Abstract
Transposable elements (TEs) have been implicated in the generation of genetic rearrangements, but their potential to mediate changes in the organization and architecture of host genomes could be even greater than previously thought. Here, we describe the naturally occurring structural and nucleotide variation around two TE insertions in the genome of Drosophila buzzatii. The studied regions correspond to the breakpoints of a widespread chromosomal inversion generated by ectopic recombination between oppositely oriented copies of a TE named Galileo. A detailed molecular analysis by Southern hybridization, PCR amplification, and DNA sequencing of 7.1 kb surrounding the inversion breakpoints in 39 D. buzzatii lines revealed an unprecedented degree of restructuring, consisting of 22 insertions of ten previously undescribed TEs, 13 deletions, 1 duplication, and 1 small inversion. All of these alterations occurred exclusively in inverted chromosomes and appear to have accumulated after the insertion of the Galileo elements, within or close to them. The nucleotide variation at the studied regions is six times lower in inverted than in noninverted chromosomes, suggesting that most of the observed changes originated in only 84,000 years. Galileo elements thus seemed to promote the transformation of these, otherwise normal, chromosomal regions in genetically unstable hotspots and highly efficient traps for transposon insertions. The particular features of two new Galileo copies found indicate that this TE belongs to the Foldback family. Together, our results strengthen the importance of TEs, and especially DNA transposons, as inducers of genome plasticity in evolution.
[The sequence data described in this paper have been submitted to the GenBank data library under accession nos. AF368842–AF368859 and AF368861–AF368900. In addition, sequences submitted under accession nos. AF162796–AF162799 were used as a basis for this study.]
Transposable elements (TEs) are intrinsic components of the genomes of all living organisms, from the simplest prokaryotes to the most complex eukaryotes (Berg and Howe 1989; Capy et al. 1998). They make up a substantial fraction of most studied genomes, although TE content varies widely in different species and tends to be positively correlated with total genome size (Hartl 2000). Current sequencing projects are revealing the precise organization of genomes and how repetitive sequences are distributed and arranged within them. In the euchromatin, TEs are usually found scattered as individual repeats interspersed with single-copy sequences. The chromosomal arms of Drosophila melanogaster, for example, contain sporadic TE insertions separated by long stretches of unique DNA (Ashburner et al. 1999; Adams et al. 2000; Benos et al. 2000). In the human genome around 35%–45% of the euchromatic portion is taken up by TEs, mainly SINEs and LINEs, more or less randomly distributed in a short period interspersion pattern (Lander et al. 2001; Venter et al. 2001). Heterochromatic regions located around centromeres and telomeres of eukaryote chromosomes, however, show a very different organization. These regions consist almost exclusively of repeated sequences and harbor a great accumulation of TE sequences. A well-known case is the pericentromeric heterochromatin of D. melanogaster, where, besides simple sequence repeats, there are many different families of mostly rearranged TEs interspersed with very little unique DNA (Gatti and Pimpinelli 1992; Pimpinelli et al. 1995; Adams et al. 2000).
Traditionally, TEs have been considered as junk DNA or mere genomic parasites, exploiting cells for their own propagation (Doolittle and Sapienza 1980; Orgel and Crick 1980). However, though probably as indirect consequences of their existence (Charlesworth et al. 1994), TEs exert a great variety of effects on the genome of their hosts and could have played a very important role in the shaping of the genetic material during evolution (Finnegan 1989; McDonald 1995; Kidwell and Lisch 1997). TEs are a major source of mutation and genetic variation by getting inserted into coding sequences or regulatory regions of genes. These insertions are generally deleterious for the organism, as happens in many Drosophila phenotypic mutants (Lindsley and Zimm 1992) and several human genetic diseases (Wallace et al. 1991; Holmes et al. 1994), but some have been involved in new gene expression patterns and even new genes with apparently beneficial effects (Britten 1996, 1997; Lander et al. 2001). Moreover, TEs possess the ability to promote genetic recombination between homologous sequences and can produce large-scale chromosomal rearrangements (Lim and Simmons 1994; Gray 2000). Specifically, TEs have been implicated in the origin of some natural chromosomal inversions in different organisms, such as bacteria (Daveran-Mingot et al. 1998), yeast (Kim et al. 1998), flies (Cáceres et al. 1999), and hominids (Schwartz et al. 1998).
One of the most outstanding examples of natural variation in chromosome structure is the extraordinarily rich inversion polymorphism in the species of the Drosophila genus. Hundreds of polymorphic inversions have been described in Drosophila, and these inversions do not distribute at random among species or among chromosomal elements within species (Krimbas and Powell 1992). Furthermore, the breakpoints of inversions are not randomly distributed along chromosomes either (Krimbas and Powell 1992; Cáceres et al. 1997). Despite the fact that not all naturally occurring inversions have TEs at their breakpoints (Wesley and Eanes 1994; Cirera et al. 1995), inversion breakpoints have been found to be associated with TE insertion sites in D. melanogaster (Lyttle and Haymer 1992; Andolfatto et al. 1999), D. willistoni (Regner et al. 1996), and the D. virilis group (Evgen'ev et al. 2000), and direct evidence for the implication of TEs in the origin of chromosomal inversions has been obtained both in the laboratory (Lim and Simmons 1994) and in nature (Cáceres et al. 1999). Therefore, it has been suggested that TEs could be responsible for the hotspots where repeated breaks have been observed (Krimbas and Powell 1992; Evgen'ev et al. 2000). However, the molecular confirmation of the existence of the hotspots and the elucidation of their anatomy have remained elusive.
Recently, we cloned and sequenced the breakpoints of a highly successful chromosomal inversion of D. buzzatii, inversion 2j, that was originated by ectopic recombination between oppositely oriented copies of a TE (Cáceres et al. 1999). This inversion inverted a central segment of the 2 standard (2st) chromosomal arrangement, the ancestral arrangement of chromosome 2 for all of the D. buzzatii cluster species (Ruiz and Wasserman 1993), comprising around one-fourth of its euchromatic fraction. In all 2j chromosomes both inversion breakpoints were found to contain large insertions that were absent from the noninverted 2st chromosomes. Because these insertions fulfilled all characteristic features of TEs (Capy et al. 1998), they were considered copies of a new transposon that was named Galileo. However, the insertion at the proximal breakpoint exhibited a very complex structure, with copies of several different internal repeats in an apparently chaotic arrangement. In addition, a preliminary study revealed that some variation in the structure of both breakpoint insertions existed among inverted chromosomes. Thus, the further characterization of the 2j breakpoints offered the opportunity to get a deeper insight into the molecular nature of inversion breakpoints and to investigate the long-term effects that TE insertions raised up to a high frequency might have on the organization of the genome.
Here, an exhaustive molecular analysis of the 2j breakpoint regions in 9 lines with 2st chromosomes and 30 lines with the 2j inversion has uncovered an amazing degree of naturally occurring structural variation among 2j chromosomes, caused by the insertion of multiple TEs inside each other, deletions, and other small DNA rearrangements. The observed structural diversity contrasts with the low level of nucleotide variation, suggesting that the structural changes have accumulated in a short period of time. Therefore, the breakpoints of inversion 2j appear to be highly variable hotspots.
RESULTS
Structural Variation at Inversion 2j Breakpoint Regions
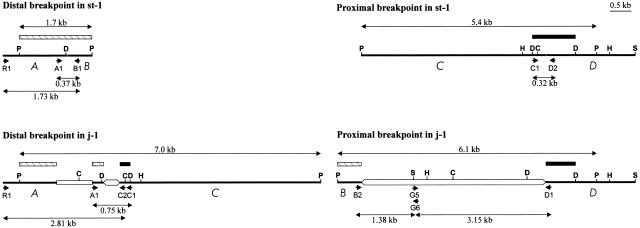
Figure 1 shows the breakpoint regions of inversion 2j in the two D. buzzatii lines that were previously characterized, st-1 and j-1 (Cáceres et al. 1999). In 2st chromosomes the breakpoint regions have been designated as AB (distal breakpoint) and CD (proximal breakpoint). Inversion 2j took place between A and B sequences and between C and D sequences, and the breakpoint regions in 2j chromosomes consist of AC (distal breakpoint) and BD (proximal breakpoint). Large insertions not present in 2st chromosomes are found in the chromosomes with the inversion between A and C sequences and between B and D sequences. In this study, several molecular techniques with increasing resolution power and accuracy were sequentially used to examine the structure of the 2j breakpoints in other 2st and 2j lines: Southern blot hybridization, PCR amplification of different segments, restriction mapping of the PCR products, and DNA sequencing.
Figure 1.
Physical map of the distal and proximal 2j breakpoint regions in the st-1 and j-1 lines. Thick lines represent the single-copy A, B, C, and D sequences. TE insertions are represented as empty boxes. Hatched and black rectangles correspond, respectively, to the AB and CD probes used for the Southern hybridization analysis. Small arrows represent primers used in the PCR amplification. Some of the restriction sites found in this region are shown: C, ClaI; D, DraI; H, HindIII; P, PstI; S, SalI.
No structural variation in the AB or CD regions was found between nine 2st lines of diverse geographic origins. Southern blot hybridization of PstI-digested genomic DNA with AB and CD probes revealed in all 2st lines the same bands of 1.7 kb and 5.4 kb, respectively, corresponding to the distal and proximal 2j breakpoint regions (Fig. 1). PCR amplification of the 1.73-kb R1–B1 and 0.37-kb A1–B1 segments (distal breakpoint) or the 0.32-kb C1–D2 segment (proximal breakpoint) did not show any size variation between the 2st lines either. Restriction mapping of the PCR products corroborated the absence of differences within each segment.
Clearly contrasting results were found in 2j chromosomes. First, variation in the restriction map of the breakpoint regions in 30 2j lines was analyzed by Southern blot hybridization. Genomic DNA of all 2j lines was digested with PstI and hybridized with a CD probe. Two hybridization bands were observed in each of the 2j lines, corresponding to the proximal and distal breakpoints with their respective insertions, and remarkable variation was detected among them: There were 11 bands of different sizes for the proximal breakpoint, whereas there were 6 different bands for the distal breakpoint (Table 1). For those lines whose PstI hybridization pattern did not coincide with that of j-1 (Fig. 1), a more detailed restriction map of the breakpoint region was elaborated by repeated Southern hybridization using additional restriction enzymes (ClaI, DraI, EcoRI, EcoRV, HindIII, SalI, and XbaI) and AB and CD probes. This resulted in the identification of nine main structural types in the proximal breakpoint and six in the distal breakpoint (Table 1).
Table 1.
Molecular Analysis by Southern Blot Hybridization and PCR Amplification of the 2j Breakpoint Regions of the 30 2j Lines Used in This Study
| Name | Geographic origin | Hybridization bands (kb) | PCR products (kb) | ||||
|---|---|---|---|---|---|---|---|
| Proximal | Distal | B2-G6 | G5-D1 | R1-C2 | A1-C1 | ||
| j-1 | Carboneras (Spain) | 6.1 | 7.0 | 1.38 | 1.92 | 2.81 | 0.75 |
| j-2 | Carboneras (Spain) | 6.1 | 7.0 | 1.38 | 1.92 | 2.81 | 0.75 |
| j-3 | Carboneras (Spain) | 6.1 | 7.0 | 1.38 | 1.92 | 2.81 | 0.75 |
| j-4 | Carboneras (Spain) | 6.1 | 7.0 | 1.38 | 1.92 | 2.81 | 0.75 |
| j-5 | Carboneras (Spain) | 6.1 | 7.0 | 1.38 | 1.92 | 2.81 | 0.75 |
| j-6 | Carboneras (Spain) | 6.1 | 7.0 | 1.38 | 1.92 | 2.81 | 0.75 |
| j-7 | Caldetas (Spain) | 6.1 | 7.0 | 1.38 | 1.92 | 2.81 | 0.75 |
| j-8 | San Luis (Argentina) | 8.5 | 7.0 | 4.15 | 2.13 | 2.83 | 0.77 |
| j-9 | Quilmes (Argentina) | 5.0 | 8.5 | 1.32 | 2.07 | 4.34 | 2.28 |
| j-10 | Palo Labrado (Argentina) | 5.1 | 9.0 | 1.38 | 2.13 | — | — |
| j-11 | Los Negros (Bolivia) | 8.8 | 7.0 | 1.32 | 2.07 | 2.83 | 0.77 |
| j-12 | Guaritas (Brazil) | 8.8 | 7.0 | 1.32 | 2.07 | 2.83 | 0.77 |
| j-13 | Guaritas (Brazil) | 8.8 | 7.0 | 1.32 | 2.07 | 2.81 | 0.75 |
| j-14 | Laboratory (Australia) | 6.1 | 7.0 | 1.38 | 1.92 | 2.83 | 0.77 |
| j-15 | Catamarca (Argentina) | 6.1 | 7.0 | 1.38 | 1.92 | 2.81 | 0.75 |
| j-16 | Salta (Argentina) | 12.1 | 7.0 | 1.38 | — | 2.83 | 0.77 |
| j-17 | Tilcara (Argentina) | 6.0 | 7.0 | 1.38 | — | 2.83 | 0.77 |
| j-18 | Termas Rio Hondo (Argentina) | 5.0 | 7.0 | 1.32 | 2.07 | 2.83 | 0.77 |
| j-19 | Ticucho (Argentina) | 10.3 | 8.9 | 1.32 | 2.25 | — | — |
| j-20 | Hemmant Australia) | 6.1 | 7.0 | 1.38 | 1.92 | 2.81 | 0.75 |
| j-21 | Hemmant (Australia) | 6.1 | 7.0 | 1.38 | 1.92 | 2.81 | 0.75 |
| j-22 | Trinkey (Australia) | 8.8 | 7.0 | 1.32 | 2.07 | 2.83 | 0.77 |
| jz3-1 | Carboneras (Spain) | 9.9 | 7.0 | 1.32 | 3.11 | 2.83 | 0.77 |
| jz3-2 | Carboneras (Spain) | 9.9 | 7.0 | 1.32 | 3.11 | 2.81 | 0.75 |
| jz3-3 | Kariouan (Tunisia) | 9.9 | 7.0 | 1.32 | 3.11 | 2.83 | 0.77 |
| jz3-4 | Tilcara (Argentina) | 8.3 | 9.2 | 1.34 | — | — | — |
| jq7-1 | Carboneras (Spain) | 7.5 | 7.0 | 1.36 | — | 2.81 | 0.75 |
| jq7-2 | Mogan, Canary Islands (Spain) | 7.5 | 11.0 | 1.36 | — | 3.62 | 1.56 |
| jq7-3 | Caldetas (Spain) | 7.5 | 7.0 | 1.36 | — | 2.81 | 0.75 |
| jq7-4 | Otamendi (Argentina) | 6.1 | 7.0 | 1.38 | 1.92 | 2.83 | 0.77 |
Hybridization bands are those obtained by Southern hybridization of PstI-digested genomic DNA of each line with the CD probe. Proximal and distal refer to the proximal and distal breakpoint, respectively. Proximal breakpoint bands indicated in boldface include a 3.8-kb extra segment due to a polymorphism in a PstI site. Products of each PCR were digested with different restriction enzymes: B2-G6, BamHI–EcoRI; G5-D1, R1-C2, and A1-C1, DraI.
In the PCR analysis of the 2j lines, smaller regions, containing just the breakpoint insertions and the adjacent single-copy DNA, were studied. Primer pairs B2–G6 and G5–D1 (proximal breakpoint) and R1–C2 and A1–C1 (distal breakpoint) were used with genomic DNA of all 2j lines (Fig. 1). The PCR products of each line were compared by gel electrophoresis and were digested with restriction enzymes to detect and map any variation existing between them (Table 1). The PCR results revealed a small difference between two lines (j-16 and jz3–4) belonging to one of the previous nine structural types defined in the proximal breakpoint and between several lines previously ascribed to the same structural type of the distal breakpoint, but otherwise confirmed the restriction maps obtained from the Southern hybridizations. However, two problems arose in the PCR amplifications. First, Taq DNA polymerase sometimes jumped between distant parts of certain DNA templates, causing an excision of the intervening segment. By sequencing the G5–D1 PCR products of lines j-1 and j-19 we showed that two different ∼1-kb deletions have occurred during the amplification. In both cases the deletions were found to take place between short homologous sequences repeated in direct orientation that were contained within long inverted repeats. Thus, the PCR excision mechanism resembles that of spontaneous deletion by slippage during DNA replication (Farabaugh et al. 1978; Albertini et al. 1982), which is stimulated by the formation of stem–loop secondary structures (Egner and Berg 1981). On the other hand, no amplification occurred in some of the 2j lines (Table 1) and other combinations of primers different of the previous ones were assayed. Nevertheless, a few breakpoint segments could not be amplified either with the new combinations of primers or with PCR conditions specially designed for the amplification of difficult templates (see Methods).
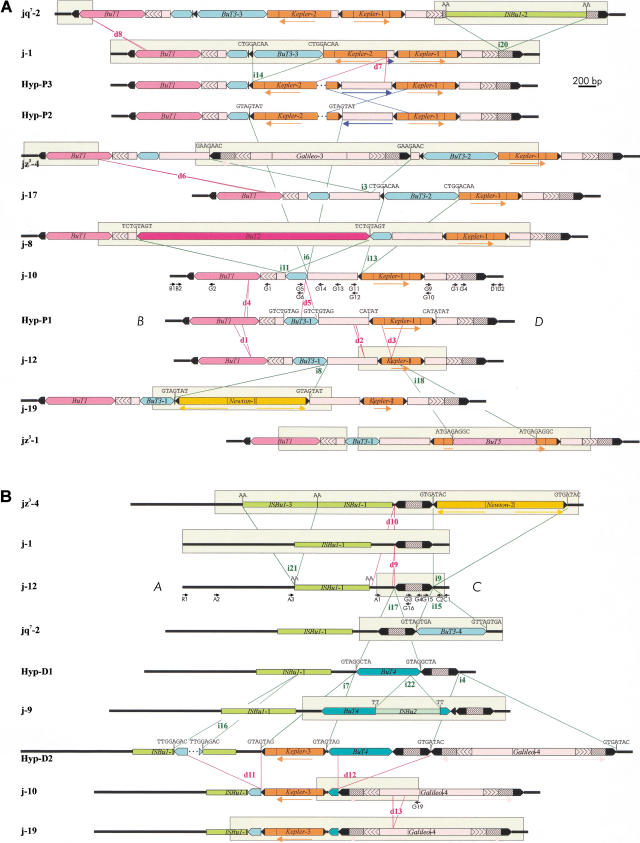
As a final step, we sequenced the regions that were found to differ between 2j lines (Fig. 2). Fragments showing varying restriction patterns were cloned and sequenced completely from the corresponding PCR products. However, when two or more 2j lines did not show any variation in the restriction map of a particular region, only the DNA of one of them was sequenced as representative. A thorough effort was made to isolate and characterize all segments in which differences have been detected. Therefore, for those segments that were not PCR-amplified or that suffered deletions during PCR, we turned to traditional cloning. Two λ genomic libraries of the j-19 and jz3–4 lines were constructed and in both lines the two breakpoints of inversion 2j were isolated. Those segments differing with regard to the other 2j lines in each breakpoint were cloned and sequenced.
Figure 2.
Schematic representation of the structures found at the proximal (A) and distal (B) breakpoints of inversion 2j in the 30 2j lines studied. All different structures are shown, except for that of j-16 in the proximal breakpoint, which differs from jz3–4 by the absence of d6 deletion. Thick lines represent the single-copy A, B, C, and D sequences. TEs are represented as colored boxes and sharp ends correspond to the ITRs. Insertions and deletions are delimited by green and red lines, respectively, and are named with an i or a d followed by a number. Target site duplications flanking the insertions are shown above them. Blue lines indicate the inversion of an internal segment. Arrows below the diagrams inform on the orientation of some homologous segments. Segments sequenced in each structure are enclosed within clear rectangles. Only the D. buzzatii lines representative of each structural variant are shown. Lines sharing the same structure in the proximal breakpoint are jq7–1, jq7–2, and jq7–3; j-1, j-2, j-3, j-4, j-5, j-6, j-7, j-14, j-15, j-20, j-21, and jq7–4; j-9, j-11, j-12, j-13, j-18, and j-22 (deletion d2 was detected during j-12 sequencing and we do not know whether it is present in other lines or not); jz3–1, jz3–2, and jz3–3. Lines sharing the same structure in the distal breakpoint are j-1, j-2, j-3, j-4, j-5, j-6, j-7, j-13, j-15, j-20, j-21, jz3–2, jq7–1, and jq7–3; j-8, j-11, j-12, j-14, j-16, j-17, j-18, j-22, jz3–1, jz3–3, and jq7–4. Hyp are hypothetical structures not found in our sample of 2j lines. Small black arrows are PCR primers used in the study.
Altogether, the Southern blot hybridization and PCR data allowed us to infer the structures present at the breakpoints of the 30 2j lines studied, and DNA sequencing let us fully identify the changes that differentiate them (Fig. 2). Ten different structural types were found in the proximal breakpoint and seven in the distal breakpoint, and most of them were related by relatively simple changes, such as insertions or deletions of DNA segments. Thus, with this information we were able to postulate a plausible evolutionary sequence of changes between the breakpoint structures. To better illustrate the changes, five hypothetical variants (Hyp) have been represented as intermediaries between the observed ones. Also, for the sake of simplicity, we have considered that all insertions occurred independently, although a few of them could have originated in a single event. In the proximal breakpoint, the simplest structure is that of Hyp–P1, which contains a Galileo insertion between B and D sequences with three other TEs inserted inside (Fig. 2A). All of the TEs inside Galileo are flanked by direct repeats, presumably generated by the duplication of the target site during the insertion event, with the only exception of BuT1. In the latter case, the absence of the outermost nucleotide of the right inverted terminal repeat (ITR), suggests that a deletion after the BuT1 insertion removed its last base pair, the right target site duplication, and part of the left long ITR of Galileo (see below). From Hyp–P1, eight large insertions of seven different TEs, eight deletions, and the inversion of an internal segment are required to generate the structural diversity actually seen in the proximal breakpoint (see Fig. 2A for details). In the distal breakpoint, the simplest structure is that of j-12, formed by a 392-bp Galileo insertion between A and C sequences and an ISBu1 insertion in A (Fig. 2B). From here, eight insertions of seven different TEs, five deletions and a small duplication should have occurred to explain the other six structural variants observed (see Fig. 2B for details).
The most important features of the 22 large insertions (named from i1 to i22) found at the breakpoints of inversion 2j are summarized in Table 2. The target site duplications flanking most insertions, the presence of multiple copies, and the variation found among lines identify the inserted DNA sequences as TEs (Capy et al. 1998). According to sequence similarities between the inserted sequences, we have recognized ten different previously undescribed TEs (that will be described in detail elsewhere). Apart from the original Galileo-1 and Galileo-2 insertions that were implicated in the generation of inversion 2j (Cáceres et al. 1999), there are two more Galileo copies inserted at the 2j breakpoints, Galileo-3 and Galileo-4. These new Galileo copies are basically composed of very long ITRs, with a relatively small and heterogeneous central region that does not seem to encode any protein involved in their transposition. Like the first two copies, they do not show homology to any known sequence in the available databases, but they display significant structural similarity to the Foldback elements described in many organisms (Bingham and Zachar 1989; Hoffman-Liebermann et al. 1989; Hankeln and Schmidt 1990; Yuan et al. 1991; Rebatchouk and Narita 1997), including the ability to form stable secondary structures when denatured (as indicated by the difficulties encountered in the PCR amplification of the segments containing these elements). Five other insertions corresponding to two closely related TEs (average sequence identity 84%) also show similarities to Foldback elements. These new elements have been named Kepler and Newton and share many of their characteristics with Galileo (average sequence identity 73%), suggesting that they belong to the same family: (1) The terminal 40 bp of their ITRs are identical (except for one single nucleotide difference); (2) all of them tend to duplicate 7 bp of the target site upon insertion (Table 2); and (3) Newton elements exhibit very long ITRs resembling those of Galileo elements. Moreover, insertions i10 to i17 correspond to four different TEs that can be ascribed to Class II (Finnegan 1989; Capy et al. 1998) and have been designated as D. buzzatii transposons or BuTs. Based on sequence homologies they have been included in the hAT superfamily (Calvi et al. 1991). BuT1 and BuT2 show similarity to the element Gandalf of D. koepferae (Marín and Fontdevila 1995), whereas BuT3 and BuT4 are related to the element Hopper of Bactrocera dorsalis (Handler and Gomez 1997). Finally, five insertions could not be neatly classified into any of the previously known TE families. BuT5 ends in ITRs of just three base pairs (followed by subterminal imperfect inverted repeats of 17 bp), generates 9-bp duplications during insertion, shows a moderately repetitive pattern by in situ hybridization to D. buzzatii polytene chromosomes (J.M. Ranz, pers. comm.), and has been tentatively considered a Class II TE. The other four insertions belong to a new class of highly repetitive mobile elements, whose members do not possess ITRs and seem to duplicate two base pairs upon insertion. We have called them ISBu elements because of their structural and sequence similarity to the IS elements of the species of the obscura group of Drosophila (Hagemann et al. 1998).
Table 2.
TE Insertions at the Breakpoint Regions of Inversion 2j of Drosophila buzzatii
| Insertion | TE | Size (bp) | ITRs (bp) | Target site (bp) | BP |
|---|---|---|---|---|---|
| Foldback-like elements | |||||
| i1 | Galileo-1 | 1589 | 228/443 | 7 | P |
| i2 | Galileo-2 | 392 | 106 | 7 | D |
| i3 | Galileo-3 | 2204 | 683/684 | 7 | P |
| i4 | Galileo-4 | 2083 | 918/916 | ND | D |
| i5 | Kepler-1 | 722 | 150 | 5 | P |
| i6 | Kepler-2 | 735 | ND | 7 | P |
| i7 | Kepler-3 | 692 | 20 | ND | D |
| i8 | Newton-1 | 1510 | 572/575 | 7 | P |
| i9 | Newton-2 | 1512 | 575/574 | 7 | D |
| hobo, Activator, Tam3 (hAT) elements | |||||
| i10 | BuT1 | 801 | 15/14 | ND | P |
| i11 | But2 | 2775 | 12 | 8 | P |
| i12 | BuT3-1 | 413 | 23 | 8 | P |
| i13 | BuT3-2 | 844 | 23 | 8 | P |
| i14 | BuT3-3 | 798 | 23 | 8 | P |
| i15 | BuT3-4 | 795 | 23 | 8 | D |
| i16 | BuT3-5 | 147 | ND | ND | D |
| i17 | BuT4 | 721 | 24/23 | 8 | D |
| Unclassified elements | |||||
| i18 | BuT5 | 1039 | 3 | 9 | P |
| i19 | ISBul-1 | 841 | — | 2 | D |
| i20 | ISBul-2 | 1467 | — | 2 | P |
| i21 | ISBul-3 | 853 | — | 2 | D |
| i22 | ISBu2 | 726 | — | 2 | D |
Elements have been classified by structural and sequence similarities with described TEs according to Capy et al. (1998). When different, the size of the left and right inverted terminal repeats (ITRs) are indicated. BP refers to the location of the element in the proximal (P) or distal (D) breakpoint.
ND, data that could not be determined due to deletions.
Several other types of genetic rearrangements besides the multiple TE insertions have been found at the 2j breakpoints. We have detected 13 deletions of more than 17 bp (Fig. 2): d1, 93 bp; d2, 24 bp; d3, 238 bp; d4, 32 bp; d5, 179 bp; d6, 41 bp; d7, >536 bp; d8, 20 bp; d9, 17 bp; d10, 248 bp; d11, >649 bp; d12, 1023 bp; and d13, 136 bp (the lengths of d7 and d11 are minimum estimates, as the real size of the deleted fragments is not known). Five of these deletions seem to have originated by the well-established mechanism of slipped-strand mispairing (Farabaugh et al. 1978; Albertini et al. 1982): d2, d3, and d6 took place between two repeated sequences of 3–4 bp, eliminating one of them and the intervening DNA; d8 and d13 removed one copy of a sequence of 20 bp and 136 bp, respectively, duplicated in tandem. A similar mechanism could also have generated the tandem duplication of the terminal 41 bp of Galileo-2 in j-9 (Fig. 2B). Finally, in some of the 2j lines we have found a change of orientation of a 55-bp Galileo-1 internal fragment, which suggests that an inversion has occurred inside the proximal breakpoint insertion (Fig. 2A). This inversion spanned ∼600 bp and was probably generated by recombination between the oppositely oriented ITRs of Kepler-1 and Kepler-2 in Hyp-P2.
Nucleotide Variation at Inversion 2j Breakpoint Regions
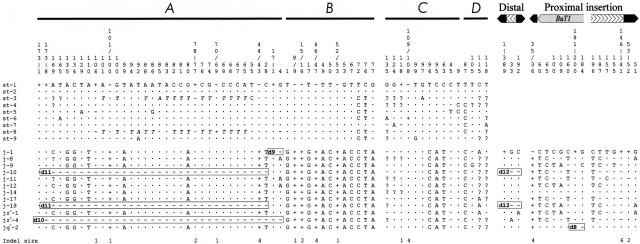
In addition to the structural variation study, we sequenced 596 bp corresponding to the A, B, C, and D single-copy sequences in the nine 2st lines and 12 2j lines representing the diversity of structural types found. For comparison, we obtained the nucleotide sequence of the same regions in D. martensis, another species of the D. buzzatii complex (Ruiz and Wasserman 1993). These are seemingly noncoding intergenic regions, located 0.5–3.7 kb apart from the rox8 (A), Pp1α-96A (C), and nAcRβ-96A (D) coding sequences (Cáceres et al. 1999). However, the last 112 bp of D show homology to a putative D. melanogaster ORF recently discovered (Adams et al. 2000) that would require further investigation. In the 12 2j lines we sequenced also 839 bp of the distal breakpoint insertion and the ends of the proximal breakpoint insertion. Figure 3 summarizes the 81 polymorphic sites found and Table 3 shows the estimates of the nucleotide diversity, π (Nei 1987), calculated ignoring sites with alignment gaps or missing data only in pairwise comparisons.
Figure 3.
Nucleotide polymorphism at the breakpoint regions of inversion 2j. Nucleotide position is represented above the sequences. The breakpoints are taken as start point of A, B, C, D, distal breakpoint insertion, and proximal breakpoint insertion sequences. Nucleotides identical to the first sequence are indicated by a dot and missing data by a question mark. Deletions and insertions are indicated by minus and plus signs, respectively, and their size in base pairs is shown below. Gross deletions affecting the sequenced regions are named as in Fig. 2 and are included in rectangles. TE insertions and target site duplications are not shown. In 2st lines there is a 18-bp stretch between A and B sequences resembling Galileo footprints (Cáceres et al. 1999) that is not represented here either. Positions A65 to A101 in st-3 and st-8 accumulate multiple nucleotide changes with regard to the other lines and are shown in italics.
Table 3.
Nucleotide Variation in the Breakpoint Regions of Inversion 2j of Drosophila buzzatii
| Region | Total (N = 21) | 2st (N = 9) | 2j (N = 12) | ||||
|---|---|---|---|---|---|---|---|
| m | S | π | S | π | S | π | |
| ABCD | 596 | 35 | 0.0197 | 15 | 0.0075 | 3 | 0.0013 |
| A | 179 | 13 | 0.0251 | 5 | 0.0063 | 2 | 0.0189 |
| B | 143 | 9 | 0.0320 | 2 | 0.0076 | 0 | 0 |
| C | 155 | 9 | 0.0167 | 6 | 0.0104 | 0 | 0 |
| D | 119 | 4 | 0.0045 | 2 | 0.0055 | 1 | 0.0015 |
| Insertions | 839 | — | — | — | — | 13 | 0.0066 |
| proximal | 447 | — | — | — | — | 11 | 0.0096 |
| distal | 392 | — | — | — | — | 2 | 0.0007 |
Positions A65 to A101 of st-3 and st-8 lines, probably originated by some sort of genetic exchange, have been excluded from the estimation of the nucleotide diversity.
N, number of sequences considered; m, maximum number of nucleotides sequenced in each region; S, number of segregating sites; π, average number of pairwise differences between sequences per nucleotide.
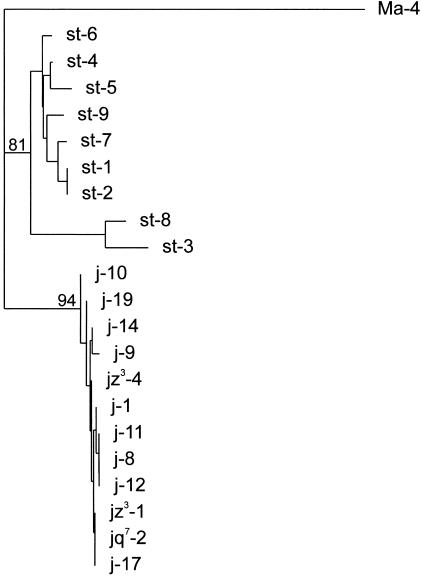
Considering the four single-copy regions together, nucleotide diversity is six times lower in 2j chromosomes than in 2st chromosomes (Table 3). We carried out computer simulations of the coalescent process using the DnaSP program (Rozas and Rozas 1999) to assess whether the nucleotide variation in each chromosomal arrangement was significantly different. Ten thousand trees were generated assuming the average number of nucleotide differences of 2st chromosomes, constant population size and no recombination, and a statistically significant probability of 0.01 of obtaining nucleotide diversity values as the one observed in 2j chromosomes or lower was found. In addition, 2st and 2j chromosomes exhibit a great number of fixed differences, including 17 nucleotide substitutions and six indels of 1–4 bp (TE insertions and target site duplications excluded). Using D. martensis as outgroup, a neighbor-joining tree (Saitou and Nei 1987) was built with the single-copy sequences of 2st and 2j lines (Fig. 4). All 2j sequences formed a monophyletic cluster of high bootstrap value, clearly separated from that of 2st sequences, confirming the proposed unique origin of the inversion (Cáceres et al. 1999).
Figure 4.
Neighbor-joining phylogenetic tree of the breakpoint sequences of inversion 2j based on the A, B, C, and D sequence data for the nine 2st and 12 2j Drosophila buzzatii lines. The Ma-4 Drosophila martensis line was used as outgroup. Bootstrap values in percentage out of 500 replicates are indicated for the main nodes.
No significant departures from the neutral model were found with the Tajima (1989) and Fu and Li (1993) tests, and nucleotide variation was used to date the origin of the inversion and of the sampled 2st and 2j alleles. The age of the inversion was estimated from the fixed differences between 2st and 2j chromosomes. The average number of nucleotide differences, dxy (Nei 1987), between 2st and 2j chromosomes is 0.0353 and between D. buzzatii and D. martensis is 0.1094. Subtracting from both figures the intraspecific polymorphism (0.0197), the net average number of nucleotide substitutions is obtained (Nei 1987). Combining the available information (Russo et al. 1995; Rodríguez-Trelles et al. 2000), we have estimated the divergence time between D. buzzatii and D. martensis as 5.8 million years (Myr) and this results in a rate of 7.7 × 10−9 nucleotide substitutions per site and per year for the breakpoint regions. Therefore, the 2j inversion should be ∼1 Myr old, which is consistent with its widespread distribution through most D. buzzatii populations. The coalescence time of 2st and 2j alleles was estimated from the average number of pairwise differences between the sequences of each chromosomal arrangement (Rozas et al. 1999). Accordingly, the sampled 2st alleles are estimated to be 485,000 years old and the sampled 2j alleles 84,000 years old.
Finally, we have used the Kreitman and Hudson's homogeneity test to detect differences in polymorphism levels between the studied regions (Kreitman and Hudson 1991). In the pooled set of 21 2st and 2j sequences no significant differences in polymorphism across A, B, C, and D regions were found (X2L = 2.86, df = 3, P = 0.41). However, the TE sequences inserted at the proximal breakpoint accumulate strikingly higher nucleotide variation between 2j chromosomes than the single-copy regions and the distal breakpoint insertion (X2L = 8.61, df = 2, P = 0.01). The difference between the polymorphism levels between 2j chromosomes at the TE insertions of each breakpoint (X2L = 4.00, df = 1, P = 0.04), which are expected to be equally selectively constrained, suggests that there could be an intrinsic increased rate of nucleotide change at the proximal breakpoint insertion.
DISCUSSION
Our detailed analysis of the breakpoints of inversion 2j has allowed us to characterize and reconstruct the evolutionary sequence of changes that has occurred in these regions. This study has revealed a great extent of genetic rearrangement at the breakpoints, consisting of 22 insertions of 10 different TEs, 13 deletions, a duplication, and an internal inversion. The low level of nucleotide variation at the single-copy sequences among 2j chromosomes suggests that the different structures in each breakpoint were generated gradually from a common ancestor in a short period of time. According to the coalescence time of the sampled 2j alleles, the changes that differentiate them, that is, 16 of the TE insertions, the 13 deletions, the duplication, and the internal inversion, are estimated to have occurred <84,000 years ago. Together with the inversion 2j itself, this represents a rapid degree of genome restructuring never found before in nature and qualifies the 2j breakpoints as genetically unstable hotspots.
Typically, the density of TE insertions in D. melanogaster euchromatin is low. The 2.9-Mb sequence from the Adh region (Ashburner et al. 1999) and the 2.6-Mb sequence from the tip of the X chromosome (Benos et al. 2000) display just one insertion every 171 kb and 155 kb on average, respectively. These values coincide with the previous observed frequencies of polymorphic insertions in particular gene regions of D. melanogaster and other Drosophila species (Table 4). The frequency of insertions found at the 2j breakpoints in D. buzzatii 2j chromosomes is, however, ∼100 times higher than the D. melanogaster average and ∼40 times bigger than the highest frequency of insertions ever found in the genus Drosophila, that of the vermilion locus of D. ananassae (Table 4). This complex array of broken and rearranged TEs accumulated in the 2j breakpoints in 2j chromosomes clearly differs from the expected organization of ordinary euchromatin and resembles more closely some D. melanogaster heterochromatic regions (Miklos et al. 1988; Vaury et al. 1989; Devlin et al. 1990; Locke et al. 1999).
Table 4.
Frequency of Naturally Occurring Insertions in Different Drosophila Species
| Species | DNA analyzed | Frequency of insertions (insertions/kb/chromosome) | Reference |
|---|---|---|---|
| D. buzzatii | |||
| 2st chromosomes | 7.1 kba | 0 | This study |
| 2j chromosomes | 7.1 kba | 0.601b | This study |
| D. melanogaster | 578 kb | 0.005 | Charlesworth and Langley 1991 |
| D. melanogaster | 229 kb | 0.004 | Aquadro 1993 |
| D. simulans | 165 kb | 0.0005 | Aquadro 1993 |
| D. pseudoobscura | 32 kb | 0 | Aquadro 1993 |
| D. ananassae | |||
| forked locus | 18 kb | 0.004 | Stephan and Langley 1989 |
| vermilion locus | 18 kb | 0.017 | Stephan and Langley 1989 |
For 2st and 2j chromosomes, the length of the single-copy region analyzed by Southern hybridization of PstI-digested DNA in 2st chromosomes was considered.
Only those insertions known to have occurred independently were computed.
What is the cause of these hotspots? The structural diversity in 2j chromosomes contrasts sharply with the lack of TE insertions and structural variation in the homologous regions of 2st chromosomes and points to an effect of the inversion or of the initial Galileo insertions as most likely explanations for the hotspots. It has been argued that TEs should accumulate around inversion breakpoints because the reduction of recombination protects them from being eliminated by deleterious ectopic exchanges (Montgomery et al. 1987; Eanes et al. 1992; Sniegowski and Charlesworth 1994), and this could in part account for the insertions at the 2j breakpoints. However, we think that the former explanation does not agree completely with our observations. First, TE insertions accumulate exclusively in very small regions around the 2j inversion breakpoints. Of the 12.3 kb corresponding to the studied region in the 2j ancestral chromosome, all TE insertions have accumulated just in the 5.1 kb comprised by the Galileo-1, Galileo-2, and ISBu1-1 elements and none in the surrounding single-copy DNA. In the two other polymorphic inversions in which variation around the breakpoints was analyzed, In(3L)P and In(2L)t of D. melanogaster, only two TE insertions were found in 2.5 kb and 5 kb studied, respectively (Hasson and Eanes 1996; Andolfatto et al. 1999). Second, although differences in mobility levels may be involved, the complete absence among the TEs inserted in the 2j breakpoints of retrotransposons, which seem to constitute the majority of TEs in Drosophila (Arkhipova et al. 1995), is noteworthy. Third, given the actual intermediate frequency of inversion 2j, the reduction in recombination is expected to affect 2st and 2j chromosomes in a similar way. Finally, the recombination reduction hypothesis does not account for deletions and other chromosomal rearrangements.
Accordingly, we favor the idea that the Galileo insertions were probably the main inducers of the generation of the hotspots. It is particularly remarkable that Galileo elements seem to belong to the Foldback family. These elements have a distinctive internally repeated structure and the FB elements of D. melanogaster are characterized by the production of extremely unstable mutations and chromosomal rearrangements at unusually high frequencies in laboratory populations (Bingham and Zachar 1989; Lovering et al. 1991). TE insertions, deletions, and the other DNA rearrangements are not distributed uniformly along the studied regions in 2j chromosomes. Instead, they appear to have occurred after Galileo-1 and Galileo-2 insertions, within or very close to them (Fig. 2). Fourteen TEs out of 20 are inserted within Galileo-1 or Galileo-2 elements and all of the observed deletions occurred inside or at the ends of pre-existing Galileo or Galileo-like elements. The fact that all 2j chromosomes share three TE insertions and one hypothetical deletion inside the Galileo-1 element and an ISBu1 insertion at the distal breakpoint is suggestive of the hotspots predating the origin of the 2j inversion, but a population bottleneck affecting 2j chromosomes could also be invoked.
There are several cases of nested insertion of TEs inside Foldback elements (Bingham and Zachar 1989; Hoffman-Liebermann et al. 1989). This sometimes has been interpreted as a mechanism to direct TE insertion outside of gene coding regions to reduce the damage inflicted to the host by their mobilization (Kidwell and Lisch 1997). Among Class II TEs, insertion site preference has been examined only for D. melanogaster P elements, which show some tendency to insert into accessible chromatin regions in the 5′ end of genes and into pre-existing P copies (Engels 1996; Liao et al. 2000). Nevertheless, many more examples are known among retrotransposons. In Saccharomyces cerevisiae, Ty1, Ty2, Ty3, and Ty4 elements are mostly located in regions upstream of tRNA genes and other genes transcribed by RNA polymerase III, whereas Ty5 prefers to integrate near silent chromatin at the telomeres (Ji et al. 1993; Zou and Voytas 1997; Boeke and Devine 1998; Kim et al. 1998). In addition, blocks of nested retrotransposons are formed in the intergenic regions of the maize genome by repeated insertion of them inside each other. In particular, 14 of the 23 retrotransposons found in the adh1-F region were inserted within other retrotransposons (SanMiguel et al. 1996, 1998). Finally, there are also retrotransposons that seem to preferentially target heterochromatic regions, such as the KERV-1 element of kangaroos (Waugh O'Neill et al. 1998) or the I element of D. melanogaster (Dimitri et al. 1997).
On the other hand, TEs, and especially DNA transposons, are largely known to mediate the production of various types of genetic rearrangements, including deletions, duplications, and inversions, with high efficiency. In laboratory studies, P elements have been found to promote deletions and duplications of the flanking genomic sequences (Preston et al. 1996) and internal deletions of P DNA (Staveley et al. 1995), whereas deletions recovered from mariner elements usually affect the ITR of the element and the DNA where is inserted (Lohe et al. 2000). In both cases, extra DNA appears sometimes between the deletion endpoints, as happens in our d4 and d5 deletions, which were accompanied by the introduction of a new nucleotide. In addition, TEs are involved in promoting genetic recombination between homologous sequences (Sved et al. 1990; McCarron et al. 1994; Lohe et al. 2000). We have already shown that recombination between Galileo copies was implicated in the generation of inversion 2j (Cáceres et al. 1999), and several other naturally occurring inversions in Diptera could have originated by a similar mechanism as well (Lyttle and Haymer 1992; Mathiopoulos et al. 1998; Andolfatto et al. 1999). At the molecular level, genetic instability might result from the presence of inverted repeats or the mechanism of transposition of the TEs inserted at the 2j breakpoints. Excluding ISBu1 and ISBu2, all of the other elements are thought to transpose by a conservative cut-and-paste mechanism (Finnegan 1989; Capy et al. 1998), in which DNA breaks induced by the transposase at the transposon ends could be aberrantly repaired by host repair functions, producing many different types of DNA alterations (Lohe et al 2000). Either an increased mutation rate attributable to repeated repair events or an increased frequency of genetic exchange with other copies of the element could account for the higher nucleotide variation observed at the TE insertion of the proximal breakpoint.
Several lessons can be drawn from this work. We have been able to follow the effects of particular TE insertions on the genome through evolutionary time and to see how these TEs seem to have altered the dynamics of ordinary euchromatic regions, transforming them into highly unstable heterochromatin-like structures. Previously, insertion and expansion of P transposon transgenes in the D. melanogaster genome was found to induce local formation of heterochromatin and this was proposed to be caused by the pairing of adjacent repeats (Dorer and Henikoff 1994). Also, the TE clustering at the 2j breakpoints is consistent with the retrotransposon associations found in D. virilis chromosomes by in situ hybridization (Evgen'ev et al. 2000) but challenges the prototypical picture of the Drosophila genome provided by D. melanogaster (Ashburner et al. 1999; Adams et al. 2000; Benos et al. 2000). An analogous disparity in TE distribution is found between two plant species with very different genome sizes, Arabidopsis thaliana and Zea mays. Similar to D. melanogaster, A. thaliana has a relatively small genome and is atypical in that most TEs are located in the pericentromeric region (Lin et al. 1999; Mayer et al. 1999). Our results are reminiscent of the explosive accumulation of 23 retrotransposons in the originally 80-kb adh-1 region of maize over the last 6 Myr that resulted in the triplication of its size (SanMiguel et al. 1996, 1998). However, the TE insertion rate observed in the 7.1-kb 2j breakpoint regions of D. buzzatii is even faster. The important effects that these blocks of TEs could have on genome evolution and the possibility that Galileo or other Foldback elements could be involved in analogous hotspots at other locations of the D. buzzatii genome are very interesting questions for further investigation.
METHODS
Drosophila Stocks
Thirty-nine lines of D. buzzatii and one of D. martensis were used in the study. The D. buzzatii lines (except jq7–3 and jq7–4) are isogenic for chromosome 2 and bear one of four different 2 chromosome arrangements: 2st, 2j, 2jz3, or 2jq7 (2jz3 and 2jq7 derive from the 2j arrangement and carry inversions 2z3 and 2q7, respectively). These lines were isolated from different natural populations covering the whole range of the species distribution. The geographic origins of the 2st lines are: st-1 and st-2, Carboneras (Spain); st-3, Vipos (Argentina); st-4, Guaritas (Brazil); st-5, Catamarca (Argentina); st-6, Salta (Argentina); st-7, Termas de Rio Hondo (Argentina); st-8, Ticucho (Argentina); and st-9, Trinkey (Australia). The geographic origin of the 2j lines is given in Table 1. The D. martensis line (Ma-4) is from Guaca (Venezuela).
Southern Hybridization and Construction of Genomic Libraries
Southern hybridization was carried out by standard methods as described previously (Ranz et al. 1999). Two probes were used for the analysis of the 2j breakpoint regions (Fig. 1). The AB probe consists of a 1.7-kb PstI fragment containing 1178 bp of A and 510 bp of B sequences, whereas the CD probe consists of a 0.9-kb DraI fragment containing 242 bp of C and 715 bp of D sequences (Cáceres et al. 1999). Two genomic libraries of the j-19 and jz3–4 D. buzzatii lines were constructed in the λGEM-11 vector (Promega) as described in Cáceres et al. (1999). To isolate the clones containing the 2j breakpoints, these libraries were screened by plaque hybridization with the AB and CD probes.
PCR Amplification
For the PCR amplification, different pairs of oligonucleotide primers covering the entire regions of study were designed (see Table 5, available as an on-line supplement at http://www.genome.org, for sequence of primers). To specifically amplify the breakpoint insertions, primers that anneal to inserted repetitive sequences were always used in combination with primers located on the flanking nonrepetitive DNA. PCRs were carried out in a volume of 50 μl, including 100–200 ng of genomic DNA of each line, 20 pmoles of the different primers, 200 μM dNTPs, 1.5 mM MgCl2, and 1–1.5 units of Taq DNA polymerase. Typical temperature cycling conditions were 30 rounds of 30 sec at 94°C, 30 sec at 50–70°C (depending on the primer pair used), and 60–180 sec at 72°C. Difficult templates that were not amplified with the normal PCR conditions were assayed with the GC-Rich PCR System (Roche), using 0.5–2 M GC-Rich resolution solution and an elongation temperature of 68°C.
DNA Sequencing and Sequence Analysis
DNA fragments of interest coming from restriction enzyme digestion or PCR amplification were cloned into Bluescript II SK (Stratagene) or pGEM-T (Promega) vectors, respectively. These fragments were sequenced on an ALFexpress (Amersham Pharmacia Biotech) or an ABI 373 A (Perkin-Elmer) automated DNA sequencer, using M13 universal and reverse primers. Nucleotide sequences were analyzed with the Wisconsin Package (Genetics Computer Group). Bestfit was used to align pairs of homologous sequences in different lines to detect inserted or deleted segments. Similarity searches through the GenBank/EMBL databases using FASTA, BLASTX, and TBLASTX were carried out to identify the inserted sequences. To analyze the nucleotide variation at the 2j breakpoints, we sequenced the same regions as in Cáceres et al. (1999) in six additional 2st lines and seven additional 2j lines. Both strands of PCR-generated templates were sequenced completely with different pairs of primers (Table 5, available as an on-line supplement at http://www.genome.org). Sequences were multiply aligned with Clustal W (Thompson et al. 1994). Polymorphism analysis was performed using the DnaSP program (Rozas and Rozas 1999). Phylogenetic analysis was performed using the PHYLIP software package (J. Felsenstein).
Acknowledgments
We are deeply indebted to J.M. Ranz for the data on the repetitive nature of BuT5 and general advice at all stages of this work. J.S.F Barker kindly provided us with 15 of the D. buzzatii stocks used. J. Rozas greatly contributed to improve the nucleotide variation analysis. We also thank A. Barbadilla for helpful discussion of results, and M. Ashburner, A. Berry, P. Capy, F. Casares, A. Navarro, and D. Petrov for valuable comments and suggestions. Work was supported by grant PB98–0900-C02–01 from the Dirección General de Investigación Científica y Técnica (Ministerio de Educación y Cultura, Spain) awarded to A.R. and a doctoral FI fellowship from the Comissionat per a Universitats i Recerca (Generalitat de Catalunya, Spain) awarded to M.C.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL caceres@salk.edu; FAX (858) 558-7454.
Article published on-line before print: Genome Res., 10.1101/gr. 174001.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.174001.
REFERENCES
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Albertini AM, Hofer M, Calos MP, Miller JH. On the formation of spontaneous deletions: The importance of short sequence homologies in the generation of large deletions. Cell. 1982;29:319–328. doi: 10.1016/0092-8674(82)90148-9. [DOI] [PubMed] [Google Scholar]
- Andolfatto P, Wall JD, Kreitman M. Unusual haplotype structure at the proximal breakpoint of In(2L)t in a natural population of Drosophila melanogaster. Genetics. 1999;153:1297–1311. doi: 10.1093/genetics/153.3.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aquadro CF. Molecular population genetics of Drosophila. In: Oakeshott J, Whitten MJ, editors. Molecular approaches to fundamental and applied entomology. New York: Springer-Verlag; 1993. pp. 222–266. [Google Scholar]
- Arkhipova IR, Lyubomirskaya NV, Ilyin YV. Drosophila retrotransposons. Heidelberg: Springer-Verlag; 1995. [Google Scholar]
- Ashburner M, Misra S, Roote J, Lewis S, Blazej R, Davis T, Doyle C, Galle R, George R, Harris N, et al. An exploration of the sequence of a 2.9-Mb region of the genome of Drosophila melanogaster: The Adh region. Genetics. 1999;153:179–219. doi: 10.1093/genetics/153.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benos PV, Gatt MK, Ashburner M, Murphy L, Harris D, Barrell B, Ferraz C, Vidal S, Brun C, Demailles J, et al. From sequence to chromosome: The tip of the X chromosome of Drosophila melanogaster. Science. 2000;287:2220–2222. doi: 10.1126/science.287.5461.2220. [DOI] [PubMed] [Google Scholar]
- Berg DE, Howe MM. Mobile DNA. Washington, D.C.: American Society for Microbiology; 1989. [Google Scholar]
- Bingham PM, Zachar Z. Retrotransposons and the FB transposon from Drosophila melanogaster. In: Berg DE, Howe MM, editors. Mobile DNA. Washington, D.C.: American Society for Microbiology; 1989. pp. 485–502. [Google Scholar]
- Boeke JD, Devine SE. Yeast retrotransposons: Finding a nice quiet neighborhood. Cell. 1998;93:1087–1089. doi: 10.1016/s0092-8674(00)81450-6. [DOI] [PubMed] [Google Scholar]
- Britten RJ. DNA sequence insertion and evolutionary variation in gene regulation. Proc Natl Acad Sci. 1996;93:9374–9377. doi: 10.1073/pnas.93.18.9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Mobile elements inserted in the distant past have taken on important functions. Gene. 1997;205:177–182. doi: 10.1016/s0378-1119(97)00399-5. [DOI] [PubMed] [Google Scholar]
- Cáceres M, Barbadilla A, Ruiz A. Inversion length and breakpoint distribution in the Drosophila buzzatii species complex: Is inversion length a selected trait? Evolution. 1997;51:1149–1155. doi: 10.1111/j.1558-5646.1997.tb03962.x. [DOI] [PubMed] [Google Scholar]
- Cáceres M, Ranz JM, Barbadilla A, Long M, Ruiz A. Generation of a widespread Drosophila inversion by a transposable element. Science. 1999;285:415–418. doi: 10.1126/science.285.5426.415. [DOI] [PubMed] [Google Scholar]
- Calvi BR, Hong TJ, Findley SD, Gelbart WM. Evidence for a common evolutionary origin of inverted repeat transposons in Drosophila and plants: hobo, Activator, and Tam3. Cell. 1991;66:465–471. doi: 10.1016/0092-8674(81)90010-6. [DOI] [PubMed] [Google Scholar]
- Capy P, Bazin C, Higuet D, Langin T. Dynamics and evolution of transposable elements. Heidelberg: Springer-Verlag; 1998. [Google Scholar]
- Charlesworth B, Langley CH. Population genetics of transposable elements in Drosophila. In: Selander RK, Clark AG, Whittam TS, editors. Evolution at the molecular level. Sunderland, MA: Sinauer; 1991. pp. 150–176. [Google Scholar]
- Charlesworth B, Sniegowski P, Stephan W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature. 1994;371:215–220. doi: 10.1038/371215a0. [DOI] [PubMed] [Google Scholar]
- Cirera S, Martín-Campos JM, Segarra C, Aguadé M. Molecular characterization of the breakpoints of an inversion fixed between Drosophila melanogaster and D. subobscura. Genetics. 1995;139:321–326. doi: 10.1093/genetics/139.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daveran-Mingot M-L, Campo N, Ritzenthaler P, le Bourgeois P. A natural large chromosomal inversion in Lactococcus lactis is mediated by homologous recombination between two insertion sequences. J Bacteriol. 1998;180:4834–4842. doi: 10.1128/jb.180.18.4834-4842.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin RH, Bingham B, Wakimoto BT. The organization and expression of the light gene, a heterochromatic gene of Drosophila melanogaster. Genetics. 1990;125:129–140. doi: 10.1093/genetics/125.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitri P, Arcà B, Berghella L, Mei E. High genetic instability of heterochromatin after transposition of the LINE-like I factor in Drosophila melanogaster. Proc Natl Acad Sci. 1997;94:8052–8057. doi: 10.1073/pnas.94.15.8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle WF, Sapienza C. Selfish genes, the phenotype paradigm, and genome evolution. Nature. 1980;284:601–603. doi: 10.1038/284601a0. [DOI] [PubMed] [Google Scholar]
- Dorer DR, Henikoff S. Expansions of transgene repeats cause heterochromatin formation and gene silencing in Drosophila. Cell. 1994;77:993–1002. doi: 10.1016/0092-8674(94)90439-1. [DOI] [PubMed] [Google Scholar]
- Eanes WF, Wesley C, Charlesworth B. Accumulation of P elements in minority inversions in natural populations of Drosophila melanogaster. Genet Res. 1992;59:1–9. doi: 10.1017/s0016672300030111. [DOI] [PubMed] [Google Scholar]
- Egner C, Berg DE. Excision of transposon Tn5 is dependent on the inverted repeats but not on the transposase function of Tn5. Proc Natl Acad Sci. 1981;78:459–463. doi: 10.1073/pnas.78.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels WR. P elements in Drosophila. In: Saedler H, Gierl A, editors. Transposable elements. Berlin: Springer-Verlag; 1996. pp. 103–123. [Google Scholar]
- Evgen'ev MB, Zelentsova H, Poluectova H, Lyozin GT, Veleikodvorskaja V, Pyatkov KI, Zhivotovsky LA, Kidwell MG. Mobile elements and chromosomal evolution in the virilis group of Drosophila. Proc Natl Acad Sci. 2000;94:7704–7711. doi: 10.1073/pnas.210386297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh PJ, Schmeissner U, Hofer M, Miller JH. Genetic studies of the lac repressor. VII. On the molecular nature of spontaneous hotspots in the lacI gene of Escherichia coli. J Mol Biol. 1978;126:847–863. doi: 10.1016/0022-2836(78)90023-2. [DOI] [PubMed] [Google Scholar]
- Finnegan DJ. Eukaryotic transposable elements and genome evolution. Trends Genet. 1989;5:103–107. doi: 10.1016/0168-9525(89)90039-5. [DOI] [PubMed] [Google Scholar]
- Fu Y-X, Li W-H. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti M, Pimpinelli S. Functional elements in Drosophila melanogaster heterochromatin. Annu Rev Genet. 1992;26:239–275. doi: 10.1146/annurev.ge.26.120192.001323. [DOI] [PubMed] [Google Scholar]
- Gray YHM. It takes two transposons to tango: Transposable-element-mediated chromosomal rearrangements. Trends Genet. 2000;16:461–468. doi: 10.1016/s0168-9525(00)02104-1. [DOI] [PubMed] [Google Scholar]
- Hagemann S, Miller WJ, Haring E, Pinsker W. Nested insertions of short mobile sequences in Drosophila P elements. Chromosoma. 1998;107:6–16. doi: 10.1007/s004120050277. [DOI] [PubMed] [Google Scholar]
- Handler AM, Gomez SP. A new hobo, Ac, Tam3 transposable element, hopper, from Bactrocera dorsalis is distantly related to hobo and Ac. Gene. 1997;185:133–135. doi: 10.1016/s0378-1119(96)00658-0. [DOI] [PubMed] [Google Scholar]
- Hankeln T, Schmidt ER. New Foldback transposable element TFB1 found in histone genes of the midge Chironomus thummi. J Mol Biol. 1990;215:477–482. doi: 10.1016/S0022-2836(05)80159-7. [DOI] [PubMed] [Google Scholar]
- Hartl DL. Molecular melodies in high and low C. Nat Rev Genet. 2000;1:145–149. doi: 10.1038/35038580. [DOI] [PubMed] [Google Scholar]
- Hasson E, Eanes WF. Contrasting histories of three gene regions associated with In(3L)Payne of Drosophila melanogaster. Genetics. 1996;144:1565–1575. doi: 10.1093/genetics/144.4.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman-Liebermann B, Liebermann D, Cohen SN. TU elements and Puppy sequences. In: Berg DE, Howe MM, editors. Mobile DNA. Washington, D.C.: American Society for Microbiology; 1989. pp. 575–592. [Google Scholar]
- Holmes SE, Dombroski BA, Krebs CM, Boehm CD, Kazazian HH. A new retrotransposable human L1 element from the LRE2 locus on chromosome 1q produces a chimaeric insertion. Nature Genet. 1994;7:143–148. doi: 10.1038/ng0694-143. [DOI] [PubMed] [Google Scholar]
- Ji H, Moore DP, Blomberg MA, Braiterman LT, Voytas DF, Natsoulis G, Boeke JD. Hotspots for unselected Ty1 transposition events on yeast chromosome III are near tRNA genes and LTR sequences. Cell. 1993;73:1007–1018. doi: 10.1016/0092-8674(93)90278-x. [DOI] [PubMed] [Google Scholar]
- Kidwell MG, Lisch D. Transposable elements as sources of variation in animals and plants. Proc Natl Acad Sci. 1997;94:7704–7711. doi: 10.1073/pnas.94.15.7704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Vanguri S, Boeke JD, Gabriel A, Voytas DF. Transposable elements and genome organization: A comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 1998;8:464–478. doi: 10.1101/gr.8.5.464. [DOI] [PubMed] [Google Scholar]
- Kreitman M, Hudson RR. Inferring the evolutionary histories of the Adh and Adh-dup loci in Drosophila melanogaster from patterns of polymorphism and divergence. Genetics. 1991;127:565–582. doi: 10.1093/genetics/127.3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimbas CB, Powell JR. Introduction. In: Krimbas CB, Powell JR, editors. Drosophila inversion polymorphism. Boca Raton, FL: CRC Press; 1992. pp. 1–52. [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Liao G-C, Rehm EJ, Rubin GM. Insertion site preferences of the P transposable element in Drosophila melanogaster. Proc Natl Acad Sci. 2000;97:3347–3351. doi: 10.1073/pnas.050017397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JK, Simmons MJ. Gross chromosome rearrangements mediated by transposable elements in Drosophila melanogaster. BioEssays. 1994;16:269–275. doi: 10.1002/bies.950160410. [DOI] [PubMed] [Google Scholar]
- Lin X, Kaul S, Rounsley S, Shea TP, Benito M-I, Town CD, Fujii CY, Mason T, Bowman CL, Barnstead M, et al. Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature. 1999;402:761–777. doi: 10.1038/45471. [DOI] [PubMed] [Google Scholar]
- Lindsley DL, Zimm GG. The genome of Drosophila melanogaster. San Diego: Academic Press; 1992. [Google Scholar]
- Locke J, Podemski L, Roy K, Pilgrim D, Hodgetts R. Analysis of two cosmid clones from chromosome 4 of Drosophila melanogaster reveals two new genes amid an unusual arrangement of repeated sequences. Genome Res. 1999;9:137–149. [PMC free article] [PubMed] [Google Scholar]
- Lohe AR, Timmons C, Beerman I, Lozovskaya ER, Hartl DL. Self-inflicted wounds, template-directed gap repair and a recombination hotspot: Effects of the mariner transposase. Genetics. 2000;154:647–656. doi: 10.1093/genetics/154.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovering R, Harden N, Ashburner M. The molecular structure of TE146 and its derivatives in Drosophila melanogaster. Genetics. 1991;128:357–372. doi: 10.1093/genetics/128.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyttle TW, Haymer DS. The role of the transposable element hobo in the origin of endemic inversions in wild populations of Drosophila melanogaster. Genetica. 1992;86:113–126. doi: 10.1007/BF00133715. [DOI] [PubMed] [Google Scholar]
- Marín I, Fontdevila A. Characterization of Gandalf, a new inverted-repeat transposable element of Drosophila koepferae. Mol Gen Genet. 1995;248:423–433. doi: 10.1007/BF02191642. [DOI] [PubMed] [Google Scholar]
- Mathiopoulos KD, della Torre A, Predazzi V, Petrarca V, Coluzzi M. Cloning of inversion breakpoints in the Anopheles gambiae complex traces a transposable element at the inversion junction. Proc Natl Acad Sci. 1998;95:12444–12449. doi: 10.1073/pnas.95.21.12444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer K, Schüller C, Wambutt R, Murphy G, Volckaert G, Pohl T, Düsterhöft A, Stiekema W, Entian K-D, Terryn N, et al. Sequence and analysis of chromosome 4 of the plant Arabidopsis thaliana. Nature. 1999;402:769–777. doi: 10.1038/47134. [DOI] [PubMed] [Google Scholar]
- McCarron M, Duttaroy A, Doughty G, Chovnick A. Drosophila P element transposase induces male recombination additively and without a requirement for P element excision or insertion. Genetics. 1994;136:1013–1023. doi: 10.1093/genetics/136.3.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JF. Transposable elements: Possible catalysts of organismic evolution. Trends Ecol Evol. 1995;10:123–126. doi: 10.1016/s0169-5347(00)89012-6. [DOI] [PubMed] [Google Scholar]
- Miklos GLG, Yamamoto M-T, Davies J, Pirrotta V. Microcloning reveals a high frequency of repetitive sequences characteristic of chromosome 4 and the β-heterochromatin of Drosophila melanogaster. Proc Natl Acad Sci. 1988;85:2051–2055. doi: 10.1073/pnas.85.7.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery EA, Charlesworth B, Langley CH. A test for the role of natural selection in the stabilization of transposable element copy number in a population of Drosophila melanogaster. Genet Res. 1987;49:31–41. doi: 10.1017/s0016672300026707. [DOI] [PubMed] [Google Scholar]
- Nei M. Molecular evolutionary genetics. New York: Columbia University Press; 1987. [Google Scholar]
- Orgel LE, Crick FHC. The ultimate parasite. Nature. 1980;284:604–607. doi: 10.1038/284604a0. [DOI] [PubMed] [Google Scholar]
- Pimpinelli S, Berloco M, Fanti L, Dimitri P, Bonaccorsi S, Marchetti E, Caizzi R, Caggese C, Gatti M. Transposable elements are stable structural components of Drosophila melanogaster heterochromatin. Proc Natl Acad Sci. 1995;92:3804–3808. doi: 10.1073/pnas.92.9.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston CR, Sved JA, Engels WR. Flanking duplications and deletions associated with P-induced male recombination in Drosophila. Genetics. 1996;144:1623–1638. doi: 10.1093/genetics/144.4.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranz JM, Cáceres M, Ruiz A. Comparative mapping of cosmids and gene clones from a 1.6 Mb chromosomal region of Drosophila melanogaster in three species of the distantly related subgenus Drosophila. Chromosoma. 1999;108:32–45. doi: 10.1007/s004120050349. [DOI] [PubMed] [Google Scholar]
- Rebatchouk D, Narita JO. Foldback transposable elements in plants. Plant Mol Biol. 1997;34:831–835. doi: 10.1023/a:1005855008823. [DOI] [PubMed] [Google Scholar]
- Regner LP, Pereira MSO, Alonso CEV, Abdelhay E, Valente VLS. Genomic distribution of P elements in Drosophila willistoni and a search for their relationship with chromosomal inversions. J Hered. 1996;87:191–198. doi: 10.1093/oxfordjournals.jhered.a022984. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Trelles F, Alarcón L, Fontdevila A. Molecular evolution and phylogeny of the buzzatii complex (Drosophila repleta group): A maximum-likelihood approach. Mol Biol Evol. 2000;17:1112–1122. doi: 10.1093/oxfordjournals.molbev.a026392. [DOI] [PubMed] [Google Scholar]
- Rozas J, Rozas R. DnaSP version 3: An integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics. 1999;15:174–175. doi: 10.1093/bioinformatics/15.2.174. [DOI] [PubMed] [Google Scholar]
- Rozas J, Segarra C, Ribó G, Aguadé M. Molecular population genetics of the rp49 gene region in different chromosomal inversions of Drosophila subobscura. Genetics. 1999;151:189–202. doi: 10.1093/genetics/151.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz A, Wasserman M. Evolutionary cytogenetics of the Drosophila buzzatii species complex. Heredity. 1993;70:582–596. doi: 10.1038/hdy.1993.85. [DOI] [PubMed] [Google Scholar]
- Russo CAM, Takezaki N, Nei M. Molecular phylogeny and divergence times of drosophilid species. Mol Biol Evol. 1995;12:391–404. doi: 10.1093/oxfordjournals.molbev.a040214. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- SanMiguel P, Tikhonov A, Jin Y-K, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer PS, Edwards KJ, Lee M, Avramova Z, Bennetzen JL. Nested retrotransposons in the intergenic regions of the maize genome. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- SanMiguel P, Gaut BS, Tikhonov A, Nakajima Y, Bennetzen JL. The paleontology of intergene retrotransposons of maize. Nat Genet. 1998;20:43–45. doi: 10.1038/1695. [DOI] [PubMed] [Google Scholar]
- Schwartz A, Chan DC, Brown LG, Alagappan R, Pettay D, Disteche C, McGillivray B, de la Chapelle A, Page DC. Reconstructing hominid Y evolution: X-homologous block, created by X-Y transposition, was disrupted by Yp inversion through LINE-LINE recombination. Hum Mol Genet. 1998;7:1–11. doi: 10.1093/hmg/7.1.1. [DOI] [PubMed] [Google Scholar]
- Sniegowski PD, Charlesworth B. Transposable element numbers in cosmopolitan inversions from a natural population of Drosophila melanogaster. Genetics. 1994;137:815–827. doi: 10.1093/genetics/137.3.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staveley BE, Heslip TR, Hodgetts RB, Bell JB. Protected P-element termini suggests a role for inverted-repeat-binding protein in transposase-induced gap repair in Drosophila melanogaster. Genetics. 1995;139:1321–1329. doi: 10.1093/genetics/139.3.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan W, Langley CH. Molecular genetic variation in the centromeric region of the X chromosome in three Drosophila ananassae populations. I. Contrasts between the vermilion and forked loci. Genetics. 1989;121:89–99. doi: 10.1093/genetics/121.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sved JA, Eggleston WB, Engels WR. Germ-line and somatic recombination induced by in vitro modified P elements in Drosophila melanogaster. Genetics. 1990;124:331–337. doi: 10.1093/genetics/124.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaury C, Bucheton A, Pelisson A. The β-heterochromatic sequences flanking the I elements are themselves defective transposable elements. Chromosoma. 1989;98:215–224. doi: 10.1007/BF00329686. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Wallace MR, Andersen LB, Saulino AM, Gregory PE, Glover TW, Collins FS. A de novo Alu insertion results in neurofibromatosis type 1. Nature. 1991;353:864–866. doi: 10.1038/353864a0. [DOI] [PubMed] [Google Scholar]
- Waugh O'Neill RJ, O'Neill MJ, Marshall Graves JA. Undermethylation associated with retroelement activation and chromosome remodelling in an interspecific mammalian hybrid. Nature. 1998;393:68–72. doi: 10.1038/29985. [DOI] [PubMed] [Google Scholar]
- Wesley CS, Eanes WF. Isolation and analysis of the breakpoint sequences of chromosome inversion In(3L)Payne in Drosophila melanogaster. Proc Natl Acad Sci. 1994;91:3132–3136. doi: 10.1073/pnas.91.8.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Finney M, Tsung N, Horvitz HR. Tc4, a Caenorhabditis elegans transposable element with an unusual fold-back structure. Proc Natl Acad Sci. 1991;88:3334–3338. doi: 10.1073/pnas.88.8.3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou S, Voytas DF. Silent chromatin determines target preference of the Saccharomyces retrotransposon Ty5. Proc Natl Acad Sci. 1997;94:7412–7416. doi: 10.1073/pnas.94.14.7412. [DOI] [PMC free article] [PubMed] [Google Scholar]