Abstract
Protein nanopores are under investigation as key components of rapid, low-cost platforms to sequence DNA molecules. Previously, it has been shown that the α-hemolysin (αHL) nanopore contains three recognition sites, capable of discriminating between individual DNA bases when oligonucleotides are immobilized within the nanopore. However, the direct sequencing of RNA is also of critical importance. Here, we achieve sharply defined current distributions that enable clear discrimination of the four nucleobases, guanine, cytosine, adenine and uracil, in RNA. Further, the modified bases, inosine, N6-methyladenosine and N5-methylcytosine, can be distinguished.
Keywords: α-hemolysin, nanopore, RNA sequencing, modified bases, protein engineering, single-nucleotide discrimination
Single-molecule nanopore technology is under development for ultra-rapid, low-cost sequencing of DNA and RNA molecules. Two types of nanopore are being investigated: solid state pores1 and protein pores, such as the heptameric α-hemolysin (αHL) pore. Protein pores have spearheaded the approach as they can be precisely manipulated by chemical and genetic engineering2, which facilitates the determination of sequence in individual DNA strands through base-dependent transitions in ionic current flow3. By immobilizing and stretching DNA strands within protein nanopores, the four canonical DNA bases and epigenetically modified bases have been individually identified by ionic current recording4–10. Individual oxidized bases and abasic sites can also be identified after chemical modification11, 12. However, ssDNA moves through protein nanopores at remarkably high velocities (e.g. ~1–3 µs per nucleobase)13, which provides insufficient signal-to-noise for individual bases to be identified. Therefore, enzymes have been used to ratchet DNA through pores14–17. By using approaches related to these published methods, Oxford Nanopore Technologies have demonstrated nanopore sequencing, achieving kilobase reads18. Additional studies of nanopore sequencing are appearing in the open literature19, 20.
Nanopore sequencing of RNA has received less attention. The direct high-throughput sequencing of RNAs (mRNA, miRNA etc) will allow the rapid identification and quantitation of functional elements of the genome and reveal important splicing patterns and post-transcriptional modifications21–23. RNA sequencing will be valuable in medical diagnosis, the selection of therapies and prognosis. For example, transcriptome sequencing has already been used to detect gene fusions in cancer24, 25. The ability to sequence extracellular RNA from plasma enhances the power of such approaches22, 26, 27. The genomes of RNA viruses and viral RNA transcripts are also accessible. RNA sequencing might also be used to monitor the levels and turnover of therapeutic RNAs28–30. Further, nanopores can directly identify modified bases, which are prevalent in RNA9, 31.
Short ssRNA homopolymer molecules have been distinguished on the basis of differences in residual currents (IRES) recorded while the RNAs are translocating through the wild-type (WT) αHL pore in an applied potential32–35. The transition between two homopolymer runs oligo(rA) and oligo(rC) within a single translocating RNA strand has also been observed, and may be accentuated by differences in the helical structures of the two regions32, 33. However, as in the case of DNA, individual RNA bases have not been identified in moving strands. Therefore, we have examined nucleobase identification in RNA strands by capturing and immobilizing them within the αHL pore with the biotin-streptavidin approach used previously for DNA base identification5, 6. The 5 nm-long β barrel of the αHL nanopore contains three recognition sites, R1, R2 and R3, capable of recognizing individual bases in ssDNA (Figure 1a). We investigated RNA base substitutions at position 9 of synthetic oligonucleotides (bases numbered from the 3' end) to probe the most promising recognition site R1 (which is located at the central constriction of the pore, comprising residues Lys-147, Glu-111 and Met-113, Figure 1b). Modification of the charge distribution within R1 has a powerful effect on IRES for DNA oligonucleotides5, 8. Therefore, we employed homoheptameric mutant pores made from αHL E111N/K147N (NN) and αHL E111N/K147N/M113Y (NNY) subunits.
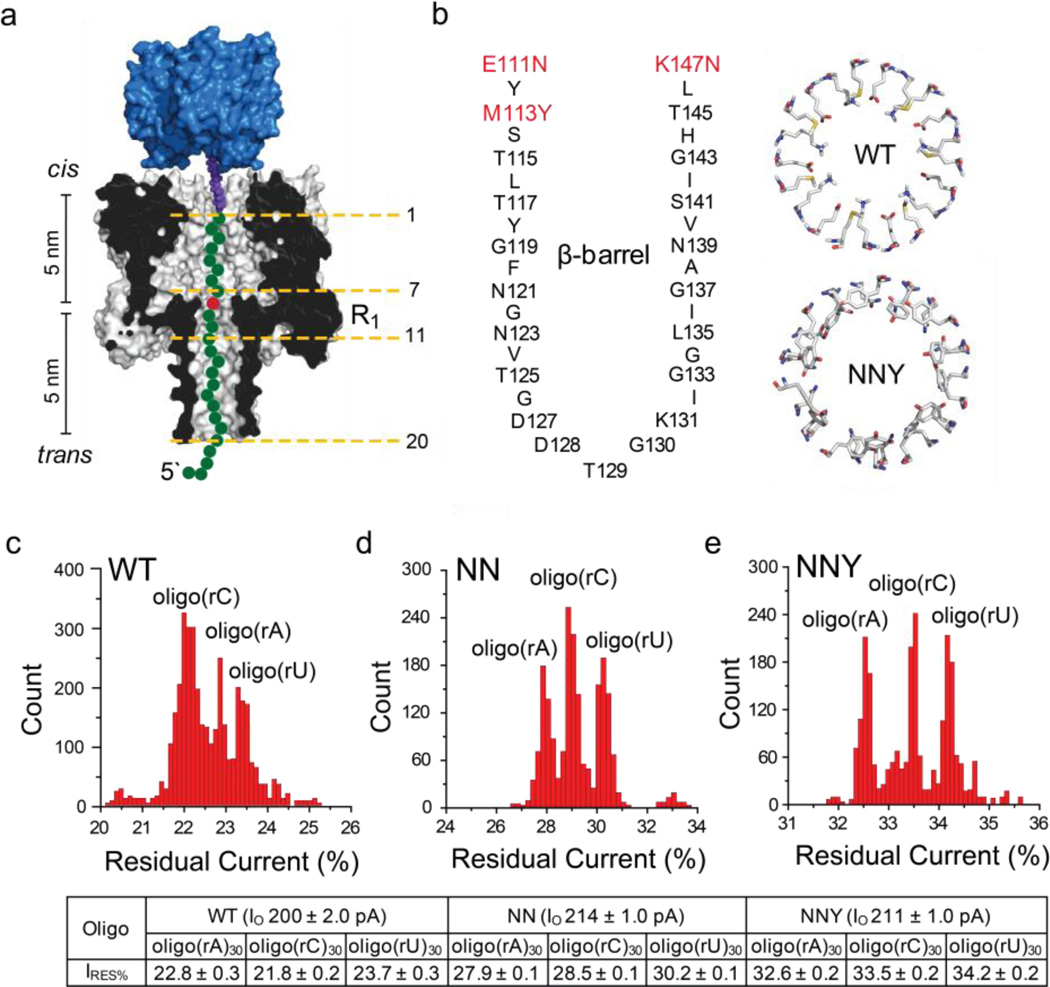
Figure 1. Interaction of homopolymeric RNA strands with αHL pores.
(a) Schematic representation of a homopolymeric RNA oligonucleotide (green circles), with a single ribonucleotide substituted at position 9 (red circle), immobilized inside an αHL pore (grey, cross-section) by using a 3'-biotin-TEG (purple)•streptavidin (blue) complex. The structure of the biotin linker is provided in Figure S1. The αHL pore can be divided into 2 parts, each 5 nm in length; an upper vestibule located between the cis entrance and the central constriction, and a 14-stranded, transmembrane, antiparallel β barrel, located between the central constriction and trans entrance. The oligonucleotide bases are numbered relative to the 3'-biotin tag and position 9 was chosen for interrogation by recognition site R1 (at the constriction). (b) The amino acid sequence of the transmembrane β barrel. The mutated residues at the top of the barrel are highlighted (red); these mutations enlarge the diameter of the pore. (Right, top) A view of the WT residues Glu-111, Lys-147 and Met-113 from the cis side of the pore. These residues form a constriction with diameter of ~13 Å. (Right, bottom) A view of the NNY residues Asn-111, Asn-147 and Tyr-113 from the cis side of the pore. These residues form a constriction with diameter of ~18 Å. The images were generated in Pymol. (c) Histogram of the residual current levels for RNA homopolymers oligo(rA)30, oligo(rC)30 and oligo(rU)30 immobilized in the WT αHL pore. (d) Histogram of the residual current levels for the homoheptameric αHL pore formed from the mutant E111N/K147N (NN). (e) Histogram of the residual current levels for the homoheptameric αHL pore formed from the mutant E111N/K147N/M113Y (NNY). Gaussian fits were performed for each peak, and the mean value of the open pore (IO), the residual currents (IRES%) and the standard deviation for each oligonucleotide are displayed in the table below the histograms.
Single nucleotide detection in homopolymeric strands
RNA base discrimination was first tested in homopolymer oligonucleotides, consisting of 30 nucleotides (the sequences of the oligonucleotides used in this paper are in Table S1). ssRNA oligonucleotides with biotin tags at the 3' end (Figure S1) were allowed to form complexes with streptavidin (SI Methods). In this state, the strands were captured and immobilized by αHL pores in an applied potential, but they were not translocated into the trans compartment (Figure 1a, Figure S2 automated voltage protocol). The one-second capture sequence was repeated for at least 400 cycles for each ssRNA added, with >90% of the cycles giving current blockades. The extended residence times of the oligonucleotides within the pore allowed reduction of the current noise by stringent filtration, thereby improving the signal-to-noise ratio and the precision of the measurements.
Once captured, the immobilized RNA molecules caused a sequence-dependent decrease in the ionic current through the pore. At +200 mV in 1 M KCl, 25 mM Tris-HCl, pH 7.5, containing 100 µM EDTA, WT αHL pores have a mean open pore current level (IO WT) of 199 ± 6 pA (n = 12), while the pores formed from NN and NNY, the mutants used in this work, gave IONN = 214 ± 7 pA (n = 8) and IONNY = 210 ± 8 pA (n = 8), respectively. Although the open pore currents carried by the WT pore, NN and NNY are similar (Figure S3), the residual currents in the presence of ssDNA5, 36, 37 and ssRNA (this work) are ~50% higher in the mutant pores (Figure 1c–e) owing to the increased cross section of the lumen after mutation. Immobilized oligo(rA)30 blocked NNY pores to a greater extent (IRES%oligo(rA) = 32.6 ± 0.2%) than oligo(rC)30 (IRES%oligo(rC) = 33.5 ± 0.2%) and oligo(rU)30 (IRES%oligo(rU) = 34.2 ± 0.2%) (Figure 1e, IRES% = IRES/IO X 100). The residual current difference between the oligo(rC)30 and the oligo(rA)30 oligonucleotide blockades (ΔIRES% = IRES%oligo(rC) - IRES%oligo(rA)) is +0.9 ± 0.2% and the ΔIRES% between oligo(rU)30 and the oligo(rA)30 oligonucleotide (ΔIRES% = IRES%oligo(rU) - IRES%oligo(rA)) is +1.6 ± 0.2%. Furthermore, the difference in residual current between the two most widely dispersed current peaks, (ΔIRES%OVERALL) between the three homopolymers also increases in the mutant pores compared to WT, providing improved levels of discrimination: αHL WT, ΔIRES%OVERALL = 1.5 ± 0.4%; αHL NN, ΔIRES%OVERALL = 2.4 ± 0.2% and αHL NNY, ΔIRES%OVERALL = 2.8 ± 0.4% (n = 3 for each pore). There is also a change in the pattern of the current blocks with oligo(rC) producing higher IRES% values than oligo(rA) in the NN and NNY pores, compared to WT (Figure 1c–e).
The IRES% values of the immobilized RNA homopolymers increased with applied positive potential for WT, NN and NNY pores (Figure S4, Table S2). A likely explanation is stretching of the RNA molecules inside the pore. Single-stranded homopolymers are highly flexible with a persistence length of ~1 nm38, 39. Therefore, the molecules will be elongated by confinement within the β barrel and further elongated by the applied potential, which generates a force of ~10 pN on the molecule (SI Methods)40–42. At higher potentials, the RNAs will become more fully extended within the pore, and blockade of the ionic current will be correspondingly reduced, resulting in higher IRES% values. The overall dispersion, ΔIRES%OVERALL , is also affected by increased potentials, which result in a tighter distribution of blockades by the three homopolymers, e.g. for αHL NNY we observed a ~64% decrease in dispersion, from ΔIRES%OVERALL = 4.4 ± 0.4% at +100 mV to 2.8 ± 0.4% at +200 mV (Table S2). The product of the sequential differences (δ) between each of the three residual current levels in the histograms, can also be determined to gauge the ability of the pore to discriminate between the three homopolymers (Table S2, SI Methods). All three pores are able to discriminate between oligo(rA), oligo(rC) and oligo(rU) within the potential range (+100 to +200 mV, δ > 0). With αHL NNY displaying the best discrimination at the highest applied potential (+200 mV), δNNY = 1.8 ± 0.2%. These investigations of voltage dependence aid in the optimization of IRES overlap.
Discrimination of all four ribobases in single-stranded oligonucleotides
We examined the ability of WT, NN and NNY αHL pores to distinguish between rG, rA, rU, and rC within individual nucleic strands. A first set of four oligonucleotides comprised oligo(dC)30 with a ribonucleotide at position-9 relative to the biotinylated 3' end. A second set of four oligonucleotides comprised oligo(rA)30 with a ribonucleotide at position-9. The oligo(dC) and oligo(rA) backgrounds were used, as both sequences are thought to have minimal secondary structure32, 43. A lack of secondary structure in our model system is advantageous, because any current differences observed between oligonucleotides can be attributed to the nucleobase sequences, rather than structural differences.
For the oligo(dC)30 oligos, WT αHL pores showed weak discrimination between rC, rA, rU and rG (in order of increasing IRES%, Figure 2c). The αHL NN pores displayed improved discrimination, clearly separating the bases in the order rC, rA, rG and rU (in order of increasing IRES%, Figure 2d). The overall dispersion of current levels was far greater for NN pores (NN-ΔIRES%OVERALL = 2.2 ± 0.4% and δNN = 0.3 ± 0.02%) than it was for WT pores (WT-ΔIRES%OVERALL = 1.2 ± 0.2% and δWT = 0.1 ± 0.02%). The αHL NNY pores displayed a twofold improvement in the dispersion of IRES% for the four nucleobases, compared to NN: NNY, ΔIRES%OVERALL = 4.5 ± 0.4%, δNNY = 2.4 ± 0.2%, along with a different order of increasing IRES% (rG<rA<rC<rU, Figures 2a and 2e). In the NNY pore, the tyrosines at position 113 may provide enhanced hydrogen bonding and aromatic stacking interactions with the immobilized bases7, 44, 45.
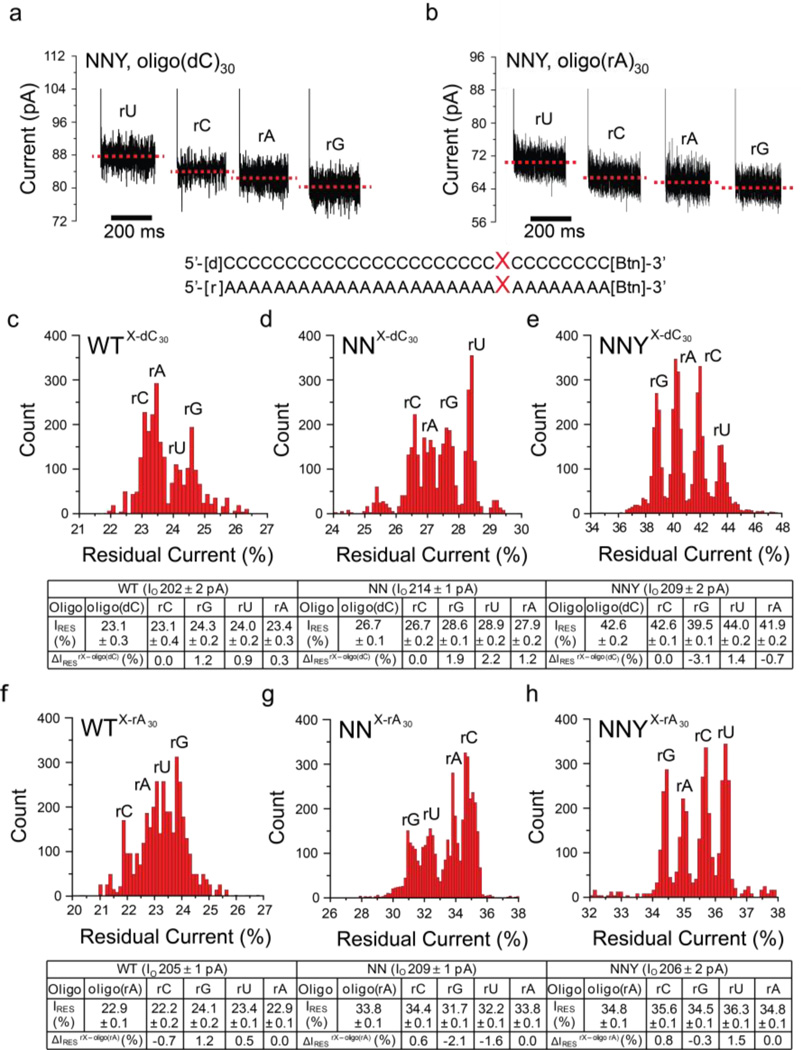
Figure 2. Discrimination of individual ribobases in ssDNA and ssRNA with the WT, NN and NNY αHL pores.
(a) Current traces for immobilized ssDNAs and (b) ssRNAs in the αHL NNY pore at +200 mV. (Below) Sequences of the oligonucleotides, biotinylated at the 3' ends. Two sets of four oligonucleotides were used, based on oligo(dC)30 and oligo(rA)30. Each set contained rG, rA, rC, or rU at position 9 (represented by X) relative to the biotin tag. Residual current (IRES%) histograms were compiled for oligonucleotide sets examined with the three αHL pores: (c) oligo(dC)30 oligonucleotides examined with the WT pore. (d) oligo(dC)30 oligonucleotides examined with the NN pore. (e) oligo(dC)30 oligonucleotides examined with the NNY pore. (f) oligo(rA)30 oligonucleotides examined with the WT pore. (g) oligo(rA)30 oligonucleotides examined with the NN pore. (h) oligo(rA)30 oligonucleotides examined with the NNY pore. Experiments were conducted at least 3 times, and the results displayed in each histogram are from a typical experiment. Gaussian fits were performed for each peak, and the mean values of the open pore currents (IO, pA), the normalized residual currents (IRES%) and the differences in the normalized residual currents (ΔIRES%) are displayed in the tables below the histograms with their standard deviations (± S.D). ΔIRES% is defined as the difference in residual current between an rX (X= rG, rC, rA or rU) oligonucleotide and either oligo(dC)30 or oligo(rA)30. ΔIRES%rX-oligo(dC) = IRES for the rX oligonucleotide - IRES for oligo(dC)30 or ΔIRES%rX-oligo(rA) = IRES% for the rX oligonucleotide - IRES% for oligo(rA)30.
Similar results were obtained for the second set of oligos in the oligo(rA)30 background. Again, the WT αHL pore showed the weakest discrimination between rC, rA, rU and rG (in order of increasing IRES%, Figure 2f), with a narrow dispersion between the residual current levels: ΔIRES%OVERALL = 1.9 ± 0.1% and δWT = 0.2 ± 0.01%. The αHL NN pore displayed discrimination that differed from that of the WT, with the increasing order of IRES% being rG, rU, rA and rC (Figure 2g), and the dispersion was again improved, ΔIRES%OVERALL = 2.7 ± 0.2% and δNN = 0.5 ± 0.01%. The αHL NNY pores gave the best dispersion in IRES% with a similar pattern between the four nucleobases as seen for the oligo(dC) background (rG, rA, rC and rU, in order of increasing IRES%, Figures 2b and 2h).
The voltage dependencies of the current blocks caused by the four RNA bases within the oligo(dC)30 and oligo(rA)30 chains were also examined (Figures S5, S6 and S7; Tables S3 and S4). We observed a similar trend to that seen in the homopolymer data, in which an increased applied positive potential gave higher IRES% levels for the DNA and RNA oligos across the three pores (Figure S5). For the NN and NNY pores, the four bases within the RNA oligo(rA) gave a higher ΔIRES%OVERALL (between +100 and +140 mV) than that observed for the DNA oligo(dC): eg. at +120 mV, within the oligo(rA) chain, NNY-ΔIRES%OVERALL = 4.9 ± 0.3%, δNNY = 4.2 ± 0.2% (n = 3); at +120 mV, within the oligo(dC) chain, NNY-ΔIRES%OVERALL = 2.8 ± 0.3%, δNNY = 0.7 ± 0.02% (n = 3) (Table S4). In addition, at each applied potential, we consistently observed lower IRES% levels for the four bases within the RNA background oligo(rA)30 in comparison to the DNA background oligo(dC)30.
Examining modified ribobases in ssDNA and ssRNA oligonucleotides
Cellular RNAs contain more than a hundred different base modifications at thousands of sites. These RNA modifications are dynamic and have critical regulatory roles31. The internal N6-methyladenosine (m6A) modification in messenger RNA (mRNA) is one of the most abundant modifications in higher eukaryotes, and is present at 3 to 5 sites on average per mRNA. The inability to carry out this modification leads to apoptosis. 5-Methylcytosine (m5C) is also widespread in cellular RNAs. While epigenetic DNA methylation has been extensively studied, the precise location of m5C in RNA remains to be elucidated. The deamination of adenosine (rA) to inosine (rI) is another important modification, which occurs in the editing of mRNA46, 47. rI base is read as rG by the ribosome, leading to amino acid substitution with functional implications48.
We therefore attempted to distinguish the riboforms of I, m6A and m5C from rG, rA, rC and rU at position-9 in ssDNA and ssRNA oligonucleotides by using the αHL NNY pore. The NNY pore had given superior discrimination between the standard bases (rG, rA, rC and rU) at recognition site R1 based on the increased ΔIRES%OVERALL values observed in the homopolymeric strands oligo(dC) and oligo(rA). The addition of the rI oligonucleotide to the standard mixture of four gave: ΔIRES%OVERALL = 5.8 ± 0.4%, δ = 2.3 ± 0.2% in the oligo(dC) background; and ΔIRES%OVERALL = 2.6 ± 0.2% and δ = 0.2 ± 0.04% in the oligo(rA) background. The difference in residual current levels between the rI oligo and oligo(rA), ΔIRES%rI-rA, was +0.9 ± 0.1% (n = 3) (Figure 3a–c, Tables S5 and S6). These results were striking, given the small chemical difference between the two bases.
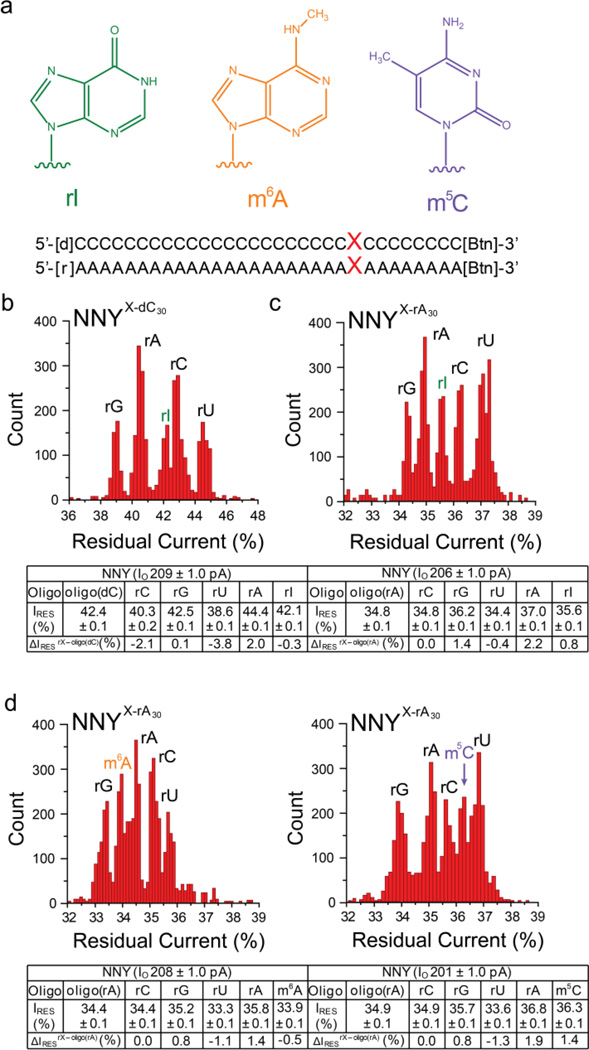
Figure 3. Discrimination of individual modified ribobases in ssDNA and ssRNA with the NNY αHL pore.
(a) Chemical structures of the modified bases: rI, inosine; m6A, N6-methyladenosine; m5C, 5-methylcytosine. (Below) Sequences of oligonucleotides biotinylated at the 3' end. Three sets of five oligo(dC)30 or five oligo(rA)30 oligonucleotides were used. Each set contained oligonucleotides with the four standard bases, rG, rA, rC, or rU at position 9 (represented by X) relative to the biotin tag, as well as an oligonucleotide with a modified base, one of rI, m6A or m5C. Histograms of the residual currents (IRES%) from oligonucleotide sets examined with the NNY pore were constructed: (b) the oligo(dC)30 set containing rI; (c) the oligo(rA)30 set containing rI; (d) the oligo(rA)30 set containing m6A; (e) the oligo(rA)30 set containing m5C oligo(rA)30. Experiments were conducted at least 3 times, and the results displayed in each panel are from a typical experiment. Gaussian fits were performed for each peak, and the mean values of the open pore currents (IO, pA), the normalized residual currents (IRES%) and the differences between the normalized residual currents (ΔIRES%) are displayed in the table below the histograms with their standard deviations (± S.D). ΔIRES% is defined as the residual current between an rX (X= rG, rC, rA, rU, rI, m6A or m5C) oligonucleotide and either oligo(dC)30 or oligo(rA)30. ΔIRES%rX-oligo(dC) = IRES% of the rX oligonucleotide - IRES% of oligo(dC)30 and ΔIRES%rX-oligo(rA) = IRES% of the rX oligonucleotide - IRES% of oligo(rA)30. The experiments were conducted at +200 mV.
m6A oligonucleotides also produced distinct current blocks (Figure 3a,d). The ΔIRES%m6A-rA between m6A and rA was −0.5 ± 0.1% (n = 3) in oligo(rA) with a similar dispersion of ΔIRES%OVERALL to that observed with rI: ΔIRES%OVERALL = 2.5 ± 0.1% and δ = 0.1 ± 0.04%. m5C could also be identified in the same manner (Figure 3a,e, Table S6): in oligo(rA), ΔIRES%m5C-rA = +1.4 ± 0.4% (n = 3), ΔIRES%OVERALL = 3.2 ± 0.2% and δ = 0.3 ± 0.02%. While the molecular bases for the small differences in IRES% levels for the modified bases are unclear, the observed current levels are distinct from all other bases and each other, demonstrating the possibility of using the αHL nanopore for mapping modified bases in the transcriptome. The ΔIRES% patterns for the WT, NN and NNY αHL pores (Figures 4a and 4b; Tables S5 and S6) demonstrate the potential to identify all seven bases within the oligo(dC) and oligo(rA) backgrounds. The αHL NNY pore displays a large dispersion of current levels for the seven bases in oligo(dC) and a more modest dispersion in oligo(rA), showing that base identification is modulated by the background7.
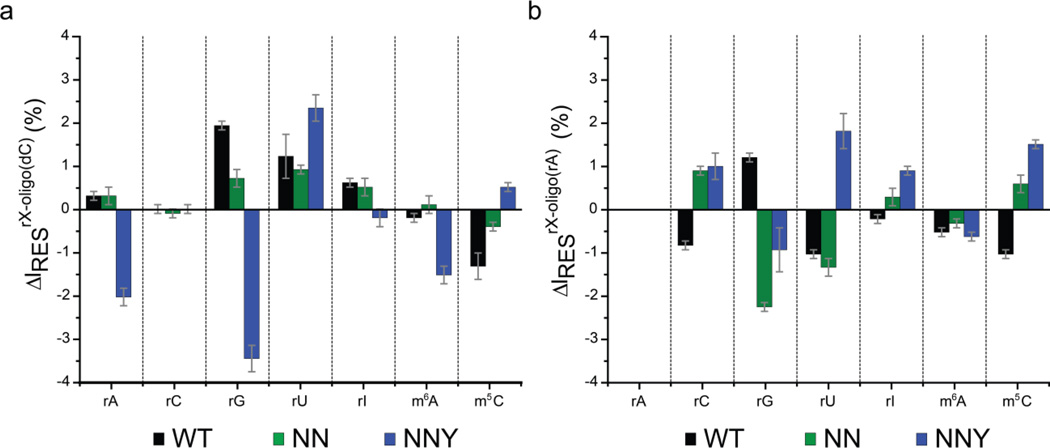
Figure 4. Plots of residual current difference for all seven bases identified in ssDNA and ssRNA oligonucleotides.
(a) ΔIRES% for each base substituted at position 9 of oligo(dC)30 for WT (black bars), NN (green bars) and NNY (blue bars) αHL pores. ΔIRES%rX-oligo(dC) = IRES% of the rX oligonucleotide - IRES% of oligo(dC)30. X = rG, rA, rC, rU, rI, m6A or m5C. (b) ΔIRES% for each base substituted at position 9 of oligo(rA)30 for WT (black bars), NN (green bars) and NNY (blue bars) αHL pores. ΔIRES%rX-oligo(rA) = IRES% of the rX oligonucleotide - IRES% of oligo(rA)30. X = rG, rA, rC, rU, rI, m6A or m5C. The errors given are standard deviations from three experiments (Table S5 and S6).
Recognition of individual nucleotides in a heteropolymeric background
The ability to sequence ssRNA would require the recognition of individual nucleotides in heteropolymeric backgrounds. To examine discrimination within a heteropolymer, we tested the R1 site in the NNY pore. The sequence we used (Figure 5) did not contain secondary structures, such as hairpins, as predicted by the mfold algorithm49, 50. At +200 mV, all four bases at position 9 were recognized with the same order of IRES% (rG, rA, rC and rU, Figure 5a), as seen in the homopolymeric backgrounds (Figure 2 and 3). The dispersion of the current levels in the histogram (ΔIRES%OVERALL = 1.7 ± 0.3%, δ = 0.2 ± 0.06%) were also similar to those seen in the homopolymeric backgrounds, suggesting that the different current levels were indeed directly due to the nucleobases changes, rather than changes in secondary structure. The voltage dependence of the currents arising from the four bases again displayed similar characteristics to the results obtained in the homopolymer backgrounds, with higher positive potentials resulting in higher IRES% levels (Figure 5b, Table S7). The span between rC and rA (Figure 5c) in the residual current histogram (ΔIRES%rC-rA = +0.7 ± 0.1%) was similar to that seen in the oligo(rA) homopolymeric background, ΔIRES%rC-rA = +0.8 ± 0.2% (Figure 2h, Table S4).
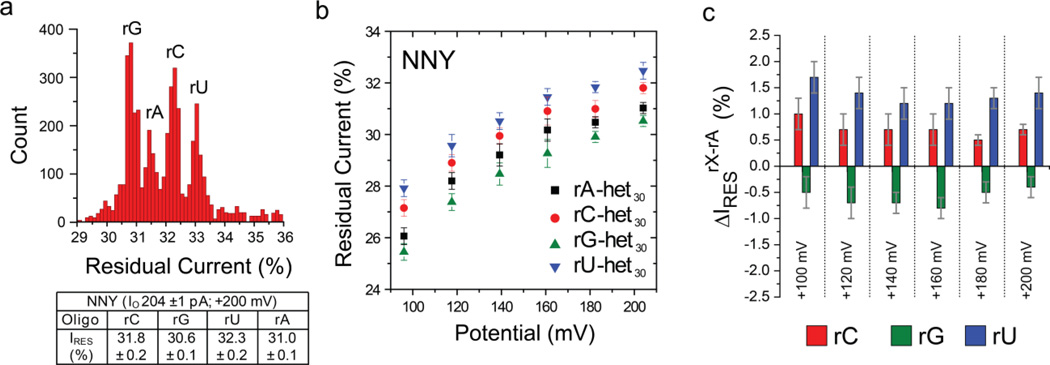
Figure 5. Discrimination of individual ribobases in heteropolymeric ssRNA with the NNY αHL pore.
(a) Histogram of the residual currents (IRES%) from a heteropolymeric ssRNA set (sequence 5’-[r]UAGCUAAACCGAUAGCUUCAGXCAUGUAAC[Btn]-3’) examined with the NNY pore. The four 3'-biotinylated RNA strands differed only at position 9 as shown. Experiments were conducted at least 3 times, and the panel displays the results from a typical experiment. Gaussian fits were performed for each peak, and the mean values of the open pore currents (IO, pA) and the normalized residual currents (IRES%) are displayed in the table below the histogram. (b) Plots of IRES% versus the applied potential for the bases at position 9: rG, rA, rC and rU. (c) Plots of residual current difference, ΔIRES%, for each base. ΔIRES% rX-rA = IRES% of the rX-het30 oligonucleotide - IRES% of rA-het30 oligonucleotide. The errors given are standard deviations from three independent experiments (Table S7).
Conclusions
We have shown that individual RNA bases can be identified in immobilized DNA and RNA strands. By using the αHL NNY mutant pore, which has superior nucleobase discrimination properties, we were able to distinguish between the standard bases (rG, rA, rC, and rU) and the modified bases rI, m6A and m5C. The ultra-rapid sequencing of RNA will ultimately require an active process to control movement of the nucleic acid through the pore14, 15, 17, 19, 20, 51. Hence, it will be necessary to combine αHL or an alternative protein nanopore with a processive RNA translocating enzyme, such as a exoribonuclease52 or a reverse transcriptase53, to ratchet RNA through the pore at a speed at which base identification is feasible. For a functional device, a ~10 ms measurement per base is feasible, and would reduce the sequencing time for a human genome to less than a day with a 104-pore array device. The data acquisition rate and signal filtering required for a 10 ms measurement time would allow the separation of the currents levels seen with NN and NNY in the present work. Additionally, integration of the protein nanopore with a processing enzyme will support the unfolding of translocating RNA molecules, thereby overcoming a major concern in the analysis of native RNA, which can form complex structures mediated by the base-pairing54, 55. For high throughput sequencing, thousands of pores must be incorporated into arrays and viable approaches towards this end are under development1, 56.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by grants from the National Institutes of Health and Oxford Nanopore Technologies. The authors thank E. Mikhailova for the preparation of proteins, and D. Stoddart and A. Heron for useful discussions.
Footnotes
ASSOCIATED CONTENT
Supporting Information. Details of experimental procedures, oligonucleotide sequences, and the data displayed in Figures 1–5. This material is available free of charge via the Internet at http://pubs.acs.org.
Hagan Bayley is the Founder, a Director and a share-holder of Oxford Nanopore Technologies, a company engaged in the development of nanopore sequencing technology.
REFERENCES
- 1.Dekker C. Nat Nanotechnol. 2007;2(4):209–215. doi: 10.1038/nnano.2007.27. [DOI] [PubMed] [Google Scholar]
- 2.Bayley H, Cremer PS. Nature. 2001;413(6852):226–230. doi: 10.1038/35093038. [DOI] [PubMed] [Google Scholar]
- 3.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Proc Natl Acad Sci U S A. 1996;93(24):13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashkenasy N, Sanchez-Quesada J, Bayley H, Ghadiri MR. Angew Chem Int Ed Engl. 2005;44(9):1401–1404. doi: 10.1002/anie.200462114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stoddart D, Heron AJ, Mikhailova E, Maglia G, Bayley H. Proc Natl Acad Sci U S A. 2009;106(19):7702–7707. doi: 10.1073/pnas.0901054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Purnell RF, Schmidt JJ. ACS Nano. 2009;3(9):2533–2538. doi: 10.1021/nn900441x. [DOI] [PubMed] [Google Scholar]
- 7.Stoddart D, Maglia G, Mikhailova E, Heron AJ, Bayley H. Angew Chem Int Ed Engl. 2010;49(3):556–559. doi: 10.1002/anie.200905483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stoddart D, Heron AJ, Klingelhoefer J, Mikhailova E, Maglia G, Bayley H. Nano Lett. 2010;10(9):3633–3637. doi: 10.1021/nl101955a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wallace EV, Stoddart D, Heron AJ, Mikhailova E, Maglia G, Donohoe TJ, Bayley H. Chem Commun (Camb) 2010;46(43):8195–8197. doi: 10.1039/c0cc02864a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manrao EA, Derrington IM, Pavlenok M, Niederweis M, Gundlach JH. PLoS One. 2011;6(10):e25723. doi: 10.1371/journal.pone.0025723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schibel AEP, An N, Jin Q, Fleming AM, Burrows CJ, White HS. J Am Chem Soc. 2010;132(51):17992–17995. doi: 10.1021/ja109501x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.An N, Fleming AM, White HS, Burrows CJ. Proceedings of the National Academy of Sciences. 2012;109(29):11504–11509. doi: 10.1073/pnas.1201669109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meller A, Nivon L, Brandin E, Golovchenko J, Branton D. Proc Natl Acad Sci U S A. 2000;97(3):1079–1084. doi: 10.1073/pnas.97.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cockroft SL, Chu J, Amorin M, Ghadiri MR. J Am Chem Soc. 2008;130(3):818–820. doi: 10.1021/ja077082c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lieberman KR, Cherf GM, Doody MJ, Olasagasti F, Kolodji Y, Akeson M. J Am Chem Soc. 2010;132(50):17961–17972. doi: 10.1021/ja1087612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olasagasti F, Lieberman KR, Benner S, Cherf GM, Dahl JM, Deamer DW, Akeson M. Nature Nanotechnology. 2010;5(11):798–806. doi: 10.1038/nnano.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chu J, Gonzalez-Lopez M, Cockroft SL, Amorin M, Ghadiri MR. Angew Chem Int Ed Engl. 2010;49(52):10106–10109. doi: 10.1002/anie.201005460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayden EC. Nature. 2012 [Google Scholar]
- 19.Cherf GM, Lieberman KR, Rashid H, Lam CE, Karplus K, Akeson M. Nat Biotechnol. 2012;30(4):344–348. doi: 10.1038/nbt.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manrao EA, Derrington IM, Laszlo AH, Langford KW, Hopper MK, Gillgren N, Pavlenok M, Niederweis M, Gundlach JH. Nat Biotechnol. 2012;30(4):349–353. doi: 10.1038/nbt.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nilsen TW, Graveley BR. Nature. 2010;463(7280):457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ozsolak F, Milos PM. Nat Rev Genet. 2011;12(2):87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pritchard CC, Cheng HH, Tewari M. Nat Rev Genet. 2012;13(5):358–369. doi: 10.1038/nrg3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitelman F, Johansson B, Mertens F. Nat Rev Cancer. 2007;7(4):233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 25.Maher CA, Kumar-Sinha C, Cao X, Kalyana-Sundaram S, Han B, Jing X, Sam L, Barrette T, Palanisamy N, Chinnaiyan AM. Nature. 2009;458(7234):97–101. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Metzker ML. Nat Rev Genet. 2010;11(1):31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 27.Wang Z, Gerstein M, Snyder M. Nat Rev Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghildiyal M, Zamore PD. Nat Rev Genet. 2009;10(2):94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davidson BL, McCray PB., Jr Nat Rev Genet. 2011;12(5):329–340. doi: 10.1038/nrg2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matzke M, Matzke AJ, Kooter JM. Science. 2001;293(5532):1080–1083. doi: 10.1126/science.1063051. [DOI] [PubMed] [Google Scholar]
- 31.He C. Nat Chem Biol. 2010;6(12):863–865. doi: 10.1038/nchembio.482. [DOI] [PubMed] [Google Scholar]
- 32.Akeson M, Branton D, Kasianowicz JJ, Brandin E, Deamer DW. Biophys J. 1999;77(6):3227–3233. doi: 10.1016/S0006-3495(99)77153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Butler TZ, Gundlach JH, Troll MA. Biophys J. 2006;90(1):190–199. doi: 10.1529/biophysj.105.068957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Butler TZ, Gundlach JH, Troll M. Biophys J. 2007;93(9):3229–3240. doi: 10.1529/biophysj.107.107003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin J, Kolomeisky A, Meller A. Phys Rev Lett. 2010;104(15):158101. doi: 10.1103/PhysRevLett.104.158101. [DOI] [PubMed] [Google Scholar]
- 36.Maglia G, Restrepo MR, Mikhailova E, Bayley H. Proc Natl Acad Sci U S A. 2008;105(50):19720–19725. doi: 10.1073/pnas.0808296105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rincon-Restrepo M, Mikhailova E, Bayley H, Maglia G. Nano Lett. 2011;11(2):746–750. doi: 10.1021/nl1038874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seol Y, Skinner GM, Visscher K, Buhot A, Halperin A. Physical Review Letters. 2007;98(15):158103. doi: 10.1103/PhysRevLett.98.158103. [DOI] [PubMed] [Google Scholar]
- 39.Seol Y, Skinner GM, Visscher K. Physical Review Letters. 2004;93(11):118102. doi: 10.1103/PhysRevLett.93.118102. [DOI] [PubMed] [Google Scholar]
- 40.Sauer-Budge AF, Nyamwanda JA, Lubensky DK, Branton D. Physical Review Letters. 2003;90(23):238101. doi: 10.1103/PhysRevLett.90.238101. [DOI] [PubMed] [Google Scholar]
- 41.Mathé J, Visram H, Viasnoff V, Rabin Y, Meller A. Biophys J. 2004;87(5):3205–3212. doi: 10.1529/biophysj.104.047274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keyser UF, Koeleman BN, Van Dorp S, Krapf D, Smeets RMM, Lemay SG, Dekker NH, Dekker C. Nature Physics. 2006;2(7):473–477. [Google Scholar]
- 43.Adler BA, Grossman L, Fasman GD. Proc Natl Acad Sci U S A. 1967;57(2):423–430. doi: 10.1073/pnas.57.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jones S, Daley DT, Luscombe NM, Berman HM, Thornton JM. Nucleic Acids Res. 2001;29(4):943–954. doi: 10.1093/nar/29.4.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mackay JP, Font J, Segal DJ. Nat Struct Mol Biol. 2011;18(3):256–261. doi: 10.1038/nsmb.2005. [DOI] [PubMed] [Google Scholar]
- 46.Gallo A, Galardi S. RNA Biol. 2008;5(3):135–139. doi: 10.4161/rna.5.3.6739. [DOI] [PubMed] [Google Scholar]
- 47.Kawahara Y, Zinshteyn B, Sethupathy P, Iizasa H, Hatzigeorgiou AG, Nishikura K. Science. 2007;315(5815):1137–1140. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bass BL. Annu Rev Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zuker M. Nucleic Acids Res. 2003;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ying X, Luo H, Luo J, Li W. Nucleic Acids Res. 2004;32(Web Server issue):W150–W153. doi: 10.1093/nar/gkh445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Olasagasti F, Lieberman KR, Benner S, Cherf GM, Dahl JM, Deamer DW, Akeson M. Nat Nanotechnol. 2010;5(11):798–806. doi: 10.1038/nnano.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raines RT. Chem Rev. 1998;98(3):1045–1065. doi: 10.1021/cr960427h. [DOI] [PubMed] [Google Scholar]
- 53.Kohlstaedt LA, Wang J, Friedman JM, Rice PA, Steitz TA. Science. 1992;256(5065):1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 54.Berman HM, Olson WK, Beveridge DL, Westbrook J, Gelbin A, Demeny T, Hsieh SH, Srinivasan AR, Schneider B. Biophys J. 1992;63(3):751–759. doi: 10.1016/S0006-3495(92)81649-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Andronescu M, Bereg V, Hoos HH, Condon A. Bmc Bioinformatics. 2008;9 doi: 10.1186/1471-2105-9-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hall AR, Scott A, Rotem D, Mehta KK, Bayley H, Dekker C. Nat Nanotechnol. 2010;5(12):874–877. doi: 10.1038/nnano.2010.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.