Abstract
Understanding the molecular basis of common traits is a primary challenge of modern genetics. One model holds that rare mutations in many genetic backgrounds may often phenocopy one another, together explaining the prevalence of the resulting trait in the population. For the vast majority of phenotypes, the role of rare variants and the evolutionary forces that underlie them are unknown. In this work, we use a population of Saccharomyces paradoxus yeast as a model system for the study of common trait variation. We observed an unusual, flocculation and invasive-growth phenotype in one-third of S. paradoxus strains, which were otherwise unrelated. In crosses with each strain in turn, these morphologies segregated as a recessive Mendelian phenotype, mapping either to IRA1 or to IRA2, yeast homologs of the hypermutable human neurofibromatosis gene NF1. The causal IRA1 and IRA2 haplotypes were of distinct evolutionary origin and, in addition to their morphological effects, associated with hundreds of stress-resistance and growth traits, both beneficial and disadvantageous, across S. paradoxus. Single-gene molecular genetic analyses confirmed variant IRA1 and IRA2 haplotypes as causal for these growth characteristics, many of which were independent of morphology. Our data make clear that common growth and morphology traits in yeast result from a suite of variants in master regulators, which function as a mutation-driven switch between phenotypic states.
Keywords: fungal evolution, common trait variation, flocculation
A primary goal of modern genetics is to understand the molecular basis of traits that segregate at high frequency in populations. Toward this end, hundreds of studies have sought to map causal genes, using tests for allele sharing among affected but unrelated individuals. Against a backdrop of recent successes in fruit fly and plant populations (Atwell et al. 2010; Brachi et al. 2010; Chan et al. 2011; Filiault and Maloof 2012; Mackay et al. 2012; Magwire et al. 2012; Weber et al. 2012; Dunn et al. 2013), association mapping in many systems has yielded loci that explain only a small part of the variation in a given trait across individuals (McCarthy et al. 2008). The latter challenges have motivated the proposal that the bulk of common phenotypic variation may be attributable to rare, highly penetrant mutations (Dickson et al. 2010; McClellan and King 2010; Veltman and Brunner 2012), including recurrent variation at hypermutable loci (Shen et al. 1996; Chan et al. 2010; Michaelson et al. 2012). As yet, the genetic architecture of most common traits remains unknown, and experimental systems in which to investigate the principles of common trait variation have been at a premium in the literature.
Saccharomyces yeasts have long been a workhorse of the molecular-genetic research community, with resources now becoming available for analyses of population-level variation (Liti et al. 2009). Among the most well-studied phenotypes in the classic yeast literature are cell-clumping and filamentation-like behaviors in certain laboratory strains, which serve as models for fungal pathogenesis and have been subject to elegant molecular dissection (Carstens and Lambrechts 1998; Jin et al. 2008; Soares 2011; Ryan et al. 2012). These morphologies can arise spontaneously in laboratory and brewing yeast, owing to variants in a number of hypermutable genes, including effectors and regulators of budding and adhesion (Halme et al. 2004; Verstrepen et al. 2005). In wild populations, the prevalence of morphological trait variation and its consequences for growth and fitness have not been well characterized. Wild yeast have, however, been the focus of phenomic profiling efforts (Liti et al. 2009; Warringer et al. 2011), which have revealed strain differences in growth phenotypes in hundreds of conditions. Although some genome-scale mapping analyses have been reported (Cubillos et al. 2011; Warringer et al. 2011; Zörgö et al. 2012), for the vast majority of growth attributes varying among yeasts, the molecular basis remains to be identified.
We set out to use yeast morphologies as a testbed for the study of common trait variation, and the role of hypermutable loci, in a system of genetically tractable, wild-collected Saccharomyces paradoxus strain backgrounds. Our goal was to establish whether and how differences in hypermutable morphology genes segregate in wild yeast populations and to what extent they underlie morphology and growth behaviors. The pursuit of these questions led us to the discovery of a suite of hundreds of common traits in wild yeast and to the mapping of their genetic determinants.
Materials and Methods
Yeast strains
Strains used in this study are listed in Supporting Information, Table S1. With the exception of wild progenitor strains, all were homothallic diploid strains obtained from the Saccharomyces Genome Resequencing Project (SGRP) collection (Liti et al. 2009) or engineered from these SGRP strains. All data were generated with these laboratory-derived strains except for the results presented in Figure S10, Figure S15, and Figure S16; the latter used homothallic progenitor strains that were obtained directly from either V. Koufopanou or G. Liti.
Hybrid strains were generated by sporulation and tetrad dissection of two strains, immediately followed by single-cell mating of progeny from each, and confirmed by PCR amplification of regions containing strain-specific indels. Homozygous deletion strains were constructed by transformation with a KanMX cassette amplified from the pUG6 plasmid (Güldener et al. 1996), followed by sporulation, tetrad dissection, and selection. Reciprocal hemizygotes were constructed by transformation of the KanMX cassette into a given hybrid background, followed by selection and PCR at the appropriate locus to determine which allele of the target gene had been deleted. Deletion of FLO10 and FLO11 in all backgrounds, as well as IRA1 in the W7 × Z1 background, was accomplished by replacing the entire ORF with the KanMX cassette. Engineered mutation of IRA1 and IRA2 in all other backgrounds, and FLO9 in all backgrounds, was accomplished by replacing 2000–3000 bp of the 5′ end of the respective gene, including the start codon, with the KanMX cassette.
Invasive growth
Cultures of each strain were grown to stationary phase, followed by normalization to an optical density of 2.0. Four microliters of a 1:100 dilution of this culture was then spotted onto a YPD plate (2% dextrose, 2% bacto-peptone, 1% yeast extract, 2% agar) and allowed to grow at room temperature for 5 days. Plates were photographed, washed under a stream of distilled water such that all nonadherent cells were removed, and photographed again. Custom Python scripts were used to quantify invasive growth from digital images by calculating the proportion of the original colony that remained after washing.
DNA sequence analysis
For the species tree in Figure 1, whole-genome alignments of laboratory derivatives of S. paradoxus strains were downloaded from the Saccharomyces Genome Resequencing Project (Liti et al. 2009) and accessed using the alicat.pl script. Sites with an error probability >0.0001 as inferred by Liti et al. (2009) were excluded from analysis. For species phylogenies, 19,695 polymorphic sites across S. paradoxus from these published data were used to infer a tree, using FastTree (Price et al. 2010), which was visualized using FigTree. These publicly available sequences were used, in Table S2, as input into custom Python scripts to calculate genetic identity between strains KPN3828 and KPN3829 and to calculate nucleotide diversity (Nei and Li 1979) within each S. paradoxus population, as
where xi and xj are the respective frequencies of the ith and jth sequences in the population and πij is the number of nucleotide differences per nucleotide site between the ith and jth sequences.
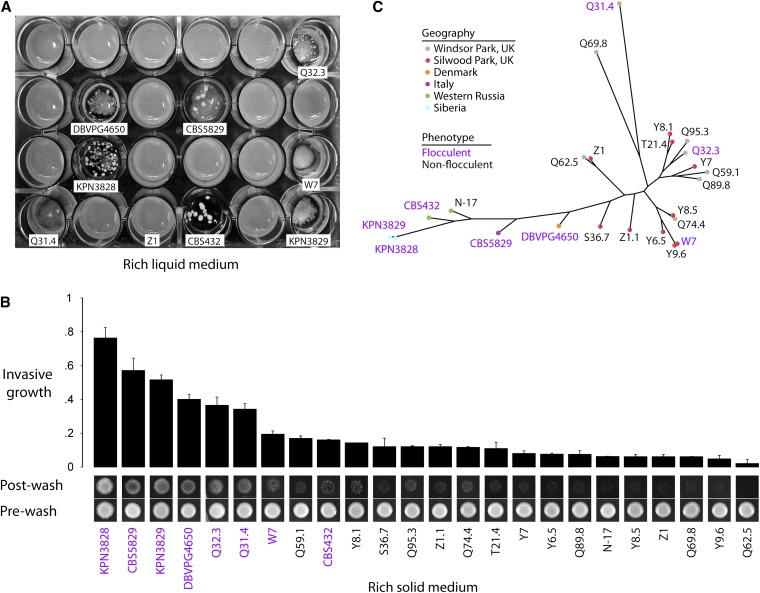
Figure 1.
Flocculation and invasive growth vary across European S. paradoxus. (A) Photograph of cultures of European S. paradoxus strains after overnight growth in rich liquid yeast peptone dextrose (YPD) medium containing 2% glucose. The eight flocculent strains and the nonflocculent strain Z1 are labeled with identifiers as in Liti et al. (2009). (B) Photographs at bottom are of invasive growth assays: a colony of each strain was grown on YPD solid medium for 5 days and then photographed before (“pre-wash”) and after (“post-wash”) removal of nonadherent cells by a water wash. At top, each bar height reports the ratio of cell density in colonies after and before a water wash as a mean of two replicate colonies; error bars represent one standard deviation. (C) Maximum-likelihood genome tree of European S. paradoxus. Branch lengths are proportional to the number of segregating sites that differentiate each pair of strains. In B and C, identifiers of flocculent strains are in purple.
In Figure 4 and Figure S9, inference of gene trees of the coding regions of IRA1 and IRA2 in laboratory derivatives of S. paradoxus strains and trees of the regions 800 bp directly upstream of the start codon of each of these loci used a combination of published and amended sequence data as follows. We used additional Sanger sequencing to fill gaps and confirm polymorphisms in the IRA1 orthologs of laboratory derivatives of KPN3829 and W7 and the IRA2 orthologs of laboratory derivatives of KPN3828 and CBS5829 from Liti et al. (2009). Reads were assembled with the PhredPhrap script (Ewing and Green 1998). For other strains analyzed in gene trees in Figure 4, additional high-coverage genomic sequences used to confirm polymorphisms in IRA1 and IRA2 were downloaded from ftp://ftp.sanger.ac.uk/pub/users/dmc/yeast/SGRP2. The complete set of gene sequences was used as input into FastTree as above.
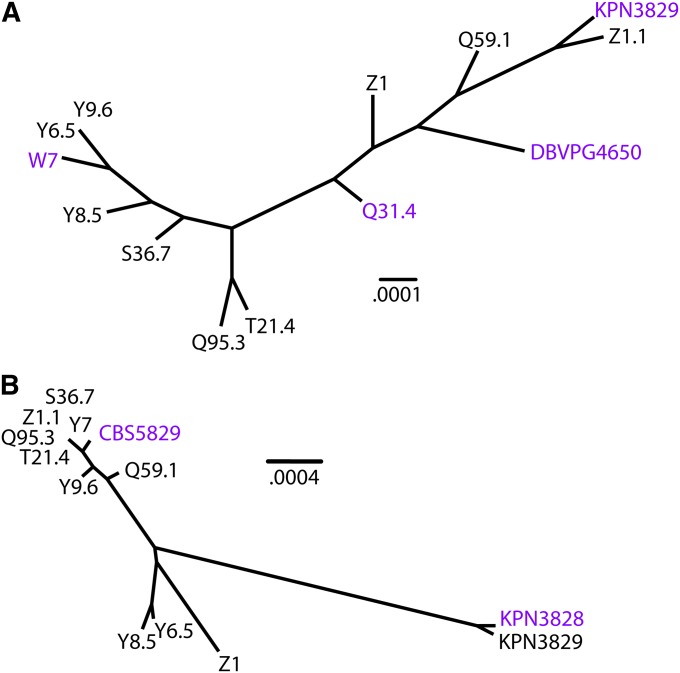
Figure 4.
IRA1 and IRA2 haplotypes from flocculent/invasive strains are not closely related. Each panel shows a maximum-likelihood phylogeny inferred from coding sequences of IRA1 (A) or IRA2 (B). Identifiers of strains in which IRA1 or IRA2 underlies flocculent and invasive growth traits are colored purple. Scale bars indicate frequencies of base pair substitutions per site.
For analysis of original isolates of Q31.4, KPN3829, KPN3828, DBVPG4650, and CBS5829 in Figure S10, we generated complete sequence data of IRA1 and IRA2 coding regions and the regions 800 bp upstream of each of these two genes by Sanger sequencing, and we visualized the results with Jalview (Waterhouse et al. 2009).
Linkage analysis
For each strain, marker SNPs were identified in IRA1 or IRA2 and strains were genotyped by Sanger sequencing of a region containing the marker. Genotypes at a given locus in a given cross were used in linkage analysis as follows. Two progeny were obtained from each of four tetrads, comprising two flocculent pairs and two nonflocculent pairs. Among the flocculent pairs, the probability pfloc of observing by chance a pairs sharing the flocculent parent’s allele, b pairs sharing the nonflocculent parent’s allele, and c pairs with distinct alleles was calculated as pfloc = n!/(a!b!c!)[(1/6)a(2/3)c(1/6)b], and the chance probability pnonfloc of observing a given pattern of allele sharing among nonflocculent pairs was calculated analogously. The final probability of the observed allele sharing under the binomial null was taken as the product of pfloc and pnonfloc.
Quantification of flocculation
Quantification of flocculation in the presence of mannose was conducted for a given strain as in Van Mulders et al. (2009) with several modifications as follows. A preculture was grown overnight in liquid medium and then pipetted vigorously to disrupt flocs. A total of 0.6 ml of this culture was then added to a well containing 0.6 ml YPD and, separately, to a well containing 0.6 ml of 0.5 M EDTA, in a 24-well plate. The plate was shaken at room temperature for 15 min to settle flocs at the bottom of the well, after which 200 µl of culture media was pipetted off the top of each well. Optical density at 600 nm (OD600) of this 200-µl aliquot of media was measured, and flocculation ability of a given strain was calculated as 1 − (OD600 in YPD/OD600 in EDTA). The final measure of the degree of flocculation for each strain in a given condition was calculated as the mean of three such assays from the same overnight culture.
Association analysis of growth fitness data
Fitness data for S. paradoxus strains were downloaded from Warringer et al. (2011). These values included fitness measurements for 5 of the strains we had determined to harbor variant IRA gene alleles and 15 nonflocculent strains. For each of the 592 environment–parameter measurements in the data set, fitness of the strains bearing variant IRA genes was grouped together and compared to fitness of the nonflocculent strains, using a Wilcoxon rank-sum test. At a given P-value threshold t, we estimated the expected number of false positives nf as the product of p0 and the number of tests, with the false discovery rate (FDR) then calculated as nf divided by the number of true tests with P-value < p0; we set p0 to attain an FDR of 0.05.
Fitness profiling
A preculture of each strain was grown for 24 hr at room temperature in complete synthetic media (CSM) (0.67% yeast nitrogen base without amino acids, 0.079% complete supplement mixture) supplemented with 2% glucose. Each strain was back-diluted to an OD of 0.02 in a 96-well plate containing 150 µl of a given treatment in CSM. Plates were vigorously shaken for 10 sec and placed in a VersaMax MicroPlate reader (Molecular Devices). OD600 was measured every 10 min for 72 hr of growth at a temperature of 27° without shaking. Each experiment that characterized a flocculent European strain included on the plate a replicate culture of the nonflocculent European strain Z1, which was used to normalize growth measurements of the respective flocculent strain in Figure 5 and Figure S12.
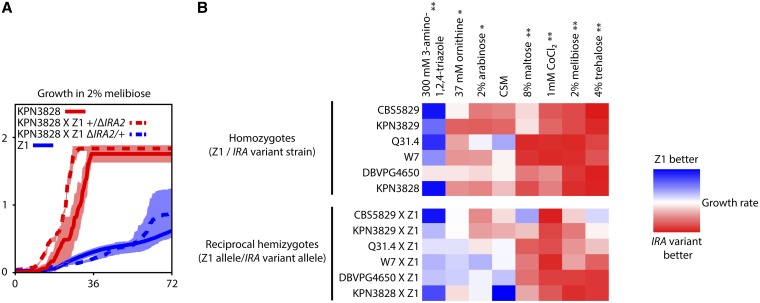
Figure 5.
Variation at IRA1 and IRA2 underlies growth rate differences in multiple conditions. (A) Shown is an example growth time course in liquid medium containing 2% melibiose as the sole carbon source, with time after inoculation on the x-axis and cell density, measured as the optical density (OD) at 600 nm, on the y-axis. Each curve reports the mean of at least two measurements of one homozygous European diploid (the flocculent strain KPN3828 or the nonflocculent strain Z1) or reciprocal hemizygote diploid constructed in the KPN3828 × Z1 hybrid background (+/ΔIRA2, bearing only the KPN3828 allele of IRA2, or ΔIRA2/+, bearing only the Z1 allele of IRA2; see Figure 3A for schematic). Error bars indicate one standard deviation. (B) Each cell reports the results of a growth experiment as in A. Color in each cell represents a ratio of the growth rates, during log-phase growth, of two strains measured in liquid culture, with each row showing data from one strain pair and each column showing data from one media condition. Top, each cell reports the ratio of growth of the homozygous nonflocculent, noninvasive European strain Z1 to growth of the indicated homozygous European strain harboring a variant IRA1 or IRA2 allele. Bottom, each row gives results from a pair of hybrid reciprocal hemizygote strains interrogating IRA1 or IRA2 in the indicated strain and Z1: each cell reports the growth of the hemizygote bearing the Z1 allele of the respective IRA gene, relative to the growth of the hemizygote bearing the variant allele at the IRA gene. Media labels at top marked by an asterisk are those in which homozygote European strains bearing variant IRA genes differed significantly from Z1 (Wilcoxon’s rank-sum test, P < 0.05 after Bonferroni correction). Media marked by ** are those inducing two significant growth effects: homozygote European strains bearing variant IRA genes differed from Z1, and hemizygote strains bearing variant IRA genes differed from hemizygotes bearing the Z1 allele at the IRA genes (Wilcoxon’s rank-sum test, P < 0.05 after Bonferroni correction). CSM, complete synthetic medium. Δ, engineered loss-of-function allele. Raw measurements and significance estimates are given in File S2.
From a given time course of OD600 measurements, custom Python scripts were used to quantify lag time, growth rate, and density change parameters as described in Warringer et al. (2011), with minor modifications as follows: the growth rate parameter was calculated in a sliding window as the slope of the line of best fit between 10 consecutive growth rate measurements corresponding to 100 min of growth. The top slope was discarded and a mean was taken of the second- to the fourth-highest slopes. Population doubling time was calculated as the ratio of ln(2) to this mean as in Warringer et al. (2011). Each fitness parameter value was calculated as the average of at least two replicate wells on the same plate inoculated from the same preculture well. For a given parameter and condition in Figure 5, Figure S12, and File S2, significance of the effect of IRA variants was assessed in parental strains by comparing the set of measurements of flocculent European strains to the set of all replicates of Z1, using the Wilcoxon rank-sum test, and for a given parameter, P-values were corrected for multiple testing across conditions by the Bonferroni method. Separately, for reciprocal hemizygotes, we estimated analogously the significance of differences between the set of measurements across all strains with variant alleles and the set of strains with wild-type alleles. In Figure S13 and File S3, for a given parameter and condition, the set of measurements across all strains with wild-type FLO9 was compared to the set of measurements in strains mutant for FLO9, using the Wilcoxon rank-sum test, and for a given parameter, P-values were corrected for multiple testing across conditions by the Bonferroni method. In each of these analyses, results were comparable, using a paired Wilcoxon test (data not shown).
Results
A polyphyletic S. paradoxus clade displays a flocculent and invasive phenotype
We used strains derived from wild isolates of S. paradoxus in a screen for flocculation, the formation of macroscopic cell aggregates during growth in liquid medium. Of the 24 strains in the well-defined European population of this yeast (Liti et al. 2009), 8 were flocculent (Figure 1A). These strains formed flocs across a range of cell densities and environmental treatments (Figure S1), in contrast to reports of condition-specific flocculation in S. cerevisiae (Smit et al. 1992; Dengis et al. 1995; Bayly et al. 2005; Sampermans et al. 2005). We also surveyed European S. paradoxus strains for invasive growth, the ability of yeast colonies to adhere to and penetrate a solid substrate (Guo et al. 2000; Palecek et al. 2000; Bester et al. 2006). Most strains invaded a solid rich-medium agar substrate to some extent (Figure 1B), again in contrast to the requirement for nutrient limitation often seen in S. cerevisiae (Vinod et al. 2008; Zaman et al. 2008). S. paradoxus strains with the most dramatic invasive phenotype were also those flocculating in liquid media, such that the two traits were tightly correlated across the population (Spearman’s rank correlation = 0.81, P = 2.14e-6).
To begin to investigate the evolutionary history of these morphology traits, we inferred the phylogeny of European S. paradoxus, using genome-scale polymorphism data (Liti et al. 2009). Surprisingly, flocculent/highly invasive strains were scattered across the phylogeny, and strains collected from neighboring locations, even those from within the same English county park, often had dissimilar morphologies (Figure 1C). Such a pattern was consistent with either of two interpretations. On the one hand, the flocculent/invasive phenotype could have been independently acquired in multiple strains, following descent from a nonflocculent, noninvasive ancestor. Alternatively, the phenotype could have been lost in multiple strains following descent from a flocculent, invasive common ancestor. Under either model, morphological variation in S. paradoxus would be a product of independent mutational events in distinct lineages.
Flocculation and invasive growth are linked Mendelian traits
To characterize further the molecular and evolutionary basis of flocculation and invasive growth in S. paradoxus, we sought to evaluate the genetic complexity of these traits. For this purpose, we carried out crosses between each flocculent, homothallic European strain and the nonflocculent European strain Z1. Seven European flocculent strains successfully formed hybrid diploids when mated as single cells with Z1. Each such hybrid strain was nonflocculent but showed a degree of invasive growth intermediate between that of its parents (see Figure 3 and Figure S6); thus, flocculation acted as a recessive phenotype and invasive growth as incompletely dominant. For each homothallic hybrid diploid, we induced sporulation, dissected the resulting tetrads, and allowed each recombinant spore to form an isogenic colony of homozygous diploid cells. Over 70% of the progeny formed colonies in each cross, except for the mating between Z1 and the flocculent European strain DBVPG4650, in which 50% of the spores dissected from each tetrad failed to yield viable colonies. For each of the remaining crosses, we cultured progeny strains in liquid culture and observed, in five cases, a 1:1 ratio of flocculent to nonflocculent morphologies (Table 1 and Figure S2). Among the progeny of each such cross, flocculation and invasive growth were coincident (Figure S2 and data not shown), indicating that for a given parent strain, a single variant locus was causal for both traits. Segregation patterns among the progeny of the flocculent, invasive European strain CBS432 suggested a polygenic model and were not investigated further.
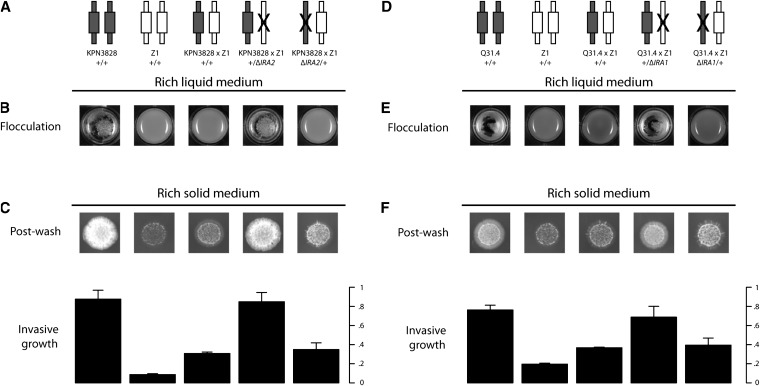
Figure 3.
Variation at IRA1 and IRA2 underlies flocculation and invasive growth. Each panel reports results of reciprocal hemizygote analysis of genetic variation at an IRA gene, between one flocculent, invasive European strain and the nonflocculent European strain Z1. Each column represents one strain, with each element in the bottom panels showing results from the strain indicated at top. (A and D) Each cartoon represents one diploid strain, with the haploid genome inherited from a flocculent parent or Z1 represented as a symbol with dark or light shading, respectively, and a solid X indicating an engineered loss-of-function of an IRA gene as indicated in labels. Δ, engineered loss-of-function allele. The fourth and fifth strains in each experiment are isogenic to one another at all loci except the IRA gene indicated, such that trait variation between them can be attributed to genetic differences at the manipulated IRA locus. (B and E) Overnight cultures in rich liquid medium as in Figure 1. (C and F) Invasive growth assays of colonies grown on solid medium as in Figure 1. Bar heights report mean invasive growth measurements of two replicate colonies and error bars represent one standard deviation.
Table 1. Two distinct Mendelian loci underlie flocculation in the S. paradoxus population.
| Straina | Proportion of flocculent recombinants from crossb | Linkage groupc |
|---|---|---|
| W7 | 0.5 (8/16) | 1 |
| Q31.4 | 0.5 (8/16) | 1 |
| KPN3829 | 0.5 (6/12) | 1 |
| KPN3828 | 0.5 (6/12) | 2 |
| CBS5829 | 0.5 (8/16) | 2 |
| CBS432 | 0.3 (6/20) | ND |
Flocculent, highly invasive European strains mated to the nonflocculent, noninvasive European strain Z1.
Results of phenotype scoring of segregants from a cross between the indicated flocculent strain and Z1. For all strains except CBS432, the invasive growth trait segregated with the flocculation trait; photographs of segregants from representative crosses are shown in Figure S2.
Results of complementation crosses among flocculent, invasive European strains. ND, not done. Photographs of segregants are shown in Figure S3.
We reasoned that if flocculation and invasive growth were a monogenic trait in a given European strain, some or all of the affected strains could share the same causal locus. To test this, we carried out complementation analyses as follows. In a cross between haploid cells of two flocculent, invasive European strains harboring variants at the same causal locus, all progeny are expected to exhibit both morphological phenotypes. In a cross between two haploids harboring causal variants at distinct loci, 25% of progeny will inherit neither causal allele and will exhibit a nonflocculent, noninvasive phenotype. From crosses of pairs of the five flocculent European strains, we inferred two linkage groups: the UK strains Q31.4 and W7 and the Siberian strain KPN3829 showed evidence for a single causative locus, distinct from that shared by a second Siberian strain, KPN3828, and the Italian strain CBS5829 (Table 1 and Figure S3). The evidence for unlinked causal loci in the two Siberian strains was particularly striking, given the >99.7% identity of these two genomes, and suggested that the determinants of flocculation and invasive growth among S. paradoxus strains could be highly mutable over short evolutionary timescales.
FLO9 and FLO11 are terminal effectors of flocculation and invasive growth
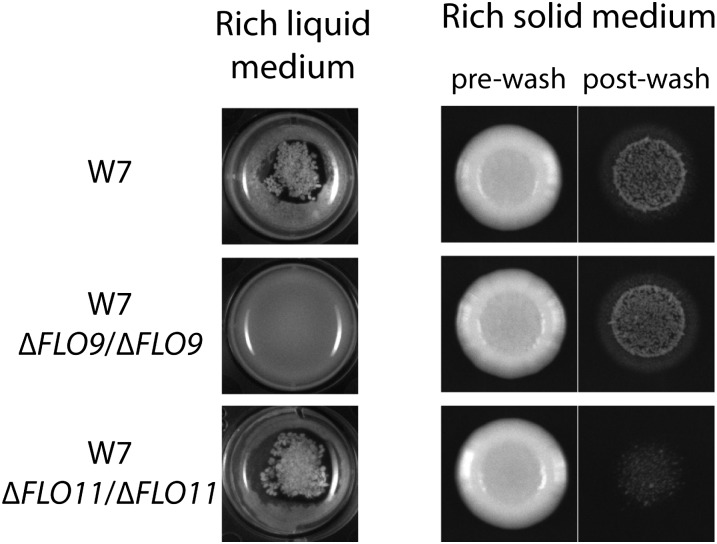
We expected that the variants underlying flocculation and invasive growth in S. paradoxus were likely to lie in genes of the adhesion or cell polarity regulatory networks, which in S. cerevisiae comprise hundreds of components (Palecek et al. 2000; Jin et al. 2008). To streamline a candidate-based search among these genes, we elected to identify terminal effectors that mediated flocculation and invasive growth in S. paradoxus. We first tested for the involvement of adhesion proteins of the flocculin (FLO) family by treating liquid cultures of several flocculent European strains with mannose, a known FLO protein inhibitor. We observed a dose-dependent inhibition of flocculation by mannose, suggesting that FLO proteins did indeed play a role in the flocculent phenotype (Figure S4). We next tested directly which members of the FLO family were required for flocculation and invasive growth by knocking out, in turn, each of the S. paradoxus orthologs of five known S. cerevisiae FLO genes in the flocculent European strain W7. Deletion of FLO9 abolished flocculation in liquid culture but had no effect on invasive growth on solid media; by contrast, a FLO11 mutant was fully flocculent but almost entirely noninvasive (Figure 2). Deletion of FLO1, FLO5, and FLO10 had no morphological effect (data not shown). We repeated these deletion experiments in the flocculent, invasive European strain CBS5829, which fell into a different linkage group from that of W7 (Table 1) and thus harbored a distinct causal variant for the morphologies. We also analyzed FLO gene deletions in a third flocculent, invasive European strain, KPN3829. The results in each case mirrored those from the W7 strain: FLO9 was required for flocculation and FLO11 for invasive growth (Figure S5). We conclude that in the context of multiple distinct genetic variants that drive morphological traits in S. paradoxus, FLO9 is an effector of flocculation and FLO11 mediates invasive growth.
Figure 2.
FLO9 and FLO11 are terminal effectors of flocculation and invasive growth. Each row reports morphologies of one homozygous derivative of the flocculent, invasive European strain W7. Δ, engineered loss-of-function allele. Left photographs show overnight cultures in rich liquid medium, and right photographs show the results of invasive growth assays of colonies grown on rich solid medium, both as in Figure 1.
Variants at IRA1 and IRA2 underlie flocculation and invasive growth
In pursuing the genetic basis of variation in flocculation and invasive growth in S. paradoxus, we reasoned that the causal polymorphisms were likely to lie in master regulators upstream of the FLO genes. In S. cerevisiae, the MAPK and PKA signaling pathways act as primary regulators of FLO gene expression and as central control points for the sensing of and response to environmental conditions (Palecek et al. 2002). To assess the role of regulators of the MAPK and PKA cascades in control of morphological phenotypes, we carried out candidate-based linkage analyses in crosses between flocculent European strains and the nonflocculent European strain Z1. In a first test of candidate genes in the flocculent European strain W7, only inheritance at IRA1, a negative regulator of Ras1/Ras2 and a homolog of the hypermutable human neurofibromatosis gene NF1 (Ballester et al. 1990; Shen et al. 1996), was correlated with flocculation and invasive growth phenotypes among progeny in a cross with Z1 (Figure S2). We likewise detected co-inheritance between IRA1 and morphology traits in the other strains of this linkage group and no evidence for linkage at other tested candidate loci (data not shown). As expected, inheritance at IRA1 was not linked to flocculation and invasive growth in KPN3828 or CBS5829, which had formed a distinct linkage group in complementation analysis (Table 1). Instead, in these two strains our candidate-based linkage analysis detected co-inheritance between both morphological traits and IRA2, an inhibitor of Ras1/Ras2 paralogous to IRA1 (Figure S2 and not shown).
To establish variants in IRA1 and IRA2 as causal for morphological phenotypes, we used the reciprocal hemizygote approach (Steinmetz et al. 2002), in which a gene of interest is deleted in each homolog in turn of a hybrid diploid to create a pair of hemizygotes that differ only at the manipulated locus. We constructed reciprocal hemizygotes at IRA genes in hybrid diploids formed by mating each flocculent, invasive European strain to Z1. Phenotyping of these diploids confirmed the IRA loci as causal for morphologies in each case: the hemizygote harboring the IRA1 or IRA2 allele from the flocculent, invasive parent strain exhibited these morphologies, and the hemizygote bearing the IRA1 or IRA2 allele from the Z1 parent was nonflocculent and minimally invasive (Figure 3 and Figure S6). This held true not only for the strains we had analyzed by linkage mapping, but also for the flocculent European strain DBVPG4650, which was refractory to linkage analyses: reciprocal hemizygote experiments revealed IRA1 to be the causal locus in this strain (Figure S6). We conclude that IRA1 underlies flocculent and invasive growth traits in strains W7, KPN3829, Q31.4, and DBVPG4650, and IRA2 underlies these traits in strains KPN3828 and CBS5829, validating our linkage and complementation analyses and highlighting IRA genes as a nexus of evolutionary change in this yeast population.
IRA1 and IRA2 variants are loss-of-function alleles of distinct evolutionary origin
Both Ira1 and Ira2 promote the GTP hydrolysis activity of Ras and facilitate the conversion of Ras1/Ras2 from a GTP-bound (active) to a GDP-bound (inactive) state (Tanaka et al. 1990). We hypothesized that the IRA1 and IRA2 alleles underlying morphological traits in S. paradoxus were likely loss-of-function variants, since such mutations in laboratory strains of S. cerevisiae deregulate Ras1/Ras2, leading to hyperactivation of the PKA signaling cascade, which in turn promotes flocculation and invasive growth (Tanaka et al. 1989; Palecek et al. 2002; Halme et al. 2004). To test this hypothesis, we disrupted the variant IRA coding region in each strain in which we had found these variants to confer flocculation and invasive growth morphologies. Phenotypes of the resulting IRA null strains differed only marginally from those of their wild-type parents (Figure S7), making clear that the IRA gene variants present in the wild S. paradoxus population are complete, or nearly complete, loss-of-function alleles. Additionally, disruption of either IRA1 or IRA2 in the nonflocculent Z1 strain resulted in a flocculent and invasive phenotype, confirming that a loss-of-function mutation in either gene is sufficient to confer these traits in S. paradoxus (Figure S8).
To investigate the evolutionary history of the IRA1 and IRA2 alleles in European S. paradoxus, we used Sanger sequencing to fill gaps and confirm polymorphisms in publicly available DNA sequence data (Liti et al. 2009), and we inferred gene trees for each IRA locus. The results revealed no consistent relationship between variant alleles in a given IRA gene; instead, the strains whose IRA gene alleles we had identified as causal for morphological phenotypes formed polyphyletic clades, reflecting the likely origin of each allele through distinct mutational events (Figure 4). Gene trees inferred from promoter regions of either gene gave similar results (Figure S9). We observed no derived alleles common to the flocculent/invasive strains and absent from the remainder of the population, further arguing against a shared evolutionary origin of the morphologies (Figure S10). Taken together, our mutational and sequence analyses supported a model in which independently derived, loss-of-function mutations in IRA1 and IRA2 have led to the convergence of six S. paradoxus strains on flocculent and invasive growth phenotypes.
Hundreds of fitness traits are associated with IRA1 and IRA2 variation
Given that the target of IRA1 and IRA2, yeast Ras, is central to metabolic and stress response signaling processes, we hypothesized that variant alleles of these genes could have wide-ranging pleiotropic effects. To investigate this, we used publicly available fitness measurements of strains derived from European S. paradoxus isolates in 199 growth conditions (Warringer et al. 2011). For each growth condition, our analyses considered lag time prior to exponential growth, doubling rate during exponential growth, and final cell density at the end of the culture, for a total of 592 environment–parameter combinations. In each case, we tested for a significant difference in fitness between European strains whose flocculent and invasive morphologies we had established to be the result of variation at IRA1 or IRA2 and nonflocculent, noninvasive European strains. This analysis identified 371 cases in which a growth trait was significantly associated with mutations in IRA1 or IRA2 (false discovery rate <0.05; Figure S11 and File S1). Associations were apparent in a variety of conditions, including alternative carbon and nitrogen sources and toxin treatments. Flocculent and invasive strains bearing variants in IRA1 and IRA2 had advantages with respect to some growth parameters and conditions and defects in others (Figure S11). Thus, IRA1 and IRA2 variants were associated with hundreds of differences in growth traits, implicating these loci as global correlates of phenotype across S. paradoxus strains.
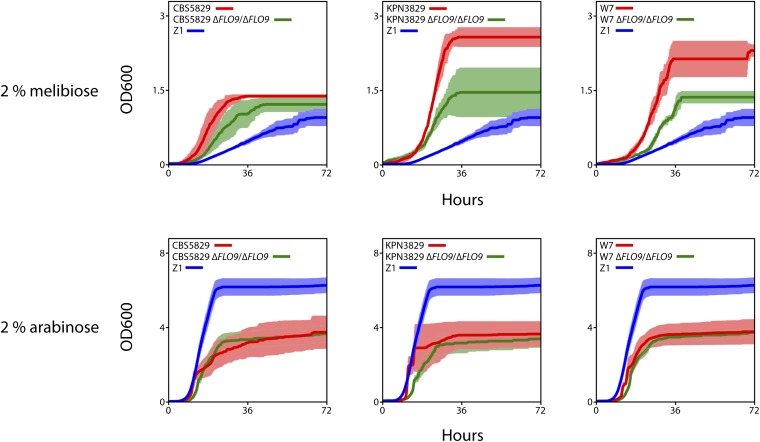
To validate the role of IRA1 and IRA2 mutations in growth phenotypes across S. paradoxus, we used the phenotypic profiles in Figure S11 to identify traits likely to be affected by IRA gene variants, and we evaluated these predictions in reciprocal hemizygote assays. We grew reciprocal hemizygote strain pairs interrogating the IRA genes, and the respective parent strains, in eight environmental conditions and measured growth parameters in each case. The results revealed extensive differences between the hemizygotes of strain pairs (Figure 5, Figure S12, and File S2), confirming sequence variation at IRA1 and IRA2 as causal for the majority of growth traits tested. In most cases, the strains of a reciprocal hemizygote pair mirrored differences between the respective parents, and the effect of an IRA gene variant in one hemizygote pair usually phenocopied the effects in other pairs (Figure 5 and Figure S12). The most marked benefits of IRA1 and IRA2 mutations were to growth rates (Figure 5), and the most uniformly deleterious effects were on cell densities reached by cultures after nutrient exhaustion (Figure S12). These results establish variants in IRA1 and IRA2 as toggles between distinct phenotypic states in S. paradoxus, underlying growth benefits and disadvantages in a range of conditions and genetic backgrounds.
Growth traits in strains bearing IRA gene mutations can be independent of morphology
Given that cell clumping can affect growth and viability (Halme et al. 2004), we asked whether variation in growth traits across S. paradoxus could be attributed to mechanisms independent of morphology. For this purpose, in each of three flocculent European strains bearing variants in IRA1 and IRA2, we eliminated flocculation by mutating the effector FLO9, and we compared growth in these nonflocculent strains to that in isogenic controls across a panel of conditions. The results identified growth traits fully or partly independent of flocculation (Figure 6, Figure S13, and File S3). For example, in all strains tested, IRA variant alleles compromised the maximal cell density attainable by cultures at stationary phase in the presence of 2% arabinose (Figure S12), and this fitness defect was unaffected by FLO9 mutation (Figure 6 and Figure S13). Likewise, in cultures using melibiose as a carbon source, IRA variant alleles conferred an advantage in several growth parameters (Figure 5 and Figure S12), which persisted, although in some cases with reduced magnitude, in nonflocculent FLO9 mutant backgrounds (Figure 6 and Figure S13). Interestingly, in a few conditions the growth effects of FLO9 mutation differed across strains, likely reflecting the action of modifier loci segregating in the population (Figure S13). We conclude that flocculation contributes to, but cannot fully explain, the growth profile of strains bearing variant alleles at IRA1 and IRA2, highlighting the pleiotropic effects of these polymorphisms on yeast growth and stress resistance.
Figure 6.
Fitness of strains bearing variant IRA gene alleles is partly or completely independent of flocculation. Each column of panels shows the results of growth experiments with one flocculent homozygote European strain, its engineered nonflocculent derivative, and the nonflocculent European strain Z1. Each row of panels shows results from cultures grown in one environmental condition. In a given panel, each trace reports the time course of growth of one strain, with solid lines reporting the mean of three replicate cultures and error bars indicating one standard deviation. Δ, engineered loss-of-function allele. OD600, optical density of cultures at 600 nm. Raw measurements and significance estimates are given in File S3.
Variant IRA1 and IRA2 alleles are present in wild yeast
In light of previous reports of rapid evolution of IRA1 and IRA2 in laboratory S. cerevisiae (Halme et al. 2004), we aimed to trace the history of these loci during the processing of S. paradoxus strains from wild collection to laboratory derivatives. For each of the six wild-derived strains in which we had identified IRA gene variants, we obtained the originally isolated wild progenitor diploid and cultured it with minimal passaging. Five of these cultures (progenitors of CBS5829, DBVPG4650, KPN3828, KPN3829, and Q31.4) displayed a flocculent and invasive growth phenotype, confirming the prevalence of these phenotypes in wild yeast populations (Figure S14). The sixth strain, the progenitor of W7, was nonflocculent and minimally invasive (Figure S14). Progeny from sporulation of this strain were nonflocculent and noninvasive (data not shown), ruling out heterozygosity of a loss-of-function allele in the wild diploid and establishing that the mutation had been acquired after introduction into the laboratory in this case.
To investigate the genetic basis of morphological traits in wild, flocculent progenitor strains, we mated a haploid from each of three such strains to the nonflocculent strain Z1, with the remaining two, KPN3828 and Q31.4, refractory to repeated mating attempts. In each of the three hybrids successfully generated between wild progenitors and Z1, sporulation revealed a pattern of Mendelian inheritance of the flocculation phenotype (Figure S15). Reciprocal hemizygote experiments implicated variation at IRA1 in the progenitor strain of KPN3829 and at IRA2 in the progenitor strain of CBS5829, consonant with our analyses of their laboratory derivatives (Figure S6 and Figure S16). Interestingly, in reciprocal hemizygote analysis of the mating between Z1 and the progenitor of DBVPG4650, we validated IRA2 as causal for flocculation, in contrast to our discovery of IRA1 as causal in the monosporic laboratory derivative of this strain (Figure S6 and Figure S16), which could reflect heterozygosity at both IRA1 and IRA2 in the wild DBVPG4650 diploid. Taken together, these data make clear that flocculation and invasive growth segregate among wild S. paradoxus and can be attributed to variation at IRA1 and IRA2, underscoring the relevance to yeast evolution of our observations in laboratory derivative strains.
Discussion
The search for the molecular basis of common trait variation dominates the modern study of genetics. In Saccharomyces yeasts, although elegant linkage studies have mapped phenotypic differences between pairs of genetically distinct strains (Steinmetz et al. 2002; Gerke et al. 2009; Kim and Fay 2009; Lee et al. 2011; Parts et al. 2011; Ehrenreich et al. 2012; Bloom et al. 2013), the determinants of common phenotypes, and the evolutionary forces that underlie them, are less well understood (Hittinger et al. 2010; Will et al. 2010; Warringer et al. 2011). In this work we have characterized a panel of flocculent and invasive, but otherwise unrelated, European S. paradoxus strains. Our results showed flocculation and invasive growth to be linked Mendelian traits, caused by loss-of-function alleles of either IRA1 or IRA2 that are of distinct evolutionary origin. Because each independent allele of IRA1 and IRA2 largely phenocopies the others, in toto they represent a suite of rare variants that underlie common traits in the S. paradoxus population.
Evolvability of IRA1 and IRA2
The independent loss-of-function alleles of IRA1 and IRA2 we have uncovered in S. paradoxus echo previous reports of mutations in these genes in wild and laboratory S. cerevisiae, including some acquired during the course of experimental culture (Halme et al. 2004; Kao and Sherlock 2008; Smith and Kruglyak 2008; Parts et al. 2011). The recurrent focus on IRA gene variants in the yeast literature a priori could reflect ascertainment bias, given the dramatic phenotypes of these alleles. However, in unbiased sequence analyses, we detected elevated nucleotide diversity at IRA1 and IRA2 in most S. paradoxus populations (Table S2), consistent with either hypermutability or relaxed selection at these loci. Because the striking fitness consequences of IRA gene mutations render the latter model unlikely, the most compelling interpretation of results in the field is as evidence for hypermutability at IRA1 and IRA2. These findings parallel the pattern of widespread de novo variation in the human homolog, NF1, which underlies susceptibility to neurofibromatosis (Ballester et al. 1990; Shen et al. 1996; Ars et al. 2000), raising the possibility that the mechanism for hypermutability at these loci may be shared between yeast and human. Our work leaves open the question of which nucleotide changes, among the dozens segregating in the population, underlie growth and morphology behaviors. Sequence analyses identified a nonsense mutation in IRA1 in both the progenitor and the laboratory derivative of Q31.4 (Figure S10), representing a plausible causal variant in this background. Likewise, in several other cases, IRA genes of wild flocculent progenitor strains harbored one or more private nonsynonymous or regulatory mutations, which may underlie the observed growth and morphology phenotypes (Figure S10).
If IRA1 and IRA2 are prone to mutation, are variants at these loci evolutionarily relevant? Our work makes clear that IRA gene polymorphisms confer fitness benefits and disadvantages in a range of environmental conditions. And as a complement to landmark studies of the advantages of morphological traits in S. cerevisiae (Smukalla et al. 2008; Koschwanez et al. 2011), our results show that growth traits in S. paradoxus strains harboring mutations at the IRA loci can be independent of cell aggregation. The strong and pleiotropic effects of IRA gene mutations set them apart from rare, recessive growth defects (Doniger et al. 2008; Zörgö et al. 2012) and weakly deleterious variants (Li et al. 2008; Liti et al. 2009; Elyashiv et al. 2010) thought to be maintained in yeast populations as a consequence of their peculiar demography. Instead, IRA1 and IRA2 exhibit the features of contingency loci (Moxon et al. 1994), whose hypermutability serves as a bet-hedging strategy. Under this model, constantly arising mutations at the IRA genes would provide short-term benefits in some environments and be eliminated or compensated for when conditions change (Halme et al. 2004). A specialist role for mutants at the IRA genes is further suggested by the loss of IRA1 function arising under laboratory selection for tolerance to low glucose (Kao and Sherlock 2008). In contrast to the classic understanding of diversifying selection (Murrell et al. 2012), the distinct wild alleles we have cataloged at the IRA genes all yield very similar, convergent phenotypes. Thus, in addition to the phenotypic switches driven by stochastic noise in biochemical processes and by protein aggregation (Acar et al. 2005; Halfmann et al. 2012; Li and Kowal 2012), our work establishes rapid evolution of master regulators as a mechanism of phenotype switching in yeast.
Mechanisms of IRA1 and IRA2 variant phenotypes
In investigating morphologies of S. paradoxus, we identified FLO11 as an effector of invasive growth, dovetailing with the roles of this gene in multicellularity behaviors of S. cerevisiae (Lo and Dranginis 1996; Lambrechts et al. 1996; Reynolds and Fink 2001). By contrast, our dissection of flocculation in S. paradoxus implicated FLO9 as the major effector, a gene whose functional role in wild S. cerevisiae is unknown (Van Mulders et al. 2009). The emerging picture from our work and that of others (Verstrepen et al. 2005; Douglas et al. 2007; Smukalla et al. 2008; Govender et al. 2011) is one in which the activity of flocculin genes and the traits they underlie are highly variable on short evolutionary timescales. Our findings make clear that morphology traits driven by the FLO genes have fitness benefits in certain growth conditions, plausibly mediated by the increased efficiency of sugar uptake observed in clumping yeast cultures (Koschwanez et al. 2011) and protection of cells in the interior of flocs from soluble chemical stressors (Smukalla et al. 2008).
Additional fitness effects of IRA gene variants are likely to be mediated by other downstream targets of the cAMP/PKA pathway. We expect that poor growth and low cell density in stationary phase, which we observe in S. paradoxus IRA1 and IRA2 variants in many stress conditions, result from the repression of protective stress responses seen in hyperactive PKA mutants (Markwardt et al. 1995; Smith et al. 1998; Budovskaya et al. 2004) as well as the disadvantages of flocculation in late phases of growth (Smukalla et al. 2008). Some growth advantages that we have mapped to IRA gene variants, including resistance to cobalt chloride, a known hypoxia mimic (Kwast et al. 1999), are likely tied to the increased respiratory capacity of hyperactive Ras mutants (Dejean et al. 2002). Our findings of rapid growth by IRA1 and IRA2 variants in some conditions after transfer from nutrient-poor stationary phase may be a consequence of the metabolic program activated by unregulated Ras1 and Ras2, which in wild-type cells is associated with starved cultures upon addition of glucose (Wang et al. 2004; Santangelo 2006). It is also tempting to speculate that the release of checks on cell growth may also underlie some of the advantages we observe in IRA gene variants, at least on the relatively short timescales we analyze here.
Mapping rare variants that underlie common traits
Whether rare variants underlie common traits is one of the most controversial questions in the current genetics literature (Dickson et al. 2010; McClellan and King 2010; Wray et al. 2011), with a few landmark studies implicating hypermutable loci as the determinants of common phenotypes (Shen et al. 1996; Chan et al. 2010; Michaelson et al. 2012). Our work establishes the hypermutable IRA genes as drivers of a broad swath of yeast phenotypes. Such a central role for mutational hotspots in yeast trait variation would be consistent with the evidence for allelic series at other loci in recent mapping studies (Ehrenreich et al. 2012). In the ongoing search for such loci in populations, our results underscore the power of a candidate-based approach, drawing on knowledge of gene networks and population-genomic data to pinpoint rare variants with sizeable fitness consequences. With the increasing availability of sequence compendia and functional-genetic resources, this strategy holds promise for application to common traits in many organisms.
Supplementary Material
Acknowledgments
We thank Jasper Rine and Sarah Bissonette for assistance with automated growth assays; Gianni Liti for generously providing sequence data before publication; Vassiliki Koufopanou, Hana Lee, Perihan Saygin, and Jonas Warringer for helpful discussions; Gianni Liti and Vassiliki Koufopanou for strains; and Chris Ellison, Leonid Kruglyak, Hilary Martin, and Joshua Schraiber for insightful comments on the manuscript.
Footnotes
Communicating editor: M. Johnston
Literature Cited
- Acar M., Becskei A., van Oudenaarden A., 2005. Enhancement of cellular memory by reducing stochastic transitions. Nature 435: 228–232. [DOI] [PubMed] [Google Scholar]
- Ars E., Serra E., García J., Kruyer H., Gaona A., et al. , 2000. Mutations affecting mRNA splicing are the most common molecular defects in patients with neurofibromatosis type 1. Hum. Mol. Genet. 9: 237–247. [DOI] [PubMed] [Google Scholar]
- Atwell S., Huang Y. S., Vilhjálmsson B. J., Willems G., Horton M., et al. , 2010. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465: 627–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballester R., Marchuk D., Boguski M., Saulino A., Letcher R., et al. , 1990. The NF1 locus encodes a protein functionally related to mammalian GAP and yeast IRA proteins. Cell 63: 851–859. [DOI] [PubMed] [Google Scholar]
- Bayly J. C., Douglas L. M., Pretorius I. S., Bauer F. F., Dranginis A. M., 2005. Characteristics of Flo11-dependent flocculation in Saccharomyces cerevisiae. FEMS Yeast Res. 5: 1151–1156. [DOI] [PubMed] [Google Scholar]
- Bester M. C., Pretorius I. S., Bauer F. F., 2006. The regulation of Saccharomyces cerevisiae FLO gene expression and Ca2+-dependent flocculation by Flo8p and Mss11p. Curr. Genet. 49: 375–383. [DOI] [PubMed] [Google Scholar]
- Bloom J. S., Ehrenreich I. M., Loo W. T., Lite T.-L. V., Kruglyak L., 2013. Finding the sources of missing heritability in a yeast cross. Nature 494: 234–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachi B., Faure N., Horton M., Flahauw E., Vazquez A., et al. , 2010. Linkage and association mapping of Arabidopsis thaliana flowering time in nature. PLoS Genet. 6: e1000940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budovskaya Y. V., Stephan J. S., Reggiori F., Klionsky D. J., Herman P. K., 2004. The Ras/cAMP-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J. Biol. Chem. 279: 20663–20671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carstens, E., and M. Lambrechts, 1998 Flocculation, pseudohyphal development and invasive growth in commercial wine yeast strains. S. Afr. J. Enol. Vitic. 19: 52–61. [Google Scholar]
- Chan E. K. F., Rowe H. C., Corwin J. A., Joseph B., Kliebenstein D. J., 2011. Combining genome-wide association mapping and transcriptional networks to identify novel genes controlling glucosinolates in Arabidopsis thaliana. PLoS Biol. 9: e1001125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y. F., Marks M. E., Jones F. C., Villarreal G., Shapiro M. D., et al. , 2010. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science 327: 302–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubillos F. A., Billi E., Zörgö E., Parts L., Fargier P., et al. , 2011. Assessing the complex architecture of polygenic traits in diverged yeast populations. Mol. Ecol. 20: 1401–1413. [DOI] [PubMed] [Google Scholar]
- Dejean L., Beauvoit B., Alonso A.-P., Bunoust O., Guérin B., et al. , 2002. cAMP-induced modulation of the growth yield of Saccharomyces cerevisiae during respiratory and respiro-fermentative metabolism. Biochim. Biophys. Acta 1554: 159–169. [DOI] [PubMed] [Google Scholar]
- Dengis P. B., Nélissen L. R., Rouxhet P. G., 1995. Mechanisms of yeast flocculation: comparison of top- and bottom-fermenting strains. Appl. Environ. Microbiol. 61: 718–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson S. P., Wang K., Krantz I., Hakonarson H., Goldstein D. B., 2010. Rare variants create synthetic genome-wide associations. PLoS Biol. 8: e1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doniger S. W., Kim H. S., Swain D., Corcuera D., Williams M., et al. , 2008. A catalog of neutral and deleterious polymorphism in yeast. PLoS Genet. 4: e1000183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas L. M., Li L., Yang Y., Dranginis A. M., 2007. Expression and characterization of the flocculin Flo11/Muc1, a Saccharomyces cerevisiae mannoprotein with homotypic properties of adhesion. Eukaryot. Cell 6: 2214–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn B., Paulish T., Stanbery A., Piotrowski J., Koniges G., et al. , 2013. Recurrent rearrangement during adaptive evolution in an interspecific yeast hybrid suggests a model for rapid introgression. PLoS Genet. 9: e1003366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenreich I. M., Bloom J., Torabi N., Wang X., Jia Y., et al. , 2012. Genetic architecture of highly complex chemical resistance traits across four yeast strains. PLoS Genet. 8: e1002570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elyashiv E., Bullaughey K., Sattath S., Rinott Y., Przeworski M., et al. , 2010. Shifts in the intensity of purifying selection: an analysis of genome-wide polymorphism data from two closely related yeast species. Genome Res. 20: 1558–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B., Green P., 1998. Base-calling of automated sequencer traces usingPhred. II. Error probabilities. Genome Res. 8: 186–194. [PubMed] [Google Scholar]
- Filiault D. L., Maloof J. N., 2012. A genome-wide association study identifies variants underlying the Arabidopsis thaliana shade avoidance response. PLoS Genet. 8: e1002589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerke J., Lorenz K., Cohen B., 2009. Genetic interactions between transcription factors cause natural variation in yeast. Science 323: 498–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govender P., Kroppenstedt S., Bauer F. F., 2011. Novel wine-mediated FLO11 flocculation phenotype of commercial Saccharomyces cerevisiae wine yeast strains with modified FLO gene expression. FEMS Microbiol. Lett. 317: 117–126. [DOI] [PubMed] [Google Scholar]
- Guo B., Styles C. A., Feng Q., Fink G. R., 2000. A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc. Natl. Acad. Sci. USA 97: 12158–12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güldener U., Heck S., Fiedler T., Beinhauer J., Hegemann J. H., 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24: 2519–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann R., Jarosz D. F., Jones S. K., Chang A., Lancaster A. K., et al. , 2012. Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature 482: 363–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halme A., Bumgarner S., Styles C., Fink G. R., 2004. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell 116: 405–415. [DOI] [PubMed] [Google Scholar]
- Hittinger C. T., Goncalves P., Sampaio J. P., Dover J., Johnston M., et al. , 2010. Remarkably ancient balanced polymorphisms in a multi-locus gene network. Nature 464: 54–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin R., Dobry C. J., McCown P. J., Kumar A., 2008. Large-scale analysis of yeast filamentous growth by systematic gene disruption and overexpression. Mol. Biol. Cell 19: 284–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao K. C., Sherlock G., 2008. Molecular characterization of clonal interference during adaptive evolution in asexual populations of Saccharomyces cerevisiae. Nat. Genet. 40: 1499–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. S., Fay J. C., 2009. A combined-cross analysis reveals genes with drug-specific and background-dependent effects on drug sensitivity in Saccharomyces cerevisiae. Genetics 183: 1141–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koschwanez J. H., Foster K. R., Murray A. W., 2011. Sucrose utilization in budding yeast as a model for the origin of undifferentiated multicellularity. PLoS Biol. 9: e1001122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwast K. E., Burke P. V., Staahl B. T., Poyton R. O., 1999. Oxygen sensing in yeast: evidence for the involvement of the respiratory chain in regulating the transcription of a subset of hypoxic genes. Proc. Natl. Acad. Sci. USA 96: 5446–5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts M. G., Bauer F. F., Marmur J., Pretorius I. S., 1996. Muc1, a mucin-like protein that is regulated by Mss10, is critical for pseudohyphal differentiation in yeast. Proc. Natl. Acad. Sci. USA 93: 8419–8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. N., Magwene P. M., Brem R. B., 2011. Natural variation in CDC28 underlies morphological phenotypes in an environmental yeast isolate. Genetics 188: 723–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Kowal A. S., 2012. Environmental regulation of prions in yeast. PLoS Pathog. 8: e1002973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. F., Costello J. C., Holloway A. K., Hahn M. W., 2008. “Reverse ecology” and the power of population genomics. Evolution 62: 2984–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G., Carter D. M., Moses A. M., Warringer J., Parts L., et al. , 2009. Population genomics of domestic and wild yeasts. Nature 458: 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo W. S., Dranginis A. M., 1996. FLO11, a yeast gene related to the STA genes, encodes a novel cell surface flocculin. J. Bacteriol. 178: 7144–7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F. C., Richards S., Stone E. A., Barbadilla A., Ayroles J. F., et al. , 2012. The Drosophila melanogaster Genetic Reference Panel. Nature 482: 173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magwire M. M., Fabian D. K., Schweyen H., Cao C., Longdon B., et al. , 2012. Genome-wide association studies reveal a simple genetic basis of resistance to naturally coevolving viruses in Drosophila melanogaster. PLoS Genet. 8: e1003057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markwardt D. D., Garrett J. M., Eberhardy S., Heideman W., 1995. Activation of the Ras/cyclic AMP pathway in the yeast Saccharomyces cerevisiae does not prevent G1 arrest in response to nitrogen starvation. J. Bacteriol. 177: 6761–6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy M. I., Abecasis G. R., Cardon L. R., Goldstein D. B., Little J., et al. , 2008. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 9: 356–369. [DOI] [PubMed] [Google Scholar]
- McClellan J., King M.-C., 2010. Genetic heterogeneity in human disease. Cell 141: 210–217. [DOI] [PubMed] [Google Scholar]
- Michaelson J. J., Shi Y., Gujral M., Zheng H., Malhotra D., et al. , 2012. Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell 151: 1431–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moxon E. R., Rainey P. B., Nowak M. A., Lenski R. E., 1994. Adaptive evolution of highly mutable loci in pathogenic bacteria. Curr. Biol. 4: 24–33. [DOI] [PubMed] [Google Scholar]
- Murrell B., Wertheim J. O., Moola S., Weighill T., Scheffler K., et al. , 2012. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 8: e1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Li W. H., 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 76: 5269–5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palecek S. P., Parikh A. S., Kron S. J., 2000. Genetic analysis reveals that FLO11 upregulation and cell polarization independently regulate invasive growth in Saccharomyces cerevisiae. Genetics 156: 1005–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palecek S. P., Parikh A. S., Kron S. J, 2002. Sensing, signalling and integrating physical processes during Saccharomyces cerevisiae invasive and filamentous growth. Microbiology 148: 893–907. [DOI] [PubMed] [Google Scholar]
- Parts L., Cubillos F. A., Warringer J., Jain K., Salinas F., et al. , 2011. Revealing the genetic structure of a trait by sequencing a population under selection. Genome Res. 21: 1131–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M. N., Dehal P. S., Arkin A. P., 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS ONE 5: e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds T. B., Fink G. R., 2001. Bakers’ yeast, a model for fungal biofilm formation. Science 291: 878–881. [DOI] [PubMed] [Google Scholar]
- Ryan O., Shapiro R. S., Kurat C. F., Mayhew D., Baryshnikova A., et al. , 2012. Global gene deletion analysis exploring yeast filamentous growth. Science 337: 1353–1356. [DOI] [PubMed] [Google Scholar]
- Sampermans S., Mortier J., Soares E. V., 2005. Flocculation onset in Saccharomyces cerevisiae: the role of nutrients. J. Appl. Microbiol. 98: 525–531. [DOI] [PubMed] [Google Scholar]
- Santangelo G. M., 2006. Glucose signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70: 253–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M. H., Harper P. S., Upadhyaya M., 1996. Molecular genetics of neurofibromatosis type 1 (NF1). J. Med. Genet. 33: 2–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit G., Straver M. H., Lugtenberg B. J., Kijne J. W., 1992. Flocculence of Saccharomyces cerevisiae cells is induced by nutrient limitation, with cell surface hydrophobicity as a major determinant. Appl. Environ. Microbiol. 58: 3709–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A., Ward M. P., Garrett S., 1998. Yeast PKA represses Msn2p/Msn4p-dependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J. 17: 3556–3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E. N., Kruglyak L., 2008. Gene–environment interaction in yeast gene expression. PLoS Biol. 6: e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smukalla S., Caldara M., Pochet N., Beauvais A., Guadagnini S., et al. , 2008. FLO1 is a variable green beard gene that drives biofilm-like cooperation in budding yeast. Cell 135: 726–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares E. V., 2011. Flocculation in Saccharomyces cerevisiae: a review. J. Appl. Microbiol. 110: 1–18. [DOI] [PubMed] [Google Scholar]
- Steinmetz L. M., Sinha H., Richards D. R., Spiegelman J. I., Oefner P. J., et al. , 2002. Dissecting the architecture of a quantitative trait locus in yeast. Nature 416: 326–330. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Matsumoto K., Toh-E A., 1989. IRA1, an inhibitory regulator of the RAS-cyclic AMP pathway in Saccharomyces cerevisiae. Mol. Cell. Biol. 9: 757–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Nakafuku M., Satoh T., Marshall M. S., Gibbs J. B., et al. , 1990. S. cerevisiae genes IRA1 and IRA2 encode proteins that may be functionally equivalent to mammalian ras GTPase activating protein. Cell 60: 803–807. [DOI] [PubMed] [Google Scholar]
- Van Mulders S. E., Christianen E., Saerens S. M. G., Daenen L., Verbelen P. J., et al. , 2009. Phenotypic diversity of Flo protein family-mediated adhesion in Saccharomyces cerevisiae. FEMS Yeast Res. 9: 178–190. [DOI] [PubMed] [Google Scholar]
- Veltman J. A., Brunner H. G., 2012. De novo mutations in human genetic disease. Nat. Rev. Genet. 13: 565–575. [DOI] [PubMed] [Google Scholar]
- Verstrepen K. J., Jansen A., Lewitter F., Fink G. R., 2005. Intragenic tandem repeats generate functional variability. Nat. Genet. 37: 986–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinod P. K., Sengupta N., Bhat P. J., Venkatesh K. V., 2008. Integration of global signaling pathways, cAMP-PKA, MAPK and TOR in the regulation of FLO11. PLoS ONE 3: e1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Pierce M., Schneper L., Güldal C. G., Zhang X., et al. , 2004. Ras and Gpa2 mediate one branch of a redundant glucose signaling pathway in yeast. PLoS Biol. 2: e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warringer J., Zörgö E., Cubillos F. A., Zia A., Gjuvsland A., et al. , 2011. Trait variation in yeast is defined by population history. PLoS Genet. 7: e1002111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A. M., Procter J. B., Martin D. M. A., Clamp M., Barton G. J., 2009. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A. L., Khan G. F., Magwire M. M., Tabor C. L., Mackay T. F. C., et al. , 2012. Genome-wide association analysis of oxidative stress resistance in Drosophila melanogaster. PLoS ONE 7: e34745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will J. L., Kim H. S., Clarke J., Painter J. C., Fay J. C., et al. , 2010. Incipient balancing selection through adaptive loss of aquaporins in natural Saccharomyces cerevisiae populations. PLoS Genet. 6: e1000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray N. R., Purcell S. M., Visscher P. M., 2011. Synthetic associations created by rare variants do not explain most GWAS results. PLoS Biol. 9: e1000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman S., Lippman S. I., Zhao X., Broach J. R., 2008. How Saccharomyces responds to nutrients. Annu. Rev. Genet. 42: 27–81. [DOI] [PubMed] [Google Scholar]
- Zörgö E., Gjuvsland A., Cubillos F. A., Louis E. J., Liti G., et al. , 2012. Life history shapes trait heredity by accumulation of loss-of-function alleles in yeast. Mol. Biol. Evol. 29: 1781–1789. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.