Abstract
We present here the description of genes coding for molluscan hemocyanins. Two distantly related mollusks, Haliotis tuberculata and Octopus dofleini, were studied. The typical architecture of a molluscan hemocyanin subunit, which is a string of seven or eight globular functional units (FUs, designated a to h, about 50 kDa each), is reflected by the gene organization: a series of eight structurally related coding regions in Haliotis, corresponding to FU-a to FU-h, with seven highly variable linker introns of 174 to 3,198 bp length (all in phase 1). In Octopus seven coding regions (FU-a to FU-g) are found, separated by phase 1 introns varying in length from 100 bp to 910 bp. Both genes exhibit typical signal (export) sequences, and in both cases these are interrupted by an additional intron. Each gene also contains an intron between signal peptide and FU-a and in the 3′ untranslated region. Of special relevance for evolutionary considerations are introns interrupting those regions that encode a discrete functional unit. We found that five of the eight FUs in Haliotis each are encoded by a single exon, whereas FU-f, FU-g, and FU-a are encoded by two, three and four exons, respectively. Similarly, in Octopus four of the FUs each correspond to an uninterrupted exon, whereas FU-b, FU-e, and FU-f each contain a single intron. Although the positioning of the introns between FUs is highly conserved in the two mollusks, the introns within FUs show no relationship either in location nor phase. It is proposed that the introns between FUs were generated as the eight-unit polypeptide evolved from a monomeric precursor, and that the internal introns have been added later. A hypothesis for evolution of the ring-like quaternary structure of molluscan hemocyanins is presented.
Hemocyanins are extracellular oxygen transport proteins found in two animal phyla—Arthropoda and Mollusca. Although the hemocyanins of both phyla use binuclear copper sites for oxygen binding, they exhibit substantial differences in tertiary and quaternary structure. Whereas arthropod hemocyanins function as multisubunit aggregates of protomers of about 75 kDa, molluscan hemocyanins are built from very much larger polypeptide chains, ranging from 350 to 450 kDa in mass. Each such molluscan polypeptide chain is comprised of seven or eight different functional units (FUs), connected by short “linker” peptides. Each FU carries a pair of copper atoms, which serve to bind an oxygen molecule. Each copper is liganded by three histidine residues, forming a copper A site and a copper B site in each FU (1–4). Recently, we and our collaborators reported the sequence of the polypeptide subunit of the cephalopod mollusk Octopus dofleini (2), the 2.3-Å tertiary structure of the C-terminal FU of this subunit (3), the polypeptide subunit sequence of the gastropod mollusk Haliotis tuberculata (4), and a 12-Å reconstruction of the quaternary structure of this hemocyanin (5). The polypeptide chain of Octopus hemocyanin (OdH) was found to contain 2,896 aa residues, divided into seven FUs, denoted OdH-a to OdH-g (from N to C terminus). The polypeptide chain of Haliotis hemocyanin (HtH) (isoform 1) consists of a corresponding set of FUs (termed HtH1-a to HtH1-g), plus an additional C-terminal unit (HtH1-h) that carries a specific tail extension of about 95 aa; in total, the protein subunit of HtH1 comprises 3,404 aa. The various FUs exhibit considerable similarity (≈45% protein sequence identity) within as well as between the two mollusks; units in corresponding positions have ≈55% identity.
On the other hand, comparison of either of these molluscan protein sequences with those determined for subunits of arthropod hemocyanins (1, 6, 7) reveals only small similarity, mainly in the region of one of the two copper binding sites, the B site. The sequence surrounding the other site (the A site) resembles more closely that found in tyrosinases (1, 2, 8, 9). A similar conclusion is reached from comparison of the tertiary structure of the Octopus FU OdH-g with the results from x-ray diffraction studies of several arthropod hemocyanin subunits (3). Nonetheless the overall geometry of the six histidine copper ligands that together hold the two copper atoms is startlingly similar in the two phyla. These observations raise important questions concerning the evolution of these two classes of respiratory proteins and of the phylogenetic relationships between arthropods and mollusks. Current views would place these two phyla (once held to be closely related) in entirely different superphyla—the arthropods as Ecdysozoans, the mollusks as Lophotrochozoans (see, for example ref. 10). The limited similarities between the hemocyanins of mollusks and arthropods then would suggest their independent evolution, following quite different paths, from a (presumably monomeric) ancestral copper protein (1, 6, 8, 9, 11), perhaps tyrosinase or phenoloxidase (12).
There is much more potential evolutionary information in genes than in the corresponding cDNA. The sequence of one arthropod hemocyanin gene has been published (13), but no corresponding published data exists for the molluscan hemocyanins. Therefore, it seemed a logical next step to follow the cDNA sequencing of HtH and OdH with determination of the sequences of their genes. In sequencing the cDNA corresponding to the HtH, the two distinct isoforms HtH1 and HtH2 previously described at the protein level were found (14, 15); counterparts termed KLH1 and KLH2 have been studied in detail in another gastropod, the keyhole limpet Megathura crenulata (16), and shown to exhibit differential regulation. The hemocyanin isoforms HtH1 and HtH2 share only ≈65% sequence identity (4), and therefore their genes could be easily distinguished. Here we present only the data on HtH1, as an example of a gastropod hemocyanin gene sequence. Also in Octopus dofleini, two variants of the hemocyanin sequence, referred to as the A and G forms (OdHA and OdHG), were found (17), but they are very similar (≈96%), in contrast to the gastropod hemocyanin isoforms. In this report, we present only the data for OdHG, as an example of a cephalopod hemocyanin gene sequence.
Materials and Methods
Animals.
The European abalone H. tuberculata (Archaeogastropoda) was a gift from the Syndicate Mixte d' Equipment du Littoral, Blainville sur Mer, France. The Pacific octopus, O. dofleini, was obtained at the Oregon State University Marine Science Center, Newport, OR.
Isolation and Analysis of Genomic DNA from Haliotis.
Genomic DNA was isolated from two mixed tissues (heart, mantle, 250 mg) of a single adult abalone by using the Stratagene genomic-DNA isolation kit according to the manufacturer's instructions. PCR was performed by using more than 50 different specific oligonucleotides (Genaxis, Spechbach, Germany) derived from the cDNA coding for HtH1. For generating PCR fragments up to 2.5 kbp, a standard three-step PCR protocol with Taq polymerase (GIBCO) was applied (94°C for 20 s; 55°C for 30 s; 72°C for 120 s; 35 cycles). For larger fragments, we used the Expand-PCR system (Roche Diagnostics) according to the manufacturer's instructions. PCR fragments were analyzed in standard agarose gels in 1× TBE (89 mM Tris/chloride/89 mM sodium borate/2 mM sodium EDTA, pH 7.5), purified by the gel extraction kit from Qiagen (Chatsworth, CA), and directly sequenced (Seqlab, Göttingen, Germany), using the PCR oligonucleotides as primers. Alternatively, cloning in pGemT easy (Promega) was performed, and positive clones were subsequently sequenced with standard and/or gene-specific primers. The sequences obtained were computer-analyzed as described (4).
Isolation and Analysis of Genomic DNA from Octopus.
Genomic DNA was isolated from O. dofleini tissues (brain, branchial gland, and gonad) of an adult octopus by using the Qiagen DNEasy kit. Fragments of the hemocyanin gene were obtained from the total genomic DNA by using PCR (PTC-150 minicycler, MJ Research, Cambridge, MA). Primers for PCR were designed whenever possible from the cDNA sequences for this hemocyanin. Oligonucleotide primers were synthesized by the Oregon State University Central Services Laboratory. The DNA used was isolated from an animal that had both A and G type hemocyanins. Because the sequence differences between these two forms are small, care was taken to have redundant overlaps. Some of the primers were A and G type-specific, but in other cases it was necessary to clone the PCR products into the cloning vectors pTrc His2-TOPO or pCR4-TOPO (Invitrogen) to obtain clones that represented homogenous (A or G) products. PCR products or sequences within vectors were sequenced in the Oregon State University Central Services Laboratory. macvector (version 5.0.2) and assemblylign (version 1.0.9b) (Oxford Molecular, Oxford) were used to produce sequence contigs and alignments.
Results
The sequences of the HtH1 gene and the OdHG gene were determined. Fig. 1 gives a schematic picture of the gene structure; for more details, see Fig. 5, which is published as supplemental material on the PNAS web site, www.pnas.org. Exon lengths are given in Table 1. The complete nucleotide and protein sequences of both genes can be found through GenBank. Both coding sequences begin with a typical “signal” peptide. A stop codon is observed near the 3′ end of the terminal exon in each gene (TAG in Haliotis and TAA in Octopus). As usually observed, the sequence AATAAA also is found before the poly(A) attachment site. As Fig. 1 shows, two types of introns can be recognized in these genes. Introns in the linker region between two functional units will be referred to as linker introns. Introns within a functional unit will be called internal introns. All but one of the splice junctions follow the GT/AG rule for donor/acceptor sites (18); the exception is the intron in the Octopus 3′ untranslated region (UTR), which begins with AT. Linker introns invariably start with a strictly conserved GT sequence, and in most cases stop with a conserved CAG triplet. Exceptions are intron HtH1(a4/b), which is TAG, and introns OdHG(c/d) and OdHG(f2/g), which are AAG. Details are illustrated in Table 2, which is published as supplemental material. All linker introns in both organisms show phase 1. In both organisms the same is true for the intron between signal peptide and FU-a and the introns found within the 3′UTR. In contrast, the nine internal introns (six in Haliotis and three in Octopus) exist in all three phases.
Figure 1.
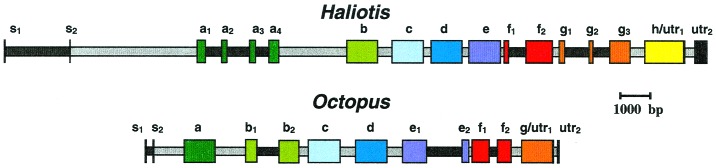
Gene structures of HtH1 and OdHG. Exons encoding FUs (a–h) are shown in different colors; introns (gray for linker introns, black for internal introns) are shown to scale. s1 and s2 correspond to the two exons encoding each signal sequence. utr indicates the 3′ UTR.
Table 1.
Size comparison of the different exons encoding HtH1 and OdHG
|
Haliotis
|
Octopus
|
||||
|---|---|---|---|---|---|
| Exon | bp | aa | Exon | bp | aa |
| s1 | 32* | s1 | 36 | ||
| s2 | 16 | s2 | 28 | ||
| s1–s2 | 48 | 15* | s1–s2 | 64 | 21 |
| a1 | 350 | ||||
| a2 | 221 | ||||
| a3 | 255 | ||||
| a4 | 407 | ||||
| a1–a4 | 1233 | 411 | a | 1233 | 411 |
| b1 | 463 | ||||
| b2 | 782 | ||||
| b | 1245 | 415 | b1–b2 | 1245 | 415 |
| c | 1242 | 414 | c | 1242 | 414 |
| d | 1239 | 413 | d | 1248 | 416 |
| e1 | 1019 | ||||
| e2 | 241 | ||||
| e | 1260 | 420 | e1–e2 | 1260 | 420 |
| f1 | 191 | f1 | 659 | ||
| f2 | 1060 | f2 | 583 | ||
| f1–f2 | 1251 | 417 | f1–f2 | 1242 | 414 |
| g1 | 219 | ||||
| g2 | 164 | ||||
| g3 | 826 | ||||
| g1–g3 | 1209 | 403 | g | 1220 | 407 |
| h | 1535 | 511 | — | — | — |
Exon size is given in bp and aa; the total numbers per FU also are shown. For sequence identities between the FUs in percent, see ref. 4. * indicates that the signal sequence is still incomplete.
Databank searches using only intron sequences showed no significant homologies to transposable elements, ORFs for (regulatory) peptides or tRNA genes. The introns HtH1(s1/s2) and HtH1(g1/g2) both contain a (GT)n-rich microsatellite. A prominent microsatellite region also is found within the internal intron OdHG(e1/e2). This microsatellite is unique in containing quite long (ACAT)n, (GT)n, and (AT)n segments, with n as large as 40 in the latter case.
Discussion
Comparison of the Two Molluscan Hemocyanin Genes: Exon-Intron Structure.
The structure of the HtH1 gene and the OdHG gene are compared schematically in Fig. 1. Here we have distinguished exons corresponding to different functional units by different colors. In both cases, the most striking feature of the gene organization is the presence of a linker intron between each pair of FUs (see Figs. 2 and 3). Additionally, in both species, an intron cuts the coding region of the signal peptide into two exons, a second intron separates the signal peptide region from the first FU, and another intron inserts in the 3′ UTR. In only three of the eight FUs of HtH (HtH1-a, HtH1-f, and HtH1-g) and in three of the seven FUs of OdH (OdHG-b, OdHG-e, and OdHG-f) do we observe a division of the FU into two or more exons by internal introns. The other nine FUs are each encoded by a single, continuous exon. Because each FU of the protein contains about 400 aa, this means that many of the exons are unusually large (see Table 1). Exons corresponding to FUs not interrupted by internal introns average around 1,250 bp, and that for HtH1-h is nearly 300 bp longer. On the other hand, most of the introns are relatively short compared to those of higher organisms (19).
Figure 2.
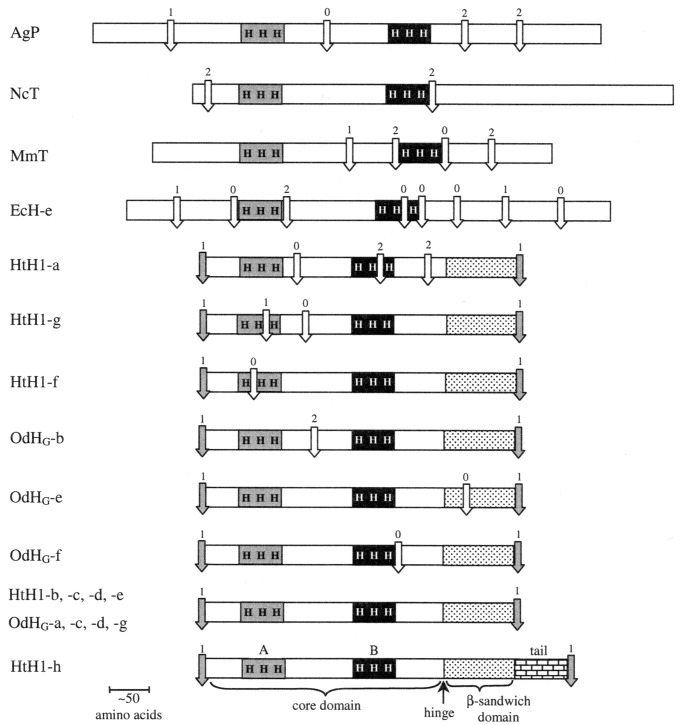
Comparison of intron locations in molluscan hemocyanins and several related proteins. Internal introns are shown by white arrows, linker introns by gray arrows. The copper-binding sites (A and B), with their histidines (H) are also schematically included. The β-sandwich domain in molluscan FUs is indicated by the dotted region, with the hinge location marked. The unique tail of FU-h is also indicated. Notation (aside from molluscan hemocyanin FUs): AgP Anopheles gambia (insect) prophenoloxidase; NcT, Neurospora crassa (fungus) tyrosinase; MmT, Mus musculus (mouse) tyrosinase, EcH-e, Eurypelma californicum (tarantula) hemocyanin subunit e.
Figure 3.
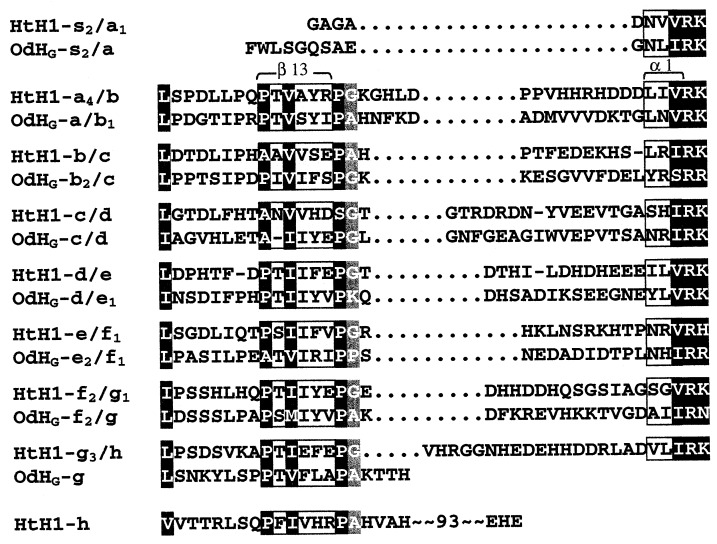
Sequence surrounding the linker introns. The protein linker regions between the various FUs (designated by the respective exon pair) have been arranged so as to (i) align conserved residues upstream from the linker intron insertion sites (indicated by dotted lines) and (ii) align the (I/V)RK consensus downstream from the insertion site. Strongly conserved residues are marked in black; there are many more of them in the β-sheet region of each preceding exon, to the left of the intron insertion site. The residues to the right of the site constitute the protein linker sequence; there is little conservation here until the (I/V)RK motif, which marks the beginning of the first α-helix in the next exon. To make both alignments, it has been necessary to introduce gaps in some of the linker sequences; their positions are not well defined. The positions of two secondary structure elements as deduced from OdH-g (3) are marked in light gray, and the positional conservation of a small amino acid is marked in dark gray.
The cephalopod hemocyanin gene especially exhibits a considerably higher exon/intron ratio than do most eukaryotic structural genes, with a consequence that the overall gene is not extremely long (about 14.8 kbp from the start of the signal peptide to the 3′ UTR), with an ORF of 8,688 bases for the secreted polypeptide, plus 63 bases for the signal peptide. The gastropod hemocyanin gene is twice as large (28.6 kbp from the signal peptide region to the 3′ UTR), with a reading frame of 10,212 bases for the secreted polypeptide.
All of the linker introns in both genes have phase 1. Each of these linker introns is inserted at a nearly equivalent point just downstream from a conserved region at the C terminus of the preceding FU, and upstream from a highly conserved sequence [(V/I)RK] in the following FU (see Fig. 3). This means that each protein linker sequence is clearly defined and lies just downstream from the preceding linker intron. The internal introns have all possible phases and are located in quite different positions within each of their FU coding sequences (see Figs. 1 and 2, and discussion below). Among the six internal introns in the HtH gene, three have phase 0, two have phase 2, and one has phase 1; none of these introns is comparable to any of the three internal introns (two of phase 0, one of phase 2) of the OdH gene. Thus, the linker introns and internal introns present a striking contrast. The regularity of the former can be explained as a reflection of the genetic mechanism that created a multiunit structure from a monomeric precursor (see below). These introns presumably have remained in nearly the same position and phase since the evolution of the multiunit structure, roughly 750 million years (4). In contrast, the internal introns appear to be scattered, both in position and phase, in a random manner within each of the hemocyanin genes, and between them. The situation stands in stark contrast to that of the globin genes, in which a few internal intron positions have been held over eons (20). How can the apparent randomness in internal intron position in hemocyanin genes be explained? Intron sliding from one or a few initial positions can be ruled out; there is a paucity of evidence for sliding in any genome, and it is very difficult to explain how phase differences, as observed here, could be generated (21, 22). In the absence of extreme sliding, we are left with two alternatives: either (i) all of the internal introns observed today are relics of a larger group of primordial introns (“introns early”), or (ii) internal introns have been added (as well as deleted?) over the evolutionary time since gastropods and cephalopods arose (“introns late”). The first option seems unlikely. If one assumes that the nine different internal introns (six in Haliotis and three in Octopus) were all present in the ancestral protomer gene (introns-early scenario), the original eight-FU gene must have contained a total of 72 internal introns. Consequently, it would be necessary to invoke a minimum of 69 independent intron losses in the OdH gene and of 66 independent losses in the HtH gene to produce the present distribution. Moreover, in all nine cases of intron survival within this hypothetical hemocyanin gene, seven of the eight original copies of each intron would have to have been wiped out, leaving no common introns in the gastropod and cephalopod lines. Such a scenario seems highly improbable. In contrast, if one follows the intron-late scenario for the internal introns, only six and three independent intron gains, perhaps accompanied by some losses of earlier introns, would have been required for Haliotis and Octopus, respectively. Thus, the genomic organization of these molluscan hemocyanins appears to support the intron-late model. In addition, it strongly suggests that: (i) The eight FU genes arose by repeated gene duplication and fusion, possibly by unequal crossing over (23), thereby producing the linker introns. (ii) In the ca. 750 million years since that time the linker introns have changed their sequence beyond recognition, but retained their phase and nearly the same position. (iii) The internal introns have been randomly inserted after the separation of gastropods and cephalopods, ca. 520 million years ago.
Finally, we may ask whether or not the internal introns define functional modules within a FU, as expected by some versions of the intron-early model (24). As Fig. 2 shows, this is not the case. According to the x-ray diffraction studies of OdH-g (3), there are two well-defined domains within the molluscan functional unit, the N-terminal, α-helical core domain, which carries the oxygen-binding site, and a β-sandwich domain rich in β-sheets; the domains are connected by a hinge. All but one of the introns recognized in this work lie in the α-helical domain, and none is found in the hinge region between domains (Fig. 2; see also Fig. 6, which is published as supplemental material). Thus, there is little evidence to relate existing internal introns to the stepwise assembly of a modular structure. Indeed, if these introns have been added after the evolution of a multi-FU structure (as we argue above) there would be no reason to expect such evidence.
Comparison with Possibly Related Genes.
Because molluscan and arthropod hemocyanins exhibit certain sequence and structural similarities (particularly in the region of the copper-binding sites) it is of interest to compare the gene structures for OdH and HtH with the only reported arthropod hemocyanin gene, that for subunit e of the hemocyanin of the tarantula Eurypelma californicum (13). As Fig. 2 shows, there is very little similarity between the genomic organization of any FU from Haliotis or Octopus and that of the Eurypelma subunit. Whereas the 15 molluscan FUs are split by at most three introns (and in nine cases by none), the gene for the arthropod subunit (which corresponds in function to a single molluscan FU) contains no less than eight introns and spans about 55 kbp, about four times the length of the OdH gene and twice the length of the HtH gene, which code for 5- to 6-fold more protein sequence, respectively. These genetic differences reinforce the idea that arthropod and molluscan hemocyanins are only distantly related. Indeed, recently Burmester (25) has argued by a convincing statistical approach that arthropod and molluscan hemocyanin sequences are not related at all. Molluscan hemocyanins do exhibit sequence similarity with tyrosinases, especially in regions surrounding both copper-binding sites (1, 2, 8, 9). However, comparison of the exon-intron structure of these two classes of proteins indicates little, if any, similarity (Fig. 2). This is perhaps not surprising, because comparison of the two molluscan genes that are clearly related shows little similarity in internal intron placement.
Evolutionary Implications.
Considering all of the available data, it seems likely that both tyrosinases and molluscan hemocyanins evolved from a polypeptide chain that contained the copper A and B regions (1, 2, 4, 8, 9). The highly regular placement of the linker introns with respect to each preceding FU exon points to a repetitive, simple process for generating the multifunctional unit structures of molluscan hemocyanins from such a primordial unit. A possible scheme is shown in Fig. 4. Three rounds of gene duplication and fusion using an identical splice site would generate an eight-unit structure as seen in HtH. This is the kind of mechanism that has been proposed for the evolution of certain multiunit invertebrate hemoglobins (23, 26–28). However, to obtain the existing gene, another step would have been required; the N-terminal signal sequence had to be added. Fungal tyrosinases (and presumably the putative precursor unit for the hemocyanin gene as well) lack signal sequences; they are cytoplasmic proteins with enzymatic, not transport, functions. It is hard to judge when the signal was added, for two extreme models are possible. On the one hand, the signal sequence may have been added only after the rounds of duplication generating the eight-unit polypeptide chain were completed. Evidence from comparison of FU sequences in both Octopus and Haliotis indicates that this series of duplication was very rapid in evolutionary time (2, 4). Alternatively, the signal sequence could have been added before gene duplications. In this model, the linker protein sequences that lie between the FUs, downstream from each linker intron, could represent the much-evolved relics of what was originally the signal sequence of a single-unit progenitor. We tend to favor the latter view, for it provides a rationale both for hemocyanin functional evolution and the development of linker peptides. Perhaps the first use of a hemocyanin for oxygen transport depended on the evolution of a signal sequence, permitting export of a monomeric oxygen-binding protein into the hemolymph. Gene duplication and fusion, building larger proteins, then could allow higher concentrations of the oxygen transporter in the blood, while maintaining low osmolality and low viscosity, and permitting high cooperativity (29).
Figure 4.
Hypothesis for the evolution of the eight-FU molluscan hemocyanin subunit from a mono-FU precursor. Although our present results strongly suggest that the eight-FU subunit evolved from three subsequent gene duplication and fusion events, a pedigree of the latter could not be constructed from the sequence data so far. Thus, for the present scenario, recently revealed details of the quaternary structure (5) and general considerations of the evolution of extracellular oxygen carriers (29, 30) have been taken into account. It is proposed that the dimeric repeating unit as well as the di-pentameric architecture of the extant molluscan hemocyanin evolved before the eight-unit polypeptide, and that the latter grew from the C-terminal to the N-terminal FU and not vice versa. ∼ denotes the signal peptide that probably gave rise to the different linker peptides.
The above scenario requires that the OdH has lost a pre-existing FU, to give the number seven. From where was the unit lost? Comparison of the cDNA sequence of the hemocyanin from Haliotis with that from Octopus shows strong pairwise similarity between FUs a–g in the two organisms; this suggests that it is the C-terminal unit (FU-h) that is missing in Octopus (4). Moreover, in the phylogenetic tree derived from aligning these sequences, HtH1-h forms a discrete branch that separates from the other seven FU branches long before the gastropod-cephalopod split (4). This observation (which is independent of whether the unique tail extension of FU-h is taken into account or not) further supports the idea that the seven-unit hemocyanin found in Octopus evolved from an eight-unit progenitor and not the other way around.
The present study reveals no homology between the linker intron sequences, which would allow one to calculate a pedigree, as in the case of Artemia and Daphnia hemoglobin (27, 28). Thus, on the basis of the available sequence data alone, the order of the evolution of the eight hemocyanin FUs remains obscure, and further speculation requires information from other structural levels. A recent high-resolution (12 Å) three-dimensional reconstruction of the HtH1 di-decamer revealed that the di-pentameric architecture of the half molecule (the decamer) is mirrored by its internal “collar,” a rather compact di-pentameric ring; this collar is composed of 10 copies of FU-h, arranged as five symmetrical dimers (5). In addition, the 12-Å reconstruction indicated an antiparallel dimer of subunits to be the repeating unit, with an “arc” piece composed of two FU-g as its stabilizing center. FU-g alone is able to stabilize the repeating unit in Octopus, and moreover, it exhibits the most conservative sequence (4), further indicating that it indeed plays a crucial role in the formation of the dimeric repeating unit. Also FU-f and FU-e participate in the dimeric core of the repeating unit, whereas FU-d to FU-a are responsible for shaping the periphery of the subunit dimer at its two ends (5). Our idea is that under the selection pressure to form a larger respiratory protein (see above), the molluscan hemocyanin started from a FU homodimer, which later oligomerized into a di-pentameric ring; comparable protein structures have been observed in invertebrate hemoglobins (30). Then the core and finally the periphery of the extant decameric cylinder evolved (Fig. 4; see also Fig. 7, which is published as supplemental material). The intriguing aspect of this idea is that it proposes that the dimeric repeating unit as well as the di-pentameric architecture of molluscan hemocyanin pre-existed the evolution of the eight-unit polypeptide. In this context it would seem unlikely that this process started from FU-a, whereas the opposite possibility, an origin from FU-h, is supported by structural considerations. We propose that an FU-h precursor equipped with a secretion signal (∼h) and able to dimerize after secretion (to form a h-h homodimer) became able to oligomerize into a di-pentameric (= decameric) ring (Fig. 4). Next, the ancient single-unit polypeptide grew by three subsequent steps of gene duplication and fusion, producing a stepwise transformation of the simple ring into the highly complex molluscan hemocyanin cylinder (Fig. 4). Specifically, we propose: (i) A first gene duplication and fusion yielded ∼h∼h, which then evolved to ∼g∼h, both still forming a pair of homodimers within the repeating unit (5). (ii) A subsequent gene duplication, fusion, and further evolution produced ∼e∼f∼g∼h, which formed a larger homodimer, further consolidating the core of the repeating unit (Fig. 4). (iii) A third duplication, fusion, and evolution gave ∼a∼b∼c∼d∼e∼f∼g∼h, with two copies of the FU-a to FU-d sequence in each dimer of subunits furnishing the two peripheral enlargements of the core (Fig. 4). The tail extension of FU-h, which is not reflected in any of the other FUs (see Fig. 2), must have occurred at a later date. As discussed above, the copies of the primordial signal peptide (∼) subsequently transformed, and independently evolved, into the different linker peptides.
Supplementary Material
Acknowledgments
The American group is especially grateful to colleague Andrew Karplus for stimulating discussion. The careful and diligent assistance of Kristin Rorrer as well as the staff in the Oregon State University Central Services Laboratory is gratefully acknowledged. The American group acknowledges the support of this research by National Science Foundation Grant MCB-9805570 to K.I.M. The work of the German group has been financially supported by the biosyn company, Fellbach, and the Deutsche Forschungsgemeinschaft (Grant Ma 843/4-3 to J.M.).
Abbreviations
- FU
functional unit
- HtH
Haliotis tuberculata hemocyanin
- OdH
Octopus dofleini hemocyanin
- UTR
untranslated region
Footnotes
References
- 1.van Holde K E, Miller K I. Adv Protein Chem. 1995;47:1–81. doi: 10.1016/s0065-3233(08)60545-8. [DOI] [PubMed] [Google Scholar]
- 2.Miller K I, Cuff M E, Lang W F, Varga-Weisz P, Field K, van Holde K E. J Mol Biol. 1998;278:827–842. doi: 10.1006/jmbi.1998.1648. [DOI] [PubMed] [Google Scholar]
- 3.Cuff M E, Miller K I, van Holde K E, Hendrickson W A. J Mol Biol. 1998;278:855–870. doi: 10.1006/jmbi.1998.1647. [DOI] [PubMed] [Google Scholar]
- 4.Lieb B, Altenhein B, Markl J. J Biol Chem. 2000;275:5675–5681. doi: 10.1074/jbc.275.8.5675. [DOI] [PubMed] [Google Scholar]
- 5.Meissner U, Dube P, Harris J R, Stark H, Markl J. J Mol Biol. 2000;298:21–34. doi: 10.1006/jmbi.2000.3631. [DOI] [PubMed] [Google Scholar]
- 6.Markl J, Decker H. Adv Comp Env Physiol. 1992;13:325–376. [Google Scholar]
- 7.Voit R, Feldmaier-Fuchs G, Schweikardt T, Decker H, Burmester T. J Biol Chem. 2000;275:39339–39344. doi: 10.1074/jbc.M005442200. [DOI] [PubMed] [Google Scholar]
- 8.Lerch K, Huber M, Scheider M-J, Drexel R, Linzen B. J Inorg Biochem. 1986;26:213–217. [Google Scholar]
- 9.Durstewitz G, Terwilliger N B. Mol Biol Evol. 1997;14:266–276. doi: 10.1093/oxfordjournals.molbev.a025762. [DOI] [PubMed] [Google Scholar]
- 10.Conway Morris S. Curr Opin Genet Dev. 1998;8:662–667. doi: 10.1016/s0959-437x(98)80034-8. [DOI] [PubMed] [Google Scholar]
- 11.van Holde K E. Zoology. 1998;100:287–297. [Google Scholar]
- 12.Decker H, Terwilliger N B. J Exp Biol. 2000;203:1777–1782. doi: 10.1242/jeb.203.12.1777. [DOI] [PubMed] [Google Scholar]
- 13.Voll W, Voit R. Proc Natl Acad Sci USA. 1990;87:5312–5316. doi: 10.1073/pnas.87.14.5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keller H, Lieb B, Altenhein B, Gebauer W, Richter S, Stricker S, Markl J. Eur J Biochem. 1999;264:27–38. doi: 10.1046/j.1432-1327.1999.00564.x. [DOI] [PubMed] [Google Scholar]
- 15.Lieb B, Altenhein B, Lehnert R, Gebauer W, Markl J. Eur J Biochem. 1999;265:134–144. doi: 10.1046/j.1432-1327.1999.00694.x. [DOI] [PubMed] [Google Scholar]
- 16.Harris J R, Markl J. Micron. 1999;30:597–623. doi: 10.1016/s0968-4328(99)00036-0. [DOI] [PubMed] [Google Scholar]
- 17.Lang W H. Biochemistry. 1988;27:7267–7282. doi: 10.1021/bi00419a015. [DOI] [PubMed] [Google Scholar]
- 18.Long M, de Souza S J, Rosenberger C, Gilbert W. Proc Natl Acad Sci USA. 1998;95:219–223. doi: 10.1073/pnas.95.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hawkins J D. Nucleic Acids Res. 1998;16:9893–9905. doi: 10.1093/nar/16.21.9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jellie A M, Tate W P, Trotman C N A. J Mol Evol. 1996;42:641–647. doi: 10.1007/BF02338797. [DOI] [PubMed] [Google Scholar]
- 21.Li W-H. Molecular Evolution. Sunderland, MA: Sinauer Associates; 1997. pp. 379–418. [Google Scholar]
- 22.Stoltzfus A, Logsdon J M, Jr, Palmer J D, Doolittle F W. Proc Natl Acad Sci USA. 1997;94:10739–10744. doi: 10.1073/pnas.94.20.10739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naito Y, Riggs C, Vandergon T L, Riggs A F. Proc Natl Acad Sci USA. 1991;88:6672–6676. doi: 10.1073/pnas.88.15.6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilbert W, Marchionni M, McKnight G. Cell. 1986;46:151–153. doi: 10.1016/0092-8674(86)90730-0. [DOI] [PubMed] [Google Scholar]
- 25.Burmester T. Mol Biol Evol. 2001;18:184–195. doi: 10.1093/oxfordjournals.molbev.a003792. [DOI] [PubMed] [Google Scholar]
- 26.Moens L, Vanfleteren J, De Baere I, Jellie A M, Tate W, Trotman C N A. FEBS Lett. 1992;312:105–109. doi: 10.1016/0014-5793(92)80915-4. [DOI] [PubMed] [Google Scholar]
- 27.Matthews C M, Trotmann C N A. J Mol Evol. 1998;47:763–771. doi: 10.1007/pl00006435. [DOI] [PubMed] [Google Scholar]
- 28.Hebert P D N, Um Y M, Prokopowitch C D, Taylor D J. J Mol Evol. 1999;49:769–779. doi: 10.1007/pl00006599. [DOI] [PubMed] [Google Scholar]
- 29.Mangum C P. Am Zool. 1998;38:1–13. [Google Scholar]
- 30.Ilan E, David M M, Daniel E. Biochemistry. 1981;20:6190–6194. doi: 10.1021/bi00524a043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.