Abstract
Humoral immunity is a critical component of the immune system that is established during fetal life and expands upon exposure to pathogens. The extensive humoral immune response repertoire is generated in large part via immunoglobulin (Ig) heavy chain variable region diversity. The horse is a useful model to study the development of humoral diversity because the placenta does not transfer maternal antibodies; therefore, Igs detected in the fetus and pre-suckle neonate were generated in utero. The goal of this study was to compare the equine fetal Ig VDJ repertoire to that of neonatal, foal, and adult horse stages of life. We found similar profiles of IGHV, IGHD, and IGHJ gene usage throughout life, including predominant usage of IGHV2S3, IGHD18S1, and IGHJ1S5. CDR3H lengths were also comparable throughout life. Unexpectedly, Ig sequence diversity significantly increased between the fetal and neonatal age, and, as expected, between the foal and adult age.
Keywords: equine, immunoglobulin, diversity, development
1. Introduction
The immense repertoire of antigen-specific immunoglobulins (Igs) is generated predominantly by diversity in variable regions of the heavy and light chains. Expressed Ig heavy chains result from recombination of discontinuous germline variable (IGHV), diversity (IGHD), joining (IGHJ), and constant (IGHC) region gene segments. The encoded VH, D, and JH domains form 3 complementarity determining regions (CDRs) that bind antigen (Kabat and Wu 1991). The 3 CDRs are separated by conserved framework regions that provide structural support (Kirkham et al. 1992). The IGHV gene encodes CDR1H and CDR2H, and framework regions 1, 2, and 3 (Tonegawa 1983). CDR3H is encoded by the 3’ end of IGHV and the junction of IGHD and IGHJ gene segments. Multiple mechanisms contribute to variable region diversity, including combinatorial diversity from IGHV, IGHD, and IGHJ gene segment usage; CDR3H junctional diversity from the insertion of non-templated (N) and palindromic (P) nucleotides; and somatic hypermutation, which allows affinity maturation (Alt and Baltimore 1982; Tonegawa 1983). Once antigen specificity is achieved, Ig function can be modulated by constant region isotype-switching.
In most species, the IGHV locus contains multiple functional IGHV genes, multiple IGHV genes with functional coding sequence but altered or missing splice or recombination signals (often denoted as VHORF), and multiple IGHV pseudogenes. The total number of IGHV genes varies considerably across species, from over 160 in mice and rats to less than 20 in cows and sheep (Das et al. 2008). Recent annotation of Ig variable region genes from the equine genome revealed 54 IGHV genes, 40 IGHD genes, and 8 IGHJ genes (Sun et al. 2010). Expressed equine IGHV sequences have been reported from IGHM and IGHE cDNA clones and from two studies characterizing horse Ig VDJ sequences (Navarro et al. 1995; Schrenzel et al. 1997; Almagro et al. 2006; Sun et al. 2010). Functional IGHV genes can be sorted into families or subgroups that share at least 75% nucleotide identity among members; the equine IGHV genes with functional coding sequence have been sorted into 7 subgroups (Giudicelli and Lefranc 1999; Sun et al. 2010). Across species, IGHV gene families can be classified into 3 phylogenetic clans (I, II, and III), and it has been suggested that they have been present for about 370 million years (Schroeder et al. 1990; Das et al. 2008). Clan II and III IGHV genes are found in most species, whereas clan I IGHV genes are less common. A comparative analysis of IGHV genes spanning 16 species reported that the evolution of IGHV genes is characterized by species-specific IGHV gene expansion and contraction, and overall, this evolution is more complex than expected (Das et al. 2008).
Biases in IGHV gene usage have been reported during fetal life in humans and mice (Feeney 1990, Schroeder and Wang 1990). Studies of Ig repertoire report biases because usage of IGHV genes from the VH3 family is strongly favored during human fetal and neonatal life, and DHQ52, JH3, and JH4 are overrepresented during human fetal life (Schroeder and Wang 1990; Bauer et al. 2002). Of note, IGHV gene usage in human neonates is not exclusive to VH3 and the overall pattern of VH usage is similar to VH usage in adults. Recent studies of antigen-specific B cell responses revealed that the rotavirus-specific repertoire is dominated by VH1 and VH4 usage in both human infants and adults, and the respiratory syncytial virus-specific repertoire is dominated by VH3 usage in both human infants over 3 months of age and adults (Weitkamp et al. 2003; Williams et al. 2009). Therefore, it seems that neonates can produce antigen-specific Ig responses with similar IGHV gene usage as adults.
We hypothesized that equine fetal and neonatal immunoglobulin repertoire diversity (before antigenic exposure) contains VH, D, JH gene usage similar to the young foal and adult horse (post antigenic exposure). The understanding of fetal and neonatal humoral competency has implications to vaccinology and immunity against pathogens in early age. The goal of this study is to compare the Ig VDJ repertoire and diversity in equine fetal life to that in neonatal, young foal, and adult horse stages of life. Our previous studies described the development of primary and secondary lymphoid tissues in the equine fetus, neonate, and young foal, including the expansion of lymphocyte populations and formation of lymphoid follicles (Tallmadge et al. 2009). The finding that fetal lymphoid tissues express mRNA for genes known to be essential for B cell development and function suggested active development and gene recombination during equine gestation, including immunoglobulin isotype switching, because a corresponding production of IgM and IgG proteins was detectable in serum in a limited scale at birth. This study extends those results by directly examining the diversity present at recombined Ig heavy chain VDJ regions from progressive developmental stages. We also used these samples in two methods for the amplification of the Ig VDJ repertoire: a) generation of 5’ rapid amplification of cDNA ends (RACE) library cDNA followed by PCR with a conserved IGHJ reverse primer and b) oligo d(T)-primed cDNA template followed by PCR using previously described degenerate primers (Almagro et al. 2006).
Materials and methods
2.1 Tissue and blood samples
These experiments were approved by the Cornell University Center for Animal Resources and Education and Institutional Animal Care and Use Committee for the use of vertebrates in research. Tissue samples from a healthy induced-abortion equine Thoroughbred x Warmblood fetus without uterine disease (102 days of gestation), a neonate Warmblood foal (pre-suckle, < 1-hour-old), a 2 month-old Warmblood foal, and a Thoroughbred adult horse were archived and available for this study from research investigations performed by us and other investigators over the years at Cornell University College of Veterinary Medicine. None of the donors were related. Tissue samples were collected within 1 hour of euthanasia, snap frozen in liquid nitrogen, and stored at −80°C until use.
2.2 RACE library construction, reverse transcriptase polymerase chain reaction (RT-PCR) and cloning
RNA was isolated from snap-frozen tissues following homogenization by QIAshredder (Qiagen, Valencia, CA) as directed by the RNeasy kit (Qiagen) including on-column digestion of contaminating genomic DNA. RNA was quantified with a Nanodrop (Thermo Scientific, Wilmington, DE) and one microgram was used to generate a 5’-RACE library with the SMARTer™ RACE cDNA Amplification Kit (Clontech Laboratories, Inc., Mountain View, CA). PCR amplification of Ig heavy chain variable regions was performed with a reverse primer designed to amplify all 8 horse IGHJ regions, 5’ CTCGCCTGAGGAGACGGTGACCAG 3’ (Supplemental figure 1). Amplification reactions contained 1X iProof high fidelity buffer, 1.5 mM MgCl2, 0.2 mM dNTPs, the provided universal forward primer with 0.5 µM gene-specific reverse primer (Integrated DNA Technologies, Coralville, IA), and 0.02 U iProof DNA polymerase (error rate of 4.4 × 10−7, Bio-Rad Laboratories, Hercules, CA). Thermal cycling parameters were 5 cycles of 98°C for 30 seconds and 72°C for 1 minute; 5 cycles of 98°C for 30 seconds, 70°C for 30 seconds and 72°C for 1 minute; and 27 cycles of 98°C for 30 seconds, 68°C for 30 seconds and 72°C for 1 minute. Amplification products were run on 1% agarose gels and stained with GelGreen nucleic acid stain (Phenix Research Products, Candler, NC) for visualization. PCR products of approximately 550 bp were excised from the gel, purified with GeneJET gel extraction kit (Thermo Scientific), and cloned with the CloneJET PCR cloning kit (Thermo Scientific). Individual colonies were expanded in LB broth with ampicillin, and plasmid DNA was purified with the GeneJET plasmid miniprep kit (Thermo Scientific). More than 30 clones were sequenced from each sample.
For reverse transcription, cDNA synthesis reactions contained 5.5 mM MgCl2, 0.5 mM dNTPs, 2.5 µM oligo d(T) (Applied Biosystems, Foster City, CA), 0.4 U RNasin Ribonuclease Inhibitor (Promega, Madison, WI), and 1 U Moloney Murine Leukemia Virus Reverse Transcriptase (MuLV RT), and 1X M-MuLV RT buffer (Applied Biosystems). To test for residual genomic DNA contamination, control samples did not receive M-MuLV RT. Amplification reactions contained 1X iProof high fidelity, 1.5 mM MgCl2, 0.2 mM dNTPs, 0.5 µM forward and reverse primer (Integrated DNA Technologies), and 0.02 U iProof DNA polymerase (error rate of 4.4 × 10−7, Bio-Rad Laboratories). The primers were previously published: HorIGHV forward primer 5’ CAGGTGCARCTGMAGGAGTCRG 3’ and HorJH5 reverse primer 5’ ACGGTGACCAGGATACCCTG 3’ (Almagro et al. 2006). Thermal cycling parameters were 98 °C for 30 seconds; 35 cycles of 98 °C for 10 seconds, 58 °C for 20 seconds, 72 °C for 30 seconds; and a final extension of 72°C for 10 minutes. Amplification product were run on 1% agarose gels and stained with GelGreen nucleic acid stain (Phenix Research Products) for visualization. PCR products of approximately 350 bp were excised from the gel, purified and cloned as described above.
2.3 Sequence analysis
Clones were sequenced at the Cornell University Life Sciences Core Laboratories Center, Ithaca, NY. Replicate sequences and sequences encoding premature stop codons (IGVDJ120 and IGVDJ121) were eliminated from the analyses. Sequences are available through GenBank with accession numbers HQ403608-HQ403643 and KC549680-KC549800. Sequence content analysis and identity matrix were performed with Geneious software (Biomatters, Ltd., Drummond et al. 2009). The Shapiro-Wilk Normality test (http://dittami.gmxhome.de/shapiro/) revealed that in most cases data were not normally distributed and hence the Wilcoxon-Mann-Whitney Rank Sum test was used to assess comparisons of pairwise nucleotide identity levels, nucleotide identity to germline IGHV genes, and the length of non-templated nucleotides (KaleidaGraph, Synergy Software). The level of amino acid polymorphism was calculated from the variability index described by Wu and Kabat (1970) and plotted against the amino acid position. Variability is determined by the number of different amino acids at a given residue divided by the frequency of the most common amino acid at that residue. IGHV, IGHD, and IGHJ gene usage was determined by comparing the obtained sequences with annotated germline sequences (Sun et al., 2010) using BLAST and manual review of each match. IGHV genes were annotated first, followed by IGHJ genes. IGHD genes were annotated by comparing the sequence between IGHV and IGHJ to the 40 germline IGHD genes using the NCBI BLAST bl2seq tool. BLAST hits were reviewed for identity level, gaps, length, and orientation; accepted hits exceeded the IGHD identification constraint of 5 consecutive nucleotide matches established by Brezinschek et al. (1995). Assigned IGHD genes spanned 7 to 32 bases and most had ≥90% nucleotide identity. If no hits were accepted, tblastn searches were used to compare the predicted amino acid sequence of the clone to translated germline IGHD genes. Hits with low identity were not assigned a germline IGHD gene and are labeled ‘not determined’ in Table 2. Bias of IGHV, IGHD, and IGHJ gene usage was assessed with the chi-square test using GraphPad Prism version 6.01 for Windows, GraphPad Software, La Jolla California USA, www.graphpad.com. Phylogenetic analysis based on amino acid sequence was performed with MEGA version 5 (Tamura et al. 2011), using p-distances and the neighbor-joining method (Saitou and Nei 1987). To determine the level of support for each node, bootstrap resampling was performed with 1,000 replications. Framework regions 1, 2, and 3 were used for the analysis as defined by Lefranc et al. (2003); complementarity-determining regions and primer sequences were excluded. For comparative phylogenetic analysis, 88 IGHV sequences were used: 40 horse IGVDJ sequences from this study, 16 horse germline IGHV and 6 IGHVORF (Sun et al. 2010), 1 cow (U49765), 1 sheep (Z49188), 1 pig (AF064686), 7 human (L22582, X62111, X92206, Z12367, M99686, X92224, Z27509), and 16 mouse (X02459, J00502, M61217, X01437, AF290966, X03398, X03256, U23020, L14362, AF064445, AC073563, M22439, AC073589, X03572, U39293, AC073563). Representative sub-group sequences from each species were randomly chosen.
Table 2.
Germline gene usage of equine immunoglobulin heavy chain VDJ sequences.
| IGHV | VD | IGHD | DJ | IGHJ | ||||
|---|---|---|---|---|---|---|---|---|
| Sequence | gene | Identity1 | junction | gene | Identity1 | junction | gene | Identity1 |
| A. Sequences obtained with RACE protocol | ||||||||
| FETUS: | ||||||||
| IGVDJ1 | 2S3 | 98 | 12 | 18S1 | 100 | 2 | 1S3 | 100 |
| IGVDJ2 | 2S3 | 98 | 3 | 17S1/2 | 100 | 0 | 1S5 | 97 |
| IGVDJ3 | 2S3 | 98 | 10 | 18S1 | 100 | 0 | 1S3 | 100 |
| IGVDJ4 | 2S3 | 98 | 0 | 9S1 | 73 | 3 | 1S3 | 100 |
| IGVDJ5 | 2S3 | 98 | 9 | 18S1 | 100 | 0 | 1S5 | 100 |
| IGVDJ6 | 2S3 | 98 | 6 | 22S1 | 89 | 0 | 1S3 | 100 |
| IGVDJ7 | 2S3 | 98 | 5 | 15S2 | 100 | 0 | 1S2 | 100 |
| IGVDJ8 | 2S3 | 98 | 4 | 13S1 | 100 | 0 | 1S2 | 100 |
| IGVDJ9 | 2S3 | 97 | 9 | 24S1 | 100 | 0 | 1S5 | 93 |
| IGVDJ10 | 2S3 | 97 | 0 | 15S2 | 100 | 0 | 1S2 | 100 |
| IGVDJ11 | 2S3 | 99 | 10 | 15S2 | 100 | 7 | 1S5 | 100 |
| IGVDJ12 | 2S3 | 99 | 13 | 22S1 | 100 | 0 | 1S3 | 100 |
| IGVDJ13 | 2S2 | 100 | 15 | 18S1 | 100 | 2 | 1S7 | 100 |
| IGVDJ14 | 2S2 | 100 | 3 | 18S1 | 100 | 6 | 1S5 | 100 |
| IGVDJ15 | 2S2 | 100 | 9 | 7S2 | 100 | 7 | 1S5 | 100 |
| IGVDJ16 | 2S2 | 100 | 4 | 11S1 | 100 | 0 | 1S5 | 100 |
| IGVDJ17 | 2S2 | 100 | ND | IGVDJ108, −115 | ND | ND | 1S3 | 100 |
| IGVDJ18 | 2S2 | 100 | 6 | 15S2 | 100 | 4 | 1S6 | 100 |
| IGVDJ19 | 2S2 | 99 | 1 | 15S1 | 100 | 0 | 1S4 | 88 |
| IGVDJ20 | 2S2 | 99 | 6 | 18S1 | 93 | 2 | 1S3 | 100 |
| IGVDJ21 | 2S4 | 99 | 11 | 11S1 | 100 | 0 | 1S5 | 100 |
| IGVDJ22 | 2S4 | 99 | 8 | 15S2 | 100 | 0 | 1S3 | 93 |
| IGVDJ23 | 2S4 | 99 | 8 | 17S1/2 | 100 | 0 | 1S2 | 100 |
| IGVDJ24 | 2S4 | 98 | 8 | 7S2 | 100 | 0 | 1S6 | 100 |
| IGVDJ25 | 2S4 | 98 | 8 | 17S1/2 | 100 | 0 | 1S4 | 100 |
| IGVDJ26 | 4S2 | 100 | 7 | 18S1 | 100 | 0 | 1S5 | 100 |
| IGVDJ27 | 4S2 | 100 | 10 | 18S1 | 93 | 5 | 1S6 | 100 |
| IGVDJ28 | 1S3 | 100 | 2 | 18S1 | 100 | 9 | 1S6 | 100 |
| IGVDJ29 | 4S17 | 99 | 9 | 7S1 | 100 | 0 | 1S3 | 92 |
| IGVDJ30 | 4S17 | 99 | 12 | 18S1 | 100 | 5 | 1S6 | 100 |
| IGVDJ31 | 4S5 | 100 | 2 | 4S1 | 100 | 0 | 1S5 | 100 |
| NEONATE: | ||||||||
| IGVDJ32 | 2S3 | 96 | 13 | 4S1 | 100 | 18 | 1S3 | 91 |
| IGVDJ33 | 2S3 | 97 | 22 | 18S1 | 100 | 2 | 1S2 | 97 |
| IGVDJ34 | 2S3 | 97 | ND | ND | ND | ND | 1S3 | 100 |
| IGVDJ35 | 2S3 | 98 | 5 | 15S1 | 100 | 17 | 1S5 | 97 |
| IGVDJ36 | 2S3 | 99 | 10 | 15S2 | 100 | 14 | 1S5 | 100 |
| IGVDJ37 | 2S3 | 97 | 8 | 4S1 | 91 | 0 | 1S5 | 88 |
| IGVDJ38 | 2S3 | 97 | 16 | 15S1 | 100 | 12 | 1S7 | 85 |
| IGVDJ39 | 2S3 | 97 | 12 | 15S1 | 95 | 0 | 1S2 | 73 |
| IGVDJ40 | 2S3 | 97 | 14 | 10S1 | 93 | 4 | 1S5 | 93 |
| IGVDJ41 | 2S3 | 96 | ND | ND | ND | ND | 1S5 | 93 |
| IGVDJ42 | 2S3 | 97 | 3 | 10S1 | 100 | 0 | 1S7 | 87 |
| IGVDJ43 | 2S3 | 96 | 11 | 20S1 | 100 | 22 | 1S3 | 100 |
| IGVDJ44 | 2S3 | 97 | 24 | 18S1 | 100 | 15 | 1S5 | 100 |
| IGVDJ45 | 2S3 | 97 | 10 | 22S1 | 100 | 0 | 1S5 | 100 |
| IGVDJ46 | 2S3 | 96 | 2 | 7S1 | 88 | 5 | 1S5 | 89 |
| IGVDJ47 | 2S2 | 99 | 16 | 7S2 | 100 | 11 | 1S7 | 91 |
| IGVDJ48 | 2S2 | 99 | 12 | 18S1 | 100 | 0 | 1S2 | 100 |
| IGVDJ49 | 2S2 | 100 | 4 | 11S1 | 100 | 0 | 1S5 | 100 |
| IGVDJ50 | 2S2 | 97 | 14 | 18S1 | 93 | 0 | 1S5 | 91 |
| IGVDJ51 | 2S2 | 95 | 12 | 9S1 | 94 | 0 | 1S2 | 95 |
| IGVDJ52 | 2S2 | 97 | 20 | 11S1 | 100 | 5 | 1S7 | 87 |
| IGVDJ53 | 2S2 | 97 | ND | ND | ND | ND | 1S3 | 96 |
| IGVDJ54 | 2S2 | 98 | 7 | 18S1 | 100 | 4 | 1S7 | 92 |
| IGVDJ55 | 2S2 | 98 | 11 | 9S1 | 100 | 16 | 1S5 | 90 |
| IGVDJ56 | 2S4 | 100 | 10 | 17S1/2 | 100 | 0 | 1S5 | 97 |
| IGVDJ57 | 2S4 | 99 | 25 | 4S1 | 100 | 6 | 1S5 | 93 |
| IGVDJ58 | 2S4 | 98 | 16 | 6S1 | 100 | 10 | 1S7 | 100 |
| IGVDJ59 | 2S4 | 97 | 11 | 15S2 | 100 | 0 | 1S4 | 100 |
| IGVDJ60 | 2S4 | 97 | 12 | 9S1 | 100 | 7 | 1S3 | 95 |
| IGVDJ61 | 2S4 | 95 | 7 | 26S1 | 94 | 0 | 1S5 | 97 |
| IGVDJ62 | 1S6 | 100 | 10 | 7S1 | 100 | 8 | 1S3 | 96 |
| FOAL: | ||||||||
| IGVDJ63 | 2S3 | 98 | 20 | 26S1 | 100 | 2 | 1S5 | 97 |
| IGVDJ64 | 2S3 | 97 | 8 | 14S1 | 100 | 0 | 1S5 | 93 |
| IGVDJ65 | 2S3 | 97 | 11 | 17S1/2 | 100 | 0 | 1S5 | 93 |
| IGVDJ66 | 2S3 | 95 | 24 | 18S1 | 100 | 5 | 1S5 | 100 |
| IGVDJ67 | 2S3 | 97 | 5 | 20S1 | 100 | 15 | 1S3 | 100 |
| IGVDJ68 | 2S3 | 95 | 26 | 18S1 | 100 | 0 | 1S6 | 96 |
| IGVDJ69 | 2S3 | 95 | ND | ND | ND | ND | 1S3 | 93 |
| IGVDJ70 | 2S3 | 97 | 11 | 18S1 | 93 | 0 | 1S5 | 84 |
| IGVDJ71 | 2S3 | 97 | 34 | 15S2 | 100 | 0 | 1S5 | 95 |
| IGVDJ72 | 2S3 | 92 | ND | ND | ND | ND | 1S3 | 80 |
| IGVDJ73 | 2S3 | 99 | 18 | 26S1 | 100 | 6 | 1S3 | 100 |
| IGVDJ74 | 2S3 | 97 | 11 | 18S1 | 100 | 5 | 1S6 | 100 |
| IGVDJ75 | 2S3 | 98 | 15 | 7S1 | 100 | 6 | 1S5 | 100 |
| IGVDJ76 | 2S3 | 96 | 2 | 20S2 | 100 | 6 | 1S7 | 83 |
| IGVDJ77 | 2S3 | 96 | 20 | 17S1/2 | 93 | 4 | 1S3 | 100 |
| IGVDJ78 | 2S3 | 97 | 23 | 17S1/2 | 100 | 6 | 1S5 | 100 |
| IGVDJ79 | 2S3 | 96 | 4 | 17S1/2 | 93 | 6 | 1S3 | 100 |
| IGVDJ80 | 2S3 | 97 | 10 | 18S1 | 93 | 12 | 1S5 | 100 |
| IGVDJ81 | 2S3 | 97 | 11 | 17S1/2 | 93 | 8 | 1S5 | 100 |
| IGVDJ82 | 2S2 | 100 | 7 | 7S2 | 100 | 8 | 1S5 | 100 |
| IGVDJ83 | 2S2 | 98 | 8 | 12S1 | 85 | 9 | 1S5 | 100 |
| IGVDJ84 | 2S2 | 100 | 10 | 4S1 | 94 | 5 | 1S3 | 96 |
| IGVDJ85 | 2S2 | 99 | 11 | 18S1 | 100 | 0 | 1S2 | 94 |
| IGVDJ86 | 2S4 | 96 | 13 | 7S1 | 100 | 11 | 1S3 | 100 |
| IGVDJ87 | 2S4 | 94 | 4 | 15S2 | 93 | 3 | 1S5 | 90 |
| IGVDJ88 | 2S4 | 96 | ND | ND | ND | ND | 1S5 | 100 |
| IGVDJ89 | 4S2 | 100 | 17 | 18S1 | 100 | 1 | 1S5 | 91 |
| IGVDJ90 | 1S3 | 100 | 17 | 22S1 | 100 | 0 | 1S3 | 100 |
| ADULT: | ||||||||
| IGVDJ91 | 2S3 | 97 | 2 | 17S1/2 | 100 | 6 | 1S5 | 74 |
| IGVDJ92 | 2S3 | 97 | 15 | 2S1 | 100 | 19 | 1S2 | 100 |
| IGVDJ93 | 2S3 | 92 | 29 | 4S1 | 100 | 17 | 1S5 | 85 |
| IGVDJ94 | 2S3 | 89 | 0 | 7S1 | 100 | 20 | 1S5 | 90 |
| IGVDJ95 | 2S3 | 92 | 23 | 2S1 | 100 | 0 | 1S5 | 79 |
| IGVDJ96 | 2S3 | 90 | 16 | 10S1 | 100 | 9 | 1S5 | 89 |
| IGVDJ97 | 2S3 | 91 | 13 | 28S1 | 100 | 1 | 1S5 | 100 |
| IGVDJ98 | 2S3 | 93 | 16 | 15S2 | 92 | 14 | 1S5 | 82 |
| IGVDJ99 | 2S3 | 89 | 20 | 5S3 | 100 | 21 | 1S7 | 81 |
| IGVDJ100 | 2S3 | 91 | 17 | 6S1 | 100 | 6 | 1S5 | 85 |
| IGVDJ101 | 2S3 | 93 | ND | ND | ND | ND | 1S5 | 94 |
| IGVDJ102 | 2S3 | 90 | 10 | 9S1 | 94 | 15 | 1S3 | 100 |
| IGVDJ103 | 2S3 | 87 | 37 | 16S1 | 100 | 3 | 1S2 | 100 |
| IGVDJ104 | 2S3 | 92 | 18 | 2S1 | 100 | 2 | 1S5 | 96 |
| IGVDJ105 | 2S3 | 90 | 22 | 1S1 | 100 | 13 | 1S5 | 93 |
| IGVDJ106 | 2S3 | 93 | 8 | 20S1 | 100 | 17 | 1S5 | 67 |
| IGVDJ107 | 2S3 | 87 | 9 | 15S2 | 91 | 0 | 1S5 | 67 |
| IGVDJ108 | 2S3 | 87 | 0 | IGVDJ17, −115 | 100 | 28 | 1S3 | 88 |
| IGVDJ109 | 2S3 | 87 | 12 | 22S1 | 100 | 17 | 1S4 | 74 |
| IGVDJ110 | 2S3 | 91 | 37 | 18S1 | 100 | 0 | 1S2 | 91 |
| IGVDJ111 | 2S3 | 91 | 16 | 16S1 | 100 | 1 | 1S5 | 77 |
| IGVDJ112 | 2S3 | 83 | ND | ND | ND | ND | 1S3 | 100 |
| IGVDJ113 | 2S2 | 92 | 1 | 26S1 | 100 | 9 | 1S5 | 75 |
| IGVDJ114 | 2S2 | 92 | 30 | 25S1 | 93 | 4 | 1S2 | 97 |
| IGVDJ115 | 2S2 | 89 | ND | IGVDJ17, −108 | ND | ND | 1S3 | 91 |
| IGVDJ116 | 2S4 | 93 | 6 | 18S1 | 100 | 11 | 1S3 | 93 |
| IGVDJ117 | 2S4 | 89 | 23 | 18S1 | 100 | 3 | 1S5 | 100 |
| IGVDJ118 | 2S4 | 86 | 30 | 14S1 | 100 | 0 | 1S3 | 82 |
| IGVDJ119 | 2S4 | 89 | 14 | 12S1 | 100 | 4 | 1S3 | 96 |
| IGVDJ120 | 2S3 | 86 | 14 | 15S1 | 100 | 9 | 1S4 | 86 |
| IGVDJ121 | 4S2 | 84 | 19 | 22S1 | 94 | 4 | 1S5 | 85 |
| B. Sequences obtained with RT-PCR protocol | ||||||||
| Fet Spln VDJ1 | 2S3 | 99 | 9 | 11S1 | 100 | 1 | 1S5 | 100 |
| Fet Spln VDJ2 | 2S3 | 99 | 6 | 4S1 | 100 | 0 | 1S3 | 100 |
| Fet Spln VDJ3 | 2S2 | 100 | 0 | 11S1 | 100 | 0 | 1S5 | 98 |
| Fet Spln VDJ4 | 2S2 | 100 | 2 | 15S2 | 100 | 6 | 1S5 | 98 |
| Fet Spln VDJ5 | 2S2 | 100 | 0 | 17S1/2 | 96 | 0 | 1S5 | 100 |
| Fet Spln VDJ6 | 2S3 | 97 | ND | ND | ND | ND | 1S3 | 100 |
| Fet Spln VDJ7 | 2S4 | 98 | 5 | 20S1 | 94 | 0 | 1S3 | 100 |
| Fet Spln VDJ8 | 2S3 | 97 | 1 | 18S1 | 100 | 4 | 1S6 | 100 |
| Fet Spln VDJ9 | 2S3 | 96 | ND | ND | ND | ND | 1S5 | 100 |
| Neo MLN VDJ1 | 2S4 | 97 | ND | ND | ND | ND | 1S7 | 88 |
| Neo MLN VDJ2 | 2S3 | 97 | 11 | 19S1 | 100 | 0 | 1S5 | 98 |
| Neo MLN VDJ3 | 2S3 | 100 | 7 | 7S1 | 100 | 2 | 1S3 | 93 |
| Neo MLN VDJ4 | 2S2 | 99 | 0 | 17S1/2 | 83 | 8 | 1S5 | 95 |
| Neo MLN VDJ5 | 2S2 | 98 | 5 | 15S2 | 100 | 4 | 1S2 | 96 |
| Neo MLN VDJ6 | 2S2 | 100 | ND | ND | ND | ND | 1S5 | 96 |
| Neo MLN VDJ7 | 2S3 or 2S4 | 95 | 12 | 15S1 | 86 | 1 | 1S5 | 90 |
| Neo MLN VDJ8 | 2S2 | 99 | 3 | 15S2 | 100 | 8 | 1S5 | 96 |
| Neo MLN VDJ9 | 2S2 | 97 | 2 | 13S1 | 86 | 0 | 1S5 | 91 |
| Foal MLN VDJ1 | 2S3 | 97 | 10 | 4S1 | 93 | 4 | 1S3 | 100 |
| Foal MLN VDJ2 | 2S2 | 99 | 5 | 10S1 | 95 | 15 | 1S5 | 100 |
| Foal MLN VDJ3 | 2S3 | 97 | 7 | 4S1 | 94 | 1 | 1S5 | 100 |
| Foal MLN VDJ4 | 2S4 | 99 | 6 | 18S1 | 92 | 0 | 1S3 | 97 |
| Foal MLN VDJ5 | 2S3 | 97 | 3 | 18S1 | 100 | 1 | 1S5 | 100 |
| Foal MLN VDJ6 | 2S4 | 97 | 12 | 7S2 | 100 | 3 | 1S7 | 100 |
| Foal MLN VDJ7 | 2S3 | 96 | ND | ND | ND | ND | 1S5 | 100 |
| Foal MLN VDJ8 | 4S1 | 99 | ND | ND | ND | ND | 1S5 | 100 |
| Foal MLN VDJ9 | 4S2 | 99 | ND | ND | ND | ND | 1S5 | 97 |
| Adult MLN VDJ1 | 2S3 | 94 | ND | ND | ND | ND | 1S5 | 92 |
| Adult MLN VDJ2 | 2S3 | 70 | ND | ND | ND | ND | 1S3 | 92 |
| Adult MLN VDJ3 | 2S3 | 87 | ND | ND | ND | ND | 1S5 | 91 |
| Adult MLN VDJ4 | 2S4 | 78 | ND | ND | ND | ND | 1S5 | 94 |
| Adult MLN VDJ5 | 2S4 | 78 | ND | ND | ND | ND | ND | ND |
| Adult MLN VDJ6 | 2S2 | 90 | ND | ND | ND | ND | 1S2 | 94 |
| Adult MLN VDJ7 | 2S4 | 92 | ND | ND | ND | ND | 1S5 | 97 |
| Adult MLN VDJ8 | 2S4 | 92 | ND | ND | ND | ND | 1S2 | 100 |
| Adult MLN VDJ9 | 2S2 | 93 | ND | ND | ND | ND | 1S5 | 97 |
‘ND’ denotes germline gene segment could not be determined
Identity indicates the percent nucleotide identity of the sequence to the germline gene
1.4 Immunoglobulin gene name nomenclature
The name of Ig heavy chain variable, diversity, and joining gene segments were assigned according to guidelines set forth by IMGT, the international ImMunoGeneTics information system (www.imgt.org), and personal communication with Y. Sun. IGHV genes were named according to subgroup (Sun et al., 2010): ‘VH1’ was renamed ‘IGHV1S1’ to designate sequence 1 of subgroup 1. Twenty-eight subgroups were established for the 40 IGHD genes based on >75% nucleotide identity, 2 subgroups were established for the 8 IGHJ genes, and the genes named accordingly. Table 1 lists the correspondent gene names assigned by Sun et al. (2010) and the new designations.
Table 1.
IGHV gene segment nomenclature.
| IGHV Subgroup |
Previous designation |
New designation |
IGHD Subgroup |
Previous designation |
New designation |
|---|---|---|---|---|---|
| 1 | VH1 | IGHV1S1 | 1 | DH1 | IGHD1S1 |
| 1 | VH6 | IGHV1S2 | 2 | DH2 | IGHD2S1 |
| 1 | VH7 | IGHV1S3 | 3 | DH3 | IGHD3S1 |
| 1 | VHORF6 | IGHV1S4 | 4 | DH4 | IGHD4S1 |
| 1 | pVH24 | IGHV1S5 | 5 | DH5 | IGHD5S1 |
| 1 | novel VH1 | IGHV1S6 | 5 | DH10 | IGHD5S2 |
| 2 | VH2 | IGHV2S1 | 5 | DH15 | IGHD5S3 |
| 2 | VH4 | IGHV2S2 | 5 | DH21 | IGHD5S4 |
| 2 | VH5 | IGHV2S3 | 5 | DH25 | IGHD5S5 |
| 2 | VH14 | IGHV2S4 | 5 | DH28 | IGHD5S6 |
| 3 | VHORF1 | IGHV3S1 | 6 | DH6 | IGHD6S1 |
| 3 | VHORF4 | IGHV3S2 | 7 | DH7 | IGHD7S1 |
| 3 | VHORF5 | IGHV3S3 | 7 | DH17 | IGHD7S2 |
| 3 | pVH8 | IGHV3S4 | 8 | DH8 | IGHD8S1 |
| 3 | pVH9 | IGHV3S5 | 8 | DH16 | IGHD8S2 |
| 3 | pVH14 | IGHV3S6 | 8 | DH19 | IGHD8S3 |
| 3 | pVH17 | IGHV3S7 | 9 | DH9 | IGHD9S1 |
| 3 | pVH21 | IGHV3S8 | 10 | DH11 | IGHD10S1 |
| 3 | pVH23 | IGHV3S9 | 11 | DH12 | IGHD11S1 |
| 3 | pVH27 | IGHV3S10 | 12 | DH13 | IGHD12S1 |
| 4 | VH3 | IGHV4S1 | 12 | DH37 | IGHD12S2 |
| 4 | VH9 | IGHV4S2 | 13 | DH14 | IGHD13S1 |
| 4 | VH11 | IGHV4S3 | 14 | DH18 | IGHD14S1 |
| 4 | VH12 | IGHV4S4 | 15 | DH20 | IGHD15S1 |
| 4 | VH13 | IGHV4S5 | 15 | DH27 | IGHD15S2 |
| 4 | VHORF2 | IGHV4S6 | 16 | DH22 | IGHD16S1 |
| 4 | pVH4 | IGHV4S7 | 17 | DH23 | IGHD17S1 |
| 4 | pVH5 | IGHV4S8 | 17 | DH26 | IGHD17S2 |
| 4 | pVH12 | IGHV4S9 | 18 | DH24 | IGHD18S1 |
| 4 | pVH13 | IGHV4S10 | 19 | DH29 | IGHD19S1 |
| 4 | pVH15 | IGHV4S11 | 20 | DH30 | IGHD20S1 |
| 4 | pVH22 | IGHV4S12 | 20 | DH40 | IGHD20S2 |
| 4 | pVH26 | IGHV4S13 | 21 | DH31 | IGHD21S1 |
| 4 | pVH28 | IGHV4S14 | 22 | DH32 | IGHD22S1 |
| 4 | pVH29 | IGHV4S15 | 23 | DH33 | IGHD23S1 |
| 4 | pVH34 | IGHV4S16 | 24 | DH34 | IGHD24S1 |
| 4 | novel VH2 | IGHV4S17 | 25 | DH35 | IGHD25S1 |
| 5 | VHORF3 | IGHV5S1 | 26 | DH36 | IGHD26S1 |
| 6 | VH8 | IGHV6S1 | 27 | DH38 | IGHD27S1 |
| 7 | VH10 | IGHV7S1 | 28 | DH39 | IGHD28S1 |
| IGHJ Subgroup |
Previous designation |
New designation |
|||
| 1 | JH1 | IGHJ1S1 | |||
| 1 | JH3 | IGHJ1S2 | |||
| 1 | JH4 | IGHJ1S3 | |||
| 1 | JH5 | IGHJ1S4 | |||
| 1 | JH6 | IGHJ1S5 | |||
| 1 | JH7 | IGHJ1S6 | |||
| 1 | JH8 | IGHJ1S7 | |||
| 2 | JH2 | IGHJ2S1 | |||
Novel IGHV1S6 was identified in clone IGVDJ62 and spans residues 38,420–38,913 of genome sequence NW_001869767
Novel IGHV4S17 was identified in clones IGVDJ29 and IGVDJ30 and spans residues 11,760–12,258 of genome sequence NW_001872990
Results
Our previous investigation demonstrated that equine neonatal B cells were able to produce all Ig isotypes at the molecular level (Tallmadge et al. 2009). Further, TdT (DNTT) mRNA expression was detected in fetal bone marrow samples, suggesting the presence of non-templated diversity. The present study directly assessed the level of Ig heavy chain variable region diversity with RACE libraries constructed from fetal spleen, neonatal, foal, and adult horse mesenteric lymph node samples. Ig transcripts were amplified with a conserved IGHJ reverse primer (supplemental Figure 1). The fetal spleen sample was collected at 102 days of gestation, approximately one-third of a full term gestation. Fetal mesenteric lymph node was not used because lymph nodes could not be recovered in sufficient amount from fetuses at this age. Maternal antibodies do not cross the equine epitheliochorial placenta and the neonatal mesenteric lymph node was collected before the ingestion of colostrum; therefore, all sequences were of fetal origin and derived independent of environmental antigen exposure.
3.1 Germline immunoglobulin heavy chain variable region gene usage over time
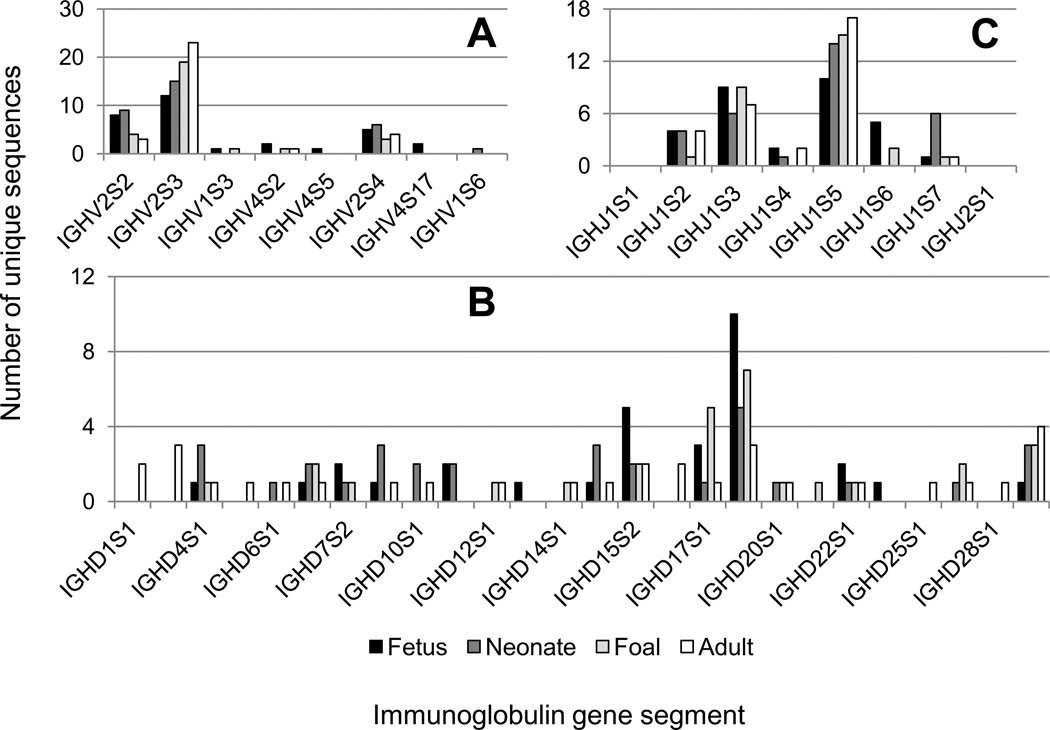
The recent annotation of IGHV, IGHD, and IGHJ genes from the horse genome sequence (Sun et al. 2010) allowed identification of the donor germline gene segments (Table 2). The fetal spleen-derived Ig VDJ sequences exhibited ≥ 97% nucleotide identity to 7 IGHV genes: IGHV2S2, IGHV2S3, IGHV1S3, IGHV4S2, IGHV4S5, IGHV2S4, and a novel IGHV gene segment, IGHV4S17 (Figure 1A). Although the newly identified IGHV4S17 gene segment was 99% identical to IGHV4S5, it is part of an unplaced genomic scaffold (NW_001872990.1, spanning positions 11,760 to 12,258), and the nucleotide differences present in the IGHV gene intron and flanking genomic sequence suggested that it was a distinct gene. Annotation of the donor IGHV gene segments on neonatal Ig VDJ sequences revealed usage of only 4 different segments: IGHV2S2, IGHV2S3, IGHV2S4, and a novel IGHV1S6 (Table 2, Figure 1A). A significant decrease in germline IGHV nucleotide identity was found in neonatal Ig VDJ sequences compared to mid-gestation fetal Ig sequences, ranging from 95 to 100% and with a median of 97% (p < 0.0001). The novel IGHV1S6 gene identified in clone IGVDJ62 was quite divergent from IGHV2S2, IGHV2S3, and IGHV2S4, resulting in pairwise nucleotide identity levels between 50 and 60%. The IGHV gene repertoire used in the foal differed from the neonate in the presence of IGHV1S3 and IGHV4S2, and the absence of IGHV1S6 (Figure 1A). The level of germline IGHV nucleotide identity was equivalent to the neonatal level (p = 0.37). Four IGHV gene segments were observed in the adult horse Ig VDJ sequences: IGHV2S2, IGHV2S3, IGHV4S2, and IGHV2S4 (Figure 1 A). The median percent identity of adult Ig VDJ sequences to germline IGHV gene segments was 90%, significantly lower than that of the foal (p < 0.0001, Table 2). Overall, IGHV2S3 usage was dominant at all ages: 39% of the fetal Ig VDJ sequences, 48% of neonatal, 68% of foal, and 74% of adult horse Ig VDJ sequences.
Figure 1. Equine immunoglobulin heavy chain gene segment usage throughout life.
The usage of immunoglobulin A) IGHV, B) IGHD, and C) IGHJ gene segments throughout life is shown and enumerated by the number of times each segment was identified in a unique IGVDJ sequence. Fetal sequences are shown in black, neonatal sequences in dark gray, foal sequences in light gray, and adult horse sequences in white. ‘ND’ represent IGHD gene segments for which the donor germline gene could not be determined.
Annotation of the germline IGHD donor genes revealed 13 different IGHD gene segments in the 31 fetal Ig VDJ sequences obtained (Figure 1B). IGHD annotation was not possible in one fetal sequence (IGVDJ17) in part because only 11 bases separated the IGHV and IGHJ regions, whereas the 40 germline IGHD genes range in length from 18 to 48 nucleotides. The usage of 17 different IGHD gene segments were identified in neonatal Ig sequences. In foal Ig VDJ sequences, 12 different annotated IGHD gene segments were identified as well as 3 others that could not be matched to the equine genome sequence. Twenty IGHD gene segments were observed in the adult horse Ig VDJ sequences and 4 were not determined. Of these 4, 2 appeared to share the same IGHD gene (IGVDJ108 and IGVDJ116); interestingly they also matched the IGHD region of the fetal IGVDJ17 sequence. In all the Ig VDJ sequences, IGHD18S1 was identified most frequently: in 30% of the fetal sequences, 16% of neonatal, 25% of foal, and 10% of the adult horse Ig VDJ sequences.
Four of the 8 IGHJ gene segments were used in all life stages: IGHJ1S2, IGHJ1S3, IGHJ1S5, and IGHJ1S7 (Figure 1C). IGHJ1S4 and IGHJ1S6 were also identified in fetal Ig VDJ sequences and used variably at later life stages. IGHJ1S5 usage was most common over time, ranging between 32 to 55% of Ig VDJ sequences.
The IGHV-IGHD-IGHJ combinations used over time were plotted to demonstrate the wide array of gene segments and combinations used at any given age (Supplemental figure 2). The usage of three IGHV gene segments (IGHV2S2, IGHV2S3 and IGHV2S4) was equivalent from fetal life to adulthood (80.7% and 96.8% respectively, p = 0.1983). Similarly, IGHJ1S5 and IGHJ1S3 accounted for 77.4% of adult horse Ig VDJ sequences and 61.3% of the fetal Ig VDJ sequences (p = 0.5952). Usage of IGHD gene segments appeared to follow the opposite trend, in which nearly twice as many gene segments were used in the adult horse compared to the fetus (23 versus 13). Comparison of the most frequently used IGHD genes IGHD18S1, IGHD17S1, and IGHD15S2 did not reveal a bias in usage over time (p = 0.8201). Together, these analyses supported the assertion that IGH variable region gene segment usage in the equine fetus was not biased.
3.2 Immunoglobulin VDJ region sequence diversity during distinct life stages
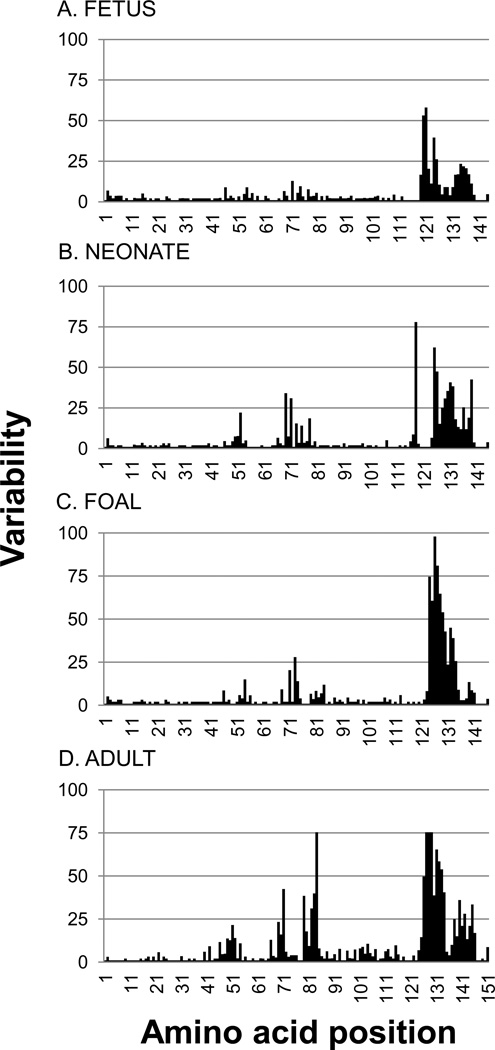
Comparison of the unique Ig VDJ sequences obtained from fetal spleen revealed pairwise nucleotide identity values that ranged from 51.1% to 96.9% with a median of 87.2% (Supplemental figure 3). Neonatal Ig VDJ sequences revealed a significant increase in Ig VDJ diversity over the last two-thirds of gestation (54 to 92.6%, median 85.8%, p < 0.01). Alignment of the predicted amino acid sequences (Supplemental figure 4) clearly displays the presence of individual residue differences among the fetal and neonatal sequences, suggesting a low level of somatic hypermutation during fetal life. The increase in diversity during the last two-thirds of gestation is also appreciable on the variability plot (Figure 2). The pairwise identity range of Ig VDJ sequences obtained from a 2-month-old foal did not differ significantly from neonatal levels (49.9 to 93.4%, median 86.3%, p = 0.06). Foal sequence IGVDJ72 showed the first signs of length variation in CDR2H in the amino acid alignment (Supplemental figure 4). The adult horse Ig VDJ sequences exhibited a large increase in sequence diversity over that of foal and earlier ages (68.8 to 86.0%, median 76.5%, p < 0.0001). The variability plot (Figure 2) and amino acid alignment (Supplemental figure 4) reveal increased sequence diversity and length variation in CDR1H and CDR2H, as well as in the intervening framework regions. The underlying nucleotide changes were consistent with somatic hypermutation, in which a trend of more frequent mutations at adenosine residues (29% vs 24%) was observed, and mutations of cytosine residues were often framed in the WRCY motif.
Figure 2. Variability plot of equine immunoglobulin heavy chain variable region sequences.
Variability plots of A) fetal, B) neonatal, C) foal, and D) adult horse immunoglobulin heavy chain variable region sequences. The variability index was determined as described by Wu and Kabat (1970).
3.3 Non-templated junctional diversity of Ig VDJ sequences
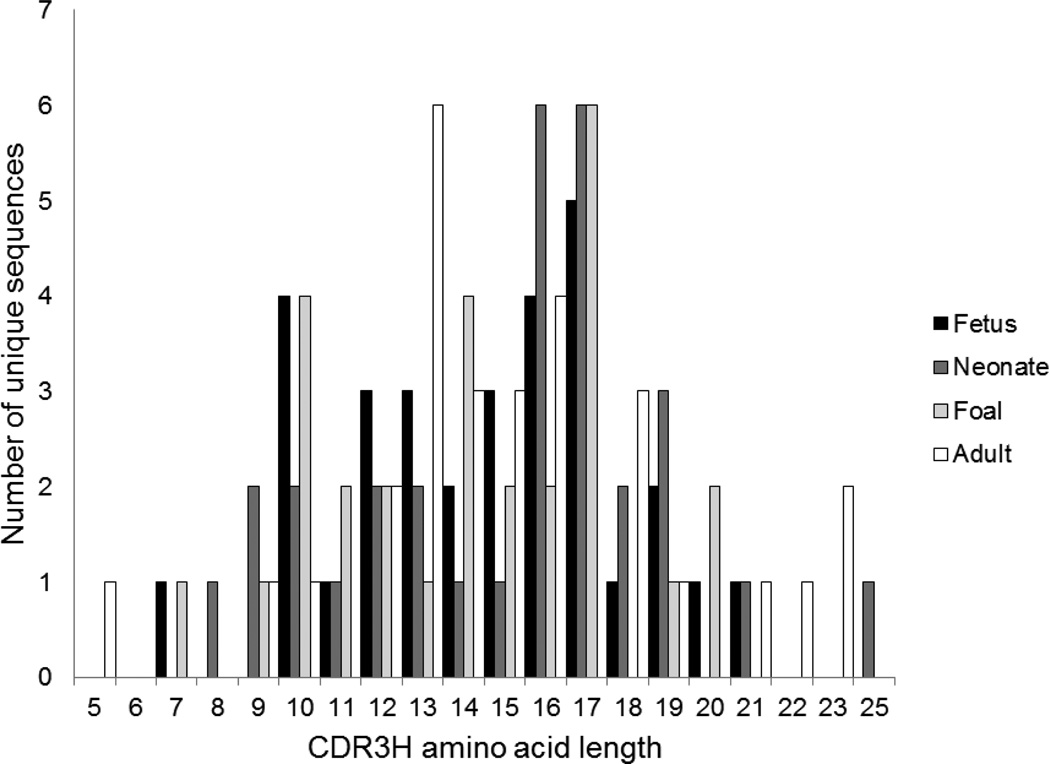
The annotation of germline gene usage also facilitated analysis of the presence of junctional diversity (Table 3). The junctional sequences often included homopolymer tracts of 2 to 5 bases, as expected for TDT-mediated N nucleotide insertions. P nucleotides were not defined because the exons were rarely full-length. During fetal life, appreciable CDR3H length variation was present, from 7 to 21 amino acids (Figure 3 and Supplemental figure 4). This was generated by a median of 8 nucleotide insertions between the IGHV and IGHD genes, and 0 nucleotide inserted between the IGHD and IGHJ genes. The length of these junctions increased significantly to 11 and 4.5, respectively, at birth, although the length of the CDR3H region remained similar. No further significant increase was detected in the length of junctional insertions of foal or adult sequences. The shortest CDR3H observed in the dataset was 5 residues, found in an adult Ig VDJ sequence.
Table 3.
Length of non-templated nucleotides inserted at junctions of Ig heavy chain gene rearrangements.
| IGHV-IGHD junction | IGHD-IGHJ junction | |||||
|---|---|---|---|---|---|---|
| Median 8 | Range | p-value* | Median | Range | p-value* | |
| Fetus | 0–15 | 0 | 0–9 | |||
| Neonate | 11 | 0–25 | 0.005 | 4.5 | 0–22 | 0.009 |
| Foal | 11 | 0–34 | 0.744 | 5 | 0–15 | 0.792 |
| Adult | 16 | 0–37 | 0.113 | 7.5 | 0–28 | 0.060 |
p-value compared to values with previous age (ie, neonate vs. fetus, foal vs. neonate).
Figure 3. CDR3H length distribution of equine immunoglobulin heavy chain sequences by age.
The range of CDR3H amino acid length is presented by age group and the number of unique sequences is plotted on the y-axis. Fetal sequences are shown in black, neonatal sequences in dark gray, foal sequences in light gray, and adult horse sequences in white.
3.4 Comparative phylogenetic analysis of immunoglobulin heavy chain variable regions
To compare our findings of IGHV gene usage to other species, we performed phylogenetic analysis. The framework regions of horse, cow, sheep, pig, human, and mouse IGHV gene families were used to generate a phylogenetic tree (Figure 4). All IGHV genes segregated into 3 clans with high bootstrap values, as previously described (Das et al. 2008; Sun et al. 2010; IMGT website). Functional horse IGHV gene segments were found in clans I and II, and only VHORF genes were found in clan III. The Ig VDJ sequences determined in this study also sorted into clans I and II and were consistent with our IGHV gene assignment based on BLAST analysis. The germline framework sequences of IGHV2S3 and IGHV2S4 clustered together because they share 100% amino acid identity when the CDRH regions are removed, and the Ig VDJ sequences derived in this study are interspersed throughout this area of the tree. Also, the framework regions of IGHV genes IGHV4S5 and IGHV4S17 could not be resolved due to their 98.8% amino acid identity; in this context, they were more similar to each other than to the fetal-derived Ig VDJ sequences, which each encoded a single amino acid residue change. Phylogenetic analysis of the horse Ig VDJ sequences obtained in this study indicates that no difference in IGHV gene usage was found with respect to developmental stage.
Figure 4. Comparative phylogenetic analysis of immunoglobulin heavy chain framework regions.
The phylogenetic tree was constructed from the amino acid sequence alignment of 88 horse, cow, sheep, pig, human and mouse IGHV gene framework regions. For species other than horse, one sequence was randomly chosen from each gene family. The tree was generated using the neighbor-joining method with p-distances model, and bootstrap values from 1,000 replicates are shown. IGHV clans are noted to the right of the respective cluster. The sequences determined in this study are indicated with the following icons: fetal - open diamond; neonatal - open triangle; foal − x; and adult horse - filled circle.
3.5 Comparison of RACE and RT-PCR methods to detect immunoglobulin heavy chain variable region diversity
A smaller dataset of 9 clones per stage was collected from the same tissues using RT-PCR with degenerate primers described by Almagro and co-workers (2006) (Table 2B). These degenerate primers detected expression of 5 IGHV genes, including IGHV4S1 that had not been detected above, and 5 IGHJ genes. Because IGHV2S3 and IGHV2S4 share 98.6% nucleotide identity, sometimes it was not possible to distinguish which was the germline donor gene. The levels of nucleotide identity and CDR3H lengths were consistent with those described above.
4. Discussion
This study assessed the developmental levels of Ig heavy chain VDJ region diversity in equine lymphoid tissues. At day 102 of a 340 day gestation, Ig VDJ diversity is present in the form of usage of multiple IGHV, IGHD, and IGHJ genes, CDR3H length variation, and modest diversity among and between the CDRH regions. Significant increases in Ig VDJ diversity were found to occur beyond the first trimester of gestation, demonstrated by sampling of mesenteric lymph node at birth prior to ingestion of colostrum. No further significant changes were observed in the 2-month postnatal period sampled, in contrast to the high diversity present in the adult horse lymphoid tissue.
Biased usage of IGHV, IGHD, or IGHJ gene segments was not found at any age in the horse, in contrast to fetal bovine, murine, and human B cells (Lawler et al. 1987; Jeong and Teale 1988; Raaphorst et al. 1992; Saini and Kaushik 2002). Horse IGHV gene segments IGHV2S2, IGHV2S3, and IGHV2S4 are used throughout life and account for >80% of each life stage dataset presented herein. The trend of several IGHV genes comprising the majority of the Ig repertoire was also found in piglets (Eguchi-Ogawa et al. 2010; Butler et al. 2011). Biased IGHV gene usage during murine fetal life has been attributed to differences in accessibility across the IGHV locus (Jeong and Teale 1988). However, assessment of the annotated equine IGHV genomic region reveals a span of more than 250,000 bases between expressed IGHV gene segments, indicating that the entire horse IGHV locus is accessible throughout life. Likewise, a study in fetal piglets found that IGHV usage is independent of genome position (Eguchi-Ogawa et al. 2010).
The most frequently identified IGHV gene segments (IGHV2S2, IGHV2S3, and IGHV2S4) in this study belong to the IGHV subgroup 2, and other annotated IGHV genes used belong to subgroups 1 (IGHV1S3) and 4 (IGHV4S2 and IGHV4S5). Most of the IGHV genes identified in this study are placed in the IGHV clan II by phylogenetic analysis, consistent with the expressed equine IGHV genes identified by other groups (Almagro et al. 2006; Sun et al. 2010). The remaining IGHV genes identified herein are placed in clan I. These findings contrast with reports of preferential utilization of clan III IGHV genes during fetal life in humans and mice (Alt et al. 1987; Tutter and Riblet 1989; Schroeder and Wang 1990). Similarly, the same set of IGHJ genes (IGHJ1S3 and IGHJ1S5) were used most frequently throughout life. Instead, a great deal of diversity throughout life appears to be derived from IGHD gene usage as many different IGHD genes were used at each time sampled, and these genes vary in sequence content and length. Perhaps the dominant usage of clan II IGHV genes is compensated for by the presence of diversity derived from IGHD/CDR3H length variation and non-templated nucleotide additions.
The composition of VD and DJ junctional sequences is consistent with N nucleotide tendencies of homopolymer tracts and guanine preference (Basu et al. 1983; Gauss and Lieber 1996). Identification of junctional diversity derived from P nucleotides was difficult because we found few full-length exons. Junctional diversity present in fetal and neonatal Ig VDJ sequences is consistent with our previous study that revealed TdT mRNA expression in equine fetal lymphoid tissues (Tallmadge et al. 2009). Antigen-independent junctional diversity due to TdT activity is also found in human, rabbit, pig, and sheep fetal tissues, but not in chick or mouse fetal tissues (Reynaud et al. 1989; Feeney 1990; Pascual et al. 1993; Reynaud et al. 1995; Tunyaplin and Knight 1995; Sinkora et al. 2003; Prabakaran et al. 2012). Interestingly, the significant increase in the length of non-templated nucleotides at junctional sites between fetal and neonatal stages was not reflected in CDR3H length, as no significant difference was detected. Presumably, the lack of correspondence can be attributed to differences in the length of IGHD genes used. CDR3H length variation in equine fetal Ig VDJ sequences is consistent with CDR3H length variation found in bovine fetal B cells, but contrasts with that during early life in humans (Shiokawa et al. 1999; Schroeder et al. 2001; Zemlin et al. 2001; Saini and Kaushik 2002; Prabakaran et al. 2012). The VD and DJ junctional diversity lengths described for adult horse Ig VDJ sequences herein are consistent with that reported by Sun et al. (2010).
We also compared the Ig diversity detected by 5’ RACE and standard RT-PCR methods with these samples. Amplification of 5’ RACE libraries is advantageous because it uses a universal forward primer and a conserved IGHJ reverse primer, amplifying all IGHJ transcripts regardless of the upstream IGHV gene, rather than an IGHV forward primer that does not detect all horse IGHV genes. The clones sequenced herein revealed that the RACE approach amplified more IGHV and IGHJ gene segments than the RT-PCR approach, including novel IGHV gene segments. Of the Ig VDJ sequences determined from 5’ RACE libraries, 15.7% do not match the degenerate IGHV forward primer and presumably would have been missed. Also, approximately 100 bases of sequence 5’ of the IGHV forward RT-PCR primer are gained with the 5’ RACE approach. This additional sequence improves IGHV gene annotation particularly in the case of IGHV2S3 and IGHV2S4 genes which share 98.6% nucleotide identity and 3 of the 6 nucleotide differences are upstream of the RT-PCR forward primer. We conclude that the additional gene segments detected justify the additional effort and cost of the RACE technique.
Overall, our study indicated that Ig heavy chain diversity in the equine fetus is not restricted with respect to germline gene usage or presence of diversity. Multiple variable region genes were used and the repertoire was comparable to that of foals and adult horses, as far as we could determine. It was unexpected to find the consistent pattern of dominant gene segment usage in all life stages (IGHV2S3, IGHD18S1, and IGHJ1S5). Limited junctional diversity and somatic hypermutation appeared to be present as early as 102 days of gestation, and similar CDR3H lengths were observed over time. The humoral system of the equine fetus exercises humoral response in the absence of pathogens for immune preparedness in early life.
Supplementary Material
Top: Schematic of immunoglobulin mRNA transcript illustrating placement of the reverse primer used for RACE RT-PCR (arrow). Bottom: Alignment of the 8 annotated IGHJ gene segments, identity to consensus indicated by dots. Reverse primer (arrow) is 100% conserved with all JH gene segments.
Immunoglobulin heavy chain V-D-J rearrangements observed in fetal (open diamond), neonatal (open triangle), foal (x), and adult horse (filled circle) lymphoid tissues. The x-axis represents the IGHD genes used with IGHV and IGHJ genes, both indicated on the left and right y-axes, respectively. The figure indicates the presence of each heavy chain V-D-J combination but does not reflect the frequency.
Pairwise nucleotide identity levels are shown for horse Ig VDJ sequences within each developmental stage. Shading indicates the level of identity: black shading with white text indicates nucleotide identity of 90–99.99%, dark gray shading with black text indicates nucleotide identity of 80–89.99%, light gray shading with black text indicates nucleotide identity of 70–79.99%, and no shading indicates nucleotide identity of ≤70%. The black diagonal represents identity to self (100%).
Alignment of fetal, neonatal, foal, and adult horse Ig VDJ sequences. Residues identical to the consensus sequence are represented by dots. Residues encoded by the reverse primer are shown in gray text, and the three complementarity-determining regions are underlined. Dashes represent gaps inserted to maximize the alignment. IGHV gene segment usage is noted after each sequence.
Highlights.
A significant increase in immunoglobulin diversity occurs during equine fetal life
No bias in Ig heavy chain gene segment usage was detected in early equine life
Predominant IGHV2S3, IGHD18S1, and IGHJ1S5 usage was identified in all life stages
Acknowledgements
This study was partially supported by the Harry M. Zweig Memorial Fund for Equine Research and National Institutes of Health grant 1R03AI079796-01.
Abbreviations
- Ig
immunoglobulin
- IGHV
immunoglobulin heavy chain variable gene
- IGHD
immunoglobulin heavy chain diversity gene
- IGHJ
immunoglobulin heavy chain joining gene
- CDR
complementarity-determining region
- RT-PCR
reverse transcription polymerase chain reaction
- mRNA
messenger RNA
- M-MuLV
Moloney Murine Leukemia Virus
- dNTPs
the four deoxynucleotide triphosphates
- cDNA
complementary DNA
- EST
expressed sequence tag
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Almagro JC, Martinez L, Smith SL, Alagon A, Estevez J, Paniagua J. Analysis of the horse V(H) repertoire and comparison with the human IGHV germline genes, and sheep, cattle and pig V(H) sequences. Mol. Immunol. 2006;43:1836–1845. doi: 10.1016/j.molimm.2005.10.017. [DOI] [PubMed] [Google Scholar]
- Alt FW, Baltimore D. Joining of immunoglobulin heavy chain gene segments: implications from a chromosome with evidence of three D-JH fusions. Proc. Natl. Acad. Sci. U S A. 1982;79:4118–4122. doi: 10.1073/pnas.79.13.4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alt FW, Blackwell TK, Yancopoulos GD. Development of the primary antibody repertoire. Science. 1987;238:1079–1087. doi: 10.1126/science.3317825. [DOI] [PubMed] [Google Scholar]
- Basu M, Hegde MV, Modak MJ. Synthesis of compositionally unique DNA by terminal deoxynucleotidyl transferase. Biochem. Biophys. Res. Commun. 1983;111:1105–1112. doi: 10.1016/0006-291x(83)91413-4. [DOI] [PubMed] [Google Scholar]
- Bauer K, Zemlin M, Hummel M, Pfeiffer S, Karstaedt J, Steinhauser G, Xiao X, Versmold H, Berek C. Diversification of Ig heavy chain genes in human preterm neonates prematurely exposed to environmental antigens. J. Immunol. 2002;169:1349–1356. doi: 10.4049/jimmunol.169.3.1349. [DOI] [PubMed] [Google Scholar]
- Brezinschek HP, Brezinschek RI, Lipsky PE. Analysis of the Heavy Chain Repertoire of Human Peripheral B Cells Using Single-Cell Polymerase Chain Reaction. J. Immunol. 1995;155:190–202. [PubMed] [Google Scholar]
- Butler JE, Sun X, Wertz N, Lager KM, Chaloner K, Urban J, Jr, Francis DL, Nara PL, Tobin GJ. Antibody repertoire development in fetal and neonatal piglets. XXI. Usage of most VH genes remains constant during fetal and postnatal development. Mol. Immunol. 2011;49:483–494. doi: 10.1016/j.molimm.2011.09.018. [DOI] [PubMed] [Google Scholar]
- Das S, Nozawa M, Klein J, Nei M. Evolutionary dynamics of the immunoglobulin heavy chain variable region genes in vertebrates. Immunogenetics. 2008;60:47–55. doi: 10.1007/s00251-007-0270-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Ashton B, Cheung M, Heled J, Kearse M, Moir R, Stones-Havas S, Thierer T, Wilson A. Geneious v4.7. 2009 [Google Scholar]
- Eguchi-Ogawa T, Wertz N, Sun X, Puimi F, Uenishi H, Wells K, Chardon P, Tobin GJ, Butler JE. Antibody repertoire development in fetal and neonatal piglets. XI. The relationship of variable heavy chain gene usage and the genomic organization of the variable heavy chain locus. J. Immunol. 2010;184:3734–3742. doi: 10.4049/jimmunol.0903616. [DOI] [PubMed] [Google Scholar]
- Feeney AJ. Lack of N regions in fetal and neonatal mouse immunoglobulin V-D-J junctional sequences. J. Exp. Med. 1990;172:1377–1390. doi: 10.1084/jem.172.5.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauss GH, Lieber MR. Mechanistic constraints on diversity in human V(D)J recombination. Mol. Cell Biol. 1996;16:258–269. doi: 10.1128/mcb.16.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudicelli V, Lefranc MP. Ontology for immunogenetics: the IMGTONTOLOGY. Bioinformatics. 1999;15(12):1047–1054. doi: 10.1093/bioinformatics/15.12.1047. [DOI] [PubMed] [Google Scholar]
- Jeong HD, Teale JM. Comparison of the fetal and adult functional B cell repertoires by analysis of VH gene family expression. J. Exp. Med. 1988;168:589–603. doi: 10.1084/jem.168.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabat EA, Wu TT. Identical V region amino acid sequences and segments of sequences in antibodies of different specificities. Relative contributions of VH and VL genes, minigenes, and complementarity-determining regions to binding of antibody-combining sites. J. Immunol. 1991;147:1709–1719. [PubMed] [Google Scholar]
- Kirkham PM, Mortari F, Newton JA, Schroeder HW., Jr Immunoglobulin VH clan and family identity predicts variable domain structure and may influence antigen binding. Embo J. 1992;11:603–609. doi: 10.1002/j.1460-2075.1992.tb05092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawler AM, Lin PS, Gearhart PJ. Adult B-cell repertoire is biased toward two heavy-chain variable-region genes that rearrange frequently in fetal pre-B cells. Proc. Natl. Acad. Sci. U S A. 1987;84:2454–2458. doi: 10.1073/pnas.84.8.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefranc MP, Pommie C, Ruiz M, Giudicelli V, Foulquier E, Truong L, Thouvenin-Contet V, Lefranc G. IMGT unique numbering for immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains. Dev. Comp. Immunol. 2003;27:55–77. doi: 10.1016/s0145-305x(02)00039-3. [DOI] [PubMed] [Google Scholar]
- Navarro P, Barbis DP, Antczak D, Butler JE. The complete cDNA and deduced amino acid sequence of equine IgE. Mol. Immunol. 1995;32:1–8. doi: 10.1016/0161-5890(94)00143-o. [DOI] [PubMed] [Google Scholar]
- Pascual V, Verkruyse L, Casey ML, Capra JD. Analysis of Ig H chain gene segment utilization in human fetal liver. Revisiting the proximal utilization hypothesis. J. Immunol. 1993;151:4164–4172. [PubMed] [Google Scholar]
- Prabakaran P, Chen W, Singarayan MG, Stewart CC, Streaker E, Feng Y, Dimitrov DS. Expressed antibody repertoires in human cord blood cells: 454 sequencing and IMGT/HighV-QUEST analysis of germline gene usage, junctional diversity, and somatic mutations. Immunogenetics. 2012;64:337–350. doi: 10.1007/s00251-011-0595-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaphorst FM, Timmers E, Kenter MJ, Van Tol MJ, Vossen JM, Schuurman RK. Restricted utilization of germ-line VH3 genes and short diverse third complementarity-determining regions (CDR3) in human fetal B lymphocyte immunoglobulin heavy chain rearrangements. Eur. J. Immunol. 1992;22:247–251. doi: 10.1002/eji.1830220136. [DOI] [PubMed] [Google Scholar]
- Reynaud CA, Dahan A, Anquez V, Weill JC. Somatic hyperconversion diversifies the single Vh gene of the chicken with a high incidence in the D region. Cell. 1989;59:171–183. doi: 10.1016/0092-8674(89)90879-9. [DOI] [PubMed] [Google Scholar]
- Reynaud CA, Garcia C, Hein WR, Weill JC. Hypermutation generating the sheep immunoglobulin repertoire is an antigen-independent process. Cell. 1995;80:115–125. doi: 10.1016/0092-8674(95)90456-5. [DOI] [PubMed] [Google Scholar]
- Saini S, Kaushik SA. Extensive CDR3H length heterogeneity exists in bovine foetal VDJ rearrangements. Scand. J. Immunol. 2002;55:140–148. doi: 10.1046/j.1365-3083.2002.01028.x. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Schrenzel MD, King DP, McKnight ML, Ferrick DA. Characterization of horse (Equus caballus) immunoglobulin mu chain-encoding genes. Immunogenetics. 1997;45:386–393. doi: 10.1007/s002510050220. [DOI] [PubMed] [Google Scholar]
- Schroeder HW, Jr, Hillson JL, Perlmutter RM. Structure and evolution of mammalian VH families. Int. Immunol. 1990;2:41–50. doi: 10.1093/intimm/2.1.41. [DOI] [PubMed] [Google Scholar]
- Schroeder HW, Jr, Wang JY. Preferential utilization of conserved immunoglobulin heavy chain variable gene segments during human fetal life. Proc. Natl. Acad. Sci. U S A. 1990;87:6146–6150. doi: 10.1073/pnas.87.16.6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder HW, Jr, Zhang L, Philips JB., 3rd Slow, programmed maturation of the immunoglobulin HCDR3 repertoire during the third trimester of fetal life. Blood. 2001;98:2745–2751. doi: 10.1182/blood.v98.9.2745. [DOI] [PubMed] [Google Scholar]
- Shiokawa S, Mortari F, Lima JO, Nunez C, Bertrand FE, 3rd, Kirkham PM, Zhu S, Dasanayake AP, Schroeder HW., Jr IgM heavy chain complementarity-determining region 3 diversity is constrained by genetic and somatic mechanisms until two months after birth. J. Immunol. 1999;162:6060–6070. [PubMed] [Google Scholar]
- Sinkora M, Sun J, Sinkorova J, Christenson RK, Ford SP, Butler JE. Antibody repertoire development in fetal and neonatal piglets. VI. B cell lymphogenesis occurs at multiple sites with differences in the frequency of in-frame rearrangements. J. Immunol. 2003;170(4):1781–1788. doi: 10.4049/jimmunol.170.4.1781. [DOI] [PubMed] [Google Scholar]
- Sun Y, Wang C, Wang Y, Zhang T, Ren L, Hu X, Zhang R, Meng Q, Guo Y, Fei J, Li N, Zhao Y. A comprehensive analysis of germline and expressed immunoglobulin repertoire in the horse. Dev. Comp. Immunol. 2010;34:1009–1020. doi: 10.1016/j.dci.2010.05.003. [DOI] [PubMed] [Google Scholar]
- Tallmadge RL, McLaughlin K, Secor E, Ruano D, Matychak MB, Flaminio MJ. Expression of essential B cell genes and immunoglobulin isotypes suggests active development and gene recombination during equine gestation. Dev. Comp. Immunol. 2009;33:1027–1038. doi: 10.1016/j.dci.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- Tunyaplin C, Knight KL. Fetal VDJ gene repertoire in rabbit: evidence for preferential rearrangement of VH1. Eur. J. Immunol. 1995;25:2583–2587. doi: 10.1002/eji.1830250927. [DOI] [PubMed] [Google Scholar]
- Tutter A, Riblet R. Conservation of an immunoglobulin variable-region gene family indicates a specific, noncoding function. Proc. Natl. Acad. Sci U S A. 1989;86:7460–7464. doi: 10.1073/pnas.86.19.7460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitkamp JH, Kallewaard N, Kusuhara K, Bures E, Williams JV, LaFleur B, Greenberg HB, Crowe JE., Jr Infant and adult human B cell responses to rotavirus share common immunodominant variable gene repertoires. J. Immunol. 2003;171:4680–4688. doi: 10.4049/jimmunol.171.9.4680. [DOI] [PubMed] [Google Scholar]
- Williams JV, Weitkamp JH, Blum DL, LaFleur BJ, Crowe JE., Jr The human neonatal B cell response to respiratory syncytial virus uses a biased antibody variable gene repertoire that lacks somatic mutations. Mol. Immunol. 2009;47:407–414. doi: 10.1016/j.molimm.2009.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TT, Kabat EA. An analysis of the sequences of the variable regions of Bence Jones proteins and myeloma light chains and their implications for antibody complementarity. J. Exp. Med. 1970;132(2):211–250. doi: 10.1084/jem.132.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemlin M, Bauer K, Hummel M, Pfeiffer S, Devers S, Zemlin C, Stein H, Versmold HT. The diversity of rearranged immunoglobulin heavy chain variable region genes in peripheral blood B cells of preterm infants is restricted by short third complementarity-determining regions but not by limited gene segment usage. Blood. 2001;97:1511–1513. doi: 10.1182/blood.v97.5.1511. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Top: Schematic of immunoglobulin mRNA transcript illustrating placement of the reverse primer used for RACE RT-PCR (arrow). Bottom: Alignment of the 8 annotated IGHJ gene segments, identity to consensus indicated by dots. Reverse primer (arrow) is 100% conserved with all JH gene segments.
Immunoglobulin heavy chain V-D-J rearrangements observed in fetal (open diamond), neonatal (open triangle), foal (x), and adult horse (filled circle) lymphoid tissues. The x-axis represents the IGHD genes used with IGHV and IGHJ genes, both indicated on the left and right y-axes, respectively. The figure indicates the presence of each heavy chain V-D-J combination but does not reflect the frequency.
Pairwise nucleotide identity levels are shown for horse Ig VDJ sequences within each developmental stage. Shading indicates the level of identity: black shading with white text indicates nucleotide identity of 90–99.99%, dark gray shading with black text indicates nucleotide identity of 80–89.99%, light gray shading with black text indicates nucleotide identity of 70–79.99%, and no shading indicates nucleotide identity of ≤70%. The black diagonal represents identity to self (100%).
Alignment of fetal, neonatal, foal, and adult horse Ig VDJ sequences. Residues identical to the consensus sequence are represented by dots. Residues encoded by the reverse primer are shown in gray text, and the three complementarity-determining regions are underlined. Dashes represent gaps inserted to maximize the alignment. IGHV gene segment usage is noted after each sequence.