Abstract
Chitin, an insoluble polymer of GlcNAc, is an abundant source of carbon, nitrogen, and energy for marine microorganisms. Microarray expression profiling and mutational studies of Vibrio cholerae growing on a natural chitin surface, or with the soluble chitin oligosaccharides (GlcNAc)2–6, GlcNAc, or the glucosamine dimer (GlcN)2 identified three sets of differentially regulated genes. We show that (i) ChiS, a sensor histidine kinase, regulates expression of the (GlcNAc)2–6 gene set, including a (GlcNAc)2 catabolic operon, two extracellular chitinases, a chitoporin, and a PilA-containing type IV pilus, designated ChiRP (chitin-regulated pilus) that confers a significant growth advantage to V. cholerae on a chitin surface; (ii) GlcNAc causes the coordinate expression of genes involved with chitin chemotaxis and adherence and with the transport and assimilation of GlcNAc; (iii) (GlcN)2 induces genes required for the transport and catabolism of nonacetylated chitin residues; and (iv) the constitutively expressed MSHA pilus facilitates adhesion to the chitin surface independent of surface chemistry. Collectively, these results provide a global portrait of a complex, multistage V. cholerae program for the efficient utilization of chitin.
The agent of Asiatic cholera, Vibrio cholerae O1, causes a dehydrating diarrheal illness and sometimes death. However, outside the human host, V. cholerae is a normal member of natural aquatic environments such as lakes, rivers, estuaries, and the ocean, which serve as the principal reservoir for this organism in nature (1). How it survives in habitats of this kind and the mechanisms by which it periodically emerges as a human pathogen are compelling questions in the ecology of infectious diseases (2). Observational studies in Bangladesh epidemiologically link seasonal phytoplankton and zooplankton blooms with cholera outbreaks. Studies of this epidemiological association identified V. cholerae attached to plankton in the environment, an observation that was confirmed by microcosm coinfection experiments. The interaction of V. cholerae with zooplankton is particularly intriguing because copepods, a subclass of crustacean zooplankton, have been incriminated in the transmission of this agent from aquatic reservoirs to susceptible human hosts (3).
Chitin, composed of β1,4-linked GlcNAc residues, is the most abundant polysaccharide in nature after cellulose. In the aquatic biosphere alone, >1011 metric tons of chitin are produced annually. This vast amount of insoluble, carbon-containing material is recycled mainly by chitinolytic bacteria, including members of the family Vibrionaceae. Chitin is found throughout all kingdoms and is the main component of the cell walls of fungi and of the exoskeletons of crustaceans. Copepods are estimated to produce billions of tons of chitin each year (4).
Many Vibrio species that live in aquatic environments are capable of using chitin as the sole carbon source. Studies of the nonpathogenic marine organism Vibrio furnissii have shown that chitin utilization is a complex process involving at least three steps: chitin sensing, attachment, and degradation (4). However, fundamental questions remain about the expression and regulation of genes involved in chitin utilization and the mechanism by which Vibrios attach to and stably colonize chitin surfaces.
Here we report microarray expression profiling studies of V. cholerae growing on a natural chitin surface. Genes within the chitin-regulated expression profile were selectively disrupted and the mutants used to functionally characterize the role of particular proteins in the efficient colonization and assimilation of chitin. The genome-wide portrait that emerges from the study of this nutritional phenotype has led to a model of the V. cholerae chitin utilization program.
Materials and Methods
Microarray Experiments. RNA was isolated by using Trizol reagent (GIBCO/BRL), treated with DNaseI (Ambion), and cleaned by using the RNeasy kit (Qiagen, Valencia, CA). Labeling of cDNA and microarray hybridizations were performed as described (5). Experiments with soluble substrates were repeated twice, and crab experiments were repeated five times. Duplicate hybridizations were performed; RNA from planktonic or noninduced bacteria was labeled with Cy3 and crab-shell attached or induced bacterial RNA was labeled with Cy5. Microarrays were scanned with a GenePix 400A instrument (Axon Instruments), using the genepix 3.0 software. To avoid fluctuations in intensity values from genes that are not expressed to a measurable level, we designated a minimum background level for each channel (5). Statistically significant changes in gene expression were identified by conducting a one-class analysis using the significance analysis of microarrays program (6) with a threshold of 2.5-fold change and a 0% false discovery rate. Hierarchical clustering was performed by using the average relative intensity values for each experiment and the cluster and treeview programs (7). Average relative expression values of each gene after adjustment of background values are shown in Table 2, which is published as supporting information on the PNAS web site. Raw data are available at http://genome-www5.stanford.edu.
Competition Experiments. Overnight cultures were diluted 1:10,000 and mixed in 1:1 ratio in 50% artificial seawater (crab experiments) or M9 media (batch culture experiments). For crab experiments, after 3 days, static incubation at 30°C the crab shell was rinsed and vortexed in fresh media. Cells were enumerated on MacConkey plates supplemented with sucrose. For batch culture experiments, after 24 h [(GlcNAc)4] or 48 h (colloidal chitin) the cultures were plated and enumerated.
Relative fitness (w) of pilA to wild type was calculated as:
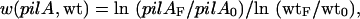 |
where pilA0 and wt0 are the initial CFU's and pilAF and wtF are the CFU's at the end of the experiment.
Quantitative Real-Time RT-PCR (qRT-PCR). Reactions were done by using the TaqMan system and an ABI Prism 7000 Sequence Detection System (Applied Biosystems). For details see Supporting Text, which is published as supporting information on the PNAS web site.
Attachment to Chitin Beads. V. cholerae expressing GFP were incubated in M9 medium containing 0.5% lactate and 0.6 mM (GlcNAc)6 for 2 h at 30°C, (GlcNAc)6 was added again, and the culture was incubated for 1.5 h. Bacteria were harvested, washed, incubated with chitin beads (New England Biolabs) in 50% artificial seawater media for 1 h, and washed twice in fresh media, and the beads were fixed in 2% formaldehyde (75 mM NaPO4-buffer/2.5 mM NaCl, pH 7.4). The number of attached bacteria was counted for 40 beads of similar size per experiment with a Zeiss Axiophot fluorescence microscope equipped with an HBO 100-W lamp. Images were acquired with a Nikon Eclipse TE300 microscope equipped for confocal microscopy with Bio-Rad MRC1000/MRC1024. The resulting z stacks were reconstructed in three dimensions by using volocity 2.0 software (Improvision, Conventry, U.K.). Bacterial strains, plasmids, and primers are listed in Tables 3 and 4, which are published as supporting information on the PNAS web site.
Results
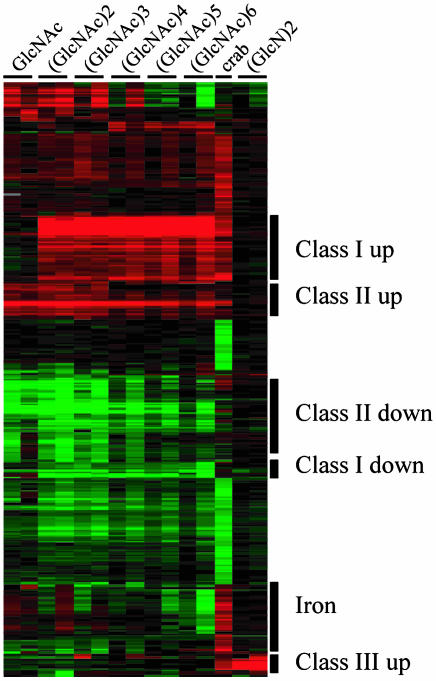
V. cholerae Expression Profiling Studies Identify Three Classes of Chitin-Regulated Genes. V. cholerae strain N16961 was incubated with a sterile crab shell fragment for 24 h, and total RNA was prepared from the crab shell-attached and free-swimming (planktonic) populations. These RNAs were labeled with Cy3 or Cy5 and applied to a DNA glass-spotted microarray representing ≈90% of the V. cholerae genome, and the differentially expressed genes were identified by statistical analysis of the fluorescence intensity values (6, 8). This analysis identified 104 genes whose expression was increased (shown in red) and 93 genes whose expression was reduced (shown in green) in the crab shell-attached compared to the planktonic population of bacteria (Fig. 1 and Fig. 6, which is published as supporting information on the PNAS web site).
Fig. 1.
Hierarchical clustering analysis of microarray expression data for 360 genes found to be significantly regulated during growth on the crab shell surface (crab) or with the soluble carbohydrates GlcNAc or (GlcNAc)n or (GlcN)2. The average fold induction values are depicted in two columns for GlcNAc, (GlcNAc)n, and (GlcN)2 (30- and 60-min time points), and in one column for the crab shell-associated population. Each of the genes is shown as a horizontal, colored stripe; individual genes are identified in Fig. 6. The locations of genes in each of the three response classes and those regulated by iron (see text) are labeled. The most intense red and green colors correspond to increased or decreased expression values of 5-fold or more, respectively.
Natural sources of chitin are insoluble composite materials containing chitin polysaccharide, protein, and minerals. Therefore, to identify chitin-regulated genes in the crab shell-attached expression profile, we performed expression profiling studies of planktonic V. cholerae exposed to the chitin monomer GlcNAc, to the soluble chitin oligosaccharides (GlcNAc)n (n = 2–6) and to the glucosamine dimer (chitosan dimer) (GlcN)2. Genes which were differentially regulated by these carbohydrates are also shown in Fig. 1.
Statistical analysis of the crab shell and soluble oligosaccharide expression profiles identified 360 differentially regulated genes (Figs. 1 and 6). When subjected to hierarchical cluster analysis, three response classes were evident.
Class I genes respond to chitin oligosaccharides, but not to GlcNAc. Within this class, exposure to chitin oligosaccharides increased the expression of 41 genes and reduced the expression of 9 (Fig. 1 and Fig. 7, which is published as supporting information on the PNAS web site).
Class II genes respond to GlcNAc. However, some of these genes also are induced by chitin oligosaccharides, an effect we attribute to the progressive degradation of chitin oligosaccharides over time, leading to the production of GlcNAc. Class II genes consist of 22 up-regulated and 51 down-regulated genes (Fig. 1 and Fig. 8, which is published as supporting information on the PNAS web site).
Class III genes are regulated by (GlcN)2, a group that contains only an up-regulated cluster of seven contiguous genes (Fig. 1 and Fig. 9, which is published as supporting information on the PNAS web site).
Many genes found to be differentially up-regulated during attachment to crab shell were not assigned to classes I, II, or III. Among these are genes encoding ribosomal proteins and translation factors, indicating that bacteria attached to the crab shell surface either grow more efficiently or are more metabolically active than bacteria in the planktonic compartment, and genes that direct the uptake and assimilation of iron (Figs. 1 and 6), indicating that crab shell-attached bacteria experience iron limitation.
Two sets of crab shell-associated, down-regulated genes were identified. Within each of these are genes encoding chemotaxis proteins. Thus, planktonic bacteria, which are suspended in a carbon-poor milieu, seem to sense and move toward the nutrient-rich chitin surface.
qRT-PCR assays of 10 genes (selected to include both chitin-regulated and chitin nonregulated genes) was used to measure transcript levels for V. cholerae grown with and without GlcNAc, (GlcNAc)2, or (GlcN)2 and in the planktonic compared to the crab shell-attached population of bacteria (Table 5, which is published as supporting information on the PNAS web site). The qRT-PCR results confirmed microarray expression results, although qRT-PCR showed higher induction values for the most strongly up-regulated genes.
The Chitin Catabolic Cascade Is Controlled by a Two-Component Sensor Histidine Kinase, ChiS. Li and Roseman (9) recently identified ChiS, a membrane-bound chitin-sensing histidine kinase in V. furnissi and V. cholerae. ChiS is encoded by VC0622, which is located upstream of a chitin-induced (GlcNAc)2 catabolic operon (VC0620–0611). Inactivation of VC0622 was found to reduce extracellular chitinase activity.
To determine whether ChiS is required for the differential regulation of genes in the V. cholerae chitin utilization program, VC0622 was disrupted and expression profiling experiments conducted with the wild type and mutant to identify genes whose (GlcNAc)2-regulated expression requires ChiS. Analysis of the resulting expression data (Fig. 2) showed that the (GlcNAc)2-dependent induction of most genes in the class I chitin-specific gene cluster requires ChiS. By contrast, the ChiS mutant responds to GlcNAc in a manner similar to wild-type bacteria. In view of these results, the ChiS-dependent genes are operationally defined to comprise the V. cholerae ChiS regulon. However, the putative cognate response regulator, “ChiR,” remains unidentified.
Fig. 2.
Expression profiles of class I induced genes. The upper row compares expression of genes in the wild-type strain after induction by (GlcNAc)2 to expression of genes in the chiS mutant after induction by (GlcNAc)2 for 30 min (chiS). The expression of genes with values depicted in green in this row are reduced in the chiS mutant compared to the wild-type strain. The lower two rows show genes (in red) that are induced in the wild-type strain after treatment with (GlcNAc)2 for 30 min or induced in the crab shell-attached population.
Functional Predictions and Chromosomal Localization of ChiS Regulon Genes. Of the 41 genes that are specifically up-regulated by (GlcNAc)2–6, 12 are annotated to encode proteins of unknown function. Five encode protein homologues of V. furnissii with biochemically defined functions: VC0972 (chitoporin) (10), VCA0027 (extracellular chitinase) (X.B.L. and S.R., unpublished data), VC0612 (N,N-diacetylchitobiose phosphorylase) (11), VC0613 (periplasmic β-N-acetylglucosminidase) (12) and VCA0700 (periplasmic chitodextrinase) (13). The functions of three V. cholerae proteins in the ChiS regulon have been biochemically defined: VC0614 (glucosamine-specific kinase) (14), VC0615 (cellobiase) (15), and VC1952 (extracellular chitinase ChiA-1) (16). Five genes encode an ABC type transporter (VC0620–0616) presumed to transport (GlcNAc)2 (9).
The V. cholerae genome is distributed between chromosome I (2.9 Mb) and chromosome II (1.07 Mb). A total of 61 of the 70 identified chitin oligosaccharide, GlcNAc, and (GlcN)2-induced genes are localized on chromosome I, pointing to the vital role this chromosome is likely to play in the survival of V. cholerae in chitin-containing aquatic habitats.
V. cholerae Produces Two Extracellular Chitinases and a Chitoporin. Microarray expression results show that crab shell chitin and the chitin oligosaccharides up-regulate, in a ChiS-dependent manner, three of the five predicted chitinase genes (VC1952, VC0769, and VCA0027) and the gene encoding chitodextrinase (VCA0700). The gene encoding a fourth predicted chitinase (VCA0811) was induced by both GlcNAc and chitin oligosaccharides, whereas expression of the fifth chitinase gene (VC1073) was unaffected by any of the tested carbohydrates (Table 2).
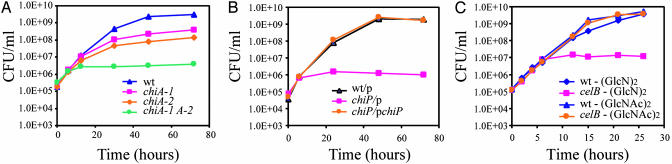
VC1952 was previously shown to encode an extracellular chitinase, ChiA-1 (16). To determine whether V. cholerae produces more than one extracellular chitinase, an in-frame deletion of chiA-1 was constructed yielding a mutant with reduced, but not abolished extracellular chitinase activity. To identify the source of the residual chitinase activity, we deleted chiA-2 (VCA0027), which is induced by chitin; the resulting mutant produced decreased extracellular chitinase activity as well (data not shown), indicating that ChiA-2 is a second V. cholerae extracellular chitinase. A double chiA-1/chiA-2 mutant was then prepared and found to grow poorly in a liquid medium supplemented with colloidal chitin (Fig. 3A). By contrast, it grew normally in a medium supplemented with GlcNAc (data not shown), showing that the growth phenotype of the double knockout is only evident when chitin provides the sole source of carbon. Taken together, these data show that V. cholerae possesses at least two extracellular chitinases, ChiA-1 and ChiA-2, and that both are regulated by chitin and chitin oligosaccharides in a ChiS-dependent manner.
Fig. 3.
Growth in media supplemented with colloidal chitin of chiA (A) and chiP (B) mutants, with or without a plasmid encoded chiP gene. (C) Growth of celB mutant in media supplemented with chitin disaccharide (GlcNAc)2 or chitosan disaccharide (GlcN)2.
Chitin oligosaccharides produced through the action of extracellular chitinases must cross the outer membrane to enter the periplasmic space. VC0972, which encodes a protein that is moderately homologous (40% identity, 66% similarity) to a previously characterized chitin-specific porin of V. furnissi (10), was found to be powerfully induced by crab shell-attached bacteria and by chitin oligosaccharides in a ChiS-dependent manner. To determine whether VC0972 also encodes a chitoporin, it was disrupted and the mutant found to grow poorly in a liquid medium containing colloidal chitin. Normal growth was restored by complementation with a plasmid-encoded copy of VC0972 (Fig. 3B). By contrast, no growth defect was observed in a medium containing GlcNAc (data not shown). We conclude that V. cholerae produces a porin in response to chitin, which enables the bacteria to sense chitin oligosaccharides and to use chitin. We suggest that this porin be named ChiP by analogy to its homologue in V. furnissii.
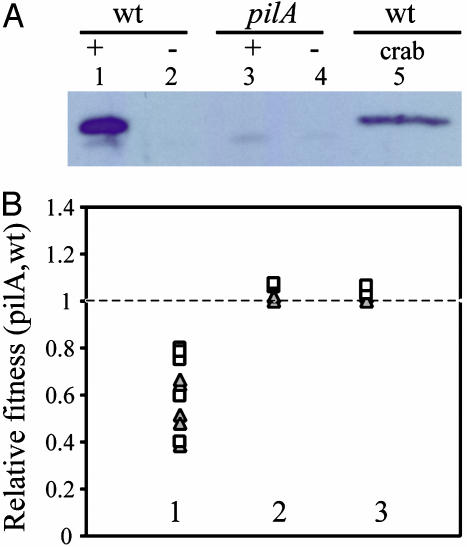
PilA, a Component of a Chitin-Regulated Pilus (ChiRP), Is Expressed by Crab Shell-Attached Bacteria and Contributes to the Colonization Process. V. cholerae produces two, functionally characterized type IV pili: the toxin coregulated pilus TCP (a virulence determinant); and the mannose-sensitive hemagglutinin MSHA (contributes to biofilm development). Chitin and chitin oligosaccharides were found to up-regulate, in a ChiS-dependent manner, 12 genes predicted to encode proteins involved in the biogenesis of a third type IV pilus (Fig. 7). These are distributed in three clusters of contiguous genes (VC0857–0861, VC2423–2426, and VC2630–2634) and one isolated gene (VC1612). One of these clusters contains a previously studied gene, pilA (VC2423), which encodes a type IV pilin of unknown function that is present in a cluster of four ORFs, pilABCD, that resembles other type IV pilus assembly operons (17). Further inspection of the 12 chitin-regulated type IV pilus biogenic genes led to the identification of two, pilB (VC2424) and VC2634, which are predicted to encode proteins with ATPase domains and which therefore might energize pilus extrusion and retraction. qRT-PCR assays confirmed the induction of pilA, showing 17- and 54-fold increases in pilA transcript abundance in crab shell-attached and chitin oligosaccharide-treated bacteria, respectively. Western blot assays using an antiserum to a PilA-6× His fusion protein showed that (GlcNAc)6-treated V. cholerae produce an ≈15-kDa protein (Fig. 4A), corresponding in size to the predicted molecular mass of PilA (17). By contrast, no such protein was produced without the inducing oligosaccharide or by a pilA mutant, prepared as a complete in-frame deletion of pilA (Fig. 4A). Western blotting of samples isolated from crab shell-associated bacteria after 24-h incubation showed that the PilA protein is expressed by crab shell-associated bacteria (Fig. 4A).
Fig. 4.
PilA is expressed by crab shell-attached bacteria and contributes to fitness during the crab shell colonization process. (A) Western blot of planktonic bacteria (lanes 1–4) with (+) or without (-) inducer and of crab shell-attached bacteria (lane 5) using a PilA antiserum. (B) Relative fitness of a pilA mutant compared to the wild-type parent in the crab shell-attached population (lane 1), in a planktonic culture containing colloidal chitin (lane 2), and in a planktonic culture containing (GlcNAc)4 (lane 3). Each data point represents one experiment in which either the wild-type strain (triangles) or the pilA mutant strain (squares) was marked by a deletion of scrA.
The expression of PilA on the crab shell surface led us to explore the possibility that it might contribute to the colonization process. This question was addressed in competition experiments in which nearly equal numbers of the wild-type parent and pilA mutant were combined and then incubated with a crab shell fragment for 72 h. Then, the crab shell was rinsed and the bacteria was detached from the crab shell surface by vigorous vortex agitation and enumerated by viable plate counting. To differentiate the two strains, we marked the wild-type strain by a deletion of scrA that encodes a IIBC component of a sucrose-specific PTS transporter, allowing the wild-type strain to be differentiated from the pilA mutant by color on MacConkey-sucrose indicator plates. To ensure that the scrA deletion did not have any effect on the marked strain's ability to colonize a chitin surface, we also deleted scrA in the pilA mutant and competed this strain against the unmarked wild-type strain. Differences in viable plate counts between wild-type and mutant were expressed as a relative fitness value, in which values <1 indicate a defect in the competitiveness of the designated strain (18) (see Materials and Methods). Relative fitness values of the pilA mutant for the 10 experimental replicates range from 0.39 to 0.80 (Fig. 4B, lane 1), indicating that deletion of pilA was associated with defective crab shell colonization or survival. The fitness loss was not caused by the scrA deletion because we obtained similar relative fitness values independent of which strain had the marker. By contrast, in liquid cultures containing either colloidal chitin or (GlcNAc)4 as the sole carbon source (Fig. 4B, lanes 2 and 3, respectively), the relative fitness value of the pilA mutant was 1.0. Together, these results and the Western blot data depicted in Fig. 4A show that PilA expression is induced by chitin and contributes to fitness during the crab shell colonization process. Moreover, this fitness phenotype is surface-dependent because the pilA mutant has no fitness disadvantage during planktonic growth on a chitin substrate. In view of its role in chitin colonization and its ChiS-dependent regulation by chitin and chitin oligosaccharides, we propose that the PilA-containing pilus be designated ChiRP, for chitin-regulated pilus.
The chitin surface dependency of the PilA colonization phenotype led us to determine whether PilA mediates adherence of V. cholerae to chitin. The wild-type parent or pilA mutant, labeled with 3H, was separately incubated with chitin particles. In five experimental replicates, wild-type bacteria attached to chitin particles in greater numbers than the pilA mutant, but the effect of the mutation was small (1.6- to 3.3-fold difference, data not shown).
V. cholerae Uses Glucosamine (GlcN) in Areas of Nonacetylated Chitin and Expresses a (GlcN)2-Specific Transporter. The crab shell microarray expression data showed that crab shell-attached bacteria express a set of genes encoding a putative cellobiose PTS transporter (VC1281–83) (Fig. 9). However, these genes are not induced by chitin oligosaccharides and V. cholerae cannot use cellobiose as a carbon source, leaving unresolved the role of genes in the VC1281–83 cluster. The purified oligosaccharides used in this study are fully acetylated, whereas in some crab shell chitins there is one nonacetylated glucosamine residue, on average, for every six GlcNAc residues (19). We speculated that the strong induction of genes comprising the putative cellobiose PTS system in crab shell-attached bacteria might be due to the presence of nonacetylated GlcN resides. To address this possibility, microarray expression profiling experiments were conducted with a soluble, nonacetylated GlcN dimer [(GlcN)2] with the same β1–4 anomeric linkage found in N,N-diacetylchitobiose. Only seven genes were found to be induced by the addition of (GlcN)2 (Figs. 1 and 9); six of the induced genes are encompassed in the operon-like cluster that encodes the putative cellobiose PTS transporter (VC1281–86), and the remaining induced gene (VC1280) is located adjacent to it, but transcribed in the opposite direction. We speculated that the substrate for the PTS transporter encoded by this operon is (GlcN)2, rather than cellobiose. To test this idea, we deleted part of the gene encoding the IIC component (celB, VC1282) and tested the mutant's capacity to use (GlcN)2. Comparison of the wild-type parent and celB mutant showed that the mutant exhibits a growth defect in a medium supplemented with (GlcN)2, whereas both strains grow equally well when the medium is supplemented with (GlcNAc)2 (Fig. 3C). Complementation of the mutant with a plasmid encoded copy of celAB restored its ability to use (GlcN)2 (data not shown). These experiments demonstrate that V. cholerae is capable of using (GlcN)2 and that this capacity requires the celB gene product.
GlcNAc Induces Expression of 22 Genes Including Genes Encoding Catabolic Enzymes, a Chemotaxis Protein, and a Chitin Adhesin. The addition of GlcNAc to a V. cholerae culture increased the expression of 22 genes (Figs. 1 and 8). Most of these genes are also up-regulated by chitin oligosaccharides and by crab shell-attached bacteria, an effect that we attribute to the release of GlcNAc from chitin oligosaccharides by the action of extracellular and/or periplasmic chitinases and β-N-acetylhexosaminidases. The expression of the 22 genes in this set does not require the ChiS protein (data not shown). Six of the 22 genes encode proteins, previously identified in V. furnissi, that are required for the uptake and metabolic conversion of GlcNAc, including NagC, NagA-1, NagB, and NagE, and enzyme I and HPr of a PTS transport system.
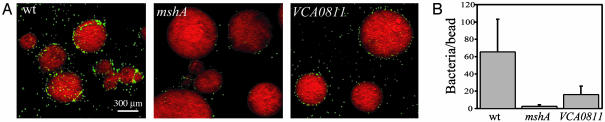
A putative chitinase (VCA0811) is also induced by GlcNAc. A search for conserved domains in VCA0811 identified a chitin-binding domain, but no glycosyl hydrolase domain, leading to the conjecture that it functions as a chitin adhesin. To address this hypothesis, VCA0811 was deleted, the mutant modified to express GFP, and tested for chitin adherence to chitin beads (Fig. 5A). The mutant attached 4-fold less than the wild-type parent (Fig. 5B). A similar result was recently reported by Kirn et al. with the classical biotype of V. cholerae O1.§ Thus, GlcNAc appears to induce a chitin adhesin, but the retention of residual chitin-binding capacity by the VCA0811 mutant indicates that the V. cholerae chitin-binding phenotype is multifactorial. GlcNAc also induced VC0449, encoding a methyl accepting chemotaxis protein; disruption of this gene reduced flagellar-dependent V. cholerae motility toward GlcNAc and (GlcNAc)2 on swarm plates (data not shown). This result and those described above suggest that GlcNAc coordinately regulates chemotaxis, adherence, and GlcNAc assimilation functions.
Fig. 5.
Attachment of GFP-labeled wild-type, mshA, and VCA0811 mutants to chitin beads. (A) Scanning confocal micrographs. (B) Average number of attached GFP-labeled bacteria.
MSHA Promotes Adherence to Chitin. mshA, encoding the principal repeating subunit of the type IV MSHA pilus, was not differentially regulated during growth of V. cholerae on the crab shell surface or in cultures containing GlcNAc, (GlcNAc)2–6 or (GlcN)2 (Tables 2 and 5). However, these data obtained during qRT-PCR experiments, showed that it was strongly, although not differentially, expressed by each growth condition. This result and recent studies showing that MSHA promotes biofilm development on borosilicate glass and adherence to zooplankton molts (20, 21) prompted us to determine whether it mediates adherence to a chitin surface. A GFP-labeled mshA mutant was found to attach ≈30-fold less to chitin beads than the GFP-labeled wild-type parent (Fig. 5).
V. cholerae Expresses Genes in the Chitin-Specific Gene Set When Associated with Living Copepods. V. cholerae has been found to be associated with copepods from natural aquatic habitats and to bind copepod molts in microcosm coinfection experiments (1). In view of these results, we used qRT-PCR to determine whether six genes in the chitin-induced gene set, one gene induced by GlcNAc, and one gene induced by (GlcN)2 were transcribed at a higher level in bacteria associated with living copepods compared to bacteria growing in a reference culture lacking chitin. Copepods of the species Tigriopus californicus, collected from Monterey Bay, CA, were incubated with V. cholerae for 6 h. Analysis of RNA from copepod-associated V. cholerae showed that four of the chitin-induced genes (VC0620, VC0972, VCA0027, and VCA0140) were transcribed at significantly higher levels than in the reference culture for each of three experiments (Table 1). The two remaining chitin-induced genes, VC1153 and VC2423, were expressed at higher levels in one of the three copepod experiments. Thus, genes in the chitin-induced gene set, found to be strongly induced by crab shell-associated bacteria 24 h after infection, are also induced in bacteria associated with living copepods 6 h after infection.
Table 1. qRT-PCR analysis of gene expression of copepod associated bacteria.
| Relative expression
|
|||
|---|---|---|---|
| Gene | Experiment 1 | Experiment 2 | Experiment 3 |
| VC0409 | 0.64 | 0.64 | 1.0 |
| VC0449 | 0.88 | 1.1 | 1.2 |
| VC0620 | 26 | 36 | 122 |
| VC0828 | 1.1 | 0.42 | 0.77 |
| VC0972 | 13 | 13 | 45 |
| VC1153 | 1.6 | 0.65 | 3.3 |
| VC1282 | 0.56 | 0.56 | 0.64 |
| VC2423 | 1.1 | 0.83 | 2.7 |
| VCA0027 | 5.3 | 9.5 | 22 |
| VCA0140 | 6.6 | 2.3 | 9.6 |
Discussion
A combination of microarray expression and genetic studies has disclosed previously unrecognized components of the V. cholerae chitin utilization program. New information includes the identification of genes belonging to the ChiS regulon; the coordinate regulation of GlcNAc-induced catabolic, adherence, and chemotaxis functions; and the identification of a previously uncharacterized gene cluster required for the assimilation of GlcN in regions of nonacetylated chitin. Taken together, these results provide a comprehensive portrait of a genetic program that governs the capacity of V. cholerae to sense, attach to, and degrade natural chitin surfaces in the aquatic biosphere.
Perhaps the most enigmatic and intriguing genes in the chitin-induced set are 12 that are predicted to direct the biogenesis and function of a type IV pilus, which we have designated ChiRP. Although located in four separate sites, the 12 genes in this group may function together as part of the same type IV pilus assembly complex because they are coordinately regulated by the same inducer (chitin oligosaccharides) and sensor (ChiS). pilA, which encodes a type IV pilin that we propose may comprise the principal repeating subunit of the ChiRP fiber, is located in a four-gene cluster, pilABCD. This cluster was previously studied by Fullner and Mekalanos (17) and by Marsh and Taylor (22), who showed that pilD codes for a prepilin peptidase required for the biogenesis of two other type IV pilus filaments (MSHA and TCP) and for the eps type II secretion system, which is required for the export of hemagglutinin/protease (HA/protease), cholera toxin, the chitinase ChiA-1, neuraminidase, and lipase. The presence of pilD in the same cluster with pilA suggests that it may also process PilA during biogenesis.
In this study, PilA was shown to contribute substantially to the fitness of V. cholerae attached to a crab shell surface, but not to the fitness of a planktonic population. One possible PilA surface-associated role is adherence, and indeed, the pilA mutant consistently attached less well to chitin than the wild-type parent. However, the average fold difference between the wild type and pilA mutant was small, particularly when compared to the effect of the mshA mutation discussed below. Accordingly, because the PilA adherence effect was small and the adherence of V. cholerae to chitin multifactorial, we doubt that the strong PilA fitness phenotype is due to its effect on adherence.
Recognizing that type IV pili perform a variety of functions, in addition to adherence, the role of PilA in f lagellar-independent surface translocation was studied. No motility phenotype for the pilA mutant was detected on colloidal chitin agar plates. Perhaps the most intriguing possible explanation for the pilA fitness phenotype comes from studies of Myxococcus xanthus. Chitin was reported to cause retraction of a polar type IV pilus which mediates social-gliding motility in this organism (23). This observation leads to a particularly attractive model for the role of the PilA-containing ChiRP in the V. cholerae chitin utilization program that could explain why type IV pilus assembly complex genes reside in the same regulon with genes that encode chitin catabolic functions. This model incorporates the fact that the eps type II chitinase secretion apparatus and most type IV pilus assembly complexes localize to the cell pole (24). In view of these topographical features, the model predicts that the assembly complex for the ChiRP localizes to the same pole as the the eps type II secretion apparatus. As a result, chitin-induced retraction of a ChiRP fiber tethered to a chitin surface would optimally orient the site of chitinase secretion with the chitin surface, enhancing chitin digestion by reducing ineffectual chitinase secretion.
PilA has also been examined for a possible role in the pathogenesis of cholera. Fullner et al. (17) showed that an in-frame deletion of pilA did not affect colonization of the infant mouse intestine. However, Hang et al. (25), using in vivo-induced antigen technology (IVIAT), showed that a PilA immunogen is expressed during human infection with V. cholerae, indicating that it might have a pathogenic function in man. This result suggests that V. cholerae might encounter GlcNAc β1–4GlcNAc-containing glyco-conjugates in the intestine, thus inducing pilA expression, but the pathogenic significance of this response is unknown.
It is natural to expect that chemotaxis toward chitin and chitin adherence will be coregulated processes. This notion received support from the induction of VCA0811 and VC0449 by GlcNAc, a chitin digestion product that is thought to diffuse away from bacteria attached to chitin surfaces (26). Deletion of VC0449, encoding a methyl accepting chemotaxis protein, resulted in decreased flagellar-mediated chemotaxis to GlcNAc and (GlcNAc)2 (data not shown), whereas deletion of VCA0811 (encoding a putative chitinase) caused ≈4-fold reduction in chitin bead adherence. Kirn et al.§, screening a V. cholerae classical biotype mutant library for entries with reduced epithelial cell adherence, discovered the same gene and showed that the mutant also adhered poorly to chitin beads. Together, these results suggest that VCA0811 might function more generally as a GlcNAc-specific lectin for a variety of surfaces displaying this common carbohydrate.
Remarkably, MSHA, a type IV pilus previously shown by Watnick et al. (21) to be important for biofilm formation on borosilicate, also functions in the adherence of V. cholerae to chitin beads, where a deletion of it was shown to cause a ≈30-fold reduction in the number of attached bacteria. Crab shells are ordinarily covered by a nonchitin surface veneer, the epicuticle (27), which V. cholerae must bind and degrade before chitin utilization can proceed. We suggest that MSHA, like its role in biofilm formation on nonnutritive surfaces, might initiate adherence to the epicuticle covering natural chitin surfaces. Because the mshA mutant also adhered less well to chitin beads, which lack an epicuticle, MshA may facilitate surface adhesion in a manner that is independent of the surface chemistry. Chiavelli et al. (20) also showed that mshA is involved in the adherence of V. cholerae O139 and the El Tor biotype of V. cholerae O1 to Daphnia pulex molts. By contrast, Watnick et al. (21) did not identify a chitin-binding phenotype for an El Tor mshA mutant using a chitin particle assay. Together, these results indicate that MSHA likely enhances chitin adherence in a surface chemistry independent manner, but does so in way that is affected by technical features of the experiment.
Analysis of the microarray expression results obtained during the course of this study has led to a stage-specific model of the V. cholerae chitin utilization program. In noncolonized, epicuticle-covered chitin surfaces, the random collision of planktonic V. cholerae with the surface leads to MSHA-mediated adherence. Later, or in surfaces already colonized with chitinolytic bacteria, gradients of GlcNAc and (GlcNAc)2 direct the bacteria to the chitin surface and induce the adhesin encoded by VCA0811. Full induction of the ChiS regulon then occurs and the coordinate expression of the chitin-regulated pilus, and chitin catabolic machinery promotes chitin colonization and the efficient digestion and assimilation of chitin. Where nonacetylated chitin residues are encountered, the (GlcN)2 gene set is induced, ensuring maximal use of the chitin substrate.
Supplementary Material
Acknowledgments
We thank Dr. Gregory Dolganov for help with primer and probe design and suggestions for quantitative real-time PCR. This work was supported by The Carlsberg Foundation, The Danish Research Council, National Institutes of Health Grants GM38759 and GM51215 (to S.R.), and an Ellison Foundation Senior Scholar Award and National Institutes of Health Grants AI053706 and AI43422 (to G.K.S.).
Abbreviations: (GlcNAc)n, chitin oligosaccharides; ChiRP, chitin-regulated pilus; qRT-PCR, quantitative real-time RT-PCR.
Footnotes
Kirn, T. J., Jude, B. A. & Taylor, R. K. (2003) in American Society for Microbiology, 103rd General Meeting, Washington Convention Center, Washington, DC, p. 115 (abstr.).
References
- 1.Lipp, E. K., Huq, A. & Colwell, R. R. (2002) Clin. Microbiol. Rev. 15, 757-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colwell, R. R. (1996) Science 274, 2025-2031. [DOI] [PubMed] [Google Scholar]
- 3.Colwell, R. R., Hug, A., Islam, M. S., Aziz, K. M. A., Yunus, M., Khan, N. H., Mahmud, A., Sack, R. B., Nair, G. B., Chakraborty, J., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 1051-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keyhani, N. O. & Roseman, S. (1999) Biochim. Biophys. Acta 1473, 108-122. [DOI] [PubMed] [Google Scholar]
- 5.Voskuil, M. I., Schnappinger, D., Visconti, K. C., Harrell, M. I., Dolganov, G. M., Sherman, D. R. & Schoolnik, G. K. (2003) J. Exp. Med. 198, 705-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tusher, V. G., Tibshirani, R. & Chu, G. (2001) Proc. Natl. Acad. Sci. USA 98, 5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. (1998) Proc. Natl. Acad. Sci. USA 95, 14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Merrell, D. S., Butler, S. M., Qadri, F., Dolganov, N. A., Alam, A., Cohen, M. B., Calderwood, S. B., Schoolnik, G. K. & Camilli, A. (2002) Nature 417, 642-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li, X. & Roseman, S. (2004) Proc. Natl. Acad. Sci. USA 101, 627-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keyhani, N. O., Li, X. B. & Roseman, S. (2000) J. Biol. Chem. 275, 33068-33076. [DOI] [PubMed] [Google Scholar]
- 11.Park, J. K., Keyhani, N. O. & Roseman, S. (2000) J. Biol. Chem. 275, 33077-33083. [DOI] [PubMed] [Google Scholar]
- 12.Keyhani, N. O. & Roseman, S. (1996) J. Biol. Chem. 271, 33425-33432. [DOI] [PubMed] [Google Scholar]
- 13.Keyhani, N. O. & Roseman, S. (1996) J. Biol. Chem. 271, 33414-33424. [DOI] [PubMed] [Google Scholar]
- 14.Park, J. K., Wang, L. X. & Roseman, S. (2002) J. Biol. Chem. 277, 15573-15578. [DOI] [PubMed] [Google Scholar]
- 15.Park, J. K., Wang, L. X., Patel, H. V. & Roseman, S. (2002) J. Biol. Chem. 277, 29555-29560. [DOI] [PubMed] [Google Scholar]
- 16.Connell, T. D., Metzger, D. J., Lynch, J. & Folster, J. P. (1998) J. Bacteriol. 180, 5591-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fullner, K. J. & Mekalanos, J. J. (1999) Infect. Immun. 67, 1393-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kerr, B., Riley, M. A., Feldman, M. W. & Bohannan, B. J. (2002) Nature 418, 171-174. [DOI] [PubMed] [Google Scholar]
- 19.Muzzarelli, R. A. A. (1973) Natural Chelating Polymers: Alginic Acid, Chitin, and Chitosan (Pergamon, Oxford).
- 20.Chiavelli, D. A., Marsh, J. W. & Taylor, R. K. (2001) Appl. Environ. Microbiol. 67, 3220-3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watnick, P. I., Fullner, K. J. & Kolter, R. (1999) J. Bacteriol. 181, 3606-3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marsh, J. W. & Taylor, R. K. (1998) Mol. Microbiol. 29, 1481-1492. [DOI] [PubMed] [Google Scholar]
- 23.Li, Y., Sun, H., Ma, X., Lu, A., Lux, R., Zusman, D. & Shi, W. (2003) Proc. Natl. Acad. Sci. USA 100, 5443-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scott, M. E., Dossani, Z. Y. & Sandkvist, M. (2001) Proc. Natl. Acad. Sci. USA 98, 13978-13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hang, L., John, M., Asaduzzaman, M., Bridges, E. A., Vanderspurt, C., Kirn, T. J., Taylor, R. K., Hillman, J. D., Progulske-Fox, A., Handfield, M., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 8508-8513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Azam, F. (1998) Science 280, 694-696. [Google Scholar]
- 27.Giraud-Guille, M. M. & Bouligand, Y. (1986) in Chitin in Nature and Technology, eds. Muzzarelli, R., Jeuniaux, C. & Gooday, G. W. (Plenum, New York), pp. 29-35.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.