, Lord CJ, Iorns E, Brough R, Swift S, Elliott R, et al. A synthetic lethal siRNA screen identifying genes mediating sensitivity to a PARP inhibitor. Embo J. 2008;27:1368–77. doi: 10.1038/emboj.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelt B, Horlings HM, Kreike B, Hayes MM, Hauptmann M, Wessels LF, et al. Refinement of breast cancer classification by molecular characterization of histological special types. J Pathol. 2008;216:141–50. doi: 10.1002/path.2407. [DOI] [PubMed] [Google Scholar]
- Yamaguchi N, Oyama T, Ito E, Satoh H, Azuma S, Hayashi M, et al. NOTCH3 signaling pathway plays crucial roles in the proliferation of ErbB2-negative human breast cancer cells. Cancer Res. 2008;68:1881–8. doi: 10.1158/0008-5472.CAN-07-1597. [DOI] [PubMed] [Google Scholar]
- Yuan B, Xu Y, Woo JH, Wang Y, Bae YK, Yoon DS, et al. Increased expression of mitotic checkpoint genes in breast cancer cells with chromosomal instability. Clin Cancer Res. 2006;12:405–10. doi: 10.1158/1078-0432.CCR-05-0903. [DOI] [PubMed] [Google Scholar]
- Zhao JJ, Liu Z, Wang L, Shin E, Loda MF, Roberts TM. The oncogenic properties of mutant p110alpha and p110beta phosphatidylinositol 3-kinases in human mammary epithelial cells. Proc Natl Acad Sci U S A. 2005;102:18443–8. doi: 10.1073/pnas.0508988102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.