Summary
Ubiquitin and ubiquitin-like (Ubl) protein modifications affect protein stability, activity and localization but we still lack broad understanding of the functions of Ubl modifications. We have profiled the protein targets of ubiquitin and six additional Ubls in mitosis using a functional assay that utilizes active mammalian cell extracts and protein microarrays and identified 1500 potential substrates; 100–200 protein targets were exclusive to each Ubl. The network structure is non-random, most targets mapping to a single Ubl. There are distinct molecular functions for each Ubl, suggesting divergent biological roles. Analysis of differential profiles between mitosis and G1 highlighted a previously under-appreciated role for the Ubl, FAT10, in mitotic regulation.
In addition to its role as a resource for Ubl modifications, our study provides a systematic approach to analyze changes in posttranslational modifications at various cellular states.
Introduction
The number of different protein isoforms in the human proteome is estimated to be about three orders of magnitude higher than the number of genes. This diversity is largely due to post-translational modifications (PTMs). Among these modifications, the Ubiquitin-Like (Ubl) molecule family appears to be very diverse in function. The Ubls comprise a class of evolutionary conserved polypeptides that can be reversibly conjugated through the formation of isopeptide bonds to lysine residues (mostly) on proteins, where they regulate activity, stability, cellular localization and interaction with other proteins (Hochstrasser, 2009). Ubiquitin and Ubl conjugation pathways have already been implicated in human diseases including numerous types of cancer(Hoeller et al., 2006), viral diseases (Edelmann and Kessler, 2008) and neurodegenerative disorders (Hattori and Mizuno, 2004). Yet, most of our knowledge stems from studies dedicated to ubiquitin and a couple of its homologs, namely SUMO and NEDD8. However, more than a dozen Ubl family members have been characterized to date, including the ones we profile here: SUMO1, SUMO2/3, NEDD8, UFM1, FAT10 and ISG15.
While the Ubl proteins share only modest primary sequence identity with ubiquitin (Figure 1A), they are closely related to it in structure, and each, like ubiquitin, requires a multi-step enzyme cascade for attachment to a target protein. It is thought that the Ubl modifiers use unique sets of the E1, E2 and E3 enzymes and Ubl-specific proteases in this cascade. Although we are still far from having a complete list of substrates for each Ubl, it is already apparent that the targets of different Ubls are not always exclusive. For instance, SUMO modification can antagonize ubiquitylation and stabilize protein substrates at the same lysine residue, both for PCNA (Papouli et al., 2005) and IkB (Lamsoul et al., 2005). In another example, the ubiquitin E3 ligase RNF4 recognizes sumoylated proteins and targets them to the proteasome (Sun et al., 2007), such ‘crosstalk’ may be conserved from yeast to humans; NEDD8, was shown to regulate the ubiquitylation efficiency of cullin E3 ligases (Morimoto et al., 2000) . While these are interesting examples, we do not know if there are general rules at work or even how common such crosstalk is. With over 500 genes that appear to be E3 ligases and over 60 E2s in the human genome, the regulatory pathways involving Ubl modifications could be of dizzying complexity. Linking the specific Ubl to their substrates is an essential first step in understanding specificity and selectivity of Ubl modifications and identifying the pathways in which they operate. However, despite their growing importance, there are remarkably few analytical tools available to analyze their behavior.
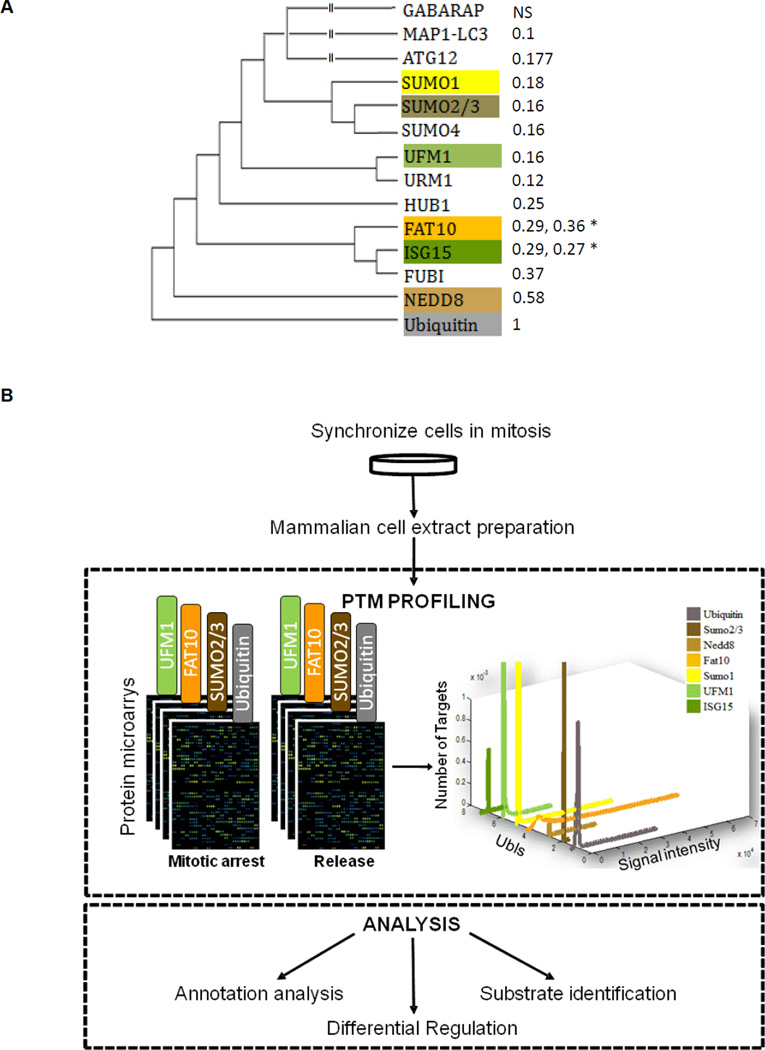
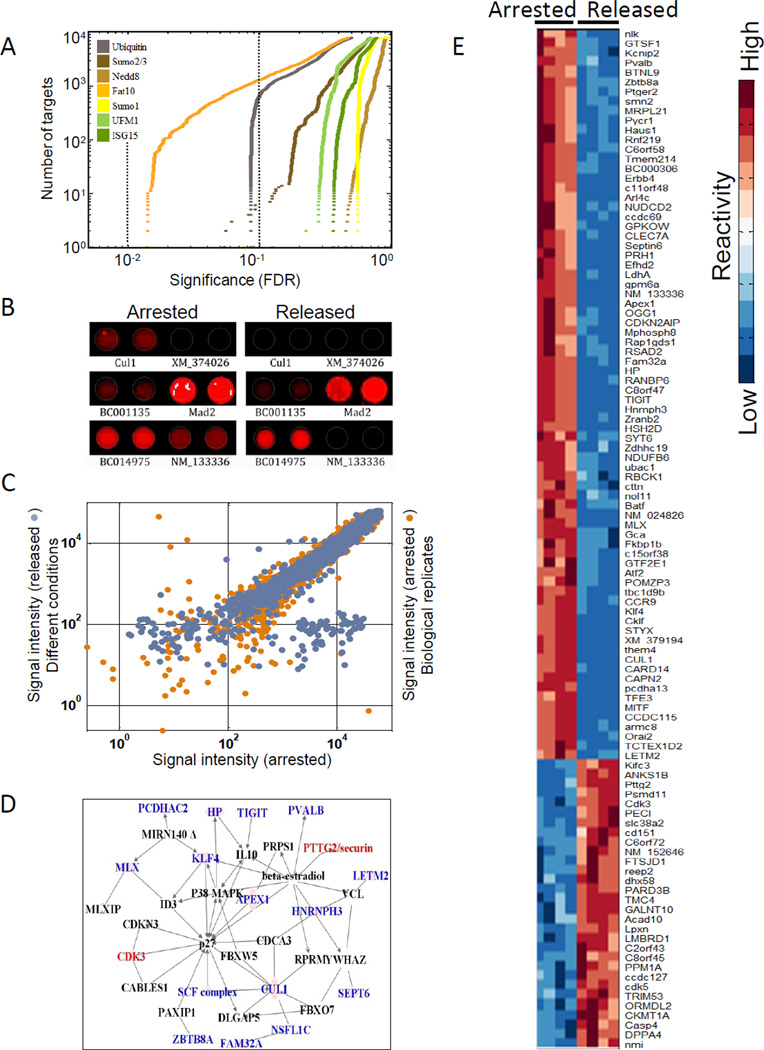
Figure 1. Global identification of ubiquitin and Ubl targets in mitosis.
(A) Phylogenetic relationship of selected members of the ubiquitin- like protein family. Branch lengths are proportional to phylogenetic distances and the percent similarity to ubiquitin sequences is given in parentheses. NS=Not Similar to ubiquitin by standard BLAST search. *Diubiquitin proteins (ISG15, FAT10) have two numbers for similarity representing each domain. (B) Experimental design. Mitotic HeLa S3 cell extracts were incubated on protein microarrays with or without the addition of Ubch10, a protein that abrogates the checkpoint arrest and allows the extracts to proceed toward mitotic exit. Ubl modifications on the spotted proteins are then measured by labeling the arrays with Ubl-specific antibodies, and fluorescently-labeled secondary antibodies are used to quantify the reactivity profile of the ~8000 proteins on the array toward each Ubl antibody.
Traditional assays on single proteins often take years before meaningful conclusions can be drawn, as was the case for several important cell cycle regulators targeted by the anaphase promoting complex, e.g. Securin, Geminin and Sororin. Though such approaches are important, it is too laborious to reach a coverage sufficient to reveal global patterns of function. Although recent advances in mass spectrometry have dramatically improved our ability to detect some modifications, it is still difficult to detect modifications that are unstable and of low abundance. We recently established a proteome-wide strategy based on functional concentrated cell extracts to monitor systematic ubiquitylation (Merbl and Kirschner, 2009a). Applying this assay we identified mitotic ubiquitylation substrates of the Anaphase-Promoting Complex (APC; we revealed more than hundred potential APC targets. Of 16 previously known Ubiquitin targets in the assay, 11 were identified, indicating that the assay was reliable with a low false negative rate. This system offered a rapid, tractable and semi-quantitative high-throughput system capable of monitoring multiple modifications on thousands of proteins in parallel. The assay makes use of the activity of cellular extracts under conditions that are relatively close to those of the complex cellular environment. It is highly reproducible, amenable to manipulation (e.g. drugs, inhibitors) and detects modifications equally for low and high abundance proteins.
We have expanded our approach to provide a comprehensive survey of the substrates of six additional Ubl modifiers (i.e. SUMO1, SUMO2/3, NEDD8, FAT10, UFM1 and ISG15) in mitosis, and to generate a Ubl interaction map. We assayed those Ubls for which there were good commercial antibodies available excluding the autophagy-related Ubls, whose activity would not be expected to be reflected in our in vitro system. The extensive identification of substrates gives us an idea not only of the degree to which the Ubl proteins target related substrates but also information about the pathways they regulate. Finally, we identified many putative Ubl targets involved in mitotic regulation and provided information about an under-appreciated role for FAT10 in mitotic progression.
Results
Global identification of ubiquitin and Ubl targets in mitosis
Concentrated cell extracts have been used extensively to study cellular processes, such as the cell cycle, nuclear membrane assembly, and cytoskeletal regulation, where they have provided insights into regulatory control at the protein level. Recently we applied such functional extracts made from mammalian cells to protein microarrays with the goal of assaying ubiquitylation in mitosis. The modification of a subset of the proteins was revealed by specific antibodies for ubiquitin and the reactivity of the target protein can be calculated from the spot intensity (see methods for image analysis and normalization). The activity of the extracts allows for the context-dependent interactions. In such cases the set of modifications is characteristic of the cellular state of the extract. For APC we were able to show that the assay is highly sensitive (spanning more than four orders of magnitude of reactivity signal) and highly reproducible (Merbl and Kirschner, 2009a). Since this approach could in principle be applied to any modification using the appropriate specific antibody, we expanded our approach to other Ubls for which less is known about their targets.
Here, we explore the role of several ubiquitin-like modifications (Ubiquitin, SUMO2/3, NEDD8, FAT10, SUMO1, UFM1 and ISG15) in mitotic regulation. For these seven modifications, we profiled the modification state of ~9000 of proteins before and after release from mitotic arrest (Figure 1B). These extracts have been shown to promote full checkpoint arrest and APC inhibition and to be relieved of that inhibition by Ubch10 (Reddy et al., 2007). Extracts were applied to duplicate microarrays for each modification under each of two conditions: ‘arrested’ (blocked in mitosis with nocodazole) and ‘released’ (released from the CP into anaphase/G1 by supplementing Ubch10).
Defining a Ubl modification network
For each Ubl, we focused on highly reactive proteins defined as having a specific reactivity greater than 2 standard deviations above the mean (when normalized to the protein abundance on the chip; see experimental procedures and Figure S1). All proteins passing these criteria have a reactivity significantly higher than the background reactivity of negative control spots (data not shown). Each Ubl reacted with 158 to 506 proteins above the threshold. We found 1543 such target proteins highly reactive to at least one of the Ubls (Table S1). For ubiquitin, which is the most investigated protein in this family, approximately 70% of the targets we identified were previously described by others either in vitro or in vivo as ubiquitylation substrates (see Table S2). Thus, the false-negative rate of the assay is tolerably low.
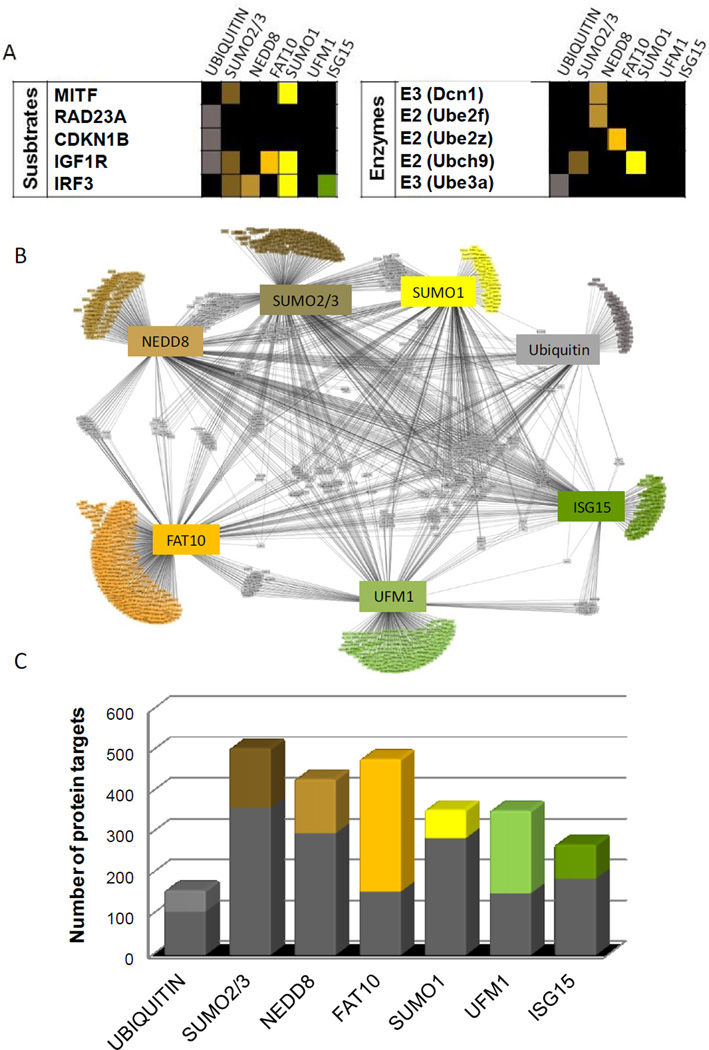
From this broad view we can ask several questions: Are there general patterns of Ubl specificity across the proteome? Specifically, are distinct pathways targeted by specific Ubls or are the modifications distributed among all biological pathways and functions? Conversely, how much overlap exists between the protein targets of the different Ubls? As a first step in our analysis, we characterized each reactive protein target’s interaction with each of the seven Ubl modifications (Figure 2A and Table S1). Some proteins are reactive to just one Ubl (e.g. Rad23a. which exhibited high reactivity only towards ubiquitin), while others react with multiple Ubls (e.g. Igf1r). Several examples of such interactions were previously reported and are shown in Figure 2A (Mitf (Murakami and Arnheiter, 2005), p27 (Carrano et al., 1999), Igf1R (Girnita et al., 2003; Sehat et al., 2010), Rad23a (Kumar et al., 1999). Among these, a few have been identified only recently. Interestingly, the assay does not only recognize putative substrates but also some of the enzymatic machinery for each of the Ubl pathways (e.g. Dcn1(Kurz et al., 2008), Ube2F(Huang et al., 2009) , Ube2z (Aichem et al., 2010b) in the cases where these enzymes were spotted on the array. Several enzymes were associated with specific Ubl reactivities for which we still do not know the underlying biology and therefore suggest open directions for investigation (see Table S1).
Figure 2. Defining a Ubl modification network.
(A) Examples of known Ubl targets and Ubl pathway enzymes identified by the assay. A color denotes reactivity toward that Ubl. The interactions reported in the literature are cited in the text. (B) The Ubl interaction network. Each protein is connected to the Ubl with which it interacts. Proteins that have multiple Ubls interactions are shown at the center and proteins that are reactive exclusively with one Ubl are shown at the rim. (C) The number of proteins targeted by each Ubl showing specificity of the Ubl pathways. The colored fractions represent the proportion of targets reacting uniquely with each Ubl whereas the grey fractions represent the targets reactive towards at least two Ubls. See related Figure S1.
To identify global patterns of Ubl modifications, we mapped the different targets for each Ubl into an “interaction network” (Figure 2B). The network consists of seven hubs, corresponding to the Ubls, and multiple nodes representing each one of the target proteins. Edges between the hubs and nodes represent the PTM interactions. The network reveals the interaction of the different Ubls with their targets, and their degree of specificity. Most of the Ubl targets (65% of 1543) map just to a single Ubl (Figure 2B, proteins assembled at the rim of the graph), whereas the remaining targets (35%) map to at least two Ubls (proteins in the center of the graph). Thus, most proteins are regulated primarily by one Ubl modification but differed for the different Ubls. For example, only 20% were unique to SUMO1, whereas 68% of FAT10 targets were unique to FAT10 (Figure 2C).
The Ubl network organization features both exclusive and preferential targeting of substrates by different Ubls
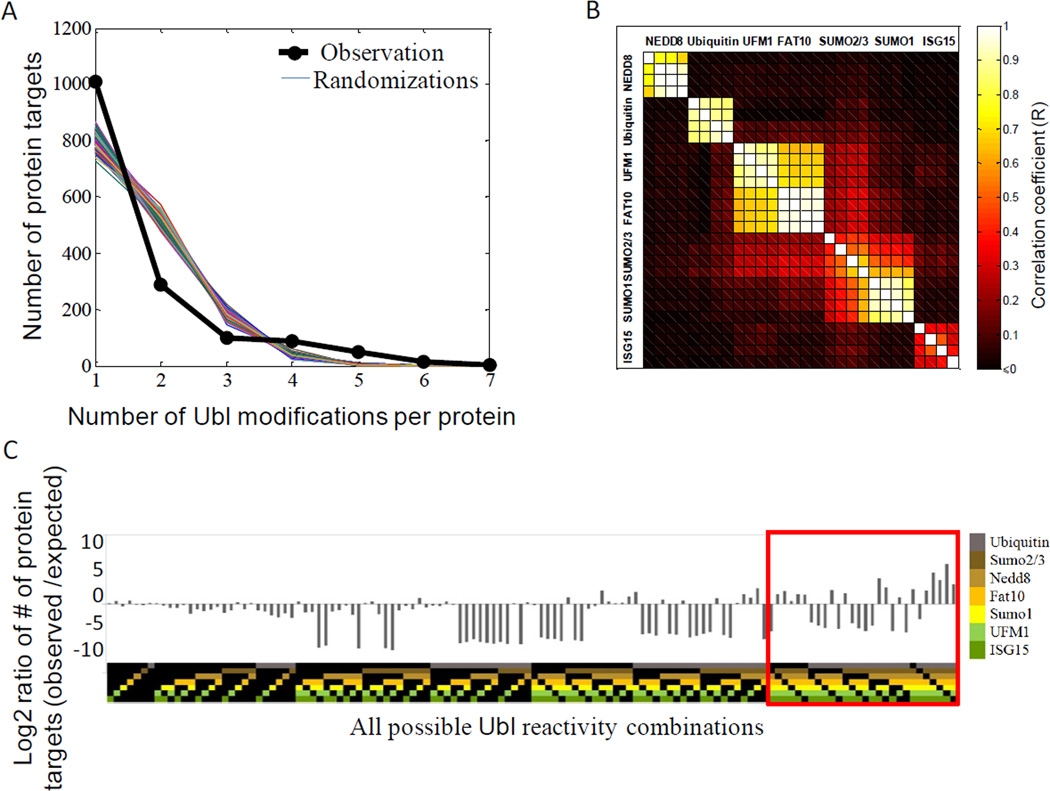
The network (Figure 2B) reveals that, a large number of target proteins interact with multiple Ubls, with a few (2.6%) interacting with five or more modifications (nodes in the center of the network). This pattern suggests that there is considerable specificity in Ubl modification, and raises the question of whether the relatively few cases of multiple reactions on a single protein have any functional significance or could merely be explained by chance. To test whether this is the case, we generated 1000 random permutations of the network that preserved the number of edges from each hub, and compared the resulting edge distribution to the empirical data (Figure 3A). The analysis shows that the number of unique targets (proteins targeted by only one Ubl), was higher than would be expected by chance (65% observed versus 52±1.4% expected), while the fraction of doubly-modified targets was much lower than expected by chance (19% observed versus 34±1% expected). Therefore, on average, a protein modified by one Ubl is less likely to be targeted by another.
Figure 3. The Ubl network features both exclusive and preferential targeting of substrates by different Ubls.
(A) Comparison of observed distribution of Ubl reactivities with expected distribution for a random Ubl network. Each of the colored lines represents the calculated frequency from 1000 random networks simulations showing exclusivity over-representation at n=1 (targets of only one Ubl) and under-representation of targets with two (n=2) or three Ubl (n=3) reactivities. The black line represents the observed frequencies in the network.
(B) Correlation analysis of Ubl profiles. Correlation coefficients between each of the Ubl reactivity profiles with all other microarrays reveals similarity among SUMO1 and SUMO2/3 targets as well as among FAT10 and UFM1 targets. Each square shows the correlation coefficient (R) between two microarray profiles, with 4 microarrays per Ubl. Black signifies no correlation or anti-correlation. Microarrays are grouped by their correlation (k-nearest neighbor clustering). (C) Ratio of observed versus expected frequencies of Ubl targets for each possible Ubl combination. The ratio represents the observed under-representation and over-representation of certain Ubl combinations when compared to a permutated network. Each Ubl combination is color coded according to the specific pattern of Ubl reactivity at the top.
The low abundance of doubly-modified targets suggests that certain Ubl combinations might be permitted while the majority is not (Figure 3A). A pair-wise correlation analysis of Ubl modifications (Figure 3B) confirmed that, indeed, many of the possible double Ubl interactions are strongly suppressed (R≤, shown in black in Figure 3B). However, certain Ubl combinations are over-represented in the correlation map, indicating that certain Ubls often co-target the same substrates. We found a high correlation between SUMO1 and SUMO2/3 targets (R=0.58), as has been previously suggested by others (Gocke et al., 2005; Matic et al., 2008), as well as between UFM1 and FAT10 modifications (R=0.63).
The enrichment of the network for exclusive Ubl targets can be studied by comparing specific Ubl reactivity combinations to the predictions of randomized permutations (Figure 3C). For single-targeted proteins, we found that only FAT10 and UFM1 exhibited a higher frequency of exclusive targets compared to random: 324 unique FAT10 targets were observed when 161 were expected, and 202 UFM1 targets were observed compared to 108 expected, suggesting that the functions of the FAT10 and UFM1 systems are largely insulated from the activity of other Ubls. The other Ubls (e.g. ubiquitin, SUMO1, SUMO2/3) exhibited the same frequency of exclusive targets as would be expected by chance, suggesting that these parallel pathways evolved independently. Thus, the over-representation of unique substrates is largely dominated by UFM1 and FAT10.
Turning to proteins subject to multiple Ubl modifications, the network exhibited a considerably higher frequency of such proteins than would be expected by chance. Looking at specific combinations of multiple Ubls (Figure 3C, red box), we find that all except one of the possible patterns of targets with 6-Ubl combination were enriched in comparison to randomized trials, with 14 proteins, including Rasl11b, Diras1, Nek4, Prkg2, Prkcb and IL15, undergoing six Ubl modifications when less than one such instance is expected by chance.
Cellular and functional classification of Ubl targets
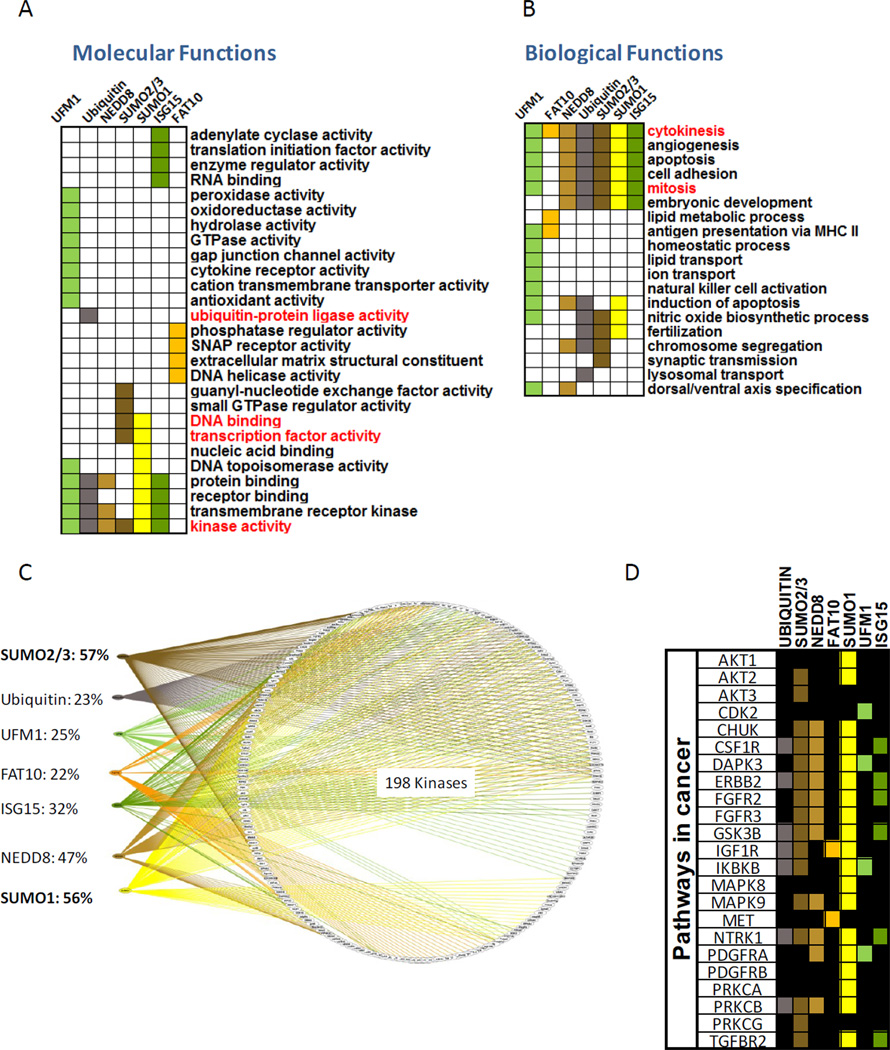
To ask whether the different Ubl pathways might be targeted to specific categories of proteins, or associated with distinct classes of biological processes, we looked for over-represented Gene Ontology (GO) terms for Ubl target protein annotations in the ‘Panther’ database. For each Ubl we calculated the enrichment for GO terms identified with its substrates. By comparing the targets of each Ubl to the list of reactive proteins only, we limit false-positive enrichments that could arise from biases in the representation of protein subsets on the chip. Figure 4A,B present a subset of the ‘molecular functions’ (A) and ‘biological processes’ (B) related to the targets of each Ubl where each column represents one Ubl (enriched terms are colored; see Table S3 for a full list).
Figure 4. Cellular and functional classification of Ubl targets.
(A) Enrichment analyses of “molecular functions” among Ubl targets, assessed by over-representation of Gene Ontology (GO) terms for the targets of each Ubl. Terms discussed in the text appear in red. (B) Same analysis as in (A) but for the “biological processes” annotations. (C) Detailed breakdown of the subset of kinases in the network, and their Ubl specificities (color-coded lines), showing an extensive crosstalk between kinases and different Ubl modifiers. See Figure S2A for a higher resolution of this image with protein names. See related figures S2 and S3.
As the assay made use of mitotic extracts, we expected to find enrichment for mitosis-related annotations. Indeed, the only over-represented categories in the cell cycle were ‘mitosis’ and ‘cytokinesis’, (Figure 4B). As a further validation, we found that known and well characterized biological functions of Ubiquitin and SUMO were enriched (Figure 4A). For example, ubiquitylation targets are the only ones enriched for the ‘Ubiquitin ligase activity’ term, with numerous targets categorized as E3 ligases or ’ring finger’ proteins. In addition, SUMO1 and SUMO2/3 targets are uniquely enriched for transcriptional-related terms and DNA binding corresponding to their established role (Gill, 2005; Girdwood et al., 2004). Interestingly, the assay also suggested a class of ‘translation initiation factors’ and RNA binding’ targeted by the ISG15 modification. Previous work suggested that ISG15 modification is involved in translation control (Okumura et al., 2007) and mainly targets newly synthesized proteins (Durfee et al., 2010) of both human and viral origin. A recent paper, suggested a role for secreted ISG15 as a cytokine inducing IFN-γ production which is key in the etiology of Mycobacterial disease (Bogunovic et al., 2012). Thus, it appears that ISG15 may exert its functions by directly targeting intracellular proteins, affecting the expression of IFN-γ target genes by acting as a secreted factor. Or putative ISG15 targets involved in translation initiation suggest that the extract system may be suitable for detection of ISG15 substrates. Our results suggest a role for ISG15 targets in various signaling pathways (Table S3 and Figure S3) and propose a few potential E3 ligases that may be involved in ISG15 conjugation.
Other findings from our analysis include the observations that UFM1 targets are enriched in transmembrane transporters, ion channels and cytokine receptors, and FAT10 targets are enriched for SNAP receptors, proteins related to extracellular matrix activity and DNA helicases. These potential targets await in vivo validation. Notably, consistent with the wide range of protein targets and biological processes ubiquitin is known to be involved in, we could not identify any unique enrichment (i.e. not shared by other Ubls) defined by the molecular function of ubiquitin targets except for the ‘ligase activity’ mentioned above. Nedd8, whose main function is to modulate E3 ligase ubiquitination activity, also did not show unique functional enrichment.
Kinases are especially enriched in the set of Ubl targets (Figure 4A, bottom row). Specifically, SUMO1 and SUMO2/3 together targeted 68% of the kinases (Figure 4C and Figure S2A) in our network. Among these, we found kinases associated with Wnt signaling pathway, MAPK signaling pathway and other pathways related to cancer (Figure 4D; see Figure S3 for a full list) as well as 20 known mitotic kinases. Indeed, we were able to confirm in vitro the SUMOylation of Polo-like kinase 1 (Plk1) and Cyclin dependent kinase-1 (Cdk1) as well as other kinases (Figure S2B). Consistent with our results, recent studies have identified both AurkB (Ban et al., 2011; Fernandez-Miranda et al., 2010) and Cdk1 (Golebiowski et al., 2009) as SUMO targets in vivo. In addition, a recent study also suggested a SUMO-modulated protein phosphorylation regulation, based on MS analysis (Yao et al., 2011). Although the biological function of these modifications is not yet clear, SUMOylation has been proposed to have both promoting and inhibitory effects on kinase activity (Yang and Sharrocks, 2006). These findings emphasize the possibility of crosstalk between the SUMOylation and phosphorylation pathways, opening a significant area for further investigation.
Many of the Ubls are implicated in a common set of biological processes (Figure 4B) that include cell cycle regulation, apoptosis, angiogenesis, cell adhesion and embryonic development. FAT10 and UFM1 are the exceptions: FAT10 targets were over-represented in ‘peroxisomal transport’, ‘lipid metabolic process’ and ‘cellular calcium homeostasis’, while UFM1 targets were implicated in pathways classified as ‘endocytosis’, ‘hemopoeisis’, ‘neurotransmitter secretion’ and ‘lipid transport’. Interestingly, both FAT10 and UFM1 appear to have enrichment for targets involved in ‘antigen presentation via MHC II’ suggesting that both may be involved in immune regulation. Yet, both FAT10 and UFM1 still have a significant number of targets in the mitosis/cytokinesis pathways, however. For example, 27 of the proteins modified by FAT10 were cell cycle regulators, 19 of which were mitosis-related. Taken together, our results suggest that the Ubl pathways evolved to regulate a wide range of cellular and physiological processes. Each Ubl has some level of discernible specialization, but they all appear to regulate multiple tasks in the cell.
Differential regulation of FAT10 on release from mitotic arrest
By comparing the modifications at prometaphase with those at anaphase/G1, we can assess the activity of Ubls at this specific transition. To detect changes in Ubl modifications under conditions that minimized variation in the extracts, we compared a nocodazole-arrested HeLa cell extract (checkpoint arrested; denoted ‘arrested’) with the same extract supplemented with the E2 enzyme, Ubch10, which relieves mitotic arrest (denoted as ‘released’), and drives the cell into anaphase and G1 (Reddy et al., 2007). The reactivity level of each protein towards each of the Ubls was calculated for these two conditions, using two microarrays per condition. An ANOVA test performed for the reactivity of each protein under these conditions (whose p-values were corrected using Storey’s false discovery rate method (Storey et al., 2007) showed that the reactivity for both ubiquitin and FAT10 had the most significant changes (q- value<0.1) upon release from the arrested into the released state (Figure 5A).
Figure 5. FAT10 targets are involved in cell cycle regulation and mitotic progression.
(A) Signal intensity of FAT10ylated protein targets were measured under nocodazole arrest and upon release from mitotic arrest. Values under the two conditions were compared using ANOVA and the resulting p-values were plotted (ascending order) for each Ubl separately. The two dotted lines indicate p-value cutoff levels of 0.1 and 0.05 (orange and brown, respectively). The y-axis denotes the cumulative number of target proteins that showed the stated significance in differential reactivity. (B) An example of duplicate spots of a FAT10 modified substrate under the two conditions, showing differences in reactivity. (C) Signal intensity of two biological replicates was compared to the signal intensity under the two different conditions for each protein. The x-axis: FAT10 signal intensity (log scale) under nocodazole arrest. Y-axis: replicate biological conditions (orange) or upon release from nocodazole arrest (blue). Dots that are off the diagonal represent the proteins that were differentially modified by FAT10 (D) Potential targets that mapped onto a known interaction network for cell cycle regulation. Color denotes increased (red) or decreased (blue) reactivity upon release from the mitotic arrest. Black names are interactors in the network that were not targeted by FAT10. (E) Proteins showing the highest differential modification under the two conditions (106) were clustered based on similarity. Signal intensity for each protein was standardized to have mean reactivity of 0 and standard deviation of 1. See related Figure S4.
We were surprised by changes in FAT10, whose role in mitosis has been little studied. Figure 5B depicts examples of FAT10 reactivity levels for several proteins under the two conditions. Our data identified Mad2 as a highly reactive substrate as previously reported (Liu et al., 1999a). However, its level was not significantly different in the two conditions. Most of the statistically significant changes in FAT10 level were dramatic, mostly decreasing two to three orders of magnitude (Figure 5C, blue dots). This is in contrast to the significant increase in ubiquitylation at the metaphase-anaphase transition as reported earlier. The striking effect on FAT10ylation could not be explained by the variability of the assay (Figure 5C orange dots) and was similar with different detection antibodies (Figure S4A). Washing with stringent conditions (3X 0.5% SDS) did not alter our results significantly suggesting that most of the interactions we detect represent covalent FAT10 conjugation (Figure S4B). 106 proteins showed a significant change in FAT10 reactivity, either an increase,30 proteins, or a decrease, 76 proteins, between the arrested and released states (q<0.002; Figure 5C and 5E).
Differential FAT10 targets are involved in cell cycle regulation and mitotic progression
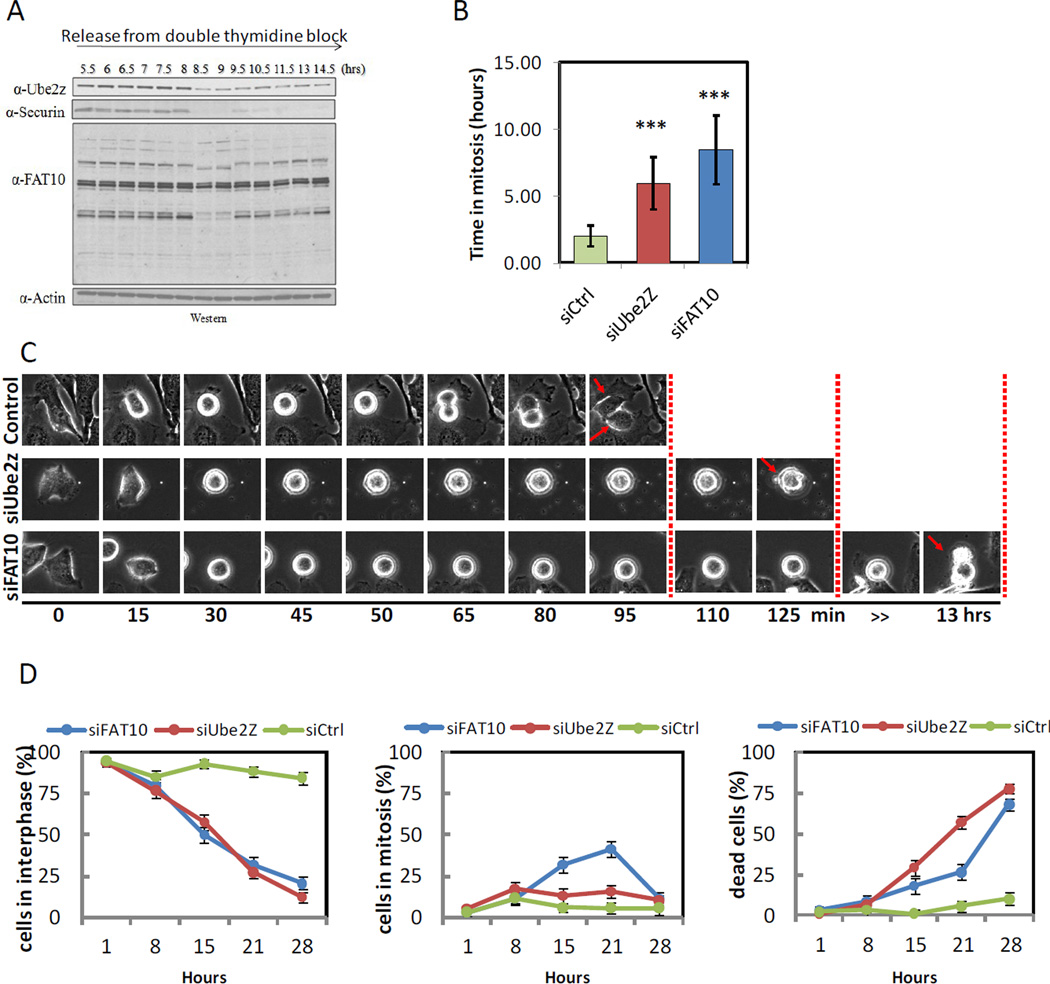
Among the proteins that showed the most striking change in FAT10 were a subset mapped onto a known cell cycle interaction network (e.g. Securin, Cul1, Septin 6 and Cdk3; see Figure 5D). Immunofluoresence showed that both FAT10 and Ube2z, the only known E2-conjugating enzyme for the FAT10 pathway, were ubiquitously highly expressed in mitosis (Figure S5A,B).We found global changes of FAT10 species at the time of mitosis (Figure 6A). The level of Ube2z was high during G2/M and dropped precipitously at the metaphase/anaphase transition, along with securin (Figure 6A), consistent with the dramatic reduction in FAT10ylation signals upon release from nocodazole arrest (see Figure S5C for cell cycle analysis). The level of Ube2z is regulated by the spindle checkpoint, since in cells released from the thymidine block into nocodazole, Ube2z was completely stable for 15 hours (Figure S5D). Thus, the timing of the degradation of Ube2z suggests that it is required for the mitotic checkpoint and that its degradation may be regulated in an APC dependent manner.
Figure 6. Inhibition of the FAT10 pathway using RNA interference leads to mitotic arrest and cell death.
(A) FAT10, Ube2z and Securin protein levels during cell cycle progression. Samples were taken at the indicated time points (see Figure S6 for FACS analysis). (B) The duration of mitosis was quantified from time lapse movies. Cells treated with siRNA for Ube2z or FAT10 spent a significantly longer time in mitosis (p<0.05) when compared to cells treated with control siRNA. Error bars depict mean +/- standard error. (C) A representative cell (n>30 per condition) undergoing mitosis for each of the different condition (siFAT10, siUbe2z and siControl) is presented. (D) Quantitation of the percentage of cells in interphase (left), mitosis (middle) and percentage of dead cells (right) in each of the conditions during the course of the experiment. Error bars depict mean +/- standard error. See related Figures S5–7 as well as Movies S1–3.
When we compared the list of targets that showed significant FAT10 changes to a database of phenotypic outcomes based on a genome-wide RNA interference screen (“Mitocheck Consortium”,(Neumann et al., 2010)) we found 8 of our candidates to have a mitotic phenotype (delay) upon knockdown and 2 additional genes to have a ‘death phenotype’ (Figure S6A). Among the 106 differentially regulated targets roughly 10% were already shown to have mitotic defects upon inhibition in vivo. We were further able to detect the in vitro FAT10ylation of four out of six proteins we tested from the FAT10 substrate list (Table 1) as well as for the currently known substrates of FAT10, p53 and Ube2z (Figure S6B). We were also able to detect FAT10ylation of 4 additional proteins by immunoprecipitation (Figure S6C). Notwithstanding the importance of establishing the role of the FAT10ylted substrates in vivo, the decrease in FAT10 modification on exiting metaphase itself suggested that the FAT10 pathway might have some regulatory role in mitotic progression.
Inhibition of the FAT10 pathway leads to prolonged mitotic arrest and cell death
To look for a regulatory role for FAT10 in mitosis we inhibited FAT10ylation via either a knockdown of FAT10, or by knockdown of its E2-conjugating enzyme, Ube2z. We transfected HeLa cells with either siRNA against FAT10 or Ube2z, or with control siRNA and allowed the cells to grow for 72 hours (Figure S7). In both knockdowns, but not in the control, there was a substantial increase in the duration of mitosis, eventually leading to cell death (Figure S7 and accompanying movies S1–3). When we quantified the average duration in mitosis (Figure 6B), we found that cells stayed in mitosis at least twice as long, on average than controls (5.93±1.96 and 8.48±2.58 hours for Ube2z and FAT10, respectively), when compared to cells transfected with control siRNA (2±0.8 hours).
We cannot at this point assess whether the effects we observe are due to FAT10 conjugation or whether FAT10 on its own or through binding interactions may exerts these effects. However, our results strongly suggest that FAT10 is involved in mitotic regulation.
Previous studies noted that FAT10 overexpression (Canaan et al., 2006b; Raasi et al., 2001; Ross et al., 2006; Snyder et al., 2009) and Ube2z overexpression (Aichem et al., 2010a; Park et al., 2007a) may lead to apoptosis. However, these studies did not link cell death to the cell cycle or mitosis. We found that either the inhibition of FAT10 or Ube2z extended mitotic arrest and triggered cell death (Figure 6C,D). The Ube2z knockdown effect may be more pronounced, since the death occurred earlier than in the FAT10 knockdown where cells arrested in mitosis for up to 30 hours before dying. Since Ube2z performs upstream to FAT10 and was shown to also mediate ubiquitinylation, its loss may be more broadly deleterious to the cell. It is interesting to note that under the FAT10 knockdown conditions all the cells that died, did so only after prolonged arrest in mitosis. Finally, by blocking the activation of the canonical apoptosis pathway via Bcl2 overexpression (McGill et al., 2002) the death phenotype could be suppressed (not shown).
Discussion
While we already know that the landscape of the proteome is largely controlled by PTM regulation, some of the most important questions about their function remain: With relatively few E3s identified for the Ubls, how is the timing and substrate specificity of these modifying activities regulated? Might the modification of one Ubl inhibitor activate another Ubl? Is there some sub-functionalization of the various Ubl modifications, either related to biological activity (kinases vs proteases), to specific pathway (eg. Hedgehog versus cytokine) or to some general type of overall biological function (transcription vs. metabolism)? The methodologies for addressing such questions on a proteome-wide scale are still limiting. In this study, we have profiled the PTMs for one cell type at the metaphase-anaphase transition. Although this choice reflects only a subset of cellular activities, mitosis involves many cellular processes that undergo structural and functional modifications. Furthermore, due to the inhibition of most transcriptional activity during mitosis, much of the regulatory burden is born by PTMs. Many modifications required for the proper progression of mitosis have been characterized to date and they include phosphorylation, acetylation, ubiquitylation and SUMOylation. Although this is already an impressive list, these modifications have usually been identified in an ad hoc manner from the study of individual proteins. Our strategy for global PTM profiling in mammalian cells is broadly applicable. It requires only the appropriate antibodies or direct labeling the small modifier itself.
Most proteins targeted by members of the Ubl group (65%) were modified by a specific Ubl; there are between 80–200 of these targets unique to each Ubl. However, the 35% of targets subject to modification by multiple Ubls may signify especially interesting regulatory processes. One pattern that emerged from the network is that the Ubl pathways appear largely independent of each other despite some potential crosstalk between them. As outliers, the FAT10 an UFM1 pathways were insular in their choice of targets, suggesting that these pathways may have more specialized uses. It was a concern of ours that given the similar chemistry of the Ubl modification reactions, there could be an appreciable background rate of non-enzymatic coupling of the Ubls to target proteins and this would genearate strong overlap in the Ubl targets. This turned out not to be the case. We found instead clear specificity in the choice of targets for the different Ubls. The sparseness of modification and the statistically few examples of overlap suggest that the examples where we did find overlap might be real and of interest. Multiple targeting is not a simple expectation of statistical overlap of independent events.
As with any high-throughput approach, the results reported here will require additional validation using more directed experiments. Though complete verification of the Ubl network we generated is impossible, we found that among the 1543 substrates we identified, between 200–500 interactions were previously documented in the literature. It is hard to give an exact number for the true positive rate as it depends on the criteria by which an interaction is defined. For example, the assessment varies depending on whether target protein homologs in different species are considered. In addition, the methods by which the modifications were identified (e.g. MS) and the nature of the interactions (e.g. immunoprecipitation under denaturing conditions versus yeast two-hybrid) may vary between different studies.
Our analysis of differential Ubl modification profiles upon release from nocodazole arrest however is largely protected from false hits that would affect each spot for each condition in the same way. In this case, differences between the arrested and released conditions may be meaningful, even when absolute measurements are not completely accurate. All but two Ubl pathways altered some of their targets during mitosis, suggesting that more PTMs may be involved in mitotic regulation than previously thought. Surprisingly, we found that FAT10, an under-investigated modification pathway, exhibited the most dramatic changes in signal intensity of modified proteins. Ubiquitin, which has already been widely investigated for its role in regulating the metaphase-anaphase transition, had the second highest change upon release from mitotic arrest.
In our analysis we found that, the reactivity pattern of FAT10 was strikingly different from that of ubiquitin in the metaphase-anaphase transition. While the poly-ubiquitylation signal increases strongly, the FAT10 signal decreased for 76 out of 106 targets. Of these, at least 10% are presumably important in mitosis, as judged by the fact that knockdowns by RNAi caused mitotic defects or death. In support of the importance of FAT10 in mitosis, we showed that inhibiting the pathway by knocking-down FAT10 or its E2-conjugating enzyme (Ube2z) resulted in a prolonged mitotic arrest, followed by cell death. Although Ube2z overexpression was shown previously to lead to apoptosis it was not linked to defects in mitosis (Park et al., 2007b).
Our observations of significantly higher FAT10ylation signal under nocodazole arrest together with the decrease of Ube2z levels at the end of mitosis provides a clue for how Ube2z functions in mitotic exit. However, though Ube2z is the only known E2-conjugating enzyme for the FAT10 pathway, it is also able to conjugate ubiquitin (Aichem et al., 2010b; Jin et al., 2007). Thus, Ube2z may regulate both ubiquitylation and FAT10ylation events in mitosis. Of potential regulatory interest is that among the FAT10 substrates were regulators of G-protein signaling family members (Rgs1, Rgs4, Rgs5, Rgs14, Rgs10, Rgs14) two of which were recently identified as ubiquitinylated Ube2z substrates (Lee et al., 2011). However, since Ube2z can conjugate ubiquitin as well as FAT10, it is not clear whether the RGS family may also be regulated via the FAT10 pathway in vivo.
Although FAT10 was discovered more than a decade ago, our knowledge of its biological function is limited. FAT10, also known as diubiquitin, has been suggested to be the only Ubl modifier that might target proteins for degradation through conjugation (Hipp et al., 2005; Pelzer and Groettrup, 2010). However, few in vivo covalent substrates of FAT10 have been identified to date (Aichem et al., 2012) including Ube2z (in cis) and p53 (Aichem et al., 2010b; Li et al., 2011). Thus, it is still unclear whether FAT10 acts as part of a signaling pathway or funnels proteins to the proteasome for degradation. In addition, it was recently shown that FAT10 itself is rapidly degraded by the ubiquitin-proteasome system (Buchsbaum et al., 2012). The known phenotypes of FAT10 misregulation tell us little about its targets. FAT10 has been implicated in at least three aspects of carcinogenesis and mitosis: Overexpression increases mitotic nondisjunction and chromosome instability (Ren et al., 2006). FAT10 is up-regulated in several epithelial tumors (Lee et al., 2003; Qing et al., 2011; Zhang et al., 2006) and overexpression of FAT10 causes apoptotic cell death (Raasi et al., 2001; Ross et al., 2006; Snyder et al., 2009). Finally, in one molecular study, overexpression of FAT10 greatly reduced the binding of Mad2, to kinetochores (Liu et al., 1999b) during prometaphase. These observations provide evidence for a link between FAT10 and mitosis. However, a key role for FAT10 in mitosis regulation might seem surprising since FAT10 knockout mice are viable (Canaan et al., 2006a). This expectation is not borne out for other seemingly critical cell cycle regulators, such as Cyclin D1, D2, D3, Cyclin E1 and E2, Cdk2, E2F, p27, suggesting that compensation in mitosis may be common. .
That FAT10, a vertebrate specific protein, has an important function in mitosis may also seem surprising on evolutionary grounds. . Early studies suggested a potential role for FAT10 in adaptive immunity, another process that arose in the vertebrate clade. Taken together, we propose that FAT10 may function in mitosis only in a context-specific manner such as specific cell types or under certain conditions (e.g. inflammation). We might imagine that the role of FAT10 in mitosis is not unlike adaptive immunity, both systems grafted onto more ancient pathways. If that is true, there may be therapeutic opportunities for FAT10 in proliferative diseases that are quite distinct from other mitotic targets.
Experimental Procedures
Tissue Culture and Cell Synchronization
For extract preparation, HeLa S3 cells were synchronized in prometaphase by treatment with nocodazole and prepared as previously described. (Merbl and Kirschner, 2009b; Rape and Kirschner, 2004; Storey et al., 2007).
PTM profiling and microarrays scanning were done as previously described (see supplementary materials).
Ube2z and FAT10 siRN
To deplete FAT10 and Ube2z, Dharmacon siGENOME SMARTpool against FAT10 or Ube2z (M-008266-03 and M-008596-02, respectively) were used in all experiments at a final concentration of 20 nM. As a control Dharmacon siGENOME Non-Targeting siRNA Pool #1 and #2 were used at 20 nM (D-001206-13-05 and D-001206-14-05, respectively). siRNA transfection was performed using Oligofectamine (Invitrogen) according to the manufacturer’s instructions. Cells were transfected for 48–72 depending on the experiment and collected for analysis by western blot, RT-PCR or followed by time-lapse microscopy. We note that the observed phenotype was dependent on the high knockdown efficiency.
RT-PCR analysis
To estimate the mRNA expression of FAT10 and Ube2z in cells treated with RNAi RT-PCR analysis was performed using TaqMan probes Hs00225039, Hs00197374 and 4333476 for Ube2z, FAT10 and GAPDH, respectively (Applied Biosystems). FAT10 and Ube2z levels were normalized to GAPDH in the same sample and then compared between the cell transfected with control non-targeting control siRNA (see above).
Microscopy and time lapse imaging
Cells were seeded in glass-bottom plates (MatTek) in CO2-independent medium (Invitrogen) supplemented with 10 FBS, 100 U/ml penicillin and 100 µg/ml streptomycin. For fluorescent time-lapse imaging cells were seeded in phenol red-free CO2-independent medium (Invitrogen). Image acquisition was performed using Nikon TE2000 automated inverted microscope with a 20X objective enclosed in a humidified incubation chamber maintained at 37°C. Images were collected every 15 minutes using a motorized stage. Images were viewed and analyzed using MetaMorph software (Molecular Dynamics).
In vitro SUMOylation
E1 (250 nM) and E2 (0.5uM) enzymes were added to a S35-radioactively labeled substrate (TNT® Quick Coupled Transcription/Translation; Promega) with 10uM recombinant SUMO1, SUMO2 or SUMO3. The reaction was supplemented with energy mix and allowed to run in room temperature for 2 hours. As a negative control, the same reaction is performed without the addition of the E1 enzyme and ran for the same amount of time (the right lane of each gel). Reactions are stopped by addition of sample buffer containing 5%β-mercaptoethanol. To identify modified substrates the samples were analyzed by SDS-PAGE and phosphorimaging.
In vitro FAT10ylation
Uba6 (250 nM), Ube2z (0.5uM) and FAT10 (10uM) were added to s35-radiactively labeled substrate (TNT® Quick Coupled Transcription/Translation; Promega). The reaction was supplemented with energy mix and allowed to run in room temperature for 1 hour at 30C. As a negative control, the same reaction is performed without the addition of FAT10 and ran for the same amount of time. Reactions are stopped by addition of sample buffer containing 5%β-mercaptoethanol.
Supplementary Material
Acknowledgments
We thank Jennifer Waters and the Nikon Imaging Center at Harvard Medical School for help with light microscope. We thank Anthony Hyman and Ina Poser for providing BAC clones. We thank Einav Laser for her support and Allon Klein, Ana Hernandez and Shai Shen-Orr for important scientific discussions and for their advice. We thank Sean Irwin, Tao Wu and Weiping Wang for technical support. We thank Becky Ward and Shay Tal for critically reading this manuscript and for their insightful comments. This work was supported by the NIH grant GM039023.
References
- Aichem A, Kalveram B, Spinnenhirn V, Kluge K, Catone N, Johansen T, Groettrup M. The proteomic analysis of endogenous FAT10 substrates identifies p62/SQSTM1 as a substrate of FAT10ylation. J Cell Sci. 2012;125:4576–4585. doi: 10.1242/jcs.107789. [DOI] [PubMed] [Google Scholar]
- Aichem A, Pelzer C, Lukasiak S, Kalveram B, Sheppard PW, Rani N, Schmidtke G, Groettrup M. USE1 is a bispecific conjugating enzyme for ubiquitin and FAT10, which FAT10ylates itself in cis. Nat Commun. 2010a;1:1–10. doi: 10.1038/ncomms1012. [DOI] [PubMed] [Google Scholar]
- Aichem A, Pelzer C, Lukasiak S, Kalveram B, Sheppard PW, Rani N, Schmidtke G, Groettrup M. USE1 is a bispecific conjugating enzyme for ubiquitin and FAT10, which FAT10ylates itself in cis. Nat Commun. 2010b;1:13. doi: 10.1038/ncomms1012. [DOI] [PubMed] [Google Scholar]
- Ban R, Nishida T, Urano T. Mitotic kinase Aurora-B is regulated by SUMO-2/3 conjugation/deconjugation during mitosis. Genes Cells. 2011;16:652–669. doi: 10.1111/j.1365-2443.2011.01521.x. [DOI] [PubMed] [Google Scholar]
- Bogunovic D, Byun M, Durfee LA, Abhyankar A, Sanal O, Mansouri D, Salem S, Radovanovic I, Grant AV, Adimi P, et al. Mycobacterial disease and impaired IFN-gamma immunity in humans with inherited ISG15 deficiency. Science. 2012;337:1684–1688. doi: 10.1126/science.1224026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchsbaum S, Bercovich B, Ciechanover A. FAT10 is a proteasomal degradation signal that is itself regulated by ubiquitination. Mol Biol Cell. 2012;23:225–232. doi: 10.1091/mbc.E11-07-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canaan A, Yu X, Booth CJ, Lian J, Lazar I, Gamfi SL, Castille K, Kohya N, Nakayama Y, Liu YC, et al. FAT10/diubiquitin-like protein-deficient mice exhibit minimal phenotypic differences. Mol Cell Biol. 2006a;26:5180–5189. doi: 10.1128/MCB.00966-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canaan A, Yu X, Booth CJ, Lian J, Lazar I, Gamfi SL, Castille K, Kohya N, Nakayama Y, Liu YC, et al. FAT10/diubiquitin-like protein-deficient mice exhibit minimal phenotypic differences. Mol Cell Biol. 2006b;26:5180–5189. doi: 10.1128/MCB.00966-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrano AC, Eytan E, Hershko A, Pagano M. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat Cell Biol. 1999;1:193–199. doi: 10.1038/12013. [DOI] [PubMed] [Google Scholar]
- Durfee LA, Lyon N, Seo K, Huibregtse JM. The ISG15 conjugation system broadly targets newly synthesized proteins: implications for the antiviral function of ISG15. Mol Cell. 2010;38:722–732. doi: 10.1016/j.molcel.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann MJ, Kessler BM. Ubiquitin and ubiquitin-like specific proteases targeted by infectious pathogens: Emerging patterns and molecular principles. Biochim Biophys Acta. 2008;1782:809–816. doi: 10.1016/j.bbadis.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Miranda G, Perez de Castro I, Carmena M, Aguirre-Portoles C, Ruchaud S, Fant X, Montoya G, Earnshaw WC, Malumbres M. SUMOylation modulates the function of Aurora-B kinase. J Cell Sci. 2010;123:2823–2833. doi: 10.1242/jcs.065565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill G. Something about SUMO inhibits transcription. Curr Opin Genet Dev. 2005;15:536–541. doi: 10.1016/j.gde.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Girdwood DW, Tatham MH, Hay RT. SUMO and transcriptional regulation. Semin Cell Dev Biol. 2004;15:201–210. doi: 10.1016/j.semcdb.2003.12.001. [DOI] [PubMed] [Google Scholar]
- Girnita L, Girnita A, Larsson O. Mdm2-dependent ubiquitination and degradation of the insulin-like growth factor 1 receptor. Proc Natl Acad Sci U S A. 2003;100:8247–8252. doi: 10.1073/pnas.1431613100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gocke CB, Yu H, Kang J. Systematic identification and analysis of mammalian small ubiquitin-like modifier substrates. J Biol Chem. 2005;280:5004–5012. doi: 10.1074/jbc.M411718200. [DOI] [PubMed] [Google Scholar]
- Golebiowski F, Matic I, Tatham MH, Cole C, Yin Y, Nakamura A, Cox J, Barton GJ, Mann M, Hay RT. System-wide changes to SUMO modifications in response to heat shock. Sci Signal. 2009;2:ra24. doi: 10.1126/scisignal.2000282. [DOI] [PubMed] [Google Scholar]
- Hattori N, Mizuno Y. Pathogenetic mechanisms of parkin in Parkinson's disease. Lancet. 2004;364:722–724. doi: 10.1016/S0140-6736(04)16901-8. [DOI] [PubMed] [Google Scholar]
- Hipp MS, Kalveram B, Raasi S, Groettrup M, Schmidtke G. FAT10, a ubiquitinindependent signal for proteasomal degradation. Mol Cell Biol. 2005;25:3483–3491. doi: 10.1128/MCB.25.9.3483-3491.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458:422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeller D, Hecker CM, Dikic I. Ubiquitin and ubiquitin-like proteins in cancer pathogenesis. Nat Rev Cancer. 2006;6:776–788. doi: 10.1038/nrc1994. [DOI] [PubMed] [Google Scholar]
- Huang DT, Ayrault O, Hunt HW, Taherbhoy AM, Duda DM, Scott DC, Borg LA, Neale G, Murray PJ, Roussel MF, et al. E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol Cell. 2009;33:483–495. doi: 10.1016/j.molcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Li X, Gygi SP, Harper JW. Dual E1 activation systems for ubiquitin differentially regulate E2 enzyme charging. Nature. 2007;447:1135–1138. doi: 10.1038/nature05902. [DOI] [PubMed] [Google Scholar]
- Kumar S, Talis AL, Howley PM. Identification of HHR23A as a substrate for E6-associated protein-mediated ubiquitination. J Biol Chem. 1999;274:18785–18792. doi: 10.1074/jbc.274.26.18785. [DOI] [PubMed] [Google Scholar]
- Kurz T, Chou YC, Willems AR, Meyer-Schaller N, Hecht ML, Tyers M, Peter M, Sicheri F. Dcn1 functions as a scaffold-type E3 ligase for cullin neddylation. Mol Cell. 2008;29:23–35. doi: 10.1016/j.molcel.2007.12.012. [DOI] [PubMed] [Google Scholar]
- Lamsoul I, Lodewick J, Lebrun S, Brasseur R, Burny A, Gaynor RB, Bex F. Exclusive ubiquitination and sumoylation on overlapping lysine residues mediate NF-kappaB activation by the human T-cell leukemia virus tax oncoprotein. Mol Cell Biol. 2005;25:10391–10406. doi: 10.1128/MCB.25.23.10391-10406.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CG, Ren J, Cheong IS, Ban KH, Ooi LL, Yong Tan S, Kan A, Nuchprayoon I, Jin R, Lee KH, et al. Expression of the FAT10 gene is highly upregulated in hepatocellular carcinoma and other gastrointestinal and gynecological cancers. Oncogene. 2003;22:2592–2603. doi: 10.1038/sj.onc.1206337. [DOI] [PubMed] [Google Scholar]
- Lee PC, Sowa ME, Gygi SP, Harper JW. Alternative ubiquitin activation/conjugation cascades interact with N-end rule ubiquitin ligases to control degradation of RGS proteins. Mol Cell. 2011;43:392–405. doi: 10.1016/j.molcel.2011.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Santockyte R, Yu S, Shen RF, Tekle E, Lee CG, Yang DC, Chock PB. FAT10 modifies p53 and upregulates its transcriptional activity. Arch Biochem Biophys. 2011;509:164–169. doi: 10.1016/j.abb.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YC, Pan J, Zhang C, Fan W, Collinge M, Bender JR, Weissman SM. A MHC-encoded ubiquitin-like protein (FAT10) binds noncovalently to the spindle assembly checkpoint protein MAD2. Proc Natl Acad Sci U S A. 1999a;96:4313–4318. doi: 10.1073/pnas.96.8.4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YC, Pan J, Zhang C, Fan W, Collinge M, Bender JR, Weissman SM. A MHC-encoded ubiquitin-like protein (FAT10) binds noncovalently to the spindle assembly checkpoint protein MAD2. Proceedings of the National Academy of Sciences of the United States of America. 1999b;96:4313–4318. doi: 10.1073/pnas.96.8.4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matic I, van Hagen M, Schimmel J, Macek B, Ogg SC, Tatham MH, Hay RT, Lamond AI, Mann M, Vertegaal AC. In vivo identification of human small ubiquitin-like modifier polymerization sites by high accuracy mass spectrometry and an in vitro to in vivo strategy. Mol Cell Proteomics. 2008;7:132–144. doi: 10.1074/mcp.M700173-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill GG, Horstmann M, Widlund HR, Du J, Motyckova G, Nishimura EK, Lin YL, Ramaswamy S, Avery W, Ding HF, et al. Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell. 2002;109:707–718. doi: 10.1016/s0092-8674(02)00762-6. [DOI] [PubMed] [Google Scholar]
- Merbl Y, Kirschner MW. Large-scale detection of ubiquitination substrates using cell extracts and protein microarrays. Proc Natl Acad Sci U S A. 2009a;106:2543–2548. doi: 10.1073/pnas.0812892106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merbl Y, Kirschner MW. Large-scale detection of ubiquitination substrates using cell extracts and protein microarrays. Proc Natl Acad Sci U S A. 2009b;106:2543–2548. doi: 10.1073/pnas.0812892106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto M, Nishida T, Honda R, Yasuda H. Modification of cullin-1 by ubiquitin-like protein Nedd8 enhances the activity of SCF(skp2) toward p27(kip1) Biochem Biophys Res Commun. 2000;270:1093–1096. doi: 10.1006/bbrc.2000.2576. [DOI] [PubMed] [Google Scholar]
- Murakami H, Arnheiter H. Sumoylation modulates transcriptional activity of MITF in a promoter-specific manner. Pigment Cell Res. 2005;18:265–277. doi: 10.1111/j.1600-0749.2005.00234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann B, Walter T, Heriche JK, Bulkescher J, Erfle H, Conrad C, Rogers P, Poser I, Held M, Liebel U, et al. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464:721–727. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura F, Zou W, Zhang DE. ISG15 modification of the eIF4E cognate 4EHP enhances cap structure-binding activity of 4EHP. Genes Dev. 2007;21:255–260. doi: 10.1101/gad.1521607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papouli E, Chen S, Davies AA, Huttner D, Krejci L, Sung P, Ulrich HD. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol Cell. 2005;19:123–133. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Park KM, Kang E, Jeon YJ, Kim N, Kim NS, Yoo HS, Yeom YI, Kim SJ. Identification of novel regulators of apoptosis using a high-throughput cell-based screen. Mol Cells. 2007a;23:170–174. [PubMed] [Google Scholar]
- Park KM, Kang E, Jeon YJ, Kim N, Kim NS, Yoo HS, Yeom YI, Kim SJ. Identification of novel regulators of apoptosis using a high-throughput cell-based screen. Mol Cells. 2007b;23:170–174. [PubMed] [Google Scholar]
- Pelzer C, Groettrup M. FAT10 : Activated by UBA6 and Functioning in Protein Degradation. Subcell Biochem. 2010;54:238–246. doi: 10.1007/978-1-4419-6676-6_19. [DOI] [PubMed] [Google Scholar]
- Qing X, French BA, Oliva J, French SW. Increased expression of FAT10 in colon benign, premalignant and malignant epithelial neoplasms. Exp Mol Pathol. 2011;90:51–54. doi: 10.1016/j.yexmp.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raasi S, Schmidtke G, Groettrup M. The ubiquitin-like protein FAT10 forms covalent conjugates and induces apoptosis. J Biol Chem. 2001;276:35334–35343. doi: 10.1074/jbc.M105139200. [DOI] [PubMed] [Google Scholar]
- Rape M, Kirschner MW. Autonomous regulation of the anaphase-promoting complex couples mitosis to S-phase entry. Nature. 2004;432:588–595. doi: 10.1038/nature03023. [DOI] [PubMed] [Google Scholar]
- Reddy SK, Rape M, Margansky WA, Kirschner MW. Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature. 2007;446:921–925. doi: 10.1038/nature05734. [DOI] [PubMed] [Google Scholar]
- Ren J, Kan A, Leong SH, Ooi LL, Jeang KT, Chong SS, Kon OL, Lee CG. FAT10 plays a role in the regulation of chromosomal stability. J Biol Chem. 2006;281:11413–11421. doi: 10.1074/jbc.M507218200. [DOI] [PubMed] [Google Scholar]
- Ross MJ, Wosnitzer MS, Ross MD, Granelli B, Gusella GL, Husain M, Kaufman L, Vasievich M, D'Agati VD, Wilson PD, et al. Role of ubiquitin-like protein FAT10 in epithelial apoptosis in renal disease. J Am Soc Nephrol. 2006;17:996–1004. doi: 10.1681/ASN.2005070692. [DOI] [PubMed] [Google Scholar]
- Sehat B, Tofigh A, Lin Y, Trocme E, Liljedahl U, Lagergren J, Larsson O. SUMOylation mediates the nuclear translocation and signaling of the IGF-1 receptor. Sci Signal. 2010;3:ra10. doi: 10.1126/scisignal.2000628. [DOI] [PubMed] [Google Scholar]
- Snyder A, Alsauskas Z, Gong P, Rosenstiel PE, Klotman ME, Klotman PE, Ross MJ. FAT10: a novel mediator of Vpr-induced apoptosis in human immunodeficiency virus-associated nephropathy. Journal of virology. 2009;83:11983–11988. doi: 10.1128/JVI.00034-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Dai JY, Leek JT. The optimal discovery procedure for large-scale significance testing, with applications to comparative microarray experiments. Biostatistics. 2007;8:414–432. doi: 10.1093/biostatistics/kxl019. [DOI] [PubMed] [Google Scholar]
- Sun H, Leverson JD, Hunter T. Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. Embo J. 2007;26:4102–4112. doi: 10.1038/sj.emboj.7601839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SH, Sharrocks AD. Interplay of the SUMO and MAP kinase pathways. Ernst Schering Res Found Workshop. 2006:193–209. doi: 10.1007/3-540-37633-x_11. [DOI] [PubMed] [Google Scholar]
- Yao Q, Li H, Liu BQ, Huang XY, Guo L. SUMOylation-regulated protein phosphorylation, evidence from quantitative phosphoproteomics analyses. J Biol Chem. 2011;286:27342–27349. doi: 10.1074/jbc.M111.220848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DW, Jeang KT, Lee CG. p53 negatively regulates the expression of FAT10, a gene upregulated in various cancers. Oncogene. 2006;25:2318–2327. doi: 10.1038/sj.onc.1209220. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.