Abstract
OBJECTIVE:
We examined whether the risk of food-allergen sensitization varied according to self-identified race or genetic ancestry.
METHODS:
We studied 1104 children (mean age: 2.7 years) from an urban multiethnic birth cohort. Food sensitization was defined as specific immunoglobulin E (sIgE) levels of ≥0.35 kilo–units of allergen (kUA)/L for any of 8 common food allergens. Multivariate logistic regression analyses were used to evaluate the associations of self-identified race and genetic ancestry with food sensitization. Analyses also examined associations with numbers of food sensitizations (0, 1 or 2, and ≥3 foods) and with logarithmically transformed allergen sIgE levels.
RESULTS:
In this predominantly minority cohort (60.9% black and 22.5% Hispanic), 35.5% of subjects exhibited food sensitizations. In multivariate models, both self-reported black race (odds ratio [OR]: 2.34 [95% confidence interval [CI]: 1.24–4.44]) and African ancestry (in 10% increments; OR: 1.07 [95% CI: 1.02–1.14]) were associated with food sensitization. Self-reported black race (OR: 3.76 [95% CI: 1.09–12.97]) and African ancestry (OR: 1.19 [95% CI: 1.07–1.32]) were associated with a high number (≥3) of food sensitizations. African ancestry was associated with increased odds of peanut sIgE levels of ≥5 kUA/L (OR: 1.25 [95% CI: 1.01–1.52]). Similar ancestry associations were seen for egg sIgE levels of ≥2 kUA/L (OR: 1.13 [95% CI: 1.01–1.27]) and milk sIgE levels of ≥5 kUA/L (OR: 1.24 [95% CI: 0.94–1.63]), although findings were not significant for milk.
CONCLUSIONS:
Black children were more likely to be sensitized to food allergens and were sensitized to more foods. African ancestry was associated with peanut sensitization.
Keywords: food allergy, sensitization, racial disparities, genetic ancestry
WHAT'S KNOWN ON THIS SUBJECT:
Racial disparities in food sensitization have been described in inner-city populations. However, clear understanding of racial disparities is difficult in US populations with significant ancestral diversity, because self-identified race is imprecise, compared with genetic ancestry.
WHAT THIS STUDY ADDS:
Black children were more likely to be sensitized to multiple foods. Only some of the differences were associated with genetic ancestry. African ancestry was particularly notable for increased risk of peanut sensitization at levels associated with clinical reactivity.
Food allergy can be a severe, potentially life-threatening illness, with an estimated prevalence of ∼3% among US children.1–3 A recent National Health and Nutrition Examination Survey–based prevalence study of food allergy showed a threefold increase in the rate of food allergy among black subjects and, in particular, racial differences for peanut allergy.1 Similarly, a recent, non–population-based study of young children at high risk, including multiple ethnicities, suggested that nonwhite subjects had a higher rate of peanut sensitization at a level more likely to be associated with clinical peanut allergy.4 Previous surveys of food allergy5,6 were not designed to evaluate food allergy in minority populations.
Although self-identified race is an important proxy for ethnocultural differences and has been used to investigate racial disparities, it is an imprecise estimate, compared with genetic ancestry.7 This is particularly true for the growing proportions of people in the US population who self-identify as members of ethnic groups with admixed ancestry, such as black and Hispanic groups.8 Genetic ancestry has been shown to be associated with a number of biological measurements, including lung function, white blood cell count, and creatinine clearance.9–11 The associations of genetic ancestry with food sensitization remain unexplored, despite recent suggestions of racial disparities.
With our urban, multiethnic, birth cohort, we sought to determine (1) whether the presence of sensitization to food or the number of foods to which a child was sensitized varied according to self-reported race/ethnicity and (2) whether these outcomes were associated with genetic ancestry, as estimated by using a set of ancestry informative markers (AIMs). Secondarily, we sought to corroborate whether there were racial disparities and ancestry associations with respect to elevated levels of specific immunoglobulin E (sIgE) for allergens (milk, egg, and peanut), which typically are associated with clinical reactivity among children 1 to 5 years of age.
METHODS
Patient Population
The Boston Birth Cohort (BBC) is a general-population, multiethnic cohort that includes subjects from a range of socioeconomic strata, from inner-city poor people to a smaller number of middle class subjects.12,13 Any mother who was admitted to the labor and delivery floor at the Boston Medical Center and delivered a live singleton infant was eligible to participate. Mother-infant pairs were recruited 24 to 72 hours after delivery. If BBC subjects planned to receive primary pediatric care at the Boston Medical Center, then the mother also was invited to have her child participate in the postnatal follow-up study, the Children's Health Study (CHS),13 with visits scheduled at 6 to 12 months and 2, 4, and 6 years. Subjects who provided consent are interviewed by using a structured questionnaire at all visits. Medical records for the mother and infant are reviewed by using a standardized abstraction form, to obtain clinical data and birth outcome data.
The current CHS enrollment includes 1794 subjects (enrollment of 91% of all subjects approached from the BBC). Of subjects enrolled in the CHS, key outcome and covariate data and genetic ancestry information were available for 1104 subjects (61.5% [1104 of 1794 subjects]) for this analysis. These people did not differ from subjects in the CHS who were not included, in terms of key sociodemographic variables including gender, self-identified race, and socioeconomic status (SES) measures, including the mother's highest level of education and annual household income (data not shown). The BBC and CHS study protocols were approved by the Boston University Medical Center institutional review board and the Children's Memorial Hospital institutional review board.
Determination of Self-identified Race
The race/ethnicity variable was based on mothers' self-reports of race, in keeping with the policy of the National Center for Health Statistics regarding reporting of infants' race.14 For this analysis, race was categorized as non-Hispanic black, non-Hispanic white, Hispanic, or “other.” More than one-half of the participants in our “other” group were of Afro-Caribbean origin. Given the small number of Asian participants (n = 21) and the wide difference in African ancestry between self-identified Asian participants and the majority of those in the “other” group, we excluded Asian subjects.
Determination of Genetic Ancestry
To determine global genetic ancestry, we genotyped 150 AIMs that were selected randomly from an admixture mapping panel15 to be highly informative among 3 ancestral populations (African, European, and Asian), with averaged δ (allele frequency difference between 2 ancestral populations) of >0.5. For our analysis, we estimated individual ancestral proportions with Structure 2.3.1 (University of Chicago, Chicago, IL) by using the 144 AIMs that had call rates of >98.0%. The variable for individual African ancestry was expressed in 10% increments, for ease of interpretation.
Assessment of Sensitization to Foods
At the 2-year visit, sIgE concentrations in plasma for each of 8 food allergens (egg white, cow's milk, peanut, soy, shrimp, walnut, wheat, and cod) were measured by using the ImmunoCAP test at Quest Diagnostics (Madison, NJ), according to the manufacturer's protocol (range: 0.35 to ≥100 kilo–units of allergen [kUA]/L). Sensitization was defined as sIgE levels of ≥0.35 kUA/L. The number of food sensitizations for each subject was categorized as 0 (reference), 1 or 2, or ≥3 foods. For allergen (egg, milk, and peanut) sIgE levels, values for children with undetectable levels were set at 0.175 kUA/L, which was one-half of the lower limit of detection (0.35 kUA/L), to allow logarithmic transformation. Peanut sIgE levels also were dichotomized at ≥5 kUA/L, a level associated with a greater likelihood of clinical reactivity among children 1 to 5 years of age.16 Milk and egg sIgE levels were dichotomized at 5 and 2 kUA/L, respectively, which corresponded to 95th percentile positive predictive values for children ≤2 years of age.16 These cutoff points, rather than those for children >5 years of age (15 kUA/L for milk and 7 kUA/L for egg), were chosen because of the ages of the children in the cohort. For the assessment of cutoff points, the control group included all children with levels below the cutoff points.
Definitions of Other Covariates
Mothers were considered atopic if they reported a physician diagnosis of 1 of the following conditions: atopic dermatitis, allergic rhinitis, or asthma. Maternal smoking during pregnancy and postnatal smoking were categorized on the basis of mothers' responses to questionnaires. Exclusive breastfeeding was determined on the basis of mothers' responses to questionnaires at the 6-month visit. SES was estimated according to the mother's highest level of education (categorized as less than high school, high school or general equivalency diploma, or some college) and annual household income (categorized as above or below $30 000 or not known).
Statistical Analyses
For comparison of baseline characteristics of the population according to self-identified race, analyses of variance and χ2 tests were used to characterize all continuous variables and all categorical variables, respectively. In parallel models, we examined the association of food sensitization with African ancestry and self-identified race by using multivariate logistic regression analyses. The basic models included the key determinant variables (race or ancestry), demographic measures (child's age and child's gender), and measures of SES (mother's age, mother's highest level of education, and annual household income). The second models evaluated other potential confounding variables from literature reports (cesarean delivery, firstborn child, breastfeeding, and maternal atopy) and confounders from our data (maternal prenatal and postnatal smoking), which were retained on the basis of significance (differentially distributed according to primary predictor; P < .2). For analysis of the number of foods to which children were sensitized, polytomous logistic regression for a categorical outcome (0 foods, 1 or 2 foods, or ≥3 foods) was used with the same covariates. For analyses of peanut, egg, and milk sIgE levels, Tobit regressions were used with the same covariates. All models for ancestry assumed 3 ancestral populations. All analyses were performed by using Stata for Windows 9.20 (Stata Corp, College Station, TX).
RESULTS
Baseline Characteristics of Cohort
For the 1104 children included in the sample, self-reported race was as follows: 60.9% black, 22.5% Hispanic, 5.9% white, and 10.8% other. This was a primarily urban sample, with 48.7% of subjects having an annual household income below $30 000 per year. There were approximately equal numbers of boys and girls, with a mean age of 2.65 years (SD: 2.23 years) at the time of assessment of sensitization through ImmunoCAP measurements. Also, as shown in Table 1, there was a wide range of African ancestry among each of the ethnic groups, with the exception of self-identified white subjects. In addition to food sensitization, a number of other characteristics varied according to race, including maternal smoking patterns, mother's highest level of education, annual household income, and child's age. Of these differentially distributed characteristics, only age was associated with food sensitization, as shown in Supplemental Table 6.
TABLE 1.
Baseline Characteristics of Cohort According to Self-reported Race
| Total Sample (N = 1104) | Black (n = 672) | Hispanic (n = 248) | White (n = 65) | Other (n = 119) | P | |
|---|---|---|---|---|---|---|
| Outcome variables | ||||||
| Food sensitization, n (%) | 393 (35.5) | 252 (37.5) | 83 (33.5) | 14 (21.5) | 43 (36.1) | .07 |
| No. of foods to which individual was sensitized, mean ± SD | 0.78 ± 1.37 | 0.85 ± 1.45 | 0.75 ± 1.39 | 0.4 ± 0.92 | 0.65 ± 1.02 | <.001 |
| Peanut sensitization, n (%) | 149 (13.4) | 106 (15.9) | 27 (11.0) | 6 (9.4) | 7 (5.9) | .009 |
| Milk sensitization, n (%) | 239 (21.9) | 154 (23.2) | 57 (23.4) | 5 (7.9) | 23 (19.5) | .04 |
| Egg sensitization, n (%) | 232 (21.4) | 141 (21.2) | 57 (23.7) | 7 (11.1) | 27 (22.9) | .18 |
| Descriptive variables | ||||||
| Preterm birth, n (%) | 314 (28.4) | 191 (28.4) | 73 (29.4) | 19 (29.2) | 31 (26.1) | .92 |
| Maternal postnatal smoking, n (%) | 172 (15.6) | 105 (15.6) | 28 (11.3) | 27 (41.5) | 12 (10.1) | <.0001 |
| Prenatal smoke exposure, n (%) | 111 (10.1) | 64 (9.5) | 14 (5.7) | 24 (36.9) | 9 (7.6) | <.0001 |
| Female, n (%) | 547 (49.6) | 332 (49.4) | 123 (49.6) | 37 (56.9) | 55 (46.2) | .58 |
| Child's age, mean ± SD, y | 2.65 ± 2.23 | 2.82 ± 2.32 | 2.36 ± 2.04 | 2.49 ± 2.39 | 2.42 ± 1.98 | .03 |
| Mother's age at delivery, mean ± SD, y | 28.5 ± 6.54 | 28.88 ± 6.69 | 27.25 ± 6.07 | 29.03 ± 5.65 | 28.69 ± 6.88 | .10 |
| Maternal atopy, n (%) | 393 (35.6) | 248 (37.1) | 81 (32.7) | 28 (43.1) | 35 (29.4) | .16 |
| Cesarean section delivery, n (%) | 366 (33.1) | 221 (32.9) | 83 (33.5) | 23 (35.4) | 39 (32.7) | .98 |
| Firstborn child, n (%) | 450 (40.8) | 273 (40.6) | 96 (38.7) | 41 (63.1) | 40 (33.6) | .001 |
| Exclusive breastfeeding, n (%) | 62 (5.6) | 29 (4.3) | 19 (7.7) | 5 (7.7) | 9 (7.6) | .14 |
| Mother's highest level of education, n (%) | ||||||
| Less than high school | 355 (30.3) | 169 (25.2) | 114 (46.0) | 13 (20.0) | 39 (32.8) | |
| High school | 404 (36.6) | 263 (39.1) | 76 (30.7) | 23 (35.4) | 42 (35.3) | |
| Some college | 365 (33.1) | 240 (35.7) | 58 (23.4) | 29 (44.6) | 38 (31.9) | <.0001 |
| Annual household income, n (%) | ||||||
| Less than $30 000 | 538 (48.7) | 336 (50.0) | 123 (49.6) | 27 (41.5) | 52 (43.7) | |
| At least $30 000 | 337 (30.5) | 197 (29.3) | 66 (26.6) | 31 (47.7) | 43 (36.1) | |
| Unknown | 229 (20.7) | 139 (20.7) | 59 (23.8) | 7 (10.8) | 24 (20.2) | .02 |
| Ancestry, median (interquartile range), % | ||||||
| European | 14.7 (0.4–46.8) | 6.9 (2.2–14.5) | 50.9 (40.3–62.1) | 94.9 (82.3–98.8) | 34.0 (5.6–51.3) | <.0001 |
| African | 77.7 (39.4–88.8) | 83.0 (78.5–93.3) | 21.7 (7.7–43.2) | 1.1 (0.6–8.2) | 58.2 (39.7–86.1) | <.0001 |
| Asian | 5.1 (2.5–10.9) | 4.4 (2.2–8.3) | 14.8 (5.5–39.3) | 2.6 (1.3–7.7) | 4.1 (2.5–8.6) | <.0001 |
All P values represent differences according to race. P values for categorical variables were determined with χ2 statistics. All P values for continuous variables were determined with analyses of variance.
Race, Ancestry, and Food Sensitization
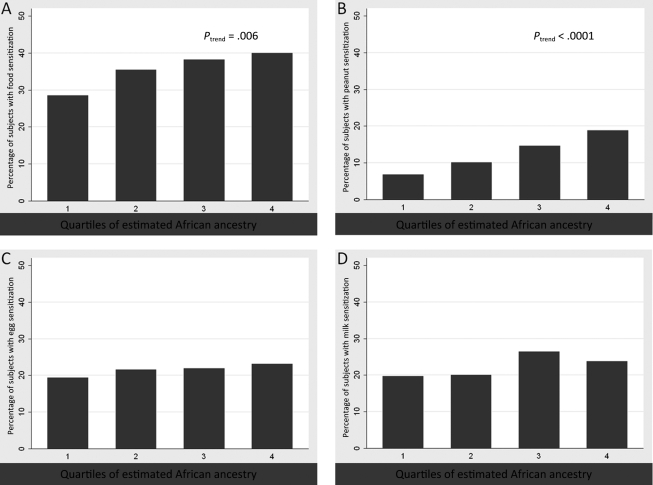
In both initial models including SES measures, self-identified black race (odds ratio [OR]: 2.22 [95% confidence interval [CI]: 1.20–4.11]) and African ancestry, expressed in 10% increments (OR: 1.07 [95% CI: 1.02–1.13]), were associated with food sensitization (Table 2). With inclusion of previously known confounders (maternal atopy, cesarean delivery, firstborn status, and exclusive breastfeeding) and potential confounders from our data (prenatal smoke exposure and maternal postnatal smoking), the ORs for self-identified race (OR: 2.34 [95% CI: 1.24–4.44]) and African ancestry (OR: 1.07 [95% CI: 1.02–1.14]) remained unchanged in magnitude and significance. The association of African ancestry and proportions of subjects with food sensitization is presented in Fig 1A. To evaluate the nature of the relationship at different levels of ancestry, African ancestry is depicted in quartiles in Fig 1. The associations with peanut, egg, and milk are shown in Fig 1, B, C, and D, respectively. It was notable that the association was strongest for sensitization to peanut.
TABLE 2.
Ancestry, Race, and Presence of Food Sensitization
| Model With Self-reported Race |
Model With Genetic Ancestry |
Model With Self-reported Race and Confoundersa |
Model With Genetic Ancestry and Confoundersa |
|||||
|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | |
| Self-reported race (reference: white) | ||||||||
| Black | 2.22 (1.20–4.11) | .01 | 2.34 (1.24–4.44) | .009 | ||||
| Hispanic | 1.84 (0.95–3.56) | .07 | 1.97 (0.99–3.92) | .05 | ||||
| Other | 2.12 (1.04–4.29) | .04 | 2.29 (1.10–4.75) | .03 | ||||
| African ancestry | 1.07 (1.02–1.13) | .01 | 1.07 (1.02–1.14) | .009 | ||||
Ancestry was expressed in 10% increments.
This model included potential confounders considered on the basis of literature reports (maternal atopy, cesarean section, firstborn status, and exclusive breastfeeding), irrespective of significance, and variables found to be potential confounders of the association of race with any food sensitization in our data (prenatal smoke exposure and maternal postnatal smoking).
FIGURE 1.
Quartiles of African ancestry and food sensitization. The y-axes represent the percentage of children with food sensitization in each group, and the x-axes represent the subjects grouped according to quartile of African ancestry, as estimated with AIMs. The quartiles for African ancestry were as follows: quartile 1, 0%–39.35%; quartile 2, 39.36%–77.70%; quartile 3, 77.71%–88.80%; quartile 4, 88.81%–100%. A, Any food sensitization; B, peanut; C, egg; D, milk.
Race, Ancestry, and Number of Food Sensitizations
In Table 3, we examine the association of race and African ancestry with the number of foods to which an child was sensitized, by using 2 separate polytomous logistic regressions. Self-identification as black was associated with ORs of 2.03 (95% CI: 1.01–4.10) for sensitization to 1 or 2 foods and 3.76 (95% CI: 1.09–12.97) for sensitization to ≥3 foods. Similarly, each 10% increment in African ancestry was associated with ORs of 1.04 (95% CI: 0.98–1.10) for 1 or 2 foods and 1.19 (95% CI: 1.07–1.32) for ≥3 foods.
TABLE 3.
Ancestry, Race, and Number of Foods to Which Individual Was Sensitized
| Models With Self-reported Race |
Models With Genetic Ancestry |
|||
|---|---|---|---|---|
| OR (95% CI) | P | OR (95% CI) | P | |
| 1 or 2 foods | ||||
| Self-reported race | ||||
| Black | 2.03 (1.01–4.10) | .047 | ||
| Hispanic | 1.77 (0.84–3.77) | .14 | ||
| Other | 2.18 (0.98–4.84) | .06 | ||
| African ancestry | 1.04 (0.98–1.10) | .19 | ||
| ≥3 foods | ||||
| Self-reported race | ||||
| Black | 3.76 (1.09–12.97) | .04 | ||
| Hispanic | 2.87 (0.77–10.66) | .12 | ||
| Other | 2.71 (0.66–11.10) | .17 | ||
| African ancestry | 1.19 (1.07–1.32) | .001 | ||
All regression analyses were based on polytomous regression with outcome categories of 0, 1 or 2, or ≥3. Ancestry was expressed in 10% increments. The models included potential confounders considered on the basis of literature reports (maternal atopy, cesarean section, firstborn status, and exclusive breastfeeding), irrespective of significance, and variables found to be potential confounders of the association of race with any food sensitization in our data (prenatal smoke exposure and maternal postnatal smoking).
Race, Ancestry, and Allergen sIgE Levels
In Table 4, we present the results of our secondary analyses, including the associations of self-identified black race and African ancestry with logarithmically transformed peanut, egg, and milk sIgE levels. Self-reported black race was associated with increased logarithmically transformed sIgE levels for egg (β: 1.90 [95% CI: 0.60–3.19]) and milk (β: 1.69 [95% CI: 0.27–3.11]) and showed a similar direction of effect for peanut, although that was not significant (β: 1.34 [95% CI: −0.63 to 3.31]). African ancestry was associated only with an increase in logarithmically transformed peanut sIgE levels (for each 10% increment in African ancestry, β: 0.48 [95% CI: 0.25–0.72]).
TABLE 4.
Associations With sIgE Levels
| Peanut |
Milk |
Egg |
||||
|---|---|---|---|---|---|---|
| β (95% CI) | P | β (95% CI) | P | β (95% CI) | P | |
| Self-reported race model | ||||||
| Black | 1.34 (−0.63 to 3.31) | .18 | 1.90 (0.60 to 3.19) | .004 | 1.69 (0.27 to 3.11) | .02 |
| Hispanic | 0.33 (−1.81 to 2.48) | .76 | 2.08 (0.71 to 3.44) | .003 | 1.91 (0.41 to 3.41) | .01 |
| Other | −1.03 (−3.52 to 1.46) | .42 | 1.48 (0.04 to 2.92) | .04 | 1.76 (0.16 to 3.35) | .03 |
| African ancestry model | 0.48 (0.25 to 0.72) | <.0001 | 0.02 (−0.07 to 0.12) | .61 | 0.08 (−0.03 to 0.19) | .15 |
All models for the logarithm of allergen sIgE levels were Tobit models based on truncation of data at the lower level of detection for sIgE. Levels of sIgE to allergen were expressed on a logarithmic scale. The model included demographic and SES variables as described for the other tables. The model also included potential confounders considered on the basis of literature reports (maternal atopy, cesarean section, firstborn status, and exclusive breastfeeding), irrespective of significance, and variables found to be potential confounders of the association of race with any food sensitization in our data (prenatal smoke exposure and maternal postnatal smoking). Ancestry was expressed in 10% increments.
In Table 5, we present the associations of self-identified race and ancestry with levels of sIgE associated with clinical reactivity. We found no association of black race with peanut sIgE levels of ≥5 kUA/L. Of note, the total number of children with this phenotype (n = 37) in the cohort was small. Given the even smaller number of white children (reference group) with levels above these cutoff points for egg and milk, we report the associations for a combined variable of sIgE level above the cutoff points for clinical reactivity for any 1 of egg, milk, or peanut. Again, there was no association of sensitization beyond these levels with self-identified race. However, African ancestry was associated with peanut sIgE levels of ≥5 kUA/L. Specifically, with each 10% increase in African ancestry in adjusted analyses, there was a 25% increase in the likelihood of peanut sIgE levels being ≥5 kUA/L. Similarly, for each 10% increase in African ancestry in adjusted analyses, there was a 17% increase in the likelihood of sIgE levels being beyond the cutoff point for any 1 of milk, egg, or peanut.
TABLE 5.
Likelihood of sIgE Levels Above Clinical Cutoff Points Suggesting Reactivity
| ≥5 kUA/L Peanut Cutoff Point |
≥5 kUA/L Milk, ≥2 kUA/L Egg, or ≥5 kUA/L Peanut Cutoff Point |
|||
|---|---|---|---|---|
| OR (95% CI) | P | OR (95% CI) | P | |
| Self reported race model | ||||
| Black | 0.68 (0.19–2.50) | .56 | 1.93 (0.66–5.66) | .23 |
| Hispanic | 0.48 (0.11–2.12) | .33 | 1.35 (0.42–4.31) | .51 |
| Other | 0.31 (0.05–2.01) | .21 | 1.16 (0.32–4.17) | .82 |
| African ancestry model | 1.25 (1.01–1.52) | .03 | 1.17 (1.06–1.31) | .003 |
All models for the logarithm of allergen sIgE levels were Tobit models based on truncation of data at the lower level of detection for sIgE. Levels of sIgE to allergen were expressed on a logarithmic scale. The model included demographic and SES variables as described for the other tables. The model also included potential confounders considered on the basis of literature reports (maternal atopy, cesarean section, firstborn status, and exclusive breastfeeding), irrespective of significance, and variables found to be potential confounders of the association of race with any food sensitization in our data (prenatal smoke exposure and maternal postnatal smoking). Ancestry was expressed in 10% increments.
Although we could not evaluate the effect of self-identified race on the likelihood of sIgE levels above those predictive of clinical reactivity for egg and milk individually, we were able to evaluate the effect of ancestry. For each 10% increase in African ancestry in adjusted analyses, the likelihoods of milk sIgE levels of ≥5 kUA/L (OR: 1.24 [95% CI: 0.94–1.63]) and egg sIgE levels of ≥2 kUA/L (OR: 1.13 [95% CI: 1.01–1.27]) were of similar direction and magnitude, compared with findings shown in Table 5 for peanut, although the results for milk were not significant.
DISCUSSION
In this urban, multiethnic, birth cohort, we found that self-identified black ethnicity was associated with a more than twofold increased likelihood of sensitization to foods, as well as sensitization to a greater number of foods, with controlling for both SES and early-life atopy-associated factors. African ancestry also was positively associated with the probability of sensitization to food and with a greater likelihood of being sensitized to multiple (≥3) foods. Lastly, although we did not corroborate recent findings in which peanut sensitization of ≥5 kUA/L was more common among black children (on the basis of self-reported race),4 we found that each 10% increment in African ancestry was associated with a 24% to 25% increase in the likelihood of having a peanut sIgE level beyond a cutoff point associated with increased likelihood of clinical reactivity.16
The results of our study are in keeping with recent findings that black children were at higher risk of both food allergy and peanut sensitization in 2 national cohorts.1,4 Although prospective studies with large, population-based, European cohorts have evaluated risk factors for food allergy,17,18 our study and the National Health and Nutrition Examination Survey study1 are the first multiethnic US studies. Furthermore, our study and the National Health and Nutrition Examination Survey were conducted with cohorts not selected according to high-risk status, which allows for better evaluation of the effects of race and ethnicity on food sensitization.
Study of the association of genetic ancestry with food sensitization is especially important for the US population. According to National Center for Health Statistics regulations,14 infant race is recorded as the mother's race. With more interracial people and marriages, traditional ethnic classifications will become increasingly imprecise, compared with genetic ancestry. Furthermore, as demonstrated in the recent US Census,8,19 there are increasing proportions of minority groups with admixed ancestral origins in the US population, particularly black20–22 and Hispanic23,24 groups. Self-identified race for these populations would underestimate the degree of genetic variation within these populations. Genetic factors influence the development of food allergy.25 As has been shown with susceptibility alleles for multiple other diseases,7 genetic factors that affect food sensitization may be distributed differently across ancestral groups. The association of ancestry with food sensitization is consistent with the findings of studies that found associations with other atopic diseases, such as asthma, and total IgE levels.26,27 The presence of ancestry associations across atopic diseases may represent population differences in genotypes associated with IgE production.28
In our sample, self-identified race was not associated with increased likelihood of peanut sensitization or peanut sIgE levels of ≥5 kUA/L. This may be attributable to several possible factors. Our sample was not selected on the basis of a history of milk or egg allergy, compared with the aforementioned study by Sicherer et al.4 It also is possible that, with an increase in sample size, particularly with more white subjects, our study might have greater power to detect an association with self-identified race and peanut allergy phenotypes.
It was interesting that African ancestry but not self-identified black race was associated with peanut sIgE levels. The opposite was seen for milk and egg. These different associations with ancestry are clearly seen in Fig 1, in which milk does not display a pattern of association similar to seen for sensitization in general or sensitization to peanut or egg. One potential explanation for these discrepancies is that there may be ethnocultural factors, such as differences in diet and early-life feeding practices, that may have more relevance than genetic factors for milk sensitization. In contrast, genetic factors associated with ancestry may have more relevance for peanut sensitization, which was found to be greater among black children in 2 other studies.1,4 However, it is important to clarify that the associations with ancestry do not necessarily represent racially distributed genetic factors.29 It is possible that these associations are attributable to environmental factors that vary with ancestry and influence sensitization, such as early-life dietary patterns and vitamin D levels.30–34
Our study has several limitations. First, it is possible that some residual confounding from subtle socioeconomic factors or unmeasured early-life exposures remains. However, it would be harder to attribute the linear association of African ancestry with sensitization rates to this type of residual confounding. Second, our ancestry variable was derived from a panel of 150 AIMs and not genome-wide association study data. Panels of AIMs smaller than ours are sufficient to control for population stratification attributable to African ancestry,35 and accuracy similar to that of ancestry estimates from genome-wide association study data may be obtained with a dramatically smaller number of AIMs.36 Furthermore, ancestry has been used as a covariate across multiple self-identified ethnicities for multiethnic populations,37,38 and the results were shown to be highly correlated with the results of a principal component-based analysis.37 Third, this study involves a young cohort. It remains to be determined whether these disparities in food sensitization will persist as the children grow older and whether they will translate into disparities in clinical reactivity to food. However, the children had a mean age of 2.7 years. Before 1 year of age there can be some cyclical variations in sIgE levels.39 The mean age of the cohort makes these findings more likely to be representative of persistent sensitization.
The clinical relevance of sensitization at a level of ≥0.35 kUA/L to any of the foods tested needs to be considered. The association with being multiply sensitized was stronger for both self-identified race and African ancestry. Another group that found African ancestry generally increased total IgE levels.27 Although it is possible that African ancestry simply may increase sIgE levels beyond the level of detection for a number of foods, we also found associations with peanut sIgE levels of ≥5 kUA/L. This suggests that race and ancestry not only are associated with minimal increases in sIgE levels but also may be associated with clinical reactivity, in keeping with the findings of 2 other studies that evaluated race and food allergy.1,4 However, it should be noted that these cutoff points (which currently are used clinically regardless of patient ethnicity) were established with primarily white populations. Other populations might have different predictive values. In National Health Interview Survey data, black people had higher rates of detectable sIgE but did not report food allergy at higher rates.40 Although this discrepancy may represent differences in reporting, additional rigorous study of food allergy in minority populations is necessary.
CONCLUSIONS
In this urban birth cohort, we found that black race and African ancestry were associated with increased risk of food sensitization, particularly to peanut. It remains to be determined whether this disparity is associated with increased rates of clinical reactivity, but the association of African ancestry with peanut sIgE levels of ≥5 kUA/L is provocative. This study emphasizes the need for continued follow-up monitoring of this cohort to determine the association of African ancestry with the development of clinical food allergies. Furthermore, additional studies to explore environmental and genetic factors that vary with African ancestry may provide new insights into the causes of racial disparities in food sensitization and food allergy.
ACKNOWLEDGMENTS
The parent study was supported in part by the March of Dimes (Perinatal Epidemiological Research Initiative grants 20-FY02-56 and 21-FY07-605) and National Institutes of Health grants R21ES011666 and R01HD041702. The follow-up study was supported in part by the Food Allergy Initiative, the Sunshine Charitable Foundation, National Institutes of Health grants R21AI079872 and U01AI090727, and Department of Defense grant W81XWH-10-1-0123. Dr Kumar was supported by National Institutes of Health grant K23HL093023. Dr Liu was supported by National Institutes of Health Clinical and Translational Science Award KL2RR025740 (to Northwestern University) and grant R21AI087888.
FINANCIAL DISCLOSURE: The authors have indicated they have no financial relationships relevant to this article to disclose.
Funded by the National Institutes of Health (NIH)
- AIM
- ancestry informative marker
- BBC
- Boston Birth Cohort
- CHS
- Children's Health Study
- UA
- units of allergen
- SES
- socioeconomic status
- IgE
- immunoglobulin E
- sIgE
- specific immunoglobulin E
- OR
- odds ratio
- CI
- confidence interval
Supplementary Material
Supplemental Table 6 (PDF)
REFERENCES
- 1. Liu AH, Jaramillo R, Sicherer SH, et al. National prevalence and risk factors for food allergy and relationship to asthma: results from the National Health and Nutrition Examination Survey 2005–2006. J Allergy Clin Immunol. 2010;126(4):798–806.e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rona RJ, Keil T, Summers C, et al. The prevalence of food allergy: a meta-analysis. J Allergy Clin Immunol. 2007;120(3):638–646 [DOI] [PubMed] [Google Scholar]
- 3. Boyce JA, Assa'ad A, Burks AW, et al. Guidelines for the diagnosis and management of food allergy in the United States: report of the NIAID-sponsored expert panel. J Allergy Clin Immunol. 2010;126(6 suppl):S1–S58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sicherer SH, Wood RA, Stablein D, et al. Maternal consumption of peanut during pregnancy is associated with peanut sensitization in atopic infants. J Allergy Clin Immunol. 2010;126(6):1191–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sicherer SH, Munoz-Furlong A, Godbold JH, Sampson HA. US prevalence of self-reported peanut, tree nut, and sesame allergy: 11-year follow-up. J Allergy Clin Immunol. 2010;125(6):1322–1326 [DOI] [PubMed] [Google Scholar]
- 6. Sicherer SH, Munoz-Furlong A, Sampson HA. Prevalence of seafood allergy in the United States determined by a random telephone survey. J Allergy Clin Immunol. 2004;114(1):159–165 [DOI] [PubMed] [Google Scholar]
- 7. Rotimi CN, Jorde LB. Ancestry and disease in the age of genomic medicine. N Engl J Med. 2010;363(16):1551–1558 [DOI] [PubMed] [Google Scholar]
- 8. US Census Bureau Annual estimates of the resident population by sex, race, and Hispanic origin for the United States: April 1, 2000 to July 1, 2008. Available at: www.census.gov/popest/national/asrh/NC-EST2008/NC-EST2008-03.xls Accessed June 6, 2011
- 9. Nalls MA, Wilson JG, Patterson NJ, et al. Admixture mapping of white cell count: genetic locus responsible for lower white blood cell count in the Health ABC and Jackson Heart studies. Am J Hum Genet. 2008;82(1):81–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kumar R, Seibold MA, Aldrich MC, et al. Genetic ancestry in lung-function predictions. N Engl J Med. 2010;363(4):321–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Peralta CA, Ziv E, Katz R, et al. African ancestry, socioeconomic status, and kidney function in elderly African Americans: a genetic admixture analysis. J Am Soc Nephrol. 2006;17(12):3491–3496 [DOI] [PubMed] [Google Scholar]
- 12. Wang X, Zuckerman B, Pearson C, et al. Maternal cigarette smoking, metabolic gene polymorphism, and infant birth weight. JAMA. 2002;287(2):195–202 [DOI] [PubMed] [Google Scholar]
- 13. Kumar R, Yu Y, Story RE, et al. Prematurity, chorioamnionitis, and the development of recurrent wheezing: a prospective birth cohort study. J Allergy Clin Immunol. 2008;121(4):878–884.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. US Office of Management and Budget Revisions to the Standards for the Classification of Federal Data on Race and Ethnicity. Washington, DC: US Office of Management and Budget; 1997 [Google Scholar]
- 15. Price AL, Patterson N, Yu F, et al. A genomewide admixture map for Latino populations. Am J Hum Genet. 2007;80(6):1024–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sicherer SH, Sampson HA. Food allergy. J Allergy Clin Immunol. 2010;125(2 suppl 2):S116–S125 [DOI] [PubMed] [Google Scholar]
- 17. Lack G, Fox D, Northstone K, Golding J; Avon Longitudinal Study of Parents and Children Study Team Factors associated with the development of peanut allergy in childhood. N Engl J Med. 2003;348(11):977–985 [DOI] [PubMed] [Google Scholar]
- 18. Tariq SM, Stevens M, Matthews S, Ridout S, Twiselton R, Hide DW. Cohort study of peanut and tree nut sensitisation by age of 4 years. BMJ. 1996;313(7056):514–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. US Census Bureau National Population Estimates, Characteristics, National Demographic Components of Change by Hispanic or Latino Origin. Washington, DC: US Census Bureau; 2003. Available at: www.census.gov/popest/archives/2000s/vintage_2002/NA-EST2002-ASRO-05.html Accessed August 20, 2011 [Google Scholar]
- 20. Halder I, Yang BZ, Kranzler HR, Stein MB, Shriver MD, Gelernter J. Measurement of admixture proportions and description of admixture structure in different U.S. populations. Hum Mutat. 2009;30(9):1299–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Parra EJ, Kittles RA, Argyropoulos G, et al. Ancestral proportions and admixture dynamics in geographically defined African Americans living in South Carolina. Am J Phys Anthropol. 2001;114(1):18–29 [DOI] [PubMed] [Google Scholar]
- 22. Yaeger R, Avila-Bront A, Abdul K, et al. Comparing genetic ancestry and self-described race in African Americans born in the United States and in Africa. Cancer Epidemiol Biomarkers Prev. 2008;17(6):1329–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Choudhry S, Seibold MA, Borrell LN, et al. Dissecting complex diseases in complex populations: asthma in Latino Americans. Proc Am Thorac Soc. 2007;4(3):226–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lai CQ, Tucker KL, Choudhry S, et al. Population admixture associated with disease prevalence in the Boston Puerto Rican Health Study. Hum Genet. 2009;125(2):199–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hong X, Tsai HJ, Wang X. Genetics of food allergy. Curr Opin Pediatr. 2009;21(6):770–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Choudhry S, Burchard EG, Borrell LN, et al. Ancestry-environment interactions and asthma risk among Puerto Ricans. Am J Respir Crit Care Med. 2006;174(10):1088–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vergara C, Caraballo L, Mercado D, et al. African ancestry is associated with risk of asthma and high total serum IgE in a population from the Caribbean coast of Colombia. Hum Genet. 2009;125(5–6):565–579 [DOI] [PubMed] [Google Scholar]
- 28. Vergara C, Tsai YJ, Grant AV, et al. Gene encoding Duffy antigen/receptor for chemokines is associated with asthma and IgE in three populations. Am J Respir Crit Care Med. 2008;178(10):1017–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Adeyemo A, Rotimi C. Genetic variants associated with complex human diseases show wide variation across multiple populations. Public Health Genomics. 2010;13(2):72–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brehm JM, Schuemann B, Fuhlbrigge AL, et al. Serum vitamin D levels and severe asthma exacerbations in the Childhood Asthma Management Program study. J Allergy Clin Immunol. 2010;126(1):52–58.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Freishtat RJ, Iqbal SF, Pillai DK, et al. High prevalence of vitamin D deficiency among inner-city African American youth with asthma in Washington, DC. J Pediatr. 2010;156(6):948–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Miyake Y, Sasaki S, Tanaka K, Hirota Y. Dairy food, calcium and vitamin D intake in pregnancy, and wheeze and eczema in infants. Eur Respir J. 2010;35(6):1228–1234 [DOI] [PubMed] [Google Scholar]
- 33. Searing DA, Zhang Y, Murphy JR, Hauk PJ, Goleva E, Leung DY. Decreased serum vitamin D levels in children with asthma are associated with increased corticosteroid use. J Allergy Clin Immunol. 2010;125(5):995–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Signorello LB, Williams SM, Zheng W, et al. Blood vitamin D levels in relation to genetic estimation of African ancestry. Cancer Epidemiol Biomarkers Prev. 2010;19(9):2325–2331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kosoy R, Nassir R, Tian C, et al. Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Hum Mutat. 2009;30(1):69–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tian C, Gregersen PK, Seldin MF. Accounting for ancestry: population substructure and genome-wide association studies. Hum Mol Genet. 2008;17(R2):R143–R150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang H, Haiman CA, Kolonel LN, et al. Self-reported ethnicity, genetic structure and the impact of population stratification in a multiethnic study. Hum Genet. 2010;128(2):165–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tsai H-J, Yu Y, Zhang S, et al. Association of genetic ancestry with preterm delivery and related traits among African American mothers. Am J Obstet Gynecol. 2009;201:94.e1–e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Holt PG, Rowe J, Kusel M, et al. Toward improved prediction of risk for atopy and asthma among preschoolers: a prospective cohort study. J Allergy Clin Immunol. 2010;125(3):653–659, 659.e1–659.e7 [DOI] [PubMed] [Google Scholar]
- 40. Branum AM, Lukacs SL. Food allergy among children in the United States. Pediatrics. 2009;124(6):1549–1555 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 6 (PDF)