Background: It remains unknown how Dot1 or the Dot1 complex specifically targets the transcribed regions.
Results: A functional interaction between hDOT1L and RNAPII targets hDOT1L and subsequent H3K79 methylations to active genes.
Conclusion: hDOT1L interacts with phosphorylated CTD of RNAPII.
Significance: This represents novel mechanistic insight into the understanding of targeting and propagation of hDOT1L along gene transcription.
Keywords: Histone Methylation, Nucleosome, RNA Polymerase II, Transcription, Ubiquitylation, CTD, hDOT1L
Abstract
Histone-modifying enzymes play a pivotal role in gene expression and repression. In human, DOT1L (Dot1-like) is the only known histone H3 lysine 79 methyltransferase. hDOT1L is associated with transcriptional activation, but the general mechanism connecting hDOT1L to active transcription remains largely unknown. Here, we report that hDOT1L interacts with the phosphorylated C-terminal domain of actively transcribing RNA polymerase II (RNAPII) through a region conserved uniquely in multicellular DOT1 proteins. Genome-wide profiling analyses indicate that the occupancy of hDOT1L largely overlaps with that of RNAPII at actively transcribed genes, especially surrounding transcriptional start sites, in embryonic carcinoma NCCIT cells. We also find that C-terminal domain binding or H3K79 methylations by hDOT1L is important for the expression of target genes such as NANOG and OCT4 and a marker for pluripotency in NCCIT cells. Our results indicate that a functional interaction between hDOT1L and RNAPII targets hDOT1L and subsequent H3K79 methylations to actively transcribed genes.
Introduction
In the nuclei of eukaryotic cells, DNA is packaged along with histones and other nuclear proteins to form chromatin. The basic unit of chromatin, the nucleosome, is composed of 147 base pairs of DNA wrapped around a histone octamer with two molecules of H2A, H2B, H3, and H4 (1, 2). The N-terminal tails of histones are subject to a variety of post-translational modifications, including acetylation, methylation, phosphorylation, and ubiquitylation (3–5).
The histone H3 residue, lysine 79, which is a conserved core residue located in a loop within the histone fold domain, is multiply methylated by Dot1 (disruptor of telomeric silencing 1) (6–8). Yeast Dot1 has been shown to affect gene expression, silencing at telomeres, DNA damage responses (6, 8–11), meiotic checkpoint control (12), and cell cycle progression (13).
Dot1 is evolutionarily conserved from yeast to humans (8, 14–16). In Drosophila melanogaster, H3K79 methylation by grappa, the Dot1 homolog, is an important euchromatin modification that has been correlated with gene activity (14). Mutational analyses of grappa in D. melanogaster have shown both Polycomb and Trithorax group mutant phenotypes, as well as telomeric silencing (17). In mammals, several studies have shown that hDOT1L (human Dot1-like) and several myeloid/lymphoid or mixed-lineage leukemia fusion partners, such as ENL/MLLT1, AF4/MLLT2, AF9/MLLT3, and AF10/MLLT10, exist in a single complex that is integral for the trimethylation of H3K79 (H3K79me3) (18–20). Murine DOT1L is involved in cell cycle progression, the differentiation of mouse embryonic stem cells, and fetal development (21, 22) and is required for leukemogenesis (20, 23).
In yeast, proper H3K79me3 requires monoubiquitylation of histone H2B on lysine 123 (H2Bub1)3 (24–26). H2Bub1 directly stimulates FACT-mediated transcription in vitro (27). Loss of the histone H2B ubiquitylation enzymes or substitution of the H2B C-terminal lysine residue with arginine leads to transcription defects, suggesting that the dynamic turnover of H2Bub1 is critical to transcription (28). In mammals, it has been reported that RAD6-mediated transcription coupled with H2Bub1 directly stimulates H3K4 methylation (29), and synthetically monoubiquitylated H2B stimulates hDOT1L-mediated H3K79 methylation events in vitro (29–31). Indeed, genome-wide analyses have shown that the H3K79 methylations co-occupied with H2Bub1 in the transcribed regions of active genes (32, 33). However, we do not yet know how H3K79 methylations globally impact chromatin structure in relation to transcription or how Dot1 or the Dot1 complex specifically targets the transcribed regions.
Enrichment of H3K79 methylations in actively transcribed regions suggests that Dot1, which methylates H3K79, plays an important role in transcription (34, 35). Because active transcription is highly related to RNAPII occupancy, we hypothesized that hDOT1L binds to RNAPII to methylate the nucleosomes of actively transcribed regions. In this study, we demonstrate that hDOT1L directly binds to the phosphorylated CTD of RNAPII from cell culture and in vitro experiments. There is a strong genome-wide correlation between the occupancies of hDOT1L and RNAPII near the transcription start sites of active genes. Depletion of hDOT1L results in reduced level of H3K79me3 and mRNA expression in pluripotency-related genes in the embryonic carcinoma NCCIT cells (36–38) that can differentiate into any of the three embryonic germ layers (i.e., ectoderm, mesoderm, and endoderm), as well as extra-embryonic cell lineages.
EXPERIMENTAL PROCEDURES
Chromatin Immunoprecipitation
ChIP assays were carried out as previously described with minor modifications (39). Briefly, harvested NCCIT cells were incubated in 1% formaldehyde for 20 min at room temperature. Cross-linked cells were then lysed in SDS lysis buffer, and DNA was sonicated. Chromatin was subjected to immunoprecipitation with a mixture of protein A- and G-agarose (GE Healthcare) at 4 °C. The chromatin-antibody-agarose complexes were then sequentially washed with the following solutions: low salt wash buffer, high salt wash buffer, LiCl buffer, and TE buffer. The chromatin complexes were eluted, 350 mm NaCl was added to the eluates, and reverse histone-DNA cross-linking was performed by heating at 68 °C. Finally, the DNA/chromatin solution was treated with proteinase K and RNase A, and the DNA was precipitated. The resulting DNA was used for either ChIP-seq or qPCR analyses.
Data Processing
The ChIP-seq results for hDOT1L and RNAPII were mapped to the human genome (UCSC hg18) using the Bowtie program (40) with default parameters. These data can be found in the NCBI GEO database under accession number GSE26598. The mapped sequence reads were extended to the average fragment length (200 bp), and the number of overlapping sequence tags was obtained at 10-bp intervals across the human genome. The ChIP-seq signal was defined as the ratio of target read count/target size divided by total read count/genome size (i.e., the average read count). The processed ChIP-seq data for IgG input, H2Bub1, and H3K79me1/2/3 were downloaded from the GEO database (accession no. GSE25882) (32). Then mRNA-seq data were downloaded from GSE25882 and mapped to the human genome using TopHat (41) with default parameters. Each gene expression level was defined by FPKM (fragments/kilobase of exon/million fragments mapped) scores generated by Cufflinks (42). Genes with FPKM scores less than 0.05 were removed, because they might be in unexpressed or unsequenced regions. All of the genome annotations were defined using the Refseq identifiers in the UCSC Genome browser (hg18 assembly). A gene body region was defined as the region spanning the transcriptional start site (TSS) to the end site, according to the Refseq identifiers.
Preparation of Nuclear Extracts, Immunoprecipitation, and Western Blot Analysis
Nuclear extracts were prepared as previously described (43) with minor modifications. HEK293T and NCCIT cells were harvested and washed twice with cold PBS. The cell pellets were resuspended in 5× volumes of extraction buffer I (10 mm HEPES, pH 7.9, 1.5 mm MgCl2, 10 mm KCl) containing protease inhibitor, mixed well, vortexed for 15 s, and incubated on ice for 10 min. Then 10% Nonidet P-40 was added to a final concentration of 0.9%, and the cells were vortexed for 15 s and incubated on ice for 5 min. Each sample was centrifuged at 5,000 rpm for 5 min at 4 °C to precipitate the cell nuclei, and the supernatant was completely removed. The pelleted cell nuclei were washed twice with extraction buffer I, mixed well with extraction buffer II (20 mm HEPES, pH 7.9, 150 mm NaCl, 1.5 mm MgCl2, 0.2 mm EDTA) containing protease inhibitor, vortexed for 20 s, and incubated on ice for 40 min. The nuclear suspension was then vortexed for 10 s, homogenized by being passed through a 23-gauge syringe needle 4–6 times, and cleared by centrifugation at 14,000 rpm for 15 min at 4 °C. The supernatant was taken as the nuclear extract and was incubated overnight with anti-FLAG M2-agarose beads (Sigma; A2220). The immunoprecipitated complexes were washed three times with extraction buffer II, mixed with SDS sample buffer, and boiled for 5 min. The samples were then separated by 8% SDS-PAGE and subjected to Western blot analysis using the appropriate antibodies.
Peptide Pulldown Assay
The peptide pulldown assay was performed as previously described (44) with minor modifications. The synthetic unphosphorylated and phosphorylated Ser-5 and Ser-2 CTD peptides were kindly provided from Drs. Bing Li and Jerry Workman, and the synthetic 6PC (a scrambled CTD-like peptide with phosphoserine) and phosphorylated Ser-5/2 CTD peptides were synthesized by AnaSpec (Fremont, CA). The biotinylated CTD peptides (∼1.5 μg) were bound to 0.1 mg of streptavidin-coated Dynabeads M280 in 100 μl of CTD coupling buffer (25 mm Tris-HCl, pH 8.0, 1 m NaCl, 1 mm dithiothreitol, 5% glycerol, 0.03% Nonidet P-40) at 4 °C for 2 h. The peptide-bound beads were washed once with CTD coupling buffer and twice with peptide pulldown buffer (25 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1 mm dithiothreitol, 5% glycerol, 0.03% Nonidet P-40) and were finally resuspended in 50 μl of peptide pulldown buffer. Proteins (500–1000 ng) were mixed with the beads and incubated at 4 °C for 1 h on a Dyna-Mixer (Dynal). Following three washes with peptide pulldown buffer, the samples were separated by SDS-PAGE and subjected to Western blot analysis.
Transfection of siRNA
Sequences that could effectively silence the expression of hDot1l were selected as follows: 5′-AGAAGCUGUUGAAGGAGAAUU-3′ (sense) and 5′-UUCUCCUUCAACAGCUUCUUU-3′ (antisense). The control siRNA sequences against Egfp were 5′-GUUCAGCGUGUCCGGCGAGUU-3′ (sense) and 5′-CUCGCCGGACACGCUGAACUU-3′ (antisense). The 21-nucleotide synthetic siRNA duplexes were prepared by Samchully Pharm (Seoul, Korea). NCCIT cells (2 × 105 cells/well) were seeded to 6-well plates. Twenty-four hours later, the cells were transfected with hDot1l or control siRNA using FuGENE HD (Roche Applied Science) according to the manufacturer's protocol. The cells were analyzed 48 h after transfection by Western blot analysis and RT-qPCR.
Accession Number
The ChIP-seq data from this study are available from NCBI GEO under accession number GSE26598.
RESULTS
hDOT1L Interacts Directly with RNAPII
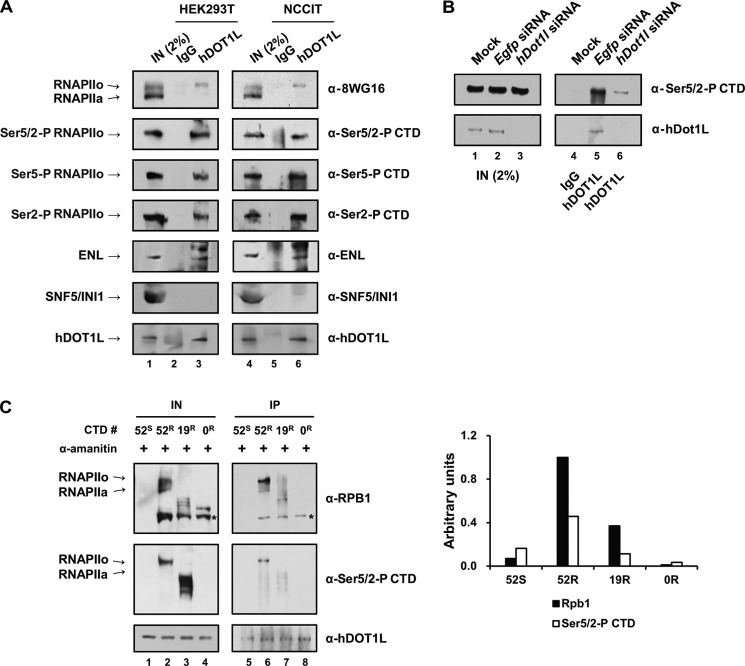
To investigate whether hDOT1L binds to RNAPII, we subjected nuclear extracts from HEK293T and NCCIT cells to co-immunoprecipitation analyses using specific antibodies. Interestingly, the immunoprecipitation of full-length hDOT1L followed by immunoblotting with antibodies against Ser5-P, Ser5/2-P, and Ser2-P CTD resulted in the detection of RNAPIIo form, with phosphorylated CTD, in both cell lines (Fig. 1A). hDOT1L did not co-immunoprecipitate with the negative control, hSNF5/INI1, which is a core component of the SWI/SNF chromatin remodeling complex. Enl/MLLT1, a component of the hDOT1L complex, was used as a positive control. Furthermore, cells that had been depleted of hDOT1L by siRNA transfection did not show co-immunoprecipitation of RNAPIIo (Fig. 1B, compare lane 5 (Egfp) with lane 6 (hDOT1L knockdown)). These results suggest that hDOT1L physically associates with the RNAPII with phosphorylated CTD in cultured cells.
FIGURE 1.
hDOT1L interacts with the RNAPII with phosphorylated CTD in cultured cells. A, nuclear proteins from HEK293T and NCCIT cells were extracted and immunoprecipitated with anti-rabbit IgG or -hDOT1L at 4 °C. Immunoprecipitated samples were resolved by SDS-PAGE and immunoblotted using the indicated antibodies on the right. B, co-immunoprecipitation was performed as described above, using nuclear proteins from HEK293T cells depleted of endogenous hDOT1L by siRNA (100 nm). C, HEK293T cells were transfected with plasmids expressing α-amanitin-sensitive wild-type hRpb1 (52S), with 52 heptad (YSPTSPS) repeats in CTD, and α-amanitin-resistant hRpb1 mutants with 52 (52R), 19 (19R), and 0 (0R) heptad repeats in CTD (45). Twenty-four hours post-transfection, the cells were treated with 5 μg/ml of α-amanitin to inhibit wild-type hRPb1. Forty-eight hours post-transfection, nuclear proteins were fractionated and immunoprecipitated with anti-hDOT1L at 4 °C. Immunoprecipitated samples were resolved by SDS-PAGE and immunoblotted using the antibodies indicated on the right. The band intensities were measured in arbitrary units of band intensity of the phosphorylated RNAPII (RNAPIIo) signals normalized by that of each immunoprecipitated hDOT1L. IN (input) represents 2% in Ser(P)-5/2 CTD and RPB1 and 100% in hDOT1L. The labeling (RNAPIIo and RNAPIIa) on the left side refers to only the 52R. Asterisks mark the degraded 180-kDa IIB form (45). IP, immunoprecipitation.
To further support the assertion that hDOT1L interacts with phosphorylated RNAPII, we employed α-amanitin-resistant human RNAPII large subunit (hRpb1) mutants (45). Our results revealed that hDOT1L interacted with the phosphorylated form of RNAPII (52R and 19R) but not the mutant without a CTD (0R) (Fig. 1C). Integrity of CTD length also appeared to play a role in the interaction because the ratio of the IP band of the phosphorylated RNAPII over that of the input of 19R (0.37 for Rpb1 and 0.10 for Ser(P)-5/2) was lower than that of 52R (1.00 for Rpb1 and 0.46 for Ser(P)-5/2). These data strongly reinforce the notion that there is a physical interaction between hDOT1L and the phosphorylated CTD of RNAPII.
Previous studies using defined biotinylated synthetic CTD peptides in yeast showed that the H3K36 histone methyltransferase, ySet2p, directly binds to CTD peptides phosphorylated at Ser-5 and Ser-2 (44, 46, 47). We adopted a similar system using CTD peptides that consisted of three repeats of the heptad sequence (YSPTSPS). hDOT1L bound to singly phosphorylated CTD peptides (CTD-Ser(P)-5 and CTD-Ser(P)-2), as well as the doubly phosphorylated CTD peptide (CTD-Ser(P)-5/2) (Fig. 2, upper panel). In contrast, hDOT1L did not bind to a scrambled CTD-like peptide with phosphoserine (6PC) (Fig. 2, upper panel), indicating that amino acid context as well as phosphoserine was important for the binding. As a positive control, yeast Set2 also bound to the phosphorylated CTD peptides as expected (Fig. 2, bottom panel) (48). Thus, we conclude that hDOT1L directly binds to the phosphorylated CTD of actively transcribing RNAPII.
FIGURE 2.
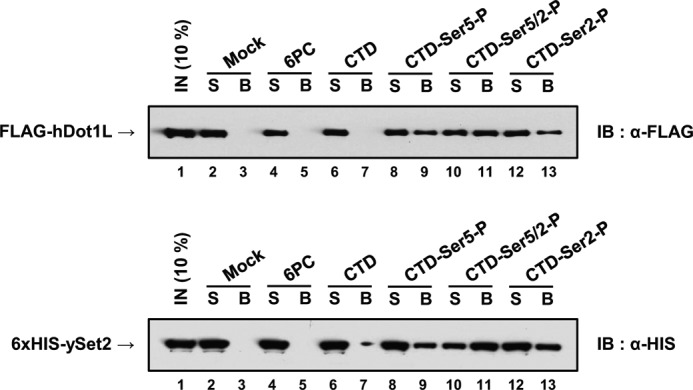
hDOT1L directly binds to the phosphorylated CTD in vitro. In vitro peptide pulldown assay. In the experiment, we used a scrambled CTD-like peptide with phosphoserine (6PC), a CTD peptide with nonphosphorylated serine residues (CTD), and phosphorylated CTD-Ser(P)-5 (CTD-Ser5-P), CTD-Ser(P)-5/2(CTD-Ser5/2-P), and CTD-Ser(P)-2 (CTD-Ser2-P) peptides (58). Synthetic biotinylated CTD peptides (1.5 μg) were adsorbed to 100 μg of streptavidin-coated magnetic beads. Full-length FLAG-hDOT1L (1,000 ng) purified from the baculovirus expression system and bacterially expressed His6-ySet2p (500 ng) (supplemental Fig. S1) were incubated at 4 °C with either magnetic beads alone (Mock) or beads immobilized with the indicated peptides. Shown are the immunoblots (IB) with anti-FLAG and -His. S and B indicate supernatant and bead fractions, respectively. IN, input.
hDOT1L Is Associated with Active Transcription and RNAPII in a Genome-wide Manner
Because our data indicate that hDOT1L interacts with RNAPII in cultured cells, we hypothesized that the genome-wide localization of hDOT1L would correlate with that of RNAPII during active transcription. To examine this point, we analyzed the genome-wide distribution of hDOT1L and RNAPII in undifferentiated human embryonic carcinoma NCCIT cells. We also compared the ChIP data of hDOT1L and RNAPII with our previous results about H3K79 mono-, di-, and trimethylation and H2Bub1 ChIP data and mRNA-seq data from NCCIT cells downloaded from the GEO database (accession number GSE25882) (32).
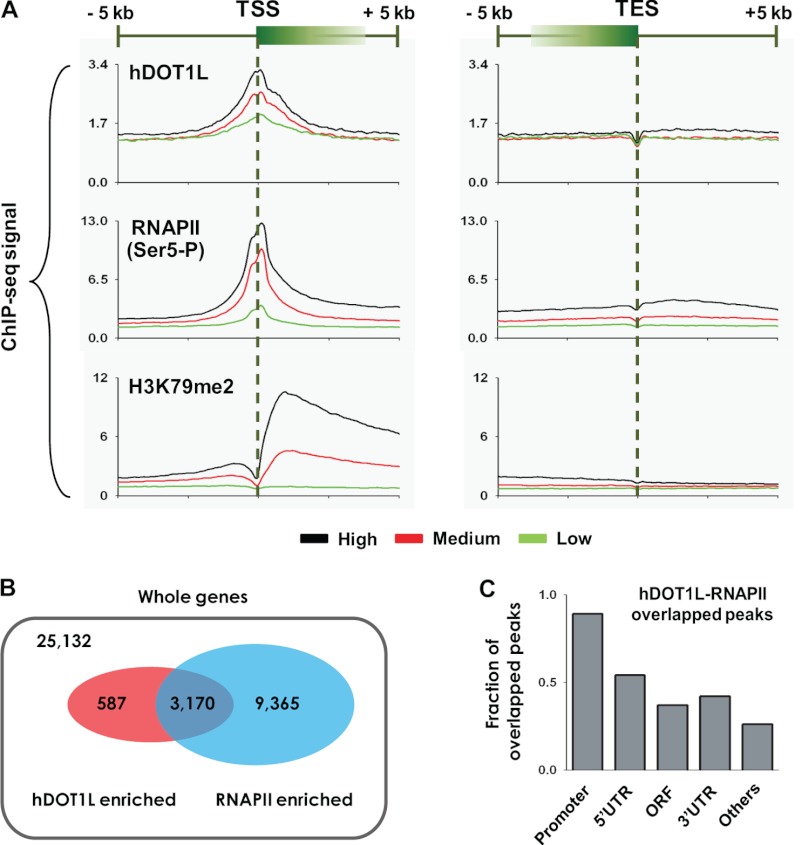
Our results revealed that hDOT1L and RNAPII manifested strong signals at and around the TSSs (5 kb upstream/downstream) (Fig. 3A, left panel), whereas the relative signal rapidly decreased along transcribed regions near the transcription end sites (5 kb upstream/downstream) (Fig. 3A, right panel). The signal patterns for H2Bub1 displayed a strong 5′ bias along the transcribed regions, as did those for all three H3K79 methylations (Fig. 3A and supplemental Fig. S2). In all cases, highly expressed genes (black lines) displayed stronger signal intensities compared with lowly expressed genes (green lines) (Fig. 3A). Among the 3,757 hDOT1L-targeted genes we identified, 3,170 were also targeted by RNAPII (Fig. 3B). Importantly, 89% of the hDOT1L peaks overlapped with those of RNAPII at the promoter regions, and 54% of the hDOT1L peaks overlapped with those of RNAPII at the 5′-UTR regions, suggesting that the specific functions of hDOT1L in association with RNAPII are concentrated in these regions (Fig. 3C).
FIGURE 3.
Genome-wide properties of hDOT1L and RNAPII. A, genome-wide profiles of hDOT1L, RNAPII, and H3K79me2 are shown for 5 kb upstream and downstream of TSSs and transcription end sites (TES) in NCCIT cells. The ChIP-seq signals are presented for high (top 30%), middle, and low (bottom 30%) level gene expression according to the FPKM scores generated by Cufflinks (42). ChIP-seq for RNAPII was performed using an antibody specific to the Ser-5-phosphorylated form. B, the targeted genes were defined from the detected peaks from the MACS peak detection program (59). Among 25,132 total genes, 3,757 and 12,535 were targeted by hDOT1L and RNAPII, respectively. C, by using MACS, 48,272 RNAPII and 12,026 hDOT1L peaks were identified in the NCCIT genome, and 4,851 peaks were found to overlap. The fraction of overlapped peaks is the value based on the peaks overlapped between RNAPII and hDOT1L versus the peaks of hDOT1L in the indicated genomic regions. For example, among the 1,411 hDOT1L peaks in promoter regions, 1,259 overlapped with RNAPII peaks. Genomic regions were defined as follows: promoter (2 kb upstream of TSS to TSS), 5′-UTR (TSS to coding start site), ORF (coding start site to coding end site), 3′-UTR (coding end site to the transcription end site), and others (remaining genomic regions).
Given the strong correlation of hDOT1L and RNAPII occupancy at the TSS, we next examined their target genes in an effort to determine whether the presence of both hDOT1L and RNAPII is associated with active transcription. We found that the top 500 high quality hDOT1L binding sites (determined using GREAT) were strongly correlated with the Ser-5-phosphorylated RNAPII (supplemental Fig. S3A). Furthermore, the expression levels of transcripts associated with the high quality hDOT1L-binding sites were significantly higher than those of all transcripts (p < 0.0001) (supplemental Fig. S3B). Interestingly, functional annotation analysis of the top 500 high quality hDOT1L binding sites revealed that hDOT1L was significantly associated with the promoters of genes encoding RNAPII-related proteins, such as TAF12, POLR2I, and GTF2H3, as well as those encoding ribosomal proteins, including RPL10A, RPL3, RPLP1, RPS3, RPS12, and RPS21 (supplemental Fig. S3C). Overall, we found that hDOT1L target genes tend to be highly expressed, and genes co-occupied by both hDOT1L and RNAPII were also highly expressed (supplemental Fig. S3D).
H3K79 Methylations Significantly Overlap with H2Bub1 at Actively Transcribed Genes
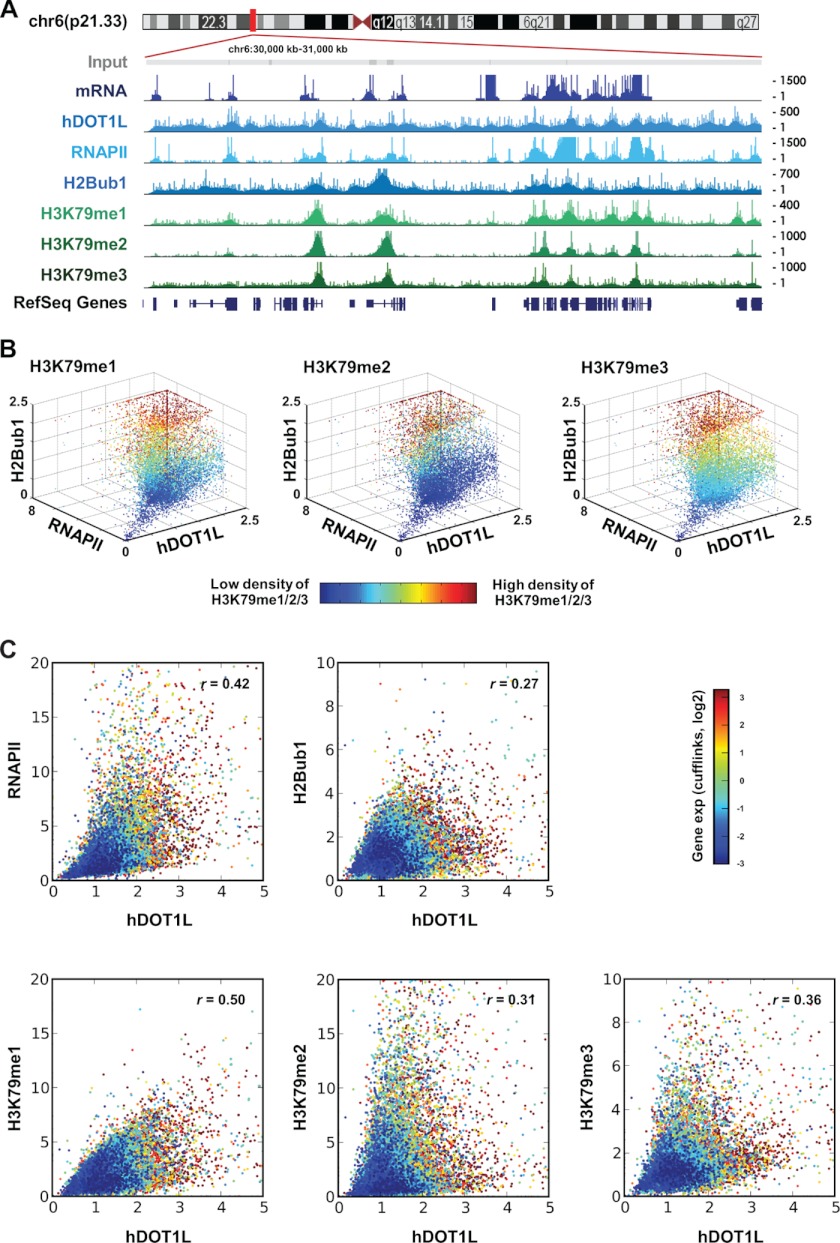
Next, we investigated the genome-wide overlap between RNAPII and the hDOT1L substrate, H3K79. hDOT1L is broadly distributed throughout the genome, showing only 2-fold enrichment at peak regions (for example, see the TSS in Fig. 3A), and relatively weak signals compared with those of RNAPII (Fig. 4A). However, we consistently observed slightly increased hDOT1L signals at RNAPII-enriched regions (Fig. 4A). In contrast to the relatively broad distribution of the hDOT1L enzyme, however, H3K79 methylations showed strong enrichment within regions of active transcription. Interestingly, the H2Bub1 signal showed a distribution very similar to that of H3K79 methylations. Therefore, the enzymatic activity of hDOT1L appears to overlap significantly with RNAPII and H2Bub1 occupancy at actively transcribed genes.
FIGURE 4.
Mapping the genome-wide co-occupancies of hDOT1L and RNAPII. A, ChIP-seq results for chr6:30,000 to 31,000 kb are shown. All of the patterns were drawn using the UCSC genome browser with a smoothing window of 10 pixels. The y axis indicates tag counts at 1-kb intervals. Mapped short read sequences without any treatment (Input) were randomly distributed throughout the genome. ChIP-seq for RNAPII was performed using an antibody specific to the Ser-5-phosphorylated form. B, each spot indicates the average ChIP-seq signals in the gene body region of each gene, and its color indicates the density of H3K79 methylations. The r values indicate Pearson correlation coefficients. C, the Pearson correlation coefficients (r values) between DOT1L and other modifications are shown.
Because the distribution patterns of H3K79 methylations overlapped with that of H2Bub1 (Fig. 4A and supplemental Fig. S2) and H3K79 methylations are catalyzed by hDOT1L, we postulated that the combination of hDOT1L, H2Bub1, and RNAPII determines H3K79 methylation levels. To test this possibility, we looked for correlations among these proteins in gene body regions across the human genome. Positive correlations were consistently found between the ChIP-seq signals of hDOT1L and RNAPII in gene body regions (Fig. 4B). Furthermore, when correlation coefficients were calculated between hDOT1L and the other features, we found a strong correlation with RNAPII (Fig. 4C). The H2Bub1-enriched genes were also likely to be simultaneously enriched for H3K79me1/2/3, hDOT1L, and RNAPII and showed high levels of transcription (Fig. 4B, red spots). These results suggest that the cooperation of RNAPII, hDOT1L, and H2Bub1 is required for the establishment of densely H3K79-methylated regions and highly active gene transcription.
We further examined the relationships among H2Bub1, hDOT1L, RNAPII, and H3K79 methylations by defining two groups of genes according to hDOT1L occupancy in the gene body region: the top 30% of genes with high hDOT1L signals versus the bottom 30% of genes with low hDOT1L signals (supplemental Fig. S4A). The hDOT1L-enriched genes showed stronger RNAPII signals (2.3-fold enrichment) compared with the hDOT1L-depleted genes. This observation was also noted for H2Bub1 and H3K79 methylation. H3K79 mono-, di-, and trimethylation were more dependent on H2Bub1 occupancy (supplemental Fig. S4B). These results further support the notion that although hDOT1L is broadly distributed throughout the genome, it is specifically enriched in RNAPII-targeted regions, where it may mediate H3K79 methylations in the presence of H2Bub1.
Identification of a Novel Interacting Region in hDOT1L Required for Interaction with the Phosphorylated CTD of RNAPII
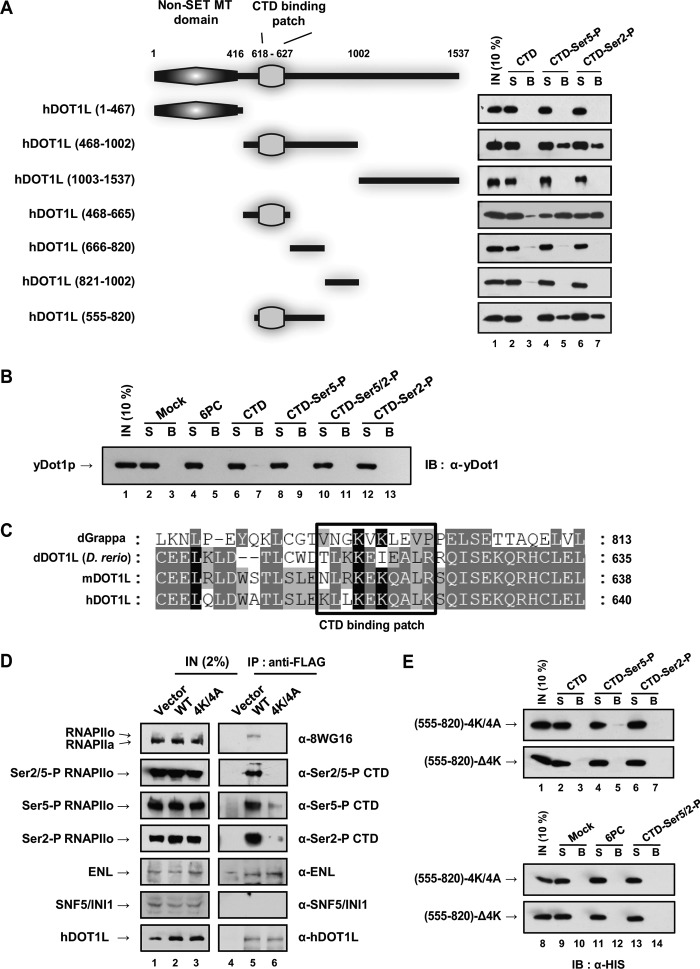
To investigate the specific region in hDOT1L required for interaction with the phosphorylated CTD of RNAPII, we generated several hDOT1L deletion constructs (supplemental Fig. S5A). The hDOT1L (1–467) construct containing an intact catalytic core domain and the hDOT1L (1003–1537) construct both failed to show any interaction with the phosphorylated CTD peptide (Fig. 5A). The hDOT1L (468–1002) construct, however, bound to Ser-5- and Ser-2-phosphorylated CTDs but not to unmodified CTD. Further investigation of the hDOT1L middle region (468–1002) through additional overlapping truncations demonstrated that the amino acid residues 555–665 were required for the interaction of hDOT1L with phosphorylated CTD peptides (Fig. 5A).
FIGURE 5.
hDOT1L mutants do not bind to the phosphorylated form of RNAPII in cultured cells or in vitro. A and B, peptide pulldown assays were performed with purified recombinant FLAG-tagged hDOT1L (1–467), (468–1002), and (1003–1537); His6-tagged hDOT1L (468–665), (666–820), (821–1002), and (555–820); and yDot1p (800 ng). C, sequence alignment of hDOT1L homologs. Alignment was performed using the ClustalW2 multiple sequence alignment program and the GENEDOC software. D, HEK293T cells were transfected with control vector and vectors expressing full-length FLAG-hDOT1L (WT) and -hDOT1L-4K/618–627/4A (4K/4A). The cells were extracted and immunoprecipitated with anti-FLAG at 4 °C. Immunoprecipitated samples were resolved by SDS-PAGE and immunoblotted using the antibodies indicated on the right. E, peptide pulldown assay was performed using mutant proteins (800 ng). (555–820)-4K/4A and (555–820)-Δ4K represent His6-hDOT1L (555–820)-4K/618–627/4A and (555–820)-Δ4K/618–627, respectively. Shown is the immunoblot with anti-His. S and B indicate supernatant and bead fractions, respectively, in A, B, and E. IN, input; IB, immunoblot; IP, immunoprecipitation.
According to structural analyses of the yeast Dot1 (yDot1) and hDOT1L (16, 49), the 360 amino acids at the N terminus of hDOT1L share significant sequence homology with yDot1 (supplemental Fig. S5B), but the long middle and C-terminal domains of hDOT1L (amino acid 431–1537) are conserved only in multicellular organisms (16, 17). As expected, full-length yDot1 purified from bacteria (supplemental Fig. S5A) failed to bind to either unmodified CTD or phosphorylated CTD in vitro (Fig. 5B).
When we aligned the full amino acid sequences of dGrappa (Drosophila homolog), dDOT1L (Danio rerio), murine DOT1L, and human DOT1L (Fig. 5C), we noted a conserved region within the region found to be required for the interaction of hDOT1L with phosphorylated CTD. We hypothesized that this region, which comprises amino acids 618–627 of hDOT1L, may be a RNAPII phosphorylated CTD-binding patch; thus, we have termed it the “CTD-binding patch.”
To determine whether the CTD-binding patch within hDOT1L is essential for the interaction between hDOT1L and the phosphorylated CTD of RNAPII in HEK293T cells, we generated a hDOT1L-4K/618–627/4A (4K/4A; four lysines substituted to alanines in the CTD-binding patch of hDOT1L) mutant and used it for co-immunoprecipitation analyses. The 4K/4A mutant did not co-immunoprecipitate with RNAPII (Fig. 5D, lane 6). To determine whether mutations in the CTD-binding patch of hDOT1L could disrupt RNAPII CTD binding in vitro, we carried out peptide pulldown analyses using highly purified hDOT1L (555–820)-4K/4A and (555–820)-Δ4K (Δ4K; four lysines deleted in the CTD-binding patch). Both of these hDOT1L mutants failed to bind phosphorylated CTD peptides (Fig. 5E). These results indicate that the CTD-binding patch is essential for the interaction of hDOT1L with the phosphorylated CTD of RNAPII.
Knockdown of hDOT1L Represses H3K79 Methylation and Actively Transcribed Genes
Because hDOT1L is the only known H3K79 methyltransferase (33), and we have shown that it interacts with the phosphorylated CTD of RNAPII, we next tested whether H3K79 methylations directly mediate the active transcription of genes enriched with both hDOT1L and RNAPII. Although the target genes of hDOT1L are not fully known, we analyzed a subset of actively transcribed genes, including Nanog, Oct4, and Sox2, in NCCIT cells. These are well known pluripotency-related genes, and our genome-wide ChIP data indicated that they were enriched with both hDOT1L and RNAPII (supplemental Fig. S6A). We also measured the relative mRNA-seq, hDOT1L, and RNAPII signals of pluripotency-related genes, stem cell development-related genes, and other genes. In the NCCIT cells, we first noted that the pluripotency maintenance-related genes were highly expressed compared with all genes (supplemental Fig. S6B, top left, and Table S3). These genes were also simultaneously enriched with both hDOT1L and RNAPII (high ChIP-seq signals were found for both hDOT1L and RNAPII) compared with development-related genes, such as those associated with the endoderm, mesoderm, and ectoderm (supplemental Fig. S6B, top right and bottom left, and Tables S4–S6). This suggests that the co-occupancy of hDOT1L and RNAPII may play a role in maintaining pluripotency-related genes. Although this result may originate from the higher expression level of pluripotency maintenance-related genes in this cell line, not the property of pluripotency itself, the significant enrichment indicates that hDOT1L may play a role in pluripotency maintenance.
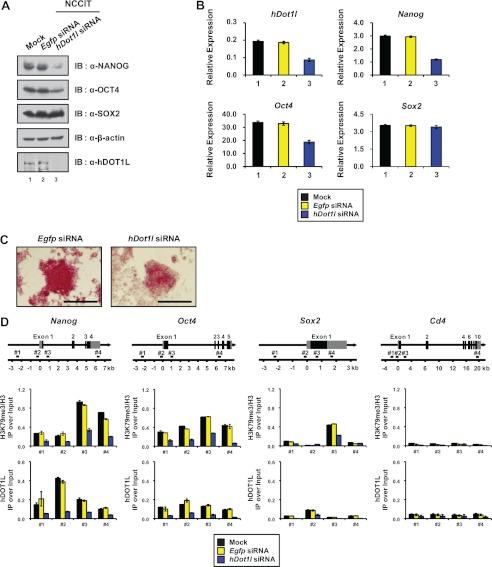
We next evaluated the importance of hDOT1L and RNAPII co-occupancy in the expression of pluripotency-related genes by siRNA-mediated knockdown of hDOT1L in NCCIT cells. After transfection with hDot1l siRNA (supplemental Fig. S7A), hDOT1L expression was reduced in both protein and mRNA levels (Fig. 6, A and B). Importantly, hDOT1L knockdown repressed the expression of the representative pluripotency-related genes, Nanog and Oct4, but not that of the control SOX2 gene, compared with cells transfected with the control Egfp siRNA (Fig. 6, A and B). In addition, the activity of alkaline phosphatase, a marker of pluripotency, was markedly reduced in hDot1l siRNA-transfected cells compared with control cells transfected with Egfp siRNA (Fig. 6C).
FIGURE 6.
Depletion of hDOT1L affects actively transcribed gene expression and H3K79 methylation. A, Egfp and hDot1l siRNAs were transiently transfected into NCCIT cells. Whole cell extracts were resolved by SDS-PAGE and immunoblotted (IB) using the indicated antibodies on the right. β-Actin was used as a loading control. B, the mRNA levels were analyzed by RT-qPCR (supplemental Table S1), with each expression value normalized with respect to that of β-actin. The error bars denote standard deviations from three biological replicates. C, Egfp (60 nm) and hDot1l (100 nm) siRNA were transfected into NCCIT cells. Three days after transfection, the cells were fixed and stained for alkaline phosphatase activity. The cells expressing AP show red colorations. The scale bar indicates 250 μm. D, NCCIT cells were transfected as in A, and ChIP analyses were carried out with the indicated antibodies. All of the signals were normalized with respect to the input and total histone H3. The error bars denote standard deviations from three biological replicates, each consisting of three qPCRs (supplemental Table S2).
To investigate that the down-regulation of NANOG and OCT4 was correlated with decreased H3K79 methylation, we performed a ChIP assay for hDOT1L and H3K79me3. After hDot1l siRNA transfection, the H3K79me3 level was reduced throughout the gene body of Nanog and Oct4, including the region around TSS (#2) where preferential binding of hDOT1L was seen (Fig. 6D). Interestingly, a significant level of H3K79me3 was also detected in a specific region within the gene body of Sox2, however, a slight reduction of H3K79me3 in the region after hDot1l siRNA transfection did not affect the Sox2 expression (Fig. 6, A, B, and D). These results show that hDOT1L is important for the proper level of H3K79me3 and the transcription of a subset of highly expressed pluripotency-related genes in human embryonic carcinoma cells.
A Functional Link between hDOT1L and the Phosphorylated RNAPII Is Important for Regulating the Expression of Actively Transcribed Genes in NCCIT Cells
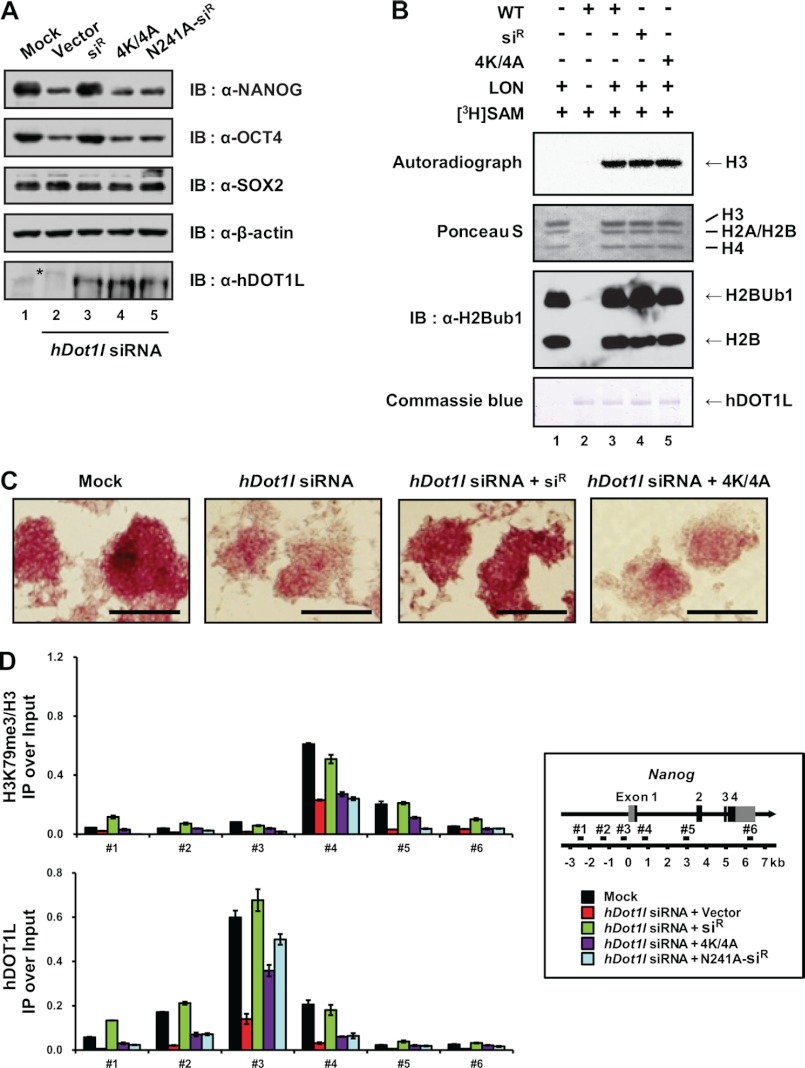
We next examined whether the decreases in H3K79 methylations and the expression levels of NANOG and OCT4 could be rescued by the expression of exogenous mutant hDOT1L resistant to hDot1l siRNA (supplemental Fig. S7A). We expressed wild-type hDOT1L-WT-siR (siR) and 4K/4A, resistant to hDot1l siRNA because of the respective silent and missense mutations in target sequences (supplemental Fig. S7), in NCCIT cells that had been subjected to siRNA-mediated knockdown of endogenous hDOT1L. As expected, the expression levels of NANOG and OCT4 increased following treatment with exogenous siR but not the RNAPII binding-defective mutant, 4K/4A (Fig. 7A). The enzymatic activities of siR and 4K/4A as histone methyltransferases were comparable with those of the WT protein (Fig. 7B).
FIGURE 7.
hDOT1L bound to RNAPII regulates the expression of actively transcribed genes. A, control vector, wild-type FLAG-hDOT1L-WT-siR (siR), −4K/618–627/4A (4K/4A), and -N241A-siR (N241A-siR) were transfected into hDOT1L knockdown NCCIT cells. Whole cell extracts were analyzed by SDS-PAGE and immunoblotted using the indicated antibodies on the right. β-Actin was used as a loading control. A nonspecific band in the hDOT1L immunoblot is indicated by an asterisk. B, histone methyltransferase assay against nucleosomes was performed using purified recombinant proteins from a baculovirus expression system. The samples were separated by SDS-PAGE and analyzed by autoradiograph. Long oligonucleosomes (LON) were resolved by SDS-PAGE and immunoblotted using anti-H2Bub1 antibody detecting H2B and H2Bub1. Long oligonucleosome and FLAG-hDOT1L proteins were stained with Ponceau S and Coomassie Blue, respectively. C, NCCIT cells were transfected as in A, and 3 days after transfection, the cells were fixed and stained for alkaline phosphatase activity. The bar indicates 250 μm. D, NCCIT cells were transfected as in A, and ChIP analyses were carried out with the indicated antibodies. All signals were normalized with respect to the input and total histone H3. The error bars denote standard deviations from three biological replicates, each consisting of three qPCRs.
Next, we checked whether the rescue of pluripotency-related gene expression by siR influenced the levels of alkaline phosphatase in NCCIT cells. Indeed, the decreased alkaline phosphatase activity seen in cells transfected with hDot1l siRNA was largely recovered following transfection with siR, but not 4K/4A (Fig. 7C). To validate that the recovery of NANOG and OCT4 expression by siR was correlated with increased H3K79 methylations, we used ChIP to examine histone modifications. The decreased incidence of H3K79me3 in the transcribed region of Nanog and Oct4 following siRNA-induced knockdown of hDOT1L was dramatically rescued by the exogenous expression of siR but not 4K/4A (Fig. 7D and supplemental Fig. S8).
To test the hypothesis that the ability of hDOT1L, in cooperation with RNAPII, to regulate pluripotency-related genes is dependent on its enzymatic activity, we introduced a silent mutation into the hDOT1L-N241A methylation-defective mutant (supplemental Fig. S7A) (49). hDOT1L-N241A-siR (N241A-siR), resistant to hDot1l siRNA, was overexpressed in hDOT1L knockdown NCCIT cells, and the H3K79me3 level was investigated. As expected, the reduced occurrence of H3K79me3 in hDOT1L knockdown cells was not rescued by the exogenous expression of N241A-siR (Fig. 7D and supplemental Fig. S8). Furthermore, the decreases in NANOG and OCT4 expression caused by hDot1l siRNA were not rescued in N241A-siR-transfected cells (Fig. 7A, lane 5). Therefore, we conclude that the functional interaction of hDOT1L with the phosphorylated CTD of RNAPII and subsequent histone H3K79 methylations by hDOT1L is a prerequisite for pluripotency-related gene expression.
DISCUSSION
Previous studies have demonstrated that the function and existence of Dot1 and global H3K79 methylations are strictly evolutionarily conserved from yeast to humans. Both Dot1 and H3K79 methylations, however, are absent from Schizosaccharomyces pombe (50), and a Dot1 homolog has not been found in Arabidopsis thaliana (51), suggesting that there are variations in the existence and functions of Dot1 and H3K79 methylations across species. Additionally, the distributions of Dot1 and H3K79 methylations during active transcription show different patterns in yeast and humans. In yeast, yDot1 and H3K79 methylations are detectable only at the coding regions of active genes (52). In humans, our present work reveals that hDOT1L is distributed at and around the transcription start sites, whereas H3K79 methylations are detected in the coding regions (similar to the pattern in yeast). Additionally, we report that the distribution of hDOT1L is much broader than that of H3K79 methylations at the gene coding sequences.
Many of the distinctions between the yDot1 and hDOT1L proteins are likely to be a consequence of structural differences. The full-length hDOT1L consists of 1,537 amino acids (16), but only the N-terminal domain shows homology with yDot1. The long middle and C-terminal domains of hDOT1L are unique to multicellular organisms, such as flies and humans. Here, we show that amino acid residues 618–627 of hDOT1L contain a CTD-binding patch that is critical for specific interactions with the phosphorylated CTD of RNAPII in cultured cells and in vitro. Additionally, we show that hDOT1L but not yDot1 functionally interacts with the phosphorylated RNAPII, indicating that the general interaction mechanism of yDot1 with RNAPII may differ from that of human hDOT1L. The CTD-binding patch may facilitate the targeting of hDOT1L to the coding region of active genes (Figs. 3A and 4A) through its interaction with elongating RNAPII.
We show that hDOT1L knockdown reduces the expression of NANOG and OCT4 and that this is correlated with decreases in hDOT1L binding and H3K79 methylation (Figs. 6 and 7). Thus, we suggest a causal relationship whereby the depletion of hDOT1L leads to reduced targeting and propagation of this enzyme across actively transcribed genes, such as Nanog and Oct4. The resulting decrease in H3K79 methylations at the whole gene body region may cause a decrease in gene expression. Although a reduction in H3K79me3 signal is observed at the 5′ end of the Sox2 gene, the expression of SOX2 is not affected (Figs. 6 and 7), suggesting the distribution pattern of H3K79 methylation may be a major determinant to regulate the expression of active genes.
When RNAPII binds to a promoter, Ser-5 of the CTD is phosphorylated (53). During the transition state from transcription initiation to elongation, the CTD is phosphorylated at both Ser-5 and Ser-2 (54–56). Elongating RNAPII loses Ser(P)-5 but retains Ser(P)-2 at the 3′ region of gene body (57). Because Dot1p directly interacts with monoubiquitylated H2B to produce H3K79 methylations (31), it seems likely that both the phosphorylation states of RNAPII during active transcription and the genome-wide pattern of H2Bub1 together may affect hDOT1L in regulating gene transcription through H3K79 methylations.
However, we do not envisage that hDOT1L would be targeted to promoters through elongating RNAPII. Rather, it is likely that hDOT1L would target the promoters of active genes via unknown factors (probably transcription factors), and the targeted hDOT1L would move to the gene body region together with elongating RNAPII through an interaction via the CTD-binding patch. Interaction between hDOT1L and monoubiquitylated H2B would further affect the production of mono-, di-, and trimethylations of H3K79. Further studies are required to examine the potential mechanism(s) responsible for the selective targeting of hDOT1L.
Supplementary Material
Acknowledgments
We thank Dr. Jerry Workman for helpful suggestions and discussions. We also thank Samantha G. Pattenden for critical reading of the manuscript. We gratefully acknowledge Drs. Bing Li and Jerry Workman for providing the synthetic unphosphorylated and phosphorylated Ser-5 and Ser-2 CTD peptides and LON (long oligonucleosomes from HeLa cells). We also acknowledge Alberto R. Kornblihtt for kindly providing the pAT7Rpb1αAm(R) constructs.
This work was supported by Epigenomic Research Program for Human Stem Cells Grant 2007-2004134, Stem Cell Research Program Grant 2012M 3A9B 4027953, and grants from the Ministry of Education, Science & Technology (South Korea) and the KRIBB/KRCF Research Initiative Program (NAP), Rural Development Administration (South Korea).

This article contains supplemental text, Tables S1–S6, and Figs. S1–S8.
- H2Bub1
- H2B monoubiquitylation
- RNAPII
- RNA polymerase II
- CTD
- C-terminal domain
- FPKM
- fragments/kilobase of exon/million fragments mapped
- TSS
- transcriptional start site
- qPCR
- quantitative PCR.
REFERENCES
- 1. Kornberg R. D. (1974) Chromatin structure. A repeating unit of histones and DNA. Science 184, 868–871 [DOI] [PubMed] [Google Scholar]
- 2. Kornberg R. D., Lorch Y. (1999) Twenty-five years of the nucleosome, review fundamental particle of the eukaryote chromosome. Cell 98, 285–294 [DOI] [PubMed] [Google Scholar]
- 3. Shilatifard A. (2006) Chromatin modifications by methylation and ubiquitination. Implications in the regulation of gene expression. Annu. Rev. Biochem. 75, 243–269 [DOI] [PubMed] [Google Scholar]
- 4. Lachner M., O'Sullivan R. J., Jenuwein T. (2003) An epigenetic road map for histone lysine methylation. J. Cell Sci. 116, 2117–2124 [DOI] [PubMed] [Google Scholar]
- 5. Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997) Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]
- 6. Lacoste N., Utley R. T., Hunter J. M., Poirier G. G., Côte J. (2002) Disruptor of telomeric silencing-1 is a chromatin-specific histone H3 methyltransferase. J. Biol. Chem. 277, 30421–30424 [DOI] [PubMed] [Google Scholar]
- 7. Ng H. H., Feng Q., Wang H., Erdjument-Bromage H., Tempst P., Zhang Y., Struhl K. (2002) Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 16, 1518–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. van Leeuwen F., Gafken P. R., Gottschling D. E. (2002) Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109, 745–756 [DOI] [PubMed] [Google Scholar]
- 9. Wysocki R., Javaheri A., Allard S., Sha F., Côté J., Kron S. J. (2005) Role of Dot1-dependent histone H3 methylation in G1 and S phase DNA damage checkpoint functions of Rad9. Mol. Cell Biol. 25, 8430–8443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Giannattasio M., Lazzaro F., Plevani P., Muzi-Falconi M. (2005) The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6-Bre1 and H3 methylation by Dot1. J. Biol. Chem. 280, 9879–9886 [DOI] [PubMed] [Google Scholar]
- 11. Ng H. H., Ciccone D. N., Morshead K. B., Oettinger M. A., Struhl K. (2003) Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells. A potential mechanism for position-effect variegation. Proc. Natl. Acad. Sci. U.S.A. 100, 1820–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. San-Segundo P. A., Roeder G. S. (2000) Role for the silencing protein Dot1 in meiotic checkpoint control. Mol. Biol. Cell 11, 3601–3615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schulze J. M., Jackson J., Nakanishi S., Gardner J. M., Hentrich T., Haug J., Johnston M., Jaspersen S. L., Kobor M. S., Shilatifard A. (2009) Linking cell cycle to histone modifications. SBF and H2B monoubiquitination machinery and cell-cycle regulation of H3K79 dimethylation. Mol. Cell 35, 626–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schübeler D., MacAlpine D. M., Scalzo D., Wirbelauer C., Kooperberg C., van Leeuwen F., Gottschling D. E., O'Neill L. P., Turner B. M., Delrow J., Bell S. P., Groudine M. (2004) The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 18, 1263–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang W., Hayashizaki Y., Kone B. C. (2004) Structure and regulation of the mDot1 gene, a mouse histone H3 methyltransferase. Biochem. J. 377, 641–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Feng Q., Wang H., Ng H. H., Erdjument-Bromage H., Tempst P., Struhl K., Zhang Y. (2002) Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 12, 1052–1058 [DOI] [PubMed] [Google Scholar]
- 17. Shanower G. A., Muller M., Blanton J. L., Honti V., Gyurkovics H., Schedl P. (2005) Characterization of the grappa gene, the Drosophila histone H3 lysine 79 methyltransferase. Genetics 169, 173–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mueller D., Bach C., Zeisig D., Garcia-Cuellar M. P., Monroe S., Sreekumar A., Zhou R., Nesvizhskii A., Chinnaiyan A., Hess J. L. (2007) A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood 110, 4445–4454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mohan M., Herz H. M., Takahashi Y. H., Lin C., Lai K. C., Zhang Y., Washburn M. P., Florens L., Shilatifard A. (2010) Linking H3K79 trimethylation to Wnt signaling through a novel Dot1-containing complex (DotCom). Genes Dev. 24, 574–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bitoun E., Oliver P. L., Davies K. E. (2007) The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum. Mol. Genet. 16, 92–106 [DOI] [PubMed] [Google Scholar]
- 21. Jones B., Su H., Bhat A., Lei H., Bajko J., Hevi S., Baltus G. A., Kadam S., Zhai H., Valdez R. (2008) The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure. PLoS Genet. 4, e1000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Barry E. R., Krueger W., Jakuba C. M., Veilleux E., Ambrosi D. J., Nelson C. E., Rasmussen T. P. (2009) ES cell cycle progression and differentiation require the action of the histone methyltransferase Dot1L. Stem Cells 27, 1538–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Okada Y., Feng Q., Lin Y., Jiang Q., Li Y., Coffield V. M., Su L., Xu G., Zhang Y. (2005) hDOT1L links histone methylation to leukemogenesis. Cell 121, 167–178 [DOI] [PubMed] [Google Scholar]
- 24. Dover J., Schneider J., Tawiah-Boateng M. A., Wood A., Dean K., Johnston M., Shilatifard A. (2002) Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 277, 28368–28371 [DOI] [PubMed] [Google Scholar]
- 25. Sun Z. W., Allis C. D. (2002) Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418, 104–108 [DOI] [PubMed] [Google Scholar]
- 26. Lee J. S., Shukla A., Schneider J., Swanson S. K., Washburn M. P., Florens L., Bhaumik S. R., Shilatifard A. (2007) Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell 131, 1084–1096 [DOI] [PubMed] [Google Scholar]
- 27. Pavri R., Zhu B., Li G., Trojer P., Mandal S., Shilatifard A., Reinberg D. (2006) Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 125, 703–717 [DOI] [PubMed] [Google Scholar]
- 28. Weake V. M., Workman J. L. (2008) Histone ubiquitination. Triggering gene activity. Mol. Cell 29, 653–663 [DOI] [PubMed] [Google Scholar]
- 29. Kim J., Guermah M., McGinty R. K., Lee J. S., Tang Z., Milne T. A., Shilatifard A., Muir T. W., Roeder R. G. (2009) RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell 137, 459–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McGinty R. K., Kim J., Chatterjee C., Roeder R. G., Muir T. W. (2008) Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature 453, 812–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Oh S., Jeong K., Kim H., Kwon C. S., Lee D. (2010) A lysine rich region in Dot1p is crucial for direct interaction with H2B ubiquitylation and high level methylation of H3K79. Biochem. Biophys. Res. Commun. 399, 512–517 [DOI] [PubMed] [Google Scholar]
- 32. Jung I., Kim S. K., Kim M., Han Y. M., Kim Y. S., Kim D., Lee D. (2012) H2B monoubiquitylation is a 5′-enriched active transcription mark and correlates with exon-intron structure in human cells. Genome Res. 22, 1026–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Steger D. J., Lefterova M. I., Ying L., Stonestrom A. J., Schupp M., Zhuo D., Vakoc A. L., Kim J. E., Chen J., Lazar M. A. (2008) DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol. Cell Biol. 28, 2825–2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kouskouti A., Talianidis I. (2005) Histone modifications defining active genes persist after transcriptional and mitotic inactivation. EMBO J. 24, 347–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Barski A., Cuddapah S., Cui K., Roh T. Y., Schones D. E., Wang Z., Wei G., Chepelev I., Zhao K. (2007) High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 [DOI] [PubMed] [Google Scholar]
- 36. Damjanov I. (1993) Pathogenesis of testicular germ cell tumours. Eur. Urol. 23, 2–5 [DOI] [PubMed] [Google Scholar]
- 37. Teshima T., Chatani M., Hata K., Inoue T. (1988) High-dose rate intracavitary therapy for carcinoma of the uterine cervix. II. Risk factors for rectal complication. Int. J. Radiat. Oncol. Biol. Phys. 14, 281–286 [DOI] [PubMed] [Google Scholar]
- 38. Damjanov I., Horvat B., Gibas Z. (1993) Retinoic acid-induced differentiation of the developmentally pluripotent human germ cell tumor-derived cell line, NCCIT. Lab. Invest. 68, 220–232 [PubMed] [Google Scholar]
- 39. Spencer V. A., Sun J. M., Li L., Davie J. R. (2003) Chromatin immunoprecipitation. A tool for studying histone acetylation and transcription factor binding. Methods 31, 67–75 [DOI] [PubMed] [Google Scholar]
- 40. Langmead B., Trapnell C., Pop M., Salzberg S. L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Trapnell C., Pachter L., Salzberg S. L. (2009) TopHat. Discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., van Baren M. J., Salzberg S. L., Wold B. J., Pachter L. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Suganuma T., Kawabata M., Ohshima T., Ikeda M. A. (2002) Growth suppression of human carcinoma cells by reintroduction of the p300 coactivator. Proc. Natl. Acad. Sci. U.S.A. 99, 13073–13078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li B., Howe L., Anderson S., Yates J. R., 3rd, Workman J. L. (2003) The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 278, 8897–8903 [DOI] [PubMed] [Google Scholar]
- 45. de la Mata M., Kornblihtt A. R. (2006) RNA polymerase II C-terminal domain mediates regulation of alternative splicing by SRp20. Nat. Struct. Mol. Biol. 13, 973–980 [DOI] [PubMed] [Google Scholar]
- 46. Li J., Moazed D., Gygi S. P. (2002) Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. J. Biol. Chem. 277, 49383–49388 [DOI] [PubMed] [Google Scholar]
- 47. Ho C. K., Shuman S. (1999) Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol. Cell 3, 405–411 [DOI] [PubMed] [Google Scholar]
- 48. Kizer K. O., Phatnani H. P., Shibata Y., Hall H., Greenleaf A. L., Strahl B. D. (2005) A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol. Cell Biol. 25, 3305–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Min J., Feng Q., Li Z., Zhang Y., Xu R. M. (2003) Structure of the catalytic domain of human DOT1L, a non-SET domain nucleosomal histone methyltransferase. Cell 112, 711–723 [DOI] [PubMed] [Google Scholar]
- 50. Hiromi N. (2009) Evolutionary conservation levels of subunits of histone-modifying protein complexes in fungi. Comp. Funct. Genomics 2009, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang K., Sridhar V. V., Zhu J., Kapoor A., Zhu J. K. (2007) Distinctive core histone post-translational modification patterns in Arabidopsis thaliana. PLoS One 2, e1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shahbazian M. D., Zhang K., Grunstein M. (2005) Histone H2B ubiquitylation controls processive methylation but not monomethylation by Dot1 and Set1. Mol. Cell 19, 271–277 [DOI] [PubMed] [Google Scholar]
- 53. Sims R. J., 3rd, Belotserkovskaya R., Reinberg D. (2004) Elongation by RNA polymerase II. The short and long of it. Genes Dev. 18, 2437–2468 [DOI] [PubMed] [Google Scholar]
- 54. Cho E. J., Kobor M. S., Kim M., Greenblatt J., Buratowski S. (2001) Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev. 15, 3319–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhou K., Kuo W. H., Fillingham J., Greenblatt J. F. (2009) Control of transcriptional elongation and cotranscriptional histone modification by the yeast BUR kinase substrate Spt5. Proc. Natl. Acad. Sci. U.S.A. 106, 6956–6961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Qiu H., Hu C., Hinnebusch A. G. (2009) Phosphorylation of the Pol II CTD by KIN28 enhances BUR1/BUR2 recruitment and Ser2 CTD phosphorylation near promoters. Mol. Cell 33, 752–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Komarnitsky P., Cho E. J., Buratowski S. (2000) Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14, 2452–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Phatnani H. P., Jones J. C., Greenleaf A. L. (2004) Expanding the functional repertoire of CTD kinase I and RNA polymerase II. Novel phosphoCTD-associating proteins in the yeast proteome. Biochemistry 43, 15702–15719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nusbaum C., Myers R. M., Brown M., Li W., Liu X. S. (2008) Model-based analysis of ChIP-seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.