Abstract
Transposon activity is known to cause chromosome rearrangements in the host genome. Surprisingly, extremely little is known about Dissociation (Ds)-induced chromosome rearrangements in Arabidopsis, where Ds is intensively used for insertional mutagenesis. Here, we describe three Arabidopsis mutants with reduced fertility and propose that excision of a hybrid Ds element induced a large genomic deletion flanking Ds. In the mutants anat and haumea, the deletion mechanism consists of a local Ds transposition from replicated into unreplicated DNA followed by Ds excision, where one end of the newly transposed element and one end of the Ds transposon at the donor site served as substrate for transposase. Excision of this hybrid element reminiscent of a macrotransposon leads to loss of the chromosomal piece located between the two ends, including one full Ds element and the flanking genomic sequence. This mechanism was found to be responsible for several other deletions and occurs at a genetically trackable frequency. Thus, it could be applied to efficiently generate deletions of various sizes in the vicinity of any existing Ds element present in the genome. In the mutant tons missing, a mechanism that involves endogenous repetitive sequences caused a large flanking deletion at a position unlinked to the starter locus. Our study of Ds transposition in Arabidopsis revealed previously undescribed mechanisms that lead to large genomic deletions flanking Ds elements, which may contribute to genome dynamics and evolution.
Keywords: genome evolution, rearrangements, transposition
Transposons are fundamental components of most prokaryotic and eukaryotic genomes and significantly contribute to their variation in size and structure. They are well known to induce genome reorganization as a result of transposition or recombination events. Transposons of various families including Ac/Ds have been associated with flanking deletions (1-8). Hybrid element insertion (HEI) and excision of Drosophila P elements, a mechanism similar to HEI for Tam3 in Antirrhinum, and nonlinear transposition and transposition of partially replicated Ac/Ds macrotransposons in maize create flanking deletions (4, 9-12). These mechanisms have in common that terminal inverted repeats (TIR) of different transposons, located on either the same or different sister chromatids, serve as substrates for transposase. For Ac/Ds, most reports associate chromosomal rearrangements such as deletions with elements of complex structure (11-17), whereas single simple Ds elements are commonly not believed to cause chromosomal rearrangements (18). Unequal homologous recombination where transposable elements serve as dispersed sites of sequence homology is the major recombination mechanism known to cause transposon-induced deletions (19, 20). Ac and P elements have also been shown to destabilize flanking direct repeats, creating a genomic deletion after double-strand break-induced repair (21, 22).
Ac/Ds elements transpose in a wide variety of heterologous plants as well as yeast, where they exhibit the same characteristics: they predominately transpose to nearby sites and usually create an eight base-pair footprint repair product (23-28). The maize Ac/Ds transposon system has been used successfully to establish large collections of insertional mutants in Arabidopsis (29-32). Although Ds is intensively used for both forward and reverse genetic approaches, few data have been reported indicating that Ds transposition can result in chromosomal rearrangements in this model plant (33). Knowledge about such events could be applied in functional genomic studies. Since the discovery of transposons, not only has their ability to transpose been exploited (e.g., for gene tagging) but so too has their ability to induce chromosomal rearrangements, for example in the development of a system to generate defined genomic deletions in Drosophila (34).
Here, we report the molecular and genetic analysis of the three Arabidopsis mutants anat (ana), haumea (hma), and tons missing (tms), which have large genomic deletions adjacent to Ds that range in size from 64 to 104 kb. We propose that the deletion mechanism involves a local transposition from replicated into unreplicated DNA, followed by an excision event where TIRs from the newly transposed and the original Ds at the donor site served as substrate for transposase. The proposed mechanism is different from chromosome-breaking events triggered by complex Ds loci in maize and does not involve homologous recombination. The described deletions occur independently of the location in the genome and at a sufficiently high frequency to be useful for the establishment of a collection of Ds-marked local deficiencies. The mechanism leading to tms-type deletions suggests that endogenous repetitive Arabidopsis sequences may interact with Ds in unequal homologous recombination events or in a mechanism similar to the one we describe for ana and hma. The finding that local transposition after chromosome replication is frequently associated with hybrid element excision deleting large pieces of flanking DNA provides another mechanism for how transposons may contribute to genome evolution.
Materials and Methods
Plant Materials. Insertional mutagenesis using enhancer detection and gene-trap Ds elements was performed as described (35). Growth conditions are described in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Isolation of Ds Flanks. Thermal asymmetric interlaced-PCR (TAIL-PCR) was performed as described (36). To isolate Ds flanks where TAIL-PCR failed, an inverse PCR procedure was used. For details, see Supporting Materials and Methods.
Southern Blot Analysis and Long-Range PCR. Genomic DNA was prepared as for inverse PCR and Southern blots performed as described (37). To amplify large fragments spanning Ds elements, the Expand Long Template PCR System (Roche Applied Science) was used. For details, see Supporting Materials and Methods.
Simple Sequence Length Polymorphism (SSLP) and Cleaved Amplified Polymorphic Sequences (CAPS) Marker Analysis. Plant DNA was prepared as described (38). SSLP and CAPS markers were designed by using the CEREON database (39). PCR was performed as described for inverse PCR. For CAPS marker analysis, a quarter of the product was digested with the respective restriction enzyme. Primer sequences, restriction enzymes, and expected product lengths have been submitted to the Arabidopsis Information Resource (TAIR) marker database under the names used here.
PCR Genotyping and Fluorescence in Situ Hybridization (FISH). DNA preparation of ana F2 siblings and PCR were carried out as described for SSLP/CAPS marker analysis. For details, see Supporting Materials and Methods. Preparation of extended chromatin fibers from leaf tissue and subsequent FISH analysis were carried out as described (40).
Results and Discussion
Selection of Candidate Mutants. Semisterility and reduced transmission of the kanamycin resistance marker present on Ds is a powerful selection strategy for the isolation of mutants defective in genes required for female gametogenesis (41, 42). We have used the enhancer detection and gene-trap system developed at Cold Spring Harbor Laboratory based on the maize Ac/Ds transposon (35) and applied this strategy to isolate genes required for normal fertility (41, 42). It is also known that mutants bearing chromosomal deficiencies can result in a reduced transmission of the mutant chromosome (1, 43, 44); if chromosomal deletions occurred upon Ds transposition in Arabidopsis, this mutant class was likely to be recovered in our screen. We checked our database for mutants with a distorted segregation ratio of the kanamycin marker and analyzed the structure of the Ds insertion. In several cases, we found that the sequences flanking Ds were from different locations on the same chromosome, indicating a chromosomal rearrangement. We focused our further analyses on the maternal-effect mutant ana and the semisterile mutant hma. The sequences flanking the 3′ and 5′ ends of Ds were separated by 104 and 83 kb, respectively. Another mutant, SGT6201 (31), was likely associated with a rearrangement because the sequence flanks were separated by 512 kb. We included this mutant in our study and named it tons missing (tms), because we expected a large number of genes to be deleted.
Deletion Mutants Have a Distorted Ds Segregation and Reduced Fertility. On self-fertilization, each of the three heterozygous mutants exhibits a characteristic seed abortion phenotype that strictly cosegregates with the kanamycin resistance marker present on Ds (Fig. 1 A-C). The ana mutant exhibits maternal-effect seed abortion. Approximately 50% of the seeds aborted during embryogenesis when the mutant allele was maternally inherited. In heterozygous hma plants, ≈40% of the ovules degenerate before fertilization. The tms mutation causes embryo lethality with ≈20% of the seeds aborting late during embryogenesis (for more details on fertility phenotypes, see Table 1, which is published as supporting information on the PNAS web site).
Fig. 1.
Reduced fertility in ana, hma, and tms mutants. (A-C) Immature siliques from ana (A), hma (B), and tms (C) heterozygous mutants. ana produces predominantly late aborting seeds (a), rare early aborted seeds (ea), and occasionally some infertile ovules (i). hma shows ≈40% infertile ovules (arrow), and tms displays close to 25% aborted seeds (arrow). (Bar = 215 μm.) (D) Southern blot analysis of genomic DNA from heterozygous ana, hma, and tms using the Ds5′ probe (see also Fig. 6).
Reduced Ds transmission through male and female gametes leads to the distorted segregation ratio of the kanamycin marker. Ds transmission was slightly reduced in the ana male gamete and entirely blocked in the female. Transmission was found strongly impaired in both male and female hma gametes: it is completely blocked through the male gametophyte and is transmitted by only ≈6% of the hma mutant female gametophytes. Ds transmission in tms was reduced to ≈80% in the female and to ≈60% in the male gamete (for details, see Table 2, which is published as supporting information on the PNAS web site).
Deletion Mutants Contain a Single Intact Ds. Perfect cosegregation of the fertility phenotypes with the kanamycin resistance marker indicated a single segregating Ds insertion in all three mutants analyzed, but the 3′ and 5′ sequences flanking Ds identified different locations on the chromosome. This suggested either (i) the presence of more than one element at the locus, (ii) that the element broke into two or more parts, or (iii) that there was a single element combined with a chromosomal rearrangement. To test whether the mutants under study contained single Ds elements, we performed Southern blot analysis with a probe hybridizing to the 5′ end of Ds (Fig. 1D). A single band was obtained for each mutant. To test whether the Ds elements were intact and complete, we amplified them by using primers based on genomic sequences identified by thermal asymmetric interlaced-PCR or inverse PCR on both flanks of Ds. For all three mutants, we could amplify the expected ≈7 kb Ds fragment. None of the primer combinations yielded any product using Landsberg erecta (Ler) WT DNA as template (data not shown). A double digestion of the PCR products with EcoRI and BamHI confirmed that the single Ds was intact in each mutant. Taken together, these experiments demonstrate that each mutant carried a complete and single Ds (see Fig. 6, which is published as supporting information on the PNAS web site, for details).
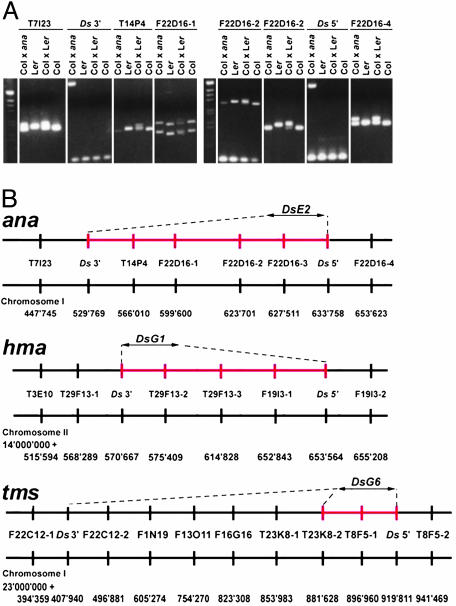
Marker and FISH Analyses Confirm Large Flanking Deletions. Despite the two Ds sequence flanks being annotated 104, 83, and 512 kb apart in ana, hma, and tms, respectively, we were able to amplify the complete Ds elements by using primers derived from these sequence flanks. The successful PCR amplification can be explained only by the proximity of the genomic sequences targeted by the primers. Because no product was detected when these primers were used on Ler WT DNA, this proximity is specific to the mutant chromosomes and suggests that the Ds transposition had induced chromosome breakage and rearrangements at the site of insertion. We could not exclude, however, that in addition to some Ds-induced rearrangements, the genomic organization in the regions studied, differed between the Ler and Columbia (Col) genome, which were used to generate the insertional mutants and the genome sequence (45), respectively.
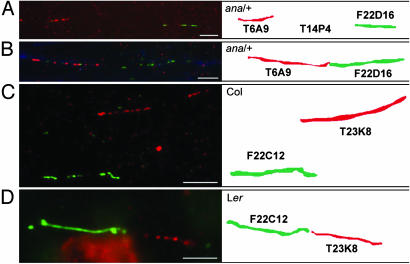
To address this question, we designed polymorphic markers covering the genomic regions expected to be deleted and/or rearranged (39). We generated mutant Ler/Col F1 hybrids and analyzed the genotype for each marker in the regions under study. We expected to obtain only a Col allele for markers where the Ler allele was deleted. This was the case for all of the markers between the Ds flanking sequences for ana and hma, suggesting that the corresponding genomic region was deleted in these mutants (Fig. 2 A and B; see also Fig. 7, which is published as supporting information on the PNAS web site). Using extended-fiber FISH (40), we could cytogenetically visualize this deletion encompassing nearly the entire BAC T14P4 in ana, where the two bacterial artificial chromosome (BAC) clones flanking T14P4 (T6A9 and F22D16) were expected to come into close proximity to each other (Fig. 3 A and B). In contrast, only two Ler markers (T23K8-2 and T8F5-1) were missing in the F1 Ler/Col hybrid in tms, suggesting that between 38 and 64 kb were deleted adjacent to the 5′ end of Ds (Fig. 2B; see also Fig. 7). However, this did not explain the predicted distance of 512 kb between the two flanking sequences of Ds. We hypothesized that the genomic organization between Ler and Col differed, because such differences had been previously observed for Ler and C24 (46). We addressed this question by using extended-fiber FISH and consistently detected F22C12 and T23K8 BAC signals adjacent to each other in the Ler genome, whereas they were separated in the Col genome as annotated (Fig. 3 C and D). This result suggests that the region between the marker T23K8-2 and the flanking sequence of the 3′ end of Ds is not a Ds-induced inversion but represents an inverted region between the Ler and the Col genomes. Taken together, our results suggest that in each of the mutants analyzed, a genomic region of 104, 83, and 33-64 kb spanning 29, 18, and 21 annotated genes, respectively, was deleted (see Table 3, which is published as supporting information on the PNAS web site, for deleted genes).
Fig. 2.
Polymorphic marker analysis of mutant Ler/Col F1 hybrids. (A) Polymorphisms at the ana locus were determined by PCR amplification by using DNA template from the mutant Ler/Col F1 hybrid (Col × ana) and the controls Ler, Col, and Col/Ler hybrid (Col × Ler) (see Fig. 7 for hma and tms). (B) The position of each marker located on distinct BAC clones and the position of the Ds element are schematically drawn (not to scale). Bars in red indicate the deleted regions on the mutant chromosomes. Ds flanking primers were used in combination with primers at the end of Ds (Ds3-1 and Ds5-1, ref. 36) to confirm the insertion position for each mutant (Ds3′ and Ds5′).
Fig. 3.
FISH on extended chromatin fibers. Extended chromatin fibers were prepared from ana (A and B), Col (C), and Ler (D) leaf tissue and hybridized with differentially labeled BAC DNA probes, as indicated in the scheme. When hybridizing extended chromatin fibers of ana heterozygous plants with the differentially labeled T6A9 and F22D16 BAC probes, two distinct hybridization patterns were obtained. We interpret A as a WT chromatin fiber showing a gap between both signals as expected from the presence of the (nonlabeled) T14P4 BAC region and B as a mutant chromatin fiber showing adjacent T6A9 and F22D16 BAC signals expected from the nearly complete deletion of T4P14 in ana. In Col (C), F22C12 and T23K8 hybridization signals are not adjacent to each other, as expected from the annotation in the public database, whereas in Ler (D), the signals are adjacent to each other. This indicates a discrepancy in genomic organization between the Ler and Col genomes in this region. (Bars = 10 μm.)
Ds-Mediated Deletions Occur at the Starter Locus or at Unlinked Loci. To gain more insight into the mechanism that led to chromosomal deletions, we analyzed the position, orientation, and sequences flanking Ds in both the starter lines (35) and the mutants. The starter lines are independent transformants harboring a T-DNA carrying the enhancer detection (DsE) or gene-trap (DsG) construct at different positions in the Arabidopsis genome. The mutants ana, hma, and tms originate from three different starter lines, DsE2, DsG1, and DsG6, respectively. Ds in ana and hma had apparently not transposed from the excision site, whereas in tms the Ds inserted at an unlinked position (Fig. 4).
Fig. 4.
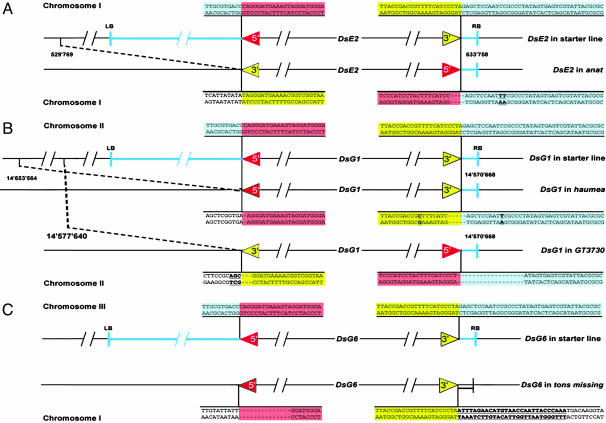
Comparison of Ds junctions in the starter lines and after transposition in the mutants ana, hma, GT3730, and tms. Schematic representation of the Ds element in the starter lines DsE2 (A), DsG1 (B), and DsG6 (C) and in the corresponding mutant. Black line, plant genomic sequence; blue line and highlighted, T-DNA fragment and sequence junction; red and yellow arrows and highlighted sequence, 5′ and 3′ Ds end, respectively. In ana (A), DsE2 retention is accompanied by a 3-bp deletion at the Ds5′ terminus, a 1-bp deletion of the T-DNA vector at the site of excision, and a 104-kb genomic deletion flanking the 3′ end of Ds. At position +11 bp counted from the Ds3′ end, an additional T was inserted, and a C→T exchange occurred (bold and underlined). In hma (B), DsG1 is excised and reinserted in the same orientation at the site of excision causing an 83-kb genomic deletion. On the Ds5′ terminus, a single base pair is deleted, whereas the four outermost bases on the Ds3′ end are missing. Aberrant transposition is accompanied with single base exchanges at position -12 (T→C) and at position + 11 (C→T), as referenced to the 5′ terminus (bold and underlined). In GT3730 DsG1 reinserted at the site of excision in opposite orientation accompanied by a 7-kb genomic deletion at the 3′ flank and a 17-bp deletion at the 5′ flank of the mutant Ds where the 3-bp GAT are substituted by CGA. In tms (C), 14 bp on the Ds5′ terminus are deleted, and 28-bp repetitive sequences from an AtREP3 annotated transposon (underlined and bold) flank the Ds3′ terminus.
Analysis of the junctions of the Ds elements in the starter lines and the mutants showed that not only were large fragments of plant genomic sequence deleted in ana and hma but also the entire T-DNA sequence flanking the 5′ end of Ds in the starter construct, including the negative selectable marker Indole Acetic Acid Hydrolase (IAAH) (35). Loss of the IAAH marker explains why these mutants were recovered after the positive/negative selection process (35). In contrast, the T-DNA sequences flanking the 3′ end of Ds were still present at the starter locus in both ana and hma. The two mutants differed, however, in that the Ds was in an inverted orientation with respect to the starter construct in hma but not in ana. Small deletions and base-pair changes were found at the junction between the end of Ds and the remaining T-DNA in both these mutants. This finding is inconsistent with the deletions originating through homologous recombination between two linked Ds elements and strongly suggests that excision of Ds was followed by double-strand break repair at the excision site, i.e., the junction between the retained Ds and T-DNA sequences from the donor site.
Ds-Induced Deletions Occur at Various Starter Loci. We were able to identify additional Ds insertion mutants with an ana-type deletion and inverted retention of the transposon at the excision site for the starter lines DsG1 (GT3730) (Fig. 4), DsG9 (GT2633), and DsE3 (ET1881). In these insertions, the junction between Ds and the remaining T-DNA shows the same characteristics of double-strand break-induced repair as described above. Although GT2633 and ET1881 also exhibit strongly reduced fertility, GT3730 has no obvious phenotype and contains a flanking deletion of only 7 kb, which can be maintained homozygously. Thus, the observed mechanism can induce flanking deletions of various sizes and induces deletions at diverse locations in the genome.
Formation of Flanking Deletions Depends on Transposase Activity. Observation of the retention of an inverted Ds accompanied by deletions at several loci suggested no other requirement for the rearrangement than a Ds element mobilized by Ac activity. However, because the mutants under study were inbred several times before molecular analysis, we wanted to investigate whether transposition and creation of the deletion occurred in consecutive steps (i.e., in different cells) or were coupled (i.e., occurred in the same cell). A two-step mechanism could involve a local transposition followed by the formation of the deletion. If these steps were uncoupled, we would expect to recover two sectors that carry either two linked Ds elements or one Ds associated with the deletion. To test whether we could recover an intermediate of a two-step mechanism, we analyzed 64 kanamycin-selected ana F2 siblings derived from the F1 plant carrying both the stable Ac transposase source and the starter Ds. We recovered 13 individuals with the ana phenotype and the molecularly confirmed deletion, but no plants with two linked Ds elements. This suggests that the formation of the ana deletion was coupled to transposition and occurred in the same cell of the F1 plant. This finding is consistent with the low frequency of Ds transposition in Arabidopsis: a high level of Ac activity is reached in only a few cells, where it can lead to both transposition and the formation of deletions. The coupled occurrence of both steps in ana provides further evidence against a mechanism where transposition is followed by homologous recombination between dispersed Ds elements, the second step of which would be independent of transposase and could occur at any position in the pedigree.
Intrachromosomal Local Transposition Followed by Immediate Excision of a Hybrid Ds Induces the Flanking Deletions. That the Ds element in ana and three other deletions of this type are inverted with respect to the starter locus implies that chromosome breakage must have occurred at both Ds ends. In hma, where Ds is in the same orientation as at the starter locus, a characteristic footprint at both ends also suggested that Ds was excised. Hybrid element insertion (4) and nonlinear transposition (11), which have been shown to cause flanking deletions in Drosophila, Antirrhinum, and maize, cannot explain this structure, because these mechanisms leave one junction of the transposon unchanged. The same is true for unequal homologous recombination between two linked Ds where one junction of the transposon is also left intact.
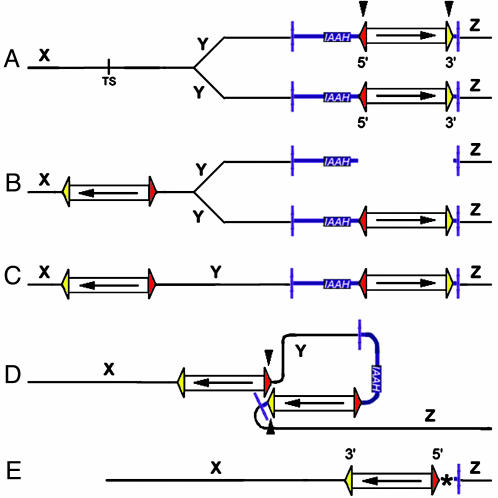
We propose a model to account for ana-type deletions where an intrachromosomal local transposition from replicated into unreplicated chromatin (Fig. 5 A and B) is followed by an excision event involving the 5′ Ds end of the newly transposed element and the 3′ Ds end of the element at the donor site (hybrid excision element) (Fig. 5 D and E). The excised element reminiscent of a macrotransposon described previously (12) consists of the starter Ds element including T-DNA sequence containing the IAAH negative selectable marker and the genomic plant sequence that was found deleted in the mutants. Because only one end of the excised element has a Ds TIR, the fragment is likely lost during the process, whereas the two other ends, the 5′ TIR of the transposed Ds and the T-DNA flanking the 3′ TIR of the donor Ds, are joined by double-strand break repair (Fig. 5E). A similar mechanism may be responsible for the hma mutation, but the formation of the hybrid excision element would involve two 3′ Ds ends in this case. Although the TIRs of Ds are nonequivalent, they do share high sequence similarity, and some deviations from the WT sequence are tolerated for transposition. Thus, an excision involving two 3′ ends, one of which is partially defective (Fig. 4), can be imagined. That we find base-pair changes typical of double-strand break repair processes at both Ds ends supports the hypothesis that the hma deletion also involved excision of a hybrid element. That we found several other mutants with an inverted Ds at the starter locus, but hma remains the only one with Ds in the original orientation, suggests that hma-type deletions occur rarely, whereas ana-type deletions are the rule.
Fig. 5.
Proposed transposition mechanism resulting in an inverted Ds and a flanking deletion at the site of excision. (A) The Ds element as present in the T-DNA context of the starter line. The donor locus has replicated, and the target site (TS) is located in unreplicated DNA. (B) Local transposition of one replicated Ds copy into the unreplicated TS produces a locally transposed Ds in inverted orientation. (C) Intermediate product with two closely linked Ds; only the relevant chromatid is shown. (D) The subsequent excision event involves the 5′ TIR of the newly transposed Ds and the 3′ TIR of the Ds at the donor locus. The hybrid element being excised contains plant genomic DNA Y, the left part of the T-DNA vector including the negative selectable marker IAAH, and the Ds element at the donor site. (E) The resulting deletion chromosome. The asterisk highlights the double-strand break repair signature between the T-DNA flanking the 3′ Ds end in the starter construct and the retained Ds. 5′ Ds TIRs are drawn in red, 3′ Ds TIRs in yellow, T-DNA in blue. X, Y, and Z indicate plant genomic DNA sequences. Arrowheads indicate DNA cuts by Ac transposase; arrows indicate the orientation of the Ds.
Endogenous Repetitive Sequences Are Involved to Create tms-Type Deletions. A transposition event from the starter locus on chromosome III to an unlinked site on chromosome I occurred in tms (Fig. 4C). Therefore, another mechanism must underlie this deletion. In contrast to ana and hma, we cannot distinguish whether excision/transposition events only or excision/transposition events in combination with illegitimate recombination caused the tms deletion. The 5′ Ds end in tms shows a truncation of 14 bp. Strikingly, we found 28 bp adjacent to the 3′ Ds flank that are identical to a sequence located internally of two direct repeats of a repetitive element (AtREP3) (47), located at 64 kb distance from the 3′ Ds flank. AtREP3 belongs to the AtREP family of 1- to 3-kb-long repetitive elements that, together with the highly similar AthE1 family (48), constitutes >1% of the Arabidopsis genome. The maximal size of the tms deletion, based on the PCR marker analysis, is 64 kb, consistent with an endogenous repetitive sequence element from Arabidopsis being involved in the formation the tms deletion. After Ds transposition to a nearby location, this sequence may have served as substrate for intrachromosomal hybrid element excision or illegitimate recombination between Ds and the endogenous repetitive sequence. Thus, endogenous repetitive sequences may lead to excision events of genomic plant DNA in combination with a nearby transposed Ds in Arabidopsis, leading to gross chromosomal rearrangements. This view is supported by the molecular analysis of the female gametophytic mutant hadad (hdd) (41), where Ds inserted in immediate proximity of an annotated endogenous DNA transposon of the hAT family, and resulted in a large chromosomal deletion adjacent to Ds (data not shown).
Deletions at Ds Excision Sites Occur at Genetically Trackable Frequencies With the exception of GT3730, the seven lines analyzed here were identified in a screen for mutants affecting fertility. Although observed at different starter loci, it was unclear at what frequencies anat- and hma-type deletions occur without preselection for reduced fertility. Thus we reanalyzed the integration pattern of 931 Ds lines generated from the same starter lines reported earlier. Parinov et al. (31) report that despite IAAH counter selection, 15% of the transposants had a Ds element inserted somewhere in the T-DNA. Only half of these were within the negative selectable IAAH gene itself, whereas the remaining 7-8% could not easily be explained. A short-range transposition into the T-DNA followed by hybrid element excision as discussed here would create small deletions removing part of the T-DNA vector including the IAAH gene, such that they would be recovered in Sundaresan's selection scheme (35).
In the same study, 9% of all transposants derived from DsG1 were found to be insertions within 1 Mb of the donor site despite counter selection. Such short-range transpositions can be recovered if the transposed Ds recombined away from the starter locus. Because the correlation between the physical and genetic maps in the area of the starter locus does not significantly differ from the average of 0.2 Mb per centiMorgan, we expect such events to occur at frequencies of only a few percent. The remainder could at least in part be explained by the mechanism outlined here where the IAAH gene as well as flanking genomic sequences are deleted by hybrid element excision. In our biased sample, we did indeed find two such events (hma and GT3730) derived from the DsG1 starter locus. Taken together, the analysis of Parinov's data suggests that Ds-induced deletions occur at a genetically trackable frequency of 5-10% at the site of excision.
Deletions at Unlinked Sites Occur at a Low Frequency. To estimate the frequency of deletions at unlinked sites (e.g., tms and hdd), we analyzed 5,616 flanking sequences of transposants from the Cold Spring Harbor Laboratory collection available at the National Center for Biotechnology Information. Using an e-value of 1e-60 to minimize wrong hits because of low sequence quality, we found 1,400 lines for which both 3′ and 5′ flanking sequences were available (see Table 4, which is published as supporting information on the PNAS web site). In 8% of the lines, 5′ and 3′ Ds flanking sequences did not match to the same chromosomal location. In 3.1% of all lines, the two ends were separated by >500 kb. These lines probably do not contain such large deletions, because they are likely not transmitted through either gametophyte. The average size deletion recovered after γ irradiation in Arabidopsis has been estimated at <160 kb (49). We found Ds flanks separated by <160 kb at a frequency of 1.3%, but some larger deletions may also be transmitted. Other rearrangments, e.g., inversions, two linked Ds elements, or differences between Ler and Col genomes, may also be present in these two classes. Among the 10 lines we have studied to date, in which 3′ and 5′ flanking sequences were derived from different chromosomal positions, nine were deletions, and one appears to be a translocation (this study and unpublished data). Although this is admittedly a biased sample, deletions appear to be rather frequent. In summary, deletions at unlinked sites, such as described here for tms, occur at a low frequency of ≈1%.
Hybrid Ds Element Excision as a Tool for Functional Genomics. That deletions flanking the excision site occur at a rather high frequency among recovered transposants makes hybrid element excision an attractive tool to generate defined Ds-marked local deficiencies in Arabidopsis. Chromosomes bearing deletions are very valuable tools in any genetic system. Based on specific properties of the hobo and P element, different strategies have been established to efficiently generate defined chromosomal deletions in Drosophila (4, 34, 50). Although attempted (29, 51) and despite the fact that transposon-induced deletions have been used successfully in plants, e.g., to delete specific loci in Antirrhinum (52), no broadly used system to create genome-wide tagged deletions has been developed. The mechanism leading to flanking deletions described here allows the use of any Ds element and does not require specially designed elements, few of which have been mapped to date (29). Given that close to 100,000 Ds lines have been generated (http://genetrap.cshl.org, www.jic.bbsrc.ac.uk/SCIENCE/cdb/exotic, http://pfgweb.gsc.riken.go.jp, and www.arabidopsis.org/abrc/ima.html), deletions can potentially be generated in virtually any region of the genome. A simple screen using nested primers flanking the Ds in combination with Ds-derived primers should allow the identification of deletions flanking the Ds in the progeny of a cross between Ac and the selected Ds element. If the primers are designed such that they produce a product only if intervening sequences are deleted, deletions can easily be identified even within DNA samples from pooled plants, making this an efficient strategy. Given that up to 50% of Ac/-;Ds/- hybrids produce transposants (35), and among these 5-10% carry an associated deletion, it should be possible to identify deletions with reasonable efficiency. Generation of a mutant collection with defined local deficiencies as proposed here promises to be of great value for the phenotypic analysis of the Arabidopsis genome, complementing other functional genomic approaches. Having available a mutant collection with defined deficiencies would provide a number of immediate advantages for the Arabidopsis researcher. Nested overlapping deficiencies could greatly facilitate the cloning of known recessive mutations that have been localized to a specific region by crossing the mutant with a set of deletions uncovering the respective region. Importantly, deletions will allow the elimination of tandem duplicated genes and even gene clusters with related function. This approach seems particularly suited for the study of the many duplicated genes where a single knockout does not lead to any phenotypic change, and double knockouts are difficult to generate due to the close linkage of the two genes.
Conclusion
Our study in Arabidopsis shows that the ability to create large genomic deletions adjacent to Ds is characteristic for any Ds and does not, as suggested by work in maize, depend on a complex molecular structure of the locus. Therefore, transposase activity alone (local transposition and hybrid element excision), not in conjunction with recombination, can mediate elimination of sequences adjacent to the insertion site, which may play an important evolutionary role. Our analysis revealed a previously undescribed mechanism by which transposons can lead to genome contraction and thus an additional feature of transposons that contributes to genome dynamics and evolution.
Supplementary Material
Acknowledgments
We thank V. Sundaresan, R. Martienssen, and coworkers for access to their Ds mutagenesis system before publication; R. Guyot for help with bioinformatics; M. Curtis and B. Keller for comments on the manuscript; V. Gagliardini and P. Kopf for technical assistance; J.-J. Pittet for help with art work; and A. Buendia for personal computer support at the University of Barcelona. D.R.P. and C.B. were supported by the Roche Research Foundation. C.K. is an Human Frontier Science Program Fellow, and J.A.d.C.-N. is supported by a scholarship from the Fundação para a Ciência e Tecnologia, Portugal. Our work on Ds-induced gametophytic mutants was supported by the University of Zürich, Grant MCB-9723948 of the National Science Foundation, and Grant 31-64061.00 of the Swiss National Science Foundation (to U.G.).
Abbreviations: FISH, fluorescence in situ hybridization; Ler, Landsberg erecta; Col, Columbia; BAC, bacterial artificial chromosome; TIR, terminal inverted repeat.
References
- 1.Robertson, D. S. & Stinard, P. S. (1987) Genetics 115, 353-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blackman, R. K., Grimaila, R., Koehler, M. M. D. & Gelbart, W. M. (1987) Cell 49, 497-505. [DOI] [PubMed] [Google Scholar]
- 3.Lister, C. & Martin, C. (1989) Genetics 123, 417-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Preston, C. R., Sved, J. A. & Engels, W. R. (1996) Genetics 144, 1623-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McClintock, B. (1951) Cold Spring Harbor Symp. Quant. Biol. 16, 13-47. [DOI] [PubMed] [Google Scholar]
- 6.Dooner, H. K. (1985) Plant Genet. 561-573.
- 7.Green, M. M. (1969) Genetics 61, 423-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reif, H. J. & Saedler, H. (1975) Mol. Gen. Genet. 137, 17-28. [DOI] [PubMed] [Google Scholar]
- 9.Lister, C., Jackson, D. & Martin, C. (1993) Plant Cell 5, 1541-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooley, L., Kelley, R. & Spradling, A. (1988) Science 239, 1121-1128. [DOI] [PubMed] [Google Scholar]
- 11.Zhang, J. & Peterson, T. (1999) Genetics 153, 1403-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dooner, H. K. & Belachew, A. (1991) Genetics 129, 855-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Courage-Tebbe, U., Doring, H. P., Fedoroff, N. & Starlinger, P. (1983) Cell 34, 383-393. [DOI] [PubMed] [Google Scholar]
- 14.Weil, C. F. & Wessler, S. R. (1993) Plant Cell 5, 515-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weck, E., Courage, U., Doring, H. P., Fedoroff, N. V. & Starlinger, P. (1984) EMBO J. 3, 175-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doring, H. P., Nelsen-Salz, B., Garber, R. & Tillmann, E. (1989) Mol. Gen. Genet. 219, 299-305. [DOI] [PubMed] [Google Scholar]
- 17.English, J. J., Harrison, K. & Jones, J. (1995) Plant Cell 7, 1235-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fedoroff, N. V. (1989) Cell 56, 181-191. [DOI] [PubMed] [Google Scholar]
- 19.Davis, P. S., Shen, M. W. & Judd, B. H. (1987) Proc. Natl. Acad. Sci. USA 84, 174-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin, C., Mackay, S. & Carpenter, R. (1988) Genetics 119, 171-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Athma, P. & Peterson, T. (1991) Genetics 128, 163-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Preston, C. R., Engels, W. & Flores, C. (2002) Genetics 161, 711-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Sluys, M. A., Tempe, J. & Fedoroff, N. (1987) EMBO J. 6, 3881-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones, J. D., Carland, F., Lim, E., Ralston, E. & Dooner, H. K. (1990) Plant Cell 2, 701-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bancroft, I. & Dean, C. (1993) Genetics 134, 1221-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Long, D., Swinburne, J., Martin, M., Wilson, K., Sundberg, E., Lee, K. & Coupland, G. (1993) Mol. Gen. Genet. 241, 627-636. [DOI] [PubMed] [Google Scholar]
- 27.Rinehart, T. A., Dean, C. & Weil, C. F. (1997) Plant J. 12, 1419-1427. [DOI] [PubMed] [Google Scholar]
- 28.Weil, C. F. & Kunze, R. (2000) Nat. Genet. 26, 187-190. [DOI] [PubMed] [Google Scholar]
- 29.Osborne, B. I., Wirtz, U. & Baker, B. (1995) Plant J. 7, 687-701. [DOI] [PubMed] [Google Scholar]
- 30.Smith, D., Yanai, Y., Liu, Y. G., Ishiguro, S., Okada, K., Shibata, D., Whittier, R. F. & Fedoroff, N. V. (1996) Plant J. 10, 721-732. [DOI] [PubMed] [Google Scholar]
- 31.Parinov, S., Sevugan, M., Ye, D., Yang, W. C., Kumaran, M. & Sundaresan, V. (1999) Plant Cell 11, 2263-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.May, B. P. & Martienssen, R. A. (2003) Crit. Rev. Plant Sci. 22, 1-35. [Google Scholar]
- 33.Oh, S.-A., Park, S. K., Jang, I., Howden, R., Moore, J. M., Grossniklaus, U. & Twell, D. (2003) Sex. Plant Reprod. 16, 99-102. [Google Scholar]
- 34.Huet, F., Lu, J. T., Myrick, K. V., Baugh, L. R., Crosby, M. A. & Gelbart, W. M. (2002) Proc. Natl. Acad. Sci. USA 99, 9948-9953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sundaresan, V., Springer, P., Volpe, T., Haward, S., Jones, J. D., Dean, C., Ma, H. & Martienssen, R. (1995) Genes Dev. 9, 1797-1810. [DOI] [PubMed] [Google Scholar]
- 36.Grossniklaus, U., Vielle-Calzada, J. P., Hoeppner, M. A. & Gagliano, W. B. (1998) Science 280, 446-450. [DOI] [PubMed] [Google Scholar]
- 37.Ausubel, F. M. (1995) Current Protocols in Molecular Biology (Wiley, New York).
- 38.Edwards, K., Johnstone, C. & Thompson, C. (1991) Nucleic Acids Res. 19, 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jander, G., Norris, S. R., Rounsley, S. D., Bush, D. F., Levin, I. M. & Last, R. L. (2002) Plant Physiol. 129, 440-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fransz, P. F., Alonso-Blanco, C., Liharska, T. B., Peeters, A. J., Zabel, P. & de Jong, J. H. (1996) Plant J. 9, 421-430. [DOI] [PubMed] [Google Scholar]
- 41.Moore, J. M., Calzada, J. P., Gagliano, W. & Grossniklaus, U. (1997) Cold Spring Harbor Symp. Quant. Biol. 62, 35-47. [PubMed] [Google Scholar]
- 42.Page, D. R. & Grossniklaus, U. (2002) Nat. Rev. Genet. 3, 124-136. [DOI] [PubMed] [Google Scholar]
- 43.McClintock, B. (1938) Genetics 23, 315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vollbrecht, E. & Hake, S. (1995) Dev. Genet. 16, 44-63. [Google Scholar]
- 45.The Arabidopsis Genome Initiative. (2000) Nature 408, 796-815. [DOI] [PubMed] [Google Scholar]
- 46.Fransz, P. F., Armstrong, S., de Jong, J. H., Parnell, L. D., van Drunen, C., Dean, C., Zabel, P., Bisseling, T. & Jones, G. H. (2000) Cell 100, 367-376. [DOI] [PubMed] [Google Scholar]
- 47.Kapitonov, V. V. & Jurka, J. (1999) Genetica 107, 27-37. [PubMed] [Google Scholar]
- 48.Surzycki, S. A. & Belknap, W. R. (1999) J. Mol. Evol. 48, 684-691. [DOI] [PubMed] [Google Scholar]
- 49.Vizir, I. Y., Anderson, M. L., Wilson, Z. A. & Mulligan, B. J. (1994) Genetics 137, 1111-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cooley, L., Thompson, D. & Spradling, A. C. (1990) Proc. Natl. Acad. Sci. USA 87, 3170-3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vizir, I. Y. & Mulligan, B. J. (1999) J. Hered. 90, 412-417. [DOI] [PubMed] [Google Scholar]
- 52.Ingram, G. C., Simon, R., Carpenter, R. & Coen, E. S. (1998) Curr. Biol. 8, 1079-1082. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.