Abstract
Double-stranded RNA-mediated interference (RNAi) is a simple and rapid method of silencing gene expression in a range of organisms. The silencing of a gene is a consequence of degradation of RNA into short RNAs that activate ribonucleases to target homologous mRNA. The resulting phenotypes either are identical to those of genetic null mutants or resemble an allelic series of mutants. Specific gene silencing has been shown to be related to two ancient processes, cosuppression in plants and quelling in fungi, and has also been associated with regulatory processes such as transposon silencing, antiviral defense mechanisms, gene regulation, and chromosomal modification. Extensive genetic and biochemical analysis revealed a two-step mechanism of RNAi-induced gene silencing. The first step involves degradation of dsRNA into small interfering RNAs (siRNAs), 21 to 25 nucleotides long, by an RNase III-like activity. In the second step, the siRNAs join an RNase complex, RISC (RNA-induced silencing complex), which acts on the cognate mRNA and degrades it. Several key components such as Dicer, RNA-dependent RNA polymerase, helicases, and dsRNA endonucleases have been identified in different organisms for their roles in RNAi. Some of these components also control the development of many organisms by processing many noncoding RNAs, called micro-RNAs. The biogenesis and function of micro-RNAs resemble RNAi activities to a large extent. Recent studies indicate that in the context of RNAi, the genome also undergoes alterations in the form of DNA methylation, heterochromatin formation, and programmed DNA elimination. As a result of these changes, the silencing effect of gene functions is exercised as tightly as possible. Because of its exquisite specificity and efficiency, RNAi is being considered as an important tool not only for functional genomics, but also for gene-specific therapeutic activities that target the mRNAs of disease-related genes.
INTRODUCTION
RNA silencing is a novel gene regulatory mechanism that limits the transcript level by either suppressing transcription (transcriptional gene silencing [TGS]) or by activating a sequence-specific RNA degradation process (posttranscriptional gene silencing [PTGS]/RNA interference [RNAi]). Although there is a mechanistic connection between TGS and PTGS, TGS is an emerging field while PTGS is undergoing an explosion in its information content. Here, we have limited our discussion to PTGS/RNAi-related phenomena.
Pioneering observations on PTGS/RNAi were reported in plants, but later on RNAi-related events were described in almost all eukaryotic organisms, including protozoa, flies, nematodes, insects, parasites, and mouse and human cell lines, as shown in Table 1. Three phenotypically different but mechanistically similar forms of RNAi, cosuppression or PTGS in plants, quelling in fungi, and RNAi in the animal kingdom, have been described. More recently, micro-RNA formation, heterochromatinization, etc., have been revealed as other facets of naturally occurring RNAi processes of eukaryotic cells.
TABLE 1.
Eukaryotic organisms exhibiting RNAi-related phenomena
| Kingdom | Species | Stage tested | Delivery method | Reference(s) |
|---|---|---|---|---|
| Protozoans | Trypanosoma brucei | Procyclic forms | Transfection | 52 |
| Plasmodium falciparum | Blood stage | Electroporation and soaking | 143, 150 | |
| Toxoplasma gondii | Mature forms in fibroblast | Transfection | 4 | |
| Paramecium | Mature form | Transfection and feeding | 14 | |
| Leishmania donovanii | Tried but not working | 183 | ||
| Invertebrates | Caenorhabditis elegans | Larval stage and adult stage | Transfection, feeding bacteria carrying dsRNA, soaking | 26, 31 |
| Caenorhabditis briggsae | Adult | Injection | 79 | |
| Brugia malayi (filarial worm) | Adult worm | Soaking | 1 | |
| Schistosoma mansoni | Sporocysts | Soaking | 23 | |
| Hydra | Adult | Delivered by micropipette | 49 | |
| Planaria | Adult | Soaking | 49 | |
| Lymnea stagnalis (snail) | Adult | Injection | 122 | |
| Drosophila melanogaster | Cell lines, adult, embryo | Injection for adult and embryonic stages, soaking and transfection for cell lines | 96, 114, 155 | |
| Cyclorrphan (fly) | Early embryonic stages | Injection | 200 | |
| Milkweed bug | Early embryonic stages | Injection | 102 | |
| Beetle | Early embryonic stages | Injection | 27 | |
| Cockroach | Larval stage | Injection | 146 | |
| Spodoptera frugiperda | Adult and cell line | Injection and soaking | 176, 215 | |
| Vertebrates | Zebra fish | Embryo | Microinjection | 224 |
| Xenopus laevis | Embryo | Injection | 162 | |
| Mice | Prenatal, embryonic stages, and adult | Injection | 31, 229 | |
| Humans | Human cell lines | Transfection | 42 | |
| Plants | Monocots/dicots | Plant | Particle bombardment with siRNA/transgenics | 88 |
| Fungi | Neurospora crassa | Filamentous fungi | Transfection | 51 |
| Schizosaccharomyces pombe | Filamentous fungi | Transgene | 178 | |
| Dictyostelium discoideum | Transgene | 147 | ||
| Algae | Chlamydomonas reinhardtii | Transfection | 231 |
During the occurrence of RNAi/PTGS, double-stranded RNA (dsRNA) molecules, which cleave the inducer molecules into smaller pieces first (16) and eventually destroy the cellular or viral cognate mRNA molecules (called the target) (17) act as inducers or activators of this process. As a result, the target mRNAs cannot accumulate in the cytosol, although they remain detectable by nuclear run-on assays (73). In certain instances, the DNA expressing the target mRNA also undergoes methylation as a by-product of the degradation process (226).
The natural functions of RNAi and its related processes seem to be protection of the genome against invasion by mobile genetic elements such as viruses and transposons as well as orchestrated functioning of the developmental programs of eukaryotic organisms. There are several excellent recent reviews which deal with different aspects of RNAi separately (95, 191). Here, we have put together the various aspects of the RNAi process known to date, identified the mechanistic similarities and differences operating in various forms of eukaryotic life, and focused on the experimental results that have led to conceptual advancements in this field.
UNRAVELING RNA SILENCING
In order to understand the process of homology-dependent RNA silencing, it would be prudent to overview the process itself and describe its important features. In the later part of this review, the genetics, biochemistry, and potential therapeutic applications of the process will be dealt with.
PTGS in Plants
In plants, the RNA silencing story unfolded serendipitously during a search for transgenic petunia flowers that were expected to be more purple. In 1990, R. Jorgensen's laboratory wanted to upregulate the activity of a gene for chalcone synthase (chsA), an enzyme involved in the production of anthocyanin pigments. Surprisingly, some of the transgenic petunia plants harboring the chsA coding region under the control of a 35S promoter lost both endogene and transgene chalcone synthase activity, and thus many of the flowers were variegated or developed white sectors (163). The loss of cytosolic chsA mRNA was not associated with reduced transcription, as demonstrated by run-on transcription tests in isolated nuclei (216). Jorgensen coined the term cosuppression to describe the loss of mRNAs of both the endo- and the transgene.
Around the same time, two other laboratories (105, 217) also reported that introduction of the transcribing-sense transgenes could downregulate the expression of homologous endogenous genes. Subsequently, many similar events of cosuppression were reported in the literature. All cases of cosuppression resulted in the degradation of endogene and transgene RNAs after nuclear transcription had occurred (120). Since posttranscriptional RNA degradation was observed in a wide range of transgenes expressing the plant, bacterial, or viral sequences, it was rechristened posttranscriptional gene silencing (PTGS). PTGS could be initiated not only by sense transgenes but also by antisense transgenes, and biochemical evidence suggests that similar mechanisms might operate in both cases (81). It is worthwhile to point out that although the cosuppression phenomenon was originally observed in plants, it is not restricted to plants and has also been demonstrated in metazoans and mammals (98).
In keeping with the times, the observed alterations in the PTGS-related phenotypes were attributed to multiple-site integrations, aberrant RNA formations, repeat structures of the transgenes, etc. Later on, it became clear that the expression of the transgene led to the formation of dsRNA, which, in turn, initiated PTGS. For example, in the case of cosuppressed petunia plants, chsA mRNA formed a partial duplex, since there are regions of self-complementarity located between chsA 3′ coding region and its 3′ untranslated region (154). This was revealed by DNA sequence analysis and experimental detection of in vitro-transcribed, RNase-resistant duplex chsA RNA. In an independent study, a p35S-ACC (1-aminocyclopropane-1-carboxylate [ACC] oxidase) sense transgene carrying a small inverted repeat in the 5′ untranslated region was introduced into tomato to test the role of dsRNA structure as an inducer of PTGS. Cosuppression of the endogenous acc gene occurred at a higher frequency in these plants than in those harboring only the p35S-ACC sense transgene without the inverted repeat (93).
Reports from several laboratories in the past few years have established that the loss in steady-state accumulation of the target mRNA is almost total if the designed transgene construct of the transgenic plant produces the nuclear transcript in the duplex conformation. Very recently it was reported that the expression of self-cRNA of plum pox virus under the control of rolC promoter caused degradation of transgenic viral RNA and as a result, the systemic disease resistance to challenge inoculum of plum pox virus occurred with a high frequency in transgenic Nicotiana benthamiana (170). This evidence points out that the production of dsRNA is required to initiate PTGS in plants. Based on this, plants carrying strongly transcribing transgenes in both the sense and antisense orientations are currently being produced that show strong PTGS features. These transgenic plants can silence endogene, invading viral RNA, or unwanted foreign genes in a sequence-specific and heritable manner.
Generally, the sense and antisense components of the above-mentioned transgenes are separated only by an intron to increase the efficacy of PTGS (43, 198). For example, Arabidopsis thaliana and Lycopersicon esculentum (tomato) plants were transformed with a transgene construct designed to generate self-complementary iaaM and ipt transcripts. iaaM and ipt are oncogenes of agrobacteria that are responsible for crown gall formation in infected plants. The transgenic lines retained susceptibility to Agrobacterium transformation but were highly refractory to tumorigenesis, providing functional resistance to crown gall disease by posttranscriptional degradation of the iaaM and ipt transcripts (72).
Quelling and RNAi
While reports of PTGS in plants were piling up, homology-dependent gene silencing phenomena were also observed independently in fungal systems. These events were called quelling. Quelling came to light during attempts to boost the production of an orange pigment made by the gene al1 of the fungus Neurospora crassa (50). An N. crassa strain containing a wild-type al1+ gene (orange phenotype) was transformed with a plasmid containing a 1,500-bp fragment of the coding sequence of the al1 gene. A few transformants were stably quelled and showed albino phenotypes. In the al1-quelled strain, the level of unspliced al1 mRNA was similar to that of the wild-type strain, whereas the native al1 mRNA was highly reduced, indicating that quelling and not the rate of transcription affected the level of mature mRNA in a homology-dependent manner.
The phenomenon of RNAi first came into the limelight following the discovery by Fire et al. (78), who unequivocally demonstrated the biochemical nature of inducers in gene silencing by introducing purified dsRNA directly into the body of Caenorhabditis elegans. The investigators injected dsRNA corresponding to a 742-nucleotide segment of unc22 into either the gonad or body cavity region of an adult nematode. unc22 encodes an abundant but nonessential myofilament protein, and the decrease in unc22 activity is supposed to produce an increasingly severe twitching phenotype. The injected animal showed weak twitching, whereas the progeny individuals were strong twitchers. The investigators showed that similar loss-of-function individuals could also be generated with dsRNAs corresponding to four other nematode genes. The phenotypes produced by interference by various dsRNAs were extremely specific.
This experiment paved the way for easy production of null mutants, and the process of silencing a functional gene by exogenous application of dsRNA was termed RNA interference (RNAi). RNAi in C. elegans was also initiated simply by soaking the worms in a solution containing dsRNAs or by feeding the worms Escherichia coli organisms that expressed the dsRNAs (209). This is a very potent method, requiring only catalytic amounts of dsRNA per cell to silence gene expression. The silencing spread not only from the gut of the worm to the remainder of the body, but also through the germ line to several generations. These phenomena of RNAi have also been demonstrated to occur in Drosophila melanogaster and many other invertebrates and vertebrates.
Insights from Virus-Infected Plants (Virus-Induced Gene Silencing)
Besides the processes mentioned above, homology-driven RNA degradation also occurs during the growth of viral genomes in infected plants (73). Viruses can be either the source, the target, or both the source and the target of silencing. PTGS mediated by viruses can occur with RNA viruses, which replicate in the cytoplasm, and also with DNA viruses, which replicate in the nucleus (71). As early as in the 1920s, it was known that plants could be protected from a severe virus by prior infection with a mild strain of a closely related virus. Although the mechanism of such cross protection in plants remained unknown for a long time, such phenomena could be explained partly in terms of PTGS that could be induced by the mild strain and targeted later against the virulent viral genome. It was also found that transforming plants with virus-derived transgenes gave protection against the challenge viruses even when no transgene protein was produced (132).
Analyses of these virus-resistant plants revealed that the transgenes were highly transcribed in the nucleus, whereas the steady-state level of cytoplasmic mRNA was very low. Further analysis suggested that some of the transgenic mRNA molecules assumed the conformation of dsRNA, which triggered sequence-specific degradation of self and other homologous or cRNA sequences in the cytoplasm. Thus, in the virus-resistant lines, not only the transgene mRNAs but also the mRNA from the homologous endogenous gene and the invading viral RNA (with homology to the transgene) were degraded.
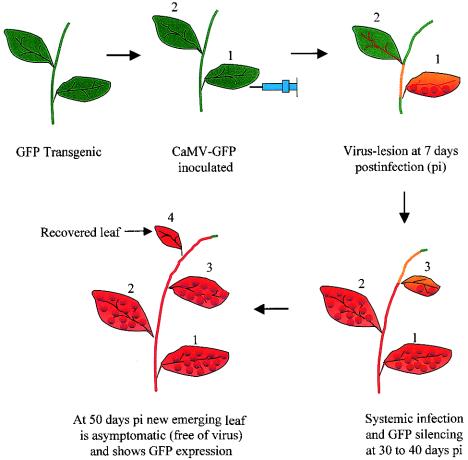
Another form of virus-induced gene silencing is the phenomenon of viral recovery itself. When Brassica napus was inoculated with cauliflower mosaic virus (a DNA virus), lesions at the site of virus entry were visible 5 to 7 days postinoculation. Symptoms of systemic infections were apparent by 10 to 14 days postinoculation. Symptoms were most prominent at 30 to 40 days postinoculation and declined thereafter (i.e., the plants recovered), with the newly emergent leaves remaining asymptomatic at 50 days postinoculation (5).
Figure 1 diagrammatically illustrates the systemic spread of RNAi in plants. Such recovery occurred by a PTGS-like mechanism because 19S and 35S RNAs encoded by the cauliflower mosaic virus were degraded while cauliflower mosaic virus DNA was still replicating in the nucleus. Induction of PTGS was visualized if the cauliflower mosaic virus infection and subsequent recovery were followed up in a transgenic B. napus expressing a p35S-GUS (β-glucuronidase) transgene. At the site of inoculation, GUS silencing associated with local lesions was first observed 7 days postinoculation. GUS silencing eventually spread systemically, and the GUS activity of the entire plant was suppressed by 50 days postinoculation. In this particular example, cauliflower mosaic virus acted as the inducer of PTGS for the transgenes sharing homology with the virus within the transcribed region. However, the virus itself was also the target of the induced PTGS, since 19S and 35S RNAs were found degraded.
FIG. 1.
Schematic illustration of systemic viral spread as well as RNAi and subsequent viral recovery in plants. Green and red indicate the presence and loss of GFP fluorescence, respectively, and orange denotes the presence of both colors. The red dots on leaves show viral lesions. The bold arrows indicate the stages of plant growth, and the leaves are numbered accordingly. An arrow with a thin line shows a newly emerged leaf recovered from viral attack.
A similar example of virus-induced gene silencing was found when Nicotiana clevelandii was infected with an RNA nepovirus, tomato black ring virus (179). RNA viruses make abundant dsRNA during intracellular replication of their genomes and thus elicit cellular PTGS degradative activity. Virus-induced gene silencing also occurs with viruses that do not undergo recovery. When a DNA geminivirus, tomato golden mosaic virus (TGMV), infected N. benthamiana, a high level of viral DNA replication in the nucleus and accumulation of viral RNA in the cytoplasm occurred. An infection by a recombinant TGMV carrying the coding sequence of the sulfur (su) gene of the host plant in either the sense or antisense orientation led to the bleaching of leaves due to PTGS of the endogenous su gene, but the DNA of the recombinant did not fail to replicate (117). Here, TGMV acted as an inducer of PTGS but was not itself a target of PTGS. Thus, plant viruses elicit PTGS but sometimes can escape the degradative PTGS activity.
Based on the principles of virus-induced gene silencing, vectors designed with the genome sequence of RNA viruses tobacco mosaic virus, potato virus X, and tobacco rattle virus are being widely used to knock down the expression of host genes. The characteristics of many plant genes were revealed by observing the loss-of-function-related phenotypic changes when the recombinant vectors incorporating the concerned host genes were introduced into plants (136). Of these vectors, the TRV-based are more promising because these are capable of inducing-meristematic gene silencing, which has not been possible to achieve with other RNA virus-based vectors. Meristematic gene silencing employing TGMV vectors has also been reported (173). Thus, virus-induced gene silencing-based techniques are extremely useful for studies related to functional genomics in plants.
IMPORTANT FEATURES OF RNA SILENCING
Independently of one another, investigations on diverse organisms, labeled variously as PTGS in plants, RNAi in animals, quelling in fungi, and virus-induced gene silencing, have converged on a universal paradigm of gene regulation. The critical common components of the paradigm are that (i) the inducer is the dsRNA, (ii) the target RNA is degraded in a homology-dependent fashion, and, as we will see later, (iii) the degradative machinery requires a set of proteins which are similar in structure and function across most organisms. In most of these processes, certain invariant features are observed, including the formation of small interfering RNA (siRNA) and the organism-specific systemic transmission of silencing from its site of initiation.
siRNA
The key insight in the process of PTGS was provided from the experiments of Baulcombe and Hamilton (92), who identified the product of RNA degradation as a small RNA species (siRNA) of ≈25 nucleotides of both sense and antisense polarity. siRNAs are formed and accumulate as double-stranded RNA molecules of defined chemical structures, as mentioned later. siRNAs were detected first in plants undergoing either cosuppression or virus-induced gene silencing and were not detectable in control plants that were not silenced. siRNAs were subsequently discovered in Drosophila tissue culture cells in which RNAi was induced by introducing >500-nucleotide-long exogenous dsRNA (96), in Drosophila embryo extracts that were carrying out RNAi in vitro (240), and also in Drosophila embryos that were injected with dsRNA (236). Thus, the generation of siRNA (21 to 25 nucleotides) turned out to be the signature of any homology-dependent RNA-silencing event.
The siRNAs resemble breakdown products of an E. coli RNase III-like digestion (13). In particular, each strand of siRNA has 5′-phosphate and 3′-hydroxyl termini and 2- to 3-nucleotide 3′ overhangs. Interestingly, in vitro-synthesized siRNAs can, in turn, induce specific RNA degradation when added exogenously to Drosophila cell extracts (69). Specific inhibition of gene expression by these siRNAs has also been observed in many invertebrate and some vertebrate systems (67). Recently, Schwarz et al. (189) provided direct biochemical evidence that the siRNAs could act as guide RNAs for cognate mRNA degradation.
Amplification and Systemic Transmission
Besides the formation of siRNAs, another intriguing characteristic of homology-dependent gene silencing is that the inducer dsRNA molecules do not act stoichiometrically. It was estimated that only two molecules of dsRNA per cell were able to induce RNAi of an abundantly expressed C. elegans gene such as unc22. In another report, injection of dsRNA into the intestine of a C. elegans hermaphrodite generated RNAi, which could be stably inherited to the F2 generation. These two findings led to the proposal that RNAi signals could be systemic and amplifiable in nature (78). The similar systemic effects of RNAi have also been demonstrated in the planarian Schmidtea mediterranea and the cnidarian Hydra magnipapillata (140).
Similar evidence is also available for plant PTGS. The new tissues growing from a GUS-expressing scion grafted onto a GUS-silenced rootstock show progressive silencing of GUS expression (168). The silencing signal seems to spread by a nonmetabolic, gene-specific diffusible signal, which travels both between cells, through plasmadesmata, and long distances via the phloem (75). In the case of virus-induced gene silencing, the systemic character has also been revealed (185). To account for the gene specificity of a systemic signal, it has been proposed that the signal could be an RNA molecule (228). However, such processes are not universal, as these are not found in flies and mammals.
COMPONENTS OF GENE SILENCING
Both genetic and biochemical approaches have been undertaken to understand the basis of silencing. Genetic screens were carried out in the fungus Neurospora crassa, the alga Chlamydomonas reinhardtii, the nematode Caenorhabditis elegans, and the plant A. thaliana to search for mutants defective in quelling, RNA interference, or PTGS. Analyses of these mutants led to the identification of host-encoded proteins involved in gene silencing and also revealed that a number of essential enzymes or factors are common to these processes. Some of the components identified serve as initiators, while others serve as effectors, amplifiers, and transmitters of the gene silencing process. In the years to come, many other components as well as their interrelations will be revealed. Here, we outline what is known so far.
Dicer
RNase III family members are among the few nucleases that show specificity for dsRNAs (164) and cleave them with 3′ overhangs of 2 to 3 nucleotides and 5′-phosphate and 3′-hydroxyl termini (69). Bernstein et al. (17) identified an RNase III-like enzyme in Drosophila extract which was shown to have the ability to produce fragments of 22 nucleotides, similar to the size produced during RNAi. These authors showed that this enzyme is involved in the initiation of RNAi. Owing to its ability to digest dsRNA into uniformly sized small RNAs (siRNA), this enzyme was named Dicer (DCR). These nucleases are evolutionarily conserved in worms, flies, fungi, plants, and mammals. Dicer has four distinct domains: an amino-terminal helicase domain, dual RNase III motifs, a dsRNA binding domain, and a PAZ domain (a 110-amino-acid domain present in proteins like Piwi, Argo, and Zwille/Pinhead), which it shares with the RDE1/QDE2/Argonaute family of proteins that has been genetically linked to RNAi by independent studies (34, 203). Cleavage by Dicer is thought to be catalyzed by its tandem RNase III domains. Some DCR proteins, including the one from D. melanogaster, contain an ATP-binding motif along with the DEAD box RNA helicase domain.
The predicted C. elegans Dicer homologue, K12H4.8, was referred as DCR1 because it was demonstrated to be the functional ortholog of the Drosophila Dicer protein (173). The 8,165-bp DCR1 protein has a domain structure similar to that of the Drosophila Dicer protein. dcr1 mutants of C. elegans showed defects in RNAi of germ line-expressed genes but no effect on the RNAi response of somatic genes. These mutants were found to be sterile, suggesting the important role of this gene in germ line development apart from RNAi (119). CAF1 has been identified as a Dicer homologue in A. thaliana, but it is not involved in PTGS activity. The structure of CAF1 shows the presence of the four distinct domains that were identified in the Drosophila Dicer protein (17, 36, 108). Dicer homologues from many different sources have been identified; some recombinant Dicers have also been examined in vitro, and phylogenetic analysis of the known Dicer-like proteins indicates a common ancestry of these proteins (83).
Complete digestion by RNase III enzyme results in dsRNA fragments of 12 to 15 bp, half the size of siRNAs (235). The RNase III enzyme acts as a dimer and thus digests dsRNA with the help of two compound catalytic centers, whereas each monomer of the Dicer enzyme possesses two catalytic domains, with one of them deviating from the consensus catalytic sequences.
Recently, the crystal structure of the RNase III catalytic domain was solved, and this led to the model for generation of 23- to 28-mer diced siRNA products (20). In this model, the dimeric Dicer folds on the dsRNA substrate to produce four compound catalytic sites so that the two terminal sites having the maximum homology with the consensus RNase III catalytic sequence remain active, while the other two internal sites bearing partial homology lose functional significance. Thus, the diced products appear as the limit digests of the RNase III enzymes and are double the size of the normal 12- to 15-mer fragments. Such a model also predicts that certain changes in Dicer structure might modify the spacing between the two active terminal sites and thus generate siRNAs of variable sizes bearing species-specific imprints (98). Clearly, the crystal structure of Dicer is necessary to authenticate this model.
Guide RNAs and RNA-Induced Silencing Complex
Hammond et al. (96) determined that the endogenous genes of Drosophila S2 cells could be targeted in a sequence-specific manner by transfection with dsRNA, and loss-of-function phenotypes were created in cultured Drosophila cells. The inability of cellular extracts treated with a Ca2+-dependent nuclease (micrococcal nuclease, which can degrade both DNA and RNA) to degrade the cognate mRNAs and the absence of this effect with DNase I treatment showed that RNA was an essential component of the nuclease activity. The sequence-specific nuclease activity observed in the cellular extracts responsible for ablating target mRNAs was termed the RNA-induced silencing complex (RISC) (96).
After partial purification of crude extracts through differential centrifugation and anion exchange chromatography, the nuclease cofractionated with a discrete ≈25-nucleotide RNA species. These results suggested that small RNAs were associated with sequence-specific nuclease and served as guides to target specific messages based upon sequence recognition. In another report, the multicomponent RNAi nuclease was purified to homogeneity as a ribonucleoprotein complex of ≈500 kDa (97). One of the protein components of this complex was identified as a member of the Argonaute family of proteins and was termed Argonaute2 (AGO2). AGO2 is homologous to RDE1, a protein required for dsRNA-mediated gene silencing in C. elegans. AGO2 is a ≈130-kDa protein containing polyglutamine residues, PAZ, and PIWI domains characteristic of members of the Argonaute gene family. The Argonaute family members have been linked both to the gene-silencing phenomenon and to the control of development in diverse species. The first link between Argonaute protein and RNAi was shown by isolation of rde1 mutants of C. elegans in a screen for RNAi-deficient mutants. Argonaute family members have been shown to be involved in RNAi in Neurospora crassa (QDE3) as well as in A. thaliana (AGO1) (75).
Recently, two independent groups identified additional components of the RISC complex. Hammond and group showed the presence of two RNA binding proteins, the Vasa intronic gene and dFMR proteins, in the RISC complex isolated from Drosophila flies (35). Of these, dFMR is a homologue of the human fragile X mental retardation protein. In a parallel study, Siomi and group also isolated a novel ribonucleoprotein complex from the Drosophila lysate that contained dFMRI, AGO2, a Drosophila homologue of p68 RNA helicase (Dmp68), and two ribosomal proteins, L5 and L11, along with 5S rRNA (106). Both of these groups showed not only the presence of these components in the RISC complex, but also interactions among these proteins in vitro. Other components of RISC have not been clearly established yet. Nevertheless, some of the proteins mentioned below could very well constitute the RISC complex.
RNA and DNA Helicases
Aberrant RNA elimination surveillance seems to be common to most eukaryotic organisms. However, a diverse array of proteins specific for each organism seem to carry out such surveillance. Broadly, they fall in the biochemically similar group of RNA-DNA helicases. A mutant strain (mut6) of C. reinhardtii was isolated in which a gene required for silencing a transgene was disrupted (232). This RNAi-resistant mutant also showed an elevated transposition activity. The mut6 gene was cloned and sequenced. The deduced MUT6 protein contains 1,431 amino acids and is a member of the DEAH box RNA helicase family. It also has a glycine-rich region that includes several RGG repeats, resembling an RGG box, a motif implicated in RNA binding and protein-protein interactions. MUT6 also has three putative nuclear localization signals and is predicted to be nuclear by PSORT analysis (161). MUT6 RNA helicase may be involved in degradation of misprocessed aberrant RNAs and thus could be a part of an RNAi-related surveillance system.
In Neurospora crassa, three classes of quelling-defective mutants (qde1, qde2, and qde3) have been isolated (46). The qde3 gene has been cloned, and the sequence encodes a 1,955-amino-acid protein (48). The protein shows homology with several polypeptides belonging to the family of RecQ DNA helicases, which includes the human proteins for Bloom's syndrome and Werner's syndrome (238). In addition, QDE3 is believed to be involved in the activation step of gene silencing. The DNA helicase activity of QDE3 may function in the DNA-DNA interaction between introduced transgenes or with a putative endogenous gene required for gene-silencing activation by unwinding the double-stranded DNA. These interactions may induce changes in methylation or chromatin structure, producing an altered state that could result in aberrant RNA production. Thus, QDE3 protein may be more important for the transcriptional part of gene silencing, i.e., TGS.
When the RNAi sensitivity of several existing C. elegans mutants was examined, two mutant strains, mut2 and mut7, that had previously shown elevated levels of transposon mobilization also showed resistance to RNAi. Ketting et al. (116) identified a mutator gene, mut7, in C. elegans and characterized it at the molecular level. MUT7 was found to be homologous to proteins with 3′-5′ exonuclease domains, such as Werner's syndrome protein and E. coli RNase D. It contained all the key catalytic residues for nuclease activity. A model was proposed in which MUT7 was speculated to play a role in repressing transposition by degrading the target mRNA with its exonuclease activity.
smg (suppressor of morphological effects on genitalia) mutants of C. elegans, defective in a process called nonsense-mediated decay, have been isolated (63). Seven smg genes which are involved in nonsense-mediated decay have been identified (29, 100). Since this process also involves RNA degradation, the function of these genes, if any, in the RNAi process was examined. Animals mutant for a subset of these genes, smg2, smg5, and smg6, were initially silenced by dsRNA but later showed rapid recovery from the effects of RNAi, unlike the wild-type worms, which remained silenced. Thus, these genes might affect the persistence of RNA interference. On the other hand, smg1, smg3, and smg4 mutant animals behaved like wild-type worms and did not recover from RNAi at all, indicating that these genes are not required for RNAi persistence. The smg5 and smg6 genes have not been cloned, but the smg2 gene shows homology to Saccharomyces cerevisiae upf1, which encodes an ATPase with RNA-binding and helicase activities.
The SMG proteins could unwind dsRNA to provide a template for amplification activity. In this way, the three SMG proteins might facilitate amplification of the silencing signal and cause persistence of the silenced state. Alternatively, SMG proteins could increase the number of dsRNA molecules by promoting endonucleolytic cleavage of existing dsRNA molecules, which has been observed in Drosophila flies. No SMG2 homologues have been identified in plants or fungi. However, a search of the A. thaliana genome sequence database revealed a number of candidates with either helicase and/or RNase domains.
In a recent report, Tijsterman et al. (208) showed that unlike sense oligomers, single-stranded oligomers of antisense polarity could induce gene silencing in C. elegans. The antisense RNA-induced gene silencing was explained by proposing that RNA synthesis was primed on the mRNA by antisense RNA, resulting in dsRNAs, which acted as substrates for Dicer-dependent degradation. Antisense RNAs showed a requirement for the mutator/RNAi genes mut7 and mut14 but acted independently of the RNAi genes rde1 and rde4 of C. elegans. The mut14 gene was cloned by genetic mapping and subsequent candidate gene approach. The MUT14 protein is a member of the family of putative RNA helicases that contain the signature DEAD box motif. These proteins are involved in diverse cellular functions. The helicase activity of MUT14 might thus act to permit de novo RNA synthesis on the target.
Dalmay et al. (54) identified an sde3 locus in A. thaliana plants which is required for the PTGS phenotype. They proposed that SDE3 protein might be involved in the production of dsRNA. SDE3 differs markedly from QDE3/MUT7 and has slight similarity to MUT6 in the helicase motif. Although it is highly similar to Upf1p and SMG2, it is unlikely that SDE3 is the functional homologue of Upf1p and SMG2 because it lacks important motifs (167). Notably, no SDE3 homologue was found in C. elegans, suggesting that SDE3-like proteins are regulators rather than essential cofactors of PTGS and are not used in C. elegans. This is further supported by the observation that sde3 mutant plants exhibit only partial loss of PTGS (55). The closest homologue of SDE3 as identified by BlastP was a mouse protein encoded by gb110 (91, 159). These SDE3 homologues have RNA helicase motifs that are quite distinct from those of the DEAD, DEAH, and Ski2p types of RNA helicase (134). It has been speculated that SDE3 and SMG2 are multifunctional RNA helicases involved in PTGS.
Translation Initiation Factor
Mutants of C. elegans showing resistance to dsRNA-mediated RNAi were selected by Tabara et al. (203). They genetically mapped seven mutant strains that were placed in four complementation groups. One of the groups, rde1, consisted of three alleles. Gene rde1 is a member of a large family which includes Drosophila homologues (piwi and sting) and Arabidopsis homologues (argonaute and zwille) and rabbit eIF2C. The full-length cDNA sequence for rde1 was determined, and the deduced protein, consisting of 1,020 amino acids, was referred to as RDE1. The RDE1 protein is homologous to the product of the quelling deficiency (qde2) gene in Neurospora crassa (75). The initiation step of RNAi might be affected in the rde1 mutant, as it completely lacks an interference response to several dsRNAs. It does not show any increase in transposon mobilization and or any effect on growth and development.
RNA-Dependent RNA Polymerase
The effects of both RNAi and PTGS are potent and systemic in nature. This has led to a proposed mechanism in which RNA-dependent RNA polymerases (RdRPs) play a role in both triggering and amplifying the silencing effect. Transgenic and virus-infected plants show an accumulation of aberrant transgenic and viral RNAs. The RdRP enzymes might recognize these aberrant RNAs as templates and synthesize antisense RNAs to form dsRNAs that are finally the targets for sequence-specific RNA degradation (45, 47, 56, 133).
Genetic screens of Neurospora crassa (QDE1) (48) and A. thaliana (SDE1/SGS2) (54, 160) led to the identification of proteins which are similar to tomato RdRP (77, 187) and are required for quelling and PTGS, respectively. This testifies to the importance of RdRP in gene silencing. Cogoni et al. (45) cloned the qde1 gene from N. crassa. It encodes a 158-kDa protein which lacks the typical signal peptide or a transmembrane domain, indicating its intracellular location. Dalmay et al. (54) found that the 113-kDa Arabidopsis RdRP is encoded by sde1. It is a plant homologue of QDE1 in N. crassa and EGO1 in C. elegans, which are required for quelling and RNAi, respectively. The SDE1 protein is required for transgene silencing but not for virus-induced PTGS, suggesting that SDE1 might be required to produce dsRNA, the initiator of PTGS (54).
The dsRNA produced as an intermediate in virus replication by virus-encoded RdRP might induce PTGS itself, and thus SDE1 may not be required for virus-induced PTGS. Plants with the sde mutation grow and develop normally, excluding a role for sde in development or basic cellular function. Two PTGS-controlling genes, sgs2 and sgs3, were identified in A. thaliana by another group of workers (160). Later, it was found that sgs2 and sde1 are different descriptions of the same gene. On comparing the protein sequence of all the RdRPs, a conserved block was identified which seems to be crucial for RdRP function in PTGS and RNAi. sgs3 mutants have the same molecular and phenotypic characteristics as sgs2 mutants, but the SGS3 protein shows no significant similarity with any known putative proteins.
In C. elegans, EGO1, a protein required for RNAi, was found to be similar to tomato RdRP and the QDE1 protein of Neurospora crassa (197), as mentioned earlier. For a number of germ line-expressed genes, ego1 mutants were resistant to RNA interference. The ego1 transcript is found predominantly in the germ line. ego1 is thus yet another example of a gene encoding an RdRP-related protein with an essential developmental function. RdRP is speculated to play a role in the amplification of the dsRNA signal, allowing its spread throughout the organism (50, 77, 168, 221). The RdRP is also perhaps responsible for sustaining PTGS at the maintenance level even in the absence of the dsRNA that initiates the RNAi effect.
In spite of its omnipresence in different kinds of eukaryotic cells, RdRP homologues are not coded by either the Drosophila or human genome. Though the systemic characteristics of RNAi have not been revealed yet in either flies or humans, the amplification of siRNAs may be an essential step of RNAi even in these systems. Hence, it is important to know how these steps of RNAi are biochemically carried out in the absence of RdRP activity.
Transmembrane Protein (Channel or Receptor)
The systemic spread of gene silencing from one tissue to another has been well established in C. elegans and plants. To investigate the mechanism of systemic RNAi, Winston et al. (231) constructed and used a special transgenic strain of C. elegans (HC57). They identified a systemic RNA interference-deficient (sid) locus required to transmit the effects of gene silencing between cells with green fluorescent protein (GFP) as a marker protein. Of the 106 sid mutants belonging to three complementation groups (sid1, sid2, and sid3), they isolated and characterized sid1 mutants. The sid1 mutants had no readily detectable mutant phenotype other than failure to show systemic RNAi. Interestingly, these mutants also failed to transmit the effect of RNAi to the progeny.
The SID1 polypeptide is predicted to be a 776-amino-acid membrane protein consisting of a signal peptide and 11 putative transmembrane domains. Based on the structure of SID1, it was suggested that it might act as a channel for the import or export of a systemic RNAi signal or might be necessary for endocytosis of the systemic RNAi signal, perhaps functioning as a receptor. No homologue of sid1 was detected in D. melanogaster, which may be consistent with the apparent lack of systemic RNAi in the organism (80, 174). However, the presence of SID homologues in humans and mice might hint at the systemic characteristics of RNAi in mammals.
Genetic Mutations with Unknown Function
The three other complementation groups identified by Tabara et al. (203) in C. elegans are rde2 and rde3, with one allele each, and rde4, with two alleles. rde4 mutants behaved like the rde1 strain in not showing any increase in transposon mobilization and no effect on growth and development. The product of rde2 remains to be identified. mut2, rde2, and rde3 exhibited high-level transposition similar to mut7. This suggests a possible biological role of RNAi in transposon silencing (203).
Mello and colleagues (87) have proposed that rde1 and rde4 respond to dsRNA by producing a secondary extragenic agent that is used by the downstream genes rde2 and mut7 to target specific mRNAs for PTGS. According to this view, rde1 and rde4 act as initiators of RNAi whereas rde2 and mut7 are effectors. Various components of gene silencing have been listed in Table 2.
TABLE 2.
Components of posttranscriptional gene silencing
| Phenomenon | Organism | Mutation causing defective silencing | Gene function | Developmental defect |
|---|---|---|---|---|
| Posttranscriptional | Plant (Arabidopsis thaliana) | sgs2/sde1 | RdRP | None |
| gene silencing | sgs3 | Unknown function | None | |
| sde3 | RecQ helicase | |||
| ago1 | Translation initiation factor | Pleiotropic effects on development & fertility | ||
| caf1 | RNA helicase & RNase III | |||
| Quelling | Fungus (Neurospora crassa) | qde-1 | RdRP | None |
| qde-2 | Translation initiation factor | None | ||
| qde-3 | RecQ DNA helicase | |||
| RNA interference | Worm (Caenorhabditis elegans) | ego-1 | RdRP | Gametogenetic defect & sterility |
| rde-1 | Translation initiation factor | None | ||
| rde-2, rde-3, rde-4, mut-2 | Unknown function | None | ||
| K12H4.8 (dcr-1) | Dicer homologue RNA helicase & RNase III | Sterility | ||
| mut-7 | Helicase & RNase D | None | ||
| mut-14 | DEAD box RNA helicase | |||
| smg-2 | Upflp helicase | |||
| smg-5 | Unknown function | |||
| smg-6 | Unknown function | |||
| sid-1 | Transmembrane protein | |||
| Alga (Chlamydomonas reinhardtii) | mut-6 | DEAH box RNA helicase |
MECHANISM OF RNA INTERFERENCE
As the various pieces of the RNAi machinery are being discovered, the mechanism of RNAi is emerging more clearly. In the last few years, important insights have been gained in elucidating the mechanism of RNAi. A combination of results obtained from several in vivo and in vitro experiments have gelled into a two-step mechanistic model for RNAi/PTGS. The first step, referred to as the RNAi initiating step, involves binding of the RNA nucleases to a large dsRNA and its cleavage into discrete ≈21- to ≈25-nucleotide RNA fragments (siRNA). In the second step, these siRNAs join a multinuclease complex, RISC, which degrades the homologous single-stranded mRNAs. At present, little is known about the RNAi intermediates, RNA-protein complexes, and mechanisms of formation of different complexes during RNAi. In addition to several missing links in the process of RNAi, the molecular basis of its systemic spread is also largely unknown.
Processing of dsRNA into siRNAs
Studies of PTGS in plants provided the first evidence that small RNA molecules are important intermediates of the RNAi process. Hamilton and Baulcombe (92), while studying transgene-induced PTGS in five tomato lines transformed with a tomato 1-aminocyclopropane-1-carboxyl oxidase (ACO), found accumulation of aco small RNAs of 25 nucleotides. More direct evidence about the generation of siRNAs in RNAi came from an in vitro cell-free system obtained from a Drosophila syncytial blastoderm embryo by Tuschl et al. (212). These authors were able to reproduce many of the features of RNAi in this system. When dsRNAs radiolabeled within either the sense or the antisense strand were incubated with Drosophila lysate in a standard RNAi reaction, 21- to 23-nucleotide RNAs were generated with high efficiency. Single-stranded 32P-labeled RNA of either the sense or antisense strand was not efficiently converted to 21- to 23-nucleotide products. The formation of the 21- to 23-nucleotide RNAs did not require the presence of corresponding mRNAs.
The role of the small RNAs in RNAi was confirmed independently by Elbashir et al. (69), who showed that synthetic 21- to 23-nucleotide RNAs, when added to cell-free systems, were able to guide efficient degradation of homologous mRNAs. To assess directly if the siRNAs were the true intermediates in an RNAi reaction, Zamore et al. (240) fractionated both the unprocessed dsRNAs and processed dsRNAs from the Renilla luc dsRNA-treated cell-free Drosophila system and showed that only the fractions containing native siRNAs were able to bring about the cognate RNA degradation and their ability to degrade RNA was lost when these fractions were treated at 95°C for 5 min. These in vivo and in vitro studies thus provided the evidence that siRNAs are the true intermediates of the RNAi reaction.
Together with the experiments to identify siRNAs as the key molecules for the RNAi effect, several investigators carried out the logical search for polypeptides that could generate such molecules. Based on the binding and cleavage properties of E. coli RNase III enzymes, Bass (13) for the first time predicted the involvement RNase III-type endonucleases in the degradation of dsRNA to siRNAs. The RNase III enzyme makes staggered cuts in both strands of dsRNA, leaving a 3′ overhang of 2 nucleotides. The first evidence for the involvement of RNase III enzyme in RNAi was provided by T. Tuschl’s group, who chemically analyzed the sequences of the 21- to 23-nucleotide RNAs generated by the processing of dsRNA in the Drosophila cell-free system. They showed the presence of 5′-phosphate, 3′-hydroxyl, and a 3′ 2-nucleotide overhang and no modification of the sugar-phosphate backbone in the processed 21- to 23-nucleotide RNAs (69).
Two groups recently identified candidate enzymes involved in degradation by scanning the genomes of D. melanogaster and C. elegans for genes encoding proteins with RNase III signatures (17, 115). Bernstein et al. (17) showed that one of these identified genes, dicer in Drosophila, codes for the RNA processing enzyme that fragments dsRNA into 22-nucleotide fragments in vitro. An antiserum raised against Dicer could also immunoprecipitate a protein from the Drosophila extract or from S2 cell lysate, and these Dicer protein immunoprecipitates were able to produce RNAs of about 22 nucleotides from the dsRNA substrate. The direct correspondence in size of these RNAs with those generated from dsRNA by cell extract suggested a role of this protein in dsRNA degradation. The role of Dicer in RNAi was further confirmed by the fact that the introduction of Dicer dsRNA into Drosophila cells diminished the ability of the transfected cells to carry out RNAi in vitro. Similar experimental studies were carried out with C. elegans extract, and an ortholog of Dicer named DCR1 was identified.
A number of in vivo and in vitro experimental studies have shown that the production of 21- to 23-nucleotide RNAs from dsRNA requires ATP. The rate of 21- to 23-nucleotide RNA formation from corresponding dsRNAs has been shown to be six times slower in the Drosophila extract depleted for ATP by treatment with hexokinase and glucose (165). Bernstein et al. (17) and Ketting et al. (115) showed that the Dicer immunoprecipitates from D. melanogaster as well as S2 cell extracts and DCR1 immunoprecipitates from C. elegans extract required ATP for the production of 22-nucleotide RNAs (17, 115). Recently, Nykanen et al. (165) reduced ATP levels in Drosophila extract by 5,000-fold with a sensitive ATP depletion strategy and showed considerable reduction in the rate of siRNA production in the Drosophila cell extract. These experiments suggest that ATP controls the rate of siRNA formation. However, it is still unclear whether ATP is absolutely rate limiting for the production of siRNAs from dsRNA.
The RNase activity and dsRNA binding of 218-kDa recombinant human Dicer have also been examined in vitro (175). The enzyme generated siRNA products from dsRNA quite efficiently in the presence of Mg2+ and the absence of ATP. The RNase activity was sensitive to ionic interactions, whereas the dsRNA binding was quite effective in presence of high salt and did not require Mg2+ at all. The dsRNA binding domain is located at the C terminus of Dicer, which is separable from the helicase and PAZ motifs. Human Dicer expressed in mammalian cells colocalized with calreticulin, a resident protein of the endoplasmic reticulum. In other systems, Dicer has also been found to complex with various other proteins (35, 106). Hence, it is possible that the Dicer RNase activity functions as a complex of proteins in vivo.
Amplification of siRNAs
One of the many intriguing features of RNA interference is the apparently catalytic nature of the phenomenon. A few molecules of dsRNA are sufficient to degrade a continuously transcribed target mRNA for a long period of time. Although the conversion of long dsRNA into many small siRNAs results in some degree of amplification, it is not sufficient to bring about such continuous mRNA degradation. Since mutations in genes encoding RNA-dependent RNA polymerase (RdRP) affect RNAi, it was proposed that this type of polymerase might replicate siRNAs as epigenetic agents, permitting their spread throughout plants and between generations in C. elegans. Recent studies by Lipardi et al. (135) and Sijen et al. (193) provided convincing biochemical and genetic evidence that RdRP indeed plays a critical role in amplifying RNAi effects.
Lipardi et al. (135), while investigating the dsRNA-dependent degradation of target mRNA in a Drosophila embryo cell extract system, showed the generation of full-length cognate dsRNAs from labeled siRNAs at early time points. Both single-stranded RNAs (equivalent to target mRNA) and dsRNAs served as templates for copying by RdRP. New full-length dsRNAs were formed rapidly and cleaved. They also showed a strict requirement for the 3′-hydroxyl group and 5′-phosphate group on siRNAs for primer extension in the RdRP-mediated reaction (135).
Sijen et al. (193) further revealed the role of RdRP activity in RNAi. In an RNAi reaction, they observed the formation of new siRNA species corresponding to target mRNAs but different from trigger dsRNAs. They named these new siRNAs secondary siRNAs. With a primary trigger dsRNA specific for the lacZ region of the target mRNA that encoded a GFP-LacZ fusion protein, these authors demonstrated the degradation of a separate GFP mRNA target. This kind of RNAi induced by secondary siRNAs was named transitive RNAi. These authors demonstrated the requirement for the rrf1 gene, a C. elegans gene with sequence homology to RdRP, in the generation of secondary siRNAs and transitive RNAi (193).
Amplification of siRNAs might occur at various stages of the RNAi reaction and has been documented in plants, C. elegans, N. crassa, and Dictyostelium discoideum but not in flies and mammals (66). Though the RdRP activity is present in Drosophila embryo extract, as mentioned earlier, it is surprising that the fly genome does not code for RdRP. Additionally, numerous experiments also suggest that RdRP is not required for RNAi in D. melanogaster (98).
Degradation of mRNA
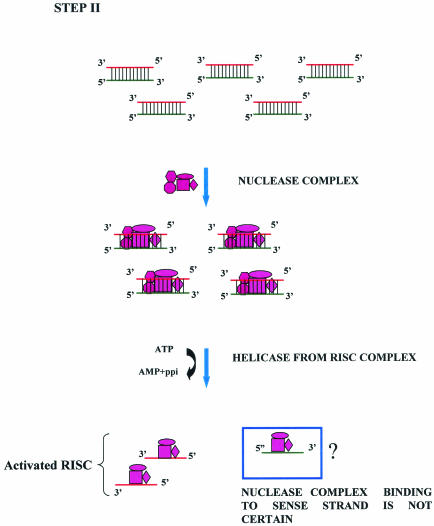
In the effector step of RNAi, the double-stranded siRNAs produced in the first step are believed to bind an RNAi-specific protein complex to form a RISC. This complex might undergo activation in the presence of ATP so that the antisense component of the unwound siRNA becomes exposed and allows the RISC to perform the downstream RNAi reaction. Zamore and colleagues (240) demonstrated that a ≈250-kDa precursor RISC, found in Drosophila embryo extract, was converted into a ≈100-kDa complex upon being activated by ATP. This activated complex cleaved the substrate. The size and constitution of the precursor as well as the activated RISC might vary depending on the choice of system (98). The antisense siRNAs in the activated RISC pair with cognate mRNAs, and the complex cuts this mRNA approximately in the middle of the duplex region.
A few independent studies demonstrated the importance of the RISC complex in this part of RNAi reactions. The mRNA-cleaving RNA-protein complexes have also been referred to as siRNP (small interfering ribonucleoprotein particles). It is widely believed that this nuclease is probably different from Dicer, judging from the substrate requirements and the nature of the end products. Since the target cleavage site has been mapped to 11 or 12 nucleotides downstream of 5′ end of the guide siRNA, a conformational rearrangement or a change in the composition of an siRNP ahead of the cleavage of target mRNA is postulated. Finally, the cleaved mRNAs are perhaps degraded by exoribonucleases (96).
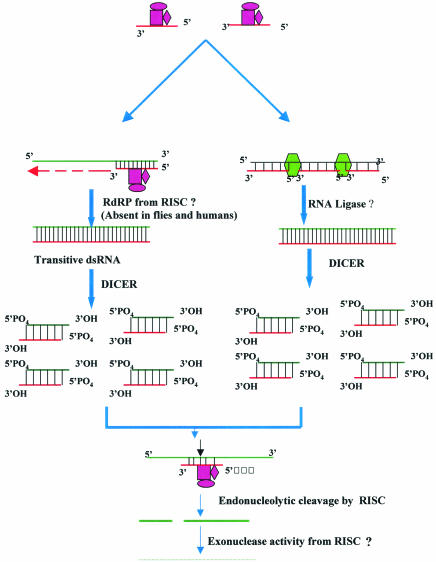
A part of cleaved fragments of mRNA at the end of step 2 might also be converted to the duplex forms by the RdRP-like activity. These forms might have siRNA-like functions and eventually enter the pool of the amplification reaction. Thus, it is likely that amplification of the RNAi reaction takes place at both step 1 and step 2 of RNAi. In another model, it has been proposed that siRNAs do not act as primers for the RdRP-like enzymes, but instead assemble along the length of the target RNA and are then ligated together by an RNA ligase to generate cRNA. The cRNA and target RNA hybrid would then be diced by the DCR protein. All these models were summarized by Schwarz et al. (189). Most of the steps involved in the mechanism of RNAi have been illustrated schematically in Fig. 2.
FIG. 2.
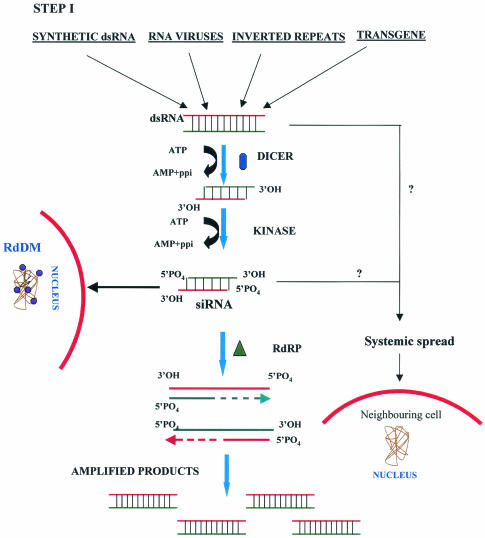
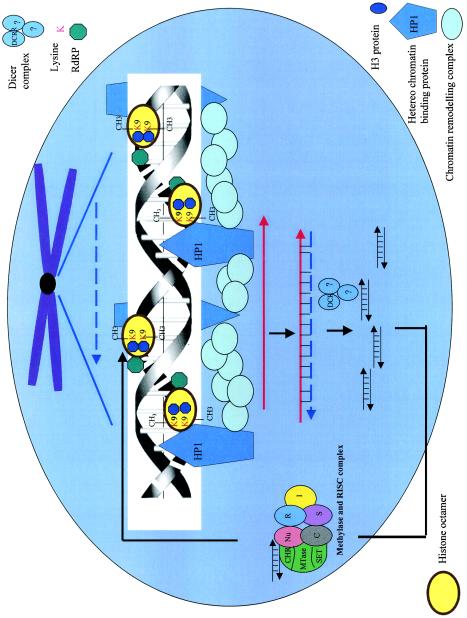
Two-step model for the mechanism of gene silencing induced by double-stranded RNA. In step I, dsRNA is cleaved by the Dicer enzyme to produce siRNAs. A putative kinase seems to maintain 5′ phosphorylation at this step. The siRNAs have also been proposed to be responsible for nuclear DNA methylation (•) and systemic spread of silencing. Amplification might occur due to the presence of RdRP (▴). In step II, the siRNAs generated in step I bind to the nuclease complex (RISC). A helicase present in the complex might activate RISC by unwinding the siRNAs. The antisense component of siRNA in the RISC guides the complex towards the cognate mRNA (—), resulting in endonucleolytic cleavage (↓) of the mRNA. RdDM, RNA-dependent DNA methylation.
RNA SILENCING FOR GENOME INTEGRITY AND DEFENSE
Considerable evidence indicates that PTGS has evolved as a protective mechanism against parasitic DNA sequences such as transposons and the RNA sequences of plant viruses. DNA methylation and transcriptional gene silencing (TGS) are mainly responsible for keeping the transposition frequency at a minimum. However, PTGS also provides additional protection against the genomic instability caused by transposons. Mutations in the C. elegans mut-7 gene increase the transposition frequency in the germ line and downregulate RNAi as well (58), implicating RNAi in the control of transposons. Recently, Djikeng et al. (61) cloned and sequenced the siRNA products of an RNA interference event occurring in Trypanosoma brucei. By sequencing over 1,300 siRNA-like fragments, they observed abundant 24- to 26-nucleotide fragments homologous to the ubiquitous retrotransposon INGI and the site-specific retroposon SLACS. Thus, they convincingly demonstrated that RNAi is involved in silencing the retroposon transcript.
In plants, PTGS has been widely linked with RNA virus resistance mechanisms (219, 227). Plant RNA viruses are, in fact, both inducers and targets for PTGS and gene-silencing-defective mutants of plants show increased sensitivity to viral infections (160). The direct role of dsRNA in inhibiting viral infection has recently been demonstrated by Tenllado and Diaz-Ruiz (207). They showed that dsRNAs derived from viral replicase sequences could interfere with virus infection in a sequence-specific manner by directly delivering the dsRNAs to leaf cells either by mechanical coinoculation with the virus or via an Agrobacterium-mediated transient-expression approach. Successful interference with the infection of plants by representative viruses belonging to the tobamovirus, potyvirus, and alfamovirus genera has been demonstrated. These results support the view that a dsRNA intermediate in virus replication acts as an efficient initiator of PTGS in natural virus infections.
The clinching support for the notion that PTGS has evolved as an antiviral mechanism has come from reports that plant viruses encode proteins that are suppressors of PTGS (8, 25, 222). These suppressors have evolved to save the viral RNA genomes from the PTGS degradative machinery of host plants. Different types of viral suppressors have been identified through the use of a variety of silencing suppression assays. Suppressors HC-PRO, P1, and AC2 are one type (encoded by potyviruses, rice yellow mottle sobemovirus, and geminiviruses of subgroup III, respectively) that is able to activate GFP expression in all tissues of previously silenced GFP-expressing plants (222). HC-PRO reduces target mRNA degradation and is thus responsible for reduced accumulation of siRNAs (137, 145). The second type of suppressors include movement proteins, i.e., p25 of potato virus X, which are involved in curbing the systemic aspect of transgene-induced RNA silencing (220). The third type includes cytomegalovirus 2b protein, which is involved in systemic signal-mediated RNA silencing (60). The cytomegalovirus 2b protein is nucleus localized and also inhibits salicylic acid-mediated virus resistance (141). Other types of viral suppressors with undefined biochemical activities are also known (128). These findings not only provide the strongest support that PTGS functions as a natural, antiviral defense mechanism, but also offer valuable tools for dissecting the biochemical pathways of PTGS (128).
The PTGS degradative machinery can both detect and inactivate repetitive DNA sequences, suggesting it controls the expansion of repetitive elements, including endogenous genes (18). Although RNAi occurs in mammals and mammalian cell cultures, its role in animal virus protection is not clear. In mammals, dsRNA induces RNAi as well as interferon-mediated nonspecific RNA degradation and other nonspecific responses leading to blockage in protein synthesis and cell death (2). Thus, mammals seem to have evolved multiple mechanisms to detect and target dsRNA and to fight viruses. These various mechanisms may have different specificities or can function in distinct tissues or during development (210). A few other roles of RNAi in development and genome maintenance will be discussed in later sections.
MECHANISTIC DIFFERENCES AMONG THE BIOSYNTHETIC PATHWAYS OF siRNA
Although the functional parallelism of gene silencing is quite apparent in plants and animals, a few unique attributes separate the pathways in these groups. For example, systemic spreading of the RNAi reaction from the site of initiation is known to occur in plants and worms (74, 79), but not in flies or mammals. The noteworthy distinct molecules that have been identified to cause differences at the pre-Dicer, Dicer, and post-Dicer stages of gene silencing pathways are mentioned below.
Pre-Dicer stage.
Plant proteins such as SGS2 (RdRP), SGS3 (coiled protein), AGO1 (responsible for plant development), and HEN1 (enhancer of floral hua1 mutation) are required for PTGS activities induced by the sense transgenes. But if the transgenes are in the form of hairpins expressing the panhandle dsRNA, the absence of or defects in the above-mentioned proteins do not play any role in altering the PTGS/cosuppression function. Hence, those proteins supposedly play a role upstream of dicing of dsRNA and may be involved in the formation and stabilization of dsRNA (22).
Homologues of SGS3 are unknown beyond the plant world. Even though HEN1 analogs are known in bacteria, yeasts, and animals, their roles in sense PTGS have not yet been identified. Likewise, SGS2 homologues are known in C. elegans, N. crassa, and Dictyostelium discoideum, but their roles at the pre-dicing stage have not been established yet in those systems. The equivalents of SGS2 in other animal systems are nonexistent both structurally and functionally (205). The role of worm AGO1 protein, i.e., RDE1, is also unique, as described earlier. AGO1 homologues are present in all eukaryotes, but they mostly function as a component (AGO2) of the animal RISC complex (32). The plant HEN1 protein is believed to be nuclear because of the presence of the nuclear localization signal at its N-terminal region (40). Since HEN1 is essential for plant PTGS (cosuppression), which is supposedly a cytoplasmic activity, the exploration of its subcellular distribution is of utmost importance. Boutet et al. (22) speculated that HEN1 could be a dsRNA stabilizing protein, and since many such proteins are known in the animal kingdom, it would be of interest to find animal analogs of the plant HEN1 protein. In fungus as well as the animal system, sense transgene-induced PTGS phenomena are known, but the machinery operative at the pre-dicing stage is still elusive.
The roles of plant SGS2, SGS3, AGO1, and HEN1 proteins may be limited at the stage of production of dsRNA from the transcript of sense transgenes, but no mechanism has been established regarding the presentation of the dsRNA to Dicer for the generation of siRNA. However, such a mechanism has been reported in C. elegans. The RDE4 and RDE1 (AGO1) proteins of C. elegans were reported as initiators of RNAi and speculated to have no mechanistic role in the downstream processes of RNAi (87, 203). Unlike the Arabidopsis AGO1 and HEN1 proteins, RDE4 and RDE1 proteins are required for RNAi even when the dsRNAs are produced intracellularly in transgenic worms (203), but the defects in RDE4 and RDE1 are of no consequence if exogenous siRNAs or short antisense RNAs drive the RNAi reaction (208). RDE4 binds tightly to dsRNA (during the RNAi reaction) by virtue of its two RNA-binding domains and is always found in a tight complex with RDE1 protein even in absence of the RNAi reaction. During RNAi, RDE4 is found in a complex with RDE1, Dicer (DCR1), and a conserved DEXH-box RNA helicase (DRH1/DRH2). Based on these observations and other genetic evidence, Tabara and coworkers postulated that RDE4 and RDE1 functioned together to detect and retain foreign dsRNA and present the dsRNA to DCR1 for processing into siRNAs (202). Analogs of the RDE4 and DRH proteins are found in many eukaryotes, including plants and humans, but their roles have not been defined yet.
Dicer stage.
The plant Dicer responsible for biosynthesis of plant siRNA is not known yet, whereas the Dicers of C. elegans, D. melanogaster, and humans as effectors for siRNA have been well characterized. The A. thaliana and rice genomes both encode at least seven RNase III-like proteins, of which at least four are putative homologues of Dicer, conveniently called DCLs (i.e., DCL1, DCL2, DCL3, and DCL4). The genetic evidence rules out that the Arabidopsis DCL1 (or CAF1) could be competent for siRNA formation (76). The roles of other DCL proteins are still to be revealed.
Interestingly, both in vivo and in vitro data suggest that the end products of plant dicing activities are different from those of the animal Dicers. When uniformly32P-labeled dsRNA was incubated with wheat germ extract, Zamore et al. (205) found that the dsRNA was chopped into siRNAs of two discrete size classes, one ≈21 nucleotides and the other 24 to 25 nucleotides long, whereas D. melanogaster and human Dicers generated only the 21-nucleotide siRNAs. Two similar size classes were also produced with cauliflower extract and were found independently in the set of 423 endogenous small RNAs cloned from A. thaliana. Thus, in plants, dicing activity leads to the generation of two distinct classes of siRNAs.
With specific synthetic siRNAs that supposedly bind tightly to and inhibit Dicer as competitors, Zamore et al. concluded that a different Dicer-like enzyme was responsible for the generation of each class of siRNA. These two distinct classes of siRNAs were reported first in vivo from transgenic plants bearing the silenced GFP sense transgenes (94). With an array of plant virus-encoded suppressors of gene silencing, Baulcombe et al. proposed that the 21-mer siRNAs controlled localized PTGS via mRNA degradation and the 24-mer siRNAs triggered systemic silencing and methylation of the homologous DNA. It remains to be seen whether this kind of dual dicing activity reflects any novel pathway intrinsic to plant RNAi. Interestingly, the longer (≈25-mer) siRNAs have also been detected in the natural RNAi biology of Trypanosoma brucei (61).
Post-Dicer stage.
RISC has been isolated from D. melanogaster, C. elegans, and humans, and only some of its components have been characterized biochemically and genetically. Both mammalian and Drosophila RISC contain AGO2 proteins, whereas the GEMIN3 (a DEAD box helicase) and GEMIN4 proteins are found only in mammalian RISC (103). Similarly, dFXR, a homologue of the human fragile X mental retardation protein, is found only in Drosophila RISC (35). However, there is no report on the isolation of RISC in plants. Hence, mechanistically little is known about postdicing activity, especially in plants. A worthwhile question to address is whether there is any anchoring site for the occurrence of RNAi in the cytoplasm. Recently, it was reported that E. coli RNase III binds to the 70S ribosome and is functionally modified after binding (6). It is widely believed that the RISC associates with eukaryotic ribosomes (96). Hence, the exploration of ribosome association of the RNAi activities, especially of dicing and postdicing leading to mRNA degradation, might shed light on RNAi mechanisms in the future. The various affinities of ribosome-binding complexes might also reveal interesting system-specific features.
RdRP-dependent siRNA amplification and systemic spreading from the site of origin is another area where many system-specific variations have been noticed. RdRP homologues are not present in many organisms, so the mechanisms by which sense transgene-mediated PTGS are effected in those organisms remain a mystery (98). In other systems where RdRP is present, the biochemical steps and details of siRNA amplification may not necessarily be the same.
In C. elegans, RRF1 (a putative homologue of RdRP), along with other proteins, is required for RNAi even when the trigger dsRNA is expressed directly from the hairpin transgene in the nuclei of somatic tissues, whereas SGS2 (Arabidopsis RdRP) is dispensable for PTGS activity if induced directly by hairpin sense transgenes in A. thaliana. This suggests that the RdRP-mediated putative amplification steps of worms are different from those of plants (37). In plants, the SGS2-dependent spreading of silencing occurs from the region homologous to the trigger dsRNA into both the adjacent nonhomologous 5′ and 3′ regions of a target transgene (214). In contrast, spreading occurs only in the 5′ region in worms and fungi, which is consistent with the primer-dependent 5′-3′ copying activity of RdRP. Hence, in plants, the spread of silencing requires other activities (such as chromatin modification) in addition to that of RdRP (37).
In worms, tissue-specific variations of RdRP-dependent RNAi have also been reported, but not in plants or other systems. EGO1 is essential for RNAi in the germ line of C. elegans, whereas another RdRP homologue, RRF1, is required for silencing in soma (193, 197). Another intriguing observation is that the loss of function of RRF3 (third putative RdRP of worms) is responsible for the enhancement of sensitivity to RNAi in several tissues of C. elegans. Here, RRF3 acts as a negative regulator of RNAi, a fact difficult to reconcile with the postulated activity of RdRP (195). For systemic transmission of gene silencing, the membrane-bound SID1 protein of C. elegans and the plasmodesmatal connections of plants are implicated, but in both cases, the molecular nature of the moving signal has not been ascertained yet. An association between Dicer and the RdRP has been suspected in the case of Dictyostelium discoideum and C. elegans, but conclusive evidence is still lacking (37).
siRNA: SYNTHESIS, DELIVERY, AND GENE KNOCKDOWN
The natural RNAi biology of eukaryotic cells offers a protection mechanism against foreign nucleic acids; however, only in the recent past has the exploitation of its mechanistic details sparked a revolution in the investigation of cellular gene functions. Transcriptional regulation with the dsRNA technology provides an easy means to identify cellular characteristics in response to both internal and external cues. However, the application of RNAi in higher eukaryotes, particularly mammalian cells, has been hampered by the presence of a number of dsRNA-triggered pathways that mediate nonspecific suppression of gene expression (152). These nonspecific responses to dsRNA are not triggered by dsRNAs shorter than 30 bp, including the siRNA duplexes. Moreover, studies in C. elegans and D. melanogaster have clearly demonstrated that synthetic siRNAs can produce effects similar to those of the long dsRNAs (69, 236). Based on these experimental analyses, siRNAs are now being optimized for systematic exploration of the function of genes in a variety of organisms.
Prior to the siRNA era, approaches such as gene targeting by homologous recombination, ribozymes, and antisense technologies were commonly used to determine gene functions. All such approaches have their limitations, and none can be applied universally (201). The dawn of siRNA-directed knockdown approaches facilitated studies of gene function in a rapid and inexpensive way. This siRNA technology has the potential to decipher the function of virtually any gene that is expressed in a cell type- or pathway-specific manner. In the span of only a few years, large-scale functional analysis of almost all the ≈19,000 genes of C. elegans has been carried out with the siRNA-directed knockdown approach. A fairly detailed account of this technology has recently been reviewed by Dykxhoorn et al. (66).
Here, some of the salient aspects of the technology are summarized. In brief, the application of siRNA for gene silencing involves a careful consideration of the following variables: (i) selecting the siRNA sequence in the target gene; (ii) synthesis of siRNAs or construction of plasmids bearing DNA sequence encoding for siRNAs; (iii) optimizing transfection of the siRNAs or the plasmids expressing siRNAs in the target cells; and (iv) monitoring the efficacy of gene silencing.
Selection and Generation of siRNA
Several siRNAs synthesized against different regions of the same target mRNA show different silencing efficiencies (101). A number of groups have analyzed several parameters for optimizing siRNA-induced gene silencing, and these include the length, secondary structure, sugar backbone, and sequence specificity of the siRNA duplex. The efficacy of these parameters has been tested on several occasions for induction of RNAi in D. melanogaster and human cells (69, 189). No consensus on choosing the siRNA sequence has evolved. A line of thinking seems to suggest the following. The sequence should be selected in the region 50 to 100 bp downstream of the start codon. The 5′ or 3′ untranslated regions and regions near the start codon should be avoided, assuming that untranslated region-binding proteins and translation initiation complexes may interfere with the binding of siRNP or RISC endonuclease complex. The GC content of the siRNAs should be kept between 30 and 70%. The computer programs developed by Lin (Jack Lin's siRNA sequence finder; www.Ic.sunysb.edu/stu/shiklin/rnai.html) and by Ambion (www.ambion.com) offer helpful guidelines to select potential siRNA sequences and determine whether these selected sequences match mRNA sequences other than those of intended target.
Based on different experimental approaches, a few guidelines have been laid for the synthesis of siRNAs. A general rule is that the sequence of one strand should be AA(N19)TT, where N is any nucleotide, i.e., these siRNAs should have a 2-nucleotide 3′ overhang of uridine residues. The siRNAs should be 21 nucleotides long. The siRNAs should have 5′-phosphate and 3′-hydroxyl group for efficiency. Compared to antisense or ribozyme technology, the secondary structure of the target mRNA does not appear to have a strong effect on silencing. The 21-nucleotide siRNAs can be chemically synthesized with appropriately protected ribonucleoside phosphoramidites and a conventional synthesizer and thus are widely available commercially. However, the use of chemically synthesized siRNA in RNAi has been restricted because of the high synthesis cost. Due to the paucity of information on the selection of siRNAs and their structures, these general guidelines are suggestive and do not guarantee the silencing effect. To overcome the siRNA selection ambiguity, Yang et al. (235) incubated dsRNA with the E. coli RNase III enzyme to generate a random array of siRNAs. The introduction of such a reaction soup resulted in the silencing of the target gene.
The exorbitant cost of synthesizing siRNAs and their lack of amplification in mammalian cells have compelled investigators to explore alternative strategies to generate a continuous supply of a battery of siRNAs. Several groups have devised strategies to synthesize short RNAs in vitro (64) or by introducing plasmids with the ability to make de novo siRNAs inside the cell (235, 239). DNA-based plasmid vectors have been designed by cloning siRNA templates downstream of an RNA polymerase III transcription unit, which normally encodes the small nuclear RNA U6 or human RNase H1.
Two approaches have been developed for expressing siRNAs. In the first, sense and antisense strands constituting the siRNA duplex are transcribed by individual promoters (64), and in the second, siRNAs are expressed as fold-back stem-loop structures that give rise to siRNAs with a small loop. A stretch of four to five thymidines is added at the end to the siRNA template that acts as a transcription termination signal. Many of these plasmid-based vectors, such as pSilencer 1.0 (Ambion) and pSuper (DNA Engine), are now commercially available. These vectors provide advantages over chemically synthesized siRNAs, but use of these plasmid vectors also remains limited due to numerous disadvantages, including the transient nature of siRNA expression and low as well as variable transfection efficiency.
To circumvent these problems. virus-based high-efficiency siRNA delivery systems are also being developed. A retrovirus-based system developed by Devree and Silver (59) is cited here as an example. The U6 promoter along with the siRNA-generating hairpin construct was cloned upstream of the 3′ long terminal repeat of the commercially available pMSCV-puro vector. The in vitro-packaged recombinant virus was allowed to transfect HeLa cells with high efficiency in the presence of puromycin selection, and a dramatic downregulation of the target gene product was observed. A downregulation of this extent was not possible with the plasmid-based delivery system. Such virus-based vectors or their improved variants hold the promise to efficiently detect the function of any gene in virtually any cell type, provided that the production of recombinant virus is not a limitation.
Most of the siRNA expression vectors produced to date use RNA polymerase III regulatory units, which do not allow tissue-specific siRNA expression. However, Shiagawa and Ishiid reported a polymerase II promoter-based plasmid encoding a dsRNA expression system that could eventually express siRNA in a tissue-specific manner (192). In their novel scheme, a pDECAP vector was used, which expressed long dsRNAs corresponding to the ski gene (encoding a transcriptional repressor) in the form of a hairpin. The engineered hairpin RNA expressed from a cytomegalovirus promoter lacked the 7-methylguanosine cap structure at its 5′ end and a poly(A) tail at its 3′ end. The transcript of such a design did not exit the nucleus to reach the cytoplasm and thus prevent the interferon pathway-mediated nonspecific antiviral response. The double-stranded transcript was diced in the nucleus, and the siRNAs were subsequently released into the cytoplasm to mediate the gene-specific silencing. The silencing was specific, since the level of a related protein, SNO, remained unaffected.
The same vector was also used to create ski knockdown mice, the phenotype of which was similar to that of ski knockout embryos, which exhibited defects in neural tube and eye formation. Later generations of such vectors may use more tissue-specific cis-acting elements in the employed promoter to stringently knock down gene functions in the animal system. It is pertinent to highlight here that because plants do not elicit an interferon-mediated antiviral response, the dsRNA/siRNA delivery system need not be as complex as the pDECAP system.
Transfection of siRNA and Detection of Gene Silencing
An attempt to understand a gene's function in diverse organisms necessitates optimization of protocols for efficient delivery of siRNAs into cells. A number of transfection reagents are being employed for transfecting siRNA into different cell lines. Lipofectamine 2000 and Oligofectamine (Invitrogen) are being routinely used for siRNA delivery. A few newer transfection reagents such as TransIT-TKO (Mirus) and Ambion's Siport Amine and Siport, have also been used successfully in cultured cell lines. Electroporation has been used to transfect siRNAs in cell lines as well as in parasites such as Trypanosoma brucei and Plasmodium falciparum (150, 213). In adult mice, naked siRNAs have been delivered by hydrodynamic transfection methods to combat hepatitis C virus infection in the intact liver (151). The transfecting siRNAs have been used successfully for studying the role of proteins in DNA damage response and cell cycle control, general cell cycle metabolism, signaling, the cytoskeleton and its rearrangement during mitosis, membrane trafficking, transcription, and DNA methylation (211). These molecules have also been used to differentiate between housekeeping and other genes (112).
The preferred way to detect specific gene knockdown by RNAi is to study the depletion of the target protein by immunofluorescence and Western blotting with the specific antibody. In addition, the knockdown phenotype and Northern blot analysis can also be used to detect the effects of siRNA. If the gene is essential, cellular growth is delayed or arrested, and [3H]thymidine uptake can also be used to assign the function of a particular gene (70).
siRNA Introduction into Plants
siRNAs have been delivered into tobacco plants by biolistic pressure to cause silencing of GFP expression. Silencing occasionally was detected as early as a day after bombardment, and it continued to potentiate up to 3 to 4 days postbombardment. Systemic spread of silencing occurred 2 weeks later to manifest in the vascular tissues of the nonbombarded leaves that were closest to the bombarded ones. After a month or so, the loss of GFP expression was seen in nonvascular tissues as well. RNA blot hybridization with systemic leaves indicated that the biolistically delivered siRNAs induced the de novo formation of siRNAs, which accumulated to cause systemic silencing (118).
MICRO-RNA
Since the RNAi machinery is present constitutively within eukaryotic cells, it is important to explore and understand the metabolic advantages that are accorded by RNAi-related proteins during the intrinsic normal growth of cells and development of organisms. The natural RNAi machinery not only keeps the mobile transposable elements from disrupting the integrity of genomes, as was suggested by analyses in lower plants, A. thaliana, C. elegans, D. melanogaster, and animals (9, 94, 138, 203, 232), but also participates in organism development. Genetic defects in C. elegans RNAi genes ego1 and dicer cause known, specific developmental errors (87, 119, 197). Similarly, the Argonaute family of genes of A. thaliana (especially the ZWILLE proteins) is also responsible for plant architecture and meristem development (32), and the Dicer homologue of A. thaliana, CAF1, is required for embryo development (83). Thus, genetic evidence illustrates the role of the RNAi machinery as a controller of development-related genes. The mechanistic details of these developmental processes are beginning to emerge.
In 1991, Ambros and coworkers first isolated a lin4 mutant of C. elegans which was arrested at the first larval stage (127). Later on, the let7 mutation was isolated in the same system, which was responsible for development through the fourth larval stage. Both lin4 and let7 encode short 22-nucleotide mature RNAs and were called short temporal RNA because they control the temporal development program of C. elegans. The mature lin4 RNA defines (negatively regulates) the mRNA expression of the lin14 and lin28 heterochronic genes with the antisense-mediated repression mechanism of translation initiation and thus specifies the fate of cells during the first three larval stages. Recent studies have revealed that the short temporal RNAs are actually members of a group of tiny RNAs (21 to 28 nucleotides) called the micro-RNAs, isolated members of which could easily run to a few hundreds. Some of the components of the RNAi machinery have also been clearly established as the effector proteins for the maturation of micro-RNAs.
Identification and Biogenesis
A range of biochemical techniques have been applied to clone the 21- to 28-nucleotide RNAs that are present during the normal cellular development of many organisms, for exploring the abundance and complexity of micro-RNA. Micro-RNAs have been found to be abundant and phylogenetically extensive in plants, flies, worms, and humans. In D. melanogaster, C. elegans, plants, and humans, more than 600 micro-RNAs have been identified (123, 125, 126, 137). Bioinformatic analyses of the complete genome sequences have been extremely useful for identification studies. The genome sequences of a variety of organisms revealed the authenticity of these micro-RNAs, the nature of the precursor RNAs, the genomic locations of micro-RNA genes, and the evolutionarily conserved character of some of these micro-RNAs. With the RNA folding program mfold (148) and Northern analyses of micro-RNA, it has been universally inferred that most micro-RNAs arise from the imperfectly annealed 70-nucleotide hairpin precursor RNA whose expression is often developmentally regulated. These micro-RNAs are thus predicted to be processed from multiple bulged and partially duplex precursors, like the short temporal RNA precursors (186).
The identification of micro-RNAs is the first major hurdle in micro-RNA-related research. The first step in computational identification of micro-RNAs from genome sequences is identification of sequences forming hairpin loops (stem-loop sequences). For this purpose, software such as srnloop (85) and RNA fold (130) is used. These are Blast-like software packages which identify short complementary sequences within a specified distance on the genome. The hairpin sequences obtained by this analysis are then evaluated as candidate micro-RNAs based on different criteria, such as GC content and minimum free energy, and by passing through different filters, such as short-repeat filters and structure quality filters (85).
Another important criterion that has been used for the identification of candidate micro-RNAs is the correspondence of a hairpin of one species with that of another species. Two hairpins are said to be in correspondence if a short sequence (>19 nucleotides) in the stem of one hairpin is also present in the stem of another hairpin, although the two hairpins may have otherwise variable sequences. If a hairpin from one species has correspondence with a hairpin from one or more other species, this strengthens its status as a candidate to be a micro-RNA. The homology of hairpins with known micro-RNAs is also considered a useful criterion to select candidate micro-RNAs.
D. P. Bartel's laboratory has developed a computational procedure, called MiRscan, to identify micro-RNAs based on their homology to known micro-RNAs with respect to the characteristic features in the stem region. MiRscan evaluates the stem-loops by passing a 21-nucleotide window along the stem region and assigning a likelihood score to each window that measures how well its attributes resemble those of the previously experimentally identified and validated micro-RNAs (129, 130). The candidate micro-RNAs identified by these procedures are experimentally validated by Northern blot assay of total small RNAs with the stem region of the candidate as a probe or by a more sensitive PCR assay of the amplified small RNA library (85, 130). Detection of a 21- to 24-nucleotide band in these assays validates a candidate micro-RNA, whereas a ≈70-nucleotide band is detected in Dicer-deficient mutants, further confirming that the micro-RNAs arise from a ≈70-nucleotide precursor.
An analysis of micro-RNA expression in cell lines and tissues suggests cell- or tissue-specific expression. For example, micro-RNA 1 (miR1) is specifically expressed in human heart tissues and stage-specifically in mouse embryogenesis (126). A. thaliana small RNA 39 is detected exclusively in inflorescence tissues and downregulates the expression of a Scarecrow-like transcription factors (137, 139). Considering the diverse functions in which micro-RNAs have been implicated, micro-RNAs have also been named variously, i.e., micro-RNAs which mediate spatial development are referred to as sdRNAs, while cell cycle micro-RNAs are referred to as ccRNAs, etc. The regulated expression patterns of these micro-RNAs are suggestive of their functions in developmental control. However, many micro-RNAs are uniformly expressed, suggesting their role in general gene regulation (186). Downregulation of micro-RNAs leads to serious developmental defects, as evidenced by isolation of various micro-RNA mutants. Recent reports reveal that miR15 and miR16 are located in human chromosome 13q14, a region which gets deleted in more than half of B-cell chronic lymphocytic leukemias. Detailed deletion and expression analyses point out that these two micro-RNAs are located within a 30-kb region of loss in chronic lymphocytic leukemias, and both genes are deleted or downregulated in a majority (≈68%) of chronic lymphocytic leukemia cases (30).
A majority of micro-RNAs occur in relatively short (≈70-nucleotide) and single stem-loop precursor structures. However, in both animals and plants, some micro-RNAs are arranged in clusters. The genes in the tandem clusters are coexpressed, for example, in the germ line and early embryos of C. elegans and D. melanogaster (123, 125) and in the inflorescence tissues of A. thaliana (137). A set of seven highly related C. elegans micro-RNA genes that are coexpressed are so tightly clustered within 1-kb region that they are predicted to form a precursor from which all the seven mature micro-RNAs are processed (186). Similarly, several micro-RNAs originate from each of the five chromosomes of A. thaliana containing clusters of two to four micro-RNAs spaced irregularly within the intergenic region. Interestingly, three of the clusters contain micro-RNA sequences of both sense and antisense polarities, a scenario not found in the animal system yet. Such variations in the precursor structures of micro-RNAs may point towards distinct mechanisms of biosynthesis of micro-RNAs, although all micro-RNAs originate by transcription events that are independent of adjacent conventional genes.
Apoptosis-Related Micro-RNA
The proliferation of tissues and organs of any organism requires careful coordination between cell proliferation and cellular death. The proliferation processes of a cell include active inhibition of the apoptotic process. Recently, two micro-RNA genes, bantam and mir14, that suppress cell death by inhibiting the translation of apoptotic messages have been isolated from D. melanogaster. Expression of the bantam 21-nucleotide micro-RNA is temporally and spatially regulated in response to patterning cues. The proapoptosis gene hid has been identified as a target for regulation by bantam micro-RNA (24). bantam deletion mutants grow poorly and die as early pupae, whereas mir14 mutants are viable but stress sensitive and cursed with a reduced life span. The mir14 suppresses death induced by expression of Rpr, Hid, Grim, or the apical caspase Dronc (234). mir14 also regulates fat metabolism by decreasing the levels of triacylglycerol and diacylglycerol. bantam is related to mir80 and mir82 of C. elegans, indicating that the mir80 family of RNAs might be involved in apoptosis in worms (33). Identification of these micro-RNAs promises discovery of similar micro-RNAs in other systems and reveals the hidden treasure of knowledge relating to micro-RNA-controlled biological functions (11).
Kinship of siRNA- and Micro-RNA-Related Pathways
Since micro-RNAs are derived from their precursor dsRNAs and are similar in size to siRNAs, the biogenesis of siRNAs and micro-RNAs is similar. In fact, both siRNAs and micro-RNAs are processed by Dicer activities in animals as well as in plants (86, 96, 104, 115, 240). Human recombinant Dicer can process pre-let7 RNA to mature let7 quite efficiently in vitro (175). Recent work by D. P. Bartel's group (181) has also shown that caf1 (dicer homologue) mutants of A. thaliana fail to process micro-RNAs. The genetic and biochemical data point toward interaction between Dicer and the Argonaute group of proteins in C. elegans and D. melanogaster for processing the micro-RNAs (86, 95). The similar interaction is possibly also present in plants between Dicer on one hand and PNH (zwille/pinhead) on the other to generate plant micro-RNAs (83). Additionally, both forms of small RNA, micro-RNAs and siRNAs, were found integrally associated with riboprotein complexes containing a member of the PIWI/PAZ domain family, siRNAs in the RISC and micro-RNAs in the microribonucleoprotein complexes (96).
In fact, recent evidence suggests that at least for some micro-RNAs, the microribonucleoprotein and the RISC complex could be the same entity (103, 137, 139, 182). Though the same or similar DCR and subsequent ribonucleocomplexes are required to process mature forms of the micro-RNAs, in some cases, such as C. elegans lin4 and let7, the ≈22-nucleotide form is processed from the 5′ part of the stem, and in other cases, such as miR1 and miR58, maturation results from the 3′ part of the precursors. Thus, there is a gene specificity of micro-RNA processing and/or stabilization (126). Since the biosynthetic pathways of micro-RNAs and siRNAs are somewhat similar, the viral suppressors that inhibit siRNA formation are also expected to interfere in the biogenesis of micro-RNAs. A detailed understanding of this suppression process may unravel the hitherto unknown molecular basis of virus-induced development-related diseases in eukaryotes, especially in plants.
An RNA-silencing suppressor, PI/HC-PRO of turnip mosaic virus, induces a number of developmental defects in the vegetative and reproductive organs of A. thaliana. Many of these defects are reminiscent of observed defects in Dicer-like mutants of A. thaliana. The PI/HC-PRO suppressor interferes with the formation of miR171, and as a result the downstream target mRNAs accumulate instead of being cleaved, causing developmental errors (113). Thus, it is interesting that the counterdefensive strategy of the viruses has evolved not only to protect the viral RNA genome from the host degradative machinery but also to subvert the cellular development program in favor of the virus.
However, it is important to mark the distinctions among the pathways leading to the formation as well as the activities of siRNAs and micro-RNAs. Although over 600 micro-RNAs from various organisms have been identified (33), only about 3% of them are fully complementary to the target mRNA sequences. All known micro-RNAs are derived in vivo from dsRNA precursors which are imperfectly annealed. Since the biosynthesis and activities of the micro-RNAs do not require perfect complementarity, noncanonical pathways of RNAi may be involved for the micro-RNAs because the usual RNAi calls for extensive complementarity of the dsRNA. It is only because of this characteristic mismatch between the sequences of micro-RNA and cognate mRNA that the in silico identification of the target mRNA is so difficult (182). The imperfect nature of annealing between the two partners is viewed as the prime cause for translational repression of the target mRNA (172).
Second, the mature micro-RNAs are always found in the single-stranded conformation in nature for some unknown reason, whereas siRNAs are double-stranded when detected. Third, unlike siRNAs, micro-RNAs enter riboprotein complexes with differing PPD proteins (PAZ and Piwi domains), depending on the specificity of the micro-RNA or its precursor with the cognate PPD proteins (86). The sequence or structure of a micro-RNA or its precursor might ensure that it functions as a translational repressor and not as a trigger of RNAi. It is widely speculated that the siRNAs and micro-RNAs are distinguished following their biosyntheses, and these two are then allowed to form related but distinct ribonucleoprotein complexes that target downstream substrates for degradation or translation repression, respectively. This hypothesis is based on the observation that siRNAs or exogenously supplied hairpin RNAs containing even a single mismatch with their substrate fail to repress the target mRNAs and do not simply shift their regulatory mode to translation inhibition (98).
Fourth, a viral suppressor of RNA silencing, the HC-PRO protein of potato virus Y, has been found to differentially regulate the accumulation of siRNAs and micro-RNAs in tobacco (144). The HC-PRO protein prevents accumulation of siRNAs of the silenced genes and thus releases silencing in a universal manner, but the same protein helps accumulation of all micro-RNAs tested, namely, miR167, miR164, and miR156 of tobacco, in vivo. This result indicates that the dicing complexes for siRNA and micro-RNA may not be exactly similar in biochemical features, and as a result the biochemical functions of the complexes are different in response to this particular HC-PRO protein. Lastly, not all RNAi pathway mutants are developmentally aberrant, whereas micro-RNA pathway mutants are expected to be defective in organism architecture and development. For example, an RNA-dependent RNA polymerase-defective mutant of A. thaliana, the sgs2/sde1 mutant, shows defects only in cosuppression phenomena (a form of RNAi) but is perfectly normal in phenotypic development (54, 160). This observation raises the question of whether the RdRP-dependent amplification step is required at all for micro-RNAs.
Functional Classifications
A number of micro-RNAs, including let7, are conserved across all organisms throughout evolution. About 12% of the micro-RNAs identified so far in animal systems are conserved at least among nematodes, D. melanogaster, and humans. Interestingly, a majority of the micro-RNAs are speculated to control development-related genes. However, the mechanisms of such control are not quite established yet. Micro-RNAs probably employ a variety of mechanisms to downregulate target genes. Micro-RNAs such as C. elegans lin4 and let7 have been shown to imperfectly anneal to the 3′ untranslated region of the target mRNA.
A vast majority of micro-RNAs probably belongs to this category. Due to imperfect complementarity, some micro-RNAs may also anneal to a host of different target mRNAs either simultaneously or in a temporally controlled manner. On the other hand, there are micro-RNAs, located mostly in the A. thaliana intergenic region, which have perfect or nearly perfect complementarity to the target mRNAs. Such micro-RNAs might trigger site-specific cleavage of the mRNA after being incorporated into a functional RISC-like complex. In such a situation, micro-RNAs act like siRNAs. The A. thaliana inflorescence-specific small RNA 39 cleaves the middle part of mRNA of the three scarecrow gene family members in a similar fashion (137).
On the basis of nearly perfect complementarity with the micro-RNAs, numerous A. thaliana mRNA targets have been predicted, and these targets have also been phylogenetically conserved in rice. Fifteen cleavage-type targets were validated recently by in vitro or in vivo micro-RNA-guided cleavage assays. The majority of these predicted mRNA targets encode members of large family of transcription factors, including Phavoluta (PHV), Phabulosa (PHB), cup-shaped cotyledon 1 (CUC1), CUC2, etc. These transcription factors are required for meristem identity, cell division, organ separation, and organ polarity (33). On the other hand, mir172 likely acts in cell fate specifications as a translational repressor of APETALA2 in Arabidopsis flower development (39).
There are other varieties of micro-RNA which also interact with target mRNAs affecting the posttranscriptional steps, such as RNA splicing, mRNA localization, and RNA turnover. In D. melanogaster, many micro-RNAs are known to be complementary to the 3′ untranslated region sequence motifs, which are responsible for mediating negative posttranscriptional regulation. These sequence motifs include the K box (CUGUGAUA), the B-rd box (AGCUUUA), and the recently found GY box (UGUCUUCC). All micro-RNAs showing complementarity to these motifs are expressed either broadly throughout development or in the narrow window of embryogenesis of D. melanogaster (124). It is possible to have even a fourth class of micro-RNAs, which may serve as guides for modification of chromosomal DNA and control the epigenetic processes of nuclear genomes (225).
Genetic Diversity in Species-Specific Biosynthesis of Micro-RNA
The siRNA and micro-RNA pathways closely parallel each other. It has been mentioned earlier that the biosynthesis of siRNAs have interesting system-specific features. Hence, system-specific features of micro-RNAs would also be of no surprise. Here, we illustrate some of those features. In C. elegans, D. melanogaster, and other animals, the Dicer proteins responsible for siRNA formation are also involved in the biosynthesis of micro-RNAs; but in A. thaliana, DCL1 (or CAF1) is responsible for micro-RNA but not for siRNA formation (76). Interestingly, the DCL1 mRNA is predicted to be a micro-RNA target, indicating that the micro-RNA-related apparatus in plants is regulated by a negative feedback loop (233). In plants, HEN1 is required for both siRNA and micro-RNA formation (22). Such a role for HEN1 orthologues in other systems is not known yet. Both CAF1 and HEN1 have nuclear localization sequence signals, raising the question of whether plant micro-RNAs are made intranuclearly.
The functions of plant micro-RNAs may be different from those of their animal counterparts in some events. The animal micro-RNAs act as translational repressors, whereas some plant micro-RNAs act on the target mRNA posttranscriptionally, like siRNAs (139). In animals, the majority of the AGO family members tightly regulate the biosynthesis of micro-RNAs (32), whereas in plants, especially A. thaliana, only one member of the 10 constituents of the AGO family, Ziwelle, alone contributes to the synthesis of micro-RNA. Surprisingly, though A. thaliana ago1 mutants show a strong hypermorphic phenotype, AGO1 protein is not responsible for plant micro-RNA formation (22). However, AGO1 is required for initiation of PTGS, whereas Zwielle is not. Recently, Vauchert et al. also isolated a few ago1 alleles of A. thaliana which were hypomorphic in nature (157).
The biogenesis of some plant micro-RNAs seems to be different from that of their animal counterparts. Most of the Arabidopsis micro-RNAs belong to group1, as their precursor forms are detected poorly or not at all. Despite the absence or greatly reduced abundance of the mature micro-RNAs, accumulations of pre-micro-RNA are never detected in Dicer-defective caf1 mutants (171). However, the pre-micro-RNAs accumulate to a higher level in C. elegans and metazoans in which Dicer activity is abolished or reduced (86). Very few Arabidopsis micro-RNAs belong to group II, including the micro-RNAs miR176, -177, -178, and 179, the pre-micro-RNA transcripts of which are, however, detectable. The levels of these precursor transcripts do not change in either the caf1 or hen1 mutant background. Such facts indicate that even within the same plant, the biosynthesis pathways of micro-RNAs might vary depending on the particular micro-RNA. The tissue specificity of micro-RNAs is well known. Hence, micro-RNAs specific to tissues that are unique either to animals (e.g., brain) or plants (roots, for example) might exemplify variant pathways of biosynthesis of micro-RNAs.
The discovery of micro-RNAs has been branded one of the top discoveries in developmental molecular biology. The survey of micro-RNAs is still at a subsaturated stage. The future will witness the discovery of hundreds of new micro-RNAs and their corresponding mRNA targets, and the mysteries of developmental pathways from embryogenesis to adulthood will be unfolded.
SMALL-RNA-MEDIATED EFFECTS ON CHROMOSOMAL DNA
The siRNAs work not only at the posttranscriptional stage but also leave their indelible marks on the genomes to repress the gene transcription activity or selectively remove portions of the genomes, especially of protozoans. These stunning discoveries have been reported only in the span of the last 2 years, the detailed mechanisms of which are still to be revealed and have been reviewed in two recent articles (57, 109). In the present review, we describe these effects briefly with special emphasis on plant systems, since the genetics and biochemistry of some of these processes are better illustrated in plants.
Broadly speaking, the siRNAs bring about three different biochemical end products with the chromatin DNA: DNA methylation, as revealed mostly in plant systems; heterochromatin formation; and programmed elimination of DNA. DNA methylation had been reckoned a major source of transcriptional gene silencing (TGS), and mechanistically TGS had been viewed very distinctively from PTGS in the past. But recent developments have caused a blurring in the identity between these two pathways (218), and some of these developments will be highlighted below. The discoveries of such epigenetic changes have ignited a revolution not only in the field of gene regulation but also in gene maintenance and gene evolution.
RNA-Dependent DNA Methylation
A role for RNA in guiding de novo cytosine methylation of homologous DNA sequences was first discovered in viriod-infected plants and subsequently also in nonpathogenic plant systems (194). When the dsRNA degradation mediated PTGS occurs in plants, the genomic DNA regions homologous to dsRNA are often found methylated at almost all the sensitive cytosine residues. This process is generally referred to as RNA-dependent-DNA methylation and the corresponding part of the genome, especially the promoter region might remain transcriptionally silent. The initiator of RNA-dependent DNA methylation/TGS could be either the transgene-derived dsRNA or the consequent siRNA (110, 111, 214). Depending on the sequence information of the dsRNA, RNA-dependent DNA methylation was found to occur at the open reading frame and/or the promoter region of the genome (10, 149). If methylation occurred only at the open reading frame, TGS did not result. However, RNA-dependent DNA methylation at the promoter sequences induced TGS, which, unlike PTGS, was stable and heritable (98). RNA-dependent DNA methylation within the host genes has also been found to occur preponderantly during virus-induced gene silencing, a type of RNAi that is generally initiated by plant virus vectors carrying portions of host genes, as has been described earlier (214).
It was demonstrated that the movement of transposons was controlled by transcriptional suppression (TGS) and that methylation also played a role in this suppression, depending on the nature of the transposon (226). In animals and lower plants, siRNAs corresponding to the transposable elements were discovered and cloned earlier (9, 232), and in A. thaliana and Nicotiana species, the siRNAs corresponding to retroelements have recently been discovered (94). These siRNAs are perhaps responsible for the methylation of the homologous DNA.
There are also conflicting data in the literature concerning the cause-and-effect relationship between PTGS and DNA methylation. In some examples, there is no correlation between PTGS and DNA methylation (153). In other events, as mentioned earlier, the correlation is strong (137). Llave et al. (137) showed that a viral protein, HC-PRO, that suppresses PTGS/RNAi, when introduced into GUS-silenced tobacco, inhibited the maintenance of small RNAs and caused a concomitant decrease in methylation of the GUS sequence in the plant genome. This study suggested that DNA methylation of the silenced gene could be directly correlated with PTGS. However, in a contrasting study carried out by Mette et al. (153), HC-PRO was found to increase the methylation of a target promoter DNA when gene silencing was induced by the promoter dsRNA. The later study also revealed that the amount of promoter siRNA was elevated fivefold in the presence of HC-PRO. Taken together, both of these studies indicate that the level of target DNA methylation is directly related to the amount of siRNA present in the cell, and thus the apparent differences between these observations can be resolved. In other words, the availability of siRNA may determine the level of RNA-directed DNA methylation. In the events of RNA-dependent DNA methylation, the chromodomain containing DNA methylases acts either alone or in combination with other proteins, such as piwi-containing proteins, to form complexes with the siRNAs and cause sequence-specific RNA-dependent DNA methylation, finally resulting in TGS (10).
Evidence of cross talk between PTGS and TGS has been obtained from the mutational analysis of A. thaliana and D. melanogaster. Two types of A. thaliana mutants, ddm1 (deficient in DNA methylation) and met1 (methyl transferase), were isolated from a screen of mutations causing a reduction in global methylation of the genome. The locus ddm1 encodes an SNF2/SW12-like chromatin-modeling protein, whereas MET1 is a major DNA methyltransferase. Both of these mutants exhibit marked reduction in PTGS activity, as measured by the accumulation of transgene transcripts (10, 218). Although the patterns of reduction are different with these mutants, these studies highlight the strong correlation between PTGS and TGS.
In D. melanogaster, polycomb protein-dependent TGS is also affected by mutations in PIWI, a family of proteins required for RNAi (169). Other evidence includes the argonaute4 gene of A. thaliana, which controls both locus-specific siRNA accumulation and DNA methylation (241); the Arabidopsis sde4 locus, which is of unknown biochemical function but is responsible for (retroelement TS SINE-specific) siRNA formation (94); and the Arabidopsis rts1 (RNA-mediated transcription silencing) mutation, which causes a ≈50% reduction in target promoter DNA methylation (10). However, not all TGS mutations affect the PTGS pathways and vice versa, suggesting that the two pathways diverge at some point (218).
RNA-dependent DNA methylation has been reported only in plants until now. Aufsatz et al. (10) have also shown that asymmetric non-CpG methylation is mostly affected by RNA-dependent DNA methylation, but the existence of non-CpG methylation in mammals has always been a contentious issue. Mammalian DNA is methylated mostly at symmetric CpG or CNG sites by various forms of DNA methyltransferases. However, using a dual-labeling nearest-neighbor technique and the bisulfite genomic sequencing methods, Ramsahoye et al. (177) found that the genomes of embryonic stem cells but not that of somatic tissues harbored non-CpG methylation, which accounted for 15 to 20% of total cytosine methylation. This methylation is perhaps caused by the methylase Dnmt 3a, which is highly expressed in embryonic stem cells but poorly expressed in somatic tissues (179). Other studies have also revealed that in D. melanogaster and mammals, non-CpG methylation is an early embryonic event (10), and this methylation can be catalyzed by Dnmt 2, which is primarily active at the initial stages of development (142).
In the above-mentioned studies, however, no connection between non-CpG methylation and any homologous RNA has been shown. Hence, if RNA-dependent DNA methylation occurs at all in animals, it might be limited to the early developmental stages when the effector proteins may be found in abundance. In contrast, RNA-dependent DNA methylation is observed throughout plant development, implying the continuous availability of the appropriate plant DNA methyltransferases. This feature also explains the ease of RNA-dependent DNA methylation detection in plants (10).
Heterochromatin Formation
Even for organisms in which RNA-dependent DNA methylation is supposedly absent, there is growing evidence that RNAi processes cause chromatin modifications leading to TGS. This evidence reveals that the connections between TGS and PTGS are strong across all layers of eukaryotic life. For example, in C. elegans, in which DNA methylation has not been detected, some PTGS mutations, namely mut7 and rde2, derepress transgenes which are affected by polycomb-dependent TGS (203). The polycomb group of proteins are known to keep the chromatin in the closed or compact conformation. Conversely, it has also been found recently that the polycomb proteins MES3, MES4, and MES6 are required for RNAi, at least under some experimental conditions (65, 121).
Generally, in eukaryotic systems, histone modifications make the chromatin structure inert to transcription by heterochromatin formation, which is modulated greatly by the RNAi processes, as recent discoveries have revealed. In almost all organisms heterochromatin formation requires that histone H3 of the chromatin be deacetylated and then methylated at lysine 9. The SET domain of a special group of histone methyltransferases carries out this function. This methylated lysine is subsequently bound by a heterochromatin binding protein, HP1. The binding of the chromodomain containing HP1 to Met H3-K9 is highly specific and of very high affinity (12). This binding may be followed by multimerization of HP1 and complex formation with other chromatin-remodeling proteins. As a result of this multicomplex formation, the chromatin becomes condensed and locked in a transcriptionally repressed heterochromatic state.
Once formed, the heterochromatin spreads a large distance due to cooperative protein-protein interactions of chromatin-remodeling factors, the components of which have not been fully identified yet. However, these structures are generally initiated at places containing repeated DNA sequences, for example, centromere, telomere, mating locus, and elsewhere in the genome containing repetitive DNA in the fission yeast Schizosaccharomyces pombe (7). These repeats are responsible for producing dsRNAs, which are processed by the RNAi machinery. D. P. Bartel's group has discovered abundant species of centromeric repeat-specific siRNAs from S. pombe (180). Volpe et al. (223) demonstrated that these siRNAs are blocked and instead, large noncoding RNAs (≈1.4 to 2.4 kb) homologous to the centromere repeats accumulate in dcr1, ago1, and rdrp1 mutants of S. pombe. These mutant cells also do not show the heterochromatin-mediated silencing of a ura4+ gene inserted into the outer and inner repeats that flank the central core of the centromeres. A corresponding reduction in Met-H3 K9 is also observed in the outer repeats of these mutant cells (223). This loss in gene silencing is phenotypically similar in cells lacking the histone methyltransferase (clr4) or the HP1-like (swi6) activity.
That the DNA repeats are central to the RNAi-like processing of dsRNA and concomitant heterochromatin formation was clearly established by the findings of Hall and colleagues (89), who inserted a 3.6-kb centromere H repeat, normally present at the silent mating type domain, in a euchromatic position (ura4 locus). The introduction of this repeat was sufficient to turn on the silencing of a linked reporter gene and induce H3-K9 methylation and recruitment of HP1-like factors (Swi6) (89). The link between the RNAi machinery and heterochromatin formation has also been established by a recent finding in A. thaliana. From a large screen of mutants, Zilberman et al. (241) found that the ago4 gene is responsible for the RNAi-related silencing of the A. thaliana superman gene, which is implicated in flower formation. The ago4-1 mutation reactivates the silent superman allele and decreases non-CpG as well as H3-K9 methylation. Significantly, the same mutation also blocks DNA methylation and the accumulation of siRNA corresponding to the retroelement at SN1 (241).
The above-mentioned facts are put together in a model (Fig. 3) showing a link between siRNA and heterochromatin formation. In the wild-type scenario, one strand of the centromeric region is constitutively expressed, whereas the complementary strand, which is subjected to heterochromatic repression, is occasionally transcribed (57). Such transcription will lead to the formation of dsRNA, which will be processed by the RNAi machinery. This processing might even be a nuclear step, since a component of this machinery, the RdRP, was found to be physically bound to the outer repeats of the centromeric region in a chromatin immunoprecipitation assay (223). The siRNA thus formed might enter a complex containing the histone methyltransferase enzyme. This complex could be a nuclear equivalent of the RISC complex (Nu.RISC of Fig. 3) lacking nuclease activity (98). Such a complex would be guided to the appropriate DNA region following the DNA-RNA base pairing rules, and the histone H3-K9 of the region might be methylated to eventually generate the heterochromatin structure. Since RdRP is found locally, the spread of the heterochromatic structure may be associated with the extension of the 3′ end of the siRNA primer. It has also been shown in N. crassa and A. thaliana that H3-K9 methylation directs DNA methylation (107, 204). The methylated DNA could be complexed further with the methyl-binding proteins. Following these binding events, the chromatin structure will be extremely compact and condensed and would remain transcriptionally inert.
FIG. 3.
Role of the RNAi process in heterochromatinization of nuclear DNA. The methylated (H3-K9) histone and many chromatin components are involved in the cross talk in several epigenetic regulatory pathways within the nucleus. The centromeric region (black oval) of the chromatin (thick purple line) might be responsible for the production of dsRNA transcripts (continuous red line and broken blue line). The transcript coming out of the DNA strand subjected to heterochromatinization is represented by broken blue lines. The siRNAs of the nucleus join the complex, of which the histone methyltransferase is a constituent. The siRNA binding the chromodomain (CHR), methyltransferase (MTase), and the SET domain of the methyltransferase are indicated.
DNA Elimination
The most dramatic effect of siRNA-mediated heterochromatin formation followed by chromosomal DNA elimination and rearrangement has been recorded in the ciliated protozoan Tetrahymena pyriformis (156, 206). Among unicellular organisms, T. pyriformis is unique because of its nuclear dimorphism. The two nuclei, the micronucleus and macronucleus, serve different functions. The polyploid macronucleus is the transcription center of the cell during vegetative growth, whereas the diploid and transcriptionally inert micronucleus acts as the germ line nucleus. During conjugation, the micronucleus gives rise to the macronucleus, and this transition is accompanied by two interesting and peculiar recombinant events. First, approximately 6,000 internal eliminated sequences of five pairs of micronucleus chromosomes, accounting for about 15% of genomic micro-DNA, are removed. Second, the remaining parts of these chromosomes are broken into 200 to 300 minichromosomes concomitant with the deletion of <50 nucleotide breakage eliminated sequences. The mechanisms of removal of internal eliminated sequences and breakage eliminated sequences remained elusive for a long time but were recently unveiled, courtesy of the awareness of the siRNA world.
Mochizuki et al. (156) showed that in wild-type cells of Tetrahymena pyriformis, siRNAs of about ≈26 to ≈31 nucleotides were produced which hybridized to micronuclear genomic DNA and not the macronuclear DNA, indicating that these siRNAs could be internal eliminated sequence/breakage eliminated sequence-specific and are referred to as scan RNAs. These scan RNAs were not made in twi1 mutants, and the production of internal eliminated sequence/breakage eliminated sequence elements was also impaired by the twi1 mutation. twi1 produces a Piwi-related protein during the sexual cycle and can transmit the RNA-encoded information from the micronucleus to the old macronucleus and finally to the new macronucleus to mark the sequences to be eliminated (57). The defect in accumulation of the scan RNAs in the twi1 mutant was similar to the case of another mutant, pdd1 (156). In a related report, Taverna et al. (206) showed that the protein PDD1 was the effector protein for DNA excision and that PDD1 along with Met H3-K9 was associated preferentially with the internal eliminated sequence/breakage eliminated sequence elements in the new macronucleus that developed from the micronucleus during the sexual cycle. PDD1 contains two chromodomains and an additional RNA-binding domain (3).
The above data and the model presented in the earlier section lead to a straightforward and interesting scheme for programmed DNA degradation in Tetrahymena pyriformis. The bidirectional transcription that occurs across the internal eliminated sequence repeats (38) may form the dsRNA, which would give rise to the scan RNAs following the action of RNAi-related Dicing complexes that perhaps also include the Twi1 and PDD proteins. These scan RNAs eventually may be associated with the nuclear equivalents of RISC factor in the new macronucleus to provide heterochromatic sites at the internal eliminated sequence/breakage eliminated sequence regions. The chromodomain containing PDD proteins may remain bound to the scan RNA and thus guide to destroying the cognate DNA. As an extension of this work, Yao et al. found that a similar RNAi process recognized and deleted a foreign neomycin resistance gene of bacterial origin which was integrated in a Tetrahymena chromosome (237). These two studies together strongly suggest an siRNA- (or scan-RNA)-based mechanism that controls genome-wide DNA arrangements and provides genomic surveillance against invading foreign DNAs.
Thus, the Tetrahymena pyriformis as well as S. pombe data show how dramatic the epigenetic consequences of the genome could be following the formation of siRNA molecules in cells. Discovery of the link between the RNAi processes and the epigenetic chromatin modification as well as chromosome behavior is probably the most fascinating and novel face of regulation of gene silencing mechanism. The RNAi machinery is reported to control many explosive features of cellular biology, namely stem cell maintenance (53), cell fate determination (21), nonrandom chromosome segregation (188), etc. A recent report established that the fission yeast RNAi-related genes ago1, dcr1, and rdp1 also control the fidelity of chromosome segregation during mitosis and meiosis. As discussed earlier, these gene products are required to maintain centromeric silencing. The report also demonstrated that the chromosome missegregation of the RNAi mutants occurred due to the loss of centromeric cohesion, suggesting a clear link between centromeric silencing and cohesion. This report broadly hinted that the regulation of chromosomal dynamics could be largely traced to the natural RNAi biology of the eukaryotic cells (90).
It is not difficult to imagine that we might witness RNAi-related unifying signals in diverse chromosome behaviors, namely X-chromosome inactivation, satellite-repeat contraction and expansion, hybrid dysgenesis in D. melanogaster, chromatin diminution in ascarid nematodes, nuclear dominance in plants, and so on in the not so distant future (57).
APPLICATIONS OF RNAi
Besides being an area of intense, upfront basic research, the RNAi process holds the key to future technological applications. Genome sequencing projects generate a wealth of information. However, the ultimate goal of such projects is to accelerate the identification of the biological function of genes. The functions of genes can be analyzed with an appropriate assay, by examining the phenotype of organisms that contain mutations in the gene, or on the basis of knowledge gained from the study of related genes in other organisms. However, a significant fraction of genes identified by the sequencing projects are new and cannot be rapidly assigned functions by these conventional methods.
RNAi technology is proving to be useful to analyze quickly the functions of a number of genes in a wide variety of organisms. RNAi has been adapted with high-throughput screening formats in C. elegans, for which the recombination-based gene knockout technique has not been established. Chromosomes I and III of C. elegans have been screened by RNAi to identify the genes involved in cell division and embryonic development (82, 84). Recently, a large-scale functional analysis of ≈19,427 predicted genes of C. elegans was carried out with RNA interference. This study identified mutant phenotypes for 1,722 genes (112). Similarly, in D. melanogaster, RNAi technology has been successfully applied to identify genes with essential roles in biochemical signaling cascades, embryonic development, and other basic cellular process (44). In plants, gene knockdown-related functional studies are being carried out efficiently when transgenes are present in the form of hairpin (or RNAi) constructs. Plant endotoxins could also be removed if the toxin biosynthesis genes are targeted with the RNAi constructs. Recently, the theobromine synthase of the coffee plant was knocked down with the hairpin construct of the transgene, leading to the production of decaffeinated coffee plants (166). Virus-induced gene silencing has also been proven to be a successful approach for plant genetics (15).
Given the fact that RNAi is easy to apply, whole-genome screens by RNAi may become a common method of choice in the near future. RNAi may facilitate drug screening and development by identifying genes that can confer drug resistance or genes whose mutant phenotypes are ameliorated by drug treatment, providing information about the modes of action of novel compounds. Although RNAi is unlikely to replace the existing knockout technology, it may have a tremendous impact for those organisms that are not amenable to the knockout strategy. It may also be a method of choice to study the simultaneous functions of a number of analogous genes in organisms in which redundancy exists with respect to a particular function, because many of these genes can be silenced simultaneously.
Given the gene-specific features of RNAi, it is conceivable that this method will play an important role in therapeutic applications. Since siRNAs direct cellular RNAi biology, these are potential therapeutic reagents because of their power to downregulate the expression pattern of mutant genes in diseased cells. However, central to this hypothesis is the assumption that the effect of exogenous siRNA applications will remain gene specific and show no nonspecific side effects relating to mismatched off-target hybridization, protein binding to nucleic acids, etc. Though it was demonstrated that mismatches of more than even one nucleotide within the 19- to 20-mer siRNAs effectively disrupted proper degradation of the target mRNA (68), the gene specificity of siRNAs needs to be confirmed on a genome-wide scale.
Recently, Chi et al. (41) reported that the GFP siRNA-induced gene silencing of transient or stably expressed GFP mRNA was highly specific in the human embryonic kidney (HEK) 293 cell background. The specific silencing did not produce secondary changes in global gene expression, as detected by the DNA microarray experiment. They also failed to detect the presence of transitive RNAi in experimentally engineered human cell lines (41). In their own experiments, Semizarov et al. (190) reached a similar conclusion while using siRNAs corresponding to akt1, rb1, and plk1 in the human non-small cell lung carcinoma cell line H1299. These experiments prove that siRNAs could be used as highly specific tools for targeted gene knockdown and can be used in high-throughput approaches and drug target validation. This exquisite sequence-specific effect of siRNAs has also been exploited in silencing the mutant allele of the diseased gene while not affecting the wild-type allele of the healthy version of the same gene (158).
siRNAs have been shown to inhibit infection by human immunodeficiency virus, poliovirus, and hepatitis C virus in cultured cell lines (152). Bitko and Barik (19) successfully used siRNAs to silence genes expressed from respiratory syncytial virus, an RNA virus that causes severe respiratory disease in neonates and infants. siRNA treatment has also been shown to reduce the expression of the BCR-ABL oncoprotein in leukemia and lymphoma cell lines, leading to apoptosis in these cells (230). With respect to future medical applications, siRNA-based therapy seems to have a great potential to combat carcinomas, myeloma, and cancer caused by overexpression of an oncoprotein or generation of an oncoprotein by chromosomal translocation and point mutations (211).
Recently, the therapeutic potential of the siRNA technique has been demonstrated in vivo in mouse models. McCaffrey et al. (151) and Song et al. (199) demonstrated effective targeting of a sequence from hepatitis C virus and the fas gene by RNA interference in mouse liver (199). An epiallelic series of p53 hypomorphs created by RNAi have been shown to produce distinct tumor phenotypes in mice in vivo, suggesting that RNAi can stably suppress gene expression (99). Song et al. (199) have shown that treatment with fas siRNA abrogated hepatocyte necrosis and inflammatory infiltration and protected mice from liver fibrosis and fulminant hepatitis. Rubinson et al. (184) showed highly specific, stable, and functional silencing of gene expression in transgenic mice with the lentivirus system for the delivery of siRNAs.
Although the delivery of siRNAs to a proper site remains problematic for gene therapy, chemical modifications of siRNAs such as changing the lipophilicity of the molecules or the methods previously developed for the application of antisense oligonucleotides or nuclease-resistant ribozymes might help the entry and stability of siRNAs within the transfected cells or tissues. The absence of specific micro-RNAs has been demonstrated in carcinoma cells, implying that cancer development could be arrested by introduction of the missing micro-RNAs. The micro-RNAs could be supplied in the form of siRNAs, since the function of micro-RNAs can be mimicked by the exogenous siRNA (62). However, independent of its biomedical applications, RNAi appears to be a forthcoming method for functional genomics.
CONCLUDING REMARKS
In the footsteps of the discovery of the double-helical structure of DNA, some outstanding discoveries have been recorded, but few of them really match the explosive content and implication of dsRNA-mediated gene silencing. This homology-dependent silencing has established a novel paradigm with far-reaching consequences in the field of transcription regulation. The regulatory mechanism offers cellular protection against parasitic nucleic acid sequences, carries out epigenetic as well as genetic alterations on the one hand, and governs organisms architecture and development on the other. Capitalizing on the basic principles of silencing, large-scale functional genomics have come into play in diverse organisms. Studies conducted at the laboratory level have revealed the tremendous power of siRNAs as therapeutics and have demonstrated the potential of micro-RNAs to reverse cellular developmental aberrations.
The new paradigm has a lot more to offer than it has delivered already. The stepwise detailed mechanism of RNAi and its related processes is waiting to be explored. The rationale for many unexplained genetic findings of RNAi in worms, plants, and other organisms will be revealed in the wake of further mechanistic discoveries. The cytoplasmic location of RNAi is evident, but the evidence of nuclear connections of RNAi and related events are also too many. Surprisingly, there are some components of RNAi, GEMIN3 and GEMIN4 of humans, which partition in both the nuclear and cytoplasmic compartments. Hence, clarification of the subcellular locations of the RNAi processes is required. Hopefully, the detailed biochemical framework of RNAi would provide such clarifications.
As we gain more insight into the mechanisms, more effective methods for analysis of gene functions may evolve. We may learn more about geriatrics, nervous diseases, genetic imprints, nuclear dominance in plants, and so on and thus might wield control over such processes in the future. Meanwhile, the knockdown technology might improve vastly with better-designed plasmid- or virus-based vectors for delivery of siRNAs to the appropriate tissues at the appropriate time. Such technology is bound to give a new shape to therapeutic gene silencing as well. The science and technology of RNAi has given us a cultural ocean of virtually bottomless depth.
.
REFERENCES
- 1.Aboobaker, A. A., and M. L. Blaxter. 2003. Use of RNA interference to investigate gene function in the human filarial nematode parasite Brugia malayi. Mol. Biochem. Parasitol. 129:41-51. [DOI] [PubMed] [Google Scholar]
- 2.Abraham, N., D. J. Stojdl, P. L. Duncan, N. Methot, T. Ishii, M. Dube, B. C. Vanderhyden, H. L. Atkins, D. A. Gray, and M. W. McBurney. 1999. Characterization of transgenic mice with targeted disruption of the catalytic domain of the double-stranded RNA-dependent protein kinase, PKR. J. Biol. Chem. 274:5953-5962. [DOI] [PubMed] [Google Scholar]
- 3.Akhtar, A., D. Zinc, and P. B. Becker. 2000. Chromodomains are protein-RNA interaction modules. Nature 407:405. [DOI] [PubMed] [Google Scholar]
- 4.Al-Anouti, F., T. Quach, and S. Ananvoranich. 2003. Double-stranded RNA can mediate the suppression of uracil phosphoribosyltransferase expression in Toxoplasma gondii. Biochem. Biophys. Res. Commun. 302:316-323. [DOI] [PubMed] [Google Scholar]
- 5.Al-Kaff, N. S., S. N. Covey, M. M. Kreike, A. M. Page, R. Pinder, and P. Dale. 1998. Transcriptional and posttranscriptional plant gene silencing in response to a pathogen. Science 279:2113-2115. [DOI] [PubMed] [Google Scholar]
- 6.Allas, U., A. Liiv, and J. Remme. 2003. Functional interaction between RNase III and the Escherichia coli ribosome. BMC Mol. Biol. 4:8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allshire, R. 2002. Molecular biology. RNAi and heterochromatin—a hushed-up affair. Science 297:1818-1819. [DOI] [PubMed] [Google Scholar]
- 8.Anandalakshmi, R., G. J. Pruss, X. Ge, R. Marathe, A. C. Mallory, T. H. Smith, and V. B. Vance. 1998. A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. USA 95:13079-13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aravin, A. A., N. M. Naumova, A. V. Tulin, V. V. Vagin, Y. M. Rozovsky, and V. A. Gvozdev. 2001. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 11:1017-1027. [DOI] [PubMed] [Google Scholar]
- 10.Aufsatz, W., M. F. Mette, J. Van der Winden, A. J. Matzke, and M. Matzke. 2002. RNA-directed DNA methylation in Arabidopsis. Proc. Natl. Acad. Sci. USA 99:16499-16506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baehrecke, E. H. 2003. miRNAs: Micro managers of programmed cell death. Curr. Biol. 13:R473-R475. [DOI] [PubMed] [Google Scholar]
- 12.Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. [DOI] [PubMed] [Google Scholar]
- 13.Bass, B. L. 2000. Double-stranded RNA as a template for gene silencing. Cell 101:235-238. [DOI] [PubMed] [Google Scholar]
- 14.Bastin, P., A. Galvani, and L. Sperling. 2001. Genetic interference in protozoa. Res. Microbiol. 302:316-323. [DOI] [PubMed] [Google Scholar]
- 15.Baulcombe, D. C. 2001. Fast forward genetics based on virus induced gene silencing. Curr. Opin. Plant Biol. 2:109-113. [DOI] [PubMed] [Google Scholar]
- 16.Bender, J. 2001. A vicious cycle: RNA silencing and DNA methylation in plants. Cell 106:129-132. [DOI] [PubMed] [Google Scholar]
- 17.Bernstein, E., A. A. Caudy, S. M. Hammond, and G. J. Hannon. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409:363-366. [DOI] [PubMed] [Google Scholar]
- 18.Bingham, P. M. 1997. Cosuppression comes to the animals. Cell 90:385-387. [DOI] [PubMed] [Google Scholar]
- 19.Bitko, V., and S. Barik. 2001. Phenotypic silencing of cytoplasmic genes with sequence specific double-stranded short interfering RNA and its application in the reverse genetics of wild-type negative strand RNA virus. BMC Microbiol. 1:34-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blaszczyk, J., J. E. Trppea, M. Bubunenko, K. M. Routzahn, D. S. Waugh, D. L. Court, and X. Ji. 2001. Crystallographic and modeling studies of RNase III suggest a mechanism for double-stranded RNA cleavage. Structure 9:1225-1236. [DOI] [PubMed] [Google Scholar]
- 21.Bohmert, K., I. Camus, C. Bellini, D. Bouchez, M. Caboche, and C. Benning. 1998. AGO 1 defines a novel class of Arabidopsis controlling leaf development. EMBO J. 17:170-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boutet, S., F. Vazquez, J. Liu, C. Beclin, M. Fagard, A. Gratias, J. B. Morel, P. Crete, X. Chen, and H. Vaucheret. 2003. Arabidopsis HEN1. A genetic link between endogenous miRNA controlling development and siRNA controlling transgene silencing and virus resistance. Curr. Biol. 13:843-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyle, J. P., X. J. Wu, C. B. Shoemaker, and T. P. Yoshino. 2003. With RNA interference to manipulate endogenous gene expression in Schistosoma mansoni sporocysts. Mol. Biochem. Parasitol. 128:205-215. [DOI] [PubMed] [Google Scholar]
- 24.Brennecke, J., D. R. Hipfner, A. Stark, R. B. Russell, and S. M. Cohen. 2003. bantam encodes a developmentally regulated micro-RNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113:25-36. [DOI] [PubMed] [Google Scholar]
- 25.Brigneti, G., O. Voinnet, W. X. Li, L. H. Ji, S. W. Ding, and D. C. Baulcombe. 1998. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17:6739-6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Brooks, D. R., and R. E. Isaac. 2002. Functional genomics of parasitic worms: the dawn of a new era. Parasitol. Int. 51:319-325. [DOI] [PubMed] [Google Scholar]
- 27.Brown, S. J., J. P. Mahaffey, M. D. Lorenzen, R. E. Denell, and J. W. Mahaffey. 1999. With RNAi to investigate orthologous homeotic gene function during the development of distantly related insects. Evol. Dev. 1:11-15. [DOI] [PubMed] [Google Scholar]
- 28.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 29.Cali, B. M., S. L. Kuchma, J. Latham, and P. Anderson. 1999. Smg-7 is required for mRNA surveillance in C. elegans. Genetics 151:605-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calin, C. A., D. Dumitru, M. Shimizu, R. Bichi, S. Zupo, S. Noch, H. Aldler, S. Rattan, M. Keating, K. Rai, L. Rassenti, T. Kipps, M. Negrini, F. Bullrich, and C. M. Croce. 2002. Frequent deletions and downregulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 99:15524-15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caplen, N. J., S. Parrish, F. Imani, A. Fire, and R. A. Morgan. 2001. Specific inhibition of gene expression by small double-stranded RNA in invertebrate and vertebrate systems. Proc. Natl. Acad. Sci. USA 98:9742-9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carmell, M. A., Z. Xuan, M. Q. Zhang, and G. J. Hannon. 2002. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 16:2733-2742. [DOI] [PubMed] [Google Scholar]
- 33.Carrington, J. C., and V. Ambros. 2003. Role of micro-RNAs in plant and animal development. Science 301:336-338. [DOI] [PubMed] [Google Scholar]
- 34.Catalanotto, C., G. Azzalin, G. Macino, and C. Cogoni. 2000. Gene silencing in worms and fungi. Nature 404:245. [DOI] [PubMed] [Google Scholar]
- 35.Caudy, A. A., M. Myers, G. J. Hannon, and S. M. Hammond. 2002. Fragile X-related protein and VIG associate with RNA interference machinery. Genes Dev. 16:2491-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cerruti, L., N. Mian, and A. Bateman. 2000. Domains in gene silencing in cell differentiation proteins: the novel PAZ domain and redefinition of the Piwi domain. Trends Biochem. Sci. 25:481-482. [DOI] [PubMed] [Google Scholar]
- 37.Cerutti, H. 2003. RNA interference: traveling in the cell and gaining functions? Trends Genet. 19:39-46. [DOI] [PubMed] [Google Scholar]
- 38.Chalker, D. L., and M. C. Yao. 2001. Nongenic, bidirectional transcription precedes and may promote developmental DNA deletion in Tetrahymena thermophila. Genes Dev. 15:1287-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen, X. 11 September 2003. A micro-RNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 10.1126/Science.1088060. [DOI] [PMC free article] [PubMed]
- 40.Chen, X., J. Liu, Y. Cheng, and D. Jia. 2002. HEN1 functions pleiotropically in Arabidopsis development and acts in C function in the flower. Development 129:1085-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chi, J. T., H. Y. Chang, N. N. Wang, D. S. Chang, N. Dunthy, and P. O. Brown. 2003. Genomewide view of gene silencing by small interfering RNAs. Proc. Natl. Acad. Sci. USA 100:6343-6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chiu, Y.-L., and T. M. Rana. 2002. RNAi in human cells and functional features of small interfering RNA. Mol. Cell 10:549-561. [DOI] [PubMed] [Google Scholar]
- 43.Chuang, C. F., and E. M. Meyerowitz. 2000. Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 97:4985-4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clemens, J. C., C. A. Worley, N. S. Leff, M. Merda, T. Maehama, B. A. Hemmings, and J. E. Dixon. 2000. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc. Natl. Acad. Sci. USA 97:6499-6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cogoni, C., and G. Macino. 1997. Conservation of transgene-induced post-transcriptional gene silencing in plants and fungi. Trends Plant Sci. 2:438-443. [Google Scholar]
- 46.Cogoni, C., and G. Macino. 1997. Isolation of quelling-defective (qde) mutants impaired in posttranscriptional transgene-induced gene silencing in Neurospora crassa. Proc. Natl. Acad. Sci. USA 94:10233-10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cogoni, C., and G. Macino. 1999. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature 399:166-169. [DOI] [PubMed] [Google Scholar]
- 48.Cogoni, C., and G. Macino. 1999. Posttranscriptional gene silencing in Neurospora by a RecQ DNA helicase. Science 286:2342-2344. [DOI] [PubMed] [Google Scholar]
- 49.Cogoni, C., and G. Macino. 2000. Post-transcriptional gene silencing across kingdoms. Curr. Opin. Genet. Dev. 10:638-643. [DOI] [PubMed] [Google Scholar]
- 50.Cogoni, C., J. T. Irelan, M. Schumacher, T. J. Schmidhauser, E. U. Selker, and G. Macino. 1996. Transgene silencing of al-1 gene in vegetative cells of Neurospora is mediated by a cytoplasmic effector and does not depend on DNA-DNA interaction or DNA methylation. EMBO J. 15:3153-3163. [PMC free article] [PubMed] [Google Scholar]
- 51.Cogoni, C., N. Romano, and G. Macino. 1994. Suppression of gene expression by homologous transgenes. Antonie Van Leeuwenhoek 65:205-209. [DOI] [PubMed] [Google Scholar]
- 52.Cottrell, T. R., and T. L. Doering. 2003. Silence of the strands: RNA interference in eukaryotic pathogens. Trends Microbiol. 11:37-43. [DOI] [PubMed] [Google Scholar]
- 53.Cox, D. N., A. Chao, J. Baker, L. Chang, D. Qiao, and H. Lin. 1998. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 12:3715-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dalmay, T., A. Hamilton, S. Rudd, S. Angell, and D. C. Baulcombe. 2000. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101:543-553. [DOI] [PubMed] [Google Scholar]
- 55.Dalmay, T., R. Horsefield, T. H. Braunstein, and D. C. Baulcombe. 2001. SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. EMBO J. 20:2069-2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Depicker, A., and M. Van Montagu. 1997. Post-transcriptional gene silencing in plants. Curr. Opin. Cell Biol. 9:373-382. [DOI] [PubMed] [Google Scholar]
- 57.Dernburg, A. F., and G. H. Karpen. 2002. A chromosome RNAissance. Cell 111:159-162. [DOI] [PubMed] [Google Scholar]
- 58.Dernburg, A. F., J. Zalevsky, M. P. Calaiacovo, and A. M. Villeneuve. 2000. Transgene-mediated cosuppression in the C. elegans germ line. Genes Dev. 14:1578-1583. [PMC free article] [PubMed] [Google Scholar]
- 59.Devree, E., and P. A. Silver. 2002. Retrovirus-delivered siRNA. BMC Biotechnol. 2:15-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ding, S. W., B. J. Anderson, H. R. Hasse, and R. H. Symons. 1994. New overlapping gene encoded by the cucumber mosaic virus genome. Virology 198:593-601. [DOI] [PubMed] [Google Scholar]
- 61.Djikeng, A., H. Shi, C. Tschudi, and E. Ullu. 2001. RNA interference in Trypanosoma bruci: cloning of small interfering RNA provides evidence for retroposon-derived 24-26 nucleotide RNA. RNA 7:1522-1530. [PMC free article] [PubMed] [Google Scholar]
- 62.Doench, J. G., C. P. Petersen, and P. A. Sharp. 2003. SiRNA can function as miRNA. Genes Dev. 17:438-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Domeier, M. E., D. P. Morse, S. W. Knight, M. Portereiko, B. L. Bass, and S. E. Mango. 2000. A link between RNA interference and nonsense-mediated decay in Caenorhabditis elegans. Science 289:1928-1930. [DOI] [PubMed] [Google Scholar]
- 64.Donze, O., and D. Picard. 2002. RNA interference in mammalian cells with siRNA synthesized with T7 RNA polymerase. Nucleic Acid Res. 30:E46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dudley, N. R., J. C. Labbe, and B. Goldstein. 2002. With RNA interference to identify genes required for RNA interference. Proc. Natl. Acad. Sci. USA 99:4191-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dykxhoorn, D. M., C. D. Novina, and P. A. Sharp. 2003. Killing the messenger: short RNAs that silence gene expression. Nat. Rev. Mol. Cell. Biol. 4:457-467. [DOI] [PubMed] [Google Scholar]
- 67.Elbashir, S. M., J. Harborth, W. Lendecknel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in mammalian cell culture. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 68.Elbashir, S. M., J. Martinez, A. Patkaniowska, W. Lendeckel, and T. Tuschl. 2001. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 20:6877-6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Elbashir, S. M., W. Lendeckel, and T. Tuschl. 2001. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15:188-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Elbashir, S. M., J. Harborth, K. Weber, and T. Tuschl. 2002. Analysis of gene functions in somatic mammalian cells with small interfering RNAs. Methods 26:199-213. [DOI] [PubMed] [Google Scholar]
- 71.English, J. J., E. Mueller, and D. C. Baulcombe. 1996. Suppression of virus accumulation in transgenic plants exhibiting silencing of nuclear genes. Plant Cell 8:179-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Escobar, M. A., E. L. Civerolo, K. R. Summerfelt, and A. M. Dandekar. 2001. RNAi-mediated oncogene silencing confers resistance to crown gall tumorigenesis. Proc. Natl. Acad. Sci. USA 98:13437-13442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fagard, M., and H. Vaucheret. 2000. (Trans) gene silencing in plants: how many mechanisms? Annu. Rev. Plant Physiol. Plant Mol. Biol. 51:167-194. [DOI] [PubMed] [Google Scholar]
- 74.Fagard, M., and H. Vaucheret. 2000. Systemic silencing signal(s). Plant Mol. Biol. 43:285-293. [DOI] [PubMed] [Google Scholar]
- 75.Fagard, M., S. Boutet, J.-B. Morel, C. Bellini, and H. Vaucheret. 2000. AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc. Natl. Acad. Sci. USA 97:11650-11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Finnegan, E. J., R. Margis, and P. M. Waterhouse. 2003. Posttranscriptional gene silencing is not compromised in the Arabidopsis CARPEL FACTORY (Dicer-LIKE1) mutant, a homolog of Dicer-1 from Drosophila. Curr. Biol. 13:236-240. [DOI] [PubMed] [Google Scholar]
- 77.Fire, A. 1999. RNA triggered gene silencing. Trends Genet. 15:358-363. [DOI] [PubMed] [Google Scholar]
- 78.Fire, A., S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver, and C. C. Mello. 1998. Potent and specific genetic interference by double-stranded RNA in C. elegans. Nature 391:806-811. [DOI] [PubMed] [Google Scholar]
- 79.Fjose, A., S. Ellingsen, A. Wargelius, and H. C. Seo. 2001. RNA interference: mechanisms and applications. Biotechnol. Annu. Rev. 7:31-57. [DOI] [PubMed] [Google Scholar]
- 80.Fortier, E., and J. M. Belote. 2000. Temperature-dependent gene silencing by an expressed inverted repeat in Drosophila. Genesis 26:240-244. [DOI] [PubMed] [Google Scholar]
- 81.Francesco, D. S., S. Hanspeter, L. Alejandro, T. Cornia, B. Estelle, and M. Frederick. 2001. Sense and anti sense mediated gene silencing in tobacco is inhibited by the same viral suppressors and is associated with accumulation of small RNAs. Proc. Natl. Acad. Sci. USA 96:6506-6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fraser, A. J. G., R. S. Kamath, P. Zipperten, M. M. Campos, M. Sohrmann, and J. Ahringer. 2000. Functional genomic analysis of C. elegans chromosome I by systemic RNA interference. Nature 408:325-330. [DOI] [PubMed] [Google Scholar]
- 83.Golden, T. A., S. E. Schauer, J. D. Lang, S. Pien, A. R. Mushegian, U. Grossniklaus, D. W. Meinke, and A. Ray. 2002. SHORT INTEGUMENTS1/SUSPENSOR1/CARPEL FACTORY, a Dicer homolog, is a maternal effect gene required for embryo development in Arabidopsis. Plant Physiol. 130:808-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gonczy, P., C. Echerverri, K. Oegema, A. Coulron, S. J. M. Jones, R. R. Copley, J. Duperon, J. Oegema, M. Brehm, E. Carrin, E. Hannak, M. Kirkham, S. Pichler, K. Flohrs, A. Gossen, S. Liedel, A. M. Alleaume, C. Martin, N. Ozlu, P. Bork, P., and A. A. Hyman. 2000. Functional genomic analysis of cell division in C. elegans with RNAi of genes on chromosome III. Nature 408:331-335. [DOI] [PubMed] [Google Scholar]
- 85.Grad, Y., J. Aach, G. D. Hayes, B. J. Reinhart, G. M. Church, G. Ruvkun, and J. Kim. 2003. Computational and experimental identification of C. elegans micro-RNAs. Mol. Cell 11:1253-1263. [DOI] [PubMed] [Google Scholar]
- 86.Grishok, A., A. E. Pasquinelli, D. Conte, N. Li, S. Parrish, I. Ha, D. L. Baillie, A. Fire, G. Ruvkun, and C. C. Mello. 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106:23-34. [DOI] [PubMed] [Google Scholar]
- 87.Grishok, A., H. Tabara, and C. C. Mello. 2000. Genetic requirements for inheritance of RNAi in C. elegans. Science 287:2494-2497. [DOI] [PubMed] [Google Scholar]
- 88.Guo, H.-S., J.-F. Fei, Qi-Xie, and N.-H. Chua. 2003. A chemical-regulated inducible RNAi in plants. Plant J. 34:383-392. [DOI] [PubMed] [Google Scholar]
- 89.Hall, I. M., G. D. Shankaranarayana, K.-I. Noma, N. Ayoub, A. Cohen, and S. L. Grewal. 2002. Establishment and maintenance of a heterochromatin domain. Science 297:2232-2237. [DOI] [PubMed] [Google Scholar]
- 90.Hall, I. M., K. Noma, and S. I. Grewal. 2003. RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc. Natl. Acad. Sci. USA 100:193-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hamann, L., K. Jensen, and K. Harbers. 1993. Consecutive inactivation of both alleles of the gb110 gene has no effect on the proliferation and differentiation of mouse embryonic stem cells. Gene 126:279-284. [DOI] [PubMed] [Google Scholar]
- 92.Hamilton, A. J., and D. C. Baulcombe. 1999. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286:950-952. [DOI] [PubMed] [Google Scholar]
- 93.Hamilton, A. J., S. Brown, H. Yuanhai, M. Ishizuka, and A. Lowe. 1998. A transgene with repeated DNA causes high frequency, post-transcriptional suppression of ACC-oxidase gene expression in tomato. Plant J. 15:737-746. [DOI] [PubMed] [Google Scholar]
- 94.Hamilton, A., O. Voinnet, L. Chappell, and D. Baulcombe. 2002. Two classes of short interfering RNA in RNA silencing. EMBO J. 21:4671-4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hammond, S. M., A. A. Caudy, and G. J. Hannon. 2001. Post-transcriptional gene silencing by double-stranded RNA. Nat. Rev. Genet. 2:110-119. [DOI] [PubMed] [Google Scholar]
- 96.Hammond, S. M., E. Berstein, D. Beach, and G. J. Hannon. 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404:293-296. [DOI] [PubMed] [Google Scholar]
- 97.Hammond, S. M., S. Boettcher, A. A. Caudy, R. Kobayashi, and G. J. Hannon. 2001. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293:1146-1150. [DOI] [PubMed] [Google Scholar]
- 98.Hannon, G. J. 2002. RNA Interference. Nature 418:244-251. [DOI] [PubMed] [Google Scholar]
- 99.Heman, M. T., J. S. Fridmann, J. T. Zilfou, E. Hernando, P. J. Paddison, C. Cordon-Cardo, G. J. Hannon, and S. W. Lowe. 2003. An epi-allelic series of p53 hypomorphs created by stable RNAi produces distinct phenotypes in vivo. Nat. Genet. 33:396-400. [DOI] [PubMed] [Google Scholar]
- 100.Hodgkin, J., A. Papp, R. Pulak, V. Ambros, and P. Anderson. 1989. A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics 123:301-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Holen, T., M. Amarzguioui, M. T. Wiiger, E. Babaie, and H. Prydz. 2002. Positional effects of short interfering RNAs targetting the human coagulation trigger tissue factor. Nucleic Acid Res. 30:1757-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hughes, C. L., and T. C. Kaufman. 2000. RNAi analysis of deformed, proboscipedia and sex combs reduced in the milkweed bug Oncopeltus fasciatus: novel roles for Hox genes in the hemipteran head. Development 127:3683-3694. [DOI] [PubMed] [Google Scholar]
- 103.Hutvagner, G., and P. D. Zamore. 2002. A micro-RNA in a multiple-turnover RNAi enzyme complex. Science 297:2056-2060. [DOI] [PubMed] [Google Scholar]
- 104.Hutvagner, G., J. McLachlan, A. E. Pasquinelli, E. Balint, T. Tuschl, P. D. Zamore. 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293:834-838. [DOI] [PubMed] [Google Scholar]
- 105.Ingelbrecht, I., H. Van Houdt, M. Van Montagu, and A. Depicker. 1994. Post-transcriptional silencing of reporter transgenes in tobacco correlates with DNA methylation. Proc. Natl. Acad. Sci. USA 91:10502-10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ischizuka, A., M. C. Siomi, and H. Siomi. 2002. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Gene Dev. 16:2497-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jackson, J. P., A. M. Lindroth, X. Cao, and S. E. Jacobsen. 2002. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416:556-560. [DOI] [PubMed] [Google Scholar]
- 108.Jacobson, S. E., M. P. Running, and E. M. Meyerowitz. 1999. Disruption of an RNA helicase/RNase III gene in Arabidopsis causes unregulated cell division in floral-meristems. Development 126:5231-5243. [DOI] [PubMed] [Google Scholar]
- 109.Jenuwein, T. 2002. Molecular biology. An RNA-guided pathway for the epigenome. Science 297:2215-2218. [DOI] [PubMed] [Google Scholar]
- 110.Jones, L., A. J. Hamilton, O. Voinnet, C. L. Thomas, A. J. Maule, and D. C. Baulcombe. 1999. RNA-DNA interactions and DNA methylation in posttranscriptional gene silencing. Plant Cell 11:2291-2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jones, L., F. Ratcliff, and D. C. Baulcombe. 2001. RNA-directed transcriptional gene silencing in plants can be inherited independently of the RNA trigger and requires Met1 for maintenance. Curr. Biol. 11:747-757. [DOI] [PubMed] [Google Scholar]
- 112.Kamath, R. S., A. G. Fraser, Y. Dong, G. Poulin, R. Durbin, M. Gotta, A. Kanapin, N. I. Bot, S. Moreno, M. Sohrmann, D. P. Welchman, P. Zipperlen, and J. Ahringer. 2003. Syst. functional analysis of the Caenorhabditis elegans genome with RNAi. Nature 421:231-237. [DOI] [PubMed] [Google Scholar]
- 113.Kasschau, K. D., Z. Xie, E. Allen, C. Llave, E. J. Chapman, K. A. Krizan, and J. C. Carrington. 2003. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell 4:205-217. [DOI] [PubMed] [Google Scholar]
- 114.Kennerdell, J. R., and R. W. Carthew. 1998. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell 95:1017-1026. [DOI] [PubMed] [Google Scholar]
- 115.Ketting, R. F., S. E. Fischer, E. Bernstein, T. Sijen, G. J. Hannon, and R. H. A. Plasterk. 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 15:2654-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ketting, R. F., T. H. A. Haverckamp, H. G. A. M. Van Luenen, and R. H. A. Plasterk. 1999. mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNase D. Cell 99:133-141. [DOI] [PubMed] [Google Scholar]
- 117.Kjemtrup, S., K. S. Sampson, C. G. Peele, L. V. Nguyen, M. A. Conkling, W. F. Thompson, and D. Robertson. 1998. Gene silencing from plant DNA carried by geminivirus. Plant J. 14:91-100. [DOI] [PubMed] [Google Scholar]
- 118.Klahre, U., P. Crete, S. A. Leuenberger, V. A. Iglesias, and F. Meins. 2002. High molecular weight RNAs and small interfering RNAs induce systemic post-transcriptional gene silencing in plants. Proc. Natl. Acad. Sci. USA 99:11981-11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Knight, S. W., and B. L. Bass. 2001. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in C. elegans. Science 293:2269-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kooter, J. M., M. A. Matzke, and P. Meyer. 1999. Listening to silent gene: transgene silencing, gene regulation and pathogen control. Trends Plant Sci. 4:340-347. [DOI] [PubMed] [Google Scholar]
- 121.Korf, I., Y. Fan, and S. Strome. 1998. The Polycomb group in Caenorhabditis elegans and maternal control of germline development. Development 125:2468-2478. [DOI] [PubMed] [Google Scholar]
- 122.Korneev, S. A., I. Kemenes, V. Straub, K. Staras, E. I. Korneeva, G. Kemenes, P. R. Benjamin, and M. OShea. 2002. Suppression of nitric oxide (NO)-dependent behavior by double-stranded RNA-mediated silencing of a neuronal NO synthase gene. J. Neurosci. 22:RC227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lagos-Quintana, M., R. Rauhut, W. Lendeckel, and T. Tuschl. 2001. Identification of novel genes coding for small expressed RNAs. Science 294:853-858. [DOI] [PubMed] [Google Scholar]
- 124.Lai, E. C. 2002. Micro RNAs are complementary to 3′ untranslated region sequence motifs that mediate negative post-transcriptional regulation. Nat. Genet. 30:363-364. [DOI] [PubMed] [Google Scholar]
- 125.Lau, N. C., L. P. Lim, E. G. Weinstein, and D. P. Bartel. 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294:858-862. [DOI] [PubMed] [Google Scholar]
- 126.Lee, R. C., and V. Ambros. 2001. An extensive class of small RNAs in Caenorhabditis elegans. Science 294:862-864. [DOI] [PubMed] [Google Scholar]
- 127.Lee, R. C., L. Feinbaum, and V. Ambros. 1993. The C. elegans heterochronic Gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75:843-854. [DOI] [PubMed] [Google Scholar]
- 128.Li, W. X., and S. W. Ding. 2001. Viral suppressors of RNA silencing. Curr. Opin. Biotechnol. 12:150-154. [DOI] [PubMed] [Google Scholar]
- 129.Lim, L. P., M. E. Glasner, S. Yekta, C. B. Burge, and D. P. Bartel. 2003a. Vertebrate micro-RNA genes. Science 299:1540. [DOI] [PubMed] [Google Scholar]
- 130.Lim, L. P., N. C. Lau, E. C. Weinstein, A. Abdelhakim, S. Yekta, M. W. Rhoades, C. B. Burge, and D. P. Bartel. 2003b. The micro-RNAs of C. elegans. Genes Dev. 17:991-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Reference deleted.
- 132.Lindbo, J. A., and W. G. Dougherty. 1992. Untranslatable transcripts of tobacco etch virus coat protein gene sequence can interfere with tobacco etch virus replication in transgenic plants and protoplasts. Virology 189:725-733. [DOI] [PubMed] [Google Scholar]
- 133.Lindbo, J. A., L. Silva-Rosales, W. M. Proebsting, and W. G. Dougherty. 1993. Induction of a highly specific antiviral state in transgenic plants: implications for regulation of gene expression and virus resistance. Plant Cell 5:1749-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Linder, L., and M.-C. Daugeron. 2000. Are DEAD-box proteins becoming respectable helicases? Nat. Struct. Biol. 7:97-99. [DOI] [PubMed] [Google Scholar]
- 135.Lipardi, C., Q. Wei, and B. M. Paterrson. 2001. RNAi as random degradation PCR: siRNA primers convert mRNA into dsRNA that are degraded to generate new siRNAs. Cell 101:297-307. [DOI] [PubMed] [Google Scholar]
- 136.Liu, Y., M. Schiff, G. Serion, X.-W. Deng, and S. P. Dinesh-kumar. 2002. Role of SCF Ubiquitin-ligase and the Cop9 signalosome in the N-gene mediated resistance response to tobacco mosaic virus. Plant Cell 14:1483-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Llave, C., K. D. Kasschau, and J. C. Carrington. 2000. Virus encoded suppressor of post-transcriptional gene silencing targets a maintenance step in silencing pathway. Proc. Natl. Acad. Sci. USA 97:13401-13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Llave, C., K. D. Kasschau, M. A. Rector, and J. C. Carrington. 2002. Endogenous and silencing-associated small RNAs in plants. Plant Cell 14:1605-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Llave, C., Z. Xie, K. D. Kasschau, and J. C. Carrington. 2002. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297:2053-2056. [DOI] [PubMed] [Google Scholar]
- 140.Lohmann, J. U., I. Endl, and T. C. G. Bosch. 1999. Silencing of developmental genes in hydra. Dev. Biol. 214:211-214. [DOI] [PubMed] [Google Scholar]
- 141.Lucy, A. P., H. S. Guo, W. X. Li, and S. W. Ding. 2000. Suppression of post transcriptional gene silencing by a plant viral protein localized in the nucleus. EMBO J. 19:1672-1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lyko, F. 2001. DNA methylation learns to fly. Trends Genet. 17:169-172. [DOI] [PubMed] [Google Scholar]
- 143.Malhotra, P., P. V. N. Dasaradhi, A. Kumar, A. Mohmmed, N. Agrawal, R. K. Bhatnagar, and V. S. Chauhan. 2002. Double-stranded RNA-mediated gene silencing of cysteine proteases (falcipain-1 and -2) of Plasmodium falciparum. Mol. Microbiol. 45:1245-1254. [DOI] [PubMed] [Google Scholar]
- 144.Mallory, A. C., B. J. Reinhart, D. Bartel, V. B. Vance, and L. H. Bowman. 2002. A viral suppressor of RNA silencing differentially regulates the accumulation of short interfering RNAs and micro-RNAs in tobacco. Proc. Natl. Acad. Sci. USA 99:15228-15233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mallory, A. C., L. Ely, T. H. Smith, R. Marathe, R. Anandalakshmi, M. Fagard, H. Vaucheret, G. Pruss, L. Bowman, and V. B. Vance. 2001. HC-Pro suppression of gene silencing eliminates the small RNA but not the transgene methylation are the mobile signal. Plant Cell 13:571-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Marie, B., J. P. Bacon, and J. M. Blagburn. 2000. Double-stranded RNA interference shows that Engrailed controls the synaptic specificity of identified sensory neurons. Curr. Biol. 10:289-292. [DOI] [PubMed] [Google Scholar]
- 147.Martens, H., J. Novotny, J. Oberstrass, T. L. Steck, P. Postlethwait, and W. Nellen. 2002. RNAi in Dictyostelium: the role of RNA-directed RNA polymerases and double-stranded RNase. Mol. Biol. Cell 13:445-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 149.Matzke, M., A. J. Matzke, and J. M. Kooter. 2001. RNA: guiding gene silencing. Science 293:1080-1083. [DOI] [PubMed] [Google Scholar]
- 150.McRobert, L., and G. A. McConkey. 2002. RNA interference (RNAi) inhibits growth of Plasmodium falciparum. Mol. Biochem. Parasitol. 119:273-278. [DOI] [PubMed] [Google Scholar]
- 151.McCaffery, A. P., L. Meuse, T.-T. Pham, D. S. Conklin, G. J. Hannon, and M. A. Kay. 2002. RNA interference in adult mice. Nature 418:38-39. [DOI] [PubMed] [Google Scholar]
- 152.McManus, M. T., and P. A. Sharp. 2002. Gene silencing in mammals by small interfering RNAs. Nat. Rev. Genet. 3:737-747. [DOI] [PubMed] [Google Scholar]
- 153.Mette, M. F., A. J. Matzke, and M. A. Matzke. 2001. Resistance of RNA-mediated TGS to Hc-Pro, a viral suppressor of PTGS, suggests alternate pathways for dsRNA processing. Curr. Biol. 11:1119-1123. [DOI] [PubMed] [Google Scholar]
- 154.Metzlaff, M., M. ODell, P. D. Cluster, and R. B. Flavell. 1997. RNA-mediated RNA degradation and chalcone synthase A silencing in Petunia. Cell 88:845-854. [DOI] [PubMed] [Google Scholar]
- 155.Misquitta, L., and B. M. Paterson. 1999. Targeted disruption of gene function in Drosophila by RNA interference (RNA-i): a role for nautilus in embryonic somatic muscle formation. Proc. Natl. Acad. Sci. USA 96:1451-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Mochizuki, K., N. A. Fine, T. Fujisawa, and M. A. Gorovsky. 2002. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in Tetrahymena. Cell 110:689-699. [DOI] [PubMed] [Google Scholar]
- 157.Morel, J. B., C. Godon, P. Mourrain, C. Beclin, S. Boutet, F. Feuerbach, F. Proux, and H. Vaucheret. 2002. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 14:629-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Moss, E. G. 2003. Silencing unhealthy alleles naturally. Trends Biotechnol. 21:185-187. [DOI] [PubMed] [Google Scholar]
- 159.Mourelatos, Z., J. Dostie, S. Paushkin, A. Sharma, B. Charroux, L. Abel, J. Rappsilber, M. Mann, and G. Dreyfuss. 2002. miRNPs: a novel class of ribonucleoproteins containing numerous micro-RNAs. Genes Dev. 16:720-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Mourrain, P., C. Beclin, T. Elmayan, F. Feuerbach, C. Godon, J.-B. Morel, D. Jouette, A. M. Lacombe, S. Nikic, N. Picault, K. Remoue, M. Sanial, T.-A. Vo, and H. Vaucheret. 2000. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101:533-542. [DOI] [PubMed] [Google Scholar]
- 161.Nakai, K., and P. Horton. 1999. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 24:34-36. [DOI] [PubMed] [Google Scholar]
- 162.Nakano, H., S. Amemiya, K. Shiokawa, and M. Taira. 2000. RNA interference for the organizer-specific gene Xlim-1 in Xenopus embryos. Biochem. Biophys. Res. Commun. 274:434-439. [DOI] [PubMed] [Google Scholar]
- 163.Napoli, C., C. Lemieux, and R. Jorgensen. 1990. Introduction of chimeric chalcone synthase gene into Petunia results in reversible cosuppression of homologous genes in trans. Plant Cell 2:279-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Nicholson, A. W. 1999. Function, mechanism and regulation of bacterial ribonucleases. FEMS Microbiol. Rev. 23:371-390. [DOI] [PubMed] [Google Scholar]
- 165.Nykanen, A., B. Haley, and P. D. Zamore. 2001. ATP requirement and small interfering RNA structure in the RNA interference pathway. Cell 107:309-321. [DOI] [PubMed] [Google Scholar]
- 166.Ogita, S., H. Uefuji, Y. Yamaguchi, N. Koizumi, and H. Sano. 2003. Producing decaffeinated coffee plants. Nature 423:823. [DOI] [PubMed] [Google Scholar]
- 167.Page, M. F., B. Carr, K. R. Anders, A. Grimson, and P. Anderson. 1999. SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol. Cell. Biol. 19:5943-5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Palauqui, J. C., T. Elmayan, J. M. Pollien, and H. Vaucheret. 1997. Systemic acquired silencing: Transgene specific posttranscriptional gene silencing is transmitted by grafting from silenced stocks to nonsilenced scions. EMBO J. 16:4738-4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Pal-Bhadra, M., U. Bhadra, and J. A. Birchler. 2002. RNAi-related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol. Cell 9:315-327. [DOI] [PubMed] [Google Scholar]
- 170.Pandolfini, T., B. Molesini, L. Avesani, A. Spena, and A. Polverari. 2003. Expression of self-complementary hairpin RNA under the control of the rolC promoter confers systemic disease resistance to plum pox virus without preventing local infection. BMC Biotechnol. 3:7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Park, W., J. Li, R. Song, J. Messing, and X. Chen. 2002. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in micro-RNA metabolism in Arabidopsis thaliana. Curr. Biol. 12:1484-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Pasquinelli, A. E. 2002. MicroRNAs: deviants no longer. Trends Genet. 18:171-173. [DOI] [PubMed] [Google Scholar]
- 173.Peele, C., C. V. Jorden, N. Muangsan, M. Turnage, E. Egelkrout, P. Eagle, L. Hanely-Bowdoin, and D. Robertson. 2001. Silencing of a-meristematic gene with geminivirus-derived vectors. Plant J. 27:357-366. [DOI] [PubMed] [Google Scholar]
- 174.Piccin, A., A. Salameh, C. Benna, F. Sandrelli, G. Mazzotta, M. Zordan, E. Rosatol, C. P. Kyriacou, and R. Costa. 2001. Efficient and heritable knockout of an adult phenotype in Drosophila with a GAL-4 driven hairpin RNA incorporating a heterologous spacer. Nucleic Acid Res. 29:55-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Provost, P., D. Dishart, J. Doucet, D. Frendewey, B. Samuelsson, and O. Radmark. 2002. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J. 21:5864-5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Rajagopal, R., S. Sivakumar, N. Agrawal, P. Malhotra, and R. K. Bhatnagar. 2002. Silencing of midgut aminopeptidase N of Spodoptera litura by double-stranded RNA establishes its role as Bacillus thuringiensis toxin receptor. J. Biol. Chem. 277:46849-46851. [DOI] [PubMed] [Google Scholar]
- 177.Ramsahoye, B. H., D. Biniszkiewicz, F. Lyko, V. Clark, A. P. Bird, and R. Jaenisch. 2000. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc. Natl. Acad. Sci. USA 97:5237-5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Raponi, M., and G. M. Arndt. 2003. Double-stranded RNA-mediated gene silencing in fission yeast. Nucleic Acid Res. 31:4481-4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ratcliff, F., B. D. Harrison, and D. C. Baulcombe. 1997. A similarity between virus defense and gene silencing in plants. Science 276:1558-1560. [DOI] [PubMed] [Google Scholar]
- 180.Reinhart, B. J., and D. P. Bartel. 2002. Small RNAs correspond to centromere heterochromatic repeats. Science 297:1831. [DOI] [PubMed] [Google Scholar]
- 181.Reinhart, B. J., E. G. Weinstein, M. W. Rhoades, B. Bartel, and D. P. Bartel. 2002. MicroRNAs in plants. Genes Dev. 16:1616-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Rhoades, M. W., B. J. Reinhart, L. P. Lim, C. B. Burge, B. Bartel, and D. P. Bartel. 2002. Prediction of plant micro-RNA targets. Cell 110:513-520. [DOI] [PubMed] [Google Scholar]
- 183.Robinson, K. A., and S. M. Beverley. 2003. Improvements in transfection efficiency and tests of RNA interference (RNAi) approaches in the protozoan parasite Leishmania. Mol. Biochem. Parasitol. 128:217-228. [DOI] [PubMed] [Google Scholar]
- 184.Rubinson, D. A., C. P. Dillon, A. V. Kwiatkowski, C. Sievers, L. Yang, J. Kopinja, M. Zhang, M. T. McManus, F. B. Gertler, M. L. Scott, and L. V. Parijs. 2003. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat. Genet. 33:401-406. [DOI] [PubMed] [Google Scholar]
- 185.Ruiz, M. T., O. Voinnet, and D. C. Baulcombe. 1998. Initiation and maintenance of virus-induced gene silencing. Plant Cell 10:937-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Ruvkun, G. 2001. Molecular biology. Glimpses of a tiny RNA world. Science 294:797-799. [DOI] [PubMed] [Google Scholar]
- 187.Schiebel, W., T. Pelissier, L. Riedel, S. Thalmier, R. Schiebel, D. Kempe, F. Lottspeich, H. L. Sanger, and M. Wassenegger. 1998. Isolation of an RNA-directed RNA polymerase-specific cDNA clone from tomato. Plant Cell 10:2087-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Schmidt, A., G. Palumbo, M. P. Bozzetti, P. Tritto, S. Pimpinelli, and V. Schafer. 1999. Genetic and molecular characterization of sting, a gene involved in crystal formation and meiotic drive in the male germ line of Drosophila melanogaster. Genetics 151:749-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Schwartz, D. S., G. Hutvagner, B. Haley, and P. D. Zamore. 2002. Evidence that siRNAs function as guides, not primers, in the Drosophila and human pathways. Mol. Cell 10:537-548. [DOI] [PubMed] [Google Scholar]
- 190.Semizarov, D., L. Frost, A. Sarthy, P. Kroeger, D. N. Halbert, and S. W. Fesik. 2003. Specificity of short interfering RNA determined through gene expression signatures. Proc. Natl. Acad. Sci. USA 100:6347-6352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Sharp, P. A. 2001. RNA interference. Genes Dev. 15:485-490. [DOI] [PubMed] [Google Scholar]
- 192.Shiagawa, T., and S. Ishii. 2003. Generation of Ski-knockdown mice by expressing a long double-strand RNA from an RNA polymerase II promoter. Genes Dev. 17:1340-1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Sijen, T., J. Fleenor, F. Simmer, K. L. Thijssen, S. Parrish, L. Timmons, R. H. Plasterk, and A. Fire. 2001. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107:465-476. [DOI] [PubMed] [Google Scholar]
- 194.Sijen, T., and J. M. Kotel. 2000. Post-transcriptional gene silencing: RNAs on the attack or the defence. Bioassays 22:520-531. [DOI] [PubMed] [Google Scholar]
- 195.Simmer, F., M. Tijsterman, S. Parrish, S. P. Koushika, M. L. Nonet, A. Fire, J. Ahringer, and R. H. Plasterk. 2002. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr. Biol. 12:1317-1319. [DOI] [PubMed] [Google Scholar]
- 196.Skipper, M. 2002. RNA interference: Interfering with infection. Nat. Rev. Genet. 3:572-573. [Google Scholar]
- 197.Smardon, A., J. M. Spoerke, S. C. Stacey, M. E. Klein, N. Mackin, and, E. M. Maine. 2000. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr. Biol. 10:169-178. [DOI] [PubMed] [Google Scholar]
- 198.Smith, N. A., S. P. Singh, M. B. Wang, P. A. Stoutjesdijk, A. G. Green, and P. M. Waterhouse. 2000. Total silencing by intron spliced hairpin RNA. Nature 407:319-320. [DOI] [PubMed] [Google Scholar]
- 199.Song, E., S. K. Lee, J. Wang, N. Ince, N. Ouyang, J. Min, J. Chen, P. Shankar, and J. Libberman. 2003. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat. Med. 9:347-351. [DOI] [PubMed] [Google Scholar]
- 200.Stauber, M., H. Taubert, and U. Schmidt-Ott. 2000. Function of bicoid and hunchback homologs in the basal cyclorrhaphan fly Megaselia (Phoridae). Proc. Natl. Acad. Sci. USA 97:10844-10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Sullenger, B. A., and E. Gilboa. 2002. Emerging clinical applications of RNA. Nature 418:252-258. [DOI] [PubMed] [Google Scholar]
- 202.Tabara, H., E. Yigit, H. Siomi, and C. C. Mello. 2002. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell 109:861-871. [DOI] [PubMed] [Google Scholar]
- 203.Tabara, H., M. Sarkissian, W. G. Kelly, J. Fleenor, A. Grishok, L. Timmons, A. Fire, and C. C. Mello. 1999. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99:123-132. [DOI] [PubMed] [Google Scholar]
- 204.Tamaru, H., and E. U. Selker. 2001. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414:277-283. [DOI] [PubMed] [Google Scholar]
- 205.Tang, G., B. J. Reinhart, D. P. Bartel, and P. D. Zamore. 2003. A biochemical framework for RNA silencing in plants. Genes Dev. 17:49-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Taverna, S. D., R. S. Coyne, and C. D. Allis. 2002. Methylation of histone h3 at lysine 9 targets programmed DNA elimination in Tetrahymena. Cell 110:701-711. [DOI] [PubMed] [Google Scholar]
- 207.Tenllado, F., and J. R. Diaz-Ruiz. 2001. Double-stranded RNA-mediated interference with plant virus infection. J. Virol. 75:12288-12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Tijsterman, M., R. F. Ketting, K. L. Okihara, T. Sijen, and R. H. A. Plasterk. 2002. RNA helicase MUT-14-dependent gene silencing triggered in C. elegans by short antisense RNAs. Science 295:694-697. [DOI] [PubMed] [Google Scholar]
- 209.Timmons, L., and A. Fire. 1998. Specific interference by ingested dsRNA. Nature 395:854. [DOI] [PubMed] [Google Scholar]
- 210.Tuschl, T. 2001. RNA interference and small interfering RNA. Chembiochem 2:239-245. [DOI] [PubMed] [Google Scholar]
- 211.Tuschl, T., and A. Borkhardt. 2002. Small interfering RNAs. Mol. Intervent. 2:158-167. [DOI] [PubMed] [Google Scholar]
- 212.Tuschl, T., P. D. Zamore, R. Lehmann, D. P. Bartel, and P. A. Sharp. 1999. Targeted mRNA degradation by double-stranded RNA in vitro. Genes Dev. 13:3191-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Ullu, E., and C. Tschudi. 2000. RNAi: applications in parasitology. New Technol. Life Sci. 6:43-46. [Google Scholar]
- 214.Vaistij, F. E., L. Jones, and D. C. Baulcombe. 2002. Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell 14:857-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Valdes, V. J., A. Sampieri, J. Sepulveda, and L. Vaca. 2003. With double-stranded RNA to prevent in vitro and in vivo viral infections by recombinant baculovirus. J. Biol. Chem. 278:19317-19324. [DOI] [PubMed] [Google Scholar]
- 216.Van Blokland, R., N. vander Geest, J. N. M. Mol, and J. M. Kooter. 1994. Transgene-mediated suppression of chalcone synthase expression in Petunia hybrida results from an increase in RNA turnover. Plant J. 6:861-877. [Google Scholar]
- 217.Vander Krol, A. R., L. A. Mur, M. Beld, J. N. M. Mol, and A. R. Stuitje. 1990. Flavanoid genes in petunia: addition of limited number of gene copies may lead to a suppression of gene expression. Plant Cell 2:291-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Vaucheret, H., and M. Fagard. 2001. Transcriptional gene silencing in plants: targets, inducers and regulators. Trends Genet. 17:29-35. [DOI] [PubMed] [Google Scholar]
- 219.Voinnet, O. 2001. RNA silencing as a plant immune system against viruses. Trends Genet. 17:449-459. [DOI] [PubMed] [Google Scholar]
- 220.Voinnet, O., C. Lederer, and D. C. Baulcombe. 2000. A viral movement protein prevents spread of gene silencing signal in Nicotiana benthamiana. Cell 103:157-167. [DOI] [PubMed] [Google Scholar]
- 221.Voinnet, O., P. Vain, S. Angell, and D. C. Baulcombe. 1998. Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell 95:177-187. [DOI] [PubMed] [Google Scholar]
- 222.Voinnet, O., Y. M. Pinto, and D. C. Baulcombe. 1999. Suppressor of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl. Acad. Sci. USA 96:14147-14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Volpe, T. A., C. Kidner, I. M. Hall, G. Teng, S. L. S. Grewal, and R. A. Martienssen. 2003. Regulation of heterochromatic silencing and histone H3 Lysine-9 methylation by RNAi. Science 297:1833-1837. [DOI] [PubMed] [Google Scholar]
- 224.Wargelius, A., S. Ellingsen, and A. Fjose. 1999. Double-stranded RNA induces specific developmental defects in zebrafish embryos. Biochem. Biophys. Res. Commun. 263:156-161. [DOI] [PubMed] [Google Scholar]
- 225.Wassenegger, M. 2000. RNA-directed DNA methylation. Plant Mol. Biol. 43:203-220. [DOI] [PubMed] [Google Scholar]
- 226.Wassenegger, M., and T. Pelissier. 1998. A model for RNA-mediated gene silencing in higher plants. Plant Mol. Biol. 37:349-362. [DOI] [PubMed] [Google Scholar]
- 227.Waterhouse, P. M., M. B. Wang, and E. J. Finnegan. 2001. Virus resistance and gene silencing: killing the messenger. Trends Plant Sci. 6:297-301. [DOI] [PubMed] [Google Scholar]
- 228.Waterhouse, P. M., M. W. Graham, and M. B. Wang. 1998. Virus resistance in gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc. Natl. Acad. Sci. USA 95:13959-13964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Wianny, F., and M. Zernicka-Goetz. 2000. Specific interference with gene function by double-stranded RNA in early mouse development. Nat. Cell Biol. 2:70-75. [DOI] [PubMed] [Google Scholar]
- 230.Wilda, M., U. Fuchs, W. Wossmann, and A. Borkhardt. 2002. Killing of leukamic cells with BCR/ABL fusion gene by RNA interference (RNAi). Oncogene. 21:5716-5724. [DOI] [PubMed] [Google Scholar]
- 231.Winston, W. M., C. Molodowitch, and C. P. Hunter. 2002. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science 295:2456-2459. [DOI] [PubMed] [Google Scholar]
- 232.Wu-Scharf, D., B. Jeong, C. Zhang, and H. Cerutti. 2000. Transgene and transposon silencing in Chlamydomonas reinhardtii by a DEAH-box RNA helicase. Science 290:1159-1162. [DOI] [PubMed] [Google Scholar]
- 233.Xie, Z., K. D. Kasschau, and J. C. Carrington. 2003. Negative feedback regulation of Dicer-like1 in Arabidopsis by micro-RNA-guided mRNA degradation. Curr. Biol. 13:784-789. [DOI] [PubMed] [Google Scholar]
- 234.Xu, P., S. Y. Vernooy, M. Guo, and B. A. Hay. 2003. The Drosophila micro-RNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr. Biol. 13:790-795. [DOI] [PubMed] [Google Scholar]
- 235.Yang, D., F. Buchholz, Z. Huang, A. Goga, C.-Y. Chen, F. M. Brodsky, and J. M. Bishop. 2002. Short RNA duplexes produced by hydrolysis with RNase III mediate effective RNA interference in mammalian cells. Proc. Natl. Acad. Sci. USA 99:9942-9947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Yang, D., H. Lu, and J. W. Erichson. 2000. Evidence that processed small dsRNA may mediate sequence specific mRNA degradation during RNAi in Drosophila embryos. Curr. Biol. 10:1191-1200. [DOI] [PubMed] [Google Scholar]
- 237.Yao, M. C., P. Fuller, and X. Xi. 2003. Programmed DNA deletion as an RNA-guided system of genome defense. Science 300:1581-1584. [DOI] [PubMed] [Google Scholar]
- 238.Yu, C. E., J. Oshima, Y. H. Fu, E. M. Wijsman, F. Hisama, R. Alisch, S. Matthews, J. Nakura, T. Miki, S. Ouais, G. M. Martin, J. Mulligan, and G. D. Schellenberg. 1996. Positional cloning of the Werners syndrome gene. Science 272:258-262. [DOI] [PubMed] [Google Scholar]
- 239.Yu, J. Y., S. L. DeRuiter, and D. L. Turner. 2002. RNA interference by expression of short interfering RNAs and hairpin RNA by mammalian cells. Proc. Natl. Acad. Sci. USA 99:6047-6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Zamore, P. D., T. Tuschl, P. A. Sharp, and D. P. Bartel. 2000. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21- to 23-nucleotide intervals. Cell 101:25-33. [DOI] [PubMed] [Google Scholar]
- 241.Zilberman, D., X. Cao, and S. E. Jacobsen. 2003. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299:716-719. [DOI] [PubMed] [Google Scholar]