Abstract
The 50th anniversary of the classic Monod–Wyman–Changeux (MWC) model provides an opportunity to survey the broader conceptual and quantitative implications of this quintessential biophysical model. With the use of statistical mechanics, the mathematical implementation of the MWC concept links problems that seem otherwise to have no ostensible biological connection including ligand–receptor binding, ligand-gated ion channels, chemotaxis, chromatin structure and gene regulation. Hence, a thorough mathematical analysis of the MWC model can illuminate the performance limits of a number of unrelated biological systems in one stroke. The goal of our review is twofold. First, we describe in detail the general physical principles that are used to derive the activity of MWC molecules as a function of their regulatory ligands. Second, we illustrate the power of ideas from information theory and dynamical systems for quantifying how well the output of MWC molecules tracks their sensory input, giving a sense of the “design” constraints faced by these receptors.
Keywords: Monod–Wyman–Changeux model, allostery, chromatin structure, bacterial chemotaxis
Introduction
Modern biology has garnered deep insights from a large collection of “model systems” ranging from specific molecules such as hemoglobin, Lac repressor and the nicotinic acetylcholine (nACh) receptor to organisms such as Escherichia coli and its phages to the fruit fly Drosophila melanogaster and beyond.1,2 Studies of “model” molecules such as hemoglobin have given rise, in turn, to very general statistical mechanical models that provide a simple link between the structural conformation of these molecules and their regulation by external ligands. One such model, the Monod–Wyman–Changeux (MWC) model,3,4 is beginning to assume similar proportions in biology to those of the Ising model in physics, which has been used to explain diverse phenomena ranging from magnetism to the liquid–gas transition.5 As we describe in this article, the MWC model sheds light on a similarly broad swath of biological phenomena.
A signature feature of any powerful model is its ability to make convincing connections between apparently unrelated phenomena. A simple search on the initials “MWC” on PubMed reveals the vast array of different molecular situations in which researchers have appealed to the MWC model as an instructive conceptual framework. A similar search on the Web of Science in December of 2012 reveals 1517 unique citations for the 1963 paper of Monod, Changeux and Jacob and 6086 unique citations for the 1965 paper of Monod, Wyman and Changeux.3,4 In speaking of the papers that introduced these ideas, Monod noted “The first paper was, really, on the idea of indirect regulation. Which I think is the really important idea. The second paper is a physical-chemical interpretation of this fact in terms of the geometry of the molecule”.6 In our paper, we hope to explain why Monod christened the idea of indirect regulation embodied in the MWC model “the second secret of life”6 by mathematically examining the general implications of the indirect regulation concept and its implementation in statistical mechanical language. It is interesting to note that, of the two papers, the second paper4 is more cited than the first,3 despite Monod's claim of the greater importance of the former.
Indirect regulation, the subject of the first of this important pair of papers and a key feature of the MWC model, arises when a macromolecule of interest has two classes of conformational states, which we will refer to generically as the inactive and active states. The molecular decision of whether or not the macromolecule is active is dictated by the binding of some regulatory ligand or ligands that bind preferentially to one state over the other, thereby tilting the balance between the inactive and active states.2,4,7,8 As we will see below, in the simplest variant of the model, there are thus four distinct states, active and inactive both with and without ligand. This simplest picture can serve as the starting point for a whole suite of more complex models involving multiple binding sites and hence, cooperativity, different sets of intermediate states and multiple ligands, for example.
The organization of the paper is as follows. In States and Weights in the MWC Setting, we show how “states-and-weights” diagrams illustrate all of the different microscopic states available to an MWC molecule and how statistical mechanics can be used to assign “statistical weights” to each such state. With these results in hand, we show how to compute the activity of an MWC molecule as a function of the concentration of its governing ligand. In Case Studies in MWC Thinking, we highlight a few of the many biological processes that can be described by an MWC model, including ligand-gated ion channels, two-component signal transduction systems and gene regulation. Some of the unexpected predictions that arise from such models are highlighted in these various examples. In An Information-Theoretic Perspective of the MWC Concept, we then consider how information theory can be used to characterize the ability of MWC molecules to “read” the state of their environment and to convert it into cellular decisions. In Dynamical MWC, we introduce a simplified model of dynamics for MWC molecules and analyze how well such molecules “read” an environment whose state is changing in time. In Discussion, we close by reflecting on the MWC concept as viewed through the prism of “toy models” in statistical mechanics and how it provides a powerful complement to more microscopically realistic perspectives that have emerged from structural biology. Some of the detailed steps of our derivations are presented in several appendices that follow the reference list. We consider these appendices an important part of the overall review since they provide details for results used in the literature that are rarely presented pedagogically, if at all.
The references cited throughout the paper are intended to provide an entry into the vast literature on the MWC concept with special emphasis on how physical scientists have exploited the model. Specifically, we place less emphasis on fitting the data for one particular molecule and more emphasis on the general features of such molecules and the physical constraints that they must face. Given the more than 7000 citations of the two papers that launched the MWC world view, it is no surprise that our list of references is representative rather than complete, and we apologize in advance to those whose important work has been neglected.
States and Weights in the MWC Setting
For the purposes of this article, we define an MWC molecule as having two classes of structural states.3,4 In the case of hemoglobin, for example, these states correspond to the famed “T” (tense) and “R” (relaxed). For ligand-gated channels such as the nACh receptor or cyclic guanosine monophosphate (cGMP)-gated ion channels such as those found in photoreceptors, these two states correspond to the closed and open states of the channel.9 These same ideas can be used even farther afield such as to describe different structural states of chromatin in which the DNA molecule is either “inaccessible” or “accessible”.10,11 In this section, we will denote the two states of the receptor by “I” and “A”, referring to the inactive state and to the active state, respectively. Our goals in this section are twofold. First, we aim to discuss indirect regulation and cooperativity in the MWC model. Second, and perhaps more importantly, we aim to provide a clear recipe for converting diagrammatic descriptions of ligand–receptor binding (“states diagrams”) into testable equations. To achieve the latter goal, we begin this section by describing the noncooperative one-site MWC molecule. Then, we focus our attention on the general n-site MWC molecule from which we can understand how the indirect regulation inherent in the MWC model can give rise to apparently cooperative interactions.
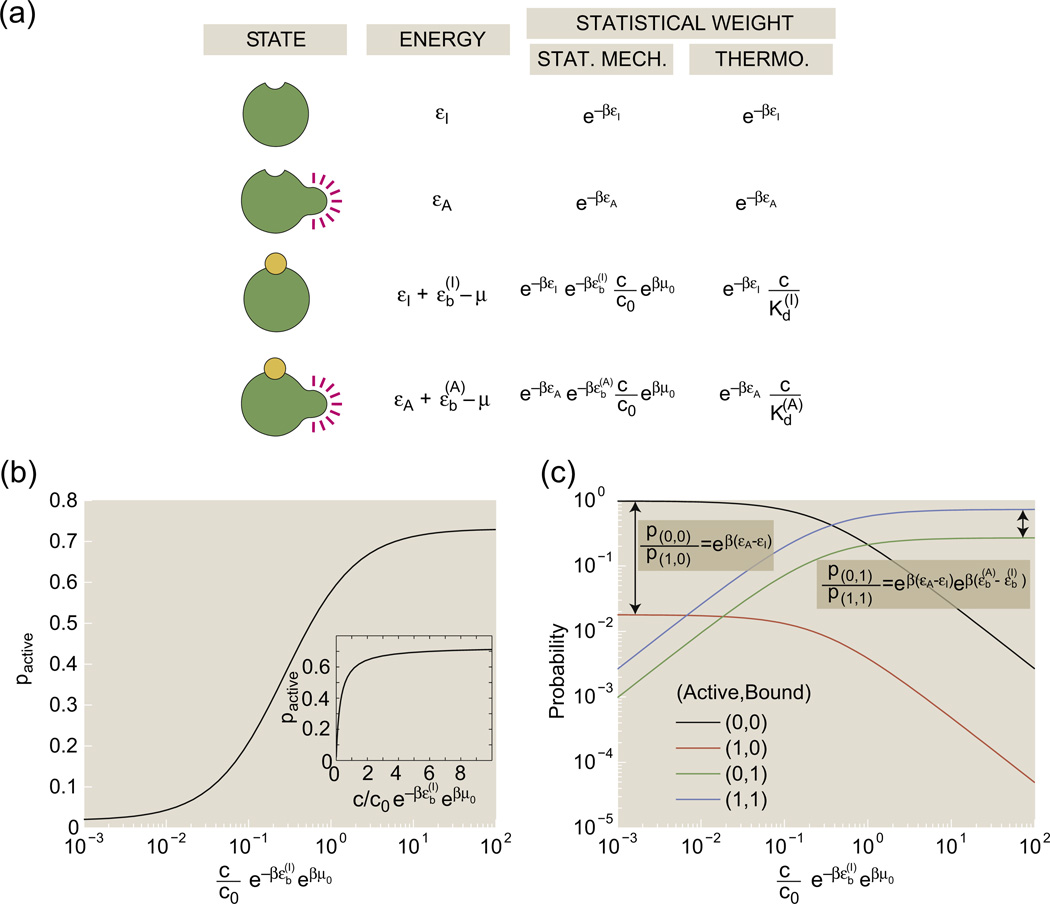
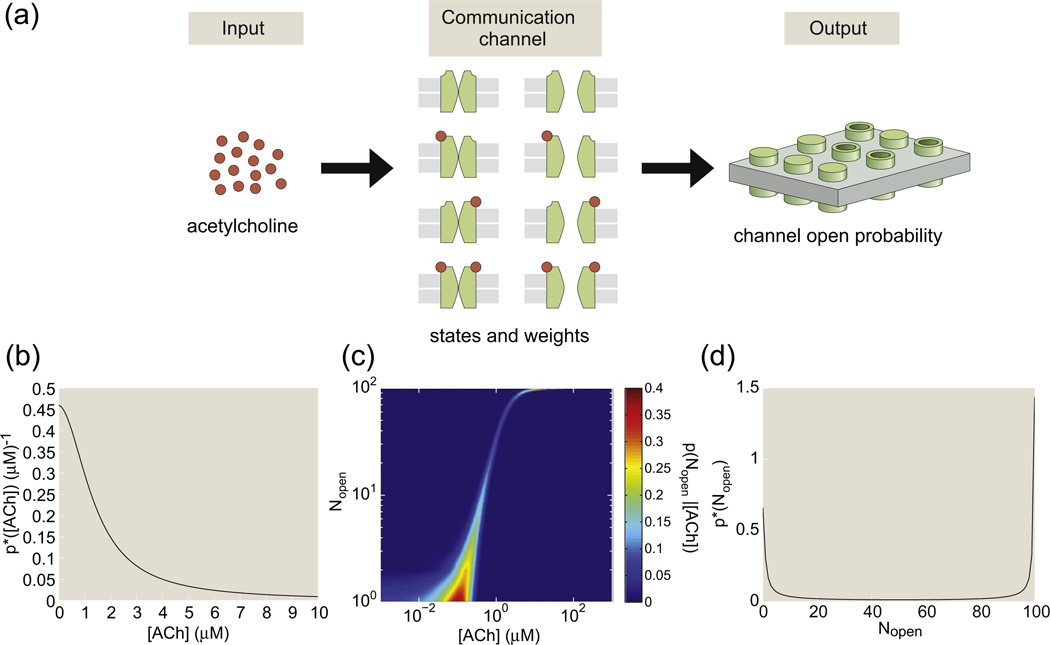
An MWC molecule with a single binding site has four possible states: the receptor can be inactive or active, and the binding site can have or cannot have a bound ligand. Figure 1a provides a generic schematic of these four distinct molecular situations and also lays the groundwork for a statistical mechanical investigation of the relative probabilities of these different states. Each of the possible microscopic states has probability proportional to its Gibbs factor, exp(−(Estate − nstate μ)/kBT), where Estate is the energy of the microscopic state of interest, nstate is the number of ligands bound to the receptor in that state and μ is the chemical potential that here encapsulates the free-energy cost of moving a ligand molecule from the solution to the receptor.2,5 For the remainder of this article, we define the variable as is typically done in the statistical mechanics literature.
Fig. 1.
States-and-weights diagram of the one-site MWC molecule. (a) Each of the four states has an associated energy, part of which is due to the conformational degrees of freedom of the molecule and part of which reflects the free energy of the binding process. (b) pactive as given in Eq. (2) as a function of concentration in units of the inactive state's dissociation constant. The activity curve is shown on a log scale in the main plot and on a linear scale in the inset. (c) The four curves show the probabilities of each of the distinct states as a function of the ligand concentration. Each state is labeled by a pair of numbers. The first number of the pair is 1 if the receptor is active and 0 if the receptor is inactive; the second number of the pair is 1 if a ligand is bound and 0 if no ligand is bound. The parameter values used in the figure are Δε = εI − εA = −4 kBT and .
We can decompose the energy of a state Estate into two different contributions: the conformational energy of the receptor and the energy of binding a ligand. We label the conformational energy of the active receptor as εA, the conformation energy of the inactive receptor as εI, the energy of binding a ligand to the active receptor as and the energy of binding a ligand to the inactive receptor as . Calculating the conformational energy of the receptor or the energy of binding a ligand from first principles could be incredibly complicated, as it depends on the details of the bonding interactions within the receptor, between the receptor and the surrounding solution and between the receptor and ligands. The free-energy cost of removing a ligand from a dilute solution is encapsulated in the chemical potential μ,
| (1) |
where μ0 and c0 are an unspecified reference chemical potential and its corresponding (unspecified) reference ligand concentration. Using the prescription that each state has a probability proportional to exp(−β(Estate − nstate μ)), we find “weights” shown in Fig. 1a for each state. From these weights, we can derive the probability of the one-site receptor being active, which can be calculated as the sum of the weights of the active receptor normalized by the total sum of all of the weights, namely,
| (2) |
Figure 1b shows that the probability of the active state increases as ligand concentration increases; this curve is often called the “activity curve”.
We can also derive the “binding curve” as the average number of bound ligands, which for the one-site receptor is the sum of the weights of the receptors with one ligand normalized by the total sum of all of the weights,
| (3) |
When the active state has a larger affinity for the ligand than the inactive state, , both pactive and 〈Nbound〉 increase with the ligand concentration c. These are consequences of the fact that an increase in ligand concentration increases the statistical weight of the bound, active state relative to the weight of the unbound, inactive state. Figure 1c plots the probabilities of each of these states as a function of concentration. As expected, the inactive, unbound state dominates at low ligand concentrations and the active, bound state dominates at high concentrations.
Some readers may find Eqs. (2) and (3) unfamiliar since activity curves and binding curves are often written in terms of a different set of parameters. Thermodynamic language is often used instead of statistical mechanical language, employing dissociation constants†,‡ and and the conformational equilibrium constant L = e−β(εA−εI).2 Note that, as commented on previously, only energy differences are meaningful. With this parameterization, the activity curve for the one-site MWC molecule has the form
| (4) |
and the binding curve has the form
| (5) |
The thermodynamic formulation is directly related to the original MWC parameters§. The activity curve and binding curve given in Eqs. (4) and (5) can in turn be fit to the MWC equation for pactive to find the microscopic parameters of the MWC model, for example, , L. For the rest of this paper, we will use a combination of statistical mechanics and thermodynamic notation: dissociation constants and will be used in preference of and , respectively, and conformational energies εA and εI will be used in preference of the conformational energy equilibrium constant L. Of course, this choice is a matter of personal taste; we find that the notation favored here combines the brevity of the original MWC notation4 with the clarity of biophysical understanding provided by statistical mechanics notation.
The concept of indirect regulation is already present in a one-site MWC molecule. Typically, the inactive receptor is more energetically favorable than the active receptor, εA > εI or L > 1. The presence of ligand shifts the balance toward the active receptor because the binding of ligand to the active receptor is more favorable than the binding of ligand to the inactive receptor, that is, . However, studying the one-site receptor cannot elucidate the phenomenon of cooperativity, in which the binding of one ligand appears to encourage or discourage the binding of the next. Thus, we turn our attention to a more general n-site MWC molecule. Some details of cooperativity calculations are confined to Supplemental Information, Appendix 1.
As for the one-site MWC molecule, the n-site MWC molecule can be in either an active state or an inactive state, and in each state, each of the n sites can either be empty or have a bound ligand. A full states-and-weights diagram would therefore have 2 × 2n states.
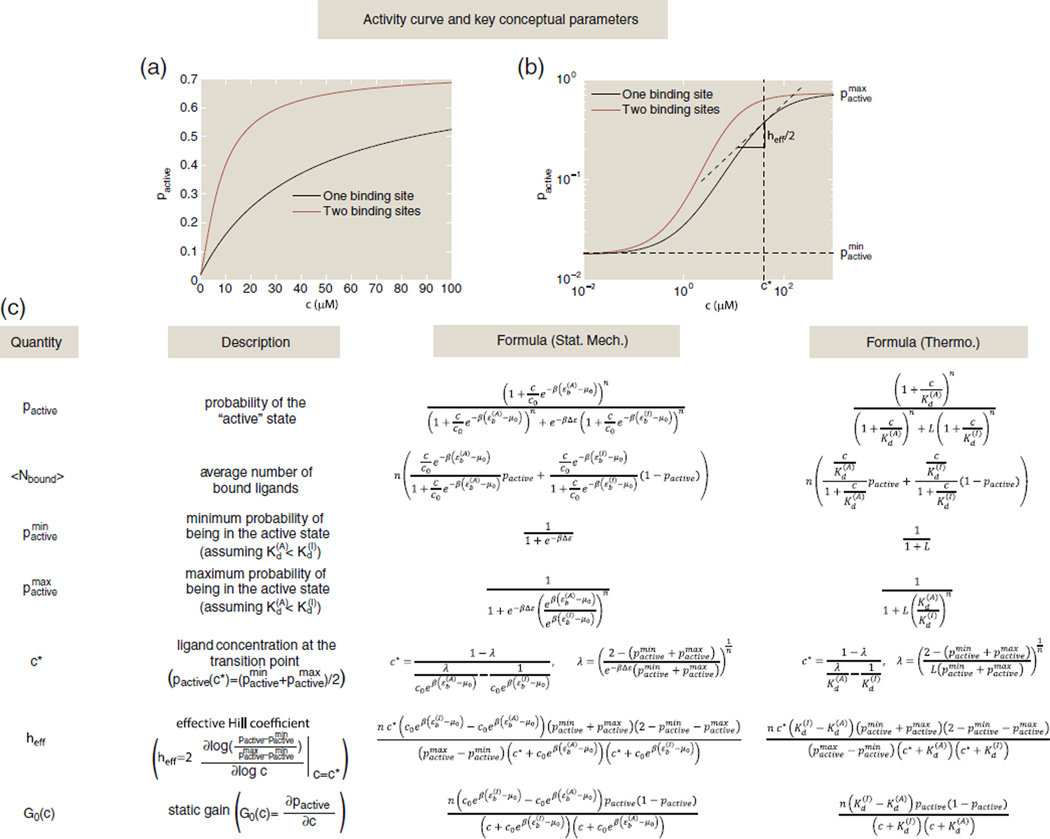
The weights assigned to each state follow the general pattern set forth in Fig. 1a, and deriving the formulas for key quantities such as pactive follows similar logic to that of the n = 1 case. For the reader's convenience, a number of these key formulas are listed for the general MWC molecule with n binding sites in Fig. 2, including formulas for the activity curve pactive and the binding curve 〈Nbound〉. Note that when faced with some new problem, we find that it is often much simpler to write down the various states and their associated statistical weights in statistical mechanical language and then to convert to a more familiar Kd language at the end. It is for this reason that we illustrate the results in both languages.
Fig. 2.
Table of key quantities that can be computed within the MWC framework. (a) The activity curve on a linear scale for two MWC molecules: a one-site receptor with Δε = εI − εA = −4 kBT, and , giving a difference in binding energy of ; and a two-site receptor with Δε = εI − εA = −4 kBT, , and , giving a binding energy difference of . (b) The activity curves from (a) with concentrations on a log scale. The transition point concentration c* = 40.6 µM and effective Hill coefficient heff = 1 are shown with vertical and horizontal lines, respectively, for the one-site receptor. (c) This table gives formulas for some of the key parameters of interest in both statistical mechanics and thermodynamic language. Here, n is the total number of binding sites on the receptor, L = e−βΔε is the conformational equilibrium constant where Δε = εI − εA is the difference in conformational energy between the inactive and active state, is the inactive state's dissociation constant for ligand binding, is the active state's dissociation constant for ligand binding and c is the ligand concentration.
As n increases, the activity curves become flatter for high and low concentrations and steeper near the “transition point”, the halfway point between a minimally and maximally active receptor denoted by c* in Fig. 2. This steepness is a signature of cooperativity, which means that the binding of one ligand seems to encourage or discourage the next.8,12,13 In the MWC model, this phenomenon is the result of indirect regulation rather than direct regulation: the presence of ligand increases the probability of the receptor existing in the state with higher ligand affinity, thereby increasing the probability of the next ligand binding. There are certainly other models that can explain cooperativity, often by postulating direct energetic interactions between bound ligands.2 One popular and simple model of cooperativity postulates that activity curves follow the Hill function2,12,13 given by
| (6) |
If the Hill function were derived from a states-and-weights diagram similar to that in Fig. 1a, then there would only be two states for this receptor with identical binding sites of dissociation constant KA: a receptor with no ligands bound and a receptor with h ligands bound. This is often not a realistic mechanistic explanation for cooperativity despite the Hill function's ubiquitous presence in the biological literature. However, h is a useful proxy for the degree of cooperativity in the system since it quantifies exactly the steepness of the activity curve at the transition point. Similarly, one can define an effective Hill coefficient, heff, for any activity curve as twice the slope of the activity curve on a log–log scale at its transition point.12 A formula for heff in the case of the MWC model is given in Fig. 2. The effective Hill coefficient is a useful heuristic for cooperativity. If |heff| > 1, then the presence of one bound ligand increases the likelihood of the next ligand binding, a signature of positive cooperativity; if |heff| < 1, then the presence of one bound ligand decreases the likelihood of the next ligand binding; and if |heff| ≃ 1, then the binding of one ligand indifferent to the binding of the next, a signature of no cooperativity. As shown in Supplemental Information, Appendix 1, the effective Hill coefficient for the MWC molecule can be summarized as follows. When the active state and the inactive state of an MWC molecule with multiple binding sites have different ligand affinities, |heff| > 1, and when there is only one binding site or when the two states have equal affinity for the ligand, then there is no signature of cooperativity, |heff| = 1. To model an MWC molecule with negative cooperativity, one needs to introduce repulsive interaction energies between ligands, as was done by Narula and Igoshin, for example.11
With the statistical mechanical preliminaries now in place, the remainder of the paper is devoted to specific case studies. Each such study is introduced either to highlight some specific twist on the statistical mechanical analysis (such as the presence of more than one binding site) or to examine modern applications of the MWC concept to problems of current interest.
Case Studies in MWC Thinking
The MWC concept presented above has been applied to a very wide array of different molecular scenarios, as evidenced by the massive citation list.
In the 1960s, the MWC concept and related models were used to great effect as the basis for thinking about several important “model” molecules,4,14,15 most famously, hemoglobin. For a beautiful review on the evolution of thinking on hemoglobin, see the work of Eaton et al.16 Recall that the hemoglobin molecule binds oxygen molecules, which are then delivered to tissues throughout the body. Hemoglobin can bind up to four oxygen molecules and therefore has a more complicated states-and-weights diagram and activity curve than that of the simpler one-site receptor in Fig. 1. One of the signature features of the binding curve of oxygen on hemoglobin is its characteristic sigmoidal shape that indicates the existence of cooperativity, described in detail in the previous section. Traditionally, one of the most well studied ways of characterizing cooperativity is through the Hill function as already discussed in Eq. (6). As we already noted in States and Weights in the MWC Setting, Hill cooperativity is very strict in the sense that, from a statistical mechanical perspective, it banishes the states of partial occupancy that are present in the MWC framework. One of the key insights of the MWC perspective is that it too can give rise to sigmoidal binding curves, but on the basis of a different underlying picture of the allowed molecular states. Models of ligand–receptor binding that include intermediate states can also give rise to sigmoidal binding curves, but unlike the MWC model, these models often posit direct energetic interactions between bound ligands. In the remainder of this section, rather than focusing on MWC classics such as hemoglobin, we highlight the spectrum of more recent examples of the MWC concept that have been applied to topics of great current research interest ranging from bacterial chemotaxis to the accessibility of chromatin to DNA binding proteins.
MWC ligand-gated channels
The cell membrane is richly decorated with a host of different molecular species, many of which detect and respond to molecules present in the external milieu. Ion channels are one of the most important examples of such membrane-bound proteins that respond to external cues resulting in changes of the cellular state such as a change in the membrane potential. One mechanism by which ion channels can detect environmental signals is through an MWC-like mechanism: when a molecule binds to the channel, it shifts its equilibrium such that the open state is more likely than the closed one. There are a number of important ligand-gated ion channels, but we will primarily focus on two examples: (i) the nACh receptors at neuromuscular junctions17 and (ii) cGMP-gated ion channels that enable photoreceptors to amplify their response to light.9,18
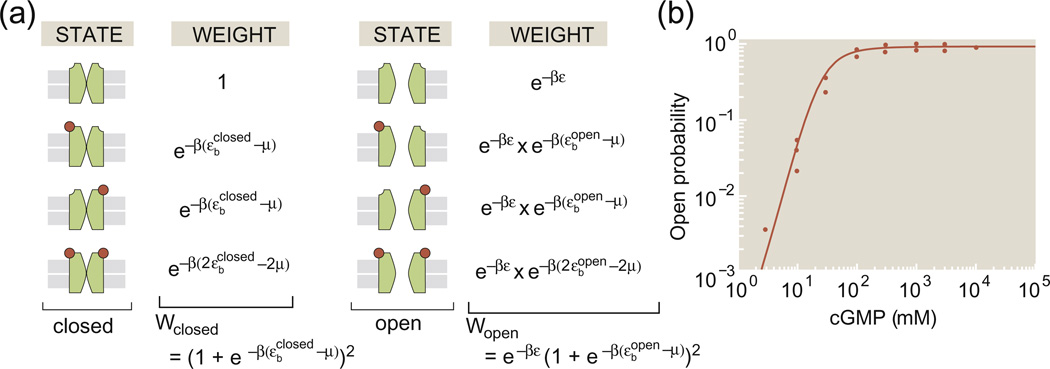
Figure 3a shows the states and weights for a toy MWC model of an ion channel in which we imagine that there are two distinct binding sites for the relevant ligand. The four states on the left of Fig. 3a correspond to all the different variants on the closed state and the four states on the right correspond to the different variants of the open state. For convenience, we have taken the conformational energy of the closed state to be zero while the conformational energy of the open state is given by ε.We can add up the statistical weights for all of the open states permitting us to compute the open probability as
| (7) |
Fig. 3.
Toy model of a ligand-gated ion channel. (a) The model ion channel has two binding sites for the control ligand and can exist in four distinct states of occupancy (i.e., empty, 2 × single occupancy, double occupancy) for both the closed and open states. (b) Open probability for a cGMP-gated channel as a function of the cGMP concentration and fit to an MWC model with four binding sites.19
Much effort on the use of MWC models has been aimed at the rigorous attempt to figure out the answers to precise questions such as how many binding sites are present in the MWC molecule of interest, whether or not those sites are heterogeneous and what are the precise values of the molecular parameters associated with the various states. Of course, these questions are all of great interest. We show an example of the kind of data that engenders these discussions in Fig. 3b, which shows the open probability of cGMP-gated ion channels as a function of the concentration of cGMP. These channels are a key part of the signal transduction pathway in the retina, undergoing a gating transition from open to closed when photoreceptors are exposed to light.2,18,20 The key point for our discussion here is to note that the MWC concept gives us a framework for thinking about how channel gating depends upon key parameters such as the ligand concentration, the number of binding sites (as revealed in the effective Hill coefficient) and a variety of other quantities of interest, including those shown in Fig. 2.2,19,21
MWC and bacterial chemotaxis
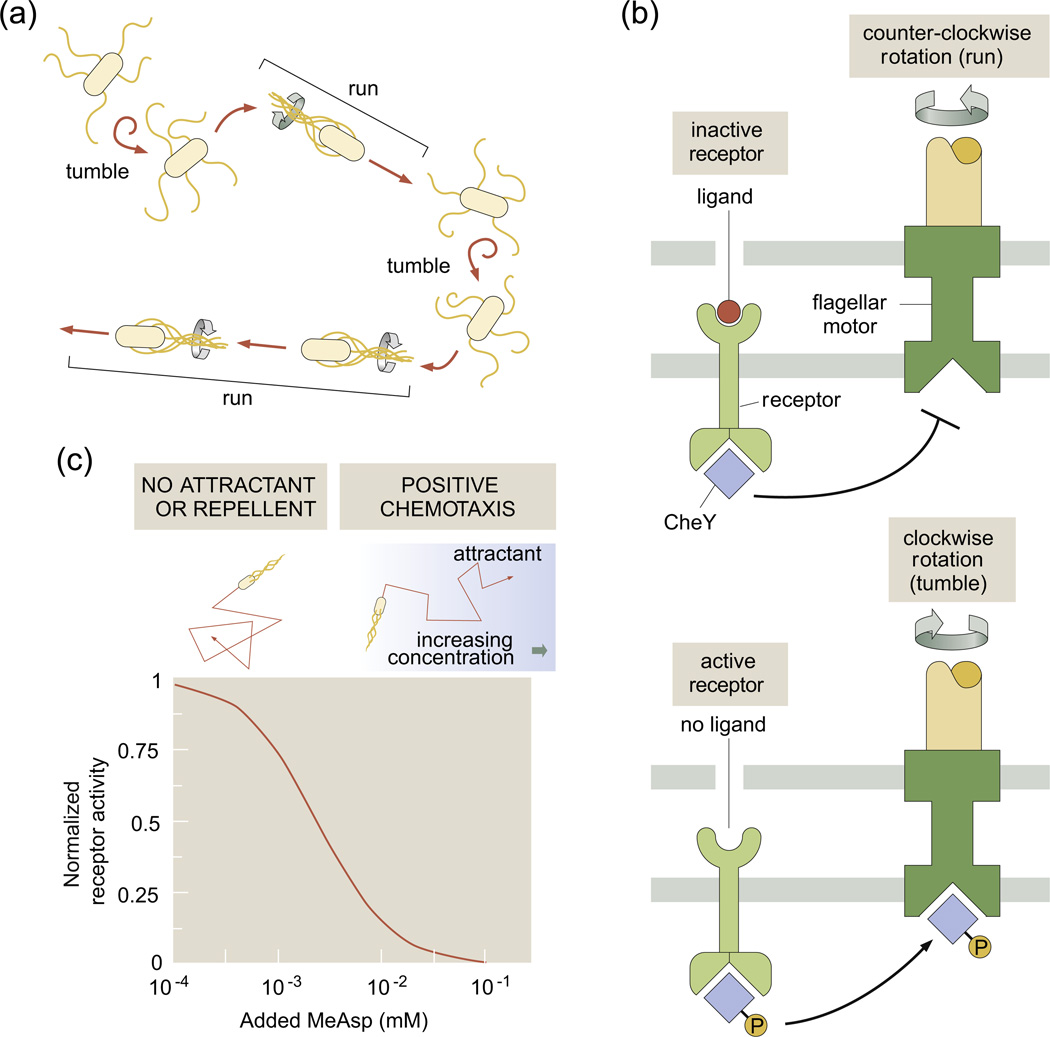
A second recent application of the MWC model that illustrates its adaptability to new experimental situations is that of bacterial chemotaxis, the process whereby bacteria are observed to move up gradients of chemoattractant.22,23 In the time since the development of the MWC concept, one of the best studied (at least in quantitative detail) examples of signal transduction in living organisms is provided by this fascinating directed motion. Specifically, bacterial motility in these situations is characterized by “runs” during which the bacterium uses its flagella to swim in a roughly straight path, punctuated by “tumbles” during which the bacterium reorients and then swims off in a new direction (see Fig. 4a).22,25
Fig. 4.
Bacterial chemotaxis. (a) A schematic showing the motion of a bacterium that consists of a series of runs and tumbles. (b) A chemoreceptor and the bacterial flagellar motor are shown in the same membrane region, although in real bacteria, they are often on opposite poles. In the presence of ligand, CheY is not phosphorylated and hence the motor is not induced to alter its rotation direction. (c) Activity of the chemoreceptor in the limits of low and high chemoattractant concentration with MWC parameters taken from Ref. 24.
The circuit that mediates this bacterial decision making has been subjected to detailed experimental scrutiny, and recent fluorescence resonance energy transfer (FRET) experiments26,27 provide precise quantitative data on the signal transduction pathways involved in bacterial chemotaxis. For our purposes here, the key point is that the bacterial surface is decorated with chemoreceptors that serve the role of detecting chemoattractants in the surrounding medium and then changing the state of phosphorylation of its diffusible response regulators (CheY). Once phosphorylated, CheY-P then induces the bacterial flagellar motor to undergo a change of rotational direction that leads to a tumble. This process is shown schematically in Fig. 4b. In this case, the chemoreceptor is actually inactive in the presence of ligands. In the presence of chemoattractants, the bacterium is what Howard Berg has dubbed an “optimist” and would like to simply keep going in the same direction (i.e., to not undergo a tumble).
The MWC concept has been applied in both clever and subtle ways to describe the response of bacteria to chemoattractants through sets of different different chemoreceptors as indicated schematically in Fig. 5.28–31 The simplest MWC description of a chemotactic receptor is identical with the scenario shown in Fig. 1 except that, in this case, binding to the inactive state has the lower Kd (i.e., or ), which means that, in the absence of ligands, the receptor is active and that binding of ligands renders the receptor inactive. This is shown in Fig. 4c. The simplest model of activity as a function of chemoattractant concentration is given by Eq. (2), but respecting the condition or described above. This is consistent with Berg's optimism principle in the sense that, with chemoattractant present, the bacteria tumble less often.
Fig. 5.
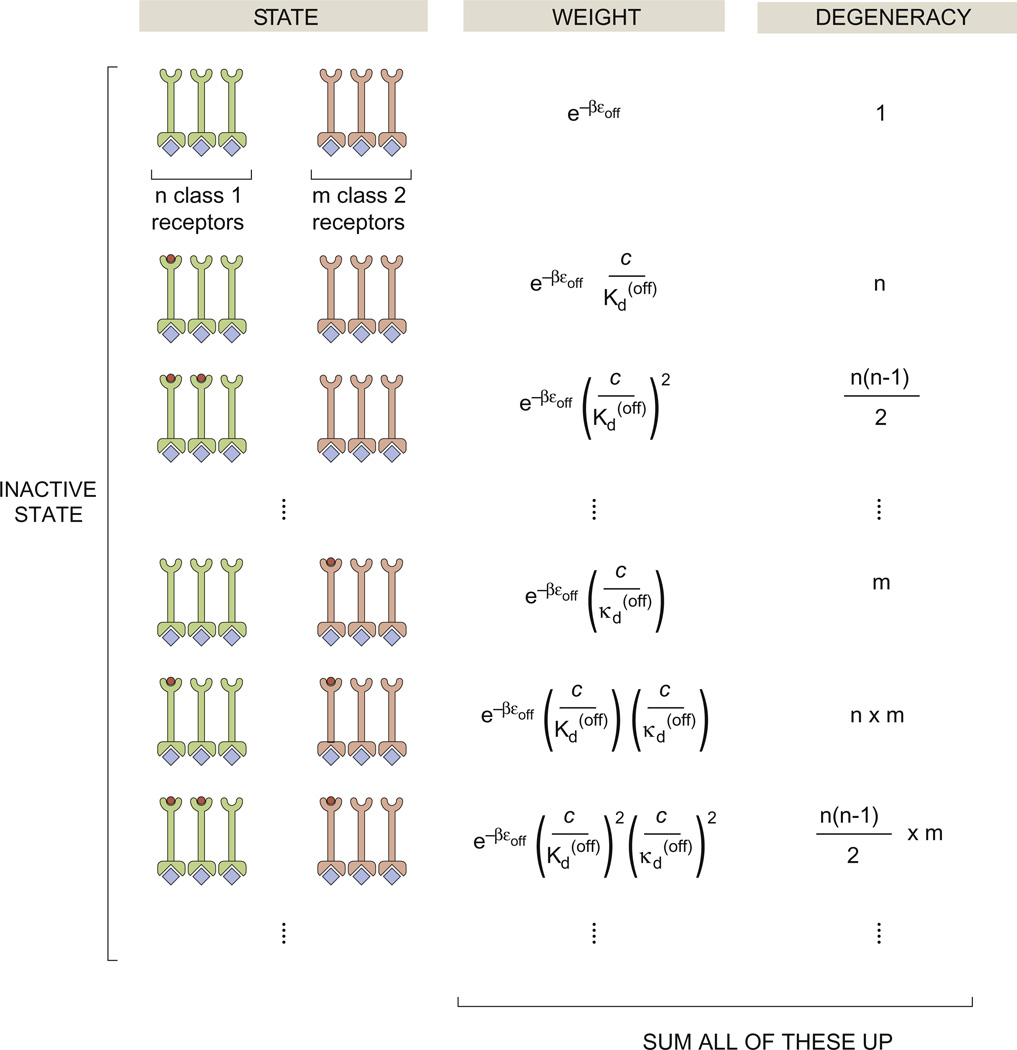
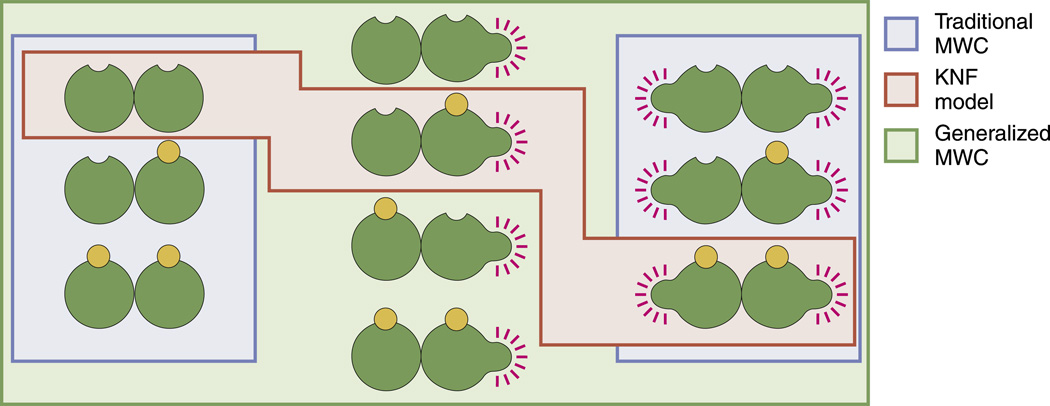
States-and-weights diagram for chemotaxis clusters. The various states shown in the figure correspond to different states of occupancy of the chemotactic receptors while in the inactive state. There is a corresponding set of diagrams (not shown) for the active state. The statistical weights of the different states reflect how many ligands have been drawn out of solution to bind the chemoreceptors. The degeneracies correspond to how many different ways there are of realizing a given state of binding. For example, in the second state shown in the figure, there is only one ligand bound on one of the n class 1 receptors. The class 1 receptors and class 2 receptors have conformational energy ε(off) in the inactive state and ε(on) in the active state. The n class 1 receptors have dissociation constant in the inactive state and in the active state; the m class 2 receptors have dissociation constants in the inactive state and in the active state.
Figure 5 shows how the simplest MWC concept can be extended to account for several particularly interesting features of chemoreceptors in bacteria. For instance, multiple receptors are clumped together into clusters, an aspect of these receptors that has been long recognized and recently studied systematically across different species.32–35 For the case of clusters of size n and all of the same species, the activity is given by
| (8) |
a simple extension of the models introduced already and which can be developed in direct analogy with the way we worked out the ion channel open probability in Fig. 3. The consequence of this clustering is an effective increase in cooperativity that sharpens the response of the chemotactic two-component signaling system to chemoattractant with respect to the response of a single chemoreceptor.
The FRET experiments of Sourjik and Berg measured the fluorescence signal change when the response regulator CheY-P interacts with downstream signaling partners, thereby effectively measuring activity curves for a number of different mutants of the receptors that mediate chemoreception. These experiments provided stringent constraints on any theoretical explanations set forth to explain chemotactic activity.26,27 Indeed, models following the MWC concept found that the only way to explain the data was to consider that the receptor clusters are chemically heterogeneous, which means that a specific cluster of chemoreceptors will contain receptors of more than one type that have different binding affinities for the same chemoattractant. Specifically, as shown in Fig. 5, if there are n copies of the first receptor type and m copies of the second receptor type, when constructing the states-and-weights diagram, we must sum over all possible states of activity and ligand occupancy. If we ascribe binding constants and to the second receptor type, the activity of the receptor cluster as a function of chemoattractant concentration is given by
| (9) |
a result used to consider the activity data coming from FRET experiments for a number of different chemoreceptor mutants in quantitative detail.28–31
As shown in this section, the MWC model has been used to great effect in a number of different situations, producing powerful predictions and insights into cellular signaling. We now describe a completely different implementation of the MWC concept in the context of the behavior of genomic DNA.
MWC and genomic accessibility
The MWC model has been recently and perhaps unexpectedly applied to transcriptional regulation.10,11,36,37
Genomic DNA can exist in a compact state (i.e., nucleosome bound or in some higher-order chromatin configuration) that is inaccessible to various molecules, for example, to transcription factors that activate some gene of interest. However, sufficiently high concentrations of transcription factors (i.e., the ligand) can increase the favorability of the chromatin open state, even though the open conformation of chromatin incurs a free energy cost. As will be discussed below, there are many variants on this basic picture in which combinations of transcription factors lead to different logic functions such as AND, OR and so on.11 Our aim here is to illustrate the overarching conceptual picture through several specific examples.
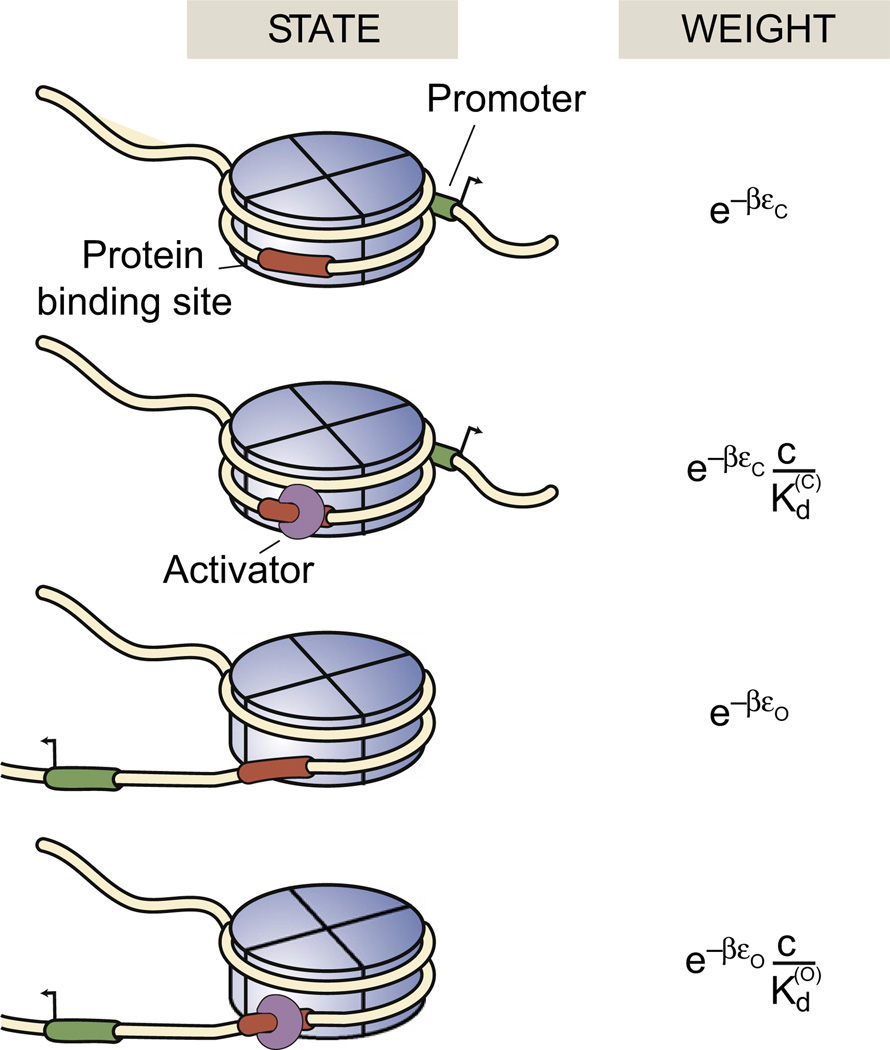
As a first foray into DNA accessibility problems from the MWC perspective, we consider the accessibility of a DNA segment wrapped within a single nucleosome. To get a first impression of the kinds of molecular states of interest and how they can be described using statistical mechanics, Fig. 6 shows a hypothetical eukaryotic promoter bound with some disposition relative to a nucleosome. We note from the outset that, because of the rules of nucleosome positioning,38–40 the real situation is more subtle than this and that this example is intended only to illustrate the “indirect regulation” that could be exercised by the presence of nucleosome-bound DNA. As seen in the figure, the DNA segment of interest harbors both a promoter and a binding site for a transcription factor. When the promoter is wrapped within the nucleosome, the gene of interest is inactive. The four states of this promoter in this simple model then correspond to inactive and active configurations of the promoter and the transcription factor binding site either unoccupied or occupied, with the transcription factor serving as the ligand in much the same way as other ligands did in previous examples. Computing the probability of the active state follows the developments described above and further details can be found elsewhere (see chapter 10 of Phillips et al.).2,41
Fig. 6.
MWC model of nucleosome accessibility. States-and-weights diagram for a toy model of nucleosome accessibility that illustrates how transcription factors could alter the equilibrium of nucleosome-bound DNA. εc and εo refer to the conformational energies of the closed and open states, respectively, and and are the dissociation constants for transcription factor binding in those two states.
One of the most compelling discoveries to emerge from the study of eukaryotic gene regulation, especially in multicellular organisms, is the existence of binding sites contained on the DNA known as enhancers that result in regulatory “action at a distance”. Interestingly, the MWC concept is also useful for characterizing these ubiquitous eukaryotic regulatory architectures. The concept of such enhancers is that there are binding sites that are not in genomic proximity to the promoter they control. Depending upon the binding of transcription factors to these enhancers, the genes will be expressed to differing extents. A particularly intriguing aspect of these enhancers from the point of view of more traditional views of gene regulation is their extreme flexibility—in some cases, there seems to be a generic indifference to the number of binding sites, their specific position and even their chemical identity.42
For example, the embryonic development of the fruit fly D. melanogaster's body plan is determined by the expression levels of a hierarchy of genes with single-cell resolution43,44 along the anterior–posterior axis of the embryo. One such gene is even-skipped, which is expressed in seven stripes along the anterior–posterior axis of the embryo. Each one of these stripes is controlled by an individual enhancer located up to 8 kb upstream or downstream of the actual eve gene.42 The enhancer that controls stripe 2, for example, is located 1.5 kb upstream from the gene and in its minimal form spans 480 bp.45 It contains several binding sites for two activators and two repressors. Specifically, it has three binding sites for the activator Bicoid despite the fact that the deletion of one of these sites does not cause any qualitative changes to the output pattern.45 Perhaps more revealing in terms of the flexibility of these regions is the fact that this enhancer sequence has undergone significant changes throughout evolution while retaining its function. For example, in Drosophila pseudoobscura, the same enhancer has lost and gained binding sites while the remaining binding sites have changed in their affinities with respect to the D. melanogaster enhancer. The spacing between some of these sites has also changed in some cases by up to 80 bp.46 Nevertheless, when the D. pseudoobscura enhancer is introduced into D. melanogaster, not only does it result in a very similar pattern of expression but also it can even rescue mutations in the eve gene.46–48
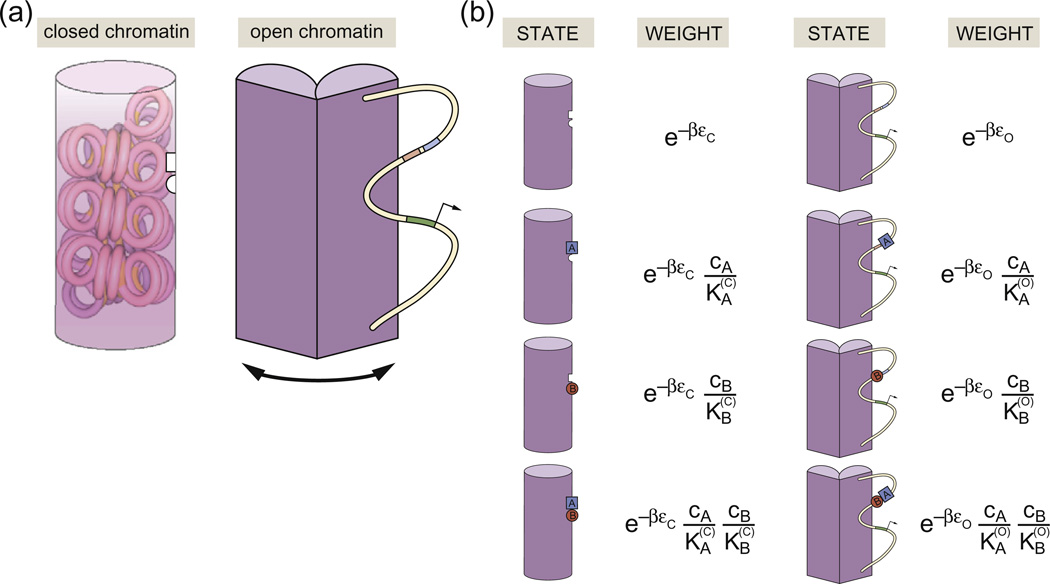
Recent quantitative models that have had some success in explaining these observations are predicated on the idea that these enhancers affect gene expression levels by controlling chromatin accessibility. This is in stark contrast to a picture in which transcriptional cooperativity is attributed to direct interactions between transcription factors and the basal transcription apparatus. Figure 7a shows a schematic example of how the MWC concept can be applied to model chromatin state. In the “closed” or inaccessible state, the DNA is wrapped up in some tight nucleosomal configuration, here indicated by one of many hypothetical higher-order chromatin structures (i.e., the putative 30-nm fiber).1 While in this state, the promoter of interest is hypothesized to be unavailable for transcription. The concept of the model is that RNA polymerase and transcription factors can bind more easily to DNA when it is in its open or accessible state, indicated schematically in Fig. 7 by DNA that is freely available in the “open chromatin” configuration.
Fig. 7.
Schematic description of MWC chromatin. (a) The genomic DNA exists in two classes of state, one of which is “off” and the other one of which is “on” and permits transcription. Transcription factor binding controls the relative probability of these different eventualities. (b) States and weights for the binding of two transcription factors, here denoted by A and B, which occupy the open and closed conformations with different affinity. The concentration of transcription factors A and B is given by cA and cB, respectively. The conformational energies of the closed and open states are given by εc and εo. The dissociation constant for A is when chromatin is in the closed state and is in the open state, and the dissociation constant for B is when chromatin is in the closed state and is when chromatin is in the open state.
Even within the relatively simple scenario depicted in Fig. 7a, there is already a great deal of conceptual and quantitative flexibility to account for a host of different regulatory architectures. For example, one can imagine situations such as shown in Fig. 7b in which the transcription factors are more favorably bound in either the closed or the open conformation, thus stabilizing one state or the other. Similarly, one can imagine both positive and negative cooperativity between the transcription factors themselves through direct physical contacts, permitting the construction of various logic functions such as AND and OR functions (and many others).11 From the perspective of the MWC model itself, the key parameters that come into play are the difference in energy between the closed and open conformations, Δε = εc − εo, the binding energies (or Kd values) for the relevant transcription factors in each of the states and the effective Hill coefficient that can be tuned by changing the number of binding sites for the DNA binding proteins in question. In Fig. 7c, we show an example of how the probability of being in the active state depends upon the concentrations of the two species of transcription factor.
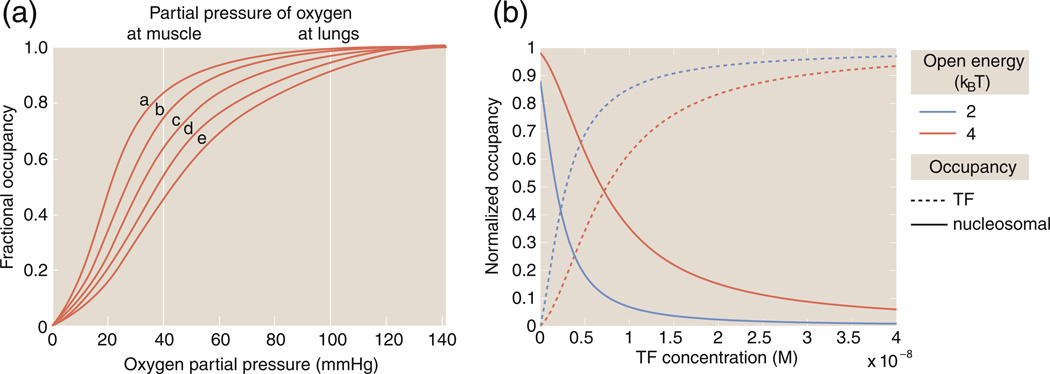
The Bohr effect generalized
A beautiful example of the unifying power of MWC models is the suggestion of an analog of the Bohr effect in the context of chromatin. The reader is reminded that the Bohr effect refers to the oxygen binding properties of hemoglobin and how the affinity for oxygen is tuned by changes in the pH, for example, as shown in Fig. 8a. Originally, the Bohr effect was an empirical observation. In the language of MWC models, however, the Bohr effect can be thought of in terms of how the binding curves are altered as the difference in energy between the two conformational states is changed. Mirny recently described an analogous, Bohr-like effect in gene regulation using the MWC model of chromatin state in which (for example) changes in the histone–DNA affinity can affect changes in the occupancy curve as shown in Fig. 8b.10
Fig. 8.
The Bohr effect and MWC models. (a) The Bohr effect and oxygen binding to hemoglobin as a function of pH. The hemoglobin binding curves are shown for five values of the pH: (a) 7.5, (b) 7.4, (c) 7.2, (d) 7.0 and (e) 6.8. The vertical lines indicate the partial pressures experienced in muscle and in the lungs. (b) The “Bohr effect” in the context of chromatin showing how the occupancy of a transcription factor on nucleosomal DNA changes as the histone–DNA affinity (for example) is changed, as described in Eq. (11). For the figure shown here, we have and . The closed state energy has been chosen as the reference energy and is taken as zero.
Specifically, we consider the example given in Fig. 7b for the case in which the two binding sites are used by the same transcription factor. In this case, we can use the states and weights highlighted in Fig. 7b to compute the probability that the DNA will be in the closed (inactive) state as
| (10) |
Note that, for the case considered here, the two enhancers bind the same transcription factor with the same affinities, although those affinities are different in the open and closed chromatin conformations. From Fig. 2, the average number of bound transcriptions factors is
| (11) |
As seen in Fig. 8b, the modulation of this binding curve as a function of the energy difference between open and closed chromatin conformations, εo − εc, reflects the chromatin Bohr effect.
Interestingly, the mutants considered in the bacterial chemotaxis setting correspond effectively to different states of methylation of the chemotaxis receptors.26,27 Like in the case of chromatin, our view is that the theoretical models using the MWC concept in that context too are yet another example of the “Bohr effect”, but now in the context of chemotaxis.28–31 This discussion provides a prime example of the unexpected biological insights that come from classifying biological topics on the basis of their conceptual proximity based on the underlying physics or mathematics, rather than on the basis of biological concepts.
An Information-Theoretic Perspective of the MWC Concept
The MWC model provides a simple conceptual mechanism whereby ligands can regulate “at a distance”. For instance, as described in Case Studies in MWC Thinking, enhancers can affect the expression of a distant gene.49 So far, we have focused on the generic features of MWC models and how to calculate molecular activity from pictures of states. However, a powerful advantage of an analytically tractable model such as the MWC model is that it can be used to calculate quantities that are difficult to measure but that still have great conceptual value. Calculating these quantities can shed light on how a regulatory system works and why it works in the way that it does. In this section, we shift away from a discussion of the MWC model itself and ask more general questions about the capacities of MWC molecules as regulators.
Specifically, we discuss the recent information-theoretic description of MWC molecules as sensors of ligand concentration.8,50,51 To see what ideas are in play, consider the case of bacterial transcription. If E. coli are grown in media rich in lactose instead of glucose, they produce an enzyme to digest the lactose. This production is mediated by transcription factors that allow information about the environmental conditions (lactose and glucose concentrations) to affect protein production (β-galactosidase enzyme) by influencing the likelihood of an RNA polymerase molecule to transcribe the relevant gene. Evolutionarily, it seems highly beneficial for an organism to excel at gathering information about environmental conditions and using that information to regulate protein production. However, such descriptions are qualitative, whereas we desire a way to quantify “how well” the output of a sensor (e.g., β-galactosidase production) tracks noisy sensory input (e.g., lactose concentration).
One quantification strategy is to make an educated guess as to how the molecule's behavior affects the organism's fitness. This approach is fraught with risk, as what initially appears to be noise often turns out to be signal in biological systems,52–54 and biological intuitions for fitness functions are often based on these guesses as to what is a signal and what is noise. For example, it is of course interesting that the sensory systems described by MWC models can be tuned to mimic the Boolean logic gates that underlie today's computers,11,55 but there is no guarantee that MWC molecules have been selected to mimic Boolean logic gates. There is a more general quantitative framework, information theory, that does not require knowing exactly what computation is being done by the cell but that still allows us to quantify how well the sensor output tracks input.56
Many have already written excellent reviews of information theory with a biological bent57–61; thus, we will just introduce the definitions that we need for this section. Given a black box system (e.g., an MWC molecule) that takes a noisy input X (ligand concentration) and returns a noisy output Y (whether or not the receptor is in the active state), then measuring Y provides information about the state of X. There is a unique function that will quantify the information content of a probability distribution subject to certain plausible assumptions about the form of this function.56,62,63 From this function, we can specify the amount of information about X gained by measuring Y as the “mutual information” I(X;Y)56,62,63 (see Supplemental Material, Appendix 2 for details). There are several ways to calculate the mutual information, but the one that we will focus on here uses the conditional probability of y given x, p(y|x), and can be written as
| (12) |
If Y tracks X well, then I(X;Y) is large; on the other hand, if Y and X are independent, then I(X;Y) = 0.62,63 What is the maximal amount of information one can expect between input and output? An answer to this question can be captured mathematically by computing the “channel capacity”, which is the mutual information for an optimal p(x), explicitly
| (13) |
See Supplemental Material, Appendix 2 for details.
The quantities described above provide principles for quantifying what is possible in a molecular signaling system, and interestingly, some biological systems seem to be operating very close to channel capacity. A spectacular example of this appears to be the expression of the Hunchback protein in the early Drosophila embryo, which is activated by the Bicoid transcription factor. It has been argued that the probability distribution of the Bicoid transcription factor is optimized so as to maximize the mutual information between this input and the output Hunchback expression level.50 Case studies such as these have motivated other investigators to study the channel capacity of MWC models more generally,8 a topic we take up below.
We stress here that not all MWC molecules are near-optimal sensors, nor should we expect them to be. For instance, hemoglobin picks up oxygen in the bloodstream and deposits that oxygen in distant tissues. This task does not necessarily require maximizing the mutual information between oxygen concentration and the number of oxygen molecules bound to hemoglobin. However, many of the MWC molecules described in Case Studies in MWC Thinking are likely to be high-performance sensors of their environment. The nACh receptor at the neuromuscular junction must turn a chemical signal in the form of diffusing acetylcholine (ACh) molecules into a corresponding electrical signal that can contract a muscle fiber. If nACh receptors misrepresent an incoming signal, the consequences could range from an inability to stimulate the motor system to an inability to stop moving. Similarly, the cGMP receptor must turn a chemical signal based on the presence of light in the environment into an electrical signal. If our cGMP receptors in our photoreceptors do not accurately represent the incoming light signal, then we will not be able to see. These and other such receptors could encode information about ligand concentration in terms of the average number of bound ligands or the probability that the receptor is in the active state. Hence, it makes sense to study the sensing properties of MWC molecules, and our motivation for doing such an analysis is inspired by a recent general analysis of the sensing properties of MWC molecules.8
In the remaining portion of this section, we use a toy model of N independent ligand-gated ion channels (inspired by the example of nACh receptors) to illustrate how to quantify the ideas presented in the previous paragraph. Though we will use the specific language of ion channels, the concepts apply much more broadly. Indeed, general reflections of this kind were analyzed comprehensively and in more generality elsewhere.8 Here, we present an abridged version of their analysis specialized to a toy model of a ligand-gated ion channel such as the nACh receptor; many of the calculational details have been relegated to Supplemental Material, Appendix 3.
Model system: nACh receptors
To illustrate the power of these information-theoretic principles, we now investigate in detail an MWC model of a ligand-gated ion channel. Again, the hope is that calculations on this toy model will provide some qualitative insight into their functionality.
To see why ligand-gated ion channels could plausibly be conceptualized as sensors, we now describe the nACh receptors that lie at neuromuscular synaptic junctions, which are a key component of the communication pathway between the nervous system and the motor system. When our brain decides that a particular muscle should contract, for example, to avoid a hot stove, a motor neuron releases vesicles of ACh molecules across a synaptic gap to a muscle fiber. On the other side of this synaptic gap are many thousands of nACh receptors, with a surface density of roughly 105 receptors per square micron. The diffusing ACh molecules bind to the nACh receptors, stochastically opening some number Nopen of the total number of receptors N. Each open channel allows for an influx of sodium ions and an outflux of potassium ions, which results in a net depolarization of the muscle fiber. When membrane potential reaches threshold, the muscle fiber contracts.64,65
Extensive studies have revealed that nACh receptors behave as MWC molecules, though the two binding sites in a nACh receptor are not necessarily identical, and there are likely more than two states of the nACh receptor.66,67 However, despite its short-comings, the two-state MWC model of the nACh receptor's response to ACh is an excellent example with which to illustrate the information-theoretic underpinnings of decision making based upon input–output functions.
It is clear that, in order to operate effectively, nACh receptors should respond quickly to commands from the nervous system. As described above, the response of nACh receptors should reflect the size of the stimulus. Ligand-gated ion channels more generally face a difficulty similar to that of nACh receptors: they convey information about some stimulus from the outside to the inside of the cell body, and they use energy by virtue of maintaining a difference between the ion concentrations inside and outside of cells. Inspired by the example of the nACh receptor, we investigate the ability of ligand-gated ion channels to turn an input, ligand concentration c, into an output, the number of open ion channels Nopen. Conceptually, we are asking how well the Nopen output tracks the c input. The calculations shown here are merely illustrative of the kinds of calculations that could be done to elucidate the functioning of a particular sensory system. There are plenty of systems, including this ensemble of nACh receptors, whose input might be best described by a magnitude other than c and whose output might be best described by a magnitude other than Nopen.
In general, this problem is challenging since c and Nopen are both fluctuating quantities, typical of any such microscopic variable in biology. Even if each receptor experienced exactly the same ligand concentration, the number of open ion channels is still subject to fluctuations. In particular, for every ligand concentration, there is a corresponding probability of being open, popen(c). Recall from Eq. (7) and Fig. 3 that this probability is given by
| (14) |
where is the dissociation constant of the open ion channel, is the dissociation constant of the closed ion channel and ε is the energy difference between the closed and open ion channels. Even when ligand molecules are absent, there is a nonzero probability of being in the active state. This limit defines the minimum, baseline probability of being open given by
| (15) |
Likewise, as the ligand concentration tends to infinity, there is a nonzero probability of the receptor being in the inactive state and the ion channel being closed. This limit as c → ∞ defines a maximum value of the probability of being in the open state, namely,
| (16) |
For example, for the nACh receptor, we have and using characteristic MWC parameters for this channel.68 Thus, there is a probability that all N nACh receptors will be closed, but this probability is very small even when [ACh] = 0.
With N identical and independent ion channels, the conditional probability that Nopen of the ion channels are open given a ligand concentration c is the binomial distribution
| (17) |
This binomial distribution has mean N̅open(c) = Npopen(c) and variance . For each value of c, the distribution p(Nopen|c) is highly peaked about its mean, as can be seen in Fig. 9, a fact that we will use later on to evaluate the channel capacity using a “small-noise approximation”. However, there are still fluctuations in Nopen that prevent the output Nopen from determining the ligand concentration c noiselessly.
Fig. 9.
Information transmission through a two-site MWC molecule. The case of nACh receptors is illustrated for concreteness. (a) ACh (input) binds to ligand-gated ion channels (communication channel), thereby influencing the number of open ion channels (output). (b) The probability distribution of ACh concentration that maximizes the mutual information between input ([ACh]) and output (Nopen) from Eq. (21). (c) The MWC ligand–receptor binding probabilities determine the conditional distribution of the total number of nACh receptors open as a function of ACh concentration, p(Nopen|[ACh]) from Eq. (17), shown here as a heat map. (d) The probability distribution of Nopen that maximizes the mutual information between input ([ACh]) and output (Nopen). (b–d) Plots assume a total of 100 nACh receptors on the synaptic cleft for visualization purposes, although this underestimates the number of nACh receptors on a typical synaptic cleft and all plots use MWC parameters characteristic of nACh receptors.68
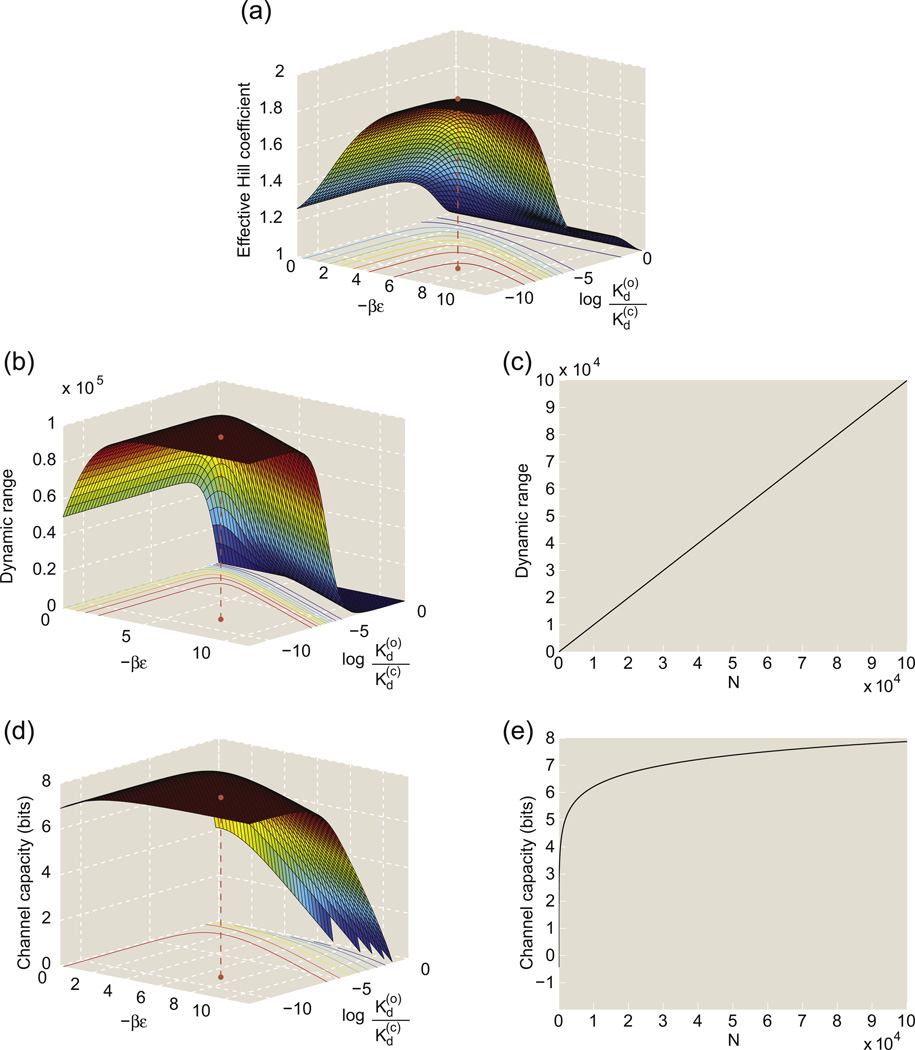
As ligand concentration varies, the most likely value of Nopen varies from to . The “dynamic range”8 captures the range of this likely output,
| (18) |
The dynamic range already provides a first glimpse into how well the output Nopen follows the input c; the larger the dynamic range, the better Nopen will be able to distinguish between different values of c despite the intrinsic fluctuations in Nopen. This quantity is shown in Fig. 10a as a function of the difference in energy between the inactive state and the active state (−ε) and the difference in ligand binding affinity between the active and inactive states () for a fixed N = 105. In this plot, we also show the point corresponding to the experimentally available data for the nACh receptor, just for comparison.68 Figure 10d shows the dynamic range of a two-site MWC molecule as a function of the number of receptors N using characteristic MWC parameters for the nACh receptor.68
Fig. 10.
Sensor properties of an ensemble of independent MWC molecules with two binding sites. In plots (a), (b) and (d), the MWC parameters are characterized by − βε, the conformational energy difference between the open and closed states (in units of kBT), and , the difference in ligand binding energies between the open and closed states (in units of kBT). (a) The effective Hill coefficient of a two-site MWC molecule plotted as a function of MWC parameters. (b) The dynamic range of 105 independent two-site MWC molecules plotted as a function of MWC parameters. (c) The dynamic range of N two-site MWC molecule with MWC parameters characteristic of a nACh receptor,68 plotted as a function of the total number of receptors N. (d) The channel capacity of 105 independent two-site MWC molecules plotted as a function of MWC parameters. (e) The channel capacity of N two-site MWC molecule with MWC parameters characteristic of a nACh receptor,68 plotted as a function of the total number of receptors N.
We are now ready to calculate the mutual information between the concentration of ligand and the number of open channels. If the joint probability distribution of c and Nopen is p(c,Nopen), then following the procedure outlined in Eq. (12), mutual information is defined as
| (19) |
As shown in Supplemental Material, Appendix 2, by invoking key approximations such as the “small-noise approximation”, this can be simplified as
| (20) |
Notice that, in this equation, we have replaced the probability distribution p(Nopen) with the value of the probability around the mean of the distribution, p(N̅open). This latter distribution is directly determined by the probability distribution of the ligand concentration p(c) since N̅open = Npopen(c) and thus the probability distributions of c andN̅open are related by .
Without measuring the probability distribution of ligand concentration p(c), we are unable to calculate the mutual information given in Eq. (20). Hence, we instead calculate the channel capacity by considering the distribution p(c) or, equivalently, the distribution p(N̅open) that maximizes the mutual information. We hope that the channel capacity can still give insight into the workings of the system, as it did for the Hunchback/Bicoid gradient.50 We note that some of the systems described by the MWC model are unable to alter the probability distribution of ligand concentrations and are therefore unlikely to operate at channel capacity. For example, bacteria cannot usually control the probability distribution of chemoattractant in the environment so that their bacterial chemotactic receptors operate constantly at channel capacity. However, the body can alter the probability distribution of ACh concentration at the neuromuscular junction by altering the size distribution and number distribution of synaptic vesicles. In short, a calculation of the channel capacity will not always be meaningful, but we suspect that this calculation can be made relevant for ligand-gated ion channels and other MWC molecules. The optimal p(N̅open) can be found using variational calculus, a step that is described in Supplemental Material, Appendix 3. The form of p(N̅open) that maximizes the mutual information is
| (21) |
where Z is a normalization constant,
| (22) |
Using Eqs. (21) and (22) to simplify Eq. (20) yields
| (23) |
indicating that the channel capacity increases as the noise of the output decreases. In Supplemental Material, Appendix 3, we compute Z explicitly, thus permitting us to write the channel capacity of the N identical uncoupled ligand-gated ion channels as
| (24) |
where and are given by Eqs. (15) and (16). A more general formula for the channel capacity of N receptors when each receptor has n binding sites is given in Ref. 8, and a more detailed derivation of the channel capacity is given elsewhere.51,58,69,70
Dynamic range and channel capacity are closely related to the previously described concept of cooperativity. A receptor with a high degree of cooperativity will have a “steeper” activity curve popen(c) near its transition point. Intuitively, increasing the cooperativity increases the ability of the system of N independent receptors to differentiate between different ligand concentrations c near the transition point. For a given , increasing cooperativity will increase , thereby increasing dynamic range and channel capacity according to Eqs. (18) and (24), respectively. The left column of Fig. 10 below shows how dynamic range, channel capacity and effective Hill coefficient vary as a function of MWC parameters, the conformational energy difference (ε) and the difference in binding energy (). Recall that the effective Hill coefficient, defined in States and Weights in the MWC Setting and Fig. 2, is a measure of the degree of cooperativity. All three quantities are highly correlated and are largest when the closed ion channel is far more energetically favorable than the open ion channel and when the open ion channel has much higher affinity for the ligand than the closed ion channel. The results of Ref. 8 also show that all three quantities increase when n increases, since increasing the number of sites increases the effective Hill coefficient.
The right column of Fig. 10 shows that the dynamic range increases linearly with the total number of ion channels N but that channel capacity increases more slowly as the logarithm of N. Increasing the total number of ion channels always increases the channel capacity, but increasing the channel capacity by n bits requires increasing the total number of ion channels (and the average number of ion channels) by a factor of 4n. However, increasing the total number of ion channels requires manufacturing proteins, which requires materials and energy.2,71 Hence, increasing the channel capacity might be potentially energetically expensive. Again, we emphasize that these calculations are merely illustrative, and we are not claiming that nACh receptors or any other receptors have this tradeoff between energy and information.
Optimization principle: Maximization of mutual information
Operating at channel capacity requires that the probability distribution of ligand concentration takes on a peculiar form shown in Fig. 9. If the measured distribution of ligand concentration matched this predicted probability distribution, then this match would provide additional support for the claim that the ensemble of ligand-gated ion channels have evolved to maximize mutual information between the input (c) and the output (Nopen) within biophysical constraints. Such a measurement was made for the Bicoid/Hunchback system in the early Drosophila embryo, and the predicted probability distribution of Hunchback gene expression was strikingly close to the empirical probability distribution.50 Sometimes, an organism cannot control the probability distribution of ligand concentration, but this does not preclude use of information theory. For instance, the chemotactic receptors described in Case Studies in MWC Thinking must use information about chemoattractant concentration to decide on whether the cell runs or tumbles. There are different information-theoretic optimal ways to move in different chemoattractant gradients, and the optimal movements can be compared to the observed movements of the organism, as was done in Ref. 72.
The idea that biological systems might have evolved to maximize the mutual information between their “input” and “output” is certainly not new. This optimality principle has previously been applied to a variety of “information bottlenecks” in the brain, in which some physical barrier prevents information in one region from being copied directly into another region, but where the second region needs the information encoded by the first region. For example, the light intensities hitting photoreceptors must be encoded as binary spikes sent by retinal ganglion neurons to the lateral geniculate nucleus, and the distribution of spike times chosen should convey maximal information about the incident light intensities.57,59 A few investigators have begun to calculate the mutual information between the input and output of molecular systems,60,73,74 in particular, for genetic regulatory circuits and signal transduction pathways.
It is important to note that mutual information calculations are suggestive and potentially helpful for understanding organism behavior but never definitive. Mutual information between the input and output could be well correlated with another quantity that the system has evolved to maximize, in the same way that cooperativity is correlated with dynamic range and channel capacity in the calculation above. Additionally, in these calculations, we might have incorrectly identified the input and output of the system.
Finally, mutual information calculations can be incredibly difficult. In particular, it is often difficult to analyze nonlinear systems subject to time-varying environmental stimuli and systems with feedback. Many biological systems, however, respond nonlinearly to time-varying environmental stimuli and are part of a feedback loop. Analyzing these systems using information theory will likely require theoretical advances in communication theory.
Dynamical MWC
Molecules that are described in terms of the MWC concept are not necessarily optimal information encoders as described in the previous section. Nevertheless, there are many processes in the life of an organism where signaling molecules should respond quickly and definitively to changes in ligand concentration. Another feature that might be expected of such molecules is that they block out high-frequency molecular noise so that they only respond to changes that occur over “long” time-scales. To explore such time-dependent properties, we need to go beyond the equilibrium statistical mechanics description exploited thus far.
Though the original formulation of the MWC model was primarily an equilibrium concept, generalization to the dynamical situation is relatively straightforward and has been undertaken by many workers in the meantime,8,75–81 sometimes under the heading of the “kinetic allosteric model”.82–84 The chemical reactions underlying the dynamical MWC model were outlined in one of the original papers,4 and an application of transition state theory85–87 to those chemical reactions yields the kinetic allosteric model. The dynamical MWC model that we will present in this review article uses only the law of mass action and does not come close to using the full power of transition state theory, which can calculate the values of rate constants from first principles.85–87
A smaller set of researchers have turned to the language of control theory88,89 to describe the kinetics of MWC molecules using transfer functions.8,80,90,91 In doing so, these researchers have described the dynamics of an MWC molecule using frequency instead of time. The two descriptions are mathematically related; high-frequency signals oscillate quickly and change on small timescales, whereas low-frequency signals oscillate slowly and change on long timescales. Transfer functions and frequency response functions are often economical ways of describing the response of a system to a change in the inputs. These functions are often used in electrical engineering and signal processing to design filters that block noise and not signal. More recent articles have calculated the transfer functions that describe how MWC molecules respond to changes in ligand concentration8,80 and showed that a general MWC molecule does not respond strongly to quickly changing ligand concentrations.8
In this section, we will describe a general approach to the dynamics of MWC molecules, once again illustrated through the special case of ligand-gated ion channels. Specifically, we will examine the response of such a channel to changes in the concentration of the relevant gating ligand.
Transition matrices and master equations
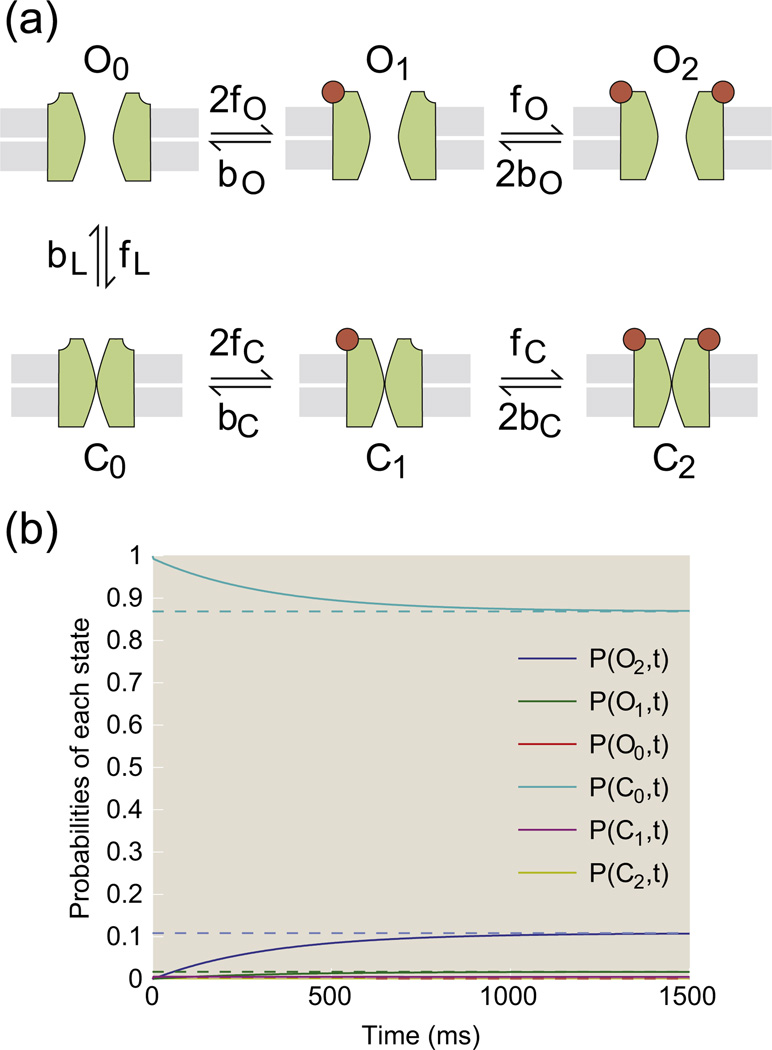
The general MWC molecule can be active or inactive and can have anywhere from 0 to n ligands bound to it in each of these states of activity, giving a total of 2 × 2n states. For even small numbers of binding sites, models of the transitions between these 2 × 2n states can become unwieldy. However, if all of the binding sites are identical, then this description can be greatly simplified. There are different ways for the receptor with n sites to bind k ligands, but all of these configurations have the same energy and therefore the same Boltzmann weight. Hence, we can describe the state of an n-site MWC molecule in terms of only two variables: the configurational state of the receptor and how many ligands are bound to the receptor in total. In this simplified description, there are 2 × (n = 1) different states for an MWC molecule with n binding sites. For example, for the toy model of the nACh receptor considered here, the possible ligand–receptor configurations are {O2,O1, O0,C0,C1,C2} where Oi denotes the open nACh ion channel with i bound ACh molecules and Ci denotes the closed nACh ion channel with i bound ACh molecules, as shown in Fig. 11. The state of the system x(t) can be described as a list of the concentrations of each of these configurations,
| (25) |
although the ordering of the states in x is completely arbitrary. A dynamical MWC model will describe how x evolves with time. If the system is Markovian (“memory-less”), then x obeys a first-order ordinary differential equation,
| (26) |
where M is a so-called “transition matrix” of size 2(n + 1) × 2(n + 1). For those unfamiliar with matrices and vectors, row i of Eq. (26) is
| (27) |
and this formulation is mathematically equivalent to that given by Eq. (26). Deriving a dynamical MWC model is therefore equivalent to specifying the elements of the transition matrix, Mij.
Fig. 11.
Dynamics of an MWC ligand-gated ion channel with two binding sites. (a) Schematic showing states and rates of transition between MWC states in a simplified kinetic allostery model. (b) Probabilities of being in each state as a function of time, starting from a nonequilibrium configuration, as calculated using Eq. (33) with c = 0.5 µM and rate constants fO = 1 µM−1 ms−1, bO = 170 ms−1, fC = 1 µM−1 ms−1, bC = 0.04 ms−1, fL = 1 µM−1 ms−1 and bL = 8 × 10−4 ms−1. The dotted lines show the equilibrium values of each of these probabilities.
The statistical mechanics of chemical reactions constrains the form of the matrix elements Mij. To see this, consider the situation shown in Fig. 11: how might we change the concentration of open ion channels with two ligands bound, [O2]? Based on the arrows in the diagram, there are two elementary reactions that can change this concentration. First, a ligand could bind to the open site of an open ion channel with one ligand bound, O1 + L → O2. Second, an open ion channel with two bound ligands could lose one ligand to the solution, O2 → O1 + L. The law of mass action implies that
| (28) |
The kinetic rates kO2→O1 and kO1→O2 are linked together by the requirement that the ratio of kinetic rates yields the equilibrium constant, . The factor of 2 in the reaction rate for O1 + L → O2 arises because of a degeneracy in state space: there are two “types” of O1 molecules, one in which the ligand is bound at the left site and one in which the ligand is bound at the right site. An alternative and equivalent viewpoint is that there are two ligands on O2 that can unbind from the receptor, but there is only one site on O1 to which a ligand can bind.8 The kinetic rates kO1→O2 and kO2→O1 reflect the height of activation energy barriers between the states, and the concentration dependence of the reaction rates encodes the frequency with which the reactants will meet. Those readers interested in calculating these rates more exactly using transition state theory should consult other references, for example, Refs. 85,86. Note that Eq. (27) when i = 1 is
| (29) |
Comparison of Eqs. (28) and (29) indicates that
| (30) |
Where c = [L], the ligand concentration. The rates kO1→O2 and kO2→O1 are denoted as the forward and backward rates fO and bO, respectively. Similar logic can be used to determine the rest of the transition matrix M, which we list in full here as
| (31) |
The rates fO, bO, fC, bC, fL and bL correspond to the forward and backward rates for binding a ligand to the receptor in its open state, the forward and backward rates for binding a ligand to the receptor in its closed state and the forward and backward rates for switching from the closed state to the open state, respectively, as shown in Fig. 11. See Supplemental Material, Appendix 4. The ratio of forward and backward kinetic rates is the equilibrium constant, yielding
| (32) |
The transition matrix M in Eq. (31) includes only a subset of the elementary reactions that could affect concentrations of the various ligand–receptor configurations. For example, another elementary reaction that is often included in dynamical MWC models is O2 → C2, which describes the phenomenon of the ion channel opening and closing while ligands are bound. It is also possible to allow for rate constants for each binding site to be different or to allow for rate constants that change as a function of the total number of ligands. However, the transition matrix in Eq. (31) has sufficient complexity to give the correct qualitative behavior of the transfer function.8 Our aim is to illustrate the general principles underlying dynamical MWC models, not to find the exact kinetic rates or transfer function for a particular receptor.
Equations (26) and (31) give us a simple framework with which to analyze the dynamics of an MWC molecule. In fact, if the concentration is a function of time c(t) and the rate parameters are constant, then there is an exact analytic solution for the state vector x(t),89 namely,
| (33) |
If M is a function of x(t) because the rate parameters are being continuously altered by some feedback mechanism, then solving Eq. (26) becomes a much more challenging proposition unless there is a separation of timescales. For instance, if the equilibration time between MWC states is much smaller than the timescale on which rate constants are altered, then x(t) ≃ xeq. This assumption can be a useful approximation for simulating systems with feedback, for example, the precise adaptation of bacterial chemotactic receptors.24
Responses to changes in ligand concentration: The frequency response of an MWC molecule
One of the most interesting properties of a ligand-gated ion channel such as the nACh receptor is that it operates outside of equilibrium. Its task is to transition to the active state (i.e., open ion channel) when vesicles of ACh are released and travel across the synaptic gap. This means that, at the time of an incoming action potential, the nACh receptor must respond to what is essentially a jump in ACh concentration. Similar arguments can be made with sudden changes in chemoattractant concentration in the setting of bacterial chemotaxis or changes in the acetylation of a histone in the context of chromatin serving as two specific examples.
To see the significance of the temporal response of an MWC molecule, consider the following two hypothetical ligand-gated ion channels that can be described by the MWC model. One receptor responds very quickly to changes in ligand concentration, tracking the changes in ligand concentration with high fidelity. The second receptor takes a longer time to respond to changes in ligand concentration. If stray ligand molecules (or some other competing ligand such as nicotine) occasionally find their way to these receptors, then the signal that the two receptors send to the motor neuron soma will be quite different. The first, quick receptor will track the quickly changing concentrations of ligand, causing a correspondingly rapid change in membrane voltage. The second, slower receptor will not necessarily be able to track the quickly changing ligand concentrations, resulting in slower but more deliberate responses of the membrane voltage.
To quantify this, we could first determine how an MWC molecule with two binding sites responds when perturbed from its rest state, and we can solve this problem exactly using Eq. (33). To find the probability of the channel being open from the state vector x, we need to compute
| (34) |
where the projection vector is defined as PR = (1 1 1 0 0 0) and the total concentration of receptor molecules [R] = [O2] + [O1] + [O0] + [C0] + [C1] + [C2] is constant. Combining Eq. (34) above with Eq. (33) that describes the evolution of the state vector, we can now write the time evolution of the open probability as
| (35) |
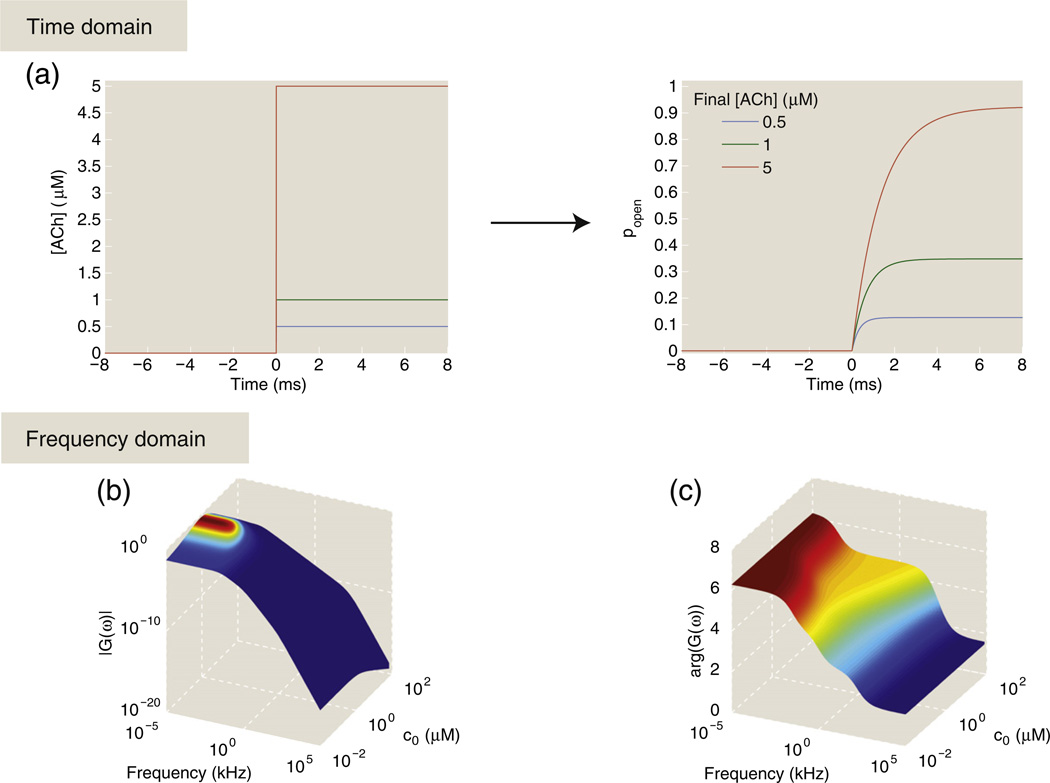
Figure 12a shows plots of popen(t) for a step function increase in ligand concentration, which could be a simple approximation for the change in ACh concentration during the transmission of an action potential, for example. The step function increase in ligand concentration leads to a slower, almost rounded step function increase in the probability of the channel opening; this curve is a linear combination of exponentials whose decay constants are the eigenvalues of the transition matrix.89
The rounded response of the ion channel to changes in the concentration suggests that the receptor has difficulty responding to quickly changing concentrations. In other words, high-frequency responses are damped relative to lower-frequency oscillations, and thus sharp changes in ligand concentration become “rounded”. A much more economical way of quantifying this idea is to calculate the frequency response function of the ion channel, which is defined as
| (36) |
where F(popen) denotes the (invertible) Fourier transform of popen(t), namely,
| (37) |
and F(c) is the Fourier transform of the ligand concentration. One of the typical assumptions of Eq. (36) is that an oscillatory ligand concentration with frequency ω will lead to an oscillatory popen at frequency ω.
The oscillatory response of the receptor is therefore characterized by its amplitude, that is, how strongly it feels the effects of that oscillatory ligand concentration, and its phase, that is, the phase delay between when ligand concentration and popen reach their respective maximum in time. The amplitude of the frequency response function |G(ω)| indicates which frequencies the system blocks and which frequencies it passes. In engineering applications, the shape of |G(ω)| is often designed to attenuate unwanted noise while passing the signal through the system largely unaltered, thereby increasing the signal-to-noise ratio. This type of filtering can substantially affect the fitness of an organism. For instance, the ability of E. coli to sense chemoattractants depends strongly on the frequency filter properties of its bacterial chemotactic receptors.92 Many biological systems act as low-pass frequency filters,8,80,90–92 which means that |G(ω)| is relatively large for small ω and that |G(ω)| trails off rapidly for ω above a certain “cutoff frequency” ωcutoff. This finding holds for MWC molecules, and the cutoff frequencies of MWC-like receptors are given by the time constants of their internal dynamics,8 as we will illustrate below for a toy model of a ligand-gated ion channel such as the nACh receptor.
To find G(ω) for a toy model of a ligand-gated ion channel, we choose a particular initial ligand concentration and consider only small fluctuations since a ligand-gated ion channel responds to changes in ligand concentration in a nonlinear way. We present only a sketch of the derivation here, but we show more mathematical details in Supplemental Material, Appendix 5. If the changes in ligand concentration are sufficiently small, then it is appropriate to deal with this system as a perturbative one and to expand the equations above about a particular ligand concentration c0, using
| (38) |
If Δc(t) < < c0, then we can similarly write
| (39) |
and
| (40) |
where |Δx(t)| < < |x0| and Δpopen(t) < < popen(c0). The entries in the transition matrix M in Eq. (31) are all linear functions of the ligand concentration c, which allows us to rewrite the transition matrix as a linear function of Δc(t),
| (41) |
where
| (42) |
and
| (43) |
This decomposition is approximately true for more complicated transition matrices whose entries are nonlinear functions of c, as can be shown using a Taylor expansion. Substituting Eqs. (41), (39) and (40) into our first-order differential equation for x in Eq. (26) yields a linear first-order differential equation
| (44) |
Recall that the quantity Δpopen can be related to the change in state vector using Eq. (34),
| (45) |
Equation (44) is easily solved in the Fourier domain because time derivatives , once Fourier transformed, turn into multiplication by iω, yielding
| (46) |
Solving for by rearranging terms in this equation above gives
| (47) |
From this, we can find the Fourier transform of Δpopen as
| (48) |
and from this, the frequency response function of this ligand-gated ion channel,
| (49) |
By diagonalizing the matrix iω − M0, we can rewrite this frequency response function as linear combinations of frequency response functions Gk(ω),
| (50) |
where
| (51) |
and ak(c0) are linear weighting coefficients that are frequency independent and ωcutoff,k are the different internal time constants of the MWC molecule. These cutoff frequencies are complicated functions of the various kinetic rates that do not have any obvious scaling relationship with the MWC parameters since equilibrium constants say nothing about how quickly the corresponding reactions occur. These filters in Eq. (51) are low-pass first-order frequency filters since
| (52) |
will be roughly constant at low frequencies, when ω < < ωcutoff,k, and will drop sharply at higher frequencies, when ω > >ωcutoff,k. Essentially, high-frequency noise in ligand concentration will be “filtered out”. A ligand-gated ion channel cannot track changes in ligand concentration that happen more quickly than the fastest time constant of its internal dynamics. The frequency response of this ligand-gated ion channel is shown in Fig. 12 as a function of both frequency and mean ligand concentration c0. The response is generally decreased when the ligand-gated ion channel is saturated, which confirms a reasonable intuition: that a ligand-gated ion channel with both binding sites filled with ligand will have a hard time responding to any changes in ligand concentration.
Fig. 12.
Characterization of the response properties of a toy model for a ligand-gated ion channel. (a) Step function increases in ligand concentration lead to smooth increases in the probability of an open channel. (b and c) Plots of the magnitude |G(ω)| and argument arg(G(ω)) of the frequency response function of the ligand-gated ion channel to small fluctuations in ligand concentration. These frequency responses are shown as a function of the mean ligand concentration c0 about which the ligand concentration fluctuations. Rate constants: fO = 1 µM−1 ms−1, bO = 170 ms−1, fC = 1 µM−1 ms−1, bC = 0.04 ms−1, fL = 1 µM−1 ms−1 and bL = 8 × 10−4 ms−1.
Here we note that a similar analysis applies to all systems whose dynamics can be described by an equation such as Eq. (26), that is, any Markovian system.89 If perturbations are larger and depend on time in a more complicated fashion, the response will not necessarily take the form of a first-order low-pass filter. A Markovian system with feedback, for instance, can act as a higher-order low-pass frequency filter.90
Discussion
In his wonderful book The Eighth Day of Creation, Horace Freeland Judson describes the work culminating in the MWC model as follows: “Two decades of work had coalesced; as the theory of the repressor had done, yet in a manner more fundamental and embracing, allostery brought diverse and apparently contradictory modes of regulation under an overarching singular vision”. In our article, we have examined the way in which this overarching singular vision can be cast in the language of statistical mechanics and used to describe a stunningly broad variety of different biological circumstances. Though we accept the proposition that science moves forward by forceful and detailed debates about very specific molecular mechanisms, we also think that it is sometimes useful to step back and take a broader view of the features that the many different molecular mechanisms have in common.
The papers from the early 1960s that introduced this important class of two-state models described both a concept and a specific class of models.3,4 One of the points of our article has been to argue that, in some cases, an emphasis on specific molecular details can lead to molecular obfuscation rather than molecular enlightenment. For example, in considering some particular molecule such as hemoglobin, the nACh receptor or some transcription factor, deep and far-reaching debates focus on very specific molecular mechanisms. An example of some of the different subsets of states that can be included when constructing a statistical mechanical or kinetic model of a given molecule is shown in Fig. 13.93,94 One can easily go even farther to include other features such as the distinction between different subunits as demonstrated clearly in the case of hemoglobin, to name but one example.16 When these mechanisms are recast in mathematical form, they lead in turn to an allied passionate discourse on the ability of this or that molecular model to “fit” particular data sets. From our perspective, the more important service of such models is to provide a unifying framework that casts completely different systems such as hemoglobin and nucleosomes in the same light and that make polarizing predictions about new classes of experiments.
Fig. 13.
Molecular cartoons showing the variety of different allowed states and subsets of states considered in different models.93,94 The states shaded in light blue correspond to the traditional MWC model. The states shaded in light pink correspond to a sequential model of the KNF form.15 The green box surrounds all of the states and generalizes the MWC scenario to include other intermediates. The version shown here is a slight variant on that presented in the excellent review by Hilser et al.94
One of the most influential tools arising from studies of biological similarity are phylogenetic trees, which succinctly capture the evolutionary history of the immense biological diversity seen in both the hidden world of microbes and macroscopic organisms. Historically, though much was learned about these questions by studying morphology and form, a powerful modern alternative is based on comparing the genome sequences of different organisms. The power of models such as the MWC model is that they serve as the basis for a different kind of phylogeny, namely, a phylogeny of concepts in which apparently completely distinct biological entities such as hemoglobin and chromatin end up being described by precisely the same physics and mathematics. This conceptual phylogeny then tells us stories about the function of one system in terms of the other in much the same way that sequence gazing allows us to understand biological function in one organism by studying another. In our view, the MWC concept should be seen as one of the key branches on the “phylogenetic tree” of fundamental concepts that tie together diverse and broad classes of molecules describing everything from enzyme action to molecular sensors (e.g., bacterial chemotactic receptors) to the physical state of genomic DNA. We have every reason to expect that the next 50 years will see many more examples of the MWC concept serving as the basis for fundamental biological insights.
Supplementary Material
Acknowledgements
We are grateful to Bill Bialek, Leonid Mirny, Oleg Igoshin, Ned Wingreen, Robert Endres, Julie Theriot, Jane Kondev, Aleksandra Walczak and Nigel Orme for insightful discussions. We are also indebted to Ron Milo, Madhav Mani, Christopher Hillar, Vanessa M. Burns, Stephanie Marzen, Qinren Zhen, Erik Madsen, Wesley Yu and the anonymous referee for their helpful comments on this paper. We are especially grateful to Prof. Jean-Pierre Changeux for many stimulating discussions and insights on the subject of allostery and the model that bears his name. Financial support from the National Institutes of Health through NIH Award Number R01 GM08421 and the Director's Pioneer Award, grant DP1 OD000217A and La Fondation Pierre Gilles de Gennes (R.P.) and the National Science Foundation Graduate Research Fellowship Program (S.M.) are gratefully acknowledged.
Abbreviations used
- MWC
Monod–Wyman–Changeux
- cGMP
cyclic guanosine monophosphate
- FRET
fluorescence resonance energy transfer
- nACh
nicotinic acetylcholine
- ACh
acetylcholine
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jmb.2013.03.013
In the more general case, this dissociation constant is the equilibrium constant for a reaction in which the ligand binds to a particular site on the receptor. As all sites of the receptor in the MWC model described here are identical, it does not matter which one we consider.
The dissociation constants and equilibrium constants are related via the equations .
The original MWC parameters measure ligand concentration α in units of the active state dissociation constant , that is, , and denote the ratio of dissociation constants as .
References
- 1.Alberts B. Molecular Biology of the Cell. 5th edit. New York, NY: Garland Science; 2008. [Google Scholar]
- 2.Phillips R, Kondev J, Theriot J, Garcia HG. In: Physical Biology of the Cell. 2nd edit. Orme N, editor. New York, NY: Garland Science; 2013. Illustrated by. [Google Scholar]
- 3.Monod J, Changeux JP, Jacob F. Allosteric proteins and cellular control systems. J. Mol. Biol. 1963;6:306–329. doi: 10.1016/s0022-2836(63)80091-1. [DOI] [PubMed] [Google Scholar]
- 4.Monod J, Wyman J, Changeux JP. On the nature of allosteric transitions: a plausible model. J. Mol. Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 5.Goldenfeld N. Lectures on Phase Transitions and the Renormalization Group. Boulder, CO: Westview Press; 1992. [Google Scholar]
- 6.Judson HF. The Eighth Day of Creation: Makers of the Revolution in Biology. Plainview, NY: Expanded Edition CSHL Press; 1996. [Google Scholar]
- 7.Graham I, Duke T. The logical repertoire of ligand-binding proteins. Phys. Biol. 2005;2:159–165. doi: 10.1088/1478-3975/2/3/003. [DOI] [PubMed] [Google Scholar]
- 8.Martins BM, Swain PS. Trade-offs and constraints in allosteric sensing. PLoS Comput. Biol. 2011;7:e1002261. doi: 10.1371/journal.pcbi.1002261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hille B. Ion Channels of Excitable Membranes. 3rd edit. Sunderland, MA: Sinauer; 2001. [Google Scholar]
- 10.Mirny LA. Nucleosome-mediated cooperativity between transcription factors. Proc. Natl Acad. Sci. USA. 2010;107:22534–22539. doi: 10.1073/pnas.0913805107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Narula J, Igoshin OA. Thermodynamic models of combinatorial gene regulation by distant enhancers. IET Syst. Biol. 2010;4:393. doi: 10.1049/iet-syb.2010.0010. [DOI] [PubMed] [Google Scholar]
- 12.Hill TL. Cooperativity Theory in Biochemistry: Steady-State and Equilibrium Systems. New York, NY: Springer-Verlag; 1985. [Google Scholar]
- 13.Dill KA, Bromberg S. Molecular Driving Forces: Statistical Thermodynamics in Biology, Chemistry, Physics, and Nanoscience. 2nd edit. London, NY: Garland Science; 2011. [Google Scholar]
- 14.Gerhart JC, Pardee AB. The enzymology of control by feedback inhibition. J. Biol. Chem. 1962;237:891–896. [PubMed] [Google Scholar]
- 15.Koshland JDE, Nemethy G, Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966;5:365–385. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- 16.Eaton WA, Henry ER, Hofrichter J, Bettati S, Viappiani C, Mozzarelli A. Evolution of allosteric models for hemoglobin. IUBMB Life. 2007;59:586–599. doi: 10.1080/15216540701272380. [DOI] [PubMed] [Google Scholar]
- 17.Changeux J-P, Edelstein SJ. Nicotinic Acetylcholine Receptors—From Molecular Biology to Cognition. New York, NY: Odile Jacob; 2005. [Google Scholar]
- 18.Rodieck RW. The First Steps in Seeing. 1st edit. Sunderland, MA: Sinauer; 1998. [Google Scholar]
- 19.Goulding EH, Tibbs GR, Siegelbaum SA. Molecular mechanism of cyclic-nucleotide-gated channel activation. Nature. 1994;372:369–374. doi: 10.1038/372369a0. [DOI] [PubMed] [Google Scholar]
- 20.Dowling JE. The Retina. 2nd edit. Cambridge, MA: The Belknap Press; 2012. [Google Scholar]
- 21.Zhong W, Gallivan JP, Zhang Y, Li L, Lester HA, Dougherty DA. From ab initio quantum mechanics to molecular neurobiology: a cation-π binding site in the nicotinic receptor. Proc. Natl Acad. Sci. USA. 1998;95:12088–12093. doi: 10.1073/pnas.95.21.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berg HC. Motile behavior of bacteria. Phys. Today. 2000;53:24–29. [Google Scholar]
- 23.Berg HC. E. coli in Motion. 1st edit. New York, NY: Springer-Verlag; 2004. [Google Scholar]
- 24.Clausznitzer D, Oleksiuk O, Lovdok L, Sourjik V, Endres RG. Chemotactic response and adaptation dynamics in Escherichia coli. PLoS Comput. Biol. 2010;6:1–11. doi: 10.1371/journal.pcbi.1000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berg HC. Random Walks in Biology. Princeton, NJ: Princeton University Press; 1993. Expanded Edition. [Google Scholar]
- 26.Sourjik V, Berg HC. Receptor sensitivity in bacterial chemotaxis. Proc. Natl Acad. Sci. USA. 2002;99:123–127. doi: 10.1073/pnas.011589998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sourjik V, Berg HC. Binding of the Escherichia coli response regulator CheY to its target measured in vivo by fluorescence resonance energy transfer. Proc. Natl Acad. Sci. USA. 2002;99:12669–12674. doi: 10.1073/pnas.192463199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mello BA, Tu Y. Quantitative modeling of sensitivity in bacterial chemotaxis: the role of coupling among different chemoreceptor species. Proc. Natl Acad. Sci. USA. 2003;100:8223–8228. doi: 10.1073/pnas.1330839100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mello BA, Tu Y. An allosteric model for heterogeneous receptor complexes: understanding bacterial chemotaxis responses to multiple stimuli. Proc. Natl Acad. Sci. USA. 2005;102:17354–17359. doi: 10.1073/pnas.0506961102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keymer JE, Endres RG, Skoge M, Meir Y, Wingreen NS. Chemosensing in Escherichia coli: two regimes of two-state receptors. Proc. Natl Acad. Sci. USA. 2006;103:1786–1791. doi: 10.1073/pnas.0507438103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Endres RG, Wingreen NS. Precise adaptation in bacterial chemotaxis through “assistance neighborhoods”. Proc. Natl Acad. Sci. USA. 2006;103:13040–13044. doi: 10.1073/pnas.0603101103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maddock JR, Shapiro L. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- 33.Greenfield D, McEvoy AL, Shroff H, Crooks GE, Wingreen NS, Betzig E, Liphardt J. Self-organization of the Escherichia coli chemotaxis network imaged with super-resolution light microscopy. PLoS Biol. 2009;7:e1000137. doi: 10.1371/journal.pbio.1000137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Briegel A, Ortega DR, Tocheva EI, Wuichet K, Li Z, Chen S, et al. Universal architecture of bacterial chemoreceptor arrays. Proc. Natl Acad. Sci. USA. 2009;106:17181–17186. doi: 10.1073/pnas.0905181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu J, Li C, Yu Z, Huang P, Wu H, Wei C, et al. Efficient and specific modifications of the Drosophila genome by means of an easy TALEN strategy. J. Genet. Genomics. 2012;39:209–215. doi: 10.1016/j.jgg.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 36.Polach KJ, Widom J. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J. Mol. Biol. 1995;254:130–149. doi: 10.1006/jmbi.1995.0606. [DOI] [PubMed] [Google Scholar]
- 37.Miller JA, Widom J. Collaborative competition mechanism for gene activation in vivo. Mol. Cell. Biol. 2003;23:1623–1632. doi: 10.1128/MCB.23.5.1623-1632.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. Genomescale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 39.Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Z, Pugh BF. High-resolution genome-wide mapping of the primary structure of chromatin. Cell. 2011;144:175–186. doi: 10.1016/j.cell.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prinsen P, Schiessel H. Nucleosome stability and accessibility of its DNA to proteins. Biochimie. 2010;92:1722–1728. doi: 10.1016/j.biochi.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 42.Davidson EH. The Regulatory Genome: Gene Regulatory Networks in Development and Evolution. Burlington, MA: Academic; 2006. [Google Scholar]
- 43.Poustelnikova E, Pisarev A, Blagov M, Samsonova M, Reinitz J. A database for management of gene expression data in situ. Bioinformatics. 2004;20:2212–2221. doi: 10.1093/bioinformatics/bth222. [DOI] [PubMed] [Google Scholar]
- 44.Gilbert SF. Developmental Biology. 9th edit. Sunderland, MA: Sinauer Associates; 2010. [Google Scholar]
- 45.Small S, Blair A, Levine M. Regulation of even-skipped stripe 2 in the Drosophila embryo. EMBO J. 1992;11:4047–4057. doi: 10.1002/j.1460-2075.1992.tb05498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ludwig MZ, Bergman C, Patel NH, Kreitman M. Evidence for stabilizing selection in a eukaryotic enhancer element. Nature. 2000;403:564–567. doi: 10.1038/35000615. [DOI] [PubMed] [Google Scholar]
- 47.Ludwig MZ, Patel NH, Kreitman M. Functional analysis of eve stripe 2 enhancer evolution in Drosophila: rules governing conservation and change. Development. 1998;125:949–958. doi: 10.1242/dev.125.5.949. [DOI] [PubMed] [Google Scholar]
- 48.Ludwig MZ, Palsson A, Alekseeva E, Bergman CM, Nathan J, Kreitman M. Functional evolution of a cis-regulatory module. PLoS Biol. 2005;3:e93. doi: 10.1371/journal.pbio.0030093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Changeux J-P. Allostery and the Monod–Wyman–Changeux model after 50 years. Annu. Rev. Biophys. 2012;41:103–133. doi: 10.1146/annurev-biophys-050511-102222. [DOI] [PubMed] [Google Scholar]
- 50.Tkačik G, Callan JCG, Bialek W. Information flow and optimization in transcriptional regulation. Proc. Natl Acad. Sci. 2008;105:12265–12270. doi: 10.1073/pnas.0806077105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walczak AM, Tkačik G, Bialek W. Optimizing information flow in small genetic networks. II. Feed-forward interactions. Phys. Rev. E. 2010;81:041905. doi: 10.1103/PhysRevE.81.041905. [DOI] [PubMed] [Google Scholar]
- 52.Blake WJ, Balazsi G, Kohanski MA, Isaacs FJ, Murphy KF, Kuang Y, et al. Phenotypic consequences of promoter-mediated transcriptional noise. Mol. Cell. 2006;24:853–865. doi: 10.1016/j.molcel.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 53.Raj A, van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell. 2008;135:216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eldar A, Elowitz MB. Functional roles for noise in genetic circuits. Nature. 2010;467:167–173. doi: 10.1038/nature09326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buchler NE, Gerland U, Hwa T. On schemes of combinatorial transcription logic. Proc. Natl Acad. Sci. USA. 2003;100:5136–5141. doi: 10.1073/pnas.0930314100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shannon CE. A mathematical theory of computation. The Bell System Technical Journal. 1948;27:379–423. [Google Scholar]
- 57.Rieke F, Warland D, de Ruyter van Steveninck R, Bialek W. Spikes: Exploring the Neural Code. Cambridge, MA: MIT Press; 1997. [Google Scholar]
- 58.Bialek W. Biophysics: Searching for Principles. Princeton, NJ: Princeton University Press; 2012. [Google Scholar]
- 59.Borst A, Theunissen FE. Information theory and neural coding. Nat. Neurosci. 1999;2:947–957. doi: 10.1038/14731. [DOI] [PubMed] [Google Scholar]
- 60.Rhee A, Cheong R, Levchenko A. The application of information theory to biochemical signaling systems. Phys. Biol. 2012;9:045011–045021. doi: 10.1088/1478-3975/9/4/045011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Adami C. Information theory in molecular biology. Phys. Life Rev. 2004;1:3–22. [Google Scholar]
- 62.Cover TM, Thomas JA. Elements of Information Theory. New York, NY: John Wiley and Sons; 2006. [Google Scholar]
- 63.Mackay DJC. Information Theory, Inference, and Leaning Algorithms. New York, NY: Cambridge University Press; 2003. [Google Scholar]
- 64.Kandel E, Schwartz J, Jessell T. Principles of Neural Science. 4th edit. New York, NY: The McGraw Hill Companies, Inc; 2000. [Google Scholar]
- 65.Galzi JL, Revah F, Bessis A, Changeux JP. Functional architecture of the nicotinic acetylcholine receptor: from electric organ to brain. Annu. Rev. Pharmacol. 1991;31:37–72. doi: 10.1146/annurev.pa.31.040191.000345. [DOI] [PubMed] [Google Scholar]
- 66.Karlin A. Emerging structure of nicotinic acetylcholine receptors. Nat. Neurosci. Rev. 2002;3:102–114. doi: 10.1038/nrn731. [DOI] [PubMed] [Google Scholar]
- 67.Corringer PJ, Novere NL, Changeux JP. Nicotinic receptors at the amino acid level. Annu. Rev. Pharmacol. Toxicol. 2000;40:431–458. doi: 10.1146/annurev.pharmtox.40.1.431. [DOI] [PubMed] [Google Scholar]
- 68.Prince RJ, Sine SM. Acetylcholine and epibatidine binding to muscle acetylcholine receptors distinguish between concerted and uncoupled models. J. Biol. Chem. 1999;274:19623–19629. doi: 10.1074/jbc.274.28.19623. [DOI] [PubMed] [Google Scholar]
- 69.Tkačik G, Callan CG, Bialek W. Information capacity of genetic regulatory elements. Phys. Rev. E. 2008;78:011910. doi: 10.1103/PhysRevE.78.011910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tkačik G, Walczak AM, Bialek W. Optimizing information flow in small genetic networks. III. A self-interacting gene. Phys. Rev. E. 2012;85:041903. doi: 10.1103/PhysRevE.85.041903. [DOI] [PubMed] [Google Scholar]
- 71.Attwell D, Laughlin SB. An energy budget for signaling in the grey matter of the brain. J. Cereb. Blood Flow Metab. 2001;21:1133–1145. doi: 10.1097/00004647-200110000-00001. [DOI] [PubMed] [Google Scholar]
- 72.Andrews BW, Iglesias PA. An information-theoretic characterization of the optimal gradient sensing response of cells. Public Library of Science Computational Biology. 2007;3:1489–1497. doi: 10.1371/journal.pcbi.0030153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Waltermann C, Klipp E. Information theory based approaches to cellular signaling. Biochim. Biophys. Acta. 2011;1810:924–932. doi: 10.1016/j.bbagen.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 74.Tkačik G, Walczak AM. Information transmission in genetic regulatory networks: a review. J. Phys.: Condens. Matter. 2011;23:153102. doi: 10.1088/0953-8984/23/15/153102. [DOI] [PubMed] [Google Scholar]
- 75.Levantino M, Spilotros A, Cammarata M, Schiro G, Ardiccioni C, Vallone B, et al. The Monod-Wyman-Changeux allosteric model accounts for the quaternary transition dynamics in wild type and a recombinant mutant human hemoglobin. Proc. Natl Acad. Sci. 2012;109:14894–14899. doi: 10.1073/pnas.1205809109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cammarata M, Levantino M, Wulff M, Cupane A. Unveiling the timescale of the R–T transition in human hemoglobin. J. Mol. Biol. 2010;400:951–962. doi: 10.1016/j.jmb.2010.05.057. [DOI] [PubMed] [Google Scholar]
- 77.Cammarata M, Levantino M, Schotte F, Anfinrud PA, Ewald F, Choi J, et al. Tracking the structural dynamics of proteins in solution using time-resolved wide-angle X-ray scattering. Nat. Methods. 2008;5:881–886. doi: 10.1038/nmeth.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Henry ER, Jones CM, Hofrichter J, Eaton WA. Can a two-state MWC allosteric model explain hemoglobin kinetics? Biochemistry. 1997;36:6511–6528. doi: 10.1021/bi9619177. [DOI] [PubMed] [Google Scholar]
- 79.Shulman RG. Spectroscopic contributions to the understanding of hemoglobin function: implications for structural biology. IUBMB Life. 2001;51:351–357. doi: 10.1080/152165401753366104. [DOI] [PubMed] [Google Scholar]
- 80.Faas GC, Schwaller B, Vergara JL, Mody I. Resolving the fast kinetics of cooperative binding: Ca2+ buffering by calretinin. PLoS Biol. 2007;5:2646–2660. doi: 10.1371/journal.pbio.0050311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hoggett JG, Kellett GL. Kinetics of the cooperative binding of glucose to dimeric yeast hexokinase P-I. J. Biochem. 1995;385:405–410. doi: 10.1042/bj3050405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cuadri-Tomé C, Barén C, Jara-Pérez V, Parody-Morreale A, Martinez JC, Cámara-Artigas A. Kinetic analysis and modelling of the allosteric behaviour of liver and muscle glycogen phosphorylases. J. Mol. Recognit. 2006;19:451–457. doi: 10.1002/jmr.772. [DOI] [PubMed] [Google Scholar]
- 83.Inobe T, Kikushima K, Makio T, Arai M, Kuwajima K. The allosteric transition of GroEL induced by metal fluoride-ADP complexes. J. Mol. Biol. 2003;329:121–134. doi: 10.1016/s0022-2836(03)00409-1. [DOI] [PubMed] [Google Scholar]
- 84.Inobe T, Arai M, Nakao M, Ito K, Kamagata K, Makio T, et al. Equilibrium and kinetics of the allosteric transition of GroEL studied by solution X-ray scattering and fluorescence spectroscopy. J. Mol. Biol. 2003;327:183–191. doi: 10.1016/s0022-2836(03)00087-1. [DOI] [PubMed] [Google Scholar]
- 85.Truhlar DG, Garrett BC, Klippenstein SJ. Current status of transition-state theory. J. Phys. Chem. 1996;100:12771–12800. [Google Scholar]
- 86.Truhlar DG, Hase WL, Hynes JT. Current status of transition-state theory. J. Phys. Chem. 1983;87:2664–2682. [Google Scholar]
- 87.Laldler KJ, King MC. The development of transition-state theory. J. Phys. Chem. 1983;87:2657–2664. [Google Scholar]
- 88.Iglesias PA, Ingalls BP. Control Theory and Systems Biology. Cambridge, MA: MIT Press; 2009. [Google Scholar]
- 89.Astrom KJ, Murray RM. Feedback Systems: An Introduction for Scientists and Engineers. Princeton, NJ: Princeton University Press; 2008. [Google Scholar]
- 90.Sakamoto N. A transfer-function representation for regulatory responses of a controlled metabolic pathway. Biosystems. 1987;20:317–327. doi: 10.1016/0303-2647(87)90050-5. [DOI] [PubMed] [Google Scholar]
- 91.Sakamoto N, Naka T. Validity of transfer-function representation of input-output relation in allosteric models. Biosystems. 1986;19:317–326. doi: 10.1016/0303-2647(86)90009-2. [DOI] [PubMed] [Google Scholar]
- 92.Andrews BW, Yi TM, Iglesias PA. Optimal noise filtering in the chemotactic response of Escherichia coli. PLoS Comput. Biol. 2006;2:e154. doi: 10.1371/journal.pcbi.0020154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Eigen M. New looks and outlooks on physical enzymology. Q. Rev. Biophys. 1968;1:3–33. doi: 10.1017/s0033583500000445. [DOI] [PubMed] [Google Scholar]
- 94.Hilser VJ, Wrabl JO, Motlagh HN. Structural and energetic basis of allostery. Annu. Rev. Biophys. 2012;41:585–609. doi: 10.1146/annurev-biophys-050511-102319. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.