Background: Galectins from peritoneal cells of conger eel contribute to the encapsulation of nematode.
Results: A new galectin from peritoneal cells, congerin P (Con-P), shows unusual sequence, specificity, and allosteric regulation by mannoside.
Conclusion: Con-P is a new type of galectin with allosteric carbohydrate-binding ability.
Significance: Con-P is the first known lectin allosterically modulated by its ligands.
Keywords: Allosteric Regulation, Carbohydrate-binding Protein, Fluorescence, Galectin, Lectin, Molecular Evolution, Protein Expression, Protein Sequence, Recombinant Protein Expression, Mannose
Abstract
Conger eel has two galectins, termed congerins I and II (Con I and II), that function in mucus as biodefense molecules. Con I and II have acquired a novel protein fold via domain swapping and a new ligand-binding site by accelerated evolution, which enables recognition of some marine bacteria. In this study, we identified a new congerin isotype, congerin P (Con-P), from the peritoneal cells of conger eel. Although Con-P displayed obvious homology with galectins, we observed substitution of 7 out of 8 amino acid residues in the carbohydrate recognition domain that are conserved in all other known galectins. To understand the structure-function relationships of this unique galectin, recombinant Con-P was successfully expressed in Escherichia coli by using a Con II-tagged fusion protein system and subsequently characterized. In the presence of d-mannose, Con-P displayed 30-fold greater hemagglutinating activity than Con I; however, no activity was observed without mannose, indicating that d-mannoside can act as a modulator of Con-P. Frontal affinity chromatography analysis showed that activated Con-P, allosterically induced by mannose, displayed affinity for oligomannose-type sugars as well as N-acetyllactosamine-type β-galactosides. Thus, Con-P represents a new member of the galectin family with unique properties.
Introduction
The galectins are a group of lectins with an affinity for β-d-galactoside that share a highly conserved carbohydrate recognition domain (CRD)2 of about 130 amino acid residues. Based on their structural features, galectins are classified into three types: prototype (monomer or homodimer containing a single CRD), tandem-repeat type (two CRDs on a single chain), and chimera type (one CRD linked to an N-terminal domain on a single chain) (1). Galectins are widely distributed among both lower invertebrates (e.g. marine sponges) and higher vertebrates, including humans, and have been proposed to participate in diverse physiological functions involving development, differentiation, morphogenesis, immunity, apoptosis, and metastasis of malignant cells (2).
The conger eel (Conger myriaster) contains two prototype galectins, congerins I and II (Con I and II), that are found in the skin mucus and frontier organs that delineate the body from the outer environment, such as the epidermal club cells of the skin, wall of the oral cavity, pharynx, esophagus, and gills (3, 4). Con I and II are prototype galectins, composed of subunits containing 135 and 136 amino acids, respectively, and display 48% amino acid sequence identity (4–6). Previous studies of Con I and Con II based on molecular evolutionary and x-ray crystallography analyses revealed that these proteins have evolved via accelerated substitutions under natural selection pressure from ancestors (4, 7–13). As a result, Con I has acquired a new strand-swap fold, which stabilizes the dimer structure that is essential for pathogen-coagulating activity. In addition, Con II has acquired a unique ligand-binding site that recognizes novel carbohydrates on pathogens (8–13).
Lectins and lectin-like molecules have been found in the skin mucus of fish (14) and may participate in innate or acquired immunity through their agglutinating activity. Indeed, Con I and II recognize some marine bacteria, including Vibrio anguillarum (15). Their localization profiles in fish tissues suggest that congerins are expressed both in the skin and also in the upper digestive tract and gill filament (16, 17). Furthermore, we demonstrated that the rate of peritoneal macrophages ingesting latex microspheres was significantly increased by recombinant Con I, suggesting that congerins act as opsonins (3). These observations indicate that congerins participate in the immune defense system, including innate immunity, on the internal and external body surface of the conger eel.
Cucullanidae nematodes are intra-alimentary canal parasites of marine and freshwater fish. Glycoconjugates, recognized by some lectins, are distributed on the surface of nematodes (19–23). Thus, we postulated that congerins in the abdominal cavity bind to glycoconjugates on nematodes, thereby facilitating their encapsulation. Indeed, we found that nematodes parasitized the abdominal cavity of Japanese conger eel intensively and were frequently encapsulated, damaged, and killed by the peritoneal leukocytes (24). Immunohistochemical staining of peritoneal cells with anti-congerin antibody showed distinct staining, which was depressed by lactose. Furthermore, Con I, Con II, and conger eel C-type lectins were detected in parasitic nematodes and peritoneal cells by MALDI-TOF-MS/MS analysis (24). These results indicate that several lectins, including congerins, exist in the peritoneal cells of the conger eel.
In this study, we identified a new isotype of congerin from the peritoneal fluid of the Japanese conger eel. The cDNA encoding the novel conger eel galectin, termed congerin P (Con-P), was cloned from the peritoneal cells by RT-PCR and rapid amplification of 5′ and 3′ complementary DNA ends (5′- and 3′-RACE). The nucleotide sequence and deduced amino acid sequence of Con-P included 130 amino acid residues, and Con-P shared 21.9 and 22.6% sequence identity with Con I and Con II, respectively. Con-P differed significantly from Con I and Con II due to the presence of a cysteine residue at position 58 and the replacement of seven amino acid residues in CRD, including Trp at position 70, which is conserved among all known galectins. The primary structure of Con-P was elucidated; however, its biological functions and carbohydrate binding activity remained unexplained due to its low expression efficiency and instability in the conger eel and consequent difficulty in the purification of native Con-P. Thus, to understand the biological functions of this unique galectin, a recombinant expression system for Con-P was developed by using the Con II-tag fusion protein system, which was recently developed for the expression of venomous proteins (25). As a result, active recombinant Con-P was prepared and characterized.
EXPERIMENTAL PROCEDURES
Materials
Restriction endonucleases and other enzymes were purchased from Takara Bio Inc. (Kyoto, Japan). Synthetic oligonucleotide primers were custom-synthesized by FASMAC Co., Ltd. (Kanagawa, Japan). Anti-Con-P polyclonal antibody was manufactured by Medical and Biological Laboratories (MBL) Co., Ltd. (Ina, Japan). All reagents used were of the purest grade commercially available.
Preparation of Peritoneal Cells and Fluid from the Conger Eel
Japanese conger eels were purchased from a commercial fish seller and reared in fiberglass-reinforced plastic tanks with running seawater until use. Fish were fed on a commercial diet for eels (Nosan Kogyo, Co., Japan) ad libitum. A total of 21 fish were fasted for 2 days, anesthetized with 2-phenoxyethanol, and sacrificed as reported previously (24). Briefly, whole blood was collected from the caudal vein using a 21-gauge needle attached to a syringe. The ventral skin was inactivated with 70% ethanol and removed before laparotomy to avoid contaminating the peritoneal fluid with skin mucus. Following longitudinal incision of the ventral muscle and peritoneum, the abdominal cavity was washed with 2 ml of ice-cold PBS (10 mm sodium phosphate (pH 7.2), containing 0.15 m NaCl), and the peritoneal cell suspension was recovered.
After washing the abdominal cavity with PBS, the lavage was centrifuged at 700 × g for 15 min. The precipitated cells were immediately frozen and stored at −80 °C for further experiments.
Molecular Cloning and Sequence Determination of Isogalectin cDNA
Total RNA was extracted from the peritoneal cells of conger eel by the guanidinium thiocyanate/phenol/chloroform extraction method (26). Poly(A)+ RNA was purified using the Micro-FastTrack kit (Invitrogen), and cDNA synthesis was performed using poly(A)+ RNA (1 μg) and the Marathon cDNA Amplification kit (Clontech). PCR was conducted using a Takara PCR Thermal Cycler, Pfu turbo DNA polymerase, and the following conditions: 94 °C for 3 min, followed by 30 cycles of 94 °C for 1 min, 50 °C for 1 min, and 72 °C for 1 min.
Oligonucleotide primers (Galco-S, 3′UTRco-A, Con2sp-S, and Con2sp-A) were designed based on the aligned nucleotide sequences of Con I, Con II, and Anguilla japonica lectin 1 (AJL1) from the Japanese eel (27). Common primers used for all three galectins were Galco-S (5′-CGTTTCTCGRTCAATGTGGG-3′) and 3′UTRco-A (5′-ATACTCWGTGAGTTTGCACAGATC-3′), which correspond to the conserved sequences at the N-terminal and 3′-untranslated regions, respectively. RT-PCR was conducted using the Access Quick Master Mix (Promega, Madison, WI) in combination with Pfu Turbo DNA polymerase (Toyobo, Japan) and the appropriate primers (Galco-S and 3′UTRco-A). PCR was conducted using a Takara PCR Thermal Cycler and the following conditions: 94 °C for 2 min, followed by 35 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min.
The 5′- and 3′-RACE method was used to determine the flanking regions and the full sequence of the novel isogalectin, Con-P. For 5′- and 3′-RACE, adaptor primers, AP1 (5′-CCATCCTAATACGACTCACTATAGGGC-3′) and AP2 (5′-ACTCACTATAGGGCTCGAGCGGC-3′) as well as gene-specific primers, GSP1-S (5′-TGGCGTAGTCCTCCATACGCAAACG-3′) and GSP2-AS (5′-TCCTGGTGGGTCTTCAGTTGATACT-3′), were used. PCR products were subcloned into the pCR-TOPO vector by using a TOPO cloning kit (Invitrogen), and the nucleotide sequences of cloned DNA fragments were determined by the dideoxy chain-termination method using universal T7 and SP6 primers and an Applied Biosystems Model 377 and 310 DNA sequencers. The nucleotide sequence data reported in this study were deposited in GenBankTM database under accession number AB109240 (cDNA encoding the novel isogalectin, Con-P, from the peritoneal cells of Japanese conger eel). Multiple sequence alignments were performed with the ClustalW program (28).
Preparation of Anti-Con-P Antibody
Anti-Con-P polyclonal antibody was manufactured by MBL Co., Ltd., using carrier protein-conjugated synthetic oligopeptide derived from Con-P sequence. Peptide sequence for Con-P antigen was assessed by total antigenic score, including secondary structure prediction (Robson and Gamier), accessibility, flexibility, surface probability, hydrophilicity, dipole and antigenicity, and consequently the peptide, HTQTETSFPFQKSRSFE(C), corresponding to the putative loop region (66th to 82nd) of Con-P was designed. Synthetic peptide was purified by reversed phase-HPLC, using Merck C18 (250 × 4.6 mm inner diameter) column, confirmed by MALDI TOF MS, and then conjugated with carrier protein, keyhole limpet hemocyanin, via C-terminal additional cysteine. After rabbits were immunized by carrier protein-conjugated Con-P peptide, anti-Con-P polyclonal antibody was obtained.
Expression of Recombinant Congerin P (rCon-P) in Escherichia coli
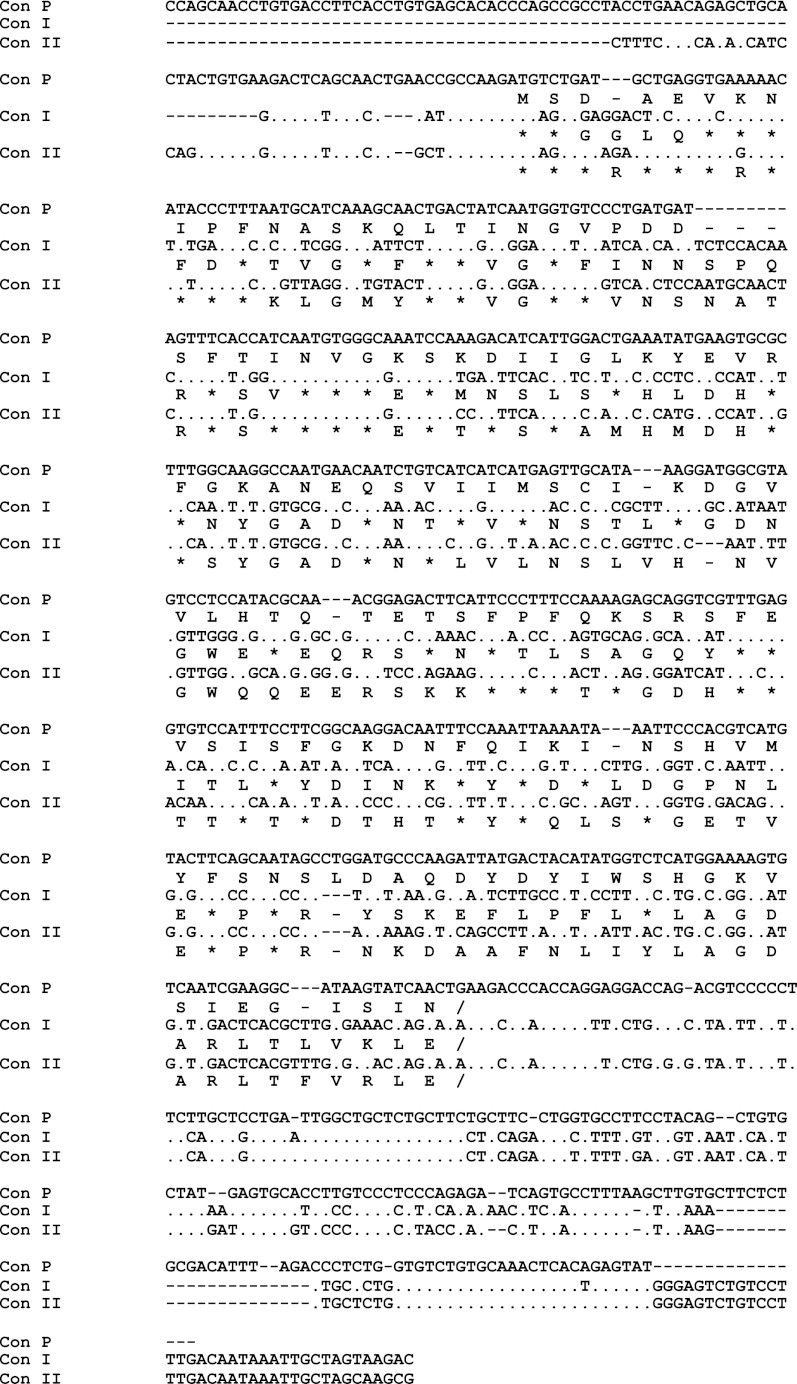
The expression plasmid for rCon-P was constructed by using the Con II-tagged fusion protein system as reported previously for Protobothrops flavoviridis lysine 49-myotoxic phospholipase A2 (BPII) (25). Briefly, the pTV-Con II-tagged plasmid vector, which contained the recognition sequence (FAGP) of the Microbacterium liquefaciens protease (MLP) isolated from the MIM-CG-9535-I strain (29) and an StuI site in the linker region, was prepared by PCR using pTV-Con II-tagged BPII as a template and the following primers: P1, 5′-CGACTAGGCCTCGGTACTAGGATTGGCCCCGCGAACCCCGCTGG-3′, and P2, 5′-TGCAAGAAGGCCTATACATGCTAA-3′. PCR was performed using KOD Plus DNA polymerase (Toyobo Co., Ltd., Osaka, Japan) and the following conditions: an initial denaturing step at 94 °C for 2 min, followed by 30 cycles of a 15-s denaturing step at 94 °C, a 30-s annealing step at 55 °C, and a final extension step at 68 °C for 4 min. The PCR product encoding the pTV-Con II-tagged vector was self-ligated after digestion with StuI, which generates blunt ends. The mature gene encoding Con-P was amplified by PCR to introduce the PstI site at the 3′-terminal end by using the following primers: P3, 5′-TCTGATGCTGAGGTGAAAAAC-3′, and P4, 5′-TCCTCCTGCAGGGTCTTCAGTTGATACT-3′. PCR was performed using KOD Plus DNA polymerase and the following conditions: an initial denaturing step at 94 °C for 2 min, followed by 30 cycles of a 15-s denaturing step at 94 °C, a 30-s annealing step at 50 °C, and a final extension step at 68 °C for 30 s. The amplified PCR product was digested with PstI and ligated into the pTV-Con II-tagged vector between StuI and PstI sites, resulting in the generation of an expression vector for Con-P, pTV-Con II tag-MLP/thrombin-Con-P.
Expression and Purification of rCon-P
The expression plasmid for Con-P was transformed into E. coli JM109 cells with or without the chaperone plasmid pG-Tf2. Transformants were cultured at 30 °C in 2× YT medium containing either 100 μg/ml ampicillin (pTV-Con II-tagged Con-P) or 100 μg/ml ampicillin plus 30 μg/ml chloramphenicol (pTV-Con II-tagged Con-P and pG-Tf2). The expression of recombinant protein was induced by adding isopropyl 1-thio-β-d-galactopyranoside (IPTG) at a final concentration of 1 mm when cells reached logarithmic phase (optical density = 0.2). After an additional 24 h of culture, the cells were harvested by centrifugation. The expression profiles of recombinant proteins were examined by SDS-PAGE on 15% (w/v) gels and detected by Coomassie Blue staining or Western blotting using anti-Con II tag or anti-Con-P antibodies. The expressed proteins were quantified by Western blotting using enhanced chemiluminescence detection reagents (GE Healthcare) and Scion Image software (Scion Co., MD).
Bacterial cells expressing recombinant proteins were resuspended in 50 mm Tris-HCl buffer (pH 7.5) containing 1 mm phenylmethylsulfonyl fluoride and lysed by sonication. The supernatant containing soluble Con II-tagged rCon-P was adsorbed to HCl-treated Sepharose 4B beads by a batch method at 4 °C. The beads washed with 50 mm Tris-HCl (pH 7.5) containing 0.15 m NaCl, and Con II-tagged rCon-P was eluted with 0.2 m lactose. Con II-tagged rCon-P was digested with MLP at a concentration of 1 μg per 500 μg of fusion protein for 10 h at 15 °C to remove the Con II tag. Subsequently, rCon-P was purified by affinity chromatography using mannose-Toyopearl AF-650 M beads by a batch method at 4 °C. Briefly, the beads absorbed to Con-P were washed with 50 mm Tris-HCl (pH 7.5) containing 0.15 m NaCl, and rCon-P was eluted with 0.2 m mannose.
MALDI-TOF MS/MS Analysis
To confirm the structure of rCon-P, purified Con II-tagged rCon-P was reduced, carboxymethylated, and digested with Achromobacter protease I (lysyl endopeptidase) (Wako, Japan) at 37 °C. The digest was desalted, applied to a Sep-Pak column (Waters Associates, Milford, MA), and then separated by using a DiNa Nano LC system equipped with a DiNa MALDI spotting device (KYA Technologies Co., Tokyo, Japan). The peptides were eluted from a reversed phase-HiQ sill C18 column by using a binary gradient of acetonitrile in 0.1% trifluoroacetic acid. The column effluent was mixed directly with the MALDI matrix solution (4 mg/ml of α-cyano-4-hydroxycinnamic acid and 80 μg/ml of ammonium citrate in 70% acetonitrile containing 0.1% trifluoroacetic acid) at a flow rate of 2.5 μl/min before spotting onto MALDI target plates (Opti-TOFTM 384-well insert; AB Sciex, Foster City, CA). The molecular mass and sequence of peptides on MALDI target plates were analyzed by using an AB SCIEX TOF/TOF 5800 Analyzer and 4000 Series Explorer software (version 3.5.1) (AB Sciex). Protein identification was performed with ProteinPilot software (version 3.0; AB Sciex) using the Paragon method. Each MS/MS spectrum was searched against a protein sequence database (NCBInr, downloaded from ftp.ncbi.nih.gov).
Dynamic Light Scattering Technique
Dynamic light scattering was employed to estimate the apparent molecular weights of Con-P and Con II-tagged Con-P by using a DynaPro 99 instrument (Protein Solutions). Protein samples were dissolved in 50 mm Tris-HCl (pH 7.5) at a final concentration of 500 μm. The sample solution was titrated against d-mannose (0.5 μm to 10 mm) at 25 °C.
Hemagglutination Assay
Hemagglutination activity was determined by a serial 2-fold dilution method on a 96-well microtiter plate using rabbit erythrocytes. Each well contained 50 μl of 2-fold serial dilutions of lectin solution, to which 50 μl of 2% (v/v) rabbit erythrocytes was added. Agglutination activity was assessed after incubation for 1 h at room temperature. The maximum dilution with hemagglutination activity was defined as the hemagglutination titer.
Frontal Affinity Chromatography (FAC)
The carbohydrate binding activity of Con-P was analyzed by FAC according to the previously reported method (30, 31). Purified rCon-P was dissolved in coupling buffer (0.2 m NaHCO3 (pH 8.3), 0.5 m NaCl, and 0.1 m mannose) and immobilized onto Hi-Trap N-hydroxysuccinimide (NHS)-activated matrix (GE Healthcare) according to the manufacturer's instructions. To deactivate excess active NHS groups, the resin was blocked and washed with 0.5 m ethanolamine (pH 8.3) in 0.5 m NaCl and 0.1 m sodium acetate buffer (pH 4.0) containing 0.5 m NaCl, respectively. The resin was equilibrated with 1 mm EDTA in 20 mm Na2PO4 (pH 7.2) and packed into a stainless steel column (inner diameter, 4.0 × 10 mm). The column was connected to an HPLC system consisting of an LC9A pump, a fluorescence detector RF-550 (Shimadzu Co., Kyoto, Japan), and a data processing system.
Various pyridylaminated (PA) oligosaccharides (10 nm) were dissolved in elution buffer and applied to the Con-P-immobilized column through a 2-ml sample injection loop at a flow rate of 0.25 ml/min at 25 °C. The elution profiles of PA oligosaccharides were monitored by a fluorescence detector connected with Dynamix Compare Module system (Rainin Instrument Co., Inc.). PA-rhamnose, which had no specific interaction with the immobilized congerins, was used as a negative control in FAC analysis.
Fluorescence Spectroscopy
The fluorescence spectra of Con-P, which contained a single Trp residue at position 117, were measured using a spectrofluorophotometer (RF-5300 PC; Shimadzu Co., Kyoto, Japan). The tryptophan excitation wavelength was set at 280 nm, and the emission spectra were recorded from 300 to 400 nm. The concentration of Con-P was determined by UV spectroscopy using an absorption value of 331 for 1% solution at 280 nm in a 1-cm path length. Samples were dissolved in 50 mm Tris-HCl (pH 7.5) at a final concentration of 250 μm. Mannose was titrated to the sample solution at a concentration ranging from 1 to 100 μm and 5 to 125 mm, respectively. The association constants Ka of Con-P toward mannose were calculated from the Scatchard plots, ΔF/F0 versus ΔF/F0/[Man], where ΔF is the difference (F − F0) of the fluorescence emission intensity, F0 and F, in the absence and presence of a ligand mannose at a concentration [Man], respectively.
RESULTS
Detection and Sequencing of a Novel Galectin from the Abdominal Cavity of Conger Eel
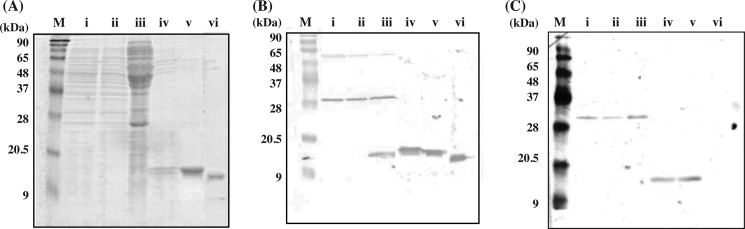
We recently demonstrated that galectins are present in the abdominal cavity of the conger eel and contribute to the encapsulation of nematode parasitic worms (24). To confirm the expression of congerins in the abdominal cavity, RT-PCR was carried out on peritoneal cells using a pair of primers (Galco-S and 3′UTRco-A) that contained sequences conserved in both Con I and Con II. As a result, a DNA band was amplified with the predicted molecular mass of 500 bp (supplemental Fig. S1A). However, the amplified DNA fragment encoded a novel galectin-like amino acid sequence, which differed from that of Con I or Con II (Fig. 1). This novel galectin from the peritoneal cells of conger eel was termed congerin P (Con-P).
FIGURE 1.
Nucleotide sequences and deduced amino acid sequences of Con-P and congerins. Con-P, congerin P; Con I, congerin I; Con II, congerin II. Dashes indicate gaps introduced to maximum sequence identity, and dots and asterisks represent the positions identical to Con-P in the nucleotide and amino acid sequences, respectively. The nucleotide sequences of Con I and Con II were from GenBankTM database with accession codes AB010276 and AB010277, respectively.
The full cDNA sequence encoding Con-P, determined by the RACE method, included 672 nucleotides with a 5′-untranslated region (UTR) of 93 bp, a protein-coding region of 393 bp, and a 3′-UTR of 186 bp. The open reading frame encoded a mature protein of 131 amino acid residues (Fig. 1). Alignment of the amino acid sequences of Con-P with those of Con I and Con II is shown in Fig. 2. Con-P shared only 21.9 and 22.6% amino acid sequence identity with Con I and Con II, respectively. Remarkably, 7 out of 8 amino acid residues in the CRD of Con-P that are conserved among all known galectins, including Trp-70, were substituted to other amino acids. Mutations observed in key amino acids included Lys-44, Ser-61, Val-70, Thr-73, and Thr-75, whereas Arg-48 remained unsubstituted. Greater sequence similarity between Con-P and Con I or Con II was seen for the UTRs compared with that for the protein-coding regions; sectional homologies between Con-P and Con I or Con II were 67 and 54% for the 5′ UTR, 47 and 48% for the coding region, and 64 and 63% for the 3′-UTR, respectively. Mathematical analysis was carried out to compare Con-P and Con I or II cDNA; specifically, the number of nucleotide substitutions per site (KN) for the UTR and the number of nucleotide substitutions per nonsynonymous site (KA) or synonymous site (KS) for the protein-coding region were analyzed. As shown in Table 1, nonsynonymous substitutions have occurred frequently in the protein-coding region, as the KA/KS values of the coding region for pairs of Con-P and Con I or Con-P and Con II were close to 1. The KN/KS values were ∼0.5–0.6, suggesting that the protein-coding region of congerins is more variable than the noncoding regions and that Con-P has evolved without any constraint or accelerated substitution.
FIGURE 2.
Amino acid sequence alignment of galectins from fish and human galectin-1. Con-P, congerin P; Con I, congerin I; Con II, congerin II; AJL1, Japanese eel galectin; Eele, electric eel galectin; hGal-1, human galectin-1. Asterisks show conserved amino acid residues in the CRD. The arrow indicates the Trp-117 residue of Con-P.
TABLE 1.
KN/KS and KA/KS values for pair of congerin P, Con I, and Con II
| Pair of genes | KN | KS | KA | KN/KS | KA/KS | KA/KN |
|---|---|---|---|---|---|---|
| Con-P vs. Con I | 0.505 | 0.956 | 0.906 | 0.53 | 0.95 | 1.79 |
| Con-P vs. Con II | 0.493 | 0.854 | 0.757 | 0.58 | 0.89 | 1.53 |
| Con I vs. Con II | 0.098 | 0.130 | 0.389 | 0.75 | 2.98 | 3.95 |
Gene Expression Analysis
Gene expression of Con-P in various tissues was examined by RT-PCR using specific primers, which distinguish Con-P expression from that of Con I and Con II (supplemental Fig. S1B). In general, Con-P was expressed in almost all of the various tissues analyzed except for spleen; however, expression of Con I and Con II was restricted to the skin and upper digestive tract as reported previously (4, 24). In some specimens, Con-P was the only congerin isotype expressed in the peritoneal cells and was also transcribed in encapsulated bodies in the abdominal cavity.
Expression and Purification of rCon-P
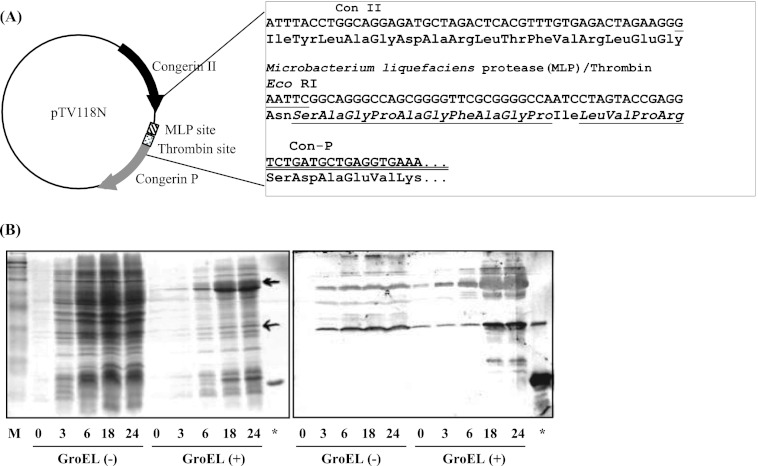
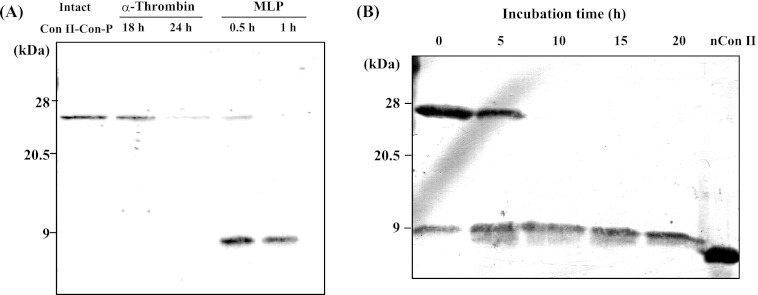
Given the drastic amino acid changes observed in the CRD of Con-P, it was of interest to investigate its biological functions and molecular properties. However, intact Con-P protein could not be obtained, which was probably due to its low expression or instability in the conger eel. Thus, rCon-P was successfully expressed as a fusion protein in E. coli by using a Con II tag expression system, which was previously developed to allow expression of the intractable snake venom phospholipase A2 protein (Fig. 3A) (25). The expression level of Con II-tagged Con-P was increased by co-expression with chaperones (GroES-GroEL-Tf) at 18–24 h post-IPTG induction, and expression reached a maximum at 24 h after IPTG induction (Fig. 3B). As a result, the Con II-tagged rCon-P fusion protein was obtained in soluble form and purified by affinity chromatography on an HCl-treated Sepharose 4B column (supplemental Fig. S2). Subsequently, purified Con II-tagged rCon-P was enzymatically cleaved, as the linker region contains recognition sites for both MLP (GPAG/FAGP) and α-thrombin (LVPR). Although α-thrombin could not cleave the Con II tag, MLP effectively cleaved the Con II tag within 30 min at the optimal temperature range of 37–42 °C (Fig. 4A). However, lectin activity was completely lost following incubation at 40 °C for 60 min. Thus, to prevent inactivation of MLP and nonspecific digestion (inactivation) of Con-P, the cleavage reaction was conducted at 15 °C for 10 h, resulting in successful cleavage of the Con II tag from fusion proteins without degradation or inactivation of Con-P (Fig. 4B). Active rCon-P was purified by affinity chromatography on a mannose-Toyopearl AF-650 M column (Fig. 5) and was obtained with a typical yield of 2.5 mg from 1 liter of culture. Using anti-Con II tag antibody, two positive bands corresponding to the Con II tag and rCon-P, respectively, were detected due to the cross-reactivity of antibody against Con-P in addition to Con II (Fig. 5B). Hence, the purification of rCon-P was also confirmed by using specific anti-Con-P antibody, indicating that rCon-P was successfully obtained as a single band (Fig. 5C). Furthermore, the structure of rCon-P was confirmed by MALDI-TOF MS/MS using ProteinPilot software with the Paragon method after digested with Achromobacter protease I and subsequently separated by a nano-LC system. As a result, the 27 peptide fragments were detected with 95% confidence intervals, and 82.3% of rCon-P sequence was covered with nonredundant 10 peptide fragments, including QLTINGVPDDSFTINVGK (16–33), YEVRFGK (42–48), ANEQSVIIMSCIK (49–61), DGVVLHTQTETSFPFQK (62–78), SRSFEVSIS (79–87), INSHVMYFSN (97–106), FSNSLDAQDYDYIWSHGK (104–121), and VSIEGISIN (122–130) (supplemental Fig. S3).
FIGURE 3.
Recombinant expression of Con II-tagged Con-P. Schematic representation of the expression plasmid for Con II-tagged Con-P (A) and time course expression levels of recombinant protein in E. coli in the absence (GroEL(−)) or presence (GroEL(+)) of chaperones (B) are shown. SDS-PAGE analysis with Coomassie Brilliant Blue staining (left) and Western blot analysis using anti-Con II antibody (right) are shown. Monomeric Con II-tagged Con-P appears at ∼28 kDa. The asterisk indicates native Con II.
FIGURE 4.
Restriction protease digestion of Con II-tagged Con-P fusion protein. Restriction digestion of Con II-tagged Con-P with α-thrombin and MLP at 37 °C (Α) and time course digestion of Con II-tagged Con-P with MLP at 15 °C (B) are shown. nCon II, native Con II. Following MLP digestion, cleaved Con II tag was detected by the presence of a 15-kDa band by using anti-Con II antibody.
FIGURE 5.
Purification of recombinant Con-P. SDS-PAGE analysis (A) and Western blot analysis of Con-P by using anti-Con II-tag antibody (B) or anti-Con-P antibody (C) for each purification step are shown. Restriction digestion of recombinant Con II-tagged Con-P with MLP was at 15 °C. Following MLP digestion, rCon-P was purified by mannose-immobilized TOYOPEARL AF-650 M column. The following are shown: E. coli lysate (lane i); soluble fraction containing Con II-tagged Con-P (lane ii); Con II-tagged Con-P purified by affinity purification on an HCl-treated Sepharose 4B column (lane iii); MLP-digested products of Con II-tagged Con-P (lane iv); purified rCon-P by mannose column (lane v), and native Con II (lane vi).
Carbohydrate Binding Activity of rCon-P
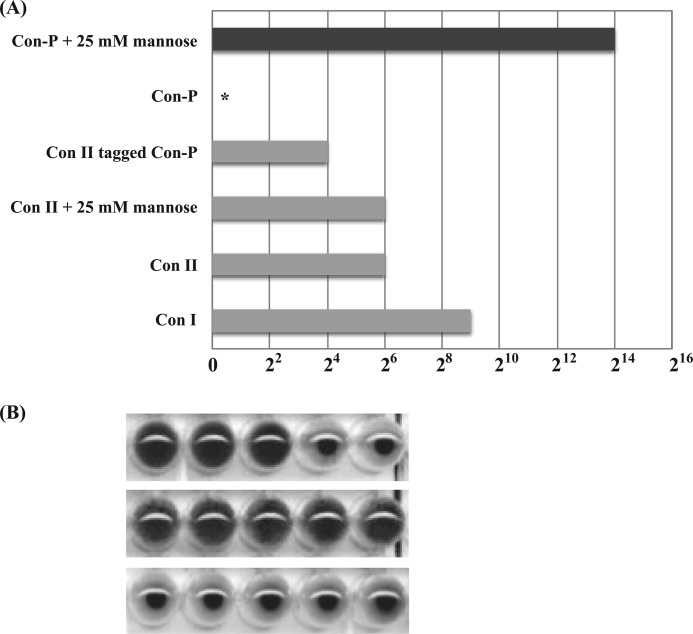
Although purified rCon-P alone showed no hemagglutination activity, it adsorbed to a d-mannose-immobilized column and could be eluted from the column with d-mannose during the purification steps. To clarify the effect of mannose on rCon-P, its hemagglutinating activity with or without 25 mm mannose was assessed. rCon-P showed strong hemagglutinating activity with 25 mm mannose, but no activity was observed in the absence of mannose (Fig. 6). Furthermore, in the presence of mannose, rCon-P displayed 32- and 256-fold greater hemagglutination activity than Con I and Con II, respectively. These results indicate that Con-P has the ability to bind to carbohydrate chains in the presence of mannose and that mannose acts as a modulator of binding. However, the hemagglutinating activity of rCon-P decreased with time and was completely lost after incubation for more than 4.5 h, even at 4 °C (supplemental Fig. S4). In contrast, Con II-tagged Con-P retained its activity. Thus, the hemagglutination assay and sugar inhibition test were carried out immediately after MLP digestion of Con II-tagged Con-P. The strong mannose-activated hemagglutinating activity of Con-P was inhibited by lactose (Fig. 6B), suggesting that Con-P contains a mannose-binding site that affects the binding of β-galactoside ligands via an allosteric mechanism.
FIGURE 6.
Mannose-induced hemagglutination activity of Con-P. Relative hemagglutination activities of congerins at 250 μg/ml (A) and hemagglutination activity of MLP-digested Con II-tagged Con-P (B) are shown. The asterisk indicates no hemagglutinating activity. Con-P showed 30-fold greater hemagglutinating activity than Con I in the presence of 25 mm mannose but not absence of mannose. MLP-digested Con II-tagged Con-P contained equal amounts of Con-P and Con II. Hemagglutination activity of Con-P was assessed in the absence and presence of 25 mm mannose, respectively, and was completely inhibited by 20 mm lactose.
To further elucidate the mechanism of instability and inactivation of Con-P, the process of inactivation of Con-P in solution and the effects of the Con II tag were evaluated by a dynamic light scattering technique (supplemental Fig. S5). Con-P formed a multimer with a high hydrodynamic radius (Rh), indicating that it was in a state of micro-aggregation. This aggregation could not be inhibited by mannose; in contrast, it was somewhat promoted by mannose (supplemental Fig. S5A). Conversely, Con II-tagged Con-P showed a much smaller apparent molecular weight, corresponding to the monomer subunit, compared with Con-P with and without mannose, indicating that the Con II tag plays a role in preventing aggregation of Con-P (supplemental Fig. S5B).
Because purified Con-P was extremely unstable and inactivated within 4.5 h, it was difficult to examine the function of Con-P when expressed alone (supplemental Fig. S4). Thus, to determine the carbohydrate specificity of Con-P by FAC, rCon-P was obtained by MLP digestion on the column after the more stable Con II-tagged Con-P was immobilized onto HiTrap NHS-activated HP resin. Although active Con II tags were contained in the Con-P-immobilized column, the effect of Con II tags on the FAC assay can be considered negligible due to the strong activity of Con-P compared with that of Con II. FAC analysis of Con-P without mannose showed that the chromatograms were similar to those of the negative control (rhamnose). In contrast, Con-P activated with mannose showed a strong interaction with the PA sugars, even at low concentrations, whereas Con II displayed no or very weak affinity with PA sugars at the same concentrations (Fig. 7 and supplemental Fig. S9). In fact, the immobilized Con-P (also Con II tag) on the column, of which concentration was estimated to be 65 pmol from the Bt value (total amount of immobilized ligands) of Woolfe-Hofstee-type plots (supplemental Fig. S7 and supplemental Table S2), was 98.5 times lower than the Con II-conjugated column (Bt, 6.4 nmol) used for FAC assay. Furthermore, no accelerated effect of mannose on Con II was observed, rather Con II showed the competitive inhibitory effects (supplemental Fig. S6). Thus, the Con II tag had no effect on the FAC analysis of Con-P.
FIGURE 7.
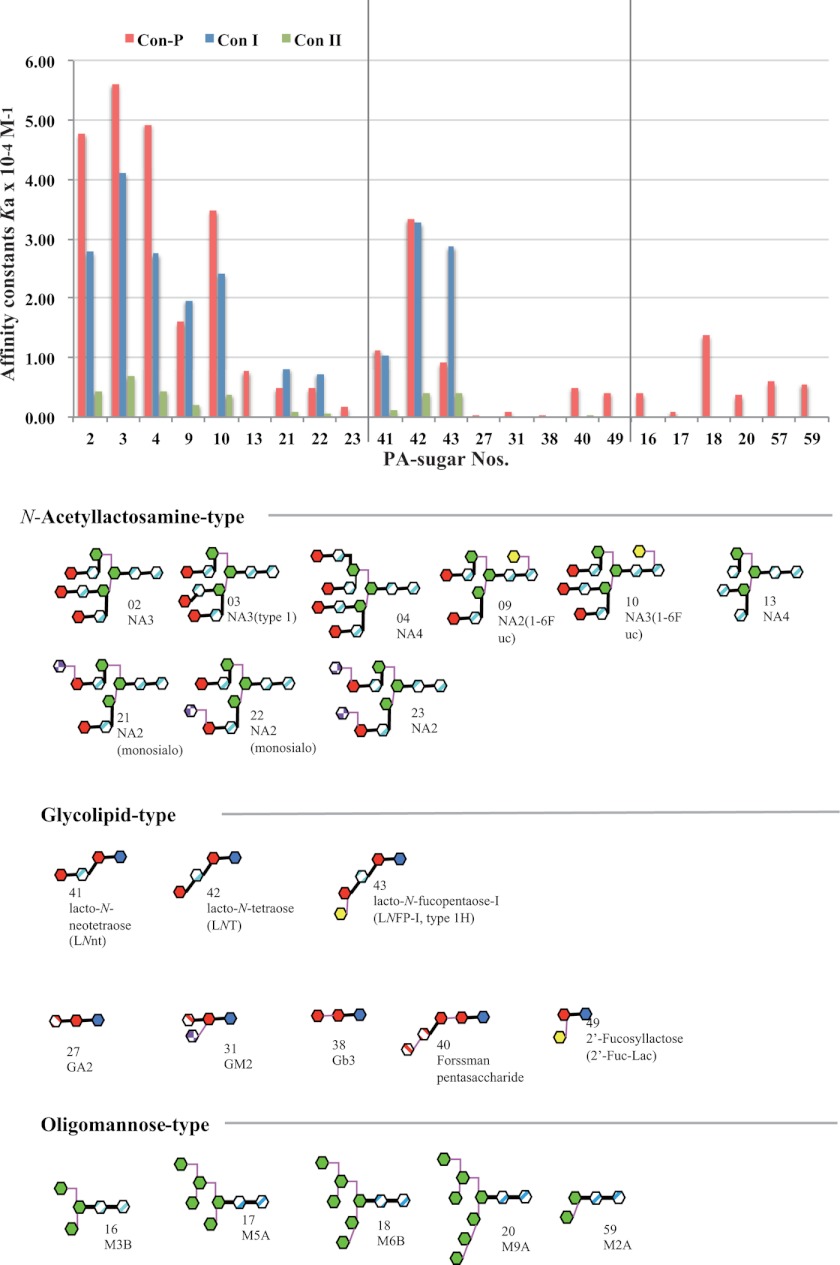
Carbohydrate specificity of activated Con-P. Affinity constants of Con-P, Con I, and Con II for 22 PA sugars were determined by frontal affinity chromatography analysis using buffer containing 20 mm mannose. No data are available for the binding of Con I and Con II to oligo-mannose-type carbohydrates.
The carbohydrate binding specificity of activated rCon-P was analyzed by FAC using 22 types of PA sugars, and the associate constants (Ka) were calculated (Fig. 7 and supplemental Table S1). Con-P displayed high affinity for complex-type N-acetyllactosamine carbohydrates, including NA3, NA3 (type 1), NA4, NA2 (1–6Fuc), NA3 (1–6Fuc), NA2 (monosialo), and NA2 (monosialo) (sugar numbers 2–4, 9, 10, 21, and 22, respectively). Similar to Con I, high affinity of Con-P for specific oligosaccharides, including lacto-N-biosyl (Galβ1–3GlcNAc) or lacto-N-neobiosyl (Galβ1–4GlcNAc) moieties such as LNnT, LNT, and LNFP-I (sugar numbers 41–43), was observed. Con-P also displayed affinity for NA4 (1–6Fuc), NA2 (disialo), GM2, Forssman pentasaccharide, and 2′-Fuc-Lac (sugar numbers 11, 23, 31, 40, and 49) but not for GA2 or Gb3 (sugar numbers 27, 38), which Con I and Con II could not recognize. Interestingly, Con-P showed affinity for oligo (high) mannose-type carbohydrates (sugar numbers 16–18, 20, and 59). In contrast, galectins, including Con I and Con II, do not recognize mannose, with the exception of the human eosinophil Charcot-Leyden crystal protein (human galectin-10) (32) and human placental protein-13 (human galectin-13) (33). Thus, the carbohydrate-binding specificities of Con-P differ significantly from those of other galectins.
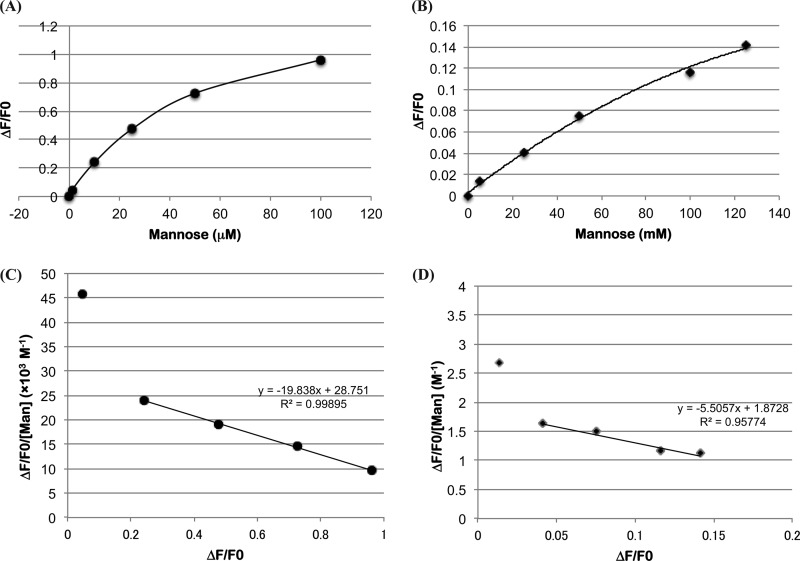
In general, Trp residues in galectins play an important role in carbohydrate binding through stacking interactions. Con-P contained a single Trp residue at position 117 instead of Trp-66 (Fig. 2). To assess the binding properties of Con-P and the effect of mannose on binding, the fluorescence spectra of Con-P in the presence of different concentrations of mannose were measured (Fig. 8). The fluorescence intensity of Con-P gradually increased in response to increasing concentrations of mannose, indicating that mannose binds to Con-P with dual affinity in the ranges of 10–100 μm and 5–125 mm mannose, respectively. The Ka values of Con-P with mannose were estimated by Scatchard plots to be 2.0 × 104 and 5.4 m−1 in the first (corresponding to a range of 10–100 μm mannose) and the second (corresponding to a range of 5–125 mm mannose), respectively (Fig. 8). Furthermore, the yeast mannan also showed the increasing fluorescence intensity in a concentration-dependent manner (supplemental Fig. S9).
FIGURE 8.
Mannose-binding ability of Con-P. Titration of mannose (A and B) with 250 μm Con-P solution and Scatchard plots (C and D) at concentrations ranging from 1 to 100 μm (A and C) and from 5 to 125 mm (B and D), respectively. Fluorescence intensities at 320 nm between the free Trp-117 of Con-P and mannose at a given concentration were measured. Tryptophan excitation wavelength was set to 280 nm, and emission spectrum was recorded from 300 to 400 nm. The association constants Ka of Con-P toward mannose were estimated from Scatchard plots displaying ΔF/F0 versus ΔF/F0/[S]. ΔF, which is the difference in fluorescence intensity at 320 nm between free Con-P and with mannose at a given concentration [S], was normalized by F0.
DISCUSSION
Previously, we isolated and characterized two galectin isotypes, congerins I and II, from conger eel (5, 6, 30). In this study, we identified a novel galectin isotype, termed Con-P, from the peritoneal cells of conger eel. Con-P shared 21.9 and 22.6% amino acid sequence identity with Con I and Con II, respectively, and 20.4% identity with human galectin-1 (Fig. 2). Although Con-P could be classified as a prototype galectin on the basis of sequence homology, it possessed unique amino acid residues in the CRD. The x-ray structural analysis of galectins co-crystallized with Gal(β1–4)GlcNAc (N-acetyllactosamine) or its derivatives has identified several amino acid residues that are important for the carbohydrate-binding ability of galectins, including His-44, Arg-48, Asn-61, Trp-70, Glu-73, and Arg-75 (marked by asterisks in Fig. 2), which are conserved among all known galectins (7, 8, 34). However, with the exception of Arg-48, the corresponding residues in Con-P were substituted with other amino acids, namely H44K, N61S, W70V, E73T, and R75T. Mutations at these key sites have previously been shown to almost completely abrogate carbohydrate binding activity (35).
Mathematical analysis to compare Con-P with Con I or Con II cDNA showed that nonsynonymous substitutions have occurred frequently in the protein-coding region and that Con-P has evolved without any constraint or via accelerated substitutions from a common ancestor gene (Table 1). Similar evolutionary behavior with accelerated evolution has been observed for Con I and Con II as well as in several gene families, including biological offense and defense systems such as snake venom isozymes, conus peptides, and reproduction systems (36). Con-P may also display unique biological activities due to its novel structural characteristics, such as the replacement of 7 out of 8 amino acid residues in the CRD, including Trp-70, which is conserved in all known galectins (Fig. 2).
Intact Con-P protein could not be obtained from conger eels probably due to its low expression or instability. The biological functions and molecular properties of Con-P, including the essential carbohydrate binding (lectin) activity, are still unknown. Thus, the expression of rCon-P protein is essential to clarify its functions and properties. However, we were unable to express recombinant Con-P in E. coli by using a variety of commercially available expression vector systems due to the instability or toxicity of the protein (data not shown). In this study, active rCon-P was expressed in E. coli by using Con II as an affinity and expression tag, which was reported previously to permit expression of toxic proteins, such as snake phospholipases A2, with the correct disulfide bonds and in the soluble form (25). This approach also resulted in successful expression of rCon-P, indicating that the Con II tag expression system is useful for the recombinant expression of toxic and instable proteins. MLP, a collagenase-like metalloprotease isolated from M. liquefaciens MIM-CG-9535-I (29), was essential for the preparation of active rCon-P, as MLP efficiently cleaved the Con II tag at 15 °C without inactivating Con-P (Figs. 4 and 5). Thus, MLP demonstrated high performance cleavage activity of unstable proteins, even at low temperatures, compared with commercially available general proteases.
To further elucidate the mechanism of inactivation of Con-P, the state of Con-P in solution and the effects of the Con II tag were analyzed by a dynamic light scattering technique (supplemental Fig. S5). It revealed that Con-P formed a multimer in a state of micro-aggregation. Conversely, Con II-tagged Con-P showed a much smaller apparent molecular weight, corresponding to the monomer subunit, compared with Con-P with and without mannose. These results indicate that the Con II tag plays a role in preventing aggregation of Con-P (supplemental Fig. S5B).
The most interesting findings in this study were that Con-P showed the greatest hemagglutinating activity (0.06 μg/ml) and carbohydrate binding activity in the presence of d-mannose but not hemagglutinating activity in the absence of mannose (Figs. 6 and 7 and supplemental Fig. S6) despite the substitution of 7 out of consensus 8 amino acids in the galectin CRD. Similarly to the human eosinophil Charcot-Leyden crystal protein (human galectin-10) (32) and the human placental protein-13 (human galectin-13) (33), Con-P displayed mannose-binding specificity, which is unusual among the galectin family. These results suggest that the carbohydrate-binding ability of Con-P is regulated by d-mannoside, which acts as an effector molecule. Thus, Con-P is a new type of galectin with allosteric carbohydrate-binding ability. To our knowledge, Con-P is the first lectin (to be described) that is allosterically modulated by its ligands. However, d-galactose, d-glucose, d-fucose, d-xylose, and l-rhamnose could not substitute for d-mannose (data not shown). Although d-glucose and d-mannose are similar in the direction of hydroxy groups, and some lectins recognize both mannose and glucose by same CRD, Con-P could not recognize glucose moiety as a modulator. It is speculated that secondary axial hydroxyl group of mannoside, which is only different between mannose and glucose, is critical for the binding to Con-P.
Con-P (with the Con II tag) for FAC analysis was immobilized at low concentrations (98.5 times lower) compared with Con I and Con II, and activated Con-P displayed more than 100-fold greater hemagglutinating activity and more than 10-fold affinity for N-acetyllactosamine-type β-galactoside than Con II (Figs. 6 and 7). Furthermore, no accelerated effect of mannose on Con II was observed (supplemental Fig. S6B). For these reasons, the FAC profiles of Con-P in this study appeared to exclude the effects of the Con II tag. Con-P possessed the broad carbohydrate-binding specificity compared with Con I and Con II, as Con-P recognized NA4 (1–6Fuc), NA2 (disialo), GM2, Forssman pentasaccharide, and 2′-Fuc-Lac carbohydrates (sugar numbers 11, 23, 31, 40, and 49, respectively) as well as oligomannose-type carbohydrates (sugar numbers 16, 17, 18, 20, and 59), which Con I and Con II could not recognize (Fig. 7). Thus, Con-P showed significant carbohydrate binding activity as a member of the galectin family, even though Con-P has mutations in 7 out of 8 amino acids in its CRD. These findings led us to question how Con-P can recognize specific carbohydrates with an unusual CRD. To address this question, we analyzed the mannose binding activity by using fluorescence spectra derived from a single tryptophan (Trp-117) near the C-terminal region (Fig. 2). Interestingly, fluorescence of the Trp residue increased in a mannose concentration-dependent manner in two phases. The binding affinity (Ka) of Con-P with mannose and the number of binding site in the first phase, corresponding to a range of 10–100 μm, were estimated to be 2.0 × 104 m−1 and 1.45, respectively (Fig. 8). Also fluorescence of the Trp residue increased in a yeast mannan concentration-dependent manner (supplemental Fig. S9). These results suggest that the two binding sites for mannose existed in Con-P, and the Trp-117 residue in Con-P contributes to the specific binding of both the effector mannose and ligand sugars. Previously, we determined the three-dimensional structure of the following: 1) Con II at 1.45 Å resolution with a molecule of MES, whose shape resembles sulfono-sugar, bound to the extended cleft; 2) the complex structure of Con II and lacto-N-fucopentaose III [Gal β1–4(Fuc α1–3)GlcNAcβ1–3Gal β1–4Glc] at 2.2 Å resolution (8, 9). The structural studies revealed that the extended sugar-binding cleft in Con II stably binds to sugars through amino acid residues such as Tyr-122, which corresponds to Trp-117 in Con-P. These observations suggest that the Trp-117 residue plays a role in the modulating effect of mannose, and mannose-induced conformational changes around the Trp-117 residue might strongly enhance the sugar-binding ability. The other examples of lectin or lectin-like proteins with changing the carbohydrate binding specificity have been found in galectin-related proteins from mushroom (CGL3) and legume lectin from Dolichos biflorus seed (DBL), respectively. In both cases, aromatic amino acid residues in CRD (stacking with pyranose ring) are replaced by other residues such as Arg (in CGL3) and Leu (in DBL) (37–39). In particular, in the case for DBL, it was reported that the specific binding of an N-acetylated sugar was achieved through a give-and-take mechanism; a lack of aromatic stacking against the sugar ring is compensated by a favorable interaction of the N-acetyl group with the lectin, which depends on the presence of a specific subsite, and on the correct positioning of the sugar ring (39). Further studies such as x-ray structural analysis are required to elucidate the detailed mechanisms of carbohydrate recognition and modulation for Con-P by unusual CRD.
Mannose-containing sugars are key molecules involved in pathogen infection and defense responses through binding to lectins such as dendritic cell-specific ICAM-3 grabbing nonintegrin and mannose-binding protein on macrophage cells (40). Both Con I and Con II recognize some marine bacteria (e.g. V. anguillarum) via a galactose or lactose sugar unit, whereas Con-P recognizes oligo-mannose sugars via allosteric regulation. Actually, some mannose-specific lectins bind strongly to parasitic nematodes and nematode eggs (18). Furthermore, Con-P was detected and cloned from the peritoneal cells and the cells that encapsulated parasitic nematodes in the abdominal cavity (24), and Con-P interacted with yeast mannan (supplemental Fig. S9) and mannose to induce lectin activity. Thus, it is possible that Con-P acts as a defense molecule by binding to mannoside on the cell surface of invading pathogens and parasites as natural ligands and activated at the internal and external body surface of the conger eel.
Supplementary Material

This article contains supplemental Figs. S1–S9, Tables S1 and S2, and Methods.
The nucleotide sequence(s) reported in this paper has been submitted to the DDBJ/GenBankTM/EBI Data Bank with accession number(s) AB109240.
- CRD
- carbohydrate recognition domain
- Con I
- congerin I
- Con II
- congerin II
- Con-P
- congerin P
- FAC
- frontal affinity chromatography
- IPTG
- isopropyl 1-thio-β-d-galactopyranoside
- MLP
- M. liquefaciens protease
- NHS
- N-hydroxysuccinimide
- PA
- pyridylaminated
- RACE
- rapid amplification of complementary DNA ends
- rCon-P
- recombinant Con-P.
REFERENCES
- 1. Kasai K., Hirabayashi J. (1996) Galectins. A family of animal lectins that decipher glycocodes. J. Biochem. 119, 1–8 [DOI] [PubMed] [Google Scholar]
- 2. Perillo N. L., Marcus M. E., Baum L. G. (1998) Galectins. Versatile modulators of cell adhesion, cell proliferation, and cell death. J. Mol. Med. 76, 402–412 [DOI] [PubMed] [Google Scholar]
- 3. Nakamura O., Matsuoka H., Ogawa T., Muramoto K., Kamiya H., Watanabe T. (2006) Opsonic effect of congerin, a mucosal galectin of the Japanese conger, Conger myriaster (Brevoort). Fish Shellfish Immunol. 20, 433–435 [DOI] [PubMed] [Google Scholar]
- 4. Ogawa T., Ishii C., Kagawa D., Muramoto K., Kamiya H. (1999) Accelerated evolution in the protein-coding region of galectin cDNAs, congerin I and congerin II, from skin mucus of conger eel (Conger myriaster). Biosci. Biotechnol. Biochem. 63, 1203–1208 [DOI] [PubMed] [Google Scholar]
- 5. Muramoto K., Kamiya H. (1992) The amino-acid sequence of a lectin from conger eel, Conger myriaster, skin mucus. Biochim. Biophys. Acta 1116, 129–136 [DOI] [PubMed] [Google Scholar]
- 6. Muramoto K., Kagawa D., Sato T., Ogawa T., Nishida Y., Kamiya H. (1999) Functional and structural characterization of multiple galectins from the skin mucus of conger eel, Conger myriaster. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 123, 33–45 [DOI] [PubMed] [Google Scholar]
- 7. Shirai T., Mitsuyama C., Niwa Y., Matsui Y., Hotta H., Yamane T., Kamiya H., Ishii C., Ogawa T., Muramoto K. (1999) High resolution structure of the conger eel galectin, congerin I, in lactose-liganded and ligand-free forms. Emergence of a new structure class by accelerated evolution. Structure 7, 1223–1233 [DOI] [PubMed] [Google Scholar]
- 8. Shirai T., Matsui Y., Shionyu-Mitsuyama C., Yamane T., Kamiya H., Ishii C., Ogawa T., Muramoto K. (2002) Crystal structure of a conger eel galectin (congerin II) at 1.45 Å resolution. Implication for the accelerated evolution of a new ligand-binding site following gene duplication. J. Mol. Biol. 321, 879–889 [DOI] [PubMed] [Google Scholar]
- 9. Shirai T., Shionyu-Mitsuyama C., Ogawa T., Muramoto K. (2006) Structure-based studies of the adaptive diversification process of congerins. Mol. Divers 10, 567–573 [DOI] [PubMed] [Google Scholar]
- 10. Konno A., Ogawa T., Shirai T., Muramoto K. (2007) Reconstruction of a probable ancestral form of conger eel galectins revealed their rapid adaptive evolution process for specific carbohydrate recognition. Mol. Biol. Evol. 24, 2504–2514 [DOI] [PubMed] [Google Scholar]
- 11. Konno A., Yonemaru S., Kitagawa A., Muramoto K., Shirai T., Ogawa T. (2010) Protein engineering of conger eel galectins by tracing of molecular evolution using probable ancestral mutants. BMC Evol. Biol. 10, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Konno A., Ogawa T., Shirai T. (2010) in Ribosomal Proteins and Protein Engineering: Design, Selection and Applications (Ortendhal V., Salchow H., eds) pp. 65–90, NOVA Publishers, Inc., Hauppauge, NY [Google Scholar]
- 13. Konno A., Kitagawa A., Watanabe M., Ogawa T., Shirai T. (2011) Tracing protein evolution through ancestral structures of fish galectin. Structure 19, 711–721 [DOI] [PubMed] [Google Scholar]
- 14. Ingram G. A. (1980) Substances involved in the natural resistance of fish to infection. A review. J. Fish Biol. 16, 23–60 [Google Scholar]
- 15. Kamiya H., Muramoto K., Goto R. (1988) Purification and properties of agglutinins from conger eel, Conger myriaster (Brevoort), skin mucus. Dev. Comp. Immunol. 12, 309–318 [DOI] [PubMed] [Google Scholar]
- 16. Nakamura O., Watanabe T., Kamiya H., Muramoto K. (2001) Galectin containing cells in the skin and mucosal tissues in Japanese conger eel, Conger myriaster. An immunohistochemical study. Dev. Comp. Immunol. 25, 431–437 [DOI] [PubMed] [Google Scholar]
- 17. Nakamura O., Ishii T., Ando Y., Amagai H., Oto M., Imafuji T., Tokuyama K. (2002) Potential role of vitamin D receptor gene polymorphism in determining bone phenotype in young male athletes. J. Appl. Physiol. 93, 1973–1979 [DOI] [PubMed] [Google Scholar]
- 18. Hillrichs K., Schnieder T., Forbes A. B., Simcock D. C., Pedley K. C., Simpson H. V. (2012) Use of fluorescent lectin binding to distinguish Teladorsagia circumcincta and Haemonchus contortus eggs, third-stage larvae and adult worms. Parasitol. Res. 110, 449–458 [DOI] [PubMed] [Google Scholar]
- 19. Page A. P., Rudin W., Maizels R. M. (1992) Lectin binding to secretory structures, the cuticle and the surface coat of Toxocara canis-infective larvae. Parasitology 105, 285–296 [DOI] [PubMed] [Google Scholar]
- 20. Araujo A. C., Souto-Padrón T., de Souza W. (1993) Cytochemical localization of carbohydrate residues in microfilariae of Wuchereria bancrofti and Brugia malayi. J. Histochem. Cytochem. 41, 571–578 [DOI] [PubMed] [Google Scholar]
- 21. Rhoads M. L., Fetterer R. H. (1994) Purification and characterization of surface-associated proteins from adult Haemonchus contortus. J. Parasitol. 80, 756–763 [PubMed] [Google Scholar]
- 22. Nayar J. K., Mikarts L. L., Chikilian M. L., Knight J. W., Bradley T. J. (1995) Lectin binding to extracellularly melanized microfilariae of Brugia malayi from the hemocoel of Anopheles quadrimaculatus. J. Invertebr. Pathol. 66, 277–286 [DOI] [PubMed] [Google Scholar]
- 23. Schallig H. D., van Leeuwen M. A. (1996) Carbohydrate epitopes on Haemonchus contortus antigens. Parasitol. Res. 82, 38–42 [DOI] [PubMed] [Google Scholar]
- 24. Nakamura O., Watanabe M., Ogawa T., Muramoto K., Ogawa K., Tsutsui S., Kamiya H. (2012) Galectins in the abdominal cavity of the conger eel Conger myriaster participate in the cellular encapsulation of parasitic nematodes by host cells. Fish Shellfish Immunol., in press [DOI] [PubMed] [Google Scholar]
- 25. Seto M., Ogawa T., Kodama K., Muramoto K., Kanayama Y., Sakai Y., Chijiwa T., Ohno M. (2008) A novel recombinant system for functional expression of myonecrotic snake phospholipase A2 in Escherichia coli using a new fusion affinity tag. Protein Expr. Purif. 58, 194–202 [DOI] [PubMed] [Google Scholar]
- 26. Chomczynski P., Sacchi N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate/phenol/chloroform extraction. Anal. Biochem. 162, 156–159 [DOI] [PubMed] [Google Scholar]
- 27. Tasumi S., Yang W. J., Usami T., Tsutsui S., Ohira T., Kawazoe I., Wilder M. N., Aida K., Suzuki Y. (2004) Characteristics and primary structure of a galectin in the skin mucus of the Japanese eel, Anguilla japonica. Dev. Comp. Immunol. 28, 325–335 [DOI] [PubMed] [Google Scholar]
- 28. Thompson J. D., Higgins D. G., Gibson T. J. (1994) CLUSTAL W. Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kanayama Y., Sakai Y. (2005) Purification and properties of a new type of protease produced by Microbacterium liquefaciens. Biosci. Biotechnol. Biochem. 69, 916–921 [DOI] [PubMed] [Google Scholar]
- 30. Hirabayashi J., Hashidate T., Arata Y., Nishi N., Nakamura T., Hirashima M., Urashima T., Oka T., Futai M., Muller W. E., Yagi F., Kasai K. (2002) Oligosaccharide specificity of galectins: a search by frontal affinity chromatography. Biochim. Biophys. Acta 1572, 232–254 [DOI] [PubMed] [Google Scholar]
- 31. Hirabayashi J., Arata Y., Kasai K. (2003) Frontal affinity chromatography as a tool for elucidation of sugar recognition properties of lectins. Methods Enzymol. 362, 353–368 [DOI] [PubMed] [Google Scholar]
- 32. Swaminathan G. J., Leonidas D. D., Savage M. P., Ackerman S. J., Acharya K. R. (1999) Selective recognition of mannose by the human eosinophil Charcot-Leyden crystal protein (galectin-10). A crystallographic study at 1.8 Å resolution. Biochemistry 38, 13837–13843 [DOI] [PubMed] [Google Scholar]
- 33. Than N. G., Pick E., Bellyei S., Szigeti A., Burger O., Berente Z., Janaky T., Boronkai A., Kliman H., Meiri H., Bohn H., Than G. N., Sumegi B. (2004) Functional analyses of placental protein 13/galectin-13. Eur. J. Biochem. 271, 1065–1078 [DOI] [PubMed] [Google Scholar]
- 34. Barondes S. H., Cooper D. N., Gitt M. A., Leffler H. (1994) Galectins. Structure and function of a large family of animal lectins. J. Biol. Chem. 269, 20807–20810 [PubMed] [Google Scholar]
- 35. Hirabayashi J., Kasai K. (1994) Further evidence by site-directed mutagenesis that conserved hydrophilic residues form a carbohydrate-binding site of human galectin-1. Glycoconj. J. 11, 437–442 [DOI] [PubMed] [Google Scholar]
- 36. Ogawa T. (2006) Molecular diversity of proteins in biological offense and defense systems. Mol. Divers 10, 511–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wälti M. A., Walser P. J., Thore S., Grünler A., Bednar M., Künzler M., Aebi M. (2008) Structural basis for chitotetraose coordination by CGL3, a novel galectin-related protein from Coprinopsis cinerea. J. Mol. Biol. 379, 146–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bouckaert J., Hamelryck T., Wyns L., Loris R. (1999) Novel structures of plant lectins and their complexes with carbohydrates. Curr. Opin. Struct. Biol. 9, 572–577 [DOI] [PubMed] [Google Scholar]
- 39. Hamelryck T. W., Loris R., Bouckaert J., Dao-Thi M. H., Strecker G., Imberty A., Fernandez E., Wyns L., Etzler M. E. (1999) Carbohydrate binding, quaternary structure, and a novel hydrophobic binding site in two legume lectin oligomers from Dolichos biflorus. J. Mol. Biol. 286, 1161–1177 [DOI] [PubMed] [Google Scholar]
- 40. Torrelles J. B., Azad A. K., Henning L. N., Carlson T. K., Schlesinger L. S. (2008) Role of C-type lectins in mycobacterial infections. Curr. Drug Targets 9, 102–112 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.