Abstract
Jak/STAT is an important signaling pathway mediating multiple events in development. We describe participation of metazoan co-activator SAYP/PHF10 in this pathway downstream of STAT. The latter, via its activation domain, interacts with the conserved core of SAYP. STAT is associated with the SAYP-containing co-activator complex BTFly and recruits BTFly onto genes. SAYP is necessary for stimulating STAT-driven transcription of numerous genes. Mutation of SAYP leads to maldevelopments similar to those observed in STAT mutants. Thus, SAYP is a novel co-activator mediating the action of STAT.
INTRODUCTION
Signaling pathways in metazoans orchestrate complex developmental events. This process usually requires elaborate transcription machinery controlling the expression of multiple genes. One of important pathways having a role in all metazoans is the Jak/Stat pathway. It has multiple functions, being responsible, in particular, for germ-cell function, morphogenesis and patterning, as well as for cell differentiation and proliferation (1). The final effector of the pathway is the family of STAT transcription activators (2). From a researcher's standpoint, Jak/Stat in Drosophila has an advantage of being simple: it consists of unique receptor dome, Janus kinase hop, and transcription factor STAT92E (below, referred to as STAT).
The recruitment of STAT onto chromatin occurs in cooperation with other factors (3). An important role in mediating the function of STAT in transcription activation is played by its C-terminal portion carrying the activation domain (3,4). Reliably identified co-activators for the STAT family are histone-modifying acetyltransferases CBP/p300 (5,6), the GCN5-containing complex (7) and chromatin-remodeling factor Brahma (8–11). In addition, some novel components of the pathway have been revealed among transcription factors (12,13). In particular, transcription factors Brahma, TFIID and SAYP have proved to be positive regulators of the pathway. SAYP was previously described as a transcription co-activator-mediating gene activation via a novel mechanism, by coupling chromatin remodeler Brahma and transcription initiation factor TFIID into one co-activator complex BTFly (14). SAYP is a conserved factor in metazoans. Its vertebrate homologue, named PHF10, shares with SAYP a conserved core consisting of the SAY domain an two PHD fingers (15).
Here, we describe the participation of SAYP in mediating STAT-driven transcription activation. Mutation in the gene encoding SAYP manifests itself similarly to those in the Jak/Stat pathway. Both SAYP and STAT co-occupy multiple loci in the genome. We have demonstrated the association of STAT with the SAYP-containing complex and revealed the domains mediating this interaction. The presence of SAYP is important for activation of STAT-dependent genes. As shown by ChIP analysis, SAYP is recruited onto STAT-dependent genes together with Brahma and TFIID.
MATERIALS AND METHODS
Experiments with S2 cell culture
Schneider cell line 2 (S2) of Drosophila were maintained at 25°C in Schneider's insect medium (Sigma) containing 10% FBS (HyClone). Conditions optimal for activation of STAT were determined experimentally. Pervanadate solution (PV) was prepared from sodium vanadate and hydrogen peroxide and then treated with catalase. Cells were treated with 100 μM PV for 2 h (for measuring mRNA level) or 30 min (for ChIP).
DNA fragments encoding SAYP with 3×FLAG epitope and STAT (form F, 761 amino acids) with HA epitope were cloned into pAc5.1/V5-HisB vector (Invitrogen). The cell line stably expressing tagged SAYP was established as described (14).
Antibodies and western blot analysis
Antibodies used in this study were described previously (14,16). Antibodies against STAT (261–456 amino acids fragment of form F) were raised in rabbits and affinity purified. These and other antibodies raised in our laboratory were used in a 1:500 dilution for western and in an amount of 5 μg for immunoprecipitation. Antibodies against fasciclin III, (obtained by C. Goodman) and beta-tubulin (obtained by M. Klymkowsky) were from the Developmental Studies Hybridoma Bank.
Genes expression analysis by reverse transcription–PCR
The following STAT-dependent genes were chosen for analysis: SOCS36E (17); dm [(Drosophila homologue of vertebrate STAT-dependent c-myc (18)]; buffy and debcl [Drosophila homologues of vertebrate STAT-dependent bcl-2 (19)]; slbo (20); eve (21); dpp (22); apontic (23); and DIAP1 (24).
For measuring gene expression, RNA was extracted with Trizol (Ambion) from five pairs of ovaries or 3 × 106 S2 cells and treated with DNase I. Reverse transcription (RT) was performed from random hexanucleotide primers and measured by qPCR. The sequences of the primers are given in the Supplementary Data. As a reference, we used the levels of actin and histone H1 mRNAs, which were stable upon PV treatment.
ChIP and Quantitative (q) PCR Analysis
The protocol for ChIP with S2 cells was described previously (25). As a negative control, measurements at rDNA and ChIP with nonspecific antibodies were used in each experiment, the signal in the latter case being at least 10 times weaker than in the former. The sequences of the primers are given in the Supplementary Data.
Immunostaining
Ovaries were stained as described (16). Polytene chromosomes were stained with rat anti-SAYP, rabbit anti-STAT, and the corresponding secondary antibodies (Molecular Probes) following the procedure described previously (16), fixation with 4% FA was performed for 2 min. Salivary glands were treated with 500 μM PV for 1 h.
Gel filtration of nuclear extract and immunoprecipitation
Preparation of the nuclear extract from Drosophila embryos, gel filtration, and immunoprecipitation were performed as described (14).
Drosophila genetic crosses
Cultivation of flies and genetic crosses were described previously (26). Females y2wae(y)3u1/FM4 and males carrying Stat92E mutation were selected for crossing. The mutation was caused by P-element insertion in the line 11681 (ry506 P{ry+t7.2=PZ}Stat92E06346/TM3, ryRK Sb1 Ser1). All genetic crosses were carried out at 25°C and repeated no less than three times. At least 50 flies of each viable genotype were screened for each strain.
RESULTS
Phenotypic manifestations of SAYP mutation
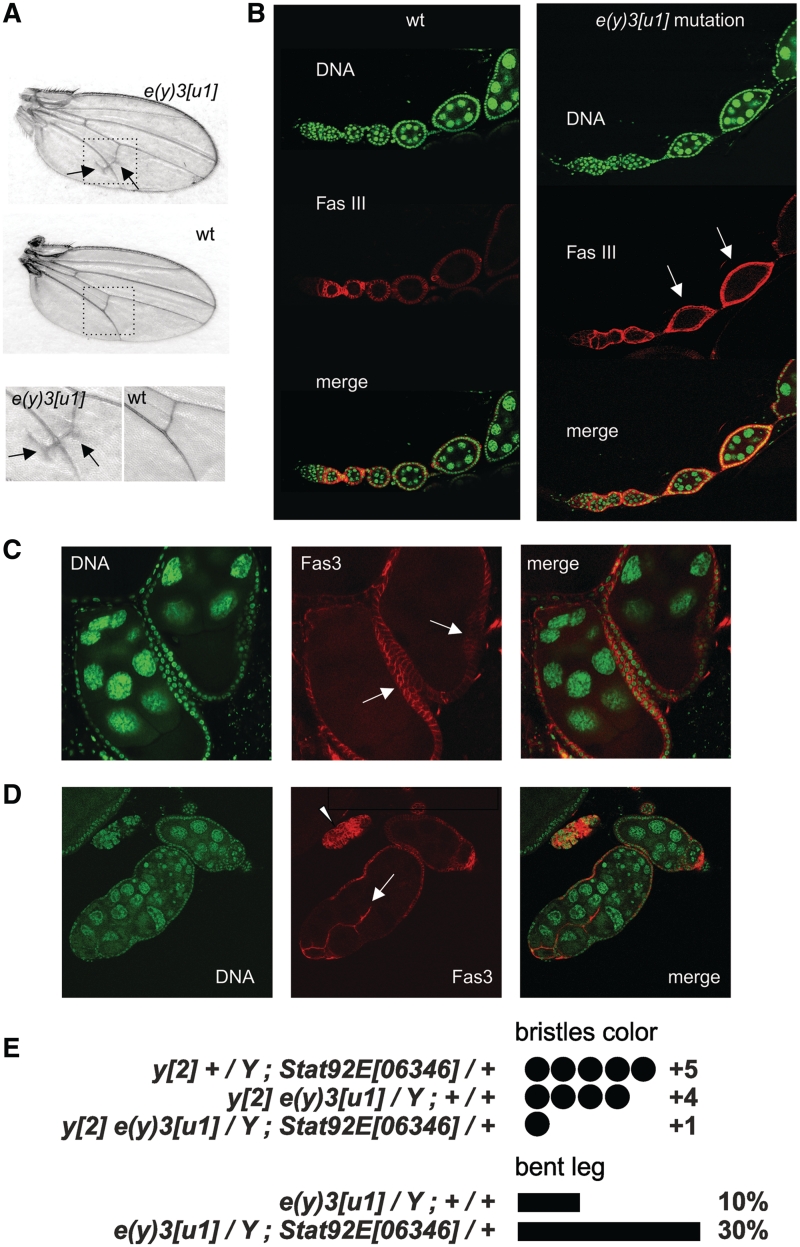
To check the possibility of SAYP participation in signaling pathways, we thoroughly analyzed the phenotype of flies with the e(y)3u1 mutation in the gene encoding SAYP (16). The main molecular manifestation of this hypomorphic mutation was a lower level of e(y)3 transcripts. Initially, e(y)3u1 was reported to suppress the expression of yellow2 allele in bristles (27,28). All flies homozygous for e(y)3u1 had lower viability, homozygous females were sterile, hemizygous males had a characteristic bent-leg phenotype and early embryos showed defects in cell cycle progression (14–16). In addition, the presence of ectopic longitudinal veins was revealed in the posterior wing blade (Figure 1A). Such a phenotype was observed in all flies carrying the e(y)3u1 allele in either homo- or hemizygous state. It is noteworthy in this context that STAT also regulates wing venation, with the hypomorphic mutation Stat92EHJ resulting in the formation of similar ectopic longitudinal veins (29).
Figure 1.
Manifestations of e(y)3u1 mutation. (A) Wing venation abnormalities in mutants, compared to wild-type (wt) flies. Arrows indicate the ectopic longitudinal vein material close to the posterior cross-vein. The bottom row shows images of the posterior cross-vein region at higher magnification. (B) Ovarioles from wild-type and homozygous mutant flies. Fas3-positive cells in wt flies are detected until stage 2, whereas in mutants they could be found in follicles of stages 3–4 (arrows). Staining with DAPI (green), Fas3 (red), and merged images are shown. (C) Egg chambers with several layers of follicle cells (arrows). (D) Fused follicles with invasive follicular epithelium (arrow) and an island of overproliferated follicular cells (arrowhead). (E) The level of bristle coloring and the frequency of bent leg phenotype in flies carrying the e(y)3 and Stat92E mutations. A combination of both mutations aggravated the mutant phenotype.
SAYP is abundant in various cells of the growing ovary, and females homozygous for e(y)3u1 are sterile (16). We checked the possible source of female sterility by inspecting the structure of ovaries in mutant flies. The ovary of an adult female consists of ovarioles, each representing an assembly line of developing ovarian follicles (egg chambers). Each follicle is covered by a monolayer of follicular cells and contains a pair of specific cells located at its anterior and posterior poles, named polar cells; adjacent follicles are connected by a column of stalk cells (30). Until Stage 2, epithelial follicular cells in the egg chambers express high levels of Fasciclin III (Fas3) (31), which marks immature proliferating follicular cells in the germarium. Progressive down-regulation of Fas3 takes place in the vitellarium after Stage 2, with the level of its expression in the stalk and polar cells remaining high.
Different degrees of disturbance were revealed in e(y)3u1 mutants. A mild mutant phenotype was characterized by maldevelopment of follicular cells: the number of Fas3-positive cells was increased, and they were detected in follicles of the vitellarium until stage 4–5 (Figure 1B). Moreover, they appeared to retain their proliferative potential, as follows from the fact that egg chambers with several layers of follicular cells and aggregates of these cells were detected (Figure 1C and D). In extreme cases, we observed the formation of fused follicles (cysts) at different stages of development, with epithelial cells within the cysts often showing disorganized, invasive growth (Figure 1D).
Jak/Stat plays an important role in follicle cells differentiation, and a similar phenotype of fused egg chambers has been reported for flies with hypomorphic mutations of hop (Janus kinase in Drosophila) and STAT92E (32,33).
Finally, we checked the effect of combining of mutation in e(y)3 and Stat92E. We observed that amorph Stat92E06346 in a heterozygous state enhances the phenotypic expression of the e(y)3u1 mutation, which is manifested in almost complete suppression of yellow2 in bristles and increased frequency of flies with the bent femur phenotype (Figure 1E). Stat92E is expressed at the stage of pupae and in larval imaginal discs (34,35) and, therefore, may have a role in the development of this phenotype.
Thus, we have found that mutations in the genes encoding SAYP and components of the Jak/Stat pathway manifest themselves similarly. However, the described phenotypes are not unique for this pathway, being also manifested in cases of mutations in other cascades (36–38). Moreover, the cross-talk of pathways in development (39) does not allow any unequivocal conclusion concerning the participation of SAYP in the Jak/STAT pathway. That is why we further checked the STAT–SAYP interaction at the molecular level.
STAT interacts with SAYP
To confirm the interaction of SAYP with STAT biochemically, antibodies against STAT were raised in rabbits, affinity purified, and their specificity was checked on both endogenous and recombinant tagged STAT (Figure 2A).
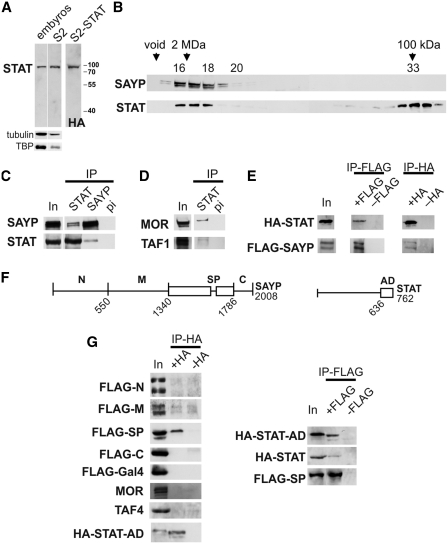
Figure 2.
STAT is associated with SAYP-containing complex. (A) Western blot analysis of total proteins from embryos or S2 cells with antibodies against STAT. To confirm their specificity, lysate of S2 cells expressing HA-tagged STAT was stained with anti-HA antibodies. Staining for tubulin and TBP is shown as a loading control. (B) Fractions after gel filtration of DNase I-treated nuclear extracts from embryos on Superose 6 were analyzed for the presence of SAYP and STAT proteins. STAT was detected not only as a free protein (fractions 32–34) but also in association with high-MW complexes, together with SAYP (fractions 16–17). (C) STAT and SAYP co-immunoprecipitated with each other from gel filtration fractions 16–17 of the extracts from embryos. IP with pre-immune IgG was used as a negative control. Equal portions of the input (In) and precipitated (IP) material were tested. (D) Antibodies against STAT co-immunoprecipitated MOR (component of Brahma) and TAF1 (component of TFIID) from gel filtration fractions 16–17. (E) Recombinant Flag-SAYP and HA-STAT co-immunoprecipitated with each other. Both proteins were co-expressed in cells, then IP with antibodies against either FLAG or HA was performed. In control IP, only one recombinant protein was expressed. Equal portions of the input (In) and precipitated (IP) material were tested. (F) Scheme of SAYP and STAT proteins. Boundaries of the tested regions and the total protein length (number of amino acids) are indicated. (G) Recombinant Flag-Gal4-fusions of SAYP fragments and the HA-tagged activation domain of STAT were tested for co-immunoprecipitation (the left column). Both proteins were co-expressed in cells, then IP with antibodies against HA was performed. In control IP, only Flag-tagged recombinant protein was expressed. Recombinant Flag-SP fragment co-immunoprecipitated HA-AD of STAT and HA-STAT (the right column). Equal amounts of the input (In) and precipitated (IP) material were tested.
Gel filtration of the nuclear extract from Drosophila embryos showed that the elution profiles of SAYP and STAT partially overlapped. SAYP migrated in fractions 16 and 17 (Figure 2B) as a component of high-molecular-weight protein complex BTFly including also Brahma and TFIID (14). A significant proportion of STAT was also found in these fractions. To confirm STAT–SAYP association in fractions 16–17, a co-immunoprecipitation experiment was performed. The results showed that anti-SAYP antibodies co-precipitated STAT, and vice versa (Figure 2C). Moreover, antibodies against STAT were capable of co-precipitating the components of Brahma and TFIID (Figure 2D), although to a lesser extent than SAYP.
To verify the SAYP–STAT interaction, both proteins with tags were co-expressed in cell culture. HA-tagged STAT was able to co-precipitate Flag-tagged SAYP, and vice versa (Figure 2E). It is noteworthy that the strength of this interaction was lower than that of SAYP–BAP170 and SAYP–TAF5 interactions in the stable BTFly complex (14), as could be expected for a transient activator–co-activator interaction.
To further study this interaction, we mapped the interacting domains within the proteins. Flag-Gal4-tagged separate domains of SAYP (described in ref. 14) and the HA-tagged C-terminal portion of STAT carrying the activation domain (AD) of STAT (40) were co-expressed in S2 cells (Figure 2F). As a result, a fairly strong interaction of STAT AD with the conserved SAY-PHD fragment of SAYP was revealed (Figure 2G).
We have previously demonstrated that the SAY domain interacts with TFIID and Brahma (14). However, the STAT AD has shown no comparable interaction with the subunits of endogenous TFIID and Brahma. Therefore, these factors are unlikely to mediate the observed STAT AD–SAY-PHD interaction. A relatively strong association of the two overexpressed proteins indicates that they probably interact with each other directly, although we cannot exclude that some other protein mediates their interaction in the lysate.
Thus, a portion of STAT is associated with the SAYP-containing protein complex, and STAT, via its activation domain, can interact with the conserved core of SAYP.
SAYP and STAT jointly control numerous genes
We assessed STAT–SAYP co-localization in the genome using preparations of polytene chromosomes from salivary glands. Both hop and STAT are expressed in this organ in larvae (35), indicating that the corresponding pathway is active. Co-immunostaining of chromosomes with antibodies against STAT and SAYP revealed a significant co-localization of these factors, although not complete (Supplementary Figure S1A). To further activate STAT in the salivary glands, they were treated with pervanadate (PV); in this case, the factors showed an even higher degree of co-localization (Figure 3, Supplementary Figure S1B). These results showed that STAT was present in multiple loci of euchromatin, which were often co-occupied by SAYP.
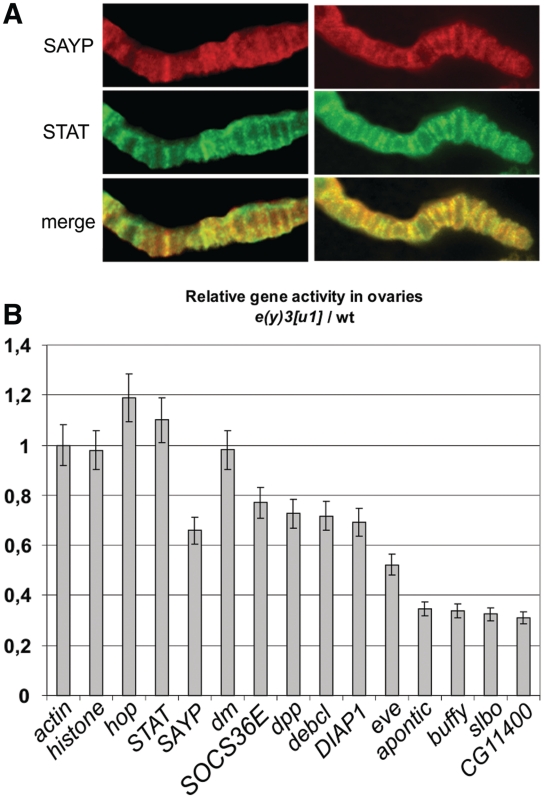
Figure 3.
SAYP controls multiple STAT-dependent genes. (A) Immunostaining of polytene chromosomes with antibodies against STAT and SAYP shows significant co-localization of these factors. (B) The levels of expression of different genes in the ovaries of homozygous e(y)3u1 mutants relative to those in wt flies, with the ratio for actin mRNA taken as 1. CG11400 is a SAYP-dependent gene (14) shown as a control.
To directly check the importance of SAYP for the activity of STAT-dependent genes in vivo, we used the ovaries of adult flies, in which the Jak/Stat pathway is active in many cells (32). Several genes were chosen which are known to be STAT-dependent (see ‘Materials and Methods’ section). Measurements of the level of gene expression showed that the reduced content of SAYP in hypomorphic e(y)3u1 mutants led to a drop in the expression of the STAT-dependent genes (Figure 3B). Importantly, the level of hop and STAT expression in these mutants was not reduced, indicating that SAYP acted downstream of STAT in the pathway.
We conclude that STAT and SAYP occur together on multiple sites genome-wide, with the presence of SAYP being important for the activity of STAT-dependent genes in the organism.
SAYP is recruited onto STAT-driven genes
To further investigate activation of STAT-dependent genes, we used Schneider (S2) cell line expressing the components of the Jak/Stat pathway (41). Western blot analysis confirmed that the level of the STAT protein (relative to the levels of tubulin and TBP) in these cells was markedly higher than in embryos (Figure 2A), while the embryo in Drosophila is the stage characterized by the highest STAT activity (35). Therefore, S2 cells are an appropriate model for studying the functioning of the pathway of interest. Treatment of S2 cells with PV, which causes the accumulation of phosphorylated STAT in them (42,43) is used to study short-term gene activation. Using this approach, we found that STAT-driven transcription took place in the untreated cell line and that PV treatment moderately stimulated STAT-dependent gene expression (see below).
The PV treatment for 2 hours resulted in induction of STAT-dependent genes, with their expression in S2 cells increasing several fold (Figure 4A) but the levels of STAT, SAYP, and BTFly components remaining unchanged (Supplementary Figure S2). To study the role of SAYP in this process, we changed its content in cells by either its RNAi knockdown or 5-fold overexpression (Supplementary Figure S3). The relative level of induction of STAT-dependent genes by PV proved to be the highest in cells overexpressing SAYP and the lowest in SAYP knockdown cells (Figure 4, Supplementary Figure S4). It should be noted that the observed changes in gene activation were not due to changes in the expression of hop and STAT in the cells (Supplementary Figure S3). Thus, SAYP has a positive effect on short-term induction of STAT-dependent genes in S2 cells.
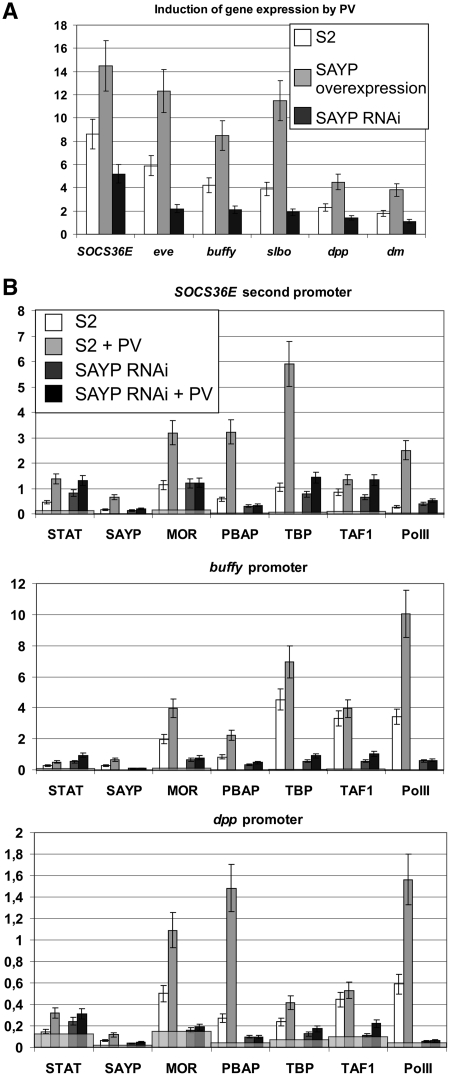
Figure 4.
SAYP is important for STAT-driven response in S2 cells. (A) The relative induction of different genes expression (the ratio of levels of expression after and before 2-h PV treatment) in normal S2 cells and cells with SAYP overexpression and SAYP knockdown. (B) The levels of different factors on promoters of genes before and after PV treatment in normal cells and cells with SAYP knockdown. The results of ChIP are shown as the percentage of input, gray staining shows the levels of the factors on rDNA.
We then directly checked whether SAYP is recruited onto STAT-dependent genes upon their activation by ChIP. After the PV treatment, an increase was observed in the contents of not only STAT but also of SAYP, TFIID and Brahma components and PolII on the promoters of several genes studied (Figure 4B).
To confirm the positive role of SAYP in this process, the recruitment of the above factors was measured upon SAYP knockdown. The results showed that the recruitment of STAT was not impaired, while the contents of TFIID, Brahma and especially PolII on the promoters dropped significantly. As shown in our previous study (14), SAYP knockdown did not affect the total content of TFIID and PolII but reduced the content of Brahma in the cells. Testing of the control promoters of housekeeping genes hsp70 (14) and actin (Supplementary Figure S5) upon SAYP knockdown showed that the recruitment of TFIID and PolII was not affected, while the level of Brahma was reduced to a lesser extent than on STAT-dependent genes. The increase of STAT signal after SAYP knockdown may be explained by higher accessibility of the STAT protein to antibodies on the promoter without co-activators.
The presence of SAYP and BTFly components on STAT-dependent genes was also checked by ChIP in early embryos (0–6 h), in which STAT is expressed at a high level (35). Since STAT was recently shown to be important for activation of numerous genes in the early Drosophila embryo (44), we checked several known targets for this protein. The contents of STAT and SAYP were found to be elevated relative to those on the promoters of housekeeping genes (Figure 5A).
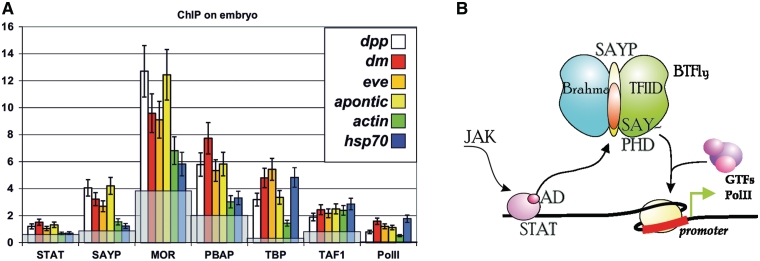
Figure 5.
SAYP is recruited onto STAT-dependent genes. (A) The presence of different factors on the promoters of indicated genes in early embryos (0–6 h). The results of ChIP are shown as the percentage of input, gray staining shows the levels of the factors on rDNA. (B) Model of SAYP participation in the Jak/Stat pathway: STAT, via its activation domain, mediates the recruitment of SAYP and associated BTFly complex, which is a prerequisite for PolII engagement.
Thus, SAYP is recruited onto STAT-dependent genes and has a role in inducing their expression. Its recruitment is also important for subsequent engagement of other co-activators and PolII.
DISCUSSION
We have analyzed the role of co-activator SAYP in the Jak/Stat pathway in Drosophila and found that SAYP interacts with STAT and mediates its activation potential (Figure 5B). SAYP operates as a component of large protein complex BTFly, which also contains Brahma and TFIID, and mediates subsequent recruitment of PolII onto the promoter (14). The results of polytene chromosome staining and measurement of multiple STAT-dependent genes testify to genome-wide cooperation of STAT and SAYP in gene activation.
The activation domain of STAT interacts with the SAY-PHD fragment of SAYP within the BTFly, and the STAT–BTFly association is not mediated by Brahma or TFIID. It is noteworthy that the interacting fragment of SAYP belongs to its conserved core, which is also found in vertebrate homologues of SAYP (16). Therefore, the above interaction may also take place in other species.
This finding broadens the known spectrum of transcription factors mediating the effect of STAT on gene expression. Such a diversity of cooperating factors appears to provide a basis for the specificity and strength of Jak/Stat-driven response in different cell types. In particular, BTFly may serve for rapid induction of transcription, as proposed previously (15). Indeed, we have observed the positive effect of SAYP content on short-term induction after STAT activation. SAYP is also important for PolII stalling (N.E.V., personal communication), which may provide for the precision of responses of target genes in the signaling pathway (44,45). One more point of interest is the putative role of SAYP in coordinating the crosstalk between Jak/Stat and other signaling pathways, which follows from the fact that SAYP is also involved in the ecdysone cascade (46) and probably in some other cascades (15).
Mutation of both STAT and SAYP leads to formation of excess numbers of ovarian follicular cells and ectopic wing veins. As shown in our previous study (15), SAYP is abundant in cells with high proliferative potential, and its mutation results in overproliferation of polar cells in embryos. Therefore, SAYP may participate in regulation of proliferation of certain cell types. Interestingly, PHF10—a vertebrate homologue of SAYP—is required for proliferation of stem/progenitor cells (47) and fibroblasts (48). On the other hand, it has been shown that STAT also has a role in proliferation and growth control (49). Therefore, it appears that SAYP and STAT are jointly involved in regulation of proliferation and differentiation of certain cell types during metazoan development.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–5 and Supplementary Materials.
FUNDING
Funding for open access charge: ‘Molecular and Cell Biology’ of the Russian Academy of Sciences; RFBR (10-04-00257 and 11-04-91976); Fellowship from University of Oslo, Centre for Medical Studies in Russia (to Y.S.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Nataliya Soshnikova and Anton Golovnin for their assistance in this study and to N.A. Gorgolyuk for his help in preparing the article.
REFERENCES
- 1.Li WX. Canonical and non-canonical JAK-STAT signaling. Trends Cell Biol. 2008;18:545–551. doi: 10.1016/j.tcb.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calo V, Migliavacca M, Bazan V, Macaluso M, Buscemi M, Gebbia N, Russo A. STAT proteins: from normal control of cellular events to tumorigenesis. J. Cell Physiol. 2003;197:157–168. doi: 10.1002/jcp.10364. [DOI] [PubMed] [Google Scholar]
- 3.Shuai K. Modulation of STAT signaling by STAT-interacting proteins. Oncogene. 2000;19:2638–2644. doi: 10.1038/sj.onc.1203522. [DOI] [PubMed] [Google Scholar]
- 4.Horvath CM. STAT proteins and transcriptional responses to extracellular signals. Trends Biochem. Sci. 2000;25:496–502. doi: 10.1016/s0968-0004(00)01624-8. [DOI] [PubMed] [Google Scholar]
- 5.Bhattacharya S, Eckner R, Grossman S, Oldread E, Arany Z, D'Andrea A, Livingston DM. Cooperation of Stat2 and p300/CBP in signalling induced by interferon-alpha. Nature. 1996;383:344–347. doi: 10.1038/383344a0. [DOI] [PubMed] [Google Scholar]
- 6.Zhang JJ, Vinkemeier U, Gu W, Chakravarti D, Horvath CM, Darnell JE., Jr Two contact regions between Stat1 and CBP/p300 in interferon gamma signaling. Proc. Natl Acad. Sci. USA. 1996;93:15092–15096. doi: 10.1073/pnas.93.26.15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paulson M, Press C, Smith E, Tanese N, Levy DE. IFN-stimulated transcription through a TBP-free acetyltransferase complex escapes viral shutoff. Nature Cell Biol. 2002;4:140–147. doi: 10.1038/ncb747. [DOI] [PubMed] [Google Scholar]
- 8.Huang M, Qian F, Hu Y, Ang C, Li Z, Wen Z. Chromatin-remodelling factor BRG1 selectively activates a subset of interferon-alpha-inducible genes. Nature Cell Biol. 2002;4:774–781. doi: 10.1038/ncb855. [DOI] [PubMed] [Google Scholar]
- 9.Ni Z, Karaskov E, Yu T, Callaghan SM, Der S, Park DS, Xu Z, Pattenden SG, Bremner R. Apical role for BRG1 in cytokine-induced promoter assembly. Proc. Natl Acad. Sci. USA. 2005;102:14611–14616. doi: 10.1073/pnas.0503070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wurster AL, Pazin MJ. BRG1-mediated chromatin remodeling regulates differentiation and gene expression of T helper cells. Mol. Cell Biol. 2008;28:7274–7285. doi: 10.1128/MCB.00835-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Cheng MB, Zhang YJ, Zhong X, Dai H, Yan L, Wu NH, Shen YF. A switch from hBrm to Brg1 at IFNgamma-activated sequences mediates the activation of human genes. Cell Res. 2010;20:1345–1360. doi: 10.1038/cr.2010.155. [DOI] [PubMed] [Google Scholar]
- 12.Baeg GH, Zhou R, Perrimon N. Genome-wide RNAi analysis of JAK/STAT signaling components in Drosophila. Genes Dev. 2005;19:1861–1870. doi: 10.1101/gad.1320705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muller P, Kuttenkeuler D, Gesellchen V, Zeidler MP, Boutros M. Identification of JAK/STAT signalling components by genome-wide RNA interference. Nature. 2005;436:871–875. doi: 10.1038/nature03869. [DOI] [PubMed] [Google Scholar]
- 14.Vorobyeva NE, Soshnikova NV, Nikolenko JV, Kuzmina JL, Nabirochkina EN, Georgieva SG, Shidlovskii YV. Transcription coactivator SAYP combines chromatin remodeler Brahma and transcription initiation factor TFIID into a single supercomplex. Proc. Natl Acad. Sci. USA. 2009;106:11049–11054. doi: 10.1073/pnas.0901801106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vorobyeva NE, Soshnikova NV, Kuzmina JL, Kopantseva MR, Nikolenko JV, Nabirochkina EN, Georgieva SG, Shidlovskii YV. The novel regulator of metazoan development SAYP organizes a nuclear coactivator supercomplex. Cell Cycle. 2009;8:2152–2156. doi: 10.4161/cc.8.14.9115. [DOI] [PubMed] [Google Scholar]
- 16.Shidlovskii YV, Krasnov AN, Nikolenko JV, Lebedeva LA, Kopantseva M, Ermolaeva MA, Ilyin YV, Nabirochkina EN, Georgiev PG, Georgieva SG. A novel multidomain transcription coactivator SAYP can also repress transcription in heterochromatin. EMBO J. 2005;24:97–107. doi: 10.1038/sj.emboj.7600508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karsten P, Hader S, Zeidler MP. Cloning and expression of Drosophila SOCS36E and its potential regulation by the JAK/STAT pathway. Mech. Dev. 2002;117:343–346. doi: 10.1016/s0925-4773(02)00216-2. [DOI] [PubMed] [Google Scholar]
- 18.Reed JC, Nowell PC, Hoover RG. Regulation of c-myc mRNA levels in normal human lymphocytes by modulators of cell proliferation. Proc. Natl Acad. Sci. USA. 1985;82:4221–4224. doi: 10.1073/pnas.82.12.4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weber-Nordt RM, Egen C, Wehinger J, Ludwig W, Gouilleux-Gruart V, Mertelsmann R, Finke J. Constitutive activation of STAT proteins in primary lymphoid and myeloid leukemia cells and in Epstein-Barr virus (EBV)-related lymphoma cell lines. Blood. 1996;88:809–816. [PubMed] [Google Scholar]
- 20.Starz-Gaiano M, Melani M, Meinhardt H, Montell D. Interpretation of the UPD/JAK/STAT morphogen gradient in Drosophila follicle cells. Cell Cycle. 2009;8:2917–2925. doi: 10.4161/cc.8.18.9547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harrison DA, McCoon PE, Binari R, Gilman M, Perrimon N. Drosophila unpaired encodes a secreted protein that activates the JAK signaling pathway. Genes Dev. 1998;12:3252–3263. doi: 10.1101/gad.12.20.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopez-Onieva L, Fernandez-Minan A, Gonzalez-Reyes A. Jak/Stat signalling in niche support cells regulates dpp transcription to control germline stem cell maintenance in the Drosophila ovary. Development. 2008;135:533–540. doi: 10.1242/dev.016121. [DOI] [PubMed] [Google Scholar]
- 23.Starz-Gaiano M, Melani M, Wang X, Meinhardt H, Montell DJ. Feedback inhibition of Jak/STAT signaling by apontic is required to limit an invasive cell population. Dev. Cell. 2008;14:726–738. doi: 10.1016/j.devcel.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 24.Betz A, Ryoo HD, Steller H, Darnell JE., Jr STAT92E is a positive regulator of Drosophila inhibitor of apoptosis 1 (DIAP/1) and protects against radiation-induced apoptosis. Proc. Natl Acad. Sci. USA. 2008;105:13805–13810. doi: 10.1073/pnas.0806291105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kopytova DV, Orlova AV, Krasnov AN, Gurskiy DY, Nikolenko JV, Nabirochkina EN, Shidlovskii YV, Georgieva SG. Multifunctional factor ENY2 is associated with the THO complex and promotes its recruitment onto nascent mRNA. Genes Dev. 2010;24:86–96. doi: 10.1101/gad.550010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Georgiev PG, Gerasimova TI. [Detection of new genes participating in the trans-regulation of locus yellow of MDG-4 in Drosophila melanogaster] Genetika. 1989;25:1409–1419. [PubMed] [Google Scholar]
- 27.Georgiev PG, Gerasimova TI. Novel genes influencing the expression of the yellow locus and mdg4 (gypsy) in Drosophila melanogaster. Mol. Gen. Genet. 1989;220:121–126. doi: 10.1007/BF00260865. [DOI] [PubMed] [Google Scholar]
- 28.Kozitsina MV, Abramova NA, Georgiev PG. [Interaction between the enhancers of yellow and the yellow gene in Drosophila melanogaster] Genetika. 1992;28:59–67. [PubMed] [Google Scholar]
- 29.Yan R, Luo H, Darnell JE, Jr, Dearolf CR. A JAK-STAT pathway regulates wing vein formation in Drosophila. Proc. Natl Acad. Sci. USA. 1996;93:5842–5847. doi: 10.1073/pnas.93.12.5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spradling AC. In: The Development of Drosophila melanogaster. Bate M, Arias AM, editors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 1–70. [Google Scholar]
- 31.Wu X, Tanwar PS, Raftery LA. Drosophila follicle cells: morphogenesis in an eggshell. Semin. Cell Dev. Biol. 2008;19:271–282. doi: 10.1016/j.semcdb.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGregor JR, Xi R, Harrison DA. JAK signaling is somatically required for follicle cell differentiation in Drosophila. Development. 2002;129:705–717. doi: 10.1242/dev.129.3.705. [DOI] [PubMed] [Google Scholar]
- 33.Baksa K, Parke T, Dobens LL, Dearolf CR. The Drosophila STAT protein, stat92E, regulates follicle cell differentiation during oogenesis. Dev. Biol. 2002;243:166–175. doi: 10.1006/dbio.2001.0539. [DOI] [PubMed] [Google Scholar]
- 34.Bach EA, Ekas LA, Ayala-Camargo A, Flaherty MS, Lee H, Perrimon N, Baeg GH. GFP reporters detect the activation of the Drosophila JAK/STAT pathway in vivo. Gene Exp. Patterns. 2007;7:323–331. doi: 10.1016/j.modgep.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 35.Daines B, Wang H, Wang L, Li Y, Han Y, Emmert D, Gelbart W, Wang X, Li W, Gibbs R, et al. The Drosophila melanogaster transcriptome by paired-end RNA sequencing. Genome Res. 2011;21:315–324. doi: 10.1101/gr.107854.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blair SS. Wing vein patterning in Drosophila and the analysis of intercellular signaling. Annu. Rev. Cell Dev. Biol. 2007;23:293–319. doi: 10.1146/annurev.cellbio.23.090506.123606. [DOI] [PubMed] [Google Scholar]
- 37.Fernandez-Minan A, Martin-Bermudo MD, Gonzalez-Reyes A. Integrin signaling regulates spindle orientation in Drosophila to preserve the follicular-epithelium monolayer. Curr. Biol. 2007;17:683–688. doi: 10.1016/j.cub.2007.02.052. [DOI] [PubMed] [Google Scholar]
- 38.Matakatsu H, Tadokoro R, Gamo S, Hayashi S. Repression of the wing vein development in Drosophila by the nuclear matrix protein plexus. Development. 1999;126:5207–5216. doi: 10.1242/dev.126.23.5207. [DOI] [PubMed] [Google Scholar]
- 39.Callus BA, Mathey-Prevot B. SOCS36E, a novel Drosophila SOCS protein, suppresses JAK/STAT and EGF-R signalling in the imaginal wing disc. Oncogene. 2002;21:4812–4821. doi: 10.1038/sj.onc.1205618. [DOI] [PubMed] [Google Scholar]
- 40.Karsten P, Plischke I, Perrimon N, Zeidler MP. Mutational analysis reveals separable DNA binding and trans-activation of Drosophila STAT92E. Cell Signal. 2006;18:819–829. doi: 10.1016/j.cellsig.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 41.Cherbas L, Willingham A, Zhang D, Yang L, Zou Y, Eads BD, Carlson JW, Landolin JM, Kapranov P, Dumais J, et al. The transcriptional diversity of 25 Drosophila cell lines. Genome Res. 2011;21:301–314. doi: 10.1101/gr.112961.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sweitzer SM, Calvo S, Kraus MH, Finbloom DS, Larner AC. Characterization of a Stat-like DNA binding activity in Drosophila melanogaster. J. Biol. Chem. 1995;270:16510–16513. doi: 10.1074/jbc.270.28.16510. [DOI] [PubMed] [Google Scholar]
- 43.Shi S, Larson K, Guo D, Lim SJ, Dutta P, Yan SJ, Li WX. Drosophila STAT is required for directly maintaining HP1 localization and heterochromatin stability. Nat. Cell Biol. 2008;10:489–496. doi: 10.1038/ncb1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsurumi A, Xia F, Li J, Larson K, LaFrance R, Li WX. STAT is an essential activator of the zygotic genome in the early Drosophila embryo. PLoS Genet. 2011;7:e1002086. doi: 10.1371/journal.pgen.1002086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Levine M. Paused RNA polymerase II as a developmental checkpoint. Cell. 2011;145:502–511. doi: 10.1016/j.cell.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vorobyeva NE, Nikolenko JV, Krasnov AN, Kuzmina JL, Panov VV, Nabirochkina EN, Georgieva SG, Shidlovskii YV. SAYP interacts with DHR3 nuclear receptor and participates in ecdysone-dependent transcription regulation. Cell Cycle. 2011;10:1821–1827. doi: 10.4161/cc.10.11.15727. [DOI] [PubMed] [Google Scholar]
- 47.Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, Wu H, Aebersold R, Graef IA, Crabtree GR. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Banga SS, Peng L, Dasgupta T, Palejwala V, Ozer HL. PHF10 is required for cell proliferation in normal and SV40-immortalized human fibroblast cells. Cytogenet. Genome Res. 2009;126:227–242. doi: 10.1159/000251960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.