Abstract
Virulence factors generally enhance a pathogen's fitness and thereby foster transmission. However, most studies of pathogen fitness have been performed by averaging the phenotypes over large populations. Here, we have analyzed the fitness costs of virulence factor expression by Salmonella enterica subspecies I serovar Typhimurium in simple culture experiments. The type III secretion system ttss-1, a cardinal virulence factor for eliciting Salmonella diarrhea, is expressed by just a fraction of the S. Typhimurium population, yielding a mixture of cells that either express ttss-1 (TTSS-1+ phenotype) or not (TTSS-1− phenotype). Here, we studied in vitro the TTSS-1+ phenotype at the single cell level using fluorescent protein reporters. The regulator hilA controlled the fraction of TTSS-1+ individuals and their ttss-1 expression level. Strikingly, cells of the TTSS-1+ phenotype grew slower than cells of the TTSS-1− phenotype. The growth retardation was at least partially attributable to the expression of TTSS-1 effector and/or translocon proteins. In spite of this growth penalty, the TTSS-1+ subpopulation increased from <10% to approx. 60% during the late logarithmic growth phase of an LB batch culture. This was attributable to an increasing initiation rate of ttss-1 expression, in response to environmental cues accumulating during this growth phase, as shown by experimental data and mathematical modeling. Finally, hilA and hilD mutants, which form only fast-growing TTSS-1− cells, outcompeted wild type S. Typhimurium in mixed cultures. Our data demonstrated that virulence factor expression imposes a growth penalty in a non-host environment. This raises important questions about compensating mechanisms during host infection which ensure successful propagation of the genotype.
Author Summary
Pathogenic bacteria require virulence factors to foster growth and survival of the pathogen within the host. Therefore, virulence factor expression is generally assumed to enhance the pathogen's fitness. However, most studies of pathogen fitness have been performed by averaging the phenotypes over large pathogen populations. Here, we have analyzed for the first time the fitness costs of virulence factor expression in a simple in vitro culture experiment using the diarrheal pathogen Salmonella enterica subspecies I serovar Typhimurium (S. Typhimurium). TTSS-1, the cardinal virulence factor for eliciting Salmonella diarrhea, is expressed by just a fraction of the clonal S. Typhimurium population. Surprisingly, time lapse fluorescence microscopy revealed that ttss-1-expressing S. Typhimurium cells grew at a reduced rate. Thus, the pathogen has to “pay” a significant “price” for expressing this virulence factor. This raises important questions about compensating mechanisms (e.g. benefits reaped through TTSS-1 driven host-interactions) ensuring successful propagation of the genotype.
Introduction
The ability to infect a host and elicit disease is dictated by the virulence factors expressed by a given pathogen. This may include, but is not limited to, protective factors neutralizing antibacterial defenses, enzymes involved in nutrient acquisition within the host, regulators of virulence factor expression and toxins or secretion systems for subverting host cell signal transduction. The coordinated expression of such virulence factors enhances colonization, growth/survival within the host and transmission. However, most studies of virulence factor function and pathogen fitness have been performed in bulk assays, averaging the phenotypes over large pathogen populations of genetically identical cells. In contrast, little is known about the potential advantages, costs or burdens arising from virulence factor expression by an individual cell of the pathogen population. Therefore, single cell analyses might be of significant interest, in particular if virulence factors, which are expressed in a bistable fashion by some but not all members of a pathogen population, e.g. the ttss-1 system of S. Typhimurium [1], [2], [3], [4], [5], as described in this paper.
Bistable gene expression is genetically encoded. In most cases, one particular genotype expresses one predictable phenotype in a given environment. However, in some cases, two different phenotypes are expressed by isogenic organisms living in the same environment. This is termed phenotypic variation, bimodal gene expression or bistability and represents a special case of gene expression [6]. The importance of bistability for pathogenic bacterial fitness and evolution is just beginning to be understood.
Like other cases of gene expression, bistability is generally observed in response to particular environmental cues. The response is driven by a dedicated (set of) regulator(s), which responds to environmental signals (operon model of Jacob [7]). This response is subject to stochastic fluctuations. In particular in the case of regulators expressed in a few copies per cell, this can significantly affect the active regulator concentration thus randomizing the corresponding phenotype in a population [8], [9]. In combination with non-linear responses (e.g. regulator multimerization, feedback loops), this can lead to formation of phenotypically distinct and stable subpopulations of isogenic bacteria [6], [8], [9], [10], [11]. In terms of evolution, two models may explain the advantage of bistability: i. in “bet hedging”, the optimally adapted phenotype will prevail and ensure the survival of the shared genotype in a changing environment [12]. ii. in “division of labor”, both phenotypes cooperate to ensure survival of the shared genotype [4]. In either way, the bistable expression of certain genes is thought to promote the survival of the genotype. However, it has remained poorly understood whether/how bistability may affect the lifestyle of pathogenic bacteria.
Salmonella enterica subspecies 1 serovar Typhimurium (S. Tm) is a pathogenic Gram-negative bacterium causing numerous cases of diarrhea, worldwide. Its' type III secretion system 1 (TTSS-1) was recently identified as an example for bistable gene expression [1], [3], [5], [13]. TTSS-1 is a well-known virulence determinant of S. Tm required for eliciting diarrheal disease [14], [15], [16]. The needle like TTSS-1 apparatus injects effector proteins into host epithelial cells, thus triggering host cell invasion and pro-inflammatory responses [17], [18], [19]. TTSS-1 is encoded on a genomic island (Salmonella pathogenicity island 1 (SPI-1)), which also harbors genes for effector proteins and for several regulators of ttss-1 expression, e.g. hilA, hilC and hilD [20], [21].
The bistable ttss-1 expression is controlled by a complex regulatory network, which includes coupled positive feedback loops, controls the threshold for ttss-1 induction and amplifies ttss-1 expression [5], [22]. Bistable ttss-1 expression is observed in “ttss-1 inducing” environments, i.e. the gut lumen of infected mice or in non-host environments, e.g. when S. Tm is grown to late logarithmic phase in LB [1], [2], [4], [5]. This yields mixed populations of isogenic S. Tm cells that express ttss-1 (TTSS-1+ phenotype), or do not (TTSS-1− phenotype), in a bimodal fashion. In the mouse gut, only the TTSS-1+ cells can actively invade the mucosal tissue and efficiently trigger inflammation [4], [18]. This inflammatory response may help to overcome the commensal microflora, thus enhancing Salmonella growth and transmission [23], [24], [25], [26], [27], [28], [29]. Experimental data indicate that bistable ttss-1 expression might represent an example of “division of labor” [4], but further data is required to settle this point. At any rate, ttss-1 expression seems to be instrumental for eliciting diarrheal disease and enhancing pathogen transmission. But the functional properties of the TTSS-1+ phenotype are not well understood.
The complex setting of the infected animal gut has hampered the analysis of the TTSS-1+ phenotype. In vitro experiments are essential for gaining detailed mechanistic insights. Here, we have analyzed the induction of ttss-1 expression and its effects on the growth rate of the TTSS-1+ phenotype by single cell reporter assays, competitive growth experiments and mathematical modeling. In such non-host environments, expression of the ttss-1 virulence system expression imposed a growth penalty on the TTSS-1+ cells. This may have important implications with respect to compensatory mechanisms during the infection of animal hosts.
Results
Single cell reporters for studying the TTSS-1+ phenotype
We started our analysis of the TTSS-1+ phenotype by probing ttss-1 expression at the single cell level. For this purpose, we chose the sicA promoter (PsicA), which controls expression of the chromosomal sicAsipBCDA operon (Fig. S1C). This operon encodes key parts of the TTSS-1 virulence system. On the one hand, we employed a transcriptional sipA-tsrvenus reporter gene cassette placing the reporter downstream of the sicAsipBCDA operon (Fig. S1; [2], [3]). Due to its localization at the bacterial poles, the tsrvenus reporter allows detecting <10 proteins per cell [30]. Thus, sipA-tsrvenus provides a highly sensitive reporter for the TTSS-1+ phenotype.
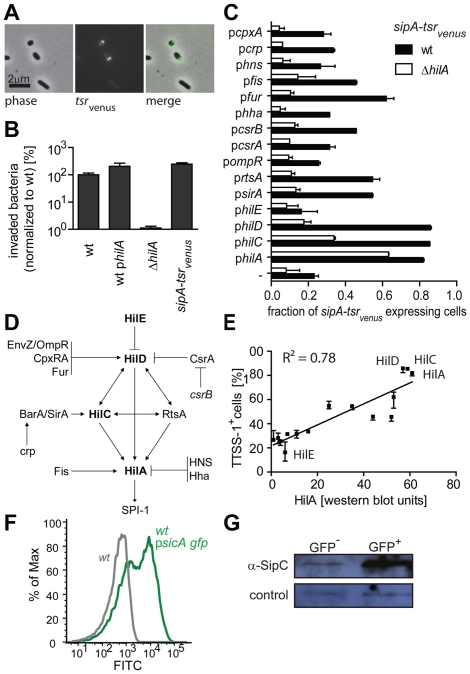
Next, we verified the performance of the sipA-tsrvenus reporter. sipA-tsrvenus expression was bistable and TTSS-1− and TTSS-1+ individuals were distinguishable by the presence/absence of Tsrvenus spots at the bacterial poles ([30]; Fig. 1A; Fig. S1D). TTSS-1 expression and virulence were not compromised (Fig. 1B). The accurate response of sipA-tsrvenus to Salmonella signaling cascades was established by disturbing known elements of the TTSS-1 gene regulation network and FACS analysis of sipA-tsrvenus expression (Fig. 1C, D). In line with the published work on ttss-1 regulation (Fig. 1D): i. Over-expression of positive TTSS-1 regulators increased the abundance of tsrvenus-expressing individuals (Fig. 1C; Fig. S1D). In particular, hilA, hilC and hilD over-expression increased the fraction of sipA-tsrvenus expressing individuals from ∼20% to 80–100%. ii. The median signal intensity per sipA-tsrvenus expressing cell increased when positive regulators were over-expressed (philA: 3.8±0.3-fold; philC: 4.0±0.1-fold; philD: 4±0.1-fold; median ± s.d.). iii. Control experiments in a ΔhilA mutant verified that expression of the TTSS-1+ phenotype depended on the ttss-1 master-regulator, HilA (Fig. 1C; open bars) and iv. The average HilA protein levels of the analyzed strains correlated positively with the fraction of tsrvenus-expressing individuals (r2 = 0.78; quantitative Western blot; Fig. 1E). These data verified the accurate performance of the sipA-tsrvenus reporter and demonstrated that hilA-dependent regulation affects both, the fraction of TTSS-1+ individuals and the level of ttss-1 expression per cell.
Figure 1. sipA-tsrvenus as a single cell reporter for ttss-1 expression.
A) Bistable expression of sipA-tsrvenus in wt S. Tm (M2001). Living bacteria (4 h in LB) were imaged by fluorescence- and phase contrast microscopy. Bar, 2 µm; B) Invasion into MDCK cells (3 indep. experiments; ±s.d.; Materials and Methods). C) Response of the sipA-tsrvenus reporter to over-expression of known ttss-1 regulators. Wt S. Tm (sipA-tsrvenus; M2001; black bars) or ΔhilA (sipA-tsrvenus; M2018; open bars) harboring the indicated regulator-expression plasmids (Table S2) were cultured for 4 h in LB and FACS-analyzed (triplicates ±s.d.). D) ttss-1 regulation cascade depicting the regulators analyzed in C) and E; adapted from [35], [36], [59], [60], [61], [62]). E) Correlation between HilA protein levels and the fraction of ttss-1 expressing individuals. The fraction of cells with the TTSS-1+ phenotype (from C) was plotted against the average HilA expression (average of ≥3 independent quantitative Western blots per regulator and strain). F) Bistable expression of psicA-gfp in S. Tm SL1344 determined and separated by FACS; G) Western blot analysis of TTSS-1- and TTSS-1+ subpopulations from F) using a polyclonal rabbit α-SipC antibody.
In addition, we employed psicA-gfp, a reporter plasmid expressing gfp under control of the sicA promoter. This construct yielded brighter fluorescence than the chromosomal sipA-tsrvenus and was better suited for FACS analysis. Again, this reporter yielded a bistable expression pattern (Fig. 1F). Using wt S. Tm psicA-gfp we separated TTSS-1+ and TTSS-1- subpopulations by FACS. Western blot analysis of the FACS-sorted subpopulations verified coincident expression of psicA-gfp and the TTSS-1 protein SipC (Fig. 1F, G). This indicated that our fluorescent reporter constructs are faithful reporters of the bistable expression of the TTSS-1+ phenotype.
Time-lapse microscopy reveals retarded growth of TTSS-1+ individuals
During our experiments, we observed that hilA, hilC and hilD over-expression led to reduced culture densities (e.g. OD600 for wt sipA-tsrvenus: 3.4±0.3 vs. wt sipA-tsrvenus philA: 2.0±0.3; mean ± s.d.). This was a first hint suggesting that retarded growth might be a general feature of the TTSS-1+ phenotype. However, it remained to be shown whether growth retardation occurs in wild type cells expressing normal levels of hilA, hilC and hilD.
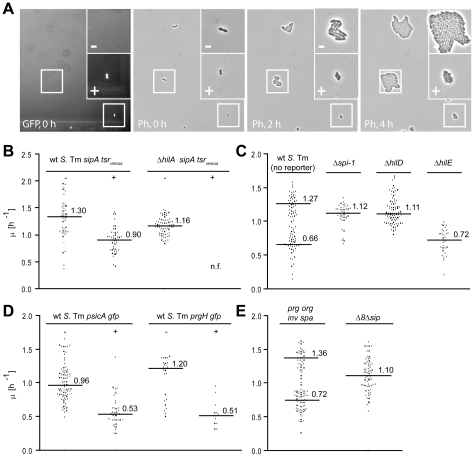
The growth rate of the TTSS-1+ individuals was analyzed by time-lapse microscopy. Wild type S. Tm harboring gfp- or tsrvenus-reporters for ttss-1 expression were placed on an agar pad (LB, 1.5% agarose), the TTSS-1+ individuals were identified by fluorescence microscopy and growth was analyzed by time-lapse phase contrast microscopy (1 frame/30 min; Fig. 2A). Imaging did not impose detectable photo damage to the bacteria, as indicated by the unaltered growth rate (Fig. S2). Strikingly, TTSS-1+ individuals grew slower than TTSS-1− individuals (wt S. Tm sipA-tsrvenus (M2001); µT1+ = 0.90 h−1 vs. µT1 − = 1.30 h−1; p = 0.027 for the factor ‘phenotype’ in a two-way ANOVA; Fig. 2B). The negative control strain ΔhilA sipA-tsrvenus yielded only TTSS-1− individuals, which grew at the “fast” rate (µT1 − = 1.16 h−1; Fig. 2B). Thus, TTSS-1+ individuals seemed to grow at a reduced rate.
Figure 2. Time-lapse microscopy reveals retarded growth of TTSS-1+ individuals.
Bacteria (4 h LB subculture, OD600 = 1), were placed on an agar pad (37°C) and imaged to detect ttss-1 expression (fluorescence) and growth (phase contrast; 1 frame/30 min). A) Sample images from a typical time-lapse microscopy experiment with wt S. Tm (SL1344, psicA-gfp). B)-D): Time-lapse microscopy experiments with wt S. Tm (M2001; sipA-tsrvenus) and an isogenic hilA mutant (M2018; sipA-tsrvenus; B); wt S. Tm (SL1344; no reporter) and mutants lacking ttss-1, hilD or hilE (no reporter); C); wt S. Tm (SL1344; psicA-gfp and an isogenic wt reporter strain (SL1344 prgH-gfp; D); mutants lacking most genes encoding the TTS apparatus (prg-org, inv-spa) or most effector proteins and the translocon (Δ8Δsip); E). Each data point represents the growth rate of an individual micro colony. Data were from ≥3 independent experiments. Black line, median; Numbers, median growth rates.
To exclude potential artifacts attributable to the sipA-tsrvenus reporter, we analyzed unmodified wild type S. Tm not harboring any reporter (Fig. 2C; Fig. S2). Using a maximum likelihood approach, we identified two populations with distinct growth rates (likelihood ratio test for two populations versus one population, p<0.001, µslow = 0.66 h−1 vs. µfast = 1.27 h−1; Fig. 2C), very similar to the ones described above (Fig. 2B). Furthermore, unmarked mutants lacking the entire SPI-1 region (Δspi-1) or the positive ttss-1 regulator hilD yielded exclusively fast growing cells, while deletion of the negative ttss-1 regulator hilE yielded only slow growing cells (Fig. 2C). Finally, wild type S. Tm harboring psicA-gfp or a chromosomal gfp-reporter for the TTSS-1 gene prgH [1] yielded slow growing TTSS-1+ and fast growing TTSS-1− cells (µT1+ = 0.51 h−1 vs. µT1 − = 1.2 h−1; p = 0.006 for the factor ‘phenotype’ in a two-way ANOVA; Fig. 2D). Bacteria expressing the psicA-gfp or prgH-gfp reporters grew even slower than the TTSS-1− sipA-tsrvenus bacteria or the slow-growing wt S. Tm subpopulation (Fig. 2BC). Presumably, this was attributable to the additional “burden” conferred by the GFP expression, as described, before [31].
Thus, the time-lapse microscopy experiments verified bistable ttss-1 expression and revealed that the TTSS-1− phenotype has a reduced growth rate, even at wild type HilA and TTSS-1 levels (µT1+ in the range of 0.7 h−1 vs. µT1 − in the range of 1.3 h−1). This was confirmed in a dye dilution assay (Fig. S3).
Our data suggested that ttss-1 expression represents a “cost” to the bacterial cell. However the mechanism explaining this growth retardation had remained unclear. We speculated that expression of the TTS apparatus itself or the sheer load of the proteins transported by the TTSS-1 (effectors, translocon proteins) might play a role. To test these hypotheses, we analyzed two additional S. Tm mutants. In the first mutant, termed Δprg-orgΔinv-spa, we deleted most apparatus-encoding genes (Table S1). This mutant formed two populations with distinct growth rates (likelihood ratio test for two populations versus one population, p<0.001, µslow = 0.72 h−1 vs. µfast = 1.36 h−1; Fig. 2E), very similar to those described for wild type S. Tm (Fig. 2C). The second mutant, termed Δ8Δsip, was lacking the genes for most TTSS-1 effector proteins and the secreted translocon components including sipB, sipC, sipD, sipA, sptP, sopE, sopE2, sopB and sopA (Tab. S1). In contrast to wild type S. Tm, we could not distinguish two subpopulations in this mutant (likelihood ratio test for two populations versus one population, p = 0.73; Fig. 2E). Instead, this mutant displayed a median growth rate of µ = 1.10 h−1, similar to the fast growing subpopulation of S. Tm wt and the mutants Δspi-1 and hilD (Fig. 2C). This data suggests, that expression of the effector proteins and translocon components is “costly” and provides at least in part a mechanistic explanation for the growth retardation of wild type S. Tm cells of the TTSS-1+ phenotype.
Retarded growth and ttss-1 induction determine the fraction of TTSS-1+ individuals: a mathematical analysis
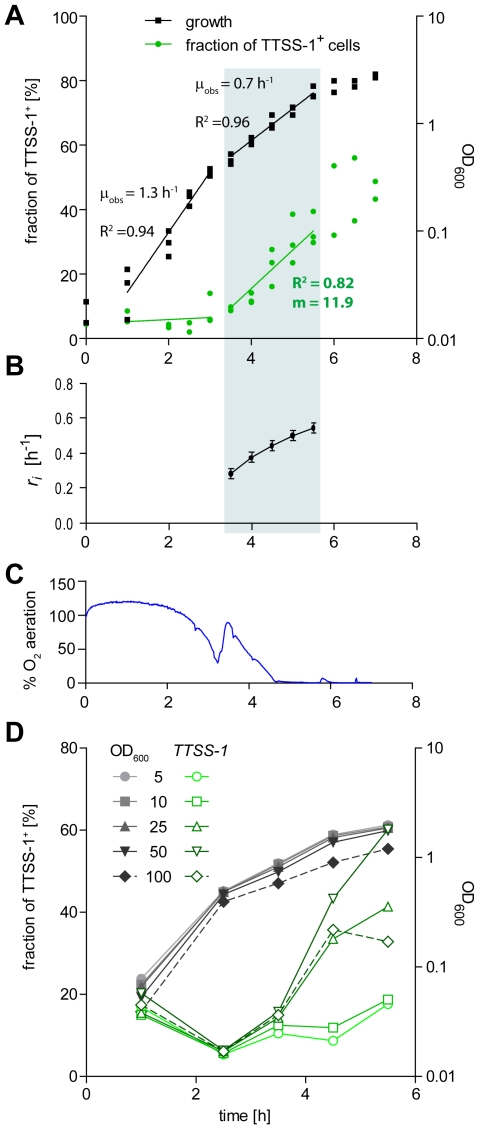
When monitoring growth and bistable ttss-1 expression in a wt S. Tm (psicA-gfp) culture, the fraction of TTSS-1+ individuals began to rise after 2.5 h as soon as the culture entered the late logarithmic phase, increased in a linear fashion, and reached approx. 60% after 7 h once the culture entered the stationary phase (Fig. 3A).
Figure 3. Time course experiment analyzing the initiation of ttss-1 expression.
A) Wt S. Tm (SL1344, psicA-gfp) was sub-cultured under mild aeration in LB. Growth (OD600, black) and ttss-1 expression (FACS, green) was analyzed and fitted separately for early and late log phase. Gray: late logarithmic phase. m: apparent initiation rate of ttss-1 expression, as determined from the slope of the fitted line. B) Calculation of the mean value of ri(t) during the late log phase using eq. 4, data from A) and 86 individual µT1 − values for S. Tm psicA gfp (from Fig. 2D); error bars depict the SEM. C) pO2 during the experiment. D) Growth (OD600, black) and ttss-1 expression (FACS, green) in 250 ml flasks (shaken 160 rpm, 37°C) harboring the indicated volume of LB (inoculation: 1/100 from a 12 h S. Tm psicA-gfp culture).
Our results implied that two different parameters affect the fraction of TTSS-1+ individuals and the overall growth progression in the late logarithmic phase: i. Competitive growth. TTSS-1+ individuals are steadily outgrown by the fast-growing TTSS-1− individuals (µT1+<µT1 −; Fig. 2); this constantly reduces the size of the TTSS-1+ subpopulation. ii. ttss-1 induction. Presumably, initiation of ttss-1 expression in TTSS-1− individuals compensates the “TTSS-1+ losses” attributable to competitive growth and explains the increasing fractions of TTSS-1+ individuals during the late logarithmic phase.
To infer the dynamic initiation rate ri of ttss-1 expression in the late logarithmic phase from our experimental data, we devised a mathematical model describing the growth of the TTSS-1+ (NT1+; growth rate µT1+) and the TTSS-1− population (NT1 −; growth rate µT1 −) as a function of time (t):
| (1) |
| (2) |
It should be noted that the model does not include a term for “switching off” ttss-1 expression. This was justified by our failure to observe “off switching” in the experiments shown in Fig. 2 and further supported by other data (Fig. S2 and data shown below). During the late logarithmic phase, the relative abundance of the TTSS-1+ individuals increased, and the fraction α of TTSS-1− individuals (NT1-) decreased in a linear fashion (Fig. 3A):
| (3) |
Equation (2) can be rearranged to calculate ri(t) (see Text S1 for details):
| (4) |
With the data from Fig. 3A and by using equation (3) we could determine NT1 − (t) and, after fitting an empirical function to NT1 − (t), also dNT1 −/dt. Using equation (4), this allowed calculating ri(t) during the late logarithmic phase (see Text S1 for details). We found that the mean initiation rate (ri) of ttss-1 expression increased continuously during the late logarithmic phase, e.g. from 0.28 h−1 at 3.5 h to 0.54 h−1 at 5.5 h (SEM = 0.03 h−1; Fig. 3B).
Environmental signals affecting ttss-1 expression in the late logarithmic phase
The initiation rate of ttss-1 expression seemed to increase upon entry into the late logarithmic growth phase (Fig. 3A). Therefore, it might be induced by growth-related environmental signals (e.g. oxygen depletion, quorum signals, nutrient depletion, metabolite accumulation). To address this, we analyzed the partial oxygen pressure (pO2) during growth. As expected, pO2 declined to <30% relative aeration during the first three hours (Fig. 3C). After approximately 3.5 h, we detected a transient rebound of the oxygen pressure followed by a steady decline to <3% relative aeration during the next hour. This undulation of oxygen pressure is indicative of a change in the growth physiology at 3.5 h and was in line with the reduced growth rate (Fig. 3A, shaded area).
The data suggested that altered metabolism, nutrient availability, waste product accumulation, the reduced growth rate or the low oxygen pressure might represent cues inducing ttss-1 expression. As a first approach to test the role of pO2, we performed batch culture growth experiments in identical 250 ml culture flasks filled with the indicated volumes of media (wt S. Tm psicA gfp grown in 5, 10, 25, 50 or 100 ml LB; Fig. 3D). This setup allowed analyzing the effect of reduced pO2 (i.e. in larger, poorly aerated culture volumes) at equivalent growth rates. We observed that the fraction of ttss-1 expressing cells increased in larger culture volumes. Therefore, low oxygen tension might represent one environmental cue directly or indirectly inducing bistable ttss-1 expression. However, the evidence is merely circumstantial at this moment and other cues might well be involved. Identification of these cues will benefit from the strategies for determining ri as described above.
Time lapse microscopy detects the emergence and the reduced growth rate of TTSS-1+ cells
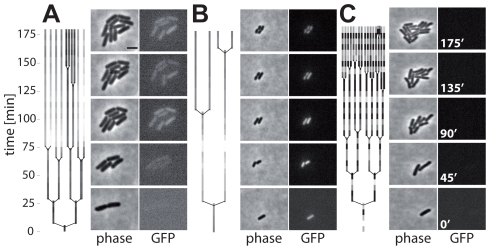
In liquid culture, the initiation of ttss-1 expression occurred in the late logarithmic phase. However, our initial time lapse microscopy data for bacteria sampled from this growth phase did not show initiation of ttss-1 expression (Fig. 2). We reasoned that this might be attributable to the lack of inducing environmental signals, as these experiments had been performed on agar pads soaked with fresh LB medium. To test this hypothesis, we modified the time lapse microscopy experiment and imaged bacteria (S. Tm psicA-gfp) placed on agar pads soaked with filter-sterilized spent medium taken from a culture at the same growth phase (OD600 = 0.9, see Materials and Methods). We analyzed growth of 191 micro colonies. At the beginning, 135 did not express ttss-1. But remarkably, we observed 15 of 135 initially TTSS-1− micro colonies, in which individual bacteria induced ttss-1 expression during the course of our imaging experiment (e.g. Fig. 4A, Fig. S4; Video S1). After induction, the TTSS-1+ cells grew at a slower rate than their TTSS-1− siblings. In addition, we observed numerous TTSS-1+ bacteria (56 micro colonies) and TTSS-1− bacteria (120 micro colonies) which did not “switch” their ttss-1 expression status. In line with the results above, ttss-1 expression and the interval between two cell divisions was negatively correlated (Fig. 4A,B,C, Spearman's rho = −0.747, p<0.0001, N = 29).
Figure 4. Time-lapse microscopy shows onset of ttss-1 expression and concomitant growth retardation.
Lineage trees with corresponding phase contrast and GFP images of S. Tm (M556; psicA-gfp) grown on agar pads with spent LB. Coloring of the lineage trees reflects the relative mean GFP intensity of individual cells (dark = low; light = high; scaled to the highest fluorescence in tree). A) On-switching of ttss-1 expression in a fraction of the micro colony. B) Micro colony uniformly expressing ttss-1 throughout the assay. C) Micro colony not expressing ttss-1 throughout the assay. Scale bar, 2 µm; see also Fig. S4.
These experiments support the stochastic initiation of ttss-1 expression. But the initiation rate of ttss-1 expression (<0.04 h−1) was lower than that predicted from the batch culture experiment shown in Fig. 3 (ri = 0.18−0.45 h−1). This might be attributable to the lack of some environmental cue, e.g. low oxygen pressure, as time lapse microscopy was performed at ambient atmosphere. Only two micro colonies showed a decrease in fluorescence as expected for “off-switching”. Hence, the rate of off-switching is not substantial. This indicated that our mathematical model, which assumed that “switching off” the ttss-1 expression would be negligible, was justified (equation (1) did not include ri(t)NT1-(t)). These experiments verified that ttss-1 expression is initiated in a stochastic fashion under “inducing” environmental conditions and that the TTSS-1+ phenotype exhibits a growth defect.
Handicap of wt S. Tm in a competitive growth experiment
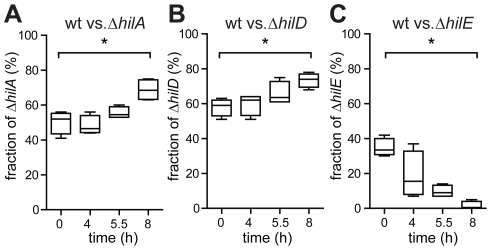
Finally, we confirmed the growth penalty attributable to ttss-1 expression in the late logarithmic phase in competition experiments. Wt S. Tm expresses ttss-1 in a bistable fashion and forms a significant fraction of slow-growing TTSS-1+ cells during the late logarithmic phase (Fig. 3). This slows down the apparent growth of the total wild type population (see above). In contrast, hilA or hilD mutants, which do not express ttss-1, yield a pure population of fast-growing TTSS-1− cells (Figs. 1 and 2). Thus, in a mixed culture, hilA or hilD mutants should outgrow wt S. Tm. Indeed, both mutants out-competed the wt strain during the late logarithmic phase of the mixed culture (ΔhilA, ΔhilD; Fig. 5A,B). In contrast, a hilE mutant, which forms a larger fraction of TTSS-1+ cells than wt S. Tm (Fig. 2), was outcompeted by wt S. Tm in this type of assay (ΔhilE, Fig. 5C). This verified the growth penalty of TTSS-1+ cells in LB batch cultures.
Figure 5. Competitive growth experiment confirming that ttss-1 expression retards growth.
A) Wt S. Tm (ATCC14028, kmS) and an isogenic hilA mutant (M2005, kmR), were used to inoculate a sub-culture at a ratio of approx. 1∶1. Growth of the mixed culture was monitored via OD600. B) Competitive growth between wt S. Tm and an isogenic hilD mutant (M2007, kmR), resp. an isogenic hilE mutant (M2008, cmR), C). The fraction of wt S. Tm was determined by differential plating on LB agar (50 µg/ml kanamycin, resp. 30 µg/ml chloramphenicol) at the indicated time points. Data were derived from four experiments (±s.d., p = 0.014).
Discussion
The effect of virulence factor expression on the fitness of an individual pathogen cell has remained unclear. We have analyzed the fitness costs associated with the expression of ttss-1, which encodes a key virulence function of S. Tm. An in vitro system was chosen for a detailed analysis of the growth phenotype of TTSS-1+ cells. We found that these cells have a reduced growth rate. This established that ttss-1 expression represents a burden (and not an advantage) at the level of the individual cell, at least in the non-host environment of our assay system. The growth penalty affects the fraction of TTSS-1+ individuals and the overall growth progression in a S. Tm culture. Mathematical modeling and experimental data demonstrated that this growth penalty and an increasing initiation rate of ttss-1 expression during the late logarithmic growth phase were sufficient to explain the dynamic abundance of TTSS-1+ and TTSS-1− individuals in a clonal S. Tm batch culture.
Evidence for bistability of ttss-1 expression has only recently been accumulated. Under inducing conditions, single cell reporters for expression of ttss-1 or effector proteins yielded cells in the “on” and cells in the “off” state [1], [2], [3], [5], [32]. The regulatory network controlling ttss-1 expression includes at least three positive feedback loops and this architecture is thought to set the threshold for initiating ttss-1 expression and to amplify the level of expression [5], [32], [33]. The TTSS-1+ phenotype can persist for several hours, even if the bacteria are shifted into environments normally not inducing ttss-1 expression (histeresis; shift to fresh LB, Fig. 2; Fig. S2). However, it should also be noted that it has not been possible to define unequivocally where stochasticity is introduced. In fact, stochastic initiation of ttss-1 expression might hinge on different regulators in different environments.
TTSS-1+ cells have at least two important characteristics. First, they express the virulence factors enabling host manipulation and elicitation of disease [13], [17], [18]. Second, as we have found here, they grow at a reduced rate. ttss-1 expression may represent a “burden” in itself. The mechanism explaining the growth defect of TTSS-1+ cells is of significant interest. A partial disruption of the proton gradient by “leaky” TTSS assembly-intermediates and/or the metabolic energy required for biosynthesis of the TTSS may offer plausible explanations. Typical TTSS-1+ cells are estimated to express 20–200 TTS apparatuses and approx. 3−10×104 effector proteins, amounting to a significant fraction of the total cellular protein [2], [3]. Indeed, deleting the translocon and most effector proteins significantly increased the growth rate of the TTSS-1+ cells (Δ8Δsip; Fig. 2E), indicating that these proteins account at least in part for the cost of ttss-1 expression. However, the growth rate of Δ8Δsip (µ = 1.10 h−1) was still lower than that of the TTSS-1− subpopulation of wt S. Tm (µfast = 1.27 h−1), suggesting that other factors do also contribute to growth retardation.
An alternative explanation for the reduced growth rate of TTSS-1+ cells might reside in coordinated expression of a complex regulon. This might be reminiscent of the prf virulence regulon of Listeria monocytogenes, which coordinates metabolism and virulence gene expression thus controlling environment-specific fitness phenotypes in vitro and in vivo [34]. Several global regulators (e.g. crp, mlc, fur; [7], [35], [36]) and silencing proteins (hns, hha; [37], [38]) can control ttss-1 expression. Moreover, HilA may control multiple loci apart from ttss-1 (25). And we have observed co-expression of ttss-1 and of fliC, which encodes a key structural component of the flagella, in the late logarithmic phase (Fig. S5). Accordingly, ttss-1 expression might be one feature of a “differentiated” state which also includes adaptations reducing the growth rate. It is tempting to speculate that this state might be particularly adapted for mucosal tissue invasion. This would be an important topic for future research.
Interestingly, similar phenomena have been observed in other ttss-expressing pathogens. In Pseudomonas aeruginosa, growth in suboptimal media was shown to result in bistable ttss expression [39]. But it remained unclear whether growth might be affected. In contrast, the plasmid-encoded TTSS of Yersinia spp. is well known to cause growth retardation in response to host cell contact or low calcium environments [40], [41]. However, in this case, ttss induction seems to be uniform even in suboptimal media [42]. Thus, bistability and growth retardation do occur in other ttss expressing bacteria, but specific adaptations may exist for each pathogen.
Which environmental cues induce ttss-1 expression in S. Tm? ttss-1 is expressed in the lumen of the host's intestine and in the late logarithmic phase in LB-batch culture. Low oxygen pressure is common to both environments and may represent an inducing signal (see Fig. 3C). In line with this hypothesis, Shigella flexneri, a closely related gut pathogen, can modulate the activity of its TTSS in response to low oxygen pressures typically observed at the gut wall [43]. Similarly, HilA-mediated ttss-1 expression is known to respond to oxygen pressure [21], [44]. In addition, numerous other internal and external cues are known to affect ttss-1 expression, including osmolarity, pH, growth rate, or the presence of short chain fatty acids like acetate [45], [46], [47], [48], [49], [50], [51]. The sum of these environmental cues seems to determine the level of ttss-1 induction. This might explain our observation of a low, but detectable initiation rate of ttss-1 expression on agar pads soaked with spent medium (Fig. 4). This environment should harbor most cues present in the late log culture medium, but lacks low oxygen pressure, which could not be established in the real time microscopy setup.
In summary, our findings indicate that the TTSS-1+ phenotype is more complex than previously anticipated. Currently, we can only speculate how this affects the real infection and transmission in vivo. Our results suggest that the TTSS-1+ subpopulation is constantly drained by the burdens inflicted by immune defenses within the infected gut mucosa [4] and by the reduced growth rate (this work). The latter should represent a competitive disadvantage against all other bacteria (commensals and TTSS-1− S. Tm cells) present in the gut lumen. Moreover, this burden should materialize even before invading the gut tissue and may explain why ttss-1 defective mutants are sometimes (though rarely) found in infected animal flocks and isolated in one case of a human outbreak [52], [53]. In order to explain the evolution and mainentance of bistable ttss-1 expression and the successful propagation of the ttss-1 genotype, one has to predict that the TTSS-1+ phenotype must confer some type of advantage. According to the “division of labor” model, the advantage might emanate from a “public good”, i.e. the TTSS-1 induced gut inflammation fostering Salmonella growth in the gut lumen and enhancing transmission. Alternatively, the TTSS-1+ phenotype might include (unidentified) features enhancing the survival and growth of the ttss-1 expressing bacteria themselves, e.g. in permissive niches of the host's intestine or by enhancing the chances of chronic infection and long-term shedding. Identifying these mechanisms will represent an important step for understanding the evolution of bistable ttss-1 expression.
Materials and Methods
Bacteria
All strains were derivatives of Salmonella Typhimurium SL1344 or ATCC14028 (see Tab. S1 and Text S2 for references). All plasmids and primers are shown in Tab. S2 and S3. Bacteria were inoculated (1∶100 in LB) from 12 h overnight cultures (LB, supplemented with the appropriate antibiotics) and grown under mild aeration for 4 h at 37°C, if not stated otherwise. In Fig. 1C,E, the medium included 0.01% arabinose.
The mutants were constructed using the lambda red recombination system [54]. The chloramphenicol or kanamycin resistance cassette of pKD3 (cat) resp. pKD4 (aphT) were amplified by PCR using the primer pairs ÄhilA::kan-fw and ÄhilA::kan-rev, ÄhilD::kan-fw and ÄhilD::kan-rev, ÄhilE::cat-fw and ÄhilE::cat-rev and electroporated into SL1344 harboring pKD46 to generate the regulator mutants M2005 (ÄhilA::aphT), M2007 (ÄhilD::aphT) and M2008 (ÄhilE::cat). Mutants were selected by plating on LB-Agar (50 µg/ml kanamycin or 30 µg/ml chloramphenicol). M2072 (termed Δprg-orgΔinv-spa in this paper) was also generated using the lambda red system using the primers invG-fw and spaS-rev as well as prgH-fw and orgC–rev and the plasmids pKD3 and pKD4 to generate prgHIJKorgABC::aphT, invGEABCIJspaOPQRS::cat, a mutant lacking most genes of the TTS apparatus. For construction of strain M2532 (termed Δ8Δsip in this paper), we transduced the ÄsipBCDA-sptP::aphT allele from SB245 (SL1344, ÄsipBCDA-sptP::aphT fliGHI::Tn10; K. Kaniga and J. E. Galan, unpublished data) via P22 into M2400 (SL1344, ÄsopE, ÄsopE2, ÄsopB, ÄsipA, ÄsptP, ÄsopA, ÄspvB, ÄspvC), which has been previously described [55]. M2532 fails to express most TTSS-1 effector proteins and the translocon components.
To create the suicide plasmid pM2002, pVS152Tsr [30] was digested with the restriction endonucleases Eco47III and XmaI. The tsrvenus encoding fragment was ligated into pM1300 (digested with MslI and XmaI, [56]) downstream of a truncated sipA fragment (nt 1156–2058 of the orf), to finally create pM2002 and introduced by homologous recombination into the genome of ATCC14028 to generate the reporter strain M2001. To obtain the tsrvenus reporter for hilA (M2076), the c-terminal region of hilA (nt 114 to 1661 of the orf) was amplified using the primer pair hilA-fw-XmaI-NcoI and hilA-rev-NheI-XbaI and cloned into pBluescriptII (Invitrogen) using the restriction endonucleases XmaI and XbaI, yielding pM2090. This plasmid was digested with NheI and NotI to introduce the tsrvenus encoding PCR fragment (template pM2002, primers: venus-NheI-fw and venus-NotI-rev, digested with NheI and NotI) to obtain pM2095. The entire region ranging from hilA to tsrvenus was cloned into pSB377 using the restriction enzymes NotI and XmaI yielding the suicide plasmid pM2080. This plasmid was used to generate the hilA reporter strain M2076 by homologous recombination into the genome of ATCC14028. To obtain the tsrvenus reporter for fliC, tsrvenus was amplified by PCR (primers: tsr-XmaI-fw and venus-XbaI-rev) and cloned into pBluescriptII using XmaI and XbaI thus yielding pM2533. After amplification of fliC by PCR using SL1344 chromosomal DNA as template and primers fliC-XhoI-fw and fliC-HindIII-rev, the fliC encoding fragment was cloned via XhoI and HindIII upstream of the tsrvenus gene into pM2533, thus yielding pM2539. Subsequently, the construct was moved via XhoI and XbaI into the suicide plasmid pGP704, thus yielding pM2819. This plasmid was used to create the fliC-tsrvenus reporter strain M2821 by homologous recombination into the genome of SL1344.
All over-expression plasmids from pM2010 to pM2042 were obtained by digesting the indicated PCR fragments (Table S2 and S3 for plasmids and primers) with EcoRI and XbaI into pBAD24.
All mutations were verified by PCR or DNA sequencing.
HilA expression was analyzed by quantitative Western blot using an affinity-purified rabbit α-HilA antiserum (Fig. 1E). Recombinant HilA was used for normalization. SipC was detected using an α-SipC serum (Fig. 1G).
For invasion, MDCK cells were grown in MEM (Invitrogen), infected for 30 min (MOI = 5; [57], washed and incubated in MEM (400 µg/ml gentamicin; 1 h). Intracellular bacteria were enumerated by plating.
FACS
Prior to analysis, fluorophore formation was ensured (2 h, RT, 30 µg/ml chloramphenicol). Tsrvenus and Gfp emission was analyzed at 530 nm (supplement; FACSCalibur 4-color, Becton Dickinson). Bacteria were identified by side scatter (SSC). Data were analyzed with FlowJo software (Tree Star, Inc.). For Tsrvenus (Fig. 1), ln-transformed fluorescence values for 40000 events were median-normalized (subtraction) and compared to the similarly normalized data from the reporterless control strain, thus yielding the fraction of TTSS-1+ individuals. For sorting bacterial cells, S. Tm (psicA-gfp) cells were sorted by FACS (Aria Becton Dickinson, FACSDiva Software).
Time-lapse microscopy
Bacteria were placed on a 1.5% agarose pad equilibrated with LB, sealed under a glass coverslip and mounted (37°C temp. control; Axioplan2; Plan-APOCHROMAT 63x/1.4 oil; Zeiss or IX81, UPlanFLN 100x/1.3 Oil, Olympus). Reporter fluorescence (Exc. 470/20 nm; BP 495 nm; Em. 505–530 nm) and micro colony growth (phase contrast) were monitored and evaluated using Axiovision software (Zeiss). The slope of the ln-tranformed bacterial numbers (t), as determined from the logarithmic growth phase, yielded the growth rate µ. For sipA-tsrvenus and prgH-gfp, the micro colonies were scored visually as TTSS-1+ or TTSS-1−. To analyze differences in growth rates between TTSS-1+ and TTSS-1− micro colonies, we performed a full-factorial analysis of variance with the two factors phenotype (fixed) and experiment (random). Variance was analyzed in SPSS 17.0 (SPSS Inc. - Chicago, IL).
Growth rates w/o reporter were analyzed via a maximum likelihood approach to test for two subpopulations with different growth rates. The growth rate measurements from five independent experiments (87 micro colonies) were combined. Using maximum likelihood, we fitted a bi-modal distribution (the sum of two normal probability density functions) and a unimodal (normal) distribution, and compared the two fits with a likelihood ratio test using R software [58].
In Fig. 4, cell growth and ttss-1 expression were analyzed using a modified version of the cell tracking software described in [9]. The first cell in each micro colony that could be observed over a whole division was used to analyze the statistical association between ttss-1 expression and the interval between two divisions (by non-parametric correlation analysis using PASW Statistics 18.0.0). 157 micro colonies were analyzed to estimate the fraction of micro colonies in which all cells, none of the cells, and a fraction of the cells expressed ttss-1. These groupings were based on visual inspection of each micro colony.
Supporting Information
Graphical maps and bistable gene expression by the gfp and venus constructs. A) Transcriptional reporters for prgH and sicA expression. The prgH and sicA promoters are driving gfp expression. The prgH-gfp reporter is integrated into the chromosomal proV locus [1]. The sicA-gfp reporter is plasmid-encoded (pM972; psicA-gfp). B) Bistable ttss-1 expression as detected using the prgH-gfp and sicA-gfp reporters. Wild type S. Tm SL1344 w/o reporter (black), harboring psicA-gfp (green), or harboring prgH-gfp (red) were cultured for 4 h in LB, gfp expression was analyzed by FACS and the results were plotted using FlowJo7.5 software (Materials and Methods). C) Transcriptional reporter for sipA expression. The sipA-tsrvenus reporter was constructed by integrating pM2002 into the S. Tm chromosome at the 3′-end of the sicAsipBCDA operon. D) Bistable ttss-1 expression profile of wild type S. Tm ATCC14028 w/o any reporter (gray), with the sipA-tsrvenus reporter (green) or with the sipA-tsrvenus reporter and philA (purple); FACS data were analyzed by using MSExcel2007 and Prism5 software.
(PDF)
Growth of individual wild type S . Tm SL1344 cells as observed by time lapse light microscopy. Primary data used to determine the growth rates of wt S. Tm (no reporter) in Fig. 2C. Bacteria grown as described in the legend to Fig. 2 were placed on a 1.5% agarose pad equilibrated with fresh LB and imaged by time-lapse microscopy. Growth of single bacteria (growing up into micro-colonies) was monitored by phase contrast time lapse microscopy and analyzed using Axiovision software (Zeiss, see also legend to Fig. 2 and Materials and Methods). The number of bacteria per micro-colony was determined every 30 minutes for a total of 3 h. A) Micro-colonies assigned to the group of “fast growing” bacteria (see Fig. 2C); curves in B) depict slow growing micro-colonies. The prominent black curves in A) and B) depict the medians. Both subpopulations display a brief lag phase followed by exponential growth throughout the rest of the imaging experiment.
(PDF)
Dye dilution assay confirmed retarded growth of TTSS-1+ individuals. A dye dilution assay served as a second, independent method for measuring growth of TTSS-1+ individuals. In this type of assay, bacteria are labeled with a stable dye which is diluted by 2-fold during each cell division. Here, we used the membrane dye PKH26 and a S. Tm wbaP strain harboring a ttss-1 reporter plasmid (SKI12, psicA-gfp). This strain lacks the LPS O-side chain and allowed efficient membrane labeling of living cells with PKH26. It should be noted that the wbaP strain grew normally in LB-broth and efficiently invaded host cells, a hallmark of TTSS-1 function [63]. A) SKI12 pM972 was sub-cultured (LB, 4 h, OD600 = 1), washed three times with 4°C PBS, and incubated for 2 min at room temperature with 5 µM PKH26 (50 mM acetate buffer pH 5; Sigma-Aldrich). Excess dye was removed by washing three times with LB. Then, the bacteria were grown in LB, aliquots were removed at the indicated times and GFP- and PKH26 fluorescence were analyzed by FACS (PKH26 = red fluorescence). B) Dye-dilution rates of the TTSS-1+ and TTSS-1- sub-populations. The median fluorescence intensity of the left (TTSS-1-) and the right (TTSS-1+) quadrants were plotted at each time point, analyzed. Line: exponential fit to the experimental data. The TTSS-1- individuals displayed an apparent PKH26 dilution rate of t1/2 = 36 min (i.e. µ = 1.1 h−1; Fig. 3B). The PKH26 dilution rate of the TTSS-1+ individuals amounted to t1/2 = 86 min (i.e. µ = 0.48 h−1; Fig. 3B). This was in line with our results from time-lapse microscopy and confirmed that the TTSS-1+ phenotype has a reduced growth rate.
(PDF)
Quantification of fluorescence intensity in time-lapse microscopy. Fluorescence was quantified over time for the growing micro colonies analyzed in Fig. 4. Each line shows fluorescence of a single cell, branching of lines indicates division events. A, B, and C correspond to A, B, and C in Fig. 4.
(PDF)
fliC is co-regulated with ttss-1 . S. Tm possessing a transcriptional reporter for ttss-1 (psicA mCherry, plasmid) and either sipA-tsrvenus or hilA-tsrvenus or fliC-tsrvenus (each on chromosome) were grown in LB to an OD600 of 1 and examined for co-expression by microscopy. The co-expression of psicA gfp and sipA-tsrvenus served as a direct positive control. We could observe a less efficient expression of mCherry, even though gfp and mCherry are driven by literally the same promoter (psicA, see also Figure S1). Most probably this is caused by the stability of the different fluorophores and the higher sensitivity of the tsrvenus reporter. In the case of hilA and fliC we could determine a co-expression of ttss-1 genes. At least all TTSS-1+ (psicA mCherry) featured hilA and fliC expression. It was recently shown that FliC, which assembles to the flagella, underlies noisy gene expression (besides phase variation [64]) and emerges FliC+ and FliC- subpopulations [65]. A) Quantification of four independent experiments; shown is the median ± s.d.; B) Representative microscopy pictures of the three strains.
(PDF)
Bacterial strains.
(PDF)
Plasmids.
(PDF)
Primer sequences.
(PDF)
Mathematical model for calculating the rate of initiation ri of ttss-1 gene expression during the late log phase.
(PDF)
References Supporting Information.
(DOC)
Heterogenous induction of ttss-1 genes in a micro-colony.
(AVI)
Acknowledgments
We are grateful to N. Freed, B. Stecher, M. Diard, P. Kaiser and M. De Lisa for thoughtful discussions, R. Regoes for help with statistics, to J. Hinton for sharing JH3010 and to Emma Slack for advice on the PKH26 FACS strategy. We thank M. Elowitz and N. Rosenfeld for cell tracking software, R. Peña-Miller for helpful comments and modifications on the cell tracking software, and T. Bergmiller for help with image analysis.
Footnotes
The authors have declared that no competing interests exist.
This work was funded by grants from the ETH research foundation (TH-10 06-1), the SNF (310000-113623/1) and the EU (INCO-2006-CT-032296) to WDH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hautefort I, Proenca MJ, Hinton JC. Single-copy green fluorescent protein gene fusions allow accurate measurement of Salmonella gene expression in vitro and during infection of mammalian cells. Appl Environ Microbiol. 2003;69:7480–7491. doi: 10.1128/AEM.69.12.7480-7491.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schlumberger MC, Muller AJ, Ehrbar K, Winnen B, Duss I, et al. Real-time imaging of type III secretion: Salmonella SipA injection into host cells. Proc Natl Acad Sci U S A. 2005;102:12548–12553. doi: 10.1073/pnas.0503407102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winnen B, Schlumberger MC, Sturm A, Schupbach K, Siebenmann S, et al. Hierarchical effector protein transport by the Salmonella Typhimurium SPI-1 type III secretion system. PLoS ONE. 2008;3:e2178. doi: 10.1371/journal.pone.0002178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ackermann M, Stecher B, Freed NE, Songhet P, Hardt WD, et al. Self-destructive cooperation mediated by phenotypic noise. Nature. 2008;454:987–990. doi: 10.1038/nature07067. [DOI] [PubMed] [Google Scholar]
- 5.Saini S, Ellermeier JR, Slauch JM, Rao CV. The role of coupled positive feedback in the expression of the SPI1 type three secretion system in Salmonella. PLoS Pathog. 2010;6:e1001025. doi: 10.1371/journal.ppat.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smits WK, Kuipers OP, Veening JW. Phenotypic variation in bacteria: the role of feedback regulation. Nat Rev Microbiol. 2006;4:259–271. doi: 10.1038/nrmicro1381. [DOI] [PubMed] [Google Scholar]
- 7.Lim S, Yun J, Yoon H, Park C, Kim B, et al. Mlc regulation of Salmonella pathogenicity island I gene expression via hilE repression. Nucleic Acids Res. 2007;35:1822–1832. doi: 10.1093/nar/gkm060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 9.Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB. Gene regulation at the single-cell level. Science. 2005;307:1962–1965. doi: 10.1126/science.1106914. [DOI] [PubMed] [Google Scholar]
- 10.Raj A, van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell. 2008;135:216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raser JM, O'Shea EK. Noise in gene expression: origins, consequences, and control. Science. 2005;309:2010–2013. doi: 10.1126/science.1105891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kussell E, Kishony R, Balaban NQ, Leibler S. Bacterial persistence: a model of survival in changing environments. Genetics. 2005;169:1807–1814. doi: 10.1534/genetics.104.035352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schlumberger MC, Hardt WD. Salmonella type III secretion effectors: pulling the host cell's strings. Curr Opin Microbiol. 2006;9:46–54. doi: 10.1016/j.mib.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 14.Watson PR, Galyov EE, Paulin SM, Jones PW, Wallis TS. Mutation of invH, but not stn, reduces Salmonella-induced enteritis in cattle. Infect Immun. 1998;66:1432–1438. doi: 10.1128/iai.66.4.1432-1438.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsolis RM, Adams LG, Ficht TA, Baumler AJ. Contribution of Salmonella typhimurium virulence factors to diarrheal disease in calves. Infect Immun. 1999;67:4879–4885. doi: 10.1128/iai.67.9.4879-4885.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barthel M, Hapfelmeier S, Quintanilla-Martinez L, Kremer M, Rohde M, et al. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun. 2003;71:2839–2858. doi: 10.1128/IAI.71.5.2839-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wallis TS, Galyov EE. Molecular basis of Salmonella-induced enteritis. Mol Microbiol. 2000;36:997–1005. doi: 10.1046/j.1365-2958.2000.01892.x. [DOI] [PubMed] [Google Scholar]
- 18.Hapfelmeier S, Hardt WD. A mouse model for S. typhimurium-induced enterocolitis. Trends Microbiol. 2005;13:497–503. doi: 10.1016/j.tim.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 19.Santos RL, Raffatellu M, Bevins CL, Adams LG, Tukel C, et al. Life in the inflamed intestine, Salmonella style. Trends Microbiol. 2009;17:498–506. doi: 10.1016/j.tim.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schechter LM, Damrauer SM, Lee CA. Two AraC/XylS family members can independently counteract the effect of repressing sequences upstream of the hilA promoter. Mol Microbiol. 1999;32:629–642. doi: 10.1046/j.1365-2958.1999.01381.x. [DOI] [PubMed] [Google Scholar]
- 21.Bajaj V, Hwang C, Lee CA. hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol Microbiol. 1995;18:715–727. doi: 10.1111/j.1365-2958.1995.mmi_18040715.x. [DOI] [PubMed] [Google Scholar]
- 22.Ellermeier JR, Slauch JM. Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr Opin Microbiol. 2007;10:24–29. doi: 10.1016/j.mib.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 23.Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007;5:2177–2189. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raffatellu M, George MD, Akiyama Y, Hornsby MJ, Nuccio SP, et al. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe. 2009;5:476–486. doi: 10.1016/j.chom.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:204. doi: 10.1016/j.chom.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 26.Stecher B, Barthel M, Schlumberger MC, Haberli L, Rabsch W, et al. Motility allows S. Typhimurium to benefit from the mucosal defence. Cell Microbiol. 2008;10:1166–1180. doi: 10.1111/j.1462-5822.2008.01118.x. [DOI] [PubMed] [Google Scholar]
- 27.Stecher B, Hardt WD. The role of microbiota in infectious disease. Trends Microbiol. 2008;16:107–114. doi: 10.1016/j.tim.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 28.Lawley TD, Bouley DM, Hoy YE, Gerke C, Relman DA, et al. Host transmission of Salmonella enterica serovar Typhimurium is controlled by virulence factors and indigenous intestinal microbiota. Infect Immun. 2008;76:403–416. doi: 10.1128/IAI.01189-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winter SE, Keestra AM, Tsolis RM, Baumler AJ. The blessings and curses of intestinal inflammation. Cell Host Microbe. 2010;8:36–43. doi: 10.1016/j.chom.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu J, Xiao J, Ren X, Lao K, Xie XS. Probing gene expression in live cells, one protein molecule at a time. Science. 2006;311:1600–1603. doi: 10.1126/science.1119623. [DOI] [PubMed] [Google Scholar]
- 31.Wendland M, Bumann D. Optimization of GFP levels for analyzing Salmonella gene expression during an infection. FEBS Lett. 2002;521:105–108. doi: 10.1016/s0014-5793(02)02834-x. [DOI] [PubMed] [Google Scholar]
- 32.Temme K, Salis H, Tullman-Ercek D, Levskaya A, Hong SH, et al. Induction and relaxation dynamics of the regulatory network controlling the type III secretion system encoded within Salmonella pathogenicity island 1. J Mol Biol. 2008;377:47–61. doi: 10.1016/j.jmb.2007.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bailly-Bechet M, Benecke A, Hardt WD, Lanza V, Sturm A, et al. J Math Biol: [Epub ahead of print]; 2010. An externally modulated, noise-driven switch for the regulation of SPI1 in Salmonella enterica serovar Typhimurium. [DOI] [PubMed] [Google Scholar]
- 34.Bruno JC, Jr, Freitag NE. Constitutive activation of PrfA tilts the balance of Listeria monocytogenes fitness towards life within the host versus environmental survival. PLoS One. 2010;5:e15138. doi: 10.1371/journal.pone.0015138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teplitski M, Goodier RI, Ahmer BM. Catabolite repression of the SirA regulatory cascade in Salmonella enterica. Int J Med Microbiol. 2006;296:449–466. doi: 10.1016/j.ijmm.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 36.Ellermeier JR, Slauch JM. Fur regulates expression of the Salmonella pathogenicity island 1 type III secretion system through HilD. J Bacteriol. 2008;190:476–486. doi: 10.1128/JB.00926-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olekhnovich IN, Kadner RJ. Role of nucleoid-associated proteins Hha and H-NS in expression of Salmonella enterica activators HilD, HilC, and RtsA required for cell invasion. J Bacteriol. 2007;189:6882–6890. doi: 10.1128/JB.00905-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rietsch A, Mekalanos JJ. Metabolic regulation of type III secretion gene expression in Pseudomonas aeruginosa. Mol Microbiol. 2006;59:807–820. doi: 10.1111/j.1365-2958.2005.04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brubaker RR, Surgalla MJ. The Effect of Ca++ and Mg++ on Lysis, Growth, and Production of Virulence Antigens by Pasteurella Pestis. J Infect Dis. 1964;114:13–25. doi: 10.1093/infdis/114.1.13. [DOI] [PubMed] [Google Scholar]
- 41.Higuchi K, Kupferberg LL, Smith JL. Studies on the nutrition and physiology of Pasteurella pestis. III. Effects of calcium ions on the growth of virulent and avirulent strains of Pasteurella pestis. J Bacteriol. 1959;77:317–321. doi: 10.1128/jb.77.3.317-321.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wiley DJ, Rosqvist R, Schesser K. Induction of the Yersinia type 3 secretion system as an all-or-none phenomenon. J Mol Biol. 2007;373:27–37. doi: 10.1016/j.jmb.2007.07.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marteyn B, West NP, Browning DF, Cole JA, Shaw JG, et al. Modulation of Shigella virulence in response to available oxygen in vivo. Nature. 2010;465:355–358. doi: 10.1038/nature08970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schiemann DA, Shope SR. Anaerobic growth of Salmonella typhimurium results in increased uptake by Henle 407 epithelial and mouse peritoneal cells in vitro and repression of a major outer membrane protein. Infect Immun. 1991;59:437–440. doi: 10.1128/iai.59.1.437-440.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lostroh CP, Lee CA. The Salmonella pathogenicity island-1 type III secretion system. Microbes Infect. 2001;3:1281–1291. doi: 10.1016/s1286-4579(01)01488-5. [DOI] [PubMed] [Google Scholar]
- 46.Huang Y, Suyemoto M, Garner CD, Cicconi KM, Altier C. Formate acts as a diffusible signal to induce Salmonella invasion. J Bacteriol. 2008;190:4233–4241. doi: 10.1128/JB.00205-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee CA, Falkow S. The ability of Salmonella to enter mammalian cells is affected by bacterial growth state. Proc Natl Acad Sci U S A. 1990;87:4304–4308. doi: 10.1073/pnas.87.11.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ernst RK, Dombroski DM, Merrick JM. Anaerobiosis, type 1 fimbriae, and growth phase are factors that affect invasion of HEp-2 cells by Salmonella typhimurium. Infect Immun. 1990;58:2014–2016. doi: 10.1128/iai.58.6.2014-2016.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee CA, Jones BD, Falkow S. Identification of a Salmonella typhimurium invasion locus by selection for hyperinvasive mutants. Proc Natl Acad Sci U S A. 1992;89:1847–1851. doi: 10.1073/pnas.89.5.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lawhon SD, Maurer R, Suyemoto M, Altier C. Intestinal short-chain fatty acids alter Salmonella typhimurium invasion gene expression and virulence through BarA/SirA. Mol Microbiol. 2002;46:1451–1464. doi: 10.1046/j.1365-2958.2002.03268.x. [DOI] [PubMed] [Google Scholar]
- 51.Galan JE, Curtiss R., 3rd Expression of Salmonella typhimurium genes required for invasion is regulated by changes in DNA supercoiling. Infect Immun. 1990;58:1879–1885. doi: 10.1128/iai.58.6.1879-1885.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hu Q, Coburn B, Deng W, Li Y, Shi X, et al. Salmonella enterica serovar Senftenberg human clinical isolates lacking SPI-1. J Clin Microbiol. 2008;46:1330–1336. doi: 10.1128/JCM.01255-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rahn K, De Grandis SA, Clarke RC, McEwen SA, Galan JE, et al. Amplification of an invA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol Cell Probes. 1992;6:271–279. doi: 10.1016/0890-8508(92)90002-f. [DOI] [PubMed] [Google Scholar]
- 54.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoffmann C, Galle M, Dilling S, Kappeli R, Muller AJ, et al. In macrophages, caspase-1 activation by SopE and the type III secretion system-1 of S. typhimurium can proceed in the absence of flagellin. PLoS One. 2010;5:e12477. doi: 10.1371/journal.pone.0012477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ehrbar K, Hapfelmeier S, Stecher B, Hardt WD. InvB is required for type III-dependent secretion of SopA in Salmonella enterica serovar Typhimurium. J Bacteriol. 2004;186:1215–1219. doi: 10.1128/JB.186.4.1215-1219.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ehrbar K, Friebel A, Miller SI, Hardt WD. Role of the Salmonella pathogenicity island 1 (SPI-1) protein InvB in type III secretion of SopE and SopE2, two Salmonella effector proteins encoded outside of SPI-1. J Bacteriol. 2003;185:6950–6967. doi: 10.1128/JB.185.23.6950-6967.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Team RDC. R: A language and environment for statistical computing. R Foundation for Statistical Computing. 2009.
- 59.Ellermeier CD, Ellermeier JR, Slauch JM. HilD, HilC and RtsA constitute a feed forward loop that controls expression of the SPI1 type three secretion system regulator hilA in Salmonella enterica serovar Typhimurium. Mol Microbiol. 2005;57:691–705. doi: 10.1111/j.1365-2958.2005.04737.x. [DOI] [PubMed] [Google Scholar]
- 60.Schechter LM, Jain S, Akbar S, Lee CA. The small nucleoid-binding proteins H-NS, HU, and Fis affect hilA expression in Salmonella enterica serovar Typhimurium. Infect Immun. 2003;71:5432–5435. doi: 10.1128/IAI.71.9.5432-5435.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Humphreys S, Rowley G, Stevenson A, Anjum MF, Woodward MJ, et al. Role of the two-component regulator CpxAR in the virulence of Salmonella enterica serotype Typhimurium. Infect Immun. 2004;72:4654–4661. doi: 10.1128/IAI.72.8.4654-4661.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Altier C, Suyemoto M, Lawhon SD. Regulation of Salmonella enterica serovar typhimurium invasion genes by csrA. Infect Immun. 2000;68:6790–6797. doi: 10.1128/iai.68.12.6790-6797.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ilg K, Endt K, Misselwitz B, Stecher B, Aebi M, et al. O-antigen-negative Salmonella enterica serovar Typhimurium is attenuated in intestinal colonization but elicits colitis in streptomycin-treated mice. Infect Immun. 2009;77:2568–2575. doi: 10.1128/IAI.01537-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bonifield HR, Hughes KT. Flagellar phase variation in Salmonella enterica is mediated by a posttranscriptional control mechanism. J Bacteriol. 2003;185:3567–3574. doi: 10.1128/JB.185.12.3567-3574.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Freed NE, Silander OK, Stecher B, Bohm A, Hardt WD, et al. A simple screen to identify promoters conferring high levels of phenotypic noise. PLoS Genet. 2008;4:e1000307. doi: 10.1371/journal.pgen.1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Graphical maps and bistable gene expression by the gfp and venus constructs. A) Transcriptional reporters for prgH and sicA expression. The prgH and sicA promoters are driving gfp expression. The prgH-gfp reporter is integrated into the chromosomal proV locus [1]. The sicA-gfp reporter is plasmid-encoded (pM972; psicA-gfp). B) Bistable ttss-1 expression as detected using the prgH-gfp and sicA-gfp reporters. Wild type S. Tm SL1344 w/o reporter (black), harboring psicA-gfp (green), or harboring prgH-gfp (red) were cultured for 4 h in LB, gfp expression was analyzed by FACS and the results were plotted using FlowJo7.5 software (Materials and Methods). C) Transcriptional reporter for sipA expression. The sipA-tsrvenus reporter was constructed by integrating pM2002 into the S. Tm chromosome at the 3′-end of the sicAsipBCDA operon. D) Bistable ttss-1 expression profile of wild type S. Tm ATCC14028 w/o any reporter (gray), with the sipA-tsrvenus reporter (green) or with the sipA-tsrvenus reporter and philA (purple); FACS data were analyzed by using MSExcel2007 and Prism5 software.
(PDF)
Growth of individual wild type S . Tm SL1344 cells as observed by time lapse light microscopy. Primary data used to determine the growth rates of wt S. Tm (no reporter) in Fig. 2C. Bacteria grown as described in the legend to Fig. 2 were placed on a 1.5% agarose pad equilibrated with fresh LB and imaged by time-lapse microscopy. Growth of single bacteria (growing up into micro-colonies) was monitored by phase contrast time lapse microscopy and analyzed using Axiovision software (Zeiss, see also legend to Fig. 2 and Materials and Methods). The number of bacteria per micro-colony was determined every 30 minutes for a total of 3 h. A) Micro-colonies assigned to the group of “fast growing” bacteria (see Fig. 2C); curves in B) depict slow growing micro-colonies. The prominent black curves in A) and B) depict the medians. Both subpopulations display a brief lag phase followed by exponential growth throughout the rest of the imaging experiment.
(PDF)
Dye dilution assay confirmed retarded growth of TTSS-1+ individuals. A dye dilution assay served as a second, independent method for measuring growth of TTSS-1+ individuals. In this type of assay, bacteria are labeled with a stable dye which is diluted by 2-fold during each cell division. Here, we used the membrane dye PKH26 and a S. Tm wbaP strain harboring a ttss-1 reporter plasmid (SKI12, psicA-gfp). This strain lacks the LPS O-side chain and allowed efficient membrane labeling of living cells with PKH26. It should be noted that the wbaP strain grew normally in LB-broth and efficiently invaded host cells, a hallmark of TTSS-1 function [63]. A) SKI12 pM972 was sub-cultured (LB, 4 h, OD600 = 1), washed three times with 4°C PBS, and incubated for 2 min at room temperature with 5 µM PKH26 (50 mM acetate buffer pH 5; Sigma-Aldrich). Excess dye was removed by washing three times with LB. Then, the bacteria were grown in LB, aliquots were removed at the indicated times and GFP- and PKH26 fluorescence were analyzed by FACS (PKH26 = red fluorescence). B) Dye-dilution rates of the TTSS-1+ and TTSS-1- sub-populations. The median fluorescence intensity of the left (TTSS-1-) and the right (TTSS-1+) quadrants were plotted at each time point, analyzed. Line: exponential fit to the experimental data. The TTSS-1- individuals displayed an apparent PKH26 dilution rate of t1/2 = 36 min (i.e. µ = 1.1 h−1; Fig. 3B). The PKH26 dilution rate of the TTSS-1+ individuals amounted to t1/2 = 86 min (i.e. µ = 0.48 h−1; Fig. 3B). This was in line with our results from time-lapse microscopy and confirmed that the TTSS-1+ phenotype has a reduced growth rate.
(PDF)
Quantification of fluorescence intensity in time-lapse microscopy. Fluorescence was quantified over time for the growing micro colonies analyzed in Fig. 4. Each line shows fluorescence of a single cell, branching of lines indicates division events. A, B, and C correspond to A, B, and C in Fig. 4.
(PDF)
fliC is co-regulated with ttss-1 . S. Tm possessing a transcriptional reporter for ttss-1 (psicA mCherry, plasmid) and either sipA-tsrvenus or hilA-tsrvenus or fliC-tsrvenus (each on chromosome) were grown in LB to an OD600 of 1 and examined for co-expression by microscopy. The co-expression of psicA gfp and sipA-tsrvenus served as a direct positive control. We could observe a less efficient expression of mCherry, even though gfp and mCherry are driven by literally the same promoter (psicA, see also Figure S1). Most probably this is caused by the stability of the different fluorophores and the higher sensitivity of the tsrvenus reporter. In the case of hilA and fliC we could determine a co-expression of ttss-1 genes. At least all TTSS-1+ (psicA mCherry) featured hilA and fliC expression. It was recently shown that FliC, which assembles to the flagella, underlies noisy gene expression (besides phase variation [64]) and emerges FliC+ and FliC- subpopulations [65]. A) Quantification of four independent experiments; shown is the median ± s.d.; B) Representative microscopy pictures of the three strains.
(PDF)
Bacterial strains.
(PDF)
Plasmids.
(PDF)
Primer sequences.
(PDF)
Mathematical model for calculating the rate of initiation ri of ttss-1 gene expression during the late log phase.
(PDF)
References Supporting Information.
(DOC)
Heterogenous induction of ttss-1 genes in a micro-colony.
(AVI)