Abstract
We investigated seed bugs of the genus Nysius (Insecta: Hemiptera: Lygaeidae) for their symbiotic bacteria. From all the samples representing 4 species, 18 populations and 281 individuals, specific bacterial 16S rRNA gene sequences were consistently identified, which formed a distinct clade in the Gammaproteobacteria. In situ hybridization showed that the bacterium was endocellularly localized in a pair of large bacteriomes that were amorphous in shape, deep red in color, and in association with gonads. In the ovary of adult females, the endosymbiont was also localized in the ‘infection zone' in the middle of each germarium and in the ‘symbiont ball' at the anterior pole of each oocyte, indicating vertical transmission of the endosymbiont through the ovarial passage. Phylogenetic analyses based on bacterial 16S rRNA, groEL and gyrB genes consistently supported a coherent monophyly of the Nysius endosymbionts. The possibility of a sister relationship to ‘Candidatus Kleidoceria schneideri', the bacteriome-associated endosymbiont of a lygaeid bug Kleidocerys resedae, was statistically rejected, indicating independent evolutionary origins of the endosymbionts in the Lygaeidae. The endosymbiont genes consistently exhibited AT-biased nucleotide compositions and accelerated rates of molecular evolution, and the endosymbiont genome was only 0.6 Mb in size. The endosymbiont phylogeny was congruent with the host insect phylogeny, suggesting strict vertical transmission and host–symbiont co-speciation over evolutionary time. Based on these results, we discuss the evolution of bacteriomes and endosymbionts in the Heteroptera, most members of which are associated with gut symbiotic bacteria. The designation ‘Candidatus Schneideria nysicola' is proposed for the endosymbiont clade.
Keywords: stinkbug, Nysius, Lygaeidae, bacteriome, endosymbiont
Introduction
Symbiotic bacteria are universally associated with a diverse array of insects (Buchner, 1965; Bourtzis and Miller, 2003). Among them, the most intimate mutualistic associations are found in obligate endosymbionts like Buchnera in aphids and Wigglesworthia in tsetse flies, wherein the symbiotic bacteria are harbored in specialized cells called bacteriocytes. In the body of these insects, the bacteriocytes often form a symbiotic organ called bacteriome, where the endosymbionts play their biological roles such as provisioning of essential nutrients (Douglas, 1998; Baumann, 2005). Early histological studies described the formation processes of the bacteriocytes and the bacteriome in aphids, tsetse flies, lice and many other insects (reviewed in Buchner, 1965). However, recent studies using modern molecular and cytological techniques are scarce, except for those on the development of bacteriocytes in the pea aphid Acyrthosiphon pisum (Braendle et al., 2003; Miura et al., 2003). The evolutionary origin of the novel organ for symbiosis is unknown.
The suborder Heteroptera is a group of insects with a sucking mouthpart, the so-called true bugs or stinkbugs. The infraorder Pentatomomorpha is placed under the Heteroptera, and contains over 12 500 species that are grouped into five superfamilies (Schuh and Slater, 1995; Henry, 1997). Besides relatively minor predacious and mycophagous species, the majority of them are plant sap feeders, and most of them possess a number of sac- or tube-like outgrowths, called crypts or caeca, in a posterior region of the midgut, whose cavity harbors specific bacterial symbionts (Buchner, 1965; Kikuchi et al., 2008). In the superfamily Pentatomoidea, their gut symbionts belong to distinct lineages of the Gammaproteobacteria, and are vertically transmitted through host generations (Fukatsu and Hosokawa, 2002; Prado et al., 2006; Hosokawa et al., 2006, 2010; Kikuchi et al., 2009; Prado and Almeida, 2009a, 2009b; Kaiwa et al., 2010, 2011). On the other hand, in the superfamilies Lygaeoidea and Coreoidea, their gut symbionts belong to the β-proteobacterial genus Burkholderia, and are acquired from the environmental bacterial pool every host generation (Kikuchi et al., 2005, 2007, 2011). In summary, extracellular symbiotic associations with specific bacteria in midgut crypts are regarded as the default symbiotic system generally found among diverse stinkbugs.
However, Schneider (1940) described a few lygaeoid stinkbugs that exceptionally host endosymbiotic bacteria within specialized bacteriomes. Nysius punctipennis, N. senecionis, N. thymi and N. lineolatus (Lygaeidae: Orsillinae) possessed paired red bacteriomes associated with the gonads, Kleidocerys (=Ischnorrhynchus) resedae and K. ericae (Lygaeidae: Ischnorrhynchinae) were with an unpaired bacteriome associated with the midgut and Ischnodemus sabuleti (Blissidae) was with a pair of bacteriomes in the fat body. Considering that most of the other lygaeoid groups are consistently associated with extracellular Burkholderia symbionts in the midgut crypts (Kikuchi et al., 2011), the exceptional lygaeoid stinkbugs harboring the bacteriome-associated endosymbiont are of great interest. These lygaeoid stinkbugs would provide novel insights into the evolutionary origin of the symbiotic organ.
Since the pioneer work by Schneider (1940), neither the bacteriomes nor the endosymbionts of the lygaeoid stinkbugs have been investigated. Recently, the endosymbiont of K. resedae was microbiologically characterized and designated as ‘Candidatus Kleidoceria schneideri' (Küchler et al., 2010). Here we report a series of comprehensive analyses on the bacteriome-associated endosymbionts of another lygaeoid group, seed bugs of the genus Nysius representing 4 species, 18 populations and 281 individuals, using multidisciplinary approaches from morphology, histology, PCR diagnosis, molecular phylogenetic and evolutionary analyses, in situ hybridization, electron microscopy and pulsed-field gel electrophoresis.
Materials and methods
Insect materials
Table 1 lists insect samples of Nysius and other lygaeid species used in this study. Most of the samples were preserved in acetone upon collection for DNA analyses (Fukatsu, 1999). Some of the insects were dissected and subjected to in situ hybridization, electron microscopy or pulsed-field gel electrophoresis.
Table 1. Samples of lygaeid stinkbugs, detection of their symbionts, and accession numbers of nucleotide sequences determined in this study.
| Taxon | Localitya | Dateb | Collector | Host plant | Symbiotic organ |
Symbiont detectionc |
Sequence accession number |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Schneideriad | Burkholderia | 16S rRNA | gyrB | groEL | COI | |||||||
| Family Lygaeidae | ||||||||||||
| Subfamily Orsillinae | ||||||||||||
| Nysius plebeius | Tsukuba, Ibaraki | 11/09 /09 | YM | Conyza sumatrensis | Paired bacteriome | ♀ ♂ | 100% (37/37) 100% (26/26) | 0% (0/37) 0% (0/26) | AB576918 | AB583935 | AB583951 | AB619226 |
| Mito, Ibaraki | 10/09 /09 | YK | C. sumatrensis | ♀ ♂ | 100% (14/14) 100% (7/7) | 0% (0/14) 0% (0/7) | AB576919 | AB583937 | AB583952 | AB619227 | ||
| Fukuyama, Hiroshima | 22/09 /09 | YM | C. sumatrensis | ♀ ♂ | 100% (5/5) 100% (7/7) | 0% (0/5) 0% (0/7) | AB576920 | AB583938 | — | AB619228 | ||
| Takamatsu, Kagawa | 16/09 /09 | YK | C. sumatrensis | ♀ ♂ | 100% (2/2) | 0% (0/2) | AB576921 | AB583939 | AB583953 | AB619229 | ||
| Koshi, Kumamoto | 28/09 /09 | YK | C. sumatrensis | ♀ ♂ | 100% (3/3) 100% (3/3) | 0% (0/3) 0% (0/3) | AB576922 | AB583940 | AB583954 | AB619230 | ||
| Tokunoshima, Kagoshima | 30/09 /09 | TH | C. sumatrensis | ♀ ♂ | 100% (4/4) 100% (6/6) | 0% (0/4) 0% (0/6) | AB576923 | AB583941 | — | AB619231 | ||
| Ishigakijima, Okinawa | 09/09 /09 | TH | Chamaesyce hirta | ♀ ♂ | 100% (15/15) 100% (15/15) | 0% (0/15) 0% (0/15) | AB576924 | AB583942 | AB583955 | AB619232 | ||
| Iriomotejima, Okinawa | 10/09 /09 | TH | C. hirta | ♀ ♂ | 100% (3/3) 100% (3/3) | 0% (0/3) 0% (0/3) | AB576925 | AB583943 | AB583956 | AB619233 | ||
| Kumejima, Okinawa | 10/04 /10 | TH | Youngia sp. | ♀ ♂ | 100% (1/1) | 0% (0/1) | AB576926 | — | — | AB619234 | ||
| Nysius sp. 1 | Tsukuba, Ibaraki | 12/09 /09 | YM | C. sumatrensis | Paired bacteriome | ♀ ♂ | 100% (23/23) 100% (20/20) | 0% (0/23) 0% (0/20) | AB576927 | AB583944 | AB583957 | AB619235 |
| Mito, Ibaraki | 10/09 /09 | YK | C. sumatrensis | ♀ ♂ | 100% (11/11) 100% (4/4) | 0% (0/11) 0% (0/4) | AB576928 | AB583945 | — | AB619236 | ||
| Takamatsu, Kagawa | 16/09 /09 | YK | C. sumatrensis | ♀ ♂ | 100% (4/4) 100% (7/7) | 0% (0/4) 0% (0/7) | AB576929 | AB583946 | AB583958 | AB619237 | ||
| Shimanto, Kochi | 16/09 /09 | YK | C. sumatrensis | ♀ ♂ | 100% (3/3) 100% (4/4) | 0% (0/3) 0% (0/4) | AB576930 | AB583947 | — | AB619238 | ||
| Koshi, Kumamoto | 28/09 /09 | YK | C. sumatrensis | ♀ ♂ | 100% (2/2) 100% (2/2) | 0% (0/2) 0% (0/2) | AB576931 | AB583948 | — | AB619239 | ||
| Nysius sp. 2 | Yakushima, Kagoshima | 14/05 /10 | TH | Erigeron philadelphicus | Paired bacteriome | ♀ ♂ | 100% (2/2) 100% (8/8) | 0% (0/2) 0% (0/8) | — | — | — | AB619240 |
| Kunigami, Okinawa | 09/06 /10 | TH | E. philadelphicus | ♀ ♂ | 100% (4/4) 100% (1/1) | 0% (0/4) 0% (0/1) | — | — | — | AB619241 | ||
| Kumejima, Okinawa | 11/04 /10 | TH | Youngia sp. | ♀ ♂ | 100% (2/2) | 0% (0/2) | AB576932 | AB583949 | AB583959 | AB619242 | ||
| Nysius expressus | Sapporo, Hokkaido | 02/09 /09 | YM | E. philadelphicus | Paired bacteriome | ♀ ♂ | 100% (16/16) 100% (17/17) | 0% (0/16) 0% (0/17) | AB576933 | AB583950 | AB583960 | AB619243 |
| Subfamily Lygaeinae | ||||||||||||
| Spilostethus hospes | Ishigakijima, Okinawa | 09/09 /09 | TH | Chamaesyce hirta | Absent | Negative | Negative | AB619244 | ||||
| Oncopeltus nigriceps | Ishigakijima, Okinawa | 05/07 /02 | KK | Calotropis gigantean | Absent | Negative | Negative | AB619246 | ||||
| Graptostethus servus | Ishigakijima, Okinawa | 02/10 /10 | TH | Carissa macrocarpa | Absent | Negative | Negative | — | ||||
| Tropidothorax belongolowi | Koshi, Kumamoto | 09/27 /09 | YK | C. sumatrensis | Absent | Negative | Negative | — | ||||
| Arocatus sericans | Takashima, Shiga | 28/08 /10 | SK | (sweeping) | Absent | Negative | Negative | AB619254 | ||||
| Arocatus melanostomus | Koka, Shiga | 29/08 /10 | SK | Clematis apiifolia | Absent | Negative | Negative | AB619255 | ||||
| Subfamily | ||||||||||||
| Ishchnorhynchinae | ||||||||||||
| Kleidocerys resedae | Sapporo, Hokkaido | 09/08 /10 | YK | Betula platyphylla | A single bacteriome | Negative | Negative | AB585972 | AB619224 | AB583961 | AB619247 | |
| Okunikko, Tochigi | 10/09 /10 | YM | B. platyphylla | Negative | Negative | AB624347 | AB619225 | — | AB619249 | |||
| Family | ||||||||||||
| Rhyparochromidae (outgroup) | ||||||||||||
| Togo hemipterus | Tsukuba, Ibaraki | 17/07 /07 | YK | Digitaria ciliaris | Midgut crypts | Negative | Positive | AB619252 | ||||
Abbreviations: COI, mitochondrial cytochrome oxidase I gene; groEL, bacterial groEL gene; gyrB, bacterial gyrB gene; KK, Katsuyuki Kouno; Negative, PCR-negative amplification; Positive, PCR-positive amplification; 16S rRNA, bacterial 16S rRNA gene; SK, Shuhei Kada; YK, Yoshitomo Kikuchi; YM, Yu Matsuura; TH, Takahiro Hosokawa.
All localities are in Japan.
For example, ‘11/09/09' means 11 September 2009.
For example, ‘0% (0/37)' means 0% infection frequency of the symbiont with 0 positive insects of 37 insects examined.
The bacteriocyte-associated endosymbiont of Nysius spp.
DNA extraction, PCR, cloning, genotyping and sequencing procedures
Adult insects were dissected in a phosphate-buffered saline with fine forceps under a dissection microscope, and their bacteriomes, ovaries and midguts were isolated. Each of the insect tissues was homogenized and digested in a 1.5 ml plastic tube with a proteinase K-containing lysis buffer, extracted with phenol–chloroform and subjected to ethanol precipitation of whole nucleic acids. Bacterial 16S rRNA, groEL and gyrB genes were amplified by PCR from the DNA samples, and subjected to cloning, restriction fragment length polymorphism genotyping and sequencing as described (Kikuchi et al, 2009). Insect mitochondrial cytochrome oxidase I (COI) gene, which has been widely used for phylogenetic characterization of insects and other organisms (Hebert et al., 2003), was also amplified by PCR and subjected to direct sequencing. The primers and PCR conditions are listed in Supplementary Table S1. The primers and PCR conditions used for diagnostic PCR are also listed in Supplementary Table S1.
Molecular phylogenetic, molecular evolutionary and genomic analyses
Multiple alignments of nucleotide sequences were generated with the program Vector NTI Advance 10.3.1 (Invitrogen, Carlsbad, CA, USA). Phylogenetic analyses were conducted by maximum likelihood, maximum parsimony and neighbor-joining methods. Maximum likelihood trees were generated with the program PhyML 3.0 (Guindon and Gascuel, 2003), whereas maximum parsimony and neighbor-joining trees were constructed with the program MEGA 4 (Tamura et al., 2007). In the maximum likelihood analyses, models were chosen on the basis of the Akaike information criterion with the programs Modeltest 3.7 (Posada and Crandall, 1998) and PAUP* Version 4.0b10 (Swofford, 2001), which selected the GTR + I + G model for each of the 16S rRNA, groEL, gyrB and COI gene phylogenies. Bootstrap tests were conducted with 100 replications. Relative rate tests were performed with the program RRTree (Robinson-Rechavi and Huchon, 2000). The phylogenetic affinity between the endosymbionts of Nysius spp. and the endosymbiont of K. resedae was evaluated by an approximately unbiased test (Shimodaira, 2002) using the concatenated alignment of 16S rRNA, groEL and gyrB gene sequences as a single data set. The nucleotide sequences from 11 endosymbionts and 2 free-living bacteria were organized into 9 topological units, the log-likelihood scores for 131 135 possible trees for the topological units were calculated and the best 1000 trees were subjected to the analysis. Log-likelihood scores for all trees and site-wise log-likelihoods for the selected 1000 trees were estimated using RAxML (Stamatakis, 2006). The data were used as input for CONSEL 0.1 g (Shimodaira and Hasegawa, 2001) to calculate P-values. Pulsed-field gel electrophoresis of the endosymbiont genome was performed with the bacteriome samples dissected from 50 adult insects of N. sp. 1 as described (Kikuchi et al., 2009).
Histological procedures
Fluorescent in situ hybridization was performed with a fluorochrome-labelled oligonucleotide probe Cy3-NpSch1274 (5′-TATACTTTTTGAGGTTCGCTTGCTC-3′) that specifically targets 16S rRNA of the endosymbionts of Nysius spp. Adult insects were dissected in phosphate-buffered saline and fixed with Carnoy's solution (ethanol/chloroform/acetic acid=6:3:1) overnight, and treated with 6% hydrogen peroxide in 80% ethanol for several weeks for quenching autofluorescence of the tissues (Koga et al., 2009). Then, the tissue samples were subjected to in situ hybridization as described (Kikuchi et al., 2009). For transmission electron microscopy, adult insects of N. plebeius were dissected with fine forceps in 0.1 sodium cacodylate buffer (pH 7.4) containing 2.5% glutaraldehyde, and isolated bacteriomes and ovaries were processed into ultrathin sections and observed as described (Fukatsu et al., 2000).
Results
General observation of symbiotic organs in Nysius spp. and allied lygaeid species
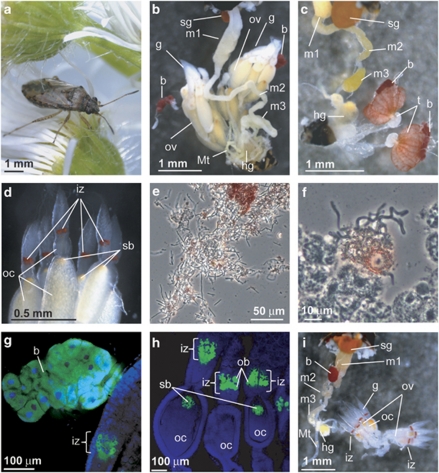
In both adult females and males of N. plebeius (Figure 1a), a pair of large bacteriomes, amorphous in shape and deep red in color, was found in association with their gonads (Figures 1b and c). Although many allied lygaeoid and coreoid species have a characteristic midgut fourth section with a number of crypts for harboring gut symbiotic bacteria (Kikuchi et al., 2011), such a midgut structure was absent in N. plebeius (Figures 1b and c). In the ovaries of adult females, a red-pigmented zone, previously designated as ‘infection zone' (Schneider, 1940), was observed in each germarium, and the anterior pole of each oocyte was marked by an orange-colored region, wherein a structure so-called ‘symbiont ball' was located (Schneider, 1940) (Figure 1d). The bacteriome contained numerous tubular bacterial cells, as did the infection zone of the ovaries (Figures 1e and f). Similar histological and symbiotic configurations were also observed in other Nysius species including N. sp. 1, N. sp. 2 and N. expressus (Supplementary Figures S1A–C). These observations agreed with the early report on European Nysius species (Schneider, 1940).
Figure 1.
(a) An adult female of Nysius plebeius. (b) A dorsal view of the dissected abdomen of an adult female of N. plebeius. (c) A dorsal view of the dissected abdomen of an adult male of N. plebeius. Note that large red bacteriomes, the symbiotic organs, are present on both sides of the gonads in both sexes. (d) An enlarged image of germaria and developing oocytes in the ovarioles of an adult female of N. plebeius. Note a red infection zone in each germarium and an orange region at the anterior pole of each oocyte, the locations of the endosymbiont in the female reproductive system. (e) A phase-contrast microscopic image of a crushed bacteriome, in which numerous tubular endosymbiont cells are observed. Note that red materials are seen outside the bacterial cells. (f) A similar image of a crushed germarium containing an infection zone. (g) Endosymbiont localization in the bacteriome visualized by in situ hybridization. (h) Endosymbiont localization in the infection zone of the germaria as well as in the symbiont ball at the anterior pole of the oocytes visualized by in situ hybridization. In (g) and (h), green and blue indicate the endosymbionts and the host insect nuclei, respectively. (i) A ventral view of the dissected abdomen of an adult female of Kleidocerys resedae collected at Sapporo, Japan. Note the presence of a single red-colored bacteriome associated with the midgut, the absence of the paired bacteriome of the Nysius type and the presence of the red-colored infection zone in each germarium. b, bacteriome; g, germarium; hg, hind gut; iz, infection zone; m1, midgut first section; m2, midgut second section; m3, midgut third section; Mt, Malpighian tubule; ob, ovarial bacteriocyte; oc, oocyte; ov, ovariole; sb, symbiont ball; sg, stink gland.
In European Kleidocerys species, namely K. resedae and K. ericae, an unpaired, red-colored bacteriome has been identified in association with the midgut (Schneider, 1940; Küchler et al., 2010). We collected samples of K. resedae from two distant populations in Japan, Sapporo and Okunikko, and confirmed the same symbiotic architecture: a reddish bacteriome was associated with the midgut, whereas no midgut fourth section with crypts was found (Figure 1i).
In the family Lygaeidae, the subfamily Orsillinae, including the genus Nysius, the subfamily Ischnorrhynchinae, including the genus Kleidocerys, and the subfamily Lygaeinae constitute a well-defined clade (Henry, 1997). However, histological inspection of lygaeine species revealed neither bacteriomes nor midgut fourth section with crypts (Supplementary Figures S1D–F).
Electron microscopy of the symbiotic organs of N. plebeius
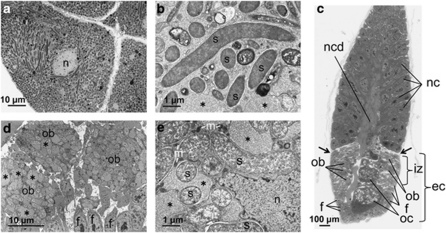
Transmission electron microscopy revealed that numerous tubular bacterial cells are harbored in the cytoplasm of bacteriocytes constituting the bacteriome (Figures 2a and b). Figure 2c shows a semi-ultrathin section of the germarium region of an ovariole dissected from an adult female. At the tip of the ovariole, a number of nurse cells were arranged in a corn-like shape, in the center of which a nutritive cord for material transport to oocytes was seen. Following the nurse cell region, an egg chamber region was present, where several incipient oocytes were located centrally, whereas a layer of many follicle cells was located peripherally. In the infection zone region, peculiar round cells, tentatively called ovarial bacteriocytes, were found between the follicle cell layer and the oocytes. Transmission electron microscopy demonstrated that in the infection zone, the endosymbiont cells were preferentially localized in the ovarial bacteriocytes (Figures 2d and e). In both the bacteriome bacteriocytes and the ovarial bacteriocytes, peculiar vacuoles were frequently associated with the endosymbiont cells (Figures 2b and e, asterisks).
Figure 2.
Transmission electron microscopy of the endosymbiotic system of Nysius plebeius. (a) Bacteriocytes in a bacteriome whose cytoplasm is filled with tubular endosymbiont cells. (b) An enlarged image of the endosymbiont cells in the bacteriome. (c) A light microscopic image of semi-ultrathin section of a germarium region of an ovariole, stained with toluidine blue. (d) An enlarged image of infection zone in an ovariole, where ovarial bacteriocytes containing many endosymbionts are surrounded by follicular cells. (e) An enlarged image of endosymbiont cells in an ovarial bacteriocyte. Asterisks indicate vacuole-like structures that are abundantly found in the ovarial bacteriocytes. b, bacteriocyte; ec, egg chamber; f, follicular cell; iz, infection zone; n, nucleus; nc, nurse cell; ncd, nutritive cord; ob, ovarial bacteriocyte; oc, oocyte; s, endosymbiont cell.
Bacterial 16S rRNA gene sequences from Nysius spp.
From the bacteriomes of five adult females of N. plebeius collected at Tsukuba, a bacterial 16S rRNA gene region was amplified by PCR and cloned, and 7–24 clones for each of the samples were subjected to restriction fragment length polymorphism genotyping. Of the 55 clones examined, 48 clones exhibited the same major pattern, whereas the other 7 clones showed two minor patterns. When three or more clones of the major type were sequenced for each of the samples, all the sequences, 1479 bp in size, were identical to each other, exhibiting the highest BLAST (Basic Local Alignment Search Tool) hit to 16S rRNA gene sequence of secondary endosymbiont of mealybug Planococcus kraunhiae (91.4% (1364/1492) sequence identity; accession number AB374417). Meanwhile, the minor clones yielded two α-proteobacterial sequences: one (accession number AB624349) with the highest BLAST hit to 16S rRNA gene sequence of Wolbachia endosymbiont of spider mite Bryobia sarothamni (99.9% (1426/1427) sequence identity; accession number EU499315), and the other (accession number AB624350) with the highest BLAST hit to 16S rRNA sequence of an uncultured bacterium of the Rickettsiales from tick Ixodes persulcatus (98.8% (1390/1407) sequence identity; accession number AF493952).
Similarly, bacteriome-derived 16S rRNA gene clones of the major restriction fragment length polymorphism genotype were sequenced for samples of N. plebeius, N. sp. 1, N. sp. 2 and N. expressus that represented 8, 5, 1 and 1 populations, respectively. The 1479 bp sequences from N. plebeius were 100% identical within the species, and so were the 1421 bp sequences from N. sp. 1. Meanwhile, the sequence identities between the bacterial sequences from different Nysius species ranged from 98.8% to 99.6%.
Furthermore, bacteriome-derived 16S rRNA gene clones were also sequenced for samples of K. resedae collected at Sapporo and Okunikko, Japan. The 1474 bp sequences were almost identical with the exception of a nucleotide site between the populations, exhibiting the highest BLAST hit to 16S rRNA gene sequence of Kleidoceria schneideri, the endosymbiont of K. resedae sampled in Germany (99.8% (1472/1475) sequence identity; accession number FN555107).
In situ hybridization of the endosymbiont in N. plebeius
Dissected tissues of N. plebeius were subjected to whole mount in situ hybridization with a fluorochrome-labelled probe specifically targeting the endosymbiont 16S rRNA. In adult females, the endosymbiont signals were specifically detected in the bacteriomes, the infection zone of germaria, and the symbiont ball located at the anterior pole of mature oocytes (Figures 1g and h). In adult males, the endosymbiont signals were found in the bacteriomes, but not in the reproductive organs (data not shown).
Prevalence of the endosymbiont infection in natural populations of Nysius spp.
In total, 151 individuals of N. plebeius from 9 populations, 80 individuals of N. sp. 1 from 5 populations, 17 individuals of N. sp. 2 from 3 populations and 33 individuals of N. expressus from 1 population were subjected to diagnostic PCR surveys of their symbiotic bacteria. The bacteriocyte-associated endosymbiont was detected from all the Nysius samples representing 4 species, 18 populations and 281 individuals (Table 1). In contrast, Burkholderia spp., known as symbiotic associates harbored in the midgut crypts of diverse lygaeoid and coreoid species (Kikuchi et al., 2011), were not detected in any of the Nysius samples (Table 1).
Molecular phylogenetic analysis of the endosymbiont of Nysius spp.
Bacterial 16S rRNA, groEL and gyrB genes were amplified by PCR, cloned and sequenced from Nysius spp. representing 4 species and 16 populations in total (Table 1) and subjected to molecular phylogenetic analyses (Supplementary Figures S2–S4). In all the phylogenies, the endosymbionts of Nysius spp. consistently formed a well-defined monophyletic group in the Gammaproteobacteria. The phylogenetic relationships reflected the systematics of the host insects: the endosymbionts of N. plebeius from different populations formed a clade, and so did the endosymbionts of N. sp. 1 from different populations. In the DNA databases, no bacterial sequences were identified as closely related to the endosymbiont clade of Nysius spp. In the γ-proteobacterial phylogenies, although statistical supports for the clusters were generally low, the following insect symbionts were placed nearby the endosymbiont clade of Nysius spp.: endosymbionts of various insects such as Blochmannia of ants, Wigglesworthia of tsetse flies, Buchnera of aphids and Baumannia of sharpshooters; Kleidoceria endosymbiont of the birch catkin bug K. resedae; and gut symbionts of various pentatomoid stinkbugs such as Rozenkranzia of acanthosomatids, Ishikawaella of plataspids and Benitsuchiphilus of parastrachiids (Supplementary Figures S2–S4).
Relationship to the endosymbiont of K. resedae
In the 16S rRNA gene phylogeny, the endosymbiont of K. resedae clustered with the endosymbionts of Nysius spp., but statistical supports for the grouping were consistently very low (Supplementary Figure S2), suggesting that the apparent clustering is dubious. In the groEL and gyrB gene phylogenies, the endosymbiont of K. resedae did not cluster with the endosymbionts of Nysius spp., but was grouped with endosymbionts of other insects such as Wigglesworthia of tsetse flies (Supplementary Figures S3 and S4). An approximately unbiased test statistically rejected the hypothesis that the endosymbiont of K. resedae constitutes a clade with the endosymbionts of Nysius spp. (Table 2).
Table 2. AU tests for clades to assess the phylogenetic relationships of the endosymbionts of Nysius and Kleidoceria schneideri.
| Clade | Δln La | P-value |
|---|---|---|
| (Ksc, Wig) | −12.2 | 0.983 |
| (Ksc, Wig, Blo) | −6.2 | 0.915 |
| (SymNys, Ksc, Wig, Blo) | −5.3 | 0.850 |
| (SymNys, Ksc, Wig) | 6.2 | 0.282 |
| (SymNys, Bau) | 6.7 | 0.094 |
| (SymNys, Blo) | 6.9 | 0.224 |
| (SymNys, Ben, Buc) | 8.1 | 0.789 |
| (SymNys, Ksc) | 20.2 | 0.024b |
| (SymNys, Wig) | 21.1 | 0.017b |
| (SymNys, Buc) | 26.3 | 0.051 |
| (SymNys, Ben) | 27.7 | 0.021b |
Abbreviations: AU test, approximately unbiased test; Bau, Baumannia cicadellinicola; Ben, Benitsuchiphilus tojoi; Blo, Blochmannia floridanus and B. pennsylvanicus; Buc, Buchnera aphidicola (aphid Acyrthosiphon pisum) and B. aphidicola (aphid Schizaphis graminum); Ksc, Kleidoceria schneideri; SymNys, endosymbionts of Nysius plebeius (Tsukuba) and N. expressus; Wig, Wigglesworthia glossinidia.
Difference of log-likelihood between the best trees among those that reject and support the monophyly of the clade.
The clade is significantly rejected (P<0.05).
Molecular phylogenetic analysis of Nysius spp. and allied lygaeid stinkbugs
Mitochondrial COI gene was amplified by PCR and sequenced from Nysius spp. and allied stinkbug species of the family Lygaeidae, and subjected to molecular phylogenetic analyses. The phylogenetic relationship agreed with the systematics of the lygaeid stinkbugs: the three subfamilies respectively formed well-defined monophyletic groups in the Lygaeidae, and congenic species also respectively constituted monophyletic groups in the phylogeny. Nysius spp. and K. resedae did not form a clade (Supplementary Figure S5).
Co-speciating pattern between Nysius spp. and their endosymbionts
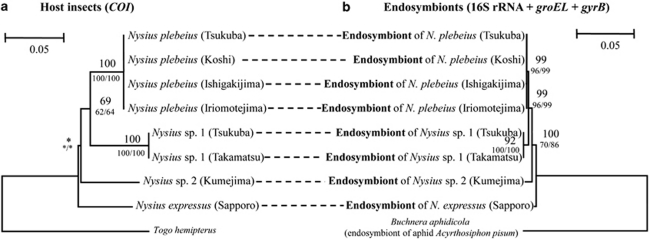
Figure 3 contrasts the phylogeny of Nysius spp. inferred from mitochondrial COI gene sequences with the phylogeny of their endosymbionts inferred from concatenated sequences of bacterial 16S rRNA, groEL and gyrB genes. The topology of the host insect phylogeny was congruent with the topology of the endosymbiont phylogeny.
Figure 3.
Phylogenetic concordance between the Nysius species and their endosymbionts. (a) A maximum likelihood phylogeny of the host insects inferred from mitochondrial COI gene sequences (558 aligned nucleotide sites). (b) A maximum likelihood phylogeny of the endosymbionts inferred from concatenated sequences of 16S rRNA, groEL and gyrB genes (2876 aligned nucleotide sites).
Molecular evolutionary and genomic aspects of the endosymbionts of Nysius spp.
Nucleotide compositions of 16S rRNA, groEL and gyrB genes of the Nysius endosymbionts were AT-rich, 51.1–51.4%, 63.3–64.5% and 70.4–71.8%, in comparison with those of free-living γ-proteobacteria at the values around 44–46%, 42–50% and 40–52%, respectively (Supplementary Figures S2–S4). Evolutionary rates of 16S rRNA, groEL and gyrB genes of the Nysius endosymbionts were significantly higher than those of free-living γ-proteobacteria such as Escherichia coli and Pantoea spp., whereas the evolutionary rates were almost equivalent to those of aphid endosymbiont Buchnera (Table 3). Pulsed-field gel electrophoresis of the bacteriomes of N. sp. 1 estimated the genome size of the endosymbiont as 0.6 Mb (Supplementary Figure S6).
Table 3. Relative rate tests for comparing the molecular evolutionary rates of 16S rRNA, groEL and gyrB gene sequences between the lineages of the endosymbiont of Nysius plebeius, its free-living relatives and Buchnera aphidicola, the obligate endosymbionts of aphids.
| Gene | Lineage 1 | Lineage 2 | Outgroup | K1a | K2b | K1–K2 | Rate ratioc | P-valued |
|---|---|---|---|---|---|---|---|---|
| 16S rRNA | Endosymbiont of N. plebeius (AB576918) | Escherichia coli (U00096) and Pantoea agglomerans (AY691545) | Vibrio cholerae (CP001485) | 0.068 | 0.028 | 0.040 | 2.4 | 3.6 × 10−5 |
| groEL | Endosymbiont of N. plebeius (AB583951) | E. coli (U00096) and P. agglomerans (AB008142) | V. cholerae (CP001485) | 0.094 | 0.030 | 0.064 | 3.1 | 1.9 × 10−7 |
| gyrB | Endosymbiont of N. plebeius (AB583935) | E. coli (U00096) and P. ananatis (NC_013956) | V. cholerae (CP001485) | 0.212 | 0.060 | 0.152 | 3.5 | 1.3 × 10−7 |
| 16S rRNA | Endosymbiont of N. plebeius (AB576918) | Buchnera aphidicola str. APS (NC_002528) and B. aphidicola str. Sg (NC_004061) | V. cholerae (CP001485) | 0.053 | 0.066 | −0.013 | 0.80 | 0.28 |
| groEL | Endosymbiont of N. plebeius (AB583951) | B. aphidicola str. APS (NC_002528) and B. aphidicola str. Sg (NC_004061) | V. cholerae (CP001485) | 0.073 | 0.069 | 0.004 | 1.1 | 0.79 |
| gyrB | Endosymbiont of N. plebeius (AB583935) | B. aphidicola str. APS (NC_002528) and B. aphidicola str. Sg (NC_004061) | V. cholerae (CP001485) | 0.133 | 0.167 | −0.034 | 0.80 | 0.21 |
Estimated mean distance between lineage 1 and the last common ancestor of lineages 1 and 2.
Estimated mean distance between lineage 2 and the last common ancestor of lineages 1 and 2.
K1/K2.
P-values are estimated by the program RRTree (Robinson-Rechavi and Huchon, 2000). The numbers of nucleotide sites used are 1224, 1010 and 640 for 16S rRNA, groEL and gyrB genes, respectively.
Discussion
From these results, we conclude that: (1) seed bugs of the genus Nysius are associated with a specific clade of γ-proteobacterial endosymbionts (Supplementary Figures S2–S4); (2) the endosymbiont is endocellularly localized in specific organs, bacteriomes, associated with host gonads of both sexes (Figures 1b, c and g); (3) in female ovaries, the endosymbiont is also localized in ‘infection zone' in the middle of germaria as well as in ‘symbiont ball' at the anterior pole of oocytes (Figures 1d and h), indicating vertical transmission of the endosymbiont via the ovarial passage; (4) the endosymbiont does not form a clade with K. schneideri, the endosymbiont of birch catkin bug K. resedae (Supplementary Figures S2–S4 and Table 2), suggesting multiple evolutionary origins of the bacteriome-associated endosymbionts in the family Lygaeidae; (5) the endosymbiont genes exhibit AT-biased nucleotide compositions and accelerated rates of molecular evolution (Supplementary Figures S2–S4 and Table 3), and the endosymbiont genome is as small as 0.6 Mb in size (Supplementary Figure S6), indicating a remarkable reductive genome evolution in the endosymbiont lineage; and (6) the phylogenetic relationship of the endosymbionts agrees with the phylogenetic relationship of the host insects (Figure 3), suggesting an intimate host–symbiont association over evolutionary time.
Host–symbiont co-evolution in the genus Nysius
Thus far, in diverse insect endosymbioses of obligate nature, it has been shown that the phylogenetic relationship of the endosymbionts generally agrees with the phylogenetic relationship of their hosts (Moran et al., 1993, 2005; Chen et al., 1999; Sauer et al., 2000; Thao et al., 2000; Thao and Baumann, 2004; Takiya et al., 2006; Conord et al., 2008). In some stinkbug lineages, such co-cladogenetic patterns with their obligate gut symbionts have been observed (Hosokawa et al., 2006; Kikuchi et al., 2009). Our results suggest that in the seed bug genus Nysius, the endosymbionts and their hosts have experienced such a co-evolutionary history: the endosymbiont was already present in the common ancestor of the Nysius species and has been subjected to strict vertical transmission and co-speciation over evolutionary time (Figure 3). However, it should be noted that we cannot rule out the possibility that the apparent phylogenetic congruence could have occurred by chance. Considering only 15 possible rooted tree topologies for four species, the chance that the symbiont tree will exactly match the host tree is expected to be 6.7%, which is not a negligible value. To confirm the validity of the host–symbiont co-speciation, therefore, more Nysius species should be added to the analysis.
Reductive genome evolution of the endosymbiont of Nysius spp.
The AT-biased nucleotide compositions and the accelerated evolutionary rates of the endosymbiont genes (Supplementary Figures S2–S4 and Table 3), and reduced genome size of the endosymbiont (Supplementary Figure S6), are also supportive of the long-lasting host–symbiont co-evolution. The estimated genome size, 0.6 Mb, is strikingly smaller than those of free-living γ-proteobacteria like E. coli (4.6 Mb) and Vibrio cholerae (4.0 Mb) (Blattner et al., 1997; Heidelberg et al., 2000), relatively smaller than those of obligate gut symbionts of other stinkbugs like Ishikawaella of plataspids (0.82–0.83 Mb) and Rosenkranzia of acanthosomatids (0.90–0.96 Mb) (Hosokawa et al., 2006; Kikuchi et al., 2009), and as small as those of obligate endosymbionts of other insects like Buchnera of aphids (0.42–0.65 Mb) and Wigglesworthia of tsetse flies (0.70 Mb) (Shigenobu et al., 2000; Akman et al., 2002). It has been argued that such evolutionary patterns may be attributed to stable and nutrition-rich endocellular habitat for the endosymbiont and also attenuated purifying selection because of the small population size and strong bottleneck associated with the lifestyle of the vertically transmitted endosymbiont (Wernegreen 2002; Moran et al. 2008).
Biological function of the endosymbiont
The biological function of the endosymbiont for the Nysius seed bugs is currently unknown. However, considering the highly developed endosymbiotic system (Figure 1), the 100% infection frequencies in natural host populations (Table 1), the presumably long-lasting host–symbiont association (Figure 3) and the essential roles of the gut symbionts in other stinkbug groups (Abe et al., 1995; Fukatsu and Hosokawa, 2002; Hosokawa et al., 2006; Kikuchi et al., 2007, 2009; Prado and Almeida, 2009a, 2009b), it appears likely that the endosymbiont also plays some important biological roles in Nysius spp. Plausibly, the endosymbiont may supply nutritional supplements for the hosts, such as essential amino acids and vitamins, as has been demonstrated for other plant-sucking insects (Douglas, 1998; Baumann, 2005).
Transmission route of the endosymbiont
The localization of the endosymbiont at the anterior pole of oocytes, which comprises the structure called ‘symbiont ball' (Schneider, 1940) (Figures 1d and h), indicates vertical transmission of the endosymbiont via the ovarial passage through host generations. At the tip of each ovariole, the endosymbiont signals were detected at a distinct, zone-like and red-colored region in the middle of germarium (Figures 1d and h), the structure referred to as ‘infection zone' (Schneider, 1940). In the fruitfly Drosophila melanogaster, a zonal region in the middle of each germarium, which represents the somatic stem cell niche, was shown to be the entry point of a Wolbachia endosymbiont into the germline (Frydman et al., 2006). Whether the infection zone observed in the ovary of Nysius species corresponds to the somatic stem cell niche in the ovary of Drosophila is of interest and deserves further studies. At present, our electron microscopic observations suggest that the endosymbiont cells are not accumulated in the presumable somatic stem cell niche (Figure 2c, arrows) but localized in the ovarial bacteriocytes (Figures 2d and e). Although vertically transmitted endosymbionts of diverse insects tend to localize at the posterior pole of host eggs where the germ plasm destined to form germline cells is located (Buchner, 1965; Miura et al., 2003; Veneti et al., 2004), some insects, including coccids, lygaeid bugs and others, exceptionally exhibit endosymbiont localization at the anterior pole of their eggs (Schneider, 1940; Buchner, 1965; Fukatsu and Nikoh, 2000; Küchler et al., 2010).
Pigmentation of the symbiotic organs
The symbiotic organs of Nysius spp., namely the bacteriome as well as the infection zone in the ovary, are vividly colored in red (Figures 1b–d). Smeared specimens of the bacteriome and the infection zone revealed that the endosymbiont cells are not colored but some materials derived from the host cytoplasm are red in color (Figures 1e and f). Electron microscopic images of the infection zone showed that amorphous vesicles are frequently associated with the endosymbiont cells (asterisks in Figures 2b and e), which might be related to the accumulation of the red pigment in the symbiotic organs. It is currently unknown whether the endosymbiont or the host produces the red pigment and what is the biological relevance of the red pigment. Interestingly, the bacteriome and the infection zone of a distinct lygaeid species K. resedae are also colored in red (Figure 1i) (Schneider, 1940; Küchler et al., 2010). The stink gland and the testis of these insects were also reddish in color (Figures 1b, c and i and Supplementary Figure S1), but diverse stinkbug groups generally exhibit these morphological traits. The colocalization of the red pigment with the endosymbiont will be useful in tracing the developmental dynamics of the bacteriome-associated endosymbiosis in these lygaeid stinkbugs.
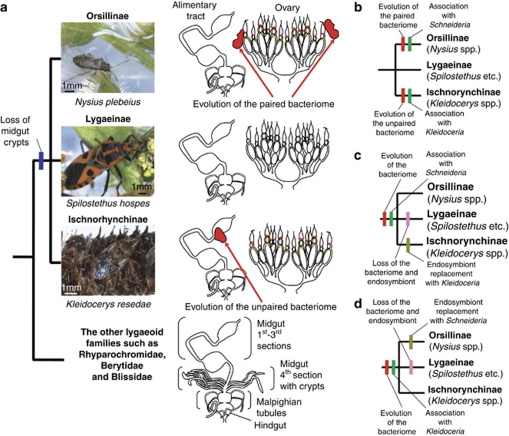
Evolution of the bacteriomes and the endosymbionts in the Lygaeidae
In the family Lygaeidae, the subfamilies Orsillinae (Nysius spp. and others), Lygaeinae (Spilostethus spp., Oncopeltus spp. and others) and Ischnorhynchiinae (Kleidocerys spp. and others) form a monophyletic group (Supplementary Figure S5 and Table 1) (Henry, 1997). All members of the subfamilies examined thus far lack the midgut fourth section with crypts (Figures 1b, c and i and Supplementary Figure S1) (Kikuchi et al., 2011), suggesting that their common ancestor had already lost the midgut crypts (Figure 4a). On the grounds that the endosymbiont clade of Nysius species is phylogenetically distinct from the endosymbiont of Kleidocerys species (Supplementary Figures S2–S4 and Table 2), and that the paired bacteriome of Nysius species anatomically look quite different from the unpaired bacteriome of Kleidocerys species (Figure 4a), it seems plausible that the bacteriomes have independently evolved in the Nysius lineage and the Kleidocerys lineage, respectively (Figure 4b). On the other hand, the red color of the bacteriomes, the red-colored infection zone in the germaria and the symbiont ball at the anterior pole of oocytes are commonly observed in the Nysius species and the Kleidocerys species (Figure 4a), which favors an alternative hypothesis that the bacteriome and the endosymbiont evolved in the common ancestor and subsequently lost in the lineage of Lygaeinae, while a bacteriome remodeling and an endosymbiont replacement have occurred either in the Nysius lineage or in the Kleidocerys lineage (Figures 4b and c). At present, it is elusive which of these or other evolutionary hypotheses are to be substantiated. In the family Blissidae that is allied to the Lygaeidae, I. sabuleti was reported to possess a pair of bacteriomes in the fat body (Schneider, 1940). Future studies on the Ischnodemus system would shed light on the endosymbiotic evolution in the Lygaeidae. Recently, from the bulrush bug Chilacis typhae (Lygaeoidea: Artheneidae), a γ-proteobacterial endosymbiont was characterized in strongly enlarged epithelial cells at an anterior region of the midgut (Küchler et al., 2011). On the grounds that (1) the endosymbiont is phylogenetically not close to the endosymbionts of Nysius spp. and Kleidocerys spp., (2) the histological configuration of the symbiotic organ is totally different from the symbiotic organs of Nysius spp. and Kleidocerys spp. and (3) the host insect is also phylogenetically not close to Nysius spp. and Kleidocerys spp. (Henry, 1997), it seems likely that the endosymbiotic system in C. typhae has an evolutionary origin distinct from the endosymbiotic systems in Nysius spp. and Kleidocerys spp.
Figure 4.
Evolution of symbiotic organs and endosymbionts in the Lygaeidae. (a) Symbiotic organs in the lygaeid subfamilies Orsillinae, Lygaeinae and Ischnorhynchinae. (b–d) Alternative hypotheses that account for the evolution of the bacteriomes and the endosymbionts in the Lygaeidae.
Proposal of candidate name
On account of these distinct and coherent microbiological, phylogenetic and evolutionary traits described in this study, we propose the designation ‘Candidatus Schneideria nysicola' for the endosymbiotic bacterial clade associated with seed bugs of the genus Nysius. The generic name honors Gerhard Schneider, who first described the bacteriome and the endosymbiont of Nysius spp. (Schneider, 1940). The specific name indicates the association with Nysius seed bugs.
Perspectives
The bacteriome, a novel organ specialized for harboring endosymbiotic microorganisms, is of evolutionary and developmental interest. To gain insights into the enigma, the Nysius seed bugs and their relatives will offer ideal study systems for the following reasons: (1) in the family Lygaeidae, the bacteriome may have evolved twice in the lineages of Nysius spp. and Kleidocerys spp.; (2) the Lygaeidae also embraces stinkbugs without the bacteriome such as Spilostethus, Graptostethus and Tropidothorax species; (3) therefore, morphological, developmental and molecular biological comparisons between allied species with and without the symbiotic organ are feasible; (4) in Nysius plebeius and Oncopeltus fasciatus, it has been shown that RNA interference works efficiently (Hughes and Kaufman, 2000; Futahashi et al., 2011), which enables molecular genetic approaches to the mechanisms as to what host genes are involved in the endosymbiosis and how the symbiotic organ is differentiated and formed; and (5) we have established rearing techniques for the Nysius, Kleidocerys, Spilostethus and Graptostethus species in Petri dishes on plant seeds, which offer tractable model systems for experimental and functional studies. Thus far, the aphid–Buchnera association has been the best-studied model endosymbiotic system with the bacteriome, wherein both the host genome and the endosymbiont genome have been sequenced (Shigenobu et al., 2000; International Aphid Genomics Consortium, 2010a), developmental processes of the bacteriome formation have been histologically documented (Braendle et al., 2003; Miura et al., 2003), the transcriptomics of the symbiotic organ have been conducted (Nakabachi et al., 2005; Hansen and Moran, 2011) and a number of physiological, ecological and molecular works on the endosymbiosis have been accumulated (reviewed in Douglas, 1998; Baumann, 2005). On the other hand, as almost all species of the family Aphididae possess the bacteriome harboring Buchnera endosymbiont and a few exceptional species without the bacteriome represent secondary losses of the symbiotic system (Fukatsu et al., 1994; Braendle et al., 2003), evolutionarily meaningful comparisons between allied aphid species with and without the symbiotic organ are difficult. In aphids, it has been reported that RNA interference generally does not work efficiently (Jaubert-Possamai et al., 2007; International Aphid Genomics Consortium, 2010b), which has hindered approaches to the molecular mechanisms underlying the endosymbiotic association. Genome sequencing of the Schneideria endosymbiont and transcriptomics of the bacteriome of the Nysius host will, in combination with the laboratory rearing protocols and the RNA interference techniques, provide a complementary and promising model system for insect symbiosis studies.
Acknowledgments
We thank Katsuyuki Kouno and Shuhei Kada for providing insect samples, Yukinobu Nakatani and Tadashi Ishikawa for identification of Nysius species and Satoshi Hanada for advice on bacterial nomenclature. This study was supported by the Program for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry (BRAIN). YM. was supported by the Japan Society for the Promotion of Science (JSPS) Predoctoral Fellowship for Young Scientists.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Abe Y, Mishiro K, Takanashi M. Symbiont of brown-winged green bug, Plautia stali Scott. Jpn J Appl Entomol Zool. 1995;39:109–115. [Google Scholar]
- Akman L, Yamashita A, Watanabe H, Oshima K, Shiba T, Hattori M, et al. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat Genet. 2002;32:402–407. doi: 10.1038/ng986. [DOI] [PubMed] [Google Scholar]
- Baumann P. Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu Rev Microbiol. 2005;59:155–189. doi: 10.1146/annurev.micro.59.030804.121041. [DOI] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G, III, Bloch CA, Perna NT, Burland V, Riley M, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Bourtzis K, Miller T. Insect Symbiosis. CRC Press: Boca Raton; 2003. [Google Scholar]
- Braendle C, Miura T, Bickel R, Shingleton AW, Kambhampati S, Stern DL. Developmental origin and evolution of bacteriocytes in the aphid–Buchnera symbiosis. PLoS Biol. 2003;1:e21. doi: 10.1371/journal.pbio.0000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchner P. Endosymbiosis of Animals with Plant Microorganisms. Interscience: New York, USA; 1965. [Google Scholar]
- Chen X, Li S, Aksoy S. Concordant evolution of a symbiont with its host insect species: molecular phylogeny of genus Glossina and its bacteriome-associated endosymbiont, Wigglesworthia glossinidia. J Mol Evol. 1999;48:49–58. doi: 10.1007/pl00006444. [DOI] [PubMed] [Google Scholar]
- Conord C, Despres L, Vallier A, Balmand S, Miquel C, Zundel S, et al. Long-term evolutionary stability of bacterial endosymbiosis in Curculionoidea: additional evidence of symbiont replacement in the Dryophthoridae family. Mol Biol Evol. 2008;25:859–868. doi: 10.1093/molbev/msn027. [DOI] [PubMed] [Google Scholar]
- Douglas AE. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu Rev Entomol. 1998;43:17–37. doi: 10.1146/annurev.ento.43.1.17. [DOI] [PubMed] [Google Scholar]
- Frydman HM, Li JM, Robson DN, Wieschaus E. Somatic stem cell niche tropism in Wolbachia. Nature. 2006;441:509–512. doi: 10.1038/nature04756. [DOI] [PubMed] [Google Scholar]
- Fukatsu T. Acetone preservation: a practical technique for molecular analysis. Mol Ecol. 1999;8:1935–1945. doi: 10.1046/j.1365-294x.1999.00795.x. [DOI] [PubMed] [Google Scholar]
- Fukatsu T, Aoki S, Kurosu U, Ishikawa H. Phylogeny of Cerataphidini aphids revealed by their symbiotic microorganisms and basic structure of their galls: implications for host-symbiont coevolution and evolution of sterile soldier castes. Zool Sci. 1994;11:613–623. [Google Scholar]
- Fukatsu T, Hosokawa T. Capsule-transmitted gut symbiotic bacterium of the Japanese common plataspid stinkbug, Megacopta punctatissima. Appl Environ Microbiol. 2002;68:389–396. doi: 10.1128/AEM.68.1.389-396.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukatsu T, Nikoh N. Endosymbiotic microbiota of the bamboo pseudococcid Antonina crawii (Insecta, Homoptera) Appl Environ Microbiol. 2000;66:643–650. doi: 10.1128/aem.66.2.643-650.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukatsu T, Nikoh N, Kawai R, Koga R. The secondary endosymbiotic bacterium of the pea aphid Acyrthosiphon pisum (Insecta: Homoptera) Appl Environ Microbiol. 2000;66:2748–2758. doi: 10.1128/aem.66.7.2748-2758.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futahashi R, Tanaka K, Matsuura Y, Tanahashi M, Kikuchi Y, Fukatsu T. Laccase2 is required for cuticular pigmentation in stinkbugs. Insect Biochem Mol Biol. 2011;41:191–196. doi: 10.1016/j.ibmb.2010.12.003. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hansen AK, Moran NA. Aphid genome expression reveals host–symbiont cooperation in the production of amino acids. Proc Natl Acad Sci USA. 2011;108:2849–2854. doi: 10.1073/pnas.1013465108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert PDN, Cywinska A, Ball SL, deWaard JR. Biological identifications through DNA barcodes. Proc R Soc London B Biol Sci. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholelae. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry TJ. Phylogenetic analysis of family groups within the infraorder Pentatomomorpha (Hemiptera: Heteroptera), with emphasis on the Lygaeoidea. Ann Entomol Soc Am. 1997;90:275–301. [Google Scholar]
- Hosokawa T, Kikuchi Y, Nikoh N, Meng XY, Hironaka M, Fukatsu T. Phylogenetic position and peculiar genetic traits of the midgut bacterial symbiont in the stinkbug Parastrachia japonensis. Appl Environ Microbiol. 2010;76:4130–4135. doi: 10.1128/AEM.00616-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa T, Kikuchi Y, Nikoh N, Shimada M, Fukatsu T. Strict host-symbiont cospeciation and reductive genome evolution in insect gut bacteria. PLoS Biol. 2006;4:e337. doi: 10.1371/journal.pbio.0040337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes CL, Kaufman TC. RNAi analysis of Deformed, proboscipedia and Sex combs reduced in the milkweed bug Oncopeltus fasciatus: novel roles for Hox genes in the hemipteran head. Development. 2000;127:3683–3694. doi: 10.1242/dev.127.17.3683. [DOI] [PubMed] [Google Scholar]
- International Aphid Genomics Consortium Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 2010a;8:e1000313. doi: 10.1371/journal.pbio.1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Aphid Genomics Consortium 2010bAphid White Paper II: proposal to complete development of the aphid model . http://www.aphidbase.com/aphidbase/news/aphid_white_paper_ii .
- Jaubert-Possamai S, Trionnaire GL, Bonhomme J, Christophides GK, Rispe C, Tagu D. Gene knockdown by RNAi in the pea aphid Acyrthosiphon pisum. BMC Biotechnol. 2007;7:63. doi: 10.1186/1472-6750-7-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiwa N, Hosokawa T, Kikuchi Y, Nikoh N, Meng XY, Kimura N, et al. Primary gut symbiont and secondary, Sodalis-allied symbiont in the scutellerid stinkbug Cantao ocellatus. Appl Environ Microbiol. 2010;76:3486–3494. doi: 10.1128/AEM.00421-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiwa N, Hosokawa T, Kikuchi Y, Nikoh N, Meng XY, Kimura N, et al. Bacterial symbionts of the giant jewel stinkbug Eucoryssus grandis (Hemiptera: Scutelleridae) Zool Sci. 2011;28:169–174. doi: 10.2108/zsj.28.169. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y, Meng XY, Fukatsu T. Gut symbiotic bacteria of the genus Burkholderia in the broad-headed bugs Riptortus clavatus and Leptocorisa chinensis (Heteroptera: Alydidae) Appl Environ Microbiol. 2005;71:4035–4043. doi: 10.1128/AEM.71.7.4035-4043.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y, Hosokawa T, Fukatsu T. Insect-microbe mutualism without vertical transmission: a stinkbug acquires beneficial gut symbiont from environment every generation. Appl Environ Microbiol. 2007;73:4308–4316. doi: 10.1128/AEM.00067-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y, Hosokawa T, Fukatsu T.2008Diversity of bacterial symbiosis in stinkbugsIn Dijk TV (ed)Nova Science Publishers, Inc.: New York, USA; 39–63. [Google Scholar]
- Kikuchi Y, Hosokawa T, Fukatsu T. An ancient but promiscuous host-symbiont association between Burkholderia gut symbionts and their heteropteran hosts. ISME J. 2011;5:446–460. doi: 10.1038/ismej.2010.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y, Hosokawa T, Nikoh N, Meng XY, Kamagata Y, Fukatsu T. Host-symbiont co-speciation and reductive genome evolution in gut symbiotic bacteria of acanthosomatid stinkbugs. BMC Biol. 2009;7:2. doi: 10.1186/1741-7007-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga R, Tsuchida T, Fukatsu T. Quenching autofluorescence of insect tissues for in situ detection of endosymbionts. Appl Entomol Zool. 2009;44:281–291. [Google Scholar]
- Küchler SM, Dettner K, Kehl S. Molecular characterization and localization of the obligate endosymbiotic bacterium in the birch catkin bug Kleidocerys resedae (Heteroptera: Lygaeidae, Ischnorhynchinae) FEMS Microbiol Ecol. 2010;73:408–418. doi: 10.1111/j.1574-6941.2010.00890.x. [DOI] [PubMed] [Google Scholar]
- Küchler SM, Dettner K, Kehl S. Characterization of an obligate intracellular bacterium in the midgut epithelium of the bulrush bug Chilacis typhae (Heteroptera, Lygaeidae, Artheneinae) Appl Environ Microbiol. 2011;77:2869–2876. doi: 10.1128/AEM.02983-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura T, Braendle C, Shingleton A, Sisk G, Kambhampati S, Stern DL. A comparison of parthenogenetic and sexual embryogenesis of the pea aphid Acyrthosiphon pisum (Hemiptera: Aphidoidea) J Exp Zool B. 2003;295:59–81. doi: 10.1002/jez.b.3. [DOI] [PubMed] [Google Scholar]
- Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008;42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- Moran MA, Munson MA, Baumann P, Ishikawa H. A molecular clock in endosymbiotic bacteria is calibrated using the insect host. Proc R Soc London B. 1993;253:167–171. [Google Scholar]
- Moran NA, Tran P, Gerardo NM. Symbiosis and insect diversification: an ancient symbiont of sap-feeding insects from the bacterial phylum Bacteroidetes. Appl Environ Microbiol. 2005;71:8802–8810. doi: 10.1128/AEM.71.12.8802-8810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabachi A, Shigenobu S, Sakazume N, Shiraki T, Hayashizaki Y, Carninci P, et al. Transcriptome analysis of the aphid bacteriocyte, the symbiotic host cell that harbors an endocellular mutualistic bacterium, Buchnera. Proc Natl Acad Sci USA. 2005;102:5477–5482. doi: 10.1073/pnas.0409034102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Prado SS, Almeida RP. Phylogenetic placement of pentatomid stink bug gut symbionts. Curr Microbiol. 2009a;58:64–69. doi: 10.1007/s00284-008-9267-9. [DOI] [PubMed] [Google Scholar]
- Prado SS, Almeida RP. Role of symbiotic gut bacteria in the development of Acrosternum hilare and Murgantia histrionica. Entomol Exp Appl. 2009b;132:21–29. [Google Scholar]
- Prado SS, Rubinoff D, Almeida RPP. Vertical transmission of a pentatomid caeca-associated symbiont. Ann Entomol Soc Am. 2006;99:577–585. [Google Scholar]
- Robinson-Rechavi M, Huchon D. RRTree: relative-rate tests between groups of sequences on a phylogenetic tree. Bioinformatics. 2000;16:296–297. doi: 10.1093/bioinformatics/16.3.296. [DOI] [PubMed] [Google Scholar]
- Sauer C, Stackebrandt E, Gadau J, Holldobler B, Gross R. Systematic relationships and cospeciation of bacterial endosymbionts and their carpenter ant host species: proposal of the new taxon Candidatus Blochmannia gen. nov. Int J Syst Evol Microbiol. 2000;50:1877–1886. doi: 10.1099/00207713-50-5-1877. [DOI] [PubMed] [Google Scholar]
- Schneider G. Beiträge zur Kenntnis der symbiontischen Einrichtungen der Heteropteren. Z Morphol Ökol Tiere. 1940;36:565–644. [Google Scholar]
- Schuh RT, Slater JA. True Bugs of the World (Hemiptera: Heteroptera) Cornell University Press: New York, USA; 1995. [Google Scholar]
- Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature. 2000;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 2002;51:492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001;17:1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–-2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Swofford DL.2001PAUP* Version 4.0b10 [computer program]4.0b10 edition. Sinauer: Sunderland, MA [Google Scholar]
- Takiya DM, Tran PL, Dietrich CH, Moran NA. Co-cladogenesis spanning three phyla: leafhoppers (Insecta: Hemiptera: Cicadellidae) and their dual bacterial symbionts. Mol Ecol. 2006;15:4175–4191. doi: 10.1111/j.1365-294X.2006.03071.x. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thao ML, Baumann P. Evolutionary relationships of primary prokaryotic endosymbionts of whiteflies and their hosts. Appl Environ Microbiol. 2004;70:3401–3406. doi: 10.1128/AEM.70.6.3401-3406.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thao ML, Moran NA, Abbot P, Brennan EB, Burckhardt DH, Baumann P. Cospeciation of psyllids and their primary prokaryotic endosymbionts. Appl Environ Microbiol. 2000;66:2898–2905. doi: 10.1128/aem.66.7.2898-2905.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veneti Z, Clark ME, Karr TL, Savakis C, Bourtzis K. Heads or tails: host-parasite interactions in the Drosophila-Wolbachia system. Appl Environ Microbiol. 2004;70:5366–5372. doi: 10.1128/AEM.70.9.5366-5372.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernegreen JJ. Genome evolution in bacterial endosymbionts of insects. Nat Rev Genet. 2002;3:850–861. doi: 10.1038/nrg931. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.