INTRODUCTION
Abscisic acid (ABA) is an optically active 15-C weak acid that was first identified in the early 1960s as a growth inhibitor accumulating in abscising cotton fruit (“abscisin II”) and leaves of sycamore trees photoperiodically induced to become dormant (“dormin”) (reviewed in Addicott, 1983). It has since been shown to regulate many aspects of plant growth and development including embryo maturation, seed dormancy, germination, cell division and elongation, and responses to environmental stresses such as drought, salinity, cold, pathogen attack and UV radiation (reviewed in Leung and Giraudat, 1998; Rock, 2000). However, despite the name, it does not appear to control abscission directly; the presence of ABA in abscising organs reflects its role in promoting senescence and/or stress responses, the processes preceding abscission. Although ABA has historically been thought of as a growth inhibitor, young tissues have high ABA levels, and ABA-deficient mutant plants are severely stunted (Figure 1) because their ability to reduce transpiration and establish turgor is impaired. Exogenous ABA treatment of mutants restores normal cell expansion and growth.
Figure 1.

Exogenous ABA suppresses growth inhibition of ABA-deficient mutants. Plants with one of three mutant alleles of aba1 were grown with (bottom) or without (top) ABA treatment (spraying twice weekly with 10 µM ABA for 8 weeks). (Photograph courtesy of J. Zeevaart.)
ABA is ubiquitous in lower and higher plants. It is also produced by some phytopathogenic fungi (Assante et al., 1977; Neill et al., 1982; Kitagawa et al., 1995) and has even been found in mammalian brain tissue (Le Page-Degivry et al., 1986). As a sesquiterpenoid, it was long thought to be synthesized directly from farnesyl pyrophosphate, as in fungi (reviewed in Zeevaart and Creelman, 1988). However, it is actually synthesized indirectly from carotenoids. As a weak acid (pKa=4.8), ABA is mostly uncharged when present in the relatively acidic apoplastic compartment of plants and can easily enter cells across the plasma membrane. The major control of ABA distribution among plant cell compartments follows the “anion trap” concept: the dissociated (anion) form of this weak acid accumulates in alkaline compartments (e.g. illuminated chloroplasts) and may redistribute according to the steepness of the pH gradients across membranes. In addition to partitioning according to the relative pH of compartments, specific uptake carriers contribute to maintaining a low apoplastic ABA concentration in unstressed plants.
Despite the ease with which ABA can enter cells, there is evidence for extracellular as well as intracellular perception of ABA (reviewed in Leung and Giraudat, 1998; Rock, 2000). Multiple receptor types are also implicated by the variation in stereospecificity among ABA responses.
Genetic studies, especially in Arabidopsis, have identified many loci involved in ABA synthesis and response and analyzed their functional roles in ABA physiology (reviewed in Leung and Giraudat, 1998; Rock, 2000). Many likely signaling intermediates correlated with ABA response (e.g. ABA-activated or -induced kinases and DNA-binding proteins that specifically bind ABA-responsive promoter elements) have also been identified by molecular and biochemical studies, but the relationships among these proteins are unclear. Cell biological studies have identified secondary messengers involved in ABA response. Ongoing studies combine these approaches in efforts to determine coherent models of ABA signaling mechanism(s).
ABA BIOSYNTHESIS AND METABOLISM
ABA is a sesquiterpenoid (C15H20O4) with one asymmetric, optically active carbon atom at C-1′ (Figure 2). The naturally occurring form is S-(+)-ABA; the side chain of ABA is by definition 2-cis,-4-trans. Trans, trans-ABA is biologically inactive, but R-(−)-ABA (a possible product of racemization via the catabolite ABA-trans-diol; Vaughn and Milborrow, 1988; Rock and Zeevaart, 1990) does have biological activities (Zeevaart and Creelman, 1988; Toorop et al., 1999) which suggests that multiple ABA receptors exist. Future studies should elucidate whether R-(−)-ABA is found in nature, since the isolation of R-(−)-ABA stereoselective response mutants (chotto, [cho]) suggests a genetic basis for its activity (Nambara et al., 2002). It is not yet clear if the cho1 and cho2 mutants identify genetically redundant factors that may only have a minor effect on germination.
Figure 2.
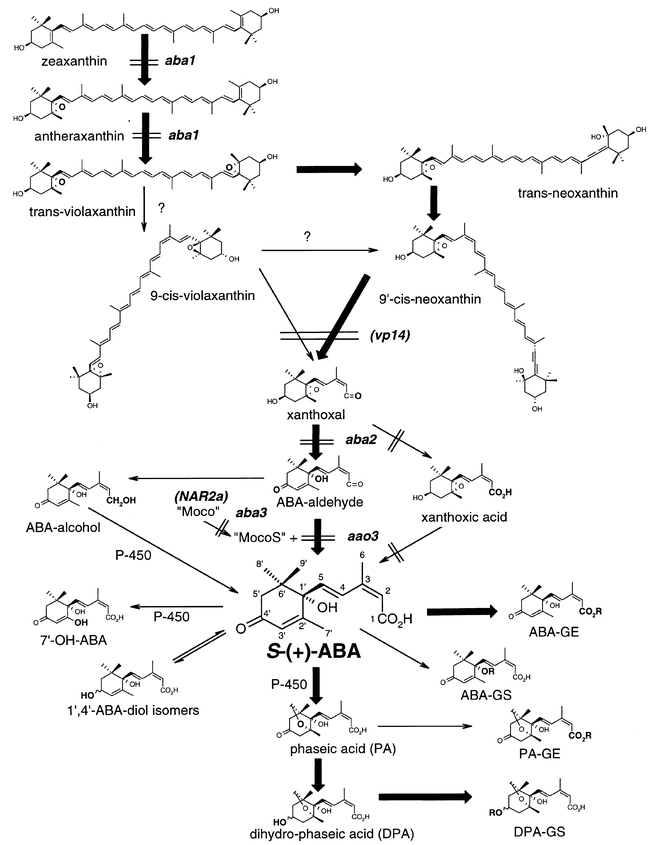
Proposed biosynthetic pathway of ABA after zeaxanthin, with positions of steps blocked in some ABA biosynthetic mutants indicated. Also shown are the major inactivation products of ABA (phaseic acid, dihydrophaseic acid, and the glucose ester of ABA) and the minor metabolites (ABA-diols, 7′-hydroxy-ABA, ABA-1′-glucoside, and glucose esters and glucosides of PA and DPA).
The regulation of physiological processes controlled by ABA is primarily at the level of de novo ABA biosynthesis and turnover. This requires de novo synthesis of the relevant enzymes rather than redistribution of existing ABA pools, although xylem transport of ABA is a drought signal from roots to shoots (Zeevaart and Creelman, 1988; Milborrow, 2001; Hartung et al., 2002). Genetic analysis in Arabidopsis of physiological processes related to ABA activity (seed germination, dormancy, osmotic stress, transpiration, gene expression) has resulted in isolation of ABA-deficient mutants, underscoring the important and direct role of ABA metabolism in plant growth and development and providing the means to elucidate the ABA biosynthetic and signaling pathway(s). Several ABA biosynthetic steps have been elucidated by characterization of maize, tomato, and Nicotinana plumbaginifolia mutants, but orthologues have yet to be uncovered by mutant analysis in Arabidopsis (and vice-versa), despite evidence that the ABA biosynthetic pathway is conserved among all plants (reviewed in Zeevaart, 1999; Liotenberg et al., 1999; Koornneef et al., 1998; Cutler and Krochko, 1999; Milborrow, 2001; Seo and Koshiba, 2002). All ABA-deficient mutants isolated to date are pleiotropic (in fact, only one of four ABA biosynthetic loci in Arabidopsis came from an ABA-based screen), but no ABA-null or ABA catabolism mutants have been uncovered. Many ABA response mutants have altered hormone levels and some ABA biosynthetic genes are regulated by ABA, suggesting that ABA metabolism is subject to feedback regulation. Taken together, these observations suggest that the processes of ABA homeostasis are complex. Genetic redundancy can account for some of the complexity, but little is known about the tissue specificity, subcellular compartmentation, or regulation of ABA metabolism. There is mounting evidence that manipulation of ABA biosynthesis or signaling can confer stress adaptation in transgenic plants, and that ABA homeostasis is part of a complex hormonal network that serves to integrate environmental inputs with intrinsic developmental programs (Chory and Wu, 2001). There is much yet to be learned about the molecular genetics of ABA metabolism, and it is anticipated that such knowledge will result in practical applications of agronomic importance.
Early, Shared Steps In ABA Biosynthesis
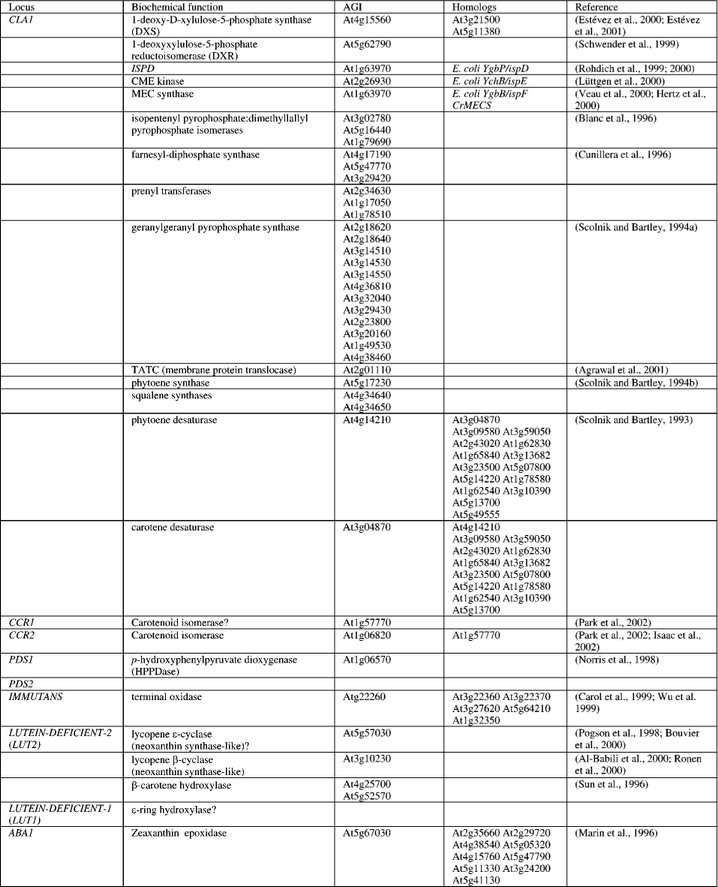
Until recently, it was thought that all isoprenoids in plants, of which there are tens of thousands including photosynthetic pigments (chlorophylls, tocopherols, carotenoids), hormones (ABA, gibberellins, cytokinins and brassinosteroids), and antimicrobial agents (phytoalexins) were synthesized from the cytoplasmic acetate/mevalonate pathway shared with animals and fungi (reviewed in DellaPenna, this edition). The plastidic MEP pathway, named for the first committed molecule (2C-methyl-D-erythritol-4-phosphate), was only recently discovered in plants and found to occur in protozoa, most bacteria, and algae (reviewed in Lichtenthaler, 1999). The MEP pathway produces isopentenyl pyrophosphate from glyceraldehyde-3-phosphate and pyruvate in the plastid for biosynthesis of isoprene, monoterpenes, diterpenes, carotenoids, plastoquinones and phytol conjugates such as chlorophylls and tocopherols. The discovery followed analysis of isotope labeling patterns in certain eubacterial and plant terpenoids that could not be explained in terms of the mevalonate pathway, which resolved a longstanding conundrum of why radiolabelled mevalonate that was fed to plants was not incorporated efficiently into ABA (reviewed in Milborrow, 2001). Prior to the elucidation of the MEP pathway, an Arabidopsis albino mutant, chloroplasts altered-1 (cla1), was described (Mandel et al., 1996) and later shown to encode 1-deoxy-D-xylulose-5-phosphate synthase; DXS (Table 1), the first enzyme of the MEP pathway (Estévez et al., 2000). Quantitation of isoprenoids, ABA, and measurement of physiological parameters in cla1 mutants and transgenic Arabidopsis plants that over- or under-express CLA1 showed that DXS was rate-limiting for isopentenyl diphosphate production and that ABA and other metabolites including GA were affected (Estévez et al., 2001). There are two conserved CLA1 homologues in Arabidopsis, 84% and 68% similar. Because deoxy-xylulose phosphate is shared by the MEP, thiamine (vitamin B1), and pyridoxine (vitamin B6) biosynthesis pathways, this shared metabolite may help explain why albino phenotypes occur in thiamine-deficient plants. Regulation of the early steps in isoprenoid biosynthesis may contribute to ABA biosynthetic rates.
Table 1.
Arabidopsis loci encoding enzymes required for ABA metabolism, listed in order of function within the biosynthetic pathways.
The subsequent four enzymatic steps of the MEP pathway have been characterized in bacteria and plants and their corresponding genes cloned (Table 1; reviewed in DellaPenna, this edition). The enzyme 1-deoxyxylulose-5-phosphate reductoisomerase, encoded by the DXR gene in Arabidopsis, produces the branched polyol MEP from 1-deoxy-D-xylulose when expressed in E. coli (Schwender et al., 1999). MEP is then converted to 4-diphosphocytidyl-2-C-methylerythritol (CME) by a CTP-dependent synthase homologous to E. coli YgbP/ispD (ISPD) with a putative plastid import sequence, consistent with its purported site of action in the plastid. Supportive evidence for this function comes from radiolabelling studies that show isolated chromoplasts of Capsicum incorporate CME into carotenoids (Rohdich et al., 1999), and the Arabidopsis ISPD cDNA when expressed in E. coli can catalyze the formation of CME from MEP (Rohdich et al., 2000).
The next step is phosphorylation of the 2-hydroxyl group of CME to CMEP by an ATP-dependent CME kinase, homologous to E. coli YchB/ispE gene and found in chromoplasts (Lüttgen et al., 2000). The Arabidopsis homologue of YchB/ispE protein is similar to that of the protein predicted by the tomato cDNA pTOM41 implicated in chromoplast biogenesis (Lüttgen et al., 2000).
The YgbB/ispF gene product of E. coli converts CMEP to 2-C-methyl-D-erythritol 2,4-cyclodiphosphate (MEC), and Capsicum chromoplasts contain this MEC synthase activity (Herz et al., 2000). There are ygbB/ispF homologues in Arabidopsis and Catharanthus roseus (CrMECS); the CrMECS transcript is up-regulated along with the DXR gene in cultured cells that produce monoterpene indole alkaloids (Veau et al., 2000). The final steps of the MEP pathway are unknown, but lead to isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP) which are substrates for isoprenoid biosynthetic enzymes. Nothing is yet known about the regulation of the MEP pathway.
There are three IPP:DMAPP isomerases in Arabidopsis (Blanc et al., 1996). The enzyme farnesyl-diphosphate synthase catalyzes the synthesis of farnesyl diphosphate (FPP) from IPP and DMAPP. Arabidopsis has at least three such genes (Cunillera et al., 1996). There are three prenyl transferases (prephytoene pyrophosphatase dehydrogenase), and 12 geranylgeranyl pyrophosphate synthase homologues (Scolnik and Bartley, 1994; Table 1).
The carotenoid biosynthetic pathway and genes are well characterized (see Cunningham and Gantt, 1998; Hirschberg, 2001; DellaPenna, this edition for reviews) and the corresponding Arabidopsis genes, along with some description of viviparous and ABA-deficient mutants of other species are briefly described here and listed in Table 1. The affected gene in a rice viviparous mutant that is pale green, wilty, and has reduced drought-induced ABA accumulation encodes a bacterial-like Sec-independent membrane protein translocase (OsTATC)(Agrawal et al., 2001) that may function in chloroplast biogenesis and therefore have indirect effects on ABA biosynthesis. EST databases indicate that a TATC homologue is expressed in Arabidopsis. A phytoene synthase and two related tandem squalene synthases (farnesyl-diphosphate farnesyltransferase) are expressed in Arabidopsis. Phytoene is subjected to four consecutive desaturation (dehydrogenation) reactions that lead to the formation of lycopene. Phytoene desaturation to ξ-carotene via phytofluene is catalyzed by phytoene desaturase (Scolnik and Bartley, 1993), and ξ-carotene desaturation to lycopene via neurosporene is catalyzed by ξ-carotene desaturase. These enzymes share significant homology to each other and a large family of flavin-containing oxidases and require a number of cofactors in plastids. The viviparous-5 mutant of maize is ABA deficient (Neill et al., 1986) and encodes a phytoene desaturase (Hable et al., 1998). Two groups have recently demonstrated by map-based cloning that carotenoid desaturation in plants requires a third distinct enzyme activity, a carotenoid isomerase (Park et al., 2002; Isaacson et al., 2002). The carotenoid and chloroplast regulation-2 (ccr2) gene was identified genetically in Arabidopsis by the partial inhibition of lutein synthesis in light and the accumulation of poly-cis-carotene precursors in dark-grown tissue. CCR2 is orthologous to the tangerine gene of tomato (Isaacson et al., 2002) and encodes the carotenoid isomerase CRTISO (Park et al., 2002). Genetic evidence for quinone and tocopherol requirements in carotenoid biosynthesis was obtained with the Arabidopsis phytoene desaturation (pds1, pds2) mutants. PDS1 encodes p-hydroxyphenylpyruvate dioxygenase (HPPDase), the first committed step in the synthesis of both plastoquinone and tocopherols (Norris et al., 1998). The pds2 mutant has yet to be characterized at the molecular level. The albino sectors of immutans (im) plants contain reduced levels of carotenoids (resulting in photo-oxidative damage to plastids) and increased levels of the carotenoid precursor phytoene. The IM gene product has amino acid similarity to the mitochondrial alternative oxidases, of which there are five structurally similar genes, suggesting that IM may function as a terminal oxidase in plastids (Carol et al., 1999; Wu et al. 1999). There are also two lycopene cyclase-like genes, β and ϵ expressed in Arabidopsis, one of which may also carry out neoxanthin biosynthesis since it has recently been shown in tomato and potato that neoxanthin synthase is a paralogue of lycopene cyclase and/or capsanthin capsorubin synthase (Bouvier et al., 2000; Al-Babili et al., 2000; Ronen et al., 2000) and there is no neoxanthin synthase homologue in Arabidopsis. Two β-carotene hydroxylase homologues are expressed in Arabidopsis (Sun et al., 1996). Two additional Arabidopsis mutants besides aba1 have been isolated that selectively eliminate and substitute a range of xanthophylls. The lutein-deficient-2 (lut2) mutation results in stoichiometric accumulation of violaxanthin and antheraxanthin at the expense of lutein and probably encodes the lycopene ϵ-cyclase (Pogson et al., 1998). The lut1 mutant accumulates the precursor of lutein, zeinoxanthin and may encode an ϵ-ring hydroxylase. The maize vivi-parous5 gene may encode phytoene desaturase (Liu et al, 1996); white-3, vp2, and vp12 genes have not yet been characterized at the molecular level but may encode phytoene desaturase components, and yellow-9 (y9) and vp9 may encode ξ-carotene desaturases, while the y3 and vp7 genes may encode lycopene cyclases (Robertson, 1961; Neill et al., 1986; Maluf et al., 1997; Janick-Buckner et al., 2001).
An allelic series of the first-described ABA-deficient mutant of Arabidopsis, aba (now designated aba1), came out of a suppressor screen of the non-germinating gibberellin-deficient ga1 mutant (Koornneef et al., 1982). The aba1 mutant alleles helped resolve a longstanding question of whether ABA biosynthesis was via a direct pathway from farnesyl pyrophosphate or through an indirect pathway from carotenoids, or both. Several lines of evidence suggested the latter:
the carotenoid-deficient viviparous mutants of maize, and plants treated with the carotenoid biosynthesis inhibitor fluridone, are ABA-deficient (Gamble and Mullet, 1986; Neill et al., 1986).
Heavy oxygen feeding studies and mass spectrometry of 18O-labeled ABA show 18O incorporation predominantly in the carboxyl group of ABA, which indicates a large precursor pool that already contains the ring oxygens for ABA (hypothesized to be a xanthophyll)(Creelman and Zeevaart, 1984).
Xanthoxal (previously called xanthoxin), an oxidative cleavage product of epoxycarotenoids found in plants, can be converted to ABA by cell-free extracts of plants (Sindhu et al., 1990).
There is a stoichiometric correlation between drought-induced ABA biosynthesis and xanthophyll changes in dark grown, water-stressed bean leaves (Li and Walton, 1990; Parry et al., 1990).
By quantitation of carotenoids and 18O-labeled ABA in the three aba1 alleles which exhibit different degrees of phenotypic severity of growth inhibition (Figure 1), Rock and Zeevaart (1991) found a correlation between the deficiencies of ABA and the epoxycarotenoids violaxanthin and neoxanthin. There was a corresponding accumulation of the epoxycarotenoid biosynthetic precursor zeaxanthin and a high percentage incorporation of 18O into the ring oxygens of ABA synthesized in the mutants (albeit small amounts of ABA, demonstrating a smaller precursor pool of epoxy-labeled ABA precursor [xanthophylls]). In addition to identifying the biochemical nature of the aba locus (zeaxanthin epoxidase, ZEP) and providing conclusive evidence for the indirect pathway of ABA biosynthesis from epoxycarotenoids (which was independently discovered by Duckham et al., 1991), the analysis of 18O labeling patterns of ABA and trans-ABA from the allelic series allowed inference about the physiological importance and source of the residual ABA in the mutants (Rock et al., 1992). It was concluded that all ABA was synthesized from carotenoids and a complete loss of ABA biosynthetic capacity in Arabidopsis would be lethal. A corollary to this hypothesis is that genetic redundancy might account for additional ABA biosynthetic capacity. The aba1 mutant has also proved a valuable resource to analyze the function of epoxycarotenoids, for example in photosynthesis and light-harvesting complex assembly, non-photochemical fluorescence quenching, and the xanthophyll cycle involved in protection of photoinhibition (Rock et al., 1992a; Pogson et al., 1998; Niyogi et al., 1998; Niyogi, 1999). Indeed, a mutant isolated on the basis of altered nonphotochemical quenching (npq2) is allelic to aba1 (Niyogi et al., 1998). The gene encoding the enzyme responsible for the reverse reaction, violaxanthin de-epoxidase, which is an important activity regulating the xanthophyll cycle, is encoded by the NPQ1 locus (Niyogi et al., 1998) and was previously cloned from lettuce (Bugos and Yamamoto, 1996).
The aba1 gene was first identified by virtue of the generation of a transposon-tagged, non-dormant wilty mutant of Nicotiana plumbaginifolia (Npaba2) that was shown to be orthologous to Arabidopsis aba1 (Marin et al., 1996). The molecular basis for two aba1 mutant alleles has been determined and the reduction in their AtZEP transcript levels correlates with the molecular defect identified (Audran et al., 2001). Arabidopsis ABA1 and NpABA2 orthologues encode a chloroplast-imported protein sharing similarities with mono-oxygenases and oxidases of bacterial origin. NpABA2 expressed in bacteria exhibits zeaxanthin epoxidase activity in vitro. The NpABA2 mRNA accumulates in all plant organs, but transcript levels are found to be higher in aerial parts (stems and leaves) than in roots and seeds. In seeds of Arabidopsis and tobacco, the ABA1/NpABA2 mRNA level peaks around the middle of development when ABA levels begin to increase. In conditions of drought stress, NpABA2/ABA1 mRNA accumulates concurrently with increases in ABA in roots but not in leaves of Arabidopsis, N. plumbaginifolia and tomato (Audran et al, 1998; 2001; Thompson et al., 2000a). Transgenic plants over-expressing NpABA2 mRNA exhibit increased ABA levels in mature seeds and delayed germination, while antisense NpABA2 expression results in a reduced ABA abundance in transgenic seeds and rapid seed germination (Frey et al., 1999). Homologues of AtABA1 have been cloned from tomato (Burbidge et al., 1997), Capsicum (Bouvier et al., 1996), and cowpea (Iuchi et al., 2000). The rice OsABA1 gene is an orthologue of ABA1 since a transposon-tagged Osaba1 mutant is viviparous, wilty, and ABA-deficient (Agrawal et al., 2001). In cowpea neither ABA nor drought stress regulate ZEP gene expression, while in tomato and Arabidopsis roots, but not leaves, drought induces ZEP mRNA accumulation (Burbidge et al., 1997; Iuchi et al., 2000; Audran et al., 2001). In tobacco and tomato leaves, ZEP expression is subject to diurnal fluctuations (Audran et al., 1998; Thompson et al., 2000a), which may be because epoxycarotenoids protect the photosynthetic apparatus from photo-oxidatative damage via the xanthophyll cycle. These results suggest that ZEP expression has a regulatory role in seeds and under some conditions ZEP may be rate-limiting for ABA and epoxy-carotenoid biosynthesis, which may be under feedback regulation. The ABA1locus is unique in Arabidopsis, but 9 other genes with significant (BLASTp E < 0.07) homology to ABA1 are present in the genome (see Table 1). However, there is no more homology between these Arabidopsis proteins and ABA1 than between ABA1 and putative flavoprotein mono-oxygenases and cyclic hydrocarbon hydroxylases in Streptomyces, Pseudomonas, E. coli and Bacillus, so their function in ABA biosynthesis is dubious. Nonetheless, they might be good candidates for reverse genetic studies of ABA metabolism (e.g. xanthoxal 4′-oxidase, ABA-8′-hydroxylase; see below).
Late, Specific Steps In ABA Biosynthesis
The well-established antagonism between ABA and gibberellin action in seed germination, originally elucidated by isolation of the aba1 mutant of Arabidopsis as a suppressor of ga1 (Koornneef et al., 1982), has been exploited by groups who have screened mutagenized seed for germination in the presence of the gibberellin biosynthesis inhibitor paclobutrazol. These screens have resulted in the isolation of two additional Arabidopsis ABA biosynthetic mutants (aba2, aba3; Leon-Kloosterziel et al., 1996). Additional alleles at aba1, aba2 and aba3 have been isolated from numerous other screens related to hormones, sugar, salt, or stress (Table 2). A molybdopterin organic cofactor (MoCo) chelates the trace element required for essential redox reactions in carbon, nitrogen and sulfur cycles such as nitrate reductase (NR), sulfite oxidase, xanthine dehydrogenase (XDH), and aldehyde oxidase (AO), and a barley mutant (nar2a) defective in MoCo synthesis and lacking NR, XDH, and AO enzyme activities is ABA deficient (Walker-Simmons et al., 1989). Protein extracts from aba2 and aba3 plants display a reduced ability to convert xanthoxal to ABA (Schwartz et al., 1997). Xanthoxal oxidase from tomato is specific for the natural S-(+)-ABA enantiomer (Yamamoto and Oritani, 1996). The next putative intermediate in ABA synthesis, ABA-aldehyde, is efficiently converted to ABA by extracts from aba2 but not by extracts from aba3 plants, indicating that the aba2 mutant is blocked in the conversion of xanthoxal to ABA-aldehyde (4′-hydroxyl oxidation and 1′,2′-epoxy isomerase activities; Figure 2) and that aba3 is impaired in the conversion of ABA-aldehyde to ABA (Schwartz et al., 1997). The ABA2 gene encodes an enzyme related to a family (>12 members) of short-chain dehydrogenase/reductases and is expressed throughout the plant, but its expression is not regulated by ABA, salt, or osmoticum (González-Guzmán et al., 2002). ABA2 can catalyze the conversion of xanthoxal to ABA-aldehyde in vitro (Cheng et al., 2002; González-Guzmán et al., 2002).
Table 2.
Arabidopsis mutants defective in ABA synthesis or response selected on the basis of other signaling defects.
ABA-aldehyde oxidase is non-specific for the natural (+) and unnatural (−) enantiomers of ABA (Yamamoto and Oritani, 1996; Schwartz et al., 1997; Seo et al., 2000). Extracts from the aba3 mutant also lack XDH activity. Treatment of aba3 extracts with Na2S restores ABA-aldehyde oxidase activity, suggesting that the genetic lesion of aba3 affects the final step of sulfurylation of the MoCo form required for AO and XDH activities (Schwartz et al., 1997; Figure 2). However, NR is unaffected in the aba3 mutant, indicating that the regulation of MoCo (or ABA-aldehyde oxidase) activity is more complex. Overexpression of NR was also seen in the ABA-deficient MoCo mutant Npaba1 of Nicotiana (Leydecker et al., 1995).
Finally, it has been recently suggested that xanthoxic acid, rather than ABA-aldehyde, is the immediate precursor to ABA, but the evidence is circumstantial (Cowan, 2001; Milborrow, 2001). Given the wide substrate specificity of aldehyde oxidases and the unknown nature of the 4′-hydroxyl oxidase and/or epoxyisomerases that could act on xanthoxic acid and xanthoxal, it may be that a metabolic matrix exists which includes both xanthoxic acid and ABA-aldehyde nodes (Figure 2). Because labelled IPP can be incorporated into ABA by washed, intact chloroplasts of spinach leaves, all three phases of ABA biosynthesis (terpenoid, carotenoid, and xanthoxal oxidation) occur within chloroplasts (Milborrow, 2001). However, Sindhu et al (1990) obtained contradictory results for the xanthoxal oxidation reactions using cell free extracts of bean that showed no xanthoxal oxidase activity by chloroplast extracts. Results with cell-free extracts from various species including Arabidopsis suggest that xanthoxal and ABA aldehyde oxidation steps are not rate-limiting for ABA biosynthesis (Sindhu et al., 1990; Schwartz et al., 1997).
Xiong et al. (2001a) have cloned the LOW OSMOTIC STRESS-5(los5)/aba3 locus by map-based methods and it has similarity to molybdopterin cofactor sulfurase. Bittner et al. (2001) simultaneously reported cloning aba3 by using degenerate PCR primers of conserved molybdenum cofactor genes from animals, and further showed that ABA3 could activate AO in vitro via a cysteine desulfurase activity. The LOS5/ABA3 gene is expressed ubiquitously in different plant parts and is induced by drought, salt, or ABA treatment. The N-terminal half of the 819 aa protein (there is evidence for splice variants (Bittner et al., 2001)) has significant homology to the class V family of pyridoxyl-phosphate-dependent aminotransferases (bacterial NifS-like), of which there are five members in Arabidopsis. There are another two Arabidopsis genes with 46% similarity to the novel C-terminus of ABA3 which are 74% similar to each other, suggesting this domain is functionally important (possibly in determining target enzyme specificity). The flacca (flc) mutant of tomato may also be orthologous to aba3 and Npaba1 because all three mutants are ABA-deficient, cannot convert xanthoxal or ABA-aldehyde to ABA, lack AO and XDH but not NR activities, and in vitro sulfurylation with Na2S reactivates preexisting XDH and AO proteins in flc, and Npaba1, as in aba3, extracts (Rock et al., 1991; Parry et al., 1991; Leydecker et al., 1995; Marin and Marion-Poll, 1997; Schwartz et al., 1997; Abaka et al., 1998; Sagi et al., 1999). The nar2a (Az34) MoCo mutant of barley lacks all AO, XDH and NR activities, suggesting an upstream lesion in the synthesis of the MoCo which all three enzyme activities require (Walker-Simmons et al., 1989; Rock, 1991). Two Arabidopsis cDNA clones (CNX2 and CNX3) encoding genes involved in early steps of molybdenum cofactor biosynthesis were obtained by functional complementation of E. coli mutants deficient in single steps of molybdenum cofactor biosynthesis (Hoff et al., 1995). The two cDNAs have significant identity to the E. coli moaA and moaC genes involved in molybdenum cofactor biosynthesis. CNX2 is expressed in all organs but most strongly in roots, while CNX3 is not expressed in abundant levels in any tissue but roots.
Only one Arabidopsis ABA mutant to date (Arabidopsis aldehyde oxidase-3, aao3) has come out of a leaf transpiration (“wilty”) screen; other screens for evaporation-enhanced “cool” mutants (e.g. Raskin and Ladyman, 1988) might also prove productive to identify ABA mutants. AAO3 encodes an aldehyde oxidase isoform, AOδ, that in conjunction with the sulfurylated MoCo form efficiently catalyzes the conversion of ABA-aldehyde to ABA (Seo et al., 2000b). The aao3 mutant is ABA-deficient in leaves and has no detectable AOδ activity, but seed dormancy is nearly normal, unlike all other ABA-deficient mutants. Analysis of growth, transpiration, and seed dormancy phenotypes of double mutant aba3/aao3 plants showed additive effects of the mutations, which could be caused by leakiness of the mutations or different tissue-specificities of AAO3 expression (ABA3 is expressed ubiquitously). Only slightly lower ABA levels were found in aao3 seeds compared to wild type, suggesting that other aldehyde oxidases may be involved in ABA biosynthesis in distinct tissues such as seeds. Three other homologous gene products, (AAO1, 74% similar; AAO2, 74% similar; and AAO4, 81% similar) are poor catalysts for ABA-aldehyde oxidation (Seo et al., 2000a; Hoff et al., 1998; Sekimoto et al., 1998). Two tandem copies of a xanthine dehydrogenase-like gene show 46% similarity with AAO3. A 58 aa gene fragment with 75% identity with AAO3 is also present in the genome. It is not clear which, if any, of these contribute to ABA biosynthesis.
The accumulation of AAO3 and ABA3 mRNAs is induced by dehydration and ABA in leaves (Seo et al., 2000a; Xiong et al 2001a; 2001b), suggesting that ABA-aldehyde oxidation is positively regulated by drought and ABA and is rate-limiting for ABA biosynthesis in leaves. However, AAO3 protein does not accumulate in drought-stressed leaves (Seo et al., 2000; M. Seo, unpublished observations). The regulatory mechanisms of the late steps of ABA biosynthesis are not known, but may involve RNA processing or turnover, or post-translational control (Xiong et al., 2001b).
The ABA-deficient mutants sitiens (sit) of tomato and droopy (dr) of potato are probably orthologs of the ABA-aldehyde oxidase apoprotein mutant aao3. This proposition is based on synteny of the mutants' map positions which are near to an aldehyde oxidase gene, TAO1, in these closely related species. In addition they both accumulate trans-ABA-alcohol, trans-ABA and catabolites (as does flacca), and are deficient in ABA-aldehyde oxidase activity (Taylor et al., 1988; Duckham et al., 1989; Rock, 1991; Rock et al., 1991; Marin and Marion-Poll, 1997; Ori et al., 1997). Although three putative aldehyde oxidase cDNAs have been cloned from tomato (Min et al., 2000), the sitiens gene has not yet been identified and it remains to be determined if sit, dr and aao3 (Seo et al., 2000b) are orthologues. It is unknown why cis and trans isomers of ABA-alcohol (likely reduced from ABA-aldehyde) accumulate in flc and sit mutants, but this phenomenon resulted in discovery of a minor shunt pathway of ABA biosynthesis from ABA-alcohol that bypasses ABA-aldehyde and is catalyzed by a P-450 mono-oxygenase (Rock et al., 1991; Figure 2). There are over 400 hypothetical monooxygenase genes in Arabidopsis (Initiative, 2000) and several steps in the carotenoid biosynthetic pathway also incorporate molecular oxygen catalyzed by this diverse and ubiquitous class of enzymes. The shunt pathway operates in all species of plants examined, including Arabidopsis (Rock et al., 1992b), but it does not appear to be regulated or physiologically significant except in those mutants which are impaired in ABA-aldehyde oxidation. Feeding studies with 18O2 and deuterium-labeled ABA-aldehyde isomers in the aba1 mutants suggested that synthesis of trans-ABA in Arabidopsis appears to be primarily via a parallel pathway from all-trans-epoxy-carotenoids, rather than isomerization at the xanthoxal or ABA-aldehyde/alcohol steps (Rock et al., 1992b).
The key ABA biosynthetic step, in terms of potential regulation by environmental and developmental cues, is the epoxy-carotenoid cleavage enzyme (termed NCED, for 9-cis-epoxy-carotenoid dioxygenase). The nature of this first committed step of ABA biosythesis was elucidated by cloning of a maize viviparous, ABA-deficient mutant vp14, which encodes a protein with homology to bacterial lignostilbene dioxygenases and whose transcript is strongly induced by water stress (Tan et al., 1997) and repressed by re-hydration. The VP14 protein is imported into chloroplasts with cleavage of a short stroma-targeting domain; the N-terminal 160 aa residues of the mature protein are necessary, but not sufficient, for thylakoid membrane targeting and were shown to associate with specific VP14-binding components (Tan et al., 2001). Schwartz et al. (1997) showed that a VP14 fusion protein could specifically cleave 9-cis-, but not all-trans-, xanthophylls to form xanthoxal and C25-apocarotenoids in a reaction that required oxygen, ferrous iron, ascorbate and a detergent. Homologous genes from several species have been isolated and characterized (Watillon et al., 1998; Qin and Zeevaart, 1999; Cherneys and Zeevaart, 2000; Iuchi et al., 2000; Thompson et al., 2000a). The de facto biosynthetic substrate(s) for the cleavage enzyme (neoxanthin, antheraxanthin, violaxanthin, and/or lutein-5,6-epoxide) are not known, but it has been assumed that 9-cis-neoxanthin is, and 9-cis-lutein-5,6-epoxide is not a substrate (Bungard et al., 1999; Milborrow, 2001). Studies on the double mutant lut2/aba1, which is deficient in all species of epoxycarotenoids, did not show a more severe ABA-deficient phenotype compared to aba1 alone (Pogson et al., 1998), suggesting that 9-cis-lutein-5,6-epoxide is not a substrate for the cleavage enzyme in planta. The 2-cis-double bond of ABA is essential for its biological activity but it is not known how the relevant trans bond in violaxanthin, neoxanthin, and/or antheraxanthin is isomerized or whether this step is regulated to modulate ABA biosynthesis. The molar amounts of 9-cis-neoxanthin substrate in leaves exceeds by about two orders of magnitude the molar amounts of ABA, whereas 9-cis-violaxanthin, antheraxanthin and lutein epoxide are present in similar molar amounts to ABA. In etiolated bean leaves, ABA levels increased up to 40-fold under stress, whereas the level of 9-cis-violaxanthin showed only a minor decrease (Li and Walton, 1990). In stressed tomato roots, a decrease in trans-neoxanthin was observed, which suggests that it is an intermediate between trans-violaxanthin and 9-cis-neoxanthin (Parry et al., 1992). Taken together, these data suggest that 9-cis-neoxanthin is probably the major substrate for the cleavage enzyme. Nothing is known about which subcellular pools of epoxy-carotenoids are available as substrates for isomerization and cleavage (e.g. those found in oil bodies, or complexed with membranes- or proteins), or whether suborganellar localization of VP14 in plastids is a regulatory mechanism in ABA biosynthesis
The ABA-deficient notabilis (not) mutant of tomato is probably orthologous to vp14 (Burbidge et al., 1999). However, it also shows some ABA-aldehyde oxidase-affected phenotypes such as relatively more trans-ABA biosynthesis than wild type, increased loss of 18O label in the ABA side chain (due to exchange with aqueous medium; Rock et al., 1991), and slightly reduced ABA-aldehyde oxidase activity (Taylor et al., 1988). Ectopic expression of a tomato NCED cDNA causes overproduction of ABA in tomato and tobacco, which suggests that NCED is rate-limiting for ABA biosynthesis (Thompson et al., 2000b). Seven predicted NCED genes (Table 1) are found in Arabidopsis, as well as genes encoding two additional distantly related proteins (37 and 41% similarity with AtNCED1, respectively). Although AtNCED1 shows sequence similarity to VP14/NOT and is drought-induced (Neill et al., 1998), this gene does not have a plastid-targeting N-terminal peptide and does not function specifically as a 9-cis-epoxycarotenoid dioxygenase, but rather has a relaxed specificity in vitro for xanthophylls and has been renamed AtCCD1 (Carotenoid Cleavage Dioxygenase; Schwartz et al., 2001). Iuchi et al (2001) cloned all seven cDNAs corresponding to AtNCED1-6&9 and showed that only AtNCED3, and to a lesser extent AtNCED9, was induced by drought stress. This apparent controversy of drought inducibility of AtNCED1 may be due to different experimental conditions between groups; it has been shown in bean that drought-induced NCED expression is dynamic, with transcripts accumulating rapidly and decreasing to pre-stress levels within 24 hr (Qin and Zeevaart, 1999). Overexpression of AtNCED3 in transgenic Arabidopsis caused an increase in endogenous ABA levels, promoted transcription of ABA-inducible genes, decreased transpiration rate of leaves, and improved plant tolerance to drought (Iuchi et al., 2001; Table 1). Similar results, including increased seed dormancy, have been reported for transgenic tobacco expressing the bean PvNCED1 (Qin and Zeevaart, 2002). By contrast, antisense supression and T-DNA knockout lines of AtNCED3 gave a drought-intolerant phenotype. These results demonstrate the key role of AtNCED3 in ABA biosynthesis under drought-stress and show that physiological processes of agronomic significance may be manipulated in crops by genetic engineering of ABA biosynthetic genes.
An amazing aspect of ABA biosynthesis is its regulation by the water status of the plant, first demonstrated by Wright and Hiron (1969). Loss of cell turgor stimulates ABA biosynthesis, but little is known about how cell wall/membrane interactions might be coupled to transcriptional events. Stretch-activated plasma membrane ion channels are one possibility (Ding and Pickard, 1993). Factors besides dehydration, such as low and high temperatures, salt and flooding have been reported to cause rises in ABA, but these factors all share a common phenomenon of water deficit. A crucial link between ABA signalling and osmotic stress perception was recently elucidated with the demonstration that an Arabidopsis transmembrane two-component histidine kinase (AtHK1; At2g17820) is a functional osmosensor (Urao et al., 1999). Expression of the AtHK1 gene is increased by ABA, drought, hypotonic solutions, cold, and salt stress. It is a member of a family of two-component sensors in Arabidopsis that include the cytokinin WOODEN LEG/CYTOKININ-RESPONSE1 (WOL/CRE1, AHK4) and ethylene (ETR, EIN, ERS) receptors (Inoue et al., 2001; Ueguchi et al., 2001; Mahonen et al., 2000). Perhaps the osmosensor is the mechanism for initiating ABA biosynthesis and other stress responses, thereby integrating ABA signalling with other overlapping stress pathways.
ABA Catabolism
Much is known about ABA catabolism in various species, and the reader is referred to recent excellent reviews on the subject (Cutler and Krochko, 1999; Zeevaart, 1999). Arabidopsis utilizes the two major pathways of ABA catabolism: (i) hydroxylation of ABA at the 8′ position by a P-450 type monoxygenase (which has been partially characterized in vitro (Krochko et al., 1998)) to give an unstable intermediate (8′-OH-ABA) that rearranges spontaneously to phaseic acid (PA), and (ii) esterification of ABA to ABA-glucose ester (ABA-GE; Fig. 2). Conjugation of ABA to ABA-GE is irreversible (Zeevaart and Boyer, 1984). The PA pathway predominates in Arabidopsis (Rock et al., 1992; Windsor and Zeevaart, 1997) and therefore the ABA-8′-hydroxylase is the rate-limiting step in catabolism and a likely target for regulation at the transcriptional level by water status (dehydration and rehydration). Other known minor catabolites such as the ABA-diols, 7′-hydroxy-ABA, ABA-1′-glucoside, 3-hydroxy-3-methylglutaryl conjugate of 8′-hydroxy-ABA, phaseic and dihydrophaseic glucose esters and glucosides have not been studied in Arabidopsis but probably occur. Little is known about the enzymes that catalyze ABA breakdown and no genes have been cloned that encode these activities.
The concentration of ABA is negatively regulated by phytochrome action such that a phytochrome-deficient (pew1) mutant of Nicotiana is hyperdormant, drought resistant, and accumulates more ABA than wild type (Krapiel et al., 1994). To determine whether this reflects phytochrome regulation of ABA biosynthesis or degradation, double mutants combining the pew1 mutation with a defect in ABA biosynthesis (the Npaba1 mutation) were analyzed. Npaba1 mutants accumulate the glucoside of trans-ABA-1-alcohol (Kraepiel et al., 1994; Schwartz et al., 1997), reflecting the increased accumulation of trans-ABA-1-alcohol due to blockage at the ABA-aldehyde oxidation step. If the pew1 mutation resulted in de-repression of ABA biosynthesis at an early (e.g. cleavage) step, more trans-ABA-alcohol-glucoside would accumulate in the double mutant, but this was not observed (Krapiel et al., 1994). The phytochrome- and ABA-deficient double mutants (pew1 Npaba1) were non-dormant due to their ABA deficiency yet accumulated no more trans-ABA-alcohol glucoside than Npaba1 alone, indicating that phytochrome regulates ABA degradation rather than ABA biosynthesis (Krapiel et al., 1994). Additional evidence consistent with a model of phytochrome regulation of ABA metabolism comes from a photoperiod mutant of tomato that overproduces ABA (Fellner et al., 2001). Because the 8′-hydroxylase activity is rate-limiting for ABA catabolism and is up-regulated by ABA and stress (Krochko et al., 1998; Windsor and Zeevaart, 1997), it is an important target for genetic engineering of ABA levels in plants since ABA accumulation triggers its own degradation. Consistent with this, abi1-1, abi2-1 and abi3-1 mutants accumulate 1.5- to nearly three-fold as much ABA as wild-type seeds (Koornneef et al., 1984), while the supersensitive sad1 mutants have decreased ABA levels (Xiong et al., 2001a).
ABA PERCEPTION
Recognition Site(s)
There is indirect evidence for both intracellular and extracellular ABA receptors. Schwartz et al (1994) tested by three different methods whether ABA can act from within guard cells. They first observed a correlation of the extent to which ABA inhibits stomatal opening and promotes stomatal closure in Commelina in proportion to radioactive ABA uptake. They then demonstrated that microinjection of ABA into the cytoplasm of Commelina guard cells triggered stomatal closure. Finally, they observed that application of ABA to the cytosol of Vicia guard-cell protoplasts via a patch-clamp electrode inhibited inward K+ currents, a stimulus sufficient to prevent stomatal opening. Similarly, Allan et al (1994) demonstrated that stomatal closure ensued after the intracellular release of microinjected “caged” ABA by photolysis. These results are consistent with, but do not prove, an intracellular site of phytohormone action. Recent patch clamp studies of Ca2+ flux across the Vicia guard cell plasma membrane show ABA induces rapid Ca2+ channel activation when added to the cytosolic side of inside-out patches, but delays activation when added in the cell-attached configuration (Hamilton et al., 2000), consistent with existence of an ABA receptor in close association with the Ca2+ channel on the cytoplasmic side of the plasma membrane. In contrast, after observing that extracellular application of 10 µM ABA inhibited stomatal opening by 98% at pH 6.15, but only by 57% at pH 8.0 when the ABA is unable to cross the membrane as an anion, Anderson et al (1994) concluded that intracellular ABA alone did not suffice to inhibit stomatal opening under the imposed conditions. This led them to suggest that a reception site for ABA-mediated inhibition of stomatal opening must exist on the extracellular side of the plasma membrane of guard cells. MacRobbie (1995) observed a correlation between buffered high external pH conditions and attenuation of ABA-induced ion efflux. Furthermore, extra-cellular ABA perception was observed in two studies using ABA-protein conjugates that cannot enter the cell, yet are biologically active, to induce ion channel activity (Jeannette et al., 1998) and gene expression (Schultz and Quatrano, 1997; Jeannette et al., 1998). Taken together, these results are consistent with a contribution of both extracellular and intracellular ABA receptors. However, other interpretations are possible, for example direct ABA action on plasma and tonoplast membranes or ion channels from the cytoplasmic side, higher affinity of an ABA receptor for the protonated form, or pH-dependent pathways.
Additional indirect in vitro evidence for an ABA receptor complex at the cell surface was provided by surface plasmon resonance studies in conjunction with a protoplast ABA-inducible gene expression assay (Desikan et al., 1999). JIM19 is one of a panel of monoclonal antibodies generated against pea guard cell protoplasts that recognizes a cell surface glycoprotein and can modulate ABA responses in barley aleurone and rice protoplasts (Wang et al., 1995; Desikan et al., 1999). Desikan et al. (1999) observed specific binding of plasma membranes to JIM19 that was antagonized significantly by ABA but not by the biologically inactive ABA catabolite phaseic acid. The in vitro interactions of plasma membranes with JIM19, ABA, and phaseic acid correlated with the biological activities of these reagents on activation of Em-GFP and Em-GUS promoter-reporter genes measured by flow cytometry and enzyme assays, respectively. Taken together, these data suggested that JIM19 interacts with a functional complex involved in ABA signalling.
Another indirect biochemical assay using plasma membrane-enriched fractions from barley aleurone protoplasts allowed Ritchie and Gilroy (2000) to observe ABA-stimulated PLD activity. The transient nature (20 min) and degree (1.5- to 2-fold) of activation in vitro were similar to that measured in vivo. The activation of PLD in vitro by ABA was dependent on the presence of GTP. Addition of GTP γS transiently stimulated PLD in an ABA-independent manner, whereas treatment with GDP βS or pertussis toxin blocked the PLD activation by ABA. Remarkably, the sole Gα subunit in Arabidopsis is required for ABA inhibition of stomatal opening and pH-independent activation of anion channels (Wang et al., 2001). These results suggest the existence of an ABA receptor system and elements (e.g. glycoproteins) at the plasma membrane linked via G proteins to PLD activation.
The use of ABA analogs in germination and gene expression bioassays has allowed the inference of multiple ABA receptors with different structural requirements for activity in different response pathways (Walker-Simmons et al., 1997; Kim et al., 1999). Given the lack of concrete leads, the search for ABA receptors should include intracellular compartments, proteins regulated by or involved in ABA responses, and non-proteinaceous molecules. It is critically important for any receptor studies to correlate the specificity of interaction with ABA analogs possessing different degrees of biological activity.
Potential Receptor(s)
Genetic approaches to ethylene, cytokinin, and brassinosteroid signaling have yielded cognate hormone receptor mutants, but to date no ABA receptor mutants have been described. A variety of ABA-binding proteins and carriers (Pédron et al., 1998; Zhang et al., 2001; Windsor et al., 1994) have been reported, but until very recently there was only an unconfirmed report of a potential receptor (Hornberg and Weiler, 1984). Immunological evidence has now been reported for an ABA-binding protein that is linked to an ABA-mediated physiological process, making this protein a prime candidate for an ABA receptor. Zhang et al. (2002) purified to apparent homogeneity by affinity chromatography a 42 kD ABA-specific binding protein from the epidermis of broad bean leaves. The protein had an equilibrium Kd of 21 nM for ABA with one apparent binding site, and R-(−)-ABA and trans-ABA were incapable of displacing 3H-(B1)-ABA bound to the protein, establishing its stereospecificity for natural ABA. Pretreatment of guard cell protoplasts of bean leaves with a monoclonal antibody raised against the 42 kD protein significantly decreased, in a dose-dependent manner, the ABA-induced PLD activity of protoplasts, providing exciting evidence for the ABA receptor-like nature of the 42 kD protein. It will be interesting to determine if the monoclonal antibody can antagonize other ABA responses.
Consistent with the notion of a cytoplasmic receptor, Zhang et al (2001) found cytosolic ABA-binding proteins from apple fruit that showed stereospecificity for R-(+)-ABA. Additional circumstantial evidence for an intracellular ABA receptor comes from results of Zheng et al. (1998), who probed a maize cDNA expression library with anti-ABA-binding-protein antibodies and isolated a clone with 60% homology to nucleic acid binding proteins. This ABA-binding protein appears to be present in a complex that includes rRNA, possibly providing a mechanism for direct ABA regulation of translation.
Polyclonal antiserum raised against an anti-ABA monoclonal antibody, such that some of the antibodies could mimic the structure of ABA and therefore bind to ABA-binding proteins, including an ABA receptor, has been used to identify an ABA-inducible gene product from barley embryos, designated aba45 (Liu et al., 1999). There is a family of aba45 homologues in Arabidopsis [At5g13200; At1g28200; At4g01600; At5g23350; At5g23360; At5g23370; At5g08350; At4g40100], two of which (At1g28200, At5g08350) were identified in a yeast two hybrid screen using an Arabidopsis formin-like protein AFH1 (At3g25500; Banno and Chua, 2000), which itself is a member of a large gene family homologous to yeast BNR1 and BNI1 genes involved in budding, cell polarity, cytokinesis, and filament formation (Drees et al., 2001). Transgenic studies have also shown that the small GTPase Rop6/AtRac1 can inhibit ABA effects on actin cytoskeleton reorganization in guard cells (Lemichez et al., 2001). However, there is yet no evidence, e.g. specific and saturable binding of ABA to the gene product, to indicate the aba45-like gene encodes an ABA receptor or that it interacts with the cytoskeleton.
ABA is known to regulate plasma membrane and tonoplast ion channel activities very rapidly (Assmann and Shimazaki, 1999) and it is plausible that ABA interacts directly with transport proteins or other metabolic factors such that enzymes or complexes may have allosteric sites for ABA binding. ABA also has direct effects on membrane fluidity and thermal behavior (Shripathi et al., 1997), which suggests that some ABA activities may not require interaction with a receptor.
It is quite possible that multiple ABA response mechanisms operate simultaneously in plants (and animals). Sutton et al (2000) used microinjection of cell-specific Vicia faba mRNA pools into Xenopus oocytes to demonstrate that an ABA signal transduction pathway exists in frog oocytes that can be coupled to a mesophyll cell-specific K+ outward-rectifying channel, but not to a co-expressed guard cell-specific K+ inward-rectifying channel whose ABA regulatory mechanism is encoded solely by a co-expressed guard cell mRNA population. The authors concluded that mesophyll cells and guard cells use distinct and different receptor types and/or signal transduction pathways in ABA regulation of K+ channels. The nature of the frog ABA perception pathway is unknown, e.g. whether it includes an ABA receptor or only downstream elements, but the system should be useful in characterizing ABA response mechanisms, for example by expression cloning of rate-limiting or autonomously functioning components. A potential shortfall of the expression cloning approach could be encountered if the response mechanism is multifactorial and/or nonlinear. To date, only one specific gene product has been identified by using an oocyte system to screen for clones contributing to an ABA-stimulated Ca2+-dependent Cl− current: a syntaxin-like protein, Nt-SYR1 (Leyman et al 1999). Although regulated by ABA and stress signals, its effect on ABA signaling is likely to be indirect. It is hoped that systematic analyses of the entire proteome, combined with biochemical, genetic, and cell biological studies will finally elucidate the enigmatic ABA receptor(s).
IDENTIFICATION OF SIGNALING INTERMEDIATES
Three major approaches have been used to identify regulatory factors controlling ABA response: genetics, biochemistry, and pharmacology/cell biology (reviewed in Rock, 2000). Genetic studies have screened for aberrant responses to ABA, based on either a physiological phenotype or aberrant expression of a marker gene. Biochemical studies have identified cis-acting regulatory regions required for “correct” expression of ABA-inducible genes, then used ligand-binding assays or yeast one-hybrid screens to isolate genes encoding proteins that specifically recognize these DNA sequences. In addition, a variety of ABA-activated or -induced kinases, phosphatases, phospholipases, and transcription factors have been analyzed to determine whether the correlations with ABA-induced gene expression or protein activation reflect any functional significance. Cell biological studies have tested the roles of candidate secondary messengers and signaling intermediates in regulating cellular responses such as stomatal closure or ABA-inducible gene expression. The completion of the Arabidopsis genome sequence (Initiative, 2000) has provided a fourth, in silico, approach to identifying potential regulatory factors. However, this approach requires functional testing by reverse genetics to identify the physiological role of any potential regulator. The fact that >60% of the genes in Arabidopsis belong to multi-gene families suggests a strong potential for functional redundancy that could mask the effects of loss of function alleles isolated by traditional forward genetic strategies. Indeed, there is conclusive evidence for such redundancy in ABA biosynthesis and signaling, suggesting that ABA mutant or engineered phenotypes may be tissue-specific and subtle.
Genetic Approach
Hormone response mutants have traditionally been defined as individuals that resemble mutants with defects in hormone biosynthesis, yet can not be restored to a wild type phenotype by addition of the relevant hormone. Identification of such mutants is complicated by the possibility that a hormone-insensitive phenotype can result from changes in a hormone-independent process. For example, a wilty phenotype may result from defects in ABA biosynthesis or response. However, wilty mutants of maize and tomato have been characterized whose primary defects appear to be abnormal vascular tissue, resulting in decreased water flow through the plant (Postlethwait and Nelson, 1957; Alldridge, 1964; Rock and Ng, 1999). In general, the classification of hormone-response mutant is reserved for those showing highly pleiotropic phenotypes. It is worth noting that many of the first-isolated hormone response mutants (e.g. abi1, abi2, axr2, gai, and etr1) have dominant phenotypes that were instrumental in elucidating genetically redundant processes.
Screens, selections, and questions
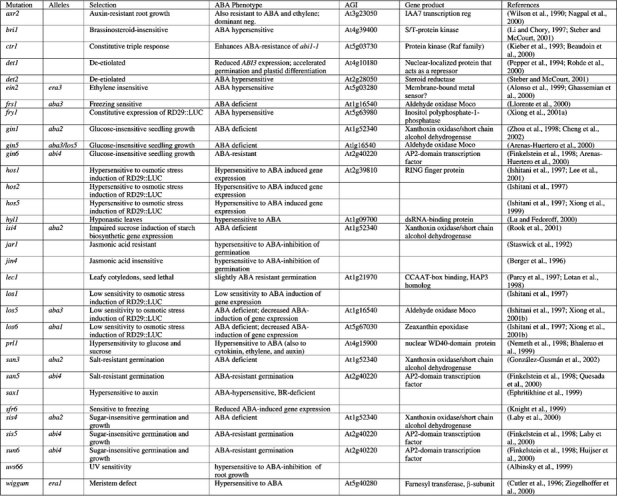
The genetic screens and selections that have been used to date include production of non-dormant seeds (Koornneef et al., 1982); loss or gain of sensitivity to ABA at germination (Koornneef et al., 1984; Finkelstein, 1994a; Cutler et al., 1996), seedling growth (Lopez-Molina and Chua, 2000), or root growth (Himmelbach et al., 1998); mis-expression of reporter genes (Ishitani et al., 1997; Foster and Chua, 1999; Delseny et al., 2001); and screens for suppressors or enhancers of GA-deficient non-germinating lines or ABA-INSENSITIVE (ABI) lines (Steber et al., 1998; Beaudoin et al., 2000; Ghassemian et al., 2000) (Table 3). Additional mutants have been isolated with defects in responses to multiple signals, including ABA, via non-ABA-based screens such as salt-resistant germination (Quesada et al., 2000), sugar-resistant seedling growth or gene expression (Arenas-Huertero et al., 2000; Huijser et al., 2000; Laby et al., 2000; Rook et al., 2001), or defects in auxin, brassinosteroid or ethylene response (Wilson et al., 1990; Alonso et al., 1999; Ephritikhine et al., 1999; Li et al., 2001) (Table 2). The fact that mutations in only some of the hormone response genes appear to affect multiple signaling pathways suggests that interactions among these pathways are relatively specific. Possible mechanisms of cross-talk are discussed in many recent reviews (McCourt, 1999; Sheen et al., 1999; Gibson, 2000; Coruzzi and Zhou, 2001; Gazzarrini and McCourt, 2001).
Table 3.
Arabidopsis mutants selected on the basis of altered ABA sensitivity.
Studies of ABA biosynthesis and response mutants have been used to address three fundamental questions: 1) what is the biological role of ABA or any given locus in regulating specific growth responses, 2) what are the products of these loci, and 3) how do they interact to regulate hormone response? The fact that screens based on altered dormancy have successfully identified ABA deficient mutants with pleiotropic effects on ABA responses indicate that ABA is required for dormancy induction, some aspects of seed maturation, and drought-induced stomatal closure (Koornneef et al., 1982). However, the specific role of ABA in germination inhibition at seed maturity and regulation of stress tolerances is less clear. For example, although effective for conferring tolerance to abiotic stresses such as drought, cold, and salinity, ABA is not essential for all responses to these stresses even though they share the common element of dehydration stress (reviewed in Shinozaki and Yamaguchi-Shinozaki, 2000). This initially led to the view that there were ABA-dependent and ABA-independent pathways of response presumed to be functioning in parallel (Nordin et al., 1991). Further analysis of specific signaling mutants provided evidence for cross-talk among these pathways (Ishitani et al., 1997), such that ABA is now considered part of an interconnected signaling network (Shinozaki and Yamaguchi-Shinozaki, 2000).
Identities, interactions, and implications of genetically defined ABA signaling loci
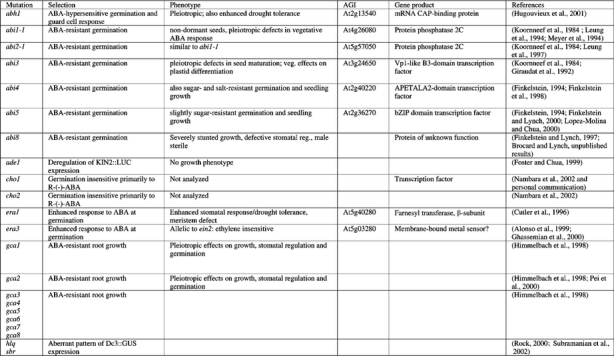
ABI1 and ABI2 were initially identified by mutations resulting in pleiotropically decreased sensitivity to ABA (Koornneef et al., 1984). Subsequent studies showed that both the abi1-1 and abi2-1 mutants are incompletely dominant, with the degree of dominance varying among responses and with unidentified environmental or developmental factors (Finkelstein, 1994b). These loci encode highly homologous members of the PP2C family of ser/thr protein phosphatases (Leung et al., 1994; Meyer et al., 1994; Leung et al., 1997; Rodriguez et al., 1998a) and the original mutations have identical amino acid substitutions in their catalytic domains, resulting in decreased phosphatase activity and a dominant negative phenotype (Leung et al., 1997; Gosti et al., 1999). Together with over-expression studies in protoplasts (Sheen, 1996, 1998), these results indicate that these PP2Cs are likely to act as negative regulators of ABA response. The inducibility of ABI1 and ABI2 gene expression by ABA may be part of a feedback loop that resets the cell to monitor ABA levels continuously, or they might act at distinct steps such that their coordinate induction has a double negative, i.e. positive, effect on ABA response. BLAST analysis of the Arabidopsis thaliana genome shows that the PP2C family contains 69 members, 25 of which contain two conserved G residues correlated with ABA signaling (Figure 3). In addition to ABI1 and ABI2, two other family members (AtPP2C and AtPP2C-HA) have been shown to repress ABA response when over-expressed (Rodriguez et al., 1998b; Sheen, 1998) (Table 4). A comprehensive reverse genetic approach should enable us to learn how many more of these PP2Cs are actually involved in ABA responses.
Figure 3.
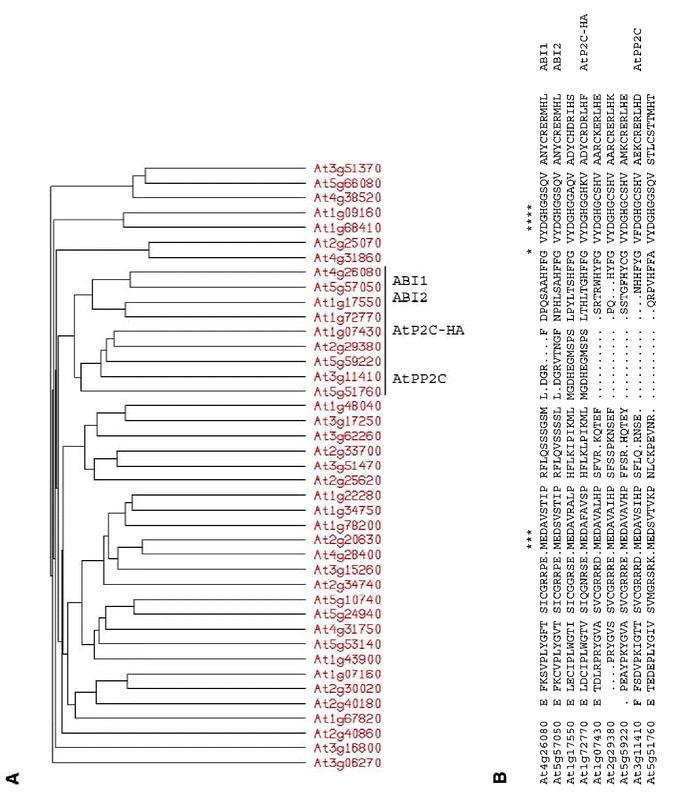
Homologies among Arabidopsis PP2Cs. (A) ABI1 and 40 most closely related predicted Arabidopsis proteins identified by a BLAST search (identified by AGI number for the corresponding genes) were analyzed by the Pileup program of GCG, using progressive pair-wise alignments. All four of the PP2Cs previously shown to affect ABA signaling are present in a subfamily of nine members (comprising approximately 13% of the PP2Cs identified by BLAST analysis), underlined and labeled on the dendrogram. (B) Comparison of a conserved region of the PP2C domain within the ABI1 subfamily. Residues labeled with * have been shown to be critical to phosphatase function and/or ABA signaling. Although this and several other conserved areas extend well beyond these critical residues, these homologies are limited to this subfamily.
Table 4.
Arabidopsis genes implicated in ABA response by reverse genetics
The molecular similarity between abi1-1 and abi2-1 has led to the suggestion that their products might act on overlapping subsets of substrates (Leung et al., 1997), but the physiological characterization of the mutants shows that the defects due to the abi1-1 mutation are more extensive. Furthermore, studies of guard cell signaling in these mutants have suggested that ABI1 and ABI2 act either at distinct steps or in parallel pathways (Pei et al., 1997). Although another PP2C has been shown to negatively regulate a mitogen-activated protein kinase (MAPK) associated with stress response in alfalfa (Meskiene et al., 1998), to date only one candidate substrate for ABI1 has been reported: a homeobox-leucine zipper transcription factor shown to interact in a yeast two-hybrid assay (Himmelbach and Grill, 2001). Yeast two-hybrid studies have also shown an interaction between ABI2 and SOS2 (Xiong and Zhu, 2001), a serine/threonine protein kinase identified on the basis of its role in salt-stress signaling (Liu et al., 2000). Both the ABI4 and ABI5 gene products (described below) contain ser/thr-rich domains that could be sites of phosphorylation (Finkelstein et al., 1998; Finkelstein and Lynch, 2000a) and recent studies have demonstrated that ABI5 protein is stabilized by ABA-induced phosphorylation (Lopez-Molina et al., 2001). Although either ABI4 or ABI5 could be a substrate for dephosphorylation by the ABI PP2Cs, consistent with a negative regulatory role for the PP2Cs, neither interacts with ABI1 in a two-hybrid assay (Nakamura et al., 2001) despite showing strong genetic interactions as digenic mutants (Finkelstein, 1994a). Recently two ABI5-related transcription factors, AREB1 and AREB2, were shown to promote ABA-activation of target gene expression (Uno et al., 2000). This activation was repressed by either protein kinase inhibitor treatment of wild-type cells or the dominant negative abi1-1 mutation. Similarly, overexpression of abi1-1 inhibits transactivation of ABA-inducible promoters by either ABI5 (Gampala et al., 2001a) or VP1/ABI3 (Hagenbeek et al., 2000). It is not known whether these results reflect direct or indirect effects on phosphorylation status of these or other transcription factors.
The three remaining cloned ABA insensitive loci, ABI3, ABI4, and ABI5, encode transcription factors of the B3-, APETALA2- (AP2), and basic leucine zipper- (bZIP) domain families, respectively, and regulate overlapping subsets of seed-specific and/or ABA-inducible genes (Giraudat et al., 1992; Finkelstein et al., 1998; Finkelstein and Lynch, 2000a; Lopez-Molina and Chua, 2000). ABI3 contains four conserved domains: an acidic activation domain and three basic domains (B1, B2 and B3). Similarities in sequence and mutant phenotype have led to the suggestion that ABI3 and maize VIVIPAROUS1 (VP1) are orthologs, which was recently validated by complementation of most abi3 defects by VP1 expression (Suzuki et al., 2001). ABI3 can activate transcription in vivo and the conserved B3 domain of VP1binds in vitro to the conserved RY element present in the C1 and Em promoters (Suzuki et al., 1997). However, this domain is not essential for ABA-regulated gene expression in the seed (Carson et al., 1997). Mutational studies have shown that the B2 domain is critical for regulation of Em and 2S albumin genes (Bies-Etheve et al., 1999), and interaction with an ABA response element (ABRE) (Ezcurra et al., 2000) and the ABRE-binding protein EmBP-1 (Hill et al., 1996). However, the intact purified protein does not specifically bind DNA in vitro, suggesting that it interacts with other proteins that mediate DNA binding (Suzuki et al., 1997). Consistent with this hypothesis, mutational analyses of VP1/ABI3-responsive promoters have shown that ABREs are sufficient but not necessary for VP1 transactivation (Vasil et al., 1995). VP1 also acts as a transcriptional repressor of some genes that are induced by GA during germination (Hoecker et al., 1995); domain mapping has shown that the VP1 repressor function is distinct from the activation domain.
Presumed DNA-binding and protein interaction domains are also present in ABI4 and ABI5: the AP2 and bZIP domains, respectively. Although ABI4 is most closely related to the Drought Response Element Binding (DREB) subfamily of the AP2-domain family, the similarity is confined to the AP2 domain. DRE cis-elements are not present in a variety of ABI4-regulated genes (Finkelstein, unpublished observations) and the target sequence for ABI4 binding is currently unknown. In contrast, ABI5 was identified independently by homology to a sunflower gene isolated via a yeast one-hybrid screen using the Dc3 promoter as bait and was designated AtDPBF1 (Arabidopsis thalianaDc3 Promoter Binding Factor 1) (Kim et al., 2002). In vitro studies with the DPBFs have demonstrated that this subfamily binds to G-box elements required for ABA regulation and consequently designated ABREs (ABA response elements) (Kim et al., 1997). However, the ABI5/DPBF subfamily has a broader consensus-binding site than the other bZIP proteins in that its members tolerate variability in the ACGT core element essential to the ABRE G-box. Analyses of transcript accumulation in abi5 mutants suggest that ABI5 also has both activator and repressor functions, but that ABI5 and ABI3 may have either synergistic or antagonistic effects on gene expression, depending on the gene (Finkelstein and Lynch, 2000a; Delseny et al., 2001). Synergistic interactions between ABI5 and the ABI3 ortholog VP1 have been demonstrated in rice protoplasts (Gampala et al., 2002). It will be interesting to learn whether ABI3/VP1 interacts with a broad range of the DPBFs.
Recent yeast two-hybrid studies have shown that ABI3 and ABI5 interact directly via the B1 domain of ABI3 (Nakamura et al., 2001), suggesting that ABI5 binding to ABRE elements may tether ABI3 to some of its target promoters and facilitate its interaction with RY elements and transcription complexes. Consistent with this, an ABI5 homolog from rice was identified in a yeast two-hybrid screen using the basic domains of rice VP1 as bait (Hobo et al., 1999). Two-hybrid screens using the B2 and B3 domains of ABI3 as bait have identified interactions with several presumed transcription factors, including a CONSTANS-related factor, the RPB5 subunit of RNA Pol II, and a homolog of the Human C1 protein involved in cell cycle control (Kurup et al., 2000). Additional interactions may involve other bZIP proteins, such as the ortholog of EmBP1, that may be indirectly linked to ABI3 via interactions with a 14-3-3 protein, as described for connections among the maize proteins EmBP1, Vp1 and GF14 (Schultz et al., 1998). Such interactions may either promote or inhibit DNA binding (Nantel and Quatrano, 1996), and may trigger chromatin remodeling to permit ABA-mediated gene activation (Li et al., 1999).
The LEC1 gene encodes another class of transcriptional regulator, a homolog of the HAP3 subunit of CCAAT-binding factors (Lotan et al., 1998), a family composed of 10 genes in Arabidopsis. While CCAAT boxes are general promoter motifs, their binding factors often show tissue- or stage-specific expression thereby providing specificity by formation of different hetero- or homodimers that bind to and activate specific sets of genes (Lekstrom-Himes and Xanthopoulos, 1998). Although the LEC1 gene is expressed primarily during early embryogenesis and mutations have very limited effects on ABA sensitivity (West et al., 1994; Lotan et al., 1998), it appears to potentiate ABA response by genetic interactions with ABI3, ABI4 and ABI5 (Meinke et al., 1994; Parcy et al., 1997; Brocard and Finkelstein, unpublished observations). The LEC2 gene has also been cloned recently and found to encode another member of the B3-domain family of transcription factors (Stone et al., 2001).
The ERA loci were identified in a screen for enhanced response to ABA inhibition of germination (Cutler et al., 1996); the recessive nature of the mutants implies defects in negative regulators of ABA signaling. The era1 mutations have pleiotropic effects including production of hyperdormant seeds, increased drought tolerance due to altered ion fluxes in guard cells (Pei et al., 1998), and abnormally large meristems due to defects in cell division control. An additional era1 allele, designated wiggum, has been isolated on the basis of the meristem defect (Ziegelhoffer et al., 2000). Digenic mutant analyses indicate that ERA1 acts epistatically (downstream) of ABI1 and ABI2, but upstream of ABI3 (Cutler et al., 1996). ERA1 encodes the β subunit of farnesyl transferase, indicating that it is likely to be involved in lipidation of possible signaling molecules, but few of its specific targets are known. ERA3 has recently been found to be allelic to EIN2 (Ghassemian et al., 2000), which encodes a membrane-bound putative divalent cation sensor that appears to represent a point of cross-talk between ethylene, ABA, auxin, jasmonic acid, and stress signaling (Alonso et al., 1999).
Three recently cloned loci affecting ABA response, ABH1 (Hugouvieux et al., 2001), HYL1 (Lu and Fedoroff, 2000), and SAD1, encode components that could affect RNA accumulation at a post-transcriptional step. The abh1 mutant, isolated on the basis of ABAhypersensitivity at germination, displays enhanced guard cell response and drought tolerance. Digenic analyses show additive effects with abi1-1 and era1-2, suggesting action in separate pathways. ABH1 encodes a homolog of the mRNA CAP-binding complex and may be involved in mRNA processing of negative regulators of ABA signaling. Transcriptional profiling comparing wild-type and abh1 plants showed only 18 genes (0.2% of those represented on the chip) were down-regulated, including some previously identified as ABA-inducible and some encoding good candidate signaling molecules, e.g. AtPP2C, a Ca2+- binding protein, and several genes implicated in response to oxidative stress. The hyl1 mutant has pleiotropic physiological defects including stunted growth, hyponastic leaves (usually refers to upward bending of petiole, but describes upward curling of leaf blade in this case), and late flowering that may reflect defects in hormonal signaling including reduced response to auxin and cytokinins and hypersensitivity to ABA (Lu and Fedoroff, 2000). HYL1 expression is repressed by ABA, consistent with a role as a negative regulator of ABA response, and it encodes a dsRNA binding protein. Although some ABA-inducible genes have higher basal levels of expression in the hyl1 mutant, the direct target(s) and mechanism of action of the HYL1 protein are not yet known. The sad1 mutant is supersensitive to ABA, drought and NaCl, possibly because it is defective in drought-induced ABA biosynthesis. SAD1 encodes an Sm-like protein (Xiong et al. 2001a) and is therefore likely to be involved in RNA processing or turnover (Fromont-Racine et al., 2000).
The growth control by ABA (gca1-gca8) mutants were isolated in a screen for ABA-resistant root growth (Himmelbach et al., 1998). While the effects of gca3-gca8 are limited to root growth control, the gca1 and gca2 mutants have pleiotropic effects reminiscent of abi1-1 and abi2-1. However, unlike these abi mutants, the gca mutants are recessive and therefore likely to affect positive regulators of ABA response. Although none of the GCA loci have been cloned, recent studies with gca2 have shown that its defect in stomatal regulation is at least partially due to altered kinetics of the ABA-induced [Ca2+]cyt oscillations required to elicit full stomatal closure (Allen et al., 2001). It is not known whether the root growth defect also reflects disrupted Ca2+ signaling.
Many loci have been identified using screens based on aberrant reporter gene expression in the presence or absence of ABA, salt, osmotic, sugar or cold stress (Ishitani et al., 1997; Foster and Chua, 1999; Rook et al., 2001). Depending on the nature of the defective expression, most of these have been designated hos (high osmotic stress response), los (low osmotic stress response), or cos (constitutive osmotic stress response). Some of those isolated on the basis of defective osmotic stress response have been shown to display aberrant response to ABA as well as to some or all of the environmental stresses listed above. Of these, the FRY1, HOS1, LOS5 and LOS6 loci have been cloned (Lee et al., 2001; Xiong et al., 2001b; Xiong et al., 2001c). The fry1 mutant displays fiery luciferase reporter expression due to constitutive activation of an RD29A promoter. In addition, fry1 plants are hypersensitive to ABA and NaCl. The FRY1 gene encodes inositol polyphosphate 1-phosphatase and functions in IP3 catabolism; the observed stress hypersensitivity appears to reflect sustained IP3 signaling (Xiong et al., 2001b). FRY1 is identical to SAL1, initially isolated on the basis of conferring salt tolerance in yeast transformants (Quintero et al., 1996). Hos1 mutants are also hyper-responsive to ABA and cold; the HOS1 gene encodes a novel RING finger protein that may participate in inactivating components of ABA signaling (Lee et al., 2001). LOS5 and LOS6 are allelic to ABA3 and ABA1, respectively (Xiong et al., 2001c), indicating that the osmotic stress-induction of RD29A expression is ABA-dependent. However, while exogenous ABA can rescue the defect in salt signaling, it is not sufficient to restore response to cold. The isi3 (impaired sucrose induction of ADP glucose pyrophosphorylase promoter) mutant is allelic to ABI4 and isi4 is allelic to ABA2. Additional mutant alleles of ABI4, designated sun6, were isolated in screens for sucrose-uncoupled expression of the plastocyanin promoter (Dijkwel et al., 1997).
In addition to the mutants with pleiotropic defects in ABA response, many mutants have been identified with pleiotropic defects in response to multiple hormones including ABA. Two independent jasmonic acid (JA) resistant mutants, jar1 and jin4, also display hypersensitivity to ABA in seed germination assays (Staswick et al., 1992; Berger et al., 1996). Mutants with defects in response to multiple hormones include axr2-1 (a dominant negative mutant resistant to auxin, ethylene, and ABA; Wilson et al., 1990), sax1 (hypersensitive to ABA and auxin, rescuable by exogenous brassinosteroids (BR); Ephritikhine et al., 1999), bri1 and bin2 (BR insensitive, ABA-hypersensitive root growth; Clouse et al., 1996; Li et al., 2001), prl1 (increased sensitivity to sugar, ethylene, ABA, auxin, cytokinin, and cold stress; Nemeth et al., 1998), ein2/era3 (decreased sensitivity to ethylene, cytokinins, auxin transport inhibitors, methyl jasomonate, and increased response to ABA; Alonso et al., 1999; Ghassemian et al., 2000), and ctr1 (constitutive ethylene signaling, enhanced resistance to ABA inhibition of germination; Kieber et al., 1993; Beaudoin et al., 2000).
Biochemical Approach
Identification of ABA-regulated genes and their ABA-responsive cis-acting sequences have constituted the starting point for many biochemical studies of ABA signaling (reviewed in Busk and Pages, 1998; Rock, 2000). In most vegetative tissues, ABA-inducible genes are presumed to be involved in response to abiotic stresses that result in cellular dehydration. In maturing seeds, ABA-regulated genes include those involved in synthesis of storage reserves as well as induction of desiccation tolerance. Overall, ABA-regulated genes encode relatively high-abundance transcripts required for adaptation to stress or for reserve synthesis, and low abundance transcripts encoding signaling components. Although the initial focus of such studies was on working backward to regulators of the high abundance transcripts by sequential identification of cis-acting regulatory regions and the transcription factors that specifically recognize these DNA sequences, recent studies have also focused on the physiological roles of ABA-regulated kinases, lipases, etc. Many of these studies have been conducted in species other than Arabidopsis, but the ABA signaling mechanisms appear to be highly conserved. The advent of genome-wide transcriptional profiling, coupled with the availability of the complete genome sequence, should facilitate identification of target genes for specific regulatory factors, as well as rapid identification of candidate cis-acting sequences of coordinately regulated genes.
Transcriptional regulation
The cis-acting sequences required or sufficient for ABA-inducibility fall into four main groups: the G-box elements designated ABREs and the functionally equivalent CE3 (coupling element)-like sequences, the RY/Sph elements, and recognition sequences for MYB and MYC class transcription factors (Table 5) (reviewed in Busk and Pages, 1998; Rock, 2000). Trans-acting factors that interact with these sequences were initially identified by ligand-binding screens of cDNA expression libraries (Guiltinan et al., 1990); more recent efforts have used one-hybrid screens in yeast with the cis-acting sequence of interest controlling reporter gene expression (Kim et al., 1997; Choi et al., 2000; Uno et al., 2000). These studies have shown that the ABREs are bound by bZIPs and the RY elements are bound by B3-domain proteins. In Arabidopsis, each of these transcription factor classes is represented by a gene family.
Table 5.
Promoter elements regulating ABA-induction and their corresponding DNA-binding proteins. The underlined sequences are most highly conserved; precise cis-acting sequences present in specific genes are detailed in (Rock, 2002). Many of the DNA-binding proteins are members of large families comprised of dozens (bZIP and B3 domain) to hundreds (Myb) of related proteins regulating diverse processes in plant growth. It is likely that specificity of the response is controlled by the “context” of the conserved binding site, the specific protein(s) bound, and the other proteins present in a complex interacting with any given promoter.
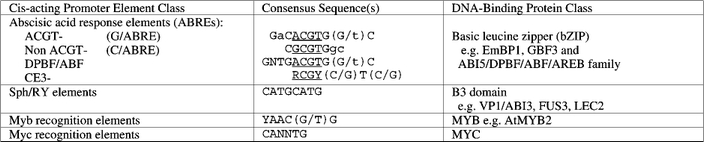
There are approximately 80 predicted bZIP factor genes in Arabidopsis (Riechmann et al., 2000; Jakoby et al., 2002), many of which are likely to participate in regulating response to cues other than ABA. The ABI5 subfamily is comprised of at least nine bZIPs, eight of which have been shown to be correlated with ABA-inducible expression (Choi et al., 2000; Uno et al., 2000; Kim et al., 2002); cDNA analyses indicate that at least one of these (ABF3/AtDPBF5/AtbZIP37) appears to undergo alternative splicing (Brocard et al., 2002). Overexpression of the ABRE-Binding Factors 1 (ABF1) and ABF3 is sufficient to transactivate the Em promoter in rice protoplasts, but these factors show different sensitivities to ABA trans-activation (Finkelstein et al., 2002), suggesting different modalities or species differences. The constitutive overexpression of ABF3 or ABF4 in Arabidopsis resulted in ABA hypersensitivity and other ABA-associated phenotypes such as sugar sensitivity, reduced germination and post-germination growth, altered stress-inducible gene expression, and drought tolerance (Kang et al., 2002). ABF3 overexpression phenotypes were ABA-dependent, whereas ABF4 overexpression lines severely affected only vegetative growth and showed some auxin and ethylene phenotypes, suggesting possible developmental aspects of ABF4 activity. These studies should be interpreted with caution since “skwelching” (titration by non-natural binding partners and shifting of endogenous factor kinetics) or abnormal hierarchical cascades due to ectopic expression may be involved. Despite the similarities among binding sites and ABA- or stress-inducible expression of the ABI5-homologous subfamily members, recent studies have shown that they are subject to cross-regulation by ABI3, ABI4 and ABI5 ranging from hyper-induction to repression (Brocard et al., 2002). These results are not consistent with a model of simple functional redundancy within this family. Studies of bZIPs from other species have shown that in vitro binding of ABREs does not necessarily reflect action in ABA signaling in vivo (Guiltinan et al., 1990; Izawa et al., 1993; Finkelstein et al., 2002). Interestingly, although first identified in connection with light-regulated gene expression and belonging to a subgroup distinct from the ABI5 subfamily (Jakoby et al., 2002), the bZIP GBF3 is ABA-inducible and may participate in ABA-regulation (Lu et al., 1996). Targeted analyses of loss- and gain-of-function lines for each of the bZIPs should identify those family members responsible for responding to specific signals in specific tissues.
Of the 43 B3-domain family members encoded in Arabidopsis, 14 are within the ABI3/VP1-related subfamily (Riechmann et al., 2000). In addition to ABI3, this subfamily includes two members of the leafy cotyledon class of regulators that control embryo maturation: FUS3 (Luerssen et al., 1998) and LEC2 (Stone et al., 2001)(see Signaling Mechanisms section).
Involvement of MYB and MYC factors in ABA-regulated gene expression was first suspected following cloning of stress-induced members of both of these families (Urao et al., 1993; Abe et al., 1997). The MYB class of transcription factors is also represented by a large gene superfamily in Arabidopsis, comprised of 190 genes (Riechmann et al., 2000). In contrast there is only a single MYC with the canonical b-HLH-ZIP domain structure, but this shares extensive homology with 139 predicted bHLH factor genes. Expression of AtMyc1 and three specific MYB family members is induced by both drought and ABA (Abe et al., 1997). Because the MYB/MYC response system requires de novo synthesis of MYB and MYC proteins, it has been suggested to participate in slow adaptive responses to dehydration stress (Shinozaki and Yamaguchi-Shinozaki, 2000). Some members of the homeodomain-leucine zipper (HD-Zip) family of transcription factors (ATHB6, ATHB7, and ATHB12) have also been shown to be induced by ABA or abiotic stress, but their roles in ABA response are not yet known (Söderman et al., 1996; Lee and Chun, 1998; Söderman et al., 1999).
Components of early steps in ABA signal transduction
Biochemical and pharmacological studies have shown that early events in ABA signaling involve participation of GTP-binding proteins, phospholipases, protein kinases and phosphatases. Due to the ubiquitous participation of these classes of proteins in a wide range of signaling events, a critical question is always that of how specificity is conferred. For those classes of regulators represented by large gene families, it is likely that individual family members perform specialized functions. However, in the case of heterotrimeric G-proteins, Arabidopsis has only one or two isoforms of each G-protein subunit and recent studies show that loss of function for the single Gα gene (GPA1) disrupts aspects of both auxin and ABA signaling (Ullah et al., 2001; Wang et al., 2001), indicating that this component is not a likely source of response specificity.
Another major class of GTPase molecular switches in plants is a plant-specific branch of the RAS superfamily, the monomeric Rops (Rho/rac-related GTPases from plants). The 11 Arabidopsis members of this subfamily, also known as Aracs or AtRacs (Yang, 2002) represent four distinct groups, whose physiological and developmental roles are being analyzed in loss- and gain-of-function (constitutively active and dominant-negative) transgenic lines. Although interpretation of the transgenic phenotypes may be complicated by ectopic action of the transgenes or disruption of closely related family members, it appears that several of the Rops inhibit various aspects of ABA response, ranging from germination control to stomatal function. Furthermore, some may act downstream of ERA1 (the farnesylase activity could act on Rops) in a signaling pathway. The pleiotropic defects of the transgenic lines suggest that Rop response specificity may depend on interactions with specific activators and targets, as well as specialization of function among the monomeric G proteins.
Secondary messengers in ABA signaling regulating stomatal function and gene expression include inositol triphosphate (IP3) and phosphatidic acid (PA), produced by phospholipase C (PLC) and phospholipase D (PLD), respectively (see following section) (Gilroy et al., 1990; Jacob et al., 1999; Ritchie and Gilroy, 1998; Gampala et al., 2001b). Arabidopsis contains 6 PLC genes; ABA induces expression of only one of these, AtPLC1 (Hirayama et al., 1995). Reverse genetic analyses have shown that AtPLC1 is required, but not sufficient, for ABA effects on germination, growth and vegetative gene expression (Sanchez and Chua, 2001). In addition, the more highly phosphorylated inositide IP6 contributes to ABA-inhibition of stomatal opening (Lemtiri-Chlieh et al., 2000). The recent discovery that a defect in phosphoinositide metabolism due to the fry1 mutation results in hypersensitivities to ABA and abiotic stresses further emphasizes the role of phosphoinositides as secondary messengers (Xiong et al., 2001b). Studies with microsomes derived from barley aleurone plasma membranes have demonstrated that stimulation of PLD activity is mediated by G-protein activity following perception of ABA at the plasma membrane (Ritchie and Gilroy, 2000). There are five subfamilies of PLD genes in Arabidopsis, representing 11 genes, many of which are induced by a variety of stresses (Wang et al., 2000) and show different tissue distributions and subcellular localizations (Fan et al., 1999). PLDα, the most prevalent phospholipase D, is the only family member whose expression and activity are increased by ABA. Consistent with a role for this family member in ABA signaling, antisense suppression of this gene slows abscisic acid- and ethylene-promoted senescence of detached Arabidopsis leaves (Fan et al., 1997) and impairs stomatal closure in response to drought stress (Sang et al., 2001). In contrast, although expression and activity of AtPLDδ are induced by dehydration, they are not significantly induced by ABA (Katagiri et al., 2001).
Many kinases representing multiple gene families have been implicated in ABA signaling affecting stomatal regulation and/or gene expression (Table 6). ABA may enhance either expression (Hwang and Goodman, 1995; Hong et al., 1997; Lee et al., 1998; Mikami et al., 1998; Gomez-Cadenas et al., 1999; Piao et al., 1999) or activity (Li and Assmann, 1996; Burnett et al., 2000) of these kinases. Although initial functional studies with pharmacological inhibitors did not discriminate among the roles of individual family members, recent studies with dominant negative alleles have assayed the role of specific kinases (Sheen, 1996; Li et al., 2000).
Table 6.
Kinases implicated in ABA signaling
Consistent with the importance of phosphorylation status in ABA signaling, several protein phosphatases have also been shown to affect ABA signaling. Several members of the PP2C family of protein phosphatases have pleiotropic negative effects on ABA signaling and inhibition of PP1/PP2A phosphatases by okadaic acid (OA) also alters both ABA-induced gene expression and stomatal closure. The effect of OA on ABA response varies among species (Kuo et al., 1996; Grabov et al., 1997; Pei et al., 1997; Wu et al., 1997); in Arabidopsis, OA partially inhibits ABA activation of S-type anion channels and stomatal closure (Pei et al., 1997). In addition, the PP2B class of calcineurin-like Ca2+ binding proteins are potential Ca2+ sensors; one of these, AtCBL1, is induced by drought (Kudla et al., 1999). However, relatively few of the potential Ca2+ sensors correlated with stress response are also ABA responsive (Takahashi et al., 2000).
Cell biological approach
Single cells are often used to test the roles of candidate secondary messengers and signaling intermediates in regulating cellular responses. Commonly used single cell systems for analyzing ABA responses are ion channel regulation in guard cells and transient gene expression assays in microinjected tissues or protoplasts. Although most of the initial studies were done on species other than Arabidopsis, most of the signaling mechanisms are conserved and can be analyzed further by studies of Arabidopsis mutants or functional studies of Arabidopsis genes in heterologous assay systems.
ABA-induced stomatal closing occurs within minutes due to ionic and osmotically-driven turgor and shape changes in specific epidermal cells, and is therefore an excellent accessible model system for early events in ABA signaling. Stomatal closure is correlated with increased [Ca2+]cyt, and ABA-induced closing has been shown to involve both Ca2+-dependent and Ca2+-independent mechanisms (reviewed in Schroeder et al., 2001). However, auxin-induced stomatal opening also appears to involve [Ca2+]cyt elevations, indicating that response specificity must be achieved via the detailed characteristics of the Ca2+ oscillation frequencies, amplitudes, and localizations, which presumably reflect the mechanism(s) of Ca2+ release and the cellular interpretation of the [Ca2+]cyt change.
[Ca2+]cyt elevations may be due to release of Ca2+ from intracellular stores and/or influx through plasma membrane channels. IP3 or cyclic ADP-ribose (cADPR) can induce Ca2+ release from intracellular stores (Gilroy et al., 1990; Leckie et al., 1998); inhibitor studies indicate that both contribute to the elevations but neither is sufficient for full response (MacRobbie, 2000). Recent studies have shown that ABA stimulates nitric oxide (NO) synthesis in guard cells, which induces stomatal closure in a cADPR- and cGMP-dependent manner, indicating that NO is an even earlier secondary messenger in this response pathway (Neill et al., 2002). Yet another calcium-mobilizing molecule in plants, sphingosine-1-phosphate, was recently implicated in linking drought-induced abscisic acid signaling to stomatal closure (Ng et al., 2001). Furthermore, [Ca2+]cyt elevations may be self-amplifying by inducing further Ca2+ release from the vacuoles (McAinsh et al., 1995). ABA appears to promote Ca2+ influx currents through Ca2+-permeable plasma membrane channels (Grabov and Blatt, 1998) by enhancing production of reactive oxygen species (ROS), e.g. H2O2, that can serve as secondary messengers leading to channel activation (Pei et al., 2000). Several stresses leading to stomatal closure result in ROS production (Lee et al., 1999), and it has been suggested that the ROS-dependent pathway of response is shared by multiple stresses. Consistent with this, enhanced tolerance of abiotic stresses results from constitutive activation of a ROS-activated MAPK cascade (Kovtun et al., 2000). However, the sequence of events involving ABA, ROS production and ABI1 function is unclear as distinct studies have demonstrated inhibition of ABI1 by H2O2 in vitro (Meinhard and Grill, 2001) and inhibition of ABA-induced ROS production in abi1-1 mutants (Murata et al., 2001). Finally, while ABA signaling can result in sustained [Ca2+]cyt elevations, continuous monitoring of [Ca2+]cyt has demonstrated that various signals affecting stomatal aperture induce [Ca2+]cyt oscillations with distinct periodicity, and that imposing the correct periodicity with exchanges of external buffer solutions can restore normal response to mutants (Allen et al., 2000; Allen et al., 2001).
ABA-induced stomatal closing also partly depends on cytosolic alkalization (Irving et al., 1992); this effect can occur in isolated membrane patches and appears to function by increasing the number of K+out channels available for activation (Miedema and Assmann, 1996). Furthermore, increased external pH decreases K+in channel activity (Hedrich et al., 1995) and increases activation of a guard cell localized K+out channel (Ache et al., 2000). Increasing pH may also be a feedback mechanism for ABA desensitization via activation of ABI1, a negative regulator of ABA response (Leube et al., 1998).
Transient gene expression assays also monitor fairly rapid responses to ABA, but over a longer time scale: several hours rather than minutes to hours. As described for guard cell signaling, ABA-induction of gene expression depends on action of Ca2+, IP3, PA and cADPR as secondary messengers (Gilroy and Jones, 1992; Heimovaara-Dijkstra et al., 1995; Wu et al., 1997; Ritchie and Gilroy, 1998; Ghelis et al., 2000b; Webb et al., 2001) and may require S-type anion channel activity (Ghelis et al., 2000a). Interactions among transcription factors and their dependence on specific secondary messengers or phosphorylation states has also been analyzed in transient assays following bombardment or electroporation of genes encoding these signaling components (Hagenbeek et al., 2000; Uno et al., 2000; Gampala et al., 2001b).
ABA SIGNALING MECHANISMS
To date, ∼50 loci affecting ABA signaling have been described in the literature (Tables 2 and 3); mutants at many more loci have been isolated but have not yet been characterized sufficiently for publication. Twenty-six of these loci have been cloned and found to encode transcription factors, protein phosphatases or kinases, putative RNA binding proteins or processing enzymes, a farnesyl transferase, enzymes of phospholipid metabolism, and GTP-binding proteins. In addition to the genetically defined transcription factors involved in seed and ABA response, many factors presumed to regulate ABA-inducible and embryonic gene expression have been identified biochemically. While it is likely that many of these transcription factors regulate some of the same genes, the majority of specific target genes for most regulatory factors are unknown. Furthermore, for most factors it is not known whether regulation of common target genes is accomplished by independent binding to distinct cis-acting sites, activation of a regulatory cascade, combinatorial action of factors, or a combination of these mechanisms. Similarly, ABA-regulated phosphatases, kinases, and lipases have been identified biochemically, but the specific roles in ABA signaling are still speculative for most of these. Many of these questions are being addressed by molecular analyses of lines with loss or gain of these specific regulatory factors resulting from mutations or ectopic expression (Table 4).
Comparison of expression patterns and mutant phenotypes has provided some surprises and demonstrated that none of the known loci act completely stage-specifically. Characterization of monogenic and some digenic mutant phenotypes have shown interactions among “ABA specific” regulators, as well as with regulators that appear to function in networks regulating response to sugars, salt, and most known hormones. Although initially selected on the basis of increased or decreased ABA response, not all mutants show consistent hyper- or hyposensitivity for all ABA-regulated responses. Thus it is most useful to consider genetic interactions in the context of specific cell types, since the participants and goals of ABA signaling vary. In the following sections, we will consider the ABA signaling networks in maturing seeds, germination, seedlings, vegetative stress responses, stomatal regulation, and flowering.
Maturing Seeds
Developing embryos enter maturation phase when they undergo a transition from growth by cell division to cell enlargement and begin to accumulate storage reserves. This growth phase transition is correlated with an increase in seed ABA content that appears to be required for cell cycle arrest at the G1/S transition (Levi et al., 1993; Liu et al., 1994). During seed maturation, there are two peaks of ABA accumulation: one of maternal and one of embryonic origin (Karssen et al., 1983). The first peak is maternally derived and occurs at 10 days after pollination (DAP), immediately preceding maturation phase. This early peak, along with FUS3 and LEC gene function, are required to prevent premature germination at the end of the cell division phase of embryogenesis (Raz et al., 2001). Although this ABA peak is reduced three-fold in fus3 mutants, only the double mutants combining fus3 with ABA deficiency are highly viviparous (Nambara et al., 2000).
In wild-type seeds, embryonic ABA accumulates later, and to only one-third the level accumulated at mid-embryogenesis; however, it is essential for induction of dormancy, which is maintained despite a ∼6 fold decrease in ABA by seed maturity (Karssen et al., 1983). The ABA content of a mature dry wild-type seed is similar to that of the peak ABA level in an ABA-deficient mutant, suggesting that dormancy maintenance in mature seeds relies on signals other than endogenous ABA. Consistent with this, several reduced dormancy mutants (ats, rdo1, rdo2, dag1, ttg) have been identified that have wild-type ABA levels and sensitivity to ABA (Leon-Kloosterziel et al., 1996; Papi et al., 2000). In some of these (ats and ttg), the decreased dormancy is attributed to testa defects. Furthermore, dormancy is controlled by only a subset of the known ABA response loci. Conversely, the hyperdormant mutant comatose (cts) has a seed-specific defect in GA response (Russell et al., 2000), but CTS function is only required when dormancy is imposed by the action of other loci. Such comparisons of genetic controls of vivipary and dormancy show that the timing and relevant regulatory factors differ, although both affect the same developmental decision, i.e. whether or not to germinate.
Reserve accumulation and late embryogenesis-abundant (LEA) gene expression during maturation are largely controlled by the combinatorial action of transcription factors. Extensive analyses of promoter sequences for storage protein and LEA genes have demonstrated the presence of elements required for hormone responsiveness, stage- and tissue-specificity. Consistent with this, some of the required factors regulate ABA response, e.g. ABI3, ABI4, and ABI5, while others primarily promote embryonic growth, e.g. LEC1, LEC2, and FUS (reviewed in Holdsworth et al., 1999, 2001). Accumulation of some ABA-inducible proteins is also subject to post-transcriptional control (Bies et al., 1998); it is possible that some of the recently identified loci encoding putative RNA processing proteins contribute to this level of regulation.
Physiological studies have shown that the ABI3, ABI4, and ABI5 loci have similar qualitative effects on seed development and ABA sensitivity, consistent with action in a common pathway, but that null mutations in ABI3 are more severe than those in ABI4 or ABI5 (Finkelstein et al., 1998; Parcy et al., 1994; Finkelstein and Lynch, 2000a). Action in a common pathway was also suggested by the similarity in the genetic interactions among these loci and with abi1 mutants (Finkelstein and Somerville, 1990; Finkelstein, 1994a). Recent studies show extensive cross-regulation of expression among ABI3, ABI4, and ABI5 (Söderman et al., 2000) and ectopic expression of either ABI3 or ABI4 results in ABA hypersensitivity of vegetative tissues, including ABA-inducible vegetative expression of several “seed-specific” genes, which is partly dependent on increased ABI5 expression (Parcy et al., 1994; Söderman et al., 2000). Taken together, these results suggest that seed-specific or ABA-inducible expression might be at least partially controlled by regulatory complexes containing these three transcription factors. Consistent with this, ABI3 (or its monocot ortholog VIVIPAROUS1) and ABI5 (or its rice homolog TRAB1) display direct and synergistic interactions in two-hybrid analyses in yeast and transient reporter activation assays in rice protoplasts (Hobo et al., 1999; Gampala et al., 2001a; Nakamura et al., 2001). However, ABI4 does not appear to interact directly with either ABI3 or ABI5 in these assays.
Although the ectopic expression studies were initially interpreted to mean that the seed-specificity of embryonic gene expression reflected seed-specific expression of key regulators (Parcy et al., 1994), most of these regulators are expressed and functional during vegetative growth (Finkelstein et al., 1998; Arenas-Huertero et al., 2000; Finkelstein and Lynch, 2000a; Huijser et al., 2000; Laby et al., 2000; Rohde et al., 2000b; Lopez-Molina et al., 2001). Alternatively, seed specificity may be conferred by repressors of embryonic characteristics, e.g. PICKLE (PKL), that repress post-germination expression of embryogenesis-promoting regulators such as LEC1 (Ogas et al., 1997; Ogas et al., 1999).
In addition to the genetically defined transcriptional regulators, several embryonically expressed homologs of ABI5 have been identified by homology to sunflower proteins identified in one-hybrid screens with the Dc3 promoter (Kim et al., 2002). These have been designated AtDPBF1- AtDPBF5 (Arabidopsis thaliana Dc3 Promoter Binding Factor); AtDPBF1 is identical to ABI5. These bZIP factors form heterodimers in some combinations, including with ABI5, suggesting that they may participate in regulation of many of the same target genes. This shared target specificity could provide functional redundancy that would explain the weakly ABA-resistant phenotype of abi5 null mutants. However, comparison of expression patterns within this subfamily shows differences that are not consistent with simple functional redundancy (Brocard et al., 2002).
More severe defects in seed maturation are observed in double mutants combining the weak abi3-1 allele with ABA deficiency (the aba-1 mutant); these plants produce seeds that have a high degree of denatured protein, and fail to lose chlorophyll, accumulate storage proteins, or attain desiccation tolerance (Koornneef et al., 1989; Wolkers et al., 1998). A similar “green seed” phenotype is observed with null alleles of ABI3 (abi3-3 and abi3-4) (Giraudat et al., 1992; Nambara et al., 1992). The aba, abi3-1 effects on desiccation tolerance and seed protein accumulation can be reversed by application of exogenous ABA (Meurs et al., 1992). These results suggest that ABI3 regulates processes in seed development that can respond to, but do not require, high endogenous ABA. However, the importance of some endogenous ABA in seed maturation has been demonstrated by the observation that severe reduction of free ABA by seed-specific expression of anti-ABA antibodies results in a “green seed” phenotype in transgenic tobacco (Phillips et al., 1997).
Some combinations of mutations in ABA biosynthesis or response (e.g.ABI1, ABI3, ABI4, or ABI5) with those in FUS3 or LEC1 result in even more severe defects; these plants produce highly pigmented seeds that fail to accumulate storage reserves or attain desiccation tolerance (Keith et al., 1994; Meinke et al., 1994; Parcy et al., 1997; Nambara et al., 2000; Brocard and Finkelstein, unpublished observations). They may also be viviparous, but the degrees of vivipary and seed resistance to exogenous ABA are not well correlated (Finkelstein et al., 2002). Even though the lec1 and fus3 mutations have little or no effect on ABA sensitivity, the double mutants are at least 10-fold less sensitive to ABA than their monogenic abi parents. Thus, ABI-dependent ABA sensitivity is potentiated by the FUS3 and LEC1 gene products. In the case of the digenics involving abi3, ABI3 protein accumulation was significantly decreased in the double mutants (Parcy et al., 1997).
In summary, many of the best-characterized loci regulating stage-specific ABA response in embryos encode transcription factors with complex patterns of cross-regulation and some direct interactions such that ectopic expression of several of these factors can confer “seed-specific” gene expression on another developmental stage or tissue. The effects of ABA and some key regulators at mid- and late embryogenesis are shown schematically in Figure 4.
Figure 4.
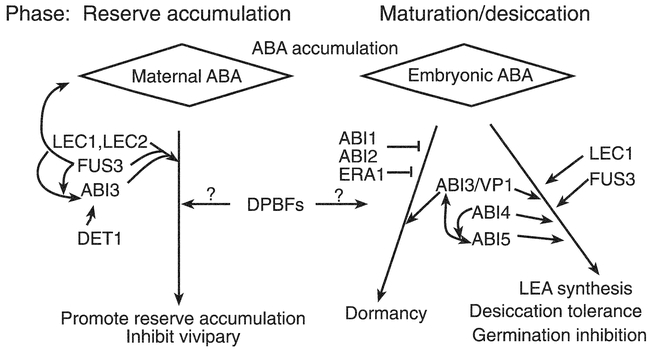
Schematic of signaling pathways in seed development. Arrows represent promotion of processes or expression of the regulators. Bars represent inhibitors of the indicated processes. Positions of loci do not imply order of gene action. Reprinted with permission from Finkelstein et al. (2002).
Germination
While endogenous ABA is essential for induction of dormancy, and dormancy maintenance is correlated with de novo synthesis of ABA during imbibition (reviewed in Finkelstein et al., 2002), ABA deficiency alone is not sufficient to cause vivipary in Arabidopsis (Karssen et al., 1983). Consequently, although as little as 3 B5M exogenous ABA is sufficient to suppress germination of mature seeds, it is not clear whether the low level of endogenous ABA remaining in Arabidopsis seeds at this stage regulates germination. Surprisingly, radicle emergence is observed even in the presence of up to 100 B5M ABA when supplemented with low concentrations of sugar (either glucose or sucrose at 30–90 mM) or peptone, but greening and subsequent seedling growth is still blocked (Garciarrubio et al., 1997; Finkelstein and Lynch, 2000b). Although the exogenous sugar might permit germination by overcoming a nutritional deficiency resulting from inhibition of reserve mobilization by exogenous ABA (Garciarrubio et al., 1997), reserve mobilization via the glyoxylate cycle is not essential for germination, and post-germinative growth can be supported by either photosynthesis or exogenous sugar in the absence of a functional glyoxylate cycle (Eastmond et al., 2000). Furthermore, ABA does not prevent mobilization of seed lipid reserves, despite inhibiting visible germination, i.e. radicle emergence (Pritchard and Graham, 2001). Other studies show that wild-type seeds quickly accumulate ABI5 protein when incubated on low sucrose and ABA for up to 5d post stratification (Lopez-Molina et al., 2001) such that germination is arrested following radicle emergence (Finkelstein and Lynch, 2000b), The ability to induce ABI5 accumulation is strongly correlated with maintenance of desiccation tolerance in these seedlings. ABA, the induced ABI5, and potentially other interacting factors may prevent the loss of desiccation tolerance by delaying escape from phase two of germination under conditions of low moisture.
The commitment to germinate is also controlled by antagonistic interactions between ABA and gibberellins, ethylene, and brassinosteroids (BR). Genetic evidence for the ABA/GA antagonism has been provided by the isolation of ABA-deficient mutations as suppressors of non-germination due to GA-deficiency (Koornneef et al., 1982), and the GA response mutant sleepy (sly) as a suppressor of abi1-1 (Steber et al., 1998). The interaction with BR was discovered when sly was shown to be rescued by BR (Steber and McCourt, 2001) and BR-deficient and insensitive lines were found to be hypersensitive to ABA (Li et al., 2001; Steber and McCourt, 2001). Interactions with ethylene signaling were implied by the isolation of new alleles of ethylene response genes such as ein2 and ctr1 in screens for suppressors and enhancers of seed sensitivity to ABA (Beaudoin et al., 2000; Ghassemian et al., 2000). This result led to careful re-examination of the monogenic phenotypes and the discovery that the single mutants have slightly altered ABA response. Comparison of mono- and digenic phenotypes show that the ctr1 and abi1-1 mutations synergistically enhance ABA-resistant germination (Beaudoin et al., 2000), suggesting action in interacting pathways. In contrast, abi3 mutations appear epistatic to the ABA hypersensitivity conferred by ein2/era3, consistent with action in the same pathway, while the ein2/era3 and abi1-1 effects are additive, suggesting action in parallel pathways.
Reverse genetic studies of monomeric Rho/rac-related GTPases suggest that Rop9 and Rop10 act redundantly in negatively regulating ABA effects on seed germination and seedling growth (reviewed in Yang, 2002). Both of these Rops contain farnesylation motifs and display ERA1-dependent localization to the plasma membrane, suggesting that they might act downstream of ERA1 in a signaling pathway, consistent with the era1-like phenotype of the double mutants. Transgenic studies have also shown that Rop2 negatively regulates seed dormancy and inhibition of germination by ABA (Li et al., 2001). However, manipulation of Rop2 activity also disrupted responses to auxins and brassinolides and a wide variety of developmental processes.
Although the biological relevance of exogenous ABA inhibition of germination is unclear, this response provides a quantitative assay for ABA sensitivity that can be dissected genetically. In addition to being regulated by the loci described above, recent studies have shown that AtPLC1 is required for ABA response and that ABA response can be overcome by high level expression of an Ins(1,4,5)P35-phosphatase (AtIP5PII) (Sanchez and Chua, 2001). Similar reductions in ABA sensitivity were observed in assays of seedling growth and gene expression; these were correlated with near-elimination of the ABA-induced rise in IP3 levels in the transgenic lines. Interestingly, transgenic lines with increased PLC or reduced Ins(1,4,5)P35-phosphatase activity did not increase IP3 levels or ABA response in the absence of added ABA. Thus, PLC1 expression is necessary but not sufficient for ABA response. However, IP3 levels and ABA sensitivity are enhanced in fry1 mutants, which are defective in phosphoinositide catabolism via an inositol polyphosphate 1-phosphatase (Xiong et al., 2001a).
Interactions among some of the hormonal and environmental signals and regulatory elements controlling germination are shown schematically in Figure 5. Additional regulators specifically controlling dormancy without altering ABA sensitivity are not included.
Figure 5.
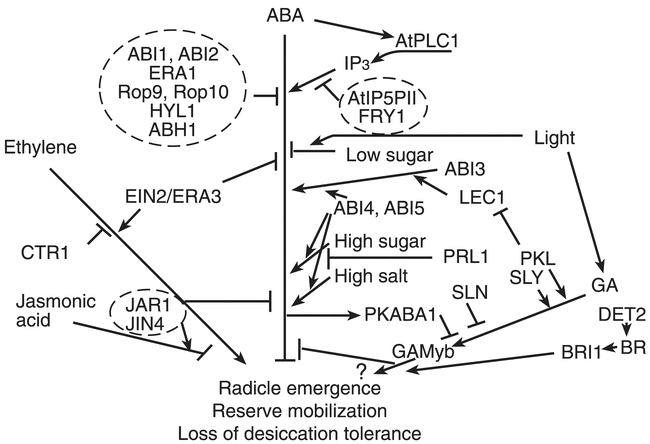
Schematic of signaling pathways that interact with ABA regulation of germination. Arrows represent promotion of processes or expression of the regulators. Bars represent inhibitors of the indicated processes. Positions of loci do not imply order of gene action. Note that PKABA1, GAMyb and SLN are barley genes; their Arabidopsis orthologs have not yet been identified. Reprinted with permission from Finkelstein et al. (2002).
Seedling growth
The relative importance of the ABI loci changes after germination. Seedling growth of abi3 plants is slightly less sensitive to ABA-inhibition than wild type, but abi1 and abi2 seedling growth is not inhibited by ABA at concentrations resulting in 50–70% inhibition of wild type seedling growth (Finkelstein and Somerville, 1990). This difference is apparent in both shoot and root growth, even at the level of individual root hair elongation (Schnall and Quatrano, 1992). While mutations in ABI3, ABI4 and ABI5 primarily affect gene expression during seed maturation, all three genes are also expressed in vegetative tissues (Finkelstein et al., 1998; Rohde et al., 1999; Finkelstein and Lynch, 2000a) suggesting they may play a role in vegetative ABA response. Consistent with this, additional abi4 mutants have been isolated on the basis of exhibiting salt-resistant germination (Quesada et al., 2000) or sugar-insensitive seedling growth or gene expression (Arenas-Huertero et al., 2000; Huijser et al., 2000; Laby et al., 2000; Rook et al., 2001) and abi5 mutants exhibit a mild sugar-insensitive phenotype. Mutations in ABI4 and ABI5 also produce mild defects in ABA-regulated vegetative gene expression (Finkelstein and Lynch, 2000a; Söderman et al., 2000). In contrast to the other ABIs, post-germination ABI3 expression is localized to the meristem and appears to regulate vegetative quiescence processes, plastid differentiation, and floral determination (Rohde et al., 1999; Kurup et al., 2000; Rohde et al., 2000a).
Several of the sugar-resistance screens are based on the observation that high concentrations of exogenous sugars (>300 mM) inhibit seedling growth. This is an ABA-dependent effect that cannot be simply attributed to the osmotic effects of sugar (reviewed in Gibson, 2000), but its physiological relevance is unclear, in part because it is not known how these conditions affect endogenous sugar pools. Although ABA and sugar could act in either parallel or intersecting pathways, two lines of evidence suggest that sugar signaling may be mediated, in part, by ABA: abi4 or aba mutants can ameliorate the sugar hypersensitivity due to hexokinase overexpression (Arenas-Huertero et al., 2000), and exposure to high glucose induces both ABA synthesis and expression of ABI4 and ABI5 (Arroyo Becerra et al., 2001; Brocard et al., 2002). These interactions appear relatively specific because abi1, abi2, and abi3 mutants show essentially normal sugar response (Arenas-Huertero et al., 2000; Huijser et al., 2000; Laby et al., 2000). However, seedlings that over-express ABI3, ABI4, or ABI5 are hypersensitive to glucose, consistent with a role for these genes in both ABA and sugar responses (Finkelstein et al., 2002); the near-normal sugar response of abi3 seedlings may reflect the relatively limited post-germination regulatory contributions of ABI3.
ABA appears to play another role in nutrient signaling in seedlings by mediating the regulatory effects of nitrate on root branching (Signora et al., 2001). Nitrate effects are also concentration dependent, with low nitrate stimulating localized lateral root elongation and high nitrate inhibiting lateral root elongation throughout the root system; these effects are antagonized by increasing sugar. As described for sugar sensing, only ABA-deficient, abi4 and abi5 seedlings displayed decreased sensitivity to nitrate inhibition, suggesting that they mediate the inhibitory effect. In contrast, the stimulatory effect of low nitrate also requires ABA, but is enhanced in all of the abi mutants tested except abi5. Although the precise roles of the ABIs in regulating lateral root growth are not understood, it is noteworthy that ABI5 is specifically expressed in root tips from emergence onward (Brocard et al., 2002).
The nature of the interactions between ABA and ethylene signaling is also complex following germination. Although these hormones have antagonistic effects on germination, both inhibit root growth (Beaudoin et al., 2000; Ghassemian et al., 2000). Disruption of ethylene signaling reduced sensitivity of root growth to inhibition by ABA, suggesting that they act in the same or parallel pathways controlling root growth. However, treatment with AVG to block ethylene synthesis results in increased ABA sensitivity and ethylene overproducing mutants have decreased ABA sensitivity, implying another antagonistic interaction. This apparent inconsistency might be explained by ABA inhibition of root growth by signaling through the ETR1 response pathway, but only in the absence of ethylene (Ghassemian et al., 2000).
ABA inhibits growth via a combination of limited cell extensibility (Kutschera and Schopfer, 1986) and inhibited cell division due to arrest at the G1 phase of the cell cycle (Levi et al., 1993; Liu et al., 1994). The effects of ABA on progression through the cell cycle might reflect ABA-induced expression of a cyclin-dependent protein kinase inhibitor that interacts with both Cdc2a and CycD3 and is correlated with decreased Cdc2-like histone H1 kinase activity (Wang et al., 1998).
The studies described above focus on the inhibitory effects of high ABA on growth. However, even well-watered ABA-deficient plants exhibit stunted growth suggesting that the low endogenous ABA levels in unstressed plants promote growth. Recent studies in maize and tomato indicate that the stunted growth of ABA-deficient plants is due to a failure to inhibit ethylene production, reflecting another antagonistic interaction between ABA and ethylene (Sharp et al., 2000; Spollen et al., 2000).
Stress responses
A critical function of ABA during vegetative growth is to optimize growth during environmental stress by maintaining osmotic homeostasis. At the whole plant level, low ABA (characteristic of mild water stress conditions) promotes root growth but inhibits shoot growth, leading to an increased root:shoot ratio. This response has been largely overlooked in most mutant characterizations. In contrast, high ABA inhibits growth of both roots and shoots, but promotes formation of arrested lateral roots, i.e. drought rhizogenesis (Vartanian et al., 1994). The inhibitory effects of high ABA on root elongation are disrupted in the abi1-1 and abi2-1 mutants, but not in the abi3, abi4 and abi5 mutants. However, ectopic expression of ABI3, ABI4 or ABI5 confers hypersensitivity to ABA-inhibition of root growth (Parcy et al., 1994; Soderman et al., 2000; Lopez-Molina et al., 2001; Brocard et al., 2002), indicating that each of these transcription factors can regulate root ABA response, but normally don, possibly because they are usually not strongly expressed in roots. Although the sax1, hyl1, and sad1 mutants show increased sensitivity to ABA-inhibition of root growth (Ephritikhine et al., 1999; Lu and Fedoroff, 2000; Xiong et al, 2001), root growth of era3/ein2 mutants is less sensitive than that of wild-type (Ghassemian et al., 2000). Drought rhizogenesis is disrupted in ABA-deficient aba1, ABA-insensitive abi1-1, and auxin/ethylene/ABA-resistant axr1-3, but not in abi2-1 or abi3 mutants (Vartanian et al., 1994).
The flow of water across cell membranes to maintain growth and within the transpiration stream is partially controlled by the aquaporins present in these membranes. ABA promotes water uptake and flow by increasing cell-to-cell water movement across roots, possibly by effects on aquaporins (Hose et al., 2000). Within the transpiration stream, the route of water movement reflects the relative resistance of apoplastic and symplastic paths, which are affected by stomatal aperture, relative humidity (R.H.) and flow across cell membranes. Under high flux conditions (open stomates, low R.H.), water movement is primarily apoplastic and osmotic permeabilities are moderate in wild-type plants and low in aba or abi mutants with defects in stomatal function leading to abnormally high transpiration rates (Morillon and Chrispeels, 2001). Consistent with a role for ABA in regulating osmotic permeability, ABA treatment induced stomatal closure, followed by an increase in osmotic permeability. However, this appears to be an indirect effect because osmotic permeability increased in all genotypes when transpiration was reduced by high R.H.
At the cellular level, ABA can promote tolerance of some abiotic stresses including drought, salinity, and cold or heat (reviewed in Rock, 2000; Shinozaki and Yamaguchi-Shinozaki, 2000; Xiong and Zhu, 2001; Larkindale and Knight, 2002). In addition, it can induce tolerance of hypoxic stress in roots, but not shoots (Ellis et al., 1999). The presumption is that these signals induce accumulation of protectants such as small hydrophilic proteins, sugars, proline, and glycine-betaine, or activate detoxifying mechanisms that confer stress tolerance. Consistent with this, constitutive expression of transcription factors or some of their target genes can increase stress tolerance (reviewed in Bartels, 2001; Thomashow, 2001). However, the enhanced signaling and stress-induced gene expression in the fry1 mutant is not protective (Xiong et al., 2001a). Surprisingly, these mutants are hypersensitive to stress-inhibition of growth even though heterologous expression of this gene confers salt tolerance in yeast (Quintero et al., 1996). This result suggests that the ability to attenuate a stress signal is important for it to be effective.
Extensive studies of stress- and ABA-induced gene expression reveal two waves of response: an early transient response, peaking at ∼3 hrs, and a late sustained response (from ∼10 hrs onward). The “early” genes include those encoding members of a MAPK cascade, some of which are induced within 1–10 min of stress or ABA treatment, as well as transcription factors and early response to dehydration (erd) genes (reviewed in Shinozaki and Yamaguchi-Shinozaki, 2000; Xiong and Zhu, 2001). It is noteworthy that many of the genes encoding presumed signaling components show relatively specific induction by drought, cold, salinity, wounding or osmotic stress, but only one of these has been shown to be ABA-responsive (Knetsch et al., 1996). Similarly although ABA signaling can be mediated by inositol 1,4,5-triphosphate, hyperosmotic stress induces a rapid and transient increase in IP3 independent of abscisic acid in Arabidopsis cell culture (Takahashi et al., 2001). The “late” genes include members of the responsive to dehydration (rd), cold-regulated (cor), low temperature induced (lti) and cold-induced (kin) gene classes; these are presumed to contribute to the adaptive aspects of induced tolerance (reviewed in Shinozaki and Yamaguchi-Shinozaki, 2000). Many of these encode proteins that are structurally similar to some of the LEA proteins that accumulate during the acquisition of desiccation tolerance in seeds, while others encode proteases, presumed chaperonins, enzymes of sugar or other compatible solute metabolism, ion and water-channel proteins, and enzymes that detoxify active oxygen species (reviewed in Ingram and Bartels, 1996). As implied by their names, many of these are induced by a variety of abiotic stresses as well as ABA treatment and several have been shown to confer enhanced cold tolerance in Arabidopsis (reviewed in Thomashow, 1999), osmotolerance in yeast (Swire-Clark and Marcotte, 1999), or general stress tolerance in tobacco (Bartels, 2001).
Comparison of stress-induced gene expression in ABA biosynthesis and response mutants has demonstrated that there are both ABA-dependent and –independent signaling pathways (reviewed in Rock, 2000). Furthermore, analysis of ABA, temperature, and osmotic stress effects on marker gene expression in wild-type plants showed a complex array of interactions (Xiong et al., 1999). While sustained ABA and cold treatment had additive effects on RD29A::LUC expression, consistent with action through independent signaling pathways, ABA and NaCl had synergistic effects. Isolation of mutants with defects in stress- and ABA-induced signaling has provided evidence for cross-talk among even the “independent” signaling pathways (Ishitani et al., 1997). A recent model to explain these interactions proposes at least four independent signaling pathways mediating response to drought stress and two additional pathways mediating cold response (Shinozaki and Yamaguchi-Shinozaki, 2000). Of these, only two are ABA-dependent and they depend on either MYC/MYB- or bZIP-regulated gene expression. The ABA-independent pathways depend on expression mediated by drought response element binding (DREB) family members or as yet unidentified factors. However, as discussed earlier, both bZIPs and MYBs are large families in Arabidopsis, some members of which have overlapping but not identical expression/activation profiles, and it is likely that they will have both redundant and discrete functions in controlling expression of stress-induced genes. Recent studies have identified 5 members of the ABI5-homologous bZIP subfamily that are induced by salt and/or drought stress (Choi et al., 2000; Uno et al., 2000). Activation of at least some of these is regulated by their phosphorylation state in an ABI1-dependent manner (Uno et al., 2000). Preliminary transcriptional profiling studies have indicated that 1–4% of Arabidopsis genes are strongly induced by drought or cold treatments, less than one-third of these are induced by both treatments, and more than two-thirds had not been previously identified as stress-induced (Seki et al., 2001). These results suggest that many of the detailed gene expression analyses to date may provide a skewed or “anecdotal” view of the relevant signaling networks.
Stomatal regulation
In another important response to drought stress, ABA regulates the transpiration rate via effects on stomatal aperture both by promoting closure and inhibiting opening (Schroeder et al., 2001). Although both effects result in closed stomata, they are not simple reversals of the same process. This view is underscored by the recent findings that loss of Gα function blocks ABA-inhibition of opening but does not affect ABA-promotion of closure (Wang et al., 2001), whereas a guard cell-specific ABA-activated serine-threonine protein kinase (AAPK) specifically functions in ABA-induced closure (Li et al., 2000).
Mutants with decreased ABA response, and consequently a wilty phenotype, include abi1-1, abi2-1 (Koornneef et al., 1984), abi8 (Finkelstein, unpublished results) and gca2 (Himmelbach et al., 1998; Pei et al., 2000), while hypersensitive lines include abh1 (Hugouvieux et al., 2001) and era1 (Pei et al., 1998). Surprisingly, the abi1 and abi2 mutants differ from the aba mutant in that exogenous ABA actually intensifies, rather than reverses, withering of stems and siliques due to long-term water stress (Koornneef et al., 1984). One possible explanation for this is that the mutants are capable of recognizing the ABA treatment and may respond non-productively by increasing turnover, decreasing synthesis, or altering redistribution of ABA to the relevant location, i.e. the guard cells.
Under drought conditions, apoplastic pH increases, resulting in greater apoplastic retention of ABA. The ABA concentration in xylem sap increases greatly and functions as a root-to-shoot signal leading to reduced transpiration in leaves (reviewed in Davies and Zhang, 1991). The magnitude of the change in xylem ABA content varies widely among species and it has been suggested that ABA may also be transported in a conjugated form, then released by hydrolysis in leaves (reviewed in Hartung et al., 1998). However, the postulated hydrolases have yet to be identified. Consistent with this long-distance signaling role for ABA, response to systemically imposed drought stress is impaired in aba1, abi1-1 and abi2-1 plants (Koornneef et al., 1982; Koornneef et al., 1984). However, these mutants show wild-type stomatal response to humidity, indicating that guard cell response to atmospheric water potential is not mediated by ABA (Assmann et al., 2000).
Upon arrival at guard cells, ABA can be perceived either intra- or extracellularly. Evidence from application of impermeant ABA derivatives suggest that extracellular perception prevents stomatal opening (Jeannette et al., 1999), while microinjection experiments with caged ABA show that intracellular ABA can induce stomatal closure (Allan et al., 1994). Although no ABA receptors have been definitively identified to date, many secondary messengers, kinases, and phosphatases involved in stomatal regulation have been identified. One of the earliest electrophysiological changes in guard cells exposed to ABA is a transient depolarization reflecting an increase in [Ca2+]cyt. Another early step in inhibition of opening involves G-protein function in a pH-independent pathway, while ABA-promotion of closure appears to involve a pH-dependent pathway (Wang et al., 2001). Analyses of mutant responses to various secondary messengers and inhibitors of signaling intermediates have permitted partial dissection of these pathways (Figure 6). The ABI PP2Cs and GCA2 all act upstream of the Ca2+ oscillations, with ABI2 and GCA2 apparently mediating response to H2O2, and ABI1 affecting ROS production (Murata et al., 2001). However, the relationship among most of the kinases and phosphatases is still obscure.
Figure 6.
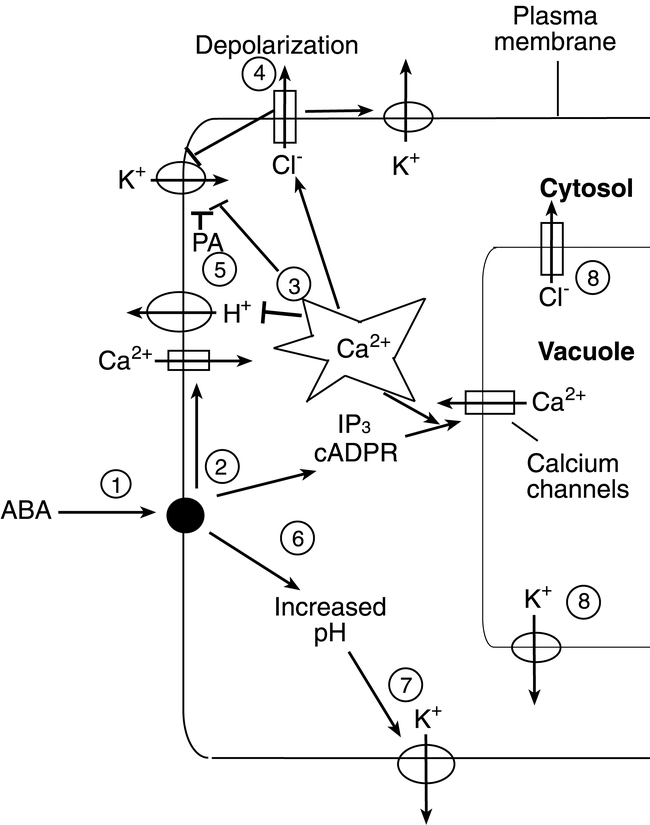
- -production of reactive oxygen species that contribute to opening of plasma membrane Ca2+in channels
- -release from internal stores through three types of Ca2+ channels regulated by IP3 (produced by phospholipase C), cyclic ADP-ribose (cADPR), and Ca2+ itself.
- -inhibits plasma membrane H+ pumps
- -inhibits K+in channels, and
- -activates Cl−out (anion) channels, resulting in depolarization of the membrane.
Following these initial ABA-induced signaling events, the ionic fluxes resulting in closed stomata are now well-defined and involve inhibition of plasma membrane proton pumps, and activation of slow/sustained (S-type) anion channels leading to depolarization and consequent deactivation of K+in channels and activation of K+out channels (reviewed in Schroeder et al., 2001). The resulting K+ and anion efflux from guard cells leads to osmotic water loss and stomatal closure. A similar chain of events occurs in response to high CO2 levels, which also induce stomatal closing, but the initial signaling events differ such that abi1-1 and abi2-1 mutants show at least 50% wild-type response to CO2 (Leymarie et al., 1998). Recent studies have shown that ABA and CO2 signals interact strongly to promote stomatal closure, suggesting that they act through converging signaling pathways (Leymarie et al., 1999).
Although ABA-induced ion efflux and water loss from guard cells reduces stomatal conductance, comparable losses in other plant cells would result in loss of turgor throughout the plant body despite the reduced transpiration rate. However, tissue- and cell-specific channels serve to maintain cellular K+ in the rest of the plant. Xylem K+ content is regulated by activity of a stelar K+ outward rectifier (SKOR) (Gaymard et al., 1998), which is repressed by ABA. A similar effect is achieved in phloem tissue by ABA-inducible expression of a weak inward K+ rectifier, AKT2 (Lacombe et al., 2000). ABA also inhibits an outward K+ current in mesophyll cells, demonstrating cell-type specific response to ABA within the leaf (Sutton et al., 2000). Another possible benefit of this differential control is that reduction of K+out-channel activity during salt stress may protect mesophyll cells against cytotoxic effects of Na+ uptake that could result from limited permeability to Na+ influx through these channels.
The guard cell shape changes that lead to changes in stomatal aperture involve a substantial change in volume due to osmotic gain or loss of water. This is accompanied by up to two-fold changes in membrane surface area, accomplished by vesicle secretion and endocytosis (reviewed in Schroeder et al., 2001). Consistent with this, ABA induces expression of an annexin-like gene (Kovacs et al., 1998), requires syntaxin function for normal ion channel response (Leyman et al., 1999) and reorganization of the actin cytoskeleton to a randomly oriented pattern (Eun and Lee, 1997).
The syntaxin Nt-Syr1 is located primarily at the plasma membrane in roots and to lesser extents in stems, leaves and flowers; its expression in leaves is transiently enhanced by ABA. Enhanced expression of Nt-Syr1 by salt stress was not observed in ABA-deficient Npaba1 mutants or plants carrying the Arabidopsis dominant negative ABA INSENSITIVE1-1 (abi1-1) gene, providing evidence that Nt-SYR1 is regulated by ABA and stress signals (Leyman et al., 2000). Arabidopsis has a large family of Nt-SYR1-like syntaxin genes, but any potential roles in ABA response are untested. At least one other SNARE, AtSNAP33, binds NtSyr1 (Kargul et al., 2001), and recent NtSyr1 overexpression and structure/function data support a role for NtSyr1 in vesicle trafficking to the plasma membrane (Geelen et al., 2002), consistent with the change in surface area of guard cell plasma membranes during stomatal movements. Syntaxin also regulates ion channel activities in neurons and similar processes may occur in guard cells (reviewed in Blatt, 2000).
The ABA-induced actin reorganization is also ABI1-dependent (Eun et al., 2001) and inhibited by AtRac1, a small GTPase protein that is subject to ABI1-dependent inactivation by ABA (Lemichez et al., 2001). Studies with Commelina have shown that ABA effects on actin reorganization are mediated by changes in [Ca2+]cyt, as well as protein kinase and phosphatase activities (Hwang and Lee, 2001).
Although the best-characterized aspects of guard cell response are electrophysiological and ultrastructural changes, some of the various ABA response loci have been shown to modify guard-cell specific gene expression. Recent studies have demonstrated that an increase in [Ca2+]cyt, mediates ABA regulated gene expression in guard cells, similar to its role in regulating guard cell turgor (Webb et al., 2001). Of particular interest are the observations that 35S::ABI3 expression acts epistatically to abi1-1 in regulating stomatal aperture (Parcy and Giraudat, 1997), 35S::ABI5 expression enhances stress-induced stomatal closure (Lopez-Molina et al., 2001), and abh1 mutants have altered guard cell expression of a small number of genes (Hugouvieux et al., 2001). Some of the regulatory targets of these transcription factors and CAP-binding protein may play critical roles in stomatal function.
Flowering
A variety of ABA synthesis or response loci have been implicated in controlling meristem function or flowering time (reviewed in Rohde et al., 2000b). The aba1 and abi1 mutants exhibit early flowering under short days (Martinez-Zapater, 1994) and abi3-4 mutants flower early regardless of daylength (Kurup et al., 2000), while the ABA hypersensitive mutant hyl1 exhibits delayed flowering (Lu and Fedoroff, 2000), consistent with an inhibitory role of ABA in the floral transition. Studies of the flowering promoting gene LEAFY showed that LFY expression is strongly induced by a combination of sucrose and GA, but that ABA fully blocks the GA-induced increase regardless of the GA3 concentration (Blazquez et al., 1998). Although ABA did not completely eliminate LFY promoter activity, the ABA effect appeared “epistatic” to the GA effect. Genetic interactions with DET1 (Rohde et al., 2000a) and CONSTANS (Kurup et al., 2000), and physical interactions with TIMING OF CAB EXPRESSION (TOC1) and a CONSTANS-related factor (Kurup et al., 2000) suggest that ABI3 affects flowering through cross-talk with light and circadian rhythm controls. However, these interactions are complex in that the epistatic relationships vary depending on photoperiodic conditions. Flowering and fruit production is also enhanced in abi3 mutants, apparently reflecting the delayed senescence and continued photosynthesis of cauline leaves (Robinson and Hill, 1999).
ERA1, initially identified as a negative regulator of ABA response in germination (Cutler et al., 1996), also affects meristem development and was consequently re-identified as the WIGGUM gene (Ziegelhoffer et al., 2000). Mutants in ERA1/WIG have unusually large meristems, possibly reflecting defects in regulating division vs. differentiation within the meristem. However, it is counter-intuitive that enhancement of ABA response, which includes suppression of progress through the cell cycle, would lead to excessive cell divisions.
CONCLUSIONS AND PERSPECTIVES
Studies of the ABA biosynthesis and response mutants of Arabidopsis have complemented similar studies in other species. The hormone deficient mutants have been valuable in providing confirmations of proposed biosynthetic pathways, such as the “indirect” carotenoid pathway of ABA synthesis and the plastidic MEP pathway of synthesis for ABA precursors. Detailed biochemical studies have identified enzymes responsible for the many steps of ABA biosynthesis and candidate genes or gene families have been identified for nearly all of these. The roles of specific family members are being tested by expression analyses and reverse genetics. Expression of several of these enzymes increases in response to drought and ABA, resulting in positive feedback regulation of ABA biosynthesis. ABA also promotes synthesis of enzymes required for its degradation, providing a mechanism for homeostasis of ABA accumulation.
Although the number of available mutants is still far from saturating the biosynthesis pathway, any mutants deficient in bioactive hormones are useful for studying the roles of those hormones in physiological and developmental processes. Such studies have confirmed the role of endogenous ABA in dormancy induction and stomatal regulation. However, they have also shown that many responses induced by either environmental stresses (e.g. drought, cold or salinity) or exogenous ABA probably do not require endogenous ABA to mediate response to the environmental cues.
The hormone response mutants provide a means of dissecting the signal transduction pathways used by each hormone. Genetic and/or molecular studies have identified approximately 50 ABA response loci, many of whose functions are conserved across species, and the list is still growing. Within Arabidopsis, ABA signaling is mediated by both redundant and independent mechanisms, some of which also appear to affect response to other signals. In fact, double mutant analyses are revealing cryptic effects of a variety of loci, implying interactions exist between ABA signaling and responses to most major classes of hormones, light, abiotic stresses, and nutrient status. Similar to signaling in response to other hormones, ABA response appears to involve the interaction of both positive and negative regulators. An additional similarity is that some of the negative regulators were initially identified on the basis of reduced response due to dominant negative mutations. In contrast to studies of other hormonal signaling mechanisms, neither biochemical nor genetic approaches have unambiguously identified an ABA receptor yet.
ABA regulates developmental events such as embryo maturation and phase transitions in seeds, as well as some responses to environmental stress. Some loci regulate both sets of processes and there are many similarities in terms of the relevant secondary messengers. However, the best- characterized embryonic regulators encode proteins that affect transcription, whereas most of the identified genes playing major roles in vegetative growth encode proteins expected to act at earlier stages in a signaling pathway, affecting processes such as protein phosphorylation or farnesylation, RNA processing, or phosphoinositide metabolism.
Although we have learned much about ABA signaling, our view of the relevant pathways is still fragmented. Some of the remaining unknowns include the identities of the receptors, the substrates of the various kinases and phosphatases, which of the known ABA response loci interact directly or indirectly, and the identities of additional signaling elements linking the known elements into complete pathways or networks. However, the genes cloned to date can be used for construction of transgenic plants with conditionally altered hormone synthesis or response. Given the efficacy of ABA in regulating such basic processes as seed development, dormancy vs. germination, transpiration and stress responses, these could have important biotechnological applications.
Acknowledgments
We thank many members of the scientific community for sharing manuscripts in press. We thank Tim Lynch and Senthil Subramanian for critical readings of the manuscript. Work in the laboratory of R.R.F. is supported by National Science Foundation grants IBN-9728297 and IBN-9982779. Work in the laboratory of C.D.R. is supported by a 2003 Research Enhancement Fund grant from the Texas Tech University Institute for University Research.
Footnotes
Citation: Finkelstein R.R., and Rock C.D. (2002) Abscisic Acid Biosynthesis and Response. The Arabidopsis Book 1:e0058. doi:10.1199/tab.0058
elocation-id: e0058
Published on: September 30, 2002
Table 1.
(Continued)
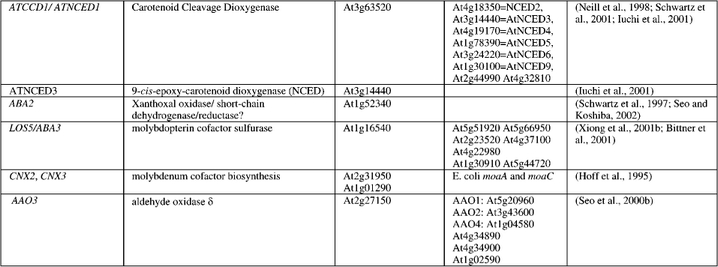
REFERENCES
- Abe H., Yamaguchi-Shinozaki K., Urao T., Iwasaki T., Hosokawa D., Shinozaki K. Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid regulated gene expression. Plant Cell. 1997;91(1):1859–1868. doi: 10.1105/tpc.9.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ache P., Becker D., Ivashikina N., Dietrich P., Roelfsema M. R. G., Hedrich R. GORK, a delayed outward rectifier expressed in guard cells of Arabidopsis thaliana, is a K+-selective, K+-sensing ion channel. FEBS Lett. 2000;4861(1):93–98. doi: 10.1016/s0014-5793(00)02248-1. [DOI] [PubMed] [Google Scholar]
- Addicott F. Abscisic acid. 1983. (New York, NY, Praeger Publishers)
- Agrawal G. K., Yamazaki M., Kobayashi M., Hirochika R., Miyao A., Hirochika H. Screening of the rice viviparous mutants generated by endogenous retrotransposon tos17 insertion, tagging of a zeaxanthin epoxidase gene and a novel OsTATC gene. Plant Physiol. 2001;1251(1):1248–1257. doi: 10.1104/pp.125.3.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akaba S., Leydecker M-T., Moureaux T., Oritani T., Koshiba T. Aldehyde oxidase in wild type and aba1 mutant leaves of Nicotiana plumbaginifolia. Plant Cell Physiol. 1998;391(1):1281–1286. [Google Scholar]
- Al-Babili S., Hugueney P., Schledz M., Welsch R., Frohnmeyer H., Laule O., Beyer P. Identification of a novel gene coding for neoxanthin synthase from Solanum tuberosum. FEBS Lett. 2000;4851(1):168–172. doi: 10.1016/s0014-5793(00)02193-1. [DOI] [PubMed] [Google Scholar]
- Albinsky D., Masson J. E., Bogucki A., Afsar K., Vass I., Nagy F., Paszkowski J. Plant responses to genotoxic stress are linked to an ABA/salinity signaling pathway. Plant J. 1999;171(1):73–82. [Google Scholar]
- Allan A. C., Fricker M. D., Ward J. L., Beale M. H., Trewavas A. J. Two transduction pathways mediate rapid effects of abscisic acid in Commelina guard cells. Plant Cell. 1994;61(1):1319–1328. doi: 10.1105/tpc.6.9.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alldridge N. Anomalous vessel elements in wilty dwarf tomato. Bot. Gazette. 1964;1251(1):138–142. [Google Scholar]
- Allen G., Chu S., Schumacher K., Shimazaki C., Vafeados D., Kemper A., Hawke S., Tallman G., Tsien R., Harper J. Alteration of stimulus-specific guard cell calcium oscillations and stomatal closing in Arabidopsis det3 mutant. Science. 2000;2891(1):2338–2342. doi: 10.1126/science.289.5488.2338. [DOI] [PubMed] [Google Scholar]
- Allen G., Chu S., Harrington C., Schumacher K., Hoffmann T., Tang Y., Grill E., Schroeder J. A defined range of guard cell calcium oscillation parameters encodes stomatal movements. Nature. 2001;4111(1):1053–1057. doi: 10.1038/35082575. [DOI] [PubMed] [Google Scholar]
- Alonso J., Hirayama T., Roman G., Nourizadeh S., Ecker J. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science. 1999;2841(1):2148–2152. doi: 10.1126/science.284.5423.2148. [DOI] [PubMed] [Google Scholar]
- Anderson B. E., Ward J. M., Schroeder J. I. Evidence for an extracellular reception site for abscisic acid in Commelina guard cells. Plant Physiol. 1994;1041(1):1177–1183. doi: 10.1104/pp.104.4.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas-Huertero F., Arroyo A., Zhou L., Sheen J., Leon P. Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar. Genes Dev. 2000;141(1):2085–2096. [PMC free article] [PubMed] [Google Scholar]
- Arroyo Becerra A., Arenas F., Jimenez A., Cantero A., Leon-Mejia P. The participation of ABA in the glucose-mediated regulation in Arabidopsis. 2001. Paper presented at: Twelfth International Conference on Arabidopsis Research (Madison, WI)
- Assante G., Merlini L., Nasini G. (+)-Abscisic acid, a metabolite of the fungus Cercospora rosicola. Experientia. 1977;331(1):1556. [Google Scholar]
- Assmann S., Shimazaki K. The multisensory guard-cell: stomatal responses to blue-light and abscisic acid. Plant Physiol. 1999;1191(1):809–815. doi: 10.1104/pp.119.3.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assmann S. M., Snyder J. A., Lee Y-R. J. ABA-deficient (aba1) and ABA-insensitive (abi1-1, abi2-1) mutants of Arabidopsis have a wild-type stomatal response to humidity. Plant Cell Envir. 2000;231(1):387–395. [Google Scholar]
- Audran C., Borel C., Frey A., Sotta B., Meyer C., Simonneau T., Marion-Poll A. Expression studies of the zeaxanthin epoxidase gene in Nicotiana plumbaginifolia. Plant Physiol. 1998;1181(1):1021–1028. doi: 10.1104/pp.118.3.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audran C., Liotenberg S., Gonneau M., North H., Frey A., Tap-Waksman K., Vartanian N., Marion-Poll A. Localisation and expression of zeaxanthin epoxidase mRNA in Arabidopsis in response to drought stress and during seed development. Aust. J. Plant Physiol. 2001;281(1):1161–1173. [Google Scholar]
- Banno H., Chua N-H. Characterization of the Arabidopsis formin-like protein AFH1 and its interacting protein. Plant Cell Physiol. 2000;411(1):617–626. doi: 10.1093/pcp/41.5.617. [DOI] [PubMed] [Google Scholar]
- Bartels D. Targeting detoxification pathways: An efficient approach to obtain plants with multiple stress tolerance? Trends Plant Sci. 2001;61(1):284–286. doi: 10.1016/s1360-1385(01)01983-5. [DOI] [PubMed] [Google Scholar]
- Beaudoin N., Serizet C., Gosti F., Giraudat J. Interactions between abscisic acid and ethylene signaling cascades. Plant Cell. 2000;121(1):1103–1115. doi: 10.1105/tpc.12.7.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S., Bell E., Mullet J. Two methyl jasmonate-insensitive mutants show altered expression of AtVsp in response to methyl jasmonate and wounding. Plant Physiol. 1996;1111(1):525–531. doi: 10.1104/pp.111.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalerao R. P., Salchert K., Bako L., Okresz L., Szabados L., Muranaka T., Machida Y., Schell J., Koncz C. Regulatory interaction of PRL1 WD protein with Arabidopsis SNF1-like protein kinases. Proc. Natl. Acad. Sci. USA. 1999;961(1):5322–5327. doi: 10.1073/pnas.96.9.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bies N., Aspart L., Carles C., Gallois P., Delseny M. Accumulation and degradation of Em proteins in Arabidopsis thaliana: Evidence for post-transcriptional controls. J. Exp. Bot. 1998;491(1):1925–1933. [Google Scholar]
- Bies-Etheve N., da Silva Conceicao A., Giraudat J., Koornneef M., Leon-Kloosterziel K., Valon C., Delseny M. Importance of the Arabidopsis ABI3 protein for Em and 2S albumin gene regulation. Plant Mol. Biol. 1999;401(1):1045–1054. doi: 10.1023/a:1006252512202. [DOI] [PubMed] [Google Scholar]
- Bittner F., Oreb M., Mendel R. R. ABA3 is a molybdenum cofactor sulfurase required for activation of aldehyde oxidase and xanthine dehydrogenase in Arabidopsis thaliana. J. Biol. Chem. 2001;2761(1):40381–40384. doi: 10.1074/jbc.C100472200. [DOI] [PubMed] [Google Scholar]
- Blanc V. M., Mullin K., Pichersky E. Nucleotide sequences of Ipi genes from Arabidopsis and Clarkia. Plant Physiol. 1996;1111(1):652. [Google Scholar]
- Blatt M. R. Ca2+ signalling and control of guard-cell volume in stomatal movements. Curr. Opin. Plant Biol. 2000;31(1):196–204. [PubMed] [Google Scholar]
- Blazquez M. A., Green R., Nilsson O., Sussman M. R., Weigel D. Gibberellins promote flowering of Arabidopsis by activating the LEAFY promoter. Plant Cell. 1998;101(1):791–800. doi: 10.1105/tpc.10.5.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier F., d'Harlingue A., Backhaus R. A., Kumagai M. H., Camara B. Identification of neoxanthin synthase as a carotenoid cyclase paralog. Eur. J. Biochem. 2000;2671(1):6346–6352. doi: 10.1046/j.1432-1327.2000.01722.x. [DOI] [PubMed] [Google Scholar]
- Bouvier F., d'Harlingue A., Hugueney P., Marin E., Marion-Poll A., Camara B. Xanthophyll biosynthesis. Cloning, expression, functional reconstitution, and regulation of beta-cyclohexenyl carotenoid epoxidase from pepper (Capsicum annuum). J. Biol. Chem. 1996;2711(1):28861–28867. doi: 10.1074/jbc.271.46.28861. [DOI] [PubMed] [Google Scholar]
- Brocard I., Lynch T., Finkelstein R. Regulation and role of the Arabidopsis ABA-insensitive (ABI)5 gene in ABA, sugar and stress response. Plant Physiol. 2002. in press. [DOI] [PMC free article] [PubMed]
- Bugos R. C., Yamamoto H. Y. Molecular cloning of violaxanthin de-epoxidase from romaine lettuce and expression in Escherichia coli. Proc. Natl. Acad. Sci. USA. 1996;931(1):6320–6325. doi: 10.1073/pnas.93.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bungard R. A., Ruban A. V., Hibberd J. M., Press M. C., Horton P., Scholes J. D. Unusual carotenoid composition and a new type of xanthophyll cycle in plants. Proc. Natl. Acad. Sci. USA. 1999;961(1):1135–1139. doi: 10.1073/pnas.96.3.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbidge A., Grieve T. M., Jackson A., Thompson A., McCarty D. R., Taylor I. B. Characterization of the ABA-deficient tomato mutant notabilis and its relationship with maize Vp14. Plant J. 1999;171(1):427–431. doi: 10.1046/j.1365-313x.1999.00386.x. [DOI] [PubMed] [Google Scholar]
- Burbidge A., Grieve T., Terry C., Corlett J., Thompson A., Taylor I. Structure and expression of a cDNA encoding zeaxanthine epoxidase, isolated from a wilt-related tomato (Lycopersicon esculentum Mill.) library. J. Exp. Bot. 1997;481(1):1749–1750. [Google Scholar]
- Burnett E. C., Desikan R., Moser R. C., Neill S. J. ABA activation of an MBP kinase in Pisum sativum epidermal peels correlates with stomatal responses to ABA. J. Exp. Bot. 2000;511(1):197–205. doi: 10.1093/jexbot/51.343.197. [DOI] [PubMed] [Google Scholar]
- Busk P. K., Pages M. Regulation of abscisic acid-induced transcription. Plant Mol. Biol. 1998;371(1):425–435. doi: 10.1023/a:1006058700720. [DOI] [PubMed] [Google Scholar]
- Carlberg C. Lipid soluble vitamins in gene regulation. Biofactors. 1999;101(1):91–97. doi: 10.1002/biof.5520100202. [DOI] [PubMed] [Google Scholar]
- Carol P., Stevenson D., Bisanz C., Breitenbach J., Sandmann G., Mache R., Creelman R. A., Zeevaart J. A. D. Incorporation of oxygen into abscisic acid and phaseic acid from molecular oxygen. Plant Physiol. 1984;751(1):166–169. doi: 10.1104/pp.75.1.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson C., Hattori T., Rosenkrans L., Vasil V. The quiescent/colorless alleles of viviparous1 show that the conserved B3 domain of VP1 is not essential for ABA-regulated gene expression in the seed. Plant J. 1997;121(1):1231–1240. doi: 10.1046/j.1365-313x.1997.12061231.x. others., a. [DOI] [PubMed] [Google Scholar]
- Cheng W-H., Endo A., Zhou L., Penney J., Chen H-C., Arroyo A., Leon P., Nambara E., Asami T., Seo M., Koshiba T., Sheen J. A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell. 2002;141(1):2723–2743. doi: 10.1105/tpc.006494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernys J. T., Zeevaart J. A. D. Characterization of the 9-cis-epoxycarotenoid dioxygenase gene family and the regulation of abscisic acid biosynthesis in avocado. Plant Physiol. 2000;1241(1):343–353. doi: 10.1104/pp.124.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H., Hong J., Ha J., Kang J., Kim S. ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 2000;2751(1):1723–1730. doi: 10.1074/jbc.275.3.1723. [DOI] [PubMed] [Google Scholar]
- Chory J., Wu D. Weaving the complex web of signal transduction. Plant Physiol. 2001;1251(1):77–80. doi: 10.1104/pp.125.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse S., Langford M., McMorris T. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 1996;1111(1):671–678. doi: 10.1104/pp.111.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coupland G., Kuntz M. Mutations in the Arabidopsis gene IMMUTANS cause a variegated phenotype by inactivating a chloroplast terminal oxidase associated with phytoene desaturation. Plant Cell. 1999;111(1):57–68. doi: 10.1105/tpc.11.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coruzzi G. M., Zhou L. Carbon and nitrogen sensing and signaling in plants: Emerging ‘matrix effects’. Curr Opin Plant Biol. 2001;41(1):247–253. doi: 10.1016/s1369-5266(00)00168-0. [DOI] [PubMed] [Google Scholar]
- Cowan A. K. Abscisic acid biosynthesis in vascular plants is a constitutive process. S. African J. Bot. 2001;671(1):497–505. [Google Scholar]
- Cunillera N., Arro M., Delourme D., Karst F., Boronat A., Ferrer A. Arabidopsis thaliana contains two differentially expressed farnesyl-diphosphate synthase genes. J. Biol. Chem. 1996;2711(1):7774–7780. doi: 10.1074/jbc.271.13.7774. [DOI] [PubMed] [Google Scholar]
- Cunningham F. X., Gantt E. Genes and enzymes of carotenoid biosynthesis in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998;491(1):557–583. doi: 10.1146/annurev.arplant.49.1.557. [DOI] [PubMed] [Google Scholar]
- Cutler A. J., Krochko J. E. Formation and breakdown of ABA. Trends Plant Sci. 1999;41(1):472–478. doi: 10.1016/s1360-1385(99)01497-1. [DOI] [PubMed] [Google Scholar]
- Cutler S., Ghassemian M., Bonetta D., Cooney S., McCourt P. A protein farnesyl transferase involved in abscisic acid signal transduction in Arabidopsis. Science. 1996;2731(1):1239–1241. doi: 10.1126/science.273.5279.1239. [DOI] [PubMed] [Google Scholar]
- Davies W., Zhang J. Root signals and the regulation of growth and development of plants in drying soil. Ann. Rev. Plant Physiol. Plant Mol. Biol. 1991;421(1):55–76. [Google Scholar]
- Delseny M., Bies-Etheve N., Carles C., Hull G., Vicient C., Raynal M., Grellet F., Aspart L. Late Embryogenesis Abundant (LEA) protein regulation during Arabidopsis seed maturation. J. Plant Physiol. 2001;1581(1):419–427. [Google Scholar]
- Desikan R., Hagenbeek D., Neill S. J., Rock C. D. Flow cytometry and surface plasmon resonance analyses demonstrate that the monoclonal antibody JIM19 interacts with a rice cell surface component involved in abscisic acid signalling in protoplasts. FEBS Lett. 1999;4561(1):257–262. doi: 10.1016/s0014-5793(99)00972-2. [DOI] [PubMed] [Google Scholar]
- Dijkwel P. P., Huijser C., Weisbeek P. J., Chua N-H., Smeekens S. C. M. Sucrose control of phytochrome A signaling in Arabidopsis. Plant Cell. 1997;91(1):583–595. doi: 10.1105/tpc.9.4.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J. P., Pickard B. G. Mechanosensory calcium-selective cation channels in epidermal cells. Plant J. 1993;31(1):83–110. doi: 10.1111/j.1365-313x.1993.tb00013.x. [DOI] [PubMed] [Google Scholar]
- Drees B. L., Sundin B., Brazeau E., Caviston J. P., Chen G. C., Guo W., Kozminski K. G., Lau M. W., Moskow J. J., Tong A., Schenkman L. R., McKenzie A., Brennwald P., Longtine M., Bi E., Chan C., Novick P., Boone C., Pringle J. R., Davis T. N., Fields S., Drubin D. G. A protein interaction map for cell polarity development. J. Cell Biol. 2001;1541(1):549–571. doi: 10.1083/jcb.200104057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckham S. C., Linforth R. S. T., Taylor I. B. Abscisic-acid-deficient mutants at the aba gene locus of Arabidopsis thaliana are impaired in the epoxidation of zeaxanthin. Plant Cell Environ. 1991;141(1):601–606. [Google Scholar]
- Duckham S. C., Taylor I. B., Linforth R. S. T., Al-Naieb R. J., Marples B. A., Bowman W. R. The metabolism of cis-ABA-aldehyde by the wilty mutants of potato, pea, and Arabidopsis thaliana. J. Exp. Bot. 1989;401(1):901–905. [Google Scholar]
- Eastmond P. J., Germain V., Lange P. R., Bryce J. H., Smith S. M., Graham I. A. Postgerminative growth and lipid catabolism in oilseeds lacking the glyoxylate cycle. Proc. Natl. Acad. Sci. USA. 2000;971(1):5669–5674. doi: 10.1073/pnas.97.10.5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis M., Dennis E., Peacock W. Arabidopsis roots and shoots have different mechanisms for hypoxic stress tolerance. Plant Physiol. 1999;1191(1):57–64. doi: 10.1104/pp.119.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephritikhine G., Fellner M., Vannini C., Lapous D., Barbier-Brygoo H. The sax1 dwarf mutant of Arabidopsis thaliana shows altered sensitivity of growth responses to abscisic acid, auxin, gibberellins and ethylene and is partially rescued by exogenous brassinosteroid. Plant Journal. 1999;181(1):303–314. doi: 10.1046/j.1365-313x.1999.00454.x. [DOI] [PubMed] [Google Scholar]
- Estévez J. M., Cantero A., Romero C., Kawaide H., Jimenez L. F., Kuzuyama T., Seto H., Kamiya Y., León P. Analysis of the expression of CLA1, a gene that encodes the 1-deoxyxylulose 5-phosphate synthase of the 2-C-methyl-D-erythritol-4-phosphate pathway in Arabidopsis. Plant Physiol. 2000;1241(1):95–103. doi: 10.1104/pp.124.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estévez J. M., Cantero A., Reindl A., Reichler S., León P. 1-Deoxy-D-xylulose-5-phosphate synthase, a limiting enzyme for plastidic isoprenoid biosynthesis in plants. J. Biol. Chem. 2001;2761(1):22901–22909. doi: 10.1074/jbc.M100854200. [DOI] [PubMed] [Google Scholar]
- Eun S-O., Lee Y. Actin filaments of guard cells are reorganized in response to light and abscisic acid. Plant Physiol. 1997;1151(1):1491–1498. doi: 10.1104/pp.115.4.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eun S-O., Bae S-H., Lee Y. Cortical actin filaments in guard cells respond differently to abscisic acid in wild-type and abi1-1 mutant Arabidopsis. Planta. 2001;2121(1):466–469. doi: 10.1007/s004250000489. [DOI] [PubMed] [Google Scholar]
- Ezcurra I., Wycliffe P., Nehlin L., Ellerstrom M., Rask L. Transactivation of the Brassica napus napin promoter by ABI3 requires interaction of the conserved B2 and B3 domains of ABI3 with different cis-elements: B2 mediates activation through an ABRE, whereas B3 interacts with an RY/G-box. Plant J. 2000;241(1):57–66. doi: 10.1046/j.1365-313x.2000.00857.x. [DOI] [PubMed] [Google Scholar]
- Fan L., Zheng S., Wang X. Antisense suppression of phospholipase Dα retards abscisic acid- and ethylene-promoted senescence of postharvest arabidopsis leaves. Plant Cell. 1997;91(1):2183–2196. doi: 10.1105/tpc.9.12.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L., Zheng S., Cui D., Wang X. Subcellular distribution and tissue expression of phospholipase Dα, Dβ, and Dγ in Arabidopsis. Plant Physiol. 1999;1191(1):1371–1378. doi: 10.1104/pp.119.4.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellner M., Zhang R. C., Pharis R. P., Sawhney V. K. Reduced de-etiolation of hypocotyl growth in a tomato mutant is associated with hypersensitivity to, and high endogenous levels of, abscisic acid. J. Exp. Bot. 2001;521(1):725–738. doi: 10.1093/jexbot/52.357.725. [DOI] [PubMed] [Google Scholar]
- Finkelstein R., Somerville C. Three classes of abscisic acid (ABA)-insensitive mutations of Arabidopsis define genes that control overlapping subsets of ABA responses. Plant Physiol. 1990;941(1):1172–1179. doi: 10.1104/pp.94.3.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R., Lynch T. Abi8: a near-lethal ABA response mutant. 1997. Paper presented at: 8th International Conference on Arabidopsis Research (Madison, WI)
- Finkelstein R., Lynch T. The Arabidopsis Abscisic Acid Response Gene ABI5 Encodes a Basic Leucine Zipper Transcription Factor. Plant Cell. 2000a;121(1):599–609. doi: 10.1105/tpc.12.4.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R., Lynch T. Abscisic acid inhibition of radicle emergence but not seedling growth is suppressed by sugars. Plant Physiol. 2000b;1221(1):1179–1186. doi: 10.1104/pp.122.4.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R., Gampala S., Rock C. Abscisic acid signaling in seeds and seedlings. Plant Cell. 2002;141(1):S15–S45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R. R. Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. Plant J. 1994a;51(1):765–771. [Google Scholar]
- Finkelstein R. R. Maternal effects govern variable dominance of two abscisic acid response mutations in Arabidopsis thaliana. Plant Physiol. 1994b;1051(1):1203–1208. doi: 10.1104/pp.105.4.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R. R., Wang M. L., Lynch T. J., Rao S., Goodman H. M. The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA2 domain protein. Plant Cell. 1998;101(1):1043–1054. doi: 10.1105/tpc.10.6.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster R., Chua N-H. An Arabidopsis mutant with deregulated ABA gene expression: implications for negative regulator function. Plant J. 1999;171(1):363–372. doi: 10.1046/j.1365-313x.1999.00384.x. [DOI] [PubMed] [Google Scholar]
- Frey A., Audran C., Marin E., Sotta B., Marion-Poll A. Engineering seed dormancy by the modification of zeaxanthin epoxidase gene expression. Plant Mol. Biol. 1999;391(1):1267–1274. doi: 10.1023/a:1006145025631. [DOI] [PubMed] [Google Scholar]
- Fromont-Racine M., Mayes A. E., Brunet-Simon A., Rain J-C., Colley A., Dix I., Decourty L., Joly N., Ricard F., Beggs J. Dao. Genome-wide protein interaction screens reveal functional networks involving Sm-like proteins. Yeast. 2000;171(1):95–110. doi: 10.1002/1097-0061(20000630)17:2<95::AID-YEA16>3.0.CO;2-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble P. E., Mullet J. E. Inhibition of carotenoid accumulation and abscisic acid biosynthesis in fluridone-treated dark-grown barley. Eur. J. Biochem. 1986;1601(1):117–121. doi: 10.1111/j.1432-1033.1986.tb09947.x. [DOI] [PubMed] [Google Scholar]
- Gampala S., Finkelstein R., Rock C. ABA INSENSITIVE-5 transactivates abscisic acid-inducible gene expression in rice protoplasts. 2001a. Paper presented at: Plant Biology 2001 (Providence, Rhode Island, USA, American Society of Plant Biologists)
- Gampala S., Hagenbeek D., Rock C. Functional interactions of lanthanum and phospholipase D with the abscisic acid signaling effectors VP1 and ABI1-1 in rice protoplasts. J. Biol. Chem. 2001b;2761(1):9855–9860. doi: 10.1074/jbc.M009168200. [DOI] [PubMed] [Google Scholar]
- Gampala S. S. L., Finkelstein R. R., Sun S. M., Rock C. D. ABA INSENSITIVE-5 interacts with ABA signaling effectors in rice protoplasts. J. Biol. Chem. 2002;2771(1):1689–1694. doi: 10.1074/jbc.M109980200. [DOI] [PubMed] [Google Scholar]
- Garciarrubio A., Legaria J. P., Covarrubias A. A. Abscisic acid inhibits germination of mature Arabidopsis seeds by limiting the availability of energy and nutrients. Planta. 1997;2031(1):182–187. doi: 10.1007/s004250050180. [DOI] [PubMed] [Google Scholar]
- Gaymard F., Pilot G., Lacombe B., Bouchez D., Bruneau D., Boucherez J., Michaux-Ferriere N., Thibaud J-B., Sentenac H. Identification and disruption of a plant shaker-like outward channel involved in K+ release into the xylem sap. Cell. 1998;941(1):647–655. doi: 10.1016/s0092-8674(00)81606-2. [DOI] [PubMed] [Google Scholar]
- Gazzarrini S., McCourt P. Genetic interactions between ABA, ethylene and sugar signaling pathways. Curr. Opin. Plant Biol. 2001;41(1):387–391. doi: 10.1016/s1369-5266(00)00190-4. [DOI] [PubMed] [Google Scholar]
- Geelen D., Leyman B., Batoko H., Di Sansabastiano G-P., Moore I., Blatt M. R. The abscisic acid-related SNARE homolog NtSyr1 contributes to secretion and growth: evidence from competition with its cytosolic domain. Plant Cell. 2002;141(1):387–406. doi: 10.1105/tpc.010328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghassemian M., Nambara E., Cutler S., Kawaide H., Kamiya Y., McCourt P. Regulation of abscisic acid signaling by the ethylene response pathway in Arabidopsis. Plant Cell. 2000;121(1):1117–1126. doi: 10.1105/tpc.12.7.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghelis T., Dellis O., Jeannette E., Bardat F., Cornel D., Miginiac E., Rona J-P., Sotta B. Abscisic acid specific expression of RAB18 involves activation of anion channels in Arabidopsis thaliana suspension cells. FEBS Lett. 2000a;4741(1):43–47. doi: 10.1016/s0014-5793(00)01574-x. [DOI] [PubMed] [Google Scholar]
- Ghelis T., Dellis O., Jeannette E., Bardat F., Miginiac E., Sotta B. Abscisic acid plasmalemma perception triggers a calcium influx essential for RAB18 gene expression in Arabidopsis thaliana suspension cells. FEBS Lett. 2000b;4831(1):67–70. doi: 10.1016/s0014-5793(00)02088-3. [DOI] [PubMed] [Google Scholar]
- Gibson S. I. Plant sugar-response pathways. Part of a complex regulatory web. Plant Physiol. 2000;1241(1):1532–1539. doi: 10.1104/pp.124.4.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilroy S., Read N. D., Trewavas A. J. Elevation of cytoplasmic calcium by caged calsium or caged inositol trisphosphate initiates stomatal closure. Nature. 1990;3431(1):769–771. doi: 10.1038/346769a0. [DOI] [PubMed] [Google Scholar]
- Gilroy S., Jones R. Gibberellic acid and abscisic acid coordinately regulate cytoplasmic calcium and secretory activity in barley aleurone protoplasts. Proc. Natl. Acad. Sci. USA. 1992;891(1):3591–3595. doi: 10.1073/pnas.89.8.3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat J., Hauge B., Valon C., Smalle J., Parcy F., Goodman H. Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell. 1992;41(1):1251–1261. doi: 10.1105/tpc.4.10.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Cadenas A., Verhey S. D., Holappa L. D., Shen Q., Ho T-H. D., Walker-Simmons M. K. An abscisic acid-induced protein kinase, PKABA1, mediates abscisic acid-suppressed gene expression in barley aleurone layers. Proc. Natl. Acad. Sci. USA. 1999;961(1):1767–1772. doi: 10.1073/pnas.96.4.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Gusmán M., Apostolova N., Bellés J. M., Barrero J. M., Piqueras P., Ponce M. R., Micol J. L., Serrano R., Rodríguez P. L. The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin into abscisic aldehyde. Plant Cell. 2002. in press. [DOI] [PMC free article] [PubMed]
- Gosti F., Beaudoin N., Serizet C., Webb A., Vartanian N., Giraudat J. ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell. 1999;111(1):1897–1909. doi: 10.1105/tpc.11.10.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabov A., Leung J., Giraudat J., Blatt M. R. Alteration of anion channel kinetics in wild-type and abi1-1 transgenic Nicotiana benthamiana guard cells by abscisic acid. Plant J. 1997;121(1):203–213. doi: 10.1046/j.1365-313x.1997.12010203.x. [DOI] [PubMed] [Google Scholar]
- Grabov A., Blatt M. R. Membrane voltage initiates Ca2+ waves and potentiates Ca2+ increases with abscisic acid in stomatal guard cells. Proc. Natl. Acad. Sci. USA. 1998;951(1):4778–4783. doi: 10.1073/pnas.95.8.4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiltinan M., WR Marcotte J., Quatrano R. A plant leucine zipper protein that recognizes an abscisic acid response element. Science. 1990;2501(1):267–271. doi: 10.1126/science.2145628. [DOI] [PubMed] [Google Scholar]
- Hable W. E., Oishi K. K., Schumaker K. S. Viviparous-5 encodes phytoene desaturase, an enzyme essential for abscisic acid (ABA) accumulation and seed development in maize. Mol. Gen. Genet. 1998;2571(1):167–176. doi: 10.1007/s004380050636. [DOI] [PubMed] [Google Scholar]
- Hagenbeek D., Quatrano R., Rock C. Trivalent ions activate abscisic acid-inducible promoters through an ABI1-dependent pathway in rice protoplasts. Plant Physiol. 2000;1231(1):1553–1560. doi: 10.1104/pp.123.4.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton D. W. A., Hills A., Kohler B., Blatt M. R. Ca2+ channels at the plasma membrane of stomatal guard cells are activated by hyperpolarization and abscisic acid. Proc. Natl. Acad. Sci. USA. 2000;971(1):4967–4972. doi: 10.1073/pnas.080068897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung W., Sauter A., Hose E. Abscisic acid in the xylem: where does it come from, where does it go to? J. Exp. Bot. 2002;531(1):27–32. [PubMed] [Google Scholar]
- Hartung W., Wilkinson S., Davies W. J. Factors that regulate abscisic acid concentrations at the primary site of action at the guard cell. J. Exp. Bot. 1998;491(1):361–367. [Google Scholar]
- Hedrich R., Moran O., Conti F., Busch H., Becker D., Gambale F., Dreyer I., Kuch A., Neuwinger K., Palme K. Inward rectifier potassium channels in plants differ from their animal counterparts in response to voltage and channel modulators. Eur. Biophys. J. 1995;241(1):107–115. doi: 10.1007/BF00211406. [DOI] [PubMed] [Google Scholar]
- Heimovaara-Dijkstra S., Mundy J., Wang M. The effect of intracellular pH on the regulation of the Rab 16A and the alpha-amylase 1/6-4 promoter by abscisic acid and gibberellin. Plant Mol. Biol. 1995;271(1):815–820. doi: 10.1007/BF00020234. [DOI] [PubMed] [Google Scholar]
- Hemerly A. S., Ferreira P., De Almeida Engler J., Van Montagu M., Engler G., Inze D. Cdc2a Expression in Arabidopsis is linked with competence for cell division. Plant Cell. 1993;51(1):1711–1723. doi: 10.1105/tpc.5.12.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz S., Wungsintaweekul J., Schuhr S. A., Hecht S., LFCttgen H., Sagner S., Fellermeier M., Eisenreich W., Zenk M. H., Bacher A., Rohdich F. Biosynthesis of terpenoids: YgbB protein converts 4-diphosphocytidyl-2C-methyl-D-erythritol 2-phosphate to 2C-methyl-D-erythritol 2,4-cyclodiphosphate. Proc. Natl. Acad. Sci. USA. 2000;971(1):2486–2490. doi: 10.1073/pnas.040554697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill A., Nantel A., Rock C., Quatrano R. A conserved domain of the Viviparous-1 gene product enhances the DNA binding activity of the bZIP protein EmBP-1 and other transcription factors. J. Biol. Chem. 1996;2711(1):3366–3374. doi: 10.1074/jbc.271.7.3366. [DOI] [PubMed] [Google Scholar]
- Himmelbach A., Iten M., Grill E. Signalling of abscisic acid to regulate plant growth. Phil. Trans. Royal Soc. London B Biol. Sci. 1998;3531(1):1439–1444. doi: 10.1098/rstb.1998.0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelbach A., Grill E. Arabidopsis ABA responses are regulated by a HD-Zip protein, a target of the protein phosphatase 2C ABI1. 2001. Paper presented at: Twelfth International Conference on Arabidopsis Research (Madison, WI, USA)
- Hirayama T., Ohto C., Mizoguchi T., Shinozaki K. A gene encoding a phosphatidylinositol-specific phospholipase C is induced by dehydration and salt stress in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 1995;921(1):3903–3907. doi: 10.1073/pnas.92.9.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschberg J. Carotenoid biosynthesis in flowering plants. Curr. Opin. Plant Biol. 2001;41(1):210–218. doi: 10.1016/s1369-5266(00)00163-1. [DOI] [PubMed] [Google Scholar]
- Hobo T., Kowyama Y., Hattori T. A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proc. Natl. Acad. Sci. U.S.A. 1999;961(1):15348–15353. doi: 10.1073/pnas.96.26.15348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U., Vasil I. K., McCarty D. R. Integrated control of seed maturation and germination programs by activator and repressor functions of Viviparous-1 of maize. Genes Dev. 1995;91(1):2459–2469. doi: 10.1101/gad.9.20.2459. [DOI] [PubMed] [Google Scholar]
- Hoff T., Frandsen G. I., Rocher A., Mundy J. Biochemical and genetic characterization of three molybdenum cofactor hydroxylases in Arabidopsis thaliana. Biochim. Biophys. Acta. 1998;13981(1):397–402. doi: 10.1016/s0167-4781(98)00085-2. [DOI] [PubMed] [Google Scholar]
- Hoff T., Schnorr K. M., Meyer C., Caboche M. Isolation of 2 Arabidopsis cDNAs involved in early steps of molybdenum cofactor biosynthesis by functional complementation of Escherichia coli mutants. J. Biol. Chem. 1995;2701(1):6100–6107. doi: 10.1074/jbc.270.11.6100. [DOI] [PubMed] [Google Scholar]
- Holdsworth M., Kurup S., McKibbin R. Molecular and genetic mechanisms regulating the transition from embryo development to germination. Trends Plant Sci. 1999;41(1):275–280. [Google Scholar]
- Holdsworth M., Lenton J., Flintham J., Gale M., Kurup S., McKibbin R., Bailey P., Larner V., Russell L. Genetic control mechanisms regulating the initiation of germination. J. Plant Physiol. 2001;1581(1):439–445. [Google Scholar]
- Hong S. W., Jon J. H., Kwak J. M., Nam H. G. Identification of a receptor-like protein kinase gene rapidly induced by abscisic acid, dehydration, high salt, and cold treatments in Arabidopsis thaliana. Plant Physiol. 1997;1131(1):1203–1212. doi: 10.1104/pp.113.4.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornberg C., Weiler E. High affinity binding sites for ABA on the plasmalemma of Vicia guard cells. Nature. 1984;3101(1):321–324. [Google Scholar]
- Hose E., Steudle E., Hartung W. Abscisic acid and hydraulic conductivity of maize roots: A study using cell- and root-pressure probes. Planta. 2000;2111(1):874–882. doi: 10.1007/s004250000412. [DOI] [PubMed] [Google Scholar]
- Hugouvieux V., Kwak J., Schroeder J. A mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell. 2001;1061(1):477–487. doi: 10.1016/s0092-8674(01)00460-3. [DOI] [PubMed] [Google Scholar]
- Huijser C., Kortstee A., Pego J., Weisbeek P., Wisman E., Smeekens S. The Arabidopsis SUCROSE UNCOUPLED-6 gene is identical to ABSCISIC ACID INSENSITIVE-4: involvement of abscisic acid in sugar responses. Plant Journal. 2000;231(1):577–585. doi: 10.1046/j.1365-313x.2000.00822.x. [DOI] [PubMed] [Google Scholar]
- Hwang I., Goodman H. M. An Arabidopsis thaliana root-specific kinase homolog is induced by dehydration, ABA, and NaCI. Plant J. 1995;81(1):37–43. doi: 10.1046/j.1365-313x.1995.08010037.x. [DOI] [PubMed] [Google Scholar]
- Hwang J-U., Lee Y. Abscisic acid-induced actin reorganization in guard cells of dayflower is mediated by cytosolic calcium levels and by protein kinase and protein phosphatase activities. Plant Physiol. 2001;1251(1):2120–2128. doi: 10.1104/pp.125.4.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram J., Bartels D. The molecular basis of dehydration tolerance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996;471(1):377–403. doi: 10.1146/annurev.arplant.47.1.377. [DOI] [PubMed] [Google Scholar]
- Initiative Arabidopsis Genome Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;4081(1):796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Inoue T., Higuchi M., Hashimoto Y., Seki M., Kobayashi M., Kato T., Tabata S., Shinozaki K., Kakimoto T. Identification of CRá as a cytokinin receptor from Arabidopsis. Nature. 2001;4091(1):1060–1063. doi: 10.1038/35059117. [DOI] [PubMed] [Google Scholar]
- Irving H., Gehring C., Parish R. Changes in cytosolic pH and calcium of guard cells precede stomatal movements. Proc. Natl. Acad. Sci. USA. 1992;891(1):1790–1794. doi: 10.1073/pnas.89.5.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacson T., Ronen G., Zamir D., Hirschberg J. Cloning of tangerine from tomato reveals a carotenoid isomerase essential for the production of β-carotene and xanthophylls in plants. Plant Cell. 2002;141(1):333–342. doi: 10.1105/tpc.010303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani M., Xiong L., Stevenson B., Zhu J-K. Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: Interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell. 1997;91(1):1935–1949. doi: 10.1105/tpc.9.11.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S., Kobayashi M., Yamaguchi-Shinozaki K., Shinozaki K. A stress-inducible gene for 9-cis-epoxycarotenoid dioxygenase involved in abscisic acid biosynthesis under water stress in drought-tolerant cowpea. Plant Physiol. 2000;1231(1):553–562. doi: 10.1104/pp.123.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S., Kobayashi M., Taji T., Naramoto M., Seki M., Kato T., Tabata S., Kakubari Y., Yamaguchi-Shinozaki K., Shinozaki K. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 2001;271(1):325–333. doi: 10.1046/j.1365-313x.2001.01096.x. [DOI] [PubMed] [Google Scholar]
- Izawa T., Foster R., Chua N-H. Plant bZIP protein DNA binding specificity. J. Mol. Biol. 1993;2301(1):1131–1144. doi: 10.1006/jmbi.1993.1230. [DOI] [PubMed] [Google Scholar]
- Jacob T., Ritchie S., Assmann S., Gilroy S. Abscisic acid signal transduction in guard cells is mediated by phospholipase D activity. Proc. Natl. Acad. Sci. USA. 1999;961(1):12192–12197. doi: 10.1073/pnas.96.21.12192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby M., Weisshaar B., Dröge-Laser W., Vicente-Carbajosa J., Tiedemann J., Kroj T., Parcy F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002;71(1):106–111. doi: 10.1016/s1360-1385(01)02223-3. [DOI] [PubMed] [Google Scholar]
- Janick-Buckner D., O'Neal J. M., Joyce E. K., Buckner B. Genetic and biochemical analysis of the y9 gene of maize, a carotenoid biosynthetic gene. Maydica. 2001;461(1):41–46. [Google Scholar]
- Jeannette E., Rona J. P., Bardat F., Cornel D., Sotta B., Miginiac E. Induction of Rab18 gene expression and activation of K+ outward rectifying channels depend on an extracellular perception of ABA in Arabidopsis thaliana suspension cells. Plant J. 1999;181(1):13–22. doi: 10.1046/j.1365-313x.1999.00423.x. [DOI] [PubMed] [Google Scholar]
- Kargul J., Gansel X., Tyrrell M., Sticher L., Blatt M. R. Protein-binding partners of the tobacco syntaxin NtSyr1. FEBS Lett. 2001;5081(1):253–258. doi: 10.1016/s0014-5793(01)03089-7. [DOI] [PubMed] [Google Scholar]
- Karssen C., Brinkhorst-van der Swan D., Breekland A., Koornneef M. Induction of dormancy during seed development by endogenous abscisic acid: studies of abscisic acid deficient genotypes of Arabidopsis thaliana (L.). Heynh. Planta. 1983;1571(1):158–165. doi: 10.1007/BF00393650. [DOI] [PubMed] [Google Scholar]
- Katagiri T., Takahashi S., Shinozaki K. Involvement of a novel Arabidopsis phospholipase D, AtPLDδ, in dehydration-inducible accumulation of phosphatidic acid in stress signalling. Plant J. 2001;261(1):595–605. doi: 10.1046/j.1365-313x.2001.01060.x. [DOI] [PubMed] [Google Scholar]
- Keith K., Kraml M., Dengler N. G., McCourt P. Fusca3: A heterochronic mutation affecting late embryo development in Arabidopsis. Plant Cell. 1994;61(1):589–600. doi: 10.1105/tpc.6.5.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber J. J., Rothenberg M., Roman G., Feldmann K. A., Ecker J. R. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the Raf family of protein kinases. Cell. 1993;721(1):427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- Kim B. T., Min Y. K., Asami T., Park N. K., Kwon O. Y., Cho K. Y., Yoshida S. 2-Fluoroabscisic acid analogs- their synthesis and biological activities. J. Agric. Food Chem. 1999;471(1):313–317. doi: 10.1021/jf980265l. [DOI] [PubMed] [Google Scholar]
- Kim S., Ma J., Perret P., Li Z., Thomas T. Arabidopsis ABI5 subfamily members have distinct DNA binding and transcriptional activities. Plant Physiol. 2002. submitted. [DOI] [PMC free article] [PubMed]
- Kim S. Y., Chung H-J., Thomas T. L. Isolation of a novel class of bZIP transcription factors that interact with ABA-responsive and embryo-specification elements in the Dc3 promoter using a modified yeast one-hybrid system. Plant J. 1997;111(1):1237–1251. doi: 10.1046/j.1365-313x.1997.11061237.x. [DOI] [PubMed] [Google Scholar]
- Kitagawa Y., Yamamoto H., Oritani T. Biosynthesis of abscisic acid in the fungus Cercospora cruenta: Stimulation of biosynthesis by water stress and isolation of a transgenic mutant with reduced biosynthetic capacity. Plant Cell Physiol. 1995;361(1):557–564. [Google Scholar]
- Knetsch M. L. W., Wang M., Snaar-Jagalska B. E., Heimovaara-Dijkstra S. Abscisic acid induces mitogen-activated protein kinase activation in barley aleurone protoplasts. Plant Cell. 1996;81(1):1061–1067. doi: 10.1105/tpc.8.6.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight H., Veale E., Warren G., Knight M. The sfr6 mutation in Arabidopsis suppresses low-temperature induction of genes dependent on the CRT/DREB1 sequence motif. Plant Cell. 1999;111(1):875–886. doi: 10.1105/tpc.11.5.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M., Hanhart C., Hilhorst H., Karssen C. In vivo inhibition of seed development and reserve protein accumulation in recombinants of abscisic acid biosynthesis and responsiveness mutants in Arabidopsis thaliana. Plant Physiol. 1989;901(1):463–469. doi: 10.1104/pp.90.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M., Jorna M. L., Brinkhorst-van der Swan D. L. C., Karssen C. M. The isolation of abscisic acid (ABA) deficient mutants by selection of induced revertants in non-germinating gibberellin sensitive lines of Arabidopsis thaliana (L. ) Heynh. Theoret Appl. Genet. 1982;611(1):385–393. doi: 10.1007/BF00272861. [DOI] [PubMed] [Google Scholar]
- Koornneef M., Léon-Kloosterziel K. M., Schwartz S. H., Zeevaart J. A. D. The genetic and molecular dissection of abscisic acid biosynthesis and signal transduction in Arabidopsis. Plant Physiol. Biochem. 1998;361(1):83–89. [Google Scholar]
- Koornneef M., Reuling G., Karssen C. The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol. Plant. 1984;611(1):377–383. [Google Scholar]
- Kovacs I., Ayaydin F., Oberschall A., Ipacs I., Bottka S., Pongor S., Dudits D., Toth E. C. Immunolocalization of a novel annexin-like protein encoded by a stress and abscisic acid responsive gene in alfalfa. Plant J. 1998;151(1):185–197. doi: 10.1046/j.1365-313x.1998.00194.x. [DOI] [PubMed] [Google Scholar]
- Kovtun Y., Chiu W-L., Tena G., Sheen J. Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc. Natl. Acad. Sci. USA. 2000;971(1):2940–2945. doi: 10.1073/pnas.97.6.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraepiel Y., Rousselin P., Sotta B., Kerhoas L., Einhorn J., Caboche M., Miginiac E. Analysis of phytochrome- and ABA-deficient mutants suggests that ABA degradation is controlled by light in Nicotiana plumbaginifolia. Plant J. 1994;61(1):665–672. [Google Scholar]
- Krochko J. E., Abrams G. D., Loewen M. K., Abrams S. R., Cutler A. J. (+)-Abscisic acid 8-hydroxylase is a cytochrome P450 monooxygenase. Plant Physiol. 1998;1181(1):849–860. doi: 10.1104/pp.118.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla J., Xu Q., Harter K., Gruissem W., Luan S. Genes for calcineurin B-like proteins in Arabidopsis are differentially regulated by stress signals. Proc. Natl. Acad. Sci. USA. 1999;961(1):4718–4723. doi: 10.1073/pnas.96.8.4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo A., Cappelluti S., Cervantes-Cervantes M., Rodriguez M., Bush D. S. Okadaic acid, a protein phosphatase inhibitor, blocks calcium changes, gene expression, and cell death induced by gibberellin in wheat aleurone cells. Plant Cell. 1996;81(1):259–269. doi: 10.1105/tpc.8.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurup S., Jones H., Holdsworth M. Interactions of the developmental regulator ABI3 with proteins identified from developing Arabidopsis seeds. Plant J. 2000;211(1):143–155. doi: 10.1046/j.1365-313x.2000.00663.x. [DOI] [PubMed] [Google Scholar]
- Kutschera U., Schopfer P. Effect of auxin and abscisic acid on cell wall extensibiltiy in maize coleoptiles. Planta. 1986;1671(1):527–535. doi: 10.1007/BF00391229. [DOI] [PubMed] [Google Scholar]
- Laby R., Kincaid M., Kim D., Gibson S. The Arabidopsis sugar-insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response. Plant J. 2000;231(1):587–596. doi: 10.1046/j.1365-313x.2000.00833.x. [DOI] [PubMed] [Google Scholar]
- Lacombe B., Pilot G., Michard E., Gaymard F., Sentenac H., Thibaud J-B. A shaker-like K+ channel with weak rectification is expressed in both source and sink phloem tissues of Arabidopsis. Plant Cell. 2000;121(1):837–851. doi: 10.1105/tpc.12.6.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkindale J., Knight M. Protection against heat stress-induced oxidative damage in Arabidopsis involves calcium, abscisic acid, ethylene, and salicylic acid. Plant Physiol. 2002;1281(1):682–695. doi: 10.1104/pp.010320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Page-Degivry M-T., Bidard J-N., Rouvier E., Bulard C., Lazdunski M. Presence of abscisic acid, a phytohormone, in the mammalian brain. Proc. Natl. Acad. Sci. USA. 1986;831(1):1155–1158. doi: 10.1073/pnas.83.4.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leckie C. P., McAinsh M. R., Allen G. J., Sanders D., Hetherington A. M. Abscisic acid-induced stomatal closure mediated by cyclic ADP-ribose. Proc. Natl. Acad. Sci. USA. 1998;951(1):15837–15842. doi: 10.1073/pnas.95.26.15837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Xiong L., Gong Z., Ishitani M., Stevenson B., Zhu J-K. The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo-cytoplasmic partitioning. Genes Dev. 2001;151(1):912–924. doi: 10.1101/gad.866801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Choi H., Suh S., Doo I-S., Oh K-Y., Choi E. J., Schroeder Taylor A. T., Low P. S., Lee Y. Oligogalacturonic acid and chitosan reduce stomatal aperture by inducing the evolution of reactive oxygen species from guard cells of tomato and Commelina communis. Plant Physiol. 1999;1211(1):147–152. doi: 10.1104/pp.121.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. H., Lee M. H., Chung W. I., Liu J. R. WAPK, a Ser/Thr protein kinase gene of Nicotiana tabacum, is uniquely regulated by wounding, abscisic acid and methyl jasmonate. Mol. Gen. Genet. 1998;2591(1):516–522. doi: 10.1007/s004380050843. [DOI] [PubMed] [Google Scholar]
- Lee Y-H., Chun J-Y. A new homeodomain-leucine zipper gene from Arabidopsis thaliana induced by water stress and abscisic acid treatment. Plant Mol. Biol. 1998;371(1):377–384. doi: 10.1023/a:1006084305012. [DOI] [PubMed] [Google Scholar]
- Lekstrom-Himes J., Xanthopoulos K. Biological role of the CCAAT/enhancer-binding protein family of transcription factors. J. Biol. Chem. 1998;2731(1):28545–28548. doi: 10.1074/jbc.273.44.28545. [DOI] [PubMed] [Google Scholar]
- Lemichez E., Wu Y., Sanchez J-P., Mettouchi A., Mathur J., Chua N-H. Inactivation of AtRac1 by abscisic acid is essential for stomatal closure. Genes Dev. 2001;151(1):1808–1816. doi: 10.1101/gad.900401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemtiri-Chlieh F., MacRobbie E. A. C., Brearley C. A. Inositol hexakisphosphate is a physiological signal regulating the K+-inward rectifying conductance in guard cells. Proc. Natl. Acad. Sci. USA. 2000;971(1):8687–8692. doi: 10.1073/pnas.140217497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léon-Kloosterziel K. M., Alvarez-Gil M., Ruijs G. J., Jacobsen S. E., Olszewski N. E., Schwartz S. H., Zeevaart J. A. D., Koornneef M. Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J. 1996;101(1):655–661. doi: 10.1046/j.1365-313x.1996.10040655.x. [DOI] [PubMed] [Google Scholar]
- Leon-Kloosterziel K., Van De Bunt G., Zeevaart J., Koornneef M. Arabidopsis mutants with a reduced seed dormancy. Plant Physiol. 1996;1101(1):233–240. doi: 10.1104/pp.110.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leube M. P., Grill E., Amrhein N. ABI1 of Arabidopsis is a protein serine/threonine phosphatase highly regulated by the proton and magnesium ion concentration. FEBS Lett. 1998;4241(1):100–104. doi: 10.1016/s0014-5793(98)00149-5. [DOI] [PubMed] [Google Scholar]
- Leung J., Bouvier-Durand M., Morris P-C., Guerrier D., Chefdor F., Giraudat J. Arabidopsis ABA response gene ABI1: Features of a calcium-modulated protein phosphatase. Science. 1994;2641(1):1448–1452. doi: 10.1126/science.7910981. [DOI] [PubMed] [Google Scholar]
- Leung J., Merlot S., Giraudat J. The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell. 1997;91(1):759–771. doi: 10.1105/tpc.9.5.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J., Giraudat J. Abscisic acid signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998;491(1):199–222. doi: 10.1146/annurev.arplant.49.1.199. [DOI] [PubMed] [Google Scholar]
- Levi M., Brusa P., Chiatante D., Sparvoli E. Cell cycle reactivation in cultured pea embryo axes: Effect of abscisic acid. In Vitro Cell. Dev. Biol. Plant. 1993;29P1(1):47–50. [Google Scholar]
- Leydecker M. T., Moureaux T., Kraepiel Y., Schnorr K., Caboche M. Molybdenum cofactor mutants, specifically impaired in xanthine dehydrogenase-activity and abscisic-acid biosynthesis, simultaneously overexpress nitrate reductase. Plant Physiol. 1995;1071(1):1427–1431. doi: 10.1104/pp.107.4.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyman B., Geelen D., Quintero F. J., Blatt M. R. A tobacco syntaxin with a role in hormonal control of guard cell ion channels. Science. 1999;2831(1):537–540. doi: 10.1126/science.283.5401.537. [DOI] [PubMed] [Google Scholar]
- Leyman B., Geelen D., Blatt M. R. Localization and control of expression of Nt-Syr1, a tobacco snare protein. Plant J. 2000;241(1):369–381. doi: 10.1046/j.1365-313x.2000.00886.x. [DOI] [PubMed] [Google Scholar]
- Leymarie J., Vavasseur A., Lasceve G. CO2 sensing in stomata of abi1-1 and abi2-1 mutants of Arabidopsis thaliana. Plant Physiol. Biochem. (Paris) 1998;361(1):539–543. [Google Scholar]
- Leymarie J., Lasceve G., Vavasseur A. Elevated CO2 enhances stomatal responses to osmotic stress and abscisic acid in Arabidopsis thaliana. Plant Cell Envir. 1999;221(1):301–308. [Google Scholar]
- Li G., Bishop K., Chandrasekharan M., Hall T. b-phaseolin gene activation is a two-step process: PvALF-facilitated chromatin modification followed by abscisic acid-mediated gene activation. Proc. Natl. Acad. Sci. USA. 1999;961(1):7104–7109. doi: 10.1073/pnas.96.12.7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Assmann S. M. An abscisic acid-activated and calcium-independent protein kinase from guard cells of fava bean. Plant Cell. 1996;81(1):2359–2368. doi: 10.1105/tpc.8.12.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;901(1):929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- Li J., Wang X-Q., Watson M. B., Assmann S. M. Regulation of abscisic acid-induced stomatal closure and anion channels by guard cell AAPK kinase. Science. 2000;2871(1):300–303. doi: 10.1126/science.287.5451.300. [DOI] [PubMed] [Google Scholar]
- Li J., Nam K. H., Vafeados D., Chory J. BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 2001;1271(1):14–22. doi: 10.1104/pp.127.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Walton D. C. Violaxanthin is an abscisic acid precursor in water-stressed dark-grown bean leaves. Plant Physiol. 1990;921(1):551–559. doi: 10.1104/pp.92.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler H. K. The 1-deoxy-D-xylulose-5-phosphate pathway of isoprenoid biosynthesis in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999;501(1):47–65. doi: 10.1146/annurev.arplant.50.1.47. [DOI] [PubMed] [Google Scholar]
- Liotenberg S., North H., Marion-Poll A. Molecular biology and regulation of abscisic acid biosynthesis in plants. Plant Physiol. Biochem. 1999;371(1):341–350. [Google Scholar]
- Liu J., Ishitani M., Halfter U., Kim C-S., Zhu J-K. The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc. Natl. Acad. Sci. USA. 2000;971(1):3730–3734. doi: 10.1073/pnas.060034197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. H., Luo M., Cheng K. J., Mohapatra S. S., Hill R. D. Identification and characterization of a novel barley gene that is ABA-inducible and expressed in embryo and aleurone. J. Exp. Bot. 1999;501(1):727–728. [Google Scholar]
- Liu Y., Bergervoet J. H. W., Ric De Vos C. H., Hilhorst H. W. M., Kraak H. L., Karssen C. M., Bino R. J. Nuclear replication activities during imbibition of abscisic acid-and gibberellin-deficient tomato (Lycopersicon esculentum Mill.) seeds. Planta. 1994;1941(1):368–373. [Google Scholar]
- Liu Z. H., Matthews P. D., Burr B., Wurtzel E. T. Cloning and characterization of a maize cDNA encoding phytoene desaturase, an enzyme of the carotenoid biosynthetic pathway. Plant Mol. Biol. 1996;301(1):269–279. doi: 10.1007/BF00020113. [DOI] [PubMed] [Google Scholar]
- Llorente F., Oliveros J. C., Martinez-Zapater J. M., Salinas J. A freezing-sensitive mutant of Arabidopsis, frs1, is a new aba3 allele. Planta. 2000;2111(1):648–655. doi: 10.1007/s004250000340. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L., Chua N-H. A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana. Plant Cell Physiol. 2000;411(1):541–547. doi: 10.1093/pcp/41.5.541. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., Chua N-H. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2001;981(1):4782–4787. doi: 10.1073/pnas.081594298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan T., Ohto M-A., Yee K. M., West M. A. L., Lo R., Kwong R. W., Yamagishi K., Fischer R. L., Goldberg R. B., Harada J. J. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell. 1998;931(1):1195–1205. doi: 10.1016/s0092-8674(00)81463-4. [DOI] [PubMed] [Google Scholar]
- Lu C., Fedoroff N. A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell. 2000;121(1):2351–2365. doi: 10.1105/tpc.12.12.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Paul A-L., McCarty D. R., Ferl R. J. Transcription factor veracity: Is GBó responsible for ABA-regulated expression of Arabidopsis Adh. Plant Cell. 1996;81(1):847–857. doi: 10.1105/tpc.8.5.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luerssen H., Kirik V., Herrmann P., Misera S. FUSCA3 encodes a protein with a conserved VP1/ABI3-like B3 domain which is of functional importance for the regulation of seed maturation in Arabidopsis thaliana. Plant J. 1998;151(1):755–764. doi: 10.1046/j.1365-313x.1998.00259.x. [DOI] [PubMed] [Google Scholar]
- Lüttgen H., Rohdich F., Herz S., Wungsintaweekul J., Hecht S., Schuhr C. A., Fellermeier M., Sagner S., Zenk M. H., Bacher A., Eisenreich W. Biosynthesis of terpenoids: YchB protein of Escherichia coli phosphorylates the 2-hydroxy group of 4-diphosphocytidyl-2C-methyl-D-erythritol. Proc. Natl. Acad. Sci. USA. 2000;971(1):1062–1067. doi: 10.1073/pnas.97.3.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRobbie E. A. C. ABA-induced ion efflux in stomatal guard cells: multiple actions of ABA inside and outside the cell. Plant J. 1995;71(1):565–576. [Google Scholar]
- MacRobbie E. A. C. ABA activates multiple Ca2+ fluxes in stomatal guard cells, triggering vacuolar K+ (Rb+) release. Proc. Natl. Acad. Sci. USA. 2000;971(1):12361–12368. doi: 10.1073/pnas.220417197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahonen A. P., Bonke M., Kauppinen L., Riikonen M., Benfey P. N., Helariutta Y. A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes Dev. 2000;141(1):2938–2943. doi: 10.1101/gad.189200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maluf M. P., Saab I. N., Wurtzel E. T., Sachs M. M. The viviparous12 maize mutant is deficient in abscisic acid, carotenoids, and chlorophyll synthesis. J Exp. Bot. 1997;481(1):1259–1268. [Google Scholar]
- Mandel M. A., Feldmann K. A., Herrera-Estrella L., Rocha-Sosa M., León P. CLA1, a novel gene required for chloroplast development, is highly conserved in evolution. Plant J. 1996;91(1):649–658. doi: 10.1046/j.1365-313x.1996.9050649.x. [DOI] [PubMed] [Google Scholar]
- Marin E., Marion-Poll A. Tomato flacca mutant is impaired in ABA aldehyde oxidase and xanthine dehydrogenase activities. Plant Physiol. Biochem. 1997;351(1):369–372. [Google Scholar]
- Marin E., Nussaume L., Quesada A., Gonneau M., Sotta B., Hugueney P., Frey A., Marion-Poll A. Molecular identification of zeaxanthin epoxidase of Nicotiana plumbaginifolia, a gene involved in abscisic acid biosynthesis and corresponding to the ABA locus of Arabidopsis thaliana. EMBO J. 1996;151(1):2331–2342. [PMC free article] [PubMed] [Google Scholar]
- Martinez-Zapater J. M. The Transition to Flowering in Arabidopsis. 1994;1(1):403–433. In Arabidopsis, E. M. Meyerowitz, and C. R. Somerville, eds (Plainview, New York: Cold Spring Harbor Laboratory Press), pp. [Google Scholar]
- McAinsh M. R., Webb A. A. R., Taylor J. E., Hetherington A. M. Stimulus-Induced Oscillations in Guard Cell Cytosolic Free Calcium. Plant Cell. 1995;71(1):1207–1219. doi: 10.1105/tpc.7.8.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCourt P. Genetic analysis of hormone signaling. Ann. Rev. Plant Physiol. Plant Mol. Biol. 1999;501(1):219–243. doi: 10.1146/annurev.arplant.50.1.219. [DOI] [PubMed] [Google Scholar]
- Meinhard M., Grill E. Hydrogen peroxide is a regulator of ABI1, a protein phosphatase 2C from Arabidopsis. FEBS Lett. 2001;5081(1):443–446. doi: 10.1016/s0014-5793(01)03106-4. [DOI] [PubMed] [Google Scholar]
- Meinke D. W., Franzmann L. H., Nickle T. C., Yeung E. C. Leafy cotyledon mutants of Arabidopsis. Plant Cell. 1994;61(1):1049–1064. doi: 10.1105/tpc.6.8.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meskiene I., Bogre L., Glaser W., Balog J., Brandstotter M., Zwerger K., Ammerer G., Hirt H. MP2C, a plant protein phosphatase 2C, functions as a negative regulator of mitogen-activated protein kinase pathways in yeast and plants. Proc. Natl. Acad. Sci. USA. 1998;951(1):1938–1943. doi: 10.1073/pnas.95.4.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurs C., Basra A., Karssen C., van Loon L. Role of abscisic acid in the induction of desiccation tolerance in developing seeds of Arabidopsis thaliana. Plant Physiol. 1992;981(1):1484–1493. doi: 10.1104/pp.98.4.1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K., Leube M., Grill E. A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science. 1994;2641(1):1452–1455. doi: 10.1126/science.8197457. [DOI] [PubMed] [Google Scholar]
- Miedema H., Assmann S. M. A membrane-delimited effect of internal pH on the K+ outward rectifier of Vicia faba guard cells. J. Memb. Biol. 1996;1541(1):227–237. doi: 10.1007/s002329900147. [DOI] [PubMed] [Google Scholar]
- Mikami K., Katagiri T., Iuchi S., Yamaguchi-Shinozaki K., Shinozaki K. A gene encoding phosphatidylinositol-4-phosphate 5-kinase is induced by water stress and abscisic acid in Arabidopsis thaliana. Plant J. 1998;151(1):563–568. doi: 10.1046/j.1365-313x.1998.00227.x. [DOI] [PubMed] [Google Scholar]
- Milborrow B. V. The pathway of biosynthesis of abscisic acid in vascular plants: a review of the present state of knowledge of ABA biosynthesis. J. Exp. Bot. 2001;521(1):1145–1164. [PubMed] [Google Scholar]
- Min X. J., Okada K., Brockmann B., Koshiba T., Kamiya Y. Molecular cloning and expression patterns of three putative functional aldehyde oxidase genes and isolation of two aldehyde oxidase pseudogenes in tomato. Biochim. Biophys. Acta. 2000;14931(1):337–341. doi: 10.1016/s0167-4781(00)00190-1. [DOI] [PubMed] [Google Scholar]
- Morillon R., Chrispeels M. J. The role of ABA and the transpiration stream in the regulation of the osmotic water permeability of leaf cells. Proc. Natl. Acad. Sci. USA. 2001;981(1):14138–14143. doi: 10.1073/pnas.231471998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata Y., Pei Z-M., Mori I. C., Schroeder J. Abscisic acid activation of plasma membrane Ca2+ channels in guard cells requires cytosolic NAD(P)H and is differentially disrupted upstream and downstream of reactive oxygen species production in abi1-1 and abi2-1 protein phosphatase 2C mutants. Plant Cell. 2001;131(1):2513–2523. doi: 10.1105/tpc.010210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal P., Walker L., Young J., Sonawala A., Timpte C., Estelle M., Reed J. AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol. 2000;1231(1):563–573. doi: 10.1104/pp.123.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S., Lynch T., Finkelstein R. Physical interactions between ABA response loci of Arabidopsis. Plant J. 2001;261(1):627–635. doi: 10.1046/j.1365-313x.2001.01069.x. [DOI] [PubMed] [Google Scholar]
- Nambara E., Naito S., McCourt P. A mutant of Arabidopsis which is defective in seed development and storage protein accumulation is a new abi3 allele. Plant J. 1992;21(1):435–441. [Google Scholar]
- Nambara E., Kawaide H., Kamiya Y., Naito S. Characterization of an Arabidopsis thaliana mutant that has a defect in ABA accumulation: ABA-dependent and ABA-independent accumulation of free amino acids during dehydration. Plant Cell Physiol. 1998;391(1):853–858. doi: 10.1093/oxfordjournals.pcp.a029444. [DOI] [PubMed] [Google Scholar]
- Nambara E., Hayama R., Tsuchiya Y., Nishimura M., Kawaide H., Kamiya Y., Naito S. The role of ABI3 and FUS3 loci in Arabidopsis thaliana on phase transition from late embryo development to germination. Dev. Biol. 2000;2201(1):412–423. doi: 10.1006/dbio.2000.9632. [DOI] [PubMed] [Google Scholar]
- Nambara E., Suzuki M., Abrams S., McCarty D. R., Kamiya Y., McCourt P. A screen for genes that function in abscisic acid signaling in Arabidopsis thaliana. Genetics. 2002. in press. [DOI] [PMC free article] [PubMed]
- Nantel A., Quatrano R. Characterization of three rice basic-leucine zipper factors, including two inhibitors of EmBP-1 DNA binding activity. J Biol. Chem. 1996;2711(1):31296–31305. doi: 10.1074/jbc.271.49.31296. [DOI] [PubMed] [Google Scholar]
- Neill S. J., Burnett E. C., Desikan R., Hancock J. T. Cloning of a wilt-responsive cDNA from an Arabidopsis thaliana suspension culture cDNA library that encodes a putative 9-cis-epoxy-carotenoid diogenase. J. Exp. Bot. 1998;491(1):1893–1894. [Google Scholar]
- Neill S. J., Desikan R., Clarke A., Hancock J. Nitric oxide is a novel component of abscisic acid signaling in stomatal guard cells. Plant Physiol. 2002;1281(1):13–16. [PMC free article] [PubMed] [Google Scholar]
- Neill S., Horgan R., Walton D., Lee T. The biosynthesis of abscisic-acid in Cercospora rosicola. Phytochemistry. 1982;211(1):61–65. [Google Scholar]
- Neill S. J., Horgan R., Parry A. D. The carotenoid and abscisic acid content of viviparous kernels and seedlings of Zea mays L. Planta. 1986;1691(1):87–96. doi: 10.1007/BF01369779. [DOI] [PubMed] [Google Scholar]
- Nemeth K., Salchert K., Putnoky P., Bhalerao R., Koncz-Kalman Z., Stankovic-Stangeland B., Bako L., Mathur J., Okresz L., Stabel S. Pleiotropic control of glucose and hormone responses by PRL1, a nuclear WD protein, in Arabidopsis. Genes Dev. 1998;121(1):3059–3073. doi: 10.1101/gad.12.19.3059. others., a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng C. K-Y., Carr K., McAinsh M. R., Powell B., Hetherington A. M. Drought-induced guard cell signal transduction involves sphingosine-1-phosphate. Nature. 2001;4101(1):596–599. doi: 10.1038/35069092. [DOI] [PubMed] [Google Scholar]
- Niyogi K. K. Photoprotection revisited: genetic and molecular approaches. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999;501(1):333–359. doi: 10.1146/annurev.arplant.50.1.333. [DOI] [PubMed] [Google Scholar]
- Niyogi K. K., Grossman A. R., Björkman O. Arabidopsis mutants define a central role for the xanthophyll cycle in the regulation of photosynthetic energy conversion. Plant Cell. 1998;101(1):1121–1134. doi: 10.1105/tpc.10.7.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordin K., Heino P., Palva E. T. Separate signal pathways regulate the expression of a low-temperature-induced gene in Arabidopsis thaliana (L.). Heynh. Plant Mol. Biol. 1991;161(1):1061–1071. doi: 10.1007/BF00016077. [DOI] [PubMed] [Google Scholar]
- Norris S. R., Shen X., DellaPenna D. Complementation of the Arabidopsis pds1 mutation with the gene encoding p-hydroxyphenylpyruvate dioxygenase. Plant Physiol. 1998;1171(1):1317–1323. doi: 10.1104/pp.117.4.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogas J., Cheng J-C., Sung Z. R., Somerville C. Cellular differentiation regulated by gibberellin in the Arabidopsis thaliana pickle mutant. Science. 1997;2771(1):91–94. doi: 10.1126/science.277.5322.91. [DOI] [PubMed] [Google Scholar]
- Ogas J., Kaufmann S., Henderson J., Somerville C. PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc. Natl. Acad. Sci. USA. 1999;961(1):13839–13844. doi: 10.1073/pnas.96.24.13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori N., Eshed Y., Pinto P., Paran I., Zamir D., Fluhr R. TAO1, a representative of the molybdenum cofactor containing hydroxylases from tomato. J. Biol. Chem. 1997;2721(1):1019–1025. doi: 10.1074/jbc.272.2.1019. [DOI] [PubMed] [Google Scholar]
- Papi M., Sabatini S., Bouchez D., Camilleri C., Costantino P., Vittorioso P. Identification and disruption of an Arabidopsis zinc finger gene controlling seed germination. Genes Dev. 2000;141(1):28–33. [PMC free article] [PubMed] [Google Scholar]
- Parcy F., Valon C., Raynal M., Gaubier-Comella P., Delseny M., Giraudat J. Regulation of gene expression programs during Arabidopsis seed development: Roles of the ABI3 locus and of endogenous abscisic acid. Plant Cell. 1994;61(1):1567–1582. doi: 10.1105/tpc.6.11.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy F., Giraudat J. Interactions between the ABI1 and the ectopically expressed ABI3 genes in controlling abscisic acid responses in Arabidopsis vegetative tissues. Plant J. 1997;111(1):693–702. doi: 10.1046/j.1365-313x.1997.11040693.x. [DOI] [PubMed] [Google Scholar]
- Parcy F., Valon C., Kohara A., Misera S., Giraudat J. The ABSCISIC ACID-INSENSITIVE3, FUSCA3, and LEAFY COTYLEDON1 loci act in concert to control multiple aspects of Arabidopsis seed development. Plant Cell. 1997;91(1):1265–1277. doi: 10.1105/tpc.9.8.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H., Kreunen S. S., Cuttriss A. J., DellaPenna D., Pogson B. J. Identification of the carotenoid isomerase provides insight into carotenoid biosynthesis, prolammellar body formation, and photomorphogenesis. Plant Cell. 2002;141(1):321–332. doi: 10.1105/tpc.010302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry A. D., Babiano M. J., Horgan R. The role of cis-carotenoids in abscisic acid biosynthesis. Planta. 1990;1821(1):118–128. doi: 10.1007/BF00239993. [DOI] [PubMed] [Google Scholar]
- Parry A. D., Blonstein A. D., Babiano M. J., King P. J., Horgan R. Abscisic acid metabolism in a wilty mutant of Nicotiana plumbaginifolia. Planta. 1991;1831(1):237–243. doi: 10.1007/BF00197794. [DOI] [PubMed] [Google Scholar]
- Parry A. D., Griffiths A., Horgan R. Abscisic-acid biosynthesis in roots .2. The effects of water-stress in wild-type and abscisic-acid-deficient mutant (notabilis) plants of Lycopersicon esculentum Mill. Planta. 1992;1871(1):192–197. doi: 10.1007/BF00201937. [DOI] [PubMed] [Google Scholar]
- Pédron J., Brault M., Näke C., Miginiac E. Detection of abscisic acid-binding proteins in the microsomal protein fraction of Arabidopsis thaliana with abscisic acid protein conjugates used as affinity probes. Eur. J. Biochem. 1998;2521(1):385–390. doi: 10.1046/j.1432-1327.1998.2520385.x. [DOI] [PubMed] [Google Scholar]
- Pei Z., Murata Y., Benning G., Thomine S., Klusener B., Allen G., Grill E., Schroeder J. Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature. 2000;4061(1):731–734. doi: 10.1038/35021067. [DOI] [PubMed] [Google Scholar]
- Pei Z-M., Kuchitsu K., Ward J. M., Schwarz M., Schroeder J. I. Differential abscisic acid regulation of guard cell slow anion channels in Arabidopsis wild-type and abi1 and abi2 mutants. Plant Cell. 1997;91(1):409–423. doi: 10.1105/tpc.9.3.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Z-M., Ghassemian M., Kwak C. M., McCourt P., Schroeder J. I. Role of farnesyltransferase in ABA regulation of guard cell anion channels and plant water loss. Science. 1998;2821(1):287–290. doi: 10.1126/science.282.5387.287. [DOI] [PubMed] [Google Scholar]
- Pepper A., Delaney T., Washburn T., Poole D., Chory J. DET1, a negative regulator of light-mediated development and gene expression in Arabidopsis, encodes a novel nuclear-localized protein. Cell. 1994;781(1):109–116. doi: 10.1016/0092-8674(94)90577-0. [DOI] [PubMed] [Google Scholar]
- Phillips J., Artsaenko O., Fiedler U., Horstmann C., Mock H-P., Muentz K., Conrad U. Seed-specific immunomodulation of abscisic acid activity induces a developmental switch. EMBO J. 1997;161(1):4489–4496. doi: 10.1093/emboj/16.15.4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao H. L., Pih K. T., Lim J. H., Kang S. G., Jin J. B., Kim S. H., Hwang I. An Arabidopsis GSK3/shaggy-like gene that complements yeast salt stress-sensitive mutants is induced by NaCl and abscisic acid. Plant Physiol. 1999;1191(1):1527–1534. doi: 10.1104/pp.119.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson B. J., Niyogi K. K., Björkman O., DellaPenna D. Altered xanthophyll compositions adversely affect chlorophyll accumulation and nonphotochemical quenching in Arabidopsis mutants. Proc. Natl. Acad. Sci. USA. 1998;951(1):13324–13329. doi: 10.1073/pnas.95.22.13324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postlethwait S., Nelson O. A chronically wilted mutant of maize. Am. J. Bot. 1957;441(1):628–633. [Google Scholar]
- Pritchard S., Graham I. A. The effect of phytohormones on storage reserve mobilisation during germination in Arabidopsis. 2001. Paper presented at: Twelfth International Conference on Arabidopsis Research (Madison, WI)
- Qin X., Zeevaart J. A. D. The 9-cis-epoxycarotenoid cleavage reaction is the key regulatory step of abscisic acid biosynthesis in water-stressed bean. Proc. Natl. Acad. Sci. USA. 1999;961(1):15354–15361. doi: 10.1073/pnas.96.26.15354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X., Zeevaart J. A. D. Overexpression of a 9-cis-epoxycarotenoid dioxygenase gene in Nicotiana plumbaginifolia increases abscisic acid and phaseic acid levels and enhances drought tolerance. Plant Physiol. 2002;1281(1):544–551. doi: 10.1104/pp.010663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V., Ponce M., Micol J. Genetic analysis of salt-tolerant mutants in Arabidopsis thaliana. Genetics. 2000;1541(1):421–436. doi: 10.1093/genetics/154.1.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero F. J., Garciadeblas B., Rodriguez-Navarro A. The SAL1 gene of Arabidopsis, encoding an enzyme with 3′(2′), 5′-bisphosphate nucleotidase and inositol polyphosphate 1-phosphatase activities, increases salt tolerance in yeast. Plant Cell. 1996;81(1):529–537. doi: 10.1105/tpc.8.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin I., Ladyman J. A. R. Isolation and characterization of a barley mutant with abscisic-acid insensitive stomata. Planta. 1988;1731(1):73–78. doi: 10.1007/BF00394490. [DOI] [PubMed] [Google Scholar]
- Raz V., Bergervoet J., Koornneef M. Sequential steps for developmental arrest in Arabidopsis seeds. Development. 2001;1281(1):243–252. doi: 10.1242/dev.128.2.243. [DOI] [PubMed] [Google Scholar]
- Riechmann J., Heard L. J., Martin G., Reuber L., Jiang C-Z., Keddie J., Adam L., Pineda O., Ratcliffe O. J., Samaha R. R., Creelman R., Pilgrim M., Broun P., Zhang J. Z., Ghandehari D., Sherman B. K., Yu G-L. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science. 2000;2901(1):2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- Ritchie S., Gilroy S. Abscisic acid signal transduction in the barley aleurone is mediated by phospholipase D activity. Proc. Natl. Acad. Sci. USA. 1998;951(1):2697–2702. doi: 10.1073/pnas.95.5.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie S., Gilroy S. Abscisic acid stimulation of phospholipase D in the barley aleurone is G-protein-mediated and localized to the plasma membrane. Plant Physiol. 2000;1241(1):693–702. doi: 10.1104/pp.124.2.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson D. S. Genetics. 1961;461(1):649–662. doi: 10.1093/genetics/46.6.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson C. K., Hill S. A. Altered resource allocation during seed development in Arabidopsis caused by the abi3 mutation. Plant Cell Envir. 1999;221(1):117–123. [Google Scholar]
- Rock C. D. Biochemical genetics of abscisic acid biosynthesis. 1991. Ph.D. thesis. Michigan State University, East Lansing, MI, USA.
- Rock C. Pathways to abscisic acid-regulated gene expression. New Phytol. 2000;1481(1):357–396. doi: 10.1046/j.1469-8137.2000.00769.x. [DOI] [PubMed] [Google Scholar]
- Rock C. D., Heath T. G., Gage D. A., Zeevaart J. A. D. Abscisic alcohol is an intermediate in abscisic acid biosynthesis in a shunt pathway from abscisic aldehyde. Plant Physiol. 1991;971(1):670–676. doi: 10.1104/pp.97.2.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock C. D., Bowlby N. R., Hoffmann-Benning S., Zeevaart J. A. D. The aba mutant of Arabidopsis thaliana (L.) Heynh. has reduced chlorophyll fluorescence yields and reduced thylakoid stacking. Plant Physiol. 1992a;1001(1):1796–1801. doi: 10.1104/pp.100.4.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock C. D., Heath T. G., Zeevaart J. A. D. 2-Trans-abscisic acid biosynthesis and metabolism of ABA-aldehyde and xanthoxin in wild type and the aba mutant of Arabidopsis thaliana. J. Exp. Bot. 1992b;431(1):249–256. [Google Scholar]
- Rock C., Ng P. Dominant Wilty mutants of Zea mays (Poaceae) are not impaired in abscisic acid perception or metabolism. Am. J. Bot. 1999;861(1):1796–1800. [PubMed] [Google Scholar]
- Rock C. D., Zeevaart J. A. D. Abscisic (ABA)-aldehyde is a precursor to, and 1′-,4′-trans-ABA-diol a catabolite of, ABA in apple. Plant Physiol. 1990;931(1):915–923. doi: 10.1104/pp.93.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock C. D., Zeevaart J. A. D. The aba mutant of Arabidopsis thaliana is impaired in epoxy-carotenoid biosynthesis. Proc. Natl. Acad. Sci. USA. 1991;881(1):7496–7499. doi: 10.1073/pnas.88.17.7496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez P., Benning G., Grill E. ABI2, a second protein phosphatase 2C involved in abscisic acid signal transduction in Arabidopsis. FEBS Letters. 1998a;4211(1):185–190. doi: 10.1016/s0014-5793(97)01558-5. [DOI] [PubMed] [Google Scholar]
- Rodriguez P., Leube M., Grill E. Molecular cloning in Arabidopsis thaliana of a new protein phosphatase 2C (PP2C) with homology to ABI1 and ABI2. Plant Mol. Biol. 1998b;381(1):879–883. doi: 10.1023/a:1006012218704. [DOI] [PubMed] [Google Scholar]
- Rohde A., Van Montagu M., Boerjan W. The ABSCISIC ACID-INSENSITIVE 3 (ABI3) gene is expressed during vegetative quiescence processes in Arabidopsis. Plant Cell Envir. 1999;221(1):261–270. [Google Scholar]
- Rohde A., De Rycke R., Beeckman T., Engler G., Van Montagu M., Boerjan W. ABI3 affects plastid differentiation in dark-grown Arabidopsis seedlings. Plant Cell. 2000a;121(1):35–52. doi: 10.1105/tpc.12.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde A., Kurup S., Holdsworth M. ABI3 emerges from the seed. Trends Plant Sci. 2000b;51(1):418–419. doi: 10.1016/s1360-1385(00)01736-2. [DOI] [PubMed] [Google Scholar]
- Rohdich F., Wungsintaweekul J., Fellermeier M., Sagner S., Herz S., Kis K., Eisenreich W., Bacher A., Zenk M. H. Cytidine 5′-triphosphate-dependent biosynthesis of isoprenoids: YgbP protein of Escherichia coli catalyzes the formation of 4-diphosphocytidyl-2-C-methylerythritol. Proc. Natl. Acad. Sci. USA. 1999;961(1):11758–11763. doi: 10.1073/pnas.96.21.11758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohdich F., Wungsintaweekul J., Eisenreich W., Richter G., Schuhr C. A., Hecht S., Zenk M. H., Bacher A. Biosynthesis of terpenoids: 4-diphosphocytidyl-2C-methyl-D-erythritol synthase of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2000;971(1):6451–6456. doi: 10.1073/pnas.97.12.6451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronen G., Carmel-Goren L., Zamir D., Hirschberg J. An alternative pathway to β-carotene formation in plant chromoplasts discovered by map-based cloning of Beta and old-gold color mutations in tomato. Proc. Natnl. Acad. Sci. USA. 2000;971(1):11102–11107. doi: 10.1073/pnas.190177497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook F., Corke F., Card R., Munz G., Smith C., Bevan M. W. Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic acid signalling. Plant J. 2001;261(1):421–433. doi: 10.1046/j.1365-313x.2001.2641043.x. [DOI] [PubMed] [Google Scholar]
- Russell L., Larner V., Kurup S., Bougourd S., Holdsworth M. The Arabidopsis COMATOSE locus regulates germination potential. Development. 2000;1271(1):3759–3767. doi: 10.1242/dev.127.17.3759. [DOI] [PubMed] [Google Scholar]
- Sagi M., Fluhr R., Lips S. H. Aldehyde oxidase and xanthine dehydrogenase in a flacca tomato mutant with deficient abscisic acid and wilty phenotype. Plant Physiol. 1999;1201(1):571–577. doi: 10.1104/pp.120.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez J-P., Chua N-H. Arabidopsis PLC1 is required for secondary responses to abscisic acid signals. Plant Cell. 2001;131(1):1143–1154. doi: 10.1105/tpc.13.5.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang Y., Zheng S., Li W., Huang B., Wang X. Regulation of plant water loss by manipulating the expression of phospholipase Da. Plant J. 2001;281(1):135–144. doi: 10.1046/j.1365-313x.2001.01138.x. [DOI] [PubMed] [Google Scholar]
- Schnall J., Quatrano R. Abscisic acid elicits the water-stress response in root hairs of Arabidopsis thaliana. Plant Physiol. 1992;1001(1):216–218. doi: 10.1104/pp.100.1.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder J., Allen G., Hugouvieux V., Kwak J., Waner D. Guard Cell Signal Transduction. Ann. Rev. Plant Phys. Plant Mol. Biol. 2001;521(1):627–658. doi: 10.1146/annurev.arplant.52.1.627. [DOI] [PubMed] [Google Scholar]
- Schultz T., Medina J., Hill A., Quatrano R. 14-3-3 proteins are part of an abscisic acid-VIVIPAROUS1 (VP1) response complex in the Em promoter and interact with VP1 and EmBP1. Plant Cell. 1998;101(1):837–847. doi: 10.1105/tpc.10.5.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz T. F., Quatrano R. Surface perception of ABA by rice cells assayed by Em gene expression. Plant Sci. 1997;1301(1):63–71. [Google Scholar]
- Schwartz A., Wu W-H., Tucker E. B., Assmann S. M. Inhibition of inward K+ channels and stomatal response by abscisic acid: an intracellular locus of phytohormone action. Proc. Natl. Acad. Sci. USA. 1994;911(1):4019–4023. doi: 10.1073/pnas.91.9.4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S. H., Léon-Kloosterziel K. M., Koornneef M., Zeevaart J. A. D. Biochemical characterization of the aba2 and aba3 mutants in Arabidopsis thaliana. Plant Physiol. 1997;1141(1):161–166. doi: 10.1104/pp.114.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S. H., Tan B. C., Gage D. A., Zeevaart J. A. D., McCarty D. R. Specific oxidative cleavage of carotenoids by VP14 of maize. Science. 1997;2761(1):1872–1874. doi: 10.1126/science.276.5320.1872. [DOI] [PubMed] [Google Scholar]
- Schwartz S. H., Qin X. Q., Zeevaart J. A. D. Characterization of a novel carotenoid cleavage dioxygenase from plants. J. Biol. Chem. 2001;2761(1):25208–25211. doi: 10.1074/jbc.M102146200. [DOI] [PubMed] [Google Scholar]
- Schwender J., Muller C., Zeidler J., Lichtenthaler H. K. Cloning and heterologous expression of a cDNA encoding 1-deoxy-D-xylulose-5-phosphate reductoisomerase of Arabidopsis thaliana. FEBS Lett. 1999;4551(1):140–144. doi: 10.1016/s0014-5793(99)00849-2. [DOI] [PubMed] [Google Scholar]
- Scolnik P. A., Bartley G. E. Phytoene desaturase from Arabidopsis. Plant Physiol. 1993;1031(1):1475. doi: 10.1104/pp.103.4.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scolnik P. A., Bartley G. E. Nucleotide sequence of an Arabidopsis cDNA for geranylgeranyl pyrophosphate synthase. Plant Physiol. 1994a;1041(1):1469–1470. doi: 10.1104/pp.104.4.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scolnik P. A., Bartley G. E. Nucleotide sequence of an Arabidopsis cDNA for phytoene synthase. Plant Physiol. 1994b;1041(1):1471–1472. doi: 10.1104/pp.104.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M., Narusaka M., Ishida J., Nanjo T., Fujita M., Oono Y., Kamiya A., Makjima M., Satou M., Sakurai T. Arabidopsis encyclopedia using full-length cDNAs and its application for expression profiling under abiotic stress conditions. 2001. Paper presented at: Twelfth International Conference on Arabidopsis Research (Madison, WI) [DOI] [PubMed]
- Sekimoto H., Seo M., Kawakami N., Komano T., Desloire S., Liotenberg S., Marion-Poll A., Caboche M., Kamiya Y., Koshiba T. Molecular cloning and characterization of aldehyde oxidases in Arabidopsis thaliana. Plant Cell Physiol. 1998;391(1):433–442. doi: 10.1093/oxfordjournals.pcp.a029387. [DOI] [PubMed] [Google Scholar]
- Seo M., Koiwai H., Akaba S., Komano T., Oritani T., Kamiya Y., Koshiba T. Abscisic aldehyde oxidase in leaves of Arabidopsis thaliana. Plant J. 2000a;231(1):481–488. doi: 10.1046/j.1365-313x.2000.00812.x. [DOI] [PubMed] [Google Scholar]
- Seo M., Koshiba T. Complex regulation of ABA biosynthesis in plants. Trends Plant Sci. 2002;71(1):41–48. doi: 10.1016/s1360-1385(01)02187-2. [DOI] [PubMed] [Google Scholar]
- Seo M., Peeters A. J., Koiwai H., Oritani T., Marion-Poll A., Zeevaart J. A. D., Koornneef M., Kamiya Y., Koshiba T. The Arabidopsis aldehyde oxidase 3 (AAO3) gene product catalyzes the final step in abscisic acid biosynthesis in leaves. Proc. Natl. Acad. Sci. USA. 2000b;971(1):12908–12913. doi: 10.1073/pnas.220426197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp R. E., LeNoble M. E., Else M. A., Thorne E. T., Gherardi F. Endogenous ABA maintains shoot growth in tomato independently of effects on plant water balance: Evidence for an interaction with ethylene. J. Exp. Bot. 2000;511(1):1575–1584. doi: 10.1093/jexbot/51.350.1575. [DOI] [PubMed] [Google Scholar]
- Shears S. B. The versatility of inositol phosphates as cellular signals. Biochim. Biophys. Acta. 1998;14361(1):49–67. doi: 10.1016/s0005-2760(98)00131-3. [DOI] [PubMed] [Google Scholar]
- Sheen J. Ca-2+-dependent protein kinases and stress signal transduction in plants. Science. 1996;2741(1):1900–1902. doi: 10.1126/science.274.5294.1900. [DOI] [PubMed] [Google Scholar]
- Sheen J. Mutational analysis of protein phosphatase 2C involved in abscisic acid signal transduction in higher plants. Proc. Natl. Acad. Sci. USA. 1998;951(1):975–980. doi: 10.1073/pnas.95.3.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J., Zhou L., Jang J. Sugars as signaling molecules. Curr. Opin. Plant Biol. 1999;21(1):410–418. doi: 10.1016/s1369-5266(99)00014-x. [DOI] [PubMed] [Google Scholar]
- Shinozaki K., Yamaguchi-Shinozaki K. Molecular responses to dehydration and low temperature: Differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 2000;31(1):217–223. [PubMed] [Google Scholar]
- Shripathi V., Swamy G. S., Chandrasekhar K. S. Microviscosity of cucumber (Cucumis sativus L.) fruit protoplast membranes is altered by triacontanol and abscisic acid. Biochim. Biophys. Acta. 1997;13231(1):263–271. doi: 10.1016/s0005-2736(96)00193-9. [DOI] [PubMed] [Google Scholar]
- Signora L., Smet I., Foyer C., Zhang H. ABA plays a central role in mediating the regulatory effects of nitrate on root branching in Arabidopsis. Plant J. 2001;281(1):655–662. doi: 10.1046/j.1365-313x.2001.01185.x. [DOI] [PubMed] [Google Scholar]
- Sindhu R. K., Griffin D. H., Walton D. C. Abscisic aldehyde is an intermediate in the enzymatic conversion of xanthoxin to abscisic acid in Phaseolus vulgaris L. leaves. Plant Physiol. 1990;931(1):689–694. doi: 10.1104/pp.93.2.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderman E., Mattsson J., Engstrom P. The Arabidopsis homeobox gene ATHB-7 is induced by water deficit and by abscisic acid. Plant J. 1996;101(1):375–381. doi: 10.1046/j.1365-313x.1996.10020375.x. [DOI] [PubMed] [Google Scholar]
- Söderman E., Hjellstrom M., Fahleson J., Engstrom P. The HD-Zip gene ATHB6 in Arabidopsis is expressed in developing leaves, roots and carpels and up-regulated by water deficit conditions. Plant Mol. Biol. 1999;401(1):1073–1083. doi: 10.1023/a:1006267013170. [DOI] [PubMed] [Google Scholar]
- Söderman E., Brocard I., Lynch T., Finkelstein R. Regulation and function of the Arabidopsis ABA-insensitive4 (ABI4) gene in seed and ABA response signaling networks. Plant Physiol. 2000;1241(1):1752–1765. doi: 10.1104/pp.124.4.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spollen W. G., LeNoble M. E., Samuels T. D., Bernstein N., Sharp R. E. Abscisic acid accumulation maintains maize primary root elongation at low water potentials by restricting ethylene production. Plant Physiol. 2000;1221(1):967–976. doi: 10.1104/pp.122.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P., Su W., Howell S. Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl. Acad. Sci. USA. 1992;891(1):6837–6840. doi: 10.1073/pnas.89.15.6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steber C., McCourt P. A role for brassinosteroids in germination in Arabidopsis. Plant Physiol. 2001;1251(1):763–769. doi: 10.1104/pp.125.2.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steber C. M., Cooney S. E., McCourt P. Isolation of the GA-response mutant sly1 as a suppressor of ABI1-1 in Arabidopsis thaliana. Genetics. 1998;1491(1):509–521. doi: 10.1093/genetics/149.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S. L., Kwong L. W., Yee K. M., Pelletier J., Lepiniec L., Fischer R. L., Goldberg R. B., Harada J. J. LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. USA. 2001;981(1):11806–11811. doi: 10.1073/pnas.201413498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S., Rajagopal B., Rock C. D. Harlequin (hlq) and short blue root (sbr), two Arabidopsis mutants that ectopically express an ABA- and auxin-inducible transgenic carrot promoter and have pleiotropic effects on morphogenesis. Plant Mol. Biol. 2002;491(1):93–105. doi: 10.1023/a:1014472417150. [DOI] [PubMed] [Google Scholar]
- Sun Z., Gantt E., Cunningham F. X., Jr. Cloning and functional analysis of the beta-carotene hydroxylase of Arabidopsis thaliana. J. Biol. Chem. 1996;2711(1):24349–24352. doi: 10.1074/jbc.271.40.24349. [DOI] [PubMed] [Google Scholar]
- Sutton F., Paul S. S., Wang X. Q., Assmann S. M. Distinct abscisic acid signaling pathways for modulation of guard cell versus mesophyll cell potassium channels revealed by expression studies in Xenopus laevis oocytes. Plant Physiol. 2000;1241(1):223–230. doi: 10.1104/pp.124.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M., Kao C., McCarty D. The conserved B3 domain of VIVIPAROUS1 has a cooperative DNA binding activity. Plant Cell. 1997;91(1):799–807. doi: 10.1105/tpc.9.5.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M., Kao C-Y., Cocciolone S., McCarty D. R. Maize VP1 complements Arabidopsis abi3 and confers a novel ABA/auxin interaction in roots. Plant J. 2001;281(1):409–418. doi: 10.1046/j.1365-313x.2001.01165.x. [DOI] [PubMed] [Google Scholar]
- Swire-Clark G. A., Marcotte W. R. The wheat LEA protein Em functions as an osmoprotective molecule in Saccharomyces cerevisiae. Plant Mol. Biol. 1999;391(1):117–128. doi: 10.1023/a:1006106906345. [DOI] [PubMed] [Google Scholar]
- Takahashi S., Katagiri T., Yamaguchi-Shinozaki K., Shinozaki K. An Arabidopsis gene encoding a Ca2+ binding protein is induced by abscisic acid during dehydration. Plant Cell Physiol. 2000;411(1):898–903. doi: 10.1093/pcp/pcd010. [DOI] [PubMed] [Google Scholar]
- Takahashi S., Katagiri T., Hirayama T., Yamaguchi-Shinozaki K., Shinozaki K. Hyperosmotic stress induces a rapid and transient increase in inositol 1,4,5-triphosphate independent of abscisic acid in Arabidopsis cell culture. Plant Cell Physiol. 2001;421(1):214–222. doi: 10.1093/pcp/pce028. [DOI] [PubMed] [Google Scholar]
- Tan B. C., Cline K., McCarty D. R. Localization and targeting of the VP14 epoxy-carotenoid dioxygenase to chloroplast membranes. Plant J. 2001;271(1):373–382. doi: 10.1046/j.1365-313x.2001.01102.x. [DOI] [PubMed] [Google Scholar]
- Tan B. C., Schwartz S. H., Zeevaart J. A. D., McCarty D. R. Genetic control of abscisic acid biosynthesis in maize. Proc. Natl. Acad. Sci. USA. 1997;941(1):12235–12240. doi: 10.1073/pnas.94.22.12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor I. B., Linforth R. S. T., Al-Naieb R. J., Bowman W. R., Marples B. A. The wilty mutants flacca and sitiens are impaired in the oxidation of ABA-aldehyde to ABA. Plant Cell Environ. 1988;111(1):739–745. [Google Scholar]
- Thomashow M. F. Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. 1999;1(1):571–599. doi: 10.1146/annurev.arplant.50.1.571. In Annual Review of Plant Physiology and Plant Molecular Biology, R. L. Jones, ed (Palo Alto, California: Annual Reviews Inc.), pp. [DOI] [PubMed] [Google Scholar]
- Thomashow M. F. So what's new in the field of plant cold acclimation? Lots!. Plant Physiol. 2001;1251(1):89–93. doi: 10.1104/pp.125.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A. J., Jackson A. C., Parker R. A., Morpeth D. R., Burbidge A., Taylor I. B. Abscisic acid biosynthesis in tomato: regulation of zeaxanthin epoxidase and 9-cis-epoxycarotenoid dioxygenase mRNAs by light/dark cycles, water stress and abscisic acid. Plant Mol. Biol. 2000a;421(1):833–845. doi: 10.1023/a:1006448428401. [DOI] [PubMed] [Google Scholar]
- Thompson A. J., Jackson A. C., Symonds R. C., Mulholland B. J., Dadswell A. R., Blake P. S., Burbidge A., Taylor I. B. Ectopic expression of a tomato 9-cis-epoxycarotenoid dioxygenase gene causes over-production of abscisic acid. Plant J. 2000b;231(1):363–374. doi: 10.1046/j.1365-313x.2000.00789.x. [DOI] [PubMed] [Google Scholar]
- Toorop P. E., Bewley J. D., Abrams S. R., Hilhorst H. W. M. Structure-activity studies with ABA analogs on germination and endo-b-mannanase activity in tomato and lettuce seeds. J. Plant Physiol. 1999;1541(1):679–685. [Google Scholar]
- Ueguchi C., Sato S., Kato T., Tabata S. The AHK4 gene involved in the cytokinin-signaling pathway as a direct recptor molecule in Arabidopsis thaliana. Plant Cell Physiol. 2001;421(1):751–755. doi: 10.1093/pcp/pce094. [DOI] [PubMed] [Google Scholar]
- Ullah H., Chen J-G., Young J., Im K-H., Sussman M., Jones A. Modulation of cell proliferation by heterotrimeric G protein in Arabidopsis. Science. 2001;2921(1):2066–2069. doi: 10.1126/science.1059040. [DOI] [PubMed] [Google Scholar]
- Ulmasov T., Hagen G., Guilfoyle T. J. Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. USA. 1999;961(1):5844–5849. doi: 10.1073/pnas.96.10.5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno Y., Furihata T., Abe H., Yoshida R., Shinozaki K., Yamaguchi-Shinozaki K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. USA. 2000;971(1):11632–11637. doi: 10.1073/pnas.190309197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urao T., Yakubov B., Satoh R., Yamaguchi-Shinozaki K., Seki M., Hirayama T., Shinozaki K. A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. Plant Cell. 1999;111(1):1743–1754. doi: 10.1105/tpc.11.9.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urao T., Yamaguchi-Shinozaki K., Urao S., Shinozaki K. An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell. 1993;51(1):1529–1539. doi: 10.1105/tpc.5.11.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartanian N., Marcotte L., Giraudat J. Drought rhizogenesis in Arabidopsis thaliana. Plant Physiology. 1994;1041(1):761–767. doi: 10.1104/pp.104.2.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasil V., Marcotte W. R. J., Rosenkrans L., Cocciolone S. M., Vasil I. K., Quatrano R. S., McCarty D. R. Overlap of viviparous 1 (VP1) and abscisic acid response elements in the Em promoter: G-Box elements are sufficient but not necessary for VP1 transactivation. Plant Cell. 1995;71(1):1511–1518. doi: 10.1105/tpc.7.9.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan G. T., Millborrow B. V. The stability of the 1′-4′-diols of abscisic acid. Phytochem. 1988;271(1):339–343. [Google Scholar]
- Veau B., Courtois M., Oudin A., Chenieux J. C., Rideau M., Clastre M. Cloning and expression of cDNAs encoding two enzymes of the MEP pathway in Catharanthus roseus. Biochim. Biophys. Acta. 2000;15171(1):159–163. doi: 10.1016/s0167-4781(00)00240-2. [DOI] [PubMed] [Google Scholar]
- Walker-Simmons M. K., Holappa L. D., Abrams G. D., Abrams S. R. ABA metabolites induce group 3 LEA mRNA and inhibit germination in wheat. Physiol. Plant. 1997;1001(1):474–480. [Google Scholar]
- Walker-Simmons M., Kudrna D. A., Warner R. L. Reduced accumulation of ABA during water stress in a molybdenum cofactor mutant of barley. Plant Physiol. 1989;901(1):728–733. doi: 10.1104/pp.90.2.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Qi Q., Schorr P., Cutler A. J., Crosby W., Fowke L. C. ICK1, a cyclin-dependent protein kinase inhibitor from Arabidopsis thaliana interacts with both Cdc2a and CycD3, and its expression is induced by abscisic acid. Plant J. 1998;151(1):501–510. doi: 10.1046/j.1365-313x.1998.00231.x. [DOI] [PubMed] [Google Scholar]
- Wang M., Heimovaara-Dijkstra S., Van der Meulen R. M., Knox J. P., Neill S. J. The monoclonal antibody JIM19 modulates abscisic acid action in barley aleurone protoplasts. Planta. 1995;1961(1):271–276. [Google Scholar]
- Wang X., Wang C., Sang Y., Zheng L., Qin C. Determining functions of multiple phospholipase Ds in stress response of Arabidopsis. Biochem. Soc. Trans. 2000;281(1):813–816. [PubMed] [Google Scholar]
- Wang X-Q., Ullah H., Jones A., Assmann S. G protein regulation of ion channels and abscisic acid signaling in Arabidopsis guard cells. Science. 2001;2921(1):2070–2072. doi: 10.1126/science.1059046. [DOI] [PubMed] [Google Scholar]
- Watillon B., Kettmann R., Arrdouani A., Hecquet J. F., Boxus P., Burny A. Apple messenger RNAs related to bacterial lignostilbene dioxygenase and plant SAUR genes are preferentially expressed in flowers. Plant Mol. Biol. 1998;361(1):909–915. doi: 10.1023/a:1005914506110. [DOI] [PubMed] [Google Scholar]
- Webb A. A. R., Larman M. G., Montgomery L. T., Taylor J. E., Hetherington A. M. The role of calcium in ABA-induced gene expression and stomatal movements. Plant J. 2001;261(1):351–362. doi: 10.1046/j.1365-313x.2001.01032.x. [DOI] [PubMed] [Google Scholar]
- West M., Yee K., Danao J., Zimmerman J., Fischer R., Goldberg R., Harada J. LEAFY COTYLEDON1 is an essential regulator of late embryogenesis and cotyledon identity in Arabidopsis. Plant Cell. 1994;61(1):1731–1745. doi: 10.1105/tpc.6.12.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A., Pickett F., Turner J., Estelle M. A dominant mutation in Arabidopsis confers resistance to auxin, ethylene and abscisic acid. Mol. Gen. Genet. 1990;2221(1):377–383. doi: 10.1007/BF00633843. [DOI] [PubMed] [Google Scholar]
- Windsor M. L., Milborrow B. V., Abrams S. R. Stereochemical requirements of the saturable uptake carrier for abscisic acid in carrot suspension culture cells. J. Exp. Bot. 1994;451(1):227–233. [Google Scholar]
- Windsor M. L., Zeevaart J. A. D. Induction of ABA 8′-hydroxylase by (+)-S-, (−)-R- and 8′,8′,8′-trifluoro-S-abscisic acid in suspension cultures of potato and Arabidopsis. Phytochem. 1997;451(1):931–934. doi: 10.1016/s0031-9422(97)00022-8. [DOI] [PubMed] [Google Scholar]
- Wolkers W. F., Alberda M., Koornneef M., Leon-Kloosterziel K. M., Hoekstra F. A. Properties of proteins and the glassy matrix in maturation-defective mutants seeds of Arabidopsis thaliana. Plant J. 1998;161(1):133–143. doi: 10.1046/j.1365-313x.1998.00277.x. [DOI] [PubMed] [Google Scholar]
- Wu D., Wright D. A., Wetzel C., Voytas D. F., Rodermel S. The IMMUTANS variegation locus of Arabidopsis defines a mitochondrial alternative oxidase homolog that functions during early chloroplast biogenesis. Plant Cell. 1999;111(1):43–56. doi: 10.1105/tpc.11.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Kuzma J., Marechal E., Graeff R., Lee H. C., Foster R., Chua N-H. Abscisic acid signaling through cyclic ADP-ribose in plants. Science. 1997;2781(1):2126–2130. doi: 10.1126/science.278.5346.2126. [DOI] [PubMed] [Google Scholar]
- Wright S. T. C., Hiron R. W. P. (+)-Abscisic acid, thee growth inhibitor induced in detached wheat leaves by a period of wilting. Nature. 1969;2241(1):719–720. [Google Scholar]
- Xiong L., Gong Z., Rock C. D., Subramanian S., Guo Y., Xu W., Galbraith D., Zhu J-K. Modulation of abscisic acid signal transduction and biosynthesis by an Sm-like protein in Arabidopsis. Devel. Cell. 2001a;11(1):771–781. doi: 10.1016/s1534-5807(01)00087-9. [DOI] [PubMed] [Google Scholar]
- Xiong L., Ishitani M., Lee H., Zhu J-K. HOS5 - a negative regulator of osmotic stress-induced gene expression in Arabidopsis thaliana. Plant J. 1999;191(1):569–578. doi: 10.1046/j.1365-313x.1999.00558.x. [DOI] [PubMed] [Google Scholar]
- Xiong L., Ishitani M., Zhu J-K. Interaction of osmotic stress, temperature, and abscisic acid in the regulation of gene expression in Arabidopsis. Plant Physiol. 1999;1191(1):205–211. doi: 10.1104/pp.119.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L., Ishitani M., Lee H., Zhang C., Zhu J-K. FIERY1 encoding an inositol polyphosphate 1-phosphatase is a negative regulator of abscisic acid and stress signaling in Arabidopsis. Genes Dev. 2001b;151(1):1971–1984. doi: 10.1101/gad.891901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L., Ishitani M., Lee H., Zhu J-K. The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stess-responsive gene expression. Plant Cell. 2001c;131(1):2063–2083. doi: 10.1105/TPC.010101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L., Zhu J-K. Abiotic stress signal transduction in plants: Molecular and genetic perspectives. Physiol. Plant. 2001;1121(1):152–166. doi: 10.1034/j.1399-3054.2001.1120202.x. [DOI] [PubMed] [Google Scholar]
- Yamamoto H., Oritani T. Stereoselectivity in the biosynthetic conversion of xanthoxin into abscisic acid. Planta. 1996;2001(1):319–325. [Google Scholar]
- Yang Z. Small GTPases: Versatile Signaling Switches in Plants. Plant Cell. 2002;141(1):S375–388. doi: 10.1105/tpc.001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart J. A. D. Abscisic acid metabolism and its regulation. 1999;1(1):189–207. In: Hooykaas, P.J.J., Hall, M.A., Libbenga, K.R., eds. Biochemistry and Molecular Biology of Plant Hormones. Amsterdam: Elsevier Science. [Google Scholar]
- Zeevaart J. A. D., Boyer G. L. Accumulation and transport of abscisic acid and its metabolites in Ricinus and Xanthium. Plant Physiol. 1984;741(1):934–939. doi: 10.1104/pp.74.4.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart J. A. D., Creelman R. A. Metabolism and physiology of abscisic acid. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1988;391(1):439–473. [Google Scholar]
- Zeevaart J., Creelman R. Metabolism and physiology of abscisic acid. Ann. Rev. Plant Physiol. Plant Mol. Biol. 1988;391(1):439–473. [Google Scholar]
- Zhang D. P., Chen S. W., Peng Y. B., Shen Y. Y. Abscisic acid-specific binding sites in the flesh of developing apple fruit. J. Exp. Bot. 2001;521(1):2097–2103. doi: 10.1093/jexbot/52.364.2097. [DOI] [PubMed] [Google Scholar]
- Zhang D. P., Wu Z-Y., Li X-Y., Zhao Z-X. Purification and identification of a 42-kilodalton abscisic acid-specific-binding protein from epidermis of broad bean leaves. Plant Physiol. 2002;1281(1):714–725. doi: 10.1104/pp.010531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z., Jin Z., Zhou X., Xia K., Ma C. An ABA-binding protein with nucleic acid-binding property. Science in China C. 1998;411(1):209–216. doi: 10.1007/BF02882729. [DOI] [PubMed] [Google Scholar]
- Zhou L., Jang J-C., Jones T. L., Sheen J. Glucose and ethylene signal transduction crosstalk revealed by an Arabidopsis glucose-insensitive mutant. Proc. Natl. Acad. Sci. (USA) 1998;951(1):10294–10299. doi: 10.1073/pnas.95.17.10294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegelhoffer E., Medrano L., Meyerowitz E. Cloning of the Arabidopsis WIGGUM gene identifies a role for farnesylation in meristem development. Proc. Natl. Acad. Sci. USA. 2000;971(1):7633–7638. doi: 10.1073/pnas.130189397. [DOI] [PMC free article] [PubMed] [Google Scholar]


