Abstract
DNA damage checkpoint and DNA repair mechanisms play critical roles in the stable maintenance of genetic information. Various forms of DNA damage that arise inside cells due to common errors in normal cellular processes, such as DNA replication, or due to exposure to various DNA damaging agents, must be quickly detected and repaired by checkpoint signaling and repair factors. Telomeres, the natural ends of linear chromosomes, share many features with undesired “broken” DNA, and are recognized and processed by various DNA damage checkpoint and DNA repair proteins. However, their modes of action at telomeres must be altered from their actions at other DNA damage sites to avoid telomere fusions and permanent cell cycle arrest. Interestingly, accumulating evidence indicates that DNA damage checkpoint and DNA repair proteins are essential for telomere maintenance. In this article, we review our current knowledge on various mechanisms by which DNA damage checkpoint and DNA repair proteins are modulated at telomeres and how they might contribute to telomere maintenance in eukaryotes.
Keywords: Telomere, Telomerase, Pot1, Shelterin, DNA repair, DNA damage checkpoint, Review
2. INTRODUCTION
The maintenance of genomic stability is essential for normal cell growth and survival. Since the integrity of genomic DNA is constantly under threat from various genotoxic agents in the environment and errors in normal cellular processes such as DNA replication, eukaryotic cells have evolved various complex mechanisms which ensure that genetic information can be propagated free of errors. One form of DNA damage that is especially challenging for cells to repair is DNA double-strand breaks (DSBs). In eukaryotic cells, DSBs are repaired by two major DNA repair mechanisms known as non-homologous end-joining (NHEJ) and homologous recombination (HR) (1). Furthermore, DSBs strongly activate DNA damage checkpoint responses to arrest cell cycle progression, so that cells have enough time to properly process and repair DSBs.
Telomeres are specialized nucleoprotein structures responsible for protecting the ends of eukaryotic chromosomes (2). Telomeric DNA ends do not fully activate DNA damage checkpoint responses and escape re-joining/repair by DNA repair enzymes. Therefore, one might predict that telomeres protect chromosome ends simply by denying access to the DNA damage response machineries. However, many DNA damage checkpoint and DNA repair proteins robustly bind telomeres and play essential roles in telomere maintenance (3, 4). While much progress has been made in recent years, the precise mechanisms by which DNA damage response factors contribute to telomere maintenance are still not fully understood. In this article, we review our current knowledge on the contributions made by DNA damage checkpoint and DNA repair proteins to maintain telomeres, and how their actions are regulated at telomeres, focusing on recent findings from studies in yeast and mammalian cells. For more comprehensive reviews on telomere maintenance mechanisms, readers are referred to many excellent reviews that have been published recently (2–7).
3. CHECKPOINT SIGNALING AT DNA BREAKS AND REPLICATION FORKS
Successful detection and repair of DSBs, much like many other biological processes, involve several proteins that work collaboratively in signaling cascades. Proteins involved in DNA damage responses are highly conserved among eukaryotes from yeasts to humans (summarized in Table 1). DNA damage checkpoint proteins have been classified into four major functional groups: sensors, mediators, transducers and effectors (8, 9). In mammalian cells, checkpoint signaling is initiated by the sensor kinases ATM (Ataxia Telangiectasia Mutated) and ATR (ATM and Rad3-related), in complex with their respective binding partners MRN (Mre11-Rad50-Nbs1) and ATRIP (ATR Interacting Protein). Additional checkpoint sensor complexes Rad17-RFC (composed of Rad17 and four small subunits of Replication Factor C) and Rad9-Rad1-Hus1 (9-1-1) are also crucial for the cell’s ability to properly respond to DSBs. These sensor complexes, with the help of mediators such as Mdc1, 53BP1, TopBP1 and Claspin, activate the transducer kinases Chk1 and Chk2, which in turn regulate Cdc25 phosphatases to inhibit cyclin-dependent kinases (effectors) and stop cell cycle progression (1, 10).
Table 1.
Checkpoint and DNA repair proteins discussed in this review
| Factors (Hs = Human, Sc = budding yeast, Sp = fission yeast) | Function in DNA damage response | Function in telomere maintenance |
|---|---|---|
| Mre11-Rad50-Nbs1 (MRN) (Hs) Mre11-Rad50-Xrs2 (MRX) (Sc) Rad32-Rad50-Nbs1 (MRN) (Sp) |
Sensor of DSBs; Nbs1/Xrs2 interacts with ATM/Tel1 via evolutionarily conserved C-terminal motif; involved in HR & NHEJ | Required for damage response to short telomeres and their preferential elongation by telomerase in Sc; important for generating telomeric G tail and preventing NHEJ at telomeres; promotes Type II telomere recombination in Sc; promotes HR-based telomere maintenance in Sp and human ALT cells; required for ATM signaling at telomeres |
| ATM (Hs) Tel1 (Sc, Sp) |
PIKK family kinase; recruited to DNA damage sites by MRN/MRX; activates CHK2 in response to DNA damage; phosphorylate many factors at DNA damage sites, including histone H2AX, to amplify DNA damage responses and facilitate DNA repair | Preferentially binds critically short telomeres in both Sc & Sp; regulates telomerase recruitment to telomeres in Sc; important for mammalian telomere length regulation; phosphorylates mammalian TRF1; inhibited by Sc Rif1/Rif2 and mammalian TRF2 |
| RPA (Hs, Sc, Sp) | Binds to ssDNA tracts; plays critical roles in DNA replication, various forms of DNA repair; provides platform to facilitate recruitment of checkpoint sensors ATR-ATRIP and 9-1-1 | Important for telomere length maintenance in Sc and Sp; regulates telomerase access to telomeres in late S phase in Sc; binds Sp telomeres in S phase in a replication-dependent manner; competes with G-tail binding proteins for binding to telomeric G-overhang |
| ATRIP (Hs) Ddc2 (Sc) Rad26 (Sp) |
Recruited to RPA-coated ssDNA at damage sites; binding partner of ATR/Mec1/Rad3; recruits ATR to damage sites via the C-terminus of ATRIP | Important for telomere length maintenance in Sp but not in Sc; binds Sp telomeres in S phase in a replication-dependent manner; binds short dysfunctional telomeres, and required for activation of senescence following telomerase loss in Sc |
| ATR (Hs) Mec1 (Sc) Rad3 (Sp) |
PIKK family kinase; recruited to DNA damage sites by ATRIP; induces cell cycle arrest following DNA damage by activating CHK1; can activate ATM in a Nbs1 C-terminus independent manner; primary sensor kinase activated by DSBs in Sc and Sp but not in mammals | Important for telomere length maintenance in Sp and mammals but not in Sc; binds short dysfunctional telomeres, and required for activation of senescence following telomerase loss in Sc; associates with human telomeres in S phase even before replication is completed; inhibited by Cdc13 in Sc and POT1 in mammals |
| Rad9-Hus1-Rad1 (9-1-1) (Hs, Sp) Ddc1-Mec3-Rad17 (Sc) |
Forms PCNA-like clamp; Loaded to telomeres and sites of DNA damage by the replication factor C (RFC)-like clamp loader Rad17-Rfc2–5 (Hs, Sp) or Rad24-Rfc2–5 (Sc) | Associates with telomerase and regulates its activity (Hs); mutations in 9-1-1 subunits generally cause telomere shortening (Sc, Sp), except in Mec3 that has been reported to cause telomere elongation (Sc) |
Depending on the type of DNA lesion being detected and the phase of the cell cycle in which the DNA lesion occurs, cells respond differently to DNA damage. DSBs, generated by genotoxic agents such as ionizing radiation, are first sensed by the MRN complex, which then recruits ATM to sites of damage via the C-terminal ATM-interaction domain of Nbs1 and promotes ATM kinase activation (11–14). Activated ATM kinase phosphorylates and regulates many proteins that are involved in cell cycle checkpoint control, apoptotic responses and DNA repair, including p53, Chk2, BRCA1, SMC1, FANCD2, Rad17, Artemis and Nbs1 (15, 16). While it was initially thought that ATR responds to DSBs independently of ATM, recent studies have found that ATR acts downstream of ATM and MRN to activate Chk1 in response to DSBs during S and G2 phases of the cell cycle (17–20) (Figure 1A). ATM and MRN are important for the activation of ATR in response to DSBs since they promote resection of DNA ends to expose long stretches of single-stranded DNA (ssDNA), which in turn will be coated by RPA (replication protein A) and serves as a recruitment and activation platform for the ATR-ATRIP complex (21, 22). Conversely, ATR-ATRIP can also function upstream of ATM following replication fork stalling or exposure to UV irradiation (Figure 1B). Interestingly, this mode of ATM activation does not require the C-terminal ATM interaction domain of Nbs1 or Mre11 (23). The 9-1-1 complex, which forms a PCNA-like ring-shaped complex (24), is also loaded to RPA-coated ssDNA with the help of Rad17-RFC, and contributes to the activation of ATR-ATRIP kinase in collaboration with TopBP1 (25, 26).
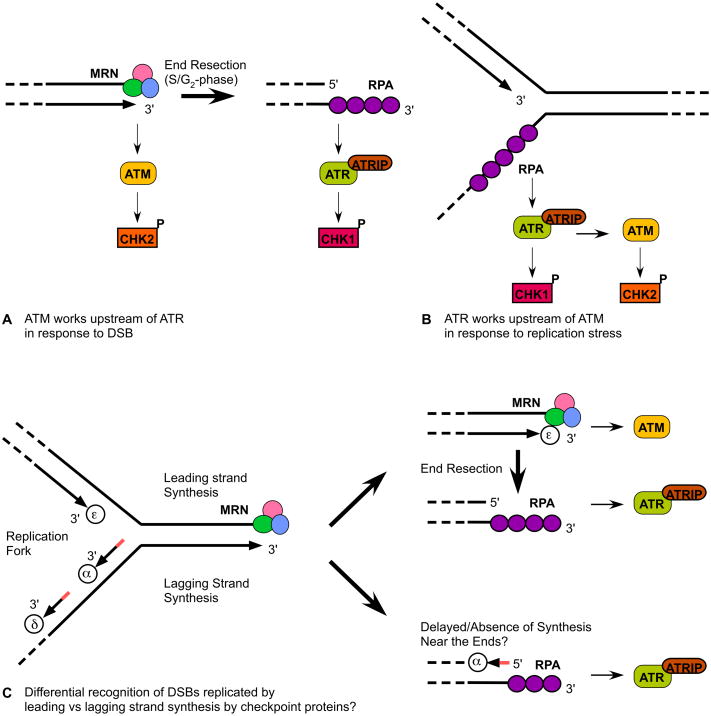
Figure 1.
DNA damage checkpoint and DNA replication checkpoint responses in mammalian cells. (A) A signaling crosstalk between ATM-MRN and ATR-ATRIP in response to DSBs. (B) A signaling crosstalk between ATR-ATRIP and ATM in response to DNA replication stress. (C) Checkpoint signaling possibilities at leading strand and lagging strand after DNA replication forks encounter DSBs. Short red lines on lagging strand represent Okazaki fragments. The lagging strand DNA polymerases (alpha and delta) and the leading strand DNA polymerase (epsilon) are also indicated as circles with the corresponding Greek letters.
DSBs generated in late G1- or S-phase may occur too late to induce proper checkpoint and repair responses prior to the arrival of DNA replication forks (Figure 1C). For example, a recent study in budding yeast has found that a single DSB generated in G1 is not able to induce Mec1 (ATR ortholog)-dependent checkpoint activation during S-phase and DNA replication continues all the way to the DSB, since the rate of DSB resection is too slow to generate enough RPA-coated ssDNA required to activate Mec1 in S-phase (27). As DNA replication forks reach DSBs, DNA strands replicated by leading strand synthesis and those replicated by lagging strand synthesis are likely to have distinct structures, and thus likely to be recognized/processed by different checkpoint and DNA repair proteins. While leading strand synthesis could theoretically continue to the very end, generating blunt end DNA, lagging strand synthesis is likely to leave behind substantial amounts of ssDNA since the final Okazaki fragment cannot be synthesized very close to the 3′ ends of DSBs (28) (Figure 1C). It was recently shown that blunt ends as well as ends with short single-stranded tails are the preferred substrates for ATM activation (22). However, as the single-stranded tail gets longer, ATM activation gets attenuated, and RPA coated long ssDNA would activate ATR (22). Thus, DSBs replicated by leading strand synthesis might be preferentially recognized by ATM-MRN, while DSBs replicated by lagging strand synthesis might be recognized primarily by ATR-ATRIP (Figure 1C). Likewise, since blunt-ended DSBs would serve as good substrates for NHEJ repair, while extended ssDNA at DSBs would promote HR repair (8), DSBs replicated by leading strand synthesis might be more suitable for NHEJ repair while DSBs replicated by lagging strand synthesis might be more likely to be repaired by HR repair.
The modes by which DNA damage checkpoint and repair proteins respond when DNA replication forks encounter DSBs are highly relevant for telomere biology, since telomeres are dependent on checkpoint and repair proteins for their stable maintenance and complete replication (29–32). Furthermore, the observation that normally dormant replication origins are activated by the presence of proximal DSBs (27) is intriguing, since short telomeres replicate earlier in S-phase than longer telomeres due to the activation of normally dormant sub-telomeric origins among critically short telomeres in budding yeast (33). Thus, while longer telomeres could repress firing of replication origins perhaps by forming heterochromatin structures, shorter telomeres seem to lose the ability to inhibit replication origin firing and behave similarly to accidental/undesired DSBs, which lose repression of replication origin firing perhaps due to changes in DNA topological constraints and/or chromatin structures upon formation of DNA breaks.
4. TELOMERE SPECIFIC FACTORS AND INIHIBITION OF DNA DAMAGE RESPONSES AT TELOMERES
What are the essential features of telomeric DNA that distinguish them from other types of undesired DSBs? In most eukaryotic cells, telomeres are made up of repetitive GT-rich sequences, which mostly consist of double-stranded DNA (dsDNA) that terminate with 3′ single-stranded tails, known as G-tails (2). A specialized reverse transcriptase, known as telomerase, ensures the stable maintenance of these telomeric GT-rich repeats by counteracting the loss of telomeric DNA caused by the inability of conventional semi-conservative DNA polymerases to fully replicate ends of linear DNA molecules (known as end-replication problem) (34). Telomerase utilizes its RNA subunit as template to synthesize a GT-rich strand beyond the end of the original genomic DNA. Both dsDNA and G-tail portions of telomeric repeat sequences play critical roles in the assembly of the telomere protection complex. Repeated cell divisions in the absence of telomerase gradually deplete GT-rich repeats and their associated telomere protection factors. Once telomeres become too short to bind sufficient telomere protection proteins, they will be treated like other undesired DSBs, leading to increased fusion and recombination events among critically short telomeres (3, 4). Studies have further shown that critically short or unprotected telomeres massively attract and activate checkpoint sensors as well as adaptors and downstream effectors of checkpoint signaling. In addition, at least in budding yeast, Tel1-MRX (ATM-MRN) appears to act upstream of Mec1-Ddc2 (ATR-ATRIP) in processing and checkpoint signaling of short or unprotected telomeres in S/G2-phase by contributing to the resection of telomeres, much like in the case of undesired/accidental DSBs (35–37) (Figure 1A).
In human cells, telomeres are bound and protected by a “shelterin” complex, consisting of TRF1, TRF2, RAP1, POT1, TPP1 and TIN2 (2) (Figure 2 and Table 2). While TRF1 and TRF2 bind specifically to dsDNA telomeric repeats, POT1 binds to G-tails. TRF2 is important for repressing ATM activation at telomeres, while POT1 is important for repressing ATR activation at telomeres (38, 39). Interestingly, TRF2 has been shown to interact with both ATM and Chk2 kinases (40, 41). Thus, TRF2 might inhibit ATM-dependent checkpoint activation by interfering with the communication between ATM and Chk2. Additional DNA repair proteins, including Apollo and the MRN complex, associate with TRF2, suggesting that their activities might be regulated by TRF2 at telomeres (42–46). POT1 is likely to inhibit activation of ATR and its downstream target Chk1 by protecting telomeres against excessive resection and accumulation of RPA at telomeres (47, 48). Mammalian RAP1 does not bind directly to telomeric DNA, but its recruitment to telomeres by TRF2 is important for inhibition of NHEJ at telomeres (49). The mammalian shelterin complex is also thought to promote insertion of 3′ telomeric G-tails into the dsDNA portion of telomeres to generate a “t-loop” structure, thereby hiding telomeric 3′ ends from DNA repair and checkpoint proteins (2, 50, 51).
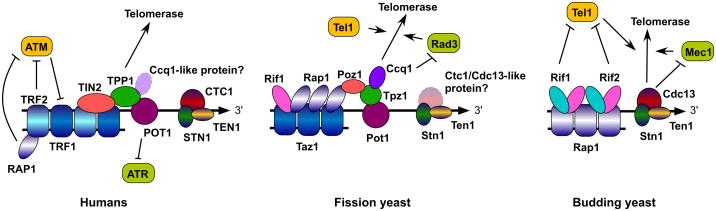
Figure 2.
Models of telomere proteins in humans, fission yeast and budding yeast. Evolutionarily related proteins are represented in the same color. See Table 2 and main text for more details on the proteins shown.
Table 2.
Telomere-specific proteins discussed in this review
| Factors Hs = human, Sc = budding yeast, Sp = fission (yeast) | Telomeric binding specificity | Function |
|---|---|---|
| CTC1-STN1-TEN1 (Hs) Cdc13-Stn1-Ten1 (Sc) Stn1-Ten1 (Sp) |
G tail-binding | Required for telomere protection; shown to closely resemble heterotrimeric RPA complex in OB-fold domain structure and function; Sc Cdc13 required for normal telomerase function and to prevent Mec1-dependent checkpoint responses at telomeres; Sc Stn1 inhibits the S-phase checkpoint; Sp Cdc13 ortholog remains to be identified |
| POT1 (Hs) Pot1 (Sp) |
G tail-binding | Required for telomere protection in both Sp and mammals; binds telomeric DNA through OB-fold domains; along with TPP1, prevents ATR signaling at mammalian telomeres; Sp Pot1 dissociates from telomeres during telomere replication in late S-phase providing an open conformation for telomerase-mediated telomere extension; Sc appears to lack Pot1 ortholog |
| TPP1(Hs) Tpz1 (Sp) |
Associated with G tail- binding proteins | Tpz1 (Sp) and mammalian TPP1 are orthologs; Associates with both Pot1 & Poz1 (Sp) or mammalian POT1 & TIN2; prevents ATR signaling at mammalian telomeres along with POT1; required for normal telomerase function; Sc appears to lack TPP1 ortholog but Sc Est3 is thought to serve analogous function |
| TIN2 (Hs) Poz1 (Sp) |
Associates with Tpz1 and Rap1 (Sp) or TPP1 and mammalian TRF1/2; negatively regulates telomerase-mediated telomere elongation in Sp | |
| Ccq1 (Sp) | Identified thus far only in Sp; component of the SHREC heterochromatin complex; associates with Tpz1, and required for telomerase recruitment and function; also required for inhibiting the Rad3-dependent checkpoint at telomeres | |
| TRF1/TRF2 (Hs) Rap1 (Sc) Taz1 (Sp) |
Double stranded telomeric DNA | Binds directly to GT-rich telomeric DNA through Myb-like domain; Sp Taz1 is related to mammalian TRF1/TRF2; TRF2 inhibits ATM signaling at telomeres, and prevents NHEJ and HR; TRF1 is required for efficient telomere replication and inhibition of ATR signaling in S phase; Sp Taz1 is required for prevention of NHEJ and HR as well as normal telomeric DNA replication; Sp Taz1 and Sc Rap1 negatively regulate telomerase-mediated telomere elongation; Sc appears to lack TRF1/TRF2 ortholog, however Sc Rap1 fulfills analogous function |
| RAP1 (Hs) Rap1 (Sp) |
Associated with double stranded telomeric DNA - binding proteins | Sc Rap1 homologs containing Myb domains, recruited by Taz1 (Sp) or mammalian TRF2; prevent NHEJ at telomeres; Sp Rap1 promotes telomere recombination, and negatively regulates telomerase-mediated telomere elongation |
| RIF1 (Hs) Rif1/Rif2 (Sc) Rif1 (Sp) |
Recruited by Rap1 (Sc) or Taz1 (Sp); Sp Rif1 & human RIF1 may have stronger affinity for dysfunctional telomeres; Both Sp and Sc Rif1 negatively regulate telomerase-mediated telomere elongation; Sc Rif1/Rif2 negatively inhibit Tel1 at telomeres |
An analogous shelterin-like complex, consisting of Taz1, Rap1, Pot1, Tpz1, Poz1 and Ccq1, is found at telomeres of fission yeast Schizosaccharomyces pombe (52) (Figure 2 and Table 2). While mammalian Rif1 protein is not associated with functional telomeres (53, 54), fission yeast Rif1 directly interacts with Taz1 and associates with functional telomeres (55). Taz1 is thought to represent a functional counterpart of the mammalian TRF1 and TRF2 proteins, and specifically binds the dsDNA portion of telomeric repeats (56, 57). Deletion of Taz1 or Rap1 leads to loss of telomere protection against NHEJ in cells arrested in G0/G1-phase of the cell cycle (58–60). Interestingly, while Taz1 is required to inhibit recombination among telomeres (59, 61), Rap1 promotes recombination-based telomere maintenance in the absence of telomerase (61). This finding is quite surprising since efficient recruitment of Rap1 to telomeres is dependent on Taz1 (55, 61). Taz1 also promotes replication of telomeric repeat sequences by the conventional DNA replication machinery (62), much like mammalian TRF1 (63). However, exponentially growing cells deleted for Taz1 are surprisingly robust in their growth with very little sign of checkpoint activation (56, 57). Therefore, fission yeast cells must posses a Taz1-independent mechanism that inhibits full activation of the DNA damage checkpoint. In fact, the G-tail binding protein Pot1, in collaboration with Tpz1 and Ccq1, provides protection against telomere fusions and Rad3 (ATR)-dependent checkpoint activation in fission yeast cells (52, 64).
In contrast to mammalian and fission yeast cells, budding yeast Saccharomyces cerevisiae lacks TRF1/TRF2-like proteins. Instead, Rap1 binds directly to the dsDNA portion of telomeric repeats and is responsible for recruiting Rif1 and Rif2 to telomeres (2). (Figure 2 and Table 2). Much like its mammalian and fission yeast counterparts, budding yeast Rap1 inhibits NHEJ at telomeres (65, 66). Budding yeast also appears to lack Pot1, but utilizes Cdc13 instead, along with its accessory factors Stn1 and Ten1 to protect telomeres (Figure 2 and Table 2) (67–70). Therefore, it was initially thought that this G-tail recognition complex, termed CST (Cdc13-Stn1-Ten1) (71), may only exist in budding yeast, while other eukaryotic species utilize evolutionarily conserved Pot1-like proteins for G-tail protection. However, this view was challenged by recent discoveries of orthologs for CST complex subunits in mammalian and plant cells (72, 73) (Figure 2). Thus, the CST complex (for CTC1-STN1-TEN1 in higher eukaryotes) may represent the most conserved telomere-capping complex among eukaryotes. Given that fission yeast orthologs of Stn1 and Ten1 have been described (74), one might anticipate that fission yeast may also carry a Cdc13/Ctc1-like protein (Figure 2) although no obvious ortholog has been identified. It is worth noting that unlike budding yeast Cdc13, mammalian CTC1 shows no preference for telomeric repeat sequences (72), and in fact may play a more global role in promoting DNA replication (75). In any case, it remains unclear how the CST and shelterin complexes work together at telomeres. In fission yeast, deletion of either Stn1 or Ten1 leads to complete loss of telomere protection and fusion of chromosomes, much like in the case of Pot1 or Tpz1 deletion (72, 74). However, it appears that Pot1-Tpz1 and Stn1-Ten1 exist as two distinct complexes in fission yeast, since no interaction has been detected between them. Furthermore, Stn1 recruitment to fission yeast telomeres can be uncoupled from Pot1 recruitment (72, 76). In contrast, mammalian Stn1 has been reported to associate with TPP1 although it is currently unclear if any of the shelterin subunits makes direct contact with the CST complex (77).
The CST complex has been proposed to represent a telomere specific ssDNA binding complex resembling RPA (71–73), and recently determined X-ray crystal structures of Stn1 and Ten1 are consistent with this hypothesis (78, 79). In addition, the CST complex interacts with subunits of the DNA polymerase alpha complex, which is involved in lagging strand synthesis (75, 80–83). Cdc13 inactivation results in excessive accumulation of long stretches of single stranded telomeric DNA and activation of a Mec1-dependent checkpoint response in budding yeast, resulting in cell death (68, 84). However, rare survivor cells, which maintain telomeres in the absence of Cdc13 and Stn1, can be isolated if the DNA damage checkpoint is abolished and nuclease activities at telomeres are all attenuated to prevent telomere resection (85). These data indicated that the essential function of the CST complex is to protect telomeres against inappropriate checkpoint activation and DNA degradation.
Besides regulating DNA repair and DNA damage checkpoint at telomeres, GT-rich specific telomere proteins play important roles in regulation of telomerase. The dsDNA telomere-binding proteins are important for the negative regulation of telomerase, since disruption of TRF1, Taz1 or Rap1 function causes massive telomerase-dependent telomere elongation (86). G-tail binding proteins show dual roles in both positive and negative regulation of telomerase action at telomeres. The negative regulation of telomerase by Pot1 in both mammalian and fission yeast cells requires the function of several shelterin subunits that connect Pot1 to dsDNA telomere-binding proteins (52, 87, 88). In mammalian cells, TPP1 and TIN2 connect POT1 to TRF1 and TRF2 to inhibit telomere elongation (87–89), but the POT1-TPP1 sub-complex has also been implicated in telomerase recruitment and enhancement of telomerase processivity (90, 91). In fission yeast, Poz1, which connects Pot1-Tpz1 to Taz1-Rap1, is critical for the negative regulation of telomerase (52). Fission yeast Ccq1, which directly interacts with Tpz1, is essential for promoting the interaction between Tpz1 and telomerase to recruit telomerase to telomeres (52, 64, 92). In budding yeast, Cdc13 contributes to the recruitment/activation of telomerase via association with Est1, the regulatory subunit of telomerase (93, 94). In collaboration with Stn1 and Ten1, Cdc13 is thought to form a protective complex, which can inhibit telomere extension by telomerase (67, 80, 95).
5. POSSIBLE ROLES OF CHECKPOINT AND DNA REPAIR PROTEINS IN TELOMERE MAINTENANCE
The structure of telomeric DNA undergoes dynamic cell cycle-regulated changes. The length of the G-tail has been shown to increase during S-phase in budding and fission yeast, as well as in mammalian cells (96–98). In budding yeast, S-phase specific long G-tails are generated independent of telomerase action, but depend partially on the MRX (Mre11-Rad50-Xrs2) complex (99, 100). In addition, various nucleases involved in resection of DSBs to generate 3′ ssDNA overhangs, such as Sae2 (related to mammalian CtIP and fission yeast Ctp1), Exo1 and Dna2 helicase/nuclease, along with RecQ-like helicase Sgs1, are likely to be involved in processing of budding yeast telomeres in S-phase, especially at critically short telomeres (101). In fission yeast, the MRN complex and Dna2 have been implicated in generation of the G-tail (96, 102). In particular, the MRN/MRX complex increases its association with telomeres during S-phase in mammalian, fission yeast and budding yeast cells (76, 103, 104). Thus, it appears that telomeres become less protected or “open” during S-phase, providing access to end processing enzymes that are responsible for resection of DSBs. Of course, telomerase could also be considered as an enzyme that “repairs” short telomeres, and like other DNA repair proteins, gains the ability to act on telomeres during S-phase when telomeres are replicated by the conventional DNA replication machinery (76, 93, 105).
If DNA repair factors gain better access to “open” telomeres during S-phase, do DNA damage checkpoint proteins also gain access to S-phase telomeres? Studies in yeasts and mammalian cells have found that ATM-MRN and ATR-ATRIP are recruited to functional telomeres during S- and G2-phases (76, 104, 106), supporting the notion that “open” telomeres are more accessible to checkpoint proteins. In fission yeast, the arrival of lagging strand DNA polymerases (alpha and delta) at telomeres is significantly delayed compared to the arrival of the leading strand DNA polymerase (epsilon), and significant quantities of RPA and Rad26 (ATRIP) transiently accumulate at replicating telomeres (76). Thus, replicating telomeres might be primarily recognized as unusual/stressed replication forks by Rad3-Rad26 (ATR-ATRIP) (76). Likewise, studies in mammalian cells have shown that replication of lagging-strand telomeres is significantly delayed compared to leading-strand telomeres (98), and that lagging strand telomeres carry significantly longer telomeric G-rich single-strand tails than leading strand telomeres (107). Thus, replicating telomeres are likely to accumulate high levels of RPA on the lagging strand and to activate ATR/Rad3 (Figure 1C). The Rad17-RFC complex also accumulates at telomeres during S-phase in mammalian cells (104). What about ATM kinase? In mammalian cells, ATM is maximally recruited to telomeres in G2-phase, significantly later than the timing of maximal recruitment for the MRN complex and ATR kinase (104). Given an established preference of ATM for binding to blunt ends or ends with only short ssDNA overhangs (22), ATM may be efficiently recruited to leading strand telomeres but not to lagging strand telomeres, unless lagging strand synthesis is fully completed or telomeres are processed to reduce G-tail length on the lagging strand (Figure 1C). Indeed, inactivation of TRF2, which results in strong activation of ATM (38), leads to preferential fusion among leading strand telomeres (38), consistent with the idea that ATM preferentially binds leading strand telomeres. Once telomeres are processed to re-generate the G-tail on leading strand telomeres, ATM binding may diminish and ATR may be recruited/activated, much like in the case of the ATM to ATR crosstalk observed at DSBs (17–20, 22) (Figure 1A).
Studies have not yet characterized in detail how leading strand and lagging strand DNA polymerases are coordinated when DNA replication forks encounter non-telomeric DSBs. In fact, the observed delay in lagging strand synthesis compared to leading strand synthesis at telomeres might not be unique to telomeres. While it is possible that telomere specific features, such as the presence of shelterin, CST, and/or recruitment of telomerase might be responsible for the delayed arrival of lagging strand DNA polymerases, it is just as likely that these telomere factors are contributing to minimize the delay or even promoting synthesis of the last Okazaki fragment near the 3′ ends of telomeres. Indeed, while lagging strand DNA polymerases leave large gaps near DSBs in vitro (28), both budding yeast and fission yeast cells lacking telomerase have been estimated to lose only a few bases from telomeres per every cell division (108, 109). Thus, native telomeres appear to possess telomerase-independent mechanism(s) that promote initiation of lagging strand synthesis very close to the 3′ ends of parental telomeres, rather than leaving large gaps. Efficient and timely synthesis of Okazaki fragments very close to the ends of lagging strand telomeres may also be critical for reducing ssDNA-bound RPA to attenuate checkpoint responses at telomeres. Since the CST complex in budding yeast and mammalian cells has been shown to interact with the DNA polymerase alpha-primase complex (75, 80, 81), the CST complex may be involved in ensuring that lagging strand synthesis can continue close to the 3′ end of parental telomeres. In addition, since TRF1 in mammalian cells and Taz1 in fission yeast promote efficient replication of telomeres (62, 63) and Taz1 prevents long G-tail formation (102), TRF1/Taz1 may play a critical role in regulating the arrival of leading and lagging strand polymerases at telomeres.
Simultaneous inactivation of ATM and ATR causes telomere maintenance defects in a wide range of organisms (29, 30, 110, 111). The phenotype of fission yeast cells lacking both Rad3 (ATR) and Tel1 (ATM) is especially dramatic, as these rad3 tel1 double deletion mutant cells show fusion of all three chromosomes due to telomere maintenance defects (30). In budding yeast, kinase activities of Tel1 (ATM) and Mec1 (ATR) are also important for their telomere function (112). Remarkably, the requirement for ATM and ATR kinases in telomere maintenance is conserved even in Drosophila, where retrotransposons have replaced telomerase and neither the shelterin complex nor the CST complex exist (111). In addition, the 9-1-1 complex and Rad17-RFC are required for telomerase-dependent telomere maintenance in Caenorhabditis elegans (113–115), and the 9-1-1 complex interacts with telomerase and modulates telomerase activity in mammalian cells (116). Fission yeast cells deleted for 9-1-1 subunits or Rad17 show significant telomere shortening (117, 118). Thus, the S/G2-phase recruitment of checkpoint sensors is in fact very important for telomere maintenance. However, mutations in mediators and effectors of canonical checkpoint signaling, such as Crb2, Chk1 and Cds1 in fission yeast and Rad9, Rad53 and Chk1 in budding yeast, cause very little or no effect on telomere length maintenance (117–119). Thus, telomere length regulation by checkpoint sensor proteins likely involves telomere-specific effectors that are distinct from effectors in DNA damage or DNA replication checkpoint regulation, but are phosphorylated by ATM and/or ATR.
What are telomere-specific substrates of ATM and ATR kinases? Budding yeast Cdc13 is phosphorylated by Tel1 (ATM) and Mec1 (ATR) kinases within its Est1 interaction domain, suggesting that Tel1 and Mec1 might promote interaction between Est1 and Cdc13 via phosphorylation of Cdc13 (120). Interestingly, a telomere maintenance defect observed in tel1 mec1 double mutant cells can be suppressed by deleting either Rif1 or Rif2 (negative regulators of telomerase, see Figure 2 and Table 2), or by reducing Rap1 accumulation at telomeres (121). Thus, the requirement for Tel1 (ATM) and Mec1 (ATR) in telomere maintenance could be bypassed simply by making telomeres more accessible to telomerase by removing inhibitors of telomerase (121). This might indicate that the most critical contributions of Tel1 and Mec1 in telomere maintenance are to remove telomerase inhibitors. In fact, mammalian ATM kinase has been shown to promote dissociation of TRF1, a negative regulator of telomerase, from telomeres by phosphorylating TRF1 (122). Thus, ATM and ATR kinases may have evolutionarily conserved roles in switching telomeres to a more accessible state for telomerase by promoting dissociation of telomeric repeat specific dsDNA-binding factors. In budding yeast, Mec1 and Tel1 are known to phosphorylate several DNA repair factors that are implicated in proper processing of critically short or unprotected telomeres, including Exo1 nuclease (123) and Pif1 helicase (124). Thus, ATM and ATR could also carry out their essential telomere function by modulating general DNA damage response factors.
Unlike in budding yeast, deletion of negative regulators of telomerase (Taz1, Rap1 or Rif1) is not sufficient to reverse chromosome fusions observed in tel1 rad3 double mutant fission yeast cells (92). Currently there is no known mutation that can suppress chromosome fusions in tel1 rad3 double mutant cells, and available evidence indicates that tel1 rad3 double mutant cells are defective not only in telomerase recruitment but also in telomere protection (92, 118). In particular, we have recently observed that recruitment of Ccq1 to telomeres is greatly reduced in tel1 rad3 double mutant cells. Ccq1 contributes to both inhibition of checkpoint activation at telomeres and telomerase recruitment (52, 64). Thus, we have proposed that Tel1/Rad3 and Ccq1 form a regulatory loop to ensure that telomeres that are transiently de-protected would preferentially recruit/activate Tel1/Rad3 to promote recruitment of Ccq1, and to re-establish proper protection of telomeres (92). Telomeres in fission yeast, much like in budding yeast or mammalian cells, undergo cell cycle regulated changes from a “closed” to an “open” state during S-phase (76). As it has been shown for budding yeast (35, 37, 106, 125), shorter telomeres in fission yeast are also likely to recruit/activate Tel1/Rad3 more strongly than longer telomeres. However, it is currently not known which protein(s) must be phosphorylated by Tel1/Rad3 to allow stable maintenance of telomeres in fission yeast. Furthermore, it is not yet clear whether ATM/ATR-dependent phosphorylation events may regulate the recruitment of telomerase and/or protection factors in a similar manner in mammalian cells, since a Ccq1 ortholog has not been identified to date (Figure 2 and Table 2).
The MRN/MRX complex and ATM kinase work in the same pathway to regulate telomere maintenance (111, 118, 122, 126). A conserved C-terminal motif in Nbs1 or Xrs2 interacts with ATM/Tel1 to promote recruitment of ATM/Tel1 to DSBs (12, 13, 127). In budding yeast, the MRX complex and Tel1 are preferentially recruited to critically short telomeres and promote recruitment of telomerase (37, 106, 128, 129). However, a recent study in budding yeast has shown that, while deleting as little as 20 amino acids from the C-terminus of Xrs2 is sufficient to disrupt the interaction between Tel1 and Xrs2, these mutant cells maintain significantly longer telomeres than mutant cells carrying a complete deletion of either xrs2 or tel1 (130). Therefore, budding yeast Xrs2 can contribute to telomere maintenance independently of its C-terminal Tel1 interaction domain. Even more surprisingly, Tel1 was able to contribute to telomere length maintenance independently of its kinase activity (130). In fission yeast, Rad3 (ATR) lacking its kinase domain contributes to telomere maintenance by promoting recruitment of Tel1 (ATM) to telomeres in the absence of the C-terminal 60 amino acid Tel1-interaction domain of Nbs1 (131). Therefore, these recent studies have begun to reveal novel non-kinase functions for ATM and ATR in telomere maintenance. Furthermore, much like in DSB processing (17, 23), telomere length regulation by ATM and ATR kinases is likely to involve complex crosstalks between the ATM and ATR pathways. Since lagging strand and leading strand telomeres are likely to have very distinct structures after DNA replication (Figure 1C) and also appear to be differentially processed or extended by nucleases and telomerase (98, 132), future studies must take into account how ATM, ATR and other checkpoint and DNA repair proteins could differentiate leading strand and lagging strand telomeres, in order to fully understand the functional contributions of these proteins at telomeres.
6. PERSPECTIVE
It has been over 70 years since Barbara McClintock and Hermann Muller realized that ends of chromosomes are protected from degradation, recombination and fusion events that can occur at other DSBs. The pioneering work of Drs. Elizabeth Blackburn, Jack Szostak and Carol Greider (winners of the Nobel Prize for Physiology and Medicine 2009) in the 1980s has allowed researchers to better understand the molecular nature of telomeres and how complete replication of telomeric DNA is accomplished in eukaryotic cells (133). It has become increasingly clear in recent years that telomere biology has human medical implications for various age related diseases, tumor progression and stem cell biology (134, 135). Studies have just begun to unravel mechanisms by which DNA damage checkpoint and DNA repair proteins balance their functions at telomeres. The existing evidence generally supports the notion that the DNA damage checkpoint kinases ATM and ATR primarily contribute to functional telomere maintenance by acting on telomere-specific substrates and controlling proper processing of telomeric DNA, rather than by regulating cell cycle progression. Coupled with the major advances made by researchers in the DNA repair and checkpoint fields, we should expect many more exciting discoveries in the telomere field, which may ultimately allow us to fully understand the molecular mechanisms by which DNA repair and checkpoint proteins are modulated to ensure stable maintenance of telomeres in coming years.
Acknowledgments
This work was supported by the NIH grant GM078253. Lakxmi Subramanian was also supported by a pre-doctoral fellowship from the American Heart Association and the Henrietta Lange Burk Fund. We thank Bettina A. Moser for critical reading of the manuscript.
Footnotes
This is an, un-copyedited, author manuscript that has been accepted for publication in the Frontiers in Bioscience.
Cite this article as appearing in the Journal of Frontiers in Bioscience. Full citation can be found by searching the Frontiers in Bioscience (http://bioscience.org/search/authors/htm/search.htm) following publication and at PubMed (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?CMD=search&DB=pubmed) following indexing. This article may not be duplicated or reproduced, other than for personal use or within the rule of "Fair Use of Copyrighted Materials" (section 107, Title 17, U.S. Code) without permission of the copyright holder, the Frontiers in Bioscience.
From the time of acceptance following peer review, the full final copy edited article of this manuscript will be made available at http://www.bioscience.org/. The Frontiers in Bioscience disclaims any responsibility or liability for errors or omissions in this version of the un-copyedited manuscript or in any version derived from it by the National Institutes of Health or other parties.
References
- 1.Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet. 2001;27(3):247–54. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 2.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–34. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 3.Lydall D. Taming the tiger by the tail: modulation of DNA damage responses by telomeres. EMBO J. 2009;28(15):2174–2187. doi: 10.1038/emboj.2009.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Longhese MP. DNA damage response at functional and dysfunctional telomeres. Genes Dev. 2008;22(2):125–40. doi: 10.1101/gad.1626908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Sullivan RJ, Karlseder J. Telomeres: protecting chromosomes against genome instability. Nat Rev Mol Cell Biol. 2010;11(3):171–81. doi: 10.1038/nrm2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bianchi A, Shore D. How telomerase reaches its end: mechanism of telomerase regulation by the telomeric complex. Mol Cell. 2008;31(2):153–65. doi: 10.1016/j.molcel.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 7.Stern JL, Bryan TM. Telomerase recruitment to telomeres. Cytogenet Genome Res. 2008;122(3–4):243–54. doi: 10.1159/000167810. [DOI] [PubMed] [Google Scholar]
- 8.Harrison JC, Haber JE. Surviving the breakup: the DNA damage checkpoint. Annu Rev Genet. 2006;40:209–35. doi: 10.1146/annurev.genet.40.051206.105231. [DOI] [PubMed] [Google Scholar]
- 9.Melo J, Toczyski D. A unified view of the DNA-damage checkpoint. Curr Opin Cell Biol. 2002;14(2):237–45. doi: 10.1016/s0955-0674(02)00312-5. [DOI] [PubMed] [Google Scholar]
- 10.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432(7015):316–23. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 11.Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308(5721):551–4. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 12.Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434(7033):605–11. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 13.You Z, Chahwan C, Bailis J, Hunter T, Russell P. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol Cell Biol. 2005;25(13):5363–79. doi: 10.1128/MCB.25.13.5363-5379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dupre A, Boyer-Chatenet L, Gautier J. Two-step activation of ATM by DNA and the Mre11-Rad50-Nbs1 complex. Nat Struct Mol Biol. 2006;13(5):451–7. doi: 10.1038/nsmb1090. [DOI] [PubMed] [Google Scholar]
- 15.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316(5828):1160–6. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 16.Shiloh Y. The ATM-mediated DNA-damage response: taking shape. Trends Biochem Sci. 2006;31(7):402–10. doi: 10.1016/j.tibs.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 17.Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol. 2006;8(1):37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- 18.Myers JS, Cortez D. Rapid activation of ATR by ionizing radiation requires ATM and Mre11. J Biol Chem. 2006;281(14):9346–50. doi: 10.1074/jbc.M513265200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cuadrado M, Martinez-Pastor B, Murga M, Toledo LI, Gutierrez-Martinez P, Lopez E, Fernandez-Capetillo O. ATM regulates ATR chromatin loading in response to DNA double-strand breaks. J Exp Med. 2006;203(2):297–303. doi: 10.1084/jem.20051923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adams KE, Medhurst AL, Dart DA, Lakin ND. Recruitment of ATR to sites of ionising radiation-induced DNA damage requires ATM and components of the MRN protein complex. Oncogene. 2006;25(28):3894–904. doi: 10.1038/sj.onc.1209426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300(5625):1542–8. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 22.Shiotani B, Zou L. Single-stranded DNA orchestrates an ATM-to-ATR switch at DNA breaks. Mol Cell. 2009;33(5):547–58. doi: 10.1016/j.molcel.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stiff T, Walker SA, Cerosaletti K, Goodarzi AA, Petermann E, Concannon P, O’Driscoll M, Jeggo PA. ATR-dependent phosphorylation and activation of ATM in response to UV treatment or replication fork stalling. EMBO J. 2006;25(24):5775–82. doi: 10.1038/sj.emboj.7601446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dore AS, Kilkenny ML, Rzechorzek NJ, Pearl LH. Crystal structure of the rad9-rad1-hus1 DNA damage checkpoint complex--implications for clamp loading and regulation. Mol Cell. 2009;34(6):735–45. doi: 10.1016/j.molcel.2009.04.027. [DOI] [PubMed] [Google Scholar]
- 25.Mordes DA, Glick GG, Zhao R, Cortez D. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev. 2008;22(11):1478–89. doi: 10.1101/gad.1666208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Majka J, Niedziela-Majka A, Burgers PM. The checkpoint clamp activates Mec1 kinase during initiation of the DNA damage checkpoint. Mol Cell. 2006;24(6):891–901. doi: 10.1016/j.molcel.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doksani Y, Bermejo R, Fiorani S, Haber JE, Foiani M. Replicon dynamics, dormant origin firing, and terminal fork integrity after double-strand break formation. Cell. 2009;137(2):247–58. doi: 10.1016/j.cell.2009.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohki R, Tsurimoto T, Ishikawa F. In vitro reconstitution of the end replication problem. Mol Cell Biol. 2001;21(17):5753–66. doi: 10.1128/MCB.21.17.5753-5766.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ritchie KB, Mallory JC, Petes TD. Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1999;19(9):6065–75. doi: 10.1128/mcb.19.9.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naito T, Matsuura A, Ishikawa F. Circular chromosome formation in a fission yeast mutant defective in two ATM homologues. Nat Genet. 1998;20(2):203–6. doi: 10.1038/2517. [DOI] [PubMed] [Google Scholar]
- 31.Bi X, Srikanta D, Fanti L, Pimpinelli S, Badugu R, Kellum R, Rong YS. Drosophila ATM and ATR checkpoint kinases control partially redundant pathways for telomere maintenance. Proc Natl Acad Sci U S A. 2005;102(42):15167–72. doi: 10.1073/pnas.0504981102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oikemus SR, Queiroz-Machado J, Lai K, McGinnis N, Sunkel C, Brodsky MH. Epigenetic telomere protection by Drosophila DNA damage response pathways. PLoS Genet. 2006;2(5):e71. doi: 10.1371/journal.pgen.0020071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bianchi A, Shore D. Early replication of short telomeres in budding yeast. Cell. 2007;128(6):1051–62. doi: 10.1016/j.cell.2007.01.041. [DOI] [PubMed] [Google Scholar]
- 34.Gilson E, Geli V. How telomeres are replicated. Nat Rev Mol Cell Biol. 2007;8(10):825–38. doi: 10.1038/nrm2259. [DOI] [PubMed] [Google Scholar]
- 35.Abdallah P, Luciano P, Runge KW, Lisby M, Geli V, Gilson E, Teixeira MT. A two-step model for senescence triggered by a single critically short telomere. Nat Cell Biol. 2009;11(8):988–93. doi: 10.1038/ncb1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vodenicharov MD, Wellinger RJ. DNA degradation at unprotected telomeres in yeast is regulated by the CDK1 (Cdc28/Clb) cell-cycle kinase. Mol Cell. 2006;24(1):127–37. doi: 10.1016/j.molcel.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 37.Viscardi V, Bonetti D, Cartagena-Lirola H, Lucchini G, Longhese MP. MRX-dependent DNA damage response to short telomeres. Mol Biol Cell. 2007;18(8):3047–58. doi: 10.1091/mbc.E07-03-0285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448(7157):1068–71. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- 39.Churikov D, Price CM. Pot1 and cell cycle progression cooperate in telomere length regulation. Nat Struct Mol Biol. 2008;15(1):79–84. doi: 10.1038/nsmb1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karlseder J, Hoke K, Mirzoeva OK, Bakkenist C, Kastan MB, Petrini JH, de Lange T. The telomeric protein TRF2 binds the ATM kinase and can inhibit the ATM-dependent DNA damage response. PLoS Biol. 2004;2(8):E240. doi: 10.1371/journal.pbio.0020240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buscemi G, Zannini L, Fontanella E, Lecis D, Lisanti S, Delia D. The shelterin protein TRF2 inhibits Chk2 activity at telomeres in the absence of DNA damage. Curr Biol. 2009;19(10):874–9. doi: 10.1016/j.cub.2009.03.064. [DOI] [PubMed] [Google Scholar]
- 42.Zhu XD, Kuster B, Mann M, Petrini JH, de Lange T. Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat Genet. 2000;25(3):347–52. doi: 10.1038/77139. [DOI] [PubMed] [Google Scholar]
- 43.van Overbeek M, de Lange T. Apollo, an Artemis-related nuclease, interacts with TRF2 and protects human telomeres in S phase. Curr Biol. 2006;16(13):1295–302. doi: 10.1016/j.cub.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 44.Lenain C, Bauwens S, Amiard S, Brunori M, Giraud-Panis MJ, Gilson E. The Apollo 5′ exonuclease functions together with TRF2 to protect telomeres from DNA repair. Curr Biol. 2006;16(13):1303–10. doi: 10.1016/j.cub.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 45.Kim H, Lee OH, Xin H, Chen LY, Qin J, Chae HK, Lin SY, Safari A, Liu D, Songyang Z. TRF2 functions as a protein hub and regulates telomere maintenance by recognizing specific peptide motifs. Nat Struct Mol Biol. 2009;16(4):372–9. doi: 10.1038/nsmb.1575. [DOI] [PubMed] [Google Scholar]
- 46.Chen Y, Yang Y, van Overbeek M, Donigian JR, Baciu P, de Lange T, Lei M. A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins. Science. 2008;319(5866):1092–6. doi: 10.1126/science.1151804. [DOI] [PubMed] [Google Scholar]
- 47.Hockemeyer D, Daniels JP, Takai H, de Lange T. Recent expansion of the telomeric complex in rodents: Two distinct POT1 proteins protect mouse telomeres. Cell. 2006;126(1):63–77. doi: 10.1016/j.cell.2006.04.044. [DOI] [PubMed] [Google Scholar]
- 48.Wu L, Multani AS, He H, Cosme-Blanco W, Deng Y, Deng JM, Bachilo O, Pathak S, Tahara H, Bailey SM, Behringer RR, Chang S. Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell. 2006;126(1):49–62. doi: 10.1016/j.cell.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 49.Bae NS, Baumann P. A RAP1/TRF2 complex inhibits nonhomologous end-joining at human telomeric DNA ends. Mol Cell. 2007;26(3):323–34. doi: 10.1016/j.molcel.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 50.Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97(4):503–14. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 51.Poulet A, Buisson R, Faivre-Moskalenko C, Koelblen M, Amiard S, Montel F, Cuesta-Lopez S, Bornet O, Guerlesquin F, Godet T, Moukhtar J, Argoul F, Declais AC, Lilley DM, Ip SC, West SC, Gilson E, Giraud-Panis MJ. TRF2 promotes, remodels and protects telomeric Holliday junctions. EMBO J. 2009;28(6):641–51. doi: 10.1038/emboj.2009.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miyoshi T, Kanoh J, Saito M, Ishikawa F. Fission yeast Pot1-Tpp1 protects telomeres and regulates telomere length. Science. 2008;320(5881):1341–4. doi: 10.1126/science.1154819. [DOI] [PubMed] [Google Scholar]
- 53.Silverman J, Takai H, Buonomo SB, Eisenhaber F, de Lange T. Human Rif1, ortholog of a yeast telomeric protein, is regulated by ATM and 53BP1 and functions in the S-phase checkpoint. Genes Dev. 2004;18(17):2108–19. doi: 10.1101/gad.1216004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu L, Blackburn EH. Human Rif1 protein binds aberrant telomeres and aligns along anaphase midzone microtubules. J Cell Biol. 2004;167(5):819–30. doi: 10.1083/jcb.200408181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kanoh J, Ishikawa F. spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr Biol. 2001;11(20):1624–30. doi: 10.1016/s0960-9822(01)00503-6. [DOI] [PubMed] [Google Scholar]
- 56.Miller KM, Cooper JP. The telomere protein Taz1 is required to prevent and repair genomic DNA breaks. Mol Cell. 2003;11(2):303–13. doi: 10.1016/s1097-2765(03)00041-8. [DOI] [PubMed] [Google Scholar]
- 57.Cooper JP, Nimmo ER, Allshire RC, Cech TR. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature. 1997;385(6618):744–7. doi: 10.1038/385744a0. [DOI] [PubMed] [Google Scholar]
- 58.Ferreira MG, Cooper JP. The fission yeast Taz1 protein protects chromosomes from Ku-dependent end-to-end fusions. Mol Cell. 2001;7(1):55–63. doi: 10.1016/s1097-2765(01)00154-x. [DOI] [PubMed] [Google Scholar]
- 59.Ferreira MG, Cooper JP. Two modes of DNA double-strand break repair are reciprocally regulated through the fission yeast cell cycle. Genes Dev. 2004;18(18):2249–54. doi: 10.1101/gad.315804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miller KM, Ferreira MG, Cooper JP. Taz1, Rap1 and Rif1 act both interdependently and independently to maintain telomeres. EMBO J. 2005;24(17):3128–35. doi: 10.1038/sj.emboj.7600779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Subramanian L, Moser BA, Nakamura TM. Recombination-based telomere maintenance is dependent on Tel1-MRN and Rap1 and inhibited by telomerase, Taz1, and Ku in fission yeast. Mol Cell Biol. 2008;28(5):1443–55. doi: 10.1128/MCB.01614-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miller KM, Rog O, Cooper JP. Semi-conservative DNA replication through telomeres requires Taz1. Nature. 2006;440(7085):824–8. doi: 10.1038/nature04638. [DOI] [PubMed] [Google Scholar]
- 63.Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138(1):90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tomita K, Cooper JP. Fission yeast Ccq1 is telomerase recruiter and local checkpoint controller. Genes Dev. 2008;22(24):3461–74. doi: 10.1101/gad.498608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pardo B, Marcand S. Rap1 prevents telomere fusions by nonhomologous end joining. EMBO J. 2005;24(17):3117–27. doi: 10.1038/sj.emboj.7600778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marcand S, Pardo B, Gratias A, Cahun S, Callebaut I. Multiple pathways inhibit NHEJ at telomeres. Genes Dev. 2008;22(9):1153–8. doi: 10.1101/gad.455108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grandin N, Damon C, Charbonneau M. Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. EMBO J. 2001;20(5):1173–83. doi: 10.1093/emboj/20.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Garvik B, Carson M, Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol Cell Biol. 1995;15(11):6128–38. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lin JJ, Zakian VA. The Saccharomyces CDC13 protein is a single-strand TG1–3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo. Proc Natl Acad Sci USA. 1996;93(24):13760–5. doi: 10.1073/pnas.93.24.13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nugent CI, Hughes TR, Lue NF, Lundblad V. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science. 1996;274(5285):249–52. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- 71.Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V. RPA-like proteins mediate yeast telomere function. Nat Struct Mol Biol. 2007;14(3):208–14. doi: 10.1038/nsmb1205. [DOI] [PubMed] [Google Scholar]
- 72.Miyake Y, Nakamura M, Nabetani A, Shimamura S, Tamura M, Yonehara S, Saito M, Ishikawa F. RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol Cell. 2009;36(2):193–206. doi: 10.1016/j.molcel.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 73.Surovtseva YV, Churikov D, Boltz KA, Song X, Lamb JC, Warrington R, Leehy K, Heacock M, Price CM, Shippen DE. Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol Cell. 2009;36(2):207–18. doi: 10.1016/j.molcel.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Martin V, Du LL, Rozenzhak S, Russell P. Protection of telomeres by a conserved Stn1-Ten1 complex. Proc Natl Acad Sci USA. 2007;104(35):14038–43. doi: 10.1073/pnas.0705497104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Casteel DE, Zhuang S, Zeng Y, Perrino FW, Boss GR, Goulian M, Pilz RB. A DNA polymerase-α primase cofactor with homology to replication protein A-32 regulates DNA replication in mammalian cells. J Biol Chem. 2009;284(9):5807–18. doi: 10.1074/jbc.M807593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Moser BA, Subramanian L, Chang YT, Noguchi C, Noguchi E, Nakamura TM. Differential arrival of leading and lagging strand DNA polymerases at fission yeast telomeres. EMBO J. 2009;28(7):810–20. doi: 10.1038/emboj.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wan M, Qin J, Songyang Z, Liu D. OB fold-containing protein 1 (OBFC1), a human homolog of yeast Stn1, associates with TPP1 and is implicated in telomere length regulation. J Biol Chem. 2009;284(39):26725–31. doi: 10.1074/jbc.M109.021105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sun J, Yu EY, Yang Y, Confer LA, Sun SH, Wan K, Lue NF, Lei M. Stn1-Ten1 is an Rpa2-Rpa3-like complex at telomeres. Genes Dev. 2009;23(24):2900–14. doi: 10.1101/gad.1851909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gelinas AD, Paschini M, Reyes FE, Heroux A, Batey RT, Lundblad V, Wuttke DS. Telomere capping proteins are structurally related to RPA with an additional telomere-specific domain. Proc Natl Acad Sci U S A. 2009;106(46):19298–303. doi: 10.1073/pnas.0909203106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Puglisi A, Bianchi A, Lemmens L, Damay P, Shore D. Distinct roles for yeast Stn1 in telomere capping and telomerase inhibition. EMBO J. 2008;27(17):2328–39. doi: 10.1038/emboj.2008.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Qi H, Zakian VA. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase α and the telomerase-associated Est1 protein. Genes Dev. 2000;14(14):1777–88. [PMC free article] [PubMed] [Google Scholar]
- 82.Petreaca RC, Chiu HC, Eckelhoefer HA, Chuang C, Xu L, Nugent CI. Chromosome end protection plasticity revealed by Stn1p and Ten1p bypass of Cdc13p. Nat Cell Biol. 2006;8(7):748–55. doi: 10.1038/ncb1430. [DOI] [PubMed] [Google Scholar]
- 83.Grossi S, Puglisi A, Dmitriev PV, Lopes M, Shore D. Pol12, the B subunit of DNA polymerase alpha, functions in both telomere capping and length regulation. Genes Dev. 2004;18(9):992–1006. doi: 10.1101/gad.300004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Weinert TA, Kiser GL, Hartwell LH. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 1994;8(6):652–65. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- 85.Zubko MK, Lydall D. Linear chromosome maintenance in the absence of essential telomere-capping proteins. Nat Cell Biol. 2006;8(7):734–40. doi: 10.1038/ncb1428. [DOI] [PubMed] [Google Scholar]
- 86.Smogorzewska A, de Lange T. Regulation of telomerase by telomeric proteins. Annu Rev Biochem. 2004;73:177–208. doi: 10.1146/annurev.biochem.73.071403.160049. [DOI] [PubMed] [Google Scholar]
- 87.Loayza D, De Lange T. POT1 as a terminal transducer of TRF1 telomere length control. Nature. 2003;423(6943):1013–8. doi: 10.1038/nature01688. [DOI] [PubMed] [Google Scholar]
- 88.Kendellen MF, Barrientos KS, Counter CM. POT1 association with TRF2 regulates telomere length. Mol Cell Biol. 2009;29(20):5611–9. doi: 10.1128/MCB.00286-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.O’Connor MS, Safari A, Xin H, Liu D, Songyang Z. A critical role for TPP1 and TIN2 interaction in high-order telomeric complex assembly. Proc Natl Acad Sci USA. 2006;103(32):11874–9. doi: 10.1073/pnas.0605303103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445(7127):506–10. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- 91.Xin H, Liu D, Wan M, Safari A, Kim H, Sun W, O’Connor MS, Songyang Z. TPP1 is a homologue of ciliate TEBP-β and interacts with POT1 to recruit telomerase. Nature. 2007;445(7127):559–62. doi: 10.1038/nature05469. [DOI] [PubMed] [Google Scholar]
- 92.Moser BA, Subramanian L, Khair L, Chang YT, Nakamura TM. Fission yeast Tel1ATM and Rad3ATR promote telomere protection and telomerase recruitment. PLoS Genet. 2009;5(8):e1000622. doi: 10.1371/journal.pgen.1000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chan A, Boule JB, Zakian VA. Two pathways recruit telomerase to Saccharomyces cerevisiae telomeres. PLoS Genet. 2008;4(10):e1000236. doi: 10.1371/journal.pgen.1000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Evans SK, Lundblad V. Est1 and Cdc13 as comediators of telomerase access. Science. 1999;286(5437):117–20. doi: 10.1126/science.286.5437.117. [DOI] [PubMed] [Google Scholar]
- 95.Pennock E, Buckley K, Lundblad V. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell. 2001;104(3):387–96. doi: 10.1016/s0092-8674(01)00226-4. [DOI] [PubMed] [Google Scholar]
- 96.Tomita K, Kibe T, Kang HY, Seo YS, Uritani M, Ushimaru T, Ueno M. Fission yeast Dna2 is required for generation of the telomeric single-strand overhang. Mol Cell Biol. 2004;24(21):9557–67. doi: 10.1128/MCB.24.21.9557-9567.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wellinger RJ, Wolf AJ, Zakian VA. Saccharomyces telomeres acquire single-strand TG1–3 tails late in S phase. Cell. 1993;72(1):51–60. doi: 10.1016/0092-8674(93)90049-v. [DOI] [PubMed] [Google Scholar]
- 98.Zhao Y, Sfeir AJ, Zou Y, Buseman CM, Chow TT, Shay JW, Wright WE. Telomere extension occurs at most chromosome ends and is uncoupled from fill-in in human cancer cells. Cell. 2009;138(3):463–75. doi: 10.1016/j.cell.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Larrivee M, LeBel C, Wellinger RJ. The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex. Genes Dev. 2004;18(12):1391–6. doi: 10.1101/gad.1199404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wellinger RJ, Ethier K, Labrecque P, Zakian VA. Evidence for a new step in telomere maintenance. Cell. 1996;85(3):423–33. doi: 10.1016/s0092-8674(00)81120-4. [DOI] [PubMed] [Google Scholar]
- 101.Bonetti D, Martina M, Clerici M, Lucchini G, Longhese MP. Multiple pathways regulate 3′ overhang generation at S. cerevisiae telomeres. Mol Cell. 2009;35(1):70–81. doi: 10.1016/j.molcel.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 102.Tomita K, Matsuura A, Caspari T, Carr AM, Akamatsu Y, Iwasaki H, Mizuno K, Ohta K, Uritani M, Ushimaru T, Yoshinaga K, Ueno M. Competition between the Rad50 complex and the Ku heterodimer reveals a role for Exo1 in processing double-strand breaks but not telomeres. Mol Cell Biol. 2003;23(15):5186–97. doi: 10.1128/MCB.23.15.5186-5197.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Takata H, Tanaka Y, Matsuura A. Late S phase-specific recruitment of Mre11 complex triggers hierarchical assembly of telomere replication proteins in Saccharomyces cerevisiae. Mol Cell. 2005;17(4):573–83. doi: 10.1016/j.molcel.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 104.Verdun RE, Karlseder J. The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell. 2006;127(4):709–20. doi: 10.1016/j.cell.2006.09.034. [DOI] [PubMed] [Google Scholar]
- 105.Jady BE, Richard P, Bertrand E, Kiss T. Cell cycle-dependent recruitment of telomerase RNA and Cajal bodies to human telomeres. Mol Biol Cell. 2006;17(2):944–54. doi: 10.1091/mbc.E05-09-0904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sabourin M, Tuzon CT, Zakian VA. Telomerase and Tel1p preferentially associate with short telomeres in S. cerevisiae. Mol Cell. 2007;27(4):550–61. doi: 10.1016/j.molcel.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chai W, Du Q, Shay JW, Wright WE. Human telomeres have different overhang sizes at leading versus lagging strands. Mol Cell. 2006;21(3):427–35. doi: 10.1016/j.molcel.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 108.Singer MS, Gottschling DE. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266(5184):404–9. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 109.Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, Lingner J, Harley CB, Cech TR. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277(5328):955–9. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 110.Vespa L, Couvillion M, Spangler E, Shippen DE. ATM and ATR make distinct contributions to chromosome end protection and the maintenance of telomeric DNA in Arabidopsis. Genes Dev. 2005;19(18):2111–5. doi: 10.1101/gad.1333805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ciapponi L, Cenci G. Telomere capping and cellular checkpoints: clues from fruit flies. Cytogenet Genome Res. 2008;122(3–4):365–73. doi: 10.1159/000167824. [DOI] [PubMed] [Google Scholar]
- 112.Mallory JC, Petes TD. Protein kinase activity of Tel1p and Mec1p, two Saccharomyces cerevisiae proteins related to the human ATM protein kinase. Proc Natl Acad Sci USA. 2000;97(25):13749–54. doi: 10.1073/pnas.250475697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ahmed S, Hodgkin J. MRT-2 checkpoint protein is required for germline immortality and telomere replication in C. elegans. Nature. 2000;403(6766):159–64. doi: 10.1038/35003120. [DOI] [PubMed] [Google Scholar]
- 114.Hofmann ER, Milstein S, Boulton SJ, Ye M, Hofmann JJ, Stergiou L, Gartner A, Vidal M, Hengartner MO. Caenorhabditis elegans HUS-1 is a DNA damage checkpoint protein required for genome stability and EGL-1-mediated apoptosis. Curr Biol. 2002;12(22):1908–18. doi: 10.1016/s0960-9822(02)01262-9. [DOI] [PubMed] [Google Scholar]
- 115.Boerckel J, Walker D, Ahmed S. The Caenorhabditis elegans Rad17 homolog HPR-17 is required for telomere replication. Genetics. 2007;176(1):703–9. doi: 10.1534/genetics.106.070201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Francia S, Weiss RS, Hande MP, Freire R, d’Adda di Fagagna F. Telomere and telomerase modulation by the mammalian Rad9/Rad1/Hus1 DNA-damage-checkpoint complex. Curr Biol. 2006;16(15):1551–8. doi: 10.1016/j.cub.2006.06.066. [DOI] [PubMed] [Google Scholar]
- 117.Dahlen M, Olsson T, Kanter-Smoler G, Ramne A, Sunnerhagen P. Regulation of telomere length by checkpoint genes in Schizosaccharomyces pombe. Mol Biol Cell. 1998;9(3):611–21. doi: 10.1091/mbc.9.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nakamura TM, Moser BA, Russell P. Telomere binding of checkpoint sensor and DNA repair proteins contributes to maintenance of functional fission yeast telomeres. Genetics. 2002;161(4):1437–52. doi: 10.1093/genetics/161.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Longhese MP, Paciotti V, Neecke H, Lucchini G. Checkpoint proteins influence telomeric silencing and length maintenance in budding yeast. Genetics. 2000;155(4):1577–91. doi: 10.1093/genetics/155.4.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tseng SF, Lin JJ, Teng SC. The telomerase-recruitment domain of the telomere binding protein Cdc13 is regulated by Mec1p/Tel1p-dependent phosphorylation. Nucleic Acids Res. 2006;34(21):6327–36. doi: 10.1093/nar/gkl786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chan SW, Chang J, Prescott J, Blackburn EH. Altering telomere structure allows telomerase to act in yeast lacking ATM kinases. Curr Biol. 2001;11(16):1240–50. doi: 10.1016/s0960-9822(01)00391-8. [DOI] [PubMed] [Google Scholar]
- 122.Wu Y, Xiao S, Zhu XD. MRE11-RAD50-NBS1 and ATM function as co-mediators of TRF1 in telomere length control. Nat Struct Mol Biol. 2007;14(9):832–40. doi: 10.1038/nsmb1286. [DOI] [PubMed] [Google Scholar]
- 123.Morin I, Ngo HP, Greenall A, Zubko MK, Morrice N, Lydall D. Checkpoint-dependent phosphorylation of Exo1 modulates the DNA damage response. EMBO J. 2008;27(18):2400–10. doi: 10.1038/emboj.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Makovets S, Blackburn EH. DNA damage signalling prevents deleterious telomere addition at DNA breaks. Nat Cell Biol. 2009;11(11):1383–6. doi: 10.1038/ncb1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hirano Y, Fukunaga K, Sugimoto K. Rif1 and rif2 inhibit localization of tel1 to DNA ends. Mol Cell. 2009;33(3):312–22. doi: 10.1016/j.molcel.2008.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ritchie KB, Petes TD. The Mre11p/Rad50p/Xrs2p complex and the Tel1p function in a single pathway for telomere maintenance in yeast. Genetics. 2000;155(1):475–9. doi: 10.1093/genetics/155.1.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Nakada D, Matsumoto K, Sugimoto K. ATM-related Tel1 associates with double-strand breaks through an Xrs2-dependent mechanism. Genes Dev. 2003;17(16):1957–62. doi: 10.1101/gad.1099003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bianchi A, Shore D. Increased association of telomerase with short telomeres in yeast. Genes Dev. 2007;21(14):1726–30. doi: 10.1101/gad.438907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Goudsouzian LK, Tuzon CT, Zakian VA. S. cerevisiae Tel1p and Mre11p are required for normal levels of Est1p and Est2p telomere association. Mol Cell. 2006;24(4):603–10. doi: 10.1016/j.molcel.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 130.Ma Y, Greider CW. Kinase-independent functions of TEL1 in telomere maintenance. Mol Cell Biol. 2009;29(18):5193–202. doi: 10.1128/MCB.01896-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Subramanian L, Nakamura TM. A kinase-independent role for the Rad3ATR-Rad26ATRIP complex in recruitment of Tel1ATM to telomeres in fission yeast. PLoS Genet. 2010;6(2):e1000839. doi: 10.1371/journal.pgen.1000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chai W, Sfeir AJ, Hoshiyama H, Shay JW, Wright WE. The involvement of the Mre11/Rad50/Nbs1 complex in the generation of G-overhangs at human telomeres. EMBO Rep. 2006;7(2):225–30. doi: 10.1038/sj.embor.7400600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Blackburn EH, Greider CW, Szostak JW. Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging. Nat Med. 2006;12(10):1133–8. doi: 10.1038/nm1006-1133. [DOI] [PubMed] [Google Scholar]
- 134.Lansdorp PM. Telomeres and disease. EMBO J. 2009;28(17):2532–40. doi: 10.1038/emboj.2009.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Blasco MA. Telomeres and human disease: ageing, cancer and beyond. Nat Rev Genet. 2005;6(8):611–22. doi: 10.1038/nrg1656. [DOI] [PubMed] [Google Scholar]