Abstract
Sphingolipid metabolism in metazoan cells consists of a complex interconnected web of numerous enzymes, metabolites and modes of regulation. At the centre of sphingolipid metabolism reside CerSs (ceramide synthases), a group of enzymes that catalyse the formation of ceramides from sphingoid base and acyl-CoA substrates. From a metabolic perspective, these enzymes occupy a unique niche in that they simultaneously regulate de novo sphingolipid synthesis and the recycling of free sphingosine produced from the degradation of pre-formed sphingolipids (salvage pathway). Six mammalian CerSs (CerS1–CerS6) have been identified. Unique characteristics have been described for each of these enzymes, but perhaps the most notable is the ability of individual CerS isoforms to produce ceramides with characteristic acyl-chain distributions. Through this control of acyl-chain length and perhaps in a compartment-specific manner, CerSs appear to regulate multiple aspects of sphingolipid-mediated cell and organismal biology. In the present review, we discuss the function of CerSs as critical regulators of sphingolipid metabolism, highlight their unique characteristics and explore the emerging roles of CerSs in regulating programmed cell death, cancer and many other aspects of biology.
Keywords: ceramide, ceramide synthase (CerS), de novo sphingolipid synthesis, salvage pathway, sphingolipid, sphingosine
INTRODUCTION
Sphingolipids are an immensely diverse class of lipids that include several molecules (e.g. ceramides, sphingoid bases, ceramide phosphate and sphingoid base phosphates) that possess important bioactive properties and control a myriad of cellular and physiological programmes [1]. The metabolism of these lipids (Figure 1) involves numerous enzymes that take ‘simple’ sphingoid bases (e.g. sphingosine) and convert them into sphingolipids of a wide range of complexity (e.g. sphingomyelin and glycosphingolipids).
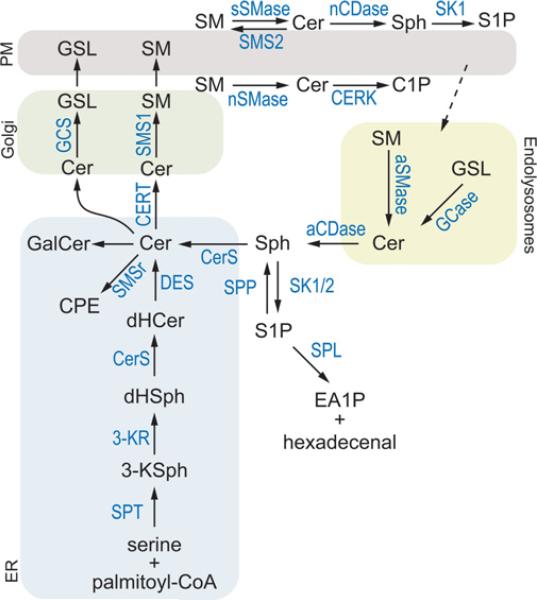
Figure 1. A ‘simplified’ view of sphingolipid metabolism.
De novo sphingolipid biosynthesis begins with the condensation of serine and palmitoyl-CoA catalysed by the serine palmitoyltransferase complex (SPT). Its product, 3-ketosphinganine (3-KSph) is enzymatically reduced to dihydrosphingosine (dHSph) by 3-ketosphinganine reductase (3-KR). dHSph is the substrate of CerSs, which produce a wide variety of dihydroceramides (dHCer) of various acyl-chain length (e.g. C14:0-dHCer to C26:0-dHCer). dHCer can be reduced to form ceramide (Cer) by dihydroceramide desaturase (DES). Ceramide can be metabolized to ceramide phosphoethanolamine (CPE) or galactosylceramide (GalCer) in the ER or transported to the Golgi via the ceramide transport protein (CERT) or through vesicular trafficking. In the Golgi, complex sphingolipids such as sphingomyelin (SM) and glycosphingolipids (GSLs) are synthesized via sphingomyelin synthases (SMS1 or SMS2), SMS-related protein (SMSr) or glycosphingolipids synthases (GCS) respectively. Ceramide may also be metabolized to ceramide 1-phosphate (C1P) via ceramide kinase (CERK) at the Golgi or plasma membrane (PM). The degradation of complex sphingolipids occurs through multiple pathways. At the outer leaflet of the plasma membrane, ceramide can be produced from sphingomyelin via secretory SMase (sSMase) and produce sphingosine via neutral CDase (nCDase). Sphingosine (Sph) can then be metabolized into S1P via SK1. Complex sphingolipids can be degraded to ceramide via the endolysosomal pathway, which contains aSMase and glycosidases (GCase). Ceramide is subsequently hydrolysed to sphingosine via acid CDase (aCDase). Free sphingosine can either be converted into S1P by SK1 or SK2 or re-synthesized into ceramide via CerS. This latter pathway is called the sphingosine salvage or recycling pathway. S1P can be hydrolysed back to sphingosine via S1P phosphatase (SPP) or degraded by S1P lyase (SPL) to ethanolamine 1-phosphate (EA1P) and hexadecenal.
One of the most important modifications of sphingoid bases is acylation of the free primary amine group to produce ceramides. This reaction occurs through at least two known mechanisms: the acyl-CoA-dependent CerS (ceramide synthase) reaction and the acyl-CoA-independent reverse CDase (ceramidase) reaction. Years of research have established the former as the most physiologically relevant means of ceramide synthesis, whereas the latter persists as an experimental curiosity with indeterminate significance. The first descriptions of the synthesis of ceramide from sphingoid bases came from studies in the 1960s showing that fractions containing a CDase activity also possessed a ‘reverse’ CDase activity, i.e. the ability to convert non-esterified ‘free’ fatty acids and sphingosine into ceramide [2–4]. Subsequently, an acyl-CoA-dependent CerS reaction was described by Sribney [5]. Additional studies showed that the substrate specificity of the acyl-CoA-dependent reaction better approximated the acyl-chain distribution of tissue sphingolipids in mammals [6,7]. It was therefore concluded and accepted that the bona fide physiological CerS reaction was acyl-CoA-dependent. Acyl-CoA-dependent ceramide, dihydroceramide and phytoceramide synthesis is largely believed to be due to the same enzymes in yeast and mammals. By convention we will therefore collectively refer to these activities as ‘ceramide synthase’, recognizing that, in some systems (e.g. yeast), dihydroceramide and phytoceramide are the predominant ceramides produced.
For many years, little more was known about CerS, but many questions persisted. Was it one enzyme or multiple enzymes? Which genes encoded its activity? What was the basis of different acyl-CoA specificities observed in different tissues? What is the biological role of the CerS reaction? Is this reaction merely an anabolic conduit for sphingolipid synthesis or does it have a role in cell signalling? Over the next decades several different lines of research would culminate in the identification of CerS genes, inhibitors and hitherto unexpected roles for these enzymes in critical aspects of biology.
IDENTIFICATION AND CHARACTERIZATION OF THE MAMMALIAN CerS FAMILY
The first gene responsible for ceramide synthesis was identified in Saccharomyces cerevisiae. This gene, LAG1, was first identified by a screen for genes differentially expressed in yeast aging [8]. Later it was described as a longevity assurance gene, because its deletion promoted longer chronological lifespan in yeast [9]. Several years later, LAG1 and its cognate LAC1 were found to be necessary for yeast ceramide synthesis and postulated to be enzymes possessing CerS activity [10,11]. Screening of cDNA libraries of S. cerevisiae, human, mouse and Caenorhabditis elegans revealed seven homologues of LAG1: one additional yeast gene LAC1, two C. elegans genes LAG1Ce-1 (also known as hyl-1) and LAG1Ce-2 (also known as hyl-2), a gene in human and mouse called UOG1 [upstream of growth and differentiation factor-1; now known as LASS1 (LAG1 homologue, ceramide synthase 1) or CERS1] and a gene in humans and dogs known to be part of the TRAM (translocating chain-associated membrane protein) family of proteins [12]. The protein sequences of the yeast, nematode and human genes showed limited similarity, except for a 52-amino-acid span that had an identity plus similarity of 52%; this sequence was called the Lag1p motif. Separate work looking for TRAM homologues also identified both LAG1 and LAC1 (then called DGT1) [13]. TRAM homologues, Lag1p homologues and some CLN (neuronal ceroid lipofuscinosis) proteins all possess a domain of five predicted transmembrane helices termed the TLC (TRAM, LAG1 and CLN8 homology) domain. This larger grouping has been termed the TLC family of proteins. CLN proteins are involved in the human neurodegenerative syndromes called CLNs [14]. Several CLN proteins have been implicated in the regulation of sphingolipid metabolism, although none has been demonstrated to possess enzymatic activity [15–17].
One of the mammalian genes identified in the screen for Lag1p homologues was an unusual bicistronic mRNA containing GDF1 (growth and differentiation factor-1) and UOG1 [12]. These genes were first identified in the early 1990s: GDF1 was known to encode a member of the TGF-β (transforming growth factor-β) family [18], but the function of the UOG1 gene product was unknown [19]. However, expression of UOG1 functionally complemented lag1Δlac1Δ yeast ceramide synthesis, cell growth and lifespan [12]. These data strongly suggested that the mammalian protein functioned similarly to Lag1p homologues in yeast [20].
Further characterization of UOG1 and its homologues led to the identification of the mammalian CerS family. Expression of UOG1 (Lass1 or CerS1) in HEK (human embryonic kidney)-293 cells conferred increased CerS activity and sphingolipid synthesis [21]. Interestingly, the increased synthesis was largely limited to sphingolipids containing C18:0 fatty acids (stearate), suggesting that this enzyme had a unique specificity for C18:0--CoA as a fatty acyl substrate. Subsequent work by the same group analysed in a similar fashion the overexpression of the previously identified TRAM homologues trh1 and trh4 (now known as CerS4 and CerS5 respectively) and found these genes also conferred increased CerS activity and sphingolipid synthesis [22]. However, overexpression of trh1/CerS4 or trh4/CerS5 showed differential effects on sphingolipid metabolism with regard to acyl-chain length and distinct from those of UOG1/CerS1. Two more mammalian homologues, Lass3 (CerS3) and Lass6 (CerS6), were subsequently identified and cloned [23,24]. Because CerS5 has been purified and shown in vitro to have CerS activity [25], there remains little doubt that each CerS family member represents a genuine enzyme with CerS activity.
GENOMIC ORGANIZATION OF CerS FAMILY MEMBERS
The genomic organization of human CerS genes is described in Table 1. As mentioned above, CerS1 is unique among CerS genes in that it is part of a bicistronic mRNA with GDF1 [19]. Both CerS1 and CerS2 have two transcript variants. The long variant of CerS1 encodes the full protein, whereas the short variant lacks the last 13 amino acids. The function of the shorter variant in unknown, but the truncation appears not to affect activity of the protein [26]. The shorter mRNA variant of CerS2 encodes the same full-length CerS2 protein as the longer variant and is the result of a splicing event in the 5′-UTR (untranslated region) of the mRNA transcript. The function of the shorter CerS2 transcript is currently unknown.
Table 1.
Genomic organization of human CerS
| CerS name | Other names | Chromosome location | Number of transcripts | Notes |
|---|---|---|---|---|
| CerS1 | Lass1/UOG1 | 19p12 | 2 | Transcript variant 1 includes GDF1 and a longer CerS1; transcript variant 1 has a shorter and distinct C-terminus |
| CerS2 | Lass2/TRH3 | 1q21.3 | 2 | Transcript variants encode the same protein, but variant 2 has an alternative splice site in the 5′-UTR |
| CerS3 | Lass3/T3I | 15q26.3 | 1 | – |
| CerS4 | Lass4/TRH1 | 19p13.2 | 1 | – |
| CerS5 | Lass5/TRH4 | 12q13.12 | 1 | – |
| CerS6 | Lass6/T1I | 2q24.3 | 1 | – |
SUBCELLULAR LOCALIZATION OF CerS PROTEINS
Early studies examining CerS activity in subcellular fractions of brain revealed that acyl-CoA-dependent ceramide synthesis occurred mostly in microsomes, which was distinct from the reverse CDase activity occurring in lysosomal and mitochondrial fractions [6]. CerS activity was characterized further as being enriched in ER (endoplasmic reticulum), but not Golgi, fractions [27,28]. Some studies have also found CerS activity in mitochondria-enriched fractions and mitochondria-associated membranes. CerS activity was partially purified from bovine brain [29] and liver [30] mitochondria-enriched fractions (although the actual organelle content of these fractions was not described). Bionda et al. [31] attempted to characterize and compare CerS activity in microsomes, mitochondria and mitochondria-associated membranes and found that, in addition to CerS activity, there may be a significant amount of reverse CDase activity in mitochondrial fractions [31]. Recent work from our group has confirmed the presence of reverse CDase activity in mitochondria and have shown that it is probably due to neutral CDase present in this fraction [32].
Since the discovery of CerS genes, many studies have used microscopy to study their localization utilizing overexpression approaches. Overexpression of N-terminal epitope-tagged mammalian CerSs in HEK-293 cells revealed these proteins as largely confined to the ER and nuclear envelope [21,22,33,34]. Interestingly, expression of a C-terminal haemagglutinin-tagged CerS1 resulted a Golgi-like localization [21]. The authors suggested that the C-terminal tag may have masked the K(X)KXX ER retrieval motif of CerS1, thus preventing its retention in the ER. However, more recent evidence has revealed that cells expressing C-terminal FLAG-tagged CerS1 do not show constitutive localization in the Golgi; instead, this protein localizes to the ER [26,34]. Interestingly, treating cells that express C-terminally tagged CerS1 with various cellular stressors caused translocation of this protein, or perhaps a C-terminal fragment of it, from the ER to the Golgi. Although not all CerSs have been tested for the effects of C-terminal tagging or stress-induced translocation, neither CerS4 nor CerS5 appears to behave in this manner [34]. No other stimulus-induced changes in the subcellular distribution of any other CerSs have been described.
STRUCTURE AND TOPOLOGY OF CerS PROTEINS
Before the identification of CerS proteins, little was known about the structure and topology of CerS. A couple of studies found that the enzymatic activity of CerS was accessible from the cytosolic leaflet of the ER membrane [27,28]. With the identification of mammalian CerS genes, much more has been elucidated (Table 2). Regarding the topology, work by Mizutani et al. [33] has elucidated several topological features of one CerS, CerS6. CerS6 has five proposed transmembrane domains, a luminal N-terminus and N-glycosylation site, and a cytosolic C-terminus. CerS2 and CerS5 were also found to be N-glycosylated. Because of the high similarity in the predicted transmembrane domains of CerS family members, other CerSs may share the same features as CerS6. However, the transmembrane domain prediction by UniProt (http://www.uniprot.org) suggests that CerS1–CerS4 possess six transmembrane domains, whereas CerS6 is the only one with five [35]. The transmembrane topologies of the CerSs other than CerS6 have not been experimentally determined.
Table 2.
Features of human CerS proteins
| CerS | Number of amino acids* | Predicted molecular mass (kDa)† | Predicted number of transmembrane domains* | Subcellular location | Notes |
|---|---|---|---|---|---|
| CerS1 | 350 (variant 1), 337 (variant 2) | 39.5 | 6 | ER/Golgi | – |
| CerS2 | 380 | 44.9 | 6 | ER | – |
| CerS3 | 383 | 46.2 | 6 | ER | – |
| CerS4 | 394 | 46.4 | 6 | ER | – |
| CerS5 | 392 | 45.8 | 6 | ER | – |
| CerS6 | 384 | 44.9 | 5 | ER | Glycosylation at Asn18 (not necessary for activity) |
Information obtained from UniProt (http://www.uniprot.org) [35,91].
Prediction by ExPASY Proteomics server (http://www.expasy.ch/tools/pi_tool.html)
FUNCTIONAL DOMAINS AND MOTIFS
CerS family members possess several identified functional domains. All Lag1p homologues contain a Lag1p motif (which is part of the larger TLC domain), and every mammalian CerS, except CerS1, contains a Hox (homeobox)-like domain [36]. Interestingly, the Hox-like domain, which is not required for activity [37,38], is only seen in vertebrate Lag1p homologues and not those of yeast, plants or worms [36]. The function of the Hox-like domain is currently unknown.
Several studies have identified specific amino acids that are required for CerS activity. Spassieva et al. [38] mutated conserved residues in the Lag1p motifs of CerS1 and CerS5 and found that several that were required for activity [38]. Interestingly, catalytic activity of CerS1 may control its CerS1 degradation and C-terminal translocation to the Golgi apparatus (see below) [26,39].
Additional domains have been identified as necessary for CerS activity. Removal of amino acids flanking the Hox-like domain and preceding the TLC domain results in a complete loss of activity [37]. More specifically, two positively charged amino acids between these two domains are necessary for the full activity of CerS5 or CerS6.
Unique among CerSs, CerS2 possesses two additional motifs that have been shown to have partial homology with S1P (sphingosine 1-phosphate) receptors [40]. Laviad et al. [40] found that S1P could inhibit CerS activity in CerS2-overexpressing cells, but not cells overexpressing other CerS isoforms. Inhibition by S1P occurred through a non-competitive mechanism and had an EC50 value of approximately 15 μM. Two S1P receptor-like motifs at amino acids 228–239 and 319–330 were identified as potential S1P-binding sites. When these domains were mutated, CerS2 lost its ability to be inhibited by S1P, suggesting that CerS2 may be regulated in vivo by S1P levels and therefore may have a unique regulatory function in sphingolipid metabolism.
SUBSTRATE SPECIFICITY OF CerS ISOFORMS
When acyl-CoA-dependent ceramide synthesis was first described in the late 1960s and early 1970s, it was suggested that this activity in mouse brain was governed by more than one enzyme [6,7]. Enzymatic work by Ullman and Radin [41] further supported this notion. It is now well appreciated that one of the most remarkable features of mammalian CerSs is that each CerS protein can regulate the synthesis of ceramides containing particular acyl-chain lengths. These preferences have been characterized by several studies and are summarized in Table 3.
Table 3. CerS substrate preferences.
dHSph, dihydrosphingosine.
| Acyl-CoA preference* |
|||||
|---|---|---|---|---|---|
| CerS | In vitro | Gain of function† | Loss of function‡ | Km towards dHSph (μM)§ | Reference(s) |
| CerS1 | C18:0 | C18:0 | C18:0 | 2.5 ± 0.7 | [21,33,38,80] |
| C20:0 | C18:1 | ||||
| CerS2 | C20:0 | C20:0 | C22:0 | 4.8 ± 0.4 | [20,33,40,92] |
| C22:0 | C22:0 | C22:1 | |||
| C24:0 | C24:0 | C24:0 | |||
| C26:0 | C24:1 | C24:1 | |||
| C26:0 | C26:0 | ||||
| C26:1 | C26:1 | ||||
| CerS3∥ | C16:0 | C18:0 | 1.7 ± 0.4 | [23] | |
| C18:0 | C20:0 | ||||
| C22:0 | C22:0 | ||||
| C24:0 | C24:0 | ||||
| CerS4 | C18:0 | C18:0 | C24:1 | 1.8 ± 0.4 | [22,33,92] |
| C20:0 | C20:0 | ||||
| C22:0 | C22:0 | ||||
| C24:0 | |||||
| C26:0 | |||||
| CerS5 | C14:0 | C16:0 | C16:0 | 1.8 ± 0.4 | [22,25,90] |
| C16:0 | C18:0 | ||||
| C18:0 | |||||
| C18:1 | |||||
| CerS6 | C14:0 | C16:0 | C16:0 | 2.0 ± 0.6 | [33,93,94] |
| C16:0 | |||||
| C18:0 | |||||
Bold chain lengths indicate the primary N-acyl length affected in each experimental situation.
Determined by overexpression of CerS proteins and measurement of ceramide or other sphingolipids.
Determined by siRNA-mediated knockdown or knockout of the indicated CerS.
Values taken from Lahiri et al. [43].
Recent results from Jennemann et al. [90a] show that CerS3-deficient animals lack ultra-long ceramides (>C26) in the epidermis.
The long-chain-base preference of CerS has also been described. The earliest studies of in vitro CerS activity showed no clear preference for dihydrosphingosine or sphingosine [6]. According to various studies, the Km values for sphingosine and dihydrosphingosine range from ~2 μM to 170 M [30,42–44]. The most recent data come from the Futerman group [43], where microsomes were prepared from HEK-293 cells overexpressing CerS proteins. In that study, only Km values for dihydrosphingosine were determined for the various CerS isoforms and were found to range from 1.7 μM (for CerS3) to 4.8 μM (for CerS2) (Table 3).
CerSs may also exhibit stereospecificity towards the sphingoid base. Naturally occurring sphingolipids contain sphingoid bases possessing the D-erythro (2S,3R) configuration, so D-erythro-sphingoid bases serve as adequate substrate for the CerS reaction. Synthetic threo-sphingoid bases are also substrates for in vitro CerS activity, although perhaps with less efficiency than erythro bases [5,6,45]. Conversely, L-erythro--dihydrosphingosine is not metabolized to dihydroceramide in vitro [44,45]. However, work by Stoffel and Bister [46] studied the in vivo metabolism of the four stereoisomers of dihydrosphingosine and found that all four isomers were converted into ceramide. Intriguingly, L-erythroand D-threo-ceramide were converted into ceramide, but not complex sphingolipids. An explanation for this phenomenon is currently lacking, but one might speculate that L-erythro- and D-threo-ceramides these researchers measured may have been 1-O-acyl sphingoid bases instead of the common N-acyl ceramides.
TISSUE DISTRIBUTION OF MAMMALIAN CerS FAMILY MEMBERS
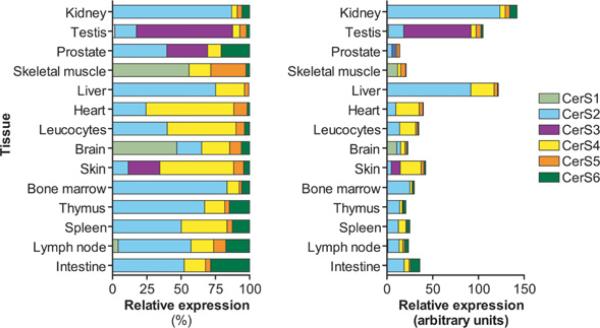
Many studies have reported the tissue distribution of mammalian CerSs and have shown that each tissue has a unique profile of CerS expression. Jiang et al. [12] analysed the human tissue expression of CerS1 by Northern blotting and found the highest expression in brain, skeletal muscle and testis. This was later confirmed in mouse tissues by other groups [22,33,40]. The tissue distribution of the other CerSs (CerS2–CerS6) in adult mouse tissue was also determined and is summarized in Figure 2 [40].
Figure 2. Tissue distribution of mouse CerSs.
The distribution of CerS1–CerS6 in mouse tissues based on data reported by Laviad et al. [40].
CerS expression changes during development. Multiple alterations in CerS transcripts have been observed during mouse brain development, including decreasing expression of CerS6 and decreased CerS2 expression, especially in myelin-producing cells [47]. CerS3 has also been linked to testicular germ cell and keratinocyte development commensurate with its high expression in skin and testes (discussed below) [48,49].
REGULATION OF CerS FAMILY MEMBERS
Our understanding of the regulation of mammalian CerSs is still in its infancy. However, several studies have suggested that these enzymes are regulated at multiple levels, including transcription, post-translation modification and degradation.
Regulation of CerS mRNA levels
The regulation of CerS expression at the mRNA level is largely uncharacterized. However, a few stimuli and experimental systems have demonstrated changes in CerS expression, including several inducers of apoptosis (Table 4). CerS5 mRNA levels were shown to be up-regulated in several models of stress, including DNA damage [50], hypoxia/re-oxygenation injury [51] and treatment with the AMPK (AMP-activated protein kinase) inhibitor Compound C [52]. In other work, CerS3 and CerS6 mRNA levels were increased by activation of cannabinoid receptors in a mantle cell lymphoma cell line [53]. CerS5 mRNA was elevated in the livers of CerS2-knockout mice, along with several enzymes involved in lipid metabolism [54]. Leptin, which increases insulin sensitivity of white adipose tissue, also decreased the mRNA levels of several sphingolipid enzymes including CerS2 and CerS4 [55]. These data support the hypothesis that CerSs can be transcriptionally regulated to control ceramide-mediated signalling.
Table 4.
Effectors of CerS mRNA levels
| CerS | Stimulus | Model system | Effect | Reference |
|---|---|---|---|---|
| CerS2 | Leptin | Rat white adipose tissue | Decrease | [55] |
| CerS3 | R(+)-Methanandamide or Win55 | Rec-1 mantle cell lymphoma cell line | Increase | [53] |
| CerS4 | Leptin | Rat white adipose tissue | Decrease | [55] |
| CerS5 | IR | Molt-4 leukaemia cells | Increase | [50] |
| Hypoxia/re-oxygenation | NT-2 neuronal precursor cells | Increase | [51] | |
| Compound C | MCF-7 breast adenocarcinoma cells | Increase | [95] | |
| CerS2-knockout mouse | Liver | Increase | [54] | |
| CerS6 | R(+)-Methanandamide or Win55 | Rec-1 mantle cell lymphoma cell line | Increase | [53] |
Post-translational regulation
Our understanding of the post-translational modifications of CerSs is also very limited. A few studies have recently suggested several modes of regulation for CerS1, including translocation and proteasome-mediated degradation [26,39]. Genotoxic or oxidative stresses, such as cisplatin, doxorubicin, dithiothreitol or UV light, caused a loss of CerS1 in a proteasome-dependent manner. Inhibition of p38 MAPK (mitogen-activated protein kinase) prevented the stress-induced degradation.
Part of CerS1 also undergoes a translocation in response to these stresses. Sridevi et al. [26] found that, in cells overexpressing C-terminally tagged CerS1, the C-terminus translocated to the Golgi in response to oxidative or genotoxic stress. The authors described a proteolytic event dependent on the proteasome that generated an ~ 17 kDa fragment containing the C-terminal tag. Furthermore, both the translocation and proteolysis were dependent on CerS1 catalytic activity and regulated by PKC (protein kinase C). This regulation by proteolysis appears to be specific for CerS1, as neither CerS4 nor CerS5 was degraded following stress.
Another mechanism of CerS1 regulation may be through phosphorylation. Sridevi et al. [39] showed that phorbol esters could increase the levels of a phosphorylated FLAG–CerS1. The authors argued that PKC activation increased the phosphorylation of FLAG–CerS1; however, the authors showed in a separate experiment that phorbol ester increases the protein levels of FLAG–CerS1 (through inhibition of degradation as mentioned above). Therefore it is difficult at this point to distinguish the effects of increased protein levels compared with increased phosphorylation. Nevertheless, the identification of a phosphorylated CerS protein invites further study of the post-translational regulation of these proteins.
Alterations in CerS activity have also been seen in fibroblasts from patients with CLN, and one of the CLN genes responsible, CLN9, has been shown to regulate CerS activity [16]. CLN9 encodes a protein that possesses a TLC domain and is part of the larger TLC family of proteins that includes yeast and mammalian CerS. Schulz et al. [16] showed that deficiency in CLN9 results in decreased CerS activity, decreased ceramide and dihydroceramide, and increased apoptosis. Expression of either Lag1p or CerS1 partially corrected the ceramide and cell death phenotype.
Regulators of CerS activity
The ability to study CerS regulation through the investigation of individual CerS proteins is a recent development. Over many years, numerous studies have demonstrated the regulation of CerS activity by various stimuli in a myriad of experimental systems, many in the context of programmed cell death. Pro-apoptotic inducers include chemotherapeutic agents, TNF (tumour necrosis factor)-α and IR (ionizing radiation) (Table 5). Only recently have individual CerSs been implicated in programmed cell death. These data are discussed later in the present review.
Table 5. Pro-apoptotic inducers of CerS activity.
bFGF, basic fibroblast growth factor; 4-HPR, 4-hydroxy(phenyl)retinamide.
| Inducer | Cell line/experimental system | Reference |
|---|---|---|
| Daunorubicin | P388 lymphocytic leukaemia cells | [60] |
| Doxorubicin | FTC-133 follicular thyroid carcinoma | [97] |
| Camptothecin | FTC-133 follicular thyroid carcinoma | [97] |
| TNF-α | Kym-1 rhabdomyosarcoma cells | [98] |
| Fenretinide (4-HPR) | CHLA-90 neuroblastoma cells | [99] |
| UV light (UV-B) | Human keratinocytes | [100] |
| Vascular endothelial growth factor receptor tyrosine kinase inhibitor SU5416 | Mouse lung | [101] |
| 125I-Deoxyuridine | Bovine aortic endothelial cells | [102] |
| Phorbol ester | CWR22-Rv1 prostate cancer cells | [103] |
| LNCaP prostate cancer cells | [104] | |
| IR | bFGF-treated C57BL6 mice | [105] |
| HeLa cervical carcinoma cells | [71] | |
| IR ± phorbol ester | LNCaP prostate cancer cells | [106] |
A few non-apoptotic regulators of CerS activity have been described. Two examples come from studies of sphingolipid metabolism in the skin. Uchida et al. [56] examined vitamin C-induced changes in sphingolipid metabolism in human keratinocytes and found multiple enzyme activities were regulated including an increase in CerS activity. Another study examining mouse skin found decreased CerS activity in chronologically aged compared with young mice [57]. Although the functional role of CerS in skin remains to be elucidated, one can speculate that CerS regulation will be important in the development and maintenance of skin functions such as the epidermal permeability barrier [49].
CerS AS REGULATORS OF DE NOVO AND SALVAGE CERAMIDE SYNTHESIS
As described above, CerSs control both de novo synthesis of sphingolipids as well as the recycling of sphingosine from the breakdown of pre-formed sphingolipids (Figure 3). The regulation of de novo compared with recycling pathways is poorly understood, and different cells may rely differentially on one pathway over another for the synthesis and maintenance of sphingolipids. For example, synthesis of glycosphingolipids in human foreskin fibroblasts was found to rely mostly on sphingosine recycling, whereas mouse C2C12 myoblasts predominantly used de novo synthesis to make these lipids [58]. On the other hand, C6 rat glioma cells used approximately 30% de novo and 60% recycling to make glycosphingolipids. In normal human fibroblasts results showed that a significant amount of sphingosine produced through the breakdown of glycosphingolipids is targeted for degradation [59].
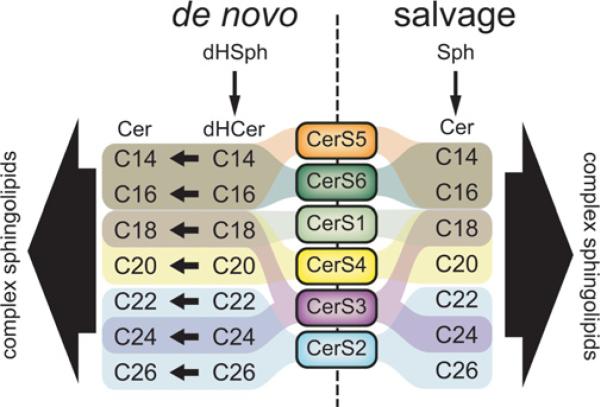
Figure 3. CerSs as regulators of de novo and salvage production of multiple ceramide species.
The major contributions of each CerS isoform to de novo and salvage synthesis of different ceramide species are depicted. Cer, ceramide; dHCer, dihydroceramide; dHSph, dihydrosphingosine; Sph, sphingosine.
Until recently, pharmacological inhibitors such as FB1 (fumonisin B1) (Figure 4) were the only tools available to probe the contribution of CerSs to sphingolipid metabolism. With the identification of individual CerS genes, the ability to study the functional importance of this reaction, or series of reactions, has expanded tremendously. Whereas FB1 has only been useful to study CerS activity as a whole, targeted manipulation of CerS has allowed the dissection of the unique contributions that each of these enzymes makes to sphingolipid metabolism.
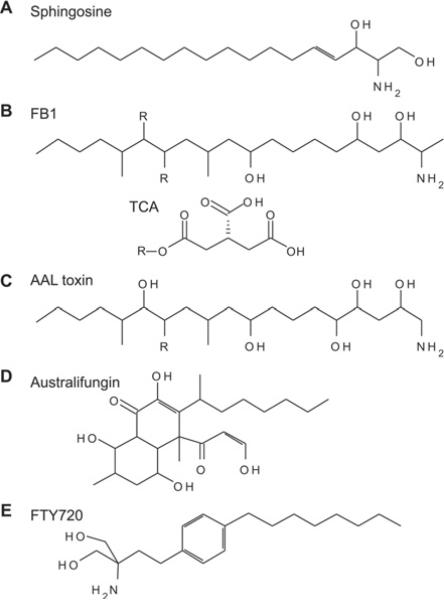
Figure 4. Inhibitors of CerS activity.
Known inhibitors of CerS include sphingosine (A) analogues such as fumonisins (B), a toxin from Alternaria alternata f. sp. lycopersici (AAL toxin) (C) and australifungin (D) [124]. Of these compounds, the FB1 (B) is the best characterized and most widely used to experimentally inhibit CerS. Fumonisins are produced by the fungus Fusarium moniliforme and were found to inhibit CerS activity in vitro and in vivo [125]. The structure of fumonisins consists of a hydrocarbon chain containing side chains including hydroxy and methyl groups and a single amino/amide group. In fumonisin B species, two of these hydroxy groups are esterified to tricarballylic acid (TCA). The TCA groups can be removed from the hydrocarbon backbone via intracellular esterases to form hydrolysed fumonisins (HF) or aminopolyols (AP). Hydrolysed FB1 (HFB1) is less potent than FB1 at inhibiting CerS and cell growth, suggesting that both the backbone and TCA groups are required for efficient binding to these enzymes [126,127]. An interesting characteristic of HFB1 is that it also is a substrate for the CerS reaction [127,128]. The resulting product, N-acyl-HFB1, is considerably more potent in cells than either HFB1 or FB1 (more than 10-fold). However, in vitro, HFB1 is less potent than FB1 [127]. AAL toxin (C) is highly similar in structure to fumonisins, and similarly inhibits CerS activity and elevates sphingoid bases [129,130]. Australifungin (D), a product of the fungus Sporormiella australis, is more potent than FB1 at inhibiting fungal CerS activity [124,131]. However, the β-ketoaldehyde moiety of australifungin can react with the free amines of sphingoid bases causing these to be sequestered, making its experimental use as an in vitro and in vivo CerS inhibitor problematic. FTY720 (fingolimod) (E), a myriocin analogue, has been shown to inhibit CerS through a complex mechanism including both uncompetitive and non-competitive modes [132,133].
BIOLOGICAL FUNCTIONS OF CerS: EMERGING THEMES
CerS and programmed cell death
For nearly two decades, CerSs have been implicated in the control of programmed cell death or apoptosis. The first description of CerS-mediated cell death was by Bose et al. [60] who demonstrated that CerS activity and ceramide generation could be induced by treatment of P388 lymphocytic leukaemia cells with the chemotherapeutic agent daunorubicin [60]. Since then, many models of programmed cell death have been shown to be sensitive to CerS inhibition (Table 6). A full discussion of this literature is beyond the scope of the present review, but the interested reader is referred to a recent extensive review by our group on the role of de novo sphingolipid synthesis and CerS in programmed cell death [61]. However, we will discuss briefly the emerging data regarding CerS-mediated cell death.
Table 6. Selected studies demonstrating inhibition of cell death by FB1.
N/D, not detected.
| Death inducer | Cell line/system | FB1 inhibits ceramide | FB1 inhibits death | Reference |
|---|---|---|---|---|
| Receptor-mediated | ||||
| TNF-α | MCF-7 breast carcinoma cells | + | + | [107] |
| L929 fibroblasts | + | + | [107] | |
| HL-60 promyelocytic leukaemia cells | + | + | [108] | |
| TNF-α/cycloheximide | Cerebral endothelial cells | + | + | [109] |
| B-cell receptor cross-linking | Ramos B-cells | + | + | [110] |
| T-cell receptor cross-linking | T-cell hybridoma | N/D | + | [111] |
| Cancer chemotherapeutic agents | ||||
| Camptothecin | 4B1 (human Fas-expressing L929 mouse fibroblasts) | N/D | + | [112] |
| FTC-133 follicular thyroid carcinoma cells | + | + | [97] | |
| Doxorubicin | MCF-7 breast carcinoma cells | N/D | + | [113] |
| Rat primary astrocytes | N/D | + | [114] | |
| FTC-133 follicular thyroid carcinoma cells | + | + | [97] | |
| Daunorubicin | P388 lymphocytic leukaemia cells | + | + | [60] |
| Hen follicular granulosa cells | N/D | + | [115] | |
| Etoposide | MOLT4 leukaemia cells | + | + | [116] |
| Fenretinide (4-HPR) | HL-60 promyelocytic leukaemia | + | + | [117] |
| Endothelial cells | + | + | [118] | |
| Fludarabine | Chronic B-cell leukaemia | + | + | [119] |
| LNCaP prostate cancer cells | + | + | [120] | |
| Inostamycin | Ms-1 small cell lung carcinoma cells | + | + | [121] |
| Oxaliplatin + SK inhibitor | RKO colon carcinoma cells | + | + | [122] |
| Vascular endothelial growth factor inhibitor SU5416 | Mouse lung | N/D | + | [101] |
| IR and non-IR | ||||
| γ-Irradiation | Mouse colonic crypts | N/D | + | [105] |
| Human lymphoblasts | + | + | [123] | |
| LNCaP prostate cancer cells | + | + | [106] | |
| 125I-Labelled 5-iodo-29-deoxyuridine | Bovine aortic endothelial cells | + | + | [102] |
| UV-B | Human keratinocytes | + | + | [100] |
| UV-C | Hen follicular granulosa cells | N/D | + | [115] |
Programmed cell death is a complex process whereby cells commit suicide in response to cellular stresses or extracellular signals. An important step in programmed cell death signalling is the MOMP (mitochondrial outer membrane permeabilization), leading to the release of pro-apoptotic factors into the cytosol. MOMP is governed by the balance of pro-apoptotic and anti-apoptotic proteins of the Bcl-2 family. Two related Bcl-2 family proteins, Bax and Bak, are necessary for MOMP to occur.
Work in our laboratory has demonstrated that Bak-deficient cells fail to induce the production of long-chain ceramide (e.g. C16:0 and C18:0 species) in response to several different apoptotic stimuli. These cells also had altered CerS activity, suggesting a specific role for Bak in regulating ceramide-mediated cell death [62]. Inhibition of de novo ceramide synthesis also inhibited caspase 3/7 activation and apoptosis, suggesting that ceramide functions downstream of mitochondria but upstream of caspase activation, possibly through controlling MOMP. Previous work also identified the ability of ceramide to independently form channels in mitochondrial membranes [63–67]. These channels could also be modified in vitro by the presence of pro- and anti-apoptotic Bcl-2 family members [68,69], suggesting that they might co-operate in vivo to promote MOMP.
Subsequent work from Kolesnick and co-workers has further supported a role for CerSs and ceramide in controlling MOMP. Lee et al. [70] found that CerS activity is necessary for Bax insertion and MOMP induced by IR. In that study, FB1 inhibited Bax insertion into mitochondrial membranes, cytochrome c release and caspase activation. Although ceramide itself did not induce cytochrome c release from isolated mouse liver mitochondria, they found that it was permissive for Bax insertion and cytochrome c release. The same group also found that IR increased in vitro C16:0-CerS activity in mitochondria-associated membrane fractions, but not in whole-cell homogenates or ER fractions [71]. These data further support a role for CerSs, particularly CerS5 and CerS6, in promoting organelle-specific ceramide generation and cell death.
Recent work from our laboratory suggests an alternative way that ceramide can function as a pro-apoptotic lipid [72]. Using a cell culture model of UV-induced cell death, we found that inhibition of CerSs or de novo sphingolipid synthesis had no effect on Bax activation and cytochrome c release in MCF-7 cells. However, inhibition of CerS with FB1 reduced caspase 3/7 activation and prevented plasma membrane permeabilization. A striking finding of the study was that, although the de novo synthesis inhibitor myriocin greatly depleted cellular ceramide levels, it had no effect on cell death. Conversely, inhibition of CerSs using FB1 inhibited ceramide generation, reduced caspase activation and prevented plasma membrane permeabilization. The only difference in ceramide generation between myriocin and FB1 was that myriocin was partially ineffective at inhibiting late ceramide generation, whereas FB1 produced a near-complete reduction in ceramide levels. Simultaneous knockdown of CerS5 and CerS6 (but not each CerS alone) also inhibited plasma membrane permeabilization. These data support a specific role for CerS5 and CerS6 (and their product C16:0-ceramide) in the regulation of post-mitochondrial cell death. Furthermore, the de novo pathway appears to be inconsequential in this mode of ceramide-mediated cell death; only sphingosine salvage through CerS5 and CerS6 was necessary for the post-mitochondrial effects. This lends further support to the hypothesis that C16:0-ceramide is an important regulator of cell death in some systems, but how CerS5/CerS6-derived ceramide mediates these effects is still unknown.
Although the studies described above fall short of coming to a unified consensus of ceramide's role in programmed cell death, an important theme is emerging. It is clear that CerSs and ceramide generation are intertwined with MOMP and its downstream events. More importantly, it appears that only a specific subset of ceramide is probably involved in the control of these events. This subset of ceramide may be defined by location (e.g. mitochondria-associated membranes), enzymes (e.g. CerS/CerS6), chain length (e.g. C16:0-ceramide) and/or biosynthetic pathway (de novo compared with salvage).
CerS as regulators of chemotherapeutic drug and radiation sensitivity in cancer
In addition to the studies mentioned above, several lines of evidence point to a role for CerSs in regulating the sensitivity to cancer chemotherapeutic agents and radiation. Min et al. [34] demonstrated that CerS1 overexpression in HEK-293 cells increased sensitivity to the chemotherapeutic agents cisplatin, carboplatin, doxorubicin and vincristine [34]. CerS5 overexpression increased sensitivity as well, but only to doxorubicin and vincristine. CerS4 expression, on the other hand, failed to significantly increase sensitivity to any of the drugs tested.
Similarly to CerS1 in HNSCC (head and neck squamous cell carcinoma) cells, CerS6 may be a key determinant in the sensitivity of colon carcinoma cells to TRAIL (TNF-related apoptosis-induced ligand), which was found to induce death selectively in cancer cells, but whose efficacy is limited by the presence of resistance mechanisms [73]. White-Gilbertson et al. [74] found decreased CerS6 expression in TRAIL-resistant SW620 cells compared with TRAIL-sensitive SW480 cells. Knockdown of CerS6 in SW480 cells promoted TRAIL resistance, whereas overexpression of CerS6 in SW620 cells restored TRAIL sensitivity.
Walker et al. [75] have described roles for CerS6 and aSMase (acid sphingomyelinase) in regulating cell death induced by the multi-kinase inhibitor sorafenib and the histone deacetylase inhibitor vorinostat in a variety of cancer cells. In this model, sorafenib and vorinostat signalled cell death via the CD95 death receptor, which was partially dependent on aSMase, CerS6 and de novo ceramide synthesis. In this same study, CerS6 overexpression sensitized SW620 cells to combined sorafenib and vorinostat. These data suggest that CerS6 is a regulator of the therapeutic efficacy of combined sorafenib and vorinostat treatment. The mechanism of this regulation remains to be determined.
CerS6 has also been identified as a mediator of cell death induced by MDA-7 (melanoma differentiation-associated gene-7)/IL-24 (interleukin-24), which is a cytokine that induces apoptosis in several cancer models but exhibits little or no toxicity towards normal cells [76]. In GBM6 glioblastoma multiforme cells, MDA-7/IL24 induced death that was dependent on ceramide, calcium release and ROS (reactive oxygen species). Knockdown of aSMase, knockdown of CerS6 or inhibition of de novo synthesis blocked cytosolic calcium elevations and blunted ROS generation. Ceramide generation was downstream of PERK (protein kinase R-like ER kinase), as cells expressing a dominant-negative form of this protein failed to induce ceramide generation. That study thus provides further evidence that components of the salvage pathway (e.g. aSMase and CerS) co-operate to produce pro-apoptotic ceramide.
In addition to identifying CerS activation in mitochondria-associated membranes, the study by Mesicek et al. [71] also showed that CerS2 and CerS5 could differentially regulate cell sensitivity to IR. Overexpression of CerS2 delayed IR-induced apoptosis in HeLa cells, whereas overexpression of CerS5 promoted it, suggesting that cell death is limited by CerS5-dependent ceramide production. Additional work by Panjarian et al. [50] found that the induction of p53 in Molt-4 cells by IR increased the expression of CerS5, but not CerS6, over 4–10 h following the irradiation. These studies highlight the emerging role of CerS5 and C16:0-ceramide as important components and mediators of the cell stress response.
CerS5 plays a role in sphingosine salvage pathway signalling and the response to cellular stress
Ceramide created through the sphingosine salvage pathway has been implicated in the regulation of several signalling pathways. The salvage pathway involves activation of a sphingomyelinase or glycosidase, leading to the generation of ceramide that is converted into free sphingosine, which is subsequently converted back into ceramide in another compartment via CerS. Although not often appreciated, the identification of the salvage pathway in sphingolipid signalling complicates the interpretation of some sphingolipid data. For example, one might observe activation of aSMase and the elevation of ceramide induced by a particular stimulus. The simple interpretation would be that the observed ceramide generation is from sphingomyelin hydrolysis. However, a more complex hypothesis should be entertained: the ceramide increase could be due to activation of the salvage pathway, leading to the production of free sphingosine which is in turn metabolized into ceramide via CerS in another compartment.
One of the first biological roles of the salvage pathway was in PKC signalling. Becker et al. [77] demonstrated that the sphingosine salvage pathway was regulated by PKC such that phorbol ester treatment caused an increase salvage-derived ceramide. This ceramide in turn regulated the translocation of PKCβII to a juxtanuclear compartment that the authors termed the ‘pericentrion’. Subsequent work by Kitatani et al. [78] showed that the salvage pathway was important in regulating phorbol-ester-induced phosphorylation and dephosphorylation of p38-MAPK. Knockdown of CerS5 recapitulated the enhanced phosphorylation of p38-MAPK, suggesting that CerS5 was the main CerS responsible for the effect.
In addition to its role in the PKC/p38-MAPK pathway, CerS5 may play an important role in stress-induced cell death. Jin et al. [51] found that hypoxia/re-oxygenation injury of NT-2 neuronal precursor cells caused aSMase activation and an increase in the mRNA levels of CerS5, and an increase in production of C14:0- and C16:0-ceramide from exogenous sphingosine (a surrogate marker of in vivo CerS activity). Knockdown of aSMase or CerS5 using siRNA (small interfering RNA) inhibited the acute increase in ceramide and partially protected the cells from Bax-mediated apoptosis. These data support a model whereby aSMase and CerS5 co-operate through the salvage pathway to produce long-chain ceramide and activate the mitochondrial cell death pathway. In another study by Jin et al. [52], the AMPK inhibitor Compound C was found to increase CerS5 mRNA levels in MCF-7 breast cancer cells. The increase in CerS5 expression was associated with C16:0-ceramide generation and translocation of Bax to the mitochondria [52].
CerS1 and head and neck cancer
Soon after the first mammalian CerSs were discovered, Ogretmen and co-workers [79] found that the ceramide profiles of head and neck tumours showed an interesting feature: tumours exhibited higher C16:0-ceramide and lower C18:0-ceramide levels than adjacent normal tissues. These ceramide profiles were also correlated with differences in CerS1 mRNA, suggesting that decreased CerS1 expression might be the cause of lower C18:0-ceramide in these tumours. Further experiments demonstrated a growth-inhibiting and pro-apoptotic effect of overexpression of CerS1 and production of C18:0-ceramide in HNSCC cells. The role of CerS1 has been extended to include an ability to control the sensitivity of HNSCC cells to combined gemcitabine/doxorubicin treatment [80]. Furthermore, lower C18:0-ceramide levels in human HNSCC samples may be associated with higher degrees of lymphovascular invasion and nodal metastasis [81]. Overall, these data suggest that CerS1 and C18:0-ceramide may be negative regulators of HNSCC cells.
CerS1 and neurodegeneration
Mutations in the mouse CerS1 gene have recently identified as the cause of a neurodegenerative phenotype in two mouse strains at the Jackson Laboratory [82]. These mice, termed flincher (fln) and toppler (to) because of their ataxic phenotypes, were found to have a frameshift and point mutation respectively that abrogated CerS1 catalytic function. Loss of CerS1 activity in the brains of fln mice was associated with decreased C18:0- and C18:1-ceramide and increased C16:0-ceramide and sphingoid base levels. Consistent with the high expression of CerS1 in Purkinje cells, these mice exhibited degeneration of the Purkinje cells and began to develop cerebellar ataxia approximately 3 weeks after birth. Furthermore, brains of fln mice accumulated lipofuscin, a lipid–protein deposit associated with neurodegeneration, more than wild-type mice. On the basis of these findings, CerS1 and its production of C18:0- and C18:1-sphingolipids are critical for maintaining neuronal function, particularly in the cerebellar Purkinje cells.
CerSs in ER stress and autophagy
CerS have been implicated in the regulation of the ER stress response in two different model systems. According to Spassieva et al. [83], down-regulation of CerS2 results in the up-regulation of long-chain sphingolipids and causes phosphorylation of eIF2α (eukaryotic initiation factor-2α) and up-regulation of CHOP (CCAAT/enhancer-binding protein-homologous protein), which are downstream effectors of the ER-stress-responsive kinase PERK. CerS2 knockdown also resulted in autophagy and cell-cycle arrest, but not apoptosis. These data suggest that disruption of sphingolipid metabolism can cause a complex cellular stress response, including ER stress and autophagy. It will be interesting to see whether ER stress and autophagy function to protect these cells from cell death.
In another study, CerS6 down-regulation produced ER stress that led to apoptosis. Senkal et al. [84] found that loss of CerS6 in HNSCC cells led to reduced C16:0-ceramide levels and the up-regulation of ATF6 (activating transcription factor 6) and CHOP, both mediators of the UPR (unfolded protein response). The up-regulation of CHOP was associated with a decrease in Bcl-2 expression and an increase in executioner caspase activity. That study demonstrates a cell-type-specific role for CerS6 in ER homoeostasis and supports the hypothesis that specific CerS may have unique roles in regulating cell fate in different cell types.
CerS and breast cancer
Several studies have examined the role of CerSs in the pathobiology of breast cancer. One study by Ruckhaberle et al. [85] found that EsR (oestrogen receptor)-negative tumours had an overall increase in SK1 (sphingosine kinase 1) expression and decreased CerS4 and CerS6 expression. The authors found that high SK1 expression predicted a decrease in survival. A subsequent study by the same group examined ceramide levels and CerS expression in more detail and found that both benign and malignant breast tumours had higher ceramide levels than normal tissue [86]. They also found that certain ceramides correlated with increased nodal metastases and/or expression of EsR. The authors also attempted to correlate CerS expression with ceramide levels and found approximate correlations of CerS2 expression with C16:0-ceramide and C24:1-ceramide, and CerS6 expression with C18:0-ceramide and C20:0-ceramide. These data are remarkable in that the only correlation that fits with the known substrate preferences for CerS is that between CerS2 and C24:1-ceramide.
A separate study by Futerman and co-workers [87] examined the expression of CerSs in paired healthy compared with cancerous breast tissues. These authors found increased CerS2 and CerS6 expression in cancerous compared with normal tissue; they also observed increased CerS4 levels in some tumours, but this was not common enough to reach statistical significance across the whole dataset. Although preliminary, these two studies indicate that CerS and the ceramides they produce may be dysregulated in breast cancers, and it is possible that they play an important role in regulating the biology of these diseases.
CerS2-knockout mice: liver and nervous system dysfunction
The generation of CerS2-knockout mice has provided an exquisite and exciting window into roles of CerSs in regulating sphingolipid metabolism and mammalian biology. CerS2-knockout mice were generated independently by two groups with each reporting similar sphingolipid and biological phenotypes [54,88,89]. According to these studies, CerS2-null mice are born at normal Mendelian ratios and no embryonic, prenatal or perinatal abnormalities have been reported besides a reduced body weight compared with wild-type animals [54,89].
Analysis of CerS2-null mice revealed major alterations in the sphingolipid composition of the several organs of these mice with specific attention paid to the effects on the liver, where CerS2 mRNA is in relatively high abundance [40]. As predicted, these organs contain only trace amounts of very-long-chain (C22-C24) sphingolipids. However, not all very-long-chain sphingolipids were absent, as the kidneys of CerS2-knockout mice still contained some hexosylceramides with chain lengths ≥22 carbons [88].
Both groups also found that C16:0-ceramide was elevated, but Imgrund et al. [88] found that total liver ceramide was decreased, whereas Pewzner-Jung et al. [54] showed that total ceramide levels were unchanged except at 7 days of age. CerS2-null livers also exhibit elevated C16:0-hexosylceramides and C16:0-sphingomyelin [54,88]. Pewnzer-Jung et al. [89] found large increases in sphingosine and dihydrosphingosine in CerS2-null livers compared with wild-type. Dihydrosphingosine 1-phosphate and S1P, however, were not significantly elevated.
The expression of several sphingolipid enzymes was also elevated in livers of CerS2-null mice [54]. CerS5 expression was increased, although C16:0-CerS activity remained the same as wild-type levels. Neutral sphingomyelinase-2 mRNA and activity were also up-regulated, as was glucosylceramide synthase activity. In addition, there were no significant changes in total glycerophospholipids or cholesterol levels in CerS2-null mice compared with wild-type animals, although there were slight differences in the acyl-chain composition of phosphatidylethanolamine.
The most striking phenotype of CerS2-null mice was the presence of hepatic abnormalities and eventually the mice developed hepatic adenomas, some of which progressed to hepatocellular carcinomas [88]. Proliferation and apoptosis were increased in these livers within the first few months of age, and progressive nodular hyperplasia was evident as early as 4 months in some mice, and livers from CerS2-null mice increased in mass relative to body weight after 6–12 months. According to Pewzner-Jung et al. [89], more than a quarter of animals older than 8 months were found to have hepatocellular carcinomas, most of which were well-differentiated. Tumours in CerS2-null mice generated by Imgrund et al. [88] also showed a loss of lobular and ultrastructural architecture. On the basis of the late onset of the tumours and presence of chronic hepatopathy, it is likely that CerS2 is not a tumour suppressor itself, but its loss causes long-term apoptosis and proliferation that promotes carcinogenesis [89].
Analysis of gene expression in livers from CerS2-null mice showed both down-regulation and up-regulation of several metabolic and cell signalling pathways compared with livers from wild-type mice [89]. Functional enrichment analysis of the up-regulated genes revealed several key regulators of the cell cycle such as p53 and p21WAF1/CIP1, and down-regulated genes included components of PPARα (peroxisome-proliferator-activated receptor α) signalling and fatty acid metabolism. How each of these alterations is related to the disruption of sphingolipid metabolism (e.g. loss of very-long-chain sphingolipids, elevated dihydrosphingosine) remains to be determined, but each metabolic change has its own sequelae.
In addition to the progressive hepatopathy, CerS2-null mice experienced multiple structural changes in the central and peripheral nervous systems [88]. Progressive cystic degeneration in both the white matter and internal granule layer IV was seen in 9-month-old CerS2-null mice. Consistent with loss of both white and grey matter, these mice exhibited difficulties with the initiation of motor activities, with also the possibility of peripheral nervous dysfunction.
CerS3 in testicular and keratinocyte function
CerS3 possesses many characteristics that make it unique among CerSs. It has a broad substrate specificity and a very limited tissue distribution (Figure 2). However, it is highly expressed in the testes and has been linked to sphingolipid production in these organs [23,48]. The germ cells of the testes contain sphingolipids with ultra long-chain (26–32 carbons) polyunsaturated (four to six double bonds) fatty acids [VLC-PUFAs (very-long-chain polyunsaturated fatty acids)]. During development, an increase in CerS3 expression is associated with the appearance of the VLCPUFA sphingolipids as well as differentiation and development of the germ cells [48]. Mice lacking germ cells failed to up-regulate CerS3 or produce VLC-PUFA-containing sphingolipids. That study suggests that CerS3 may play a key role in the regulation of testicular sphingolipids and male fertility. CerS3 also has a high expression in the skin and may play an important role in the synthesis of skin lipids [23,40] (Figure 2). Mizutani et al. [90] found that CerS3 expression was increased in keratinocytes undergoing differentiation and hypothesized that CerS3 may contribute to the formation of the epidermal permeability barrier. A more mechanistic understanding of CerS3 function in these tissues is awaited. Recent results from Jennemann et al. [90a] have demonstrated a critical role for CerS3 in epidermal ceramide production and epidermal permeability barrier.
SUMMARY AND PERSPECTIVE
CerSs clearly play a critical role in regulating the biosynthesis of ceramide, a class of lipids that have clear metabolic and biological importance in a variety of settings (Figure 5). From a metabolic perspective, these enzymes represent a critical node where a relatively small set of substrates (e.g. dihydrosphingosine, sphingosine or phytosphingosine) are transformed into a dazzling array of lipid species of varying acyl-CoA compositions. Through the generation of ceramide and dihydroceramide, these enzymes have the potential to modify all aspects of sphingolipid metabolism and signalling. Although it is experimentally convenient to focus only on the proximal products of these enzymes (i.e. ceramides), we must remind ourselves of the far-reaching metabolic consequences of changes in CerS activity. CerSs are not merely ceramide factories, they also control the levels of bioactive sphingoid bases and sphingoid base phosphates (e.g. S1P). Moreover, these enzymes serve to regulate the composition of complex sphingolipids. These issues also have significant implications for the development of CerSs as potential therapeutic targets.
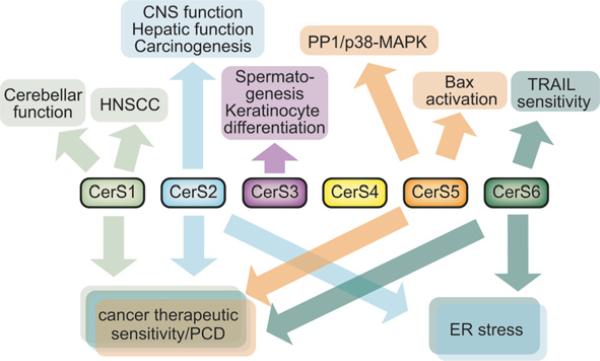
Figure 5. Multiple unique and shared biological roles of CerSs.
Through the generation of multiple different ceramide species, each CerS can potentially regulate a variety of biological phenomena. PCD, programmed cell death; PP1, protein phosphatase 1.
As more and more roles of particular sphingolipid species are described and our understanding of sphingolipids continues to transition from structural to function, questions arise as to the relative significance of individual lipids. Are there some sphingolipids that are structurally different but functionally redundant? For example, does sphingomyelin have acyl-chain-specific functions or is it merely a repository for C16:0-ceramide (and sphingosine)? Is ceramide the only chain-length-specific signalling molecule or do many sphingolipid species function in a chain-length-specific manner? Can CerSs function redundantly to maintain complex sphingolipid synthesis despite differential effects on acyl-chain length? We know from the CerS2-knockout mouse that very-long-chain sphingolipids are largely dispensable in some tissues for overall viability [54,88,89]. However, the development of hepatic and CNS (central nervous system) dysfunction later in adulthood supports an important role for this large subset of sphingolipids. An understanding of the specific contribution of ceramides, sphingomyelins and glycosphingolipids to the pathology in CerS2-knockout mice will probably take many years to elucidate. Data from additional CerS-knockout mice, particularly those implicated in cell signalling (e.g. CerS1, CerS5 and CerS6), will help to clarify the importance of these enzymes in both the regulation of sphingolipid levels and cellular function.
A major limitation in the field of CerS research is the lack of pharmacological inhibitors for individual CerS isoforms. FB1, although effective, has several drawbacks. First, it is a relatively expensive fungal product and requires micromolar concentrations for effective inhibition of CerS in vivo. Secondly, it has a complex mechanism of action (see the legend to Figure 4). Finally, it does not appear to be specific for any particular CerS. Until specific inhibitors of these proteins are developed, CerS-mediated metabolism and function will remain confined to cell culture models and knockout animals. CerS-specific inhibitory compounds would certainly accelerate the transition from the bench to pre-clinical and clinical studies.
Now is an exciting time for sphingolipid research and for the study of CerS in particular. Tremendous and significant gains in our understanding of CerS have been achieved in the past decade. However, as we have elaborated above, many important unanswered questions remain. With the right tools and judicious use of our intellectual (and monetary) capital, the study of CerS will probably transform these enzymes from biochemical curiosities into clinically relevant controllers of human biology.
Acknowledgments
FUNDING
This work is supported by the MUSC Department of Pharmaceutical Sciences Training Program in Environmental Stress Signaling [National Institutes of Health (NIH)/National Institute of Environmental Health Sciences (NIEHS)] [grant number 5T32ES012878] and a Ruth L. Kirschstein National Research Service Award (NIH/NIEHS) [grant number 1 F30 ES016975-01] (to T.D.M.), and was supported in part by the NIH [grant numbers R01AG016583, R01GM062887] and a MERIT Award by the Office of Research and Development, Department of Veterans Affairs, Ralph H. Johnson Veterans Affairs Medical Center (to L.M.O.), and by the NIH/National Cancer Institute (NCI) [grant number P01 CA097132 (to Y.A.H.)].
Abbreviations
- aSMase
acid sphingomyelinase
- CDase
ceramidase
- CerS
ceramide synthase
- CHOP
CCAAT/enhancer-binding protein-homologous protein
- CLN
neuronal ceroid lipofuscinosis
- ER
endoplasmic reticulum
- EsR
oestrogen receptor
- FB1
fumonisin B1
- fln
flincher
- GDF1
growth and differentiation factor-1
- HEK
human embryonic kidney
- HNSCC
head and neck squamous cell carcinoma
- Hox
homeobox
- IL-24
interleukin-24
- IR
ionizing radiation
- LAC1
longevity assurance gene cognate 1
- LAG1
longevity assurance gene 1
- MAPK
mitogen-activated protein kinase
- MDA-7
melanoma differentiation-associated gene-7
- MOMP
mitochondrial outer membrane permeabilization
- PERK
protein kinase R-like ER kinase
- PKC
protein kinase C
- ROS
reactive oxygen species
- S1P
sphingosine 1-phosphate
- siRNA
small interfering RNA
- SK
sphingosine kinase
- TNF
tumour necrosis factor
- TRAIL
TNF-related apoptosis-induced ligand
- TRAM
translocating chain-associated membrane protein
- TLC
TRAM, LAG1 and CLN8 homology
- UOG1
upstream of growth and differentiation factor-1
- UTR
untranslated region
- VLC-PUFA
very-long-chain polyunsaturated fatty acid
REFERENCES
- 1.Hannun YA, Obeid LM. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 2008;9:139–150. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- 2.Gatt S. Enzymic hydrolysis and synthesis of ceramides. J. Biol. Chem. 1963;238:3131–3133. [PubMed] [Google Scholar]
- 3.Gatt S. Enzymatic hydrolysis of sphingolipids. I. Hydrolysis and synthesis of ceramides by an enzyme from rat brain. J. Biol. Chem. 1966;241:3724–3730. [PubMed] [Google Scholar]
- 4.Yavin E, Gatt S. Enzymatic hydrolysis of sphingolipids. 8. Further purification and properties of rat brain ceramidase. Biochemistry. 1969;8:1692–1698. doi: 10.1021/bi00832a052. [DOI] [PubMed] [Google Scholar]
- 5.Sribney M. Enzymatic synthesis of ceramide. Biochim. Biophys. Acta. 1966;125:542–547. doi: 10.1016/0005-2760(66)90042-7. [DOI] [PubMed] [Google Scholar]
- 6.Morell P, Radin NS. Specificity in ceramide biosynthesis from long chain bases and various fatty acyl coenzyme A's by brain microsomes. J. Biol. Chem. 1970;245:342–350. [PubMed] [Google Scholar]
- 7.Morell P, Braun P. Biosynthesis and metabolic degradation of sphingolipids not containing sialic acid. J. Lipid Res. 1972;13:293–310. [PubMed] [Google Scholar]
- 8.Egilmez NK, Chen JB, Jazwinski SM. Specific alterations in transcript prevalence during the yeast life span. J. Biol. Chem. 1989;264:14312–14317. [PubMed] [Google Scholar]
- 9.D'Mello NP, Childress AM, Franklin DS, Kale SP, Pinswasdi C, Jazwinski SM. Cloning and characterization of LAG1, a longevity-assurance gene in yeast. J. Biol. Chem. 1994;269:15451–15459. [PubMed] [Google Scholar]
- 10.Guillas I, Kirchman PA, Chuard R, Pfefferli M, Jiang JC, Jazwinski SM, Conzelmann A. C26-CoA-dependent ceramide synthesis of Saccharomyces cerevisiae is operated by Lag1p and Lac1p. EMBO J. 2001;20:2655–2665. doi: 10.1093/emboj/20.11.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schorling S, Vallee B, Barz WP, Riezman H, Oesterhelt D. Lag1p and Lac1p are essential for the acyl-CoA-dependent ceramide synthase reaction in Saccharomyces cerevisae. Mol. Biol. Cell. 2001;12:3417–3427. doi: 10.1091/mbc.12.11.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang JC, Kirchman PA, Zagulski M, Hunt J, Jazwinski SM. Homologs of the yeast longevity gene LAG1 in Caenorhabditis elegans and human. Genome Res. 1998;8:1259–1272. doi: 10.1101/gr.8.12.1259. [DOI] [PubMed] [Google Scholar]
- 13.Barz WP, Walter P. Two endoplasmic reticulum (ER) membrane proteins that facilitate ER-to-Golgi transport of glycosylphosphatidylinositol-anchored proteins. Mol. Biol. Cell. 1999;10:1043–1059. doi: 10.1091/mbc.10.4.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winter E, Ponting CP. TRAM, LAG1 and CLN8: members of a novel family of lipid-sensing domains? Trends Biochem. Sci. 2002;27:381–383. doi: 10.1016/s0968-0004(02)02154-0. [DOI] [PubMed] [Google Scholar]
- 15.Jalanko A, Braulke T. Neuronal ceroid lipofuscinoses. Biochim. Biophys. Acta. 2009;1793:697–709. doi: 10.1016/j.bbamcr.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 16.Schulz A, Mousallem T, Venkataramani M, Persaud-Sawin DA, Zucker A, Luberto C, Bielawska A, Bielawski J, Holthuis JC, Jazwinski SM, et al. The CLN9 protein, a regulator of dihydroceramide synthase. J. Biol. Chem. 2006;281:2784–2794. doi: 10.1074/jbc.M509483200. [DOI] [PubMed] [Google Scholar]
- 17.Rusyn E, Mousallem T, Persaud-Sawin DA, Miller S, Boustany RM. CLN3p impacts galactosylceramide transport, raft morphology, and lipid content. Pediatr. Res. 2008;63:625–631. doi: 10.1203/PDR.0b013e31816fdc17. [DOI] [PubMed] [Google Scholar]
- 18.Lee SJ. Identification of a novel member (GDF-1) of the transforming growth factor-β superfamily. Mol. Endocrinol. 1990;4:1034–1040. doi: 10.1210/mend-4-7-1034. [DOI] [PubMed] [Google Scholar]
- 19.Lee SJ. Expression of growth/differentiation factor 1 in the nervous system: conservation of a bicistronic structure. Proc. Natl. Acad. Sci. U.S.A. 1991;88:4250–4254. doi: 10.1073/pnas.88.10.4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guillas I, Jiang JC, Vionnet C, Roubaty C, Uldry D, Chuard R, Wang J, Jazwinski SM, Conzelmann A. Human homologues of LAG1 reconstitute acyl-CoA-dependent ceramide synthesis in yeast. J. Biol. Chem. 2003;278:37083–37091. doi: 10.1074/jbc.M307554200. [DOI] [PubMed] [Google Scholar]
- 21.Venkataraman K, Riebeling C, Bodennec J, Riezman H, Allegood JC, Sullards MC, Merrill AH, Jr, Futerman AH. Upstream of growth and differentiation factor 1 (uog1), a mammalian homolog of the yeast longevity assurance gene 1 (LAG1), regulates N-stearoyl-sphinganine (C18-(dihydro)ceramide) synthesis in a fumonisin B1-independent manner in mammalian cells. J. Biol. Chem. 2002;277:35642–35649. doi: 10.1074/jbc.M205211200. [DOI] [PubMed] [Google Scholar]
- 22.Riebeling C, Allegood JC, Wang E, Merrill AH, Jr, Futerman AH. Two mammalian longevity assurance gene (LAG1) family members, trh1 and trh4, regulate dihydroceramide synthesis using different fatty acyl-CoA donors. J. Biol. Chem. 2003;278:43452–43459. doi: 10.1074/jbc.M307104200. [DOI] [PubMed] [Google Scholar]
- 23.Mizutani Y, Kihara A, Igarashi Y. LASS3 (longevity assurance homologue 3) is a mainly testis-specific (dihydro)ceramide synthase with relatively broad substrate specificity. Biochem. J. 2006;398:531–538. doi: 10.1042/BJ20060379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weinmann A, Galle PR, Teufel A. LASS6, an additional member of the longevity assurance gene family. Int. J. Mol. Med. 2005;16:905–910. [PubMed] [Google Scholar]
- 25.Lahiri S, Futerman AH. LASS5 is a bona fide dihydroceramide synthase that selectively utilizes palmitoyl CoA as acyl donor. J Biol Chem. 2005;280:33735–33738. doi: 10.1074/jbc.M506485200. [DOI] [PubMed] [Google Scholar]
- 26.Sridevi P, Alexander H, Laviad EL, Min J, Mesika A, Hannink M, Futerman AH, Alexander S. Stress-induced ER to Golgi translocation of ceramide synthase 1 is dependent on proteasomal processing. Exp. Cell Res. 2010;316:78–91. doi: 10.1016/j.yexcr.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mandon EC, Ehses I, Rother J, van Echten G, Sandhoff K. Subcellular localization and membrane topology of serine palmitoyltransferase, 3-dehydrosphinganine reductase, and sphinganine N-acyltransferase in mouse liver. J. Biol. Chem. 1992;267:11144–11148. [PubMed] [Google Scholar]
- 28.Hirschberg K, Rodger J, Futerman AH. The long-chain sphingoid base of sphingolipids is acylated at the cytosolic surface of the endoplasmic reticulum in rat liver. Biochem. J. 1993;290:751–757. doi: 10.1042/bj2900751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimeno H, Soeda S, Yasukouchi M, Okamura N, Nagamatsu A. Fatty acyl-Co A:sphingosine acyltransferase in bovine brain mitochondria: its solubilization and reconstitution onto the membrane lipid liposomes. Biol. Pharm. Bull. 1995;18:1335–1339. doi: 10.1248/bpb.18.1335. [DOI] [PubMed] [Google Scholar]
- 30.Shimeno H, Soeda S, Sakamoto M, Kouchi T, Kowakame T, Kihara T. Partial purification and characterization of sphingosine N-acyltransferase (ceramide synthase) from bovine liver mitochondrion-rich fraction. Lipids. 1998;33:601–605. doi: 10.1007/s11745-998-0246-2. [DOI] [PubMed] [Google Scholar]
- 31.Bionda C, Portoukalian J, Schmitt D, Rodriguez-Lafrasse C, Ardail D. Subcellular compartmentalization of ceramide metabolism: MAM (mitochondria-associated membrane) and/or mitochondria? Biochem. J. 2004;382:527–533. doi: 10.1042/BJ20031819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Novgorodov SA, Wu BX, Gudz TI, Bielawski J, Ovchinnikova TV, Hannun YA, Obeid LM. Novel pathway of ceramide production in mitochondria: thioesterase and neutral ceramidase produce ceramide from sphingosine and acyl-CoA. J. Biol. Chem. 2011;286:25352–25362. doi: 10.1074/jbc.M110.214866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mizutani Y, Kihara A, Igarashi Y. Mammalian Lass6 and its related family members regulate synthesis of specific ceramides. Biochem. J. 2005;390:263–271. doi: 10.1042/BJ20050291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Min J, Mesika A, Sivaguru M, Van Veldhoven PP, Alexander H, Futerman AH, Alexander S. (Dihydro)ceramide synthase 1 regulated sensitivity to cisplatin is associated with the activation of p38 mitogen-activated protein kinase and is abrogated by sphingosine kinase 1. Mol. Cancer Res. 2007;5:801–812. doi: 10.1158/1541-7786.MCR-07-0100. [DOI] [PubMed] [Google Scholar]
- 35.Jain E, Bairoch A, Duvaud S, Phan I, Redaschi N, Suzek BE, Martin MJ, McGarvey P, Gasteiger E. Infrastructure for the life sciences: design and implementation of the UniProt website. BMC Bioinformatics. 2009;10:136. doi: 10.1186/1471-2105-10-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Venkataraman K, Futerman AH. Do longevity assurance genes containing Hox domains regulate cell development via ceramide synthesis? FEBS Lett. 2002;528:3–4. doi: 10.1016/s0014-5793(02)03248-9. [DOI] [PubMed] [Google Scholar]
- 37.Mesika A, Ben-Dor S, Laviad EL, Futerman AH. A new functional motif in Hox domain-containing ceramide synthases: identification of a novel region flanking the Hox and TLC domains essential for activity. J. Biol. Chem. 2007;282:27366–27373. doi: 10.1074/jbc.M703487200. [DOI] [PubMed] [Google Scholar]
- 38.Spassieva S, Seo JG, Jiang JC, Bielawski J, Alvarez-Vasquez F, Jazwinski SM, Hannun YA, Obeid LM. Necessary role for the Lag1p motif in (dihydro)ceramide synthase activity. J. Biol. Chem. 2006;281:33931–33938. doi: 10.1074/jbc.M608092200. [DOI] [PubMed] [Google Scholar]
- 39.Sridevi P, Alexander H, Laviad EL, Pewzner-Jung Y, Hannink M, Futerman AH, Alexander S. Ceramide synthase 1 is regulated by proteasomal mediated turnover. Biochim. Biophys. Acta. 2009;1793:1218–1227. doi: 10.1016/j.bbamcr.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laviad EL, Albee L, Pankova-Kholmyansky I, Epstein S, Park H, Merrill AH, Jr, Futerman AH. Characterization of ceramide synthase 2: tissue distribution, substrate specificity, and inhibition by sphingosine 1-phosphate. J. Biol. Chem. 2008;283:5677–5684. doi: 10.1074/jbc.M707386200. [DOI] [PubMed] [Google Scholar]
- 41.Ullman MD, Radin NS. Enzymatic formation of hydroxy ceramides and comparison with enzymes forming nonhydroxy ceramides. Arch. Biochem. Biophys. 1972;152:767–777. doi: 10.1016/0003-9861(72)90272-x. [DOI] [PubMed] [Google Scholar]
- 42.Merrill AH, Jr, van Echten G, Wang E, Sandhoff K. Fumonisin B1 inhibits sphingosine (sphinganine) N-acyltransferase and de novo sphingolipid biosynthesis in cultured neurons in situ. J. Biol. Chem. 1993;268:27299–27306. [PubMed] [Google Scholar]
- 43.Lahiri S, Lee H, Mesicek J, Fuks Z, Haimovitz-Friedman A, Kolesnick RN, Futerman AH. Kinetic characterization of mammalian ceramide synthases: determination of Km values towards sphinganine. FEBS Lett. 2007;581:5289–5294. doi: 10.1016/j.febslet.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 44.Nikolova-Karakashian MN, Wales TR, Wang E, Menaldino DS, Goh J, Liotta DC, Merrill AH., Jr . Ceramide synthase and ceramidases in the regulation of sphingoid base metabolism. In: Hannun YA, editor. Sphingolipid-Mediated Signal Transduction. R. G. Landes; Georgetown, TX: 1997. pp. 159–172. [Google Scholar]
- 45.Venkataraman K, Futerman AH. Comparison of the metabolism of l-erythro- and l-threo-sphinganines and ceramides in cultured cells and in subcellular fractions. Biochim. Biophys. Acta. 2001;1530:219–226. doi: 10.1016/s1388-1981(01)00085-3. [DOI] [PubMed] [Google Scholar]
- 46.Stoffel W, Bister K. Stereospecificities in the metabolic reactions of the four isomeric sphinganines (dihydrosphingosines) in rat liver. Hoppe–Seylers Z. Physiol. Chem. 1973;354:169–181. doi: 10.1515/bchm2.1973.354.1.169. [DOI] [PubMed] [Google Scholar]
- 47.Becker I, Wang-Eckhardt L, Yaghootfam A, Gieselmann V, Eckhardt M. Differential expression of (dihydro)ceramide synthases in mouse brain: oligodendrocyte-specific expression of CerS2/Lass2. Histochem. Cell. Biol. 2008;129:233–241. doi: 10.1007/s00418-007-0344-0. [DOI] [PubMed] [Google Scholar]
- 48.Rabionet M, van der Spoel AC, Chuang CC, von Tumpling-Radosta B, Litjens M, Bouwmeester D, Hellbusch CC, Korner C, Wiegandt H, Gorgas K, et al. Male germ cells require polyenoic sphingolipids with complex glycosylation for completion of meiosis: a link to ceramide synthase-3. J. Biol. Chem. 2008;283:13357–13369. doi: 10.1074/jbc.M800870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mizutani Y, Mitsutake S, Tsuji K, Kihara A, Igarashi Y. Ceramide biosynthesis in keratinocyte and its role in skin function. Biochimie. 2009;91:784–790. doi: 10.1016/j.biochi.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 50.Panjarian S, Kozhaya L, Arayssi S, Yehia M, Bielawski J, Bielawska A, Usta J, Hannun YA, Obeid LM, Dbaibo GS. De novo N-palmitoylsphingosine synthesis is the major biochemical mechanism of ceramide accumulation following p53 up-regulation. Prostaglandins Other Lipid Mediat. 2008;86:41–48. doi: 10.1016/j.prostaglandins.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 51.Jin J, Hou Q, Mullen TD, Zeidan YH, Bielawski J, Kraveka JM, Bielawska A, Obeid LM, Hannun YA, Hsu YT. Ceramide generated by sphingomyelin hydrolysis and the salvage pathway is involved in hypoxia/reoxygenation-induced Bax redistribution to mitochondria in NT-2 cells. J. Biol. Chem. 2008;283:26509–26517. doi: 10.1074/jbc.M801597200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jin J, Mullen TD, Hou Q, Bielawski J, Bielawska A, Zhang X, Obeid LM, Hannun YA, Hsu YT. AMPK inhibitor Compound C stimulates ceramide production and promotes Bax redistribution and apoptosis in MCF7 breast carcinoma cells. J. Lipid Res. 2009;50:2389–2397. doi: 10.1194/jlr.M900119-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gustafsson K, Sander B, Bielawski J, Hannun YA, Flygare J. Potentiation of cannabinoid-induced cytotoxicity in mantle cell lymphoma through modulation of ceramide metabolism. Mol. Cancer Res. 2009;7:1086–1098. doi: 10.1158/1541-7786.MCR-08-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pewzner-Jung Y, Park H, Laviad EL, Silva LC, Lahiri S, Stiban J, Erez-Roman R, Brugger B, Sachsenheimer T, Wieland F, et al. A critical role for ceramide synthase 2 in liver homeostasis: I. Alterations in lipid metabolic pathways. J. Biol. Chem. 2010;285:10902–10910. doi: 10.1074/jbc.M109.077594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bonzon-Kulichenko E, Schwudke D, Gallardo N, Molto E, Fernandez-Agullo T, Shevchenko A, Andres A. Central leptin regulates total ceramide content and sterol regulatory element binding protein-1C proteolytic maturation in rat white adipose tissue. Endocrinology. 2009;150:169–178. doi: 10.1210/en.2008-0505. [DOI] [PubMed] [Google Scholar]
- 56.Uchida Y, Behne M, Quiec D, Elias PM, Holleran WM. Vitamin C stimulates sphingolipid production and markers of barrier formation in submerged human keratinocyte cultures. J. Invest. Dermatol. 2001;117:1307–1313. doi: 10.1046/j.0022-202x.2001.01555.x. [DOI] [PubMed] [Google Scholar]
- 57.Jensen JM, Forl M, Winoto-Morbach S, Seite S, Schunck M, Proksch E, Schutze S. Acid and neutral sphingomyelinase, ceramide synthase, and acid ceramidase activities in cutaneous aging. Exp. Dermatol. 2005;14:609–618. doi: 10.1111/j.0906-6705.2005.00342.x. [DOI] [PubMed] [Google Scholar]
- 58.Gillard BK, Clement RG, Marcus DM. Variations among cell lines in the synthesis of sphingolipids in de novo and recycling pathways. Glycobiology. 1998;8:885–890. doi: 10.1093/glycob/8.9.885. [DOI] [PubMed] [Google Scholar]
- 59.Smith ER, Merrill AH., Jr Differential roles of de novo sphingolipid biosynthesis and turnover in the “burst” of free sphingosine and sphinganine, and their 1-phosphates and N-acyl-derivatives, that occurs upon changing the medium of cells in culture. J. Biol. Chem. 1995;270:18749–18758. doi: 10.1074/jbc.270.32.18749. [DOI] [PubMed] [Google Scholar]
- 60.Bose R, Verheij M, Haimovitz-Friedman A, Scotto K, Fuks Z, Kolesnick R. Ceramide synthase mediates daunorubicin-induced apoptosis: an alternative mechanism for generating death signals. Cell. 1995;82:405–414. doi: 10.1016/0092-8674(95)90429-8. [DOI] [PubMed] [Google Scholar]
- 61.Mullen TD, Obeid LM. Ceramide and apoptosis: exploring the enigmatic connections between sphingolipid metabolism and programmed cell death. Anticancer Agents Med. Chem. 2011 doi: 10.2174/187152012800228661. the press. [DOI] [PubMed] [Google Scholar]
- 62.Siskind LJ, Mullen TD, Rosales KR, Clarke CJ, Hernandez-Corbacho MJ, Edinger AL, Obeid LM. The BCL-2 protein BAK is required for long-chain ceramide generation during apoptosis. J. Biol. Chem. 2010;285:11818–11826. doi: 10.1074/jbc.M109.078121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Colombini M. Ceramide channels and their role in mitochondria-mediated apoptosis. Biochim. Biophys. Acta. 2010;1797:1239–1244. doi: 10.1016/j.bbabio.2010.01.021. [DOI] [PubMed] [Google Scholar]
- 64.Siskind LJ, Colombini M. The lipids C2- and C16-ceramide form large stable channels. Implications for apoptosis. J. Biol. Chem. 2000;275:38640–38644. doi: 10.1074/jbc.C000587200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Siskind LJ, Kolesnick RN, Colombini M. Ceramide channels increase the permeability of the mitochondrial outer membrane to small proteins. J. Biol. Chem. 2002;277:26796–26803. doi: 10.1074/jbc.M200754200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Siskind LJ, Kolesnick RN, Colombini M. Ceramide forms channels in mitochondrial outer membranes at physiologically relevant concentrations. Mitochondrion. 2006;6:118–125. doi: 10.1016/j.mito.2006.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Samanta S, Stiban J, Maugel TK, Colombini M. Visualization of ceramide channels by transmission electron microscopy. Biochim. Biophys. Acta. 2011;1808:1196–1201. doi: 10.1016/j.bbamem.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Siskind LJ, Feinstein L, Yu T, Davis JS, Jones D, Choi J, Zuckerman JE, Tan W, Hill RB, Hardwick JM, Colombini M. Anti-apoptotic Bcl-2 family proteins disassemble ceramide channels. J. Biol. Chem. 2008;283:6622–6630. doi: 10.1074/jbc.M706115200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ganesan V, Perera MN, Colombini D, Datskovskiy D, Chadha K, Colombini M. Ceramide and activated Bax act synergistically to permeabilize the mitochondrial outer membrane. Apoptosis. 2010;15:553–562. doi: 10.1007/s10495-009-0449-0. [DOI] [PubMed] [Google Scholar]
- 70.Lee H, Rotolo JA, Mesicek J, Penate-Medina T, Rimner A, Liao WC, Yin X, Ragupathi G, Ehleiter D, Gulbins E, et al. Mitochondrial ceramide-rich macrodomains functionalize bax upon irradiation. PLoS ONE. 2011;6:e19783. doi: 10.1371/journal.pone.0019783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mesicek J, Lee H, Feldman T, Jiang X, Skobeleva A, Berdyshev EV, Haimovitz-Friedman A, Fuks Z, Kolesnick R. Ceramide synthases 2, 5, and 6 confer distinct roles in radiation-induced apoptosis in HeLa cells. Cell. Signalling. 2010;22:1300–1307. doi: 10.1016/j.cellsig.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mullen TD, Jenkins RW, Clarke CJ, Bielawski J, Hannun YA, Obeid LM. Ceramide synthase-dependent ceramide generation and programmed cell death: involvement of salvage pathway in regulating postmitochondrial events. J. Biol. Chem. 2011;286:15929–15942. doi: 10.1074/jbc.M111.230870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thorburn A, Behbakht K, Ford H. TRAIL receptor-targeted therapeutics: resistance mechanisms and strategies to avoid them. Drug Resist. Update. 2008;11:17–24. doi: 10.1016/j.drup.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.White-Gilbertson S, Mullen T, Senkal C, Lu P, Ogretmen B, Obeid L, Voelkel-Johnson C. Ceramide synthase 6 modulates TRAIL sensitivity and nuclear translocation of active caspase-3 in colon cancer cells. Oncogene. 2009;28:1132–1141. doi: 10.1038/onc.2008.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Walker T, Mitchell C, Park MA, Yacoub A, Graf M, Rahmani M, Houghton PJ, Voelkel-Johnson C, Grant S, Dent P. Sorafenib and vorinostat kill colon cancer cells by CD95-dependent and -independent mechanisms. Mol. Pharmacol. 2009;76:342–355. doi: 10.1124/mol.109.056523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lebedeva IV, Emdad L, Su ZZ, Gupta P, Sauane M, Sarkar D, Staudt MR, Liu SJ, Taher MM, Xiao R, et al. mda-7/IL-24, novel anticancer cytokine: focus on bystander antitumor, radiosensitization and antiangiogenic properties and overview of the phase I clinical experience. Int. J. Oncol. 2007;31:985–1007. [PubMed] [Google Scholar]
- 77.Becker KP, Kitatani K, Idkowiak-Baldys J, Bielawski J, Hannun YA. Selective inhibition of juxtanuclear translocation of protein kinase CβII by a negative feedback mechanism involving ceramide formed from the salvage pathway. J. Biol. Chem. 2005;280:2606–2612. doi: 10.1074/jbc.M409066200. [DOI] [PubMed] [Google Scholar]
- 78.Kitatani K, Idkowiak-Baldys J, Bielawski J, Taha TA, Jenkins RW, Senkal CE, Ogretmen B, Obeid LM, Hannun YA. Protein kinase C-induced activation of a ceramide/protein phosphatase 1 pathway leading to dephosphorylation of p38 MAPK. J. Biol. Chem. 2006;281:36793–36802. doi: 10.1074/jbc.M608137200. [DOI] [PubMed] [Google Scholar]
- 79.Koybasi S, Senkal CE, Sundararaj K, Spassieva S, Bielawski J, Osta W, Day TA, Jiang JC, Jazwinski SM, Hannun YA, et al. Defects in cell growth regulation by C18:0-ceramide and longevity assurance gene 1 in human head and neck squamous cell carcinomas. J. Biol. Chem. 2004;279:44311–44319. doi: 10.1074/jbc.M406920200. [DOI] [PubMed] [Google Scholar]
- 80.Senkal CE, Ponnusamy S, Rossi MJ, Bialewski J, Sinha D, Jiang JC, Jazwinski SM, Hannun YA, Ogretmen B. Role of human longevity assurance gene 1 and C18-ceramide in chemotherapy-induced cell death in human head and neck squamous cell carcinomas. Mol. Cancer Ther. 2007;6:712–722. doi: 10.1158/1535-7163.MCT-06-0558. [DOI] [PubMed] [Google Scholar]
- 81.Karahatay S, Thomas K, Koybasi S, Senkal CE, Elojeimy S, Liu X, Bielawski J, Day TA, Gillespie MB, Sinha D, et al. Clinical relevance of ceramide metabolism in the pathogenesis of human head and neck squamous cell carcinoma (HNSCC): attenuation of C18-ceramide in HNSCC tumors correlates with lymphovascular invasion and nodal metastasis. Cancer Lett. 2007;256:101–111. doi: 10.1016/j.canlet.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhao L, Spassieva SD, Jucius TJ, Shultz LD, Shick HE, Macklin WB, Hannun YA, Obeid LM, Ackerman SL. A deficiency of ceramide biosynthesis causes cerebellar purkinje cell neurodegeneration and lipofuscin accumulation. PLoS Genet. 2011;7:e1002063. doi: 10.1371/journal.pgen.1002063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Spassieva SD, Mullen TD, Townsend DM, Obeid LM. Disruption of ceramide synthesis by CerS2 down-regulation leads to autophagy and the unfolded protein response. Biochem. J. 2009;424:273–283. doi: 10.1042/BJ20090699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Senkal CE, Ponnusamy S, Bielawski J, Hannun YA, Ogretmen B. Antiapoptotic roles of ceramide-synthase-6-generated C16-ceramide via selective regulation of the ATF6/CHOP arm of ER-stress-response pathways. FASEB J. 2010;24:296–308. doi: 10.1096/fj.09-135087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ruckhaberle E, Rody A, Engels K, Gaetje R, von Minckwitz G, Schiffmann S, Grosch S, Geisslinger G, Holtrich U, Karn T, Kaufmann M. Microarray analysis of altered sphingolipid metabolism reveals prognostic significance of sphingosine kinase 1 in breast cancer. Breast Cancer Res. Treat. 2008;112:41–52. doi: 10.1007/s10549-007-9836-9. [DOI] [PubMed] [Google Scholar]
- 86.Schiffmann S, Sandner J, Birod K, Wobst I, Angioni C, Ruckhaberle E, Kaufmann M, Ackermann H, Lotsch J, Schmidt H, et al. Ceramide synthases and ceramide levels are increased in breast cancer tissue. Carcinogenesis. 2009;30:745–752. doi: 10.1093/carcin/bgp061. [DOI] [PubMed] [Google Scholar]
- 87.Erez-Roman R, Pienik R, Futerman AH. Increased ceramide synthase 2 and 6 mRNA levels in breast cancer tissues and correlation with sphingosine kinase expression. Biochem. Biophys. Res. Commun. 2010;391:219–223. doi: 10.1016/j.bbrc.2009.11.035. [DOI] [PubMed] [Google Scholar]
- 88.Imgrund S, Hartmann D, Farwanah H, Eckhardt M, Sandhoff R, Degen J, Gieselmann V, Sandhoff K, Willecke K. Adult ceramide synthase 2 (CERS2)-deficient mice exhibit myelin sheath defects, cerebellar degeneration, and hepatocarcinomas. J. Biol. Chem. 2009;284:33549–33560. doi: 10.1074/jbc.M109.031971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pewzner-Jung Y, Brenner O, Braun S, Laviad EL, Ben-Dor S, Feldmesser E, Horn-Saban S, Amann-Zalcenstein D, Raanan C, Berkutzki T, et al. A critical role for ceramide synthase 2 in liver homeostasis: II. Insights into molecular changes leading to hepatopathy. J. Biol. Chem. 2010;285:10911–10923. doi: 10.1074/jbc.M109.077610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mizutani Y, Kihara A, Chiba H, Tojo H, Igarashi Y. 2-Hydroxy-ceramide synthesis by ceramide synthase family: enzymatic basis for the preference of FA chain length. J. Lipid Res. 2008;49:2356–2364. doi: 10.1194/jlr.M800158-JLR200. [DOI] [PubMed] [Google Scholar]
- 90a.Jennemann R, Rabionet M, Gorgas K, Epstein S, Dalpke A, Rothermel U, Bayerle A, vander Hoeven F, Imgrund S, Kirsch J, et al. Loss of ceramide synthase 3 causes lethal skin barrier disruption. Hum. Mol. Genet. 2011 doi: 10.1093/hmg/ddr494. doi:10.1093/hmg/ddr494. [DOI] [PubMed] [Google Scholar]
- 91.UniProt Consortium The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mullen TD, Spassieva S, Jenkins RW, Kitatani K, Bielawski J, Hannun YA, Obeid LM. Selective knockdown of ceramide synthases reveals complex interregulation of sphingolipid metabolism. J. Lipid Res. 2011;52:68–77. doi: 10.1194/jlr.M009142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Baran Y, Salas A, Senkal CE, Gunduz U, Bielawski J, Obeid LM, Ogretmen B. Alterations of ceramide/sphingosine-1-phosphate rheostat involve in the regulation of resistance to imatinib-induced apoptosis in K562 human chronic myeloid leukemia (CML) cells. J. Biol. Chem. 2007;282:10922–10934. doi: 10.1074/jbc.M610157200. [DOI] [PubMed] [Google Scholar]
- 94.Wooten-Blanks LG, Song P, Senkal CE, Ogretmen B. Mechanisms of ceramide-mediated repression of the human telomerase reverse transcriptase promoter via deacetylation of Sp3 by histone deacetylase 1. FASEB J. 2007;21:3386–3397. doi: 10.1096/fj.07-8621com. [DOI] [PubMed] [Google Scholar]
- 95.Jin J, Mullen TD, Hou Q, Bielawski J, Bielawska A, Zhang X, Obeid LM, Hannun YA, Hsu YT. AMPK inhibitor compound C stimulates ceramide production and promotes Bax redistribution and apoptosis in MCF7 breast carcinoma cells. J. Lipid Res. 2009;50:2389–2397. doi: 10.1194/jlr.M900119-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Reference deleted
- 97.Rath G, Schneider C, Langlois B, Sartelet H, Morjani H, Btaouri HE, Dedieu S, Martiny L. De novo ceramide synthesis is responsible for the anti-tumor properties of camptothecin and doxorubicin in follicular thyroid carcinoma. Int. J. Biochem. Cell Biol. 2009;41:1165–1172. doi: 10.1016/j.biocel.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 98.Bourteele S, Hausser A, Doppler H, Horn-Muller J, Ropke C, Schwarzmann G, Pfizenmaier K, Muller G. Tumor necrosis factor induces ceramide oscillations and negatively controls sphingolipid synthases by caspases in apoptotic Kym-1 cells. J. Biol. Chem. 1998;273:31245–31251. doi: 10.1074/jbc.273.47.31245. [DOI] [PubMed] [Google Scholar]
- 99.Wang H, Maurer BJ, Reynolds CP, Cabot MC. N-(4-hydroxyphenyl) retinamide elevates ceramide in neuroblastoma cell lines by coordinate activation of serine palmitoyltransferase and ceramide synthase. Cancer Res. 2001;61:5102–5105. [PubMed] [Google Scholar]
- 100.Uchida Y, Nardo AD, Collins V, Elias PM, Holleran WM. De novo ceramide synthesis participates in the ultraviolet B irradiation-induced apoptosis in undifferentiated cultured human keratinocytes. J. Invest. Dermatol. 2003;120:662–669. doi: 10.1046/j.1523-1747.2003.12098.x. [DOI] [PubMed] [Google Scholar]
- 101.Petrache I, Natarajan V, Zhen L, Medler TR, Richter AT, Cho C, Hubbard WC, Berdyshev EV, Tuder RM. Ceramide upregulation causes pulmonary cell apoptosis and emphysema-like disease in mice. Nat. Med. 2005;11:491–498. doi: 10.1038/nm1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liao WC, Haimovitz-Friedman A, Persaud RS, McLoughlin M, Ehleiter D, Zhang N, Gatei M, Lavin M, Kolesnick R, Fuks Z. Ataxia telangiectasia-mutated gene product inhibits DNA damage-induced apoptosis via ceramide synthase. J. Biol. Chem. 1999;274:17908–17917. doi: 10.1074/jbc.274.25.17908. [DOI] [PubMed] [Google Scholar]
- 103.Truman JP, Gueven N, Lavin M, Leibel S, Kolesnick R, Fuks Z, Haimovitz-Friedman A. Down-regulation of ATM protein sensitizes human prostate cancer cells to radiation-induced apoptosis. J. Biol. Chem. 2005;280:23262–23272. doi: 10.1074/jbc.M503701200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Garzotto M, White-Jones M, Jiang Y, Ehleiter D, Liao WC, Haimovitz-Friedman A, Fuks Z, Kolesnick R. 12-O-tetradecanoylphorbol-13-acetate-induced apoptosis in LNCaP cells is mediated through ceramide synthase. Cancer Res. 1998;58:2260–2264. [PubMed] [Google Scholar]
- 105.Ch'ang HJ, Maj JG, Paris F, Xing HR, Zhang J, Truman JP, Cardon-Cardo C, Haimovitz-Friedman A, Kolesnick R, Fuks Z. ATM regulates target switching to escalating doses of radiation in the intestines. Nat. Med. 2005;11:484–490. doi: 10.1038/nm1237. [DOI] [PubMed] [Google Scholar]
- 106.Garzotto M, Haimovitz-Friedman A, Liao WC, White-Jones M, Huryk R, Heston WD, Cardon-Cardo C, Kolesnick R, Fuks Z. Reversal of radiation resistance in LNCaP cells by targeting apoptosis through ceramide synthase. Cancer Res. 1999;59:5194–5201. [PubMed] [Google Scholar]
- 107.Dbaibo GS, El-Assaad W, Krikorian A, Liu B, Diab K, Idriss NZ, El-Sabban M, Driscoll TA, Perry DK, Hannun YA. Ceramide generation by two distinct pathways in tumor necrosis factor α-induced cell death. FEBS Lett. 2001;503:7–12. doi: 10.1016/s0014-5793(01)02625-4. [DOI] [PubMed] [Google Scholar]
- 108.Laouar A, Glesne D, Huberman E. Involvement of protein kinase C-β and ceramide in tumor necrosis factor-α-induced but not Fas-induced apoptosis of human myeloid leukemia cells. J. Biol. Chem. 1999;274:23526–23534. doi: 10.1074/jbc.274.33.23526. [DOI] [PubMed] [Google Scholar]
- 109.Xu J, Yeh CH, Chen S, He L, Sensi SL, Canzoniero LM, Choi DW, Hsu CY. Involvement of de novo ceramide biosynthesis in tumor necrosis factor-α/cycloheximide-induced cerebral endothelial cell death. J. Biol. Chem. 1998;273:16521–16526. doi: 10.1074/jbc.273.26.16521. [DOI] [PubMed] [Google Scholar]
- 110.Kroesen BJ, Pettus B, Luberto C, Busman M, Sietsma H, de Leij L, Hannun YA. Induction of apoptosis through B-cell receptor cross-linking occurs via de novo generated C16-ceramide and involves mitochondria. J. Biol. Chem. 2001;276:13606–13614. doi: 10.1074/jbc.M009517200. [DOI] [PubMed] [Google Scholar]
- 111.Tonnetti L, Veri MC, Bonvini E, D'Adamio L. A role for neutral sphingomyelinase-mediated ceramide production in T cell receptor-induced apoptosis and mitogen-activated protein kinase-mediated signal transduction. J. Exp. Med. 1999;189:1581–1589. doi: 10.1084/jem.189.10.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Suzuki A, Iwasaki M, Kato M, Wagai N. Sequential operation of ceramide synthesis and ICE cascade in CPT-11-initiated apoptotic death signaling. Exp. Cell Res. 1997;233:41–47. doi: 10.1006/excr.1997.3498. [DOI] [PubMed] [Google Scholar]
- 113.Cuvillier O, Nava VE, Murthy SK, Edsall LC, Levade T, Milstien S, Spiegel S. Sphingosine generation, cytochrome c release, and activation of caspase-7 in doxorubicin-induced apoptosis of MCF7 breast adenocarcinoma cells. Cell Death Differ. 2001;8:162–171. doi: 10.1038/sj.cdd.4400793. [DOI] [PubMed] [Google Scholar]
- 114.Mercier C, Decleves X, Masseguin C, Fragner P, Tardy M, Roux F, Gabrion J, Scherrmann JM. P-glycoprotein (ABCB1) but not multidrug resistance-associated protein 1 (ABCC1) is induced by doxorubicin in primary cultures of rat astrocytes. J. Neurochem. 2003;87:820–830. doi: 10.1046/j.1471-4159.2003.02034.x. [DOI] [PubMed] [Google Scholar]
- 115.Witty JP, Bridgham JT, Johnson AL. Induction of apoptotic cell death in hen granulosa cells by ceramide. Endocrinology. 1996;137:5269–5277. doi: 10.1210/endo.137.12.8940345. [DOI] [PubMed] [Google Scholar]
- 116.Perry DK, Carton J, Shah AK, Meredith F, Uhlinger DJ, Hannun YA. Serine palmitoyltransferase regulates de novo ceramide generation during etoposide-induced apoptosis. J. Biol. Chem. 2000;275:9078–9084. doi: 10.1074/jbc.275.12.9078. [DOI] [PubMed] [Google Scholar]
- 117.DiPietrantonio AM, Hsieh TC, Olson SC, Wu JM. Regulation of G1/S transition and induction of apoptosis in HL-60 leukemia cells by fenretinide (4HPR). Int. J. Cancer. 1998;78:53–61. doi: 10.1002/(sici)1097-0215(19980925)78:1<53::aid-ijc10>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 118.Erdreich-Epstein A, Tran LB, Bowman NN, Wang H, Cabot MC, Durden DL, Vlckova J, Reynolds CP, Stins MF, Groshen S, Millard M. Ceramide signaling in fenretinide-induced endothelial cell apoptosis. J. Biol. Chem. 2002;277:49531–49537. doi: 10.1074/jbc.M209962200. [DOI] [PubMed] [Google Scholar]
- 119.Biswal SS, Datta K, Acquaah-Mensah GK, Kehrer JP. Changes in ceramide and sphingomyelin following fludarabine treatment of human chronic B-cell leukemia cells. Toxicology. 2000;154:45–53. doi: 10.1016/s0300-483x(00)00296-1. [DOI] [PubMed] [Google Scholar]
- 120.Sumitomo M, Ohba M, Asakuma J, Asano T, Kuroki T, Hayakawa M. Protein kinase Cδ amplifies ceramide formation via mitochondrial signaling in prostate cancer cells. J. Clin. Invest. 2002;109:827–836. doi: 10.1172/JCI14146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kawatani M, Simizu S, Osada H, Takada M, Arber N, Imoto M. Involvement of protein kinase C-regulated ceramide generation in inostamycin-induced apoptosis. Exp. Cell Res. 2000;259:389–397. doi: 10.1006/excr.2000.4986. [DOI] [PubMed] [Google Scholar]
- 122.Nemoto S, Nakamura M, Osawa Y, Kono S, Itoh Y, Okano Y, Murate T, Hara A, Ueda H, Nozawa Y, Banno Y. Sphingosine kinase isoforms regulate oxaliplatin sensitivity of human colon cancer cells through ceramide accumulation and Akt activation. J. Biol. Chem. 2009;284:10422–10432. doi: 10.1074/jbc.M900735200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Vit JP, Rosselli F. Role of the ceramide-signaling pathways in ionizing radiation-induced apoptosis. Oncogene. 2003;22:8645–8652. doi: 10.1038/sj.onc.1207087. [DOI] [PubMed] [Google Scholar]
- 124.Mandala SM, Harris GH. Isolation and characterization of novel inhibitors of sphingolipid synthesis: australifungin, viridiofungins, rustmicin, and khafrefungin. Methods Enzymol. 2000;311:335–348. doi: 10.1016/s0076-6879(00)11094-8. [DOI] [PubMed] [Google Scholar]
- 125.Wang E, Norred WP, Bacon CW, Riley RT, Merrill AH., Jr Inhibition of sphingolipid biosynthesis by fumonisins. Implications for diseases associated with Fusarium moniliforme. J. Biol. Chem. 1991;266:14486–14490. [PubMed] [Google Scholar]
- 126.Schmelz EM, Dombrink-Kurtzman MA, Roberts PC, Kozutsumi Y, Kawasaki T, Merrill AH., Jr Induction of apoptosis by fumonisin B1 in HT29 cells is mediated by the accumulation of endogenous free sphingoid bases. Toxicol. Appl. Pharmacol. 1998;148:252–260. doi: 10.1006/taap.1997.8356. [DOI] [PubMed] [Google Scholar]
- 127.Humpf HU, Schmelz EM, Meredith FI, Vesper H, Vales TR, Wang E, Menaldino DS, Liotta DC, Merrill AH., Jr Acylation of naturally occurring and synthetic 1-deoxysphinganines by ceramide synthase. Formation of N-palmitoyl-aminopentol produces a toxic metabolite of hydrolyzed fumonisin, AP1, and a new category of ceramide synthase inhibitor. J. Biol. Chem. 1998;273:19060–19064. doi: 10.1074/jbc.273.30.19060. [DOI] [PubMed] [Google Scholar]
- 128.Seiferlein M, Humpf HU, Voss KA, Sullards MC, Allegood JC, Wang E, Merrill AH., Jr Hydrolyzed fumonisins HFB1 and HFB2 are acylated in vitro and in vivo by ceramide synthase to form cytotoxic N-acyl-metabolites. Mol. Nutr. Food Res. 2007;51:1120–1130. doi: 10.1002/mnfr.200700118. [DOI] [PubMed] [Google Scholar]
- 129.Merrill AH, Jr, Wang E, Gilchrist DG, Riley RT. Fumonisins and other inhibitors of de novo sphingolipid biosynthesis. Adv. Lipid Res. 1993;26:215–234. [PubMed] [Google Scholar]
- 130.Abbas HK, Tanaka T, Duke SO, Porter JK, Wray EM, Hodges L, Sessions AE, Wang E, Merrill AH, Jr, Riley RT. Fumonisin- and AAL-toxin-induced disruption of sphingolipid metabolism with accumulation of free sphingoid bases. Plant Physiol. 1994;106:1085–1093. doi: 10.1104/pp.106.3.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mandala SM, Thornton RA, Frommer BR, Curotto JE, Rozdilsky W, Kurtz MB, Giacobbe RA, Bills GF, Cabello MA, Martin I, et al. The discovery of australifungin, a novel inhibitor of sphinganine N-acyltransferase from Sporormiella australis. Producing organism, fermentation, isolation, and biological activity. J. Antibiot. 1995;48:349–356. doi: 10.7164/antibiotics.48.349. [DOI] [PubMed] [Google Scholar]
- 132.Berdyshev EV, Gorshkova I, Skobeleva A, Bittman R, Lu X, Dudek SM, Mirzapoiazova T, Garcia JG, Natarajan V. FTY720 inhibits ceramide synthases and up-regulates dihydrosphingosine 1-phosphate formation in human lung endothelial cells. J. Biol. Chem. 2009;284:5467–5477. doi: 10.1074/jbc.M805186200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lahiri S, Park H, Laviad EL, Lu X, Bittman R, Futerman AH. Ceramide synthesis is modulated by the sphingosine analog FTY720 via a mixture of uncompetitive and noncompetitive inhibition in an acyl-CoA chain length-dependent manner. J. Biol. Chem. 2009;284:16090–16098. doi: 10.1074/jbc.M807438200. [DOI] [PMC free article] [PubMed] [Google Scholar]