Abstract
Motivation:Granzyme B (GrB) and caspases cleave specific protein substrates to induce apoptosis in virally infected and neoplastic cells. While substrates for both types of proteases have been determined experimentally, there are many more yet to be discovered in humans and other metazoans. Here, we present a bioinformatics method based on support vector machine (SVM) learning that identifies sequence and structural features important for protease recognition of substrate peptides and then uses these features to predict novel substrates. Our approach can act as a convenient hypothesis generator, guiding future experiments by high-confidence identification of peptide-protein partners.
Results:The method is benchmarked on the known substrates of both protease types, including our literature-curated GrB substrate set (GrBah). On these benchmark sets, the method outperforms a number of other methods that consider sequence only, predicting at a 0.87 true positive rate (TPR) and a 0.13 false positive rate (FPR) for caspase substrates, and a 0.79 TPR and a 0.21 FPR for GrB substrates. The method is then applied to ∼25 000 proteins in the human proteome to generate a ranked list of predicted substrates of each protease type. Two of these predictions, AIF-1 and SMN1, were selected for further experimental analysis, and each was validated as a GrB substrate.
Availability: All predictions for both protease types are publically available at http://salilab.org/peptide. A web server is at the same site that allows a user to train new SVM models to make predictions for any protein that recognizes specific oligopeptide ligands.
Contact: craik@cgl.ucsf.edu; sali@salilab.org
Supplementary information: Supplementary data are available at Bioinformatics online
1 INTRODUCTION
Protein–peptide specificity is an important biological phenomenon. There are many systems in which a protein recognizes a specific amino acid sequence, often falling on a flexible, solvent-exposed region of another protein. Such systems include the modular scaffolding PDZ domains, which recognize specific sequences on the C-terminal tails of their substrates (Jemth and Gianni, 2007); multifunctional SH3 domains, which recognize a linear motif of the form Pro-Xaa-Xaa-Pro (Kaneko et al., 2008); and class I MHC proteins, which bind nine-residue peptides with specificity varying across different MHC molecules (Sieker et al., 2009).
Here, we focus on the protein–peptide specificity of the pro-apoptotic proteases granzyme B (GrB) and caspases interacting with their respective protein substrates. GrB is a serine protease delivered by natural killer cells into virally infected and tumor cells (Pardo et al., 2009; Russell and Ley, 2002). The caspases are a family of endogenous cysteine proteases activated by extracellular death ligands and environmental stresses (Nicholson and Thornberry, 2003). Both protease types recognize and cleave specific peptide sequences containing an aspartic acid residue on their target substrates, activating different pathways that lead to cell death. Identifying these substrates has led to a wealth of knowledge about how the proteases contribute to apoptosis, how the cleavage events lead to cell death, and which substrates to target for therapeutic purposes.
Substrates of the two protease types have been discovered with a variety of experimental techniques, ranging from low-throughput gel-based methods to proteomic efforts that can identify hundreds of cleaved proteins (Bredemeyer et al., 2005; Casciola-Rosen et al., 1999; Dix et al., 2008; Mahrus et al., 2008). However, different datasets overlap only partially, indicating that many substrates remain to be identified. For example, two proteomics studies, respectively, reported 261 and 292 caspase cleavage sequences, although the high-confidence overlap between the two sets was only 64 [Fig. 1A in Johnson and Kornbluth (2008)].
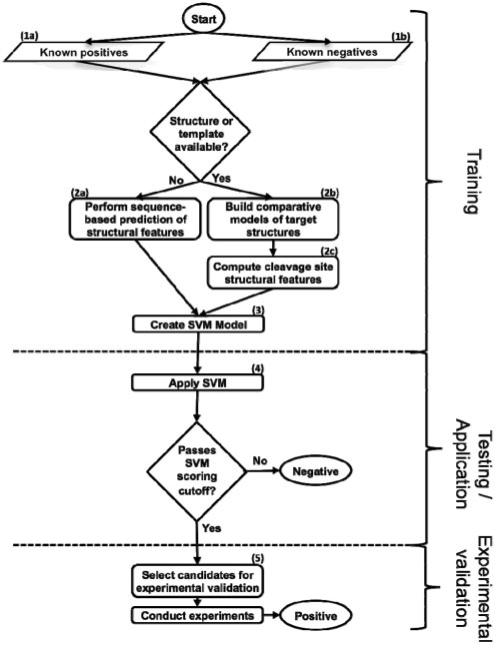
Fig. 1.
Flowchart of procedure. Peptides are scored with the SVM trained on sequence and structure features; the peptides that pass the cutoffs derived from benchmarking are the final candidates for experimental validation.
To reduce this gap, accurate computational techniques could be used to predict protein–peptide interactions for guiding further focused experiments. A number of approaches have been taken in the systems described. For PDZ interactions, examples of such methods include position-specific scoring matrices (PSSMs; Stiffler et al., 2007) and Bayesian inference (Chen et al., 2008). SH3 binding partners have been predicted with neural networks (Ferraro et al., 2005; Zhang et al., 2006) and MHC class I interactions have been predicted with support vector machines (SVMs; Jacob and Vert, 2008). Finally, molecular docking methods have been developed to analyze both systems (Bui et al., 2006; Hou et al., 2009).
Computational methods have also been applied to predict substrates recognized by GrB and caspases. These methods take advantage of both protease types having a near-absolute requirement for Asp at the P1 position, while allowing degenerate preference for different residue types in the positions immediately surrounding P1. These studies rely on fixed sequence searches. (Wilkins et al., 1999), PSSMs based on frequencies of residue types in known cleavage sites (Garay-Malpartida et al., 2005; Lohmüller et al., 2003; Verspurten et al., 2009) and positional-scanning combinatorial substrate libraries (PS-SCLs; Backes et al., 2005; Boyd et al., 2005), SVMs using residue composition around the cleavage site (Wee et al., 2006), and Bayesian neural networks (Yang, 2005).
Cleavage sequences for both GrB and caspases are generally thought to occur on flexible, disordered regions of substrates (Hubbard, 1998). However, it was previously shown in an analysis of caspase substrate structures that many of these known cleavage sites are in α-helices and even occasionally on β-strands (Mahrus et al., 2008; Timmer et al., 2009). This observation motivates the choice of a machine learning algorithm that relies on the structure as well as sequence information. Here, we describe such a protocol incorporating SVM learning. The method is trained and benchmarked on separate pools consisting of known GrB and caspase cleavage sequences. It is then applied to the human proteome to generate a list of high-confidence predictions for experimental validation. Two such candidates are the proteins AIF-1 and SMN1, which are experimentally validated as being cleaved by GrB. The approach has the potential to provide greater coverage of substrates for both GrB and caspases, and can be easily adapted to other protein–peptide systems through our web server that can learn from any user-supplied protein–peptide training set.
2 METHODS
2.1 Structural characteristics of sequences
Datasets of known cleavage sequences were compiled for benchmarking, and all human proteome octapeptides with Asp in the fourth position were processed for the application step (Section 3). Comparative models were generated by the automated modeling pipeline ModPipe (Pieper et al., 2009), and only good quality models [those predicted to have >80% of their C-α atoms within 3.5 Å of the native state, as assessed by the model evaluation algorithm TSVMod (Eramian et al., 2008)] were considered (Fig. 1, step 2b). It has been previously shown that secondary structure features computed from accurate comparative models are similar to those for crystallographic structures (Chakravarty and Sanchez, 2004). For a solved structure or a comparative model, the DSSP program was used to assess secondary structure (mapping results ‘H’, ‘G’ and ‘I’ to α-helix; ‘B’ and ‘E’ to β-sheet; and ‘S’, ‘T’ and ‘L’ to loop) and solvent accessibility (Kabsch and Sander, 1983; Fig. 1, step 2c). When a structure or model was not available, sequence-based algorithms were used to predict secondary structure (Fig. 1, step 2a; Jones, 1999). A sequence-based algorithm was also used to predict disorder on all known substrates regardless of whether a structure or model was available (Jones and Ward, 2003). A cleavage sequence was defined as being in a loop if four or more of its residues were predicted to be in this conformation, devoid of regular secondary structure; similarly, a cleavage sequence was defined to be solvent accessible if four or more of its residues were >16% exposed to solvent (Kabsch and Sander, 1983). Error bars represent two times an SD, which is calculated for a binomial experiment with (n* p* (1−p))1/2; values for n can be found in Supplementary Figure 1a. Training on octapeptides spanning P4 to P4′ gave the best performance relative to peptides of other lengths and positions (data not shown).
2.2 Scoring of potential cleavage sites by an SVM
SVMs are machine learning algorithms that can be used for classification. They create a kernel function hypersurface that maximally separates two sets of n-dimensional training set (i.e. classified) vectors, followed by predicting an unclassified vector as falling on one side or the other of the separation. Each dimension in the vector is a feature number, which has a corresponding value. Here, a single cleavage sequence had eight features representing its oligopeptide sequence. Each residue was assigned a feature number by the formula n*20+i, where n represents the zero-based position in the peptide sequence of the residue and i represents the position of the residue in a zero-based alphabetical ordering of all residues. Thus, a glutamate (i = 3) in the second position (n = 1) would have the feature number 23. The value for all sequence features was 1.
The outputs of the structural assessment algorithms were used to create additional features for each cleavage sequence. Each of these algorithms assigned a value to each residue in the cleavage sequence. The program Disopred outputs values from 0 to 1 that correspond to the predicted degrees of disorder. DSSP outputs a calculated solvent accessibility fraction and both DSSP and PSI-PRED output a predicted structure type of loop, α-helix or β-sheet. These algorithms each added eight features to a cleavage sequence, where the structure types were assigned the values 1, 2 and 3 corresponding to loop, helix and sheet, respectively, and the other values were the raw score outputs of the algorithms.
The SVM-light software was used to execute the SVM algorithm (Joachims, 1999; Fig. 1, step 3). A radial basis kernel function was used, sampling different values of the parameters C (selecting from 1, 10, 100 and 1000) and γ (0.01, 0.1, 1, 10 and 100) to find those that performed best in the assessment, as has been done previously (Wee et al., 2006).
2.3 Benchmarking of scoring by jackknifing
A jackknife procedure was employed to test different scoring functions, in which 90% of the positives for each type of protease were randomly selected into a training set, and the remaining 10% were placed in a test set, along with the known negatives. The ratio of negatives to positives in the test set was 39 : 1 for the GrB benchmark and 35 : 1 for the caspase procedure, reflecting the ratio of negatives to positives observed in respective known substrates. Scores for the peptides were ranked and the false positive rate (FPR) against the true positive rate (TPR) was assessed at different score thresholds (Fig. 1, step 4). The jackknife procedure was repeated 1000 times and the results were averaged. Error bars for the averaged FPR μ at each TPR represent two times an SD, which is calculated over the distribution of FPRs for all iterations (x from i to N) by ((1/N)∑(xi−μ)2)1/2.
To ensure that random assignment of all experimentally identified peptides into different training and testing sets did not artificially influence predictive accuracy due to some similarities between the two sets, a separate jackknifing procedure was performed and compared the original to random assignment. Here, for each peptide x in the test set, no other peptide y was included in the training set if y was derived from a protein with >25% sequence identity to the protein from which x was derived. These included other peptides on x's protein itself. We describe this restriction as ‘homolog-filtering’.
2.4 Comparison of the protocol to other approaches
We applied to the datasets the following published methods: (i) an SVM trained on sequence information, using the original encoding and parameter sampling scheme (Wee et al., 2006); (ii) the GrabCas method, which incorporates in vitro PS-SCLs into a PSSM, using default parameters; (iii) a PSSM based on the frequency of residue types appearing in each position in the training set, incorporating the generalized PoPS algorithm to score a sequence (Boyd et al., 2005).
2.5 Experimental validation on select substrates
The method was applied to all octapeptides in the human proteome with Asp in the fourth position. Certain peptides were selected for experimental validation using the following procedure. The expression of a predicted substrate at the mRNA level was determined by consulting the BioGPS database (https://biogps.gnf.org/; Fig. 1, step 5). The availability of a literature-validated antibody was determined by consulting http://www.labome.com. K562 cells were grown in Iscove's modified Dulbecco's medium, 10% FBS, 1 × Glutamax, 1× Penn/Strep to a density of ∼ 5 × 105 cells/ml. K562 cells were harvested by centrifugation, washed in PBS, and lyzed in MPER™ (Thermo Scientific, Rockford, IL) at 1 × 107 cells/ml according to the manufacturer's instructions. Protein concentration was determined by BCA assay (Thermo Scientific, Rockford, IL).
Pichia-expressed human GrB (Thornberry et al., 1997) and Escherichia coli-expressed human caspase-3 and -8 (Stennicke and Salvesen, 1999) were purified as previously described. K562 MPER™ lysates were diluted 1 : 2 into 500 mM HEPES pH 8.0, 100 mM NaCl, 0.01% Tween-20 to raise the pH for optimal GrB activity and diluted 1 : 2 into MPER and 20 mM DTT for optimal caspase activity. GrB or a mixture of caspase-3 and -8 were added for either 1 h or ∼19 h before quenching proteolysis by adding LDS sample buffer (Invitrogen, Carlsbad, CA) and incubating at 70○C for 10 min. The final concentration of exogenous protease (GrB or total caspase) was 1 μ M, 500, 250, 100, 50 and 25 nM. Untreated lysate was incubated for 19 h to account for the activity of endogenous proteases. Caspase-inhibited lysates were pretreated with 100 μ M z-VAD-FMK (Bachem, Torrance, CA) and 100μ M z-DEVD-FMK (Bachem, Torrance, CA) for at least 1 h at 37○C and then treated with GrB as described. To verify that the exogenous protease added to the lysate was active, immunoblots against validated substrates were performed as described: pro-caspase-3 for GrB, PARP for caspase-3 and BID for caspase-8 (Supplementary Fig. 3).
7μg of total protein from each protease-treated and -untreated sample were subjected to electrophoresis on denaturing and reducing NuPAGE Bis-Tris gels (Invitrogen, Carlsbad, CA). Proteins were then transferred to Polyvinylidene Fluoride (PVDF) membranes and blocked in Tris buffered saline Triton X-100 (TBST) containing 5% (w/v) milk. Membranes were then incubated with substrate-specific antibodies, washed and incubated with HRP-conjugated secondary antibodies (BioRad, Hercules, CA). Immunoblots were developed on film with the ECL Plus detection system (GE Healthcare, Piscataway, NJ). To verify that equal amounts of protein were being compared across samples, GAPDH levels were quantified in parallel with either a rabbit anti-GAPDH or mouse anti-GAPDH antibody and appropriate Cy3 or Cy5 conjugated secondary antibody (GE Healthcare, Piscataway, NJ). Fluorescence was quantified on Typhoon Scanner (GE Healthcare, Piscataway, NJ). A representative GAPDH immunoblot is shown in Supplementary Figure 3. All primary antibodies were from either (Cell Signaling, Beverly, MA) or (Santa Cruz Biotechnology, Santa Cruz, CA).
3 RESULTS
3.1 Benchmark sets
For each protease type, two sets of octapeptides were compiled to benchmark the method (Fig. 1, steps 1a and 1b). These sets included peptides cleaved (‘positives’) and not cleaved (‘negatives’) by the proteases, respectively (Supplementary Fig. 1a). For GrB, the positives include 54 cleavage sequences from literature (i.e. our ‘GrBah’ dataset; Supplementary Table 1) and 305 cleavage sequences from a proteomics experiment that used combined fractional diagonal chromatography for isolating peptides (Van Damme et al., 2008). These positives spanned the P4 to P4′ positions using the traditional protease nomenclature (Schechter and Berger, 1968). Positives for caspase substrates were drawn from the literature-curated Casbah dataset (Lüthi and Martin, 2007) as well as a separate proteomics dataset obtained in experiments with the Jurkat cell line (Mahrus et al., 2008). The negatives for both protease types were all octapeptides in known protein substrates that are outside of the experimentally identified cleavage site and contain Asp in the fourth position (Supplementary Fig. 1b). While it is possible that some of these negatives are in fact cut by the protease and were missed experimentally, many of the positives in the benchmark sets were confirmed by studies that afford a high degree of coverage. The use of octapeptides outside the cleavage site is therefore a suitable source for a statistical description of the negatives' properties.
3.2 Difference in peptide sequence between positives and negatives
The frequencies of amino acid residue types appearing at each position in the peptides were calculated for positives of both protease types and the combined set of negatives. Instead of the qualitative sequence logos commonly used to plot residue-type frequencies (Crooks et al., 2004), we created a representation allowing for a more quantitative comparison of residue characteristics and identity (Fig. 2a). A large degree of degeneracy is observed in the positives, with both GrB and caspase substrates allowing for six or more residue types appearing at frequencies >5% at six of the eight subsites in the peptide. Aside from the requirement for Asp at the P1 site, the most stringent specificities are for large hydrophobic residues at the GrB P4 site (occurring in 62% of all substrates), and for small non-polar residues at the caspase P1′ site (occurring in 74% of all substrates). Residue-type frequencies in the positives for both protease types differ from those in the negatives.
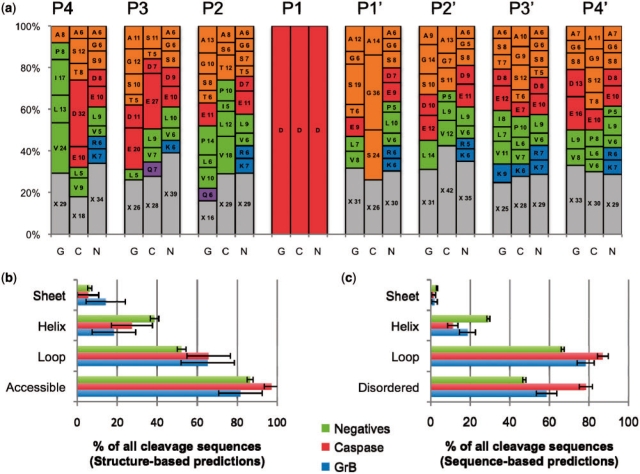
Fig. 2.
Sequence and structural properties of cleavage sequences. (a) A stacked histogram showing the relative frequency of each residue type at each position in the cleavage sequence for substrates of GrB (G) and caspases (C), and negatives (N). The numbering spans positions from P4 to P4′. Letters on the plots represent the one-letter code for each amino acid residue type, followed by its percentage at that position. X (gray) represents the total percentage for all residue types that are present in the position at <5% relative frequency. Amino acid residue types are grouped by general characteristic (Green: hydrophobic; orange: small non-polar; red: charged acidic; blue: basic; purple: polar). (b) Structural properties of protease cleavage sequence positives and negatives as assessed by DSSP for substrates where a solved structure or good quality comparative model was available. Numbers may not add to 100% as some peptides did not have more than four residues in any one of the three secondary structure conformations. (c) Structural properties as assessed by predictive methods that consider the protein primary sequence only. Disopred predicted disorder in all substrates. PSI-PRED predicted secondary structure, in cases where a structure or model of the substrate was not available.
3.3 Enrichment of structural features in cleavage sequences
Structural features were assessed for enrichment in known cleavage sequences compared with the negatives (Fig. 2b and c). Previous reviews of protease substrates (Hubbard, 1998) show that the cleaved sequence is more likely to be exposed to solvent, flexible, disordered and lacking secondary structure. In solved structures and comparative models, cleavage sequences are indeed more likely to be in a loop than the negatives, with 65.3% ± 13.3% of GrB sites and 65.0% ± 10.9% of caspase sites being in such a conformation compared with 52.2% ± 2.1% of the negatives. Solvent accessibility was greatest in the caspase substrates (97.3% ± 3.7% of cleavage sequences), followed by the negative set (86.5% ± 1.5%), and then by the GrB substrates (81.6% ± 10.8%). When structures or comparative models were not available, predictions gave a similar enrichment, although the magnitude of cleavage sequences in a loop conformation for all three sets was increased between 12% and 20%. This agreement in the relative distributions (Fig. 2b and c) suggests that any errors in PSI-PRED are generally not limiting in predicting the secondary structure of cleavage sites in the substrates to which it was applied. Finally, the amount of predicted disorder (i.e. sequences that are flexible, dynamic and unresolved in an electron density map obtained by X-ray crystallography) was also greater by 12% for GrB substrates and by 37% for caspase cleavage sequences than in the negatives.
3.4 Benchmarking of scoring functions
Using a jackknifing procedure and the datasets, we benchmarked a scoring function for predicting whether or not an octapeptide is a substrate of a given protease type, incorporating an SVM trained on both structure and sequence (Section 2). Receiver operator characteristic (ROC) plots were generated to assess the ability of the scoring functions to distinguish between positives and negatives (Fig. 3). The critical point of the ROC plot represents the optimal tradeoff between coverage and accuracy (i.e. the minimal combined false positive and false negative rates) and was used to compare the performance of different methods.
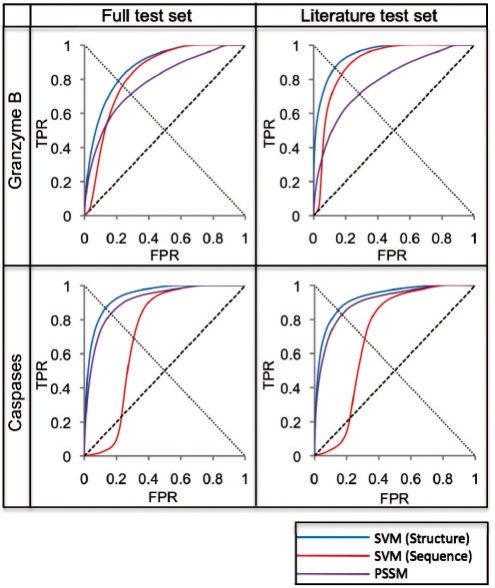
Fig. 3.
Benchmark results. Results from different methods applied to four different datasets, represented by ROC curves. The line from (0,0) to (1,1) represents a random predictor; a perfect classifier would go from (0,0) to (0,1) and then to (1,1). The critical point of the ROC curve is where each curve intersects the line from (1,0) to (0,1). Full test sets included all known substrates for the respective protease type, and Literature test sets excluded the large proteomic datasets, retaining only the GrBah and Casbah substrates. SVM (Structure) was developed in the current study; SVM (Sequence) was taken from a previous study that trained on cleavage sequence residue type only (Wee et al., 2006); PSSM implemented the GrabCas method for GrB substrates (Backes et al., 2005), while for caspases it was trained on frequency of residue types at each position in known cleavage sequences, using the PoPS (Boyd et al., 2005) algorithm. All ROC plots were interpolated through a number of points equal to the number of test set positives in each dataset (Supplementary Fig. 1a).
Due to preferences of these proteases for specific residue types around cleavage sites, as well as the enrichment of certain structural features at these sites, we hypothesized that the best classifier would incorporate these aspects of proteolysis. Indeed, the SVM trained on these features did well to discriminate between positives and negatives in the benchmark sets [Fig. 3; ‘SVM (Structure)’]. The GrB benchmark set was classified with a 0.79 TPR at a 0.21 FPR at its critical point. Furthermore, these rates improved (0.87 TPR at 0.14 FPR) when the SVM was trained on all known GrB substrates but assessed on a test set consisting of only the literature-curated GrBah dataset. The caspase benchmark produced similar results on both datasets. Error bars for the FPRs across 1000 iterations were assessed and calculated as less than 0.002 for all points; these are omitted from the figure as they are smaller than the width of the curve itself.
Due to the potential for biasing an estimate of prediction accuracy by including peptides from similar proteins in both the training and testing set, we performed the jackknifing procedure with homolog filtering (Section 2). When this condition was imposed, the TPRs and FPRs did not change significantly (Supplementary Fig. 2). This observation implies that including peptides from related proteins across the two sets does not significantly influence the estimate of the prediction accuracy. The likely reason is that the features used by the classifier depend on the peptides themselves and not on the proteins from which they were derived.
3.5 Comparison with other methods
The results of the method were compared with those obtained by two previously described methods tested on the same datasets. An SVM trained on sequence only predicted GrB substrates with a 0.76 TPR at a 0.25 FPR at its critical point when assessed on the full test set [Fig. 3, ‘SVM (Sequence)’]. GrabCas achieved a 0.71 TPR at a 0.29 FPR on the same test set (Fig. 3, ‘PSSM’). Similar discrepancies were observed on the GrBah test set and on both caspase test sets, here using the PoPS algorithm as the basis for the PSSM (Section 2).
3.6 Criteria for selecting targets for experimental validation
The method was applied to all human proteome octapeptides with Asp in the fourth position to produce a score for each potential cleavage sequence. Two proteins, Apoptosis Inducing Factor 1 (AIF-1) and Survival Motor Neuron 1 (SMN1), fulfilled the following criteria for experimental followup: (i) they were not in any benchmark dataset, (ii) the corresponding mRNA was expressed in the K562 cell line (highly susceptible to granzyme-induced cell death), (iii) a validated antibody was available and (iv) evidence supported a role in apoptosis. To test whether these candidates were cleaved by GrB or caspases, K562 lysates were treated with varying concentrations of exogenous protease for either 1 h or 19 h. As the benchmark set contains substrates of both initiator and executioner caspases, a mixture composed of caspase-8 and -3 was chosen. To determine if exogenously added GrB was the causative protease, K562 lysates were pretreated with broad-spectrum caspase inhibitors before GrB addition.
3.7 Cleavage of AIF-1 by GrB
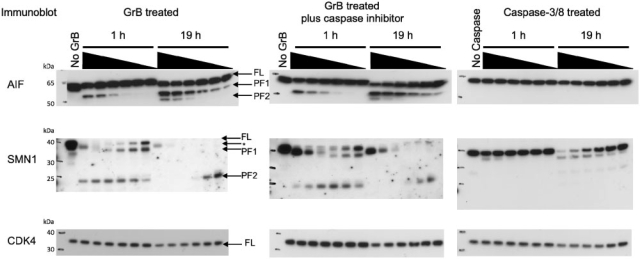
AIF-1 is a mitochondrial flavoprotein that translocates to the nucleus during apoptosis and facilitates DNA fragmentation. Interestingly, AIF-1 has a high-scoring GrB cleavage sequence (VPQD126KAPS) that is partially solvent exposed and in a loop conformation, as determined in its X-ray structure. Addition of GrB to K562 lysates results in the appearance of a ∼55 kDa proteolytic product that is both time and concentration dependent [labeled as proteolytic fragment 1 (PF1) in Fig. 4]. A second ∼50 kDa proteolytic fragment [labeled as proteolytic fragment 2 (PF2) in Fig. 4] is detected only at the highest concentrations of GrB after 19 h. The anti-AIF1 antibody was raised against a peptide sequence derived from the C-terminus of the protein. The antibody will therefore recognize both full-length protein and any proteolytic product containing this C-terminal epitope, making cleavage at VQPD126 the most likely explanation for the observed 55 kDa product. AIF-1 did not contain high-scoring caspase cleavage sites. In agreement with this prediction, the same proteolysis pattern is observed when GrB is added to K562 lysates pretreated with caspase inhibitors (Fig. 4). Furthermore, addition of exogenous caspase to K562 lysates resulted in no detectable proteolysis of AIF-1. These data indicate that proteolysis of AIF-1 is directly dependent on GrB.
Fig. 4.
Immunoblots of predicted GrB substrates. K562 lysates were treated with increasing concentrations of GrB or a mixture of caspase-3 and caspase-8 for either 1 h or 19 h. The final concentration of exogenously-added protease was 1 μ M, 500, 250, 100, 50 and 25 nM. For caspases, the final concentration refers to the concentration of total caspase (caspase-3 plus caspase-8). The no protease controls were incubated at 37○C for 19 h to account for the activity of endogenous proteases. The caspase-inhibited lysates were pretreated with 100 μM z-VAD-FMK and 100 μM z-DEVD-FMK at 37○C and then treated with GrB. Bands corresponding to full-length (FL) protein, proteolytic fragment 1 (PF1) and proteolytic fragment 2 (PF2) are indicated with arrows. Controls showed that the SMN1 antibody cross-reacts with GrB (Supplementary Fig. 4). The GrB band is indicated by an arrow and asterisk.
3.8 Cleavage of SMN1 by GrB
Proteolysis of SMN1 is observed during apoptosis in neurons; one study demonstrated that cleavage occurs at ICPD252SLDD and suggested a caspase as the causative protease (Kerr et al., 2000). When evaluated with our method, this site instead scored poorly with the caspase SVM model but scored well with the GrB SVM model (Fig. 5). To determine if SMN1 is a GrB substrate, GrB-treated K562 lysates in the presence and absence of caspase inhibitors were immunoblotted for SMN1. Both the appearance of the ∼37 kDa and ∼23 kDa proteolytic products (labeled PF1 and PF2 in Fig. 4) are caspase independent. SMN1 did contain a high-scoring caspase cleavage sequence, located six residues C-terminal to the predicted GrB cleavage site. Addition of exogenous caspase to K562 lysate resulted in the appearance of a ∼37 kDa proteolytic product, consistent with cleavage near the predicted GrB site (Fig. 4).
Fig. 5.
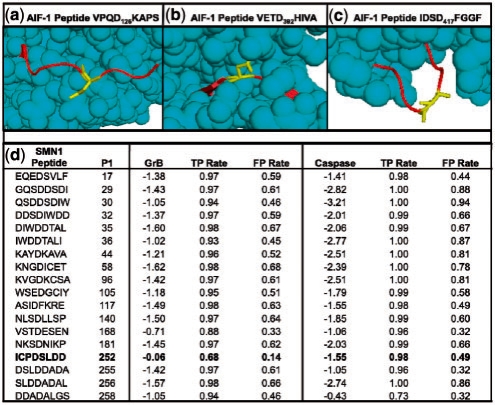
Details of novel GrB substrates. (a) Solved structure of AIF-1 (PDB ID 1M6I), highlighting Asp126. Cleavage at this site is consistent with the observed banding patterns on the immunoblot. (b) A peptide on AIF-1 centered on Asp392 that scores well when examining sequence only, but poorly when structure is considered, likely due to being largely inaccessible to solvent. (c) Another high scoring site on AIF-1 at Asp417; it is unclear why this site is not cleaved despite favorable sequence and structure properties. (d) Scores for the SMN1 protein for all octopeptides with Asp in the fourth position, as assessed by SVMs trained on GrB and caspase substrates, respectively. The ‘GrB’ and ‘Caspase’ columns indicate the scores that the respective SVMs assigned to each peptide, and TPR and FPR signify rates that these scores would fall on in the benchmark set.
3.9 CDK4 is not cleaved by GrB
Proteins were predicted to be negatives if all candidate cleavage sequences did not score higher than a threshold defined by the SVM critical point. To determine if a predicted negative is cleaved by GrB and caspases, immunoblotting for CDK4 in protease-treated lysates was performed. In all cases, a slight reduction in the amount of full-length protein is evident only after 19 h at 37○C and at high concentration of exogenous protease (Fig. 4), validating our negative predictions.
4 DISCUSSION
4.1 Overview
In an effort to increase the coverage, accuracy and efficiency of identifying protease substrates, we developed and benchmarked a bioinformatics method that takes advantage of the current knowledge about known substrates as well as general rules of protein structure (Fig. 1). Its predictive power was quantified by the degree to which it distinguishes between positives and negatives in a benchmark set. To demonstrate the utility of the approach, we applied it to predict novel substrates of the GrB protease and caspases, followed by experimental validation of two biologically important predictions, AIF-1 and SMN1. These results thus benefited from the synergy of sequence- and structure-based predictions combined with biological intuition to select targets for validation. The computational method has two main benefits. First, it acts as a hypothesis generator; when applied to all proteins in a proteome of interest, it produces a list of high-confidence predictions suitable for a focused and efficient experimental followup. Second, the computational method lends insight into the structural aspects that determine whether a site can be cleaved.
4.2 Proteome-wide prediction of protease substrates
The method was applied to all proteins in the human proteome to identify those most likely cleaved by GrB and caspases, resulting in many predictions made with high confidence. For example, the top 500 predicted caspase substrates with Gene Ontology (GO; Ashburner et al., 2000) annotation received a score corresponding to a 0.002 FPR and a 0.110 TPR in the ROC plot (Fig. 3). GO assignments for these sequences suggest their role in apoptosis (21 proteins), signaling (53), transcription regulation (51) and proteolysis (18), all of which are hallmarks of many known substrates targeted by caspases to induce cell death. Similar results are observed for predicted GrB substrates (Supplementary Table 2).
Once experimentally validated, these substrates lend critical insight into apoptosis. A case in point are the two GrB substrates validated in this study, AIF-1 and SMN1, which are potentially involved in two novel apoptotic pathways initiated by GrB cleavage. All predictions are available at http://salilab.org/peptide. Each predicted substrate site is annotated with the structural assignments that were used to make the predictions, the TPRs and FPRs for their scores, and links to the MODBASE database of comparative protein structure models to view any known structures or models of the substrate.
4.3 Cleavage of SMN1 and AIF-1 by GrB
The high-confidence predictions generated by this method are valuable for both streamlining experimental validation (Fig. 4) and generating novel hypotheses regarding the roles of substrates in cell death. AIF-1 is tethered to the inner mitochondrial membrane (IMM); therefore, its translocation to the nucleus requires both mitochondrial outer membrane permeabilization (MOMP) and proteolysis of the IMM tether. The cathepsins B, S and L have been shown to proteolyze AIF-1 around residue 100, 26 residues N-terminal to the predicted GrB cleavage site (Yuste et al., 2005). The redundancy of multiple proteases liberating AIF-1 from the mitochondria might represent a strategy to overcome anti-apoptotic resistance mechanisms, such as Hsp70 overexpression. Hsp70 has been shown to inhibit import of AIF-1 to the nucleus (Ravagnan et al., 2001). GrB cleaves and inactivates Hsp70 (Loeb et al., 2006) and therefore might facilitate AIF-1 nuclear import.
SMN1 cleavage was first observed during neuronal apoptosis induced by viral infection and ischemic injury in mice (Kerr et al., 2000). Mutation of Asp252 to Ala abolished cleavage, leading to the speculation that caspase was the causative protease. Interestingly, SMN1 cleavage was induced by adding brain extracts from either ischemically injured or virally infected mice, raising the possibility that cytotoxic T lymphocyte (CTLs) and therefore GrB was present in the extracts.
In a separate study, SMN1 cleavage has been observed in a differentiated neuronal cell line during growth factor withdrawal. CTLs are absent in this ex vivo study, thereby excluding GrB and implying a caspase as the causative protease (Vyas et al., 2002). Interestingly immunoblotting for SMN1 in the neuronal lysate suggested that proteolysis is inefficient, consistent with our observation that SMN1 is proteolyzed far more efficiently by GrB than the caspases. In light of evidence for a role of CTLs in both ischemic brain injury (Yilmaz and Granger, 2010) and virally infected neurons (Neumann et al., 2002), GrB should be examined as the causative protease for SMN1 cleavage in vivo.
4.4 Benefit of incorporating structural features in classifier training
The method was compared with several previous approaches benchmarked on the same datasets. One study using an SVM trained on sequence features did well to discriminate between positives and negatives (Wee et al., 2006), but was still outperformed by the current SVM that incorporates structure as well as sequence features (Fig. 3). This improvement shows that structural features of the cleavage sequence can add predictive value to a substrate identification method. Additionally, the method outperformed two other methods based on PSSMs. The first method, GrabCas, uses the results of in vitro small peptide libraries to predict GrB substrates (Backes et al., 2005; Thornberry et al., 1997). These in vitro libraries often do not fully reflect the observed protein-peptide specificity in known biological substrates. In contrast, our SVM training set does include biological substrates. The second method, PoPS, was trained only on the observed frequencies of residue types at each position in the caspase training set (Boyd et al., 2005). This PSSM does not take into account cooperativity across residue pairs. In contrast, the pair correlations can be encoded in our SVM.
It was shown previously that caspase cleavage sites can occur in regions of regular secondary structure (Mahrus et al., 2008). Here, we show that GrB substrates display the same tendency. Indeed, > 35% of known cleavage sequences in both GrB and caspase substrates fall on a region that has regular secondary structure (Fig. 2b). One possibility is that these regions undergo local unfolding prior to cleavage by the protease. These observations demonstrate the limitations of making predictions based on sequence and then filtering for expected secondary structure, as opposed to using a machine learning algorithm that makes unbiased predictions by combining sequence and structure in an integrated fashion.
An example of the power of incorporating structure into prediction is shown by comparing two potential cleavage sequences in AIF-1, VPQD126KAPS (Fig. 5a) and VETD392HIVA (Fig. 5b). Both sites were evaluated with the sequence-based SVM (Wee et al., 2006) as well as our SVM that includes structural information. VPQD126KAPS, which was suggested experimentally as the GrB cleavage site (Fig. 4), was scored with the sequence-based SVM corresponding to a 0.73 FPR. When structural features were incorporated, this site scored with much higher confidence at a 0.17 FPR. The site is on a fully exposed, flexible portion of the solved AIF-1 structure. VETD392HIVA, on the other hand, evaluates at a 0.05 FPR when scored with the sequence-based SVM, but falls to a lower confidence 0.34 FPR when structural features are included. This site is almost completely buried and portions of it fall on a β-strand. The difference between these two sites demonstrates the importance of considering structural information when predicting protease cleavage sites. Interestingly, a third sequence at IDSD417FGGF is not cleaved despite having favorable sequence and structure features (Fig. 5c); further understanding of the dynamics of GrB-substrate recognition is needed to determine why this is the case.
4.5 General applicability of the approach
The protocol presented in this study was applied to predict substrates for GrB and caspases, two types of proteases that recognize extended, specific oligopeptide sequences possessing certain structural features. However, the approach is generally applicable to predict interaction partners for any protein that recognizes its peptide partners based on the features encoded in our method. Thus, we provide a web server (http://salilab.org/peptide) that allows users: (i) to construct and apply a new SVM based on a user-provided training set; (ii) to benchmark the ability of the SVM to predict interaction partners for a protein of interest; (iii) to use the newly generated SVM to make proteome-wide predictions; and (iv) to make the SVM and its predictions publically available for use by others. As a result, our approach may become a widely useful hypothesis generator that can increase the pace of biological discovery by guiding future experiments in a variety of protein–peptide systems.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Julie Zorn for providing us with purified recombinant caspase-3 and caspase-8; Dr Benjamin Webb for software support; and Dr Avner Schlessinger and Dr Kaveh Maghsoudi for helpful discussions. We also thank Ron Conway, Mike Homer, Intel, IBM, Hewlett-Packard and NetApp for computer hardware gifts to A.S.
Funding: National Institutes of Health (R01 GM54762 to A.S., NIHCA72006 and CA128765 to C.S.C.); Leukemia and Lymphoma Society Fellowship (5552-06 to D.R.H.); University of California Cancer Research Coordinating Committee fellowship and the Chih Foundation Award (to D.T.B.).
Conflict of Interest: none declared.
REFERENCES
- Ashburner M, et al. Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backes C, et al. GraBCas: a bioinformatics tool for score-based prediction of Caspase- and Granzyme B-cleavage sites in protein sequences. Nucleic Acids Res. 2005;33:W208–W213. doi: 10.1093/nar/gki433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd SE, et al. PoPS: a computational tool for modeling and predicting protease specificity. J. Bioinform. Comput. Biol. 2005;3:551–585. doi: 10.1142/s021972000500117x. [DOI] [PubMed] [Google Scholar]
- Bredemeyer AJ, et al. Use of protease proteomics to discover granzyme B substrates. Immunol. Res. 2005;32:143–153. doi: 10.1385/IR:32:1-3:143. [DOI] [PubMed] [Google Scholar]
- Bui HH, et al. Structural prediction of peptides binding to MHC class I molecules. Proteins. 2006;63:43–52. doi: 10.1002/prot.20870. [DOI] [PubMed] [Google Scholar]
- Casciola-Rosen L, et al. Cleavage by granzyme B is strongly predictive of autoantigen status: implications for initiation of autoimmunity. J. Exp. Med. 1999;190:815–826. doi: 10.1084/jem.190.6.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty S, Sanchez R. Systematic analysis of added-value in simple comparative models of protein structure. Structure. 2004;12:1461–1470. doi: 10.1016/j.str.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Chen JR, et al. Predicting PDZ domain-peptide interactions from primary sequences. Nat. Biotechnol. 2008;26:1041–1045. doi: 10.1038/nbt.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, et al. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix MM, et al. Global mapping of the topography and magnitude of proteolytic events in apoptosis. Cell. 2008;134:679–691. doi: 10.1016/j.cell.2008.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eramian D, et al. How well can the accuracy of comparative protein structure models be predicted? Protein Sci. 2008;17:1881–1893. doi: 10.1110/ps.036061.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraro E, et al. A neural strategy for the inference of SH3 domain-peptide interaction specificity. BMC Bioinformatics. 2005;6(Suppl. 4):S13. doi: 10.1186/1471-2105-6-S4-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garay-Malpartida HM, et al. CaSPredictor: a new computer-based tool for caspase substrate prediction. Bioinformatics. 2005;21(Suppl. 1):i169–i176. doi: 10.1093/bioinformatics/bti1034. [DOI] [PubMed] [Google Scholar]
- Hou T, et al. Characterization of domain-peptide interaction interface: a generic structure-based model to decipher the binding specificity of SH3 domains. Mol. Cell Proteomics. 2009;8:639–649. doi: 10.1074/mcp.M800450-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard SJ. The structural aspects of limited proteolysis of native proteins. Biochim. Biophys. Acta. 1998;1382:191–206. doi: 10.1016/s0167-4838(97)00175-1. [DOI] [PubMed] [Google Scholar]
- Jacob L, Vert JP. Efficient peptide-MHC-I binding prediction for alleles with few known binders. Bioinformatics. 2008;24:358–366. doi: 10.1093/bioinformatics/btm611. [DOI] [PubMed] [Google Scholar]
- Jemth P, Gianni S. PDZ domains: folding and binding. Biochemistry. 2007;46:8701–8708. doi: 10.1021/bi7008618. [DOI] [PubMed] [Google Scholar]
- Joachims T. Making large-scale SVM learning practical. In: Scholkopf B, et al., editors. Advances in Kernel Methods - Support Vector Learning. Boston: MIT-Press; 1999. pp. 169–184. [Google Scholar]
- Johnson CE, Kornbluth S. Caspase cleavage is not for everyone. Cell. 2008;134:720–721. doi: 10.1016/j.cell.2008.08.019. [DOI] [PubMed] [Google Scholar]
- Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- Jones DT, Ward JJ. Prediction of disordered regions in proteins from position specific score matrices. Proteins. 2003;53(Suppl. 6):573–578. doi: 10.1002/prot.10528. [DOI] [PubMed] [Google Scholar]
- Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Kaneko T, et al. The SH3 domain–a family of versatile peptide- and protein-recognition module. Front. Biosci. 2008;13:4938–4952. doi: 10.2741/3053. [DOI] [PubMed] [Google Scholar]
- Kerr DA, et al. Survival motor neuron protein modulates neuron-specific apoptosis. Proc. Natl Acad. Sci. USA. 2000;97:13312–13317. doi: 10.1073/pnas.230364197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb CRK, et al. Granzyme B proteolyzes receptors important to proliferation and survival, tipping the balance toward apoptosis. J. Biol. Chem. 2006;281:28326–28335. doi: 10.1074/jbc.M604544200. [DOI] [PubMed] [Google Scholar]
- Lohmüller T, et al. Toward computer-based cleavage site prediction of cysteine endopeptidases. Biol. Chem. 2003;384:899–909. doi: 10.1515/BC.2003.101. [DOI] [PubMed] [Google Scholar]
- Lüthi AU, Martin SJ. The CASBAH: a searchable database of caspase substrates. Cell Death Differ. 2007;14:641–650. doi: 10.1038/sj.cdd.4402103. [DOI] [PubMed] [Google Scholar]
- Mahrus S, et al. Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell. 2008;134:866–876. doi: 10.1016/j.cell.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann H, et al. Cytotoxic T lymphocytes in autoimmune and degenerative CNS diseases. Trends Neurosci. 2002;25:313–319. doi: 10.1016/s0166-2236(02)02154-9. [DOI] [PubMed] [Google Scholar]
- Nicholson DW, Thornberry NA. Apoptosis. Life and death decisions. Science. 2003;299:214–215. doi: 10.1126/science.1081274. [DOI] [PubMed] [Google Scholar]
- Pardo J, et al. The biology of cytotoxic cell granule exocytosis pathway: granzymes have evolved to induce cell death and inflammation. Microbes Infect. 2009;11:452–459. doi: 10.1016/j.micinf.2009.02.004. [DOI] [PubMed] [Google Scholar]
- Pieper U, et al. MODBASE, a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2009;37:D347–354. doi: 10.1093/nar/gkn791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravagnan L, et al. Heat-shock protein 70 antagonizes apoptosis-inducing factor. Nat. Cell Biol. 2001;3:839–843. doi: 10.1038/ncb0901-839. [DOI] [PubMed] [Google Scholar]
- Russell JH, Ley TJ. Lymphocyte-mediated cytotoxicity. Annu. Rev. Immunol. 2002;20:323–370. doi: 10.1146/annurev.immunol.20.100201.131730. [DOI] [PubMed] [Google Scholar]
- Schechter I, Berger A. On the active site of proteases. 3. Mapping the active site of papain; specific peptide inhibitors of papain. Biochem. Biophys. Res. Commun. 1968;32:898–902. doi: 10.1016/0006-291x(68)90326-4. [DOI] [PubMed] [Google Scholar]
- Sieker F, et al. Predicting affinity and specificity of antigenic peptide binding to major histocompatibility class I molecules. Curr. Protein Pept. Sci. 2009;10:286–296. doi: 10.2174/138920309788452191. [DOI] [PubMed] [Google Scholar]
- Stennicke HR, Salvesen GS. Caspases: preparation and characterization. Methods. 1999;17:313–319. doi: 10.1006/meth.1999.0745. [DOI] [PubMed] [Google Scholar]
- Stiffler MA, et al. PDZ domain binding selectivity is optimized across the mouse proteome. Science. 2007;317:364–369. doi: 10.1126/science.1144592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornberry NA, et al. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J. Biol. Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- Timmer J, et al. Structural and kinetic determinants of protease substrates. Nat. Struct. Mol. Biol. 2009;16:1101–1108. doi: 10.1038/nsmb.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme P, et al. Analysis of protein processing by N-terminal proteomics reveals novel species-specific substrate determinants of granzyme B orthologs. Mol. Cell Proteomics. 2008;8:258–272. doi: 10.1074/mcp.M800060-MCP200. [DOI] [PubMed] [Google Scholar]
- Verspurten J, et al. SitePredicting the cleavage of proteinase substrates. Trends Biochem. Sci. 2009;34:319–323. doi: 10.1016/j.tibs.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Vyas S, et al. Involvement of survival motor neuron (SMN) protein in cell death. Hum. Mol. Genet. 2002;11:2751–2764. doi: 10.1093/hmg/11.22.2751. [DOI] [PubMed] [Google Scholar]
- Wee LJ, et al. SVM-based prediction of caspase substrate cleavage sites. BMC Bioinformatics. 2006;7(Suppl. 5):S14. doi: 10.1186/1471-2105-7-S5-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins MR, et al. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999;112:531–552. doi: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- Yang ZR. Prediction of caspase cleavage sites using Bayesian bio-basis function neural networks. Bioinformatics. 2005;21:1831–1837. doi: 10.1093/bioinformatics/bti281. [DOI] [PubMed] [Google Scholar]
- Yilmaz G, Granger DN. Leukocyte recruitment and ischemic brain injury. Neuromolecular Med. 2010;12:193–204. doi: 10.1007/s12017-009-8074-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuste VJ, et al. Cysteine protease inhibition prevents mitochondrial apoptosis-inducing factor (AIF) release. Cell Death Differ. 2005;12:1445–1448. doi: 10.1038/sj.cdd.4401687. [DOI] [PubMed] [Google Scholar]
- Zhang L, et al. An integrated machine learning system to computationally screen protein databases for protein binding peptide ligands. Mol. Cell Proteomics. 2006;5:1224–1232. doi: 10.1074/mcp.M500346-MCP200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.