Abstract
The protein folding reaction carries great significance for cellular function and hence continues to be the research focus of a large interdisciplinary protein science community. Single-molecule methods are providing new and powerful tools for dissecting the mechanisms of this complex process by virtue of their ability to provide views of protein structure and dynamics without associated ensemble averaging. This review briefly introduces common FRET and force methods, and then explores several areas of protein folding where single-molecule experiments have yielded insights. These include exciting new information about folding landscapes, dynamics, intermediates, unfolded ensembles, intrinsically disordered proteins, assisted folding and biomechanical unfolding. Emerging and future work is expected to include advances in single-molecule techniques aimed at such investigations, and increasing work on more complex systems from both the physics and biology standpoints, including folding and dynamics of systems of interacting proteins and of proteins in cells and organisms.
1. Introduction
The proper folding of proteins is critically important in cellular activities, and involves substantial interesting physics and complexity. Hence, this physical chemical process has generated tremendous interest in the multidisciplinary field of protein science. The protein folding field has advanced greatly over the past 50 years due to knowledge acquired through a multitude of experimental, theoretical and computational approaches [1–8]. Experimental strategies have included ensemble methodologies such as NMR, fluorescence and CD spectroscopy, small-angle x-ray scattering, x-ray crystallography and protein engineering, among many others. In comparison, the application of single-molecule techniques in protein folding studies is relatively new, with many early single-molecule protein folding experiments being proof of principle studies or direct validations of previous ensemble observations. As we discuss here, important new insights are now increasingly being obtained from single-molecule folding studies, highlighting the potential impact and unique contributions that single-molecule applications can provide to this area of research that has engaged several generations of protein scientists (Fig. 1).
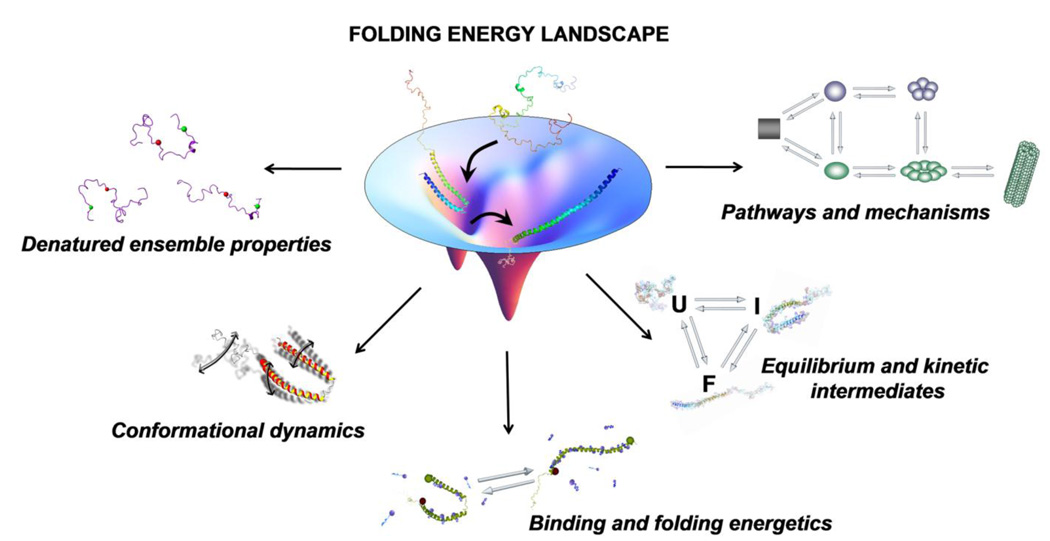
Fig. 1.
Several areas of interest in the protein folding field linked to the concept of the protein folding funnel.
1.1 Protein folding problems
The field of protein folding can be thought of in terms of three related but separate problems [3]. The first is the conformational thermodynamics issue, i.e., for a given amino acid sequence and protein environment, what determines a protein’s native structure? The second deals with the kinetics, mechanisms and energy landscapes of folding. And last is the task of predicting protein native structures. Along these lines, several more specific questions (Fig. 1) are addressed in the field, including:
What are the thermodynamic states involved as a protein folds? Are there intermediates in the folding pathway? Are the intermediates on- or off-pathway? What are the structural properties of each thermodynamic state? What does the transition state look like?
What are the sizes of the denatured state ensemble? Does it resemble a random coil, or is there significant residual structure, resulting in more collapsed conformations?
What are the dynamics or fluctuations involved in each of the thermodynamic states? How much conformational fluctuations are present in the native state ensemble?
What is the shape of the protein free energy landscape associated with folding? How rugged or smooth are the contours in the funnel-shaped landscape? What energy barriers are involved in the folding transitions? Can fast protein folders achieve global downhill folding?
What is the nature of the interactions involved in protein folding? What are the enthalpic and entropic contributions to folding?
How does interaction with ligands, molecular chaperones, protein partners or small molecule solutes such as salts and osmolytes change the energy landscape? How do parameters and conditions relevant to the cell affect protein folding?
The answers to many of these questions are critically important to function in the cell, and are encoded in a generalized energy landscape for protein binding and folding. Importantly, they will shed deep insight into fundamental biology, nanotechnology and protein design, and provide new ideas for strategies in combating protein misfolding diseases.
1.2 Early events in the history of single-molecule protein folding studies
Single-molecule techniques have the advantage of allowing the detailed examination of heterogeneous populations without ensemble averaging. Single-molecule measurements also eliminate the necessity of synchronization, which is required in some ensemble experiments. Research into single-molecule protein folding is a natural by-product of the recent technological advancements in single-molecule instrumentation, particularly in single-molecule fluorescence (especially FRET) and force techniques (i.e., atomic force microscopy (AFM), and optical and magnetic tweezers). Applications of these single-molecule techniques range from the study of protein and nucleic acid folding and dynamics to structural investigations of protein complexes and other biomolecular machines (including the nucleosome, ribosome and motor proteins), and investigations in live cells [9–19].
The first reported single-molecule ‘unfolding’ measurements were performed using optical tweezers and AFM on the muscle protein titin, whose physiological function naturally involves unfolding/stretching [20–22]. Single-molecule fluorescence experiments on denaturant-induced protein unfolding soon followed [23, 24]. Early work on protein folding that employed single-molecule FRET (smFRET) were performed using simple systems [23, 24]. For example, an early study on chymotrypsin inhibitor 2 (CI2) demonstrated the direct observation of folded and unfolded populations for a two-state system, and also showed that data acquired at the single-molecule level are consistent with bulk or ensemble data [23]. Early single-molecule unfolding experiments on a membrane protein were performed using AFM by Oesterhelt et al. [25], and presented the interesting observation that the transmembrane α-helices of individual bacteriorhodopsin molecules unfold and dissociate from the membrane in a pairwise and sequential manner. More recently, there has been a surge in single-molecule protein folding studies. This review is not meant to be comprehensive; rather it discusses several examples of information about the protein folding problem that are available from single-molecule studies, providing brief descriptions of common single-molecule and supporting techniques to orient the reader. The reader is also directed to several other reviews and original papers in the field for more information, several of which are cited throughout this review.
2. Single-molecule techniques
Here, we briefly introduce a couple of techniques that have been used in several single-molecule protein folding studies, and also discuss the use of computation and supporting ensemble methods. For additional technical details of instrumentation and theoretical aspects of single-molecule fluorescence and force techniques, the reader is referred to the following reviews: [9–19, 26–32].
2.1 Single-molecule FRET
Förster/fluorescence resonance energy transfer (FRET) is the non-radiative transfer of singlet excitation energy from donor to acceptor chromophores. This transfer has a strong distance dependence in the 30–70 Å range, making it well-suited for probing conformational properties of proteins and other biomolecules. There are two methods for applying FRET at single-molecule resolution (smFRET): experiments can be carried out on “immobilized” or “freely diffusing” molecules [26]. In the latter technique, numerous single molecules freely diffusing in solution are observed one at a time, from which histograms and population distributions are generated. In comparison, experiments on immobilized proteins follow signals from single molecules for longer times (e.g., seconds), from which equilibrium and kinetic information can be derived. The free diffusion method has the advantage over immobilized methods in minimizing the potential for surface interaction artifacts. On the other hand, the immobilized methods have the ability to monitor longer time trajectories of conformational fluctuations. To circumvent surface interaction problems with the immobilized methods, Rhoades et al. [33, 34] have used encapsulation of protein molecules in lipid vesicles. And recently, Chung et al. [35] have tackled significant issues in the immobilized methods for protein folding studies through careful analysis of fluorescent dye properties. smFRET methods provide information about several key molecular characteristics of particular interest to understanding protein folding, including conformational fluctuations, structural distributions and stochastic transitions of the protein within and between unfolded, intermediate and native ensembles. Furthermore, a range of timescales may be probed ranging from millisecond (or slower) to nanosecond (or faster) times (using correlation-type analyses as described later), covering regions of importance for local to global fluctuations and structural transitions. In addition, smFRET measurements can be carried out both in vitro and in cells, adding to its wide applicability.
2.2 Force methods (AFM and tweezers)
A number of single-molecule protein folding studies have been performed using atomic force microscopy (AFM), with a few key ones using optical tweezers (Fig. 2) [27, 36]. With AFM, proteins are held between a flexible cantilever and a surface, are pulled on, and the response of the molecule in terms of extension can be studied. Similarly, with optical tweezers, the molecule is held between two beads and the extension can be studied while applying forces to the molecule. These methods have the advantage of having more direct control on the reaction coordinate (which can be defined as the end-to-end distance), and the potential disadvantage in the necessity for the attachment of bulky “handle” surfaces or molecules. Changing pulling geometries can result in a high degree of anisotropy in the unfolding energy landscape [37], allowing a detailed exploration of folding energy landscapes [38]. The pulling force exhibits directionality, which can be controlled to mimic that which can naturally occur in vivo, for example, during the unfolding of muscle proteins or the action of the ATPase motors [39]. The use of force-clamp AFM instrumentation has opened the way for protein folding studies because it has enabled protein folding to be studied in reversible conditions, which is necessary for equilibrium thermodynamics investigations [22, 40]. More recently, the use of lock-in single-molecule force spectroscopy has increased the force sensitivity to sub-pN regime [41, 42]. There are fewer studies on protein folding using optical tweezers, but it has the advantage over AFM in that low forces and loading rates can often be used, resulting in increased spatial resolution.
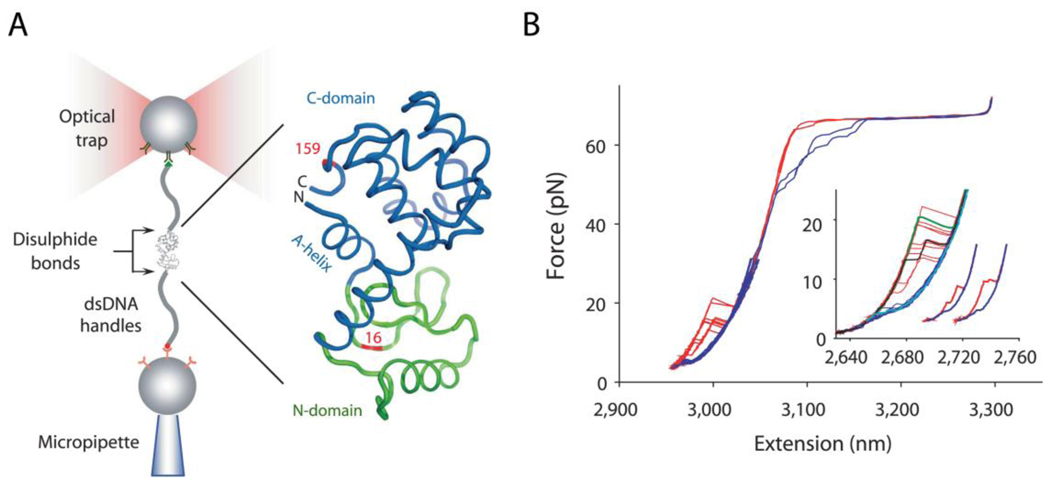
Fig. 2.
T4 lysozyme folding cooperativity investigated using optical tweezers. A. The experimental setup, with the protein attached to beads via long DNA handles, and using optical trap for manipulation. B. Unfolding (red) and refolding (blue) force-extension curves, with the inset showing data at higher resolution, showing variability in folding-unfolding behavior. Adapted by permission from Macmillan Publishers Ltd: Nature, Shank et al. [36] 465:637 (2010), copyright 2010.
2.3 In silico single-molecule protein folding
Progress in single-molecule protein folding research was accelerated by insights from computation and molecular dynamics (MD) simulations [6, 43–48]. For example, steered molecular dynamics (SMD) simulations have validated and predicted many of the results from force manipulation of biomolecules [49]. By default, these simulations already start off using single-molecule proteins. In most single-molecule studies, the assignments or characterization of the structural or conformational changes down to atomic resolution could only be done in silico. In some instances, these simulations have been predictive, such as in the importance of the linkers in spectrin where mutations in these regions were found to cause disease [49]. In another recent example, collaboration between single-molecule experimental and computational groups has resulted in new insight into the folding of the dimeric Rop protein [50].
2.4 Essential experimental controls
Because single-molecule techniques are still relatively new, evolving and non-standard in most laboratories, it is necessary to use controls to check the reliability of the results acquired using single-molecule methods. In most cases, standard ensemble measurements should be used as controls for comparison and can give hints as to whether something is amiss [23, 27]. In other cases, protein engineering methods can be used to advantage, such as in studying the different modules of titin and the corresponding AFM sawtooth curves [51] and the use of point mutations to validate models and methods [23]. For example, the presence of an intermediate can be further validated by making a point mutation to alter the population of the intermediate [52, 53]. It is also sometimes necessary to perform ensemble measurements to separate out the contribution of artifacts related to dye properties, such as that performed in Citrine unfolding to check for the presence of an intermediate [54].
3. Equilibrium and kinetic information from single-molecule protein folding experiments
3.1 Thermodynamic and kinetic parameters
Protein energy landscapes can be fully described by the structural states involved, by the rates of folding and unfolding, and the transition activation barriers. Population distributions can be examined without significant model-dependent analysis and the free energy of unfolding (ΔG°N-D) can be obtained directly from smFRET population histograms of the native and the denatured ensembles as a function of increasing denaturant, as first demonstrated by Deniz et al. for CI2 [23]. This aspect is most useful in the study of more complex proteins, as is the case when multiple coexisting states complicate simple ensemble analyses [55–57]. Also in an early study, Schuler et al. [58] presented limits to the activation energy barrier to folding from the initial estimates of polypeptide reconfiguration time according to Kramers theory, providing experimental information about the elusive pre-exponential factor [28]. A clever aspect of this study was the use of polyproline as a well-defined distance standard for more facile analysis of the data; however, even these molecules were later reported by these and other authors to be somewhat conformationally heterogeneous [59, 60], which underlines the difficulty of obtaining “ideal” distance standards.
Unfolded states show a large degree of conformational fluctuations occurring in a wide range of timescales. For example, the natively unfolded proteins Sup35-NM prion and α-synuclein were demonstrated to exhibit high degrees of conformational fluctuations in the ns-µs timescales, as measured by FCS and FCS-FRET techniques [55, 61]. The width of the transfer efficiency histogram peaks can reveal the underlying chain dynamics involved [28], taking into account contributions from shot noise. In the work by Nettels et al. [62], a reconfiguration time of ~50 ns was extracted from photon statistics for unfolded Csp, which places the free energy barrier at ~10 kBT. Furthermore, using two parameters (shape of the end-to-end distance distributions and the effective end-to-end diffusion coefficient as a function of the denaturant concentration), Nettels et al. were able to describe a more quantitative free energy surface of the collapse of Csp.
Lipman et al. [63] initiated the use of microfluidic devices combined with smFRET to study protein folding kinetics, and observed the same folding rates as those observed in ensemble measurements. Orte et al. [54] also implemented a homemade nanomanipulation system to obtain the kinetic parameters for a triangular kinetic scheme (with the presence of intermediate) of unfolding of the β-barrel protein Citrine (a variant of GFP). More recent reports have continued this work, and such non-equilibrium measurements of folding will continue to be important in the future.
Thermodynamic parameters can also be obtained using force methods. For example, the Crooks fluctuation theorem [64, 65] was employed to extract the equilibrium ΔG° from non-equilibrium force vs. distance curves detailed in [38]. Using optical tweezers, Gebhardt et al. [38] obtained the kinetic folding and unfolding rates, as well as the intermediates and transition state locations from the distances to generate a full distance-resolved energy landscape for GCN4 leucine zipper. In another AFM study, Rief and co-workers obtained the folding, unfolding rates (and related to kon, koff binding rates), stoichiometry and cooperativity involved in calmodulin binding to a variety of peptide partners [66, 67]. It is important to note that thermodynamic parameters from mechanical unfolding by force techniques could be significantly different from smFRET chemical unfolding experiments. Along these lines, Best et al. have shown using ϕ–analysis to characterize transition states that different unfolding pathways are explored by mechanical and denaturant unfolding [44].
3.2 Folding intermediates and the physics of folding landscapes
Many smFRET studies to date have been with simple two-state folders, e.g., CI2 [23], CspTm [58], GCN4 [24], protein L [68], and protein G B1 domain [35]. Additional peaks in smFRET histograms of proteins under partially folding conditions signal the existence of folding intermediates. In some cases, different states are difficult to resolve and the presence of broad distributions will require substantial modeling. For example, recent studies on SNase [69] showed two-state folding behavior for protein variants that have been previously demonstrated as three-state folders. The authors indicate that it is most likely that they were not able to resolve the intermediate states. This illustrates the importance of having multiple probes, as was the case for studies on CspTm and the four-helical Im7 (immunity protein). Data from multiple probes confirmed the two-state nature of CspTm [58, 70] and that Im7 folds via an on-pathway intermediate under appropriate conditions [71]. In another study using two-color coincidence detection (TCCD) coupled to a nano-mixer, Orte et al. were able to detect an on-pathway intermediate [54]. Recent work has revealed details of a complex, multistate folding-binding landscape for the neuronal protein α-synuclein [55] (see below – section 4.1 and Fig. 3). In another example, the protein dimer Rop was studied using smFRET by Gambin et al.[50]. Based on computational analysis and previous ensemble data, Rop was predicted to have a dual-funneled native state. smFRET experiments directly validated this prediction and also provided interesting insight into the mechanism by which the system interconverts between the two minima.
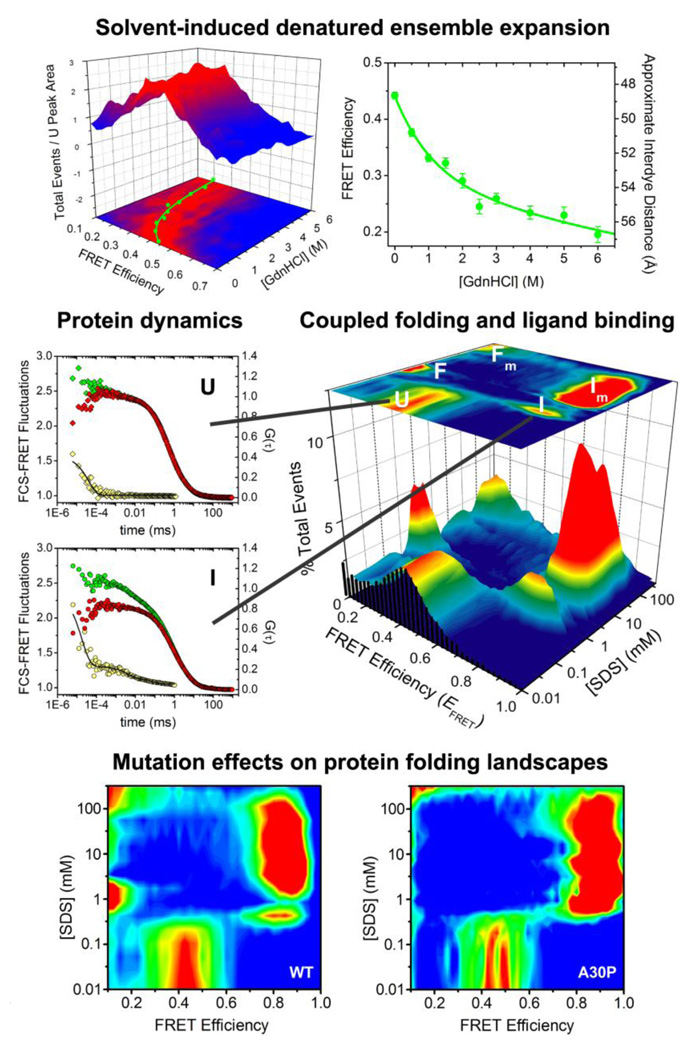
Fig. 3.
Equilibrium, dynamic and binding-folding behavior of an intrinsically disordered protein studied using single-molecule fluorescence. α-Synuclein denaturation demonstrates non-cooperative expansion (top, adapted from [26], Ferreon et al. Methods Enzymol. (2010) 472:179). SDS binding reveals multistate folding and varied dynamics (middle, adapted from [55], Ferreon et al. Proc. Natl. Acad. Sci. USA, (2009) 106:5645). The Parkinson’s disease-linked mutation A30P substantially alters α-synuclein binding-folding landscape (bottom, adapted from [56], Ferreon et al. Angew. Chem. Int. Ed. (2010) 49:3469).
Similarly, with AFM, the presence of additional features in the extension trace suggests the presence of intermediates [49]. Early AFM work on titin showed seemingly simple unfolding in an all-or-none fashion, although subsequent work showed a reversible intermediate that causes an extension of ~7Å [53, 72]. For this protein, an unfolding intermediate was suggested to function as a kinetic trap to buffer and prevent the module from completely unfolding [73]. The existence of intermediates in the fibronectin domain FnIII1 is believed to have relevance for the protein’s strength and formation of fibrils [74]. In several protein folding scenarios, intermediates have been suggested as obligate precursors in the folding pathway to the native state and are characterized by an ensemble of minimum energy collapsed structures (MECS) [75]. Ceconi et al. [52] have shown using optical tweezers that E.coli RNase H unfolds in a two-state manner, and refolds through an intermediate that is similar to that observed in ensemble measurements. This on-pathway intermediate was characterized by ‘hopping’ movements from intermediate to unfolded states, transient non-local contacts and non-specific tertiary interactions. This study also demonstrated how single-molecule methods could directly prove connectivity in different states involved in protein folding studies. With increased spatial resolution for optical tweezers, Gebhardt et al. [38] resolved two intermediate states in the GCN4 based leucine zipper unfolding. In the mechanical unfolding of green fluorescent protein, Dietz and Rief [76] showed the presence of intermediate states that are structurally different from denaturation experiments.
The case of barrierless or continuous protein folding is predicted by the folding landscape theory. Here, folding approaches the ‘speed limit’ or the maximum rate of protein folding [28, 77, 78], and the observed rates approaches the diffusion coefficient [79]. Several proteins with fast folding kinetics are believed to be within or near a downhill regime [77, 79], and there has been significant discussion about this folding regime [80–84] for the case of BBL. Single-molecule experiments can in principle distinguish between barrierless or global downhill folding and barrier-limited folding. However, this requires that the observation time be in the µs regime for most fast folders [84]. In this context, Chung et al. [35] have continued to make progress towards measuring transition path times from smFRET equilibrium trajectories. These types of studies point to the need for continued progress towards more rapid and higher signal/noise smFRET measurements.
3.3 Sizes of unfolded states
smFRET has been used for several studies of the unfolded states of proteins. These heterogeneous ensembles have been of substantial interest because of their intrinsic complexity, links to polymer physics and the distinct possibility of residual structure that guides the subsequent folding of proteins. Most proteins studied by smFRET showed that native states are fairly constant in dimensions but the unfolded states expand with increasing denaturant concentration. Ziv et al. [85] explained this behavior using polymer theory. The authors noted an interesting correlation between slopes of collapse and folding free energies vs. denaturant concentration, pointing to the mediatory role played by the denatured state in denaturant effects on folding. Recent data in FynSH3 using multiple pairwise probes showed that the denatured ensemble dimensions are in agreement with a random coil ensemble, with potentially significant deviations that can suggest residual structure, as seen in NMR studies [86]. Studies on Staphylococcal nuclease (SNase) showed that in 4M GdmCl, the protein has an Rg of 42–44 Å, close to what is expected for a random coil (45Å), but significantly larger than the 37Å measured by SAXS in 6M GdmCl [69]. Recently, Nettels et al. [70] studied the temperature-induced collapse of CspTm using smFRET and molecular simulations, and concluded that the “hydrophobic collapse” is not the main mechanism for the observed collapse, and that secondary structure formation and hydrogen bonding plays significant roles in the collapse. This was also verified by using an extremely hydrophilic intrinsically disordered protein, prothymosin α, which also showed collapse. The “unfolded states” of intrinsically disordered proteins (IDP; see section on IDPs below) can be directly studied under native conditions as discussed below.
4. Other novel insights into protein biophysics and biology
4.1 Intrinsically disordered proteins
More than 50% of the cellular proteome consists of proteins with long disordered regions [87], and >70% of cancer-related proteins have significant disorder [88]. Uversky et al. introduced the ‘D2 concept’ to show that there is a widespread occurrence of disordered proteins in diseases [89]. The advantages of intrinsic disorder in cellular processes include increased flexibility, potential for high binding specificity coupled with lower affinity, and interaction with multiple binding partners [90, 91]. With IDPs, the native states have a wider conformational space that is accessible, which could be relevant to protein function through formation of transient encounter complexes via a ‘flycasting mechanism’ [92, 93], although the corresponding slower diffusion would also influence the binding kinetics.
In an early study, Mukhopadhyay et al. [61] showed that for the amyloid determining region of the yeast prion Sup35, the natively unfolded ensemble is more compact than the denaturant-induced unfolded ensembles of most proteins, and observed a gradual expansion of the natively unfolded state upon increasing denaturant concentration, indicating a collapsed unfolded state. Moreover, the ensemble is rapidly interconverting (on the sub-µs timescale). This behavior may have relevance for the aggregation of this protein, which is thought of as an example of the class of functional amyloids. With another IDP, α-synuclein, a similar collapsed state is observed in the absence of denaturants, and a non-cooperative transition of the natively unfolded protein with denaturant titration was observed (Fig. 3) [26]. A recent combination computation and FCS study provided new insight into the effects of charge content and distribution on IDP conformational ensembles [48]; the role of charge was also explored in a recent smFRET study [94]. Overall, the disordered states of IDPs encode interesting features that differ from those of unfolded globular proteins, and that may play key roles in their function and dysfunction.
Another exciting feature of IDPs is that most undergo disorder-to-order transitions upon binding to partners, which could be key to their cellular function. Using single-molecule FRET, Ferreon et al. [55] studied the coupled folding and ligand binding equilibria of the Parkinson’s disease-associated IDP α-synuclein. In the presence of the lipid mimetic SDS and small unilamellar phospholipid vesicles, a remarkable conformational plasticity of α-synuclein was revealed as the protein cycles through multiple conformational states as a function of ligand properties (Fig. 3). The observed conformations exhibited different conformational fluctuations in various timescales. The reported single-molecule results could be of relevance to the in vivo function of α-synuclein, the protein’s natural membrane interactions, and its propensity to misfold and aggregate. smFRET data in a subsequent paper [56] showed that the mutation A30P associated with early-onset Parkinson’s disease has a pronounced effect on the protein’s folding-binding landscape. Other smFRET studies have provided similar interesting information about some aspects of wild-type α-synuclein folding-binding properties [95, 96]. AFM studies have also provided new information about this protein [97, 98], including revealing the presence of significant intermolecular interactions between α-synuclein molecules, which may be important during aggregation, and residual structure that varies between wild-type and mutant proteins.
4.2 Assisted protein folding
4.2.1 Molecular chaperones
Many cellular proteins are prone to misfolding and aggregation during the folding reaction. To counter these adverse pathways, there is a class of protein molecular chaperones whose primary cellular function is to aid the protein folding process. The best-studied example is the bacterial GroEl-GroES system. The mechanisms of GroEl-assisted folding have been extensively investigated in ensemble experiments [99, 100]. Several ensemble studies suggest that GroEl-GroES behaves like a passive Anfinsen cage that captures aggregation-prone folding intermediates [99], although other studies have suggested that GroEl-GroES may play a more active role in assisted folding such as in changing the conformation of substrates inside the cage [100].
Recent studies using smFRET indicates that substrates (maltose binding protein [MBP] and rhodanese) inside the GroEl cavities in the early stages prior to GroES binding and ATP hydrolysis are characterized by a broadened conformational distribution [101, 102]. More recently, Chakraborty et al. [103] presented smFRET data as supportive of GroEl-GroES rescuing a protein from a kinetically trapped state. Also in recent work, Hofmann et al. [104] reported smFRET results that show differential effects of this chaperonin system on folding of the different domains of MBP. In a different chaperonin system, Bechtluft et al. [105] used optical tweezers to investigate the mechanism of SecB in the translocation of newly synthesized polypeptides across membranes. Also using MBP as the test substrate, they showed that SecB interacts with the molten globule-like state and extended states of MBP, protecting the hydrophobic regions and preventing aggregation. Thus, in these examples, by binding to chaperones, the folding energy landscape is re-shaped, with previously easily accessible conformational states such as the aggregated states now exhibiting higher energy barriers to formation.
4.2.2 Chemical chaperones - osmolytes
Nature employs naturally occurring small molecules collectively known as osmolytes to modulate the conformational properties of proteins in response to a wide variety of environmental stresses [106]. Ensemble studies by Bolen and co-workers have shed light on the mechanism of protein action for a variety of these osmolytes [107–112]. From single-molecule AFM studies, Garcia-Manyes et al. [113] showed that ethanol and the osmolyte glycerol affect the activation energy barrier to unfolding of ubiquitin, and both molecules have varying effects on the protein native, transition and unfolded states. While glycerol has an effect on the native and extended states, it had no effect on the ensemble of minimum energy collapsed structures that are necessary intermediates prior to the native state form.
4.3 Co-translational folding
Co-translational folding incorporates additional interesting elements into the folding process [114]. Folding occurs before the entire protein chain has been synthesized. Additionally, the ribosome plays a significant role in protein folding through a co-translational folding process. Co-translational folding narrows the available configurations that a protein can access. Recently, Katranidis et al. [115] used single-molecule fluorescence microscopy to follow the co-translational folding of a fast folding and maturing variant of green fluorescent protein (GFP), GFP Emerald. They found that the entire process of translation and folding occurs with a time constant of 5.3 min. This time also reflects the maturation of the chromophore, which is the limiting step to the actual detection of the process. They also observed appearance of the fastest GFP molecules within one min for a significant fraction of proteins.
4.4 Bio-mechanical forces
Mechanical forces govern many cellular processes. Studies in AFM and force manipulation methods have given valuable insights into these biological processes [42]. The protein titin acts as an efficient entropic spring, and the purpose of the force hysteresis is to keep efficiency high due to the repetitive mechanical loading in muscles [52]. In a detailed AFM study by Li et al. [73] on the different force-extension properties of the sub-domains of the elastic region of cardiac titin, the investigators were able to reconstitute the function of the whole from the sum of its parts. In particular, the unfolding of the proximal Ig modules was shown to protect cardiac sarcomeres from being damaged at forces beyond the physiological range. However, recent studies by Williams et al. [116] argue against this conclusion because I27 was found to withstand higher forces and under physiological forces, the partially unfolded intermediate does not contribute to mechanical strength. However, titin may also have other biological functions, e.g., as a mechanical signal transducer [42, 117]. Stacked helical ankyrin repeats were found to behave like a mechanical nanospring [118]. The amazing reversible force generated by the ankyrin repeats has relevance in mechanotransduction signaling in cells such as the mechanoreceptors in hair cells. Studies on the extracellular matrix protein tenascin, which consists of fibronectin type III domains, suggest that their elasticity is similar to titin and that they aid in mechanical interactions necessary in cell rolling and migration [74].
An interesting aspect of mechanical unfolding of proteins was observed in studies of variably linked ubiquitin chain either in N-C or Lys48 position, demonstrating a difference in mechanical stability with respect to the direction of the pulling geometry. The lower force necessary to unfold the Lys48-Ub provides a possible mechanism for which proteins will be targeted for proteosomal degradation, where proteins attached to the Lys48-polyubiquitin chain is required to unfold before the polyubiquitin-proteosome interaction breaks [39].
5. Emerging areas and future directions
Several questions in the area of protein folding were posed in the introductory sections of this article. We hope the reader has obtained a feel for how single-molecule methods are contributing to answering several of these key questions. Continued progress in the field will however necessitate improvements in multiple areas including instrumentation and physical, chemical and in silico analyses tools, such as by use of microfluidics [63, 119–122], improved dyes and sample conditions, smaller cantilevers, enhanced imaging modes [123–125], improved measurements of biomolecular thermodynamics [126], multicolor FRET [127], and extension to shorter single-molecule distance rulers [128–130]. In addition, the theoretical understanding of single-molecule data can build upon excellent existing work in the area [45, 131–140]. This is important to permit improved means to uncover thermodynamic and time-dependent information from complex single-molecule data. While most single-molecule folding studies have been on relatively simple systems, the majority of the cellular proteome is composed of large, multi-domain proteins that have complex folding behavior and sometimes a significant degree of irreversibility [141]. Single-molecule measurements will continue to target these types of systems, affording substantial advantages in measuring details of the complexity and also minimizing the side-reaction of aggregation. In addition to probing systems with direct relevance to biology, single-molecule methods will also increasingly target protein systems encoding novel physics, such as downhill folders, knotted proteins and systems of interacting proteins. More complex protein systems will particularly benefit from the development of new chemical biology methods of sample preparation, for example for site-specific labeling or attachment [142]. In addition to folding, studies are also beginning to target misfolding and aggregation [55, 143, 144], the latter of which has been closely linked to disease but also has potential functional implications. Single-molecule methods should prove even more valuable for mechanistic studies of this extremely heterogeneous reaction. Another key point is that most folded proteins fold in the unique environment of the cell, presenting additional features such as macromolecular crowding and a huge number of possible binding partners. Understanding the influence of such parameters will also continue to be at the forefront by use of both in vitro and cell-based experiments. As the field matures further, the strengths of single-molecule analyses will also be increasingly combined with results of other experimental, computational and theoretical analyses to probe more complex systems with better resolution in many dimensions, and to directly relate folding and dynamics with function at various levels of organization.
Acknowledgements
We thank contributing coworkers and collaborators, and gratefully acknowledge the NSF (PHY0750049 to A.A.D.) and NIH (Grant GM066833 to A.A.D.) for support for the Deniz laboratory work reviewed in this article.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Allan Chris M. Ferreon, Email: aferreon@scripps.edu.
Ashok A. Deniz, Email: deniz@scripps.edu.
References
- 1.Frauenfelder H, Sligar SG, Wolynes PG. The energy landscapes and motions of proteins. Science. 1991;254:1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 2.Leopold PE, Montal M, Onuchic JN. Protein folding funnels: A kinetic approach to the sequence-structure relationship. Proc. Natl. Acad. Sci. U S A. 1992;89:8721–8725. doi: 10.1073/pnas.89.18.8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dill KA, Ozkan SB, Shell MS, Weikl TR. The protein folding problem. Annu. Rev. Biophys. 2008;37:289–316. doi: 10.1146/annurev.biophys.37.092707.153558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fersht AR. From the first protein structures to our current knowledge of protein folding: Delights and scepticisms. Nat. Rev. Mol. Cell Biol. 2008;9:650–654. doi: 10.1038/nrm2446. [DOI] [PubMed] [Google Scholar]
- 5.Lindorff-Larsen K, Rogen P, Paci E, Vendruscolo M, Dobson CM. Protein folding and the organization of the protein topology universe. Trends Biochem. Sci. 2005;30:13–19. doi: 10.1016/j.tibs.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 6.Thirumalai D, O'Brien EP, Morrison G, Hyeon C. Theoretical perspectives on protein folding. Annu. Rev. Biophys. 2010;39:159–183. doi: 10.1146/annurev-biophys-051309-103835. [DOI] [PubMed] [Google Scholar]
- 7.Bartlett AI, Radford SE. An expanding arsenal of experimental methods yields an explosion of insights into protein folding mechanisms. Nat. Struct. Mol. Biol. 2009;16:582–588. doi: 10.1038/nsmb.1592. [DOI] [PubMed] [Google Scholar]
- 8.Baldwin AJ, Kay LE. NMR spectroscopy brings invisible protein states into focus. Nat. Chem. Biol. 2009;5:808–814. doi: 10.1038/nchembio.238. [DOI] [PubMed] [Google Scholar]
- 9.Deniz AA, Mukhopadhyay S, Lemke EA. Single-molecule biophysics: At the interface of biology, physics and chemistry. J. R. Soc. Interface. 2008;5:15–45. doi: 10.1098/rsif.2007.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moerner WE. A dozen years of single-molecule spectroscopy in physics, chemistry, and biophysics. J. Phys. Chem. B. 2002;106:910–927. [Google Scholar]
- 11.Weiss S. Measuring conformational dynamics of biomolecules by single molecule fluorescence spectroscopy. Nat. Struct. Biol. 2000;7:724–729. doi: 10.1038/78941. [DOI] [PubMed] [Google Scholar]
- 12.Woodside MT, García-García C, Block SM. Folding and unfolding single RNA molecules under tension. Curr. Opin. Chem. Biol. 2008;12:640–646. doi: 10.1016/j.cbpa.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bustamante C. In singulo biochemistry: When less is more. Annu. Rev. Biochem. 2008;77:45–50. doi: 10.1146/annurev.biochem.012108.120952. [DOI] [PubMed] [Google Scholar]
- 14.Joo C, Balci H, Ishitsuka Y, Buranachai C, Ha T. Advances in single-molecule fluorescence methods for molecular biology. Annu. Rev. Biochem. 2008;77:51–76. doi: 10.1146/annurev.biochem.77.070606.101543. [DOI] [PubMed] [Google Scholar]
- 15.Kapanidis AN, Strick T. Biology, one molecule at a time. Trends Biochem. Sci. 2009;34:234–243. doi: 10.1016/j.tibs.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 16.Perkins TT. Optical traps for single molecule biophysics: A primer. Laser Photon. Rev. 2009;3:203–220. [Google Scholar]
- 17.Peters R. Single-molecule fluorescence analysis of cellular nanomachinery components. Annu. Rev. Biophys. Biomol. Struct. 2007;36:371–394. doi: 10.1146/annurev.biophys.36.040306.132715. [DOI] [PubMed] [Google Scholar]
- 18.Michalet X, Weiss S, Jager M. Single-molecule fluorescence studies of protein folding and conformational dynamics. Chem. Rev. 2006;106:1785–1813. doi: 10.1021/cr0404343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakon JJ, Weninger KR. Detecting the conformation of individual proteins in live cells. Nat. Methods. 2010;7:203–205. doi: 10.1038/nmeth.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kellermayer MS, Smith SB, Granzier HL, Bustamante C. Folding-unfolding transitions in single titin molecules characterized with laser tweezers. Science. 1997;276:1112–1116. doi: 10.1126/science.276.5315.1112. [DOI] [PubMed] [Google Scholar]
- 21.Tskhovrebova L, Trinick J, Sleep JA, Simmons RM. Elasticity and unfolding of single molecules of the giant muscle protein titin. Nature. 1997;387:308–312. doi: 10.1038/387308a0. [DOI] [PubMed] [Google Scholar]
- 22.Rief M, Gautel M, Oesterhelt F, Fernandez JM, Gaub HE. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 23.Deniz AA, Laurence TA, Beligere GS, Dahan M, Martin AB, Chemla DS, Dawson PE, Schultz PG, Weiss S. Single-molecule protein folding: Diffusion fluorescence resonance energy transfer studies of the denaturation of chymotrypsin inhibitor 2. Proc. Natl. Acad. Sci. U S A. 2000;97:5179–5184. doi: 10.1073/pnas.090104997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Talaga DS, Lau WL, Roder H, Tang J, Jia Y, DeGrado WF, Hochstrasser RM. Dynamics and folding of single two-stranded coiled-coil peptides studied by fluorescent energy transfer confocal microscopy. Proc. Natl. Acad. Sci. U S A. 2000;97:13021–13026. doi: 10.1073/pnas.97.24.13021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oesterhelt F, Oesterhelt D, Pfeiffer M, Engel A, Gaub HE, Muller DJ. Unfolding pathways of individual bacteriorhodopsins. Science. 2000;288:143–146. doi: 10.1126/science.288.5463.143. [DOI] [PubMed] [Google Scholar]
- 26.Ferreon AC, Moran CR, Gambin Y, Deniz AA. Single-molecule fluorescence studies of intrinsically disordered proteins. Methods Enzymol. 2010;472:179–204. doi: 10.1016/S0076-6879(10)72010-3. [DOI] [PubMed] [Google Scholar]
- 27.Borgia A, Williams PM, Clarke J. Single-molecule studies of protein folding. Annu. Rev. Biochem. 2008;77:101–125. doi: 10.1146/annurev.biochem.77.060706.093102. [DOI] [PubMed] [Google Scholar]
- 28.Schuler B. Single-molecule fluorescence spectroscopy of protein folding. Chemphyschem. 2005;6:1206–1220. doi: 10.1002/cphc.200400609. [DOI] [PubMed] [Google Scholar]
- 29.Haustein E, Schwille P. Single-molecule spectroscopic methods. Curr. Opin. Struct. Biol. 2004;14:531–540. doi: 10.1016/j.sbi.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 30.Michalet X, Kapanidis AN, Laurence T, Pinaud F, Doose S, Pflughoefft M, Weiss S. The power and prospects of fluorescence microscopies and spectroscopies. Annu. Rev. Biophys. Biomol. Struct. 2003;32:161–182. doi: 10.1146/annurev.biophys.32.110601.142525. [DOI] [PubMed] [Google Scholar]
- 31.Ha T. Single-molecule fluorescence resonance energy transfer. Methods. 2001;25:78–86. doi: 10.1006/meth.2001.1217. [DOI] [PubMed] [Google Scholar]
- 32.Deniz AA, Laurence TA, Dahan M, Chemla DS, Schultz PG, Weiss S. Ratiometric single-molecule studies of freely diffusing biomolecules. Annu. Rev. Phys. Chem. 2001;52:233–253. doi: 10.1146/annurev.physchem.52.1.233. [DOI] [PubMed] [Google Scholar]
- 33.Rhoades E, Gussakovsky E, Haran G. Watching proteins fold one molecule at a time. Proc. Natl. Acad. Sci. U S A. 2003;100:3197–3202. doi: 10.1073/pnas.2628068100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rhoades E, Cohen M, Schuler B, Haran G. Two-state folding observed in individual protein molecules. J. Am. Chem. Soc. 2004;126:14686–14687. doi: 10.1021/ja046209k. [DOI] [PubMed] [Google Scholar]
- 35.Chung HS, Louis JM, Eaton WA. Experimental determination of upper bound for transition path times in protein folding from single-molecule photon-by-photon trajectories. Proc. Natl. Acad. Sci. U S A. 2009;106:11837–11844. doi: 10.1073/pnas.0901178106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shank EA, Cecconi C, Dill JW, Marqusee S, Bustamante C. The folding cooperativity of a protein is controlled by its chain topology. Nature. 2010;465:637–640. doi: 10.1038/nature09021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schlierf M, Rief M. Surprising simplicity in the single-molecule folding mechanics of proteins. Angew. Chem. Int. Ed. Engl. 2009;48:820–822. doi: 10.1002/anie.200804723. [DOI] [PubMed] [Google Scholar]
- 38.Gebhardt JC, Bornschlogl T, Rief M. Full distance-resolved folding energy landscape of one single protein molecule. Proc. Natl. Acad. Sci. U S A. 2010;107:2013–2018. doi: 10.1073/pnas.0909854107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carrion-Vazquez M, Li H, Lu H, Marszalek PE, Oberhauser AF, Fernandez JM. The mechanical stability of ubiquitin is linkage dependent. Nat. Struct. Biol. 2003;10:738–743. doi: 10.1038/nsb965. [DOI] [PubMed] [Google Scholar]
- 40.Samori B, Zuccheri G, Baschieri R. Protein unfolding and refolding under force: Methodologies for nanomechanics. Chemphyschem. 2005;6:29–34. doi: 10.1002/cphc.200400343. [DOI] [PubMed] [Google Scholar]
- 41.Schlierf M, Berkemeier F, Rief M. Direct observation of active protein folding using lock-in force spectroscopy. Biophys. J. 2007;93:3989–3998. doi: 10.1529/biophysj.107.114397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galera-Prat A, Gomez-Sicilia A, Oberhauser AF, Cieplak M, Carrion-Vazquez M. Understanding biology by stretching proteins: Recent progress. Curr. Opin. Struct. Biol. 2010;20:63–69. doi: 10.1016/j.sbi.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Onuchic JN, Wang J, Wolynes PG. Analyzing single molecule trajectories on complex energy landscapes using replica correlation functions. Chem. Phys. 1999;247:175–184. [Google Scholar]
- 44.Best RB, Fowler SB, Herrera JL, Steward A, Paci E, Clarke J. Mechanical unfolding of a titin Ig domain: Structure of transition state revealed by combining atomic force microscopy, protein engineering and molecular dynamics simulations. J. Mol. Biol. 2003;330:867–877. doi: 10.1016/s0022-2836(03)00618-1. [DOI] [PubMed] [Google Scholar]
- 45.Lenz P, Cho SS, Wolynes PG. Analysis of single molecule folding studies with replica correlation functions. Chem. Phys. Lett. 2009;471:310–314. doi: 10.1016/j.cplett.2009.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ensign DL, Pande VS. Bayesian single-exponential kinetics in single-molecule experiments and simulations. J. Phys. Chem. B. 2009;113:12410–12423. doi: 10.1021/jp903107c. [DOI] [PubMed] [Google Scholar]
- 47.Guzman DL, Randall A, Baldi P, Guan Z. Computational and single-molecule force studies of a macro domain protein reveal a key molecular determinant for mechanical stability. Proc. Natl. Acad. Sci. U S A. 2010;107:1989–1994. doi: 10.1073/pnas.0905796107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mao AH, Crick SL, Vitalis A, Chicoine CL, Pappu RV. Net charge per residue modulates conformational ensembles of intrinsically disordered proteins. Proc. Natl. Acad. Sci. U S A. 2010;107:8183–8188. doi: 10.1073/pnas.0911107107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sotomayor M, Schulten K. Single-molecule experiments in vitro and in silico. Science. 2007;316:1144–1148. doi: 10.1126/science.1137591. [DOI] [PubMed] [Google Scholar]
- 50.Gambin Y, Schug A, Lemke EA, Lavinder JJ, Ferreon AC, Magliery TJ, Onuchic JN, Deniz AA. Direct single-molecule observation of a protein living in two opposed native structures. Proc. Natl. Acad. Sci. U S A. 2009;106:10153–10158. doi: 10.1073/pnas.0904461106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H, Oberhauser AF, Fowler SB, Clarke J, Fernandez JM. Atomic force microscopy reveals the mechanical design of a modular protein. Proc. Natl. Acad. Sci. U S A. 2000;97:6527–6531. doi: 10.1073/pnas.120048697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cecconi C, Shank EA, Bustamante C, Marqusee S. Direct observation of the three-state folding of a single protein molecule. Science. 2005;309:2057–2060. doi: 10.1126/science.1116702. [DOI] [PubMed] [Google Scholar]
- 53.Marszalek PE, Lu H, Li H, Carrion-Vazquez M, Oberhauser AF, Schulten K, Fernandez JM. Mechanical unfolding intermediates in titin modules. Nature. 1999;402:100–103. doi: 10.1038/47083. [DOI] [PubMed] [Google Scholar]
- 54.Orte A, Craggs TD, White SS, Jackson SE, Klenerman D. Evidence of an intermediate and parallel pathways in protein unfolding from single-molecule fluorescence. J. Am. Chem. Soc. 2008;130:7898–7907. doi: 10.1021/ja709973m. [DOI] [PubMed] [Google Scholar]
- 55.Ferreon AC, Gambin Y, Lemke EA, Deniz AA. Interplay of α-synuclein binding and conformational switching probed by single-molecule fluorescence. Proc. Natl. Acad. Sci. U S A. 2009;106:5645–5650. doi: 10.1073/pnas.0809232106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ferreon AC, Moran CR, Ferreon JC, Deniz AA. Alteration of the α-synuclein folding landscape by a mutation related to Parkinson's disease. Angew. Chem. Int. Ed. Engl. 2010;49:3469–3472. doi: 10.1002/anie.201000378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ferreon AC, Deniz AA. α-Synuclein multistate folding thermodynamics: Implications for protein misfolding and aggregation. Biochemistry. 2007;46:4499–4509. doi: 10.1021/bi602461y. [DOI] [PubMed] [Google Scholar]
- 58.Schuler B, Lipman EA, Eaton WA. Probing the free-energy surface for protein folding with single-molecule fluorescence spectroscopy. Nature. 2002;419:743–747. doi: 10.1038/nature01060. [DOI] [PubMed] [Google Scholar]
- 59.Doose S, Neuweiler H, Barsch H, Sauer M. Probing polyproline structure and dynamics by photoinduced electron transfer provides evidence for deviations from a regular polyproline type II helix. Proc. Natl. Acad. Sci. U S A. 2007;104:17400–17405. doi: 10.1073/pnas.0705605104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Best RB, Merchant KA, Gopich IV, Schuler B, Bax A, Eaton WA. Effect of flexibility and cis residues in single-molecule FRET studies of polyproline. Proc. Natl. Acad. Sci. U S A. 2007;104:18964–18969. doi: 10.1073/pnas.0709567104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mukhopadhyay S, Krishnan R, Lemke EA, Lindquist S, Deniz AA. A natively unfolded yeast prion monomer adopts an ensemble of collapsed and rapidly fluctuating structures. Proc. Natl. Acad. Sci. U S A. 2007;104:2649–2654. doi: 10.1073/pnas.0611503104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nettels D, Gopich IV, Hoffmann A, Schuler B. Ultrafast dynamics of protein collapse from single-molecule photon statistics. Proc. Natl. Acad. Sci. U S A. 2007;104:2655–2660. doi: 10.1073/pnas.0611093104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lipman EA, Schuler B, Bakajin O, Eaton WA. Single-molecule measurement of protein folding kinetics. Science. 2003;301:1233–1235. doi: 10.1126/science.1085399. [DOI] [PubMed] [Google Scholar]
- 64.Crooks GE. Entropy production fluctuation theorem and the nonequilibrium work relation for free energy differences. Phys. Rev. E. 1999;60:2721–2726. doi: 10.1103/physreve.60.2721. [DOI] [PubMed] [Google Scholar]
- 65.Collin D, Ritort F, Jarzynski C, Smith SB, Tinoco I, Jr, Bustamante C. Verification of the Crooks fluctuation theorem and recovery of RNA folding free energies. Nature. 2005;437:231–234. doi: 10.1038/nature04061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Junker JP, Ziegler F, Rief M. Ligand-dependent equilibrium fluctuations of single calmodulin molecules. Science. 2009;323:633–637. doi: 10.1126/science.1166191. [DOI] [PubMed] [Google Scholar]
- 67.Junker JP, Rief M. Single-molecule force spectroscopy distinguishes target binding modes of calmodulin. Proc. Natl. Acad. Sci. U S A. 2009;106:14361–14366. doi: 10.1073/pnas.0904654106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Merchant KA, Best RB, Louis JM, Gopich IV, Eaton WA. Characterizing the unfolded states of proteins using single-molecule FRET spectroscopy and molecular simulations. Proc. Natl. Acad. Sci. U S A. 2007;104:1528–1533. doi: 10.1073/pnas.0607097104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu P, Meng X, Qu P, Zhao XS, Wang CC. Subdomain-specific collapse of denatured staphylococcal nuclease revealed by single molecule fluorescence resonance energy transfer measurements. J. Phys. Chem. B. 2009;113:12030–12036. doi: 10.1021/jp809825x. [DOI] [PubMed] [Google Scholar]
- 70.Nettels D, Muller-Spath S, Kuster F, Hofmann H, Haenni D, Ruegger S, Reymond L, Hoffmann A, Kubelka J, Heinz B, Gast K, Best RB, Schuler B. Single-molecule spectroscopy of the temperature-induced collapse of unfolded proteins. Proc. Natl. Acad. Sci. U S A. 2009 doi: 10.1073/pnas.0900622106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pugh SD, Gell C, Smith DA, Radford SE, Brockwell DJ. Single-molecule studies of the Im7 folding landscape. J. Mol. Biol. 2010;398:132–145. doi: 10.1016/j.jmb.2010.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhuang X, Rief M. Single-molecule folding. Curr. Opin. Struct. Biol. 2003;13:88–97. doi: 10.1016/s0959-440x(03)00011-3. [DOI] [PubMed] [Google Scholar]
- 73.Li H, Linke WA, Oberhauser AF, Carrion-Vazquez M, Kerkvliet JG, Lu H, Marszalek PE, Fernandez JM. Reverse engineering of the giant muscle protein titin. Nature. 2002;418:998–1002. doi: 10.1038/nature00938. [DOI] [PubMed] [Google Scholar]
- 74.Oberhauser AF, Marszalek PE, Erickson HP, Fernandez JM. The molecular elasticity of the extracellular matrix protein tenascin. Nature. 1998;393:181–185. doi: 10.1038/30270. [DOI] [PubMed] [Google Scholar]
- 75.Garcia-Manyes S, Dougan L, Badilla CL, Brujic J, Fernandez JM. Direct observation of an ensemble of stable collapsed states in the mechanical folding of ubiquitin. Proc. Natl. Acad. Sci. U S A. 2009;106:10534–10539. doi: 10.1073/pnas.0901213106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dietz H, Rief M. Exploring the energy landscape of GFP by single-molecule mechanical experiments. Proc. Natl. Acad. Sci. U S A. 2004;101:16192–16197. doi: 10.1073/pnas.0404549101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schuler B, Eaton WA. Protein folding studied by single-molecule FRET. Curr. Opin. Struct. Biol. 2008;18:16–26. doi: 10.1016/j.sbi.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kubelka J, Hofrichter J, Eaton WA. The protein folding 'speed limit'. Curr. Opin. Struct. Biol. 2004;14:76–88. doi: 10.1016/j.sbi.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 79.Munoz V. Conformational dynamics and ensembles in protein folding. Annu. Rev. Biophys. Biomol. Struct. 2007;36:395–412. doi: 10.1146/annurev.biophys.36.040306.132608. [DOI] [PubMed] [Google Scholar]
- 80.Garcia-Mira MM, Sadqi M, Fischer N, Sanchez-Ruiz JM, Munoz V. Experimental identification of downhill protein folding. Science. 2002;298:2191–2195. doi: 10.1126/science.1077809. [DOI] [PubMed] [Google Scholar]
- 81.Sadqi M, Fushman D, Munoz V. Atom-by-atom analysis of global downhill protein folding. Nature. 2006;442:317–321. doi: 10.1038/nature04859. [DOI] [PubMed] [Google Scholar]
- 82.Huang F, Sato S, Sharpe TD, Ying L, Fersht AR. Distinguishing between cooperative and unimodal downhill protein folding. Proc. Natl. Acad. Sci. U S A. 2007;104:123–127. doi: 10.1073/pnas.0609717104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li P, Oliva FY, Naganathan AN, Munoz V. Dynamics of one-state downhill protein folding. Proc. Natl. Acad. Sci. U S A. 2009;106:103–108. doi: 10.1073/pnas.0802986106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huang F, Ying L, Fersht AR. Direct observation of barrier-limited folding of BBL by single-molecule fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. U S A. 2009;106:16239–16244. doi: 10.1073/pnas.0909126106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ziv G, Thirumalai D, Haran G. Collapse transition in proteins. Phys. Chem. Chem. Phys. 2009;11:83–93. doi: 10.1039/b813961j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McCarney ER, Werner JH, Bernstein SL, Ruczinski I, Makarov DE, Goodwin PM, Plaxco KW. Site-specific dimensions across a highly denatured protein; A single molecule study. J. Mol. Biol. 2005;352:672–682. doi: 10.1016/j.jmb.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 87.Xie H, Vucetic S, Iakoucheva LM, Oldfield CJ, Dunker AK, Uversky VN, Obradovic Z. Functional anthology of intrinsic disorder. 1. Biological processes and functions of proteins with long disordered regions. J. Proteome Res. 2007;6:1882–1898. doi: 10.1021/pr060392u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. J. Mol. Biol. 2002;323:573–584. doi: 10.1016/s0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- 89.Uversky VN, Oldfield CJ, Dunker AK. Intrinsically disordered proteins in human diseases: Introducing the D2 concept. Annu. Rev. Biophys. 2008;37:215–246. doi: 10.1146/annurev.biophys.37.032807.125924. [DOI] [PubMed] [Google Scholar]
- 90.Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Curr. Opin. Struct. Biol. 2002;12:54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
- 91.Tompa P. Intrinsically unstructured proteins. Trends Biochem. Sci. 2002;27:527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 92.Wright PE, Dyson HJ. Linking folding and binding. Curr. Opin. Struct. Biol. 2009;19:31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shoemaker BA, Portman JJ, Wolynes PG. Speeding molecular recognition by using the folding funnel: the fly-casting mechanism. Proc. Natl. Acad. Sci. U S A. 2000;97:8868–8873. doi: 10.1073/pnas.160259697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Muller-Spath S, Soranno A, Hirschfeld V, Hofmann H, Ruegger S, Reymond L, Nettels D, Schuler B. Charge interactions can dominate the dimensions of intrinsically disordered proteins. Proc. Natl. Acad. Sci. U S A. 2010;107:14609–14614. doi: 10.1073/pnas.1001743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Trexler AJ, Rhoades E. α-Synuclein binds large unilamellar vesicles as an extended helix. Biochemistry. 2009;48:2304–2306. doi: 10.1021/bi900114z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Veldhuis G, Segers-Nolten I, Ferlemann E, Subramaniam V. Single-molecule FRET reveals structural heterogeneity of SDS-bound α-synuclein. Chembiochem. 2009;10:436–439. doi: 10.1002/cbic.200800644. [DOI] [PubMed] [Google Scholar]
- 97.Sandal M, Valle F, Tessari I, Mammi S, Bergantino E, Musiani F, Brucale M, Bubacco L, Samori B. Conformational equilibria in monomeric α-synuclein at the single-molecule level. PLoS biology. 2008;6:e6. doi: 10.1371/journal.pbio.0060006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yu J, Malkova S, Lyubchenko YL. α-Synuclein misfolding: Single molecule AFM force spectroscopy study. J. Mol. Biol. 2008;384:992–1001. doi: 10.1016/j.jmb.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 99.Horwich AL, Fenton WA. Chaperonin-mediated protein folding: Using a central cavity to kinetically assist polypeptide chain folding. Q. Rev. Biophys. 2009;42:83–116. doi: 10.1017/S0033583509004764. [DOI] [PubMed] [Google Scholar]
- 100.Lin Z, Rye HS. GroEL-mediated protein folding: Making the impossible, possible. Crit. Rev. Biochem. Mol. Biol. 2006;41:211–239. doi: 10.1080/10409230600760382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hillger F, Hanni D, Nettels D, Geister S, Grandin M, Textor M, Schuler B. Probing protein-chaperone interactions with single-molecule fluorescence spectroscopy. Angew. Chem. Int. Ed. Engl. 2008;47:6184–6188. doi: 10.1002/anie.200800298. [DOI] [PubMed] [Google Scholar]
- 102.Sharma S, Chakraborty K, Muller BK, Astola N, Tang YC, Lamb DC, Hayer-Hartl M, Hartl FU. Monitoring protein conformation along the pathway of chaperonin-assisted folding. Cell. 2008;133:142–153. doi: 10.1016/j.cell.2008.01.048. [DOI] [PubMed] [Google Scholar]
- 103.Chakraborty K, Chatila M, Sinha J, Shi Q, Poschner BC, Sikor M, Jiang G, Lamb DC, Hartl FU, Hayer-Hartl M. Chaperonin-catalyzed rescue of kinetically trapped states in protein folding. Cell. 2010;142:112–122. doi: 10.1016/j.cell.2010.05.027. [DOI] [PubMed] [Google Scholar]
- 104.Hofmann H, Hillger F, Pfeil SH, Hoffmann A, Streich D, Haenni D, Nettels D, Lipman EA, Schuler B. Single-molecule spectroscopy of protein folding in a chaperonin cage. Proc. Natl. Acad. Sci. U S A. 2010;107:11793–11798. doi: 10.1073/pnas.1002356107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bechtluft P, van Leeuwen RG, Tyreman M, Tomkiewicz D, Nouwen N, Tepper HL, Driessen AJ, Tans SJ. Direct observation of chaperone-induced changes in a protein folding pathway. Science. 2007;318:1458–1461. doi: 10.1126/science.1144972. [DOI] [PubMed] [Google Scholar]
- 106.Yancey PH, Clark ME, Hand SC, Bowlus RD, Somero GN. Living with water stress: Evolution of osmolyte systems. Science. 1982;217:1214–1222. doi: 10.1126/science.7112124. [DOI] [PubMed] [Google Scholar]
- 107.Bolen DW, Rose GD. Structure and energetics of the hydrogen-bonded backbone in protein folding. Annu. Rev. Biochem. 2008;77:339–362. doi: 10.1146/annurev.biochem.77.061306.131357. [DOI] [PubMed] [Google Scholar]
- 108.Liu Y, Bolen DW. The peptide backbone plays a dominant role in protein stabilization by naturally occurring osmolytes. Biochemistry. 1995;34:12884–12891. doi: 10.1021/bi00039a051. [DOI] [PubMed] [Google Scholar]
- 109.Qu Y, Bolen CL, Bolen DW. Osmolyte-driven contraction of a random coil protein. Proc. Natl. Acad. Sci. U S A. 1998;95:9268–9273. doi: 10.1073/pnas.95.16.9268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Auton M, Bolen DW. Additive transfer free energies of the peptide backbone unit that are independent of the model compound and the choice of concentration scale. Biochemistry. 2004;43:1329–1342. doi: 10.1021/bi035908r. [DOI] [PubMed] [Google Scholar]
- 111.Auton M, Bolen DW. Predicting the energetics of osmolyte-induced protein folding/unfolding. Proc. Natl. Acad. Sci. U S A. 2005;102:15065–15068. doi: 10.1073/pnas.0507053102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Auton M, Ferreon AC, Bolen DW. Metrics that differentiate the origins of osmolyte effects on protein stability: A test of the surface tension proposal. J. Mol. Biol. 2006;361:983–992. doi: 10.1016/j.jmb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 113.Garcia-Manyes S, Dougan L, Fernandez JM. Osmolyte-induced separation of the mechanical folding phases of ubiquitin. Proc. Natl. Acad. Sci. U S A. 2009;106:10540–10545. doi: 10.1073/pnas.0902090106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cabrita LD, Dobson CM, Christodoulou J. Protein folding on the ribosome. Curr. Opin. Struct. Biol. 2010;20:33–45. doi: 10.1016/j.sbi.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 115.Katranidis A, Atta D, Schlesinger R, Nierhaus KH, Choli-Papadopoulou T, Gregor I, Gerrits M, Buldt G, Fitter J. Fast biosynthesis of GFP molecules: A single-molecule fluorescence study. Angew. Chem. Int. Ed. Engl. 2009;48:1758–1761. doi: 10.1002/anie.200806070. [DOI] [PubMed] [Google Scholar]
- 116.Williams PM, Fowler SB, Best RB, Toca-Herrera JL, Scott KA, Steward A, Clarke J. Hidden complexity in the mechanical properties of titin. Nature. 2003;422:446–449. doi: 10.1038/nature01517. [DOI] [PubMed] [Google Scholar]
- 117.Kruger M, Linke WA. Titin-based mechanical signalling in normal and failing myocardium. J. Mol. Cell Cardiol. 2009;46:490–498. doi: 10.1016/j.yjmcc.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 118.Lee G, Abdi K, Jiang Y, Michaely P, Bennett V, Marszalek PE. Nanospring behaviour of ankyrin repeats. Nature. 2006;440:246–249. doi: 10.1038/nature04437. [DOI] [PubMed] [Google Scholar]
- 119.Hamadani KM, Weiss S. Nonequilibrium single molecule protein folding in a coaxial mixer. Biophys. J. 2008;95:352–365. doi: 10.1529/biophysj.107.127431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pfeil SH, Wickersham CE, Hoffmann A, Lipman EA. A microfluidic mixing system for single-molecule measurements. Rev. Sci. Instrum. 2009;80:055105. doi: 10.1063/1.3125643. [DOI] [PubMed] [Google Scholar]
- 121.Lemke EA, Gambin Y, Vandelinder V, Brustad EM, Liu HW, Schultz PG, Groisman A, Deniz AA. Microfluidic device for single-molecule experiments with enhanced photostability. J. Am. Chem. Soc. 2009;131:13610–13612. doi: 10.1021/ja9027023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gambin Y, Simonnet C, VanDelinder V, Deniz A, Groisman A. Ultrafast microfluidic mixer with three-dimensional flow focusing for studies of biochemical kinetics. Lab Chip. 2010;10:598–609. doi: 10.1039/b914174j. [DOI] [PubMed] [Google Scholar]
- 123.Abbondanzieri EA, Greenleaf WJ, Shaevitz JW, Landick R, Block SM. Direct observation of base-pair stepping by RNA polymerase. Nature. 2005;438:460–465. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.King GM, Carter AR, Churnside AB, Eberle LS, Perkins TT. Ultrastable atomic force microscopy: Atomic-scale stability and registration in ambient conditions. Nano Lett. 2009;9:1451–1456. doi: 10.1021/nl803298q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kodera N, Yamamoto D, Ishikawa R, Ando T. Video imaging of walking myosin V by high-speed atomic force microscopy. Nature. 2010;468:72–76. doi: 10.1038/nature09450. [DOI] [PubMed] [Google Scholar]
- 126.Huguet JM, Bizarro CV, Forns N, Smith SB, Bustamante C, Ritort F. Single-molecule derivation of salt dependent base-pair free energies in DNA. Proc. Natl. Acad. Sci. U S A. 2010;107:15431–15436. doi: 10.1073/pnas.1001454107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gambin Y, Deniz AA. Multicolor single-molecule FRET to explore protein folding and binding. Mol. Biosyst. 2010;6:1540–1547. doi: 10.1039/c003024d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhu P, Clamme JP, Deniz AA. Fluorescence quenching by TEMPO: A sub-30 A single-molecule ruler. Biophys. J. 2005;89:L37–L39. doi: 10.1529/biophysj.105.071027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bates M, Blosser TR, Zhuang X. Short-range spectroscopic ruler based on a single-molecule optical switch. Phys. Rev. Lett. 2005;94:108101. doi: 10.1103/PhysRevLett.94.108101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Doose S, Neuweiler H, Sauer M. Fluorescence quenching by photoinduced electron transfer: A reporter for conformational dynamics of macromolecules. Chemphyschem. 2009;10:1389–1398. doi: 10.1002/cphc.200900238. [DOI] [PubMed] [Google Scholar]
- 131.Dudko OK, Hummer G, Szabo A. Theory, analysis, and interpretation of single-molecule force spectroscopy experiments. Proc. Natl. Acad. Sci. U S A. 2008;105:15755–15760. doi: 10.1073/pnas.0806085105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Talaga DS. Information-theoretical analysis of time-correlated single-photon counting measurements of single molecules. J. Phys. Chem. A. 2009;113:5251–5263. doi: 10.1021/jp8082908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gopich IV, Szabo A. Decoding the pattern of photon colors in single-molecule FRET. J. Phys. Chem. B. 2009;113:10965–10973. doi: 10.1021/jp903671p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Liu Y, Park J, Dahmen KA, Chemla YR, Ha T. A comparative study of multivariate and univariate hidden Markov modelings in time-binned single-molecule FRET data analysis. J. Phys. Chem. B. 2010;114:5386–5403. doi: 10.1021/jp9057669. [DOI] [PubMed] [Google Scholar]
- 135.Suzuki Y, Dudko OK. Single-molecule rupture dynamics on multidimensional landscapes. Phys. Rev. Lett. 2010;104:048101. doi: 10.1103/PhysRevLett.104.048101. [DOI] [PubMed] [Google Scholar]
- 136.Gopich IV, Szabo A. Single-molecule FRET with diffusion and conformational dynamics. J. Phys. Chem. B. 2007;111:12925–12932. doi: 10.1021/jp075255e. [DOI] [PubMed] [Google Scholar]
- 137.Hummer G, Szabo A. Free energy profiles from single-molecule pulling experiments. Proc. Natl. Acad. Sci. U S A. 2010;107:21441–21446. doi: 10.1073/pnas.1015661107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kalinin S, Valeri A, Antonik M, Felekyan S, Seidel CAM. Detection of structural dynamics by FRET: A photon distribution and fluorescence lifetime analysis of systems with multiple states. J. Phys. Chem. B. 2010;114:7983–7995. doi: 10.1021/jp102156t. [DOI] [PubMed] [Google Scholar]
- 139.Nir E, Michalet X, Hamadani KM, Laurence TA, Neuhauser D, Kovchegov Y, Weiss S. Shot-noise limited single-molecule FRET histograms: Comparison between theory and experiments. J. Phys. Chem. B. 2006;110:22103–22124. doi: 10.1021/jp063483n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Palo K, Mets Ü, Loorits V, Kask P. Calculation of photon-count number distributions via master equations. Biophys. J. 2006;90:2179–2191. doi: 10.1529/biophysj.105.066084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Fitter J. The perspectives of studying multi-domain protein folding. Cell Mol. Life Sci. 2009;66:1672–1681. doi: 10.1007/s00018-009-8771-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Brustad EM, Lemke EA, Schultz PG, Deniz AA. A general and efficient method for the site-specific dual-labeling of proteins for single molecule fluorescence resonance energy transfer. J. Am. Chem. Soc. 2008;130:17664–17665. doi: 10.1021/ja807430h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hillger F, Nettels D, Dorsch S, Schuler B. Detection and analysis of protein aggregation with confocal single molecule fluorescence spectroscopy. J. Fluoresc. 2007;17:759–765. doi: 10.1007/s10895-007-0187-z. [DOI] [PubMed] [Google Scholar]
- 144.Orte A, Birkett NR, Clarke RW, Devlin GL, Dobson CM, Klenerman D. Direct characterization of amyloidogenic oligomers by single-molecule fluorescence. Proc. Natl. Acad. Sci. U S A. 2008;105:14424–14429. doi: 10.1073/pnas.0803086105. [DOI] [PMC free article] [PubMed] [Google Scholar]