Abstract
Human immunodeficiency virus type 1 (HIV-1) is characterized by sequence variability. The third variable region (V3) of the HIV-1 envelope glycoprotein gp120 plays a key role in determination of viral coreceptor usage (tropism) and pathogenesis. This report describes a novel denaturing heteroduplex tracking assay (HTA) to analyze the genetic variation of HIV-1 V3 DNA. It improved upon previous non-denaturing HTA approaches to distinguish HIV-1 CCR5 and CXCR4 tropic viruses in mixed populations. The modifications included the use of a single-stranded fluorescent probe based on the consensus V3 sequence of HIV-1 CCR5 tropic viruses, Locked Nucleic Acid (LNA) “clamps” at both ends of heteroduplex DNA, and denaturing gel electrophoresis using Mutation Detection Enhancement (MDE®) as matrix. The analysis demonstrated that the LNA “clamps” increased its melting temperature (Tm) and the thermal stability of heteroduplex DNA. The partially denaturing gel used a defined concentration of formamide, and significantly induced mobility shifts of heteroduplex DNA that was dependent on the number and patterns of DNA mismatches and insertions/deletions. This new technique successfully detected tropisms of 53 HIV-1 V3 clones of known tropism, and was able to separate and detect multiple V3 DNA variants encoding tropisms for CCR5 or CXCR4 in a mixture. The assay had the sensitivity to detect 0.5% minority species. This method may be useful as a research tool for analysis of viral quasispecies and for genotypic prediction of HIV-1 tropism in clinical specimens.
Keywords: HIV-1 gp120 V3, denaturing heteroduplex tracking assay, Locked Nucleic Acid clamps, viral tropism
1. Introduction
Viral diversity is a hallmark of human immunodeficiency virus type 1(HIV-1) infection (Coffin, 1995). Molecular approaches to analyze HIV-1 genetic variation have the potential to contribute to research and clinical care. HIV-1 viral entry to the host CD4+ cell begins with attachment of the HIV-1 gp120 to the CD4 receptor, which induces a conformational change in gp120 and exposes the third variable region (V3). HIV-1 gp120 V3 then binds to either CCR5 (R5) or CXCR4 (X4) as coreceptor, thereby triggering envelope glycoprotein 41 (gp41) mediated fusion between the viral envelope and the cell membrane, and delivery of nucleocapsid into the host cell (Huang et al., 2005; Feng et al., 1996). The V3 loop is the major determinant of HIV-1 co-receptor usage (Alkhatib et al., 1996; Deng et al. 1996; Berger, 1997). HIV-1 tropism, which is whether a virus uses R5 or X4 as coreceptor for entry, plays a critical role in disease progression and pathogenesis (Connor et al., 1997; Berger et al., 1999; Philpott et al., 2001; Weiser et al., 2008).
HIV-1 stains transmitted in vivo generally use the R5 coreceptor for entry (Samson et al., 1996; Bjorndal et al., 1997; Freel et al. 2003). Years after infection is established, however, X4 strains are detected in approximately 50% of infected individuals, with X4 and R5 viruses usually coexisting in the viral swarm. The emergence of X4 variants in patients infected with HIV-1 is associated with rapid depletion of CD4 cells and acceleration of HIV-1 disease progression (Koot et al. 1993; Shankarappa et al., 1999; Connor et al., 1997;). The introduction of the new class of antiretroviral drugs, entry inhibitors, increases the options of antiretroviral regimens, however, patients should be tested for HIV tropism prior to initiating CCR5 antagonist-based therapy. A rapid, sensitive assay that is able to detect minority strains in a sample would be very useful for research and clinical management.
Previous work has led to the identification of putative sequence motifs distinguishing the HIV-1 X4 phenotypes from the R5 phenotypes. The common genotypic predictive algorithms for HIV-1 tropism include basic residue at 11/25 of V3 (Chesebro et al., 1992; Fouchier et al., 1995), total positive charge of V3 (Bhattacharyya et al., 1996), structure modeling (Cardozo et al., 2007) and bioinformatics (Jensen et al., 2003; Resch et al., 2001; Sing et al., 2007). However, the heterogeneous nature of the HIV viral population from infected patients creates difficulty in accurately determining tropism with regular sequencing approaches (Low et al., 2007; McGovern et al., 2010). Direct sequencing of PCR products is problematic for detection of minority strains, and yields complicated results when samples contain a mixture of variants that have different point mutations or insertion/deletions. Cell-based HIV-1 phenotypic tropism assays are alternative methods of tropism detection, but they are labor-intensive, slow, and expensive.
A gel based electrophoresis method, known as the heteroduplex tracking assay (HTA), has the merits of detecting various virus strains with different mutations in a mixture (Delwart et al., 1993; Delwart and Gordon, 1997). In the HTA, the desired target gene region is amplified by PCR, and a labeled probe is allowed to anneal to the PCR products to form duplex DNA. In a polyacrylamide gel electrophoresis, DNA heteroduplexes migrate more slowly than DNA homoduplexes due to kinks or bends formed by mismatches and deletions/insertions (Bhattacharyya and Lilley 1989;). The HTA has been used for HIV-1 tropism detection previously (Weiser et al., 2008; Nelson et al., 1997). The sensitivity of the HTA allows rare variants with as little as 1–3% of the total viral population to be detected and quantified (Weiser et al., 2008; Schnell et al., 2008; Upchurch et al., 2000), but its ability to distinguish DNA with minor sequence differences is uncertain.
Because subtle changes in the HIV-1 V3 sequence may cause a switch in tropism between some HIV R5 and X4-using viruses, the use of the HTA for HIV-1 tropism detection requires that the shift in mobility of a heteroduplex be strictly dependent on the extent of divergence between the hybridizing sequences. This report describes modifications to the previously reported HTA method that significantly increases its ability to differentiate DNA heteroduplexes containing slight sequence divergence. The improvements include: 1) A single-stranded fluorescent probe based on a V3 consensus sequence of HIV-1 R5 tropic viruses. 2) Denaturing gel electrophoresis using formamide as solvent to enhance the structural conformation change of heteroduplexes. 3) Use of Mutation Detection Enhancement (MDE®) gel as matrix to detect the mobility shift of heteroduplex DNA based on their subtle structural conformation changes (Børresen 1994; Glavac and Dean, 1995). 4) Application of Locked Nucleic Acids (LNA) (Vester and Wengel, 2004; Koshkin et al., 1998) “clamps” at both 5′ and 3′ end regions of the heteroduplex to prevent the two single strands of DNA from totally separating from each other. The modified HIV-1 V3 denaturing HTA tropism assay was able to detect accurately tropisms of a total of 53 HIV-1 V3 clones of known tropism, and had the sensitivity to detect 0.5 % of minor species from the mixture.
2. Materials and Methods
2.1. Viral strains
HIV-1 strains with well characterized tropism were selected in this study. These strains were derived from two sources: 1) previous studies in the Women’s Interagency HIV Study (WIHS) cohort, a multi-center, longitudinal investigation of HIV-1 infection of women (Weiser et al., 2008; Shi et al., 2010; Anastos et al., 2000; Kemal et al., 2003, 2007b; Philpott et al., 2001) and 2) the NIH AIDS Research and Reference Reagent Program (https://www.aidsreagent.org). A few plasma samples were obtained from the WIHS (New York City site). The institutional review boards at Montefiore Medical Center, Bronx, NY and the New York State Department of Health approved the study, and the patients have provided informed consent.
2.2. Amplification of HIV-1 V3 DNA from Clones
2.2.1 Cloning of HIV-1 gp120 V3 DNA from virus strains of known tropism
Clones of the HIV-1 gp120 V3 region were made from the selected HIV-1 viruses. The tropisms of these viruses were genotypically predicted by Geno2pheno algorithms (Sing et al., 2007) (Table 1). Among these 53 V3 clones, 27 had phenotypes confirmed by cell-based tropism assays (https://www.aidsreagent.org; Philpott et al., 2001; Weiser et al., 2008). To construct these clones, RT-PCR was performed to amplify a 140bp V3 DNA fragment from 7093nt to 7232nt according to HIV-1 HXB2 position as described previously (Philpott et al., 2001; Weiser et al., 2008). Briefly, after reverse transcription, the gp120 V3 DNA was amplified by a 35 cycle PCR by using V3 forward primer V3F 5′ AATCTGTAGAAATTAATTGTACAAGAC 3′ (Integrated DNA Technologies, Coralville, USA) and reverse primer V3R 5′ TGCTCTACTAATGTTACAATGTGCTTGTCTTAT 3′ (Integrated DNA Technologies, Coralville, USA). The GeneAmp Gold PCR Reagent Kit (Life Technologies, Grand Island, USA) was used in the PCR. The product V3 DNA was gel purified by using QIAquick Gel Extraction Kit (Qiagen, Valencia, USA), and cloned into vector pSC-A-amp/kan (Agilent Technologies, Santa Clara, USA) following the kit instructions. A total of 53 V3 clones were obtained, and each of them was confirmed by sequencing.
Table 1.
Tropism prediction of HIV-1 V3 clones by the V3 denaturing HTA
| V3 Clone ID | Mismatches and Insertions/Deletions* | RMP** (%) | Tropism by HTA *** | Tropism by Genotype† | Tropism by Phenotype †† | V3 Clone ID | Mismatches and Insertions/Deletions | RMP (%) | Tropism by HTA | Tropism by Genotype | Tropism by Phenotype | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | WCM-19 | 6 | 167 | R5 | R5 | R5 | 1 | WC51P X4 | 12 | 128 | X4 | X4 | X4 |
| 2 | WC28PR5 | 6 | 167 | R5 | R5 | NA‡ | 2 | WC2P X4 | 10 | 128 | X4 | X4 | X4 |
| 3 | WC203PR5 | 9 | 167 | R5 | R5 | NA | 3 | WC9PX4 | 13 | 126 | X4 | X4 | X4 |
| 4 | SW87 | 7 | 166 | R5 | R5 | NA | 4 | WC205PX4 | 11 | 125 | X4 | X4 | NA |
| 5 | BaL | 3 | 166 | R5 | R5 | R5 | 5 | WC203PX4 | 11 | 125 | X4 | X4 | NA |
| 6 | WCM-28 | 5 | 165 | R5 | R5 | R5 | 6 | MN | 11 | 125 | X4 | X4 | X4 |
| 7 | JRCSF | 5 | 160 | R5 | R5 | R5 | 7 | WC28PX4a | 14 | 118 | X4 | X4 | NA |
| 8 | JRFL | 5 | 164 | R5 | R5 | R5 | 8 | WC207PX4 | 12 | 113 | X4 | X4 | NA |
| 9 | WCM-13 | 6 | 164 | R5 | R5 | R5 | 9 | WC28PX4b | 15 | 112 | X4 | X4 | NA |
| 10 | WCM-1 | 6 | 164 | R5 | R5 | R5 | 10 | SPL-3 | 15 Δ | 109 | X4 | X4 | X4 |
| 11 | WC9PR5 | 7 | 164 | R5 | R5 | R5 | 11 | WC209PX4 | 13 | 100 | X4 | X4 | NA |
| 12 | WC202PR5b | 8 | 163 | R5 | R5 | NA | 12 | WC28PX4c | 16 | 95 | X4 | X4 | NA |
| 13 | SW54 | 6 | 162 | R5 | R5 | NA | 13 | WC26PX4 | 20 | 95 | X4 | X4 | NA |
| 14 | WC201PR5 | 8 | 162 | R5 | R5 | NA | 14 | W212PX4 | 13 | 87 | X4 | X4 | NA |
| 15 | WCM-4 | 7 | 161 | R5 | R5 | R5 | 15 | WC21PX4 | 11 ΔΔΔ | 87 | X4 | X4 | NA |
| 16 | WCM-23 | 7 | 160 | R5 | R5 | R5 | 16 | WC4PX4 | 15 | 85 | X4 | X4 | X4 |
| 17 | WC211PR5 | 10 | 159 | R5 | R5 | NA | 17 | WC202PX4 | 14 | 82 | X4 | X4 | NA |
| 18 | SF162 | 8 | 157 | R5 | R5 | R5 | 18 | LAV | 12 ΔΔ+Δ | 80 | X4 | X4 | X4 |
| 19 | 91US005 | 9 | 154 | R5 | R5 | R5 | 19 | TYBE | 12 Δ+ Δ | 79 | X4 | X4 | X4 |
| 20 | WC202PR5f | 4 Δ | 152 | R5 | R5 | NA | 20 | WC213PX4 | 18 Δ | 72 | X4 | X4 | NA |
| 21 | WC206PR5 | 6 Δ | 143 | R5 | R5 | NA | 21 | WC41PX4 | 17 Δ | 69 | X4 | X4 | X4 |
| 22 | WC204PR5 | 4 Δ | 143 | R5 | R5 | NA | 22 | WC211PX4 | 18 | 67 | X4 | X4 | NA |
| 23 | WCM-7 | 5 Δ | 142 | R5 | R5 | R5 | 23 | ELI | 26 Δ | 63 | X4 | X4 | X4 |
| 24 | QZ4598 | 4 Δ | 136 | R5 | R5 | R5 | 24 | 92HT599 | 18 Δ Δ Δ+Δ | 62 | X4 | X4 | X4 |
| 25 | WC214PR5 | 10 Δ | 136 | R5 | R5 | NA | |||||||
| 26 | 92US727 | 10 | 135 | R5 | R5 | R5 | |||||||
| 27 | WC208PR5 | 6 Δ | 134 | R5 | R5 | NA | |||||||
| 28 | WC4PR5 | 9 Δ | 133 | R5 | R5 | R5 | |||||||
| 29 | WC210PR5 | 8 Δ | 125 | X4 | R5 | NA |
Mismatches and Insertions/Deletions: number indicates number of mismatches; Δ indicates a three base insertion/deletion; Δ+ Δ indicate two separated three base insertions/deletions; ΔΔΔ indicates three consecutive three base insertions/deletions. ΔΔ+Δ indicates two consecutive three base insertions/deletions and another separated three base insertion/deletion. ΔΔΔ+Δ indicates three consecutive three base insertions/deletions and another separated three base insertion/deletion.
Relative Mobility Percentage (RMP) = (Migration distance of a heteroduplex band/migration diatance of the single stranded-probe band in the same lane) × 100%.
Tropism prediction by HTA: RMP of ≥ 130% was set as a cut-off value for R5 virus prediction. RMP of < 130% was set as a cut-off value for X4 virus prediction.
Genotypic Tropism: tropism prediction by using Geno2pheno coreceptor prediction (false positive rate at 10%).
Phenotypes were determined by cell-based assays. Strains obtained from the NIH AIDS Research and Reference Reagent Program were phenotypically characterized (www.aidsreagent.org). Other HIV strains studied were phenotypically characterized as described (15, 16).
NA: not available
2.2.2 Amplification and purification of V3 DNA for HTA
To produce HIV-1 V3 DNA for HTA from these clones with known tropism, a PCR was performed in 35 cycles. The reaction contained 20μM of both LNA incorporated forward primer V3FL 5′ AAT+CTG+TAG+AAATTAATTG+TACAAGAC 3′ (Exiqon, Vedbaek, Denmark) (LNA™ bases is indicated as +G, +A, +T, +C) and reverse primer V3RL 5′ TG+CT+CT+ACTAATGTTA+CAATGTGCTTGTCTTAT 3′ (Exiqon, Vedbaek, Denmark) (LNA™ bases is indicated as +G, +A, +T, +C), and 5ng of plasmid DNA. The GeneAmp Gold PCR Reagent Kit (Life Technologies, Grand Island, USA) was used in PCR and the manufacturer’s instructions were followed. All amplified V3 DNA products were purified by using the QIAquick PCR Purification Kit (Qiagen, Valencia, USA), and quantified by using the Quant-iT™ dsDNA HS Assay (Life Technologies, Grand Island, USA). In control experiments, V3 DNA samples without LNA were amplified by the regular V3 primer V3F and V3R.
2.3. Analysis of melting temperature (Tm) of LNA primers
V3 primer reverse complementary oligonucleotides V3FRC 5′ GTCTTGTACAATTAATTTCTACAGATT 3′ (Integrated DNA Technologies, Coralville, USA) and V3RRC 5′ ATAAGACAAGCACATTGTAACATTAGTAGAGCA 3′ (Integrated DNA Technologies, Coralville, USA) were synthesized. Both LNA primer (V3FL and V3RL), and control DNA primer (V3F and V3R) were paired correspondingly with their reverse complementary oligonucleotides (V3FL/V3FRC, V3F/V3FRC; V3RL/V3RRC, V3R/V3RRC). A 25μl mixture was made that included 0.1 M NaCl, 10 mM Tris-HCl pH 7.4, and 2 mM EDTA, 50 μM of paired primer and its reverse complementary oligonucleotide, and 1 x high resolution melting dye EvaGreen (Biotium, Hayward, USA). Samples were run on an Applied Biosystems 7500 Fast Instrument by using the following thermocycling program: 95°C for 1 min, 55°C for 1 min, ramp to 95°C in 0.1°C increments every 30 sec. Derivative melt curves were created and Tm was calculated by using Applied Biosystems 7500 Software, version 2.0.1. Tm is defined as the temperature at which the maximum change in fluorescence occurs (peak of derivative curve).
2.4. Preparation of R5V3 single-stranded fluorescent probe for HTA
2.4.1. Probe design
An alignment of 425 sequences was retrieved from the Los Alamos HIV database (http://www.hiv.lanl.gov) by using the following criteria: HIV-1, Subtype B, V3 sequence of only R5 using viruses, and non-syncytium-inducing (NSI) phenotype. After eliminating identical V3 sequences and adding together other V3 sequences that had exclusively the R5 using phenotype by Philpott and Weiser (Philpott et al., 2001; Weiser et al., 2008), an alignment of 104 exclusive R5 using V3 sequences was made. By using the tool “consensus maker” (http://www.hiv.lanl.gov), the HIV-1 V3 consensus sequence of only R5 using viruses (R5V3) was generated as: 5′-TGTACAAGACCCAACAACAATACAAGAAAAGGTATACATATAG GACCAGGCAGAGCATTTTATGCAACAGGAGATATAATAGGAGATATAAGACAAGCACATTG T-3′.
2.4.2. Construction of the plasmid pSC-R5V3
To construct a clone that has the inserted R5V3 consensus sequence for probe preparation, a 140bp DNA fragment was produced. It included the R5V3 sequence and both forward and reverse primer binding sequences, and was constructed by using a modified recombinant PCR method (Innis et al., 1990). Briefly, a 79 base R5V3 probe forward oligonucleotide (R5V3-OF) 5′ AAATTAATTGTACAAGACCCAACAACAATACAAGAAAAGGTATACATATAGGACCAGGCAGA GCATTTTATGCAACAGG 3′ and a 77 base R5V3 probe reverse oligonucleotide 5′ GTTACAATGTGCTTGTCTTATATCTCCTATTATATCTCCTGTTGCATAAAATGCTCTGCCTGG TCCTATATGTATAC 3′ were synthesized (Integrated DNA Technologies, Coralville, USA). Because there is a 40bp complementary sequence overlap between the 3′ end regions of R5V3-OF and R5V3-OR oligonucleotides, an overlapping extention reaction was performed with 1 X HF buffer, 200μM dNTPs, 20μM of each R5V3-OF and R5V3-OR, and 0.5 unit of Phusion® High-Fidelity DNA Polymerase (New England Biolabs, Ipswich, USA) in a 25μl reaction. The reaction was heated at 98°C for 30sec, followed by 5 cycles of 98°C 10sec, 50°C 30sec, and 72°C 30sec; and ended with an extension 72°C for 5min. Then, another 30 cycle PCR including primer pair V3F and V3R and 2 μl of product from the first step was carried out by using Phusion® High-Fidelity DNA Polymerase following the manufacturer’s instructions. Finally, after a 2% agarose gel electrophoresis, the 140bp DNA PCR product was gel purified by using QIAEX II Gel Extraction Kit (Qiagen, Valencia, USA), and then cloned into vector pSC-A-amp/kan (Agilent Technologies, Santa Clara, USA). A recombinant plasmid was confirmed by sequencing and named as pSC-R5V3.
3.1.1. Preparation of R5V3 single-stranded fluorescent probe
An asymmetric PCR was used to make the R5V3 single-stranded fluorescent probe (Innis et al., 1990). The 100μl reaction consisted of 1X PCR Gold Buffer, 2.5mM Mg2SO4, 200μM each dNTPs, 5.0 μM excess primer V3FL-FAM 5′ 6-FAM™-AAT+CTG+TAG+AAATTAATTG+TACAAGAC 3′ (Exiqon, Vedbaek, Denmark) (LNA™ bases is indicated as +G, +A, +T, +C) and 0.1 μM limiting primer V3R, 10ng of plasmid pSC-R5V3, and 2 units of AmpliTaq Gold DNA Polymerase (Life Technologies, Grand Island, USA). PCR started with a holding cycle of 95°C for 5 min, followed by 40 cycles of 95°C 15sec, 55°C 30sec, 72°C 30sec; then extension at 72°C for 5min, and the reaction was finally kept at 4°C. The fluorescent gel image under the Spectroline TVR-312R UV transilluminator (312nm wavelength) (Spectronics, Westbury, USA) was found to be the same as that scanned by PharosFX Systems (BioRad, Hercules, USA) set the wavelength for FITC, but with weaker fluorescence. The main product of the asymmetric PCR, the 6-FAM end-labeled single-stranded probe migrated much slower than the double-stranded V3 DNA in an 8 % native polyacrylamide gel (Sigma, St. Louis, USA) electrophoresis. The band of slow migration was cut out from the gel under the UV transilluminator, and purified by using QIAEX II Gel Extraction Kit (Qiagen, Valencia, USA) following the manufacturer’s instructions. To determine the amount of probe needed in a single HTA test, the purified probe was diluted serially and mixed with 200ng R5V3 target DNA in an HTA performed as described below. One dose of probe is defined as the quantity of probe that is needed for an excessive single strand band to be clearly visible after the gel was electrophoresed and scanned by PharosFX (BioRad, Hercules, USA). In control experiments, regular R5V3 probe without LNA was produced in an asymmetric PCR using V3F-FAM 5′ 6-FAM™-AATCTGTAGAAATTAATTGTACAAGAC 3′ (Integrated DNA Technologies, Coralville, USA) as excess primer.
2.5. Denaturing HTA
2.5.1. Denaturing MDE gel
A 1mm thick gel was made with 16 × 10 cm glass plates. The gel had two layers; a layer of 20ml gel consisted of 0.6 x TBE (Lonza, Basel, Switzerland), 0.5 X MDE® gel solution (Lonza, Basel, Switzerland), 5% ethylene glycol(Sigma, St. Louis, USA), and deionized formamide (Life Technologies, Grand Island, USA) with varying concentrations determined by individual experiments. On top of the MDE gel, 5ml of 8% native polyacrylamide gel (acrylamide: bis = 49: 1) (Sigma, St. Louis, USA) was made to form the sample loading well and to seal formamide from diffusion. Ammonium persulfate (Sigma, St. Louis, USA) and TEMED (N, N, N, N′-tetramethylethylenediamine) (Sigma, St. Louis, USA) were used to initiate polymerization.
2.5.2 Formation of Heteroduplex DNA
The heteroduplex formation between R5V3 probe and V3 DNA sample was prepared in a 20 μl reaction that consisted of 2μl 10x annealing buffer (1 M NaCl, 100 mM Tris-HCl pH 7.4, and 20 mM EDTA), 50 to 100ng purified V3 DNA, and 1 dose of probe as quantified previously. The reaction mixture was heated for 5 minutes at 95°C, then immediately chilled to 4°C for 2min. The heteroduplex DNA was mixed with 6 x gel loading buffer (New England Biolabs, Ipswich, USA), and then loaded in the sample wells on the casted MDE gel.
2.5.3. Electrophoresis
Electrophoresis was carried out at 40V for 16h at room temperature by applying the Dcode Mutation Detection System (BioRad, Hercules, USA). Buffer 0.6 x TBE (Lonza, Basel, Switzerland) was pumped to cycle between upper and lower electrophoresis buffer tanks. After electrophoresis the gel was analyzed by laser scanner PharosFX (BioRad, Hercules, USA) using the FITC low density setting and 50 micrometer resolution. The gel image was analyzed by “Quantity One” (BioRad, Hercules, USA) software. Bands in each lane were located automatically, and migration distances of each band were measured by the software.
2.5.4. Calculation of relative mobility percentage (RMP)
In order to compare heteroduplex bands from different lanes more precisely, the mobility of each heteroduplex band relative to that of the single-stranded probe found in the same lane was calculated. The relative mobility percentage (RMP) was defined as a unit to measure heterduplex DNA mobility. RMP = (migration distance of a heteroduplex DNA band/migration distance of single-stranded probe band in the same lane) × 100%.
2.6. Sequence alignment of HIV-1 V3 clones
GenBank does not allow submission of sequences shorter than 200bp. Therefore, the V3 sequence alignment of HIV-1 clones in this study is provided in Supplementary Material-1.
3. Results
3.1. The application of LNA clamps increase thermal stability of the V3 heteroduplex DNA
3.1.1. LNA increases the melting temperature (Tm) of DNA
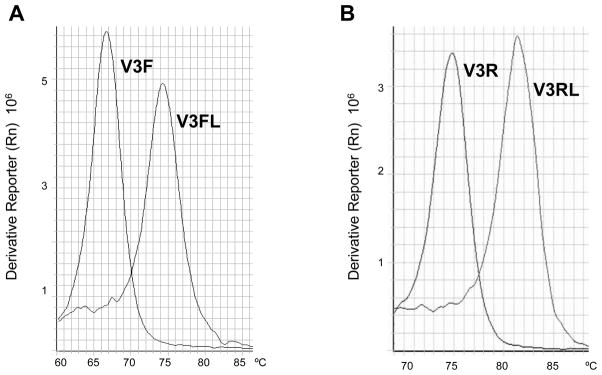
LNA and DNA primers and their complementary oligonucleotides were denatured and then annealed to form double stranded DNA respectively. Melt curve analysis was performed, and Tm was calculated. As shown in Fig. 1, the Tm of LNA primer V3FL to its complementary oligo V3FRC was 74.3°C, while the Tm of regular DNA primer V3F to V3FRC was 66.6°C (Fig. 1(A)). The Tm was increased by 7.7C after LNAs were introduced. The Tm for LNA primer V3RL to its complementary oligonucleotide V3RRC was 81.3°C, while the Tm of regular DNA primer V3F to its V3FRC was 74.7°C (Fig. 1(B)). The Tm of V3RL was increased by 6.6 °C compared to the V3R primer. In both LNA primer V3FL and V3RL there were 4 LNA nucleotides incorporated. These data showed that the incorporation of LNA nucleotides in primers V3FL and V3RL significantly increased their Tm, and thus increased thermal stability of DNA duplexes.
Fig. 1.
Analysis of Tm of LNA primers by melt curve analysis. Paired forward or reverse primers and their complementary oligonucleotides (V3FL/V3FRC, V3F/V3FRC; V3RL/V3RRC, V3R/V3RRC), were denatured and then annealed to form double stranded DNA. Melt curve analysis was performed and Tm was calculated. (A) Derivative melt curves of V3 forward primer. V3F, standard primer; V3FL, LNA primer. (B) Derivative melt curves of V3 reverse primer. V3R, standard primer; V3RL, LNA primer.
3.1.1. The introduction of LNA clamps increased the stability of V3 heteroduplex DNA in denaturing HTA
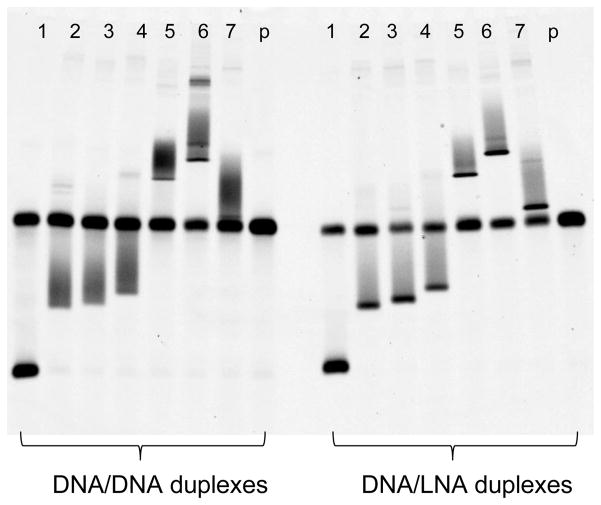
By using the LNA primer V3FL-FAM as the excess primer in asymmetric PCR, 4 LNA nucleotides were incorporated into the 5′ end of the R5V3 single-stranded fluorescent probe. Another LNA primer, V3RL, was used as reverse primer for target V3 DNA amplification, and then four LNA nucleotides were introduced to the 5′ end of the reverse strand of the V3 DNA. When heteroduplex DNA formed between the R5V3 probe and target V3 DNA, both its 3′ end and 5′ end had 4 LNA/DNA base parings. These LNA/DNA base parings served as “clamps” to create regions with high melting temperature at both 5′ and 3′ end regions of the heteroduplex DNA. To test the effect of LNA clamps in the HTA, heteroduplex DNAs representing different patterns of mismatches and insertions/deletions were chosen, and a denaturing MDE gel with 30% formamide was performed. As shown in Fig. 2, the V3 heteroduplex DNA sample in lane 2 (WC2PX4), lane 3 (WC205PX4), and lane 4 (WC207PX4) had ten, eleven, and twelve multiple mismatches individually. V3 heteroduplex DNA in lane 5 (WC41P), lane 6 (ELI), lane 7 (LAV) had both multiple mismatches and deletions/insertions. A sample DNA sequence alignment is shown in Supplementary Material-1. On the left side of the electrophoresis gel, regular DNA duplexes were used as control. All bands were found to have substantial smearing; some of them, as in lanes 2, 4 and 6, even had minor second bands. The right side of the same gel had DNA/LNA duplexes samples with the application of LNA clamps. All heteroduplexes with LNA clamps showed a sharper and higher intensity centered band, with only slight smearing. The data demonstrated that the application of LNA clamps resulted in an increase of the thermal stability of heteroduplex DNA, and thereby improved the performance of the HTA in denaturing MDE® gel electrophoresis.
Fig. 2.
Comparison of performance between heteroduplex DNA with and without LNA clamps in HTA using denaturing MDE® gel with 30% formamide. On the left side of the electrophoresis gel, regular DNA/DNA duplexes were used. In the right side of the same gel, DNA/LNA duplexes with LNA clamps were used. The following cloned V3 DNA was used to the HTA experiment: 1. R5V3. 2. WC2PX4. 3. WC205PX4. 4. WC207PX4. 5. WC41P. 6. ELI. 7. LAV. p. Probe only. The sample sequence alignments can be found in Supplementary material-1.
3.2. Denaturing HIV-1 V3 HTA increases the ability to detect heteroduplex DNA formed between the R5V3 probe and V3 DNA from either CCR5 or CXCR4 tropic viruses
3.2.1. Denaturing HTA increased the ability to differentiate heteroduplex DNA depending on the number of mismatches
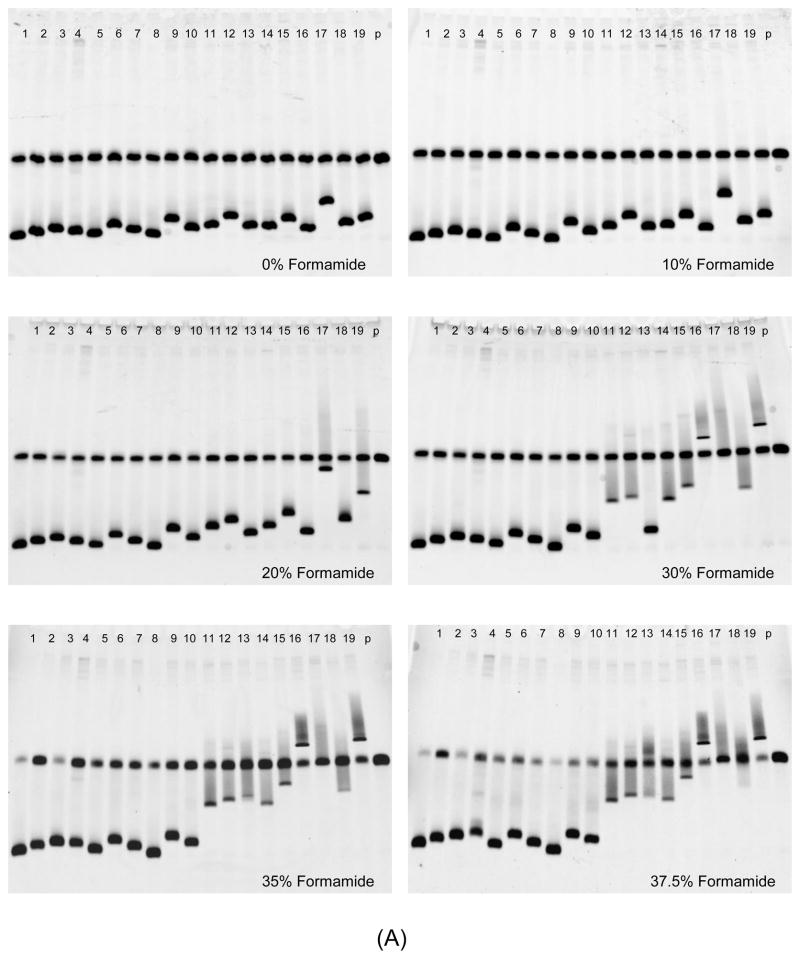
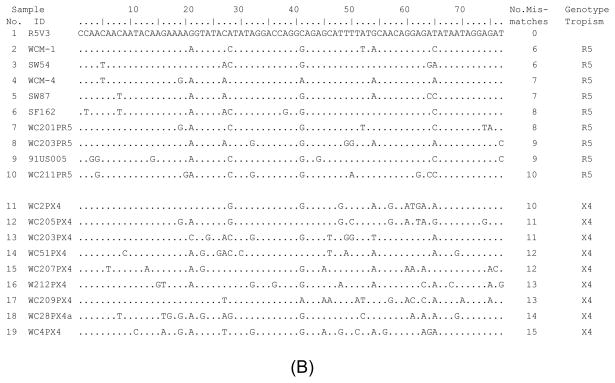
A group of heteroduplex DNAs with different numbers of mismatches was made by denaturing and re-annealing the R5V3 probe with HIV-1 V3 DNA amplified from clones of known tropism. HTAs with different concentrations of formamide were performed, and their ability to differentiate different heteroduplex DNA was analyzed. As shown in Fig. 3. sample 1 was a homoduplex control. Samples 2 to 10 were V3 heteroduplexes produced from R5 tropic viruses, which have six to ten mismatches individually. Samples 11 to 19 were V3 heteroduplex DNA from X4 tropic viruses, which have ten to fifteen mismatches respectively. The HTA gel images are shown in Fig. 3(A), and the alignment of V3 DNA sequences tested is shown in Fig. 3(B). In the HTA with 0% and 10% formamide the migration distances were similar among heteroduplex DNA samples 2 to 19, except that the heteroduplex of sample 17 had a relative slower mobility. When the formamide concentration in a denaturing gel was increased to 20% and 30%, the mobility shift was observed starting from heteroduplex DNA that had higher numbers of mismatches, including V3 heteroduplex DNA samples 11, 12, and 14 to 19. Further gel mobility shifts of heteroduplex DNA of all X4 tropic viruses (samples from 11 to 19) were observed in denaturing HTAs with 35% and 37.5% formamide. However, the migrating behavior of heteroduplex DNA from R5 viruses did not experience noticeable changes in HTAs with 0% to 37.5% formamide.
Fig. 3.
Analysis of DNA heteroduplexes with multiple mismatches using HTA with denaturing MDE® gel under different formamide concentrations. (A) HTA gel pictures. Formamide concentration is indicated in each gel. Samples 2 to 19 were heteroduplexes with six to fifteen mismatches, whose sequence alignment is shown as (B). (B) Alignment of target HIV-1 V3 DNA sequences. The sequence starts after primer V3F and ends before primer V3R, corresponding to nucleotide position of HIV-1 V3 11 to 87.
3.2.2. Denaturing HTA increased the ability to differentiate heteroduplex DNA depending on the distribution pattern of mismatches
Both heteroduplexes of samples 10(R5) and 11(X4) had ten mismatches, but displayed a totally different level of mobility shift in V3 HTA when the formamide concentration was raised to 30% and higher (Fig. 3(A)). The sequence alignment in Fig. 3(B) showed that sample 11 had four consecutive mismatches in a row and all ten mismatches tended to be located toward the 3′ end of the heteroduplex molecule, while the ten mismatches found in sample 10 were evenly distributed. The heteroduplex DNA sample 17 had thirteen mismatches. Most of its mismatches were closely clustered together and located at the 3′ end region of the heteroduplex. An increasing mobility shift was observed in sample 17 in the HTA even with a low formamide concentration. These observations suggested that the distribution pattern of mismatches in the heteroduplex was one of the factors that influenced the vulnerability of the heteroduplex to formamide and resulted in the subsequent mobility shift.
3.2.3. Denaturing HTA differentiated heteroduplex DNA of either R5 tropic or X4 tropic viruses with mismatches and insertions/deletions
Approximately 10% to 15% of isolated HIV-1 V3 sequences contain insertions/deletions. Therefore, another set of samples, HIV-1 V3 heteroduplex DNA contained both insertions/deletions and multiple mismatches were tested. The result showed that the heteroduplex DNA, formed with the R5V3 probe and either R5 or X4 specific V3 DNA with insertions/deletions, could also be successfully distinguished by the mobility shift in denaturing HTAs with appropriate formamide concentrations (see Supplementary Material-2).
3.3. The development and characterization of an HIV-1 V3 denaturing HTA for tropism detection
Based on the above results, an HIV-1 V3 denaturing HTA for tropism detection was developed with the following features: the application of LNA clamps to heteroduplex DNA; use of the R5V3 single-stranded fluorescent probe; and use of denaturing MDE® gel electrophoresis with 36% formamide and 5% ethylene glycol. The HIV-1 V3 denaturing HTA was characterized by experiments under the following different settings.
3.3.1. Tropism prediction of clone DNA by HIV-1 V3 denaturing HTA
The ability of the V3 denaturing HTA to predict tropism was tested by examining a group of 53 HIV-1 V3 clones with known tropisms. HIV-1 V3 DNA was amplified from each individual clone, and an HTA was performed. Migration distances of each heteroduplex DNA were measured by the “Quantity One” software, and the RMP of each sample was calculated accordingly. As listed in Table 1, all 24 V3 clones predicted as X4 tropic by algorithms Geno2pheno had an RPM value from 62 to128; while all 29 clones predicted as R5 tropic by genotype had an RPM (%) from 125 to 167. If an RMP of < 130 was set as a cut-off value for X4 virus prediction, then all the X4 viruses would be accurately predicted. Using this cut-off, 28 out of 29 R5 tropic samples would be predicted by HTA as R5. There was only one sample, WC210PR5, which had a disparity between the HTA result and Geno2pheno prediction. It would have been called as an X4 tropic virus by the HTA with an RMP of 125, but was predicted as R5 tropic by Geno2pheno. Sample WC210PR5 formed a heteroduplex with both a three base deletion and eight multiple mismatches, but its phenotypic tropism was unavailable. The HTA results were also compared with cell-based phenotypes of 16 R5-tropic clones and 11 X4-tropic clones (Table 1). In all cases the HTA result agreed with the cell-based phenotypic assay. The cell-based phenotypes agreed with the genotypic sequence-based predictions in all cases.
3.3.2. Tropism detection of multiple variants in a mixture by HIV-1 V3 denaturing HTA
A mixed sample was created by combining equal amounts of five different V3 DNAs from V3 clones of SW87, 91US005, WC205PX4, WC207PX4 and W212PX4. Each of them forms seven, nine, eleven, twelve and thirteen mismatches individually in heteroduplex DNA after being denatured and annealed with the R5V3 probe. Fig. 4 shows the results of the V3 denaturing HTA using this mixed V3 DNA sample (lane 1) along with V3 DNA from individual clones (lane 2–6). The HTA successfully separated and resolved five individual bands of heteroduplex DNA from the mixture. SW87 (lane 2) and 91US005 (lane 3) are R5 tropic viruses, and WC205PX4 (lane 4), WC207PX4 (lane 5), and W212PX4 (lane 6) are predicted to be X4 tropic viruses.
Fig. 4.
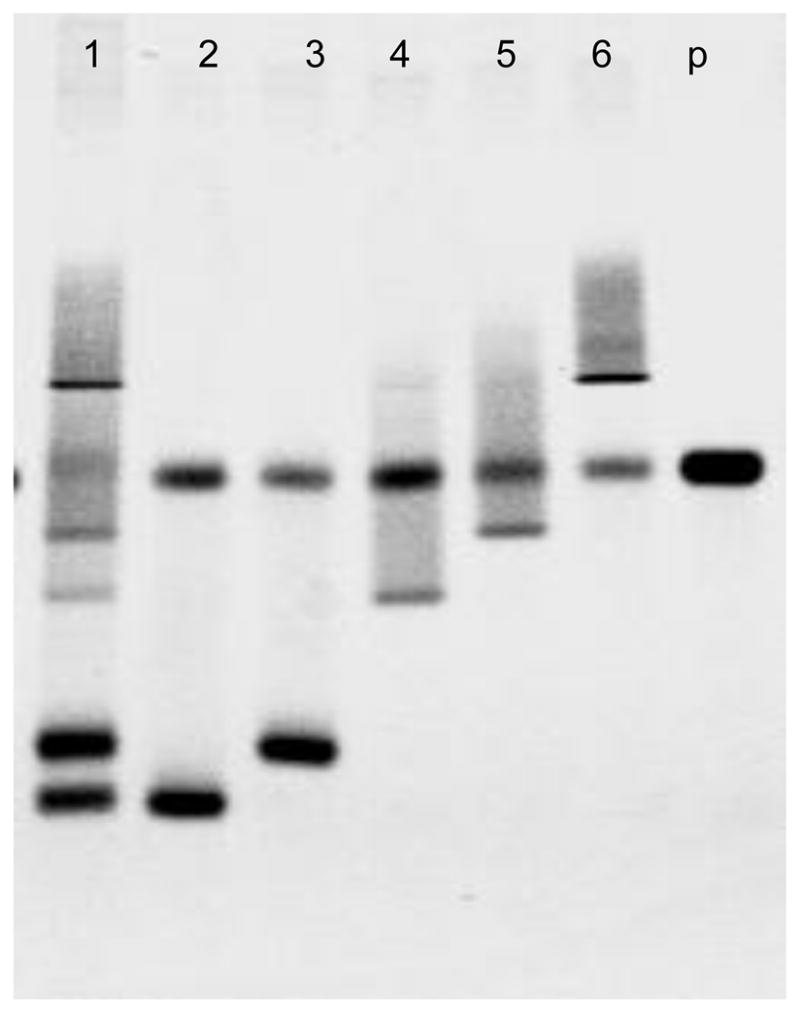
Detection of different V3 DNA in a mixture by the HIV-1 V3 denaturing HTA. 1. Mixture of 50ng each V3 DNA from clone SW87, 91US005, WC205PX4, WC207PX4 and W212PX4. 2. 50ng DNA from V3 clone SW87. 3. 50ng DNA from V3 clone 91US005. 4. 50ng DNA from V3 clone WC205PX4. 5. 50ng DNA from V3 clone WC207PX4. 6. 50ng DNA from V3 clone W212PX4. p. probe only. Sequence alignment of V3 clone SW87, 91US005, WC205PX4, WC207PX4 and W212PX4 can be found at Fig. 3(B).
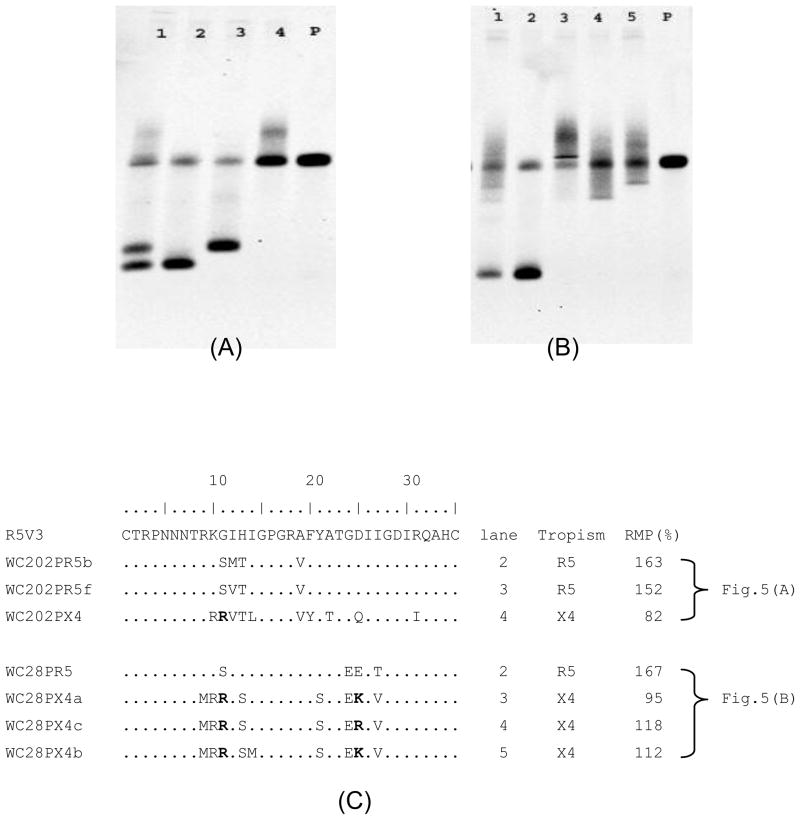
3.3.3. Tropism detection of HIV-1 from plasma samples with a mixture of R5 and X4 tropic viruses
The HIV-1 V3 denaturing HTA tropism assay was used to test some clinical samples. Fig. 5 shows two examples using plasma samples from patients infected with HIV-1. In Fig. 5(A), lane 1 shows the HTA results of the HIV-1 V3 RT PCR product directly amplified from plasma from patient WC202. Three heteroduplex DNA bands were detected in lane 1, indicating the sample contained a mixture of virus populations. Cloning of the same V3 RT PCR product found three different clones WC202PR5b, WC202PR5f and WC202PX4. Lanes 2, 3 and 4 show the HTA results by using V3 DNA from individual clones, whose heteroduplex DNA band (lanes 2, 3, and 4) matched the position of the corresponding heteroduplex bands from the original sample (lane 1). If using the RMP value (cut-off value < 130) for tropism prediction, V3 clones of WC202PR5b (RMP=163) and WC202PR5f (RMP=152) were predicted as R5 tropic, and the V3 clone of WC202PX4 (RMP=82) was predicted to be X4 tropic (Fig. 5(C)).
Fig. 5.
Tropism determination of HIV-1 from patient plasma samples by the HIV-1 V3 denaturing HTA. (A) HIV-1 V3 denaturing HTA result using plasma sample of patient WC202. 1. 100ng of purified HIV-1 V3 RT PCR product from plasma sample of patient WC202. 2. 50ng V3 DNA from clone WC202PR5b. 3. 50ng V3 DNA from clone WC202PR5f. 4. 50ng V3 DNA from clone WC202PX4. p. probe only. (B) HIV-1 V3 denaturing HTA result using plasma sample of patient WC28. 1. 100ng of purified HIV-1 V3 RT PCR product from plasma sample of patient WC28. 2. 50ng V3 DNA from clone WC28PR5. 3. 50ng V3 DNA from clone WC28PX4a. 4. 50ng V3 DNA from clone WC28PX4c. 5. 50ng V3 DNA from clone WC28PX4b. p. probe only. (C) Amino acid sequence alignment of V3 clones from samples WC202 and WC28.
Another example shown in Fig. 5(B) was the result of an HIV-1 V3 denaturing HTA using plasma from patient WC28. Lane 1 shows the HTA result using the HIV-1 V3 RT PCR product. There was one major band and three detectable minor shifted bands in lane 1. The cloning of the same V3 RT PCR product obtained four different clones. The denaturing HTA results of each cloned V3 DNA were shown in lanes 2, 3, 4, and 5. In the HTA, each heteroduplex band from cloned DNA was able to find a matched band at the same position in lane 1 in which the original V3 RT PCR product from patient WC28 was used. If using the RMP (%) value to predict their tropism, clone WC28PR5 (RMP=167) (lane2) was R5 tropic, and the clones WC28PXc (RMP=95) (lane 3), WC28PXa (RMP=118) (lane 4), WC28PXb (RMP=112) (lane 5) were X4 tropic.
The amino acids sequence variations of cloned V3 DNA isolated from both sample WC202P and WC28P are shown in Fig. 5(C). The tropism predictions of each clone based on V3 sequences by using the Geno2pheno algorithm are also shown, and matched the prediction by the HIV-1 V3 denaturing HTA in terms of RMP values.
3.3.4. The detection sensitivity of the denaturing HIV-1 V3 HTA
The detection sensitivity of the denaturing HTA assay was also examined. It detected 1ng of X4 tropic virus (HIV-1 LAV) V3 DNA in a mixture with 200ng R5 tropic virus (HIV-1 JRCSF) V3 DNA; and 1ng of R5 tropic virus (JRCSF) DNA in a mixture with 200ng X4 tropic virus (LAV) V3 DNA. In both cases, the minority species was detected when it represented on 0.5% of the total mixture. The original gel picture of the sensitivity assay can be found in Supplementary Material 3.
4. Discussion
The original HTA is a nondenaturing polyacrylamide gel-based assay that separates viral variants based on mobility shift of their heteroduplexes after electrophoresis. Delwart et al. reported that the greater the divergence in the heteroduplex DNA molecules the slower the heteroduplex migrated, as long as the overall divergence between the two strands was less than 20–25% (Delwart et al. 1993; 1994; 1997). However, it was reported that shifts in mobility do not reflect precisely the total number of mismatches in the heteroduplex in a regular HTA (Schnell et al., 2008). Another report showed that the mobility of heteroduplexes was proportional to the level of mismatch only when that level exceeded 4.5% (Upchurch et al., 2000). The result in Fig. 3 showed that among 140bp heteroduplex DNAs formed with the R5V3 probe and either R5 or X4 cloned V3 DNAs, which had numbers from six to fifteen mismatches (4% to 11%, over all percentage of mismatches), there were no significant differences in the HTA mobility shift if using nondenaturing and low formamide denaturing gel electrophoresis.
However, results in this study substantiated that formamide of appropriate concentration can induce mobility shift of heteroduplex DNA in denaturing MDE® gel electrophoresis depending on the extent of sequence divergence between the two strands of the heteroduplex. The number and distribution pattern of mismatches in the heteroduplex were important factors in determining its degree of mobility shift induced by formamide. This result agrees with previous reported observations under native gel electrophoresis that the overall number of mismatches and clustering of several mismatches together in heteroduplexes appeared to be necessary for the major shift in mobility (Bhattacharyya and Lilley, 1989; Nelson et al., 1997; Upchurch et al., 2000).
Formamide is a commonly used denaturing agent for DNA. It lowers the Tm of DNAs linearly depending on the sequence composition, such as (G+C) content (Blake and Delcourt, 1996). Mismatches are another major factor that influence the Tm of the regional heteroduplex. Therefore, heteroduplex DNA with higher number of mismatches and clustered mismatches tended to be more sensitive to formamide, causing partial denaturation and regional conformation change, ultimately resulting in mobility shifts in the HTA. The application of LNA clamps created higher Tm at the ends of a DNA duplex, which prevented two single strands of DNA from totally melting. The LNA clamps made it possible for the partially denatured DNA between the two end clamps to be analyzed by HTA using denaturing gels with elevated formamide concentrations.
Because the R5V3 consensus sequence as probe was used in HTA, the heteroduplex DNA formed with R5 tropic viruses was most likely to have fewer mismatches than heteroduplex DNA of X4 tropic viruses. Scattered patterns of mismatch distribution were expected to be seen in heteroduplex DNA formed with R5 V3 samples. It was reported that V3 region of X4 variants had much variation especially around amino acids positions 19 to 25 (Nelson et al., 1997, Milich et al., 1993; 1997), which would result in more clustered mismatches in heteroduplex DNA produced with V3 DNA of X4 variants. The V3 sequence alignments in Supplementary Material-1 showed that in most cases heteroduplex DNA of X4 tropic viruses had more mismatches; they also tended to form more closely clustered mismatches than heteroduplexes of R5 strains.
It was estimated from published sequences that about 10%–15% of isolated HIV-1 V3 sequences were found containing deletions/insertions, which occurred in both R5 and X4 tropic viruses. Deletions/insertions result in large heteroduplex mobility shifts in an HTA (Bhattacharyya and Lilley, 1989; Nelson et al., 1997; Upchurch et al., 2000). A similar result was observed using the denaturing HTA. As shown in Table 1, most V3 DNA samples that had deletions/insertions had lower mobility (RMP) values than those with only mismatches. The characteristics of DNA structural changes in heteroduplexes caused by insertions/deletions and mismatches are distinct. Insertions/deletions form bulges in the heteroduplex; while mismatches result in bubble-like structures (Bhattacharyya and Lilley, 1989). Formamide may have had a different impact on the DNA structural changes between heteroduplexes containing deletions/insertions and heteroduplexes containing multiple mismatches. In the development of HIV-1 V3 HTA tropism assay efforts were made to optimize the formamide concentration, and electrophoresis conditions so as to differentiate V3 DNA heteroduplexes of R5 and X4 viruses that involved both mismatches and insertions/deletions.
In conclusion, the data reported demonstrated that the denaturing HIV-1 V3 HTA has a sensitivity of approximately 0.5% to detect X4 tropic strains in mixed samples. Since single-stranded R5V3 probe was end-labeled with fluorescence, the data will be suitable for quantitative analysis. It also has the potential to be automated and performed by using a capillary electrophoresis system. Moreover, the denaturing HTA provides a novel and useful molecular tool to study other viral infections characterized by quasispecies diversity.
Supplementary Material
A novel denaturing heteroduplex tracking assay
The use of a single-stranded fluorescent probe based on the consensus V3 sequence of HIV-1 CCR5 tropic viruses
The use of Locked Nucleic Acid (LNA) “clamps” at both ends of heteroduplex DNA.
The partially denaturing gel electrophoresis with formamide induced significant mobility shifts of heteroduplex DNA based on patterns of mutations.
A useful research method for analysis of viral quasispecies and for genotypic prediction of HIV-1 tropism.
Acknowledgments
We thank the study subjects for their participation. We thank the Wadsworth Center Applied Genomics Technologies Core for DNA sequencing. We thank the NIH AIDS Research and Reference Reagent Program for HIV-1 strains. This study was supported by the National Institutes of Health (grants RO1-AI52015 to B.W., UO1-AI35004 to K.A., and an ARRA supplement to UO1-AI35004 to B.W.) and by Health Research Inc (grant to H.B.)
Footnotes
Conflict of Interest statement: H. Burger and B. Weiser are inventors of 6 patents for the determination of HIV tropism for clinical monitoring. The patents are owned by Health Research Inc., which has licensed the technologies to Quest Diagnostics for clinical use. B. Shi, H. Burger and B. Weiser are coinventors of a pending patent application for detection of X4 strains of HIV-1 by Heteroduplex Tracking Assay.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anastos K, Gange SJ, Lau B, Weiser B, Detels R, Giorgi JV, Margolick JB, Cohen M, Phair J, Melnick S, Rinaldo CR, Kovacs A, Levine A, Landesman S, Young M, Muñoz A, Greenblatt RM. Association of race and gender with HIV-1 RNA levels and immunologic progression. J Acquir Immune Defic Syndr. 2000;24:218–226. doi: 10.1097/00126334-200007010-00004. [DOI] [PubMed] [Google Scholar]
- Alkhatib G, Combadiere C, Broder CC, Feng Y, Kennedy PE, Murphy PM, Berger EA. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- Berger EA. HIV entry and tropism: the chemokine receptor connection. AIDS. 1997;11(Suppl A):S3–S16. [PubMed] [Google Scholar]
- Berger EA, Murphy PM, Farber JM. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Ann Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya A, Lilley DM. The contrasting structures of mismatched DNA sequences containing looped-out bases (bulges) and multiple mismatches (bubbles) Nucleic Acids Res. 1989;17:6821–6840. doi: 10.1093/nar/17.17.6821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya D, Brooks BR, Callahan L. Positioning of positively charged residues in the V3 loop correlates with HIV type 1 syncytium-inducing phenotype. AIDS Res Hum Retroviruses. 1996;12:83–90. doi: 10.1089/aid.1996.12.83. [DOI] [PubMed] [Google Scholar]
- Bjorndal A, Deng H, Jansson M, Fiore JR, Colognesi C, Karlsson A, Albert J, Scarlatti G, Littman DR, Fenyö EM. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J Virol. 1997;71:7478–7487. doi: 10.1128/jvi.71.10.7478-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake RD, Delcourt SG. Thermodynamic effects of formamide on DNA stability. Nucleic Acids Res. 1996;24:2095–2103. doi: 10.1093/nar/24.11.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Børresen AL. Current Protocols in Human Genetics. New York: John Wiley & Sons; 1994. Mismatch detection using heterodplex analysis; pp. 7.3.1–7.3.3. [Google Scholar]
- Cardozo T, Kimura T, Philpott S, Weiser B, Burger H, Zolla-Pazner S. Structural basis for coreceptor selectivity by the HIV type 1 V3 loop. AIDS Res Hum Retroviruses. 2007;23:415–426. doi: 10.1089/aid.2006.0130. [DOI] [PubMed] [Google Scholar]
- Chesebro B, Wehrly K, Nishio J, Perryman S. Macrophagetropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-celltropic isolates: definition of critical amino acids involved in cell tropism. J Virol. 1992;66:6547–6554. doi: 10.1128/jvi.66.11.6547-6554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin JM. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- Connor RI, Sheridan KE, Ceradini D, Choe S, Landau NR. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delwart EL, Shpaer EG, Louwagie J, McCutchan FE, Grez M, Rübsamen-Waigmann H, Mullins JI. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science. 1993;262:1257–1261. doi: 10.1126/science.8235655. [DOI] [PubMed] [Google Scholar]
- Delwart EL, Sheppard HW, Walker BD, Goudsmit J, Mullins JI. Human immunodeficiency virus type 1 evolution in vivo tracked by DNA heteroduplex mobility assays. J Virol. 1994;68:6672–6683. doi: 10.1128/jvi.68.10.6672-6683.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delwart EL, Gordon CJ. Tracking changes in HIV-1 envelope quasispecies using DNA heteroduplex analysis. Methods. 1997;12:348–354. doi: 10.1006/meth.1997.0489. [DOI] [PubMed] [Google Scholar]
- Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di MP, Marmon S, Sutton RE, Hill CM, Davis CB, Peiper SC, Schall TJ, Littman DR, Landau NR. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- Fouchier RA, Brouwer M, Broersen SM, Schuitemaker H. Simple determination of human immunodeficiency virus type 1 syncytium-inducing V3 genotype by PCR. J Clin Microbiol. 1995;33:906–911. doi: 10.1128/jcm.33.4.906-911.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freel SA, Fiscus SA, Pilcher CD, Menezes P, Giner J, Patrick E, Lennox JL, Hicks CB, Eron JJ, Shugars DC. Envelope diversity, coreceptor usage and syncytiuminducing phenotype of HIV-1 variants in saliva and blood during primary infections. AIDS. 2003;17:2025–2033. doi: 10.1097/00002030-200309260-00003. [DOI] [PubMed] [Google Scholar]
- Glavac D, Dean M. Applications of heteroduplex analysis for mutation detection in disease genes. Human Mutation. 1995;6:281–287. doi: 10.1002/humu.1380060402. [DOI] [PubMed] [Google Scholar]
- Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, Stanfield RL, Dimitrov DS, Korber B, Sodroski J, Wilson IA, Wyatt R, Kwong PD. Structure of a V3-containing HIV-1 gp120 core. Science. 2005;310:1025–1028. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innis M, Gelfand D, Sninsky J, White T. PCR Protocols: A Guide to Methods and Applications. Academic Press Inc; 1990. pp. 76–83. [Google Scholar]
- Jensen MA, Li FS, van ‘t Wout AB, Nickle DC, Shriner D, He HX, McLaughlin S, Shankarappa R, Margolick JB, Mullins JI. Improved coreceptor usage prediction and genotypic monitoring of R5-to-X4 transition by motif analysis of human immunodeficiency virus type 1 env V3 loop sequences. J Virol. 2003;77:13376–13388. doi: 10.1128/JVI.77.24.13376-13388.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemal KS, Foley B, Burger H, Anastos K, Minkoff H, Kitchen C, Philpott SM, Gao W, Robison E, Holman S, Dehner C, Beck S, Meyer WA, 3rd, Landay A, Kovacs A, Bremer J, Weiser B. HIV-1 in genital tract and plasma of women: compartmentalization of viral sequences, coreceptor usage, and glycosylation. Proc Natl Acad Sci USA. 2003;100:12972–12977. doi: 10.1073/pnas.2134064100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemal KS, Burger H, Mayers D, Anastos K, Foley B, Kitchen C, Huggins P, Schroeder T, Picchio G, Back S, Gao W, Meyer WA, 3rd, Weiser B. HIV-1 drug resistance in variants from the female genital tract and plasma. J Infect Dis. 2007;195:535–545. doi: 10.1086/510855. [DOI] [PubMed] [Google Scholar]
- Koot M, I, Keet P, Vos AH, de Goede RE, Roos MT, Coutinho RA. Prognostic value of HIV-1 syncytium-inducing phenotype for rate of CD4R cell depletion and progression to AIDS. Ann Intern Med. 1993;118:681–688. doi: 10.7326/0003-4819-118-9-199305010-00004. [DOI] [PubMed] [Google Scholar]
- Koshkin AA, Nielsen P, Meldgaard M, Rajwanshi VK, Singh SK, Wengel J. LNA (Locked Nucleic Acid): An RNA Mimic Forming Exceedingly Stable LNA:LNA Duplexes. J Am Chem Soc. 1998;120:13252–13253. [Google Scholar]
- Low AJ, Dong W, Chan D, Sing T, Swanstrom R, Jensen M, Pillai S, Good B, Harrigan PR. Current V3 genotyping algorithms are inadequate for predicting X4 co-receptor usage in clinical isolates. AIDS. 2007;21:F17–24. doi: 10.1097/QAD.0b013e3282ef81ea. [DOI] [PubMed] [Google Scholar]
- Milich L, Margolin B, Swanstrom R. V3 loop of the human immunodeficiency virus type 1 Env protein: interpreting sequence variability. J Virol. 1993;67:5623–5634. doi: 10.1128/jvi.67.9.5623-5634.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milich L, Margolin BH, Swanstrom R. Patterns of amino acid variability in NSI-like and SI-like V3 sequences and a linked change in the CD4-binding domain of the HIV-1 Env protein. Virology. 1997;239:108–118. doi: 10.1006/viro.1997.8821. [DOI] [PubMed] [Google Scholar]
- McGovern RA, Thielen A, Mo T, Dong W, Woods CK, Chapman D, Lewis M, James I, Heera J, Valdez H, Harrigan PR. Population-based V3 genotypic tropism assay: a retrospective analysis using screening samples from the A4001029 and MOTIVATE studies. AIDS. 2010;24:2517–2525. doi: 10.1097/QAD.0b013e32833e6cfb. [DOI] [PubMed] [Google Scholar]
- Nelson JA, Fiscus SA, Swanstrom R. Evolutionary variants of the human immunodeficiency virus type 1 V3 region characterized by using a heteroduplex tracking assay. J Virol. 1997;71:8750–8758. doi: 10.1128/jvi.71.11.8750-8758.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott S, Weiser B, Anastos K, Kitchen CM, Robison E, Meyer WA, 3rd, Sacks HS, Mathur-Wagh U, Brunner C, Burger H. Preferential suppression of CXCR4-specific strains of HIV-1 by antiviral therapy. J Clin Invest. 2001;107:431–438. doi: 10.1172/JCI11526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resch W, Hoffman N, Swanstrom R. Improved success of phenotype prediction of the human immunodeficiency virus type 1 from envelope variable loop 3 sequence using neural networks. Virology. 2001;288:51–62. doi: 10.1006/viro.2001.1087. [DOI] [PubMed] [Google Scholar]
- Shankarappa R, Margolick JB, Gange SJ, Rodrigo AG, Upchurch D, Frazadegan H. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol. 1999;73:10489–10502. doi: 10.1128/jvi.73.12.10489-10502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell G, Ince WL, Swanstrom R. Identification and recovery of minor HIV-1 variants using the heteroduplex tracking assay and biotinylated probes. Nucleic Acids Res. 2008;36:e146. doi: 10.1093/nar/gkn713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi B, Kitchen C, Weiser B, Mayers D, Foley B, Kemal K, Anastos K, Suchard M, Parker M, Brunner C, Burger H. Evolution and recombination of genes encoding HIV-1 drug resistance and tropism during antiretroviral therapy. Virology. 2010;404:5–20. doi: 10.1016/j.virol.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sing T, Low AJ, Beerenwinkel N, Sander O, Cheung PK, Domingues F, Büch J, Däumer M, Kaiser R, Lengauer T, Harrigan PR. Predicting HIV co-receptor usage based on genetic and clinical covariates. Antiviral Therapy. 2007;12:1097–1106. [PubMed] [Google Scholar]
- Samson M, Libert F, Doranz BJ, Rucker J, Liesnard D, Farber CM. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- Weiser B, Philpott S, Klimkait T, Burger H, Kitchen C, Bürgisser P, Gorgievski M, Perrin L, Piffaretti JC, Ledergerber B Swiss HIV Cohort Study. HIV-1 coreceptor usage and CXCR4-specific viral load predict clinical disease progression during combination antiretroviral therapy. AIDS. 2008;22:469–479. doi: 10.1097/QAD.0b013e3282f4196c. [DOI] [PubMed] [Google Scholar]
- Upchurch DA, Shankarappa R, Mullins JI. Position and degree of mismatches and the mobility of DNA heteroduplexes. Nucleic Acids Res. 2000;28:E69. doi: 10.1093/nar/28.12.e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vester B, Wengel J. LNA (locked nucleic acid): high-affinity targeting of complementary RNA and DNA. Biochemistry. 2004;43:13233–13241. doi: 10.1021/bi0485732. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.