Abstract
Genetic association studies, pharmacological investigations, and analysis of mice lacking individual genes have made it clear that cocaine administration and withdrawal have a profound impact on multiple neurotransmitter systems. The GABAergic medium spiny neurons of the nucleus accumbens (NAc) exhibit changes in the expression of genes encoding receptors for glutamate and in the signaling pathways triggered by dopamine binding to G-protein coupled dopamine receptors. Deep sequence analysis provides a sensitive, quantitative and global analysis of the effects of cocaine on the NAc transcriptome. RNA prepared from the NAc of adult male mice receiving daily injections of saline or cocaine, or cocaine followed by a period of withdrawal, was used for high-throughput sequence analysis. Changes were validated by qPCR or Western blot. Based on pathway analysis, a preponderance of the genes affected by cocaine and withdrawal were involved in the cadherin, heterotrimeric G-protein, and Wnt signaling pathways. Distinct subsets of cadherins and protocadherins exhibited a sustained increase or decrease in expression. Sustained down-regulation of several heterotrimeric G-protein β- and γ-subunits was observed. In addition to altered expression of receptors for small molecule neurotransmitters, neuropeptides and endocannabinoids, changes in the expression of plasma membrane transporters and vesicular neurotransmitter transporters were also observed. The effects of chronic cocaine and withdrawal on the expression of genes essential to cholinergic, glutamatergic, GABAergic, peptidergic, and endocannabinoid signaling are as profound as their effects on dopaminergic transmission. Simultaneous targeting of multiple withdrawal-specific changes in gene expression may facilitate development of new therapeutic approaches that are better able to prevent relapse.
Keywords: RNA-Seq, pathway analysis, Wnt/cadherin signaling, heterotrimeric G-protein, glutamate, neuropeptide, acetylcholine, GABA
Introduction
Chronic drug exposure induces persistent changes in the brain that underlie the addiction-associated behavioral abnormalities seen in human addicts and rodent models of addiction. Data from broad expression studies using microarray technology and from single-gene approaches suggest that cocaine exposure elicits widespread modifications to the transcriptional landscape, including drug-induced changes in epigenetics, RNA processing, microRNAs, and gene transcription (McClung and Nestler, 2008; Eipper-Mains et al., 2011). These alterations are believed to be integral to the neural plasticity seen in addiction.
Dopaminergic, glutamatergic, and peptidergic signaling in the mesolimbic system are altered in response to cocaine. Repeated exposure to cocaine leads to sensitization, the progressive and persistent amplification of behavioral and motivational responses to a fixed dose of the same drug (Hyman et al., 2006). Sensitization may persist for weeks to years after cessation of drug taking, presumably contributing to a reformed addict’s risk for relapse (Saunders and Robinson, 2011), which occurs in up to ninety percent of addicted individuals (Wee and Koob, 2010; Kalivas and Volkow, 2011).
A majority of current research focuses on the effects of cocaine on the mesolimbic dopaminergic system, identified in prior studies as the reward center of the brain (Nestler, 2005a). Dopaminergic projections from the ventral tegmental area as well as glutamatergic projections from the prefrontal cortex synapse on inhibitory GABAergic medium spiny neurons in the ventral striatum (nucleus accumbens; NAc) (Kauer and Malenka, 2007). This convergence of dopaminergic and glutamatergic projections, as well as accompanying biochemical and morphological changes, identified the NAc as a key integration point in the rewarding and addictive effects of drugs of abuse (Nestler, 2005b; Hyman et al., 2006; Kalivas and Volkow, 2011; Saunders and Robinson, 2011).
High-throughput sequencing has a low background signal, facilitates identification and absolute quantification of low-abundance and novel transcripts (Mortazavi et al., 2008; Wang et al., 2008a; Wang et al., 2008b; Wahlstedt et al., 2009; Wang et al., 2009; Metzker, 2010; Eipper-Mains et al., 2011). We used next-generation sequencing of mRNAs purified from the NAc to obtain a more complete picture of the molecular response to chronic cocaine exposure and withdrawal. Bioinformatic analysis of cocaine-regulated transcripts revealed substantial involvement of the Wnt/cadherin and heterotrimeric G-protein signaling pathways. Evaluation of the synthetic enzymes, transporters, and receptors involved in catecholaminergic, glutamatergic, GABAergic, cholinergic, and lipid signaling identified complex responses unique to cocaine and withdrawal.
Methods and Materials
Cocaine Treatment of Mice
Adult male C57BL/6 mice (Jackson Laboratories, Bar Harbor, Maine; 2–5 months old) were group-housed in the animal facility at the University of Connecticut Health Center on a 12 h light/dark cycle (lights, 7:00am–7:00pm) and handled in accordance with Institutional Animal Care and Use Committee guidelines. After acclimation for one week, animals were handled for 1 min/day for 2–3 days prior to injections. On each treatment day, animals were allowed to acclimate to the behavior room for 45–60 minutes before beginning injections.
Cocaine treatment was performed as described (Eipper-Mains et al., 2011). Animals were given saline (10 ml/kg/day) or cocaine (10 or 20 mg/kg/day) intraperitoneally for 7 days and locomotor activity was monitored daily using a 15”×15” Plexiglas chamber (PAS Open Field System, San Diego Instruments); after the 7th injection, locomotor activity was three-fold higher in mice receiving cocaine than in mice receiving saline (Figure S1) (Kiraly et al., 2010; Eipper-Mains et al., 2012). Administering 10 mg/kg cocaine on the first and last days produces more reliable locomotor sensitization (locomotor ambulations on day 7 divided by locomotor ambulations on day 1), as originally demonstrated in rats (Pierce et al., 1996), and was used in later experimental sets in this work. Animals were sacrificed by decapitation 24 hours after the final injection with the exception of mice in the withdrawal groups, which were kept in the home cage for 8 or 28 days after the final cocaine dose prior to tissue harvest; elevated locomotor responding was maintained for the 28 day withdrawal period (Figure S1). Quantitative polymerase chain reaction (qPCR) validation was performed on RNA samples used for library preparation (8 animals analyzed separately in each treatment group) and on two biological replicate groups of mice sensitized in a similar manner (Kiraly et al., 2010): 8 each saline and cocaine; 4 each saline, cocaine, and 28 days withdrawal. Pairwise comparisons were performed and subjected to t-test (p<0.05 taken as significant).
RNA and Library Preparation and Sequencing
NAc punches were taken from individual mice (at least 8 animals/treatment group). Total RNA was prepared using TRIzol (Invitrogen) (Eipper-Mains et al., 2011). Average yield of total RNA per NAc punch was 6.3±0.6 µg. Total RNA used for NAc library preparation consisted of equal amounts of RNA from 8 individual mice for each treatment. One Saline, one Cocaine, and one 7-day Withdrawal mRNA-Seq library was prepared from each pooled RNA sample according to manufacturer’s specifications (Illumina). Briefly, total RNA was treated with DNase I; poly(A)+ RNA purified using Dynal magnetic beads (Invitrogen) was fragmented by partial alkaline hydrolysis (Ambion) and reverse-transcribed using random primers and SuperScript II (Invitrogen). cDNA was size-selected (~200 bp insert) on 2% agarose gels. Libraries were prepared for sequencing using the Paired-End DNA Sample Prep Kit (Illumina). NAc libraries were sequenced in nine lanes (3 technical replicates per sample) on an Illumina GAIIx using a 37-cycle paired-end sequencing protocol. Replicates were analyzed for intra-sample disparity and read data from all three lanes were then merged into one composite data file per sample; intra-sample coefficient of determination, R2 ≥ 0.98. The composite file was used for subsequent analyses.
mRNA-Seq Data Analysis
Sequences and quality scores were extracted from image files using Firecrest, Bustard, and GERALD software. Sequences were aligned to the Mus musculus genome (2007, mm9; NCBI Build 37) using Bowtie (http://bowtie-bio.sourceforge.net; version 0.12.7) (Langmead et al., 2009) permitting multi-alignment and allowing up to 2-mismatches. Ambiguous alignments were resolved using the Spliced Paired-end Aligner Perl script (Brooks et al., 2011). Robust assembly of novel transcripts and a complete analysis of cocaine-induced alternative splicing requires greater transcript coverage and read depth than we obtained (Trapnell et al., 2010).
Normalized mRNA expression was calculated as RPKM (reads per kilobase gene model per million mapped reads) (Mortazavi et al., 2008) from aligned sequence reads; Z-scores were computed from frequency data using the following equation: (x - μ)/σ, x, sample frequency; μ, mean across all samples; σ, standard deviation across all samples. Gene expression data (Z-scores) were hierarchically clustered using Gene Cluster 3.0 (http://rana.lbl.gov/EisenSoftware.htm); heat maps were generated using Java Treeview (http://www.sourceforge.net/projects/jtreeview/files). Gene lists were submitted to the Panther website (http://www.pantherdb.org) for analysis of Pathway and Protein Class enrichment compared to the Mus musculus reference gene list. Western and qPCR analyses were performed as described (Eipper-Mains et al., 2011); primer sequences are in Table S1.
Results
Genome coverage
To characterize the effects of chronic cocaine and withdrawal on mRNA expression in the NAc, we prepared mRNA-Seq libraries from RNA pooled from mice subjected to daily saline injections (Saline), daily cocaine injections (Cocaine) or daily cocaine injections followed by a week with no injections (Withdrawal). Paired-end sequencing yielded approximately 20 million uniquely mapped reads for each library (Table 1A). The current annotation of the Mus musculus genome (2007, mm9) spans ~1.6 billion bases; 3.2% of the genome encodes mature mRNAs and 40.6% encodes primary transcripts. We detected 5–6% of the genome in the poly(A)+ RNA-Seq data from each NAc library. Approximately 80% of the reads in each library mapped to annotated exons; 15–16% of reads mapped to introns and 5% of reads aligned to intergenic regions (Table 1B). Mitochondrial reads accounted for roughly 2% of all mapped sequences in these libraries (Figure S2). Scatter plots of mitochondrial (Figure S3) and ribosomal (Figure S4) gene expression demonstrated no evidence for cocaine-regulation, so subsequent analyses were performed on a filtered gene list that excluded both ribosomal and mitochondrial genes.
Table 1. Read mapping and base coverage for RNA-Seq data.
Sequences from NAc libraries were aligned to the Mus musculus reference genome (2007, mm9; NCBI Build 37) using Bowtie (Langmead et al., 2009) and allowing up to 2-mismatches. (A) Aligning read numbers and percent coverage of the genome. (B) Reads were characterized as aligning to exonic, intronic, or intergenic regions based on the existing annotation.
| A. High-throughput sequencing alignment information | |||
|---|---|---|---|
| Sample | Treatment | Number aligning reads | Total genome coverage |
| NAc lysate | Saline | 26,563,428 | 5.8% |
| Cocaine | 24,696,981 | 5.8% | |
| Withdrawal | 19,767,382 | 5.1% | |
| B. Read mapping | ||||
|---|---|---|---|---|
| Sample | Treatment | Exonic | Intronic | Intergenic |
| NAc lysate | Saline | 79.4% | 15.5% | 5.1% |
| Cocaine | 79.0% | 16.1% | 4.9% | |
| Withdrawal | 79.0% | 16.2% | 4.8% | |
Genome-wide assessment of the effect of cocaine on gene expression
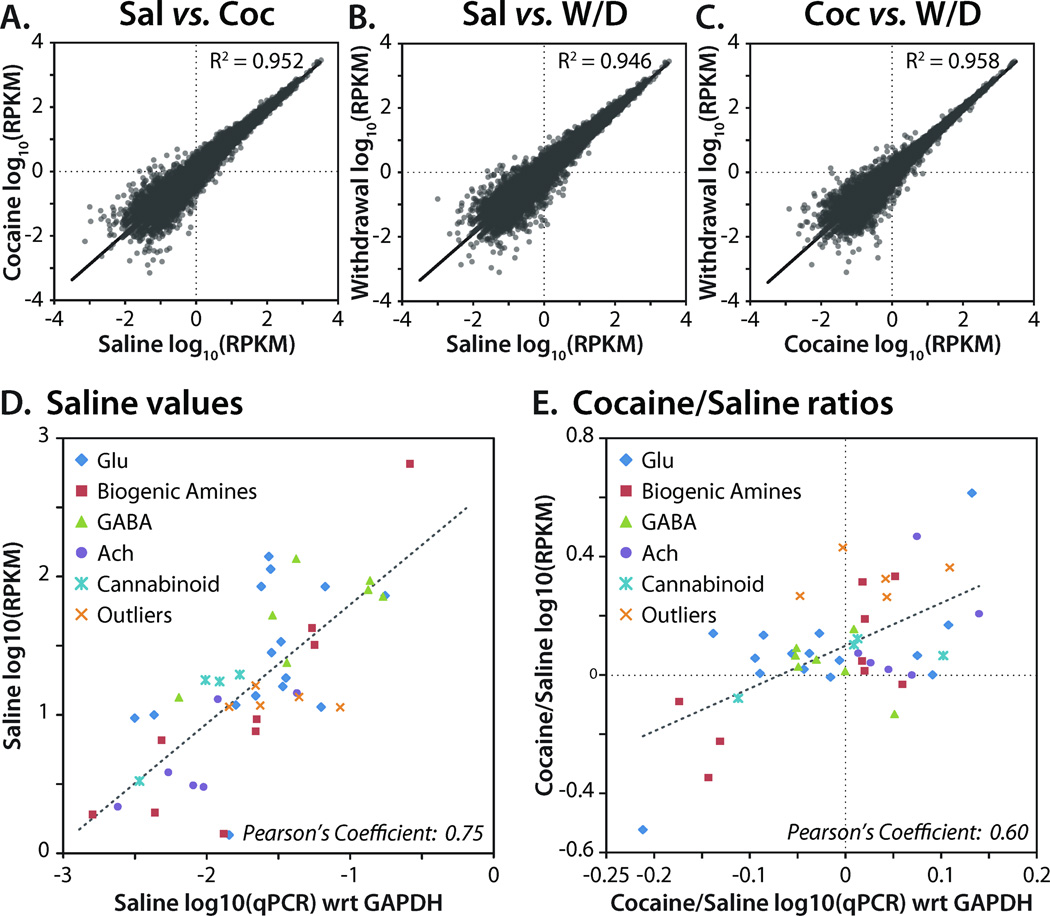
Gene expression over a broad (106) dynamic range was evaluated in each library. Expression was quantified in reads per kilobase of exon per million mapped sequence reads (RPKM), a normalized measure of exonic read density (Mortazavi et al., 2008). For the three NAc libraries, pairwise scatter plots revealed similarly varied expression between the Saline/Cocaine, Saline/Withdrawal and Cocaine/Withdrawal libraries (Figure 1A–C). The variability of gene expression between samples was highest for genes expressed at lower levels. For this reason, a lower limit of expression was applied before examining regulation. Since biogenic amine transporter (Slc6a2, NET; Slc6a3, DAT; Slc6a4, SERT) transcripts are not expressed in the NAc (http://mouse.brain-map.org/), we used their RPKM values to set this lower limit; for inclusion in subsequent analyses, a gene had to have an RPKM of > 1.0 in at least one of the three treatment groups. We used qPCR to validate the RNA-Seq data (Figure 1D,E). The RNA-Seq and qPCR expression levels of 47 transcripts from five neurotransmitter systems from the Saline sample showed strong agreement across the two methods (Pearson’s r = 0.75) over a greater than 1000-fold range of expression (Figure 1D). A comparison of the ratios of Cocaine/Saline measured by RNA-Seq and qPCR was also carried out (Figure 1E). Cocaine-mediated changes in gene expression estimated from the RNA-Seq data were consistently larger than the changes observed with qPCR; nevertheless, the ratios show good qualitative agreement (Pearson’s r = 0.60). Differences presumably reflect the much larger dynamic range of RNA-Seq and quantification across entire transcripts rather than between single primer pairs.
Figure 1. High-throughput sequence analysis of NAc mRNA.
(A-C) Pairwise scatter plots of mRNA expression data for NAc libraries; shown as log10(RPKM) per sample. Ribosomal and mitochondrial genes are not included in the scatter plots. R2 values were calculated by least squares best fit. Abbreviations: Sal, Saline; Coc, Cocaine; W/D, Withdrawal; RPKM, reads per kilobase gene model per million mapped reads (Mortazavi et al., 2008). (D) RPKM data for 47 transcripts in the Saline library are compared to qPCR data (normalized to Glyceraldehyde 3-phosphate dehydrogenase, GAPDH) for the same transcripts; Pearson’s Coefficient represents the linear correlation coefficient, r. (E) Cocaine/Saline ratios for the same 47 transcripts were calculated using RPKM data and qPCR data. Pearson’s Coefficient represents the linear correlation coefficient, r, between two samples, X and Y. Pearson’s r = covariance(X,Y)/σXσY, where σ is the standard deviation.
Regulation of NAc transcript levels by cocaine and withdrawal
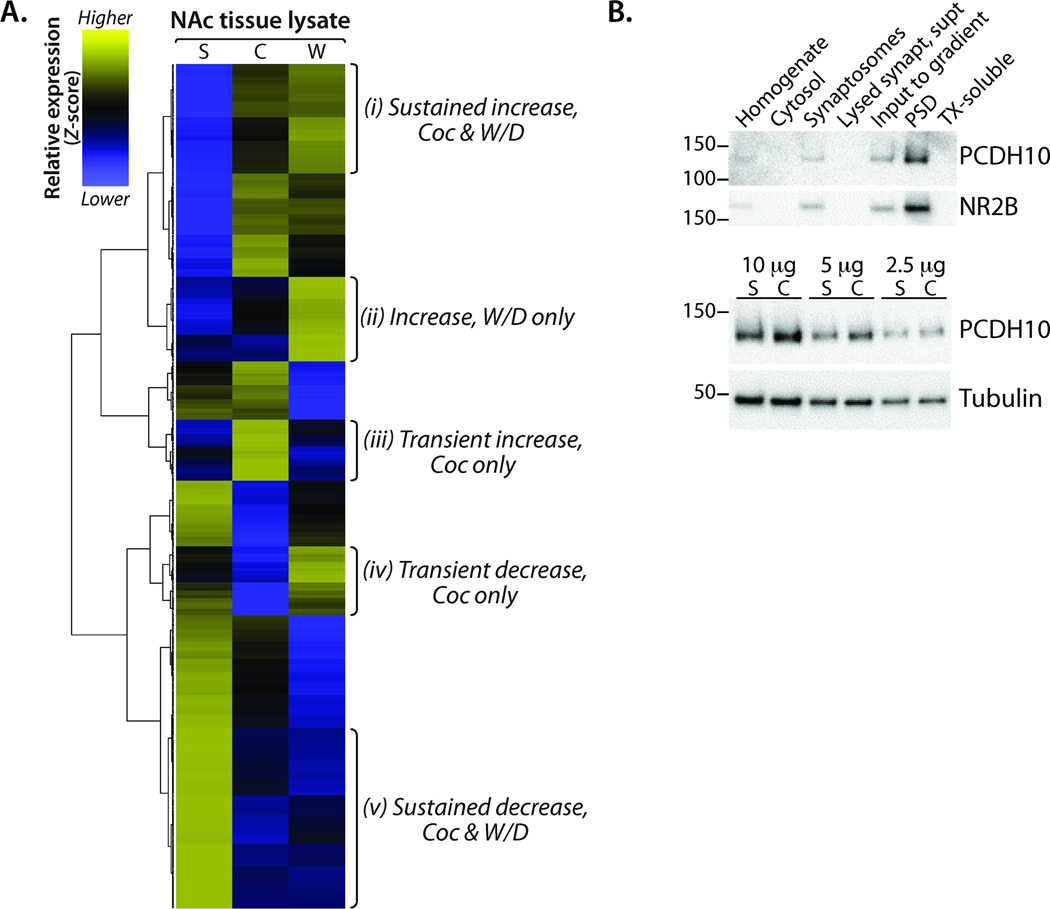
To evaluate enrichment and depletion of transcripts in NAc lysates prepared from mice in the Saline, Cocaine, and Withdrawal groups, we calculated the relative abundance of each mRNA in the three treatment groups, computed Z-scores across samples, and hierarchically clustered genes for visualization in a heat map. Of the 19,890 detected genes, 13,184 were expressed in at least one library at an RPKM > 1.0 and were used in downstream analyses. Several patterns of expression emerged from hierarchical clustering, including transcripts showing (i) sustained increases in both Cocaine and Withdrawal, (ii) increases only in Withdrawal, (iii) transient increases only in Cocaine, (iv) transient decreases only in Cocaine, and (v) sustained decreases in Cocaine and Withdrawal (Figure 2A). The full gene list for each cluster is available in Table S2.
Figure 2. Differential regulation of clusters of genes in the NAc following cocaine and withdrawal.
(A) Normalized mRNA frequency as a function of total reads for NAc lysate libraries (Saline, Cocaine, Withdrawal) was calculated for all genes in which one sample had an RPKM ≥ 1.0 (n = 13,184). Hierarchical gene expression clustering using Gene Cluster 3.0 and Java TreeView generated the dendrogram and heat map. Z-score was computed on normalized mRNA frequency across all samples. Blue, low expression; yellow, high expression; grey, no expression. Gene lists from the indicated Clusters (i-v) were used in subsequent protein class and pathway analyses. (B) The effects of Cocaine on one of the protocadherins identified as a target were verified. Subcellular fractionation of striatal tissue revealed enrichment of PCDH10 at the PSD. PSDs purified from the striata of Saline and Cocaine treated mice were analyzed for PCDH10; after normalization to βIII tubulin, the Coc/Sal ratio for PCDH10 rose 24% (P < 0.01; t-test).
Since transcripts exhibiting a sustained change in response to cocaine are included in Clusters i (sustained increase), ii (increase, Withdrawal), and v (sustained decrease), we used Panther Pathway (Table 2A and Table S3) analysis to identify common pathways. Three pathways were enriched (P < 0.005) in these three Clusters: cadherin signaling, heterotrimeric G-protein signaling (Gαi and Gαs-mediated), and Wnt signaling; since there was a great deal of overlap in the cadherin and Wnt signaling pathways, they are discussed together. Other pathways were unique to specific clusters (Table S3). For example, components of the ionotropic glutamate receptor, metabotropic glutamate receptor group III, and nicotinic acetylcholine receptor pathways were enriched only in Cluster i (sustained increase) while components of the muscarinic acetylcholine receptor 1 and 3 pathway were enriched in Cluster ii (increase, Withdrawal).
Table 2. Panther Pathways from Cocaine- and Withdrawal-regulated genes in the NAc.
Gene lists from Clusters i-v (Figure 2A) were used in Panther Pathway and Protein Class analysis (http://www.pantherdb.org); pathways enriched (P < 0.005) in all three lists relative to the Mus musculus reference genome are indicated. Complete pathway lists are provided in Table S3.
| PANTHER Pathway | Mus musculus genes (26,185) |
(i) Sustained Increase, Coc & W/D (1,387) |
(i) P- value |
(ii) Increase, W/D only (1,001) |
(ii) P- value |
(v) Sustained Decrease, Coc & W/D (2,670) |
(v) P- value |
|---|---|---|---|---|---|---|---|
| Cadherin signaling | 167 | 19 | 1.94E-03 | 16 | 9.25E-04 | 30 | 2.70E-03 |
| Heterotrimeric G-protein signaling – Gαi & Gαs |
169 | 19 | 2.21E-03 | 18 | 1.32E-04 | 32 | 8.89E-04 |
| Wnt signaling | 348 | 46 | 3.67E-08 | 27 | 5.77E-04 | 64 | 9.02E-06 |
Abbreviations: Coc, Cocaine; W/D, Withdrawal.
The pathways exhibiting sustained changes were examined in more detail. Unique sets of cadherins and protocadherins appeared in each Cluster (Table 3). While many cadherins and protocadherins showed a sustained increase in expression (Cluster i), others increased only in Withdrawal (Cluster ii) or showed a transient response to Cocaine (Clusters iii and iv). The clustered families of protocadherins, Pcdhα, -β, and γ, are important in neuronal development and are synaptically enriched in mature myelinated neurons (Morishita and Yagi, 2007). The Pcdhβ gene cluster includes 22 genes; three showed a sustained increase (Cluster i), five were increased only after Withdrawal (Cluster ii), five exhibited an increase only after Cocaine (Cluster iii), and one showed a sustained decrease in expression (Cluster v). In contrast, the Pcdhγ cluster of 23 genes exhibited a more homogeneous expression pattern in which expression of all 21 detected isoforms showed a sustained decrease (Cluster v).
Table 3. Clustered genes from the Cadherin/Wnt and Heterotrimeric G-Protein Signaling Pathways.
The cadherin and protocadherin genes appearing in Clusters i-v are identified. Genes in the heterotrimeric G-protein signaling pathway (Gαi and Gαs) that fell into Clusters i-v are grouped by function; GPCRs for dopamine, GABA, glutamate and acetylcholine are discussed separately and GPCRs without known ligands are not listed. Intracellular signaling proteins are grouped by function.
| (i) Sustained Increase, Coc & W/D |
(ii) Increase, W/D only | (iii) Transient Increase, Coc only |
(iv) Transient Decrease, Coc only |
(v) Sustained Decrease, Coc & W/D |
||
|---|---|---|---|---|---|---|
| Cadherin / Wnt Signaling | ||||||
| Cadherins |
Cdh4, Cdh5, Cdh6, Cdh8, Cdh11 |
Cdh7, Cdh19, Cdh22 | Cdh24 | Cdh13 | ||
| Protocadherins |
Pcdh1, Pcdh10, Pcdh18, Pcdhb13, Pcdhb14, Pcdhb16 |
Pcdhb2, Pcdhb6, Pcdhb7, Pcdhb8, Pcdhb9 |
Pcdh20, Pcdhb5, Pcdhb10, Pcdhb11, Pcdhb19, Pcdhb20 |
Pcdha4, Pcdhb12, Pcdhga1, Pcdhga2, Pcdhga3, Pcdhga4, Pcdhga5, Pcdhga6, Pcdhga7, Pcdhga8, Pcdhga9, Pcdhga10, Pcdhga11, Pcdhga12, Pcdhgb1, Pcdhgb2, Pcdhgb4, Pcdhgb5, Pcdhgb6, Pcdhgb7, Pcdhgb8, Pcdhgc3, Pcdhgc4, Pcdhgc5 |
||
| Heterotrimeric G-Protein Signaling (Gαi & Gαs) | ||||||
| Type of GPCR: | ||||||
| Neuropeptide binding |
Fzd5, Fzd8, Hcrtr2, Npffr1, Oprk1, Oprm1, Prlhr, Tacr1 |
Fzd6, Fzd7, Mc3r, Oprd1, Pth1r, Smo, Trhr2 |
Hcrtr1, Npbwr1, Npy2r, Oxtr, Sstr3, Tacr3 |
Fzd9, Glpr, Nmbr, Npy5r | Mc4r, Mchr1, Mtnr1a, Sstr2 | |
| Lipid binding | S1pr3 | S1pr4 | Lpar1 | |||
| Cadherin type | Celsr2, Celsr3 | Celsr1 | ||||
| Intracellular signaling proteins: | ||||||
| GPCR kinases | Grk6 | Grk1 | ||||
| Arrestins | Arrb1 | Arrb2 | ||||
| Gβ subunits | Gnb4 |
Gnb1, Gnb1l, Gnb2, Gnb2l1, Gnb5 |
||||
| Gγ subunits | Gng7 | Gng2 |
Gng3, Gng8, Gng10, Gng11, Gng13 |
|||
| RGSs | Rgs6 | Rgs8, Rgs11 | Rgs17 | Rgs9, Rgs19 |
Rgs2, Rgs4, Rgs10, Rgs16, Rgs20 |
|
| Adenylate cycases | Adcy1, Adcy4, Adcy6, Adcy9 | Adcy2, Adcy5 | Adcy8 | |||
| Phosphorylase b kinases | Phka2 | Phkg1, Phkg2 | ||||
| CREBs | Creb3l2, Crebbp | Creb3 | ||||
Abbreviations: Coc, Cocaine; W/D, Withdrawal.
We used an antibody to protocadherin 10 (PCDH10), which is known to play a role in the development of striatal axons and thalamocortical projections (Uemura et al., 2007), to evaluate its subcellular localization and quantify its levels in postsynaptic densities (PSDs) purified from the striata of saline- and cocaine-treated mice. PCDH10 was enriched in PSD preparations (Figure 2B); furthermore, PSDs purified from striatal extracts of cocaine-treated mice contained more PCDH10 than striatal PSDs from saline controls. A role for extracellular matrix proteins, including cadherins and protocadherins, is consistent with previous studies of cocaine-regulated gene expression in mice (Eipper-Mains et al., 2011) and postmortem human brain (Mash et al., 2007; Lull et al., 2008).
Consistent with the literature, heterotrimeric G-protein signaling (Gαi and Gαs-mediated) was identified as a major pathway targeted by cocaine. In addition to G-protein coupled receptors (GPCRs) for the major neurotransmitters (discussed below), neuropeptide (opioid, galanin, somatostatin), lipid, and cadherin-type GPCRs showed cocaine-related changes in expression (Table 3).
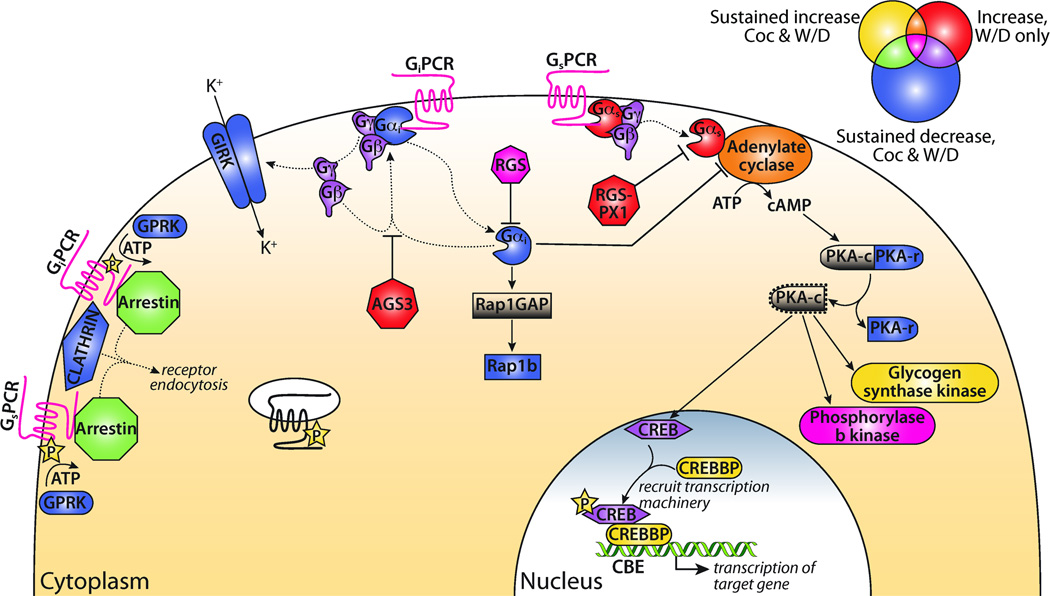
Panther Pathway analysis identified cocaine-regulated changes in the repertoire of intracellular signaling proteins downstream of the regulated Gαi and Gαs-coupled GPCRs (Table 3 and Figure 3). The diagram in Figure 3 depicts the complex interplay of intracellular signaling proteins. For example, only two members of the Mus arrestin gene family are expressed outside of the retina (Kovacs et al., 2009); Arrb1 showed a sustained increase in expression (Cluster i) while Arrb2 exhibited a sustained decrease (Cluster v). Transcripts encoding the β- and γ-subunits of heterotrimeric G-proteins were primarily identified in Cluster v (sustained decrease). In contrast, Regulators of G-protein signaling (RGS genes) appeared in all clusters. Transcripts encoding four adenylate cyclase genes showed a sustained increase in expression (Cluster i) while the transcripts encoding two additional adenylate cyclase genes demonstrated increased expression only in the Withdrawal group (Cluster ii).
Figure 3. Gαi and Gαs heterotrimeric G-protein signaling schematic.
Model based on Panther Pathway diagram (http://www.pantherdb.org). Venn diagram depicts color key for signaling components. Genes labeled yellow showed a sustained increase in Coc and W/D (cluster i), red genes increased in W/D only (cluster ii), and blue genes had decreased expression in both Coc and W/D (cluster v); overlap colors indicate contribution of > 1 cluster to signaling group. For example, GPCRs, which appear in all three clusters, are shown in magenta. Genes from Clusters i, ii, and v were used for this analysis. Abbreviations: Coc, Cocaine; W/D, Withdrawal.
Panther Protein Class analysis (Table S4) identified Cytoskeletal Proteins, Transcription Factors, and Transferases as the three Protein Classes enriched in genes exhibiting a sustained response (Clusters i, ii, and v; P < 0.0001). Sustained up-regulation of G-protein modulators and guanyl-nucleotide exchange factors (Cluster i) was also observed. Several Protein Classes involved in interactions with Ca2+ (calcium-binding proteins, calmodulin and intracellular calcium-sensing proteins) were enriched only in Cluster v (sustained down-regulation). GPCRs, ligand-gated ion channels, membrane trafficking proteins and neuropeptides were also enriched in Cluster v.
Widespread effects of cocaine and withdrawal on catecholaminergic, GABAergic, cholinergic, glutamatergic, and endocannabinoid pathways
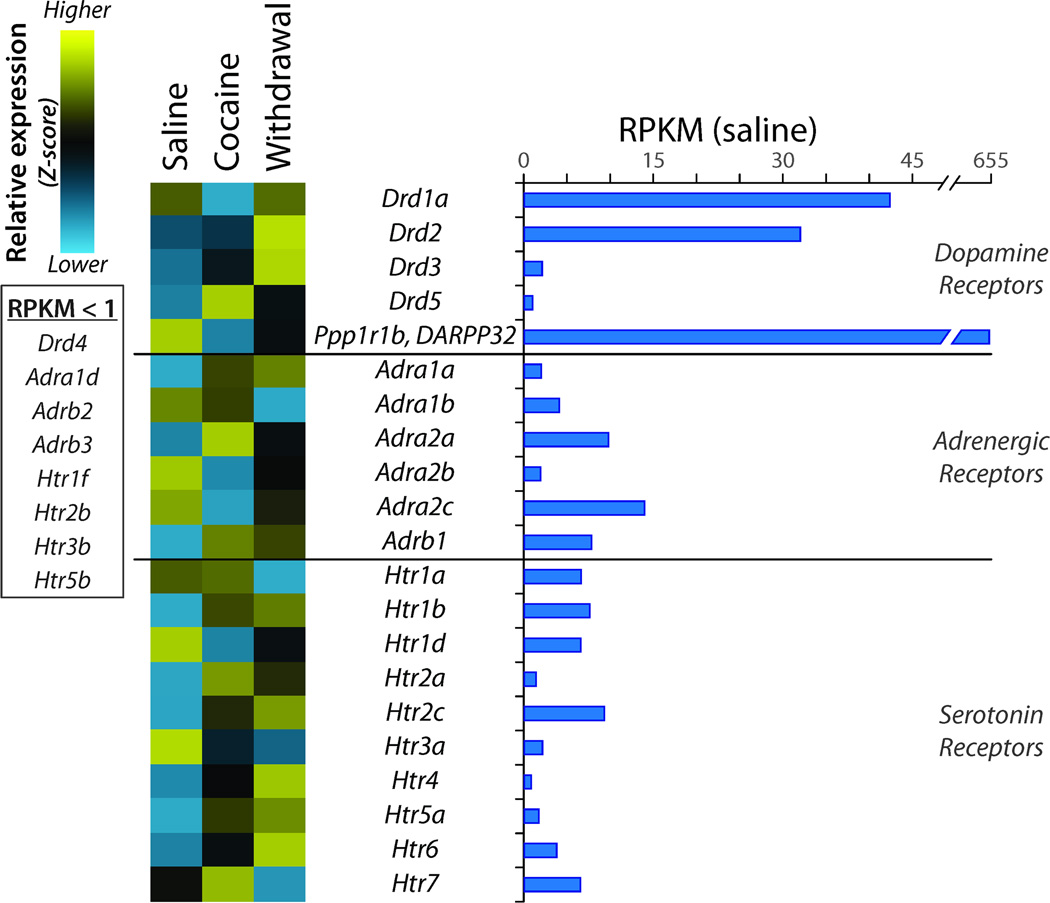
Dopamine, norepinephrine, and serotonin receptors expressed in the NAc play a critical role in addiction. Transcripts encoding the dopamine receptors, Drd1a, Drd2, Drd3, and Drd5, exhibited different patterns of response to cocaine and withdrawal (Figure 4). Drd1a was known to decrease in response to cocaine treatment, while Drd2 was known to increase (Hoffmann et al., 2012). Adra1a, Adra2a and Adrb1 adrenergic receptor expression rose in response to cocaine treatment, while expression of Adra1b, Adra2b, and Adra2c fell. Six of the twelve serotonin receptors (Htr1b, Htr2a, Htr2c, Htr4, Htr5a, Htr6) exhibited increased expression in response to cocaine and/or withdrawal. The long-term changes during withdrawal were verified by qPCR for Drd1a, Drd2, Adra1a, Adra2b, Htr1d, Htr2a, Htr2c, and Ppp1r1b (DARPP32).
Figure 4. Regulation of Receptors for Biogenic Amines.
RPKM data are shown for dopamine, adrenergic and serotonin receptor transcripts (RPKM > 1) in the Saline library. Z-scores were calculated as described in Methods. Transcripts included in this group but expressed at an RPKM < 1 are listed to the side. Ppp1r1b (DARPP32), a multiply phosphorylated phosphatase inhibitor highly expressed in medium spiny neurons, is included for comparison.
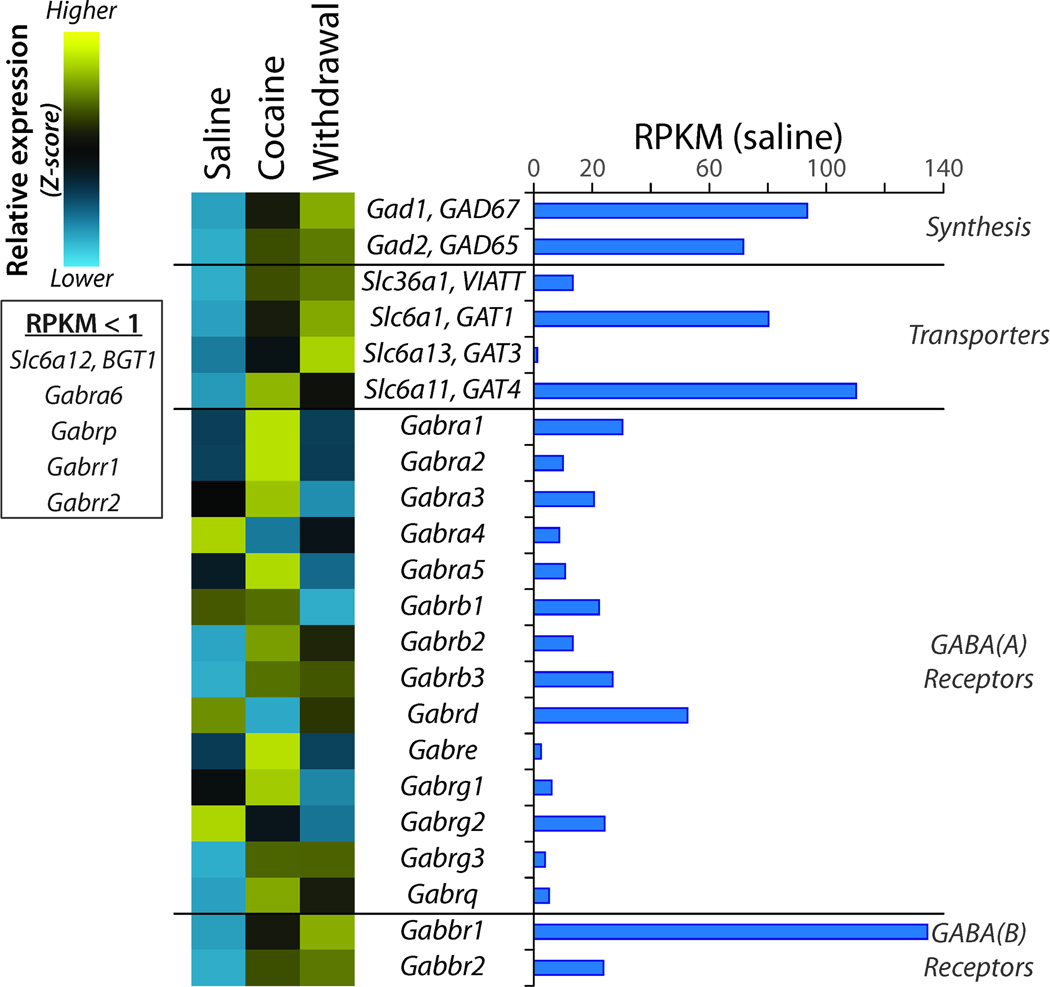
Over 90% of the neurons in the NAc are GABAergic medium spiny interneurons (Kemp and Powell, 1971); the genes involved in synthesizing, storing and retrieving GABA (γ-aminobutyric acid) are expressed in the NAc, along with ionotropic and metabotropic GABA receptors (Figure 5). Expression of the GABA-synthesizing enzymes (Gad65, Gad67), the vesicular inhibitory amino acid transmitter transporter (Viatt) and the plasma membrane transporters (Gat1, Gat3 and Gat4) rose in response to cocaine and withdrawal. The three Na+/Cl--dependent GABA transporters are expressed by astrocytes and retrieve GABA after secretion (Bolteus and Bordey, 2004; Schousboe et al., 2004; Beenhakker and Huguenard, 2010); neurons also express Gat1 (Schousboe et al., 2011). Levels of Gat4 were highest after Cocaine while levels of Gat1 were highest in Withdrawal. Multiple GABAA receptor subunits are expressed at similar levels; while some showed a sustained increase in expression (Gabrb2, Gabrb3, Gabrg3, Gabrq), Gabrb1 expression decreased in Withdrawal and Gabrd expression decreased after Cocaine but recovered in Withdrawal. Expression of GABAB receptor subunits (Gabbr1, Gabbr2) increased after Cocaine and increased further during Withdrawal.
Figure 5. Regulation of Genes Affecting GABAergic Transmission.
Data were analyzed as described for Figure 4. The transcripts encoding glutamic acid decarboxylase 65 and 67 (Gad1 and Gad2) are expressed at similar levels. GABA transport into synaptic vesicles requires Slc36a1; retrieval of GABA from extracellular space requires Slc6a1, Slc6a13, or Slca11. Multiple ionotropic GABAA receptor subunits are expressed in the NAc; expression of metabotropic GABAB receptor Gabbr1 greatly exceeds expression of Gabbr2.
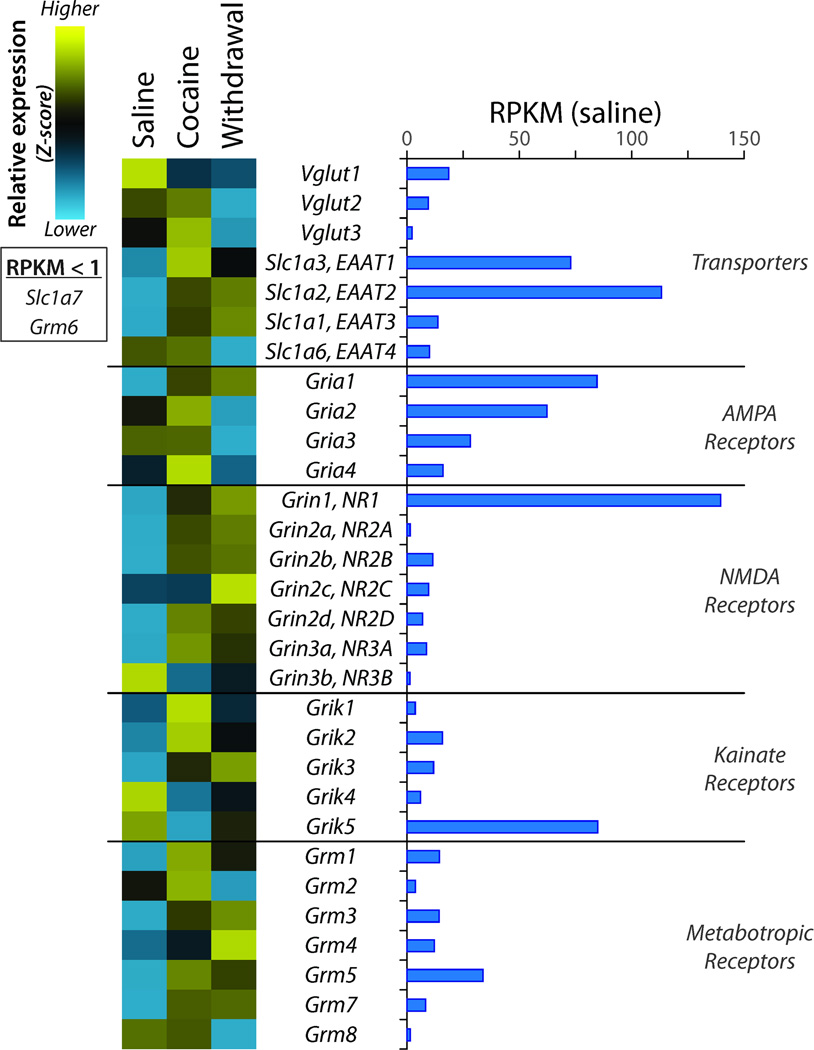
Glutamatergic signaling plays an essential role in drug abuse, with major glutamatergic inputs onto NAc medium spiny neurons (MSNs) coming from amygdala, hippocampus, and prefrontal cortex (Kalivas, 2009; Wolf, 2010). The effects of cocaine on expression of ionotropic and metabotropic glutamate receptors have been studied in detail (Kalivas, 2009; Kalivas et al., 2009; Kalivas and Volkow, 2011) and were largely confirmed in our analysis (Figure 6). Glutamatergic inputs acutely excite NAc MSNs primarily via activation of AMPA receptors, a step considered necessary for initiation of drug seeking behavior (Wolf and Ferrario, 2010). AMPA receptor expression showed only small changes during Cocaine administration; Gria1 increased substantially while Gria3 decreased during Withdrawal (confirmed by qPCR). Expression of several NMDA receptor subunits increased during Cocaine administration (Grin1, Grin2a, Grin2b, Grin2d, Grin3a) (several confirmed by qPCR). Expression of kainate receptors Grik1, Grik2, and Grik3 showed a sustained increase while expression of Grik4 and Grik5 showed a sustained decrease. Expression of the five most-highly expressed metabotropic receptors for glutamate (Grm1, Grm3, Grm4, Grm5, and Grm7) showed a sustained increase. Vesicular glutamate transporters (Vglut) can be expressed in GABAergic and cholinergic neurons as well as in glial cells (El Mestikawy et al., 2011). Transcripts encoding all three Vgluts were detectable in the NAc, which contains no primarily glutamatergic neurons; expression of Vglut1 mRNA exceeded that of Vglut2. Na+-dependent plasma membrane glutamate transporters (Eaat1 through Eaat5), which remove glutamate from the synaptic cleft and perisynaptic area, also play a critical role in glutamatergic transmission. Three of the four Eaats expressed in the NAc (Eaat1, Eaat2, and Eaat3) exhibited a sustained increase in expression while Eaat4 exhibited reduced expression in Withdrawal. Eaat1 and Eaat2 are primarily expressed in glia, while Eaat3, Eaat4 and Eaat5 show widespread neuronal expression throughout the brain (Tzingounis and Wadiche, 2007).
Figure 6. Regulation of Genes Affecting Glutamatergic Transmission.
Data were analyzed as described for Figure 4. Expression of all three vesicular glutamate transporters (Vglut) and four of the plasma membrane glutamate transporters (Slc1a1, Slc1a2, Slc1a3, Slc1a6) was above the cutoff level. Multiple subunits of the ionotropic glutamate receptors (AMPA, NMDA, and Kainate) and multiple metabotropic glutamate receptors are expressed at similar levels.
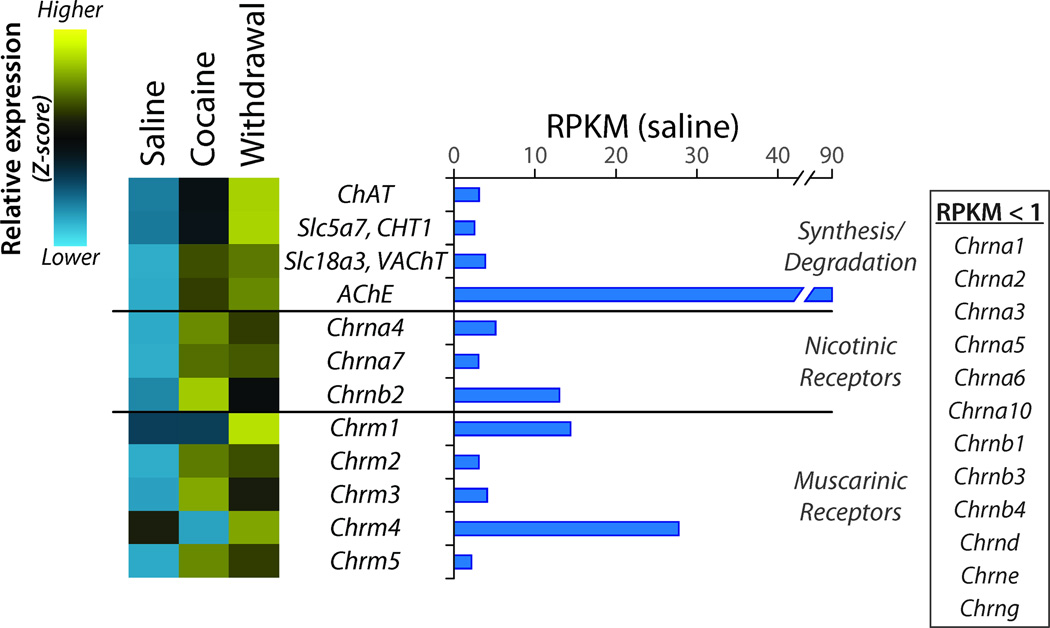
A small population of giant aspiny cholinergic interneurons is found in the NAc (Threlfell and Cragg, 2011); expression of both the plasma membrane choline transporter (Cht1), which provides the rate-limiting substrate, and choline acetyltransferase (ChAT) was increased by Cocaine and further increased by Withdrawal (Figure 7). Expression of each of the major nicotinic ACh receptor subunits found in the NAc (Chrna4, Chrna7, and Chrnb2) and three muscarinic ACh receptors (Chrm2, Chrm3, and Chrm5) showed sustained increases, many verified by qPCR.
Figure 7. Regulation of Genes Affecting Cholinergic Transmission.
Data were analyzed as described for Figure 4. Although only a small percentage of the neurons in the NAc are cholinergic, choline acetyltransferase (ChAT), high affinity plasma membrane choline transporter (Slc5a7) and vesicular acetylcholine transporter (Slc18a3) transcript levels all fall above the 1 RPKM cutoff. Only three of the many nicotinic acetylcholine receptor subunits are expressed in the NAc while all five muscarinic receptors are expressed. Expression of each of the genes involved in cholinergic transmission is elevated in withdrawal.
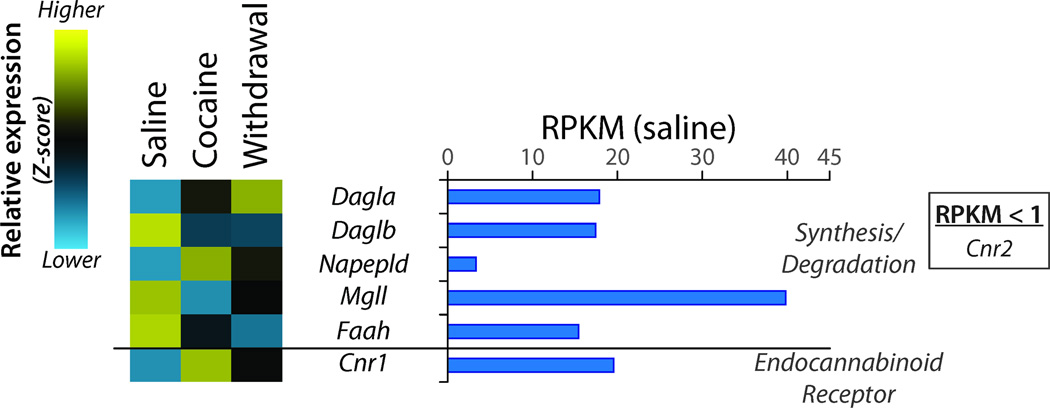
The endocannabinoid system is heavily involved in many reward-seeking responses to drugs of abuse (Orio et al., 2009). Levels of transcripts encoding Cnr1 (the CB1 endocannabinoid receptor) and the cannabinoid synthetic enzymes Dagla and Napepld showed sustained increases in Cocaine and Withdrawal, while Daglb showed a sustained decrease (Figure 8). The effect of Cocaine expression on each of these genes was confirmed by qPCR analysis of two separate sets of animals.
Figure 8. Regulation of Genes Affecting Endocannabinoid Signaling.
RPKM data were analyzed as described for Figure 4. While transcripts encoding the two enzymes that synthesize the endocannabinoid 2-arachidonoylglycerol (diacylglycerol lipase α and β; Dagla and Daglb) are expressed at similar levels in the NAc, they respond to cocaine in an opposite manner. Expression of transcripts encoding cannabinoid receptor 1 (Cnr1) greatly exceeds that of cannabinoid receptor 2 (Cnr2) and is responsive to cocaine.
Discussion
Single-gene, proteomic, and microarray analyses demonstrated that cocaine produces widespread, multi-faceted responses in the NAc (McClung and Nestler, 2008; Eipper-Mains et al., 2011). Next-generation sequencing identified pathways not previously thought to play any role in addiction and facilitated systematic cataloging of changes in all components of known signaling pathways. With greater sequencing depth, changes in editing, splicing, imprinting and allele-specific expression can be quantified (Mortazavi et al., 2008; Wang et al., 2008a; Wang et al., 2008b; Wahlstedt et al., 2009; Wang et al., 2009; Metzker, 2010; Eipper-Mains et al., 2011). In situ hybridization and immunostaining will be required to localize changes in gene expression to specific cell types in the core and shell regions of the NAc. Components of the Wnt, cadherin, and heterotrimeric G-protein signaling pathways were over-represented among cocaine-responsive transcripts; cross-talk between these signaling cascades is extensive (Force et al., 2007).
Global comparisons of gene expression identify signaling pathways targeted by cocaine
Cadherin/Wnt pathways
By sequencing PSD-localized, cocaine-regulated striatal microRNAs, we previously identified miR-8 family members as key players in the response to cocaine (Eipper-Mains et al., 2011). Although the miR-8 family has primarily been studied for its role in cancer progression and metastasis, we verified cocaine regulation of two bioinformatically-predicted cadherin family targets (Eipper-Mains et al., 2011). We identified over forty cocaine-regulated cadherin and protocadherin transcripts; genes from the Pcdhβ cluster were primarily up-regulated following cocaine in both a transient and sustained fashion (Clusters i-iii), whereas genes from the Pcdhγ cluster were exclusively found to undergo a sustained down-regulation following cocaine exposure (Cluster v).
A few studies have suggested a role for cell adhesion and extracellular matrix proteins in the response to cocaine. Disturbances in cortical cytoarchitecture in animal models of prenatal cocaine exposure resembled changes observed when Wnt/cadherin pathway function was altered; this lead to the prediction that these pathways play a role in the response to cocaine (Novikova et al., 2005). As observed here (Cluster i), Cdh6 and Cdh11 expression increased in the frontal cortex of E18 pups taken from dams treated with cocaine for 10 days (Novikova et al., 2005). Also contributing to alterations in the extracellular milieu is cocaine-mediated down-regulation of glycosyltransferase expression; these enzymes are crucial for the synthesis of cell surface glycoproteins. In a compilation documenting changes in gene expression in postmortem tissue from cocaine addicts, cell adhesion molecules were identified as major targets (Lehrmann et al., 2003; Mash et al., 2007).
Heterotrimeric G-protein pathways
Transcripts encoding GPCRs, heterotrimeric G-protein subunits, regulators of G-protein signaling (RGS), GPCR kinases (GRK) and adenylate cyclases (Adcy) responded differently to cocaine and withdrawal. Genes encoding GPCRs and RGSs were identified in all five clusters. Particularly striking was the prevalence of GPCRs for neuropeptides, with different receptor subtypes in different clusters (e.g., Wnt, galanin, somatostatin, and opioid receptors). The μ-opioid (Oprm1) receptor, associated with increased sensations of reward and pleasure, showed a sustained increase in expression in response to cocaine, as expected (Di Chiara and Imperato, 1988; Wee and Koob, 2010), as did the endocannabinoid (Cnr1) receptor (Adamczyk et al., 2012a; Adamczyk et al., 2012b). The σ-opioid receptor (Oprd1), also involved in positive reinforcement of drugs of abuse (Le Merrer et al., 2009), increased expression only in Withdrawal. The κ-opioid receptor (Oprk1), crucial to the negative emotional state of withdrawal (Wee and Koob, 2010), showed a sustained increase, as also observed in postmortem tissue from cocaine overdose victims (Mash and Staley, 1999). Transcripts encoding several neuropeptides were substantially down-regulated by cocaine: Pomc (proopiomelanocortin),Tac1 (tachykinin), Oxt (oxytocin) and Cartpt (CART; cocaine and amphetamine regulated peptide).
Consistent with a major role for cadherins in the response to cocaine, seven transmembrane cadherins (Celsr) were identified in clusters i and ii. Genes encoding adenylate cyclases appeared in Clusters i, ii, and iii. On the other hand, the majority of genes encoding the β- and γ-subunits of the heterotrimeric G-proteins fell into cluster v (sustained decrease). The few genes whose expression is uniquely regulated in Withdrawal are of special interest; these transcripts may provide insight into the biochemical and cell biological alterations underlying reinstatement of drug-seeking behavior, ultimately helping identify potential therapeutic approaches to prevent relapse (Wolf, 2010).
Neurotransmitter-specific effects
Expression of the D3 dopamine receptor (Drd3) increased in response to Cocaine and remained elevated after Withdrawal (cluster i); in contrast, D2 dopamine receptor (Drd2) expression increased only after Withdrawal. An increase in D3 receptor expression was observed in the NAc of cocaine overdose victims (Mash and Staley, 1999) and in rats 45 days after cocaine self-administration ceased (Conrad et al., 2010). Recent work suggests that dopamine receptor dysregulation, mediated by decreased D2 and increased D3 receptor expression, contributes to increased cocaine-seeking behavior after prolonged withdrawal (Conrad et al., 2010). Alterations in serotonin signaling are also recognized as important in drug addiction (Dreyer, 2010), and we observed cocaine-responsive changes in gene expression across many of the serotonin receptors.
Expression of many of the genes responsible for GABA synthesis, transport into synaptic vesicles, and retrieval from the synaptic cleft rose following cocaine treatment and increased further during withdrawal. Daily cocaine injections decreased GABAA receptor-mediated maximal evoked currents and miniature inhibitory postsynaptic currents on dopamine neurons in the VTA (Liu et al., 2005); furthermore, GABAA receptor function has been implicated in dopamine-mediated alcohol reward (Heilig et al., 2011). GABAB receptor agonists attenuated cocaine reinforcement and reduced cocaine craving in a study of human cocaine users (Brebner et al., 2002); while the precise mechanism of these behavioral effects is unknown, activation of GABAB receptors in the NAc reduces dopamine release from dopaminergic projections from the VTA (Erhardt et al., 2002). Changes in GABAA receptor subunit expression, which fell during withdrawal, and in GABAB receptor expression, which rose, could contribute significantly to altered synaptic transmission.
A growing body of literature has focused on the role of AMPA, NMDA and kainate receptor-mediated glutamatergic transmission into the NAc in the development of addiction and addictive-like behaviors (Kalivas et al., 2009). Increased glutamate receptor mRNA expression parallels increases in cell surface expression of GluR1-containing AMPA receptors following cocaine withdrawal (Boudreau et al., 2007), along with a specific population of GluR2-lacking calcium-permeable AMPA receptors (Wolf, 2010). We saw sustained up-regulation of Gria1 (GluR1) transcripts, a transient increase in Gria4 (GluR4) transcripts, and no change in Gria2 (GluR2) transcripts. While expression of kainate receptor 3 (Grik3) rose substantially, expression of Grik4 and Grik5 fell. NMDA receptors are critical for the behavioral response to drugs of abuse (Lee et al., 2010; Pascoli et al., 2011); new spines appearing in the NAc following cocaine are enriched in NMDA receptors and are not formed if NMDA receptor currents are blocked (Huang et al., 2009; Ren et al., 2010). Correspondingly, transcripts encoding several NMDA receptor subunits rose in response to cocaine. Expression of the more prevalent metabotropic glutamate receptors also increased. Perhaps the most significant finding is that expression of Slc17a7 and Slc17a6 (Vglut1 and Vglut2) exhibited sustained down-regulation after cocaine treatment and withdrawal.
Cholinergic transmission in the NAc contributes to addiction through the modulation of dopamine signaling and reward processing (Mark et al., 2011). Increased expression of both choline acetyltransferase and the plasma membrane choline transporter in the giant aspiny cholinergic interneurons could increase cholinergic signaling at both nicotinic and muscarinic ACh receptors expressed on the GABAergic output neurons.
Conclusion
We used next-generation sequencing to generate a comprehensive catalog of transcriptional changes that occur in response to cocaine treatment and withdrawal in the primary reward center of the brain. Pathway analysis enabled the complete categorization of transcriptional changes in signaling pathways known to be important in addiction along with the identification of pathways not previously considered important. These data should serve as a resource for development of new targets for cocaine therapy and facilitate selection of protein interaction and pathway targets for future study. With more information about their normal physiological roles, the extracellular domains of the many cadherins and protocadherins that respond to cocaine may be useful therapeutic targets. Pharmaceutical cocktails targeted to a subset of the specific GPCRs whose expression is altered during withdrawal may prove to be more effective than treatments targeting a single pathway.
Supplementary Material
Acknowledgements
We thank Darlene D’Amato for indefatigable laboratory support. This work was supported by National Institutes of Health Grants DA-15464 and DA-23082 (to BAE and REM), DA-26706 (to DDK), and GM-62516 (to BRG).
Abbreviation list
- NAc
nucleus accumbens
- PCDH
protocadherin
- PSD
postsynaptic density
- qPCR
quantitative polymerase chain reaction
- RGS
regulator of G-protein signaling
- RPKM
reads per kilobase gene model per million mapped reads
Footnotes
None of the authors have any conflicts of interest.
References
- Adamczyk P, Faron-Górecka A, Kuśmider M, Dziedzicka-Wasylewska M, Papp M, Filip M. Long-lasting increase in [(3)H]CP55,940 binding to CB1 receptors following cocaine self-administration and its withdrawal in rats. Brain Res. 2012a doi: 10.1016/j.brainres.2012.02.052. [DOI] [PubMed] [Google Scholar]
- Adamczyk P, Miszkiel J, McCreary AC, Filip M, Papp M, Przegaliński E. The effects of cannabinoid CB1, CB2 and vanilloid TRPV1 receptor antagonists on cocaine addictive behavior in rats. Brain Res. 2012b;1444:45–54. doi: 10.1016/j.brainres.2012.01.030. [DOI] [PubMed] [Google Scholar]
- Beenhakker MP, Huguenard JR. Astrocytes as gatekeepers of GABAB receptor function. J Neurosci. 2010;30:15262–15276. doi: 10.1523/JNEUROSCI.3243-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolteus AJ, Bordey A. GABA release and uptake regulate neuronal precursor migration in the postnatal subventricular zone. J Neurosci. 2004;24:7623–7631. doi: 10.1523/JNEUROSCI.1999-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau AC, Reimers JM, Milovanovic M, Wolf ME. Cell surface AMPA receptors in the rat nucleus accumbens increase during cocaine withdrawal but internalize after cocaine challenge in association with altered activation of mitogen-activated protein kinases. J Neurosci. 2007;27:10621–10635. doi: 10.1523/JNEUROSCI.2163-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brebner K, Childress AR, Roberts DCS. A potential role for GABA(B) agonists in the treatment of psychostimulant addiction. Alcohol Alcohol. 2002;37:478–484. doi: 10.1093/alcalc/37.5.478. [DOI] [PubMed] [Google Scholar]
- Brooks AN, Yang L, Duff MO, Hansen KD, Park JW, Dudoit S, Brenner SE, Graveley BR. Conservation of an RNA regulatory map between Drosophila and mammals. Genome Res. 2011;21:193–202. doi: 10.1101/gr.108662.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad KL, Ford K, Marinelli M, Wolf ME. Dopamine receptor expression and distribution dynamically change in the rat nucleus accumbens after withdrawal from cocaine self-administration. Neuroscience. 2010;169:182–194. doi: 10.1016/j.neuroscience.2010.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Chiara G, Imperato A. Opposite effects of mu and kappa opiate agonists on dopamine release in the nucleus accumbens and in the dorsal caudate of freely moving rats. J Pharmacol Exp Ther. 1988;244:1067–1080. [PubMed] [Google Scholar]
- Dreyer J-L. New insights into the roles of microRNAs in drug addiction and neuroplasticity. Genome Med. 2010;2:92. doi: 10.1186/gm213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eipper-Mains JE, Eipper BA, Mains RE. Global Approaches to the Role of miRNAs in Drug-Induced Changes in Gene Expression. Front Genet. 2012;3:109. doi: 10.3389/fgene.2012.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eipper-Mains JE, Kiraly DD, Palakodeti D, Mains RE, Eipper BA, Graveley BR. microRNA-Seq reveals cocaine-regulated expression of striatal microRNAs. RNA. 2011;17:1529–1543. doi: 10.1261/rna.2775511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Mestikawy S, Wallén-Mackenzie Å, Fortin GM, Descarries L, Trudeau L-E. From glutamate co-release to vesicular synergy: vesicular glutamate transporters. Nat Rev Neurosci. 2011;12:204–216. doi: 10.1038/nrn2969. [DOI] [PubMed] [Google Scholar]
- Erhardt S, Mathé JM, Chergui K, Engberg G, Svensson TH. GABA(B) receptor-mediated modulation of the firing pattern of ventral tegmental area dopamine neurons in vivo. Naunyn Schmiedebergs Arch Pharmacol. 2002;365:173–180. doi: 10.1007/s00210-001-0519-5. [DOI] [PubMed] [Google Scholar]
- Force T, Woulfe K, Koch WJ, Kerkelä R. Molecular scaffolds regulate bidirectional crosstalk between Wnt and classical seven-transmembrane-domain receptor signaling pathways. Sci STKE. 2007;2007:pe41. doi: 10.1126/stke.3972007pe41. [DOI] [PubMed] [Google Scholar]
- Heilig M, Goldman D, Berrettini W, O'Brien CP. Pharmacogenetic approaches to the treatment of alcohol addiction. Nat Rev Neurosci. 2011;12:670–684. doi: 10.1038/nrn3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann HM, Nadal R, Vignes M, Ortiz J. Chronic cocaine self-administration modulates ERK1/2 and CREB responses to dopamine receptor agonists in striatal slices. Addict Biol. 2012;17:565–575. doi: 10.1111/j.1369-1600.2011.00353.x. [DOI] [PubMed] [Google Scholar]
- Huang YH, Lin Y, Mu P, Lee BR, Brown TE, Wayman G, Marie H, Liu W, Yan Z, Sorg BA, Schlüter OM, Zukin RS, Dong Y. In vivo cocaine experience generates silent synapses. Neuron. 2009;63:40–47. doi: 10.1016/j.neuron.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman SE, Malenka RC, Nestler EJ. Neural mechanisms of addiction: the role of reward-related learning and memory. Annu Rev Neurosci. 2006;29:565–598. doi: 10.1146/annurev.neuro.29.051605.113009. [DOI] [PubMed] [Google Scholar]
- Kalivas PW. The glutamate homeostasis hypothesis of addiction. Nat Rev Neurosci. 2009;10:561–572. doi: 10.1038/nrn2515. [DOI] [PubMed] [Google Scholar]
- Kalivas PW, Volkow ND. New medications for drug addiction hiding in glutamatergic neuroplasticity. Mol Psychiatry. 2011;16:974–986. doi: 10.1038/mp.2011.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalivas PW, Lalumiere RT, Knackstedt L, Shen H. Glutamate transmission in addiction. Neuropharmacology. 2009;56 Suppl 1:169–173. doi: 10.1016/j.neuropharm.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauer JA, Malenka RC. Synaptic plasticity and addiction. Nat Rev Neurosci. 2007;8:844–858. doi: 10.1038/nrn2234. [DOI] [PubMed] [Google Scholar]
- Kemp JM, Powell TP. The structure of the caudate nucleus of the cat: light and electron microscopy. Philos Trans R Soc Lond B Biol Sci. 1971;262:383–401. doi: 10.1098/rstb.1971.0102. [DOI] [PubMed] [Google Scholar]
- Kiraly DD, Ma X-M, Mazzone CM, Xin X, Mains RE, Eipper BA. Behavioral and morphological responses to cocaine require kalirin7. Biol Psychiatry. 2010;68:249–255. doi: 10.1016/j.biopsych.2010.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs JJ, Hara MR, Davenport CL, Kim J, Lefkowitz RJ. Arrestin development: emerging roles for beta-arrestins in developmental signaling pathways. Dev Cell. 2009;17:443–458. doi: 10.1016/j.devcel.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Merrer J, Becker JAJ, Befort K, Kieffer BL. Reward processing by the opioid system in the brain. Physiol Rev. 2009;89:1379–1412. doi: 10.1152/physrev.00005.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M-C, Yasuda R, Ehlers MD. Metaplasticity at single glutamatergic synapses. Neuron. 2010;66:859–870. doi: 10.1016/j.neuron.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrmann E, Oyler J, Vawter MP, Hyde TM, Kolachana B, Kleinman JE, Huestis MA, Becker KG, Freed WJ. Transcriptional profiling in the human prefrontal cortex: evidence for two activational states associated with cocaine abuse. Pharmacogenomics J. 2003;3:27–40. doi: 10.1038/sj.tpj.6500146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q-s, Pu L, Poo M-m. Repeated cocaine exposure in vivo facilitates LTP induction in midbrain dopamine neurons. Nature. 2005;437:1027–1031. doi: 10.1038/nature04050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lull ME, Freeman WM, Vrana KE, Mash DC. Correlating human and animal studies of cocaine abuse and gene expression. Ann N Y Acad Sci. 2008;1141:58–75. doi: 10.1196/annals.1441.013. [DOI] [PubMed] [Google Scholar]
- Mark GP, Shabani S, Dobbs LK, Hansen ST. Cholinergic modulation of mesolimbic dopamine function and reward. Physiol Behav. 2011;104:76–81. doi: 10.1016/j.physbeh.2011.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mash DC, Staley JK. D3 dopamine and kappa opioid receptor alterations in human brain of cocaine-overdose victims. Ann N Y Acad Sci. 1999;877:507–522. doi: 10.1111/j.1749-6632.1999.tb09286.x. [DOI] [PubMed] [Google Scholar]
- Mash DC, Ffrench-Mullen J, Adi N, Qin Y, Buck A, Pablo J. Gene expression in human hippocampus from cocaine abusers identifies genes which regulate extracellular matrix remodeling. PLoS One. 2007;2:e1187. doi: 10.1371/journal.pone.0001187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung CA, Nestler EJ. Neuroplasticity mediated by altered gene expression. Neuropsychopharmacology. 2008;33:3–17. doi: 10.1038/sj.npp.1301544. [DOI] [PubMed] [Google Scholar]
- Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- Morishita H, Yagi T. Protocadherin family: diversity, structure, and function. Curr Opin Cell Biol. 2007;19:584–592. doi: 10.1016/j.ceb.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Nestler EJ. Is there a common molecular pathway for addiction? Nat Neurosci. 2005a;8:1445–1449. doi: 10.1038/nn1578. [DOI] [PubMed] [Google Scholar]
- Nestler EJ. The neurobiology of cocaine addiction. Sci Pract Perspect. 2005b;3:4–10. doi: 10.1151/spp05314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikova SI, He F, Bai J, Lidow MS. Neuropathology of the cerebral cortex observed in a range of animal models of prenatal cocaine exposure may reflect alterations in genes involved in the Wnt and cadherin systems. Synapse. 2005;56:105–116. doi: 10.1002/syn.20134. [DOI] [PubMed] [Google Scholar]
- Orio L, Edwards S, George O, Parsons LH, Koob GF. A role for the endocannabinoid system in the increased motivation for cocaine in extended-access conditions. J Neurosci. 2009;29:4846–4857. doi: 10.1523/JNEUROSCI.0563-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascoli V, Besnard A, Hervé D, Pagès C, Heck N, Girault J-A, Caboche J, Vanhoutte P. Cyclic adenosine monophosphate-independent tyrosine phosphorylation of NR2B mediates cocaine-induced extracellular signal-regulated kinase activation. Biol Psychiatry. 2011;69:218–227. doi: 10.1016/j.biopsych.2010.08.031. [DOI] [PubMed] [Google Scholar]
- Pierce RC, Bell K, Duffy P, Kalivas PW. Repeated cocaine augments excitatory amino acid transmission in the nucleus accumbens only in rats having developed behavioral sensitization. J Neurosci. 1996;16:1550–1560. doi: 10.1523/JNEUROSCI.16-04-01550.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Z, Sun WL, Jiao H, Zhang D, Kong H, Wang X, Xu M. Dopamine D1 and N-methyl-D-aspartate receptors and extracellular signal-regulated kinase mediate neuronal morphological changes induced by repeated cocaine administration. Neuroscience. 2010;168:48–60. doi: 10.1016/j.neuroscience.2010.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders BT, Robinson TE. Individual variation in the motivational properties of cocaine. Neuropsychopharmacology. 2011;36:1668–1676. doi: 10.1038/npp.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schousboe A, Madsen KK, White HS. GABA transport inhibitors and seizure protection: the past and future. Future Med Chem. 2011;3:183–187. doi: 10.4155/fmc.10.288. [DOI] [PubMed] [Google Scholar]
- Schousboe A, Sarup A, Larsson OM, White HS. GABA transporters as drug targets for modulation of GABAergic activity. Biochem Pharmacol. 2004;68:1557–1563. doi: 10.1016/j.bcp.2004.06.041. [DOI] [PubMed] [Google Scholar]
- Threlfell S, Cragg SJ. Dopamine signaling in dorsal versus ventral striatum: the dynamic role of cholinergic interneurons. Front Syst Neurosci. 2011;5:11. doi: 10.3389/fnsys.2011.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzingounis AV, Wadiche JI. Glutamate transporters: confining runaway excitation by shaping synaptic transmission. Nat Rev Neurosci. 2007;8:935–947. doi: 10.1038/nrn2274. [DOI] [PubMed] [Google Scholar]
- Uemura M, Nakao S, Suzuki ST, Takeichi M, Hirano S. OL-Protocadherin is essential for growth of striatal axons and thalamocortical projections. Nat Neurosci. 2007;10:1151–1159. doi: 10.1038/nn1960. [DOI] [PubMed] [Google Scholar]
- Wahlstedt H, Daniel C, Ensterö M, Ohman M. Large-scale mRNA sequencing determines global regulation of RNA editing during brain development. Genome Res. 2009;19:978–986. doi: 10.1101/gr.089409.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W-X, Wilfred BR, Baldwin DA, Isett RB, Ren N, Stromberg A, Nelson PT. Focus on RNA isolation: obtaining RNA for microRNA (miRNA) expression profiling analyses of neural tissue. Biochim Biophys Acta. 2008a;1779:749–757. doi: 10.1016/j.bbagrm.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Sun Q, McGrath SD, Mardis ER, Soloway PD, Clark AG. Transcriptome-wide identification of novel imprinted genes in neonatal mouse brain. PLoS One. 2008b;3:e3839. doi: 10.1371/journal.pone.0003839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee S, Koob GF. The role of the dynorphin-kappa opioid system in the reinforcing effects of drugs of abuse. Psychopharmacology (Berl) 2010;210:121–135. doi: 10.1007/s00213-010-1825-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf ME. The Bermuda Triangle of cocaine-induced neuroadaptations. Trends Neurosci. 2010;33:391–398. doi: 10.1016/j.tins.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf ME, Ferrario CR. AMPA receptor plasticity in the nucleus accumbens after repeated exposure to cocaine. Neurosci Biobehav Rev. 2010;35:185–211. doi: 10.1016/j.neubiorev.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.