Abstract
In the barley (Hordeum vulgare) Hooded (Kap) mutant, the duplication of a 305-bp intron sequence leads to the overexpression of the Barley knox3 (Bkn3) gene, resulting in the development of an extra flower in the spikelet. We used a one-hybrid screen to identify four proteins that bind the intron-located regulatory element (Kap intron-binding proteins). Three of these, Barley Ethylene Response Factor1 (BERF1), Barley Ethylene Insensitive Like1 (BEIL1), and Barley Growth Regulating Factor1 (BGRF1), were characterized and their in vitro DNA-binding capacities verified. Given the homology of BERF1 and BEIL1 to ethylene signaling proteins, we investigated if these factors might play a dual role in intron-mediated regulation and ethylene response. In transgenic rice (Oryza sativa), constitutive expression of the corresponding genes produced phenotypic alterations consistent with perturbations in ethylene levels and variations in the expression of a key gene of ethylene biosynthesis. In barley, ethylene treatment results in partial suppression of the Kap phenotype, accompanied by up-regulation of BERF1 and BEIL1 expression, followed by down-regulation of Bkn3 mRNA levels. In rice protoplasts, BEIL1 activates the expression of a reporter gene driven by the 305-bp intron element, while BERF1 can counteract this activation. Thus, BEIL1 and BERF1, likely in association with other Kap intron-binding proteins, should mediate the fine-tuning of Bkn3 expression by ethylene. We propose a hypothesis for the cross talk between the KNOX and ethylene pathways.
Organogenesis in plants is a continuous process involving meristems, populations of pluripotent cells. The shoot apical meristem (SAM) is formed during embryogenesis and initiates organ primordia in postembryonic growth. SAM activity entails two contrasting processes: the slow proliferation of cells in the central zone, necessary for meristem maintenance, and the recruitment of cells at the meristem flanks, where lateral organ primordia are initiated (Scofield and Murray, 2006).
Knotted1-like homeobox (KNOX) genes represent an ancient class of transcription factors present in all examined plant species as well as in single-cell green and red algae (Mukherjee et al., 2009). A subset of this gene family (class I KNOX genes) regulates SAM functions in both monocots and dicots. KNOX I genes play pivotal roles not only in SAM formation and maintenance but also in morphogenetic processes throughout plant development (Hake et al., 2004): they contribute to cell fate and cell differentiation of vegetative (Vollbrecht et al., 1991; Becraft and Freeling, 1994) and reproductive (Müller et al., 1995; Williams-Carrier et al., 1997) tissues.
KNOX I genes encode transcription factors belonging to the TALE-homeobox superfamily (Bürglin, 1997), including maize (Zea mays) KNOTTED1 (KN1; Vollbrecht et al., 1991) and the closely related Oryza sativa Homeobox1 (OSH1; Matsuoka et al., 1993), Barley Knox3 (BKN3; Müller et al., 1995), and Arabidopsis (Arabidopsis thaliana) SHOOTMERISTEMLESS (STM; Long et al., 1996).
STM is first expressed in a single apical cell of Arabidopsis globular stage embryos (Long and Barton, 1998), whereas OSH1 (together with OSH15, OSH43, and OSH71) is transcribed at 3 d after pollination, in a cellular domain where the SAM will subsequently form in the (late globular) rice (Oryza sativa) embryo (Sentoku et al., 1999). Loss of KNOX I function causes defects in apical meristem formation (Long et al., 1996; Aida et al., 1999) and maintenance (Kerstetter et al., 1997; Vollbrecht et al., 2000).
In both monocot and dicot species with simple leaves, KNOX I genes are expressed in the shoot meristem and stem and repressed in differentiating leaf primordia (for review, see Hay and Tsiantis, 2010). In grasses, for example, leaf founder cells are organized in a ring-like domain where KNOX I genes are down-regulated (Schneeberger et al., 1998; Sentoku et al., 1999). Gain-of-function mutants with ectopic KNOX I gene expression exhibit knot-like meristematic structures in the vicinity of leaf veins in maize (Smith et al., 1992) and ectopic floral structures on barley (Hordeum vulgare) lemmas (Müller et al., 1995).
Correct spatial and temporal expression of KNOX I genes is ensured by different regulatory mechanisms involving a number of trans-acting factors. The MYB transcription factor ROUGH SHEATH2 (RS2) negatively regulates Kn1 during maize leaf initiation (Schneeberger et al., 1998; Timmermans et al., 1999; Tsiantis et al., 1999): rs2 mutants have abnormal leaf morphology and fail to repress KNOX I expression. Antirrhinum and Arabidopsis RS2 orthologs, PHANTASTICA and ASYMMETRIC LEAVES1 (AS1), repress the expression of AmSTM1 and KNOTTED-LIKE FROM ARABIDOPSIS THALIANA1 (KNAT1)/BREVIPEDICELLUS genes, respectively (Waites et al., 1998; Byrne et al., 2000). Among additional loci reviewed by Hake et al. (2004), AS2 encodes a plant-specific Lateral Organ Boundary protein that promotes leaf differentiation through the repression of KNOX I genes (Ori et al., 2000; Lin et al., 2003). RS2/AS1 proteins, in conjunction with AS2 and a homolog of the chromatin-remodeling protein HIRA (Phelps-Durr et al., 2005), interact with KNOX I cis-regulatory sequences, suggesting that epigenetic silencing of KNOX genes plays a role in the maintenance of determinacy throughout organogenesis (Guo et al., 2008).
Furthermore, chromatin immunoprecipitation experiments also provide evidence for the epigenetic control of KNOX gene expression. In Arabidopsis, STM is a direct target of histone H3K27 trimethylation mediated by CURLY LEAF (Schubert et al., 2006) as part of a silencing mechanism involving a POLYCOMB REPRESSIVE COMPLEX1-like complex (Xu and Shen, 2008). In maize, manipulation of the expression of hda101, a gene encoding an Rpd3-type histone deacetylase, is associated with morphological defects and alterations of Rs2 and Kn1 expression (Rossi et al., 2007).
Intronic cis-acting elements can also condition KNOX gene transcription. An insertion in a 5′ intron of the snapdragon (Antirrhinum majus) hirzina gene induces changes in leaf shape and the formation of a tube resembling an ectopic spur on the ventral petal (Golz et al., 2002). Insertions in intron III of maize Kn1 (Greene et al., 1994) are responsible for misexpression of the gene outside the shoot meristem. In barley, the dominant Hooded (Kap) phenotype is associated with a direct tandem duplication of 305 bp in intron IV, resulting in ectopic expression of Bkn3 at the distal end of the lemma and the development of an extra flower in place of the awn present in wild-type spikelets (Müller et al., 1995). In transgenic tobacco (Nicotiana tabacum) lines, the barley 305-bp element can drive reporter gene expression at the base of the flower, in contrast to the Bkn3 promoter, whose activity is confined to the SAM (Santi et al., 2003). The 305-bp intron element, therefore, can act as a floral-specific regulatory element, presumably recruiting transcription factors that modulate the expression of genes involved in floral evocation.
To gain insight into intron-mediated KNOX I gene regulation, a one-hybrid screen, using the 305-bp element as the DNA target, was used to identify four candidate barley Kap intron-binding proteins (KIBPs). The first of these proteins, Barley B Recombinant (BBR), has been shown to bind the 305-bp element in vitro and in vivo and to cause morphological alterations in leaves and flowers when overexpressed in tobacco (Santi et al., 2003).
Here, we report the isolation and molecular and functional characterization of Barley Ethylene Response Factor1 (BERF1), Barley Ethylene Insensitive Like1 (BEIL1), and Barley Growth Regulating Factor1 (BGRF1), three other genes encoding proteins that bind the Bkn3 305-bp intronic sequence. We focus mainly on the role of BERF1 and BEIL1, as some aspects of BGRF1 characterization will be described elsewhere. Functional studies in rice support a role for these genes in the transcriptional regulation of developmental processes. We propose a model for interactions between ethylene and intron-mediated regulation of KNOX I genes involved in grass spike development.
RESULTS
Isolation and Organization of KIBP Genes
A yeast one-hybrid screen facilitated the identification of genes encoding proteins that bind the 305-bp intron-located region of the Bkn3 gene (Müller et al., 1995): besides BBR (Santi et al., 2003), three additional putative plant transcription factors, collectively designated as KIBP, were isolated. To further characterize the corresponding genes, a barley genomic library was screened, yielding sequences of 7,834 bp for BERF1, 6,870 bp for BEIL1, and 5,184 bp for BGRF1 (GenBank accession numbers are reported in Table I). The putative structures of the three genes were determined based on comparisons of longest available cDNA and genomic sequences (Fig. 1). BLAST searches of the barley EST collection (Barley HarvEST database; http://harvest.ucr.edu/) recovered EST clusters showing particular similarity with BERF1 (TA38707_4513, TA33959_4513, and TA33465_4513) and BEIL1 (U35 15803 and U35 12078). Sequence information was used to position BERF1, BEIL1, and BGRF1 in a linkage map of the barley genome, following Castiglioni et al. (1998; Table I; Supplemental Figs. S1 and S2). Comparisons of map positions and allele sequences led to the exclusion of associations of these genes with previously characterized mutants (Castiglioni et al., 1998; Roig et al., 2004; data not shown). Hence, to date, no barley phenotypic data are available for loss- or gain-of-function mutations for BERF1, BEIL1, and BGRF1.
Table I.
Accession numbers, barley map positions, and colinear rice genomic regions for the three KIBP genes studied
| KIBP Gene | Genomic Sequence Accession No. | Barley Map Position (Chromosome, Sublinkage Groupsa) | Rice Colinear Regions (Chromosome, Coordinates) | Supplemental Figures with Original Data |
| BGRF1 | HQ328942 | 2H, 19–20 | 2, 24–27.1 Mb | S1A, S2A |
| 4, 24.7–31 Mb | ||||
| BERF1 | HQ328941 | 5H, 63–64 | 9, 16–19.1 Mb | S1B, S2B |
| BEIL1 | HQ328940 | 7H, 6 | 8, 24.7–27.2 Mb | S1C, S2C |
As defined by Castiglioni et al. (1998) and Pozzi et al. (2003).
Figure 1.
Genomic organization of BGRF1, BERF1, and BEIL1 genes. The reconstructed BGRF1 gene includes a transcribed region of 3,613 bp with four exons and three introns. The BERF1 gene is organized in a transcribed region of 2,227 bp consisting of four exons and three introns. The BEIL1 gene includes a transcribed region composed of three exons (3,256 bp) and two introns (in the 5′ and 3′ untranslated regions). White boxes, thick lines, and thin lines represent exons, introns, and 5′ and 3′ genomic regions (identified from barley genomic clones), respectively. The positions of start and stop codons are indicated. The putative start of transcription is indicated based on the longest available cDNA clones.
KIBP Genes: Sequence and Phylogenetic Analyses
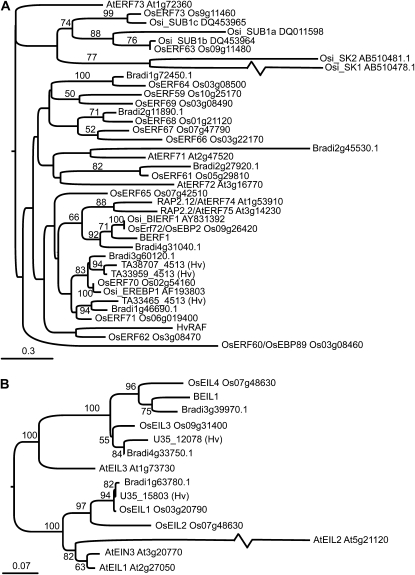
To help infer possible functions of KIBP genes, comparative sequence analyses were conducted on the Arabidopsis, rice, and Brachypodium distachyon genomes as well as the barley EST database. BERF1 contains a stretch of 60 amino acids typical of the APETALA2 superfamily and conserved in all known plant ERF proteins, known to be sufficient for sequence-specific DNA binding (Ohme-Takagi and Shinshi, 1995). The ERF gene family includes 122 members in Arabidopsis and 139 in rice, assigned to 12 and 15 groups, respectively (Nakano et al., 2006). BERF1 is closely related to group VII ERFs from rice and Arabidopsis (Okamuro et al., 1997; Nakano et al., 2006; Fig. 2A; Supplemental Table S1; Supplemental Fig. S3A) and shares distinctive motifs of this group, including the N-terminal MCGGIA(I/L) motif. Rice, Arabidopsis, Brachypodium, and barley ERF VII sequences were aligned with BERF1. The BERF1 sequence clusters with sequences that show overall colinearity outside of the ERF domain (OsERF72, O. sativa indica benzothiadiazole-induced ERF1 [OsiBIERF1], OsERF70, O. sativa indica ethylene-responsive element-binding protein 1 [OsiEREBP1], and OsERF71 and Arabidopsis RAP2.12 and RAP2.2). The analysis, including the positions of Brachypodium and Arabidopsis sequences, supports the orthology of BERF1 with OsERF72/O. sativa ethylene-responsive element-binding protein 2 gene (OsEBP2) (Lin et al., 2007) of japonica rice and its indica counterpart OsiBIERF1 (Cao et al., 2006). OsERF72/OsEBP2 maps on rice chromosome 9, around the chromosomal region colinear to the BERF1 locus (Supplemental Fig. S2B). A Brachypodium counterpart (Bradi4g31040) to these genes is discernible, while Arabidopsis RAP2.2 and RAP2.12 are recovered as moderately supported co-orthologs to this cluster of monocot genes. Deeper relationships, even within the ERF VII subfamily, are hard to ascertain due to the limited number of unambiguously alignable amino acid positions outside the DNA-binding domain. ERF proteins play a role in the transcriptional regulation of diverse biological processes related to growth and development as well as in responses to hormones, various stresses, and environmental stimuli (for review, see Nakano et al., 2006). In some indica rice varieties, the ERF VII gene Submergence1A and related SNORKEL1 (SK1) and SK2 genes have been shown to play important (and contrasting) functions in ethylene-mediated internode growth in response to submergence (Hattori et al., 2009; Bailey-Serres and Voesenek, 2010; Hinz et al., 2010; Jung et al., 2010). Transcriptional regulation by ethylene has been reported for other ERF VII genes, including OsEBP89/OsERF60 and OsEBP2/OsERF72 in rice (Yang et al., 2002; Mao et al., 2006; Lin et al., 2007), RAP2.2 in Arabidopsis (Hinz et al., 2010), and HvRAF (for H. vulgare root abundant factor), the only characterized member of this group in barley (Jung et al., 2007).
Figure 2.
Phylogenetic trees of ERFs and EIL proteins from barley, rice, Brachypodium, and Arabidopsis. A, Maximum likelihood phylogenetic tree showing the relationships between group VII ERF proteins from barley (Hv), rice subspecies japonica (Os) and indica (Osi), Brachypodium distachyon (Bradi), and Arabidopsis (At). Bootstrap proportions are shown where over 50%, and the scale bar represents substitutions per site. B, Phylogenetic tree showing relationships between BEIL1 and EIL proteins.
BEIL1 is related to Ethylene Insensitive (EIN) 3-Like (EIL) transcription factors from Arabidopsis (Chao et al., 1997) and rice (Mao et al., 2006; Supplemental Table S2; Supplemental Fig. S3B). The BEIL1 sequence emerges as the well-supported ortholog of OsEIL4, and one of the previously identified barley EST clusters (U35 12078) is likely to correspond to the ortholog of OsEIL3, while U35 15803 is the well-supported ortholog of OsEIL1 (Fig. 2B). The orthology of BEIL1 and OsEIL4 is consistent with the synteny between the barley and OsEIL4 loci (Supplemental Fig. S3C). OsEIL4 and OsEIL3 (as well as the corresponding barley sequences BEIL1 and U35 12078 and their uncontroversial Brachypodium orthologs) are co-orthologs of AtEIL3, and the synteny evidence is consistent with their derivation from a whole genome duplication event preceding the divergence of rice and barley (Thiel et al., 2009). In Arabidopsis, EIN3 and EIL1 act as downstream regulators of the ethylene signaling pathway (Chao et al., 1997), a role attributed also to OsEIL1 in rice (Mao et al., 2006) and related proteins in other species (for review, see Guo and Ecker, 2004). EIL genes have been implicated in other plant processes, as in the case of AtEIL3, which regulates sulfate uptake and metabolism in Arabidopsis (Maruyama-Nakashita et al., 2006).
BGRF1 shows most similarity to OsGRF3, OsGRF4, and OsGRF5, members of the subfamily A of rice GRFs (Choi et al., 2004). BGRF and GRF proteins share common features in their N-terminal half. The QLQ domain is similar to the one described by Treich et al.(1995), involved in protein-protein interaction; the WRC domain contains a stretch of basic amino acids considered a nuclear localization signal, as well as the C3H motif, CX9CX10CX2H (van der Knaap et al., 2000), suggested to function in DNA binding. The C3H motif is a component of a group of atypical zinc finger transcription factors, the first example of which was found in Hordeum Repressor of Transcription, a protein that binds the GA Response Element (Raventós et al., 1998). A study of rice GRF genes including detailed sequence comparisons with BGRF1 will be presented elsewhere. OsGRF3 maps on rice chromosome 4 within the region colinear with the BGRF1 locus, while OSGRF4 is located on chromosome 2, in the vicinity of the second synteny interval (Supplemental Fig. S2A). In Arabidopsis, GRF proteins are known to regulate cell proliferation in leaf primordia and influence the development of appropriate leaf size and shape (Kim and Kende, 2004; Lee et al., 2009). In rice, OsGRF1 has been proposed to play a role in GA-induced stem elongation through activity in the intercalary meristem at the base of the growing internode, a type of meristematic tissue present only in monocots (van der Knaap et al., 2000; Choi et al., 2004).
KIBPs Are Nucleus Localized and Able to Bind the 305-bp Element in Vitro
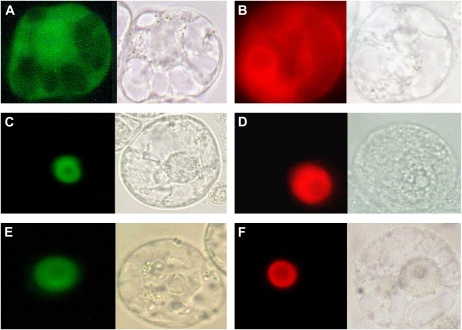
The presence of DNA-binding domains and localization to the nucleus of BERF1 and BEIL1 (Fig. 3) support their putative function as transcription factors. BGRF1 also contains a putative nuclear localization signal (K and R residues within the WRC domain, amino acids 121–165), which should function as in the related OsGRF1 protein, also shown to localize to the nucleus (van der Knaap et al., 2000). However, Red Fluorescent Protein (RFP) fusion experiments show a less clear subcellular localization for BGRF1 compared with BERF1 and BEIL1, with a fainter and more diffuse fluorescent signal, also present in the nucleus (data not shown).
Figure 3.
Subcellular localization of BEIL1 and BERF1 in transiently transformed tobacco protoplasts. A and B, Cytoplasmic and nuclear localization of GFP and RFP, respectively. C and D, Nuclear localization of BEIL1 fused to GFP and RFP, respectively. E and F, Nuclear localization of BERF1 fused to GFP and RFP, respectively. To the right of each image, the corresponding bright-field image is shown.
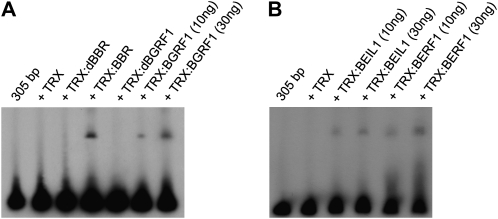
Recombinant KIBPs were produced in prokaryotic systems and tested in vitro for binding to the intron IV 305-bp element of the Bkn3 gene. Coding sequences were expressed in Escherichia coli to produce N-terminal fusions with a modified thioredoxin (His-Patch thioredoxin [TRX]), and the tagged proteins were purified by affinity chromatography (Supplemental Fig. S4). In these experiments, the BBR protein, previously studied by Santi et al. (2003), was used as a control. When TRX:KIBP proteins were used in electrophoretic mobility shift assays (EMSAs), binding to the 305-bp element was evident (Fig. 4). Truncated versions of BBR and BGRF1, lacking the zinc finger domains, were not able to shift the 305-bp element (Fig. 4A). Deletions within the 305-bp element allowed the localization of the binding region for each KIBP. Three overlapping fragments were produced by PCR (Supplemental Fig. S5): sequences spanning the region from 80 to 305 bp were retarded by TRX:BEIL1, TRX:BERF1, and TRX:BGRF1 (data not shown). This region was subsequently divided into 11 overlapping fragments (numbered 1–11), produced by annealing 34-bp-long complementary oligonucleotides (Fig. 5D; Supplemental Table S5).
Figure 4.
Interaction of KIBPs with the 305-bp element in vitro. A, Binding of full-length TRX:BBR and TRX:BGRF1 to the 305-bp element, detected as a band shift. B, Binding of increasing amounts of full-length TRX:BEIL1 and TRX:BERF1 to the 305-bp element, detected as a band shift. TRX and deleted versions TRX:dBBR and TRX:dBGRF1 are negative controls.
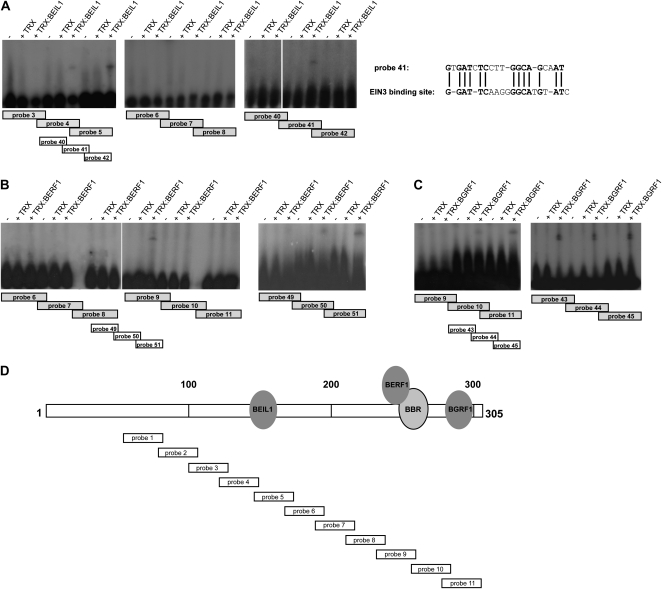
Figure 5.
Mapping of KIBP-binding sites within the 305-bp regulatory element in vitro. A, Left, in vitro interaction between TRX:BEIL1 and the region 100 to 241 bp of the regulatory element. Right, binding of TRX:BEIL1 to the region 122 to 175 bp. The positions of probes 40 to 42 relative to probes 4 and 5 are indicated. Bases similar to a DNA fragment recognized by EIN3 are in boldface. B, Left, in vitro interaction between TRX:BERF1 and the region 168 to 305 bp of the regulatory element. Right, binding of TRX:BERF1 to the region 200 to 262 bp. The positions of probes 49 to 51 relative to probes 8 and 9 are indicated. C, In vitro interaction between TRX:BGRF1 and the region 236 to 305 bp of the regulatory element, covered by 34-bp probes (left), and the region 257 to 305 bp, covered by 24-bp probes (right). The positions of probes 43 to 45 relative to probes 10 and 11 are indicated. −, Free probe. D, Schematic representation of the 305-bp intronic element. Eleven overlapping 34-bp fragments (1–11) spanning the region from 80 to 305 bp were tested in gel retardation assays. The positions of the KIBP-binding sites are indicated.
BEIL1 bound probes 4 and 5, covering the DNA region from 122 to 175 bp (Fig. 5A), whereas BERF1 was able to recognize probe 9 (Fig. 5B), spanning the region from 236 to 265 bp of the 305-bp element. These sequences were dissected into overlapping 24-bp oligonucleotides (Fig. 5; Supplemental Table S6). Sequence GTGATCTCCTTGGCAGCAAT (fragment 41) contained the BEIL1-binding site and displayed similarities with a sequence present in the AtERF1 promoter bound by AtEIN3 (Fig. 5A; Solano et al., 1998). BERF1 was able to bind fragments 50 and 51, spanning the region 220 to 262 bp (Fig. 5B).
BGRF1 has affinity for the region from 257 to the end of the 305-bp element (covered by probes 10 and 11; Fig. 5C), which contains the TTGAC motif, considered to be the core element of the W-box recognized by plant zinc finger transcription factors of the WRKY family (PLACE database; Higo et al., 1999). Subsequent band shift experiments, performed with shorter synthetic oligonucleotides spanning the region corresponding to probes 10 and 11, displayed binding of TRX:BGRF1 to all probes used (Fig. 5C).
Competition assays show that binding of BGRF1, BEIL1, and BERF1 to specific radiolabeled fragments is increasingly displaced by 5×, 25×, and 50× molar excess of unlabeled fragments with the same sequence (Supplemental Fig. S4, D–F). A schematic representation of the regions within the intron IV 305-bp element that mediate KIBP binding is shown in Figure 5D.
KIBPs Are Able to Interact with the 305-bp Element in Rice Protoplasts
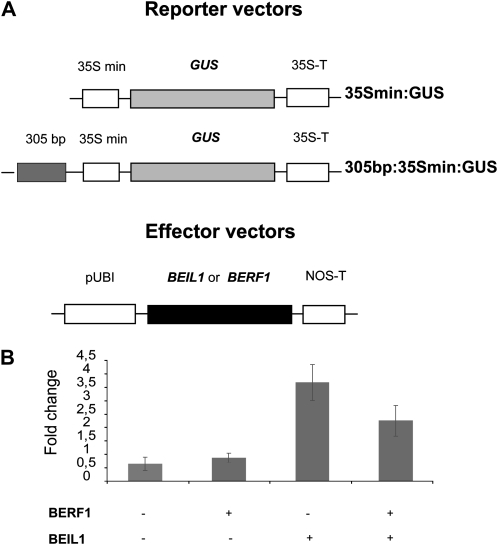
In order to confirm yeast one-hybrid and EMSA DNA-binding results and to analyze interactions between BERF1 and BEIL1 proteins and the 305-bp element in vivo, we transfected rice protoplasts with a GUS reporter construct driven by the 35S minimal promoter (35Smin:GUS) or a similar GUS construct where the 305-bp element was inserted upstream of the 35S minimal promoter (305:35Smin:GUS). These reporter constructs were used in combination with one or both of the effector plasmids, PUbi:BERF1 and PUbi:BEIL1 (Fig. 6A). In all cases, a 35S:luciferase construct was cotransfected as a control for transformation efficiency. Activity of the effectors was derived from GUS activity normalized on luciferase activity.
Figure 6.
Interaction of KIBPs with the 305-bp element in rice protoplasts. Transactivation activity of BERF1 and BEIL1 was tested on 305-bp 35S minimal-driven expression of the GUS gene by transient transformation. A, Schematic representation of reporter and effector plasmids. In the two effector plasmids, BERF1 and BEIL1 coding sequences were inserted downstream of the constitutive ubiquitin promoter. B, GUS activity was normalized on LUC activity, which served as a control for transformation efficiency. Each histogram indicates the GUS-LUC activity ratio of the effector with 305:35Smin:GUS after subtraction of the value obtained with the same effector and the control 35Smin:GUS. The graph reports mean values of six independent transformations of each construct combination, and error bars represent sd of biological replicates.
The expression of the reporter gene when 305:35Smin:GUS was cotransformed with BEIL1 was increased around 3.9-fold compared with the expression of 35Smin:GUS with BEIL1 (Fig. 6B). Reporter expression did not differ significantly between BERF1 with 305:35Smin:GUS and BERF1 with 35Smin:GUS (Fig. 6B). When both BERF1 and BEIL1 were cotransfected with 305:35Smin:GUS, GUS expression was reduced to about half the activity obtained with BEIL1 alone (Fig. 6B). These results support the ability of both BERF1 and BEIL1 to bind the 305-bp element in vivo, consistent with EMSA experiments. These results suggest that BERF1 and BEIL1 play antagonistic roles in intron-mediated regulation of Bkn3.
KIPB Genes Have Broader Expression Patterns Than Bkn3 during Barley Development
In order to survey the expression of KIBP genes and Bkn3 during the development of wild-type Atlas and of its Hooded version Kap-Atlas, reverse transcription (RT)-PCR analyses were performed on roots, stems, and leaves of 5-d-old seedlings (Fig. 7A), on developing leaves and immature inflorescences of 4-week-old plants (Fig. 7B), and on flag leaves and developing spikelets of 7-week-old plants (Fig. 7C). In all these experiments, the Ubiquitin gene was used as a control. BBR and BERF1 genes were expressed in all analyzed organs throughout development. For BERF1, two distinct transcripts were identified, likely deriving from alternative splicing: in the shorter transcript, the three introns were spliced out, producing a protein of 359 amino acids, whereas in the long version, intron II (78 bp) was retained, resulting in a protein of 385 amino acids. The two forms seem to be equally represented.
Figure 7.
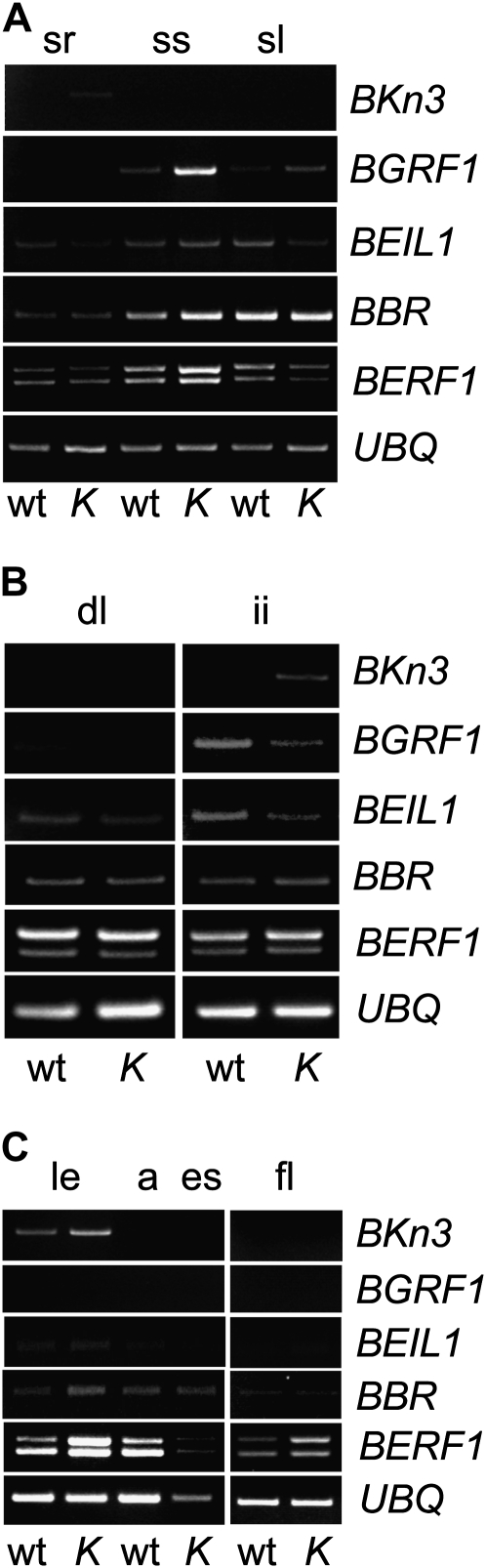
Bkn3 and KIBP gene expression profiles throughout barley development. A, RT-PCR analysis of total RNA extracted from seedling roots (sr), seedling stems (ss), and seedling leaves (sl). B, RT-PCR analysis of total RNA extracted from developing leaves (dl) and immature inflorescences (ii) of 4-week-old plants. C, RT-PCR analysis of total RNA extracted from lemmas (le), wild-type awns (a) or Hooded ectopic structures (es), and flag leaves (fl) of 7-week-old plants. The Ubiquitin (UBQ) gene was used as control. wt, Cultivar Atlas; K, Kap mutant in background Atlas.
In 5-d-old seedlings (Fig. 7A), BEIL1 and BGRF1 transcripts were detected after 35 PCR cycles in all tissues, with the exception of BGRF1, which is not expressed in roots. Bkn3 transcripts were detected, although at low levels, in roots of Kap seedlings.
In 4-week-old plants (Fig. 7B), BEIL1 transcripts were present in developing leaves and inflorescences, while those of BGRF1 were detected in immature inflorescences. Bkn3 transcripts were absent in developing leaves and wild-type developing spikes. According to Kirby and Appleyard (1987), in 4-week-old plants at the awn primordium stage, the meristematic dome of the ear has already ceased its activity and the shoot apex has already formed all leaf and spikelet primordia. Bkn3 expression is evident in Kap developing spikes (Fig. 7B): at this stage, ectopic meristems are formed in the mutant at the tip of lemmas (Yagil and Stebbins, 1969) as a result of the reactivation of Bkn3 activity (Müller et al., 1995). In wild-type immature inflorescences, no signal could be detected for Bkn3 with this technique after 30 PCR cycles.
In 7-week-old plants (Fig. 7C), BGRF1 and BEIL1 transcripts were not detected in any of the examined organs. Bkn3 transcripts were absent in flag leaves, in differentiating wild-type awns, and in the hoods of Kap spikelets. Bkn3 transcription was evident in green developing lemmas in both the wild type and Kap; at this stage, before anthesis takes place, lemma and palea are actively dividing (Kirby and Appleyard, 1987).
In immature inflorescences, corresponding to the critical stage of awn versus hood formation, the expression of BERF1, BEIL1, and BGRF1 was detected, implying the possibility for interactions of the respective proteins in the regulation of Bkn3.
KIPB genes thus show broader expression patterns than Bkn3, suggesting that they may play additional roles besides the regulation of Bkn3.
Ethylene Is Able to Partially Suppress the Hooded Phenotype in Barley
The finding that BERF1 and BEIL share sequence similarity with components of the ethylene cascade suggested that they may also regulate Bkn3 expression in response to ethylene. To investigate if ethylene affects the expression of genes involved in Bkn3 modulation, the potential effect of ethylene on the wild type and Kap-Atlas was addressed by treating 4-week-old barley plants with ethephon, a compound that releases ethylene. As detailed in the section describing developmental expression patterns of KIBP genes, this is the critical stage for awn versus hood formation. Following ethephon treatment, the development of plants was examined and the expression levels of KIBP and Bkn3 genes were analyzed in young leaves and immature inflorescences of both genotypes considered.
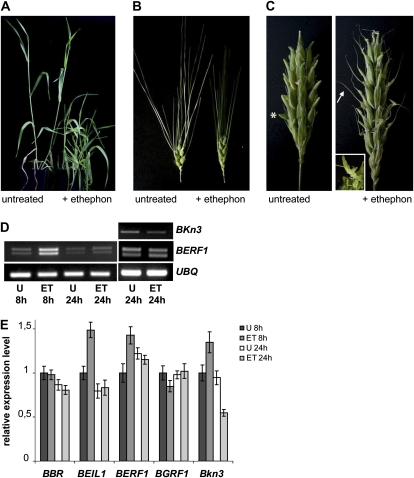
Two weeks after treatment, both wild-type and Kap untreated plants produced on average one tiller, and the main stem brought a developing spike at the booting stage (the growth stage of grasses at the time when the spike is enclosed by the sheath of the uppermost leaf [Kirby and Appleyard 1987]; Fig. 8A; data not shown). In contrast, in both wild-type and Kap ethephon-treated plants, growth of the main stem was arrested and apical dominance was affected, resulting in reduced height and increased tillering: two to three tillers became precociously senescent (Fig. 8A; data not shown). In treated plants, booting took place 2 weeks after untreated plants (data not shown). In wild-type spikes, ethephon treatment induced a shortening of awn length and a decrease in kernel size (Fig. 8B; Lauer, 1991). In Kap, 80% of treated spikes displayed a partial change toward the awned condition with varying degrees of penetrance, depending on the position of the spikelet in the ear (Fig. 8C). This is consistent with the developmental gradient of spikelet differentiation, starting from the mid part of the ear and progressing toward the base and the tip (Kirby and Appleyard, 1987). In some spikelets from ethephon-treated Kap plants, additional ovaries were produced: this phenotype is reminiscent of those of kn1 loss-of-function mutants reported by Vollbrecht et al. (2000), in which recessive kn1 mutations result in extra carpels. Similar modifications were never observed in untreated Kap plants under our growth conditions. These results suggest that ethylene may antagonize the Kap phenotype through the repression of Bkn3.
Figure 8.
Ethephon-induced alterations in barley. A, Left, untreated Kap plants carrying the main culm at the booting stage. Right, ethephon-treated Kap plants displaying three to four tillers each. B, Left, untreated wild-type spike. Right, ethephon-treated wild-type spike. C, Left, untreated Kap spike carrying the typical ectopic structures (developing hoods, indicated by an asterisk). Right, ethephon-treated Kap spike displaying rudimental awns (indicated by an arrow). The inset shows an abnormal ovary in closeup. D, Left, RT-PCR results of RNA extracted from Kap developing leaves 8 and 24 h after ethephon treatment. Right, RT-PCR results of RNA extracted from Kap immature inflorescences. Ubiquitin (UBQ) was used as an amplification control in PCR. E, Relative expression levels of Bkn3 and KIBP genes in immature inflorescences of the Kap mutant 8 and 24 h after ethephon treatment. The ratio between each gene level and the Ubiquitin gene from untreated plants was arbitrarily set at 1. Three biological replicates were performed independently, and mean values with sd are indicated. U, Untreated; ET, ethephon treated. [See online article for color version of this figure.]
In order to gain insight into the gene expression changes that accompany these phenotypic modifications, we determined the levels of expression of BERF1, BEIL1, and Bkn3 at two different time points in untreated and ethephon-treated wild-type and Kap plants. RNA samples were extracted from the fifth developing leaves and inflorescences. In leaves of both genotypes, BERF1 reached a maximum up-regulation of 3-fold compared with the control 8 h after ethephon treatment, with unaltered relative amounts of its two alternative transcripts (Fig. 8D; results from quantitative [q]RT-PCR not shown); at 24 h, BERF1 expression in the wild type and Kap aligned to untreated plants (Fig. 8D). Bkn3 expression was already shown to be absent in leaves (Fig. 7B). In wild-type and Kap immature inflorescences, 8 h after treatment, BERF1 and BEIL1 were up-regulated 1.5-fold, whereas at 24 h, BERF1 and BEIL1 expression levels were not significantly different from those of untreated plants (Fig. 8, D and E). In Kap inflorescences, levels of Bkn3 transcripts were reduced by 50% at 24 h after ethephon treatment. This is consistent with RT-PCR data based on a different Bkn3-specific primer pair (Fig. 8D): this down-regulation can be correlated with the partial reversion toward awn formation in ethephon-treated spikes. Taken together, these results are consistent with a possible repressive role of BERF1 and/or BEIL1 in the expression of Bkn3. RNA from ethephon-treated plants was additionally tested for the levels of expression of BGRF1 and BBR, thus producing further control data: expression levels of these two KIBP genes were unaffected by ethylene (Fig. 8E).
BERF1 and BEIL1 upstream genomic sequences were scanned for the presence of boxes mediating the ethylene response, as described by Lin et al. (2007). Multiple copies of the GCC box, considered to be the core motif of the ethylene-responsive elements (Ohme-Takagi and Shinshi, 1995), are present in both BERF1 and BEIL1 upstream sequences (Supplemental Fig. S6).
Transgenic Rice Overexpressing KIBP Genes Displays Ethylene-Related Alterations
Considering the lack of barley mutants defective in BERF1 and BEIL1 gene function, the morphological effects of the constitutive expression of these genes under the control of the Ubiquitin promoter were studied in transgenic rice, allowing further investigation of their involvement in the ethylene pathway.
To date, few transcription factors involved in the ethylene signal transduction pathway have been characterized in rice. Among these, OsEIL1 was shown to act as a positive regulator of the ethylene response (Mao et al., 2006), as overexpressor rice plants displayed an increase in the expression levels of the ERF gene OsEBP89 and of aminocyclopropane carboxylic acid (ACC) oxidase gene 1 (OsACO1), encoding a key enzyme in ethylene biosynthesis.
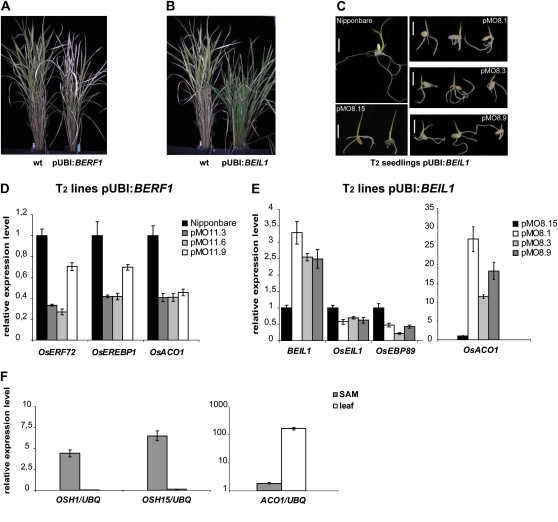
Several pUBI:BERF1 T0 lines expressing the transgene were selected and plants with the highest levels of expression (pMO11) were reproduced to T1 (Supplemental Fig. S7C). Analyses of segregating progeny from other T0 lines were not possible, as plants were sterile. Southern-blot analysis of T1 and T2 plants (Supplemental Fig. S7A) indicated that at least two copies of the transgene were present in the rice genome, most probably organized in tandem. Morphological analysis of T1 plants showed that they were shorter (plant height to the base of the panicle was −10%) and developed significantly fewer tillers (−30%) compared with Nipponbare (Fig. 9A).
Figure 9.
Transgenic rice plants overexpressing BERF1 and BEIL1. A and B, BERF1 and BEIL1 top overexpressor plants (T1 generation, pMO11 and pMO8 lines, respectively) compared with wild-type (wt) rice (Nipponbare). C, Five-day-old seedlings from Nipponbare and T2 segregation progeny of BEIL1 overexpression plants (pMO8). Bars = 1 cm. D, Relative expression levels of endogenous OsERF72, OsEREBP1, and OsACO1 genes in BERF1 expressors (T2 generation). Total RNA was extracted from a pool of five 10-d-old seedlings. The ratio between each gene level and the OsUBQ gene level obtained from wild-type plants was arbitrarily set at 1. Nipponbare was used as a wild-type control. E, Relative expression levels of the exogenous BEIL1 gene and endogenous OsEIL1, OsEBP89, and OsACO1 genes. Total RNA was extracted from a pool of five 7-d-old seedling leaves of BEIL1 overexpression plants (T2 generation). The ratio between each gene level and the OsUBQ gene level obtained from mild overexpressor plants (pMO8.15) was arbitrarily set at 1. F, Relative expression levels of endogenous OSH1, OSH15, and OsACO1 genes in 1-week-old wild-type seedlings. Total RNA was extracted from a 1-cm region of the stem containing the SAM and the first true leaf from a pool of five Nipponbare plants. qRT-PCR was performed in triplicate, and mean values with sd are shown. Two independent biological replicates gave similar results. [See online article for color version of this figure.]
Five T0 lines carrying the pUBI:BEIL1 construct were selected that expressed the transgene. BEIL1 overexpressors (pMO8) were self-pollinated, and the T1 plants obtained were subjected to Southern-blot analysis (Supplemental Fig. S7B). Plants carrying two copies of the transgenic cassette displayed a stronger expression of BEIL1 than plants carrying three copies (Supplemental Fig. S7D). Macroscopic analysis of T1 plants showed both a decrease in overall height and a bushy phenotype associated with a higher level of BEIL1 expression and of the OsACO1 gene (Fig. 9B; Supplemental Fig. S7D). In addition, booting was delayed by about 3 to 4 weeks in top overexpressor plants (data not shown). Consistent with a potential role of BEIL1 in the ethylene pathway, delayed flowering has also been observed following ACC (an ethylene precursor) treatment in Arabidopsis (Achard et al., 2007).
T2 rice plants overexpressing BERF1 and BEIL1 were analyzed by qRT-PCR to assess the expression levels of endogenous genes involved in the ethylene pathway. OsACO1 is up- and down-regulated in pUBI:BEIL1 and pUBI:BERF1 plants, respectively (Fig. 9, D and E): the positive effect of BEIL1 mimics the outcome of OsEIL1 overexpression (Mao et al., 2006), whereas repression of OsACO1 by BERF1 may be due to the cosuppression of related endogenous genes OsERF72/OsEBP2 and OsEREBP1, previously shown to be responsive to ethylene and to bind the GCC box, respectively, implying potential roles in the ethylene cascade (Cheong et al., 2003; Lin et al., 2007). BEIL1 top overexpressors display a slight decrease in endogenous OsEIL1 (and of OsEBP89) compared with BEIL1 mild overexpressors: this result appears to be in contrast with previous findings (Mao et al., 2006). However, the increase of OsACO1 transcription could be explained by the BEIL1-mediated transactivation of other ERF genes (different from OsEBP89) that may act as nuclear modulators of the ethylene cascade.
When T2 seedlings were grown on Murashige and Skoog medium, BERF1 expressors did not show any visible alteration (data not shown), whereas BEIL1 expressors displayed a short-shoot and short-root phenotype (Fig. 9C) similar to that of rice transgenic plants overexpressing OsEIL1 and wild-type plants treated with high concentration of ACC (Mao et al., 2006). However, BEIL1 expressors did not show the coiled primary root like OsEIL1 expressors.
Together, data from rice transgenic lines support the link between BEIL1, BERF1, and the ethylene pathway, consistent with their homology to transcription factors involved in the response to this hormone.
In addition, we monitored the relative expression levels of two rice homeobox genes, OSH1 and OSH15, and of OsACO1 in 1-week-old wild-type rice by qRT-PCR analysis (Fig. 9F). OSH1 and OSH15 expression could not be detected in the first true leaf, but transcripts of these two homeobox genes were present in a 1-cm region of the developing stem comprising the SAM. On the contrary, OsACO1 was highly expressed in the first true leaf, its transcripts being 100 times more represented in developing leaves than in developing stems containing the SAM. These findings strongly support a mutual exclusion pattern between KNOX I genes and OsACO1, an ethylene biosynthesis gene.
DISCUSSION
Regulation of KNOX Genes
The correct spatial and temporal expression of class I KNOX genes is crucial for the initiation of organ primordia at the flanks of the SAM (for review, see Hake et al., 2004). Besides the transcriptional and epigenetic mechanisms reviewed in the introduction, posttranslational control occurs through specific protein-protein interactions and protein trafficking (Müller et al., 2001; Hake et al., 2004). Despite these insights, we still lack a thorough understanding of either the mechanisms underlying the establishment and maintenance of patterns of KNOX I expression or the metabolic pathways leading from gene expression to final phenotypes.
In barley, a link has been established between the level of expression of a class I KNOX gene, Bkn3, and the Kap phenotype. The molecular lesion underlying this dominant mutant is a duplication of a 305-bp intron sequence within the Bkn3 gene itself, causing its overexpression at the distal end of the lemma (Müller et al., 1995). Interestingly, transposon insertions disrupting intron structure of the orthologous intron of maize kn1 result in gene overexpression (Greene et al., 1994). Despite our fragmentary knowledge of the mechanisms underlying the phenotype, Kap can be employed to study the regulation of KNOX I and other genes. Here, we have attempted to associate expression levels with altered mutant morphologies as well as characterizing proteins that interact with Bkn3 intron IV sequences (KIBPs). In vitro binding of these gene products to defined regions within the 305-bp element is consistent with results from yeast one-hybrid and rice protoplast assays. Although further work is needed to define the minimal binding site of each KIBP protein, available data support sequence-specific binding of the proteins to the intronic element and suggest a complex regulatory mechanism, potentially involving interactions between different KIBPs.
The similarity of BERF1 and BEIL1 to previously characterized proteins involved in ethylene signaling (Chao et al., 1997; Solano et al., 1998; Mao et al., 2006) prompted us to investigate whether the two proteins play a role in both intron-mediated regulation and ethylene response. The reduced height and increased tillering of transgenic rice plants that express BEIL1 constitutively resemble phenotypic alterations produced by ethephon treatment in barley. BEIL-overexpressing rice plants exhibit phenotypes consistent with perturbations in ethylene levels (Mao et al., 2006) and significantly increased levels of mRNA from OsACO1, a key gene in ethylene biosynthesis. Taken together, data from rice overexpression suggest that BEIL1 may act as a positive regulator of ethylene biosynthesis. A link between BERF1 and the ethylene pathway is also indicated by alterations in OsACO1 expression in rice transgenics. However, interpretation of these results is complicated due to effects of the transgene on the expression of rice homologs and the sterility of most of the transgenic lines.
In barley, cold and short-day treatments can cause the reversion of Kap toward the awned phenotype, possibly by altering the hormonal balance of lemma primordia (Yagil and Stebbins, 1969). We have used the Kap mutant as a tool to evaluate the effect of different hormones on meristematic activity during the reproductive phase in barley. We investigated the effect of ethephon treatment on the Kap phenotype and monitored the expression of the genes under study. The partial suppression of the Kap phenotype is accompanied by 50% down-regulation of Bkn3 transcript levels, suggesting that ethylene may antagonize the Kap phenotype through the repression of Bkn3 mediated by BERF1 and/or BEIL1. As a basal level of Bkn3 expression was previously detected by poly(A+) RNA-blot analysis in wild-type immature inflorescences at the awn primordium stage, where up-regulation is seen in Kap (Müller et al., 1995), we may hypothesize that quantitative rather than qualitative changes in Bkn3 expression are implicated in awn versus hood development. As the duplication of the 305-bp enhancer element of the Bkn3 gene is responsible for the Kap phenotype (Müller et al., 1995), we propose that there is a connection between ethylene and Bkn3 regulation. Considering evidence implicating BEIL1 and BERF1 in ethylene responses, we infer that the connection between ethylene and Bkn3 may be mediated by these transcription factors, in agreement with their up-regulation in response to ethephon treatment.
The interplay between BERF1 and BEIL1 is likely to be complex, as indicated by reporter assays in rice protoplasts: while BEIL1 can activate the expression of a reporter gene driven by the 305-bp intron element, BERF1 can counteract this activation, suggesting that in planta, the two proteins, likely in association with other proteins such as BGRF1 and BBR, may interact to mediate the ethylene-supported fine-tuning of Bkn3 expression. This idea is supported by the comparison of developmental patterns of KIBP and Bkn3 gene expression, showing partially overlapping domains of transcript accumulation. Based on this interpretation, fine-tuning of Bkn3 expression is expected to depend on the balance of action between different KIBPs interacting with the cis-regulatory element. While efforts to demonstrate protein-protein interactions by in vitro approaches have been unsuccessful to date, in the future it will be interesting to investigate the possible formation of a complex. Analogously to BEIL1 and BERF1, kiwifruit (Actinidia deliciosa) EIL and ERF proteins were recently demonstrated to have contrasting effects in reporter gene expression studies: AdEIL2 and AdEIL3 activated transcription of the ripening-related gene AdXET5 (for xyloglucan endotransglycosylase), while the potential repressor AdERF9 suppressed the activity of this promoter (Yin et al., 2010).
Interactions between KNOX Genes and Hormones
Most studies of interactions between KNOX I genes and hormones have been performed in Arabidopsis or rice (Hay et al., 2004; Sakamoto et al., 2006). Transgenic plants that overexpress KNOX I genes show changes in the levels of endogenous auxin, cytokinin (CK), and GA (Kusaba et al., 1998; Hay et al., 2004). In rice, KNOX I proteins increase CK biosynthesis (Sakamoto et al., 2006) and decrease GA biosynthesis (Sakamoto et al., 2001): the resulting hormonal regime favors SAM formation and maintenance (Sakamoto et al., 2006). It has been proposed that KNOX I genes and CK act synergistically to trigger cell proliferation and antagonistically in cell expansion mediated by elevated GA levels (Jasinski et al., 2005).
The interaction of KNOX I with ethylene has been less studied. In Arabidopsis, ethylene and the KNAT2 gene antagonistically regulate meristematic activity (Hamant et al., 2002): overexpression of KNAT2 antagonizes ethylene, and ethylene regulates KNAT2 expression in the SAM. Treatment with ACC, an ethylene precursor, spatially restricts KNAT2 expression in the SAM and reduces cell number in the L3 layer. These results, while indicating the existence of an interaction between KNOX I genes and ethylene, do not clarify the mechanism underlying this cross talk.
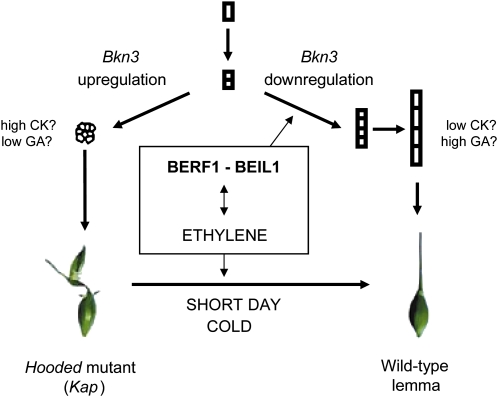
In this paper, we provide evidence for a role of ethylene in the regulation of a barley KNOX I gene. Based on our data, a model illustrating interactions among ethylene, BEIL1, and BERF1 in Bkn3 regulation can be formulated (Fig. 10). BEIL1 and BERF1 act in the ethylene pathway, as their expression is modulated by ethephon and regulates a key gene in ethylene biosynthesis (central box). BEIL1 and BERF1 also play a role in controlling Bkn3 expression by interacting with the 305-bp intronic element: the dynamics of their opposing actions may be delicate and complex, as suggested by protoplast reporter-effector assays. Consistent with the data presented here, we propose that ethylene acts as a negative upstream regulator of Bkn3. However, interactions between the hormone, Bkn3, and KIBP genes may also occur at other levels. Finally, although the model focuses on BEIL1 and BERF1, intron-mediated Bkn3 expression likely depends on interactions of these proteins with BBR, BGRF1, and possibly other factors. The proposed model may be validated in Arabidopsis and rice, where functional genomics tools are better developed than in barley.
Figure 10.
Model of interactions between Bkn3 and ethylene in barley plants (modified from Stebbins and Yagil, 1966). Right, in wild-type lemma primordia, cells divide and elongate to form the awn. Left, in the Kap mutant lemma primordia, cells divide without elongating and form the hood, due to the up-regulation of Bkn3 via the rearrangement of intron IV. Ethylene treatment is linked to the down-regulation of Bkn3 and to the partial restoration of awn formation. BERF1 and BEIL1 are up-regulated in response to ethylene and regulate both ethylene biosynthesis and Bkn3 expression through binding to the intronic element. CK and GA pathways may also be involved in the determination of lemma primordia cell fate toward differentiation (wild type) or proliferation (Kap). [See online article for color version of this figure.]
A possible implication of our model is that KNOX I genes like Bkn3 may, in part, mediate the cross talk between ethylene and GA and CK hormones. The cross talk is complex and also influenced by environmental factors (for review, see Weiss and Ori, 2007), consistent with earlier findings (Yagil and Stebbins, 1969). Reinforcing the idea of a KNOX I-ethylene-GA link, BGRF1 shares sequence similarity with OsGRF1, a rice gene implicated in GA-mediated internode growth in deepwater rice varieties, characterized by the ability to elongate rapidly upon submergence during the monsoon season (van der Knaap et al., 2000). This growth response is mediated by GAs that act to promote cell proliferation in the intercalary meristem and to stimulate cell expansion in the internodal elongation zone (Choi et al., 2004). Interestingly, this response is known to also involve ethylene through the action of the ERF genes SK1 and SK2 (for review, see Bailey-Serres and Voesenek, 2010). Our results suggest that similar interactions between ethylene, GA, ERF, and GRF transcription factors may affect cell proliferation/elongation during barley spikelet development through the regulation of KNOX I activity.
MATERIALS AND METHODS
Hybond-P and -N membranes, the ECL Western Blotting Analysis System, Hyperfilm MP autoradiography film, and radioisotopes were purchased from Amersham Biosciences (GE Healthcare). Oligonucleotides were purchased from Sigma-Genosys and are listed in Supplemental Tables S3 to S8.
Plant Material
Barley (Hordeum vulgare) seeds were vernalized at 4°C for 3 weeks in the dark. Seedlings of the wild type (var Atlas) and Hooded mutant (Kap-Atlas) were transferred to soil and grown until maturity in long-day conditions (16 h of light/18°C, 8 h of dark/13°C).
To analyze the expression profiles of genes of interest throughout barley development, three biological replicates were used. To assess the effect of ethylene on KIBP and Bkn3 genes, plants were treated with 10 mm ethephon (Sigma-Aldrich), previously dissolved in 50 mm phosphate buffer, pH 7, with 0.1% Tween 20, when the fifth leaf emerged as stated by Yagil and Stebbins (1969). Each treatment consisted of 60 plants, and three biological replicates were performed. Controls consisted of untreated plants as well as plants sprayed with buffer without ethephon. Ethephon was sprayed twice at a time interval of 18 h, and plants were covered with a plastic bag; samples were harvested at 8 and 24 h from the second treatment as pools of five developing leaves and immature inflorescences. RNA samples were extracted from the fifth developing leaves and inflorescences. qRT-PCR analyses (see below), carried out on untreated plants and plants treated with Tween-phosphate buffer, indicated that in leaf and inflorescence, Ubiquitin and KIBP gene expression levels were not altered in the wild type or Kap (data not shown).
Genetic Mapping of BGRF1, BERF1, and BEIL1 Genes
Map positions of KIBP genes were determined from linkage analysis between polymorphic gene fragments amplified with the specific primers reported in Supplemental Table S3 and subjected to standard single-strand conformation polymorphism analysis and amplified fragment length polymorphism markers previously mapped on the Proctor × Nudinka population. Segregation analysis used MAPMAKER/EXP3.0 with a logarithm of the odds score of 3 and maximum distance of 50 centimorgan. Map positions of genes were entered into the linkage map of Castiglioni et al. (1998). Details on mapping populations and markers used for each gene are given in the legend of Supplemental Figure S2, A to C.
Phylogenetic Analyses
Rice (Oryza sativa), Arabidopsis (Arabidopsis thaliana), and Brachypodium distachyon BERF1- and BEIL1-related sequences were identified based on similarity searches of the respective genome sequences (http://blast.ncbi.nlm.nih.gov/Blast.cgi and http://www.tigr.org/plantProjects.shtml). Pair-wise and multiple sequence alignments and similarity values were obtained using BioEdit (Hall, 1999). Conceptually translated sequences were aligned with MUSCLE (Edgar, 2004), and alignments were manually refined using Se-Al (http://tree.bio.ed.ac.uk/software/seal/). Regions of the alignment not unambiguously alignable were excluded using GBlocks (Castresana, 2000) to yield data sets of 14 sequences, 242 positions for EIL and 39 sequences, 96 positions for ERF (ERF DNA-binding domain). PhyML (Guindon and Gascuel, 2003) was used to perform bootstrap analyses (100 replicates) using the WAG substitution model (Whelan and Goldman, 2001) with four γ-distributed substitution rate categories and an invariable rate category).
Barley-Rice Synteny Analyses
Based on previous analyses, the regions hosting KIBP genes share colinearity with defined rice genomic intervals (Rossini et al., 2006): RFLP marker sequences deriving from this work and flanking the KIBP gene of interest were obtained from the GrainGenes database (http://wheat.pw.usda.gov/GG2/index.shtml). BLASTn searches against the HarvEST barley database (http://harvest.ucr.edu/) allowed in many cases the recovery of unigene clusters corresponding to the RFLP probe. These were then mapped on the current assembly of the rice genome sequence by tBLASTx searches through the Gramene server (http://www.gramene.org/multi/blastview), confirming previous results (Rossini et al., 2006), except for slight changes in chromosome coordinates. Details of the markers used for comparative mapping are given in Supplemental Figure S3, A to C.
Cloning
Yeast one-hybrid and barley library screens were as described by Santi et al. (2003). A genomic λEMBL3 library from the barley genotype Calcaroides-C15 was screened with the longest cDNA clones obtained from the one-hybrid screen. Among 2 million λ clones plated, three positives were obtained for BERF1, three for BEIL1, and six for BGRF1. The longest cDNA clones from the screens were introduced in pBluescript KS− (Agilent Technologies) as EcoRI fragments and used as templates in PCR-based clonings. All constructs were verified by sequencing, and gene-specific primers are listed in Supplemental Table S4.
To produce fusions with RFP and GFP, full-length coding sequences for BERF1 and BEIL1 were amplified with primers NcoI-BERF1 f/NcoI-BERF1 r and NcoI-BEIL1 f/NcoI-BEIL1 r and cloned as NcoI fragments in pGJ1425 and pGFP-JS, respectively. Nuclear localization was assessed in transiently transformed tobacco (Nicotiana tabacum) protoplasts as reported by Santi et al. (2003).
In order to produce N-terminal fusions with a modified thioredoxin, KIBP coding sequences were subcloned in pENTR4 (Invitrogen) or pGEM-T Easy (Promega) vectors as described in Supplemental Materials and Methods S1 and then introduced into pThioHisB expression vector (Invitrogen). BBR and BERF1 coding sequences were excised from pENTR4 constructs and in-frame cloned in pThioHisB as NcoI/XhoI fragments. BGRF1 and BEIL1 coding sequences were excised from pGEM-T Easy constructs and in-frame cloned in pThioHisB as KpnI/PstI and BglII/PstI fragments, respectively. Partial versions of BBR and BGRF1, deleted in their DNA-binding domain, were obtained by cloning NcoI/SacI and NcoI/PstI restriction fragments in pThioHisB vector.
To overexpress BERF1 and BEIL1 in rice, binary constructs were obtained by Gateway technology. attB-flanked PCR products, produced by the amplification of BERF1 and BEIL1 cDNAs with attB-specific primers, were recombined in a modified version of the pCAMBIA-5300 vector containing attP sites downstream of the ubiquitin promoter. To test the transactivation activity of BERF1 and BEIL1 in rice protoplasts, HindIII-KpnI fragments carrying the ubiquitin promoter and BERF1 and BEIL1 coding sequences were excised from pCAMBIA constructs obtained previously and cloned into a pCRII-TOPO vector (Invitrogen) containing the NOS terminator.
All clones aimed at the expression of BERF1 contain the short version of the coding sequence where the second intron is spliced out.
Recombinant Protein Production and Purification
Escherichia coli (TOP10 strain) cells were transformed with pThioHis constructs, grown at 24°C, and induced with 0.5 mm isopropyl β-d-1-thiogalactopyranoside for 3 h. TRX fusions were purified via affinity chromatography on ProBond resin (Invitrogen) and eluted with a step gradient of imidazole (50, 100, and 250 mm). TRX fusions were analyzed by SDS-PAGE, transferred to Hybond-P membranes by western blotting, and immunodetected with anti-thioredoxin-specific antibody (Sigma-Aldrich) and the ECL Western Blotting Analysis System, as suggested by the manufacturer (GE Healthcare).
DNA-Protein Interactions
Purified recombinant proteins (TRX fusions, 50 mm imidazole elution) were tested in EMSAs. The 305-bp element was obtained by XhoI digestion of (3× 305):GUS plasmid, and protruding ends were labeled as described by Santi et al. (2003). To map binding sites, synthetic oligonucleotides spanning the whole regulatory element were designed (Supplemental Tables S5 and S6). Specific complementary primers were denatured for 10 min at 100°C and allowed to anneal overnight at room temperature; the annealing reaction was diluted to a final concentration of 100 ng μL−1 prior to end labeling using T4 Polynucleotide Kinase (Invitrogen) and [γ32P]ATP (10 μCi μL−1). Unincorporated nucleotides were removed from radiolabeled probes with the QIAquick Nucleotide Removal kit (Qiagen). Binding reactions were performed in 1× G1 buffer (Santi et al., 2003) with 10 to 30 ng of recombinant protein and 5 pmol of specific probe for 20 min at room temperature. Double-stranded poly(dI-dC)-poly(dI-dC), bovine serum albumin, and heparin (Sigma-Aldrich) were used as nonspecific competitors in DNA-protein binding assays [1 μg of poly(dI-dC), 1 μg of bovine serum albumin, and 500 ng of heparin each reaction for bigger fragments; 100 ng of poly(dI-dC), 100 ng of bovine serum albumin, and 30 ng of heparin each reaction for smaller fragments]. The 305-bp KIBP complexes were resolved onto native 5% polyacrylamide (37.5:1) gels, whereas complexes with smaller probes were electrophoresed onto native 10% polyacrylamide gels. Gels were dried for 45 min at 80°C and exposed to autoradiography film at −80°C with intensifying screens.
Transgenic Rice Production and Analysis
Transformed Agrobacterium tumefaciens was cocultivated with rice calli (cv Nipponbare), and plants were regenerated as described by Sallaud et al. (2003). Transformed plantlets resistant to hygromycin were transferred to test tubes and subjected to RT-PCR screening: Ubiquitin was considered the reference, and barley gene-specific primers were used to discriminate different degrees of transgene expression. Lines overexpressing BEIL1 and BERF1 were selected based on RT-PCR carried out on retrotranscribed RNA samples extracted from developing leaves (Supplemental Fig. S7). Selected regenerants were transferred to soil pots and grown until maturity in a containment greenhouse at the Centre de Coopération Internationale en Recherche Agronomique pour le Développement, Montpellier, France. T0 overexpressor plants were self-pollinated, and T1 and T2 progeny were further characterized.
Rice genomic DNA was extracted from young leaves of 4-week-old transgenic plants with the mixed alkyl trimethyl ammonium bromide method (Sallaud et al., 2003). For this, 10 μg of purified DNA was digested with EcoRI and separated on 0.8% agarose gels. DNA was transferred onto Hybond-N membranes using a standard Southern-blot protocol, and transgene copy number was assessed by hybridization with a radiolabeled probe related to the Hygromycin Phosphotransferase gene (Sallaud et al., 2003).
Gene Expression Analysis
KIBP gene expression profiles during barley development and in transgenic rice plants were determined by RT-PCR, whereas the expression of genes of interest in hormone-treated barley and in T2 transgenic rice lines was analyzed by qRT-PCR. Total RNA was isolated from different organs using TRI-Reagent (Sigma-Aldrich) and treated with deoxyribonuclease I, amplification grade (Invitrogen). A total of 1.5 μg of DNaseI-treated total RNA was primed with oligo(dT) and random hexamers for RT-PCR and qRT-PCR applications, respectively, and retrotranscribed with SuperScript III Reverse Transcriptase (Invitrogen). cDNAs were diluted 1:5, and 2 μL was used for further analyses.
Standard PCR was performed with specific primers (Supplemental Table S7) and GoTaq polymerase in 1× Green GoTaq reaction buffer (Promega). Amplicons were electrophoresed on 3% agarose gels supplemented with SYBR Safe DNA gel stain (Invitrogen). Quantitative analysis of gene expression in hormone-treated barley plants and T2 transgenic rice lines was by qRT-PCR in the 7300 Real-Time PCR System (Applera) by using SYBR Green Master Mix and specific primers reported in Supplemental Table S8.
Transient Expression Experiments
Effector constructs described in the cloning section and reporter vectors described by Santi et al. (2003) were used in transient transformations of protoplasts derived from 2-week-old rice seedlings of the Chinese cultivar Minghui86 (indica subspecies) grown in the dark at 28°C and 85% humidity. Protoplast isolation and polyethylene glycol-mediated transfection were performed as described by Chen et al. (2006). In each experiment, vectors carrying the firefly luciferase gene under the control of a constitutive promoter were introduced as transformation controls.
After transfection, protoplasts were incubated in W5 medium (154 mm NaCl, 125 mm CaCl2, 5 mm KCl, and 2 mm MES, pH 5.7) at 25°C for 16 h in the dark and used for both GUS assay and protein quantification. GUS activity was estimated at 0.5, 3, 6, and 24 h and was expressed as pmol 4-methylumbelliferone mg−1 min−1 referred to luciferase activity. For each experiment, all the transfections were performed in triplicate, and GUS assays were performed in triplicate for each transfection.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: BGRF1 (HQ328942), BERF1 (HQ328941), and BEIL1 (HQ328940).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Assignment of the BGRF1 locus to barley chromosome 2H (A), the BERF1 locus to barley chromosome 5H (B), and the BEIL1 locus to barley chromosome 7H (C).
Supplemental Figure S2. Colinearity between barley chromosome 2H and rice chromosomes 2 and 4 (A), barley chromosome 5H and rice chromosome 9 (B), and barley chromosome 7H and rice chromosome 8 (C).
Supplemental Figure S3. Sequence alignment of class VII ERFs (A) and EIL proteins (B).
Supplemental Figure S4. Analysis of purified thioredoxin fusions and EMSA experiments.
Supplemental Figure S5. KIBP-specific binding to the 305-bp element.
Supplemental Figure S6. Nucleotide sequences and analysis of BERF1 and BEIL1 upstream genomic sequences.
Supplemental Figure S7. Analysis of transgenic rice plants overexpressing BERF1 and BEIL1.
Supplemental Table S1. Information on ERF genes from Arabidopsis and japonica and indica rice closely related to BERF1.
Supplemental Table S2. Information on EIL genes from Arabidopsis and rice.
Supplemental Table S3. Primers used in single-strand conformation polymorphism analysis for mapping of KIBP genes.
Supplemental Table S4. Modified primers used in PCR-based cloning of KIBP genes.
Supplemental Table S5. Specific oligonucleotides used in EMSA to assess the interaction between KIBP and 34-bp double-stranded fragments.
Supplemental Table S6. Specific oligonucleotides used in EMSA to map KIBP-binding sites.
Supplemental Table S7. Specific primers used in standard RT-PCR experiments
Supplemental Table S8. Specific primers used in qRT-PCR experiments.
Supplementary Material
Acknowledgments
We gratefully acknowledge the help of Jean Cristophe Breitler for providing technical help in the production of constructs for rice transformation and Soraya Pelaz and the staff at the Centre for Research in Agricultural Genomics for their advice and support on real-time PCR analyses, rice, and protoplast experiments.
References
- Achard P, Baghour M, Chapple A, Hedden P, Van Der Straeten D, Genschik P, Moritz T, Harberd NP. (2007) The plant stress hormone ethylene controls floral transition via DELLA-dependent regulation of floral meristem-identity genes. Proc Natl Acad Sci USA 104: 6484–6489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M, Ishida T, Tasaka M. (1999) Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development 126: 1563–1570 [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J, Voesenek LA. (2010) Life in the balance: a signaling network controlling survival of flooding. Curr Opin Plant Biol (in press) [DOI] [PubMed] [Google Scholar]
- Becraft PW, Freeling M. (1994) Genetic analysis of Rough sheath1 developmental mutants of maize. Genetics 136: 295–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürglin TR. (1997) Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res 25: 4173–4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne ME, Barley R, Curtis M, Arroyo JM, Dunham M, Hudson A, Martienssen RA. (2000) Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408: 967–971 [DOI] [PubMed] [Google Scholar]
- Cao Y, Song F, Goodman RM, Zheng Z. (2006) Molecular characterization of four rice genes encoding ethylene-responsive transcriptional factors and their expressions in response to biotic and abiotic stress. J Plant Physiol 163: 1167–1178 [DOI] [PubMed] [Google Scholar]
- Castiglioni P, Pozzi C, Heun M, Terzi V, Müller KJ, Rohde W, Salamini F. (1998) An AFLP-based procedure for the efficient mapping of mutations and DNA probes in barley. Genetics 149: 2039–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17: 540–552 [DOI] [PubMed] [Google Scholar]
- Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker JR. (1997) Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89: 1133–1144 [DOI] [PubMed] [Google Scholar]
- Chen S, Tao L, Zeng L, Vega-Sanchez ME, Umemura K, Wang GL. (2006) A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol Plant Pathol 7: 417–427 [DOI] [PubMed] [Google Scholar]
- Cheong YH, Moon BC, Kim JK, Kim CY, Kim MC, Kim IH, Park CY, Kim JC, Park BO, Koo SC, et al. (2003) BWMK1, a rice mitogen-activated protein kinase, locates in the nucleus and mediates pathogenesis-related gene expression by activation of a transcription factor. Plant Physiol 132: 1961–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi D, Kim JH, Kende H. (2004) Whole genome analysis of the OsGRF gene family encoding plant-specific putative transcription activators in rice (Oryza sativa L.). Plant Cell Physiol 45: 897–904 [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golz JF, Keck EJ, Hudson A. (2002) Spontaneous mutations in KNOX genes give rise to a novel floral structure in Antirrhinum. Curr Biol 12: 515–522 [DOI] [PubMed] [Google Scholar]
- Greene B, Walko R, Hake S. (1994) Mutator insertions in an intron of the maize knotted1 gene result in dominant suppressible mutations. Genetics 138: 1275–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704 [DOI] [PubMed] [Google Scholar]
- Guo H, Ecker JR. (2004) The ethylene signaling pathway: new insights. Curr Opin Plant Biol 7: 40–49 [DOI] [PubMed] [Google Scholar]
- Guo M, Thomas J, Collins G, Timmermans MC. (2008) Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. Plant Cell 20: 48–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake S, Smith HM, Holtan H, Magnani E, Mele G, Ramirez J. (2004) The role of knox genes in plant development. Annu Rev Cell Dev Biol 20: 125–151 [DOI] [PubMed] [Google Scholar]
- Hall TA. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41: 95–98 [Google Scholar]
- Hamant O, Nogué F, Belles-Boix E, Jublot D, Grandjean O, Traas J, Pautot V. (2002) The KNAT2 homeodomain protein interacts with ethylene and cytokinin signaling. Plant Physiol 130: 657–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori Y, Nagai K, Furukawa S, Song XJ, Kawano R, Sakakibara H, Wu J, Matsumoto T, Yoshimura A, Kitano H, et al. (2009) The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 460: 1026–1030 [DOI] [PubMed] [Google Scholar]
- Hay A, Barkoulas M, Tsiantis M. (2004) PINning down the connections: transcription factors and hormones in leaf morphogenesis. Curr Opin Plant Biol 7: 575–581 [DOI] [PubMed] [Google Scholar]
- Hay A, Tsiantis M. (2010) KNOX genes: versatile regulators of plant development and diversity. Development 137: 3153–3165 [DOI] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz M, Wilson IW, Yang J, Buerstenbinder K, Llewellyn D, Dennis ES, Sauter M, Dolferus R. (2010) Arabidopsis RAP2.2: an ethylene response transcription factor that is important for hypoxia survival. Plant Physiol 153: 757–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinski S, Piazza P, Craft J, Hay A, Woolley L, Rieu I, Phillips A, Hedden P, Tsiantis M. (2005) KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr Biol 15: 1560–1565 [DOI] [PubMed] [Google Scholar]
- Jung J, Won SY, Suh SC, Kim H, Wing R, Jeong Y, Hwang I, Kim M. (2007) The barley ERF-type transcription factor HvRAF confers enhanced pathogen resistance and salt tolerance in Arabidopsis. Planta 225: 575–588 [DOI] [PubMed] [Google Scholar]
- Jung KH, Seo YS, Walia H, Cao P, Fukao T, Canlas PE, Amonpant F, Bailey-Serres J, Ronald PC. (2010) The submergence tolerance regulator Sub1A mediates stress-responsive expression of AP2/ERF transcription factors. Plant Physiol 152: 1674–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstetter RA, Laudencia-Chingcuanco D, Smith LG, Hake S. (1997) Loss-of-function mutations in the maize homeobox gene, knotted1, are defective in shoot meristem maintenance. Development 124: 3045–3054 [DOI] [PubMed] [Google Scholar]
- Kim JH, Kende H. (2004) A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc Natl Acad Sci USA 101: 13374–13379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby EJM, Appleyard M. (1987) Cereal Development Guide. National Agricultural Centre Cereal Unit, Stoneleigh, UK [Google Scholar]
- Kusaba S, Kano-Murakami Y, Matsuoka M, Tamaoki M, Sakamoto T, Yamaguchi I, Fukumoto M. (1998) Alteration of hormone levels in transgenic tobacco plants overexpressing the rice homeobox gene OSH1. Plant Physiol 116: 471–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer JG. (1991) Barley tiller response to plant density and ethephon. Agron J 83: 968–973 [Google Scholar]
- Lee BH, Ko JH, Lee S, Lee Y, Pak JH, Kim JH. (2009) The Arabidopsis GRF-INTERACTING FACTOR gene family performs an overlapping function in determining organ size as well as multiple developmental properties. Plant Physiol 151: 655–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Zhao W, Meng X, Peng Y. (2007) Molecular cloning and characterization of a rice gene encoding AP2/EREBP-type transcription factor and its expression in response to infection with blast fungus and abiotic stresses. Physiol Mol Plant Pathol 70: 60–68 [Google Scholar]
- Lin WC, Shuai B, Springer PS. (2003) The Arabidopsis LATERAL ORGAN BOUNDARIES-domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial-abaxial patterning. Plant Cell 15: 2241–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JA, Barton MK. (1998) The development of apical embryonic pattern in Arabidopsis. Development 125: 3027–3035 [DOI] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK. (1996) A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69 [DOI] [PubMed] [Google Scholar]
- Mao C, Wang S, Jia Q, Wu P. (2006) OsEIL1, a rice homolog of the Arabidopsis EIN3 regulates the ethylene response as a positive component. Plant Mol Biol 61: 141–152 [DOI] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Tohge T, Saito K, Takahashi H. (2006) Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell 18: 3235–3251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka M, Ichikawa H, Saito A, Tada Y, Fujimura T, Kano-Murakami Y. (1993) Expression of a rice homeobox gene causes altered morphology of transgenic plants. Plant Cell 5: 1039–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee K, Brocchieri L, Bürglin TR. (2009) A comprehensive classification and evolutionary analysis of plant homeobox genes. Mol Biol Evol 26: 2775–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J, Wang Y, Franzen R, Santi L, Salamini F, Rohde W. (2001) In vitro interactions between barley TALE homeodomain proteins suggest a role for protein-protein associations in the regulation of Knox gene function. Plant J 27: 13–23 [DOI] [PubMed] [Google Scholar]
- Müller KJ, Romano N, Gerstner O, Garcia-Maroto F, Pozzi C, Salamini F, Rohde W. (1995) The barley Hooded mutation caused by a duplication in a homeobox gene intron. Nature 374: 727–730 [DOI] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H. (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140: 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohme-Takagi M, Shinshi H. (1995) Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7: 173–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamuro JK, Caster B, Villarroel R, Van Montagu M, Jofuku KD. (1997) The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc Natl Acad Sci USA 94: 7076–7081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori N, Eshed Y, Chuck G, Bowman JL, Hake S. (2000) Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127: 5523–5532 [DOI] [PubMed] [Google Scholar]
- Phelps-Durr TL, Thomas J, Vahab P, Timmermans MC. (2005) Maize rough sheath2 and its Arabidopsis orthologue ASYMMETRIC LEAVES1 interact with HIRA, a predicted histone chaperone, to maintain knox gene silencing and determinacy during organogenesis. Plant Cell 17: 2886–2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozzi C, di Pietro D, Halas G, Roig C, Salamini F. (2003) Integration of a barley (Hordeum vulgare) molecular linkage map with the position of genetic loci hosting 29 developmental mutants. Heredity 90: 390–396 [DOI] [PubMed] [Google Scholar]
- Raventós D, Skriver K, Schlein M, Karnahl K, Rogers SW, Rogers JC, Mundy J. (1998) HRT, a novel zinc finger, transcriptional repressor from barley. J Biol Chem 273: 23313–23320 [DOI] [PubMed] [Google Scholar]
- Roig C, Pozzi C, Santi L, Müller J, Wang Y, Stile MR, Rossini L, Stanca M, Salamini F. (2004) Genetics of barley hooded suppression. Genetics 167: 439–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi V, Locatelli S, Varotto S, Donn G, Pirona R, Henderson DA, Hartings H, Motto M. (2007) Maize histone deacetylase hda101 is involved in plant development, gene transcription, and sequence-specific modulation of histone modification of genes and repeats. Plant Cell 19: 1145–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossini L, Vecchietti A, Nicoloso L, Stein N, Franzago S, Salamini F, Pozzi C. (2006) Candidate genes for barley mutants involved in plant architecture: an in silico approach. Theor Appl Genet 112: 1073–1085 [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Kobayashi M, Itoh H, Tagiri A, Kayano T, Tanaka H, Iwahori S, Matsuoka M. (2001) Expression of a gibberellin 2-oxidase gene around the shoot apex is related to phase transition in rice. Plant Physiol 125: 1508–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Sakakibara H, Kojima M, Yamamoto Y, Nagasaki H, Inukai Y, Sato Y, Matsuoka M. (2006) Ectopic expression of KNOTTED1-like homeobox protein induces expression of cytokinin biosynthesis genes in rice. Plant Physiol 142: 54–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallaud C, Meynard D, van Boxtel J, Gay C, Bès M, Brizard JP, Larmande P, Ortega D, Raynal M, Portefaix M, et al. (2003) Highly efficient production and characterization of T-DNA plants for rice (Oryza sativa L.) functional genomics. Theor Appl Genet 106: 1396–1408 [DOI] [PubMed] [Google Scholar]
- Santi L, Wang Y, Stile MR, Berendzen K, Wanke D, Roig C, Pozzi C, Müller KJ, Müller J, Rohde W, et al. (2003) The GA octodinucleotide repeat binding factor BBR participates to the transcriptional regulation of selected plant homeobox genes. Plant J 34: 813–826 [DOI] [PubMed] [Google Scholar]
- Schneeberger RG, Tsiantis M, Freeling M, Langdale JA. (1998) The rough sheath2 gene negatively regulates homeobox gene expression during maize leaf development. Development 125: 2857–2865 [DOI] [PubMed] [Google Scholar]
- Schubert D, Primavesi L, Bishopp A, Roberts G, Doonan J, Jenuwein T, Goodrich J. (2006) Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J 25: 4638–4649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield S, Murray JAH. (2006) KNOX gene function in plant stem cell niches. Plant Mol Biol 60: 929–946 [DOI] [PubMed] [Google Scholar]
- Sentoku N, Sato Y, Kurata N, Ito Y, Kitano H, Matsuoka M. (1999) Regional expression of the rice KN1-type homeobox gene family during embryo, shoot, and flower development. Plant Cell 11: 1651–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LG, Greene B, Veit B, Hake S. (1992) A dominant mutation in the maize homeobox gene, Knotted-1, causes its ectopic expression in leaf cells with altered fates. Development 116: 21–30 [DOI] [PubMed] [Google Scholar]
- Solano R, Stepanova A, Chao Q, Ecker JR. (1998) Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev 12: 3703–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins GL, Yagil E. (1966) The morphogenetic effects of the hooded gene in barley. I. The course of development in hooded and awned genotypes. Genetics 54: 727–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel T, Graner A, Waugh R, Grosse I, Close TJ, Stein N. (2009) Evidence and evolutionary analysis of ancient whole-genome duplication in barley predating the divergence from rice. BMC Evol Biol 9: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermans MC, Hudson A, Becraft PW, Nelson T. (1999) ROUGH SHEATH2: a Myb protein that represses knox homeobox genes in maize lateral organ primordia. Science 284: 151–153 [DOI] [PubMed] [Google Scholar]
- Treich I, Cairns BR, de los Santos T, Brewster E, Carlson M. (1995) SNF11, a new component of the yeast SNF-SWI complex that interacts with a conserved region of SNF2. Mol Cell Biol 15: 4240–4248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiantis M, Schneeberger R, Golz JF, Freeling M, Langdale JA. (1999) The maize rough sheath2 gene and leaf development programs in monocot and dicot plants. Science 284: 154–156 [DOI] [PubMed] [Google Scholar]
- van der Knaap E, Kim JH, Kende H. (2000) A novel gibberellin-induced gene from rice and its potential regulatory role in stem growth. Plant Physiol 122: 695–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollbrecht E, Reiser L, Hake S. (2000) Shoot meristem size is dependent on inbred background and presence of the maize homeobox gene, knotted1. Development 127: 3161–3172 [DOI] [PubMed] [Google Scholar]
- Vollbrecht E, Veit B, Sinha N, Hake S. (1991) The developmental gene Knotted-1 is a member of a maize homeobox gene family. Nature 350: 241–243 [DOI] [PubMed] [Google Scholar]
- Waites R, Selvadurai HR, Oliver IR, Hudson A. (1998) The PHANTASTICA gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum. Cell 93: 779–789 [DOI] [PubMed] [Google Scholar]
- Weiss D, Ori N. (2007) Mechanisms of cross talk between gibberellin and other hormones. Plant Physiol 144: 1240–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan S, Goldman N. (2001) A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol 18: 691–699 [DOI] [PubMed] [Google Scholar]
- Williams-Carrier RE, Lie YS, Hake S, Lemaux PG. (1997) Ectopic expression of the maize kn1 gene phenocopies the Hooded mutant of barley. Development 124: 3737–3745 [DOI] [PubMed] [Google Scholar]
- Xu L, Shen WH. (2008) Polycomb silencing of KNOX genes confines shoot stem cell niches in Arabidopsis. Curr Biol 18: 1966–1971 [DOI] [PubMed] [Google Scholar]
- Yagil E, Stebbins GL. (1969) The morphogenetic effects of the Hooded gene in barley. II. Cytological and environmental factors affecting gene expression. Genetics 62: 307–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HJ, Shen H, Chen L, Xing YY, Wang ZY, Zhang JL, Hong MM. (2002) The OsEBP-89 gene of rice encodes a putative EREBP transcription factor and is temporally expressed in developing endosperm and intercalary meristem. Plant Mol Biol 50: 379–391 [DOI] [PubMed] [Google Scholar]
- Yin XR, Allan AC, Chen KS, Ferguson IB. (2010) Kiwifruit EIL and ERF genes involved in regulating fruit ripening. Plant Physiol 153: 1280–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.