Abstract
A large body of literature provides compelling evidence for the role of evolutionarily conserved core histone residues in various biological processes. However, site-directed mutagenesis of individual residues that are known to be sites of posttranslational modifications often does not result in clear phenotypic defects. In some cases, the combination of multiple mutations can give rise to stronger phenotypes, implying functional redundancy between distinct residues on histones. Here, we examined the “histone redundancy hypothesis” by characterizing double deletion of all pairwise combinations of amino-terminal tails (N-tails) from the four core histones encoded in budding yeast. First, we found that multiple lysine residues on the N-tails of both H2A and H4 are redundantly involved in cell viability. Second, simultaneous deletion of N-tails from H2A and H3 leads to a severe growth defect, which is correlated with perturbed gross chromatin structure in the mutant cells. Finally, by combining point mutations on H3 with deletion of the H2A N-tail, we revealed a redundant role for lysine 4 on H3 and the H2A N-tail in hydroxyurea-mediated response. Altogether, these data suggest that the N-tails of core histones share previously unrecognized, potentially redundant functions that, in some cases are different from those of the widely accepted H2A/H2B and H3/H4 dimer pairs.
Keywords: acetylation, methylation
The basic repeating unit of chromatin is a nucleosome, consisting of ≈147 base pairs of DNA wrapped around an octamer consisting of two copies of each of the four core histone proteins, H2A, H2B, H3, and H4. This octamer is formed by pairwise associations of the histones, a more stable [H3:H4]2 tetramer (tetrasome) bound by two H2A:H2B dimers (1). Core histones represent some of the most evolutionarily conserved proteins and can be structurally divided into two structural configurations—well-structured histone-fold domains that govern histone:histone and histone:DNA interactions and unstructured amino-terminal tails (N-tails) protruding from the surface of nucleosomal core (1).
Regulation of posttranslational modifications (PTMs) on histones is one of the well-documented mechanisms to modulate chromatin structure and function. Histone N-tails are well endowed with an extensive array of multiple potentially modifiable residues (Fig. 1A; ref. 2). A number of studies have shown that specific modifications within these N-tails are associated with diverse DNA-templated processes. Site-specific acetylation (ac) on the N-tails of newly synthesized H3 and H4, for example, is correlated with DNA replication-dependent chromatin assembly (3, 4). However, methylation (me) on lysine 4 of H3 (H3K4) in concert with acetylation of lysines within the N-tail of H4 is strongly correlated with transcriptional activation, whereas methylation of other lysines, such as H3K9 and H3K27, are linked to silencing in many organisms (5). Furthermore, phosphorylation (ph) of serine 10 and 28 of H3 (H3S10 and H3S28) are enriched in mitotic cells and H4S1ph is induced in response to DNA double-strand breaks in yeast cells (6–8).
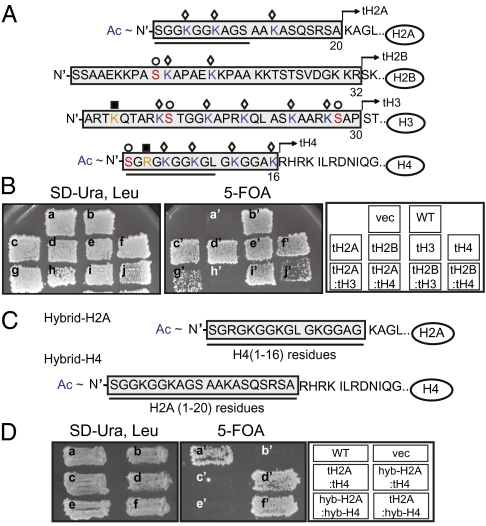
Fig. 1.
Amino-terminal tails (N-tails) from H2A and H4 play a redundant role in cell viability. (A) Amino acid sequences within N-tails of yeast core histones, H2A, H2B, H3, and H4. Residues deleted from each histone are in shaded boxes. Each tailless histone lacking N-tail is designated as tH2A, tH2B, tH3, and tH4. Identified modifications in yeast are marked with the following symbols: ◇, acetylation on lysines (blue); ■, methylation on lysines and arginines (orange); ○, phosphorylation on serines (red). Acetylation on N-termini of H2A and H4 are marked as Ac. The sequences displaying strong similarity between the H2A and H4 N-tails are underlined. (B) Viability of histone shuffle strains with tailless histones. Cells on the SD-Ura, Leu plate contain tailless histones empty vector alone (vec) (a), wild-type H2A/H2B/H3/H4 genes (WT) (b), tH2A/H2B/H3/H4 (tH2A) (c), H2A/tH2B/H3/H4 (tH2B) (d), H2A/H2B/tH3/H4 (tH3) (e), H2A/H2B/H3/tH4 (tH4) (f), tH2A/H2B/tH3/H4 (tH2A:tH3) (g), tH2A/H2B/H3/H4 (tH2A:tH4) (h), tH2A/H2B/tH3/H4 (tH2B:tH3) (i), H2A/tH2B/H3/tH4 (tH2B/tH4) (j) as well as wild-type counterparts in separate plasmids. Cells on the 5-FOA plate lacking wild-type histone genes in the URA3 plasmid were marked with a′–j′. (C) Amino acid sequences of N-terminal residues of hybrid H2A and hybrid H4. Residues replaced for hybrid histones are underlined and in shaded boxes. (D) Viability of histone shuffle strains. Cells on the SD-Ura, Leu plate carry mutant histones: wild-type H2A/H2B/H3/H4 genes (WT) (a), empty vector alone (vec) (b), tH2A/H2B/H3/tH4 (tH2A:tH4) (c), hybrid-H2A/H2B/H3/tH4 (hyb-H2A:tH4) (d), hybrid-H2A/H2B/H3/hybrid-H4 (hyb-H2A:hyb-H4) (e), tH2A/H2B/H3/hybrid-H4 (tH2A:hyb-H4) (f). Cells on the 5-FOA plate lacking wild-type histone genes in the URA3 plasmid were marked with a′–f′.
However, despite the intimate correlation of the above-mentioned modifications with various aspects of nuclear function, individual mutations of these residues often do not display prominent phenotypes in cells. For example, neither the alanine substitution mutation of H3S10 or of H3S28 nor the double mutation at these sites exhibit clear mitotic defects in budding yeast or in Xenopus (9–11). Similarly, cells containing a mutation of any single lysine on the H4 N-tail do not display an obvious DNA replication or chromatin assembly defect (12, 13), and prominent transcriptional phenotypes, with the exception of H4K16 (10, 14). Furthermore, recent systematic mutagenesis studies demonstrate that, despite the extremely well conserved nature of histone residues throughout different organisms, only a few mutations on the individual residues (including nonmodifiable sites) bring about prominent phenotypic defects (10, 15, 16).
One possible explanation for the lack of clear phenotypes by single mutations is a functional redundancy of multiple residues in a given biological process. Previous studies showing more striking phenotypes caused by combinations of different histone mutations lend support to this idea. For example, the lethal phenotype caused by quadruplet mutations of all four lysine residues (H4K5,8,12,16) within the H4 N-tail, which cannot be recapitulated with any combination of triple mutations on the lysines, indicates the redundancy of all four lysine residues for cell viability (17). Furthermore, triple mutations on lysines within H4 N-tail (H4K5,8,12) in combination with deletion of H3 N-tail, which carries five acetylatable lysines (H3K9,14,18,23,27), causes cellular lethality (12). Similarly, double deletion of the H2A/H2B or of the H3/H4 N-tails is lethal (18, 19). These results suggest redundant roles for multiple residues both within and between histone N-tails.
The elegant genetic studies cited above have largely been focused on cooperative and/or overlapping roles for N-tails of either H2A/H2B or H3/H4 pairs, likely because of their well-known, pairwise association during nucleosome assembly/disassembly and structural features of assembled nucleosomal octamers (1, 20). Interestingly, however, sequence similarity exists on the N-tails of H2A and H4, notably the extreme amino-terminal residues are nearly identical between the two (underlined in Fig. 1A). Moreover, acetylation of both H2A and H4 N-tails in yeast can be mediated by the same histone acetylatransferase (HAT) complex, NuA4 (21), whereas, in contrast, H2B and H3 are acetylated by another HAT complex, SAGA (22, 23). Interestingly, in mammalian cells, methylation of arginine 3 in the H4 tail (H4R3) is a preferred site of the PRMT1 methyltransferase, but if this site is mutated to glutamine (H4R3Q), methylation switches to R3 in the H2A tail (24). Collectively, these observations suggest that N-tails of “nonconventionally” paired histones, such as H2A/H4 and H2B/H3, could play redundant functions in a given biological context, perhaps as targets of common enzyme systems. These reports led us hypothesize that the modest phenotype by a single mutation on histone residues may be attributed to functional redundancy of different histone residues: “histone redundancy hypothesis.”
In this study, we sought to determine whether redundant or cooperative roles are played by the N-tails of histones in different combinations in addition to the better-studied H2A/H2B or H3/H4 pairs. To address this question, we characterized mutant yeast cells either lacking histone N-tails in various combinations or carrying substitution mutations within relevant tails. First, we show that double deletion of N-tails from H2A and H4 and simultaneous mutations on lysines on both the H2A and the H4 N-tails is lethal. This result alone underscores the point that certain N-tail elements may provide redundant functions that lie outside of the more conventional and widely accepted view of their roles in chromatin assembly and nucleosome structure (e.g., H3 with H4). We also find that cells lacking double N-tails from H2A and H3 are defective in cell growth without displaying dramatic cell cycle defects and display defects in global chromatin structure. Lastly, we demonstrate that H3K4 and H2A N-tails are involved in hyrdoxyurea (HU)-mediated checkpoint activation in a redundant manner. Taken together, our study reveals potential redundant functions across novel pairwise histone N-tail combinations, lending support to the histone redundancy hypothesis.
Results and Discussion
Acetylatable Lysines on the N-Tails of both H2A and H4 Play Redundant Roles in Cell Viability.
To investigate potential redundant functions for the N-tail domains of the four core histones, we constructed histone-shuffle strains carrying tailless core histones in different combinations (Fig. 1A and Fig. S1A; ref. 25). Viability of histone tail-delete mutant cells was then examined by growing the histone shuffle strains under selective conditions where wild-type histone genes were lost (25). Consistent with previous literature (18, 25, 26), cells lacking an N-tail from each single histone (designated as tH2A, tH2B, tH3, and tH4) were viable (Fig. 1B, c′–f′). In addition, cells lacking two N-tails from the following pairs of histones, H2A/H3 (tH2A:tH3), H2B/H3 (tH2B:tH3), and H2B/H4 (tH2B:tH4), were viable, although the growth of tH2A:tH3 and tH2B:tH4 cells under selective conditions was slower than those of other mutant cells (Fig. 1B, g′, i′, and j′). By contrast, and quite unexpectedly, tH2A:tH4 double-tail deletion mutant cells were inviable (Fig. 1B, h′). We note that high copy expression of tailless H2A and H4, genes of which were delivered into cells by 2μ plasmid, did not restore cellular viability (Fig. S1B); ruling out the possibility that reduced levels of tailless H2A and H4 as the sole cellular H2A and H4 might cause the lethality of tH2A:tH4 mutant cells. These results suggest that the N-tails of H2A and H4 are redundantly involved in cell viability, the underlying cause of which is unclear.
The N-tails of H2A and H4, especially the first 10 residues from the N terminus (underlined in Fig. 1A), exhibit strong sequence similarity to each other. Previous studies showed that lysine residues embedded in the N-tails of H2A (K4, K7, K13) and H4 (K5, K8, K12) can be acetylated by common enzymatic activities such as the NuA4 complex in vitro (21, 27). Importantly, not only acetylated H4 but also at least two acetylated lysines of H2A (H2AK4ac and H2AK7ac) have been detected in yeast cells by mass spectrometry analyses (28). To determine whether acetylatable lysines within the N-tails of H2A and H4 are redundantly involved in cell viability, we combined triple lysine point mutations of H2A (H2AK4,7,13R: H2A-3KR) either with triple-site point mutations of H4 (H4K5,8,12R: H4-3KR) or with truncation of H4 N-tail (tH4). As with the double-tail deletion, either group of combinatorial mutations were lethal (Fig. S1C, f′ and g′). This result implies that the lysine residues within the N-tails of H2A and H4 share an essential function for cell viability that likely involves acetylation of the lysines. To determine further which specific lysine residues on the H2A and H4 N-tails played the redundant role in cell viability, we generated different sets of combinatory mutations on the lysines. However, none of the other combinatorial mutations recapitulated the lethal phenotype caused by H2A-3KR:H4-3KR mutation (Table S1). These results demonstrate that all six lysine residues on H2A (K4,7,13) and H4 (K5,8,12) are functionally redundant to maintain cell viability. Independently, Altaf et al. (27) reported that H2A-3KQ (H2AK4,7,13Q) combined with H4-3KQ (H4K5,8,12Q) also resulted in cellular lethality. Taken together, we postulate that the lethality caused by simultaneous mutations on lysines of H2A and H4 N-tails is likely due to the lack of acetylation on the N-tails of H2A or H4. It remains to be determined which specific biological process requires the essential histone acetylation for viability. Unlike H4, acetylation of lysines on the H2A N-tail is less well-linked to defined biological processes, perhaps due to their lower abundance. However, our genetic analysis with combinatorial mutations within the H2A and H4 tails clearly revealed that mutations of acetylatable lysines on the H2A N-tail can be buffered by acetylatable lysines within the H4 N-tail, which share a function in cell viability, highlighting the biological importance of acetylated H2A N-tail. An alternative possibility, namely that the lysines themselves, rather than their acetylation, are critical for viability is also plausible.
Given the uncovered redundancy of H2A and H4 N-tails, we next wondered whether fusing the acetylatable H2A N-tail to a tailless H4 could rescue viability or vice versa. To address this question, we generated yeast strains carrying hybrid-H2A or -H4, such that the H4 N-tail was fused to tailless H2A and the H2A N-tail was fused to tailless H4 (Fig. 1C). Cells carrying a pair of hybrid-H2A/tH4 or of hybrid-H4/tH2A were viable (Fig. 1D, d′ and f′). Surprisingly, however, the copresence of hybrid-H2A and hybrid-H4 resulted in the cellular lethality (Fig. 1D, e′). This result indicates that, although the N-tails of either H2A or H4 can be interchanged to retain cell viability, simultaneous mispositioning of both N-tails is not tolerable in cells. Therefore, we suggest that not only the presence of histone N-tails, but also the spatial context of the tails in chromatin, have implications for biological processes.
Double Deletion of the Histone N-Tails Uncovers Unidentified Phenotypes.
To uncover other possible redundancy of pairwise N-tails of the four core histones in specific biological processes beyond cellular viability, we sought to explore different genetic assays to search for potential phenotypic defects in the viable tail-delete mutant cells. First, we characterized the growth of viable single- and double-delete N-tail mutant cells under different temperatures. The doubling time of mutant cells in nutrient-rich media (YPD) at 30 °C showed that tH2A:tH3 cells grew the most slowly among the mutant and wild-type cells (Fig. 2A and Table S2). When grown under different temperatures, all mutants except tH2B cells displayed growth defects to variable degrees. Among them, tH2A:tH3 cells exhibit the most striking and synergistic sensitivities to both high (37 °C) and low (16 °C) temperatures compared with that of either single tail-delete tH2A or tH3 cells (Fig. 2A), demonstrating a redundant role for N-tails of H2A and H3 in cell growth.
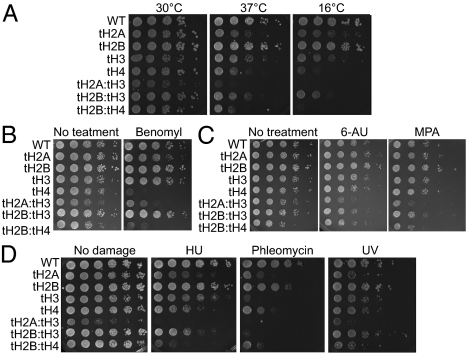
Fig. 2.
N-tails of core histones function in diverse biological processes in a redundant manner. (A) Histone tail deletion mutant strains were grown to exponential phase. Ten-fold serially diluted cells were spotted onto YPD and grown at different temperatures, 16 °C, 30 °C, and 37 °C. (B) Different histone tail deletion mutant cells were tested for benomyl sensitivity by spotting serially diluted cells on YPD containing 10 μg/mL benomyl. (C) Serially diluted, different histone tail deletion mutant cells were spotted on YPD containing 10 μg/mL 6-AU or 10 μg/mL MPA. (D) DNA damage sensitivities of histone tail deletion mutant cells were tested by spotting serially diluted cells on YPD containing 100 mM HU and 1 μg/mL phleomycin. To test UV sensitivity, a plate with spotted cells was irradiated with 2 J/m2. The photos were taken 3–4 d after spotting.
Second, we checked the sensitivity of mutant cells to benomyl, a microtubule depolymerizing reagent, to assay for mitotic defects of the mutant cells. Consistent with the previous report that cells lacking the N-tail of H4 displayed hypersensitivity to benomyl because of mitotic defects (28), both tH4 and tH2B:tH4 cells were hypersensitive to benomyl in our assays (Fig. 2B). In addition, double deletion of the H2A and H3 N-tails also showed benomyl hypersensitivity, whereas the other single tail deletes (tH2A, tH2B, and tH3) and tH2B:tH3 cells were not benomyl-sensitive under the same condition. This result suggests a redundant involvement of N-tails from H2A and H3 in mitotic processes, which is not shared with N-tail of H2B.
Third, we determined the sensitivities of viable histone tail deletes mutant cells in response to 6-azauracil (6-AU) and mycophenolic acid (MPA), which perturb transcriptional elongation by depleting the intracellular level of ribonucleotides (10). Although none of the single-tail deletion mutant cells displayed sensitivities, double-tail deletes, tH2A:tH3 and tH2B:tH3 cells were sensitive to 6-AU and MPA, moderately and slightly, respectively (Fig. 2C). We note that, consistent with the observation above (Fig. 1B), tH2B:tH4 cells displayed growth defect on synthetic media used in this assay even without 6-AU or MPA. We speculate that the gene expression patterns in tH2B:tH4 cells under restricted nutrients may be different from those in nutrient-rich YPD media, affecting the cell growth. However, no further growth defect in tH2B:tH4 cells was observed in the presence of 6-AU or MPA, demonstrating that tH2B:tH4 cells were not sensitive to transcriptional elongation inhibitors. This result raises a possibility that chromatin regulation during transcriptional elongation in cells is defective in the simultaneous absence of N-tails either from H2A/H3 or from H2B/H3.
Lastly, we examined whether double deletion of histone N-tails mediates the sensitivity of mutant cells to diverse DNA damaging reagents. Single deletion of the N-tail from each histone mediated cellular sensitivities to HU, phleomycin, and UV irradiation (UV) to varying extents, consistent with previous reports (Fig. 2D; refs. 10 and 29). tH2B:tH3 cells showed more sensitivity to HU than either single tail-delete mutant, tH2B, or tH3 cells, whereas the sensitivity of tH2B:tH3 cells to phleomycin and UV was similar to that of tH3 cells. However, tH2A:tH3 cells exhibited the dramatically enhanced sensitivities to all three reagents compared with either single tail deletes. Therefore, these data suggest that the H2A N-tail is involved in DNA damage response in a redundant manner with that of H3. In addition, because tH2B:tH4, but not tH2B:tH3 cells, exhibited more enhanced sensitivity to all three damaging reagents than each single tail deletes, we postulate that the H2B N-tail can compensate for the loss of H4 N-tail but not of the H3 N-tail in the damage-induced response. Altogether, the phenotypic characterizations above revealed roles for histone N-tails in different biological processes (summarized in Table S3).
Global Chromatin Structure Is Altered in the Double N-Tail Delete tH2A:tH3 Cells.
Because tH2A:tH4 cells are not viable, it is difficult to examine further the mechanistic nature of a potentially redundant function(s) for the N-tails of H2A and H4. Instead, we sought to investigate further the redundancy between the H2A and H3 N-tails in cell growth and diverse stress sensitivities. To examine whether modifiable residues within the N-tails of H2A and H3 play redundant roles in cell growth, we first used the H2A-3KR (K4,7,13R) mutant cells discussed previously. Combining this mutant with five simultaneous mutations on acetylatable lysines within H3 N-tail (H3K9,14,18,23,27R: H3-5KR) did not recapitulate growth defects observed in tH2A:tH3 cells. Moreover, mutating methylatable H3K4 to alanine (H3K4A) in combination of H2A-3KR and H3-5KR did not aggravate the growth of mutant cells (Fig. S2A). Therefore, it is unlikely that the absence of specific modifications on H2A and H3 N-tails, notably acetylation and methylation, leads to slow cell growth phenotypes seen when both these N-tails are missing.
Intriguingly, tH2A:tH3 cells, but not tH2B:tH3 cells, displayed abnormally elongated cell shapes, which is reminiscent of the morphology of the H4 N-tail deletion mutant cells, cell cycle defects in which are known to mediate the growth defect (17). We therefore wondered whether the slow growth of tH2A:tH3 cells was also correlated with cell cycle defects, like that of the H4 N-tail delete mutant cell. However, the cell cycle distribution of asynchronous tH2A:tH3 mutant cells was not dramatically different from wild-type cells (Fig. S2B).
Because tH2A:tH3 mutant cells were defective in growth under various conditions, we next wondered whether cells lacking the N-tails from both H2A and H3 could have a defect in global chromatin structure, which might potentially lead to problems in multiple chromatin-related processes. To determine gross chromatin structure, we isolated nuclei from tH2A:tH3 and wild-type cells and performed micrococcal nuclease (MNase) digestion. The MNase-digested chromatin from tH2A:tH3 cells exhibited a less distinct nucleosomal “ladder” (i.e., more smeared background) than that from wild-type cells (Fig. 3 A and B), which suggests that this mutant has a more open chromatin structure.
Fig. 3.
Double N-tail delete, tH2A:tH3 mutant cells have more open chromatin structure. (A) Bulk chromatin from wild-type and tH2A:tH3 cells was digested with different amounts of MNase (lanes 1 and 5, 0 U; lanes 2 and 6, 1 U; lanes 3 and 7, 2 U; lanes 4 and 8, 4 U) and visualized with ethidium bromide on agarose gel. M, DNA ladders; U, relative units of Mnase. (B) Densitometric analysis of MNase-digested DNA shown in A was measured by plotting profile with Image J (National Center for Biotechnology Information, NCBI). Dashed lines mark mono- (n1), di- (n2), and tri- (n3) nucleosomes. Labels for plotted profile (1–8) are matched with lanes in A. (C) Chromatin assembly, as assayed by superhelical density analysis of 2-μm plasmids (ectopically introduced pRS426), is shown in histone-tail deletion mutants and wild-type cells. Samples from histone shuffle strain-derived cells were run from lanes 1–6 (lane 1, wild type; lane 2, tH2A; lane 3, tH2B; lane 4, tH3; lane 5, tH2A:tH3; lane 6, tH2B:tH3). Lanes 7 and 8 contain samples from BY4741 (wild type with two endogenous copies of each wild-type core histones) and cac1Δ derived from BY4741, respectively. N, nicked plasmids and the distributed topoisomers of the plasmid are marked with a bracket. The midpoint of topoisomer distribution in each lane (marked with a closed circle) was found by profiling the toposiomers with Image J (NCBI).
Compared with more closed chromatin forms, open chromatin structures tend to have lower occupancy of nucleosomes and/or less regularly positioned nucleosomes (30). Because incorporation of a nucleosome into a circular DNA introduces a negative supercoil, a difference in the number of nucleosomes on the episomal DNA affects distribution of the DNA topoisomers, which, in turn, can be separated in agarose gels (31). To examine whether the more open chromatin structure in tH2A:tH3 cells was correlated with changes in the number of nucleosomes, we measured supercoiling of a 2μ plasmid in cells. Among the different histone tail deletes and wild-type cells examined in this assay, tH2A:tH3 cells displayed the most prominent difference in topoisomer distribution (Fig. 3C, lane 5). To determine whether the change in topoisomer distribution was caused either by depletion of nucleosomes or by enrichment of nucleosomes, we compared the topoisomers from wild-type cells containing two endogenous histone genes (BY4741) and from its derivative lacking CAC1 (BY4741-cac1Δ), the largest subunit of CAF-1, in parallel with those from histone shuffle strains (Fig. 3C, lanes 7 and 8). Because a defect in chromatin assembly factor, CAF-1, leads to a lower number of nucleosomes into episomal plasmid (32), cac1Δ cells was served as a control for nucleosome depletion in this study. We note that the topoisomer distribution in the wild-type histone shuffle strain was different from that of BY4741 strain (Fig. 3C, lane 1 vs. lane 7), suggesting a structural difference in chromatin from the wild-type histone shuffle strain compared with that in the BY4741 strain. Nonetheless, the same directional shift of topoisomer distribution in tH2A:tH3 mutant cells as that in cac1Δ cells compared with their wild-type counterparts (Fig. 3C, lane 1 vs. lane 5 and lane 7 vs. lane 8) indicates that double deletion of N-tails from H2A and H3 alters chromatin structure, at least in part, by lowering the number of nucleosomes assembled on chromatin. However, we do not rule out the possibility that a more open chromatin structure in tH2A:tH3 cells may also be partially attributable to the possibility of weakened interactions between DNA and tailless H2A/H3.
The N-tails of core histones are known to be dispensable for in vitro nucleosome assembly (30, 33). Other studies of in vitro assembled nucleosomal arrays revealed that individual histone N-tails (except the H4 N-tail) play only moderate effects on high-order chromatin structure (30, 34). However, by revealing a specific requirement for the H2A or the H3 N-tail, our results raise the intriguing possibility that these tails play a redundant role in maintaining intact cellular chromatin. The presence of other physiological factors, such as multiple histone modifications and/or chromatin remodeling activities, might further constrain the cellular requirement for the H2A or the H3 N-tails.
H2A N-Tail and H3K4 Play a Redundant Role in HU-Mediated Response.
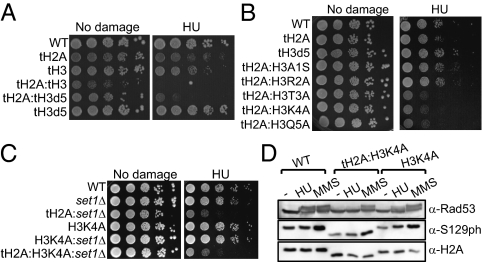
We then asked whether shorter truncations of H2A and H3 N-tails could recapitulate the synergistic phenotypic defects observed in tH2A:tH3 cells (Figs. 2 and 3). Interestingly, deletion of the first five residues from the N terminus of H3 in combination of the H2A N-tail (tH2A:tH3d5) was sufficient to recapitulate the HU sensitivity seen in tH2A:tH3 cells, whereas the other synergistic phenotypes observed required deletion of entire N-tails both from H2A and from H3, (Fig. 4A and Fig. S3A). A superhelical density assay showed no difference in bulk chromatin structure from tH2A:tH3d5 cells compared with that in wild-type cells (Fig. S3B), indicating that the HU sensitivity of the mutant cells is not correlated with a defect in gross chromatin structure. To examine what, if any, residue or potential modification in the first five N-terminal residues of H3 might be important for HU sensitivity, the H2A N-tail deletion was combined with individual H3 point mutations (H3S1A, H3R2A, H3T3A, H3K4A, and H3Q5A). Among these, three point mutations at or immediately surrounding H3K4 (H3T3A, H3K4A, and H3Q5A) enhanced HU sensitivity of cells lacking the H2A N-tail, whereas H3S1A and H3R2A did not affect the HU sensitivity of the H2A N-tail deletion mutant cells (Fig. 4B).
Fig. 4.
Lysine 4 on H3 and its adjacent residues are involved in HU-mediated response in a redundant manner with the H2A N-tail. (A) HU sensitivities of mutant strains carrying tailless H2A (tH2A) and truncated H3 (tH3d5: H3Δ1–5). (B) HU sensitivities of mutants carrying tH2A and a point mutation on H3 (H3S1A, H3R2A, H3T3A, H3K4A, H3Q5A). (C) HU sensitivities of set1 deletion mutant cells combined with histone mutations (tH2A, tH3K4A, or tH2A:tH3K4A). (D) Exponentially growing histone mutant, tH2A:H3K4A, H3K4A, and wild-type cells (labeled as WT) were treated with 100 mM HU or 0.2% MMS for 2 h. Whole-cell extracts from the mutant cells were subjected to immunoblot analyses with indicated antibodies against Rad53, H2AS129ph (labeled as S129ph), and H2A.
H3K4 is methylated by Set1 in yeast cells (35), and Set1 has been suggested to be involved in DNA damage response because deletion of SET1 alleviated the DNA damage sensitivity of checkpoint-defective MEC3 deletion strain by derepressing the expression of DNA repair genes (36, 37). In contrast, a more recent study showed that either SET1 deletion or H3K4A mutation enhances the DNA damage sensitivity of DNA repair-defective RAD50 deletion mutant cells (38). To determine whether methylation on H3K4 is involved in HU sensitivity, we knocked out SET1 in the H2A N-tail delete mutant strains (tH2A and tH2A:tH3K4A). Double deletion of the H2A N-tail and SET1 (tH2A:set1Δ) phenocopied the HU sensitivity of tH2A:tH3K4A mutant cells (Fig. 4C). In contrast, SET1 deletion did not aggravate the HU sensitivity of tH2A:tH3K4A. This result suggests that H3K4 is epistatic to SET1; implying the involvement of Set1-mediated methylation of H3K4 in HU response when the H2A N-tail is absent. Taken together, our data suggest that the H2A tail, in combination with K4 in the H3 tail, likely through methylation of H3K4, play a redundant role in mediating a response to HU. In addition, we speculate that the HU sensitivities of tH2A:H3T3A and tH2A:H3Q5A mutant cells could be due to the defects either in Set1-mediated methylation on H3K4 and/or in binding of effector proteins to methylated H3K4, given the fact that these residues are right next to H3K4. We note a recent report showing that H3K4 can be acetylated in yeast (39). Therefore, it is still possible that H3K4ac may also play a role in HU sensitivity.
To further characterize redundancy between the methylatable lysine 4 of H3 and the H2A N-tail in HU-mediated response, we tested the checkpoint activation in response to HU and methylmethansulfonate (MMS) by immunoblotting for hyperphosphorylated Rad53 (Rad53ph) and H2AS129ph, the presence of which is indicative of activation of the checkpoint signaling (40, 41). A primary cellular effect of HU treatment is depletion of dNTPs, which, in turn, mediates the stalling of DNA replication forks, followed by activation of the replication checkpoint without the formation of DNA breaks (42). The activation of replication checkpoint can be monitored by the appearance of Rad53ph (40). However, cellular treatment of MMS induces alkylation on DNA, which leads to the formation of DNA breaks, followed by activation of the DNA damage checkpoint (43). In response to MMS, robust inductions of Rad53ph and H2AS129ph were detected in H3K4A, tH2A:H3K4A, and wild-type cells (Fig. 4D). By contrast, HU treatment did not generate Rad53ph in tH2A:H3K4A cells, whereas strong and moderate induction of Rad53ph was detected in wild-type and H3K4A cells, respectively. In addition, the MMS- and HU-induced Rad53ph was not compromised in tH2A cells (Fig. S3C). These results indicate that the activation of the HU-mediated replication checkpoint but not of the DNA damage checkpoint is defective in tH2A:H3K4A cells. A single point mutation on H3K4 fails to bring about a prominent HU sensitivity (10, 38). However, when H3K4A mutation or SET1 deletion was introduced into cells lacking RAD50, a DNA double-strand break repair factor, a more enhanced HU sensitivity was observed (38). Our results suggest that the combination of H3K4A point mutation with deletion of H2A N-tail leads to defects in the activation of HU-mediated response, likely without affecting the DNA damage checkpoint. We anticipate that elucidating the function for methylatable H3K4 in a redundant manner with the H2A N-tail in HU-mediated response will improve our understanding of the replication checkpoint.
By characterizing combinations of deletion and point mutations of the budding yeast core histones, our study lends experimental support to the histone redundancy hypothesis, especially as regulatory information carried or embedded in the unstructured histone N-tails. Notably, our findings call attention to redundancy between nonconventional pairs of histones such the H2A and H3 pair. One caveat to our studies is that phenotypic defects observed in the histone mutant cells examined might be mediated by secondary, indirect effects through, for example, perturbed gene expression. Alternatively, subtle defects in general chromatin structure may be introduced by given combinatory histone mutations to varying degrees, resulting in different phenotypes. Future studies will be required to address these and other issues, but our work calls attention to which histone termini and residues should be focused on in several important downstream biological processes.
Materials and Methods
Yeast Strains.
Yeast strains used in this study are described in Table S4.
MNase Analysis.
MNase analysis of bulk chromatin from yeast cells were performed as described (44).
Superhelical Density Analysis.
Plasmid DNA extracted as described in the literature (45) was subject to Southern blot analysis by using a probe specific to URA3 on the plasmid.
Western Blot Analysis.
Whole-cell extract was prepared by bead-beating in trichloroacetic acid. Anti-H2A (Active Motif; AM39235), anti-H2AS129ph (Active Motif; AM39271), and anti-Rad53 (Santa Cruz; sc-6749) were used for immunoblotting analyses.
Supplementary Material
Acknowledgments
We thank members of the C.D.A. laboratory for critical comments on the manuscript. We are grateful for support from National Institutes of Health Grants GM 40922 (to C.D.A., J.-A.K., and the Rockefeller University) and GM28920 (to M.M.S. and University of Virginia). J.-A.K. was also supported by Rockefeller University's Women and Science Initiative fellowship.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1203453109/-/DCSupplemental.
References
- 1.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Wang GG, Allis CD, Chi P. Chromatin remodeling and cancer, Part I: Covalent histone modifications. Trends Mol Med. 2007;13:363–372. doi: 10.1016/j.molmed.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 3.Sobel RE, Cook RG, Perry CA, Annunziato AT, Allis CD. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc Natl Acad Sci USA. 1995;92:1237–1241. doi: 10.1073/pnas.92.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Q, et al. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell. 2008;134:244–255. doi: 10.1016/j.cell.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 6.Goto H, et al. Identification of a novel phosphorylation site on histone H3 coupled with mitotic chromosome condensation. J Biol Chem. 1999;274:25543–25549. doi: 10.1074/jbc.274.36.25543. [DOI] [PubMed] [Google Scholar]
- 7.Wei Y, Yu L, Bowen J, Gorovsky MA, Allis CD. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell. 1999;97:99–109. doi: 10.1016/s0092-8674(00)80718-7. [DOI] [PubMed] [Google Scholar]
- 8.Cheung WL, et al. Phosphorylation of histone H4 serine 1 during DNA damage requires casein kinase II in S. cerevisiae. Curr Biol. 2005;15:656–660. doi: 10.1016/j.cub.2005.02.049. [DOI] [PubMed] [Google Scholar]
- 9.Hsu JY, et al. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 2000;102:279–291. doi: 10.1016/s0092-8674(00)00034-9. [DOI] [PubMed] [Google Scholar]
- 10.Dai J, et al. Probing nucleosome function: A highly versatile library of synthetic histone H3 and H4 mutants. Cell. 2008;134:1066–1078. doi: 10.1016/j.cell.2008.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de la Barre AE, Angelov D, Molla A, Dimitrov S. The N-terminus of histone H2B, but not that of histone H3 or its phosphorylation, is essential for chromosome condensation. EMBO J. 2001;20:6383–6393. doi: 10.1093/emboj/20.22.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma XJ, Wu J, Altheim BA, Schultz MC, Grunstein M. Deposition-related sites K5/K12 in histone H4 are not required for nucleosome deposition in yeast. Proc Natl Acad Sci USA. 1998;95:6693–6698. doi: 10.1073/pnas.95.12.6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith MM, et al. A novel histone H4 mutant defective in nuclear division and mitotic chromosome transmission. Mol Cell Biol. 1996;16:1017–1026. doi: 10.1128/mcb.16.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dion MF, Altschuler SJ, Wu LF, Rando OJ. Genomic characterization reveals a simple histone H4 acetylation code. Proc Natl Acad Sci USA. 2005;102:5501–5506. doi: 10.1073/pnas.0500136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakanishi S, et al. A comprehensive library of histone mutants identifies nucleosomal residues required for H3K4 methylation. Nat Struct Mol Biol. 2008;15:881–888. doi: 10.1038/nsmb.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsubara K, Sano N, Umehara T, Horikoshi M. Global analysis of functional surfaces of core histones with comprehensive point mutants. Genes Cells. 2007;12:13–33. doi: 10.1111/j.1365-2443.2007.01031.x. [DOI] [PubMed] [Google Scholar]
- 17.Megee PC, Morgan BA, Mittman BA, Smith MM. Genetic analysis of histone H4: Essential role of lysines subject to reversible acetylation. Science. 1990;247:841–845. doi: 10.1126/science.2106160. [DOI] [PubMed] [Google Scholar]
- 18.Schuster T, Han M, Grunstein M. Yeast histone H2A and H2B amino termini have interchangeable functions. Cell. 1986;45:445–451. doi: 10.1016/0092-8674(86)90330-2. [DOI] [PubMed] [Google Scholar]
- 19.Morgan BA, Mittman BA, Smith MM. The highly conserved N-terminal domains of histones H3 and H4 are required for normal cell cycle progression. Mol Cell Biol. 1991;11:4111–4120. doi: 10.1128/mcb.11.8.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eitoku M, Sato L, Senda T, Horikoshi M. Histone chaperones: 30 years from isolation to elucidation of the mechanisms of nucleosome assembly and disassembly. Cell Mol Life Sci. 2008;65:414–444. doi: 10.1007/s00018-007-7305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boudreault AA, et al. Yeast enhancer of polycomb defines global Esa1-dependent acetylation of chromatin. Genes Dev. 2003;17:1415–1428. doi: 10.1101/gad.1056603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suka N, Suka Y, Carmen AA, Wu J, Grunstein M. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol Cell. 2001;8:473–479. doi: 10.1016/s1097-2765(01)00301-x. [DOI] [PubMed] [Google Scholar]
- 23.Grant PA, et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: Characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 24.An W, Kim J, Roeder RG. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell. 2004;117:735–748. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 25.Ahn SH, et al. Sterile 20 kinase phosphorylates histone H2B at serine 10 during hydrogen peroxide-induced apoptosis in S. cerevisiae. Cell. 2005;120:25–36. doi: 10.1016/j.cell.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 26.Ling X, Harkness TA, Schultz MC, Fisher-Adams G, Grunstein M. Yeast histone H3 and H4 amino termini are important for nucleosome assembly in vivo and in vitro: Redundant and position-independent functions in assembly but not in gene regulation. Genes Dev. 1996;10:686–699. doi: 10.1101/gad.10.6.686. [DOI] [PubMed] [Google Scholar]
- 27.Altaf M, et al. NuA4-dependent acetylation of nucleosomal histones H4 and H2A directly stimulates incorporation of H2A.Z by the SWR1 complex. J Biol Chem. 2010;285:15966–15977. doi: 10.1074/jbc.M110.117069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang L, et al. Global assessment of combinatorial post-translational modification of core histones in yeast using contemporary mass spectrometry. LYS4 trimethylation correlates with degree of acetylation on the same H3 tail. J Biol Chem. 2007;282:27923–27934. doi: 10.1074/jbc.M704194200. [DOI] [PubMed] [Google Scholar]
- 29.Qin S, Parthun MR. Histone H3 and the histone acetyltransferase Hat1p contribute to DNA double-strand break repair. Mol Cell Biol. 2002;22:8353–8365. doi: 10.1128/MCB.22.23.8353-8365.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gordon F, Luger K, Hansen JC. The core histone N-terminal tail domains function independently and additively during salt-dependent oligomerization of nucleosomal arrays. J Biol Chem. 2005;280:33701–33706. doi: 10.1074/jbc.M507048200. [DOI] [PubMed] [Google Scholar]
- 31.Morse RH. Topoisomer heterogeneity of plasmid chromatin in living cells. J Mol Biol. 1991;222:133–137. doi: 10.1016/0022-2836(91)90198-f. [DOI] [PubMed] [Google Scholar]
- 32.Adkins MW, Tyler JK. The histone chaperone Asf1p mediates global chromatin disassembly in vivo. J Biol Chem. 2004;279:52069–52074. doi: 10.1074/jbc.M406113200. [DOI] [PubMed] [Google Scholar]
- 33.Shibahara K, Verreault A, Stillman B. The N-terminal domains of histones H3 and H4 are not necessary for chromatin assembly factor-1- mediated nucleosome assembly onto replicated DNA in vitro. Proc Natl Acad Sci USA. 2000;97:7766–7771. doi: 10.1073/pnas.97.14.7766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson PJ, et al. 30 nm chromatin fibre decompaction requires both H4-K16 acetylation and linker histone eviction. J Mol Biol. 2008;381:816–825. doi: 10.1016/j.jmb.2008.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Briggs SD, et al. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 2001;15:3286–3295. doi: 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schramke V, et al. The set1Delta mutation unveils a novel signaling pathway relayed by the Rad53-dependent hyperphosphorylation of replication protein A that leads to transcriptional activation of repair genes. Genes Dev. 2001;15:1845–1858. doi: 10.1101/gad.193901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Corda Y, et al. Interaction between Set1p and checkpoint protein Mec3p in DNA repair and telomere functions. Nat Genet. 1999;21:204–208. doi: 10.1038/5991. [DOI] [PubMed] [Google Scholar]
- 38.Faucher D, Wellinger RJ. Methylated H3K4, a transcription-associated histone modification, is involved in the DNA damage response pathway. PLoS Genet. 2010;6:e1001082. PubMed. doi: 10.1371/journal.pgen.1001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guillemette B, et al. H3 lysine 4 is acetylated at active gene promoters and is regulated by H3 lysine 4 methylation. PLoS Genet. 2011;7:e1001354. doi: 10.1371/journal.pgen.1001354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nyberg KA, Michelson RJ, Putnam CW, Weinert TA. Toward maintaining the genome: DNA damage and replication checkpoints. Annu Rev Genet. 2002;36:617–656. doi: 10.1146/annurev.genet.36.060402.113540. [DOI] [PubMed] [Google Scholar]
- 41.Keogh MC, et al. A phosphatase complex that dephosphorylates gammaH2AX regulates DNA damage checkpoint recovery. Nature. 2006;439:497–501. doi: 10.1038/nature04384. [DOI] [PubMed] [Google Scholar]
- 42.Pellicioli A, et al. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 1999;18:6561–6572. doi: 10.1093/emboj/18.22.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwartz JL. Monofunctional alkylating agent-induced S-phase-dependent DNA damage. Mutat Res. 1989;216:111–118. doi: 10.1016/0165-1161(89)90011-3. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Z, Reese JC. Isolation of yeast nuclei and micrococcal nuclease mapping of nucleosome positioning. Methods Mol Biol. 2006;313:245–255. doi: 10.1385/1-59259-958-3:245. [DOI] [PubMed] [Google Scholar]
- 45.Morse RH. Analysis of DNA topology in yeast chromatin. Methods Mol Biol. 2009;523:93–108. doi: 10.1007/978-1-59745-190-1_7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.