Abstract
The plant immune system is activated by microbial patterns that are detected as nonself molecules. Such patterns are recognized by immune receptors that are cytoplasmic or localized at the plasma membrane. Cell surface receptors are represented by receptor-like kinases (RLKs) that frequently contain extracellular leucine-rich repeats and an intracellular kinase domain for activation of downstream signaling, as well as receptor-like proteins (RLPs) that lack this signaling domain. It is therefore hypothesized that RLKs are required for RLPs to activate downstream signaling. The RLPs Cf-4 and Ve1 of tomato (Solanum lycopersicum) mediate resistance to the fungal pathogens Cladosporium fulvum and Verticillium dahliae, respectively. Despite their importance, the mechanism by which these immune receptors mediate downstream signaling upon recognition of their matching ligand, Avr4 and Ave1, remained enigmatic. Here we show that the tomato ortholog of the Arabidopsis thaliana RLK Suppressor Of BIR1-1/Evershed (SOBIR1/EVR) and its close homolog S. lycopersicum (Sl)SOBIR1-like interact in planta with both Cf-4 and Ve1 and are required for the Cf-4– and Ve1-mediated hypersensitive response and immunity. Tomato SOBIR1/EVR interacts with most of the tested RLPs, but not with the RLKs FLS2, SERK1, SERK3a, BAK1, and CLV1. SOBIR1/EVR is required for stability of the Cf-4 and Ve1 receptors, supporting our observation that these RLPs are present in a complex with SOBIR1/EVR in planta. We show that SOBIR1/EVR is essential for RLP-mediated immunity and propose that the protein functions as a regulatory RLK of this type of cell-surface receptors.
Keywords: plant innate immunity, receptor complex, development, defense signaling, plant–microbe interaction
Plants rely on an innate immune system, which is activated upon recognition of pathogen-derived nonself molecules, or host-derived damage products (1, 2). Conserved microbe-associated molecular patterns (MAMPs) are perceived by pattern recognition receptors (PRRs) that activate MAMP-triggered immunity (MTI). Pathogenic microbes promote virulence by secretion of effector proteins, many of which suppress MTI (3, 4). In resistant plants, these effector proteins are detected by resistance proteins that activate effector-triggered immunity (ETI), frequently resulting in the hypersensitive response (HR), a localized programmed host cell death response (1). Conceptually, MTI and ETI function in a similar fashion by using immune receptors that mount a suitable defense response to halt pathogen ingress upon recognition of appropriate ligands that betray pathogen presence (5).
Most PRRs have been identified as transmembrane receptor-like kinases (RLKs) that frequently contain an extracellular leucine-rich repeat (LRR) domain or lysin-motif (LysM) for ligand recognition and an intracellular kinase domain for activation of downstream signaling (6). The LysM–RLK Chitin Elicitor Receptor Kinase 1 mediates immunity against fungi by recognizing fungal chitin (7, 8), whereas the LRR–RLKs Flagellin Sensing 2 (FLS2) and EF-Tu Receptor (EFR) are involved in recognition of bacterial flagellin and the elongation-factor Tu protein, respectively (9, 10). Upon ligand recognition, both FLS2 and EFR form a receptor complex with the LRR–RLK Somatic Embryogenesis Receptor Kinase 3/Brassinosteroid Insensitive 1 (BRI1)-Associated receptor Kinase 1 (SERK3/BAK1) and its close paralog BAK1-like 1 (BKK1) (11–13). The transphosphorylation events that follow, together with the dissociation of the cytoplasmic kinase Botrytis-Induced Kinase 1 from the receptor complex, subsequently activate downstream defense signaling (14, 15). SERK3/BAK1 is not involved in ligand binding to FLS2 and EFR but, rather, plays a role in downstream signaling upon its recruitment by FLS2 and EFR after ligand binding (2). Hence, SERK3/BAK1 and BKK1 likely function as signal enhancers and can be regarded as coregulatory RLKs in FLS2- and EFR-mediated immunity (6, 16).
Receptor-like proteins (RLPs) form a second major class of cell-surface receptors in plants. RLPs are structurally similar to RLKs but lack a cytoplasmic kinase domain (17, 18). RLPs function in defense, such as the Cf proteins and Ve1, as well as in development (18). Examples of the latter are Clavata2 (CLV2), which plays a role in meristem maintenance, and Too Many Mouths (TMM), which regulates stomatal patterning (18). Because RLPs lack a cytoplasmic kinase domain, it is anticipated that proteins containing such a domain are recruited to activate downstream signaling (19, 20). Indeed, Arabidopsis thaliana CLV2 forms a complex with the transmembrane kinase Coryne and the LRR–RLK CLV1 (21–23), whereas TMM requires the LRR–RLK Erecta to activate downstream signaling (24).
In tomato (Solanum lycopersicum), resistance to specific races of the fungal pathogens Cladosporium fulvum (causing leaf mold disease) and Verticillium dahliae (causing vascular wilt disease) is mediated by LRR-containing RLPs (25, 26). Cf proteins confer immunity upon recognition of C. fulvum race-specific secreted effectors [also referred to as avirulence (Avr) proteins] (27), whereas Ve1 recognizes the Ave1 effector protein secreted by race 1 V. dahliae strains (28). Cf-9 was the first identified RLP (19), and since its discovery several attempts have been made to understand Cf-mediated defense signaling by identifying Cf-interacting proteins. Yeast two-hybrid analyses resulted in the isolation of several potential interactors of the cytoplasmic C terminus of Cf-9 (29–31). However, an RLK or Coryne-like protein, recruited by Cf proteins and providing a cytoplasmic kinase domain through which Cf-mediated signaling would occur, remained to be identified (20). Recently, by immunopurification of a functional Cf-4–enhanced green fluorescent protein (eGFP) fusion protein from plants, we identified endoplasmic reticulum (ER)-resident chaperones as in planta interactors of Cf proteins that are required for Cf protein biogenesis (32). Here, following a similar approach, we describe the identification of the tomato ortholog of the Arabidopsis RLK Suppressor Of BIR1-1/Evershed (SOBIR1/EVR; hereafter referred to as SOBIR1) (33, 34) and its close homolog SOBIR1-like as Cf interactors. Interestingly, both tomato homologs and Arabidopsis SOBIR1 interact with Ve1, in addition to Cf-4, and we show that SOBIR1 is required for Cf-2–, Cf-4–, and Ve1-mediated immunity. Our work reveals an essential role for SOBIR1 in the plant immune response activated by two distinct RLPs involved in resistance to fungal pathogens and suggests that SOBIR1 functions as an essential regulatory RLK of this type of cell-surface receptors.
Results
Identification of Tomato SOBIR1 and SOBIR1-like as Interactors of Cf-4 and Ve1.
To identify Cf-interacting proteins, we stably transformed Money Maker (MM)–Cf-0 tomato, lacking Cf resistance genes to C. fulvum, with a construct driving constitutive expression of a Cf-4–eGFP fusion protein (32). Transgenic line (TL) TL3 showed recognition resulting in a specific HR upon infiltration with the C. fulvum Avr4 effector, whereas TL21 did not show a response to Avr4 (Fig. S1A). Cf-4–eGFP was successfully immunopurified from TL3, whereas the fusion protein could not be detected in TL21 (Fig. S1B). To identify proteins copurifying with Cf-4, tryptic on-bead digestion of the purified proteins was performed, and the generated peptides were analyzed by mass spectrometry. Interestingly, in the sample originating from TL3, but not in the one from TL21, in addition to peptides originating from Cf-4–eGFP itself, peptides matching to two tomato RLKs encoded by Solyc06g071810.1.1 and Solyc03g111800.2.1 were identified (Table S1). The alignments presented in Fig. S2A show that the amino acid sequences of these tomato RLKs are highly homologous to each other (∼74% identical) and are closely related to the Arabidopsis RLK SOBIR1 (∼60% identity). Both tomato RLKs are more distantly related to S. lycopersicum (Sl)SERK3a/BAK1 (∼25% identical) (33, 34). Fig. S2B also shows that the nucleotide sequences of both tomato RLKs and A. thaliana (At)SOBIR1 are very similar throughout their coding regions. Hence, we named the genes encoding the two tomato RLKs SlSOBIR1 and SlSOBIR1-like. Similar to AtSOBIR1, SlSOBIR1 and SlSOBIR1-like have five predicted LRRs, in contrast to SlSERK3a/BAK1, which has only four LRRs. The SOBIR1 sequences of tomato and Arabidopsis are highly similar, both in their extracellular LRR and cytoplasmic kinase domains, whereas the homology of SOBIR1 to SlSERK3a/BAK1 is mostly restricted to their kinase domains (Fig. S2A). No peptides originating from any other RLKs were identified in the peptide sample originating from TL3.
Cf-4–eGFP is also functional in Nicotiana benthamiana (32), and immunopurification of transiently expressed Cf-4–eGFP from this plant also yielded peptides from copurifying RLKs potentially matching SOBIR1 and SOBIR1-like (Table S2). The presence of SlSOBIR1 orthologs in N. benthamiana and Nicotiana tabacum was assessed by searching public databases, indeed revealing two candidate N. benthamiana homologs, referred to as NbSOBIR1 and NbSOBIR1-like, and one N. tabacum homolog (NtSOBIR1) (Fig. S2C). To also identify proteins interacting with Ve1, eGFP-tagged Ve1 (35) was immunopurified upon its transient expression in N. benthamiana. Also for this RLP, peptides matching NbSOBIR1 and NbSOBIR1-like were identified, whereas again no peptides from other RLKs were detected (Table S3).
Tomato SOBIR1 and SOBIR1-like and Arabidopsis SOBIR1 Interact with Cf-4 and Ve1.
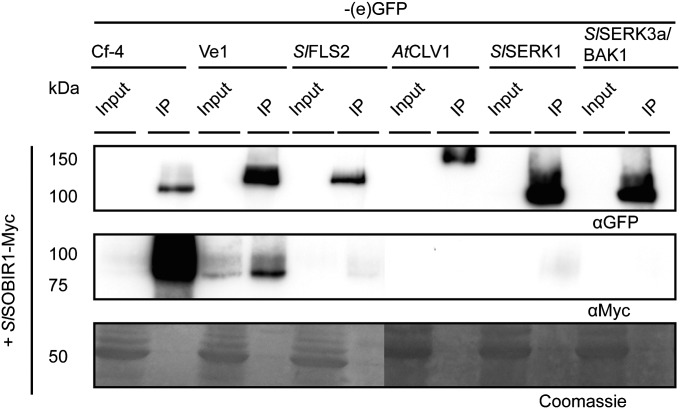
C-terminally Myc epitope-tagged versions of the tomato and Arabidopsis SOBIR1 RLKs (SlSOBIR1–Myc, SlSOBIR1-like–Myc, and AtSOBIR1–Myc) were generated to perform coimmunopurification experiments with Cf and Ve1. Transient coexpression in N. benthamiana revealed that all three SOBIR1 proteins interact with Cf-4 and Ve1 (Fig. 1 and Fig. S1C). Coexpression of constructs encoding SlSOBIR1–eGFP and Cf-4–Myc similarly revealed interaction of Cf-4–Myc with SlSOBIR1–eGFP (Fig. S3A). We then examined whether the SOBIR1 proteins also interact with RLKs known to be involved in defense and/or development. Interestingly, C-terminally (e)GFP-tagged SlSERK1, SlSERK3a/BAK1 (36), SlFLS2 (37), or AtCLV1 (38), did not copurify with SOBIR1 (Fig. 1 and Fig. S1C).
Fig. 1.
Tomato SlSOBIR1 interacts with Cf-4 and Ve1, but not with various RLKs. Tagged versions of Cf-4, Ve1, AtCLV1, SlSERK1, SlSERK3a/BAK1, and SlFLS2 (all fused to eGFP, except for SlFLS2, which was fused to GFP) were coexpressed with SlSOBIR1–Myc in N. benthamiana. Total protein extracts of transiently transformed leaf tissue were subjected to immunopurification by using GFP-affinity beads. Total proteins (Input) and immunopurified proteins (IP) were subjected to SDS/PAGE and blotted. Blots were incubated with α-GFP antibody to detect the immunopurified (e)GFP fusion proteins and incubated with α-Myc antibody to detect coimmunopurifying SOBIR1–Myc proteins. Coomassie-stained blots showing the 50-kDa Rubisco band present in the input samples confirm equal loading. Representative results for three independent experiments are shown.
To determine whether SOBIR1 requires a functional kinase domain for interaction with Cf-4, the core catalytic aspartate (D) of its conserved RD kinase motif was substituted to an asparagine (N) residue. For all tested RLKs containing the catalytic D, among which is SERK3a/BAK1, this mutation causes a loss of kinase activity (39). Interestingly, C-terminally Myc-tagged SlSOBIR1D473N, SlSOBIR1-likeD486N, and AtSOBIR1D489N all still interact with Cf-4–eGFP, showing that kinase activity of SOBIR1 is not required for interaction with the RLP (Fig. S3B). It was subsequently tested whether the presence of the Cf-4 ligand, Avr4, would lead to loss of the interaction between SOBIR1 and Cf-4. Cf-4–eGFP and SlSOBIR1–Myc were transiently coexpressed with Avr4 or the nonrecognized effector Avr9 infiltrated at two different optical densities. Interaction between Cf-4 and SlSOBIR1 was still observed in the presence of Avr4 and Avr9, indicating that the Cf-4/SlSOBIR1 complex does not dissociate upon recognition of Avr4 by Cf-4 (Fig. S3C). We further studied whether SlSOBIR1 forms homodimers and/or heterodimerizes with SlSOBIR1-like or AtSOBIR1. For this experiment, SlSOBIR1–eGFP was coexpressed with SlSOBIR1–Myc, SlSOBIR1-like–Myc, or AtSOBIR1–Myc, whereas coexpression with Cf-4–Myc was used as a control. Upon pull-down of SlSOBIR1–eGFP, Cf-4–Myc strongly copurified with the RLK. However, we did not observe copurification of SlSOBIR1–Myc, SlSOBIR1-like–Myc, or AtSOBIR1–Myc, indicating that SOBIR1 does not form homo- or heterodimers with SlSOBIR1-like or AtSOBIR1 (Fig. S3D).
SlSOBIR1 Localizes to the Plasma Membrane and Cytoplasmic Vesicles.
It has been reported that AtSOBIR1–YFP, when expressed under control of its own promoter in Arabidopsis, localizes to the plasma membrane and internal membrane compartments of epidermal leaf petiole cells and epidermal root cells (33). Confocal-laser scanning microscopy performed on N. benthamiana epidermal leaf cells transiently expressing SlSOBIR1–eGFP under control of the 35S promoter revealed that SlSOBIR1 mainly localizes to the plasma membrane (Fig. S4A). In addition, fluorescence signals were observed in mobile cytoplasmic vesicles (Fig. S4A). As previously shown, the GFP–HA control protein localizes to the cytoplasm and nucleus, whereas SlFLS2–GFP localizes to the plasma membrane (37) (Fig. S4 B–D).
Targeting SOBIR1 Compromises the Cf-4/Avr4–Induced and Ve1/Ave1-Induced HR.
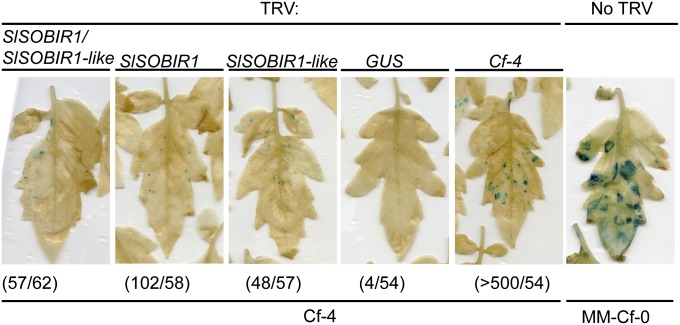
The observation that the two SOBIR1 homologs from tomato and N. benthamiana interact with Cf-4 and Ve1 (Fig. 1, Fig. S1C, and Tables S1–S3) suggests that both proteins play a role in Cf-4– and Ve1-mediated defense signaling in Solanaceous plants. Therefore, recombinant tobacco rattle virus (TRV)-based virus-induced gene silencing (VIGS) constructs were generated to target expression of the NbSOBIR1 homologs, either individually or simultaneously (Fig. S2C), and transgenic N. benthamiana expressing Cf-4 was inoculated with the different TRV constructs. Three weeks after viral inoculations, plants were transiently transformed to express Avr4 (40). Inoculation with TRV:NbSOBIR1/NbSOBIR1-like resulted in a severely compromised Avr4-triggered HR, similar to inoculation with a TRV construct targeting Cf-4 itself (TRV:Cf-4) (Fig. 2). The Avr4-triggered HR was also strongly compromised when NbSOBIR1 was targeted. When NbSOBIR1-like was targeted, the HR was affected to a much lesser extent (Fig. 2). Quantitative RT-PCRs (qRT-PCRs) revealed that expression of NbSOBIR1 was strongly reduced upon inoculation with TRV:NbSOBIR1/NbSOBIR1-like or TRV:NbSOBIR1, compared with inoculation with TRV:β-glucuronidase (GUS) (Fig. S5 A and B). Interestingly, we did not detect transcripts of NbSOBIR1-like in TRV:GUS-inoculated or TRV:NbSOBIR1/NbSOBIR1-like-inoculated plants, suggesting that NbSOBIR1-like is not expressed or is at a very low level. We therefore reasoned that the slight reduction of the Avr4-triggered HR upon inoculation of N. benthamiana:Cf-4 with TRV:NbSOBIR1-like (Fig. 2) could be attributed to cross-silencing of NbSOBIR1 by the TRV:NbSOBIR1-like construct. Indeed, qRT-PCR confirmed that NbSOBIR1 expression levels were ∼30% reduced upon inoculation with TRV:NbSOBIR1-like (Fig. S5B). Together these results indicate that NbSOBIR1 is the RLK that is required for the Cf-4–mediated HR in N. benthamiana. The Cf homolog Peru2 from Solanum peruvianum is autoactive in N. benthamiana, causing an effector-independent HR when transiently expressed (41). Interestingly, the Peru2–eGFP-triggered HR was also strongly compromised upon expression in TRV:NbSOBIR1/NbSOBIR1-like–inoculated N. benthamiana plants (Fig. S6A). To check whether the silenced plants were still able to mount programmed cell death, fully expanded leaves were also transiently transformed to express an autoactive variant of the Nucleotide Binding (NB)–LRR immune receptor Rx (RxD460V) (42) and the proapoptotic factor Bcl2-Associated protein X (BAX) (43). Because RxD460V and BAX still triggered a strong cell death, we concluded that the ability of the plants to mount programmed cell death was not compromised (Fig. 2).
Fig. 2.
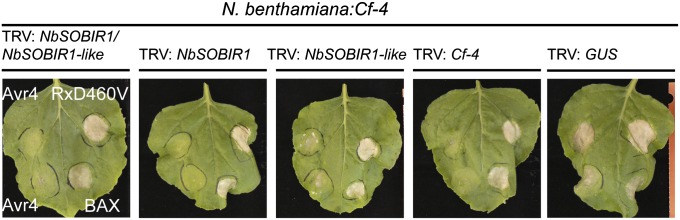
SOBIR1 is required for Cf-4–mediated HR. Transgenic N. benthamiana:Cf-4 plants were subjected to VIGS by inoculation with the TRV constructs indicated above each image. TRV:NbSOBIR1/NbSOBIR1-like targets NbSOBIR1 and NbSOBIR1-like simultaneously. TRV:Cf-4 and TRV:GUS served as controls. Three weeks after TRV inoculation, Avr4 (in duplicate), autoactive Rx (RxD460V), and BAX were transiently expressed in the order indicated in the left image, and leaves were photographed 3 d later. The experiment was performed three times with three plants for each TRV construct, and representative pictures for the experiment are shown.
Unlike in N. benthamiana, coexpression of Ve1 with Ave1 triggers an HR in N. tabacum, a plant for which TRV-based VIGS was recently established (28, 35). N. tabacum plants (cultivar Samsun) were inoculated with TRV:NbSOBIR1/NbSOBIR1-like, which also targets the NtSOBIR1 homolog (Fig. S2C), and TRV:Enhanced Disease Susceptibility 1 (EDS1) as a positive control, because EDS1 is required for Ve1-mediated immunity (26). Inoculation with TRV:GFP was included as a negative control. We used the TRV:NbSOBIR1/NbSOBIR1-like construct because we anticipated that N. tabacum, of which the currently available genome sequence is very similar to that of N. benthamiana, may contain an NtSOBIR1-like homolog in addition to NtSOBIR1, although we did not identify an NtSOBIR1-like candidate in public databases. Three weeks after inoculation with the different recombinant TRV constructs, Ve1 and Ave1 were coexpressed, revealing that plants inoculated with the VIGS constructs targeting NtSOBIR1/NtSOBIR1-like and EDS1 did not mount an HR, in contrast to the TRV:GFP-inoculated plants (Fig. S6B). Together, these results show that SOBIR1 is required for Cf-4– and Peru2-mediated HR in N. benthamiana and Ve1-mediated HR in N. tabacum.
Kinase Activity of SOBIR1 Is Required for Cf-4–Mediated HR.
To determine whether SOBIR1 requires a functional kinase domain for the Cf-4–mediated HR, we inoculated N. benthamiana:Cf-4 with TRV:NbSOBIR1/NbSOBIR1-like. These plants were then spot-infiltrated to transiently express the combinations Avr4 and AtSOBIR1–Myc or Avr4 and AtSOBIR1D489N–Myc. As a control, GUS was expressed in combination with Avr4. We reasoned that AtSOBIR1 would not be targeted by this RNA silencing because there is not sufficient sequence homology between the NbSOBIR1 genes and AtSOBIR1, and therefore AtSOBIR1, being a functional homolog of NbSOBIR1, would complement the loss of NbSOBIR1 and reconstitute the Avr4-triggered HR. However, if SOBIR1 kinase activity is required for Cf-4–mediated HR, AtSOBIR1D489N–Myc would not be able to complement.
Coexpression of GUS with Avr4 in the NbSOBIR1-silenced plants did not restore the Cf-4–mediated HR (Fig. S5C). When AtSOBIR1–Myc was coexpressed with Avr4, an HR was observed. However, when the kinase-dead mutant AtSOBIR1D489N–Myc was coexpressed with Avr4, the Avr4-triggered HR was strongly compromised, indicating that SOBIR1 kinase activity is required for Cf-4–mediated HR (Fig. S5C). RT-PCR analysis showed that full-length AtSOBIR1–Myc and AtSOBIR1D489N–Myc transcripts were present in the plants inoculated with TRV:NbSOBIR1/NbSOBIR1-like (Fig. S5D), confirming that Arabidopsis SOBIR1 is indeed not targeted by the VIGS construct. These results show that AtSOBIR1 complements NbSOBIR1 and the C-terminal Myc epitope tag does not appear to affect AtSOBIR1 function with respect to its role in Cf-4–mediated HR. Importantly, these results show that SOBIR1 kinase activity is required for the Cf-4–mediated HR.
SOBIR1 Is Required for Cf- and Ve1-Mediated Resistance to C. fulvum and V. dahliae, Respectively.
To determine whether SOBIR1 is required for Cf-4–mediated resistance of tomato to C. fulvum, TRV constructs targeting tomato SlSOBIR1 and SlSOBIR1-like individually or both genes simultaneously were generated (Fig. S2B). As a positive control, plants were inoculated with TRV:Cf-4, whereas TRV:GUS-inoculation served as a negative control. Cf-4–expressing tomato was inoculated with the different TRV constructs, and 3 wk later, plants were inoculated with a race 5 strain of C. fulvum, expressing Avr4 and the GUS reporter gene. To detect fungal colonization, leaflets were GUS-stained after 2 wk. Inoculation with constructs targeting the two SlSOBIR1 homologs either individually or simultaneously, resulted in increased fungal colonization as indicated by the much higher number of successful colonization attempts compared with the TRV:GUS-inoculated plants. This result shows that both tomato SOBIR1 homologs contribute to Cf-4–mediated resistance (Fig. 3). We also targeted both SlSOBIR1 homologs in tomato expressing Cf-2.2 and inoculated these plants with the same C. fulvum strain as used above, because this race 5 strain also expresses Avr2. Also in this case, increased fungal colonization was observed compared with the TRV:GUS control (Fig. S6C).
Fig. 3.
Targeting SOBIR1 and SOBIR1-like suppresses Cf-4–mediated resistance of tomato. Cf-4 tomato was inoculated with the indicated TRV constructs, and 3 wk later plants were inoculated with an Avr4-secreting, GUS-transgenic strain of C. fulvum. A non TRV-inoculated susceptible MM-Cf-0 plant was included as control. Two weeks later, leaflets were stained for GUS activity to detect C. fulvum colonization. For the Cf-4 tomato plants, the amount of successful colonization attempts (blue spots) vs. the total amount of leaflets analyzed for that particular experiment is indicated between parentheses. The experiment was performed three times, and representative pictures are shown.
To test the role of the SlSOBIR1 homologs in resistance to V. dahliae, tomato cultivar Motelle that carries the Ve1 gene was also inoculated with TRV:SlSOBIR1, TRV:SlSOBIR1-like, and TRV:SlSOBIR1/SlSOBIR1-like. As controls, plants were inoculated with TRV:Ve1 and TRV:GFP. Three weeks after TRV inoculation, plants were either inoculated with a race 1 strain of V. dahliae expressing Ave1 or mock-treated and subsequently monitored for development of disease symptoms (e.g., stunted growth and reduced canopy area). Targeting of the two SlSOBIR1 homologs either individually or simultaneously, as well as Ve1 itself, resulted in clear stunting and a strongly reduced canopy area compared with the mock-treated plants. These disease symptoms were not observed in plants inoculated with TRV:GFP (Fig. S6D).
SOBIR1 Is Required for Ve1-Mediated Resistance to V. dahliae in Arabidopsis.
Ve1 provides resistance to V. dahliae when introduced in Arabidopsis (44). To study the requirement of AtSOBIR1 for Ve1-mediated resistance in this plant, we tested whether Ve1 still mediates resistance to V. dahliae in an Arabidopsis sobir1-1 mutant (34). Similar to the Columbia 0 (Col-0) wild-type, the sobir1-1 mutant is susceptible to V. dahliae race 1, as shown by the stunted appearance and chlorosis upon fungal infection (Fig. 4). When transformed with the Ve1 gene, the Col-0 wild-type gains resistance to V. dahliae race 1 (Fig. 4). Strikingly, when the Ve1 gene was introduced into the sobir1-1 mutant background, the plants did not gain resistance to the pathogen, because stunting and chlorosis were still observed after inoculation with the fungus (Fig. 4). Quantitative measurement of fungal biomass confirmed these results, because only in the Col-0 wild-type plants transformed with Ve1 fungal colonization was very limited (Fig. S7A). This result indicates that in addition to its requirement in tomato, SOBIR1 is required for Ve1-mediated resistance to V. dahliae in Arabidopsis.
Fig. 4.

Ve1 is not functional in an Arabidopsis sobir1-1 mutant background. Arabidopsis ecotype Col-0 is susceptible to V. dahliae race 1 expressing Ave1 (Col-0). When transformed with the Ve1 gene, expressed under control of the 35S promoter, Col-0 gains resistance to the fungus (Ve1). Similar to the Col-0 wild-type, sobir1 mutants are susceptible (sobir1-1), whereas sobir1-1 mutant plants transformed with Ve1 remain susceptible to the fungus (Ve1 sobir1-1). The inoculation experiments and qRT-PCR quantifications (Fig. S7A) were performed three times, with similar results. A representative picture is shown.
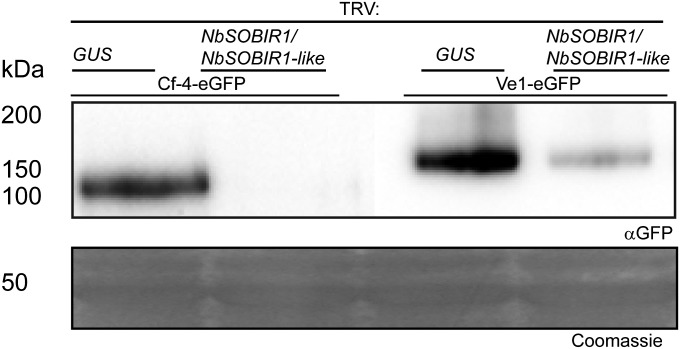
Targeting SOBIR1 in N. benthamiana Leads to Reduced Cf-4 and Ve1 Protein Levels.
To investigate whether targeting SOBIR1 affects Cf-4 and Ve1 protein levels, we inoculated N. benthamiana with TRV:NbSOBIR1/NbSOBIR1-like or the control TRV:GUS, and after 3 wk fully expanded leaves were transiently transformed to individually express eGFP-tagged Cf-4 or Ve1. Subsequently, the steady-state levels of the RLPs were determined by their immunopurification and detection by immunoblotting. Both Cf-4 and Ve1 protein levels were strongly reduced upon targeting SOBIR1, compared with the TRV:GUS-inoculated plants, indicating that SOBIR1 is required for the accumulation of Cf-4 and Ve1, and thus stabilizes these RLPs (Fig. 5). RT-PCRs revealed that Cf-4 and Ve1 are normally expressed in plants inoculated with TRV:NbSOBIR1/NbSOBIR1-like and TRV:GUS, indicating that reduced accumulation of the Cf-4 and Ve1 proteins is not due to reduced expression levels (Fig. S7B).
Fig. 5.
SOBIR1 is required for the accumulation of Cf-4 and Ve1 proteins. Cf-4 and Ve1, fused to eGFP, were expressed in leaves of N. benthamiana subjected to VIGS by inoculation with the indicated TRV constructs. Transiently expressed fusion proteins were immunopurified and subjected to SDS/PAGE, and blots were incubated with αGFP antibody for detection of the expressed proteins. The Coomassie-stained blot shows the 50-kDa Rubisco band present in the input samples to confirm equal loading. The experiment was repeated three times with similar results, and a representative picture is shown.
Tomato SOBIR1 Homologs Interact with a Broad Range of RLPs.
To test whether the tomato SOBIR1 homologs interact with additional RLPs, Cf-2.2, Cf-4E, Cf-9, and the Cf-like protein Peru2 from S. peruvianum were coexpressed as eGFP fusions with SlSOBIR1–Myc or SlSOBIR1-like–Myc in N. benthamiana. This experiment revealed that both SOBIR1 homologs copurify with the various Cf proteins (Fig. S8A). We expanded our study and examined whether more distantly related tomato RLPs also interact with the tomato SOBIR1 homologs. We fused SlEIX2 (45), mediating perception of the ethylene-inducing xylanase from Trichoderma viride, and the closest tomato orthologs of Arabidopsis CLV2 (Solyc04g056640.1), TMM (Solyc12g042760.1), and the Suppressor of Non-expressor of pathogenesis-related genes 1-1 (Npr1-1), Constitutive 2 (SNC2; Solyc02g072250.1) (46) to eGFP and coexpressed them with the Myc-tagged SOBIR1 homologs in N. benthamiana. Immunopurification of the RLPs revealed that SlEIX2, SlCLV2, and SlTMM, but not SlSNC2, interact with SlSOBIR1 and SlSOBIR1-like (Fig. S8B).
Discussion
For signal initiation by Cf proteins, a mechanistic model was proposed based on the early model of the Clavata1 (CLV1) signaling pathway, in which the RLP CLV2 interacts with the RLK CLV1. This RLK acts as a coreceptor that allows binding of the extracellular endogenous ligand CLV3 and subsequently mediates downstream signaling through its kinase domain (20, 47). Here, we report that the RLK SOBIR1 interacts with various RLPs of tomato, including the Cf proteins, Ve1 and SlEIX2, which are all involved in immunity, as well as the tomato homologs of Arabidopsis SlTMM and SlCLV2, which are involved in development (Fig. 1 and Figs. S1C and S8). However, not all RLPs interact with SOBIR1, as is exemplified by SlSNC2 (Fig. S8B). In addition, no interaction of SOBIR1 with any of the tested RLKs was found (Fig. 1 and Fig. S1C). We show that SOBIR1 is required for Cf-2.2–, Cf-4–, and Ve1-mediated immune responses (Figs. 2–4 and Figs. S5 and S6).
SOBIR1 was initially identified in a suppressor screen of the Arabidopsis bak1-interacting receptor kinase 1-1 (bir1-1) mutant and was referred to as Suppressor Of BIR1-1, 1 (34). BIR1 encodes another RLK, which interacts with SERK3/BAK1, and the bir1-1 mutant shows a constitutive defense phenotype, indicating that BIR1 is a negative regulator of defense responses. The bir1-1 phenotype is suppressed by the sobir1-1 mutation, suggesting that SOBIR1 is a positive regulator of defense signaling (34). In line with this finding, overexpression of SOBIR1 in Arabidopsis leads to constitutive defense activation (34). Although no direct interaction between SOBIR1 and BIR1 was observed, it was hypothesized that BIR1 functions in a signal transduction pathway that is dependent on SOBIR1 and which promotes pathogen resistance and cell death (34). As mentioned above, a mutation in AtSOBIR1 suppresses the bir1-1 phenotype, whereas an additional mutation in At Phytoalexin Deficient 4 (PAD4) fully reverts the bir1-1 sobir1-1 mutant phenotype back to that of wild-type plants. It was suggested that BIR1 regulates two parallel pathways—one involving resistance proteins that are dependent on PAD4, such as the Toll-Interleukin 1 Receptor (TIR)–NB–LRRs, and one involving another class of resistance proteins requiring SOBIR1 (34). We propose that the RLPs are members of this latter class of resistance proteins.
We also observed in planta interaction of SOBIR1 with RLPs involved in development. Indeed, a role of SOBIR1 in development has been described. Arabidopsis mutants in the gene encoding the ADP ribosylation factor GTPase-activating protein Nevershed (NEV) show impaired floral organ shedding after flowering (48). A screen for mutations in nev plants that restore organ shedding identified a mutation in SOBIR1 resulting in premature floral organ shedding. Hence, the name Evershed (EVR) was coined as a synonym for this RLK, which in this case functions as an inhibitor of abscission (33). Because SOBIR1/EVR was found to localize to the plasma membrane and cytoplasmic vesicles, it was proposed that the RLK regulates the signaling and internalization of other ligand-binding RLKs involved in floral organ shedding (33). Interestingly, when transiently expressed in N. benthamiana, we likewise found SlSOBIR1–eGFP to localize to the plasma membrane and mobile, cytoplasmic vesicles (Fig. S4). Similar to SOBIR1, SERK3/BAK1 also plays a role both in development and defense, and this RLK was initially identified as an interactor of the RLK BRI1, which is involved in brassinosteroid (BR) perception and signaling (49, 50). SERK3/BAK1 was also identified to act as a regulator of the RLK-type PRRs FLS2 (11, 13), EFR (12), and PEP1 Receptor protein-1, an RLK involved in perceiving endogenous peptides (51). Because Cf and Ve1 interact with SOBIR1 in planta and require SOBIR1 for mediating HR and resistance, it is tempting to speculate that SOBIR1 is involved in signaling and possible internalization of RLP-containing immune receptor complexes, similar to the function of SERK3/BAK1 in relation to RLKs involved in defense (52).
The current paradigm for several LRR–RLK-type PRRs is their rapid heterodimerization with SERK3/BAK1 upon ligand perception (11–13). By contrast, interaction between SOBIR1 and the various RLPs studied here is ligand-independent, because we did not coexpress the corresponding ligands in most of our coimmunopurification experiments and still detected copurification of SOBIR1 with the RLPs (Fig. 1 and Figs. S1C and S8). In addition, the presence of Avr4 did not affect the interaction of Cf-4 with SOBIR1 (Fig. S3C). Through mutation of its highly conserved RD motif, we showed that a functional SOBIR1 kinase domain is required for Cf-4–dependent HR (Fig. S5C), but not for interaction with Cf-4 (Fig. S3B). Possibly, the phosphorylation status of SOBIR1 changes upon ligand perception by Cf proteins, thereby allowing additional proteins to associate with the complex. Such proteins could be the previously identified Cf interactors Cf-9–Interacting Thioredoxin (CITRX) (31), the protein kinase Avr9/Cf-9–Induced Kinase 1 (ACIK1) (29), the Soluble N-ethylmaleimide-sensitive factor Adaptor protein Receptor (SNARE) protein Vesicle-Associated Protein 27 (VAP27) (30), and RLKs that reside in the active Cf-containing receptor complex. For example, recently it was shown that SERK1 is also required for Cf-4–mediated resistance of tomato. Furthermore, SERK1 and SERK3/BAK1 are both required for full Ve1-mediated resistance (26, 44). Because SOBIR1 constitutively interacts with a broad range of RLPs, either involved in defense or in development, it may be that SOBIR1 functions as a scaffold protein stabilizing receptor complexes in which RLPs take part. Alternatively, SOBIR1 could play a role as an integral part of the signaling pathway triggered by RLPs involved in different processes. In that case, downstream signaling specificity might be determined by the particular phosphorylation status of the cytoplasmic kinase domain of this regulatory RLK. For example, recent characterization of the bak1-5 mutation in Arabidopsis revealed that the function of SERK3/BAK1 in MTI, the BR response and cell death control can be mechanistically uncoupled (39). The bak1-5 mutation is in the kinase domain of SERK3/BAK1 and results in strongly impaired FLS2- and EFR-mediated immune signaling but does not affect BR signaling and the control of the cell death response (39). Such a situation might also hold for SOBIR1 in relation to signaling triggered by the different RLPs.
Together, our studies support the existence of a SOBIR1/RLP complex in planta, in which SOBIR1 is required for RLP-mediated immunity against two fungal pathogens that exhibit a different lifestyle. SOBIR1 appears to function as a regulatory RLK for RLP-containing immune receptor complexes in plants. Future experiments focusing on the cell biology of SOBIR1 and determination of its phosphorylation status and downstream interactors, in the presence and absence of the ligand that is perceived by the interacting RLP, should specify the precise role of SOBIR1 in RLP-containing signaling complexes.
Materials and Methods
Plant Materials and Growth.
Growth conditions for N. benthamiana, A. thaliana, and S. lycopersicum (tomato) are described in Sl Materials and Methods.
Primers and Vector Construction.
Sequences of primers and corresponding targets can be found in Table S4. Construction of plasmids containing Cf-2.2, -4, -4E, -9, Peru2, and Ve1, C-terminally fused to either eGFP or the Myc epitope-tag, has been described (32, 35). The construction of additional vectors for A. tumefaciens-mediated transient transformation and VIGS is described in Sl Materials and Methods.
Plant Transformations.
Plasmid pBIN-KS-35S::Cf-4–eGFP (Sol 2701) (32) was used for transformation of tomato MM–Cf-0, which does not carry a functional Cf-4 gene. Transformations and plant selections were performed as described in Sl Materials and Methods.
Protein Immunopurification and Identification.
Immunopurifications were essentially performed following the protocol described with minor modifications (32). Immunopurifications from stable transgenic tomato expressing Cf-4–eGFP were performed as described in Sl Materials and Methods.
VIGS and Disease Assays.
VIGS experiments in N. benthamiana, tobacco, and tomato were performed as described (32, 35). C. fulvum disease assays were performed as described (32), and V. dahliae disease assays were performed as described in Sl Materials and Methods.
Further experimental details can be found in Sl Materials and Methods.
Supplementary Material
Acknowledgments
We thank Prof. Pierre de Wit for critically reading the manuscript; Dr. Norbert de Ruijter for assistance with confocal microscopy; Dr. Yuelin Zhang for providing the sobir1-1 mutant seeds; Ali Ahmed and Ronnie Lubbers for assistance with experiments; and Unifarm personnel for excellent plant care. This work was supported by the Centre for BioSystems Genomics (part of the Netherlands Genomics Initiative and the Netherlands Organization for Scientific Research). P.S. and W.I.L.T. are supported by a Netherlands Organization for Scientific Research VENI grant, and B.P.H.J.T. is supported by a Netherlands Organization for Scientific Research VIDI grant. J.S., A.M.E.J., and S.R. are supported by the Gatsby Charitable Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1220015110/-/DCSupplemental.
References
- 1.Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444(7117):323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 2.Boller T, Felix G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 3.Göhre V, Robatzek S. Breaking the barriers: Microbial effector molecules subvert plant immunity. Annu Rev Phytopathol. 2008;46:189–215. doi: 10.1146/annurev.phyto.46.120407.110050. [DOI] [PubMed] [Google Scholar]
- 4.de Jonge R, Bolton MD, Thomma BPHJ. How filamentous pathogens co-opt plants: The ins and outs of fungal effectors. Curr Opin Plant Biol. 2011;14(4):400–406. doi: 10.1016/j.pbi.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 5.Thomma BPHJ, Nürnberger T, Joosten MHAJ. Of PAMPs and effectors: The blurred PTI-ETI dichotomy. Plant Cell. 2011;23(1):4–15. doi: 10.1105/tpc.110.082602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Monaghan J, Zipfel C. Plant pattern recognition receptor complexes at the plasma membrane. Curr Opin Plant Biol. 2012;15(4):349–357. doi: 10.1016/j.pbi.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 7.Miya A, et al. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci USA. 2007;104(49):19613–19618. doi: 10.1073/pnas.0705147104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shimizu T, et al. Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 2010;64(2):204–214. doi: 10.1111/j.1365-313X.2010.04324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gómez-Gómez L, Boller T. FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell. 2000;5(6):1003–1011. doi: 10.1016/s1097-2765(00)80265-8. [DOI] [PubMed] [Google Scholar]
- 10.Zipfel C, et al. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125(4):749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 11.Chinchilla D, et al. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature. 2007;448(7152):497–500. doi: 10.1038/nature05999. [DOI] [PubMed] [Google Scholar]
- 12.Roux M, et al. The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell. 2011;23(6):2440–2455. doi: 10.1105/tpc.111.084301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heese A, et al. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci USA. 2007;104(29):12217–12222. doi: 10.1073/pnas.0705306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu D, et al. A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc Natl Acad Sci USA. 2010;107(1):496–501. doi: 10.1073/pnas.0909705107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J, et al. Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe. 2010;7(4):290–301. doi: 10.1016/j.chom.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 16.Schwessinger B, Ronald PC. Plant innate immunity: Perception of conserved microbial signatures. Annu Rev Plant Biol. 2012;63:451–482. doi: 10.1146/annurev-arplant-042811-105518. [DOI] [PubMed] [Google Scholar]
- 17.Wang G, et al. A genome-wide functional investigation into the roles of receptor-like proteins in Arabidopsis. Plant Physiol. 2008;147(2):503–517. doi: 10.1104/pp.108.119487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang G, et al. The diverse roles of extracellular leucine-rich repeat-containing receptor-like proteins in plants. Crit Rev Plant Sci. 2010;29(5):285–299. [Google Scholar]
- 19.Jones DA, Thomas CM, Hammond-Kosack KE, Balint-Kurti PJ, Jones JDG. Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science. 1994;266(5186):789–793. doi: 10.1126/science.7973631. [DOI] [PubMed] [Google Scholar]
- 20.Joosten MHAJ, de Wit PJGM. The tomato-Cladosporium fulvum interaction: A versatile experimental system to study plant-pathogen interactions. Annu Rev Phytopathol. 1999;37:335–367. doi: 10.1146/annurev.phyto.37.1.335. [DOI] [PubMed] [Google Scholar]
- 21.Müller R, Bleckmann A, Simon R. The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell. 2008;20(4):934–946. doi: 10.1105/tpc.107.057547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu YF, et al. Analysis of interactions among the CLAVATA3 receptors reveals a direct interaction between CLAVATA2 and CORYNE in Arabidopsis. Plant J. 2010;61(2):223–233. doi: 10.1111/j.1365-313X.2009.04049.x. [DOI] [PubMed] [Google Scholar]
- 23.Bleckmann A, Weidtkamp-Peters S, Seidel CAM, Simon R. Stem cell signaling in Arabidopsis requires CRN to localize CLV2 to the plasma membrane. Plant Physiol. 2010;152(1):166–176. doi: 10.1104/pp.109.149930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee JS, et al. Direct interaction of ligand-receptor pairs specifying stomatal patterning. Genes Dev. 2012;26(2):126–136. doi: 10.1101/gad.179895.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rivas S, Thomas CM. Molecular interactions between tomato and the leaf mold pathogen Cladosporium fulvum. Annu Rev Phytopathol. 2005;43:395–436. doi: 10.1146/annurev.phyto.43.040204.140224. [DOI] [PubMed] [Google Scholar]
- 26.Fradin EF, et al. Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1. Plant Physiol. 2009;150(1):320–332. doi: 10.1104/pp.109.136762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stergiopoulos I, de Wit PJGM. Fungal effector proteins. Annu Rev Phytopathol. 2009;47:233–263. doi: 10.1146/annurev.phyto.112408.132637. [DOI] [PubMed] [Google Scholar]
- 28.de Jonge R, et al. Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc Natl Acad Sci USA. 2012;109(13):5110–5115. doi: 10.1073/pnas.1119623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rivas S, et al. CITRX thioredoxin interacts with the tomato Cf-9 resistance protein and negatively regulates defence. EMBO J. 2004;23(10):2156–2165. doi: 10.1038/sj.emboj.7600224. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Laurent F, Labesse G, de Wit PJGM. Molecular cloning and partial characterization of a plant VAP33 homologue with a major sperm protein domain. Biochem Biophys Res Commun. 2000;270(1):286–292. doi: 10.1006/bbrc.2000.2387. [DOI] [PubMed] [Google Scholar]
- 31.Nekrasov V, Ludwig AA, Jones JDG. CITRX thioredoxin is a putative adaptor protein connecting Cf-9 and the ACIK1 protein kinase during the Cf-9/Avr9- induced defence response. FEBS Lett. 2006;580(17):4236–4241. doi: 10.1016/j.febslet.2006.06.077. [DOI] [PubMed] [Google Scholar]
- 32.Liebrand TWH, et al. Endoplasmic reticulum-quality control chaperones facilitate the biogenesis of Cf receptor-like proteins involved in pathogen resistance of tomato. Plant Physiol. 2012;159(4):1819–1833. doi: 10.1104/pp.112.196741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leslie ME, Lewis MW, Youn JY, Daniels MJ, Liljegren SJ. The EVERSHED receptor-like kinase modulates floral organ shedding in Arabidopsis. Development. 2010;137(3):467–476. doi: 10.1242/dev.041335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao M, et al. Regulation of cell death and innate immunity by two receptor-like kinases in Arabidopsis. Cell Host Microbe. 2009;6(1):34–44. doi: 10.1016/j.chom.2009.05.019. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Z, et al. Optimized agroinfiltration and virus-induced gene silencing to study Ve1-mediated Verticillium resistance in tobacco. Mol Plant Microbe Interact. 2013;26(2):182–190. doi: 10.1094/MPMI-06-12-0161-R. [DOI] [PubMed] [Google Scholar]
- 36.Mantelin S, et al. The receptor-like kinase SlSERK1 is required for Mi-1-mediated resistance to potato aphids in tomato. Plant J. 2011;67(3):459–471. doi: 10.1111/j.1365-313X.2011.04609.x. [DOI] [PubMed] [Google Scholar]
- 37.Robatzek S, et al. Molecular identification and characterization of the tomato flagellin receptor LeFLS2, an orthologue of Arabidopsis FLS2 exhibiting characteristically different perception specificities. Plant Mol Biol. 2007;64(5):539–547. doi: 10.1007/s11103-007-9173-8. [DOI] [PubMed] [Google Scholar]
- 38.Clark SE, Williams RW, Meyerowitz EM. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell. 1997;89(4):575–585. doi: 10.1016/s0092-8674(00)80239-1. [DOI] [PubMed] [Google Scholar]
- 39.Schwessinger B, et al. Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet. 2011;7(4):e1002046. doi: 10.1371/journal.pgen.1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van der Hoorn RAL, Laurent F, Roth R, De Wit PJGM. Agroinfiltration is a versatile tool that facilitates comparative analyses of Avr9/Cf-9-induced and Avr4/Cf-4-induced necrosis. Mol Plant Microbe Interact. 2000;13(4):439–446. doi: 10.1094/MPMI.2000.13.4.439. [DOI] [PubMed] [Google Scholar]
- 41.Wulff BBH, et al. Gene shuffling-generated and natural variants of the tomato resistance gene Cf-9 exhibit different auto-necrosis-inducing activities in Nicotiana species. Plant J. 2004;40(6):942–956. doi: 10.1111/j.1365-313X.2004.02268.x. [DOI] [PubMed] [Google Scholar]
- 42.Bendahmane A, Farnham G, Moffett P, Baulcombe DC. Constitutive gain-of-function mutants in a nucleotide binding site-leucine rich repeat protein encoded at the Rx locus of potato. Plant J. 2002;32(2):195–204. doi: 10.1046/j.1365-313x.2002.01413.x. [DOI] [PubMed] [Google Scholar]
- 43.Lacomme C, Santa Cruz S. Bax-induced cell death in tobacco is similar to the hypersensitive response. Proc Natl Acad Sci USA. 1999;96(14):7956–7961. doi: 10.1073/pnas.96.14.7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fradin EF, et al. Interfamily transfer of tomato Ve1 mediates Verticillium resistance in Arabidopsis. Plant Physiol. 2011;156(4):2255–2265. doi: 10.1104/pp.111.180067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ron M, Avni A. The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell. 2004;16(6):1604–1615. doi: 10.1105/tpc.022475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, et al. Arabidopsis snc2-1D activates receptor-like protein-mediated immunity transduced through WRKY70. Plant Cell. 2010;22(9):3153–3163. doi: 10.1105/tpc.110.074120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clark SE. Cell signalling at the shoot meristem. Nat Rev Mol Cell Biol. 2001;2(4):276–284. doi: 10.1038/35067079. [DOI] [PubMed] [Google Scholar]
- 48.Liljegren SJ, et al. Regulation of membrane trafficking and organ separation by the NEVERSHED ARF-GAP protein. Development. 2009;136(11):1909–1918. doi: 10.1242/dev.033605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nam KH, Li J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell. 2002;110(2):203–212. doi: 10.1016/s0092-8674(02)00814-0. [DOI] [PubMed] [Google Scholar]
- 50.Li J, et al. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. 2002;110(2):213–222. doi: 10.1016/s0092-8674(02)00812-7. [DOI] [PubMed] [Google Scholar]
- 51.Postel S, et al. The multifunctional leucine-rich repeat receptor kinase BAK1 is implicated in Arabidopsis development and immunity. Eur J Cell Biol. 2010;89(2-3):169–174. doi: 10.1016/j.ejcb.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 52.Beck M, Zhou J, Faulkner C, MacLean D, Robatzek S. Spatio-temporal cellular dynamics of the Arabidopsis flagellin receptor reveal activation status-dependent endosomal sorting. Plant Cell. 2012;24(10):4205–4219. doi: 10.1105/tpc.112.100263. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.