SUMMARY
Flap endonuclease (FEN1), essential for DNA replication and repair, removes RNA and DNA 5′-flaps. FEN1 5′ nuclease superfamily members acting in nucleotide excision repair (XPG), mismatch repair (EXO1) and homologous recombination (GEN1) paradoxically incise structurally distinct bubbles, ends or Holliday junctions, respectively. Here structural and functional analyses of human FEN1:DNA complexes show structure-specific, sequence-independent recognition for nicked dsDNA bent 100° with unpaired 3′- and 5′-flaps. Above the active site, a helical cap over a gateway formed by two helices enforces ssDNA threading and specificity for free 5′-ends. Crystallographic analyses of product and substrate complexes reveal that dsDNA binding and bending, the ssDNA gateway, and double-base unpairing flanking the scissile phosphate control precise flap incision by the two-metal-ion active site. Superfamily conserved motifs bind and open dsDNA, direct the target region into the helical gateway permitting only non-base-paired oligonucleotides active site access, and support a unified understanding of superfamily substrate specificity.
INTRODUCTION
Flap endonucleases (FENs) play key roles in cells in all domains of life by acting in lagging strand DNA replication and long-patch base excision repair (LP-BER). Both processes generate 5′-single stranded (ss) DNA or RNA in bifurcated structures known as 5′-flaps. FEN1 removes 5′-flaps without regard to sequence using its divalent metal ion-dependent phosphodiesterase activity (Tomlinson et al., 2010). Consistent with its crucial role in DNA replication, human FEN1 is highly expressed in all proliferative tissues. Yet, many cancers show even higher levels of FEN1 expression, in several cases correlated with tumor aggressiveness (Finger and Shen, 2010). Thus, FEN1 specific inhibitors have chemotherapeutic potential (Tumey et al., 2005). Although FEN1 deletion (FEN1-/-) is embryonically lethal in mice (Larsen et al., 2003), insights into its biology come from yeast FEN1 nulls (Reagan et al., 1995), which are viable but have mutator phenotypes indicating severe genomic instability (Liu et al., 2004; Navarro et al., 2007). Homozygous FEN1 null DT40 chicken cells are viable but sensitive to oxidative DNA damaging agents (Matsuzaki et al., 2002), consistent with FEN1's role in LP-BER. FEN1 is also implicated in mitochondrial genome maintenance (Kalifa et al., 2009). Furthermore, mutations that decrease expression levels or alter FEN1 biochemical properties predispose humans and mice to cancers (Finger and Shen, 2010).
FEN1 efficiency and specificity is critical as human DNA replication generates ~50 million Okazaki primers each cell cycle. Failure to precisely remove these primers creates gaps or overlaps that would prevent efficient ligation, delay cell division and initiate post-replicative repair mechanisms, endangering genomic fidelity (Debrauwere et al., 2001). FENs possess multiple nucleic acid hydrolytic activities in vitro including cleavage of double flaps (5′- and 3′-flaps) that mimic equilibrating replication intermediates (Lyamichev et al., 1999), nicked DNA with no 5′-flap both with and without a 3′-flap, and structures where the 5′-flap can form a fold back hairpin or gapped flaps (Finger et al., 2009). Other FEN1 activities may reduce repeat expansions (Singh et al., 2007). The cellular substrate for PCNA-scaffolded FEN1 action is an equilibrating double flap with a short 5′-ss DNA flap. Critical to replication efficiency, the FEN1 product from a double flap can be ligated without further processing (Liu et al., 2004). Although RNA and DNA phosphodiester bonds are exceptionally stable to attack by water or hydroxide (Schroeder et al., 2006), FEN1s enhance the hydrolysis rate of targeted phosphodiester bonds ~1017 fold. Indeed, the catalytic efficiency of FEN1 on its optimal substrate approaches enzyme:substrate association rates in solution (Finger et al., 2009). Thus, FEN1:DNA reactions may be diffusion controlled, as seen for other charged interactions like plastocyanin with cytochrome c (Roberts et al., 1991). Yet, the basis of FEN1's specificity and catalytic power are unknown.
FEN1:DNA recognition models generally focus on the 5′-ssDNA flap portion of the substrate, but whether the FEN1 threads the free end through the helical arch or the arch clamps the 5′-flap ssDNA without threading is debated (Chapados et al., 2004; Devos et al., 2007; Liu et al., 2004). After initial interaction with the free 5′ terminus, both models propose that FEN1 slides or tracks down the ssDNA to the ss-dsDNA junction whereupon incision occurs. Functional data involving modifications to the 5′-flap both support and contradict the tracking hypothesis showing that bulky alterations of the 5′-flap are in some cases tolerated but in others drastically retard reaction (Finger et al., 2009).
Paradoxically, the diverse specificity of sequence-related enzymes in the 5′ nuclease superfamily for substrates without 5′-ss termini does not fit the ssDNA-based mechanisms. EXOI, XPG, and GEN1, whose activities span multiple DNA repair pathways, are members of the 5′ nuclease superfamily with FEN1 (Lieber, 1997; Tomlinson et al., 2010). However, XPG and GEN1 cleave DNA bubbles and four-way junctions, respectively: substrates with ss-dsDNA junctions but lacking 5′-termini for threading (Ip et al., 2008; O'Donovan et al., 1994). So a general model that explains FEN1 specificity for 5′-ss termini (whether ssDNA or a 5′-gapped flap) while reconciling substrate diversity of the 5′ nuclease superfamily is lacking.
To help elucidate the molecular basis for FEN1 functions and resolve paradoxes regarding 5′ nuclease superfamily specificities, we solved three crystal structures of human FEN1 with DNA, allowing comparative analyses of complexes with substrate and product DNA. Coupled with mutational and biochemical analyses, these FEN1:DNA complex structures reveal an unexpected sophistication to the recognition mechanism. Two separate DNA binding sites located ~1 dsDNA turn apart impose severe substrate bending, a feature likely to be superfamily conserved. These DNA binding sites coupled to helical wedges require that the 5′-flap DNA, located adjacent to the DNA bend, enter through a gateway formed between two superfamily-conserved α-helices. Substantial conformational changes in the FEN1 regions that bind the 3′- and 5′-DNA flaps enforce structure-specific binding. The combined enzyme-DNA structures show that in the reactive conformation of a FEN1 substrate, three nucleotides (nts) of the double flap DNA are unpaired, one at the 3′-terminus and two flanking the scissile phosphodiester. The collective results support a mechanism for FEN1 that resolves apparently contradictory biochemical data regarding the 5′-ssDNA terminus, explains FEN1 substrate specificity, and accommodates the diverse biological specificities of the 5′-nuclease superfamily.
RESULTS
FEN1:DNA Structure Determinations and Overall Architecture
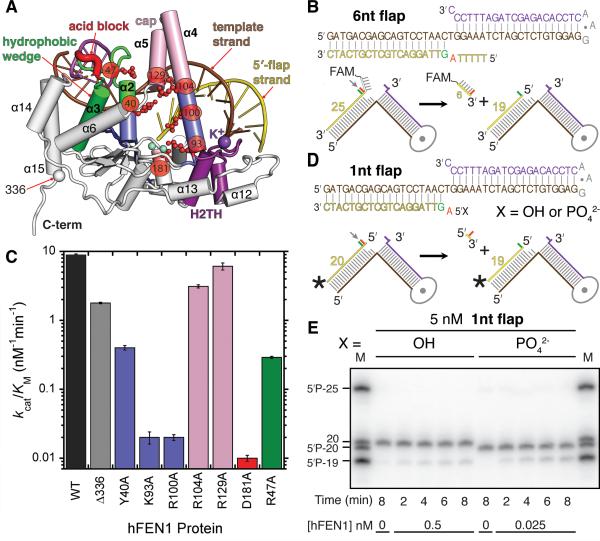
To characterize FEN1 recognition and incision of flap DNA substrates, we crystallized human FEN1 truncated after residue 336 (Δ336) and Δ336 combined with a D181A mutation as complexes with double flap DNA having either 1 or 4 nt 5′-flaps and with metal ion cofactors (Figure 1). FEN1 Δ336, which removes only the flexible, protruding PCNA binding motif and encompasses the entire catalytic domain, is termed WT for brevity. The active site D181A mutation used here severely retards incision (Shen et al., 1996). The DNA contains competing base-pairing purine-pyrimidine pairs at the DNA junction, but the C-A mismatch favors 1 nt 3′-flap formation (Figure 1A). Based on lagging-strand DNA replication, we term the strand paired to the flaps “template DNA”. We solved three structures for distinct FEN1:DNA complexes (Table S1): 1) D181A crystallized with a 1-nt flap DNA and K+ at 2.6 Å, 2) WT with a 1-nt flap DNA, K+, and Sm3+ at 2.3Å, and 3) WT with a 4-nt flap DNA, K+, Sm3+ and Mg2+ at 2.2 Å.
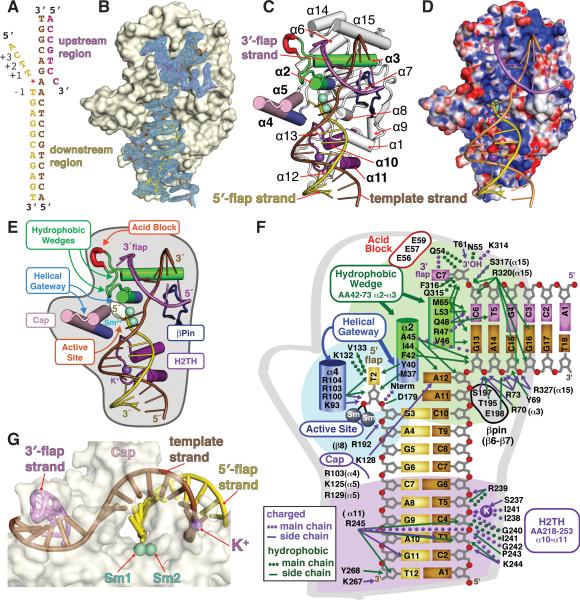
Figure 1. Binding of 5′-Flap DNA in FEN1:Sm3+:Product DNA Complex.
(All panels) FEN1-bound DNA includes 3′-flap strand (magenta), 5′-flap strand (purple), and template strand (brown).
(A) The 4 nt flap crystallization substrate numbered relative to scissile phosphate. The flap strand (+1 to +4 nts) is absent in the WT:Sm3+: product complex. The 1 nt flap substrate lacks the +2 to +4 nts.
(B) FEN1 surface and Fo-Fc electron density of DNA from PHENIX kick map shows clear density for 100° bent DNA bound by FEN1 (1.6σ map within 1.9 Å of DNA). Crystallographic statistics are in Table S1. FEN1 preferentially interacts with the template strand and the 3′- and 5′ -flap terminal nucleotides (Figure S1A)
(C) FEN1:DNA showing the structural elements involved in DNA binding and the bound DNA distortion. Key elements include the hydrophobic wedge that breaks DNA path and forms the 3′-flap binding site (green), acid block that inhibits longer 3′-flaps (red), helical gateway that permits only ssDNA and forms the active site (dark blue), helical cap that imposes the preference for 5′-termini (pink), and H2TH that binds to dsDNA upstream from 5′-flap (purple). Sm3+ (cyan) and K+ (purple) ions are shown as spheres. Comparison with FEN1:substrate complexes is shown in Figures S1B-D.
(D) Electrostatic surface (-52 mV to +52 mV) of FEN1 and cations shows how the template strand traverses the path of basic residues.
(E) Key structural elements in FEN1 involved in DNA recognition and incision.
(F) Schematic of all direct protein-DNA interactions shows that binding is concentrated to template strand and are not base-specific. Numbering is based on the pdb.
(G) A transparent FEN1 surface reveals binding sites to template dsDNA strands are spaced ~1 helical turn apart. Fixed by the bending point of DNA and the 3′ -flap binding, binding of the template strand positions the complementary strand (the 5′ -flap) at the gateway under the cap.
In both WT and D181A FEN1 with 1-nt ‘equilibrating’ flap structures, the DNA was uncleaved. In the highest resolution structure (WT with 4-nt flap, Sm3+ and Mg2+), the 5′-flap was incised 1 nt into the DNA duplex (Figure 1), consistent with FEN1 specificity. In the WT structures, two Sm3+ ions bind in the active site. In all three FEN1:DNA structures, the overall shape resembles a left-handed boxing glove (Figures 1 and S1). FEN1 is a mixed α/β structure containing a seven-stranded twisted β-sheet core plus 15 α-helices. The fold is interwoven. Multiple loops extend 12 to 50 Å across the domain, allowing residues throughout the sequence to act in DNA interactions. Helices α4 and α5 (pink, Figures 1C and 2), disordered in DNA-free FEN1 structures (Sakurai et al., 2005), cap the thumb of the glove. Flexible α2-α3 loop also orders in the DNA-bound form (green). The DNA (bent ~100°) contacts the palm and fingers along a positively-charged patch extending the length of the boxing glove (Figure 1D). The DNA bases and backbone atoms are well defined in the electron density (Figure 1B), with FEN1 binding ~16 basepairs of dsDNA plus 3′- and 5′-flap nts.
Figure 2. Sequence, Secondary Structure and Residue Function for FEN1 and the 5′ Nuclease Superfamily.
Map of FEN1 secondary structure, structural elements, and mutants to a sequence alignment of the FEN1 superfamily human members. XPG residues 121-750 were removed (dotted line) to facilitate alignment.
Recognition of the sharply bent DNA junction is mediated by four separate regions (Figure 1E): 1) the hydrophobic wedge composed of α2 and the α2-α3 loop at the junction between the upstream and downstream dsDNA portions of the two-way junction, 2) the 3′-flap binding pocket at the tip of the glove that interacts with the upstream dsDNA, 3) the Helix-2Turn-Helix (H2TH) motif at the at the glove base (“wrist”) with bound K+ ion and charged side chains that bind downstream dsDNA, and 4) the two-metal ion active site between thumb and forefinger that binds the 5′-flap strand.
FEN1 contacts the DNA primarily with helix and loop structures. Most interactions are to template strand and terminal flap nts (Figures 1F and S1A). FEN1 with metal ions has a 1,828 Å2 interface with product DNA. The terminal nts of the flaps form a third of the interface: 221 Å2 for 3′-flap and 388 Å2 for 5′-flap. Over half the interface is to the template strand. Notably, there are few interactions to the 5′-flap strand, except at the active site. Mapping the key structural elements observed in human FEN1 to a sequence alignment of superfamily members EXO1, XPG, and GEN1 shows that key residues in the gateway (Lys93, Arg100, Arg104) and H2TH (Ile238 and Ile241) are conserved, while the cap sequence is FEN1- and EXO1-specific (Figure 2).
Template Strand Binding by H2TH:K+ Bridge and Hydrophobic Wedges
The primary FEN1 interaction with DNA is to the template strand in two physically separated regions. The H2TH:K+ ion and four extended basic residues (Lys267, Arg239, Arg245, Lys244) comprise the largest FEN1:DNA interface (Figures 1F and 3A). The K+ ion is coordinated by Ile238 and Ile241 backbone carbonyl oxygens and Ser237 hydroxyl in turns formed between α10 and α11 of the H2TH motif (purple). In archaeal FEN1, this motif has three turns (H3TH) (Hosfield et al., 1998). As ion-ligand distances for Na+, typically ~2.3 Å (Kuppuraj et al., 2009), would prevent intimate DNA contact, the closest DNA-K+ distance of 2.7 Å suggests specificity for K+. The four basic residues form a track for the DNA minor groove with Arg245 acting as a rail to position the 5′-flap strand. The DNA interface is not continuous, and the template strand arcs away from the protein surface between the H2TH and near the active site and back down to the second binding region (Figure 1G). There, the template makes an abrupt ~100° bend. The bend is positioned between a track formed by two protruding regions: the α2 helix and α2-α3 loop hydrophobic wedges and the β6-β7 loop, which we term the β-pin (Figure 1E).
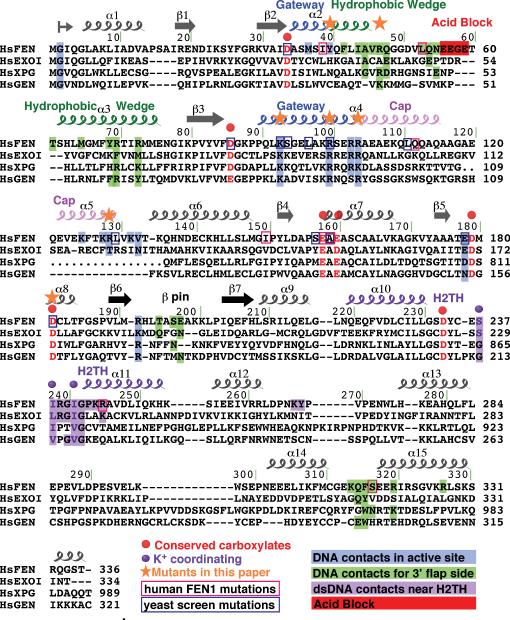
Figure 3. Key FEN1 Structural Elements in the FEN:Sm3+:Product Complex.
(A) The H2TH:K+ motif and surrounding basic residues (purple) forms an electrostatic track for the downstream dsDNA minor groove.
(B) The 3′-flap binding pocket (green) binds the 3′-hydroxyl and the unpaired nt sugar moiety.
(C) Rear view of the active site, opposite the template DNA interacting region, shows the helical gateway (blue), active site residues and product DNA. The cleaved 5′-nt sits in the helical gateway formed by α2 and α4 and under the cap. Figures S2A-C show C-capping in the gateway.
(D) Front view of the active site shows the gateway and 5′-flap strand approach to the active site.
(E) FEN1 binds to the -1 to -3 phosphates of the 5′ -flap product strand.
(F) Four conserved FEN1 carboxylate residues and two phosphate oxygens of the +1 5′-phosphate in the DNA directly coordinate the two Sm3+ ions. Distances are 2.3 – 2.5 Å. Figure S2D shows all seven highly conserved carboxylates and bound waters.
Helix α2 blocks the dsDNA path on either side, packing against the template strand unpaired A12 base (Figures 1C and 1F). The α2-α3 loop is wedged against the first basepair of the upstream dsDNA adjacent to the 3′-flap. This region was disordered in the DNA-free protein (Sakurai et al., 2005), so our structures indicate a substrate-induced, disorder-to-order transition. The β-pin, positioned closer to the active site in the DNA-free enzyme, moved to accommodate the template strand in the complex. Arg320 intercalates into the minor groove near the 3′-flap, ratcheting the upstream dsDNA in place (Figure 3B).
3′-Flap Binding
Binding of the unpaired 3′-flap nt comprises one-eighth of the FEN1:DNA binding surface, explaining the 33-fold increase in nuclease activity seen for 5′-flap substrates with an unpaired 3′-flap compared to substrates without a 3′ -flap (Finger et al., 2009). Ten residues surround the 3′-flap, similar to the A. fulgidus FEN structure with only 3′-flap upstream dsDNA (Chapados et al., 2004), indicating this is a conserved FEN1 feature from archaea to humans and extending biochemical results (Friedrich-Heineken and Hubscher, 2004) (Figures 1F and 3B). The 3′-hydroxyl forms a bifurcated hydrogen bond with the Lys314 backbone carbonyl and Thr61 hydroxyl. Five main chain and side chain residues form van der Waals interactions to the 2-deoxyribose sugar plus two interactions with the 3′-flap phosphate. The Gln54 amide nitrogen hydrogen bonds to cytosine O2, a base-independent minor groove hydrogen bond acceptor. The upstream dsDNA conforms to B-DNA characteristics. A eukaryotic FEN1-specific loop (the EEGE “acid block”, residues 56-59), blocks DNA from passing beyond the one base pocket by charge repulsion (red, Figures 1C and 1F).
5′-Flap Binding
In the product DNA complex, the -1 terminal nt on the 5′-flap strand is unpaired from the template DNA and inside the helical gateway formed by α2 and α4 (Figures 3C and 3D). This same -1 nt unpairing was also seen in a low resolution product:Mn2+ structure (Supplemental Methods). The ordered cap over the helical arch leaves the gateway open but limits active site access to ends. The α2 and α4 gateway helices are ~13-15 Å apart (Cβ-Cβ distances), permitting ssDNA passage while prohibiting entry by dsDNA, which requires ~20 Å. The 5′-flap DNA position is fixed relative to the 100° DNA bend and by the two template strand binding sites a helical turn apart. The bound template strand forms an arc under which the 5′-flap strand must pass (Figure 1G).
In the active site, the 5′-monophosphate of the cleaved product nt (-1) is coordinated by two Sm3+ ions and interacts via salt bridges with α4 Lys93 and Arg100 (Figures 3C and 3D). These basic residues are positioned to facilitate electrostatic rate acceleration of the phosphodiester hydrolysis, as seen in T5 Flap Endonuclease (Sengerova et al., 2010). The -1 base stacks with Tyr40, which is conserved in FENs but not in the superfamily. Hydrophobic contacts from Arg100, Lys132, and Val133 orient the -1 nt, but are base independent. The phosphate 3′ to the cleaved 5′-phosphate (-2 phosphate) is coordinated by the N-terminus (Gly2) (NT, Figures 1F and 3E). Met1 is removed due to glycine at position 2 (Sherman et al., 1985). Gly2 is conserved in the eukaryotic 5′ nuclease superfamily (Figure 2), but a similar role may be played by an additional metal ion in phage members (Syson et al., 2008). The -3 phosphate forms a salt bridge with Arg192, also superfamily-conserved. Overall, downstream dsDNA of the enzyme-product complex adopts B-DNA helical parameters except for nt -2, which has a C3′-endo sugar pucker and distortions in backbone dihedral angles due to the sharp bend between nts -1 and -2.
The two Sm3+ ions (Sm1 and Sm2) occupy Mg2+ ion sites seen in the FEN1 DNA-free structure (Sakurai et al., 2005). Sm1 is coordinated by Asp86, Glu160, and two oxygens of the cleaved 5′-monophosphate (Figure 3F). Sm2 is coordinated by Glu160, Asp179, Asp181 and one phosphate oxygen. Asp34, Glu158, and Asp233 interact with Sm3+ via waters (Figure S2). Seven conserved active site carboxylates are characteristic of the FEN superfamily (Figure 2).
Above the active site, the so-called helical arch or clamp region (α4 and α5) is ordered. In DNA-free structures, this region was mostly disordered. To distinguish the superfamily-conserved gateway from the non-conserved arch region, we define the C-terminal part of α4 and all α5 as the helical cap (pink, Figure 1). Three interactions appear to stabilize and help nucleate an ordered conformation of the helical cap. 1) α5 basic residues (Lys125, Lys128, and Arg129) bind the template strand (Figure 1F). 2) Arg47 and Arg104 guanidiniums C-cap the C-terminal ends of α2 and α5, respectively (Figures S2A-C). 3) Arg47 links occupation of the 3′-flap pocket to ordering α4-α5. Arg47 main chain, in the hydrophobic wedge, packs against the dsDNA adjacent to the 3′-flap, while its side chain stacks with α5 Lys128.
Double Base Unpairing Flanking the Scissile Phosphate
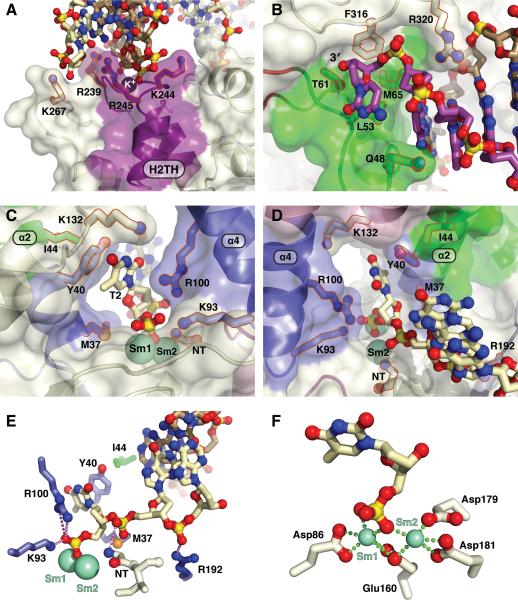
In the WT and D181A substrate structures, the two flap nts +1 and -1 on either side of the scissile phosphate are basepaired to the template strand, unlike the product complex (Figures 4A-D). Tyr40 is stacked with the +1 base in the substrate complexes, but rotates 25° to stack with the -1 base in the product complex. Ordering of the α4-α5 helical cap and the α2-α3 hydrophobic wedges in our substrate complexes (Figures S1B-D) indicates that placement of the scissile phosphate into the active site pocket is not a prerequisite for cap formation. The scissile phosphate in the WT:Sm3+-substrate complex is 5.2 Å from the cleaved 5′-phosphate in the product complex. In the D181A:substrate complex, the scissile phosphate is even further away from the active site (7.7 Å), likely due to the absence of electrostatic forces provided by active site metal ions. The template strand is displaced only slightly between the product and substrate complexes due to its interaction with α2, the hydrophobic wedge, and the β pin. As one flap base and two template strand bases are unpaired in the product complex, two bases of substrate must unpair to position the scissile phosphate between the divalent metal ions. These results reveal an important, hitherto unknown specificity step in the mechanism between binding and catalysis (Figure 4E). In the substrate complexes, the region bound by the H2TH and K+ ion conforms to B-DNA parameters, whereas the dsDNA near the active site deviates from B-DNA geometry. In fact, the +1 basepair (A-T) in the substrate forms only one of two possible H-bonds due to a pronounced base pair opening of ~20° and stagger of 0.61 Å towards the major groove. Collectively, these deviations suggest that FEN1 induces deformations in the downstream dsDNA base pairs closest to the active site to promote the requisite unpairing to provide specific incision one base into the dsDNA region.
Figure 4. Substrate and Product Complex DNA Structure Comparison Reveals a Novel Double Base Unpairing Mechanism for Scissile Phosphate Placement.
(A) DNA from WT:substrate and product complexes show the +1 and -1 nts are paired in the substrate but the -1 nt in the product is unpaired with Sm3+ ions (green spheres, panels A, C-E).
(B) Close-up of the DNA and Tyr40 from the D181A:substrate complex showing the basepairing of the +1 and -1 nts and Tyr40 stacking with the +1 nt. The overlay with the product structure outline highlights the 7.7 Å movement of the scissile phosphate of the -1 nt (green) needed for catalysis.
(C) As in (B), the +1 and -1 nt are basepaired and Tyr40 stacks with the +1 nt in the Wt:Sm3+:substrate. The scissile phosphate of the -1 nt would need to move 5.2 Å into the active site.
(D) Close-up view of the DNA and Tyr40 from the WT:Sm3+:product DNA complex. Unlike the substrate complexes, the +1 nt has been cleaved off, the -1 nt is unpaired, and Tyr40 stacks with the -1 nt.
(E) Model of double nt unpairing to move substrate into a catalytic position for incision.
Implications for Location of the Uncleaved 5′-Portion of Substrates and Mechanism for Superfamily Phosphodiesterase Activity
Two Sm3+ ions coordinate the cleaved phosphate in the product complex (Figure 3F), supporting the role for two cofactor ions in FEN1 catalysis implied by functional data (Syson et al., 2008). To examine the catalytic mechanism, we modeled substrate in catalytic position at the metals. Assuming that the scissile phosphodiester of FEN1 substrates was positioned equivalently to 5′-phosphate product and that in-line attack of the hydroxide must occur, the phosphate oxygen between the two Sm3+ ions is probably the oxygen of the attacking nucleophile that is now covalently linked to the phosphate. Models of the +1 nt based on an in-line attack from the other two phosphate oxygens show a steric clash between the protein and modeled DNA. A similar model with two metals activating an attacking water is proposed for Zn2+-dependent EndoIV (Garcin et al., 2008; Ivanov et al., 2007).
Mutational Analyses
To further test functional implications from our structures, we made key mutants in the active site, the gateway, the cap, and the hydrophobic wedge. We postulated that mutation of residues that directly bind the cleaved phosphate in the product structure (Lys93, Arg100) or that coordinate the catalytic metals (Asp181) would severely retard activity, whereas those proposed to nucleate disorder-to-order transitions in the helical cap (Arg104, Arg47 and Arg129) or interact with the -1 base would also significantly reduce activity, but to a lesser extent. To test this, we measured the catalytic efficiency (kcat/KM) of full-length WT and mutant FEN1s using a double flap substrate in the presence of K+ and Mg2+ ions under conditions where [S] < KM (Figures 5A-C and S3). Overall, mutations cluster into three classes differing in degrees of severity.
Figure 5. Structure–Guided Site-Directed Mutagenesis of Active Site, Helical Gateway, Hydrophobic Wedge and Helical Cap Residues.
(A) FEN1:Sm3+:product complex fold (ribbons) marking the position of mutations in this study (colored as in Figure 1).
(B) Substrate used in (C) with schematic of the incision.
(C) Catalytic efficiency of WT and mutant FEN1 as bar graph showing the relative severity of mutations. Controls and examples of data are in Figure S3.
(D) 5′ -phosphate substrates used in (E) with schematic of the incision.
(E) Denaturing PAGE gel showing increased incision activity on substrates with a 5′-phosphate at the +1 position.
Mutations that severely impair catalysis include residues conserved throughout the FEN1 superfamily (Figure 2). Lys93 and Arg100 interact with the product 5′-phosphate monoester in our structures and probably stabilize the unpaired conformation of DNA substrates and act as electrostatic catalysts during hydrolysis (Figure 3C). K93A and R100A drastically reduced the rate of the FEN1 catalyzed reaction by >400-fold (Figure 5C). Asp181 coordinates Sm2 in our structures (Figure 3F). D181A mutation severely reduced activity by >800-fold. In the D181A structure, Ca2+ present during crystallization was not observed, suggesting that the D181A affects divalent metal sequestration.
Two mutations (Y40A and R47A) have an intermediate effect on FEN1 activity, likely by interfering with substrate binding. Tyr40 stacks against the +1 or -1 base in the substrate and product complexes, respectively (Figures 4B-D). The functional significance of Tyr40, conserved in archaeal, yeast and mammalian FEN1 has not been noted previously. Y40A reduced the reaction rate 20-fold, supporting a significant role in activity likely through substrate positioning. Arg47 acts in multiple interactions: contacting the first base pair on the 3′-flap side, C-capping α2, and making van der Waals interactions with Lys128 on α5 (Figure S2A). In studies without K+ and a single flap substrate, a FEN1 R47A mutation altered site specificity and reduced reaction rate 2-fold (Qiu et al., 2002). Here, with higher activity and specificity enforced by a double flap substrate and K+, the reaction rate catalyzed by R47A decreased 30-fold. A similar decrease is observed when comparing second order rate constants of substrates with or without a 3′-flap (Finger et al., 2009), supporting the idea that Arg47 links 3′-flap binding to the active site by facilitating helical cap ordering.
Other arch mutations in α4 and α5 show a modest effect on FEN1 catalytic efficiency. The R104A and R129A mutations have a decrease in kcat/KM of 3 and 1.5-fold, respectively. Arg104, conserved in most superfamily members, likely contributes to FEN1 action by C-capping α5 and facilitating unpairing of the flap strand by electrostatic pulling of the flap strand +1 and -1 phosphates (Figures S2B and S2C). Arg129 on α5, conserved in all FENs but not in the superfamily, interacts with the template strand in DNA-bound structures.
The incision activity of FEN1 Δ336, used for crystallography, was only reduced 5-fold in apparent second order rate constant compared to full-length. As reduction in activity may have been due to decrease in substrate affinity from loss of non-specific electrostatic interactions mediated by the Lys rich C-terminus (Friedrich-Heineken et al., 2003), we determined the steady state kinetic parameters. For Δ336, kcat was increased 1.7-fold and KM 7.5-fold compared to WT (Table S2), showing that the catalytic core used for crystallization is fully competent for catalysis and that the C-terminus aids non-specific DNA binding. Biologically, the reduction of the FEN1 off-rate by its C-terminus may aid handoff, with product release promoted upon the C-terminus binding PCNA:ligase I.
Unexpectedly, the 1 nt flap substrate (also called an EXO substrate) is neither cleaved nor unpaired in the WT:Sm3+ crystals, despite being a viable FEN1 substrate (Finger et al., 2009). Yet, FEN1 has greater activity on nicked substrates with a 5′-phosphate than those without (Wu et al., 1996), and the crystallization substrate had a 5′-hydroxyl. So we postulated that a negatively-charged phosphate at the +1 position was needed to promote unpairing, through DNA backbone repulsion and/or pulling by positive charges from the metal ions and active site basic residues. To test this, we crystallized FEN1 with Sm3+ and a 1 nt flap with a 5′-phosphate, and the electron density showed that the DNA was cleaved and coordinated to the Sm3+ ions (Figure S3H). We also determined that, in the presence of K+ and Mg2+, the normalized initial rate of reaction on a 1 nt flap (5 nM) with a 5′-phosphate was 2.13 min-1, 8.2-fold faster than on a flap with a 5′-hydroxyl (0.26 min-1) (Figures 5D and 5E). As a similar effect was reported for the related EXO1 enzyme (Lee and Wilson, 1999), the importance of the 5′-phosphate at the +1 position may be a general superfamily feature.
FEN1 Structural Elements and the 5′ Nuclease Superfamily
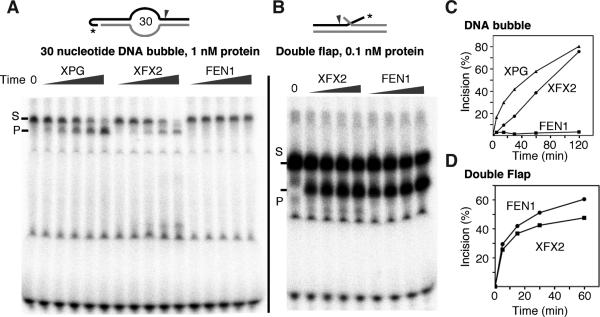
Many FEN1 structural elements appear conserved in the 5′ nuclease superfamily, but the helical cap is a FEN1 and EXO1 specific element that selects for 5′-termini. To test if we could transfer FEN1-specificity (imposed by the cap) to another family member, we designed a chimeric XFX2 construct, in which the 650 residue R-domain (also called spacer domain) of XPG is replaced by α4 and α5 sequence from an archaeal FEN arch (Sarker et al., 2005). As XPG can act on DNA bubbles, addition of a cap over the gateway in XFX2 should reduce or eliminate ability to cleave DNA bubbles.
We compared the cleavage efficiency of XPG, XFX2, and FEN1, on a 30 nt DNA bubble substrate (Figures 6 and S4). XPG but not FEN1 incised DNA bubbles, consistent with the known substrate specificities. As hypothesized, XFX2 had reduced (but not absent) activity on a bubble substrate compared to XPG. As a control, we determined that XFX2 is as proficient as FEN1 on flap DNA, indicating no inherent loss in catalytic activity. The retention of some DNA bubble incision activity by XFX2 suggests that although the helical cap motif contributes to substrate selectivity, it can be pushed back to allow some activity on DNA bubbles. Also XPG lacks an acid block that we predict provides a second direct obstacle to DNA bubbles in FEN1.
Figure 6. FEN1 Implications for XPG.
(A) Denaturing PAGE gel showing reduced bubble incision activity by XFX2. XPG, XFX2, and FEN1 incision activity of 30 nt DNA bubbles at 1 nM protein concentration. Incubation time was 5, 15, 30, 60, 120 min. Substrate sequences are in Figure S4.
(B) Denaturing PAGE gel showing XFX2 is active on 5′-flap DNA. FEN1 and XFX2 were incubated for 15, 30, and 60 min.
(C) Quantification of (A)
(D) Quantification of (B)
DISCUSSION
FEN1 Substrate Recognition and Catalysis
The results presented here reveal the extraordinary sophistication used by FEN1 to achieve specificity for its substrates, which in cells are usually short (1-6 nt) 5′-ss flaps on dsDNA (Burgers, 2009). First, FEN1 recognizes 5′ -flap DNA by its ability to form a sharp 100° bend with dsDNA on either side. Only dsDNA with a flap or break can bend at a single phosphodiester link to that degree. Second, a 3′ -flap binding pocket encloses a single unpaired nt. Biologically, this ensures that the FEN1 product is suitable for direct ligation. Mechanistically, the 3′ -flap and the DNA bend help register the incision position. Third, FEN1 requires the 5′ -flap to pass under the cap to enter the helical gateway and active site. Only DNA with free ends can do so. Fourth, FEN1 binds the template strand at the DNA bend and at the K+:H2TH, thereby positioning the strand complementary to it, the 5′ -flap strand, toward the active site. FEN1 thus employs the helical properties of dsDNA with twin binding sites one-helical turn apart (Figure 1G). Biologically, template-based positioning of the 5′ -flap prevents inadvertent incision of ssDNA or 3′- flaps and enforces specificity for ss-dsDNA junctions. Fifth, the entrance to the FEN1 active site is guarded on two sides by the α2-α4 helical gateway, which only permits passage of ssDNA and prevents dsDNA from entering the active site. The cap and gateway are separable elements: the FEN-specific cap selects for free ends and the superfamily-conserved gateway permits ssDNA/RNA access to the active site.
Importantly for replication, FEN1 must ensure incision takes place efficiently and specifically at the correct location needed to produce ligatable DNA. Specificity is achieved by a unique mechanism: double-base unpairing of the 5′-flap strand. The scissile phosphodiester is only positioned at the active site divalent metals after other specificity criteria are met and a disorder-to-order transition has occurred. The ordering of the cap and gateway upon substrate binding ensures that α4 Lys93 and Arg100 are positioned to capture the -1 phosphate and to facilitate electrostatic rate acceleration of a two-metal-ion catalyzed phosphodiester hydrolysis. The importance of Asp181, Lys93 and Arg100 shown here by in vitro studies (Figure 5), extends FEN1 screens for toxic yeast variants in vivo, which repeatedly isolated Lys93, Arg100 and active site carboxylate mutations ((Storici et al., 2002), Figure 2).
Our three structures of FEN1 with bound DNA substrate or product display protein folds within 1.4 Å RMSD and similar binding to the template and 3′-flap strands (Figures S1B-D). Yet, these differ substantially from previous structures of DNA-free hFEN1, particularly in the α2-α3 hydrophobic wedge, helical cap and α4 gateway regions, implicating substrate-induced conformational change in the FEN1 mechanism. Ordering of α4 and α5 explains FTIR results showing increased helicity of 14 residues upon DNA binding (Kim et al., 2001). The concept of a disordered cap explains previous biochemical observations of activity on bulky 5′-flaps that appeared contradictory (Finger et al., 2009; Liu et al., 2004). The disordered α4 and α5 region is ~34 residues in the DNA-free structures. Assuming that disordered residues stretch ~3.8 Å, the unfolded arch could contain three ~42 Å sides. This open conformation would accommodate bulky modified DNA flaps, even those with hairpin structure that can form in repeat sequences (Movie S1). The significance of key structural elements in FEN1 that confer specificity (3′ -flap binding site, gateway, cap) is underscored by identification of mutations in these regions that occur in human cancer specimens ((Zheng et al., 2007), Figure 2).
dsDNA Binding, Double Base Unpairing and Hand Offs
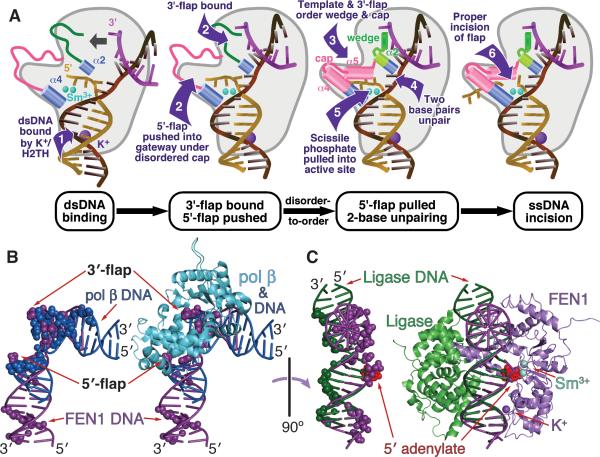
Our dsDNA-binding, double-base-flipping, ssDNA-incision model (Figure 7) explains exquisite FEN1 specificity for incision one base into dsDNA, which is not explained by existing ssDNA-binding/dsDNA-incision models (Liu et al., 2004). Our FEN1:DNA structures show that the major DNA interaction is to the template strand and dsDNA components adjacent to the 3′- and 5′-flaps, in conflict with models where FEN1 interacts first with the 5′-ss flap. Our analyses suggest FEN1 initially binds to substrate dsDNA (Figure 7), extending an idea from bacterial Pol I studies (Xu et al., 2001). FEN1 can incise both RNA and DNA flaps because it does not primarily recognize the flap but rather binds dsDNA junctions. We propose a ‘bind-push-pull-unpair’ mechanism that is based on the structures and biochemistry presented here and that explains dsDNA-based specificity. Furthermore, the observation that substrate binding affinity does not depend on 5′- flap accessibility (Gloor et al., 2010), is explained by our template strand-based binding mechanism.
Figure 7. FEN1 Primarily Binds and Bends dsDNA to Detect dsDNA Junctions and Undergoes Disorder-To-Order Transitions to Recognize 5′-flaps.
(A) FEN1 binds primarily to downstream dsDNA by the K+:H2TH, allowing FEN1 to search for target DNA structure. The second juxtaposed binding site selects for dsDNA structures that can sharply bend ~100°. Coincident with DNA template strand binding, the 5′-ss flap is directed under the disordered cap domain and through the helical gateway selecting for ssDNA nearing the active site. Assembly of the 3′-flap site, cap ordering and double base unpairing promote correct positioning of the scissile bond and rapid two-metal-ion catalyzed incision. Movie S1 shows morphing between models of three DNA-free FEN1 structures and the product DNA-bound structure.
(B) Overlay of DNA from complexes with FEN1 (product) and Pol β (1TV9) (Krahn et al., 2004) reveals that Pol β binding does not block the FEN1 downstream dsDNA binding region, suggesting a baton passing mechanism for direct handoff. (Left) DNA atoms shown within 4 Å of protein in the complex (FEN1, purple; Pol β, blue, colored spheres). (Right) Pol β shown with DNA overlay.
(C) Both FEN1 and the Ligase I DBD can bind simultaneously to the DNA, based on an overlay of 5′ -flap DNA from complexes with FEN1 (product) and Ligase I DBD (1X9N) (Pascal et al., 2004). (Left) DNA atoms shown within 4 Å of protein in the complex (FEN1, purple; Ligase I DBD, green, colored spheres). (Right) Overlaid complexes.
Biologically, this new double-strand DNA binding and double base unpairing model has implications for the mechanism by which FEN1 accepts flap substrates from DNA polymerases, which obscure the 3′-flap from FEN1 binding (Pelletier et al., 1994). Analysis of the DNA binding sites shows that FEN1 can bind downstream dsDNA exposed in the Pol β complex (Figure 7B). To access the 5′- and 3′-flaps, FEN1 would then have to displace Pol β, resulting in a direct handoff. Similarly, handoff to ligase is structurally possible: the minor grooves of the dsDNA, which are bound by Ligase I DNA binding domain (DBD) (Pascal et al., 2004), are exposed in the FEN1:DNA complex. The only steric clash occurs near the cap, suggesting an order-to-disorder mechanism for pushing FEN1 off its product DNA that reverses the binding process. Coordinated handoff of dangerous DNA intermediates by interface exchange as supported here by the way FEN1 binds DNA, and as hypothesized for APE1 in base excision repair (Mol et al., 2000) and for Rad51 in dsDNA break repair (Shin et al., 2003), may generally apply to 5' nuclease superfamily activities in DNA replication and repair processes, as aided by accessory proteins, such as PCNA (Chapados et al., 2004). Specifically, the ‘passing-the-baton’ concept proposed for base excision repair (Wilson and Kunkel, 2000) is consistent with the buried interfaces of Pol β and Ligase I and with dsDNA binding directing FEN1 and 5′ nuclease specificity, but not with FEN1 binding directed by the 5′-flap, which is partly buried in the Pol β complex for short flaps.
Unified Understanding of FEN1 Superfamily Members
Analyses of conserved and non-conserved regions in the FEN1:DNA structures provide insights into the paradox how 5′ nuclease superfamily members enforce structure specific nucleolytic activity, yet exhibit strikingly different substrate specificity. FEN1, EXO1, XPG and GEN1 are junction specific enzymes that require three separate binding sites for DNA and that incise one basepair into the duplex region of the substrate. As a group they conserve many DNA-interacting residues in the K+:H2TH motif, the active site, and the helical gateway formed by α2 and α4 (Figures 1 and 2). Most are invariantly conserved. In α2, the least conserved, residues retain helical propensity and provide a potential stacking equivalent to Tyr40 in FEN1. The conservation of α2 is supported by the robust flap incision activity of XFX2, which has an XPG α2 and a FEN α4 (Figure 6). Features such as non-incised-strand-based binding, severe bending at the ss-dsDNA junction, helical gateway selection for ssDNA, and strand unpairing at the active site are likely maintained in the superfamily. Preference of protein-DNA contacts similar to FEN for the non-incised strand was observed in foot-printing studies of XPG-DNA complexes (Hohl et al., 2003). A minimum dsDNA length of ~1 dsDNA turn, defined by the distance between the H2TH and active site, is likely needed for efficient cutting by all superfamily families.
Features such as the cap and the electronegative acid block are not conserved in the superfamily. Their absence explains the substrate diversity. The helical cap, seen is only in EXO1 and FEN1, decreased the XPG nuclease activity on bubbles in XFX2 (Figure 6). In XPG this cap region, known as the spacer or R-domain, is ~650 residues, but in GEN1 it is shorter than in FEN1. These regions likely do not cap the helical gateway allowing bubble and Holliday junction ssDNA substrates to slot through the gateway and into the active site without threading. Indeed, adding the R-domain between FEN1 residues 104 and 105 (FXF) allowed a weak but significant incision on DNA bubbles (Hohl et al., 2007). Analysis of our FEN1 structures suggests the XPG bubble could traverse the back surface of the enzyme from the active site to the second DNA binding site (used for 3′-flap binding by FEN1). Notably, this 3′-flap pocket including the electronegative acid block, which prevents binding of longer 3′-flaps, is only conserved in FENs. A specific feature in the FEN1 substrate is the duplex and 3′ -flap that we propose promotes the disorder-to-order transition. For XPG and EXOI, this substrate region would be ssDNA. So XPG and EXOI may have an analogous disorder-to-order transition promoted by ssDNA or a partner. The latter is suggested for XPG, which binds the 3′ -junction early in nucleotide excision repair (NER), but does not incise until later (Staresincic et al., 2009).
Collectively, our structural and biochemical analyses reveal a novel and unified model for FEN1 superfamily substrate recognition and incision that resolves paradoxes regarding observed specificities. Positioning of the target DNA strand through primary binding to its complementary strand and enforced double base unpairing for the scissile strand to access the two-metal-ion active site are unexpected elements identified in our FEN1-DNA complexes. Combined with the disorder-to-order transitions in the gateway and cap, our findings explain how FEN selects for ssDNA but can accommodate bulky adducts at the 5′ -flap termini. Moreover, our structurally-informed sequence and biochemical comparisons suggest that these features are the hallmark of the 5′ nuclease superfamily. Binding and bending the dsDNA, the ss gateway, and double-base unpairing would ensure that incision depends upon the nature of the dsDNA and not the ss flap, avoiding potentially damaging non-specific reactions within the 5′ -ss region in flaps in DNA replication and LP-BER (FEN1), bubbles in NER (XPG), ends in mismatch repair (EXO1), and Holliday junctions in homologous recombination (GEN1).
EXPERIMENTAL PROCEDURES
Proteins
For crystallization, WT and D181A FEN1 were cloned with a prescission protease site and (His)6tag after residue 336 (removing 44 residues at the C-terminus) and expressed in E. coli. For biochemical analysis, WT and mutant FEN1 proteins were cloned with a (His)6tag and bacterially expressed. The gene for XFX2 was synthesized by Codon Devices. Residues 86-766 in human XPG were replaced with P. furiosus FEN residues 89-128. XPG and XFX2 were expressed in insect cells. Details are in the Supplemental Information.
Crystallization and Data Collection
WT FEN1 was crystallized with 1 nt and 4 nt flap substrates, while D181A FEN1 was crystallized with 1 nt flap substrate. Designed to prevent hydrolysis, the scissile phosphodiester of the 5′-flap oligonucleotide was replaced by a 3′-phosphoramidate. However the DNA was still incised during crystallization. Details of the crystallization are provided in Supplemental Information. All X-ray diffraction data were measured from single crystals cooled to 90 K.
Structure Determination and Refinement
Data processing and refinement are detailed in Supplemental Information. Deposited coordinates are 3Q8K.pdb (WTFEN1:Product) , 3Q8L.pdb (WTFEN1:substrate), and 3Q8M.pdb (D181A:substrate).
FEN1 Incision Assays
Rates of WT and mutated FEN1-catalysed reactions (50 mM HEPES pH 7.5, 0.05 mg/mL BSA, 2.5 mM THP, 10 mM Mg(OAc)2 100 mM KCl) were determined at 37°C using a 5′-fluorescein (FAM) double flap substrate (2.5-10 nM). Reactions were quenched at the appropriate time with 250 mM EDTA. For second order rate constant determination, initial rates (v) were determined as described (Finger et al. 2009). The apparent second order rate constant (kcat/KM) was determined from a plot of normalized initial rate (v/[E]) versus substrate concentration ([S]). For steady state measurements, kcat and KM were derived from generalized nonlinear least squares using a Michaelis-Menten model. For incision assays with and without 5′-phosphate, substrate labeling, assays, and PAGE were conducted with 25 pM and 500 pM FEN1, respectively, as previously described (Finger et al., 2009).
XFX2 Activity Assays
Radiolabeled bubble DNA or flap substrate (6.6 nM) was incubated with XPG, XFX2 or FEN1 for the specified time at 37°C in an incubation buffer containing 25 mM Hepes-KOH (pH 7.5), 80 mM KCl, 4 mM MgCl2, 1 mM DTT and 5% glycerol. A hairpin on the labeled strand of the DNA bubble was added to reduce a competing end-binding activity that removes the 5′-label. A longer flap substrate used here is more comparable in size to the bubble. Substrate and product DNA were separated by denaturing PAGE and visualized by phosphorimager analysis
Supplementary Material
ACKNOWLEDGEMENTS
Work on FEN1 is supported by the NIH/NCI through RO1CA081967, R01CA073764, and P01 CA092584 (SBDR) and by BBSRC (BBF0147321). Crystal data was collected at the SIBYLS beamline 12.3.1 (ALS, Contract DE-AC02-05CH11231) and beamline 11-1 (SSRL, supported by DOE, OBER, NIH, NCRR, Biomedical Technology Program, and the NIGMS). Morphing movies were produced with CHIMERA (NIH P41 RR-01081). We thank Hong Xu and Chiharu Hitomi for technical assistance; and James Holton, Mike Pique, Elizabeth Getzoff, David Moiani, Cliff Ng and Peter Burgers for aiding structural analyses.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Burgers PM. Polymerase dynamics at the eukaryotic DNA replication fork. J Biol Chem. 2009;284:4041–4045. doi: 10.1074/jbc.R800062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapados BR, Hosfield DJ, Han S, Qiu J, Yelent B, Shen B, Tainer JA. Structural basis for FEN-1 substrate specificity and PCNA-mediated activation in DNA replication and repair. Cell. 2004;116:39–50. doi: 10.1016/s0092-8674(03)01036-5. [DOI] [PubMed] [Google Scholar]
- Debrauwere H, Loeillet S, Lin W, Lopes J, Nicolas A. Links between replication and recombination in Saccharomyces cerevisiae: a hypersensitive requirement for homologous recombination in the absence of Rad27 activity. Proc Natl Acad Sci U S A. 2001;98:8263–8269. doi: 10.1073/pnas.121075598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos JM, Tomanicek SJ, Jones CE, Nossal NG, Mueser TC. Crystal structure of bacteriophage T4 5' nuclease in complex with a branched DNA reveals how flap endonuclease-1 family nucleases bind their substrates. J Biol Chem. 2007;282:31713–31724. doi: 10.1074/jbc.M703209200. [DOI] [PubMed] [Google Scholar]
- Finger LD, Blanchard MS, Theimer CA, Sengerova B, Singh P, Chavez V, Liu F, Grasby JA, Shen B. The 3′-flap pocket of human flap endonuclease 1 is critical for substrate binding and catalysis. J Biol Chem. 2009;284:22184–22194. doi: 10.1074/jbc.M109.015065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger LD, Shen B. FEN1 (flap structure-specific endonuclease 1). In: Huret JL, editor. Atlas Genet Cytogenet Oncol Haematol. INIST; Poitiers, France: 2010. http://AtlasGeneticsOncology.org/Genes/FEN1ID40543ch11q12.html. [Google Scholar]
- Friedrich-Heineken E, Henneke G, Ferrari E, Hubscher U. The acetylatable lysines of human Fen1 are important for endo- and exonuclease activities. J Mol Biol. 2003;328:73–84. doi: 10.1016/s0022-2836(03)00270-5. [DOI] [PubMed] [Google Scholar]
- Friedrich-Heineken E, Hubscher U. The Fen1 extrahelical 3′-flap pocket is conserved from archaea to human and regulates DNA substrate specificity. Nucleic Acids Res. 2004;32:2520–2528. doi: 10.1093/nar/gkh576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcin ED, Hosfield DJ, Desai SA, Haas BJ, Bjoras M, Cunningham RP, Tainer JA. DNA apurinic-apyrimidinic site binding and excision by endonuclease IV. Nat Struct Mol Biol. 2008;15:515–522. doi: 10.1038/nsmb.1414. [DOI] [PubMed] [Google Scholar]
- Gloor JW, Balakrishnan L, Bambara RA. Flap endonuclease 1 mechanism analysis indicates flap base binding prior to threading. J Biol Chem. 2010;285:34922–31. doi: 10.1074/jbc.M110.165902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohl M, Dunand-Sauthier I, Staresincic L, Jaquier-Gubler P, Thorel F, Modesti M, Clarkson SG, Scharer OD. Domain swapping between FEN-1 and XPG defines regions in XPG that mediate nucleotide excision repair activity and substrate specificity. Nucleic Acids Res. 2007;35:3053–3063. doi: 10.1093/nar/gkm092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohl M, Thorel F, Clarkson SG, Scharer OD. Structural determinants for substrate binding and catalysis by the structure-specific endonuclease XPG. J Biol Chem. 2003;278:19500–19508. doi: 10.1074/jbc.M213155200. [DOI] [PubMed] [Google Scholar]
- Hosfield DJ, Mol CD, Shen B, Tainer JA. Structure of the DNA repair and replication endonuclease and exonuclease FEN-1: coupling DNA and PCNA binding to FEN-1 activity. Cell. 1998;95:135–146. doi: 10.1016/s0092-8674(00)81789-4. [DOI] [PubMed] [Google Scholar]
- Ip SC, Rass U, Blanco MG, Flynn HR, Skehel JM, West SC. Identification of Holliday junction resolvases from humans and yeast. Nature. 2008;456:357–361. doi: 10.1038/nature07470. [DOI] [PubMed] [Google Scholar]
- Ivanov I, Tainer JA, McCammon JA. Unraveling the three-metal-ion catalytic mechanism of the DNA repair enzyme endonuclease IV. Proc Natl Acad Sci U S A. 2007;104:1465–1470. doi: 10.1073/pnas.0603468104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalifa L, Beutner G, Phadnis N, Sheu SS, Sia EA. Evidence for a role of FEN1 in maintaining mitochondrial DNA integrity. DNA Repair (Amst) 2009;8:1242–1249. doi: 10.1016/j.dnarep.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CY, Park MS, Dyer RB. Human flap endonuclease-1: conformational change upon binding to the flap DNA substrate and location of the Mg2+ binding site. Biochemistry. 2001;40:3208–3214. doi: 10.1021/bi002100n. [DOI] [PubMed] [Google Scholar]
- Krahn JM, Beard WA, Wilson SH. Structural insights into DNA polymerase beta deterrents for misincorporation support an induced-fit mechanism for fidelity. Structure. 2004;12:1823–1832. doi: 10.1016/j.str.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Kuppuraj G, Dudev M, Lim C. Factors Governing Metal-Ligand Distances and Coordination Geometries of Metal Complexes. J Phys Chem B. 2009 doi: 10.1021/jp807972e. [DOI] [PubMed] [Google Scholar]
- Larsen E, Gran C, Saether BE, Seeberg E, Klungland A. Proliferation failure and gamma radiation sensitivity of Fen1 null mutant mice at the blastocyst stage. Mol Cell Biol. 2003;23:5346–5353. doi: 10.1128/MCB.23.15.5346-5353.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BI, Wilson DM., 3rd The RAD2 domain of human exonuclease 1 exhibits 5' to 3′ exonuclease and flap structure-specific endonuclease activities. J Biol Chem. 1999;274:37763–37769. doi: 10.1074/jbc.274.53.37763. [DOI] [PubMed] [Google Scholar]
- Lieber MR. The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. Bioessays. 1997;19:233–240. doi: 10.1002/bies.950190309. [DOI] [PubMed] [Google Scholar]
- Liu Y, Kao HI, Bambara RA. Flap endonuclease 1: a central component of DNA metabolism. Annu Rev Biochem. 2004;73:589–615. doi: 10.1146/annurev.biochem.73.012803.092453. [DOI] [PubMed] [Google Scholar]
- Lyamichev V, Brow MA, Varvel VE, Dahlberg JE. Comparison of the 5' nuclease activities of taq DNA polymerase and its isolated nuclease domain. Proc Natl Acad Sci U S A. 1999;96:6143–6148. doi: 10.1073/pnas.96.11.6143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki Y, Adachi N, Koyama H. Vertebrate cells lacking FEN-1 endonuclease are viable but hypersensitive to methylating agents and H2O2. Nucleic Acids Res. 2002;30:3273–3277. doi: 10.1093/nar/gkf440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mol CD, Izumi T, Mitra S, Tainer JA. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination. Nature. 2000;403:451–456. doi: 10.1038/35000249. [DOI] [PubMed] [Google Scholar]
- Navarro MS, Bi L, Bailis AM. A mutant allele of the transcription factor IIH helicase gene, RAD3, promotes loss of heterozygosity in response to a DNA replication defect in Saccharomyces cerevisiae. Genetics. 2007;176:1391–1402. doi: 10.1534/genetics.107.073056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donovan A, Davies AA, Moggs JG, West SC, Wood RD. XPG endonuclease makes the 3′ incision in human DNA nucleotide excision repair. Nature. 1994;371:432–435. doi: 10.1038/371432a0. [DOI] [PubMed] [Google Scholar]
- Pascal JM, O'Brien PJ, Tomkinson AE, Ellenberger T. Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature. 2004;432:473–478. doi: 10.1038/nature03082. [DOI] [PubMed] [Google Scholar]
- Pelletier H, Sawaya MR, Kumar A, Wilson SH, Kraut J. Structures of ternary complexes of rat DNA polymerase beta, a DNA template-primer, and ddCTP. Science. 1994;264:1891–1903. [PubMed] [Google Scholar]
- Qiu J, Bimston DN, Partikian A, Shen B. Arginine residues 47 and 70 of human flap endonuclease-1 are involved in DNA substrate interactions and cleavage site determination. J Biol Chem. 2002;277:24659–24666. doi: 10.1074/jbc.M111941200. [DOI] [PubMed] [Google Scholar]
- Reagan MS, Pittenger C, Siede W, Friedberg EC. Characterization of a mutant strain of Saccharomyces cerevisiae with a deletion of the RAD27 gene, a structural homolog of the RAD2 nucleotide excision repair gene. J Bacteriol. 1995;177:364–371. doi: 10.1128/jb.177.2.364-371.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts VA, Freeman HC, Olson AJ, Tainer JA, Getzoff ED. Electrostatic orientation of the electron-transfer complex between plastocyanin and cytochrome c. J Biol Chem. 1991;266:13431–13441. [PubMed] [Google Scholar]
- Sakurai S, Kitano K, Yamaguchi H, Hamada K, Okada K, Fukuda K, Uchida M, Ohtsuka E, Morioka H, Hakoshima T. Structural basis for recruitment of human flap endonuclease 1 to PCNA. EMBO J. 2005;24:683–693. doi: 10.1038/sj.emboj.7600519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarker AH, Tsutakawa SE, Kostek S, Ng C, Shin DS, Peris M, Campeau E, Tainer JA, Nogales E, Cooper PK. Recognition of RNA polymerase II and transcription bubbles by XPG, CSB, and TFIIH: insights for transcription-coupled repair and Cockayne Syndrome. Mol Cell. 2005;20:187–198. doi: 10.1016/j.molcel.2005.09.022. [DOI] [PubMed] [Google Scholar]
- Schroeder GK, Lad C, Wyman P, Williams NH, Wolfenden R. The time required for water attack at the phosphorus atom of simple phosphodiesters and of DNA. Proc Natl Acad Sci U S A. 2006;103:4052–4055. doi: 10.1073/pnas.0510879103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengerova B, Tomlinson C, Atack JM, Williams R, Sayers JR, Williams NH, Grasby JA. Bronsted analysis and rate-limiting steps for the T5 flap endonuclease catalyzed hydrolysis of exonucleolytic substrates. Biochemistry. 2010;49:8085–8093. doi: 10.1021/bi100895j. [DOI] [PubMed] [Google Scholar]
- Shen B, Nolan JP, Sklar LA, Park MS. Essential amino acids for substrate binding and catalysis of human flap endonuclease 1. J Biol Chem. 1996;271:9173–9176. doi: 10.1074/jbc.271.16.9173. [DOI] [PubMed] [Google Scholar]
- Sherman F, Stewart JW, Tsunasawa S. Methionine or not methionine at the beginning of a protein. Bioessays. 1985;3:27–31. doi: 10.1002/bies.950030108. [DOI] [PubMed] [Google Scholar]
- Shin DS, Pellegrini L, Daniels DS, Yelent B, Craig L, Bates D, Yu DS, Shivji MK, Hitomi C, Arvai AS, et al. Full-length archaeal Rad51 structure and mutants: mechanisms for RAD51 assembly and control by BRCA2. EMBO J. 2003;22:4566–4576. doi: 10.1093/emboj/cdg429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Zheng L, Chavez V, Qiu J, Shen B. Concerted action of exonuclease and Gap-dependent endonuclease activities of FEN-1 contributes to the resolution of triplet repeat sequences (CTG)n- and (GAA)n-derived secondary structures formed during maturation of Okazaki fragments. J Biol Chem. 2007;282:3465–3477. doi: 10.1074/jbc.M606582200. [DOI] [PubMed] [Google Scholar]
- Staresincic L, Fagbemi AF, Enzlin JH, Gourdin AM, Wijgers N, Dunand-Sauthier I, Giglia-Mari G, Clarkson SG, Vermeulen W, Scharer OD. Coordination of dual incision and repair synthesis in human nucleotide excision repair. EMBO J. 2009;28:1111–1120. doi: 10.1038/emboj.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storici F, Henneke G, Ferrari E, Gordenin DA, Hubscher U, Resnick MA. The flexible loop of human FEN1 endonuclease is required for flap cleavage during DNA replication and repair. EMBO J. 2002;21:5930–5942. doi: 10.1093/emboj/cdf587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syson K, Tomlinson C, Chapados BR, Sayers JR, Tainer JA, Williams NH, Grasby JA. Three metal ions participate in the reaction catalyzed by T5 flap endonuclease. J Biol Chem. 2008;283:28741–28746. doi: 10.1074/jbc.M801264200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson CG, Atack JM, Chapados BR, Tainer JA, Grasby JA. Substrate recognition and catalysis by flap endonucleases and related enzymes. Biochem Soc Trans. 2010;38:433–437. doi: 10.1042/BST0380433. [DOI] [PubMed] [Google Scholar]
- Tumey LN, Bom D, Huck B, Gleason E, Wang J, Silver D, Brunden K, Boozer S, Rundlett S, Sherf B, et al. The identification and optimization of a N-hydroxy urea series of flap endonuclease 1 inhibitors. Bioorg Med Chem Lett. 2005;15:277–281. doi: 10.1016/j.bmcl.2004.10.086. [DOI] [PubMed] [Google Scholar]
- Wilson SH, Kunkel TA. Passing the baton in base excision repair. Nat Struct Biol. 2000;7:176–178. doi: 10.1038/73260. [DOI] [PubMed] [Google Scholar]
- Wu X, Li J, Li X, Hsieh CL, Burgers PM, Lieber MR. Processing of branched DNA intermediates by a complex of human FEN-1 and PCNA. Nucleic Acids Res. 1996;24:2036–2043. doi: 10.1093/nar/24.11.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Potapova O, Leschziner AE, Grindley ND, Joyce CM. Contacts between the 5' nuclease of DNA polymerase I and its DNA substrate. J Biol Chem. 2001;276:30167–30177. doi: 10.1074/jbc.M100985200. [DOI] [PubMed] [Google Scholar]
- Zheng L, Dai H, Zhou M, Li M, Singh P, Qiu J, Tsark W, Huang Q, Kernstine K, Zhang X, et al. Fen1 mutations result in autoimmunity, chronic inflammation and cancers. Nat Med. 2007;13:812–819. doi: 10.1038/nm1599. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.