Abstract
This report of independent genome sequences of two natural populations of Drosophila melanogaster (37 from North America and 6 from Africa) provides unique insight into forces shaping genomic polymorphism and divergence. Evidence of interactions between natural selection and genetic linkage is abundant not only in centromere- and telomere-proximal regions, but also throughout the euchromatic arms. Linkage disequilibrium, which decays within 1 kbp, exhibits a strong bias toward coupling of the more frequent alleles and provides a high-resolution map of recombination rate. The juxtaposition of population genetics statistics in small genomic windows with gene structures and chromatin states yields a rich, high-resolution annotation, including the following: (1) 5′- and 3′-UTRs are enriched for regions of reduced polymorphism relative to lineage-specific divergence; (2) exons overlap with windows of excess relative polymorphism; (3) epigenetic marks associated with active transcription initiation sites overlap with regions of reduced relative polymorphism and relatively reduced estimates of the rate of recombination; (4) the rate of adaptive nonsynonymous fixation increases with the rate of crossing over per base pair; and (5) both duplications and deletions are enriched near origins of replication and their density correlates negatively with the rate of crossing over. Available demographic models of X and autosome descent cannot account for the increased divergence on the X and loss of diversity associated with the out-of-Africa migration. Comparison of the variation among these genomes to variation among genomes from D. simulans suggests that many targets of directional selection are shared between these species.
Keywords: natural selection; polymorphism; divergence; frequency spectrum; linkage disequilibrium (LD); hitchhiking, background selection; geographic differentiation; mutation; recombination; chromatin states and copy number variation (CNV)
ACCESS to sequenced genomes from natural, outbreeding populations (Begun et al. 2007; Li and Durbin 2011) places our theoretical understanding of the forces that determine patterns of genomic variation within and between taxa in a new empirical light. Alignment of the predictions of classical evolutionary genetic models with richly annotated population genomic survey data is an exciting challenge. Descriptions of the patterns of variation in these first sets of population genomic data can foster efficient sieving of hypotheses and serve as a foundation for the design of subsequent studies. Here we present the description of the genomic sequence assemblies from two collections of natural populations of Drosophila melanogaster. The polymorphism, divergence, and copy-number variation revealed in these data are presented at several scales that all support the hypothesis by Maynard Smith and Haigh (1974) that linked selection can dominate genetic drift as the driver of stochastic allele-frequency dynamics in large natural populations such as D. melanogaster. Discerning the contributions and interactions of hitchhiking (impact of linked adaptive substitutions) vs. more complex selective dynamics and background selection [i.e., the impact of selection against linked deleterious alleles (Charlesworth 1996)] remains a clear challenge.
Natural populations of D. melanogaster are found today in virtually all tropical and temperate locations, typically commensal with humans. Biogeographic analyses and population genetics surveys have identified eastern sub-Saharan Africa as the center of diversity of D. melanogaster and its likely ancestral range (Tsacas and Lachaise 1974; Veuille et al. 2004 ; Pool and Aquadro 2006). The smaller of the two population samples we surveyed is from a population in Malawi, Africa (MW), representing that biogeographic center. The second and larger sample of sequenced genomes is derived from Raleigh, North Carolina (RAL) (Jordan et al. 2007) and represents a relatively recently (≈200 years) established North American extension (Lintner 1882) of the older (≈10,000 years) “Old World” or “out-of-Africa diaspora” (Lachaise et al. 1988; Li and Stephan 2006; Thorton and Andolfatto 2006). Although populations of D. melanogaster in the Western Hemisphere appear to have a predominantly European origin, evidence of admixture from Africa into American populations has been reported (Caracristi and Schlötterer 2003; Nunnes et al. 2008).
The study of genetic variation in natural populations of D. melanogaster has played an important role in the development of evolutionary theory, largely because of the central role of the species in the advancement of knowledge of genetic inheritance. Our fundamental understanding of the biology of D. melanogaster, as well as the advanced methods and unique resources available for its study, has fueled research into the evolutionary forces shaping quantitative, cytogenetic, and molecular genetic variation. In this same context the design of experiments and interpretation of data in this study leverage new and unique resources, including recent results from the modENCODE Project (Roy et al. 2010).
While genomic annotation and descriptions and contrasts of polymorphism and divergence on different scales show our central results, other population genetics statistics were calculated and interpreted, including an estimate of the rate of recombination, the scale and direction of linkage disequilibrium, and geographic differentiation. Together these analyses provide a richly detailed new view and interpretation of population genomic variation in natural populations of D. melanogaster (Mackay et al. 2012).
Materials and Methods
Drosophila stocks
The genomes sequenced and analyzed here are derived from two sources. The first source is a collection of 37 inbred lines provided by T. F. C. Mackay. The details of their provenance and breeding are in Jordan et al. (2007). The lines listed in Table 1 are part of a larger collection established by the Mackay laboratory and available in the Bloomington Drosophila Stock Center. Briefly, inseminated females collected at the Raleigh, North Carolina Farmer’s Market in 2003 were cultured independently. For ≈20 generations single sib-pairs of progeny were mated. Thus independent inbred stocks were established from each isofemale line. The MW genomes were derived by classical balancer extractions from independent isofemale lines collected in Mwanza, Malawi by William Ballard in 2001. Isogenic X chromosome lines were established using FM7a, nod4/C(1)DX/Dp(1;Y)y+; svspa-pol as a balancer stock. The three types (second, third, or both) of autosomal inbred lines were extracted using CyO/wgSp-1; TM3, Ser1/Sb1 as the balancer stock. Independent isogenic stocks of seven X chromosomes, six second chromosomes, and five third chromosomes from the MW population were established and resequenced.
Table 1 . Stock name, Bloomington Drosophila Stock Center number, DNA preparation protocol, libraries/assemblies, GC content, mean read depth, target chromosomes, inversions, and SRA accession numbers (see text).
| DPGP stock | BDSC | DNA preparation | Library/assembly | % GC | Mean depth | Target Chrs | Cosmopolitan inversions: PCR | SRA accession |
|---|---|---|---|---|---|---|---|---|
| MW11-1 | 30858 | CsCl | MW11-1_1 | 42.0 | 9.53 | X | SRX022256 | |
| MW27-3 | 37290 | BPC | MW27-3_1 | 42.3 | 11.99 | 3 | SRX019049 | |
| MW28-1 | 30859 | CsCl | MW28-1_1 | 43.3 | 10.36 | X | SRX019104 | |
| MW28-2-3 | 30860 | CsCl | MW28-2-3_1 | 40.8 | 9.65 | 2, 3 | In(2L)t; In(2R)NS; In(3R)K | SRX000484 |
| MW38-1 | 30861 | CsCl | MW38-1_1 | 43.5 | 9.88 | X | SRX019107 | |
| MW38-2 | 30862 | CsCl | MW38-2_1 | 41.3 | 11.69 | 2 | SRX019109 | |
| MW46-1 | 36919 | BPC | MW46-1_1 | 42.3 | 12.14 | X | SRX019110 | |
| MW56-2-3 | 30863 | CsCl | MW56-2-3_1 | 43.7 | 9.88 | 2, 3 | In(3R)K | SRX000440 |
| MW6-1 | 30854 | CsCl | MW6-1_1 | 42.3 | 11.86 | X | SRX022257 | |
| MW6-2 | 30855 | BPC | MW6-2_1 | 43.0 | 11.63 | 2 | In(2L)t | SRX022258 |
| MW6-3 | 37289 | BPC | MW6-3_1 | 43.0 | 11.62 | 3 | In(3R)K | SRX022259 |
| MW63-1 | 30864 | CsCl | MW63-1_1 | 42.7 | 11.79 | X | In(X)A | SRX019022 |
| MW63-2-3 | 32046 | CsCl | MW63-2-3_2 | 42.2 | 11.41 | 2, 3 | In(2L)t; In(2R)NS | SRX000439 |
| MW9-1 | 30856 | CsCl | MW9-1_1 | 42.7 | 13.41 | X | SRX022262 | |
| MW9-2 | 30857 | BPC | MW9-2_1 | 42.8 | 11.86 | 2, 3 | SRX022263 | |
| RAL-301 | 25175 | CsCl | RAL-301_1 | 42.3 | 15.79 | X, 2, 3 | In(2L)t/+ | SRX000530 |
| RAL-303 | 25176 | CsCl | RAL-303_1 | 41.6 | 10.42 | X, 2, 3 | SRX000529 | |
| RAL-304 | 25177 | CsCl | RAL-304_1 | 42.4 | 11.22 | X, 2, 3 | In(2R)NS | SRX000531 |
| RAL-306 | 37525 | CsCl | RAL-306_1 | 43.2 | 10.24 | X, 2, 3 | SRX000532 | |
| RAL-307 | 25179 | CsCl | RAL-307_2 | 42.7 | 9.71 | X, 2, 3 | SRX000533 | |
| RAL-313 | 25180 | CsCl | RAL-313_1 | 39.7 | 10.54 | X, 2, 3 | In(2L)t | SRX022270 |
| RAL-315 | 25181 | CsCl | RAL-315_1 | 42.6 | 9.85 | X, 2, 3 | SRX000535 | |
| RAL-324 | 25182 | CsCl | RAL-324_1 | 42.7 | 11.83 | X, 2, 3 | In(3R)Mo | SRX010933 |
| RAL-335 | 25183 | CsCl | RAL-335_2 | 42.1 | 10.84 | X, 2, 3 | SRX022273 | |
| RAL-357 | 25184 | CsCl | RAL-357_1 | 41.6 | 10.94 | X, 2, 3 | SRX022274 | |
| RAL-358 | 25185 | CsCl | RAL-358_1 | 41.1 | 9.74 | X, 2, 3 | In(2L)t; In(3R)Mo | SRX000536 |
| RAL-360 | 25186 | CsCl | RAL-360_1 | 40.7 | 9.44 | X, 2, 3 | SRX000534 | |
| RAL-362 | 25187 | CsCl | RAL-362_2 | 41.6 | 10.61 | X, 2, 3 | SRX022277 | |
| RAL-365 | 25445 | CsCl | RAL-365_1 | 43.1 | 10.06 | X, 2, 3 | SRX000537 | |
| RAL-375 | 25188 | CsCl | RAL-375_1 | 43.5 | 10.15 | X, 2, 3 | SRX000538 | |
| RAL-379 | 25189 | CsCl | RAL-379_1 | 40.1 | 10.31 | X, 2, 3 | SRX000539 | |
| RAL-380 | 25190 | CsCl | RAL-380_2 | 42.8 | 9.21 | X, 2, 3 | SRX000556 | |
| RAL-391 | 25191 | CsCl | RAL-391_2 | 43.6 | 10.54 | X, 2, 3 | SRX000557 | |
| RAL-399 | 25192 | CsCl | RAL-399_1 | 41.3 | 9.55 | X, 2, 3 | SRX000558 | |
| RAL-427 | 25193 | NIBPC | RAL-427_1 | 42.6 | 10.32 | X, 2, 3 | SRX000528 | |
| RAL-437 | 25194 | NIBPC | RAL-437_1 | 42.7 | 11.32 | X, 2, 3 | In(3R)Mo | SRX010938 |
| RAL-486 | 25195 | CsCl | RAL-486_1 | 41.0 | 11.5 | X, 2, 3 | SRX022286 | |
| RAL-514 | 25196 | BPC | RAL-514_1 | 42.6 | 9.63 | X, 2, 3 | SRX022287 | |
| RAL-517 | 25197 | BPC | RAL-517_1 | 41.8 | 11.93 | X, 2, 3 | SRX022288 | |
| RAL-555 | 25198 | CsCl | RAL-555_1 | 42.8 | 11.72 | X, 2, 3 | In(3R)Mo | SRX022289 |
| RAL-639 | 25199 | CsCl | RAL-639_1 | 42.0 | 11.86 | X, 2, 3 | SRX022290 | |
| RAL-705 | 25744 | CsCl | RAL-705_1 | 43.0 | 11.66 | X, 2, 3 | SRX022291 | |
| RAL-707 | 25200 | CsCl | RAL-707_1 | 42.7 | 11.6 | X, 2, 3 | In(3R)Mo | SRX022292 |
| RAL-707 | 25201 | NIBPC | RAL-707_2 | 43.3 | 11.47 | X, 2, 3 | In(3R)Mo | SRX022293 |
| RAL-714 | 25745 | CsCl | RAL-714_1 | 42.0 | 11.25 | X, 2, 3 | In(3R)Mo | SRX022294 |
| RAL-730 | 25202 | NIBPC | RAL-730_1 | 43.2 | 11.38 | X, 2, 3 | SRX022295 | |
| RAL-732 | 25203 | CsCl | RAL-732_1 | 42.5 | 11.27 | X, 2, 3 | In(3R)K/+ | SRX022296 |
| RAL-765 | 25204 | CsCl | RAL-765_1 | 42.8 | 10.7 | X, 2, 3 | SRX022297 | |
| RAL-774 | 25205 | CsCl | RAL-774_1 | 41.1 | 10.7 | X, 2, 3 | SRX022298 | |
| RAL-786 | 25206 | CsCl | RAL-786_1 | 42.4 | 10.32 | X, 2, 3 | In(3R)P | SRX022299 |
| RAL-799 | 25207 | BPC | RAL-799_1 | 42.2 | 12.43 | X, 2, 3 | SRX022300 | |
| RAL-820 | 25208 | CsCl | RAL-820_1 | 41.6 | 10.92 | X, 2, 3 | In(3R)Mo | SRX022301 |
| RAL-852 | 25209 | CsCl | RAL-852_1 | 40.6 | 11.44 | X, 2, 3 | In(2R)NS | SRX022302 |
| ycnbwsp | 2057 | CsCl | ycnbwsp_0 | 42.8 | 11.64 | X, 2, 3 | SRX027154 | |
| ycnbwsp | 2057 | CsCl | ycnbwsp_1 | 41.6 | 11.4 | X, 2, 3 | SRX010957 |
Genomic DNA
Three genomic DNA isolation protocols were used as indicated in Table 1. Most DNAs were prepared from adults, using the nuclear-isolation/CsCl protocol in Bingham et al. (1981). “NIBPC” refers to genomic DNA preparations that followed the nuclear isolation in Bingham et al. (1981) but resuspended the nuclei in 5 ml of 100 mM NaCl, 200 mM sucrose, 100 mM Tris-HCL, 50 mM EDTA, and 0.5% SDS. In the case of “BPC” 25 adults were homogenized in 500 μl of this Tris-EDTA-SDS buffer. In both cases 0.25 vol of cold KOAc was added, mixed, and placed on ice for >30 min. These were then centrifuged at high speed and the supernatant was extracted with phenol-chloroform, ethanol precipitated, and resuspended in H2O. The BPC samples were treated with RNAse. The genomic DNA preparations of the MW chromosome X lines and RAL-365, RAL-379, RAL-391, RAL-437, RAL-514, RAL-555, RAL-730, and RAL-799 started with only adult females, while the remainder are unselected, i.e., adult females and males.
The construction of libraries, preparation of the flow cell, and 36 cycles of synthesis imaging followed the Illumina protocols described in Bentley et al. (2008). Our initial DNA concentrations were 5 μg and the target insert size was 150–200 bp. The PCR enrichment of the libraries ranged between 15 and 18 cycles. All the 36-bp reads analyzed were processed through Illumina pipeline V0226 or V030 that includes feature extraction plus parameter-matrix estimation (Firecrest module), basecalling (Bustard module), subsequent Eland alignment to BDGP Release 5, and first-pass quality score calibration. Only reads that passed the Illumina pipeline’s quality control (QC) filters were used for subsequent analysis.
Library QC and titration
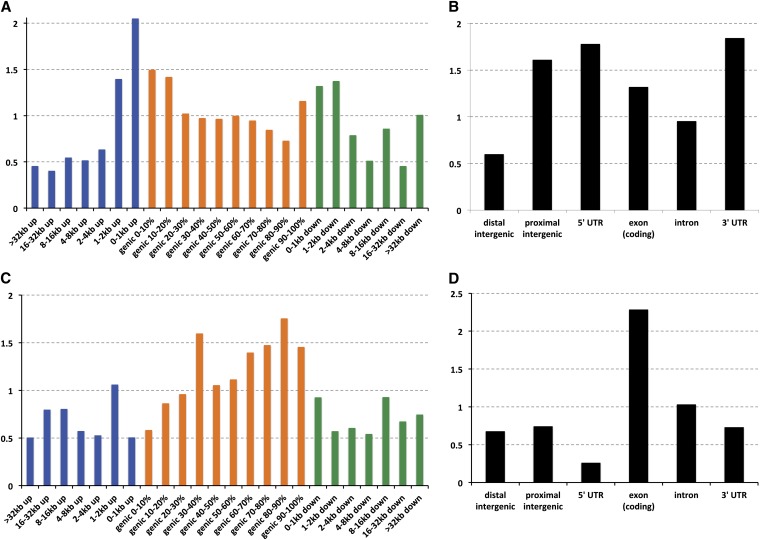
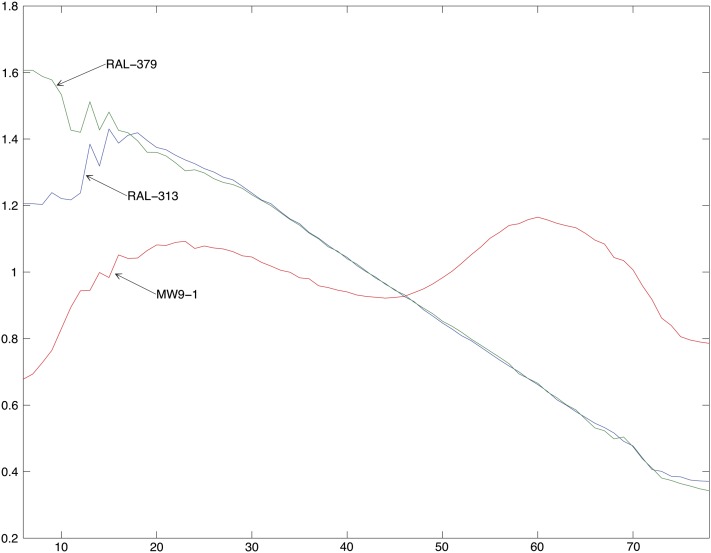
Evaluation of eight new libraries occurred on a “titration flow cell.” A serviceable library exhibited adequate intensity and a cluster density that could be adjusted on subsequent runs to the target values in subsequent lanes. The target G and C content was between 19% and 21% in each. If these three metrics were not met, a new library was prepared. The sample flow cells were generated from libraries that pass these QC and titer criteria. We found that eight lanes at the target cluster density resulted in ≥10× mean coverage of the unique portion of the genome. This was chosen as our production goal for each genome (see Table 1). Earlier studies and our own experience suggested the error increases with deviation in the GC content of the reads (Bentley et al. 2008; Ossowski et al. 2008). However, our criteria yielded a data set with only a mild dependence of apparent SNP rate (relative to the reference sequence) on GC content (see Figure 1).
Figure 1 .
SNP rate (differences from the reference sequence per base pair) of the RAL lines for base pairs with different Illumina quality scores, ≥Q10, ≥Q20, ≥Q30, and ≥Q40 (light to dark blue) plotted with depth-weighted mean GC content at unique base pairs (large solid circles). The whiskers show the range of depth-weighted GC content over lanes. The gray bars show depth-weighted %GC of the median lane. Note the apparent increase in nonreference basecalls (SNP rate) in the lines with the lowest GC content.
Table 2 . Definitions and symbols used in the methods and analyses.
| Symbol | Definitions | Equation |
|---|---|---|
| δw | Estimate of the average nucleotide substitution divergence at polarized sites in a window, weighted by (allele) sampling depth. | (1) |
| πw | Estimate of the expected heterozygosity for nucleotide substitutions per site in a random sample from a randomly mating population. Weighting is by allele sampling depth and the standard bias correction is applied to each. | (2) |
| ρ | Population recombination parameter: ρ = 2Nr/bp for both autosomes and the X chromosome. For local genomic estimates of 2Nr/bp, is determined via statistical fitting to an approximation of the equilibrium between mutation to selectively equivalent alleles and genetic drift in a single, stable outbreeding population (McVean et al. 2004). | |
| rω | Linkage disequilibrium oriented by the allele frequencies. Let p and q be the frequencies of the more common alleles at two loci, p > 1/2 and q > 1/2 (Langley and Crow 1974). And let g be the frequency of the gametotype composed of those two more common alleles. Then . | |
| HKAl | Hudson–Kreitman–Aguadé-like test statistic reflecting the significance of the deviation of the observed proportions of segregating and diverged sites in a window to the chromosome-arm averages under a model for the equilibrium between mutation to selectively equivalent alleles and genetic drift in a single stable outbreeding population (Hudson et al. 1987; Ford and Aquadro 1996). | (3) |
| TsD | A test statistic for either an excess (+) or a deficiency (−) of common alleles compared to the predictions of a model for the equilibrium between mutation to selectively equivalent alleles and genetic drift in a single stable outbreeding population (Tajima 1989). | (4) |
| χ[log(p)] | “+” or “−” the log10 of the P-value for a test statistics such as HKAl and TsD. The sign reflects the sign of the deviation from expectation: the number of segregating sites in the case of HKAl and the frequency spectrum for TsD. | |
| π/min(divl, divg) | A simple metric of reduced diversity in a window where the denominator is the lesser of local divergence and global average divergence. | |
| s | The difference in relative fitness of homozygotes for alternative alleles at a locus. The heterozygote’s relative fitness is 1 − hs, where h is the dominance coefficient. | |
| r | The rate of recombination between two closely linked genomic sites, usually adjacent base pairs unless otherwise indicated. | |
| Estimated rate of recombination per base pair based on local smoothing of incremental change in the standard genetic map. | ||
| ρS | Spearman’s rank correlation coefficient. |
Genomic regions excluded from the analyses
Because this short read resequencing technology is ineffective in repetitive genomic regions, this study focuses only on the five large euchromatic “chromosomes” of the melanogaster reference sequence (BDGP 5). The study of genomic variation in the highly repetitive chr4, chrXhet, chr2Lhet, chr2Rhet, chr3Lhet, chr3Rhet, and sparse chrY contigs are left to another technology at another time. Even within the large euchromatic arms there are many repetitive regions that are not assembled in these data, left as “N” with no quality value. As discussed below, specific genomic regions of particular genomes are excluded if there is evidence that they are not random samples of the genomes in the natural populations.
Assembly and quality calibration
The genome sequences were assembled using the MAQ program described in Li et al. (2008). We carefully investigated the error properties of such assemblies based on independent data from the reference sequence strain (ycnbwsp). An assembly-based error model was formulated that quantitatively captured the main sources of error. Application of this model allowed us to assign recalibrated quality values (similar to Phred scores) for each nucleotide in each assembly. These more realistic values allow quality to become an effective parameter in downstream population genetics analyses. The rationale, implementation, and evaluation of this approach are more thoroughly presented in Appendix A.
Background and residual heterozygosity
The sib-mating inbreeding process is, of course, not expected to be completely or uniformly successful across the genome. Regions in which closely linked recessive deleterious mutations are segregating in repulsion will resist close inbreeding and remain heterozygous (Falconer 1989, p. 101). Additionally, the balancer-chromosome method of inbreeding used with the MW lines could fail because of chance sampling of such recessive lethals. Furthermore, simple technical shortcoming such as low depth or poor primary sequence quality can yield increased levels of heterozygous base calls. Thus we routinely created both “diploid” and “haploid” MAQ assemblies for QC purposes. Plots of heterozygosity along each chromosome arm were generated by parsing the output from MAQ cns2snp. This command calls all SNPs occurring within the MAQ consensus (prior heterozygosity rate = 0.001 for the diploid assemblies). In 100-kbp windows incremented every 5 kbp along each arm, heterozygosity was calculated as the proportion of called sites that are heterozygous (see Supporting Information, Figure S1). The tendency of MAQ to call the reference sequence base when coverage and/or quality are low means that these plots are conservative in their detection of regions of residual heterozygosity. Nevertheless, they proved to be sensitive, robust, and interpretable indicators of QC problems at many levels, including the failure of inbreeding.
A specific augmentation of this method was developed to identify and delineate regions of “residual heterozygosity.” First, if a window exhibited heterozygosity >0.0075, a region of residual heterozygosity was enucleated. The region of residual heterozygosity was extended in both directions until a window with heterozygosity <0.001 was reached in each direction. This sliding-window method was conducted twice, starting at each end of the chromosome arm and proceeding to the other. Overlapping regions from different enucleation sites were merged. Second, regions of residual heterozygosity <150 kbp apart were merged and the intervening formerly “normal” heterozygosity regions were considered to be part of a larger region of excess residual heterozygosity regions. Additionally, any regions of excess residual heterozygosity within 500 kbp of either end of a chromosome arm were extended to the end of the arm.
All regions of called residual heterozygosity were verified by examining the QC plots of heterozygosity (above) with the called regions highlighted and typically masked from the subsequent analyses. In a few cases, adjustments were made to the parameters to produce calls in better agreement with the plotted heterozygosity. Figure S1 show the QC plots and coordinates of regions of residual heterozygosity found. Note the two regions (chr2L:1,677,628–1,890,473 and chrX:21,409,827–21,732,469) found to have high heterozygosity in a large portion of the assemblies. Such regions are listed in Table S1 for each assembly. These were masked in the subsequent analyses.
Regions of identity by descent
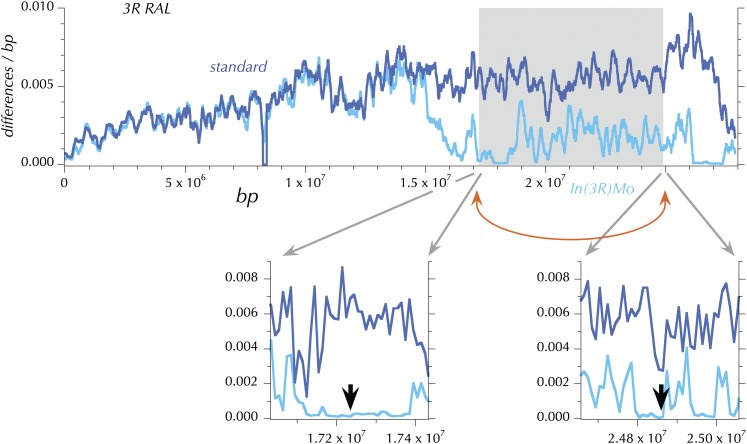
Both as a matter of quality control and to identify the potential impact of polymorphic local recombination suppressors (e.g., cosmopolitan inversions) the genomic distribution of large regions of extremely high sequence similarity between pairs of genomes was systematically determined. Each assembly was compared to all other assemblies in nonoverlapping windows of 100 kbp for the proportion of differences per base pair. Exceptional pairs of assemblies, exhibiting large numbers of consecutive windows with near zero divergence, were flagged as potentially containing identical-by-descent (IBD) segments. Plots of these measures were examined to confirm that large segments identified in a few comparisons were truly empirical outliers as well as being far beyond the theoretical expectation, assuming a large randomly mating and sampled population. These exceptions fell into two small groups, one apparently attributable to the sampling of close relatives and the second apparently associated with inversions (see below and in Corbett-Detig et al. 2012). Three genomes (RAL-303_1, RAL-304_1, and RAL-306_1) share extensive regions, including whole chromosome arms that are nearly identical. These genomes were filtered in subsequent analyses such that only one copy of each of the apparently IBD regions was included (see Table S2).
Cosmopolitan chromosome inversions
PCR-based assays for In(2L)t (Andolfatto et al. 1999), In(3L)P (Wesley and Eanes 1994), and In(3R)P (Sezgin et al. 2004) and five new assays for In(X)A, In(X)Be, In(2R)NS, In(3R)K, and In(3R)Mo were performed as described in Corbett-Detig et al. (2012). The results are presented in Table 1.
Allele sampling depth
Many local features of the genome are difficult to resequence with the approach used here. For example, sites near repetitive sequences and within highly diverged segments are less likely to be covered by uniquely mapping reads and thus more likely to have low-quality scores or be missing altogether from the sequence of an individual genome. The average sampling depth, or the average number of genomes in which a site was sequenced (above a particular quality value) in at least one genome in the sample, is presented in Table A1, Table S3, and Table S4 for each chromosome arm (and the total) in the RAL and MW samples [D. simulans genome (SIM) data are also presented for comparison in Table S5]. The median sampling depth and the total number of base pairs called are also presented. The average sampling was always close to the actual sample size. For the total data in RAL and MW the average numbers of sample genomes are 32.11 and 4.63, respectively (medians 33 and 5), not far from the values predicted for complete sampling, 33.95 and 5.76, respectively (corrected for filtered regions of residual heterozygosity and IBD, see Table S6). Thus the average numbers of quality score (Q)30 (Q40) base pairs in each assembly of the RAL and MW samples are 6.182 × 107 (5.582 × 107) bp and 5.960 × 107 (5.338 × 107) bp, respectively, indicating the size of the “unique” portion of the D. melanogaster genome that can be resequenced with these technologies.
Table A1 . Numbers of assembled Q30 and Q40 base pairs and allelic depths for the MW and RAL samples on the X and autosomes.
| Q ≥ 30 | Q ≥ 40 | |||
|---|---|---|---|---|
| MW | RAL | MW | RAL | |
| All chromosomes | ||||
| bp | 109,883,720 | 110,963,890 | 108,943,964 | 110,681,459 |
| Maximum allelic depth | 7 | 36 | 7 | 36 |
| Mean allelic depth | 5.35 | 31.76 | 4.82 | 28.69 |
| Coding maximum allelic depth | 7 | 36 | 7 | 36 |
| Coding mean allelic depth | 5.40 | 31.99 | 4.73 | 27.70 |
| Autosomes | ||||
|---|---|---|---|---|
| bp | 89,468,274 | 90,351,443 | 88,727,797 | 90,144,978 |
| Maximum allelic depth | 6 | 36 | 6 | 36 |
| Mean allelic depth | 5.10 | 31.73 | 4.61 | 28.99 |
| Coding maximum allelic depth | 6 | 36 | 6 | 36 |
| Coding mean allelic depth | 5.19 | 32.05 | 4.55 | 28.15 |
| X chromosome | ||||
|---|---|---|---|---|
| bp | 20,415,446 | 20,612,447 | 20,216,167 | 20,536,481 |
| Maximum allelic depth | 7 | 35 | 7 | 35 |
| Mean allelic depth | 6.45 | 31.91 | 5.75 | 27.37 |
| Coding maximum allelic depth | 7 | 35 | 7 | 35 |
| Coding mean allelic depth | 6.66 | 31.61 | 5.81 | 24.95 |
Local genomic regions of high polymorphism and divergence are expected to have lower sampling depth. This is borne out in Figure S6, which shows a consistent trend of higher expected heterozygosity and divergence (both defined below) among Q30 sites with lower sampling coverage for RAL, MW, and SIM. Restricting the analysis to Q40 sites reduces this trend somewhat but this also reduces the overall sampling depth (see Figure S6, Table A1, Table S7, Table S8, Table S9, and Table 10). As expected, both expected heterozygosity and divergence in the RAL, MW, and SIM samples are correlated on the local genomic scale (see Table S11). Table A1 also shows that the average sampling depth of coding base pairs is quite comparable to all unique portions of the genome. The largest discrepancy is the Q40 X chromosome where the average sampling depth of all unique base pairs is 27.37, while that for coding base pairs is 24.95. Furthermore the achieved sampling depth at Q30 is within 10% of the maximum possible (see Table S6). Still it must be acknowledged that a proportion of this association between allelic sampling depth and sequence variation could be due to the fact that base-calling errors and depth can be correlated with systematic variation in assembly quality (e.g., read depth or unannotated paralogs).
Table 10 . The proportions of significant MK tests, of significant MK tests with excess of nonsynonymous fixations, and, among significant MK tests, of genes with excess of nonsynonymous fixations for each recombination category.
| Recombination categories | χ2 P-value | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Very low (%) | Low (%) | Intermediate (%) | High (%) | “All” category | Without “very low” category | |||||||
| Proportion | Autosome | X | Autosome | X | Autosome | X | Autosome | X | Autosome | X | Autosome | X |
| MK tests with P < 0.05 | 7.20 | 10.53 | 6.56 | 10.15 | 7.67 | 9.80 | 7.24 | 13.56 | 0.8 | 0.66 | 0.61 | 0.47 |
| MK tests with an excess of nonsynonymous fixations | 1.77 | 9.21 | 2.70 | 9.14 | 6.26 | 9.15 | 5.65 | 12.49 | 1.82 × 10−7 | 0.67 | 3 × 10−4 | 0.5 |
| MK tests with an excess of nonsynonymous fixations among those with P < 0.05 | 24.56 | 87.50 | 41.18 | 90.00 | 81.61 | 93.33 | 78.05 | 91.67 | 1.12 × 10−14 | 0.95 | 4 × 10−8 | 0.94 |
Multispecies alignments
To make estimates and inferences about nucleotide substitutional divergence on the D. melanogaster and D. simulans lineages the reference sequences for these two species (BDGP R5/dm3, WUGSC mosaic 1.0/droSim1) were aligned with those of D. yakuba (WUGSC 7.1/droYak2) and D. erecta (Agencourt prelim/droEre1) in Berkeley Drosophila Genome Project’s D. melanogaster Release 5 (BDGPr5) coordinates. Alignments were produced using a combination of the Mercator (Dewey 2007) and FSA programs (Bradley et al. 2009). Mercator was used to build a one-to-one colinear orthology map between the four genomes and FSA was run on the resulting colinear blocks to produce nucleotide-level alignments. The input to Mercator consisted of all coding exon annotations for the four genomes available from the University of California, Santa Cruz (UCSC) Genome Browser (Karolchik 2003) as well as the results from running BLAT (Kent 2002) on the coding exon sequences in an all-vs.-all fashion. Mercator was run with its default parameters and the “breakpoint finding” utility included with Mercator was used to refine the coordinates of the endpoints of the collinear blocks. FSA was run on the nucleotide sequences of the colinear blocks with options “–mercator cons –exonerate –softmasked –maxram 1000”. Since the focus of our analyses is the polymorphism and divergence within the D. melanogaster lineage, insertions relative to D. melanogaster were ignored and deletions were simply treated as N’s. This multispecies genomic alignment is publicly available at www.dpgp.org.
The syntenic assemblies of the six D. simulans genomes (SIM) presented in Begun et al. (2007) were remapped to the D. melanogaster Release 5 coordinates and used throughout the analyses presented here that involve polymorphism within D. simulans.
Nucleotide-substitution polymorphism and divergence
A fundamental aspect of the way we have assembled these data is to associate each base call with a realistic estimate of the statistical confidence (as described in Appendix A). This readily affords the opportunity to check any observed and interesting pattern at increasing levels of minimum quality. This approach and other inherent properties of the technology lead to missing data. Thus at any particular site in any one of the sampled genomes the called nucleotide may or may not have sufficient quality to be included in a calculation; i.e., it may be “missing data.” The statistics described below incorporate this variation in (allele) sampling depths.
Average divergence in windows
Unless otherwise indicated, divergence was estimated as the average across sites in a segment or a domain of the proportion of “derived states.” As in Begun et al. (2007), we defined the average lineage-specific divergence as
| (1) |
where L is the number of sites in the window or domain, jl is the number of diverged alleles among the cl observed alleles at site l, n is the number of sampled genomes, c is the number of these for which there are data, and kcj is the number of sites in the window or domain at which j of the observed c sampled genomes are “derived” (diverged) from the inferred ancestral state. Ancestral states were inferred as the shared state in the aligned outgroup genomes. For D. melanogaster the ancestral state was assumed to be that in the simulans sequence if either the yakuba or the erecta sequence was aligned and shared that state. For simulans the inference was the reciprocal, melanogaster matching yakuba or erecta. Otherwise the ancestral state was not inferred and the site not included in the estimation of divergence. This parsimony-based estimate is inherently biased under virtually all models as an estimator of divergence at a particular site. However, the magnitude of this bias is likely small for the short timescales relevant for our analyses (Zuckerkandl and Pauling 1962). A more substantial bias across the genome arises from variation in rates of divergence and particularly the clustering of rapidly evolving sites that are much more likely to be excluded from any analyses incorporating divergence. The implications of this bias for particular analyses are discussed in this context.
Expected heterozygosity in windows
The most intuitive measure of population genetic variation is the estimate of the expected (under random mating) heterozygosity at a single nucleotide site (hereafter “heterozygosity” or π). We use the following estimate of heterozygosity over a range (or domain) of nucleotide sites (unless otherwise indicated),
| (2) |
where n is the number of genomes sampled, c ≤ n is the sampling depth, and kcj is the number of sites with exactly c sampling depth and j “derived alleles” (Begun et al. 2007). The designation of derived in the estimation of expected heterozygosity is, of course, not relevant. But note that in specific analyses we limited our attention to those sites at which the ancestral state can be inferred (as for δw above), while in others, all sites are considered, including those lacking useful outgroup data. The estimates of the “average” heterozygosity and divergence for chromosome arms were calculated simply as the weighted average of π in 1000-bp windows in which at least 100 bp had sampling depth >2. Weighting was by number of base pairs in the window with allelic depth >2.
HKA-like analysis (HKAl)
Powerful analyses of evolutionary genetic models can occur when the same process is observed in the same units on different scales of time or space. The most fundamental of these situations is the comparison of within-population sequence polymorphism to divergence between distinct taxa. The Hudson–Kreitman–Aguadé test assesses the prediction of the neutral model (equilibrium between selectively neutral mutation and genetic drift) by comparing the numbers of segregating sites and the average number of diverged sites in two or more genomic regions to their expectations based on estimates of the pertinent parameters of the model (see Hudson et al. 1987). Ford and Aquadro (1996) modified this approach (their “FS” test) by comparing the numbers of fixed differences and segregating sites between species. Formal applications of these tests depend on the choice of the genomic segments being compared, on the assumed rate of recombination, and on simulated distributions of the χ2-like test statistic. A more empirical and practical approach applied here is to simply compute the comparable expected values for the numbers of segregating and fixed diverged sites in a window from the chromosome-wide proportions of such sites at various sampling depths and to calculate the analogous χ2-like statistic as in Begun et al. (2007). Specifically, the proportion of all variant sites that are segregating,
| ; |
the proportion of all variant sites that are fixed,
| ; |
the observed number of segregating sites,
| ; |
the observed number of fixed sites,
| ; |
the total number of variant sites,
| ; |
the expected number of segregating sites,
| ; |
the expected number of fixed sites,
| ; |
and
| (3) |
where kcj is the number of sites with exactly c sampling depth and j derived alleles in the reference segments, e.g., the whole chromosome arm or the “trimmed” (see below) portion. And w in kwcj, pw, dw, Sw, Δw, and Tw refers to the particular window. The window size is adaptively variable such that adjacent base pairs are sequentially included in the window until O(Tw), the total number of variant sites is greater than a fixed parameter. When overlapping windows are displayed, the indicated overlap is in these units of numbers of segregating and fixed sites.
Since all the chromosome arms display a marked reduction in πw proximal to the centromeres and telomeres, these regions (see Table S14) were trimmed from the chromosome arm in the calculation of kcj. Finally, the display of the results of this HKA-like test for each window is in terms of the ±log10 of the nominal P-value associated with the ordinary χ2 with 1 d.f. and the sign of O(Sw ) − E(pw)O(Tw). χ[log(pHKAl)] associated with HKAl will be positive when the observed proportion of segregating sites is greater the trimmed chromosome arm average, given the distribution of sampling depths and average divergence at the sites in the window.
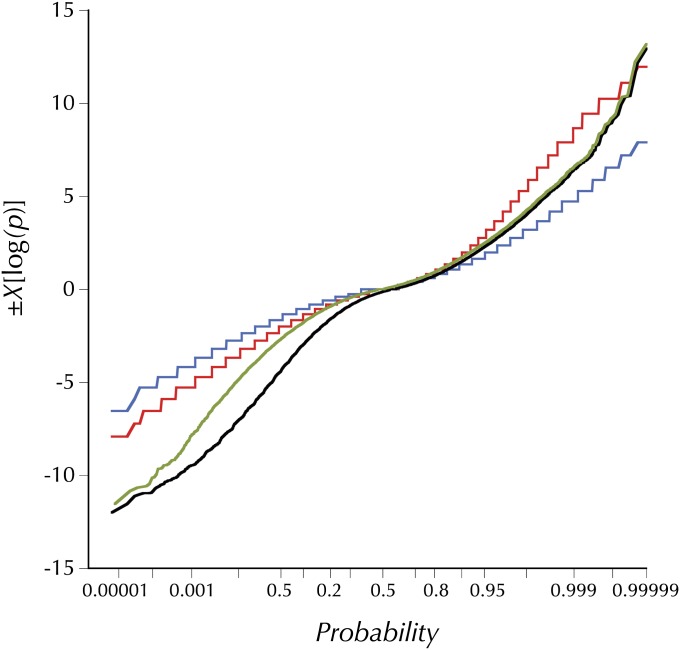
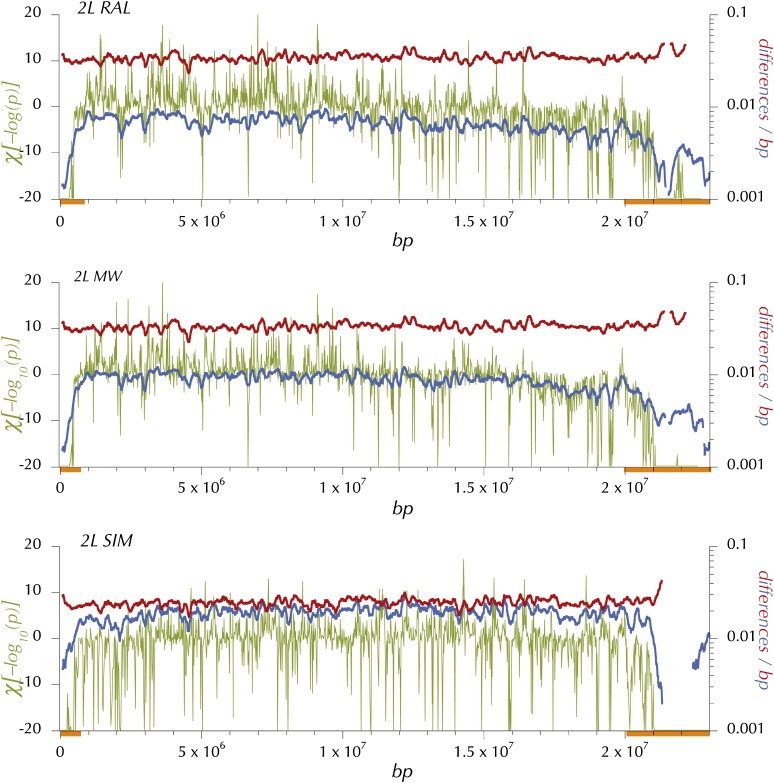
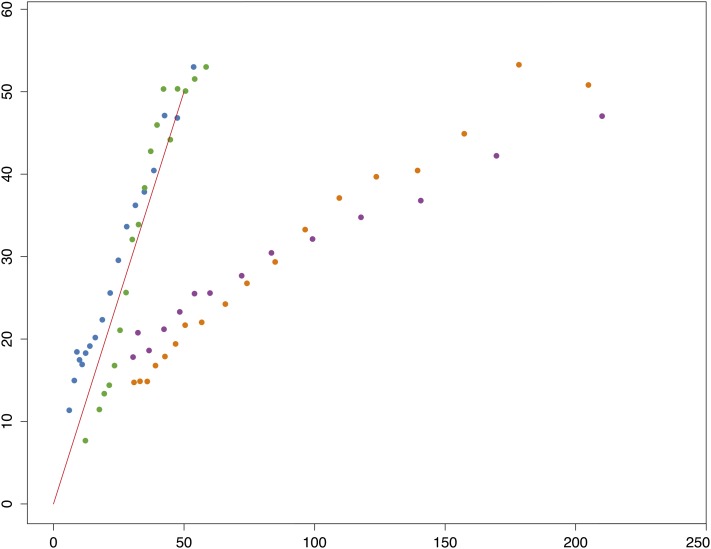
On a finer scale HKAl was calculated using Equation 3 described above. To choose a window size for fine-scaled HKAl it was first necessary to put different possible window sizes on a common basis. The false discovery approach of Benjamini and Yekutieli (2001) was applied to a geometric series of window sizes from 16 to 512 variant (polymorphic or divergent) sites. The number of windows, k with nominal P < k*0.05/n (where n is the total number of windows on the chromosome arm), was determined for each window size and is plotted in Figure S5. Despite the variation in depth of sampling and even the sequencing technologies (i.e., Sanger and Illumina sequencing for D. simulans and D. melanogaster, respectively), a maximum emerges between 20 and 70 variant base pairs per window for each of the chromosome arms for all three samples. In the interest of economical and transparent presentation a window size of 50 segregating and lineage-specific, fixed divergent sites was chosen. Windows in this range with a P ≈ 0.01 or less would be formally significant (under the naive binomial assumption) at the 0.05 level on a chromosome-arm–wide basis. Typically, less than one-quarter of windows with 50 variant base pairs reach this threshold. While the primary purpose of this approach was simply to settle on a small window size that would serve to simply and transparently annotate those regions of the genome with highly deviant divergence-relative polymorphism, it is instructive to compare the observed distributions of χ[log(pHKAl)] with naive neutral theory predictions with different assumed levels of recombination (see Figure 2). As expected, the simulations with no “intralocus” (i.e., within the 50-SNP window) recombination exhibit wider variation in both positive and negative χ[log(pHKAl)]s. Note this difference is larger in the positive domain. But as Figure 2 clearly shows, the distribution of observed χ[log(pHKAl)]s falls far below the simulated values in the negative domain and well above for the more relevant 2Nr = 8 in the positive domain. Removing the centromere- and telomere-proximal regions substantially reduces the magnitude of the deviation in the medial portion of the negative domain. But many of the most the extreme 0.05%, with nominal P-values <10−11, are outside these regions of extremely low crossing over per physical length.
Figure 2 .
The distributions of observed and simulated HKAl (signed chi-square) values. The olive line is the distribution of observed HKAl values for all adjacent windows of 50 variant sites (segregating or fixations on the melanogaster lineage) for which the expected numbers derive from the observed averages in the large subset of these windows outside the designated (“trimmed”) centromere- and telomere-proximal regions with low crossing over. The black line shows the distribution of HKAl for the same windows, using the observed averages for all windows to derive the expected numbers. The blue and red lines are the theoretical distributions for high and no recombination derived from simulations using Hudson’s ms program with the commands ms 35 1000000 -s 50 -r 8 500 -I 2 1 34 -ej 2.5 1 2 or -r 0, respectively. The parameter -ej 2.5 relates the outgroup divergence time to 4N0 and yields the observed proportion of segregating sites (0.44), both averaged over all sample sizes and only at sites with 34 observed alleles, the simulated number.
Frequency spectrum in windows
To evaluate genomic patterns of variation in the frequency spectrum of segregating sites within windows, a simple extension of the familiar Tajima’s D statistic (Tajima 1989) is used to accommodate the variation in sampling depth. Since this test statistic is constructed to approximate a N(0, 1) normalization of the difference between the expected heterozygosity and Waterson’s estimator of 4Nμ, it is natural to simply sum the D values for each of the observed sampling depths in a window and divide by the square root of the number of these observed sampling depths. Of course, this statistic, TsD, is only ∼N(0,1), but it does allow the comparison of different windows,
| (4) |
where D(πi, Si) is Tajima’s D for the sites in the window with sampling depth i and χ(Si > 3) = 0 if there are fewer than three SNPs at sampling depth i and 1 otherwise.
To assess the statistical significance of each observed value, 1000 samples with observed numbers of segregating sites at the observed pattern of sampling depths were generated using Hudson’s ms (Hudson 1990, 2002). The recombination rate was set to zero, producing conservative estimates of the critical values (pTsD) for both the positive and the negative deviations. Windows of the signed logarithm of the depend on the sign of Dw and are plotted and labeled as TsD. The window sizes for TsD were set such that the sum of the numbers of segregating sites over observed sampling depths was a constant 50.
Estimating the rate of recombination per base pair
A unified, high-resolution genetic map based on the segregation of a high density of physically mapped SNPs, such as is available in humans, has not yet been reported for D. melanogaster. The genetic mapping data available at flybase.org comprise a highly edited and rectified summation of a vast, heterogeneous and sometimes conflicting literature of genetic, cytogenetic, and physical mapping in melanogaster. To date the only available estimate of distribution of the rate of crossing over per physical length across the whole genome is that of Singh et al. (2005), which was recently updated (Fiston-Lavier et al. 2010). Their approach is to fit a third-degree polynomial of the genomic positions to the FlyBase reported genetic map positions for each chromosome arm (after removing a few obvious outliers). The derivative of the fitted functions is then their estimate of local rates of recombination (crossing over) per base pair across each of the major arms. Begun et al. (2007) presented locally smoothed interval estimates from such data for the X and noted higher-resolution parallels to the distribution of π (notably) in D. simulans. Here we pursued such local smoothing and higher-resolution maps with the following simple approach based on selected data compiled at FlyBase. We start with the genetic and genomic map positions in the “Map Conversion Table,” which is organized around the cytogenetic “lettered subdivisions.” While the specifics of each curatorial decision are not available, the general method is documented at FlyBase. The reported genetic map positions in that table for each of some 100+ lettered cytogenetic subdivisions (≈200 kbp) on each arm appear to correspond to the map position of a reported locus physically localized in that subdivision. But clearly conflicts between the genetic and physical maps have been rectified. The physical boundaries and reported genetic map positions in this FlyBase table are included in Table S12.
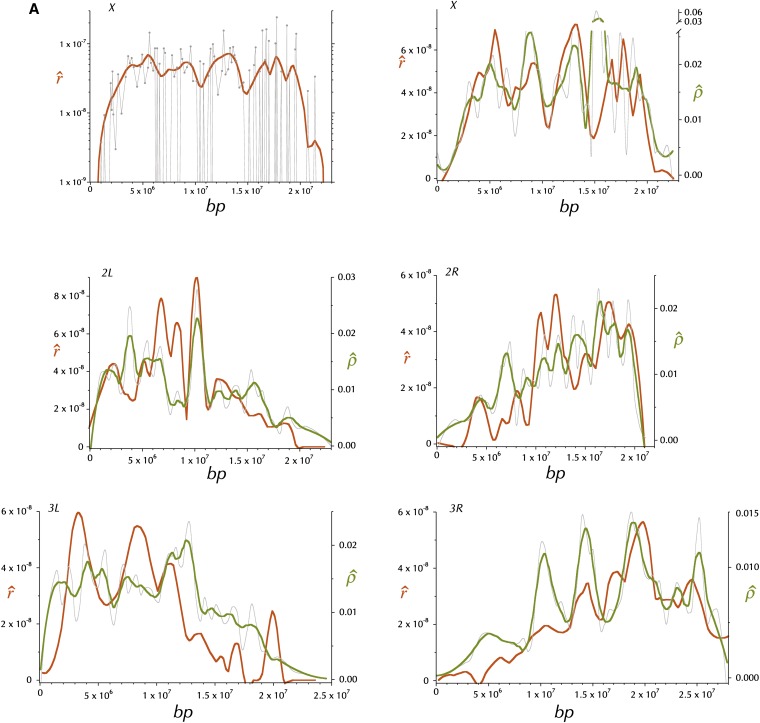
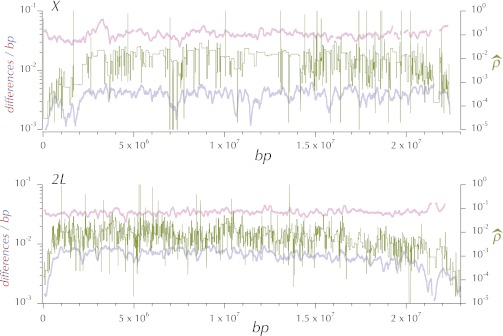
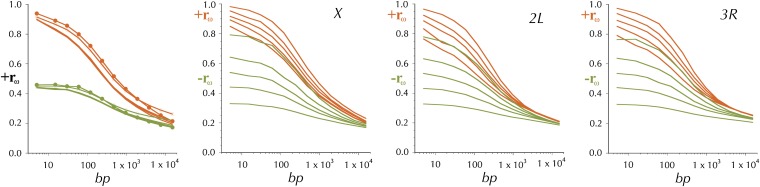
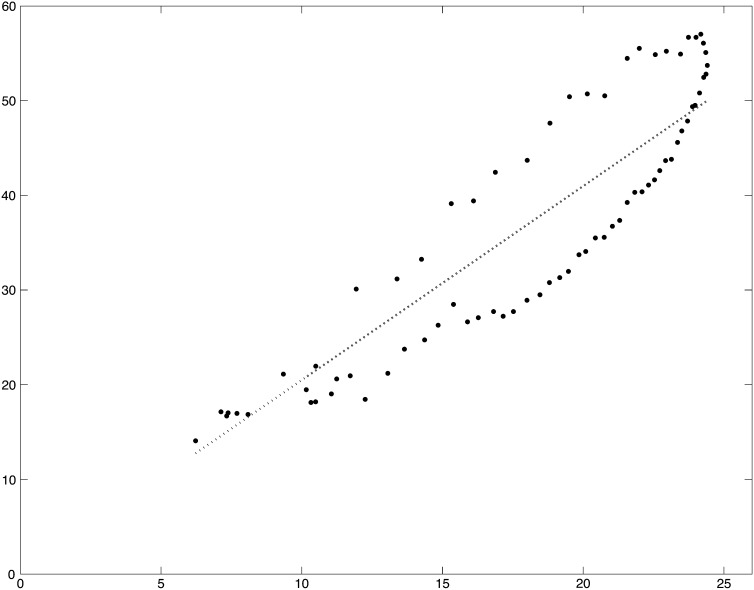
The estimate of the rate of crossing over per base pair (M/bp), (gene conversion contributes little to the underlying data) is the increment in the reported genetic map divided by the length in base pairs of the subdivision. Inset A in Figure 3 shows the distribution of these estimates, and the smoothed fit (hereafter referred to as ) for the X chromosomes plotted against the midpoint of the subdivision. The smoothing is locally weighted regression and smoothing scatterplots (loess) (Cleveland 1979) implemented in the function loess in R (R Development Core Team 2010) with the span parameter = 15% and the default tricubic weighting. This plot is logarithmic to accommodate the wide range of values. The large numbers of segments with an estimated rate of zero are obviously off the bottom of this plot. The comparably smoothed fit for each chromosome arm is plotted in the remaining 5 panels of Figure 3 on a linear scale. The data plotted in Figure 3 are in Table S12.
Figure 3 .
Distributions of estimates of the rates of recombination per base pair, r (M/bp) and ρ = 2Nr, where N is the populations size (see text). is the (orange) loess-smoothed (span = 15%) per genome per generation estimate of the rate of recombination between adjacent base pairs, derived from curated FlyBase genetic map positions. (A) and the estimated r for each subdivision on the X, plotted on a logarithmic scale. is the (olive) comparably (span = 15%) loess-smoothed per population per generation estimate of the rate of recombination (2Nr) between adjacent base pairs (see text). The gray line is the higher-resolution (span = 8%) smoothed estimate of 2Nr, .
Fine-scale recombination rate estimation
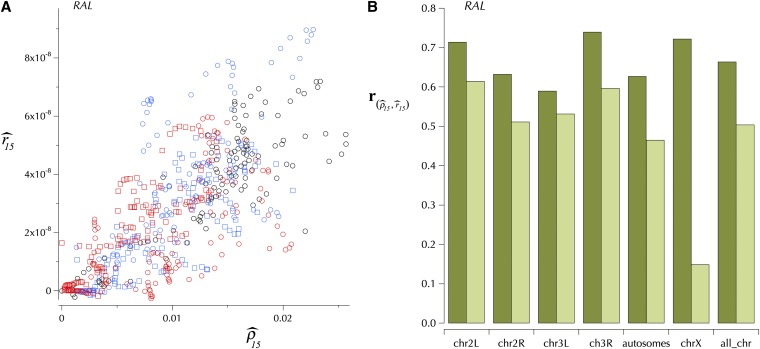
We used the program package LDhat version 2.1 (McVean et al. 2004) to estimate the fine-scale population recombination rate variation in Q30 assemblies of the 37 RAL lines. In LDhat, missing data are handled by marginalizing over the unknown allelic values in the likelihood computation. The computational complexity of this procedure scales exponentially with the number of missing entries. To avoid this hurdle in our genome-wide fine-scale recombination analysis, i.e., to create a complete data set (with no missing data), we removed missing data using the approach described below. For several reasons some assemblies have long intervals of contiguous missing alleles. For each chromosome, we found the set of missing intervals for each sample and used the resulting collection of end points to partition the chromosome into a set of nonoverlapping blocks. Then, within each block, we removed completely missing haplotypes. Finally, we removed the sites containing at least one missing entry. The data resulting from this procedure had the properties listed in Table S13.
We used LDhat’s subprogram complete to generate two-locus likelihood lookup tables with the population-scaled recombination rate ranging from 0 to 500, with an increment of 0.5 (McVean et al. 2004). The population-scaled mutation rate was set to 0.006 for autosomes and 0.004 for the X. For each sample configuration, we ran complete until either the minimum effective sample size reached 1000 or 1.0 million genealogies were sampled, whichever came first. To estimate genome-wide fine-scale population recombination rates, ρ = 2Nr, we adopted a sliding-window approach, with each window containing 1000 SNPs and consecutive windows overlapping by 250 SNPs. N is the population size and r is the recombination rate between base pairs per generation. Because of the lack of crossing over in male Drosophila meiosis, N and r are multiplied by 2 rather than by 4. LDhat’s subprogram interval was used to estimate variable recombination rates. For each window, we ran the reversible-jump MCMC for 5 million iterations, with a burn-in of 200,000 iterations, and subsequently took a sample every 2000 iterations. To stitch together the estimates in the overlapping region of consecutive windows, we discarded the estimates for 125 SNPs from the ends of each window. In each interval, the prior for the number of recombination rate changes is taken to be a Poisson distribution with mean (S − 2)e−ξ, where S is the number of SNPs in a window (in our case 1000) and ξ is a block penalty. Because of the evident spikes in the estimates (see below), we tried a range of ξ, including 15, 25, 35, and 45. Note that changing the penalty from 15 to 45 decreases the mean of the prior by a factor of ∼9.4 × 10−14. For the reversible-jump MCMC to successfully sample recombination maps with several rate changes when the penalty is as high as 45, the data must strongly support the rate changes so that an increase in the likelihood compensates for a decrease in the prior.
Because the estimated fine-scale recombination rates exhibit considerable variation on several scales, especially spikes (see below), we report here the following conservative estimate based on two independent runs with ξ = 45. In 50-kbp windows the ρ map with the fewest changes was selected to thus remove “unreplicated” spikes. Table 5 shows properties of these two maps and this combination. Spikes with a width >2 kbp are unlikely to be artifacts of the LDhat estimation.
Table 5 . Comparison of recombination per base pair based on the genetic map, , and the population genomic estimate of 2Nr, (see text).
| Genetic map: | Population genomic map: | Correlation between and | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Arm | Euchromatic (bp) | M | M/bp | Relative | Relative | Run 1 | Run 2 | Combined | |
| 2L | 22,590,693 | 0.54 | 2.39 × 10−8 | 1.0244 | 1.0755 | 0.00884 | 0.00885 | 0.00883 | 0.73 (0.80) |
| 2R | 20,972,991 | 0.53 | 2.53 × 10−8 | 1.0830 | 1.0779 | 0.00892 | 0.00884 | 0.00885 | 0.78 (0.74) |
| 3L | 24,148,966 | 0.49 | 2.03 × 10−8 | 0.8696 | 1.1985 | 0.00993 | 0.01012 | 0.00984 | 0.66 (0.78) |
| 3R | 28,652,412 | 0.56 | 1.95 × 10−8 | 0.8376 | 0.7162 | 0.00589 | 0.00597 | 0.00588 | 0.81 (0.73) |
| X | 22,775,017 | 0.66 | 2.90 × 10−8 | 1.2419 | 1.8136 | 0.01474 | 0.01465 | 0.01489 | 0.57 (0.86) |
The base pair-weighted correlation between and is shown (their logarithm in parentheses; see text and Table S15).
For the purpose of comparing this map of linkage disequilibrium-based estimates of of 2Nr with the map of , a comparably smoothed map, , and a more fine-scale were also created (see Figure 3).
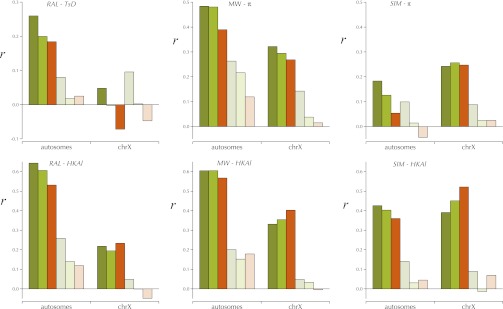
Differentiation between Africa and North America in windows
The patterns of differentiation of allele frequencies between populations can be a powerful means of discerning the impact of geographically variable selection or other processes generating allele-frequency differences. To depict genomic differentiation between the RAL and MW samples in windows, Fisher’s exact test (FET) was computed for each segregating site in the window. The statistical significance of the ensemble was gauged by Fisher’s combined probability test (FCPT). To deal with the obvious fact that such closely linked segregating sites do not meet the assumption of independence, a simple shuffling test was used to generate the null distribution of the FCPT χ2 (Hudson et al. 1992). The assignment of genomes to RAL and MW was randomly permuted and the FCPT χ2 was calculated. This was repeated until either 100 permutations had a FCPT χ2-value greater than that for the sample or 500,000 permutations were tested. From this distribution of a more reliable critical value, pHBK for the observed χ2 can be estimated; χ[log(pHBK)] is the quantity plotted and labeled Hudson–Boos–Kaplan-like (HBKl). The size of these HBKl windows was set to a specific sum of expected heterozygosity, 1.0 across contiguous segregating sites, to normalize the statistical power among windows.
Shared polymorphism
To assess the pattern of shared polymorphism between D. melanogaster and D. simulans across the genome, a simple extension of the HKA-like test was calculated that follows the approach in Wakeley and Hey (1997), except that no attempt is made to evaluate the statistical significance of deviations from the neutral model. Instead the goal was to detect genomic regions that harbored extreme amounts of shared polymorphisms. The expected proportion of shared polymorphisms was estimated for each sampling depth across each chromosome arm (ignoring the trimmed regions near centromeres and telomeres). A goodness-of-fit χ2 was calculated for each nonoverlapping window containing a total of 100 polymorphic and divergent sites. Each test has four cells, the number of sites polymorphic in the RAL and MW combined sample but monomorphic in the SIM sample, the number of sites polymorphic in the SIM sample but monomorphic in the combined D. melanogaster sample, the number of sites monomorphic in both samples, and the number of sites polymorphic in both samples. Variation in polymorphism and divergence is addressed by the HKAl analyses. To evaluate the patterns of shared polymorphism the usual (expected – observed)2/expected for the polymorphic-in-both-samples cell was treated as a 1-d.f. χ2 and the ±χ log(pWHl)] is plotted only in the genome browser [see below, labeled Wakeley–Hey-like (WHl)]. Large positive values indicate an excess and negative values a deficiency of shared polymorphisms. There is little power to detect regions deficient in shared polymorphisms given the overall low rate of shared polymorphism.
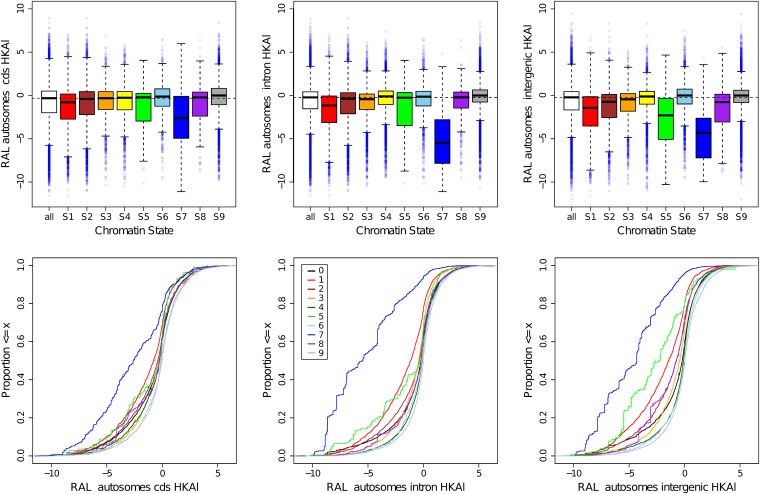
Correlations between chromatin states
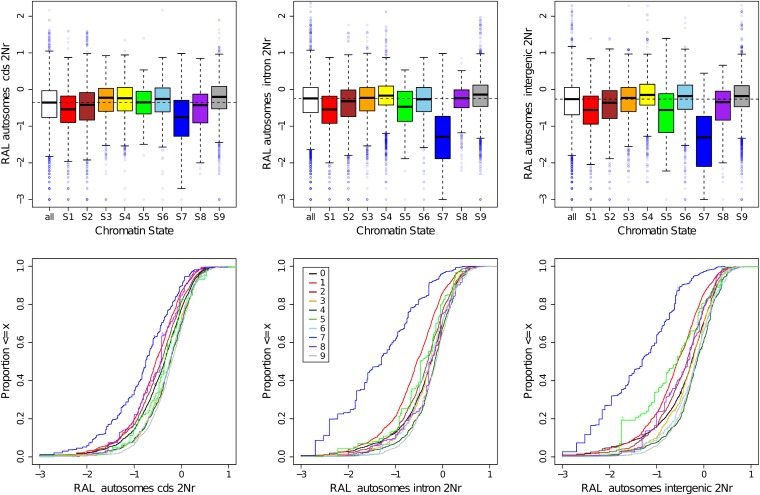
The fine-scale windows for πw, δw, HKAl, HBKl, and were intersected with the chromatin state “windows” described in Kharchenko et al. (2010), using the unionBedGraph command in the BEDTools v2.12.0 (Quinlan and Hall 2010). The distributions of resultant values were examined in two ways: box plots that capture the central tendencies and empirical cumulative distributions that display the differences in the tails more effectively (available in Figure S11 and Figure S12). Because of the variation in the length of partitioned windows, the analyses were based on weighting by number of base pairs in each window, using the Hmisc and Enmisc R packages (R Development Core Team 2010).
Gene-based methods
We defined a gene set for analyses by including only genes whose gene models (initiation codons, splice junctions, and termination codons) are either canonical or the same as the reference 5.16 annotations for every sampled genome. To ensure a minimum amount of data available for analyses, we restricted our analyses to genes having three or more alleles (lines) with at least 100 bp of data in both D. melanogaster and D. simulans samples. To have appropriate outgroup sequences to perform polarized analyses, genes for which neither D. yakuba nor D. erecta alleles had the same gene model as D. melanogaster reference annotations or had <100 bp were excluded. If more than one isoform met the above criteria, only the longest isoform was used. All statistical tests in gene-based analysis were performed with R version 2.8.0 (R Development Core Team 2010).
When analyzing bases with quality score ≥Q30, there were 9328 genes of 13,693 annotated genes that were included in the “golden gene set.” When considering bases with quality score ≥Q40, we restricted our analyses to 9258 genes that are shared with the Q30 data set (note that a few genes may be excluded from the Q30 gene set because of a premature stop codon that is not supported at Q40). Unless stated in the text, patterns observed with Q30 data were also observed with the more stringent Q40 data.
Expected nonsynonymous and synonymous heterozygosity was estimated as average pairwise differences. We include only sites with a sampling depth of ≥20 alleles in the D. melanogaster RAL sample and with ≥3 alleles in the D. simulans SIM and D. melanogaster MW samples. The numbers of nonsynonymous and synonymous sites were counted using the procedure in Nei and Gojobori (1986). Numbers of nonsynonymous and synonymous changes between two codons are calculated by averaging over all possible pathways between the pairs.
Lineage-specific divergences were estimated on branches leading to D. melanogaster and D. simulans by using D. yakuba (or D. erecta when the D. yakuba allele was not available) as the outgroup. We excluded polymorphic sites when estimating lineage-specific divergence to avoid the inflation of divergence with polymorphism (“polymorphism-adjusted divergence”). To accomplish this, we used two alleles each from D. melanogaster and D. simulans with the following criteria to capture the most of within-species polymorphism while ensuring enough statistical power for estimations. Each allele in either species was first ranked from high to low according to the proportion of bases that were not missing data (coverage). For D. melanogaster, two MW alleles with lowest rank (highest coverage) were picked. However, if any one of the MW alleles had rank ≥20, the MW allele with lower coverage was replaced by the RAL allele with the highest coverage. The two D. simulans alleles with highest coverage were included in the analyses. Lineage-specific divergence was estimated using maximum-likelihood methods implemented in PAML version 4 (Yang 2007). We used codeml with HKY as the nucleotide substitution model. The tree was assigned as [outgroup, (D. melanogaster allele 1, D. melanogaster allele 2), (D. simulans allele 1, D. simulans allele 2)], and the species-specific dN and dS were obtained from the estimates of the shared branch between two individuals in either D. melanogaster or D. simulans. Genes with <100 sites included in the PAML analysis were not included in downstream analyses.
Genetic differentiation between African and North American populations was tested by estimating averaged FST (Wright 1949; Weir and Cockerham 1984) of amino acid polymorphism. Only amino acid positions with sampling depth of at least 20 in the RAL sample and at least 3 in the MW sample were included. P-values associated with each FST were estimated using 1000 random permutations of the samples with respect to population identity (Hudson et al. 1992).
Polarized McDonald–Kreitman (MK) tests (McDonald and Kreitman 1991) were applied to D. melanogaster MW polymorphism data, using the alleles of the D. simulans mosaic assembly genome and D. yakuba or D. erecta (when the D. yakuba allele is unavailable) to count fixed differences on the D. melanogaster lineage. D. simulans polarized MK tests used D. simulans polymorphism data and the reference D. melanogaster genome and either the D. yakuba or the D. erecta allele. Codons with sampling depth greater than or equal to three in D. simulans and D. melanogaster MW samples are included in the analysis. When none of the polymorphic states was the same as those of the outgroups, we counted the site as both polymorphic (counting the differences between two ingroup alleles) and divergent (summing the differences between the outgroup state and each of the ingroup states). Polymorphic codons with more than two states within species are not included in the analysis. When two alternative codons differ at >1 bp, pathways between codons that minimized the number of nonsynonymous substitutions were used. To ensure at least modest statistical power, genes for which expectations of each of the four cells of the MK tables were less than one were removed. Statistical significance of the 2 × 2 contingency table was determined by Fisher’s exact test. Excess of nonsynonymous fixations (NSfix) and excess of nonsynonymous of polymorphisms (NSpoly) were calculated as the observations subtracted by the expectations from the 2 × 2 tables. Polarized MK tests were calculated using three different data sets: Q30 minimum data, Q40 minimum data, and Q30 minimum data with singleton alleles removed. The proportion of adaptive amino acid fixations (α) was estimated according to Smith and Eyre-Walker (2002) for individual genes.
Evidence of enrichment of statistical association in particular Gene Ontology (GO) categories was investigated for the critical values in the MK test. We combined the full GO list and the GO slim list (from the gene ontology website http://www.geneontology.org/) to annotate the GO categories of each golden gene. We considered only GO terms associated with at least five golden genes for which an MK test was calculated (after filtering criteria). For each GO term, we calculated the proportion of genes having MK tests P < 0.05 and rejecting the null hypothesis in the direction of excess amino acid fixation. The P-value associated with each GO term was determined by sampling without replacement n (the number of golden genes associated with a GO term) MK test P-values and calculating the proportion of significant MK tests. This process was repeated 10,000 times to get the empirical P-values associated with each GO term.
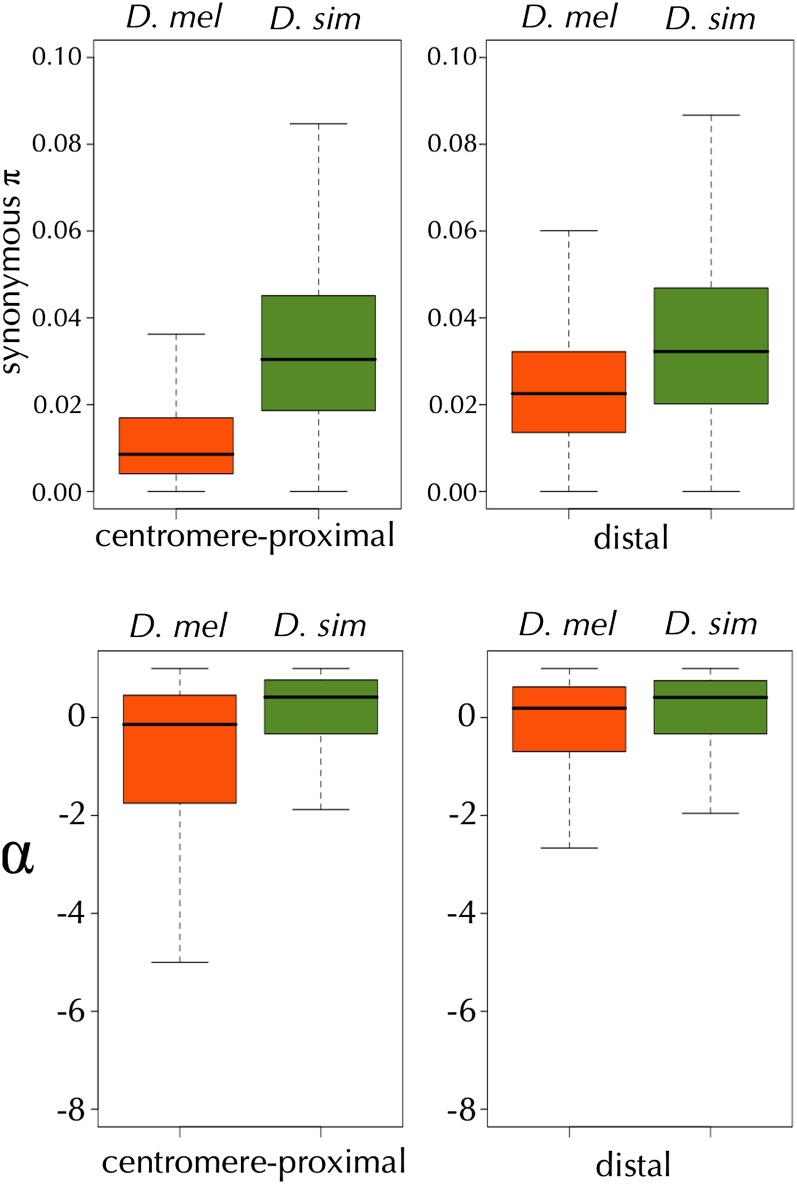
GC content of each gene was estimated as the proportion of G and C bases of the fourfold degenerate sites of the D. melanogaster reference allele. Recombination rate of the midpoint of each gene was estimated according to the genetic-map–based recombination rate estimates described above. We categorized genes into four equal bins according to genetic-map–based recombination rate, , separately for the autosomes and the X: “very low recombination” (0–2.98 cM/Mbp for X-linked genes and 0–1.07 cM/Mbp for autosomal genes), “low recombination” (2.98–4.15 cM/Mbp for X-linked genes and 1.07–2.57 cM/Mbp for autosomal genes), “intermediate recombination” (4.15–5.17 cM/Mbp for X-linked genes and 2.57–3.84 cM/Mbp for autosomal genes), and “high recombination” (>5.17 cM/Mbp for X-linked genes and >3.84 for autosomal genes) (see Figure S13 for distributions of numbers of genes with within the indicated intervals). With this binning, there are ∼300 X-linked and 2000 autosomal genes in each recombination category. Alternative binning criteria classified genes into no recombination (0 cM/Mbp), low recombination (0–3.6 cM/Mbp for X-linked genes and 0–1.89 cM/Mbp for autosomal genes), intermediate recombination (3.6–4.79 cM/Mbp for X-linked genes and 1.89–3.72 cM/Mbp for autosomal genes), and high recombination (>4.79 cM/Mbp for X-linked genes and >3.72 cM/Mbp for autosomal genes). This resulted in 37 X-linked and 468 autosomal genes in the no recombination category and ∼400 X-linked and 2300 autosomal genes in other categories. As most analyses were not sensitive to the choice of binning methods, we present only the results using the first categorizing methods to ensure equal statistical power of each bin. When investigating the effect of recombination rates on polymorphism, we used linear regression with the linear model “synonymous π ~ recombination rate.” The linear model “α ~ recombination rate” was used when analyzing the effect of recombination rates on adaptive protein evolution. P-values associated with each regression coefficient were calculated by 1000 random permutations.
Shared polymorphism in genes:
A codon that has the same two alternative states segregating in both D. melanogaster and D. simulans is considered a codon with shared ancestral polymorphism. To be conservative, codons with more than two alternative states segregating in either species were not considered. Also, if a codon was segregating for two alternative states in both D. melanogaster and D. simulans but only one state was shared between the two species or if there was no state shared when one species is monomorphic while the other is segregating for two alternative states, the codon was excluded from the analyses. The nonsynonymous and synonymous differences between pairs of codon states were calculated by the path that minimizes the number of nonsynonymous changes. We used Fisher’s exact test to test whether, in a gene, the ratio of shared polymorphic sites to all variable sites (including fixed differences between D. melanogaster and D. simulans, sites that are polymorphic within one of the two species, and shared polymorphic sites) is significantly different from the golden gene totals. Genes without any variations (both between species and within species) are removed from the calculation of the proportion of shared ancestral polymorphism for overall golden genes. Because we are interested in shared polymorphism that may have functional importance, we mainly present analysis of nonsynonymous variation.
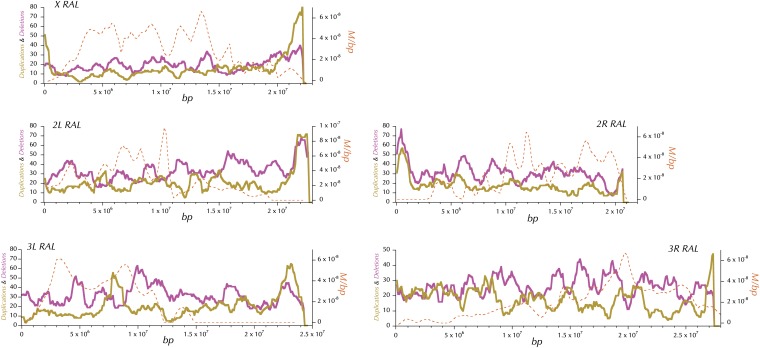
Identification and analysis of copy-number variation
To detect copy-number differences among inbred lines we examined the depth of sequence reads at each position of the genome in each sequenced line. Duplications were detected as regions of significantly increased depth, while deletions were inferred based on significantly decreased depth. We used a hidden Markov model (HMM) to segment the genome of each line into regions of euploidy and aneuploidy. The model calculates the expected read depth at each position based on the depth in the resequenced reference genome, GC content, number of SNPs, and number of small indels and then detects stretches of positions having read depth deviating from this expectation (see Appendix B). We set the minimum length of duplications and deletions to 295 bp to minimize false positive calls. Because our HMM was quite conservative, we used a second step to genotype copy-number variants (CNVs) identified in at least one line in all other lines. This genotyping step uses a likelihood-ratio test to score every line as either a duplication or a deletion based on the length and type of CNV identified by the HMM (see Appendix B).
CNVs and origins of replication
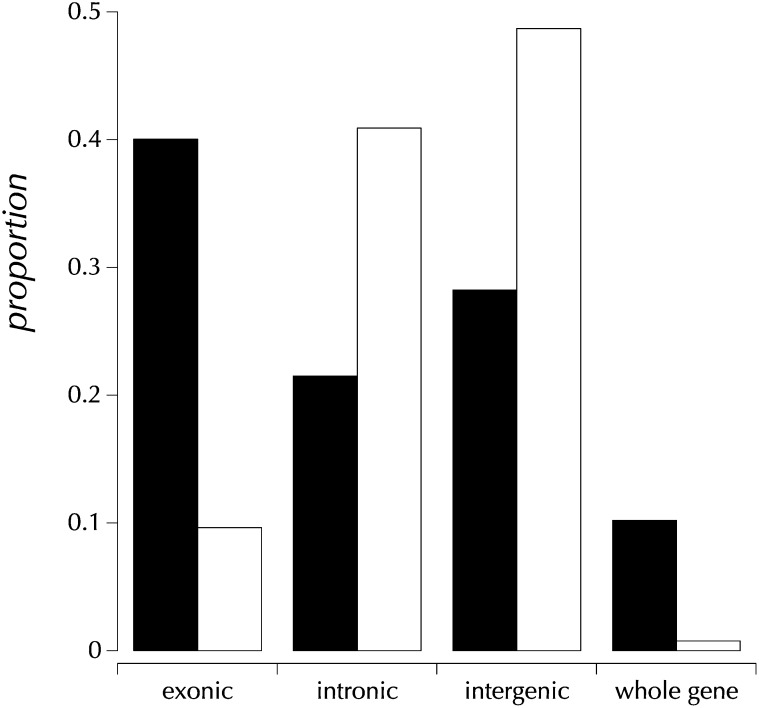
To investigate the relationship between the genomic distribution of origins of replication and CNVs, the origin of replication complex (ORC) meta-peaks ChIP-chip data based on immunoprecipitation of the replication initiation complex from three cell lines at modencode.org/ were analyzed (Roy et al. 2010). This data set is composed of 7084 annotated intervals covering a total of 1.98 million bp. Approximately 33% of these were annotated in all three cell lines, 23% in two, and 44% in one. The scoring of genomic regions reflects the number of cell lines that were positive. The annotated ORC intervals were extended in both directions by 500 bp. A null or background data set with a similar chromosomal distribution and the same sizes was generated. Each shuffled interval was placed 10 kbp away randomly 5′ or 3′ of an annotated ORC. If exactly one of the two random locations overlapped an annotated ORC, the other location was preferred. Importantly, this Monte Carlo data set preserved the total count, cell line weight, and size distribution. The numbers, sizes, and heterozgosities of CNVs overlapping these two data sets are the bases of the analyses presented.
CNVs and replication time
To examine potential associations between CNVs and replication time we looked at the replication time map of Schwaiger et al. (2009) obtained from the author’s website. This map includes replication time data for two cell lines, Kc and Cl8, which were analyzed independently for comparison. We used our CNVs called for the RAL lines for this analysis. To compare replication times of individual deletions and duplications events, the midpoint of each CNV was computed and the replication time for both cell lines was obtained from aforementioned map of Schwaiger et al. Each CNV was also classified as early, middle, and late replicating based on a clustering of the Kc replication time data also done by Schwaiger et al. and used in Cardoso-Moreira et al. (2011). To examine the potential association between the local density of copy-number variation and replication time as reported in Cardoso-Moreira and Long (2010), the numbers of deletion and duplication events were summarized in 100-kbp independent nonoverlapping windows. For each window, we also computed the corresponding expected replication time. This was done separately for the Kc and Cl8 cell lines for comparison.
Results
Polymorphism and divergence
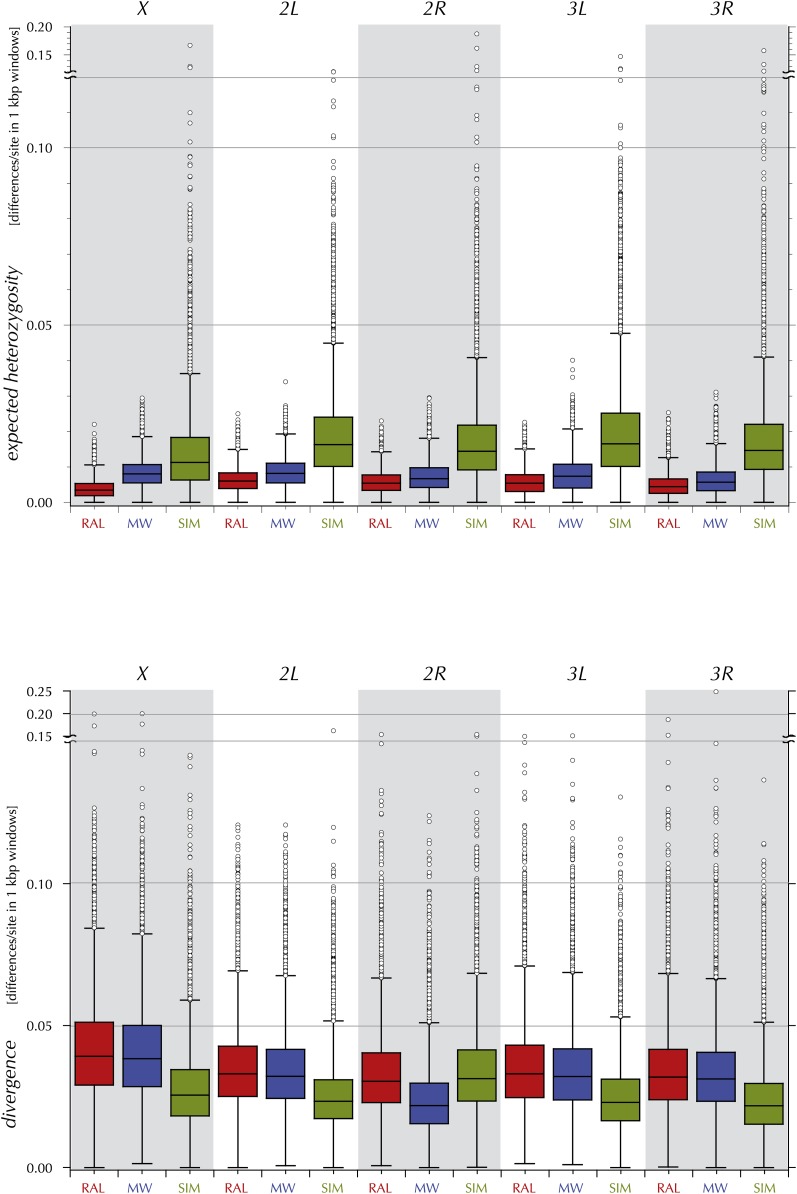
The expected heterozygosities for each of the major chromosome arms in the RAL and MW samples are in Table 3. Figure 4 (top) shows box plots of the expected heterozygosity in 1-kbp windows in each chromosome arm. The comparable estimates for the D. simulans sample (Begun et al. 2007) are presented for comparison. Clearly the expected heterozygosity of D. simulans is much greater than that of D. melanogaster (both RAL and MW). Note also that the divergence on the simulans lineage is less than on the melanogaster lineage, 0.024 vs. 0.030 respectively. Furthermore, divergence on the X is greater than that of the autosomes on both the simulans and melanogaster lineages (see Figure 4, bottom). The X-to-autosome ratios of expected heterozygosity are notably variable among the samples; the ratio for the Africa sample (MW) is well above the naively expected 0.75, at 1.10. Consistent with earlier studies (Andolfatto et al. 2001; Kauer et al. 2003; Hutter et al. 2007), the ratio in the North American sample (RAL) is far below, at 0.67. These deviations have been detected in earlier surveys of small parts of the genome and have motivated the investigation of complex demographic models (Hutter et al. 2007; Pool and Nielsen 2008). The increased scale and scope of the present data invite a reanalysis of these earlier interpretations (discussed below). At this point though, the focus is on a thorough empirical description of the genomic variation at all scales.
Table 3 . The expected heterozygosity, π and average lineage-specific divergence, δw on the chromosome arms in the RAL, MV, and SIM samples.
| Expected heterozygosity, π | Divergence, δ | |||||
|---|---|---|---|---|---|---|
| Chromosome arm | RAL | MW | SIM | RAL | MW | SIM |
| X | 0.00385 | 0.00822 | 0.01366 | 0.04137 | 0.04051 | 0.02744 |
| 2L | 0.00634 | 0.00846 | 0.01847 | 0.03497 | 0.03409 | 0.02503 |
| 2R | 0.00588 | 0.00733 | 0.01706 | 0.03370 | 0.03279 | 0.02379 |
| 3L | 0.00576 | 0.00783 | 0.01920 | 0.03504 | 0.03397 | 0.02484 |
| 3R | 0.00486 | 0.00631 | 0.01698 | 0.03395 | 0.03313 | 0.02352 |
| Genome | 0.00531 | 0.00752 | 0.01714 | 0.03084 | 0.03017 | 0.02432 |
Figure 4 .
The distributions of estimates of expected heterozygosity and of divergence in 1000-bp windows on chromosome arms (X, 2L, 2R, 3L, and 3R) for RAL, MW, and SIM.
Polymorphism and divergence across the chromosome arms
Expected heterozygosity, πw:
A striking feature of DNA sequence polymorphism in D. melanogaster and D. simulans (Begun et al. 2007) and indeed in a number of other species, e.g., tomato (Stephan and Langley 1998; Roselius et al. 2005), D. ananassae (Stephan and Langley 1989; Stephan et al. 1998), and humans (Hellmann et al. 2008; Cai et al. 2009), is the systematic reduction in centromere- and telomere-proximal regions where crossing over per physical length also declines (Hahn 2008 and references therein). Figures 5 and 6 and Figure S2, Figure S3, and Figure S4 show that π declines near the centromeres and telomeres and reveal other large-scale (≈105 bp) peaks and troughs in expected heterozygosity in overlapping 150-kbp windows incremented every 10 kbp. While the X in the RAL sample does show lower average π, it also exhibits more large-scale variation than does the X from the MW sample. Also obvious in these large-scale plots of π are the strong parallels between the two melanogaster samples and between D. melanogaster and D. simulans (see Table 4).
Figure 5 .
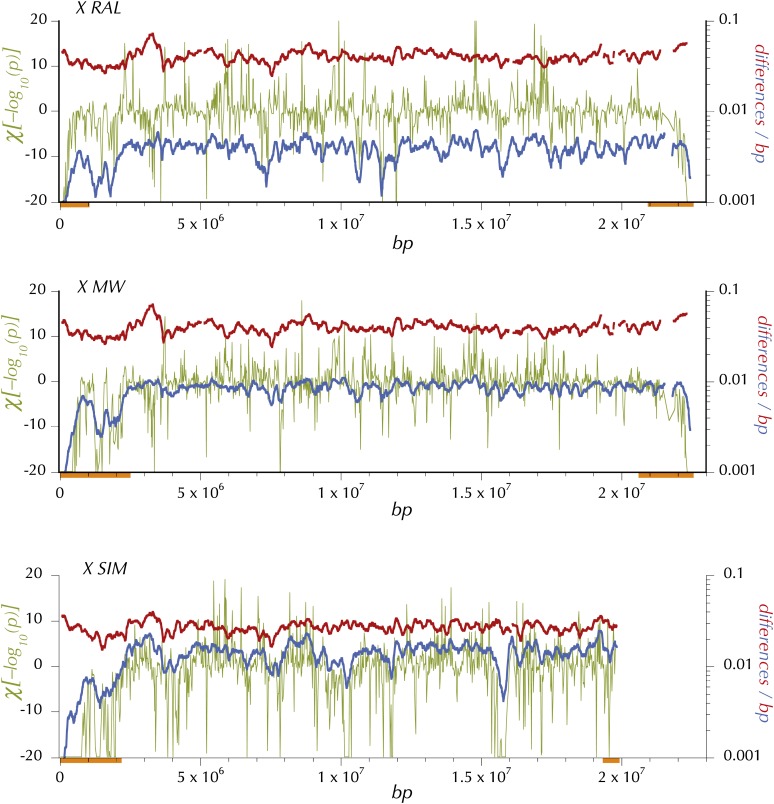
Expected heterozygosity, divergence, and HKAl on the X for the North American (RAL), African (MW), and simulans (SIM) samples. Blue shows expected heterozygosity, π at the midpoint of 150-kbp windows (incremented every 10 kbp, minimum coverage = 0.25 and Q30 sequence). Red shows lineage-specific, average Q30 divergence in 150-kbp windows (incremented every 10 kbp and minimum coverage of 0.25). A preliminary application of HKAl on the Q30 data in windows of 4096 contiguous polymorphic or divergent sites identified centromere- and telomere-proximal regions (orange bars) in which the each window exhibited a deficiency of polymorphic sites relative to the chromosome-arm average. Then HKAl was applied again on the Q30 data in windows of 512 contiguous polymorphic or divergent sites (excluding these centromere- and telomere-proximal regions from calculation of the chromosome-arm–wide expected proportions, pc and dc). χ[log(pHKAl)] (olive) is the log of the P-value associated with HKAl plotted with the sign of the difference between the observed number and the expected number of polymorphic sites in the window.
Figure 6 .
Expected heterozygosity, πw, divergence, δw, and HKAl on 2L for the North American (RAL), African (MW), and simulans (SIM) samples. Blue shows expected heterozygosity, πw at the midpoint of 150-kbp windows (incremented every 10 kbp, minimum coverage = 0.25 and Q30 sequence). Red shows lineage-specific, Q30 divergence, δw in 150-kbp windows (incremented every 10 kbp and minimum coverage of 0.25). A preliminary application of HKAl on the Q30 data in windows of 4096 contiguous polymorphic or divergent sites identified centromere- and telomere-proximal regions (orange bars) in which the each window exhibited a deficiency of polymorphic sites relative to the chromosome-arm average. Then HKAl was applied to the Q30 data in windows of 512 contiguous polymorphic or divergent sites (excluding these centromere-and telomere-proximal regions from calculation of the chromosome-arm–wide expected proportions, pc and dc). χ[log(pHKAl)] (olive) is the logarythim of the P-value associated with the HKAl plotted with the sign of the difference between the observed number and the expected number of polymorphic sited in nonoverlapping windows of 512 variable sites.
Table 4 . The correlation in πw between D. melanogaster (MW or RAL) and D. simulans (SIM) in 1000-bp windows.
| Chromosome | MW-SIM | RAL-SIM |
|---|---|---|
| X | 0.469 | 0.298 |
| Chr2L | 0.450 | 0.393 |
| Chr2R | 0.385 | 0.361 |
| Chr3L | 0.328 | 0.338 |
| Chr3R | 0.336 | 0.328 |
| All | 0.375 | 0.367 |
Average divergence, δw:
Figures 5 and 6 as well as Figure S2, Figure S3, and Figure S4 show the average lineage-specific divergence, δw, in the same overlapping 150-kbp windows incremented every 10 kbp. In contrast to πw the distribution of δw is remarkably uniform across each of the arms, although as noted above the X does consistently exhibit an overall higher amount of divergence. One can also note another subtle but consistent pattern of decreasing divergence from the centromere to the telomere (Begun et al. 2007). The generality of this observation of divergence suggests that quantitative modeling of the forces that shape polymorphism and divergence should strive to address the cause of this chromosome-arm pattern.
Contrasting polymorphism with divergence at the chromosome level
Arguably, the comparison of levels of polymorphism to divergence in different genomic regions is the most fundamental analysis directly relevant to models proposed to explain the maintenance of genetic variation and the divergence between species. The simple empirical analog of the HKA test (Hudson et al. 1987) as modified by Ford and Aquadro (1996), HKAl, identifies local genomic regions in which the relative polymorphism (numbers of segregating sites) and divergence (numbers of fixed differences) exhibit strong deviation from the chromosome arm average. In Figures 5 and 6 and Figure S2, Figure S3, and Figure S4 are plotted χ[log(pHKAl)], the signed log of the P-value for a simple 1-d.f. χ2 at the midpoint of nonoverlapping windows of 512 variable (polymorphic or diverged) sites. The expectations of numbers of segregating sites are based on the observed proportions at each sampling depth across the chromosome arm (excluding the centromere- and telomere-proximal regions in Table S14 and demarcated by the orange bars in Figures 5–6 and Figure S2, Figure S3, and Figure S4. Of course, the most striking features of the distribution of πw and this HKAl statistic are the reductions in the regions adjacent to the centromere and telomere. As is apparent in the bottom (SIM) panels of Figures 5 and 6, and Figure S2, Figure S3, and Figure S4. and as reported by Begun et al. (2007), simulans also exhibits reductions in these regions. But note that the regions of reduced polymorphism in simulans are smaller than in melanogaster (RAL and MW) on chromosome arms 2R, 3L, and 3R.
A prediction of models of linked strong directional selection is a positive correlation of the variation in polymorphism with variation in recombination per physical length, especially as the rate of recombination approaches zero (Maynard Smith and Haigh 1974; Kaplan et al. 1989; Charlesworth et al. 1993). Indeed these genomic regions do exhibit much lower levels of crossing over per physical length (Figure 3) and of , an estimate of 2Nr (Figure 7 and Figure S7). At the telomere the proportion of segregating sites plummets most precipitously in parallel with and π. On the X of melanogaster the telomere-associated suppression extends over a wider region than on the autosomes (Figure 3). At the centromeric ends of chromosome arms χ[log(pHKAl)] is extremely negative over even broader physical regions, especially on several of the autosomes in melanogaster. As Figure 3 shows, crossing over per base pair is low in these regions.
Figure 7 .
, estimates of 2Nr across chromosome arms X and 2L, generated by LDhat. Also shown for comparison are estimates of πw (blue) and δw (red) as in Figures 5 and 6 (see Figure S7 for this type of plot for all five chromosome arms).
The second striking feature of these patterns of polymorphism and divergence is the large number of extreme χ[log(pHKAl)] windows across the whole euchromatic arm between these centromere- and telomere-proximal regions. The pattern is more variable for χ[log(pHKAl)] in Figures 5 and 6 and Figure S2, Figure S3, and Figure S4 than for πw and δw because the windows of these latter two statistics are larger (150 kbp vs. ≈20 kbp) and densely overlapping (10-kbp increments). These remarkable deviations in χ[log(pHKAl)] occur both in the direction of excess polymorphism and in that of excess divergence, positive and negative χ[log(pHKAl)], respectively. The broad (>150 kbp) peaks and especially troughs in πw and χ[log(pHKAl)] often harbor a number of apparently disjunct smaller windows in which the proportions of segregating sites are quite deviant.
The view of these statistics at a finer scale (in smaller windows and in coding elements) is discussed below, but at this gross resolution the large number of windows on each arm that are associated with extremely small HKAl test P-values is remarkable and is most simply interpreted as evidence of a great deal of recent and (by deduction) recurrent selective substitution of rare variants (newly arising adaptive mutations).
Patterns of polymorphism within cosmopolitan gene arrangements
Perhaps the most striking population genomic feature of D. melanogaster is the high level of large paracentric inversion heterozygosity in tropical populations. D. simulans and its sibling species show very low levels of segregating karyotypic variation. This appears to be the conserved ancestral state, since simulans and the melanogaster “standard” euchromatic karyotype differ by only one large inversion in chromosome 3R fixed on the melanogaster lineage (Lemeunier and Ashburner 1976; Begun et al. 2007). One or more cosmopolitan, large, derived paracentric inversions are found at intermediate frequencies in each of the chromosome arms in African (Lemeunier and Ashburner 1976; Aulard et al. 2002) and other tropical populations (Mettler et al. 1977; Knibb et al. 1981). Studies of the sequence polymorphism within samples of several of these cosmopolitan inversions reinforce the conclusion that all these chromosome rearrangements arose independently and recently from the standard arrangement (Wesley and Eanes 1994; Andolfatto and Kreitman 2000; Matzkin et al. 2005). As Table 1 shows, inversion polymorphism in the MW sample is high, as expected. The overall levels of inversion polymorphism in the RAL sample are typical of a temperate population and previous sampling of this population (Langley et al. 1977; Mettler et al. 1977; Voelker et al. 1978). Much higher frequencies of the standard arrangements are observed, except on chromosome 3R. In the numerous prior surveys in Raleigh, In(3R)P was consistently found at ∼12% frequency. However, we found this inversion in only a single line. The high-inversion polymorphism on 3R in the RAL sample is now due to In(3R)Mo (∼18%), previously very rare in this and other temperate eastern North American populations (Langley et al. 1977; Mettler et al. 1977). This dramatic change in the frequencies of these two inversions is unexpected and deserves additional investigation.
Most inversions are at low frequencies in the RAL sample and counts in the MW sample are inherently few; however, the prevalence of In(3R)Mo in RAL invites a more careful population genomic analysis of the pattern of polymorphism relative to the inversion breakpoints. Due to suppression of recombination in inversion heterozygotes, especially immediately proximal to the inversion breakpoints, the inversion can remain associated with the single haplotype in which the inversion arose. Consistent with earlier observations in melanogaster of In(3R)P and In(2L)t by Wesley and Eanes (1994) and Andolfatto et al. (1999), respectively, Figure 8 shows that heterozygosity among In(3R)Mo chromosomes immediately surrounding both breakpoints is almost completely lacking. This feature is consistent with the hypothesis that single In(3R)Mo haplotypes were “captured” by the unique rearrangement event. While the lack of polymorphism relative to the standard in these small breakpoint regions indicates a recent origin for In(3R)Mo on the timescale of the mutation rate [similar to In(2L)t and In(3R)P], the fact that π throughout most of the inverted segment is much higher (yet not as high as a standard) indicates there may have been many double exchanges with standard since In(3R)Mo’s origin. Similarly there is evidence of exchanges just outside the two breakpoints.
Figure 8 .
Expected heterozygosity, πw on 3R for the North American (RAL) sample. The dark blue line shows expected heterozygosity, πw at the midpoint of 150-kbp windows (incremented every 10 kbp and Q30 sequence) for the “standard” arrangements in the RAL sample. The light blue line is the comparable plot for the In(3R)Mo arrangements of chromosome 3R in the RAL sample. The gray rectangle demarcates the inverted region in In(3R)Mo. The two insets below are blow-ups of the genomic regions surrounding the two breakpoints marked by arrows (Corbett-Detig et al. 2012).
In addition to the small regions surrounding the breakpoints, we found two other large regions of decreased polymorphism. The first is ∼0.5 Mbp and is located within the inversion near the centromere-proximal breakpoint. Note that the inversion places this region now telomere-proximal. The second region, ∼1.5 Mbp, is actually between the distal breakpoint and the telomere. Between the distal breakpoint and this large block is >1 Mbp in which we observed increased polymorphism. While crossing over outside each breakpoint of the inversion is no doubt suppressed in the In(3R)Mo heterokaryotypes, as with other rearrangements, the expected higher frequency of single crossovers than of the double crossovers required within the inversion and the intervening region of high polymorphism demands a more complex explanation. Recent hitchhiking events (Maynard Smith and Haigh 1974) restricted to this gene arrangement are perhaps more probable than an equilibrium between epistatic selection, recombination, and genetic drift. In any case the replacement of In(3R)P by In(3R)Mo and this unusual pattern of restricted polymorphism are surprising. Obviously, considerable additional investigation of the population distribution and recombination patterns is needed to support any robust analysis and interpretation.
Linkage disequilibrium
A number of previous studies in D. melanogaster concluded that the scale over which the magnitude (+ or −) of linkage disequilibrium decays is hundreds of base pairs (Miyashita and Langley 1988; Long et al. 1998), although clear exceptions have also been noted (Aquadro et al. 1992; De Luca et al. 2003; Takano-Shimizu 2004; Tatarenkov and Ayala 2007; Itoh et al. 2009). The North American (RAL) sample studied here is the first opportunity to examine this question on a full genomic scale. The average squared correlation coefficient, r2, among pairs of SNPs within the indicated intervals of separation (≤20,640 bp) on the five chromosome arms in this sample is presented in Figure 9A. Note that all but the first of these intervals increase twofold in midpoint and width at each step. The midpoints of these decay curves are indeed near 100 bp. The average r2 is initially greatest for the closest pairs on the X, and it dissipates to a level comparable to that of the other chromosome arms by 10 kbp while that of 3R remains higher. Also note that all but the first three intervals involve pairs of sites >40 bp apart, which are thus not likely to be attributable to spurious correlations induced by read mapping or basecalling.
Figure 9 .
(A–C) The decay of linkage disequilibrium with physical distance on the chromosome arms in the North American sample (RAL) of D. melanogaster. The average r2 between all pairs of Q30 SNPs with minor allele frequency (MAF) ≥0.167 separated by contiguous ranges of base pairs is plotted in A against the midpoint of each range (indicated by the dots on the abscissa). The ranges are [1, 10] and (si−1, si], for 1 < i ≤ 12, where si = 10, 20, 40, 80, 160, 320, 640, 1280, 2560, 5120, 10,240, and 20,480. The lines for the X and 3R are distinct from those for 2L, 2R, and 3L and are labeled. B shows the distribution of the average allele-frequency–oriented correlation coeffficent, rω over the same intervals for each chromosome arm (see Table 1 for the definition of rω). And C shows that much of the shift toward more positive rω is attributable to an increase in the proportion of pairs of SNPs with rω > 0.
Linkage disequilibrium can also be oriented based on allele frequencies. Here, “positive” linkage disequilibrium occurs when the alleles with frequencies >50% at a pair of sites are positively correlated, while “negative” linkage disequilibrium indicates that the more common allele at one site is associated with the less common allele at the second site (Langley and Crow 1974; Langley et al. 1974). The direction of linkage disequilibrium can vary systematically under particular models: genetic drift with little or no recombination (Golding 1984), equilibrium epistatic selection (Langley and Crow 1974), and hitchhiking (Stephan et al. 2006). Figure 9B shows that the average correlation coefficient between alleles with frequencies >0.5 (rω, see Table 1) is ≫0 for the same intervals of distance. Note that at the most proximal distances the X and 3R exhibit exceptionally high average rω . On 3R, rω merges with the other autosomal arms but on the X average rω remains higher. Figure 9C shows that this effect is at least partially attributable to an increased proportion of SNP pairs with rω > 0, not just increased magnitude of association in the positive direction. Also note that while the average rω is near 0.0 for all but the X (Figure 9B) for pairs of SNPs >1000 bp apart, the proportion of positive rω in Figure 9C dips below 50% for the last three intervals, 2560 to 5120 to 10,340 to 20,640. It is unclear whether this result is related to the original observations of negative rω between allozyme loci and polymorphic inversions in Langley et al. (1974) and the model of natural selection proffered in Langley and Crow (1974).
In Figure 10A, the decay of averages of positive and of negative rω at different ranges of distance and allele frequencies is plotted for each chromosome arm. The X chromosome is observed to have higher levels of linkage disequilibrium than the autosomes at all allele frequencies. Arms 2L, 2R, and 3L have very similar patterns of decay with distance. 3R is generally intermediate between the other autosomal arms and the X chromosome. However, the magnitude of linkage disequilibrium at long distances (>5000 bp) is greater for 3R than for any other arm. This pattern can be attributed to 3R’s high levels of inversion polymorphism (see below) that can suppress crossing over.
Figure 10 .
The distribution of positive and negative average rω at different distances (bp) for pairs of Q30 SNPs in the North American (RAL) sample. The left plot shows the pattern for the five large chromosome arms, orange is the average rω > 0, and olive is the average rω < 0, where the MAF ≥ 0.167. Solid circles mark the X, while open circles are 3R. The other three chromosome arms are hardly distinguishable. The right three panels are plots similar to the first (left) but for chromosome arms X, 3R, and 2L. The different lines of rω > 0 (orange) and rω < 0 (olive) correspond to subsets in which p(1 − p)q(1 − q), the product of the SNP allele frequencies at each locus (p and q) fall in the following intervals, respectively (increasing in rω): 0.0255–0.0383, 0.0383–0.0468, 0.0468–0.0531, 0.0531–0.0582, and 0.0582–0.0625.
The observation of high levels of linkage disequilibrium over short distances, >100 bp, is consistent with that in earlier studies. But the strong tendency toward rω > 0 has not been noted in studies of fewer sites in selected genomic regions. Only two theoretical hypotheses can be proffered for this pattern. The first is recent admixture (Cavalli-Sforza and Bodmer 1971, p. 69). The western hemispheric populations of D. melanogaster are believed to be founded by colonizing flies from both Europe and West Africa in the 18th and early 19th centuries. Assuming an asymmetric admixture contribution and a loss of ancestral polymorphisms in the recognized bottleneck associated with the out-of-Africa diaspora, linked SNPs with highly differentiated frequencies may still exhibit rω > 0. Simulation analyses (data not shown) demonstrate that demographic processes such as admixture and population bottlenecks may account for a portion of the observed skew in rω.
The alternative theoretical explanation is based on the analysis in Stephan et al. (2006) of the impact of hitchhiking on linkage disequilibrium. While the most striking result was the demonstration that the linkage disequilibrium between sites on opposites sides of the selected locus is destroyed, they also established a wide parameter domain in which rω > 0. In addition to the recognized loss of variation associated with hitchhiking, there are drastic and correlated changes in the frequencies of SNPs in the flanking regions. In particular, rare SNPs that happen to be associated with the rare, selectively favored variant (mutant) will rise in frequency together. If tightly linked (and if on the same side of the selected locus), they tend to recombine as a haplotype away from the selected site and remain at similar frequencies (>0.5 or <0.5), yielding rω > 0. Therefore, the observed bias in rω may be attributable (at least partially) to the hitchhiking effect.
Linkage disequilibrium along the chromosome arms and , an estimate of the population recombination parameter 2Nr
Studies of linkage disequilibrium in human population genomic surveys have successfully identified and quantitatively mapped systematic patterns in the rate of recombination (McVean et al. 2004; Myers et al. 2010). This advance is based on deep genotyping (>400,000 SNPs) in large samples and on the application of practical statistical approximations (Hudson 2001) in computationally scaleable software (McVean et al. 2004). These maps of estimated recombination in the ancestry of the sampled human genomes revealed a striking pattern of punctate recombination that has been independently verified in both sperm-typing experiments (Jeffreys et al. 2001; Tiemann-Boege et al. 2006) and pedigree-based investigations (Coop et al. 2008; Kong et al. 2010).
Figure 7 shows the fine-scale estimates of the population recombination parameter across chromosomes X (top) and 2L (bottom; also see Figure S7 for the patterns on all the chromosome arms) along with those for average πw and δw in 150-kbp windows (note highly smoothed versions of are shown in Figure 3 for comparison to ). The genomic pattern of these local estimates is expected to reflect via linkage disequilibrium the actual per meiosis recombination rates in a randomly mating, finite population at equilibrium for selectively neutral mutation and genetic drift. Comparable SNP genotyping-based analyses of larger human samples indicate that the fit to an equilibrium neutral model is remarkably good for most of that genome (McVean et al. 2004). And even other demographic sources of linkage disequilibrium such as population size fluctuations, geographic differentiation, and subsequent admixture fit well into this selection-free modeling of human population genomics.
But the pattern of linkage disequilibrium in a much more abundant species such as D. melanogaster may be determined more by linked selection than by genetic drift (Maynard Smith and Haigh 1974; Stephan et al. 2006). Because of the larger species population size of D. melanogaster, one must consider that while the genomic scale of an individual hitchhiking event will be on the order of the selection coefficient s (Maynard Smith and Haigh 1974; Stephan et al. 2006), the rate of adaptive substitutions may increase rapidly with population size. Two mechanisms affect this increase: proportionate increase in the rate at which favorable mutations arise and the numbers of favored variants immune from the impact of genetic drift after the first few generations (2Ns ≫ 1) in generations other than the initial few (Fisher 1922; Wahl 2011).
Clearly the estimated values, , of 2Nr in the centromere- and telomere-proximal regions are substantially lower, consistent with the highly smoothed (low-resolution) recombination rate maps based on the genetic map (see the next section). Hitchhiking and background selection (Charlesworth 1994) in these large regions of reduced recombination may also contribute to this relative reduction in values of . The analysis of the site-frequency spectrum below can address this issue further. Outside these regions, a lower than average (per base pair) estimate of 2Nr on the scale of ≥100 kbp might well reflect lower meiotic crossing over per physical distance, or it could reflect bias in caused by the contractions and distortions of the gene genealogies due to recent hitchhiking. A striking feature of these maps is the large spikes of high and low that have withstood two restrictive filters, a high penalty in the prior for the number of rate changes and replication in two long independent chains. These are analyzed and interpreted in the context of much smaller nonoverlapping windows below.
Comparison of the and the maps
To ask whether variation in the linkage disequilibrium-based estimate of recombination reflects variation in the per generation recombination rate, our estimates of the recombination rate per base pair from the standard genetic map can be compared with . Table 5 shows the chromosome-arm-wide average rates for and . The smaller, acrocentric X has a 24% higher than the average autosome, while the longer chromosome 3 has a 15% lower . The chromosome-arm-wide average varies a great deal more. The average estimate for the X is 1.8 times the autosomal average (note the smaller centromere-proximal region of lower in Figure 7), while the four autosomes range between 0.72 and 1.20 of the autosomal average. Because the genomic resolution of our estimates of is so much lower than that for , we smoothed both and to the same scale (loess span parameter = 15%). Figure 3 shows the distribution of these smoothed versions and across the five large chromosome arms. The coincidence of many features of these maps indicates that these independent approaches identify common variation in the rate of recombination at this coarse scale. Indeed Figure 11A, a scatterplot of the two estimates, and Figure 11B, the weighted (by base pairs) correlations between logarithms of these two measures, clearly indicate that they are capturing a strong pattern of covariation, which is most parsimoniously interpreted as reflecting a common factor in recombination per base pair (see Table 5; note these correlations are among non–log-transformed estimates of and ). This conclusion suggests that the variation revealed by the higher-resolution in Figure 3 and unsmoothed in Figure 7 and Figure S7 may also reflect the pattern of recombination per base pair on even finer resolutions. However, it remains possible that hitchhiking or other forces may interfere with local estimates of and that these distortions are averaged out on the scale of Figure 3.
Figure 11 .
Common patterns of the genomic distribution of the two estimates of the rate of recombination per base pair, and . (A) A scatterplot of these two estimates in windows of variable sizes (all >104 bp) for each of the untrimmed chromosome arms. Eight outliers are off this plot (2R, −0.0027, −6.2 × 10−9; X, 0.0321, 1.9 × 10−8; 0.0375, 2.2 × 10−8; 0.0400, 2.5 × 10−8; 0.0404, 2.7 × 10−8; 0.0373, 3.0 × 10−8; 0.0314, 3.3 × 10−8; 0.0275, 3.7 × 10−8). B compares the base pair-weighted correlation logarithms of and , among chromosome arms for both trimmed (light green) and untrimmed (dark green). The parallels between the two measures are high, but reduced slightly when only the trimmed regions are considered. This is not true for chromosome X, which shows much less correlation between the two estimates on the trimmed data.
Polymorphism and divergence at higher resolution
The densities of segregating and divergent sites in both the RAL and the MW samples are sufficiently great that statistically meaningful analyses can be made on a much smaller scale than depicted above at the 150-kbp resolution or that of or . The average scale of linkage disequilibrium (midpoint of decay over base pairs) in D. melanogaster is ≪500 bp and most annotated gene elements are closer to this scale. Smaller windows for χ[log(pHAKl)] and other statistics can provide higher resolution of detected outliers but the statistical power ultimately declines as the total number of variants in a window decreases. On the other hand, very large windows (≫1000 variant base pairs) may exhibit one pattern of deviation from the chromosome arm average because of the aforementioned larger scale and systematic variation along the chromosome arms, thus obscuring even strong local deviations. Table 6 compiles the basic information on available fine-scaled statistics, including πw, δw, and .
Table 6 . Fine-scale statistics.
| Name | Symbol | Window size | Range | Link |
|---|---|---|---|---|
| Expected heterozygosity | πw | ≥1000 bp (filter:coverage >250 bp) | 0.0 to 0.5 | URL |
| Average lineage-specific divergence | δw | ≥1000 bp (filter:coverage >250 bp) | 0.0 to 1.0 | URL |
| Population recombination parameter, 2Nr | Variable (see text) | 10−5 to 10 | URL | |
| Test: segregating to divergent sites | HKAl = ±χ[log(pHKAl)] | Segregating and divergent sites = 50 bp | −20 to +20 | URL |
| Test: frequency spectrum | TsD = ±χ[log(pTsD)] | Segregating sites = 50 | Depends on the nos. of sampling depths | URL |
| Test: geographic differentiation | HBKl = −log(pHKAl) | Expected no. heterozygous sites in window = 1.0 | 0 to 5 | URL |
| Variation in shared polymorphism | WHl = −log(pWHl) | Polymorphic and divergent sites = 100 | 0 ≤ | URL |
While diverse approaches to many interesting questions can start with these (or similar) high-resolution population genomic statistics (annotations), we present three distinct types here. The first is a view of these fine-scaled population genomic statistics as additional annotations in the context of a genome browser. The second is a systematic search for associations of HKAl, TsD, and HKAl with annotated elements of gene structure and chromatin “states.” And the third addresses the correlation across the genome of the finer-scaled estimate of the rate of recombination, with these deviations in the ratio of polymorphism to divergence (as measured by HKAl), in the allele frequency spectrum (as measured by TsD), in differentiation between populations (HBKl), and in the proportion of shared polymorphism between species (WHl).
Genome browser annotations, πw, δw, , HKAl, TsD, HBKl, and WHl
Arguably one of the most valuable uses of the measures and test statistics derived from small windows is via their visual juxtaposition in specific genomic regions with the already rich and high-quality structural and functional genomic annotations of D. melanogaster. The choice of genome browser (applications) and specific settings used can vary in a myriad of ways. Here we will show as examples two views of these statistics on chromosome (chr)2R in the UCSC genome browser using a track data hub, but the reader is encouraged to download and display them via other tools, e.g., ENSEMBL and IGB 6.5 (Nicol et al. 2009). The coordinates and values of windows of πw, δw, , HKAl TsD, HBKl, and WHl are available in BedGraph file format at the URLs indicated in Table 6. Also note that Ensembl provides a complete population genomic presentation of the Q30 sequence data (reported here) in the context of Ensembl’s full gene-oriented annotation of the D. melanogaster genome (FlyBase derived) that can be of great value to those interested in the functionally annotated sequence variation at particular loci, e.g., Cullin-2.
Figure S9 shows a snapshot of the UCSC Genome Browser based on the track data hub containing these fine-scale statistics. While the large-scale patterns (describe above) are apparent, here we want to emphasize the potential interest and value of these estimates and “tests” as annotations for the specific genes in and around each window. For example, Figure S10 is a blowup of the 287 kbp beginning at 8 Mbp on chr2R, also accessible as a session. In the center of this part of chr2R is a cluster of 31 protein-coding and 6 noncoding genes. It exhibits relatively low values, reduced numbers of segregating sites relative to diverged sites (reflected in the pattern of πw and in HKAl), and a strong skew in the site-frequency spectrum in the RAL sample. The RAL and MW samples exhibit a high amount of differentiation in this region, which is reflected in the high proportion of HBKl windows that have the maximum value. The πw, δw, HKAl, and TsD tracks for the MW sample are not shown in Figure S10, but they are available and can be displayed from the track data hub. Their distribution in this region is similar to that of the RAL sample. Also included in Figure S10 are annotations from the simulans sample. πw in this same gene cluster in the SIM sample is also reduced and is consistent with the parallel clustering of negative HKAl and lack of positive WHl values. Among the 31 coding genes in this cluster are 2 for which patterns of polymorphism have previously been interpreted as evidence of recent strong selection, Cyp6g1 (Schlenke and Begun 2004; Schmidt et al. 2010) and Hen1 (Kolaczkowski et al. 2011a and references therein). Many other small, but richly annotated regions of these genomes exhibit clusters of deviant HKAl, TsD, HBKl, and WHl values. Indeed the systematic analysis below demonstrates clustering of deviant population genetics statistics near gene and chromatin annotations. Specific biologically interesting hypotheses supported by these apparent associations can be addressed using the available stocks from which these sequences were derived and via gene-focused replications in the large remainder of the RAL sample as well as in additional independent population samples and phenotyping experiments (Ayroles et al. 2009; Clowers et al. 2010).
Evidence of linked selection in the large-scale genomic associations with , and
Parallels between statistics that can reflect the impact of linked selection (e.g., HKAl and TsD) and large-scale patterns of recombination per base pair can be examined in terms of overall correlations and from the perspective of specific genomic regions. The centromere- and telomere-proximal regions of strongly depressed recombination per base pair (see Figures 3 and 7 and Figure S7) correspond broadly to the regions of reduced πw and HKAl in Figures 5 and 6 and Figure S2, Figure S3, and Figure S4. The site-frequency spectrum at this large scale also shows a strong skew toward rare variants in regions of very low crossing over per physical length; this is especially evident in the larger RAL sample near the telomeres and proximal to the centromeres of 2R, 3L, and 3R. And these patterns are quite evident in the base pair-weighted correlations between or and πw , HKAl, and TsD. Of course, the strong and consistent skew of the site-frequency spectrum (TsD ≪ 0) in these regions of low crossing over is more consistent with the predictions of hitchhiking (Braverman et al. 1995) than background selection (Hudson and Kaplan 1994; Charlesworth et al. 1995).
The contrast of polymorphism with divergence summarized in HKAl exhibited the strongest association with or (see Figure 12 and Table S15). Considering the whole (untrimmed) chromosome arm, more than one-quarter of the variation in HKAl can be explained by either of these two measures. As can be seen in Figures 5 and 6 and Figure S2, Figure S3, and Figure S4, HKAl plummets in the low- to no-recombination regions proximal to the centromere and telomere. While this result is expected from conclusions of many articles in the last two decades, the magnitude and genomic footprint of the effect is striking. For the two (most certainly nonindependent) melanogaster samples, the correlations of πw with or are almost as great as for HKAl. The correlations of HKAl with or are large and comparable to each other in simulans (SIM), while the correlations of πw with or are much weaker than in melanogaster (see Figure 12). This is likely due to the large differences in overall statistical power in the two species. The power of the HKA test is sensitive to disparities in the overall proportions of diverged and polymorphic sites (see Table 3); these are more balanced in simulans, especially on the X. Most noteworthy is the fact that the largest chromosome-arm–wide correlation of HKAl with or in simulans (SIM) is on the X (note that both recombination statistics are estimated in melanogaster), while the X shows the weakest correlation with these measures of the rate of recombination in the melanogaster samples, RAL and MW. This is at least partially attributable to the more drastic depression of HKAl near the telomere of the simulans X and the lower overall π on the melanogaster X relative to that on the autosomes.
Figure 12 .
Correlations between recombination rates and HKAl, πw, and TsD. r is the base pair-weighted Pearson’s correlation coefficient between TsD, πw, and HKAl and the logarithm of (olive), (light olive), and (orange) across the autosomes and the X chromosome. The three lower columns to the right (lighter shades) are the corresponding correlations for the “trimmed” euchromatic regions (see text and Table S15 and Figure S8).
For the entire (untrimmed) autosomal arms, TsD measured in small (50 adjacent segregating sites) nonoverlapping windows exhibits ≈20% correlation with both large-scale estimates of the rates of recombination. Such a strong association is not as evident on the X. The correlation of TsD with or is much weaker if the centromere- and telomere-proximal regions are trimmed (see the stippled columns of Figure 12 and Table S15). Because of the strong dependence of Tajima’s D on the sample size, the correlations in the MW sample are substantially weaker. Still all the chromosome arms exhibit a positive correlation of with TsD (untrimmed and trimmed). This, along with consistently higher overall correlations of TsD with than with , suggests that there may be considerably more information about genomically local variation in rates of recombination using and that this variation may influence patterns of polymorphisms via hitchhiking (Braverman et al. 1995) and perhaps even background selection (Charlesworth et al. 1993, 1995). Alternatively, recurrent linked selection may systematically bias LDhat estimation of 2Nr (McVean et al. 2004; Stephan et al. 2006)
If hitchhiking does indeed play a significant role in determining the pattern of polymorphism in D. melanogaster, variation in the rate of recombination could interact with the selection and demography. For example, if we assume that the establishment of the out-of-Africa diaspora involved a severe bottleneck and subsequent local adaptation, the Hill–Robertson effect may have led to more effective selection in regions of high recombination. On the other hand, strong selection for locally adaptive rare variants would yield a bigger impact in regions of low crossing over. While HBKl exhibits weak and inconsistent correlation with or in Table S15, it is noteworthy that the more fine-scale correlates consistently with increased evidence of SNP frequency differentiation between RAL and MW. This supports the view that contains additional information about local variation in recombination rates and interactions with natural selection. This evidence of a correlation between geographic differentiation and local variation in the rate of recombination is revisited in the gene-based analysis below. But it must be noted that this pattern of weak association of geographic differentiation with recombination contrasts starkly with the clear negative correlation observed by Keinan and Reich (2010) among continental samples of humans. The large differences between humans and D. melanogaster in the genomic scale and pattern of linkage disequilibrium and in ranges of recombination per base pair are no doubt relevant to this apparent contradiction and deserve deeper investigation.
πw, δw, HKAl, HBKl, and by chromatin states and gene elements
The structural and functional annotation of the Drosophila genome provides a basis for the inference of natural selection as a primary mechanism shaping genomic patterns of polymorphism and divergence. Here we consider the patterns of polymorphism and divergence in the context of the recent annotation of the D. melanogaster chromatin states by the modENCODE Project members (Kharchenko et al. 2010; Roy et al. 2010; Riddle et al. 2011). These states identify functionally relevant, well-established combinatorial patterns of histone modifications and chromatin proteins shared by diverse organisms, including the epigenetic signatures associated with active transcription start sites (state 1), transcriptional elongation (state 2), and Polycomb Group (PcG) regulation (state 6). States 3 and 4 are enriched in noncoding sequence in and around transcribed genes with long introns. State 3 is distinguished by its association with enhancers and potential role in gene regulation. State 4 is similarly enriched in the noncoding sequences of genes expressed at low levels. In addition, they identify chromatin domains with specific importance in D. melanogaster, such as the α- and β- heterochromatic domains flanking the centromeres (state 7) and state 8, a “heterochromatin-like”pattern that also occurs in apparently tissue-specific interstitial blocks. Regions of MSL-mediated dosage compensation on the X are referred to as state 5. State 9, which represents segments of the genome with no enrichment (of the 18 examined marks), covers approximately half the genomic sequence included in the analysis, including large intergenic stretches and many genes with very low expression.
Chromatin-mapping analysis was conducted in two male tissue-culture lines: S2 cells, isolated from embryos, and neuronally-derived BG3 cells. These array-based studies of cross-linked chromatin have a resolution determined both by the distributions of the sizes of DNA in the chromatin fragments used for the chromatin immunoprecipitation and variation in the observed intensities. Genome-wide intensity maps of the 18 histone marks were partitioned into 200-bp bins, and K-means clustering was applied to generate the nine-state model (Kharchenko et al. 2010). There are limitations to the utility of data derived from cell culture lines in the investigation of patterns of polymorphism and divergence. Cultured cells are somatic, whereas mutations leading to polymorphism arise in the germline. Cell lines can be distinct from the tissues from which they are derived, exhibiting varying levels of aneuploidy, copy-number variation, and altered transcriptional programs (Cherbas et al. 2011). These two lines share in common the expression of a large set of genes necessary for growth/proliferation and common to all dividing cell types (Cherbas et al. 2011). However, it has been observed that cultured cells often retain much of the expression profile of their progenitors (Cherbas et al. 2011). Further, many chromatin domains are very similar across cell types (Kharchenko et al. 2010). For example, heterochromatin, dosage compensation, and PcG-regulated regions are largely overlapping in the S2 and BG3 cell lines and presumably in the diverse cells of the animal including the meiotic germline. The chromatin state of different genomic regions is obviously correlated strongly with function and thus natural selection. But it is equally significant that the chromatin states may strongly influence the fidelity of DNA repair (Wellinger and Thoma 1997), recombination (Alexeev et al. 2003; Heyer 2007), and the distribution and pathways of meiotic recombination events (Wu and Lichten 1994; Fan and Petes 1996; Baudat et al. 2010; Berg et al. 2010; Myers et al. 2010; Pan et al. 2011) and are thus central to population mechanisms.
An important property of these new chromatin annotations is their relationship to the detailed gene annotation already available. While chromatin state shows clear correlation with the structure and expression patterns of individual genes, the overlap of chromatin and gene annotation is complex, consistent with the widely held view that chromatin properties add an important and fundamental dimension to genome function. These annotations of chromatin states assign functionally relevant information to many previously unannotated genomic regions (see below) and provide a new resource for population genomic inference.
As presented above, the most prominent large-scale genomic feature of the population statistics is the relative reduction in expected heterozygosity, πw , especially in the large centromere-proximal regions that parallel the reductions in levels of recombination per base pair [see Figure 7 and Figure S7 as well as the tracks pi_RAL and log(2Nr/100 bp) in Figure S9 and the corresponding UCSC genome browser at the track data hub page zoomed out to the entire chromosome arm]. Within those same regions are in fact the most obvious concentrations of chromatin state windows annotated with state 7 or state 8, bearing the histone marks associated with α- and β-heterochromatin. This strong association between levels of polymorphism and crossing over per physical length is attributed to the impact of linked strong selection and is also apparent in Figures 13 and 14, which show the box plots and empirical cumulative distribution functions (respectively) of HKAl and in windows of the nine states partitioned by coding, intronic, and intergenic. Clearly genomic segments annotated as state 7 in S2 cells are highly enriched for low values of HKAl and . State 8, which is not limited to the regions flanking centromeres and differs by tissue, does not exhibit such clear enrichment. The full sets of such plots (box plots and ecdfs based on the inferred states for S2 cells) for πw (RAL, MW, and SIM), δw (RAL, MW, and SIM), HKAl (RAL, MW, and SIM), Dw (RAL), HBKl (RAL ↔ MW), and (RAL) are available in Figure S11 and Figure S12.
Figure 13 .
The distribution of (“untrimmed”) HKAl values in windows partitioned by chromatin state (inferred from S2 cells) and gene structure (coding, intron, and intergenic). The top row shows box plots (boxes are the central two quartiles, the whiskers are 1.5 times those, and the dots with light shading represent the outliers beyond the whiskers). The empirical cumulative distribution functions (ecdfs) in the bottom row give a perhaps clearer alternative view of the differences in the distributions of HKAl between chromatin states in the coding, intronic, and intergenic regions. Also see Figure S11 and Figure S12 for the box plots and ecdfs (respectively) for πw (RAL, MW, and SIM), δw (RAL, MW, and SIM), HKAl (RAL, MW, and SIM), HBKl, and . “all” in the box plots and “0” in the ecdfs refer to the sum of the chromatin states, 1–9.
Figure 14 .
The distribution of -values in windows partitioned by chromatin state inferred in S2 cells and gene structure (coding, intron, and intergenic). The top row shows box plots (boxes are the central two quartiles, the whiskers are 1.5 times those, and the dots with light shading represent the outliers beyond the whiskers). The empirical cumulative distribution functions (ecdfs) in the bottom row give a perhaps clearer alternative view of the differences in the distributions of between chromatin states in the coding, intronic, and intergenic regions. Also see Figure S11 and Figure S12 for the box plots and ecdfs (respectively) for πw (RAL, MW, and SIM), δw (RAL, MW, and SIM), HKAl (RAL, MW, and SIM), HBKl, and . “all” in the box plots and “0” in the ecdfs refer to the sum of the chromatin states, 1–9.
The molecular and evolutionary mechanism(s) responsible for the strong suppression of meiotic crossing over in the centromere- and telomere-proximal regions remains unclear (Charlesworth et al. 1986; Westphal and Reuter 2002 and references therein; Chiolo et al. 2011). Nevertheless the associations of the density of repetitive sequences with reduced crossing over and of these with characteristic heterochromatic histone marks are clear here and are commonly observed in other species. An interesting observation in this respect is the apparent association of large β-heterochromatic regions only with the most extreme reductions in recombination. In particular, neither chrX nor chr3L have large β-heterochromatic blocks proximal to their centromeres in the “arm” assemblies (distinct from the adjacent “Het” assemblies). The maps in Figure 3 of show evidence of recombination in the centromere-proximal regions on these two arms, even though the remaining three arms have no evidence of recombination in the corresponding β-heterochromatic regions in the arm assemblies. It is also worth noting that while the suppression of recombination proximal to the telomeres is comparably strong (see Figure 3), it extends over much smaller regions; state 7 (shown in Figure S9) is not obviously concentrated in these regions of suppressed crossing over. This discordance might be attributable to a difference in the distribution of chromatin state 7 during female meiosis.
Similar to state 7 but on a finer scale is state 1, which shows increased relative δw, decreased relative πw, consequent negative HKAl, and local decreases in . This elevated divergence (see Figure S11 and Figure S12) can be parsimoniously attributed to either increased mutation or more positive directional selection in these regions. Assays of chromatin openness (nuclease sensitivity) indicate that state 1 (and state 3) DNA is more exposed (Kharchenko et al. 2010) and thus potentially more accessible to DNA-damaging agents. Contradicting this attractive interpretation of the increased divergence is the reduced relative πw and consequent strongly negative distribution of HKAl in state 1 (Figure 13). Recurrent hitchhiking would yield a local depletion in polymorphism and a skew in the site-frequency spectrum that is evident in the TsD (labeled “D_w”) in the autosomal panels of Figure S11 and Figure S12. Further evidence in support of this explanation can be found in the skew toward high HBKl windows overlapping with states 1 (and 2) on the autosomes. States 1 and 2 are enriched over 5′ regions of actively transcribed genes, many of which are essential housekeeping genes with broad developmental expression (Kharchenko et al. 2010; Cherbas et al. 2011). These intergenic and intronic regions of elevated HBKl include core promoters and other regulatory elements of these genes. Thus strong directional selection for adaptive regulatory variants could account for these state 1- and 2-associated patterns in HKAl and HBKl albeit on potentially quite different timescales. Consistent with this hypothesis of increased hitchhiking in the state 1 regions is the relative reduction in , since the genomic footprint of hitchhiking scales with the reciprocal of recombination rate. While McVean et al. (2004) failed to find evidence that the LDhat estimator itself is biased in a particular parameter range of a recurrent hitchhiking model, it remains possible that the contraction and distortion of the genomic genealogies in these regions lead not only to fewer recombination events but also to a biased estimate. In any case, these state 1-associated coldspots require further experimental and theoretical investigation.
Approximately 5% of the Drosophila genome is in state 6. These blocks are enriched for the trimethylation of H3K27 and are bound by proteins from the Polycomb Group. Most significantly the gene content of these special regions of animals and plants is largely composed of fundamental developmental regulators, usually transcription factors. These often exhibit low and exquisitely determined, cell-specific expression affecting developmental fate. The cumulative distributions (Figure 13, bottom) show that polymorphism and divergence in state 6 regions are highly consistent (also see Figure S11 and Figure S12). It is difficult to even speculate about the possible genetic and population genetic mechanisms that would predict this rather conservative pattern other than the narrowest application of the neutral theory (see discussion in Ohta 1992 and references therein). Perhaps a solid clue, consistent with the hitchhiking hypothesis, for states 1 and 7 is the fact that SNPs in state 6 exhibit markedly less differentiation between the RAL and MW samples (see HBKl panels of both autosomes and X in Figure S11 and Figure S12). If temporal and spatial changes in specific environmental factors drive the directional selection shaping genomic polymorphism and divergence, the PCG-regulated genes, because of their deep positions in the developmental and physiological pathways, may be most sheltered and thus show less evolutionary response (Schwartz and Pirrotta 2008 and references therein). Interestingly, state 9 regions also exhibit relatively higher levels of polymorphism and estimated recombination (see Figures 13 and 14, Figure S11, and Figure S12). These regions include tissue-specific and inducible genes, as well as noncoding sequence outside of active gene clusters and PcG domains.
Gene-based analyses
Gene heterozygosity and divergence:
Gene-based analyses were focused on protein-coding regions. We used all sites exceeding Q30 in this analysis. All major patterns in Q30 data were also observed at the more stringent Q40 quality threshold. We restricted our analysis to genes for which all alleles matched the gene model annotated in reference D. melanogaster version 5.16 (see Materials and Methods). This yielded a list of 9328 genes with Q30 data, which is referred to as “golden genes” in the following analysis. Analysis of a subset of genes excluded due to variation in stop codon position is presented in Lee and Reinhardt (2012).
Expected heterozygosity was estimated for nonsynonymous and synonymous sites from the MW and RAL samples of D. melanogaster and from D. simulans (Begun et al. 2007). Results are shown in Table 7, and all comparisons between populations, between species, and between synonymous vs. nonsynonymous sites are highly significant by Mann–Whitney U-tests (UMW, P < 10−16). These results support the well-known patterns that African populations of D. melanogaster are more variable than U.S. populations (Begun and Aquadro 1993; Caracristi and Schlötterer 2003; Haddrill et al. 2005), that D. simulans is more variable than D. melanogaster (Aquadro et al. 1988; Andolfatto 2001; Andolfatto et al. 2011; but see Nolte and Schlötterer 2008), and that synonymous variation is at least an order of magnitude greater than nonsynonymous variation (Kreitman 1983; Begun et al. 2007; Sackton et al. 2009).
Table 7 . Nonsynonymous and synonymous polymorphism estimates for North American (RAL), African (MW), and D. simulans (SIM) samples.
| Nonsynonymous | Synonymous | |||||
|---|---|---|---|---|---|---|
| All | Autosomal | X-linked | All | Autosomal | X-linked | |
| MW | 0.0015 | 0.0015 | 0.0013 | 0.019 | 0.019 | 0.021 |
| RAL | 0.0012 | 0.0012 | 0.0007 | 0.013 | 0.014 | 0.009 |
| SIM | 0.0023 | 0.0024 | 0.0017 | 0.033 | 0.035 | 0.020 |
Nonsynonymous π and synonymous π were highly correlated, ρS = 0.28 (MW), ρS = 0.31 (RAL), and ρS = 0.30 (D. simulans) (for each, P < 10−16), consistent with the larger-than-gene size scale of variance in heterozygosity along chromosome arms. π was also significantly correlated between species, ρS = 0.55 (nonsynonymous, P < 10−16) and ρS = 0.37 (synonymous; P < 10−16). As lineage-specific divergences are both correlated between species and between nonsynonymous and synonymous estimates, we repeated the analysis using divergence-adjusted polymorphism (polymorphism estimates divided by maximum-likelihood estimated lineage-specific divergence; see Materials and Methods). We observed similar levels of correlations. If the variation across genes in divergence-adjusted standing nonsynonymous π reflects patterns of average deleterious selections while that for synonymous π is purely neutral (Ohta 1992), such a correlation might arise, although its expected magnitude is unclear. Alternatively if the variation in divergence-adjusted synonymous π reflects the impact of recent lineage-specific linked directional selection and the variation in nonsynonymous π is due to random environment (or other balancing) selection (Gillespie 1994), the observed difference would also be expected. This striking result deserves further investigation.
Consistent with previous reports (Begun 1996; Andolfatto 2001), the ratio of nonsynonymous to synonymous π is higher for autosomal genes than for X-linked genes in the African D. melanogaster sample (UMW, P < 10−8) but not statistically different in D. simulans. Comparing the ratios of nonsynonymous to synonymous π between species, D. melanogaster showed a higher value for the autosomes (UMW, P < 10−16) but not for the X chromosome (P > 0.05). This discrepancy of between-species differences for autosomes and X chromosomes could be attributed to the combined effects of assumed smaller effective population size of D. melanogaster and hemizygosity of the X chromosome (McVean and Charlesworth 1999; Andolfatto 2001) and/or a greater impact of linked selection on D. melanogaster autosomes due to the suppression of crossing over associated with polymorphic autosomal inversions (Begun 1996). However, analyses contrasting autosomal genes within 100 kbp distal and proximal to the inversion breakpoints to other autosomal genes did not reveal significant differences in the ratio of nonsynonymous to synonymous π (UMW > 0.05). The small number of genes in these inversion-breakpoint regions no doubt reduces the statistical power to detect such an effect. Note also that the young age of these polymorphic inversions (Wesley and Eanes 1994; Andolfatto and Kreitman 2000) severely restricts the possible scenarios under which the impact of linked selection might greatly reduce standing polymorphism.
In Table 8 are estimates of “polymorphism-adjusted” synonymous (dS) and nonsynonymous divergence (dN) (taking into account intraspecific variation; see Materials and Methods), showing strong correlations between dN and dS ρS = 0.11 (D. melanogaster) and ρS = 0.28 (D. simulans) (for both, P < 10−16), and between estimates on the D. melanogaster and D. simulans lineages, ρS = 0.62 (dN) and 0.10 (dS) (for both, P < 10−16). The correlation between the two lineages is stronger for dN than for dS even though correlation with dN is expected to have lower statistical power given its much smaller value than dS. In this context, the systematic divergence in codon bias on the two lineages is relevant; in particular, codon bias on the melanogaster lineage is by several measures weaker than on the simulans lineage (Akashi 1995). The dN/dS ratio, which is used to detect accelerated rates of amino acid replacement and can be used as an index for adaptive protein evolution, is also strongly correlated between D. melanogaster and D. simulans (ρS = 0.45, P < 10−16). Maximum-likelihood estimates of dN/dS have large uncertainty when synonymous divergence is low. Restricting the analysis to genes with dS estimates >0.005 on both the D. melanogaster and D. simulans lineages, we observed an even stronger correlation in relative rates of protein evolution between these two species (ρS = 0.55, P < 10−16), implying similar selective pressures between species in terms of constraint and/or directional selection. Even though previous studies pointed to the higher levels of polymorphism in suggesting that the effective population size of D. simulans may be greater than that of D. melanogaster (Aquadro et al. 1988; Andolfatto 2001; Eyre-Walker et al. 2002), in a recent study Andolfatto et al. (2011), using X-linked genes, concluded that rates of adaptive evolution on the two lineages do not differ. Our dN/dS estimates were greater on the D. simulans lineage than on the D. melanogaster lineage (Wilcoxon’s paired rank test, P < 10−11 for the three comparisons, all genes, X-linked genes, or autosomal genes). This apparently higher rate of protein evolution on the D. simulans lineage is consistent with the hypothesis of a larger effective population size. Our investigation using the McDonald–Kreitman test to detect adaptive protein evolution (see below) reached the same conclusion that there has been more adaptive evolution on the simulans lineage.
Table 8 . Maximum-likelihood estimates of divergence excluding within-species polymorphism on D. melanogaster and D. simulans lineages (see text).
| dN | dS | |||||
|---|---|---|---|---|---|---|
| Lineage | All | Autosomal | X-linked | All | Autosomal | X-linked |
| melanogaster | 0.0056 | 0.0055 | 0.0064 | 0.059 | 0.059 | 0.065 |
| simulans | 0.0051 | 0.0049 | 0.0063 | 0.037 | 0.036 | 0.041 |
The connection between base composition and synonymous divergence or heterozygosity in Drosophila has been extensively investigated (Sharp and Li 1989; Moriyama and Hartl 1993; Moriyama and Powell 1996). Those studies interpreted their analyses as consistent with a mutation–selection–drift model of codon bias (Bulmer 1991), in which genes showing more codon usage bias are more functionally constrained at synonymous sites, although the effect must be weak, 2Ns of order ≈1. Our observations with D. simulans data support this idea, as there are significant negative correlations between GC content and synonymous π (partial correlation controlling for lineage-specific dS which is correlated with both variables; ρS = −0.14, P < 10−16). However, we observed the opposite pattern in D. melanogaster. There were slight but significant positive correlations between GC content and synonymous π (partial correlation controlling for dS ρS = 0.07 and P < 10−10 for MW; ρS = 0.029 and P = 0.005 for RAL). One interpretation of this difference is that the selection on codon bias is weaker or even absent in D. melanogaster (Akashi 1995, 1996; McVean and Charlesworth 1999; Nielsen et al. 2007). Alternatively, codon preference may have shifted in particular away from strict GC bias in the D. melanogaster lineage.
The analyses of polymorphism and divergence across the genome in windows presented above confirm the emerging picture in Drosophila that polymorphism across chromosome arms is correlated with variation in crossing over per base pair. The widely acknowledged interpretation of this pattern is that the dynamic interactions between rare but strongly selected variants and closely linked, selectively neutral polymorphisms lead to a relative reduction in the standing levels of this latter category. But the relative contributions of linked, strongly selective adaptive substitutions (the hitchhiking effect) and the linked selective effect of the much more common deleterious mutations (background selection) to the levels of selectively neutral polymorphism remain unclear. Among the various potential approaches available to address this issue is the comparison of synonymous and nonsynonymous polymorphism and divergence. Clearly, the substantial difference in the average effects of newly arising synonymous and nonsynonymous mutations could provide a gauge for the relative impacts of linked selection. We first investigated the association of synonymous and nonsynonymous π with variation in the estimated rate of crossing over per base pair, M/bp (see Materials and Methods). The apparent differences in the pattern and impact of crossing over on the X led us to consider X-linked and autosomal genes separately. Consistent with the overall genomic analyses (above) and previous reports (Begun and Aquadro 1992; Ometto et al. 2005; Presgraves 2005), we observed significant correlations between the M/bp rate and both πS and πN. The correlations for synonymous π [MW, ρS = 0.27 (X chromosome, X) and 0.45 (autosomes, A); RAL, ρS = 0.18 (X) and 0.41 (A); simulans, 0.25 (X) and 0.11 (A), each P < 10−8] were of a similar magnitude to those observed for the correlation of HKAl and 2Nr (above) and uniformly greater than for nonsynonymous π [MW, ρS = 0.08 (X) and 0.13 (A); RAL, ρS = 0.11 (A); each P < 0.01 except for RAL X and D. simulans, which were nonsignificant]. The stronger correlations for πS point to the substantial impacts of linked selection on these presumably more weakly selected variants, while the weaker correlations for πN could reflect mildly deleterious nonsynonymous variants reaching higher frequencies in regions of low recombination. Alternatively, the lower nonsynonymous polymorphism and thus lower statistical power may have contributed to the pattern. Linear regression analysis (see Materials and Methods) suggested that a twofold increase of recombination rate (M/bp) at the average autosomal locus yields a 38% increase in MW πS [regression coefficients β = 1.98 × 105 (MW X), 2.71 × 105 (MW A), 7.49 × 104 (RAL X), 1.86 × 105 (RAL A), 1.94 × 105 (SIM X), and 9.15 × 104 (SIM A); permutation-based P < 0.001 for all]. Obviously, many other factors (such as average mutation rate and average functional constraint) could create substantial variation among genes in the rates of synonymous and nonsynonymous divergence as well as π. We also adopted the convenient normalization for each gene of dividing π by the lineage-specific divergence and found similar observations (not shown).
Previous studies suggested that the influence of linked selection may be most prominent in genomic regions with strongly suppressed crossing over, e.g., near centromeres and telomeres and on the neo-Y (Aguadé et al. 1989, 1994; Stephan and Langley 1989; Begun and Aquadro 1992; Langley et al. 1993, 2000; Bachtrog and Charlesworth 2002; Betancourt and Presgraves 2002; Bachtrog 2003; Braverman et al. 2005; Presgraves 2005; Begun et al. 2007; Haddrill et al. 2007), which might be the major factor driving our observations. To investigate such an effect, we restricted the analysis to genes with recombination rates above the 25th percentile [2.98 × 10−8 M/bp (X) and 1.07 × 10−8 M/bp (A), corresponding to removal of genes in the very low-recombination bin in the below analysis]. We observed weaker, yet significant correlations between πS and M/bp for D. melanogaster autosomal genes and D. simulans X-linked genes [MW, ρS = 0.26 (A); RAL, ρS = 0.24 (A); simulans, ρS = 0.13 (X); each P < 10−4], supporting the idea that recombination rate variation also has an appreciable effect on genes in genomic regions with normal rates of crossing over. For D. simulans autosomal and D. melanogaster X-linked genes, the significant correlations seem to be driven by genes with exceptionally low recombination rates. The discrepancy between D. melanogaster X-linked and autosomal genes may be attributable to the overall high-recombination environment on the X, within which the effects of crossing over level off (see Figure S13). An opposite pattern observed in D. simulans might be attributed to the fact that these recombination rates were estimated in D. melanogaster, and D. simulans may lack the degree of suppression of crossing over in the centromere-proximal regions (True et al. 1996).
Strong selection can quantitatively impede the selection of more weakly selected variants at linked sites (Hill and Robertson 1966). The difference in the impact of such linked selection on synonymous and nonsynonymous polymorphisms may be reduced in regions of lower crossing over per base pair, thus creating an opportunity for quantitative investigation of the stochastic impact of linked selection in the population genomic dynamics of D. melanogaster. The ratio of nonsynonymous to synonymous π can be interpreted as an indication of the effectiveness of purifying selection at removing slightly deleterious amino acid mutations with the assumption of no fitness impacts of synonymous variants. A higher value suggests weaker effectiveness of purifying selection. We found such a ratio is significantly correlated with recombination rate (M/bp) in all comparisons [MW, ρS = −0.077 (X) and −0.12 (A); RAL, ρS = −0.12 (X) and −0.18 (A); simulans, ρS = −0.062 (X) and −0.033 (A); each P < 10−16 except simulans X (P = 0.03)]. Excluding genes with low recombination rates (below the 25th percentile) yielded a similar significant result for D. melanogaster autosomal comparison [MW, ρS = −0.076 (A); RAL, ρS = −0.096 (A); both P < 10−16].
As shown in previous studies (Begun et al. 2007; Shapiro et al. 2007) and below (see next section), directional selection is a significant factor in protein sequence divergence in D. melanogaster and D. simulans. One therefore expects a negative correlation between nonsynonymous divergence and synonymous polymorphism (Maynard Smith and Haigh 1974; Kaplan et al. 1989; Andolfatto 2007; Begun et al. 2007; Sattath et al. 2011). We estimated partial correlations between dN and synonymous π by controlling for mutation rate (using dS on the D. melanogaster or D. simulans lineage as a proxy) and found weak but highly significant correlations for autosomal genes [ρS = −0.081 (MW), −0.088 (RAL), −0.43 (SIM); P < 10−3]. The reduction in statistical power associated with the relatively small number of X-linked genes and the reduction in overall level of polymorphism, especially in RAL, may result in the absence of such patterns on the X. The effect of hitchhiking also depends on the recombination environment around selected sites. We investigated the effects of recombination by categorizing genes into four recombination categories with the same number of genes according to M/bp, very low, low, intermediate, and high for X-linked and autosomal genes separately (see Materials and Methods and Figure S13), and found a slightly stronger negative correlation between dN and synonymous π for the very low-recombination category [MW, ρS = −0.099 (very low), −0.048 (low), −0.090 (intermediate), and −0.071 (high); RAL, ρS = −0.11 (very low), −0.044 (low), −0.11 (intermediate), and −0.075 (high); each P < 0.05]. Our observations of significant correlations within the other three autosomal recombination categories suggest that this hitchhiking effect associated with amino acid substitutions extends to all recombination environments and that at least a subset of the associated selection coefficients is of the magnitude of the map size of a typical gene (Kern et al. 2002). But note that selection coefficients driving recent hitchhiking events that are much larger than the map size of a gene will actually weaken this correlation.
Adaptive protein divergence:
We used contrasts of polymorphic and fixed synonymous and nonsynonymous variants to investigate recurrent adaptive protein divergence (McDonald and Kreitman 1991). As we wish to infer such divergence specifically in D. melanogaster, we carried out polarized tests, using parsimony to infer the variants that fixed in D. melanogaster since the split from D. simulans. We used all sites exceeding Q30 in this analysis, as all major patterns in Q30 data were also observed at the more stringent quality threshold Q40. Only genes with sufficient variation (expected value for each cell of the 2 × 2 table greater than one) were included in this analysis.
The source population of the MW sample is less likely (than that of the RAL sample) to have been perturbed by the strong selection and demographic phenomena associated with recent colonization of temperate environments. We thus focused on the MW sample. However, we also obtained MK results for the RAL sample (File S1), to include genes that lacked sufficient coverage in the MW sample (see Materials and Methods for thresholds). Of the 4774 genes that meet filtering criteria in the MW sample, 7.73% are significantly heterogeneous in the 2 × 2 contingency table at the critical value of P < 0.05. Under the premise that synonymous sites experience considerably weaker selection than nonsynonymous sites, we can determine whether the significant genes reject the null hypothesis as a result of excess amino acid divergence: 66.4% of significant genes reject in this direction. This is highly unusual (P < 0.001, by simulation with the observed marginal counts), suggesting an important role for recurrent directional selection on proteins in this species. The 20 most significant genes in D. melanogaster showing excess protein divergence are shown in Table 9; named genes in this list of 20 genes are Trc8 (negative regulation of growth), apt (organ and neuromuscular system development), Sk1 (phosphorylation), and inx7 (gap junction channel). Similar analyses using D. simulans data had more genes (n = 6011) passing filtering criteria, which can be explained by the higher level of polymorphism and thus statistical power in D. simulans. Of these, 10.65% of genes rejected the null hypothesis (at P < 0.05) and a majority of these significant genes (96.21%) exhibited an excess of amino acid fixation.
Table 9 . The top 20 genes with most significant evidence of adaptive protein evolution (significant MK tests and excess of nonsynonymous fixations).
| FlyBase ID | Symbol | Chr | P-value |
|---|---|---|---|
| FBgn0032136 | CG15828 | 2L | 1.57 × 10−09 |
| FBgn0029697 | CG15570 | X | 2.56 × 10−09 |
| FBgn0031078 | CG11943 | X | 2.89 × 10−06 |
| FBgn0030594 | CG9509 | X | 7.12 × 10−06 |
| FBgn0031868 | Rat1 | 2L | 1.12 × 10−05 |
| FBgn0028887 | CG3491 | 2L | 1.20 × 10−05 |
| FBgn0039207 | CG5789 | 3R | 1.37 × 10−05 |
| FBgn0039668 | Trc8 | 3R | 2.59 × 10−05 |
| FBgn0015903 | apt | 2R | 4.25 × 10−05 |
| FBgn0030320 | CG2247 | X | 8.15 × 10−05 |
| FBgn0030091 | CG7065 | X | 2.62 × 10−04 |
| FBgn0030300 | Sk1 | X | 3.08 × 10−04 |
| FBgn0259168 | mnb | X | 3.31 × 10−04 |
| FBgn0005617 | msl-1 | 2L | 3.51 × 10−04 |
| FBgn0030504 | CG2691 | X | 4.18 × 10−04 |
| FBgn0035657 | CG10478 | 3L | 4.91 × 10−04 |
| FBgn0052654 | Sec16 | X | 4.97 × 10−04 |
| FBgn0027106 | inx7 | X | 5.24 × 10−04 |
| FBgn0033955 | CG12866 | 2R | 6.36 × 10−04 |
| FBgn0031377 | CG15356 | 2L | 6.56 × 10−04 |
An excess of low-frequency slightly deleterious amino acid polymorphisms can lead to overly conservative conclusions regarding the prevalence of adaptive protein divergence using the McDonald–Kreitman framework (Fay et al. 2001; Charlesworth and Eyre-Walker 2008). To address this issue, we analyzed the data after removing all singleton variants. In this reduced data set (n = 2428 MW genes), 5.3% of the tests had P < 0.05. This is approximately the number expected under the null hypothesis. However, of these 5.3%, almost 83% reject the null hypothesis in the direction of excess protein divergence, which is highly unusual (P < 0.001, by simulation with the observed marginal counts), again pointing to an important role of directional selection on protein sequences.
Several models of natural selection predict an excess of adaptive divergence on the X chromosome (Charlesworth et al. 1987). Enrichment of significant tests (P < 0.05) on the X chromosome in the MW sample for the entire data set was not observed. However, among the top 1% (n = 477) of genes with the smallest MK test P-value (singletons included), there was an enrichment of X-linked genes (FET, P < 0.003), suggesting a chromosomal influence on the most rapidly adapting protein-coding regions. If most of the slightly deleterious amino acid variants are partially recessive, there should be fewer such polymorphisms segregating on the X chromosome because of its hemizygosity. Indeed, we observed the proportion of significant MK tests having excess amino acid replacement is higher on the X chromosome than on autosomes (FET, P < 0.0001). Restricting our comparison to significant MK tests with excess nonsynonymous fixations, we again observe X-linked enrichment of genes under adaptive protein evolution (FET, P < 0.03).
Genes experiencing recurrent directional selection but that retain sufficient polymorphism for carrying out MK tests are expected to be biased toward smaller selection coefficients of beneficial mutations. If most new beneficial mutations are weakly selected, a greater proportion of such new mutations may fix in regions of higher recombination (Hill and Robertson 1966), in which average linkage to other selected variants is reduced. There was no significant correlation observed between MK P-values and recombination rates. However, as mentioned above, a gene can have an excess of either nonsynonymous substitutions or nonsynonymous polymorphisms. Indeed, we observed significant negative correlations between recombination rates and MK P-values of genes with excess nonsynonymous fixations (ρS = −0.10, P < 10−7) and positive correlations for genes with excess nonsynonymous polymorphisms (ρS = 0.15, P < 10−10), consistent with the idea that selection is facilitated by recombination. To further investigate the effect of recombination on adaptive protein evolution, we categorized genes according to recombination category using the above methods and compared the proportion of genes with significant MK tests in each recombination category. While the proportion of autosomal genes with significant MK tests is not statistically different between recombination categories, the proportion of genes with significant MK tests due to excess of amino acid divergence was strongly influenced by recombination rate (Table 10). These results are consistent with previous reports suggesting recombination facilitates the spread of weakly selected favorable alleles (Betancourt and Presgraves 2002; Presgraves 2005). Previous investigations of the impact of the recombination environment on MK tests focused primarily on the contrast of genes in regions with no evidence of meiotic exchange with those in the remainder of the genome with normal levels of crossing over (Bachtrog and Charlesworth 2002; Bachtrog 2003, 2005; Haddrill et al. 2007; Betancourt et al. 2009). Even with the genes in the very low-recombination category excluded, there remains a positive relationship over the remaining three categories with the proportion of MK tests exhibiting a significant excess of nonsynonymous fixations (χ2-test, P < 0.01). These results suggest that not only the “presence” but also the extent of crossing over increases the effectiveness of selection for advantageous amino acid variants. Parallel analysis with X-linked genes did not show any significant heterogeneity for these comparisons, potentially due to many fewer genes in each recombination category (∼300) and thus lower statistical power.
Using the related approach of Smith and Eyre-Walker (2002), to obtain an estimate for the proportion of adaptive amino acid substitutions, α, for each gene, the median values on the autosomes and the X chromosome were 0.128 and 0.463, respectively. α exhibited a positive correlation with the recombination rate, for autosomal genes [ρS = 0.14 (all genes) and 0.10 (genes with very low-recombination regions removed); P < 10−9]. Parallel analysis with X-linked genes found similar, though statistically nonsignificant, trends.
Turning for a moment to the differences between melanogaster and simulans in the amount of centromere-proximal suppression of crossing over mentioned above (True et al. 1996), Figure 15 (top) shows that the difference in πs between the two species is significantly greater in centromere-proximal regions (Wilcoxon’s rank test, P < 10−16). The between-species differences in the estimated proportion of adaptive amino acid fixations (α) are also greater for genes in regions with centromeric suppression (Wilcoxon’s rank test, P < 10−9; see Figure 15, bottom). D. melanogaster genes located in genomic regions of centromeric suppression show greater effects of linked selection and may contribute disproportionately to the between-species differences in overall levels of polymorphism.
Figure 15 .
Comparisons of the levels of synonymous polymorphism (πs) and the proportions of amino acid fixations driven by positive selection (α) in centromere-proximal and distal regions of the autosomes of D. melanogaster (shown in olive) and D. simulans (shown in orange). Using the -smoothed map of the recombination rate, the boundaries between the centromere-proximal and distal regions were chosen to be the first interval where > 1.8 × 10−8 M/bp, 4,714,046 for 2L, 9,716,832 for 2R, 2,350,586 for 3L, and 13,015,705 for 3R.
To investigate general biological patterns associated with MK tests showing a significant excess of amino acid fixations, we used GO enrichment analysis. Retaining only GO categories that contained at least five genes with MK tests, we obtained P-values by permutation. Table 11 shows those biological process terms most strongly enriched for significant MK tests in the MW sample, including cystoblast division, ubiquitin moieties addition, sodium ion transport, male meiosis, protein import into nucleus, chromatin organization, and downregulation of translation. Consistent with previous findings, GO categories associated with reproduction [male meiosis, spermatogenesis, spermatid development, oocyte fate determination, and oogenesis (Swanson et al. 2001, 2004)] and stem cell maintenance [germ cell development, germ cell fate determination, and germline stem cell self-renewal (Bauer Dumont et al. 2007)] include a large number of genes showing adaptive protein evolution. Several GO terms related to neural and neuromuscular development (neural muscular synaptic transmission, regulation of synaptic growth at neuromuscular junctions, axon genesis, and axon guidance) are also enriched for genes with significant MK tests. Significant molecular function terms in Table 11 included adenylate cyclase activity, sodium ion channel activity, lipid binding, nucleotide binding, translation repression, and calmodulin binding. Significant cellular location terms (see Table 11) included chromatin, complex for import/export of cells, peroxisomal membrane, proteasome complex, spindle, mitochondrial envelope, neural–neural or neural–muscular junctions, and nuclear pore.
Table 11 . The top 10 biological, molecular, and cellular GO categories enriched with genes having evidence of adaptive protein evolution.
| GO category | Proportion of significant genes | P-value | Description |
|---|---|---|---|
| Biological | |||
| GO:0007282 | 0.600 | 1.00E-04 | Cystoblast division |
| GO:0016567 | 0.600 | 1.00E-04 | Ubiquitin moieties addition |
| GO:0006814 | 0.231 | 4.00E-04 | Sodium ion transport |
| GO:0007140 | 0.300 | 7.00E-04 | Male meiosis |
| GO:0007281 | 0.300 | 1.40E-03 | Germ cell development |
| GO:0007274 | 0.333 | 2.40E-03 | Neuromuscular synaptic transmission |
| GO:0007283 | 0.211 | 3.40E-03 | Spermatogenesis |
| GO:0007294 | 0.286 | 3.40E-03 | Germarium-derived oocyte fate determination |
| GO:0006606 | 0.286 | 3.70E-03 | Protein import into nucleus |
| GO:0051056 | 0.286 | 4.10E-03 | Regulation of GTPase-mediated signal transduction. |
| Molecular | |||
| GO:0004016 | 0.500 | 1.00E-04 | Adenylate cyclase activity |
| GO:0005272 | 0.294 | 1.00E-04 | Sodium channel activity |
| GO:0008289 | 0.267 | 6.00E-04 | Lipid binding |
| GO:0000166 | 0.175 | 1.60E-03 | Nucleotide binding |
| GO:0000900 | 0.400 | 1.60E-03 | Translation repressor activity, nucleic acid binding |
| GO:0005516 | 0.286 | 3.50E-03 | Calmodulin binding |
| GO:0015450 | 0.286 | 4.20E-03 | Active carrier-mediated of protein transportation across a membrane |
| GO:0004842 | 0.182 | 5.20E-03 | Ubiquitin-protein ligase activity |
| GO:0016887 | 0.200 | 6.10E-03 | ATP hydrolysis |
| GO:0003729 | 0.136 | 8.30E-03 | mRNA binding |
| Cellular | |||
| GO:0000785 | 0.200 | 7.00E-03 | Chromatin |
| GO:0043190 | 0.167 | 1.94E-02 | ATP-binding cassette (ABC) transporter complex |
| GO:0005778 | 0.200 | 2.26E-02 | Peroxisomal membrane |
| GO:0000502 | 0.200 | 2.65E-02 | Proteasome complex |
| GO:0005819 | 0.167 | 3.25E-02 | Spindle |
| GO:0005740 | 0.143 | 3.33E-02 | Mitochondrial envelope |
| GO:0005741 | 0.167 | 3.51E-02 | Mitochondrial outer membrane |
| GO:0016020 | 0.077 | 3.55E-02 | Lipid bilayer |
| GO:0045202 | 0.167 | 3.60E-02 | Neural–neural or neural–muscular junctions |
| GO:0005643 | 0.133 | 3.91E-02 | Nuclear pore |
A comparable GO enrichment analysis of D. simulans showed some interesting overlaps. Due to the fact that many genes have an associated MK test in only one of the species, we restricted the analysis to 2467 genes with both D. melanogaster and D. simulans MK test results (see Table 12). Overlapping significant terms included germ cell development, male meiosis, spermatogenesis, ovum development, and microtubule-based movement (biological); microtubule motor activity, lipid binding, and chromatin binding (molecular); and chromatin and spindle (cellular). Among these genes, 6.1% (n = 150) and 6.6% (n = 163) showed evidence of adaptive protein evolution (with excess of amino acid replacement) on the D. melanogaster and D. simulans lineages, respectively. There were 126 genes significant in both species (MK P < 0.05 and an excess of nonsynonymous fixations), which is significantly greater than the expected number of overlapping genes [0.061(0.066)(2467) = 10; χ2-test, P < 10−16]. There is also a slight but significantly positive correlation in species MK P-values (ρS = 0.06, P < 0.003). Restricting analysis to genes with an excess of amino acid fixations showed even stronger correlations of MK P-values (ρS = 0.13, P < 10−4). Consistent with results based on windowing analysis (see below), we observed evidence supporting the idea that certain biological functions are likely under persistent directional selection in multiple linages.
Table 12 . The top biological, molecular, and cellular GO categories enriched with genes having evidence of adaptive protein evolution in both D. melanogaster and D. simulans.
| GO category | Description |
|---|---|
| Biological | |
| GO:0007140 | Male meiosis |
| GO:0007281 | Germ cell development |
| GO:0007283 | Spermatogenesis |
| GO:0009790 | Embryonic development |
| GO:0048477 | Ovum development |
| GO:0007018 | Microtubule-based movement |
| GO:0030097 | Hemopoiesis |
| GO:0006333 | Chromatin assembly or disassembly |
| GO:0009190 | Cyclic nucleotide biosynthetic process |
| GO:0035071 | Salivary gland cell death |
| GO:0006511 | Ubiquitin-dependent protein catabolic process |
| Cellular | |
| GO:0000785 | Chromatin |
| GO:0043190 | ATP-binding cassette (AB) transporter complex |
| GO:0005819 | Spindle |
| Molecular | |
| GO:0003777 | Microtubule motor activity |
| GO:0008289 | Lipid binding |
| GO:0042626 | ATP hydrolysis |
| GO:0003682 | Chromatin binding |
| GO:0003774 | Motor activity |
Genetic differentiation between temperate and tropical D. melanogaster populations:
The observations of differences between MW and RAL D. melanogaster populations can be attributed to demographic changes associated with the expansion out of Africa. Alternatively, the colonization of the new temperate habitat by the RAL population could have led to strong selective forces and result in a greater extent of selection in the RAL population. Consistent with the window-based analysis (see below), a large proportion of genes have significant amino acid FST; 46% of the genes have permutation-based FST P-values <0.05. X-linked genes tend to be more strongly differentiated than autosomal genes (UMW test, P = 10−16 for FST value and P < 10−16 for P-values associated with FST). It is worth noting that there is a strong correlation between amino acid FST and nonsynonymous π, presumably due to the fact that genes with higher polymorphism have more statistical power to detect nonzero values and thus showed more differentiation [ρS = 0.47 (with FST) and −0.39 (with FST P-values), P < 10−16 for both comparisons]. Accordingly, the contrast between X and autosome should be conservative given the lower level of polymorphism on the X before correction for population size (UMW test, P < 10−16). The stronger differentiation on the X is consistent with the predicted (Charlesworth et al. 1987) and observed excess of adaptive evolution on the X chromosomes (see above). But note that similar predictions are also made by a range of demographic models (e.g., Hutter et al. 2007; Pool and Nielsen 2008). To further confirm that the observed geographic differentiation of amino acid sequences is the result of selection instead of solely driven by demography, we compared the per-site FST for synonymous and nonsynonymous sites. To ensure equal statistical power, we binned synonymous and nonsynonymous SNPs into categories with marginal frequencies of [0, 0.15), [0.15, 0.3), and [0.3, 0.5]. For the category that has the greatest statistical power and showed the greatest differentiation ([0.3, 0.5]), nonsynonymous sites have significantly larger Fst than synonymous sites (Wilcoxon’s rank test, P < 10−16). This result is robust to using either weighted (by sample size) or unweighted marginal allele frequency.
Because of the large number of genes having significant amino acid FST, we restricted our GO enrichment analysis to genes having FST P-value <0.001 (15.7% of all the golden genes). There are 80 biological GO categories highly enriched (P < 0.05) with genes that are highly differentiated (see Table S16). Top biological GO categories include response to damaged tissue, metabolism of nitrogen compounds, and exocytosis. Interestingly, several GO categories related to immunity, hemocyte development, recombination, and perception of and reactions to sensory cues show significant enrichment of highly differentiated genes. Significant cellular GO categories include chromosome condensin complex, nuclear plasm, and nuclear pore (see Table S17). The most enriched molecular categories include metal ion binding, taste receptor activity, helicase activity, and phospholipid binding (see Table S18).
It is interesting to investigate whether genes under recurrent adaptive evolution (long-term adaptive evolution) are also the target of selection for the local adaptation to temperate habitat (short-term adaptive evolution). We compared between-population amino acid FST and MW MK tests of 4758 genes that have results for both tests. Almost 1% of genes (0.7%) had both significant excess of amino acid fixations and between-population differentiation (FST, P < 0.001), which is not significantly different from the expected 0.9% [multiplication of 5.8% (genes have significant MK test) and 15.7% (genes have FST P-values <0.001); χ2-test, P > 0.05]. Our observations could be explained by both the differences in timescale that can be detected by these two tests and/or the fact that genes that are under local adaptation to the temperate habitat are fundamentally different from genes that are under recurrent directional selection in the tropics. Consistent with this, we observed little overlap of GO terms enriched with either set of the significant genes. Only two biological GO terms (negative regulation of Notch signaling pathway and meiotic recombination) showed both enrichment among genes with evidence of adaptive evolution and strong African–North American differentiations. Nucleoplasm is the only significant cellular GO term, while there are no molecular GO terms that have significant enrichment in both MK and FST analyses. Window-based analysis can reflect these observations as well if rare variants are the primary substrate of such adaptation, thus producing a hitchhiking effect (see below). On the other hand, if much of the selective geographic differentiation involved frequency changes in preexisting (amino acid) polymorphisms at linkage equilibrium with surrounding SNPs, the detectable changes might well be limited to actual selected sites (Hermisson and Pennings 2005).
Shared nonsynonymous polymorphisms:
Shared ancestral amino acid polymorphism maintained by balancing selection has been well documented in several cases, such as the MHC locus of vertebrates (Figueroa et al. 1988 and references therein) and the S locus of plants (Ioerger et al. 1990 and references therein). Examples of such polymorphisms have not yet been reported for D. melanogaster and D. simulans. We looked for genes with exceptional levels of shared ancestral polymorphism between D. melanogaster and D. simulans. Specifically, we compared the ratio of the number of sites that have two alternative states shared between the two species to the total number of variable sites of a focal gene with those of the overall golden gene set (see Materials and Methods). In total, we observed 539 nonsynonymous and 6886 synonymous shared polymorphisms, which are 0.98% and 3.98% of the observed corresponding polymorphic sites in D. melanogaster genes. As we are mainly interested in shared polymorphisms that are more likely to have functional and/or phenotypic effects, we considered only nonsynonymous changes in the following analysis. Assuming that the proportion of nonsynonymous polymorphisms is constant, 133 genes exhibit a significant (P < 0.05; see Materials and Methods) excess. Thirty-nine of these genes (all among the significant set) have two or more specific amino acid replacement polymorphisms shared between D. melanogaster and D. simulans. It is highly unlikely to observe 39 genes with more than one shared nonsynonymous polymorphic site (simulation-based P < 0.007).
Eight of the 39 genes also have an excess of shared synonymous polymorphism, which might have been maintained by balancing selection, perhaps due to selection on the shared nonsynonymous polymorphisms. Consistent with the overall short scale of linkage disequilibrium (see above), we did not observe any haplotypic structures in any of the 39 genes (results not shown) as has been described for the vertebrate MHC or S locus of plants. Sequencing errors are unlikely to be the sole source of these 39 genes, since 37 of these genes have at least one nonsynonymous shared polymorphism that is not a singleton in D. melanogaster.
Consistent with a biological explanation for this unusual set, the levels of both nonsynonymous and synonymous polymorphism of these 39 genes are higher than those of other genes (Wilcoxon’s rank test, P < 0.001 for both tests). We cannot exclude the possibility that the excess of shared nonsynonymous variations observed is due to conserved locus-specific higher mutation rates, especially to specific alleles. Accordingly, instead of arguing that the observed shared nonsynonymous polymorphism is truly ancestral and maintained by balancing selection, our following analysis briefly identifies and discusses the gene-specific biology of named genes likely to be relevant (see Table 13). These observed shared polymorphisms across species could potentially be the result of systematic, correlated mismapping in both species. This may especially be a problem for genes with several ancestral paralogs. We searched in the D. melanogaster genome (using BLAT) in the 1-kbp region surrounding each of the shared nonsynonymous polymorphic sites in each the 39 genes. For most genes, the “noncanonical” BLAT hits are short (20 bp) and far from the shared sites. Exceptions are Ugt36Ba, rhi, and Ino80. Yet, in these genes, the noncanonical BLAT hit that spanned the shared sites had low sequence identity (<70%); reads are unlikely to be mismapped. Additionally, if most of the observed shared polymorphisms were the result of mismapping, we expect to see increased linkage disequilibrium among these sites. However, the r2 between pairs of shared sites (either nonsynonymous or synonymous) do not have a significantly higher level of linkage disequilibrium than those of other pairs of SNPs in the same set of genes. Thus, mismapping can be excluded as the probable cause of the identified shared polymorphisms.
Table 13 . Named genes in the golden set with two or more nonsynonymous shared polymorphisms in D. melanogaster and D. simulans (see text).
| FBgn | Gene name | No. shared nonsynonymous polymorphisms | No. shared nonsingleton nonsynymous polymorphisms | No. synonymous polymorphisms | Shared amino acid coded with different codon | One of the shared nonsynonymous states present in yakuba and/or erecta | Alternative shared nonsynonymous states present in yakuba and erecta | FET P-value for nonsynonymous polymorphisms | FET P-value for synonymous polymorphisms |
|---|---|---|---|---|---|---|---|---|---|
| FBgn0040102 | Lectin-24Db | 4 | 2 | 3 | 1 | 3 | 0 | 0.001 | 0.080 |
| FBgn0261714 | Cpn | 4 | 3 | 1 | 1 | 3 | 1 | 0.002 | 1 |
| FBgn0043841 | vir-1 | 3 | 3 | 1 | 0 | 2 | 1 | 0.0001 | 0.583 |
| FBgn0050169 | Brca2 | 3 | 2 | 0 | 1 | 0 | 0 | 0.024 | 0.652 |
| FBgn0000928 | fs(1)Yb | 3 | 1 | 2 | 0 | 0 | 0 | 0.019 | 0.655 |
| FBgn0015663 | Dot | 2 | 2 | 6 | 0 | 0 | 0 | 0.005 | 0.001 |
| FBgn0052656 | Muc11A | 2 | 1 | 3 | 0 | 0 | 0 | 0.009 | 0.019 |
| FBgn0036463 | Reck | 2 | 2 | 6 | 0 | 0 | 0 | 0.008 | 0.030 |
| FBgn0031592 | Art2 | 2 | 2 | 2 | 0 | 0 | 0 | 0.006 | 0.166 |
| FBgn0032728 | Tango | 2 | 2 | 0 | 1 | 2 | 0 | 0.026 | 0.412 |
| FBgn0040262 | Ugt36Ba | 2 | 2 | 0 | 0 | 2 | 0 | 0.007 | 0.631 |
| FBgn0004400 | rhi | 2 | 1 | 0 | 0 | 0 | 0 | 0.040 | 1 |
| FBgn0032840 | sNPF | 2 | 1 | 0 | 0 | 1 | 0 | 0.001 | 1 |
| FBgn0036032 | Ino80 | 2 | 1 | 1 | 0 | 1 | 0 | 0.014 | 1 |
| FBgn0011297 | l(2)not | 2 | 2 | 1 | 1 | 2 | 0 | 0.002 | 1 |
| FBgn0000140 | asp | 2 | 1 | 2 | 0 | 2 | 0 | 0.043 | 1 |
Genes with the largest numbers (four) of shared polymorphic sites are Lectin-24Db (codes for a mannose and fucose binding protein) and the RNA-edited Cpn (codes for a photoreceptor-specific calcium-binding protein (Stapleton et al. 2006). Several aspects of their shared nonsynonymous variants suggest a long-term role of natural selection. For both Lectin-24Db and Cpn, at least two nonsynonymous polymorphisms have their two shared states present in multiple individuals, suggesting they are neither rare nor sequencing errors. The alternative states of one of the Cpn shared sites were present in D. yakuba and D. erecta. Furthermore, these two genes both possess one shared amino acid polymorphism encoded by different codons in the two species, consistent with parallel balancing selection for specific alleles.
Based on the functions of these named genes in Table 13, there are three potential scenarios in which shared amino acid polymorphisms could be attributed to balancing selection. The antagonistic interaction between Drosophila and pathogens/parasites is an obvious one. Muc11A is a gene predicted to be involved in the metabolism of chitin. Chitin is an essential component of the exo- and endoskeleton of Drosophila, including the gut and trachea, which are entry points for fungal pathogens via their degradation of the chitin (Lemaitre and Hoffmann 2007 and references therein). vir-1, which is upregulated during the infection of Drosophila C virus via the Jak-STAT pathway (Dostert et al. 2005), is another good candidate for balancing selection. The two alternative states at the three shared nonsynonymous sites of vir-1 are present in multiple individuals and in both MW and RAL populations. In addition, for one shared nonsynonymous site, one of the states was observed in D. yakuba while the other was in D. erecta.
In addition to external parasites, the interactions between Drosophila and genomic parasites, such as transposable elements and Wolbachia, can be another cause for the observed shared ancestral polymorphism. Most transposable-element families annotated in D. melanogaster are also detected in D. simulans (Begun et al. 2007; Bartolomé et al. 2009) as is Wolbachia. rhi and perhaps Brca2 have a role for host–transposable-element interactions. Brca2 (Breast cancer 2, early onset homolog), a homolog of human tumor suppressor Brca2, is essential for double-strand break repair via homologous recombination and the activation of the meiotic recombination checkpoint (Klovstad et al. 2008). Thus Brca2 may interact with double-strand breaks associated with transposable-element activity. Two of the three shared nonsynonymous polymorphic sites of Brca2 have nonsingleton alleles that are present in both MW and RAL populations. Apparent partial loss-of-function mutations of rhi (rhino), a HP1 paralog, have recently been shown to influence the generation of piRNAs, which regulates the transposition of transposable elements (Klattenhoff et al. 2009). Evidence of positive selection in rhi has been reported for both the D. melanogaster and the D. simulans lineages (Vermaak et al. 2005). The different fates of germline and somatic cell lineages of Drosophila gonads create an opportunity for an arms race between host and pathogens/parasites (Blumenstiel 2011 and references therein). For example, pathogens/parasites that can target the oocyte rather than nurse cells of the female germline have a higher chance of being vertically transmitted and, thus, greater fitness advantage. Genetic variation in fs(1)Yb [female sterile (1) Yb, responsible for female germline stem cell maintenance] and asp (abnormal spindle, a microtubule-associated protein that is involved in spindle pole organization in both mitosis and meiosis) may interact with transposable elements and other germline transmitted pathogens, leading to balancing selection.
Another interesting finding is that among the named genes in Table 13 are two predicted UDP-glycosyltransferases (UGTs), Ugt36Ba and Dot (Dorothy). The transfer of glycosyl group to hydrophobic molecules increases their hydrophilicity, thus enhancing their secretion. In insects, UGTs participate in detoxication of plant chemicals, cuticle formation, pigmentation, and olfactions (Luque and O’Reilly 2002 and references therein). The specific mechanisms of possible interactions of Drosophila UGTs with plants and pathogens are not known, but it is interesting to note that in both Ugt36Ba and Dot, there is at least one shared polymorphic site located in the domain with glycosyltransferase activity.
Copy-number variation
In addition to single-nucleotide polymorphisms, individual Drosophila differ by large duplications and deletions of DNA. Bridges (1936) was the first to identify one such duplication at the Bar gene. Subsequent genetic and molecular genetic analyses (Tsubota 2009 and references therein) implicated unequal crossing over as the main underlying mechanism and estimated such events as a major component of spontaneous mutation (Montgomery et al. 1991 and references therein). Recently whole-genome array studies have identified many more naturally occurring copy-number variants (Dopman and Hartl 2007; Emerson et al. 2008; Turner et al. 2008; Cridland and Thornton 2010).
Across all lines we detected 3631 duplications and 3953 deletions. Largely due to slight differences in the location of breakpoints called by the HMM when run on different lines, identical structural variants may have been called as distinct CNVs. We therefore devised a simple algorithm to combine overlapping calls believed to represent the same polymorphism for the purposes of analyses of CNV length and frequency presented in this section (See Appendix B). This resulted in a set of 2588 duplications and 3336 deletions. The average size of duplicates was 2069 bp and the average size of deletions was 617 bp; the longest duplication detected was 121,910 bp and the longest deletion was 31,885 bp. Overall, we were able to confirm 85–86% of deletions and 82–85% of duplications (the ranges are defined by values for the two lines). The confirmation rate for duplications is similar to that found in previous studies (86% in Emerson et al. 2008), while our deletion confirmation rate is higher than they reported, 53% (see Appendix B).
We did observe a deficit of duplications on the X chromosome, but this could be due in part to lower average coverage on the X vs. the autosomes. We observed no significant difference between the lengths of CNVs on the X and those on the autosomes. Within each chromosome, there was also obvious variation in the density of duplications and deletions, with both types of events found in larger numbers in subcentromeric regions (see Figure 16).
Figure 16 .
The distribution of duplications and deletions in 1-Mbp windows spaced every 100 kbp across the X and autosomes. To smooth the underlying count data from independent nonoverlapping 100-kbp windows, windows >3 standard deviations from the mean and with <40% consensus sequence coverage were filtered. These nonoverlapping windows were then averaged using a 1-Mb sliding window spaced every 100 kbp. The genomic distribution of is added for comparison.
Between any two lines from MW there is an average of 738.5 CNV differences; equivalent comparisons between RAL lines gave 657.2 CNV differences. This difference in copy-number heterozygosity between populations mirrors the difference in SNP diversity between MW and RAL. Given the average length of CNVs in our data set, this number of CNV differences implies a total of 764 kbp (0.64% of the genome) that vary in copy number between any two inbred lines from RAL, compared to 869 kbp (0.73% of the genome) in MW. There are a number of reasons why these values may underestimate the true extent of copy-number heterozygosity, including the fact that derived deletions and duplications in the reference genome are not queried (see Materials and Methods) and that we have used a minimum-length cutoff to define CNVs. Nonetheless, we estimate that the total number of base pair differences between individuals contained within CNVs found here is on the same order as the total number of SNP differences (∼1 Mb across the genome). This result is in stark contrast to studies in humans that suggest that the total number of bases contained within genomic regions varying in copy number between any two individuals is roughly an order of magnitude larger than the total number of SNP differences between individuals (McCarroll et al. 2008; Conrad et al. 2010). We note, however, that π for SNPs is much higher in Drosophila and that the CNV ascertainments may not be sufficiently comparable.
We examined the distribution of both duplication and deletion CNVs with respect to genes in the Drosophila genome. Similar to the results found by Emerson et al. (2008), we found an excess of deletions in intergenic regions and introns (Figure 17). Deletions that contain entire genes are especially rare, likely due to the deleterious effects of such mutations. Nevertheless, we do find many CNVs overlapping genes (29 deleted genes and 301 duplicated genes), with an average of 41.48 whole genes differing in copy number between any two inbred lines from RAL. Using GOEAST (Zheng and Wang 2008) to find biological processes overrepresented in genes varying in copy number, we find that deleted genes are enriched for response to chemical stimulus and nuclear mRNA splicing via the spliceosome. We find that duplicated regions are enriched for the following biological processes: fatty acid beta-oxidation, regulation of hormone levels, response to insecticide, protein amino acid glycosylation negative regulation of the cellular catabolic process, cellular response to hydrogen peroxide, detection of chemical stimulus involved in sensory perception of smell, and detection of pheromone. Significant expansions in gene families involved response to toxins, response to hydrogen peroxide, and olfactory perception were also found in an interspecific study of copy number in the genus Drosophila (Hahn et al. 2007). Also notable is a common deletion that was found in the mthl-8 gene, although this may be due to recurrent mutational events (Kern and Begun 2008).
Figure 17 .
The proportion of duplications (solid bars) and deletions (open bars) containing at least one entire gene, containing a portion of an exon (coding or noncoding), containing an intronic segment, or containing only intergenic regions. Note that deletions are far less likely than duplications to contain whole genes or exonic segments. Instead, deletions are disproportionately found within intergenic regions. Duplications, on the other hand, do not appear to be biased toward intergenic regions (which make up ∼35% of the genome).
When examining regions most commonly found to be aneuploid in the RAL inbred lines, we found a large region on chromosome 3L identified as elevated in copy number in >75% of the lines. This region was found to contain several chorion genes (Cp15, Cp16, Cp18, and Cp19) as well as several other genes (CG13306, CG6511, CG32022, SrpRbeta, and Prm). We concluded that, rather than being a large genomic duplication, this stretch of aneuploidy corresponds to a genomic region known to be amplified in follicle cells in association with increased chorion gene expression and eggshell development (Spradling 1981). This amplified region was also detected in the genome-wide CNV scan conducted by Dopman and Hartl (2007), implying that read-depth or array-based CNV scans will often detect such amplifications. Indeed, we also detect a less common region elevated in copy number, containing the genes CP7Fa, CP7Fb, and CP7Fc, corresponding to a different chorion cluster that is also known to be amplified (Spradling 1981; Turner et al. 2008). These results show that read-depth or array-based CNV detection methods can identify regions of the genome that are aneuploid due to amplification or to underreplication in terminally differentiated cell types (Sher et al. 2011).
We examined the allele frequencies of CNVs and found that duplications were skewed toward lower frequencies than are deletions (P < 2.2 × 10−16, UMW), a result that runs counter to the expectation that deletions are on average more deleterious than duplications and therefore subject to stronger negative selection, which has received support in a study of humans CNVs (Locke et al. 2006). This result is most likely an artifact of the lower power to genotype duplications than deletions.
We noted a significant negative correlation between CNV length and frequency for both duplications (ρS = −0.259, P < 2.2 × 10−16) and deletions (ρS = −0.366, P < 2.2 × 10−16), implying that larger duplications and deletions are likely to be deleterious, as also appears to be the case in humans (Itsara et al. 2009). Next, we compared the allele frequencies of CNVs containing entire genes, containing some exonic sequence, containing only intronic sequence, or located within intergenic regions, notably finding that whole-gene duplications were constrained to lower allele frequencies than duplications in exonic or intergenic regions (P = 0.036 for duplications and P = 0.0071 for deletions, UMW). Interestingly, the frequencies of gene duplications were not significantly lower than those of intronic duplications (P = 0.097). This may be due to the lower frequencies of intronic duplications relative to intergenic duplications, a trend that is only marginally significant in our data (P < 0.083) but has been observed previously (Emerson et al. 2008). These results suggest that whole-gene duplications, and perhaps intron duplications as well, are subject to stronger purifying selection than intergenic duplications. Although the same trend has previously been observed for exonic duplications in a previous study (Emerson et al. 2008), we find no such pattern here. When examining deletions, we found that intronic deletions had lower allele frequencies than intergenic deletions (P = 4.8 × 10−8). Strangely, this was not the case with exonic deletions, which appear at higher frequencies than intronic deletions, although this difference is not significant. Our data set contained too few whole-gene deletions to compare their frequencies to other classes of deletions. Finally, we compared the allele frequencies of CNVs on the X chromosome to those on the autosomes and found no significant difference for either duplications or deletions.
While most new duplicates in Drosophila are located in proximity (i.e., tandem) and in head-to-tail orientation relative to the locus they are copied from, a significant number of gene duplicates are located far away on the same chromosome or even on different chromosomes (Meisel et al. 2009). We used two methods to try to identify nontandem, “dispersed” duplication events. First, we compared levels of linkage disequilibrium between flanking SNPs and CNVs due to both duplications and deletions. Because duplication polymorphisms may be located a long distance from the parental locus, SNPs flanking this locus will not be in linkage disequilibrium (LD) with the CNV; the same is not true for deletions, as the location of the CNV polymorphism is known exactly (Schrider and Hahn 2010). Results for 50 SNPs flanking each CNV (or, more precisely, the location of the identified polymorphism in the reference genome) showed that LD was in fact lower around duplications than around deletions: the average maximum-r2 value among the 50 SNPS for duplications was 0.35 while for deletions it was 0.56. These results suggest that some duplications are located far from their parental loci, but they do not tell us how far away. Second, we developed a novel method for identifying polymorphic retrotransposed duplicates (“retroCNVs”) from the Illumina data (Schrider et al. 2011). Because of the crenellated patterns of read depth associated with retroCNVs—only the exons will show excess read depth, but not introns—these polymorphisms will be missed by the HMM. We identified 34 retroCNVs among the RAL lines, some at quite high frequency (Schrider et al. 2011). Because retrogenes are inserted in a seemingly random pattern across the genome, this class of duplicative CNV also represents a source of dispersed duplicates. We also detected nine intron deletion polymorphisms among the RAL lines, which are described in detail by Schrider et al. (2011).
CNVs and recombination rates:
Recombination is expected to be an important element in both the origin (Montgomery et al. 1991) of CNVs and their population dynamics via both linked selection (Hill and Robertson 1966) and gene conversion (Johnson-Schlitz and Engels 1993; Presgraves 2006). Thus we investigated the distributions of deletions and duplications among the RAL genomes in regions with different estimates of the crossing over per base pair (quartiles of the logarithm of ). The mean count of duplications per window increases by >75% when comparing the highest to the lowest recombination rate categories (see Table 14). Deletions also increase by 30% when comparing the same two quartiles. At face value this would seem to support the hypothesis that both types of CNVs are more effectively eliminated by natural selection with increasing crossing over. Also supporting this hypothesis is the observation that heterozygosity showed similar trends. There is an 88% increase of expected heterozygosity for duplications when comparing the highest to the lowest recombination quartiles and a 15% increase for deletions. The mean count of duplications per window drops by more than one-third between the lowest and the highest recombination rate categories (see Table 14). Deletions also decline by >50%.
Table 14 . Mean counts and heterozygosities of duplications and deletions in 100-kbp windows with estimated crossing-over rates in one of four categories (see text).
| Recombination quartile | ||||
|---|---|---|---|---|
| Very low | Low | High | Very high | |
| Mean duplication count | 2.04 | 1.28 | 0.90 | 1.15 |
| Mean deletion count | 2.65 | 2.36 | 2.42 | 2.03 |
| Mean duplication heterozygosity | 0.040 | 0.023 | 0.024 | 0.021 |
| Mean deletion heterozgosity | 0.069 | 0.058 | 0.063 | 0.060 |
CNVs and origins of replication:
Functional origins of replication initiate at a subset of ORC binding sites (MacAlpine et al. 2010 and references therein). And the regulation of interactions among the arising bidirectional forks is complex, with multiple pathways functioning on the scale of both the genomic neighborhood and the cell (Natsume and Tanaka 2009; Blow et al. 2011). Stalled adjacent replication forks pose a particularly serious challenge to genome replication, give rise to error-prone recovery, and have been proposed as a major source of human CNVs (Lee et al. 2007). Thus we tested whether our detected deletions and duplications were enriched near (±500 bp) annotated ORC binding sites. Table 15 compares the number of deletions overlapping the annotated ORC binding regions with a randomly derived control set of genomic regions (see Materials and Methods). The relative number of deletions increases with the number of cell lines exhibiting ORC binding at a site. Pooling all three categories, there is a 1.5-fold increase (P < 0.0001) of deletions near ORC binding sites. Similarly Table 16 shows the relationship between numbers of identified duplications and ORC binding sites. The overall proportion of ORC binding regions overlapping with duplications is 1.34 times that in the control regions (P < 0.001). These results support the hypothesis that the distribution of origins of replication contributes to genomic variation in CNV density. With much less power we failed to detect any difference in the expected heterozygosity of CNVs or the relative enrichment of deletions vs. duplications near ORC binding sites.
Table 15 . Numbers of deletions overlapping ORC binding sites compared to a comparable control region 10 kbp away (see text).
| ORC score | Deletions | Control regions | Ratio | ||
|---|---|---|---|---|---|
| 0 | 2615 | 0.91 | 2699 | 0.94 | 1.50 |
| 1/3 | 118 | 0.04 | 68 | 0.02 | 1.73 |
| 2/3 | 52 | 0.02 | 37 | 0.01 | 1.40 |
| 1 | 82 | 0.03 | 63 | 0.02 | 1.30 |
| Total | 2867 | 2867 | |||
The “ORC score” indicates in how many of three studied cell lines the ORC binding site was called. The first value in the ratio column (in italics) is the ratio for the union of three ORC score categories.
Table 16 . Numbers of duplications overlapping ORC binding sites compared to a comparable control region 10 kbp away (see text).
| ORC score | Duplications | Control regions | Ratio | ||
|---|---|---|---|---|---|
| 0 | 1459 | 0.85 | 1526 | 0.89 | 1.34 |
| 1/3 | 91 | 0.05 | 67 | 0.04 | 1.35 |
| 2/3 | 73 | 0.04 | 47 | 0.03 | 1.55 |
| 1 | 98 | 0.06 | 81 | 0.05 | 1.21 |
| Total | 1721 | 1721 | |||
The “ORC score” indicates in how many of three studied cell lines the ORC binding site was called. The first value in the ratio column (in italics) is the ratio for the union of three ORC score categories.
CNVs and replication time:
It has also recently been proposed that the timing of DNA replication affects the probability of a CNV arising, although this relationship may differ for duplications and deletions (Cardoso-Moreira and Long 2010; Cardoso-Moreira et al. 2011). These authors found that regions dense in duplication CNVs tended to be associated with later replication times, while regions dense in deletion CNVs were associated with earlier replication times (Cardoso-Moreira and Long 2010). We used our high-quality set of CNVs in RAL lines and the replication timing data from Schwaiger et al. (2009) to further test the generality of this relationship. Following Cardoso-Moreira and Long (2010), we compared the replication time of the 100-kbp windows containing the greatest number of deletions and duplications to the remaining windows in each category. Our analyses alternatively included (and excluded) the pericentromere regions and consistently excluded those CNVs associated with the chorion genes. Another notable difference between our approach and the approach of Cardoso-Moreira and Long (2010) is the smaller number of windows considered in the analysis; we targeted the maximum resolution at which our data would support independent windows. We did find that windows within the 90% quantile for duplication count tended to be later replicating compared to the rest of the data set. This observation was significant for the Cl8 cell line for all duplications (P = 0.0048) and when excluding those within the pericentromere (P = 0.022). In contrast to Cardoso-Moreira and Long (2010), we observed that windows within the 90% quantile for deletion count also tended to be later replicating. This was consistent for both cell lines regardless of whether the pericentromere was excluded. However, the difference was not significantly different from the rest of the genome for any of these four cases. We also compared the replication time of deletions to that of duplications. We observed mixed outcomes. The only significant observed difference in replication time was for the Kc cell line when including all CNVs in the analysis. In this case, deletions tended to be slightly later replicating than duplications (Wilcoxon’s P = 0.0015). This observation maintained directionality, but was not significant when the pericentromeric CNVs were excluded. Similarly, for state classifications, we found that deletions were more often classified as later replicating over earlier replicating than deletions using the HMM classification of Schwaiger et al. (2009) for the Kc cell line (see Materials and Methods). This observation was significant for all CNVs (FET P = 0.0148) but lost significance when the pericentromeric regions were excluded from the analysis. Given our earlier reported results on the higher average deleterious effects of deletions relative to duplications, we think that the biases in the location and timing of CNVs we observe are likely due to the fact that there also happens to be a higher density of genes in early-replicating portions of the genome. This implies a selective rather than a mutational cause for the observed bias in the location of CNVs.
Genes and functional sequences affected by natural selection:
As the gene-based analysis presented above focused on the coding regions, window-based analysis along the chromosomes is complementary in several ways. While the window-based analyses capture none of the powerful inference associated with the distinction between nonsynonymous and synonymous changes, the more uniform statistical power associated with the equal numbers of polymorphic-plus-diverged sites in the case of the HKAl and comparable normalizations in the HBKl and TsD analyses should not suffer from the same potential biases of gene-based analysis, in which genes with longer coding regions tend to have larger statistical power. Furthermore, window-based analyses are likely to identify the target of selection if it is located away from coding regions or if only part of a gene is under adaptive protein evolution. This is because the signals of directional selection are highly likely to be diluted by other nonselected variation in the same gene in gene-based analysis, while the window-based statistics leverage the linkage-mediated impacts of selection and/or demography. Thus the following analysis based on the intersection of the high-resolution window-based statistics and the structural and functional annotation presented here approaches a similar set of questions from a distinct, complementary angle.
The HKAl statistic was calculated for windows of 50 segregating or divergent sites in the MW sample. Although the MW sample size is small, it should still provide reasonable local estimates of sequence diversity, and its relatively stable demographic history may allow the effects of selection to be clearly observed. Windows among the lowest 2.5% of HKAl values (evaluated separately for the autosomes and the X chromosome) were classified as outliers with sweep-like patterns of diversity.
Low HKAl outliers might reflect either low diversity due to a recent sweep or elevated divergence due to many ancient recurrent sweeps. The strongest signals of selection in the genome often span multiple genes, and their targets are thus difficult to ascertain. Cases where an extended sweep signal appeared to center on a specific gene included CENP-meta (involved in chromosome segregation during mitosis and male meiosis I) and Dicer-2 (antiviral function via RNA interference), the latter being consistent with the findings of Obbard et al. (2006). A full list of HKAl outlier regions is presented in Table S19.
For each HKAl outlier region, the closest gene was defined as the gene with an exon (including UTRs) closest to the center of the HKAl outlier window. Outlier windows <10 kbp apart were considered jointly, and the signal was deemed to center in the middle of the window with the lowest χ[log(pHKAl)] value. Note that these are imprecise localizations of the targets of directional selection (especially with regard to functional elements, see below), yet they may be sufficiently informative to reveal genome-wide patterns. GO analysis was undertaken to identify functional categories overrepresented among these outlier genes. Statistical significance was assessed by randomly permuting the windows identified as low HKAl outliers (thus accounting for the effect of gene length on the likelihood of random detection). Cellular components implicated by this analysis included the nucleus, chromatin, spliceosome, and polytene chromosome puff (Table S20). Molecular functions included many related to nucleic acid binding, along with kinase activities, ATP binding, zinc ion binding, and microtubule motor activity. Biological functions with the lowest GO P-values included neuron development, chromatin silencing, mRNA splicing, and centrosome organization. Other biological functions with P < 0.05 pertained to transcription and translation, along with RNA interference, oogenesis, and spermatogenesis (Table S20).
Using the low HKAl outlier central positions and closest genes identified above, a striking pattern was noted with regard to the positions of putative sweep targets along gene regions. It was found that, relative to random expectations, sequences near the beginning and ending of genes were highly enriched for sweep signals (Figure 18A). On average, exons were targeted by outliers considerably more often than introns or distal intergenic regions (those >2 kbp from a gene). Notably, 5′-UTRs and 3′-UTRs, along with proximal intergenic regions, were more likely to be targeted by HKAl outliers than protein-coding exons. As noted above, no single localization by this method can be taken with confidence, and some UTR-centered windows could instead result from adaptive substitutions in nearby exons or noncoding regions. However, given that the strongest genome-wide HKAl enrichments in Figure 18A are for gene position bins outside exonic regions, it seems unlikely that adaptive protein-coding substitutions are the primary drivers of the HKAl enrichment observed in UTRs and nearby regions. These results are parallel to the chromatin state 1 patterns presented above and could indicate a particular importance of UTRs and nearby functional elements in the recent adaptive history of D. melanogaster (Kolaczkowski et al. 2011b).
Figure 18 .
The distribution of HKAl outliers along gene regions and among functional classes of sites. The number of outlier windows centering on each gene position bin (A and C) or functional element (B and D) was compared against the total number of analyzed windows centering on each of these categories for MW HKAl, for low outliers (A and B) and high outliers (C and D). Over- and underrepresentation of each category was calculated such that a value of 1 matches the random genome-wide expectation, while a value of 2 indicates twice as many HKAl outliers in this category as expected randomly. Gene positions (A and C) were defined with regard to the beginning of the 5′-UTR and the end of the 3′-UTR. “kbp” bins indicate distance upstream or downstream of these limits, while genic bins indicate relative position between these limits and encompass 5′-UTR, coding exon, intron, and 3′-UTR sequences. These categories are depicted separately, along with proximal intergenic (within 2 kbp of a gene) and distal intergenic regions, in B and D.
UTRs may contain sequences that interact with regulatory proteins or micro-RNAs to regulate mRNA stability or translation (Kuersten and Goodwin 2003; Pickering and Willis 2005) and hence have the potential to influence gene regulation. Genome-wide, 87% of HKAl outliers appeared to center on introns (43%), intergenic regions (38%), or UTRs (6%). Despite the inherent uncertainty of the localizations, these results suggest ample potential for adaptive cis-regulatory changes in the D. melanogaster genome. With regard to the types of genes affected by directional selection, GO enrichment analysis of HKAl outliers implicated a striking number of biological processes related to gene regulation at multiple levels, including transcription, translation, and splicing (Table S20). Processes related to RNA interference and chromatin silencing showed similar enrichments and may offer additional avenues for regulatory evolution (Levine and Begun 2008; Kolaczkowski et al. 2011a). Functional changes to genes involved in any of these processes could alter the regulation of many other genes, suggesting the possibility that relatively “higher-order” trans-regulatory changes may have a strong importance in Drosophila evolution.
Aside from implicating genes involved in gene regulation, GO enrichment analyses of HKAl and MK outliers provided further evidence for the adaptive importance of several previously suggested biological processes, including male and female reproduction. “Neuron development,” on the other hand, has not been a major focus of research on Drosophila adaptive evolution, and yet this GO category was the most significantly enriched biological process for HKAl outliers. Furthermore, the most enriched biological process for diversity ratio outliers (identifying candidates for recent adaptation in non-African populations, see below) was “negative regulation of neuron apoptosis.” One biological explanation for these results would be the evolution of resistance to insecticides, many of which target the nervous system. However, the prevalence of neural genes among MK outliers (a signal that is unlikely to be driven by extremely recent adaptation alone) suggests that other selective pressures may be at work. An alternative explanation would be a strong adaptive importance of behavioral evolution via modification of the nervous system. Detailed molecular and evolutionary studies will be needed to evaluate this and other hypotheses motivated by the data and analyses presented here.
Similar analyses were undertaken with regard to high HKAl outliers, which represent regions of excess polymorphism relative to divergence. High HKAl outlier regions were generally narrower than low HKAl windows (Table S21) and concentrated in coding exons but not UTRs (Figure 18, C and D). GO categories enriched for high HKAl outliers included terms related to myosin and muscle attachment, transmembrane and vesicle transport, and cellular junctions (Table S22). Further investigation is needed to determine whether these results indicate the selective maintenance of protein-coding variation at some genes or simply the presence of low-frequency deleterious variants.
Potential signals of adaptation specific to the temperate sample:
Both D. melanogaster and D. simulans have expanded their ranges from tropical Africa and Madagascar to inhabit temperate environments (Lachaise et al. 1988; Dean and Ballard 2004). Regions of the genome with unusually low diversity in the RAL sample, relative to the MW sample, may contain targets of adaptation to temperate environments (Schlötterer and Dieringer 2005). Diversity ratios (πRAL/πMW) were calculated for 5-kbp windows, moved in 1-kbp increments. Outliers and putative target genes were identified as described above for MW HKAl. A comparable analysis excluding outliers for πMW gave very similar results to those described below (not shown).
By far the strongest signal of non-African adaptation in the highly recombining portions of the genome comes from the region that includes Cyp6g1. This selective sweep, which has been linked to insecticide resistance (Daborn et al. 2002; Schmidt et al. 2010), has strongly reduced nucleotide diversity across ∼85 kbp in the RAL sample and was also associated with a large differentiated region in the Australian latitudinal cline (Kolaczkowski et al. 2011b). The previously described signal covering unc-119 (Glinka et al. 2006) is also apparent, covering at least 35 kbp. Other broad sweep signals (≥20 kbp) include the genes jaguar (multifunctional, with roles in oogenesis and sperm motility), Pcf11 (mRNA cleavage), CG5278 (regulation of alternative splicing), Pi3K92E (regulation of growth), PGRP-SA (regulation of antimicrobial peptides), and Stat92E (multifunctional, with roles in immunity and oogenesis), among others (Table S23).
GO analysis was conducted for diversity-ratio outliers in the same manner as described above. Many biological terms with P < 0.05 pertained to metabolism, regulation of growth, or behavior (see Table S24). Other terms included negative regulation of neuron apoptosis, oogenesis, and response to DNA damage stimulus. The set of GO terms implicated by this analysis (Table S24) was largely different from the MW HKAl results. Among the terms implicated by both analyses were the biological processes nuclear mRNA splicing via spliceosome and chromatin silencing, the cellular components precatalytic spliceosome and nuclear pore, and the molecular processes nucleic acid binding and microtubule motor activity.
Signals of reduced diversity shared between populations and species:
A simple metric was employed to scan the genome for regions of reduced diversity that may have resulted from recent selective sweeps: π/min(divl, divg), where the denominator indicates the lesser of local divergence and global average divergence (evaluated separately for the X chromosome and autosomes). This statistic is expected to yield low values after a recent selective sweep, but not due to selective constraint or elevated ancestral polymorphism. The statistic was evaluated for 5-kbp windows in 1-kbp increments. Windows within the bottom 2.5% quantile of the empirical distribution were noted, and these “valleys” of diversity were merged for outliers <10 kbp apart. A more restrictive set of cutoffs was identified for centromeric and telomeric regions (visually, based on nucleotide diversity along the chromosome in each sample) to exclude an influence of these regions on the windows identified (boundaries are listed with the relevant tables below).
Of the diversity valleys observed in the RAL sample, 44% overlapped with a valley in the MW sample. Many of these genomic regions may have experienced selective sweeps in Africa prior to the worldwide expansion of the species. More surprising was the proportion of diversity valleys shared between D. melanogaster and D. simulans (data from Begun et al. 2007). Of the RAL sample’s valleys, 24% overlapped with a valley inferred from the D. simulans data. And the MW D. melanogaster sample shared 28% of its diversity valleys with D. simulans. Based on random permutation of valley locations within the genome, the expected overlap due to chance for each of these comparisons is ∼0.3%, and each result cited above significantly exceeds this threshold (P < 0.0001).
While regions of reduced recombination were not included in the above outlier analyses, we nevertheless tested for a correlation between recombination rate and the locations of shared outliers between species. Shared valleys of diversity between MW and D. simulans did not show lower recombination rates than the full analyzed regions, as estimated by broad-scale mapping data via (1.88 × 10−8 vs. 1.69 × 10−8). Although Drosophila is not known to have a hotspot-like pattern of recombination along chromosomes, it is possible that recombination or gene conversion rates are locally reduced across some of the shared diversity valleys, increasing the influence of linked positive or negative selection and potentially influencing fine-scale estimates of . Although we cannot formally exclude background selection as a contributor to these results, this process has not been predicted to strongly reduce diversity in higher-recombination regions of the Drosophila genome. And in light of our divergence correction, selective constraint is not expected to account for the above pattern.
Instead, genes that fall within diversity valleys in both D. melanogaster and D. simulans may have been affected by recent directional selection in both species. Such genes might have contributed to the recent adaptation of both species to a human commensal ecological role. Alternatively they could simply represent isolated loci or dense gene clusters of frequent adaptive importance in general, analogous to the “hotspots of positive selection” detected in primates by Enard et al. (2010). As might be predicted from the single-species results, the 116 diversity valleys shared between MW D. melanogaster and D. simulans include genes related to RNA interference, male and female reproduction, chromatin organization, and regulation of transcription and splicing (Table S25).
Four genes involved in the nuclear pore complex (CG8219, Nup153, Nup214, and Nup358) were identified, consistent with previous data from Nup153 and other nuclear pore genes (Presgraves and Stephan 2007) in a population genetic survey of interactors of the hybrid incompatibility gene Nup96. Another hybrid incompatibility gene, Lhr (Brideau et al. 2006), also contained overlapping diversity valleys between species. Additionally, Hmr (Barbash et al. 2003) appeared on the list of MW HKAl outliers, and OdsH (Ting et al. 1998) was among the diversity ratio outliers. These data are in agreement with the hypothesis that hybrid incompatibilities in Drosophila often result from genes subject to recurrent positive selection.
Genes that contributed to the adaptation of temperate populations to cooler climates may show a low ratio of diversity between temperate and tropical populations. “Diversity ratio valleys” were defined in the same manner as described above. For D. melanogaster, the ratio of nucleotide diversity between the RAL sample and the MW sample was assessed. For D. simulans, the ratio of diversity was compared between the two U.S. strains on one hand (sim 4/6 and w501) and the three lines from Africa/Madagascar (MD106TS, MD199S, and C167.4) on the other. The D. simulans data thus consisted of very small samples and relatively low coverage. However, most 5-kbp windows met the threshold of having at least 2500 sites covered by two or more lines in each set.
Despite the limitations of the divided D. simulans data set, this species shared 6% of the diversity ratio valleys observed in D. melanogaster (P < 0.0001 compared to random expectation, calculated by permutation as described above). Thus, some of the same genes may contribute to temperate adaptation in D. melanogaster and D. simulans (Table S26). Cyp6g1 appears on this list, in agreement with the conclusion of Schlenke and Begun (2004) and Schmidt et al. (2010) that the evolution of insecticide resistance has occurred at this locus in D. simulans as well. The 28 genes flagged by shared diversity-ratio valleys between species also included nompC (perception of sound), shep (gravitaxis), Sirt2 (determination of adult life span), and klingon (olfactory learning, long-term memory).
X-linked vs. autosomal polymorphism and divergence:
With regard to X-linked vs. autosomal polymorphism and divergence, we begin by focusing on the ancestral-range population (MW). In an equilibrium population with equal numbers of males and females, equal X-linked and autosomal mutation rates, and no natural selection, the expected ratio of X-linked vs. autosomal diversity is 3/4. In the African sample of D. melanogaster, however, we observed X-linked expected heterozygosity to be ∼10% higher than the autosomal average (Table 3), consistent with previous results from sub-Saharan populations (e.g., Kauer et al. 2003; Hutter et al. 2007). The X chromosome shows an even greater excess of divergence (∼20% higher than the autosomes; Table 3). Together, these observations might be taken as evidence for a higher X-linked mutation rate. However, this hypothesis is not supported by empirical data: Keightley et al. (2009) found no significant difference between X-linked and autosomal mutation rates, and the point estimate for the X-linked mutation rate was actually ∼30% lower than the autosomal estimate. Thus, we are left without a plausible interpretation for a large portion of X-linked divergence in D. melanogaster based solely on the strong assumption that substitutions have no fitness effects.
The elevated X-linked polymorphism in African D. melanogaster also remains unexplained by mutational factors. One conceivable demographic explanation for this pattern is an extreme excess of females over males in the breeding population (Charlesworth 2001). However, a simple calculation indicates that ∼40 females per male would be required to generate the observed X-to-autosome (X/A) diversity ratio. In principal, background selection might also lead to elevated X/A diversity ratios (Charlesworth 1996). Deleterious mutations may reach higher frequencies on the autosomes (being purged more efficiently on the X chromosome due to male hemizygosity), and this may lead to a greater autosomal diversity reduction under background selection. However, background selection has not been predicted to strongly influence diversity in regions of the Drosophila genome subject to moderate or high rates of recombination. Perhaps the most plausible explanation for elevated X-linked diversity in African D. melanogaster is the one advanced by Vicoso and Charlesworth (2006). These authors found that differences in recombination rates (generally higher on the X) could account for observed deviations from a 3/4 X/A diversity ratio. Under this view, the autosomes’ lower recombination rates lead to a greater diversity reduction due to linked selection. Importantly, the high levels of autosomal inversion polymorphism observed in many D. melanogaster populations, particularly in Africa (Aulard et al. 2002), may exacerbate recombination rate differences between the X and the autosomes in natural populations, beyond what would be indicated by laboratory mapping experiments.
Relative to the African population, the North American sample showed a very different pattern of X-linked vs. autosomal variation. Here, the X/A diversity ratio was 0.67, with the North American sample retaining only 47% of the X-linked diversity present in the ancestral range population, but retaining 76% of the autosomal diversity observed in the African population [consistent with previous findings (Kauer et al. 2003; Hutter et al. 2007)].
The difference between the North American and African populations in the ratio of X-linked vs. autosomal variation was assessed using the ratio of X/A diversity ratios, defined as (RAL X-linked π/RAL autosomal π)/(MW X-linked π/MW autosomal π). This statistic will be equal to one if both populations have the same ratio of X-linked vs. autosomal nucleotide diversity. Instead, this ratio was equal to 0.61 for the empirical data. Demographic events such as population bottlenecks or founder events may lead to a disproportionate reduction in X-linked diversity (Wall et al. 2002; Pool and Nielsen 2008). Pool and Nielsen (2008) examined a model of founder events with multiple mating, finding that such histories could produce lower X/A diversity ratios than population bottlenecks. However, the lowest ratio observed in that study for any demographic parameter combination was 0.67. Further, the effect of African admixture, if selectively neutral, would be to bring the RAL sample’s X/A diversity ratio closer to 1. Hence, proposed neutral demographic models cannot account for X-linked and autosomal variation in the non-African sample.
One explanation for the X chromosome’s stronger diversity reduction in the non-African population is a stronger effect of hitchhiking on the X chromosome relative to the autosomes. The X chromosome’s hemizygosity may allow more recessive (and even underdominant) beneficial mutations to become visible to selection (Charlesworth et al. 1987; Orr and Betancourt 2001; Betancourt et al. 2004). Alternatively, even if beneficial mutations have fixed at similar frequencies on the X chromosome and the autosomes, a higher proportion of X-linked sweeps may have been driven by newly occurring variants (hard sweeps), whereas relatively more autosomal adaptation may have occurred via natural selection on standing genetic variation (“soft sweeps,” with more than one haplotype linked to the beneficial mutation). An additional possibility is that African introgression into the U.S. population may have been more extensive on the autosomes than on the X chromosome, potentially due to stronger or more efficient selection against African X chromosomes entering temperate American environments, mirroring the interpretation of Kauer et al. (2003) in their study of gene flow into Africa.
Conclusion
Our results are consistent with a significant role for positive selection in shaping patterns of polymorphism in Drosophila. This threatens to undermine the rationale for conducting standard demographic inference in this species, since the effects of natural selection may skew demographic parameters estimated under the assumption of selective neutrality. Indeed, a strong case could be made to place natural selection and population history on equal footing in modeling the dynamics shaping patterns of genomic variation in Drosophila. For species with large population sizes, it may prove essential to estimate parameters of selection and demography jointly, to have plausible inferences regarding either of these processes. The recurrent evidence of nonneutral evolution in our data stands in stark contrast to the view that human genomic variation can largely be explained without invoking hitchhiking (Hernandez et al. 2011 and references therein). One important objective for comparative population genomic analyses (along with theoretical and simulation studies) is investigation of the extent to which differences among species in the genome-wide prevalence of recurrent hitchhiking can be explained by population size (Maynard Smith and Haigh 1974; Gillespie 1999). Such investigations offer promising opportunities to address classic questions concerning the nature and impact of selection in shaping levels and patterns of genetic variation. The expansion of population genomic surveys to include geographic sampling on the relevant demographic and ecological scales allows even broader population biological investigations. The merger of the deepening knowledge of structures and functions of genomes (annotation) with full descriptions of genomic variation in natural populations represented by our analyses is an important expansion of the scope and approach of biological research.
Supplementary Material
Acknowledgments
Y.S.S. was supported by National Institutes of Health (NIH) grants R00-GM080099 and R01-GM094402, A.D.K. was supported by the Neukom Institute and Dartmouth College, and C.H.L. was supported by NIH grant HG02942.
Note added in proof: See Corbett-Detig et al. 2012 (pp. 131–137) for a related work. In addition, subsequent to the submission of this article a related paper, Mackay et al. (2012), appeared. While the scopes of that and this article precluded a systematic analysis and integration during the prepublication period, we note two notable differences. We report a finer genomic scale for the correlation between rates of crossing over and polymorphism. And Mackay et al. (2012) report a greater amount of between-species divergence in introns. We think both of these differences are likely attributable to differences in methodology.
Appendix A: Genome Assemblies
Creating MAQ Assemblies
Lanes that passed quality-control filters were subsequently aligned to the BDGP 5 reference genome, using MAQ v 0.6.8 (Li et al. 2008). For nonpaired reads MAQ uses a Bayesian calculation to determine a maximal a posteriori ungapped alignment of each read to the reference genome. To accomplish this task effectively for large problem instances, MAQ implements an approximate matching heuristic based on hash indexes. By default, MAQ indexes the first 24 bp of the read, which are typically the most accurate. The indexing scheme guarantees all alignments with at most two mismatches to the reference genome in the first 24 bp of the read will be evaluated. The hashing scheme MAQ implements allows for the possibly of additional mismatches in the first 24 bp, but the probability that they are evaluated drops off steeply. MAQ identifies and will retain the multiplicity and alignment locations of reads that align with 0 or 1 mismatch to multiple loci. We considered a locus to be unique if all reads used to determine its consensus nucleotide were mapped to only one location. All nonunique loci were masked at this stage from our subsequent analyses of the consensus sequence.
Two MAQ consensus sequences
For each genome, we used MAQ to determine two consensus sequences, diploid and a haploid, from the aligned reads. More specifically, the diploid consensus sequence was created solely to evaluate genetic anomalies and annotate regions of residual heterozygosity for subsequent masking from population genomics analyses. For our diploid consensus sequences, we left the MAQ consensus sequence parameters at their default settings. Since the diploid assemblies are an intermediate datatype used only for genetic quality control, we did not spend time optimizing these parameters.
Ultimately we resolved to create a haploid consensus sequence of each genome for subsequent population genetics analyses. The haploid model is representative of the single genome of a successfully inbred strain. We implemented a haploid consensus sequence or “assembly” by using the prior probability distribution to eliminate the heterozygous outcome from the model selection.
Two postprocessing steps are applied specifically to the haploid consensus sequences. Under the haploid model, true heterozygosity obviously leads to lower quality scores and higher consensus error rates. To prevent this, regions of residual heterozygosity are identified and filtered from the haploid consensus sequences. We also performed a postprocessing of the quality scores to improve both their accuracy and their ability to discriminate. These are described in more detail below. The results across all D. melanogaster lines are shown in Table A1.
Evaluating the Consensus Quality of Haploid MAQ Assemblies
To evaluate the quality of the Drosophila genome assemblies produced by this sequencing method and the MAQ assembly software, we sequenced and assembled two replicate libraries of the reference D. melanogaster strain, ycnbwsp (Adams et al. 2000) (see Table 1). In principle, evaluating the assembly of the genome with known sequence, such as the reference D. melanogaster strain, can provide information regarding the accuracy of the assembly process.
The fact that the sequenced genomes targeted by our project for population genetic analysis are different from the reference genome sequence means that the act of assembling the reference genome to the reference genome’s sequence provides little information on the effect of natural variation on assembly quality. In other words, assembling the reference strain against its own genome does not address all issues and sources of error.
We wanted to simulate the sequencing and assembly of novel D. melanogaster genomes, but in a controlled environment. To accomplish this we introduced “evolutionary divergence” into the reference sequence at rates consistent with expected pairwise divergence values, using the MAQ module fakemut (Li et al. 2008).
An alternative, but flawed, approach would be to introduce the evolutionary divergence into the reads to change the haplotype of the DNA sample. This can be done only by knowing precisely where the reads are located with respect to each other and consequently within their genome. Since this involves an additional layer of simulation and assumption, we deemed the simpler approach to be the best. Rates of divergence were chosen based on average heterozygosity and divergence reported in previous surveys (Pool and Aquadro 2006; Andolfatto 2007; Begun et al. 2007): 0.009 nucleotide substitutional variants, 0.0005 small insertions, and 0.0005 small deletions.
Recall that two ycnbwsp genomic DNA preparations were independently sequenced to our target read length of 36 bp and approximate target coverage of 10-fold redundancy (Table 1). For each ycnbwsp DNA sample we used the method described above to align and assemble the reads into a haploid assembly.
Formally, for each ycnbwsp sample the resulting haploid consensus sequence is denoted s = (s1, … , sn), where s ε (A, C, G, T). For each consensus sequence s there is a corresponding quality-score sequence Q = (Q1, Q2, . . . , Qn), where Q ε Z+ produced by MAQ. Each quality score Qi is defined in terms of the probability that the nucleotide si is incorrect. If the MAQ predicted error probability is εi, then Qi = −10 log10(εi).
These data have two remarkable features: first, the reference sequence is no longer the same as the genome being sequenced. This allows for a greater possibility of alignment error. Second, the sequence of the DNA sample is known, allowing us to evaluate assembly errors. We analyzed the accuracy of the consensus quality scores of these two assemblies against the “diverged” reference sequence upon which MAQ built them.
Empirical quality analysis
A mismatch to this modified or diverged high-quality BDGP 5 reference genome was considered an error. For each assembled ycnbwsp genome we ranked the consensus nucleotides by their MAQ quality scores and defined quantiles on this ranking. Within a quantile K, let errorK be the number of observed errors. The size of each quantile was expanded until errorK was greater than a threshold parameter t chosen to provide a good nonzero estimate of the quantile-specific error rate estimate . An empirical consensus quality score is computed from the observed error rate in each quantile, using . Similarly, the MAQ predicted consensus quality score Q for a quantile K was determined from the expected value of the MAQ predicted error rate for that quantile εK. More specifically and . Figure A5 plots the empirical quality score vs. the MAQ predicted consensus quality score Q for each quantile.
Figure A5 .
Empirical quality (y-axis) vs. the consensus quality score Q (x-axis) from the MAQ assemblies of both ycnbwsp assemblies (one purple and one orange). The ideal relationship between the idealized quality score and the empirical quality is shown with the red line. The final recalibrated consensus quality scores for both ycnbwsp assemblies are plotted in green and blue. The recalibrated consensus quality scores show improved accuracy as they are much closer to the ideal relationship. See text.
The comparison in Figure A5 indicates that the MAQ consensus quality scores are optimistic in this setting. MAQ includes a model parameter designed to compensate for dependent errors in the aligned data at a locus. The observed result, however, is likely a combination of multiple factors: optimistic quality scores from the Illumina basecaller, nonindependence of errors in the aligned data at a locus, and alignment errors not modeled by MAQ. To address these issues, we implemented an empirical approach to recalibrate the raw MAQ consensus quality scores.
Quality Score Recalibration
Following Ewing and Green (1998; Ewing et al. 1998), we can characterize quality scores in two ways. The accuracy of a quality score assignment measures how different the observed error rate is from the error rate predicted by a quality score such as Q. While a single metric is sufficient to characterize the overall accuracy of the quality scores across an assembly, we characterize the quantitative relationship between these over their domains. In Figure A5 we examine the accuracy of MAQ quality scores Q by comparing them to the observed error rate in quantiles.
The utility of an assignment of quality scores in the discrimination of correctly and incorrectly called bases is perhaps more important than accuracy. This discrimination of a quality score assignment measures how effective the quality score’s ranking is of correct basecalls over erroneous basecalls. This characterization is formally discussed in Ewing and Green (1998). As an example, if our assembly has an average error rate of 1 in 100, assigning a quality score of 20 to each nucleotide would be a highly accurate quality score assignment but leave the user of the sequence little ability to select a subset of the data with lower error rates.
Our goal in recalibrating the quality score assignments was to achieve better accuracy while at the same time maintaining, if not improving, the discrimination. Increased accuracy could be achieved by recalibrating the MAQ consensus sequence quality scores, using a monotonic transformation of the values. However, since we were also interested in better discrimination, we developed a richer model of consensus sequence errors specific to our experimental design.
Applying a generalized linear model
We used a generalized linear model (GLM) to determine the recalibrated quality scores. The GLM, as introduced by Nelder and Wedderburn (1972), consists of a single response variable and one or more predictor variables. The response variable is free to change in response to the predictor variables or predictors of the model. The response variable is modeled as a random variable and the predictor variables are nonrandom observations. When the distribution of the response variable can be modeled by a member of the exponential family of probability distributions, maximum-likelihood parameter estimates can be obtained via a standard linear regression technique. In contrast to the more general class of nonlinear models, the GLM is appropriate for large data sets because parameter estimation has the same computational complexity as linear regression.
Two components are needed to adapt the iterative weighted least-squares regression algorithm to perform maximum-likelihood estimation of the model parameters (Charnes et al. 1976). The link function describes the relationship between the expected value of the response variable and additive linear predictor terms. The variance function describes the variance of the predictor variable as a function of its mean. The variance function is completely specified by choosing the probability distribution for modeling the response variable. There is typically a natural link function for most exponential probability distributions.
In our case, we modeled the error rate, for a particular class of base pair, as the parameter of a Poisson distribution. We fitted the model on a table of error counts by classes determined by the predictor variables. The Poisson distribution is frequently chosen for modeling count data of this type (Chambers and Hastie 1992; Dobson 2001).
Modeling a Poisson-distributed response variable:
Our model follows a standard form for a Poisson GLM. Below we demonstrate that the observed data fits this model well. A map s → 1, … , n assigns each base pair to a class. The number of base pair classes is determined by the resolution of the m predictor variables and the structure of the model. Each predictor variable has a finite number of states. Each class represents a unique combination of the states of the m predictor variables (interaction terms were not considered). The error rate εi for the class of nucleotides indexed by the subscript i can be described by the following function of the predictor variables:
| (A1) |
It is typical to represent this in a linear form by applying the link function to both sides of the model:
| (A2) |
Relationship to the null hypothesis:
Our null hypothesis can be written in the form of Equation A2. Under the null hypothesis, that a consensus quality score Qi is correct, the error rate εi for all base pairs labeled with i is a function of Qi:
| (A3) |
If εi is modeled as a Poisson random variable, then we have
| (A4) |
| (A5) |
where β is a proportionality constant. If the quality scores are “correct,” then β = ln(10)/10 and a estimated by maximum likelihood from data would not be significantly different from its expectation, β = ln(10)/10.
Constructing a conservative alternative hypothesis:
The primary predictor of the error rate was the MAQ consensus quality score Q, which by definition is proportional to the log of the “predicted” error rate. We note that the GLM link function for the Poisson error rate parameter is also the log, and thus no transformation of Q is required. A Poisson GLM using Q as the sole predictor variable results in a linear transformation of the quality scores, improving accuracy but, of course, not their ability to discriminate.
Let counti be the number of nucleotides with quality score Qi and errori be the number of those that are errors. Let εi be the error rate of all nucleotides with quality score Qi and note that it denotes the actual error rate. For data based on counts of rare events, we model the number of errors observed in all base pairs with quality score Qi as a draw from a Poisson distribution specified with parameter εi (Dobson 2001). This implies our beginning model has the form
| (A6) |
| (A7) |
where
| (A8) |
Equation A6 represents the model, where Equation A7 is a restatement of the model in terms of the actual observed data. Equation A8 computes the recalibrated quality score Q’ after model parameters have been estimated. This model was considered along with richer ones during the selection process.
Richer alternative hypotheses:
We recognized, as have others, that local assembly errors are also a driver of consensus sequence errors. We evaluated additional predictor variables to extend the model of Equation A6. These are defined in the following list:
Depth is the number of aligned reads covering the nucleotide. We expect lower-depth parts of the assembly to be associated with higher error rates (Bentley et al. 2008; Keightley et al. 2009).
InDel is either 0 or 1 for a nucleotide depending on whether MAQ’s indel detector (indelsoa) covers the nucleotide with a predicted insertion or deletion event.
MinQ1 for a nucleotide is the minimum MAQ quality score of the nucleotide and its two adjacent neighbors.
MinQ5 for a nucleotide is the minimum MAQ quality score in the window within a distance including five nucleotides in either direction along the consensus sequence.
An interior score is defined for each position in a read as the distance to the nearest edge. It follows that each nucleotide in the consensus sequence has a corresponding set of interior scores that overlap it. Three summaries of the overlapping interior scores were computed for each nucleotide. They are illustrated in Figure A1.
MaxIS is the maximum interior score.
AveIS is the average interior score.
SumIS is the sum of interior scores.
Figure A1 .
Interior score MaxIS. An interior score is defined for each position in a read (open boxes) as the distance to the nearest edge. It follows that each nucleotide in the consensus sequence has a corresponding set of interior scores that overlap it. To compute MaxIS, the consensus sequence (shaded boxes) inherits the maximum max() of the interior scores of the overlapping reads. SumIS and AveIS are defined by replacing the max() function with sum() and ave(), respectively.
Parameter estimation, model selection, and model adequacy
For a proposed model, the parameters were estimated using the glm module of the R package for statistical computing (R Development Core Team 2010). Our training data consisted of the two independent ycnbwsp assemblies described above. When evaluating the adequacy of the model during the model selection phase, a cross-validation approach was used whereby the model was trained on one of the two ycnbwsp assemblies and tested on the other.
The base model considered was the initial model using only the MAQ quality score, Q (Equation A7). More descriptive models of the data were considered incrementally, evaluating additional predictor variables and retaining those that significantly improved the fit as well as the models’ ability to discriminate between nucleotides with correct and incorrect assignments. A number of methods are available to determine whether a model adequately explains the data (Chambers and Hastie 1992; Dobson 2001; Cohen and Cohen 2008). Because of the extremely large amount of data in the training set, all of the candidate predictor variables showed significant changes in residual deviance. The remaining criterion used to determine whether a predictor variable should be included was a quantitative improvement in discrimination.
Receiver operator characteristic (ROC) curve:
To evaluate the ability of a proposed quality score assignment to discriminate between nucleotides with correct and incorrect assignments, we used an implementation of the ROC curve. We are ultimately interested in choosing a quality score cutoff to separate high-quality (mostly correct) nucleotides from low-quality (likely incorrect) nucleotides. Our classification of the data is parameterized by quality score cutoff. Two concepts are important in evaluating the effectiveness of a binary classification. In our context, sensitivity measures the ability of our classifier to identify correct nucleotides, while specificity measures the ability of our classifier to identify incorrect nucleotides so they can be filtered from the correct nucleotides. A ROC curve is used to simultaneously evaluate the sensitivity and specificity of a classifier over a range of cutoff parameters (Cohen and Cohen 2008; Hastie et al. 2009). Our ROC curve plots the cumulative coverage vs. the cumulative error rate. Each point on this parametric curve is a quality score cutoff. The area under an ROC curve is interpreted as a quantitative summary in which better classifiers have a larger area. A perfect classification of the data occurs when incorrect nucleotides all have lower quality scores than correct nucleotides. Similarly, in our ROC curve, maximal area is achieved by a quality score assignment that maximizes coverage without errors.
In Figure A2 we assess the base model, using only the MAQ quality score Q, and all of the single variable extensions, using the ROC curve. Surprisingly, the only candidate predictor variable that did not improve the discrimination ability of the MAQ consensus quality score alone was the consensus sequence depth. This is consistent with Figure SA1, which shows a large correlation between depth and MAQ quality score Q.
Figure A2 .
ROCs for the simple models (see text). Cumulative coverage (y-axis) vs. observed error rate (x-axis) is plotted for the model based only on Q (red) and the seven extensions based on the addition of a single (indicated) predictor variable. Note the improvement of the discrimination achieved with several of the extensions.
In using an ROC to evaluate model performance, it is possible that an additional predictor variable will have a significant fit, but will not significantly affect the ROC (Cook 2007). All of the predictor variables evaluated significantly reduced residual deviance from the glm analysis; however, Depth does not improve the ROC. Our objective was to add only additional predictors that improved discrimination ability; hence we did not consider Depth.
Interpretation of parameter estimates:
For a binary predictor variable xj, a fitted model parameter βj can be interpreted in terms of the following ratio of expectations for the Poisson-distributed response variable errori:
| (A9) |
The interpretation is similar when xj is an integer. For each unit increase in the observed value of xj, there is a quantitative increase of exp(βj).
The model correctly represents the binary predictor variable InDel because the ratio of expectations in Equation A9 is well defined and expected to be greater than one. For the integer predictor variables on our list we confirmed the linear relationship between the predictor and the log error rate. The log-linearity property is clearly demonstrated for Q in Figure A5. Figure A3 additionally demonstrates that this property is well reflected in the observed data for the other predictor variables chosen for the final model (see below).
Figure A3 .
Quantile error rates for predictor variables: right, vs. depth; center, vs. MinQ5; and left, vs. MaxIS. The mean empirical quality score (y-axis) was calculated in quantiles large enough to estimate (see text). The quantile mean of predictor variables is plotted on the x-axis.
Improved consensus quality scores
The final form of our Poisson GLM is as follows:
| (A10) |
Given the high correlation with the two groups of predictors (MaxIS, AveIS, SumIS) and (MinQ1, MinQ5), only one was chosen from each group to avoid overfitting. Of the models considered, our final model maximized the area under the curve. Once the final model was determined, the two reference libraries were combined and divided in half for the final training and cross-validation.
As expected, the final model parameters indicated that the detected indel events (InDel) were positively correlated with error rate. It predicted the probability of an error was >14 times greater in the context of an InDel (see Equation A9). All other model parameters were negatively correlated with the error rate. The model chose to weight the consensus quality score (Q) and the minimum proximal quality score (MinQ5) similarly. This penalizes abrupt decreases in depth and proximal decreases in quality. This is consistent with our hypothesis that context, in this case the nominal quality, is associated with assembly errors.
In Figure A4 we evaluate our final model alongside the previously considered models. When comparing two quality score assignments, the one that gives the lower error rate for the same amount of coverage is clearly better in the practical range. The inclusion of simpler models provides details of the relative contribution of each of the predictors to the improved discrimination. MaxIS gives the largest contribution in discrimination. The striking improvement in discrimination of our model over the unprocessed MAQ consensus quality scores Q (Figure A4) validates our approach and indicates that it is possible to use additional assembly information to significantly improve MAQ consensus quality scores.
Figure A4 .
ROC for selected model (cumulative coverage vs. cumulative error rate, parameterized by the quality score cutoff). The final quality score model and three earlier models incorporating the individual components are compared to the initial MAQ quality scores Q.
The improved accuracy of our model is evaluated in Figure A5, where we plot the empirical quality score computed from the estimated error rate determined from the validation data vs. our improved quality scores along with the unprocessed MAQ consensus quality scores Q. Under the ideal scenario of perfect accuracy, the estimated quality score should be equal to the empirical quality score (red line in Figure A5).
Application to RAL and MW genomes
In Figure SA1 we illustrate the linear relationship between depth and quality score. All lines sequenced demonstrated this linear relationship, which is due to the additive effect of evidence on the consensus quality score. We define a quality score depth profile for a DNA sample as the two parameters describing the best fit of Q = α0 + α1 depth. The RAL and MW genomes were normalized to the training profile in a parametric manner by computing each quality score depth profile and applying a linear transformation to the consensus quality scores to give them a profile identical to the training profile. This was done using the glm module of R. Finally, the recalibrated quality score Q’ is computed from the model-predicted εi, using the estimated model parameters β0, … , β4, and predictor variables are computed on the consensus sequence derived from each sampled genome. The standard Phred definition of a quality score Q′ = −log10(ε) is used to assign a recalibrated quality score from the predicted ε. The mean quality score for all genomes decreased upon recalibration.
Implications of improved quality scores
These improved aspects of the data enable more robust downstream population genetics analysis. In any standard population genomic analysis, sequencing errors can critically bias estimations of important population genetic parameters and invalidate tests of statistical hypotheses; e.g., consider the impact of singleton errors on Tajima’s D (Tajima 1989). Therefore, a critical aspect of most genomic analyses, including population genomic analyses, is the ability to censor based on accurate estimates of error. Since its debut (Ewing and Green 1998; Ewing et al. 1998), the Phred scaled quality score has been the most direct and widely accepted way to do this. To better establish this utility on our genomic assemblies, we recalibrated the MAQ consensus quality scores to correctly reflect the error properties. The resulting increased accuracy means that the error properties of the data above a particular Q value are estimated better. Furthermore, by increasing the discrimination of our recalibrated quality scores using additional assembly information, we have increased the amount of data that can be included in analyses at a specified error tolerance. While the specific model developed here is specialized in terms of its technological and biological assumptions, we think the modeling procedure should be useful in different settings.
Appendix B: Annotating Segmental Aneuploidy
Our goal was to determine intervals of segmental aneuplody along the genomes of the sequenced D. melanogaster stocks. We examined the depth of sequenced reads at each position of the genome in each sequenced line to detect copy-number differences among inbred lines. Characteristically, duplications are detected as regions of significantly increased depth, while deletions are inferred based on significantly decreased depth. Following similar results for hybridization-based studies using microarray technology, we used a HMM to segment the genome of each line into regions of euploidy and aneuploidy as we describe below. Our approach was an adaptation of the ergodic model used by Colella et al. (2007) for our specific technology and genetic scenarios.
Compositional Characteristics of Illumina Libraries
In our characterization of segmental aneuplody we considered the ycnbwsp reference genome as euploid or normal copy number, and we were interested in determining deleted or duplicated intervals relative to the reference genome. Our approach was to detect the changes in read depth or expected fold coverage that arise from such an event. An important initial step in annotating regions of aneuplody was to develop a model for euploid read depth.
It has been previously observed that the representation of the library is not uniform across the range of the genomic G+C content. We observed that genomic DNA with %G+C content in the range of 20–60% was well represented in the library. However, as the %G+C gets below 20 or above 65, there is a sharp decrease in the representation of the genomic DNA in the library. This overall effect can be observed in Figure B1. Variation in the library’s compositional profile across samples can also be observed in the differences in the overall %G+C content of the library (Figure SB1).
Figure B1 .
PL(γ) vs γ (see Equation B1). The solid line represents the mean of the G+C profiles of all stocks in the CNV analysis. The dashed lines are ±1 standard deviation.
Library-Specific Sequence Composition Profiles
Since we observed that %G+C content has such a large effect on the expected fold coverage, it is an important factor to consider in modeling expected fold coverage or read depth. To better quantify the local %G+C content effect we estimated representational enrichment in windows as a function of %G+C.
In our setting, all reads have length l = 36 bp. Read depth was measured as the number of reads in windows of width w = 100 bp. A read contributes depth only to the window overlapping its leftmost coordinate. Windows overlapped by l − 1 = 35 bp. The counting scheme and overlap were chosen to make depth measurements conditionally independent given the model parameters observed in the window.
To characterize representational enrichment for each library, we computed a library-specific G+C profile that maps the local G+C content to a rate ratio indicative of how much the local fold coverage is expected to be higher or lower. The specific rate ratio is the ratio of the expected read depth in the window given the local %G+C content γ to the overall expected read depth in a window marginalizing %G+C. Formally, for a library L, its library-specific compositional profile PL maps the local %G+C content γ to the following ratio of expectations:
| (B1) |
The G+C profile for each line was computed using only the autosomal chromosomes to avoid the expected reduction in fold coverage due to a lower representation of the X chromosome in the library (due to mixtures of male and female individuals). The library-specific ratio of X chromosomes to autosomes is estimated in a subsequent phase. Figure B2 shows our library-specific autosomal compositional profiles for all libraries.
Figure B2 .
PL(γ) vs γ (see Equation B1). The lines represent the library specific G+C profiles of all stocks in the CNV analysis.
Modeling Read Depth
To model read depth, it was natural to appeal again to GLMs that utilize the analytical methods of linear regression to obtain maximum-likelihood parameter estimates for models with a single response variable, in our case read depth, and one or more predictor variables (Nelder and Wedderburn 1972). The theoretical distribution for the number of reads falling in an interval along the genome is modeled as a Poisson random variable (Lander and Waterman 1988). It is also standard practice to use the Poisson distribution in the GLM framework to model count data (Dobson 2001). Our final model (see below) incorporates estimated deviations from this ideal Poisson model.
Utilizing our library-specific compositional profile PL, we constructed a simple model to explain the windowed read count d = {d1, … , dn}. We modeled the dependence of the Poisson parameter λi from which di is drawn on the explanatory variables γ and X as
| (B2) |
Here we introduce the Boolean explanatory variable X that is 1 if the window falls on the X chromosome and 0 if the window falls on an autosomal chromosome. Recall that PL(γi) is the rate ratio for window i with %G+C = γi as given by Equation B1. The estimated parameter β0 should be interpreted as the natural log of the expected windowed read depth across the entire sample; this is also the denominator of Equation B1. Thus, the expected read depth for a window is simply the overall expected read depth multiplied by a %G+C enrichment factor and potentially an X chromosome depletion factor. The resulting GLM for this model is
| (B3) |
The model parameters of Equation B3 were estimated for each library, using the glm package in R.
The X/A Ratios
For the binary predictor variable X, the fitted model parameter βX can be interpreted in terms of the following ratio of rates for the Poisson-distributed response variable windowed read depth on the X chromosome and on the autosomes (A).
| (B4) |
We can use Equation B4 as an estimate for the library-specific X/A ratio. Figure B3 shows our estimates of the X/A ratio across all stocks; as expected, the values fell between 1 (all females) and 0.75 (one-half females).
Figure B3 .
X/A ratios. Here we plot estimated for all stocks. Values fall in the expected range between 1 (all females) and 3/4 (50% males).
Additional Features
We developed a more descriptive model by including additional terms to incorporate information about the alignment as well as the reference libraries. This more descriptive model adds two alignment features computed from the MAQ output. We considered the number of single-nucleotide differences from the reference genome sequence in the window (SNPs). We also considered the number small indels computed by the MAQ indelsoa program (indels). These are summarized in Table B1.
Table B1 . Estimates of additional model parameters across all stocks.
| Description of explanatory variable V | Median estimate of βV | Median exp(βV) |
|---|---|---|
| MAQ SNP count | −0.02 | 0.98 |
| MAQ indel | −0.15 | 0.86 |
| Log reference depth | 0.16 | NA |
| Log sample depth | 0.81 | NA |
We noted that SNPs had a small but noticeable effect. The median estimate across all stocks was a 2% reduction for each SNP when there were fewer than four of them in a window. For the few windows where there were four or more SNPs read depth was low and the windows were treated as missing data. For windows with indels predicted by MAQ the median estimate across all stocks was a 14% reduction in read depth per indel per window.
Aberrant Stocks and Overdispersion
Two quality-control steps were used to remove stocks from subsequent analysis of segmental aneuploidy. The first criterion was to identify stocks by eye with compositional profiles that were very atypical. This yielded the three stocks shown in Figure B4 that all showed a significant departure from the canonical profile of Figure B1.
Figure B4 .
Shown here are the aberrant library compositional profiles PL(γ) vs γ (see Equation B1) for RAL-379, RAL-313, and MW9-1. These were removed from subsequent analysis of copy-number variation.
The second phase involved estimating a dispersion parameter η for each stock (Figures B5 and B6). Under the ideal model, the mean λ and variance σ2 of the windowed read depth should be the same. This follows from the Poisson distribution. Unfortunately, this is not the case. However, we do find that the variance in read depth can be approximated well for a library as a function of the mean using σ2 = ηλ, as shown in Figure B5. For each particular library, the dispersion parameter η was then estimated by regression through the origin. Our estimates for η ranged from 1.6 on the low end to 5.0 on the high end. As we describe in more detail later, in addition to being an important quality control indicator, these dispersion parameters can be useful for implementing robust emission probability distributions for the HMM.
Figure B5 .
Variance in windowed read depth (y-axis) vs the predicted mean read depth (x-axis) for RAL-689 using Equation B3. Overdispersion was quantified by estimating a dispersion parameter η where σ2 = ηλ. The dispersion parameter η was estimated by regression through the origin. In the case of RAL-689, η = 2.03.
Figure B6 .
Dispersion parameter (η) estimates for all stocks. The stocks removed due to aberrant G+C profiles are labeled and cluster above η = 4. Using that information as an empirical guideline, all stocks with η > 4 were removed from subsequent analyses on the basis of overdispersion.
Our second criterion for removing stocks from the analysis was to define a threshold η = 4. This threshold was chosen because it was the lowest cutoff required to eliminate all stocks identified by phase one. As shown in Figure B6, the threshold of η = 4 eliminated the three phase-one stocks as well as four stocks that were not identified by phase one. These additional four stocks were also removed from the analysis on the basis of overdispersion.
Segmentation Using a Hidden Markov Model
To find and annotate regions of segmental aneuploidy, a four-state HMM was used. The path of hidden states through this HMM that maximized the probability of the observed read depth under the model is the basis for the annotation segmenting the genome into regions of euploidy and aneuploidy.
Operationally, we defined euploidy as copy-number 1. While two copies of each chromosome exist, they have been inbred to identity. Aneuploidy in this context refers to deletions and duplications of a segment on both copies of the inbred chromosome. Further aneuploidy is defined with respect to euploid regions of the reference.
Hidden states
Our HMM has four hidden states (Table B2). State 1 corresponds to the default state of euploidy, or single copy number. State 0 corresponds to a deletion, state 2 corresponds to a duplication of the interval, and state 3 was a catchall for segments of high copy number.
Table B2 . The hidden states of the HMM, their biological interpretation, and the mean and variance parameters used for the negative binomially distributed emission probabilities of observed read count.
| Hidden state (copy no.) | Emission probability | Description |
|---|---|---|
| 1 | NB(λ, ηλ) | Normal (euploid) |
| 0 | NB(ελ, εηλ) | Deletion |
| 2 | NB(2λ, 2ηλ) | Duplication |
| 3 | NB(3λ, 3ηλ) | High copy no. |
Emission probabilities
Rather than using the theoretically ideal Poisson distribution, a negative binomial distribution was used to account for the additional dispersion observed in the read depth. It is convenient to specify the two-parameter negative binomial distribution in terms of a mean λ and the ratio of the variance to the mean η = σ2/λ. The probability mass function for this parameterization is
| (B5) |
where the classic parameters p and r are stated in terms of the mean λ and a dispersion parameter η:
| (B6) |
The particular utility of the negative binomial distribution for approximating read depth and why we preferred it to the Poisson distribution is illustrated in Figure SB2. We compare the histogram of read depth for the ycnbwsp1 reference library to two probability density functions estimated by maximum likelihood from the same data. The negative binomial more closely approximates the observed distribution. Figure B7 shows the linear relationship between mean and variance for all lanes sequenced in the data set.
Figure B7 .
Scatterplot showing the association of the variance with the mean of read depth.
In the context of our HMM, the mean read depth λ for a particular stock is modeled as a window-specific parameter, using the observable dependent variables for that window. The dispersion parameter ν is assumed to be constant for a stock across all windows.
Transition probabilities
The transition matrix for our HMM is completely specified by two parameters, the frequency of an aneuploidy fa and the mean length of an aneuploidy la. The transition matrix of hidden states between adjacent windows i and j is given by
| (B7) |
This transition matrix implies that the length distributions of excursions from the euploid state will be modeled by the geometric distribution where la is the expected length of a segmental aneuploidy.
Determining an optimal sequence of states
We are interested in finding the sequence of hidden states π* that maximizes the probability of the observed read-depth data r under the model with parameters θ:
The standard Viterbi algorithm, a tabular approach to computing the optimum probability using dynamic programming, was applied to each stock. A subsequent traceback of the dynamic programming table yielded the optimal sequence of hidden states π*.
Scoring segmental aneuploidy:
Potential aneuploidy events were defined as excursions from the reference state 1 with a duration greater than a minimum length threshold lmin observed in the state sequence π* given by the Viterbi algorithm. Thus, each potential aneuploidy event is characterized by an interval from i to j with copy number k. We associated each aneuploidy event with the following likelihood ratio:
| (B8) |
This posterior probability pa compares the evidence for the region uniformly being in the hidden state k to the possibility that the region has a uniform copy number other than k. To determine a convenient score by which to filter and rank aneuploidy events we computed a Phred-scaled quality score, assuming that the error probability is the probability that one of the other models is correct:
| (B9) |
This allowed us to ignore certain events below a user-defined threshold. It is also used to determine the frequency of an aneuploidy event across multiple samples.
Model calibration:
The parameter lmin was adjusted for an acceptable predicted rate of false discovery. The first methodology employed was to use Monte Carlo sampling of observed read depth from the HMM and subsequent scoring to determine a false discovery rate for the chosen parameter settings. We increased lmin to 4 at which no false positive deletions were reported and the expected rate of false positive duplications was <0.5 per genome. This false positive rate was further decreased by more than one-third by conditioning on a minimum Q of 30.
The reference genome also allowed us to provide calibration of our user-settable model parameters in an attempt to minimize the number of false positives. We applied the HMM to the reference genome and then examined the detected aneuploidies. In this manner we determined a small number of apparent positives on the reference sequence with lmin set at 4, including chorion genes that are expected to be quantitatively amplified in our libraries constructed from genomic DNAs isolated from collections of both females and males (Spradling 1981).
Genotyping CNVs Across Lines
For frequency-spectrum analyses, we genotyped the discovered CNVs using a more sensitive method. For each CNV interval, we calculated the most probable copy number (0, 1, 2, or 3) in each line. Equation B9 was used to quantify our confidence in the genotype as a quality score Q. When any individual line was assigned a genotype with Q < 30, this line was considered missing data for this CNV. Thus, when determining the minor allele frequency of a discovered CNV, genotyping calls that had a quality score <30 were ignored in both the numerator and the denominator of the calculation.
Clustering independently discovered CNVs
Due to the fact that CNVs were discovered independently in each inbred line, the same polymorphism may not always be given identical breakpoints. To summarize individual events, we clustered the discovered CNVs, using a single linkage clustering. Two CNVs in different lines were identified if at least 50% of one interval overlapped with the other and vice versa. We believe that this process was effective at combining CNV calls corresponding to the same polymorphism, since the genotypes agreed ∼97% of the time for identified pairs. To facilitate length and frequency analyses of this data set, each clustered CNV was assigned the median length and the median minor allele frequency (as calculated above) of its respective component CNVs.
Overlap with earlier studies
We compared our set of CNVs to three prior data sets collected using different D. melanogaster strains (Dopman and Hartl 2007; Emerson et al. 2008; Cridland and Thornton 2010). Two of these previous studies used microarray technologies to detect CNVs, one with a spotted cDNA array (Dopman and Hartl 2007) and one with an Affymetrix tiling array (Emerson et al. 2008). While the comparison of CNV ascertainment on different strains with different platforms is problematic, we did expect that our data would have greater resolution because the entire genome is covered. We identified 76 of the 438 duplications (17.4%) and 67 of the 1107 deletions (6.1%) called by Dopman and Hartl (2007) in our own data and 280 of the 2211 duplications (12.7%) and 312 of the 1427 deletions (21.9%) called by Emerson et al. (2008) in our own data. The third data set used low-coverage paired-end data to detect possible duplications and inversions (Cridland and Thornton 2010). We looked for overlap between our duplication calls and tandem duplications detected in the Cridland and Thornton data, as nontandem duplication breakpoints could not be inferred from the data, and this data set does not contain any deletion calls. To remove likely false positives or nonduplication events from the Cridland and Thornton data, we examined only putative tandem duplications <10 kbp in length. Duplications in our data set overlapped with 173/477 (39.9%) of these putative tandem duplications in the Cridland and Thornton set—a considerably higher number than in the two array data sets.
Validating (discovered) CNVs using paired-end data
For two of the lines, RAL-437 and RAL-765, we had ∼3.5× and ∼2.8× of paired-end coverage, respectively, with 45-bp reads and average insert sizes of ∼250 bp. Since paired-end reads provide an alternative method of detecting CNVs (Korbel et al. 2007), these data were used to provide independent validation of discovered duplication and deletion polymorphisms. Paired ends were mapped to the reference genome using MAQ. Since few paired ends (<2%) mapped >350 bp away from one another, read pairs mapping at least this far apart and spanning a putative deletion were considered to confirm the deletion, while pairs having one read mapping outside of the putative deletion and one read mapping within the deletion were considered to refute it. Of the putative deletions meeting these criteria, 159 of 177 events discovered in RAL-437 were confirmed; in RAL-765, 133 of 150 were confirmed. To correct for spurious confirmation, we repeated this process 1000 times, randomizing locations, and found that on average 6.4/226 and 5.9/205 of the permuted deletions were confirmed by paired ends in RAL-437 and RAL-765, respectively. Subtracting the estimated number of spurious confirmations, we estimate that ∼86% (152.6/177) and ∼85% (127.1/150) of the discovered deletions called in RAL-437 and RAL-765 are true deletions. We also sought to confirm discovered duplications by examining read pairs exhibiting an abnormal inferred insert size [for nontandem duplications (Korbel et al. 2007)] or orientation [for tandem duplications (Cooper et al. 2008)]. Using the data, we confirmed 66 of 95 duplications discovered in RAL-437 and 64 of 87 duplications in RAL-765. We then randomized the coordinates of these putative duplications 1000 times, confirming 5.25/95 and 9.77/87 permuted duplications on average and yielding corrected confirmation rates of 64% (60.75/95) and 62% (54.23/87). As with deletions, we wished to determine the fraction of rejected duplications. However, because there is no way to reject a duplication using paired-end data, we assumed that the same fraction of duplications could be confirmed or rejected as deletions (78% for RAL-437 and 73% for RAL-765). After correcting the number of confirmable duplications, we estimate the true positive rates of duplications discovered in RAL-437 and RAL-765 to be 82% and 85%, respectively—nearly as accurate as the putative deletions. If we increase our minimum-length cutoff to l = 7, or ∼500 bp, the confirmation rate increases to ∼90% for both duplications and deletions. Thus this data set provides a largely independent and highly accurate representation of the CNVs in natural populations of D. melanogaster.
Footnotes
Communicating editor: R. Nielsen
Sequence data from this article have been deposited in the Bloomington Drosophila Stock Center (see Table 1).
Literature Cited
- Adams M. D., Celniker S. E., Holt C. A., Evans J. D., Gocayne J. D., et al. , 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195 [DOI] [PubMed] [Google Scholar]
- Aguadé M., Miyashita N., Langley C. H., 1989. Reduced variation in the yellow-achaete-scute region in natural populations of Drosophila melanogaster. Genetics 122: 607–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguadé M., Meyers W., Long A. D., Langley C. H., 1994. Single-strand conformation polymorphism analysis coupled with stratified DNA sequencing reveals reduced sequence variation in the su(s) and su(wa) regions of the Drosophila melanogaster X chromosome. Proc. Natl. Acad. Sci. USA 91: 4658–4662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi H., 1995. Inferring weak selection from patterns of polymorphism and divergence at “silent” sites in Drosophila DNA. Genetics 139: 1067–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi H., 1996. Molecular evolution between Drosophila melanogaster and D. simulans: reduced codon bias, faster rates of amino acid substitution, and larger proteins in D. melanogaster. Genetics 144: 1297–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexeev A., Mazin A., Kowalczykowski S. C., 2003. Rad54 protein possesses chromatin-remodeling activity stimulated by the Rad51-ssDNA nucleoprotein filament. Nat. Struct. Biol. 10: 182–186 [DOI] [PubMed] [Google Scholar]
- Andolfatto P., 2001. Contrasting patterns of X–Linked and autosomal nucleotide variation in Drosophila melanogaster and Drosophila simulans. Mol. Biol. Evol. 18: 279–290 [DOI] [PubMed] [Google Scholar]
- Andolfatto P., 2007. Hitchhiking effects of recurrent beneficial amino acid substitutions in the Drosophila melanogaster genome. Genome Res. 17: 1755–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto P., Kreitman M., 2000. Molecular variation at the In(2L)t proximal breakpoint site in natural populations of Drosophila melanogaster and D. simulans. Genetics 154: 1681–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto P., Wall J. D., Kreitman M., 1999. Unusual haplotype structure at the proximal breakpoint of In(2L)t in a natural population of Drosophila melanogaster. Genetics 153: 1297–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto P., Depaulis F., Navarro A., 2001. Inversion polymorphisms and nucleotide variability in Drosophila. Genet. Res. 77: 1–8 [DOI] [PubMed] [Google Scholar]
- Andolfatto P., Wong K. M., Bachtrog D., 2011. Effective population size and the efficacy of selection on the X chromosomes of two closely related Drosophila species. Genome Biol. Evol. 3: 114–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aquadro C. F., Lado K. M., Noon W. A., 1988. The rosy region of Drosophila melanogaster and Drosophila simulans. I. Contrasting levels of naturally occurring DNA restriction map variation and divergence. Genetics 119: 875–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aquadro C. F., Jennings R. M., Bland M. M., Laurie C. C., Langley C. H., 1992. Patterns of naturally occurring restriction map variation, dopa decarboxylase activity variation and linkage disequilibrium in the Ddc gene region of Drosophila melanogaster. Genetics 132: 443–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulard S., David J. R., Lemeunier F., 2002. Chromosomal inversion polymorphism in Afrotropical populations of Drosophila melanogaster. Genet. Res. 79: 49–63 [DOI] [PubMed] [Google Scholar]
- Ayroles J. F., Carbone M. A., Stone E. A., Jordan K. W., Lyman R. F., et al. , 2009. Systems genetics of complex traits in Drosophila melanogaster. Nat. Genet. 41: 299–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D., 2003. Adaptation shapes patterns of genome evolution on sexual and asexual chromosomes in Drosophila. Nat. Genet. 34: 215–219 [DOI] [PubMed] [Google Scholar]
- Bachtrog D., 2005. Sex chromosome evolution: molecular aspects of Y-chromosome degeneration in Drosophila. Genome Res. 15: 1393–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D., Charlesworth B., 2002. Reduced adaptation of a non-recombining neo-Y chromosome. Nature 416: 323–326 [DOI] [PubMed] [Google Scholar]
- Barbash D. A., Siino D. F., Aaron M., Roote J., 2003. A rapidly evolving MYB-related protein causes species isolation in Drosophila. Proc. Natl. Acad. Sci. USA 100: 5302–5307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomé C., Bello X., Maside X., 2009. Widespread evidence for horizontal transfer of transposable elements across Drosophila genomes. Genome Biol. 10: R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F., Buard J., Grey C., Fledel-Alon A., Ober C., et al. , 2010. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327: 836–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer DuMont V. L., Flores H. A., Wright M. H., Aquadro C. F., 2007. Recurrent positive selection at Bgcn, a key determinant of germ line differentiation, does not appear to be driven by simple coevolution with its partner protein Bam. Mol. Biol. Evol. 24: 182–191 [DOI] [PubMed] [Google Scholar]
- Begun D. J., 1996. Population genetics of silent and replacement variation in Drosophila simulans and D. melanogaster: X/autosome differences? Mol. Biol. Evol. 13: 1405–1407 [DOI] [PubMed] [Google Scholar]
- Begun D. J., Aquadro C. F., 1992. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature 356: 519–520 [DOI] [PubMed] [Google Scholar]
- Begun D. J., Aquadro C. F., 1993. African and North American populations of Drosophila melanogaster are very different at the DNA level. Nature 365: 548–550 [DOI] [PubMed] [Google Scholar]
- Begun D. J., Holloway A. K., Stevens K., Hillier L. W., Poh Y.-P., et al. , 2007. Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 5: e310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Yekutieli D., 2001. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 29: 1165–1188 [Google Scholar]
- Bentley D. R., Balasubramanian S., Swerdlow H. P., Smith G. P., Milton J., et al. , 2008. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456: 53–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg I. L., Neumann R., Lam K.-W. G., Sarbajna S., Odenthal-Hesse L., et al. , 2010. PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat. Genet. 42: 859–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt A. J., Presgraves D. C., 2002. Linkage limits the power of natural selection in Drosophila. Proc. Natl. Acad. Sci. USA 99: 13616–13620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt A. J., Kim Y., Orr H. A., 2004. A pseudohitchhiking model of X vs. autosomal diversity. Genetics 168: 2261–2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt A. J., Welch J. J., Charlesworth B., 2009. Reduced effectiveness of selection caused by a lack of recombination. Curr. Biol. 19: 655–660 [DOI] [PubMed] [Google Scholar]
- Bingham P. M., Levis R., Rubin G. M., 1981. Cloning of DNA sequences from the white locus of D. melanogaster by a novel and general method. Cell 25: 693–704 [DOI] [PubMed] [Google Scholar]
- Blow J. J., Ge X. Q., Jackson D. A., 2011. How dormant origins promote complete genome replication. Trends Biochem. Sci. 36: 405–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenstiel J. P., 2011. Evolutionary dynamics of transposable elements in a small RNA world. Trends Genet. 27: 23–31 [DOI] [PubMed] [Google Scholar]
- Bradley R. K., Roberts A., Smoot M., Juvekar S., Do J., et al. , 2009. Fast statistical alignment. PLoS Comput. Biol. 5: e1000392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braverman J. M., Hudson R. R., Kaplan N. L., Langley C. H., Stephan W., 1995. The hitchhiking effect on the site frequency spectrum of DNA polymorphisms. Genetics 140: 783–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braverman J. M., Lazzaro B. P., Aguadé M., Langley C. H., 2005. DNA sequence polymorphism and divergence at the erect wing and suppressor of sable loci of Drosophila melanogaster and D. simulans. Genetics 170: 1153–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brideau N. J., Flores H. A., Wang J., Maheshwari S., Wang X., et al. , 2006. Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science 314: 1292–1295 [DOI] [PubMed] [Google Scholar]
- Bridges C. B., 1936. The Bar “gene” a duplication. Science 83: 210–211 [DOI] [PubMed] [Google Scholar]
- Bulmer M., 1991. The selection-mutation-drift theory of synonymous codon usage. Genetics 129: 897–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J. J., Macpherson J. M., Sella G., Petrov D. A., 2009 Pervasive hitchhiking at coding and regulatory sites in humans. PLoS Genet. 5: e1000336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caracristi G., Schlötterer C., 2003. Genetic differentiation between American and European Drosophila melanogaster populations could be attributed to admixture of African alleles. Mol. Biol. Evol. 20: 792–799 [DOI] [PubMed] [Google Scholar]
- Cardoso-Moreira M., Emerson J. J., Clark A. G., Long M., 2011. Drosophila duplication hotspots are associated with late-replicating regions of the genome. PLoS Genet. 7: e1002340 [Google Scholar]
- Cardoso-Moreira M. M., Long M., 2010. Mutational bias shaping fly copy number variation: implications for genome evolution. Trends Genet. 26: 243–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza L. L., Bodmer W. F., 1971. The Genetics of Human Populations. W. H. Freeman, San Francisco [Google Scholar]
- Chambers J. M., Hastie T. J., 1992. Statistical Models. Wadsworth & Brooks/Cole, Pacific Grove, CA [Google Scholar]
- Charlesworth B., 1994. The effect of background selection against deleterious mutations on weakly selected, linked variants. Genet. Res. 63: 213–227 [DOI] [PubMed] [Google Scholar]
- Charlesworth B., 1996. Background selection and patterns of genetic diversity in Drosophila melanogaster. Genet. Res. 68: 131–149 [DOI] [PubMed] [Google Scholar]
- Charlesworth B., 2001. The effect of life-history and mode of inheritance on neutral genetic variability. Genet. Res. 77: 153–166 [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Langley C. H., Stephan W., 1986. The evolution of restricted recombination and the accumulation of repeated DNA sequences. Genetics 112: 947–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., Coyne J. A., Barton N. H., 1987. The relative rates of evolution of sex chromosomes and autosomes. Am. Nat. 130: 113–146 [Google Scholar]
- Charlesworth B., Morgan M. T., Charlesworth D., 1993. The effect of deleterious mutations on neutral molecular variation. Genetics 134: 1289–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D., Charlesworth B., Morgan M. T., 1995. The pattern of neutral molecular variation under the background selection model. Genetics 141: 1619–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth J., Eyre-Walker A., 2008. The McDonald–Kreitman test and slightly deleterious mutations. Mol. Biol. Evol. 25: 1007–1015 [DOI] [PubMed] [Google Scholar]
- Charnes A., Frome E. L., Yu P. L., 1976. The equivalence of generalized least squares and maximun likelihood estimates in the exponential family. J. Am. Stat. Assoc. 71: 169–171 [Google Scholar]
- Cherbas L., Willingham A., Zhang D., Yang L., Zou Y., et al. , 2011. The transcriptional diversity of 25 Drosophila cell lines. Genome Res. 21: 301–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiolo I., Minoda A., Colmenares S. U., Polyzos A., Costes S. V., et al. , 2011. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell 144: 732–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland W. S., 1979. Robust locally weighted regression and smoothing scatterplots. J. Am. Stat. Assoc. 74: 829 [Google Scholar]
- Clowers K. J., Lyman R. F., Mackay T. F. C., Morgan T. J., 2010. Genetic variation in senescence marker protein-30 is associated with natural variation in cold tolerance in Drosophila. Genet. Res. 92: 103–113 [DOI] [PubMed] [Google Scholar]
- Cohen Y., Cohen J. Y., 2008. Statistics and Data with R an Applied Approach Through Examples. Wiley, Chichester, UK [Google Scholar]
- Colella S., Yau C., Taylor J. M., Mirza G., Butler H., et al. , 2007. QuantiSNP: an objective Bayes hidden-Markov model to detect and accurately map copy number variation using SNP genotyping data. Nucleic Acids Res. 35: 2013–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad D. F., Bird C., Blackburne B., Lindsay S., Mamanova L., et al. , 2010. Mutation spectrum revealed by breakpoint sequencing of human germline CNVs. Nat. Genet. 42: 385–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook N. R., 2007. Use and misuse of the receiver operating characteristic curve in risk prediction. Circulation 115: 928–935 [DOI] [PubMed] [Google Scholar]
- Coop G., Wen X., Ober C., Pritchard J. K., Przeworski M., 2008. High-resolution mapping of crossovers reveals extensive variation in fine-scale recombination patterns among humans. Science 319: 1395–1398 [DOI] [PubMed] [Google Scholar]
- Cooper G. M., Zerr T., Kidd J. M., Eichler E. E., Nickerson D. A., 2008. Systematic assessment of copy number variant detection via genome-wide SNP genotyping. Nat. Genet. 40: 1199–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett-Detig R. B., Cardeno C., Langley C. H., 2012. Sequence-based detection and breakpoint assembly of polymorphic inversions. Genetics 192: 131–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cridland J. M., Thornton K. R., 2010. Validation of rearrangement break points identified by paired-end sequencing in natural populations of Drosophila melanogaster. Genome Biol. Evol. 2: 83–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daborn P. J., Yen J. L., Bogwitz M. R., Le Goff G., Feil E., et al. , 2002. A single p450 allele associated with insecticide resistance in Drosophila. Science 297: 2253–2256 [DOI] [PubMed] [Google Scholar]
- Dean M. D., Ballard J. W. O., 2004. Linking phylogenetics with population genetics to reconstruct the geographic origin of a species. Mol. Phylogenet. Evol. 32: 998–1009 [DOI] [PubMed] [Google Scholar]
- De Luca M., Roshina N. V., Geiger-Thornsberry G. L., Lyman R. F., Pasyukova E. G., et al. , 2003. Dopa decarboxylase (Ddc) affects variation in Drosophila longevity. Nat. Genet. 34: 429–433 [DOI] [PubMed] [Google Scholar]
- Dewey C. N., 2007. Aligning multiple whole genomes with Mercator and MAVID. Methods Mol. Biol. 395: 221–236 [DOI] [PubMed] [Google Scholar]
- Dobson A. J., 2001. An Introduction to Generalized Linear Models. Chapman & Hall/CRC, London [Google Scholar]
- Dopman E. B., Hartl D. L., 2007. A portrait of copy-number polymorphism in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 104: 19920–19925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostert C., Jouanguy E., Irving P., Troxler L., Galiana-Arnoux D., et al. , 2005. The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of Drosophila. Nat. Immunol. 6: 946–953 [DOI] [PubMed] [Google Scholar]
- Emerson J. J., Cardoso-Moreira M., Borevitz J. O., Long M., 2008. Natural selection shapes genome-wide patterns of copy-number polymorphism in Drosophila melanogaster. Science 320: 1629–1631 [DOI] [PubMed] [Google Scholar]
- Enard D., Depaulis F., Roest Crollius H., 2010. Human and non-human primate genomes share hotspots of positive selection. PLoS Genet. 6: e1000840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B., Green P., 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8: 186–194 [PubMed] [Google Scholar]
- Ewing B., Hillier L., Wendl M. C., Green P., 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8: 175–185 [DOI] [PubMed] [Google Scholar]
- Falconer D. S., 1989. Introduction to Quantitative Genetics. Longmans Green/John Wiley & Sons, Harlow, Essex, UK/New York [Google Scholar]
- Fan Q. Q., Petes T. D., 1996. Relationship between nuclease-hypersensitive sites and meiotic recombination hot spot activity at the HIS4 locus of Saccharomyces cerevisiae. Mol. Cell. Biol. 16: 2037–2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay J. C., Wyckoff G. J., Wu C.-I., 2001. Positive and negative selection on the human genome. Genetics 158: 1227–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa F., Günther E., Klein J., 1988. MHC polymorphism pre-dating speciation. Nature 335: 265–267 [DOI] [PubMed] [Google Scholar]
- Fisher R. A., 1922. On the dominance ratio. Proc. R. Soc. Edinb. 42: 321–341 [Google Scholar]
- Fiston-Lavier A.-S., Singh N. D., Lipatov M., Petrov D. A., 2010. Drosophila melanogaster recombination rate calculator. Gene 463: 18–20 [DOI] [PubMed] [Google Scholar]
- Ford M. J., Aquadro C. F., 1996. Selection on X-linked genes during speciation in the Drosophila athabasca complex. Genetics 144: 689–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie J. H., 1994. The Causes of Molecular Evolution. Oxford University Press, New York [Google Scholar]
- Gillespie J. H., 1999. The role of population size in molecular evolution. Theor. Popul. Biol. 55: 145–156 [DOI] [PubMed] [Google Scholar]
- Glinka S., De Lorenzo D., Stephan W., 2006. Evidence of gene conversion associated with a selective sweep in Drosophila melanogaster. Mol. Biol. Evol. 23: 1869–1878 [DOI] [PubMed] [Google Scholar]
- Golding G. B., 1984. The sampling distribution of linkage disequilibrium. Genetics 108: 257–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill P. R., Charlesworth B., Halligan D. L., Andolfatto P., 2005. Patterns of intron sequence evolution in Drosophila are dependent upon length and GC content. Genome Biol. 6: R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill P. R., Halligan D. L., Tomaras D., Charlesworth B., 2007. Reduced efficacy of selection in regions of the Drosophila genome that lack crossing over. Genome Biol. 8: R18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M. W., 2008. Toward a selection theory of molecular evolution. Evolution 62: 255–265 [DOI] [PubMed] [Google Scholar]
- Hahn M. W., Han M. V., Han S.-G., 2007. Gene family evolution across 12 Drosophila genomes. PLoS Genet. 3: e197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie T., Tibshirani R., Friedman J., 2009. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. Springer-Verlag, New York [Google Scholar]
- Hellmann I., Mang Y., Gu Z., Li P., de la Vega F. M., et al. , 2008. Population genetic analysis of shotgun assemblies of genomic sequences from multiple individuals. Genome Res. 18: 1020–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermisson J., Pennings P. S., 2005. Soft sweeps. Genetics 169: 2335–2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez R. D., Kelley J. L., Elyashiv E., Melton S. C., Auton A., et al. , 2011. Classic selective sweeps were rare in recent human evolution. Science 331: 920–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer W.-D., 2007. Biochemistry of eukaryotic homologous recombination. Top. Curr. Genet. 17: 95–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W. G., Robertson A., 1966. The effect of linkage on limits to artificial selection. Genet. Res. 8: 269–294 [PubMed] [Google Scholar]
- Hudson R. R., 1990. Gene genealogies and the coalescent process, pp. 1–44 in Oxford Surveys in Evolutionary Biology, edited by D. Futuyma and J. Antonovics. Oxford University Press, Oxford, pp. 1–44 [Google Scholar]
- Hudson R. R., 2001. Two-locus sampling distributions and their application. Genetics 159: 1805–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. R., 2002. Generating samples under a Wright–Fisher neutral model of genetic variation. Bioinformatics 18: 337–338 [DOI] [PubMed] [Google Scholar]
- Hudson R. R., Kaplan N. L., 1994. Gene trees with background selection. Non-Neutral Evolution: Theories and Data, edited by Golding G. B. Chapman & Hall, New York [Google Scholar]
- Hudson R. R., Kreitman M., Aguade M., 1987. A test of neutral molecular evolution based on nucleotide data. Genetics 116: 153–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. R., Boos D. D., Kaplan N. L., 1992. A statistical test for detecting geographic subdivision. Mol. Biol. Evol. 9: 138–151 [DOI] [PubMed] [Google Scholar]
- Hutter S., Li H., Beisswanger S., De Lorenzo D., Stephan W., 2007. Distinctly different sex ratios in African and European populations of Drosophila melanogaster inferred from chromosomewide single nucleotide polymorphism data. Genetics 177: 469–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioerger T. R., Clark A. G., Kao T. H., 1990. Polymorphism at the self-incompatibility locus in Solanaceae predates speciation. Proc. Natl. Acad. Sci. USA 87: 9732–9735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M., Nanba N., Hasegawa M., Inomata N., Kondo R., et al. , 2009. Seasonal changes in the long-distance linkage disequilibrium in Drosophila melanogaster. J. Hered. 101: 26–32 [DOI] [PubMed] [Google Scholar]
- Itsara A., Cooper G. M., Baker C., Girirajan S., Li J., et al. , 2009. Population analysis of large copy number variants and hotspots of human genetic disease. Am. J. Hum. Genet. 84: 148–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys A. J., Kauppi L., Neumann R., 2001. Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nat. Genet. 29: 217–222 [DOI] [PubMed] [Google Scholar]
- Johnson-Schlitz D. M., Engels W. R., 1993. P-element-induced interallelic gene conversion of insertions and deletions in Drosophila melanogaster. Mol. Cell. Biol. 13: 7006–7018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan K., Carbone M., Yamamoto A., Morgan T., Mackay T., 2007. Quantitative genomics of locomotor behavior in Drosophila melanogaster. Genome Biol. 8: R172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N. L., Hudson R. R., Langley C. H., 1989. The “hitchhiking effect” revisited. Genetics 123: 887–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D., 2003. The UCSC Genome Browser database. Nucleic Acids Res. 31: 51–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauer M. O., Dieringer D., Schlotterer C., 2003. A microsatellite variability screen for positive selection associated with the “Out of Africa” habitat expansion of Drosophila melanogaster. Genetics 165: 1137–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley P. D., Trivedi U., Thomson M., Oliver F., Kumar S., et al. , 2009. Analysis of the genome sequences of three Drosophila melanogaster spontaneous mutation accumulation lines. Genome Res. 19: 1195–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinan A., Reich D., 2010. Human population differentiation is strongly correlated with local recombination rate. PLoS Genet. 6: e1000886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W. J., 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern A. D., Begun D. J., 2008. Recurrent deletion and gene presence/absence polymorphism: telomere dynamics dominate evolution at the tip of 3L in Drosophila melanogaster and D. simulans. Genetics 179: 1021–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern A. D., Jones C. D., Begun D. J., 2002. Genomic effects of nucleotide substitutions in Drosophila simulans. Genetics 162: 1753–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko P. V., Alekseyenko A. A., Schwartz Y. B., Minoda A., Riddle N. C., 2010. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 471: 480–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff C., Xi H., Li C., Lee S., Xu J., et al. , 2009. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell 138: 1137–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klovstad M., Abdu U., Schüpbach T., 2008. Drosophila brca2 is required for mitotic and meiotic DNA repair and efficient activation of the meiotic recombination checkpoint. PLoS Genet. 4: e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knibb W. R., Oakeshott J. G., Gibson J. B., 1981. Chromosome inversion polymorphisms in Drosophila melanogaster. I. Latitudinal clines and associations between inversions in Australasian populations. Genetics 98: 833–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolaczkowski B., Hupalo D. N., Kern A. D., 2011a Recurrent adaptation in RNA interference genes across the Drosophila phylogeny. Mol. Biol. Evol. 28: 1033–1042 [DOI] [PubMed] [Google Scholar]
- Kolaczkowski B., Kern A. D., Holloway A. K., Begun D. J., 2011b Genomic differentiation between temperate and tropical Australian populations of Drosophila melanogaster. Genetics 187: 245–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A., Thorleifsson G., Gudbjartsson D. F., Masson G., Sigurdsson A., et al. , 2010. Fine-scale recombination rate differences between sexes, populations and individuals. Nature 467: 1099–1103 [DOI] [PubMed] [Google Scholar]
- Korbel J. O., Urban A. E., Affourtit J. P., Godwin B., Grubert F., et al. , 2007. Paired-end mapping reveals extensive structural variation in the human genome. Science 318: 420–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitman M., 1983. Nucleotide polymorphism at the alcohol dehydrogenase locus of Drosophila melanogaster. Nature 304: 412–417 [DOI] [PubMed] [Google Scholar]
- Kuersten S., Goodwin E. B., 2003. The power of the 3′ UTR: translational control and development. Nat. Rev. Genet. 4: 626–637 [DOI] [PubMed] [Google Scholar]
- Lachaise D., Carious M. L., David J. R., Lemeunier F., Tsacas L., et al. , 1988. Historical biogeography of the Drosophila melanogaster species subgroup. Evol. Biol. 22: 159–225 [Google Scholar]
- Lander E. S., Waterman M. S., 1988. Genomic mapping by fingerprinting random clones: a mathematical analysis. Genomics 2: 231–239 [DOI] [PubMed] [Google Scholar]
- Langley C. H., Crow J. F., 1974. The direction of linkage disequilibrium. Genetics 78: 937–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley C. H., Tobari Y. N., Kojima K.-I., 1974. Linkage disequilibrium in natural populations of Drosophila melanogaster. Genetics 78: 921–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley C. H., Ito K., Voelker R. A., 1977. Linkage disequilibrium in natural populations of Drosophila melanogaster. Seasonal variation. Genetics 86: 447–454 [PMC free article] [PubMed] [Google Scholar]
- Langley C. H., MacDonald J., Miyashita N., Aguadé M., 1993. Lack of correlation between interspecific divergence and intraspecific polymorphism at the suppressor of forked region in Drosophila melanogaster and Drosophila simulans. Proc. Natl. Acad. Sci. USA 90: 1800–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley C. H., Lazzaro B. P., Phillips W., Heikkinen E., Braverman J. M., 2000. Linkage disequilibria and the site frequency spectra in the su(s) and su(wa) regions of the Drosophila melanogaster X chromosome. Genetics 156: 1837–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. C. G., Reinhardt J., 2012. Widespread polymorphism in the positions of stop codons in Drosophila melanogaster. Genome Biol. Evol. 4: 533–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. A., Carvalho C. M. B., Lupski J. R., 2007. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell 131: 1235–1247 [DOI] [PubMed] [Google Scholar]
- Lemaitre B., Hoffmann J., 2007. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25: 697–743 [DOI] [PubMed] [Google Scholar]
- Lemeunier F., Ashburner M., 1976. Relationships within the melanogaster species subgroup of the genus Drosophila (Sophophora). II. Phylogenetic relationships between six species based upon polytene chromosome banding sequences. Proc. R. Soc. Lond. B Biol. Sci. 193: 275–294 [DOI] [PubMed] [Google Scholar]
- Levine M. T., Begun D. J., 2008. Evidence of spatially varying selection acting on four chromatin-remodeling loci in Drosophila melanogaster. Genetics 179: 475–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R., 2011. Inference of human population history from individual whole-genome sequences. Nature 475: 493–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Stephan W., 2006. Inferring the demographic history and rate of adaptive substitution in Drosophila. PLoS Genet. 2: e166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Ruan J., Durbin R., 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18: 1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lintner J. A., 1882. First Annual Report on the Injurious and Other Insects of the State of New York. Weed, Parsons & Co., Albany, NY [Google Scholar]
- Locke D. P., Sharp A. J., McCarroll S. A., McGrath S. D., Newman T. L., et al. , 2006. Linkage disequilibrium and heritability of copy-number polymorphisms within duplicated regions of the human genome. Am. J. Hum. Genet. 79: 275–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long A. D., Lyman R. F., Langley C. H., Mackay T. F., 1998. Two sites in the Delta gene region contribute to naturally occurring variation in bristle number in Drosophila melanogaster. Genetics 149: 999–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luque T., O’Reilly D. R., 2002. Functional and phylogenetic analyses of a putative Drosophila melanogaster UDP-glycosyltransferase gene. Insect Biochem. Mol. Biol. 32: 1597–1604 [DOI] [PubMed] [Google Scholar]
- MacAlpine H. K., Gordân R., Powell S. K., Hartemink A. J., MacAlpine D. M., 2010. Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Res. 20: 201–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F. C., Richards S., Stone E. A., Barbadilla A., Ayroles J. F., et al. , 2012. The Drosophila melanogaster Genetic Reference Panel. Nature 482: 173–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzkin L. M., Merritt T. J. S., Zhu C.-T., Eanes W. F., 2005. The structure and population genetics of the breakpoints associated with the cosmopolitan chromosomal inversion In(3R)Payne in Drosophila melanogaster. Genetics 170: 1143–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J., Haigh J., 1974. The hitch-hiking effect of a favourable gene. Genet. Res. 23: 23–35 [PubMed] [Google Scholar]
- McCarroll S. A., Huett A., Kuballa P., Chilewski S. D., Landry A., et al. , 2008. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn’s disease. Nat. Genet. 40: 1107–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald J. H., Kreitman M., 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351: 652–654 [DOI] [PubMed] [Google Scholar]
- McVean G. A. T., Charlesworth B., 1999. A population genetic model for the evolution of synonymous codon usage: patterns and predictions. Genet. Res. 74: 145–158 [Google Scholar]
- McVean G. A. T., Myers S. R., Hunt S., Deloukas P., Bentley D. R., et al. , 2004. The fine-scale structure of recombination rate variation in the human genome. Science 304: 581–584 [DOI] [PubMed] [Google Scholar]
- Meisel R. P., Han M. V., Hahn M. W., 2009. A complex suite of forces drives gene traffic from Drosophila X chromosomes. Genome Biol. Evol. 1: 176–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettler L. E., Voelker R. A., Mukai T., 1977. Inversion clines in populations of Drosophila melanogaster. Genetics 87: 169–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita N., Langley C. H., 1988. Molecular and phenotypic variation of the white locus region in Drosophila melanogaster. Genetics 120: 199–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery E. A., Huang S. M., Langley C. H., Judd B. H., 1991. Chromosome rearrangement by ectopic recombination in Drosophila melanogaster: genome structure and evolution. Genetics 129: 1085–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama E. N., Hartl D. L., 1993. Codon usage bias and base composition of nuclear genes in Drosophila. Genetics 134: 847–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama E. N., Powell J. R., 1996. Intraspecific nuclear DNA variation in Drosophila. Mol. Biol. Evol. 13: 261–277 [DOI] [PubMed] [Google Scholar]
- Myers S., Bowden R., Tumian A., Bontrop R. E., Freeman C., et al. , 2010. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science 327: 876–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsume T., Tanaka T. U., 2009. Spatial regulation and organization of DNA replication within the nucleus. Chromosome Res. 18: 7–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Gojobori T., 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3: 418–426 [DOI] [PubMed] [Google Scholar]
- Nelder J. A., Wedderburn R. W. M., 1972. Generalized linear models. J. R. Stat. Soc. Ser. A 135: 370–384 [Google Scholar]
- Nicol J. W., Helt G. A., Blanchard S. G., Raja A., Loraine A. E., 2009. The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics 25: 2730–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R., Bauer DuMont V. L., Hubisz M. J., Aquadro C. F., 2007. Maximum likelihood estimation of ancestral codon usage bias parameters in Drosophila. Mol. Biol. Evol. 24: 228–235 [DOI] [PubMed] [Google Scholar]
- Nolte V., Schlötterer C., 2008. African Drosophila melanogaster and D. simulans populations have similar levels of sequence variability, suggesting comparable effective population sizes. Genetics 178: 405–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes M. D. S., Neumeier H., Schlötterer C., 2008. Contrasting patterns of natural variation in global Drosophila melanogaster populations. Mol. Ecol. 17: 4470–4479 [DOI] [PubMed] [Google Scholar]
- Obbard D. J., Jiggins F. M., Halligan D. L., Little T. J., 2006. Natural selection drives extremely rapid evolution in antiviral RNAi genes. Curr. Biol. 16: 580–585 [DOI] [PubMed] [Google Scholar]
- Ohta T., 1992. The nearly neutral theory of molecular evolution. Annu. Rev. Ecol. Syst. 23: 263–286 [Google Scholar]
- Ometto L., Glinka S., De Lorenzo D., Stephan W., 2005. Inferring the effects of demography and selection on Drosophila melanogaster populations from a chromosome-wide scan of DNA variation. Mol. Biol. Evol. 22: 2119–2130 [DOI] [PubMed] [Google Scholar]
- Orr H. A., Betancourt A. J., 2001. Haldane’s sieve and adaptation from the standing genetic variation. Genetics 157: 875–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S., Schneeberger K., Clark R. M., Lanz C., Warthmann N., et al. , 2008. Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res. 18: 2024–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J., Sasaki M., Kniewel R., Murakami H., Blitzblau H. G., et al. , 2011. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell 144: 719–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering B. M., Willis A. E., 2005. The implications of structured 5′ untranslated regions on translation and disease. Semin. Cell Dev. Biol. 16: 39–47 [DOI] [PubMed] [Google Scholar]
- Pool J. E., Aquadro C. F., 2006. History and structure of sub-Saharan populations of Drosophila melanogaster. Genetics 174: 915–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool J. E., Nielsen R., 2008. The impact of founder events on chromosomal variability in multiply mating species. Mol. Biol. Evol. 25: 1728–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves D. C., 2005. Recombination enhances protein adaptation in Drosophila melanogaster. Curr. Biol. 15: 1651–1656 [DOI] [PubMed] [Google Scholar]
- Presgraves D. C., 2006. Intron length evolution in Drosophila. Mol. Biol. Evol. 23: 2203–2213 [DOI] [PubMed] [Google Scholar]
- Presgraves D. C., Stephan W., 2007. Pervasive adaptive evolution among interactors of the Drosophila hybrid inviability gene, Nup96. Mol. Biol. Evol. 24: 306–314 [DOI] [PubMed] [Google Scholar]
- Quinlan A. R., Hall I. M., 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team , 2010. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna [Google Scholar]
- Riddle N. C., Minoda A., Kharchenko P. V., Alekseyenko A. A., Schwartz Y. B., et al. , 2011. Plasticity in patterns of histone modifications and chromosomal proteins in Drosophila heterochromatin. Genome Res. 21: 147–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roselius K., Stephan W., Städler T., 2005. The relationship of nucleotide polymorphism, recombination rate and selection in wild tomato species. Genetics 171: 753–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S., Ernst J., Kharchenko P. V., Kheradpour P., Negre N., et al. , 2010. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330: 1787–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackton T. B., Kulathinal R. J., Bergman C. M., Quinlan A. R., Dopman E. B., et al. , 2009. Population genomic inferences from sparse high-throughput sequencing of two populations of Drosophila melanogaster. Genome Biol. Evol. 1: 449–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattath S., Elyashiv E., Kolodny O., Rinott Y., Sella G., 2011. Pervasive adaptive protein evolution apparent in diversity patterns around amino acid substitutions in Drosophila simulans. PLoS Genet. 7: e1001302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenke T. A., Begun D. J., 2004. Strong selective sweep associated with a transposon insertion in Drosophila simulans. Proc. Natl. Acad. Sci. USA 101: 1626–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlötterer C., Dieringer D., 2005. A novel test statistic for the identification of local selective sweeps based on microsatellite gene diversity, pp. 55–64 Selective Sweep, edited by Nurminsky D. Landes Bioscience, Georgetown, TX [Google Scholar]
- Schmidt J. M., Good R. T., Appleton B., Sherrard J., Raymant G. C., et al. , 2010. Copy number variation and transposable elements feature in recent, ongoing adaptation at the Cyp6g1 locus. PLoS Genet. 6: e1000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider D. R., Hahn M. W., 2010. Lower linkage disequilibrium at CNVs is due to both recurrent mutation and transposing duplications. Mol. Biol. Evol. 27: 103–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider D. R., Stevens K., Cardeño C. M., Langley C. H., Hahn M. W., 2011. Genome-wide analysis of retrogene polymorphisms in Drosophila melanogaster. Genome Res. 21: 2087–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaiger M., Stadler M. B., Bell O., Kohler H., Oakeley E. J., et al. , 2009. Chromatin state marks cell-type- and gender-specific replication of the Drosophila genome. Genes Dev. 23: 589–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz Y. B., Pirrotta V., 2008. Polycomb complexes and epigenetic states. Curr. Opin. Cell Biol. 20: 266–273 [DOI] [PubMed] [Google Scholar]
- Sezgin E., Duvernell D. D., Matzkin L. M., Duan Y., Zhu C.-T., et al. , 2004. Single-locus latitudinal clines and their relationship to temperate adaptation in metabolic genes and derived alleles in Drosophila melanogaster. Genetics 168: 923–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J. A., Huang W., Zhang C., Hubisz M. J., Lu J., et al. , 2007. Adaptive genic evolution in the Drosophila genomes. Proc. Natl. Acad. Sci. USA 104: 2271–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Li W.-H., 1989. On the rate of DNA sequence evolution in Drosophila. J. Mol. Evol. 28: 398–402 [DOI] [PubMed] [Google Scholar]
- Sher N., Bell G. W., Li S., Nordman J., Eng T., et al. , 2011. Developmental control of gene copy number by repression of replication initiation and fork progression. Genome Res. 22: 64–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N. D., Davis J. C., Petrov D. A., 2005. Codon bias and noncoding GC content correlate negatively with recombination rate on the Drosophila X chromosome. J. Mol. Evol. 61: 315–324 [DOI] [PubMed] [Google Scholar]
- Smith N. G. C., Eyre-Walker A., 2002. Adaptive protein evolution in Drosophila. Nature 415: 1022–1024 [DOI] [PubMed] [Google Scholar]
- Spradling A. C., 1981. The organization and amplification of two chromosomal domains containing Drosophila chorion genes. Cell 27: 193–201 [DOI] [PubMed] [Google Scholar]
- Stapleton M., Carlson J. W., Celniker S. E., 2006. RNA editing in Drosophila melanogaster: new targets and functional consequences. RNA 12: 1922–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan W., Langley C. H., 1989. Molecular genetic variation in the centromeric region of the X chromosome in three Drosophila ananassae populations. I. Contrasts between the vermilion and forked loci. Genetics 121: 89–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan W., Langley C. H., 1998. DNA polymorphism in Lycopersicon and crossing-over per physical length. Genetics 150: 1585–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan W., Xing L., Kirby D. A., Braverman J. M., 1998. A test of the background selection hypothesis based on nucleotide data from Drosophila ananassae. Proc. Natl. Acad. Sci. USA 95: 5649–5654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan W., Song Y. S., Langley C. H., 2006. The hitchhiking effect on linkage disequilibrium between linked neutral loci. Genetics 172: 2647–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson W. J., Clark A. G., Waldrip-Dail H. M., Wolfner M. F., Aquadro C. F., 2001. Evolutionary EST analysis identifies rapidly evolving male reproductive proteins in Drosophila. Proc. Natl. Acad. Sci. USA 98: 7375–7379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson W. J., Wong A., Wolfner M. F., Aquadro C. F., 2004. Evolutionary expressed sequence tag analysis of Drosophila female reproductive tracts identifies genes subjected to positive selection. Genetics 168: 1457–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano-Shimizu T., 2004. Interlocus nonrandom association of polymorphisms in Drosophila chemoreceptor genes. Proc. Natl. Acad. Sci. USA 101: 14156–14161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatarenkov A., Ayala F. J., 2007. Nucleotide variation at the dopa decarboxylase (Ddc) gene in natural populations of Drosophila melanogaster. J. Genet. 86: 125–137 [DOI] [PubMed] [Google Scholar]
- Thornton K., Andolfatto P., 2006. Approximate Bayesian inference reveals evidence for a recent, severe bottleneck in a Netherlands population of Drosophila melanogaster. Genetics 172: 1607–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiemann-Boege I., Calabrese P., Cochran D. M., Sokol R., Arnheim N., 2006. High-resolution recombination patterns in a region of human chromosome 21 measured by sperm typing. PLoS Genet. 2: e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting C. T., Tsaur S. C., Wu M. L., Wu C. I., 1998. A rapidly evolving homeobox at the site of a hybrid sterility gene. Science 282: 1501–1504 [DOI] [PubMed] [Google Scholar]
- True J. R., Mercer J. M., Laurie C. C., 1996. Differences in crossover frequency and distribution among three sibling species of Drosophila. Genetics 142: 507–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsacas L., Lachaise D., 1974. Quatre nouvelles especes de la Cote-d’Ivoire du genre Drosophila, groupe melanogaster, et discussion de l’origine du sous-groupe melanogaster (Diptera: Drosophilidae). Ann. Univ. Abidjan E. Ecol. 7: 193–211 [Google Scholar]
- Tsubota S. I., 2009. Unequal crossing-over within the B duplication of Drosophila melanogaster: a molecular analysis. Genet. Res. 57: 105. [DOI] [PubMed] [Google Scholar]
- Turner T. L., Levine M. T., Eckert M. L., Begun D. J., 2008. Genomic analysis of adaptive differentiation in Drosophila melanogaster. Genetics 179: 455–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermaak D., Henikoff S., Malik H. S., 2005. Positive selection drives the evolution of rhino, a member of the heterochromatin protein 1 family in Drosophila. PLoS Genet. 1: 96–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veuille M., Baudry E., Cobb M., Derome N., Gravot E., 2004. Historicity and the population genetics of Drosophila melanogaster and D. simulans. Genetica 120: 61–70 [DOI] [PubMed] [Google Scholar]
- Vicoso B., Charlesworth B., 2006. Evolution on the X chromosome: unusual patterns and processes. Nat. Rev. Genet. 7: 645–653 [DOI] [PubMed] [Google Scholar]
- Voelker R. A., Cockerham C. C., Johnson F. M., Schaffer H. E., Mukai T., et al. , 1978. Inversions fail to account for allozyme clines. Genetics 88: 515–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl L. M., 2011. Fixation when N and s vary: classic approaches give elegant new results. Genetics 188: 783–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeley J., Hey J., 1997. Estimating ancestral population parameters. Genetics 145: 847–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall J. D., Andolfatto P., Przeworski M., 2002. Testing models of selection and demography in Drosophila simulans. Genetics 162: 203–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir B. S., Cockerham C. C., 1984. Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370 [DOI] [PubMed] [Google Scholar]
- Wellinger R. E., Thoma F., 1997. Nucleosome structure and positioning modulate nucleotide excision repair in the non-transcribed strand of an active gene. EMBO J. 16: 5046–5056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley C. S., Eanes W. F., 1994. Isolation and analysis of the breakpoint sequences of chromosome inversion In(3L)Payne in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 91: 3132–3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal T., Reuter G., 2002. Recombinogenic effects of suppressors of position-effect variegation in Drosophila. Genetics 160: 609–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S., 1949. The genetical structure of populations. Ann. Hum. Genet. 15: 323–354 [DOI] [PubMed] [Google Scholar]
- Wu T. C., Lichten M., 1994. Meiosis-induced double-strand break sites determined by yeast chromatin structure. Science 263: 515–518 [DOI] [PubMed] [Google Scholar]
- Yang Z., 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24: 1586–1591 [DOI] [PubMed] [Google Scholar]
- Zheng Q., Wang X.-J., 2008. GOEAST: a web-based software toolkit for Gene Ontology enrichment analysis. Nucleic Acids Res. 36: W358–W363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerkandl E., Pauling L., 1962. Molecular disease, evolution and genetic heterogeneity, pp. 189–225 Horizons in Biochemistry, edited by Kasha M., Pullman B. Academic Press, New York [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.