Abstract
Nuclear Factor Y (NF-Y) is a heterotrimeric complex formed by NF-YA/NF-YB/NF-YC subunits that binds to the CCAAT-box in eukaryotic promoters. In contrast to other organisms, in which a single gene encodes each subunit, in plants gene families of over 10 members encode each of the subunits. Here we report that five members of the Arabidopsis thaliana NF-YA family are strongly induced by several stress conditions via transcriptional and miR169-related post-transcriptional mechanisms. Overexpression of NF-YA2, 7 and 10 resulted in dwarf late-senescent plants with enhanced tolerance to several types of abiotic stress. These phenotypes are related to alterations in sucrose/starch balance and cell elongation observed in NF-YA overexpressing plants. The use of transcriptomic analysis of transgenic plants that express miR169-resistant versions of NF-YA2, 3, 7, and 10 under an estradiol inducible system, as well as a dominant-repressor version of NF-YA2 revealed a set of genes, whose promoters are enriched in NF-Y binding sites (CCAAT-box) and that may be directly regulated by the NF-Y complex. This analysis also suggests that NF-YAs could participate in modulating gene regulation through positive and negative mechanisms. We propose a model in which the increase in NF-YA transcript levels in response to abiotic stress is part of an adaptive response to adverse environmental conditions in which a reduction in plant growth rate plays a key role.
Introduction
Nuclear factor Y (NF-Y) (also termed Heme Activator Protein or CCAAT Binding Factor) is a heterotrimeric transcription factor (TF) composed of NF-YA (HAP2/CBF-B), NF-YB (HAP3/CBF-A) and NF-YC (HAP5/CBF-C) subunits, which is conserved from yeast to mammals. In yeast (Saccharomyces cerevisiae) there is a fourth subunit, HAP4 necessary for complex formation and transcriptional activation [1], [2]. NF-Y activates transcription by recognizing the CCAAT- box cis-acting element present in about 30% of eukaryotic promoters in a highly specific manner via the DNA binding domain present in the NF-YA subunit [3]. The activator activity of NF-Y by binding to the CCAAT boxes has been documented for cytochrome and nitrogen metabolism-related genes in yeast [4], [5] and in genes involved in penicillin biosynthesis in Aspergillus nidulans [6]. In animals a multitude of genes, including housekeeping and inducible genes, developmentally controlled and cell cycle-related genes [7], [8], [9] contain CCAAT boxes in its promoter regions. In the case of plants, the CCAAT-box is present mainly in the promoters of genes involved in photosynthesis [10], [11].
Unlike fungi and mammals, which have only a single gene for each subunit, in plants there are more than 10 different loci encoding each subunit [12], suggesting a vast potential for the formation of different NF-Y heterotrimeric complexes that could be involved in a wide range of developmental processes and/or responses to environmental cues. In Arabidopsis, 10 NF-YA, 13 NF-YB and 13 NF-YC genes encode the different subunits of NF-Y. The expression of NF-YA family members in Arabidopsis has been reported to be regulated at the transcriptional level [12], [13] and some NF-YA family members, such as NF-YA2 and NF-YA5, are targets of the miR169 family [13], [14]. At least other six Arabidopsis NY-FA transcripts are predicted to be targets of mir169 [14].
The fact that the mir169 family comprises 14 members in Arabidopsis thaliana [14] suggests a scenario in which a complex set of regulatory combinations between several NF-YA transcripts and the numerous members of the mir169 family might control the expression of diverse groups of genes in response to developmental and/or environmental cues. Indeed, Arabidopsis NF-YA5, which is involved in drought tolerance [13], is a target for miR169 and miR169 overexpressing (OE) lines showed a similar phenotype to nf-ya5 [13], demonstrating the participation of this microRNA in drought stress responses.
Recent studies also suggest the involvement of miR169-dependent NF-YA regulation in response to nutrient stress, since some members of this microRNA family are down-regulated by nitrate and phosphate starvation [15] and the expression of some genes encoding NF-YA subunits is up-regulated by phosphate (Pi) and nitrate deprivation as well as during leaf senescence [16]–[18].
Another biological process that depends on the mir169-NF-YA interaction is plant growth. Overexpression of Arabidopsis NF-YA4 causes growth reduction [19]; while mimicry lines, with a reduced level of miR169 and increased NF-YA transcript levels, display reduced growth [20]. Other NF-Y subunits are also involved in growth control, since Arabidopsis NF-YB1 overexpression arrests growth of the shoot apical meristem [21], and rice OsHAP3E (a NF-YB9 orthologue) overexpression produces a dwarf phenotype [22]. However, the mechanism by which NF-Y modulates plant growth remains to be elucidated.
Besides the mir169-NF-YA regulatory mechanism, several studies have highlighted the importance of the NF-Y complex in plant developmental processes. For example, Arabidopsis thaliana mutants in the genes encoding NF-YB9 and NF-YB6, denominated LEAFY COTYLEDON 1 (LEC1) and LEAFY COTYLEDON 1 LIKE (L1L), respectively, produce defective embryos, which fail to acquire desiccation tolerance [23]. The notion that NF-Y complexes regulate large groups of genes involved in developmental processes is illustrated by the finding that LEC1 overexpression leads to the formation of embryo like structures in adult leaves [24] whereas lec1 produces ectopic development of trichomes on cotyledons [25]. Emerging evidence also indicates that NF-Y plays an important role in the flowering process. In Arabidopsis, CONSTANS (CO) interacts in vivo with NF-YB and NF-YC subunits, replacing a NF-YA partner, to promote flowering by regulating FLOWERING LOCUS T (FT) [21]. Additionally, NF-YA1 OE plants are late flowering, without altering CO mRNA levels, suggesting that NF-YA1 impairs the formation of the CO/NF-YB/NF-YC complex [21].
Several reports demonstrate the participation of NF-Y in abiotic stress responses. In maize (Zea mays) and Arabidopsis, overexpression of NF-YB1 and NF-YB2, respectively, promote drought resistance [26]. However, microarray analysis of the transcriptional alterations caused by NF-YB2 overexpression in Arabidopsis did not reveal a clear effect on genes directly related to stress tolerance, but rather on those involved in polysaccharide metabolism.
Here we show that the expression of 5 members of the Arabidopsis NF-YA family is induced by nutrients stress and, in contrast to the majority of stress-related transcription factors, its expression window is long-term. Overexpression of NF-YA members confers tolerance to diverse abiotic stresses, including drought, flooding, cold and heat. Unexpectedly, we found that at least several members of the NF-YA family act as negative regulators of early stress response genes and modulate overall plant growth through modification of carbohydrate metabolism and cell elongation. We propose that this genetic response conduces the plant to a state of acclimatization. We also propose that NF-YA modulates stress responses mainly by sequestering the NF-YB/NF-YC heterodimers preventing its interaction with other transcription factors that induce the expression stress-responsive genes.
Results
Members of the NF-YA family are strongly up-regulated by different abiotic stresses
It has been shown that some members of the NF-YA family are up-regulated in Pi-deprived plants [16], [27] and analysis of public microarray databases suggests that they are also deregulated by other abiotic stresses, including low nitrogen (N), drought, cold, heat, high glucose and by treatment with the stress-related hormone ABA (https://www.genevestigator.com/gv/plant.jsp). These data indicate that the members of the NF-YA family might play a general role in stress responses.
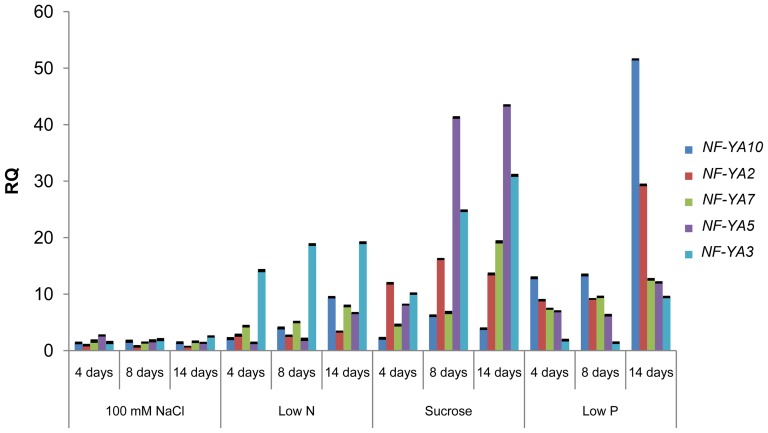
In the past years our lab has focused on the study of plant responses to nutrient stress, specifically to Pi availability. Therefore, we explore the behavior of several of these NF-YA members in response to diverse nutrient stresses. As a starting point we performed a time-course qRT-PCR analysis of the changes in transcript levels of five members of the NF-YA family (NF-YA2, 3, 5, 7, 10), which were up-regulated in our microarray analysis of the Arabidopsis response to low Pi availability [20]. Besides low Pi (5 µM), we also analyzed its response to low N (50 µM), high salinity (100 mM NaCl) and high sucrose (Suc) (9.4%). Transcript levels for these NF-YA family members increased between 5 and 50-fold, under all conditions tested, with the exception of high salinity, where only NF-YA5 and NF-YA3 transcript levels increased by 2-fold at 4 and 14 days of treatment, respectively (Figure 1). An increase in NF-YA2, NF-YA7 and NF-YA10 transcript levels was detected 4 days after germination (dag) in low P, low N and high Suc treatments, NF-YA5 and NF-YA3 showed a similar response in high Suc but no change in low N and P conditions, respectively (Figure 1). Transcript levels were maintained 8 dag and showed a further increase at 14 dag for low P and low N. In high Suc, the level of transcript induction was similar at 8 and 14 dag. This experiment also showed that even though all 5 NF-YAs tested were up-regulated in response to low P, low N and high Suc conditions, the degree of responsiveness of each NF-YA gene to each condition differed, for example, under low N NF-YA3 was the most responsive (19-fold induction at 14dag), whereas NF-YA5 was most responsive in high Suc (43-fold induction at 14dag) and NF-YA10 under low P (51-fold induction at 14 dag).
Figure 1. Expression profile of Arabidopsis NF-YA genes in seedlings exposed to different stress conditions.
Seedlings were germinated in solid media for each treatment (100 mM NaCl, low N, 9.4% Sucrose or low P). Total RNA was extracted after 4, 8 and 14 days of treatment and transcript levels for NF-YA2, 3, 7, 5 and 10 determined by qRT-PCR. Expression level of ACTIN 2 (ACT2) was used as internal reference. Values represent the means and error bars indicate the standard error (SE) of 3 independent amplification replicates.
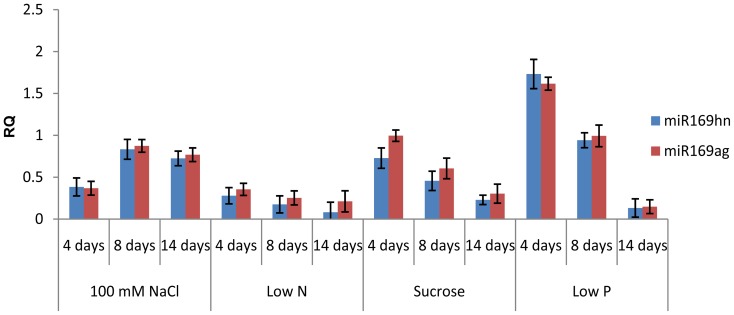
Since the expression of several members of the Arabidopsis miR169 family is repressed by low Pi and low N [15], and NF-YA5 is a target for this microRNA family [14], it was important to determine whether the expression of the miR169 family is altered by the same nutrient stress conditions under which NF-YAs mRNA levels increased. For this, we used a qRT-PCR approach similar to that previously described [15] to examine the mature forms of miR169ag and miR169hn. A more specific analysis was not possible because of the high identity between the mature miR169 sequences. A substantial decrease in the level of the mature miR169ag and miR169hn subgroups was observed in seedlings exposed to low N, low P and high Suc (Figure 2). Reduction in miR169 expression was more pronounced after 14 days of treatment, correlating with the increase in NF-YA transcript levels in response to the same treatments. Despite no clear change in expression level was observed for the miR169 subgroups under high salinity conditions, we cannot exclude the possibility that one or few specific members of the miR169 family could be controlling the low but consistent increase in transcript levels detected for NF-YA3 and NF-YA5 at 4 and 14 dag (Figure 1). This fact could be due to, the qRT-PCR analysis we used to assess miR169 levels is only able to detect subgroups of several mature miR169s but does not have specificity to detect subtle changes in the expression of a single member of the miR169 family.
Figure 2. Expression profile of mature miR169 Arabidopsis in seedlings exposed to different stress conditions.
Seedlings were germinated in solid media for each treatment (100 mM NaCl, low N, 9.4% Sucrose or low P). Total RNA was extracted after 4, 8 and 14 days of treatment and transcript levels for miR169 determined by qRT-PCR. Because of high sequence identity quantification was made for two groups of miR169, a to g and h to n. Expression level of ACT2 was used as internal reference. Values represents the means and error bars indicate SE of 3 independent amplification replicates.
Changes in NF-YA mRNA levels and miRNA169 abundance fit well with the classical behavior of miRNAs and their targets showing an inverse pattern of expression in which the NF-YA transcript level increases as the corresponding miR169 decreases.
The induction response of NF-YAs under Low Pi conditions has both a transcriptional and a post-transcriptional component
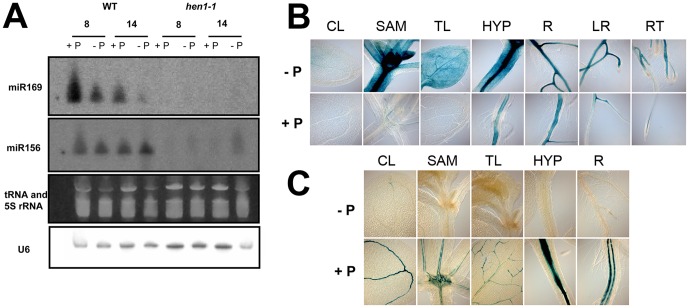
If the inverse expression patterns of miR169s and its NF-YA targets are integrated into stress response regulatory networks, and this obeys a post-transcriptional regulatory program, it would be expected that in mutants affected in microRNA biogenesis, the expression of the NF-YAs should be altered. To explore this possibility, we analyzed NF-YA transcript levels in hen 1-1 grown under contrasting Pi conditions. Since HEN1 is involved in transferring a methyl group to the 3′ end of miRNAs that confers stability to the mature miRNA [28], we expect that, in the mutant, the amount of mature miR169 is reduced leading to an increase in the uncleaved NF-YA mRNAs. Northern Blot analysis showed that the level of miR169 decreases in WT seedlings subjected to Pi-deprivation for 8 and 14 days, whereas in hen1-1 the level of mature miR169 was undetectable in both high and low Pi media (Figure 3A). qRT-PCR was performed to determine the effect of hen1-1 on NF-YA transcript levels in media containing sufficient and low Pi. As previously shown for WT seedlings, transcript levels for NF-YA2, 3, 5, 7 and 10 were higher in Pi-deprived seedlings than in those grown in Pi-sufficient conditions (Table 1). In 8-day-old hen1-1 seedlings grown in Pi sufficient conditions, NF-YA5, NF-YA3, NF-YA2 and NF-YA10 transcript levels were 9-, 5.4-, 15.7- and 5.3-fold higher than those present in WT seedlings, respectively, confirming NF-YA post-transcriptional regulation by microRNAs and suggesting that NF-YA2 is the most tightly post-transcriptionally regulated member of the tested NF-YAs (Table 1). In contrast, transcript levels for NF-YA7, which lacks the binding site for miR169, was similar in hen1-1 to that present in WT seedlings (Table 1). In 8-day-old hen1-1 seedlings grown in low Pi media a further increase in transcript levels was observed for NF-YA3 and NF-YA10 in comparison to hen1-1 seedlings grown in Pi-sufficient conditions (Table 1), suggesting that these genes are also subjected to transcriptional regulation. In the case of NF-YA2, transcript levels were similar in Pi-deprived and Pi-sufficient seedlings suggesting regulation mainly at the post-transcriptional level.
Figure 3. Temporal and Spatial Expression Patterns of miR169 gene and NF-YA10 in response to Pi availability.
(A) Northern blot analysis of miR169 in response to Pi availability (+P, 1 mM and −P, 5 µM) in 8 and 14-day-old hen 1-1 and in WT seedlings. miR156 probe was used as a control to confirm that mature microRNAs in general have decreased levels or are absent in hen1-1. U6 probe was used as a loading control. Ethidium bromide-stained tRNA and rRNA is shown below the blots to indicate the relative amount of total low molecular weight RNA loaded per lane. (B) and (C) Tissue specific expression directed by the NF-YA10 (B) and miR169ij (C) promoters as revealed by GUS histochemical assays in seedlings grown in Pi contrasting conditions 14 days after germination (dag). CL, SAM, TL, HYP, R, LR and RT, are cotyledonary leaf, shoot apical meristem, true leaf, hypocotyl, root, lateral root and root tip, respectively.
Table 1. qRT-PCR analysis of the NF-YA subfamily en Pi contrasting conditions in WT and hen 1-1 backgrounds.
| Gene | 8 days | 14 days | ||||||
| WT | hen 1-1 | WT | hen 1-1 | |||||
| +P | −P | +P | −P | +P | −P | +P | −P | |
| NF-YA5 | 1 | 6 | 9 | 1.2 | 1 | 12 | 3.3 | 4 |
| NF-YA3 | 1 | 1.4 | 5.4 | 12.4 | 1 | 9.5 | 13.6 | 11.9 |
| NF-YA2 | 1 | 9 | 15.7 | 14.2 | 1 | 29 | 21 | 62 |
| NF-YA10 | 1 | 13 | 5.3 | 32.5 | 1 | 51 | 8.9 | 84.7 |
| NF-YA7 | 1 | 9.5 | 0.7 | 2.5 | 1 | 12 | 0.9 | 6.8 |
| SPX1 | 1 | 52 | 0.5 | 65 | 1 | 169 | 0.97 | 73 |
Transcript levels that reflect a major component of post-transcriptional regulation are in bold. +P, optimum P (1 mM) −P, low P (5 µM).
Similar results were obtained for 14-day-old seedlings for NF-YA2 and 10, and for NF-YA3 and 5 for which transcript levels seem to be mainly regulated post-transcriptionally. To confirm that the effect HEN1 loss of function on NF-YA transcript levels was due to a direct effect on microRNA biogenesis and not an indirect general effect on Pi-deprivation responses, we examined the expression of SPX1, a Pi-responsive gene [27]. It was found that SPX1 transcript levels in the WT and hen1-1 were similar in both Pi-sufficient and limiting conditions, showing that hen1-1 does not have a general effect on the Arabidopsis response to Pi-availability (Table 1).
To corroborate that NF-YA2, 3, 5, 7 and 10 are also regulated at the transcriptional level by Pi-availability we fused the promoter region of these genes to the β-glucuronidase (GUS) reporter gene. In Pi-sufficient conditions, a similar expression pattern to that previously reported for PNF-YAs:GUS [12] was observed. In contrast, in Pi-deprived seedlings a significant increase of GUS activity was observed for PNF-YA2, 3, 7 and 10 lines with different patterns of tissue specific responses (Figure 3B and Figure S1A to S1D). We also produced promoter:GUS transcriptional gene fusions for some of the miR169 family members, namely miR169i/j, miR169k/l, miR169m/n, which are arranged in tandem, and for miR169h and miR169a which are monocistronic units. In seedlings grown in Pi-sufficient media we observed a pattern of expression similar to that previously reported [29], with predominant expression in the vascular tissue. Under Pi-limiting conditions it was observed that GUS staining decreased for miR169 promoters, in particular for miR169h, miR169k/l, and miR169i/j as compared to that observed for seedlings grown in Pi-sufficient media (Figure 3C and Figure S1E to S1H). There is a clear overlap between the expression patterns of PNF-YA2:GUS, PNF-YA5:GUS and PNF-YA10:GUS with PmiR169h:GUS, PmiR169i/j:GUS and PmiR169k/l:GUS in vascular tissues of leaf and hypocotyl; showing a space-time coincidence between the miRNAs and their targets suggesting that, under Pi limiting conditions, the decrease in the transcript levels of these miR169s is, at least in part, responsible for the increase in transcript levels of NF-YA2, NFYA5 and NF-YA10.
To confirm that miR169 modulates the level of NF-YA2, 3, 5, and 10 mRNAs, we generated transgenic Arabidopsis lines that overexpress the mature sequence of miR169 h to n, which is identical for all these miRNAs. For this we cloned a fragment of DNA that encodes for miR169m and miR169n precursors arranged in tandem, hereafter called miR169nm, under the control of the CaMV 35S promoter.
Two homozygous overexpressor lines were characterized by qRT-PCR, one showed a 2-fold and the other a 3.5-fold higher level of mature miR169hn when compared to WT plants (Figure S2A). qRT-PCR analysis showed that transcript levels of NF-YA2, 3, 5 and 10 were reduced in both miR169/OE lines, whereas transcript levels for NF-YA7, which lacks the target site for miR169, were unaffected by the overexpression of miR169 (Figure S2B).
Taken together, these results confirm that the post-transcriptional regulation of several members of the NF-YA family is mediated by miR169hn, which in turn are transcriptionally regulated by Pi availability.
NF-YA overexpression causes a dwarf phenotype
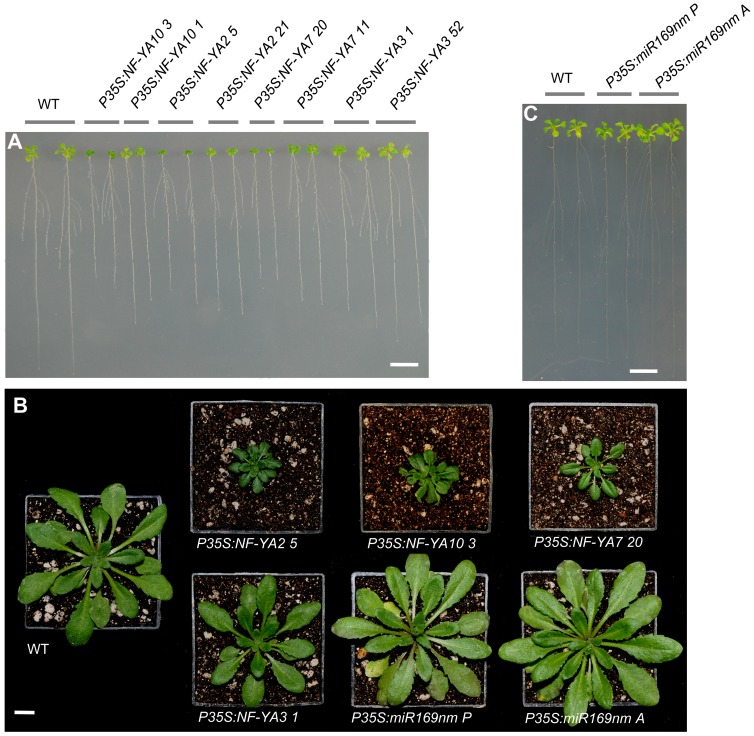
To characterize the functional role of NF-YAs, we produced transgenic plants that express miR169-resistant versions of NF-YA2, 3 and 10 genes and the native form of the NF-YA7 gene, under the control of the CaMV 35S promoter. At least 5 independent overexpressing lines were generated for each construct. After an initial phenotypic evaluation (Figure 4) and qRT-PCR analysis of two representative lines (Figure S3), one OE line for each construct was selected for further detailed evaluation. qRT-PCR analysis showed that these transgenic lines had from 20- up to 1000-fold higher transcript levels than that of the corresponding NF-YA in WT plants (Figure S3). No NF-YA5 OE lines were produced since the effect of its overexpression in Arabidopsis was previously reported [13]
Figure 4. Phenotype of transgenic lines overexpressing NF-YAs and miR169nm.
(A) and (C) Phenotype of 14-day-old seedlings overexpressing NF-YAs and miR169nm, respectively. (B) Phenotype of three-week-old plants grown under greenhouse conditions. Bars = 10 mm in (A) to (C).
Phenotypic analyses of the selected NF-YA OE lines showed that P35S:NF-YA3 plants developed in a similar fashion to WT controls, whereas P35S:NF-YA2, P35S:NF-YA7 and P35S:NF-YA10 plants showed a dwarf phenotype depending on the expression level of the transgene (Figure 4A and Figure S3). The dwarf phenotype of P35S:NF-YA2, 7 and 10 lines was observed both at the seedling and adult stages (Figure 4B), smaller siliques and seeds were also observed in these OE lines compared to those of the WT (Figure S4). The dwarf phenotype observed in NF-YA OE lines was reflected in biomass reduction, for example P35S:NF-YA2 plants had almost 2.5-fold less biomass than the WT (Figure S5). A dark green color was observed in P35S:NF-YA2, 7, and 10 leaves and to a lesser extent in leaves of P35S:NF-YA3 plants, which correlated with a higher chlorophyll content in these lines as compared to that of WT plants (Figure S6). P35S:miR169nm plants had a contrasting phenotype to that observed for NF-YA OE lines, as they produced 20% larger seeds, accumulated approximately 25% more biomass and less chlorophyll than WT plants (Figure 4B to 4C, Figure S4, Figure S5, and Figure S6). The relatively mild phenotype observed for the miR169 overexpressing lines could be explained by the fact that repression of NF-YAs containing the miR169 target sequences is incomplete and/or because the expression of NF-YA7 is not affected.
Transcriptomic analysis of genes downstream of NF-YA2, 3, 7, and 10
To identify genes regulated by NF-YAs while avoiding the potential indirect effects in gene expression related to the dwarf phenotype caused by constitutive NF-YA expression, we used the XVE inducible system (LexA-VP16-estrogen receptor) [30], [31] to drive the expression of miR169-resistant forms of NF-YA2, 3, 10 and the native form of NF-YA7 or the genomic region comprising the mir169nm. Representative transgenic lines containing these gene constructs showed WT phenotype when germinated in media lacking estradiol, whereas in estradiol-containing media, their phenotypes were similar to those observed in their constitutively expressed counterparts (data not shown). Estradiol-inducible NF-YA and miRNA169 overexpressing lines were used to analyze the effect of NF-YAs and miR169 overexpression on global RNA profiles.
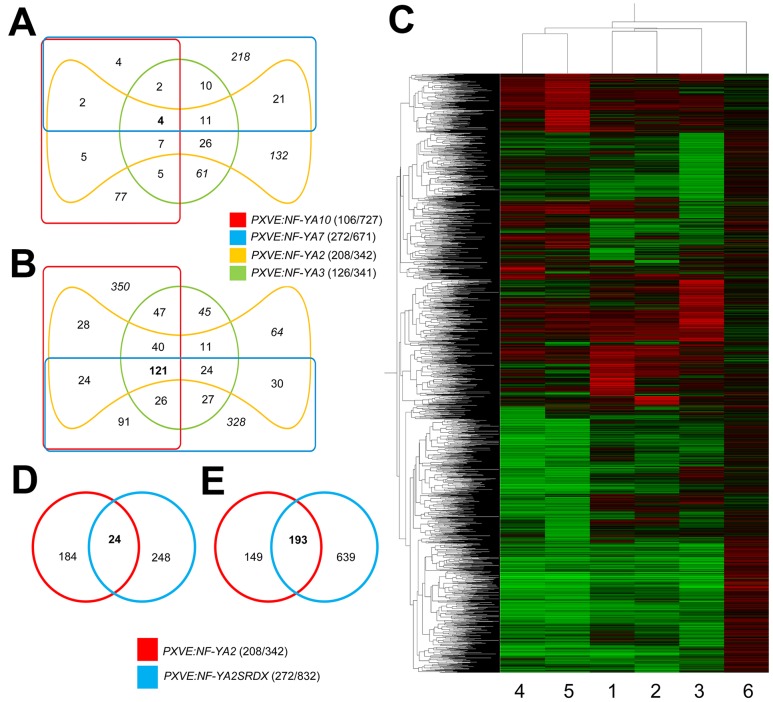
The microarray data corresponding to the overexpression of NF-YAs showed a strikingly small overlap between up-regulated genes (Figure 5A), with only 4 genes shared by the four transgenic lines. Interestingly, down-regulated genes showed a greater overlap with 121 genes common to all lines (Figure 5B). In contrast to the results obtained for PXVE:NF-YA lines, our data from the PXVE:miR169nm array showed that 270 of the 359 differentially expressed genes were induced, including 32 genes that are down-regulated in the four PXVE:NF-YAs lines (Dataset S1). These results suggest that NF-YA overexpression has a mainly negative effect on gene expression and that this repressor activity is modulated by miR169.
Figure 5. Global gene expression profile in NF-YA overexpressing lines.
(A) and (B), (D) and (E) Venn diagrams showing genes with differential expression for PXVE:NF-YAs and PXVE:NF-YA2SRDX, respectively. (A) and (D) represent up-regulated genes; (B) and (E) down-regulated genes. The number of up-regulated/down-regulated genes is given in parenthesis in each case. The number of shared genes in the corresponding data set is indicated in bold. (C) Overall picture of the hierarchical clustering analysis of differentially expressed transcripts. Expression patterns of differential genes identified in at least one of the evaluated lines (PXVE:NF-YA2, 3, 7, 10; PXVE:NF-YA2SRDX and PXVE:miRNA169nm; 1, 2, 3, 4, 5, 6, respectively. Green indicates down-regulated values, red, up-regulated values and black unchanged values.
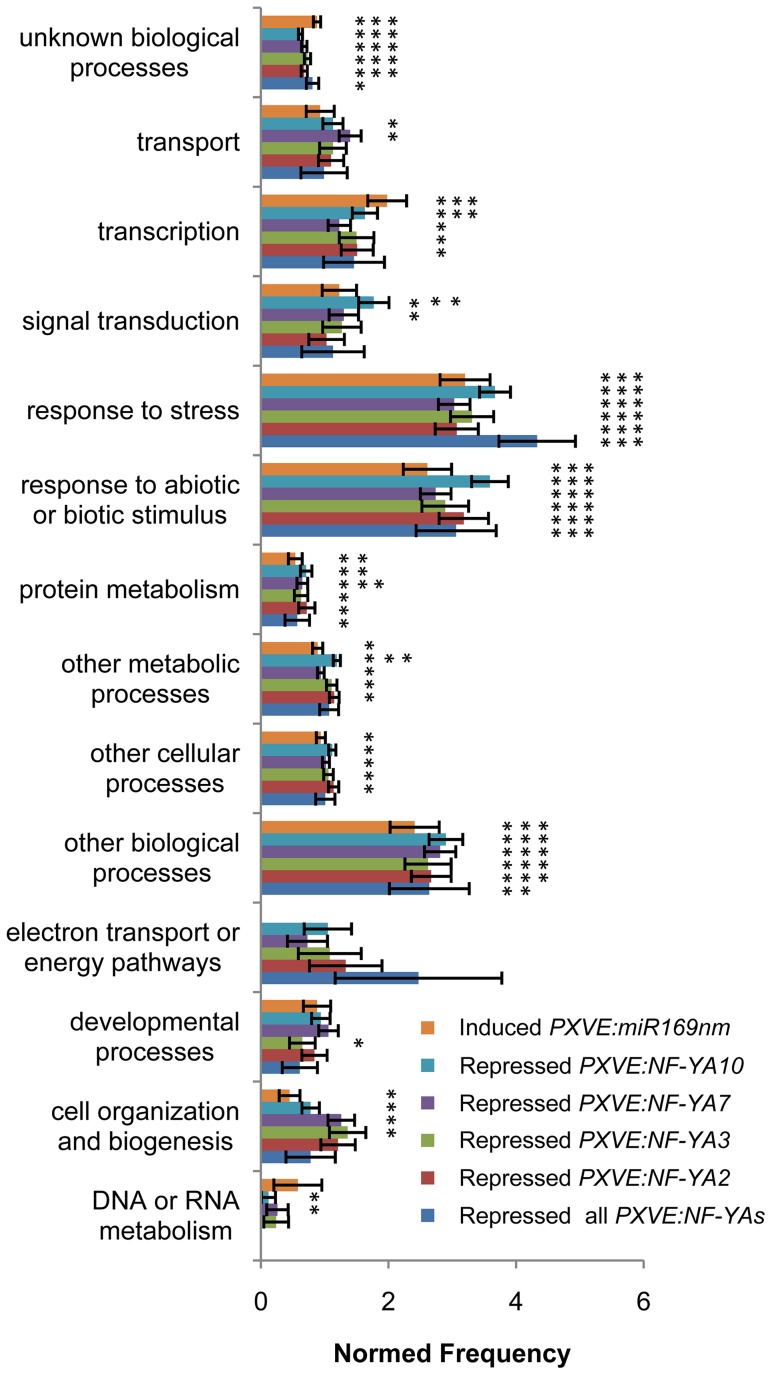
To gain insight into the co-regulatory network downstream of NF-YAs, differentially expressed genes in NF-YA overexpressing lines were clustered using the Pearson correlation and average linkage clustering. This analysis confirmed that the expression profiles of PXVE:NF-YA lines were similar and inverse to those observed for the PXVE:miR169nm line (Figure 5C, Dataset S2). Analysis of the microarray data of PXVE:NF-YA lines using the web-tool Superviewer [32], showed that the expression of stress-related genes is modulated by the NF-Y/miR169 system, since these genes were down-regulated as a consequence of NF-YA overexpression and induced in the PXVE:miR169nm line (Figure 6). Using the MapMan functional category classification we found that differentially expressed genes in NF-YA overexpressing lines include genes whose function is related to carbon metabolism, starch breakdown, glycolysis, cell wall biosynthesis, senescence, fermentation, ABA and auxin sensing, circadian clock, among others, were repressed in NF-YA OE lines (Figure S7 and Dataset S3). Moreover, using the web-tool Genesect at VirtualPlant (http://virtualplant.bio.nyu.edu/cgi-bin/vpweb/), we determined that the set of general stress-induced genes [33], [34] have a significant overlap with genes repressed in PXVE:NF-YA lines (Table 2).
Figure 6. Functional classification of differentially expressed transcripts in PXVE:NF-YAs and PXVE:miR169nm lines.
Gene ontology classification of the transcripts according to classical gene ontology categories using the web-based tool Classification Superviewer (http://bar.utoronto.ca) with normalized class score option. One, two and three asterisks indicate a p-value<0.05, 0.001 and 0.0001, respectively.
Table 2. Z-score for overlap between early general stress response genes and those repressed in PXVE:NF-YA transgenic lines.
| Experiment | Line | |||||||
| PXVE:NF-YA2 | PXVE:NF-YA3 | PXVE:NF-YA7 | PXVE:NF-YA10 | |||||
| Z-score | S | Z-score | S | Z-score | S | Z-score | S | |
| Ma and Bohnert (2007) | 18.0 | 29 | 13.4 | 23 | 13.9 | 34 | 23.7 | 58 |
| Walley et al. (2007) | 11.2 | 17 | 12.3 | 18 | 14.1 | 30 | 15.7 | 35 |
The overlap of early general stress response genes reported in two previous studies [33], [34] and the repressed genes by NF-YAs (cutoff≤2-fold, p-value≤0.05) is given. Data obtained using the Genesect web tool available through VirtualPlant 1.2 software platform [35]. S, size of intersection. Significant differences (p-value<0.001) based on a Z-score overrepresented (Z-score>11) are highlighted in bold.
NF-YA expression impairs cell elongation and has a strong impact on carbon metabolism
In coincidence with the dwarf phenotype of the NF-YA OE lines, we found that among the differentially expressed genes in PXVE:NF-YA lines, a number of genes strongly down-regulated encode for enzymes involved in cell wall modification processes such as xyloglucan endotransglucosylase-hydrolases (XTHs) and expansins (EXPs), suggesting that the dwarf phenotype observed in these lines could be due to defects in cell elongation (Table S1). To determine whether cell elongation was impaired in P35S:NF-YA lines, we measured hypocotyl elongation in etiolated P35S:NF-YA seedlings. These lines showed an important decrease in elongation capacity in this assay; P35S:NF-YA2, 3, 7 and 10 seedlings had a 49, 33, 41 and 39%, reduction in hypocotyl elongation as compared to that achieved by WT plants (Figure S8A to S8B).
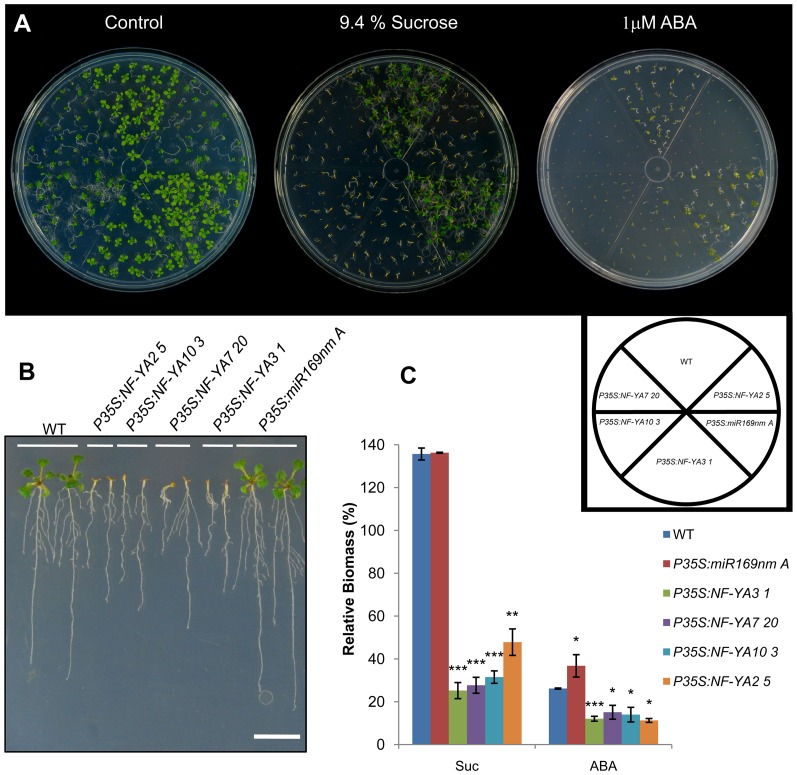
In PXVE:NF-YA seedlings we also identified several differentially expressed genes involved in carbon metabolism, such as sucrose synthases (SUS1 and SUS4), qua-quine starch (QQS), β-amylase 3 (BAM3) and α-AMYLASE 2. Interestingly, P35S:NF-YA lines treated with 9.4% Suc showed a Suc-hypersensitive phenotype, showing limited cotyledon expansion and being unable to develop true leaves as compared to the WT (Figure 7A to 7C). Moreover, we found that P35S:NF-YA2 plants had a 2-fold higher Suc content than WT whereas miR169/OE lines showed a 20% reduction (Table S2). P35S:NF-YA lines also had a higher starch content than WT plants, whereas the P35S:miR169nm line showed lower starch accumulation (Figure S9). Together these results suggest that NF-YAs can modulate plant growth (i.e. cell expansion) through the regulation of carbon metabolism related genes.
Figure 7. P35S:NF-YA lines display an increased sensitivity to sucrose and ABA.
(A) Images of twelve-day-old P35S:NF-YA, P35Smir169nm and WT, seedlings grown on 0.1× MS control media and on media supplemented with 9.4% sucrose or 1 µM ABA under standard light/dark conditions (16/8). A schematic representation of the distribution of the different lines on the plate is shown. (B) Phenotype of 14-day-old WT, P35S:NF-YA and P35Smir169nm seedlings grown in plates with medium supplemented with 9.4% sucrose and grown in vertical position in a growth chamber. Bar = 10 mm. (C) Relative biomass of wild-type and overexpressing plants grown on media supplemented with 9.4% sucrose (Suc) or 1 µM ABA (ABA) for twelve days. Total biomass was recorded and used to calculate the relative biomass (expressed as a percent of the value for the same line growing on control medium, set to 100%). Values are means and SD of three biological replicates statistically treated using a student t-test (*P<0.05, **P<0.01, ***P<0.001).
P35S:NF-YA lines have an abiotic stress tolerance phenotype related to altered ABA perception and senescence
Our microarray data also showed that the expression of genes involved in ABA perception was strongly affected in all PXVE:NF-YA lines (Table 3). Since the crosstalk between sugar and ABA signaling has been well documented [36], we tested whether NF-YA overexpression also alters ABA sensitivity. Seedlings of P35S:NF-YA lines grown on plates with 1 µM ABA showed hypersensitivity to this hormone, while the P35S:miR169nm line was slightly less sensitive than the WT (Figure 7A and 7C).
Table 3. ABA signaling pathway related genes with altered transcript level in PXVE:NF-YA transgenic lines.
| Gene ID | Description | Fold Change | |||
| PXVE:NF-YA2 | PXVE:NF-YA3 | PXVE:NF-YA7 | PXVE:NF-YA10 | ||
| At4g17870 | PYR1/RCAR11 | - | - | 0.56 | 0.61 |
| At1g01360 | PYL9/RCAR1 | 0.61 | 0.58 | 0.51 | 0.50 |
| At2g26040 | PYL2/RCAR14 | 0.52 | 0.56 | 0.61 | 0.58 |
| At2g38310 | PYL4/RCAR10 | - | - | 0.78 | 0.67 |
| At5g53160 | PYL8/RCAR3 | 0.62 | 0.56 | 0.51 | 0.62 |
| At5g05440 | PYL5/RCAR8 | - | 0.71 | 0.48 | - |
| At2g40330 | PYL6/RCAR9 | 0.66 | 0.64 | 0.60 | 0.59 |
| At1g72770 | HAB1 | - | - | - | 0.32 |
| At4g33950 | OST1 | 0.61* | 0.54 | 0.59 | 0.31 |
| At3g50500 | SNRK2.2 | - | - | 0.72 | - |
| At2g17290 | CPK6 | - | - | 0.64 | 0.67 |
| At1g35670 | CPK11 | 0.46 | - | 0.50 | 0.45 |
| At3g57530 | CPK32 | - | - | - | 0.46 |
| At3g51920 | CML9 | - | - | 0.58 | 0.55 |
| At5g37770 | CML24 | 0.62 | 0.69 | 0.62 | 0.53 |
Gene expression values shown represent fold change (estradiol treatment vs. control), p-value≤0.05. PYR, PYRABACTIN RESISTANCE; PYL, PYR1-Like; RCAR, regulatory component of ABA receptor; HAB1, Hypersensitive to ABA1; OST1, Open stomata 1; SNRK, Sucrose non-fermenting1 related kinase; CPK, calcium-dependent protein kinase; CML, Calmodulin like. Asterisk (*) indicates p-value 0.056. (−) not statistically significant.
A direct link between ABA and abiotic stress responses has been reported, i.e., transgenic plants hypersensitive to ABA are tolerant to drought stress [37]. According to our microarray data, NF-YA overexpression down-regulates the expression of the general stress response genes, including most of the 48 genes of the “anaerobic cluster” [38] (Table S3). These findings prompted us to evaluate the response of P35S:NF-YA lines to abiotic stress conditions, namely flooding, drought, heat and cold. P35S:NF-YA lines showed higher tolerance to submergence when compared to WT plants, in which leaves showed the typical yellowing characteristic of the senescence induced by hypoxia (Figure 8A, 8C to 8F); P35S:miR169nm plants behaved similar to WT (Figure 8B). We also observed that stress-induced damage by drought (Figure S10A and S10B), heat and cold (Figure S11A and S11B) was significantly less severe in P35S:NF-YA seedlings than in WT and P35S:miR169nm seedlings.
Figure 8. NF-YA overexpression produces a flooding tolerant phenotype.
(A) to (F) represent plants belonging to WT, P35S:miR169nm and P35S:NF-YA3, 7, 10, 2, respectively, after 14 days of recovery. Arrows indicate senescent leaves. Inset in (F) is a magnification of P35S:NF-YA2 rosette. Two-week-old plants of each transgenic line were completely submerged in water-filled tanks in standard light/dark cycle (16/8) for 5 days and photographed after a recovery period of 14 days. The depth of the water column during submergence was kept at 6 cm above the pot surface.
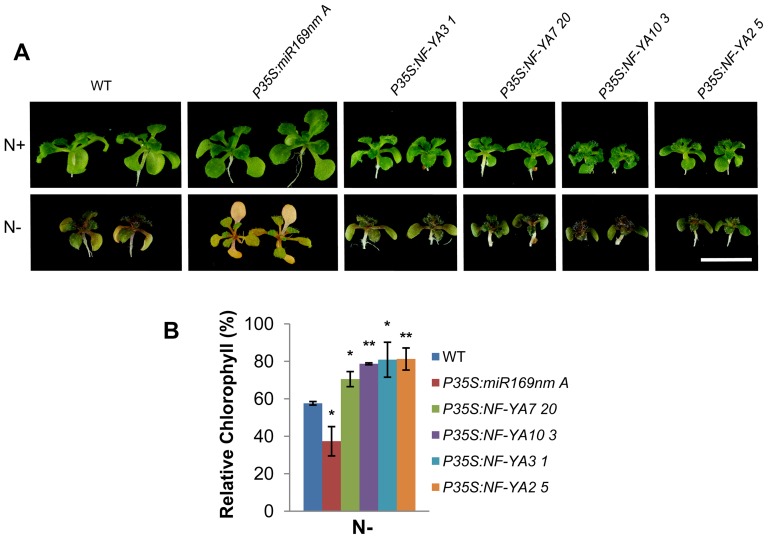
Several senescence-related genes, including SENESCENCE 1 (SEN1), POLYUBIQUITIN 10 (UBQ10), SALT TOLERANT ZINC FINGER (STZ) and WRKY6 are repressed in P35S:NF-YA plants (Table 4), suggesting that the increased expression of NF-YAs could lead to reduced senescence in Arabidopsis. We confirmed the role of NF-YA TFs in controlling senescence using an assay based on nitrogen starvation-induced senescence [39]. After two weeks of N starvation, WT plants showed clear senescence symptoms such as leaf yellowing, particularly in cotyledons. Leaf senescence was less drastic in P35S:NF-YA2, 3, 7 and 10 seedlings than that observed for the WT. In contrast, leaf senescence was more drastic in P35S:miR169nm seedlings than for the WT (Figure 9A). Chlorophyll content analysis confirmed the observed phenotypes under N starvation conditions, as chlorophyll content in WT seedlings was reduced 40%, whereas that of P35S:NF-YA lines decreased only 20 to 30% and in P35S:miR169nm seedlings reduction was about 60% (Figure 9B). Similar results were obtained using another senescence assay, by transferring plants to darkness for 7 days (Figure S12). Under greenhouse conditions, a delay between one to three weeks in developmental senescence was also observed for NF-YA overexpressing plants, as compared to the WT at the end of their life cycle (Figure S13A to S13L).
Table 4. Expression changes in genes involved in senescence related processes in PXVE:NF-YA and PXVE:miR169nm transgenic lines.
| Gene ID | Description | Fold Change | ||||
| PXVE:NF-YA2 | PXVE:NF-YA3 | PXVE:NF-YA7 | PXVE:NF-YA10 | PXVE:miR169nm | ||
| At4g35770 | SENESCENCE 1 | - | 0.48190 | 0.38967 | 0.16077 | 2.21905 |
| At4g30270 | SENESCENCE 4 | - | - | 0.46831 | 0.37010 | 1.44568 |
| At4g02380 | SENESCENCE-ASSOCIATED GENE 21 | 0.36220 | 0.54261 | 0.41296 | - | - |
| At4g35985 | Senescence/dehydration-associated protein-related | - | - | 0.41765 | 0.48309 | 1.71738 |
| At5g62000 | ORESARA 14 | - | - | 0.60483 | 0.57416 | - |
| At4g05320 | POLYUBIQUITIN 10 | 0.64522 | 0.68095 | 0.54658 | 0.60331 | - |
| At2g17850 | Rhodanese | 0.25176 | 0.11597 | 0.09108 | 0.10600 | - |
| At1g27730 | SALT TOLERANT ZINC FINGER | 0.39096 | 0.42403 | - | 0.35361 | - |
| At1g62300 | WRKY6 | 0.62249 | 0.51744 | 0.65640 | 0.39123 | 1.29049* |
| At4g01250 | WRKY22 | - | - | 0.49892 | 0.44510 | 2.13443 |
| At4g23810 | WRKY53 | 0.44315 | 0.55789 | 0.58980 | - | - |
| At5g46700 | TETRASPANIN 1 | - | 1.41474 | 1.50586 | - | - |
| At2g19580 | TETRASPANIN 2 | - | - | 0.59429 | 0.61269 | - |
| At2g23810 | TETRASPANIN 8 | - | - | 0.45217 | 0.52539 | 1.52715 |
| At5g66040 | SULFURTRANSFERASE PROTEIN 16 | 0.66635 | 0.69143 | 0.43626 | 0.64451 | - |
Gene expression values shown represent fold change (estradiol treatment vs. control), p-value≤0.05. (−), not statistically significant. Asterisk (*) indicates p-value 0.053.
Figure 9. Plants overexpressing NF-YA display reduced N starvation induced senescence.
(A) Photograph of WT, P35S:NF-YA and P35S:miR169nm plants grown for 2 weeks on N+ (N optimum) or N− (N limited) media under standard light/dark cycle. Bar = 10 mm. (B) Relative chlorophyll contents of WT and overexpressing plants grown on N contrasting conditions. Total chlorophyll content was measured, normalized per gram fresh weight of sample and used to calculate the relative content (expressed as a percent of the value for the same line growing on N optimum, set to 100%). Values are means and SD of three independent experiments. Statistically significant differences using the student t-test between WT and transgenic lines are indicated (*P<0.05, **P<0.01).
NF-YAs are part of an acclimatization strategy to cope with adverse conditions
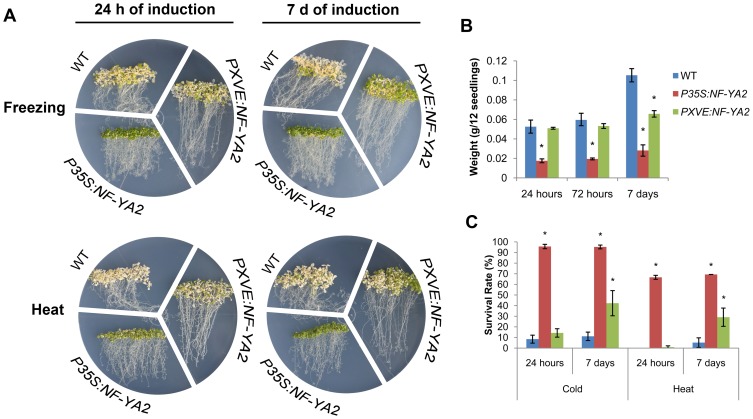
Although there is substantial evidence indicating that growth reduction is part of an adaptive strategy in plants to face abiotic stresses [40], it is also possible that a dwarf phenotype per se promotes tolerance to abiotic stresses because smaller plants have lower water and nutrient requirements. To assess whether the stress tolerance phenotype observed in P35S:NF-YA is a direct effect of NF-YA overexpression per se or indirectly by reducing plant growth, we carried out freezing and heat assays using WT, P35S:NF-YA2 and PXVE:NF-YA2 lines. Seeds from these three lines were germinated in MS media and 8 dag the seedlings were transferred to MS supplemented with 7 µM β-estradiol for short (24 h) and long (7 days) induction times prior to exposure to freezing and heat stress. The idea is that if overexpression of NF-YAs per se is sufficient to confer tolerance, a short induction time should be enough, whereas if growth reduction is responsible for tolerance, a longer induction time will be needed.
At the short induction time, freezing and heat conditions imposed a similar stress on PXVE:NF-YA2 and WT seedlings, (Figure 10A to 10D). After 7 days of induction, when PXVE:NF-YA2 seedlings show a decrease in growth rate, seedlings showed a better survival to heat and freezing stress compared to WT. The reduction in growth rate was determined by monitoring the repression of EXL (Figure S14A and S14B), a gene involved in growth promotion process [41]. A control line constitutively overexpressing NF-YA2 (P35S:NF-YA2) performed significantly better than the WT at both short and long exposure times (Figure 10E).
Figure 10. Long-term expression of NF-YA2 confers tolerance to freezing and heat.
(A) Images of eight-day-old WT, P35S:NF-YA2 and PXVE:NF-YA2 seedlings grown on 0.1× MS that were transferred to 0.1× MS+ β-estradiol (7 µM) for 24 h or 7 days prior to stress. Photographs were taken after four and seven days of recovery time for freezing (−7°C/4 h, lids off) and heat (45°C/2 h, lids on) treatments, respectively. (B) Weight of 12 seedlings of each line was determined at the indicated induction times. Data represent means and SD of three replicates statistically treated using a student t-test (*P<0.05). (C) Average survival rate of each plant line subsequent to the stress assay (n>50). Data represent means and SD of three replicates statistically treated using a student t-test (*P<0.05).
Promoters of down-regulated genes in NF-YA overexpressing lines are enriched in DNA motifs related to stress responses
Since induced ectopic expression of the NF-YAs caused changes in the expression of a large number of genes, we searched, using the Athena Web tools [42], for enriched cis-regulatory elements in the promoters of the differentially expressed genes in each line. No clear motif enrichment was detected in up-regulated genes, whereas in down-regulated genes a number of enriched motifs was found, including ABA signaling-related (ABF binding site motif, ABRE binding site motif, ACGT ABRE motif A2OSE), dehydration responses (ABRE-like binding site motif, MYB1AT), drought stress responses (AtMYC2BS in RD22), unfolded protein response (UPRMOTIFIAT), WRKY transcription factor binding sites (W-box), TFIID binding site (TATA-box motif) and motifs related to circadian rhythm (Table S4). Intriguingly, no enrichment of the CCAAT motif in the promoter of differentially expressed genes in NF-YA OE lines was observed using either the Athena or the Promomer DNA motif searching tools [43] (Table S5). Therefore, it is unlikely that the complete NF-YA/NF-YB/NF-YC complex, which recognizes the CCAAT-box, regulates the expression of genes that are repressed in NF-YA OE lines.
Transcriptional activities of NF-YAs
Down-regulation of stress related genes in NF-YA overexpressing lines could be due to the NF-YA dependent activation of one or several transcriptional repressors or to a direct repressor activity of NF-YAs. In an attempt to elucidate whether NF-YAs act as positive and/or negative regulators of gene expression we used the CRES-T system (Chimeric repressor silencing technology) [44], [45] to convert NF-YA2 into a repressor. We reasoned that if NF-YAs indirectly down-regulated gene expression by activating the expression of one or several transcriptional repressors, conversion of NF-YAs into a repressor would result in the opposite effect (transcriptional activation of the same set of genes in the absence of the repressor(s)). If NF-YAs have a direct negative effect on gene expression, conversion of NF-YAs into repressors should have a similar effect on the expression of genes that are down-regulated in NF-YA overexpressing lines. In the CRES-T system, the fusion of a TF to the EAR-motif repression domain (SRDX) causes the repression of the genes whose promoters are normally recognized by the native TF. To avoid possible lethality caused by strong repression of the NF-YAs targets, we cloned the NF-YASRDX fusions under the control of the inducible XVE system.
We obtained at least 5 independent transgenic lines harboring the PXVE:NF-YA2SRDX construct. After an initial evaluation of the phenotype of these lines, seeds of two representative lines were germinated in media supplemented with different estradiol concentrations (20 nM to 20 µM). Eight day-old PXVE:NF-YASRDX2 seedlings showed a strong dwarf phenotype which was proportional to the estradiol concentration (Figure S15A to S15D), while untreated seeds from this line showed normal morphology (data not shown). The phenotype of these NF-YA repressor lines was similar but more drastic than that observed for NF-YA overexpressing lines.
Microarray analysis of the PXVE:NF-YA2SRDX line showed that the repressor version of NF-YA2 caused a strong down-regulation in gene expression since 832 of the 1104 differentially expressed genes suffered a strong repression (Dataset S1). Comparison of differentially expressed genes between the PXVE:NF-YA2 and PXVE:NF-YA2SRDX lines showed little overlap among up-regulated genes (Figure 5D), whereas 56% of repressed genes in PXVE:NF-YA2 were also repressed in the chimeric repressor line (Figure 5E). Moreover, of 121 down-regulated genes common among the four PXVE:NF-YA lines, 107 were also repressed in PXVE:NF-YA2SRDX (Dataset S1). Clustering array data from the NF-YA2 repressor line using the Pearson correlation and average linkage clustering confirmed that the expression profiles of PXVE:NF-YA2SRDX and PXVE:NF-YA lines were similar (Figure 5C, Dataset S2). The phenotype of NF-YA2 repressor lines and the similarity of the effect of overexpression of NF-YA2 and its repressor version on global gene expression suggests that NF-YAs could act as repressors. Therefore, it is possible that NF-YAs could act both as transcriptional activators of genes, whose promoters contain the CCAAT-box (as previously documented in other systems) and as repressors of a subset of genes that lack the CCAAT-box. However, formal demonstration of these activator and repressor activities requires additional experimentation.
Potential NF-YA target genes
Comparison of the microarray data for the NF-YA2 OE line with that of the chimeric NF-YA2 repressor would allow the identification of the genes that are directly regulated by NF-YA2. We expected that genes whose expression is deregulated (both positively and negatively) in the NF-YA2 OE line and down-regulated in the PXVE:NFYA2SRDX line, would represent direct targets of NF-YA2. With this idea in mind we inspected the list of the differentially expressed genes, whose level of expression varied by at least two-fold (p-value≤0.05), for both PXVE:NF-YA2 and PXVE:NFYA2SRDX lines, and found that out of 832 down-regulated genes in the PXVE:NFYA2SRDX line, 202 genes were differentially expressed in PXVE:NF-YA2, of which 9 were induced and 193 were repressed. A search for cis-acting elements using the Athena and Promomer web tools in the 193 common genes, showed a similar motif overrepresentation to that obtained for the complete lists of differentially expressed genes (Table S6 and S7).
Because many of the genes that are down-regulated by the chimeric NF-YA2 repressor did not pass the cut off value in the NF-YA2 OE line, it is possible that these genes represent a group that is directly regulated by the NF-Y complex, but whose expression is not affected by overexpression of only one of the subunits of the complex, considering that the complete NF-YA/NF-YB/NF-YC heterotrimer is necessary to bind CCAAT-box containing promoters [4], [46]. A survey of conserved motifs in the promoters of 322 down-regulated genes in the PXVE:NF-YA2SRDX line, whose expression was not significantly altered in PXVE:NF-YA2 (Dataset S4), revealed that the CCAAT motif was overrepresented in this set of genes (Z-score = 3.4, significance value = 0.001) and that 69% of them (223) have one or more CCAAT boxes (Dataset S4), which represents more than a 2-fold increase compared to the 30% normally found in eukaryotic promoters [3]. These results suggest that NF-YA positively regulates the expression of promoters containing the CCAAT-box and when converted into a repressor down-regulates their expression. Additional experiments, such as transactivation using protoplast systems will be required to provide direct evidence of the activator and repressor activities suggested by our data.
Discussion
Expression of NF-YAs is regulated at the transcriptional and post-transcriptional level
NF-Y activity is regulated at the transcriptional, post-transcriptional and post-translational levels, with some variation throughout the different kingdoms. In human cells, where the biochemistry of the NF-Y complex is well understood, NF-Y activity is regulated through NF-YA subunit by differential splicing [47], by ubiquitination and acetylation [48] and by long noncoding RNAs [49]. In Aspergillus nidulans, NF-Y activity is controlled by the redox status through two closely positioned cysteine residues, Cys74 and Cys78, of the NF-YB subunit [50], but in plants NF-YB subunits lack the first cysteine residue suggesting that this mechanism of regulation does not operate in plants [51].
Our results show that, in Arabidopsis, NF-YA expression is regulated both at the transcriptional and post-transcriptional levels. Analysis of NF-YAs in hen1-1 and in miR169/OE lines revealed that a post-transcriptional component participates in regulating the expression of Arabidopsis NF-YA2, 3, 5 and 10. In agreement with the importance of a post-transcriptional mechanism, the expression of miR169 family members showed an inverse expression pattern in response to the same stress conditions that promote an increase in the NF-YA transcript levels. Analysis of promoter-GUS gene fusions confirmed the transcriptional component of NF-YA expression at least in contrasting Pi conditions. This transcriptional induction was not surprising since NF-YA7 has recently been identified as a direct target of the central regulator of the Pi stress response PHR1 [52].
In plants, the NF-Y complex activity could be controlled by the NF-YA subunit as previously shown for the mammalian system [48], [53]. In agreement with this hypothesis, we found that when NF-YAs are converted into repressor using the CRES-T system the expression of a large set of genes whose promoter contain one or more copies of the CCAAT-box are repressed.
NF-YA overexpression has a severe impact on growth control processes
Previous reports [17], [54], [55] and our own results show that the expression of several members of the NF-YA family is induced under abiotic stress conditions that reduce plant growth. The phenotype and changes in global gene expression observed in our Arabidopsis NF-YA OE lines suggest that several NF-YAs could regulate plant growth by modulating the expression of genes involved in carbohydrate metabolism and cell expansion. Carbohydrate metabolism has been proposed to be a key regulator of plant growth, since an imbalance between Suc and starch leads to growth reduction in plants subjected to abiotic stress [54], [56]–[59], also starch content has been proposed as a major integrator of growth control in Arabidopsis [60]. The finding that NF-YA overexpression leads to a Suc-hypersensitive phenotype, an increase in starch accumulation and a 2-fold increase in Suc content, supports the notion that reduced growth in P35S:NF-YA lines is related to alterations in carbohydrate metabolism. We speculate that Suc hyper-accumulation in P35S:NF-YA2 could be responsible for the repression of genes encoding Suc degrading enzymes such as Sucrose Synthases 1 and 4 observed in all PXVE:NF-YA lines.
In this study we found that transcript levels of BAM3, QQS and α-AMYLASE 2 are reduced in PXVE:NF-YA lines. Interestingly, it has been reported that reduced transcript levels of the starch degrading enzymes qua-quine starch (QQS) and β-amylase 3 (BAM3) result in a sex (starch excess) and dwarf phenotype [61], [62]. These evidences support the notion that NF-YA plays a key role in regulating part of the genetic program that modulates carbon metabolism and plant growth.
Plant growth depends in part on cell elongation, a process that requires re-arrangements of cell wall components where XTHs and EXPs play an active role in cell wall loosening [63]. Several XTH and EXP encoding genes such as XTH6, XTH12, XTH14, EXP8, EXP14 are repressed in several PXVE:NF-YA lines, (Table S1) and the seedlings of these lines showed reduced cell elongation (Figure S8A and S8B). When we search for CCAAT enrichment in the promoters of this set of cell wall remodeling genes (Table S1), instead of CCAAT overrepresentation, these promoters displayed an enrichment in TATA box motifs [Promomer: z-score = 4.5 (top), significance = 0.001; Athena: p-value<10−3 (the unique highly enriched)], which suggest that cell elongation process would not be directly controlled by the heterotrimeric NF-Y complex.
We provide evidence to suggest that NF-YAs modulate a transcriptional program that links abiotic environmental signals with carbon metabolism and cell expansion, two key elements for plant growth rate. We speculate that the Arabidopsis NF-YAs controls a genetic program that, in response to abiotic stresses, adjusts metabolic expenditure associated with cell elongation and carbon metabolism as a strategy to adapt to adverse conditions. This strategy would involve the action of NF-YA factors independently of the capacity of this factor to drive the expression, together with NF-YB and NF-YC, of the CCAAT-containing genes.
NF-YA has positive and negative effects on gene expression
It has been proposed that NF-Y modulates gene expression by two distinct mechanisms, the best characterized involves the association of the heterotrimeric complex NF-YA/YB/YC to the CCAAT-box motif via the DNA-binding domain of NF-YA, inducing the expression of CCAAT-containing genes [53]. Several studies have shown the requirement of the whole NF-YA/YB/YC complex to bind the CCAAT-box and activate the transcription of promoters containing this cis-acting element [4], [19], [46]. The second mechanism proposes that NF-YA acts as a repressor by sequestering the NF-YB/YC heterodimer and preventing its association with other transcription factors that mediate binding to DNA motifs other than the CCAAT-box [21], [64]. Therefore by titrating the NF-YB/YC heterodimer, NF-YA indirectly represses the expression of NF-YB/YC/TF targets.
The finding that the overexpression of the repressor version of NF-YA2 results in the negative regulation of genes whose promoters contain the CCAAT-box, suggests that, as in yeast and animal cells, NF-Y activates the expression of CCAAT-box promoters. However, since overexpression of NF-YA2 also results in the negative regulation of a similar subset of genes that are negatively regulated in the repressor version of NF-YA2, it is likely that NF-YA also acts as a negative regulator of genes expression. NF-YA repressor activity generated by sequestering the NF-YB/YC heterodimer has been previously reported in Arabidopsis. It was shown that NF-YB and NF-YC subunits interact with CO to promote flowering and that overexpression of NF-YA1 and NF-YA4 delay flowering by preventing the interaction of CO with NF-YB/NF-YC [21]. We found that when NF-YA2, 3, 7 and 10 are ectopically expressed, there is a delayed flowering (Figure S16), suggesting that these NF-YA subunits can act redundantly to NF-YA1 and NF-YA4 and prevent the interaction of CO with the NF-YB/NF-YC heterodimer and alter flowering time. In plants, another clear example of the negative effect of NF-YA on gene expression was reported by Yamamoto and co-workers (2009). These authors showed that NF-YB/NF-YC forms an heterotrimeric complex with bZIP67 to activate the expression of the CRUCIFERIN C (CRC) promoter via binding to an ABRE motif and demonstrated that overexpression of NF-YA4, 5, 7, and 9 prevents the interaction with bZIP67 and blocks the activation of CRC [64]. We found 99, 91, 175 and 285 repressed genes in the microarray analyses of PXVE:NF-YA2, 3, 7, 10, respectively, that contain the ABRE or ABRE-like motifs in their promoter sequence (Table S4). This suggests that NF-YA could regulate a large subset of ABA-responsive genes by interfering with the binding of the NF-YB/NF-YC complex to some members of the bZIP family. However, additional experimentation is required to directly demonstrate this general activity of NF-YA as a transcriptional repressor.
NF-YAs role in controlling a general abiotic stress response
The most conspicuous phenotypes of NF-YA OE plants are: a significant reduction in growth rate, delayed senescence and increased tolerance to different abiotic stresses. These phenotypes together with the finding that transcript levels of NF-YAs are induced by different abiotic stress conditions, suggest an important general role for NF-Y in the response to adverse environmental conditions in Arabidopsis. The relatively late and prolonged induction window of NF-YAs by abiotic stress found by others [65] and in this work, contrasts with that observed for more specific regulators of stress responses, which generally have early and transient expression patterns [66].
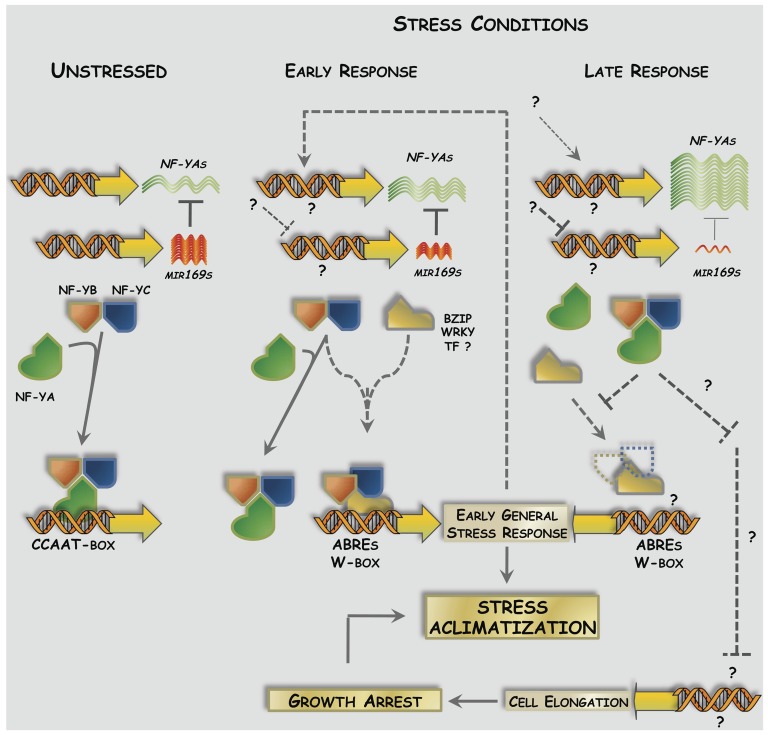
Based on our results, we propose a model (Figure 11) in which NF-YAs act as key components of a stress tolerance adaptive mechanism. NF-YAs expression is induced in response to prolonged periods of stress. After an initial period of stress, NF-YAs repress the expression of early stress response genes and activate mechanisms that reduce cell elongation and plant growth by repressing genes involved in cell elongation. Reduction in plant growth translates into a reduction in metabolic and energetic costs and water requirements related to cell expansion and plant growth. We also speculate that two different, but not mutually exclusive mechanisms could cause the reduction in cell elongation and growth. One of them related to alterations in carbon partitioning as indicated by the repression of genes involved in these processes and the increased content of Suc and starch in NF-YA OE plants and the other related to the repression of genes involved in cell elongation.
Figure 11. Hypothetical Molecular Model of NF-YA action mode.
In WT plants growing under non-stress conditions, the expression of NF-YAs is low due miR169-mediated post-transcriptional down-regulation, but sufficient to activate the transcription of CCAAT-box containing promoters. Upon exposure to abiotic stress, NF-YA levels increase due to the transcriptional activation of NF-YA expression (early) and to the repression of miR169 (late). NF-YAs repress early general stress response genes sequestering NF-YB/NF-YC heterodimer avoiding its interaction probably with bZIPs, and on the other hand, participating in the late down-regulation of cell wall remodeling genes. This last step could be responsible of growth arrest mediated by these TFs when plants face for long time a stressful environment.
Several lines of evidence support our hypothesis: 1) NF-YA overexpression represses the early general stress response, which includes the early ABA-mediated induction of gene expression [65], 2) genes involved in cell wall remodeling such as XTH and EXP are repressed in NF-YA OE plants, 3) NF-YA OE plants have increased tolerance to senescence induced by different types of environmental stress and 4) NF-YA OE plants phenocopy WT plants subjected to abiotic stress in terms of growth rate reduction. Our results suggest that NF-YA OE plants enter a kind of quiescent state that leads to an increased tolerance to different types of abiotic stress. This could be similar to the growth reduction already proposed as an adaptive trait that confers tolerance to flooding, in which “the quiescent strategy” functions as a mechanism that protects plants against an unfavorable environment by reducing the cost related to cell expansion and plant growth [67]. The role of growth arrest in abiotic stress tolerance was confirmed with NF-YA estradiol-inducible lines, since stress tolerance is not observed upon NF-YA induction but rather at later stages when growth arrest is observed.
Mechanistically our hypothetical model proposes that in WT plants growing under non stress conditions, NF-YAs expression is low, due to the presence of high levels of miR169, but sufficient to activate the transcription of CCAAT-box containing promoters. Upon exposure to abiotic stress, NF-YA levels increase due to their transcriptional activation and to the reduction in miR169 levels. Increased levels of NF-YAs repress early abiotic stress response genes, probably by sequestering the NF-YB/YC heterodimer, creating a regulatory loop to arrest early responses that represent high energy and carbon costs, as well as participating in the activation of late responses. Late abiotic response involves the repression of genes involved in cell wall remodeling and cell elongation, leading to a quiescent state that confers general abiotic resistance. Our hypothetical model is based on solid evidences and led us to generate several interesting working hypothesis for which further experimental work is needed in order to elucidate in detail the molecular mechanisms in which NF-YA seems to play a central role.
Methods
Plant Materials and Growth Conditions
Wild-type Arabidopsis thaliana (ecotype Col-0) was used to produce all transgenic lines. The hen1-1 mutant was previously reported [68]. Seeds were surface-sterilized and germinated on agar plates with limiting (5 µM) or sufficient (1 mM) NaH2PO4 in 0.1× MS medium as previously described [69]. For contrasting nitrogen conditions, N optimum medium contained 2.0 mM NH4NO3 and 1.9 mM KNO3, whereas for N limited conditions, NH4NO3 was left out of the medium and KNO3 was adjusted to 50 µM. For nitrogen starvation induced senescence 1-week-old seedlings grown on N optimum medium were transferred to either N optimum or nitrogen-deficient medium and grown under for 2 weeks. All media (pH 5.7, 1% [w/v] agar) contained 1% Suc, unless stated otherwise.
Expression Analysis
For qRT-PCR analysis, total RNA was extracted using the RNeasy Plant kit from Qiagen according to the manufacturer's instructions. cDNA templates for PCR amplification were prepared using reverse specific primers (Table S8) and SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer's instructions. qRT-PCR was performed in an ABI PRISM 7500 sequence detection system (Applied Biosystems), using SYBR Green PCR Master Mix according to the manufacturer's protocol. Data shown represent mean values obtained from at least three independent amplification reactions and the error bars indicate ±SE. Gene expression was normalized to that of ACTIN 2 (ACT2) gene by subtracting the CT value of ACT2 from the CT value of the gene of interest (NF-YA2, 3, 5, 7, 10; SPX1). Relative quantification number (RQ) was obtained from the 2−ΔΔCT method, [70] where ΔΔCT represents ΔCT (treatment sample) - ΔCT (control condition). ΔCT was calculated [CT(NF-YA2, 3, 5, 7, 10 or SPX1)*E]-[CT(ACT2)*E] and E is the PCR efficiency according to the protocol reported by Czechowski et al. (2004) [71].
Mature miR169ag and miR169hn levels were determined by qRT-PCR as previously described [72]. Total RNA was extracted using Trizol reagent (Invitrogen) and reverse-transcribed using the stem-loop primers describe by Pant et al., (2009) [15]. Small-RNA Northern blot analysis was performed as described previously [73]. Membranes containing low molecular weight RNA were probed with U6 small nucleolar RNA, miR169 and miR156 oligonucleotides (Table S8) end-labeled in the presence of [γ-32P] ATP. The hybridization signals were scanned using a Storm phosphorimager.
Histological Assays
NF-YA2, 3, 5, 7, 10 promoter fusions, and GUS histochemical analysis were carried out as described previously [74]. A genomic region upstream of each ORF was amplified by PCR. For miR169a, miR169h, miR169i/j, miR169k/l and miR169m/n promoter fusions, the promoter sequence immediately upstream to the predicted precursor sequences was amplified and cloned using the protocol mentioned above, except for miR169a which was cloned in the pCR8/GW/TOPO (Invitrogen) vector instead of pDONR221 (Invitrogen).
Generation of Overexpressing Transgenic Lines
To generate the P35S:miR169nm construct, the fragment surrounding the miRNA sequences including both fold-back structures was amplified from genomic DNA. For P35S::NF-YA constructs the coding sequence of NF-YA3, 10 and 7 was amplified with the primers indicated (Table S8) in order to remove the miR169 target site located at the 3′UTR. The OE lines under the 35S promoter were produced as previously reported [69] except that NF-YA7 and 10 that were cloned in pCR8/GW/TOPO vector instead of pDONR221. For OE lines under the estradiol inducible system, the same protocol was carried out, but the pMDC7 plasmid was used instead of pb7WG2D (Invitrogen). For NF-YA2, a CDS clone [75] (stock # G85154) was obtained from the ABRC and used in the subsequent steps as described [69] using pb7WG2D and pMDC7 vectors to generate 35S and estradiol inducible lines, respectively. Chimeric repressor version for NF-YA2 was produced by amplifying the coding sequence before the stop codon for each gene by PCR with reverse primers containing the SRDX coding sequence and a stop codon coding sequence (Table S8). Transgenic estradiol inducible lines were produced as described early using the pCR8/GW/TOPO and pMDC7 vectors.
Microarray Analysis
Estradiol inducible lines were grown on liquid media for 7 days in standard light/dark cycle conditions (16/8 h) and on the 8th day, five hours after the light cycle had started, plants were exposed for 24 h to 7 µM β-estradiol or DMSO (mock treatment). Whole seedlings were collected and total RNA was extracted using the RNeasy Kit according to the manufacturer's instructions (Qiagen). Spotted glass microarray slides were obtained from the University of Arizona (http://ag.arizona.edu/microarray/). Three biological replicates for each line were used for RNA isolation and hybridized as two technical replicates (in swap) to the two channel microarrays. Labelling, hybridization, and image processing was performed as described previously [76]. Statistical tests using the Limma software [77] were used to identify differentially expressed genes with at least two-fold change in expression at the end of the estradiol treatment for PXVE:NF-YAs, PXVE:NF-YAsSRDX and 1.5 -fold in the case of PXVE:miR169nm with an adjusted p-value≤0.05 in all cases. The p- value was adjusted with FDR.
Pigment quantification
Chlorophyll was extracted with DMSO from 100 mg of shoots and Chlorophyll a and b content measured as previously described [78].
Sucrose analysis
Sucrose amount was quantified by UPLC-MS analysis. 12-day-old shoots were ground in liquid nitrogen, soluble sugar extracted in ethanol 80% (v/v) twice at 65°C for 15 min, samples were centrifuged at 5000 rpm for 10 min. Supernatant was dried under vacuum and resuspended in acetonitrile∶water (1∶1). Sucrose amount was quantified by UPLC-MS analysis performed on an LC-MS system composed of the Waters Acquity UPLC system (Waters Corporation) equipped with an Acquity photodiode array detector interfaced with the ThermoFisher LTQ mass spectrometer (ThermoFisher). Conditions for LC were as follows: column, ACQUITY UPLC BEH Amide 1.7 mm (2.1 mm×100 mm; Waters); solvent A, MeCN∶H2O (80∶20); solvent B, MeCN∶H2O (30∶70); gradient, 0 to 25 min, initial, 88% A and 12% B; 10 min, 30% A and 70% B; 15 min 88% A and 12% B; flow rate, 0.13 mL·min−1; injection volume 5 mL and column temperature, 35°C. Conditions for MS under negative mode were as follows, capillary voltage, 2.4 kV; cone voltage, 100 V; source temperature, 120°C; desolvation temperature, 350°C; cone gas flow, 10 L·h−1; desolvation gas flow, 450 L·h−1; nebulizer and curtain gas, N2.The amount of sucrose was determined by spectrometer software (MassLynx™ v. 4.1, Micromass) using calibration curves prepared with sucrose purchased from Sigma-Aldrich.
Dark induced elongation assay
Seeds were germinated on 0.1× MS medium for 24 hours and then transferred to darkness for 5 days. Hypocotyl length was determined and Scanning Electron Microscopy was performed in an EVO® 40 Series microscope (Carl Zeiss AG).
Accession Numbers
Sequence data from this article can be found in the EMBL/GenBank data libraries under the following accession numbers: NF-YA2 (At3g05690), NF-YA3 (At1g72830), NF-YA5 (At1g54160), NF-YA7 (At1g30500), NF-YA10 (At5g06510), SPX1 (At5g20150), HEN1 (At4g20910), MIR169a (At3g13405), MIR169h (At1g19371), MIR169i (At3g26812), MIR169j (At3g26813), MIR169k (At3g26815), MIR169l (At3g26816), MIR169m (At3g26818), MIR169n (At3g26819), MIR156g (At2g19425), MIR164a (At2g47585), MIR164b (At5g01747), MIR164c (At5g27807), ACTIN 2 (At3g18780). The microarray data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession number GSE36092.
Supporting Information
Temporal and Spatial expression pattern of NF-YAs and miR169s in response to Pi availability.
(PDF)
qRT-PCR analysis of miR169nm overexpressing lines.
(PDF)
NF-YA expression level in transgenic Arabidopsis lines overexpressing NF-YAs .
(PDF)
Ectopic expression of NF-YAs and miR169nm affects seed weight and silique size.
(PDF)
Biomass accumulation is affected in P35S:NF-YA and P35S:miR169nm lines.
(PDF)
Chlorophyll content of wild-type, P35S:NF-YA and P35S:miR169nm lines.
(PDF)
MAPMAN-based functional classification of differentially expressed transcripts in PXVE:NF-YA and PXVE:miR169nm lines.
(PDF)
Dark-induced cell elongation is affected in NF-YA overexpressing plants.
(PDF)
NF-YA overexpression causes a starch excess phenotype.
(PDF)
NF-YA overexpressing lines display increased drought tolerance.
(PDF)
NF-YA overexpression enhances heat and freezing tolerance.
(PDF)
NF-YA overexpression delays dark-induced senescence.
(PDF)
NF-YA overexpression prolongs plant longevity.
(PDF)
qRT-PCR analysis of WT, PXVE:NF-YA2 and P35S:NF-YA2 lines subjected to freezing.
(PDF)
Dose-dependent effect of estradiol on seedling phenotype of homozygote and heterozygote PXVE:NF-YA2SRDX transgenic lines.
(PDF)
NF-YA overexpression delays flowering time.
(PDF)
Expression changes in genes involved in cell wall remodeling in PXVE:NF-YA transgenic lines.
(PDF)
Sucrose content in WT, P35S:NF-YA2 and P35S:miR169nm lines.
(PDF)
Expression changes in genes belonging to the “anaerobic cluster” in PXVE:NF-YA and PXVE:miR169nm transgenic lines.
(PDF)
Motifs enrichment in deregulated genes in estradiol treated PXVE transgenic lines.
(PDF)
Z-score for TATA-box and CCAAT-box motifs in the promoters of differentially expressed genes in PXVE transgenic lines.
(PDF)
Motifs enrichment in the promoters of repressed genes in PXVE:NF-YA2SRDX lines that change in its PXVE:NF-YA2 counterpart, putative indirect targets.
(PDF)
Z-score for TATA-box and CCAAT-box motifs in the promoters of repressed genes in PXVE:NF-YA2SRDX line that change in its PXVE:NF-YA2 counterpart, putative indirect targets.
(PDF)
Sequences of primers and probes used in this study.
(PDF)
Transcripts differentially expressed in PXVE:NF-YA , PXVE:miR169nm and PXVE:NF-YA2SRDX lines.
(XLS)
Expression profiles of differential genes identified in at least once of the evaluated lines, PXVE:NF-YA , PXVE:miR169nm and PXVE:NF-YA2SRDX , clustered using the Pearson correlation and average linkage clustering.
(XLS)
Functional classification of differentially expressed transcripts in estradiol-inducible lines according to MAPMAN categories using the web-based tool Classification Superviewer.
(XLS)
Putative direct targets of NF-YA2.
(XLS)
Acknowledgments
We thank Edgar A. Rodriguez-Negrete, Marco A. Garcia-Neria, Susana Fuentes-Guerra, Flor Zamudio-Hernandez, María de Jesús Ortega-Estrada, Paulina Lozano-Sotomayor and Anne-Laure Chauvin for technical support. We thank Dr June Simpson for valuable help in reviewing the manuscript. We are grateful to Dr Nam Hai Chua for providing the pMDC7 vector and to Dr Joe Ecker for providing the NF-YA2 clone.
Funding Statement
This work was supported in part by grants from the Howard Hughes Medical Institute www.hhmi.org/ (Grant 55005946) and CONACyT (Mexico) www.conacyt.mx/ (299/43979) to L.H.-E. M. A. L.-G. is indebted to CONACyT for a PhD fellowship (176135). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Forsburg SL, Guarente L (1989) Identification and characterization of HAP4: a third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes Dev 3: 1166–1178. [DOI] [PubMed] [Google Scholar]
- 2. McNabb DS, Tseng KA, Guarente L (1997) The Saccharomyces cerevisiae Hap5p homolog from fission yeast reveals two conserved domains that are essential for assembly of heterotetrameric CCAAT-binding factor. Mol Cell Biol 17: 7008–7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bucher P (1990) Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J Mol Biol 212: 563–578. [DOI] [PubMed] [Google Scholar]
- 4. McNabb DS, Xing Y, Guarente L (1995) Cloning of yeast HAP5: a novel subunit of a heterotrimeric complex required for CCAAT binding. Genes Dev 9: 47–58. [DOI] [PubMed] [Google Scholar]
- 5. Dang VD, Bohn C, Bolotin-Fukuhara M, Daignan-Fornier B (1996) The CCAAT-box binding factor stimulate ammonium assimilation in Saccaromyces cerevisiae, defining a mew cross-pathway regulation between nitrogen and carbon metabolisms. J Bacteriol 178: 1842–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Litzka OK, Bergh T, Brakhage AA (1996) The Aspergillus nidulans penicillin biosynthesis gene aat (penDE) is controlled by a CCAAT- containing DNA element. Eur J Biochem 238: 675–682. [DOI] [PubMed] [Google Scholar]
- 7. Roy B, Lee A (1995) Transduction of calcium stress through interaction of the human transcription factor CBF with the proximal CCAAT regulatory element of the grp/Bip promoter. Mol Cell Biol 15: 2263–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Berry M, Grosveld F, Dillon N (1992) A single point mutation is the cause of the Greek form of hereditary persistence of fetal haemoglobin. Nature 358: 499–502. [DOI] [PubMed] [Google Scholar]
- 9. Linhart C, Elkon R, Shiloh Y, Shamir R (2005) Deciphering transcriptional regulatory elements that encode specific cell cycle phasing by comparative genomics analysis. Cell Cycle 4: 1788–1797. [DOI] [PubMed] [Google Scholar]
- 10. Argüello-Astorga G, Herrera-Estrella L (1998) Evolution of light-regulated plant promoters. Annu Rev Plant Physiol Plant Mol Biol 49: 525–555. [DOI] [PubMed] [Google Scholar]
- 11. Stephenson TJ, McIntyre CL, Collet C, Xue GP (2010) TaNF-YC11, one of the light-upregulated NF-YC members in Triticum aestivum, is co-regulated with photosynthesis-related genes. Funct Integr Genomics 10: 265–276. [DOI] [PubMed] [Google Scholar]
- 12. Siefers N, Dang KK, Kumimoto RW, Bynum WE, Tayrose G, et al. (2009) Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol 149: 625–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li WX, Oono Y, Zhu J, He XJ, Wu JM, et al. (2008) The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 20: 2238–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jones-Rhoades MW, Bartel DP (2004) Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14: 787–799. [DOI] [PubMed] [Google Scholar]
- 15. Pant BD, Musialak-Lange M, Nuc P, May P, Buhtz A, et al. (2009) Identification of nutrient-responsive Arabidopsis and rapeseed microRNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant Physiol 150: 1541–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Misson J, Raghothama KG, Jain A, Jouhet J, Block MA, et al. (2005) A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci USA 102: 11934–11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Scheible WR, Morcuende R, Czechowski T, Fritz C, Osuna D, et al. (2004) Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol 136: 2483–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, et al. (2005) Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis . Plant J 42: 567–585. [DOI] [PubMed] [Google Scholar]
- 19. Liu JX, Howell SH (2010) bZIP28 and NF-Y transcription factors are activated by ER stress and sssemble into a transcriptional complex to regulate stress response genes in Arabidopsis . Plant Cell 22: 782–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Todesco M, Rubio-Somoza I, Paz-Ares J, Weigel D (2010) A Collection of Target Mimics for Comprehensive Analysis of MicroRNA Function in Arabidopsis thaliana . PLoS Genet 6 (7) e1001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wenkel S, Turck F, Singer K, Gissot L, Le Gourrierec J, et al. (2006) CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis . Plant Cell 18: 2971–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ito Y, Thirumurugan T, Serizawa A, Hiratsu K, Ohme-Takagi M, et al. (2011) Aberrant vegetative and reproductive development by overexpression and lethality by silencing of OsHAP3E in rice. Plant Sci 181: 105–110. [DOI] [PubMed] [Google Scholar]
- 23. Kwong RW, Bui AQ, Lee H, Kwong LW, Fischer RL, et al. (2003) LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell 15: 5–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lotan T, Ohto M, Yee KM, West MA, Lo R, et al. (1998) Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93: 1195–1205. [DOI] [PubMed] [Google Scholar]
- 25. West M, Yee KM, Danao J, Zimmerman JL, Fischer RL, et al. (1994) LEAFY COTYLEDON1 Is an Essential Regulator of Late Embryogenesis and Cotyledon Identity in Arabidopsis . Plant Cell 6: 1731–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nelson DE, Repetti PP, Adams TR, Creelman RA, Wu J, et al. (2007) Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc Natl Acad Sci USA 104: 16450–16455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Müller R, Morant M, Jarmer H, Nilsson L, Nielsen TH (2007) Genome-wide analysis of the Arabidopsis leaf transcriptome reveals interaction of phosphate and sugar metabolism. Plant Physiol 143: 156–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li J, Yang Z, Yu B, Liu J, Chen X (2005) Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis . Curr Biol 15: 1501–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li Y, Fu Y, Ji L, Wu C, Zheng C (2010) Characterization and expression analysis of the Arabidopsis mir169 family. Plant Sci 178: 271–280. [Google Scholar]
- 30. Zuo J, Niu QW, Chua NH (2000) Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J 24: 265–273. [DOI] [PubMed] [Google Scholar]
- 31. Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Provart N, Zhu T (2003) A browser-based functional classification superviewer for Arabidopsis genomics. Curr Comput Mol Biol 2003: 271–272. [Google Scholar]
- 33. Ma S, Bohnert HJ (2007) Integration of Arabidopsis thaliana stress-related transcript profiles, promoter structures, and cell-specific expression. Genome Biol 8: R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Walley JW, Coughlan S, Hudson ME, Covington MF, Kaspi R, et al. (2007) Mechanical stress induces biotic and abiotic stress responses via a novel cis-element. PLoS Genet 3: 1800–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Katari MS, Nowicki SD, Aceituno FF, Nero D, Kelfer J, et al. (2010) VirtualPlant: a software platform to support systems biology research. Plant Physiol 152: 500–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gibson SI (2004) Sugar and phytohormone response pathways: navigating a signalling network. J Exp Bot 55: 253–264. [DOI] [PubMed] [Google Scholar]
- 37. Kang JY, Choi HI, Im MY, Kim SY (2002) Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14: 343–357 Meinke DW, Franzmann LH, Nickle TC, Yeung EC (1994) Leafy Cotyledon Mutants of Arabidopsis. Plant Cell 6: 1049–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Licausi F, van Dongen JT, Giuntoli B, Novi G, Santaniello A, et al. (2010) HRE1 and HRE2, two hypoxia-inducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana . Plant J 62: 302–315. [DOI] [PubMed] [Google Scholar]
- 39. Xiao S, Gao W, Chen QF, Chan SW, Zheng SX, et al. (2010) Overexpression of Arabidopsis acyl-CoA binding protein ACBP3 promotes starvation-induced and age-dependent leaf senescence. Plant Cell 22: 1463–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Achard P, Gong F, Cheminant S, Alioua M, Hedden P, et al. (2008) The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 20: 2117–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schröder F, Lisso J, Müssig C (2011) EXORDIUM-LIKE1 promotes growth during low carbon availability in Arabidopsis . Plant Physiol 156: 1620–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. O' Connor TR, Dyreson C, Wyrick JJ (2005) Athena: A resource for rapid visualization and systematic analysis of Arabidopsis promoter sequences. Bioinformatics 21: 4411–4413. [DOI] [PubMed] [Google Scholar]
- 43. Toufighi K, Brady SM, Austin R, Ly E, Provart NJ (2005) The Botany Array Resource: e-Northerns, Expression Angling, and promoter analyses. Plant J 43: 153–163. [DOI] [PubMed] [Google Scholar]
- 44. Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M (2003) Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain in Arabidopsis . Plant J 34: 733–739. [DOI] [PubMed] [Google Scholar]
- 45. Koyama T, Furutani M, Tasaka M, Ohme-Takagi M (2007) TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis . Plant Cell 19: 473–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sinha S, Maity SN, Lu JF, Decrombrugghe B (1995) Recombinant rat CBF-C, the 3rd subunit of CBF/NF-Y, allows formation of a protein-DNA complex with CBF-A and CBF-B and with yeast HAP2 and HAP3. Proc Natl Acad Sci USA 92: 1624–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li XY, Hooft van Huijsduijnen R, Mantovani R, Benoist C, Mathis D (1992) Intron-exon organization of the NF-Y genes. Tissue-specific splicing modifies an activation domain. J Biol Chem 267: 8984–8990. [PubMed] [Google Scholar]
- 48. Manni I, Caretti G, Artuso S, Gurtner A, Emiliozzi V, et al. (2008) Posttranslational regulation of NF-YA modulates NF-Y transcriptional activity. Mol Biol Cell 19: 5203–5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, et al. (2011) Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet 43: 621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Thön M, Abdallah QA, Hortschansky P, Scharf DH, Eisendle M, et al. (2010) The CCAAT-binding complex coordinates the oxidative stress response in eukaryotes. Nucleic Acids Res 38: 1098–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hackenberg D, Wu Y, Voigt A, Adams R, Schramm P, et al. (2011) Studies on differential nuclear translocation mechanism and assembly of the three subunits of the Arabidopsis thaliana transcription factor NF-Y. Mol Plant doi:10.1093/mp/ssr107 [DOI] [PubMed] [Google Scholar]
- 52. Bustos R, Castrillo G, Linhares F, Puga MI, Rubio V, et al. (2010) A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis . PLoS Genet 6: e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mantovani R (1999) The molecular biology of the CCAAT-binding factor NF-Y. Gene 239: 15–27. [DOI] [PubMed] [Google Scholar]
- 54. Rizhsky L, Liang H, Shuman J, Shulaev V, Davletova S, et al. (2004) When defense pathways collide: the response of Arabidopsis to a combination of drought and heat stress. Plant Physiol 134: 1683–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Morcuende R, Bari R, Gibon Y, Zheng W, Pant BD, et al. (2007) Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ 30: 85–112. [DOI] [PubMed] [Google Scholar]
- 56. Strand A, Hurry V, Gustafsson P, Gardestrom P (1997) Development of Arabidopsis thaliana leaves at low temperatures releases the suppression of photosynthesis and photosynthetic gene expression despite the accumulation of soluble carbohydrates. Plant J 12: 605–614. [DOI] [PubMed] [Google Scholar]
- 57. Hoekstra FA, Golovina EA, Buitink J (2001) Mechanisms of plant desiccation tolerance. Trends Plant Sci 6: 431–438. [DOI] [PubMed] [Google Scholar]
- 58. Krapp A, Berthome R, Orsel M, Mercey-Boutet S, Yu A, et al. (2011) Arabidopsis roots and shoots show distinct temporal adaptation patterns toward nitrogen starvation. Plant Physiol 157: 1255–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lei M, Liu Y, Zhang B, Zhao Y, Wang X, et al. (2011) Genetic and genomic evidence that sucrose is a global regulator of plant responses to phosphate starvation in Arabidopsis . Plant Physiol 156: 1116–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sulpice R, Pyl ET, Ishihara H, Trenkampa S, Steinfathb M, et al. (2009) Starch as a major integrator in the regulation of plant growth. Proc Natl Acad Sci USA 106: 10348–10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Fulton DC, Stettler M, Mettler T, Vaughan CK, Li J, et al. (2008) Beta-AMYLASE4, a noncatalytic protein required for starch breakdown, acts upstream of three active beta-amylases in Arabidopsis chloroplasts. Plant Cell 20: 1040–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Li L, Foster CM, Gan Q, Nettleton D, James MG, et al. (2009) Identification of the novel protein QQS as a component of the starch metabolic network in Arabidopsis leaves. Plant J 58: 485–498. [DOI] [PubMed] [Google Scholar]
- 63. Cosgrove DJ (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6: 850–861. [DOI] [PubMed] [Google Scholar]
- 64. Yamamoto A, Kagaya Y, Toyoshima R, Kagaya M, Takeda S, et al. (2009) Arabidopsis NF-YB subunits LEC1 and LEC1-LIKE activate transcription by interacting with seed-specific ABRE-binding factors. Plant J 58: 843–856. [DOI] [PubMed] [Google Scholar]
- 65. Harb A, Krishnan A, Ambavaram MM, Pereira A (2010) Molecular and physiological analysis of drought stress in Arabidopsis reveals early responses leading to acclimation in plant growth. Plant Physiol 154: 1254–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mahajan S, Tuteja N (2005) Cold, salinity and drought stresses: an overview. Arch Biochem Biophys 444: 139–158. [DOI] [PubMed] [Google Scholar]
- 67. Bailey-Serres J, Voesenek LA (2008) Flooding stress: acclimations and genetic diversity. Annu Rev Plant Biol 59: 313–339. [DOI] [PubMed] [Google Scholar]
- 68. Chen X, Liu J, Cheng Y, Jia D (2002) HEN1 functions pleiotropically in Arabidopsis development and acts in C function in the flower. Development 129: 1085–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Perez-Torres CA, Lopez-Bucio J, Cruz-Ramirez A, Ibarra-Laclette E, Dharmasiri S, et al. (2008) Phosphate availability alters lateral root development in Arabidopsis by modulating auxin sensitivity via a mechanism involving the TIR1 auxin receptor. Plant Cell 20: 3258–3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 71. Czechowski T, Bari RP, Stitt M, Scheible WR, Udvardi MK (2004) Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant J 38: 366–379. [DOI] [PubMed] [Google Scholar]
- 72. Pant BD, Buhtz A, Kehr J, Scheible WR (2008) MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J 53: 731–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rodriguez-Negrete EA, Carrillo-Tripp J, Rivera-Bustamante RF (2009) RNA silencing against geminivirus: complementary action of posttranscriptional gene silencing and transcriptional gene silencing in host recovery. J Virol 83: 1332–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Cruz-Ramirez A, Oropeza-Aburto A, Razo-Hernandez F, Ramirez-Chavez E, Herrera-Estrella L (2006) Phospholipase DZ2 plays an important role in extraplastidic galactolipid biosynthesis and phosphate recycling in Arabidopsis roots. Proc Natl Acad Sci USA 103: 6765–6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yamada K, Lim J, Dale JM, Chen HM, Shinn P, et al. (2003) Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302: 842–846. [DOI] [PubMed] [Google Scholar]
- 76. Calderon-Vazquez C, Ibarra-Laclette E, Caballero-Perez J, Herrera-Estrella L (2008) Transcript profiling of Zea mays roots reveals gene responses to phosphate deficiency at the plant- and species-specific levels. J Exp Bot 59: 2479–2497. [DOI] [PubMed] [Google Scholar]
- 77. Smyth GK (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article 3. [DOI] [PubMed] [Google Scholar]
- 78. Richardson AD, Duigan SP, Berlyn GP (2002) An evaluation of noninvasive methods to estimate foliar chlorophyll content. New Phytol 153: 185–194. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Temporal and Spatial expression pattern of NF-YAs and miR169s in response to Pi availability.
(PDF)
qRT-PCR analysis of miR169nm overexpressing lines.
(PDF)
NF-YA expression level in transgenic Arabidopsis lines overexpressing NF-YAs .
(PDF)
Ectopic expression of NF-YAs and miR169nm affects seed weight and silique size.
(PDF)
Biomass accumulation is affected in P35S:NF-YA and P35S:miR169nm lines.
(PDF)
Chlorophyll content of wild-type, P35S:NF-YA and P35S:miR169nm lines.
(PDF)
MAPMAN-based functional classification of differentially expressed transcripts in PXVE:NF-YA and PXVE:miR169nm lines.
(PDF)
Dark-induced cell elongation is affected in NF-YA overexpressing plants.
(PDF)
NF-YA overexpression causes a starch excess phenotype.
(PDF)
NF-YA overexpressing lines display increased drought tolerance.
(PDF)
NF-YA overexpression enhances heat and freezing tolerance.
(PDF)
NF-YA overexpression delays dark-induced senescence.
(PDF)
NF-YA overexpression prolongs plant longevity.
(PDF)
qRT-PCR analysis of WT, PXVE:NF-YA2 and P35S:NF-YA2 lines subjected to freezing.
(PDF)
Dose-dependent effect of estradiol on seedling phenotype of homozygote and heterozygote PXVE:NF-YA2SRDX transgenic lines.
(PDF)
NF-YA overexpression delays flowering time.
(PDF)
Expression changes in genes involved in cell wall remodeling in PXVE:NF-YA transgenic lines.
(PDF)
Sucrose content in WT, P35S:NF-YA2 and P35S:miR169nm lines.
(PDF)
Expression changes in genes belonging to the “anaerobic cluster” in PXVE:NF-YA and PXVE:miR169nm transgenic lines.
(PDF)
Motifs enrichment in deregulated genes in estradiol treated PXVE transgenic lines.
(PDF)
Z-score for TATA-box and CCAAT-box motifs in the promoters of differentially expressed genes in PXVE transgenic lines.
(PDF)
Motifs enrichment in the promoters of repressed genes in PXVE:NF-YA2SRDX lines that change in its PXVE:NF-YA2 counterpart, putative indirect targets.
(PDF)
Z-score for TATA-box and CCAAT-box motifs in the promoters of repressed genes in PXVE:NF-YA2SRDX line that change in its PXVE:NF-YA2 counterpart, putative indirect targets.
(PDF)
Sequences of primers and probes used in this study.
(PDF)
Transcripts differentially expressed in PXVE:NF-YA , PXVE:miR169nm and PXVE:NF-YA2SRDX lines.
(XLS)
Expression profiles of differential genes identified in at least once of the evaluated lines, PXVE:NF-YA , PXVE:miR169nm and PXVE:NF-YA2SRDX , clustered using the Pearson correlation and average linkage clustering.
(XLS)
Functional classification of differentially expressed transcripts in estradiol-inducible lines according to MAPMAN categories using the web-based tool Classification Superviewer.
(XLS)
Putative direct targets of NF-YA2.
(XLS)