Abstract
Protein folding and disaggregation are crucial processes for survival of cells under unfavorable conditions. A network of molecular chaperones supports these processes. Collaborative action of Hsp70 and Hsp100 proteins is an important component of this network. J-proteins/DnaJ members as co-chaperones assist Hsp70. As against 22 DnaJ sequences noted in yeast, rice genome contains 104 J-genes. Rice J-genes were systematically classified into type A (12 sequences), type B (9 sequences), and type C (83 sequences) classes and a scheme of nomenclature of these proteins is proposed. Transcript expression profiles revealed that J-proteins are possibly involved in basal cellular activities, developmental programs, and in stress. Ydj1 is the most abundant J-protein in yeast. Ydj1 deleted yeast cells are nonviable at 37 °C. Two rice ortholog proteins of yeast Ydj1 protein namely OsDjA4 and OsDjA5 successfully rescued the growth defect in mutant yeast. As Hsp70 and J-proteins work in conjunction, it emerges that rice J-proteins can partner with yeast Hsp70 proteins in functioning. It is thus shown that J-protein machine is highly conserved.
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-012-0384-9) contains supplementary material, which is available to authorized users.
Keywords: J-proteins, Hsp70, Rice, Transcript expression, Yeast complementation
Introduction
Proper folding of proteins is central to cell functioning. This is true for the nascent proteins synthesized on ribosomes under optimal cellular conditions as well as for the pre-existing proteins when cells face stress conditions. The adverse environmental conditions are particularly more threatening to plants because of their sessile nature. Network of chaperones monitor and ensure protein quality control and homeostasis in the cells. The chaperones have been classified into various families like Hsp100, Hsp90, Hsp70/Hsp40, Hsp60/Hsp10, and sHsp. In the integrated model of protein surveillance system, Hsp70 chaperone machine collaborates with Hsp100 (Sielaff and Tsai 2010; Miot et al. 2011). Hsp70 is assisted by co-chaperone J-proteins and nucleotide exchange factor (Miot et al. 2011). Together, these proteins constitute a chaperone machine that participates in protein folding, prevention of protein aggregation, translocation of proteins across membranes, targeting proteins towards degradation, and regulation of translation initiation (Kelley 1999; Qiu et al. 2006).
The conserved signature sequence of all J-proteins is the 70 amino acid long J-domain which is mostly present near the N-terminus (Cyr et al. 1992). A highly conserved HPD tripeptide is the characteristic feature of J-domain (Kampinga and Craig 2010). Based on the presence of specific conserved regions, J-proteins are classified into three types. Type I J-proteins are characterized by an N-terminus J-domain followed by a stretch of G/F-rich region, a cysteine rich Zn finger-like motif and may contain a loosely conserved C-terminal region involved in dimerization and substrate binding (Lu and Cyr 1998). Type II J-proteins are similar to type I except that they lack the Zn-finger domain. A universally conserved tripeptide of Asp-Ile/Val-Phe referred as DIF motif in the G/F region is proposed to be critical for J-proteins of Escherichia coli (Cajo et al. 2006). Type III J-proteins are the most diverse group, defined by the presence of the J-domain which may be present anywhere along the length of the protein. The proteins which contain a J-like domain but lack the critical HPD tripeptide are classified as type IV J-proteins (Kampinga and Craig 2010). Some J-proteins contain additional domains like protein disulphide isomerase domain and ubiquitin interacting motifs that impart specific functional roles to these diverse protein family members (Kampinga and Craig 2010; Chapple et al. 2004). J-proteins bind to the ATPase domain of Hsp70 that initiates ATP hydrolytic cycle, which is central to the functioning of Hsp70 (Misselwitz et al. 1998). It has been suggested that the substrate specificity of Hsp70 proteins is largely determined by their J-protein co-chaperones. J-proteins deliver the substrate to Hsp70, an event during which a ternary complex between J-protein, the substrate and Hsp70 is probably formed and subsequently the J-protein component is released (Misselwitz et al. 1998).
Detailed information on J-proteins has accrued from studies on E. coli and Saccharomyces cerevisiae. S. cerevisiae contains 22 J-proteins (Walsh et al. 2004) and there are 6 J-domain proteins (called DnaJ) in E. coli (Mayer and Bukau 2005). Ydj1 (YNL064C) is the most abundant cytosolic J-protein and is reportedly required for the normal growth of yeast cells (Caplan et al. 1992). Null mutations of this gene lead to slow growth at 30 °C and inviability at elevated temperatures (Caplan et al. 1992). Ydj1 co-operates with Hsp70 protein Ssa1 in cellular processes like stress response, maintenance of protein quality by protein folding, suppression and rescue of protein aggregates, and translocation of proteins to endoplasmic reticulum (ER) and mitochondria (Caplan et al. 1992; Glover and Lindquist 1998). Miernyk (2001) reported 89 J-proteins in Arabidopsis. Recent analysis has reported that Arabidopsis genome has 120 J-proteins (Rajan and D’Silva 2009). In plants, J-proteins have been implicated in protection against environmental stresses (Yang et al. 2009; Qi et al. 2011; Zhou et al. 2012; Sung and Guy 2003). J-proteins have also been implicated in developmental programs of the plants. J-proteins TMS1 and EDA3 are implicated in thermotolerance of pollen tubes and development and function of the female gametophyte respectively in Arabidopsis (Valencia-Morales et al. 2012; Yang et al. 2009). Specific J-proteins are also found to be essential for Arabidopsis growth and their absence has been associated with gametophytic defect and embryo lethality (Yamamoto et al. 2008). J-proteins are shown to act by modulating activity of transcription factors as well. The flowering time in Arabidopsis is regulated by AtJ3 protein through its direct binding with a MADS-box transcription factor (Shen et al. 2011). Crystal structures of Arabidopsis JAC1 and Nicotiana NtCPIP have been determined (Takano et al. 2010; Griessl et al. 2012). CPIP interacts with capsid protein of potyvirus and mutation in CPIP leads to loss in viral infectivity suggesting that J-protein function in plant virus infection and replication (Hofius et al. 2007). Takano et al. (2010) showed that J-domain of JAC1 possesses a positive charge surface that forms a putative interface with Hsp70. Mutation in HPD tripeptide of JAC1 hampered its functional involvement in chloroplast photo-relocation movement (Takano et al. 2010).
We have aimed at developing a model for the J-proteins in higher plant rice, in this study. In earlier attempts, J-protein of rice was shown to play a role in UV-induced DNA damage (Yamamoto et al. 2005) and in interaction with viral movement protein facilitating cell-to-cell movement of virus (Lu et al. 2009). We identified 104 J-protein coding genes in rice genome. Transcript expression analysis revealed that various J-genes are regulated under different stress conditions and the genes expressed constitutively or regulated developmentally. Yeast mutant of Ydj1 protein was employed for functional complementation by J-proteins of rice.
Materials and methods
Identification of J-proteins in rice genome
Hidden Markov model-based DnaJ domain (chaperone J-domain superfamily) entries of Oryza sativa (var. japonica) genome were retrieved from Superfamily1.75 (http://supfam.org/). As the genome information in Superfamily1.75 was based on Rice Genome Annotation Project (RGAP) 5.0, the retrieved genes were subsequently analyzed in the RGAP release 6.1 (http://rice.plantbiology.msu.edu/). After removing the redundant entries, protein sequences of 125 DnaJ domain containing genes identified in rice genome were downloaded from RGAP. Subsequent analysis of these genes in SMART database to confirm the presence of DnaJ domain led to 123 DnaJ domain containing sequences and two genes (Os01g73020 and Os10g33910) containing PAM16 domain. PAM16 domain is considered as DnaJ-like protein. PAM16 sequences were excluded in tree analysis. Following the strict criteria of HPD motif containing J-domain, 104 genes qualified as J-proteins of rice. Os10g42439 was manually curated based on the available FL-cDNA sequence. Os03g12236 in RGAP coding for 256 amino acid protein showed partial domains of type I DnaJ suggesting incomplete or truncated protein. FGENESH analysis of genomic sequence of Os03g12236 at softberry (www.softberry.com/) revealed presence of one gene having 10 exons coding for 462 amino acid protein. The amino acid sequence deduced from softberry, on analysis in SMART database showed presence of all characteristic domains of type I J-proteins (Electronic supplementary material (ESM) Fig. 1).
Multiple sequence alignment of J-proteins was performed using the Clustal X 2.0 (Larkin et al. 2007) with default parameters. The NJ tree with 1,000 bootstrap was constructed in Clustal X 2.0 and viewed using tree drawing tool FigTree (http://tree.bio.ed.ac.uk/software/figtree/). Subcellular localization of J-proteins was analyzed at WoLFPSORT, Predotar, PSORT, and TargetP database. Consensus localization in two algorithms was considered as the subcellular compartment for the specific entries (ESM Table 1).
Isolation of RNA, RT-PCR analysis, and cloning of FL-cDNA
Total RNA was isolated from stressed and nonstressed (control), 3-week-old rice seedlings [O. sativa L; cultivar Pusa basmati 1 (PB1)] using TRI reagent (Sigma, USA) as per the manufacturer’s instructions. RNA quantification was carried out spectrophotometrically and the quality of RNA was analyzed by gel electrophoresis. For cDNA synthesis, 5 μg of total RNA of each sample was reverse transcribed using reverse transcriptase (RevertAid H Minus Reverse transcriptase, MBI, Fermentas) and reaction was performed according to manufacturers’ protocol. Reverse transcription polymerase chain reaction (RT-PCR) amplification was performed using gene specific primers listed in ESM Table 2.
For yeast mutant complementation and yeast-2-hybrid (Y2H) analysis, J-genes were amplified using FL-cDNA clones procured from Rice Genome Resource Centre, Japan (primers used are listed in ESM Table 2). PCR was carried out using PhusionTMHi-Fi DNA polymerase in presence of 1 % DMSO in a 50 μl reaction. The inserts digested with specific enzymes (ESM Table 2) were cloned in pAD-GAL4 or pBD vectors. For Y2H, the vectors were transformed in YRG2 cells. For yeast mutant complementation, the genes were cloned in p426 vector (Mumberg et al. 1995) under the control of glyceraldehydes-3-phosphate dehydrogenase (GPD) promoter and transformed in mutant cells. The phenotype of the yeast was scored by incubating the plates spotted with tenfold serial dilutions of yeast cells. Prior to spotting the OD600 of yeast cells was normalized to 0.2.
Results
In silico characterization of rice J-proteins
Rice genome contains a large number of J-domain containing proteins. The size of these proteins ranges from 112 (12 kD) to 1,507 (164 kD) amino acids (Table 1). Unlike other Hsp gene families, the molecular mass range of J-proteins is significantly large. This study identified 104 potential J-proteins belonging to 12 type I, 9 type II, and 83 type III (Table 1). Apart from the above 104 J-proteins, rice genome contains 21 sequences which possess J-domain but lack the crucial HPD motif present in the turn between helixII and helixIII (ESM Table 3 and ESM Fig. 2). Out of these 21 proteins, two proteins have PAM16 domain. PAM16 domain proteins are considered as type IV J-proteins or J-like proteins. Rajan and D’Silva (2009) reported four such type IV genes in Arabidopsis genome.
Table 1.
J-proteins of rice
| Locus id | Protein name | Length (aa) | DnaJ Domain | E valuea | Additional domains | Localization |
|---|---|---|---|---|---|---|
| Type A | ||||||
| Os02g43930 | OsDjA1 | 422 | 8–108 | 1.96e-34 | ZN, DnaJ_C | C/N |
| Os02g56040 | OsDjA2 | 488 | 65–176 | 5.23e-34 | ZN, DnaJ_C | CP |
| Os03g12236 | OsDjA3 | 257 | 62–166 | 7.07e-32 | ZN, DnaJ_C, TMD | CP |
| Os03g44620 | OsDjA4 | 418 | 8–108 | 6.28e-36 | ZN, DnaJ_C | C/N |
| Os03g57340 | OsDjA5 | 418 | 8–109 | 6.28e-37 | ZN, DnaJ_C | C/N |
| Os04g46390 | OsDjA6b | 417 | 8–105 | 3.27e-35 | ZN, DnaJ_C | C/N |
| Os05g26902 | OsDjA7c | 448 | 87–189 | 3.79e-33 | ZN, DnaJ_C | NP |
| Os05g26926 | OsDjA8c | 448 | 87–189 | 3.79e-33 | ZN, DnaJ_C | NP |
| Os06g02620 | OsDjA9 | 443 | 72–178 | 9.29e-32 | ZN, DnaJ_C | M |
| Os06g11440 | OsDjD10 | 1293 | 932–1,001 | 3.27e-18 | ZN, DnaJ_C | C/N |
| Os12g07060 | OsDjA11 | 420 | 65–170 | 2.22e-31 | ZN, DnaJ_C | CP |
| Os12g42440 | OsDjA12 | 468 | 10–88 | 3.79e-28 | ZN, DnaJ_C | C/N |
| Type B | ||||||
| Os01g13760 | OsDjB1 | 350 | 3–97 | 1.7e-31 | DnaJ_C | C/N |
| Os01g65480 | OsDjB2 | 328 | 8–69 | 5.63e-17 | DnaJ_C | C/N |
| Os02g03600 | OsDjB3 | 390 | 3–95 | 5.89e-31 | DnaJ_C | C/N |
| Os02g20394 | OsDjB4 | 350 | 3–73 | 1.02e-32 | DnaJ_C | C/N |
| Os05g03630 | OsDjB5 | 323 | 4–98 | 1.44e-31 | DnaJ_C | C/N |
| Os05g06440 | OsDjB6 | 348 | 22–125 | 7.07e-34 | DnaJ_C | ER |
| Os05g48810 | OsDjB7 | 363 | 3–73 | 3.14e-32 | DnaJ_C | C/N |
| Os08g06460 | OsDjB8 | 343 | 3–113 | 1.44e-31 | DnaJ_C | C/N |
| Os08g28700 | OsDjB9 | 345 | 1–105 | 6.41e-32 | DnaJ_C | C/N |
| Type C | ||||||
| Os01g01160 | OsDjC1 | 191 | 50–128 | 3.93e-23 | CP | |
| Os01g06454 | OsDjC2 | 113 | 43–104 | 2.36e-16 | SP, PAM18 | CP |
| Os01g17030 | OsDjC3 | 151 | 6–77 | 2.36e-19 | TMD | C/N |
| Os01g17040 | OsDjC4 | 212 | 4–75 | 4.32e-20 | TMD | C/N |
| Os01g25320 | OsDjC5 | 949 | 782–946 | 3.79e-37 | C/N | |
| Os01g27740 | OsDjC6 | 1009 | 57–133 | 1.83e-19 | C/N | |
| Os01g32870 | OsDjC7 | 404 | 17–98 | 3.01e-27 | cc | C/N |
| Os01g33800 | OsDjC8 | 604 | 295–389 | 2.09e-20 | cc | C/N |
| Os01g37560 | OsDjC9 | 381 | 59–190 | 2.22e-27 | DUF1977 | C/N |
| Os01g42190 | OsDjC10 | 198 | 9–100 | 2.09e-25 | C/N | |
| Os01g44310 | OsDjC11 | 1473 | 1,323–1,471 | 1.05e-35 | C/N | |
| Os01g50700 | OsDjC12 | 653 | 6–102 | 3.14e-28 | dehydrin | C/N |
| Os01g53020 | OsDjC13 | 343 | 77–174 | 6.54e-20 | Fe–S cluster | CP |
| Os01g69930 | OsDjC14 | 745 | 62–161 | 1.31e-22 | cc | C/N |
| Os01g74580 | OsDjC15 | 472 | 347–457 | 5.23e-31 | 5 TPR, SP | M |
| Os02g10180 | OsDjC16 | 477 | 351–461 | 3.4e-31 | 5 TPR, SP | NP |
| Os02g10220 | OsDjC17 | 283 | 26–105 | 9.81e-27 | C/N | |
| Os02g30620 | OsDjC18 | 735 | 56–132 | 1.06e-23 | C/N | |
| Os02g35000 | OsDjC19 | 378 | 2–102 | 7.59e-28 | C/N | |
| Os02g46640 | OsDjC20 | 122 | 10–99 | 9.68e-28 | C/N | |
| Os02g50760 | OsDjC21 | 443 | 28–99 | 2.62e-26 | C/N | |
| Os02g52270 | OsDjC22 | 133 | 29–117 | 1.96e-27 | C/N | |
| Os02g54130 | OsDjC23 | 272 | 19–91 | 1.3e-23 | C/N | |
| Os03g04400 | OsDjC24 | 297 | 7–83 | 1.7e-15 | RRM | C/N |
| Os03g10180 | OsDjC25 | 607 | 491–601 | 2.36e-26 | C/N | |
| Os03g15480 | OsDjC26 | 298 | 23–147 | 1.05e-20 | CP | |
| Os03g18200 | OsDjC27 | 664 | 25–118 | 4.58e-28 | cc | ER |
| Os03g18870 | OsDjC28 | 167 | 21–92 | 1.22e-24 | CP | |
| Os03g20730 | OsDjC29 | 166 | 54–124 | 1.02e-22 | CP | |
| Os03g28310 | OsDjC30 | 749 | 62–133 | 1.83e-23 | C/N | |
| Os03g36160 | OsDjC31 | 293 | 72–133 | 1.96e-15 | C/N | |
| Os03g51830 | OsDjC32 | 239 | 165–230 | 3.66e-13 | TMD | C/N |
| Os03g54150 | OsDjC33 | 612 | 503–603 | 9.81e-07 | C/N | |
| Os03g55360 | OsDjC34 | 506 | 44–111 | 1.96e-18 | SP | M |
| Os03g56540 | OsDjC35 | 129 | 43–103 | 4.58e-17 | PAM18 | M |
| Os03g60790 | OsDjC36 | 269 | 47–136 | 2.62e-25 | CP | |
| Os03g61550 | OsDjC37 | 261 | 31–104 | 1.57e-18 | CP | |
| Os03g61730 | OsDjC38 | 726 | 429–511 | 1.57e-18 | 5 TMD | CP |
| Os03g62120 | OsDjC39 | 478 | 65–130, 252–317 | 5.76e-12 | C/N | |
| Os03g62130 | OsDjC40 | 277 | 69–136 | 2.36e-11 | C/N | |
| Os03g62140 | OsDjC41 | 288 | 71–136 | 3.4e-12 | C/N | |
| Os03g62150 | OsDjC42 | 263 | 64–126 | 1.83e-13 | C/N | |
| Os04g24180 | OsDjC43 | 682 | 96–197 | 1.09e-24 | TMD, Sec63, SP | ER |
| Os04g31940 | OsDjC44 | 730 | 55–122 | 4.32e-23 | C/N | |
| Os04g57880 | OsDjC45 | 487 | 60–147 | 5.63e-19 | Fe–S cluster, TMD | CP |
| Os04g59060 | OsDjC46 | 275 | 167–269 | 2.09e-11 | SP | M |
| Os05g01590 | OsDjC47 | 231 | 162–224 | 1.22e-11 | SP | M |
| Os05g30130 | OsDjC48 | 368 | 100–173 | 5.23e-27 | DUF1977 | C/N |
| Os05g31062 | OsDjC49 | 395 | 176–322 | 1.98e-22 | 4TPR | C/N |
| Os05g45350 | OsDjC50 | 351 | 11–140 | 2.75e-21 | Fe–S cluster | CP |
| Os05g46620 | OsDjC51 | 339 | 2–100 | 8.64e-31 | C/N | |
| Os05g50370 | OsDjC52 | 1424 | 1,269–1,422 | 4.32e-36 | C/N | |
| Os06g09560 | OsDjC53 | 236 | 13–107 | 3.01e-22 | C/N | |
| Os06g13060 | OsDjC54 | 436 | 21–93 | 1.83e-27 | C/N | |
| Os06g34440 | OsDjC55 | 1019 | 55–132 | 1.57e-19 | C/N | |
| Os06g44160 | OsDjC56 | 143 | 44–102 | 1.57e-15 | CP | |
| Os07g03270 | OsDjC57 | 238 | 166–229 | 5.76e-14 | cc | C/N |
| Os07g09450 | OsDjC58 | 113 | 43–104 | 9.42e-17 | SP, PAM18 | NP |
| Os07g28800 | OsDjC59 | 270 | 149–264 | 6.8e-13 | M | |
| Os07g43330 | OsDjC60 | 271 | 68–146 | 8.11e-23 | SP | CP |
| Os07g44310 | OsDjC61 | 135 | 5–70 | 9.81e-13 | TMD | C/N |
| Os08g35160 | OsDjC62 | 159 | 7–86 | 6.15e-23 | C/N | |
| Os08g36980 | OsDjC63 | 175 | 7–83 | 5.23e-18 | ZnF_CSL | C/N |
| Os08g37270 | OsDjC64 | 397 | 63–141 | 5.5e-08 | AT_hook | C/N |
| Os08g41110 | OsDjC65 | 395 | 2–101 | 6.28e-30 | C/N | |
| Os08g43490 | OsDjC66 | 147 | 44–117 | 2.88e-20 | CP | |
| Os09g20320 | OsDjC67 | 330 | 53–125 | 9.42e-20 | C/N | |
| Os09g28590 | OsDjC68 | 197 | 8–83 | 1.57e-17 | ZnF_CSL | C/N |
| Os09g28890 | OsDjC69 | 372 | 63–141 | 9.42e-08 | AT_hook | C/N |
| Os09g32050 | OsDjC70 | 396 | 3–101 | 4.97e-30 | C/N | |
| Os10g03610 | OsDjC71 | 254 | 57–138 | 3.27e-10 | NP | |
| Os10g11012 | OsDjC72 | 373 | 71–126 | 0.000389 | C/N | |
| Os10g36370 | OsDjC73 | 541 | 9–84 | 8.11e-24 | C/N | |
| Os10g42439 | OsDjC74 | 1508 | 1,591–1,631 | 3.14e-12 | C/N | |
| Os11g36530 | OsDjC75 | 291 | 153–224 | 7.33e-17 | CP | |
| Os11g36960 | OsDjC76 | 1053 | 52–131 | 1.83e-20 | C/N | |
| Os11g37000 | OsDjC77 | 625 | 57–129 | 4.19e-16 | C/N | |
| Os11g43950 | OsDjC78 | 889 | 740–886 | 1.22e-36 | C/N | |
| Os12g15590 | OsDjC79 | 310 | 38–115 | 4.19e-25 | 2 TMD, SP | C/N |
| Os12g27070 | OsDjC80 | 261 | 100–173 | 7.33e-16 | M | |
| Os12g31840 | OsDjC81 | 608 | 7–81 | 8.64e-22 | 2 ZnF_C2H2 | C/N |
| Os12g36180 | OsDjC82 | 925 | 785–923 | 4.45e-37 | cc | C/N |
| Os12g41820 | OsDjC83 | 545 | 219–351 | 4.58e-19 | 4 TMD | PM |
aE value of DnaJ domain
bRNB8 in Lu et al. (2009)
cBAB70509, accession number of protein according to Yamamoto et al. (2005) and mapped to chromosome 5. Both DnaJ genes present on chromosome 5 code for exactly same protein, therefore, BAB70509 represents both the genes
DnaJ_C- C-terminal domain, RRM RNA recognition motif, SP signal peptide, TMD transmembrane domain, cc coiled coil domain, TPR tetratricopeptide region, PAM Presequence translocase-associated protein import motor, DUF domain of unknown function, C/N cytosol/nuclear, CP chloroplast, ER endoplasmic reticulum, M mitochondria, PM plasma membrane, NP not predicted
Notably, rice J-proteins were spread throughout the rice genome on all the 12 chromosomes. In addition to the signature J-domain, other diverse domains like TPR domain, Fe–S cluster or AT-hook domain provide further specificity to type III J-proteins (Table 1). According to the domain features of the J-proteins, rice types I and II proteins grouped in two separate clades along with Arabidopsis J-proteins in the phylogenetic tree drawn on aligned amino acid sequences (Fig. 1). On the other hand, type III J-proteins were scattered on several clades due to the varied arrangement of domains. One of the J-protein, i.e., Os03g62120 was found to have two DnaJ domains. There were 12 pairs of segmental duplicates (Fig. 1) and four genes were tandem duplicates arranged in same orientation on chromosome 3 (ESM Fig. 3) suggesting that duplication event has little contribution in expansion of such a large gene family. The duplicated pairs of genes were present on the same clades and were found to localize to same cell compartments. J-proteins of each type were localized to various cellular compartments (Table 1). In type I, six proteins were localized to cytosol/nuclear, three to chloroplast, one to mitochondria, and consensus localization could not be predicted for two proteins. No J-protein of type I was localized to ER in rice and Arabidopsis. In type II, eight proteins were cytosol/nuclear and 1 was ER localized and interestingly no protein was localized to mitochondria or chloroplast, the two endosymbiont organelles. In type III, 55 proteins were cytosol/nuclear, 15 proteins chloroplastic, 7 mitochondrial, and 2 ER localized.
Fig. 1.
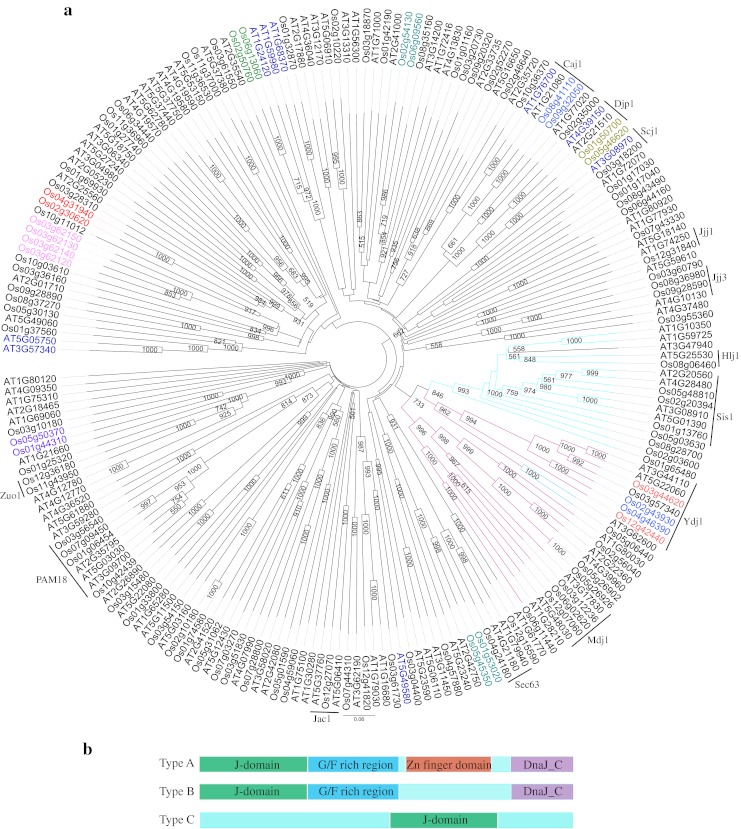
Phylogenetic tree of J-proteins of rice and Arabidopsis. a Amino acid sequences of J-proteins of rice and Arabidopsis (Rajan and D’Silva 2009) were aligned in Clustal X (2.0). The bootstrap NJ tree was generated in clustal X (2.0) and viewed in FigTree v1.1.1.1. The bootstrap support value (>50 %) is shown at the nodes. Clade in purple depicts type I J-proteins, clade in blue depicts type II J-proteins and black line clades are type III J-proteins. Arabidopsis J-proteins in blue font are type II (in Rajan and D’Silva 2009), but cluster in this tree with type III proteins. The segmental duplicated pairs are marked in same color font. b Diagrammatic representation of different types of J-domain proteins. In type C, the J-domain can be located anywhere in the protein sequence. In type D, a J-like domain is present
Nomenclature of rice J-genes
J-protein family has not been extensively studied in rice. As random nomenclature of the J-genes could lead to redundant situation, rice J-proteins identified in this study were designated a new nomenclature which is essentially adopted from Ohtsuka and Hata (2000) proposed for mammalian J-proteins. Accordingly, J-domain proteins were classified into OsDjA, OsDjB, and OsDjC where Dj denotes DnaJ domain and A, B, and C represent types I, II, and III, respectively, and prefix “Os” is the source of DnaJ protein, i.e., O. sativa. In the original scheme of Ohtsuka and Hata (2000), the type of DnaJ is followed by an Arabic numeral and a lowercase alphabet to denote the chronological order in which the sequence data is deposited in the database and the splice variants of the gene, respectively. There are only two reports in literature on J-proteins of rice (Table 1). In this study, the Arabic numbers are depicted on the basis of order in which the J-genes are arranged on the chromosomes (Table 1). Accordingly, the first gene in Table 1 is OsDjA1, where “Os” is source organism O. sativa, “Dj” is DnaJ/Hsp40/J-protein, “A” denotes type I, “1” denotes the first protein in type I on chromosome.
Rice J-proteins are induced by heat and other abiotic stresses
Transcript expression profiles of various J-proteins in response to abiotic stresses were analyzed in silico from microarray data at Genevestigator (https://www.genevestigator.com/gv/). In addition, expression of selective genes was assessed by semiquantitative RT-PCR. The results indicated diverse expression patterns for the J-genes (Fig. 2 and ESM Fig. 4). A large number of J-genes were constitutively expressed. The expression profiles as seen by RT-PCR experiments were largely similar to the microarray meta-analysis results (Fig. 2 and ESM Fig. 4). This was especially noticeable in the case of Os01g53020, Os03g18200, Os05g48810, Os06g02620, Os06g09560, and Os11g10990 genes. Several J-genes like Os03g57340, Os06g02620, Os05g48810, Os03g15033, Os01g50700, Os03g18200, Os06g09560, and Os06g44160 were heat stress (HS) inducible. Moreover, a positive correlation between transcript expression levels and the duration of stress was distinctly observed for Os01g25320, Os03g18200 and Os06g09560 in RT-PCR. Most of the type A genes were upregulated under anoxia. In type B, Os05g48810 gene showed a highly induced expression level in almost all the stress conditions except cold stress (CS). Os3g18200 was significantly downregulated in response to CS as evident from RT-PCR as well as microarray data (Fig. 2 and ESM Fig. 4). Os03g56540 and Os06g09560 genes of type C showed an increased expression level under all stress conditions except CS. The genes like Os08g03380, Os09g21250, and Os11g10990 without HPD domain were specifically induced by HS (Fig. 2).
Fig. 2.
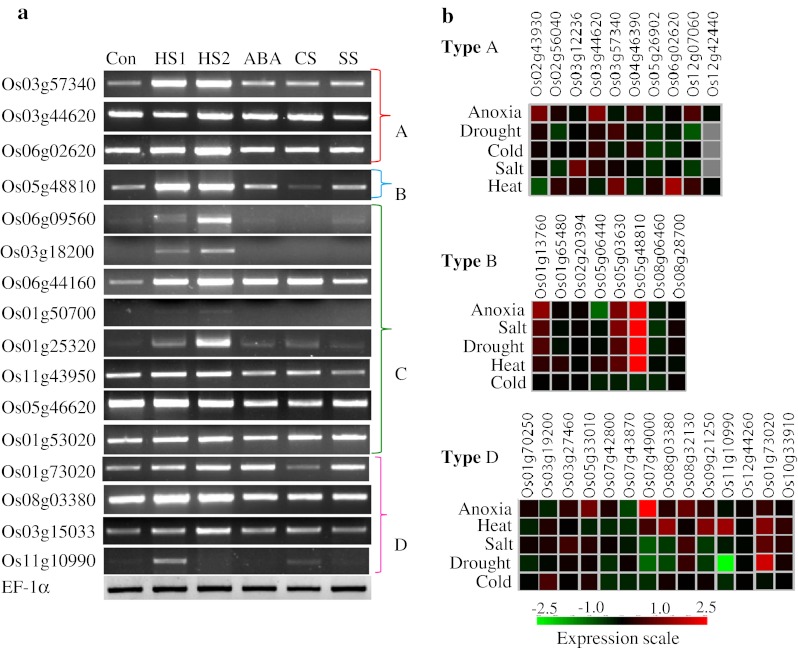
Expression analysis of selective J-proteins of rice by semiquantitative RT-PCR. a For RT-PCR cDNA used was synthesized using 5 μg RNA isolated from rice seedlings stressed as follows: HS1 heat shock at 42 °C for 10 min, HS2 heat shock at 42 °C for 60 min, ABA 100 μM abcissic acid for 3 h, CS cold stress at 6 °C for 6 h, SS salt stress in 150 mM NaCl for 6 h. Con unstressed control maintained at 26 °C. bIn silico expression analysis of types A, B, and D J-proteins of rice under stress conditions. Expression profiles of type C J-proteins are presented in ESM Fig. 4. Primers used for RT-PCR are listed in ESM Table 2
Developmental regulation of J-proteins
Transcripts for Os02g43930, Os03g44620, and Os04g46390 were highly expressed in the dough stage, indicating possible importance of these proteins at the time of seed setting (ESM Fig. 5). Os02g43930 transcript showed relatively high expression in most of the tissues and throughout the development whereas Os04g46390 transcripts showed high expression levels in vegetative tissues. Os03g62120 and Os03g62140 transcripts showed high expression at all the developmental stages and in almost all the tissues (ESM Fig. 4). Os08g06460 and Os02g20394 genes appear to be important specifically in the pollen tissues (ESM Fig. 5). Drawing parallels from a report of a J-protein ortholog that is crucial for thermotolerance in pollen and pollen tube in Arabidopsis and thus maintaining male fertility (Yang et al. 2009), similar roles for the pollen-associated J-genes of rice may be proposed.
Rice J-protein functionally complement corresponding yeast mutants
An important approach for functional analysis of stress genes is through analysis of mutants. However, genetic resources of rice are not exhaustively available. In yeast, Ydj1 (YNL064C) is the most abundant J-protein and is reportedly required for the normal growth of yeast cells (Caplan et al. 1992). It is reported that though Ydj1 expression is not heat inducible, mutation in this gene leads to slow growth at 30 °C and inviability at elevated temperatures (Caplan et al. 1992). Orthologous genes of Ydj1 in rice and in Arabidopsis (AtJ3; At3g44110) are also strongly expressed. The mutant of this gene in Arabidopsis is reported to have late flowering phenotype (Shen et al. 2011). However, no phenotypic aberration was observed in the mutant Arabidopsis lines in comparison to wild type after heat stress at various temperatures and stress duration (Fig. 3). Seven rice J-proteins (two type A: DjA4 and DjA5; one type B: DjB7; and four type C: DjC9, DjC18, DjC19, and DjC51) were expressed under the control of GPD promoter in ydj1 cells to analyze if they could complement the growth defect of ydj1 mutant. Notably, the growth defect phenotype of ydj1 at 37 °C was rescued by expression of five out of seven rice J-proteins (Fig. 3). Two genes, DjB7 and DjC51 failed to complement the growth defect at 37 °C. DjB7 expressing yeast cells showed slow growth even at 26 °C. The rice DjA4 and DjA5 were more efficient in rescuing the growth defect at 37 °C as compared to other three genes (Dj18, Dj19, and Dj51). A tree drawn on the aligned amino acid sequences of seven rice J-proteins with yeast J-proteins showed that DjA4 and DjA5 are closest orthologues of yeast Ydj1 (Fig. 3). Hence, selective rice J-proteins can function in yeast and can interact with yeast chaperone machinery.
Fig. 3.
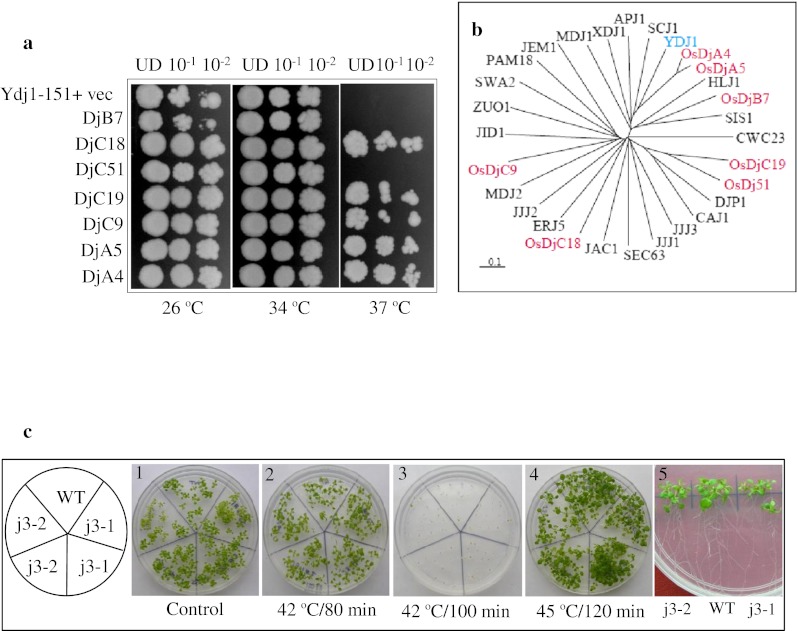
a Complementation of yeast mutant (ydj1Δ) by rice J-proteins. ydj1Δ (MATα ade2-1 leu2-3,112 his3-11, 15 trp1-1 ura3-1can1-100 ydj1-2::HIS3 LEU2::ydj1-151) mutant cells were transformed with specified J-proteins of rice as shown. The cells were grown to log phase, growth was normalized to 0.2 OD600, tenfold serial dilutions were spotted on selection medium and plates were incubated at specified temperatures. UD denotes cells normalized to 0.2 OD600. b Phylogeny of yeast J-proteins and rice J-proteins. The tree was generated in clustalX2.0 from aligned amino acid sequences of rice J-proteins used for ydj1 mutant complementation and yeast J-proteins. The tree was viewed in Treeview1.6.6. c Phenotype analysis of j3 mutant of Arabidopsis. Seven-day old seedlings of T-DNA insertion mutant lines j3-1 (Salk_132923) and j3-2 (Salk_141625) of Arabidopsis J3 protein (At3g44110) were given heat stress at 42 °C for different duration (1 and 5 unstressed, 2 and 3 stressed; 4 seeds heat stressed). The growth response was scored after 10 days of treatments
Physical interaction of rice Hsp70 and J-proteins
Pairwise interactions of J-proteins and Hsp70 were analyzed by yeast-two hybrid assays. Considering that the probability of interaction of proteins localized in the same compartment would be higher, the nucleocytoplasmic localized J-proteins and Hsp70 proteins were analyzed in this study. It is shown that yeast Ydj1 interacts with yeast Hsp70, Ssa1 (Caplan et al. 1992). Hsp70 proteins analyzed in this study included Os03g16880, Os02g60620, Os03g16920, and Os11g47760, and J-proteins included Os01g37560 (DjC9), Os02g35000 (DjC19), Os02g30620 (DjC18), Os05g46620 (DjC51), and Os05g48810 (DjB7). Notably, none of the J-proteins and Hsp70 proteins from the above range yielded positive interaction (results not shown).
Discussion
Previous studies divulged the complexity and functional diversity of J-protein family of yeast and Arabidopsis (Walsh et al. 2004; Rajan and D’Silva 2009). Our analysis highlights the diversity of J-proteins at sequence as well as at expression levels in rice. Interestingly, we found that larger genome of rice contains 104 J-proteins as against smaller genome of Arabidopsis which has 116 J-proteins. Both in prokaryotic and eukaryotic organisms, J-proteins and Hsp70 chaperones work in conjunction in controlling various cellular processes. Constitutive as well as stress modulated expression of J-proteins in this study suggests that Hsp70/J-protein bichaperone machine of rice may be involved in basal as well as stress-related cellular functions. Some functions of these proteins can be envisioned based on studies in other systems. J-proteins localized to ER may be crucial as proper folding of secretory proteins in ER is essential for the maintenance of protein quality, homeostasis and cell viability (Vembar et al. 2009). Defect in protein translocation in T-DNA insertion lines in ER proteins, AtERDJ2A and AtERDJ5C has a probable effect on pollen germination (Yamamoto et al. 2008; Yang et al. 2009). While cytosolic, ER and mitochondrial Hsp70 and J-proteins functions are conserved in all organisms, functions of chloroplastic J-proteins are unique to plants. In rice, 18 J-proteins are predicted to be localized in chloroplast. Functional association of J-proteins and Hsp70 was revealed in the biogenesis of thylakoid membranes in Chlamydomonas (CDJ2/Hsp70B; Liu et al. 2005) and movement of chloroplast in Arabidopsis that has implication in photorelocation of chloroplast (Suetsugu et al. 2010). JAC1, the J-protein involved in movement of chloroplast, was functionally impaired if its HPD tripeptide was mutated to AAA (Suetsugu et al. 2010). It will be worthwhile to assess the role of 21 J-proteins in rice that do not contain HPD motif. Further, Fe–S cluster containing proteins in Chlamydomonas function as redox switches (Dorn et al. 2010). Three Fe–S cluster containing J-proteins are present in rice (OsDjC13, OsDjC45, and OsDjC50) and like in Chlamydomonas, expression of these genes was downregulated under heat stress. Based on similarity in the expression pattern and localization, it may be argued that Fe–S cluster containing J-proteins in rice are also involved in redox sensing. The expression patterns of J-genes in specific cellular compartments differed drastically, suggesting their functional diversity. Functional diversity of J-proteins is further noted from the expression profiles of 4 J-proteins of type A containing –CAQQ sequence at the C-terminus. All these four proteins (DjA1, DjA4, DjA5, and DjA6) showed distinct developmental and stress regulation. However, expression of only DjA5 was elevated at high temperature. The spatial expression of all 4 above proteins was differential during developmental stages.
The functionality of J-protein machinery of rice was analyzed in this study by expressing 7 rice J-domain proteins in ydj1 mutant. Both E. coli and yeast mutants have been used for functional analysis of J-proteins from evolutionary diverse species (Nicoll et al. 2007; Vembar et al. 2009). This study shows that the functions of yeast cytosolic Ydj1 could be performed by expression of diverse J-proteins of rice. Ydj1 protein of yeast belongs to Type A class that contains an N-terminal J-domain, a Zn-finger domain and a C-terminal domain. In addition, Ydj1 contains a conserved -CAAX box motif at C-terminal end, which is reportedly a site for farnesylation. The latter reaction promotes association of these proteins to the cytosolic side of the ER membrane (Caplan et al. 1992). Caplan et al. (1992) have suggested that this post-translational modification is crucial for the function of yeast Ydj1 protein at elevated temperature. Rice DjA4 and DjA5, structurally similar to Ydj1 performed more efficiently than other J-proteins tested in this study in ameliorating the growth defect of ydj1 mutant at high temperature. Two rice J-proteins, DjB7 and DjC51, however failed to rescue the growth defect of yeast Ydj1 mutant. This failure in functional complementation could be due to specificity of J-proteins for their partner client proteins (Vembar et al. 2009). The other plausible reason could be difference in cellular localization of the above proteins (Sahi and Craig 2007). The rescuing of growth defect by selective J-proteins of rice indicates their capability to stimulate the ATPase activity of yeast Hsp70 as reported for other systems (Nicoll et al. 2007; Vembar et al. 2009). Moreover, it shows that sequences other than J-domains can also dictate the functional specificity of J-proteins (Sahi and Craig 2007; Hennessy et al. 2005). In addition, domain swapping studies revealed that J-domains of all J-proteins cannot functionally substitute for each other (Hennessy et al. 2000; Kluck et al. 2002; Nicoll et al. 2007). Further experiments are required to narrow down on the sequences other than J-domains for function of J-protein.
Like in other organisms, rice J-domain proteins are one of the largest and most diverse family of chaperones. It is thus suggested that multiple J-proteins can pair with single Hsp70. The pairwise interaction analysis of selected cytosolic J-proteins and Hsp70 by yeast-2 hybrid analysis did not show positive interaction in this study. It is possible that Hsp70 and J-proteins do not interact directly. There are different views proposed on the interaction of Hsp70 with J-proteins. DnaK and DnaJ were demonstrated to bind to different sites of the same substrate with substrate binding domain forming a ternary complex (Han and Christen 2003). Terada and Oike (2010) confirmed independent binding of Hsp70 and J-proteins to unfolded proteins to distinct segments of the peptide. On the contrary, direct physical interaction of Hsp70 to J-protein (both mitochondrial) was reported recently (Zhou et al. 2012). Furthermore, we predict that ∼80 J-proteins (this study) and 11 Hsp70 genes (unpublished data) are localized to cytosol/nucleus compartment. Considering such high numbers of cytosolic J-proteins and Hsp70 members, the appropriate pairing may have been missed with limited J-proteins used in pairwise interactions carried out in this study. We aim to study Hsp70 and J-protein interaction by screening cDNA library of rice with different rice Hsp70 as baits in future course.
Electronic supplementary information
FGENESH analysis of Os03g12236 (PDF 27 kb)
Multiple sequence alignment of J-domain of rice J-proteins showing consensus sequences and four helices characteristic of J-domain. The sequences were aligned in Clustal and edited in Jalview. The consensus sequence and the Logo were derived using Jalview. The amino acid residues are colored by default color scheme of ClustalX. Conserved tripeptide HPD is present between helix II and III. J-proteins in which HPD tripeptide was absent are depicted in red color font (EPS 4,581 kb)
Tandem arrangement of J-proteins on chromosome (PPTX 387 kb)
Microarray based expression meta-analysis of type C J-genes in abiotic stresses (a), types C and D during development stages of rice plant (b) and in various tissues (c) (PPTX 346 kb)
Microarray based expression meta-analysis of types A and B J-genes in development stages (a) and various tissues of rice plant (b) (PPTX 175 kb)
Prediction of localization of J-proteins of rice (XLSX 16 kb)
List of primers used in this study (XLSX 10 kb)
Details of J-proteins of rice without HPD tripeptide in J-domain (XLSX 12.2 kb)
Acknowledgement
We thank Jeffrey L. Brodsky of the University of Pittsburgh, USA for DnaJ yeast mutants and Hao Yu of the National University of Singapore, Singapore for kindly providing the seeds of T-DNA insertion mutants of Arabidopsis J3 protein (j3-1 and j3-2). We thank Centre for Plant Molecular Biology and Indo-Finland project, Department of Biotechnology, Govt. of India, for financial support.
References
- Cajo GC, Horne BE, Kelley WL, Schwager F, Georgopoulos C, Genevaux P. The role of the DIF motif of the DnaJ (Hsp40) co-chaperone in the regulation of the DnaK (Hsp70) chaperone cycle. J Biol Chem. 2006;281:12436–12444. doi: 10.1074/jbc.M511192200. [DOI] [PubMed] [Google Scholar]
- Caplan AJ, Cyr DM, Douglas MG. YDJ1p facilitates polypeptide translocation across different intracellular membranes by a conserved mechanism. Cell. 1992;71:1143–1155. doi: 10.1016/S0092-8674(05)80063-7. [DOI] [PubMed] [Google Scholar]
- Chapple JP, van der Spuy J, Poopalasundaram S, Cheetham ME. Neuronal DnaJ proteins HSJ1a and HSJ1b: a role in linking the Hsp70 chaperone machine to the ubiquitin–proteasome system? Biochem Soc Trans. 2004;32:640–642. doi: 10.1042/BST0320640. [DOI] [PubMed] [Google Scholar]
- Cyr DM, Lu X, Douglas MG. Regulation of Hsp70 function by a eukaryotic DnaJ homolog. J Biol Chem. 1992;267:20927–20931. [PubMed] [Google Scholar]
- Dorn KV, Willmund F, Schwarz C, Henselmann C, Pohl T, Hess B, Veyel D, Usadel B, Friedrich T, Nickelsen J, Schroda M. Chloroplast DnaJ-like proteins 3 and 4 (CDJ3/4) from Chlamydomonas reinhardtii contain redox-active Fe–S clusters and interact with stromal HSP70B. Biochem J. 2010;427:205–215. doi: 10.1042/BJ20091412. [DOI] [PubMed] [Google Scholar]
- Glover JR, Lindquist S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/S0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- Griessl MH, Jungkunz I, Sonnewald U, Muller YA. Purification, crystallization and preliminary X-ray diffraction analysis of the Hsp40 protein CPIP1 from Nicotiana tabacum. Acta Crystallogr F Struct Biol Crystallogr Commun. 2012;68:236–239. doi: 10.1107/S1744309111055928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W, Christen P. Mechanism of the targeting action of DnaJ in the DnaK molecular chaperone system. J Biol Chem. 2003;278:19038–19043. doi: 10.1074/jbc.M300756200. [DOI] [PubMed] [Google Scholar]
- Hennessy F, Cheetham ME, Dirr HW, Blatch GL. Analysis of the levels of conservation of the J domain among the various types of DnaJ-like proteins. Cell Stress Chaperones. 2000;5:347–358. doi: 10.1379/1466-1268(2000)005<0347:AOTLOC>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessy F, Nicoll WS, Zimmermann R, Cheetham ME, Blatch GL. Not all J domains are created equal: implications for the specificity of Hsp40–Hsp70 interactions. Protein Sci. 2005;14:1697–1709. doi: 10.1110/ps.051406805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofius D, Maier AT, Dietrich C, Jungkunz I, Bornke F, Maiss E, Sonnewald U. Capsid protein-mediated recruitment of host DnaJ-like proteins is required for Potato virus Y infection in tobacco plants. J Virol. 2007;81:11870–11880. doi: 10.1128/JVI.01525-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley WL. Molecular chaperones: how J domains turn on Hsp70s. Curr Biol. 1999;9:R305–R308. doi: 10.1016/S0960-9822(99)80185-7. [DOI] [PubMed] [Google Scholar]
- Kluck CJ, Patzelt H, Genevaux P, Brehmer D, Rist W, Schneider-Mergener J, Bukau B, Mayer MP. Structure–function analysis of HscC, the Escherichia coli member of a novel subfamily of specialized Hsp70 chaperones. J Biol Chem. 2002;277:41060–41069. doi: 10.1074/jbc.M206520200. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Liu C, Willmund F, Whitelegge JP, Hawat S, Knapp B, Lodha M, Schroda M. J-domain protein CDJ2 and HSP70B are a plastidic chaperone pair that interacts with vesicle-inducing protein in plastids 1. Mol Biol Cell. 2005;16:1165–1177. doi: 10.1091/mbc.E04-08-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Cyr DM. The conserved carboxyl terminus and zinc finger-like domain of the co-chaperone Ydj1 assist Hsp70 in protein folding. J Biol Chem. 1998;273:5970–5978. doi: 10.1074/jbc.273.10.5970. [DOI] [PubMed] [Google Scholar]
- Lu L, Du Z, Qin M, Wang P, Lan H, Niu X, Jia D, Xie L, Lin Q, Wu Z. Pc4, a putative movement protein of Rice stripe virus, interacts with a type I DnaJ protein and a small Hsp of rice. Virus Genes. 2009;38:320–327. doi: 10.1007/s11262-008-0324-z. [DOI] [PubMed] [Google Scholar]
- Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miernyk JA. The J-domain proteins of Arabidopsis thaliana: an unexpectedly large and diverse family of chaperones. Cell Stress Chaperones. 2001;6:209–218. doi: 10.1379/1466-1268(2001)006<0209:TJDPOA>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miot M, Reidy M, Doyle SM, Hoskins JR, Johnston DM, Genest O, Vitery MC, Masison DC, Wickner S. Species-specific collaboration of heat shock proteins (Hsp) 70 and 100 in thermotolerance and protein disaggregation. Proc Natl Acad Sci U S A. 2011;108:6915–6920. doi: 10.1073/pnas.1102828108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misselwitz B, Staeck O, Rapoport TA. J proteins catalytically activate Hsp70 molecules to trap a wide range of peptide sequences. Mol Cell. 1998;2:593–603. doi: 10.1016/S1097-2765(00)80158-6. [DOI] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- Nicoll WS, Botha M, McNamara C, Schlange M, Pesce ER, Boshoff A, Ludewig MH, Zimmermann R, Cheetham ME, Chapple JP, Blatch GL. Cytosolic and ER J-domains of mammalian and parasitic origin can functionally interact with DnaK. Int J Biochem Cell Biol. 2007;39:736–751. doi: 10.1016/j.biocel.2006.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka K, Hata M. Mammalian HSP40/DNAJ homologs: cloning of novel cDNAs and a proposal for their classification and nomenclature. Cell Stress Chaperones. 2000;5:98–112. doi: 10.1379/1466-1268(2000)005<0098:MHDHCO>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Wang H, Zou Y, Liu C, Liu Y, Wang Y, Zhang W. Over-expression of mitochondrial heat shock protein 70 suppresses programmed cell death in rice. FEBS Lett. 2011;585:231–239. doi: 10.1016/j.febslet.2010.11.051. [DOI] [PubMed] [Google Scholar]
- Qiu XB, Shao YM, Miao S, Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol Life Sci. 2006;63:2560–2570. doi: 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan VB, D’Silva P. Arabidopsis thaliana J-class heat shock proteins: cellular stress sensors. Funct Integr Genomics. 2009;9:433–446. doi: 10.1007/s10142-009-0132-0. [DOI] [PubMed] [Google Scholar]
- Sahi C, Craig EA. Network of general and specialty J protein chaperones of the yeast cytosol. Proc Natl Acad Sci U S A. 2007;104:7163–7168. doi: 10.1073/pnas.0702357104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Kang YG, Liu L, Yu H. The J-domain protein J3 mediates the integration of flowering signals in Arabidopsis. Plant Cell. 2011;23:499–514. doi: 10.1105/tpc.111.083048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sielaff B, Tsai FT. The M-domain controls Hsp104 protein remodeling activity in an Hsp70/Hsp40-dependent manner. J Mol Biol. 2010;402:30–37. doi: 10.1016/j.jmb.2010.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu N, Takano A, Kohda D, Wada M. Structure and activity of JAC1 J-domain implicate the involvement of the cochaperone activity with HSC70 in chloroplast photorelocation movement. Plant Signal Behav. 2010;5:1602–1606. doi: 10.4161/psb.5.12.13915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung DY, Guy CL. Physiological and molecular assessment of altered expression of Hsc70-1 in Arabidopsis. Evidence for pleiotropic consequences. Plant Physiol. 2003;132:979–987. doi: 10.1104/pp.102.019398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano A, Suetsugu N, Wada M, Kohda D. Crystallographic and functional analyses of J-domain of JAC1 essential for chloroplast photorelocation movement in Arabidopsis thaliana. Plant Cell Physiol. 2010;51:1372–1376. doi: 10.1093/pcp/pcq089. [DOI] [PubMed] [Google Scholar]
- Terada K, Oike Y. Multiple molecules of Hsc70 and a dimer of DjA1 independently bind to an unfolded protein. J Biol Chem. 2010;285:16789–16797. doi: 10.1074/jbc.M110.101501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia-Morales MD, Camas-Reyes JA, Cabrera-Ponce JL, Alvarez-Venegas R. The Arabidopsis thaliana SET-domain-containing protein ASHH1/SDG26 interacts with itself and with distinct histone lysine methyltransferases. J Plant Res. 2012;125(5):679–692. doi: 10.1007/s10265-012-0485-7. [DOI] [PubMed] [Google Scholar]
- Vembar SS, Jin Y, Brodsky JL, Hendershot LM. The mammalian Hsp40 ERdj3 requires its Hsp70 interaction and substrate-binding properties to complement various yeast Hsp40-dependent functions. J Biol Chem. 2009;284:32462–32471. doi: 10.1074/jbc.M109.000729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh P, Bursac D, Law YC, Cyr D, Lithgow T. The J-protein family: modulating protein assembly, disassembly and translocation. EMBO Rep. 2004;5:567–571. doi: 10.1038/sj.embor.7400172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Mori Y, Ishibashi T, Uchiyama Y, Ueda T, Ando T, Hashimoto J, Kimura S, Sakaguchi K. Interaction between proliferating cell nuclear antigen (PCNA) and a DnaJ induced by DNA damage. J Plant Res. 2005;118:91–97. doi: 10.1007/s10265-005-0197-3. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Maruyama D, Endo T, Nishikawa S. Arabidopsis thaliana has a set of J proteins in the endoplasmic reticulum that are conserved from yeast to animals and plants. Plant Cell Physiol. 2008;49:1547–1562. doi: 10.1093/pcp/pcn119. [DOI] [PubMed] [Google Scholar]
- Yang KZ, Xia C, Liu XL, Dou XY, Wang W, Chen LQ, Zhang XQ, Xie LF, He L, Ma X, Ye D. A mutation in Thermosensitive Male Sterile 1, encoding a heat shock protein with DnaJ and PDI domains, leads to thermosensitive gametophytic male sterility in Arabidopsis. Plant J. 2009;57:870–882. doi: 10.1111/j.1365-313X.2008.03732.x. [DOI] [PubMed] [Google Scholar]
- Zhou W, Zhou T, Li MX, Zhao CL, Jia N, Wang XX, Sun YZ, Li GL, Xu M, Zhou RG, Li B. The Arabidopsis J-protein AtDjB1 facilitates thermotolerance by protecting cells against heat-induced oxidative damage. New Phytol. 2012;194:364–378. doi: 10.1111/j.1469-8137.2012.04070.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FGENESH analysis of Os03g12236 (PDF 27 kb)
Multiple sequence alignment of J-domain of rice J-proteins showing consensus sequences and four helices characteristic of J-domain. The sequences were aligned in Clustal and edited in Jalview. The consensus sequence and the Logo were derived using Jalview. The amino acid residues are colored by default color scheme of ClustalX. Conserved tripeptide HPD is present between helix II and III. J-proteins in which HPD tripeptide was absent are depicted in red color font (EPS 4,581 kb)
Tandem arrangement of J-proteins on chromosome (PPTX 387 kb)
Microarray based expression meta-analysis of type C J-genes in abiotic stresses (a), types C and D during development stages of rice plant (b) and in various tissues (c) (PPTX 346 kb)
Microarray based expression meta-analysis of types A and B J-genes in development stages (a) and various tissues of rice plant (b) (PPTX 175 kb)
Prediction of localization of J-proteins of rice (XLSX 16 kb)
List of primers used in this study (XLSX 10 kb)
Details of J-proteins of rice without HPD tripeptide in J-domain (XLSX 12.2 kb)


