Abstract
Determining the rate of forming the truly folded conformation of ultrafast folding proteins is an important issue for both experiments and simulations. The double-norleucine mutant of the 35-residue villin subdomain is the focus of recent computer simulations with atomistic molecular dynamics because it is currently the fastest folding protein. The folding kinetics of this protein have been measured in laser temperature-jump experiments using tryptophan fluorescence as a probe of overall folding. The conclusion from the simulations, however, is that the rate determined by fluorescence is significantly larger than the rate of overall folding. We have therefore employed an independent experimental method to determine the folding rate. The decay of the tryptophan triplet-state in photoselection experiments was used to monitor the change in the unfolded population for a sequence of the villin subdomain with one amino acid difference from that of the laser temperature-jump experiments, but with almost identical equilibrium properties. Folding times obtained in a two-state analysis of the results from the two methods at denaturant concentrations varying from 1.5–6.0 M guanidinium chloride are in excellent agreement, with an average difference of only 20%. Polynomial extrapolation of all the data to zero denaturant yields a folding time of 220 (+100,-70) ns at 283 K, suggesting that under these conditions the barrier between folded and unfolded states has effectively disappeared—the so-called “downhill scenario.”
Keywords: tryptophan triplet lifetime, villin headpiece subdomain, downhill protein folding, laser temperature jump
The introduction of a variety of experimental methods to study protein folding with much improved time resolution has resulted in the discovery of proteins that fold on a microsecond timescale (1–15). The investigation of these so-called ultrafast folding proteins is important for several reasons. They are small single-domain proteins, containing less than 100 amino acid residues, and are therefore expected to fold by relatively simple mechanisms. Their small size makes them attractive for investigation using statistical mechanical models that enumerate conformations and therefore provide a comprehensive description of the kinetics by solving the set of differential equations for the time course of each conformation (16–19). Finally, with recent improvements in computing power and the application of distributed computing (20–22), their size and folding speed allows the kinetics and mechanisms of folding to be studied by atomistic molecular dynamics simulations.
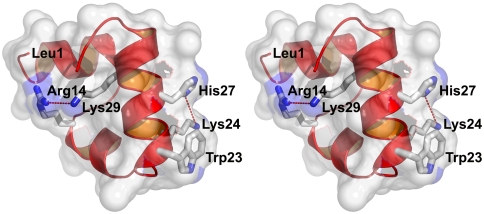
The 35-residue subdomain of the villin headpiece, called HP35, is one of the smallest naturally occurring protein domains. It folds without metals or disulfide bridges and is the most intensively studied ultrafast folder by experiment, theory, and simulations (SI Text). It has a nontrivial structure with three helices surrounding a hydrophobic core (Fig. 1). To use fluorescence of the lone tryptophan in position 23 as a probe for folding kinetics, Asn27 of the wild-type sequence, one turn away in the C-terminal helix, was replaced by a protonated histidine [HP35(His27)] to quench the fluorescence in the folded state (Fig. 1). In temperature-jump (T-jump) experiments two relaxations are observed. The faster one at approximately 70 ns is nearly temperature-independent and has been interpreted as a helix-coil transition on the folded side of the major barrier (19, 26). The slower relaxation has been interpreted as global unfolding/refolding, with a rate that increases with temperature in the range 300–360 K from ∼(10 μs)-1 to ∼(1 μs)-1 (19, 26). In a two-state analysis the protein folds in 5 µs at the unfolding temperature of 70 °C. A more stable mutant in which two lysines are replaced by norleucine [HP35(Nle24,His27,Nle29)] (Fig. 1), folds in 700 ns (24).
Fig. 1.
X-ray structure of villin headpiece subdomain, HP35(His27), (Protein Data Bank ID code 1YRF) showing key residues (sequence: LSDED FKAVF GMTRS AFANL PLWKQ QNLKK EKGLF) (23). Trp23 is the fluorescence probe; His27 replaces Asn27 of the wild-type sequence, and when protonated reduces the fluorescence of Trp23 upon folding; Lys24 and Lys29 make repulsive electrostatic interactions with protonated His27 and Arg14, respectively. The ϵ-amino nitrogen of Lys24 is 6.1 Å from the nearest protonated nitrogen of the imidazole ring of His27 and the ϵ-amino nitrogen of Lys29 is 5.3 Å from the nearest charged amino atom of Arg14. Removal of the ϵ-amino nitrogen groups of Lys24 and Lys29 to form norleucines in the mutant called HP35(Nle24,His27,Nle29) eliminates the repulsive interaction, thereby stabilizing the protein and increasing the folding rate (24, 25).
Because it is currently the fastest folding protein, this double-norleucine mutant is rapidly becoming a favorite molecule for atomistic molecular dynamics simulations (27–30). From an analysis of a large number of trajectories started from unfolded conformations, Pande and coworkers suggested that the tryptophan fluorescence change does not monitor folding of the entire molecule, which occurs more slowly in their simulations, but just the local tertiary structure surrounding this tryptophan (27, 29). Freddolino and Schulten reached the same conclusion from their simulations of both HP35(His27) and [HP35(Nle24,His27,Nle29)] (30). This alternative interpretation of the experimental results highlights an important general question for studies of ultrafast folding proteins that use a single probe to monitor the kinetics.
Experimentalists have been very much aware of this problem, as no available experimental method can unambiguously measure the formation of the native tertiary structure on the microsecond timescale (31, 32). We previously compared the folding rate for HP35(His27) obtained from fluorescence T-jump experiments with the rate for HP35 determined by an independent method based on quenching of the tryptophan triplet state upon close contact with a cysteine added to the N terminus (Cys-HP35). Quenching of tryptophan fluorescence probes the change in population of the folded state by monitoring the formation of native structure in the environment of Trp23, whereas quenching of the triplet state in the photoselection experiment probes the change in population of the unfolded state. There are two properties of the triplet-lifetime experiment that allow determination of the folding rate. First, upon contact formation cysteine quenches the triplet state 2,500 times faster than any other amino acid in the sequence of this study (33). Second, in the folded conformation the cysteine is on the opposite side of the molecule from the tryptophan. To make the contact a large part of the N-terminal portion of the protein must be unfolded (Fig. 1). Thus we can safely assume that for this polypeptide the decrease in the lifetime of the triplet state arises exclusively from triplet quenching by close contact of the cysteine with the tryptophan in a nonnative conformation. These properties allow the method to investigate the kinetics of specific intramolecular contact formation (11, 34–37) and other processes that occur on a timescale less than or comparable to the intrinsic triplet lifetime of approximately 100 µs and that also affect the quenching rate (35, 38).†
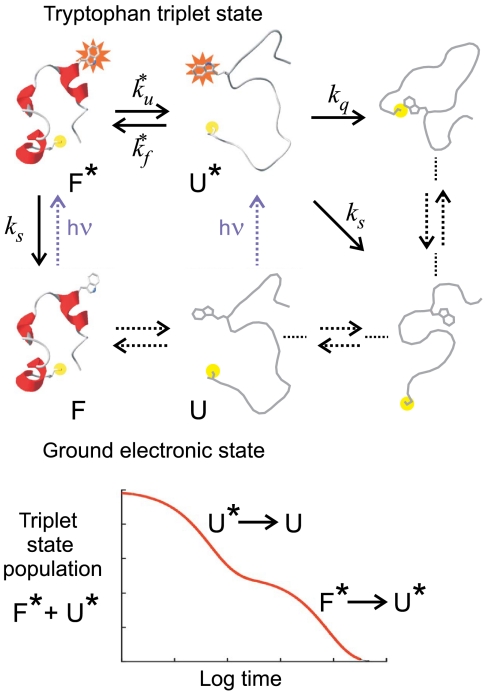
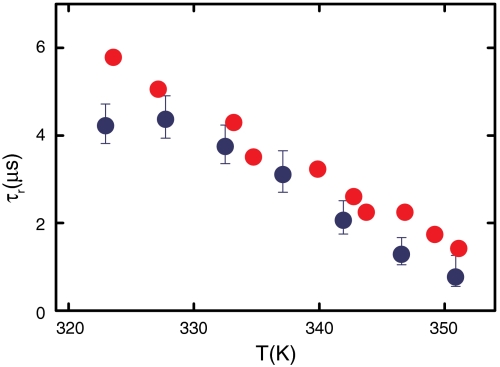
A schematic of the model used to measure the folding and unfolding rates for Cys-HP35 from the decay in the triplet-state population is shown in Fig. 2. The decay of the triplet state for Cys-HP35, analyzed in terms of a two-state model, yielded folding and unfolding rates for the protein that agreed to within the, albeit large, experimental uncertainties with the rates obtained from the T-jump experiments for HP35(His27) (Table S1) (38). Confirmation of fluorescence as an accurate probe of folding came with measurement of the kinetics using temperature jump with detection by IR spectroscopy by Gai and coworkers (39), which probes the global formation of secondary structure. They determined relaxation times for HP35(His27) in excellent agreement with those determined by tryptophan fluorescence (Fig. 3). The situation is not as clear with Met-HP35 studied extensively by Raleigh and coworkers (40), where the relaxation (exchange) rates determined in dynamic nuclear magnetic resonance experiments appear to be smaller than those obtained in infrared T-jump experiments (seeTable S1 for a detailed comparison).
Fig. 2.
Schematic of tryptophan triplet-lifetime experiment. (Upper) A UV laser pulse excites tryptophan to its lowest excited singlet state that undergoes intersystem crossing in a few nanoseconds to the triplet state, which then lives for up to 100 µs. Diffusion of the unfolded polypeptide chain to form a close contact between the N-terminal cysteine and the tryptophan depopulates the triplet state, presumably via electron transfer from the tryptophan to the cysteine. The solid arrows are processes that contribute to the observed kinetics for the decay of the triplet-state population. (Lower) In the case of a two-state protein where (i) the spontaneous decay of the triplet state (ks) is so slow that the triplet state only returns to the ground state via cysteine quenching in the unfolded state and (ii) the quenching rate is much faster than the unfolding and refolding rates (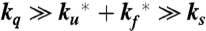 ), the decay of the triplet-state population monitored by triplet–triplet optical absorption is biphasic, with the fast phase corresponding to the triplet-quenching rate in the unfolded state (kq), the slow phase to the unfolding rate (
), the decay of the triplet-state population monitored by triplet–triplet optical absorption is biphasic, with the fast phase corresponding to the triplet-quenching rate in the unfolded state (kq), the slow phase to the unfolding rate (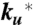 ), and the two amplitudes for the fast and slow phases given by the equilibrium fractions of the unfolded and folded states:
), and the two amplitudes for the fast and slow phases given by the equilibrium fractions of the unfolded and folded states: 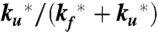 and
and 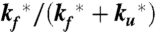 , respectively (11, 34–36, 38). In the more realistic situation that we encounter with the double-norleucine mutant, Cys-HP35(Nle24,Nle29), where the timescales are not so clearly separable, the relaxation rates and amplitudes become complex functions of the four rate coefficients, which can be obtained by fitting the data with the solution of a simple differential equation model (see Eqs. 1–5).
, respectively (11, 34–36, 38). In the more realistic situation that we encounter with the double-norleucine mutant, Cys-HP35(Nle24,Nle29), where the timescales are not so clearly separable, the relaxation rates and amplitudes become complex functions of the four rate coefficients, which can be obtained by fitting the data with the solution of a simple differential equation model (see Eqs. 1–5).
Fig. 3.
Comparison of relaxation times for HP35(His27) measured in temperature-jump experiments using IR absorption (39) (red circles) or fluorescence (25) (blue circles) detection.
Because of its importance to simulation studies, the primary objectives of this work are to determine whether tryptophan fluorescence accurately determines the global folding rate for the double-norleucine mutant HP35(Nle24,His27,Nle29), as demonstrated for HP35(His27), and to critically compare the experimental results with the results of molecular dynamics simulations for this mutant (27, 30). Moreover, the free energy barriers separating folded and unfolded states for HP35(His27) have been estimated by both calorimetric and kinetic criteria to be very low (< 2 kcal/mol) (43); thus there is the possibility that the barrier disappears in the faster-folding mutant. In this so-called downhill scenario (44), we would expect to observe probe-dependent kinetics (45, 46). On the other hand, if the kinetic properties are independent of the probe, then we can validate the use of fluorescence as a probe of global folding and obtain folding and unfolding rate coefficients from a two-state analysis. We therefore made a detailed comparison of the kinetics of the double-norleucine mutant in T-jump experiments monitored by tryptophan fluorescence with the kinetics measured using the triplet-lifetime method over a wide range of denaturant concentrations.
Results
Equilibrium: Chemical Denaturant Unfolding Curves.
The true wild-type sequence of HP35 has an asparagine in position 27. To obtain a measurable change in the tryptophan quantum yield this residue was replaced in all of the laser T-jump experiments with histidine (Fig. 1) and the measurements made at pH 4.9 to ensure that the histidine is protonated (19, 24, 26, 47, 48). The positive charge of the protonated histidine results in a decreased fluorescence of Trp23 in the third helix (Fig. 1). The problem for the triplet-lifetime experiment is that protonated histidine severely reduces the population of the triplet state and also complicates the analysis by quenching both the singlet and triplet states [protonated histidine quenches the tryptophan triplet at a rate only 25-fold slower than cysteine (33)]. On the other hand, raising the pH decreases the amplitude of the fluorescence change by > 4-fold, making it impractical to obtain accurate kinetic data from the T-jump experiments. Consequently, the double-norleucine mutant with asparagine in position 27 and an N-terminal cysteine, which we call Cys-HP35(Nle24,Nle29), was used to compare with the fluorescence temperature jump with protonated histidine in position 27 and an N-terminal cysteine, which we call Cys-HP35(Nle24,His27,Nle29). To obtain the most accurate determination of rates, the comparison was carried out at 283 K, because the population of the triplet state, like the fluorescence quantum yield (19, 24, 26), decreases markedly with increasing temperature.
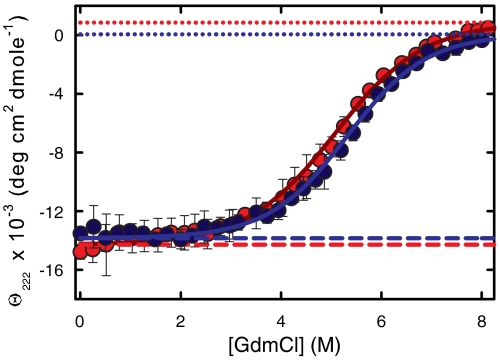
The sine qua non of such a comparison is that the two sequences have the same fractional populations of folded and unfolded states at equilibrium under the same solution conditions. The comparison of the denaturant-induced unfolding curves shows that these populations are indeed very similar with almost superimposable data (Fig. 4). Moreover, these data are well fit by a two-state model. The ratio of the equilibrium constants (Keq) for the two sequences at 5.2 M GdmCl, close to the midpoint denaturant concentrations for both sequences where the equilibrium constants are most accurately determined, is 1.27 [Keq = 1.12 for Cys-HP35(Nle24,His27,Nle29) and Keq = 0.88 for Cys-HP35(Nle24,Nle29)], with Cys-HP35(Nle24,Nle29) being the more stable; the ratios in the range 1–6.5 M GdmCl are all less than 2.2. Thus, if all of the difference in equilibrium constant appears in the folding rate, the two rates would differ by less than 2.2-fold at all GdmCl concentrations, assuming a linear-free energy relation [i.e., Φ values between zero and one, as observed for all other mutants (19, 24, 26, 47, 48)].
Fig. 4.
Equilibrium unfolding curves. (A) Comparison of denaturant unfolding curves for Cys-HP35(Nle24,Nle29) and Cys-HP35(Nle24,His27,Nle29). Equilibrium CD data collected at 10 °C (20 mM sodium acetate, 1 mm Tris(2-carboxyethyl)phosphine (TCEP), 75 μm protein, pH = 4.9) as a function of GdmCl concentration. The experimentally measured ellipticities are the dark-blue points [Cys-HP35(Nle24,Nle29)] and the red points [Cys-HP35(Nle24,His27,Nle29)]. The blue and red solid lines are the two-state fits to the data. The dashed lines are the native baselines and the dotted lines are the unfolded baselines. The denaturation midpoint concentrations, [D]mid, are 5.1( ± 0.2)M for Cys-HP35(Nle24,His27,Nle29) and 5.3( ± 0.1)M for Cys-HP35(Nle24,Nle29), whereas the m-values are 640 (+149,-112) cal mol-1 M-1 for Cys-HP35(Nle24,His27,Nle29) and 714(+88, -62) cal mol-1 M-1 for Cys-HP35(Nle24,Nle29). The error bars represent standard deviations from the average of at least three measurements on separately prepared solutions.
Kinetics: Laser T-Jump with Tryptophan Fluorescence Detection.
Fig. 5 shows the time course of the tryptophan fluorescent quantum yield at 3 GdmCl concentrations following a 5 K temperature jump. The data are well fit with a single exponential function. Assuming a two-state model, the folding and unfolding rate coefficients can be obtained from the observed relaxation rate (= kf + ku) (Fig. S1) and the equilibrium constants (Keq = ku/kf) determined from the equilibrium unfolding data in Fig. 4.
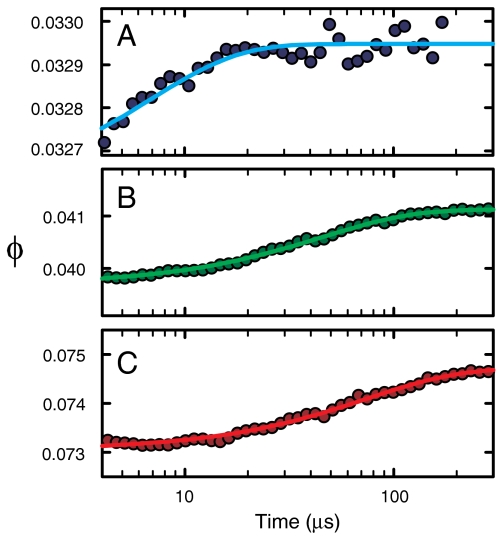
Fig. 5.
Tryptophan quantum yield (ϕ) as a function of time at 10 °C after a 5 K laser-induced T-jump for 300 μM solutions of Cys-HP35(Nle24,His27,Nle29) containing 20 mM sodium acetate, 1 mM TCEP, and either (A) 2.25 M GdmCl (blue), (B) 4 M GdmCl (green), or (C) 6 M GdmCl (red). The circles are the experimental data and the lines are fits with a single exponential function.
Kinetics: Tryptophan Triplet Lifetime.
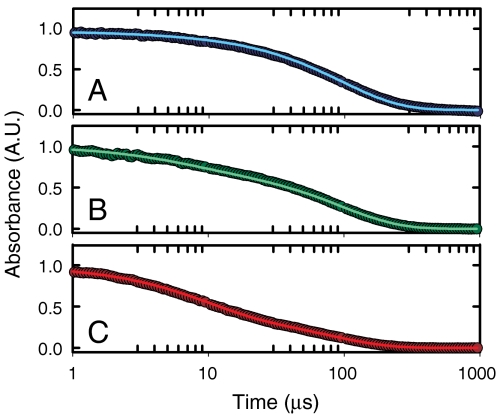
Fig. 6 shows the kinetic progress curves at three denaturant concentrations for decay of the tryptophan triplet state formed by a 5-ns UV laser pulse (curves at other denaturant concentrations are shown in Fig. S2). At 1.5 M GdmCl, where about 1% of the molecules are unfolded according to the equilibrium data in Fig. 4, there is very little contribution from quenching in the unfolded state and the decay rate is very close to ks of the model in Fig. 2. As the unfolded population increases with increasing denaturant, the decays become faster and biphasic due to both quenching of the tryptophan triplet in the unfolded molecules and to unfolding and refolding during the lifetime of the triplet.
Fig. 6.
Triplet-lifetime results. Normalized tryptophan triplet–triplet absorbance at 440 nm as a function of time on a log scale at 10 °C for 100 μM solutions of Cys-HP35(Nle24,Nle29) containing 20 mM sodium acetate, 1 mM TCEP, and either (A) 2.25 M GdmCl (blue), (B) 4.5 M GdmCl (green), or (C) 6 M GdmCl (red). The absorbance is proportional to the sum of the populations of the triplet state in the folded and unfolded states. The circles are the experimental data and the lines are the fits with the kinetic model.
The continuous curves through the data points are the fits using the kinetic model in Fig. 2 for the decay of the triplet-state population. The differential equations that describe the model and the details of the fitting are given in Materials and Methods.
Comparison of Folding Rates from Laser T-Jump with Fluorescence Detection and Triplet-Lifetime Experiments.
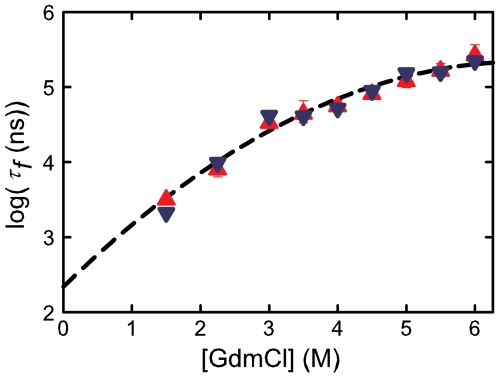
Fig. 7 compares the folding rates from the laser T-jump with fluorescence detection and triplet-lifetime experiments at 10 °C and pH = 4.9. The rates agree to within a very small experimental error at all concentrations, with an average difference between the results of approximately 20%. A polynomial extrapolation of all the data to zero denaturant yields a folding time of 220 (+100,-70) ns.
Fig. 7.
Comparison of folding times measured by temperature jump and tryptophan triplet lifetime at 283 K. (A) Folding times (points) as a function of GdmCl concentration and a quadratic fit to all the data (dashed line). The triangles (red points) correspond to folding times measured by temperature jump from experiments on Cys-HP35(Nle24,His27,Nle29), whereas the inverted triangles (blue points) are folding rates from the triplet-quenching experiment performed on Cys-HP35(Nle24,Nle29). It was not possible to obtain measurable amplitudes in temperature-jump measurements at 1.5 M GdmCl at 283 K; this data point was obtained by extrapolation from higher temperatures of observed rates by using an Arrhenius temperature dependence (Fig. S3 and Fig. S4). The parameters of the quadratic fitting function, log(τf) = α + βx + γx2, are α = 2.34 ± 0.17, β = 0.89 ± 0.10, and γ = -0.066 ± 0.013. In this polynomial fit using the program MLAB the relative weights of the T-jump points were determined by the standard deviation from the average of three measurements at each denaturant concentration, whereas for the triplet-lifetime data, the relative weight was determined by the rms residuals between the data and the two-state model fit. The relative weights at the denaturant concentrations 1.5, 2.25, 3,3.5, 4, 4.5, 5.5, and 6 M for the triplet lifetime data are 0.218, 0.311, 0.608, 0.370, 0.337, 0.430, 1.0, 0.430, and 0.508 and for the T-jump data are 1.0, 0.414, 0.817, 0.146, 0.297, 0.673, 0.236, 0.259, and 0.178 . Because of the very small amplitude for the faster relaxation at 1.5 M GdmCl in the triplet-lifetime experiment (Table S2), this point was not included in the extrapolation to zero denaturant.
Discussion
Summary of Experimental Results.
We employed two different experimental methods to determine the folding rate of the double-norleucine mutant of the ultrafast folding villin subdomain. The T-jump method with tryptophan fluorescence detection monitors the change in the population of the folded state, and is based on the increased quenching of the singlet state of Trp23 upon folding that results from the decrease in the distance from the protonated histidine, one turn away on the helix (Fig. 1). The triplet-lifetime method, on the other hand, monitors the change in the population of the unfolded state, and is based on quenching of the triplet state of tryptophan resulting from contact formation with a cysteine added to the N terminus, a contact that cannot form in the folded structure. Although an amino acid difference at position 27 between the sequences (Asn, His) was required to compare rates from the T-jump and triplet-lifetime measurements (Results), the equilibrium denaturant unfolding curves (Fig. 4) are almost identical, indicating that this difference should produce less than about a factor of two difference in the folding rates, assuming that there is a negligible difference in the interaction of the ground and triplet electronic states of Trp23 with the surrounding residues.
The populations of both singlet and triplet states of tryptophan decrease markedly with temperature, so the experiments were carried out at 283 K to maximize the signal-to-noise for the most precise comparison of the two methods, yet still be close to the temperatures of the molecular dynamics simulations of 300 K.
The data were analyzed in terms of a two-state model. There is an additional relaxation in the T-jump measurements, which results from a conformational rearrangement in the folded well (19, 26). However, it occurs at approximately 70 ns, which is sufficiently separated in time to not influence the folding time obtained in a two-state analysis where the folding time is longer than 1 μs. The folding times from the two methods are compared in Fig. 7. The agreement is excellent over the entire range of denaturant concentrations, with an average difference in the folding time between the two methods of approximately 20%, providing strong evidence that the slower of the two fluorescence phases does indeed monitor folding. Moreover, the agreement of the two methods provides a posteriori justification for the use of the two-state model in the analysis. Unlike most slower-folding two-state proteins, however, the logarithm of the folding time is not linearly dependent on the denaturant concentration. To obtain a folding time in the absence of denaturant, we therefore employed an empirical polynomial fit to all the data that yields an extrapolated value of 220 (+100,-70) ns (Fig. 7). Although the uncertainty in this number may be larger because of an assumed extrapolation function, this folding time is even shorter than the estimated theoretical speed limit for folding of N/100 μs ≈ 350 ns (14), suggesting that the double-norleucine mutant has reached the barrierless regime in the absence of denaturant at 283 K.
To further confirm our determination of the folding time, it would be important to carry out additional experiments such as laser T-jump with monitoring by infrared spectroscopy to compare with fluorescence experiments. Relaxation rates obtained by fluorescence and infrared detection can then be directly compared on the identical sequence without any assumptions or modeling, as done for the wild type (Fig. 3) (39).
Comparison of Experimental Results with Simulations.
Among the most extensive molecular dynamics simulations of folding to date for any protein are those performed by Ensign et al. for HP35(Nle24,His27,Nle29). A total of 410 trajectories, starting from nine different unfolded conformations, with a total simulation time of 354 µs, carried out at 300 K in explicit water produced approximately 70 successful folding trajectories (27). The authors tried to make contact with experiments by investigating the behavior of the property being measured in the experiments, in this case the fluorescence of the lone tryptophan. They employed a simple model for the quantum yield, similar to what was used in the successful fitting of equilibrium and kinetics data for HP35(His27) using an Ising-like statistical mechanical model (19, 47, 48). In both the theoretical model and the simulations, the lower quantum yield for Trp23 fluorescence in the folded state was implicitly assumed to result from an increased electron transfer rate in the excited singlet state solely due to the decreased distance from the protonated histidine, one turn away on the C-terminal helix. Ensign et al. assumed that there are only two quantum yields for the tryptophan, corresponding to conformations where the Trp23-His27 distance is either less than or greater than 0.75 nm.
To compare with the experimental results, Ensign et al. (27) plotted the fraction of molecules with a Trp23-His27 distance less than 0.75 nm, and found biexponential kinetics. For the majority of the trajectories (the so-called “gamma group”) they found significant differences (2- to10-fold) between the slower fluorescence phase and the times for forming the completely folded conformation using a stringent criterion in which a structure was considered folded if all three helices are intact and all three phenylalanine residues in the core are making their native contacts. To explain this discrepancy, Ensign et al. suggested that fluorescence is completely missing slowly folding conformations because it is insensitive to the structural changes associated with global folding from these starting structures. Freddolino and Schulten (30) also concluded that the fluorescence does not monitor folding. Their conclusion was based on three trajectories for HP35(His27) (Fig. 1) and six for HP35(Nle24,His27,Nle29), where in the four successful folding trajectories the Trp23-His27 distance reached the native state value approximately fivefold earlier than the folding of the entire structure.
Differences in rates such as those observed in the simulations should give rise to readily detectable differences between rates determined by fluorescence and an independent method that does not depend on the Trp23-His27 distance, in sharp contrast to our experimental results. The difference between the simulations and the experiments might be related to problems in calculating experimental quantities from the molecular dynamics trajectories. The use of the tryptophan–histidine distance as a metric for the fluorescence quantum yield may be an oversimplification. According to the quantum chemical calculations of Callis and coworkers, the rate of electron transfer from tryptophan to the amide backbone group of the tryptophan is sensitive to the local electrostatic environment (49–51). In this mechanism, the quantum yield is not simply determined by the formation of the helical turn that brings the protonated histidine within 0.75 nm.
Another source of differences between the experiments and simulations could be the inaccuracies in the force field. Bowman et al. (29, 52) used a Markov state model to analyze the trajectories of Ensign et al. (27). A biexponential fit to the (blue) folded population vs. time plot in figure 6 of ref. 29 (which is apparently replotted as unfolded population vs. time and rescaled in figure S6 of ref. 52) yields a fraction folded at infinite time of 0.12, compared to the experimental value of > 0.99 (24). These fractions correspond to equilibrium constants that differ by almost four orders of magnitude. Because the protein is so unstable the relaxation times from these populations plots correspond closely to unfolding, and should therefore not be compared with experimental relaxation times at 300 K as done in ref. 52, where the slower phase in the experiment corresponds to folding. The plots are nonexponential, so to obtain a rough estimate of folding and unfolding times, we assume a two-state model and use the mean relaxation time of approximately 330 ns from the integral of the plot in figure S6 of ref. 52, to calculate folding and unfolding times of 2 µs and 380 ns, respectively, compared to approximately 0.7 µs and 1 ms determined experimentally (24). Determination of rates unambiguously from simulations would not be an issue if trajectories were sufficiently long that many unfolding and folding transitions are observed in individual trajectories. If the waiting times observed in the trajectories are the same for a number of different order parameters, then the mean waiting time would determine the rate coefficients unambiguously, as was found for a small all-beta protein in the recent study of Shaw and coworkers (53).
Materials and Methods
Materials.
The 36-residue villin subdomain, referred to throughout as Cys-HP35(Nle24,His27,Nle29) or Cys-HP35(Nle24,Nle29) has the sequence C LSDED FKAVF GMTRS AFANL PLW(Nle)Q QH(or N)L(Nle)K EKGLF, where a cysteine has been added to the N terminus and we retained the numbering of HP35 with leucine in position 1. Norleucines (Nle) replaced the lysines in positions 24 and 29. The peptides were synthesized by standard solid-phase fluorenylmethoxycarbonyl chemistry on an Applied Biosystems peptide synthesizer AB433A, and purified to better than 95% by HPLC, as judged by mass spectrometry.
Kinetic Model for Triplet-Lifetime Experiments.
The differential equations that describe the model in Fig. 2 are:
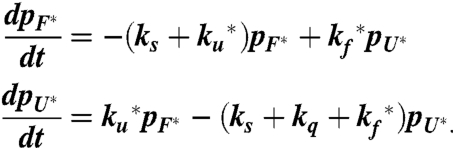 |
[1] |
The solution of these equations for the total fractional population of the triplet state as a function of time, which is proportional to the measured triplet–triplet absorbances, A(t)/A(0), with the initial conditions (t = 0)
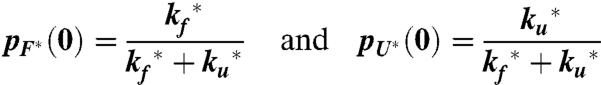 |
[2] |
is
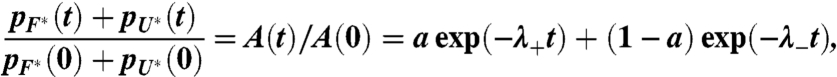 |
[3] |
where
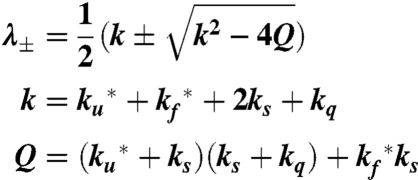 |
[4] |
and
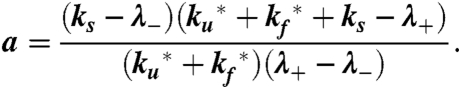 |
[5] |
Notice that for  , and
, and 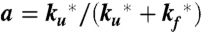 , so combining the amplitude a and slower rate λ- yields both folding and unfolding rate coefficients (Fig. 2).
, so combining the amplitude a and slower rate λ- yields both folding and unfolding rate coefficients (Fig. 2).
The model contains four adjustable parameters: the rate coefficients for folding and unfolding (with tryptophan in the triplet state), 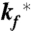 and
and 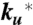 ; the spontaneous rate of decay of the tryptophan triplet to the ground electronic state in the absence of quenching by the N-terminal cysteine, ks; and the rate of decay of the tryptophan triplet state to the ground electronic state in unfolded molecules resulting from quenching by the cysteine, kq. The experimental data were modeled by calculating the total fractional population of the triplet excited state as a function of time and optimizing the parameters of the model to minimize the residuals between the calculated and observed progress curves. To reduce the interdependency of the parameters in the fit, the ratio
; the spontaneous rate of decay of the tryptophan triplet to the ground electronic state in the absence of quenching by the N-terminal cysteine, ks; and the rate of decay of the tryptophan triplet state to the ground electronic state in unfolded molecules resulting from quenching by the cysteine, kq. The experimental data were modeled by calculating the total fractional population of the triplet excited state as a function of time and optimizing the parameters of the model to minimize the residuals between the calculated and observed progress curves. To reduce the interdependency of the parameters in the fit, the ratio 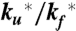 was constrained by a ground electronic state equilibrium constant (Keq = ku/kf) to which was assigned an explicit two-state dependence on denaturant concentration of the form Keq = exp{m([D] - [D]mid)/RT}. The adjustable parameters m and [Dmid] were tied to the measured equilibrium properties by simultaneously fitting a circular dichroism profile synthesized from the computed equilibrium constants (with an adjustable baseline offset) to the circular dichroism data in Fig. 4 (Fig. S5). The model therefore assumes that the folding and unfolding rates are unaffected by having tryptophan in its excited triplet state, and that the triplet–triplet absorption coefficient at 440 nm is the same in the folded and unfolded states. An additional assumption is that the spontaneous decay rate (ks) scales with viscosity, as observed for tryptophan alone, using the relation ks = ks0 η/η0, where ks0 and η0 are the values in the absence of denaturant, and the relative viscosity, η/η0, was assumed to be the same as that determined at 25 °C by Kawahara and Tanford (54). The fits were significantly improved by further assuming for the quenching rate kq a dependence on denaturant concentration of the form kq = kq0 + αq[D], with negative values for the coefficient αq.
was constrained by a ground electronic state equilibrium constant (Keq = ku/kf) to which was assigned an explicit two-state dependence on denaturant concentration of the form Keq = exp{m([D] - [D]mid)/RT}. The adjustable parameters m and [Dmid] were tied to the measured equilibrium properties by simultaneously fitting a circular dichroism profile synthesized from the computed equilibrium constants (with an adjustable baseline offset) to the circular dichroism data in Fig. 4 (Fig. S5). The model therefore assumes that the folding and unfolding rates are unaffected by having tryptophan in its excited triplet state, and that the triplet–triplet absorption coefficient at 440 nm is the same in the folded and unfolded states. An additional assumption is that the spontaneous decay rate (ks) scales with viscosity, as observed for tryptophan alone, using the relation ks = ks0 η/η0, where ks0 and η0 are the values in the absence of denaturant, and the relative viscosity, η/η0, was assumed to be the same as that determined at 25 °C by Kawahara and Tanford (54). The fits were significantly improved by further assuming for the quenching rate kq a dependence on denaturant concentration of the form kq = kq0 + αq[D], with negative values for the coefficient αq.
Supplementary Material
Acknowledgments.
We thank Wai-Ming Yau for peptide synthesis, Thang Chiu for preparation of Fig. 1, David Shaw for a preprint of his work, and Patrick Callis for helpful discussion. This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1019552108/-/DCSupplemental.
†The idea of determining the kinetics of fast processes, other than intramolecular contact formation, by monitoring triplet-state populations was first applied to the helix-coil transition by Lapidus et al. (35) and to protein folding by Buscaglia et al. (38) using intrinsic probes (tryptophan and cysteine), and has subsequently been used by Kiefhaber and coworkers using extrinsic probes (xanthone and naphthylalanine) to further investigate the helix-coil transition (41), as well as submicrosecond processes in the folding of the villin subdomain (42).
References
- 1.Jones CM, et al. Fast events in protein folding initiated by nanosecond laser photolysis. Proc Natl Acad Sci USA. 1993;90:11860–11864. doi: 10.1073/pnas.90.24.11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang GS, Oas TG. Submillisecond folding of monomeric lambda repressor. Proc Natl Acad Sci USA. 1995;92:6878–6882. doi: 10.1073/pnas.92.15.6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Phillips CM, Mizutani Y, Hochstrasser RM. Ultrafast thermally-induced unfolding of RNAse-A. Proc Natl Acad Sci USA. 1995;92:7292–7296. doi: 10.1073/pnas.92.16.7292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nolting B, Golbik R, Fersht AR. Submillisecond events in protein folding. Proc Natl Acad Sci USA. 1995;92:10668–10672. doi: 10.1073/pnas.92.23.10668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams S, et al. Fast events in protein folding: Helix melting and formation in a small peptide. Biochemistry. 1996;35:691–697. doi: 10.1021/bi952217p. [DOI] [PubMed] [Google Scholar]
- 6.Ballew RM, Sabelko J, Reiner C, Gruebele M. A single-sweep, nanosecond time resolution laser temperature-jump apparatus. Rev Sci Instrum. 1996;67:3694–3699. [Google Scholar]
- 7.Thompson PA, Eaton WA, Hofrichter J. Laser temperature jump study of the helix reversible arrow coil kinetics of an alanine peptide interpreted with a “kinetic zipper” model. Biochemistry. 1997;36:9200–9210. doi: 10.1021/bi9704764. [DOI] [PubMed] [Google Scholar]
- 8.Chan CK, et al. Submillisecond protein folding kinetics studied by ultrarapid mixing. Proc Natl Acad Sci USA. 1997;94:1779–1784. doi: 10.1073/pnas.94.5.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eaton WA, Munoz V, Thompson PA, Chan CK, Hofrichter J. Submillisecond kinetics of protein folding. Curr Opin Struct Biol. 1997;7:10–14. doi: 10.1016/s0959-440x(97)80003-6. [DOI] [PubMed] [Google Scholar]
- 10.Gruebele M. The fast protein folding problem. Annu Rev Phys Chem. 1999;50:485–516. doi: 10.1146/annurev.physchem.50.1.485. [DOI] [PubMed] [Google Scholar]
- 11.Lapidus LJ, Eaton WA, Hofrichter J. Measuring the rate of intramolecular contact formation in polypeptides. Proc Natl Acad Sci USA. 2000;97:7220–7225. doi: 10.1073/pnas.97.13.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eaton WA, et al. Fast kinetics and mechanisms in protein folding. Annu Rev Biophys Biomol Struct. 2000;29:327–359. doi: 10.1146/annurev.biophys.29.1.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferguson N, Fersht AR. Early events in protein folding. Curr Opin Struct Biol. 2003;13:75–81. doi: 10.1016/s0959-440x(02)00009-x. [DOI] [PubMed] [Google Scholar]
- 14.Kubelka J, Hofrichter J, Eaton WA. The protein folding “speed limit”. Curr Opin Struct Biol. 2004;14:76–88. doi: 10.1016/j.sbi.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 15.Dyer RB. Ultrafast and downhill protein folding. Curr Opin Struct Biol. 2007;17:38–47. doi: 10.1016/j.sbi.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 16.Munoz V, Henry ER, Hofrichter J, Eaton WA. A statistical mechanical model for beta-hairpin kinetics. Proc Natl Acad Sci USA. 1998;95:5872–5879. doi: 10.1073/pnas.95.11.5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Munoz V, Eaton WA. A simple model for calculating the kinetics of protein folding from three-dimensional structures. Proc Natl Acad Sci USA. 1999;96:11311–11316. doi: 10.1073/pnas.96.20.11311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henry ER, Eaton WA. Combinatorial modeling of protein folding kinetics: Free energy profiles and rates. Chem Phys. 2004;307:163–185. [Google Scholar]
- 19.Kubelka J, Henry ER, Cellmer T, Hofrichter J, Eaton WA. Chemical, physical, and theoretical kinetics of an ultrafast folding protein. Proc Natl Acad Sci USA. 2008;105:18655–18662. doi: 10.1073/pnas.0808600105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snow CD, Sorin EJ, Rhee YM, Pande VS. How well can simulation predict protein folding kinetics and thermodynamics? Annu Rev Biophys Biomol Struct. 2005;34:43–69. doi: 10.1146/annurev.biophys.34.040204.144447. [DOI] [PubMed] [Google Scholar]
- 21.Suits F, Pitman MC, Pitera JW, Swope WC, Germain RS. Overview of molecular dynamics techniques and early scientific results from the Blue Gene project. IBM J Res Dev. 2005;49:475–487. [Google Scholar]
- 22.Shaw DE, et al. Anton, a special-purpose machine for molecular dynamics simulation. Commun ACM. 2008;51:91–97. [Google Scholar]
- 23.Chiu TK, et al. High-resolution X-ray crystal structures of the villin headpiece subdomain, an ultrafast folding protein. Proc Natl Acad Sci USA. 2005;102:7517–7522. doi: 10.1073/pnas.0502495102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kubelka J, Chiu TK, Davies DR, Eaton WA, Hofrichter J. Submicrosecond protein folding. J Mol Biol. 2006;359:546–553. doi: 10.1016/j.jmb.2006.03.034. [DOI] [PubMed] [Google Scholar]
- 25.Bi Y, et al. Rational design, structural and thermodynamic characterization of a hyperstable variant of the villin headpiece helical subdomain. Biochemistry. 2007;46:7497–7505. doi: 10.1021/bi6026314. [DOI] [PubMed] [Google Scholar]
- 26.Kubelka J, Eaton WA, Hofrichter J. Experimental tests of villin subdomain folding simulations. J Mol Biol. 2003;329:625–630. doi: 10.1016/s0022-2836(03)00519-9. [DOI] [PubMed] [Google Scholar]
- 27.Ensign DL, Kasson PM, Pande VS. Heterogeneity even at the speed limit of folding: Large-scale molecular dynamics study of a fast-folding variant of the villin headpiece. J Mol Biol. 2007;374:806–816. doi: 10.1016/j.jmb.2007.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lei HX, Deng XJ, Wang ZX, Duan Y. The fast-folding HP35 double mutant has a substantially reduced primary folding free energy barrier. J Chem Phys. 2008;129:155104. doi: 10.1063/1.2995987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bowman GR, Beauchamp KA, Boxer G, Pande VS. Progress and challenges in the automated construction of Markov state models for full protein systems. J Chem Phys. 2009;131:124101. doi: 10.1063/1.3216567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freddolino PL, Schulten K. Common structural transitions in explicit-solvent simulations of villin headpiece folding. Biophys J. 2009;97:2338–2347. doi: 10.1016/j.bpj.2009.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang F, Settanni G, Fersht AR. Fluorescence resonance energy transfer analysis of the folding pathway of engrailed homeodomain. Protein Eng, Des Sel. 2008;21:131–146. doi: 10.1093/protein/gzm069. [DOI] [PubMed] [Google Scholar]
- 32.Mayor U, et al. The complete folding pathway of a protein from nanoseconds to microseconds. Nature. 2003;421:863–867. doi: 10.1038/nature01428. [DOI] [PubMed] [Google Scholar]
- 33.Gonnelli M, Strambini GB. Phosphorescence lifetime of tryptophan in proteins. Biochemistry. 1995;34:13847–13857. doi: 10.1021/bi00042a017. [DOI] [PubMed] [Google Scholar]
- 34.Lapidus LJ, Steinbach PJ, Eaton WA, Szabo A, Hofrichter J. Effects of chain stiffness on the dynamics of loop formation in polypeptides. Appendix: Testing a 1-dimensional diffusion model for peptide dynamics. J Phys Chem B. 2002;106:11628–11640. [Google Scholar]
- 35.Lapidus LJ, Eaton WA, Hofrichter J. Measuring dynamic flexibility of the coil state of a helix-forming peptide. J Mol Biol. 2002;319:19–25. doi: 10.1016/S0022-2836(02)00193-6. [DOI] [PubMed] [Google Scholar]
- 36.Buscaglia M, Schuler B, Lapidus LJ, Eaton WA, Hofrichter J. Kinetics of intramolecular contact formation in a denatured protein. J Mol Biol. 2003;332:9–12. doi: 10.1016/s0022-2836(03)00891-x. [DOI] [PubMed] [Google Scholar]
- 37.Buscaglia M, Lapidus LJ, Eaton WA, Hofrichter J. Effects of denaturants on the dynamics of loop formation in polypeptides. Biophys J. 2006;91:276–288. doi: 10.1529/biophysj.105.071167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buscaglia M, Kubelka J, Eaton WA, Hofrichter J. Determination of ultrafast protein folding rates from loop formation dynamics. J Mol Biol. 2005;347:657–664. doi: 10.1016/j.jmb.2005.01.057. [DOI] [PubMed] [Google Scholar]
- 39.Bunagan MR, Gao JM, Kelly JW, Gai F. Probing the folding transition state structure of the villin headpiece subdomain via side chain and backbone mutagenesis. J Am Chem Soc. 2009;131:7470–7476. doi: 10.1021/ja901860f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang MH, et al. Dynamic NMR line-shape analysis demonstrates that the villin headpiece subdomain folds on the microsecond timescale. J Am Chem Soc. 2003;20:6032–6033. doi: 10.1021/ja028752b. [DOI] [PubMed] [Google Scholar]
- 41.Fierz B, Reiner A, Kiefhaber T. Local conformational dynamics in alpha-helices measured by fast triplet transfer. Proc Natl Acad Sci USA. 2009;106:1057–1062. doi: 10.1073/pnas.0808581106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reiner A, Henklein P, Kiefhaber T. An unlocking/relocking barrier in conformational fluctuations of villin headpiece subdomain. Proc Natl Acad Sci USA. 2010;107:4955–4960. doi: 10.1073/pnas.0910001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Godoy-Ruiz R, et al. Estimating free energy barrier heights for an ultrafast folding protein from calorimetric and kinetic data. J Phys Chem B. 2008;112:5938–5949. doi: 10.1021/jp0757715. [DOI] [PubMed] [Google Scholar]
- 44.Bryngelson JD, Onuchic JN, Socci ND, Wolynes PG. Funnels, pathways, and the energy landscape of protein-folding—a synthesis. Proteins: Struct Funct Genet. 1995;21:167–195. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]
- 45.Liu F, et al. A one-dimensional free energy surface does not account for two-probe folding kinetics of protein alpha D-3. J Chem Phys. 2009;130:061101. doi: 10.1063/1.3077008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lia P, Oliva FY, Naganathan AN, Munoz V. Dynamics of one-state downhill protein folding. Proc Natl Acad Sci USA. 2009;106:103–108. doi: 10.1073/pnas.0802986106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cellmer T, Henry ER, Hofrichter J, Eaton WA. Measuring landscape roughness in ultrafast folding kinetics. Proc Natl Acad Sci USA. 2008;105:18320–18325. doi: 10.1073/pnas.0806154105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cellmer T, Henry ER, Kubelka J, Hofrichter J, Eaton WA. Relaxation rate for an ultrafast folding protein is independent of chemical denaturant concentration. J Am Chem Soc. 2007;129:14564–14565. doi: 10.1021/ja0761939. [DOI] [PubMed] [Google Scholar]
- 49.Callis PR, Liu TQ. Quantitative prediction of fluorescence quantum yields for tryptophan in proteins. J Phys Chem B. 2004;108:4248–4259. [Google Scholar]
- 50.Callis PR, Petrenko A, Muino PL, Tusell JR. Ab initio prediction of tryptophan fluorescence quenching by protein electric field enabled electron transfer. J Phys Chem B. 2007;111:10335–10339. doi: 10.1021/jp0744883. [DOI] [PubMed] [Google Scholar]
- 51.Muino PL, Callis PR. Solvent effects on the fluorescence quenching of tryptophan by amides via electron transfer. Experimental and computational studies. J Phys Chem B. 2009;113:2572–2577. doi: 10.1021/jp711513b. [DOI] [PubMed] [Google Scholar]
- 52.Bowman GR, Pande VS. Protein folded states are kinetic hubs. Proc Natl Acad Sci USA. 2010;107:10890–10895. doi: 10.1073/pnas.1003962107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shaw DE, et al. Atomic-level characterization of the structural dynamics of proteins. Science. 2010;330:341–346. doi: 10.1126/science.1187409. [DOI] [PubMed] [Google Scholar]
- 54.Kawahara K, Tanford C. Viscosity and density of aqueous solutions of urea and guanidine hydrochroide. J Biol Chem. 1966;241:3228–3232. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.