Abstract
The classification of protein folds is necessarily based on the structural elements that distinguish domains. Classification of protein domains consists of two problems: the partition of structures into domains and the classification of domains into sets of similar structures (or folds). Although similar topologies may arise by convergent evolution, the similarity of their respective folding pathways is unknown. The discovery and the characterization of the majority of protein folds will be followed by a similar enumeration of available protein folding pathways. Consequently, understanding the intricacies of structural domains is necessary to understanding their collective folding pathways. We review the current state of the art in the field of protein domain classification and discuss methods for the systematic and comprehensive study of protein folding across protein fold space via atomistic molecular dynamics simulation. Finally, we discuss our large-scale Dynameomics project, which includes simulations of representatives of all autonomous protein folds.
Keywords: protein folds, protein folding, structural classification, molecular dynamics simulations
Introduction
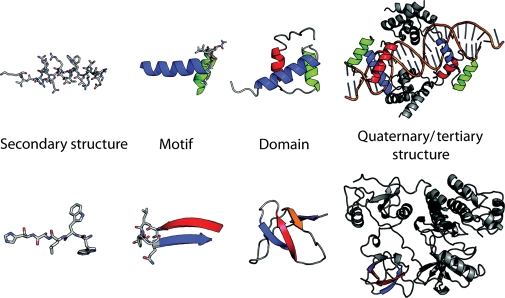
Protein structure is inherently hierarchical. Proteins have steric constraints due to the chemistry of their amino acids. Secondary structure in proteins is formed due to the hydrogen-bonding properties of the peptide backbone, the interaction of the side-chain atoms with the backbone and the chiral nature of amino acids. Secondary structure can be identified from both the backbone torsion angles (Φ and Ψ) and the hydrogen-bonding patterns observed between carbonyl and amide groups in the peptide backbone. Secondary structure (generally α-helices and β-sheets) can be built up into small repeating patterns in protein structures; these can be called ‘motifs’ or ‘supersecondary structure’ (Levitt and Chothia, 1976). Motifs are important in describing protein structure because they can be repeated within many structures (Fig. 1).
Fig. 1.
Structural elements from Pit-1 homeodomain (PDB:1AU7) and Src (PDB:1FMK). Motifs are made up of secondary structure elements but do not necessarily make up a hydrophobic core. Domains are the smallest self-contained unit within a structure. Structures may be made up of multiple domains, sometimes with repeats of the same domain.
Protein motifs assemble into larger subunits of structure called ‘domains'. Although some proteins contain only a single domain, it is often the case that proteins are composed of multiple domains. The definition of a domain can depend on multiple criteria and is not necessarily convergent. Independent of (but not necessarily distinct from) structural considerations, domains are often defined as a unit of conserved sequence (Marchler-Bauer et al., 2007; Finn et al., 2008). Domains can be defined as a unit of conserved functional behavior (e.g. a serine protease). A domain can be described as the smallest structural unit, without necessarily taking the compactness or potential stability of that unit into account. Finally, we prefer to think of domains as the smallest cooperatively folding unit within a protein structure, or as autonomous folded structures.
Classification of protein structures into domains and folds
Proteins can share similarity, in both sequence and/or structure. Although the methodologies for comparison and quantification of sequence similarity are generally agreed upon, the conventions for quantifying and classifying structural similarity are still up for debate (Sippl, 2009; Sadowski and Taylor, 2010). To classify and compare a new protein structure with existing examples, two difficult problems must be addressed: (i) identification and partitioning of multiple domains in a protein structure and (ii) comparison of those domains with existing characterized domains. The clustering of those comparisons into discrete similar groups of structures (or ‘folds') is complicated by the possibility of regions of significant structural similarity in a conserved core bordering non-conserved regions with little in common (Fig. 2). Different empirical approaches to these two problems result in different classification systems (or ‘domain dictionaries'); some of which are more suited to the study of protein folding than others.
Fig. 2.
Structurally similar domains and their structural elements. Some structurally similar proteins contain nearly identical secondary structure elements in similar orientations. Hemoglobin, chain A (PDB:2MHB, chain A) and myoglobin (1A6N) are two such members of the globin fold. Chemotaxis protein Y (PDB:3CHY), Histamine N-methyltransferase (PDB:2AOT) and Catechol O-methyltransferase (1VID) have a conserved structural core surrounded by some non-conserved regions.
Levitt and Chothia (1976) performed the first classification of protein structures into folds. Although based solely on the 31 known protein structures, they made several interesting observations that have persisted over time and growth of the Protein Data Bank (PDB): protein structures partition into four distinct classes, proteins contain repeating motifs, and there is a preference for ‘handedness’ in motifs. The original classification relied primarily on the topological arrangement of secondary structure elements. Later classifications would focus more closely on the precise geometric arrangement of these elements. Notably absent was the consideration of specific angles of interaction between secondary structure elements. For example, this consideration is an important factor in distinguishing between orthogonal and parallel helical bundles.
The structural patterns described by Levitt and Chothia were incorporated into the development of the first publicly accessible and consistently updated domain dictionary, SCOP (Structural Classification of Proteins) (Murzin et al., 1995). The initial versions of SCOP relied heavily on the visual inspection of structures to identify domains and to group domains together into folds and superfamilies in the SCOP hierarchy. The SCOP hierarchy has four levels: the family, wherein proteins are clustered together primarily on the basis of sequence identity (>30%) and/or functional similarity; the superfamily, wherein families with low sequence identity but similar functions and structural features suggest a common origin; and the fold, wherein families and superfamilies with conserved core topological arrangement are grouped. SCOP folds are also grouped by secondary structure class: all-α, all-β, α/β (where helices and sheets are mingled) and α + β (where helices and sheets are separate). The rapid increase in the rate of structure determination prompted changes to the update methodology of SCOP between versions 1.63 and 1.73 (Andreeva et al., 2008). The large populations of some superfamilies resulted in some component families that did not necessarily possess any given level of sequence similarity despite their structural similarity. Also, given the dependence on functional classification for the superfamily level, proteins determined by structural genomics initiatives that were functionally uncharacterized could only receive a provisional classification. Finally, the rate of structural determination required an introduction of an automatic PSI-BLAST protocol for screening of new structures. Following release v1.71, complete classification of all known PDB structures could no longer be guaranteed, so priority was assigned to potentially novel fold representatives. The intensive nature of expert curation led to the development of other approaches to domain classification and fold assignment.
The development and release of CATH (Class, Architecture, Topology, Homologous superfamily) was a response to this ever-increasing rate of structure determination (Orengo et al., 1997). Specifically, CATH includes automatic sequence and structure comparisons, and automatic domain detection within its classification process. The domain-partitioning algorithm in CATH underwent several revisions (Greene et al., 2007). The structural comparison algorithm used in the initial releases of CATH, Sequential Structure Alignment Program (SSAP), was later augmented by a graph-based prescreening method, GRATH (Orengo et al., 1996; Pearl et al., 2001). The hierarchal levels in CATH are as follows; the class, where proteins are grouped by their secondary structure content (the distinction between α + β and α/β is not made at the class level in CATH); the architecture, where proteins are grouped by their secondary structure arrangement, regardless of chain connectivity; the topology, where proteins are grouped by their architecture and chain connectivity; and the homologous superfamily, where proteins are grouped by possessing >35% sequence identity. Functional considerations are not an explicit component of the CATH classification. Also, domains in CATH are not required to be continuous in sequence. The architecture levels are derived by manual inspection of domains and reflect the arrangement and number of secondary structure elements regardless of chain connectivity.
When applied to known protein structures, the classification system in CATH describes a ‘fold space’ that is unevenly populated. A small number of topologies in CATH contain a disproportionately large number of domains. These highly populated topologies are referred to as ‘superfolds’. Additionally, multiple architecture levels are disproportionately populated, these are referred to as ‘superarchitectures’ (Orengo et al., 1994). While the cause of this population bias may be biological in origin, it is difficult to disentangle from potential bias in targeting proteins for structural determination (or the ease with which some folds crystallize). Buchan et al. (2002) have suggested that the overpopulation of these fold families is related to their ability to evolve and adapt new structure and function. Where architectures are highly populated, structural similarity can exist that leads to structural continuity across multiple topologies. This continuity of similarity across multiple topologies has been used as evidence for continuity within some regions of fold space and as an example of the difficulties of representing fold space as discrete folds (Kolodny et al., 2006).
An approach to domain classification that reduces or removes manual inspection is desirable both due to the rate of structure determination and the potential inconsistencies introduced by expert curation. Version 3.1β of the Dali Domain Dictionary relied on the hierarchal clustering of domains compared by the DALI structural alignment method coupled with a feature vector that contained enzymatic, functional, and keyword annotation for each domain (Holm and Sander, 1999; Dietmann et al., 2001). Domains were identified from structures using ‘compactness’ and recurrence criteria (Holm and Sander, 1998a). Folds were assigned by hierarchal clustering of the DALI Z-scores from an all versus all structural alignment of domains and using empirical similarity thresholds such that the resulting dendrogram was discretized into clusters with shared topologies. The DALI structural alignment program relied on the identification of a set of substructures (usually secondary structure elements) shared between two structures and then optimization of the alignment of the substructures’ Cα–Cα distance matrices (Holm and Sander, 1996). A neural network optimization used a vector of functional annotations to distinguish homologues from analogues within a fold and to derive ‘superfamilies’ (Dietmann and Holm, 2001). Ultimately, although the Dali Domain Dictionary was fully automated, it was hampered by the computational power required to perform all versus all comparisons at the current rate of structure release (Hasegawa and Holm, 2009).
Problems with domain classifications
The manual inspection used by SCOP and CATH during the chain partition and domain classification process allows for powerful structural and biological insights to be built into these dictionaries; it also potentially allows unintended consequences from these choices to arise within the classification system. Since both hierarchies rely on the abstraction of topology and/or functional considerations, the structural similarity of domains within families and superfamilies can be lower than expected. This problem can be exaggerated when sequence alignment becomes a component for the automatic assignment of domains to homologues (Greene et al., 2007; Andreeva et al., 2008). Although high-sequence identity often implies high structural similarity, at the borders of folds, there is sufficient overlap that unexpected structural diversity can accumulate (Reeves et al., 2006; Cuff et al., 2009a) (Fig. 3). The ‘bottom-up’ approach of structural comparison relies on purely geometric metrics of structure comparison as a means of avoiding overlapping regions that arise from the assumption that high sequence identity implies structural similarity (Valas et al., 2009). However, domain classification that relies purely on structural comparison algorithms is often blind to potentially useful distant biological relationships and determined by an arbitrary threshold of structural significance (Adam, 1996). An exhaustive review of current structural alignment algorithms has recently been published (Hasegawa and Holm, 2009). Any discrete domain classification system needs to account for the presence of observable intermediate forms between certain distinct structural topologies. The difficult choice is whether to split intermediate forms into a separate fold or to merge them into one of the structurally similar but still distinct folds.
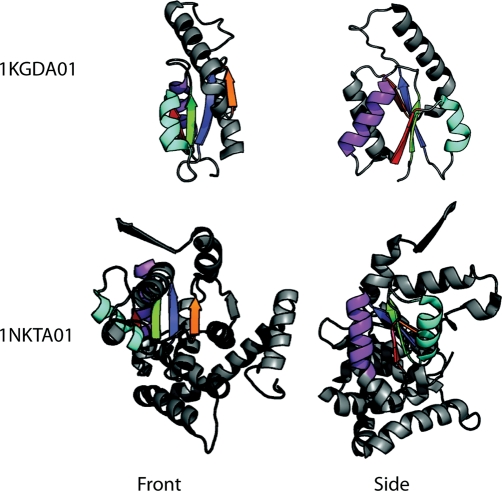
Fig. 3.
Domains of guanylate kinase (CATH: 1KGDA01) and translocation ATPase (CATH:1NGDA01), two structurally diverse domains from the CATH v1.73 superfamily 3.40.50.300, the P-loop nucleotide hydrolases. Adapted from Cuff et al. (2009a).
Fold classification is dependent on the prior problem of partitioning multi-domain chains. Results of domain-partitioning algorithms and domains partitioned by expert curation have been compared both with each other and with domain boundaries generated by the crystallographers for the structure in question (Islam et al., 1995). In an exhaustive study by Holland et al. (2006), each domain's partitioning method was analyzed with respect to: (i) the distribution of single and multiple domain chains, (ii) the ratio of continuous to discontinuous domains, (iii) the distribution of domain sizes and (iv) the distribution of fragment sizes in discontinuous domains. Although domain classification methods tend to agree on boundaries in general, they vary on their tendency to partition chains into multiple domains; both with respect to the number of domains they partition into and whether those domains are continuous or discontinuous. The domain boundaries of SCOP and CATH show the best agreement with the crystallographers’ annotations and DALI the least. Owing to the larger proportion of bona fide single-domain chains in most structure test sets, domain assignment methods can disagree on the assignment of boundaries in multi-domain chains and still agree on a majority of domains by correctly identifying single-domain chains. Religa et al. (2007) have even found that engrailed homeodomain (EnHD) can be truncated into a stable HTH motif similar to the folding intermediate of the full structure, suggesting that even in minimal single-domain proteins, the boundary of folding domains may be more complex than expected. Furthermore, as structures become more complex, domain classifications diverge and agreement drops (Fig. 3).
SCOP tends to leave large structures uncut, identifying the largest recurrent subunit rather than the smallest independent subunit, which is correlated with less discontinuous domains resulting from domain crossover. The presence of discontinuous domains and the consequent presence of domain crossover (where another domain is inserted into the region between two fragments of a discontinuous domain) have troubling implications for the definition of a domain as an independent folding unit. CATH, on the other hand, agrees more closely with algorithmic domain assignment methods (such as DALI) while maintaining agreement with SCOP on many smaller domains. DALI tends to partition chains into multiple domains, most often by emphasizing compact domains, and is the most likely to create discontinuous domains (Fig. 4). The primary conclusions of Veretnik et al. (2004) were that domain partitioning methods tend to disagree on: (i) the definition of very small domains; (ii) splitting secondary structures between domains; (iii) the size and number of discontinuous domains; (iv) closely packed or convoluted domain–domain interfaces; (v) structures with large and complex architectures; and (vi) the role of structural, functional and evolutionary concepts in the determination of domain definitions.
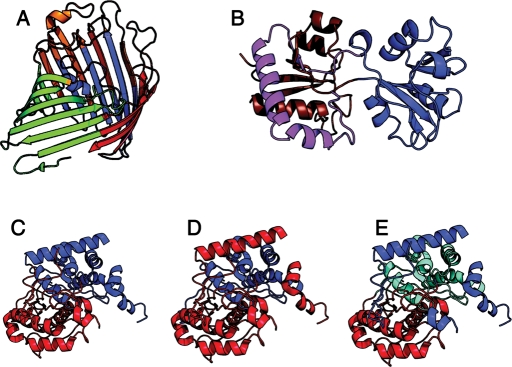
Fig. 4.
Examples of difficult domain partitions. (A) Escherichia coli phosphorin (PDB:1PHO); SCOP and CATH do not partition this structure, DALI partitions it into four separate domains. (B) Periplasmic lysine/arginine/ornithine-binding protein (PDB:2LAO); SCOP does not partition this domain (disfavors discontinuous domains), CATH and DALI partition it into two domains; domain 1 (red/brown), domain 2 (blue). (C) 3-ketoacyl-CoA thiolase (PDB:1PXT), CATH assigns two domains, (D) SCOP assigns two different domains, (E) and the AUTHORS database (Islam et al., 1995) assigns three. Adapted from Veretnik et al. (2004).
Despite the varying strategies for the derivation of domain dictionaries, most approaches agree on a large number of domain classifications. Multiple studies have compared the consensus domain assignments in expert-curated and automated methods (Hadley and Jones, 1999; Day et al., 2003; Csaba et al., 2009). Holland et al. (2006) performed a comparison of automated domain assignment methods on a curated multi-domain protein set and found that no single automatic method could consistently and accurately generate domain partitions while optimizing for continuous or minimally discontinuous domains. They suggest that this lack of consistency is the result of competing structural and functional definitions of the domain implicit in boundary definitions. Despite disagreements on some multi-chain domains, a consensus method can accurately treat a majority of the domains captured by SCOP, CATH and Dali/FSSP (Hadley and Jones, 1999; Day et al., 2003), largely due to the prevalence of single-domain protein chains.
The number of unique protein folds
Important goals of structural biology include the identification of all families of sequence domains, structures with sequences homologous to those families, folds containing those structures and functions conveyed by those folds. By identifying these elements, we hope to largely sidestep the difficult process of structurally determining the individual proteins within an organism and instead understand its components and their interactions purely from its genetic code. For these reasons, the number of total folds (and the number of associated functions) is a matter of interest. Protein folds are not equally populated, some folds are either more structurally or sequentially diverse, and these effects are not necessarily correlated (Orengo et al., 1997; Holm and Sander, 1998b). This difference in the distribution of domains complicates estimates of the total number of protein folds. In addition, families of sequences with no detectable similarity can adopt the same structural fold. Estimates of the number of protein folds have varied over two orders of magnitude (103–105) in the past two decades (Table I). The presence of highly populated folds, some sequence diverse, in domain classification systems is consistent, but these folds may arise for different reasons. The degree to which population differences are biased by the targeting of structure determination efforts is difficult to estimate, but structural prediction of whole genomes suggests that it is small (Wolf et al., 2000; Buchan et al., 2002). It is unclear whether highly populated folds arise because of convergent evolution from multiple origins, which might suggest that these folds are more easily evolved to be stable; or divergent evolution from a single origin, which might suggest that these folds are more easily evolved to adapt different functions. Furthermore, it is difficult to assess how the total number of observed folds is related to the total number of naturally occurring folds. At least one topology has been designed that has not yet been observed in nature (Kuhlman et al., 2003). Is there a significant set of folds that are physically possible but that have not been evolved? Interestingly, Govindarajan et al. (1999) estimated that there are ∼4000 unique protein folds and that ∼2200 are likely in nature.
Table I.
Estimates for the number of protein folds and superfamilies by year
| Year | Folds | Superfamilies | Reference |
|---|---|---|---|
| 1992 | <1000 | 1500 | Chothia (1992) |
| 1994 | <7700 | 23 100 | Orengo et al. (1994) |
| 1994 | 6727 | — | Alexandrov and Go (1994) |
| 1996 | 455 | — | Zhi-Xin (1996) |
| 1997 | <920 | 920 | Brenner et al. (1997) |
| 1997 | ≤5200 | 17 175 | Zhang (1997) |
| 1998 | 650 | 1150 | Zhi-Xin (1998) |
| 1998 | 836 | — | Zhang and DeLisi (1998) |
| 1999 | 3756 | — | Govindarajan et al. (1999) |
| 2000 | ∼1000 | 4000–7000 | Wolf et al. (2000) |
| 2002 | 10 000 | 50 000 | Coulson and Moult (2002) |
| 2007 | 1613 | — | Levitt (2007) |
| 2009 | ∼1700 ± 400 | ∼4000 | Sadreyev et al. (2009) |
Protein folds are islands of discrete structural similarity within which structures share some level of sequence similarity. As a corollary, sequences with high identity usually share the same structural fold. This principle is widely used to automate domain assignments to folds and in comparative modeling of sequences with no known structure. However, multiple cases of domains with detectable sequence similarity to members of multiple structurally distinct folds have been described (Grishin, 2001; Alva et al., 2008). Another complication is that domains can be classified as having the same fold because of substructure similarity while possessing significant overall structural dissimilarity (Cuff et al., 2009b). The effect of this loss of transitivity of domain similarity has been studied in both SCOP and CATH folds (Csaba et al., 2009; Pascual-Garcia et al., 2009). Both of these effects indicate that regions of fold space exist that are difficult or improper to discretize by assuming a correlation between sequence homology and structural similarity (i.e. regions of ‘continuous' fold space). That these regions exist does not negate the fact that the majority of structurally characterized domains are sourced from single-domain chains and that these chains seem to exist in consistent, thermodynamically stable islands in fold space (Cuff et al., 2009a,b; Sadreyev et al., 2009). Are these regions of fold space simply more capable of evolving into new structures, or are we simply observing the effects of higher sampling within these regions of fold space? Finally, the boundary cases in any empirical fold classification that relies on heuristic criteria not directly related to the biophysical and biological origin of folds (i.e. their folding pathways and evolutionary origins) will have boundary conditions. Any replacement for current domain classification systems will need to be generated with these criteria in mind.
Computational simulation of protein folding
Proteins fold from a partially structured denatured state through any number of increasingly structured intermediate states into the folded native ensemble. Although we can experimentally characterize the native ensemble at the atomic level, generally speaking, the atomic details of partially folded states are inaccessible. If the rules of protein structural classification are hierarchical, as discussed in the previous section, then it may also be the case that the folding of these domains displays a similar hierarchical behavior, such that small well-behaved domains studied in isolation will also reflect their behavior in the context of larger multidomain structures.
All-atom molecular dynamic (MD) simulations model states along the folding/unfolding pathway that cannot be directly observed by experiment. Recent increases in computer power have translated into increases in the length and complexity of MD simulations. Using such simulations, more sampling of the partially structured states along the folding pathway can be achieved. The structural heterogeneity of partially folded states complicates their experimental structural analysis. Spectroscopic signals from these states may originate from many different structures or from a small fraction of folded structures in a large ensemble of unfolded structures. Therefore, MD simulations can provide a theoretical model with which to interpret experimental signals. Here briefly describe three proteins studied by combining MD in the Daggett group and experimental studies in the Fersht group. These three proteins fall into 3-fold classes: mixed α/β, all-α, and all-β.
A mixed α/β-protein
Our representative mixed α/β protein is chymotrypsin inhibitor 2 (CI2). CI2 is a 64-residue protein with a single α helix and a three-stranded β-sheet, and it was the first protein whose folding was mapped in detail and extensively validated. The packing of the helix against the sheet forms the hydrophobic core. CI2 was the first protein demonstrated to fold by a two-state mechanism. The TS of CI2 was first characterized at the atomic level by MD simulation and Φ-value analysis (Li and Daggett, 1994; Otzen et al., 1994; Daggett et al., 1996; Li and Daggett, 1996). The MD-generated TS structures were further validated by the design of faster folding variants by targeting interactions in the MD structures that could not be predicted from the native state (Ladurner et al., 1998). The redesigns were successful, leading to the fastest folding CI2 variant thus far. The TS of CI2 contains partial secondary and tertiary structure and the final event during folding involves their simultaneous consolidation (Fig. 5A).
Fig. 5.
Major conformational states sampled during thermal unfolding MD simulations. (A) Native (N), transition (TS) and denatured state (D) of CI2. The TS of CI2 is characterized by the packing of the still nascent helix against the partially formed β-sheet. The denatured state of CI2 is particularly denatured, containing little secondary structure. (B) Native (N), transition (TS), intermediate (I) and denatured (D) states of the engrailed homedomain. The TS of EnHD is characterized by essentially native helices condensing into their native topology. The EnHD intermediate contains fully formed helices I and III with a partially denatured helix II. (C) The WW domain does not contain a hydrophobic core. Instead, two small hydrophobic clusters are found on either side of the β-sheet. Residues in cluster 1 (purple) and cluster 2 (cyan), associate in strands 1 (red) and 2 (blue) of FBP28 in the TS. These residues nucleate the folding of the WW domain despite the plasticity of the precise turn residues in the TS.
CI2 served as a workbench for establishing many of the early postulates of the study of protein folding by simulations of high-temperature unfolding. Through simulations of CI2, it was first established that a plausible TS could be selected from a high-temperature MD simulation and that the properties of the TS mirrored those probed experimentally at lower temperature (Li and Daggett, 1994). Microscopic reversibility was first observed in simulations of CI2, such that folding and unfolding were observed in a single continuous simulation and they followed the same steps, but in reverse (Day and Daggett, 2007). In addition, through a set of 100 independent simulations, it was demonstrated that a small number of simulations (5–10) could capture the overall properties of the full set (Day and Daggett, 2005). Furthermore, NMR studies of the denatured state of CI2 showed that long-range residual structure detected in MD simulations could be similarly detected by NMR (Kazmirski et al., 2001). Early work on CI2 established the benchmarks by which future work on EnHD and small all-β model proteins (such as the WW domains) would be measured. Furthermore, the synergy between the simulation and experiment became apparent: experiment benefits from the detailed structural information that can be obtained from MD, and MD needs experiment to establish its validity.
An all α-protein
EnHD is a 56-residue three-helix bundle that folds through an intermediate. MD simulations have provided structural models for the major states along the folding pathway of EnHD (Mayor et al., 2000). These states were then validated by experiment. For example, the MD-predicted transition state ensemble (TS) (Mayor et al., 2000) was found to agree with experimental Φ-values determined several years later (Gianni et al., 2003). Similarly, the MD-predicted intermediate (Mayor et al., 2000) was validated by direct structural determination by NMR (Religa et al., 2005). Finally, the unfolding pathway of EnHD was directly observed to be reversible, both via simulations near its melting temperature (325 K) (McCully et al., 2008) and by temperature-quenched simulations conducted on the intermediate state (McCully et al., 2010).
The MD-generated TS structures of EnHD at different temperatures are similar and are native-like with all native helices essentially fully formed (Mayor et al., 2000). The estimated half-life of unfolding by experiment was found to be very similar to the MD predictions (Mayor et al., 2003). The prediction that the TS of EnHD contains essentially native helices that are only partially packed was also confirmed by experiment (Gianni et al., 2003) (Fig. 5B). A fundamental assumption of high-temperature simulations is that the folding pathway at low temperatures contains the same states as the unfolding pathway at high temperatures. Given good agreement between MD simulations probing unfolding and experiments on both folding and unfolding, this assumption seems to be reasonable, but it was also shown directly that the overall TS properties are insensitive to temperature (Mayor et al., 2000; Mayor et al., 2003; DeMarco et al., 2004). In addition, as with CI2, this assumption was directly tested and the principle of microscopic reversibility was shown to hold for continuous trajectories of EnHD at its melting temperature (McCully et al., 2008).
In addition to the simulations around the melting point, quench simulations of thermally denatured structures of EnHD were performed (McCully et al., 2010). A series of 46 refolding simulations was started from a single thermally denatured structure (the intermediate state) from a 498K simulation of EnHD near the temperature of the maximal folding rate (310, 314 and 319 K). A single simulation, run for 700 ns, refolded. However, 45 of the 46 simulations did not refold and instead were confined in the intermediate state primarily via non-native electrostatic interactions. The intermediate state of this protein is very stable by experiment and the rate-determining step for folding is not folding to the intermediate but the transition from the intermediate to the native state. The simulations provide examples of specific non-native interactions that slow folding by stabilizing the intermediate state. Altogether these results demonstrate that the folding pathway of EnHD is well understood and demonstrates the predictive power of the MD simulations and the strength, or even necessity, of combining experimental and computational methods.
The lessons we learn from closely analyzing individual proteins must be applicable to other proteins in order to be useful. Then the question becomes, does the mechanism of a single model protein necessarily represent that of structurally similar proteins (Daggett and Fersht, 2003)? To address this question, the folding behavior of EnHD was compared with other three-helix bundles; c-Myb transforming protein (c-Myb) and human telomeric repeat factor 1 (hTRF1) were simulated and analyzed (Gianni et al., 2003; White et al., 2005). Although all three proteins belong to the same fold, hTRF1 and c-Myb folded by different apparent mechanisms than EnHD by experiment (Gianni et al., 2003). Whereas EnHD folds via the framework mechanism with formation of the helices followed by docking of the preformed helices, both hTRF1 and c-Myb displayed properties that were consistent with the nucleation–condensation mechanism, whereby secondary and tertiary interactions are closely coupled, as with CI2. MD simulations of EnHD, c-Myb and hTRF1 provided TS ensembles consistent with the experimental Φ-value analysis (White et al., 2005). Whereas the TS of EnHD contained fully formed native helices, in both c-Myb and hTRF1 helices were only partially formed. Interestingly, an intermediate was observed in MD simulations of c-Myb that was not observed by experiment. A c-Myb mutant was designed to stabilize the proposed intermediate and its population increased such that it became visible by experiment (White et al., 2005). These studies showed that these family members are linked by a common folding mechanism that is modulated in different family members by the helical propensities and strength of specific tertiary interactions.
An all β-protein
WW domains have been popular model systems for studying the formation of β-structure. They are small proteins consisting of a three-stranded β-sheet (a double hairpin) lacking a conventional hydrophobic core (Macias et al., 2000). Owing to the topology of WW domains, particular interest has focused on the role of the β-turns. The WW double hairpin fold is found as a substructure in other larger proteins. Consequently, generalizing the behavior of these domains could provide a general model for β-sheet folding. Previous combined experimental and theoretical work demonstrated that the precise main-chain hydrogen bonding of the first β-turn of FBP28 is disrupted in the TS (Petrovich et al., 2006). In the follow-up work, three WW domains, hYAPtm, FBP28 and PIN1, were studied both by experiment and by MD simulation (Sharpe et al., 2007). Using NMR relaxation methods, the S2 order parameters, reflecting motion of the NH bond vectors, were measured for both hYAPtm and FBP28 and compared with values calculated from MD. In both proteins, the S2 values compared well between experiment and simulation and showed that the first turn was highly mobile in the native state. These results, combined with the Φ-value analysis, yield a model for β-sheet folding in which the early formation of the turn per se is not necessary for folding, but side-chain interactions in the turn region are important in the nucleation of sheet formation (Fig. 5C).
Toward general principles of protein folding
Given the utility of the combined experimental/theoretical approach to the study of protein folding, a quandary remains. To what degree are these results representative of the protein universe? Even within structural families that are well studied, variation in folding behavior can be observed between family members, although the folding behavior is often conserved within a fold family (Gunasekaran et al., 2001; Zarrine-Afsar et al., 2005; Nickson and Clarke, 2010). Despite 20 years of active study on various model proteins, we have only begun to scratch the surface of available protein structures and their folding pathways. At this point, the increase in computer power and the confidence in simulation methods have advanced to a sufficient point where we must expand our horizons beyond the well-studied proteins over which we have labored in the past. A broad scale, data-driven approach to protein modeling can bring about the discovery and enumeration of general rules to protein folding only hinted at by previous studies.
Mass simulation of all known globular protein folds
The identification of domains in protein structures and the classification of these domains into folds are fairly well understood. We have demonstrated that native states and folding pathways can be fleshed out using a combination of experiment and theory. The next logical step is to combine this knowledge into a broad-based high-throughput study of folding across representatives of all known protein folds. The recent increases in available computer power have made high-throughput simulation initiatives possible and the first and most extensive of these is the Dynameomics project (Day et al., 2003; Beck et al., 2008; van der Kamp et al., 2010). The Dynameomics project entails systematic simulation of representatives of all autonomous protein folds. These representatives are taken from a consensus domain dictionary (CDD), where the consensus relates to consensus within the SCOP, CATH and Dali domain dictionaries (Day et al., 2003; Schaeffer et al., 2010). From a total of 1695 consensus folds (or metafolds), 807 individual domains were found to be autonomous folded domains. For many metafolds no suitable domains exist, for one or more reasons. Domains may contain cofactors that are structurally significant or contain large gaps or other quality factors that prohibit simulation. Interestingly, we found that the single largest reason for the rejection of metafolds was that they contained only discontinuous domains or domains were not autonomous when excised from their structure. Over 40% of the metafolds in the v2009 CDD were rejected for this reason (Schaeffer et al., 2010). Nevertheless, 807 metafolds representing a majority of the domains in the CDD (due to the differential population of metafolds) were selected and simulated. The Dynameomics database currently contains over 11 000 simulations of over 2000 systems, representing the largest collection of protein simulations and structures in the world. The native state simulations of the Top 100 targets are publicly accessible at http://www.dynameomics.org.
This large data set has then been used as a foundation for studies into native state flexibility (Benson and Daggett, 2008), transition state data mining (Jonsson et al., 2009) and a property-based reaction coordinate of folding (Benson and Daggett, 2008; Jonsson et al., 2009; Toofanny et al., 2010). In the first case, a large-scale assessment of native protein dynamics was performed. Proteins fluctuate in their native state, potentially altering the apparent accessibility of functional sites from static structures, as shown some time ago for transient cleft formation in cytochrome b5 (Storch and Daggett, 1995) that was later validated experimentally and shown to be linked to function (Storch et al., 1999a,b; Hom et al., 2000). Protein flexibility in the native state was analyzed for a subset of 253 Dynameomics targets (Benson and Daggett, 2008). The flexibility of the main chain was calculated using the principal components of the Cα atoms (Teodoro et al., 2004). The hydrophilic and polar residues were slightly more flexible than hydrophobics, as is expected given their position on the protein surface. The flexibility of α-helices was more localized toward their termini, whereas for β-strands, there was no such tendency. In comparing the native and unfolding simulations, one sees that sites involved in early unfolding tend to have high flexibility, suggesting a strong link between native dynamics and folding behavior.
Interestingly, 21 highly inflexible loops were identified, loops that were more inflexible than helices and sheets. No significant deviation in amino acid populations was identified in the inflexible loops. This finding suggests that there are structural units that outside of the conventional secondary structure definitions. In addition to the metafold representatives, a series of additional targets was selected in the fold families of EnHD, ubiquitin and Fyn SH3. The flexibility of fold family members was found to be highly correlated along their shared secondary structure. Where a member deviated from the average behavior of the fold, it was associated with low sequence and structural identity to other members of the fold.
Identification and characterization of the properties of TS ensembles is one of the principal strengths of studying protein folding by MD simulation. As the TS is an unstable species, it can only be indirectly characterized by experiment. Jonsson et al. (2009) identified a set of 183 TS ensembles from an earlier subset of Dynameomics data (Beck et al., 2008). Global properties were calculated over the set of selected TS ensembles and analyzed by fold class, experimental source and secondary structure content. Structures were analyzed based on 27 structural properties, including contacts, secondary structure content, solvent accessible surface area (SASA) and radius of gyration (Rg). No significant difference was observed for the identified TS ensembles when separated by fold class (all-α, all-β and mixed α/β), SASA or starting secondary structure. Contacts were analyzed for each residue in each TS ensemble and aggregated by residue type. Residues with the highest number of contacts in the starting structure tended to lose the most contacts (and become fractionally more exposed) in the TS ensemble. Helical residues tended to have slightly higher burial than residues starting in other or no secondary structure. This work represents the first major mining of the general properties of the unfolding pathways in Dynameomics. The general properties of the TS found here cluster around a small number of averages that are relatively invariant with respect to protein fold, residue type and secondary structure. With more extensive data mining of the native and denatured states over the now complete 807 fold set, a general picture of the entire folding pathway may emerge.
Conclusions
Ultimately, the study of protein folding must move beyond model systems. By coupling the broad-based knowledge of protein domain partitioning and fold classification with high-throughput MD simulations, the Dynameomics project provides a foundation for a new era of data-driven protein folding research. With these data and new mining tools, we are moving beyond the study of individual systems to obtain a broader, more general view of protein folding.
Funding
We are grateful for the financial support from the National Institutes of Health (GM 50789) and the External Research Program at Microsoft Research. In addition, the simulations for Dynameomics were performed using computer time through the DOE Office of Biological Research as provided by the National Energy Research Scientific Computing Center, which is supported by the Office of Science of the US Department of Energy under contract DE-AC02-05CH11231.
Acknowledgements
Dynameomics would not have been possible without ∼20 years of extensive benchmarking of MD simulations of protein folding/unfolding against experiment for well-behaved protein systems with Professor Alan Fersht and his group at the MRC Cambridge Center for Protein Engineering.
References
- Adam G. Protein Sci. 1996;5:1325–1338. doi: 10.1002/pro.5560050711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov N.N., Go N. Protein Sci. 1994;3:866–875. doi: 10.1002/pro.5560030601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alva V., Koretke K.K., Coles M., Lupas A.N. Curr. Opin. Struct. Biol. 2008;18:358–365. doi: 10.1016/j.sbi.2008.02.006. [DOI] [PubMed] [Google Scholar]
- Andreeva A., Howorth D., Chandonia J.M., Brenner S.E., Hubbard T.J., Chothia C., Murzin A.G. Nucleic Acids. Res. 2008;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck D.A., Jonsson A.L., Schaeffer R.D., Scott K.A., Day R., Toofanny R.D., Alonso D.O., Daggett V. Protein Eng. Des. Sel. 2008;21:353–368. doi: 10.1093/protein/gzn011. [DOI] [PubMed] [Google Scholar]
- Benson N.C., Daggett V. Protein Sci. 2008;17:2038–2050. doi: 10.1110/ps.037473.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S.E., Chothia C., Hubbard T.J. Curr. Opin. Struct. Biol. 1997;7:369–376. doi: 10.1016/s0959-440x(97)80054-1. [DOI] [PubMed] [Google Scholar]
- Buchan D.W.A., Shepherd A.J., Lee D., Pearl F.M.G., Rison S.C.G., Thornton J.M., Orengo C.A. Genome Res. 2002;12:503–514. doi: 10.1101/gr.213802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C. Nature. 1992;357:543–544. doi: 10.1038/357543a0. [DOI] [PubMed] [Google Scholar]
- Coulson A.F., Moult J. Proteins. 2002;46:61–71. doi: 10.1002/prot.10011. [DOI] [PubMed] [Google Scholar]
- Csaba G., Birzele F., Zimmer R. BMC Struct. Biol. 2009;9:23. doi: 10.1186/1472-6807-9-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuff A., Redfern O.C., Greene L., et al. Structure. 2009a;17:1051–1062. doi: 10.1016/j.str.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuff A.L., Sillitoe I., Lewis T., Redfern O.C., Garratt R., Thornton J., Orengo C.A. Nucleic Acids Res. 2009b;37:D310–D314. doi: 10.1093/nar/gkn877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daggett V., Fersht A.R. Trends Biochem. Sci. 2003;28:18–25. doi: 10.1016/s0968-0004(02)00012-9. [DOI] [PubMed] [Google Scholar]
- Daggett V., Li A., Itzhaki L.S., Otzen D.E., Fersht A.R. J. Mol. Biol. 1996;257:430–440. doi: 10.1006/jmbi.1996.0173. [DOI] [PubMed] [Google Scholar]
- Day R., Daggett V. Proc. Natl Acad. Sci., USA. 2005;102:13445–13450. doi: 10.1073/pnas.0501773102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day R., Daggett V. J. Mol. Biol. 2007;366:677–686. doi: 10.1016/j.jmb.2006.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day R., Beck D.A., Armen R.S., Daggett V. Protein Sci. 2003;12:2150–2160. doi: 10.1110/ps.0306803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMarco M.L., Alonso D.O.V., Daggett V. J. Mol. Biol. 2004;341:1109–1124. doi: 10.1016/j.jmb.2004.06.074. [DOI] [PubMed] [Google Scholar]
- Dietmann S., Holm L. Nat. Struct. Biol. 2001;8:953–957. doi: 10.1038/nsb1101-953. [DOI] [PubMed] [Google Scholar]
- Dietmann S., Park J., Notredame C., Heger A., Lappe M., Holm L. Nucleic Acids Res. 2001;29:55–57. doi: 10.1093/nar/29.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R.D., Tate J., Mistry J., et al. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianni S., Guydosh N.R., Khan F., Caldas T.D., Mayor U., White G.W., DeMarco M.L., Daggett V., Fersht A.R. Proc. Natl Acad. Sci., USA. 2003;100:13286–13291. doi: 10.1073/pnas.1835776100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindarajan S., Recabarren R., Goldstein R.A. Proteins. 1999;35:408–414. [PubMed] [Google Scholar]
- Greene L.H., Lewis T.E., Addou S., et al. Nucleic Acids Res. 2007;35:D291–D297. doi: 10.1093/nar/gkl959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishin N.V. J. Struct. Biol. 2001;134:167–185. doi: 10.1006/jsbi.2001.4335. [DOI] [PubMed] [Google Scholar]
- Gunasekaran K., Eyles S.J., Hagler A.T., Gierasch L.M. Curr. Opin. Struct. Biol. 2001;11:83–93. doi: 10.1016/s0959-440x(00)00173-1. [DOI] [PubMed] [Google Scholar]
- Hadley C., Jones D.T. Structure. 1999;7:1099–1112. doi: 10.1016/s0969-2126(99)80177-4. [DOI] [PubMed] [Google Scholar]
- Hasegawa H., Holm L. Curr. Opin. Struct. Biol. 2009;19:341–348. doi: 10.1016/j.sbi.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Holland T.A., Veretnik S., Shindyalov I.N., Bourne P.E. J. Mol. Biol. 2006;361:562–590. doi: 10.1016/j.jmb.2006.05.060. [DOI] [PubMed] [Google Scholar]
- Holm L., Sander C. Science. 1996;273:595–603. doi: 10.1126/science.273.5275.595. [DOI] [PubMed] [Google Scholar]
- Holm L., Sander C. Proteins. 1998a;33:88–96. doi: 10.1002/(sici)1097-0134(19981001)33:1<88::aid-prot8>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Holm L., Sander C. Nucleic Acids Res. 1998b;26:316–319. doi: 10.1093/nar/26.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L., Sander C. Nucleic Acids Res. 1999;27:244–247. doi: 10.1093/nar/27.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hom K., Ma Q.F., Wolfe G., Zhang H., Storch E.M., Daggett V., Basus V.J., Waskell L. Biochemistry. 2000;46:14025–14039. doi: 10.1021/bi001129o. [DOI] [PubMed] [Google Scholar]
- Islam S.A., Luo J., Sternberg M.J.E. Protein Eng. Des. Sel. 1995;8:513–526. doi: 10.1093/protein/8.6.513. [DOI] [PubMed] [Google Scholar]
- Jonsson A.L., Scott K.A., Daggett V. Biophys. J. 2009;97:2958–2966. doi: 10.1016/j.bpj.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmirski S.L., Wong K.-B., Freund S.M.V., Tan Y.-J., Fersht A.R., Daggett V. Proc. Natl Acad. Sci. USA. 2001;98:4349–4354. doi: 10.1073/pnas.071054398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodny R., Petrey D., Honig B. Curr. Opin. Struct. Biol. 2006;16:393–398. doi: 10.1016/j.sbi.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Kuhlman B., Dantas G., Ireton G.C., Varani G., Stoddard B.L., Baker D. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- Ladurner A.G., Itzhaki L.S., Daggett V., Fersht A.R. Proc. Natl Acad. Sci. USA. 1998;95:8473–8478. doi: 10.1073/pnas.95.15.8473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt M. Proc. Natl Acad. Sci. USA. 2007;104:3183–3188. doi: 10.1073/pnas.0611678104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt M., Chothia C. Nature. 1976;261:552–558. doi: 10.1038/261552a0. [DOI] [PubMed] [Google Scholar]
- Li A., Daggett V. Proc. Natl Acad. Sci. USA. 1994;91:10430–10434. doi: 10.1073/pnas.91.22.10430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A., Daggett V. J. Mol. Biol. 1996;257:412–429. doi: 10.1006/jmbi.1996.0172. [DOI] [PubMed] [Google Scholar]
- Macias M.J., Gervais V., Civera C., Oschkinat H. Nat. Struct. Biol. 2000;7:375–379. doi: 10.1038/75144. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A., Anderson J.B., Derbyshire M.K., et al. Nucleic Acids Res. 2007;35:D237–D240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor U., Johnson C.M., Daggett V., Fersht A.R. Proc. Natl Acad. Sci. USA. 2000;97:13518–13522. doi: 10.1073/pnas.250473497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor U., Guydosh N.R., Johnson C.M., et al. Nature. 2003;421:863–867. doi: 10.1038/nature01428. [DOI] [PubMed] [Google Scholar]
- McCully M.E., Beck D.A.C., Daggett V. Biochemistry. 2008;47:7079–7089. doi: 10.1021/bi800118b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCully M.E., Beck D.A.C., Fersht A.R., Daggett V. Biophys. J. 2010;99:1628–1636. doi: 10.1016/j.bpj.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzin A.G., Brenner S.E., Hubbard T., Chothia C. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- Nickson A.A., Clarke J. Methods. 2010;52:38–50. doi: 10.1016/j.ymeth.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orengo C.A., Jones D.T., Thornton J.M. Nature. 1994;372:631–634. doi: 10.1038/372631a0. [DOI] [PubMed] [Google Scholar]
- Orengo C.A., Taylor W.R. Methods Enzymol. 1996;266:617–635. doi: 10.1016/s0076-6879(96)66038-8. [DOI] [PubMed] [Google Scholar]
- Orengo C.A., Michie A.D., Jones S., Jones D.T., Swindells M.B., Thornton J.M. Structure. 1997;5:1093–1108. doi: 10.1016/s0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- Otzen D.E., Itzhaki L.S., elMasry N.F., Jackson S.E., Fersht A.R. Proc. Natl Acad. Sci. USA. 1994;91:10422–10425. doi: 10.1073/pnas.91.22.10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual-Garcia A., Abia D., Ortiz A.R., Bastolla U. PLoS. Comput. Biol. 2009;5:e1000331. doi: 10.1371/journal.pcbi.1000331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl F.M.G., Martin N., Bray J.E., et al. Nucleic Acids Res. 2001;29:223–227. doi: 10.1093/nar/29.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovich M., Jonsson A.L., Ferguson N., Daggett V., Fersht A.R. J. Mol. Biol. 2006;360:865–881. doi: 10.1016/j.jmb.2006.05.050. [DOI] [PubMed] [Google Scholar]
- Reeves G.A., Dallman T.J., Redfern O.C., Akpor A., Orengo C.A. J. Mol. Biol. 2006;360:725–741. doi: 10.1016/j.jmb.2006.05.035. [DOI] [PubMed] [Google Scholar]
- Religa T.L., Markson J.S., Mayor U., Freund S.M., Fersht A.R. Nature. 2005;437:1053–1056. doi: 10.1038/nature04054. [DOI] [PubMed] [Google Scholar]
- Religa T.L., Johnson C.M., Vu D.M., Brewer S.H., Dyer R.B., Fersht A.R. Proc. Natl Acad. Sci. USA. 2007;104:9272–9277. doi: 10.1073/pnas.0703434104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski M.I., Taylor W.R. J. Phys. Condens. Matter. 2010;22:033101. doi: 10.1088/0953-8984/22/3/033103. [DOI] [PubMed] [Google Scholar]
- Sadreyev R.I., Kim B.-H., Grishin N.V. Curr. Opin. Struct. Biol. 2009;19:321–328. doi: 10.1016/j.sbi.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer R.D., Jonsson A.L., Simms A.M., Daggett V. Bioinformatics. 2010 doi: 10.1093/bioinformatics/btq625. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe T., Jonsson A.L., Rutherford T.J., Daggett V., Fersht A.R. Protein Sci. 2007;16:2233–2239. doi: 10.1110/ps.073004907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sippl M.J. Curr. Opin. Struct. Biol. 2009;19:312–320. doi: 10.1016/j.sbi.2009.03.010. [DOI] [PubMed] [Google Scholar]
- Storch E.M., Daggett V. Biochemistry. 1995;30:9682–9693. doi: 10.1021/bi00030a005. [DOI] [PubMed] [Google Scholar]
- Storch E.M., Daggett V., Atkins W.M. Biochemistry. 1999a;16:5054–5064. doi: 10.1021/bi982158q. [DOI] [PubMed] [Google Scholar]
- Storch E.M., Grinstead J.S., Campbell A.P., Daggett V., Atkins W.M. Biochemistry. 1999b;16:5065–5075. doi: 10.1021/bi982159i. [DOI] [PubMed] [Google Scholar]
- Teodoro M.L., Phillips G.N., Kavraki L.E. J. Comp. Biol. 2004;10:617–634. doi: 10.1089/10665270360688228. [DOI] [PubMed] [Google Scholar]
- Toofanny R.D., Jonsson A.L., Daggett V. Biophys. J. 2010;98:2671–2681. doi: 10.1016/j.bpj.2010.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valas R.E., Yang S., Bourne P.E. Curr. Opin. Struct. Biol. 2009;19:329–334. doi: 10.1016/j.sbi.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Kamp M.W., Schaeffer R.D., Jonsson A.L., et al. Structure. 2010;18:423–435. doi: 10.1016/j.str.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veretnik S., Bourne P.E., Alexandrov N.N., Shindyalov I.N. J. Mol. Biol. 2004;339:647–678. doi: 10.1016/j.jmb.2004.03.053. [DOI] [PubMed] [Google Scholar]
- White G.W., Gianni S., Grossmann J.G., Jemth P., Fersht A.R., Daggett V. J. Mol. Biol. 2005;350:757–775. doi: 10.1016/j.jmb.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Wolf Y.I., Grishin N.V., Koonin E.V. J. Mol. Biol. 2000;299:897–905. doi: 10.1006/jmbi.2000.3786. [DOI] [PubMed] [Google Scholar]
- Zarrine-Afsar A., Larson S.M., Davidson A.R. Curr. Opin. Struct. Biol. 2005;15:42–49. doi: 10.1016/j.sbi.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Zhang C.T. Protein Eng. 1997;10:757–761. doi: 10.1093/protein/10.7.757. [DOI] [PubMed] [Google Scholar]
- Zhang C., DeLisi C. J. Mol. Biol. 1998;284:1301–1305. doi: 10.1006/jmbi.1998.2282. [DOI] [PubMed] [Google Scholar]
- Zhi-Xin W. Proteins: Structure, Function, and Genetics. 1996;26:186–191. [Google Scholar]
- Zhi-Xin W. Protein Eng. Sel. Des. 1998;11:621–626. [Google Scholar]