Abstract
Summary: Flagellar and translocation-associated type III secretion (T3S) systems are present in most Gram-negative plant- and animal-pathogenic bacteria and are often essential for bacterial motility or pathogenicity. The architectures of the complex membrane-spanning secretion apparatuses of both systems are similar, but they are associated with different extracellular appendages, including the flagellar hook and filament or the needle/pilus structures of translocation-associated T3S systems. The needle/pilus is connected to a bacterial translocon that is inserted into the host plasma membrane and mediates the transkingdom transport of bacterial effector proteins into eukaryotic cells. During the last 3 to 5 years, significant progress has been made in the characterization of membrane-associated core components and extracellular structures of T3S systems. Furthermore, transcriptional and posttranscriptional regulators that control T3S gene expression and substrate specificity have been described. Given the architecture of the T3S system, it is assumed that extracellular components of the secretion apparatus are secreted prior to effector proteins, suggesting that there is a hierarchy in T3S. The aim of this review is to summarize our current knowledge of T3S system components and associated control proteins from both plant- and animal-pathogenic bacteria.
INTRODUCTION
Higher eukaryotes such as plants, animals, and humans are permanently exposed to the risk of bacterial infections, which often lead to severe and even lethal diseases. Major infectious agents are Gram-negative bacteria, which utilize at least six different protein secretion systems (type I to type VI secretion systems) to transport bacterial virulence factors into the surrounding milieu or directly into the host cell. Protein secretion systems from Gram-negative bacteria differ significantly in structure, regulation, and substrate specificity and are summarized in recent review articles (198, 215, 218, 247, 253, 258, 453, 582). Similar systems are employed by Gram-positive bacteria, but they also contain an additional type of protein secretion system, designated type VII, that was identified in mycobacteria (558). Most pathogens use a combination of several protein secretion systems to successfully conquer their respective host organisms. Although the impact of secretion systems on bacterial virulence can vary in different pathogens, an essential role in pathogenicity has often been assigned to the type III secretion (T3S) system, which delivers bacterial proteins, so-called effector proteins, into the cytosol of eukaryotic cells (107, 465, 466, 512). This transkingdom protein transport enables the pathogen to interfere with host cellular pathways for its own benefit.
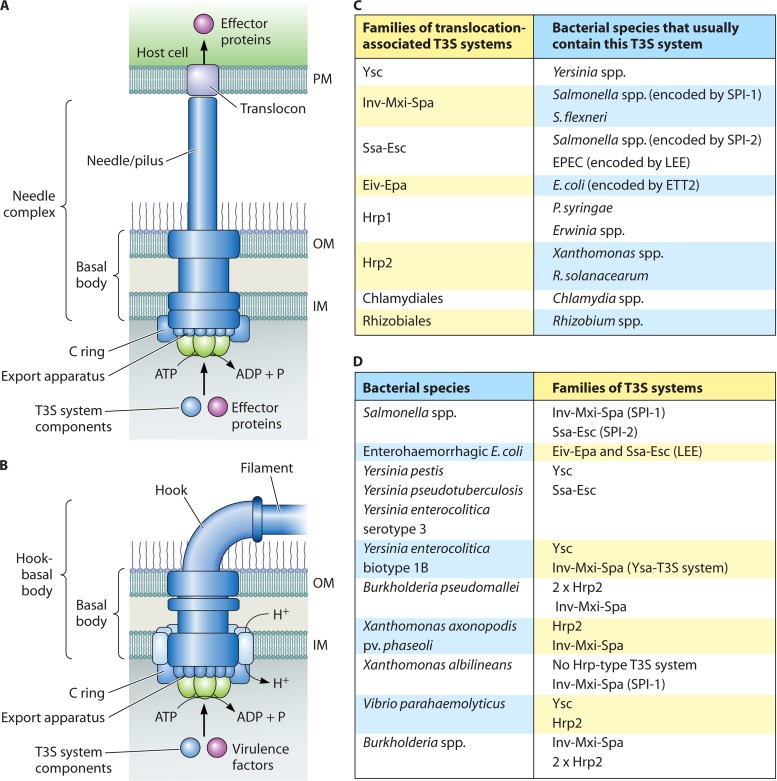
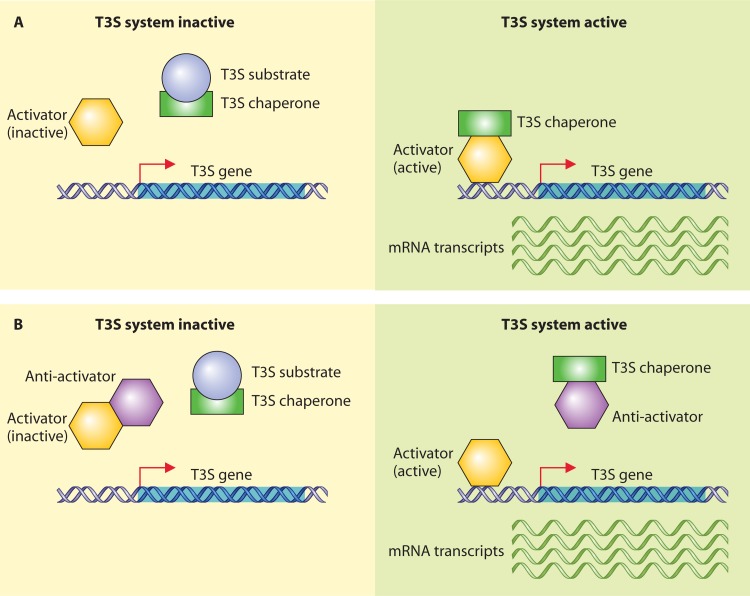
T3S systems are highly complex nanomachines that consist of more than 20 components. The membrane-spanning core apparatus is associated with an extracellular pilus-like appendage that is assumed to serve as a channel for transport of secreted proteins to the host-pathogen interface. The translocation of effector proteins into eukaryotic cells is probably mediated by a bacterial channel-like translocon that inserts into the host plasma membrane (Fig. 1A). Notably, the term “T3S system” does not refer only to secretion systems that translocate effector proteins (so-called translocation-associated T3S systems) but also to the bacterial flagellum, which is a key motility organelle and is connected via a hook to the bacterial filament (Fig. 1B). In contrast to translocation-associated T3S systems, flagellar T3S systems mainly secrete extracellular components of the flagellum, such as hook and filament proteins. However, the secretion of virulence factors by flagellar T3S systems has also been observed (617, 618).
Fig 1.
Overview of translocation-associated and flagellar T3S systems from animal- and plant-pathogenic bacteria. (A) Model of the translocation-associated T3S system. The basal body of the T3S system spans the bacterial IM and OM and consists of ring structures that are presumably connected by a periplasmic rod. The basal body is associated via an extracellular needle (animal-pathogenic bacteria) or pilus (plant-pathogenic bacteria) with a channel-like translocon in the host plasma membrane. The basal body and the needle from animal-pathogenic bacteria are referred to as the needle complex. The energy for the docking and unfolding of T3S substrates, including extracellular components of the T3S system and effector proteins, is probably provided by a cytoplasmic ATPase (shown in green) associated with the T3S system. Note that the cytoplasmic C ring is predicted only for translocation-associated T3S systems. A more detailed representation of single components of translocation-associated T3S systems is given in Fig. 2. (B) Model of the flagellar T3S system. The flagellar basal body is associated via an extracellular hook with the flagellar filament, which is 10 to 20 μm long and is the main bacterial motility organelle. The basal body is surrounded by 8 to 11 stator complexes that drive flagellar rotation and contain proton-conducting channels. The flagellar basal body and the hook are referred to as the hook-basal body. A detailed description of individual components of the flagellar T3S systems is provided in Fig. 3. (C) Summary of different families of translocation-associated T3S systems from bacterial pathogens and symbionts of plants or animals. The SPI-1-like Eiv-Epa T3S system encoded by the ETT2 gene cluster from E. coli is active in only a few strains. Note that P. syringae strains belonging to the phylogenetic subgroup 2c appear to encode an unusual T3S system that is only distantly related to the Hrp1-type T3S system (97). (D) Examples of bacterial species that possess more than one translocation-associated T3S system. Please note that most species contain an additional flagellar T3S system that is not listed in this table.
Given the architecture of T3S systems, it is assumed that T3S is a hierarchical process and that extracellular components of the secretion apparatus are secreted prior to effector proteins. Similarly, the secretion of hook components of flagellar T3S systems probably precedes the secretion of filament proteins. In the past 5 years, significant progress has been made in the analysis of the structures and functions of many core components of T3S systems as well as of T3S-associated control proteins. The aim of this review is to summarize our current knowledge of the architecture of T3S systems and the control mechanisms underlying T3S in plant- and animal-pathogenic bacteria. For a detailed description of individual proteins or regulatory mechanisms, the reader is also referred to excellent previous overview articles that provide summaries on the following topics: translocation-associated T3S systems (29, 72, 105, 161, 199, 217, 557), flagellar T3S systems (92, 161, 343, 377, 428, 549), T3S chaperones (175, 431), structures and functions of individual components of T3S systems (46, 70, 243, 281, 283, 349, 353, 389, 395, 482), and control mechanisms underlying T3S and gene expression (64, 106, 129, 212, 370, 421, 547, 555, 588).
VARIATIONS ON A THEME—DIFFERENCES AND SIMILARITIES OF T3S SYSTEMS
The structural components of T3S systems are encoded by chromosomal or plasmid-borne gene clusters that were probably acquired during evolution by horizontal gene transfer. According to phylogenetic differences in amino acid sequences, T3S systems from animal- and plant-pathogenic or symbiotic bacteria have been classified into different families, including flagellar, Ysc, Inv-Mxi-Spa, Ssa-Esc, Hrp1, and Hrp2 T3S systems as well as T3S systems of the Chlamydiales and Rhizobiales families (Fig. 1C). Ysc, Inv-Mxi-Spa, and Ssa-Esc-T3S systems have been analyzed intensively in species of the animal-pathogenic bacteria Yersinia, Salmonella, and Shigella and in enteropathogenic Escherichia coli (EPEC), while Hrp1 and Hrp2 T3S systems have been studied mainly in the plant-pathogenic bacteria Xanthomonas spp., Ralstonia solanacearum, and Pseudomonas syringae. The Inv-Mxi-Spa T3S system from Salmonella spp. and the Ssa-Esc T3S system from EPEC and Salmonella spp. are also referred to by the genomic loci that encode them (Salmonella pathogenicity island 1 [SPI-1] and SPI-2 for Salmonella spp. and locus of enterocyte effacement [LEE] for EPEC) (Fig. 1C).
Many bacteria contain more than one T3S system, including a flagellar T3S system and one or several translocation-associated T3S systems of the same or different families that might be of importance at different stages of the infection process (Fig. 1D). The SPI-1 T3S system of Salmonella spp., for instance, promotes bacterial pathogenicity before the invasion of host cells, and the corresponding genes are expressed during the initial bacterial contact with the intestinal epithelium. In contrast, the SPI-2 T3S system is activated only after bacterial entry into the eukaryotic cell cytosol. The different functional requirements of both systems might explain why Salmonella spp. possess approximately 10 to 100 SPI-1 T3S systems per cell but only 1 or a few T3S systems of the SPI-2 family type (83, 291). Different translocation-associated T3S systems have also been identified in the animal-pathogenic bacteria Yersinia spp. and Burkholderia spp. Interestingly, Burkholderia spp. contain not only a SPI-1 T3S system but also Hrp-type T3S systems that are usually specific for plant-pathogenic bacteria (449, 522) (Fig. 1D). In most cases, it is still unclear whether the different types of T3S system are required for interactions with different hosts.
Interestingly, translocation-associated T3S systems not only are linked exclusively to bacterial pathogenicity but also can contribute to symbiotic interactions, as shown for the Rhizobium-legume symbiosis (173, 574). Genes encoding components of T3S systems have also been identified in other symbiotic (e.g., Photorhabdus luminescens, Sodalis glossinidius, and the Sitophilus zeamais primary endosymbiont) and nonpathogenic bacteria (e.g., E. coli, Pseudomonas fluorescens, Desulfovibrio vulgaris, Myxococcus xanthus, and Verrucomicrobium spinosum). The precise role of T3S genes during the life cycle of these bacteria remains to be investigated.
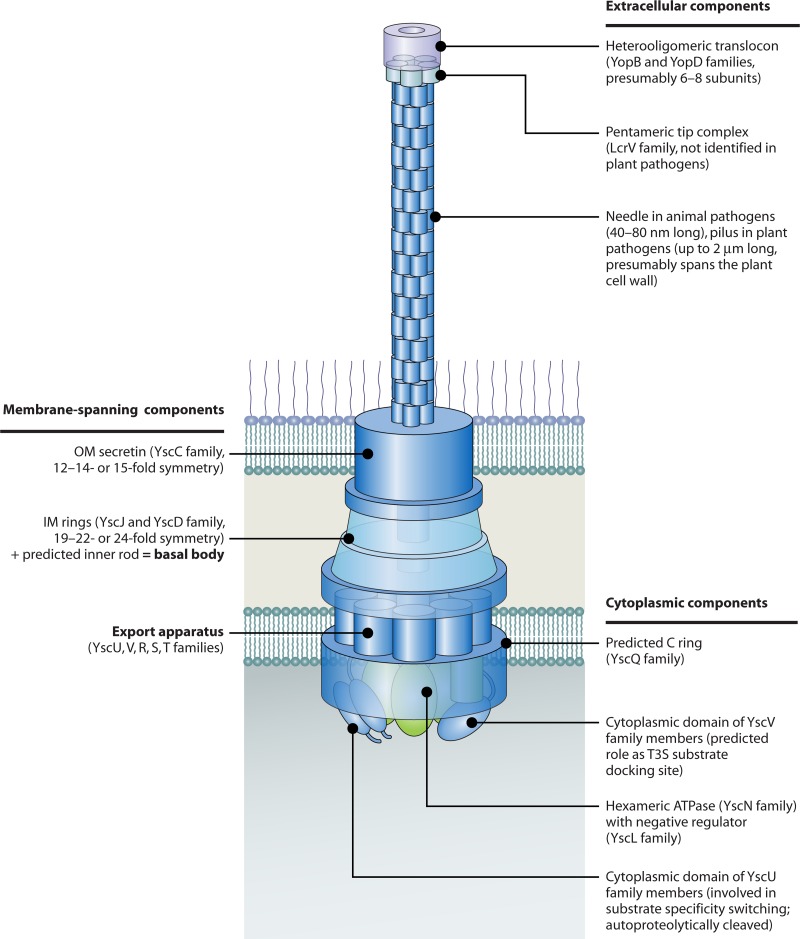
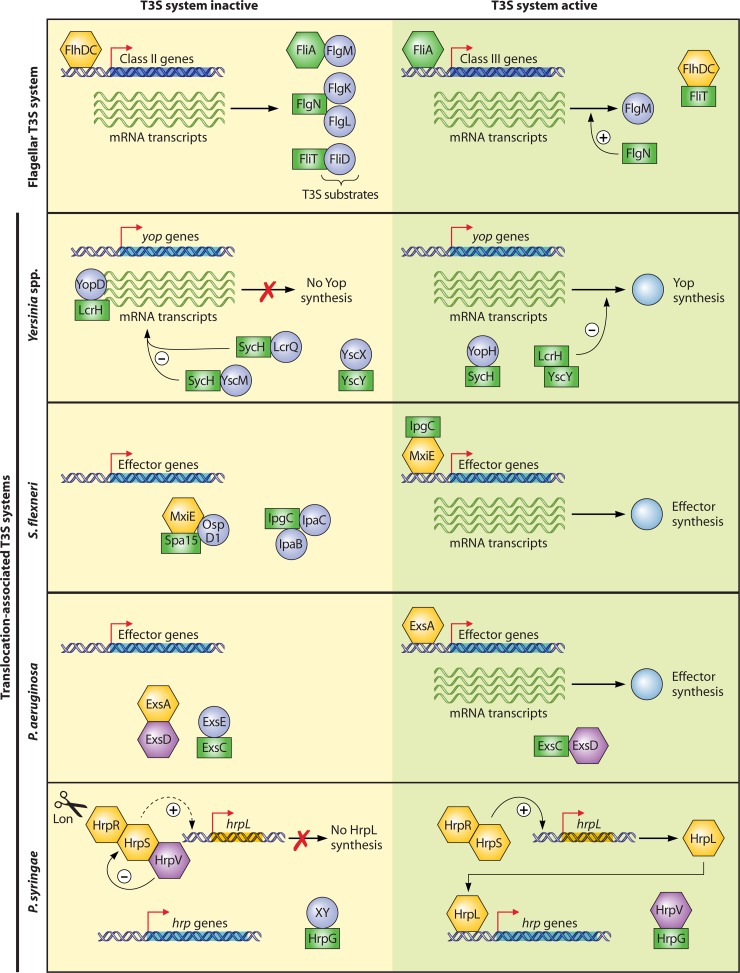
Comparative sequence analyses revealed that at least 9 of the more than 20 components of translocation-associated T3S systems are conserved among plant- and animal-pathogenic bacteria. They likely constitute the core components of the secretion apparatus in the inner membrane (IM) and outer membrane (OM). The nomenclature of these proteins refers to the Ysc proteins from the animal-pathogenic bacterium Yersinia (48). Eight components are also conserved in the flagellar T3S system, suggesting that the membrane-spanning secretion apparatuses of flagellar and translocation-associated T3S systems share a similar overall architecture. This assumption was confirmed by electron microscopy (EM) studies of isolated flagellar and translocation-associated T3S systems from Salmonella spp. and Shigella flexneri, respectively. Both systems consist of ring structures in the IM and OM that enclose a transport channel with an inner diameter of 2 to 3 nm (45, 139, 179, 291, 348, 478) (Fig. 2 and 3). The IM rings are associated with the export apparatus, which is built by members of the YscR, YscS, YscT, YscU, and YscV families and is connected to a predicted cytoplasmic C ring and an ATPase complex (see below). According to a commonly used nomenclature, the IM and OM rings that are linked by a central periplasmic rod structure are also termed the “basal body.” In contrast, the term “needle complex” refers to basal bodies of translocation-associated T3S systems that are associated with the extracellular needle (Fig. 1A). In flagellar T3S systems, the corresponding structure is called the “hook-basal body” (Fig. 1B). Individual components of the needle complex, the hook-basal body, and the export apparatus are discussed in this review and are presented in more detail in Fig. 2 and 3.
Fig 2.
Schematic representation of individual components of translocation-associated T3S systems from animal- and plant-pathogenic bacteria. Conserved membrane-spanning components of the T3S system include the OM secretin (YscC family) and constituents of the IM ring (YscD and YscJ family) and the export apparatus (YscU, -V, -R, -S, and -T families). The IM ring and the export apparatus are associated with the predicted C ring (presumably a multimer of members of the YscQ family) and the hexameric ATPase (depicted in green), which might provide the energy to facilitate the docking and entry of T3S substrates into the inner channel of the secretion system. Additional cytoplasmic components of the T3S system are the predicted regulator of the ATPase (YscL family) and the cytoplasmic domains of YscU and YscV family members, which are probably involved in substrate docking. Note that the composition of the export apparatus and the cytoplasmic parts of the secretion system is speculative and that multiple copies of a single substituent (e.g., members of the YscV protein family) can be involved in the assembly of the T3S system. Extracellular components of the T3S system include the needle (animal-pathogenic bacteria) and pilus (plant-pathogenic bacteria), which differ in length and serve as transport channels for secreted proteins at the host-pathogen interface. Translocation of effector proteins across the host plasma membrane is mediated by the channel-like translocon, which is a hetero-oligomeric protein complex and is connected to the needle via a tip complex that consists of members of the LcrV protein family. Tip complexes have so far been identified and/or characterized only for animal-pathogenic bacteria.
Fig 3.
Schematic representation of components of the flagellar T3S system. The membrane-spanning basal body consists of two OM rings (L and P rings) that are connected via a distal and proximal rod to the IM ring (MS ring). The MS ring is surrounded by 8 to 11 stator complexes in the IM that provide proton-conducting channels and is associated with the export apparatus, the C ring, the ATPase, and the regulator of the ATPase. The architecture of the export apparatus and of the cytoplasmic components of the flagellar T3S system is probably similar to that in translocation-associated T3S systems (see Fig. 2). Structures that are different from those in translocation-associated T3S systems are indicated.
OUTER APPEARANCE—EXTRACELLULAR APPENDAGES OF FLAGELLAR AND TRANSLOCATION-ASSOCIATED T3S SYSTEMS
Needles, Pili, and Flagella—Protein Transport Channels and Motility Organelles
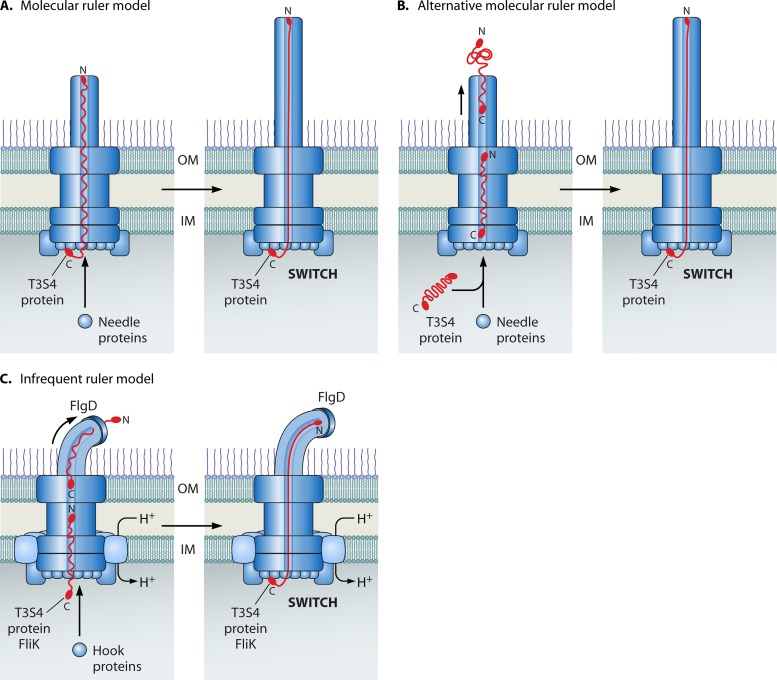
Translocation-associated and flagellar T3S systems are associated with extracellular appendages that differ significantly in their structures and compositions. The basal body of translocation-associated T3S systems is linked to an extracellular pilus (plant pathogens) or a needle (animal pathogens) (Fig. 2) that contains an inner channel through which secreted proteins might be transported. The pilus from plant pathogens is up to 2 μm long and presumably spans the plant cell wall, which is a major obstacle for the transport of bacterial effector proteins into plant cells (59, 233, 248, 315, 463, 568, 594). The T3S needle from animal pathogens is significantly shorter than the pilus from plant pathogens and often has a determined length, which varies from 40 to approximately 80 nm in different pathogens (45, 224, 257, 291, 540). Needle assembly is probably initiated in the periplasm and occurs at the tip of the needle (444, 478). Since purified needle proteins (members of the YscF family) build structures of several micrometers, the regulation of needle length likely depends on the context of the T3S system (444, 447) (see below).
Nuclear magnetic resonance (NMR) and crystal structure analyses of needle proteins suggest that they form a hairpin-like structure with a central head region that connects the helical N- and C-terminal regions (131, 448, 530, 585, 630). While the central head region is presumably located at the needle surface, the C-terminal helix is buried in the needle wall and might undergo a conformational change upon polymerization (131, 444). Interestingly, needle components share structural similarities with components of the flagellar filament, although they do not share sequence similarities at the amino acid level. Thus, the needle subunit MxiH from Shigella spp. assembles into a helical structure with 5.6 subunits per turn, which is similar to the helical symmetry of the flagellar filament (approximately 5.5 subunits per turn) (103, 616). Furthermore, crystal structure analysis of the needle protein PrgI and the needle tip protein (see below) SipD from Salmonella spp. revealed that five molecules of PrgI assemble with five molecules of SipD to form the needle tip (337). However, the symmetry of the needle from Salmonella spp. is apparently highly variable, with an average of approximately 6.2 subunits per turn (191). Structural rearrangements in the needle might therefore occur and could contribute to the transmission of signals such as host cell sensing from the tip of the needle to the base (see below). This hypothesis is supported by the finding that several mutations in needle proteins lead to constitutive T3S (102, 131, 268, 552, 569).
In addition to the needle, T3S systems from some gastrointestinal pathogens (enterohemorrhagic E. coli, EPEC, and Citrobacter rodentium) contain a filament structure on top of the needle that is composed of the filament protein EspA. The EspA filament might connect the needle to the translocon (see below) and encloses an inner channel with a diameter of 2 to 2.5 nm (117, 277, 497). Interestingly, EspA has a similar helical structure to that of the flagellar filament protein FliC (116, 277, 616). However, the EspA filament has a smaller external diameter (12 nm versus 24 nm for the flagellar filament), which could be caused by the smaller size of EspA (192 amino acids versus 494 amino acids for FliC) (116, 616). In addition to EspA filaments from gastrointestinal pathogens, a sheath-like surface appendage with a diameter of 30 to 70 nm and a highly variable length has also been observed for the SPI-2-encoded T3S system from Salmonella enterica serovar Typhimurium (83). The precise function and composition of this surface structure, however, have not yet been investigated.
In contrast to translocation-associated T3S systems, the flagellar T3S system is associated with an extracellular hook, which is composed of approximately 120 molecules of FlgE and has a length of 55 ± 6 nm (222). The hook is connected to the flagellar filament via the hook-filament junction proteins FlgK and FlgL, which are incorporated at the tip of the hook (228, 237) (Fig. 3). The filament consists of around 20,000 subunits of FliC and is 10 to 15 μm long. It terminates with a pentameric cap structure that is built by the filament cap protein FliD (616). fliD mutants are deficient in filament formation, suggesting that the filament cap is required for the assembly of FliC monomers to form a helical structure (227). EM studies revealed that the filament cap contains five leg-like anchor regions and provides a docking site for one FliC molecule. Rotation of the cap allows the entry of the next FliC molecule and thus could promote the folding and insertion of FliC monomers into the growing filament structure (615).
Port of Entry for Effector Proteins—the Translocon and the Tip Complex
The translocon.
Translocation of effector proteins into the eukaryotic cell cytosol is mediated by a bacterial channel-like translocon, which is inserted into the host plasma membrane and usually consists of two hydrophobic proteins that are referred to as major (e.g., YopB, IpaB, SipB, and EspD, with two transmembrane helices) and minor (e.g., YopD, IpaC, SipC, and EspB, with one transmembrane helix) translocon proteins (Table 1). Translocon proteins act outside the bacterial cell; however, a Yersinia yopB null mutant cannot be trans-complemented upon coinfection with another Yersinia strain that delivers YopB (473). Similar observations were reported for the translocon proteins PopB and PopD from Pseudomonas aeruginosa, suggesting that translocon proteins act in cis (95). It is therefore assumed that the membrane insertion of translocon proteins is closely linked to the activity of the corresponding secretion apparatus.
Table 1.
Conserved components of flagellar and translocation-associated T3S systemsa
| Flagellar T3S system from Salmonella spp. | Translocation-associated T3S systems |
Predicted functions/characteristics | Published structures |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Animal-pathogenic bacteria |
Plant-pathogenic bacteria |
Protein(s) | Reference(s) | ||||||
| Yersinia spp. | Salmonella spp. (SPI-1) | EPEC | S. flexneri | Xanthomonas spp. | P. syringae | ||||
| Cytoplasmic components | |||||||||
| FliI | YscN | InvC | EscN | Spa47 | HrcN | HrcN | ATPase | EscN, FliI | 239, 627 |
| FliH | YscL | — | EscL (Orf5) | MxiN | HrcL | HrpE | Regulator of the ATPase | ||
| FliM, FliN, FliG | YscQ | SpaO | EscQ (SepQ) | Spa33 | HrcQ | HrcQA + HrcQB | Predicted C ring component, possibly involved in substrate docking | HrcQB, FliM, FliG-FliM, FliG, FliN | 60, 61, 170, 429, 436 |
| IM components of the export apparatus | |||||||||
| FlhB | YscU | SpaS | EscU | Spa40 | HrcU | HrcU | Autocatalytic cleavage at the NPTH motif; cytoplasmic domain is presumably involved in the substrate specificity switch and in substrate recognition | EscUC, SpaSC, YscUC, Spa40C | 130, 334, 597, 626 |
| FlhA | YscV | InvA | EscV | MxiA | HrcV | HrcV | Cytoplasmic domain is possibly involved in substrate docking | FlhAC, InvAC | 23, 320, 388, 474, 606 |
| FliP | YscR | InvL/SpaP | EscR | Spa24 | HrcR | HrcR | |||
| FliQ | YscS | SpaQ | EscS | Spa9 | HrcS | HrcS | |||
| FliR | YscT | InvN/SpaR | EscT | Spa29 | HrcT | HrcT | |||
| YscD | PrgH | EscD | MxiG | HrcD | HrcD | Not highly conserved; forms multimeric ring structures | PrgH, MxiG, YscDN | 25, 335, 348, 356, 516 | |
| FliO | — | — | — | — | — | — | |||
| Periplasmic/IM-associated components | |||||||||
| FliF (MS ring) | YscJ | PrgK | EscJ | MxiJ | HrcJ | HrcJ | Forms multimeric ring structures; lipoprotein | EscJ, PrgK | 112, 348, 614 |
| FlgB, FlgC, FlgF, FlgG | YscI | PrgJ | EscI (rOrf8) | MxiI | HrpB2 | — | Predicted inner rod protein | ||
| OM components | |||||||||
| — | YscC | MxiD | EscC | InvG | HrcC | HrcC | Secretin; forms a multimeric channel in the OM | YscC, EscC, InvG | 67, 108, 516 |
| — | YscW | MxiM | — | InvH | — | — | Pilotin | MxiM, MxiD-MxiM | 305, 417 |
| FlgI | — | — | — | — | — | — | P ring | ||
| FlgH | — | — | — | — | — | — | L ring | ||
| Extracellular components | |||||||||
| FlgJ | — | — | — | — | — | Rod cap protein | FlgJ | 214, 270 | |
| FlgE | — | — | — | — | — | Hook protein | |||
| FlgD | — | — | — | — | — | Hook cap | |||
| FlgL, FlgK | — | — | — | — | — | Hook-filament junction proteins | |||
| FliC | — | — | — | — | — | Filament protein | FliC | 616 | |
| FliD | — | — | — | — | — | Filament cap | FliD | 615 | |
| — | YscF | PrgI | EscF | MxiH | HrpE | HrpA | Pilus or needle proteins | YscF-YscE-YscG complex, PrgI, MxiH, PrgI-SipD, PscF-PscE-PscG complex, BsaL (Burkholderia pseudomallei) | 103, 131, 337, 444, 448, 530, 585, 630 |
| — | LcrV | SipD | — | IpaD | — | — | Needle tip proteins | LcrV, SipD, IpaD, BipD | 87, 140, 252, 337, 424 |
| — | YopB, YopD | SipB, SipC | EspD, EspB | IpaB, IpaC | HrpF, XopA | HrpK | Translocon proteins | EspB, EspD, IpaB, IpaB-IpgC, PcrH-PopD, SipB, PrgI-SipD | 26, 163, 236, 249, 274, 337, 338 |
| — | — | — | EspA | — | — | — | Filament on top of the needle | EspA | 613 |
Proteins from flagellar and translocation-associated T3S systems with similar functions and/or homologous sequences are given in the same row. Alternative protein names are given in parentheses. —, no homologous or functionally equivalent proteins have been identified.
Translocon proteins form a hetero-oligomeric protein complex of presumably six to eight subunits with an internal diameter of approximately 1.2 to 3.5 nm (44, 113, 236, 339, 387, 403, 490, 569). Recent crystal structure analysis of the translocon proteins IpaB from S. flexneri and SipB from Salmonella spp. revealed structural similarities to bacterial pore-forming toxins, which could indicate a common evolutionary origin of both protein families and similar mechanistics underlying membrane insertion (26). Experimental evidence reported for translocon proteins from animal-pathogenic bacteria suggests that the formation of a functional translocation channel depends on the composition of the host cell membrane. Thus, infection studies with P. aeruginosa revealed that some cell lines are resistant against T3S-mediated protein injection, indicating the requirement of certain host cell properties for efficient effector protein translocation (357, 471). This observation was supported by the finding that the alteration of the host plasma membrane composition renders cells insensitive against T3S by P. aeruginosa (55). It was already previously proposed that the formation of a functional T3S translocon occurs preferentially in specific microdomains of the host cell membrane that are rich in cholesterol and glycosphingolipids (490). These microdomains, also known as lipid rafts, are often involved in the attachment of invading bacterial pathogens, bacterial cytotoxicity, and contact-mediated T3S, as shown, for example, for Bordetella spp. and S. flexneri (158, 187, 563). Notably, depletion of cholesterol from host cell membranes impairs bacterial entry of S. flexneri (301) and affects effector protein translocation by P. aeruginosa and EPEC (13, 570). It is therefore assumed that cholesterol is required for the efficient formation of a functional translocation channel, while it is apparently dispensable for the membrane insertion of translocon proteins per se, as shown for P. aeruginosa (490, 570).
In agreement with the predicted contribution of cholesterol to channel formation is the finding that the translocon proteins PopB, SipB, and IpaB, from P. aeruginosa, Salmonella spp., and S. flexneri, respectively, can bind cholesterol (172, 216). Furthermore, IpaB from S. flexneri also associates with raft-containing liposomes (563) and binds to the transmembrane protein CD44, which preferentially localizes to lipid rafts and was previously shown to be recruited to the site of bacterial attachment during infection by EPEC (207, 301, 506). CD44 is a receptor for hyaluronic acid and other ligands and contains a cytoplasmic domain that interacts with actin cytoskeleton-associated proteins. In this context, it is interesting that the host actin cytoskeleton was proposed to contribute to translocon channel formation. Thus, the actin-depolymerizing agents cytochalasin D and latrunculin B were shown to inhibit the translocation of ExoS by the T3S system from P. aeruginosa (570). Furthermore, it was previously reported that deletion of the T3S effector gene yopE in Yersinia pseudotuberculosis resulted in increased amounts of translocated effector proteins and increased pore-forming activity that was dependent on actin polymerization (3, 572). The inhibition of pore formation by YopE was linked to its GTPase-activating activity, which modulates the activity of Rho GTPases and is required for the YopE-mediated depolymerization of actin filaments (361, 572, 576).
Inhibition of pore formation was also shown for the cysteine protease YopT, which cleaves Rho GTPases, as well as for the effector protein YopK (141, 225, 361, 500, 572, 640). Furthermore, an increase in the size of the translocation pore was observed for mutants of Salmonella spp. that were deprived of individual effector genes (95). This indicates that some effectors could have an anti-pore-formation activity after being translocated into the host cell. The predicted inhibitory activity on pore formation could impose a feedback regulation on effector protein translocation and might ensure that all infected cells contain similar levels of effector proteins and are not killed too rapidly by an effector overdose (562). Furthermore, the negative control of pore formation by translocated effector proteins might also counteract the proinflammatory responses of the host cell that are activated in the presence of the translocation pore. Previous mutant studies of Yersinia spp. revealed that it is the presence of the T3S translocon that triggers proinflammatory responses, which are in turn suppressed by translocated effector proteins (503). This predicted feedback regulation might itself be controlled by the degradation of effector proteins inside the host cell, as shown for YopE from Yersinia spp., which is degraded by the eukaryotic ubiquitination machinery. Because the accumulation of a degradation-resistant YopE mutant derivative leads to reduced translocation of effector proteins into the host cell, it was proposed that the pathogen exploits the host proteasome to indirectly regulate effector protein delivery (194, 470). Ubiquitination and proteasome-mediated degradation were also reported for the effector proteins SopE and SopB from Salmonella spp. (276, 290).
Compared to the case for animal-pathogenic bacteria, translocon proteins from most plant-pathogenic bacteria have been studied less intensively, and the precise composition of the translocation channel still remains to be investigated. In Xanthomonas spp., effector protein translocation depends on HrpF, which is a predicted component of a channel-like protein complex (74). Interestingly, translocation of individual effector proteins appears to be reduced upon recognition of the effector protein AvrBs2 by the corresponding pepper resistance protein Bs2, which initiates plant defense responses (631). The molecular mechanisms underlying this apparent feedback control are unknown. In contrast to Xanthomonas campestris pv. vesicatoria, several predicted translocon proteins from other plant-pathogenic bacteria are not essential for pathogenicity, suggesting that additional proteins such as harpins are involved in effector protein translocation (47, 300, 364, 438). Harpins are small, heat-stable T3S substrates from plant-pathogenic bacteria that are rich in glycine and can elicit plant defense responses when infiltrating the plant apoplast at high concentrations. In line with the predicted role of harpins in effector protein translocation is the finding that the harpin protein HrpZ from P. syringae forms transmembrane channels and assembles into oligomeric structures that consist of at least 16 molecules of HrpZ (155, 211, 307, 308). Alternatively, however, some harpin proteins can also target the plant cell wall (84, 174, 317).
The tip complex.
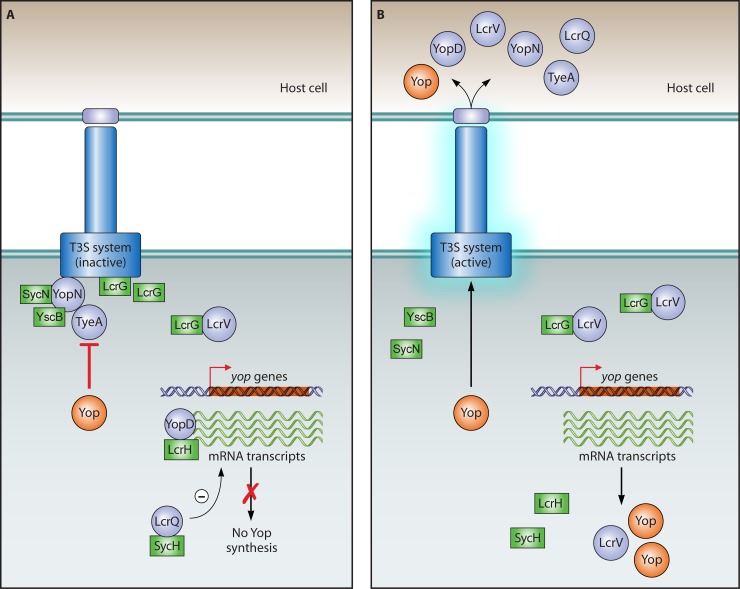
The T3S translocon from animal-pathogenic bacteria is presumably connected to the needle by a tip complex that was initially visualized by scanning transmission EM studies of needles from Yersinia enterocolitica (396). The tip complex might serve different purposes, including sensing of the host cell contact, control of T3S, and insertion of the translocon into the host plasma membrane.
The tip complex from Yersinia spp. consists of five molecules of the hydrophilic LcrV protein that oligomerize in vitro and form ring-like structures with an internal diameter of 3 to 4 nm (63, 197, 393, 395, 396). The hydrophobic translocon proteins YopB and YopD from Yersinia spp. probably do not participate in tip complex formation. This is in contrast to the tip complex from S. flexneri, which contains one molecule of the translocon protein IpaB, which forms a complex with four hydrophilic molecules of the tip protein IpaD (46, 164, 252, 418, 569). It was proposed that IpaB and IpaD plug the needle prior to host cell contact and are thus involved in the regulation of T3S (362). In agreement with this model, deletion of ipaB and ipaD leads to constitutive T3S in vitro (432, 442, 461, 501, 569). Similar findings were reported for a P. aeruginosa mutant lacking the predicted tip complex protein PcrV (310, 407). However, the deregulation of T3S in the absence of a tip protein does not appear to be a general phenomenon, because it was not observed for a Yersinia sp. lcrV mutant (32, 481). It is assumed that a conformational change in the tip complex upon completion of the translocon is transduced via the needle subunits to the base of the T3S system and activates the secretion of effector proteins (131, 569). Alternatively, recent experimental evidence reported for Salmonella spp. suggests that the secretion of effector proteins could also be activated upon a shift in the extracellular pH that is sensed by the needle (623) (see below).
ARCHITECTURE OF THE BASAL BODY AND EXPORT APPARATUS IN TRANSLOCATION-ASSOCIATED AND FLAGELLAR T3S SYSTEMS
Translocation-Associated and Flagellar T3S Systems Contain Different OM Ring Components
The OM rings of translocation-associated T3S systems are built by proteins belonging to the secretin family (Table 1), whose members also participate in the assembly of type II secretion systems and type IV pili but are absent from flagellar T3S systems. Secretins consist of an N-terminal domain with a cleavable signal sequence that directs the protein for Sec-dependent transport across the IM into the periplasm. The N-terminal region of T3S secretins is not highly conserved among different species and might form a periplasmic neck structure that connects the secretin channel to components of the IM ring (223, 492). The C-terminal membrane-spanning region of secretins multimerizes to form OM rings with a diameter of approximately 11 nm and a 12- to 14-fold symmetry (66, 108, 223, 286, 516). A 15-fold symmetry was recently reported for the OM ring of the translocation-associated T3S system from S. Typhimurium (493). Oligomerization and channel formation by secretins are often mediated by pilotins, which are small OM lipoproteins with limited sequence homology that have been identified in animal-pathogenic bacteria (e.g., see references 66, 108, 114, and 495). In the absence of their cognate pilotins, secretins localize to the IM, as shown for InvG from Salmonella spp. (114) and YscC from Yersinia spp. (66). Interestingly, experimental evidence for the presence of pilotins in plant-pathogenic bacteria is missing.
In contrast to the secretins of translocation-associated T3S systems, flagellar T3S systems contain an L (lipopolysaccharide) ring in the OM, consisting of the lipoprotein FlgH (254, 491) (Fig. 3 and Table 1). The L ring is associated with a periplasmic P (peptidoglycan) ring, which is composed of 26 copies of FlgI (229, 254, 255). L and P rings form a stiff structure that serves as bushing for the rotating rod of the flagellar T3S system and is absent from flagellar T3S systems of Gram-positive bacteria that do not possess an OM.
Is the Predicted Periplasmic Rod Structure a Building Platform for the Needle or the Pilus?
EM studies of isolated needle complexes from S. Typhimurium revealed the presence of an internal channel of the basal body, localized in the periplasm, which was referred to as the inner rod and is composed of PrgJ (347, 348). The inner rod of the T3S system from Salmonella spp. is connected by a socket-like structure to the IM rings and might be required for stable anchoring of the extracellular needle, which probably protrudes into the periplasm as revealed by single-particle EM (347, 348, 478, 605). Experimental evidence for an inner rod structure was also reported for the T3S system from EPEC as well as for flagellar T3S systems (410), but the presence of this structure has not yet been confirmed.
It is assumed that the predicted inner rod of translocation-associated T3S systems is composed of multiple copies of a single subunit (e.g., YscI from Yersinia spp.; note that predicted inner rod proteins are not highly conserved). In contrast, the inner rod of flagellar T3S systems consists of four different components that build up the proximal rod (composed of FlgB, FlgC, and FlgF) and the distal rod (composed of FlgG) (230). The latter is surrounded by the P and L rings. The assembly of the predicted rod depends on FlgJ, a protein with a dual function. The N-terminal domain of FlgJ serves as a rod-capping protein that probably assists in the formation of the inner rod, while the C-terminal domain of FlgJ acts as a muramidase. The muramidase activity of FlgJ might be involved in the degradation of peptidoglycan and thus could be required for the efficient assembly of the rod structure in the periplasm (220, 401). Since the diameter of the flagellar T3S system-associated ring structures has been estimated to be approximately 11 nm (514) or 7.5 nm (534), the system is too large to pass through the natural pores of peptidoglycan, which are approximately 2 nm wide (135). The assistance of peptidoglycan-degrading enzymes is therefore often required to facilitate the assembly of membrane-spanning high-molecular-weight protein complexes such as flagellar or translocation-associated T3S systems (also see below).
The IM Ring Components of Translocation-Associated and Flagellar T3S Systems Differ in Their Complexity
It is assumed that the predicted inner rod is associated with IM rings of the T3S system that differ in complexity. While the IM rings (or MS rings [membrane and supramembranous rings]) of flagellar T3S systems are composed of FliF (Table 1) (34, 535, 560), the IM rings of translocation-associated T3S systems consist of at least two proteins, including members of the YscJ family of lipoproteins and the less conserved YscD family (Table 1). YscJ family members form a ring structure that is located at the periplasmic site of the IM and is presumably attached to the membrane by the N-terminal lipid moiety of YscJ and homologs. Several YscJ family members also contain a predicted C-terminal transmembrane helix (10, 492, 504). YscD and homologs are lipoproteins with an N-terminal cytoplasmic and a C-terminal periplasmic domain and might form a multimeric ring structure next to the YscJ ring (25, 142, 293, 492, 516). Protein-protein interaction studies revealed that YscJ and YscD family members interact not only with each other but also with members of the YscC family of OM secretins (142, 410, 467, 479) (Table 2). Interestingly, the YscJ and YscD homologs EscJ and EscD from EPEC were also shown to interact with the needle protein EscF, suggesting that the IM rings might provide a connection not only to the secretin channel but also to the needle, which possibly sits atop the predicted periplasmic inner rod structure and thus protrudes into the periplasm (410) (see above and Table 2). In agreement with this hypothesis, needle-like structures were observed in isolated T3S needle complexes from S. Typhimurium that lacked the OM secretin (492).
Table 2.
Interaction partners of T3S system components
| Type of component and protein family | Homolog(s) (organism)a | Interacting T3S system component(s) | Interacting T3S substrate(s) and/or chaperone(s) | Reference(s) |
|---|---|---|---|---|
| Predicted C ring components | ||||
| YscQ family | YscQ (Yersinia spp.) | YscL, YscK, YscN, YscUC, YscQC | YscP (T3S4 protein) | 77, 245, 457, 458 |
| SpaO (Salmonella spp., SPI-1) | OrgA, OrgB (probably required for efficient assembly and/or stability of SpaO complex), InvC | Translocon (SipB, SipC, SipD) and effector proteins (SipA, SptP) associate with SpaO-OrgA-OrgB complex; association depends on the YopN homolog InvE and the chaperone SicA (for translocon proteins) as well as on the CBD of SptP | 304, 515 | |
| SsaQ (Salmonella spp., SPI-2) | SsaQS (generated by tandem translation of ssaQL) | 622 | ||
| Spa33 (S. flexneri) | Spa47, MxiN, MxiK, MxiG, MxiJ | Effector proteins VirA, IcsB, IpaC, and IpgB1; T3S4 protein Spa32 | 251, 256, 390 | |
| EscQ (EPEC) | EscN, EscL | 33 | ||
| CdsQ (Chlamydia trachomatis) | CdsD, CdsS, CdsT | Mcsc (chaperone of effector Cap1), complex of Cap1 and Mcsc | 304 | |
| CdsQ (Chlamydia pneumoniae) | CdsQ, CdsD, CdsL, CdsN, FlhA* | 250, 515, 524, 525 | ||
| HrcQA (P. syringae) | HrcQB | 170, 550 | ||
| HrcQB (P. syringae) | HrcQA | 170, 550 | ||
| FliM* | FliJ (chaperone-binding protein)* | 206 | ||
| FliN* | FliH* | 206, 358 | ||
| FliG* | FliF* | 419 | ||
| ATPases | ||||
| YscN family | YscN (Yersinia spp.) | YscL, YscK, YscQ | YscP (T3S4 protein that interacts with YscN-YscL-YscQ complex), YscF (needle protein), YscE and YscG (chaperones of YscF), YopR (effector protein) | 42, 123, 245, 458, 459, 510 |
| InvC (Salmonella SPI-1) | SicP (chaperone of SptP), SopD (effector) | 4, 49 | ||
| SsaN (Salmonella SPI-2) | SrcA (chaperone of SseL and PipB2), SsaE (chaperone of SseB) | 101, 369 | ||
| Spa47 (S. flexneri) | Spa33, MxiK, MxiN | MxiC (secreted regulator, YopN homolog; the N terminus of MxiC is required for the interaction) | 51, 251, 256 | |
| EscN (EPEC) | EscL, EscQ | Tir (effector), CesT (class IB chaperone) | 33, 195, 546 | |
| CdsN (C. pneumoniae) | CdsL, CdsD, CdsQ | CopN (YopN homolog and effector protein) | 250, 515, 525 | |
| HrcN (X. campestris pv. vesicatoria) | HrcN, HrcL, HrcUC, HpaC (T3S4 protein) | HpaB (class IB chaperone) | 329 | |
| HrcN (P. syringae) | HrcN, HrpE (YscL family member) | 550 | ||
| FliI* (C. pneumoniae) | CopN, CdsL, FlhAC* | 524 | ||
| FliI* (Salmonella spp.) | FlhA*, FlhB*, FliI*, FliH* | FliE*, FlgB*, FlgE*, FlgD* (rod/hook proteins), FlgK*, FlgL*, FliC* (filament proteins), FliJ* (chaperone-binding protein), FlgN* (chaperone; binds to FliI in complex with the substrate FlgK), FliT* (chaperone of filament-capping protein FliD) | 205, 238, 382, 383, 544, 638 | |
| Putative negative regulators of ATPases | ||||
| YscL family | YscL (Yersinia spp.) | YscN, YscQ, YscUC | YscP (T3S4 protein; interacts with YscN-YscL-YscQ complex) | 42, 245, 457, 458 |
| EscL (EPEC) | EscN, EscQ | EspA (filament protein) | 33, 288 | |
| CdsL (C. pneumoniae) | CdsD, CdsN, CdsQ, FliI*, FlhA* | 250, 515, 524, 525 | ||
| HrcL (X. campestris pv. vesicatoria) | HrcN, HrcU | 329 | ||
| HrpE (P. syringae) | HrcN | 550 | ||
| FliH* (Salmonella spp.) | FlhA*,FlhB*, FliI*, FliN* | FlgB*, FlgE*, FlgD* (rod/hook proteins), FlgK*, FlgL*, FliC* (filament proteins), FliJ* (chaperone escort protein) | 185, 205, 358, 382, 383, 638 | |
| Components of the export apparatus | ||||
| YscU family | YscUC (Yersinia spp.) | YscL, YscK, YscQ | 457 | |
| Spa40 (S. flexneri) | Spa32 (T3S4 protein) | 50 | ||
| EscU (EPEC) | EspR | EspD (translocon protein), EscI (predicted inner rod protein) | 110, 475 | |
| CdsU (C. pneumoniae) | FlhA* | 524 | ||
| HrcU (X. campestris pv. vesicatoria) | HrcL | HpaB (class IB chaperone) | 329 | |
| HrcUC (X. campestris pv. vesicatoria) | HrcN, HpaC (T3S4 protein) | HrpB2 (early T3S substrate) | 329, 330, 332 | |
| HrcU (P. syringae) | HopAH1 | 550 | ||
| FlhBC* (Salmonella spp.) | FliI*, FliH*, FlhAC* | FliE*, FlgB*, FlgE*, FlgD* (rod/hook proteins), FlgK* (filament-type protein, weak interaction), FliK* (T3S4 protein) | 381, 383, 386, 392 | |
| FlhBCN* (Salmonella spp.) | FlhBCC* | 381 | ||
| YscV family | YscV (Yersinia spp.) | YscC (the interaction with YscC is reduced in the absence of YscD or YscJ), YscD (YscC is required for the interaction with YscD), YscJ (YscR, YscS, and YscT are required for the interaction with YscJ) | 143 | |
| FlhAC* (Bacillus subtilis) | FliC (flagellin), FliD (filament cap; binds in complex with chaperone FliT)*, FliJ (chaperone-binding protein)* | 23 | ||
| FlhA* (C. pneumoniae) | FliF*, CdsU, CdsL | 524 | ||
| FlhA* (Salmonella spp.), FlhAC* (Salmonella spp.) | FliH*, FliI*, FliF*, FlhBC*, FlhAC* | FlgE*, FlgD* (rod/hook proteins), FlgK*, FlgL* (hook-filament junction proteins; interaction with FlhAC is enhanced by the chaperone FlgN*), FliC* (filament protein), FliJ* (chaperone-binding protein), FlgN* (chaperone) | 185, 359, 379, 383, 638 | |
| FlhA* (C. pneumoniae) | CdsL, CdsU, CdsQ | SepZ (effector) | 515, 524 | |
| YscR family | EscR (EPEC) | EscR, EscS, EscU | EspD (translocon protein) | 110 |
| YscS family | EscS (EPEC) | EscR, EscS | EspD (translocon protein) | 110 |
| IM ring components | ||||
| YscJ family | YscJ (Yersinia spp.) | YscJ, YscC, YscD | 142, 467 | |
| EscJ (EPEC) | EscC | EscF (needle protein) | 410 | |
| PrgK (S. enterica) | PrgH | 479 | ||
| YscD family | YscD (Yersinia spp.) | YscD, YscJ, YscC | 142, 467 | |
| PrgH (S. enterica) | PrgK | 479 | ||
| CdsD (C. pneumoniae) | CdsQ, CdsL, CdsN | 250, 525 | ||
| EscD (EPEC) | EscC | EscF (needle protein) | 110, 410 | |
| MxiG (S. flexneri) | Spa33 (MxiG interacts with phosphorylated peptide of Spa33) | 25 | ||
| MS ring components | FliF* | FlhA*, FliG | 419, 524 | |
| OM ring components | ||||
| YscC family | YscC (Yersinia spp.) | YscJ, YscD | 142, 467 | |
| EscC (EPEC) | EscD | EscF (needle protein), EscI (predicted inner rod protein) | 110, 410, 475 | |
| Needle/pilus components | ||||
| YscF family | YscF (Yersinia spp.) | YscN | 123 | |
| EscF (EPEC) | EscC, EscJ, EscD | EscF (needle protein) | 410 | |
| MxiH (S. flexneri) | IpaD (translocon protein) | 629 | ||
| PrgI (Salmonella spp.) | SipD (translocon protein) | 451 |
Proteins from flagellar T3S systems are marked with asterisks.
Cryo-EM studies revealed that the symmetry of the IM rings of isolated translocation-associated T3S systems from S. enterica and S. flexneri ranges from 19- to 22- or 24-fold (19, 347, 478, 493). Alternatively, a 12-fold symmetry was proposed for IM rings from S. flexneri (223), whereas a 24- to 26-fold symmetry was observed for the IM rings of flagellar T3S systems (535, 542). The model of a 24-subunit ring model for IM rings of translocation-associated T3S systems was supported by the results of crystal structure analyses of the YscJ homolog EscJ from EPEC and the YscD homolog MxiG from S. flexneri (356, 614).
Although the constituents of IM and OM rings, including members of the YscC, YscJ, and YscD families, do not share significant amino acid similarities, crystal structure analyses of EscC, EscJ, and PrgH (Table 1) revealed a common α2β3 fold that was also identified in the OM secretins GspD and DotD, from type II and type IV secretion systems, respectively, and was proposed to act as a ring-building motif (284, 400, 516, 614). However, deletion of the predicted ring-building motif in the PrgH homolog YscD did not affect the activity of the T3S system (467). The α2β3 motif was also found in the C-terminal domain of the YscV homolog InvA, which is a component of the export apparatus (320). It is therefore conceivable that ring formation is a common characteristic of IM- and OM-associated components of the T3S system. Notably, YscV and its flagellar homolog FlhA were reported to oligomerize, and it was assumed that approximately 20 subunits of FlhA are incorporated into the flagellar export apparatus, where they might form a ring structure outside the MS ring (143, 316). In future studies, it remains to be investigated whether the assembly of the export apparatus in the IM does indeed involve the formation of ring structures.
Transmembrane Components of the Export Apparatus Are Involved in Substrate Recognition
The IM rings of the needle complex most likely provide a scaffold for the assembly of the transmembrane components of the export apparatus that enclose the transport channel for secreted proteins. The export apparatus of translocation-associated T3S systems is composed of members of the YscR, YscS, YscT, YscU and YscV families, which presumably form a multimeric protein complex. In flagellar T3S systems, these proteins include members of the FlhA, FlhB, FliO, FliP, FliQ, and FliR protein families (summarized in Table 1). Components of the export apparatus contain one to eight transmembrane helices and differ in size and in the presence of cytoplasmic domains. Members of the YscU/FlhB and YscV/FlhA families of IM proteins contain two large cytoplasmic domains that were proposed to be involved in the recognition of secreted proteins (11, 23, 31, 379, 381, 383). In agreement with this hypothesis, the C-terminal domains of FlhB and FlhA from flagellar T3S systems were shown to interact with extracellular components of the flagellum (Table 2). Furthermore, an interaction was reported between the C-terminal domain of the YscU/FlhB homolog HrcU, from the plant-pathogenic bacteria Xanthomonas campestris pv. vesicatoria and P. syringae, and secreted proteins (332, 550). The contribution of YscU/FlhB family members to the substrate specificity switch is discussed below. It should be noted that in addition to the cytoplasmic domains of members of the export apparatus, the presence of substrate docking sites was also described for the ATPase and the predicted C ring (Table 2; see below). It therefore cannot be excluded that T3S systems contain multiple substrate docking sites and that different acceptor sites recognize different types of T3S substrates.
Power Supplies—the Cytoplasmic ATPase and the Flagellar Motor
The ATPase of the T3S system—key player or useful substituent?
The export apparatus of flagellar and translocation-associated T3S systems is associated with a cytoplasmic ATPase which is a member of the YscN protein family (YscN/InvC/Spa47/EscN) and forms homo- or double-hexameric ring structures with an internal diameter of approximately 2.5 to 3 nm (96, 235, 267, 397, 445). Oligomerization and membrane contact of YscN family members lead to an increase of the ATPase activity, which is predicted to provide the energy needed for the secretion process (18, 21, 96, 378, 445, 627). Oligomerization of ATPases can also be induced upon binding of a T3S chaperone, as shown for the multicargo T3S chaperone SrcA from Salmonella spp., which interacts with the SPI-2-encoded ATPase SsaN (101) (Table 2). T3S chaperones are cytoplasmic proteins that bind to one or several T3S substrates and promote their stability and/or secretion (see below). Since ATPases of flagellar and translocation-associated T3S systems interact with effectors and/or effector-chaperone complexes, they were proposed to be involved in T3S substrate recognition (4, 195, 329, 510, 544, 546). Experimental evidence suggests that the ATPase dissociates T3S substrates from their cognate chaperones (4) and contributes to the unfolding of secreted proteins prior to their entry into the secretion apparatus (4). This is probably important for efficient secretion, because the inner channel of the T3S system has a diameter of 2 to 3 nm, which is too narrow to allow the passage of fully folded proteins (4, 45, 600).
Interestingly, experimental evidence reported for Yersinia spp. suggests that T3S can also occur in the absence of a functional ATPase, albeit in reduced amounts, and might then be driven by the proton motive force (PMF) (599). The PMF refers to the electrochemical potential difference of protons across a membrane and consists of the electrical potential difference (ΔΨ) and the proton concentration difference (ΔpH). PMF was also shown to contribute to flagellar T3S in Salmonella spp. in the absence of the ATPase FliI and its regulator FliH (376, 384, 435). It was therefore proposed that the PMF drives protein transport across both membranes, whereas the ATPase is required for the efficient initial docking of T3S substrates to the secretion channel. Notably, however, evidence for ATPase-independent secretion could not be observed for the plant-pathogenic bacterium X. campestris pv. vesicatoria (329), suggesting that the contributions of different energy sources to T3S can vary among plant- and animal-pathogenic bacteria.
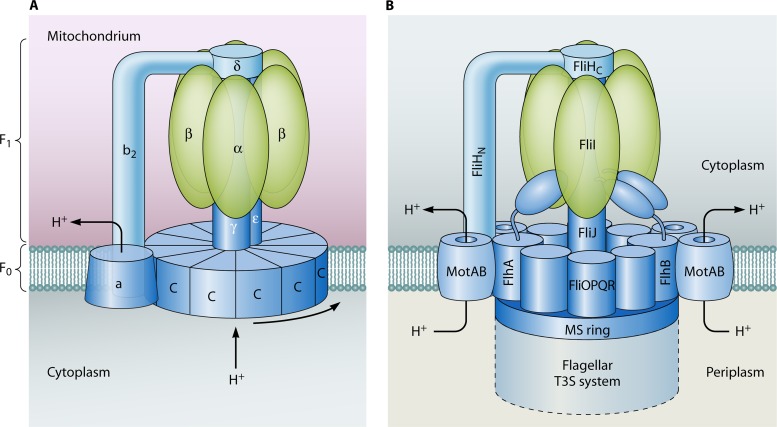
Crystal structure analyses of the T3S-associated ATPase EscN from EPEC and the flagellar T3S-associated ATPase FliI revealed a structural similarity with the α/β subunits of FoF1-ATPases (239, 627). FoF1-ATPases consist of a membrane-spanning Fo domain and a solvent-exposed F1 domain that rotate in opposite directions (Fig. 4). The F1 domain is composed of a hexamer of α and β subunits arranged around a central stalk. A second peripheral stalk, which contains b and δ subunits, connects the Fo and F1 domains. Interestingly, components of the second stalk share sequence homology with members of the YscL protein family that interact with the ATPase and are predicted regulators of its enzymatic activity (425) (Table 2; Fig. 4). A negative regulator might prevent ATP hydrolysis prior to the activation of the secretion system (382). FliH forms a FliH2-FliI complex with the ATPase and presumably promotes the docking of FliI to the secretion apparatus at the IM (21, 205, 382). The absence of FliH leads to a loss of bacterial motility, but the phenotype of fliH mutants can be suppressed upon overexpression of FliI or upon introduction of point mutations into the cytoplasmic domain of FlhA or FlhB (373). Since FlhA and FlhB interact with FliI and FliH (Table 2), mutations in the cytoplasmic domains of both proteins probably promote the docking of the ATPase complex to the export apparatus in the absence of FliH (358, 359, 383, 638). These findings suggest that FliH per se is not essential for flagellar T3S. Taken together, the data indicate that the docking of the ATPase and its enzymatic activity are important for T3S but are probably not the only energy source of the T3S system.
Fig 4.
Similarities between FoF1-ATPases and T3S-associated ATPases. (A) Model of the FoF1-ATPase. The FoF1-ATPase consists of a membrane-embedded Fo domain and a catalytic F1 domain. The F1 domain is composed of an α3β3 hexamer and is associated via the central stalk (consisting of the γ and ε subunits) and the peripheral stalk (composed of δ, b2, and a subunits) with the Fo domain. The Fo domain contains a and b (not shown) subunits and 12 c subunits that form proton-conducting channels. The energy provided by the proton influx drives the rotation of the Fo domain and ATP synthesis. (B) Model of the flagellar T3S-associated ATPase FliI and its interaction partners. FliI presumably forms a hexameric complex that is associated with the regulator of the ATPase, FliH, which shares structural similarity with the peripheral stalk of the FoF1-ATPase. Structural similarity was also reported for the chaperone-binding protein FliJ and components of the central stalk of the FoF1-ATPase (235). FliI associates with the export apparatus of the T3S system, which is connected to the MS ring in the IM and is surrounded by stator complexes (also see Fig. 2 and 3). Note that the organization of the ATPase complex is speculative and that the central position of FliJ in the ATPase ring has not been confirmed experimentally. The cytoplasmic C ring is not shown in this model.
The flagellar ATPase complex interacts with the chaperone-binding protein FliJ.
FliH, the regulator of the ATPase of flagellar T3S systems, interacts not only with the ATPase FliI but also with the soluble FliJ protein, which is an essential cytoplasmic component of the secretion machinery and contributes to the secretion of rod, hook, and filament proteins (185, 205, 372). Interestingly, analysis of the crystal structure of FliJ from S. enterica revealed a structural similarity with the γ subunit of the FoF1-ATPase, which is part of the central stalk of the soluble F1 domain (235) (see above and Fig. 4). Furthermore, the results of cryo-EM and protein-protein interaction studies suggest that FliJ inserts into the central channel of the ATPase ring and promotes the formation of a hexameric ATPase ring when mixed with FliI at a ratio of 6:1 (FliI:FliJ) (235) (Table 2). Given the previous finding that ring formation by FliI increases the ATPase activity (96, 378), FliJ could be involved in the activation of ATPase-driven T3S. However, as mentioned above, flagellar T3S can also occur in the absence of the ATPase and is then driven by the PMF, which consists of ΔΨ and ΔpH. A recent publication revealed a contribution of FliJ to the ΔΨ-driven export of flagellar T3S substrates that probably depends on the interaction of FliJ with the linker region of the cytoplasmic domain of the IM protein FlhA (FlhAC) (384). In agreement with this hypothesis, the binding sites of FliJ in FlhA were shown to be required for the functioning of both proteins, and vice versa. The authors therefore proposed that FliJ alters the conformation of FlhAC to activate ΔΨ-driven protein export (384).
Notably, however, the role of FliJ appears to be more complex. An alternative function of FliJ as a chaperone-binding protein was described because FliJ also interacts with the chaperones FlgN (chaperone of the hook-filament junction proteins FlgK and FlgL) and FliT (chaperone of the filament cap protein FliD) (23, 166). Similarly, the FliJ orthologs InvI and YscO, from Shigella spp. and Yersinia spp., respectively, interact with T3S chaperones (165). In the case of FliJ, experimental evidence suggests that FliJ promotes the interaction of the FliT-FliD complex with FlhAC (23). The interaction of the hook-filament junction protein FlgK with FlhAC, however, appears to depend on its cognate chaperone, FlgN, rather than on the presence of FliJ (379). It therefore remains to be clarified whether FliJ and homologs play a general role in the docking of chaperone-substrate complexes to components of the export apparatus or if they are proteins with multiple functions that are also involved in the activation of ATPase- and/or PMF-driven T3S.
Flagellar rotation depends on membrane-embedded stator complexes.
In addition to the ATPase, the activity of flagellar T3S systems depends on 8 to 11 membrane-embedded stator complexes, which consist of MotA and MotB and are absent from translocation-associated T3S systems. MotA contains four transmembrane helices and a cytoplasmic domain and interacts with the single-pass IM protein MotB, which is anchored by a peptidoglycan-binding domain in the bacterial cell wall (94, 128, 136). MotA and MotB form a hetero-oligomeric MotA4MotB2 complex that provides a channel for proton influx into the bacterial cytoplasm (41, 53, 483, 523) and converts the energy of the proton flux into a mechanical force that drives flagellum rotation (40, 279, 637) (Fig. 3). It has been estimated that a flow of approximately 1,200 protons is required for each rotation of the flagellar filament (360).
MotA interacts with the cytoplasmic FliG protein, which forms a ring of 26-fold symmetry on the cytoplasmic side of the MS ring and is directly involved in torque generation (183, 309, 325, 326, 535, 637). FliG is part of the switch complex that is required for flagellar rotation and the switching between clockwise and counterclockwise rotation. The switch complex (which corresponds to the predicted C ring [see below]) also consists of FliM and FliN, which form a pentameric FliM-FliN4 complex (61, 179, 242, 434, 609, 632, 633). FliM is presumably located between FliG and FliN and contains a binding site for the signaling molecule phospho-CheY, which promotes clockwise rotation of the flagellum (62, 375, 429, 434, 508, 595). A binding site for phospho-CheY was also identified in FliN, but FliN has a relatively minor role in flagellar switching and rotation (480). It is assumed that the proton flux across the MotA4MotB2 complexes induces a conformational change in the cytoplasmic domain of MotA and applies a force on the switch complex, most likely via electrostatic interactions between MotA and FliG (39, 279, 637). These electrostatic forces cause the rotation of the flagellar rotor, which consists of the predicted C ring, the IM rings, the periplasmic rod, the hook, and the flagellar filament and might rotate as one unit.
The Predicted Cytoplasmic C Ring of the T3S System Is a Potential Substrate Docking Site
FliM and FliN of flagellar T3S systems not only are involved in flagellar rotation but also form most of the cytoplasmic C ring that is associated with the IM ring complexes of the T3S system and contributes to the secretion process. The C ring is a cup-like structure with a diameter of approximately 40 nm that has been visualized by EM of isolated flagellar hook-basal body complexes and has a symmetry varying between 31- and 38-fold (179, 269, 542, 543, 619). The C ring of flagellar T3S systems is estimated to be composed of 34 copies of FliM and approximately 100 copies of FliN (436, 543, 633). The third component, FliG, interacts not only with the pentameric FliM-FliN4 complex and MotA but also with the MS ring component FliF and could therefore couple the C ring to the transmembrane components of the flagellar T3S system (61, 179, 419, 633) (Table 2). Interactions have also been observed between FliN and the ATPase regulator FliH, as well as between FliM and the ATPase-associated chaperone-binding protein FliJ (see above), suggesting that the C ring is involved in the docking of the ATPase complex (206, 358) (Table 2). Notably, the phenotype of C ring mutants can be suppressed by enhanced levels of the ATPase FliI or the master regulator FlhDC. It was therefore concluded that the C ring per se is not essential for flagellum formation (160, 280).
Predicted C ring components of translocation-associated T3S systems include members of the YscQ protein family, which share amino acid sequence similarities with FliM and FliN. YscQ and homologs interact with effector proteins or effector-chaperone complexes and were therefore proposed to act as a recruitment platform for secreted proteins (390, 515) (Table 2). The purification of recombinant YscQ revealed that it exists as a complex of two proteins, including full-length YscQ and a shorter protein corresponding to the C-terminal portion of YscQ, designated YscQ-C and synthesized from an internal translation initiation codon in YscQ (77). A similar tandem translation was recently reported for the YscQ homolog SsaQ from the Salmonella sp. SPI-2 (622). Crystal structure analysis revealed that YscQ-C forms a homodimer and shares structural similarities with the C ring component FliN and the FliN homolog HrcQB from the plant-pathogenic bacterium P. syringae (77). The C-terminal domain of HrcQB is itself structurally similar to FliN and was shown to interact with HrcQA, which shares similarities with FliM (61, 170). In agreement with a predicted function as a cytoplasmic component of the T3S system, the YscQ homolog Spa33 from S. flexneri localizes to the cytoplasmic side of the T3S system and interacts with the IM ring components MxiG and MxiJ as well as with the ATPase Spa47 (390). Furthermore, YscQ and the homologous CdsQ protein from Chlamydia spp. interact with the ATPase of the T3S system and its predicted regulators, i.e., YscL and CdsL, respectively (245, 250, 515) (Table 2). It was therefore postulated that the C ring is also present in translocation-associated T3S systems. Notably, however, in contrast to those of flagellar T3S systems, predicted C rings of translocation-associated T3S systems have not yet been visualized by EM studies (223, 347, 348, 493). The existence of these specialized cytoplasmic ring structures in translocation-associated T3S systems therefore remains to be proven.
THE CONSTRUCTION PHASE—HOW THE BASAL BODY AND EXPORT APPARATUS ARE ASSEMBLED
Stepwise Assembly of the Membrane-Spanning Basal Body
Experimental evidence suggests that there is a hierarchy in the assembly of the membrane-spanning basal body. An analysis of the translocation-associated T3S system from S. Typhimurium suggested that the ring structures in the IM and OM are assembled prior to the inner rod (529). Since IM rings and needle-like structures were observed in the absence of the OM secretin (492), the OM ring is probably dispensable for the assembly of the IM structures and the needle. In agreement with this finding, overexpression of the IM ring components PrgH and PrgK in E. coli led to the formation of ring structures even in the absence of other components of the basal body (273). Mutant studies with Salmonella spp. revealed that the IM and OM ring structures of the needle complex are dispensable for the formation of the export apparatus in the IM. Thus, it was shown that SpaP, SpaQ, and SpaR (YscR, YscS, and YscT family members) can assemble into a stable complex even in the absence of the needle complex (577). It is therefore likely that the assembly of the export apparatus precedes needle complex formation in Salmonella spp.
An inside-outside assembly was proposed not only for the translocation-associated T3S system but also the flagellar T3S system from Salmonella spp. The assembly of the flagellar basal body presumably initiates with the insertion of the MS ring into the IM and is followed by the attachment of the C ring and the stator complexes. After C ring formation, the export apparatus, the periplasmic rod, and the P and L rings are built (292). A recent study suggested, however, that the formation of the MS ring is preceded by oligomerization of the IM component FlhA, which is part of the export apparatus and might thus be the first component of the flagellar T3S system that is inserted into the IM (316).
In line with the predicted inside-outside assembly of T3S systems from Salmonella spp., it was previously reported that the localization of the OM secretin EscC of the translocation-associated T3S system from EPEC depends on the ATPase EscN and the IM protein EscV. In the absence of EscV or EscN, EscC accumulates in the periplasm, suggesting that the OM localization of the secretin depends not only on the Sec pathway but also on the assembly of IM-associated components of the T3S system (196). This hypothesis is further supported by a recent publication on the mechanisms underlying the assembly of the translocation-associated T3S system from Yersinia spp. While the formation of the T3S system in Yersinia spp. was earlier proposed to be initiated by the insertion of the OM secretin (142), experimental evidence now suggests the existence of two independent assembly pathways. One assembly platform involves the insertion of the secretin into the OM followed by the assembly of the YscD and YscJ rings. The second assembly platform probably consists of members of the export apparatus, including YscR, YscS, and YscT, that are required for the subsequent assembly of YscV (143). The export apparatus and the basal body are probably later joined together by the periplasmic YscJ protein (see above and Tables 1 and 2), which can directly bind to the export apparatus (143). It remains to be investigated whether a two step-assembly process is also applicable to the formation of translocation-associated and flagellar T3S systems of other bacterial species.
Contribution of Peptidoglycan-Degrading Enzymes
Macromolecular transport systems such as flagellar and translocation-associated T3S systems often require peptidoglycan-degrading enzymes, including lytic transglycosylases (LTs; also referred to as “specialized LTs”), for their efficient assembly because the natural pores of peptidoglycan are too narrow to allow the formation of these complex secretion systems (Table 3) (625; reviewed in references 282, 486, and 487). LTs are usually small proteins (150 to 250 amino acids) that cleave the beta-1,4-glycosidic bond between N-acetylglucosamine and N-acetylmuramic acid of peptidoglycan. Since LTs are ubiquitous in most peptidoglycan-containing eubacteria, they provide a potential target for new antibacterial drugs. LTs can be associated physically with components of protein secretion systems, as shown for VirB1 of the type IV secretion systems of Agrobacterium tumefaciens and Brucella suis (145, 586). This might ensure that peptidoglycan is degraded only locally.
Table 3.
Characteristics of T3S-associated LTs from animal- and plant-pathogenic bacteria
| LT | Organism | Characteristics or contribution to T3S and/or virulence | Reference(s) |
|---|---|---|---|
| IpgF | S. flexneri | No effect on virulence; LT activity demonstrated | 9, 625 |
| IagB | S. enterica | No effect on virulence; LT activity demonstrated | 529, 625 |
| l0045 | Enterohemorrhagic E. coli (EHEC) | Contributes to T3S and expression of the filament protein EspA; weakly expressed; localizes mainly to the periplasm | 624 |
| EtgA | EPEC | Contributes to T3S and bacterial hemolytic activity; localizes to the periplasm; N-terminally processed; degrades peptidoglycan | 192 |
| rOrf3 | Citrobacter rodentium | Contributes to virulence and T3S | 138 |
| HpaH | X. campestris pv. vesicatoria | Contributes to virulence and T3S; specifically promotes secretion and translocation of selected effector proteins | 75 |
| Xanthomonas axonopodis pv. glycines | Contributes to virulence and HR induction; weakly expressed | 272 | |
| Hpa2 | X. oryzae pv. oryzae | (No) influence on virulence (contradictory data are published); lyses the bacterial cell wall | 628, 639 |
| X. oryzae pv. oryzicola | Contributes to virulence and translocation of effector proteins; interacts with the translocon protein HrpF; secreted by the T3S system | 319 | |
| HrpH | P. syringae | Contributes to effector protein translocation; overexpression in E. coli leads to an arrest of bacterial growth; suppresses basal plant defense responses; secreted and translocated by the T3S system | 412 |
| HopP1 | P. syringae | Might contribute to effector protein translocation; suppresses basal plant defense responses; secreted and translocated by the T3S system | 412 |
| HopAJ1 | P. syringae | Might contribute to effector protein translocation | 412 |
To date, the contribution of predicted LTs to T3S and/or pathogenicity has been studied in both animal- and plant-pathogenic bacteria (75, 192, 412, 413, 624, 625, 628) (summarized in Table 3). Notably, it was observed that single LTs do not contribute significantly to T3S and virulence, presumably due to functional redundancies. Some predicted LTs that are involved in T3S, including Hpa2 from the plant-pathogenic bacterium Xanthomonas oryzae pv. oryzicola as well as HrpH and HopP1 from P. syringae, are themselves secreted, possibly to prevent further LT-mediated peptidoglycan degradation after the assembly of the secretion apparatus (319, 412) (Table 3). Interestingly, Hpa2 from X. oryzae pv. oryzicola contributes to effector protein secretion and interacts with the translocon protein HrpF, suggesting that it not only acts as an LT but also plays a role at the host-pathogen interface (319). The predicted LTs HrpH and HopP1 from P. syringae are even translocated by the T3S system into the plant cell and were shown to suppress basal plant defense responses in Nicotiana benthamiana (412, 413). Given the finding that peptidoglycan from animal-pathogenic bacteria is transported into the host cell, where it can be recognized by so-called pattern recognition receptors and cytoplasmic NOD proteins, it is tempting to speculate that translocated bacterial LTs might prevent the recognition of peptidoglycan by the host immune system (202, 203, 554, 571). Thus, it is possible that T3S-associated LTs have a dual activity as periplasmic proteins to promote the assembly of the T3S system and outside the bacterium after the assembly process.
RECOGNITION OF SECRETED PROTEINS
T3S Signals Are Not Conserved and Interchangeable among T3S Substrates
Substrates of T3S systems are targeted to the secretion system by a signal that is usually located within the N-terminal 20 to 30 amino acids (367, 485, 511). Although the N-terminal regions of T3S substrates are not conserved at the amino acid level, they often share specific amino acid compositions or patterns (20, 210, 336, 437, 477, 484). Furthermore, the analysis of several effector proteins from animal-pathogenic bacteria suggests that the region which harbors the N-terminal T3S signal is structurally disordered, i.e., lacks a unique tertiary structure. Intrinsically disordered protein regions can undergo structural alterations upon binding to their cognate folded partners, as was shown for the effector protein YopE from Yersinia spp., which binds to the cognate T3S chaperone SycE (459) (see below). The structural flexibility provided by the disordered protein regions that harbor the T3S signal might facilitate the recognition of effector proteins by components of the T3S system, including the cytoplasmic ATPase, the predicted C ring, or the cytoplasmic domains of members of the YscU and YscV protein families (Table 2; see above) (65).
The presence of an N-terminal T3S signal is not strictly conserved—although it is frequently observed—in all T3S substrates. For example, a T3S signal has been identified in the C-terminal region of the T3S effector Tir from EPEC (12). Central or C-terminal regions of T3S substrates were also shown to contribute to the secretion of the effector protein SipB from Salmonella spp. and the translocon protein EspB from enterohemorrhagic E. coli (93, 271). As an alternative to the amino acid-based T3S signal, a signal in the corresponding mRNAs of several effector proteins from Yersinia was proposed, suggesting a cotranslational secretion of these proteins (16, 17). However, mRNA-based T3S signals probably do not account for the high secretion rates observed for T3S substrates from animal-pathogenic bacteria. Real-time analysis of effector protein translocation revealed transport of several thousand effector protein molecules within the first few minutes of the infection process (156, 371, 488, 566, 603).
Interestingly, experimental evidence suggests that T3S signals are interchangeable (14, 63, 188, 466, 469), even between substrates of flagellar and translocation-associated T3S systems (151, 312, 321, 365, 366, 531, 587, 617). This suggests that the mechanisms underlying substrate recognition are conserved in both systems. Pathway specificity during T3S is probably conferred by the N-terminal or central region of T3S substrates, which provides the binding sites for specific T3S chaperones (see below). In this context, it is interesting that translocation-associated T3S systems can secrete and also translocate flagellin into the eukaryotic cell cytosol, where it might be recognized by the host immune system and can induce defense responses (365, 531). Recently, an interaction between flagellin and eukaryotic Nod-like receptors was demonstrated (278, 634). Experimental evidence reported for the animal-pathogenic bacterium P. aeruginosa revealed that the translocation-associated T3S system suppresses the expression of genes that encode components of the flagellar T3S system and vice versa (513). Similarly, the flagellar regulators FlhDC and FliA appear to repress the expression of ysc genes from Yersinia spp. (43, 232). The alternative sigma factor FliA is also required for the temperature-regulated expression of ysc genes, which are preferentially expressed at 37°C, while flagellar T3S gene expression is activated at temperatures below 30°C (104, 262, 263, 302, 462). These findings suggest an antagonistic expression of flagellar and translocation-associated T3S systems. The downregulation of flagellar T3S systems under conditions that lead to the activation of translocation-associated T3S systems, i.e., upon host cell contact, might be required to minimize host defense responses that are activated in response to flagellin.
Crossing the Borders—Translocation of Effector Proteins
The lack of amino acid sequence similarities of T3S signals significantly hampered the identification of effector proteins in plant- and animal-pathogenic bacteria. Several computational approaches that use machine-learning programs were therefore developed to identify T3S substrates from both plant- and animal-pathogenic bacteria, based on common features such as N-terminal amino acid biases in T3S signal sequences and structural elements (20, 336, 477). Additional characteristics used for the identification of effector proteins include homologies to already known effectors, the presence of typical eukaryotic protein motifs, the neighborhood of T3S chaperone genes, specific promoter elements that allow coexpression with the T3S system, and a low G+C content, which is indicative of horizontal gene transfer. Together, these approaches have led to the identification of novel effector proteins in both plant- and animal-pathogenic bacteria.
In many cases, the translocation of effector proteins into the eukaryotic cell cytosol was confirmed experimentally by the use of suitable reporter assays. For this purpose, fusion proteins between N-terminal regions of effectors and reporter proteins, such as the adenylate cyclase domain (CyaA) of the adenylate cyclase toxin of Bordetella pertussis, the TEM β-lactamase, or derivatives of avirulence proteins from plant-pathogenic bacteria that induce a cell death reaction inside resistant plant cells, were generated. CyaA is specifically activated in the presence of calmodulin in eukaryotic cells. The translocation of CyaA fusion proteins can therefore be determined by the measurement of intracellular cyclic AMP (cAMP) levels (485, 511, 512). In contrast to CyaA, the TEM β-lactamase cleaves the fluorescent substrate CCF2 and thus leads to a shift in the fluorescence spectrum, which can be detected in living cells (85, 345). In the last 6 years, additional assays, based on split-green fluorescent protein (split-GFP) systems, the recruitment of translocated effectors by GFP-chaperone fusion proteins inside the host cell, or the detection of translocated tetracysteine-tagged effectors by a specific fluorescing FlAsH reagent, have been developed and have allowed real-time imaging of effector protein arrival in the host cell (152, 156, 157, 488, 505, 567).
These assays revealed that in addition to the N-terminal T3S signal, translocation depends on a second protein region that is usually located within the N-terminal 50 to 100 amino acids of effector proteins and provides the binding site for a cognate T3S chaperone (52, 485, 512, 604) (see below). Furthermore, it was observed that effector protein translocation in animal-pathogenic bacteria starts within seconds after host cell contact and that the translocation kinetics of effector proteins can vary (156, 371, 488, 566, 603). Different translocation rates are indicative of a hierarchy in effector protein translocation that might guarantee the efficient manipulation of host cellular pathways by effector proteins. Furthermore, a temporal regulation of effector protein translocation might prevent an interference of effector proteins with antagonistic activities, as shown for SopE, SopE2, SipA, and SptP from Salmonella spp. SopE, SopE2, and SipA trigger actin polymerization, whereas SptP disrupts the changes in the actin cytoskeleton 1 to 2 h after infection (190, 521, 635, 636). While the translocation kinetics of these effector proteins differ during the initial stages of the infection process (566, 603), their cellular levels at later time points might also be regulated by other mechanisms, such as a differential proteasome-mediated degradation. It was shown that SopE and SptP are both present in the host cell about 15 min after infection and that SopE is rapidly degraded afterwards by the host cell proteasome. In contrast, SptP remains stable (290). Interestingly, the different sensitivities of SopE and SptP toward proteasomal degradation appear to depend on the N-terminal secretion and translocation signals of both proteins. Thus, a hybrid protein consisting of the N-terminal domain of SopE and the effector domain of SptP is rapidly degraded, while a fusion between the effector domain of SopE and the N-terminal region of SptP has an increased half-life (290). The precise molecular mechanisms that underlie the proteasome-dependent degradation of SopE and SptP remain to be elucidated.
Protein translocation by the T3S system has long been assumed to be a one-step transport process that is specific for effector proteins. However, a recent study reported translocation of effector proteins from Yersinia spp. that localized to the bacterial cell surface prior to translocation. Translocation was dependent on the translocon, suggesting that secretion and translocation of effector proteins can be uncoupled (5). The translocation of surface-localized YopH from Yersinia spp. was independent of the N-terminal T3S signal (amino acids 1 to 18) but required the presence of a translocation signal within amino acids 18 to 49 (5). Interestingly, translocation has been observed not only for surface-localized effector proteins but also for extracellular proteins that are usually not transported by the T3S system. Thus, the autotransporter EspC from EPEC, which is secreted by the type V secretion system, can be translocated into eukaryotic cells in a type III secretion-dependent manner (573). The corresponding targeting signal in EspC is unknown. Taken together, these findings suggest that the mechanisms underlying translocation and recognition of translocated proteins by the translocon are more complex than initially anticipated. Future experiments will have to clarify how translocation signals target effector proteins across the host plasma membrane.
Guides and Bodyguards—the T3S Chaperones
Role of T3S chaperones in T3S substrate targeting.
In addition to the secretion and translocation signal, several secreted proteins depend on cytoplasmic T3S chaperones for their efficient secretion (589, 590) (summarized in Table 4). T3S chaperones often interact as homo- or heterodimers with their cognate substrates and presumably promote the recognition of secreted proteins by components of the T3S system. Furthermore, binding of the chaperone can also prevent the premature degradation of T3S substrates. While most known T3S chaperones are cytoplasmic proteins, Spa15 from S. flexneri was shown to be secreted and translocated by the T3S system, suggesting that it has a second activity inside the host cell (171).
Table 4.
Known T3S chaperones from selected animal- and plant-pathogenic bacteria
| Organisms and chaperonea | Interaction partner(s)b | Location of CBDc | Description/comments | Reference(s) |
|---|---|---|---|---|
| Yersinia spp. | ||||
| SycD (LcrH) (Ysc T3SS) | YopB (T), YopD (T), YscY (C), YscM1 (R), YscM2 (R) | aa 53 to 149 and 278 to 292 of YopD; no discrete CBD in YopB | Dimerizes; contains three TPRs; stabilizes YopB and YopD; regulatory function (see Fig. 7) | 69, 146, 147, 180, 181, 404, 489, 537 |
| SycE (Ysc T3SS) | YopE (E), YscM1 (R), YscM2 (R) | aa 15 to 50 of YopE | Dimerizes; promotes a disorder-to-order transition in the CBD of YopE; alanine substitutions in the CBD of YopE do not affect SycE binding and YopE secretion but lead to severely reduced translocation of YopE | 36, 37, 90, 169, 459, 460, 485, 489, 537, 604 |
| SycH (Ysc T3SS) | YopH (E), YscM1 (R), YscM2 (R) | aa 20 to 69 of YopH; N-terminal region of YscM2 | Dimerizes; YscM1 and YscM2 share aa similarity with the CBD of YopH; regulatory function (see Fig. 7) | 80, 168, 441, 604 |
| SycN, YscB (Ysc T3SS) | YopN (R/E) | aa 30 to 76 of YopN | SycN and YscB form a heterodimer and stabilize YopN | 126, 244, 494 |
| SycO (Ysc T3SS) | YopO (E), YscM1 (R) | aa 20 to 77 of YopO | Dimerizes; masks membrane localization domain of YopO; overproduction of SycO leads to reduced Yop secretion | 144, 314 |
| SycT (Ysc T3SS) | YopT (E) | At least aa 52 to 103 of YopT | Dimerizes; binding to catalytically inactive YopT(C139S) is reduced | 68, 240, 327 |
| YscE, YscG (Ysc T3SS) | YscF (N) | C-terminal region of YscF | YscE and YscG form a heterodimer; YscG contains TPRs and shares a similar fold with LcrH; YscE shares structural similarity with the needle protein MxiH from S. flexneri | 125, 440, 530 |
| YscY (Ysc T3SS) | YscX (EC) | aa 50 to 110 of YscX | Regulatory function (see Fig. 7) | 127, 241 |
| SycP (Ysa T3SS) | YspP (E) | Dimerizes; stabilizes YspP | 352 | |
| SycB (Ysa T3SS) | YspB (T), YspC (T) | Dimerizes; stabilizes YspB; together with the AraC-type regulator YscE regulates the expression of ysa T3S genes | 28, 178, 581 | |
| Shigella spp. | ||||
| IpgA | IcsB (E) | aa 171 to 247 of IcsB | Stabilizes IcsB; icsB and ipgA can be translated as a single fusion protein | 409 |
| IpgC | IpaB (T), IpaC (T), MxiE (R) | aa 15 to 45 and 48 to 74 of IpaB; aa 50 to 80 (213), 73 to 122 (422) and/or 33 to 73 (328) of IpaC | Dimerizes; contains TPR motifs; stabilizes IpaB and IpaC; probably acts as a coactivator of the AraC-type transcriptional activator MxiE (see Fig. 7) | 27, 35, 213, 328, 338, 354, 362, 422, 443 |
| IpgE | IpgD (E) | Stabilizes IpgD | 405 | |
| Spa15 | IpaA(E), IpgB1 (E), OspC3 (E), OspB (E), OspD1 (R) | aa 26 to 141 of OspC3; aa 263 to 365 of IpaA; aa 23 to 190 of IpgB1 | Dimerizes; is secreted; stabilizes IpgB but not IpaA; binds to the secreted antiactivator OspD1 and acts as coantiactivator (see Fig. 7) | 171, 422, 423, 565 |
| IpaD | IpaD (T) | Possesses self-chaperoning activity | 252 | |
| Salmonella spp. | ||||
| InvB (SPI-1) | SipA (SspA) (E), SopA (SipF) (E), SopE (E), SopE2 (E) | aa 1 to 45 of SopA; aa 1 to 158 of SipA; aa 30 to 45 of SopE | Dimerizes; contributes to stability of SopE2 and SipA; CBD of SopA is required for translocation; CBD of SopE prevents secretion by SPI-1 or flagellum in the absence of InvB | 58, 149–151, 219, 311, 321 |
| SicA (SPI-1) | SipB (T), SipC (T), InvF (R) | aa 80 to 100 of SipB | Self-interacts; coactivator of the AraC-type transcriptional regulator InvF; stabilizes SipB and SipC; SipB is stable and secreted in a sicA sipC mutant; CBD of SipB is not sufficient to target the protein to the translocation-associated T3SS | 119, 120, 271, 559 |
| SicP (SPI-1) | SptP (E) | aa 35 to 139 of SptP | Dimerizes; stabilizes SptP; translation of SicP is required for translation of SptP (translational coupling) | 76, 189, 520 |
| SigE (SPI-1) | SopB (SigD) (E) | Dimerizes; stabilizes SopB | 121, 231, 275, 340 | |
| SrcA (SPI-2) | SseL (E), PipB2 (E) | Dimerizes; multicargo T3S chaperone; the srcA gene is unlinked to the T3S system genomic region | 101 | |
| SscB (SPI-2) | SseF (E) | Stabilizes SseF | 115 | |
| SseA (SPI-2) | SseB (T), SseD (T) | aa 147 to 169 of SseB; aa 138 to 194 of SseD (but aa 32 to 82 of SseD also contribute to the binding of SseA) | Contributes to stability of SseB | 99, 472, 641, 642 |
| SsaE (SPI-2) | SseB (T) | Also contributes to secretion of the effector PipB | 369 | |
| SsaQS (SPI-2) | SsaQL (YscQ homolog) | Generated by tandem translation of ssaQL | 622 | |
| EPEC/EHEC | ||||
| CesA2 (L0017) | EspA (F) | Inhibits polymerization of EspA; stabilizes EspA | 527 | |
| CesAB (CesA) | EspA (F), EspB (T) | Dimerizes; stabilizes EspA | 111, 613 | |
| CesD | EspD (T) | Also contributes to secretion of EspB | 580 | |
| CesD2 | EspD (T) | Stabilizes EspD | 402 | |
| CesF | EspF (E) | 154, 575 | ||
| CesL | SepL (E) | 620 | ||
| CesT | Tir (E), Map (E), NleA (E), EspF (E), EspG (E), EspZ (E), NleG (E), NleH (E), NleH2 (E) | N-terminal 50 to 100 aa of Tir | Dimerizes; contributes to stability of Map; also contributes to secretion of effector NleI | 1, 109, 134, 153, 318, 340, 545, 546 |
| P. syringae | ||||
| HrpG | Unknown | Acts as suppressor of the negative regulator HrpV (see Fig. 7) | 593 | |
| ShcA | HopPsyA (HopA1) (E) | N-terminal 166 aa of HopPsyA | 564 | |
| ShcM | HopPtoM (HopM1) (E) | aa 100 to 400 of HopPtoM | Protects HopPtoM from Lon-mediated degradation | 22, 333 |
| ShcF | HopPtoF (HopF2) (E) | Stabilizes HopPtoF | 499 | |
| ShcV | HopPtoV (HopV1) (E) | aa 76 to 125 of HopPtoV | 592 | |
| ShcO1 | HopO1-1 (E), HopS1 (E), HopS2 (E) | Central part of HopO1-1 | Homologous to ShcS1; can interact with ShcS1 | 209, 259 |
| ShcS1 | HopO1-1 (E), HopS1 (E), HopS2 (E) | Central part of HopO1-1 | Can interact with ShcO1; forms homodimers | 209, 259 |
| ShcS2 | HopO1-1 (E), HopS1 (E), HopS2 (E) | Central part of HopO1-1 | Homologous to ShcS1 | 209 |
| X. campestris pv. vesicatoria | ||||
| HpaB | AvrBs1 (E), AvrBs3 (E), HpaA (E), HpaC (R) | aa 1 to 50 of AvrBs3; aa 225 to 275 of HpaA | Essential for pathogenicity; contributes to the efficient T3S and translocation of multiple effector proteins | 71, 73, 331 |
| Erwinia amylovora | ||||
| DspB/F | DspA/E (E) | aa 51 to 100 and C-terminal region of DspA/E | N-terminal CBD is required for DspA/E translocation (411); minimal translocation signal does not comprise CBD (556) | 193, 411, 556 |
T3S chaperones of translocation-associated T3S systems. For pathogens with more than one T3S system, the type of the respective T3S system is written in italics. Alternative protein names are given in parentheses. T3SS, T3S system.
Known interaction partners are categorized into effectors (E), translocon proteins (T), needle proteins (N), filament proteins (F), regulators (R), chaperones (C), and extracellular proteins of the T3S system (EC). Alternative protein names are given in parentheses.
aa, amino acids.
Depending on their substrate specificities, T3S chaperones have been categorized into different classes: class IA chaperones are specific for one or several homologous effector proteins, while class IB chaperones bind to different effectors with unrelated sequences (431). Known class IB chaperones include Spa15 from S. flexneri, InvB from Salmonella spp., CesT from EPEC, and HpaB from X. campestris pv. vesicatoria (71, 73, 149, 150, 311, 423, 546). Class II chaperones interact with translocon proteins (589), whereas chaperones of flagellar T3S systems are referred to as class III chaperones (431). Chaperones of translocon proteins usually contain tandem tetratricopeptide repeats (TPRs), which are imperfect 34-amino-acid repeats that are also present in eukaryotic chaperones and are often involved in protein-protein interactions (57, 249, 427). TPRs were also identified in the T3S chaperone YscG from Yersinia spp., which binds together with its cochaperone YscE to the needle protein YscF (530) (Table 4). Similarly, TPRs are present in the T3S chaperones PscG and AscG, from P. aeruginosa and Aeromonas hydrophila, respectively, which form heterotrimeric PscG-PscE-PscF and AscG-AscE-AscF complexes (86, 447, 448).
It is assumed that T3S chaperones facilitate the binding of their cognate interaction partners to components of the secretion apparatus at the IM, such as the ATPase (see above). They might thus increase the local concentrations of secretion substrates at the base of the secretion apparatus and promote their transport to the T3S system (452). As described for S. flexneri, the inner channels of the ATPase and the secretion apparatus, as well as the surfaces of effector proteins, have an electronegative potential which creates a repulsive force and thus a need for energy to transport secreted proteins into the T3S system (452).
CBDs in T3S substrates.
Despite their moderate amino acid sequence similarities, several class I chaperones share a conserved mixed α/β fold and form dimeric structures, as revealed by crystal structure analyses (36, 37, 68, 169, 321, 327, 340, 441, 494, 520, 553, 565). Conserved structural features were also described for the chaperone-binding domains (CBDs) of T3S substrates, which are often located within the N-terminal 50 to 100 amino acids and are wrapped around the chaperone dimer in an extended conformation (37, 321, 441, 494, 520, 598). Chaperone-bound CBDs were therefore proposed to serve as three-dimensional targeting signals that are recognized by components of the T3S system at the IM (37, 167, 321). In line with this model are the findings that the CBD is often required for efficient translocation (Table 4) and that binding of the chaperone SycE to the effector protein YopE from Yersinia spp. induces a disorder-to-order transition in the CBD of YopE (459). Interestingly, it was shown that single amino acid substitutions in the CBD of YopE result in reduced translocation of YopE but do not affect YopE secretion or the interaction of YopE with SycE (460). This suggests that a postulated three-dimensional targeting signal in the CBD is required for the efficient translocation of effector proteins. Notably, however, it is not yet known whether T3S systems harbor specific recognition sites for translocated proteins. Given the finding that most T3S chaperones are not secreted and that mutations in the CBD specifically affect translocation but not secretion of YopE, it is possible that the specific targeting of T3S substrates to the translocon is controlled in the bacterial cytoplasm. The molecular mechanisms underlying the potential cross talk between the translocon and components of the T3S system in the IM remain to be investigated.
Notably, the N-terminal protein region including the CBD is not always sufficient to target T3S substrates for efficient secretion by the translocation-associated T3S systems. This was shown, for instance, for the T3S effectors SipB and Tir, from Salmonella spp. and EPEC, respectively, which depend on C-terminal protein regions for efficient secretion (12, 271) (see above). Furthermore, secretion and/or translocation of some effector proteins was also observed in the absence of the CBD, suggesting that the CBD per se is not always essential for protein export (52, 151, 314, 321, 556, 604). However, in many cases it was not analyzed whether the observed secretion of effector derivatives deprived of their CBDs was still mediated by the translocation-associated T3S system. As shown for the effector protein SptP from Salmonella, the absence of the CBD can lead to a loss of pathway specificity and thus to a secretion of SptP by the flagellar T3S system (312). It was therefore proposed that the binding of T3S chaperones to the CBD confers the specific secretion of proteins by the translocation-associated T3S system (312). However, a later study revealed that it is the CBD itself which determines the recognition specificity of flagellar or translocation-associated T3S systems (151).
In addition to their roles in protein export, CBDs might also provide a membrane-targeting signal, as shown for the CBD of the effector protein YopO from Yersinia spp. Binding of the chaperone SycO to the CBD of YopO could prevent the membrane localization of YopO inside the bacterium. In agreement with this model, deletion of the CBD in YopO did not interfere with the secretion and translocation of the protein by the wild-type strain but abolished membrane localization of YopO inside the host cell (314). Similar to SycO, the T3S chaperones SycE and SycT might mask a membrane localization domain in their corresponding interaction partners, i.e., YopE and YopT (287, 314). These observations suggest that the CBD of effector proteins not only is essential for translocation per se but also could exert an inhibitory influence on translocation by promoting membrane localization of the effector protein inside the bacterium. Taken together, these findings imply that T3S chaperones fulfill multiple functions that can vary in different pathogens. Thus, T3S chaperones not only promote stability and secretion of their cognate interaction partners but also could prevent membrane localization of effector proteins inside the bacterial cytosol. Furthermore, experimental evidence suggests that several T3S chaperones can be involved in the regulation of T3S gene expression (see below). Given that T3S chaperones often do not share significant sequence similarities with each other, the term “T3S chaperone” therefore refers to a rather heterogeneous group of proteins with structural similarities but various functions.
Contribution of T3S chaperones to the establishment of a secretion hierarchy.
In addition to their contribution to the docking and recognition of T3S substrates, T3S chaperones might also impose a hierarchy on the translocation of effector proteins. It was shown that deletion of the CBD in YopE abolishes the translocation of YopE by the wild-type strain but not by a polyeffector mutant, suggesting that the binding of the chaperone helps the effector to compete with other effectors for translocation (52). A chaperone-dependent hierarchy in effector protein translocation was also observed in EPEC. In this case, the class IB chaperone CesT from EPEC promoted the secretion of the effector protein Tir, which is itself required for the efficient secretion of additional CesT-dependent effectors. In the absence of Tir, effector protein secretion was severely reduced, presumably because of enhanced levels of free CesT that blocked the T3S system (545) (Fig. 5A). It was proposed that CesT regulates effector protein secretion after being released from Tir, presumably by binding to components of the T3S system at the IM. A negative regulation of effector protein secretion by a potentially uncomplexed chaperone was also observed for the class IB chaperone HpaB from X. campestris pv. vesicatoria, which interacts with the effector protein HpaA. Secretion of HpaA is probably required to liberate HpaB and thus to allow the efficient HpaB-mediated secretion and translocation of additional effector proteins. In the absence of HpaA, increased amounts of free HpaB lead to reduced T3S of pilus, translocon, and effector proteins (331) (Fig. 5B). The additional deletion of hpaB in an hpaA mutant therefore restores the efficient secretion of pilus and translocon proteins (331) (Fig. 5B). Notably, HpaB not only imposes a hierarchy on effector protein translocation but also appears to prevent the translocation of extracellular components of the T3S system, such as components of the translocon (Fig. 5B). Thus, it was shown that the N-terminal regions of translocon and pilus proteins can target a reporter protein for translocation in an hpaB mutant but not in the wild-type strain. This suggests that pilus and translocon proteins harbor a translocation signal that is suppressed during HpaB-mediated effector protein translocation (71; my unpublished data).
Fig 5.
Predicted functions of class IB chaperones during control of effector protein secretion in EPEC and X. campestris pv. vesicatoria. (A) Model for the function of the class IB T3S chaperone CesT from EPEC. CesT promotes the secretion and translocation of Tir, which is the first effector protein (E) that is translocated into the host cell. It is assumed that CesT binds preferentially to Tir to promote Tir secretion after assembly of the T3S system. In the absence of Tir, uncomplexed CesT might block the secretion of effector proteins by the T3S system. The question mark indicates that it is still unknown whether the inhibitory activity of CesT is linked to its potential association with components of the T3S system. Dashed arrows indicate reduced secretion and/or translocation rates. (B) Hypothetical mode of action of the class IB T3S chaperone HpaB from X. campestris pv. vesicatoria. HpaB binds to and promotes the secretion of the effector protein HpaA and additional effector proteins (E) after the secretion of translocon proteins (T). Similar to Tir, HpaA is presumably the first effector protein that is bound to the chaperone and travels the T3S system. In the absence of HpaA, the efficient secretion of effector, pilus, and translocon proteins is suppressed, probably by uncomplexed HpaB that binds to components of the secretion apparatus. The question mark indicates that it is unknown whether the association of HpaB with the T3S system contributes to its inhibitory activity. Interestingly, in the absence of HpaB, translocon proteins are secreted and even translocated, suggesting that they contain a functional translocation signal that is suppressed by HpaB. Translocon proteins are also efficiently secreted in an hpaAB double deletion mutant, in which T3S is not suppressed by uncomplexed HpaB.
FEEDBACK CONTROL—HOW GENE EXPRESSION IS COUPLED TO THE SECRETORY ACTIVITY OF THE T3S SYSTEM
T3S is controlled not only on the posttranslational level but also by transcriptional regulators, which often couple the expression of genes that encode components and substrates of the T3S system to the secretory activity of the system. A common regulatory principle involves the interaction of a T3S chaperone with either its cognate T3S substrate or a cytoplasmic regulatory protein. Upon activation of T3S, the chaperone is liberated from its secreted binding partner and can bind to regulatory proteins inside the cytoplasm, including transcriptional activators or antiactivators (Fig. 6A). Binding of the T3S chaperone can positively regulate transcriptional activators or counteract antiactivators that act as suppressors of transcriptional activators. The interaction of a regulatory chaperone with an antiactivator thus relieves the inhibitory effect of the antiactivator on the activity of the transcriptional activator and leads to the induction of T3S gene expression (Fig. 6B). Known transcriptional regulators that are involved in the control of T3S gene expression and corresponding co-, anti-, or antiantiactivators are summarized in Table 5 and briefly described below.
Fig 6.
Control of T3S gene expression by regulatory chaperones. (A) Proposed mode of action of T3S chaperones that act as coactivators of transcriptional regulators. When the T3S system is inactive, T3S chaperones are bound by their cognate substrates in the bacterial cytosol. The activation of the T3S system leads to the secretion of T3S substrates and thus to the liberation of corresponding T3S chaperones, which can subsequently interact with transcriptional activators and promote T3S gene expression. T3S chaperones are represented by green rectangles, and T3S substrates are represented by circles. Transcriptional activators are depicted in yellow. (B) Model for the activity of T3S chaperones that act as antiantiactivators. When the T3S system is inactive, T3S chaperones are bound by their corresponding T3S substrates. The induction of T3S leads to the liberation of T3S chaperones, which can subsequently bind to an antiactivator that suppresses the activity of a transcriptional activator. Upon interaction with the T3S chaperone, however, the antiactivator is released from the transcriptional activator, and the latter can induce T3S gene expression.
Table 5.
Transcriptional and posttranscriptional control proteins that link T3S gene expression with the secretory activity of flagellar and translocation-associated T3S systems
| Organism and control proteina | Predicted functions/characteristicsb |
|---|---|
| Salmonella spp. | |
| Transcriptional control proteins (flagellar T3S system) | |
| FlhDC | Transcriptional activators for class II genes; form an FlhD4-FlhC2 complex with DNA-binding activity; promote the σ70-dependent transcription of class II genes; also act as autoinhibitors of their own transcription |
| FlgM* | Anti-σ28 factor; inhibits the σ28-dependent RNA polymerase that is specific for class III promoters; expressed from class II and class III promoters; class II-expressed FlgM interacts with FliA (σ28); secreted upon hook-basal body completion |
| FliA | σ28 factor; associates with the RNA polymerase and induces the expression of class III genes after secretion of the anti-σ28 factor FlgM; expressed from class II and class III promoters; also acts as a chaperone for FlgM |
| FlgN | Chaperone of hook-filament junction proteins FlgK and FlgL; promotes the translation of class III-expressed flgM |
| FliT | Chaperone of filament cap protein FliD; inhibits the FlhDC-dependent activation of class II gene expression and FlhDC autoinhibition (leads to reinitiation of class I gene expression) when liberated from FliD |
| Posttranscriptional control proteins (translocation-associated T3S system) | |
| InvE (SPI-1) | YopN homolog; negative regulator of effector protein secretion; deletion of invE leads to reduced secretion of translocon proteins |
| SsaL* (SPI-2) | YopN homolog; negative regulator of effector protein secretion; deletion of ssaL leads to reduced secretion of translocon proteins; interacts with the SsaM-SpiC complex; dissociation of the SsaL-SsaM-SpiC complex at pH 7 activates effector protein secretion |
| SsaM (SPI-2) | Interacts with SpiC; a SpiC-SsaM complex interacts with SsaL at pH 7; negative regulator of effector protein secretion; deletion of ssaM leads to reduced secretion of translocon proteins |
| SpiC (SPI-2) | Interacts with SsaM; a SpiC-SsaM complex interacts with SsaL at pH 7; negative regulator of effector protein secretion; deletion of spiC leads to reduced secretion of translocon proteins |
| Yersinia spp. (translocation-associated T3S system) | |
| Transcriptional control proteins | |
| YopD* | Translocon protein; negative regulator of yop gene expression; binds in complex with the chaperone LcrH to the 5′-untranslated region of yop mRNAs |
| LcrH | Chaperone of YopD; negative regulator of yop gene expression; regulatory activity also requires interaction with the predicted T3S chaperone YscY |
| LcrQ* | Transcriptional repressor; blocks yop gene expression in complex with the chaperone SycH; regulatory activity depends on YopD |
| SycH | Chaperone of LcrQ and YopH; promotes YopH secretion after export of LcrQ |
| Posttranscriptional control proteins | |
| YopN* | Negative regulator of Yop and translocon protein secretion; regulatory activity depends on interaction with TyeA, SycN, and YscB; a YopN-TyeA complex acts as an internal plug of the T3S system; secretion of YopN under T3S-permissive conditions relieves the negative regulatory effect |
| TyeA* | Interacts with and prevents secretion of YopN |
| SycN | Chaperone of YopN |
| YscB | Chaperone of YopN |
| LcrG | Negative regulator of Yop secretion; inhibitory influence is counteracted by binding of LcrV; LcrG also promotes the secretion of LcrV |
| LcrV* | Tip complex protein; interacts with LcrG and counteracts the LcrG-mediated repression of Yop secretion |
| P. aeruginosa (translocation-associated T3S system) | |
| Transcriptional control proteins | |
| ExsA | AraC-type transcriptional activator; interacts with the antiactivator ExsD under T3S-nonpermissive conditions |
| ExsD | Antiactivator of ExsA; binds to ExsA and inhibits ExsA-dependent transcription |
| ExsC | Antiantiactivator; binds to ExsD and counteracts the negative regulatory activity of ExsD; also acts as a T3S chaperone of ExsE; secretion of ExsE liberates ExsC and allows its interaction with ExsD |
| ExsE* | Inhibitor of ExsC |
| Posttranscriptional control proteins | |
| PopN* | Blocks T3S of effector proteins from inside the cytosol similarly to YopN; does not interfere with secretion of translocon proteins |
| PcrG | Internal plug of T3S system; interacts with PcrV; inhibitory activity of PcrG is independent of its interaction with PcrV |
| PcrV* | Tip complex protein; interacts with PcrG; presumably acts as external plug of the T3S system |
| S. flexneri (translocation-associated T3S system) | |
| Transcriptional control proteins | |
| MxiE | AraC-type transcriptional activator; required for effector gene expression; activity of MxiE depends on its interaction with IpgC |
| IpgC | Coactivator of MxiE; interacts with MxiE and OspD1; chaperone of the translocon proteins IpaB and IpaC; binding of IpaB and IpaC to IpgC inhibits MxiE/IpgC-dependent activation of effector gene expression |
| OspD1* | Antiactivator of MxiE; binds to MxiE and to the chaperone Spa15 |
| Spa15* | Class IB T3S chaperone; promotes secretion of OspD1; coantiactivator that inhibits MxiE activity in complex with OspD1; T3S-inducing conditions lead to the secretion of OspD1 and Spa15 and thus to the activation of MxiE/IpgC-dependent effector gene expression |
| Posttranscriptional control proteins | |
| IpaB*, IpaD* | Tip complex proteins; might serve as an external plug to control effector protein secretion |
| MxiC* | YopN homolog; negative regulator of Yop secretion; deletion of mxiC leads to reduced secretion of translocon proteins in response to Congo red induction |
| Enterohemorrhagic E. coli (translocation-associated T3S system) | |
| Posttranscriptional control proteins | |
| SepL | YopN homolog; deletion of sepL leads to reduced secretion of translocon and enhanced secretion of effector proteins; SepL interacts with SepD, Tir, and the T3S chaperone CesL |
| SepD | Interacts with SepL; deletion of sepD leads to reduced secretion of translocon proteins and enhanced secretion of effectors |
Secreted proteins are indicated by asterisks, transcriptional regulators are shown in bold, and T3S chaperones are underlined.
References are given in the text.
Hierarchical Control of Gene Expression in Flagellar T3S Systems
In flagellar T3S systems, genes encoding components and substrates of the secretion apparatus are not expressed simultaneously but activated at different stages of the secretion process. According to their temporal expression patterns, flagellar genes are organized into three different classes (299). Class I genes encode the transcriptional activators FlhD and FlhC, which initiate the assembly of the flagellum. FlhD and FlhC activate genes that are expressed under the control of class II promoters and encode components of the MS, C, P, and L rings, the inner rod, and the extracellular hook (299, 322). The class II gene product FlgM binds to the sigma factor FliA (σ28) inside the bacterial cytoplasm and thus prevents the association of FliA with the RNA polymerase (8, 81, 82, 200, 414, 415) (Fig. 7). Upon completion of the hook-basal body, FlgM is secreted and liberates FliA, which can subsequently activate class III genes that encode proteins involved in the formation of the flagellar filament and the stator complexes (234, 266, 294, 299, 323, 414) (Fig. 7). Notably, FliA acts not only as a sigma factor but also as a T3S chaperone for FlgM. Since FliA and FlgM are encoded by both class II and class III genes, they can autoregulate their own expression levels (201, 324). Translation of the flgM class III mRNA is enhanced by the T3S chaperone FlgN, which binds to the hook-filament junction proteins FlgK and FlgL (6, 30, 184, 264). Thus, upon secretion of FlgK and FlgL, liberated FlgN promotes the translation of FlgM (encoded by the class III flgM gene), which can bind to and inhibit FliA (Fig. 7).
Fig 7.
Schematic representation of the regulatory mechanisms that control T3S gene expression in flagellar and translocation-associated T3S systems. The expression of effector genes or genes encoding substrates and components of the flagellar T3S system is controlled by transcriptional regulators and regulatory chaperones and depends on the activity of the T3S system. Flagellar gene expression is regulated by the transcriptional activators FlhDC, which are encoded by class I genes and activate the expression of class II genes. Class II gene products include the anti-σ28 factor FlgM, the hook-filament junction proteins FlgK and FlgL, and the filament cap protein FliD, which bind to the corresponding chaperones FliA, FlgN, and FliT, respectively. The secretion of FlgM leads to the release of the σ28 factor FliA, which activates the expression of class III genes. The liberated T3S chaperone FliT binds to FlhC and thus inhibits the expression of class II genes, whereas FlgN positively regulates the translation of flgM class III mRNA. In Yersinia spp., translation of yop mRNAs is suppressed by a YopD-LcrH complex when the T3S system is inactive. LcrQ-SycH and YscM-SycH complexes might act as additional repressors. Activation of Yop secretion leads to relief of the YopD-LcrH-mediated repression of yop mRNA translation and the liberation of SycH and LcrH upon secretion of YopD. The SycH chaperone presumably promotes YopH secretion after its release from the secreted regulator YscM. LcrH might suppress yop gene expression when bound to the T3S chaperone YscY. Effector gene expression in S. flexneri depends on the transcriptional activator MxiE and its coactivator, IpgC. Upon activation of T3S, MxiE and IpgC are released from their secreted interaction partners, i.e., Spa15, OpsD1, IpaC, and IpaD, and can activate effector gene expression. In P. aeruginosa, effector gene expression is controlled by the activator ExsA, which interacts with the antiactivator ExsD when the T3S system is inactive. The T3S chaperone ExsC, which binds to the T3S substrate ExsE, acts as an antiantiactivator when released from ExsE after activation of the T3S system. The interaction of ExsC with ExsD leads to the liberation of ExsA, which subsequently activates effector gene expression. In the plant-pathogenic bacterium P. syringae, expression of hrp (hypersensitive response and pathogenicity) genes that encode the T3S system is induced by HrpR, HrpS, and HrpL. HrpR and HrpS interact with each other and activate the expression of the alternative sigma factor HrpL, which binds to the promoters of hrp genes. HrpR-, HrpS-, and HrpL-dependent activation of hrp gene expression is counteracted by the negative regulator HrpV, which interacts with HrpS, and by the Lon protease, which degrades HrpR. Under T3S-inducing conditions, HrpV interacts with the chaperone-like protein HrpG, which interferes with the negative regulatory activity of HrpV and might also bind to a secreted but so far unknown interaction partner (XY). T3S chaperones are represented in green, and T3S substrates are represented by circles. Transcriptional regulators are depicted in yellow.
In addition to FlgN, expression of class II genes can also be repressed by FliT, which acts as a T3S chaperone of the filament cap protein FliD and also binds to the regulatory FlhDC proteins (184, 297, 610). Secretion of FliD after hook formation liberates FliT, which subsequently binds to FlhC and thus suppresses the FlhDC-dependent activation of class II gene expression (30, 610) (Fig. 7). FliT also interferes with the autoinhibitory activity of FlhDC on their own expression and thus restores the expression of class I genes (295, 610).
Control of yop Gene Expression in Yersinia spp.
The coupling of transcriptional gene regulation and T3S has been described not only for flagellar T3S systems but also for translocation-associated T3S systems from animal-pathogenic bacteria. In Yersinia spp., the expression of effector (yop) genes is specifically activated after host cell contact upon secretion of the translocon protein YopD, which acts a negative regulator of yop gene expression inside the bacterial cytoplasm. YopD presumably binds to the 5′-untranslated regions of yop mRNAs in complex with its chaperone, LcrH (also known as SycD) (15, 181). Binding of YopD-LcrH to the yop mRNA might prevent the access of ribosomes and facilitate mRNA degradation, thus leading to reduced Yop levels (88) (Fig. 7). Furthermore, LcrH might have an additional regulatory role in the suppression of yop gene expression that is independent of its binding to YopD and involves an interaction of LcrH with YscY, a predicted chaperone of the secreted YscX protein (56, 127, 181).
A second transcriptional repressor from Yersinia spp. is the secreted LcrQ protein, which was identified in Y. pseudotuberculosis. LcrQ blocks yop gene expression in complex with the chaperone SycH. The LcrQ homologs YscM1 and YscM2 from Y. enterocolitica might act in a similar manner (79, 439) (Fig. 7). Although the molecular mechanisms underlying the LcrQ/YscM-mediated control of yop gene expression are not yet understood, both proteins likely act on the transcriptional level (519). Secretion and translocation of LcrQ/YscM upon activation of the T3S system relieve the negative effect on yop gene expression and also liberate SycH, which is a common T3S chaperone of LcrQ/YscM and YopH and presumably promotes secretion and translocation of YopH after LcrQ/YscM export (78, 456, 607) (Fig. 7). LcrQ-YscM-mediated yop gene repression is linked to the action of YopD, but the mechanisms underlying the functional interplay between LcrQ/YscM and YopD are still unclear.
Regulation of T3S Genes in S. flexneri, Salmonella spp., and P. aeruginosa
Similar to the case of Yersinia spp., the expression of effector genes in S. flexneri is triggered upon host cell contact. Gene induction depends on the AraC-type transcriptional activator MxiE and its interacting coactivator, IpgC (354, 443). Under noninducing conditions, MxiE interacts with the antiactivator OspD1 and the coantiactivator Spa15, which both prevent MxiE-dependent gene expression (430) (Fig. 7). Spa15 is a class IB T3S chaperone that is itself secreted by the T3S system (171, 423, 430) (see above). Secretion of OspD1 (and Spa15) upon activation of the T3S system leads to the liberation of MxiE, which subsequently activates effector gene expression in complex with IpgC (430) (Fig. 7). IpgC also acts as a T3S chaperone that promotes the secretion of the translocon proteins IpaB and IpaC (363). Under T3S-inducing conditions, IpgC is released from its secreted interaction partners and binds to MxiE, thus promoting MxiE-dependent gene expression (430). A similar mechanism might underlie the function of the MxiE and IpgC homologs InvF and SicA, respectively, from Salmonella spp. (118, 120).
An AraC-type transcriptional activator (designated ExsA) that controls effector gene expression has also been described for P. aeruginosa. When the T3S system is inactive, ExsA is bound by the antiactivator ExsD and does not induce gene expression (355, 608) (Fig. 7). The inhibitory activity of ExsD is counteracted by the antiantiactivator ExsC, which interacts with ExsD but also acts as a T3S chaperone for the T3S substrate ExsE (122). ExsC is therefore complexed with ExsE in the cytoplasm when the T3S system is inactive (Fig. 7). Activation of the T3S system, however, results in the secretion of ExsE and thus in the liberation of ExsC, which can subsequently interact with the antiactivator ExsD. This leads to the release of ExsD-bound ExsA and to the activation of ExsA-dependent gene expression (455, 561).
Regulation of T3S Gene Expression in P. syringae by the Regulatory T3S Chaperone-Like Protein HrpG and the Lon Protease
In P. syringae, the expression of T3S genes is specifically induced when the bacterium enters the plant apoplast, by the two regulatory proteins HrpR and HrpS, which belong to the NtrC family of two-component regulators and interact with each other (54). HrpR and HrpS induce the expression of the alternative sigma factor HrpL, which binds to conserved elements (termed hrp [hypersensitive response and pathogenicity] boxes) in the promoter regions of T3S genes (541). The HrpL-dependent activation of T3S gene expression is counteracted by the negative regulator HrpV, which presumably acts upstream of HrpL and interacts with HrpS (446, 593) (Fig. 7). The activity of HrpV can be suppressed by the cytoplasmic HrpG protein, which shares typical features of a T3S chaperone and probably binds not only to HrpV but also to a secreted interaction partner that has not yet been identified (593). It is assumed that the activation of T3S leads to the release of HrpG from its predicted secreted interaction partner and thus to the subsequent interaction of HrpG with HrpV, which counteracts the negative effect of HrpV on T3S gene expression (Fig. 7).
In addition to HrpR, HrpS, HrpL, HrpV, and HrpG, the Lon protease was identified as another player in the control of T3S gene expression in P. syringae. The Lon protease is an ATP-dependent serine protease that is involved in the degradation of unstable or misfolded proteins and can contribute to the regulation of T3S genes (464, 551). In P. syringae, the Lon protease acts as a negative regulator by degrading HrpR, specifically under T3S-repressing conditions (54, 303, 420, 612). Notably, the Lon protease is also involved in the degradation of effector proteins from P. syringae. However, most effector proteins are protected from Lon-mediated degradation by the binding of their corresponding T3S chaperones (333). Lon-mediated degradation of regulatory proteins was also shown for the flagellar sigma factor FliA from E. coli, which can be protected from Lon-dependent degradation by interaction with the anti-sigma factor FlgM (see above) (24). Furthermore, in Salmonella spp., the Lon protease degrades the transcriptional activators HilC and HilD, which are involved in the regulation of the SPI-1-encoded T3S system (539). In Yersinia spp., the Lon protease and the ATP-dependent ClpXP protease degrade a small histone-like protein designated YmoA, which represses the expression of T3S genes (246). In conclusion, these findings suggest that the Lon protease is involved in the regulation of T3S gene expression in both plant- and animal-pathogenic bacteria.
ORCHESTRATION OF T3S—HOW SUBSTRATE SPECIFICITY IS CONTROLLED
Components of the extracellular needle or pilus are most likely the first proteins that travel the secretion apparatus and are therefore also referred to as “early” T3S substrates. In animal-pathogenic bacteria, the formation of T3S needles is a tightly controlled process that ensures a defined distribution of the lengths of needle structures, with a peak at 45 nm (Shigella spp. [45, 540]), 60 nm (Yersinia spp. [224, 257]), or 80 nm (Salmonella spp. [291]) (44, 257, 293, 578, 579) (Table 6 and see below). Since needles have to be sufficiently long to bridge the extracellular space between the bacterium and the host cell, needle length control is probably essential for the efficient translocation of effector proteins. In agreement with this model, a correlation between needle length and the length of the adhesin YadA from Y. enterocolitica has been observed (394). Shorter YadA molecules allowed effector protein translocation by short needles that would not be translocation competent in the context of the wild-type YadA protein (394). Notably, additional studies of Shigella and Salmonella spp. also revealed an influence of the length of extracellular lipopolysaccharide molecules on effector protein translocation (226, 596).
Table 6.
Characteristics and functions of T3S4 proteins
| T3S4 protein (organism) | Protein length (aa) | Needle/pilus/hook length (nm [unless stated otherwise]) | T3S of T3S4 protein | Interaction with YscUC/FlhBC or homologs | Mutant phenotype | References |
|---|---|---|---|---|---|---|
| Translocation-associated T3S systems of animal-pathogenic bacteria | ||||||
| YscP (Yersinia spp.) | 515 | 55–60 | Yes | Not shown | Reduced secretion of effector proteins; increased amounts of surface-localized YscF; increased secretion of the predicted inner rod protein YscI; lack of needle length control | 148, 257, 518, 605 |
| InvJ (Salmonella spp.) | 336 | ≈80 | Yes | Not shown | Lack of inner rod structures in isolated injectisomes; wild-type secretion of PrgI (needle protein) and PrgJ (inner rod protein); needles are loosely attached to the base; lack of needle length control | 98, 204, 293, 347, 528 |
| Spa32 (S. flexneri) | 292 | ≈45 | Yes | Yes | Reduced secretion of IpaB, IpaC, and IpaD; lack of needle length control | 50, 341 |
| Translocation-associated T3S systems of plant-pathogenic bacteria | ||||||
| HpaC (X. campestris pv. vesicatoria) | 212 | ≈1–2 μm | No | Yes | Reduced secretion of translocon and effector proteins; increased secretion of HrpB2 | 332, 496 |
| Flagellar T3S systems of animal-pathogenic bacteria | ||||||
| FliK (Salmonella spp.) | 405 | ≈55 | Yes | Yes | Elongated hook structures; no filaments | 374, 386, 392, 433, 536 |
T3S4 Proteins and Their Interplay with YscU/FlhB Family Members
Since the formation of the pilus/needle is a prerequisite for T3S, it presumably precedes the secretion of intermediate (translocon proteins) and late (effector proteins) substrates, suggesting that the substrate specificity of the T3S system switches. The predicted switch in T3S substrate specificity from early to late substrates is mediated by T3S substrate specificity switch (T3S4) proteins, which have been studied intensively in animal-pathogenic bacteria. T3S4 proteins are often themselves secreted by the T3S system and are involved not only in the substrate specificity switch but also in length control of the extracellular needle (Table 6). Lack of a functional T3S4 protein usually results in increased needle length and a reduced secretion of late substrates. To date, T3S4 proteins have been described for translocation-associated T3S systems from several animal-pathogenic bacteria (YscP from Yersinia spp., Spa32 from Shigella spp., and InvJ from Salmonella spp.) and the plant-pathogenic bacterium X. campestris pv. vesicatoria (HpaC) (Table 6). Furthermore, in flagellar T3S systems, the T3S4 protein FliK has been identified, which switches the substrate specificity from early (hook components) to late (filament proteins) substrates after the hook has reached a length of approximately 55 nm (374, 386).
T3S4 proteins share little amino acid sequence identity with each other but contain a structurally conserved C-terminal domain (termed the T3S4 domain) that is probably essential for the substrate specificity switch and harbors a P-X-L-G amino acid motif (2). T3S4 proteins from both translocation-associated and flagellar T3S systems interact with the C-terminal cytoplasmic domains of members of the conserved YscU/FlhB family of IM proteins, as shown for the T3S4 proteins FliK, Spa32, and HpaC (50, 332, 381, 392) (Tables 2 and 6). Binding of T3S4 proteins to the C-terminal domains of FlhB, YscU, and homologs might induce a conformational change in these domains that alters the substrate specificity of the T3S system. This hypothesis is corroborated by the finding that single point mutations in the C-terminal regions of FlhB, YscU, and homologs can restore the wild-type phenotype in T3S4 mutants of S. Typhimurium, Y. pseudotuberculosis, EPEC, and X. campestris pv. vesicatoria (148, 298, 330, 602, 626) (Table 7). It is assumed that the introduction of point mutations into the C-terminal domains of YscU/FlhB family members leads to a conformational change that is permissive for the substrate specificity switch.
Table 7.
Effects of point mutations or deletions in YscU/FlhB family members
| YscU/FlhB homolog (organism) | Mutation (expression in trans or in cis) | Characteristics of mutated YscU/FlhB homologs |
Reference(s) | |
|---|---|---|---|---|
| Cleavagea | Effect on T3S | |||
| YscU (Yersinia spp.) | Mutations in the NPTH motif | |||
| N263A (in cis) | Reduced/alternative cleavage | Secretion of Yops and LcrQ in the presence of Ca2+; reduced secretion of Yops and LcrV in the absence of Ca2+; increased amounts of surface- exposed YscF; secretion of YscP is not significantly altered | 38, 509 | |
| N263A (in trans) | Alternative/partial cleavage | Reduced secretion of LcrV and translocon and effector proteins; longer needles | 457, 509 | |
| P264A (in cis) | Reduced/alternative cleavage | Secretion of Yops in the presence and absence of Ca2+; reduced secretion of effector and translocon proteins in the absence of Ca2+; reduced secretion and expression of LcrV; increased amounts of surface-exposed YscF | 38, 509 | |
| P264A (in trans) | No/alternative cleavage | Reduced secretion of effector and translocon proteins; longer needles | 306, 509 | |
| T265A (in cis) | Wild type | Wild-type phenotype | 38 | |
| T265A (in trans) | Wild type | Wild-type phenotype | 509 | |
| H266A (in trans) | Reduced cleavage | Secretion of LcrV; wild-type needle length | 597 | |
| ΔNPTH (in cis/in trans) | No cleavage | No secretion of effectors; low level of effector gene expression; no surface-exposed YscF | 38, 306 | |
| Extragenic mutations that suppress the yscP mutant phenotype | ||||
| A268F (in trans) | NA | Restores effector protein secretion in a yscU mutant, but not secretion of the needle protein YscF | 148 | |
| Y287G (in trans) | NA | Partially restores effector protein secretion in a yscU mutant; leads to reduced amounts of surface-localized YscF in a yscP mutant; does not restore wild-type levels of YscI secretion in a yscP mutant | 148, 605 | |
| V292T (in trans) | NA | Restores effector protein secretion in a yscU mutant | 148 | |
| Y317D (in trans) | NA | Does not restore effector protein secretion in a yscU mutant but does so in a yscP yscU double mutant; leads to reduced amounts of surface-localized YscF in a yscP mutant; leads to reduced YscI secretion in a yscP mutant | 148, 605 | |
| EscU (EPEC) | Mutations in the NPTH motif | |||
| N262A (in trans) | No cleavage | Reduced/no secretion of effector and translocon proteins | 626 | |
| N262D (in trans) | Reduced cleavage | Reduced/no secretion of effector and translocon proteins | 626 | |
| P263A (in trans) | No cleavage | Reduced/no secretion of effector and translocon proteins | 626 | |
| H265A (in trans) | Cleavage | Reduced/no secretion of effector and translocon proteins | 626 | |
| SpaS (Salmonella spp.) | Mutations in the NPTH motif | |||
| N258A (in cis) | NA | Reduced secretion of needle, translocon, and effector proteins; secretion of the T3S4 protein InvJ is not significantly altered | 626 | |
| P259A (in cis) | NA | Reduced secretion of needle, translocon, and effector proteins; secretion of the T3S4 protein InvJ is not significantly altered | 626 | |
| HrcU (X. campestris pv. vesicatoria) | Mutations in the NPTH motif | |||
| N264A (in trans) | No cleavage | No secretion of translocon and effector proteins; wild-type secretion of the early T3S substrate HrpB2; interacts with HrpB2; reduced interaction with the T3S4 protein HpaC | 330 | |
| P265A (in trans) | Reduced cleavage | Reduced secretion of translocon and effector proteins; wild-type secretion of the early T3S substrate HrpB2; interacts with HrpB2; reduced interaction with the T3S4 protein HpaC | 330 | |
| P265G (in trans) | No cleavage | No secretion of translocon proteins, effectors, and the early T3S substrate HrpB2; reduced interaction with HrpB2 and the T3S4 protein HpaC | 330 | |
| P265G (in cis) | No cleavage | No secretion of translocon and effector proteins and the early T3S substrate HrpB2 | 330 | |
| T266A (in trans) | Cleavage | Wild-type secretion of translocon and effector proteins and the early T3S substrate HrpB2 | 330 | |
| H267A (in trans) | Cleavage | Reduced secretion of translocon and effector proteins; wild-type secretion of the early T3S substrate HrpB2 | 330 | |
| Extragenic suppressor mutation that suppresses the hpaC mutant phenotype | ||||
| Y318D (in cis) | Reduced cleavage | Wild-type secretion of translocon and effector proteins; oversecretion of the early T3S substrate HrpB2 in the hpaC deletion mutant is not restored; reduced interaction with the T3S4 protein HpaC and with HrpB2 | 330 | |
| FlhB (Salmonella spp.) | Mutations in the NPTH motif | |||
| N269A (in trans) | Alternative cleavage | Polyhooks | 186 | |
| P279A (in trans) | Alternative cleavage | Polyhooks | 186 | |
| Extragenic suppressor mutations that suppress the fliK mutant phenotype | ||||
| A298V (in trans) | Reduced cleavage | Partially restores motility of fliK mutant (to less than 25% of wild-type motility) | 381, 602 | |
| G293R (in trans) | Reduced cleavage | Partially restores motility of fliK mutant (to less than 25% of wild-type motility) | 381, 602 | |
| G293V (in trans) | Reduced cleavage | Partially restores motility of fliK mutant (to less than 25% of wild-type motility) | 381, 602 | |
| 348 frameshift (in trans) | Reduced cleavage | Partially restores motility of fliK mutant (to less than 25% of wild-type motility) | 381, 602 | |
| 358 frameshift (in trans) | Reduced cleavage | Partially restores motility of fliK mutant (to less than 25% of wild-type motility) | 381, 602 | |
| W353stop (in trans) | Reduced cleavage | Partially restores motility of fliK mutant (to less than 25% of wild-type motility) | 381, 602 | |
| FlhB (H. pylori) | Mutations in the NPTH motif | |||
| N265A (in cis) | No cleavage | No secretion of filament proteins | 507 | |
| P266G (in cis) | No cleavage | No secretion of filament proteins | 507 | |
NA, not analyzed.
The C-terminal domains of YscU/FlhB family members are cleaved autoproteolytically between the asparagine and proline residues of a conserved NPTH motif (letters refer to amino acids) (38, 130, 177, 306, 334, 509, 597, 626). The C-terminal cleavage products probably remain associated with the membrane-associated portions of the proteins. Since the motility of a flagellar flhBCC mutant, which lacks the C-terminal cleavage product of FlhB, can be restored partially upon ectopic expression of flhBCC, it was suggested that the cleavage product can be provided in trans and that the cleavage event per se is not crucial for protein function (381, 583). Similar findings were observed for an X. campestris pv. vesicatoria hrcUCC mutant (330). Crystal structure analyses of several YscU/FlhB family members revealed that the cleavage leads to an altered orientation of the PTH loop, while the rest of the C-terminal protein domain remains structurally unchanged (38, 130, 148, 177, 334, 597, 602, 626). Mutant derivatives of YscU/FlhB family members which carry point mutations in the conserved N, P, or T residue of the NPTH motif are no longer efficiently cleaved but still promote the secretion of early substrates and needle formation (Table 7). Furthermore, the lack of YscU/FlhB cleavage does not significantly compromise the secretion of T3S4 proteins (509, 626) (Table 7). In contrast, secretion of intermediate and late substrates is often suppressed in YscU/FlhB cleavage mutants, suggesting that the cleavage of YscU/FlhB family members, and thus the reorientation of the PTH loop, is required for the substrate specificity switch (186, 330, 507, 509, 548, 626) (Table 7). In EPEC and X. campestris pv. vesicatoria, the presence of noncleavable EscU and HrcU derivatives, respectively, leads to a significantly reduced secretion of effector proteins (330, 548) (Table 7). Since the IM association of the T3S chaperone CesT is reduced in EscU cleavage mutants, it was proposed that the cleavage is required for substrate docking (548). In agreement with this hypothesis, mutations in the NPTH motif of HrcU interfere with the interaction of HrcU with the early T3S substrate HrpB2 (330).
In contrast to EPEC and X. campestris pv. vesicatoria, Yersinia sp. mutants that are deficient in YscU cleavage still secrete effector proteins as well as hybrid proteins consisting of the N-terminal T3S signal of the effector protein YopE and the tip protein LcrV (38, 509) (Table 7). Secretion of the wild-type LcrV protein, in contrast, was severely reduced in the absence of YscU cleavage (509). YscU cleavage might therefore be required to activate the secretion of translocon proteins (intermediate substrates) but not effector proteins (late substrates). Furthermore, these data indicate that the classification of T3S substrates as early, intermediate, or late substrates depends on the N-terminal T3S signal, which might determine the time point of secretion. In agreement with this hypothesis, the results of domain swapping experiments with substrates of the flagellar T3S system revealed that hybrid proteins that contain the N-terminal regions of early substrates are also secreted as early substrates (517). Similarly, the secretion behavior of translocon and effector proteins in sepD or sepL mutants (see below) of EPEC was determined by the N-terminal T3S signal (398). To date, different models for the substrate specificity switch in translocation-associated and flagellar T3S systems have been proposed, and these are discussed below.
T3S Substrate Specificity Switching in Translocation-Associated T3S Systems
The molecular ruler model proposed for the T3S4 protein YscP from Yersinia spp.
Given the finding that T3S4 proteins from animal-pathogenic bacteria determine needle length and are secreted by the T3S system, they were proposed to act as molecular rulers that measure needle length. This so-called molecular ruler model was based on the finding that deletions and insertions in the T3S4 protein YscP result in shorter and longer needle structures, respectively (257). The simultaneous production of a short and a long version of YscP led to two different needle populations with corresponding lengths but not intermediate sizes. It was therefore assumed that the length of each needle is controlled by only one, not several, YscP molecules (single ruler model) (579). The molecular ruler model assumes that the C-terminal region of T3S4 proteins remains attached to the base of the secretion apparatus while the N-terminal portion travels the inner channel of the needle (Fig. 8A). Since an interaction between YscP and the needle protein YscF or the needle tip protein LcrV has not yet been demonstrated, the identity of YscP docking sites at the tip of the growing needle still remains to be identified. Once the ruler is stretched, the C-terminal T3S4 domain signals the switch in T3S substrate specificity, probably via interaction with the C-terminal domain of YscU, and thus activates the secretion of translocon proteins. Notably, the molecular ruler model predicts that YscP and the needle protein YscF are secreted at the same time. Since the average width of YscP in an extended alpha-helical conformation was calculated to be 1 to 1.3 nm and the inner channel of the secretion apparatus has a diameter of approximately 2 to 3 nm, it might be just sufficiently wide enough to allow the passage of two partially (YscP) or completely (YscF?) unfolded proteins (578).
Fig 8.
Proposed modes of action of T3S4 proteins from animal-pathogenic bacteria. (A) Molecular ruler model. According to the molecular ruler model, the N terminus of the T3S4 protein is attached to the tip of the growing needle. Once the T3S4 protein is stretched, the C-terminal region signals the substrate specificity switch via interaction with the C-terminal domain of a member of the YscU/FlhB family. N, N-terminal region; C, C-terminal region. (B) Alternative molecular ruler model. This model predicts that T3S4 proteins are constantly secreted during needle assembly and thereby measure needle length. The interaction between the C-terminal region of the T3S4 protein and the C-terminal domains of members of the YscU/FlhB family leads to a switch in the substrate specificity and occurs only when the needle has reached a certain length. (C) Infrequent ruler model proposed for flagellar T3S systems. During hook assembly, the T3S4 protein FliK is intermittently secreted and temporarily interacts with hook components such as the hook-capping protein FlgD. However, the rapid secretion of FliK does not allow a productive interaction of the C-terminal domain of FliK with the C-terminal domain of FlhB. After the hook has reached its physiological length of approximately 55 nm, the N-terminal region of FliK interacts more frequently with hook subunits and the reduced secretion rate of FliK allows an interaction of the C-terminal region of FliK with the C-terminal domain of FlhB, and thus the induction of the substrate specificity switch.
Possible contribution of the predicted inner rod to the substrate specificity switch.
An alternative mechanism to the molecular ruler model was proposed and considers a possible contribution of the predicted inner rod structure to the substrate specificity switch in Yersinia spp. It was shown that the absence of the T3S4 protein YscP leads to oversecretion of the putative inner rod protein YscI (605). Since the introduction of point mutations into YscU not only suppresses the yscP mutant phenotype but also restores wild-type levels of YscI secretion, it was suggested that the substrate specificity switch is linked to the control of YscI secretion (605). YscP might therefore control the assembly of the predicted inner rod, which could be required for the T3S substrate specificity switch (605). The potential contribution of T3S4 proteins to inner rod formation is supported by the finding that the absence of the T3S4 protein InvJ in Salmonella spp. leads to structural differences in the base of the T3S system that are presumably caused by a disturbance of the inner rod formation (347). Furthermore, the amounts of the inner rod protein PrgJ associated with the needle complex are significantly reduced in invJ mutants (528). Although the possible contribution of the predicted inner rod assembly to the T3S substrate specificity switch is at odds with the molecular ruler model, it cannot be excluded that a combination of both mechanisms is involved in the control of T3S.
Substrate specificity switching by the T3S4 protein Spa32 from S. flexneri.
The different sizes of T3S4 proteins from animal-pathogenic bacteria do not always correlate with the observed differences in needle length (Table 6). The T3S4 protein Spa32 from S. flexneri, for instance, is 292 amino acids long, compared with 515 amino acids for YscP from Yersinia spp. However, T3S needles from S. flexneri are only approximately 20% shorter than the needles from Yersinia spp. (Table 6). Furthermore, deletions within Spa32 do not lead to a reduction in needle length (50). Notably, however, Spa32 is functionally interchangeable with YscP from Yersinia spp. and InvJ (336 amino acids) from Salmonella spp. (50). In contrast, a Spa32-YscP hybrid protein containing the central ruler domain of YscP flanked by the N- and C-terminal regions of Spa32 led to a 2-fold increase in needle length in S. flexneri compared with that observed with the native YscP or Spa32 protein (50). It is still unclear why the ruler region of YscP leads to longer needles in the context of a Spa32-YscP hybrid but not in the wild-type protein. Since it was shown that not only the length but also the helical structure of YscP might contribute to needle length control (578), it remains to be investigated whether differences in the secondary structures of YscP and Spa32-YscP hybrid proteins could account for the differences in needle length.
The analysis of Spa32 truncation derivatives revealed that the N- and C-terminal protein regions are required for protein function. The C-terminal protein region harbors the binding site for the YscU/FlhB homolog Spa40, while the N-terminal protein portion travels the inner channel of the needle (50). It was therefore proposed that Spa32 is constantly secreted during needle assembly and signals the switch via the interaction with the C-terminal domain of Spa40 once the needle has reached its final length (50) (Fig. 8B).
Substrate specificity switching during T3S in the plant-pathogenic bacterium X. campestris pv. vesicatoria.
While T3S4 proteins have been studied intensively in animal-pathogenic bacteria, less is known about the molecular mechanisms underlying T3S substrate specificity switching in plant pathogens. Functional studies of T3S4 proteins have so far been performed only with X. campestris pv. vesicatoria (332). In contrast to T3S4 proteins from animal pathogens, HpaC from X. campestris pv. vesicatoria is a cytoplasmic protein and therefore does not act as a secreted molecular ruler (73). So far, there is no experimental evidence for length control of the extracellular T3S pilus for plant-pathogenic bacteria. Since the T3S pilus is significantly longer (up to 2 μm) than the needle from animal-pathogenic bacteria, it probably cannot be bridged by a single molecular ruler molecule, suggesting that T3S substrate specificity switching in plant-pathogenic bacteria is not linked to length control of the pilus.
The T3S4 protein HpaC switches the substrate specificity of the T3S system from the early T3S substrate HrpB2 to translocon and effector proteins (332, 468). HrpB2 is essential for pilus formation and interacts with HpaC and the C-terminal domain of the YscU/FlhB homolog HrcU (HrcUC), which also provides a binding site for HpaC (330, 332, 496, 591). Experimental evidence suggests that the HrcUC-HrpB2 interaction is required for the efficient secretion of HrpB2 prior to the substrate specificity switch, which is in agreement with the predicted role of HrcUC as a substrate acceptor site (330). Since the NPTH motif of HrcU appears to be essential for the interaction of HrcUC with HpaC and HrpB2, both proteins might compete for the same binding site in HrcUC (330, 332, 496). It is therefore possible that HpaC prevents the efficient binding of HrpB2 to HrcUC and thus promotes the recognition of effector and translocon proteins by HrcUC. However, it is still unknown whether HrcUC also acts as a substrate acceptor site for late substrates, because an interaction between HrcUC and effector proteins has not yet been observed (332).
Interestingly, the lack of substrate specificity switching in the absence of HpaC can be restored upon introduction of a point mutation into HrcUC (330). As mentioned above, point mutations in the C-terminal domains of YscU/FlhB family members could mimic a conformational change in these domains that is permissive for the secretion of late substrates. Notably, however, the increased secretion of the early substrate HrpB2 in the hpaC deletion mutant was unaltered in the presence of the suppressor mutation in HrcU, suggesting that secretion of early and late substrates from X. campestris pv. vesicatoria is controlled by independent mechanisms (330). Taken together, these studies reveal differences and similarities in the control mechanisms underlying T3S in plant- and animal-pathogenic bacteria. One major difference is the apparent lack of a secreted molecular ruler in X. campestris pv. vesicatoria. Furthermore, the finding that secretion of early and late T3S substrates from X. campestris pv. vesicatoria is controlled by different mechanisms that can be uncoupled has not been reported for animal-pathogenic bacteria.
T3S Substrate Specificity Switching in Flagellar T3S Systems
The flagellar T3S4 protein FliK presumably acts as an infrequent ruler.
The length of the flagellar hook varies from 35 to 75 nm, with a peak at 55 nm, and is controlled by the T3S4 protein FliK, which is secreted during hook assembly (222, 374). Mutation of fliK results in elongated rod and hook structures and in a loss of filament formation (222, 433) (Table 6). Since insertions and deletions outside the C-terminal T3S4 domain of FliK lead to increased and reduced hook lengths, respectively, FliK most likely acts as a molecular ruler, as proposed for YscP from Yersinia spp. (502). Interestingly, experimental evidence suggests that FliK is involved not only in hook length control but also in length control of the inner rod of the flagellar T3S system (538).
Similar to the case in translocation-associated T3S systems, the substrate specificity switch in flagellar T3S systems is induced when the C-terminal T3S4 domain of FliK (FliKC) interacts with the C-terminal domain of FlhB. As mentioned above, this interaction is probably favored at a hook length of approximately 55 nm (159, 381, 383, 392). Prior to hook completion, binding of FliKC to FlhBC is suppressed by an additional regulatory protein, termed RflH/Flk or Fluke (to clearly distinguish it from FliK), which prevents the premature secretion of filament components and is anchored in the IM (7, 265, 296, 385).
Interaction studies revealed that FliK binds to the hook-capping protein FlgD, which might provide the docking site for FliK within the growing hook structure (385, 391). It is still unclear whether the inner diameter of the hook, which is smaller than 2 nm, allows the simultaneous passage of FliK and the hook protein FlgE (498). It was therefore proposed that FliK acts as a more flexible ruler molecule that is constantly secreted through the growing hook structure and switches the substrate specificity when the hook has reached its final length (159) (Fig. 8C). This so-called infrequent ruler model, which predicts temporal measurements of the hook length by intermittently secreted FliK molecules, was recently supported by experiments in S. enterica in which fliK expression and hook polymerization were uncoupled. It was shown that the substrate specificity switch occurred immediately when fliK expression was induced in a strain with elongated hook structures (162). Furthermore, the simultaneous production of a short and a long FliK derivative resulted in short hooks corresponding to the short FliK molecule, in agreement with the infrequent ruler model (162). Thus, since the substrate specificity switch depends on the interaction between the C-terminal regions of FliK and FlhB, the first FliK molecule that travels a hook with an appropriate length will signal the switch to the secretion of filament proteins. In contrast, as mentioned above, a similar experiment performed with Yersinia spp. led to two populations, with short and long needles, suggesting that needle length in Yersinia spp. is controlled by a single static ruler molecule (579).
Interestingly, it was previously reported that overexpression of a secretion-deficient FliK derivative lacking the N-terminal 99 amino acids still allows secretion of the filament protein FliC but results in elongated hooks and severely reduced bacterial motility (221, 385). It was therefore speculated that increased amounts of an N-terminally truncated FliK derivative enable FliKC to interact with FlhBC and thus to signal the switch even in the absence of FliK secretion. In agreement with this model, substrate specificity switching by FliKΔ1–99 was increased in the absence of Fluke (385). Notably, deletions in the central part (amino acids 208 to 278) of FliK, outside the T3S4 domain (amino acids 265 to 405), did not abolish filament formation and hook length control (502). Since these FliK derivatives were initially not detected in the culture supernatant, FliK was proposed to act as an internal ruler (502). However, secretion of FliK derivatives with deletions in the central protein region was shown in a later study by the use of a more sensitive FliK-specific antibody (159).
The measuring cup model.
While the molecular ruler or tape measure model is now a widely accepted working hypothesis for FliK function, hook length control was initially considered to be controlled by the capacity of the C ring to be filled with a defined amount of the hook protein FlgE (measuring cup model). This theory was based on the observation that the lack of the C-ring component FliG, FliM, or FliN led to shorter hooks (342). According to the measuring cup model, emptying the C ring would allow access of FliK to FlhBC and thus would allow the switch to occur. However, the C ring can be filled with 50 FlgE hook subunits at most, while at least 120 subunits are required to reach the average hook length (342, 476). This implies that the C ring would have to be emptied several times before the substrate specificity switch occurs. Furthermore, the measuring cup model does not explain the finding that mutations in the hook-capping protein FlgD abolish the switch in T3S substrate specificity. flgD mutants are nonmotile and do not assemble the hook, but they secrete the hook protein FlgE (380, 416). According to the measuring cup model, the substrate specificity switch should occur when FlgE is secreted, even in the absence of FlgD. Since this is not the case, the predicted measuring device provided by the C ring is not sufficient to account for the switch in substrate specificity that activates filament formation (and thus motility) after hook assembly. In agreeement with this is the observation that hook length control and filament formation are not abolished in the absence of the C ring (159).
The molecular clock model.
Besides the molecular ruler and measuring cup models, yet another mechanism involving a molecular timing device was proposed to explain substrate specificity switching in flagellar T3S systems. According to this so-called molecular clock model, initiation of hook assembly activates a countdown which signals the switch in substrate specificity after the hook has reached a length of approximately 55 nm (391). This model was based on the observation that the length of polyhooks in fliK mutants still peaks at 55 nm, suggesting that hook length control does not depend solely on the molecular ruler function of FliK (285, 391). Since mutant derivatives of the hook protein FlgE that had a defect in polymerization resulted in shorter hooks, it was suggested that the hook polymerization rate determines the time point of the substrate specificity switch (391).
One potential mechanism that was proposed to serve as a molecular clock was the proteolytic cleavage of FlhB, which has a half-life of approximately 7 min (177, 391). Notably, however, the cleavage event itself is required but presumably not essential for the substrate specificity switch, because coproduction of both FlhB cleavage products can partially restore the motility of flhB mutants, as shown for Salmonella spp. and Helicobacter pylori (381, 583) (see above). Furthermore, the finding that polymerization-deficient FlgE mutant derivatives result in shorter hooks can be explained not only by a predicted molecular clock mechanism but also by the enhanced FliK secretion that was observed in these strains (391). According to the infrequent molecular ruler model, a high secretion rate of FliK increases the probability of an interaction between FliK and FlhBC. Since this interaction induces the substrate specificity switch, the switch might occur earlier in strains that oversecrete FliK, thus leading to shorter hooks (162). In agreement with this model, overexpression of FliK was previously shown to result in shorter hooks (399). In conclusion, the experimental data published to date on mechanisms underlying flagellar hook length control support the infrequent molecular ruler model rather than the molecular cup or molecular clock model.
A SECOND SWITCH ACTIVATES EFFECTOR PROTEIN SECRETION
In translocation-associated T3S systems, secretion is controlled not only by T3S4 proteins and transcriptional regulators but also by posttranscriptional mechanisms that might impose a hierarchy on the secretion of intermediate and late substrates. Since the insertion of the translocon is a prerequisite for effector protein translocation, there is probably a second switch in the T3S substrate specificity that activates effector protein secretion after translocon assembly. This hypothesis is supported by the finding that the secretion of translocon proteins requires a different trigger (e.g., serum albumin in Y. enterocolitica [313] and bile salts in S. flexneri [418]) from that for the secretion of effector proteins (e.g., 37°C or a low calcium level in Yersinia spp. [368, 526] and Congo red in S. flexneri [432]). Although far from being understood, several control mechanisms that underlie the secretion of late substrates have been described for translocation-associated T3S systems from animal-pathogenic bacteria. In several cases, the transit of effector proteins through the secretion channel is physically blocked by a gatekeeper protein until the formation of the translocon is completed. The trigger that activates effector protein secretion could be the contact with the host cell, which is sensed by the needle or the needle tip complex. Transduction of the signal to the base of the secretion apparatus by the needle subunits might relieve the inhibitory effect of the gatekeeper protein and activate effector protein secretion. A brief summary of known gatekeeper and control proteins that are involved in the regulation of effector protein secretion is given below.
Control of Effector Protein Secretion in Yersinia spp. and P. aeruginosa by YopN Family Members
The secretion of Yops in Yersinia spp. is controlled by at least six different proteins, including the negative regulator YopN, its T3S chaperones SycN and YscB, the YopN-interacting protein TyeA, and the additional negative regulator LcrG, which binds to the tip complex protein LcrV (Table 5; Fig. 9) (212). YopN interacts with TyeA and a SycN-YscB complex in the bacterial cytosol and blocks translocon and effector protein secretion, presumably by preventing the transit of these proteins through the inner channel of the T3S system (89, 91, 126, 176) (Fig. 9A). Deletion of tyeA, yopN, sycN, or yscB therefore results in constitutive Yop secretion (212). The cytosolic TyeA protein prevents the secretion of YopN and thus the activation of effector protein secretion (89, 532). Upon host cell contact, however, secretion and translocation of YopN are triggered by a signal that might be transmitted via the needle to the secretion apparatus and thus abolishes the inhibitory effect of YopN on T3S (176, 552) (Fig. 9B).
Fig 9.
Model of the YopN-mediated control of effector protein secretion in Yersinia spp. (A) A complex of YopN, TyeA, and the YopN-specific chaperones SycN and YscB blocks the transit of T3S substrates (represented by circles) through the secretion apparatus. The T3S chaperone LcrG, which interacts with the tip protein LcrV, acts as an additional negative regulator of Yop secretion. YopD-LcrH and LcrQ-SycH complexes suppress yop gene expression when the T3S is inactive (see Fig. 7). (B) Activation of Yop secretion. The activation of the T3S system leads to the secretion of YopD, YopN, TyeA, LcrQ, and LcrV. This relieves the negative effect of YopN and TyeA on Yop secretion as well as of YopD and LcrQ on yop gene expression and thus leads to increased synthesis of Yops, including LcrV. Increased numbers of LcrV proteins bind to LcrG and presumably suppress the negative influence of LcrG on T3S.
A similar regulatory role was proposed for LcrG, the second negative regulator of Yop secretion in Yersinia spp. (407, 454). The regulatory activity of LcrG is counteracted by the tip complex protein LcrV, which interacts with LcrG in the bacterial cytoplasm (133, 351, 406, 407). According to the LcrG titration model, the induction of T3S leads to the activation of lcrV expression (see above) and thus to increased levels of LcrV that bind to LcrG and counteract its inhibitory effect on Yop secretion (351) (Fig. 9B). Although LcrG can block Yop secretion in the absence of YopN, there might be a molecular cross talk between both regulators. It was proposed that the negative influence of YopN on the secretion of LcrV indirectly regulates the function of LcrG (212).
Control of effector protein secretion by a negative regulator that might serve as a specific “plug” of the secretion channel was also reported for P. aeruginosa. The YopN homolog PopN from P. aeruginosa blocks the secretion of effector proteins before it is itself secreted and translocated (533, 611). Notably, however, in contrast to effector proteins, translocon proteins are still secreted under conditions that do not favor PopN secretion, suggesting that the negative influence of PopN on T3S is specific for effector proteins (95). In this context, it is interesting that the secretion of translocon proteins by the T3S system from P. aeruginosa appears to be constitutive (95).
The LcrG homolog PcrG from P. aeruginosa presumably blocks effector protein secretion from inside the cytoplasm by an unknown mechanism (533). PcrG interacts with PcrV, which is a homolog of LcrV and is secreted in a PcrG-dependent manner. Notably, however, the PcrG-mediated suppression of effector protein secretion is independent of its interaction with PcrV (310). This is different from the anticipated interplay between LcrV and LcrG (see above) and suggests that despite the presence of sequence-related control proteins, the regulatory mechanisms underlying effector protein export in Yersinia spp. and P. aeruginosa can vary significantly. Different regulatory mechanisms might also explain the finding that in contrast to the mutation of lcrV, which does not lead to deregulated secretion (32, 481), deletion of pcrV leads to constitutive effector protein secretion in P. aeruginosa (310, 407) (Tables 5 and 8). The PcrV-mediated control of effector protein secretion is probably linked to the role of PcrV as a tip complex protein, because PcrV has to be secreted to regulate T3S (310). However, it is unlikely that PcrV acts simply as an external plug, because translocon proteins are secreted prior to host cell contact, when effector protein secretion is probably off (95). It was therefore proposed that the assembly of the tip complex stabilizes an “off” conformation for effector protein secretion. A signal upon host cell contact that is transduced to the base of the secretion apparatus via subunits of the needle might be required to activate the secretion of effector proteins (310). A role of the needle subunits in the sensing and transduction of the activation signal was supported by the finding that individual point mutations in needle proteins from Yersinia spp. and S. flexneri lead to alterations in the secretion profile (Table 8).
Table 8.
Contributions of selected mutations in needle, translocon, and tip complex proteins to the control of T3S
| Needle, translocon, or tip complex protein mutant(s)a | Organism | Effect on T3S | Reference(s) |
|---|---|---|---|
| Needle protein mutants | |||
| YscFD28A, YscFD46A | Yersinia spp. | Constitutive secretion of effectors, translocon proteins, and YopN; larger amounts of extracellular Ca2+ are required to block T3S | 552 |
| YscFK58R, YscFA72V, YscFN31A, YscFT70A | Yersinia spp. | WT secretion of YopE and YscF; reduced translocation of YopE and reduced pore formation | 124 |
| YscFD82G, YscFN47S, YscFN47S, N68S | Yersinia spp. | WT secretion of YopE and YscF; reduced translocation of YopE; WT pore formation | 124 |
| MxiHP44A, MxiHP51A | S. flexneri | Constitutive secretion of translocon and effector proteins; altered composition of IpaB, IpaC, and IpaD | 268, 569 |
| MxiHD73A | S. flexneri | Constitutive secretion of translocon and effector proteins; T3S is not inducible by Congo red; needles lack IpaB, IpaC, and IpaD | 268, 569 |
| MxiHK69A, MxiHR83A | S. flexneri | WT secretion of translocon proteins, but no secretion of effector proteins upon induction with Congo red; no secretion of MxiC | 268, 350 |
| Translocon protein mutants | |||
| ΔyopD mutant | Yersinia spp. | Deregulated Yop synthesis and secretion (secretion of LcrV and Yops in the presence of Ca2+); no translocation of Yops | 182, 465, 601 |
| ΔipaB mutant | S. flexneri | Unregulated secretion of translocon and effector proteins (no responsiveness to Congo red) | 362, 461, 501, 569 |
| Tip complex protein mutants | |||
| ΔlcrV mutant | Yersinia spp. | Reduced secretion of effector proteins; unaltered secretion of YopB and YopD; no pore formation | 133, 344 |
| ΔpcrV mutant | P. aeruginosa | Increased secretion of effector proteins in the presence of Ca2+; increased secretion of ExoS in the presence of eukaryotic cells; unaltered secretion of PopB and PopD; no assembly of the translocation pore | 208, 310, 533 |
| ΔipaD mutant | S. flexneri | Unregulated secretion of Ipa and effector proteins (no responsiveness to Congo red) | 362, 442, 461, 501, 569 |
| ΔsipD mutant | Increased T3S, no translocation of effector proteins | 260, 261 |
Only selected point mutant derivatives are listed.
Control of Effector Protein Export in S. flexneri
Control of effector protein secretion by an extracellular plug was also proposed for S. flexneri. Since the absence of the needle tip proteins results in constitutive secretion, it is assumed that the tip complex serves as an external plug that blocks secretion of effector proteins prior to host cell contact (569) (Tables 5 and 8). An additional protein involved in the control of effector protein secretion in S. flexneri is the secreted MxiC protein, which shares sequence and structural similarity with the YopN-TyeA complex from Yersinia spp. (132) and might act similarly to YopN. Notably, however, in contrast to the case for yopN mutants, which constitutively secrete translocon and effector proteins (176), deletion of mxiC leads to constitutive secretion of effector proteins but reduced secretion of translocon proteins in response to Congo red induction (350). This suggests that MxiC not only acts as a negative regulator of effector protein secretion but also promotes the secretion of translocon proteins. Interestingly, the formation of the tip complex is unaltered in the mxiC mutant. Since the mxiC mutant constitutively secretes effector proteins, it was proposed that the tip complex does not act as an external plug for effector protein export in the absence of MxiC (350).
Control of T3S in EPEC by SepL and SepD
In EPEC, T3S of translocon and effector proteins is controlled by SepL and SepD, which both localize to the bacterial membranes and interact with each other (137, 138, 408) (Table 5). Deletion of sepL leads to an increase in the secretion of effector proteins and to a reduced secretion of translocon proteins (137, 138). Since SepL shares homology with YopN and TyeA (see above) (426, 584), it might act together with SepD as a gatekeeper that promotes secretion of translocon proteins and prevents effector protein secretion prior to host cell contact. Interestingly, the C-terminal region of SepL interacts with the T3S chaperone CesL and the effector protein Tir but not with other effector proteins (584, 620). The SepL-Tir interaction appears to be required for the SepL-mediated control of effector protein secretion while being dispensable for the efficient secretion of translocon proteins (584). So far, the molecular mechanisms that link the SepL-Tir interaction to the control of effector protein secretion are not understood. However, given that Tir is the first effector protein that travels the T3S channel (see above), SepL-bound Tir might block the efficient access of additional effectors to components of the secretion apparatus, such as the ATPase EscN.
Control of Effector Protein Translocation by pH Sensing and a Sorting Platform in Salmonella spp.
As mentioned above, Salmonella spp. contain two translocation-associated T3S systems that are encoded by SPI-1 and SPI-2 and operate during bacterial entry into the host cell (SPI-1) and inside the host vacuole (SPI-2), respectively (Fig. 10). SPI-1- and SPI-2-mediated effector protein translocation is presumably controlled by the YopN homologs InvE (SPI-1) and SsaL (SPI-2). Deletion of invE and ssaL results in reduced secretion of translocon proteins and oversecretion of effector proteins, respectively, suggesting that InvE and SsaL not only act as negative regulators of effector secretion but also promote the secretion of translocon proteins (100, 289, 623). Notably, a similar phenotype was observed for Salmonella mutants lacking either SsaM or SpiC, two cytoplasmic regulatory proteins that interact with each other (621) (Table 5). SsaM and SpiC are involved in the control of SPI-2-mediated T3S, which is activated after acidification of the Salmonella-containing vacuole inside the host cell (450). It was previously shown that SsaM and SpiC bind to the YopN homolog SsaL at pH 5.0. The resulting SsaM-SpiC-SsaL complex promotes the secretion of translocon proteins and suppresses effector protein secretion. It might thus act as a gatekeeper for effector proteins when bacteria reside in the host vacuole (623). A shift to pH 7.2, however, leads to dissociation of the SsaL-SsaM-SpiC complex and to the activation of effector protein secretion. This pH shift might occur in nature when bacteria leave the host vacuole to enter the cytoplasm (623) (Fig. 10). The sensor for the pH shift is probably not the translocon, because a translocon mutant secretes wild-type levels of effector proteins at pH 7.2. It was therefore speculated that the extracellular pH is sensed by components of the needle (623). Alternatively, the tip complex could also serve as a sensor of the external pH. In this context, it is interesting that the tip complex protein SipD undergoes a conformational change at pH 5 to 6 (346). It remains to be investigated whether pH sensing is also involved in the control of effector protein secretion in other animal-pathogenic bacteria.
Fig 10.
Control of SPI-1- and SPI-2-mediated T3S in Salmonella spp. (A) Infection of epithelial eukaryotic cells by Salmonella spp. The SPI-1-encoded T3S system injects effector proteins into epithelial cells, which leads to cytoskeletal rearrangements and membrane ruffling. Bacteria enter the host cell cytosol via endocytosis and activate the SPI-2-encoded T3S system inside the Salmonella-containing vacuole. (B) Predicted function of the SPI-1-encoded SpaO-OrgA-OrgB complex during control of T3S. The SpaO-OrgA-OrgB complex serves as a docking platform for translocon and effector proteins that are sequentially targeted to this complex by a process that presumably requires the presence of corresponding T3S chaperones. According to the predicted hierarchy in T3S, effector proteins form a queue for docking to the SpaO-OrgA-OrgB complex, while translocon proteins are secreted. (C) Control of SPI-2-dependent secretion of translocon and effector proteins. A complex of SpiC, SsaL, and SsaM controls the secretion of translocon and effector proteins dependent on differences in the external pH. At pH 5 (inside the Salmonella-containing vacuole), the complex blocks the efficient secretion of effector proteins, while translocon proteins are secreted. A shift in the extracellular pH to pH 7 leads to the dissociation of the SpiC-SsaL-SsaM complex and thus induces effector protein secretion. The pH shift is probably sensed by the extracellular components of the T3S system. Note that the architecture of the SPI-2 T3S system is speculative and is proposed according to amino acid sequence similarities between predicted components of SPI-2 T3S systems and known translocation-associated T3S systems. The dashed arrow indicates a reduced secretion and/or translocation rate.
Control of translocon and effector protein secretion in Salmonella spp. not only depends on YopN-like gatekeeper proteins but also involves the predicted C ring of the T3S system. Experimental evidence suggests that the putative C ring provides a sorting platform for early, intermediate, and late substrates. Thus, the predicted C ring component SpaO was shown to associate with several proteins, including OrgA and -B (required for complex stability), the ATPase InvC, and translocon proteins (Table 2). Effector proteins are largely absent from the complex. However, they associate with SpaO-OrgA-OrgB in the absence of translocon proteins. Since translocon proteins are probably secreted prior to effector proteins, these findings suggest that effectors form a queue for docking to the predicted C ring (304). In the absence of the T3S4 protein InvJ (which leads to the constitutive secretion of needle and inner rod proteins but not of translocon proteins), translocon or effector proteins do not associate with the SpaO-OrgA-OrgB complex (304) (Fig. 10). It was therefore proposed that the selective binding of T3S substrates to the predicted C ring allows their sequential delivery. Interestingly, the binding of the SpaO-OrgA-OrgB complex to T3S substrates requires the presence of their cognate T3S chaperones, which might target their interaction partners to the predicted C ring (304). In conclusion, the published data on T3S in Salmonella spp. suggest that T3S is controlled not only by the gatekeeper proteins InvE and SsaL but also by SsaL-associated proteins that are released from SsaL upon a shift in the extracellular pH. Furthermore, the transport of effector proteins might also depend on the regulated binding of these proteins to the predicted C ring. Once again, this reveals the high complexity of T3S-associated control mechanisms.
CONCLUDING REMARKS
During the past 3 years, significant progress has been made in our understanding of the molecular mechanisms that underlie the assembly and control of T3S systems from Gram-negative pathogenic bacteria. While T3S systems have long been known as membrane-spanning nanomachines, the detailed characterization of several conserved components of T3S systems, including the analysis of protein crystal structures, has now shed more light on their contribution to substrate recognition and their function during the secretion process. Furthermore, recent studies revealed that the assembly of the T3S system occurs sequentially and probably involves two assembly platforms that are later joined together. The aims of future studies will certainly be to determine the location of every component of the T3S system and to generate a complete atomic model of the secretion apparatus.
In addition to the analysis of core constituents of the secretion apparatus, research on animal-pathogenic bacteria has also focused on the characterization of extracellular components of the T3S system, such as needle, translocon, and tip proteins. Experimental evidence suggests that these proteins are involved in the sensing and transduction of external signals such as the pH or host cell contact. In contrast, not much is known about the identities and functions of translocon and potential tip complex proteins from plant-pathogenic bacteria. Future research should help to functionally characterize these proteins, because it cannot be assumed that the contributions of individual proteins to the assembly and activity of the T3S system are similar in different pathogens. We also still know very little about the functions of the nonconserved components of T3S systems from plant- and animal-pathogenic bacteria. Since these proteins might reflect adaptions of the T3S system to different host organisms or extracellular environments, they should be included in future studies.
Given the architecture of T3S systems, it has been assumed that T3S is a hierarchical process. Recently, various regulatory proteins of T3S systems that are involved in transcriptional and posttranscriptional control mechanisms or the switch in substrate specificity have been characterized intensively. However, the precise mechanisms that lead to the activation of T3S and guarantee the hierarchical secretion of early, middle, and late T3S substrates are far from being understood and so far have been studied mainly in animal-pathogenic bacteria. Future research should therefore focus on the detailed characterization of individual T3S control proteins and on the analysis of their interactions with substrates and components of the T3S system. Since T3S control proteins and substrate recognition sites are not highly conserved and the experimental findings reported to date have already revealed genus- and species-specific differences in the control mechanisms underlying T3S, the characterization of individual species- and even pathovar-specific proteins will be crucial for a complete understanding of the complex T3S-associated regulatory mechanisms. Furthermore, this knowledge will help in the design of inhibitors of T3S that may be used as therapeutic agents and in development of biotechnological approaches that will employ the T3S system as a tool for the targeted and controlled delivery of foreign proteins into eukaryotic cells.
ACKNOWLEDGMENTS
I am indebted to the reviewers of the manuscript for their numerous helpful comments and to Patrick Lane for his assistance in preparing the illustrations. I am grateful to members of my research group for lively discussions and to Ulla Bonas for critical comments on the manuscript.
Work in my group is supported by grants from the Deutsche Forschungsgemeinschaft and the Sonderforschungsbereich (SFB 648).
REFERENCES
- 1. Abe A, et al. 1999. Enteropathogenic Escherichia coli translocated intimin receptor, Tir, requires a specific chaperone for stable secretion. Mol. Microbiol. 33:1162–1175 [DOI] [PubMed] [Google Scholar]
- 2. Agrain C, et al. 2005. Characterization of a type III secretion substrate specificity switch (T3S4) domain in YscP from Yersinia enterocolitica. Mol. Microbiol. 56:54–67 [DOI] [PubMed] [Google Scholar]
- 3. Aili M, et al. 2008. Regulation of Yersinia Yop-effector delivery by translocated YopE. Int. J. Med. Microbiol. 298:183–192 [DOI] [PubMed] [Google Scholar]
- 4. Akeda Y, Galan JE. 2005. Chaperone release and unfolding of substrates in type III secretion. Nature 437:911–915 [DOI] [PubMed] [Google Scholar]
- 5. Akopyan K, et al. 2011. Translocation of surface-localized effectors in type III secretion. Proc. Natl. Acad. Sci. U. S. A. 108:1639–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Aldridge P, Karlinsey J, Hughes KT. 2003. The type III secretion chaperone FlgN regulates flagellar assembly via a negative feedback loop containing its chaperone substrates FlgK and FlgL. Mol. Microbiol. 49:1333–1345 [DOI] [PubMed] [Google Scholar]
- 7. Aldridge P, Karlinsey JE, Becker E, Chevance FF, Hughes KT. 2006. Flk prevents premature secretion of the anti-sigma factor FlgM into the periplasm. Mol. Microbiol. 60:630–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Aldridge PD, et al. 2006. The flagellar-specific transcription factor, sigma28, is the type III secretion chaperone for the flagellar-specific anti-sigma28 factor FlgM. Genes Dev. 20:2315–2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Allaoui A, Menard R, Sansonetti PJ, Parsot C. 1993. Characterization of the Shigella flexneri ipgD and ipgF genes, which are located in the proximal part of the mxi locus. Infect. Immun. 61:1707–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Allaoui A, Sansonetti PJ, Parsot C. 1992. MxiJ, a lipoprotein involved in secretion of Shigella Ipa invasins, is homologous to YscJ, a secretion factor of the Yersinia Yop proteins. J. Bacteriol. 174:7661–7669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Allaoui A, Woestyn S, Sluiters C, Cornelis GR. 1994. YscU, a Yersinia enterocolitica inner membrane protein involved in Yop secretion. J. Bacteriol. 176:4534–4542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Allen-Vercoe E, Toh MC, Waddell B, Ho H, DeVinney R. 2005. A carboxy-terminal domain of Tir from enterohemorrhagic Escherichia coli O157:H7 (EHEC O157:H7) required for efficient type III secretion. FEMS Microbiol. Lett. 243:355–364 [DOI] [PubMed] [Google Scholar]
- 13. Allen-Vercoe E, Waddell B, Livingstone S, Deans J, DeVinney R. 2006. Enteropathogenic Escherichia coli Tir translocation and pedestal formation requires membrane cholesterol in the absence of bundle-forming pili. Cell. Microbiol. 8:613–624 [DOI] [PubMed] [Google Scholar]
- 14. Anderson DM, Fouts DE, Collmer A, Schneewind O. 1999. Reciprocal secretion of proteins by the bacterial type III machines of plant and animal pathogens suggests universal recognition of mRNA targeting signals. Proc. Natl. Acad. Sci. U. S. A. 96:12839–12843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Anderson DM, Ramamurthi KS, Tam C, Schneewind O. 2002. YopD and LcrH regulate expression of Yersinia enterocolitica YopQ by a posttranscriptional mechanism and bind to yopQ RNA. J. Bacteriol. 184:1287–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Anderson DM, Schneewind O. 1997. A mRNA signal for the type III secretion of Yop proteins by Yersinia enterocolitica. Science 278:1140–1143 [DOI] [PubMed] [Google Scholar]
- 17. Anderson DM, Schneewind O. 1999. Yersinia enterocolitica type III secretion: an mRNA signal that couples translation and secretion of YopQ. Mol. Microbiol. 31:1139–1148 [DOI] [PubMed] [Google Scholar]
- 18. Andrade A, Pardo JP, Espinosa N, Perez-Hernandez G, Gonzalez-Pedrajo B. 2007. Enzymatic characterization of the enteropathogenic Escherichia coli type III secretion ATPase EscN. Arch. Biochem. Biophys. 468:121–127 [DOI] [PubMed] [Google Scholar]
- 19. Andre I, Bradley P, Wang C, Baker D. 2007. Prediction of the structure of symmetrical protein assemblies. Proc. Natl. Acad. Sci. U. S. A. 104:17656–17661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Arnold R, et al. 2009. Sequence-based prediction of type III secreted proteins. PLoS Pathog. 5:e1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Auvray F, Ozin AJ, Claret L, Hughes C. 2002. Intrinsic membrane targeting of the flagellar export ATPase FliI: interaction with acidic phospholipids and FliH. J. Mol. Biol. 318:941–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Badel JL, et al. 2003. Pseudomonas syringae pv. tomato DC3000 HopPtoM (CEL ORF3) is important for lesion formation but not growth in tomato and is secreted and translocated by the Hrp type III secretion system in a chaperone-dependent manner. Mol. Microbiol. 49:1239–1251 [DOI] [PubMed] [Google Scholar]
- 23. Bange G, et al. 2010. FlhA provides the adaptor for coordinated delivery of late flagella building blocks to the type III secretion system. Proc. Natl. Acad. Sci. U. S. A. 107:11295–11300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Barembruch C, Hengge R. 2007. Cellular levels and activity of the flagellar sigma factor FliA of Escherichia coli are controlled by FlgM-modulated proteolysis. Mol. Microbiol. 65:76–89 [DOI] [PubMed] [Google Scholar]
- 25. Barison N, Lambers J, Hurwitz R, Kolbe M. 2012. Interaction of MxiG with the cytosolic complex of the type III secretion system controls Shigella virulence. FASEB J. 26:1717–1726 [DOI] [PubMed] [Google Scholar]
- 26. Barta ML, et al. 2012. The structures of coiled-coil domains from type III secretion system translocators reveal homology to pore-forming toxins. J. Mol. Biol. 417:395–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Barta ML, Zhang L, Picking WL, Geisbrecht BV. 2010. Evidence for alternative quaternary structure in a bacterial type III secretion system chaperone. BMC Struct. Biol. 10:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Basu A, Chatterjee R, Datta S. 2012. Expression, purification, structural and functional analysis of SycB: a type three secretion chaperone from Yersinia enterocolitica. Protein J. 31:93–107 [DOI] [PubMed] [Google Scholar]
- 29. Beeckman DS, Vanrompay DC. 2010. Bacterial secretion systems with an emphasis on the chlamydial type III secretion system. Curr. Issues Mol. Biol. 12:17–41 [PubMed] [Google Scholar]
- 30. Bennett JC, Thomas J, Fraser GM, Hughes C. 2001. Substrate complexes and domain organization of the Salmonella flagellar export chaperones FlgN and FliT. Mol. Microbiol. 39:781–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Berger C, Robin GP, Bonas U, Koebnik R. 2010. Membrane topology of conserved components of the type III secretion system from the plant pathogen Xanthomonas campestris pv. vesicatoria. Microbiology 156:1963–1974 [DOI] [PubMed] [Google Scholar]
- 32. Bergman T, et al. 1991. Analysis of the V antigen lcrGVH-yopBD operon of Yersinia pseudotuberculosis: evidence for a regulatory role of LcrH and LcrV. J. Bacteriol. 173:1607–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Biemans-Oldehinkel E, Sal-Man N, Deng W, Foster LJ, Finlay BB. 2011. Quantitative proteomic analysis reveals formation of an EscL-EscQ-EscN type III complex in enteropathogenic E. coli. J. Bacteriol. 193:5514–5519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bigot A, et al. 2005. Role of FliF and FliI of Listeria monocytogenes in flagellar assembly and pathogenicity. Infect. Immun. 73:5530–5539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Birket SE, et al. 2007. Preparation and characterization of translocator/chaperone complexes and their component proteins from Shigella flexneri. Biochemistry 46:8128–8137 [DOI] [PubMed] [Google Scholar]
- 36. Birtalan S, Ghosh P. 2001. Structure of the Yersinia type III secretory system chaperone SycE. Nat. Struct. Biol. 8:974–978 [DOI] [PubMed] [Google Scholar]
- 37. Birtalan SC, Phillips RM, Ghosh P. 2002. Three-dimensional secretion signals in chaperone-effector complexes of bacterial pathogens. Mol. Cell 9:971–980 [DOI] [PubMed] [Google Scholar]
- 38. Björnfot AC, Lavander M, Forsberg A, Wolf-Watz H. 2009. Auto-proteolysis of YscU of Yersinia pseudotuberculosis is important for regulation of expression and secretion of Yop proteins. J. Bacteriol. 191:4259–4267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Blair DF. 2003. Flagellar movement driven by proton translocation. FEBS Lett. 545:86–95 [DOI] [PubMed] [Google Scholar]
- 40. Blair DF, Berg HC. 1988. Restoration of torque in defective flagellar motors. Science 242:1678–1681 [DOI] [PubMed] [Google Scholar]
- 41. Blair DF, Berg HC. 1990. The MotA protein of E. coli is a proton-conducting component of the flagellar motor. Cell 60:439–449 [DOI] [PubMed] [Google Scholar]
- 42. Blaylock B, Riordan KE, Missiakas DM, Schneewind O. 2006. Characterization of the Yersinia enterocolitica type III secretion ATPase YscN and its regulator, YscL. J. Bacteriol. 188:3525–3534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bleves S, Marenne MN, Detry G, Cornelis GR. 2002. Up-regulation of the Yersinia enterocolitica yop regulon by deletion of the flagellum master operon flhDC. J. Bacteriol. 184:3214–3223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Blocker A, et al. 1999. The tripartite type III secreton of Shigella flexneri inserts IpaB and IpaC into host membranes. J. Cell Biol. 147:683–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Blocker A, et al. 2001. Structure and composition of the Shigella flexneri “needle complex,” a part of its type III secreton. Mol. Microbiol. 39:652–663 [DOI] [PubMed] [Google Scholar]
- 46. Blocker AJ, et al. 2008. What's the point of the type III secretion system needle? Proc. Natl. Acad. Sci. U. S. A. 105:6507–6513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bocsanczy AM, Nissinen RM, Oh CS, Beer SV. 2008. HrpN of Erwinia amylovora functions in the translocation of DspA/E into plant cells. Mol. Plant Pathol. 9:425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bogdanove A, et al. 1996. Unified nomenclature for broadly conserved hrp genes of phytopathogenic bacteria. Mol. Microbiol. 20:681–683 [DOI] [PubMed] [Google Scholar]
- 49. Boonyom R, Karavolos MH, Bulmer DM, Khan CM. 2010. Salmonella pathogenicity island 1 (SPI-1) type III secretion of SopD involves N- and C-terminal signals and direct binding to the InvC ATPase. Microbiology 156:1805–1814 [DOI] [PubMed] [Google Scholar]
- 50. Botteaux A, Sani M, Kayath CA, Boekema EJ, Allaoui A. 2008. Spa32 interaction with the inner-membrane Spa40 component of the type III secretion system of Shigella flexneri is required for the control of the needle length by a molecular tape measure mechanism. Mol. Microbiol. 70:1515–1528 [DOI] [PubMed] [Google Scholar]
- 51. Botteaux A, Sory MP, Biskri L, Parsot C, Allaoui A. 2009. MxiC is secreted by and controls the substrate specificity of the Shigella flexneri type III secretion apparatus. Mol. Microbiol. 71:449–460 [DOI] [PubMed] [Google Scholar]
- 52. Boyd AP, Lambermont I, Cornelis GR. 2000. Competition between the Yops of Yersinia enterocolitica for delivery into eukaryotic cells: role of the SycE chaperone binding domain of YopE. J. Bacteriol. 182:4811–4821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Braun TF, Blair DF. 2001. Targeted disulfide cross-linking of the MotB protein of Escherichia coli: evidence for two H(+) channels in the stator complex. Biochemistry 40:13051–13059 [DOI] [PubMed] [Google Scholar]
- 54. Bretz J, Losada L, Lisboa K, Hutcheson SW. 2002. Lon protease functions as a negative regulator of type III protein secretion in Pseudomonas syringae. Mol. Microbiol. 45:397–409 [DOI] [PubMed] [Google Scholar]
- 55. Bridge DR, Novotny MJ, Moore ER, Olson JC. 2010. Role of host cell polarity and leading edge properties in Pseudomonas type III secretion. Microbiology 156:356–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bröms JE, Edqvist PJ, Carlsson KE, Forsberg A, Francis MS. 2005. Mapping of a YscY binding domain within the LcrH chaperone that is required for regulation of Yersinia type III secretion. J. Bacteriol. 187:7738–7752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bröms JE, Edqvist PJ, Forsberg A, Francis MS. 2006. Tetratricopeptide repeats are essential for PcrH chaperone function in Pseudomonas aeruginosa type III secretion. FEMS Microbiol. Lett. 256:57–66 [DOI] [PubMed] [Google Scholar]
- 58. Bronstein PA, Miao EA, Miller SI. 2000. InvB is a type III secretion chaperone specific for SspA. J. Bacteriol. 182:6638–6644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Brown IR, Mansfield JW, Taira S, Roine E, Romantschuk M. 2001. Immunocytochemical localization of HrpA and HrpZ supports a role for the Hrp pilus in the transfer of effector proteins from Pseudomonas syringae pv. tomato across the host plant cell wall. Mol. Plant Microbe Interact. 14:394–404 [DOI] [PubMed] [Google Scholar]
- 60. Brown PN, Hill CP, Blair DF. 2002. Crystal structure of the middle and C-terminal domains of the flagellar rotor protein FliG. EMBO J. 21:3225–3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Brown PN, Mathews MA, Joss LA, Hill CP, Blair DF. 2005. Crystal structure of the flagellar rotor protein FliN from Thermotoga maritima. J. Bacteriol. 187:2890–2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Brown PN, Terrazas M, Paul K, Blair DF. 2007. Mutational analysis of the flagellar protein FliG: sites of interaction with FliM and implications for organization of the switch complex. J. Bacteriol. 189:305–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Broz P, et al. 2007. Function and molecular architecture of the Yersinia injectisome tip complex. Mol. Microbiol. 65:1311–1320 [DOI] [PubMed] [Google Scholar]
- 64. Brutinel ED, Yahr TL. 2008. Control of gene expression by type III secretory activity. Curr. Opin. Microbiol. 11:128–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Buchko GW, et al. 2010. A multi-pronged search for a common structural motif in the secretion signal of Salmonella enterica serovar Typhimurium type III effector proteins. Mol. Biosyst. 6:2448–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Burghout P, et al. 2004. Role of the pilot protein YscW in the biogenesis of the YscC secretin in Yersinia enterocolitica. J. Bacteriol. 186:5366–5375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Burghout P, et al. 2004. Structure and electrophysiological properties of the YscC secretin from the type III secretion system of Yersinia enterocolitica. J. Bacteriol. 186:4645–4654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Büttner CR, Cornelis GR, Heinz DW, Niemann HH. 2005. Crystal structure of Yersinia enterocolitica type III secretion chaperone SycT. Protein Sci. 14:1993–2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Büttner CR, Sorg I, Cornelis GR, Heinz DW, Niemann HH. 2008. Structure of the Yersinia enterocolitica type III secretion translocator chaperone SycD. J. Mol. Biol. 375:997–1012 [DOI] [PubMed] [Google Scholar]
- 70. Büttner D, Bonas U. 2002. Port of entry—the type III secretion translocon. Trends Microbiol. 10:186–192 [DOI] [PubMed] [Google Scholar]
- 71. Büttner D, Gürlebeck D, Noel LD, Bonas U. 2004. HpaB from Xanthomonas campestris pv. vesicatoria acts as an exit control protein in type III-dependent protein secretion. Mol. Microbiol. 54:755–768 [DOI] [PubMed] [Google Scholar]
- 72. Büttner D, He SY. 2009. Type III protein secretion in plant pathogenic bacteria. Plant Physiol. 150:1656–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Büttner D, Lorenz C, Weber E, Bonas U. 2006. Targeting of two effector protein classes to the type III secretion system by a HpaC- and HpaB-dependent protein complex from Xanthomonas campestris pv. vesicatoria. Mol. Microbiol. 59:513–527 [DOI] [PubMed] [Google Scholar]
- 74. Büttner D, Nennstiel D, Klüsener B, Bonas U. 2002. Functional analysis of HrpF, a putative type III translocon protein from Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 184:2389–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Büttner D, Noel L, Stuttmann J, Bonas U. 2007. Characterization of the non-conserved hpaB-hrpF region in the hrp pathogenicity island from Xanthomonas campestris pv. vesicatoria. Mol. Plant Microbe Interact. 20:1063–1074 [DOI] [PubMed] [Google Scholar]
- 76. Button JE, Galan JE. 2011. Regulation of chaperone/effector complex synthesis in a bacterial type III secretion system. Mol. Microbiol. 81:1474–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Bzymek KP, Hamaoka BY, Ghosh P. 2012. Two translation products of Yersinia yscQ assemble to form a complex essential to type III secretion. Biochemistry 51:1669–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Cambronne ED, Cheng LW, Schneewind O. 2000. LcrQ/YscM1, regulators of the Yersinia yop virulon, are injected into host cells by a chaperone-dependent mechanism. Mol. Microbiol. 37:263–273 [DOI] [PubMed] [Google Scholar]
- 79. Cambronne ED, Schneewind O. 2002. Yersinia enterocolitica type III secretion: yscM1 and yscM2 regulate yop gene expression by a posttranscriptional mechanism that targets the 5′ untranslated region of yop mRNA. J. Bacteriol. 184:5880–5893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Cambronne ED, Sorg JA, Schneewind O. 2004. Binding of SycH chaperone to YscM1 and YscM2 activates effector yop expression in Yersinia enterocolitica. J. Bacteriol. 186:829–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Chadsey MS, Hughes KT. 2001. A multipartite interaction between Salmonella transcription factor sigma28 and its anti-sigma factor FlgM: implications for sigma28 holoenzyme destabilization through stepwise binding. J. Mol. Biol. 306:915–929 [DOI] [PubMed] [Google Scholar]
- 82. Chadsey MS, Karlinsey JE, Hughes KT. 1998. The flagellar anti-sigma factor FlgM actively dissociates Salmonella typhimurium sigma28 RNA polymerase holoenzyme. Genes Dev. 12:3123–3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Chakravortty D, Rohde M, Jager L, Deiwick J, Hensel M. 2005. Formation of a novel surface structure encoded by Salmonella pathogenicity island 2. EMBO J. 24:2043–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Charkowski AO, et al. 1998. The Pseudomonas syringae pv. tomato HrpW protein has domains similar to harpins and pectate lyases and can elicit the plant hypersensitive response and bind to pectate. J. Bacteriol. 180:5211–5217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Charpentier X, Oswald E. 2004. Identification of the secretion and translocation domain of the enteropathogenic and enterohemorrhagic Escherichia coli effector Cif, using TEM-1 beta-lactamase as a new fluorescence-based reporter. J. Bacteriol. 186:5486–5495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Chatterjee C, et al. 2011. Crystal structure of the heteromolecular chaperone, AscE-AscG, from the type III secretion system in Aeromonas hydrophila. PLoS One 6:e19208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Chatterjee S, et al. 2011. The crystal structures of the Salmonella type III secretion system tip protein SipD in complex with deoxycholate and chenodeoxycholate. Protein Sci. 20:75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Chen Y, Anderson DM. 2011. Expression hierarchy in the Yersinia type III secretion system established through YopD recognition of RNA. Mol. Microbiol. 80:966–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Cheng LW, Kay O, Schneewind O. 2001. Regulated secretion of YopN by the type III machinery of Yersinia enterocolitica. J. Bacteriol. 183:5293–5301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Cheng LW, Schneewind O. 1999. Yersinia enterocolitica type III secretion. On the role of SycE in targeting YopE into HeLa cells. J. Biol. Chem. 274:22102–22108 [DOI] [PubMed] [Google Scholar]
- 91. Cheng LW, Schneewind O. 2000. Yersinia enterocolitica TyeA, an intracellular regulator of the type III machinery, is required for specific targeting of YopE, YopH, YopM, and YopN into the cytosol of eukaryotic cells. J. Bacteriol. 182:3183–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Chevance FFV, Hughes KT. 2008. Coordinating assembly of a bacterial macromolecular machine. Nature 6:455–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Chiu HJ, Lin WS, Syu WJ. 2003. Type III secretion of EspB in enterohemorrhagic Escherichia coli O157:H7. Arch. Microbiol. 180:218–226 [DOI] [PubMed] [Google Scholar]
- 94. Chun SY, Parkinson JS. 1988. Bacterial motility: membrane topology of the Escherichia coli MotB protein. Science 239:276–278 [DOI] [PubMed] [Google Scholar]
- 95. Cisz M, Lee PC, Rietsch A. 2008. ExoS controls the cell contact-mediated switch to effector secretion in Pseudomonas aeruginosa. J. Bacteriol. 190:2726–2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Claret L, Calder SR, Higgins M, Hughes C. 2003. Oligomerization and activation of the FliI ATPase central to bacterial flagellum assembly. Mol. Microbiol. 48:1349–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Clarke CR, Cai R, Studholme DJ, Guttman DS, Vinatzer BA. 2010. Pseudomonas syringae strains naturally lacking the classical P. syringae hrp/hrc locus are common leaf colonizers equipped with an atypical type III secretion system. Mol. Plant Microbe Interact. 23:198–210 [DOI] [PubMed] [Google Scholar]
- 98. Collazo CM, Zieler MK, Galan JE. 1995. Functional analysis of the Salmonella typhimurium invasion genes invL and invJ and identification of a target of the protein secretion apparatus encoded in the inv locus. Mol. Microbiol. 15:25–38 [DOI] [PubMed] [Google Scholar]
- 99. Coombes BK, Brown NF, Kujat-Choy S, Vallance BA, Finlay BB. 2003. SseA is required for translocation of Salmonella pathogenicity island-2 effectors into host cells. Microbes Infect. 5:561–570 [DOI] [PubMed] [Google Scholar]
- 100. Coombes BK, Brown NF, Valdez Y, Brumell JH, Finlay BB. 2004. Expression and secretion of Salmonella pathogenicity island-2 virulence genes in response to acidification exhibit differential requirements of a functional type III secretion apparatus and SsaL. J. Biol. Chem. 279:49804–49815 [DOI] [PubMed] [Google Scholar]
- 101. Cooper CA, et al. 2010. Structural and biochemical characterization of SrcA, a multi-cargo type III secretion chaperone in Salmonella required for pathogenic association with a host. PLoS Pathog. 6:e1000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Cordes FS, et al. 2005. Helical packing of needles from functionally altered Shigella type III secretion systems. J. Mol. Biol. 354:206–211 [DOI] [PubMed] [Google Scholar]
- 103. Cordes FS, et al. 2003. Helical structure of the needle of the type III secretion system of Shigella flexneri. J. Biol. Chem. 278:17103–17107 [DOI] [PubMed] [Google Scholar]
- 104. Cornelis G, Sluiters C, de Rouvroit CL, Michiels T. 1989. Homology between virF, the transcriptional activator of the Yersinia virulence regulon, and AraC, the Escherichia coli arabinose operon regulator. J. Bacteriol. 171:254–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Cornelis GR. 2010. The type III secretion injectisome, a complex nanomachine for intracellular ‘toxin’ delivery. Biol. Chem. 391:745–751 [DOI] [PubMed] [Google Scholar]
- 106. Cornelis GR, Agrain C, Sorg I. 2006. Length control of extended protein structures in bacteria and bacteriophages. Curr. Opin. Microbiol. 9:201–206 [DOI] [PubMed] [Google Scholar]
- 107. Cornelis GR, Wolf-Watz H. 1997. The Yersinia Yop virulon: a bacterial system for subverting eukaryotic cells. Mol. Microbiol. 23:861–867 [DOI] [PubMed] [Google Scholar]
- 108. Crago AM, Koronakis V. 1998. Salmonella InvG forms a ring-like multimer that requires the InvH lipoprotein for outer membrane localization. Mol. Microbiol. 30:47–56 [DOI] [PubMed] [Google Scholar]
- 109. Creasey EA, et al. 2003. CesT is a bivalent enteropathogenic Escherichia coli chaperone required for translocation of both Tir and Map. Mol. Microbiol. 47:209–221 [DOI] [PubMed] [Google Scholar]
- 110. Creasey EA, Delahay RM, Daniell SJ, Frankel G. 2003. Yeast two-hybrid system survey of interactions between LEE-encoded proteins of enteropathogenic Escherichia coli. Microbiology 149:2093–2106 [DOI] [PubMed] [Google Scholar]
- 111. Creasey EA, et al. 2003. CesAB is an enteropathogenic Escherichia coli chaperone for the type-III translocator proteins EspA and EspB. Microbiology 149:3639–3647 [DOI] [PubMed] [Google Scholar]
- 112. Crepin VF, et al. 2005. Structural and functional studies of the enteropathogenic Escherichia coli type III needle complex protein EscJ. Mol. Microbiol. 55:1658–1670 [DOI] [PubMed] [Google Scholar]
- 113. Dacheux D, Goure J, Chabert J, Usson Y, Attree I. 2001. Pore-forming activity of type III system-secreted proteins leads to oncosis of Pseudomonas aeruginosa-infected macrophages. Mol. Microbiol. 40:76–85 [DOI] [PubMed] [Google Scholar]
- 114. Daefler S, Russel M. 1998. The Salmonella typhimurium InvH protein is an outer membrane lipoprotein required for the proper localization of InvG. Mol. Microbiol. 28:1367–1380 [DOI] [PubMed] [Google Scholar]
- 115. Dai S, Zhou D. 2004. Secretion and function of Salmonella SPI-2 effector SseF require its chaperone, SscB. J. Bacteriol. 186:5078–5086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Daniell SJ, et al. 2003. 3D structure of EspA filaments from enteropathogenic Escherichia coli. Mol. Microbiol. 49:301–308 [DOI] [PubMed] [Google Scholar]
- 117. Daniell SJ, et al. 2001. The filamentous type III secretion translocon of enteropathogenic Escherichia coli. Cell. Microbiol. 3:865–871 [DOI] [PubMed] [Google Scholar]
- 118. Darwin KH, Miller VL. 1999. InvF is required for expression of genes encoding proteins secreted by the SPI1 type III secretion apparatus in Salmonella typhimurium. J. Bacteriol. 181:4949–4954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Darwin KH, Miller VL. 2000. The putative invasion protein chaperone SicA acts together with InvF to activate the expression of Salmonella typhimurium virulence genes. Mol. Microbiol. 35:949–960 [DOI] [PubMed] [Google Scholar]
- 120. Darwin KH, Miller VL. 2001. Type III secretion chaperone-dependent regulation: activation of virulence genes by SicA and InvF in Salmonella typhimurium. EMBO J. 20:1850–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Darwin KH, Robinson LS, Miller VL. 2001. SigE is a chaperone for the Salmonella enterica serovar Typhimurium invasion protein SigD. J. Bacteriol. 183:1452–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Dasgupta N, Lykken GL, Wolfgang MC, Yahr TL. 2004. A novel anti-anti-activator mechanism regulates expression of the Pseudomonas aeruginosa type III secretion system. Mol. Microbiol. 53:297–308 [DOI] [PubMed] [Google Scholar]
- 123. Davis AJ, Diaz DA, Mecsas J. 2010. A dominant-negative needle mutant blocks type III secretion of early but not late substrates in Yersinia. Mol. Microbiol. 76:236–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Davis AJ, Mecsas J. 2007. Mutations in the Yersinia pseudotuberculosis type III secretion system needle protein, YscF, that specifically abrogate effector translocation into host cells. J. Bacteriol. 189:83–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Day JB, Guller I, Plano GV. 2000. Yersinia pestis YscG protein is a Syc-like chaperone that directly binds yscE. Infect. Immun. 68:6466–6471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Day JB, Plano GV. 1998. A complex composed of SycN and YscB functions as a specific chaperone for YopN in Yersinia pestis. Mol. Microbiol. 30:777–788 [DOI] [PubMed] [Google Scholar]
- 127. Day JB, Plano GV. 2000. The Yersinia pestis YscY protein directly binds YscX, a secreted component of the type III secretion machinery. J. Bacteriol. 182:1834–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Dean GE, Macnab RM, Stader J, Matsumura P, Burks C. 1984. Gene sequence and predicted amino acid sequence of the motA protein, a membrane-associated protein required for flagellar rotation in Escherichia coli. J. Bacteriol. 159:991–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Deane JE, Abrusci P, Johnson S, Lea SM. 2010. Timing is everything: the regulation of type III secretion. Cell. Mol. Life Sci. 67:1065–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Deane JE, et al. 2008. Crystal structure of Spa40, the specificity switch for the Shigella flexneri type III secretion system. Mol. Microbiol. 69:267–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Deane JE, et al. 2006. Molecular model of a type III secretion system needle: implications for host-cell sensing. Proc. Natl. Acad. Sci. U. S. A. 103:12529–12533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Deane JE, Roversi P, King C, Johnson S, Lea SM. 2008. Structures of the Shigella flexneri type 3 secretion system protein MxiC reveal conformational variability amongst homologues. J. Mol. Biol. 377:985–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. DeBord KL, Lee VT, Schneewind O. 2001. Roles of LcrG and LcrV during type III targeting of effector Yops by Yersinia enterocolitica. J. Bacteriol. 183:4588–4598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Delahay RM, et al. 2002. Functional analysis of the enteropathogenic Escherichia coli type III secretion system chaperone CesT identifies domains that mediate substrate interactions. Mol. Microbiol. 43:61–73 [DOI] [PubMed] [Google Scholar]
- 135. Demchick P, Koch AL. 1996. The permeability of the wall fabric of Escherichia coli and Bacillus subtilis. J. Bacteriol. 178:768–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. De Mot R, Vanderleyden J. 1994. The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both Gram-positive and Gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol. Microbiol. 12:333–334 [DOI] [PubMed] [Google Scholar]
- 137. Deng W, et al. 2005. Regulation of type III secretion hierarchy of translocators and effectors in attaching and effacing bacterial pathogens. Infect. Immun. 73:2135–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Deng W, et al. 2004. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc. Natl. Acad. Sci. U. S. A. 101:3597–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. DePamphilis ML, Adler J. 1971. Fine structure and isolation of the hook-basal body complex of flagella from Escherichia coli and Bacillus subtilis. J. Bacteriol. 105:384–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Derewenda U, et al. 2004. The structure of Yersinia pestis V-antigen, an essential virulence factor and mediator of immunity against plague. Structure 12:301–306 [DOI] [PubMed] [Google Scholar]
- 141. Dewoody R, Merritt PM, Houppert AS, Marketon MM. 2011. YopK regulates the Yersinia pestis type III secretion system from within host cells. Mol. Microbiol. 79:1445–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Diepold A, et al. 2010. Deciphering the assembly of the Yersinia type III secretion injectisome. EMBO J. 29:1928–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Diepold A, Wiesand U, Cornelis GR. 2011. The assembly of the export apparatus (YscR,S,T,U,V) of the Yersinia type III secretion apparatus occurs independently of other structural components and involves the formation of an YscV oligomer. Mol. Microbiol. 82:502–514 [DOI] [PubMed] [Google Scholar]
- 144. Dittmann S, et al. 2007. The Yersinia enterocolitica type three secretion chaperone SycO is integrated into the Yop regulatory network and binds to the Yop secretion protein YscM1. BMC Microbiol. 7:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Draper O, Middleton R, Doucleff M, Zambryski PC. 2006. Topology of the VirB4 C terminus in the Agrobacterium tumefaciens VirB/D4 type IV secretion system. J. Biol. Chem. 281:37628–37635 [DOI] [PubMed] [Google Scholar]
- 146. Edqvist PJ, Aili M, Liu J, Francis MS. 2007. Minimal YopB and YopD translocator secretion by Yersinia is sufficient for Yop-effector delivery into target cells. Microbes Infect. 9:224–233 [DOI] [PubMed] [Google Scholar]
- 147. Edqvist PJ, et al. 2006. Tetratricopeptide repeats in the type III secretion chaperone, LcrH: their role in substrate binding and secretion. Mol. Microbiol. 59:31–44 [DOI] [PubMed] [Google Scholar]
- 148. Edqvist PJ, et al. 2003. YscP and YscU regulate substrate specificity of the Yersinia type III secretion system. J. Bacteriol. 185:2259–2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Ehrbar K, Friebel A, Miller SI, Hardt WD. 2003. Role of the Salmonella pathogenicity island 1 (SPI-1) protein InvB in type III secretion of SopE and SopE2, two Salmonella effector proteins encoded outside of SPI-1. J. Bacteriol. 185:6950–6967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Ehrbar K, Hapfelmeier S, Stecher B, Hardt WD. 2004. InvB is required for type III-dependent secretion of SopA in Salmonella enterica serovar Typhimurium. J. Bacteriol. 186:1215–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Ehrbar K, Winnen B, Hardt WD. 2006. The chaperone binding domain of SopE inhibits transport via flagellar and SPI-1 TTSS in the absence of InvB. Mol. Microbiol. 59:248–264 [DOI] [PubMed] [Google Scholar]
- 152. Ehsani S, Rodrigues CD, Enninga J. 2009. Turning on the spotlight—using light to monitor and characterize bacterial effector secretion and translocation. Curr. Opin. Microbiol. 12:24–30 [DOI] [PubMed] [Google Scholar]
- 153. Elliott SJ, et al. 1999. Identification of CesT, a chaperone for the type III secretion of Tir in enteropathogenic Escherichia coli. Mol. Microbiol. 33:1176–1189 [DOI] [PubMed] [Google Scholar]
- 154. Elliott SJ, et al. 2002. A gene from the locus of enterocyte effacement that is required for enteropathogenic Escherichia coli to increase tight-junction permeability encodes a chaperone for EspF. Infect. Immun. 70:2271–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Engelhardt S, et al. 2009. Separable roles of the Pseudomonas syringae pv. phaseolicola accessory protein HrpZ1 in ion-conducting pore formation. Plant J. 57:706–717 [DOI] [PubMed] [Google Scholar]
- 156. Enninga J, Mounier J, Sansonetti P, Tran Van Nhieu G. 2005. Secretion of type III effectors into host cells in real time. Nat. Methods 2:959–965 [DOI] [PubMed] [Google Scholar]
- 157. Enninga J, Rosenshine I. 2009. Imaging the assembly, structure and activity of type III secretion systems. Cell. Microbiol. 11:1462–1470 [DOI] [PubMed] [Google Scholar]
- 158. Epler CR, Dickenson NE, Olive AJ, Picking WL, Picking WD. 2009. Liposomes recruit IpaC to the Shigella flexneri type III secretion apparatus needle as a final step in secretion induction. Infect. Immun. 77:2754–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Erhardt M, et al. 2010. The role of the FliK molecular ruler in hook-length control in Salmonella enterica. Mol. Microbiol. 75:1272–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Erhardt M, Hughes KT. 2010. C-ring requirement in flagellar type III secretion is bypassed by FlhDC upregulation. Mol. Microbiol. 75:376–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Erhardt M, Namba K, Hughes KT. 2010. Bacterial nanomachines: the flagellum and type III injectisome. Cold Spring Harb. Perspect. Biol. 2:a000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Erhardt M, Singer HM, Wee DH, Keener JP, Hughes KT. 2011. An infrequent molecular ruler controls flagellar hook length in Salmonella enterica. EMBO J. 30:2948–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Erskine PT, et al. 2006. High resolution structure of BipD: an invasion protein associated with the type III secretion system of Burkholderia pseudomallei. J. Mol. Biol. 363:125–136 [DOI] [PubMed] [Google Scholar]
- 164. Espina M, et al. 2006. IpaD localizes to the tip of the type III secretion system needle of Shigella flexneri. Infect. Immun. 74:4391–4400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Evans LD, Hughes C. 2009. Selective binding of virulence type III export chaperones by FliJ escort orthologues InvI and YscO. FEMS Microbiol. Lett. 293:292–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Evans LD, Stafford GP, Ahmed S, Fraser GM, Hughes C. 2006. An escort mechanism for cycling of export chaperones during flagellum assembly. Proc. Natl. Acad. Sci. U. S. A. 103:17474–17479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Evdokimov AG, et al. 2003. Similar modes of polypeptide recognition by export chaperones in flagellar biosynthesis and type III secretion. Nat. Struct. Biol. 10:789–793 [DOI] [PubMed] [Google Scholar]
- 168. Evdokimov AG, Tropea JE, Routzahn KM, Copeland TD, Waugh DS. 2001. Structure of the N-terminal domain of Yersinia pestis YopH at 2.0 A resolution. Acta Crystallogr. D Biol. Crystallogr. 57:793–799 [DOI] [PubMed] [Google Scholar]
- 169. Evdokimov AG, Tropea JE, Routzahn KM, Waugh DS. 2002. Three-dimensional structure of the type III secretion chaperone SycE from Yersinia pestis. Acta Crystallogr. D Biol. Crystallogr. 58:398–406 [DOI] [PubMed] [Google Scholar]
- 170. Fadouloglou VE, et al. 2004. Structure of HrcQB-C, a conserved component of the bacterial type III secretion systems. Proc. Natl. Acad. Sci. U. S. A. 101:70–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Faherty CS, Maurelli AT. 2009. Spa15 of Shigella flexneri is secreted through the type III secretion system and prevents staurosporine-induced apoptosis. Infect. Immun. 77:5281–5290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Faudry E, Vernier G, Neumann E, Forge V, Attree I. 2006. Synergistic pore formation by type III toxin translocators of Pseudomonas aeruginosa. Biochemistry 45:8117–8123 [DOI] [PubMed] [Google Scholar]
- 173. Fauvart M, Michiels J. 2008. Rhizobial secreted proteins as determinants of host specificity in the rhizobium-legume symbiosis. FEMS Microbiol. Lett. 285:1–9 [DOI] [PubMed] [Google Scholar]
- 174. Fauvart M, et al. 2009. Rhizobium etli HrpW is a pectin-degrading enzyme and differs from phytopathogenic homologues in enzymically crucial tryptophan and glycine residues. Microbiology 155:3045–3054 [DOI] [PubMed] [Google Scholar]
- 175. Feldman MF, Cornelis GR. 2003. The multitalented type III chaperones: all you can do with 15 kDa. FEMS Microbiol. Lett. 219:151–158 [DOI] [PubMed] [Google Scholar]
- 176. Ferracci F, Schubot FD, Waugh DS, Plano GV. 2005. Selection and characterization of Yersinia pestis YopN mutants that constitutively block Yop secretion. Mol. Microbiol. 57:970–987 [DOI] [PubMed] [Google Scholar]
- 177. Ferris HU, et al. 2005. FlhB regulates ordered export of flagellar components via autocleavage mechanism. J. Biol. Chem. 280:41236–41242 [DOI] [PubMed] [Google Scholar]
- 178. Foultier B, et al. 2003. Identification of substrates and chaperone from the Yersinia enterocolitica 1B Ysa type III secretion system. Infect. Immun. 71:242–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Francis MR, Sosinsky GE, Thomas D, DeRosier DJ. 1994. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J. Mol. Biol. 235:1261–1270 [DOI] [PubMed] [Google Scholar]
- 180. Francis MS, Aili M, Wiklund ML, Wolf-Watz H. 2000. A study of the YopD-LcrH interaction from Yersinia pseudotuberculosis reveals a role for hydrophobic residues within the amphipathic domain of YopD. Mol. Microbiol. 38:85–102 [DOI] [PubMed] [Google Scholar]
- 181. Francis MS, Lloyd SA, Wolf-Watz H. 2001. The type III secretion chaperone LcrH co-operates with YopD to establish a negative, regulatory loop for control of Yop synthesis in Yersinia pseudotuberculosis. Mol. Microbiol. 42:1075–1093 [DOI] [PubMed] [Google Scholar]
- 182. Francis MS, Wolf-Watz H. 1998. YopD of Yersinia pseudotuberculosis is translocated into the cytosol of HeLa epithelial cells: evidence of a structural domain necessary for translocation. Mol. Microbiol. 29:799–813 [DOI] [PubMed] [Google Scholar]
- 183. Francis NR, Irikura VM, Yamaguchi S, DeRosier DJ, Macnab RM. 1992. Localization of the Salmonella typhimurium flagellar switch protein FliG to the cytoplasmic M-ring face of the basal body. Proc. Natl. Acad. Sci. U. S. A. 89:6304–6308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Fraser GM, Bennett JC, Hughes C. 1999. Substrate-specific binding of hook-associated proteins by FlgN and FliT, putative chaperones for flagellum assembly. Mol. Microbiol. 32:569–580 [DOI] [PubMed] [Google Scholar]
- 185. Fraser GM, Gonzalez-Pedrajo B, Tame JR, Macnab RM. 2003. Interactions of FliJ with the Salmonella type III flagellar export apparatus. J. Bacteriol. 185:5546–5554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Fraser GM, et al. 2003. Substrate specificity of type III flagellar protein export in Salmonella is controlled by subdomain interactions in FlhB. Mol. Microbiol. 48:1043–1057 [DOI] [PubMed] [Google Scholar]
- 187. French CT, et al. 2009. The Bordetella type III secretion system effector BteA contains a conserved N-terminal motif that guides bacterial virulence factors to lipid rafts. Cell. Microbiol. 11:1735–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Frithz-Lindsten E, et al. 1998. Functional conservation of the effector protein translocators PopB/YopB and PopD/YopD of Pseudomonas aeruginosa and Yersinia pseudotuberculosis. Mol. Microbiol. 29:1155–1165 [DOI] [PubMed] [Google Scholar]
- 189. Fu Y, Galan JE. 1998. Identification of a specific chaperone for SptP, a substrate of the centisome 63 type III secretion system of Salmonella typhimurium. J. Bacteriol. 180:3393–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Fu Y, Galan JE. 1999. A Salmonella protein antagonizes Rac-1 and Cdc42 to mediate host-cell recovery after bacterial invasion. Nature 401:293–297 [DOI] [PubMed] [Google Scholar]
- 191. Galkin VE, Schmied WH, Schraidt O, Marlovits TC, Egelman EH. 2010. The structure of the Salmonella typhimurium type III secretion system needle shows divergence from the flagellar system. J. Mol. Biol. 396:1392–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Garcia-Gomez E, Espinosa N, de la Mora J, Dreyfus G, Gonzalez-Pedrajo B. 2011. The muramidase EtgA from enteropathogenic Escherichia coli is required for efficient type III secretion. Microbiology 157:1145–1160 [DOI] [PubMed] [Google Scholar]
- 193. Gaudriault S, Paulin JP, Barny MA. 2002. The DspB/F protein of Erwinia amylovora is a type III secretion chaperone ensuring efficient secretion of the DspA/E essential pathogenicity factor. Mol. Plant Pathol. 3:313–320 [DOI] [PubMed] [Google Scholar]
- 194. Gaus K, et al. 2011. Destabilization of YopE by the ubiquitin-proteasome pathway fine-tunes Yop delivery into host cells and facilitates systemic spread of Yersinia enterocolitica in host lymphoid tissue. Infect. Immun. 79:1166–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Gauthier A, Finlay BB. 2003. Translocated intimin receptor and its chaperone interact with ATPase of the type III secretion apparatus of enteropathogenic Escherichia coli. J. Bacteriol. 185:6747–6755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Gauthier A, Puente JL, Finlay BB. 2003. Secretin of the enteropathogenic Escherichia coli type III secretion system requires components of the type III apparatus for assembly and localization. Infect. Immun. 71:3310–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Gebus C, Faudry E, Bohn YS, Elsen S, Attree I. 2008. Oligomerization of PcrV and LcrV, protective antigens of Pseudomonas aeruginosa and Yersinia pestis. J. Biol. Chem. 283:23940–23949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Gerlach RG, Hensel M. 2007. Protein secretion systems and adhesins: the molecular armory of Gram-negative pathogens. Int. J. Med. Microbiol. 297:401–415 [DOI] [PubMed] [Google Scholar]
- 199. Ghosh P. 2004. Process of protein transport by the type III secretion system. Microbiol. Mol. Biol. Rev. 68:771–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Gillen KL, Hughes KT. 1991. Negative regulatory loci coupling flagellin synthesis to flagellar assembly in Salmonella typhimurium. J. Bacteriol. 173:2301–2310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Gillen KL, Hughes KT. 1993. Transcription from two promoters and autoregulation contribute to the control of expression of the Salmonella typhimurium flagellar regulatory gene flgM. J. Bacteriol. 175:7006–7015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Girardin SE, et al. 2003. Nod1 detects a unique muropeptide from Gram-negative bacterial peptidoglycan. Science 300:1584–1587 [DOI] [PubMed] [Google Scholar]
- 203. Girardin SE, et al. 2003. Peptidoglycan molecular requirements allowing detection by Nod1 and Nod2. J. Biol. Chem. 278:41702–41708 [DOI] [PubMed] [Google Scholar]
- 204. Gong H, et al. 2010. Differential expression of Salmonella type III secretion system factors InvJ, PrgJ, SipC, SipD, SopA and SopB in cultures and in mice. Microbiology 156:116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Gonzalez-Pedrajo B, Fraser GM, Minamino T, Macnab RM. 2002. Molecular dissection of Salmonella FliH, a regulator of the ATPase FliI and the type III flagellar protein export pathway. Mol. Microbiol. 45:967–982 [DOI] [PubMed] [Google Scholar]
- 206. Gonzalez-Pedrajo B, Minamino T, Kihara M, Namba K. 2006. Interactions between C ring proteins and export apparatus components: a possible mechanism for facilitating type III protein export. Mol. Microbiol. 60:984–998 [DOI] [PubMed] [Google Scholar]
- 207. Goosney DL, DeVinney R, Finlay BB. 2001. Recruitment of cytoskeletal and signaling proteins to enteropathogenic and enterohemorrhagic Escherichia coli pedestals. Infect. Immun. 69:3315–3322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Goure J, et al. 2004. The V antigen of Pseudomonas aeruginosa is required for assembly of the functional PopB/PopD translocation pore in host cell membranes. Infect. Immun. 72:4741–4750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Guo M, et al. 2005. Pseudomonas syringae type III chaperones ShcO1, ShcS1, and ShcS2 facilitate translocation of their cognate effectors and can substitute for each other in the secretion of HopO1-1. J. Bacteriol. 187:4257–4269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Guttman DS, et al. 2002. A functional screen for the type III (Hrp) secretome of the plant pathogen Pseudomonas syringae. Science 295:1722–1726 [DOI] [PubMed] [Google Scholar]
- 211. Haapalainen M, et al. 2011. Functional mapping of harpin HrpZ of Pseudomonas syringae reveals the sites responsible for protein oligomerization, lipid interactions and plant defence induction. Mol. Plant Pathol. 12:151–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Hamad MA, Nilles ML. 2007. Roles of YopN, LcrG and LcrV in controlling Yops secretion by Yersinia pestis. Adv. Exp. Med. Biol. 603:225–234 [DOI] [PubMed] [Google Scholar]
- 213. Harrington AT, et al. 2003. Structural characterization of the N terminus of IpaC from Shigella flexneri. Infect. Immun. 71:1255–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Hashimoto W, et al. 2009. Crystal structure of the glycosidase family 73 peptidoglycan hydrolase FlgJ. Biochem. Biophys. Res. Commun. 381:16–21 [DOI] [PubMed] [Google Scholar]
- 215. Hayes CS, Aoki SK, Low DA. 2010. Bacterial contact-dependent delivery systems. Annu. Rev. Genet. 44:71–90 [DOI] [PubMed] [Google Scholar]
- 216. Hayward RD, et al. 2005. Cholesterol binding by the bacterial type III translocon is essential for virulence effector delivery into mammalian cells. Mol. Microbiol. 56:590–603 [DOI] [PubMed] [Google Scholar]
- 217. He SY, Nomura K, Whittam TS. 2004. Type III protein secretion mechanism in mammalian and plant pathogens. Biochim. Biophys. Acta 1694:181–206 [DOI] [PubMed] [Google Scholar]
- 218. Henderson IR, Navarro-Garcia F, Desvaux M, Fernandez RC, Ala'Aldeen D. 2004. Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol. Rev. 68:692–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Higashide W, Zhou D. 2006. The first 45 amino acids of SopA are necessary for InvB binding and SPI-1 secretion. J. Bacteriol. 188:2411–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Hirano T, Minamino T, Macnab RM. 2001. The role in flagellar rod assembly of the N-terminal domain of Salmonella FlgJ, a flagellum-specific muramidase. J. Mol. Biol. 312:359–369 [DOI] [PubMed] [Google Scholar]
- 221. Hirano T, Shibata S, Ohnishi K, Tani T, Aizawa S. 2005. N-terminal signal region of FliK is dispensable for length control of the flagellar hook. Mol. Microbiol. 56:346–360 [DOI] [PubMed] [Google Scholar]
- 222. Hirano T, Yamaguchi S, Oosawa K, Aizawa S. 1994. Roles of FliK and FlhB in determination of flagellar hook length in Salmonella typhimurium. J. Bacteriol. 176:5439–5449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Hodgkinson JL, et al. 2009. Three-dimensional reconstruction of the Shigella T3SS transmembrane regions reveals 12-fold symmetry and novel features throughout. Nat. Struct. Mol. Biol. 16:477–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Hoiczyk E, Blobel G. 2001. Polymerization of a single protein of the pathogen Yersinia enterocolitica into needles punctures eukaryotic cells. Proc. Natl. Acad. Sci. U. S. A. 98:4669–4674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Holmström A, et al. 1997. YopK of Yersinia pseudotuberculosis controls translocation of Yop effectors across the eukaryotic cell membrane. Mol. Microbiol. 24:73–91 [DOI] [PubMed] [Google Scholar]
- 226. Hölzer SU, Schlumberger MC, Jackel D, Hensel M. 2009. Effect of the O-antigen length of lipopolysaccharide on the functions of type III secretion systems in Salmonella enterica. Infect. Immun. 77:5458–5470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Homma M, Fujita H, Yamaguchi S, Iino T. 1984. Excretion of unassembled flagellin by Salmonella typhimurium mutants deficient in hook-associated proteins. J. Bacteriol. 159:1056–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Homma M, Iino T. 1985. Locations of hook-associated proteins in flagellar structures of Salmonella typhimurium. J. Bacteriol. 162:183–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Homma M, Komeda Y, Iino T, Macnab RM. 1987. The flaFIX gene product of Salmonella typhimurium is a flagellar basal body component with a signal peptide for export. J. Bacteriol. 169:1493–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Homma M, Kutsukake K, Hasebe M, Iino T, Macnab RM. 1990. FlgB, FlgC, FlgF and FlgG. A family of structurally related proteins in the flagellar basal body of Salmonella typhimurium. J. Mol. Biol. 211:465–477 [DOI] [PubMed] [Google Scholar]
- 231. Hong KH, Miller VL. 1998. Identification of a novel Salmonella invasion locus homologous to Shigella ipgDE. J. Bacteriol. 180:1793–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Horne SM, Pruss BM. 2006. Global gene regulation in Yersinia enterocolitica: effect of FliA on the expression levels of flagellar and plasmid-encoded virulence genes. Arch. Microbiol. 185:115–126 [DOI] [PubMed] [Google Scholar]
- 233. Hu W, Yuan J, Jin QL, Hart P, He SY. 2001. Immunogold labeling of Hrp pili of Pseudomonas syringae pv. tomato assembled in minimal medium and in planta. Mol. Plant Microbe Interact. 14:234–241 [DOI] [PubMed] [Google Scholar]
- 234. Hughes KT, Gillen KL, Semon MJ, Karlinsey JE. 1993. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science 262:1277–1280 [DOI] [PubMed] [Google Scholar]
- 235. Ibuki T, et al. 2011. Common architecture of the flagellar type III protein export apparatus and F- and V-type ATPases. Nat. Struct. Mol. Biol. 18:277–282 [DOI] [PubMed] [Google Scholar]
- 236. Ide T, et al. 2001. Characterization of translocation pores inserted into plasma membranes by type III-secreted Esp proteins of enteropathogenic Escherichia coli. Cell. Microbiol. 3:669–679 [DOI] [PubMed] [Google Scholar]
- 237. Ikeda T, Homma M, Iino T, Asakura S, Kamiya R. 1987. Localization and stoichiometry of hook-associated proteins within Salmonella typhimurium flagella. J. Bacteriol. 169:1168–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Imada K, Minamino T, Kinoshita M, Furukawa Y, Namba K. 2010. Structural insight into the regulatory mechanisms of interactions of the flagellar type III chaperone FliT with its binding partners. Proc. Natl. Acad. Sci. U. S. A. 107:8812–8817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Imada K, Minamino T, Tahara A, Namba K. 2007. Structural similarity between the flagellar type III ATPase FliI and F1-ATPase subunits. Proc. Natl. Acad. Sci. U. S. A. 104:485–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Iriarte M, Cornelis GR. 1998. YopT, a new Yersinia Yop effector protein, affects the cytoskeleton of host cells. Mol. Microbiol. 29:915–929 [DOI] [PubMed] [Google Scholar]
- 241. Iriarte M, Cornelis GR. 1999. Identification of SycN, YscX, and YscY, three new elements of the Yersinia yop virulon. J. Bacteriol. 181:675–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Irikura VM, Kihara M, Yamaguchi S, Sockett H, Macnab RM. 1993. Salmonella typhimurium fliG and fliN mutations causing defects in assembly, rotation, and switching of the flagellar motor. J. Bacteriol. 175:802–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Izore T, Job V, Dessen A. 2011. Biogenesis, regulation, and targeting of the type III secretion system. Structure 19:603–612 [DOI] [PubMed] [Google Scholar]
- 244. Jackson MW, Day JB, Plano GV. 1998. YscB of Yersinia pestis functions as a specific chaperone for YopN. J. Bacteriol. 180:4912–4921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Jackson MW, Plano GV. 2000. Interactions between type III secretion apparatus components from Yersinia pestis detected using the yeast two-hybrid system. FEMS Microbiol. Lett. 186:85–90 [DOI] [PubMed] [Google Scholar]
- 246. Jackson MW, Silva-Herzog E, Plano GV. 2004. The ATP-dependent ClpXP and Lon proteases regulate expression of the Yersinia pestis type III secretion system via regulated proteolysis of YmoA, a small histone-like protein. Mol. Microbiol. 54:1364–1378 [DOI] [PubMed] [Google Scholar]
- 247. Jani AJ, Cotter PA. 2010. Type VI secretion: not just for pathogenesis anymore. Cell Host Microbe 8:2–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248. Jin Q, He SY. 2001. Role of the Hrp pilus in type III protein secretion in Pseudomonas syringae. Science 294:2556–2558 [DOI] [PubMed] [Google Scholar]
- 249. Job V, Mattei PJ, Lemaire D, Attree I, Dessen A. 2010. Structural basis of chaperone recognition of type III secretion system minor translocator proteins. J. Biol. Chem. 285:23224–23232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Johnson DL, Stone CB, Mahony JB. 2008. Interactions between CdsD, CdsQ, and CdsL, three putative Chlamydophila pneumoniae type III secretion proteins. J. Bacteriol. 190:2972–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Johnson S, Blocker A. 2008. Characterization of soluble complexes of the Shigella flexneri type III secretion system ATPase. FEMS Microbiol. Lett. 286:274–278 [DOI] [PubMed] [Google Scholar]
- 252. Johnson S, et al. 2007. Self-chaperoning of the type III secretion system needle tip proteins IpaD and BipD. J. Biol. Chem. 282:4035–4044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Johnson TL, Abendroth J, Hol WGJ, Sandkvist M. 2006. Type II secretion: from structure to function. FEMS Microbiol. Lett. 255:175–186 [DOI] [PubMed] [Google Scholar]
- 254. Jones CJ, Homma M, Macnab RM. 1989. L-, P-, and M-ring proteins of the flagellar basal body of Salmonella typhimurium: gene sequences and deduced protein sequences. J. Bacteriol. 171:3890–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Jones CJ, Macnab RM, Okino H, Aizawa S. 1990. Stoichiometric analysis of the flagellar hook-(basal-body) complex of Salmonella typhimurium. J. Mol. Biol. 212:377–387 [DOI] [PubMed] [Google Scholar]
- 256. Jouihri N, et al. 2003. MxiK and MxiN interact with the Spa47 ATPase and are required for transit of the needle components MxiH and MxiI, but not of Ipa proteins, through the type III secretion apparatus of Shigella flexneri. Mol. Microbiol. 49:755–767 [DOI] [PubMed] [Google Scholar]
- 257. Journet L, Agrain C, Broz P, Cornelis GR. 2003. The needle length of bacterial injectisomes is determined by a molecular ruler. Science 302:1757–1760 [DOI] [PubMed] [Google Scholar]
- 258. Juhas M, Crook DW, Hood DW. 2008. Type IV secretion systems: tools of bacterial horizontal gene transfer and virulence. Cell. Microbiol. 10:2377–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259. Kabisch U, Landgraf A, Krause J, Bonas U, Boch J. 2005. Type III secretion chaperones ShcS1 and ShcO1 from Pseudomonas syringae pv. tomato DC3000 bind more than one effector. Microbiology 151:269–280 [DOI] [PubMed] [Google Scholar]
- 260. Kaniga K, Trollinger D, Galan JE. 1995. Identification of two targets of the type III protein secretion system encoded by the inv and spa loci of Salmonella typhimurium that have homology to the Shigella IpaD and IpaA proteins. J. Bacteriol. 177:7078–7085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261. Kaniga K, Tucker S, Trollinger D, Galan JE. 1995. Homologs of the Shigella IpaB and IpaC invasins are required for Salmonella typhimurium entry into cultured epithelial cells. J. Bacteriol. 177:3965–3971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262. Kapatral V, Minnich SA. 1995. Co-ordinate, temperature-sensitive regulation of the three Yersinia enterocolitica flagellin genes. Mol. Microbiol. 17:49–56 [DOI] [PubMed] [Google Scholar]
- 263. Kapatral V, Olson JW, Pepe JC, Miller VL, Minnich SA. 1996. Temperature-dependent regulation of Yersinia enterocolitica class III flagellar genes. Mol. Microbiol. 19:1061–1071 [DOI] [PubMed] [Google Scholar]
- 264. Karlinsey JE, Lonner J, Brown KL, Hughes KT. 2000. Translation/secretion coupling by type III secretion systems. Cell 102:487–497 [DOI] [PubMed] [Google Scholar]
- 265. Karlinsey JE, Pease AJ, Winkler ME, Bailey JL, Hughes KT. 1997. The flk gene of Salmonella typhimurium couples flagellar P- and L-ring assembly to flagellar morphogenesis. J. Bacteriol. 179:2389–2400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266. Karlinsey JE, et al. 2000. Completion of the hook-basal body complex of the Salmonella typhimurium flagellum is coupled to FlgM secretion and fliC transcription. Mol. Microbiol. 37:1220–1231 [DOI] [PubMed] [Google Scholar]
- 267. Kazetani K, Minamino T, Miyata T, Kato T, Namba K. 2009. ATP-induced FliI hexamerization facilitates bacterial flagellar protein export. Biochem. Biophys. Res. Commun. 388:323–327 [DOI] [PubMed] [Google Scholar]
- 268. Kenjale R, et al. 2005. The needle component of the type III secreton of Shigella regulates the activity of the secretion apparatus. J. Biol. Chem. 280:42929–42937 [DOI] [PubMed] [Google Scholar]
- 269. Khan IH, Reese TS, Khan S. 1992. The cytoplasmic component of the bacterial flagellar motor. Proc. Natl. Acad. Sci. U. S. A. 89:5956–5960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270. Kikuchi Y, Matsunami H, Yamane M, Imada K, Namba K. 2009. Crystallization and preliminary X-ray analysis of a C-terminal fragment of FlgJ, a putative flagellar rod cap protein from Salmonella. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 65:17–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271. Kim BH, Kim HG, Kim JS, Jang JI, Park YK. 2007. Analysis of functional domains present in the N-terminus of the SipB protein. Microbiology 153:2998–3008 [DOI] [PubMed] [Google Scholar]
- 272. Kim JG, et al. 2003. Characterization of the Xanthomonas axonopodis pv. glycines Hrp pathogenicity island. J. Bacteriol. 185:3155–3166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273. Kimbrough TG, Miller SI. 2000. Contribution of Salmonella typhimurium type III secretion components to needle complex formation. Proc. Natl. Acad. Sci. U. S. A. 97:11008–11013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274. Knight MJ, et al. 2006. Crystallization and preliminary X-ray diffraction analysis of BipD, a virulence factor from Burkholderia pseudomallei. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 62:761–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275. Knodler LA, Bertero M, Yip C, Strynadka NC, Steele-Mortimer O. 2006. Structure-based mutagenesis of SigE verifies the importance of hydrophobic and electrostatic residues in type III chaperone function. Mol. Microbiol. 62:928–940 [DOI] [PubMed] [Google Scholar]
- 276. Knodler LA, Winfree S, Drecktrah D, Ireland R, Steele-Mortimer O. 2009. Ubiquitination of the bacterial inositol phosphatase, SopB, regulates its biological activity at the plasma membrane. Cell. Microbiol. 11:1652–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277. Knutton S, et al. 1998. A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J. 17:2166–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278. Kofoed EM, Vance RE. 2011. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature 477:592–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279. Kojima S, Blair DF. 2001. Conformational change in the stator of the bacterial flagellar motor. Biochemistry 40:13041–13050 [DOI] [PubMed] [Google Scholar]
- 280. Konishi M, Kanbe M, McMurry JL, Aizawa S. 2009. Flagellar formation in C-ring-defective mutants by overproduction of FliI, the ATPase specific for flagellar type III secretion. J. Bacteriol. 191:6186–6191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281. Koo J, Burrows LL, Lynne Howell P. 2012. Decoding the roles of pilotins and accessory proteins in secretin escort services. FEMS Microbiol. Lett. 328:1–12 [DOI] [PubMed] [Google Scholar]
- 282. Koraimann G. 2003. Lytic transglycosylases in macromolecular transport systems of Gram-negative bacteria. Cell. Mol. Life Sci. 60:2371–2388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283. Korotkov KV, Gonen T, Hol WG. 2011. Secretins: dynamic channels for protein transport across membranes. Trends Biochem. Sci. 36:433–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284. Korotkov KV, Pardon E, Steyaert J, Hol WG. 2009. Crystal structure of the N-terminal domain of the secretin GspD from ETEC determined with the assistance of a nanobody. Structure 17:255–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285. Koroyasu S, Yamazato M, Hirano T, Aizawa SI. 1998. Kinetic analysis of the growth rate of the flagellar hook in Salmonella typhimurium by the population balance method. Biophys. J. 74:436–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286. Koster M, et al. 1997. The outer membrane component, YscC, of the Yop secretion machinery of Yersinia enterocolitica forms a ring-shaped multimeric complex. Mol. Microbiol. 26:789–797 [DOI] [PubMed] [Google Scholar]
- 287. Krall R, Zhang Y, Barbieri JT. 2004. Intracellular membrane localization of Pseudomonas ExoS and Yersinia YopE in mammalian cells. J. Biol. Chem. 279:2747–2753 [DOI] [PubMed] [Google Scholar]
- 288. Ku CP, Lio JC, Wang SH, Lin CN, Syu WJ. 2009. Identification of a third EspA-binding protein that forms part of the type III secretion system of enterohemorrhagic Escherichia coli. J. Biol. Chem. 284:1686–1693 [DOI] [PubMed] [Google Scholar]
- 289. Kubori T, Galan JE. 2002. Salmonella type III secretion-associated protein InvE controls translocation of effector proteins into host cells. J. Bacteriol. 184:4699–4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290. Kubori T, Galan JE. 2003. Temporal regulation of Salmonella virulence effector function by proteasome-dependent protein degradation. Cell 115:333–342 [DOI] [PubMed] [Google Scholar]
- 291. Kubori T, et al. 1998. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280:602–605 [DOI] [PubMed] [Google Scholar]
- 292. Kubori T, Shimamoto N, Yamaguchi S, Namba K, Aizawa S. 1992. Morphological pathway of flagellar assembly in Salmonella typhimurium. J. Mol. Biol. 226:433–446 [DOI] [PubMed] [Google Scholar]
- 293. Kubori T, Sukhan A, Aizawa SI, Galan JE. 2000. Molecular characterization and assembly of the needle complex of the Salmonella typhimurium type III protein secretion system. Proc. Natl. Acad. Sci. U. S. A. 97:10225–10230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294. Kutsukake K. 1994. Excretion of the anti-sigma factor through a flagellar substructure couples flagellar gene expression with flagellar assembly in Salmonella typhimurium. Mol. Gen. Genet. 243:605–612 [DOI] [PubMed] [Google Scholar]
- 295. Kutsukake K. 1997. Autogenous and global control of the flagellar master operon, flhD, in Salmonella typhimurium. Mol. Gen. Genet. 254:440–448 [DOI] [PubMed] [Google Scholar]
- 296. Kutsukake K. 1997. Hook-length control of the export-switching machinery involves a double-locked gate in Salmonella typhimurium flagellar morphogenesis. J. Bacteriol. 179:1268–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297. Kutsukake K, Ikebe T, Yamamoto S. 1999. Two novel regulatory genes, fliT and fliZ, in the flagellar regulon of Salmonella. Genes Genet. Syst. 74:287–292 [DOI] [PubMed] [Google Scholar]
- 298. Kutsukake K, Minamino T, Yokoseki T. 1994. Isolation and characterization of FliK-independent flagellation mutants from Salmonella typhimurium. J. Bacteriol. 176:7625–7629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299. Kutsukake K, Ohya Y, Iino T. 1990. Transcriptional analysis of the flagellar regulon of Salmonella typhimurium. J. Bacteriol. 172:741–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300. Kvitko BH, Ramos AR, Morello JE, Oh HS, Collmer A. 2007. Identification of harpins in Pseudomonas syringae pv. tomato DC3000, which are functionally similar to HrpK1 in promoting translocation of type III secretion system effectors. J. Bacteriol. 189:8059–8072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301. Lafont F, Tran Van Nhieu G, Hanada K, Sansonetti P, van der Goot FG. 2002. Initial steps of Shigella infection depend on the cholesterol/sphingolipid raft-mediated CD44-IpaB interaction. EMBO J. 21:4449–4457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302. Lambert de Rouvroit C, Sluiters C, Cornelis GR. 1992. Role of the transcriptional activator VirF and temperature in the expression of the pYV plasmid genes of Yersinia enterocolitica. Mol. Microbiol. 6:379–388 [PubMed] [Google Scholar]
- 303. Lan L, Deng X, Xiao Y, Zhou JM, Tang X. 2007. Mutation of Lon protease differentially affects the expression of Pseudomonas syringae type III secretion system genes in rich and minimal media and reduces pathogenicity. Mol. Plant Microbe Interact. 20:682–696 [DOI] [PubMed] [Google Scholar]
- 304. Lara-Tejero M, Kato J, Wagner S, Liu X, Galan JE. 2011. A sorting platform determines the order of protein secretion in bacterial type III systems. Science 331:1188–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305. Lario PI, et al. 2005. Structure and biochemical analysis of a secretin pilot protein. EMBO J. 24:1111–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306. Lavander M, et al. 2002. Proteolytic cleavage of the FlhB homologue YscU of Yersinia pseudotuberculosis is essential for bacterial survival but not for type III secretion. J. Bacteriol. 184:4500–4509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307. Lee J, Klessig DF, Nürnberger T. 2001. A harpin binding site in tobacco plasma membranes mediates activation of the pathogenesis-related gene hin1 independent of extracellular calcium but dependent on mitogen-activated protein kinase activity. Plant Cell 13:1079–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308. Lee J, et al. 2001. HrpZPsph from the plant pathogen Pseudomonas syringae pv. phaseolicola binds to lipid bilayers and forms an ion-conducting pore in vitro. Proc. Natl. Acad. Sci. U. S. A. 98:289–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309. Lee LK, Ginsburg MA, Crovace C, Donohoe M, Stock D. 2010. Structure of the torque ring of the flagellar motor and the molecular basis for rotational switching. Nature 466:996–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310. Lee PC, Stopford CM, Svenson AG, Rietsch A. 2010. Control of effector export by the Pseudomonas aeruginosa type III secretion proteins PcrG and PcrV. Mol. Microbiol. 75:924–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311. Lee SH, Galan JE. 2003. InvB is a type III secretion-associated chaperone for the Salmonella enterica effector protein SopE. J. Bacteriol. 185:7279–7284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312. Lee SH, Galan JE. 2004. Salmonella type III secretion-associated chaperones confer secretion-pathway specificity. Mol. Microbiol. 51:483–495 [DOI] [PubMed] [Google Scholar]
- 313. Lee VT, Mazmanian SK, Schneewind O. 2001. A program of Yersinia enterocolitica type III secretion reactions is activated by specific signals. J. Bacteriol. 183:4970–4978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314. Letzelter M, et al. 2006. The discovery of SycO highlights a new function for type III secretion effector chaperones. EMBO J. 25:3223–3233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315. Li CM, et al. 2002. The Hrp pilus of Pseudomonas syringae elongates from its tip and acts as a conduit for translocation of the effector protein HrpZ. EMBO J. 21:1909–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316. Li H, Sourjik V. 2011. Assembly and stability of flagellar motor in Escherichia coli. Mol. Microbiol. 80:886–899 [DOI] [PubMed] [Google Scholar]
- 317. Li JG, et al. 2010. PopW of Ralstonia solanacearum, a new two-domain harpin targeting the plant cell wall. Mol. Plant Pathol. 11:371–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318. Li M, et al. 2006. Identification and characterization of NleI, a new non-LEE-encoded effector of enteropathogenic Escherichia coli (EPEC). Microbes Infect. 8:2890–2898 [DOI] [PubMed] [Google Scholar]
- 319. Li YR, et al. 2011. Hpa2 required by HrpF to translocate Xanthomonas oryzae transcriptional activator-like effectors into rice for pathogenicity. Appl. Environ. Microbiol. 77:3809–3818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320. Lilic M, Quezada CM, Stebbins CE. 2010. A conserved domain in type III secretion links the cytoplasmic domain of InvA to elements of the basal body. Acta Crystallogr. D Biol. Crystallogr. 66:709–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321. Lilic M, Vujanac M, Stebbins CE. 2006. A common structural motif in the binding of virulence factors to bacterial secretion chaperones. Mol. Cell 3:653–664 [DOI] [PubMed] [Google Scholar]
- 322. Liu X, Matsumura P. 1994. The FlhD/FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J. Bacteriol. 176:7345–7351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323. Liu X, Matsumura P. 1995. An alternative sigma factor controls transcription of flagellar class-III operons in Escherichia coli: gene sequence, overproduction, purification and characterization. Gene 164:81–84 [DOI] [PubMed] [Google Scholar]
- 324. Liu X, Matsumura P. 1996. Differential regulation of multiple overlapping promoters in flagellar class II operons in Escherichia coli. Mol. Microbiol. 21:613–620 [DOI] [PubMed] [Google Scholar]
- 325. Lloyd SA, Blair DF. 1997. Charged residues of the rotor protein FliG essential for torque generation in the flagellar motor of Escherichia coli. J. Mol. Biol. 266:733–744 [DOI] [PubMed] [Google Scholar]
- 326. Lloyd SA, Tang H, Wang X, Billings S, Blair DF. 1996. Torque generation in the flagellar motor of Escherichia coli: evidence of a direct role for FliG but not for FliM or FliN. J. Bacteriol. 178:223–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327. Locher M, et al. 2005. Crystal structure of the Yersinia enterocolitica type III secretion chaperone SycT. J. Biol. Chem. 280:31149–31155 [DOI] [PubMed] [Google Scholar]
- 328. Lokareddy RK, Lunelli M, Eilers B, Wolter V, Kolbe M. 2010. Combination of two separate binding domains defines stoichiometry between type III secretion system chaperone IpgC and translocator protein IpaB. J. Biol. Chem. 285:39965–39975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329. Lorenz C, Büttner D. 2009. Functional characterization of the type III secretion ATPase HrcN from the plant pathogen Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 191:1414–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330. Lorenz C, Büttner D. 2011. Secretion of early and late substrates of the type III secretion system from Xanthomonas is controlled by HpaC and the C-terminal domain of HrcU. Mol. Microbiol. 79:447–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331. Lorenz C, et al. 2008. HpaA from Xanthomonas is a regulator of type III secretion. Mol. Microbiol. 69:344–360 [DOI] [PubMed] [Google Scholar]
- 332. Lorenz C, et al. 2008. HpaC controls substrate specificity of the Xanthomonas type III secretion system. PLoS Pathog. 4:e1000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333. Losada LC, Hutcheson SW. 2005. Type III secretion chaperones of Pseudomonas syringae protect effectors from Lon-associated degradation. Mol. Microbiol. 55:941–953 [DOI] [PubMed] [Google Scholar]
- 334. Lountos GT, Austin BP, Nallamsetty S, Waugh DS. 2009. Atomic resolution structure of the cytoplasmic domain of Yersinia pestis YscU, a regulatory switch involved in type III secretion. Protein Sci. 18:467–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335. Lountos GT, Tropea JE, Waugh DS. 2012. Structure of the cytoplasmic domain of Yersinia pestis YscD, an essential component of the type III secretion system. Acta Crystallogr. D Biol. Crystallogr. 68:201–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336. Löwer M, Schneider G. 2009. Prediction of type III secretion signals in genomes of Gram-negative bacteria. PLoS One 4:e5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337. Lunelli M, Hurwitz R, Lambers J, Kolbe M. 2011. Crystal structure of PrgI-SipD: insight into a secretion competent state of the type three secretion system needle tip and its interaction with host ligands. PLoS Pathog. 7:e1002163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338. Lunelli M, Lokareddy RK, Zychlinsky A, Kolbe M. 2009. IpaB-IpgC interaction defines binding motif for type III secretion translocator. Proc. Natl. Acad. Sci. U. S. A. 106:9661–9666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339. Luo W, Donnenberg MS. 2011. Interactions and predicted host membrane topology of the enteropathogenic Escherichia coli translocator protein EspB. J. Bacteriol. 193:2972–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340. Luo Y, et al. 2001. Structural and biochemical characterization of the type III secretion chaperones CesT and SigE. Nat. Struct. Biol. 8:1031–1036 [DOI] [PubMed] [Google Scholar]
- 341. Magdalena J, et al. 2002. Spa32 regulates a switch in substrate specificity of the type III secreton of Shigella flexneri from needle components to Ipa proteins. J. Bacteriol. 184:3433–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342. Makishima S, Komoriya K, Yamaguchi S, Aizawa SI. 2001. Length of the flagellar hook and the capacity of the type III export apparatus. Science 291:2411–2413 [DOI] [PubMed] [Google Scholar]
- 343. Manson MD. 2010. Dynamic motors for bacterial flagella. Proc. Natl. Acad. Sci. U. S. A. 107:11151–11152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344. Marenne MN, Journet L, Mota LJ, Cornelis GR. 2003. Genetic analysis of the formation of the Ysc-Yop translocation pore in macrophages by Yersinia enterocolitica: role of LcrV, YscF and YopN. Microb. Pathog. 35:243–258 [DOI] [PubMed] [Google Scholar]
- 345. Marketon MM, DePaolo RW, DeBord KL, Jabri B, Schneewind O. 2005. Plague bacteria target immune cells during infection. Science 309:1739–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346. Markham AP, Birket SE, Picking WD, Picking WL, Middaugh CR. 2008. pH sensitivity of type III secretion system tip proteins. Proteins 71:1830–1842 [DOI] [PubMed] [Google Scholar]
- 347. Marlovits TC, et al. 2006. Assembly of the inner rod determines needle length in the type III secretion injectisome. Nature 441:637–640 [DOI] [PubMed] [Google Scholar]
- 348. Marlovits TC, et al. 2004. Structural insights into the assembly of the type III secretion needle complex. Science 306:1040–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349. Marlovits TC, Stebbins CE. 2010. Type III secretion systems shape up as they ship out. Curr. Opin. Microbiol. 13:47–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350. Martinez-Argudo I, Blocker AJ. 2010. The Shigella T3SS needle transmits a signal for MxiC release, which controls secretion of effectors. Mol. Microbiol. 78:1365–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351. Matson JS, Nilles ML. 2001. LcrG-LcrV interaction is required for control of Yops secretion in Yersinia pestis. J. Bacteriol. 183:5082–5091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352. Matsumoto H, Young GM. 2009. Essential role of the SycP chaperone in type III secretion of the YspP effector. J. Bacteriol. 191:1703–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353. Mattei PJ, et al. 2010. Membrane targeting and pore formation by the type III secretion system translocon. FEBS J. 278:414–426 [DOI] [PubMed] [Google Scholar]
- 354. Mavris M, et al. 2002. Regulation of transcription by the activity of the Shigella flexneri type III secretion apparatus. Mol. Microbiol. 43:1543–1553 [DOI] [PubMed] [Google Scholar]
- 355. McCaw ML, Lykken GL, Singh PK, Yahr TL. 2002. ExsD is a negative regulator of the Pseudomonas aeruginosa type III secretion regulon. Mol. Microbiol. 46:1123–1133 [DOI] [PubMed] [Google Scholar]
- 356. McDowell MA, et al. 2011. Structural and functional studies on the N-terminal domain of the Shigella type III secretion protein MxiG. J. Biol. Chem. 286:30606–30614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357. McGuffie EM, Fraylick JE, Hazen-Martin DJ, Vincent TS, Olson JC. 1999. Differential sensitivity of human epithelial cells to Pseudomonas aeruginosa exoenzyme S. Infect. Immun. 67:3494–3503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358. McMurry JL, Murphy JW, Gonzalez-Pedrajo B. 2006. The FliN-FliH interaction mediates localization of flagellar export ATPase FliI to the C ring complex. Biochemistry 45:11790–11798 [DOI] [PubMed] [Google Scholar]
- 359. McMurry JL, Van Arnam JS, Kihara M, Macnab RM. 2004. Analysis of the cytoplasmic domains of Salmonella FlhA and interactions with components of the flagellar export machinery. J. Bacteriol. 186:7586–7592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360. Meister M, Lowe G, Berg HC. 1987. The proton flux through the bacterial flagellar motor. Cell 49:643–650 [DOI] [PubMed] [Google Scholar]
- 361. Mejia E, Bliska JB, Viboud GI. 2008. Yersinia controls type III effector delivery into host cells by modulating Rho activity. PLoS Pathog. 4:e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362. Menard R, Sansonetti P, Parsot C. 1994. The secretion of the Shigella flexneri Ipa invasins is activated by epithelial cells and controlled by IpaB and IpaD. EMBO J. 1354:5293–5302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363. Menard R, Sansonetti P, Parsot C, Vasselon T. 1994. Extracellular association and cytoplasmic partitioning of the IpaB and IpaC invasins of S. flexneri. Cell 7984:515–525 [DOI] [PubMed] [Google Scholar]
- 364. Meyer D, et al. 2006. PopF1 and PopF2, two proteins secreted by the type III protein secretion system of Ralstonia solanacearum, are translocators belonging to the HrpF/NopX family. J. Bacteriol. 188:4903–4917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365. Miao EA, et al. 2006. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat. Immunol. 7:569–575 [DOI] [PubMed] [Google Scholar]
- 366. Miao EA, et al. 2010. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc. Natl. Acad. Sci. U. S. A. 107:3076–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367. Michiels T, Cornelis GR. 1991. Secretion of hybrid proteins by the Yersinia Yop export system. J. Bacteriol. 173:1677–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368. Michiels T, Wattiau P, Brasseur R, Ruysschaert JM, Cornelis G. 1990. Secretion of Yop proteins by yersiniae. Infect. Immun. 58:2840–2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369. Miki T, Shibagaki Y, Danbara H, Okada N. 2009. Functional characterization of SsaE, a novel chaperone protein of the type III secretion system encoded by Salmonella pathogenicity island 2. J. Bacteriol. 191:6843–6854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370. Miller VL. 2002. Connections between transcriptional regulation and type III secretion? Curr. Opin. Microbiol. 5:211–215 [DOI] [PubMed] [Google Scholar]
- 371. Mills E, Baruch K, Charpentier X, Kobi S, Rosenshine I. 2008. Real-time analysis of effector translocation by the type III secretion system of enteropathogenic Escherichia coli. Cell Host Microbe 3:104–113 [DOI] [PubMed] [Google Scholar]
- 372. Minamino T, Chu R, Yamaguchi S, Macnab RM. 2000. Role of FliJ in flagellar protein export in Salmonella. J. Bacteriol. 182:4207–4215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373. Minamino T, Gonzalez-Pedrajo B, Kihara M, Namba K, Macnab RM. 2003. The ATPase FliI can interact with the type III flagellar protein export apparatus in the absence of its regulator, FliH. J. Bacteriol. 185:3983–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374. Minamino T, Gonzalez-Pedrajo B, Yamaguchi K, Aizawa SI, Macnab RM. 1999. FliK, the protein responsible for flagellar hook length control in Salmonella, is exported during hook assembly. Mol. Microbiol. 34:295–304 [DOI] [PubMed] [Google Scholar]
- 375. Minamino T, et al. 2011. Structural insight into the rotational switching mechanism of the bacterial flagellar motor. PLoS Biol. 9:e1000616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376. Minamino T, Imada K, Namba K. 2008. Mechanisms of type III protein export for bacterial flagellar assembly. Mol. Biosyst. 4:1105–1115 [DOI] [PubMed] [Google Scholar]
- 377. Minamino T, Imada K, Namba K. 2008. Molecular motors of the bacterial flagella. Curr. Opin. Struct. Biol. 18:693–701 [DOI] [PubMed] [Google Scholar]
- 378. Minamino T, et al. 2006. Oligomerization of the bacterial flagellar ATPase FliI is controlled by its extreme N-terminal region. J. Mol. Biol. 360:510–519 [DOI] [PubMed] [Google Scholar]
- 379. Minamino T, et al. 2012. Interaction of a bacterial flagellar chaperone FlgN with FlhA is required for efficient export of its cognate substrates. Mol. Microbiol. 83:775–788 [DOI] [PubMed] [Google Scholar]
- 380. Minamino T, Macnab RM. 1999. Components of the Salmonella flagellar export apparatus and classification of export substrates. J. Bacteriol. 181:1388–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381. Minamino T, Macnab RM. 2000. Domain structure of Salmonella FlhB, a flagellar export component responsible for substrate specificity switching. J. Bacteriol. 182:4906–4914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382. Minamino T, MacNab RM. 2000. FliH, a soluble component of the type III flagellar export apparatus of Salmonella, forms a complex with FliI and inhibits its ATPase activity. Mol. Microbiol. 37:1494–1503 [DOI] [PubMed] [Google Scholar]
- 383. Minamino T, MacNab RM. 2000. Interactions among components of the Salmonella flagellar export apparatus and its substrates. Mol. Microbiol. 35:1052–1064 [DOI] [PubMed] [Google Scholar]
- 384. Minamino T, Morimoto YV, Hara N, Namba K. 2011. An energy transduction mechanism used in bacterial flagellar type III protein export. Nat. Commun. 2:475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385. Minamino T, Moriya N, Hirano T, Hughes KT, Namba K. 2009. Interaction of FliK with the bacterial flagellar hook is required for efficient export specificity switching. Mol. Microbiol. 74:239–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386. Minamino T, et al. 2004. Domain organization and function of Salmonella FliK, a flagellar hook-length control protein. J. Mol. Biol. 341:491–502 [DOI] [PubMed] [Google Scholar]
- 387. Montagner C, Arquint C, Cornelis GR. 2011. Translocators YopB and YopD from Yersinia form a multimeric integral membrane complex in eukaryotic cell membranes. J. Bacteriol. 193:6923–6928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388. Moore SA, Jia Y. 2010. Structure of the cytoplasmic domain of the flagellar secretion apparatus component FlhA from Helicobacter pylori. J. Biol. Chem. 285:21060–21069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389. Moraes TF, Spreter T, Strynadka NC. 2008. Piecing together the type III injectisome of bacterial pathogens. Curr. Opin. Struct. Biol. 18:258–266 [DOI] [PubMed] [Google Scholar]
- 390. Morita-Ishihara T, et al. 2006. Shigella Spa33 is an essential C-ring component of type III secretion machinery. J. Biol. Chem. 281:599–607 [DOI] [PubMed] [Google Scholar]
- 391. Moriya N, Minamino T, Hughes KT, Macnab RM, Namba K. 2006. The type III flagellar export specificity switch is dependent on FliK ruler and a molecular clock. J. Mol. Biol. 359:466–477 [DOI] [PubMed] [Google Scholar]
- 392. Morris DP, et al. 2010. Kinetic characterization of Salmonella FliK-FlhB interactions demonstrates complexity of the type III secretion substrate-specificity switch. Biochemistry 49:6386–6393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393. Mota LJ. 2006. Type III secretion gets an LcrV tip. Trends Microbiol. 14:197–200 [DOI] [PubMed] [Google Scholar]
- 394. Mota LJ, Journet L, Sorg I, Agrain C, Cornelis GR. 2005. Bacterial injectisomes: needle length does matter. Science 307:1278. [DOI] [PubMed] [Google Scholar]
- 395. Mueller CA, Broz P, Cornelis GR. 2008. The type III secretion system tip complex and translocon. Mol. Microbiol. 68:1085–1095 [DOI] [PubMed] [Google Scholar]
- 396. Mueller CA, et al. 2005. The V-antigen of Yersinia forms a distinct structure at the tip of injectisome needles. Science 310:674–676 [DOI] [PubMed] [Google Scholar]
- 397. Müller SA, et al. 2006. Double hexameric ring assembly of the type III protein translocase ATPase HrcN. Mol. Microbiol. 61:119–125 [DOI] [PubMed] [Google Scholar]
- 398. Munera D, Crepin VF, Marches O, Frankel G. 2010. N-terminal type III secretion signal of enteropathogenic Escherichia coli translocator proteins. J. Bacteriol. 192:3534–3539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399. Muramoto K, Makishima S, Aizawa SI, Macnab RM. 1998. Effect of cellular level of FliK on flagellar hook and filament assembly in Salmonella typhimurium. J. Mol. Biol. 277:871–882 [DOI] [PubMed] [Google Scholar]
- 400. Nakano N, Kubori T, Kinoshita M, Imada K, Nagai H. 2010. Crystal structure of Legionella DotD: insights into the relationship between type IVB and type II/III secretion systems. PLoS Pathog. 6:e1001129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401. Nambu T, Minamino T, Macnab RM, Kutsukake K. 1999. Peptidoglycan-hydrolyzing activity of the FlgJ protein, essential for flagellar rod formation in Salmonella typhimurium. J. Bacteriol. 181:1555–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402. Neves BC, et al. 2003. CesD2 of enteropathogenic Escherichia coli is a second chaperone for the type III secretion translocator protein EspD. Infect. Immun. 71:2130–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403. Neyt C, Cornelis GR. 1999. Insertion of a Yop translocation pore into the macrophage plasma membrane by Yersinia enterocolitica: requirement for translocators YopB and YopD, but not LcrG. Mol. Microbiol. 33:971–981 [DOI] [PubMed] [Google Scholar]
- 404. Neyt C, Cornelis GR. 1999. Role of SycD, the chaperone of the Yersinia Yop translocators YopB and YopD. Mol. Microbiol. 31:143–156 [DOI] [PubMed] [Google Scholar]
- 405. Niebuhr K, et al. 2000. IpgD, a protein secreted by the type III secretion machinery of Shigella flexneri, is chaperoned by IpgE and implicated in entry focus formation. Mol. Microbiol. 38:8–19 [DOI] [PubMed] [Google Scholar]
- 406. Nilles ML, Fields KA, Straley SC. 1998. The V antigen of Yersinia pestis regulates Yop vectorial targeting as well as Yop secretion through effects on YopB and LcrG. J. Bacteriol. 180:3410–3420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407. Nilles ML, Williams AW, Skrzypek E, Straley SC. 1997. Yersinia pestis LcrV forms a stable complex with LcrG and may have a secretion-related regulatory role in the low-Ca2+ response. J. Bacteriol. 179:1307–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408. O'Connell CB, et al. 2004. SepL, a protein required for enteropathogenic Escherichia coli type III translocation, interacts with secretion component SepD. Mol. Microbiol. 52:1613–1625 [DOI] [PubMed] [Google Scholar]
- 409. Ogawa M, Suzuki T, Tatsuno I, Abe H, Sasakawa C. 2003. IcsB, secreted via the type III secretion system, is chaperoned by IpgA and required at the post-invasion stage of Shigella pathogenicity. Mol. Microbiol. 48:913–931 [DOI] [PubMed] [Google Scholar]
- 410. Ogino T, et al. 2006. Assembly of the type III secretion apparatus of enteropathogenic Escherichia coli. J. Bacteriol. 188:2801–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411. Oh CS, Carpenter SC, Hayes ML, Beer SV. 2010. Secretion and translocation signals and DspB/F-binding domains in the type III effector DspA/E of Erwinia amylovora. Microbiology 156:1211–1220 [DOI] [PubMed] [Google Scholar]
- 412. Oh HS, Kvitko BH, Morello JE, Collmer A. 2007. Pseudomonas syringae lytic transglycosylases coregulated with the type III secretion system contribute to the translocation of effector proteins into plant cells. J. Bacteriol. 189:8277–8289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413. Oh HS, Park DH, Collmer A. 2010. Components of the Pseudomonas syringae type III secretion system can suppress and may elicit plant innate immunity. Mol. Plant Microbe Interact. 23:727–739 [DOI] [PubMed] [Google Scholar]
- 414. Ohnishi K, Kutsukake K, Suzuki H, Iino T. 1990. Gene fliA encodes an alternative sigma factor specific for flagellar operons in Salmonella typhimurium. Mol. Gen. Genet. 221:139–147 [DOI] [PubMed] [Google Scholar]
- 415. Ohnishi K, Kutsukake K, Suzuki H, Lino T. 1992. A novel transcriptional regulation mechanism in the flagellar regulon of Salmonella typhimurium: an antisigma factor inhibits the activity of the flagellum-specific sigma factor, sigma F. Mol. Microbiol. 6:3149–3157 [DOI] [PubMed] [Google Scholar]
- 416. Ohnishi K, Ohto Y, Aizawa S, Macnab RM, Iino T. 1994. FlgD is a scaffolding protein needed for flagellar hook assembly in Salmonella typhimurium. J. Bacteriol. 176:2272–2281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417. Okon M, et al. 2008. Structural characterization of the type-III pilot-secretin complex from Shigella flexneri. Structure 16:1544–1554 [DOI] [PubMed] [Google Scholar]
- 418. Olive AJ, et al. 2007. Bile salts stimulate recruitment of IpaB to the Shigella flexneri surface, where it colocalizes with IpaD at the tip of the type III secretion needle. Infect. Immun. 75:2626–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419. Oosawa K, Ueno T, Aizawa S. 1994. Overproduction of the bacterial flagellar switch proteins and their interactions with the MS ring complex in vitro. J. Bacteriol. 176:3683–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420. Ortiz-Martin I, Thwaites R, Mansfield JW, Beuzon CR. 2010. Negative regulation of the Hrp type III secretion system in Pseudomonas syringae pv. phaseolicola. Mol. Plant Microbe Interact. 23:682–701 [DOI] [PubMed] [Google Scholar]
- 421. Osborne SE, Coombes BK. 2011. Expression and secretion hierarchy in the nonflagellar type III secretion system. Future Microbiol. 6:193–202 [DOI] [PubMed] [Google Scholar]
- 422. Page AL, Fromont-Racine M, Sansonetti P, Legrain P, Parsot C. 2001. Characterization of the interaction partners of secreted proteins and chaperones of Shigella flexneri. Mol. Microbiol. 42:1133–1145 [DOI] [PubMed] [Google Scholar]
- 423. Page AL, Sansonetti P, Parsot C. 2002. Spa15 of Shigella flexneri, a third type of chaperone in the type III secretion pathway. Mol. Microbiol. 43:1533–1542 [DOI] [PubMed] [Google Scholar]
- 424. Pal M, Erskine PT, Gill RS, Wood SP, Cooper JB. 2010. Near-atomic resolution analysis of BipD, a component of the type III secretion system of Burkholderia pseudomallei. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 66:990–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425. Pallen MJ, Bailey CM, Beatson SA. 2006. Evolutionary links between FliH/YscL-like proteins from bacterial type III secretion systems and second-stalk components of the FoF1 and vacuolar ATPases. Protein Sci. 15:935–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426. Pallen MJ, Beatson SA, Bailey CM. 2005. Bioinformatics analysis of the locus for enterocyte effacement provides novel insights into type-III secretion. BMC Microbiol. 5:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427. Pallen MJ, Francis MS, Futterer K. 2003. Tetratricopeptide-like repeats in type-III-secretion chaperones and regulators. FEMS Microbiol. Lett. 223:53–60 [DOI] [PubMed] [Google Scholar]
- 428. Pallen MJ, Penn CW, Chaudhuri RR. 2005. Bacterial flagellar diversity in the post-genomic era. Trends Microbiol. 13:143–149 [DOI] [PubMed] [Google Scholar]
- 429. Park SY, Lowder B, Bilwes AM, Blair DF, Crane BR. 2006. Structure of FliM provides insight into assembly of the switch complex in the bacterial flagella motor. Proc. Natl. Acad. Sci. U. S. A. 103:11886–11891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430. Parsot C, et al. 2005. A secreted anti-activator, OspD1, and its chaperone, Spa15, are involved in the control of transcription by the type III secretion apparatus activity in Shigella flexneri. Mol. Microbiol. 56:1627–1635 [DOI] [PubMed] [Google Scholar]
- 431. Parsot C, Hamiaux C, Page AL. 2003. The various and varying roles of specific chaperones in type III secretion systems. Curr. Opin. Microbiol. 6:7–14 [DOI] [PubMed] [Google Scholar]
- 432. Parsot C, Ménard R, Gounon P, Sansonetti PJ. 1995. Enhanced secretion through the Shigella flexneri Mxi-Spa translocon leads to assembly of extracellular proteins into macromolecular structures. Mol. Microbiol. 16:291–300 [DOI] [PubMed] [Google Scholar]
- 433. Patterson-Delafield J, Martinez RJ, Stocker BAD, Yamaguchi S. 1973. A new fla gene in Salmonella typhimurium—flaR—and its mutant phenotype—superhooks. Arch. Microbiol. 90:107–120 [DOI] [PubMed] [Google Scholar]
- 434. Paul K, Brunstetter D, Titen S, Blair DF. 2011. A molecular mechanism of direction switching in the flagellar motor of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 108:17171–17176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435. Paul K, Erhardt M, Hirano T, Blair D, Hughes KT. 2008. Energy source of flagellar type III secretion. Nature 451:489–492 [DOI] [PubMed] [Google Scholar]
- 436. Paul K, Gonzalez-Bonet G, Bilwes AM, Crane BR, Blair D. 2011. Architecture of the flagellar rotor. EMBO J. 30:2962–2971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437. Petnicki-Ocwieja T, et al. 2002. Genomewide identification of proteins secreted by the Hrp type III protein secretion system of Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. U. S. A. 99:7652–7657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438. Petnicki-Ocwieja T, van Dijk K, Alfano JR. 2005. The hrpK operon of Pseudomonas syringae pv. tomato DC3000 encodes two proteins secreted by the type III (Hrp) protein secretion system: HopB1 and HrpK, a putative type III translocator. J. Bacteriol. 187:649–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439. Pettersson J, et al. 1996. Modulation of virulence factor expression by pathogen target cell contact. Science 273:1231–1233 [DOI] [PubMed] [Google Scholar]
- 440. Phan J, Austin BP, Waugh DS. 2005. Crystal structure of the Yersinia type III secretion protein YscE. Protein Sci. 14:2759–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441. Phan J, Tropea JE, Waugh DS. 2004. Structure of the Yersinia pestis type III secretion chaperone SycH in complex with a stable fragment of YscM2. Acta Crystallogr. D Biol. Crystallogr. 60:1591–1599 [DOI] [PubMed] [Google Scholar]
- 442. Picking WL, et al. 2005. IpaD of Shigella flexneri is independently required for regulation of Ipa protein secretion and efficient insertion of IpaB and IpaC into host membranes. Infect. Immun. 73:1432–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443. Pilonieta MC, Munson GP. 2008. The chaperone IpgC copurifies with the virulence regulator MxiE. J. Bacteriol. 190:2249–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444. Poyraz O, et al. 2010. Protein refolding is required for assembly of the type three secretion needle. Nat. Struct. Mol. Biol. 17:788–792 [DOI] [PubMed] [Google Scholar]
- 445. Pozidis C, et al. 2003. Type III protein translocase: HrcN is a peripheral ATPase that is activated by oligomerization. J. Biol. Chem. 278:25816–25824 [DOI] [PubMed] [Google Scholar]
- 446. Preston G, Deng WL, Huang HC, Collmer A. 1998. Negative regulation of hrp genes in Pseudomonas syringae by HrpV. J. Bacteriol. 180:4532–4537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447. Quinaud M, et al. 2005. The PscE-PscF-PscG complex controls type III secretion needle biogenesis in Pseudomonas aeruginosa. J. Biol. Chem. 280:36293–36300 [DOI] [PubMed] [Google Scholar]
- 448. Quinaud M, et al. 2007. Structure of the heterotrimeric complex that regulates type III secretion needle formation. Proc. Natl. Acad. Sci. U. S. A. 104:7803–7808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449. Rainbow L, Hart CA, Winstanley C. 2002. Distribution of type III secretion gene clusters in Burkholderia pseudomallei, B. thailandensis and B. mallei. J. Med. Microbiol. 51:374–378 [DOI] [PubMed] [Google Scholar]
- 450. Rappl C, Deiwick J, Hensel M. 2003. Acidic pH is required for the functional assembly of the type III secretion system encoded by Salmonella pathogenicity island 2. FEMS Microbiol. Lett. 226:363–372 [DOI] [PubMed] [Google Scholar]
- 451. Rathinavelan T, Tang C, De Guzman RN. 2011. Characterization of the interaction between the Salmonella type III secretion system tip protein SipD and the needle protein PrgI by paramagnetic relaxation enhancement. J. Biol. Chem. 286:4922–4930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452. Rathinavelan T, et al. 2010. A repulsive electrostatic mechanism for protein export through the type III secretion apparatus. Biophys. J. 98:452–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453. Records AR. 2011. The type VI secretion system: a multi-purpose delivery system with a phage-like machinery. Mol. Plant Microbe Interact. 24:751–757 [DOI] [PubMed] [Google Scholar]
- 454. Reina LD, O'Bryant DM, Matson JS, Nilles ML. 2008. LcrG secretion is not required for blocking of Yops secretion in Yersinia pestis. BMC Microbiol. 8:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455. Rietsch A, Vallet-Gely I, Dove SL, Mekalanos JJ. 2005. ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 102:8006–8011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456. Rimpilainen M, Forsberg A, Wolf-Watz H. 1992. A novel protein, LcrQ, involved in the low-calcium response of Yersinia pseudotuberculosis shows extensive homology to YopH. J. Bacteriol. 174:3355–3363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457. Riordan KE, Schneewind O. 2008. YscU cleavage and the assembly of Yersinia type III secretion machine complexes. Mol. Microbiol. 68:1485–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458. Riordan KE, Sorg JA, Berube BJ, Schneewind O. 2008. Impassable YscP substrates and their impact on the Yersinia enterocolitica type III secretion pathway. J. Bacteriol. 190:6204–6216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459. Rodgers L, Gamez A, Riek R, Ghosh P. 2008. The type III secretion chaperone SycE promotes a localized disorder-to-order transition in the natively unfolded effector YopE. J. Biol. Chem. 283:20857–20863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460. Rodgers L, Mukerjea R, Birtalan S, Friedberg D, Ghosh P. 2010. A solvent-exposed patch in chaperone-bound YopE is required for translocation by the type III secretion system. J. Bacteriol. 192:3114–3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461. Roehrich AD, Martinez-Argudo I, Johnson S, Blocker AJ, Veenendaal AK. 2010. The extreme C terminus of Shigella flexneri IpaB is required for regulation of type III secretion, needle tip composition, and binding. Infect. Immun. 78:1682–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462. Rohde JR, Fox JM, Minnich SA. 1994. Thermoregulation in Yersinia enterocolitica is coincident with changes in DNA supercoiling. Mol. Microbiol. 12:187–199 [DOI] [PubMed] [Google Scholar]
- 463. Roine E, et al. 1997. Hrp pilus: a hrp-dependent bacterial surface appendage produced by Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. U. S. A. 94:3459–3464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464. Rosen R, et al. 2002. Protein aggregation in Escherichia coli: role of proteases. FEMS Microbiol. Lett. 207:9–12 [DOI] [PubMed] [Google Scholar]
- 465. Rosqvist R, Forsberg A, Wolf-Watz H. 1991. Intracellular targeting of the Yersinia YopE cytotoxin in mammalian cells induces actin microfilament disruption. Infect. Immun. 59:4562–4569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466. Rosqvist R, Hakansson S, Forsberg A, Wolf-Watz H. 1995. Functional conservation of the secretion and translocation machinery for virulence proteins of yersiniae, salmonellae, and shigellae. EMBO J. 14:4187–4195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467. Ross JA, Plano GV. 2011. A C-terminal region of Yersinia pestis YscD binds the outer membrane secretin YscC. J. Bacteriol. 193:2276–2289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468. Rossier O, Van den Ackerveken G, Bonas U. 2000. HrpB2 and HrpF from Xanthomonas are type III-secreted proteins and essential for pathogenicity and recognition by the host plant. Mol. Microbiol. 38:828–838 [DOI] [PubMed] [Google Scholar]
- 469. Rossier O, Wengelnik K, Hahn K, Bonas U. 1999. The Xanthomonas Hrp type III system secretes proteins from plant and mammalian pathogens. Proc. Natl. Acad. Sci. U. S. A. 96:9368–9373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470. Ruckdeschel K, et al. 2006. The proteasome pathway destabilizes Yersinia outer protein E and represses its antihost cell activities. J. Immunol. 176:6093–6102 [DOI] [PubMed] [Google Scholar]
- 471. Rucks EA, Olson JC. 2005. Characterization of an ExoS type III translocation-resistant cell line. Infect. Immun. 73:638–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472. Ruiz-Albert J, Mundy R, Yu XJ, Beuzon CR, Holden DW. 2003. SseA is a chaperone for the SseB and SseD translocon components of the Salmonella pathogenicity-island-2-encoded type III secretion system. Microbiology 149:1103–1111 [DOI] [PubMed] [Google Scholar]
- 473. Ryndak MB, Chung H, London E, Bliska JB. 2005. Role of predicted transmembrane domains for type III translocation, pore formation, and signaling by the Yersinia pseudotuberculosis YopB protein. Infect. Immun. 73:2433–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474. Saijo-Hamano Y, et al. 2010. Structure of the cytoplasmic domain of FlhA and implication for flagellar type III protein export. Mol. Microbiol. 76:260–268 [DOI] [PubMed] [Google Scholar]
- 475. Sal-Man N, Deng W, Finlay BB. 2012. EscI: a crucial component of the type III secretion system forms the inner rod structure in enteropathogenic Escherichia coli. Biochem. J. 442:119–125 [DOI] [PubMed] [Google Scholar]
- 476. Samatey FA, et al. 2004. Structure of the bacterial flagellar hook and implication for the molecular universal joint mechanism. Nature 431:1062–1068 [DOI] [PubMed] [Google Scholar]
- 477. Samudrala R, Heffron F, McDermott JE. 2009. Accurate prediction of secreted substrates and identification of a conserved putative secretion signal for type III secretion systems. PLoS Pathog. 5:e1000375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478. Sani M, et al. 2007. Structural organization of the needle complex of the type III secretion apparatus of Shigella flexneri. Micron 38:291–301 [DOI] [PubMed] [Google Scholar]
- 479. Sanowar S, et al. 2010. Interactions of the transmembrane polymeric rings of the Salmonella enterica serovar Typhimurium type III secretion system. mBio 1:e00458–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480. Sarkar MK, Paul K, Blair D. 2010. Chemotaxis signaling protein CheY binds to the rotor protein FliN to control the direction of flagellar rotation in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 107:9370–9375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481. Sarker MR, Neyt C, Stainier I, Cornelis GR. 1998. The Yersinia Yop virulon: LcrV is required for extrusion of the translocators YopB and YopD. J. Bacteriol. 180:1207–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482. Sato H, Frank DW. 2011. Multi-functional characteristics of the Pseudomonas aeruginosa type III needle-tip protein, PcrV; comparison to orthologs in other Gram-negative bacteria. Front. Microbiol. 2:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483. Sato K, Homma M. 2000. Multimeric structure of PomA, a component of the Na+-driven polar flagellar motor of Vibrio alginolyticus. J. Biol. Chem. 275:20223–20228 [DOI] [PubMed] [Google Scholar]
- 484. Schechter LM, Roberts KA, Jamir Y, Alfano JR, Collmer A. 2004. Pseudomonas syringae type III secretion system targeting signals and novel effectors studied with a Cya translocation reporter. J. Bacteriol. 186:543–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 485. Schesser K, Frithz-Lindsten E, Wolf-Watz H. 1996. Delineation and mutational analysis of the Yersinia pseudotuberculosis YopE domains which mediate translocation across bacterial and eukaryotic cellular membranes. J. Bacteriol. 178:7227–7233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 486. Scheurwater E, Reid CW, Clarke AJ. 2008. Lytic transglycosylases: bacterial space-making autolysins. Int. J. Biochem. Cell Biol. 40:586–591 [DOI] [PubMed] [Google Scholar]
- 487. Scheurwater EM, Burrows LL. 2011. Maintaining network security: how macromolecular structures cross the peptidoglycan layer. FEMS Microbiol. Lett. 318:1–9 [DOI] [PubMed] [Google Scholar]
- 488. Schlumberger MC, et al. 2005. Real-time imaging of type III secretion: Salmonella SipA injection into host cells. Proc. Natl. Acad. Sci. U. S. A. 102:12548–12553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489. Schmid A, et al. 2006. Yersinia enterocolitica type III secretion chaperone SycD: recombinant expression, purification and characterization of a homodimer. Protein Expr. Purif. 49:176–182 [DOI] [PubMed] [Google Scholar]
- 490. Schoehn G, et al. 2003. Oligomerization of type III secretion proteins PopB and PopD precedes pore formation in Pseudomonas. EMBO J. 22:4957–4967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491. Schoenhals GJ, Macnab RM. 1996. Physiological and biochemical analyses of FlgH, a lipoprotein forming the outer membrane L ring of the flagellar basal body of Salmonella typhimurium. J. Bacteriol. 178:4200–4207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492. Schraidt O, et al. 2010. Topology and organization of the Salmonella typhimurium type III secretion needle complex components. PLoS Pathog. 6:e1000824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 493. Schraidt O, Marlovits TC. 2011. Three-dimensional model of Salmonella's needle complex at subnanometer resolution. Science 331:1192–1195 [DOI] [PubMed] [Google Scholar]
- 494. Schubot FD, et al. 2005. Three-dimensional structure of a macromolecular assembly that regulates type III secretion in Yersinia pestis. J. Mol. Biol. 346:1147–1161 [DOI] [PubMed] [Google Scholar]
- 495. Schuch R, Maurelli AT. 2001. MxiM and MxiJ, base elements of the Mxi-Spa type III secretion system of Shigella, interact with and stabilize the MxiD secretin in the cell envelope. J. Bacteriol. 183:6991–6998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 496. Schulz S, Büttner D. 2011. Functional characterization of the type III secretion substrate specificity switch protein HpaC from Xanthomonas. Infect. Immun. 79:2998–3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 497. Sekiya K, et al. 2001. Supermolecular structure of the enteropathogenic Escherichia coli type III secretion system and its direct interaction with the EspA-sheath-like structure. Proc. Natl. Acad. Sci. U. S. A. 98:11638–11643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498. Shaikh TR, et al. 2005. A partial atomic structure for the flagellar hook of Salmonella typhimurium. Proc. Natl. Acad. Sci. U. S. A. 102:1023–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 499. Shan L, et al. 2004. The HopPtoF locus of Pseudomonas syringae pv. tomato DC3000 encodes a type III chaperone and a cognate effector. Mol. Plant Microbe Interact. 17:447–455 [DOI] [PubMed] [Google Scholar]
- 500. Shao F, Dixon JE. 2003. YopT is a cysteine protease cleaving Rho family GTPases. Adv. Exp. Med. Biol. 529:79–84 [DOI] [PubMed] [Google Scholar]
- 501. Shen DK, Saurya S, Wagner C, Nishioka H, Blocker AJ. 2010. Domains of the Shigella flexneri type III secretion system IpaB protein involved in secretion regulation. Infect. Immun. 78:4999–5010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502. Shibata S, et al. 2007. FliK regulates flagellar hook length as an internal ruler. Mol. Microbiol. 64:1404–1415 [DOI] [PubMed] [Google Scholar]
- 503. Shin H, Cornelis GR. 2007. Type III secretion translocation pores of Yersinia enterocolitica trigger maturation and release of pro-inflammatory IL-1beta. Cell. Microbiol. 9:2893–2902 [DOI] [PubMed] [Google Scholar]
- 504. Silva-Herzog E, Ferracci F, Jackson MW, Joseph SS, Plano GV. 2008. Membrane localization and topology of the Yersinia pestis YscJ lipoprotein. Microbiology 154:593–607 [DOI] [PubMed] [Google Scholar]
- 505. Simpson N, Audry L, Enninga J. 2010. Tracking the secretion of fluorescently labeled type III effectors from single bacteria in real time. Methods Mol. Biol. 619:241–256 [DOI] [PubMed] [Google Scholar]
- 506. Skoudy A, et al. 2000. CD44 binds to the Shigella IpaB protein and participates in bacterial invasion of epithelial cells. Cell. Microbiol. 2:19–33 [DOI] [PubMed] [Google Scholar]
- 507. Smith TG, Pereira L, Hoover TR. 2009. Helicobacter pylori FlhB processing-deficient variants affect flagellar assembly but not flagellar gene expression. Microbiology 155:1170–1180 [DOI] [PubMed] [Google Scholar]
- 508. Sockett H, Yamaguchi S, Kihara M, Irikura VM, Macnab RM. 1992. Molecular analysis of the flagellar switch protein FliM of Salmonella typhimurium. J. Bacteriol. 174:793–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 509. Sorg I, et al. 2007. YscU recognizes translocators as export substrates of the Yersinia injectisome. EMBO J. 26:3015–3024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510. Sorg JA, Blaylock B, Schneewind O. 2006. Secretion signal recognition by YscN, the Yersinia type III secretion ATPase. Proc. Natl. Acad. Sci. U. S. A. 103:16490–16495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 511. Sory MP, Boland A, Lambermont I, Cornelis GR. 1995. Identification of the YopE and YopH domains required for secretion and internalization into the cytosol of macrophages, using the cyaA gene fusion approach. Proc. Natl. Acad. Sci. U. S. A. 92:11998–12002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512. Sory MP, Cornelis GR. 1994. Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol. Microbiol. 14:583–594 [DOI] [PubMed] [Google Scholar]
- 513. Soscia C, Hachani A, Bernadac A, Filloux A, Bleves S. 2007. Cross talk between type III secretion and flagellar assembly systems in Pseudomonas aeruginosa. J. Bacteriol. 189:3124–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514. Sosinsky GE, Francis NR, Stallmeyer MJ, DeRosier DJ. 1992. Substructure of the flagellar basal body of Salmonella typhimurium. J. Mol. Biol. 223:171–184 [DOI] [PubMed] [Google Scholar]
- 515. Spaeth KE, Chen YS, Valdivia RH. 2009. The Chlamydia type III secretion system C-ring engages a chaperone-effector protein complex. PLoS Pathog. 5:e1000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 516. Spreter T, et al. 2009. A conserved structural motif mediates formation of the periplasmic rings in the type III secretion system. Nat. Struct. Mol. Biol. 16:468–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 517. Stafford GP, et al. 2007. Sorting of early and late flagellar subunits after docking at the membrane ATPase of the type III export pathway. J. Mol. Biol. 374:877–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 518. Stainier I, et al. 2000. YscP, a Yersinia protein required for Yop secretion that is surface exposed, and released in low Ca2+. Mol. Microbiol. 37:1005–1018 [DOI] [PubMed] [Google Scholar]
- 519. Stainier I, Iriarte M, Cornelis G. 1997. YscM1 and YscM2, two Yersinia enterocolitica proteins causing downregulation of yop transcription. Mol. Microbiol. 26:833–843 [DOI] [PubMed] [Google Scholar]
- 520. Stebbins CE, Galan JE. 2001. Maintenance of an unfolded polypeptide by a cognate chaperone in bacterial type III secretion. Nature 414:77–81 [DOI] [PubMed] [Google Scholar]
- 521. Stender S, et al. 2000. Identification of SopE2 from Salmonella typhimurium, a conserved guanine nucleotide exchange factor for Cdc42 of the host cell. Mol. Microbiol. 36:1206–1221 [DOI] [PubMed] [Google Scholar]
- 522. Stevens MP, et al. 2002. An Inv/Mxi-Spa-like type III protein secretion system in Burkholderia pseudomallei modulates intracellular behaviour of the pathogen. Mol. Microbiol. 46:649–659 [DOI] [PubMed] [Google Scholar]
- 523. Stolz B, Berg HC. 1991. Evidence for interactions between MotA and MotB, torque-generating elements of the flagellar motor of Escherichia coli. J. Bacteriol. 173:7033–7037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 524. Stone CB, Bulir DC, Gilchrist JD, Toor RK, Mahony JB. 2010. Interactions between flagellar and type III secretion proteins in Chlamydia pneumoniae. BMC Microbiol. 10:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 525. Stone CB, Johnson DL, Bulir DC, Gilchrist JD, Mahony JB. 2008. Characterization of the putative type III secretion ATPase CdsN (Cpn0707) of Chlamydophila pneumoniae. J. Bacteriol. 190:6580–6588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 526. Straley SC, Plano GV, Skrzypek E, Haddix PL, Fields KA. 1993. Regulation by Ca2+ in the Yersinia low-Ca2+ response. Mol. Microbiol. 8:1005–1010 [DOI] [PubMed] [Google Scholar]
- 527. Su MS, Kao HC, Lin CN, Syu WJ. 2008. Gene l0017 encodes a second chaperone for EspA of enterohaemorrhagic Escherichia coli O157:H7. Microbiology 154:1094–1103 [DOI] [PubMed] [Google Scholar]
- 528. Sukhan A, Kubori T, Galan JE. 2003. Synthesis and localization of the Salmonella SPI-1 type III secretion needle complex proteins PrgI and PrgJ. J. Bacteriol. 185:3480–3483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 529. Sukhan A, Kubori T, Wilson J, Galan JE. 2001. Genetic analysis of assembly of the Salmonella enterica serovar Typhimurium type III secretion-associated needle complex. J. Bacteriol. 183:1159–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 530. Sun P, Tropea JE, Austin BP, Cherry S, Waugh DS. 2008. Structural characterization of the Yersinia pestis type III secretion system needle protein YscF in complex with its heterodimeric chaperone YscE/YscG. J. Mol. Biol. 377:819–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 531. Sun YH, Rolan HG, Tsolis RM. 2007. Injection of flagellin into the host cell cytosol by Salmonella enterica serotype Typhimurium. J. Biol. Chem. 23:33897–33901 [DOI] [PubMed] [Google Scholar]
- 532. Sundberg L, Forsberg A. 2003. TyeA of Yersinia pseudotuberculosis is involved in regulation of Yop expression and is required for polarized translocation of Yop effectors. Cell. Microbiol. 5:187–202 [DOI] [PubMed] [Google Scholar]
- 533. Sundin C, Thelaus J, Broms JE, Forsberg A. 2004. Polarisation of type III translocation by Pseudomonas aeruginosa requires PcrG, PcrV and PopN. Microb. Pathog. 37:313–322 [DOI] [PubMed] [Google Scholar]
- 534. Suzuki H, et al. 1998. A structural feature in the central channel of the bacterial flagellar FliF ring complex is implicated in type III protein export. J. Struct. Biol. 124:104–114 [DOI] [PubMed] [Google Scholar]
- 535. Suzuki H, Yonekura K, Namba K. 2004. Structure of the rotor of the bacterial flagellar motor revealed by electron cryomicroscopy and single-particle image analysis. J. Mol. Biol. 337:105–113 [DOI] [PubMed] [Google Scholar]
- 536. Suzuki T, Iino T. 1981. Role of the flaR gene in flagellar hook formation in Salmonella spp. J. Bacteriol. 148:973–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 537. Swietnicki W, et al. 2004. Novel protein-protein interactions of the Yersinia pestis type III secretion system elucidated with a matrix analysis by surface plasmon resonance and mass spectrometry. J. Biol. Chem. 279:38693–38700 [DOI] [PubMed] [Google Scholar]
- 538. Takahashi N, et al. 2009. Autonomous and FliK-dependent length control of the flagellar rod in Salmonella enterica. J. Bacteriol. 191:6469–6472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 539. Takaya A, Kubota Y, Isogai E, Yamamoto T. 2005. Degradation of the HilC and HilD regulator proteins by ATP-dependent Lon protease leads to downregulation of Salmonella pathogenicity island 1 gene expression. Mol. Microbiol. 55:839–852 [DOI] [PubMed] [Google Scholar]
- 540. Tamano K, et al. 2000. Supramolecular structure of the Shigella type III secretion machinery: the needle part is changeable in length and essential for delivery of effectors. EMBO J. 19:3876–3887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 541. Tang X, Xiao Y, Zhou JM. 2006. Regulation of the type III secretion system in phytopathogenic bacteria. Mol. Plant Microbe Interact. 19:1159–1166 [DOI] [PubMed] [Google Scholar]
- 542. Thomas DR, Francis NR, Xu C, DeRosier DJ. 2006. The three-dimensional structure of the flagellar rotor from a clockwise-locked mutant of Salmonella enterica serovar Typhimurium. J. Bacteriol. 188:7039–7048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 543. Thomas DR, Morgan DG, DeRosier DJ. 1999. Rotational symmetry of the C ring and a mechanism for the flagellar rotary motor. Proc. Natl. Acad. Sci. U. S. A. 96:10134–10139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 544. Thomas J, Stafford GP, Hughes C. 2004. Docking of cytosolic chaperone-substrate complexes at the membrane ATPase during flagellar type III protein export. Proc. Natl. Acad. Sci. U. S. A. 101:3945–3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 545. Thomas NA, Deng W, Baker NT, Puente JL, Finlay BB. 2007. Hierarchical delivery of an essential host colonization factor in enteropathogenic Escherichia coli. J. Biol. Chem. 282:29634–29645 [DOI] [PubMed] [Google Scholar]
- 546. Thomas NA, et al. 2005. CesT is a multi-effector chaperone and recruitment factor required for the efficient type III secretion of both LEE- and non-LEE-encoded effectors of enteropathogenic Escherichia coli. Mol. Microbiol. 57:1762–1779 [DOI] [PubMed] [Google Scholar]
- 547. Thomas NA, Finlay BB. 2003. Establishing order for type III secretion substrates—a hierarchical process. Trends Microbiol. 11:398–403 [DOI] [PubMed] [Google Scholar]
- 548. Thomassin JL, He X, Thomas NA. 2011. Role of EscU auto-cleavage in promoting type III effector translocation into host cells by enteropathogenic Escherichia coli. BMC Microbiol. 11:205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 549. Thormann KM, Paulick A. 2010. Tuning the flagellar motor. Microbiology 156:1275–1283 [DOI] [PubMed] [Google Scholar]
- 550. Tian F. 2010. Pseudomonas syringae type III secretion system: secretion signals and putative docking stations. Ph.D. thesis University of Nebraska, Lincoln, NE [Google Scholar]
- 551. Tomoyasu T, Mogk A, Langen H, Goloubinoff P, Bukau B. 2001. Genetic dissection of the roles of chaperones and proteases in protein folding and degradation in the Escherichia coli cytosol. Mol. Microbiol. 40:397–413 [DOI] [PubMed] [Google Scholar]
- 552. Torruellas J, Jackson MW, Pennock JW, Plano GV. 2005. The Yersinia pestis type III secretion needle plays a role in the regulation of Yop secretion. Mol. Microbiol. 57:1719–1733 [DOI] [PubMed] [Google Scholar]
- 553. Trame CB, McKay DB. 2003. Structure of the Yersinia enterocolitica molecular-chaperone protein SycE. Acta Crystallogr. D Biol. Crystallogr. 59:389–392 [DOI] [PubMed] [Google Scholar]
- 554. Travassos LH, et al. 2005. Nod1 participates in the innate immune response to Pseudomonas aeruginosa. J. Biol. Chem. 280:36714–36718 [DOI] [PubMed] [Google Scholar]
- 555. Tree JJ, Wolfson EB, Wang D, Roe AJ, Gally DL. 2009. Controlling injection: regulation of type III secretion in enterohaemorrhagic Escherichia coli. Trends Microbiol. 17:361–370 [DOI] [PubMed] [Google Scholar]
- 556. Triplett LR, Melotto M, Sundin GW. 2009. Functional analysis of the N terminus of the Erwinia amylovora secreted effector DspA/E reveals features required for secretion, translocation, and binding to the chaperone DspB/F. Mol. Plant Microbe Interact. 22:1282–1292 [DOI] [PubMed] [Google Scholar]
- 557. Troisfontaines P, Cornelis GR. 2005. Type III secretion: more systems than you think. Physiol. Plant. 20:326–339 [DOI] [PubMed] [Google Scholar]
- 558. Tseng TT, Tyler BM, Setubal JC. 2009. Protein secretion systems in bacterial-host associations, and their description in the Gene Ontology. BMC Microbiol. 9:S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 559. Tucker SC, Galan JE. 2000. Complex function for SicA, a Salmonella enterica serovar Typhimurium type III secretion-associated chaperone. J. Bacteriol. 182:2262–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 560. Ueno T, Oosawa K, Aizawa S. 1992. M ring, S ring and proximal rod of the flagellar basal body of Salmonella typhimurium are composed of subunits of a single protein, FliF. J. Mol. Biol. 227:672–677 [DOI] [PubMed] [Google Scholar]
- 561. Urbanowski ML, Lykken GL, Yahr TL. 2005. A secreted regulatory protein couples transcription to the secretory activity of the Pseudomonas aeruginosa type III secretion system. Proc. Natl. Acad. Sci. U. S. A. 102:9930–9935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 562. Urbanowski ML, Yahr TL. 2008. Limiting too much of a good thing: a negative feedback mechanism prevents unregulated translocation of type III effector proteins. J. Bacteriol. 190:2643–2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 563. van der Goot FG, Tran van Nhieu G, Allaoui A, Sansonetti P, Lafont F. 2004. Rafts can trigger contact-mediated secretion of bacterial effectors via a lipid-based mechanism. J. Biol. Chem. 279:47792–47798 [DOI] [PubMed] [Google Scholar]
- 564. van Dijk K, Tam VC, Records AR, Petnicki-Ocwieja T, Alfano JR. 2002. The ShcA protein is a molecular chaperone that assists in the secretion of the HopPsyA effector from the type III (Hrp) protein secretion system of Pseudomonas syringae. Mol. Microbiol. 44:1469–1481 [DOI] [PubMed] [Google Scholar]
- 565. van Eerde A, Hamiaux C, Perez J, Parsot C, Dijkstra BW. 2004. Structure of Spa15, a type III secretion chaperone from Shigella flexneri with broad specificity. EMBO Rep. 5:477–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 566. Van Engelenburg SB, Palmer AE. 2008. Quantification of real-time Salmonella effector type III secretion kinetics reveals differential secretion rates for SopE2 and SptP. Chem. Biol. 15:619–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 567. Van Engelenburg SB, Palmer AE. 2010. Imaging type-III secretion reveals dynamics and spatial segregation of Salmonella effectors. Nat. Methods 7:325–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 568. Van Gijsegem F, Vasse J, Camus JC, Marenda M, Boucher C. 2000. Ralstonia solanacearum produces Hrp-dependent pili that are required for PopA secretion but not for attachment of bacteria to plant cells. Mol. Microbiol. 36:249–260 [DOI] [PubMed] [Google Scholar]
- 569. Veenendaal AK, et al. 2007. The type III secretion system needle tip complex mediates host cell sensing and translocon insertion. Mol. Microbiol. 63:1719–1730 [DOI] [PubMed] [Google Scholar]
- 570. Verove J, et al. 2012. Injection of Pseudomonas aeruginosa Exo toxins into host cells can be modulated by host factors at the level of translocon assembly and/or activity. PLoS One 7:e30488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 571. Viala J, et al. 2004. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat. Immunol. 5:1166–1174 [DOI] [PubMed] [Google Scholar]
- 572. Viboud GI, Bliska JB. 2001. A bacterial type III secretion system inhibits actin polymerization to prevent pore formation in host cell membranes. EMBO J. 20:5373–5382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 573. Vidal JE, Navarro-Garcia F. 2008. EspC translocation into epithelial cells by enteropathogenic Escherichia coli requires a concerted participation of type V and III secretion systems. Cell. Microbiol. 10:1975–1986 [DOI] [PubMed] [Google Scholar]
- 574. Viprey V, Del Greco A, Golinowski W, Broughton WJ, Perret X. 1998. Symbiotic implications of type III protein secretion machinery in Rhizobium. Mol. Microbiol. 28:1381–1389 [DOI] [PubMed] [Google Scholar]
- 575. Viswanathan VK, et al. 2004. Comparative analysis of EspF from enteropathogenic and enterohemorrhagic Escherichia coli in alteration of epithelial barrier function. Infect. Immun. 72:3218–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 576. Von Pawel-Rammingen U, et al. 2000. GAP activity of the Yersinia YopE cytotoxin specifically targets the Rho pathway: a mechanism for disruption of actin microfilament structure. Mol. Microbiol. 36:737–748 [DOI] [PubMed] [Google Scholar]
- 577. Wagner S, et al. 2010. Organization and coordinated assembly of the type III secretion export apparatus. Proc. Natl. Acad. Sci. U. S. A. 107:17745–17750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 578. Wagner S, et al. 2009. The helical content of the YscP molecular ruler determines the length of the Yersinia injectisome. Mol. Microbiol. 71:692–701 [DOI] [PubMed] [Google Scholar]
- 579. Wagner S, Stenta M, Metzger LC, Dal Peraro M, Cornelis GR. 2010. Length control of the injectisome needle requires only one molecule of Yop secretion protein P (YscP). Proc. Natl. Acad. Sci. U. S. A. 107:13860–13865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 580. Wainwright LA, Kaper JB. 1998. EspB and EspD require a specific chaperone for proper secretion from enteropathogenic Escherichia coli. Mol. Microbiol. 27:1247–1260 [DOI] [PubMed] [Google Scholar]
- 581. Walker KA, Miller VL. 2004. Regulation of the Ysa type III secretion system of Yersinia enterocolitica by YsaE/SycB and YsrS/YsrR. J. Bacteriol. 186:4056–4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 582. Wallden K, Rivera-Calzada A, Waksman G. 2010. Type IV secretion systems: versatility and diversity in function. Cell. Microbiol. 12:1203–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 583. Wand ME, et al. 2006. Helicobacter pylori FlhB function: the FlhB C-terminal homologue HP1575 acts as a “spare part” to permit flagellar export when the HP0770 FlhBCC domain is deleted. J. Bacteriol. 188:7531–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 584. Wang D, Roe AJ, McAteer S, Shipston MJ, Gally DL. 2008. Hierarchal type III secretion of translocators and effectors from Escherichia coli O157:H7 requires the carboxy terminus of SepL that binds to Tir. Mol. Microbiol. 69:1499–1512 [DOI] [PubMed] [Google Scholar]
- 585. Wang Y, et al. 2007. Differences in the electrostatic surfaces of the type III secretion needle proteins PrgI, BsaL, and MxiH. J. Mol. Biol. 371:1304–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 586. Ward DV, Draper O, Zupan JR, Zambryski PC. 2002. Peptide linkage mapping of the Agrobacterium tumefaciens vir-encoded type IV secretion system reveals protein subassemblies. Proc. Natl. Acad. Sci. U. S. A. 99:11493–11500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 587. Warren SM, Young GM. 2005. An amino-terminal secretion signal is required for YplA export by the Ysa, Ysc, and flagellar type III secretion systems of Yersinia enterocolitica biovar 1B. J. Bacteriol. 187:6075–6083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 588. Waters RC, O'Toole PW, Ryan KA. 2007. The FliK protein and flagellar hook-length control. Protein Sci. 16:769–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 589. Wattiau P, Bernier B, Deslee P, Michiels T, Cornelis GR. 1994. Individual chaperones required for Yop secretion by Yersinia. Proc. Natl. Acad. Sci. U. S. A. 91:10493–10497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 590. Wattiau P, Cornelis GR. 1993. SycE, a chaperone-like protein of Yersinia enterocolitica involved in the secretion of YopE. Mol. Microbiol. 8:123–131 [DOI] [PubMed] [Google Scholar]
- 591. Weber E, et al. 2005. The type III-dependent Hrp pilus is required for productive interaction of Xanthomonas campestris pv. vesicatoria with pepper host plants. J. Bacteriol. 187:2458–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 592. Wehling MD, Guo M, Fu ZQ, Alfano JR. 2004. The Pseudomonas syringae HopPtoV protein is secreted in culture and translocated into plant cells via the type III protein secretion system in a manner dependent on the ShcV type III chaperone. J. Bacteriol. 186:3621–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 593. Wei CF, Deng WL, Huang HC. 2005. A chaperone-like HrpG protein acts as a suppressor of HrpV in regulation of the Pseudomonas syringae pv. syringae type III secretion system. Mol. Microbiol. 57:520–536 [DOI] [PubMed] [Google Scholar]
- 594. Wei W, et al. 2000. The gene coding for the Hrp pilus structural protein is required for type III secretion of Hrp and Avr proteins in Pseudomonas syringae pv. tomato. Proc. Natl. Acad. Sci. U. S. A. 97:2247–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 595. Welch M, Oosawa K, Aizawa S, Eisenbach M. 1993. Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc. Natl. Acad. Sci. U. S. A. 90:8787–8791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 596. West NP, et al. 2005. Optimization of virulence functions through glucosylation of Shigella LPS. Science 307:1313–1317 [DOI] [PubMed] [Google Scholar]
- 597. Wiesand U, et al. 2009. Structure of the type III secretion recognition protein YscU from Yersinia enterocolitica. J. Mol. Biol. 385:854–866 [DOI] [PubMed] [Google Scholar]
- 598. Wilharm G, Dittmann S, Schmid A, Heesemann J. 2007. On the role of specific chaperones, the specific ATPase, and the proton motive force in type III secretion. Int. J. Med. Microbiol. 297:27–36 [DOI] [PubMed] [Google Scholar]
- 599. Wilharm G, et al. 2004. Yersinia enterocolitica type III secretion depends on the proton motive force but not on the flagellar motor components MotA and MotB. Infect. Immun. 72:4004–4009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 600. Wilharm G, Lehmann V, Neumayer W, Trcek J, Heesemann J. 2004. Yersinia enterocolitica type III secretion: evidence for the ability to transport proteins that are folded prior to secretion. BMC Microbiol. 4:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 601. Williams AW, Straley SC. 1998. YopD of Yersinia pestis plays a role in negative regulation of the low-calcium response in addition to its role in translocation of Yops. J. Bacteriol. 180:350–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 602. Williams AW, et al. 1996. Mutations in fliK and flhB affecting flagellar hook and filament assembly in Salmonella typhimurium. J. Bacteriol. 178:2960–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 603. Winnen B, et al. 2008. Hierarchical effector protein transport by the Salmonella typhimurium SPI-1 type III secretion system. PLoS One 3:e2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 604. Woestyn S, Sory MP, Boland A, Lequenne O, Cornelis GR. 1996. The cytosolic SycE and SycH chaperones of Yersinia protect the region of YopE and YopH involved in translocation across eukaryotic cell membranes. Mol. Microbiol. 20:1261–1271 [DOI] [PubMed] [Google Scholar]
- 605. Wood S, Jin J, Lloyd SA. 2008. YscP and YscU switch the substrate specificity of the Yersinia type III secretion system by regulating export of the inner rod protein YscI. J. Bacteriol. 190:4252–4262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 606. Worrall LJ, Vuckovic M, Strynadka NC. 2010. Crystal structure of the C-terminal domain of the Salmonella type III secretion system export apparatus protein InvA. Protein Sci. 19:1091–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 607. Wulff-Strobel CR, Williams AW, Straley SC. 2002. LcrQ and SycH function together at the Ysc type III secretion system in Yersinia pestis to impose a hierarchy of secretion. Mol. Microbiol. 43:411–423 [DOI] [PubMed] [Google Scholar]
- 608. Yahr TL, Wolfgang MC. 2006. Transcriptional regulation of the Pseudomonas aeruginosa type III secretion system. Mol. Microbiol. 62:631–640 [DOI] [PubMed] [Google Scholar]
- 609. Yamaguchi S, et al. 1986. Genetic evidence for a switching and energy-transducing complex in the flagellar motor of Salmonella typhimurium. J. Bacteriol. 168:1172–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 610. Yamamoto S, Kutsukake K. 2006. FliT acts as an anti-FlhD2C2 factor in the transcriptional control of the flagellar regulon in Salmonella enterica serovar Typhimurium. J. Bacteriol. 188:6703–6708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 611. Yang H, et al. 2007. Regulatory role of PopN and its interacting partners in type III secretion of Pseudomonas aeruginosa. J. Bacteriol. 189:2599–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 612. Yang HJ, Lee JS, Cha JY, Baik HS. 2011. Negative regulation of pathogenesis in Pseudomonas syringae pv. tabaci 11528 by ATP-dependent lon protease. Mol. Cell 32:317–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 613. Yip CK, Finlay BB, Strynadka NC. 2005. Structural characterization of a type III secretion system filament protein in complex with its chaperone. Nat. Struct. Mol. Biol. 12:75–81 [DOI] [PubMed] [Google Scholar]
- 614. Yip CK, et al. 2005. Structural characterization of the molecular platform for type III secretion system assembly. Nature 435:702–707 [DOI] [PubMed] [Google Scholar]
- 615. Yonekura K, et al. 2000. The bacterial flagellar cap as the rotary promoter of flagellin self-assembly. Science 290:2148–2152 [DOI] [PubMed] [Google Scholar]
- 616. Yonekura K, Maki-Yonekura S, Namba K. 2003. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature 424:643–650 [DOI] [PubMed] [Google Scholar]
- 617. Young BM, Young GM. 2002. YplA is exported by the Ysc, Ysa, and flagellar type III secretion systems of Yersinia enterocolitica. J. Bacteriol. 184:1324–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 618. Young GM, Schmiel DH, Miller VL. 1999. A new pathway for the secretion of virulence factors by bacteria: the flagellar export apparatus functions as a protein-secretion system. Proc. Natl. Acad. Sci. U. S. A. 96:6456–6461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 619. Young HS, Dang H, Lai Y, DeRosier DJ, Khan S. 2003. Variable symmetry in Salmonella typhimurium flagellar motors. Biophys. J. 84:571–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 620. Younis R, et al. 2010. SepL resembles an aberrant effector in binding to a class 1 type III secretion chaperone and carrying an N-terminal secretion signal. J. Bacteriol. 192:6093–6098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 621. Yu XJ, Liu M, Holden DW. 2004. SsaM and SpiC interact and regulate secretion of Salmonella pathogenicity island 2 type III secretion system effectors and translocators. Mol. Microbiol. 54:604–619 [DOI] [PubMed] [Google Scholar]
- 622. Yu XJ, Liu M, Matthews S, Holden DW. 2011. Tandem translation generates a chaperone for the Salmonella type III secretion system protein SsaQ. J. Biol. Chem. 286:36098–36107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 623. Yu XJ, McGourty K, Liu M, Unsworth KE, Holden DW. 2010. pH sensing by intracellular Salmonella induces effector translocation. Science 328:1040–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 624. Yu YC, et al. 2010. A putative lytic transglycosylase tightly regulated and critical for the EHEC type three secretion. J. Biomed. Sci. 17:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 625. Zahrl D, et al. 2005. Peptidoglycan degradation by specialized lytic transglycosylases associated with type III and type IV secretion systems. Microbiology 151:3455–3467 [DOI] [PubMed] [Google Scholar]
- 626. Zarivach R, et al. 2008. Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS. Nature 453:124–127 [DOI] [PubMed] [Google Scholar]
- 627. Zarivach R, Vuckovic M, Deng W, Finlay BB, Strynadka NC. 2007. Structural analysis of a prototypical ATPase from the type III secretion system. Nat. Struct. Mol. Biol. 14:131–137 [DOI] [PubMed] [Google Scholar]
- 628. Zhang J, Wang X, Zhang Y, Zhang G, Wang J. 2008. A conserved HpaII protein has lytic activity against the bacterial cell wall in phytopathogenic Xanthomonas oryzae. Appl. Microbiol. Biotechnol. 79:605–616 [DOI] [PubMed] [Google Scholar]
- 629. Zhang L, et al. 2007. Identification of the MxiH needle protein residues responsible for anchoring invasion plasmid antigen D to the type III secretion needle tip. J. Biol. Chem. 282:32144–32151 [DOI] [PubMed] [Google Scholar]
- 630. Zhang L, Wang Y, Picking WL, Picking WD, De Guzman RN. 2006. Solution structure of monomeric BsaL, the type III secretion needle protein of Burkholderia pseudomallei. J. Mol. Biol. 359:322–330 [DOI] [PubMed] [Google Scholar]
- 631. Zhao B, Dahlbeck D, Krasileva KV, Fong RW, Staskawicz BJ. 2011. Computational and biochemical analysis of the Xanthomonas effector AvrBs2 and its role in the modulation of Xanthomonas type three effector delivery. PLoS Pathog. 7:e1002408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 632. Zhao R, Amsler CD, Matsumura P, Khan S. 1996. FliG and FliM distribution in the Salmonella typhimurium cell and flagellar basal bodies. J. Bacteriol. 178:258–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 633. Zhao R, Pathak N, Jaffe H, Reese TS, Khan S. 1996. FliN is a major structural protein of the C-ring in the Salmonella typhimurium flagellar basal body. J. Mol. Biol. 261:195–208 [DOI] [PubMed] [Google Scholar]
- 634. Zhao Y, et al. 2011. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature 477:596–600 [DOI] [PubMed] [Google Scholar]
- 635. Zhou D, Chen LM, Hernandez L, Shears SB, Galan JE. 2001. A Salmonella inositol polyphosphatase acts in conjunction with other bacterial effectors to promote host cell actin cytoskeleton rearrangements and bacterial internalization. Mol. Microbiol. 39:248–259 [DOI] [PubMed] [Google Scholar]
- 636. Zhou D, Mooseker MS, Galan JE. 1999. Role of the S. typhimurium actin-binding protein SipA in bacterial internalization. Science 283:2092–2095 [DOI] [PubMed] [Google Scholar]
- 637. Zhou J, Lloyd SA, Blair D. 1998. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc. Natl. Acad. Sci. U. S. A. 95:6436–6441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 638. Zhu K, Gonzalez-Pedrajo B, Macnab RM. 2002. Interactions among membrane and soluble components of the flagellar export apparatus of Salmonella. Biochemistry 41:9516–9524 [DOI] [PubMed] [Google Scholar]
- 639. Zhu WG, Magbanua MM, White FF. 2000. Identification of two novel hrp-associated genes in the hrp gene cluster of Xanthomonas oryzae pv. oryzae. J. Bacteriol. 182:1844–1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 640. Zumbihl R, et al. 1999. The cytotoxin YopT of Yersinia enterocolitica induces modification and cellular redistribution of the small GTP-binding protein RhoA. J. Biol. Chem. 274:29289–29293 [DOI] [PubMed] [Google Scholar]
- 641. Zurawski DV, Stein MA. 2003. SseA acts as the chaperone for the SseB component of the Salmonella pathogenicity island 2 translocon. Mol. Microbiol. 47:1341–1351 [DOI] [PubMed] [Google Scholar]
- 642. Zurawski DV, Stein MA. 2004. The SPI2-encoded SseA chaperone has discrete domains required for SseB stabilization and export, and binds within the C-terminus of SseB and SseD. Microbiology 150:2055–2068 [DOI] [PubMed] [Google Scholar]