Abstract
The fission yeast Schizosaccharomyces pombe has been widely used to study eukaryotic cell biology, but almost all of this work has used derivatives of a single strain. We have studied 81 independent natural isolates and 3 designated laboratory strains of Schizosaccharomyces pombe. Schizosaccharomyces pombe varies significantly in size but shows only limited variation in proliferation in different environments compared with Saccharomyces cerevisiae. Nucleotide diversity, π, at a near neutral site, the central core of the centromere of chromosome II is approximately 0.7%. Approximately 20% of the isolates showed karyotypic rearrangements as detected by pulsed field gel electrophoresis and filter hybridization analysis. One translocation, found in 6 different isolates, including the type strain, has a geographically widespread distribution and a unique haplotype and may be a marker of an incipient speciation event. All of the other translocations are unique. Exploitation of this karyotypic diversity may cast new light on both the biology of telomeres and centromeres and on isolating mechanisms in single-celled eukaryotes.
Keywords: pombe, karyotype, diversity, fission yeast
Variation segregating in natural populations of model organisms is now widely studied to address questions about the populations structure (Ruderfer et al. 2006; Tsai et al. 2008), geographic differentiation (Johnson et al. 2004), and phylogeny, as well as to provide new sources of variation for the investigation of traits that were previously studied using either experimentally induced mutations or a small set of natural variants (Nieduszynski and Liti 2011). In the case of the budding yeast Saccharomyces cerevisiae, variation has, on one hand, been used to establish the origins and relatedness of the many yeast strains (Fay and Benavides 2005; Schacherer et al. 2009; Liti et al. 2009) that have been exploited by humans and, on the other, to identify new components of pathways and processes that have been exhaustively studied by laboratory methods (Torabi and Kruglyak 2011; Parts et al. 2011). In contrast; variation in Arabidopsis thaliana (Nordborg and Weigel 2008) has been studied to understand the population structure of a predominantly self-fertilizing plant (Kim et al. 2007; Fournier-Level et al. 2011), to identify variation in traits of fundamental and agricultural interest, such as disease resistance (Atwell et al. 2010; Nemri et al. 2010); and to serve as an ecological model that allows a unified understanding of trait variation at the population and nucleotide levels (Todesco et al. 2010).
The fission yeast Schizosaccharomyces pombe (Egel 2003) is a powerful complement to S. cerevisiae for the study of eukaryotic cell autonomous processes. S. pombe was originally isolated from East African beer, but subsequently it has been found in many parts of the world in indigenous fermentations, fruit, molasses, and industrial glucose. There have been two large efforts to isolate S. pombe from fruit, nectar, or fermentations: one by Florenzano et al. (1977) in the vineyards of Western Sicily, and the other by Gomes et al. (2002) in four regions of Southeast Brazil. Thus, while S. pombe has played only a minor role in biotechnology in comparison to S. cerevisiae, isolates of S. pombe are present in many of the major yeast strain collections. Although S. cerevisiae was intensively studied by brewers, oenologists, and bakers prior to its exploitation as a laboratory model, S. pombe has only been studied in any depth as a model of eukaryotic cell biology. Almost all of this work exploits derivatives of the strain 968, which was first identified in a French wine by Osterwalder and then developed as a genetic system by Leupold (1950) (Jürg Kohli, personal communication). Little is known, therefore, about the extent or nature of the variation of this yeast in nature. We assembled a collection of 81 isolates from many regions of the world. We analyzed the isolates at both the phenotypic, genotypic, and karyotypic level. We identified inherited variation in cell size but only limited variability in the proliferative ability in various environments. There are extensive karyotypic differences between many of the strains. The level of nucleotide variation, π, at neutral sites is about 0.7%, which is higher than S. cerevisiae (Liti et al. 2009). Our data suggest that S. pombe exists in small, incompletely isolated populations and that these occupy a limited range of environments. Although mechanistic analysis of the phenotypic diversity of S. pombe will require structural analysis of the different karyotypes, the diverse karyotypes may themselves provide new insights into centromere and telomere function, isolation mechanisms, and speciation in single-celled eukaryotes (Gordon et al. 2011).
Materials and Methods
Handling of strains
Isolates typically arrived on agar slopes or as freeze-dried samples. If they were freeze dried, then the yeast was reconstituted with water and streaked onto supplemented yeast extract agar plates; (YES; 5% yeast extract, 3% glucose, 225 mg/L histidine, 225 mg/L adenine, 225 mg/L leucine, and 225 mg/L uracil), 2% agar Bacto agar (Becton Dickinson). Strains were maintained either on YES agar or cultured in liquid YES.
Restriction site mapping and PCR
Filter transfer, hybridization analysis, and pulsed field gel electrophoresis were carried out as previously described (Brown 1988; Brown et al. 1990). PCR was carried using Taq polymerase (homemade or from Yorkshire Biosciences). Sanger sequencing was carried out using BigDye v3.1 (Applied Biosystems). Primers used to construct probes for filter hybridization are given in supporting information, Table S3.
DNA extraction, sequencing, and analysis
DNA for PCR was extracted from the 5 mL of yeast cultures using a protocol kindly supplied by Jacob Dalgaard of the Marie Curie Research Institute. Five milliliters of saturated culture was concentrated by centrifugation; spheroplasted using zymolyase 20T in 100 μl 1M sorbitol and 50 mM EDTA; concentrated by centrifugation once again; resuspended in 0.2 mL of DNAzol; and vortex mixed. The DNA was precipitated with an equal volume of cold ethanol. The crude DNA was treated with ribonuclease and then pronase in 10 mM Tris-HCl (pH 8.0), 1 mM EDTA, and 0.1% SDS; extracted between three and five times with a 1:1 mixture of phenol and chloroform; and finally precipitated with ethanol prior to use. Primers used to amplify and sequence DNA for diversity analysis are listed in Table S4.
DNA was amplified with Taq polymerase in an ammonium chloride buffer containing 2 mM MgCl2 using the following conditions: an initial denaturation step of 92° for 30 sec was followed by 33 cycles of 92° for 10 sec, 56° for 10 sec, and 65° for 2 min. Primers and unincorporated dNTP were removed from the reactions using Ampure (Agencourt, Beckman Coulter), the products were sequenced on each strand using the primers described above using BigDye (v2 or v3.1; Applied Biosystems), and then purified prior to electrophoretic analysis by Cleanseq (Agencourt, Beckman Coulter). Sequences were aligned and edited using Bioedit and collapsed into haplotypes using FaBox (http://www.birc.au.dk/~biopv/php/fabox/). Sequences that defined unique haplotypes at any one locus were reamplified and resequenced. Standard summary statistics were extracted with Arlequin (Excoffier et al. 2005) and DNASp (Librado and Rozas 2009). DNA for pulsed field gel electrophoresis was embedded in agarose plugs and extracted as described (Smith et al. 1987).
Microarray analysis
For microarray analysis, chromosomal DNA was size-fractionated by pulsed field gel electrophoresis, electroeluted from the gel into dialysis tubing, concentrated using Butan-2-ol, amplified by the Qiagen REPL1-g mini kit before labeling using the Agilent DNA ULS labeling kit (5190-0419), and then purified and hybridized to the Agilent S. pombe 4 × 44K ChIP-on-chip array (G4810) using unfractionated CRUK 972 DNA as competitor. In some experiments, unfractionated DNA from natural isolates was used as target. All steps were carried out according to the manufacturer’s instructions. Arrays were scanned using an Agilent Scanner, and the data were analyzed using the Agilent Genomic Workbench v5.0.14.
Quantifying natural trait variation
Strains were subjected to high-throughput phenotyping by microcultivation (n = 2) in an array of environments essentially as described (Warringer et al. 2008; Warringer and Blomberg 2003). Briefly, strains were inoculated in 350 µl of YES medium (5% yeast extract, 3% glucose, 225 mg/L histidine, 225 mg/L adenine, 225 mg/L leucine, and 225 mg/L uracil) and incubated in two serial rounds of precultivation for 48–72 h at 30°. For experimental runs, strains were inoculated to an OD of 0.05–0.1 in 350 µl of YES medium (3% glucose was replaced by 3% of alternative carbon sources where indicated) and microcultivated for 48 h or 72 h in a Bioscreen analyzer C (Oy Growth Curves, Finland). Optical density was measured every 20 min using a wide band (450–580 nm) filter. The mitotic proliferation rate (population doubling time), lag (population adaptation time), and efficiency (total change in population density) were extracted from high-density growth curves and LN (natural logarithm)–transformed (data set in Table S5). Relative fitness variable for each strain and trait, LSCij, was calculated as:
where wtkj is the fitness variable of the kth measurement of the wild-type for trait j; xij is the measure of strain i for trait j; and r indicates the run. The measure for proliferation efficiency was inverted to maintain directionality between fitness components.
Cell size at septation
Cells were cultured over night in YES containing glucose at 0.5% weight per volume until they reached mid-log phase, concentrated by centrifugation, and visualized using phase microscopy using a Zeiss Axioscop microscope with a Plan NeoFluor 40× objective. Images were collected and sizes were quantitated using Metamorph (Universal Imaging) v6.1.
Population structure revealed by Fst analysis of concatenated sequences of noncoding DNA
The SPBC660.16 large intron (which encodes phosphogluconate dehydrogease), TER1, and CEN2 sequences were stripped of indels and, in the case of the large intron of SPBC 660.16, the microsatellite; concatenated; and then grouped according to geographic origin. Fst values (Cockerham and Weir 1984) were estimated using the Arlequin package (Excoffier et al. 2005).
Linkage disequilibrium calculation
Informative SNPs were identified both in the centromere of chromosome 2 (Figure S6) and in the flanking DNA (Figure S7) and in the TER1 gene and in the flanking DNA (Figure S8) and used to estimate linkage disequilibrium (LD) statistics using DNAsp (Table 5).
Table 5 . Regions of CRUK 972, CBS 356, CBS 2777, Nott 138, 141, 143, and 145 showing copy number variation.
| Strain | Chromosome | Rearrangement | Start Breakpoint | Finish Breakpoint | Size (kb) | Comments |
|---|---|---|---|---|---|---|
| NOTT 141 | I | Duplication | 3031645–3031834 | 3211591–3211839 | 180 | |
| NOTT 143, NOTT 145 | I | Duplication | 3031645–3031834 | 3233014–3233274 | 201 | |
| NOTT 138 | I | Deletion | 124442–124568 | 124866–125001 | 0.3 | No annotated gene |
| NOTT 138 | I | Duplication | 5499761–5500081 | 5500384–5502054 | 2 | Part of a galactosidase (mel1) gene |
| CRUK 972 | II | Triplication | 1598396–161093 | 1603682–1620842 | 19 | Dgdh repeats |
| NOTT 141, NOTT 143, CBS 2777 | II | Duplication | 2036773–2037231 | 2041544–2041813 | 4 | Includes ccc2-Menkes disease protein |
| NOTT 138, NOTT 141, NOTT 143, NOTT 145, CBS 2777 | II | Deletion | 356108–356292 | 359569–360000 | 3 | SPBC1271.07E, SPBC1271.08E, mug96 |
| NOTT 138 | II | Loss of copy number | 2114026–2115566 | 2116080–2116407 | 0.5 | Mating type locus |
| CBS 356, NOTT 141, NOTT 143, NOTT 145, CBS2777 | III | deletion | 1493999–1494480 | 1499706–1500311 | 5 | Deletion of pseudogene SPCC188.10c-1 |
| NOTT 141, NOTT 143, NOTT 145 (4 copies) | III | Duplication | 1893874–1894205 | 1926082–1926689 | 32 | SPCC737.05-08, hmt1 (ABC transporter involved in response to Cd++), mug24, sly1 |
| NOTT 141, NOTT 143, NOTT 145 | III | Duplication | 1174389–1174518 | 1180220–1180872 | 6 | Includes SPCC4B3.01-1 putative 3-mercaptopyruvate sulfurtransferase |
| NOTT 141, NOTT 143 | III | Duplication | 381359–383288 | 383347–383709 | SPCC1682.06 | |
| NOTT 138 | III | Deletion | 969398–970117 | 971646–971940 | 1 | No annotated gene |
| CBS 2777, NOTT 138, NOTT 141, NOTT 143, NOTT 145 | III | Deletion | 1645637–1646096 | 1648511–1649231 | 2 | Deletion of pseudogene SPCC663.07c-1 |
| CBS 356, CBS 2777, NOTT 138, NOTT 141, NOTT 143 | III | CNV polymorphic | 2108532–2109334 | 2109565–2111026 | 1 | wtf22; listed as a pseudogene |
The indicated strains were analyzed by comparative genome hybridization using Agilent 4 × 44K ChiP-on-chip arrays, and the indicated regions of copy number variation were detected.
Results
A collection of natural isolates of S. pombe
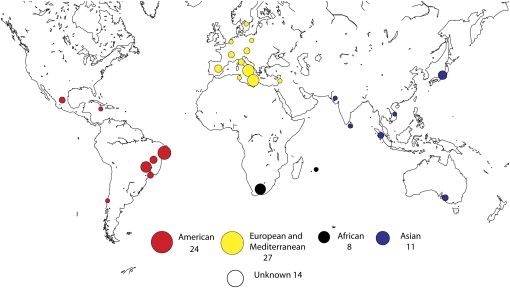
We started our project by assembling a collection of 84 S. pombe strains (Figure 1, Table 1, and Table S1). Of these strains, 81 were natural isolates and 3 were listed as laboratory strains. We first checked the ploidy by DNA staining with Cytox green and fluorescence flow cytometry; this showed that all of the strains but one (NCYC 2355) were haploid. We therefore subcloned NCYC 2355 and isolated three haploid clones. We analyzed two of these clones using the methods described below; they were identical, so we refer to one: NCYC 2355-1. We characterized the 84 haploid strains by sequence and phenotype. We initially sequenced three segments of the genome that did not code for proteins: a segment of the central core of chromosome II (II: 1,621,085–1,621,800); the gene encoding the RNA component of telomerase, TER1 (I: 3,084,446–3,086,143); and the second intron (the largest intron in the S. pombe genome, II: 230,740–231,501) of gene SPBC660.16-1, which encodes phosphogluconate dehydrogenase. To establish a preliminary estimate of the levels of outcrossing (see below), we also sequenced two loci that flanked the centromere of chromosome II (II: 1,572,330–1,572,988 and 1,658,033–1,658,751) and two loci centromere distal of the TER1 gene (I: 3,113,975–3,114,540 and I: 3,194,538–3,195,201). In total, we sequenced and analyzed 5,777 bp in 84 strains. The results established that many of the isolates were closely related and that the entire collection could be reduced to 40 identical haplotypes (Table 1). The strains with identical haplotypes either had a widespread distribution or were collected close to one another. Haplotypes with a widespread distribution could have spread around the world by natural mechanisms or as result of human activity. The geographically restricted haplotypes may represent clones that had reached a high frequency because of founder effects or population bottlenecks, or it may have arisen from restricted collection activity.
Figure 1 .
Geographic origin of the 84 strains of Schizosaccharomyes pombe used in this study. The area of the circles is proportionate to the numbers of strains from the respective areas.
Table 1 . Assorting 84 strains of S. pombe into 40 groups with shared haplotypes at seven loci.
| Strain | Haplotype Number | Origin Where Known |
|---|---|---|
| UWOPS 92.229.4 | 1 | Mexico |
| UWOPS 94.422.2 | 2 | Mexico |
| UFMG A529, UFMG 790, UFMG A826 | 3 | Brazil, Belo Horizonte and Viçosa |
| UFMG R416, UFMG R418, UFMG R420, UFMG R424, UFMG R435 | 4 | Brazil; Aracaju |
| UFMG R427 | 5 | Brazil; Aracaju |
| UFMG A1263 | 6 | Brazil, Viçosa |
| UFMG A521, UFMG A571, UFMG A602 | 7 | Brazil, Belo Horizonte |
| UFMG A1000, UFMG A1153, | 8 | Brazil, Belo Horizonte and Salinas |
| UFMG R434 | 9 | Brazil; Aracaju |
| UFMG R428 | 10 | Brazil; Aracaju |
| UFMG A1152 | 11 | Brazil, Salinas |
| UFMG R437 | 12 | Brazil; Aracaju |
| UFMG A738 | 13 | Brazil, Belo Horizonte |
| NCYC 683, NCYC 2387, DBVPG4435, AWRI 442 | 14 | Spain, Italy, South Australia |
| NCYC 936, NCYC 2355-1, | 15 | Sri Lanka, Japan |
| CBS 356, NCYC 132, NCYC535, DBVPG2817, DBVPG4437, AWRI 141 | 16 | Eastern Mediterranean, Africa, Italy, Australia |
| NCYC 380, CBS 1063, DBVPG 6281, CBS 355, DBVPG 6417 | 17 | Sicily, Spain |
| DBVPG 4433, DBVPG 6279, DBVPG 6610, DBVPG 6699, Y0036, Y0037, CRUK 972, CRUK 975, Y 468, Y 469, | 18 | Germany, Indo-China, South Africa, France, |
| CBS 2628 | 19 | Pakistan |
| CBS 2775, CBS 2776, CBS 2777 | 20 | Japan (all) |
| CBS 5680 | 21 | Poland |
| CBS 5682 | 22 | South Africa |
| CBS 7335 | 23 | Spain |
| DBVPG 2801 | 24 | Tunisia |
| DBVPG 2805 | 25 | Malta |
| DBVPG 2804, DBVPG 2806, DBVPG 2807, DBVPG 2808, DBVPG 2809 | 26 | Malta (all) |
| DBVPG 2810 | 27 | Malta |
| DBVPG 2811, DBVPG 2812, DBVPG 2814, DBVPG 2815, DBVPG 2816, DBVPG 2818 | 28 | Sicily (all) |
| Y470 | 29 | — |
| Y 831, Y 832 | 30 | South Africa (both) |
| CBS 374 | 31 | Delft |
| DBVPG 6447, DBVPG 6449 | 32 | — |
| CBS 358 | 33 | — |
| CBS 1058 | 34 | Java |
| CBS 357 | 35 | Jamaica |
| CBS 352 | 36 | Indonesia |
| CBS 1057 | 37 | Sweden |
| CBS 1059 | 38 | Mauritius |
| CBS 1044 | 39 | — |
| L2470 | 40 | Chile |
Each of the 84 strains in the original collection was sequenced using conventional Sanger methodology at seven individual loci as described in the text and in File S1. Strains were grouped according to 40 compound haplotypes. For the details of the sequences of the individual loci and the haplotype structures, see File S1.
Population genetics of S. pombe
We used sequence data to analyze the diversity and population structure of S. pombe. To minimize the consequences of biased collecting activities or human trade, we carried out this analysis using the diversity contained within the 40 different haplotypes identified in Table 1 and Table S2. The diversity π (Table 2) detected at each of the three nonprotein-coding loci varies with the highest value being observed at the locus most likely to approach neutrality, the central core of the centromere of chromosome II. A negative value of Tajima’s D suggests recent selection at or close to the region of the SPBC 660.16 intron. The value of π of 7 × 10−3 seen at the central core of chromosome II is consistent with the variation seen in pairwise comparison of 4-fold degenerate sites between the laboratory strain and strains NCYC 132 and SPK 1820, which gave values of 8.9 × 10−3 and 6.7 × 10−3 (Rhind et al. 2011), respectively, so we concluded that this estimate is correct. This is slightly higher than in the budding yeast S. cerevisiae, which has a value of 5.65 × 10−3 (Liti et al. 2009). The fact that all but one of the strains are haploid and the assumption of a neutral coalescent allows estimation of the effective mitotic population size Ne by use of relationship π = 2Neu, where u is the mutation rate per nucleotide per mitotic generation (Tsai et al. 2008). The neutral mutation rate in S. pombe is not known, so we assumed the rate that has been used for S. cerevisiae of 0.33 × 10−9/bp/generation (Lynch et al. 2008), leading to an estimate of the global effective population size of 1 × 107. It is also of interest to know how much variation is endemic to particular population. There was too little sequence variation to be able to estimate population structure a priori using the Bayesian approaches implemented in the STRUCTURE (Pritchard et al. 2000) and BAPS (Corander et al. 2008) packages, so we grouped the haplotypes according to their geographic origin, where this was possible, and then analyzed them using the F statistic (Cockerham and Weir 1984). These results (Table 3) demonstrated little by way of population structure but suggested that the American strains were the most highly differentiated. Network analysis of the CEN2, TER, and 660.16 sequences was also consistent with extensive mixing between the different haplotypes (Figure S6, Figure S8, Figure S9, and Figure S10). Two loci include sufficient data to estimate the extent of linkage disequilibrium, which is inversely proportional to the out-crossing rate. To do this, we identified informative SNPs in and around the centromere of chromosome II and within and centromere distal of the TER1 gene (Figure S6, Figure S7, Figure S8, Figure S9, and Figure S10). A four gamete test of the sequence variation around the centromere does not indicate recombination. Significant linkage disequilibrium is also detectable 30 kb from the telomerase RNA gene using both the D' and R statistics (Table 4). The extent of linkage disequilibrium would seem, therefore, to be greater than in S. paradoxus (Tsai et al. 2008), which out-crosses once in every 1000 asexual generations and which shows lower levels of nucleotide diversity. Network analysis (Figure S6, Figure S9, and Figure S10) of the haplotypes at centromere II, TER1, and SPBC660.16, however, demonstrate that there is extensive mixing between different strains. More detailed analysis will be required to measure the rate of out-crossing in S. pombe and to correlate it with the karyotypic diversity described below.
Table 2 . Nucleotide diversity at three noncoding loci in the genome of S. pombe.
| Sequence | Number of Residues | π ± SD × 10−3 | θ ± SD × 10−3 | Tajima’s D (per Sequence) | Number of Segregating Sites |
|---|---|---|---|---|---|
| CC CEN2-indels | 719 | 6.997 ± 3.863 | 6.866 ± 2.520 | 0.01131 | 21 |
| TER1-indels | 1702 | 4.329 ± 2.298 | 4.420 ± 1.472 | −0.08316 | 32 |
| 660.16 intron-minisat | 743 | 1.603 ± 1.165 | 3.798 ± 1.51 | −1.77965 | 12 |
Diversity statistics were calculated based on the sequences at the three indicated loci using Arlequin. The sequences were cleared of indels and microsatellite sequences prior to analysis.
Table 3 . Population differentiation as measured by Fst.
| Population | American | African | European | Asian | Unknown |
|---|---|---|---|---|---|
| American (15) | 0 | ||||
| African (5) | 0.075 | 0 | |||
| European (11) | 0.087 | −0.021 | 0 | ||
| Asian (7) | 0.009 | 0.02 | 0.035 | 0 | |
| Unknown origin (9) | 0.044 | −0.01 | 0.051 | 0.022 | 0 |
The sequence data from the 40 compound haplotypes were used to estimate pairwise Fst measures of population differentiation using the Arlequin package. Haplotypes (e.g. haplotypes 16 and 18) containing strains of different origins were considered as being of unknown origin. The numbers of strains in the respective populations are indicated in parentheses. Haplotypes containing strains of different known origins were assigned multiply to different populations and thus the sum of the numbers in brackets exceeds 40.
Table 4 . Linkage disequilibrium around centromere 2 and telomerase RNA gene (TER1).
| Chromosome II centromere | |||||
|---|---|---|---|---|---|
| R/chi | 1572349 | 1621091 | 1658392 | 1658455 | |
| 1572349 | −0.396 | −0.597 | −0.396 | ||
| 1621091 | 6.271* | 0.664 | 1 | ||
| 1658392 | 14.235*** | 17.622*** | 0.664 | ||
| 1658455 | 6.271* | 40*** | 17.622*** | ||
| D' | 1572349 | 1621091 | 1658392 | 1658455 | |
| 1572349 | −1 | −1 | −1 | ||
| 1621091 | 1 | 1 | |||
| 1658392 | 1 | ||||
| 1658455 | |||||
| Chromosome I TER1 region | |||||
| R/chi | 3084762 | 3086123 | 3114239 | 3114431 | 3194949 |
| 3084762 | 0.555 | 0.267 | 0.434 | −0.248 | |
| 3086123 | 12.31*** | 0.339 | 0.236 | −0.089 | |
| 3114239 | 2.857 | 4.596* | 0.85 | −0.0207 | |
| 3114431 | 7.519** | 2.222 | 28.9*** | −0.333 | |
| 3194949 | 2.462 | 0.32 | 1.709 | 1.778 | |
| D' | 3084762 | 3086123 | 3114239 | 3114431 | 3194949 |
| 3084762 | 0.826 | 0.331 | 0.457 | −0.0373 | |
| 3086123 | 0.407 | 0.236 | −0.2 | ||
| 3114239 | 1 | −0.385 | |||
| 3114431 | −0.333 | ||||
| 3194949 | |||||
Haplotypes at individual loci were identified to identify informative SNPs, and these were used to measure linkage disequilibrium by the indicated metrics using the DNAsp package. The significance of the chi squared values are indicated by asterisks: *, at the 5% level; **, at the 1% level; ***, at the 0.1% level. The numbers in bold at the margins of the table refer to the coordinates along the chromosome of the respective, informative SNPs.
Trait variation in S. pombe is defined by population structure and geographic boundaries
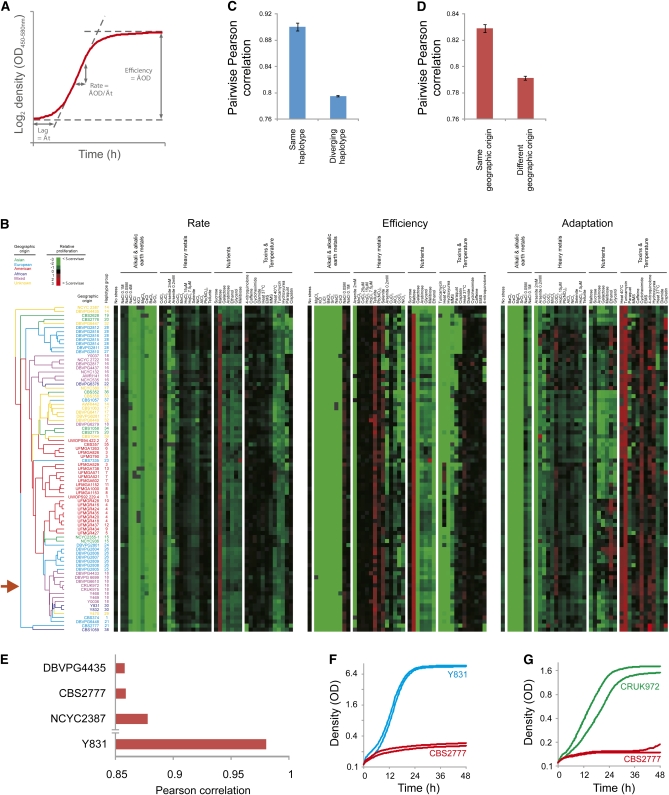
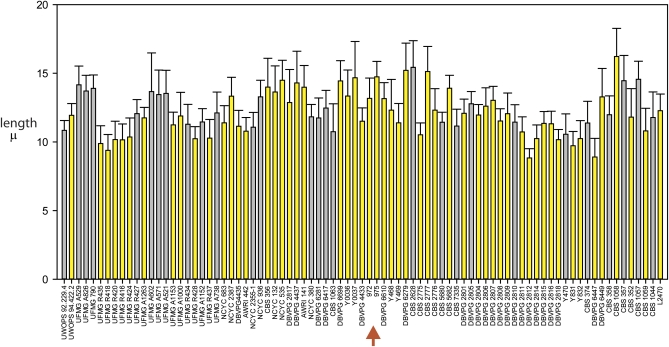
To survey natural trait variation in S. pombe, isolates were subjected to high-resolution quantification of proliferative ability in an array of environments representing variations in nutrient availability, temperature, and exposure to toxic metals and drugs. From high-density mitotic growth curves, the fitness components lag of proliferation, rate of proliferation (population doubling time), and efficiency of proliferation (population density change) were extracted, providing >120 distinct measures of organism-environment interactions (Figure 2). These fitness components were compared with those of the S. cerevisiae universal type strain BY4741. In optimal conditions, S. pombe featured a delay in the time to initiate population growth and a marginal reduction in proliferation rate relative S. cerevisiae (Figure S1, A and B). In contrast, the performance of S. pombe was very severely impaired in many stress-inducing environments (Figure 2B). In particular, S. pombe proliferation in the presence of alkali and alkaline earth metals, DNA damaging agents, and respiratory or partially respiratory carbon sources was reduced (Figure 2B and Figure S1D). Utilization of maltose, the main storage carbohydrate of barley, represented an exception, being more efficient in S. pombe (Figure S1, D and E). Overall, trait variation in S. pombe was lower than what has been reported both for the variable S. cerevisiae and its less variable wild relative S. paradoxus (Warringer et al. 2011) (Figure S2A). This suggests that S. pombe is adapted for proliferating in a more constrained ecological range than the baker’s yeasts. Nevertheless, trait variation largely followed phylogenetic boundaries as defined by haplotype structure and geographic origin: strains with the same haplotype or the same geographic origin were significantly (Student t-test, P < E−34) more similar than other strains (Figure 2B–D). In addition, we surveyed cell length at septation and found the influence of the haplotype structure to prevail for this trait (Figure 3). The most phenotypically distinct S. pombe populations were the American population, with an impaired proliferation at high temperatures, and the European population, with a prolonged lag phase when initiating respiratory proliferation as well as elevated tolerance to DNA damage (Figure S2B). The strain CBS 2777 stands out in terms of mitotic fitness traits, and it emerges as one of the most atypical strains (Figure 2E and Figure S3A). CBS 2777 is a strain with the unusual karyotype of four chromosomes (see below). CBS 2777 deviated from the S. pombe mean in more than 14% of all traits (Student t-test, FDR 5%). Most of these abnormalities represented severe proliferation deficiencies, notably the inability to tolerate DNA-damaging drugs, such as cisplatin and 4-nitroquinolone, that strain DNA replication and repair (Figure 2, G and F and Figure S3B). These abnormalities were evident also in relation to CBS2775 and CBS2776, which share haplotype, but not karyotype, structure with CBS2777.
Figure 2 .
Trait variation in S. pombe is defined by population structure and geographic origin. (A) The proliferative lag (time to initiate proliferation), proliferative rate (population doubling time), and proliferative efficiency (change in population density) were extracted from high-density growth curves (n = 2) of natural S. pombe isolates (N = 2) over 42 environments. (B) Hierarchical clustering of S. pombe natural isolates was performed using trait profiles based on all traits, a centered Pearson correlation metric, and average linkage mapping. Numbers indicate haplotype groups and color indicates geographic origin. Heat map depicts proliferation relative the S. cerevisiae universal type strain BY4741 (Log2 [isolate/BY4741]). Green = inferior proliferation, red = superior proliferation, black = BY4741 proliferation, gray = missing data. The red arrow indicates the S. pombe reference strains 972 h− and 975 h+. (C) Pearson correlation coefficients were calculated between all pairs of strains belonging to the same and to different haplotype groups. Means and standard errors of the means are displayed. (D) Pearson correlation coefficients were calculated between all pairs of strains with similar and diverging geographic origins. Means and standard errors of the means are displayed. (E) A S. pombe mean trait profile was calculated and the similarity (Pearson correlation) between the mean trait profile and the trait profile of each individual isolate was calculated. Isolates were ranked according to degree of similarity; the bottom three (most atypical) S. pombe isolates are displayed. The most typical S. pombe isolate, Y 831, is shown for comparison. (F, G) Proliferation of the S. pombe karyotype extreme CBS 2777 in presence of the DNA-damaging drugs cisplatin (F) and 4-nitroquinolone (G). Strains showing typical S. pombe behavior in these environments are included for comparison.
Figure 3 .
Variation in length at septation among natural isolates of S. pombe. S. pombe isolates were cultured to mid-exponential phase of growth, harvested, and then analyzed by phase contrast microscopy. The individual isolates are arranged on the abscissa according to their respective haplotypes, with isolates of the same haplotype placed adjacent to one another and accorded the same color (alternating gray or yellow). The red arrow indicates the S. pombe reference strains 972 h− and 975 h+.
Extensive karyotypic diversity in S. pombe
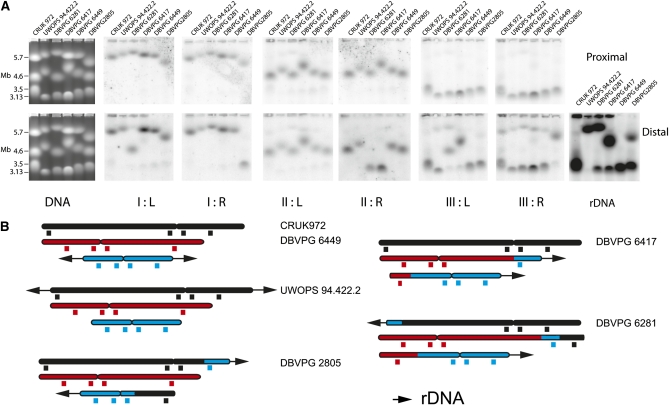
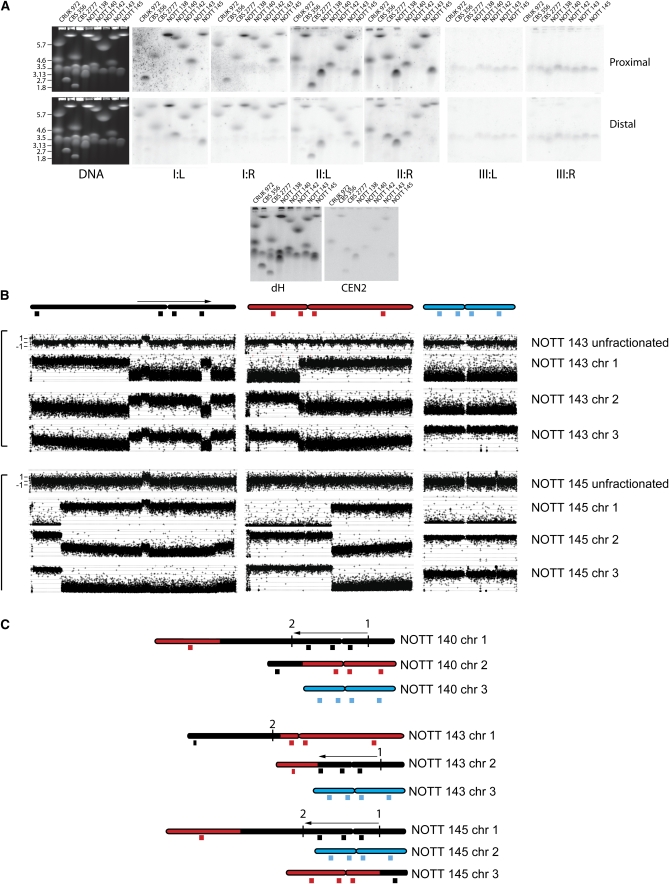
The laboratory strain of S. pombe possesses three chromosomes. Each centromere includes two blocks of tandemly repeated heterochromatic sequences that are arranged palindromically around an A+T rich central core. There is extensive sequence homology between the six blocks of tandemly repeated centromeric DNA, suggesting the possibility of polymorphism arising either from exchange between the arrays of repeats at different centromeres or from instability of individual centromeric palindromes. In light of this potential polymorphism, we analyzed the chromosomes by pulsed field gel electrophoresis, filter transfer, and hybridization using a set of single-copy probes that lie centromere proximal and distal on the six arms of the laboratory strain karyotype and with a probe for the ribosomal DNA (rDNA), which occupies a subtelomeric position at the ends of the two arms of chromosome III. Simple ethidium bromide staining of the pulsed field gels showed that four African isolates (Y468, Y470, Y831, and Y832) were mixtures of different karyotypes and were subcloned. One of the isolates (CBS 374), which gave ambiguous results upon first pulsed field gel analysis, was also subcloned. Together these subclones defined 15 additional strains, which we define as NOTT 133–145, NOTT 147, and NOTT 148. We subsequently sequenced these subclones at the most informative (as defined by number of haplotypes) of the seven loci that we had sequenced in the original set of 84 strains (SPBC660.0.16, TER1, and the central core of the centromere of chromosome II), and we confirmed that they were isogenic with respect to the original uncloned isolates. The pulsed field gel and filter hybridization analysis identified three types of gross chromosome rearrangement. Four isolates (UWOPS 94.422.2, DBVPG 2805, DBVPG 6417, and DBVPG 6281) had karyotypes in which the rDNA was rearranged. The nature of these rearrangements was analyzed as shown in Figure 4A to produce the ideograms shown in Figure 4B. Each of these rearrangements was unique in the collection. The conclusion we draw is that S. pombe shows extensive exchange of subtelomeric DNA sequences, including rDNA. It is striking that the sequences that are exchanged between the chromosomes extend hundreds of kilobases into single-copy, chromosome-specific DNA. It is also notable that there is extensive size variation between chromosomes not detectably involved in the translocation of the rDNA arrays. The cause of this variation remains to be determined but may include translocations and exchanges of subtelomeric DNA that our analyses failed to detect. The second type of rearrangement involved sequences normally present on chromosomes I and II of the laboratory strain. To facilitate discussion, we refer to these types of rearranged chromosomes using Arabic numerals, with the largest chromosome in any one strain termed chromosome 1. We identified six different rearrangements of this type. One of these rearrangements was present in what is referred to as the S. pombe type strain CBS 356 (Figure 5A) and in five other strains (NCYC 132, NCYC 535, DBVPG 2817, DBVPG 4437, and AWRI 141) that have a widespread distribution over the old world and Australia (Table 1, Figure S4). These strains together defined haplotype 16, which includes specific locus haplotypes at the three loci in and around CEN2 and at the large intron of SPBC 660.16. The widespread distribution of the strains with this haplotype suggests that the karyotype variant is long standing relative to the other rearrangements, and thus, haplotype 16 karyotype may define an incipient species. The other translocations were detected in single strains (NOTT 138, NOTT 140, NOTT 142, NOTT 143, and NOTT 145), all of which were derived by subcloning three African isolates. We analyzed all of these rearranged karyotypes first by pulsed field gel electrophoresis, then by filter transfer and hybridization (Figure 5A), and then for two of the karyotypes, NOTT 143 and NOTT 145, by comparative genomic hybridization (CGH) of gel-purified chromosomes to microarrays (Figure 5B), followed by PCR analysis and sequencing (File S1) across hypothetical breakpoints to produce the maps shown in Figure 5C. These demonstrate features held in common by the two rearrangements, first they are both translocations, and second, they both share a 2,227,883 bp pericentric inversion with respect to chromosome I of the laboratory strain. PCR showed that this inversion is present in all of the strains in the collection with the exception of DBVPG 2805, DBVPG 6610, DBVPG 4433, DBVPG 6279, DBVPG 6699, CRUK 972, and CRUK 975. These strains belong to haplotype 18 or the closely related haplotype 25. However, these strains have a widespread distribution, which suggested that the laboratory strain arrangement and possibly other rearrangements on a shared background are acting as barriers to fertile mating. Sequencing across the inversion and translocation breakpoints (File S1, and Figure S5) showed only microhomology in two of five sequences involved in the exchanges. Repetitive sequences were absent. This fact that the 2.23 Mb inversion was present in a minority of strains also suggested that the sequence arrangement in the laboratory strain was a derived trait, which we confirmed by comparison with the arrangement of the homologous sequences in S. octosporus and S. cryophilus (not shown). Pulsed field gel and filter hybridization analysis (Figure 5A) indicated that the rearrangements in NOTT 140 and 142 were also simple reciprocal translocations, and they were not characterized further. Filter hybridization and CGH microarray analysis showed that CBS 356 and NOTT 138 also contained translocations or transpositions, but in both cases, one of the two partner chromosomes was not detectably rearranged when analyzed by CGH on microarrays (not shown). The simplest explanation for these results is that one of the breakpoints is close to a telomere.
Figure 4 .
Rearrangement of the S. pombe karyotype by translocations of the rDNA and associated sequences. (A) DNA extracted from five different natural isolates (UWOPS 94.422.2, DBVPG 6281, DBVPG 6417, DBVPG 6449, and DBVPG 2805) and from laboratory strain CRUK 972 was size-fractionated by pulsed field gel electrophoresis and then analyzed by filter hybridization with centromere proximal and distal probes from each of the six arms of the three chromosomes and with a probe derived from rDNA. The figure illustrates the original ethidium bromide-stained gels and the results of the filter hybridization. Four isolates had karyotypes in which the rDNA was rearranged. In UWOPS 94.422.2, the rDNA is translocated in its entirety onto chromosome I. In DBVPG 2805, the rDNA on the right arm of the laboratory strain chromosome III is translocated together with the distal IIIR sequence onto chromosome I, and distal IR sequence is translocated onto chromosome III. In DBVPG6417, distal IIIL sequences are translocated onto chromosome II, and distal IIR sequences are translocated onto chromosome III. DBVPG 6281 appears to be a derivative of DBVPG 6417 as the rDNA is now present on chromosome I and distal IL sequences are present on chromosome II. (B) An ideogrammatic interpretation of these results together with an indication of the approximate positions of the probes used in the filter hybridization.
Figure 5 .
Rearrangement of the S. pombe karyotype by translocations between chromosomes I and II. (A) DNA extracted from seven different natural isolates (CBS 356, CBS2777, NOTT 136, NOTT 140, NOTT 142, NOTT 143, and NOTT 145) and laboratory strain CRUK 972 was size-fractionated by pulsed field gel electrophoresis and then analyzed after transfer by filter hybridization with centromere proximal and distal probes from each of the six arms of the three chromosomes and with probes for the centromeric dGdH repeat and the central core of chromosome II. The figure illustrates the original ethidium bromide-stained gels and the results of the filter hybridization. (B) DNA was extracted from strains NOTT 143 and NOTT 145, size fractionated by pulsed field gel electrophoresis, and analyzed together with unfractionated DNA by CGH using Agilent 44K ChIP on CHIP arrays using unfractionated DNA from the laboratory strain CRUK 975 as competitor. In the case of NOTT 145, chromosomes 2 and 3 are very similar in size; there is cross contamination in the CGH experiments, but this does not obscure the details of the translocation between sequences that normally reside on chromosomes I and II. (C) Ideogrammatic representation of the CGH results together with an indication of the approximate positions of the probes used in the filter hybridization. Also indicated is the position of the 2.3 Mb pericentric inversion of sequences with respect to laboratory strain chromosome I. This inversion is present in both NOTT 143, as indicated by the results of the CGH experiments, and NOTT 143, as indicated by the results of PCR analysis. For a detailed discussion of the experimental approaches needed to define the rearrangements, see File S1.
In addition to the analyses of the chromosomal rearrangements in CBS 356, NOTT 138, NOTT 140, and NOTT 142, we carried out comparative genomic hybridization (CGH) and microarray analysis CGH on unfractionated DNA of strains CRUK 972, NOTT 141, NOTT 143, NOTT 145, and CBS 2777 using CRUK 975 DNA as a competitor. This CGH analysis detected extensive regions of copy number variation (Table 5). Three of the eight regions that are amplified include genes that are involved in resistance to heavy metals, consistent with exposure to Bordeaux mixture (CuSO4/Ca(OH)2) used as a fungicide in vineyards.
The strain CBS 2777 contained four chromosomes (Figure 5A). Preliminary pulsed field gel and filter hybridization analysis of intact chromosomal DNA demonstrated a complex pattern of sequence rearrangements. Detailed functional and structural analysis of this strain will be described elsewhere.
Discussion
In many organisms, natural isolates have been a source of variation for the analysis of pathways and processes of basic biological interest. In the short term, the aim of such studies is to increase knowledge of the components of these pathways and processes, whereas in the long term, the aim is to understand the architecture of a trait or feature at all levels from DNA to phenotype in terms of the ecology of the respective organism (Mackay et al. 2009). We initiated the study of the variation of S. pombe because this organism is easy and cheap to manipulate and analyze and because much is known about the cell biology of S. pombe. We showed that the amount of phenotypic variation segregating in S. pombe is small in comparison to that seen, for example, in S. cerevisiae and that the existence of karyotypic rearrangements may limit access to some of it. We conclude that the variation that does exist (in size, for example) will need to be studied in a focused way. Our pulsed field gel-based approach is likely to have revealed the presence of only a subset of the translocations, inversions, and deletions segregating in the collection, and thus, a subset of strains, segregating a trait of particular interest, will need to be completely sequenced and characterized in terms of rearrangements. In light of the expense required to carry out such work, it would seem prudent to start with the most significant of the variable traits and characterize the strains segregating this variation first.
Cell size is a feature of the biology of S. pombe that has been intensively studied, is of general biological importance, and shows clear evidence of variation among the different isolates. Such variation is also observed in budding yeast (Nogami et al. 2007). It may be that this variation is adaptively neutral, and the correlation between the size of particular haplotype and the source from which it is isolated that is observed in haplotypes 3 and 4 of Brazilian origin has arisen by drift. Alternatively, the variation may be adaptively significant and related to the fact that the strains were isolated from different environments: caçhaca must (haplotype 3) or the frozen pulp of Eugenia uniflora (haplotye 4). A combination of experimental and ecological studies should discriminate between these alternative explanations and improve our understanding of this important aspect of cell physiology.
Although karyotypic diversity will make it necessary to adapt and apply the approaches used to exploit diversity in other model organisms to S. pombe, karyotype diversity is itself of interest. Thus, an understanding of the karyotype diversity is essential if we are to understand the nucleotide diversity because inversion and translocations will limit exchange between segments of DNA and strains. Rearranged chromosomes have been useful as both genetic reagents and in terms of their ability to cast new and often unexpected perspectives on fundamental problems. Thus the discovery of strain CBS 2777 with four chromosomes poses interesting questions about the mechanism of formation of additional centromeres and telomeres. The unexpectedly high levels of karyotypic diversity that characterize S. pombe worldwide also require explanation in population genetic and mechanistic terms. The simplest explanation, at the population genetic level, is that S. pombe exists in small, rarely out-crossing populations that favor the accumulation of weakly deleterious mutations, including karyotypic rearrangements. This explanation is consistent with both the pattern of linkage distribution around CEN2 and TER1 and with the observation that most of the rearrangements occur once in the collection or are confined to single haplotypes (Table S2). Note, however, that our perspective on out-crossing is based on only two regions of the genome and the patterns of LD within these regions will in turn be sensitive to whether they include any common rearrangements. Genome-wide estimates of LD will thus also require a knowledge of the karyotype diversity. A second, and not exclusive, adaptive explanation for the high levels of karyotypic diversity at the population genetic level is suggested by the isogenic rearrangements isolated by subcloning the African strains Y468, Y470, Y831, and Y832. The fact that the substrains are isogenic at the sequence level, vary karyotypically, and are derived from the same isolates raises the possibility that adaptive radiation may be occurring within in a limited geographic area and that the rearrangements are acting to isolate variants adapted to different, specific subniches. Although it may be possible to investigate such hypothetical microspeciation, a potential limitation of this line of work is that the traits under selection may be significant only for yeast ecology (e.g. sugar utilization) and not of general interest as regards eukaryotic cell biology. However, it seems worthwhile to study this aspect of the biology of S. pombe because speciation is a problem of general interest that remains incompletely understood, and S. pombe offers the possibility of rigorous analysis and understanding. In this respect, our population level approach extends the comparative genomics analysis of four different Schizosaccharomyces species by Rhind et al. (2011). Deeper understanding at both levels should help cast light on the mode and tempo of speciation in Schizosaccharomyces spp. and, in particular, on the relative significance of karyotypic and gene incompatibility as isolating mechanisms in this genus (Greig 2009).
It is clear from our analysis that our understanding of the origins and spread of S. pombe would be improved not only by more sequence data and data about the rearrangements segregating in the population but also by the collection and analysis of many more strains from around the world. S. pombe will grow at high concentrations of glucose and at low pH. An enrichment medium for S. pombe has been described (Florenzano et al. 1977). Thus, it may be possible to collect many more strains, although the ease with which this might be done is not yet clear. In particular, more strains from Africa and South and East Asia will be necessary to establish the geographic origins and phylogeny of the species. The success of the Brazilian collection expeditions suggests that it should be possible to isolate such strains. This work should also help define the range of environments that harbor S. pombe, provide additional karyotypic variants, and perhaps identify convenient locations for field work.
Supplementary Material
Acknowledgments
We thank André Lachance, Gert Marais, Jacky Hayles, and Eveline Bartowsky for strains; Jürg Kohli for information about the origin of the laboratory strain; and Sheila McCormick (PGEC, Albany, California) and John Armour (Nottingham University) for comments on the manuscript. This work was supported by grants from the Genetics Society (UK) and Nottingham University.
Literature Cited
- Atwell S., Huang Y. S., Vilhjalmsson B. J., Willems G., Horton M., et al. , 2010. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465: 627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown W. R. A., 1988. A physical map of the human pseudoautosomal region. EMBO J. 7: 2377–2385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown W. R. A., MacKinnon P. J., Villasante A., Spurr N., Buckle V. J., et al. , 1990. Structure and polymorphism of human telomere-associated DNA. Cell 63: 119–132 [DOI] [PubMed] [Google Scholar]
- Cockerham C. C., Weir B. S., 1984. Covariances of relatives stemming from a population undergoing mixed self and random mating. Biometrics 40: 157–164 [PubMed] [Google Scholar]
- Corander J., Marttinen P., Siren J., Tang J., 2008. Enhanced Bayesian modelling in BAPS software for learning genetic structures of populations. BMC Bioinformatics 9: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egel R. (Editor), 2003. The Molecular Biology of Schizosaccharomyces pombe. Springer-Verlag, Berlin [Google Scholar]
- Excoffier L., Laval G., Schneider S., 2005. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. Online 1: 47–50 [PMC free article] [PubMed] [Google Scholar]
- Fay J. C., Benavides J. A., 2005. Evidence for domesticated and wild populations of Saccharomyces cerevisiae. PLoS Genet. 1: 66–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florenzano G., W. Balloni, and R. Materasisi, 1977. Contributo alla acologia dei lieviti Schizosaccharomyces sulle uve. Vitis 16: 38–44 [Google Scholar]
- Fournier-Level A., Korte A., Cooper M. D., Nordborg M., Schmitt J., et al. , 2011. A map of local adaptation in Arabidopsis thaliana. Science 334: 86–89 [DOI] [PubMed] [Google Scholar]
- Gomes F. C., Pataro C., Guerra J. B., Neves M. J., Correa S. R., et al. , 2002. Physiological diversity and trehalose accumulation in Schizosaccharomyces pombe strains isolated from spontaneous fermentations during the production of the artisanal Brazilian cachaca. Can. J. Microbiol. 48: 399–406 [DOI] [PubMed] [Google Scholar]
- Gordon J. L., Byrne K. P., Wolfe K. H., 2011. Mechanisms of chromosome number evolution in yeast. PLoS Genet. 7: e1002190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig D., 2009. Reproductive isolation in Saccharomyces. Heredity 102: 39–44 [DOI] [PubMed] [Google Scholar]
- Johnson L. J., Koufopanou V., Goddard M. R., Hetherington R., Schafer S. M., et al. , 2004. Population genetics of the wild yeast Saccharomyces paradoxus. Genetics 166: 43–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Plagnol V., Hu T. T., Toomajian C., Clark R. M., et al. , 2007. Recombination and linkage disequilibrium in Arabidopsis thaliana. Nat. Genet. 39: 1151–1155 [DOI] [PubMed] [Google Scholar]
- Leupold U., 1950. Die Vererbung von Homothallie und Heterothallie bei Schizosaccharomyces pombe. Cr. Trav. Lab. Carlsberg Ser. Physiol. 24: 381–480 [Google Scholar]
- Librado P., Rozas J., 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452 [DOI] [PubMed] [Google Scholar]
- Liti G., Carter D. M., Moses A. M., Warringer J., Parts L., et al. , 2009. Population genomics of domestic and wild yeasts. Nature 458: 337–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Sung W., Morris K., Coffey N., Landry C. R., et al. , 2008. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc. Natl. Acad. Sci. USA 105: 9272–9277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F., Stone E. A., Ayroles J. F., 2009. The genetics of quantitative traits: challenges and prospects. Nat. Rev. Genet. 10: 565–577 [DOI] [PubMed] [Google Scholar]
- Nemri A., Atwell S., Tarone A. M., Huang Y. S., Zhao K., et al. , 2010. Genome-wide survey of Arabidopsis natural variation in downy mildew resistance using combined association and linkage mapping. Proc. Natl. Acad. Sci. USA 107: 10302–10307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieduszynski C. A., Liti G., 2011. From sequence to function: insights from natural variation in budding yeasts. Biochim. Biophys. Acta 1810: 959–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogami S., Ohya Y., Yvert G., 2007. Genetic complexity and quantitative trait loci mapping of yeast morphological traits. PLoS Genet. 3: e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M., Weigel D., 2008. Next-generation genetics in plants. Nature 456: 720–723 [DOI] [PubMed] [Google Scholar]
- Parts L., Cubillos F. A., Warringer J., Jain K., Salinas F., et al. , 2011. Revealing the genetic structure of a trait by sequencing a population under selection. Genome Res. 21: 1131–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., Stephens M., Donnelly P., 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N., Chen Z., Yassour M., Thompson D. A., Haas B. J., et al. , 2011. Comparative functional genomics of the fission yeasts. Science 332: 930–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruderfer D. M., Pratt S. C., Seidel H. S., Kruglyak L., 2006. Population genomic analysis of outcrossing and recombination in yeast. Nat. Genet. 38: 1077–1081 [DOI] [PubMed] [Google Scholar]
- Schacherer J., Shapiro J. A., Ruderfer D. M., Kruglyak L., 2009. Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature 458: 342–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. L., Matsumoto T., Niwa O., Klco S., Fan J. B., et al. , 1987. An electrophoretic karyotype for Schizosaccharomyces pombe by pulsed field gel electrophoresis. Nucleic Acids Res. 15: 4481–4489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todesco M., Balasubramanian S., Hu T. T., Traw M. B., Horton M., et al. , 2010. Natural allelic variation underlying a major fitness trade-off in Arabidopsis thaliana. Nature 465: 632–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torabi N., Kruglyak L., 2011. Variants in SUP45 and TRM10 underlie natural variation in translation termination efficiency in Saccharomyces cerevisiae. PLoS Genet. 7: e1002211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai I. J., Bensasson D., Burt A., Koufopanou V., 2008. Population genomics of the wild yeast Saccharomyces paradoxus: quantifying the life cycle. Proc. Natl. Acad. Sci. USA 105: 4957–4962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warringer J., Anevski D., Liu B., Blomberg A., 2008. Chemogenetic fingerprinting by analysis of cellular growth dynamics. BMC Chem. Biol. 8: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warringer J., Blomberg A., 2003. Automated screening in environmental arrays allows analysis of quantitative phenotypic profiles in Saccharomyces cerevisiae. Yeast 20: 53–67 [DOI] [PubMed] [Google Scholar]
- Warringer J., Zorgo E., Cubillos F. A., Zia A., Gjuvsland A., et al. , 2011. Trait variation in yeast is defined by population history. PLoS Genet. 7: e1002111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.