Abstract
Autism spectrum disorders (ASDs) are a group of commonly occurring, highly-heritable developmental disabilities. Human genes c3orf58 or Deleted In Autism-1 (DIA1) and cXorf36 or Deleted in Autism-1 Related (DIA1R) are implicated in ASD and mental retardation. Both gene products encode signal peptides for targeting to the secretory pathway. As evolutionary medicine has emerged as a key tool for understanding increasing numbers of human diseases, we have used an evolutionary approach to study DIA1 and DIA1R. We found DIA1 conserved from cnidarians to humans, indicating DIA1 evolution coincided with the development of the first primitive synapses. Nematodes lack a DIA1 homologue, indicating Caenorhabditis elegans is not suitable for studying all aspects of ASD etiology, while zebrafish encode two DIA1 paralogues. By contrast to DIA1, DIA1R was found exclusively in vertebrates, with an origin coinciding with the whole-genome duplication events occurring early in the vertebrate lineage, and the evolution of the more complex vertebrate nervous system. Strikingly, DIA1R was present in schooling fish but absent in fish that have adopted a more solitary lifestyle. An additional DIA1-related gene we named DIA1-Like (DIA1L), lacks a signal peptide and is restricted to the genomes of the echinoderm Strongylocentrotus purpuratus and cephalochordate Branchiostoma floridae. Evidence for remarkable DIA1L gene expansion was found in B. floridae. Amino acid alignments of DIA1 family gene products revealed a potential Golgi-retention motif and a number of conserved motifs with unknown function. Furthermore, a glycine and three cysteine residues were absolutely conserved in all DIA1-family proteins, indicating a critical role in protein structure and/or function. We have therefore identified a new metazoan protein family, the DIA1-family, and understanding the biological roles of DIA1-family members will have implications for our understanding of autism and mental retardation.
Introduction
Autism spectrum disorder (ASD) is a neurodevelopmental condition commonly diagnosed in early childhood. ASD is characterized by deficits in verbal and non-verbal communication, social interaction, and by displays of restricted and/or repetitive behaviours (Mendelian Inheritance in Man accession number 209850). In the absence of definitive neuropathological markers, these deficits remain the sole diagnostic indicators of autism. ASD has the greatest heritable basis of any developmental cognitive disorder, based on twin and family studies, with heritability estimates of around 90% [1]–[9]. In addition, spontaneous genetic alterations cause around 10% of cases [10]–[12]. As expected, many of the genes implicated in ASD have direct or indirect roles in synapse formation and function [13]–[19]. The secretory pathway plays a key role in neuron function, and abnormalities in secretion and secretory cargo have been found in increasing numbers of ASD patients [20]–[23]. Genes affecting secretory pathway traffic are also reported to be affected in those with ASD [12], [24]. Furthermore, post-translational modification of proteins in the secretory pathway via phosphorylation and sulphation are abnormal in ASD patients, as are genes involved in glycosylation events within the Golgi apparatus lumen [12], [25]–[28].
The wide phenotypic presentation of ASD is reflected in the increasing number of different gene changes identified in those affected [29]. Many genes implicated in ASD are of known function, while for other genes a role in brain function is yet to be identified. Many of the characterized and uncharacterized genes have orthologues in model organisms, which can be used to study biological function. The mouse model is the most widely used to experimentally manipulate candidate genes for ASD susceptibility [30]. Zebrafish, which have highly complex social behaviours, are also emerging as useful models for vertebrate neurodevelopment and autism research [31]–[34]. Increasing use is also being made of smaller organisms to further our understanding of neurobiology and neurological disorders. For example, the fruit fly Drosophila melanogaster encodes many homologues of human neuronal genes, including significant numbers implicated in neurological disease [35], [36]. Indeed, the first specific therapeutic treatment for a condition with co-morbid ASD symptoms was reported using a Drosophila model of fragile X syndrome [37]. This study [37] demonstrated for the first time that genetic defects such as ASD and mental retardation might be treatable after birth using drugs, rather than more complex gene therapy-based approaches.
Recently, in a study of consanguineous families, an uncharacterized gene was implicated in the etiology of ASD [38]. Deletion of this gene, known as c3orf58, was hemizygous in the unaffected parents and an unaffected sibling, but homozygous in a child with ASD. The gene was therefore renamed DIA1, for Deleted In Autism 1 [38]. We have recently identified a second human gene, closely related to DIA1, which we have named DIA1R, for DIA1-Related [39]. DIA1R mutations and deletions are associated with X-linked mental retardation and/or ASD-like syndromes [39]. In order to further understand the biological role of DIA1 and DIA1R, we have used an in silico approach to study the wider DIA1 gene family. We report that DIA1 is restricted to metazoans, while the closely-related homologue, DIA1R, is restricted to vertebrates, with the latter being strikingly absent from fish with a solitary lifestyle. An additional DIA1 family member was found in echinoderm and cephalochordate genomes, which we name DIA1L (DIA1-Like). By contrast to the DIA1 and DIA1R gene products, which encode signal peptides, DIA1L gene products are predicted to be cytosolic. Unexpectedly, Caenorhabditis elegans lacks an identifiable DIA1 homologue, suggesting gene loss in the nematode lineage. Homologues of DIA1 could not be detected prior to evolution of the Cnidaria, coinciding with the development of the first primitive synapses [40]. These findings provide us with evolutionary support for a role of DIA1 homologues in neuronal function. We have therefore identified a new gene family, the DIA1-family, where there is increasing evidence two members, DIA1 and DIA1R, play a vital role in normal human brain function.
Results
Identification of DIA1 orthologues
The human (Homo sapiens) gene c3orf58, at chromosome position 3q24, has recently been renamed DIA1 on the basis of its deletion in ASD [38]. Human DIA1 has known orthologues in the genome of ten animal species [39], [41]. To characterize the DIA1 family in detail, we used Basic Local Alignment Search Tool (BLAST) and keyword searches (search term: c3orf58) of publicly-available databases to identify further sequences orthologous to human DIA1. In total, thirty five full-length (Tables 1, S1, and S3) and forty five partial (Table S2) DIA1 orthologues were identified. ‘Partial DIA1 orthologues’ are those where the full-length gene or cDNA sequence is currently incomplete (partial), due to gaps in genomic sequence or because of the limited read-length of expressed sequence tag (EST) sequencing data. Protein length varied from 345 (Drosophila willistoni and D. yakuba DIA1) to 477 (Ciona intestinalis DIA1) amino acids, equating to between 40 and 54 kDa in predicted molecular mass (Tables 1 and S3). The average length of DIA1 proteins identified was 376 amino acids, with insect gene products, especially those of Drosophila species, being shorter than those from most other species (Table 1 and Table S3). Accession numbers of all DIA1 orthologues are provided in Tables S1 and S2.
Table 1. Physical characteristics of DIA1 proteins and similarity to orthologues from key species.
| BLASTPd similarity toe: | ||||||||||
| Metazoan speciesa | Length protein (amino acids) | pIb | Molecular massc (kDa) | H. sapiens | G. gallus | D. rerio (a) | C. intestinalis | S. purpuratus | D. melanogaster | N. vectensis |
| Cnidaria | ||||||||||
| Nematostella vectensis | 402 | 5.6 | 44.7 | 8e-46 | 4e-44 | 6e-49 | 1e-15 | 6e-47 | 7e-08 | - |
| Echinodermata | ||||||||||
| Strongylocentrotus purpuratus | 431 | 7.9 | 49.7 | 6e-80 | 3e-81 | 5e-84 | 3e-17 | - | 2e-08 | 2e-42 |
| Arthropoda | ||||||||||
| Hexapoda | ||||||||||
| Aedes aegypti | 395 | 6.0 | 45.7 | 6e-16 | 6e-15 | 2e-14 | 1e-04 | 5e-16 | 6e-14 | 5e-15 |
| Anopheles gambiae | 412 | 5.4 | 47.3 | 2e-06 | 3e-08 | 9e-09 | 0.10 | 5e-12 | 4e-10 | 0.018 |
| Culex pipiens | 400 | 5.1 | 45.2 | 2e-13 | 4e-13 | 8e-16 | 5e-04 | 1e-15 | 2e-13 | 2e-18 |
| Drosophila melanogaster | 348 | 5.3 | 40.0 | 1e-06 | 7e-07 | 2e-08 | >10 | 2e-08 | - | 5e-08 |
| Nasonia vitripennis | 404 | 5.1 | 46.2 | 3e-53 | 7e-53 | 2e-54 | 1e-11 | 9e-47 | 2e-08 | 4e-36 |
| Chordata | ||||||||||
| Urochordata | ||||||||||
| Ciona intestinalis | 477 | 9.3 | 54.5 | 2e-29 | 1e-31 | 3e-34 | - | 3e-17 | >10 | 4e-14 |
| Cephalochordata | ||||||||||
| Branchiostoma floridae | 398 | 5.2 | 46.1 | 3e-101 | 5e-98 | 9e-104 | 2e-31 | 5e-81 | 7e-12 | 6e-43 |
| Vertebrata | ||||||||||
| Neopterygii | ||||||||||
| Danio rerio (a) | 432 | 8.6 | 50.0 | 0.0 | 0.0 | - | 6e-32 | 8e-82 | 2e-07 | 1e-46 |
| Danio rerio (b) | 429 | 8.7 | 49.4 | 0.0 | 0.0 | 0.0 | 6e-31 | 4e-80 | 2e-06 | 3e-44 |
| Gasterosteus aculeatus | 434 | 8.9 | 50.0 | 0.0 | 0.0 | 0.0 | 6e-33 | 5e-81 | 3e-06 | 2e-44 |
| Oryzias latipes | 434 | 8.8 | 50.1 | 0.0 | 0.0 | 0.0 | 2e-32 | 3e-83 | 3e-06 | 2e-44 |
| Takifugu rubripes | 435 | 8.8 | 50.1 | 0.0 | 0.0 | 0.0 | 3e-34 | 8e-84 | 1e-07 | 6e-47 |
| Tetraodon nigroviridis | 425 | 8.7 | 49.2 | 0.0 | 0.0 | 0.0 | 6e-32 | 6e-82 | 1e-05 | 9e-45 |
| Tetrapoda | ||||||||||
| Gallus gallus | 429 | 8.6 | 49.0 | 0.0 | - | 0.0 | 3e-29 | 4e-79 | 7e-06 | 9e-42 |
| Xenopus tropicalis | 429 | 8.7 | 49.3 | 0.0 | 0.0 | 0.0 | 7e-32 | 3e-80 | 1e-06 | 7e-45 |
| Bos taurus | 430 | 8.8 | 49.5 | 0.0 | 0.0 | 0.0 | 1e-29 | 6e-80 | 2e-06 | 3e-41 |
| Canis familiaris | 430 | 8.8 | 49.5 | 0.0 | 0.0 | 0.0 | 1e-29 | 6e-80 | 2e-06 | 3e-41 |
| Homo sapiens | 430 | 8.8 | 49.4 | - | 0.0 | 0.0 | 1e-29 | 7e-80 | 2e-06 | 3e-41 |
| Macaca mulatta | 430 | 8.8 | 49.5 | - | 0.0 | 0.0 | 1e-29 | 7e-80 | 2e-06 | 3e-41 |
| Monodelphis domestica | 430 | 8.8 | 49.3 | 0.0 | 0.0 | 0.0 | 9e-30 | 6e-80 | 8e-07 | 2e-42 |
| Mus musculus | 430 | 8.9 | 49.5 | 0.0 | 0.0 | 0.0 | 2e-30 | 5e-79 | 9e-06 | 8e-41 |
| Pan troglodytes | 430 | 8.8 | 49.5 | 0.0 | 0.0 | 0.0 | 1e-29 | 6e-80 | 2e-06 | 3e-41 |
| Pongo pygmaeus | 430 | 8.8 | 49.5 | 0.0 | 0.0 | 0.0 | 1e-29 | 7e-80 | 2e-06 | 3e-41 |
| Pteropus vampyrus | 430 | 8.9 | 49.5 | 0.0 | 0.0 | 0.0 | 4e-29 | 7e-80 | 2e-05 | 8e-41 |
| Rattus norvegicus | 430 | 8.9 | 49.5 | 0.0 | 0.0 | 0.0 | 2e-30 | 5e-79 | 9e-06 | 8e-41 |
| Tursiops truncatus | 430 | 8.8 | 49.5 | 0.0 | 0.0 | 0.0 | 1e-29 | 7e-80 | 2e-06 | 3e-41 |
DIA1 homologues were restricted to the Metazoa, but were not found in the phylum Porifera or Nematoda. Partial homologues, including those found in the phylum Mollusca and Annelida (Table S2), are not shown. See Table S1 for accession numbers of full length DIA1 proteins. Only a single Drosophila species DIA1 is included here. Table S3 is an expanded version of this Table.
Isoelectric point calculated using the assumption that all residues have pKa values equivalent to that of isolated residues, and so may not accurately represent the value for the folded protein.
Isotopically-averaged molecular weight prediction in kiloDaltons.
The BLASTP E-value measures the statistical significance threshold for protein sequence matches. The smaller the number, the better the match. Computer shorthand nomenclature is used to present E-values when values are small. For example, 5e-01 = 0.5 and 5e-04 = 0.0005. Values below 1e-250 are indicated as zero, and details of those greater than 10 are not provided. A dash is used when protein alignments have 100% identity.
Proteins were compared to DIA1from H. sapiens, C. intestinalis, S. purpuratus, or D. melanogaster, or N. vectensis by protein BLAST. The DIA1 proteins used for comparison were chosen as representives from Class Mammalia, Class Aves, and Class Neopterygii within the subphylum Vertebrata, subphylum Urochordata, phylum Echinodermata, phylum Arthropoda, and phylum Cnidaria, respectively.
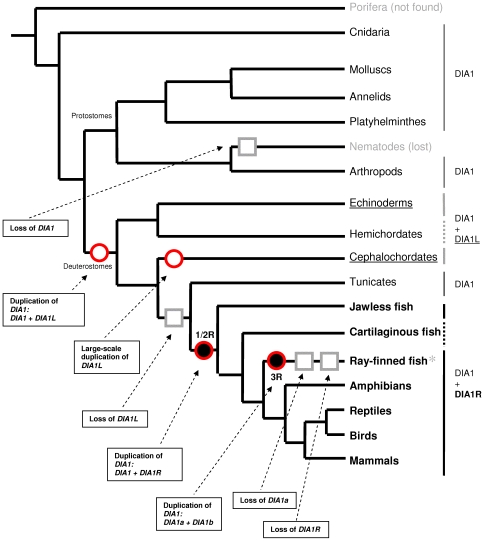
Species containing DIA1 orthologues were restricted to a single eukaryotic supergroup: the Opisthokonta (Tables 1, S1-S3). DIA1 orthologues were not found in any other eukaryotic supergroup (Plantae, Amoebozoa, Chromalveolata, Excavata, or Rhizaria) despite the availability of complete genome sequences from all supergroups, except the Rhizaria. Within the Opisthokonta, DIA1 orthologues were not found in any fungal species and were restricted to the genomes of metazoan species. Within the Metazoa, a DIA1 orthologue was not detected in sequence from the phylum Placozoa or phylum Porifera, possibly due to a scarcity of data. By contrast, orthologues were identified in species within the phylum Cnidaria, Mollusca, Annelida, Platyhelminthes, Echinodermata, Arthropoda, and Chordata (Tables S1 and S2). Within the phylum Chordata, DIA1 orthologues were present in the subphylum Urochordata (sea squirt, C. intestinalis), Cephalochordata (lancelet, Branchiostoma floridae) and Vertebrata. DIA1 orthologues were strikingly absent from the phylum Nematoda, including the completed genomes of Caenorhabditis elegans and C. briggsae. These findings are summarized in Figure 1.
Figure 1. DIA1-family superimposed on a simplified metazoan phylogeny.
DIA1 is absent from the genome sequences of nematodes (grey font) as well as fungi, plants, amoebozoa and chromalveolates (not shown). Due to a paucity of sequence data, it is unclear whether a DIA1 homologue is absent from the Porifera (grey font). DIA1L was exclusively found in echinoderm and cephalochordate genomes (underlined), which also encode DIA1. DIA1L is absent from tunicates, but a current dearth of sequence data precludes evaluation of hemichordate genomes for DIA1L homologues (indicated by a dotted bold grey line on right hand side, and a lack of underline). A bold dotted black line (right-hand side) indicates that the presence of DIA1R has been confirmed in cartilaginous fish but, probably due to a lack of sequence data, DIA1 has yet to be identified in this class of chordates. Both a DIA1 and DIA1R gene are present in vertebrate genomes (bold font), with a notable absence of DIA1R in acanthopterygian fish (asterisk). Furthermore, two DIA1 paralogues were identified in the genomes of fish from the superorder Ostariophysi, but not in fish from other superorders (see Figure 3). Data for the schematic metazoan phylogeny were from numerous sources [157]–[163]. Proposed rounds of whole-genome duplication (WGD) are indicated by filled black spheres, where two WGDs occurred early in the vertebrate lineage (1R/2R) and a third WGD (3R) in the ray-finned fish lineage before the diversification of teleosts [43], [164], [165]. Proposed duplications of DIA1-family genes are indicated by red circles, and ‘loss’ of DIA1-family genes by grey squares. Dashed arrows are used to annotate events occurring in our current model of DIA1-family evolution. Further details of two different models of DIA1-family duplication and ‘loss’ events in the fish lineage (*) can be found in Figure 3, where some fish species encode DIA1 paralogues, while others lack DIA1R. Accession numbers of DIA1, DIA1R, and DIA1L sequences can be found in Tables S1-S5, Table S7 and Table S9.
DIA1 paralogues in zebrafish
The genome of each metazoan species encode only a single DIA1 gene, with the exception of the zebrafish (Danio rerio) and minnow (Pimephales promelas) genomes, which encode two closely related DIA1 genes (Tables 1, S1 and S2). We will refer to the closely related DIA1 paralogues in these two fish species as DIA1a and DIA1b. ESTs for both DIA1 paralogues were detected in the D. rerio and P. promelas EST databases (Tables S1 and S2), indicating expression of both paralogues, and arguing against one copy being a pseudogene of the other. By contrast, the ‘completed’ genomes sequence of pufferfish Takifugu rubripes (Fugu), Tetraodon nigroviridis, and the medaka Oryzias latipes, encode only a single DIA1 orthologue (Tables S1 and S2).
Amino acid alignments of the D. rerio DIA1a and DIA1b gene products reveal an overall 88/98% amino acid identity/similarity (Figures 2 and S1). At the nucleotide level, the mRNA sequences are 77% identical (Table S1). While only partial sequence is available for the P. promelas DIA1 paralogues, amino acid alignments of the available sequences (see Table S2) reveal that DIA1a of P. promelas has greater similarity to DIA1a from D. rerio (than to DIA1b from D. rerio), while DIA1b of P. promelas is more similar to DIA1b from D. rerio (than to DIA1a from D. rerio). The available data support a model where ostariophysan fish, but not fish from other superorders, encode two functional, closely related DIA1 paralogues. These findings, superimposed on a simplified fish phylogeny, are summarized in Figure 3.
Figure 2. Amino acid sequence comparison of DIA1 from key species.
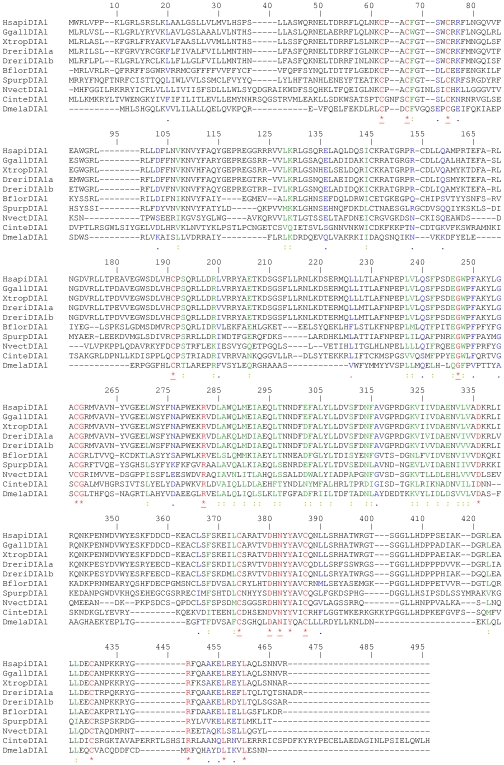
The sequence alignment was generated using CLUSTALW [47]. Identical amino acids are highlighted in red font and indicated below the alignment with an asterisk (*). Strongly similar amino acids are highlighted in green font and indicated below the alignment with a colon (:). Weakly similar amino acids are highlighted in blue font and indicated below the alignment with a full stop (.). Dissimilar amino acids are in black font. Amino acids conserved in all DIA1 proteins, as determined by alignment of DIA1 gene products from all species (Figure S2), are underlined (*). Amino acid numbering is provided above the alignment. Gaps required for optimal alignment are indicated by dashes. Standard single-letter amino acid abbreviations are used. Organism abbreviations use the first letter of the genus name, followed by the first four letters of the species (e.g. Homo sapiens DIA1 is abbreviated to HsapiDIA1). The two D. rerio DIA1 paralogues are abbreviated as DreriDIA1a and DreriDIA1b. Full species names and accession numbers can be found in Table S1.
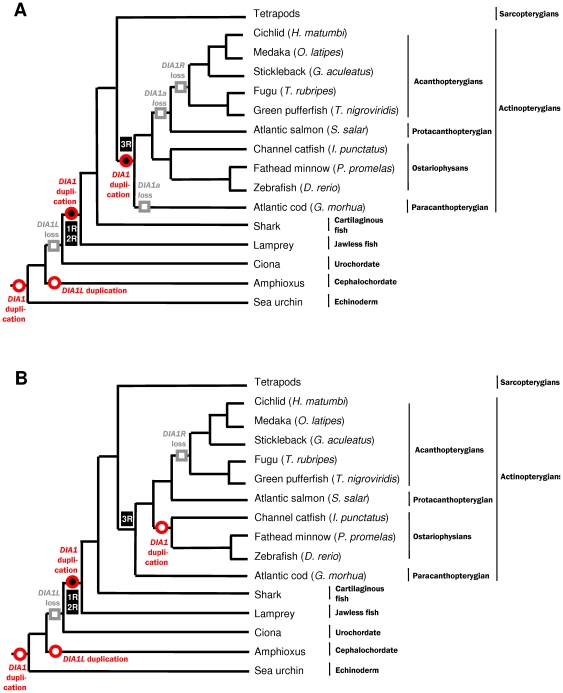
Figure 3. Fish-centric models of DIA1-family evolution.
In both models (A and B), the genome of the hypothetical chordate ancestor encodes two DIA1-family genes: DIA1 and DIA1L. The DIA1L gene has been ‘lost’ in the urochordate/vertebrate lineage, preceding the 1/2R whole genome duplications (WGDs). A duplicated copy of DIA1, which we have called DIA1R, was retained subsequent to the 1/2R WGD event, with both DIA1 and DIA1R identified in lamprey, fish, and tetrapod genomes. In the fish lineage, however, two different models, (A) and (B), could account for our current knowledge of DIA1 family members. In model (A), the DIA1 duplication generating DIA1a and DIA1b coincides with the 3R WGD. Two lineage-specific ‘losses’ of DIA1a have then occurred: the first in the G. morhua lineage, and the second in the Protacanthopterygian/Acathopterygian lineage. There are too few data available to determine whether the channel catfish encodes DIA1a, DIA1b, both, or neither. In model (B), the DIA1 duplication leading to DIA1a and DIA1b in ostariophysans does not coincide with 3R but, instead, is specific to the ostariophysan lineage. Both model (A) and (B) both predict DIA1R gene loss in the acanthopterygian lineage. Proposed rounds of WGD [43], [164], [165] are indicated by filled black spheres: numbering of the WGDs is provide in black boxes: those occurring early in the vertebrate lineage marked as 1R/2R and that in the ray-finned fish lineage marked as 3R. Proposed duplications of DIA1-family genes are indicated by red circles, and ‘loss’ of DIA1-family genes by grey squares. Data for the schematic fish phylogeny were from numerous sources [162], [166]–[169].
Comparison of DIA1 proteins
To compare DIA1 proteins with each other, we used three methods: (i) BLAST analyses, (ii) amino acid alignments, and (iii) phylogenetic analyses (see later). First, we used pair-wise protein BLAST analyses to generate ‘expect values’ (E-values) as a means of comparing the similarity between DIA1 orthologues (Table S3). All full-length DIA1 gene products were compared to DIA1 from five key species. The DIA1 sequences used for comparison of all identified DIA1 proteins were those from a species representative of the phyla Cnidaria (Nematostella vectensis), Echinodermata (Strongylocentrotus purpuratus), Arthropoda (D. melanogaster), and Chordata, where a representative of both the subphyla Urochordata (C. intestinalis) and Vertebrata (H. sapiens) were included, with a representative from the class Mammalian, class Aves, and class Neopterygii for the latter subphylum. When proteins are compared using this method, the smaller the E-value, the greater the similarity between two compared proteins. E-values of greater than 1.0 are generally considered insignificant. Our analyses found significant pair-wise E-values between all of the DIA1 gene products examined (Table S3). The only exceptions (where E-values were greater than 1.0) were in pair-wise comparisons between DIA1 from C. intestinalis and Drosophila species. By contrast, significant similarity (less than 1.0) was found between C. intestinalis DIA1 and DIA1 from all other insect species. We evaluate and discuss these similarities and differences in more detail later. Our comparisons therefore support the hypothesis that the DIA1 orthologues form a metazoan protein family.
Secondly, amino acid alignments were employed to compare DIA1 gene products at the amino acid level. While in pair-wise comparisons vertebrate DIA1 proteins were between 78–100% identical and 97–100% similar (Table S4), an amino acid alignment of DIA1 proteins from all species revealed only 10 absolutely conserved amino acids (Figure S2): 3 cysteines in the amino-terminal portion; a cysteine glycine and arginine in the central region; and an asparagine, an aspartate and two further cysteines in the carboxy-terminal portion. Blocks of highly conserved regions of amino acids within the DIA1 gene products were also identified, with the extreme amino-terminal region showing the greatest diversity (Figures 2 and S2). These alignments provide evidence that conserved regions of DIA1 are vital to the core biological function of the protein, and support the notion that this biological role is conserved in all metazoan species with a DIA1 orthologue.
Identification of DIA1R orthologues
The human gene currently annotated as cXorf36 is closely related to DIA1, and has recently been renamed DIA1R [39]. The human DIA1R gene is X-linked, located on the short arm of the X chromosome at position Xp11.3, and has orthologues in at least four vertebrate species [39]. We carried out BLAST searches using the human DIA1R sequence and keyword searches (search term: cXorf36) to identify further DIA1R orthologues. We use the term ‘orthologue’, rather than ‘paralogue’, to describe those genes most similar to DIA1 or to DIA1R, as DIA1R and DIA1 proteins differ substantially at the amino acid level, and are predicted to have evolved specific, yet-related, cellular functions. Reciprocal BLAST searches were used to confirm that newly identified DIA1R sequences had greatest similarity to DIA1R, rather than to DIA1. Reciprocal BLAST searches also ensured all currently available DIA1 and DIA1R sequences were identified, and no false positives were included. Our findings are highlighted below.
Unlike DIA1 orthologues, which were found in most metazoan phyla, DIA1R orthologues were restricted to the phylum Chordata. Furthermore, within the Chordata, DIA1R expression is restricted to the subphylum Vertebrata, including the early-branching lamprey species Petromyzon marinus (Tables S5 and S6). DIA1R was not found in subphylum Urochordata or Cephalochordata genomes. No evidence for DIA1R pseudogenes or closely-related paralogues was detected in any of the available sequence databases. All genomes encoding a DIA1R gene also had a DIA1 gene, with the exception of the EST databases for cartilaginous fish species and a single ostariophysan species (Ictalurus punctatus), where partial DIA1R sequences (Table S6), but not DIA1 sequence(s) was detected. Due to limited EST and genomic data for these species, we currently assume this absence is due to a lack of sequence data, rather than gene loss. Together, these data indicate that DIA1R arose from a DIA1 gene duplication event occurring prior to the expansion of the vertebrates, which diverged from the cephalochordates around 520 million years ago [42], and this coincides with the well-documented whole-genome duplication events in the ancestral vertebrate, prior to vertebrate expansion [43].
Absence of DIA1R in acanthopterygian fish
While DIA1R was detected in several fish species (Tables S5 and S6), there was a striking absence of DIA1R orthologues from the ‘completed’ genomes of acanthopterygian fish species including the genomes of Gasterosteus aculateus (stickleback), O. latipes (medaka fish), T. nigroviridis and T. rubripes (both pufferfish). By contrast, DIA1R was found in the genomes of fish from the superorders Ostariophysi (D. rerio and I. punctatus) and Protacanthopterygii (Salmo salar), although current EST data does not support the presence of DIA1R in the genome of another ostariophysan species, P. promelas. Conversely, while a DIA1R EST from the channel catfish (I. punctatus) was identified, a corresponding DIA1 EST was not found in this species. At present, it is mostly likely that the apparent absence of DIA1R in P. promelas, and DIA1 in I. punctatus, is due to limitations in EST data, rather than because of gene loss. Therefore, two paralogues of DIA1 are present in ostariophysan fish, but only a single DIA1R orthologue; while in the protacanthopterygian fish, we only find evidence for a single DIA1 and DIA1R gene (Tables S1, S2, S5 and S6). By contrast, in acanthopterygian genomes, only a single DIA1 gene is detected and DIA1R is absent. The timing of the DIA1 gene duplication (to form DIA1a and DIA1b) and the loss of DIA1R in acanthopterygians can only be further delineated when additional fish sequence data becomes available. Nonetheless, these data provide evidence for both DIA1 gene duplication and DIA1R gene loss events during evolution of teleost fish. Two models for DIA1 and DIA1R evolution in teleost fish, are discussed later, and are summarized in Figure 3.
Comparison of DIA1R proteins
To compare DIA1R proteins with each other, we used three methods: (i) BLAST analyses, (ii) amino acid alignments, and (iii) phylogenetic analyses. First, we used pair-wise protein BLAST analyses to generate ‘expect values’ (E-values) to assess the similarity between DIAR1 orthologues (Table 2). All full-length DIA1R gene products were compared with DIA1R from three key vertebrate species. The DIA1R sequences used for comparison with all identified full-length DIA1Rs were representative species from the subphylum Vertebrate; one each from Class Mammalia, Class Aves, and Class Neopterygii. Our analyses found significant pair-wise similarity (i.e. E-values less than 1.0) between the all of DIA1R gene products examined (Table 2). The highest pair-wise E-value (indicating the least similarity) was obtained when DIA1R from S. salar (Atlantic salmon) was compared to that of Gallus gallus (chicken), and was a value of 7e-77. As expected, each DIA1R protein was more similar to other DIA1R proteins, than to DIA1 proteins from the same species (Table 2). For example, the pair-wise E-value for DIA1R from S. salar compared to DIA1 from G. gallus was 1e-44, indicating less similarity to DIA1 than to DIA1R from the same species. Our comparisons therefore support the existence of a DIA1R-subfamily within the DIA1-family, where the subfamily genes are exclusive to the subphylum Vertebrata. Comparison of DIA1 to DIA1R gene products is discussed in detail in the section below.
Table 2. Physical characteristics of DIA1R proteins and similarity to orthologues from key species.
| BLASTPd similarity toe: | |||||||||
| DIA1R from- | DIA1 from- | ||||||||
| Speciesa | Length (amino acids) | pIb | Molecular massc (kDa) | H. sapiens | G. gallus | D. rerio | H. sapiens | G. gallus | D. rerio (a) |
| METAZOA f | |||||||||
| Chordata | |||||||||
| Vertebrata | |||||||||
| Neopterygii | |||||||||
| Danio rerio | 417 | 8.7 | 47.2 | 1e-105 | 7e-81 | - | 2e-42 | 3e-44 | 1e-43 |
| Salmo salar | 453 | 8.7 | 50.5 | 6e-101 | 7e-77 | 1e-130 | 8e-41 | 1e-44 | 1e-42 |
| Tetrapoda | |||||||||
| Aves | |||||||||
| Gallus gallus | 430 | 8.7 | 49.0 | 3e-142 | - | 2e-85 | 1e-34 | 6e-35 | 1e-35 |
| Mammalia | |||||||||
| Bos taurus | 433 | 8.4 | 48.1 | 0.0 | 9e-128 | 8e-108 | 3e-40 | 5e-43 | 1e-43 |
| Dipodomys ordii | 434 | 7.8 | 48.5 | 0.0 | 2e-127 | 5e-102 | 2e-40 | 5e-42 | 1e-43 |
| Equus caballus | 433 | 8.8 | 48.7 | 0.0 | 3e-132 | 9e-109 | 2e-41 | 2e-42 | 3e-43 |
| Homo sapiens | 433 | 8.1 | 48.6 | - | 8e-136 | 9e-106 | 3e-41 | 2e-44 | 7e-45 |
| Macaca mulatta | 433 | 8.1 | 48.6 | 0.0 | 4e-134 | 1e-104 | 7e-42 | 4e-44 | 4e-45 |
| Monodelphis domestica | 432 | 8.6 | 49.0 | 0.0 | 1e-139 | 1e-110 | 8e-46 | 2e-47 | 1e-48 |
| Ornithorhynchus anatinus | 432 | 8.9 | 48.8 | 2e-168 | 3e-131 | 5e-103 | 8e-40 | 5e-40 | 7e-44 |
| Mus musculus | 435 | 8.1 | 49.0 | 0.0 | 4e-129 | 2e-104 | 4e-38 | 3e-40 | 3e-40 |
| Rattus norvegicus | 435 | 8.5 | 48.8 | 0.0 | 6e-129 | 4e-106 | 4e-42 | 2e-44 | 1e-44 |
| Sorex araneus | 431 | 8.8 | 48.1 | 0.0 | 6e-123 | 1e-100 | 2e-38 | 7e-41 | 1e-42 |
See Table S5 for accession numbers of full length DIA1R proteins.
Isoelectric point calculated using the assumption that all residues have pKa values equivalent to that of isolated residues, so may not accurately represent the value for the folded protein.
Isotopically averaged molecular weight prediction in kiloDaltons.
The BLASTP E-value (Expect value) measures the statistical significance threshold for protein sequence matches. The smaller the number, the better the match. Computer shorthand nomenclature is used to present E-values when values are small. For example, 5e-01 = 0.5 and 5e-04 = 0.0005. Values lower than 1e-250 are treated as zero. A dash is used when alignments have 100% identity.
Proteins were compared to DIA1 (or DIA1a when paralogues were present) and DIA1R from Homo sapiens, Gallus gallus or Danio rerio by BLASTP. The proteins used for comparison were chosen as representatives from the Class Mammalia, Class Aves, and Class Neopterygii within the subphylum Vertebrata.
DIA1R orthologues are only found in the subphylum Vertebrata and not in other subphyla or phyla. For example, DIA1R orthologues are not found in the phylum Nematoda, Platyhelminthes, Cnidaria, Echinodermata, or Arthropoda.
Secondly, we carried out amino acid alignments of all DIA1R orthologues (Figure S3 and Table S4). This comparison revealed a high level of amino acid identity and similarity between DIA1R proteins from all species, with an overall 27% amino acid identity and 53% similarity across gene products from all species. Poorest conservation was within the extreme amino-terminal portion, which forms a signal peptide (see later). Comparison between DIA1R gene products in a pair-wise manner revealed 45–97% identity and 76–97% similarity between each pair (Table S4). Further comparison of DIA1R proteins was carried using phylogenetic analyses and this data will be presented and discussed later.
Comparison of DIA1 and DIA1R proteins
To compare DIA1R orthologues with their DIA1 counterparts, three approaches were used: (i) pair-wise amino acid alignments, (ii) amino acid alignments of all full length DIA1 and DIA1R proteins identified, and (iii) phylogenetic analyses. The phylogenetic analyses are provided later.
First, a detailed pair-wise comparison of DIA1 and DIA1R gene products was performed, using amino acid sequence from all species where both genes had been identified (Table S4). These analyses lead to two notable findings. (i) All DIA1R amino acid sequences are around 30% identical and 60% similar to their DIA1 counterpart of the same species (Table S4). (ii) The pair-wise amino acid identity between DIA1 homologues of two given species was always greater than that of the DIA1R proteins of those same species (Table S4). For example, while DIA1 from H. sapiens and Macaca mulatta (rhesus macaque) are 100% identical at the amino acid level, the DIA1R proteins from these same species are only 97% identical. Similarly, DIA1 from Rattus norvegicus (rat) and Mus musculus (mouse) are 100% identical at the amino acid level, while the DIA1R proteins from these same species are only 91% identical. The greater divergence of DIA1R, compared to DIA1, is more apparent when sequences from more evolutionary distant species are compared. For example, DIA1 from H. sapiens and Gallus gallus (fowl) are 90% identical (96% similar), but DIA1R proteins from these two species are only 65% identical (87% similar). These findings indicate greater evolutionary pressures favouring the conservation of DIA1, compared to DIA1R. More rapid evolution of one copy of a duplicated gene is a well-documented phenomenon [44], [45] and greater DIA1R divergence, than DIA1 divergence, is therefore expected.
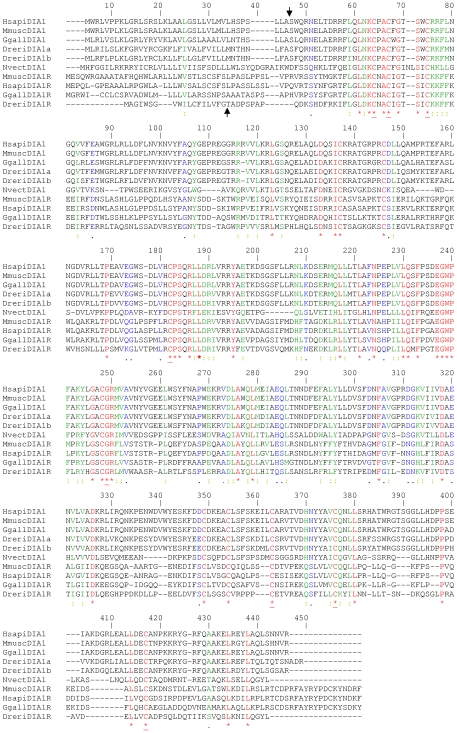
Secondly, an amino acid alignment of all DIA1 and DIA1R proteins was created (Figure S4). This comparison found a total of eight amino acids that were absolutely conserved in DIA1 and DIA1R from all species, and these are highlighted in Figure 4. The conserved residues are: three cysteine residues in the amino-terminal portion, a cysteine and glycine residue in the central portion, and three cysteine residues in the C-terminal portion. Overall, there was greater amino acid conservation between DIA1 and DIA1R proteins in the central portion, which suggests this region mediates a key, conserved protein function.
Figure 4. Amino acid sequence alignment of DIA1 and DIA1R proteins from key species.
Gene products from species with known full-length DIA1 and DIA1R orthologues were aligned using CLUSTALW [47], with DIA1 from the cnidarian species Nematostella vectensis (NvectDIA1), included for comparative purposes. Identical amino acids are highlighted in red font and indicated below the alignment with an asterisk (*). Strongly similar amino acids are highlighted in green font and indicated below the alignment with a colon (:). Weakly similar amino acids are highlighted in blue font and indicated below the alignment with a full stop (.). Dissimilar amino acids are in black font. Amino acids conserved in all DIA1 and DIA1R proteins, as determined by alignment of the DIA1 and DIA1R gene products from all species (Figure S4), are underlined (*). Amino acid numbering is provided above the alignment. Gaps required for optimal alignment are indicated by dashes. Standard single-letter amino acid abbreviations are used. Organism abbreviations use the first letter of the genus name, followed by the first four letters of the species (e.g. Homo sapiens DIA1R is abbreviated to HsapiDIA1R). Full species names and accession numbers can be found in Tables S1 and S4. Predicted signal peptide cleavage sites for human DIA1 and DIA1R (Figure S5) are indicated by arrows above or below the alignment, respectively.
Tunicate DIA1 is similar to both DIA1 and DIA1R from other species
On examination of amino acid alignments of DIA1 and DIA1R gene products, it was noticeable that the C. intestinalis (a tunicate) sequence contained frequent amino acid insertions of various lengths, when compared to the DIA1 and DIA1R sequence from all other species (Figure S4). These insertions were most prominent in the amino- and carboxy-terminal portions of the C. intestinalis DIA1 protein, rather than the central region. We therefore investigated the relationship of C. intestinalis DIA1 with other DIA1 family members in greater detail.
First, pair-wise amino acid alignments of DIA1 of C. intestinalis with DIA1 and DIA1R of key vertebrate species (H. sapiens, M. musculus, G. gallus, D. rerio), were carried out (Table S7). These analyses revealed that C. intestinalis DIA1 was only marginally more similar to DIA1 than to DIA1R from other species, being on average 23.5% identical (55% similar) to the representative vertebrate DIA1 proteins, and 22% identical (55% similar) to the DIA1R proteins.
Secondly, reciprocal pair-wise E-values between C. intestinalis DIA1, and DIA1 and DIA1R of the same key vertebrate species (H. sapiens, M. musculus, G. gallus, D. rerio), were generated (Table S7). These analyses revealed that C. intestinalis DIA1 showed greater similarity (i.e. lower E-values) to DIA1, than DIA1R orthologues using the BLAST algorithm (Table S7), with E-values of around e-32 when compared to the vertebrate DIA1 proteins, but only e-25 when compared to the DIA1R proteins from the same species. These findings suggest that C. intestinalis DIA1 indels are causing difficulties when comparisons are made with other protein family members. Close examination of the C. intestinalis DIA1 gene structure did not reveal any annotation errors contributing to the indels in the C. intestinalis DIA1 sequence, nor were any pseudogenes, or other DIA1-related sequences, detected in the C. intestinalis genome (data not shown). These results highlight the differing results that can be obtained using different alignment algorithms, such as BLAST [46] or CLUSTALW [47], and the intrinsic difficulties in obtaining optimal amino acid alignments for sequences with insertions. Our conclusions are that C. intestinalis contains a single copy of a divergent DIA1 (not a DIA1R) gene, and that this divergence will impact on the phylogenetic relationships we later derive.
Signal peptides in DIA1 and DIA1R orthologues
Human DIA1 and DIA1R, have amino-terminal signal peptides for protein targeting to the secretory pathway [39], [48]. Signal peptide functionality is supported by localization of DIA1 to the lumen of the Golgi apparatus [48]. We therefore analyzed all available DIA1 and DIA1R gene products for signal peptides, and predicted signal peptide cleavage sites, using three of the best-performing algorithms [49], [50]. The methods used were: (i) the neural network algorithm of SignalP v3.0 [51], (ii) the hidden Markov model of SignalP v3.0 [51], and (iii) the Sigcleave algorithm at EMBOSS [52]. Use of multiple algorithms also gives us the maximal possible confidence about the results, if they concur. No trans-membrane domains were predicted, nor ER-retrieval or retention motifs detected (data not shown).
Our analyses of 35 full length DIA1 orthologues revealed signal peptides (SPs) in gene products from all species, using all three prediction methods (Figure S5). Furthermore, concordant SP cleavage sites were predicted by all three methods for DIA1 proteins from 6 species, with very similar cleavage sites predicted for DIA1s from a further 11 species (i.e. where 2 of the 3 methods predicted a cleavage site aligning with the concordant site, and the remaining method predicted the adjacent amino acid as the cleavage site). As similar cleavage site prediction results were obtained for DIA1 proteins from all tetrapod species, the cleavage site for tetrapod DIA1 cleavage is predicted to be conserved, and to occur after alanine-37 (using H. sapiens DIA1 numbering). By contrast, the signal peptide cleavage site for DIA1 from Drosophila species is predicted to be after proline-24 (using D. melanogaster DIA1 numbering). While there is a consensus for SP presence in DIA1s from other species, there was no consensus for the cleavage site (Figure S5). Less concordance in prediction of the cleavage site for these species may reflect a current lack of information about the SP recognition process in such species. The majority of model proteins on which the prediction algorithms are based are from tetrapods, and less is known about cleavage of signal peptides in proteins from other species.
Our analyses of DIA1R orthologues likewise revealed the presence of SPs in DIA1R gene products from all species using all three prediction methods (Figure S5). While there was a consensus for SP presence in all DIA1Rs, there was no consensus for the position of the cleavage site, as only two species showed concordance in cleavage sites prediction from all three methods. Based on our data, the best prediction for the SP cleavage site for mammalian DIA1R is after serine-31 (using H. sapiens DIA1R numbering). Overall, we show SPs are predicted for both DIA1 and DIA1R from all species, however the exact amino acid composition of luminal DIA1 and DIA1R gene products will require experimental validation. Such data will be of benefit for refining the algorithms used for signal peptide cleavage sites, particularly in non-tetrapod species. Our data indicate that translocation into the ER and transportation to the Golgi apparatus will be a common property of all DIA1 and DIA1R gene products.
DIA1-like homologues in amphioxus and echinoderm
BLAST searches of the non-redundant (NR) database revealed the presence of additional, DIA1-like genes in some species, which we refer to as DIA1L, for DIA1-like gene. These gene products were restricted to the genomes of S. purpuratus (an echinoderm) and B. floridae (a cephalochordate). Initially, we thought these products may be due to annotation errors or be pseudogenes. Indeed, analysis of the annotated S. purpuratus DIA1L gene did reveal a splicing error in the gene model, which we corrected (Table S8 and Figure S10). The EST database provides evidence that the S. purpuratus DIA1L gene is expressed (Table S8).
By contrast to the echinoderm genome, which has a single DIA1L gene, three full-length DIA1L paralogues were identified in the B. floridae genome, which we have called DIA1La, DIA1Lb, and DIA1Lc (Table S8). Of the three full-length B. floridae DIA1L genes, two contain introns, indicating they are not processed pseudogenes (Table S8). EST data supports the expression of DIA1Lb and the intron-less B. floridae DIA1Lc gene (Table S8). For the latter, this indicates the lack of introns is due to intron-loss events, rather than DIA1Lc being a pseudogene. For B. floridae, EST sequencing is still very much a work in progress, and this may explain the current lack of expression data for the amphioxus DIA1La gene. Together these data indicate a duplication of DIA1 early in the deuterostome lineage, generating what is now DIA1L (Figure 1). However, we cannot rule out an alternative hypothesis: that the duplication event generating DIA1L preceded protostome-deuterostome divergence, and that DIA1L was lost early in the protostome lineage. Strikingly, while the echinoderm genome encodes a single copy of DIA1 and of DIA1L, the cephalochordate genome encodes a single copy of DIA1 and multiple copies of DIA1L. This indicates cephalochordate lineage-specific duplication of DIA1L. A lack of DIA1L in later-branching deuterostomes, indicates DIA1L homologues were ‘lost’ prior to tunicate divergence.
Comparison of DIA1L and DIA1 gene products
Amino acid alignments reveal S. purpuratus DIA1 and DIA1L have approximately 15% identical amino acids while, overall, 40% of aligned amino acids are similar (Table S9 and Figure S7). The corrected S. purpuratus DIA1L gene product is 636 amino acids in length (Table S8), compared to 431 residues for the homologous S. purpuratus DIA1 gene product (Table 1; Table S1). The B. floridae DIA1La gene product is 483 amino acids in length and the B. floridae DIA1Lb gene product 539 residues, again both longer than DIA1 (418 amino acids) of the same species. By contrast, B. floridae DIA1Lc is shorter (398 amino acids) than the parental DIA1 gene product. B. floridae DIA1La, DIA1Lb and DIA1Lc are all approximately 20% identical (55% similar) to each other at the amino acid level (Figure S7; Table S9) and S. purpuratus DIA1L is most similar to B. floridae DIA1Lc (∼60% similar/20% identical). By contrast to the variability between DIA1L gene products, the DIA1 gene products of S. purpuratus and B. floridae show greater similarity to each other (∼75% similar/40% identical). This finding indicates greater evolutionary pressure favouring the conservation of DIA1 sequences, compared to that favouring sequence conservation in the duplicated DIA1L gene(s), over the same evolutionary time-period.
Conservation within the DIA1 protein family
To determine key amino acids conserved across all members of the DIA1 protein family, amino acid alignments of DIA1, DIA1R, and DIA1L gene products were performed (Figure S7). Absolute conservation of four residues across the whole DIA1-protein family was found. These residues were: two cysteines in the amino-terminal region, a central glycine residue, and a further cysteine in the carboxy-terminal region of the proteins (Figure S7). In total, 15 highly conserved motifs, can be delineated (Table 3), however hydrophobic motif 1 is weakly conserved in DIA1L proteins and Drosophila DIA1 proteins, motif 12 is absent from DIA1L proteins, motifs 4 and 14 are absent from Drosophila DIA1 proteins, and motif 14 is only weakly conserved in DIA1R and DIA1L proteins (Figures S7–S9). We are currently investigating further the structure and function of the identified DIA1-family motifs. Overall, greatest amino acid similarity is found in the central portion of the extended DIA1 family (Figure 5), indicating a core role for this region in function, not only of DIA1 and DIA1R, but also of DIA1L. The relationship between DIA1-family gene products is examined further below, using phylogenetic methods.
Table 3. DIA1-family motifs.
| Motif* | Alignment consensus** | Human DIA1 sequence*** | Comment |
| 1 | F-L-x-L | FLqL | This motif is conserved in DIA1 and DIA1R proteins, but less-strongly conserved in DIA1L proteins (which are not targeted to the secretory pathway). The penultimate leucine in the motif is absent from Drosophila DIA1 proteins. |
| 2 | C-x-A-C-x-G-x(3,5)-C | CpACfGtswC | Motif contains two of the absolutely conserved DIA1-family residues. |
| 3 | A-x-Y-x(6,15)-L | AqYgepreggrrrvvklrL | The tyrosine is conserved in 73% of DIA1-family members. In some family members it is replaced by a tryptophan or phenylalanine, or there is an adjacent (or nearby) residue that is a tyrosine. |
| 4 | I-C-x(8,10)-C | ICkratgrprC | Absent from DIA1 from Drosophila species. |
| 5 | V-x(4,11)-C-x-S-x(6,10)-Y-x-E | VegwsdlvhCpSqrlldrlvrrYaE | The tyrosine is conserved in 85% of DIA1-family proteins. |
| 6 | L-x(3)-L-x-x-N-x-x-P-L-V-L-Q | LlltLafNpePLVLQ | The proline and glutamine residues of this motif are absent from DIA1L proteins from amphioxus. |
| 7 | G-W-P-x(5)-G-x-C-G | GWPfakylGaCG | Motif contains one of the absolutely conserved DIA1-family residues. |
| 8 | L-x-x-Y | LwsY | The tyrosine is conserved in 77% of DIA1-family members. In some family members it is replaced by a tryptophan or phenylalanine, or there is an adjacent (or nearby) tyrosine residue. |
| 9 | R-x-D-L-A-x-Q-L-M-x-I-x(3)-L | RvDLAwQLMeIaeqL | The first amino acid (R) of this motif is poorly conserved in DIA1R proteins. |
| 10 | F-x-L-Y-x-x-D-x(5)-F-A-V | FaLYlldvsfdnFAV | The tyrosine is conserved in 73% of DIA1-family members. In some family members it is replaced by a tryptophan or phenylalanine, or there is an adjacent (or nearby) residue that is a tyrosine. The aspartate of this motif is absent from DIA1R proteins. |
| 11 | K-V-x-I-D-x-E-x-V-x-V-x-D | KViIvDaEnVlVaD | The central glutamate is not conserved in DIA1R proteins. |
| 12 | C-x(3,4)-A-C-x(6,8)-C | CdkeAClsfskeilC | The final cysteine is well-conserved, but the remainder of motif is poorly conserved in insect and tunicate DIA1. Motif absent from DIA1L proteins. |
| 13 | [D-x-N-x-Y-x-x]-C-x-x-L-L | [DhNyYav]CqnLL | Motif contains one of the absolutely conserved DIA1-family residues. An expanded, tyrosine-containing motif [in square brackets] is found in this position in DIA1 and DIA1L, but not DIA1R, proteins (see Figures S8 and S9). |
| 14 | G-x-L-H-x(3,4)-E | GlLHDPPsE | Motif found in DIA1 only, with the exception of Drosophila DIA1 proteins. Absent from DIA1R and DIA1L. |
| 15 | L-x-E-x(16,18)-L | LdEcanpkkrygrfqaakeL | Consensus is conserved in more than 80% of DIA1 family but, while the final leucine is highly conserved, the first leucine is absent from 75% of DIA1R proteins. The charged residue is poorly conserved in DIA1 of insects. |
*Motifs numbered in amino- to carboxy-terminal direction.
**Consensus motif is that from the Boxshade consensus line using 80% similarity threshold (Figure S8), unless otherwise indicated. Underlined residues = 100% conserved. Standard single-letter amino acid abbreviations are used, where x = any amino acid, and x(6,8) indicates 6, 7, or 8 poorly/non-conserved amino acids present in that position.
***Motif-conforming residues are in upper case; poorly or non-conserved amino acids in lower case.
Figure 5. Amino acid sequence alignment of DIA1-family proteins from key species.
All full-length DIA1, DIA1R, and/or DIA1L gene products were aligned using CLUSTALW (Figure S7), and this figure represents excerpts from this master alignment, where proteins from the following phyla only are represented: Cnidaria (N. vectensis DIA1: NvectDIA1), Arthopoda (D. melanogaster DIA1: DmelaDIA1), Echinodermata (S. purpuratus DIA1 and DIA1L: SpurpDIA1 and SpurpDIA1L), Cephalochordata (B. floridae DIA1 and DIA1L paralogues: BflorDIA1, BflorDIA1La, b, and c), and Chordata. The latter includes representatives of the subphylum Urochordata (C. intestinalis DIA1: CinteDIA1) and subphylum Vertebrata (H. sapiens DIA1 and DIA1R: HsapiDIA1 and HsapiDIA1R). Amino acid numbering from the master alignment (Figure S7) is provided above the alignment. Gaps required for optimizing the master alignment (Figure S7) are indicated by dashes. Standard single-letter amino acid abbreviations are used. Organism abbreviations use the first letter of the genus name, followed by the first four letters of the species (e.g. Homo sapiens DIA1R is abbreviated to HsapiDIA1R). Full species names and accession numbers can be found in Tables S1, S4 and S7. The predicted location of the DIA1 and DIA1R signal peptides (SP) are indicated above the alignment (Figure S5). Conserved amino acid motifs detected in the master alignment (Table 3, Figure S7) are indicated in numbered boxes above the alignment. Consensus amino acids for each motif are indicated below the alignment. Amino acids absolutely conserved across the whole DIA1-family are indicated in red upper-case letters, those strongly conserved across the whole DIA1-family are in green, and those weakly conserved across the whole DIA1-family in blue (Figure S7). In addition, black lower-case letters indicate amino acids conserved in over 80% of DIA1-family sequences (Figure S8), while grey lower-case letters indicate conservation in 50–80% of DIA1-family sequences (Figure S9).
DIA1L proteins lack signal peptides
By contrast with both DIA1 and DIA1R gene products (Figure S5), DIA1L proteins do not encode predicted signal peptides (data not shown). As for DIA1 and DIA1R, we analyzed DIA1L gene products for the presence of trans-membrane domains and, similarly, none were detected (data not shown). We therefore conclude that while DIA1 and DIA1R can enter the endoplasmic reticulum, DIA1L proteins cannot, and will fulfil a cytosolic function. The altered sub-cellular localization of DIA1L compared to the parental DIA1 may contribute to the sequence divergence of DIA1L proteins compared to that of DIA1 proteins (Table S9). Subcellular relocalization has previously been described as a mechanism for duplicate gene retention [53]–[55] and as a factor contributing to asymmetric sequence divergence [56].
Identification of further DIA1L homologues in amphioxus
While a single DIA1L homologue was found in the echinoderm genome, and three full-length DIA1L homologues in the cephalochordate B. floridae (Table S8), we also found evidence that further DIA1L homologues may exist in the B. floridae genome. In the first assembly (August, 2009) of the B. floridae genome, 14 incomplete DIA1L genes were annotated. In the most recent assembly (October, 2009), we found evidence for five partial DIA1L genes (Table S10), in addition to the 3 full-length amphioxus DIA1L homologues. Our analyses (below) suggest the number of DIA1L homologues, however, will be eight, as some of the partial DIA1L genes represent redundant allelic copies (Table S10). Further analysis is hampered by gaps in the genomic sequence.
The currently annotated five incomplete DIA1L genes in the B. floridae genome were identified by BLAST searches of the NR database (Table S10). We have numbered the partial (DIA1L-pt) genes DIA1L-pt1–DIA1L-pt5, for discussion purposes. Two of the DIA1L partial genes, DIA1L-pt3 and DIA1L-pt4 are similar to each other and share synteny (Table S10), and will most likely prove to be alleles of each other (and be re-annotated in a future assembly of the B. floridae genome). Expression of DIA1L-pt3 and its proposed allele DIA1L-pt4 is supported by EST data (Table S10) but, at present, expression data for DIA1L-pt5 is lacking. DIA1L-pt1 and DIA1L-pt2 are annotated as adjacent genes encoded on opposite DNA strands. A single EST with 93% identity to both DIA1L-pt1 and DIA1L-pt2 suggests expression of at least one of these genes (Table S10). However, the significance of a lack of expression data for DIA1L-pt5 is unclear, as it must be appreciated that, for B. floridae, EST sequencing is still limited. Therefore, while a duplication of DIA1 early in the deuterostome lineage generated DIA1L, there has been large-scale lineage-specific expansion of the DIA1L in the cephalochordate lineage. Strikingly, despite the retention of DIA1L homologues in echinoderms and cephalochordates subsequent to this gene duplication event, DIA1L is absent in all later-branching deuterostomes, indicating DIA1L was ‘lost’ or diverged dramatically (precluding detection) prior to tunicate divergence (see Figure 1).
A non-processed DIA1 pseudogene in the mosquito C. pipiens?
The C. pipiens genome contains a second DIA1-like gene (Table S1) classified as putatively translated on the NR-database (accession number XP_001867819). However, there is some evidence for re-classification of the C. pipiens gene as a non-processed pseudogene (Table S1) and we are currently investigating this possibility in more detail. The presence of non-processed pseudogene provides evidence of a past lineage-specific DIA1 gene duplication event in C. pipiens. No non-processed or processed pseudogenes were detected in other species using genomic BLAST searches.
DIA1 family phylogeny
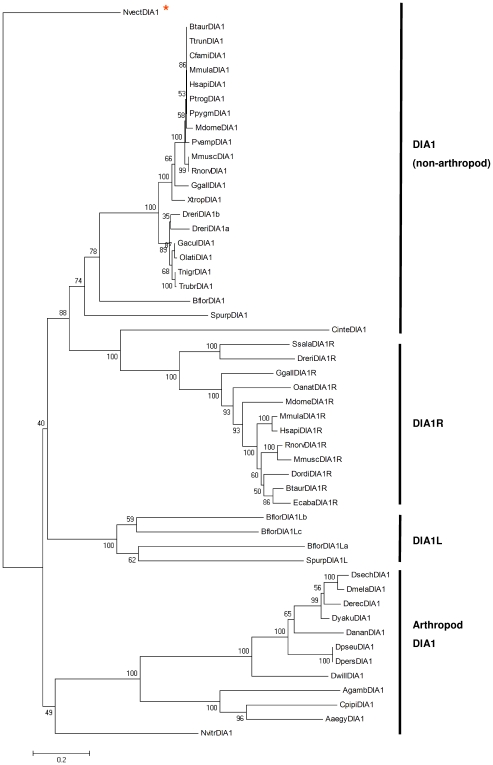
To assess the evolutionary relatedness of the DIA1 family genes, a phylogenetic tree, based on the alignment of the amino acid sequences of all DIA1 homologues (Figure S7) was generated, using a distance-based neighbour-joining method [57]. Figure 6 illustrates our current knowledge of the evolutionary relationship between DIA1, DIA1R, and DIA1L. Four key features were highlighted by these analyses. (i) DIA1R orthologues cluster with each other, supporting the hypothesis of a gene duplication, and subsequent divergence of the duplicated gene. (ii) Evidence for a second DIA1 duplication in the teleost lineage is highlighted by the clustering of the two DIA1 paralogues from D. rerio first with each other, rather than with DIA1 from other vertebrates. By contrast, the D. rerio DIA1R orthologue clusters with DIA1R of other vertebrates, not with the D. rerio DIA1 paralogues. (iii) Clustering of the DIA1L gene products of sea urchin (S. purpuratus) and amphioxus (B. floridae) together, supports the scenario of a DIA1 gene duplication early in the deuterostome lineage, followed by a lineage-specific expansion of DIA1L in B. floridae. (iv) DIA1 from the sea squirt, C. intestinalis, did not branch in the position expected from the known phylogenetic relationship of species (Figure 2). Our neighbourhood-joining analysis found C. intestinalis DIA1 clustering with the DIA1R orthologues, rather than the DIA1 orthologues, of other species (Figure 6). However, C. intestinalis does not encode a DIA1R gene, and has only one DIA1 gene, although the resulting gene product has characteristics intermediate to DIA1 and DIA1R of other species (see above). The unusual branching of C. intestinalis DIA1 may be due to one or more of the following factors: loss of a duplicated gene, convergent evolution, and/or limitations in the phylogenetic reconstruction method. Use of other amino acid alignment methods (data not shown) and different methods of phylogenetic reconstruction, such as maximum-likelihood (Figure S11) or Bayesian analysis (Figure S12), did not alter the branching of the C. intestinalis DIA1 gene to that consistent with the current evolutionary model of species.
Figure 6. Evolutionary relationships between DIA1-family members.
The evolutionary history of the DIA1 family was inferred using the neighbour-joining method [57]. The optimal tree is shown, with statistical reliability of branching assessed using 1000 bootstrap replicates [155], where percentage values are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method [170] and units are the number of amino acid substitutions per site. All positions containing gaps were eliminated from the dataset (Figure S10). There were a total of 258 positions in the final dataset. Phylogenetic analyses were conducted in MEGA4 [150]. The tree was rooted on the cnidarian N. vectensis DIA1 sequence (NvectDIA1), as highlighted with an asterisk. Organism abbreviations use the first letter of the genus name, followed by the first four letters of the species. Full species names and accession numbers can be found in Tables S1, S4 and S7.
In summary (Figures 1 and 6), the earliest-branching metazoan in which DIA1 was detected was the cnidarian, N. vectensis. The origin of DIA1L can be traced back to early in the deuterostome lineage, where a DIA1 gene duplication event and subsequent divergence occurred. In the cephalochordate lineage, large-scale, lineage-specific duplication of DIA1L has occurred, leading to an estimated 7 copies of DIA1L in the B. floridae genome. DIA1L loss (or divergence precluding detection) then occurred prior to urochordate branching, and DIA1L homologues are not detected in urochordates or vertebrates. The origin of DIA1R (via DIA1 duplication) coincides with the two WGDs occurring early in vertebrate evolution, and humans and other vertebrates have two DIA1 homologues: DIA1 and DIA1R. By contrast, further DIA1 gene duplication has occurred in the fish lineage which may have coincided with the teleost WGD, or alternatively an event specific to the ostariophysan lineage (Figure 3). Loss of DIA1R has occurred in the acanthopterygian fish lineage. Together, our data support the hypothesis that multiple gene duplication and gene ‘loss’ events have occurred during the evolution of the DIA1 family. Three subfamilies of expressed extant genes exist that share a number of common motifs: DIA1, DIA1R, and DIA1L. The DIA1 and DIA1R gene products from all species are targeted to the lumen of the secretory pathway and DIA1L gene products to the cytoplasm. Animal models may be useful in understanding why defective DIA1 and DIA1R gene products cause ASD and/or mental retardation.
Discussion
Defective human DIA1 and DIA1R genes, despite their ubiquitous tissue expression, are implicated in the etiology of autism, autism-like syndromes, and/or mental retardation [38], [39]. Little is known about DIA1 or DIA1R, except that the genes are ubiquitously expressed and they encode signal peptides for targeting to the secretory pathway, with DIA1 localizing to the lumen of the Golgi apparatus [39], [48]. Here we confirmed the presence of a signal peptide for secretory pathway targeting in DIA1 and DIA1R gene products from all species, and have studied the evolutionary history of the DIA1 gene, finding its emergence coincided with the development of the early nervous system. By contrast, the closely related DIA1R gene is exclusive to vertebrate genomes. We further identify a related gene DIA1L in echinoderm and amphioxus genomes only, where the gene products are predicted to be cytoplasmically-targeted. Therefore, the recently identified human DIA1 gene is part of a larger, evolutionarily-related gene family.
DIA1 evolution coincided with early nervous tissue development
We found DIA1 to be conserved from cnidarians to humans, but we found no evidence for DIA1 in the currently available poriferan sequence databases. Cnidarians have a basic nervous system, but are constructed on principles similar to those of complex metazoans, including the formation of organized nerve networks [58]. The cnidarian apical pole is considered a primitive head, and it is currently thought that nervous tissue first evolved in cnidarians or a closely-related ancestor [58], [59]. Therefore, the detection of DIA1 coincides with the evolution of a neuronal network, providing circumstantial evidence for a key role of DIA1 in neuronal function.
Absence of DIA1 in nematodes
Unexpectedly, DIA1 was absent in nematodes, including the completed genomes of C. elegans and C. briggsae. There are two possible explanations for this finding: (i) nematodes have lost the DIA1 gene and have no need for an equivalent gene; or (ii) a nematode DIA1 is still present, but it is undetectable due to evolutionary divergence. This apparent nematode-specific gene loss is not without precedent: it has been found that, while 20% of S. purpuratus genes are found in fruit fly, only 15% are present in nematode. Furthermore, while S. purpuratus has members of 97% of the human kinase subfamilies, while Drosophila lacks 20%, and nematodes 32% [60]. Indeed, more than half of the putative nematode genes are unique to the phylum, with 23% being species-specific [61]. Even within the nematode phylum, organisms only share only around 60% of their genes [61]. Finally, a study of ESTs from the Cnidarian Acropora millepora showed that over 10% of the Acropora ESTs with clear human homologs, have no representatives in the Drosophila or Caenorhabditis genomes [62]. Clearly, secondary gene loss in the nematode lineage is becoming an increasingly well-documented phenomenon.
The hypothesis that the lack of detection of DIA1 in nematode genomes is due to divergence, however, is also supported by the rapid evolutionary rates reported in the nematode lineage [63]–[66], with genome rearrangements occurring approximately four times faster in the worm than in the fly [67] and 50-fold the rate in vertebrates [68]. Our finding of DIA1 in arthropod genomes, but its absence in nematode genomes, is not without precedent as only 35% of genes in C. elegans and D. melanogaster are considered orthologous [69]. Therefore, while nematodes have provided insights into many areas of neurobiology, there are also nematode-specific phenomena. A lack of a DIA1 homologue in nematodes may be related to some of the differences in neurobiology found in nematodes compared to other metazoans [70]–[74]. Our overall model is that DIA1 evolved in an ancestor of the cnidarians and, despite being ubiquitously expressed in mammalian tissues [39], [41], [75], gene loss has a marked impact on neurological function in humans, thereby probably causing the symptoms of ASD. Whatever the secretory-pathway role of DIA1 is, it is now defunct or significantly different, in nematodes.
Evolution of the DIA1-related gene DIA1R
We have recently described a ubiquitously-expressed DIA1-related gene in the human genome, DIA1R, where deletion or mutation is linked to ASD-like syndromes and/or X-linked mental retardation [39]. Unlike DIA1, which we found present in the genome of most metazoans, we found DIA1R to be restricted to the subphylum Vertebrata within the phylum Chordata, and DIA1R is absent from urochordate and cephalochordate genomes. Therefore, the emergence of DIA1R coincides with the timing of the known two whole genome duplication (WGD) events, which occurred early in the vertebrate lineage around 500 million years ago [43], [76]–[79]. These WGDs preceded the dramatic rise of vertebrate life that occurred during the Cambrian explosion and coincided with the development of larger brains, a neural crest, and cranial placodes. Indeed, genome duplications often precede species-expansion, although links between genome duplication and increased species diversity remains correlative [80]. Certain genes were preferentially maintained after the vertebrate WGD events including transporters and kinases, with many being involved in signal transduction and development [81]–[83]. This provides circumstantial evidence for the involvement of DIA1-family genes in such processes. Together, these data indicate that DIA1R is a post-2R gene with a role in brain function.
Identification of a cytosolic homologue of DIA1: DIA1L
DIA1-like homologues, termed DIA1L, were detected in the genomes of S. purpuratus (an echinoderm) and B. floridae (a cephalochordate). Our data suggest an origin of DIA1L by DIA1 duplication early in the deuterostome lineage. Other gene families known to have diversified by gene duplication subsequent to the protostome-deuterostome divergence include the myosin light chain family [84], and the ST8Sia family of sialyltransferases [85]. Unlike both DIA1 and DIA1R, DIA1L proteins were unexpectedly found to lack signal peptides, and are therefore predicted to have a cytosolic location and function. There are a number of possible explanations for the restriction of DIA1L to echinoderm and cephalochordate genomes: (i) S. purpuratus and B. floridae are unique in their requirement for a DIA1-family role in both the cytoplasm and secretory pathway, resulting in subsequent loss of the gene in other species; (ii) Other species require cytoplasmic DIA1-family activity and achieve this by currently uncharacterized splicing events; or (iii) The DIA1L gene diversified rapidly after divergence of cephalochordates and is no longer recognizable as a DIA1L homologue in extant chordate genomes. While we currently cannot differentiate between these hypotheses, we favour the latter hypothesis, as the central portion of B. floridae DIA1La shows weak amino acid similarity to an as-yet-uncharacterized human gene product (data not shown).
Intensive gene duplication pre-dating vertebrate DIA1 duplication
While the echinoderm, S. purpuratus, encodes a single DIA1 and a single DIA1L gene, the amphioxus genome encodes a single DIA1 gene, and multiple DIA1L genes. While three of the DIA1L genes in B. floridae are well-documented, the current genome assembly also provides evidence for a further 5 DIA1L genes (although two of these may be allelic). These data provide evidence for a large-scale expansion of DIA1L specific to the amphioxus lineage. The genome of B. floridae was previously considered relatively unduplicated [86], [87], however more recent evidence documents large-scale duplication of genes from certain functional categories in the amphioxus lineage. These include nuclear hormone receptors [88], opsins [42], tyrosine-kinase superfamily genes [89], and receptors and receptor-adaptors of the innate immune system [90]. It has therefore been suggested that the vertebrate WGD events, occurring subsequent to amphioxus divergence, were symptoms of a pre-existing predisposition toward genomic structural change [78]. Our data support the hypothesis that the early vertebrate WGDs were preceded by remarkable gene-family expansion and genome rearrangements.
Duplication and loss of DIA1-family genes in teleost fish
Two closely-related DIA1 paralogues were found in the genome of the ostariophysan fish D. rerio and P. promelas: DIA1a and DIA1b. There are two possible explanations for this finding: (i) origin of the duplicated DIA1 gene during the fish-specific WGD (also known as ‘3R’), with retention in the ostariophysan lineage, but loss of the duplicated in gene in both the paracanthopterygian and acanthopterygian/protanthopterygian fish lineages, or (ii) a DIA1 gene duplication event occurring early after divergence of the ostariophysan lineage, with maintenance of the duplicated gene in this lineage. The two possible evolutionary scenarios are superimposed on a fish-centric phylogenetic tree in Figure 3. There are current precedents for complicated evolutionary pathways for other teleost genes. Both the Elopomorpha, Ostariophysi, Salmoniformes, and Acanthipterygii have lineage-specific duplications of the vitellogenin (Vtg) genes occurring after the 3R WGD event (our unpublished data; [91]). By contrast, two independent losses of the androgen receptor-B (AR-B) gene, subsequent to the 3R WGD, have occurred, one at the base of the Otophysi and another at the base of the Salmoniformes [92]. The precise evolutionary history of the DIA1 paralogues in fish cannot be reconstructed until significantly more fish sequence data become available.
Why ostariophysan fish have retained three DIA1-family genes is unclear. Features specific to ostariophysan fish include: small, horny projections called unculi; a bony Weberian apparatus; the release of a pheromone known as the alarm substance, when frightened; and highly social behaviour [93]. As we propose the DIA1 and DIA1R gene products play a ubiquitous role in the secretory pathway [39], it is possible that DIA1-family homologues play a role in generating these ostariophysan-specific features. Zebrafish are a widely-used model organism for studying vertebrate development in both normal and pathologic conditions [33] and are also being used to study the etiology of neurological disorders including Alzheimer disease [94] and schizophrenia [34], [95]–[97]. Indeed, due to overlapping genes, risk factors and neurological findings, methodology applicable to studies of schizophrenia is also highly relevant to autism [98]–[106]. The ability to manipulate the genome and quantitate effects on behavioural phenotypes, including social skills, makes the zebrafish an attractive model organism to study the etiology of autism [31], [107], [108].
Another unexpected finding was the absence of a DIA1R gene from the genomes of acanthopterygian fish, including the ‘completed’ genomes sequence of pufferfishes T. rubripes (Fugu) and T. nigroviridis, and the medaka O. latipes. It would be unusual to have gaps encompassing the same gene in all three species, and unprecedented to also have the same gene unrepresented in the EST databases of all acanthopterygian fish, unless it is not encoded by those species. It is possible DIA1R is present and functional, but has diverged beyond recognition in acanthopterygian fish. However, there is a precedent for ‘loss’ of other genes during acanthopterygian fish evolution, and therefore we consider this a more likely phenomenon. For example, loss of Hox-family genes, and melanocortin receptor genes have occurred in pufferfish when compared to zebrafish [109], [110]. Indeed, while 5,918 orthologous genes are found between the medaka and pufferfish, only 1,365 are found between medaka and zebrafish [111].
Given the proposed ubiquitous role of DIA1R in secretion, with specific effects on brain function [39], loss of DIA1R in acanthopterygian fish would be expected to relate to structural and/or functional differences in acanthopterygian fish compared to fish from other superorders. Indeed, differences in brain structure and function have been reported between species from the superorder Ostariophysi and Acanthopterygii [112]–[116]. Of possible relevance to the etiology of ASD, the fish lacking DIA1R are considered solitary in nature, while those with DIA1R are considered schooling fish [117].
Evolution of urochordate DIA1
Comparative analyses of the DIA1 gene product of the urochordate C. intestinalis revealed both DIA1- and DIA1R-like characteristics. The unusual phylogenetic placement of C. intestinalis DIA1 is not without precedent. It is well-known that the Ciona lineage is fast-evolving, making topologies unreliable [118]. Indeed, it has been reported that both the sea urchin and amphioxus genomes are more representative of the ancestral deuterostome than that of C. intestinalis, due to its considerable evolutionary changes [42]. In the future, the use of further sequence data from other urochordate species may resolve this issue. The unique characteristics of tunicate DIA1 and its sequence divergence may relate to traits specific to that lineage. For example, tunicates are the only animals capable of producing cellulose, which is a major component of the characteristic ‘tunic’ which is a defining feature of the subphylum Tunicata [119]. Evolutionary divergence of DIA1, and modification of its role within the secretory pathway, may be required for tunic secretion. We are currently investigating the evolution of C. intestinalis DIA1 in more detail.
Conservation within the DIA1 protein family
Amino acid sequence alignments revealed a number of amino acids conserved across the DIA1 protein family. Absolutely conserved amino acids found were: two cysteine residues in the amino-terminal region, a centrally-located glycine residue, and a further cysteine residue in the carboxy-terminal region. Conserved cysteine residues have been implicated in the dimerisation of some proteins [120] and can be essential components of metal- or calcium-ion binding sites [121]–[124]. However, none of the known consensus sequences for such motifs are found in DIA1 or DIA1R. We identified 15 unique motifs characteristic of the DIA1-family, and suggest that hydrophobic motif-1 may be a Golgi-retention motif in DIA1 and DIA1R proteins. While the retention of Golgi-resident proteins with TM domains is well-studied [125], retention of fully-luminal Golgi proteins is sparse. However, an amino-terminal leucine-rich region is essential for Golgi retention of the NEFA/NUC family of Ca2+-binding EF-hand/leucine zipper proteins [126]. None of the remaining DIA1-family motifs have a predicted function in proteins localized to the lumen of the ER or Golgi apparatus.
Arthropod DIA1 proteins were found to differ from those from other species and form their own phylogenetic clade. DIA1-motif 1 was poorly conserved in arthropod DIA1 proteins, while motifs 4 and 14 were absent in DIA1 from Drosophila species. As a result of these differences, most of the Drosophila DIA1 proteins did not have significant similarity to that of the divergent C. intestinalis DIA1 by BLAST, although there was sufficient similarity to have significant matches to DIA1 proteins from all other species. By contrast, DIA1 from the wasp species Nasonia vitripennis was the most similar to non-arthropod DIA1 proteins. This finding was not unexpected, as around 25% of Nasonia genes are more similar to human genes than to their Drosophila counterparts, reflecting the derived nature of many Drosophila genes [127]. Overall, amino acid similarity was greatest in central part of the DIA1 family sequences, compared to the amino- and carboxy-terminal portions, indicating the central part of DIA1 contains the core functional domain of this protein.
Conclusion
We have demonstrated that DIA1 and DIA1R, but not DIA1L, encode signal peptides for targeting to the secretory pathway. Together, with the finding that DIA1 or DIA1R mutations occur in patients with ASD and/or mental retardation [38], [39], a role for DIA1 and DIA1R within the secretory pathway of cells is suggested as causative. While we propose that both DIA1 and DIA1R encode a hydrophobic amino-terminal Golgi-retention motif, in the absence of known functional protein motifs and domains, ongoing studies on human DIA1 and DIA1R are required to determine the exact role(s) of these genes in cellular function, particularly the effects on cognitive function.
Materials and Methods
Detection of DIA1-family homologues
The human DIA1 amino acid sequence [38], [48] or human DIA1R amino acid sequence [39] were retrieved from the National Center for Biotechnology Information (NCBI) Entrez Protein database, and used in BLAST and keyword searches (search terms: c3orf58 or cXorf36) of the NR protein database at the NCBI [46], [128], [129]. EST and genomic databases at the NCBI were searched using the TBLASTN algorithm [46]. Additional BLAST and keyword searches were carried out on: the Ensembl database, including preliminary genome assemblies [130], [131]; the Joint Genome Institute (JGI) database [132], including the placozoan species Trichoplax adhaerens [133]; and SilkDB, the silkworm sequence database [134]. Databases were last searched in August 2009, with the exception of B. floridae, where information was updated using assembly data from October 2009.
Protein alignments and analyses
Protein sequence alignments were generated with CLUSTALW (version 1.8) [47] at NPS@ [135] with manual alignment of some positions. Boxshade (version 3.21), available at EMBnet [136]–[137], was used to format some amino acid alignments. The ExPASy Compute pI/MW tool was used to calculate theoretical molecular weights and isoelectric points [138]. Three trans-membrane prediction methods were used to analyze protein sequences: TMpred [139], TMAP [140], and HMMTOP version 2.0 [141]. Signal peptides were evaluated using SignalP version 3.0 [51], [142] or the SigCleave algorithm [52], which is part of the EMBOSS software suite [143]. Amino acid motifs and domains were investigated using the following resources: MOTIF at GenomeNet [144]; PSORT-II [145]; the Conserved Domain Database at the NCBI, which also contains data from Pfam, SMART and COG [146]; and the ELM resource [147].
Phylogeny
Gblocks version 0.91b was used to eliminate poorly aligned positions and divergent regions in aligned protein sequences [148], [149]. The evolutionary history of the DIA1 family was inferred using the neighbour-joining method [57] in MEGA4 [150], or using the PhyML maximum-likelihood algorithm [151]–[152] or Bayesian inference [153] via the ‘Phylogeny.fr’ web-server [154]. Statistical reliability of branching was assessed using either bootstrap replicates [155] or approximate likelihood ratio testing [156].
Supporting Information
Taxa, accession numbers, and chromosome location of DIA1 orthologues.
(0.05 MB PDF)
Taxa, accession numbers and chromosome location of partial DIA1 orthologues.
(0.04 MB PDF)
Physical characteristics of all available full-length DIA1 proteins and their similarity to orthologues from key species.
(0.04 MB PDF)
Pairwise comparison of DIA1 and DIA1R proteins.
(0.02 MB PDF)
Taxa, accession numbers and chromosome location of full-length DIA1R orthologues.
(0.03 MB PDF)
Taxa, accession numbers and chromosome location of partial DIA1R orthologues.
(0.03 MB PDF)
Comparison of Ciona intestinalis DIA1 to DIA1 and DIA1R from other species.
(0.02 MB PDF)
Taxa and accession numbers of full-length DIA1L genes.
(0.03 MB PDF)
Amino acid comparisons between DIA1L and DIA1 proteins.
(0.02 MB PDF)
Taxa, accession numbers and chromosome location of partial DIA1L paralogues.
(0.03 MB PDF)
Amino acid sequence comparison of DIA1a and DIA1b from zebrafish. The sequence alignment was generated using CLUSTALW [47]. Identical amino acids are highlighted in red font and indicated below the alignment with an asterisk (*). Strongly similar amino acids are highlighted in green font and indicated below the alignment with a colon (:). Weakly similar amino acids are highlighted in blue font and indicated below the alignment with a full stop (.). Dissimilar amino acids are in black font. Amino acid numbering is provided above the alignment. Gaps are indicated by dashes. The alignment shows 88% identical amino acids, and a further 10% similar amino acids, providing an overall similarity of 98%. Standard single-letter amino acid abbreviations are used. Organism abbreviation uses the first letter of the genus name, followed by the first four letters of the species (i.e., Danio rerio DIA1a is abbreviated to DreriDIA1a). Accession numbers can be found in Table S1.
(0.01 MB PDF)
Amino acid sequence comparison of DIA1 proteins. The sequence alignment of all full-length DIA1 proteins was generated using CLUSTALW [47]. A consensus amino acid sequence is presented below the alignment, where uppercase letters indicate absolutely conserved amino acids. Regions of greater than 50% conservation are shaded (identical amino acids are in black boxes and similar amino acids in grey boxes). Amino acid numbering is provided on the left-hand side of the alignment. Gaps required for optimal alignment are indicated by dashes. The alignment reveals that 10 amino acids are absolutely conserved (2% identity), with a further 6% amino acids being similar, providing an overall similarity of 8%. Standard single-letter amino acid abbreviations are used. Organism abbreviation uses the first letter of the genus name, followed by the first four letters of the species (e.g., Strongylocentrotus purpuratus DIA1 is abbreviated to SpurpDIA1). Accession numbers and full species names can be found in Table S1.
(0.08 MB PDF)
Amino acid sequence comparison of DIA1R proteins. The sequence alignment was generated using CLUSTALW [47]. Identical amino acids are highlighted in red font and indicated below the alignment with a red asterisk (*). Strongly similar amino acids are highlighted in green font and indicated below the alignment with a green colon (:). Weakly similar amino acids are highlighted in blue font and indicated below the alignment with a blue full stop (.). Dissimilar amino acids are in black font. Amino acid numbering is provided above the alignment. Gaps are indicated by dashes. The alignment shows 124 identical amino acids (27% identity), with a further 27% similar amino acids, providing an overall similarity of 54%. Standard single-letter amino acid abbreviations are used. A consensus line is provided below the alignment where the number “2” is inserted when no consensus amino acid is found. Organism abbreviations use the first letter of the genus name, followed by the first four letters of the species (e.g., Monodelphis domestica DIA1R is abbreviated to MdomeDIA1R). Accession numbers and full species names can be found in Table S5.
(0.03 MB PDF)
Amino acid sequence alignment of DIA1 and DIA1R proteins. The sequence alignment of all full-length DIA1 and DIA1R proteins was generated using CLUSTALW [47]. A consensus amino acid sequence is presented below the alignment, where uppercase letters indicate absolutely conserved amino acids. Regions of greater than 50% conservation are shaded (identical amino acids are in black boxes and similar amino acids in grey boxes) and 56% of aligned positions showed conservation in >50% of sequences. Only 8 amino acids are absolutely conserved (∼2% identity), with a further 23 amino acids being similar (∼5%) in all DIA1 and DIA1R proteins. Amino acid numbering is provided on the left-hand side of the alignment. Gaps required for optimal alignment are indicated by dashes. Standard single-letter amino acid abbreviations are used. Organism abbreviations use the first letter of the genus name, followed by the first four letters of the species (e.g., Gallus gallus DIA1R is abbreviated to GgallDIA1R). Accession numbers and full species names can be found in Tables S1 and S5.
(0.13 MB PDF)
Signal peptide localization in DIA1 and DIA1R proteins. The sequence alignment and consensus sequence of the amino terminal region of all full-length DIA1 and DIA1R proteins is from Figure S4. Abbreviations are as in Figure S4. DIA1R proteins were grouped together in the top portion of the figure, with aligned DIA1 proteins below, where arthropod sequences are placed above, nonvertebrate/nonarthropod sequences below, and vertebrate DIA1 sequences in the middle (see annotation on right-hand side of alignment). Initiation methiones were not manually aligned. Bold red amino acids represent the last amino acid of the amino-terminal signal peptide predicted using the NN algorithm [51]. Bold blue amino acids represent the last amino acid of the signal peptide predicted using the HMM prediction method [51]. Bold purple amino acids represent the last amino acid of the signal peptide predicted by both NN and HMM prediction methods. Bold underlined amino acids represent the last amino acid of the signal peptide predicted by the Sigcleave algorithm [52]. Lack of an underlined residue indicates no signal peptide cleavage site was predicted within the aligned region by Sigcleave, and lack of a red residue (or purple) indicates no signal peptide cleavage site predicted by the NN algorithm. The most commonly predicted site for cleavage of DIA1R signal peptides or vertebrate and arthropod DIA1 signal peptides are indicated with arrows above the alignment.
(0.03 MB PDF)
Amino acid sequence comparison of DIA1L proteins. A single DIA1L protein from S. purpuratus (SpurpDIA1L) and three DIA1L paralogues from B. floridae (BflorDIA1La, BflorDIA1Lb, BflorDIA1Lc) were aligned using CLUSTALW [47]. Identical amino acids shared between all four proteins are highlighted in red font and indicated below the alignment with a red asterisk (*). Strongly similar amino acids are highlighted in green font and indicated below the alignment with a green colon (:). Weakly similar amino acids are highlighted in blue font and indicated below the alignment with a blue full stop (.). Dissimilar amino acids are in black font. Amino acid numbering is provided above the alignment. Gaps are indicated by dashes. The alignment shows 7% identical amino acids, with a further 20% similar amino acids, providing an overall similarity of 27%. Amino acid similarity and identity shared at the amino-terminal ends of the three longer DIA1L proteins are indicated using the coloured fonts described above, but the corresponding annotations (*, : and .) are in grey font below the alignment. Standard single-letter amino acid abbreviations are used. Accession numbers can be found in Table S8.
(0.02 MB PDF)
Amino acid sequence comparison of DIA1-family proteins. The sequence alignment of all full-length DIA1, DIA1R and DIA1L proteins was generated using CLUSTALW [47]. Identical amino acids are highlighted in red font and indicated below the alignment with a red asterisk (*). Strongly similar amino acids are highlighted in green font and indicated below the alignment with a green colon (:). Weakly similar amino acids are highlighted in blue font and indicated below the alignment with a blue full stop (.). Dissimilar amino acids are in black font. Amino acid numbering is provided above the alignment. Gaps are indicated by dashes. The alignment shows only 4 identical amino acids and 18 similar amino acids conserved across the entire protein family (3% similarity). Standard single-letter amino acid abbreviations are used. Organism abbreviations use the first letter of the genus name, followed by the first four letters of the species (e.g., Pteropus vampyrus DIA1 is abbreviated to PvampDIA1). Accession numbers and full species names can be found in Tables S1, S5 and S8.
(0.05 MB PDF)
Amino acids conserved in at least 80% of DIA1-family members. The sequence alignment of all full-length DIA1, DIA1R and DIA1L proteins was generated using CLUSTALW [47]. A consensus amino acid sequence is presented below the alignment, where uppercase letters indicate absolutely conserved amino acids. Regions of greater than 80% conservation are shaded (identical amino acids are in black boxes and similar amino acids in grey boxes), and 10% of aligned positions showed conservation in >80% of sequences. Amino acid numbering is provided on the left-hand side of the alignment. Gaps required for optimal alignment are indicated by dashes. Standard single-letter amino acid abbreviations are used. Organism abbreviations use the first letter of the genus name, followed by the first four letters of the species (e.g., Salmo salar DIA1R is abbreviated to SsalaDIA1R). Accession numbers and full species names can be found in Tables S1, S5 and S8.
(0.10 MB PDF)
Amino acids conserved in at least 50% of DIA1-family members. The sequence alignment of all full-length DIA1, DIA1R and DIA1L proteins was generated using CLUSTALW [47]. A consensus amino acid sequence is presented below the alignment, where uppercase letters indicate absolutely conserved amino acids. Regions of greater than 50% conservation are shaded (identical amino acids are in black boxes and similar amino acids in grey boxes), and 38% of aligned positions showed conservation in >50% of sequences. Amino acid numbering is provided on the left-hand side of the alignment. Gaps required for optimal alignment are indicated by dashes. Standard single letter amino acid abbreviations are used. Organism abbreviations use the first letter of the genus name, followed by the first four letters of the species (e.g., Tursiops truncatus DIA1 is abbreviated to TtrunDIA1R). Accession numbers and full species names can be found in Tables S1, S5 and S8.
(0.14 MB PDF)
DIA1-family amino acid sequences in FASTA format. Each DIA1-family amino acid sequence starts with a “>” (greater-than) symbol followed by the species abbreviation and protein type (e.g., Oryzias latipes DIA1 is abbreviated to OlatiDIA1 and Pongo pygmaeus DIA1R to PpygmDIA1R). Following the initial title line is the actual amino acid sequence, in standard single-letter code. Accession numbers, full species names, and differences to current database sequence data (due to corrections) can be found in Tables S1, S5 and S8.
(0.04 MB PDF)
Maximum-likelihood tree of DIA1-family proteins. Proteins encoded by each full-length DIA1-family gene were aligned using CLUSTALW [47] and subjected to maximum-likelihood analysis [151] using PhyML phylogeny software [135]. Approximate likelihood-ratio test for branch-support statistics [156] was carried out, and percentage values are shown next to branches. Branch lengths are proportional to the number of amino acid substitutions per site (see scale bar). G-blocks were used to eliminate poorly aligned positions and divergent regions, since they may not be homologous or may have been saturated by multiple substitutions [148]. The tree was rooted on the cnidarian N. vectensis DIA1 sequence (NvectDIA1), as highlighted with an asterisk. Organism abbreviations use the first letter of the genus name, followed by the first four letters of the species. Full species names and accession numbers can be found in Tables S1, S4 and S7.
(0.12 MB PDF)
Phylogeny of the DIA1-family reconstructed using a Bayesian phylogenetic approach. A subset of DIA1-family gene products were aligned using CLUSTALW [47] and subjected to Bayesian inference of phylogeny using the MrBayes programme [153]. The number above each branch refers to the Bayesian posterior probability of the node, given as a percentage (e.g., 77 represents a posterior probability of 0.77). Branch lengths are proportional to the number of amino acid substitutions per site (see scale bar). Gblocks were used to curate the alignment [148]. The tree was rooted on the cnidarian N. vectensis DIA1 sequence (NvectDIA1), as highlighted with an asterisk. Organism abbreviations use the first letter of the genus name, followed by the first four letters of the species. Full species names and accession numbers can be found in Tables S1, S4 and S7.
(0.07 MB PDF)
Acknowledgments
We thank Paul Fisher and Sandra Accari for helpful discussions on evolutionary biology and the use of model organisms for studying neurological disorders, and researchers at the Olga Tennison Autism Research Centre and the Swinburne Autism Bio-Research Initiative for lively discussions on the etiology of autism spectrum disorders. Our research relies heavily on the large amounts of sequence data, and gene and protein analysis software, made available by large research centres for the use of other research scientists. We also thank the three reviewers of this paper for their helpful comments, which led to an improved final manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: NEB and SPH were supported by La Trobe University and AA by La Trobe University, the Malaysian Ministry of Higher Education, and the Islamic Science University of Malaysia. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, et al. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med. 1995;25:63–77. doi: 10.1017/s0033291700028099. [DOI] [PubMed] [Google Scholar]
- 2.Folstein SE, Rosen-Sheidley B. Genetics of autism: complex aetiology for a heterogeneous disorder. Nat Rev Genet. 2001;2:943–955. doi: 10.1038/35103559. [DOI] [PubMed] [Google Scholar]
- 3.Veenstra-VanderWeele J, Cook EH., Jr Molecular genetics of autism spectrum disorder. Mol. Psychiatry. 2004;9:819–832. doi: 10.1038/sj.mp.4001505. [DOI] [PubMed] [Google Scholar]
- 4.Rutter M. Genetic studies of autism: from the 1970s into the millennium. J Abnorm Child Psychol. 2000;28:3–14. doi: 10.1023/a:1005113900068. [DOI] [PubMed] [Google Scholar]
- 5.Ronald A, Happé F, Bolton P, Butcher LM, Price TS, et al. Genetic heterogeneity between the three components of the autism spectrum: a twin study. J Am Acad Child Adolesc Psychiatry. 2006;45:691–699. doi: 10.1097/01.chi.0000215325.13058.9d. [DOI] [PubMed] [Google Scholar]
- 6.Freitag CM. The genetics of autistic disorders and its clinical relevance: a review of the literature. Mol Psychiatry. 2007;12:2–22. doi: 10.1038/sj.mp.4001896. [DOI] [PubMed] [Google Scholar]
- 7.Losh M, Sullivan PF, Trembath D, Piven J. Current developments in the genetics of autism: from phenome to genome. J Neuropathol Exp Neurol. 2008;67:829–837. doi: 10.1097/NEN.0b013e318184482d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taniai H, Nishiyama T, Miyachi T, Imaeda M, Sumi S. Genetic influences on the broad spectrum of autism: study of proband-ascertained twins. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:844–849. doi: 10.1002/ajmg.b.30740. [DOI] [PubMed] [Google Scholar]
- 9.Lichtenstein P, Carlström E, Råstam M, Gillberg C, Anckarsäter H. The genetics of autism spectrum disorders and related neuropsychiatric disorders in childhood. Am J Psychiatry. 2010;167:1357–1363. doi: 10.1176/appi.ajp.2010.10020223. [DOI] [PubMed] [Google Scholar]
- 10.Zhao X, Leotta A, Kustanovich V, Lajonchere C, Geschwind DH, et al. A unified genetic theory for sporadic and inherited autism. Proc Natl Acad Sci U S A. 2007;104:12831–12836. doi: 10.1073/pnas.0705803104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jamain S, Betancur C, Quach H, Philippe A, Fellous M, et al. Linkage and association of the glutamate receptor 6 gene with autism. Mol Psychiatry. 2002;7:302–310. doi: 10.1038/sj.mp.4000979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jamain S, Quach H, Betancur C, Råstam M, Colineaux C, et al. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma DQ, Whitehead PL, Menold MM, Martin ER, Ashley-Koch AE, et al. Identification of significant association and gene-gene interaction of GABA receptor subunit genes in autism. Am J Hum Genet. 2005;77:377–388. doi: 10.1086/433195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng J, Schroer R, Yan J, Song W, Yang C, et al. High frequency of neurexin 1beta signal peptide structural variants in patients with autism. Neurosci Lett. 2006;409:10–13. doi: 10.1016/j.neulet.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 17.Talebizadeh Z, Lam DY, Theodoro MF, Bittel DC, Lushington GH, et al. Novel splice isoforms for NLGN3 and NLGN4 with possible implications in autism. J Med Genet. 2006;43:e21. doi: 10.1136/jmg.2005.036897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sadakata T, Washida M, Iwayama Y, Shoji S, Sato Y, et al. Autistic-like phenotypes in Cadps2-knockout mice and aberrant CADPS2 splicing in autistic patients. J Clin Invest. 2007;117:931–943. doi: 10.1172/JCI29031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chapleau CA, Larimore JL, Theibert A, Pozzo-Miller L. Modulation of dendritic spine development and plasticity by BDNF and vesicular trafficking: fundamental roles in neurodevelopmental disorders associated with mental retardation and autism. J Neurodev Disord. 2009;1:185–196. doi: 10.1007/s11689-009-9027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sadakata T, Furuichi T. Developmentally regulated Ca2+-dependent activator protein for secretion 2 (CAPS2) is involved in BDNF secretion and is associated with autism susceptibility. Cerebellum. 2009;8:312–22. doi: 10.1007/s12311-009-0097-5. [DOI] [PubMed] [Google Scholar]
- 23.Castermans D, Volders K, Crepel A, Backx L, De Vos R, et al. SCAMP5, NBEA and AMISYN: three candidate genes for autism involved in secretion of large dense-core vesicles. Hum Mol Genet. 2010;19:1368–1378. doi: 10.1093/hmg/ddq013. [DOI] [PubMed] [Google Scholar]
- 24.Giannandrea M, Bianchi V, Mignogna ML, Sirri A, Carrabino S, et al. Mutations in the small GTPase gene RAB39B are responsible for X-linked mental retardation associated with autism, epilepsy, and macrocephaly. Am J Hum Genet. 2010;86:185–195. doi: 10.1016/j.ajhg.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waring R. Sulphation and autism – what are the links? 2001. Available: http://www.autismfile.com/papers/Rosemary_Waring_Sulphation.asp. Accessed April 2010.
- 26.Waring R, Phoenix J. TPST-assay for diagnosis of autism and related disorders. SHS International Ltd: International patent WO/2001/077681 2001 [Google Scholar]
- 27.Castagnola M, Messana I, Inzitari R, Fanali C, Cabras T, et al. Hypo-phosphorylation of salivary peptidome as a clue to the molecular pathogenesis of autism spectrum disorders. J Proteome Res. 2008;7:5327–5332. doi: 10.1021/pr8004088. [DOI] [PubMed] [Google Scholar]
- 28.van der Zwaag B, Franke L, Poot M, Hochstenbach R, Spierenburg HA, et al. Gene-network analysis identifies susceptibility genes related to glycobiology in autism. PLoS One. 2009;4:e5324. doi: 10.1371/journal.pone.0005324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar RA, Christian SL. Genetics of autism spectrum disorders. Curr Neurol Neurosci Rep. 2009;9:188–197. doi: 10.1007/s11910-009-0029-2. [DOI] [PubMed] [Google Scholar]
- 30.Moy SS, Nadler JJ. Advances in behavioral genetics: mouse models of autism. Mol Psychiatry. 2008;13:4–26. doi: 10.1038/sj.mp.4002082. [DOI] [PubMed] [Google Scholar]
- 31.Tropepe V, Sive HL. Can zebrafish be used as a model to study the neurodevelopmental causes of autism? Genes Brain Behav. 2003;2:268–281. doi: 10.1034/j.1601-183x.2003.00038.x. [DOI] [PubMed] [Google Scholar]
- 32.Best JD, Alderton WK. Zebrafish: An in vivo model for the study of neurological diseases. Neuropsychiatr Dis Treat. 2008;4:567–576. doi: 10.2147/ndt.s2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Veldman MB, Lin S. Zebrafish as a developmental model organism for pediatric research. Pediatr Res. 2008;64:470–476. doi: 10.1203/PDR.0b013e318186e609. [DOI] [PubMed] [Google Scholar]
- 34.Mathur P, Guo S. Use of zebrafish as a model to understand mechanisms of addiction and complex neurobehavioral phenotypes. Neurobiol Dis. 2010;40:66–72. doi: 10.1016/j.nbd.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lloyd TE, Verstreken P, Ostrin EJ, Phillippi A, Lichtarge O, Bellen HJ. A genome-wide search for synaptic vesicle cycle proteins in Drosophila. Neuron. 2000;26:45–50. doi: 10.1016/s0896-6273(00)81136-8. [DOI] [PubMed] [Google Scholar]
- 36.Yoshihara M, Ensminger AW, Littleton JT. Neurobiology and the Drosophila genome. Funct Integr Genomics. 2001;1:235–240. doi: 10.1007/s101420000029. [DOI] [PubMed] [Google Scholar]
- 37.McBride SM, Choi CH, Wang Y, Liebelt D, Braunstein E, et al. Pharmacological rescue of synaptic plasticity, courtship behavior, and mushroom body defects in a Drosophila model of fragile X syndrome. Neuron. 2005;45:753–764. doi: 10.1016/j.neuron.2005.01.038. [DOI] [PubMed] [Google Scholar]
- 38.Morrow EM, Yoo SY, Flavell SW, Kim TK, Lin Y, et al. Identifying autism loci and genes by tracing recent shared ancestry. Science. 2008;321:218–223. doi: 10.1126/science.1157657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aziz A, Harrop SP, Bishop NE. DIA1R is an X-linked gene related to Deleted In Autism-1. PLoS One. In press. 2010 doi: 10.1371/journal.pone.0014534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller G. Origins. On the origin of the nervous system. Science. 2009;325:24–26. doi: 10.1126/science.325_24. [DOI] [PubMed] [Google Scholar]
- 41.Takatalo M, Järvinen E, Laitinen S, Thesleff I, Rönnholm R. Expression of the novel Golgi protein GoPro49 is developmentally regulated during mesenchymal differentiation. Dev Dyn. 2008;237:2243–2255. doi: 10.1002/dvdy.21646. [DOI] [PubMed] [Google Scholar]
- 42.Holland LZ, Albalat R, Azumi K, Benito-Gutiérrez E, Blow MJ, et al. The amphioxus genome illuminates vertebrate origins and cephalochordate biology. Genome Res. 2008;18:1100–1111. doi: 10.1101/gr.073676.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dehal P, Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005;3:e314. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van de Peer Y, Taylor JS, Braasch I, Meyer A. The ghost of selection past: rates of evolution and functional divergence of anciently duplicated genes. J Mol Evol. 2001;53:436–446. doi: 10.1007/s002390010233. [DOI] [PubMed] [Google Scholar]
- 45.Zhang J. Evolution by gene duplication: an update. Trends Ecol Evol. 2003;18:292–298. [Google Scholar]
- 46.Altshul SF, Madden, TL, Schaffer AA, Zhang J, Zhang Z, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1990;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takatalo MS, Kouvonen P, Corthals G, Nyman TA, Rönnholm RH. Identification of new Golgi complex specific proteins by direct organelle proteomic analysis. Proteomics. 2006;6:3502–3508. doi: 10.1002/pmic.200500516. [DOI] [PubMed] [Google Scholar]
- 49.Menne KM, Hermjakob H, Apweiler R. A comparison of signal sequence prediction methods using a test set of signal peptides. Bioinformatics. 2000;16:741–742. doi: 10.1093/bioinformatics/16.8.741. [DOI] [PubMed] [Google Scholar]
- 50.Klee EW, Ellis LB. Evaluating eukaryotic secreted protein prediction. BMC Bioinformatics. 2005;6:256. doi: 10.1186/1471-2105-6-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 52.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marques AC, Vinckenbosch N, Brawand D, Kaessmann H. Functional diversification of duplicate genes through subcellular adaptation of encoded proteins. Genome Biol. 2008;9:R54. doi: 10.1186/gb-2008-9-3-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rosso L, Marques AC, Weier M, Lambert N, Lambot MA, et al. Birth and rapid subcellular adaptation of a hominoid-specific CDC14 protein. PLoS Biol. 2008;6:e140. doi: 10.1371/journal.pbio.0060140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qian W, Zhang J. Protein subcellular relocalization in the evolution of yeast singleton and duplicate genes. Genome Biol Evol. 2009;2009:198–204. doi: 10.1093/gbe/evp021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Conant GC, Wagner A. Asymmetric sequence divergence of duplicate genes. Genome Res. 2003;13:2052–2058. doi: 10.1101/gr.1252603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 58.Galliot B, Quiquand M, Ghila L, de Rosa R, Miljkovic-Licina M, et al. Origins of neurogenesis, a cnidarian view. Dev Biol. 2009;332:2–24. doi: 10.1016/j.ydbio.2009.05.563. [DOI] [PubMed] [Google Scholar]
- 59.Grimmelikhuijzen CJ, Westfall JA. The nervous systems of cnidarians. EXS. 1995;72:7–24. doi: 10.1007/978-3-0348-9219-3_2. [DOI] [PubMed] [Google Scholar]
- 60.Sea Urchin Genome Sequencing Consortium. The genome of the sea urchin Strongylocentrotus purpuratus. Science. 2006;314:941–952. doi: 10.1126/science.1133609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parkinson J, Mitreva M, Whitton C, Thomson M, Daub J, et al. A transcriptomic analysis of the phylum Nematoda. Nat Genet. 2004;36:1259–1267. doi: 10.1038/ng1472. [DOI] [PubMed] [Google Scholar]
- 62.Kortschak RD, Samuel G, Saint R, Miller DJ. EST analysis of the cnidarian Acropora millepora reveals extensive gene loss and rapid sequence divergence in the model invertebrates. Curr Biol. 2003;13:2190–2195. doi: 10.1016/j.cub.2003.11.030. [DOI] [PubMed] [Google Scholar]
- 63.Winnepenninckx B, Backeljau T, De Wachter R. Phylogeny of protostome worms derived from 18S rRNA sequences. Mol Biol Evol. 1995;12:641–649. doi: 10.1093/oxfordjournals.molbev.a040243. [DOI] [PubMed] [Google Scholar]
- 64.Mushegian AR, Garey JR, Martin J, Liu LX. Large-scale taxonomic profiling of eukaryotic model organisms: a comparison of orthologous proteins encoded by the human, fly, nematode, and yeast genomes. Genome Res. 1998;8:590–598. doi: 10.1101/gr.8.6.590. [DOI] [PubMed] [Google Scholar]
- 65.Ledent V, Vervoort M. The basic helix-loop-helix protein family: comparative genomics and phylogenetic analysis. Genome Res. 2001;11:754–770. doi: 10.1101/gr.177001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kiontke K, Gavin NP, Raynes Y, Roehrig C, Piano F, et al. Caenorhabditis phylogeny predicts convergence of hermaphroditism and extensive intron loss. Proc Natl Acad Sci U S A. 2004;101:9003–9008. doi: 10.1073/pnas.0403094101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Coghlan A, Wolfe KH. Fourfold faster rate of genome rearrangement in nematodes than in Drosophila. Genome Res. 2002;12:857–867. doi: 10.1101/gr.172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mitreva M, Blaxter ML, Bird DM, McCarter JP. Comparative genomics of nematodes. Trends Genet. 2005;21:573–581. doi: 10.1016/j.tig.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 69.Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kawano T, Takuwa K, Nakajima T. Structure and activity of a new form of the glutamate transporter of the nematode Caenorhabditis elegans. Biosci Biotechnol Biochem. 1997;61:927–929. doi: 10.1271/bbb.61.927. [DOI] [PubMed] [Google Scholar]
- 71.Bargmann CI. Neurobiology of the Caenorhabditis elegans genome. Science. 1998;282:2028–2033. doi: 10.1126/science.282.5396.2028. [DOI] [PubMed] [Google Scholar]
- 72.Brockie PJ, Mellem JE, Hills T, Madsen DM, Maricq AV. The C. elegans glutamate receptor subunit NMR-1 is required for slow NMDA-activated currents that regulate reversal frequency during locomotion. Neuron. 2001;31:617–630. doi: 10.1016/s0896-6273(01)00394-4. [DOI] [PubMed] [Google Scholar]
- 73.Brockie PJ, Madsen DM, Zheng Y, Mellem J, Maricq AV. Differential expression of glutamate receptor subunits in the nervous system of Caenorhabditis elegans and their regulation by the homeodomain protein UNC-42. J Neurosci. 2001;21:1510–1522. doi: 10.1523/JNEUROSCI.21-05-01510.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li C. The ever-expanding neuropeptide gene families in the nematode Caenorhabditis elegans. Parasitology. 2005;131:S109–127. doi: 10.1017/S0031182005009376. [DOI] [PubMed] [Google Scholar]
- 75.Takatalo MS, Tummers M, Thesleff I, Rönnholm R. Novel Golgi protein, GoPro49, is a specific dental follicle marker. J Dent Res. 2009;88:534–538. doi: 10.1177/0022034509338452. [DOI] [PubMed] [Google Scholar]
- 76.Blomme T, Vandepoele K, De Bodt S, Simillion C, Maere S, et al. The gain and loss of genes during 600 million years of vertebrate evolution. Genome Biol. 2006;7:R43. doi: 10.1186/gb-2006-7-5-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kasahara M. The 2R hypothesis: an update. Curr Opin Immunol. 2007;19:547–552. doi: 10.1016/j.coi.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 78.Hufton AL, Groth D, Vingron M, Lehrach H, Poustka AJ, et al. Early vertebrate whole genome duplications were predated by a period of intense genome rearrangement. Genome Res. 2008;18:1582–1591. doi: 10.1101/gr.080119.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Putnam NH, Butts T, Ferrier DE, Furlong RF, Hellsten U, et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- 80.Van de Peer Y, Maere S, Meyer A. The evolutionary significance of ancient genome duplications. Nat Rev Genet. 2009;10:725–732. doi: 10.1038/nrg2600. [DOI] [PubMed] [Google Scholar]
- 81.Taylor JS, Raes J. Duplication and divergence: the evolution of new genes and old ideas. Annu Rev Genet. 2004;38:615–643. doi: 10.1146/annurev.genet.38.072902.092831. [DOI] [PubMed] [Google Scholar]
- 82.Holland LZ, Short S. Gene duplication, co-option and recruitment during the origin of the vertebrate brain from the invertebrate chordate brain. Brain Behav Evol. 2008;72:91–105. doi: 10.1159/000151470. [DOI] [PubMed] [Google Scholar]
- 83.Conant GC, Wolfe KH. Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet. 2008;9:938–950. doi: 10.1038/nrg2482. [DOI] [PubMed] [Google Scholar]
- 84.Oota S, Saitou N. Phylogenetic relationship of muscle tissues deduced from superimposition of gene trees. Biol Evol. 1999;16:856–867. doi: 10.1093/oxfordjournals.molbev.a026170. [DOI] [PubMed] [Google Scholar]
- 85.Harduin-Lepers A, Petit D, Mollicone R, Delannoy P, Petit JM, et al. Evolutionary history of the alpha 2,8-sialyltransferase (ST8Sia) gene family: tandem duplications in early deuterostomes explain most of the diversity found in the vertebrate ST8Sia genes. BMC Evol Biol. 2008;8:258. doi: 10.1186/1471-2148-8-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Holland LZ, Laudet V, Schubert M. The chordate amphioxus: an emerging model organism for developmental biology. Cell Mol Life Sci. 2004;61:2290–2308. doi: 10.1007/s00018-004-4075-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Permanyer J, Albalat R, Gonzàlez-Duarte R. Getting closer to a pre-vertebrate genome: the non-LTR retrotransposons of Branchiostoma floridae. Int J Biol Sci. 2006;2:48–53. doi: 10.7150/ijbs.2.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schubert M, Brunet F, Paris M, Bertrand S, Benoit G, et al. Nuclear hormone receptor signaling in amphioxus. Dev Genes Evol. 2008;218:651–665. doi: 10.1007/s00427-008-0251-y. [DOI] [PubMed] [Google Scholar]
- 89.D'Aniello S, Irimia M, Maeso I, Pascual-Anaya J, Jiménez-Delgado S, et al. Gene expansion and retention leads to a diverse tyrosine kinase superfamily in amphioxus. Mol Biol Evol. 2008;25:1841–1854. doi: 10.1093/molbev/msn132. [DOI] [PubMed] [Google Scholar]
- 90.Huang S, Yuan S, Guo L, Yu Y, Li J, et al. Genomic analysis of the immune gene repertoire of amphioxus reveals extraordinary innate complexity and diversity. Genome Res. 2008;18:1112–1126. doi: 10.1101/gr.069674.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Finn RN, Kristoffersen BA. Vertebrate vitellogenin gene duplication in relation to the “3R hypothesis”: correlation to the pelagic egg and the oceanic radiation of teleosts. PLoS One. 2007;2:e169. doi: 10.1371/journal.pone.0000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Douard V, Brunet F, Boussau B, Ahrens-Fath I, Vlaeminck-Guillem V, et al. The fate of the duplicated androgen receptor in fishes: a late neofunctionalization event? BMC Evol Biol. 2008;8:336. doi: 10.1186/1471-2148-8-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nelson JS. New York: John Wiley and Sons; 2006. Fishes of the world, 4th edition. [Google Scholar]
- 94.Newman M, Musgrave IF, Lardelli M. Alzheimer disease: amyloidogenesis, the presenilins and animal models. Biochim Biophys Acta. 2007;1772:285–297. doi: 10.1016/j.bbadis.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 95.Morris JA. Zebrafish: a model system to examine the neurodevelopmental basis of schizophrenia. Prog Brain Res. 2009;179:97–106. doi: 10.1016/S0079-6123(09)17911-6. [DOI] [PubMed] [Google Scholar]
- 96.Sawa A. Genetic animal models for schizophrenia: advantages and limitations of genetic manipulation in drosophila, zebrafish, rodents, and primates. Prog Brain Res. 2009;179:3–6. doi: 10.1016/S0079-6123(09)17901-3. [DOI] [PubMed] [Google Scholar]
- 97.Wood JD, Bonath F, Kumar S, Ross CA, Cunliffe VT. Disrupted-in-schizophrenia 1 and neuregulin 1 are required for the specification of oligodendrocytes and neurones in the zebrafish brain. Hum Mol Genet. 2009;18:391–404. doi: 10.1093/hmg/ddn361. [DOI] [PubMed] [Google Scholar]
- 98.Tordjman S, Drapier D, Bonnot O, Graignic R, Fortes S, et al. Animal models relevant to schizophrenia and autism: validity and limitations. Behav Genet. 2007;37:61–78. doi: 10.1007/s10519-006-9120-5. [DOI] [PubMed] [Google Scholar]
- 99.Flint J, Shifman S. Animal models of psychiatric disease. Curr Opin Genet Dev. 2008;18:235–240. doi: 10.1016/j.gde.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 100.Laviola G, Ognibene E, Romano E, Adriani W, Keller F. Gene-environment interaction during early development in the heterozygous reeler mouse: clues for modelling of major neurobehavioral syndromes. Neurosci Biobehav Rev. 2009;33:560–572. doi: 10.1016/j.neubiorev.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 101.Rapoport J, Chavez A, Greenstein D, Addington A, Gogtay N. Autism spectrum disorders and childhood-onset schizophrenia: clinical and biological contributions to a relation revisited. J Am Acad Child Adolesc Psychiatry. 2009;48:10–18. doi: 10.1097/CHI.0b013e31818b1c63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Burbach JP, van der Zwaag B. Contact in the genetics of autism and schizophrenia. Trends Neurosci. 2009;32:69–72. doi: 10.1016/j.tins.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 103.Sebat J, Levy DL, McCarthy SE. Rare structural variants in schizophrenia: one disorder, multiple mutations; one mutation, multiple disorders. Trends Genet. 2009;25:528–535. doi: 10.1016/j.tig.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Weiss LA. Autism genetics: emerging data from genome-wide copy-number and single nucleotide polymorphism scans. Expert Rev Mol Diagn. 2009;9:795–803. doi: 10.1586/erm.09.59. [DOI] [PubMed] [Google Scholar]
- 105.Cheung C, Yu K, Fung G, Leung M, Wong C, et al. Autistic disorders and schizophrenia: related or remote? An anatomical likelihood estimation. PLoS One. 2010;5:e12233. doi: 10.1371/journal.pone.0012233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Crespi B, Stead P, Elliot M. Evolution in health and medicine Sackler colloquium: Comparative genomics of autism and schizophrenia. Proc Natl Acad Sci U S A. 2010;107(Suppl 1):1736–1741. doi: 10.1073/pnas.0906080106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Elsen GE, Choi LY, Prince VE, Ho RK. The autism susceptibility gene met regulates zebrafish cerebellar development and facial motor neuron migration. Dev Biol. 2009;335:78–92. doi: 10.1016/j.ydbio.2009.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kabashi E, Brustein E, Champagne N, Drapeau P. Zebrafish models for the functional genomics of neurogenetic disorders. Biochim Biophys Acta. In press. 2010 doi: 10.1016/j.bbadis.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 109.Kurosawa G, Yamada K, Ishiguro H, Hori H. Hox gene complexity in medaka fish may be similar to that in pufferfish rather than zebrafish. Biochem Biophys Res Commun. 1999;260:66–70. doi: 10.1006/bbrc.1999.0834. [DOI] [PubMed] [Google Scholar]
- 110.Schiöth HB, Haitina T, Fridmanis D, Klovins J. Unusual genomic structure: melanocortin receptors in Fugu. Ann N Y Acad Sci. 2005;1040:460–463. doi: 10.1196/annals.1327.090. [DOI] [PubMed] [Google Scholar]
- 111.Kasahara M, Naruse K, Sasaki S, Nakatani Y, Qu W, et al. The medaka draft genome and insights into vertebrate genome evolution. Nature. 2007;447:714–719. doi: 10.1038/nature05846. [DOI] [PubMed] [Google Scholar]
- 112.Vanegas H, Ito H. Morphological aspects of the teleostean visual system: a review. Brain Res. 1983;287:117–137. doi: 10.1016/0165-0173(83)90036-x. [DOI] [PubMed] [Google Scholar]
- 113.Wullimann MF, Meyer DL, Northcutt RG. The visually related posterior pretectal nucleus in the non-percomorph teleost Osteoglossum bicirrhosum projects to the hypothalamus: a DiI study. J Comp Neurol. 1991;312:415–435. doi: 10.1002/cne.903120309. [DOI] [PubMed] [Google Scholar]
- 114.Rupp B, Wullimann MF, Reichert H. The zebrafish brain: a neuroanatomical comparison with the goldfish. Anat Embryol (Berl) 1996;194:187–203. doi: 10.1007/BF00195012. [DOI] [PubMed] [Google Scholar]
- 115.Furutani-Seiki M, Wittbrodt J. Medaka and zebrafish, an evolutionary twin study. Mech Dev. 2004;121:629–637. doi: 10.1016/j.mod.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 116.Xue HG, Yang CY, Yamamoto N, Ito H, Ozawa H. An indirect trigeminocerebellar pathway through the nucleus lateralis valvulae in a perciform teleost, Oreochromis niloticus. Neurosci Lett. 2005;390:104–108. doi: 10.1016/j.neulet.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 117.Paxton JR, Eschmeyer WN. San Diego: Academic Press; 1998. Encyclopedia of fishes, 2nd Ed. [Google Scholar]
- 118.Holland LZ, Gibson-Brown JJ. The Ciona intestinalis genome: when the constraints are off. Bioessays. 2003;25:529–532. doi: 10.1002/bies.10302. [DOI] [PubMed] [Google Scholar]
- 119.Satoh N, Kawashima T, Shoguchi E, Satou Y. Urochordate genomes. Genome Dyn. 2006;2:198–212. doi: 10.1159/000095105. [DOI] [PubMed] [Google Scholar]
- 120.Fukuda M, Kanno E, Mikoshiba K. Conserved N-terminal cysteine motif is essential for homo- and heterodimer formation of synaptotagmins III, V, VI, and X. J Biol Chem. 1999;274:31421–31427. doi: 10.1074/jbc.274.44.31421. [DOI] [PubMed] [Google Scholar]
- 121.Zimmer DB, Cornwall EH, Landar A, Song W. The S100 protein family: history, function, and expression. Brain Res Bull. 1995;37:417–429. doi: 10.1016/0361-9230(95)00040-2. [DOI] [PubMed] [Google Scholar]
- 122.McCall KA, Huang C, Fierke CA. Function and mechanism of zinc metalloenzymes. J Nutr. 2000;130(5S Suppl):1437S–1446S. doi: 10.1093/jn/130.5.1437S. [DOI] [PubMed] [Google Scholar]
- 123.Marenholz I, Heizmann CW, Fritz G. S100 proteins in mouse and man: from evolution to function and pathology (including an update of the nomenclature). Biochem Biophys Res Commun. 2004;322:1111–1122. doi: 10.1016/j.bbrc.2004.07.096. [DOI] [PubMed] [Google Scholar]
- 124.Bitto E, Bingman CA, Bittova L, Kondrashov DA, Bannen RM, et al. Structure of human J-type co-chaperone HscB reveals a tetracysteine metal-binding domain. J Biol Chem. 2008;283:30184–30192. doi: 10.1074/jbc.M804746200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Glick BS, Nakano A. Membrane traffic within the Golgi apparatus. Annu Rev Cell Dev Biol. 2009;25:113–132. doi: 10.1146/annurev.cellbio.24.110707.175421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nesselhut J, Jurgan U, Onken E, Götz H, Barnikol HU, et al. Golgi retention of human protein NEFA is mediated by its N-terminal Leu/Ile-rich region. FEBS Lett. 2001;509:469–475. doi: 10.1016/s0014-5793(01)03187-8. [DOI] [PubMed] [Google Scholar]
- 127.Werren JH, Richards S, Desjardins CA, Niehuis O, Gadau J, et al. Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science. 2010;327:343–348. doi: 10.1126/science.1178028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pruitt KD, Tatusova T, Maglott DR. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2010;38:D5–16. doi: 10.1093/nar/gkp967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Birney E, Andrews TD, Bevan P, Caccamo M, Chen Y, et al. An overview of Ensembl. Genome Res. 2004;14:925–928. doi: 10.1101/gr.1860604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Flicek P, Aken BL, Ballester B, Beal K, Bragin E, et al. Ensembl's 10th year. Nucleic Acids Res. 2010;38:D557–D562. doi: 10.1093/nar/gkp972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Markowitz VM, Chen IM, Palaniappan K, Chu K, Szeto E, et al. The integrated microbial genomes system: an expanding comparative analysis resource. Nucleic Acids Res. 2010;38:D382–D390. doi: 10.1093/nar/gkp887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Srivastava M, Begovic E, Chapman J, Putnam NH, Hellsten U, et al. The Trichoplax genome and the nature of placozoans. Nature. 2008;454:955–960. doi: 10.1038/nature07191. [DOI] [PubMed] [Google Scholar]
- 134.Duan J, Li R, Cheng D, Fan W, Zha X, et al. SilkDB v2.0: a platform for silkworm (Bombyx mori) genome biology. Nucleic Acids Res. 2010;38:D453–D456. doi: 10.1093/nar/gkp801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Combet C, Blanchet C, Geourjon C, Deléage G. NPS@: network protein sequence analysis. Trends Biochem Sci. 2000;25:147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- 136.Harper RA. EMBnet: an institute without walls. Trends Biochem Sci. 1996;21:150–152. doi: 10.1016/0968-0004(96)30012-1. [DOI] [PubMed] [Google Scholar]
- 137.D'Elia D, Gisel A, Eriksson NE, Kossida S, Mattila K, et al. The 20th anniversary of EMBnet: 20 years of bioinformatics for the Life Sciences community. BMC Bioinformatics. 2009;10(Suppl 6):S1. doi: 10.1186/1471-2105-10-S6-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, et al. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol. 1999;112:531–552. doi: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- 139.Hofmann K, Stoffel W. TMbase - a database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler. 1993;374:166–169. [Google Scholar]
- 140.Milpetz F, Argos P, Persson B. TMAP: a new email and WWW service for membrane-protein structural predictions. Trends Biochem Sci. 1995;20:204–205. doi: 10.1016/s0968-0004(00)89009-x. [DOI] [PubMed] [Google Scholar]
- 141.Tusnády GE, Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17:849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- 142.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 143.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 144.Kanehisa M. Linking databases and organisms: GenomeNet resources in Japan. Trends Biochem Sci. 1997;22:442–444. doi: 10.1016/s0968-0004(97)01130-4. [DOI] [PubMed] [Google Scholar]
- 145.Horton P, Nakai K. Better prediction of protein cellular localization sites with the k nearest neighbors classifier. Proc Int Conf Intell Syst Mol Biol. 1997;5:147–152. [PubMed] [Google Scholar]
- 146.Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, et al. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009;37:D205–D210. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Gould CM, Diella F, Via A, Puntervoll P, Gemünd C, et al. ELM: the status of the 2010 eukaryotic linear motif resource. Nucleic Acids Res. 2010;38:D167–D180. doi: 10.1093/nar/gkp1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 149.Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 150.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 151.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 152.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 153.Huelsenbeck JP, Ronquist F. MrBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 154.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 156.Anisimova M, Gascuel O. Approximate likelihood ratio test for branches: A fast, accurate and powerful alternative. Syst Biol. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- 157.Adoutte A, Balavoine G, Lartillot N, Lespinet O, Prud'homme B, et al. The new animal phylogeny: reliability and implications. Proc Natl Acad Sci U S A. 2000;97:4453–4456. doi: 10.1073/pnas.97.9.4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Halanych KM, Passamaneck Y. A brief review of metazoan phylogeny and future prospects in Hox-research. Am Zoologist. 2001;41:629–639. [Google Scholar]
- 159.Fontanillas E, Welch JJ, Thomas JA, Bromham L. The influence of body size and net diversification rate on molecular evolution during the radiation of animal phyla. BMC Evol Biol. 2007;7:95. doi: 10.1186/1471-2148-7-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gerlach D, Wolf M, Dandekar T, Müller T, Pokorny A, Rahmann S. Deep metazoan phylogeny. In Silico Biol. 2007;7:151–154. [PubMed] [Google Scholar]
- 161.Wägele JW, Mayer C. Visualizing differences in phylogenetic information content of alignments and distinction of three classes of long-branch effects. BMC Evol Biol. 2007;7:147. doi: 10.1186/1471-2148-7-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ponting CP. The functional repertoires of metazoan genomes. Nat Rev Genet. 2008;9:689–698. doi: 10.1038/nrg2413. [DOI] [PubMed] [Google Scholar]
- 163.Kuraku S, Meyer A, Kuratani S. Timing of genome duplications relative to the origin of the vertebrates: did cyclostomes diverge before or after? Mol Biol Evol. 2009;26:47–59. doi: 10.1093/molbev/msn222. [DOI] [PubMed] [Google Scholar]
- 164.Wang Y, Gu X. Evolutionary patterns of gene families generated in the early stage of vertebrates. J Mol Evol. 2000;51:88–96. doi: 10.1007/s002390010069. [DOI] [PubMed] [Google Scholar]
- 165.Meyer A, Van de Peer Y. From 2R to 3R: evidence for a fish-specific genome duplication (FSGD). Bioessays. 2005;27:937–945. doi: 10.1002/bies.20293. [DOI] [PubMed] [Google Scholar]
- 166.Cossins AR, Crawford DL. Fish as models for environmental genomics. Nat Rev Genet. 2005;6:324–333. doi: 10.1038/nrg1590. [DOI] [PubMed] [Google Scholar]
- 167.Froschauer A, Braasch I, Volff JN. Fish genomes, comparative genomics and vertebrate evolution. Curr Genomics. 2006;7:43–57. [Google Scholar]
- 168.Hoegg S, Boore JL, Kuehl JV, Meyer A. Comparative phylogenomic analyses of teleost fish Hox gene clusters: lessons from the cichlid fish Astatotilapia burtoni. BMC Genomics. 2007;8:317. doi: 10.1186/1471-2164-8-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Li C, Lu G, Ortí G. Optimal data partitioning and a test case for ray-finned fishes (Actinopterygii) based on ten nuclear loci. Syst Biol. 2008;57:519–539. doi: 10.1080/10635150802206883. [DOI] [PubMed] [Google Scholar]
- 170.Zuckerkandl E, Pauling L. Evolutionary divergence and convergence in proteins. In: Bryson V, Vogel H, editors. Evolving genes and proteins. New York: Academic Press; 1965. pp. 97–166. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Taxa, accession numbers, and chromosome location of DIA1 orthologues.
(0.05 MB PDF)
Taxa, accession numbers and chromosome location of partial DIA1 orthologues.
(0.04 MB PDF)
Physical characteristics of all available full-length DIA1 proteins and their similarity to orthologues from key species.
(0.04 MB PDF)
Pairwise comparison of DIA1 and DIA1R proteins.
(0.02 MB PDF)
Taxa, accession numbers and chromosome location of full-length DIA1R orthologues.
(0.03 MB PDF)
Taxa, accession numbers and chromosome location of partial DIA1R orthologues.
(0.03 MB PDF)
Comparison of Ciona intestinalis DIA1 to DIA1 and DIA1R from other species.
(0.02 MB PDF)
Taxa and accession numbers of full-length DIA1L genes.
(0.03 MB PDF)
Amino acid comparisons between DIA1L and DIA1 proteins.
(0.02 MB PDF)
Taxa, accession numbers and chromosome location of partial DIA1L paralogues.
(0.03 MB PDF)
Amino acid sequence comparison of DIA1a and DIA1b from zebrafish. The sequence alignment was generated using CLUSTALW [47]. Identical amino acids are highlighted in red font and indicated below the alignment with an asterisk (*). Strongly similar amino acids are highlighted in green font and indicated below the alignment with a colon (:). Weakly similar amino acids are highlighted in blue font and indicated below the alignment with a full stop (.). Dissimilar amino acids are in black font. Amino acid numbering is provided above the alignment. Gaps are indicated by dashes. The alignment shows 88% identical amino acids, and a further 10% similar amino acids, providing an overall similarity of 98%. Standard single-letter amino acid abbreviations are used. Organism abbreviation uses the first letter of the genus name, followed by the first four letters of the species (i.e., Danio rerio DIA1a is abbreviated to DreriDIA1a). Accession numbers can be found in Table S1.
(0.01 MB PDF)
Amino acid sequence comparison of DIA1 proteins. The sequence alignment of all full-length DIA1 proteins was generated using CLUSTALW [47]. A consensus amino acid sequence is presented below the alignment, where uppercase letters indicate absolutely conserved amino acids. Regions of greater than 50% conservation are shaded (identical amino acids are in black boxes and similar amino acids in grey boxes). Amino acid numbering is provided on the left-hand side of the alignment. Gaps required for optimal alignment are indicated by dashes. The alignment reveals that 10 amino acids are absolutely conserved (2% identity), with a further 6% amino acids being similar, providing an overall similarity of 8%. Standard single-letter amino acid abbreviations are used. Organism abbreviation uses the first letter of the genus name, followed by the first four letters of the species (e.g., Strongylocentrotus purpuratus DIA1 is abbreviated to SpurpDIA1). Accession numbers and full species names can be found in Table S1.
(0.08 MB PDF)
Amino acid sequence comparison of DIA1R proteins. The sequence alignment was generated using CLUSTALW [47]. Identical amino acids are highlighted in red font and indicated below the alignment with a red asterisk (*). Strongly similar amino acids are highlighted in green font and indicated below the alignment with a green colon (:). Weakly similar amino acids are highlighted in blue font and indicated below the alignment with a blue full stop (.). Dissimilar amino acids are in black font. Amino acid numbering is provided above the alignment. Gaps are indicated by dashes. The alignment shows 124 identical amino acids (27% identity), with a further 27% similar amino acids, providing an overall similarity of 54%. Standard single-letter amino acid abbreviations are used. A consensus line is provided below the alignment where the number “2” is inserted when no consensus amino acid is found. Organism abbreviations use the first letter of the genus name, followed by the first four letters of the species (e.g., Monodelphis domestica DIA1R is abbreviated to MdomeDIA1R). Accession numbers and full species names can be found in Table S5.
(0.03 MB PDF)
Amino acid sequence alignment of DIA1 and DIA1R proteins. The sequence alignment of all full-length DIA1 and DIA1R proteins was generated using CLUSTALW [47]. A consensus amino acid sequence is presented below the alignment, where uppercase letters indicate absolutely conserved amino acids. Regions of greater than 50% conservation are shaded (identical amino acids are in black boxes and similar amino acids in grey boxes) and 56% of aligned positions showed conservation in >50% of sequences. Only 8 amino acids are absolutely conserved (∼2% identity), with a further 23 amino acids being similar (∼5%) in all DIA1 and DIA1R proteins. Amino acid numbering is provided on the left-hand side of the alignment. Gaps required for optimal alignment are indicated by dashes. Standard single-letter amino acid abbreviations are used. Organism abbreviations use the first letter of the genus name, followed by the first four letters of the species (e.g., Gallus gallus DIA1R is abbreviated to GgallDIA1R). Accession numbers and full species names can be found in Tables S1 and S5.
(0.13 MB PDF)
Signal peptide localization in DIA1 and DIA1R proteins. The sequence alignment and consensus sequence of the amino terminal region of all full-length DIA1 and DIA1R proteins is from Figure S4. Abbreviations are as in Figure S4. DIA1R proteins were grouped together in the top portion of the figure, with aligned DIA1 proteins below, where arthropod sequences are placed above, nonvertebrate/nonarthropod sequences below, and vertebrate DIA1 sequences in the middle (see annotation on right-hand side of alignment). Initiation methiones were not manually aligned. Bold red amino acids represent the last amino acid of the amino-terminal signal peptide predicted using the NN algorithm [51]. Bold blue amino acids represent the last amino acid of the signal peptide predicted using the HMM prediction method [51]. Bold purple amino acids represent the last amino acid of the signal peptide predicted by both NN and HMM prediction methods. Bold underlined amino acids represent the last amino acid of the signal peptide predicted by the Sigcleave algorithm [52]. Lack of an underlined residue indicates no signal peptide cleavage site was predicted within the aligned region by Sigcleave, and lack of a red residue (or purple) indicates no signal peptide cleavage site predicted by the NN algorithm. The most commonly predicted site for cleavage of DIA1R signal peptides or vertebrate and arthropod DIA1 signal peptides are indicated with arrows above the alignment.
(0.03 MB PDF)
Amino acid sequence comparison of DIA1L proteins. A single DIA1L protein from S. purpuratus (SpurpDIA1L) and three DIA1L paralogues from B. floridae (BflorDIA1La, BflorDIA1Lb, BflorDIA1Lc) were aligned using CLUSTALW [47]. Identical amino acids shared between all four proteins are highlighted in red font and indicated below the alignment with a red asterisk (*). Strongly similar amino acids are highlighted in green font and indicated below the alignment with a green colon (:). Weakly similar amino acids are highlighted in blue font and indicated below the alignment with a blue full stop (.). Dissimilar amino acids are in black font. Amino acid numbering is provided above the alignment. Gaps are indicated by dashes. The alignment shows 7% identical amino acids, with a further 20% similar amino acids, providing an overall similarity of 27%. Amino acid similarity and identity shared at the amino-terminal ends of the three longer DIA1L proteins are indicated using the coloured fonts described above, but the corresponding annotations (*, : and .) are in grey font below the alignment. Standard single-letter amino acid abbreviations are used. Accession numbers can be found in Table S8.
(0.02 MB PDF)
Amino acid sequence comparison of DIA1-family proteins. The sequence alignment of all full-length DIA1, DIA1R and DIA1L proteins was generated using CLUSTALW [47]. Identical amino acids are highlighted in red font and indicated below the alignment with a red asterisk (*). Strongly similar amino acids are highlighted in green font and indicated below the alignment with a green colon (:). Weakly similar amino acids are highlighted in blue font and indicated below the alignment with a blue full stop (.). Dissimilar amino acids are in black font. Amino acid numbering is provided above the alignment. Gaps are indicated by dashes. The alignment shows only 4 identical amino acids and 18 similar amino acids conserved across the entire protein family (3% similarity). Standard single-letter amino acid abbreviations are used. Organism abbreviations use the first letter of the genus name, followed by the first four letters of the species (e.g., Pteropus vampyrus DIA1 is abbreviated to PvampDIA1). Accession numbers and full species names can be found in Tables S1, S5 and S8.
(0.05 MB PDF)
Amino acids conserved in at least 80% of DIA1-family members. The sequence alignment of all full-length DIA1, DIA1R and DIA1L proteins was generated using CLUSTALW [47]. A consensus amino acid sequence is presented below the alignment, where uppercase letters indicate absolutely conserved amino acids. Regions of greater than 80% conservation are shaded (identical amino acids are in black boxes and similar amino acids in grey boxes), and 10% of aligned positions showed conservation in >80% of sequences. Amino acid numbering is provided on the left-hand side of the alignment. Gaps required for optimal alignment are indicated by dashes. Standard single-letter amino acid abbreviations are used. Organism abbreviations use the first letter of the genus name, followed by the first four letters of the species (e.g., Salmo salar DIA1R is abbreviated to SsalaDIA1R). Accession numbers and full species names can be found in Tables S1, S5 and S8.
(0.10 MB PDF)
Amino acids conserved in at least 50% of DIA1-family members. The sequence alignment of all full-length DIA1, DIA1R and DIA1L proteins was generated using CLUSTALW [47]. A consensus amino acid sequence is presented below the alignment, where uppercase letters indicate absolutely conserved amino acids. Regions of greater than 50% conservation are shaded (identical amino acids are in black boxes and similar amino acids in grey boxes), and 38% of aligned positions showed conservation in >50% of sequences. Amino acid numbering is provided on the left-hand side of the alignment. Gaps required for optimal alignment are indicated by dashes. Standard single letter amino acid abbreviations are used. Organism abbreviations use the first letter of the genus name, followed by the first four letters of the species (e.g., Tursiops truncatus DIA1 is abbreviated to TtrunDIA1R). Accession numbers and full species names can be found in Tables S1, S5 and S8.
(0.14 MB PDF)
DIA1-family amino acid sequences in FASTA format. Each DIA1-family amino acid sequence starts with a “>” (greater-than) symbol followed by the species abbreviation and protein type (e.g., Oryzias latipes DIA1 is abbreviated to OlatiDIA1 and Pongo pygmaeus DIA1R to PpygmDIA1R). Following the initial title line is the actual amino acid sequence, in standard single-letter code. Accession numbers, full species names, and differences to current database sequence data (due to corrections) can be found in Tables S1, S5 and S8.
(0.04 MB PDF)
Maximum-likelihood tree of DIA1-family proteins. Proteins encoded by each full-length DIA1-family gene were aligned using CLUSTALW [47] and subjected to maximum-likelihood analysis [151] using PhyML phylogeny software [135]. Approximate likelihood-ratio test for branch-support statistics [156] was carried out, and percentage values are shown next to branches. Branch lengths are proportional to the number of amino acid substitutions per site (see scale bar). G-blocks were used to eliminate poorly aligned positions and divergent regions, since they may not be homologous or may have been saturated by multiple substitutions [148]. The tree was rooted on the cnidarian N. vectensis DIA1 sequence (NvectDIA1), as highlighted with an asterisk. Organism abbreviations use the first letter of the genus name, followed by the first four letters of the species. Full species names and accession numbers can be found in Tables S1, S4 and S7.
(0.12 MB PDF)
Phylogeny of the DIA1-family reconstructed using a Bayesian phylogenetic approach. A subset of DIA1-family gene products were aligned using CLUSTALW [47] and subjected to Bayesian inference of phylogeny using the MrBayes programme [153]. The number above each branch refers to the Bayesian posterior probability of the node, given as a percentage (e.g., 77 represents a posterior probability of 0.77). Branch lengths are proportional to the number of amino acid substitutions per site (see scale bar). Gblocks were used to curate the alignment [148]. The tree was rooted on the cnidarian N. vectensis DIA1 sequence (NvectDIA1), as highlighted with an asterisk. Organism abbreviations use the first letter of the genus name, followed by the first four letters of the species. Full species names and accession numbers can be found in Tables S1, S4 and S7.
(0.07 MB PDF)