Abstract
A global approach was used to analyze protein synthesis and stability during the cell cycle of the bacterium Caulobacter crescentus. Approximately one-fourth (979) of the estimated C. crescentus gene products were detected by two-dimensional gel electrophoresis, 144 of which showed differential cell cycle expression patterns. Eighty-one of these proteins were identified by mass spectrometry and were assigned to a wide variety of functional groups. Pattern analysis revealed that coexpression groups were functionally clustered. A total of 48 proteins were rapidly degraded in the course of one cell cycle. More than half of these unstable proteins were also found to be synthesized in a cell cycle-dependent manner, establishing a strong correlation between rapid protein turnover and the periodicity of the bacterial cell cycle. This is, to our knowledge, the first evidence for a global role of proteolysis in bacterial cell cycle control.
Bacteria have the potential to rapidly multiply and spread in an environment that provides all of the nutrients for growth. Rapid growth of virulent bacteria can be fatal for both plant and animal hosts. To understand the ability of these simple cells to quickly grow and divide while faithfully passing on their genetic information, it is critical to unravel the regulatory circuits that control the bacterial cell cycle. In particular, cells have to control DNA replication, chromosome segregation, and cytokinesis temporally and spatially and coordinate these events with growth. The periodicity of these cell cycle events is accompanied by oscillations of gene expression both in eukaryotic cells and in bacteria. In the unicellular eukaryote Saccharomyces cerevisiae, a total of 800 genes, representing ≈13% of the yeast genome, are differentially expressed during the cell cycle, many being involved in DNA replication, cell division, mitosis, and mating (1, 2). Similarly, temporal control of gene expression is an important regulatory element of cell cycle progression and development in the bacterium Caulobacter crescentus (3). In accordance, a recent study analyzing gene expression on a global scale identified 590 genes that are differentially expressed during the Caulobacter cell cycle. A large subset of these genes encode proteins involved in the execution of cell cycle events, whereby the time of gene expression reflected the time of the function of their products (4). Thus, studying cell cycle-dependent gene expression in a global manner not only catalogs periodically expressed genes but can also help to identify genes with novel cell cycle functions. Gene expression studied globally by monitoring changes of the cell's mRNA levels must be complemented by proteome analysis, because the protein rather than the mRNA is the biologically active, and thus more relevant, molecule, and because the relationship between mRNA levels and the rates of protein synthesis can be nonlinear (5). Moreover, proteomics makes it possible to study critical posttranslational control mechanisms such as modification and protein stability, which may contribute greatly to the ultimate activity of a given protein.
In eukaryotic cells, a complex regulatory network has been elucidated that authorizes faithful progression and coordination of the cell cycle by acting not only at the level of protein synthesis but also at the level of protein phosphorylation and degradation (6). Recent discoveries in C. crescentus have revealed similar multilayered regulatory mechanisms for cell cycle propagation in bacteria (7). A prime example is the transcriptional regulator CtrA, which acts as a timing device for several cell cycle events, including DNA replication, DNA methylation, and cell division. CtrA activity is temporally and spatially controlled by differential expression, phosphorylation, and protein degradation (8, 9). CtrA degradation is catalyzed by the essential ClpXP protease complex, a structural homolog of the 26S proteasome of eukaryotic cells (10). Genetic evidence suggests that ClpXP is required for additional, so far unrecognized, protein degradation events, which are critical for cell cycle progression (10). However, in contrast to the situation in eukaryotic cells, the significance and scope of specific protein degradation for cell cycle control has not been elucidated in bacteria so far. Although the total protein turnover has been estimated to be about 3% of the Escherichia coli protein mass per hour (11), only a small number of E. coli proteins with a short half life have been identified (12, 13). To address this issue, we have used two-dimensional (2-D) gel electrophoresis combined with peptide mass fingerprinting to investigate both protein synthesis and degradation during the C. crescentus cell cycle. The identification of a large fraction of proteins that are both differentially synthesized and rapidly degraded opens new entry points into analyzing the role of these proteins in directing cell cycle progression through controlled proteolysis.
Materials and Methods
Cell Cycle Synchronization and Protein Labeling.
The synchronizable C. crescentus strain NA1000 was grown at 30°C in M2 minimal glucose medium (M2G) (14). Swarmer cells were isolated by density gradient centrifugation (15) and released into fresh M2G medium. Cell cycle progression was monitored by light microscopy. To monitor differential protein expression, cells were pulse labeled at 0, 0.3, 0.6, 0.8, and 1 cell cycle units (corresponding to G1, early S, late S, G2 phase, and cell division) (16, 17) by adding 20 μCi of a [35S]methionine/cysteine mix (NEG 772, New England Nuclear) to 1 ml of culture for 4 min, followed by a 2-min chase with 0.2% tryptone, 1 mM methionine, and 0.02 mM cysteine before harvesting. To analyze protein stability, asynchronous cultures were labeled as described above and chased for up to 120 min. To assess the influence of the synchronization procedure on protein synthesis, expression patterns were compared in asynchronous cultures before and after the synchronization procedure. Swarmer, stalked, and predivisional cells were repooled after separation by density gradient centrifugation and pulse-labeled as described above. The protein expression pattern was then compared with a labeled culture that had not experienced the synchronization procedure. Proteins that were significantly up- or down-regulated as a consequence of the synchronization procedure were subtracted from the cell cycle expression data set (see below).
Protein Preparation.
For analytical gels, 1 ml of pulse labeled cells was washed twice in 20 mM phosphate buffer, pH 7, and then lysed in 200 μl lysis buffer containing 8 M urea, 4% cholamidopropyl-dimethyl-ammonio-propane sulfonate, 0.8% ampholytes, pH 3–10 (Pharmalyte, Amersham Pharmacia), 65 mM DTT, and a few grains of bromophenole blue. The incorporated radioactivity was determined by scintillation counting of trichloroacetic acid precipitated proteins.
For preparative gels, 250 ml of cells was washed in 20 mM phosphate buffer, pH 7, and resuspended in 8 ml of breaking buffer (20 mM phosphate buffer, pH 7/5% sucrose) containing a protease inhibitor mixture (Complete, Roche Diagnostics, Rotkreuz, Switzerland), 4 μg/ml of RNase, and 16 μg/ml DNase. The suspension was passed twice through a precooled French pressure cell at 1,000 psi and centrifuged at 120,000 × g. The soluble proteins were concentrated and washed twice with H2O in an Amicon filtration cell by using a membrane with a molecular weight cutoff of 10,000. Solid urea and concentrated lysis buffer were added to the protein solution to give the same final concentration as described above.
2-D Gel Electrophoresis.
2-D gel electrophoresis was performed as described (18). The proteins were separated with 18-cm Immobiline DryStrips, pH 3–10 (Amersham Pharmacia) in the first dimension and on continuous 12% SDS gels in the second dimension. Analytical gels were loaded with 106 cpm and preparative gels with 2 mg of protein. Radioactivity was detected by storage phosphor imaging, and preparative gels were stained with colloidal Coomassie blue (NOVEX, San Diego). Protein size (10–100 kDa) and isoelectric point range (pH 3–10) of the 2-D gels were determined by using 2-D gel marker proteins (Bio-Rad).
Data Processing and Analysis.
Samples from six independent labeling experiments were resolved on six independent 2-D gels for each time point investigated. The 2-D gel autoradiographs were matched and quantified by image analysis by using pdquest (Version 5.0.1, PDI Imageware Systems, Huntington Station, NY). The quantified data were then analyzed with s-plus (MathSoft, Cambridge, MA) and Excel (Microsoft) as follows: (i) The spot intensities were converted into parts per million of the total gel intensity and normalized as described in ref. 19. (ii) Spots were removed from the data set of a given time point if they were detected in less than three of the six repeats. (iii) Single outliers were removed by a standard t-significance test. (iv) All spots were removed that were present on gels of the nonsynchronous cultures but could not be detected on any of the gels of the cell cycle time points and vice versa. (v) Spots with the highest intensity below 200 ppm had high experimental variation and were removed. Finally, 979 highly reproducible protein spots were left and were used as the minimal reproducible data set for statistical analyses.
Spot intensity changes were considered statistically significant and relevant if the ANOVA or t-test confidence level was higher than 99% and if the ratio between two mean values was at least two. All spots identified by these criteria were manually investigated on the original gels, and spots with low quality or clear mismatches were removed. Spots that were significantly induced or repressed by the synchronization procedure (see above) and whose cell cycle expression patterns could be attributed only to this synchronization effect were removed from the cell cycle expression data set. Spots with differential expression patterns were sorted by hierarchical clustering with an agglomerative nesting algorithm (20) (see Fig. 4, which is published as supplemental data on the PNAS web site, www.pnas.org).
Protein Identification.
Protein spots were identified by peptide-mass fingerprinting (21). Spots were cut out from Coomassie-stained preparative gels, destained, and digested with endoproteinase Lys-C or Trypsin (21). The masses of the peptides after the proteolytic digest were determined with a matrix-assisted laser desorption ionization time-of-flight mass spectrometer (Reflex3, Bruker, Billerica, MA). All translated putative ORFs larger than 80 amino acids of the Caulobacter prerelease genome sequence version of September 1999 (22) were used to search for protein sequences that matched the measured peptides as described in ref. 23. A protein identification was considered a hit if it was detected as the only candidate in two independent experiments or if at least five peptide masses matched the best hit. Protein homology searches were done by blast-2 (24). A function was assigned to an identified protein if the similarity had an expectancy value e < 10−8 and if the function was supported by experimental data for at least one of the homologs. Antibodies against PleD (25), CcrM (26), flagellins, FtsZ (27), FliF (28), and McpA (29) were used for immunoblot analysis (25). Expression profiles, identities, and 2-D gel coordinates of all protein spots that were differentially expressed during the cell cycle, repressed, or induced by the synchrony procedure, or exhibited a change in intensity during chase are shown in Table 2 and Fig. 5 in the supplemental material, www.pnas.org.
Results and Discussion
Fifteen Percent of the C. crescentus Proteins Detected Are Differentially Expressed During the Cell Cycle.
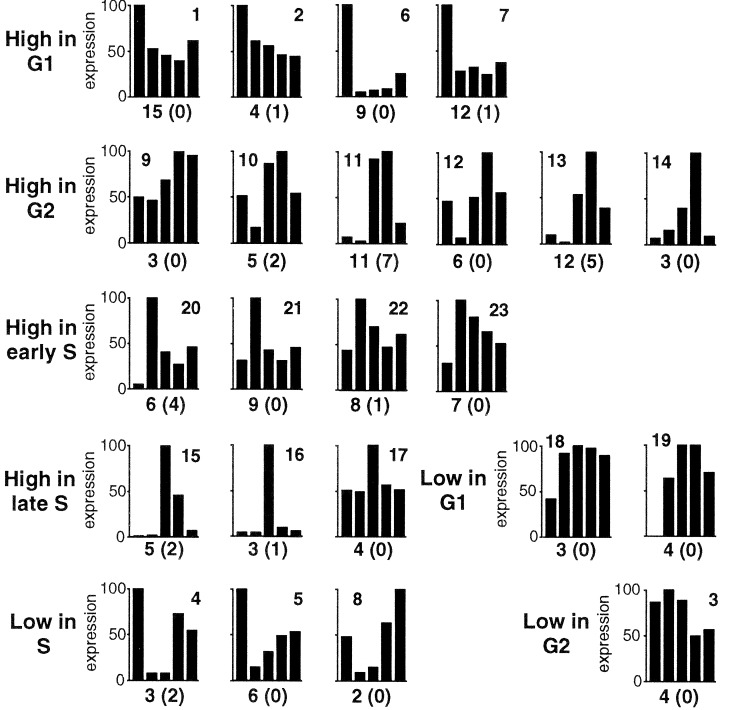
We used a global proteomics approach to independently analyze the timing of protein synthesis and decay during the C. crescentus cell cycle. First, protein synthesis was monitored by pulse labeling synchronized Caulobacter cultures at five different time points of the cell cycle (Fig. 1). A total of 979 protein spots were reproducibly detected on 2-D gels (see Materials and Methods; Fig. 5A in the supplemental material, www.pnas.org), corresponding to about one-fourth of the estimated total number of Caulobacter genes (22). Because membrane integral proteins generally cannot be well separated on 2-D gels, and low abundant proteins are difficult to visualize (30), these 979 spots mainly represent soluble highly abundant proteins. Taking into account that about 25% of all bacterial proteins are membrane-integral proteins (31), these spots represent about 35% of all soluble proteins encoded by the Caulobacter genome. The expression of 234 spots of this minimal reproducible data set oscillated significantly during the progression of synchronized cultures. To assess the influence of the synchronization protocol on protein expression, extracts of pulse-labeled nonsynchronous cells were compared before and after the synchronization procedure (see Materials and Methods). The expression of 90 protein spots was significantly affected either positively or negatively by the steps of the synchronization protocol, and the corresponding spots were removed from subsequent analysis. The remaining cell cycle-variable data set contained 144 spots (15% of all spots detected) that were randomly distributed among the resolved spots. These spots were then sorted by cluster analysis into 23 groups of proteins with distinct cell cycle expression profiles (Fig. 2). Clustering expression data have been shown to compile members of synexpression groups, which represent sets of genes that share a complex expression pattern under different conditions and that function in the same process (32, 33). In agreement with this observation, we found possible synexpression groups for several proteins involved in riboflavin synthesis (four proteins in cluster 6), energy metabolism (four proteins in the similar clusters 22 and 23), redox reactions (three proteins in cluster 1), amino acid biosynthesis (two proteins in cluster 19), carbohydrate metabolism (three proteins in the similar clusters 1 and 7), protein degradation (five proteins in the similar clusters 1, 2, and 7), and motility and chemotaxis (five proteins in the similar clusters 11 and 13) (Table 1, Fig. 2).
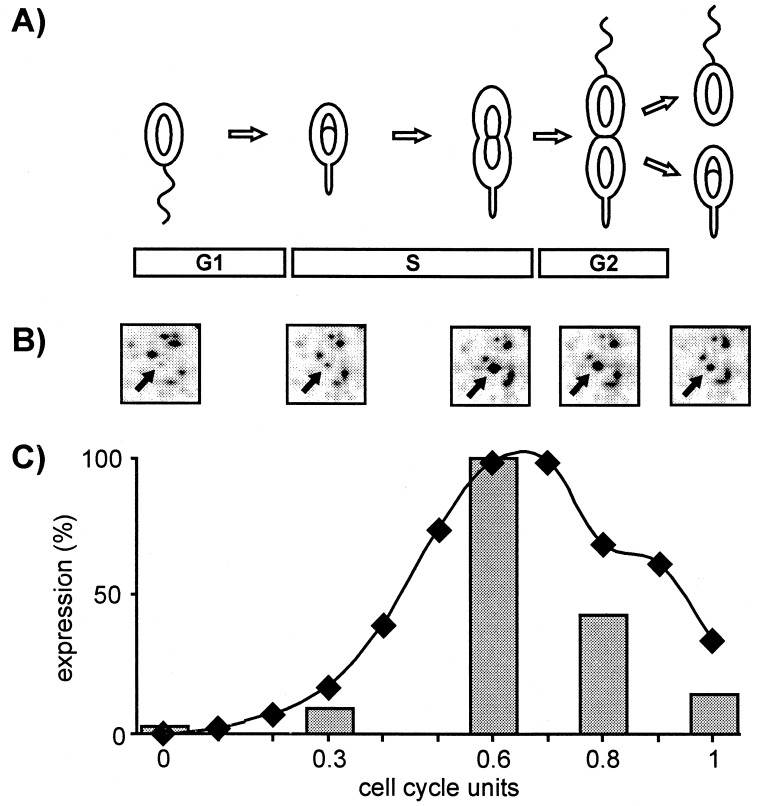
Figure 1.
Cell cycle of C. crescentus and cell cycle-dependent protein expression of the CtrA regulator. (A) Motile replication silent swarmer cells (G1 phase) differentiate into stalked cells by shedding the polar flagellum and growing a stalk at the same pole. DNA replication is initiated in stalked cells and continues as cells elongate and increase in mass during S phase. A new flagellum is assembled in the predivisional cells at the pole opposite the stalk. On completion of DNA replication, the newly synthesized chromosomes segregate to the poles, and an asymmetric cell division generates two new daughter cells (G2 phase). (B) Protein synthesis was measured during the cell cycle by pulse labeling cells of a synchronized culture with [35S]methionine in G1 (0 cell cycle units), early S (0.3), late S (0.6), G2 phase (0.8), and immediately after cell division (1.0). The labeled extracts were separated on 2-D gels, and fluctuations were determined by quantifying and comparing the spot intensities. The example shows a small area of the 2-D gels with the arrows marking the CtrA protein. (C) Oscillation of ctrA expression during the C. crescentus cell cycle. Relative levels of ctrA mRNA (diamonds) were taken from ref. 8, and CtrA protein synthesis (bars) was quantified from the 2-D gel spots shown in B for each time point investigated.
Figure 2.
Coexpression groups with distinct cell cycle expression profiles are contained within 23 clusters. The bars represent the mean relative synthesis levels (as percentage of the maximum value) of all members of a cluster at the five cell cycle time points indicated in Fig. 1C. The values of each individual member of a cluster were calculated from six independent repeats and the changes were significant at the 99% confidence level. The clusters are grouped according to the timing of the highest or lowest expression value. The number on top of each chart indicates the cluster number. The total number of protein spots assigned to each cluster is indicated at the bottom of each chart with the number of unstable spots in parenthesis.
Table 1.
Identification of C. crescentus proteins with differential cell cycle expression profiles
| ORF | CL | Function |
|---|---|---|
| Amino acid metabolism | ||
| 03246* | 16 | Tetrahydropteroyltriglutamate methyltransferase (Met) |
| 04899* | 3 | Glutamyl-tRNA(Gln)-amidotransferase subunit A (Glu) |
| 05974 | 19 | Indole-3-glycerol-phosphate synthase (Trp) |
| 06643 | 19 | Acetylornithine aminotransferase (Arg) |
| Carbohydrate metabolism | ||
| 03310† | 1 | Acetyl-CoA-acetyltransferase |
| 03861† | 7 | β-d-glucoside glucohydrolase |
| 06912* | 7 | UDP-glucose-4-epimerase |
| Cofactor metabolism | ||
| 03206 | 6 | GTP cyclohydrolase I (tetrahydrofolate synthesis) |
| 04038* | 6 | Riboflavin specific deaminase |
| 04041* | 6 | Riboflavin synthase α-chain |
| 04042 | 6 | GTP cyclohydrolase II (riboflavin synthesis) |
| 04043* | 6 | Riboflavin synthase β-chain |
| Lipid metabolism | ||
| 02302 | 3 | Enoyl-CoA-hydratase |
| 02436† | 7 | Fatty acid oxidation complex α-subunit |
| Energy metabolism | ||
| 01686 | 22 | ATP-synthase-α-subunit |
| 02110 | 22 | Aconitase |
| 06056* | 23 | NADH dehydrogenase I chain G |
| 06255 | 22 | Pyruvate kinase |
| Redox reactions | ||
| 00532 | 21 | Thioredoxin reductase |
| 00952† | 3 | Glutathione-S-transferase |
| 01378* | 1 | NADPH-dependent quinone oxidoreductase |
| 02058* | 11 | Glutathione-S-transferase |
| 02406* | 1 | NAD(P)H Nitroreductase |
| 02506* | 1 | Thioredoxin |
| RNA and protein synthesis | ||
| 02595 | 23 | Ribonuclease PH |
| 03553† | 14 | 50S Ribosomal protein L1 |
| 04420 | 9 | Peptide chain release factor 3 |
| 04755† | 6 | 50S Ribosomal protein L4 |
| 05538* | 23 | 50S Ribosomal protein L9 |
| Protein folding and degradation | ||
| 00474 | 1, 22 | Aminopeptidase |
| 00833* | 21 | Peptidase |
| 01270† | 19 | Peptidase |
| 01800* | 1, 7 | Endopeptidase |
| 01944 | 1 | Zinc-metalloprotease |
| 02143† | 2 | Prolyl-endopeptidase |
| 02145 | 3 | Prolyl-endopeptidase |
| 02174* | 7 | Peptidyl-dipeptidase |
| 03639* | 8 | Chaperone GroEL |
| 03641* | 8 | Chaperone GroES |
| 06085* | 12 | Protease ClpP |
| 06128* | 7 | Dipeptidyl-peptidase |
| 07050* | 23 | Amidohydrolase |
| Cell envelope synthesis and structure | ||
| 01771 | 2 | Penicillin binding protein |
| 05095† | 1 | 2-Dehydro-3-deoxyphosphooctonate aldolase |
| Transport protein | ||
| 01077* | 13 | TonB-dependent iron uptake receptor |
| 01388* | 23 | ABC transporter ATP binding protein |
| 01790 | 7 | TonB-dependent iron uptake receptor |
| 05691† | 2 | TonB-dependent receptor |
| Motility and chemotaxis | ||
| 03149* | 13 | Chemotaxis protein CheYI |
| 03156* | 11 | Chemotaxis protein CheR |
| 03157* | 13 | Chemotaxis protein CheB |
| 03161* | 13 | Chemotaxis protein CheD |
| 05156* | 13 | Flagellin FljL |
| 05158* | 4 | Flagellin FljK |
| Cell division | ||
| 07201* | 20 | Cell division protein FtsZ |
| DNA metabolism | ||
| 02197† | 23 | dUTP nucleotidohydrolase |
| 03051* | 13 | DNA methyltransferase CcrM |
| 05175* | 21 | Single strand binding protein SSB |
| Regulatory protein | ||
| 00839* | 15 | Cell cycle transcriptional regulator CtrA |
| 01137* | 23 | Transcriptional regulator of AsnC/Lrp family |
| 02400† | 22 | Ferric uptake regulation protein Fur |
| 02881* | 1 | Phosphate transport regulator PhoU |
| 05154* | 10 | Regulator of flagellin expression FlbT |
| 05981* | 17 | SOS response repressor LexA |
| 06096* | 5 | Nitrogen regulatory protein PII |
| 07059* | 12 | Response regulator PleD |
| 07061* | 13 | Response regulator DivK |
| Unknown function | ||
| 01570* | 1 | Conserved unknown |
| 02168* | 21 | Unknown |
| 02609* | 21 | Unknown |
| 03189* | 12 | Conserved unknown |
| 03651* | 10 | Unknown |
| 03910† | 22 | Conserved unknown |
| 04449 | 21 | Unknown |
| 05475† | 10 | Unknown |
| 05886* | 12 | Conserved unknown |
| 05929* | 7 | Conserved unknown |
| 06393* | 1 | Conserved unknown |
| 06615* | 2 | Conserved unknown |
| 07094 | 12 | Unknown |
| 07198 | 22 | Conserved unknown |
ORF, Caulobacter genome ORF number (see Materials and Methods); *, similar; †, no or inverse cell cycle oscillations were detected for the corresponding mRNAs in ref. 4; CL, cluster number of coexpression groups (Fig. 2); Function, putative function determined by blast homology searches. Proteins with a half life shorter than one cell cycle are shown in bold.
Ninety-one protein spots with differential cell cycle expression patterns were identified by peptide-mass fingerprinting or immunodetection (see Materials and Methods). Several of the identified spots represented separable isoforms of the same proteins, reducing the total number of identified proteins to 81 (Table 1). These belonged to a wide variety of functional groups such as metabolism (23%), redox reactions (7%), transcription and translation (6%), protein folding and degradation (14%), cell envelope and transport (7%), motility and chemotaxis (7%), DNA synthesis (4%), cell division (1%), and regulation (11%). No function could be assigned to 20% of the identified proteins.
Most of the genes coding for proteins listed in Table 1 were also identified in a parallel study, which used DNA microarrays to analyze the variation of mRNA levels as a function of the C. crescentus cell cycle (4). Although 18 of the corresponding 81 genes were not represented on the microarrays, 49 genes showed cell cycle expression patterns that were identical or very similar to their pulse-labeled products on 2-D gels, and four genes showed an inverse cell cycle expression pattern. Ten of the eighty-one proteins identified in our proteome study showed no fluctuation on the mRNA level, implying translational or posttranslational cell cycle control (see Fig. 6, in the supplemental material, www.pnas.org). This important finding leads to several conclusions. First, the strong correlation between data from microarrays and 2-D gels makes assaying changes in mRNA levels a valuable approximation for changes in protein synthesis. Second, differential cell cycle expression of bacterial genes seems to be regulated mainly on the transcriptional level. Third, comparable kinetics of fluctuating mRNA levels and protein expression throughout the cell cycle suggests that the majority of the corresponding mRNA species has a very short half life. Fourth, oscillating protein expression during the cell cycle can be regulated posttranscriptionally, which has not been observed in bacteria so far and indicates new regulatory mechanisms for cell cycle control.
Functional Diversity of Differentially Expressed Proteins.
Several of the differentially expressed proteins identified in this study were involved in DNA metabolism, cell division, or development (Table 1). Despite the fact that a number of replication or cell division genes have been shown to be under cell cycle control (16, 27, 34, 35), only few of the corresponding proteins were identified in this study (SSB, cluster 21; CcrM, cluster 13; FtsZ, cluster 20). Plausible explanations for this result are that most replication and cytokinesis proteins are present at very low concentrations in the cell (36, 37) and that the membrane-integral or membrane-associated cell division components were lost during the extraction for the preparative gels.
Although DNA synthesis and cell division proteins were up-regulated in early S phase, proteins required for motility and chemotaxis were induced late in the cell cycle. A single flagellum and a chemotaxis machinery are assembled during each division cycle in the C. crescentus predivisional cell (Fig. 1). Several proteins required for directed motility were found to be synthesized predominantly during this phase of development, including chemotaxis proteins CheR, CheB, CheYI and CheD, the flagellins FljL and FljK (38), and a regulator of flagellin expression, FlbT (39) (clusters 4, 10, 11, 13). These proteins account for only a small fraction of the motor and chemotaxis components identified in C. crescentus (40, 41). This result might be because of the low abundance of most flagellar components in this uniflagellated organism and because a large fraction of flagellar components and all chemoreceptors are membrane-integral proteins, which, because of their low solubility, could not be resolved in the first dimension of the 2-D gels.
A surprisingly large number of proteins were involved in metabolic functions not typically thought of as cell cycle regulated (Table 1). Enzymes of energy metabolism were up-regulated as cells entered S phase and initiated growth. Similarly, enzymes involved in the production of several amino acids (tryptophane, arginine, methionine) were synthesized predominantly in S and G2 phase, which are associated with cell mass increase. A transcriptional regulator of the Lrp/AsnC family was up-regulated in early S phase. Homologs of this protein act as global metabolic regulators and are involved in the control of amino acid metabolism in a number of different bacteria, making this protein a candidate for cell cycle control of amino acid biosynthesis (42–45). Several enzymes involved in riboflavin and tetrahydrofolate biosynthesis had a sharp expression peak in G1 (Table 1). Riboflavin and tetrahydrofolate are growth factors involved in redox reactions and in the synthesis of building blocks like purines and certain amino acids, respectively. Their induction in swarmer cells might reflect the need of this cell type to prepare for the upcoming proliferation phase of the division cycle.
Several proteins engaged in oxidative stress response were also under cell cycle control. These are the GTP cyclohydrolase II, thioredoxin, thioredoxin reductase, glutathione S-transferase, and the ferric uptake regulator Fur (46, 47). Oxidative stress can result from hydroxide radicals that are generated in the presence of Fe2+ and O2 (47). To avoid DNA damage, the cell tightly controls its iron metabolism. Fur negatively controls iron acquisition and import genes (47) and is induced in early S phase, whereas two iron uptake proteins were repressed in S phase and induced in G1 or G2 (Table 1, Fig. 2). Thus, one could speculate that the cell needs to keep the iron concentration low in S phase to prevent DNA damage during ongoing replication. Alternatively, induction of iron uptake in G1 and G2 could reflect a metabolic peculiarity of the planktonic swarmer cell type as having a specialized role in nutrient scavenging. This suggestion is in agreement with the observation that a large group of proteolytic enzymes (6 of 13) were predominantly synthesized in the swarmer cell (Table 1, Fig. 2). All of these enzymes have predicted export signal sequences (48), implying that they are involved in the degradation of extracellular polypeptides and thereby contribute to nutrient scavenging in the planktonic swarmer cell.
It is interesting to note that a large group of nutritional genes involved in amino acid and sugar metabolism as well as iron uptake are also under cell cycle control in yeast (1). The requirement for periodic induction of key metabolic pathways during growth is apparently conserved in both prokaryotic and eukaryotic cells.
Cell Cycle Expression and Protein Stability.
The concentration of several important modulators of the Caulobacter cell cycle progression such as the transcriptional regulator CtrA, the cell division protein FtsZ, and the DNA methyltransferase CcrM fluctuate during the cell cycle as a consequence of timed synthesis and degradation. In all three cases, this oscillation has important functional implications for the timing and control of cell cycle events (9, 27, 49). To identify similar fluctuations, we globally investigated protein stability in a pulse–chase experiment. Exponentially growing asynchronous cells were pulse labeled and chased for up to 120 min, equivalent to one cell cycle length. Proteins degraded in the course of one cell cycle should partially or completely disappear during the chase period. Chased extracts were separated and analyzed on 2-D gels, allowing comparison of this data set with the cell cycle expression data. For 72 protein spots, a significant change was observed during the chase period (see Materials and Methods), with 48 spots decreasing and 24 increasing in intensity (Fig. 7 in the supplemental material, www.pnas.org). The latter are either modification or processing products that appear very slowly or in response to changing conditions during the chase period. In support of this conclusion, isoforms were found for 8 of the 10 identified protein spots accumulating during chase. In contrast, isoforms were found for only 4 of the 15 identified protein spots that decreased during the chase. This result argues that most of these proteins are not substrates for modification or processing reactions. Rather, the majority of the 48 spots, which disappear during the chase, represent proteins that are degraded in the course of one division unit. By extrapolating this conclusion to the entire C. crescentus proteome, an estimated 4% of the cell's proteins have a half life of one cell cycle equivalent or shorter. Most importantly, 26 of the 48 unstable protein spots were also differentially synthesized during the cell cycle. In contrast, oscillating expression was found only for one of the 24 spots that increased during the chase period (Fig. 7 in the supplemental material, www.pnas.org). A χ2 test for independence revealed a P value of 2 × 10−18 and thus a very strong correlation between protein instability and cell cycle expression. Interestingly, most of the proteins, which are rapidly degraded, group in clusters 4, 10, 11, 13, 15, and 20, all of which are characterized by sharp peaks of expression in G1, S, or G2 phase (Figs. 2 and 3). One can assume that narrow windows of expression, in combination with rapid degradation, result in distinct periodic changes of protein concentrations during the cell cycle.
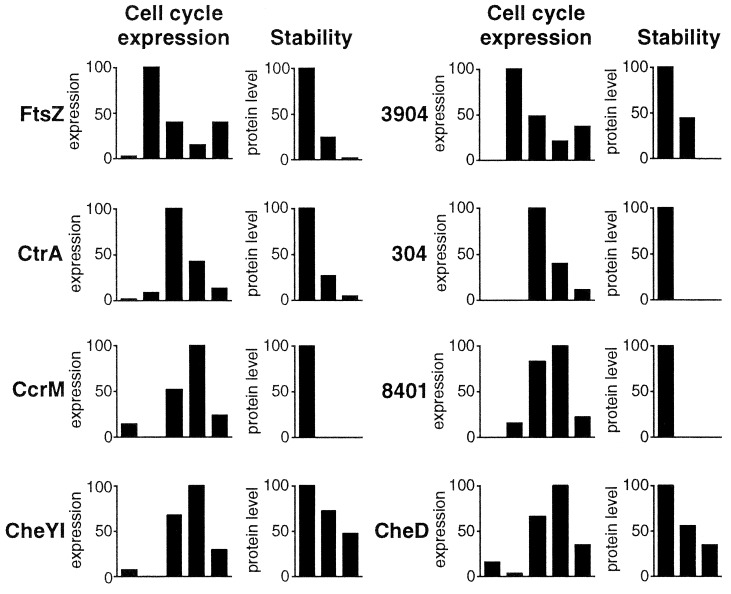
Figure 3.
Selected examples of protein spots with distinct cell cycle expression and stability profiles. The bars represent the mean relative protein synthesis levels (as percentage of the maximum value) at the five cell cycle time points indicated in Fig. 1C (Left chart) or the mean relative concentrations of the pulse-labeled protein (as percentage of the value at time 0) after chasing for 0, 60, and 120 min (Right chart). The expression and degradation profiles of the cell division protein FtsZ, the CtrA regulator, and the CcrM DNA methyltransferase are indicated and are in good agreement with patterns reported earlier for these proteins (8, 9, 26, 27, 49). In addition, the expression and degradation profiles for CheYI, CheD, and three so far unidentified proteins are shown.
Although 63% of the differentially synthesized protein spots could be identified by mass spectrometry, the unstable proteins (33%), because of their relatively low abundance, were underrepresented in the pool of identified proteins (Table 1). As expected, among the 26 spots found to be differentially expressed and rapidly degraded were CtrA, FtsZ, and CcrM (Table 1, Fig. 3). Two additional proteins, the flagellar anchor protein FliF and the chemoreceptor McpA, have recently been shown to be subject to cell cycle-dependent proteolysis in Caulobacter (28, 29). Neither was found in the pool of unstable proteins identified here, most likely because as membrane-integral proteins, they could not be resolved on the 2-D gels. Degradation of FliF and McpA is used by the cell to eliminate motility and chemotaxis during the G1 to S transition. The finding that two additional soluble chemotaxis proteins, CheYI and CheD (Table 1, Fig. 3), were rapidly degraded implies that the cell removes a major fraction of the chemotaxis components during the transition from the planktonic to the sessile stage of its life cycle. In contrast, the decline of the flagellin proteins FljL and FljK during the chase period reflects the ejection of the polar Caulobacter flagellum into the supernatant during each swarmer cell differentiation (Table 1; Fig. 1).
Conclusions
Studying cell cycle-regulated protein expression and degradation by using a global proteomics approach has provided new insights into the complexity of the bacterial cell cycle. The unexpectedly large number of proteins synthesized at a specific stage of the reproductive cycle suggests that periodic protein expression is critical for the cell either to guarantee the optimal utilization of resources or to maintain the proper order and functioning of the cell cycle. Differentially expressed proteins were found to be involved in many different aspects of the cell's metabolism. A large subgroup of periodically synthesized proteins were of unknown function (Table 1) and represent candidates for novel regulators of the C. crescentus cell cycle. The strong correlation between protein turnover and differential synthesis indicates that one of the main reasons for rapid protein degradation in bacteria is to maintain the periodicity required for ordered cell cycle progression. This is, to our knowledge, the first evidence for a global role of proteolysis in bacterial cell cycle control. In eukaryotic cells, the controlled proteolysis of key proteins at specific time points plays an essential role in promoting irreversible steps during cell cycle progression (50). On the basis of our results, we postulate that specific and controlled proteolysis plays a similar role in bacteria. It will be of particular interest to determine the identity of all proteins found to be both unstable and differentially expressed, as some might have critical functions in cell cycle progression. Characterization of these proteins will deepen our understanding of the molecular basis of bacterial growth. In an age of reemerging bacterial diseases, detailed knowledge of all regulatory processes involved in bacterial proliferation will be indispensable for the development of novel antimicrobial strategies.
Supplementary Material
Acknowledgments
We thank the Institute for Genomic Research (TIGR) for providing genome sequence data before publication and members of the Jenal lab, C. Thompson, and A. Kralli for critical reading of the manuscript. This work was supported by Swiss National Science Foundation fellowships 31–46764.96 and 31–59050.99 to U.J. and by a grant from F. Hoffmann–LaRoche, Ltd., to U.J.
Abbreviation
- 2-D
two-dimensional
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Spellman P T, Sherlock G, Zhang M Q, Iyer V R, Anders K, Eisen M B, Brown P O, Botstein D, Futcher B. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cho R J, Campbell M J, Winzeler E A, Steinmetz L, Conway A, Wodicka L, Wolfsberg T G, Gabrielian A E, Landsman D, Lockhart D J, Davis R W. Mol Cell. 1998;2:65–73. doi: 10.1016/s1097-2765(00)80114-8. [DOI] [PubMed] [Google Scholar]
- 3.Jenal U. FEMS Microbiol Rev. 2000;24:177–191. doi: 10.1016/S0168-6445(99)00035-2. [DOI] [PubMed] [Google Scholar]
- 4.Laub M T, McAdams H H, Feldblyum T, Fraser C M, Shapiro L. Science. 2000;290:2144–2148. doi: 10.1126/science.290.5499.2144. [DOI] [PubMed] [Google Scholar]
- 5.VanBogelen R A, Greis K D, Blumenthal R M, Tani T H, Matthews R G. Trends Microbiol. 1999;7:320–328. doi: 10.1016/s0966-842x(99)01540-1. [DOI] [PubMed] [Google Scholar]
- 6.Nasmyth K. Science. 1996;274:1643–1645. doi: 10.1126/science.274.5293.1643. [DOI] [PubMed] [Google Scholar]
- 7.Østerås M, Jenal U. Curr Opin Microbiol. 2000;3:171–176. doi: 10.1016/s1369-5274(00)00071-0. [DOI] [PubMed] [Google Scholar]
- 8.Quon K C, Marczynski G T, Shapiro L. Cell. 1996;84:83–93. doi: 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- 9.Domian I J, Quon K C, Shapiro L. Cell. 1997;90:415–424. doi: 10.1016/s0092-8674(00)80502-4. [DOI] [PubMed] [Google Scholar]
- 10.Jenal U, Fuchs T. EMBO J. 1998;17:5658–5669. doi: 10.1093/emboj/17.19.5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pine M. J Bacteriol. 1970;103:207–215. doi: 10.1128/jb.103.1.207-215.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larrabee K L, Phillips J O, Williams G J, Larrabee A R. J Biol Chem. 1980;255:4125–4130. [PubMed] [Google Scholar]
- 13.Gottesman S. Annu Rev Genet. 1996;30:465–506. doi: 10.1146/annurev.genet.30.1.465. [DOI] [PubMed] [Google Scholar]
- 14.Johnson R C, Ely B. Genetics. 1977;86:25–32. doi: 10.1093/genetics/86.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stephens C M, Shapiro L. Mol Microbiol. 1993;9:1169–1179. doi: 10.1111/j.1365-2958.1993.tb01246.x. [DOI] [PubMed] [Google Scholar]
- 16.Winzeler E, Shapiro L. J Mol Biol. 1995;251:346–365. doi: 10.1006/jmbi.1995.0439. [DOI] [PubMed] [Google Scholar]
- 17.Marczynski G T. J Bacteriol. 1999;181:1984–1993. doi: 10.1128/jb.181.7.1984-1993.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bjellqvist B, Pasquali C, Ravier F, Sanchez J C, Hochstrasser D. Electrophoresis. 1993;14:1357–1365. doi: 10.1002/elps.11501401209. [DOI] [PubMed] [Google Scholar]
- 19.Vohradsky J, Li X M, Thompson C J. Electrophoresis. 1997;18:1418–1428. doi: 10.1002/elps.1150180817. [DOI] [PubMed] [Google Scholar]
- 20.Kaufman L, Rousseeuw P J. Finding Groups in Data. New York: Wiley; 1990. [Google Scholar]
- 21.Fountoulakis M, Langen H. Anal Biochem. 1997;250:153–156. doi: 10.1006/abio.1997.2213. [DOI] [PubMed] [Google Scholar]
- 22.Nierman, W. C., Feldblyum, T. V., Laub, M. T., Paulsen, I. T., Nelson, K. E., Eisen J., Heidelberg, J. F., Alley, M. R. K., Ohta, N., Maddock, J. R., et al. (2001) Proc. Natl. Acad. Sci. USA 98, in press.
- 23.Berndt P, Hobohm U, Langen H. Electrophoresis. 1999;20:3521–3526. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3521::AID-ELPS3521>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 24.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aldridge P, Jenal U. Mol Microbiol. 1999;32:379–391. doi: 10.1046/j.1365-2958.1999.01358.x. [DOI] [PubMed] [Google Scholar]
- 26.Stephens C, Reisenauer A, Wright R, Shapiro L. Proc Natl Acad Sci USA. 1996;93:1210–1214. doi: 10.1073/pnas.93.3.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelly A J, Sackett M J, Din N, Quardokus E, Brun Y V. Genes Dev. 1998;12:880–893. doi: 10.1101/gad.12.6.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jenal U, Shapiro L. EMBO J. 1996;15:2393–2406. [PMC free article] [PubMed] [Google Scholar]
- 29.Alley M R, Maddock J R, Shapiro L. Science. 1993;259:1754–1757. doi: 10.1126/science.8456303. [DOI] [PubMed] [Google Scholar]
- 30.Wilkins M R, Gasteiger E, Sanchez J C, Bairoch A, Hochstrasser D F. Electrophoresis. 1998;19:1501–1505. doi: 10.1002/elps.1150190847. [DOI] [PubMed] [Google Scholar]
- 31.Wallin E, von Heijne G. Protein Sci. 1998;7:1029–1038. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eisen M B, Spellman P T, Brown P O, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niehrs C, Pollet N. Nature (London) 1999;402:483–487. doi: 10.1038/990025. [DOI] [PubMed] [Google Scholar]
- 34.Zweiger G, Shapiro L. J Bacteriol. 1994;176:401–408. doi: 10.1128/jb.176.2.401-408.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wortinger M, Sackett M J, Brun Y V. EMBO J. 2000;19:4503–4512. doi: 10.1093/emboj/19.17.4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kornberg A, Baker T A. DNA Replication. New York: Freeman; 1992. [Google Scholar]
- 37.Rothfield L, Justice S, Garcia-Lara J. Annu Rev Genet. 1999;33:423–448. doi: 10.1146/annurev.genet.33.1.423. [DOI] [PubMed] [Google Scholar]
- 38.Minnich S A, Newton A. Proc Natl Acad Sci USA. 1987;84:1142–1146. doi: 10.1073/pnas.84.5.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mangan E K, Malakooti J, Caballero A, Anderson P, Ely B, Gober J W. J Bacteriol. 1999;181:6160–6170. doi: 10.1128/jb.181.19.6160-6170.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ely B, Ely T W. Genetics. 1989;123:649–654. doi: 10.1093/genetics/123.4.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu J, Newton A. Mol Microbiol. 1997;24:233–239. doi: 10.1046/j.1365-2958.1997.3281691.x. [DOI] [PubMed] [Google Scholar]
- 42.Newman E B, Lin R. Annu Rev Microbiol. 1995;49:747–775. doi: 10.1146/annurev.mi.49.100195.003531. [DOI] [PubMed] [Google Scholar]
- 43.Beloin C, Ayora S, Exley R, Hirschbein L, Ogasawara N, Kasahara Y, Alonso J C, Hegarat F L. Mol Gen Genet. 1997;256:63–71. doi: 10.1007/s004380050546. [DOI] [PubMed] [Google Scholar]
- 44.Inoue H, Inagaki K, Eriguchi S I, Tamura T, Esaki N, Soda K, Tanaka H. J Bacteriol. 1997;179:3956–3962. doi: 10.1128/jb.179.12.3956-3962.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peekhaus N, Tolner B, Poolman B, Kramer R. J Bacteriol. 1995;177:5140–5147. doi: 10.1128/jb.177.17.5140-5147.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Storz G, Imlay J A. Curr Opin Microbiol. 1999;2:188–194. doi: 10.1016/s1369-5274(99)80033-2. [DOI] [PubMed] [Google Scholar]
- 47.Touati D. Arch Biochem Biophys. 2000;373:1–6. doi: 10.1006/abbi.1999.1518. [DOI] [PubMed] [Google Scholar]
- 48.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 49.Wright R, Stephens C, Zweiger G, Shapiro L, Alley M R. Genes Dev. 1996;10:1532–1542. doi: 10.1101/gad.10.12.1532. [DOI] [PubMed] [Google Scholar]
- 50.King R W, Deshaies R J, Peters J M, Kirschner M W. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.