Summary
To reconstruct modern human evolutionary history and identify loci that have shaped hunter-gatherer adaptation, we sequenced the whole-genomes of five individuals in each of three different hunter-gatherer populations at > 60x coverage: Pygmies from Cameroon and Khoesan-speaking Hadza and Sandawe from Tanzania. We identify 13.4 million variants, substantially increasing the set of known human variation. We found evidence of archaic introgression in all three populations and the distribution of time to most recent common ancestors from these regions is similar to that observed for introgressed regions in Europeans. Additionally, we identify numerous loci that harbor signatures of local adaptation, including genes involved in immunity, metabolism, olfactory and taste perception, reproduction, and wound healing. Within the Pygmy population, we identify multiple highly differentiated loci that play a role in growth and anterior pituitary function and are associated with height.
Introduction
Due to recent advances in sequencing technologies, whole-genome sequencing of multiple individuals in multiple populations is now feasible (Henn et al., 2010). This burgeoning field of population genomics allows inference of demographic history and natural selection that is free of SNP ascertainment bias (Luikart et al., 2003; Teo et al., 2010). Initial efforts of the 1000 Genomes Project have already identified millions of variants in diverse populations (1000 Genomes Project Consortium, 2010). However, whole-genome sequencing in the 1000 Genomes Project has generally been at low coverage, and genetic diversity in many ethnically diverse populations is yet to be characterized, particularly with respect to Africa, the ancestral homeland of all modern humans (Campbell and Tishkoff, 2010). Furthermore, differences in diet, climate, and exposure to pathogens among ethnically and geographically diverse African populations are likely to have produced distinct selection pressures, resulting in local genetic adaptations. Despite the important role that African populations have played in human evolutionary history, they remain one of the most understudied groups in human genomics. To date, few high coverage African genomes have been analyzed in published studies: four Yoruba, one !Xhosa, and one San (1000 Genomes Project Consortium, 2010; Bentley et al., 2008; Drmanac et al., 2010; Schuster et al., 2010).
A comprehensive knowledge of levels and patterns of variation in African genomes is critical for a deeper understanding of human genetic diversity, the allelic spectrum of functionally important genetic variation, including disease susceptibility, the genetic basis of adaptation to diverse environments, and the origins of modern humans. Previous analyses of mtDNA, Y-chromosome, and autosomal genetic lineages in African hunter-gatherer populations indicate that they have some of the deepest divergence times of our species (Tishkoff et al., 2007; Tishkoff et al., 2009; Veeramah et al., 2012). The forest-dwelling Pygmies of Cameroon include the Baka, Bakola, and Bedzan populations. As a result of the expansion of Bantu speaking agriculturalists within the last few thousand years into Pygmy territories, Cameroonian Pygmy populations are highly admixed (Jarvis et al., 2012) and the ancestral pygmy language has been lost (Berniell-Lee et al., 2009; Verdu et al., 2009). The Hadza and Sandawe populations live in Tanzania and speak languages that contain click-consonants and are classified as Khoesan, but these languages are highly divergent from each other and from southern African San Khoesan-speaking populations (Sands, 1998). At present, little is known about the genetic relationships between click-speaking populations (Henn et al., 2011; Tishkoff et al., 2007; Tishkoff et al., 2009; Veeramah et al., 2012). The Sandawe currently number greater than 30,000 individuals, and following the Bantu expansion, many Sandawe have switched from nomadic hunting to an agricultural subsistence pattern (Newman, 1970). By contrast, the Hadza have a current population size of ~1000 individuals, the vast majority of which still practice a hunter-gatherer lifestyle (Blurton Jones et al., 1992; Marlowe, 2010). Data from autosomes, mtDNA, and the Y chromosome indicate that although the Hadza, Sandawe, Pygmy, and San populations have an ancient divergence, they also share lineages that are rare in other populations (Behar et al., 2008; Henn et al., 2011; Tishkoff et al., 2007; Tishkoff et al., 2009).
To extend our knowledge of genomic variation in ethnically diverse Africans, we sequenced the genomes of five males from each of three African hunter-gatherer populations (Western Pygmy, Hadza, and Sandawe) at high coverage (Figure 1A). We then compared these genome sequences to a previously published genome sequence from a San hunter-gatherer (Schuster et al., 2010) and to publically available whole sequence data from other ethnically, linguistically, and geographically diverse African populations (http://www.completegenomics.com/sequence-data/download-data/).
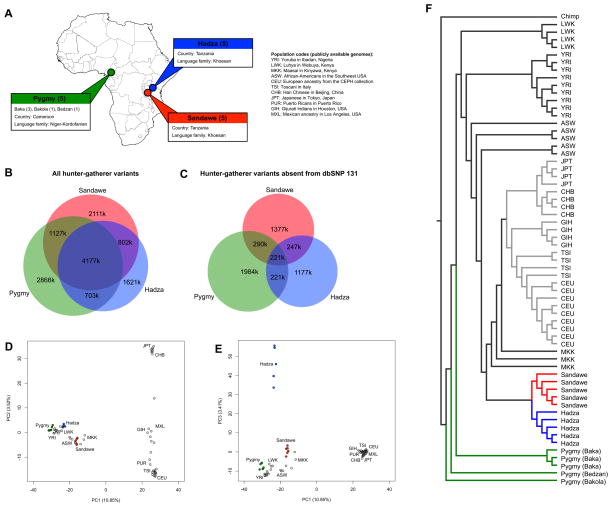
Figure 1. Genomic variation in African hunter-gatherers and other global populations.
A) Hunter-gatherer populations sequenced in our study (five males per population). HapMap abbreviations of publicly available genomes are also listed. A and B) Numbers indicate how many variants belong to each subset of populations. C and D) Principal component analysis of 68 high-coverage genomes. Pygmy genomes are indicated by green, Hadza by blue, Sandawe by red, and non-hunter-gatherer genomes by gray circles. E) Neighbor joining tree based on pairwise identity-by-state matrix distances using high-coverage whole-genome sequences from 68 individuals. See also Table S1, Table S2, Figure S2, and Figure S4.
Results
Whole-genome Sequencing
Whole-genome sequencing from 15 African hunter-gatherers was performed by Complete Genomics (Drmanac et al., 2010). We obtained approximately 60-fold coverage per genome, resulting in high confidence calls of variants from 95% of each genome (Table 1, see Supplemental Information). These genomes were compared to publically available high-coverage genomes sequenced and analyzed using the same technology and software in a diverse panel of 53 unrelated individuals (including 4 Luhya from Kenya, 4 Maasai from Kenya, 10 Yoruba from Nigeria, and 51 non-Africans, see Table S1), allowing the genomes of African hunter-gatherers to be placed within a global context.
Table 1. Summary statistics for whole-genome sequences.
Mean values ± one standard deviation are listed for each population. See also Figure S1.
| Statistic | Pygmy | Hadza | Sandawe |
|---|---|---|---|
| Number of individuals | 5 | 5 | 5 |
| Median coverage per genome | 63.4x ± 3.4x | 67.3x ± 17.8x | 61.6x ± 6.2x |
| Called genome fraction | 94.8% ± 0.7% | 96.1% ± 0.8% | 95.1% ± 1.0% |
| Ti/Tv ratio | 2.16 ± 0.01 | 2.14 ± 0.01 | 2.16 ± 0.01 |
| Variant SNPs per genome | 3.75×106 ± 0.04×106 | 3.54×106 ± 0.06×106 | 3.53×106 ± 0.05×106 |
| Homozygous variant SNP fraction | 34.04% ± 0.39% | 37.7% ± 2.7% | 33.2% ± 0.60% |
| Exonic SNP fraction | 0.621% ± 0.007% | 0.612% ± 0.008% | 0.617% ± 0.011% |
| Insertions | 1.26×105 ± 0.02×105 | 1.23×105 ± 0.03×105 | 1.19×105 ± 0.3×105 |
| Deletions | 1.49×105 ± 0.34×105 | 1.43×105 ± 0.03×105 | 1.39×105 ± 0.04×105 |
| Coding indel fraction | 0.116% ± 0.003% | 0.115% ± 0.006% | 0.118% ± 0.005% |
After applying stringent quality control filters (Supplemental Information, Figure S1) we identified 13,407,517 variants (SNPs, insertions, and deletions that differ from the human genome reference sequence, GRCh37/hg19) in the 15 hunter-gatherer genomes. In addition, we sequenced two Hadza genomes as technical replicates, and found approximately 28,000 discordant calls per genome pair, corresponding to less than one error per 100 kb, consistent with previous estimates (Drmanac et al., 2010; Lam et al., 2012). We also assessed sequence accuracy by comparing calls between whole-genome sequencing and the Illumina1M-Duo BeadChip genotyping array. Over 99.96% of calls were identical between platforms, and a large proportion (>34%) of discordant SNPs involved known tri-allelic loci.
The majority of variants are SNPs (90.9%), and we observe a greater number of deletions than insertions relative to the human reference genome (Table 1). We operationally define novel variants as those that are absent from build 131 of dbSNP. A considerable proportion of hunter-gatherer variants are absent from dbSNP 131 (41.1%, or 5,516,366 variants). Cross-referencing with the October 2011 release of the 1000 genomes project reveals that 3,062,541 variants remain absent, and thus our sequence data substantially expands the catalog of human genetic variation. Approximately half (50.8%) of the variants we identify are shared among multiple hunter-gatherer populations, and genomic regions that contain a large number of variants in one population also contain a large number of variants in other populations (Figure 1B, Figure S2). However, despite shared language and geographic proximity, we do not observe an excess of shared variants between Hadza and Sandawe Khoesan speakers, consistent with an ancient divergence and diverse mtDNA haplogroups (Table S2).
Functional Classification of Variants
Of the 13.4 million variants, 3.69 million are intronic, 37,797 are nonsynonymous, and 35,747 are synonymous variants. Moreover, 674,808 variants were located in DNase I footprints (Rosenbloom et al., 2012), and 149,072 variants occurred in cis-regulatory motifs within footprints. At the individual level, each hunter-gatherer genome contains approximately 11,500 nonsynonymous variants, 12,400 synonymous variants, and 256,000 variants in DNase I footprints. Using PolyPhen-2 classifications (Adzhubei et al., 2010), we find that ~60% of amino acid-changing variants identified in the hunter-gatherer genomes are classified as benign, ~25% as possibly damaging, and ~15% as probably damaging. In addition, benign amino acid changes are >2.7 times more likely to be found in all three hunter-gatherer populations than possibly damaging or probably damaging changes (p < 10−10, Z-test).
Comparing African and non-African genomes, we confirm that non-African populations contain a slight excess in the proportion of probably damaging nonsynonymous variants (Figure S3), consistent with population bottlenecks due to migration out-of-Africa and with prior studies (Lohmueller et al., 2008). By contrast, proportions of synonymous and nonsynonymous SNPs and the predicted number of probably damaging sites are similar for African hunter-gatherers and other African populations (Figure S3). Thus, at this broad genomic scale, natural selection appears to shape the genomes of hunter-gatherers similarly to the genomes of other African populations. However, analyses of larger sample sizes will be required to distinguish subtle differences that may exist.
Population Genetics of Functionally Important Regions of the Human Genome
The proportion of polymorphic sites, as estimated by θ, are lowest for exons in all three populations, suggesting that natural selection acts on coding sequences to reduce genetic variation (Table 2). θ is lower for introns than intergenic regions, a finding that is consistent with both background selection and positive selective sweeps. In all three hunter-gatherer populations, the mean value of Tajima’s D is lower for genic regions. This observation reflects allele frequency distributions that are shifted towards rare alleles in genic regions, a pattern that can be explained by both selective sweeps and purifying selection. Consistent with findings from HapMap Phase II data (Barreiro et al., 2008), we find that mean values of FST, a measure of allele frequency differentiation between populations, are lower for exons and higher for introns and intergenic regions. We also find that 5′ and 3′ untranslated regions have intermediate derived allele frequencies (DAF), θ, Tajima’s D, and FST, consistent with evolutionary constraint in regulatory regions. Furthermore, consistent with purifying selection against slightly deleterious mutations, the distribution of derived allele frequencies for exon SNPs are skewed towards low frequency alleles relative to intergenic SNPs, and the magnitude of this shift was similar for African and non-African populations (Figure S3, see Supplemental Information).
Table 2. Population genetic statistics for different types of variants.
Statistics are for fully called autosomal variants. Mean DAF refers to mean derived allele frequency after correcting for CpG hypermutability and biased gene conversion. See also Table S4, Figure S3, Figure S6, and Figure S5.
| Statistic | Intergenic | 10kb Upstream | 5′ UTR | Exon | Intron | 3′ UTR | 10kb Downstream | Overall |
|---|---|---|---|---|---|---|---|---|
| Within-population | ||||||||
| θper base pair (Pygmy) | 0.001132 | 0.000783 | 0.000722 | 0.000503 | 0.000860 | 0.000800 | 0.000845 | 0.000966 |
| θper base pair (Hadza) | 0.000921 | 0.000630 | 0.000564 | 0.000390 | 0.000691 | 0.000639 | 0.000681 | 0.000782 |
| θper base pair (Sandawe) | 0.001056 | 0.000728 | 0.000663 | 0.000463 | 0.000797 | 0.000743 | 0.000789 | 0.000899 |
| Mean DAF (Pygmy) | 0.2785 | 0.2756 | 0.2701 | 0.2527 | 0.2728 | 0.2648 | 0.2748 | 0.2760 |
| Mean DAF (Hadza) | 0.3239 | 0.3204 | 0.3092 | 0.3015 | 0.3191 | 0.3198 | 0.3222 | 0.3219 |
| Mean DAF (Sandawe) | 0.2912 | 0.2883 | 0.2791 | 0.2700 | 0.2855 | 0.2814 | 0.2889 | 0.2888 |
| Tajima’s D (Pygmy) | −0.4124 | −0.4350 | −0.4977 | −0.5955 | −0.4411 | −0.5010 | −0.4328 | −0.4272 |
| Tajima’s D (Hadza) | −0.0175 | −0.0145 | −0.0347 | −0.0918 | −0.0126 | −0.0300 | −0.0129 | −0.0148 |
| Tajima’s D (Sandawe) | −0.3285 | −0.3587 | −0.4549 | −0.5030 | −0.3627 | −0.4172 | −0.3567 | −0.3453 |
| Between-population | ||||||||
| FST (Pygmy, Hadza) | 0.0727 | 0.0723 | 0.0713 | 0.0681 | 0.0727 | 0.0714 | 0.0722 | 0.0726 |
| FST (Pygmy,Sandawe) | 0.0485 | 0.0482 | 0.0465 | 0.0448 | 0.0483 | 0.0473 | 0.0478 | 0.0483 |
| FST (Hadza,Sandawe) | 0.0659 | 0.0654 | 0.0643 | 0.0627 | 0.0653 | 0.0647 | 0.0625 | 0.0657 |
In addition, we investigated functional constraint for different site types by calculating the neutrality index (NI) which contrasts the levels of polymorphism and divergence of a putatively neutral class of sites to a class of sites that may be subject to selection (Rand and Kann, 1996). We used intergenic sites that were at least 50 kb from protein-coding genes as our neutral class, and calculated the NI with respect to nonsynonymous, synonymous, intronic, and DNase I footprint sites. Weak purifying selection was found for all of these sites (as defined by a NI significantly greater than 1; Table S4). Strikingly, sites classified as DNAse I footprints were more constrained (NI = 1.302, 95% CI = 1.298 – 1.306) than nonsynonymous sites (NI = 1.153, 95% CI = 1.140 – 1.167), consistent with the hypothesis that DNAse I footprints are enriched for functionally important regulatory variants.
Demographic History of African Hunter-Gatherers
Principal component analysis (PCA) reveals both continental and population-specific patterns of genetic variation. PC1 distinguishes Africans from non-Africans (with East African populations being closer to non-Africans), and PC2 differentiates Asian and European populations (Figure 1D). The Hadza are differentiated by PC3, and subsequent principal components differentiate Pygmies (PC4) and Sandawe (PC5) from other African populations (Figure S4).
To assess shared ancestry between diverse African hunter-gatherer populations we examined the percent of shared variants between Pygmy, Hadza, and Sandawe genomes and a previously sequenced San genome (Schuster et al., 2010). The percent of San variants that are shared with one other hunter-gatherer population is similar for Pygmy-, Hadza-, and Sandawe-specific variants (5.6–5.7%), suggesting that that the San diverged before other hunter-gatherer populations. However, the D-test of admixture (Green et al., 2010) indicates that the San genome shares more derived alleles with Pygmies than with the Hadza or Sandawe (p < 0.01, Table S3). This result suggests that the ancestors of the Tanzanian click-speakers (the Hadza and Sandawe) may have diverged more recently in the past than the Pygmy/San split. However, additional possibilities involve gene flow between the ancestors of Pygmies and the San or stochastic loss of shared derived alleles among the ancestors of the Hadza and Sandawe.
A neighbor joining tree indicates that Pygmies diverged before the Hadza and Sandawe split (Figure 1F), and lack of monophyly among Pygmy genomes reveals population substructure involving Baka, Bakola, and Bedzan individuals. Hadza and Sandawe genomes are nested within a cluster that also includes the Maasai, possibly due to recent shared gene flow with neighboring East African populations. With the exception of Pygmies, clustering patterns reflect shared language families: Khoesan-speaking Hadza and Sandawe individuals cluster together, as do Niger-Kordofanian speaking Yoruba and Luhya individuals.
We also observe differences in the number and cumulative size of long runs of homozygosity in each population. Of the 15 hunter-gatherer genomes analyzed in this paper, the five genomes with the most runs of homozygosity all belong to the Hadza (Figure S5). While some of these differences may be due to a population bottleneck in the Hadza (Henn et al., 2011), an additional cause may be cryptic inbreeding (Pemberton et al., 2012), as indicated by the large variance in cumulative size of runs of homozygosity within the Hadza (Figure S5). Indeed, cumulative runs of homozygosity in three Hadza genomes are more than double the size of other hunter-gatherers analyzed in this paper (Figure S5).
Consistent with an historic bottleneck and/or inbreeding in the Hadza, we find that the proportion of polymorphic sites, as quantified by θ, is lowest for the Hadza and highest for Pygmies (Table 2). Depending on mutation rates, this translates to effective population sizes of 11,300–25,700 (Pygmy), 9,200–20,900 (Hadza), and 10,600–24,000 individuals (Sandawe). Genome-wide estimates of Tajima’s D are lower for Pygmies and Sandawe compared to the Hadza (mean values of Tajima’s D are -0.4273 for Pygmies, −0.0148 for Hadza, and −0.3453 for Sandawe). These results are consistent with the observation that low frequency derived alleles (DAF ≤ 0.1) are over-represented in Pygmy and Sandawe populations and under-represented in the Hadza (Figure S6, p < 0.0001, χ2 tests of independence). Together, these results suggest that Pygmy and Sandawe populations have recently expanded in size whereas the Hadza population has recently decreased in size.
Hunter-gatherer Genomes Possess Signatures of Archaic Admixture
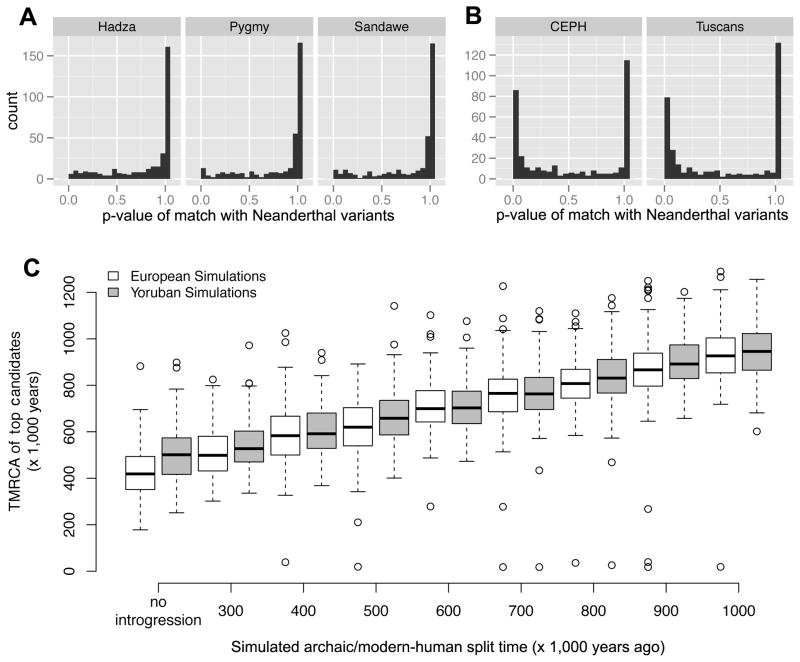
Gene flow between anatomically modern humans and archaic species has been described for European, Melanesian, and African populations (Hammer et al., 2011; Plagnol and Wall, 2006; Wall et al., 2009; Reich et al., 2010). To detect putatively introgressed regions in the Pygmy, Hadza, and Sandawe hunter-gatherer populations, we modified the summary statistic S*, which searches for clusters of population specific SNPs in near complete LD, to be suitable for genome-scale analyses. S* has previously been used to detect archaic admixture in individuals of European and African descent (Hammer et al., 2011; Plagnol and Wall, 2006; Wall et al., 2009). We first verified that our implementation of S* could accurately identify introgressed regions by performing extensive coalescent simulations (Supplemental Information, Figure S7) and analyzing publicly available whole-genome sequences from 9 CEPH and 4 Tuscan individuals sequenced by Complete Genomics. We calculated S* in 50kb sliding windows, and identified the top 350 regions (top ~0.4%) in each population with unusually large values of S* as high-confidence candidates for introgression. TMRCA distributions for these regions are significantly larger than the distribution for all loci (p < 10−16), consistent with the hypothesis that they are enriched for introgression (Figure 2A). Moreover, non-African genomic regions with high values of S* were significantly enriched for Neanderthal-specific SNPs (p < 10−16, Figure 3B). Thus, S* can robustly detect genomic regions inherited from archaic ancestors.
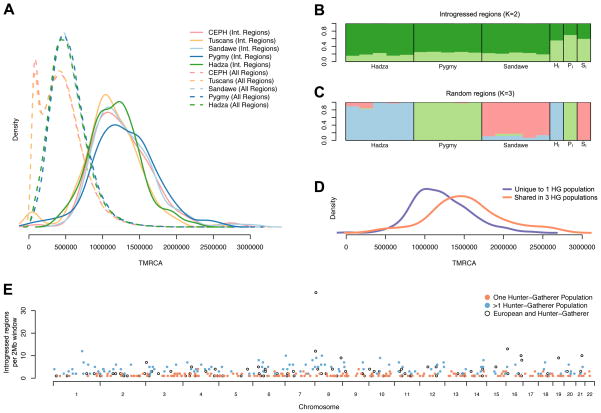
Figure 2. Times until most recent common ancestry and evidence of archaic introgression.
A) TMRCA of top candidate regions (solid lines), and of all regions (dotted lines) for the Pygmy, Hadza, and Sandawe hunter-gatherer populations and two European populations. Note, TMRCA represents the estimated time of divergence between the anatomically modern human and candidate introgressed sequences (Supplemental Information). TMRCA for top candidate regions is significantly older than random genomic regions (Kruskal-Wallis test, p < 2.2×10−16), but TMRCA for top candidate regions from each population are not significantly different (Kruskal-Wallis test, p=1). B and C) STRUCTURE plots showing the proportion of ancestry for each individual based on the most likely number of subpopulations (K = 2 for putatively introgressed regions in Panel B and K =3 for random regions in Panel C). For each population, a ‘virtual’ genome was constructed by concatenating sequence from individuals containing the putatively introgressed sequence (B) or from arbitrary individuals (C). Pi, Hi, and Si denote the virtual genomes constructed for the Pygmy, Hadza, and Sandawe samples, respectively. D) TMRCA of top candidate regions for introgression unique to a single hunter-gatherer population is significantly lower than TMRCA of regions shared between all hunter-gatherer populations (Wilcoxon rank sum test, p = 2.2×10−5). E) Genomic distribution of the top 350 introgressed regions for the Pygmy, Hadza, and Sandawe populations and two European populations, in 2Mb windows. Colors indicate whether windows contain introgressed regions from a single hunter-gatherer population (orange), multiple hunter-gatherer populations (blue), or hunter-gatherer and European populations (open black circle). Counts are for hunter-gatherer regions only. See also Figure S7.
Figure 3. Characteristics of S* in real and simulated data.
A–B) Neanderthal variants are not enriched in top candidate regions for three hunter-gatherer populations (A), but are enriched in top candidate regions from two European populations (B). C) TMRCA estimates for top 0.5% of 50kb regions in simulated data, varying time of split with the archaic population from 300kya to 1000kya; introgression was simulated into Europeans (white boxes) and Yorubans (gray boxes). See also Figure S7.
We next used S* to identify putatively introgressed regions in the African hunter-gatherer samples. In all three African hunter-gatherer samples, we found evidence of introgression from at least one archaic population. Strikingly, the median TMRCA for putatively introgressed haplotypes in the hunter-gatherer samples is similar to the median TMRCA for introgressed haplotypes in Europeans (1.2–1.3 Mya vs 1.1–1.2 Mya, respectively; Figure 2A), suggesting that the archaic African population diverged from anatomically modern humans in the same time frame as Neanderthals (simulations suggest that relative time of split with archaic populations can be recovered via TMRCA; Figure 3C). Additionally, we performed a STRUCTURE analysis of the putatively introgressed regions and of 350 random regions. If candidate regions identified by unusually large values of S* are enriched for genuine introgressed sequence, then we would expect STRUCTURE to identify two populations, as introgressed regions primarily consist of individuals carrying one archaic and one anatomically modern haplotype. In contrast, we would expect STRUCTURE to identify three populations in the randomly selected regions corresponding to the Pygmy, Hadza and Sandawe populations. Indeed, this is precisely what we find (Figure 2B and 2C), further demonstrating that top ranked S* regions are enriched for putatively introgressed sequence.
There is significant overlap (p < 10−16) among putatively introgressed regions in the three hunter-gatherer populations, consistent with either gene flow among the hunter-gatherer populations or introgression events that predate population splitting of these populations. In addition, the TMRCA of introgressed regions shared between all three populations is significantly older compared to introgressed regions observed in only one population (Wilcoxon rank-sum test, p = 2.2×10−5, Figure 2D), consistent with an introgression event predating the divergence of these populations. In contrast, we observed few introgressed regions that overlap with those observed outside of Africa. One exception is a 2 Mb window on chromosome 8 (Figure 2E, chr8:3Mb-5Mb) that contains introgressed regions in all global populations. However, we note that because the chr8:3Mb-5Mb region is enriched for CNVs, it may be more prone to false positives (Supplemental Information).
Identification of Genomic Regions with Extreme Times to Most Recent Common Ancestry
We scanned the genomes of African hunter-gatherers to identify regions with extremely long or short coalescence times, which are likely to be enriched for targets of natural selection. To this end, we calculated the time to most recent common ancestor (TMRCA) for 50kb sliding windows in the 15 hunter-gatherer genomes. The mean autosomal TMRCA across all windows is 796kya. As expected, windows spanning the HLA region, which exhibits strong signatures of balancing selection (Barreiro and Quintana-Murci, 2010), are the most ancient in the genome, with a maximum TMRCA of 5.1 million years for a 50kb window encompassing HLA-G. The oldest genic regions outside of the HLA locus include NSUN4, HCG9, MYO3A, and APOBEC4. Conversely, we also found multiple genomic regions with short TMRCA times (< 10 kya) including multiple tripartite motif containing genes (TRIM53P, TRIM64, and TRIM64B), the SPAG11A gene that is involved in sperm maturation, and NCF1, which is a subunit of neutrophil NADPH oxidase.
Evidence of Local Adaptation in African Hunter-Gatherer Populations
To identify signatures of geographically restricted adaptation, we used two complimentary approaches. First, we identified genomic regions in each hunter-gatherer population that differ the most from agricultural (Yoruba) and pastoral (Maasai) populations. This involved calculating locus-specific branch lengths (LSBL; (Shriver et al., 2004)) for each polymorphic site in the genome, and using sliding windows to identify 100kb regions that are enriched for LSBL outliers (sites with the highest values of LSBL statistics). We then focus on the top 268 (1%) most divergent 100kb windows in each population (Figure 4A–C, Table S5). Many highly-divergent 100kb windows do not contain any genes (101/268 Pygmy, 105/268 Hadza, and 119/268 Sandawe windows), and these windows may contain regulatory sequences that are important targets of adaptation. Divergent 100kb windows shared between pairs of populations include regions containing olfactory receptors (Pygmy and Hadza), major histocompatibility complex genes (Hadza and Sandawe), a gene which regulates lipid content in human breast milk (BTN1A1, Pygmy and Sandawe), and a gene involved in vascular injury repair (FLNB, Pygmy and Sandawe). We find only a single 100kb window that is highly divergent in all three populations (located 41 kb downstream of the PRDM5, a gene involved in bone development (Galli et al., 2012)), suggesting that each African hunter-gatherer population has been subject to different local selective pressures.
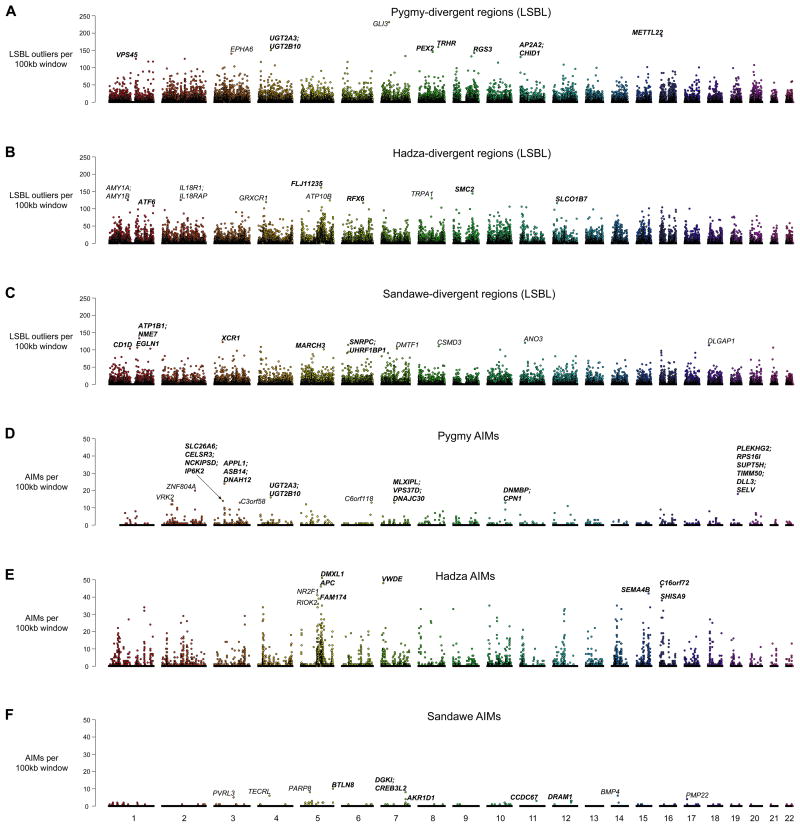
Figure 4. Divergent genomic regions between hunter-gatherers and non-hunter-gatherers and genomic distributions of ancestry informative markers (AIMs).
Each dot represents a non-overlapping 100kb window. Colors correspond to different chromosomes. For each population, genes found in the top 10 windows are listed in bold. If no genes are present in a top 10 window, the nearest gene is listed in normal font. A-C) number of LSBL outliers (top 1%) per 100kb window. D-F) number of AIMs per 100kb window. See also Table S5 and Table S6.
Genes in the top Pygmy-divergent regions of potential interest based on function include TRHR (thyrotropin-releasing hormone receptor involved in regulation of thyroid function), IFIH1 (involved in viral immunity), HESX1 (anterior pituitary development), CYBRD1 (iron absorption), UGT2B10 (breakdown of toxic endobiotic and xenobiotic compounds), and RGS3 (a GTP-ase activating protein gene that has been identified in other scans of Pygmy-specific selection (Jarvis et al., 2012; Pickrell et al., 2009)). Additionally, multiple genes in Pygmy-divergent regions are involved in spermatogenesis and fertility (ODF3, FSHR and SLC9A10). Functionally interesting genes in Hadza-divergent regions include IL18R1/IL18RAP (interleukin 18 receptor and accessory protein), CYCS (cytochrome c), CNR2 (cannabinoid receptor), and VWF (a blood glycoprotein involved in hemostasis and wound healing). Functionally interesting genes in Sandawe-divergent regions include ALDH2 (aldehyde dehydrogenase), EGLN1 (cellular response to hypoxia), HLA-DOB (MHC class II protein), and ZPBP (zona pellucida binding protein).
To determine shared functional characteristics of genes within 200kb of genomic regions enriched for high LSBL SNPs, we performed pathway analysis using DAVID, which includes KEGG and PANTHER databases (Huang da et al., 2009), for each hunter-gatherer population. These analyses revealed that immune related pathways were over-represented near Hadza-divergent and Sandawe-divergent regions (p < 0.05 after Bonferroni corrections), but not for Pygmy-divergent regions. However, these signals in the Hadza and Sandawe were almost entirely driven by the HLA-region at 6p21 (Table S6). Additionally, both Pygmy- and Hadza-divergent regions are significantly enriched for genes involved in olfactory transduction. Pathway analysis also pointed towards overrepresented retinol and porphyrin/chlorophyl metabolism pathways in Pygmies (p < 0.02 after Bonferroni corrections) and a taste transduction pathway in the Sandawe (p < 3.3×10−5 after Bonferroni corrections). The observation that highly divergent regions in three different hunter-gatherer populations are enriched for genes involved in smell or taste suggests the potential evolutionary importance of these loci with respect to local dietary adaptations.
Second, we identified high frequency population-specific variants in hunter-gatherer genomes, referred to as ancestry informative markers (AIMs, defined here as sites with variant allele frequencies >50% in a single hunter-gatherer sample and absent from the other two hunter-gatherer samples and dbSNP131). We identified < 15,000 variants per population that meet these stringent criteria (Figure 4D-F). These AIMs are not randomly distributed throughout hunter-gatherer genomes (p < 10−5 for each population); instead, we observe multiple clusters of AIMs in each population. AIM clusters may result from population-specific adaptation as well as demographic factors (Burger and Akerman, 2011; Falush et al., 2003). The number of AIMs is greatest for the Hadza (Figure 4D-F), a pattern that is consistent with population bottlenecks and greater genetic isolation relative to the other hunter-gatherer populations. Pathway analyses of genes within 50kb of AIMs suggest that there is enrichment for starch and sucrose metabolism in Pygmy genomes (p = 0.002; p = 0.247 after Bonferroni corrections) and enrichment for melanogenesis in Sandawe genomes (p = 0.0022; p = 0.168 after Bonferroni corrections, Table S6).
Identification of Candidate Genes For Short Pygmy Stature
Analyses of LSBL and AIM clusters in the Pygmies are particularly informative for identifying variants that play a role in population specific traits, including short stature. The largest Pygmy AIM cluster spans 170kb at 3p14.3 and contains a high (70%) frequency haplotype in our sample of 10 genomes, with 44 Pygmy-AIMs in 100% linkage disequilibrium (chr3:57,211,368–57,383,846, Figure 5A). Four genes lie within this cluster: HESX1 (which encodes a homeobox containing transcriptional repressor that plays a critical role in development of the anterior pituitary, the site of growth hormone synthesis and secretion), APPL1 (which is involved in crosstalk between adiponectin and insulin signaling pathways), ASB14 (which encodes a SOCS box protein), and the sperm-motility gene DNAH12. Mutations within HESX1 in humans cause septo-optic dysplasia, combined pituitary hormone disease, and/or isolated growth hormone deficiency, resulting in short stature (Dattani, 2005). Although HESX1 is expressed early in embryonic development in mouse and plays a critical role in forebrain and pituitary development, it continues to be expressed in adult pituitary in humans where it may play a role in the maintenance of anterior pituitary cell types and function (Mantovani et al., 2006). Analysis of the October 2011 release of the 1000 genomes database (1000 Genomes Project Consortium, 2010) indicates that the 44 SNPs encompassing the Pygmy 3p14.3 AIM haplotype are in complete LD and at very low frequency (<5%) in Yoruba and Luhya populations and are absent from non-African populations. Additionally, the Pygmy 3p14.3 AIM haplotype includes a previously identified non-synonymous SNP within HESX1 (rs9878928; Asn125Ser), which has been shown to be associated with pituitary developmental defects and growth hormone deficiency (Brickman et al., 2001; Gat-Yablonski et al., 2009). This non-synonymous SNP is at moderate frequency (Pickrell et al., 2009) but on distinct haplotype backgrounds in other African populations. Interestingly, this Pygmy AIM cluster lies within a 15 Mb region that shows high levels of differentiation between Pygmies and neighboring Bantu populations and is associated with height in Pygmies (Jarvis et al., 2012).
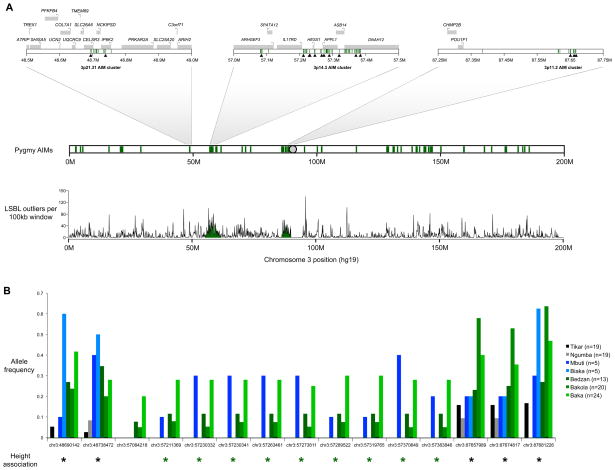
Figure 5. Pygmy AIMs, allele frequencies, and height associations.
A) Pygmy AIMs (green lines) located on chromosome 3. Green shading in the LSBL plot indicates genomic regions with an excess of LSBL outliers near the 3p14.3 and 3p11.2 AIM clusters. B) Allele frequencies of Pygmy AIMs genotyped in a broad sample of African Pygmy and Bantu individuals. Significant associations (p < 0.05) with height are indicated for males (green asterisks) and for both sexes pooled together (black asterisks). Sample sizes are also listed (n=the number of genotyped individuals per population). See also Table S7.
An additional cluster of 11 Pygmy AIMs was identified at 3p11.2 (chr3:87,657,988–87,681,226, Figure 5A). The closest gene, located ~330 kb upstream, is POU1F1 (also known as PIT1) which encodes a pituitary specific transcription factor that plays an important role in anterior pituitary development and regulation of growth hormone gene expression (Hunsaker et al., 2012) and mutations within this gene cause growth hormone deficiency and short stature (Kiess et al., 2011). These SNPs, referred to as the Pygmy 3p11.2 AIM cluster, are at >60% frequency in sequenced Pygmies and at <12% frequency in African and non-African populations from the 1000 genomes project. Both the 3p14.3 (HESX1) and 3p11.2 (POU1F1) AIM clusters are embedded within 4.2Mb and 3.3Mb blocks, respectively, where every 100kb window contains an excess of LSBL outliers (the largest continuous runs of increased LSBL in Pygmy genomes, Figure 5A).
To examine the geographic distribution and frequency of Pygmy AIMs, we genotyped a panel of 58 Pygmies (Baka, Bakola, and Bedzan ancestry) and 38 neighboring Bantu individuals (Tikar and Ngumba ancestry), as well as 5 Mbuti Pygmies and 5 Biaka Pygmies, at 15 AIM SNPs on chromosome 3 (Figure 5; Table S7). We find that the HESX1-containing AIM cluster at 3p14.3 is common (>5% frequency) in Baka, Bakola, and Bedzan Pygmies from Cameroon and Mbuti Pygmies from the Democratic Republic of the Congo, and absent from Cameroonian Bantu populations and Biaka Pygmies from the Central African Republic. Among Baka, Bakola, Bedzan Pygmies, and a single Mbuti Pygmy, the 3p14.3 AIM haplotype extends 172kb (complete LD), but in three other Mbuti Pygmies this haplotype is broken down (with 100% LD extending 97kb from chr3:57273811 to chr3:57370649). The presence of a highly divergent AIM haplotype in multiple Pygmy populations suggests that this haplotype predates the divergence of Eastern and Western Pygmies.
Additionally, we tested for genetic associations between 15 SNPs located in the 3p21.31, 3p14.3, and 3p.11.AIM clusters and height in a panel of Cameroonian Pygmy and Bantu individuals (Figure 5A). Treating sex as a covariate, we find significant associations between height and two SNPs located at 48.7M and three SNPs located at 87.6M, ~330kb from POU1F1 (p <0.03, Figure 5B and Table S7). Two of these SNPs remain significant after conservative Bonferroni corrections (chr3:48738472, and chr3:87681226). In addition, we find a significant association between the Pygmy HESX1 AIM haplotype at 3p14.3 and shorter height in males (p < 0.02, Figure 5B). When significant associations are found, the alleles associated with shorter height are all Pygmy AIM variants.
Discussion
The deluge of data from next-generation sequencing has begun, with massively large datasets of low coverage whole-genome sequences (1000 Genomes Project Consortium, 2010) and high coverage exome sequences (Tennessen et al., 2012) being reported in thousands of individuals. Here, we described high coverage whole-genome sequencing of individuals from three African hunter-gatherer populations, which harbor large amounts of previously unknown genetic diversity that is inaccessible by studying individuals of non-African ancestry or focusing only on protein-coding regions. Despite evidence of inbreeding and a population bottleneck in the Hadza, high levels of genetic diversity are maintained in all three hunter-gatherer populations. Additionally, we found significant genetic divergence among the three African hunter-gatherer populations, including between the Hadza and Sandawe who are geographically close (~150km apart) and have languages that contain click consonants, demonstrating the continued need to broadly sample human populations in order to comprehensively assess the spectrum of human genomic diversity.
We find evidence of selective constraint near genes, and these patterns are replicated in each hunter-gatherer population. We also observe signatures of local adaptation in Pygmy, Hadza, and Sandawe populations, including high locus-specific branch lengths for genes involved in taste/olfactory perception, pituitary development, reproduction, and immune function. These genetic differences reflect differences in local diets, pathogen pressures, and environments. Thus, Pygmies, Hadza and Sandawe have continued to adapt to local conditions while sustaining their own unique cultures of hunting and gathering.
Evidence of Archaic Introgression
A striking finding in our dataset is that compelling evidence exists that extant hunter-gatherer genomes contain introgressed archaic sequence, consistent with previous studies (Hammer et al., 2011; Plagnol and Wall, 2006; Reich et al., 2010; Shimada et al., 2007; Wall et al., 2009). We note that unambiguous evidence of introgression is difficult to obtain in the absence of an archaic reference sequence, which currently does not exist and may never be feasible given the rapid decay of fossils in Africa. Although we carefully filtered our data set in an attempt to analyze only high-quality sequences (Supplementary Information), it is possible that unrecognized structural variants or other alignment errors could generate a spurious signature similar to introgression. Encouragingly, we did not see an enrichment of structural variation calls in our candidate introgression regions. Additionally, through extensive simulations and analysis of European whole-genome sequences (Supplementary Information), we have demonstrated that the signatures of introgression we observed are unlikely to be entirely accounted for due to other aspects of population demographic history, natural selection, or sequencing errors. Moreover, we did not find strong evidence that introgressed regions were clustered in the genome more often than expected by chance (p > 0.05; Supplemental Information). Nor did we find significant evidence that introgressed regions were enriched in genic regions (p > 0.05); rather, genic regions were significantly depleted for introgression in several populations (Supplemental Information). Therefore, the simplest interpretation of these data is that introgressed regions in extant human populations represent neutrally evolving vestiges of archaic sequences. In short, we find that low levels of introgression from an unknown archaic population, or populations, occurred in the three African hunter-gatherer samples examined, consistent with findings of archaic admixture in non-Africans (Reich et al., 2010).
Short Stature, Pituitary Function, and Local Adaptation in Western African Pygmies
Short stature in African Pygmies is thought to be an adaptation to a tropical forest environment. Several possible fitness advantages of short height have been proposed including thermo-regulation, early cessation of growth as a trade-off for early reproduction to compensate for shorter life expectancy, easier mobility in a dense forest environment, and reduced caloric requirements (Migliano et al., 2007; Perry and Dominy, 2009). Although stature in Europeans is a highly complex trait (Lango Allen et al., 2010), the genetic architecture of this trait in Pygmies may differ (Pygmy LSBL hits are not enriched for height genes found in largely European GWAS, p = 0.888 for the top 268 LSBL windows, confirming (Jarvis et al., 2012)). AIMs within and near HESX1 and POU1F1 are strong candidates for the short stature phenotype in Pygmies, together with previously identified (chr3:45Mb-60Mb region (Jarvis et al., 2012)) and other, as yet undiscovered, loci. The observation of long-range LD maintained in diverse populations at these loci raises the possibility that undetected inversions in these chromosome 3 regions play a role in population differentiation and adaptation. Additionally, the observation that third chromosome AIM clusters exist at very low frequency in other African populations suggests that if selection has altered the frequency of AIM haplotypes in Pygmies, then it may have acted on standing variation which existed prior to the divergence of Eastern and Western Pygmies from other African populations. Furthermore, AIM variants are not included in commercially available genome-wide SNP arrays, emphasizing the critical importance of whole-genome sequencing for identifying variants of potential functional significance that may be geographically or ethnically restricted due to distinct selection pressures and/or demographic histories.
In addition to the 3p14.3 (HESX1) and 3p11.2 (POU1F1) AIM clusters, we have identified other candidate loci that may play a role in local adaptation, height, and pituitary function in Pygmies. These loci include TRHR (thyrotropin-releasing hormone receptor), APPL1, FSHR, and genes associated with Williams Syndrome (Supplemental Information). Overall, we find that highly divergent regions of Pygmy genomes (as identified by LSBL scans) are enriched for genes that play a role in pituitary function (p = 0.0082, χ2 test of independence).
Together, these results point towards the possibility that development and expression of hormones produced by the anterior pituitary may play a central role in the Pygmy phenotype, potentially influencing a number of traits including growth, reproduction, metabolism, and immunity. Further studies of pituitary function and development in vitro and using transgenic animal models will be necessary to elucidate the importance of this system in Pygmy development and physiology, and to clarify the role of variants within the 3p14.3 and 3p11.2 Pygmy AIM clusters.
Conclusion
In summary, this is one of the first population genomics analyses to use high coverage whole-genome sequencing. Our results indicate the importance of whole-genome data for reconstructing human origins, identifying targets of local adaptation, reconstructing demographic history, and identifying functionally important variants for complex traits like height. We have identified many novel targets of natural selection that play a role in immunity, reproduction, metabolism, and height in diverse hunter-gatherer populations. As sequencing costs continue to decrease, it will become feasible to do whole-genome sequencing of increasingly larger sample sizes across ethnically diverse global populations, and to integrate genomic data with functional studies using in vitro and in vivo models. Such studies will shed light on human evolutionary history and the origin of traits that make each of us unique.
Experimental procedures
Samples Used for Whole-genome Sequencing
Prior to sample collection, informed consent was obtained from all research participants, and permits were received from the Ministry of Health and National Committee of Ethics in Cameroon and from COSTECH and NIMR in Dar es Salaam, Tanzania. In addition, appropriate IRB approval was obtained from both the University of Maryland and the University of Pennsylvania. The genomes of five Pygmy (three Baka, one Bakola, one Bedzan), five Hadza (plus two technical replicates), and five Sandawe individuals were sequenced using the combinatorial probe-anchor ligation and DNA nanoarray technology of Complete Genomics (Drmanac et al., 2010). The standard Complete Genomics bioinformatics pipeline (Assembly Pipeline version 1.10 and CGA™ Tools 1.4) was used for sequence alignment, read mapping, assembly, and data analysis. As additional quality control filters, we eliminated variants found in poorly called regions (<20% missing data required), and sites heterozygous in every individual (using a departure from Hardy-Weinberg proportions test).
Population Genetic Statistics and Functional Classification of Variants
Considering only fully called sites, θ and Tajima’s D were calculated. Derived alleles were obtained using ancestral states inferred via maximum likelihood (chimpanzee, orangutan, and rhesus macaque genomes were used as outgroups), and FST was calculated using corrections for small sample size (Weir and Cockerham, 1984). DNase I hypersensitive sites and footprints were obtained from the ENCODE project (Rosenbloom et al., 2012). The neutrality index for different site types was calculated as previously described (Rand and Kann, 1996). Strength of purifying selection was analyzed in nine different populations: African hunter-gatherers (Pygmies, Hadza, and Sandawe), African farmers and pastoralists (YRI, MKK, and LWK) and non-Africans (CEU, CHB, and JPT; three letter HapMap population codes are listed in Figure 1). To control for sample size differences we randomly chose four genomes per population, and as per (Lohmueller et al., 2008) we calculated mean derived allele frequencies, proportion of “probably deleterious” variants (PolyPhen-2), and relative numbers of nonsynonymous and synonymous variants per genome for each population.
Principal Components Analysis (PCA) and Hierarchal Clustering of Genomic Data (Neighbor Joining Tree)
After randomly selecting 50,000 unlinked autosomal SNPs at fully called sites, the prcomp() function in R was used for PCA. Individuals chosen for PCA include 15 hunter-gatherers and 53 unrelated individuals from the Complete Genomics public data release (because of cryptic relatedness, one Maasai individual was excluded). We constructed a neighbor joining tree using Phylip from 1,260,982 autosomal SNPs using the chimpanzee genome as an outgroup. The 61 individuals chosen for the neighbor joining tree were a subset of those analyzed by PCA (PUR and MXL individuals excluded).
Tests of Ancient Admixture
To detect putatively introgressed archaic sequence, we employed a modified version of the S* statistic (Plagnol and Wall, 2006). To mitigate potential confounding effects of mutation rate heterogeneity and paralogous variants, CpGs and repetitive sequences (as defined by RepeatMasker) were removed, and only fully called sites were considered. After filtering, 1.2Gb of sequence was retained. To enable comparisons between windows, we modify S* by dividing by the square of the sequence length after filtering of each window. S* also requires a target population (in which we search for introgressed sequence) and a reference population. For African populations, we used 13 European genomes (9 CEPH and 4 Tuscan) as the reference, and for non-African populations we used 9 unrelated Yoruban genomes (each genome sequenced by Complete Genomics).
We calculated S* in 50kb windows, using a step size of 20kb. Windows were removed from consideration if they did not contain at least 20kb of unfiltered sequence, leaving ~2Gb of sequence. The top 350 windows (~0.4%) as ranked by S* were considered as high-confidence candidates for introgression, which is likely a very conservative threshold as determined by extensive coalescent simulations and STRUCTURE analyses.
TMRCA Estimates
TMRCA was calculated using a previously described approach (Hudson, 2007; Thomson et al., 2000), which computes the average coalescent time from nucleotide substitutions assuming that mutations are Poisson distributed. This value is then converted to an estimate of TMRCA in years by computing the divergence between chimpanzee and humans for this region (D), and setting the molecular clock to 12My/D (assuming the divergence time between humans and chimpanzee is 6 million years). Human/chimpanzee alignments were downloaded from the UCSC Genome Browser (reference versions GRCb37 and panTro2, http://hgdownload.cse.ucsc.edu/goldenPath/hg19/vsPanTro2/syntenicNet/).
Tests of Local Adaptation (LSBL Analyses) and Identification of Ancestry Informative Markers (AIMs)
Locus-specific branch lengths (LSBL) were obtained by constructing evolutionary trees for each polymorphic site in the genome, calculating genetic distances (FST) between pairs of populations for each fully called site, and calculating LSBL statistics for each hunter-gatherer population. LSBL outliers are defined here as sites with LSBL statistics in the top 1% of the empirical distribution for each population. The 268 (top 1%) most divergent 100kb windows in each population were found using a χ2 test statistic (comparing observed and expected numbers of LSBL outliers for each autosomal 100kb window). AIM clusters were operationally defined as sets of at least 10 AIMs that are no more than 25kb distant from other AIMs. For each population, DAVID 6.7 was used to run pathway analyses (KEGG and PANTHER) on the set of genes located near highly-divergent LSBL windows (within 200kb of the of the top 1% windows) and on the set of genes located within 50kb of AIMs.
Tests of Association of Pygmy AIM SNPs and Height
15 Pygmy AIM SNPs were genotyped using TaqMan® assays (Applied Biosystems) in 95 Pygmy and Bantu samples analyzed in (Jarvis et al., 2012) and 10 additional Pygmy samples (5 Biaka and 5 Mbuti) from the Coriell Institute for Medical Research. Height data and identity by state data were available for 94 of the samples from (Jarvis et al., 2012). Association between AIM SNPs and height was determined using EMMAX, a mixed-model linear regression approach that corrects for both relatedness within populations and structure between them via a pair-wise matrix of genetic relationships among individuals (Kang et al., 2010). Ancestry was treated as a covariate (using an identity by state matrix generated from Illumina1M-duo genotyping) and SNP-height associations were calculated for males, females, and both sexes pooled together (with sex as a covariate).
Supplementary Material
Research Highlights.
13.4 million variants identified in African hunter-gatherers, many of which are novel
Evidence of archaic admixture found in the genomes of African hunter-gatherers
Selection scans implicate loci involved in taste perception, metabolism, and immunity
Genetic associations with height are found for Pygmy variants located on chromosome 3
Acknowledgments
This work was supported by NSF (BCS-0827436) and NIH (R01GM076637, 8 DP1 ES022577-04) grants to S.A.T., an NIH NRSA postdoctoral fellowship (F32HG006648-01) to J.L., Rubicon Grants of the Netherlands Organization of Scientific Research to C.E. and B.F., and support from the Center of Excellence in Environmental Toxicology at the University of Pennsylvania, P30-ES013508-07. We thank J. Hirbo, J. Jarvis, A. Rawlings, L. Scheinfeld, and S. Soi, for their critical feedback and advice, and K. Addya, D. Baldwin, and B. Beggs for assistance in genotyping the samples. We also thank the 15 individuals who graciously supplied their DNA. Data reported in this paper will be available by request and at the dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/) and dbGaP (http://www.ncbi.nlm.nih.gov/gap/) websites.
Footnotes
Supplemental Information contains additional experimental procedures and data, 7 supplemental figures, 7 supplemental tables, and can be found with this article online at doi:####.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreiro LB, Laval G, Quach H, Patin E, Quintana-Murci L. Natural selection has driven population differentiation in modern humans. Nat Genet. 2008;40:340–345. doi: 10.1038/ng.78. [DOI] [PubMed] [Google Scholar]
- Barreiro LB, Quintana-Murci L. From evolutionary genetics to human immunology: how selection shapes host defence genes. Nat Rev Genet. 2010;11:17–30. doi: 10.1038/nrg2698. [DOI] [PubMed] [Google Scholar]
- Behar DM, Villems R, Soodyall H, Blue-Smith J, Pereira L, Metspalu E, Scozzari R, Makkan H, Tzur S, Comas D, et al. The dawn of human matrilineal diversity. Am J Hum Genet. 2008;82:1130–1140. doi: 10.1016/j.ajhg.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berniell-Lee G, Calafell F, Bosch E, Heyer E, Sica L, Mouguiama-Daouda P, van der Veen L, Hombert JM, Quintana-Murci L, Comas D. Genetic and demographic implications of the Bantu expansion: insights from human paternal lineages. Mol Biol Evol. 2009;26:1581–1589. doi: 10.1093/molbev/msp069. [DOI] [PubMed] [Google Scholar]
- Blurton Jones NG, Smith LC, O’Connell JF, Hawkes K, Kamuzora CL. Demography of the Hadza, an increasing and high density population of Savanna foragers. Am J Phys Anthropol. 1992;89:159–181. doi: 10.1002/ajpa.1330890204. [DOI] [PubMed] [Google Scholar]
- Brickman JM, Clements M, Tyrell R, McNay D, Woods K, Warner J, Stewart A, Beddington RS, Dattani M. Molecular effects of novel mutations in Hesx1/HESX1 associated with human pituitary disorders. Development. 2001;128:5189–5199. doi: 10.1242/dev.128.24.5189. [DOI] [PubMed] [Google Scholar]
- Burger R, Akerman A. The effects of linkage and gene flow on local adaptation: A two-locus continent-island model. Theor Popul Biol. 2011;80:272–288. doi: 10.1016/j.tpb.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MC, Tishkoff SA. The evolution of human genetic and phenotypic variation in Africa. Curr Biol. 2010;20:R166–173. doi: 10.1016/j.cub.2009.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dattani MT. Growth hormone deficiency and combined pituitary hormone deficiency: does the genotype matter? Clin Endocrinol (Oxf) 2005;63:121–130. doi: 10.1111/j.1365-2265.2005.02289.x. [DOI] [PubMed] [Google Scholar]
- Drmanac R, Sparks AB, Callow MJ, Halpern AL, Burns NL, Kermani BG, Carnevali P, Nazarenko I, Nilsen GB, Yeung G, et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli GG, Honnens de Lichtenberg K, Carrara M, Hans W, Wuelling M, Mentz B, Multhaupt HA, Fog CK, Jensen KT, Rappsilber J, et al. Prdm5 Regulates Collagen Gene Transcription by Association with RNA Polymerase II in Developing Bone. PLoS genetics. 2012;8:e1002711. doi: 10.1371/journal.pgen.1002711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gat-Yablonski G, Lazar L, Bar M, de Vries L, Weintrob N, Phillip M. Growth without growth hormone and similar dysmorphic features in three patients with sporadic combined pituitary hormone deficiencies. Horm Res. 2009;72:302–309. doi: 10.1159/000245932. [DOI] [PubMed] [Google Scholar]
- Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer MF, Woerner AE, Mendez FL, Watkins JC, Wall JD. Genetic evidence for archaic admixture in Africa. Proc Natl Acad Sci U S A. 2011;108:15123–15128. doi: 10.1073/pnas.1109300108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn BM, Gignoux CR, Jobin M, Granka JM, Macpherson JM, Kidd JM, Rodriguez-Botigue L, Ramachandran S, Hon L, Brisbin A, et al. Hunter-gatherer genomic diversity suggests a southern African origin for modern humans. Proc Natl Acad Sci U S A. 2011;108:5154–5162. doi: 10.1073/pnas.1017511108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn BM, Gravel S, Moreno-Estrada A, Acevedo-Acevedo S, Bustamante CD. Fine-scale population structure and the era of next-generation sequencing. Hum Mol Genet. 2010;19:R221–226. doi: 10.1093/hmg/ddq403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hudson RR. The variance of coalescent time estimates from DNA sequences. J Mol Evol. 2007;64:702–705. doi: 10.1007/s00239-006-0261-1. [DOI] [PubMed] [Google Scholar]
- Hunsaker TL, Jefferson HS, Morrison JK, Franklin AJ, Shewchuk BM. POU1F1-mediated activation of hGH-N by deoxyribonuclease I hypersensitive site II of the human growth hormone locus control region. J Mol Biol. 2012;415:29–45. doi: 10.1016/j.jmb.2011.11.001. [DOI] [PubMed] [Google Scholar]
- Jarvis JP, Scheinfeldt LB, Soi S, Lambert C, Omberg L, Ferwerda B, Froment A, Bodo JM, Beggs W, Hoffman G, et al. Patterns of Ancestry, Signatures of Natural Selection, and Genetic Association with Stature in Western African Pygmies. PLoS genetics. 2012;8:e1002641. doi: 10.1371/journal.pgen.1002641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HM, Sul JH, Service SK, Zaitlen NA, Kong SY, Freimer NB, Sabatti C, Eskin E. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiess W, Kratzsch J, Kruis T, Muller E, Wallborn T, Odeh R, Schlicke M, Klammt J, Pfaffle R. Genetics of human stature: Insight from single gene disorders. Horm Res Paediatr. 2011;76(Suppl 3):11–13. doi: 10.1159/000330140. [DOI] [PubMed] [Google Scholar]
- Lam HY, Clark MJ, Chen R, Natsoulis G, O’Huallachain M, Dewey FE, Habegger L, Ashley EA, Gerstein MB, Butte AJ, et al. Performance comparison of whole-genome sequencing platforms. Nat Biotechnol. 2012;30:78–82. doi: 10.1038/nbt.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, Willer CJ, Jackson AU, Vedantam S, Raychaudhuri S, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmueller KE, Indap AR, Schmidt S, Boyko AR, Hernandez RD, Hubisz MJ, Sninsky JJ, White TJ, Sunyaev SR, Nielsen R, et al. Proportionally more deleterious genetic variation in European than in African populations. Nature. 2008;451:994–997. doi: 10.1038/nature06611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luikart G, England PR, Tallmon D, Jordan S, Taberlet P. The power and promise of population genomics: from genotyping to genome typing. Nat Rev Genet. 2003;4:981–994. doi: 10.1038/nrg1226. [DOI] [PubMed] [Google Scholar]
- Mantovani G, Asteria C, Pellegrini C, Bosari S, Alberti L, Bondioni S, Peverelli E, Spada A, Beck-Peccoz P. HESX1 expression in human normal pituitaries and pituitary adenomas. Mol Cell Endocrinol. 2006;247:135–139. doi: 10.1016/j.mce.2005.12.036. [DOI] [PubMed] [Google Scholar]
- Marlowe F. The Hadza : hunter-gatherers of Tanzania. Berkeley: University of California Press; 2010. [Google Scholar]
- Migliano AB, Vinicius L, Lahr MM. Life history trade-offs explain the evolution of human pygmies. Proc Natl Acad Sci U S A. 2007;104:20216–20219. doi: 10.1073/pnas.0708024105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JL. The ecological basis for subsistence change among the Sandawe of Tanzania. Washington: National Academy of Sciences; 1970. [Google Scholar]
- Pemberton TJ, Absher D, Feldman MW, Myers RM, Rosenberg NA, Li H. Genomic patterns of homozygosity in worldwide human populations. Am J Hum Genet. 2012 doi: 10.1016/j.ajhg.2012.06.014. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry GH, Dominy NJ. Evolution of the human pygmy phenotype. Trends Ecol Evol. 2009;24:218–225. doi: 10.1016/j.tree.2008.11.008. [DOI] [PubMed] [Google Scholar]
- Pickrell JK, Coop G, Novembre J, Kudaravalli S, Li JZ, Absher D, Srinivasan BS, Barsh GS, Myers RM, Feldman MW, et al. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 2009;19:826–837. doi: 10.1101/gr.087577.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plagnol V, Wall JD. Possible ancestral structure in human populations. PLoS genetics. 2006;2:e105. doi: 10.1371/journal.pgen.0020105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand DM, Kann LM. Excess amino acid polymorphism in mitochondrial DNA: contrasts among genes from Drosophila, mice, and humans. Molecular biology and evolution. 1996;13:735–748. doi: 10.1093/oxfordjournals.molbev.a025634. [DOI] [PubMed] [Google Scholar]
- Reich D, Green RE, Kircher M, Krause J, Patterson N, Durand EY, Viola B, Briggs AW, Stenzel U, Johnson PL, et al. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 2010;468:1053–1060. doi: 10.1038/nature09710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbloom KR, Dreszer TR, Long JC, Malladi VS, Sloan CA, Raney BJ, Cline MS, Karolchik D, Barber GP, Clawson H, et al. ENCODE whole-genome data in the UCSC Genome Browser: update 2012. Nucleic Acids Res. 2012;40:D912–917. doi: 10.1093/nar/gkr1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sands B. The linguistic relationship between the Hadza and Khoisan. In: Schladt M, editor. Language, Identity, and Conceptualization among the Khoisan. Köln (Germany): Rudiger Kupper Verlag; 1998. pp. 265–283. [Google Scholar]
- Schuster SC, Miller W, Ratan A, Tomsho LP, Giardine B, Kasson LR, Harris RS, Petersen DC, Zhao F, Qi J, et al. Complete Khoisan and Bantu genomes from southern Africa. Nature. 2010;463:943–947. doi: 10.1038/nature08795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada MK, Panchapakesan K, Tishkoff SA, Nato AQ, Jr, Hey J. Divergent haplotypes and human history as revealed in a worldwide survey of X-linked DNA sequence variation. Molecular biology and evolution. 2007;24:687–698. doi: 10.1093/molbev/msl196. [DOI] [PubMed] [Google Scholar]
- Shriver MD, Kennedy GC, Parra EJ, Lawson HA, Sonpar V, Huang J, Akey JM, Jones KW. The genomic distribution of population substructure in four populations using 8,525 autosomal SNPs. Hum Genomics. 2004;1:274–286. doi: 10.1186/1479-7364-1-4-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen JA, Bigham AW, O’Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun G, et al. Evolution and Functional Impact of Rare Coding Variation from Deep Sequencing of Human Exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo YY, Small KS, Kwiatkowski DP. Methodological challenges of genome-wide association analysis in Africa. Nat Rev Genet. 2010;11:149–160. doi: 10.1038/nrg2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson R, Pritchard JK, Shen P, Oefner PJ, Feldman MW. Recent common ancestry of human Y chromosomes: evidence from DNA sequence data. Proc Natl Acad Sci U S A. 2000;97:7360–7365. doi: 10.1073/pnas.97.13.7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Gonder MK, Henn BM, Mortensen H, Knight A, Gignoux C, Fernandopulle N, Lema G, Nyambo TB, Ramakrishnan U, et al. History of click-speaking populations of Africa inferred from mtDNA and Y chromosome genetic variation. Mol Biol Evol. 2007;24:2180–2195. doi: 10.1093/molbev/msm155. [DOI] [PubMed] [Google Scholar]
- Tishkoff SA, Reed FA, Friedlaender FR, Ehret C, Ranciaro A, Froment A, Hirbo JB, Awomoyi AA, Bodo JM, Doumbo O, et al. The genetic structure and history of Africans and African Americans. Science. 2009;324:1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeramah KR, Wegmann D, Woerner A, Mendez FL, Watkins JC, Destro-Bisol G, Soodyall H, Louie L, Hammer MF. An early divergence of KhoeSan ancestors from those of other modern humans is supported by an ABC-based analysis of autosomal resequencing data. Mol Biol Evol. 2012;29:617–630. doi: 10.1093/molbev/msr212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdu P, Austerlitz F, Estoup A, Vitalis R, Georges M, Thery S, Froment A, Le Bomin S, Gessain A, Hombert JM, et al. Origins and genetic diversity of pygmy hunter-gatherers from Western Central Africa. Curr Biol. 2009;19:312–318. doi: 10.1016/j.cub.2008.12.049. [DOI] [PubMed] [Google Scholar]
- Wall JD, Lohmueller KE, Plagnol V. Detecting ancient admixture and estimating demographic parameters in multiple human populations. Mol Biol Evol. 2009;26:1823–1827. doi: 10.1093/molbev/msp096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-Statistics for the Analysis of Population Structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.