Abstract
Background
Population structure and admixture have strong confounding effects on genetic association studies. Discordant frequencies for age-related macular degeneration (AMD) risk alleles and for AMD incidence and prevalence rates are reported across different ethnic groups. We examined the genomic ancestry characterizing 538 Latinos drawn from the Los Angeles Latino Eye Study [LALES] as part of an ongoing AMD-association study. To help assess the degree of Native American ancestry inherited by Latino populations we sampled 25 Mayans and 5 Mexican Indians collected through Coriell's Institute. Levels of European, Asian, and African descent in Latinos were inferred through the USC Multiethnic Panel (USC MEP), formed from a sample from the Multiethnic Cohort (MEC) study, the Yoruba African samples from HapMap II, the Singapore Chinese Health Study, and a prospective cohort from Shanghai, China. A total of 233 ancestry informative markers were genotyped for 538 LALES Latinos, 30 Native Americans, and 355 USC MEP individuals (African Americans, Japanese, Chinese, European Americans, Latinos, and Native Hawaiians). Sensitivity of ancestry estimates to relative sample size was considered.
Results
We detected strong evidence for recent population admixture in LALES Latinos. Gradients of increasing Native American background and of correspondingly decreasing European ancestry were observed as a function of birth origin from North to South. The strongest excess of homozygosity, a reflection of recent population admixture, was observed in non-US born Latinos that recently populated the US. A set of 42 SNPs especially informative for distinguishing between Native Americans and Europeans were identified.
Conclusion
These findings reflect the historic migration patterns of Native Americans and suggest that while the 'Latino' label is used to categorize the entire population, there exists a strong degree of heterogeneity within that population, and that it will be important to assess this heterogeneity within future association studies on Latino populations. Our study raises awareness of the diversity within "Latinos" and the necessity to assess appropriate risk and treatment management.
Background
Recent years have seen great advances in discovering genetic variants associated with the biogenesis and progression of a variety of complex diseases (e.g., [1-8]). Despite the relative success of mapping susceptible loci, we are still faced with a frequent lack of replication across different populations. One possible cause is our relatively poor understanding of the degree of genetic diversity between populations. Besides the variation in genetic make-up across ethnicities, we often observe a wide range in incidence and prevalence rates across populations, for any given disease; it is likely that this range is largely due to that variation.
On the other hand, population substructure may inflate positive associations and cause hidden confounding effects due to an underlying difference in the distribution of ancestry between cases and controls [9-19]. If a particular ancestral group has relatively lower disease prevalence rates, this will result in an under-representation of that subgroup in cases versus controls. Loci with dissimilar allele frequencies across populations may induce spurious associations with phenotype. For example, the CY3A4-V gene variant and prostate cancer are reported to be substantially less common among European American than African American (AA) men; Kittles et al. studied 688 AAs and found that a strongly significant association at CYP3A4-V for prostate cancer became a non-significant signal after including ten ancestry informative markers (AIMs) [19]. Several discrepancies in both disease prevalence rates and genetic susceptibility loci have been confirmed in Latino studies. For instance, Salari et al. [20] found a higher level of European ancestry among Mexican Americans to be strongly associated with increased asthma severity, while a higher proportion of Native American ancestry was protective. Also, Choudhry et al. (2006) observed a significant difference in allele frequencies between asthma cases and controls (P = 0.0002) in Puerto Ricans, but not in Mexicans.
As Latinos form the largest minority ethnic group in the US, with close to 100 million individuals projected by 2050 [21], a growing number of genome-wide association studies will involve that population. It is therefore essential to understand the specifics of genetic structure within Latino populations, and to design association studies with reference to that structure. Thus, we examine the ancestral landscape of Latinos ascertained through the Los Angeles Latino Eye Study (LALES), the largest visual impairment epidemiologic cohort of Latinos in the US [22]. As such, this cohort represents a unique opportunity to better decipher the demographics of Latinos.
The LALES study is a population-based cohort composed of 6,357 Latinos residing in 6 census tracts of the Los Angeles County, who originated mainly in the US, Mexico, Guatemala, or El Salvador. Preliminary evidence suggests that there are differences for risk of AMD between various populations [23-32]. While prevalence rates for early AMD among Latinos are similar to those found in Caucasians [9.4% LALES vs. 7.2% Blue Mountains Eye Study (BMES) vs. 15.6% Beaver Dam] and in individuals of African descent (12.6% BES) [27,29,31,32], incidence data indicates that only 1.5% of early AMD cases advance into late AMD in Latinos, while 3.4% of cases progress in Caucasian cohorts. Despite the growing evidence for the role of complement pathway in development of AMD, discordant frequencies for a series of AMD risk alleles have been reported between different ethnic groups [24,31-35].
The difficulty in defining Latino admixture rests in our relatively poor historical understanding of the demographic events that converged into shaping the modern Latinos from the source populations of the Americas, Europe, Asia and Africa. However, the history of any population is written in its genetic make-up, and that version is forgotten much more slowly than any language-based version of the same history. While a number of studies defined the admixed nature of Latinos to be mostly composed of Native American and European descent [20,36-39], there is a considerable degree of heterogeneity within Native Americans. Wang et al. examined genetic diversity in 29 Native American populations from North, Central, and South America, and compared them to Siberian populations [40]. They depicted gradients of decrease in both genetic diversity and similarity to immigrant Siberians as a function of geographic distance from the Bering Strait. Unfortunately, the relative paucity of available genome-wide data for the Native American populations has made even the genetic data hard to interpret. Consequently, in addition to the data inherent in the LALES study, we have also generated genotype data for a number of Native American individuals.
Previous studies identified ancestry informative marker (AIM) polymorphisms that exhibit large differences in allele frequencies across populations of European, Asian, and African descent, and therefore confer increased power for detecting levels of population stratification [38,41-44]. A series of projects have since followed, describing the effects these ancestries have on numerous genetic risk factors [18,45-55]. However, such AIMs are liable to be less powerful when describing the ethnicity of Latinos. For example, Mexican Americans contain a rather small percentage of African heritages and are mostly composed of a mixture of European and Native American ancestry [20,36,47,50-52]. The historical focus on the HapMap has meant that a clear and comprehensive description of genetic admixture among American Latinos has been lacking, and has only recently started to emerge [20,37,38,56]. Our analysis uses AIMs genotyped for 6 population samples: (1) LALES Latinos, (2) Native Americans selected through Coriell's institute for medical research laboratory http://ccr.coriell.org, (3) Yoruba Africans (YRI) from the HapMap II database, (4) Asian, African and European descent individuals from the USC Multiethnic Panel (USC MEP), consisting of samples from the Multiethnic Cohort (MEC) [57,58], and (5-6) two additional Chinese cohorts [59,60]. We use this set of marker data to infer the important demographic characteristics of Latinos. This will enable investigators to increase the power of future association studies based on Latino populations.
Results
LALES demographics
A total of 500 out of 538 genotyped subjects were included in the final analysis after a sample call rate test was performed at the 0.80 level. Age, gender, and self-reported geographic birthplace distributions for the 500 LALES subjects are given in Table 1. Recent Latino-based population studies reported various ancestry estimates between Puerto Ricans and Mexican Americans [20,36,39,61]. Overall, LALES birth locations were dispersed as 68.4% Mexico, 18.2% USA, 5.4% El Salvador, 3.4% Guatemala, and 4.6% from other places. There is little difference between cases and controls in this respect, as would be expected given that the inclusion criteria for cases and controls in the original LALES cohort (n = 6357) study design required a matched frequency for birthplace location.
Table 1.
LALES sample demographics
| LALES Demographics | Cases | Controls | |
|---|---|---|---|
| Age Average (S.D.) | All (n = 500) | 60.34 (11.37) | 60.04 (11.54) |
| Males (n = 227) | 59.62 (11.37) | 60.74 (11.55) | |
| Females (n = 273) | 61.12 (12.40) | 59.61 (11.54) | |
| Birthplace % | Mexico | 68.0 | 68.8 |
| USA | 18.0 | 18.4 | |
| EL Salvador | 5.6 | 5.2 | |
| Guatemala | 3.6 | 3.2 | |
| Other | 4.8 | 4.4 | |
Note.
S.D. = Standard Deviation
n = Number
Other = Other birthplace locations
Estimation of LALES Population Structure and Admixture
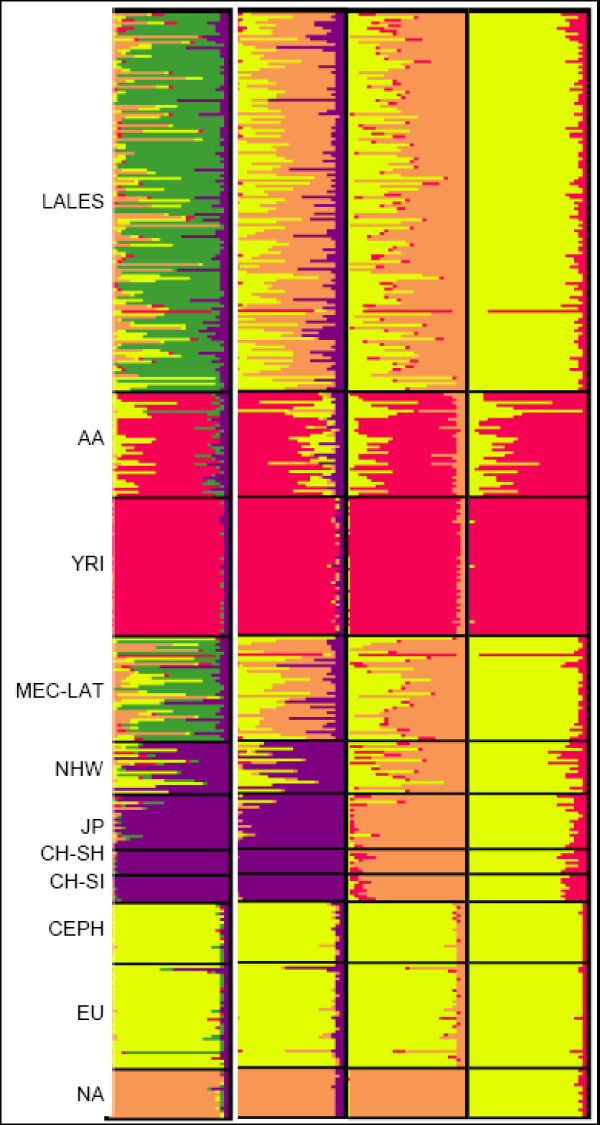
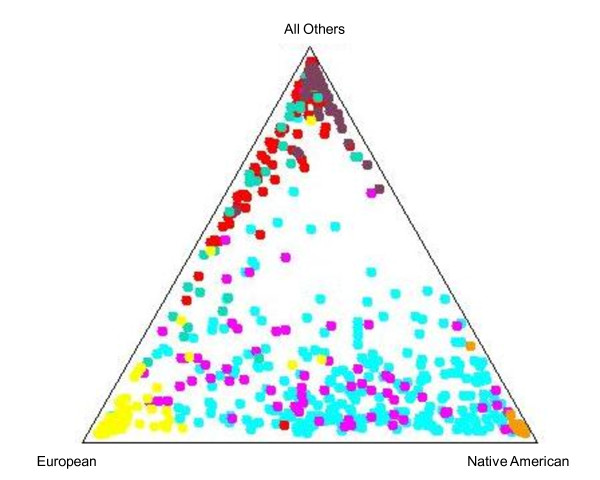
Population structure for the LALES, YRI, USC MEP, and NA samples for each of the K = {2, ..., 5} cluster models are illustrated in Figure 1. Reported results represent an average from 3 different runs, all of which gave consistent results, reflecting proper MCMC convergence. For STRUCTURE analysis estimates see Additional file 1, Table S1; the log likelihood of the data, lnPr(X|K), and the corresponding allele frequency difference measure FK are summarized for each K = {2, ..., 5}. Previous studies suggest that Latinos are a mixture of three main source populations (Native American, European, and Asian), with rather little African descent [20,36,40,47,61]. For this reason, we focus on the modeling results of K = 4 for which the second largest likelihood [lnPr(X|K = 4) = -116312.20] where the average LALES Latino admixture is partitioned as 45.2 - 54.3% Native American, 32.1 - 40.1% European, 9.7 - 11.5% Asian, and 4.0 - 5.2% African-American (Tables 2 and 3). We estimated Latino admixture proportions from the inclusion of LALES controls only (n = 250). Nucleotide distance dispersions of individual ancestry vectors for K = 4 are plotted in Figure 2, where each individual is mapped on the triangular coordinates between Native American, European and 'Other' ethnicities
Figure 1.
Individual ancestry proportions for the LALES, Multiethnic Cohort, and Native American sampled populations. Population Number Code: AA - MEC African Americans; MEC-LAT - MEC Latinos; JP - MEC Japanese (AS); NH - MEC Native Hawaiians; CH-Sh - China Shanghai (AS); CH-Si - China Singapore (AS); CEPH - CEPH (EU); MEC-EU - MEC Europeans (EU); LALES - LALES Latinos; NA - Native American; YRI - Yoruba Africans
Table 2.
Estimation of ancestry proportions for the LALES, MEC/Chinese/CEPH, and Native American populations
| Population | AF | EU | AS | NA |
|---|---|---|---|---|
| MEC African American | 0.721 | 0.167 | 0.066 | 0.047 |
| MEC Native Hawaiian | 0.031 | 0.329 | 0.591 | 0.049 |
| MEC Japanese | 0.012 | 0.034 | 0.892 | 0.061 |
| Chinese - Shanghai | 0.013 | 0.018 | 0.916 | 0.052 |
| Chinese - Singapore | 0.014 | 0.02 | 0.934 | 0.032 |
| MEC European | 0.009 | 0.939 | 0.029 | 0.022 |
| CEPH | 0.015 | 0.891 | 0.056 | 0.038 |
| Native American | 0.005 | 0.013 | 0.032 | 0.949 |
| MEC Latinos | 0.059 | 0.453 | 0.116 | 0.373 |
| LALES Latinos | 0.049 | 0.401 | 0.098 | 0.452 |
Note.
Estimates for the LALES Latino population are based on the LALES controls.
European - EU; African - AF; Asian - AS; Native American - NA.
Table 3.
Estimation of ancestry proportions for the LALES Latinos based on 111 SNPs from two admixture models: (1) African American, European, Native American, and Asian source populations, and (2) Yoruba African, European, Native American, and Asian source populations.
| Source Populations | LALES Ancestry Estimates | NA | EU | AF | AS |
|---|---|---|---|---|---|
| AA, AS, NA, EU | All | 0.480 | 0.352 | 0.052 | 0.115 |
| El Salvador/Guatemala | 0.484 | 0.259 | 0.095 | 0.163 | |
| Mexico | 0.444 | 0.369 | 0.050 | 0.136 | |
| USA | 0.320 | 0.458 | 0.051 | 0.171 | |
| YRI, AS, NA, EU | All | 0.543 | 0.321 | 0.040 | 0.097 |
| El Salvador/Guatemala | 0.596 | 0.230 | 0.084 | 0.091 | |
| Mexico | 0.539 | 0.332 | 0.034 | 0.095 | |
| USA | 0.453 | 0.394 | 0.025 | 0.128 | |
Note.
Estimates for the LALES Latino populations are based on the LALES controls.
African American - AA; African - AF; Asian - AS; European - EU;
Native American - NA; Yoruba African - YRI.
Figure 2.
Cluster ancestry distribution for the LALES, Multiethnic Panel, and Native American samples. Each individual is positioned proportional to his/her ancestral similarity to each the three reference groups. Individuals placed at a particular corner are completely assigned to the corresponding population, whereas those in the centroid area are equidistant from each of the three group lineages. LALES Latinos - turquoise; MEC Latinos - pink; African - red; Asian - purple; European - yellow; Native Hawaiian - green; Native American - orange.
In comparison to LALES Latinos, those ascertained through the MEC cohort show a stronger relatedness to Europeans (~40.1% vs. ~45.3%) with correspondingly lower Native American ancestry (~45.2% vs. ~37.3%) (Table 2). This discrepancy is likely to be a consequence of differentiation in selection of individuals for the two cohorts from the different birth places. Roughly 18% of the LALES Latinos were born within the US and 68% within Mexico, with smaller proportions born in Guatemala and El Salvador (Table 1). For the MEC sample these proportions are somewhat different, with 47% of Latinos born in the US, 34% in Mexico, 10% in Central/South American, and 4% in Cuba. Three MEC Latino individuals were of unknown birth origin.
When we split the data by birth origin (i.e. US vs. Mexico vs. Central/South America or El Salvador/Guatemala), even though there are some differences in EU and NA proportions between MEC and LALES Latinos, we detect in both cohorts a gradient of linear increase in NA ancestry from North (US) to South (El Salvador for LALES or South America for MEC) with a corresponding decrease in European descent (Table 4).
Table 4.
Estimation of ancestry proportions for the LALES and MEC Latinos by birthplace location
| Latinos | Birth Region | NA | EU | AS | AF |
|---|---|---|---|---|---|
| LALES | El Salvador + Guatemala | 0.52 | 0.30 | 0.12 | 0.07 |
| Mexico | 0.49 | 0.37 | 0.11 | 0.04 | |
| USA | 0.42 | 0.42 | 0.12 | 0.04 | |
| Other | 0.35 | 0.42 | 0.10 | 0.14 | |
| MEC | Central/South America | 0.51 | 0.37 | 0.06 | 0.05 |
| Mexico | 0.39 | 0.39 | 0.08 | 0.13 | |
| USA | 0.35 | 0.47 | 0.04 | 0.14 | |
Note.
LALES = Los Angeles Latino Eye Study
MEC = Multi Ethnic Cohort
EU - European; AF - African; AS - Asian; NA - Native American
Moreover, individual NA and EU ancestry distributions between Salvadorans/Guatemalans and the rest of the LALES cohort were significantly different (Wilcoxon signed P-values = 0.012 and 0.009, respectively). Since relatively few individuals were born in El Salvador and Guatemala, we included both LALES cases and controls for the computation of Wilcoxon tests. We note however that separate analyses of LALES cases or controls gave very similar ancestry estimates (Additional file 1, Table S2), resulting in non-significant differences (P-values > 0.5) for any of the NA, EU, AF, or AS proportions.
All 223 MEC markers were selected from the admixture map panel developed by Smith et al. (2004). The authors estimate these markers (3,011) to be optimal for distinguishing European, West African, Amerindian, and East Asian mixtures. Recent studies have identified extensive heterogeneity across African populations [62,63]; the STRUCTURE analysis depicted 14 ancestral clusters across Africa. This issue is also relevant for Native American populations; Wang et al. [40] and Tishkoff et al. [62] both report high variation among Native Americans. For this reason we sought to include Native Americans that co-inhibit the same regions as most of our LALES cohort. We note that the MEC study ascertained African Americans rather than Yoruban Africans (YRI). To compare Latino ancestry estimates derived from AAs vs. YRIs we performed parallel Structure analyses for a subset of 111 AIMs identified in the YRI HapMap II database; this resulted in an overall increase in NA ancestry of ~6% (54.3% vs. 48.0%) and a corresponding decrease in EU origin of ~3% (35.2% vs. 32.1%) when YRIs rather than AAs were set as founders (Table 3). However, some degree in variation will result from using the smaller set of 111. To examine the potential extent of this variation we selected random samples of 111 SNPs from the total of 176 SNPs that passed the call rate threshold of 0.98. The average ancestry estimates across LALES Latinos ranged from 42.2% to 51.4% NA and from 32.1% to 37.9% EU (Additional file 1, Table S3). However, regardless of the admixture model or the set of markers analyzed, the North to South trend among Latino populations for NA and EU mixtures remains the consistent; lowest NA heritage within US born Latinos, and highest within El-Salvador/Guatemala.
While, for ease of interpretation we focus our results on the assumption of four source populations, the strongest log-likelihood was obtained at K = 5 for both the AA and YRI based analyses [lnPr(X|K = 5) = -116186.10 and -80348.2, respectively vs. lnPr(X|K = 4) = -116312.2 and - 81256.7, respectively]. The 5th cluster explains in both analyses approximately 63.0% of LALES and 47.6% of MEC Latino ancestry, though this substructure is found in none of the founder populations (Figure 1; Additional file 1, Table S4).
Selection of markers informative for distinguishing between Native American and European ethnicity
It would clearly be useful to determine a set of SNPs that might be helpful in untangling admixture in Latinos, but the HapMap data contains no Native American individuals. With this in mind, Table S5 (see Additional file 1) summarizes the chromosomal positions and allele frequencies of 42 SNPs for which we detected at least 30% difference in allele frequencies (δ > 0.3) between NA and EU populations. This set of markers offers an addition to the previously reported Latino population admixture map markers provided by Price et al. (2007) [37].
Tests for population structure and recent admixture
The HWE test was used as a means of detecting population structure and/or recent admixture. While none of the 176 AIMs failed HWE, the overall distribution of genotype homozygosity showed a greater shift to the right (higher homozygosity) in the LALES Latinos than in any of the founder populations (Additional file 2, Figure S1). This tendency is reduced in the MEC Latinos. Additional Figure S2 (see Additional file 3) reveals a potential explanation for this. We examined the distribution of homozygosity within the LALES population for those born within vs. outside the US. Given that the MEC Latino population contains a larger proportion of individuals born within the US, a smaller signature of increased homozygosity might be expected.
Finally, from a total of 15,931 pair-wise SNP combinations we obtained a subset of 15,163 pairs formed by SNPs positioned on different chromosomes; 10.0% of the unlinked pairs were significantly associated in the LALES cohort compared to 6.7% in MEC Latinos. These results point towards evidence for recent population admixture in Latinos that have recently populated the US, as they compose ~82% of the LALES vs. 50% of the MEC cohort.
Effect of Sample Size on Admixture Estimation
We used two sampling techniques to explore the effect of relative sample size on inferred ancestry. In a first approach, we sub-sampled the LALES cohort to produce a sample of size 70, broadly consistent with the other samples in our data. Despite the wide variation of estimated NA and EU admixture proportions within LALES individuals, this approach typically resulted in estimates broadly similar to those resulting from the initial dataset analysis (Additional file 1, Table S6). Estimated NA and EU ancestries had a mean (s.d.) over 100 sampled datasets of 45.0% (2.0%) and 42.0% (2.0%), respectively, compared to original estimates of 45.2% and 40.1%. Using a second bootstrapping approach (sampling with replacement) we increased smaller datasets to 250 individuals each, matching the size of the LALES control set. We report average ancestry estimates over 100 samples (Additional file 1, Table S6; Additional file 4, Figure S3). Mean EU ancestry in LALES Latinos increased to 44.3% (s.d. = 0.6%), with a correspondingly lower NA percentage (42.2% (0.7%)). While this outcome is only suggestive, it does seem that a sample size of 70 individuals per ethnic group is sufficient to obtain reliable estimates, at least in the present context. However, if there is a perceived need to increase the size of smaller samples by using boot-strapping, somewhat altered estimates of admixture proportions may result.
Discussion
Association studies of recently admixed populations may produce spurious allelic associations for markers that are in linkage disequilibrium with a causal gene, a reason for replication failures in other populations [9,16,18,64]. It is therefore necessary to first assess the extent of admixture when designing association studies that involve populations such as Latinos. The degree of genetic variation within 'Latino' populations is not well understood, so in this paper we evaluated admixture in Latinos ascertained through the Los Angeles Latino Eye Study, the most comprehensive eye disease study in the US. Our paper raises awareness of the diversity within "Latinos" themselves and provides a resource for future invasive examination of ancestry-specific AMD mechanisms or other related biological pathways. A distinctive characteristic of the LALES study is the ascertainment of Latinos from different geographic regions, an aspect that allowed us to better characterize the extent of Native American and European variation.
Depending on the details of which SNPs were incorporated in our analysis and, correspondingly, which African populations were used as a reference, the LALES Latinos were estimated to inherit in the region of 50% NA and 40% EU ancestry. This reflects the importance of structure within reference populations, such as the Africans here, as well. However, whichever set of Africans was used as a reference, we observed a consistent trend for Native American ancestry to increase on a north (lowest) to south (highest) gradient within the Americans. It is also important to note that our study focused on using K = 4 clusters (AF, AS, EU, and NA) in the STRUCTURE analysis, whereas earlier studies used K = 3 (AF, EU, and NA) [20,38]. When we replicate the approach of Salari et al. (2005) and of Collins et al. (2004), by excluding Asians and running an analysis with K = 3 we recover broadly the same estimated ancestry proportions in both Mexican LALES Latinos (53.4% NA and 40.3% EU) and the overall LALES cohort (49.3% NA and 41.1% EU).
Increased homozygosity is a commonly-used signature for admixture. We observe elevated levels of homozygosity in Latinos. The increase is higher in the LALES Latinos than in those from the MEC cohort, an indicator of more recent population admixture among Latinos that have migrated recently to the US. Indeed, when we compared US with non-US born LALES Latinos, we observed an increase in the level of homozygosity in the latter. Another indicator of recent admixture and/or population structure is the degree of allelic association between markers positioned on different chromosomes. 10% vs. 6.7% of unlinked locus pairs were associated in LALES vs. MEC Latinos, an additional confirmation of heterogeneity within Latinos. Finally, in an attempt to aid the design of future studies involving Latinos, we reported a set of SNPs with high differences in allele frequencies between Native Americans and Europeans.
The issue of whether the results from a STRUCTURE analysis are affected by discrepancies between sample sizes across ethnic groups is not typically addressed. Our results suggest two things. First, unequal sample sizes do not appear to bias estimates of ancestry, at least in the context of the present paper. Second, they support the belief that sample sizes of 25 or great are typically sufficient to give meaningful estimates of ancestry. Finally, when we tried another common strategy, inflating sample sizes by boot-strapping, ancestry estimates did appear to change from those found in the original sample. While these results are clearly only suggestive, they do imply that caution should be exercised before employing such an approach. However, we also note that the standard deviation of the estimates appears to decrease as sample-size increases, as would be expected. The relative merits in the trade-off between the apparent change in ancestry estimates in the boot-strapped samples and the decrease in standard deviation of those estimates, remains to be assessed in future studies.
Conclusion
In summary, we found strong evidence for recent population admixture in Latinos ascertained through the LALES cohort. By specifically incorporating, and in some cases collecting genotype data for each of the likely source populations, we were able to identify the ethnicity related to each component of the Latino genetic make-up. The highest ancestral component was Native American, with gradients of increasing NA ancestry as a function of birth origin from North to South (US, Mexico, Guatemala, El Salvador). These findings reflect the historic migration patterns of the NA population and suggest that while the 'Latino' label is used to categorize the entire population, there exists a strong degree of heterogeneity within that population, and that it will be imperative to assess this heterogeneity and control for it within future association studies using Latino populations.
Methods
Selection of ancestry informative markers (AIMs)
We used a set of 233 AIMs, dispersed throughout the genome, and chosen from a set of high-density admixture map markers described in Smith et al. [65]. These SNPs exhibit a substantial difference in allele frequencies across ethnicities [66]. In addition, AIMs are specifically chosen to lack linkage with any known human disease candidate. These SNPs had been previously genotyped among the USC MEP. Given the existence of this data, and our desire to incorporate it within our study, we ourselves genotyped the LALES sample and the NA collection of individuals at the same set of AIMs.
Study Subjects
Six datasets were compiled for the estimation of Latino ancestry for the ongoing ocular disease study of the LALES cohort: LALES, NA, YRI, and a multiethnic panel comprised of subjects from the MEC and two Chinese cohorts. We genotyped two distinct datasets for the same set of AIMs described above: (1) 538 LALES subjects and (2) 30 Native Americans. A brief description of the LALES, NA, and MEC datasets is provided below. Ninety YRI samples from the HapMap II project were incorporated in the population admixture models.
LALES Subjects
538 LALES participants (268 cases: 268 controls) with an average age (s.d.) of 56.7 (11.2) years were genotyped for this study (Table 1). All LALES cases were diagnosed with early AMD through the detection of bilateral, intermediate to large soft drusen deposits. Controls lacked drusen in either eye and were matched with cases based on age and birthplace location. Details of the LALES cohort design are described elsewhere [22,67,68]. All procedures followed the Declaration of Helsinki for research involving human subjects. The Los Angeles County/University of Southern California Medical Center Institutional Review Board approved the project, and informed consent was obtained from all participants.
Native American Subjects
In order to establish a reference set for the NA lineage in Latinos, we genotyped 25 Mayan Amerindian and 5 Mexican Indian DNA samples from Coriell's human population repository collection http://ccr.coriell.org/. The Mayan samples were specifically chosen because they represent ancient Native American civilizations that lived before the arrival of Europeans in what nowadays are eastern and southern Mexico, El Salvador, Guatemala, Belize, and Honduras. Since the dispersion of geographic regions for the LALES cohort covers Mexico and most of Central America, the Mayan and Mexican Indian samples overlap the birth locations for most of the LALES cohort.
MEC Subjects
The Multiethnic Cohort (MEC) study is a prospective cohort of approximately 215,000 individuals from California and Hawaii [57]. This study was established between 1993-1996 and includes men and women primarily from five racial and ethnic populations in Hawaii and California (African Americans, European Americans, Latinos, Japanese Americans and Native Hawaiians). The USC MEP sample includes 355 individuals; 18 Chinese males from a prospective cohort from Shanghai, China [59], 17 females from the Singapore Health Study [60], 40 parents from 20 CEU trios from HapMap [69], and 280 MEC women without a history of cancer, namely, 70 Europeans, 70 African Americans, 70 Latinos from the Los Angeles area, 35 Japanese, and 35 Hawaiians. This multiethnic panel has been reported previously in de Bakker et al. [70] and Haiman et al. [69].
Genotyping
The 538 LALES and 30 Native American subjects were genotyped using the Illumina GoldenGate platform for the 233 AIMs (USC Genomics Core Laboratory, Los Angeles, CA). The MEP panel samples were genotyped using the same platform (USC Genomics Core Laboratory, Los Angeles, CA). 176 SNPs out of 233 had genotype call rates > 0.98 and were chosen for the present analysis. Samples with an overall genotype call rate ≤ 0.8 were removed from analysis, resulting in a total of 500 LALES (250 cases, 250 controls) and 30 Native American individuals being included in the downstream analyses.
Statistical Analysis
We employed a series of methods to evaluate the level of admixture among Latinos, to estimate the relative proportions of AF, AS, EU, and NA background in both LALES and MEC Latinos, and to assess the correlation of NA and EU ancestry with the LALES AMD case-control status. Ethnic proportions were inferred through the Markov chain Monte Carlo (MCMC) algorithm of Falush and Pritchard using the STRUCTURE 2.2 software package [71-73].
Assessment of Latino population admixture was performed using three different statistics: (1) the Pearson chi-square test to identify SNPs in Hardy Weinberg disequilibrium, (2) an overall assessment across all AIMs of the distribution of homozygous genotypes within each sampled population and also of that within US-born vs. non-US born Latinos, and (3) a measure for excess association between physically unlinked loci in LALES and in MEC Latinos.
Estimation of Population Ancestry
The genetic make-up of LALES Latinos was inferred using the admixture modeling implemented in STRUCTURE 2.2 [71-73], and allowing for correlation between allele frequencies among populations. The ALPHA Dirichlet parameter for degree of admixture was inferred, starting at an initial value of 1.0 and a standard deviation of proposal for updating ALPHA of 0.025. We ran 45,000 burn-in repetitions and a further 50,000 iterations after the burn-in period. When using STRUCTURE, accurately deciding the number of clusters K that best describes a population's substructure is a rather difficult task [71-75]. Our solution was to focus on the value of K which not only captures most of the structure in a population, but also offers an experimentally relevant interpretation. We ran the analysis using different values of K and obtained the estimated log-likelihood of the data (lnPr(X|K)) at each run. For each K-value three independent analyses were completed to ensure that lnPr(X|K) estimates were consistent across runs. The average likelihood from the three independent runs is reported for each K, where the posterior probability of K can be computed as 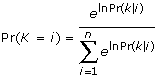 .
.
A second parameter of interest is the divergence in allele frequencies between the K clusters, traditionally referred to as Wright's Fst measure [76]. The current STRUCTURE implementation reports FK, an analogue of Fst, proposed by Falush et al. (2003) [73]. The FK-based model allows for variation in drift rates between populations, computing a different FK measure for each of the K populations rather than assessing an overall Fst measure across all populations.
STRUCTURE analyses were performed first on the final set of 176 AIMs for the merged dataset of the LALES, NA, and USC MEP. These AIMs were selected from the high-density admixture map for disease gene discovery in African Americans (Smith et al., 2004); the STRUCTURE model integrates this information in estimating Latino ancestry. However, given the high heterogeneity among African populations (Tishkoff et al., 2009), we compared these estimates with those obtained from an additional analysis based on a subset of 111 SNPs for which 90 Yoruba Africans from the HapMap II database were also included in the ancestry model.
Since AIMs were selected for their lack of linkage with loci known to be associated with human diseases, the inclusion of cases would be unlikely to affect overall approximations. However, to avoid any potential biases we report the population structure results based only on the inclusion of LALES controls. In addition, as part of our continuing LALES Latino eye study we also completed a separate STRUCTURE analysis using only the 250 AMD cases. This additional step allowed us to further examine potential differences in ethnic background between AMD cases and controls by using the Wilcoxon signed test. Lastly, association between any of the AIMs and AMD status was tested using an additive genetic model. Allelic regression analysis was also conducted by including individual EU and NA ancestry estimates as model covariates for assessing the strength of association between any of the AIMs and AMD. Final p-values were corrected for multiple comparisons through Bonferroni adjustment at the 2.84*10-4 (or 0.05/176) threshold.
Identification of population structure and recent admixture
In a random-mating population we expect genotypes to be in Hardy-Weinberg equilibrium (HWE) [77]. Deviations from this equilibrium are typically thought to be due to population structure, selection or genotyping errors. For example, admixture will cause a modification of genotype frequencies in a population due to the influx of alleles from other populations [78]. Deviations due to selection are unlikely in the present context given that the AIMs were chosen to be optimal for distinguishing large scale population mixtures and for making precise ancestry estimates (Smith et al. 2004) [65]. Given this, we checked among Latinos for deviations from HWE in the set of 176 AIM SNPs using a Pearson's chi-square test with one degree of freedom. In addition, we tested for excess of homozygosity, a trademark of recent admixture. Choudhry and Siegmund implemented the T statistic measure for estimating the amount of deviation from HWE and the trend in homozygosity across all markers, where 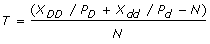 , N is the total number of individuals, PD and Pd denote estimated allele frequencies, and XDD and Xdd are the homozygote genotypic counts [36]. Under the assumption of HWE and based on the selection of randomly chosen genome-wide loci, a standard normal distribution is expected to fit the frequencies of the T-statistic [61], with heterozygote frequencies distributed towards the left, homozygote counts towards the right. The observed distribution of this T-statistic was contrasted between the LALES, MEC and Native American populations. We further searched for potential variation within Latinos themselves by evaluating regional specific homozygosity trends of individuals originating in different birthplace locations. A final analysis of population admixture was conducted by assessing the degree of allelic association between physically unlinked markers [16,61,79]. Any associations between AIM pairs from these SNP pairs would most likely be due to recent admixture or population substructure.
, N is the total number of individuals, PD and Pd denote estimated allele frequencies, and XDD and Xdd are the homozygote genotypic counts [36]. Under the assumption of HWE and based on the selection of randomly chosen genome-wide loci, a standard normal distribution is expected to fit the frequencies of the T-statistic [61], with heterozygote frequencies distributed towards the left, homozygote counts towards the right. The observed distribution of this T-statistic was contrasted between the LALES, MEC and Native American populations. We further searched for potential variation within Latinos themselves by evaluating regional specific homozygosity trends of individuals originating in different birthplace locations. A final analysis of population admixture was conducted by assessing the degree of allelic association between physically unlinked markers [16,61,79]. Any associations between AIM pairs from these SNP pairs would most likely be due to recent admixture or population substructure.
Bootstrap Methods for Assessing the Effect of Sample Size on Population Structure Inference
An emerging concern when assessing ancestral proportions is the size of the genotyped samples within a given study. Two issues surface when inferring population structure: (1) the minimum sample size requirement for a given population, and (2) the difference in the size of the analyzed sub-populations. There is a danger that estimates of population ancestry might be influenced by the size of the (sub)population being analyzed. For example, it is plausible to imagine that it is easier to identify a population for which we have a large number of representatives than one with relatively few members. This is a particular concern in our study, given the discrepancies between sample sizes across ethnic groups, and this issue is not generally addressed in the literature. To guard against this issue we employed two commonly-used techniques for adjusting sample sizes. First, smaller samples were inflated by Boot-strapping (i.e. sampling at random with replacement) until they reached the LALES control sample size (n = 24 controls). Chinese and Japanese subjects were merged and categorized as 'Asians', while White and CEPH samples were grouped into a single 'European' population. We applied this scheme to inflate each of the following samples: 70 African Americans, 70 Latinos from LA (non-LALES), 35 Native Hawaiians, 70 Asians (35 Japanese, 18 Chinese from Shangai, and 17 Chinese from Singapore), and 110 Caucasians (40 CEPHs and 70 Europeans). Through a second approach we reduced the size of the LALES control cohort by selecting 70 individuals through random sampling without replacement. Unselected individuals were excluded from the subsequent STRUCTURE analysis. Each of the two schemes were repeated 100 times, and every resulting data-set was analyzed with STRUCTURE 2.2 under the K = 4 model parameterization used on the original data. We then reanalyzed the data to see if our earlier conclusions remained true.
Authors' contributions
All authors read and approved the final manuscript. CJS contributed to the proposal of the study design, performed the statistical analysis, interpretation, and writing of the manuscript; PM contributed to the study design, statistical, and interpretative coordination of the project. He was involved in the revision and final approval of the manuscript; SA is co-investigator of the LALES cohort. He participated in the design, reviewing, and final approval of the manuscript; DVC contributed with the genotyping of the MEC AIMs data, advised on methods to be used, and gave critical reviews final approval of the manuscript; LLM participated in the genotyping of the MEC - Hawaiian data, methodology and review of the manuscript; CAH took part in the acquisition of the MEC AIMs data, advised on methodology and final manuscript review; RV is the main PI of the LALES cohort study and of the current population admixture project. He led and coordinated the acquisition of the LALES and Native American data, contributed to the merging of the LALES/Native American and MEC cohorts, proposed and guided this study, and gave interpretation, critical review, and final approval of the manuscript.
Author's Information
CJS is a post-doctoral scholar in the department of Human Genetics and Neuroscience at the University of California - Los Angeles (UCLA); PM, DVC, and CAH are associate professors in the department of Biostatistics at USC, with research interests in the fields of Population Genetics, Statistical Genetics, and Biostatistics. DC and CAH are co-investigators of the MEC cohort; SA is dean of Biostatistics at USC, and co-investigator of the LALES study; LLM is professor of epidemiology at the University of Hawaii, Honolulu. He is principal investigator of the MEC cohort; RV is the director and PI of the LALES cohort study, and a Research to Prevent Blindness Sybil B. Harrington Scholar. Dr. Varma is director of the Glaucoma Service, Ocular Epidemiology Center and Clinical Trials, and professor in the department of Ophthalmology at USC.
Supplementary Material
Additional Tables. Table S1. Simulation summary statistics of ancestry clustering models. Table S2. Comparison of ancestry proportion medians (1st : 3rd quartile) among LALES Latinos by birth location and case-control status. Table S3. Range of ancestry proportion estimates (low - high) for LALES Latinos for random sets of 111 SNPs from the total 176 AIMs genotyped for the LALES, NA, and MEC cohorts. Table S4. Proportion of membership of each pre-defined population in each of the 5 clusters. Table S5. Ancestry informative markers with difference in allele frequency (δ) greater than 0.3 between Native American and European ancestry among Latinos. Table S6. Bootstrap simulation results for the increased and decreased sample size methods
Figure S1. Distribution of T-values for testing overall homozygosity and heterozygosity trends
Figure S2. Distribution of T-values for testing overall homozygosity and heterozygosity trends in LALES Latinos born within the US versus LALES Latinos born outside the US
Figure S3. Bootstrap re-sampling: distribution of European and Native American ancestry frequencies in LALES Latinos
Contributor Information
Corina J Shtir, Email: stir@usc.edu.
Paul Marjoram, Email: pmarjora@usc.edu.
Stanley Azen, Email: sazen@usc.edu.
David V Conti, Email: dconti@usc.edu.
Loic Le Marchand, Email: loic@crch.hawaii.edu.
Christopher A Haiman, Email: haiman@usc.edu.
Rohit Varma, Email: rvarma@usc.edu.
Acknowledgements
Supported by the National Eye Institute, National Institutes of Health, Bethesda, MD (grant nos. EY11753 and EY03040), and by an unrestricted grant from the Research to Prevent Blindness, New York, NY. Dr. Varma is a Research to Prevent Blindness Sybil B. Harrington Scholar. The Multiethnic Cohort Study was supported by National Cancer Institute (NCI) grants CA63464 and CA54281. Dr. Marjoram was supported by the NIH grant HG004049.
We thank Jeffrey Wall for discussions and comments on the manuscript.
References
- Gudmundsson J, Sulem P, Manolescu A, Amundadottir LT, Gudbjartsson D, Helgason A, Rafnar T, Bergthorsson JT, Agnarsson BA, Baker A. et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39(5):631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, Hinds DA, Pennacchio LA, Tybjaerg-Hansen A, Folsom AR. et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316(5830):1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, Green T, Kuballa P, Barmada MM, Datta LW. et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39(5):596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ. et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316(5829):1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S. et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445(7130):881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU. et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316(5829):1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinthorsdottir V, Thorleifsson G, Reynisdottir I, Benediktsson R, Jonsdottir T, Walters GB, Styrkarsdottir U, Gretarsdottir S, Emilsson V, Ghosh S. et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat Genet. 2007;39(6):770–775. doi: 10.1038/ng2043. [DOI] [PubMed] [Google Scholar]
- Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, Timpson NJ, Perry JR, Rayner NW, Freathy RM. et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316(5829):1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardon LR, Palmer LJ. Population stratification and spurious allelic association. Lancet. 2003;361(9357):598–604. doi: 10.1016/S0140-6736(03)12520-2. [DOI] [PubMed] [Google Scholar]
- Wakeley J, Lessard S. Theory of the effects of population structure and sampling on patterns of linkage disequilibrium applied to genomic data from humans. Genetics. 2003;164(3):1043–1053. doi: 10.1093/genetics/164.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DC, Witte JS. Point: population stratification: a problem for case-control studies of candidate-gene associations? Cancer Epidemiol Biomarkers Prev. 2002;11(6):505–512. [PubMed] [Google Scholar]
- Wacholder S, Rothman N, Caporaso N. Population stratification in epidemiologic studies of common genetic variants and cancer: quantification of bias. J Natl Cancer Inst. 2000;92(14):1151–1158. doi: 10.1093/jnci/92.14.1151. [DOI] [PubMed] [Google Scholar]
- Altshuler D, Kruglyak L, Lander E. Genetic polymorphisms and disease. N Engl J Med. 1998;338(22):1626. doi: 10.1056/NEJM199805283382214. [DOI] [PubMed] [Google Scholar]
- Lander ES, Schork NJ. Genetic dissection of complex traits. Science. 1994;265(5181):2037–2048. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- Balding DJ. A tutorial on statistical methods for population association studies. Nat Rev Genet. 2006;7(10):781–791. doi: 10.1038/nrg1916. [DOI] [PubMed] [Google Scholar]
- Helgason A, Yngvadottir B, Hrafnkelsson B, Gulcher J, Stefansson K. An Icelandic example of the impact of population structure on association studies. Nat Genet. 2005;37(1):90–95. doi: 10.1038/ng1492. [DOI] [PubMed] [Google Scholar]
- Hinds DA, Stokowski RP, Patil N, Konvicka K, Kershenobich D, Cox DR, Ballinger DG. Matching strategies for genetic association studies in structured populations. Am J Hum Genet. 2004;74(2):317–325. doi: 10.1086/381716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell CD, Ogburn EL, Lunetta KL, Lyon HN, Freedman ML, Groop LC, Altshuler D, Ardlie KG, Hirschhorn JN. Demonstrating stratification in a European American population. Nat Genet. 2005;37(8):868–872. doi: 10.1038/ng1607. [DOI] [PubMed] [Google Scholar]
- Kittles RA, Chen W, Panguluri RK, Ahaghotu C, Jackson A, Adebamowo CA, Griffin R, Williams T, Ukoli F, Adams-Campbell L. et al. CYP3A4-V and prostate cancer in African Americans: causal or confounding association because of population stratification? Hum Genet. 2002;110(6):553–560. doi: 10.1007/s00439-002-0731-5. [DOI] [PubMed] [Google Scholar]
- Salari K, Choudhry S, Tang H, Naqvi M, Lind D, Avila PC, Coyle NE, Ung N, Nazario S, Casal J. et al. Genetic admixture and asthma-related phenotypes in Mexican American and Puerto Rican asthmatics. Genet Epidemiol. 2005;29(1):76–86. doi: 10.1002/gepi.20079. [DOI] [PubMed] [Google Scholar]
- U.S. Census Bureau. http://www.census.gov/population/www/socdemo/hispanic/files/Internet_Hispanic_in_US_2006 Accessed May-2009.
- Varma R, Paz SH, Azen SP, Klein R, Globe D, Torres M, Shufelt C, Preston-Martin S. The Los Angeles Latino Eye Study: design, methods, and baseline data. Ophthalmology. 2004;111(6):1121–1131. doi: 10.1016/j.ophtha.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Allikmets R, Shroyer NF, Singh N, Seddon JM, Lewis RA, Bernstein PS, Peiffer A, Zabriskie NA, Li Y, Hutchinson A. et al. Mutation of the Stargardt disease gene (ABCR) in age-related macular degeneration. Science. 1997;277(5333):1805–1807. doi: 10.1126/science.277.5333.1805. [DOI] [PubMed] [Google Scholar]
- Klein R, Klein BE, Knudtson MD, Meuer SM, Swift M, Gangnon RE. Fifteen-year cumulative incidence of age-related macular degeneration: the Beaver Dam Eye Study. Ophthalmology. 2007;114(2):253–262. doi: 10.1016/j.ophtha.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Klein R, Knudtson MD, Klein BE. Statin use and the five-year incidence and progression of age-related macular degeneration. Am J Ophthalmol. 2007;144(1):1–6. doi: 10.1016/j.ajo.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein R, Peto T, Bird A, Vannewkirk MR. The epidemiology of age-related macular degeneration. Am J Ophthalmol. 2004;137(3):486–495. doi: 10.1016/j.ajo.2003.11.069. [DOI] [PubMed] [Google Scholar]
- Leske MC, Wu SY, Hennis A, Nemesure B, Yang L, Hyman L, Schachat AP. Nine-year incidence of age-related macular degeneration in the Barbados Eye Studies. Ophthalmology. 2006;113(1):29–35. doi: 10.1016/j.ophtha.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Zareparsi S, Buraczynska M, Branham KE, Shah S, Eng D, Li M, Pawar H, Yashar BM, Moroi SE, Lichter PR. et al. Toll-like receptor 4 variant D299G is associated with susceptibility to age-related macular degeneration. Hum Mol Genet. 2005;14(11):1449–1455. doi: 10.1093/hmg/ddi154. [DOI] [PubMed] [Google Scholar]
- Varma R, Fraser-Bell S, Tan S, Klein R, Azen SP. Prevalence of age-related macular degeneration in Latinos: the Los Angeles Latino eye study. Ophthalmology. 2004;111(7):1288–1297. doi: 10.1016/j.ophtha.2004.01.023. [DOI] [PubMed] [Google Scholar]
- Klein ML, Francis PJ. Genetics of age-related macular degeneration. Ophthalmol Clin North Am. 2003;16(4):567–574. doi: 10.1016/S0896-1549(03)00063-4. [DOI] [PubMed] [Google Scholar]
- Klein R, Klein BE, Linton KL. Prevalence of age-related maculopathy. The Beaver Dam Eye Study. Ophthalmology. 1992;99(6):933–943. doi: 10.1016/s0161-6420(92)31871-8. [DOI] [PubMed] [Google Scholar]
- Mitchell P, Smith W, Attebo K, Wang JJ. Prevalence of age-related maculopathy in Australia. The Blue Mountains Eye Study. Ophthalmology. 1995;102(10):1450–1460. doi: 10.1016/s0161-6420(95)30846-9. [DOI] [PubMed] [Google Scholar]
- Grassi MA, Fingert JH, Scheetz TE, Roos BR, Ritch R, West SK, Kawase K, Shire AM, Mullins RF, Stone EM. Ethnic variation in AMD-associated complement factor H polymorphism p.Tyr402His. Hum Mutat. 2006;27(9):921–925. doi: 10.1002/humu.20359. [DOI] [PubMed] [Google Scholar]
- Tedeschi-Blok N, Buckley J, Varma R, Triche TJ, Hinton DR. Population-based study of early age-related macular degeneration: role of the complement factor H Y402H polymorphism in bilateral but not unilateral disease. Ophthalmology. 2007;114(1):99–103. doi: 10.1016/j.ophtha.2006.07.043. [DOI] [PubMed] [Google Scholar]
- Klein R, Klein BE, Knudtson MD, Wong TY, Cotch MF, Liu K, Burke G, Saad MF, Jacobs DR Jr. Prevalence of age-related macular degeneration in 4 racial/ethnic groups in the multi-ethnic study of atherosclerosis. Ophthalmology. 2006;113(3):373–380. doi: 10.1016/j.ophtha.2005.12.013. [DOI] [PubMed] [Google Scholar]
- Choudhry S, Coyle NE, Tang H, Salari K, Lind D, Clark SL, Tsai HJ, Naqvi M, Phong A, Ung N. et al. Population stratification confounds genetic association studies among Latinos. Hum Genet. 2006;118(5):652–664. doi: 10.1007/s00439-005-0071-3. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson N, Yu F, Cox DR, Waliszewska A, McDonald GJ, Tandon A, Schirmer C, Neubauer J, Bedoya G. et al. A genomewide admixture map for Latino populations. Am J Hum Genet. 2007;80(6):1024–1036. doi: 10.1086/518313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins-Schramm H, Chima B, Morii T, Wah K, Figueroa Y, Criswell L, Hanson R, Knowler W, Silva G, Belmont J. et al. Mexican American ancestry-informative markers: examination of population structure and marker characteristics in European Americans, Mexican Americans, Amerindians and Asians. Human Genetics. 2004;114:263–271. doi: 10.1007/s00439-003-1058-6. [DOI] [PubMed] [Google Scholar]
- Krueger SK, Siddens LK, Martin SR, Yu Z, Pereira CB, Cabacungan ET, Hines RN, Ardlie KG, Raucy JL, Williams DE. Differences in FMO2*1 allelic frequency between Hispanics of Puerto Rican and Mexican descent. Drug Metab Dispos. 2004;32(12):1337–1340. doi: 10.1124/dmd.104.001099. [DOI] [PubMed] [Google Scholar]
- Wang S, Lewis CM, Jakobsson M, Ramachandran S, Ray N, Bedoya G, Rojas W, Parra MV, Molina JA, Gallo C. et al. Genetic variation and population structure in native Americans. PLoS Genet. 2007;3(11):e185. doi: 10.1371/journal.pgen.0030185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Butler J, Patterson N, Capelli C, Pascali VL, Scarnicci F, Ruiz-Linares A, Groop L, Saetta AA, Korkolopoulou P. et al. Discerning the Ancestry of European Americans in Genetic Association Studies. PLoS Genetics. 2008;4(1):e236. doi: 10.1371/journal.pgen.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaff CL, Parra EJ, Bonilla C, Hiester K, McKeigue PM, Kamboh MI, Hutchinson RG, Ferrell RE, Boerwinkle E, Shriver MD. Population structure in admixed populations: effect of admixture dynamics on the pattern of linkage disequilibrium. Am J Hum Genet. 2001;68(1):198–207. doi: 10.1086/316935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA. Algorithms for selecting informative marker panels for population assignment. J Comput Biol. 2005;12(9):1183–1201. doi: 10.1089/cmb.2005.12.1183. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW. Genetic structure of human populations. Science. 2002;298(5602):2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- Li M, Atmaca-Sonmez P, Othman M, Branham KE, Khanna R, Wade MS, Li Y, Liang L, Zareparsi S, Swaroop A. et al. CFH haplotypes without the Y402H coding variant show strong association with susceptibility to age-related macular degeneration. Nat Genet. 2006;38(9):1049–1054. doi: 10.1038/ng1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, Abraham C, Regueiro M, Griffiths A. et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314(5804):1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv E, John EM, Choudhry S, Kho J, Lorizio W, Perez-Stable EJ, Burchard EG. Genetic Ancestry and Risk Factors for Breast Cancer among Latinas in the San Francisco Bay Area. Cancer Epidemiol Biomarkers Prev. 2006;15(10):1878–1885. doi: 10.1158/1055-9965.EPI-06-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner AP, Ziv E, Lind DL, Nievergelt CM, Schork NJ, Cummings SR, Phong A, Burchard EG, Harris TB, Psaty BM. et al. Population structure, admixture, and aging-related phenotypes in African American adults: the Cardiovascular Health Study. Am J Hum Genet. 2005;76(3):463–477. doi: 10.1086/428654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv E, John EM, Choudhry S, Kho J, Lorizio W, Perez-Stable EJ, Burchard EG. Genetic ancestry and risk factors for breast cancer among Latinas in the San Francisco Bay Area. Cancer Epidemiol Biomarkers Prev. 2006;15(10):1878–1885. doi: 10.1158/1055-9965.EPI-06-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, McElree C, Roth MP, Shanahan F, Targan SR, Rotter JI. Familial empirical risks for inflammatory bowel disease: differences between Jews and non-Jews. Gut. 1993;34(4):517–524. doi: 10.1136/gut.34.4.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menotti A, Lanti M, Puddu PE, Kromhout D. Coronary heart disease incidence in northern and southern European populations: a reanalysis of the seven countries study for a European coronary risk chart. Heart. 2000;84(3):238–244. doi: 10.1136/heart.84.3.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi F, Arcieri P, Bertina RM, Chiarotti F, Corral J, Pinotti M, Prydz H, Samama M, Sandset PM, Strom R. et al. Contribution of factor VII genotype to activated FVII levels. Differences in genotype frequencies between northern and southern European populations. Arterioscler Thromb Vasc Biol. 1997;17(11):2548–2553. doi: 10.1161/01.atv.17.11.2548. [DOI] [PubMed] [Google Scholar]
- Shaffer J, Kammerer C, Reich D, McDonald G, Patterson N, Goodpaster B, Bauer D, Li J, Newman A, Cauley J. et al. Genetic markers for ancestry are correlated with body composition traits in older African Americans. Osteoporosis International. 2007;18:733–741. doi: 10.1007/s00198-006-0316-6. [DOI] [PubMed] [Google Scholar]
- Castellano M. Geographic ancestry, angiotensinogen gene polymorphism, and cardiovascular risk. Hypertension. 2006;48(4):562–563. doi: 10.1161/01.HYP.0000239226.62314.31. [DOI] [PubMed] [Google Scholar]
- Azofeifa J, Hahn M, Ruiz E, Hummerich L, Morales AI, Jimenez G, Barrantes R. The STR polymorphism (AAAAT)n within the intron 1 of the tumor protein 53 (TP53) locus in 17 populations of different ethnic groups of Africa, America, Asia and Europe. Rev Biol Trop. 2004;52(3):645–657. doi: 10.15517/rbt.v1i2.15352. [DOI] [PubMed] [Google Scholar]
- Conrad DF, Jakobsson M, Coop G, Wen X, Wall JD, Rosenberg NA, Pritchard JK. A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nat Genet. 2006;38(11):1251–1260. doi: 10.1038/ng1911. [DOI] [PubMed] [Google Scholar]
- Kolonel LN, Henderson BE, Hankin JH, Nomura AM, Wilkens LR, Pike MC, Stram DO, Monroe KR, Earle ME, Nagamine FS. A multiethnic cohort in Hawaii and Los Angeles: baseline characteristics. Am J Epidemiol. 2000;151(4):346–357. doi: 10.1093/oxfordjournals.aje.a010213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike MC, Kolonel LN, Henderson BE, Wilkens LR, Hankin JH, Feigelson HS, Wan PC, Stram DO, Nomura AM. Breast cancer in a multiethnic cohort in Hawaii and Los Angeles: risk factor-adjusted incidence in Japanese equals and in Hawaiians exceeds that in whites. Cancer Epidemiol Biomarkers Prev. 2002;11(9):795–800. [PubMed] [Google Scholar]
- Yuan JM, Ross RK, Wang XL, Gao YT, Henderson BE, Yu MC. Morbidity and mortality in relation to cigarette smoking in Shanghai, China. A prospective male cohort study. JAMA. 1996;275(21):1646–1650. doi: 10.1001/jama.275.21.1646. [DOI] [PubMed] [Google Scholar]
- Wu AH, Seow A, Arakawa K, Van Den Berg D, Lee HP, Yu MC. HSD17B1 and CYP17 polymorphisms and breast cancer risk among Chinese women in Singapore. Int J Cancer. 2003;104(4):450–457. doi: 10.1002/ijc.10957. [DOI] [PubMed] [Google Scholar]
- Choudhry S, Taub M, Mei R, Rodriguez-Santana J, Rodriguez-Cintron W, Shriver MD, Ziv E, Risch NJ, Burchard EG. Genome-wide screen for asthma in Puerto Ricans: evidence for association with 5q23 region. Hum Genet. 2008;123(5):455–468. doi: 10.1007/s00439-008-0495-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Reed FA, Friedlaender FR, Ehret C, Ranciaro A, Froment A, Hirbo JB, Awomoyi AA, Bodo JM, Doumbo O. et al. The Genetic Structure and History of Africans and African Americans. Science. 2009;324(5930):1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auton A, Bryc K, Boyko AR, Lohmueller KE, Novembre J, Reynolds A, Indap A, Wright MH, Degenhardt JD, Gutenkunst RN. et al. Global distribution of genomic diversity underscores rich complex history of continental human populations. Genome Res. 2009;19(5):795–803. doi: 10.1101/gr.088898.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton DG, Walker NM, Smyth DJ, Pask R, Cooper JD, Maier LM, Smink LJ, Lam AC, Ovington NR, Stevens HE. et al. Population structure, differential bias and genomic control in a large-scale, case-control association study. Nat Genet. 2005;37(11):1243–1246. doi: 10.1038/ng1653. [DOI] [PubMed] [Google Scholar]
- Smith MW, Patterson N, Lautenberger JA, Truelove AL, McDonald GJ, Waliszewska A, Kessing BD, Malasky MJ, Scafe C, Le E. et al. A high-density admixture map for disease gene discovery in african americans. Am J Hum Genet. 2004;74(5):1001–1013. doi: 10.1086/420856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA, Li LM, Ward R, Pritchard JK. Informativeness of genetic markers for inference of ancestry. Am J Hum Genet. 2003;73(6):1402–1422. doi: 10.1086/380416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser-Bell S, Wu J, Klein R, Azen SP, Varma R. Smoking, alcohol intake, estrogen use, and age-related macular degeneration in Latinos: the Los Angeles Latino Eye Study. Am J Ophthalmol. 2006;141(1):79–87. doi: 10.1016/j.ajo.2005.08.024. [DOI] [PubMed] [Google Scholar]
- Klein R, Davis MD, Magli YL, Segal P, Klein BE, Hubbard L. The Wisconsin age-related maculopathy grading system. Ophthalmology. 1991;98(7):1128–1134. doi: 10.1016/s0161-6420(91)32186-9. [DOI] [PubMed] [Google Scholar]
- Haiman CA, Stram DO, Pike MC, Kolonel LN, Burtt NP, Altshuler D, Hirschhorn J, Henderson BE. A comprehensive haplotype analysis of CYP19 and breast cancer risk: the Multiethnic Cohort. Hum Mol Genet. 2003;12(20):2679–2692. doi: 10.1093/hmg/ddg294. [DOI] [PubMed] [Google Scholar]
- de Bakker PI, Burtt NP, Graham RR, Guiducci C, Yelensky R, Drake JA, Bersaglieri T, Penney KL, Butler J, Young S. et al. Transferability of tag SNPs in genetic association studies in multiple populations. Nat Genet. 2006;38(11):1298–1303. doi: 10.1038/ng1899. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164(4):1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Molecular Ecology Notes. 2007;7(4):574–578. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Donnelly P. Case-control studies of association in structured or admixed populations. Theor Popul Biol. 2001;60(3):227–237. doi: 10.1006/tpbi.2001.1543. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Rosenberg NA, Donnelly P. Association mapping in structured populations. Am J Hum Genet. 2000;67(1):170–181. doi: 10.1086/302959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. The genetical structure of populations. Ann Eugen. 1951. pp. 323–354. [DOI] [PubMed]
- Hartl DL. A Primer of Population Genetics. 2. Sunderland, MA.: Sinauer Associates, Inc; 1988. [Google Scholar]
- Law B, Buckleton JS, Triggs CM, Weir BS. Effects of Population Structure and Admixture on Exact Tests for Association Between Loci. Genetics. 2003;164(1):381–387. doi: 10.1093/genetics/164.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai HJ, Kho JY, Shaikh N, Choudhry S, Naqvi M, Navarro D, Matallana H, Castro R, Lilly CM, Watson HG. et al. Admixture-matched case-control study: a practical approach for genetic association studies in admixed populations. Hum Genet. 2006;118(5):626–639. doi: 10.1007/s00439-005-0080-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Tables. Table S1. Simulation summary statistics of ancestry clustering models. Table S2. Comparison of ancestry proportion medians (1st : 3rd quartile) among LALES Latinos by birth location and case-control status. Table S3. Range of ancestry proportion estimates (low - high) for LALES Latinos for random sets of 111 SNPs from the total 176 AIMs genotyped for the LALES, NA, and MEC cohorts. Table S4. Proportion of membership of each pre-defined population in each of the 5 clusters. Table S5. Ancestry informative markers with difference in allele frequency (δ) greater than 0.3 between Native American and European ancestry among Latinos. Table S6. Bootstrap simulation results for the increased and decreased sample size methods
Figure S1. Distribution of T-values for testing overall homozygosity and heterozygosity trends
Figure S2. Distribution of T-values for testing overall homozygosity and heterozygosity trends in LALES Latinos born within the US versus LALES Latinos born outside the US
Figure S3. Bootstrap re-sampling: distribution of European and Native American ancestry frequencies in LALES Latinos