Abstract
PTEN is a tumor suppressor gene mutated in various advanced human neoplasias, including glioblastomas and prostate, breast, endometrial, and kidney cancers. This tumor suppressor is a lipid phosphatase that negatively regulates cell survival and proliferation mediated by phosphatidylinositol 3-kinase/protein kinase B signaling. Using the Cre–loxP system, we selectively inactivated Pten in murine tissues in which the MMTV-LTR promoter is active, resulting in hyperproliferation and neoplastic changes in Pten-null skin and prostate. These phenotypes had early onset and were completely penetrant. Abnormalities in Pten mutant skin consisted of mild epidermal hyperplasia, whereas prostates from these mice exhibited high-grade prostatic intraepithelial neoplasia (HGPIN) that frequently progressed to focally invasive cancer. These data demonstrate that Pten is an important physiological regulator of growth in the skin and prostate. Further, the early onset of HGPIN in Pten mutant males is unique to this animal model and implicates PTEN mutations in the initiation of prostate cancer. Consistent with high PTEN mutation rates in human prostate tumors, these data indicate that PTEN is a critical tumor suppressor in this organ.
Prostate cancer is the most common cancer among North American and European men, affecting 1 in 9 men over 65 years of age (1, 2). Malignancy of the prostate is the second-leading cause of male cancer deaths and there is currently no effective cure for advanced stages of this disease (1, 2).
One of the most frequent genetic alterations in prostate tumors occurs at chromosomal region 10q (1, 3). PTEN is a tumor-suppressor gene residing within 10q23 that is frequently deleted in various advanced human cancers, including glioblastomas and prostate, breast, and endometrial tumors (4, 5). PTEN mutations have been identified in 10–15% of all prostate tumors and in up to 60% of advanced prostate cancers and cell lines (1, 3, 5, 6).
Germ-line mutations of PTEN have also been shown to cause Cowden's disease (CD), an autosomal dominant multiple hamartoma syndrome believed to occur at an incidence of at least 1/200,000 (7). Hamartomas are benign tumors that develop in multiple organs of CD patients such as the skin and breast. Consistent with PTEN′s role as a tumor suppressor, individuals with CD have an increased susceptibility to developing malignancies of the breast, endometrium, and thyroid (7).
PTEN is a lipid phosphatase that dephosphorylates phosphoinositides at the 3 position of the inositol ring (8, 9). The membrane phosphoinositide and second messenger phosphatidylinositol 3,4,5-trisphosphate [PI(3,4,5)P3] has been identified as a physiological substrate of PTEN. Growth factor-stimulated production of PI(3,4,5)P3 results in activation of cell survival and proliferation mediated by phosphatidylinositol 3-kinase/protein kinase B (PI3K–PKB/Akt) signaling. PTEN suppresses tumor growth by negatively regulating this signal transduction pathway (4). Elevated PKB/Akt activity and PI(3,4,5)P3 levels in Pten-null murine cells are associated with a decreased sensitivity to various apoptotic stimuli and accelerated G1/S progression (9, 10).
Targeted disruption of murine Pten results in hyperproliferation and embryonic lethality by day 9.5 (4, 9, 11). Young Pten heterozygous mice are highly susceptible to thymic lymphomas, whereas carcinomas of the endometrium, breast, and prostate predominate late in life (12). Tumors from Pten+/- mice have lost the wild-type Pten allele, implying that tumor formation requires loss of both Pten alleles. Analysis of tissue-specific Pten-knockout mice has demonstrated that deletion of both Pten alleles results in tumorigenesis of germ cells, T lymphocytes, and mammary epithelium (13–15).
To specifically investigate the role of Pten in homeostasis of the prostate and skin epithelium, we used the Cre-loxP system to generate mice with tissue-specific deletion of Pten in these tissues. Data in this study demonstrate an important physiological role for Pten as a negative regulator of growth in the skin and a critical tumor suppressor in the prostate.
Methods
Genotype and Southern Blot Analysis. Mouse genotyping of the Pten-loxP locus and detection of Cre-mediated recombination of Pten by Southern blot analysis and PCR were performed as described in ref. 16. A 280-bp PCR product was amplified for MMTV-cre genotyping by using MMTV-LTR primer (5′-GGT TCT GAT CTG AGC TCT GAG TG-3′) and Cre primer (5′-CAT CAC TCG TTG CAT CGA CCG G-3′).
Histology and Immunohistochemistry. Upon dissection, tissues were fixed in 10% formalin or 4% paraformaldehyde. Immunohistochemistry and coimmunohistochemistry were carried out as described in ref. 16 with antibodies to phospho-PKB/Akt (Cell Signaling Technology, Beverly, MA), Ki67 and SMA (DAKO), androgen receptor (Affinity Bioreagents, Golden, CO), p63 (Santa Cruz Biotechnology), pan-cadherin (NeoMarkers, Fremont, CA), and loricrin, filaggrin, and keratins 14 and 10 (Babco, Richmond, CA) and counterstained with hematoxylin. For details on measurements of epidermal thickness, cell number, and surface area, see Supporting Methods, which is published as supporting information on the PNAS web site.
Keratinocyte Cultures. Keratinocytes were derived from the epidermis of newborn mice, plated in Eagle's minimal essential medium containing 0.2 mM calcium, and switched to 0.05 mM calcium-containing medium 18 h later (see Supporting Methods for details). For growth curves, keratinocytes plated at 2 × 105 cells per 3.5-cm dish were trypsinized, and viable cells were counted twice for each sample by using the trypan blue exclusion assay. For crystal violet staining, a 0.05% staining solution was applied to cells for 1 min, and the cells were rinsed and dried for analysis. All keratinocyte experiments were done at least in duplicate and repeated at least three times from newly derived keratinocytes.
Western Blot Analysis. Preparation of whole cell protein lysates, protein resolution by SDS/8% PAGE and immunoblotting were performed as in ref. 9. Pten (Cascade, Winchester, MA) and β-tubulin (Santa Cruz Biotechnology) antibodies were used at 1:1,000.
Results
Deletion of Pten in the Skin, Prostate, and Thymus. A Pten flox mouse was previously generated by flanking exons 4 and 5 of the Pten gene with loxP sites (13). Pten was deleted in several tissues by breeding homozygous Pten flox mice with MMTV (mouse mammary tumor virus)-cre transgenic mice [transgenic line A (17)]. Cre reporter assays of MMTV-cre mice have demonstrated Cre-mediated recombination in several tissues, including mammary epithelial cells, skin, lymphocytes, oocytes, seminal vesicles, and salivary epithelial cells (17). Cre-mediated recombination in the prostate of the MMTV-cre mice has recently been documented and is consistent with the ability of hormones to up-regulate the MMTV-LTR promoter (18, 19).
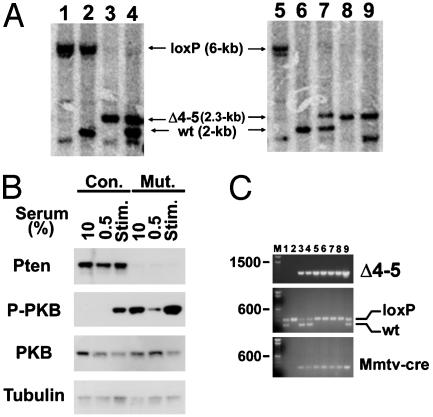
Southern blot analysis using genomic DNA from tissues and cells derived from PtenloxP/loxP; MMTV-cre mice demonstrate that Cre-mediated recombination occurred in the majority of cells in the thymus and epidermis (Fig. 1A). Pten was strongly expressed in keratinocytes derived from skin of Ptenwt/loxP; MMTV-cre control animals, but absent in the majority of PtenloxP/loxP; MMTV-cre keratinocytes (Fig. 1B). Consistent with Pten's ability to negatively regulate PKB/Akt activation, levels of activated and phosphorylated PKB/Akt were increased in Pten mutant keratinocytes (Fig. 1B). Cre-mediated recombination in the prostate of Pten mutant males was investigated by PCR of the Pten-loxP locus. Amplification of an 849-bp PCR product specific for Pten deletion was detected in prostate tissue from PtenloxP/loxP; MMTV-cre and Ptenwt/loxP; MMTV-cre mice (Fig. 1C). Together, these data indicate that Cre-mediated recombination of the Pten allele occurred in the epidermis, prostate, and thymus of PtenloxP/loxP; MMTV-cre mice.
Fig. 1.
Targeted conditional deletion of Pten in murine skin, thymus, and prostate. (A) Southern-blot genotypic analysis for deletion of Pten in keratinocytes and thymus. Genomic DNA extracted from cultured keratinocytes and thymus tissue was digested with HindIII and hybridized with a probe situated upstream of exon 4 to show bands for wild-type (wt, 2-kb), nonrecombined (loxP, 6-kb), and recombined (Δ4-5, 2.3-kb) Pten alleles. DNA from keratinocytes and thymi is shown in Left and Right, respectively. Genotypes for each lane are as follows: 1 and 5, PtenloxP/loxP;2, Ptenwt/loxP;3,8,and9, PtenloxP/loxP; MMTV-cre; and 4 and 7, Ptenwt/loxP; MMTV-cre mice. Lane 9 is DNA from a thymic lymphoma. (B) Western blot analysis of cultured keratinocytes. Pten expression is lost in PtenloxP/loxP; MMTV-cre keratinocytes (Mut.) compared with Ptenwt/loxP; MMTV-cre control (Con.) keratinocytes. Weak expression of Pten in mutant samples indicates incomplete Cre-mediated recombination within the keratinocyte population. Phospho-PKB levels are elevated in PtenloxP/loxP; MMTV-cre keratinocytes compared with Ptenwt/loxP; MMTV-cre control keratinocytes under basal (10% FCS), serum-starved (0.5% FCS), and serum-stimulated conditions (Stim., starved and stimulated with 10% FCS). (C) PCR analysis for Cre-mediated recombination of the Pten locus (Δ4-5, Top), genotyping for Pten-loxP (Middle), and MMTV-cre (Bottom). Genomic DNA was isolated from the prostate of mice for PCR analysis, and products were run on an agarose gel. For Δ4-5 PCR, primers situated upstream of the 5′ loxP site and downstream of the 3′ loxP site amplified an 849-bp product only upon Cre-mediated deletion of Pten4-5. The Pten-loxP and wt alleles yielded products of 335 and 228 bp, respectively. Genotypes for each lane are as follows: 1, Ptenwt/loxP; 2, PtenloxP/loxP; 3–4, Ptenwt/loxP; MMTV-cre; and 5–8, PtenloxP/loxP; MMTV-cre mice. Positive control in lane 9 corresponds to PCR products from genomic DNA of Ptenwt/loxP; Gfap-cre brain tissue (16). Numbers on the left indicate size of DNA marker in base pairs.
Deletion of Pten was also detected in mammary tissue from PtenloxP/loxP; MMTV-cre mice (data not shown). A severe developmental defect was observed in mammary epithelium from these females and will be described elsewhere.
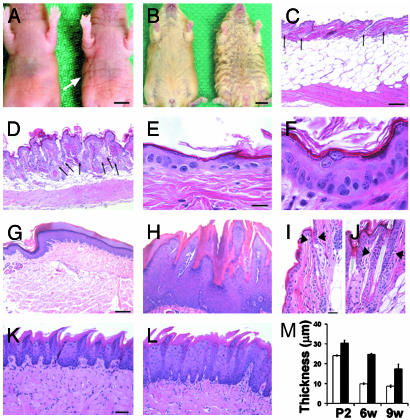
Epidermal Hyperplasia and Coat Abnormalities in PtenloxP/loxP; MMTV-cre Mice. Ptenlox/loxP; MMTV-cre mutant mice were viable and newborns appeared normal and healthy. However, by 7 days of age, skin on the ventral surface of PtenloxP/loxP; MMTV-cre animals appeared thickened and wrinkled compared with heterozygous Ptenwt/loxP; MMTV-cre controls (Fig. 2A). By P21, the coat of PtenloxP/loxP; MMTV-cre mice was shaggy and ruffled compared with the smooth coat on control mice (Fig. 2B). Appendages of Pten mutant mice such as ears and eyelids were thicker than those of control mice (data not shown).
Fig. 2.
Deletion of Pten in the skin results in skin, coat abnormalities, and hamartomas. (A) Skin on the abdomen of PtenloxP/loxP; MMTV-cre mice is more wrinkled (right, white arrow) than skin on Ptenwt/loxP; MMTV-cre control littermates (left) at postnatal day 7 (P7). (B) Coat of PtenloxP/loxP; MMTV-cre mice (right) is shaggy and ruffled compared with the smooth coat of Ptenwt/loxP; MMTV-cre littermate (left) at P21. (C–L) Hematoxylin and eosin staining. Epidermis of PtenloxP/loxP; MMTV-cre mice is papillary in appearance (D). Acanthosis with hyperkeratosis and hypergranulosis are apparent in Pten mutant skin (F). Skin from control mice is shown in C and E. Black arrows denote sebaceous glands, which were 61% more abundant in the dermis of Pten mutants compared with control skin by 3 wk (n = 4, P = 0.01). (G–L) Staining of paws (G and H), hair follicles (I and J), and tongue epithelium (K and L) of Ptenwt/loxP; MMTV-cre (G, I, and K) and PtenloxP/loxP; MMTV-cre (H, J, and L) animals. Epithelium of hair follicles and tongue of PtenloxP/loxP; MMTV-cre mice is several cell layers thicker (J and L) than controls (I and K). Arrowheads denote hair follicle epithelium. Acanthosis with hypergranulosis, hyperkeratosis, and parakeratosis are present within hamartomatous growths on paws of Pten mutant mice (H). Tissues are from 3-wk-old (C–F, I, and J) and 9- to 10-wk-old (G, H, K, and L) animals. (M) Average epidermal thickness in P2, 6- and 9-wk-old PtenloxP/loxP; MMTV-cre mice (black bars) was increased compared with Ptenwt/loxP; MMTV-cre mice (white bars, n = 3 for each genotype and age, P < 0.03). (Scale bar for A = 0.5 cm; B = 1 cm; C, D, G, and H = 100 μm; E and F = 10 μm; I and J = 20 μm; and K and L = 50 μm.)
Abnormalities in Pten mutant skin were further investigated by sectioning and histological staining. Epidermis from Pten mutant animals was acanthotic (hyperplastic) by P7 and appeared papillary by P21 (Fig. 2 D and F). Epidermis of Pten mutant skin was >2-fold thicker than controls by 6 wk (Fig. 2M). Hyperplastic changes in the Pten-null epidermis included hyperkeratosis and hypergranulosis (Fig. 2F and Fig. 5, which is published as supporting information). Sebaceous glands were more abundant in the dermis of Pten mutant compared with control skin (Fig. 2D). Whereas cells occasionally appeared hypertrophied in the Pten mutant epidermis (Fig. 2F and Fig. 5 D and F), cultured keratinocytes lacking Pten were only mildly enlarged (<10%, data not shown).
Acanthosis with hyperkeratosis and hypergranulosis were observed on the palmoplantar regions of paws from Pten mutant mice that were similar to the benign tumors on the hands and feet of patients with CD (Fig. 2H) (20, 21). Whereas no benign tumors were observed in hair follicles or tongue of Pten mutant mice, these tissues were mildly hyperplastic in the absence of Pten (Fig. 2 J and L). Thus, loss of Pten leads to development of palmoplantar hamartomas and hyperplasia of the hair follicle and tongue epithelium in mice.
A failure of keratinocytes to undergo terminal differentiation could result in excessive cell divisions and epidermal hyperplasia. The differentiation program of keratinocytes lacking Pten was intact in vivo (Fig. 5) and in vitro (data not shown). However, keratinocytes in the proliferative and differentiated cell layers of Pten mutant epidermis were each increased in number by >50% compared with controls (Fig. 5K). These data indicate that the epidermal hyperplasia in Pten mutant skin is composed of both proliferative and differentiated keratinocytes and is not caused by a failure to initiate terminal differentiation.
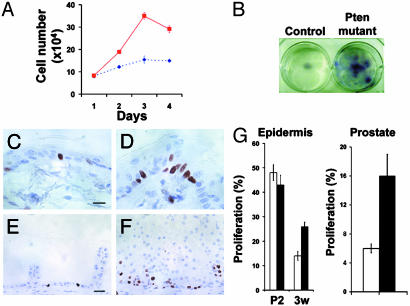
Increased Proliferation in Pten-Null Epidermis. The increased cellularity in the PtenloxP/loxP; MMTV-cre epidermis suggested an increase in proliferation. To determine whether loss of Pten conferred a growth advantage in these cells, primary keratinocytes were derived from PtenloxP/loxP; MMTV-cre and Ptenwt/loxP; MMTV-cre animals, seeded at low density, and grown in culture for several days. Pten mutant keratinocytes grew more rapidly than control cells (Fig. 3A). The majority of control keratinocytes have exhausted their proliferative potential and die by 21 days. However, many foci of Pten-null keratinocytes remained attached to the culture dish by this time point (Fig. 3B), indicating that the enhanced growth seen in Pten mutant keratinocytes is associated with a significant expansion of these cells over time. We investigated the proportion of proliferating and dead cells but were not able to detect a statistically significant difference in proliferation or viability in the presence or absence of Pten (data not shown). Therefore, we cannot determine the relative contributions of either proliferation or death to the enhanced growth of Pten-deficient cultured keratinocytes, and it is possible that effects on both of these phenomena contribute to this phenotype.
Fig. 3.
Increased proliferation of Pten-null keratinocytes and prostate. (A) Pten-null keratinocytes (solid line) outgrew Ptenwt/loxP; MMTV-cre keratinocytes (dotted line) (trypan blue exclusion). (B) Increased expansion of Pten-null keratinocytes compared with heterozygous controls over 21 days in culture (crystal violet stain). (C–F) Ki67 immunohistochemistry in epidermis (C and D) and prostate (E and F) of Ptenwt/loxP; MMTV-cre (C and E) and PtenloxP/loxP; MMTV-cre (D and F) mice. (G) Percentage of Ki67-positive basal keratinocytes (Left) is greater in Pten mutant mice (black bars, n = 3) by 3 wk compared with controls (white bars, n = 3, P = 0.01). Proliferation of basal keratinocytes from 2-day-old Pten mutant animals (black bar, n = 3) is not significantly different from controls (white bar, n = 3, P = 0.39). Pten mutant animals have a greater percentage of Ki67-positive prostate epithelial cells (Right; black bar, n = 3) compared with glands from control animals (white bar, n = 3, P = 0.05). (Scale bar for C and D = 10 μm; E and F = 20 μm.)
The ability of Pten to regulate keratinocyte proliferation in vivo was investigated by immunohistochemistry for proliferation marker Ki67 in the epidermis of Pten mutant and control animals. Coimmunohistochemistry for K14 and Ki67 demonstrate that proliferation is restricted to the basal layer in both control and Pten mutant mice (Fig. 5 I and J). However, basal keratinocytes of the Pten mutant epidermis had a near 2-fold increase in proliferation compared with control cells at 3 wk (Fig. 3G).
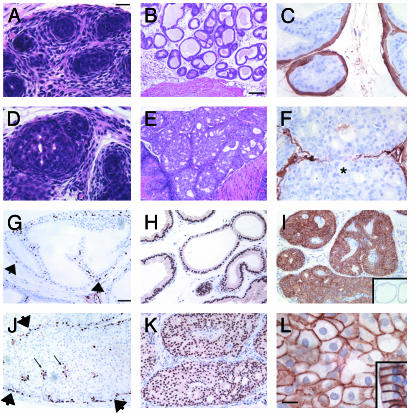
Complete Penetrance and Early Onset of High-Grade Neoplasia in Pten Mutant Prostate. To investigate the consequences of Pten loss in the prostate, developing and mature tissue was collected from Pten mutant males and histologically analyzed. Whereas development of the murine prostate gland begins late in embryogenesis, the majority of ductal morphogenesis in this organ occurs within the first 2 postnatal weeks (22, 23). Prostatic epithelium from six of nine PtenloxP/loxP; MMTV-cre animals was hyperplastic by 5 days of age compared with controls (Fig. 4D). By 2 wk, high-grade prostate intraepithelial neoplasia (HGPIN) and prostate carcinoma in situ (PCIS) were present in all Pten mutant males (n = 4/4, Table 1). HGPIN lesions were composed of a papillary proliferation of epithelial cells with cellular atypia. In PCIS, epithelial cells formed a solid bridge across the lumen that filled the glandular structure (Fig. 4E). Features of atypia evident in HGPIN and PCIS as early as 2 wk include hyperchromasia, prominent nucleoli, nuclear enlargement, and loss of polarity. From 3 wk onwards, additional features of atypia, including pleiomorphism and cell enlargement, were observed in these prostate lesions (Fig. 6B, which is published as supporting information). Surface area measurements demonstrated that prostate cells lacking Pten were >50% larger than control cells at 3 wk (Fig. 4L). Necrosis was frequently observed within the lumen of PCIS lesions (Fig. 6D), indicative of comedocarcinoma and higher grade of disease. Neoplasms originated from all regions of the prostate in Pten mutant animals, including the ventral, anterior, and dorsolateral lobes.
Fig. 4.
Prostate neoplasias in PtenloxP/loxP; MMTV-cre mice. Prostate glands from PtenloxP/loxP; MMTV-cre animals (D) are more cellular than control glands (A) by P5. (B) Glands from the prostate of a 3-wk control male are one to two cells thick with a hollow lumen. (E) PCIS from 3-wk Pten mutant animal. Neoplastic growth with cellular atypia (see text and Fig. 6B) fills the lumen. Smooth muscle forms a continuous layer around control glands (C). Neoplasm in Pten mutant prostate gland invades through smooth muscle (F, above asterisk). p63 is expressed in the nucleus of basal cells from control (G) and Pten mutant (J) glands (arrowheads). A minority of cells within the neoplastic growths of Pten-null prostate were p63-positive (arrows). Androgen receptor is expressed in luminal epithelial cells from Ptenwt/loxP; MMTV-cre prostate tissue (H) and in the majority of cells within prostate neoplasms of Pten mutant animals (K). (I) Up-regulation of phospho-PKB expression in Pten-null prostate epithelium compared with Ptenwt/loxP; MMTV-cre mice (I Inset). Surface area of Pten-null prostate cells (L; 62.3 ± 5.2 μm2, n = 4) was larger than control cells (L; Inset, 40.5 ± 1.5 μm2, n = 4, P = 0.007). Hematoxylin and eosin staining is shown in A, B, D, and E. Immunohistochemistry for smooth muscle actin is shown in C and F; p63 is shown in G and J. Androgen receptor is shown in H and K. Phospho-PKB is shown in I. Pan-cadherin is shown in L. (Scale bar for A and D = 20 μm, B and E = 100 μm, G–K = 50 μm, and L = 10 μm.)
Table 1. Neoplasias in prostate of PtenloxP/loxP; MMTV-cre mice.
| Control (Ptenwt/loxP; MMTV-cre)
|
PtenloxP/loxP; MMTV-cre
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Age, wk | Neoplasia | HGPIN | PCIS | Invasion | Neoplasia | HGPIN | PCIS | Invasion |
| 2* | 0/4 | 0/4 | 0/4 | 0/4 | 4/4 | 4/4 | 4/4 | 0/4 |
| 3 | 0/7 | 0/7 | 0/7 | 0/7 | 7/7 | 4/7 | 7/7 | 2/7 |
| 4–6 | 0/5 | 0/5 | 0/5 | 0/5 | 8/8 | 7/8 | 7/8 | 3/8 |
| 7–10 | 2/9 | 2/9 | 2/9 | 0/9 | 16/16 | 10/16 | 16/16 | 7/16 |
| 11–19 | 1/2 | 1/2 | 0/2 | 0/2 | 3/3 | 2/3 | 3/3 | 3/3 |
| Total | 3/27 | 3/27 | 2/27 | 0/27 | 38/38 | 27/38 | 37/38 | 15/38 |
| % | 11 | 11 | 7 | 0 | 100 | 71 | 97 | 39 |
Neoplasia includes all lesions showing HGPIN, PCIS, and/or invasion.
Genotypes of control mice 2 wk of age were Ptenwt/loxP; MMTV-cre, Ptenwt/loxP, or PtenloxP/loxP
Growths within the prostate of Pten mutant animals invaded surrounding tissues as early as 3 wk. Between 7 and 14 wk, focal invasion was observed in more than half (10/19) of PtenloxP/loxP; MMTV-cre males (Table 1 and Figs. 4F and 6C), signifying disease progression to cancer. Significantly, prostate neoplasias were completely penetrant in PtenloxP/loxP; MMTV-cre males (n = 38/38, Table 1). As expected from a previous study reporting a low incidence of prostate neoplasias in young Pten+/- males, prostate lesions were detected in 3/23 Ptenwt/loxP; MMTV-cre males 3–19 wk old (11).
Consistent with Pten's ability to negatively regulate PKB/Akt activity, phospho-PKB expression was elevated in Pten-null prostate compared with control glands in which expression was undetectable (Fig. 4I). Up-regulation of PKB activity in this organ was detected as early as P5 in PtenloxP/loxP; MMTV-cre males (data not shown).
Differentiation in Pten-Null Prostate. Prostatic epithelium consists mainly of basal and luminal cells (1). Whereas prostatic stem cells are believed to reside in the proliferative basal cell population, luminal cells are highly differentiated secretory cells. To determine the nature of neoplastic cells in the Pten mutant prostate, expression of differentiation markers was investigated by immunohistochemistry. The p63 gene product is selectively expressed in basal cells of epithelial tissues, including the prostate (24). Basal cells from the prostate of both Ptenwt/loxP; MMTV-cre and PtenloxP/loxP; MMTV-cre mice expressed p63 (Fig. 4 G and J). The vast majority of neoplastic cells within the lumen of Pten mutant glands did not express p63. However, a small percentage (<2%) of p63-positive cells was observed within these neoplastic foci. The androgen receptor (AR) is expressed in the nucleus of luminal epithelial cells from control prostate glands (Fig. 4H) (1). Cells within neoplastic lesions of PtenloxP/loxP; MMTV-cre prostates expressed nuclear AR (Fig. 4K). Together, these findings demonstrate that prostate epithelium can differentiate in the absence of Pten and that the prostate tumors were luminal in nature. In addition, basal and luminal cells were each increased by >3-fold in the Pten mutant glands compared with heterozygous controls (Fig. 6E). The increased cellularity in both the stem cell and differentiated populations of Pten mutant glands suggests that, similarly to the Pten-null epidermis, neoplasia results from expansion of the basal cell population.
Increased Proliferation in Pten-Null Prostate Epithelium. To determine whether increased proliferation contributes to neoplasia in the Pten mutant prostate, immunohistochemistry for Ki67 was performed. Similarly to keratinocytes in Pten mutant skin, the percentage of Ki67-positive epithelial cells was >2-fold greater in the prostates of PtenloxP/loxP; MMTV-cre males compared with heterozygous controls (Fig. 3 E–G). These findings indicate that Pten negatively regulates proliferation of the mouse prostate.
Thymic Lymphomas in PtenloxP/loxP; MMTV-cre Mice. All PtenloxP/loxP; MMTV-cre mice were sick or dead by 14 wk (Fig. 7, which is published as supporting information). Internal organs of Pten mutant mice were collected and analyzed for potential abnormalities. Thymic lymphomas were found in the majority of PtenloxP/loxP; MMTV-cre mice by 14 wk (Table 2, which is published as supporting information), and it is believed that these malignancies were responsible for the premature mortality of these animals. The development of lymphomas upon loss of Pten in T-lymphocytes by this age is in agreement with previous studies (13, 25). The early mortality of Pten mutant mice due to the incidence of lymphomas precluded analyses of these mice for progression of malignancies affecting the skin and prostate past 14 wk. Skin, breast, and thyroid tumors were not observed in PtenloxP/loxP; MMTV-cre mice before they succumbed to lymphomas. In addition, prostate tumors did not metastasize to the internal organs of the Pten mutant males, and bone metastases are highly unlikely, given the focally invasive nature of these tumors by the time these animals died of lymphoid malignancy.
Discussion
Pten heterozygous mice are highly susceptible to tumor development (4, 12). Loss of heterozygosity for Pten in these tumors implies that homozygous inactivation of Pten is necessary for development of neoplasias. Analysis of mice with tissue-specific deletion of both Pten alleles revealed that homozygous inactivation of Pten results in hyperproliferation and neoplasia in lymphoid tissues, gonads, breast, and neural stem cells (13–15, 26–28). Neoplastic changes in Pten-null lymphocytes, keratinocytes, neural cells, and mammary epithelial cells are also associated with a resistance to apoptotic stimuli. Consequently, a high frequency of tumors arise from Pten-null T lymphocytes, breast, and germ cells. These studies demonstrate a physiological role for Pten in regulation of proliferation and apoptosis. However, Pten nullizygosity does not result in neoplastic proliferation of all cell types. Deletion of Pten in terminally differentiated brain and heart cells causes enlargement of these organs as a consequence of increased cell size (28, 29). Thus, consistent with earlier studies in Drosophila, Pten functions as an important regulator of cell and organ size (28).
In the current study, we report that inactivation of Pten in mouse skin and prostate results in neoplasia. Overgrowth of these organs was the result of increased proliferation and, in the prostate, increased cell size. Potential defects in apoptosis are unlikely to contribute to the phenotypes described in Pten mutant skin and prostate because basal rates of apoptosis are negligible in these tissues at the tested times (30, 31). Significantly, the prostate and skin lesions in PtenloxP/loxP; MMTV-cre mice have early onset and are completely penetrant, indicating that loss of Pten is sufficient for neoplastic proliferation of these tissues.
Development and growth of the epidermis and prostate are controlled by components of PI3K signaling. Overexpression of transforming growth factor α (TGF-α) or insulin-like growth factor 1 (IGF-1) in mouse keratinocytes results in wrinkled skin and epidermal hyperplasia as well as coat abnormalities comparable to those of Pten mutant mice (32, 33). Significantly, these transgenic mice develop benign skin tumors with age. In contrast, mice lacking EGF receptor, IGF-1 receptor, Fgfr2-IIIB, or both Akt1 and Akt2 have a thin epidermis because of a reduction in proliferation (34–36). In the prostate, overexpression experiments have implicated Igf-1, TGF-α, Fgf8b, and PKB/Akt in development and tumorigenesis of this gland (1, 37, 38). Further, growth of Igf-1-null prostate epithelium is impaired (37). Given Pten's ability to antagonize growth factor-mediated PI3K signaling, up-regulation of PI3K-PKB/Akt signaling likely contributes to neoplasias in Pten mutant skin and prostate. Consistent with this notion, increased PKB phosphorylation was observed in both Pten-null keratinocytes and prostate.
Whereas PTEN mutations are rare in human basal and squamous cell carcinomas, loss of PTEN activity and up-regulation of PI3K signaling occurs during mouse skin carcinogenesis (21, 39). Cutaneous tumors were not observed in PtenloxP/loxP; MMTV-cre mice by the time they died of thymic lymphomas at 3 months of age. The absence of skin tumors in young Pten mutant mice is in agreement with a recent study in which Pten was deleted specifically in keratinocytes by the use of the keratin 5 promoter-driven Cre (40). These mice developed squamous papillomas and squamous cell carcinomas only by 4–8 months of age.
The breast, endometrial, and thyroid tumors that arise in Pten heterozygous mice overlap with malignancies of certain CD patients (7, 11, 12). Hamartomas characteristic of CD are present in some organs of Pten heterozygous mice such as the intestine, but not in the skin or brain (11, 12). We and others previously reported that loss of both Pten alleles is sufficient for development of a mouse phenocopy of Lhermitte–Duclos disease, a rare neural manifestation of CD characterized by macrocephaly and hypertrophied cerebellar neurons (16, 41). Hamartomas originating from mucocutaneous tissues are found in >90% of CD patients, and there is limited evidence that PTEN loss of heterozygosity occurs in these growths (7, 42). It is therefore possible that loss of the wild-type PTEN allele is sufficient for development of these growths. Indeed, benign keratoses similar to those that develop on the hands and feet of CD patients were found on the paws and feet of PtenloxP/loxP; MMTV-cre mice (20, 21). Whereas benign tumors were not found in other mucocutaneous tissues of Pten mutant mice such as the hair follicle and oral cavity, hyperplasia was detected at these sites. These findings indicate that, in mice, loss of Pten is sufficient for hyperproliferation of mucocutaneous tissues affected in CD patients. Together, these data imply that loss of both PTEN alleles is involved in mucocutaneous hamartoma formation in humans with CD and suggest that PtenloxP/loxP; MMTV-cre mice may offer valuable insight into this manifestation of CD. Interestingly, the spectrum of tissues predisposed to tumor development in CD patients does not include the thymus or prostate as is seen in the PtenloxP/loxP; MMTV-cre mice. However, PTEN mutations have been described in sporadic human prostate tumors (3).
Despite the high rate of loss of heterozygosity on various chromosomes in prostate cancer, few loss-of-function animal models of this disease exist. Mice deficient in homeobox gene Nkx3.1 or retinoid receptor RXRα develop HGPINs by 1 year of age (37). The long latency period for tumor development implies that additional genetic mutations are required for neoplasia to occur in the prostate in these models. The high frequency of chromosome 10 deletions and PTEN mutations in human prostate cancers suggests that inactivation of this tumor suppressor is an important step during prostate tumorigenesis. In mice, Pten haploinsufficiency accelerates tumorigenesis in various prostate cancer-prone backgrounds (43–45). The age of onset of prostate neoplasias in Nkx3.1-/-, p27-/-, and transgenic adenocarcinoma of mouse prostate (TRAMP) mice is reduced in a Pten heterozygous background. However, prostate lesions still take several months to develop in these compound mutants.
In this study, we report that inactivation of Pten alone results in high-grade prostate neoplasia within only 2 wk of age. Prostate tissue from PtenloxP/loxP; MMTV-cre neonates is hyperplastic and progresses to carcinoma in situ before development of this gland is complete (23), and these lesions frequently become invasive. The only other existing animal model with a prostate phenotype comparable to that of the Pten mutant mice is the TRAMP transgenic mice, which rely on dysregulation of multiple signaling molecules (37). In contrast to the TRAMP mice, the secretory nature of prostate tumors in the Pten mutant animals resembles that of human prostate adenocarcinomas (24).
Importantly, the early onset and complete penetrance of prostate lesions in the PtenloxP/loxP; MMTV-cre mice indicate that inactivation of Pten alone is sufficient for the initiation of prostate cancer. The incidence of thymic lymphomas within the first 3 months of life in the PtenloxP/loxP; MMTV-cre mice limits the study of prostate tumor metastases in this model. Conditional mutants in which deletion of Pten is targeted specifically to the prostate have recently been reported (46), and these Pten mutant animals develop metastatic prostate lesions with age. Prostate-specific Pten conditional mutants of this nature will also be applicable for therapy and prevention trials. Together, analyses of the various conditional Pten-knockout mice indicate that this gene is a critical regulator of proliferation in the mouse prostate and that maintenance of Pten signaling is of paramount importance to prevent initiation and progression of prostate tumorigenesis.
The phenotypes described in the PtenloxP/loxP; MMTV-cre mice demonstrate an important role for Pten in tissue homeostasis of the skin and prostate. Further study of conditional Pten mutant animals will help to advance our understanding of Pten signaling, growth control, and tumorigenesis in these organs.
Supplementary Material
Acknowledgments
We thank G. Melino, E. Candi, C. Abate-Shen, K. Economides, C. Cheung, D. Sauder, I. Freed, B. Wong, and C. Rosen for helpful comments and thoughtful discussions; D. Wakeham, D. Bouchard, and K. Watson for technical assistance; and M. Cully for critical review of the manuscript. This work was supported by the Canadian Breast Cancer Initiative. S.A.B. holds an Ontario Graduate Scholarship.
Abbreviations: CD, Cowden's disease; PI3K, phosphatidylinositol 3-kinase; PKB, protein kinase B; Pn, postnatal day n; HGPIN, high-grade prostate intraepithelial neoplasia; PCIS, prostate carcinoma in situ.
References
- 1.Abate-Shen, C. & Shen, M. M. (2000) Genes Dev. 14, 2410-2434. [DOI] [PubMed] [Google Scholar]
- 2.Parker, S. L., Tong, T., Bolden, S. & Wingo, P. A. (1996) CA Cancer J. Clin. 46, 5-27. [DOI] [PubMed] [Google Scholar]
- 3.Ittmann, M. M. (1998) Oncol. Rep. 5, 1329-1335. [DOI] [PubMed] [Google Scholar]
- 4.Stambolic, V., Mak, T. W. & Woodgett, J. R. (1999) Oncogene 18, 6094-6103. [DOI] [PubMed] [Google Scholar]
- 5.Ali, I. U., Schriml, L. M. & Dean, M. (1999) J. Natl. Cancer Inst. 91, 1922-1932. [DOI] [PubMed] [Google Scholar]
- 6.Vlietstra, R. J., van Alewijk, D. C., Hermans, K. G., van Steenbrugge, G. J. & Trapman, J. (1998) Cancer Res. 58, 2720-2723. [PubMed] [Google Scholar]
- 7.Eng, C. (2003) Hum. Mutat. 22, 183-198. [DOI] [PubMed] [Google Scholar]
- 8.Maehama, T. & Dixon, J. E. (1998) J. Biol. Chem. 273, 13375-13378. [DOI] [PubMed] [Google Scholar]
- 9.Stambolic, V., Suzuki, A., de la Pompa, J. L., Brothers, G. M., Mirtsos, C., Sasaki, T., Ruland, J., Penninger, J. M., Siderovski, D. P. & Mak, T. W. (1998) Cell 95, 29-39. [DOI] [PubMed] [Google Scholar]
- 10.Sun, H., Lesche, R., Li, D. M., Liliental, J., Zhang, H., Gao, J., Gavrilova, N., Mueller, B., Liu, X. & Wu, H. (1999) Proc. Natl. Acad. Sci. USA 96, 6199-6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Podsypanina, K., Ellenson, L. H., Nemes, A., Gu, J., Tamura, M., Yamada, K. M., Cordon-Cardo, C., Catoretti, G., Fisher, P. E. & Parsons, R. (1999) Proc. Natl. Acad. Sci. USA 96, 1563-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stambolic, V., Tsao, M. S., Macpherson, D., Suzuki, A., Chapman, W. B. & Mak, T. W. (2000) Cancer Res. 60, 3605-3611. [PubMed] [Google Scholar]
- 13.Suzuki, A., Yamaguchi, M. T., Ohteki, T., Sasaki, T., Kaisho, T., Kimura, Y., Yoshida, R., Wakeham, A., Higuchi, T., Fukumoto, M., et al. (2001) Immunity 14, 523-534. [DOI] [PubMed] [Google Scholar]
- 14.Kimura, T., Suzuki, A., Fujita, Y., Yomogida, K., Lomeli, H., Asada, N., Ikeuchi, M., Nagy, A., Mak, T. W. & Nakano, T. (2003) Development (Cambridge, U.K.) 130, 1691-1700. [DOI] [PubMed] [Google Scholar]
- 15.Li, G., Robinson, G. W., Lesche, R., Martinez-Diaz, H., Jiang, Z., Rozengurt, N., Wagner, K. U., Wu, D. C., Lane, T. F., Liu, X., et al. (2002) Development (Cambridge, U.K.) 129, 4159-4170. [DOI] [PubMed] [Google Scholar]
- 16.Backman, S. A., Stambolic, V., Suzuki, A., Haight, J., Elia, A., Pretorius, J., Tsao, M. S., Shannon, P., Bolon, B., Ivy, G. O. & Mak, T. W. (2001) Nat. Genet. 29, 396-403. [DOI] [PubMed] [Google Scholar]
- 17.Wagner, K. U., McAllister, K., Ward, T., Davis, B., Wiseman, R. & Hennighausen, L. (2001) Transgenic Res. 10, 545-553. [DOI] [PubMed] [Google Scholar]
- 18.Bierie, B., Nozawa, M., Renou, J. P., Shillingford, J. M., Morgan, F., Oka, T., Taketo, M. M., Cardiff, R. D., Miyoshi, K., Wagner, K. U., et al. (2003) Oncogene 22, 3875-3887. [DOI] [PubMed] [Google Scholar]
- 19.Beato, M. (1991) Crit. Rev. Oncog. 2, 195-210. [PubMed] [Google Scholar]
- 20.Brownstein, M. H., Mehregan, A. H., Bikowski, J. B., Lupulescu, A. & Patterson, J. C. (1979) Br. J. Dermatol. 100, 667-673. [DOI] [PubMed] [Google Scholar]
- 21.Fistarol, S. K., Anliker, M. D. & Itin, P. H. (2002) Eur. J. Dermatol. 12, 411-421. [PubMed] [Google Scholar]
- 22.Sugimura, Y., Cunha, G. R. & Donjacour, A. A. (1986) Biol. Reprod. 34, 961-971. [DOI] [PubMed] [Google Scholar]
- 23.Marker, P. C., Donjacour, A. A., Dahiya, R. & Cunha, G. R. (2003) Dev. Biol. 253, 165-174. [DOI] [PubMed] [Google Scholar]
- 24.Signoretti, S., Waltregny, D., Dilks, J., Isaac, B., Lin, D., Garraway, L., Yang, A., Montironi, R., McKeon, F. & Loda, M. (2000) Am. J. Pathol. 157, 1769-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki, A., de la Pompa, J. L., Stambolic, V., Elia, A. J., Sasaki, T., del Barco Barrantes, I., Ho, A., Wakeham, A., Itie, A., Khoo, W., et al. (1998) Curr. Biol. 8, 1169-1178. [DOI] [PubMed] [Google Scholar]
- 26.Anzelon, A. N., Wu, H. & Rickert, R. C. (2003) Nat. Immunol. 4, 287-294. [DOI] [PubMed] [Google Scholar]
- 27.Suzuki, A., Kaisho, T., Ohishi, M., Tsukio-Yamaguchi, M., Tsubata, T., Koni, P. A., Sasaki, T., Mak, T. W. & Nakano, T. (2003) J. Exp. Med. 197, 657-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Backman, S., Stambolic, V. & Mak, T. (2002) Curr. Opin. Neurobiol. 12, 516-522. [DOI] [PubMed] [Google Scholar]
- 29.Crackower, M. A., Oudit, G. Y., Kozieradzki, I., Sarao, R., Sun, H., Sasaki, T., Hirsch, E., Suzuki, A., Shioi, T., Irie-Sasaki, J., et al. (2002) Cell 110, 737-749. [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez-Villanueva, J., Greenhalgh, D., Wang, X. J., Bundman, D., Cho, S., Delehedde, M., Roop, D. & McDonnell, T. J. (1998) Oncogene 16, 853-863. [DOI] [PubMed] [Google Scholar]
- 31.Bruckheimer, E. M., Brisbay, S., Johnson, D. J., Gingrich, J. R., Greenberg, N. & McDonnell, T. J. (2000) Oncogene 19, 5251-5258. [DOI] [PubMed] [Google Scholar]
- 32.Vassar, R. & Fuchs, E. (1991) Genes Dev. 5, 714-727. [DOI] [PubMed] [Google Scholar]
- 33.Bol, D. K., Kiguchi, K., Gimenez-Conti, I., Rupp, T. & DiGiovanni, J. (1997) Oncogene 14, 1725-1734. [DOI] [PubMed] [Google Scholar]
- 34.Fuchs, E. & Raghavan, S. (2002) Nat. Rev. Genet. 3, 199-209. [DOI] [PubMed] [Google Scholar]
- 35.Liu, J. P., Baker, J., Perkins, A. S., Robertson, E. J. & Efstratiadis, A. (1993) Cell 75, 59-72. [PubMed] [Google Scholar]
- 36.Peng, X. D., Xu, P. Z., Chen, M. L., Hahn-Windgassen, A., Skeen, J., Jacobs, J., Sundararajan, D., Chen, W. S., Crawford, S. E., Coleman, K. G. & Hay, N. (2003) Genes Dev. 17, 1352-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Powell, W. C., Cardiff, R. D., Cohen, M. B., Miller, G. J. & Roy-Burman, P. (2003) Curr. Drug Targets 4, 263-279. [DOI] [PubMed] [Google Scholar]
- 38.Majumder, P. K., Yeh, J. J., George, D. J., Febbo, P. G., Kum, J., Xue, Q., Bikoff, R., Ma, H., Kantoff, P. W., Golub, T. R., Loda, M. & Sellers, W. R. (2003) Proc. Natl. Acad. Sci. USA 100, 7841-7846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Segrelles, C., Ruiz, S., Perez, P., Murga, C., Santos, M., Budunova, I. V., Martinez, J., Larcher, F., Slaga, T. J., Gutkind, J. S., et al. (2002) Oncogene 21, 53-64. [DOI] [PubMed] [Google Scholar]
- 40.Suzuki, A., Itami, S., Ohishi, M., Hamada, K., Inoue, T., Komazawa, N., Senoo, H., Sasaki, T., Takeda, J., Manabe, M., et al. (2003) Cancer Res. 63, 674-681. [PubMed] [Google Scholar]
- 41.Kwon, C. H., Zhu, X., Zhang, J., Knoop, L. L., Tharp, R., Smeyne, R. J., Eberhart, C. G., Burger, P. C. & Baker, S. J. (2001) Nat. Genet. 29, 404-411. [DOI] [PubMed] [Google Scholar]
- 42.Trojan, J., Plotz, G., Brieger, A., Raedle, J., Meltzer, S. J., Wolter, M. & Zeuzem, S. (2001) J. Invest. Dermatol. 117, 1650-1653. [DOI] [PubMed] [Google Scholar]
- 43.Kwabi-Addo, B., Giri, D., Schmidt, K., Podsypanina, K., Parsons, R., Greenberg, N. & Ittmann, M. (2001) Proc. Natl. Acad. Sci. USA 98, 11563-11568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Di Cristofano, A., De Acetis, M., Koff, A., Cordon-Cardo, C. & Pandolfi, P. P. (2001) Nat. Genet. 27, 222-224. [DOI] [PubMed] [Google Scholar]
- 45.Kim, M. J., Cardiff, R. D., Desai, N., Banach-Petrosky, W. A., Parsons, R., Shen, M. M. & Abate-Shen, C. (2002) Proc. Natl. Acad. Sci. USA 99, 2884-2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, S., Gao, J., Lei, Q., Rozengurt, N., Pritchard, C., Jiao, J., Thomas, G. V., Li, G., Roy-Burman, P., Nelson, P. S., et al. (2003) Cancer Cell 4, 209-221. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.