Using powerful tools to investigate the regulation of global gene expression in the model microalga Chlamydomonas reinhardtii, we observed an impact of CO2 and CIA5, a key transcription regulator, on expression of almost 25% of all the genes. We also discovered an array of gene clusters with distinctive expression patterns that provide insight into the regulatory interaction between CIA5 and CO2.
Abstract
We used RNA sequencing to query the Chlamydomonas reinhardtii transcriptome for regulation by CO2 and by the transcription regulator CIA5 (CCM1). Both CO2 and CIA5 are known to play roles in acclimation to low CO2 and in induction of an essential CO2-concentrating mechanism (CCM), but less is known about their interaction and impact on the whole transcriptome. Our comparison of the transcriptome of a wild type versus a cia5 mutant strain under three different CO2 conditions, high CO2 (5%), low CO2 (0.03 to 0.05%), and very low CO2 (<0.02%), provided an entry into global changes in the gene expression patterns occurring in response to the interaction between CO2 and CIA5. We observed a massive impact of CIA5 and CO2 on the transcriptome, affecting almost 25% of all Chlamydomonas genes, and we discovered an array of gene clusters with distinctive expression patterns that provide insight into the regulatory interaction between CIA5 and CO2. Several individual clusters respond primarily to either CIA5 or CO2, providing access to genes regulated by one factor but decoupled from the other. Three distinct clusters clearly associated with CCM-related genes may represent a rich source of candidates for new CCM components, including a small cluster of genes encoding putative inorganic carbon transporters.
INTRODUCTION
The photosynthetic conversion of inorganic carbon (Ci) into organic form is responsible for the abundance of biomass on earth. In this process, ribulose-1,5-bis-phosphate carboxylase/oxygenase (Rubisco) catalyzes the initial incorporation of CO2 via the carboxylation of ribulose bisphosphate by CO2 (reviewed in Andersson, 2008). Although critically important, the catalytic activity of Rubisco is slow compared with many other enzymes and also cannot discriminate completely between CO2 and O2; the oxygenation of ribulose bisphosphate is competitive with the carboxylation reaction. Under present atmospheric conditions, CO2 assimilation rates often are limited by the CO2 concentration, and in many photosynthetic species, ranging from cyanobacteria and algae to C4 vascular plants, an active CO2-concentrating mechanism (CCM) has evolved to help offset the deficiencies of Rubisco (Raven et al., 2008). CCMs are especially prevalent in aquatic photosynthetic organisms.
Chlamydomonas reinhardtii, a unicellular green alga that serves as a reference organism, also exhibits acclimations to varied CO2 levels (reviewed in Spalding, 2009). C. reinhardtii must overcome the 10,000-fold slower diffusion of CO2 in water relative to air. Thus, active transport and accumulation of Ci, either as CO2 or as HCO3−, plays a critical role in the C. reinhardtii CCM (Moroney and Ynalvez, 2007; Spalding, 2008). Internal accumulation of Ci occurs against a large concentration gradient, so accumulation must occur as HCO3− because its permeability across lipid membranes is 1000-fold lower than that of CO2. However, Rubisco uses CO2 as substrate, so, along with Ci transporters, carbonic anhydrases (CAs), which catalyze interconversion of CO2 and HCO3−, also play important roles in the CCM, (Spalding et al., 1983a; Coleman and Grossman, 1984; Moroney et al., 2011).
The C. reinhardtii CCM is induced by low CO2 concentrations, and the discovery of CCM-related genes has been based on identifying genes with elevated expression under limiting CO2 (lower than 0.05%) compared with high CO2 (1 to 5% CO2) (Spalding and Jeffrey, 1989; Chen et al., 1997; Somanchi and Moroney, 1999; Miura et al., 2004; Yamano and Fukuzawa, 2009). Many CAs and putative transporters or other LCI (for low CO2 inducible) genes have been discovered by this criterion and have been hypothesized to relate to the CCM of C. reinhardtii (Miura et al., 2004; Yamano and Fukuzawa, 2009).
The detailed regulatory mechanisms of the CCM remain unclear, but two important transcription regulators have been identified and characterized based on their relationship to the CCM. A zinc-finger type transcription regulator, CIA5 (or CCM1), was identified by complementation of the cia5 mutant (Moroney et al., 1989), which is unable to acclimate to limiting CO2 conditions, and, independently, by cloning of a tagged allele of cia5, ccm1 (Fukuzawa et al., 2001; Xiang et al., 2001). Expression of most putative Ci transporters and induced CAs requires CIA5, even though the expression of CIA5 itself does not depend on the CO2 level, so posttranslational activation of CIA5 in low CO2 apparently is required for CIA5 to regulate these genes (Fukuzawa et al., 2001; Xiang et al., 2001; Miura et al., 2004). Another transcription regulator, low-CO2 response regulator1 (LCR1), has a Myb domain and appears to regulate the expression of at least three limiting CO2 induced genes, carbonic anhydrase1 (CAH1), low-CO2-induced gene1 (LCI1), and LCI6. LCR1 itself also is induced by limiting CO2, and this induction requires Ci accumulation5 (CIA5) (Yoshioka et al., 2004). Because of the extensive connection of CIA5 to regulation of the CCM-related genes, including LCR1, CIA5 is often called the master regulator of the CCM.
Regarding the mechanism of Ci transport and accumulation in the CCM, the first barrier to Ci uptake is the plasma membrane. Two CIA5-regulated genes encoding candidate transporters have been implicated in Ci transport across the plasma membrane: high light–induced gene3 (HLA3) encodes a putative ATP binding cassette type transporter and is induced under low CO2 conditions, and knockdown of its expression impairs photosynthesis, Ci uptake, and growth in alkaline conditions (Duanmu et al., 2009a). LCI1 encodes a plasma membrane protein reported to increase Ci uptake in LCR1 mutants when expressed transgenically (Ohnishi et al., 2010). Two Rhesus-like proteins, RHP1 and RHP2, also are predicted to be plasma membrane located (Yoshihara et al., 2008). The RHP1 protein has been proposed as a CO2 channel to facilitate CO2 influx under high CO2 conditions (Soupene et al., 2002, 2004), and its expression is reportedly upregulated in high CO2.
Some chloroplast envelope proteins also are candidates to transport Ci into the stroma. The low CO2–induced gene A (LCIA [NAR1.2]) gene, which encodes a Formate/Nitrite Transporter family protein targeted to the chloroplast envelope, is induced in low CO2 and requires CIA5 for expression (Galván et al., 2002; Miura et al., 2004). LCIA has been reported to increase HCO3− transport when transfected into Xenopus laevis oocytes (Mariscal et al., 2006), and its product has been implicated in Ci transport in HLA3-LCIA coknockdown C. reinhardtii strains (Duanmu et al., 2009a). RNA interference knockdown of chloroplast carrier protein1 (CCP1) and CCP2, which encode nearly identical, LCI chloroplast envelope proteins (Spalding and Jeffrey, 1989; Ramazanov et al., 1993; Chen et al., 1997) resulted in poor growth under low CO2 conditions, although no direct evidence for a defect in Ci transport or photosynthesis was demonstrated (Pollock et al., 2004).
The combined transport of HCO3− across the plasma membrane and the chloroplast envelope results in the accumulation of HCO3− in the chloroplast stroma. Since Rubisco, located in the pyrenoid, cannot use HCO3−, a specific CA, carbonic anhydrase3 (CAH3), dehydrates the accumulated HCO3− to CO2 in the thylakoid lumen, taking advantage of the acidic lumen environment to drive nearly complete conversion of HCO3− to CO2 (Spalding, 2008; Moroney et al., 2011). This essential role of CAH3 also mandates the transport or facilitated diffusion of HCO3− across the thylakoid membrane, but this has not yet been demonstrated.
Another set of low CO2–induced genes, low CO2–induced gene B (LCIB) and three related genes, LCIC, LCID, and LCIE, also have been implicated in Ci transport and accumulation even though they are predicted to be soluble chloroplast proteins (Wang and Spalding, 2006; Moroney and Ynalvez, 2007; Spalding, 2008). LCIB mutants fail to accumulate internal Ci in low CO2 conditions and are thus unable to grow in air levels of CO2 (Spalding et al., 1983b; Wang and Spalding, 2006). Notably, LCIB mutants have revealed the existence of a third acclimation state at very low CO2 concentrations (<0.02%): Both LCIB allelic mutants pmp1 and ad1 die under low CO2 conditions (<0.05 and >0.02%) but are able to grow slowly under very low CO2 (<0.02%) conditions (Wang and Spalding, 2006).
Because transmembrane domains are not evident, these LCIB family proteins cannot be stand-alone Ci transporters. It has been suggested that they might serve as Ci transport regulators or as Ci transport complex subunits (Wang and Spalding, 2006), but LCIB either distributes through the stroma or concentrates around the pyrenoid (Duanmu et al., 2009b; Yamano et al., 2010), making interaction with Ci transporters unlikely. Mutants defective in the thylakoid lumen CA, CAH3, suppress the LCIB mutation phenotype (Duanmu et al., 2009b), suggesting a role for LCIB and LCIC, with which LCIB forms a heteromeric complex (Yamano et al., 2010), in preventing the leakage of CO2 from the stroma. CAH6, a putative chloroplast stromal CA, also may be involved in CO2-to-HCO3− conversion in the stroma to reduce diffusive loss of CO2 from the chloroplast (Mitra et al., 2004).
Even though the C. reinhardtii CCM has been extensively studied in recent years, we still know little about the limiting CO2 acclimation process, and the potential for discovery of new genes involved in this process is very high. The acclimation to limiting CO2 and induction of the CCM in C. reinhardtii appear to be regulated by the master regulator, CIA5 (or CCM1) (Miura et al., 2004). The cia5 mutant appears to completely lack induction of the CCM, although it is viable under high CO2 conditions and grows more slowly than the wild type in air levels of CO2. Also, most identified LCI genes remain uninduced when cia5 is exposed to low CO2 (Moroney et al., 1989; Spalding et al., 2002). Aside from being a critical upstream regulator of the CCM and other low CO2 acclimation responses and likely requiring posttranslational activation in low CO2, the details of CIA5 function remain undiscovered. CIA5 has been proposed to be a transcription regulator (Fukuzawa et al., 2001; Xiang et al., 2001), but we know very little about sequences recognized by its putative DNA binding domain or the genes it directly regulates downstream.
To better understand the CCM and low CO2 acclimation of C. reinhardtii in general, as well as the function of CIA5, we conducted RNA sequencing (RNA-Seq) experiments employing the Illumina Genome Analyzer II because of its superiority over the traditional microarray methods (González-Ballester et al., 2010; Wang et al., 2010; Castruita et al., 2011) using two C. reinhardtii strains: the 137c wild type (cc125) and cia5 (cc2702), a mutant in the 137c background with a point mutation in CIA5. To also gain insight into the multiple acclimation states, the strains were grown at three different CO2 concentrations as quantified below: high CO2 (H-CO2), low CO2 (L-CO2), and very low CO2 (VL-CO2). Our transcriptome comparison identified a massive impact of CIA5 and CO2 on the transcriptome and revealed an array of gene clusters with distinctive expression patterns that provide insight into the regulatory interaction between CIA5 and CO2. Individual gene clusters responded primarily to CIA5, to CO2, or to an interaction between the two. This study of transcriptome-wide gene expression patterns provides insight into the massive impact of these two factors and their interaction on C. reinhardtii gene expression in addition to identifying compelling new candidates for CCM functional components.
RESULTS
Identification of Differentially Expressed Genes
This transcriptome study was designed employing three CO2 acclimation states, H-CO2 (5% CO2), L-CO2 (0.033 to 0.041%), and VL-CO2 (0.011 to 0.015%), and two strains (genotypes): the cia5 mutant and its original wild-type progenitor, 137c. Processing of RNA samples on the Illumina Genome Analyzer system yielded more than 12 million reads mapped to the transcriptome for each sample, and more than 90% of these were uniquely mapped to the C. reinhardtii genome (see Supplemental Table 1 online). We detected expression for 15,649 of 15,818 filtered Augustus 5.0 gene models (>99% coverage). Since Augustus 5.0 predictions were based on the Chlamydomonas Version 4 genome assembly, we also acquired annotation information from the filtered Version 4 model set available from the Joint Genome Initiative database as user annotation references and sources for the common gene names.
As an aid to examining gene expression level distributions, we calculated the reads per kilobase of exon model per million of aligned reads (RPKM) values as normalized expression estimates for each gene model in each sample. The shape of distributions for the average RPKM values are very similar among the six conditions, as are the 5th, 50th, and 95th percentiles of these distributions (see Supplemental Figure 1 online). Also, the calculated correlation coefficients, based on the log-transformed RPKM values after eliminating genes with zero count in either of the two replicates, between the two biological replicates for each condition range from 0.935 to 0.983, indicating high correlation between replicates.
To evaluate the reliability of our RNA-Seq results, we performed quantitative PCR (qPCR) on eight previously studied genes (CAH1, CAH3, CAH6, CIA5/CCM1, HLA3, LCIB, LCIE, and RHP1) using the same RNA samples as those used for RNA-Seq. These genes were selected to represent a wide range of expression levels and expression patterns under the conditions used. For all eight genes, the expression patterns from RNA-Seq and qPCR agree very well visually and also are highly correlated, with correlation coefficients ranging from 0.92 to 0.995 (see Supplemental Figure 2 online).
After validating our RNA-Seq results with qPCR, we applied a generalized linear model analysis based on a negative binomial distribution and conducted an overall test to determine which genes vary in expression among any of the six treatment groups, where a treatment group is defined by a strain-by-induction condition combination (see Methods for details). While controlling the false discovery rate (FDR) at 2.5% using Benjamini and Hochberg’s method (Benjamini and Hochberg 1995), we identified 3678 genes as differentially expressed (DE) among the six treatment groups (see Supplemental Data Set 1 online). This number is similar in scale to the 5884 DE genes at 30, 60, or 180 min after CO2 deprivation in wild-type C. reinhardtii cells reported in the companion publication (Brueggeman et al., 2012).
The overall test identified genes with differential expression in any of the six treatment groups. The transcript levels of these genes might be affected by: (1) the CO2 concentration, (2) the presence/absence of functional CIA5, and/or (3) the interaction of CO2 concentration and the presence/absence of functional CIA5. To provide more detailed information about how the CO2 level or the presence/absence of CIA5 affects gene expression, we used a C/S impact model. Under this model, we separately tested for a CO2 effect (due to varied CO2 levels; C-effect), a strain effect (due to varied genotypes; S-effect), and an interaction effect between CO2 levels and genotype (CS-effect) using the generalized linear model (C/S impact test) as described in Methods. When we control the FDR level at 2.5%, this C/S impact test identifies most of the DE genes, with only 165 of the 3678 DE genes identified from the overall test failing to show significance for any one of the three possible effects. Among the other 3513 genes, 2230 exhibit significant C-effect, 2787 exhibit significant S-effect, and 372 exhibit significant CS-effect (see Supplemental Data Set 1 online).
To facilitate a closer comparison of previously reported LCI genes with our results, we also conducted a pairwise comparison of our expression data for the wild type in H-CO2, L-CO2, and VL-CO2 conditions using the DESeq package (Anders and Huber, 2010), which was reported to be one of the best methods for identifying DE genes between two treatment groups (Kvam et al., 2012). When we controlled FDR at level 2.5% using Benjamini and Hochberg’s method (Benjamini and Hochberg, 1995), we identified 345 genes DE for the L-CO2 versus H-CO2 pairwise comparison and 696 genes DE for the VL-CO2 versus H-CO2 pairwise comparison (see Supplemental Data Set 2 online). Surprisingly, no genes were identified as DE for the VL-CO2 versus L-CO2 pairwise comparison.
Reproducibility across Laboratories
The companion study by Brueggeman et al. (2012) focused exclusively on the effects of CO2 deprivation on gene expression. Their focus on the time course for induction from 0 to 3 h nicely complements our study, which compares the impact of CO2 deprivation and CIA5 on gene expression following a 4-h induction in limiting CO2. Their findings support many of our observations and conclusions regarding transcriptome changes associated with CO2 deprivation. Nonetheless, the two studies were conducted completely independently and involved significant differences in experimental conditions (e.g., light and temperature) and in the C. reinhardtii strains used. Therefore, the differences found in the patterns and the magnitude of gene expression between these two studies are not unexpected (i.e., considerably lower interlaboratory reproducibility than intralaboratory reproducibility is expected). Further details about the reproducibility of our expression estimates and their correlation with results from our companion article can be found at the end of Methods and in Supplemental Figures 3 to 5 online.
Clusters of Genes with Similar Expression Patterns
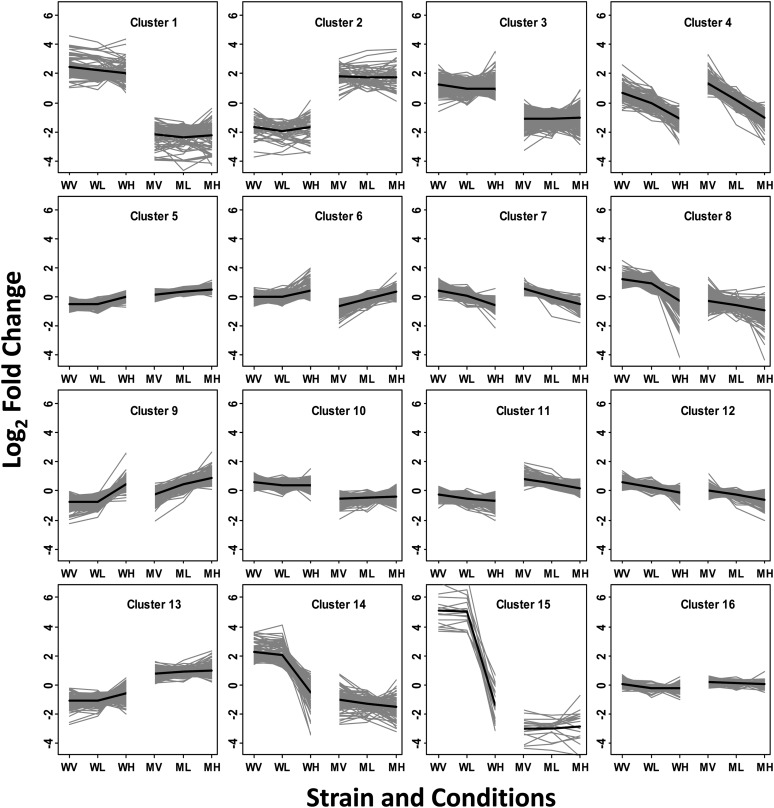
We applied a model-based clustering algorithm to identify distinct gene expression profiles among identified DE genes and chose a total of 16 clusters to maintain as few tight clusters as possible while including most of the distinct expression patterns (Figure 1). Each gray line in Figure 1 represents the expression pattern for an individual gene, and the single black line indicates the average behavior for all genes in that cluster.
Figure 1.
Clustering.
The expression patterns of 3678 selected DE genes for 16 clusters. The horizontal axis indicates each strain and CO2 induction condition: WV, the wild type under VL-CO2 induction; WL, the wild type under L-CO2 induction; WH, the wild type under H-CO2 induction; MV, cia5 under VL-CO2 induction; ML, cia5 under L-CO2 induction; MH, cia5 under H-CO2 induction. The vertical axis indicates the log2 fold change calculated between each condition and the average across all six conditions. Each gray line symbolizes the expression pattern of one gene, and the bold back line illustrates the average expression pattern of all genes in each cluster.
When sorted by cluster, the C/S impact test results (see Supplemental Figure 6 online) confirmed many of the visually observed patterns in the clusters. For example, gene expression patterns in clusters 1, 2, 3, 10, 11, and 13, the CIA5 clusters, appear to be affected mainly by the presence/absence of CIA5 but only minimally by CO2. In agreement with this, 1060 (∼76%) of the 1396 genes in these six CIA5 clusters exhibit only a significant S-effect (no C-effect or CS-effect).
By contrast, the expression patterns in clusters 4, 7, and 9, the CO2 clusters, appear to respond to variation in CO2, with little apparent difference between the genotypes. Accordingly, 415 of the 764 genes in these three CO2 clusters exhibit only a significant C-effect (no S-effect or CS-effect). Like those in the CO2 clusters, genes in clusters 6 and 12 also exhibit visually parallel changes in response to changing CO2 between the genotypes but also show a slightly larger expression shift between the genotypes (Figure 1). Many genes in these two pseudo-CO2 clusters exhibit C+S effects or only C-effect, but the larger proportion of genes exhibiting S-effect distinguishes them from the CO2 clusters (see Supplemental Figure 6 online).
Clusters 8, 14, and 15, the CCM clusters, exhibit a pattern of induction or upregulation under limiting CO2 and repression by the absence of CIA5. These three CCM clusters also contain a considerable number of genes exhibiting significant C+S-effects (both C- and S-effects) and C+S+CS-effects (all C-, S-, and CS-effects) as well as a large number of genes exhibiting only S-effects and few or no genes exhibiting only C-effects (see Supplemental Figure 6 online).
Genes in clusters 5 and 16 show the mildest changes over the six strain-by-treatment combinations (see Supplemental Figure 6 online) and exhibit a mix of genes with all three effects (C-effect, S-effect, and CS-effect). These two clusters also include the largest proportion of genes that were detected by the overall test but not the individual test for any one of the three possible effects from the C/S impact model.
Functional Implications of the Gene Expression Clusters
In addition to the distribution of genes into clusters based on similar expression patterns, we used two complementary methods to examine the DE genes within each cluster for commonalities of function: the Algal Functional Annotation Tool (Lopez et al., 2011) and manual curation. In employing the Algal Functional Annotation Tool, we used the Gene Ontology (GO) terms based on orthology to Arabidopsis thaliana to overcome the limitation of available annotations for C. reinhardtii. We compiled all GO terms that showed statistical significance (P < 0.01) in at least one gene cluster and generated a summary heat map to visualize an overview of the resulting functional information by clusters (see Supplemental Figure 7 online). A detailed list of GO terms identified for the clusters can be found in Supplemental Data Set 3 online. In the heat map, GO terms were subjected to hierarchical clustering so that gene clusters with common significant ontology terms are placed close to each other in the tree. Although details of the identified GO terms and associated genes corresponding to the heat map are found in Supplemental Data Set 3 online, Supplemental Figure 7 online illustrates that very few of the significant GO terms (20 out of 210) overlap among any of the 16 cluster entries, which suggests that the genes separated into clusters based on distinctive expression patterns also tend to be involved in varied biological processes, providing independent support for our clustering results.
In addition to tabulating the significant GO terms associated with each gene cluster, the total number of unique genes represented within all of the significant GO categories for each cluster was determined. For example, Table 1 indicates that 22 GO terms were identified by the Algal Functional Annotation Tool to be associated with cluster 1, but these 22 GO terms represent only three unique genes, since each of the three genes is associated with multiple GO terms. This example is not unique; in many of the clusters, the significant GO category hits represented only a small number of individual genes, even if the number of GO category hits was high. On the other hand, 60 unique genes (32.1% of the genes) in cluster 5 were included among the significant GO term hits. However, only in six of the clusters, (4, 5, 6, 9, 10, and 11) were at least 8% of the genes in the cluster identified among the GO hits.
Table 1. GO Categories of Clusters.
| Clusters |
Functional Annotation Tool |
Manual Curation |
||||||
|---|---|---|---|---|---|---|---|---|
| Cluster | Total Genes | Cluster Group | GO Terms | Unique Genes | Unique Genes (%) | Main GO Termsa | Primary Functional Category | Total (%) |
| 1 | 124 | CIA5 | 22 | 3 | 2.4% | N/S | Signaling | 16% |
| 2 | 95 | CIA5 | 9 | 4 | 4.2% | N/S | Metabolism | 19% |
| 3 | 387 | CIA5 | 25 | 8 | 2.1% | N/S | Signaling | 14% |
| 4 | 150 | CO2 | 45 | 15 | 10.0% | Catabolic processes | Metabolism | 24% |
| 5 | 187 | – | 33 | 60 | 32.1% | Biosynthetic processes | Metabolism | 29% |
| 6 | 240 | – | 10 | 30 | 12.5% | RNA modification; protein localization | Gene expression regulation | 23% |
| 7 | 243 | CO2 | 4 | 7 | 2.9% | N/S | Metabolism | 16% |
| 8 | 360 | CCM | 6 | 8 | 2.2% | N/S | Metabolism | 12% |
| 9 | 371 | CO2 | 23 | 49 | 13.2% | RNA processes, nitrogen metabolism | Gene expression regulation | 28% |
| 10 | 460 | CIA5 | 34 | 63 | 13.7% | Intracellular trafficking, proteolysis, regulation of processes | Signaling | 19% |
| 11 | 183 | CIA5 | 10 | 15 | 8.2% | Small molecule metabolic processes | Metabolism | 22% |
| 12 | 463 | – | 1 | 3 | 0.7% | N/S | Signaling | 13% |
| 13 | 147 | CIA5 | 2 | 3 | 2.0% | N/S | Metabolism | 32% |
| 14 | 138 | CCM | 6 | 3 | 2.2% | N/S | Signaling | 14% |
| 15 | 35 | CCM | 3 | 1 | 2.9% | N/S | Metabolism | 20% |
| 16 | 95 | – | 1 | 3 | 3.2% | N/S | Metabolism | 20% |
Summary of the identified GO terms and primary functional categories within the 16 clusters using the Algal Functional Annotation tool and manual curation, respectively.
N/S indicates that significant GO term hits included less than 5% of the unique genes in the cluster.
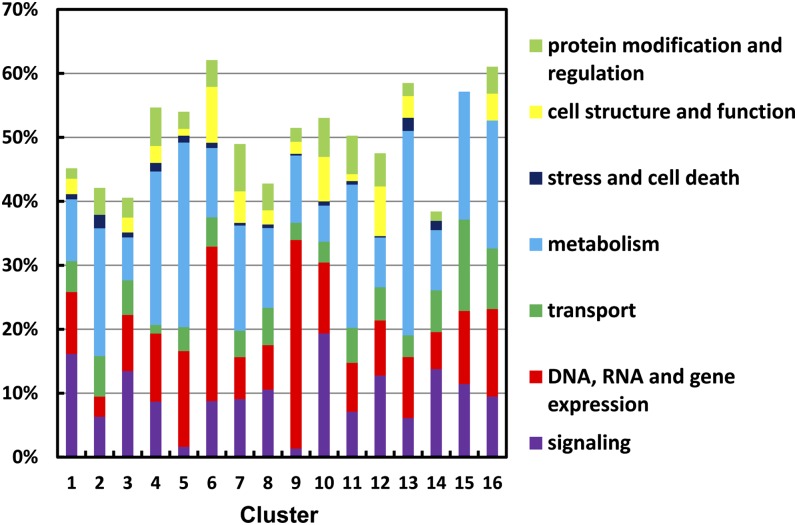
Because of the relative paucity of functional annotation in the C. reinhardtii genome, the Algal Functional Annotation Tool was unable to provide much functional information for more than a few gene clusters. Therefore, we also employed manual curation to place DE genes into eight broad functional categories (see Supplemental Data Set 4 online). Not surprisingly, the most abundant manual functional category of DE genes in all 16 clusters was “unknown,” which is represented by the difference between 100% and the sum of all other functional categories for each cluster in Figure 2 and accounts for 38 to 62% of the genes in each. Among the 16 clusters, the most abundant of the eight manual categories after “unknown,” are “metabolism,” “signaling,” and “gene expression and regulation.” The relative distribution of DE genes among these eight manually curated categories is illustrated for each gene cluster in Figure 2, and a compilation of the primary functional category in each cluster identified as including the largest proportion of genes (excluding the “unknown” category) is summarized in Table 1.
Figure 2.
Distribution of Genes in Manual Functional Categories within Each Cluster.
The functional categories (protein modification and regulation, cell structure and function, stress and cell death, metabolism, transport, gene expression and regulation, and signaling) were determined manually based on a combination of existing annotation and automated identification of functional domains. Percentages indicate the sum of genes in each functional category (indicated by color). The difference between the summed percentage and 100% represents the “unknown” category, which is not included.
No single functional category among the genes in each major cluster group (CIA5, CO2, and CCM clusters) was consistently apparent by either method. However, those CIA5 clusters (1, 3, and 10) with higher gene expression in the wild type all have signaling as the primary functional category, whereas those clusters (2, 11, and 13) with higher gene expression in cia5 have metabolism as the primary functional category. Furthermore, the functional categories of signaling and gene expression together accounted for more than half of the genes in CIA5 clusters 1, 3, and 10, excluding the unknown category. Of these CIA5 clusters, only clusters 10 and 11 contained more than 8% of the genes identified as GO category hits, but these GO hits agreed with the primary functional category of signaling for cluster 10, in that they fell in the general areas of intracellular trafficking, proteolysis, and regulation of processes, and of metabolism for cluster 11, in that they fell in the general area of metabolic processes (see Supplemental Data Set 3 online).
Similarly, the CO2 cluster 9 with increased transcript abundance at higher CO2 concentrations has gene expression as the primary functional category, whereas those clusters (4 and 7) with increased gene expression at lower CO2 concentration have metabolism as the primary functional category. Furthermore, the functional categories of gene expression and signaling combined accounted for more than half of the genes in CO2 cluster 9, excluding the unknown category. Of the three CO2 clusters, only 4 and 9 each contained more than 8% of the genes identified as GO category hits, and in both cases, these GO hits agreed with the primary functional category identified. In CO2 cluster 9, which had gene expression as its primary functional category, the GO category hits fell in the general areas of RNA processes and nitrogen metabolism (see Supplemental Data Set 3 online). For CO2 cluster 4, which had metabolism identified as its primary functional category, the GO category hits fell in the general area of catabolic processes (see Supplemental Data Set 3 online), and a large proportion of the cluster 4 genes in the manually curated metabolism category were putative catabolic genes (see Supplemental Data Set 4 online).
Among the CCM clusters, the various functional categories appeared to be relatively evenly dispersed, with the exception of cluster 15. Although cluster 15 had the somewhat common category of metabolism as its primary functional category, it is notable in having, among all the clusters, the highest proportion (∼14%) of manually curated genes in the transport functional category.
Key CO2 Assimilation-Related Genes and Pathways
In addition to the segregation of genes into broad functional categories, we also analyzed the distribution of specific groups of genes among the gene expression pattern clusters, such as previously reported LCI genes, Calvin cycle genes, photorespiratory pathway genes, and CA genes.
Of 2274 genes exhibiting a C-effect and/or a CS-effect (see Supplemental Data Set 1 online), and thus indicating a statistically significant response to CO2 concentration, 1350 were upregulated in wild-type L-CO2 versus H-CO2, and 418 had a fold change of 2 or greater. This selection of genes is the most comparable to classic LCI genes reported previously (Chen et al., 1996; Somanchi and Moroney, 1999; Miura et al., 2004; Wang et al., 2005; Wang and Spalding, 2006; Yamano and Fukuzawa, 2009). We selected 106 of these previously reported LCI genes for a direct comparison with genes identified as having a C-effect or CS-effect. Among these 106 previously reported LCI genes, 49 exhibit a C-effect or CS-effect, and 45 of these were upregulated in either L-CO2 or VL-CO2 conditions compared with H-CO2 in the wild-type strain (see Supplemental Data Set 5 online). We also used a recently proposed statistical method implemented in the Bioconductor package DESeq (Anders and Huber, 2010) to perform a direct, pairwise comparison of gene expression for H-CO2 versus either L-CO2 or VL-CO2, which identified a highly overlapping but slightly different list of 40 previously reported LCI genes as upregulated in our experiment. In combination with the C/S impact model, the DESeq analysis supports 53 of the previously reported LCI genes as upregulated in either L-CO2 or VL-CO2 (see Supplemental Data Set 5 online).
Data included in our companion article (Brueggeman et al., 2012) demonstrate a low-CO2 upregulation of 40 of the 106 previously identified LCI genes, and 35 of these overlap with the 53 genes identified here as being upregulated by L-CO2 or VL-CO2. In combination, our data and the data from Brueggeman et al. (2012) provide support for upregulation of 60 of the 106 previously reported LCI genes. In addition to showing downregulation for three of the same four previously reported LCI genes that our data identify as downregulated, our companion article identifies an additional two previously reported LCI genes that exhibit downregulation under their experimental conditions (see Supplemental Data Set 5 online).
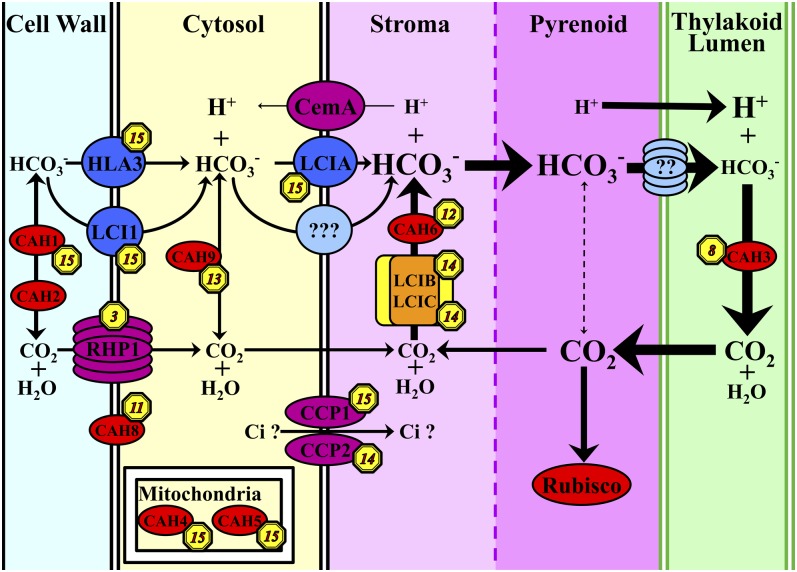
From our list of 49 previously reported LCI genes showing a C-effect or CS-effect, 36 genes fall into the CCM clusters (8, 14, and 15), and an additional seven genes in the CCM clusters were identified as upregulated in L-CO2 or VL-CO2 based on the pairwise DESeq analysis (see Supplemental Data Set 5 online). To explore this relationship further, we selected 10 intensively studied, CIA5-regulated, LCI genes (CAH1, CAH3, CCP1, CCP2, HLA3, LCIA, LCIB, LCIC, LCI1, and LCR1; highlighted in Supplemental Data Set 5 online) implicated as functionally involved in the CCM (Spalding, 2008; Wang et al., 2011) and found all to be contained in CCM clusters 8, 14, and 15, and all except CAH3 were identified as DE by our companion article (Brueggeman et al., 2012). Figure 3 provides a schematic model of the proposed C. reinhardtii CCM, including Ci uptake and accumulation processes (modified from Wang et al., 2011), including identified and proposed locations of the various CAs (Moroney et al., 2011). A major proportion of the CCM/CA genes in this model were included in CCM clusters 15, 14, or 8, providing additional validation of our clustering results.
Figure 3.
Gene Cluster Distribution within a Hypothetical CCM/Ci Transport Model.
The cluster label for each gene is indicated by a yellow octagon containing a cluster number. In this schematic representation of the CCM, most of the putative CCM elements and many previously identified CO2-responsive genes, including the putative Ci transporters HLA3, LCI1, LCIA, CCP1, and CCP2 and CCM-related CAs CAH1 and CAH3, and other indentified CCM genes like LCIB and LCIC are indicated as having expression patterns categorized in the “CCM clusters (clusters 8, 14, and 15).” This figure is modified from Wang et al. (2011), with the addition of the LCI chloroplast membrane proteins CCP1 and CCP2 (Chen et al., 1997) and CA locations (Moroney et al., 2011). Transport proteins with demonstrated CCM function are shown in dark blue, known transport proteins with suspected CCM function are shown in purple, predicted but unknown transport proteins are shown in light blue, known soluble enzymes are shown in red, and the soluble LCIB/LCIC complex is shown in orange and yellow.
Although a large proportion of the previously reported LCI genes identified here as DE genes were found to be associated with CCM clusters 8, 14, and 15, a substantial number also were associated with CO2 clusters 7 and 9. Of the 53 previously reported LCI genes supported by our data, seven fell into CO2 cluster 7, four genes fell into CO2 cluster 9, and one each into CIA5 clusters 11 and 12 (see Supplemental Data Set 5 online). Six previously reported LCI genes that were not supported as LCI genes by our data (i.e., no C-effect or CS-effect and not DE based on the DESeq analysis) were identified as DE genes but fell mostly into CIA5 clusters 3, 10, 11, and 13.
By visual inspection, the gene expression pattern in CCM cluster 15 shows very low expression in H-CO2 and induction in VL-CO2 and L-CO2 conditions for the wild type and very low expression in any CO2 conditions for the cia5 mutant. CCM clusters 8 and 14, on the other hand, show only upregulation of expression under VL-CO2 and L-CO2 conditions for the wild type, relative to the modest expression in H-CO2 conditions, and almost equally low expression under any CO2 conditions for the cia5 mutant. Thus, the patterns for CCM clusters 8, 14, and 15 progress from mild upregulation of expression to high-level induction, respectively. Only 35 genes showed the high-level induction and were grouped in CCM cluster 15, so every gene in this cluster is listed in Table 2, and all genes in clusters 8, 14, and 15 are listed in Supplemental Data Set 6 online.
Table 2. Genes in Cluster 15.
| Name | Protein IDa | Descriptionb | Average RPKMc | Significant Effectsd | Primary Functional Categorye |
|---|---|---|---|---|---|
| CAH1 | 522126 | CA, periplasmic, α type | 1223.8 | C+S+CS | Metabolism |
| CAH4 | 522732 | Mitochondrial CA, β type | 226.5 | C+S+CS | Metabolism |
| CAH5 | 522733 | Mitochondrial CA, β type | 157.3 | C+S+CS | Metabolism |
| CCP1 | 522130 | LCI chloroplast envelope protein | 162.0 | C+S+CS | Transport |
| CGL28 | 510019 | RNA binding protein | 200.7 | C+S+CS | Unknown |
| CYC6 | 516039 | Cytochrome c6 | 1.2 | S | Metabolism |
| DNJ15 | 514023 | DnaJ-like protein | 12.8 | S | Gene Expression |
| DNJ31 | 518238 | DnaJ-like protein | 13.1 | S | Gene Expression |
| *HFO7 | 523344 | Histone H4 | <0.1 | – | Gene Expression |
| HLA3 | 518934 | ATP binding cassette transporter | 289.5 | C+S | Transport |
| *KIR1 | 526069 | Keto acid isomerase-like protein | <0.1 | – | Unknown |
| LCI1 | 520703 | LCI membrane protein | 224.0 | C+S | Transport |
| LCI23 | 523507 | LCI protein | 75.8 | C+S | Unknown (TM) |
| LCIE | 522129 | LCIB-like gene | 1.6 | S | Metabolism |
| LCR1 | 519760 | Low-CO2 response regulator | 79.0 | C+S | Gene Expression |
| LHCSR2 | 525378 | Stress-related chlorophyll a/b binding protein 2 | 45.7 | C+S+CS | Metabolism |
| LHCSR3 | 525376 | Stress-related chlorophyll a/b binding protein 3 | 51.7 | C+S+CS | Metabolism |
| NAR1.2 | 524076 | Anion transporter; LCIA | 209.7 | C+S | Transport |
| – | 516770 | PRLI-interacting factor L | 44.9 | S | Signaling |
| – | 509757 | Acetyltransferase | 27.5 | C+S | Unknown |
| – | 519249 | Ser/Thr protein kinase | 53.5 | S | Signaling |
| – | 522781 | ND | 4.85 | S | Unknown (TM) |
| – | 516290 | ND | 51.5 | C+S | Transport |
| – | 510680 | ND | 44.6 | C+S | Unknown (TM) |
| – | 520458 | ND | 20.7 | S+CS | Unknown |
| – | 512353 | ND | 16.8 | C+S+CS | Unknown (TM) |
| – | 522486 | ND | 8.7 | C+S | Signaling |
| – | 524386 | ND | 6.4 | S | Unknown (TM) |
| – | 524387 | ND | 2.3 | S | Unknown |
| *– | 512735 | ND | <0.1 | S | Unknown |
| *– | 519540 | ND | <0.1 | – | Unknown |
| *– | 522103 | ND | <0.1 | – | Unknown |
| *– | 510710 | ND | <0.1 | – | Signaling |
| *– | 518019 | ND | <0.1 | – | Unknown |
| *– | 511100 | ND | <0.1 | – | Unknown |
Asterisks indicate genes with an average expression level lower than 0.05 RPKM and “–” indicates unnamed gene.
Augustus 5.0 gene model protein ID.
ND means no description available.
Average RPKM across all six treatment conditions.
Individual effect having a q-value <0.025 by C/S impact test, where “C” means CO2 effect, “S” means strain effect, “CS” means interaction effect, and “–” means no significant C/S impact effect.
Result from our manual curation, where “unknown (TM)” indicates gene of unknown function containing at least one putative transmembrane domain.
Based on our manual functional curation, CCM cluster 15 contains a relatively large proportion of genes in the manually annotated transport functional category (Figure 2) and includes essentially all the genes for which there is either compelling evidence for a Ci transport role for the gene product in the CCM (LCIA, LCI1, and HLA3) or a strong argument for the gene product as a good candidate for Ci transport (CCP1). As with all the clusters, a large proportion (15/35) of the genes in CCM cluster 15 falls into the “unknown” functional category. However, it is notable that five of the 15 genes of unknown function in cluster 15 are putative transmembrane proteins.
Within CCM cluster 15, all 17 genes with significant C-effects also exhibit S-effects (includes CAH1, CAH4, CAH5, LCI1, LCR1, CCP1, HLA3, and LCIA), and eight of these (includes CAH1, CAH4, CAH5, and CCP1) also have a significant CS-effect. Seven genes that did not exhibit any significant C/S impact effects have expression levels in the lowest 3% of genes, with a mean RPKM lower than 0.023 for all seven genes. Of the remaining 11 genes in CCM cluster 15 with significant expression levels, all exhibit only S-effect except one (protein ID 520458; also shows CS-effect).
CAs catalyze the reversible hydration of CO2 to HCO3− and serve critical roles for the CCM (Moroney et al., 2011) (Figure 3). Among the nine identified α and β CA genes (Table 3), CAH1, CAH4, and CAH5 fell into CCM cluster 15 and have all three significant C+S+CS-effects, as described above. These three CA genes are strongly induced in low CO2 and thus may be directly involved in the Ci transport and accumulation process of the CCM or at least in the acclimation to low CO2. CAH3, the thylakoid lumen CA required for dehydration of stromal HCO3 (Moroney et al., 2011), exhibits both significant C+S-effects and was placed in cluster 8, which contains genes whose visual expression patterns indicate modest upregulation in response to limiting CO2. CA genes CAH8 and CAH9 showed mainly S-effects and fell into CIA5 clusters 11 and 13, respectively, and CAH6 showed both C+S-effects and fell into cluster 12. CAH2 and CAH7 were not identified as DE genes.
Table 3. CAs.
| Name | Protein IDa | Description and Subcellular Location | q-Valuesb | Clusterc | Significant Effectsd |
|---|---|---|---|---|---|
| CAH1 | 522126 | α-CA, periplasm | 3.4E-04 | 15 | C+S+CS |
| CAH2 | 522125 | α-CA, periplasm | 3.2E-01 | – | – |
| CAH3 | 526413 | α-CA, thylakoid lumen | 1.5E-02 | 8 | C+S |
| CAH4 | 522732 | β-CA, mitochondria | 2.7E-03 | 15 | C+S+CS |
| CAH5 | 522733 | β-CA, mitochondria | 3.4E-03 | 15 | C+S+CS |
| CAH6 | 512520 | β-CA, chloroplast stroma | 5.4E-03 | 12 | C+S |
| CAH7 | 515107 | β-CA, unknown | 2.5E-02 | – | – |
| CAH8 | 526207 | β-CA, plasma membrane | 1.6E-02 | 11 | S |
| CAH9 | 522626 | β-CA, cytoplasm | 2.3E-02 | 13 | S |
Augustus 5.0 gene model protein ID.
q-values calculated by overall test.
A “–” indicates the gene was not identified as DE in overall test.
Individual effect having a q-value <0.025 by C/S impact test, where “C” means CO2 effect, “S” means strain effect, “CS” means interaction effect, and “–” indicates the gene was not identified as DE in overall test so was not included in C/S impact test.
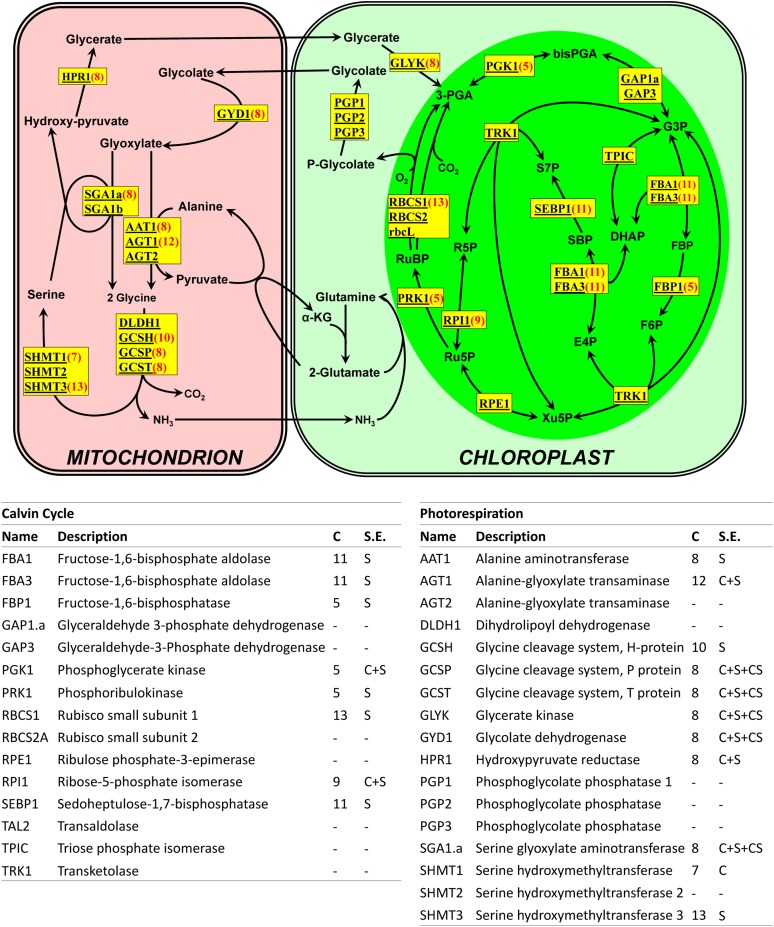
We also scrutinized the genes encoding enzymes of the Calvin cycle and the photorespiratory pathway (Spalding, 2009), since these important carbon metabolism pathways are expected to respond to CO2 concentration (Figure 4). Eight of the 15 genes involved in the Calvin cycle were DE in our experiment. Two fructose bisphosphate aldolase genes FBA1 and FBA3, the sedoheptulose bisphosphatase gene SEBP1, and one of the two Rubisco small subunit genes, RBCS1, were found in CIA5 clusters 11 and 13, both of which show increased gene expression in the cia5 mutant but relatively little effect of CO2 concentration. Two critical kinase-encoding genes, phosphoglycerate kinase1 (PGK1) and phosphoribulokinase1 (PRK1), and the fructose bisphosphatase gene FBP1 were included in cluster 5, which shows a pattern of mildly increasing gene expression with increasing CO2 concentration as well as mildly increased gene expression in the cia5 mutant. The ribose-5-phosphate isomerase1 (RPI1) gene was in CO2 cluster 9, which shows significantly increased expression under higher CO2 concentration but only modest expression increase in the cia5 mutant. Thus, aside from PGK1 and RPI1, in which both C-effect and S-effect were detected, all DE genes from the Calvin cycle show only S-effects and increased expression in cia5.
Figure 4.
Genes Involved in the Calvin Cycle and Photorespiration.
(A) Schematic of the Calvin cycle and photorespiration pathway in C. reinhardtii (Spalding, 2009). Yellow boxes indicate genes involved in each reaction, and the red numbers in parentheses indicate the cluster in which each gene was found.
(B) Summary of detailed information about each gene. C, cluster number (a “-” indicates the gene was not identified as DE in overall test and not assigned a cluster); S.E., significant effects, which are individual effects having a q-value <0.025 by the C/S impact test, where C is CO2 effect, S is strain effect, CS is CO2 and strain interaction effect, and “-” indicates the gene was not identified as DE in overall test so was not included in C/S impact test.
On the other hand, the expression of photorespiratory pathway genes was strongly affected by the CO2 concentration; many of the genes were upregulated in L-CO2 and VL-CO2 (Figure 4). Accordingly, the photorespiratory genes alanine aminotransferase1 (AAT1), glycerate kinase (GLYK), glycolate dehydrogenase (GYD1), hydroxypyruvate reductase1 (HPR1), and serine glyoxylate aminotransferase1 (SGA1) and all Gly decarboxylase complex subunit genes, except glycine cleavage system, H-protein (GCSH) and dihydrolipoyl dehydrogenase1 (DLDH1), fell into CCM cluster 8, even though the Algal Functional Annotation Tool only identified two genes, AAT1 and HPR, among the GO hits for photorespiration in cluster 8 (see Supplemental Data Set 3 online). These cluster 8 photorespiratory genes, which encode enzymes spanning the entire pathway from glycolate to phosphoglycerate, appear to be regulated by both CIA5 and CO2; accordingly, all exhibited C-effects and S-effects, and all, except AAT1 and HPR1, exhibited CS-effects.
Of those photorespiratory pathway genes not in CCM cluster 8, GCSH and serine hydroxymethyltransferase3 (SHMT3) were found in CIA5 clusters 10 and 13, respectively, with S-effects only, SHMT1 was captured in CO2 cluster 7 with only significant C-effect, and alanine-glyoxylate transaminase1 (AGT1) was found in cluster 12 with a significant C+S-effect. Some photorespiratory genes, such as the three phosphoglycolate phosphatase genes PGP1, PGP2, and PGP3, were not identified as being DE in our experiment even though phosphoglycolate phosphatase activity was reported to increase in response to limiting CO2 (Marek and Spalding, 1991; Tural and Moroney, 2005). One isoform of Ala-glyoxylate transaminase (AGT2), one isoform of Ser hydroxymethyltransferase (SHMT2), and the Gly decarboxylase complex subunit DLDH1 also were not identified as DE genes under the conditions used.
DISCUSSION
Identification of DE Genes
In this article, our primary objective was to gain insight into the transcriptome-wide changes in the patterns of gene expression that occur in response to the interaction between CO2 concentration and the transcription regulator CIA5. An additional benefit expected was the identification of candidate genes that may play significant roles in the CCM. To address these objectives, we analyzed the gene expression profiles of two genotypes, the wild type (137c) and cia5, under three different CO2 concentrations using an overall test to identify 3678 genes that showed differential expression in at least one of the six treatments (two genotypes × three conditions). This identification of over 3600 DE genes, which represents almost 20% of the C. reinhardtii transcriptome, revealed massive changes in gene expression in response to the combination of CO2 concentration changes and the presence/absence of CIA5.
Further detailed analysis of the 3678 DE genes was performed using two additional methods: (1) C/S impact tests for C-effects, S-effects, and CS-effects, for each gene; and (2) a cluster analysis of the gene expression patterns across the six conditions. Whereas cluster analysis grouped DE genes with similar expression patterns, the C/S impact test provided quantitative evaluations of individual environmental induction and strain effects. The majority of genes identified as DE genes by the overall test showed one or more significant C/S impact effects when tested for C-, S-, and CS-effects. Only ∼5% (165 out of 3678) of the DE genes identified by the overall test were not identified as having significant individual effects in the C/S impact test, possibly due to different power of detection inherent in the overall test and the C/S impact test.
Cluster analysis, in combination with identification of individual C-effects, S-effects, and CS-effects, revealed clusters of genes regulated primarily by CIA5 (predominantly S-effects; CIA5 clusters), regulated primarily by CO2 (predominantly C-effects; CO2 clusters), and regulated by interaction of CO2 and CIA5 (predominately CS-effects and combinations of C-effects, S-effects, and CS-effects). The delineation of these clusters directly addressed our overall objective of gaining insight into the patterns of gene expression in response to interaction between CO2 and CIA5, as well as revealing specific genes regulated by CO2, by CIA5, and by the interaction of CO2 and CIA5. Based on reports of induction or upregulation of CCM-related genes in low CO2, genes functionally involved in the C. reinhardtii CCM were expected to be among the third general group of genes, those regulated by both CO2 and CIA5.
Comparison with Previously Reported LCI Genes
Although our major objective was to discover a spectrum of gene expression patterns in response to the interaction between CO2 and CIA5, we also performed direct pairwise comparisons in the wild-type strain between H-CO2 and either L-CO2 or VL-CO2 to provide a more detailed analysis of the differential expression of genes in response to low or limiting CO2. Because of the historical connection between low-CO2 upregulated genes and the CCM, we included these DESeq analyses to enrich the comparisons between previously reported LCI genes and DE genes identified in this study.
Because of the interest in the CCM specifically, at least 106 genes have been reported as LCI genes, many of which also reportedly require CIA5 for differential expression (Chen et al., 1996; Somanchi and Moroney, 1999; Miura et al., 2004; Wang et al., 2005; Wang and Spalding, 2006; Yamano and Fukuzawa, 2009). Some of these LCI genes, such as CAH3, LCI1, LCIA, LCIB, and HLA3, reportedly play important roles in the C. reinhardtii CCM (Galván et al., 2002; Wang and Spalding, 2006; Duanmu et al., 2009a, 2009b; Ohnishi et al., 2010), and the function of others, such as CCP1, CCP2, and LCIC, in the CCM also has been implicated (Pollock et al., 2004; Wang and Spalding, 2006; Yamano et al., 2010). However, only 53 of 106 previously reported LCI genes were supported as L-CO2 or VL-CO2 upregulated DE genes in this study. This discrepancy is not unexpected because the different strains and different light, CO2 concentration, and other environmental conditions used among the various studies almost certainly will result in variations in the genes responding and because DE gene identification may be impacted by a shifting in the population distribution among the cell division cycle phases in response to a shift from H-CO2 to L-CO2 conditions (Dillard et al., 2011). In addition, the 4-h induction time used here may not identify genes that are DE only earlier or later than 4 h. Indeed, the companion article by Brueggeman et al. (2012) documents significant changes in gene expression during a 3-h time course following CO2 depletion but also reports the lack of induction of several previously reported LCI genes. Only 40 previously reported LCI genes were supported by Brueggeman et al. (2012) as low-CO2 upregulated. In combination with those supported by our data, 60 previously reported LCI genes are supported as upregulated under the conditions used in the two studies combined.
Many previously reported LCI genes have not been further characterized or confirmed beyond initial observations, which in many cases used no statistical procedure to control FDRs (Miura et al., 2004; Yamano and Fukuzawa, 2009). The greater sensitivity of RNA-Seq and our more reliable statistical approach provide significant advantages over previous studies. Therefore, in addition to the impact of environmental and strain differences on the absence of some LCI genes from our list of DE genes, some previously reported LCI genes may not represent bona fide LCI genes. Our data indicate that four previously reported LCI genes are actually downregulated by L-CO2 or VL-CO2, and, in addition to supporting the downregulation of three of these four genes, our companion article (Brueggeman et al., 2012) identified two more previously reported LCI genes as downregulated by low CO2.
The results reported here and in our companion article (Brueggeman et al., 2012) complement and extend past reports of differential expression by supporting 60 previously reported LCI genes, directly contradicting six others, and leaving the remaining 40 as not clearly supported under the conditions used in the two studies. In addition, the two companion studies identified a large number of additional genes as regulated by CO2, CIA5, or both.
CCM Clusters
Of 57 previously reported LCI genes identified in this study as DE genes, 43 fell into the CCM clusters 8, 14, and 15, all of which exhibited expression patterns expected for classic LCI genes (i.e., high expression for the wild type in L-CO2 and VL-CO2 but lower expression in H-CO2 and consistently lower expression in cia5 under all CO2 concentrations). In addition to those in the CCM clusters, 11 of the previously reported LCI genes, including four that were downregulated by low CO2, fell into CO2 clusters 7 and 9, which exhibit little impact of the presence/absence of CIA5, suggesting that the genes included are not likely to be involved in the CCM. This illustrates an important value of sorting gene expression patterns into clusters, which provide richer insight into the identification of likely functional CCM genes than provided by the LCI approach alone.
The CCM cluster 15 contains only 35 of the 3678 DE genes but may be a rich source of candidate functional CCM genes. Of the 35 genes in cluster 15, eight have RPKM expression levels lower than 0.05 across all conditions, making them unlikely candidates for a significant role in the CCM. The remaining 27 genes in CCM cluster 15, which includes all the genes that encode transport proteins strongly implicated or suspected as Ci transporters as well as the CCM regulatory gene LCR1, the LCIB-like gene LCIE, and the well-studied CA genes CAH1, CAH4, and CAH5 must be good candidates for a functional or regulatory role in the C. reinhardtii CCM. Considering the burden of transporting one of the highest flux inorganic nutrients, as well as the selection against wasting energy on transport when CO2 is abundant, such an expression pattern is not surprising for genes encoding Ci transporters and other conditionally critical CCM components.
Among the remaining genes of CCM cluster 15, four encoding stress-induced light harvesting chlorophyll proteins, stress-related light harvesting complex protein2 (LHCSR2) and LHCSR3, and DnaJ-like, putative chaperonins, DNJ15 and DNJ31, may represent general stress response elements, and a few genes, such as 522486 (putative guanylate cyclase), 519249 (putative protein kinase), and 516770 (putative PRLI interacting factor), encode potential signaling elements. However, the most intriguing group of cluster 15 genes may be the six unknown or little known transmembrane protein–encoding genes, including 524386, 512353, 510680, 522781, 516290, and 523507, since their expression patterns parallel those of all likely transporter-encoding genes so far identified. Therefore, these genes rank high as possible undiscovered Ci transporters.
Key CO2 Assimilation-Related Genes and Pathways outside the CCM
CAs are expected to play important roles in microalgae because of the poor solubility and diffusion rate of CO2 in water and the critical importance of interconversion of these Ci forms internally. Of the nine confirmed α-CA and β-CA genes (Moroney et al., 2011), seven were identified as DE genes in our experiment (Table 3, Figure 3), which is not unexpected given the importance of Ci uptake and accumulation in the CCM and the well-known differential expression of the three CCM cluster 15 CA genes. S-effect appeared to influence more CA genes than C-effect and CS effect, which reinforces our thoughts about the extent of CIA5 influence. Also, this study confirms CAs as important functional targets for further study regarding the dynamics of the CCM.
The expression of a number of Calvin cycle genes, including one Rubisco small subunit gene, was impacted by the cia5 mutation. DE genes encoding Calvin cycle enzymes were found in clusters 5, 9, 11, and 13, all of which exhibit increased expression in cia5 relative to the wild type but which vary in their responses to changes in CO2 concentration. Reinforcing the implication that the Calvin cycle DE genes respond primarily to CIA5, all these genes exhibited a significant S-effect, with only two, PGK1 (cluster 5) and RPI1 (CO2 cluster 9), exhibiting a significant C-effect. Although we have no clear explanation for a CIA5 role in regulation of several Calvin cycle genes, the increased expression of these genes in the cia5 mutant argues for a role of CIA5 in carbon assimilation independent of the CO2 concentration. This observation may reflect a role for CIA5 as a repressor of Calvin cycle genes under as yet unidentified conditions, where the absence of CIA5 activity might result in a modest increase in expression of these Calvin cycle genes.
Photorespiration results from the low specificity of Rubisco and competes with CO2 assimilation via the Calvin cycle, so lower CO2 concentrations increase the Rubisco oxidase reaction relative to the carboxylase reaction and increase the demand for photorespiratory enzymes. The apparent regulation of a number of these genes by CIA5 and CO2 is consistent with previous reports of low-CO2 induced expression of photorespiratory pathway genes (Marek and Spalding, 1991; Tural and Moroney, 2005), but, contrary to previous reports, we did not see differential expression of any PGP genes. This discrepancy might be explained if PGP activity is regulated at the post transcriptional level or if the change in PGP gene expression occurs in a time frame not captured by our 4-h induction time point. Unlike the DE Calvin cycle genes, most of the DE photorespiratory pathway genes exhibited significant C-, S-, and CS-effects, rather than just significant S-effects, demonstrating that many of the photorespiratory pathway DE genes are regulated by both CO2 and CIA5.
Not all photorespiratory genes appear to be regulated by CIA5, but the apparent regulation of a large fraction of them by this protein argues that CIA5 plays a significant role in regulation of the photorespiratory pathway. Notably, those photorespiratory pathway genes that exhibit significant S-effect show a differential expression opposite to that of the Calvin cycle genes (i.e., they have increased expression in the wild type relative to cia5). This expression pattern across the genotypes is consistent with CIA5 acting as an inducer of photorespiratory pathway genes in contrast with its putative role as a mild repressor of several Calvin cycle genes.
CIA5 Clusters and the Impact of CIA5
CIA5 appears to serve much broader and more extensive roles than indicated by the phenotype of cia5, which grows similar to the wild type either heterotrophically or mixotrophically in acetate or photoautotrophically in H-CO2 and even grows slowly in L-CO2 (Spalding, 2008). Most genes in CIA5 clusters 1, 2, 3, 10, 11, and 13 show clear regulation by CIA5 but little regulation by CO2 concentration, indicating that low-CO2 activation of CIA5 is not always required for function of CIA5. More than 76% of the 1396 genes in these six clusters exhibit only a significant S-effect (no C-effect or CS-effect), and, including genes that also show a significant C-effect or CS-effect, almost 95% exhibit a significant S-effect. Furthermore, of 3678 identified DE genes, over 62% show a significant S-effect, including those that also exhibit a significant C-effect and/or CS-effect, and more than half of those genes regulated directly or indirectly by CIA5 show only a significant S-effect. Thus, almost 15% of all C. reinhardtii genes are regulated in some way by CIA5, and almost 7.5% of all genes are regulated by CIA5 independent of any changes in the CO2 concentration.
CIA5 is very likely involved in the upstream regulation of multiple physiological processes, and, although its own transcript abundance does not change, the presence/absence of CIA5 (and its potential activation/inactivation) may have a major impact on the expression of genes encoding many secondary regulatory genes, including those encoding transcription factors and signal transduction components involved in regulation of a number of processes. Consistent with this expectation, manual curation and, in most cases, the Functional Annotation Tool, identified signaling or signaling plus gene expression as key functional categories for the three CIA5 clusters with increased expression in the wild type, arguing that when the presence of CIA5 increases transcript abundance for specific genes, it appears to do so as an upstream activator of positive signaling pathways and/or other gene expression activators. On the other hand, both manual curation and the Functional Annotation Tool pointed to metabolism as the primary functional category for the three CIA5 clusters in which the presence of CIA5 resulted in decreased expression of specific genes (decreased expression in the wild type). Thus, when the presence of CIA5 coincides with the repression of specific genes, CIA5 appears to act as an upstream repressor of specific metabolic functions. Notably, half of the Calvin cycle genes identified as DE fall into these three CIA5 clusters, and most of the others fall in cluster 5, which has somewhat similar characteristics, including decreased gene expression in the wild type and metabolism as primary functional category.
CO2 Clusters and the Impact of CO2
Regulation of gene expression by CO2 concentration also appears to be more extensive than expected. Most genes in the CO2 clusters 4, 7, and 9 show clear CO2 concentration regulation but little or no apparent effect of CIA5. Almost 55% of the 764 genes in these three clusters exhibit only a significant C-effect (no S-effect or CS-effect), and, when including those genes that also show a significant S-effect or CS-effect, over 90% exhibit a significant C-effect. Furthermore, of 3678 identified DE genes, over 60% show a significant C-effect, including those that also exhibit a significant S-effect and/or CS-effect, and ∼30% of those genes regulated directly or indirectly by CO2 concentration show only a significant C-effect. Thus, CO2 concentration significantly affects the expression of over 60% of the DE genes, and more than 14% of all genes detected in this experiment. This means that ∼14% of all C. reinhardtii genes are regulated by CO2 concentration, most of which (over 10% of the genes) also exhibit some form of CIA5 regulation. However, almost 4% of all genes are regulated by CO2 apparently independent of CIA5.
Manual curation and the Functional Annotation Tool both identified gene expression as the key functional category for the CO2 cluster (cluster 9) with decreased expression of genes in L-CO2 or VL-CO2 relative to H-CO2, which is consistent with either limiting CO2 acting as an upstream repressor or elevated CO2 acting as an upstream activator, respectively, of genes involved in regulation of gene expression. On the other hand, manual curation and, in one case, the Functional Annotation Tool, identified metabolism as the primary functional category for the two CO2 clusters (clusters 4 and 7) with increased expression of genes in L-CO2 or VL-CO2 relative to H-CO2, which is consistent with either limiting CO2 acting as an upstream activator or elevated CO2 acting as an upstream repressor, respectively, of genes involved in specific metabolic functions. Notably, both functional characterizations of cluster 4 revealed enrichment in putative catabolic genes, suggesting that low CO2 concentrations cause starvation and stimulate the expression of genes involved in degrading and remobilizing existing molecules.
More generally, the abundance of metabolism as a primary functional category in nine of the 16 gene clusters, including CIA5 clusters 2, 11, and 13, CO2 clusters 4 and 7, and CCM clusters 8 and 15 suggests that a major impact of changes in CO2 and CIA5 is on the expression of genes encoding metabolic enzymes. This seems reasonable, since large changes in metabolism may well accompany substantial changes in CO2 concentration. These conjectures also are strongly supported by Brueggeman et al. (2012), who report a marked decrease in expression of numerous genes involved in anabolic processes following CO2 deprivation. Alternatively, the high frequency of metabolism as a primary functional category also may reflect an annotation bias; metabolism-related genes may be easier to annotate, resulting in their disproportionate representation among the manually annotated genes.
Multiple Acclimation States
Although there is compelling evidence demonstrating a distinction between the VL-CO2 and L-CO2 acclimation states of C. reinhardtii, none of our transcriptome analyses identified any genes DE between the VL-CO2 and L-CO2 induction conditions for the wild type. Since a large number of DE genes were identified for L-CO2 or VL-CO2 versus H-CO2 conditions, the apparent absence of DE genes in the VL-CO2 versus L-CO2 comparison likely reflects at least a paucity of DE genes distinguishing these two acclimation states under the conditions used. Based on our data, we suggest at least two possible conclusions regarding the distinction between the L-CO2 and VL-CO2 acclimation states: (1) These two acclimation states are controlled at levels beyond transcript abundance, or (2) differential expression of genes distinguishing these two acclimation states is evident only earlier or later than the 4-h acclimation time used in our experiment. On the other hand, if we assume a very limited number of genes are DE in VL-CO2 versus L-CO2 under our experimental conditions, our one-time test of >15,000 genes may have elevated the problem of multiple testing and reduced our power of detection. To test this assumption, future experiments could increase the number of biological replicates and/or sequence to a greater depth of coverage.
Summary
This transcriptome study resulted in a number of new insights regarding the global regulation of genes by CO2 concentration, by CIA5, and by the combination or interaction of CO2 and CIA5. Gene expression patterns were classified into distinct clusters, many of which could be characterized as responding primarily to CIA5 or CO2 based on the C/S impact test and visual inspection of the expression patterns. Regulation by CIA5 independent of CO2 demonstrates that low-CO2 activation of CIA5 is not essential for its function. Three distinct gene expression clusters with response to both CIA5 and CO2 were clearly associated with CCM-related genes and may prove to represent a rich source of candidates for new CCM components, especially cluster 15, which may contain a significant number of putative CCM-related transporter genes. These expression pattern clusters should also represent a more robust source of insight than the LCI gene approach with regard to the role of CIA5 and CO2 regulation on limiting CO2 acclimation responses in general and on the function of the CCM specifically. An example of this is the indication, based on expression patterns observed with Calvin cycle genes and photorespiratory genes, that CIA5 may act as an upstream activator of photorespiratory genes and a mild upstream repressor of Calvin cycle genes. Thus, this study of transcriptome-wide patterns of gene expression related to CO2 and CIA5 provides insight into the massive impact of these two factors and their interaction on gene expression in C. reinhardtii, in addition to identifying compelling new candidates for functional CCM components and highlighting new questions to be addressed in subsequent work.
METHODS
Chlamydomonas reinhardtii Strains and Culture Conditions
C. reinhardtii wild-type strain cc125 was obtained from the Chlamydomonas Resource Center (University of Minnesota, Minneapolis, MN). The cia5 mutant (strain cc2702) was a gift from Donald P. Weeks (University of Nebraska, Lincoln, NE). Media and growth conditions for C. reinhardtii were described previously (Wang and Spalding, 2006). All strains were maintained on CO2 minimal plates in high CO2 (air enriched with 5% CO2) chambers at room temperature, under continuous illumination (50 μmol photons m−2 s−1). Liquid cultures were grown on a gyratory shaker (speed 200 rpm) under white light (∼100 μmol photons m−2 s−1) at room temperature.
Induction and RNA Isolation
Liquid cultures of all strains were grown under H-CO2 (5% [v/v] CO2) to a concentration of 1.0 to 2.0 million cells per mL. Cell cultures were then equally distributed into nine flasks, with three flasks each aerated with H-CO2 (5%), L-CO2 (nominally 300 to ∼500 ppm; actually 330 to 410 ppm), or VL-CO2 (nominally 100 to ∼200 ppm; actually 110 to 152 ppm). Selection of gas flow lines and position on the shaker were completely random. After 4 h induction, cultures aerated with the same CO2 concentrations were combined and centrifuged to collect cells. Two biological replicates of each strain and of each induction condition were processed for RNA isolation as previously described (Wang and Spalding, 2006), and crude RNA samples were cleaned with DNase I and the RNeasy MinElute Cleanup kit (Qiagen).
Sequencing and Alignment
The DNA Facility at Iowa State University processed the cleaned RNA samples, prepared the libraries, and generated sequences (one sample per lane on the flow cell) on a Genome Analyzer II system (Illumina). No less than 12 million reads were obtained for each sample (details can be found in Supplemental Table 1 online). Raw and processed sequence files are available at the National Center for Biotechnology Information Gene Expression Omnibus (accession number GSE33927).
Raw reads were aligned to the version 4.0 assembly of the C. reinhardtii genome (http://genome.jgi-psf.org/chlamy/chlamy.info.html) using gmap (Wu and Watanabe, 2005), which tries to align every single read to the genome as an mRNA (possibly spliced) read without previous knowledge of the genome annotation or sequencing coverage. Alignment files were processed to retrieve unique alignments with an alignment score higher than r-15, where r is the read length. This choice allows us to (1) keep two-block alignments (reads that span two different exons) where only one block is reported by gmap (that typically fails to provide alignment blocks smaller than 16 because of its indexing strategy) and (2) keep alignments for trimmed reads with sequencing errors at the 3′ end. Only high-similarity (less than three mismatches) and intron-like alignments (defined here as those with up to one gap smaller than 5 kb) were used for expression estimation.
Counts per gene were estimated by requiring complete overlap between each alignment and the transcript genomic coordinates and after normalization of transcript sequence coverage by read length for each sample. This strategy attempts to remove the impact that different read lengths could have on the final results, but almost identical results were found with alternative methods (see the end of Methods). Augustus 5.0 gene models (http://augustus.gobics.de/predictions/chlamydomonas/) were used as the reference annotation in this work. The original annotation was filtered to keep only the first prediction for each locus (“_t1” transcripts). Total number of sequences, percentage of uniquely aligned reads, and total gene counts per sample are provided in Supplemental Table 1 online. Gene counts were used for differential expression analysis. Expression estimates for each sample are provided in units of RPKM (reads per kilobase of exon model per million of aligned reads; Mortazavi et al., 2008).
Normalization and Statistical Analysis
The primary statistical analysis was performed in the R statistical programming language (version 2.10.1 from http://www.r-project.org/). A generalized linear model with negative binomial distribution and logarithm link function was fitted to the counts of reads separately for each gene, while the sequencing depths were used as the offsets for normalization purpose. Fixed factors in the model included CO2 condition, strain, and their interaction. An overall test was conducted to identify genes with differential expression in any of the six treatment groups. This test was performed by comparing the full model with six separate means and a reduced model with the same mean for all six groups using quasi-likelihood-based approach. The set of P values from the overall test for each gene was adjusted for FDR control as described by Benjamini and Hochberg (1995). For each of the genes identified as DE by the overall test while FDR was controlled at 2.5%, we further tested (C/S impact test) for CO2 effect (C-effect), strain effect (S-effect), and CO2 and strain interaction effect (CS-effect). These C/S impact tests were conducted by comparing the full model with the appropriate reduced models using quasi-likelihood-based F-test with the R function “drop1.” FDR was controlled for each set of P values using Benjamini and Hochberg’s method.
The DE genes identified by the overall test were clustered by a model-based clustering method implemented in an R package, MBCluster.Seq (Y. Si, P. Liu, P. Li, and T.P. Brutnell, unpublished, http://www.stat.iastate.edu/preprint/articles/2011-11.pdf), assuming the observed counts following negative binomial distributions. Results were evaluated for variations in the number of clusters from 10 to 30. To maintain the clusters as few and tight as possible while including most of the patterns, 16 was chosen to be the total number of clusters for further analysis by visual inspection of the clustering results.
Pairwise comparisons between VL-CO2 and L-CO2 states, VL-CO2 and H-CO2 states, or L-CO2 and H-CO2 states were performed using the Bioconductor package DESeq (Anders and Huber, 2010), which performs variance stabilization by borrowing information across genes (Anders and Huber, 2010). The set of P values for each test was adjusted for FDR control as described by Benjamini and Hochberg (1995).
Quantitative Real-Time PCR Analysis
SYBR green one-step quantitative PCR system was used for qPCR analysis (Quanta Biosciences). All experiments were performed on a Bio-Rad iCycler iQ real-time PCR detection system using primers described in Supplemental Table 2 online. The RNA samples used as templates for qPCR were the same as those used for RNA-Seq. The CBLP gene was used as internal control for normalization of qPCR data. Pearson correlations were calculated for each gene across six strain treatment conditions between RNA-Seq and qPCR methods, based on average log2 fold change of two biological replicates.
Functional Categorization for DE Genes Using the Pathways Tool
The Algal Functional Annotation Tool (Lopez et al., 2011) was used to investigate the biological processes associated with each cluster. To address the limitation of annotations availability for C. reinhardtii, we used the GO terms based on orthology to Arabidopsis thaliana as the framework for GO term selection. After inputting the gene list for each cluster separately, P values were generated for each GO term for each cluster entry. GO terms lacking any hits were assigned a P value of 1, and other terms not statistically significant (P > 0.01) for any cluster entry were excluded in generating the summary heat map (see Supplemental Figure 7 online).
Manual Curation for DE Genes
The principles we followed when manually curating these genes were as follows: (1) manual annotation was used if available; (2) if there was no manual annotation, the automated annotation domain information based on the Augustus 5.0 gene model was used to guide curation of the gene; (3) if there was no manual annotation and no identified domains from the automated annotation, the gene was marked as “unknown,” or as “unknown transmembrane,” if one or more transmembrane regions were predicted; (4) any domain information provided by automated annotation of the gene models was used to place the genes into broad functional categories. The categories used included general biological pathways and general protein functions, such as “metabolism” or “signaling.” From this process, we placed all genes into eight general categories reflective of putative function: (1) signaling (including protein kinases, cyclic nucleotide synthesis, and metabolism, etc.); (2) gene expression and regulation (including transcription, translation, RNA processing, chromatin structure, and dynamics, etc.); (3) transport (including Ci transport, ion transport, and metabolite transport, etc.); (4) metabolism (including amino acid, photosynthesis, photorespiration, carbohydrate, acetate, lipid, and macronutrients, etc.); (5) stress and cell death (including oxidative stress, autophagy, and programmed cell death, etc.); (6) cell structure and function (including cell wall, cytoskeleton, vesicular trafficking, protein trafficking, cell division, and cell motility, etc.); (7) protein modification and regulation (including proteases and protein modifications other than kinases, etc.); and (8) unknown.
An Alternative RNA-Seq Data Evaluation for Reproducibility and Comparative Analysis
Here, we provide additional data regarding the quality and reproducibility of our RNA-Seq libraries, along with a comparative analysis with the results from our companion article (Brueggeman et al., 2012). To this end, we used a different, simplified pipeline for all data to highlight the biological differences and remove any potential differences due to the computational methods. Specifically, we performed the steps below.
To remove any dependency with the various parameters involved in gapped-alignment algorithms, we first obtained nongapped alignments to Augustus 5 transcript sequences using bowtie. The potential issues introduced by this approach (slightly worse alignment rates and missing annotations in the genome with sequence similarity to the annotated genome) are not expected to make a difference in the expression and differential expression results for most genes.
Trimming sequences: To remove the impact that different read length and error rates could have in comparing different RNA-Seq libraries, the results below correspond to trimmed libraries (60 bp) showing very similar error rate profiles (around 1% at the 3′ end; data not shown). The rate of unique alignment to Augustus 5 transcripts is in the range of 70 to 80% for all libraries (see Supplemental Table 1 online).
Unique hits from bowtie were compiled to build the count matrix per gene per sample, for both our libraries and those from our companion article. The previous matrix was normalized to compute expression estimates in unit of RPKMs. This normalization was performed after imputation of missing values (0s were imputed a value of 1 count to regularize fold changes and differential expression estimates) and filtering those genes with no counts in any sequencing lane (absent or nonmappable genes).
Some remarks that can be made from these normalized values follow:
High-expression tails: as normalized RPKM values provide relative expression measures, one of the main sources of ambiguity when comparing two different RNA-Seq data sets comes from the distribution of the high-expression genes. Very small changes in the high-expression tail of the distribution changes significantly the estimates for moderately and slightly expressed genes. As an example, we can compute the fraction of the total RPKMs corresponding to the top 100 highly expressed transcripts. For those conditions that are similar in both articles, the numbers are as follows: Fang/Spalding data set high CO2 #1 = 0.4117; Fang/Spalding data set high CO2 #2 = 0.4650; Fang/Spalding data set very low CO2 #1 = 0.3849; Fang/Spalding data set very low CO2 #2 = 0.4445; Brueggeman/Ladunga data set 0 h #1 = 0.4528; Brueggeman/Ladunga data set 0 h #2 = 0.5357; Brueggeman/Ladunga data set 3 h #1 = 0.6786; and Brueggeman/Ladunga data set 3 h #2 = 0.7031.
For instance, the last number means that 70% of the RPKMs come from the top 100 genes in the 3-h sample (second replicate) of the Brueggeman et al. (2012) data set. This observation, in its turn, will clearly impact the mean variance distributions so that the number of reported DE genes is potentially different. The biological interpretation of this difference in the expression distributions can be found in both articles and can be easily understood from the differences in experimental conditions and/or genetic background.
Reproducibility. A high correlation between fold change estimates from both RNA-Seq and PCR is shown and discussed in this article for a number of relevant genes. Regarding the reproducibility of our RNA-Seq expression estimates for different replicates, Supplemental Figure 3 online shows mean difference scatterplots for replicates of the same condition in our article. The x axis values are geometric means of the expression of the two replicates, while the y axis shows the fold change between replicates. To assist with the visualization of these graphs, shown are line plots with the 90th percentile (red), mean (green), and 10th percentile (cyan) of the fold changes. This means that above the red line fall the 10% higher fold changes between replicates and below the cyan is the 10% higher negative fold changes between replicates. These plots show both that the replicates fold change distributions are centered around 0 as expected (green lines) and that the expression estimates from both replicates area consistent for a majority of the transcriptome. Very similar results were found for the experiments from our companion article (Brueggeman et al., 2012). It is worth mentioning that the highest reproducibility is observed for the mutant samples, most likely due to a lower sensitivity to fluctuations in CO2 levels.
Fold change comparative plots: Fold changes between control and experiment were obtained from the mean expression estimates across replicates for each condition. We focus here on those comparisons that are common to both articles (very low/high CO2 for our data set and 0 versus 3 h on the Brueggeman et al. [2012] data set). Supplemental Figure 4 online compares both fold change estimates. The left panel shows a scatterplot along with results from a linear fit. The black line is a guide to the eye, representing an ideal linear relationship (slope = 1, no offset, expected if both experiments were completely equivalent). A linear fit provides the results highlighted in red font, with slope 1.13 and offset = −0.56. The correlation between both fold change distributions is 0.60. Together, these results indicate that both data sets have the same whole-transcriptome trends in fold expression. The same data are shown in the right panel of Supplemental Figure 4 online as a smoothed density plot. It is clear that fold changes for a majority of genes are in close agreement between both data sets, in accordance with the discussion in the articles regarding the similarity between the reported regulated genes.
Fold-change comparative plots for different analysis pipelines: Supplemental Figure 5 online shows scatterplots of log2 fold changes estimated for data in this article with two different analysis pipelines. The x axis corresponds to the fold change estimates presented in the main article, while the y axis plots estimates from the pipeline introduced above. Red lines represent a perfect linear relationship. The agreement between both estimates is apparent for the whole fold change dynamic range.
Accession Numbers
All sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers listed in Supplemental Data Set 2 online. Accession numbers for specifically discussed genes are as follows: AAT1, EDP08011; AGT1, EDO97315; AGT2, EDO96807; CAH1, EDP04241; CAH3, EDP00852; CAH4, EDO96058; CAH5, EDP07024; CAH6, EDO96552; CAH7, EDO99006; CAH8, EDO99999; CAH9, EDP07163; CCM1/CIA5, EDP07542; CCP1, EDP04147; CCP2, EDP04238; DLDH1, EDP01871; DNJ15, EDP03107; DNJ31, EDO98634; FBA1, EDO98285; FBA3, EDO97897; FBP1, EDP05318; GCSH, EDP08614; GLYK, EDP03009; GYD1, EDP01639; HLA3, EDP07736; HPR1, EDP05213; LCI1, EDP06069; LCI6, EDP02960; LCIA/NAR1.2, EDP04946; LCIB, EDP07837; LCIC, EDP04956; LCID, EDP04142; LCIE, EDP04243; LCR1, BAD13492; LHCSR2, EDP01013; LHCSR3, EDP01087; PGK1, EDO98586; PGP1, EDP06184; PGP2, EDP05829; PGP3, EDP08194; PRK1, EDP02974; RBCS1, EDO96904; RHP1, EDP01722; RHP2, EDP01723; RPI1, EDP04506; SEBP1, EDP04487; SGA1, EDO97196; SHMT1, EDO97448; SHMT2, EDO97351; and SHMT3, EDP00905. C. reinhardtii strains (available from the Chlamydomonas Stock Center; http://chlamycollection.org/) used in this work are as follows: the 137c wild type (strain cc125) and the cia5 mutant (strain cc2702).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Gene Expression Level Distributions for Each Treatment Condition.
Supplemental Figure 2. Validation of RNA-Seq by qPCR.
Supplemental Figure 3. Mean Difference Scatterplots for Biological Replicates.
Supplemental Figure 4. Comparison of Log2 Fold Change Estimates between Different Data Sets.
Supplemental Figure 5. Comparison of Log2 Fold Change Estimates between Different Analysis Pipelines.
Supplemental Figure 6. Distribution of C/S Impact Test Results by Cluster.
Supplemental Figure 7. Heat Map for GO Category Hits Based on the Algal Functional Annotation Tool.
Supplemental Table 1. Alignment Statistics for the Transcriptome Sequencing Experiment.
Supplemental Table 2. List of qPCR Primers.
Supplemental Data Set 1. Overall and C/S Impact Test.
Supplemental Data Set 2. DESeq Summary.
Supplemental Data Set 3. Gene Ontology Analysis.
Supplemental Data Set 4. Manual Curation of Genes.
Supplemental Data Set 5. Previously Reported LCI Genes.
Supplemental Data Set 6. Genes in CCM clusters 8, 14, and 15.
Supplementary Material
Acknowledgments
This work was supported by USDA National Research Initiative Competitive grants (2007-35318-18433), by the National Science Foundation (MCB-0952323), and by the Department of Energy Advanced Research Projects Agency-Energy Program (DEAR0000010) to M.H.S., as well as by the Institute of Genomics and Proteomics (Department of Energy Cooperative Agreement DE-FC02-02ER63421 to David Eisenberg) and the National Institutes of Health (R24GM092473) to S.S.M. and M.H.P.
AUTHOR CONTRIBUTIONS
W.F., P.L., and M.H.S. designed the research. W.F. performed the research. Y.S. and P.L. contributed new analytical tools. All authors analyzed data, and all authors contributed to the writing of the article.
Glossary
- Ci
inorganic carbon
- Rubisco
ribulose-1,5-bis-phosphate carboxylase/oxygenase
- CCM
CO2-concentrating mechanism
- CA
carbonic anhydrase
- RPKM
reads per kilobase of exon model per million of aligned reads
- qPCR
quantitative PCR
- FDR
false discovery rate
- DE
differentially expressed
- GO
Gene Ontology
References
- Anders S., Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson I. (2008). Catalysis and regulation in Rubisco. J. Exp. Bot. 59: 1555–1568 [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300 [Google Scholar]
- Brueggeman A.J., Gangadharaiah D.S., Cserhati M.F., Casero D., Weeks D.P., Ladunga I. (2012). Activation of the carbon-concentrating mechanism by CO2 deprivation coincides with massive transcriptional and metabolic restructuring in Chlamydomonas reinhardtii. Plant Cell 24: 1860–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castruita M., Casero D., Karpowicz S.J., Kropat J., Vieler A., Hsieh S.I., Yan W., Cokus S., Loo J.A., Benning C., Pellegrini M., Merchant S.S. (2011). Systems biology approach in Chlamydomonas reveals connections between copper nutrition and multiple metabolic steps. Plant Cell 23: 1273–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.-Y., Burow M.D., Mason C.B., Moroney J.V. (1996). A low-CO2-inducible gene encoding an alanine: alpha-ketoglutarate aminotransferase in Chlamydomonas reinhardtii. Plant Physiol. 112: 677–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.Y., Lavigne L.L., Mason C.B., Moroney J.V. (1997). Cloning and overexpression of two cDNAs encoding the low-CO2-inducible chloroplast envelope protein LIP-36 from Chlamydomonas reinhardtii. Plant Physiol. 114: 265–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman J.R., Grossman A.R. (1984). Biosynthesis of carbonic anhydrase in Chlamydomonas reinhardtii during adaptation to low CO(2). Proc. Natl. Acad. Sci. USA 81: 6049–6053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillard S.R., Van K., Spalding M.H. (2011). Acclimation to low or limiting CO2 in non-synchronous Chlamydomonas causes a transient synchronization of the cell division cycle. Photosynth. Res. 109: 161–168 [DOI] [PubMed] [Google Scholar]
- Duanmu D., Miller A.R., Horken K.M., Weeks D.P., Spalding M.H. (2009a). Knockdown of limiting-CO2-induced gene HLA3 decreases HCO3- transport and photosynthetic Ci affinity in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 106: 5990–5995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duanmu D., Wang Y., Spalding M.H. (2009b). Thylakoid lumen carbonic anhydrase (CAH3) mutation suppresses air-Dier phenotype of LCIB mutant in Chlamydomonas reinhardtii. Plant Physiol. 149: 929–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuzawa H., Miura K., Ishizaki K., Kucho K.I., Saito T., Kohinata T., Ohyama K. (2001). Ccm1, a regulatory gene controlling the induction of a carbon-concentrating mechanism in Chlamydomonas reinhardtii by sensing CO2 availability. Proc. Natl. Acad. Sci. USA 98: 5347–5352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galván A., Rexach J., Mariscal V., Fernández E. (2002). Nitrite transport to the chloroplast in Chlamydomonas reinhardtii: Molecular evidence for a regulated process. J. Exp. Bot. 53: 845–853 [DOI] [PubMed] [Google Scholar]
- González-Ballester D., Casero D., Cokus S., Pellegrini M., Merchant S.S., Grossman A.R. (2010). RNA-seq analysis of sulfur-deprived Chlamydomonas cells reveals aspects of acclimation critical for cell survival. Plant Cell 22: 2058–2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvam V.M., Liu P., Si Y. (2012). A comparison of statistical methods for detecting differentially expressed genes from RNA-seq data. Am. J. Bot. 99: 248–256 [DOI] [PubMed] [Google Scholar]
- Lopez D., Casero D., Cokus S.J., Merchant S.S., Pellegrini M. (2011). Algal Functional Annotation Tool: A web-based analysis suite to functionally interpret large gene lists using integrated annotation and expression data. BMC Bioinformatics 12: 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek L.F., Spalding M.H. (1991). Changes in photorespiratory enzyme activity in response to limiting CO2 in Chlamydomonas reinhardtii. Plant Physiol. 97: 420–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariscal V., Moulin P., Orsel M., Miller A.J., Fernández E., Galván A. (2006). Differential regulation of the Chlamydomonas Nar1 gene family by carbon and nitrogen. Protist 157: 421–433 [DOI] [PubMed] [Google Scholar]
- Mitra M., Lato S.M., Ynalvez R.A., Xiao Y., Moroney J.V. (2004). Identification of a new chloroplast carbonic anhydrase in Chlamydomonas reinhardtii. Plant Physiol. 135: 173–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Yamano T., Yoshioka S., Kohinata T., Inoue Y., Taniguchi F., Asamizu E., Nakamura Y., Tabata S., Yamato K.T., Ohyama K., Fukuzawa H. (2004). Expression profiling-based identification of CO2-responsive genes regulated by CCM1 controlling a carbon-concentrating mechanism in Chlamydomonas reinhardtii. Plant Physiol. 135: 1595–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroney J.V., Husic H.D., Tolbert N.E., Kitayama M., Manuel L.J., Togasaki R.K. (1989). Isolation and characterization of a mutant of Chlamydomonas reinhardtii deficient in the CO2 concentrating mechanism. Plant Physiol. 89: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroney J.V., Ma Y., Frey W.D., Fusilier K.A., Pham T.T., Simms T.A., DiMario R.J., Yang J., Mukherjee B. (2011). The carbonic anhydrase isoforms of Chlamydomonas reinhardtii: Intracellular location, expression, and physiological roles. Photosynth. Res. 109: 133–149 [DOI] [PubMed] [Google Scholar]
- Moroney J.V., Ynalvez R.A. (2007). Proposed carbon dioxide concentrating mechanism in Chlamydomonas reinhardtii. Eukaryot. Cell 6: 1251–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Ohnishi N., Mukherjee B., Tsujikawa T., Yanase M., Nakano H., Moroney J.V., Fukuzawa H. (2010). Expression of a low CO2-inducible protein, LCI1, increases inorganic carbon uptake in the green alga Chlamydomonas reinhardtii. Plant Cell 22: 3105–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock S.V., Prout D.L., Godfrey A.C., Lemaire S.D., Moroney J.V. (2004). The Chlamydomonas reinhardtii proteins Ccp1 and Ccp2 are required for long-term growth, but are not necessary for efficient photosynthesis, in a low-CO2 environment. Plant Mol. Biol. 56: 125–132 [DOI] [PubMed] [Google Scholar]
- Ramazanov Z., Mason C.B., Geraghty A.M., Spalding M.H., Moroney J.V. (1993). The low CO2-inducible 36-kilodalton protein is localized to the chloroplast envelope of Chlamydomonas reinhardtii. Plant Physiol. 101: 1195–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven J.A., Cockell C.S., De La Rocha C.L. (2008). The evolution of inorganic carbon concentrating mechanisms in photosynthesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363: 2641–2650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somanchi A., Moroney J.V. (1999). As Chlamydomonas reinhardtii acclimates to low-CO2 conditions there is an increase in cyclophilin expression. Plant Mol. Biol. 40: 1055–1062 [DOI] [PubMed] [Google Scholar]
- Soupene E., Inwood W., Kustu S. (2004). Lack of the Rhesus protein Rh1 impairs growth of the green alga Chlamydomonas reinhardtii at high CO2. Proc. Natl. Acad. Sci. USA 101: 7787–7792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soupene E., King N., Feild E., Liu P., Niyogi K.K., Huang C.H., Kustu S. (2002). Rhesus expression in a green alga is regulated by CO(2). Proc. Natl. Acad. Sci. USA 99: 7769–7773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding M.H. (2008). Microalgal carbon-dioxide-concentrating mechanisms: Chlamydomonas inorganic carbon transporters. J. Exp. Bot. 59: 1463–1473 [DOI] [PubMed] [Google Scholar]
- Spalding M.H. (2009). CO2-concentrating mechanism and carbon assimilation. In The Chlamydomonas Sourcebook: Organellar and Metabolic Processes, 2nd ed, Vol. 2, D. Stern and E Harris, eds (Amsterdam: Elsevier Publishers), pp. 257–301 [Google Scholar]
- Spalding M.H., Jeffrey M. (1989). Membrane-associated polypeptides induced in Chlamydomonas by limiting CO2 concentrations. Plant Physiol. 89: 133–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding M.H., Spreitzer R.J., Ogren W.L. (1983a). Carbonic anhydrase-deficient mutant of Chlamydomonas reinhardii requires elevated carbon dioxide concentration for photoautotrophic growth. Plant Physiol. 73: 268–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding M.H., Spreitzer R.J., Ogren W.L. (1983b). Reduced inorganic carbon transport in a CO2-requiring mutant of Chlamydomonas reinhardii. Plant Physiol. 73: 273–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding M.H., Van K., Wang Y., Nakamura Y. (2002). Acclimation of Chlamydomonas to changing carbon availability. Funct. Plant Biol. 29: 221–230 [DOI] [PubMed] [Google Scholar]
- Tural B., Moroney J.V. (2005). Regulation of the expression of photorespiratory genes in Chlamydomonas reinhardtii. Can. J. Bot. 83: 810–819 [Google Scholar]
- Wang L., Li P., Brutnell T.P. (2010). Exploring plant transcriptomes using ultra high-throughput sequencing. Brief Funct Genomics 9: 118–128 [DOI] [PubMed] [Google Scholar]
- Wang Y., Duanmu D., Spalding M.H. (2011). Carbon dioxide concentrating mechanism in Chlamydomonas reinhardtii: Inorganic carbon transport and CO2 recapture. Photosynth. Res. 109: 115–122 [DOI] [PubMed] [Google Scholar]
- Wang Y., Spalding M.H. (2006). An inorganic carbon transport system responsible for acclimation specific to air levels of CO2 in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 103: 10110–10115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Sun Z., Horken K.M., Im C.S., Xiang Y., Grossman A.R., Weeks D.P. (2005). Analyses of CIA5, the master regulator of the carbon-concentrating mechanism in Chlamydomonas reinhardtii, and its control of gene expression. Can. J. Bot. 83: 765–779 [Google Scholar]
- Wu T.D., Watanabe C.K. (2005). GMAP: A genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21: 1859–1875 [DOI] [PubMed] [Google Scholar]
- Xiang Y., Zhang J., Weeks D.P. (2001). The Cia5 gene controls formation of the carbon concentrating mechanism in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 98: 5341–5346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano T., Fukuzawa H. (2009). Carbon-concentrating mechanism in a green alga, Chlamydomonas reinhardtii, revealed by transcriptome analyses. J. Basic Microbiol. 49: 42–51 [DOI] [PubMed] [Google Scholar]
- Yamano T., Tsujikawa T., Hatano K., Ozawa S.I., Takahashi Y., Fukuzawa H. (2010). Light and low-CO2-dependent LCIB-LCIC complex localization in the chloroplast supports the carbon-concentrating mechanism in Chlamydomonas reinhardtii. Plant Cell Physiol. 51: 1453–1468 [DOI] [PubMed] [Google Scholar]
- Yoshihara C., Inoue K., Schichnes D., Ruzin S., Inwood W., Kustu S. (2008). An Rh1-GFP fusion protein is in the cytoplasmic membrane of a white mutant strain of Chlamydomonas reinhardtii. Mol. Plant 1: 1007–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka S., Taniguchi F., Miura K., Inoue T., Yamano T., Fukuzawa H. (2004). The novel Myb transcription factor LCR1 regulates the CO2-responsive gene Cah1, encoding a periplasmic carbonic anhydrase in Chlamydomonas reinhardtii. Plant Cell 16: 1466–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.