Abstract
The energy landscape approach has played a fundamental role in advancing our understanding of protein folding. Here, we quantify protein folding energy landscapes by exploring the underlying density of states. We identify three quantities essential for characterizing landscape topography: the stabilizing energy gap between the native and nonnative ensembles δE, the energetic roughness ΔE, and the scale of landscape measured by the entropy S. We show that the dimensionless ratio between the gap, roughness, and entropy of the system 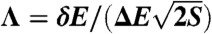 accurately predicts the thermodynamics, as well as the kinetics of folding. Large Λ implies that the energy gap (or landscape slope towards the native state) is dominant, leading to more funneled landscapes. We investigate the role of topological and energetic roughness for proteins of different sizes and for proteins of the same size, but with different structural topologies. The landscape topography ratio Λ is shown to be monotonically correlated with the thermodynamic stability against trapping, as characterized by the ratio of folding temperature versus trapping temperature. Furthermore, Λ also monotonically correlates with the folding kinetic rates. These results provide the quantitative bridge between the landscape topography and experimental folding measurements.
accurately predicts the thermodynamics, as well as the kinetics of folding. Large Λ implies that the energy gap (or landscape slope towards the native state) is dominant, leading to more funneled landscapes. We investigate the role of topological and energetic roughness for proteins of different sizes and for proteins of the same size, but with different structural topologies. The landscape topography ratio Λ is shown to be monotonically correlated with the thermodynamic stability against trapping, as characterized by the ratio of folding temperature versus trapping temperature. Furthermore, Λ also monotonically correlates with the folding kinetic rates. These results provide the quantitative bridge between the landscape topography and experimental folding measurements.
Keywords: energy landscape theory, biomolecular dynamics
Understanding how the amino acid sequence (i.e., primary structure) of each protein enables the native three-dimensional structure to be reached is one of the major challenges in molecular biophysics. In 1969, the arguments of Levinthal led to the suggestion of an apparent kinetic paradox (1). That is, if proteins were to randomly explore all possible states, cosmological timescales would be required for each protein to find the folded configuration. However, naturally occurring proteins fold in milliseconds to seconds. Protein folding theory has resolved this paradox by demonstrating that the underlying energy landscape is “funneled” towards the native state (2–9), however, local traps may be encountered during folding. To ensure that the folding occurs on biologically relevant timescales, the steepness of the protein folding funnel should be large, compared with the roughness due to local traps. Although this theory has provided the conceptual framework for interpreting folding experiments, both qualitatively and quantitatively (2, 5–18), it has yet to be explicitly demonstrated how the shape of the underlying landscape governs the thermodynamic stability and speed of folding, as measured experimentally (19). Here, we meet this challenge by quantifying the landscape topography and establishing the connection between the thermodynamics and kinetics of protein folding.
Naturally selected proteins differ from random sequences in that they fold into unique three-dimensional functional configurations. This indicates that the information necessary to fold is embedded in the amino acid sequence. This intrinsic information manifests in the form of energetic interactions and entropic contributions, which together define the temperature-dependent free-energy landscape. For describing folding, we are interested in the solvent-averaged effective energy, rather than the direct interactions between atoms (20, 21). Whereas detailed solvent–solute interactions contribute to the overall energetics, hydrophobic and hydrophilic interactions are described as solvent-averaged effects that drive folding. Similarly, the entropy of interest is the solvent-averaged configurational entropy of the polymer. Accordingly, the energy funnel describes the funnel-like character of the effective energy and configurational entropy.
We quantify the landscape by calculating the density of states in configuration and energy space. The density of states gives the probability of each energy/configuration and it does not depend explicitly on temperature. Accordingly, it describes the intrinsic energy landscape. The density of states is obtained from the microcanonical ensemble, whereas simulations of protein folding are usually performed for the canonical ensemble, which yields information on the free energy. In order to obtain the density of states, we transform the simulated canonical ensemble to the microcanonical ensemble.
To identify a robust metric that measures the “degree of funneledness” and overall topography of the landscape, we have used structure-based models with and without explicit energetic roughness (details in Materials and Methods). For both, funneled and rough landscapes, we explicitly demonstrate that three quantities are essential for characterizing the effective folding landscape topography: the energy gap (bias or slope of the funnel) (22, 23) between the native state and the average nonnative states 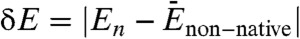 , the roughness in the energies ΔE, and the size of the funnel measured by the configurational entropy of the system S = kB ln Ω (where Ω is the number of states). We show that the dimensionless ratio between the gap, roughness and configurational entropy of the system
, the roughness in the energies ΔE, and the size of the funnel measured by the configurational entropy of the system S = kB ln Ω (where Ω is the number of states). We show that the dimensionless ratio between the gap, roughness and configurational entropy of the system 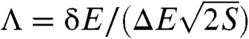 measures the landscape topography, and quantifies the degree of the funnel. Consistent with previous work (24), Λ is strongly correlated with the thermodynamic stability against traps (characterized by the ratio of the folding temperature versus the trapping temperature) and the folding kinetic rates. These results provide the quantitative bridge between the landscape topography and experimental folding measurements.
measures the landscape topography, and quantifies the degree of the funnel. Consistent with previous work (24), Λ is strongly correlated with the thermodynamic stability against traps (characterized by the ratio of the folding temperature versus the trapping temperature) and the folding kinetic rates. These results provide the quantitative bridge between the landscape topography and experimental folding measurements.
Results and Discussions
Density of States and the Energy Landscape of Protein Folding.
We explored the effective energy landscape and quantified the underlying topography to establish the link between the thermodynamics and kinetics of protein folding. We first performed molecular dynamics simulations, using a structure-based model, with and without energetic frustration included. The canonical ensemble can be efficiently sampled with replica (temperature) exchange methods (25–32). This provides a temperature-dependent distribution of the energies, which we then transform into the microcanonical ensemble through use of the weighted histogram analysis method (WHAM) algorithm (33). Because size effects can have a large influence on dynamics, it is crucial that we partition size-scaling and structural (or topological) effects. To do so, we studied two sets of proteins: The first group is formed by 13 proteins that vary in size, and the second is composed of nine proteins with similar size and different structural motifs (see SI Appendix for full analysis of each protein).
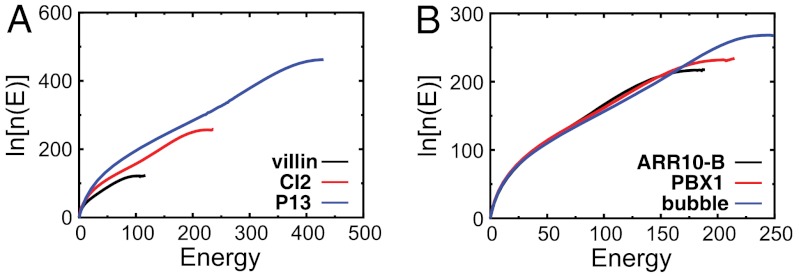
To visualize the energy landscape of folding, we projected the density of states onto specific order parameters, or reaction coordinates. The simplest representation of the landscape is the energy spectrum (Fig. 1) (34–36). From this, one may extract the stability gap δE and roughness ΔE (indicated by vertical arrows for each protein). The energy gap between the native and nonnative ensembles measures the bias, or slope, towards the native state. In contrast, the dispersion in energies describes the energetic roughness of the landscape. As shown in Fig. 1, there is a distinct gap between the native energy and the average energy of the nonnative states. With these models, all of the fast folding proteins that we studied have significant gaps, or biases, towards the native state. However, for each protein, the degree of roughness and scale of the gap are unique. As we will show, these differences allow one to classify proteins according to their thermodynamic and kinetic properties, and thereby provide a quantitative bridge between the funnel diagram and the physical–chemical properties of protein folding.
Fig. 1.
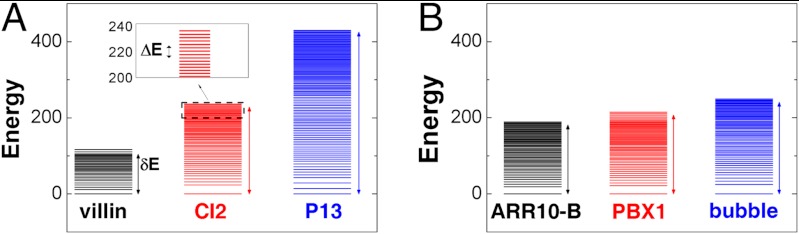
Folding energy landscapes in zero dimensions. The distribution of the energy levels (spectrum) for (A) three (of 13 studied. See SI Appendix.) proteins with different sizes and (B) three (of nine) proteins with the same size. The lowest (native) energy En is set to 0 for visualization purposes. The stability gap δE between the native state and the average nonnative states is indicated by vertical arrows. Each energy level of the distribution represents the sum of a cluster of states, except for the native band. The inset is a magnification of the energy levels of CI2, with the scale of the energetic roughness ΔE indicated by a vertical arrow.
Size, Steepness, and Roughness of Folding Funnels.
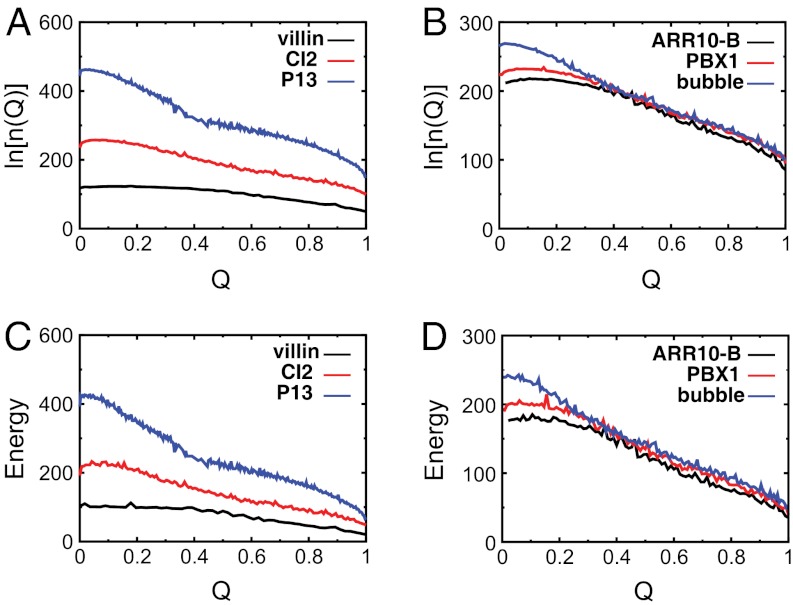
For an energetically unfrustrated model, the number of states (i.e., the configurational entropy S = ln[n(E)]) decreases monotonically with decreasing energy. In other words, the states are more sparsely distributed as the energy is lowered. This is different from the canonical ensemble distribution n(E,T), where T is the temperature and low energies have higher populations [n(E,T) ∼ n(E) exp[E/kBT]]. The distribution of energy states, or density of states n(E), is intrinsic to the system (i.e., not temperature dependent), and thus reflects the underlying landscape of the protein folding funnel. The configurational entropy S(E) measures how many states are accessible and, therefore, describes the size of the state space. Initially, the size of the funnel is large for the unfolded states. As expected, the size of the funnel decreases as the native ensemble is approached. Additionally, the rate of decrease, or the slope, of the configurational entropy towards the native state in energy is different for different proteins (Fig. 2).
Fig. 2.
Folding energy landscapes in one dimension. Logarithm of the density of states as a function of energy for (A) three (of 13) proteins with different sizes and (B) three (of nine) proteins of the same size. The lowest (native) energy En is set to 0 for visualization purposes.
To estimate the glassy-like trapping temperature Tg, which describes the temperature at which the configurational entropy associated with nonnative states vanishes, the density of states was partitioned into native and nonnative ensembles. This was achieved by classifying the simulated configurations by their structural proximity to the native state (SI Appendix, Fig. S5). Using the microcanonical ensemble, Tg was determined according to the thermodynamic Maxwell relationship ∂S/∂E = 1/T, and ∂S/∂E was calculated for the unfolded ensemble. Tg corresponds to the temperature below which the system becomes trapped or “frozen.” Consistent with these measures of Tg, in our simulations, as T approaches Tg, the kinetics significantly slows down, has increased dispersion in the folding time, and becomes nonself-averaging (SI Appendix, Figs. S14 and S20). From Tg, the roughness of the underlying energy landscape of folding ΔE is determined from the relation 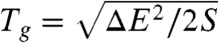 , where S is the configurational entropy of nonnative states (22). In contrast to Tg, the folding temperature Tf is obtained directly from the heat capacity curve, which is experimentally accessible.
, where S is the configurational entropy of nonnative states (22). In contrast to Tg, the folding temperature Tf is obtained directly from the heat capacity curve, which is experimentally accessible.
Because experiments do not report on the configurational entropy and enthalpy of each configuration directly, it is important that we understand the connection between energetic and structural metrics of protein folding. Fig. 3 A and B show the density of states as a function of the order parameter Q, which is defined here as the fraction of native contacts formed (37). The configurational entropy, or density of states, decreases monotonically with Q. In other words, as folding proceeds, the size of the funnel [measured by S(Q)] decreases, where the the slope is distinct for each protein. The average energy also monotonically decreases as the system becomes more folded (Fig. 3 C and D). This depicts the funneled nature of the simulated landscapes. For an analytical mean field theory of protein folding (22), the energy gap is closely linked to the slope of E(Q). Here, there is also a strong correlation between the average slope of the energy landscape E(Q) and the energy gap (SI Appendix, Figs. S12 and S18), explicitly confirming that the models describe the landscape as being funneled.
Fig. 3.
Folding energy landscapes in one dimension. Logarithm of the density of states as a function of the fraction of native contacts Q for (A) three (of 13) proteins of different sizes and (B) three of the nine proteins of same size. Average energy as a function of Q for (C) three of the 13 different-sized proteins and (D) three of the nine proteins with same size.
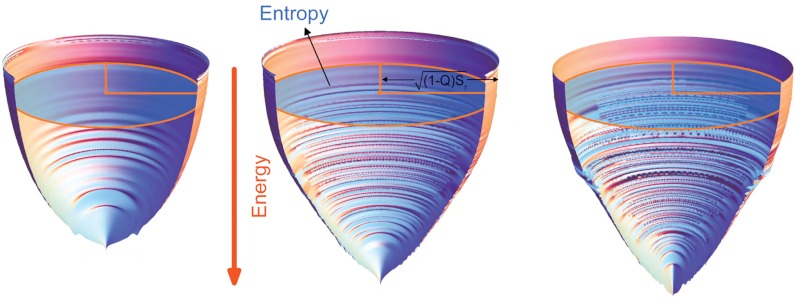
There are many qualitative folding funnel illustrations available, though none have been quantitatively derived from experimental or theoretical measurements. In Fig. 4, we show three quantified folding funnels of villin headpiece, CI2 and P13, obtained directly from simulations. These funnels encapsulate the data described in Figs. 1–3. The horizontal axis is 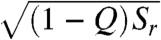 , where the area in the ellipsoid is equal to the configurational entropy S(E), which provides the size of the funnel. The picture illustrates that as folding proceeds towards the native state, S(E) and the effective energy E(Q) (vertical axis) decrease. The steepness of the funnel is determined by the slope of the energy (with respect to Q), as well as the slope of the configurational entropy. The depicted roughness in the funnel is due to the imperfect correlation between Q and E (details in SI Appendix). As the protein folds, the radius of the funnel [related to (1 - Q)], the steepness of landscape, and the size of the funnel all decrease. The overall shape of the funnel is then determined by the steepness over the roughness and the size (entropy) of the unfolded ensemble.
, where the area in the ellipsoid is equal to the configurational entropy S(E), which provides the size of the funnel. The picture illustrates that as folding proceeds towards the native state, S(E) and the effective energy E(Q) (vertical axis) decrease. The steepness of the funnel is determined by the slope of the energy (with respect to Q), as well as the slope of the configurational entropy. The depicted roughness in the funnel is due to the imperfect correlation between Q and E (details in SI Appendix). As the protein folds, the radius of the funnel [related to (1 - Q)], the steepness of landscape, and the size of the funnel all decrease. The overall shape of the funnel is then determined by the steepness over the roughness and the size (entropy) of the unfolded ensemble.
Fig. 4.
Quantified funneled landscapes for villin headpiece (Left), CI2 (Center) and P13 (Right) obtained from simulation. The depth of the funnel is the energy and the cross-section perpendicular to the energy axis is an ellipsoid with an area equal to the configurational entropy S(E). The semimajor axis of the ellipsoid at each stratum is 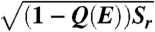 , which is a function of the fraction of native contacts Q(E), Sr is a constant for each protein. The other semiaxis is given by
, which is a function of the fraction of native contacts Q(E), Sr is a constant for each protein. The other semiaxis is given by 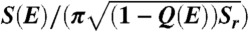 . Both energy and configurational entropy are normalized by the protein size.
. Both energy and configurational entropy are normalized by the protein size.
Free-Energy Profile of Protein Folding.
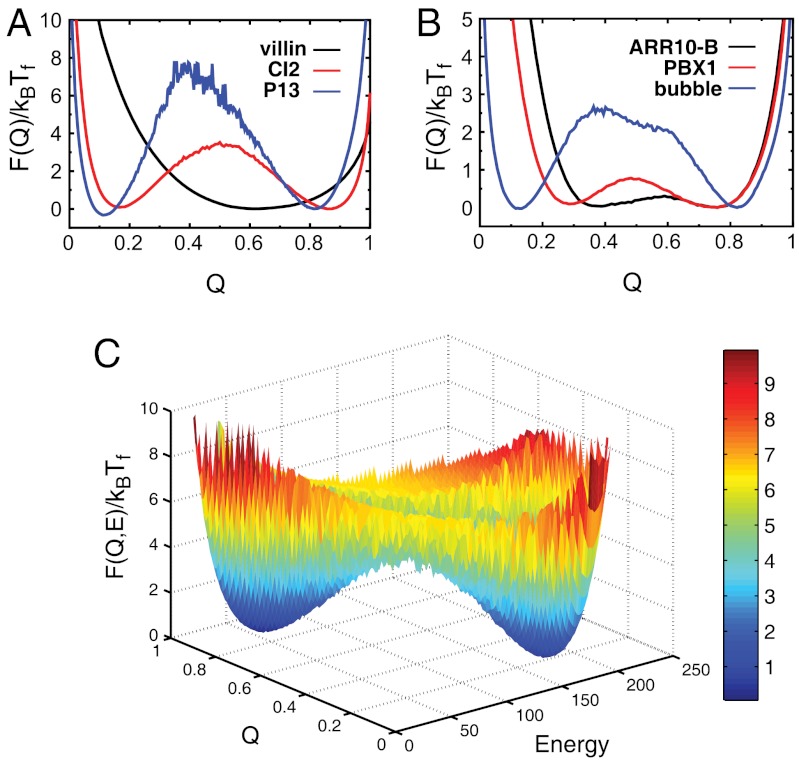
In contrast to the density of states, the free-energy landscape is dependent on temperature, which can give the impression that a particular landscape is not funneled, even when the underlying density of states is. Fig. 5 A and B show the free energy as a function of the folding coordinate F(Q), at the folding temperature Tf. The profiles were calculated from a Cα structure-based model for downhill folders (villin headpiece), two-state folders (CI2 and P13) with different sizes, and proteins with the same size but different structural topologies. The shape of the free-energy landscape results from a balance between energy and configurational entropy, leading to its explicit dependence on temperature. For two-state folders, at Tf the free energies of the folded and unfolded ensembles are equal. Below Tf, the free energy is dominated by the effective enthalpy and the system is biased towards the native ensemble. Above Tf, the free energy is dominated by the configurational entropy and is heavily weighted towards the nonnative ensemble. For comparison to other common measures of folding, we also calculated the free energy at Tχ (38), which is the temperature at which the native ensemble accounts for 80% of the population (SI Appendix, Fig. S28 A–C). At T0, the folding kinetics is the fastest (SI Appendix, Fig. S28 D–F). Because Tχ is lower than Tf, the free-energy profiles are biased towards the native state. As temperature approaches T0, the folding of each protein becomes downhill-like. Two-dimensional free-energy profiles at Tf, Tχ, T0 are shown in Fig. 5C and SI Appendix, Fig. S28 G and H. The relative free energy of the native ensemble and nonnative ensemble provides a measure of thermodynamics (or equilibrium constants), linking to thermodynamic folding experiments.
Fig. 5.
The free energy at folding temperature in one dimension as a function of fraction of native contacts Q and in two dimension as a function of Q and energy. Free-energy profile, at folding temperature Tf, as a function of Q for (A) three (of 13) proteins with different sizes and (B) three (of nine) proteins with the same size. (C) Two-dimensional free-energy profile for CI2 as a function of Q and energy at Tf normalized by the corresponding kBTf.
Some of the studied proteins are strong folders, with one basin of attraction in the free-energy landscape. The slope of the free-energy landscape for the one-state folders measures how easy it is to fold. For other proteins, two basins of attraction emerge, leading to the two-state behavior of the proteins, as is often seen in protein folding experiments. The barrier heights of the two-state folders measure how difficult it is to fold. The often nonfunnel-like folding free-energy landscape is the result of a competition and imperfect cancelation between energy and configurational entropy. This is the view from the canonical ensemble. For the microcanonical ensemble, whereas the underlying energy landscape is also determined by the competition between energy, roughness, and configurational entropy, there is only one basin of attraction that shapes the energy landscape.
Energy Landscape Topography Determines the Thermodynamics of Folding.
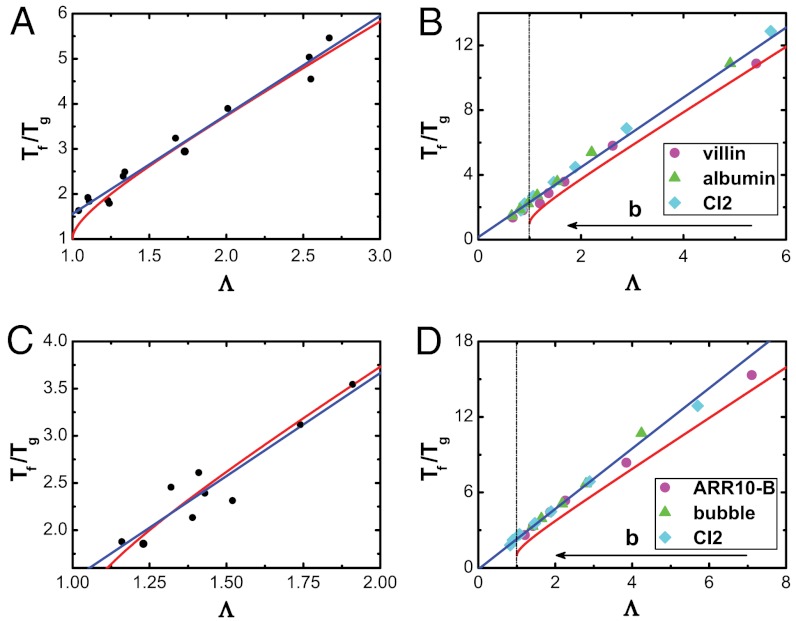
The landscape topographic measure Λ correlates with the thermodynamic folding stability against trapping for several classes of proteins and energetic models (Fig. 6): (a) proteins of different sizes with different structural topologies that are modeled without energetic roughness [i.e., completely funneled landscape (39)]; (b) proteins of different sizes, modeled with energetic roughnesses; (c) proteins of the same size, but different topologies, without energetic roughness included; (d) proteins of the same size with different structural topologies and energetic roughness included. For each group, we find a strong positive correlation between thermodynamic stability [characterized by the dimensionless ratio of folding transition temperature to the trapping temperature Tf/Tg (40–42)] and Λ, which is consistent with expectations from mean field theory (2, 7, 22, 43). That is, the steeper and less rugged (or the smaller the size of) the funnel, the more thermodynamically stable the protein is against energetic traps. This demonstrates that the thermodynamic stability of proteins that is measured from experiments will reflect the the underlying topography of the landscape. Specifically, these results demonstrate that Tf and Tg (44) measurements will allow us to infer the characteristics of the underlying landscape.
Fig. 6.
The correlation between folding against trapping Tf/Tg and landscape topography Λ. The red line is the analytical mean field theory prediction of the relationship between Tf/Tg and Λ: Tf/Tg = Λ + (Λ2 - 1)1/2. The blue line is a linear fit of Tf/Tg versus Λ. For all cases, the correlation coefficient (c.c) is greater than 0.9. (A) Folding of proteins of different sizes and different structural topologies, but no energetic roughness. (B) Folding of different-sized proteins with energetic roughness. (C) Folding of proteins of similar size with different structural topologies but no energetic roughness included. (D) Folding of same-size proteins with energetic roughness included. In B and D, b sets the energy variance of the non-native interactions (details in SI Appendix). Λ monotonically correlates with b.
For each class of proteins, the origin of the correlation between Λ and Tf/Tg may be attributed to different features. Because the slope, roughness, and configurational entropy of the landscape, as well as the folding and trapping temperatures, all depend on protein size, the correlation across different-sized proteins (Fig. 6A) is expected from scaling considerations (SI Appendix, Figs. S11 and S13). This is inconsistent with mean field approximations because finite sizes and the capillarity of protein folding are accounted for here (45). When energetic roughness is included (Fig. 6B) the trapping temperature and Λ are altered, and the correlation is due to the energetic roughness dependence of both. For proteins with different structural topologies (Fig. 6C), size-scaling arguments cannot account for the correlation, which implicates the residue–residue connectivity as leading to changes in the underlying slope, roughness, and size of the landscape. When introducing energetic roughness to proteins of the same size (Fig. 6D) the origin of the correlation is again due to concomitant changes in landscape topography and thermodynamic stability. Thus, these results demonstrate that Λ is a measure of landscape topography that can accurately predict the thermodynamic stability against trapping, whether or not they are size-scaling or energetic in origin.
Energy Landscape Topography Determines the Kinetics of Folding.
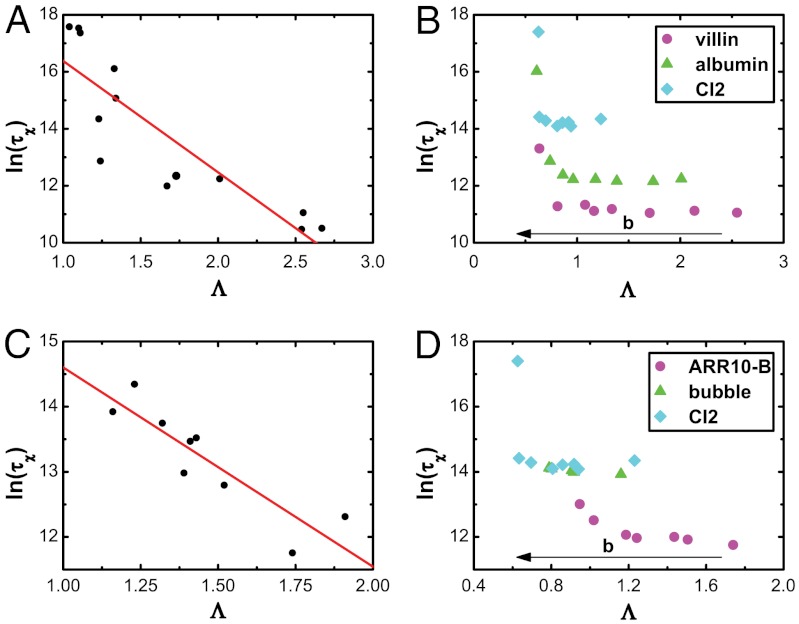
To explore the relationship between the landscape topography and kinetics, we have simulated folding and calculated the kinetic rates. We first investigated proteins of different sizes, using an energetically unfrustrated model that has been shown to produce folding kinetics that are correlated with experimental measurements (11, 45–58). Kinetic simulations were performed at the temperature Tχ, where 80% of the population occupies the native ensemble (38). We find that the rates are strongly negatively correlated with the landscape topographic measure Λ (Fig. 7). This illustrates that the steeper, less rugged, or smaller size (i.e., less entropy) of the folding funnel, the faster folding occurs (59, 60). This strong correlation indicates that experimental measures of the folding speed can be used to obtain quantitative measures of the underlying energy landscape.
Fig. 7.
The correlation between the kinetic rates of folding and landscape topography Λ, shown for: (A) proteins of different sizes and topologies, without energetic roughness included (c.c. = -0.87); (B) proteins of different sizes with varying degrees of energetic roughness included; (C) proteins of similar size and different structural topologies but no energetic roughness included (c.c. = -0.89); (D) proteins of the same size with energetic roughness included. The landscape topography Λ is strongly correlated with folding time. The folding time is in reduced units.
Because the kinetic rate and landscape topography measure Λ depend on size, the correlation for proteins of different sizes (in the energetically unfrustrated model) may result from size-dependent effects. To address this potential limitation in our description of kinetics and topography, we calculated the folding kinetics for proteins of different sizes, with a variable degree of energetic roughness included (Fig. 7B). We find a comparable correlation when using models with energetic roughness. Because the landscape topography is directly related to the energetic roughness, and the kinetic rates also depend on the energetic roughness (rougher landscape slows down kinetics), the correlation between the landscape topography and folding speed reflects the influence of energetic roughness. It is worth noting that, when the nonnative interaction strength is increased from zero, the correlation between folding rate and Λ is negligible. This is consistent with theoretical considerations and simulations that have shown that a small degree of nonnative interactions (relative to a completely unfrustrated landscape) can decrease the free-energy barrier and accelerate folding rates (39, 61). In the present study, the model is structure based, where large values of Λ correspond to a very small degree of energetic roughness, whereas proteins must have a finite degree of roughness in their landscapes. Thus, for proteins in solution, one does not expect to observe such accelerating effects because the landscapes have a finite degree of roughness already. Therefore, the correlation between folding rates and Λ is likely to be monotonic in real proteins, where larger Λ leads to larger folding rate.
To remove size-scaling effects and identify the structural contributions to the folding kinetics, we performed simulations of nine proteins that are comparable in size (Fig. 7C). We first explored the folding kinetics for proteins of the same size and different structural topologies, with a model that lacks energetic roughness. For those cases, the structural topologies determine the configurational entropy, landscape slope, and connectivity of substates. The correlation between the landscape topography and folding speed, therefore, demonstrates the structural-topology dependence of the landscape. To ensure that these results are not model dependent, we repeated these calculations with a model that included stabilizing nonnative interactions (Fig. 7D). We again find that the energetic roughness has a considerable impact on the landscape topography and leads to a correlation between the underlying topography and folding speed.
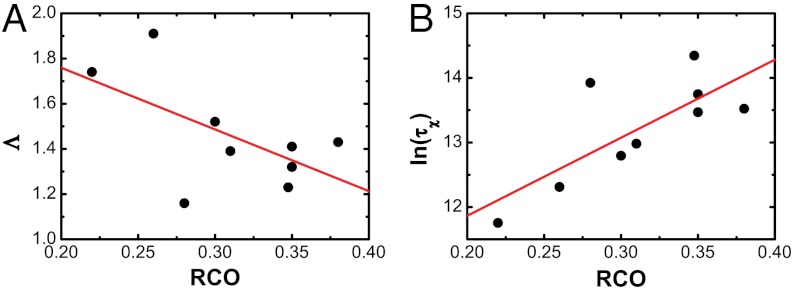
Because other metrics, such as contact order [CO, an entropic measure based on the native structure of proteins (53, 61–64)], are found to be correlated with the kinetics of folding, Λ should be correlated with those quantities. The correlation between the relative contact order (RCO) and Λ for proteins of the same size with the energetically unfrustrated model implies that Λ contains the structural topological information of each protein (Fig. 8A). In addition, our results show that RCO is correlated with the folding rate for the nine same-sized energetically unfrustrated proteins (Fig. 8B). Compared with RCO, the energy landscape topography measure Λ shows a stronger correlation with the folding rates. Although such observations reinforce the notion that native-state topology influences the kinetics, the presented landscape topography measure Λ captures the topological and energetic contributions to the landscape and is capable of predicting the thermodynamics and kinetics of folding.
Fig. 8.
The correlation between landscape topography Λ, kinetic rates of folding and RCO. (A) Λ versus RCO for nine proteins of the same size but different structural topologies (c.c. = -0.59). (B) Folding time ln(τχ) as a function of RCO (c.c. = 0.76).
Conclusions
We have quantified the energy landscape of protein folding by calculating the underlying density of states. Based on the density of states, we have shown that the dimensionless ratio between the gap, roughness, and configurational entropy of the system [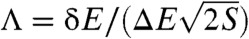 ] quantifies the topography of the underlying landscape and measures the degree of its funneledness. Λ represents a universal metric by which the overall landscape topography may be extracted from thermodynamic or kinetic measurements. Further, we have demonstrated that Λ is not only appropriate for particular subsets of proteins (such as fast folding proteins) but also for proteins of different sizes and energetic character.
] quantifies the topography of the underlying landscape and measures the degree of its funneledness. Λ represents a universal metric by which the overall landscape topography may be extracted from thermodynamic or kinetic measurements. Further, we have demonstrated that Λ is not only appropriate for particular subsets of proteins (such as fast folding proteins) but also for proteins of different sizes and energetic character.
Each protein studied has a folding funnel associated with a unique slope, roughness, and size. The topography measure of the underlying folding landscape, Λ, is shown to be correlated strongly (monotonically) with the thermodynamic stability against traps, characterized by the ratio of thermodynamic stability temperature versus trapping temperature. In addition, the landscape topography measure Λ also monotonically correlates with the folding kinetic rates. These demonstrate how the energy landscape topography determines the thermodynamics and kinetics of protein folding.
Energy landscape theory has guided our understanding of protein folding and has led to progress in the study of folding thermodynamics and kinetics. In our studies, because the landscape topography, thermodynamics, and kinetics are so strongly correlated, Λ represents the meeting point for theoretical quantification of the landscape topography and experimental evaluation of stability and kinetics. In addition, this quantitative energy landscape framework can be extended to multidomain proteins and folding in vivo. It is worth noting that our studies have used a coarse-grained structure-based model to explore the energy landscape. However, as computing hardware and software continue to advance, the presented framework may be utilized in conjunction with all-atom molecular dynamic simulations. Such efforts will allow one to partition the effects of solvent interactions on protein folding and identify how they contribute to the global topology of the energy landscape. In closing, the current study bridges the gap between the quantification of the underlying folding energy landscape topography and experiments (or simulations) on thermodynamics and kinetics of protein folding.
Materials and Methods
A structure-based model (13), coarse grained at the residue level, was used to explore the density of states. To ensure efficient sampling, the density of states was calculated from replica exchanged molecular dynamics simulations (25), performed at 48 temperatures for each protein. To obtain a single measure of the density of states, WHAM (33) was used. The deviation of each configuration from the native structure was used to divide sampled configurations into native and nonnative ensembles. With the thermodynamic relation 1/T = ∂S/∂E, we calculated the slope of the density of states for the nonnative ensemble to estimate the temperature at which the configurational entropy of the nonnative states vanishes (1/Tg). From the trapping temperature Tg, we directly obtained the landscape topological roughness: 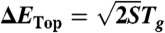 . S is the configurational entropy of the nonnative states. The energy gap δE is defined as the difference between the average energy of the nonnative states and the energy of native state. S and δE were calculated from the density of states.
. S is the configurational entropy of the nonnative states. The energy gap δE is defined as the difference between the average energy of the nonnative states and the energy of native state. S and δE were calculated from the density of states.
To explore the role of the energetic roughness on the landscape topographical measure, we introduced energetic frustration into the structure-based model by including nonnative interactions (39, 65). The weight of each nonnative interaction was assigned according to a Gaussian distribution with a variance of b2. We modulated the scale of the energetic roughness by changing b2. b2 = 0 corresponds to a structure-based model. The energetic roughness ΔEEne introduced by nonnative interactions is expressed as 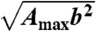 , where Amax is the maximum number of nonnative contacts the protein can form.
, where Amax is the maximum number of nonnative contacts the protein can form.
The total landscape roughness of proteins ΔETotal is due to both topological and energetic roughness contributions. In purely structure-based models (13, 66), there are no nonnative interactions, and the energetic contribution to the roughness is ΔEEne = 0. When nonnative interactions are included, the roughness of the energy landscape is the sum of the topological roughness and energetic roughness. To calculate ΔETotal, we make the assumption that modulating the variance of nonnative interactions does not significantly alter the topological contribution to the roughness. This assumption is valid because the topological roughness arises from constraints on the geometry and shape of the native structure, which is not significantly perturbed when the energetic roughness is sufficiently low (67,68). In other words, the nonnative interactions in real proteins can be approximated as energetic heterogeneity perturbations to the background roughness introduced by the topology. Accordingly, we use the expression: 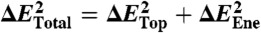 to calculate the total roughness of each protein. The trapping transition temperature (Tg) will be:
to calculate the total roughness of each protein. The trapping transition temperature (Tg) will be: 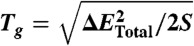 . The folding landscape measure is
. The folding landscape measure is 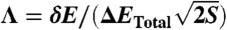 .
.
Supplementary Material
ACKNOWLEDGMENTS.
X.C. and E.W. are supported by the National Natural Science Foundation (NSF) of China (Grants 21190040 and 11174105) and the 973 project 2009CB930100 and 2010CB933600. J.W. is supported by the National Science Foundation. R.J.O., J.C., and V.B.P.L. were supported by Brazilian Agencies Fundação de Amparo à Pesquisa do Estado de São Paulo, Coordenação de Aperfeiçoamento de Pessoal de Nível Superior and Conselho Nacional de Desenvolvimento Científico e Tecnológico. This research was supported by resources supplied by the Núcleo de Computação Científica/Grid Universidade Estadual Paulista—Universidade Estadual Paulista. This work was supported by the Center for Theoretical Biological Physics sponsored by the NSF (Grant PHY-0822283) and by NSF-MCB-1214457. J.N.O. is a CPRIT Scholar in Cancer Research sponsored by the Cancer Prevention and Research Institute of Texas.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1212842109/-/DCSupplemental.
References
- 1.Levinthal C. In: Debrunner P, Tsibris J, Munck E, editors. Proceedings in Mossbauer Spectroscopy in Biological Systems; Urbana, IL: University of Illinois Press; 1969. pp. 22–24. [Google Scholar]
- 2.Bryngelson JD, Wolynes PG. Spin-glasses and the statistical-mechanics of protein folding. Proc Natl Acad Sci USA. 1987;84:7524–7528. doi: 10.1073/pnas.84.21.7524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bryngelson JD, Wolynes PG. Intermediates and barrier crossing in a random energy-model (with applications to protein folding) J Phys Chem. 1989;93:6902–6915. [Google Scholar]
- 4.Leopold PE, Montal M, Onuchic JN. Protein folding funnels—A kinetic approach to the sequence structure relationship. Proc Natl Acad Sci USA. 1992;89:8721–8725. doi: 10.1073/pnas.89.18.8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan HS, Dill KA. Transition-states and folding dynamics of proteins and heteropolymers. J Chem Phys. 1994;100:9238–9257. [Google Scholar]
- 6.Abkevich VI, Gutin AM, Shakhnovich EI. Free-energy landscape for protein-folding kinetics—Intermediates, traps, and multiple pathways in theory and lattice model simulations. J Chem Phys. 1994;101:6052–6062. [Google Scholar]
- 7.Bryngelson JD, Onuchic JN, Socci ND, Wolynes PG. Funnels, pathways, and the energy landscape of protein-folding—A synthesis. Proteins. 1995;21:167–195. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]
- 8.Wang J, Onuchic J, Wolynes P. Statistics of kinetic pathways on biased rough energy landscapes with applications to protein folding. Phys Rev Lett. 1996;76:4861–4864. doi: 10.1103/PhysRevLett.76.4861. [DOI] [PubMed] [Google Scholar]
- 9.Brooks CL, Onuchic JN, Wales DJ. Statistical thermodynamics—Taking a walk on a landscape. Science. 2001;293:612–613. doi: 10.1126/science.1062559. [DOI] [PubMed] [Google Scholar]
- 10.Itzhaki LS, Otzen DE, Fersht AR. The structure of the transition-state for folding of chymotrypsin inhibitor-2 analyzed by protein engineering methods—Evidence for a nucleation-condensation mechanism for protein-folding. J Mol Biol. 1995;254:260–288. doi: 10.1006/jmbi.1995.0616. [DOI] [PubMed] [Google Scholar]
- 11.Klimov DK, Thirumalai D. Linking rates of folding in lattice models of proteins with underlying thermodynamic characteristics. J Chem Phys. 1998;109:4119–4125. [Google Scholar]
- 12.Gruebele M, Sabelko J, Ervin J. Observation of strange kinetics in protein folding. Proc Natl Acad Sci USA. 1999;96:6031–6036. doi: 10.1073/pnas.96.11.6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clementi C, Nymeyer H, Onuchic JN. Topological and energetic factors: What determines the structural details of the transition state ensemble and “en-route” intermediates for protein folding? An investigation for small globular proteins. J Mol Biol. 2000;298:937–953. doi: 10.1006/jmbi.2000.3693. [DOI] [PubMed] [Google Scholar]
- 14.Nymeyer H, Garcia AE, Onuchic JN. Folding funnels and frustration in off-lattice minimalist protein landscapes. Proc Natl Acad Sci USA. 1998;95:5921–5928. doi: 10.1073/pnas.95.11.5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Onuchic JN, Nymeyer H, Garcia AE, Chahine J, Socci ND. The energy landscape theory of protein folding: Insights into folding mechanisms and scenarios. Adv Protein Chem. 2000;53:87–152. doi: 10.1016/s0065-3233(00)53003-4. [DOI] [PubMed] [Google Scholar]
- 16.Eaton WA, Schuler B, Lipman EA. Probing the free-energy surface for protein folding with single-molecule fluorescence spectroscopy. Nature. 2002;419:743–747. doi: 10.1038/nature01060. [DOI] [PubMed] [Google Scholar]
- 17.Stell G, Lee CL, Wang J. First-passage time distribution and non-Markovian diffusion dynamics of protein folding. J Chem Phys. 2003;118:959–968. doi: 10.1103/PhysRevE.67.041905. [DOI] [PubMed] [Google Scholar]
- 18.Zhou YQ, Zhang C, Stell G, Wang J. Temperature dependence of the distribution of the first passage time: Results from discontinuous molecular dynamics simulations of an all-atom model of the second beta-hairpin fragment of protein g. J Am Chem Soc. 2003;125:6300–6305. doi: 10.1021/ja029855x. [DOI] [PubMed] [Google Scholar]
- 19.Englander SW, Milne JS, Xu YJ, Mayne LC. Experimental study of the protein folding landscape: Unfolding reactions in cytochrome c. J Mol Biol. 1999;290:811–822. doi: 10.1006/jmbi.1999.2924. [DOI] [PubMed] [Google Scholar]
- 20.Wolynes PG. Biomolecular folding in vacuo. Proc Natl Acad Sci USA. 1995;92:2426–2427. doi: 10.1073/pnas.92.7.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolynes PG, Onuchic JN, Thirumalai D. Navigating the folding routes. Science. 1995;267:1619–1620. doi: 10.1126/science.7886447. [DOI] [PubMed] [Google Scholar]
- 22.Plotkin SS, Wang J, Wolynes PG. Statistical mechanics of a correlated energy landscape model for protein folding funnels. J Chem Phys. 1997;106:2932–2948. [Google Scholar]
- 23.Shakhnovich EI. Proteins with selected sequences fold into unique native conformation. Phys Rev Lett. 1994;72:3907–3910. doi: 10.1103/PhysRevLett.72.3907. [DOI] [PubMed] [Google Scholar]
- 24.Goldstein R, Luthey-Schulten Z, Wolynes PG. Optimal protein-folding codes from spin-glass theory. Proc Natl Acad Sci USA. 1992;89:4918–4922. doi: 10.1073/pnas.89.11.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okamoto Y, Sugita Y. Replica-exchange molecular dynamics method for protein folding. Chem Phys Lett. 1999;314:141–151. [Google Scholar]
- 26.Garcia AE, Onuchic JN. Folding a protein in a computer: An atomic description of the folding/unfolding of protein A. Proc Natl Acad Sci USA. 2003;100:13898–13903. doi: 10.1073/pnas.2335541100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodinger T, Pomes R. Enhancing the accuracy, the efficiency and the scope of free energy simulations. Curr Opin Struct Biol. 2005;15:164–170. doi: 10.1016/j.sbi.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 28.Rodinger T, Howell PL, Pomes R. Distributed replica sampling. J Chem Theor Comput. 2006;2:725–731. doi: 10.1021/ct050302x. [DOI] [PubMed] [Google Scholar]
- 29.Zhang C, Ma J. Comparison of sampling efficiency between simulated tempering and replica exchange. J Chem Phys. 2008;129:134112. doi: 10.1063/1.2988339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang C, Ma J. Enhanced sampling in generalized ensemble with large gap of sampling parameter: Case study in temperature space random walk. J Chem Phys. 2009;130:194112. doi: 10.1063/1.3139192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang C, Ma J. Enhanced sampling and applications in protein folding in explicit solvent. J Chem Phys. 2010;132:244101. doi: 10.1063/1.3435332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rauscher S, Neale C, Pomes R. Simulated tempering distributed replica sampling, virtual replica exchange, and other generalized-ensemble methods for conformational sampling. J Chem Theor Comput. 2010;5:2640–2662. doi: 10.1021/ct900302n. [DOI] [PubMed] [Google Scholar]
- 33.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. The weighted histogram analysis method for free-energy calculations on biomolecules 1. The method. J Comput Chem. 1992;13:1011–1021. [Google Scholar]
- 34.Honeycutt JD, Thirumalai D. Metastability of the folded states of globular proteins. Proc Natl Acad Sci USA. 1990;87:3526–3529. doi: 10.1073/pnas.87.9.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sali A, Shakhnovich E, Karplus M. How does a protein fold. Nature. 1994;369:248–251. doi: 10.1038/369248a0. [DOI] [PubMed] [Google Scholar]
- 36.Shakhnovich EI, Gutin AM. Implications of thermodynamics of protein folding for evolution of primary sequences. Nature. 1990;346:773–775. doi: 10.1038/346773a0. [DOI] [PubMed] [Google Scholar]
- 37.Cho SS, Levy Y, Wolynes PG. P versus q: Structural reaction coordinates capture protein folding on smooth landscapes. Proc Natl Acad Sci USA. 2006;103:586–591. doi: 10.1073/pnas.0509768103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klimov DK, Thirumalai D. Criterion that determines the foldability of proteins. Phys Rev Lett. 1996;76:4070–4073. doi: 10.1103/PhysRevLett.76.4070. [DOI] [PubMed] [Google Scholar]
- 39.Clementi C, Plotkin SS. The effects of nonnative interactions on protein folding rates: Theory and simulation. Protein Sci. 2004;13:1750–1766. doi: 10.1110/ps.03580104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eastwood M, Hardin C, Luthey-Schulten Z, Wolynes PG. Evaluating protein structure prediction schemes using energy landscape theory. IBM Res Dev. 2001;45:475–497. [Google Scholar]
- 41.Hardin C, Eastwood MP, Prentiss M, Luthey-Schulten Z, Wolynes PG. Folding funnels the key to robust protein structure prediction. J Comput Chem. 2002;23:138–146. doi: 10.1002/jcc.1162. [DOI] [PubMed] [Google Scholar]
- 42.Fujitsuka Y, Takada S, Luthey-Schulten Z, Wolynes PG. Optimizing physical energy functions for protein folding. Prot Struct Func Gen. 2004;54:88–103. doi: 10.1002/prot.10429. [DOI] [PubMed] [Google Scholar]
- 43.Wang J, Verkhivker GM. Energy landscape theory, funnels, specificity, and optimal criterion of biomolecular binding. Phys Rev Lett. 2003;90:188101. doi: 10.1103/PhysRevLett.90.188101. [DOI] [PubMed] [Google Scholar]
- 44.Iben IET, et al. Glassy behavior of a protein. Phys Rev Lett. 1989;62:1916–1919. doi: 10.1103/PhysRevLett.62.1916. [DOI] [PubMed] [Google Scholar]
- 45.Wolynes PG. Folding funnels and energy landscapes of larger proteins within the capillarity approximation. Proc Natl Acad Sci USA. 1997;94:6170–6175. doi: 10.1073/pnas.94.12.6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cieplak M, Hoang TX, Li MS. Scaling of folding properties in simple models of proteins. Phys Rev Lett. 1999;83:1684–1687. [Google Scholar]
- 47.Thirumalai D. From minimal models to real proteins—Time scales for protein-folding kinetics. J Phys I. 1995;5:1457–1467. [Google Scholar]
- 48.Finkelstein AV, Badretdinov AY. Rate of protein folding near the point of thermodynamic equilibrium between the coil and the most stable chain fold. Fold Des. 1997;2:115–121. doi: 10.1016/s1359-0278(97)00016-3. [DOI] [PubMed] [Google Scholar]
- 49.Cieplak M, Hoang TX. Scaling of folding properties in go models of proteins. J Biol Phys. 2000;26:273–294. doi: 10.1023/A:1010359024559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takada S, Koga N. Roles of native topology and chain-length scaling in protein folding: A simulation study with a go-like model. J Mol Biol. 2001;313:171–180. doi: 10.1006/jmbi.2001.5037. [DOI] [PubMed] [Google Scholar]
- 51.Li MS, Klimov DK, Thirumalai D. Dependence of folding rates on protein length. J Phys Chem B. 2002;106:8302–8305. [Google Scholar]
- 52.Finkelstein AV, et al. Contact order revisited: Influence of protein size on the folding rate. Protein Sci. 2003;12:2057–2062. doi: 10.1110/ps.0302503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Galzitskaya OV, Garbuzynskiy SO, Ivankov DN, Finkelstein AV. Chain length is the main determinant of the folding rate for proteins with three-state folding kinetics. Proteins. 2003;51:162–166. doi: 10.1002/prot.10343. [DOI] [PubMed] [Google Scholar]
- 54.Li MS, Klimov DK, Thirumalai D. Thermal denaturation and folding rates of single domain proteins: Size matters. Polymer. 2004;45:573–579. [Google Scholar]
- 55.Munoz V, Naganathan AN, Sanchez-Ruiz JM. Direct measurement of barrier heights in protein folding. J Am Chem Soc. 2005;127:17970–17971. doi: 10.1021/ja055996y. [DOI] [PubMed] [Google Scholar]
- 56.Naganathan AN, Munoz V. Scaling of folding times with protein size. J Am Chem Soc. 2005;127:480–481. doi: 10.1021/ja044449u. [DOI] [PubMed] [Google Scholar]
- 57.Kouza M, Li MS, O’Brien EP, Hu CK, Thirumalai D. Effect of finite size on cooperativity and rates of protein folding. J Phys Chem A. 2006;110:671–676. doi: 10.1021/jp053770b. [DOI] [PubMed] [Google Scholar]
- 58.Klimov DK, Thirumalai D. Linking rates of folding in lattice models of proteins with underlying thermodynamic characteristics. J Chem Phys. 1998;109:4119–4125. [Google Scholar]
- 59.Saven JG, Wang J, Wolynes PG. Kinetics of protein folding: The dynamics of globally connected rough energy landscapes with biases. J Chem Phys. 1994;101:11037–11043. [Google Scholar]
- 60.Socci ND, Onuchic JN, Wolynes PG. Diffusive dynamics of the reaction coordinate for protein folding funnels. J Chem Phys. 1996;104:5860–5868. [Google Scholar]
- 61.Plotkin SS. Speeding protein folding beyond the go model: How a little frustration sometimes helps. Proteins: Struct Funct Bioinform. 2001;45:337–345. doi: 10.1002/prot.1154. [DOI] [PubMed] [Google Scholar]
- 62.Baker D, Plaxco KW, Simons KT. Contact order, transition state placement and the refolding rates of single domain proteins. J Mol Biol. 1998;277:985–994. doi: 10.1006/jmbi.1998.1645. [DOI] [PubMed] [Google Scholar]
- 63.Raleigh DP, Kuhlman B, Luisi DL, Evans PA. Global analysis of the effects of temperature and denaturant on the folding and unfolding kinetics of the n-terminal domain of the protein l9. J Mol Biol. 1998;284:1661–1670. doi: 10.1006/jmbi.1998.2246. [DOI] [PubMed] [Google Scholar]
- 64.Dinner AR, Karplus M. The roles of stability and contact order in determining protein folding rates. Nat Struct Biol. 2001;8:21–22. doi: 10.1038/83003. [DOI] [PubMed] [Google Scholar]
- 65.Clementi C, Das P, Matysiak S. Balancing energy and entropy: A minimalist model for the characterization of protein folding landscapes. Proc Natl Acad Sci USA. 2005;102:10141–10146. doi: 10.1073/pnas.0409471102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Whitford PC, et al. An all-atom structure-based potential for proteins: Bridging minimal models with all-atom empirical forcefields. Proteins: Struct Funct Bioinform. 2009;75:430–41. doi: 10.1002/prot.22253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thirumalai D, Klimov DK, Woodson SA. Kinetic partitioning mechanism as a unifying theme in the folding of biomolecules. Theor Chem Acc. 1997;96:14–22. [Google Scholar]
- 68.Thirumalai D, Klimov DK. Deciphering the timescales and mechanisms of protein folding using minimal off-lattice models. Curr Opin Struct Biol. 1999;9:197–207. doi: 10.1016/S0959-440X(99)80028-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.