Abstract
Anopheles triannulatus s.l. is a species complex, however in Colombia its taxonomic status is unclear. This study was conducted to understand the level of genetic differentiation or population structure of specimens of An. triannulatus s.l. from northwestern and southeastern Colombia. Cytochrome oxidase subunit I (COI) and internal transcribed spacer (ITS2) sequence analyses suggested high genetic differentiation between the NW and SE populations. A TCS network and Bayesian inference analysis based on 814 bp of COI showed two main groups: group I included samples from the NW and group II samples from the SE. Two main ITS2-polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) patterns were found. Pattern I is present in both the NW and SE, and pattern II is found in the SE specimens. To further elucidate the taxonomic status of An. triannulatus s.l. in Colombia and how these COI lineages are related to the Triannulatus Complex species, the evaluation of immature stages, male genitalia, and additional mitochondrial and nuclear markers will be needed.
Introduction
Anopheles triannulatus s.l. Neiva and Pinto (1922) of the Oswaldoi subgroup is widely distributed in Central America and east of the Andes in South America,1 and in 2005 it was also reported from Trinidad.2 This species complex is considered a local or secondary vector in Brazil,3–5 an important vector in Loreto Department, Peru6; it was implicated as the possible vector during a malaria epidemic in Venezuela,7 and samples from San Carlos, Cojedes were discovered with natural Plasmodium oocysts.8
The first evidence for the existence of a species complex within An. triannulatus s.l. came from the observation of morphologic variations of male genitalia, eggs, and larvae.9 Based on morphology, allozyme, and random amplified polymorphic DNA, three species were proposed: Anopheles triannulatus s.s., Anopheles halophylus, and Anopheles triannulatus C.10,11 In Latin America, several population studies have been conducted using various markers in an attempt to establish the taxonomic status of An. triannulatus s.l.; for example, isozymes separated four Brazilian An. triannulatus populations into two groups, with Macapa-Amapa state in one group and Janauri Lake Manaus-Amazonas state, Ji-Paraná-Rondonia state and Aripuanã-Mato Grosso state in the other; the estimated low gene flow probably influenced the sub-population structure.12 Cytochrome oxidase subunit I (COI) gene sequence analyses of specimens of An. triannulatus from northeast of the Amazon river, central and southern Brazil showed five groups with a significant signal of population expansion, suggesting that these populations were not at equilibrium.13 Additional analyses of partial fragments of both timeless and cpr genes of specimens of the Triannulatus Complex to assess their genetic variability and divergence showed partial separation between An. halophylus and An. triannulatus C with timeless, whereas cpr sequences showed a clear separation among the three species of this complex.14 In Colombia, a study reported polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) patterns for the internal transcribed spacer (ITS2) in seven species collected in a locality in the northwestern (NW) region of the country; among them, An. triannulatus s.l. presented a distinctive pattern15 that was congruent with An. triannulatus s.l. specimens from other NW localities.16 These results suggested that the PCR-RFLP-ITS2 assay could be used to differentiate An. triannulatus s.l. specimens from the other Anopheles species included in the test, especially among those in the Oswaldoi Group.
Considering that An. triannulatus s.l. constitutes a complex of species that may differ in their ability to transmit Plasmodium spp., and in Colombia this species is widely distributed displaying anthropophilic and zoophilic behavior,17–20 the aim of this study was to assess the genetic variability of An. triannulatus s.l. specimens from the NW and SE regions. Mitochondrial DNA (mtDNA) COI sequences were used to address the following questions: 1) What is the level of genetic differentiation or population structure among An. triannulatus s.l. populations from NW and SE Colombia? 2) Is there more than one An. triannulatus COI lineage in these two regions? 3) What is the phylogenetic relatedness between An. triannulatus s.l. lineages from Colombia and Brazil? Additionally, in this study the nuclear ITS2 marker was used to confirm the taxonomic classification of the An. triannulatus s.l. specimens and to determine whether this marker supported the COI results.
Materials And Methods
Mosquito collection and processing.
Adult female An. triannulatus s.l. were collected from January 2009 to March 2011 in six localities, three in NW Colombia: El Bagre-BAG (07°35′39″N, 74°49′42″W) and San Pedro de Uraba-SPU (8°16′60″N, 76°22′60″W) in Antioquia, and Puerto Libertador-PLT (07°43′36″N, 75°51′16″W) in Cordoba Department. Furthermore, in this study were included previously collected and identified specimens of An. triannulatus s.l. from Santa Rosa de Lima-SRL, Bolivar Department and Zaragoza-ZAR, Antioquia Department.18,19 The three localities in the SE were: Leticia-LET (4°12′54″S, 69°55′58″W), Puerto Nariño-PNA (3°46′24″S, 70°22′55″W), and Tarapaca-TAR (2°53′44″S, 69°45′29″W) in Amazonas Department (Figure 1). Specimens were collected using human landing catches under a protocol, along with informed consent forms, approved by the Bioethics Committee for Human Research (Sede de Investigación Universitaria-SIU) of University of Antioquia. In SPU and PLT some specimens were also collected resting on animals such as dogs or cattle. Anopheles triannulatus s.l. mosquitoes were identified based on morphological features.21 For each specimen, a photographic record was obtained, and one hind leg and two wings were removed and fixed on a glass slide with Euparal mountant and conserved as entomological support of the Anopheles species collection of the Molecular Microbiology Group, University of Antioquia. The DNA was extracted from individual mosquito abdomens using a salt precipitation protocol.22
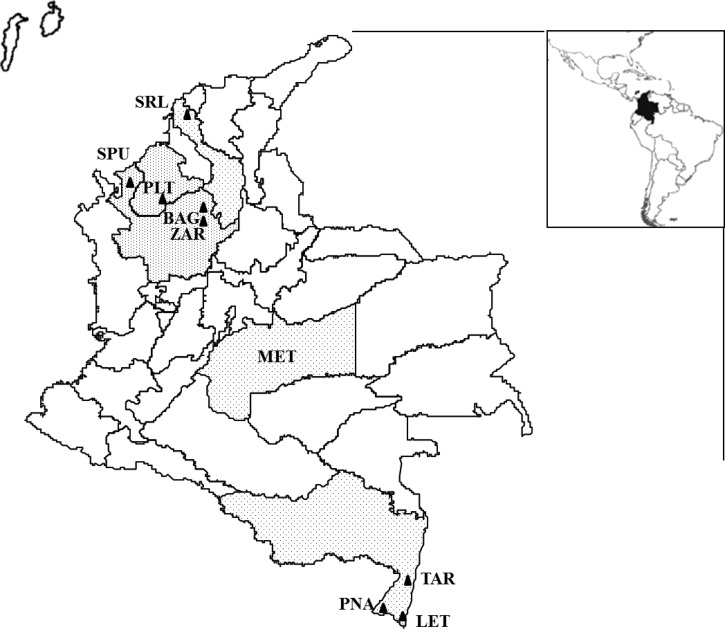
Figure 1.
Distribution of collection localities for Anopheles triannulatus s.l. NW region: El Bagre-BAG, Zaragoza-ZAR, and San Pedro de Uraba-SPU in Antioquia Department, Puerto Libertador-PLT in Córdoba, and Santa Rosa de Lima-SRL in Bolívar. SE region: Leticia-LET, Tarapaca-TAR and Puerto Nariño-PNA in Amazonas Department. The Cytochrome oxidase subunit I (COI) sequences from Meta-MET Department obtained in GenBank were included in the analyses.
ITS2 analyses.
All specimens used in subsequent analyses were confirmed as An. triannulatus s.l. by a PCR-RFLP-ITS2 assay.15,16 Furthermore, an in silico AluI restriction enzyme analysis using NEBcutter software23 was performed using 10 ITS2 sequences obtained in this work representative of NW and SE specimens, three from Meta-MET Department available in GenBank (accession nos. HM022422, HM022423, and HM022426) and one sequence from Brazil (accession no. AF462377).24 This procedure allowed PCR-RFLP-ITS2 pattern prediction and comparison with laboratory results. The PCR-RFLP-ITS2 band sizes were calculated with FragSize 1.0.3 software.25 Twenty ITS2 PCR products that included the two RFLP patterns obtained were cloned into CloneJET PCR Cloning kit (Fermentas, St. Leon, Germany) or pGEM-T easy vector plasmid (Promega, Madison, WI) and used to transform Escherichia coli strain DH5α, following the manufacturer's instructions. To evaluate intra-individual ITS2 variability, three to five clones from each specimen were sequenced and the obtained sequences were edited with Geneious 5.2 software.26 All consensuses of the forward and reverse sequences were aligned with the MUSCLE algorithm27 in Geneious. The ITS2 was annotated for each individual sequence/clone following parameters available in the ITS2 database28 and the guanine-cytosine content evaluated in Geneious.
MtDNA COI amplification and scheme to rule out co-amplification of numts.
For the COI analysis, a 1,300 bp fragment was amplified using primers UEA3 and UEA10.29 The PCR was performed in a 25-μL reaction volume using 4 uL of DNA and PCR conditions and cycles as previously described.30 The PCR products were purified using the QIAquick PCR Purification Kit (QIAGEN, Düsseldorf, Germany) or ExoSAP-IT (USB Corporation, Cleveland, OH). All fragments were sequenced in both directions. Sequences were visually edited and the consensuses of the forward and reverse sequences were aligned with MUSCLE in Geneious26 and imported into programs for sequence analyses. After editing, sequences were again visually reviewed to check for base calls and confirm variations in nucleotide positions. To rule out for the presence of numts, a MegaBLAST was performed with each sequence to search for identity to sequences available in the databases. The PCR products and chromatograms were carefully observed to detect the potential presences of ghost bands or ambiguities, respectively. Finally, translation of nucleotides to amino acids was carried out to detect stop codons.31–33
MtDNA COI phylogenetic analysis.
Phylogenetic relatedness and haplotype network analyses were performed in two ways. One used the 814-bp sequences obtained, located at positions 1,900–2,714 on the Anopheles darlingi mitochondrial genome RefSeq GQ918272,34 to understand the phylogenetic relationships with three An. triannulatus s.s. COI sequences from Meta Department35 available in GenBank (accession nos. HM022387, HM022388, and HM022389) and one An. triannulatus s.l. sequence from Mato Grosso, Salobra, Brazil36 (accession no. AF417702). A second analysis was performed to include 50 shorter An. triannulatus s.l. COI sequences from Brazil available in GenBank (accession no. GU445849-GU445898). A 337 bp overlapping region located at positions 2,377–2,714 on the An. darlingi mitochondrial genome RefSeq GQ918272,34 was used to analyze the phylogenetic relationships of the Colombian and Brazilian COI sequences. Anopheles albimanus (FJ015203) and Anopheles nuneztovari s.l. (Naranjo and others, unpublished data) COI sequences were included as outgroups. A statistical parsimony-based analysis that calculates the maximum number of mutational connections between pairs of COI sequences with a 95% parsimony criterion was conducted in TCS 1.21 software.37 This method accepts the existence of ancestral haplotypes, which according to the coalescence theory would be the most frequent in a population.38 To resolve some ambiguous loops in the networks resulting from 814 and 337 bp sequences, several recommendations were followed.38 For example, if one haplotype was connected to various geographically separated groups, the haplotype was assigned to the group showing the highest frequency of sequences from the same locality. This is because a singleton is more likely to be connected to haplotypes from the same population than to haplotypes from different ones.38 Additionally, a median-joining haplotype network run in Network version 4.6.1.039,40 and a neighbor-net network in SplitsTree4 version 4.1041,42 were estimated. The networks were compared with the Bayesian inference (BI) analysis43 performed with the COI sequences and also with the concatenated (COI + ITS2) dataset using the Geneious software and for both datasets the model of molecular evolution of Hasegawa-Kishino-Yano, 85 (HKY85) was run using Modeltest 0.1.1 software.44 For the BI analysis the Markov Chain Monte Carlo algorithm was allowed to run for 20,000,000 generations with a sampling every 1,000 generations after a burn-in of 5,000,000 generations (50,000 trees).
Indices of genetic variation and population structure.
Intrapopulation (intra-localities) and interpopulation (inter-localities) genetic diversity was estimated in DnaSP 5.045 and Arlequin 3.11 software,46 taking into account the two possible mitochondrial lineages defined by the previous COI and concatenated (COI + ITS2) BI analyses and COI haplotype network. The three An. triannulatus s.s. sequences from MET were excluded because the small sample size would not produce reliable results. The following estimates of variability were calculated: number of mtDNA haplotypes, nucleotide diversity (π), haplotype diversity (h), and number of polymorphic sites (S). The number of migrants per generation (Nm) among localities and the level of genetic differentiation between the NW and SE lineages measured by the fixation index FST47 were calculated in Arlequin and the significance was estimated by permutation tests (10,000 replicates). Analysis of molecular variance (AMOVA) was used to examine variation within, between and among collection sites for each possible lineage. The statistical significance for AMOVA was evaluated using > 10,000 permutations. Uncorrected pairwise genetic distances were calculated between and within COI lineages in MEGA software version 5.0.48
Neutrality tests and parameters of population expansion.
To measure deviations from the null hypothesis of constant population size and random mating, neutrality tests of COI sequences were conducted in DnaSP and Arlequin software. Fu's Fs values49 and Tajima's D50 were estimated by comparing the differences between the number of segregating sites and the average number of nucleotide differences. Fu and Li's D and F statistics51 were used to compare estimates of θ based on mutations in internal and external branches of a genealogy. Fu's Fs values49 were used to assess haplotype structure on the basis of haplotype frequency distribution. Mismatch distribution, a frequency distribution of the observed number of pairwise sequence differences, was used to differentiate between a smooth unimodal distribution and a ragged or multimodal distribution.52,53 The shape of the distribution is highly informative and helps to determine whether a population is in expansion or equilibrium. The raggedness statistic (r) to quantify the smoothness of the mismatch distribution54 was calculated in DnaSP. To evaluate the signal of population expansion, three parameters were calculated, τ (expansion age), θ0 and θ1 (before and after expansion, respectively) with Arlequin. Time since the population expansion was estimated from t = τ/2 μ, where τ (tau) is the date of the growth or decline measured in units of mutational time (τ = 2 μt; t is the time in generations and μ is the mutation rate per site [sequence size] and per generation).55
Results
ITS2 analyses.
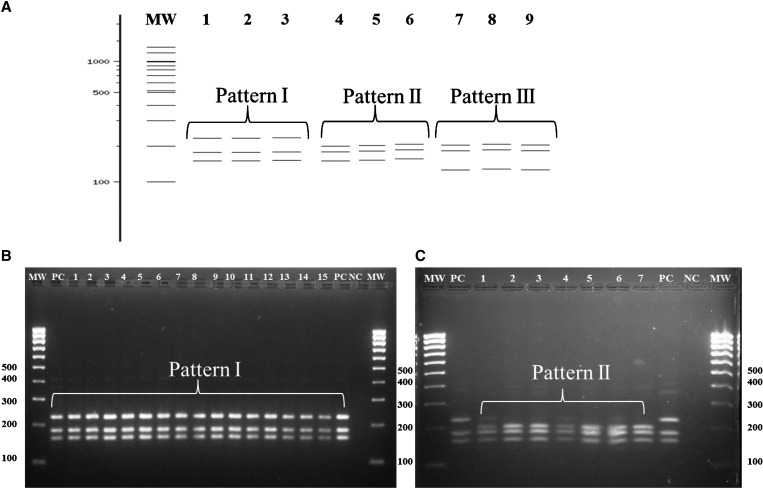
Specimens identified as An. triannulatus s.l. was confirmed as such by PCR-RFLP-ITS2. From the ITS2 products cloned from five specimens from LET, six from PNA, one from TAR, four from PLT, three from SPU, and one from BAG, three to five clones were obtained per specimen (Table 1). In silico RFLP-ITS2 analysis showed three patterns. Patterns I and II were observed in sequences obtained in this work (accession nos. JX852283–JX852320) and pattern III corresponded to ITS2 sequences downloaded from GenBank of An. triannulatus s.s. from MET (Figure 2A). When performing the PCR-RFLP-ITS2 in the laboratory the in silico results were confirmed (Figure 2B and C). Pattern I is the same as previously described for An. triannulatus s.l. from NW Colombia,15,16 with three bands (226, 180, and 156 bp), and was present in both the NW and SE regions (Figure 2, Table 1). Pattern II of 198, 180, and 155 bp, was found in specimens of the three SE localities (Figure 2, Table 1), however, one specimen from TAR showed a pattern of 218, 177, and 155 bp that, when compared with the larger NW sequences, reveled several deletions and one nucleotide insertion (Table 1). In general, the small differences in ITS2 patterns are caused by the presence of indels of distinct lengths and substitutions. Specifically, a deletion of 24 bp present in the sequences with pattern II is the main cause of its differentiation from pattern I (Table 1). In addition, the pattern I sequences were similar to the only ITS2 sequence reported in GenBank for An. triannulatus s.l. specimens from Rondonia State, Brazil.24 The ITS2 GC contents varied from 57.4% in pattern I sequences to 57.8% in those of pattern II; these values are similar to previously reports (58.2%) for the specimens from Rondonia State.24 There was heterogeneity in ITS2 sizes among specimens. The analysis in the ITS2 database28 showed that the ITS2 lengths were 434 bp with pattern I, 405 bp with pattern II, and 422 bp for the TAR specimens. Intra-individual variations were detected in An. triannulatus s.l. clones from SPU (2), PLT (3), LET (4), and PNA (8) and were mainly caused by substitutions, whereas insertions and deletions were observed at the interpopulation level (Table 1).
Table 1.
ITS2 PCR-RFLP pattern allocation and sequence features for Anopheles triannulatus s.l. from northwestern and southeastern Colombia
| Department locality (abbreviation) | RFLP pattern | Specimen code-clone substitution, deletion, or insertion (base position) | |
|---|---|---|---|
| I Specimens/no. cloned/sequences obtained | II Specimens/no. cloned/sequences obtained | ||
| Antioquia | |||
| El Bagre (BAG) | 81/1/5 | NA | |
| San Pedro de Urabá (SPU) | 30/3/9 | NA | Substitutions: SPU726-3: G/A (286), SPU708-3: G/A (286) |
| Córdoba | |||
| Puerto Libertador (PLT) | 71/4/13 | NA | Substitutions: PLT3657-1: A/G (203, 286), PL3756-5: T/C (57), PLT3687-5: C/T (159) |
| Total northwestern | 182/8/27 | NA | TGTGCGATAGTCCACACACGCACA (306-329)* |
| Amazonas | |||
| Leticia (LET) | 9/1/5 | 2/4/15 | Substitutions: LET26-2: G/A (32), C/T (161), LET62-5: G/A (83), LET13-5: T/C (285) |
| Puerto Nariño (PNA) | 2/1/4 | 63/5/20 | Substitutions: PNA15-2: A/G (136), PNA122-3: G/A (402), PNA156-4: C/T (192), PNA 122-1: A/G (372), PNA39-1: T/C (213), PNA20-4: A/T (130), PNA20-1: T/C (285) |
| Tarapacá (TAR) | 10/1/3 | 91/0/0 | Substitutions: TAR337: A/G (154), TAR337-1: C/T (275), Insertion: C (346). Deletions: T (37), GAC (241–243), TTGGA (278–282), GAGA (414–417) |
| Total southeastern | 21/3/12 | 226/9/35 | Deletion in ITS2 sequences showing pattern II: GTGCGATAGTCCACACACGCACA (306-329) |
Sequence present in all specimens with pattern I and not observed in those with pattern II.
ITS2 = internal transcribed spacer; PCR-RFLP = polymerase chain reaction-restriction fragment length polymorphism; NA = does not apply.
Figure 2.
Polymerase chain reaction-restriction fragment length polymorphism-internal transcribed spacer (PCR-RFLP-ITS2) patterns corresponding to Anopheles triannulatus s.l. from Colombia. (A) Scheme representing in silico PCR-RFLP-ITS2 patterns. Lanes: 1–3 Pattern I (226, 180, and 156 bp), 4–6 Pattern II (198, 180, and 155 bp), 7–9 Pattern III (198, 180, and 126 bp). (B and C) 2% agarose gel showing ITS2-PCR-RFLP patterns obtained from An. triannulatus s.l. specimens from NW and SE Colombia. (B) Lanes: 1–15 Pattern I. (C) Lanes: 1–7 Pattern II. MW = Molecular weight; PC = Positive control-An. triannulatus s.l. ITS2 clone; NC = RFLP negative control.
Genetic diversity and numts analysis.
An 814 bp COI sequence was analyzed for 286 An. triannulatus s.l. specimens from NW and SE localities yielding 141 haplotypes (accession nos. JX852142–JX852282) (Table 2). No insertion/deletion events were observed. The adenine and thymine content (A + T) was 71.2%, similar to other Neotropical anophelines such as An. nuneztovari s.l. (70.2%)56 and An. albimanus (69.2%)30 and (70.0%).57 For the diversity measurements, COI sequences from ZAR and BAG were analyzed together because these sites are only 11 km apart and the Nm estimate detected infinite migrants between them. Mutations were all transitions or transversions, resulting in 115 synonymous changes and four amino acid changes. There were 135 polymorphic sites. The h and π values were very similar for all NW and SE populations, h values were high and ranged from 0.972 to 0.976, and π values were 0.0513 and 0.0518, in the NW and SE, respectively. Intra-population diversity varied in the different localities but was higher in SPU and TAR (Table 2). The methodology implemented to rule out the co-amplification of numts showed no double peaks in the chromatograms and no ghost bands in the electrophoresis gels. The MegaBLAST showed > 96% identity of the sequences with those of An. triannulatus s.s. from Meta (GenBank accession nos. HM022387, HM022388, and HM022389), and no stop codons were detected during the translation of each sequence into protein with the 814 bp corresponding to 271 amino acids of the COI protein.
Table 2.
COI sequence analyses for Anopheles triannulatus s.l. from northwestern and southeastern Colombia
| Department (state) locality (abbreviation) | Haplotype designation (frequency) | COI sequences analyzed | Genetic polymorphisms | |||
|---|---|---|---|---|---|---|
| S | H | h (SD) | π (SD) | |||
| Antioquia | ||||||
| El Bagre (BAG) | A(2), B(2), C(7), D(7), E(2), G(2), N(1), O(3), P(2), Q(2), R(2), S(2), T(2), CC(1), DD(1) | 51 | 35 | 32 | 0.950 ± 0.015 | 0.0438 ± 0.0240 |
| Zaragoza (ZAR)* | A(2), C(1), D(2), E(1), G(2), DD(1) | 11* | NA | NA | NA | NA |
| San Pedro de Urabá (SPU) | B(3), C(2), E(1), H(2), K(1), N(1), Y(2), Z(2), AA(2), BB(1), CC(1) | 31 | 71 | 23 | 0.987 ± 0.013 | 0.0698 ± 0.0373 |
| Córdoba | ||||||
| Puerto Libertador (PLT) | A(10), B(8), E(4), H(2), I(4), J(3), K(2), L(3), M(3), N(1), U(2), V(2), W(2), X(2), BB(1) | 62 | 31 | 29 | 0.947 ± 0.015 | 0.0488 ± 0.0265 |
| Total northwestern | A(14), B(13), C(10), D(9), E(8), F(6)†, G(5), H(4), I(4), J(3), K(3), L(3), M(3), N(3), O(3), P(2), Q(2), R(2), S(2), T(2), U(2), V(2), W(2), X(2), Y(2), Z(2), AA(2), BB(2), CC(2), DD(2) | 155 | 95 | 67 | 0.972 ± 0.005 | 0.0513 ± 0.0274 |
| Amazonas | ||||||
| Leticia (LET) | EE(8), FF(3), GG(1), HH(3), II(4), JJ(2), KK(1), LL(2), MM(2), NN(1), OO(1), PP(1)-RR(1), SS(1), VV(1) | 55 | 93 | 37 | 0.970 ± 0.013 | 0.0476 ± 0.0259 |
| Puerto Nariño (PNA) | EE(2), FF(6), GG(4), II(1), JJ(1), KK(1), LL(1), MM(1), OO(1), SS(1), TT(2), UU(2), VV(1) | 42 | 34 | 30 | 0.970 ± 0.015 | 0.0395 ± 0.0221 |
| Tarapacá1 (TAR) | EE(3), FF(2), GG(2), HH(2), JJ(1), KK(2), NN(1), PP(1), QQ(1)-RR(1) | 34 | 52 | 28 | 0.990 ± 0.001 | 0.0694 ± 0.0340 |
| Total southeastern | EE(13), FF(11), GG(7), HH(5), II(5), JJ(4), KK(4), LL(3), MM(3), NN(2), OO(2), PP(2), QQ(2)-RR(2), SS(2), TT(2), UU(2), VV(2) | 131 | 116 | 74 | 0.976 ± 0.006 | 0.0518 ± 0.0277 |
ZAR COI sequences (11) were analyzed together with those from BAG.
Haplotypes corresponding to specimens from Santa Rosa de Lima (SRL) in Bolivar were not included in the genetic diversity analyses caused by the low number of specimens available.
S = number of segregating sites, H = haplotype number, h = haplotype diversity, π = nucleotide diversity; NA = not apply.
Phylogenetic analysis.
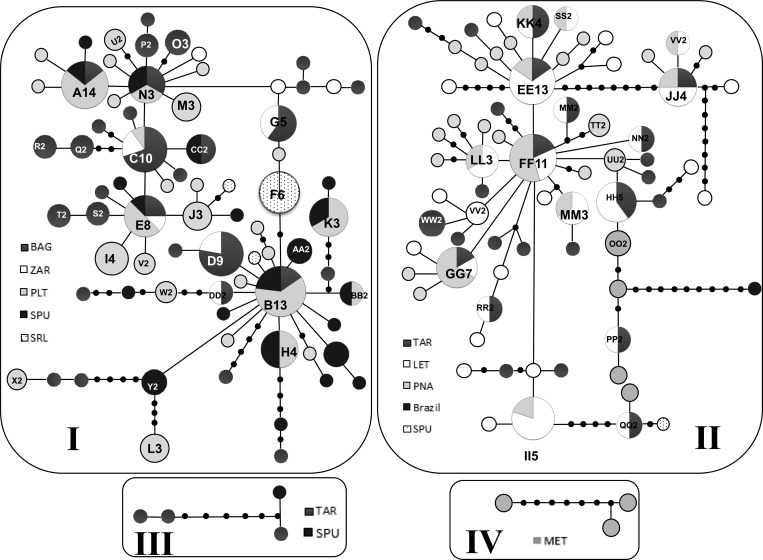
The two networks, based on 814 bp (Figure 3) and on 337 bp (not shown), were star shaped with many singletons at the tips. These tip alleles are considered to be more recently derived and geographically restricted suggesting a demographic expansion.38,58 No shared haplotypes between the two regions/clades were observed in these networks (Figure 3).
Figure 3.
Parsimony-based haplotype network of 141 Cytochrome oxidase subunit I (COI) haplotypes (814 bp) representing 286 Anopheles triannulatus s.l. specimens from NW and SE Colombia. Each circle represents a haplotype and the color and letters depict the origin, and numbers the frequency of each haplotype. The smallest circles denote unique haplotypes. Each black circle represents one mutational step. Roman numbers represent the region or location: I. NW, II. SE, III. TAR-SE (TAR337, TAR338, and TAR349) and SPU-NW haplotype (SPU718) and IV. MET (MG40-3 and MGMV05-6).
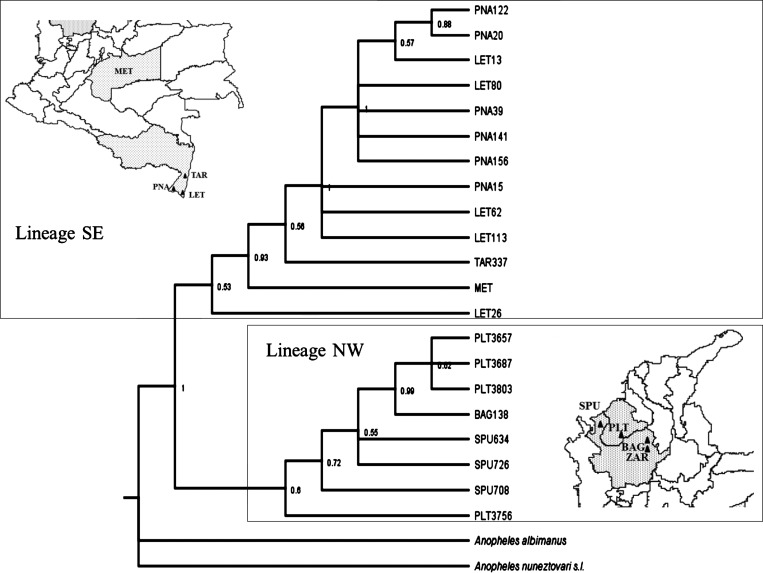
The TCS, Network, and SplitsTree4 networks and Bayesian performed with the 814 bp COI sequences showed four equivalent separate clades (Figure 3 and Supplemental file). Clade I included most NW haplotypes, clade II contained SE haplotypes and one An. triannulatus s.l. sequence from Mato Grosso, Salobra, Brazil (the only one available in GenBank with the appropriate size to allow comparison with sequences from this work), although separated from the SE sequences by 10 mutational steps; clade III contained three TAR (SE) and one SPU (NW) haplotypes and clade IV included the three An. triannulatus s.s. haplotypes from MET (Figure 3). Haplotypes within each clade differed by one or more mutational steps. The most frequent and ancestral haplotype detected in the NW clade contained 14 sequences, 10 from PLT, and two each from BAG and SPU. Of the eight sequences obtained from SRL specimens, six formed a single haplotype (F6) (Figure 3) and two constituted single haplotypes. For the SE clade the most ancestral haplotype was detected in LET (N = 8). There were also multiple haplotypes from PNA (N = 3) and TAR (N = 2) (Figure 3). In the BI analyses based on the 814 bp COI sequences, the same four groups that corresponded to clades I-IV of the TCS network were detected; however, the polytomies present did not allow the resolution of intra-group phylogenetic relationships (data not shown). Results of the concatenated (COI + ITS2) Bayesian analysis showed two monophyletic groups, both with fairly modest levels of support (0.60 and 0.53 for NW and SE lineages, respectively) (Figure 4 ). The MET and TAR sequences that correspond to Clades III and IV of the network (Figure 3) grouped together with the SE sequences. As in the previous results, the concatenated BI analysis also indicated the presence of two distinct An. triannulatus lineages.
Figure 4.
Bayesian topology generated under the molecular evolution model HKY, employing the combined Cytochrome oxidase subunit I + internal transcribed spacer (COI + ITS2) sequence dataset for Anopheles triannulatus s.l. from NW and SE Colombia. Three COI + ITS2 sequences from MET were included. Anopheles nuneztovari s.l. and Anopheles albimanus were used as outgroups. Numbers in the branches represent posterior probabilities.
In the alignment of the 814 bp COI sequences of this study with the 475 bp COI sequences from Brazil,13 a 337 bp overlapping region was obtained. The haplotype network built with these sequences showed all haplotypes connected in two clades because with the shorter sequences several haplotypes collapsed together. Clade I included two major subclades, one contained mainly sequences from SE and the other sequences from Brazil, and within this, the three TAR (SE), one SPU (NW) haplotype, and the three MET-Colombian haplotypes were included. Clade II contained exclusively NW haplotypes (not shown). However, the BI analysis with the 337 bp sequences lacked resolution and unlike the TCS network, only one group was detected containing all haplotypes mixed together (not shown).
Population genetic differentiation and neutrality tests.
The FST values greater than 0.739 between the NW and SE populations indicated high genetic differentiation and subdivision into two groups. The highest FST value was observed between PNA and BAG/ZAR (0.791) and the lowest, between PNA and LET (0.005). The Nm values were higher among populations within each lineage, ∞ to 28.8 in NW and ∞ to 86.2 in SE, and lower when comparing the two lineages (0.18), indicating high gene flow within NW and SE populations but restricted between the two regions. The AMOVA based on COI haplotype frequencies revealed 73.5% of the total variance was among populations and 25.9% within populations. The COI uncorrected pairwise genetic distances between lineages ranged from 2.3% to 3.3%. The within lineage distances were < 1.5% for the NW and < 2% for the SE.
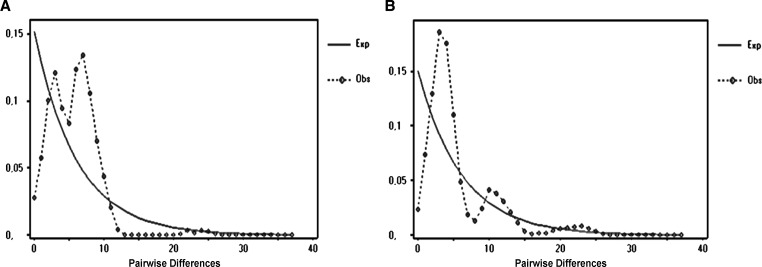
The τ estimates and the current and historical population parameters θ implied a historical expansion episode for all populations. There were τ-values greater than zero and a high difference between the initial and final effective population sizes found, with θ0 = 0.000 and θ1 = 17.446 for the NW lineage and θ0 = 0.068 and θ1 = 99999.000 for the SE lineage. The τ estimates for the NW and SE lineages were in the range of 2.373–7.740, suggesting population expansion.59,60 Expansions were estimated to have started ~36,827 y.a. for the NW and 13,591 y.a. for the SE lineage. These expansions likely occurred during the late Pleistocene Epoch.60–62 Significant negative values for Fu's Fs and Tajima's D neutrality tests were found for all populations implying population expansion or genetic hitchhiking,49 and a departure from neutral equilibrium. Mismatch distribution analysis for the NW and SE lineages showed a bimodal distribution (Figure 5), that suggests that the populations may have undergone a bottleneck or colonization events as previously found for An. nuneztovari s.l. subclade II-B.56
Figure 5.
Mismatch distribution of Anopheles triannulatus Cytochrome oxidase subunit I (COI) sequences. (A) NW COI sequences. (B) SE COI sequences (r = 0.023; P = 0.08).
Discussion
This study was conducted to understand the level of genetic differentiation or population structure of specimens An. triannulatus s.l. from NW and SE Colombia. The two main PCR-RFLP-ITS2 patterns found for An. triannulatus s.l. specimens were not able to precisely differentiate lineages; however, they are still useful to discriminate An. triannulatus s.l. from other anopheline species. In this work, pattern I was found in the NW and SE specimens, but pattern II was only found in those from SE; however, this pattern does not seem to be exclusive to this region because in previous work with An. triannulatus s.l. from NW Colombia a similar pattern has been observed (Correa M, personal communication). The different patterns are the result of insertions, deletions, or substitutions in the sequences that do not affect the AluI recognition site, therefore, conserving the three band pattern typically observed for An. triannulatus s.l.15,16
By PCR-RFLP-ITS2 or COI analysis, it was not possible to determine whether An. triannulatus s.l. from NW and SE were An. triannulatus s.s., An. halophylus, or An. triannulatus C. However, the COI and COI + ITS2 based BI analyses, haplotype network, and population genetic differentiation analyses indicated a substantial genetic division in two lineages that coincide with their geographical locations. Various criteria for the identification of lineages have been proposed,63 among these, monophyly is a criterion for species delimitation under the general lineage concept of monophyletic groups with distinct evolutionary lineages.64,65 Thus, results of this work suggest the presence of two basal lineages without gene flow between them, supporting the division between NW and SE populations. These results are important because lineage identification is a necessary precursor to species discovery.63 It is possible that An. triannulatus s.l. populations from NW and SE Colombia are undergoing speciation, as has been demonstrated for the Anopheles gambiae Complex molecular forms M and S.66 In addition, the values of intra-population diversity detected (h: 0.972–0.976), indicated that An. triannulatus s.l. populations from the NW and SE Colombia have high genetic diversity. Similar values have been found for other Neotropical anopheline species complex in the Brazilian Amazonian, for example for Anopheles marajoara lineage 1 (h: 0.828), lineage 2 (h: 0.780),67 and An. nuneztovari s.l. (h: 0.9560).56 High variability in the sequences may also indicate co-amplification of numts.68 In DNA barcoding studies, numts co-amplification have been reported in 11 taxa of insects leading to misidentification and an overestimation of species.68 Few studies on anophelines from South America have determined the presence/absence of pseudogenes or their influence on the genetic diversity estimates.57,69 In this study, the analysis conducted to detect numts showed that the high variability detected is caused by genetic differences and not the result of numts co-amplification.31–33
Mitochondrial DNA shows ample signatures of genomic events such as gene flow, migration, bottlenecks, and speciation.70 In this study, the high level of differentiation and lack of shared mitochondrial haplotypes between An. triannulatus s.l. NW and SE populations indicated an absence of, or very little gene flow between them. However, despite the clear restrictions to gene flow, there was insufficient evidence to define these two groups as distinct species or to relate them to one of the three described species in the Triannulatus Complex (An. triannulatus s.s., An. halophylus or An. triannulatus C). The substantial genetic variation between NW and SE lineages (73.5%) was higher than the one found for the two proposed An. marajoara lineages (61.9%),67 results that also support the existence of two An. triannulatus s.l. lineages in the studied regions. In addition, FST values between both populations (0.74) supported the genetic separation of two An. triannulatus lineages and indicated that they are genetically distinct. Lower FST values (0.47–0.63), based on COI sequences of Anopheles scanloni Sallum and Peyton, 2005 suggested high levels of differentiation that separated four mtDNA lineages.71 Similarly, FST values (0.68–0.79) between species C and D of the Anopheles dirus complex suggested a very different population history for the species.72 Therefore, the FST values for the NW and SE An. triannulatus lineages may also suggest separate evolutionary histories. Similarly, results of the uncorrected pairwise genetic analysis supported the presence of two lineages in these Colombian regions. A 3% value is used as the speciation threshold,73 however, a lower threshold was set to separate all of the Annulipes Complex species.74 It is argued that the 3% assessment would minimizing false positives, but it may also generate false negatives, and that intraspecific variation is well constrained in the 2–3% range,75 values that are comparable to the ones found for the NW and SE lineages.
Results of the neutrality tests, mismatch distribution, parameters of population expansion, and the star-like network showed that NW and SE lineages and populations within each lineage were not in equilibrium, most likely as a result of a population expansion. Similarly, a significant signal of population expansion was found for An. triannulatus populations collected northeast of the Amazon River, central and south Brazil.13 Fu's Fs values in this study were also higher than those reported for An. darlingi populations of south, central and northeastern Brazil60 and for An. nuneztovari s.l. clade I and subclade II-C of South America.56 In addition, a signal of population expansion was found with the τ estimate and the value was higher (2.373–7.740) than the ones for An. darlingi in the Brazilian Amazonia (1.091–5.628).60 The estimated time of divergence for the An. triannulatus populations suggested that lineage diversification occurred very recently, during the Pleistocene Epoch. Similar divergence times have been found for other Neotropical anophelines such as An. albimanus,30,57,76 An. darling,77 and An. nuneztovari s.l.56,78 and it had been suggested that climatic changes may be a common force driving Neotropical speciation.
The differentiation detected in An. triannulatus s.l. populations from Colombia may have several explanations. First, the genetic variation observed in COI sequences between the An. triannulatus lineages may be influenced by allopatric distribution (potential isolation by distance), or possible reproductive isolation.79 Furthermore, different lineages/species of the Triannulatus Complex may be present in these regions of Colombia. Geographic barriers have been shown to contribute to the genetic structure of other Latin American anophelines, for example, the Amazon River is a substantial barrier to gene flow among An. triannulatus13 and An. darlingi populations of the Brazilian Amazon.60 Geography is widely recognized as a key factor for speciation,80 it is possible that the Andean mountains that separate the NW and SE An. triannulatus populations are acting as a barrier to gene flow and contributing to the genetic structure detected.
Furthermore, given the wide geographic distribution of An. triannulatus s.l. in Colombia, it will be important to expand the collections to cover localities between the two regions evaluated to search for the connections between the NW and SE lineages. As demonstrated in species phylogenetic analysis, a wide range of sampling can help to address the role of geography in speciation80; it is also necessary to carry out work with immature stages and male genitalia to identify whether different species of this complex are present in Colombia and compare mtDNA and nuclear DNA data to further assess its population structure.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to G. Gómez, N. Naranjo, and M. Altamiranda who cooperated in specimen collection and identification.
Footnotes
Financial support: This study was supported by Comité para el Desarrollo de la Investigación-CODI, Universidad de Antioquia, grant 8700-033 to DAR, the United States National Institutes of Health grants R03 AI076710-02 to MMC and R01 AI54139-02 to JEC.
Authors' addresses: Doris A. Rosero, Luz M. Jaramillo, Lina A. Gutiérrez, and Margarita M. Correa, Grupo de Microbiología Molecular, Escuela de Microbiología, Universidad de Antioquia, Medellín, Colombia, E-mails: roserodoris@hotmail.com, luz.montanito@gmail.com, liangutibui@gmail.com, and mcorrea@quimbaya.udea.edu.co. Jan E. Conn, Griffin Laboratory, Wadsworth Center, New York State Department of Health, Slingerlands, New York, E-mail: jconn@wadsworth.org.
References
- 1.Faran M. Mosquito studies (Diptera: Culicidae) XXXIV. A revision of the Albimanus section of the subgenus Nyssorhynchus of Anopheles. Contrib Am Entomol Inst. 1980;15:1–214. [Google Scholar]
- 2.Chadee DD, Wilkerson RC. Anopheles triannulatus (Neiva and Pinto): a new Anopheles record from Trinidad, West Indies. J Am Mosq Control Assoc. 2005;21:316–317. doi: 10.2987/8756-971X(2005)21[316:ATNAPA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 3.Oliveira-Ferreira J, Lourenco-de-Oliveira R, Teva A, Deane LM, Daniel-Ribeiro CT. Natural malaria infections in anophelines in Rondonia State, Brazilian Amazon. Am J Trop Med Hyg. 1990;43:6–10. [PubMed] [Google Scholar]
- 4.de Arruda M, Carvalho MB, Nussenzweig RS, Maracic M, Ferreira AW, Cochrane AH. Potential vectors of malaria and their different susceptibility to Plasmodium falciparum and Plasmodium vivax in northern Brazil identified by immunoassay. Am J Trop Med Hyg. 1986;35:873–881. doi: 10.4269/ajtmh.1986.35.873. [DOI] [PubMed] [Google Scholar]
- 5.Tadei WP, Dutary-Thatcher B. Malaria vectors in the Brazilian Amazon: Anopheles of the subgenus Nyssorhynchus. Rev Inst Med Trop Sao Paulo. 2000;42:87–94. doi: 10.1590/s0036-46652000000200005. [DOI] [PubMed] [Google Scholar]
- 6.Aramburú J, Ramal C, Witzig R. Malaria reemergence in the Peruvian Amazon Region. Emerg Infect Dis. 1999;5:209–215. doi: 10.3201/eid0502.990204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benarroch EI. Studies on malaria in Venezuela. Am J Epidemiol. 1931;14:690–693. [Google Scholar]
- 8.Gabaldon A, Cova Garcia P. Zoogeografìa de los anofelinos en Venezuela. I. Los dos vectores principales. Tijeretazos Malar. 1946;10:78–127. [Google Scholar]
- 9.Rosa-Freitas MG, Lourenco-de-Oliveira R, de Carvalho-Pinto CJ, Flores-Mendoza C, Silva-do-Nascimento TF. Anopheline species complexes in Brazil. Current knowledge of those related to malaria transmission. Mem Inst Oswaldo Cruz. 1998;93:651–655. doi: 10.1590/s0074-02761998000500016. [DOI] [PubMed] [Google Scholar]
- 10.Silva-do-Nascimento TF, Lourenço-de-Oliveira R. Anopheles halophylus, a new species of the subgenus Nyssorhynchus (Diptera: Culicidae) from Brazil. Mem Inst Oswaldo Cruz. 2002;97:801–811. doi: 10.1590/s0074-02762002000600010. [DOI] [PubMed] [Google Scholar]
- 11.Silva-do-Nascimento TF, Wilkerson RC, Lourenço-de-Oliveira R, Monteiro FA. Molecular confirmation of the specific status of Anopheles halophylus (Diptera: Culicidae) and evidence of a new cryptic species within An. triannulatus in central Brazil. J Med Entomol. 2006;43:455–459. doi: 10.1603/0022-2585(2006)43[455:mcotss]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 12.Santos JM, Maia JF, Tadei WP. Differentiation and genetic variability in natural populations of Anopheles (N.) triannulatus (Neiva & Pinto, 1922) of Brazilian Amazonia. Braz J Biol. 2004;64:327–336. doi: 10.1590/s1519-69842004000200019. [DOI] [PubMed] [Google Scholar]
- 13.Pedro PM, Uezu A, Sallum MA. Concordant phylogeographies of 2 malaria vectors attest to common spatial and demographic histories. J Hered. 2010;101:618–627. doi: 10.1093/jhered/esq054. [DOI] [PubMed] [Google Scholar]
- 14.Silva-do-Nascimento TF, Rona LD, Peixoto AA, Lourenço-de-Oliveira R. Molecular divergence in the timeless and cpr genes among three sympatric cryptic species of the Anopheles triannulatus complex. Mem Inst Oswaldo Cruz. 2011;106((Suppl I)):218–222. doi: 10.1590/s0074-02762011000900027. [DOI] [PubMed] [Google Scholar]
- 15.Zapata MA, Cienfuegos AV, Quiros OI, Quiñones ML, Luckhart S, Correa MM. Discrimination of seven Anopheles species from San Pedro de Uraba, Antioquia, Colombia, by polymerase chain reaction-restriction fragment length polymorphism analysis of ITS sequences. Am J Trop Med Hyg. 2007;77:67–72. [PubMed] [Google Scholar]
- 16.Cienfuegos AV, Rosero DA, Naranjo N, Luckhart S, Conn JE, Correa MM. Evaluation of a PCR-RFLP-ITS2 assay for discrimination of Anopheles species in northern and western Colombia. Acta Trop. 2011;118:128–135. doi: 10.1016/j.actatropica.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brochero H, Pareja PX, Ortiz G, Olano VA. Breeding places and biting activity of Anopheles species in the municipality of Cimitarra, Santander, Colombia. Biomedica. 2006;26:269–277. [PubMed] [Google Scholar]
- 18.Gutiérrez LA, Gonzalez JJ, Gomez GF, Castro MI, Rosero DA, Luckhart S, Conn JE, Correa MM. Species composition and natural infectivity of anthropophilic Anopheles (Diptera:Culicidae) in the states of Cordoba and Antioquia, northwestern Colombia. Mem Inst Oswaldo Cruz. 2009;104:1117–1124. doi: 10.1590/s0074-02762009000800008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gutiérrez LA, Naranjo N, Jaramillo LM, Muskus C, Luckhart S, Conn JE, Correa MM. Natural infectivity of Anopheles species from the Pacific and Atlantic Regions of Colombia. Acta Trop. 2008;107:99–105. doi: 10.1016/j.actatropica.2008.04.019. [DOI] [PubMed] [Google Scholar]
- 20.Rodríguez M, Pérez L, Caicedo JC, Prieto G, Arroyo JA, Kaur H, Suarez-Mutis M, de La Hoz F, Lines J, Alexander N. Composition and biting activity of Anopheles (Diptera: Culicidae) in the Amazon region of Colombia. J Med Entomol. 2009;46:307–315. doi: 10.1603/033.046.0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.González R, Carrejo N. Second edition. Programa Editorial Universidad de Valle; Cali: 2009. (Introducción al estudio taxonómico de Anopheles de Colombia: claves y notas de distribución). [Google Scholar]
- 22.Rosero DA, Gutiérrez LA, Cienfuegos AV, Jaramillo LM, Correa MM. Optimización de un procedimiento de extracción de ADN para mosquitos anofelinos. Rev Chil Entomol. 2010;36:260–263. [Google Scholar]
- 23.Vincze T, Posfai J, Roberts RJ. NEBcutter: a program to cleave DNA with restriction enzymes. Nucleic Acids Res. 2003;31:3688–3691. doi: 10.1093/nar/gkg526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marrelli MT, Floeter-Winter LM, Malafronte RS, Tadei WP, Lourenco-de-Oliveira R, Flores-Mendoza C, Marinotti O. Amazonian malaria vector anopheline relationships interpreted from ITS2 rDNA sequences. Med Vet Entomol. 2005;19:208–218. doi: 10.1111/j.0269-283X.2005.00558.x. [DOI] [PubMed] [Google Scholar]
- 25.Oliveira de Carvalho M. 2002. (FragSize: DNA Band Size Determination).http://www.bioinformatics.org Available at. Accessed October 2011.
- 26.Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Duran C, Field M, Heled J, Kearse M, Markowitz S, Moir R, Stones-Havas S, Sturrock S, Thierer T, Wilson A. Research Software for Biologists, Not Computer Scientists. 2011. http://www.geneious.com Available at. Accessed October 2011.
- 27.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keller A, Schleicher T, Schultz J, Müller T, Dandekar T, Wolf M. 5.8S-28S rRNA interaction and HMM-based ITS2 annotation. Gene. 2009;430:50–57. doi: 10.1016/j.gene.2008.10.012. http://its2.bioapps.biozentrum.uni-wuerzburg.de/ Available at. Accessed October 2011. [DOI] [PubMed] [Google Scholar]
- 29.Lunt DH, Zhang DX, Szymura JM, Hewitt GM. The insect cytochrome oxidase I gene: evolutionary patterns and conserved primers for phylogenetic studies. Insect Mol Biol. 1996;5:153–165. doi: 10.1111/j.1365-2583.1996.tb00049.x. [DOI] [PubMed] [Google Scholar]
- 30.Gutiérrez LA, Naranjo NJ, Cienfuegos AV, Muskus CE, Luckhart S, Conn JE, Correa MM. Population structure analyses and demographic history of the malaria vector Anopheles albimanus from the Caribbean and the Pacific regions of Colombia. Malar J. 2009;8:259. doi: 10.1186/1475-2875-8-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hlaing T, Tun-Lin W, Somboon P, Socheat D, Setha T, Min S, Chang MS, Walton C. Mitochondrial pseudogenes in the nuclear genome of Aedes aegypti mosquitoes: implications for past and future population genetic studies. BMC Genet. 2009;10:11. doi: 10.1186/1471-2156-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buhay JE. “COI-like” sequences are becoming problematic in molecular systematic and DNA barcoding studies. J Crustac Biol. 2009;29:96–110. [Google Scholar]
- 33.Song H, Buhay JE, Whiting MF, Crandall KA. Many species in one: DNA barcoding overestimates the number of species when nuclear mitochondrial pseudogenes are coamplified. Proc Natl Acad Sci USA. 2008;105:13486–13491. doi: 10.1073/pnas.0803076105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moreno M, Marinotti O, Krzywinski J, Tadei WP, James AA, Achee NL, Conn JE. Complete mtDNA genomes of Anopheles darlingi and an approach to anopheline divergence time. Malar J. 2010;9:127. doi: 10.1186/1475-2875-9-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahumada M, Quiñones ML. Universidad Nacional de Colombia, Facultad de Medicina; Infecciones y Salud en el Trópico: 2009. (Determinación del papel como vector de malaria de las especies de Anopheles presentes en el Departamento del Meta, Colombia. Tesis de Maestría). [Google Scholar]
- 36.Sallum MA, Schultz TR, Foster PG, Aronstein K, Wirtz RA, Wilkerson RC. Phylogeny of Anophelinae (Diptera: Culicidae) based on nuclear ribosomal and mitochondrial DNA sequences. Syst Entomol. 2002;27:361–382. [Google Scholar]
- 37.Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Mol Ecol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- 38.Crandall KA, Templeton AR. Empirical tests of some predictions from coalescent theory with applications to intraspecific phylogeny reconstruction. Genetics. 1993;134:959–969. doi: 10.1093/genetics/134.3.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 40.Fluxus Technology . 2012. www.fluxus-engineering.com/network_terms.htm (Network 4.6.1.0. Copyright 2011 Fluxus Technology Ltd. All rights reserved). Available at. [Google Scholar]
- 41.Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 42.Bryant D, Moulton V. Neighbor-net: an agglomerative method for the construction of phylogenetic networks. Mol Biol Evol. 2004;21:255–265. doi: 10.1093/molbev/msh018. [DOI] [PubMed] [Google Scholar]
- 43.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 44.Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 45.Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- 46.Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 47.Wright S. The genetical structure of populations. Ann Eugen. 1951;15:323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
- 48.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fu YX. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rogers AR, Harpending H. Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol. 1992;9:552–569. doi: 10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed] [Google Scholar]
- 53.Rogers AR. Genetic evidence for a Pleistocene population explosion. Evolution. 1995;49:608–615. doi: 10.1111/j.1558-5646.1995.tb02297.x. [DOI] [PubMed] [Google Scholar]
- 54.Harpending HC. Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Hum Biol. 1994;66:591–600. [PubMed] [Google Scholar]
- 55.Schneider S, Excoffier L. Estimation of past demographic parameters from the distribution of pairwise differences when the mutation rates vary among sites: application to human mitochondrial DNA. Genetics. 1999;152:1079–1089. doi: 10.1093/genetics/152.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scarpassa VM, Conn JE. MtDNA tracks a complex evolutionary history with Pleistocene divergence for the neotropical malaria vector Anopheles nuneztovari sensu lato. Am J Trop Med Hyg. 2011;85:857–867. doi: 10.4269/ajtmh.2011.11-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Loaiza JR, Scott ME, Bermingham E, Rovira J, Conn JE. Evidence for pleistocene population divergence and expansion of Anopheles albimanus in Southern Central America. Am J Trop Med Hyg. 2010;82:156–164. doi: 10.4269/ajtmh.2010.09-0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Castelloe J, Templeton AR. Root probabilities for intraspecific gene trees under neutral coalescent theory. Mol Phylogenet Evol. 1994;3:102–113. doi: 10.1006/mpev.1994.1013. [DOI] [PubMed] [Google Scholar]
- 59.Hasan AU, Suguri S, Fujimoto C, Itaki RL, Harada M, Kawabata M, Bugoro H, Albino B. Genetic diversity in two sibling species of the Anopheles punctulatus group of mosquitoes on Guadalcanal in the Solomon Islands. BMC Evol Biol. 2008;8:318. doi: 10.1186/1471-2148-8-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pedro PM, Sallum MA. Spatial expansion and population structure of the neotropical malaria vector, Anopheles darlingi (Diptera: Culicidae) Biol J Linn Soc Lond. 2009;97:854–866. [Google Scholar]
- 61.Hewitt G. The genetic legacy of the Quaternary ice ages. Nature. 2000;405:907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- 62.Matthews SD, Meehan LJ, Onyabe DY, Vineis J, Nock I, Ndams I, Conn JE. Evidence for late Pleistocene population expansion of the malarial mosquitoes, Anopheles arabiensis and Anopheles gambiae in Nigeria. Med Vet Entomol. 2007;21:358–369. doi: 10.1111/j.1365-2915.2007.00703.x. [DOI] [PubMed] [Google Scholar]
- 63.de Queiroz K. Species concepts and species delimitation. Syst Biol. 2007;56:879–886. doi: 10.1080/10635150701701083. [DOI] [PubMed] [Google Scholar]
- 64.Wiens JJ, Penkrot TA. Delimiting species using DNA and morphological variation and discordant species limits in spiny lizards (Sceloporus) Syst Biol. 2002;51:69–91. doi: 10.1080/106351502753475880. [DOI] [PubMed] [Google Scholar]
- 65.Reeves PA, Richards CM. Species delimitation under the general lineage concept: an empirical example using wild North American hops (Cannabaceae: Humulus lupulus) Syst Biol. 2011;60:45–59. doi: 10.1093/sysbio/syq056. [DOI] [PubMed] [Google Scholar]
- 66.Lehmann T, Diabate A. The molecular forms of Anopheles gambiae: a phenotypic perspective. Infect Genet Evol. 2008;8:737–746. doi: 10.1016/j.meegid.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McKeon SN, Lehr MA, Wilkerson RC, Ruiz JF, Sallum MA, Lima JB, Povoa MM, Conn JE. Lineage divergence detected in the malaria vector Anopheles marajoara (Diptera: Culicidae) in Amazonian Brazil. Malar J. 2010;9:271. doi: 10.1186/1475-2875-9-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moulton MJ, Song H, Whiting MF. Assessing the effects of primer specificity on eliminating numt coamplification in DNA barcoding: a case study from Orthoptera (Arthropoda: Insecta) Mol Ecol Resour. 2010;1:615–627. doi: 10.1111/j.1755-0998.2009.02823.x. [DOI] [PubMed] [Google Scholar]
- 69.Ruiz F, Wilkerson R, Conn JE, McKeon SN, Levin DM, Quiñones M, Povoa M, Linton YM. DNA barcoding reveals both known and novel taxa in the Albitarsis Group (Anopheles: Nyssorhynchus) of Neotropical malaria vectors. Parasites & Vectors. 2012;5:44. doi: 10.1186/1756-3305-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Avise JC. Molecular Markers, Natural History, and Evolution. Second edition. Sunderland, MA: Sinauer; 2004. [Google Scholar]
- 71.O'Loughlin SM, Somboon P, Walton C. High levels of population structure caused by habitat islands in the malarial vector Anopheles scanloni. Heredity. 2007;99:31–40. doi: 10.1038/sj.hdy.6800959. [DOI] [PubMed] [Google Scholar]
- 72.Walton C, Handley JM, Tun-Lin W, Collins FH, Harbach RE, Baimai V, Butlin RK. Population structure and population history of Anopheles dirus mosquitoes in Southeast Asia. Mol Biol Evol. 2000;17:962–974. doi: 10.1093/oxfordjournals.molbev.a026377. [DOI] [PubMed] [Google Scholar]
- 73.Hebert PD, Cywinska A, Ball SL, deWaard JR. Biological identifications through DNA barcodes. Proc Biol Sci. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Foley DH, Wilkerson RC, Cooper RD, Volovsek ME, Bryan JH. A molecular phylogeny of Anopheles annulipes (Diptera: Culicidae) sensu lato: the most species-rich anopheline complex. Mol Phylogenet Evol. 2007;43:283–297. doi: 10.1016/j.ympev.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 75.Meyer CP, Paulay G. DNA barcoding: error rates based on comprehensive sampling. PLoS Biol. 2005;3:e422. doi: 10.1371/journal.pbio.0030422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Loaiza JR, Scott ME, Bermingham E, Sanjur OI, Wilkerson R, Rovira J, Gutierrez LA, Correa MM, Grijalva MJ, Birnberg L, Bickersmith S, Conn JE. Late Pleistocene environmental changes lead to unstable demography and population divergence of Anopheles albimanus in the northern Neotropics. Mol Phylogenet Evol. 2010;57:1341–1346. doi: 10.1016/j.ympev.2010.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mirabello L, Conn JE. Molecular population genetics of the malaria vector Anopheles darlingi in Central and South America. Heredity. 2006;96:311–321. doi: 10.1038/sj.hdy.6800805. [DOI] [PubMed] [Google Scholar]
- 78.Mirabello L, Conn JE. Population analysis using the nuclear white gene detects Pliocene/Pleistocene lineage divergence within Anopheles nuneztovari in South America. Med Vet Entomol. 2008;22:109–119. doi: 10.1111/j.1365-2915.2008.00731.x. [DOI] [PubMed] [Google Scholar]
- 79.Gutiérrez LA, Gomez GF, Gonzalez JJ, Castro MI, Luckhart S, Conn JE, Correa MM. Microgeographic genetic variation of the malaria vector Anopheles darlingi root (Diptera: Culicidae) from Cordoba and Antioquia, Colombia. Am J Trop Med Hyg. 2010;83:38–47. doi: 10.4269/ajtmh.2010.09-0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Barraclough TG, Vogler AP. Detecting the geographical pattern of speciation from species-level phylogenies. Am Nat. 2000;155:419–434. doi: 10.1086/303332. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.