Abstract
Senescence represents a state of indefinite growth arrest in cells that have reached their replicative life span, have become damaged, or express aberrant levels of cancer-related proteins. While senescence is widely considered to represent tumor-suppressive mechanism, the accumulation of senescent cells in tissues of older organisms is believed to underlie age-associated losses in physiologic function and age-related diseases. With the emergence of microRNAs (miRNAs) as a major class of molecular regulators of senescence, we review the transcriptional and post-transcriptional factors that control senescence-associated microRNA biosynthesis. Focusing on their enhancement or repression of senescence, we describe the transcription factors that govern the synthesis of primary (pri-)miRNAs, the proteins that control the nuclear processing of pri-miRNAs into precursor (pre-)miRNAs, including RNA editing enzymes, RNases, and RNA helicases, and the cytoplasmic proteins that affect the final processing of pre-miRNAs into mature miRNAs. We discuss how miRNA biogenesis proteins enhance or repress senescence, and thus influence the senescent phenotype that affects normal tissue function and pathology.
1. Introduction
As first described by Hayflick almost 50 years ago (Hayflick 1965), most untransformed cells divide in culture for a finite number of times, after which they reach a state of long-term, G1-phase growth arrest named senescence. Although senescent cells do not divide, they can remain viable and metabolically active for a long time, displaying a senescence-associated secretory phenotype (SASP) (Coppé et al., 2010). Cellular senescence is triggered by many factors, including DNA damage caused by telomere erosion (the loss of chromosome ends as they shorten progressively with each round of division), oxidative damage, and excessive mitogenic signals from activated oncoproteins or inactivated tumor suppressors (Kuilman et al., 2010). With advancing age, senescent cells accumulate in tissues and are believed to contribute to age-related changes (Rodier and Campisi 2011).
In vivo senescence
Senescence is widely recognized as a tumor suppressor mechanism, as evidenced by several facts: i) some factors that promote tumor growth can also trigger senescence, and thus senescence is viewed as a safeguard mechanism against tumorigenesis following oncogenic stimulation, ii) that senescence depends on two major tumor suppressor pathways (p53 and pRB/p16), and iii) that defects in these pathways fail to trigger senescence and lead to tumorigenesis (Narita and Lowe, 2005; Kang et al., 2011). However, there is mounting evidence that senescent cells accumulate in tissues over time and can promote carcinogenesis in older individuals. As part of the SASP, senescent cells secrete factors with promalignant effects, such as the growth-related oncogene (GRO), and factors that promote degradation and invasion of the basement membrane, such as interleukins (IL-6 and IL-8), vascular endothelial growth factor (VEGF), and matrix metalloproteases (reviewed in Rodier and Campisi 2011, Coppé et al., 2010). Together, these lines of evidence indicate that senescence could function as a tumor-suppressive mechanism in young individuals and a tumor-promoting mechanism in older individuals.
Besides cancer, cellular senescence may also contribute to other age-related diseases. For example, overexpression of p53 in mice triggered the accumulation of senescent cells in vivo, linked to the premature appearance of age-associated changes, including loss of fertility and subcutaneous fat, osteoporosis, sarcopenia, thinning of the skin, and reduced hair growth (Tyner et al., 2002; Maier et al., 2004). In another example, a mouse model of a premature aging syndrome (Hutchinson-Gilford progeria, HGPS) showed signs of chronic DNA damage response, persistent p53 activation, and cellular senescence (Varela et al., 2005). Recently, a mouse model in which senescent cells were genetically removed showed a delayed onset of age-related manifestations in skeletal muscle, adipose tissue, and eye (Baker et al., 2011). Senescence may affect age-related diseases by depleting progenitor stem cells in various tissues and by secreting factors that compromise tissue architecture or function, although these possibilities await experimental support.
Posttranscriptional regulation of senescence
With rising interest in the molecular mechanisms that drive cellular senescence, post-transcriptional processes have emerged as pivotal players. Two main groups of factors control senescence-associated gene expression beyond transcription: mRNA-binding proteins and noncoding RNAs, particularly microRNAs (miRNAs). The specific mRNA-binding proteins implicated in senescence are discussed in two excellent reviews in this issue (Wang 2012, and Masuda and Rokutan 2012). The specific microRNAs implicated in senescence were also reviewed recently (Gorospe and Abdelmohsen 2011); they include microRNAs that elicit their actions by acting upon the p53 pathway (e.g., miR-34) or the pRB/p16 pathway (e.g., miR-130, miR-106, miR-24), miRNAs that affect SASP (e.g., miR146), and miRNAs affecting other senescence-regulatory proteins (e.g., miR-519, miR-29, and miR-30).
MicroRNAs have emerged as key effectors of senescence-triggering stimuli. Here, we will discuss the senescence-associated changes in transcription of primary (pri-)miRNAs, the editing and nuclear processing of pri-miRNAs into hairpin-loop precursor (pre-)miRNAs, and the further cytoplasmic processing of pre-miRNAs into mature miRNAs. The target mRNAs, whose stability and/or translation are lowered by the miRNA, will also be indicated. The influence of these regulatory events on cellular senescence, as well as the roles of the proteins that govern each step in miRNA biogenesis will be described in the corresponding sections. Comprehensive reviews of microRNA biosynthesis and the influence of miRNAs on target mRNAs are available (Krol et al., 2010; Winter et al., 2009; Siomi and Siomi MC, 2010).
2. Regulators of pri-miRNA transcription linked to senescence
Transcription of the primary microRNA, the first step in microRNA biogenesis, is tightly regulated. Like other mRNA transcripts, pri-miRNAs are transcribed by RNA polymerase II, although some are transcribed by RNA polymerase III (Borchert et al., 2006; Carthew and Sontheimer 2009, Kim et al., 2009b). Several transcription factors (TFs) recruit the transcription machinery to the appropriate promoters to drive senescence-associated pri-miRNA transcription.
TP53
The TP53 family comprises three TFs, p53, p63 and p73, implicated in microRNA transcription (Boominathan, 2010; Dotsch et al., 2010). Among them, the tumor suppressor protein p53, which is upregulated in senescent cells and can trigger senescence (Itahana et al., 2001; Nardella et al., 2011), has gained much attention as a transcriptional regulator of a subset of miRNAs. Global miRNA expression analyses following DNA damage revealed the p53-dependent upregulation of several miRNAs, notably the tumor suppressor microRNA miR-34a (Chang et al., 2007). Activated p53 upregulated the expression of miR-34a, miR-34b, and miR-34c and induced cellular senescence, while overexpression of miR-34a alone induced senescence in cancer cells (Tazawa et al., 2007; Kumamoto et al., 2008). In turn, the miR-34 family of microRNAs downregulates the expression of several proteins that promote cell proliferation and survival including E2F3, Bcl-2, Met, Cdk6, Cyclin E2, and Sirt1 (Kaller et al; 2011; reviewed by Hermeking, 2010). Another microRNA, miR-200c, was also transcriptionally upregulated by p53 under oxidative stress conditions that cause both apoptosis and senescence in human umbilical vein endothelial cells (Magenta et al., 2011; Schubert and Brabletz, 2011).
Collectively, these three TFs regulated several additional miRNAs, including let-7, miR-143, miR-107, miR-16, miR-145, miR-134, miR-449a, miR-503, and miR-21 (Boominathan, 2010; Knouf et al., 2011). The tumor suppressor microRNA let-7 reduced lung cancer cell proliferation by repressing the production of the high-mobility group protein (HMG)A2 and triggered senescence, in keeping with the upregulation of let-7 family members in senescent cells (Lee and Dutta 2007; Boyerinas et al., 2010; Tzatsos et al., 2011). Perhaps representing a negative regulatory loop, p63 directly transactivated miR-130b, which in turn reduced expression of the cyclin-dependent kinase (cdk) inhibitor p21 and blocked Ras-induced senescence (Su et al., 2010; Borgdorff et al., 2010). In addition to miRNAs, p63 also regulates transcription of Dicer, an essential factor in miRNA biogenesis, as discussed below (Boominathan 2010).
c-Myc
This proto-oncogenic TF is generally downregulated in senescence cells, although it is capable of inducing senescence (Hydbring and Larsson, 2010). Activation of c-Myc broadly decreased the levels of several miRNAs, including miR-15a, miR-16-a, miR-34a and let-7 (Chang et al., 2008). c-Myc and p53 have opposite effects on miR-34a expression: c-Myc suppresses miR-34a expression and helps cell to evade senescence, while p53 promotes it and enhances senescence. c-Myc upregulates the transcription of the genomic locus of miR-17-92 and by doing this it can induce tumorigenesis and inhibit the oncogene-induced senescence triggered by c-Myc (O’Donnell et al., 2005; Hong et al., 2010). Consistent with this paradigm, microRNA members of the miR-17-92 cluster are downregulated in senescent cells, and cancer cells undergo growth arrest or senescence upon the inhibition of these miRNAs (Li et al., 2009; Takakura et al., 2008). Importantly, some miRNAs belonging to this cluster are altered by human aging, age-related cardiac failure, and cellular senescence (Li et al., 2009; Hackl et al., 2010; van Almen et al., 2011), further highlighting the role of c-Myc as a regulator of miRNAs implicated in evading cellular senescence. Downregulation of c-Myc together with upregulation of p53 in senescent cells appears necessary for establishing an effective tumor suppressor mechanism.
E2F1
The E2F family consists of eight members (E2F1-8), including activators and repressors of transcription (DeGregori & Johnson 2006). Together, they influence cell cycle progression, DNA replication, DNA repair, differentiation, autophagy, and apoptosis (Dimova and Dyson 2005, Polager and Ginsberg 2008). E2F1 binds the retinoblastoma protein (RB) in a cell-cycle dependent manner and promotes proliferation while, paradoxically, it also increases the expression of genes that induce senescence and apoptosis such as p16 and p53 (Emmrich and Putzer, 2010; Collado and Serrano, 2010). Independently of pRB, E2F1 also induces the expression of the p14ARF tumor suppressor, which is highly expressed in senescent cells (Good et al., 1996). Moreover, while E2F1 is downregulated in senescent WI-38 fibroblasts, it induces senescence in IMR90 fibroblasts (Dimri et al., 1994, 2000). This dual function of E2F1 is further reflected in the subset of microRNAs it regulates. E2F1-3 directly activate oncogenic microRNA (oncomiR) clusters miR17-92, miR106b-25 and miR106a-363 (Sylvestre et al., 2007, Woods et al., 2007); the miR-17–92 cluster prevents Ras-induced senescence in primary human fibroblasts (Hong et al., 2010), suggesting that E2F1 can suppress cellular senescence at least in part by enhancing the expression of oncomiRs. However, E2F1 also transcriptionally upregulates the expression levels of other microRNAs such as miR-449a and miR-449b, which lower the levels of oncogenes CDK6 and CDC25A, causing cell cycle arrest and a negative feedback regulation of the pRb-E2F1 pathway (Yang et al., 2009). In sum, E2F1 is capable of enhancing and repressing senescence through the subsets of microRNAs it regulates. Whether the divergent activities of E2F1 depend on the specific tissue, the relative abundance of proteins competing with E2F1 (such as other E2Fs) or the levels of E2F1 cofactors remains to be studied.
C/EBPα
The CCAAT/enhancer-binding protein (C/EBP) α is essential for granulopoiesis and is downregulated in acute myeloid leukemia (AML). Recently, CEBPα was reported to upregulate the transcription of miR-34a during granulopoiesis (Pulikkan et al., 2010a). Since miR-34a suppresses cell proliferation and induces senescence, the C/EBPα-enhanced miR-34a might be essential for directing cells into growth arrest or senescence to suppress AML.
ERα
Although the TF estrogen receptor (ER)α promotes carcinogenesis, it can bind at the transcription start site of the miR-221/miR-222 cluster and represses its transcription (Di Leva et al., 2010). These miRNAs inhibit the production of cdk inhibitor p27(Kip1), which increases in senescent cells and is required for senescence (Alexander and Hinds 2001,; Galardi et al., 2007; Marasa et al., 2010). The fact that miR-221 and miR-222 levels are lower in senescent fibroblasts (Marasa et al., 2010) suggest that ERα may paradoxically also promote senescence.
3. microRNA Editing and Senescence
RNA editing is a posttranscriptional process in which cytidine (C) and adenosine (A) are modified to uridine (U) and inosine (I), respectively. The more frequent editing (A-to-I) is carried out by the ADAR (adenosine deaminases acting on RNA) family of proteins, which comprises the widely expressed ADAR 1 and 2, and the predominantly neuronal ADAR 3 (Chen et al., 2000; Kim et al., 2004a; Eisenberg et al., 2005). A-to-I editing has been reported in 6% to 16% of pri-miRNAs and a result, the pri-miRNAs are not further processed (Blow et al., 2006; Kawahara et al., 2008; Dupuis and Maas, 2010). For example, editing of pri-miR-142 by ADAR results in suppression of its processing by Drosha, and therefore miR-142 expression levels in ADAR1 null or ADAR2 null mice was higher than in wild type animals (Yang et al., 2006). It is not known if miR-142 plays a role in senescence, but it does attenuate the proliferation of hematopoietic cells (Sun et al., 2010). In another example, the precursor of miR-22 (a tumor suppressor and senescence inducer) is also subject to A-to-I RNA editing which impairs its processing (Luciano et al., 2004; Xu et al., 2011)
A-to-I editing was also detected in pri-miR-223, pri-miR-1, and pri-miR-143 (Yang et al., 2006). miR-233 affects senescence by downregulating E2F1 and thereby blocks cell cycle progression, miR-1 has tumor suppressive activity in head and neck squamous cell carcinoma, and miR-143 is capable of inducing fibroblast senescence, likely by targeting K-RAS (Chen et al., 2009; Bonifacio and Jarstfer, 2010; Pulikkan et al., 2010b; Nohata et al., 2011). Several other miRs were found to be subject to editing including miR-151, -197, -376a, -379 and -99a; miR-197 is downregulated in senescent fibroblasts and miR-376a is upregulated in senescence induced by the B-RAF oncogene (Bonifacio and Jarstfer, 2010). Since all types of RNAs can be edited, the process may also potentially change the mRNA target sites, suggesting that RNA editing might be a general regulatory mechanism during senescence (Mehler 2008). The implication of RNA editing on the aging process, as influenced by ADAR specifically, was recently reviewed (Montano and Long, 2011).
4. Nuclear processing of senescence-associated miRNAs
DGCR8 and Drosha
A pri-miRNA may give rise to multiple pre-miRNA precursor hairpins, each ~70 nucleotides long. Processing requires the recognition of the pri-miRNA by the microprocessor complex, which includes the nuclear RNA-binding protein DiGeorge Syndrome Critical Region 8 (DGCR8, also known as Pasha). DGCR8 facilitates RNA cleavage by another component of the microprocessor complex, the ribonuclease III-type protein Drosha (Newman and Hammond, 2010). DGCR8 is essential for miRNA biogenesis and for suppressing the self-renewal ability of embryonic stem (ES) cells (Wang et al., 2007). DGCR8 expression is negatively regulated by the ING (Inhibitor of Growth) tumor suppressor proteins, which participate in cellular senescence by interacting with p53 and enhancing p53 stability and translation (Soliman & Riabowol 2007). High ING levels in senescent cells lower DGCR8 levels and thus decrease miRNA processing, an observation that agrees with the lower levels of many miRNAs in senescent cells, in aged individuals, and in cancer cells (Marasa et al., 2009, 2010; Noren Hooten et al., 2010; Melo and Esteller, 2011). Interestingly, Drosha and DGCR8 crossregulate each other: DGCR8 stabilizes the Drosha protein via protein-protein interaction, while Drosha-DGCR8 complex cleaves hairpin structures embedded in the DGCR8 mRNA and thereby destabilizes the mRNA (Han et al., 2009). This crossregulation may play a role in maintaining miRNA levels in the cell. While Drosha is downregulated in senescent cells, its general role in senescence is unclear, as reduction of Drosha levels in carcinoma cells did not itself trigger cellular senescence (Srikantan et al., 2011).
Smad
The Smad group of proteins transduce extracellular signals from transforming growth factor (TGF) β ligands to the nucleus, where they act as TFs (Massague et al., 2005). After phosphorylation of the Smads receptor (R-Smads) the R-Smads-Smad4 complex translocates to the nucleus, where it interacts with pri-miRNA transcripts and the microprocessor complex. This interaction stimulates the production of pre-miRNAs and mature miRNAs such as miR-21 and miR-199a (Davis et al., 2008). All-trans-retinoic acid (ATRA) increases the levels of Smad proteins in mouse embryonic neural tissue and induces miR-21 expression in estrogen receptor-positive breast carcinoma cells (Zhang et al., 2009; Terao et al., 2011). miR-21 is downregulated in senescent cells, and is often upregulated in cancer cells, where it targets tumor suppressors such as the programmed cell death 4 (PDCD4), PTEN, and the Ras homolog RhoB (Marasa et al., 2010; reviewed by Feliciano et al., 2011). Recently, miR-199a was found to promote proliferation and survival of breast cancer cells (Shatseva et al., 2011). In sum, Smad signaling promotes proliferation (Bruna et al., 2007) and by inducing miR-21 and miR-199a it may prevent senescence. In agreement with this hypothesis, silencing of miR-21 promotes the growth inhibition and senescence induced by ATRA (Terao et al., 2011).
DDX
DEAD-box (DDX) proteins are RNA helicases involved in diverse cellular processes, including proliferation, translation initiation, splicing, ribosome assembly, and microRNA processing (Fuller-Pace and Moore, 2011; van Kouwenhove et al., 2011). DDX5 (p68) and DDX17 (p72) associated with the microprocessor complex and regulated the expression of a subset of miRNAs (reviewed by van Kouwenhove et al., 2011). DDX5 was found to unwind the pre-let-7, a step required for the function of let-7 in tumor suppression and senescence (Salzman et al., 2007; Boyerinas et al., 2010). However, DDX5 and DDX17 are likely involved in promoting proliferation and suppressing senescence, given their high expression in tumors and the reduced cell growth and increased apoptosis of mouse embryo fibroblasts (MEFs) deficient in DDX proteins (Fukuda et al., 2007; Fuller-Pace and Moore, 2011). It remains to be studied in detail if they modulate senescence specifically by modulating the processing of miRNAs.
hnRNPs
Many heterogeneous nuclear ribonucleoproteins (hnRNPs) are downregulated in senescent cells (Zhu et al., 2002). HnRNPs control broad areas of RNA metabolism, including pre-mRNA splicing and mRNA stability and translation. Some hnRNPs are also implicated in microRNA biogenesis; for example, hnRNP A1 recognizes a hairpin within the pri-miR-18a transcript and assists Drosha with its processing (Guil and Caceres 2007). miR-18a is downregulated during stress-induced senescence in both HDF and HTM cells (Li et al., 2009) and it is also lower in aged cardiomyocytes and in the hearts of old mice prone to heart failure (Kim et al., 2004b; van Almen et al., 2011). While hnRNP A1 facilitates pri-miR-18a processing, hnRNP A1 inhibits the processing of pri-let-7 (Michlewski and Caceres, 2010). Phosphorylation of hnRNP A1 by p38 was proposed to influence microRNA biogenesis and processing, thus impacting upon cellular senescence (Shimada et al., 2009). Further work is needed to elucidate if other hnRNPs affect pri-microRNA processing and senescence.
KSRP
The RNA-binding protein KSRP (KH-type splicing regulatory protein) is best known for its binding to AU-rich elements (AREs) to promote the decay of target mRNAs (Gherzi et al., 2010). Recently, however, KSRP was also reported to participate in the processing of a subset of microRNAs, including miR-20, let-7, and miR-26b (Trabucchi et al., 2009; Briata et al., 2011). Although miR-20 is a member of an oncogenic cluster, it is involved in MEF senescence in culture, is upregulated in senescent cells, suppresses tumorigenesis, and induces senescence (Marasa et al., 2010; reviewed by Rizzo et al., 2010). KSRP binds to pri-let-7a-1 to promote let-7a biogenesis, antagonizing hnRNP A1, which inhibits let-7a processing (Michlewski and Caceres, 2010). KSRP knockdown increases cell proliferation, in part because KSRP is essential for production of the anti-proliferative miRNA let-7 (Legesse-Miller et al., 2009). KSRP may also affect the production of other anti-proliferative miRNAs such as miR-1 and miR-20a (Gherzi et al., 2010; Trabucchi et al., 2009). Thus, KSRP may regulate cell fate by a unique mechanism that includes an interplay with other RNA-binding proteins (e.g., hnRNP A1) to determine the expression levels of microRNAs that directly regulate cell proliferation and senescence.
Lin28
Lin28 (lineage protein 28) was the first RNA-binding protein identified as essential for embryonic development (Viswanathan et al., 2010). In Caenorhabditis elegans, Lin28 co-transcriptionally binds to pri-let-7 to regulate its maturation (Van Wynsberghe et al., 2011). The mammalian homologs, Lin28 and Lin28b, associate with the terminal loop of pre-let-7, blocking the processing of mature let-7. This negative regulatory effect of Lin28 on microRNA biogenesis is linked to pre-miR uridylation by TUTase4 (TUT4) (Heo et al., 2009). Lin28 may influence the establishment of the pluripotent state, as let-7 expression is low or absent in different stem and progenitor cell populations. In addition, the decreased self-renewal capacity of aging tissues and terminally differentiated stem cells has been associated to the reduced levels of Lin28 and increased levels of mature let-7, which in turn suppresses the expression of self-renewal proteins, inhibits stem cell growth, and promotes senescence (Viswanathan and Daley, 2010; Tzatsos et al., 2011). The above-described effects of let-7 on senescence and the emerging role of Lin28 in suppressing cell senescence and promoting tumorigenesis are highlighted in several cancer studies (Yang et al., 2010; King et al., 2011; Xue et al., 2011).
SNIP1
The Smad nuclear-interacting protein 1 (SNIP1) is highly expressed in tumor cells, promotes cell proliferation and tumorigenesis, and its downregulation causes G1 cell cycle arrest (Roche et al., 2004; Fujii et al., 2006). These effects are at least in part mediated by its association with different TFs such as c-Myc and by its binding to cyclin D1 mRNA causing its stabilization (Larsson, 2006; Bracken et al., 2008). SNIP1 is also involved in inducing mature miRNA by binding to either the pri-miRNA or Drosha (reviewed by van Kouwenhove et al., 2011). For example, miR-21 expression is significantly lower in SNIP1-depleted cells, and this reduction was attributed to the impaired processing of pri-miR-21, which in turn influenced PDCD4 expression levels (Asangani et al., 2008; Yu et al., 2008).
SRF1
The serine/arginine-rich splicing factor 1 (SRF1, also known as SF2 and ASF) is essential for alternative splicing, directly influencing splice site selection. SRF1 recognizes the stem region of a subset of pri-miRNAs and enhances the processing by Drosha of microRNAs such as miR-221 and miR-222 (Wu et al., 2010). As explained above, these miRNAs regulate the expression of the cdk inhibitor p27(Kip1) (Kedde et al., 2010) which increases with and directly promotes senescence (Alexander and Hinds 2001, Marasa et al., 2010). Thus, SRF1 and SRF1-dependent microRNA processing may repress senescence.
Nucleolin
Nucleolin is a multifunctional protein involved in several processes including proliferation, cell survival, ribosomal assembly, and viral infection. It binds DNA to modulate transcription and it also binds RNA, stabilizing or altering the translation of target mRNAs (Mongelard and Bouvet, 2007; Abdelmohsen et al., 2011). Nucleolin expression is regulated by the competitive interaction of the RNA-binding protein HuR and miR-494 with the nucleolin (NCL) mRNA (Tominaga et al., 2011). The fact that senescent cells show high levels of miR-494 and low levels of HuR is in keeping with the low expression levels of nucleolin in senescent human diploid fibroblasts (Wang et al., 2001; Faraonio et al., 2011; and our unpublished findings). Recently, nucleolin was found to interact with the microprocessor complex to enhance the biogenesis of miR-15a and miR-16 (Pickering et al., 2011); consistent with the low levels of nucleolin in senescent cells, miR-15a abundance declines with senescence (Marasa et al., 2010). As miR-15a represses the production of the anti-apoptotic protein Bcl-2 (Cimmino et al., 2005), downregulation of miR-15a might be essential for maintaining high Bcl-2 levels, which promotes premature senescence in human carcinoma cells expressing oncogenic Ras (Crescenzi et al., 2003; Tombor et al., 2003). Cells need to maintain a delicate balance of nucleolin, miR-15a and Bcl-2 levels in order to both avoid excessive apoptosis and aberrant proliferation.
NF90, NF45
The RNA-binding protein NF90 (nuclear factor of 90 kDa) is involved in processes such as transcription, DNA repair, mRNA stability, mRNA export, and protein translation (Kuwano et al., 2010; Shamanna et al., 2011). It can heterodimerize with NF45 and negatively regulates miRNA processing by interacting with pri-miRNA. Reducing NF90 decreased pri-let-7a-1 levels and increased mature let-7a levels, inhibited DNA synthesis, and lowered cell growth accompanied by the accumulation of multinucleated cells, often seen in senescent cells (Guan et al., 2008, Santarosa et al., 2009, Sakamoto et al., 2009; Singh et al., 2010; Tzatsos et al., 2011). Thus, the NF90/NF45 complex may promote cell division by helping cells to evade cellular senescence.
5. Cytoplasmic microRNA processing implicated in senescence
Dicer
A member of the RNase III family of endoribonucleases, Dicer binds to exported microRNA precursors (pre-miRNAs) and aids in the formation of the mature miRNAs that target mRNAs for silencing (Jaskiewicz and Filipowicz, 2008). Dicer is essential for the maturation of the vast majority of miRNAs, including microRNAs that promote or suppress cellular senescence and modulate tumorigenesis. However, Dicer ablation and loss of miRNAs were found to trigger senescence in primary cells, accompanied by high levels of both p53 and p19ARF (Mudhasani et al., 2008; Srikantan et al., 2011). TAp63 [which contains the N-terminal transactivation (TA) domain of p63] was found to regulate metastasis by coordinately regulating Dicer and miRNAs: TAp63 transcriptionally regulated Dicer expression and influenced miRNA levels, while TAp63 deficiency induced senescence and inhibited the metastatic potential of osteosarcomas (Kimura et al., 2010).
Argonaute
After Dicer releases the mature miRNA from a double-stranded pre-miRNA, Argonaute proteins (Ago) bind the mature miRNA and become part of the RNA-induced silencing complex (RISC), which contains additional proteins such as the dsRNA-binding protein TRBP [human immunodeficiency virus (HIV) transactivating response RNA (TAR) binding protein], and PACT (protein activator of the interferon induced protein kinase). The complex Ago/miRNA/RISC is then directed to specific subcellular locations to cleave or suppress the translation of target RNAs (reviewed by Cenik and Zamore, 2011). Although eight Ago proteins exist in human cells, only Ago1 and Ago2 mediate the silencing effect of microRNAs, with Ago1 primarily suppressing mRNA translation and Ago2 promoting target mRNA cleavage (Forstemann et al., 2007). Ago2 is essential for development, since Ago2-null mice are embryonic lethal, and Ago proteins are highly expressed in cancer cells (Carmell et al., 2002; Liu et al., 2004, Morita et al., 2007; Adams et al., 2009). Although Dicer is required for miRNA processing, Ago2 can process a subset of miRNAs independently of Dicer (Cheloufi et al., 2010). Besides the cytoplasmic roles of Ago proteins, nuclear Ago2 was recently found to be critical for stem cells to escape senescence by downregulating miR-10b and miR-23b (Kim et al., 2011). Further studies are required to understand the role of Ago2 in senescence and aging.
6. Auxiliary factors linked to Senescence
HuR
Human antigen R (HuR) is an RNA-binding protein expressed ubiquitously and capable of stabilizing and modulating the translation of target mRNAs. HuR promotes carcinogenesis by enhancing proliferation, survival, angiogenesis, tissue invasion, while at the same time blocking senescence (Wang et al., 2001; Abdelmohsen and Gorospe, 2010; Srikantan and Gorospe, 2012). HuR can compete or cooperate with miRNA-loaded RISC to regulate target mRNA expression (reviewed by Srikantan et al., submitted). As examples of competition, HuR relieved cationic amino acid transporter 1 (CAT1) mRNA from miR-122-mediated repression under stress conditions in human liver cells (Bhattacharyya et al., 2006), enhanced TOP2A mRNA translation by antagonizing miR-548c-3p, and enhanced NCL mRNA expression by competing with its negative regulator, miR-494 (Srikantan et al., 2011; Tominaga et al., 2011). Instances of cooperation include HuR’s promotion of the loading of let-7-RISC to regulate MYC mRNA, and of miR-19-RISC to regulate RHOB mRNA (Kim et al., 2009; Glorian et al., 2011).
FMRP
The RNA-binding protein fragile X mental retardation protein (FMRP) is involved in essential processes such as brain development and female reproductive function. Altered or mutated FMRP causes mental retardation, premature ovarian failure, Parkinson’s disease, and aberrant expression of the amyloid precursor protein (APP), linked to Alzheimer’s disease and neuronal senescence (Charlesworth 1996; Raina et al., 2001; Golde & Miller 2009; Sokol et al., 2011). FMRP associate with the RISC to regulate gene expression (Jin et al., 2004); for example, it associates with microRNAs miR-125b and miR-132 to regulate synaptic structure and function (Edbauer et al., 2010). Upon cell cycle arrest, Ago and fragile X-related protein 1 (FXR1), a paralog of FMRP, were recruited to the RISC to enhance tumor necrosis factor α (TNFA) mRNA translation (Vasudevan et al., 2007). Further studies are needed to elucidate the role FMRP and FMRP-associated miRNAs in cellular senescence, particularly neuronal senescence.
TRBP
TRBP is implicated in cytoplasmic miRNA processing with Dicer and Ago2, enhances Dicer protein stability and serves as a platform for RISC assembly (Chendrimada et al., 2005; Heo and Kim 2009). TRBP was found to be mutated in some human cancers and the resulting truncated protein had an impaired ability to process microRNA (Melo et al., 2009). Interestingly, TRBP phosphorylation by the extracellular signal-regulated kinase (ERK) led to differential expression of growth-related miRNAs: pro-growth miRNAs (e.g., miR-17) increased and anti-growth miRNAs (e.g., let-7) decreased (Paroo et al., 2009). The ERK phosphorylation pathway is attenuated in old animals and in senescent cells (Ikeyama et al., 2003; Bose et al., 2004), likely representing a beneficial advantage of senescent cells to suppress the oncogenic activity of TRBP (Benkirane et al., 1997).
Gemins
As members of the DEAD-box putative RNA helicases, gemins are involved in cellular processes such as translation initiation, nuclear and mitochondrial splicing, and ribosome and spliceosome assembly, as well as the assembly of small nuclear ribonucleoproteins (snRNPs) (Battle et al., 2006). Members of this family include DDX20 (gemin 3) and DDX42 (gemin 4), which interact with Ago2 and miRNAs in the RISC (Mourelatos et al., 2002). The aging-related disease spinal muscular atrophy (SMA) is mainly caused by deletion or mutations in the gene ‘survival of motor neuron’ (SMN). Gemins are involved in SMA by interacting with SMN. The enzyme telomerase, closely linked to senescence, was found to be associated with SMN, influencing the functional telomerase holoenzyme (Bachand et al., 2002).
7. Concluding remarks
The recognition that senescent cells critically affect many pathologies, particularly those associated with the aging process, has fueled efforts to identify the molecular regulators of senescence. Given that miRNAs constitute a major class of senescence regulators (Gorospe and Abdelmohsen, 2011), many studies have sought to understand the mechanisms that control their expression and function. As described in this review, the senescence-associated biogenesis of microRNAs is regulated on multiple levels. At the level of transcription, several TFs have been identified as regulators of the production of senescence-associated pri-miRNAs, although genome-wide analyses of this process are still needed. In this regard, it is plausible that another class of noncoding RNAs [long intergenic noncoding (linc) RNAs] may help to coordinate the transcription of senescence-associated miRNAs. Senescence-associated miRNAs are also tightly regulated by proteins that process pri-miRNAs and pre-miRNAs, including RNA editing enzymes, RNA-binding proteins, RNases, and RNA helicases. Since many of these regulatory events have only been described for one or a few miRNAs thus far, the identification of miRNA subsets regulated in each manner also awaits systematic study. To this end, several methodologies based on the immunoprecipitation of RNA-binding proteins (Keene et al., 2006; Hafner et al., 2010; Darnell 2010) will be particularly informative.
Finally, a general challenge in the area of senescence is to extend the studies in cultured senescent cells to in vivo settings. Although some animal models to study senescence in physiologic settings are becoming available [for example (Baker et al., 2011)], there are still many limitations to the analysis of senescence in vivo. As appropriate genetic models become available, the influence of miRNA biogenesis regulators should be tested in the context of senescence and the influence of senescence on physiologic changes and pathologies. These molecular and animal studies will broaden and deepen our understanding of the complex role of microRNAs in senescence, and will point to promising therapeutic targets in disease processes arising from aberrant senescence.
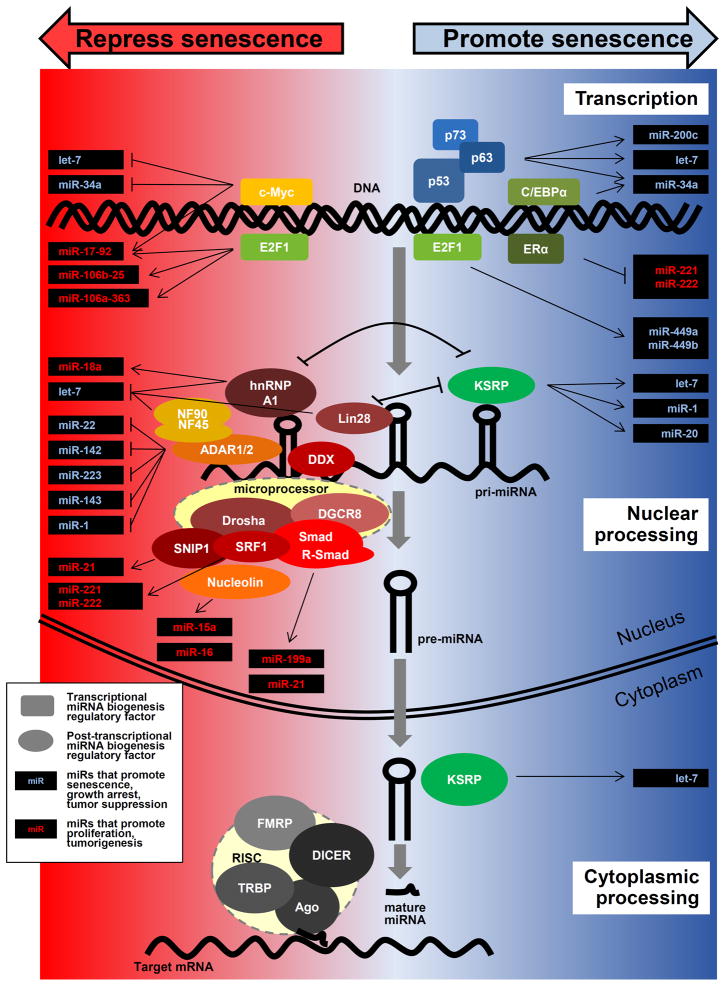
Figure 1.
Schematic representation of microRNA biogenesis, depicting the senescence-associated factors involved. Top, transcriptional regulators of senescence-associated primary (pri)-miRNA biosynthesis (rectangles). Middle, regulators of pri-miRNA processing into pre-mRNAs, including components of the microprocessor complex (Drosha, DGCR8 and accessory factors, ovals). Bottom, regulators of pre-miRNA processing into mature miRNAs, particularly the components of the RISC and accessory proteins (ovals). On the right part of the schematic (blue background) are factors that mainly promote senescence, either by increasing the expression of growth-inhibitory, senescence-enhancing microRNAs (e.g., miR-34a or let-7, blue text) or by inhibiting the production of proliferative, growth-promoting, oncogenic miRNA (e.g., miR-221/miR-222, red text). On the left part of the schematic (red background) are factors that mainly repress senescence, either by promoting the expression of proliferative, oncogenic miRNAs (e.g., miR-21, the miR-17-92 cluster, red text) or by lowering the production of senescence-promoting miRNAs (e.g., miR-34a, let-7, miR-1, blue text).
Hightlights.
MicroRNAs (miRNAs) play a pivotal role in cellular senescence
MiRNA biosynthesis is tightly regulated during senescence
Senescence modulates the transcription factors expressing primary (pri-)miRNAs
Senescence affects the nuclear processing of pri-miRNA to precursor (pre-)miRNAs
Senescence influences the cytoplasmic processing of pre-miRNAs to mature miRNAs
Acknowledgments
This work was supported in its entirety by the National Institute on Aging-Intramural Research Program or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdelmohsen K, Gorospe M. Posttranscriptional regulation of cancer traits by HuR. Wiley Interdiscip Rev RNA. 2010;1:214–229. doi: 10.1002/wrna.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelmohsen K, Tominaga K, Lee EK, Srikantan S, Kang MJ, et al. Enhanced translation by Nucleolin via G-rich elements in coding and non-coding regions of target mRNAs. Nucleic Acids Res. 2011;39:8513–8530. doi: 10.1093/nar/gkr488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams BD, Claffey KP, White BA. Argonaute-2 expression is regulated by epidermal growth factor receptor and mitogen-activated protein kinase signaling and correlates with a transformed phenotype in breast cancer cells. Endocrinology. 2009;150:14–23. doi: 10.1210/en.2008-0984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander K, Hinds PW. Requirement for p27(KIP1) in retinoblastoma protein-mediated senescence. Mol Cell Biol. 2001;21:3616–31. doi: 10.1128/MCB.21.11.3616-3631.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–36. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- Bachand F, Boisvert FM, Cote J, Richard S, Autexier C. The product of the survival of motor neuron (SMN) gene is a human telomerase-associated protein. Mol Biol Cell. 2002;13:3192–202. doi: 10.1091/mbc.E02-04-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DJ, Wijshake T, Tchkonia T, LeBrasseur NK, Childs BG, van de Sluis B, Kirkland JL, van Deursen JM. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479:232–236. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battle DJ, Kasim M, Yong J, Lotti F, Lau CK, Mouaikel J, Zhang Z, Han K, Wan L, Dreyfuss G. The SMN complex: an assembly machine for RNPs. Cold Spring Harb Symp Quant Biol. 2006;71:313–320. doi: 10.1101/sqb.2006.71.001. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Blow MJ, Grocock RJ, van Dongen S, Enright AJ, Dicks E, Futreal PA, Wooster R, Stratton MR. RNA editing of human microRNAs. Genome Biol. 2006;7:R27. doi: 10.1186/gb-2006-7-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacio LN, Jarstfer MB. MiRNA profile associated with replicative senescence, extended cell culture, and ectopic telomerase expression in human foreskin fibroblasts. PLoS One. 2010;5:e12519. doi: 10.1371/journal.pone.0012519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boominathan L. The tumor suppressors p53, p63, and p73 are regulators of microRNA processing complex. PLoS One. 2010;5:e10615. doi: 10.1371/journal.pone.0010615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- Borgdorff V, Lleonart ME, Bishop CL, Fessart D, Bergin AH, Overhoff MG, Beach DH. Multiple microRNAs rescue from Ras-induced senescence by inhibiting p21(Waf1/Cip1) Oncogene. 2010;29:2262–2271. doi: 10.1038/onc.2009.497. [DOI] [PubMed] [Google Scholar]
- Bose C, Bhuvaneswaran C, Udupa KB. Altered mitogen-activated protein kinase signal transduction in human skin fibroblasts during in vitro aging: differential expression of low-density lipoprotein receptor. J Gerontol A Biol Sci Med Sci. 2004;59:126–135. doi: 10.1093/gerona/59.2.b126. [DOI] [PubMed] [Google Scholar]
- Boyerinas B, Park SM, Hau A, Murmann AE, Peter ME. The role of let-7 in cell differentiation and cancer. Endocr Relat Cancer. 2010;17:F19–36. doi: 10.1677/ERC-09-0184. [DOI] [PubMed] [Google Scholar]
- Bracken CP, Wall SJ, Barre B, Panov KI, Ajuh PM, Perkins ND. Regulation of cyclin D1 RNA stability by SNIP1. Cancer Res. 2004;68:7621–7628. doi: 10.1158/0008-5472.CAN-08-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briata P, Chen CY, Giovarelli M, Pasero M, Trabucchi M, Ramos A, Gherzi R. KSRP, many functions for a single protein. Front Biosci. 2011;16:1787–1796. doi: 10.2741/3821. [DOI] [PubMed] [Google Scholar]
- Bruna A, Darken RS, Rojo F, Ocana A, Penuelas S, et al. High TGFbeta-Smad activity confers poor prognosis in glioma patients and promotes cell proliferation depending on the methylation of the PDGF-B gene. Cancer Cell. 2007;11:147–160. doi: 10.1016/j.ccr.2006.11.023. [DOI] [PubMed] [Google Scholar]
- Carmell MA, Xuan Z, Zhang MQ, Hannon GJ. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 2002;16:2733–2742. doi: 10.1101/gad.1026102. [DOI] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenik ES, Zamore PD. Argonaute proteins. Curr Biol. 2011;21:R446–9. doi: 10.1016/j.cub.2011.05.020. [DOI] [PubMed] [Google Scholar]
- Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B. Evolution of senescence: Alzheimer’s disease and evolution. Curr Biol. 1996;6:20–22. doi: 10.1016/s0960-9822(02)00411-6. [DOI] [PubMed] [Google Scholar]
- Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CX, Cho DS, Wang Q, Lai F, Carter KC, Nishikura K. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA. 2000;6:755–767. doi: 10.1017/s1355838200000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Guo X, Zhang H, Xiang Y, Chen J, et al. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene. 2009;28:1385–1392. doi: 10.1038/onc.2008.474. [DOI] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppé JP, Desprez PY, Krtolica A, Campisi J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu Rev Pathol. 2010;5:99–118. doi: 10.1146/annurev-pathol-121808-102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crescenzi E, Palumbo G, Brady HJ. Bcl-2 activates a programme of premature senescence in human carcinoma cells. Biochem J. 2003;375:263–274. doi: 10.1042/BJ20030868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell RB. HITS-CLIP: panoramic views of protein-RNA regulation in living cells. Wiley Interdiscip Rev RNA. 2010;1:266–286. doi: 10.1002/wrna.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Leva G, Gasparini P, Piovan C, Ngankeu A, Garofalo M, Taccioli C, Iorio MV, Li M, Volinia S, Alder H, Nakamura T, Nuovo G, Liu Y, Nephew KP, Croce CM. MicroRNA cluster 221-222 and estrogen receptor alpha interactions in breast cancer. J Natl Cancer Inst. 2010;102:706–721. doi: 10.1093/jnci/djq102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimova DK, Dyson NJ. The E2F transcriptional network: old acquaintances with new faces. Oncogene. 2005;24:2810–2826. doi: 10.1038/sj.onc.1208612. [DOI] [PubMed] [Google Scholar]
- Dimri GP, Hara E, Campisi J. Regulation of two E2F-related genes in presenescent and senescent human fibroblasts. J Biol Chem. 1994;269:16180–16186. [PubMed] [Google Scholar]
- Dimri GP, Itahana K, Acosta M, Campisi J. Regulation of a senescence checkpoint response by the E2F1 transcription factor and p14(ARF) tumor suppressor. Mol Cell Biol. 2000;20:273–285. doi: 10.1128/mcb.20.1.273-285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dötsch V, Bernassola F, Coutandin D, Candi E, Melino G. p63 and p73, the ancestors of p53. Cold Spring Harb Perspect Biol. 2010;2:a004887. doi: 10.1101/cshperspect.a004887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis DE, Maas S. MiRNA editing. Methods Mol Biol. 2010;667:267–279. doi: 10.1007/978-1-60761-811-9_18. [DOI] [PubMed] [Google Scholar]
- Edbauer D, Neilson JR, Foster KA, Wang CF, Seeburg DP, Batterton MN, Tada T, Dolan BM, Sharp PA, Sheng M. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 2010;65:373–384. doi: 10.1016/j.neuron.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg E, Adamsky K, Cohen L, Amariglio N, Hirshberg A, et al. Identification of RNA editing sites in the SNP database. Nucleic Acids Res. 2005;33:4612–4617. doi: 10.1093/nar/gki771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraonio R, Salerno P, Passaro F, Sedia C, Iaccio A, et al. A set of miRNAs participates in the cellular senescence program in human diploid fibroblasts. Cell Death Differ. 2011 doi: 10.1038/cdd.2011.143. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feliciano A, Sanchez-Sendra B, Kondoh H, Lleonart ME. MicroRNAs Regulate Key Effector Pathways of Senescence. J Aging Res. 2011;2011:205378. doi: 10.4061/2011/205378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1. Cell. 2007;130:287–297. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda T, Yamagata K, Fujiyama S, Matsumoto T, Koshida I, et al. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat Cell Biol. 2007;9:604–611. doi: 10.1038/ncb1577. [DOI] [PubMed] [Google Scholar]
- Fujii M, Lyakh LA, Bracken CP, Fukuoka J, Hayakawa M, et al. SNIP1 is a candidate modifier of the transcriptional activity of c-Myc on E box-dependent target genes. Mol Cell. 2006;24:771–783. doi: 10.1016/j.molcel.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Fuller-Pace FV, Moore HC. RNA helicases p68 and p72: multifunctional proteins with important implications for cancer development. Future Oncol. 2011;7:239–251. doi: 10.2217/fon.11.1. [DOI] [PubMed] [Google Scholar]
- Gherzi R, Chen CY, Trabucchi M, Ramos A, Briata P. The role of KSRP in mRNA decay and microRNA precursor maturation. Wiley Interdiscip Rev RNA. 2010;1:230–239. doi: 10.1002/wrna.2. [DOI] [PubMed] [Google Scholar]
- Glorian V, Maillot G, Poles S, Iacovoni JS, Favre G, Vagner S. HuR-dependent loading of miRNA RISC to the mRNA encoding the Ras-related small GTPase RhoB controls its translation during UV-induced apoptosis. Cell Death Differ. 2011;18:1692–1701. doi: 10.1038/cdd.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golde TE, Miller VM. Proteinopathy-induced neuronal senescence: a hypothesis for brain failure in Alzheimer’s and other neurodegenerative diseases. Alzheimers Res Ther. 2009;1:5. doi: 10.1186/alzrt5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good L, Dimri GP, Campisi J, Chen KY. Regulation of dihydrofolate reductase gene expression and E2F components in human diploid fibroblasts during growth and senescence. J Cell Physiol. 1996;168:580–588. doi: 10.1002/(SICI)1097-4652(199609)168:3<580::AID-JCP10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Gorospe M, Abdelmohsen K. MicroRegulators come of age in senescence. Trends Genet. 2011;27:233–241. doi: 10.1016/j.tig.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan D, Altan-Bonnet N, Parrott AM, Arrigo CJ, Li Q, et al. Nuclear factor 45 (NF45) is a regulatory subunit of complexes with NF90/110 involved in mitotic control. Mol Cell Biol. 2008;28:4629–4641. doi: 10.1128/MCB.00120-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guil S, Caceres JF. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol. 2007;14:591–596. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- Hackl M, Brunner S, Fortschegger K, Schreiner C, Micutkova L, Mück C, Laschober GT, Lepperdinger G, Sampson N, Berger P, Herndler-Brandstetter D, Wieser M, Kühnel H, Strasser A, Rinnerthaler M, Breitenbach M, Mildner M, Eckhart L, Tschachler E, Trost A, Bauer JW, Papak C, Trajanoski Z, Scheideler M, Grillari-Voglauer R, Grubeck-Loebenstein B, Jansen-Dürr P, Grillari J. miR-17, miR-19b, miR-20a, and miR-106a are down-regulated in human aging. Aging Cell. 2010;9:291–296. doi: 10.1111/j.1474-9726.2010.00549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayflick L. The limited in vitro lifetime of human diploid cell strains. Exp Cell Res. 1965;37:614–636. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Heo I, Kim VN. Regulating the regulators: posttranslational modifications of RNA silencing factors. Cell. 2009;139:28–31. doi: 10.1016/j.cell.2009.09.013. [DOI] [PubMed] [Google Scholar]
- Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17:193–199. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- Hong L, Lai M, Chen M, Xie C, Liao R, Kang YJ, Xiao C, Hu WY, Han J, Sun P. The miR-17-92 cluster of microRNAs confers tumorigenicity by inhibiting oncogene-induced senescence. Cancer Res. 2010;70:8547–8557. doi: 10.1158/0008-5472.CAN-10-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hydbring P, Larsson LG. Tipping the balance: Cdk2 enables Myc to suppress senescence. Cancer Res. 2010;70:6687–6691. doi: 10.1158/0008-5472.CAN-10-1383. [DOI] [PubMed] [Google Scholar]
- Ikeyama S, Kokkonen G, Martindale JL, Wang XT, Gorospe M, Holbrook NJ. Effects of aging and calorie restriction of Fischer 344 rats on hepatocellular response to proliferative signals. Exp Gerontol. 2003;38:431–439. doi: 10.1016/s0531-5565(02)00239-5. [DOI] [PubMed] [Google Scholar]
- Itahana K, Dimri G, Campisi J. Regulation of cellular senescence by p53. Eur J Biochem. 2001;268:2784–2791. doi: 10.1046/j.1432-1327.2001.02228.x. [DOI] [PubMed] [Google Scholar]
- Jaskiewicz L, Filipowicz W. Role of Dicer in posttranscriptional RNA silencing. Curr Top Microbiol Immunol. 2008;320:77–97. doi: 10.1007/978-3-540-75157-1_4. [DOI] [PubMed] [Google Scholar]
- Jin P, Zarnescu DC, Ceman S, Nakamoto M, Mowrey J, Jongens TA, Nelson DL, Moses K, Warren ST. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat Neurosci. 2004;7:113–117. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- Kaller M, Liffers ST, Oeljeklaus S, Kuhlmann K, Röh S, Hoffmann R, Warscheid B, Hermeking H. Genome-wide characterization of miR-34a induced changes in protein and mRNA expression by a combined pulsed SILAC and microarray analysis. Mol Cell Proteomics. 2011;10:M111. doi: 10.1074/mcp.M111.010462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, Megraw M, Kreider E, Iizasa H, Valente L, Hatzigeorgiou AG, Nishikura K. Frequency and fate of microRNA editing in human brain. Nucleic Acids Res. 2008;36:5270–5280. doi: 10.1093/nar/gkn479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedde M, van Kouwenhove M, Zwart W, Oude Vrielink JA, Elkon R, Agami R. A Pumilio-induced RNA structure switch in p27-3′ UTR controls miR-221 and miR-222 accessibility. Nat Cell Biol. 2010;12:1014–1020. doi: 10.1038/ncb2105. [DOI] [PubMed] [Google Scholar]
- Keene JD, Komisarow JM, Friedersdorf MB. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat Protoc. 2006;1:302–307. doi: 10.1038/nprot.2006.47. [DOI] [PubMed] [Google Scholar]
- Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009a;23:1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009b;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Kim K, Nose K, Shibanuma M. Significance of nuclear relocalization of ERK1/2 in reactivation of c-fos transcription and DNA synthesis in senescent fibroblasts. J Biol Chem. 2000;275:20685–20692. doi: 10.1074/jbc.M908723199. [DOI] [PubMed] [Google Scholar]
- Kim BS, Jung JS, Jang JH, Kang KS, Kang SK. Nuclear Argonaute 2 regulates adipose tissue-derived stem cell survival through direct control of miR10b and selenoprotein N1 expression. Aging Cell. 2011;10:277–91. doi: 10.1111/j.1474-9726.2011.00670.x. [DOI] [PubMed] [Google Scholar]
- Kim DD, Kim TT, Walsh T, Kobayashi Y, Matise TC, et al. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004a;14:1719–1725. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH, Park GT, Lim YB, Rue SW, Jung JC, et al. Expression of connective tissue growth factor, a biomarker in senescence of human diploid fibroblasts, is up-regulated by a transforming growth factor-beta-mediated signaling pathway. Biochem Biophys Res Commun. 2004b;318:819–25. doi: 10.1016/j.bbrc.2004.04.108. [DOI] [PubMed] [Google Scholar]
- Kimura S, Naganuma S, Susuki D, Hirono Y, Yamaguchi A, Fujieda S, Sano K, Itoh H. Expression of microRNAs in squamous cell carcinoma of human head and neck and the esophagus: miR-205 and miR-21 are specific markers for HNSCC and ESCC. Oncol Rep. 2010;23:1625–1633. doi: 10.3892/or_00000804. [DOI] [PubMed] [Google Scholar]
- King CE, Cuatrecasas M, Castells A, Sepulveda AR, Lee JS, Rustgi AK. LIN28B promotes colon cancer progression and metastasis. Cancer Res. 2011;71:4260–4268. doi: 10.1158/0008-5472.CAN-10-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knouf EC, Garg K, Arroyo JD, Correa Y, Sarkar D, Parkin RK, Wurz K, O’Briant KC, Godwin AK, Urban ND, Ruzzo WL, Gentleman R, Drescher CW, Swisher EM, Tewari M. An integrative genomic approach identifies p73 and p63 as activators of miR-200 microRNA family transcription. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr731. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- Kuilman T, Michaloglou C, Mooi WJ, Peeper DS. The essence of senescence. Genes Dev. 2010;24:2463–2479. doi: 10.1101/gad.1971610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto K, Spillare EA, Fujita K, Horikawa I, Yamashita T, et al. Nutlin-3a activates p53 to both down-regulate inhibitor of growth 2 and up-regulate mir-34a, mir-34b, and mir-34c expression, and induce senescence. Cancer Res. 2008;68:3193–3203. doi: 10.1158/0008-5472.CAN-07-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson LG. SNIP1: Myc’s new helper in transcriptional activation. Mol Cell. 2006;24:811–812. doi: 10.1016/j.molcel.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legesse-Miller A, Elemento O, Pfau SJ, Forman JJ, Tavazoie S, Coller HA. let-7 overexpression leads to an increased fraction of cells in G2/M, direct down-regulation of Cdc34, and stabilization of Wee1 kinase in primary fibroblasts. J Biol Chem. 2009;284:6605–6609. doi: 10.1074/jbc.C900002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Luna C, Qiu J, Epstein DL, Gonzalez P. Alterations in microRNA expression in stress-induced cellular senescence. Mech Ageing Dev. 2009;130:731–741. doi: 10.1016/j.mad.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Luciano DJ, Mirsky H, Vendetti NJ, Maas S. RNA editing of a miRNA precursor. RNA. 2004;10:1174–1177. doi: 10.1261/rna.7350304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier B, Gluba W, Bernier B, Turner T, Mohammad K, Guise T, Sutherland A, Thorner M, Scrable H. Modulation of mammalian life span by the short isoform of p53. Genes Dev. 2004;18:306–319. doi: 10.1101/gad.1162404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magenta A, Cencioni C, Fasanaro P, Zaccagnini G, Greco S, Sarra-Ferraris G, Antonini A, Martelli F, Capogrossi MC. miR-200c is upregulated by oxidative stress and induces endothelial cell apoptosis and senescence via ZEB1 inhibition. Cell Death Differ. 2011;18:1628–1639. doi: 10.1038/cdd.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marasa BS, Srikantan S, Martindale JL, Kim MM, Lee EK, Gorospe M, Abdelmohsen K. MicroRNA profiling in human diploid fibroblasts uncovers miR-519 role in replicative senescence. Aging. 2010;2:333–343. doi: 10.18632/aging.100159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marasa BS, Srikantan S, Masuda K, Abdelmohsen K, Kuwano Y, Yang X, Martindale JL, Rinker-Schaeffer CW, Gorospe M. Increased MKK4 abundance with replicative senescence is linked to the joint reduction of multiple microRNAs. Sci Signal. 2009;2:ra69. doi: 10.1126/scisignal.2000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- Masuda K, Rokutan K. General RBP expression in human tissues as a function of age. Ageing Res Rev. 2012 doi: 10.1016/j.arr.2012.01.005. This issue. [DOI] [PubMed] [Google Scholar]
- Mehler MF. Epigenetic principles and mechanisms underlying nervous system functions in health and disease. Prog Neurobiol. 2008;86:305–341. doi: 10.1016/j.pneurobio.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michlewski G, Cáceres JF. Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nat Struct Mol Biol. 2010;17:1011–1018. doi: 10.1038/nsmb.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo SA, Esteller M. Dysregulation of microRNAs in cancer: playing with fire. FEBS Lett. 2011;585:2087–2099. doi: 10.1016/j.febslet.2010.08.009. [DOI] [PubMed] [Google Scholar]
- Melo SA, Ropero S, Moutinho C, Aaltonen LA, Yamamoto H, et al. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat Genet. 2009;41:365–370. doi: 10.1038/ng.317. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Mongelard F, Bouvet P. Nucleolin: a multiFACeTed protein. Trends Cell Biol. 2007;17:80–86. doi: 10.1016/j.tcb.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Montano M, Long K. RNA surveillance-an emerging role for RNA regulatory networks in aging. Ageing Res Rev. 2011;10:216–224. doi: 10.1016/j.arr.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita S, Horii T, Kimura M, Goto Y, Ochiya T, Hatada I. One Argonaute family member, Eif2c2 (Ago2), is essential for development and appears not to be involved in DNA methylation. Genomics. 2007;89:687–696. doi: 10.1016/j.ygeno.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, et al. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002;16:720–728. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudhasani R, Zhu Z, Hutvagner G, Eischen CM, Lyle S, et al. Loss of miRNA biogenesis induces p19Arf-p53 signaling and senescence in primary cells. J Cell Biol. 2008;181:1055–1063. doi: 10.1083/jcb.200802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardella C, Clohessy JG, Alimonti A, Pandolfi PP. Pro-senescence therapy for cancer treatment. Nat Rev Cancer. 2011;11:503–511. doi: 10.1038/nrc3057. [DOI] [PubMed] [Google Scholar]
- Narita M, Lowe SW. Senescence comes of age. Nature Med. 2005;11:920–922. doi: 10.1038/nm0905-920. [DOI] [PubMed] [Google Scholar]
- Newman MA, Hammond SM. Emerging paradigms of regulated microRNA processing. Genes Dev. 2010;24:1086–1092. doi: 10.1101/gad.1919710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohata N, Sone Y, Hanazawa T, Fuse M, Kikkawa N, Yoshino H, Chiyomaru T, Kawakami K, Enokida H, Nakagawa M, Shozu M, Okamoto Y, Seki N. miR-1 as a tumor suppressive microRNA targeting TAGLN2 in head and neck squamous cell carcinoma. Oncotarget. 2011;2:29–42. doi: 10.18632/oncotarget.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noren Hooten N, Abdelmohsen K, Gorospe M, Ejiogu N, Zonderman AB, Evans MK. microRNA expression patterns reveal differential expression of target genes with age. PLoS One. 2010;5:e10724. doi: 10.1371/journal.pone.0010724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- Paroo Z, Ye X, Chen S, Liu Q. Phosphorylation of the human microRNA-generating complex mediates MAPK/Erk signaling. Cell. 2009;139:112–122. doi: 10.1016/j.cell.2009.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering BF, Yu D, Van Dyke MW. Nucleolin interacts with the microprocessor complex to affect microRNAs 15a and 16 biogenesis. J Biol Chem. 2011 doi: 10.1074/jbc.M111.265439. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polager S, Ginsberg D. E2F - at the crossroads of life and death. Trends Cell Biol. 2008;18:528–535. doi: 10.1016/j.tcb.2008.08.003. [DOI] [PubMed] [Google Scholar]
- Pulikkan JA, Dengler V, Peramangalam PS, Peer Zada AA, Müller-Tidow C, Bohlander SK, Tenen DG, Behre G. Cell-cycle regulator E2F1 and microRNA-223 comprise an autoregulatory negative feedback loop in acute myeloid leukemia. Blood. 2010b;115:1768–1778. doi: 10.1182/blood-2009-08-240101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulikkan JA, Peramangalam PS, Dengler V, Ho PA, Preudhomme C, Meshinchi S, Christopeit M, Nibourel O, Müller-Tidow C, Bohlander SK, Tenen DG, Behre G. C/EBPα regulated microRNA-34a targets E2F3 during granulopoiesis and is down-regulated in AML with CEBPA mutations. Blood. 2010a;116:5638–5649. doi: 10.1182/blood-2010-04-281600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina AK, Pardo P, Rottkamp CA, Zhu X, Pereira-Smith OM, Smith MA. Neurons in Alzheimer disease emerge from senescence. Mech Ageing Dev. 2001;123:3–9. doi: 10.1016/s0047-6374(01)00333-5. [DOI] [PubMed] [Google Scholar]
- Rizzo M, Mariani L, Pitto L, Rainaldi G, Simili M. miR-20a and miR-290, multi-faceted players with a role in tumourigenesis and senescence. J Cell Mol Med. 2010;14:2633–2640. doi: 10.1111/j.1582-4934.2010.01173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche KC, Wiechens N, Owen-Hughes T, Perkins ND. The FHA domain protein SNIP1 is a regulator of the cell cycle and cyclin D1 expression. Oncogene. 2004;23:8185–8195. doi: 10.1038/sj.onc.1208025. [DOI] [PubMed] [Google Scholar]
- Rodier F, Campisi J. Four faces of cellular senescence. J Cell Biol. 2011;192:547–556. doi: 10.1083/jcb.201009094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto S, Aoki K, Higuchi T, Todaka H, Morisawa K, et al. The NF90-NF45 complex functions as a negative regulator in the microRNA processing pathway. Mol Cell Biol. 2009;29:3754–3769. doi: 10.1128/MCB.01836-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman DW, Shubert-Coleman J, Furneaux H. P68 RNA helicase unwinds the human let-7 microRNA precursor duplex and is required for let-7-directed silencing of gene expression. J Biol Chem. 2007;282:32773–32779. doi: 10.1074/jbc.M705054200. [DOI] [PubMed] [Google Scholar]
- Santarosa M, Del Col L, Tonin E, Caragnano A, Viel A, Maestro R. Premature senescence is a major response to DNA cross-linking agents in BRCA1-defective cells: implication for tailored treatments of BRCA1 mutation carriers. Mol Cancer Ther. 2009;8:844–854. doi: 10.1158/1535-7163.MCT-08-0951. [DOI] [PubMed] [Google Scholar]
- Schubert J, Brabletz T. p53 Spreads out further: suppression of EMT and stemness by activating miR-200c expression. Cell Res. 2011;21:705–707. doi: 10.1038/cr.2011.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamanna RA, Hoque M, Lewis-Antes A, Azzam EI, Lagunoff D, et al. The NF90/NF45 complex participates in DNA break repair via nonhomologous end joining. Mol Cell Biol. 2011;31:4832–4843. doi: 10.1128/MCB.05849-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatseva T, Lee DY, Deng Z, Yang BB. MicroRNA miR-199a-3p regulates cell proliferation and survival by targeting caveolin-2. J Cell Sci. 2011;124:2826–2836. doi: 10.1242/jcs.077529. [DOI] [PubMed] [Google Scholar]
- Shimada N, Rios I, Moran H, Sayers B, Hubbard K. p38 MAP kinase-dependent regulation of the expression level and subcellular distribution of heterogeneous nuclear ribonucleoprotein A1 and its involvement in cellular senescence in normal human fibroblasts. RNA Biol. 2009;6:293–304. doi: 10.4161/rna.6.3.8497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, George J, Shukla Y. Role of senescence and mitotic catastrophe in cancer therapy. Cell Div. 2010;5:4. doi: 10.1186/1747-1028-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Siomi H, Siomi MC. Posttranscriptional regulation of microRNA biogenesis in animals. Mol Cell. 2010;38:323–332. doi: 10.1016/j.molcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- Sokol DK, Maloney B, Long JM, Ray B, Lahiri DK. Autism, Alzheimer disease, and fragile X: APP, FMRP, and mGluR5 are molecular links. Neurology. 2011;76:1344–1352. doi: 10.1212/WNL.0b013e3182166dc7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soliman MA, Riabowol K. After a decade of study-ING, a PHD for a versatile family of proteins. Trends Biochem Sci. 2007;32:509–519. doi: 10.1016/j.tibs.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Srikantan S, Tominaga K, Gorospe M. Functional interplay between RNA-binding protein HuR and microRNAs. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikantan S, Abdelmohsen K, Lee EK, Tominaga K, Subaran SS, Kuwano Y, Kulshrestha R, Panchakshari R, Kim HH, Yang X, Martindale JL, Marasa BS, Kim MM, Wersto RP, Indig FE, Chowdhury D, Gorospe M. Translational control of TOP2A influences doxorubicin efficacy. Mol Cell Biol. 2011;31:3790–3801. doi: 10.1128/MCB.05639-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Chakravarti D, Cho MS, Liu L, Gi YJ, et al. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature. 2010;467:986–990. doi: 10.1038/nature09459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Shen W, Yang S, Hu F, Li H, Zhu TH. miR-223 and miR-142 attenuate hematopoietic cell proliferation, and miR-223 positively regulates miR-142 through LMO2 isoforms and CEBPβ. Cell Res. 2010;20:1158–1169. doi: 10.1038/cr.2010.134. [DOI] [PubMed] [Google Scholar]
- Sylvestre Y, De Guire V, Querido E, Mukhopadhyay UK, Bourdeau V, et al. An E2F/miR-20a autoregulatory feedback loop. J Biol Chem. 2007;282:2135–2134. doi: 10.1074/jbc.M608939200. [DOI] [PubMed] [Google Scholar]
- Takakura S, Mitsutake N, Nakashima M, Namba H, Saenko VA, et al. Oncogenic role of miR-17-92 cluster in anaplastic thyroid cancer cells. Cancer Sci. 2008;99:1147–1154. doi: 10.1111/j.1349-7006.2008.00800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazawa H, Tsuchiya N, Izumiya M, Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci USA. 2007;104:15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terao M, Fratelli M, Kurosaki M, Zanetti A, Guarnaccia V, Paroni G, Tsykin A, Lupi M, Gianni M, Goodall GJ, Garattini E. Induction of miR-21 by retinoic acid in estrogen receptor-positive breast carcinoma cells: biological correlates and molecular targets. J Biol Chem. 2011;286:4027–4042. doi: 10.1074/jbc.M110.184994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga K, Srikantan S, Lee EK, Subaran SS, Martindale JL, et al. Competitive regulation of nucleolin expression by HuR and miR-494. Mol Cell Biol. 2011;31:4219–4231. doi: 10.1128/MCB.05955-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyner SD, Venkatachalam S, Choi J, Jones S, Ghebranious N, Igelmann H, Lu X, Soron G, Cooper B, Brayton C, et al. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415:45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- Tzatsos A, Paskaleva P, Lymperi S, Contino G, Stoykova S, Chen Z, Wong KK, Bardeesy N. Lysine-specific demethylase 2B (KDM2B)-let-7-enhancer of zester homolog 2 (EZH2) pathway regulates cell cycle progression and senescence in primary cells. J Biol Chem. 2011;286:33061–33069. doi: 10.1074/jbc.M111.257667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Almen GC, Verhesen W, van Leeuwen RE, van de Vrie M, Eurlings C, Schellings MW, Swinnen M, Cleutjens JP, van Zandvoort MA, Heymans S, Schroen B. MicroRNA-18 and microRNA-19 regulate CTGF and TSP-1 expression in age-related heart failure. Aging Cell. 2011;10:769–779. doi: 10.1111/j.1474-9726.2011.00714.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kouwenhove M, Kedde M, Agami R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer. 2011;11:644–656. doi: 10.1038/nrc3107. [DOI] [PubMed] [Google Scholar]
- Van Wynsberghe PM, Kai ZS, Massirer KB, Burton VH, Yeo GW, Pasquinelli AE. LIN-28 co-transcriptionally binds primary let-7 to regulate miRNA maturation in Caenorhabditis elegans. Nat Struct Mol Biol. 2011;18:302–308. doi: 10.1038/nsmb.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela I, Cadiñanos J, Pendás AM, Gutiérrez-Fernández A, Folgueras AR, Sánchez LM, Zhou Z, Rodríguez FJ, Stewart CL, Vega JA, et al. Accelerated ageing in mice deficient in Zmpste24 protease is linked to p53 signalling activation. Nature. 2005;437:564–568. doi: 10.1038/nature04019. [DOI] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ. Lin28: A microRNA regulator with a macro role. Cell. 2010;140:445–449. doi: 10.1016/j.cell.2010.02.007. [DOI] [PubMed] [Google Scholar]
- Wang W. Regulatory RNA-binding proteins in senescence. Ageing Res Rev. 2012 doi: 10.1016/j.arr.2012.02.006. This issue. [DOI] [PubMed] [Google Scholar]
- Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Yang X, Cristofalo VJ, Holbrook NJ, Gorospe M. Loss of HuR is linked to reduced expression of proliferative genes during replicative senescence. Mol Cell Biol. 2001;21:5889–5898. doi: 10.1128/MCB.21.17.5889-5898.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem. 2007;282:2130–2134. doi: 10.1074/jbc.C600252200. [DOI] [PubMed] [Google Scholar]
- Wu H, Sun S, Tu K, Gao Y, Xie B, Krainer AR, Zhu J. A splicing-independent function of SF2/ASF in microRNA processing. Mol Cell. 2010;38:67–77. doi: 10.1016/j.molcel.2010.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Takeshita F, Hino Y, Fukunaga S, Kudo Y, Tamaki A, Matsunaga J, Takahashi RU, Takata T, Shimamoto A, Ochiya T, Tahara H. miR-22 represses cancer progression by inducing cellular senescence. J Cell Biol. 2011;193:409–424. doi: 10.1083/jcb.201010100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue D, Peng Y, Wang F, Allan RW, Cao D. RNA-binding protein LIN28 is a sensitive marker of ovarian primitive germ cell tumours. Histopathology. 2011;59:452–459. doi: 10.1111/j.1365-2559.2011.03949.x. [DOI] [PubMed] [Google Scholar]
- Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, et al. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Feng M, Jiang X, Wu Z, Li Z, et al. miR-449a and miR-449b are direct transcriptional targets of E2F1 and negatively regulate pRb-E2F1 activity through a feedback loop by targeting CDK6 and CDC25A. Genes Dev. 2009;23:2388–2393. doi: 10.1101/gad.1819009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Lin X, Zhong X, Kaur S, Li N, Liang S, Lassus H, Wang L, Katsaros D, Montone K, Zhao X, Zhang Y, Bützow R, Coukos G, Zhang L. Double-negative feedback loop between reprogramming factor LIN28 and microRNA let-7 regulates aldehyde dehydrogenase 1-positive cancer stem cells. Cancer Res. 2010;70:9463–9472. doi: 10.1158/0008-5472.CAN-10-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Bi L, Zheng B, Ji L, Chevalier D, et al. The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc Natl Acad Sci USA. 2008;105:10073–10078. doi: 10.1073/pnas.0804218105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D, Xu G, Ghandhi S, Hubbard K. Modulation of the expression of p16INK4a and p14ARF by hnRNP A1 and A2 RNA binding proteins: implications for cellular senescence. J Cell Physiol. 2002;193:19–25. doi: 10.1002/jcp.10147. [DOI] [PubMed] [Google Scholar]