Abstract
Previous studies have revealed that mouse primordial germ cells (PGCs) undergo genome-wide DNA methylation reprogramming to reset the epigenome for totipotency. However, the precise 5-methylcytosine (5mC) dynamics and its relationship with the generation of 5-hydroxymethylcytosine (5hmC) are not clear. Here we analyzed the dynamics of 5mC and 5hmC during PGC reprograming and germ cell development. Unexpectedly, we found a specific period (E8.5-9.5) during which both 5mC and 5hmC levels are low. Subsequently, 5hmC levels increase reaching its peak at E11.5 and gradually decrease until E13.5 likely by replication-dependent dilution. Interestingly, 5hmC is enriched in chromocenters during this period. While this germ cell-specific 5hmC subnuclear localization pattern is maintained in female germ cells even in mature oocytes, such pattern is gradually lost in male germ cells as mitotic proliferation resumes during the neonatal stage. Pericentric 5hmC plays an important role in silencing major satellite repeat, especially in female PGCs. Global transcriptome analysis by RNA-seq revealed that the great majority of differentially expressed genes from E9.5 to 13.5 are upregulated in both male and female PGCs. Although only female PGCs enter meiosis during the prenatal stage, meiosis-related and a subset of imprinted genes are significantly upregulated in both male and female PGCs at E13.5. Thus, our study not only reveals the dynamics of 5mC and 5hmC during PGC reprogramming and germ cell development, but also their potential role in epigenetic reprogramming and transcriptional regulation of meiotic and imprinted genes.
Keywords: DNA demethylation, 5-hydroxymethylcytosine, primordial germ cells, reprogramming, pericentric heterochromatin
Introduction
Unlike most other species such as Caenorhabditis elegans, Drosophila melanogaster, and Xenopus laevis, whose germ cells are specified at fertilization, mammalian germ cells originate from the pluripotent epiblast1. Therefore, mammalian germ cells need to erase their epigenetic features that are characteristic of somatic cells to acquire an epigenetic state compatible with the germ cell program. Primordial germ cells (PGCs) are identified as alkaline phosphatase- and Prdm1-positive cells at the base of the allantois at E7.251. During gastrulation, PGCs migrate along the hindgut and colonize into the genital ridge around E10.5. In the genital ridge, both male and female PGCs stop proliferation around E14.5. At this time, female PGCs enter the meiotic prophase and arrest at the diplotene stage, thus defining a limited pool of gametes called the ovarian reservoir. Arrested oocytes will only resume meiosis during adulthood after hormonal stimulation. On the other hand, mitotic arrest takes place in male germ cell from E14.5 to neonatal stage. Male germ cells resume mitotic proliferation soon after birth, and subsequently enter meiosis during puberty2,3.
During PGC migration and colonization from E8.5 to E13.5, genome-wide epigenetic reprogramming takes place, including reactivation of the inactive X chromosome, global changes in histone modifications (eg., loss of H3K9me2, H3K9ac, H4/H2AR3me2, and linker histone H1; gain of H3K27me3), and global loss of DNA methylation3,4,5,6,7,8,9. Global DNA demethylation is known to be essential for germ cells to establish allele-specific DNA methylation pattern that is necessary for appropriate genomic imprinting10. Methylation at the 5-position of cytosine of the CpG dinucleotide (5mC) plays an important role in regulating gene expression, including the regulation of imprinted genes. Although the enzymes responsible for DNA methylation has been well studied, how DNA demethylation is achieved has been elusive for a long time11. Recent studies have demonstrated that the Tet family proteins are capable of iterative oxidation of 5mC into 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC)12,13,14,15. Since 5fC and 5caC can be further processed by Tdg and base excision repair (BER)15,16, conversion of 5mC to 5hmC by Tet proteins is believed to be the first step in the DNA demethylation process. In addition, because 5hmC cannot serve as a substrate for Dnmt117, generation of 5hmC can lead to passive demethylation in a replication-dependent manner. Although several lines of evidence suggest that PGCs go through global demethylation during their reprogramming, the dynamics and the mechanism of demethylation are still controversial7,16,18,19. Additionally, the biological significance of PGC reprogramming is also unclear.
In this study, we investigated the dynamics of 5mC and its oxidative derivatives during PGC reprogramming. Using immunohistochemistry, immunocytochemistry and mass spectrometric analyses, we demonstrate that PGC reprogramming involves three global demethylation steps: 1) loss of bulk 5mC in an oxidation-independent manner, 2) oxidation of the remaining 5mC to 5hmC, and 3) dilution of 5hmC by a replication-dependent process. Additionally, we identified a novel germ cell-specific epigenetic signature, the pericentric region-enriched 5hmC, and characterized its replication-dependent dynamics. Finally, we analyzed the transcriptome and found that gene transcription is globally activated during PGC reprogramming.
Results
Decrease of 5mC level correlates with increase in 5hmC during PGC reprogramming
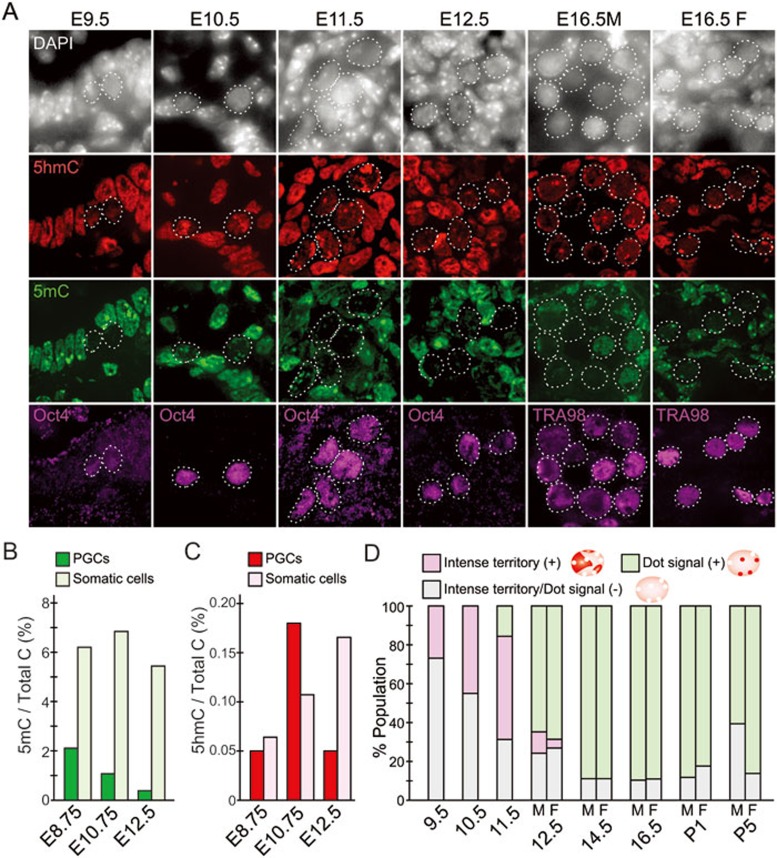
Although several studies have analyzed the dynamics of DNA methylation during germ cell development6,8,10,19,20,21,22, it is not clear whether loss of 5mC correlates with the generation of 5hmC. To address this question, we determined the 5mC and 5hmC dynamics during PGC reprogramming by immunostaining of E9.5 to E16.5 embryos with 5mC and 5hmC antibodies. Consistent with previous reports8, PGCs are already hypomethylated compared with surrounding somatic cells at E9.5 (Figure 1A). While the reemergence of 5mC in male germ cell is observed around E16.5, its level is consistently lower compared to the adjacent somatic cells during the reprogramming period. In contrast, 5hmC exhibited a dynamic change in both signal intensity and staining pattern. While 5hmC levels in PGCs are also low compared to those of neighboring somatic cells at E9.5, it becomes markedly increased between E10.5-11.5 and then the level is gradually decreased.
Figure 1.
Dynamics of 5mC and 5hmC during PGC reprogramming. (A) Representative images of reprogramming PGCs and adjacent somatic cells co-stained with 5mC (green), 5hmC (red), and germ cell markers (Oct4 and TRA98) of the cryosection of mouse embryos (E9.5-11.5) or embryonic gonads (E12.5 and E16.5, male and female). Germ cells are indicated by dashed circles. (B, C) Mass spectrometric quantification of 5mC (B) and 5hmC (C). PGCs and somatic cells from 4-10 embryos were FACS sorted based on the expression of Oct4-GFP transgenes in gonads. (D) Quantification of the 5hmC staining patterns demonstrates its dynamic change during PGC reprogramming and germ cell development. M, male; F, female.
To evaluate the dynamics of 5mC and 5hmC levels during PGC reprogramming quantitatively, we purified PGCs and somatic cells using transgenic mice carrying an Oct4 promoter-driven GFP reporter. After isolation, the genomic DNA from these cells was subjected to mass spectrometric analysis. We found that the 5mC levels in somatic cells are relatively stable, around 6% of total C, from E8.75 to E12.5 (Figure 1B). However, the 5mC level in PGCs is already low (∼2%) at E8.75 and continues to decrease to less than 1% at E12.5 (Figure 1B and Supplementary information, Figure S1), consistent with immunostaining results shown in Figure 1A. In contrast to 5mC, the 5hmC levels appear to be more dynamic. The level of 5hmC gradually increases from E8.75 to E12.5 and reaches about 0.17% of total C in somatic cells (Figure 1C). In contrast, 5hmC peaks at E10.75 in PGCs (Figure 1C and Supplementary information, Figure S1). This dynamic change in 5mC and 5hmC levels in PGCs is compatible with a mechanism by which global DNA demethylation involves Tet-mediated oxidation of 5mC23,24,25.
5fC and 5caC are relatively stable during germ cell reprogramming
Since Tet proteins can iteratively oxidize 5hmC to 5fC and 5caC, a decrease in 5hmC level after E11.5 could be caused by further oxidation of 5hmC to 5fC and 5caC. To explore this possibility, we immunostained the genital ridge sections of E9.5 to E12.5 embryos with 5fC and 5caC antibodies. Unlike the obvious change in 5hmC levels, no dramatic change in 5fC or 5caC levels was observed during PGC reprogramming (Supplementary information, Figure S2). Although further oxidation by Tet proteins cannot be excluded, the results suggest that it is unlikely a major reason for the reduction of 5hmC in E12.5 PGCs.
Reprogramming of subnuclear localization pattern of 5hmC in PGCs
Immunostaining pictures presented in Figure 1A indicate that not only the signal intensity but also the subnuclear localization pattern of 5hmC in PGCs change during PGC reprogramming. Intense territories of 5hmC in the PGC nucleus increased by E11.5 and disappeared around E12.5. Concomitant with loss of intense 5hmC mark, 5hmC foci appeared at E11.5 and became evident at later stages. Based on the nuclear 5hmC staining pattern, PGCs can be divided into three groups (Figure 1D). Quantification of the percentage of each group during PGC reprogramming and germ cell development is presented in Figure 1D.
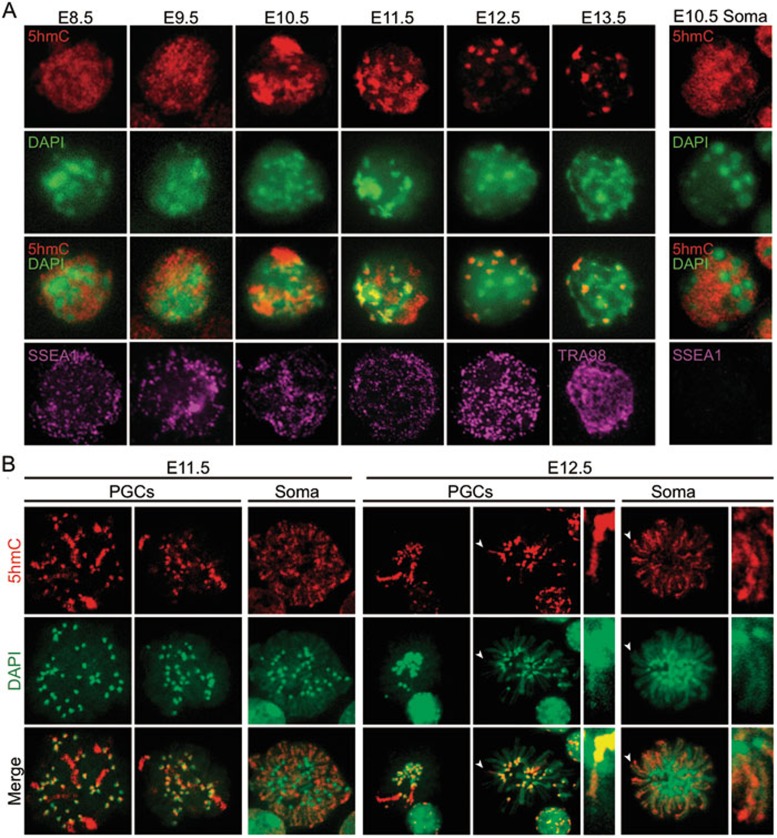
To analyze 5hmC subnuclear localization in more detail, we performed immunostaining of the surface spread of PGCs and surrounding somatic cells. Similar to previous studies on embryonic stem cells26,27, 5hmC signal in somatic cells is mainly enriched in euchromatic regions but excluded from DAPI-dense chromocenters (Figure 2A, right panel). Although a similar staining pattern is observed in E8.5 and E9.5 PGCs, intense 5hmC territories are observed in E10.5 and E11.5 PGCs, and the intense 5hmC is further restricted to limited foci in E12.5 and E13.5 PGCs (Figure 2A, left panels). To determine the chromosome location of the dense 5hmC staining foci, we stained chromosome spreads of purified PGCs and found that 5hmC is mainly localized to the DAPI-heavy pericentric heterochromatin at E11.5 and E12.5 (Figure 2B). In addition, we also observed the enrichment of 5hmC in several chromosomes at E11.5 and this number decreased in E12.5 PGCs (Figure 2B). When each chromosome is scrutinized, only one of the sister chromatids is enriched with 5hmC in E12.5 PGCs, while both chromatids are 5hmC positive in somatic cells (Figure 2B). This staining pattern suggests that replication-dependent dilution might be responsible for the loss of 5hmC at the late stage of PGC reprogramming, which is similar to that observed in the paternal genome of preimplantation embryos28,29.
Figure 2.
Subnuclear localization dynamics of 5hmC during PGC reprogramming. (A) Representative images of reprogramming PGC nuclear staining with 5hmC (red), DAPI (green), and germ cell markers (SSEA1 and TRA98) of surface spreads. The highest signal intensity was detected in the E10.5 PGC nucleus. Enrichment of 5hmC at the chromocenters starts from E10.5 and became obvious from E11.5. Since neighboring somatic cells show no change in 5hmC pattern through development, only a representative image of E10.5 embryo is shown (right panel). (B) Representative images of chromosome spreads of E11.5 and E12.5 PGCs and somatic cells co-stained with 5hmC antibody and DAPI. PGCs and somatic cells were purified by FACS sorting based on the expression of an Oct4-GFP transgene. Note that only a few chromosomes have intense signal in the chromosome body in PGCs, while all chromosomes are broadly stained in somatic cells. Arrowheads indicate enlarged chromatids shown in the adjacent panels.
Fate of 5hmC in pericentric heterochromatin of PGCs during germ cell development
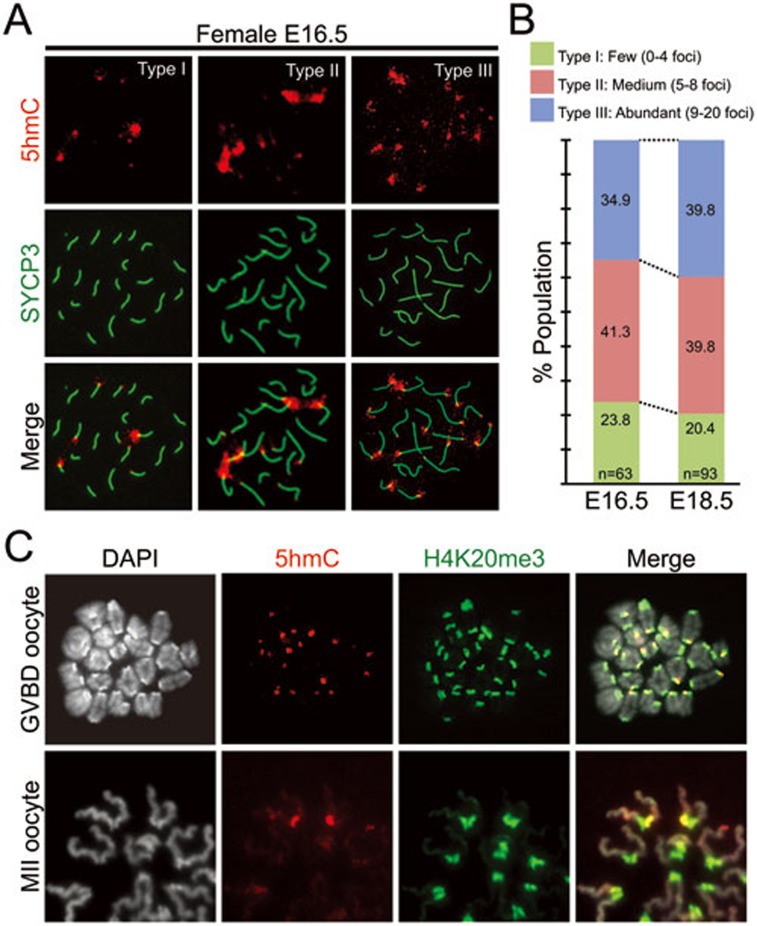
To investigate the fate of 5hmC present in pericentric heterochromatin of PGCs, we first focused on female germ cells as they enter the meiotic cell cycle at the late embryonic stage. Immunostaining of E16.5 and E18.5 meiotic germ cells revealed the presence of variable 5hmC foci at the chromocenter (Figure 3A). When grouped by foci numbers, the distribution of this pattern is very similar between E16.5 and E18.5 PGCs, indicating that 5hmC is relatively stable in these germ cells (Figure 3B). Indeed, 5hmC foci can still be observed in some of the H4K20me3-marked pericentric region of germinal vesicle breakdown (GVBD), and MII oocytes in adult females (Figure 3C). These results suggest that some of the pericentric 5hmC deposited in early gonadal stages can be maintained through female germ cell development. Interestingly, 5hmC is only observed in one of the two centric regions of GVBD and MII oocytes (Figure 3C), indicating that 5hmC in pericentric region is not maintained after DNA replication. Given that female germ cells have already entered meiosis at E14.5, the variation in the intensity and the number of 5hmC foci appears to be generated before E14.5 as explained in Supplementary information, Figure S3.
Figure 3.
Pericentric 5hmC is maintained through female germ cell development. (A) Representative images of E16.5 pachytene stage oocytes co-stained with 5hmC (red) and a synaptonemal complex marker SYCP3 (green). Germ cells are classified into three types based on the number of large 5hmC foci. Type I, less than 4 large foci; type II, five to eight foci; type III, more than nine foci. (B) Percentages of each group in E16.5 and E18.5 PGCs. Note that the distribution of the three groups is not significantly altered between E16.5 and E18.5 PGCs. (C) Representative images of 5hmC (red) and H4K20me3 (green), a pericentric heterochromatin marker, in germinal vesicle breakdown (GVBD), and metaphase II (MII) oocytes.
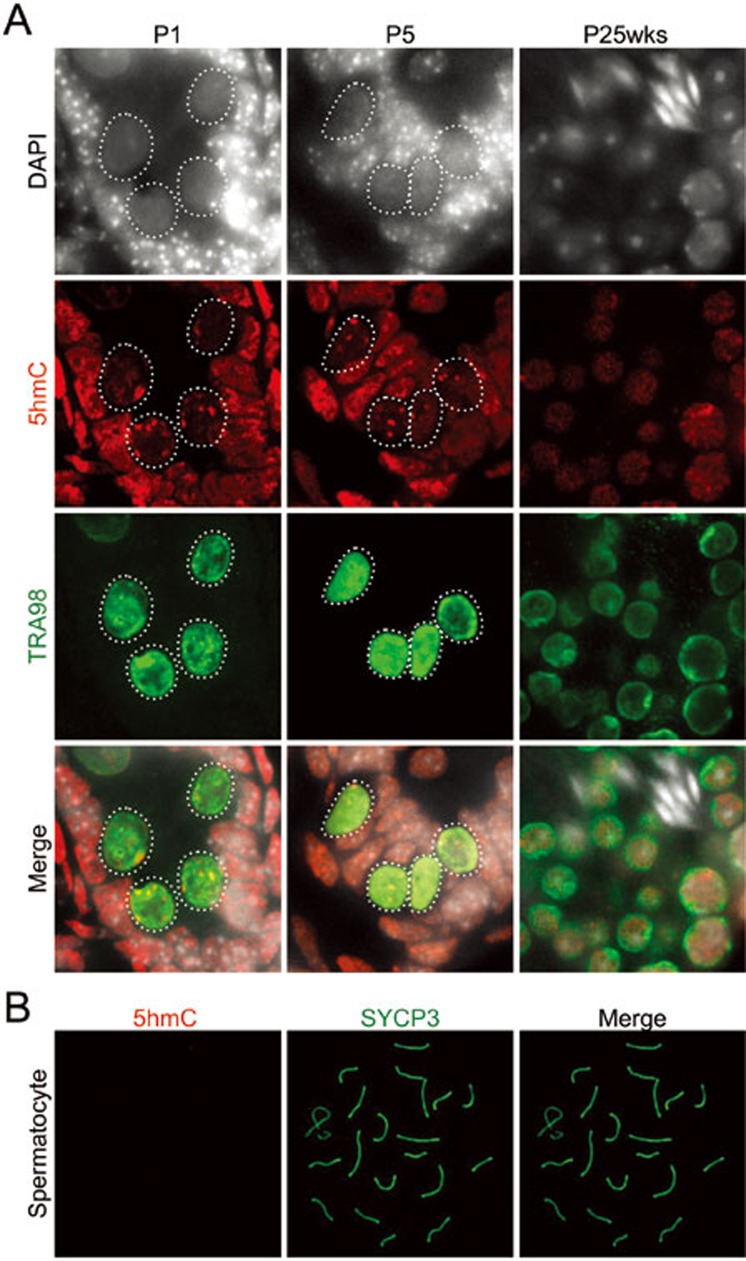
We next analyzed the 5hmC fate in male germ cells. While 5hmC foci are observed in E16.5 through P1 and greatly decreased in P5 testis (Figures 1D and 4A), no such foci are observed in adult male germ cells such as spermatocytes and spermatids (Figure 4A and 4B). Since the decrease in 5hmC foci coincides with the period in which male germ cells are mitotically active, one likely explanation for the loss of pericentric 5hmC is replication-dependent dilution.
Figure 4.
Loss of pericentric 5hmC during postnatal male germ cell development. (A) Representative images of cryosections of P1, P5, and 25-week-old testis co-stained with 5hmC (red) and germ cell marker TRA98 (green) antibodies. Dashed circles indicate germ cells. (B) Representative images of pachytene stage spermatocyte co-stained with 5hmC (red) and a synaptonemal complex marker SYCP3 (green). No 5hmC signal was detected at this stage.
To evaluate the relationship between the expression of the major satellite repeat and the accumulation of pericentric 5hmC, we analyzed its expression in wild-type and Tet1-deficient E13.5 PGCs. We found that loss of function of Tet1 increased the major satellite expression by 8-12 folds in female and 1.5-5 folds in male PGCs, respectively (Supplementary information, Figure S4). Since pericentric 5hmC is generated by Tet124, this result suggests that pericentric 5hmC is functionally important for major satellite repeat silencing. Interestingly, major satellite expression level is much lower in male than female PGCs in Tet1-KO, although 5hmC is undetectable in both sexes24. This suggests that 5hmC-independent mechanism for silencing major satellite expression may also exist and this mechanism appears to work in a sex-dependent manner. Indeed, even in wild-type mice, the expression level of satellite repeat in female PGCs is about three times higher than that in male PGCs (Supplementary information, Figure S4).
Gene expression analysis during PGC reprogramming
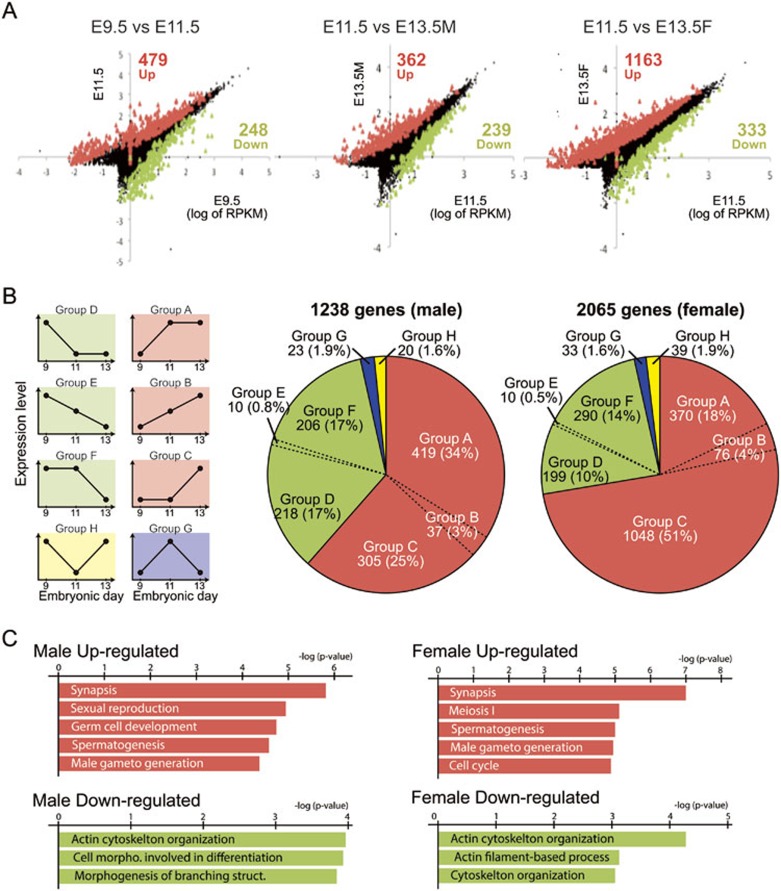
DNA methylation has been implicated in transcriptional regulation. We therefore asked whether PGC reprogramming is accompanied with gene expression change. To this end, PGCs from E9.5, E11.5, and E13.5 male and female embryos were purified and their transcriptomes were profiled using a recently developed Smart-Seq method30. This analysis generated 22-38 million unique reads for each sample, allowing detection of more than 10 000 expressed transcripts for each stage of PGC development (Supplementary information, Table S1). The sample and library preparation procedures were reliable given that the sequencing reads were highly reproducible among biological replicates (r = 0.97-0.99) (Supplementary information, Figure S5). Compared with E9.5 PGCs, we found that 479 genes are significantly upregulated and 248 genes are downregulated in E11.5 PGCs (Figure 5A and Supplementary information, Table S2). When compared with E11.5 PGCs, male E13.5 PGCs have 362 upregulated, and 239 downregulated genes, whereas female E13.5 PGCs have 1 163 upregulated and 333 downregulated genes (Figure 5A and Supplementary information, Tables S3 and S4). Overall, the number of upregulated genes is greater than that of the downregulated genes in every comparison, suggesting that gene expression is generally activated during PGC reprogramming. This notion is supported by a distribution shift of the gene groups that are classified by expression value (Supplementary information, Figure S6). The number of genes that are expressed at a very low level (RPKM < −4) is gradually decreased from E9.5 to E13.5, and the number of genes expressed at a low level (RPKM between −4 and 0) is increased.
Figure 5.
Transcriptional change during PGC reprogramming. (A) Scatter plot comparing transcriptome between E9.5 and E11.5 (left), E11.5 and E13.5 male (middle), and E11.5 and E13.5 female (right) PGCs. Red and green dots represent significantly up- and downregulated genes, respectively. The numbers of up- and downregulated genes are indicated. (B) Pie chart illustration of the percentage in each group that is categorized by the trend of change from E9.5 to E13.5 PGCs. Expression patterns of each group are illustrated in left panels. (C) Gene Ontology analysis of upregulated (Group A+B+C) and downregulated (Group D+E+F) genes in E13.5 compared with that in E9.5 PGCs with a cut-off FDR < 0.05. The most enriched biological processes based on their P-values are shown.
To analyze the expression change in more detail, we classified the differentially expressed (DE) genes based on their change trend from E9.5 to E13.5. Among them, more than 60% (761 out of 1 238) of DE genes in male PGCs and 70% (1 494 out of 2 065) in female PGCs are upregulated from E9.5 to E13.5 (Figure 5B). In male PGCs, the number of genes upregulated at an early phase (from E9.5 to 11.5: Group A) and late phase (from E11.5 to E13.5: Group C) are roughly equal, but genes that are upregulated in both stages are relatively rare (Group B; Supplementary information, Table S5). An overall similar pattern is also observed in male downregulated genes (Figure 5B, Groups D, E, F), as well as female up- and downregulated genes (Figure 5B, Group A-F). This gene expression trend suggests that there are two major phases in the regulation of gene expression during PGC reprogramming. The first phase takes place from E9.5 to E11.5 when PGCs enter the genital ridges. The second phase starts at E11.5 and ends at E13.5 when epigenetic reprogramming completes. Interestingly, a little over half of the differentially regulated genes in female PGCs are upregulated at the late stage (Group C; Supplementary information, Table S6).
Since female PGCs enter meiosis around E13.5, massive activation of gene expression at this stage may be important for PGCs to go through meiosis. Indeed, we found that many genes critical for meiosis are upregulated in this stage (Supplementary information, Figure S7A). Consistently, gene ontology (GO) analysis also revealed the enrichment of genes involved in synapsis and meiosis in this gene groups (Figure 5C and Supplementary information, Figure S7, and Table S7). Reverse transcriptase quantitative PCR (RT-qPCR) analysis confirmed upregulation of meiosis-related genes, including Sycp3, Syce1, Hormad1, Mael, and Sohlh2 (Supplementary information, Figure S7B). In addition, we found that a subset of meiosis-related genes is significantly upregulated even in male PGCs (Figure 5C and Supplementary information, Figure S7B and Table S8). These results suggest that activation of meiotic genes is likely mediated by a common mechanism during PGC reprogramming such as DNA demethylation. On the other hand, downregulated genes in both male and female PGCs (eg., Claudin7) include genes important for cytoskeleton organization and activity (Figure 5C and Supplementary information, Figure S7B), suggesting that PGCs become less motile once they have migrated into genital ridges.
Given that erasure of genomic imprinting is believed to be one of the major purposes of PGC reprogramming, we asked whether imprinted genes are present in the DE genes. Indeed, we found that a total of 25 out of 143 known imprinted genes are differentially expressed in both or either male or female PGCs (Supplementary information, Figure S8). Differential expression of a subset of imprinted genes during PGC reprogramming indicates that epigenetic reprogramming not only erases DNA methylation, but also regulates expression of at least a subset of imprinted genes.
Discussion
Global DNA demethylation in PGCs takes place in three steps
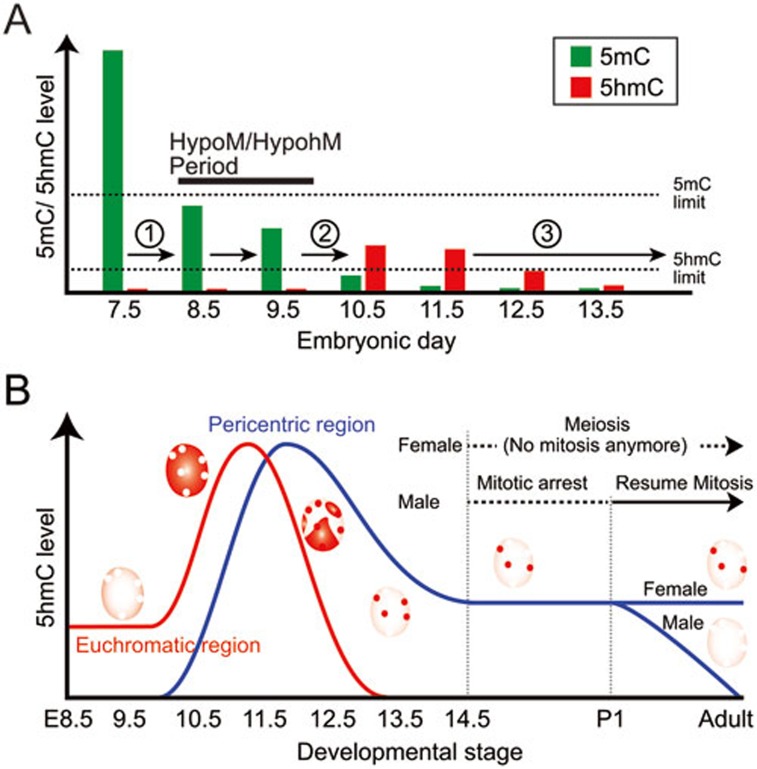
In this study, we analyzed the dynamics of 5mC and 5hmC during PGC reprogramming. Based on our results, DNA demethylation in PGCs appears to occur in three steps (Figure 6A). The first step involves a massive loss of 5mC around E8.5 as 5mC levels in PGCs at this time is significantly lower than that in somatic cells. This step appears to be Tet independent as both Tet proteins and 5hmC are at very low levels at this stage24. Since Uhrf1 (also called NP95), the essential partner of Dnmt1, is downregulated in E8.5 PGCs, this first step is likely mediated by replication-dependent dilution31. The second step involves oxidation of the remaining 5mC to 5hmC by Tet proteins around E10.5 as 5hmC level is specifically increased in PGCs at this time. The final step features the loss of 5hmC in a replication-dependent manner that takes place from E10.5 to E13.5. As massive loss of 5mC takes place about two days before the increase of 5hmC, the hypomethyl/hypohydroxymethylation (hypoM/hypohM) period, when both 5mC and 5hmC are at extremely low levels, spans E8.5 to E9.5. Both immunostaining and mass spectrometry analyses demonstrate that the bulk of 5mC has already disappeared at E8.5. Since 5hmC can only be generated from 5mC, the detected upregulation of 5hmC at E10.5 can only be derived from the remaining 5mC after the first demethylation step. While we currently do not have a good explanation for the hypoM/hypohM period, it might be caused by the different sensitivity of the 5mC and 5hmC antibodies (Figure 6A).
Figure 6.
Diagrammatic illustration of the 5mC and 5hmC dynamics and distribution pattern during germ cell development. (A) Diagrammatic illustration of the three steps of DNA demethylation during PGC reprogramming. (1) Loss of bulk DNA methylation takes place in a Tet-independent manner. (2) Oxidation of remaining 5mC to 5hmC by Tet proteins. (3) Replication-dependent dilution of 5hmC. Detection limit of 5mC and 5hmC by immunohistochemistry is indicated by dashed line. HypoM/hypohM, hypomethyl/hypohydroxymethylation. (B) Diagrammatic illustration of 5hmC subnuclear localization pattern during germ cell development. 5hmC level in euchromatic (red line) and pericentric (blue line) region is indicated. Note that the fates of pericentric 5hmC in late stage male and female germ cells are different.
The observation of a hypoM/hypohM period is consistent with a previous study demonstrating that a rapid and massive reduction of 5mC takes place in E8.0 to E8.5 PGCs8. Similarly, a recent study demonstrated that 5mC in E9.5 PGCs drops to 30%32. Using bisulfite sequencing, this study demonstrated that certain genomic regions such as imprinted regions, CpG islands in the X chromosome, and meiotic genes remain to be methylated until E11.5. As bisulfite sequencing cannot differentiate 5hmC from 5mC, the methylation detected in these regions is likely 5hmC. Consistent with this notion, Tet1 has been shown to play an important role in promoter demethylation and activation of a subset of meiotic genes during PGC reprogramming24. However, other demethylation pathways, such as AID-mediated deamination16,19, may also contribute to this process.
Replication-dependent loss of 5hmC during PGC reprogramming
One of the most interesting observations in this study is that the loss of 5hmC in an entire chromosome or a pericentric heterochromatin appears to go through a replication-dependent dilution process, although the timing of 5hmC loss is different (Figure 6B). We have previously reported that a replication-dependent dilution of the 5mC oxidation products, 5hmC/5fC/5caC, is responsible for the loss of 5mC during preimplantation development28,29. Considering that Tdg expression level dramatically decreases in E8.5 to E12.5 PGCs33, it seems that 5hmC in PGCs is passively removed without further oxidation as we failed to detect accumulation of 5fC or 5caC when 5hmC levels drop between E10.5 to E12.5. This data suggests that the processivity of Tet proteins may be regulated. Although the regulatory mechanism of Tet enzymatic activity is currently unclear, Tet-mediated 5mC oxidation and replication-dependent dilution appear to be a common mechanism in both PGCs and preimplantation embryos. During the preparation of this manuscript, three important reports with similar conclusions were published32,33,34. Together, these studies support that replication-dependent dilution is part of the mechanism responsible for the loss of 5mC during PGC reprogramming.
Germ cell-specific pericentric 5hmC enrichment
In this study, we uncovered 5hmC as a novel germ cell-specific pericentric epigenetic mark. Previous studies in several other cell types have shown that 5hmC is excluded from the DAPI dense heterochromatin, including pericentric heterochromatin26,27. Thus, the enrichment of 5hmC in pericentric heterochromatin appears to be germ cell specific. Our recent study suggests that the pericentric 5hmC is generated by Tet1 as deletion of Tet1 resulted in loss of pericentric 5hmC in PGCs24. Despite dynamic changes in histone modifications of constitutive heterochromatin during germ cell development35, 5hmC appears to be stable in both male and female PGCs unless they undergo mitosis (Figure 6B). We found that PGCs derived from ESCs through in vitro differentiation also show pericentric 5hmC enrichment (data not shown). We found that Tet1-KO female PGCs show significant upregulation of major satellite expression. Although the biological function of the expression of major satellite in germ cell development is unclear, meiotic phenotype in Tet1-KO female PGCs suggests that it may have an important role in germ cell development24.
Epigenetic reprogramming and gene expression in PGCs
By performing RNA-seq analysis, we identified a significant number of genes whose expression are dynamically regulated during PGC reprogramming. Overall, we found that gene expression is preferentially activated during the reprogramming process, and genes involved in meiosis are significantly enriched in the activated gene group. This general trend is consistent with previous studies demonstrating that transcription repression-associated epigenetic marks, such as DNA methylation and H3K9me2, disappear during the reprogramming process8. Although only female PGCs enter meiosis at the embryonic stage, our observation that meiotic genes are also upregulated in male PGCs suggests that epigenetic reprogramming has a common transcriptional effect in both male and female germ cells. In addition, we also identified a subset of imprinted genes whose expression is dynamically regulated during PGC reprogramming. It has been shown that loss of DNA methylation in some imprinted genes takes place around E10.5-13.56,32. Given that this time window correlates with the gene expression change, DNA demethylation at the differentially methylated region (DMR) of these genes is likely the cause of the gene expression change.
Overall, our study reveals the dynamics of 5mC and 5hmC during PGC reprogramming that supports a model in which DNA demethylation in PGCs occurs through multiple steps and with both active and passive mechanisms. In addition, our transcriptome studies suggest that PGC reprogramming may have an important role in the activation of a subset of meiotic and imprinted genes. However, the details regarding whether a specific genomic locus is demethylated, when it is demethylated, and through which mechanism it is demethylated remain to be determined.
Materials and Methods
Animals and purification of PGCs
All animal studies were performed in accordance with guidelines of the Institutional Animal Care & Use Committee at the Harvard Medical School. Purification of PGCs using FACS sorting is performed as described previously24. Briefly, homozygous male of transgenic mouse bearing the GOF18ΔPE-EGFP (Tg(Pou5f1-EGFP), Jackson Lab) gene were timely mated with C57BL/6J. Noon of the vaginal plug appearance was defined as embryonic day (E) 0.5. The parts of embryos (E8.5-10.5) or genital ridges (E11.5-13.5) containing PGCs were dissociated by treating with trypsin followed by hyarulonidase. Germ cells were purified based on the expression of GFP using FACS Aria II flow cytometry (BD Bioscience).
Immunohistochemistry
Post-implantation embryos were dissected from pregnant females at the day indicated. Following fixation, embedding, sectioning, and staining for the immunohistochemistry were performed as described previously24. Briefly, embryos fixed with 4% paraformaldehyde (PFA)/PBS were embedded in OCT compound (Sakura) and sliced to 10 μm thickness using a cryostat. After several washes and permeabilization, sections were treated with hydrochloric acid solution (4N hydrochloric acid, 0.1% Triton-X 100 in distilled water), followed by washing and incubation with blocking buffer (3% BSA, 2% donkey serum, PBS containing 0.1% Tween 20 (PBST)). Then, sections were incubated with primary antibodies, followed by washes and incubation with appropriate secondary antibodies.
Chromosome spread and immunocytochemistry for PGCs and oocytes
Fully-grown GV-stage oocytes were obtained from 8-week-old BDF1 mice 44-48 h after injection with 7.5 I.U PMSG. The ovaries were removed from the mice and transferred to M2 media (Millipore) containing 0.2 mM 3-isobutyl-1-methylxanthine (IBMX; Sigma-Aldrich). The ovarian follicles were punctured with a 27-gauge needle, and the cumulus cells were gently removed from the cumulus-oocyte complexes using a narrow-bore glass pipette. GVBD oocytes were obtained by incubation of GV-stage oocytes with IBMX-free α-MEM medium supplemented with 5% FBS for 4 h. MII-stage oocytes were collected from superovulated females. Zona pellucida was removed by treatment with acidic tyrode's solution (Sigma-Aldrich). For chromosome spread of MII oocytes, oocytes were treated with a low osmotic solution (0.9% sodium citrate and 0.3% PVP) for 10 min before fixation. To prepare chromosome spreads, purified PGCs and oocytes were put on a glass slide dipped in the fixation solution of 1% PFA in DW (pH 9.2) containing 0.15% Triton X-100 and 3 mM dithiothreitol (Sigma-Aldrich)29. The slides were then incubated overnight at 4 °C. After fixation, the slides were washed in 0.4% Photoflo (Kodak) in DW and dried for 30 min at room temperature. The samples were washed with PBST and treated with 0.5% Triton X-100 for 15 min. After washing with PBST, they were denatured with 4N hydrochloric acid for 10 min, neutralized with 100 mM Tris-HCl (pH 8.5) for 15 min, and incubated in PBS/BSA overnight at 4 °C. The samples were then incubated with primary antibodies, followed by washes and incubation with appropriate secondary antibodies.
Antibodies
Primary antibodies used in this study include rabbit anti-5hmC (Active Motif #39791), rabbit anti-5fC and -5caC28, mouse anti-5mC (Eurogenetec #BI-MECY-1000), mouse anti-H4K20me3 (Abcam #78517), mouse anti-SYCP3 (Abcam #ab97672), mouse anti-SSEA1 (Millipore #MAB4301), rat anti-TRA98 (BioAcademia 73-003), and goat anti-Oct3/4 (Santa Cruz #sc-8628). Secondary antibodies used in this study include fluorescein isothiocyanate-conjugated anti-mouse IgG (Jackson Immuno-Research), Alexa Flour 546 donkey anti-rabbit IgG, Alexa Flour 488 donkey anti-mouse IgG, Alexa Flour 488 donkey anti-mouse IgM, Alexa Flour 647 donkey anti-goat IgG, Alexa Flour 647 chick anti-rat IgG (Life technologies).
RNA-seq and gene ontology analysis
Total RNA was purified from 300-5 000 sorted PGCs using RNA microprep kit (Zymo Research, USA). The cDNA synthesis and amplification was performed with the SMARTer ultra low input RNA kit (Clontech, USA). The amplified cDNA was then fragmented by S2 sonicator (Covaris, USA) and converted to sequencing libraries following the Illumina's construction protocol for low input DNA (Illumina, USA). Barcoded libraries were pooled and sequenced in three lanes of Illumina Hiseq 2000 instrument.
mRNA-seq reads generated from each sample were aligned to the mouse genome (mm9, NCBI build 37) with Bowtie/Tophat v1.3.1 (http://tophat.cbcb.umd.edu), which allows mapping across splice sites by read segmentation. All programs were used with default setting unless otherwise specified. Mapped reads (> 90% of total reads) were subsequently assembled into transcripts guided by reference annotation (mm9, USCS gene annotation) with Cufflinks v1.2.1 (http://cufflinks.cbcb.umd.edu). Expression level of each transcript was quantified with normalized RPKM (reads per kilobase of exon per million mapped reads). Two or three biological replicates were used in each stage to identify transcripts that showed significant differences at a FDR cutoff < 0.05 between developmental stages (E9.5, E11.5 and E13.5) by Cuffdiff v1.2.1. Functional annotation of significantly different transcripts and enrichment analysis was performed with DAVID (http://david.abcc.ncifcrf.gov).
RT-qPCR of purified PGCs and somatic cells
RT-qPCR were performed following previously described method24. Briefly, total RNA was isolated from the purified E9.5, 11.5 and 13.5 PGCs using RNeasy Mini Kit (Qiagen), followed by a treatment of TURBO DNase (Life Technologies) and cDNA synthesis by Superscript III First-Strand synthesis system (Invitrogen). Real-time quantitative PCR reactions were performed on an ABI ViiA7 Sequence Detection System (Applied Biosystems) using SYBR Green (Applied Biosystems). Relative gene expression levels were analyzed using comparative Ct methods, where Ct is the cycle threshold number, and normalized to Gapdh. The primers used for RT-qPCR were summarized in Supplementary information, Table S9.
Mass spectrometry analysis
For mass spectrometry analysis, around 4 000 PGCs from about sixty E8.75 embryos, around 3 500 PGCs from about forty E10.75 genital ridges, and around 6 000 PGCs from about ten E12.5 genital ridges were used. Genomic DNAs were extracted from the sorted cells and applied to ultra-performance liquid chromatography coupled with a TSQ-Quantum Ultra triple-quadrupole mass analyzer (ThermoFinnigan)13,14.
Accession number
The RNA-seq data has been deposited in the Gene Expression Omnibus under accession number GSE41908.
Acknowledgments
We thank Jinzhao Wang and Wei Jiang for FACS sorting; Ho-Lim Fung for Illumina sequencing; Susan Wu and Gustavo German for critical reading of the manuscript. This work was partially supported by NIH R01GM097253 and CIRM RB3-05083 (to KZ); and GM68804 and U01DK089565 (to YZ). YZ is an Investigator of the Howard Hughes Medical Institute.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
LC-MS/MS chromatograms of C, 5mC, and 5hmC of the DNA purified from E8.75, E10.75, and E12.5 PGCs
No accumulation of 5fC or 5caC during PGC reprogramming
Diagrammatic illustration of how variations in the number and level of pericentric 5hmC may be generated
Tet1 mediated pericentric 5hmC generation suppresses major satellite expression
Biological replicates of RNA-seq data is highly reproducible
Global activation of gene expression during PGC reprogramming
Meiotic genes are significantly up-regulated in both male and female E13.5 PGCs
List of imprinted genes whose expression was significantly changed in both male and female E13.5 PGCs
Mapping statistics for RNAseq
Differentially expressed genes between E9.5 and E11.5 PGCs
Differentially expressed genes between E11.5 and male E13.5 PGCs
Differentially expressed genes between E11.5 and female E13.5 PGCs
Groups of 1238 differentially expressed genes based on their expression changes from E9.5, E11.5 to E13.5 male PGCs.
Groups of 2065 differentially expressed genes based on their expression changes from E9.5, E11.5 to E13.5 female PGCs.
Functional annotation of 2065 genes whose expression changed from E9.5, E11.5 to E13.5 female PGCs.
Functional annotation of 1238 genes whose expression changed from E9.5, E11.5 to E13.5 male PGCs.
List of primer sequences for RT-qPCR
References
- Saitou M, Yamaji M. Germ cell specification in mice: signaling, transcription regulation, and epigenetic consequences. Reproduction. 2010;139:931–942. doi: 10.1530/REP-10-0043. [DOI] [PubMed] [Google Scholar]
- Ewen KA, Koopman P. Mouse germ cell development: from specification to sex determination. Mol Cell Endocrinol. 2010;323:76–93. doi: 10.1016/j.mce.2009.12.013. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat Rev Genet. 2008;9:129–140. doi: 10.1038/nrg2295. [DOI] [PubMed] [Google Scholar]
- Ancelin K, Lange UC, Hajkova P, et al. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat Cell Biol. 2006;8:623–630. doi: 10.1038/ncb1413. [DOI] [PubMed] [Google Scholar]
- Hajkova P, Ancelin K, Waldmann T, et al. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature. 2008;452:877–881. doi: 10.1038/nature06714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P, Erhardt S, Lane N, et al. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev. 2002;117:15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- Saitou M, Kagiwada S, Kurimoto K. Epigenetic reprogramming in mouse pre-implantation development and primordial germ cells. Development. 2012;139:15–31. doi: 10.1242/dev.050849. [DOI] [PubMed] [Google Scholar]
- Seki Y, Hayashi K, Itoh K, Mizugaki M, Saitou M, Matsui Y. Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev Biol. 2005;278:440–458. doi: 10.1016/j.ydbio.2004.11.025. [DOI] [PubMed] [Google Scholar]
- Sugimoto M, Abe K. X chromosome reactivation initiates in nascent primordial germ cells in mice. PLoS Genet. 2007;3:e116. doi: 10.1371/journal.pgen.0030116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Inoue K, Ono R, et al. Erasing genomic imprinting memory in mouse clone embryos produced from day 11.5 primordial germ cells. Development. 2002;129:1807–1817. doi: 10.1242/dev.129.8.1807. [DOI] [PubMed] [Google Scholar]
- Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortellino S, Xu J, Sannai M, et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valinluck V, Sowers LC. Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer Res. 2007;67:946–950. doi: 10.1158/0008-5472.CAN-06-3123. [DOI] [PubMed] [Google Scholar]
- Hackett JA, Zylicz JJ, Surani MA. Parallel mechanisms of epigenetic reprogramming in the germline. Trends Genet. 2012;28:164–174. doi: 10.1016/j.tig.2012.01.005. [DOI] [PubMed] [Google Scholar]
- Popp C, Dean W, Feng S, et al. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guibert S, Forné T, Weber M. Global profiling of DNA methylation erasure in mouse primordial germ cells. Genome Res. 2012;22:633–641. doi: 10.1101/gr.130997.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett JA, Reddington JP, Nestor CE, et al. Promoter DNA methylation couples genome-defence mechanisms to epigenetic reprogramming in the mouse germline. Development. 2012;139:3623–3632. doi: 10.1242/dev.081661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maatouk DM, Kellam LD, Mann MRW, et al. DNA methylation is a primary mechanism for silencing postmigratory primordial germ cell genes in both germ cell and somatic cell lineages. Development. 2006;133:3411–3418. doi: 10.1242/dev.02500. [DOI] [PubMed] [Google Scholar]
- Hajkova P, Jeffries SJ, Lee C, Miller N, Jackson SP, Surani MA. Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science. 2010;329:78–82. doi: 10.1126/science.1187945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Hong K, Liu R, et al. Tet1 controls meiosis by regulating meiotic gene expression. Nature. 2012;492:443–447. doi: 10.1038/nature11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011;25:2436–2452. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubiura M, Okano M, Kimura H, Kawamura F, Tada M. Chromosome-wide regulation of euchromatin-specific 5mC to 5hmC conversion in mouse ES cells and female human somatic cells. Chromosome Res. 2012;20:837–848. doi: 10.1007/s10577-012-9317-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Li Y, et al. Integrating 5-Hydroxymethylcytosine into the epigenomic landscape of human embryonic stem cells. PLoS Genet. 2011;7:e1002154. doi: 10.1371/journal.pgen.1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Shen L, Dai Q, He C, Zhang Y. Generation and replication-dependent dilution of 5fC and 5caC during mouse preimplantation development. Cell Res. 2011;21:1670–1676. doi: 10.1038/cr.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011;334:194. doi: 10.1126/science.1212483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsköld D, Luo S, Wang YC, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nature Biotechnol. 2012;30:777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimoto K, Yabuta Y, Ohinata Y, Shigeta M, Yamanaka K, Saitou M. Complex genome-wide transcription dynamics orchestrated by Blimp1 for the specification of the germ cell lineage in mice. Genes Dev. 2008;22:1617–1635. doi: 10.1101/gad.1649908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seisenberger S, Andrews S, Krueger F, et al. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol Cell. 2012;48:849–862. doi: 10.1016/j.molcel.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett JA, Sengupta R, Zylicz JJ, et al. Germline DNA demethylation dynamics and imprint erasure through 5-Hydroxymethylcytosine Science 2012. Dec 13. doi: 10.1126/science.1229277 [DOI] [PMC free article] [PubMed]
- Kagiwada S, Kurimoto K, Hirota T, Yamaji M, Saitou M.Replication-coupled passive DNA demethylation for the erasure of genome imprints in mice EMBO J 2012. Dec 14. doi: 10.1038/emboj.2012.331 [DOI] [PMC free article] [PubMed]
- Yoshioka H, McCarrey JR, Yamazaki Y. Dynamic nuclear organization of constitutive heterochromatin during fetal male germ cell development in mice. Biol Reprod. 2009;80:804–812. doi: 10.1095/biolreprod.108.072603. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
LC-MS/MS chromatograms of C, 5mC, and 5hmC of the DNA purified from E8.75, E10.75, and E12.5 PGCs
No accumulation of 5fC or 5caC during PGC reprogramming
Diagrammatic illustration of how variations in the number and level of pericentric 5hmC may be generated
Tet1 mediated pericentric 5hmC generation suppresses major satellite expression
Biological replicates of RNA-seq data is highly reproducible
Global activation of gene expression during PGC reprogramming
Meiotic genes are significantly up-regulated in both male and female E13.5 PGCs
List of imprinted genes whose expression was significantly changed in both male and female E13.5 PGCs
Mapping statistics for RNAseq
Differentially expressed genes between E9.5 and E11.5 PGCs
Differentially expressed genes between E11.5 and male E13.5 PGCs
Differentially expressed genes between E11.5 and female E13.5 PGCs
Groups of 1238 differentially expressed genes based on their expression changes from E9.5, E11.5 to E13.5 male PGCs.
Groups of 2065 differentially expressed genes based on their expression changes from E9.5, E11.5 to E13.5 female PGCs.
Functional annotation of 2065 genes whose expression changed from E9.5, E11.5 to E13.5 female PGCs.
Functional annotation of 1238 genes whose expression changed from E9.5, E11.5 to E13.5 male PGCs.
List of primer sequences for RT-qPCR