Abstract
Aims: The prototypical protein disulfide bond (Dsb) formation and protein refolding pathways in the bacterial periplasm involving Dsb proteins have been most comprehensively defined in Escherichia coli. However, genomic analysis has revealed several distinct Dsb-like systems in bacteria, including the pathogen Salmonella enterica serovar Typhimurium. This includes the scsABCD locus, which encodes a system that has been shown via genetic analysis to confer copper tolerance, but whose biochemical properties at the protein level are not defined. The aim of this study was to provide functional insights into the soluble ScsC protein through structural, biochemical, and genetic analyses. Results: Here we describe the structural and biochemical characterization of ScsC, the soluble DsbA-like component of this system. Our crystal structure of ScsC reveals a similar overall fold to DsbA, although the topology of β-sheets and α-helices in the thioredoxin domains differ. The midpoint reduction potential of the CXXC active site in ScsC was determined to be −132 mV versus normal hydrogen electrode. The reactive site cysteine has a low pKa, typical of the nucleophilic cysteines found in DsbA-like proteins. Deletion of scsC from S. Typhimurium elicits sensitivity to copper (II) ions, suggesting a potential involvement for ScsC in disulfide folding under conditions of copper stress. Innovation and Conclusion: ScsC is a novel disulfide oxidoreductase involved in protection against copper ion toxicity. Antioxid. Redox Signal. 19, 1494–1506.
Introduction
Thioredoxin (TRX)2-like proteins are central to the redox biology of the cell. These proteins catalyze the thiol–disulfide exchange reactions that are essential for the maintenance of the correct thiol redox state of proteins. Such systems are of particular importance in the periplasm of pathogenic bacteria, where disulfide bond (Dsb) formation is essential for motility, type III secretion systems, and for folding a variety of toxins and secreted enzymes (24, 52).
The formation of Dsb in the periplasm of most γ-Proteobacteria is facilitated by two processes: (i) oxidative disulfide folding, and (ii) disulfide isomerization. The most extensively studied example of oxidative disulfide folding is that of the DsbAB system of Escherichia coli K-12, where periplasmic thiols are oxidized by the soluble protein DsbA, and the electrons are shuttled to the electron transport chain by the membrane-bound DsbB (4). Misfolded disulfides are then corrected by the soluble disulfide isomerase DsbC, driven by reducing power from the integral membrane protein DsbD [reviewed in (14)]. However, a wide diversity of oxidizing machineries exists among bacteria (15), exemplified by the existence of three functionally distinct DsbA proteins in Neisseria meningitidis (52).
Innovation.
Many bacterial pathogens overcome the biocidal action of copper during infection. The scsABCD locus of Salmonella encodes four proteins with thioredoxin-like motifs that restore tolerance to copper in copper-sensitive mutants of Escherichia coli. Here we have determined the structural and biochemical properties of the soluble, periplasmically located ScsC protein. We also demonstrate that ScsC contributes to the survival of Salmonella during conditions of copper stress. Our findings have implications on the mechanism by which Salmonella circumvents the toxic effects of redox-active copper (II) ions. Scs proteins are encoded by several pathogens, suggesting this system may constitute a broad protective mechanism.
In addition to the oxidative DsbAB system, S. Typhimurium also encodes an alternative DsbLI redox pair similar to that of Uropathogenic E. coli (UPEC) strain CFT073 (57), as well as a plasmid-encoded DsbA-like protein called SrgA (6). S. Typhimurium DsbA (StDsbA) is required for disulfide folding of the P-ring flagellar protein FlgI (58), DsbL is likely to be involved in the assembly of arylsulfate sulfotransferase (AssT) (18), and SrgA is necessary for the assembly of the major structural subunit of the plasmid-encoded fimbriae, PefA (6). In addition to these TRX-like proteins, the scs (suppressor of copper sensitivity) locus of S. Typhimurium encodes four proteins with TRX-like CXXC motifs (19). The expression of all four genes, denoted scsA-D, was shown to restore tolerance to copper in copper-sensitive mutants of E. coli, hence the scs nomenclature (19). The ScsA protein was predicted to be a copper-binding integral membrane complex, while the ScsB protein was reported to exhibit homology to several proteins, including the membrane-spanning disulfide interchange protein DsbD. The ScsC protein was predicted to be an outer membrane protein involved in disulfide folding, although work presented herein indicates that ScsC is a soluble periplasmic protein. ScsD was predicted to be a periplasmically located, membrane-anchored, TRX-like protein with similarities to TlpA of Bradyrhizobium japonicum or Neisseria gonorrhoeae (1). A recent study in Caulobacter crescentus has demonstrated that CcScsB maintains ScsC protein of C. crescentus (CcScsC) in a reduced form; it therefore appears that these proteins interact in a similar way to the DsbC/DsbD system. Reduced CcScsC was then shown to facilitate disulfide folding of periplasmic RNase I (of C. crescentus) and RcsF (an outer membrane protein of E. coli K12) via an isomerization mechanism (9). CcScsB was also shown to directly reduce TlpA (TRX-like-protein A), which subsequently reduces the peroxiredoxins PprX and PrxL (9). The CGYC active site motif of CcScsC is identical to that of EcDsbC (Table 1) and like this E. coli isomerase, CcScsC also forms a homodimer in solution. However, dimerization in CsScsC appears to be mediated by a long alpha helix, whereas the dimerization interface in EcDsbC is formed by β-strand interactions.
Table 1.
Redox Midpoints and pKa Values of Selected Thioredoxin-Like Proteins
| Protein | Motif | Redox midpoint Eo′ (mV) [Reference] | pKa of active cysteine [Reference] |
|---|---|---|---|
| Escherichia coli thioredoxin 1 | CGPC | −284 (20) | 7.1 (20) |
| E. coli thioredoxin 2 | CGPC | −221 (20) | 5.1 (20) |
| E. coli glutaredoxin 1 | CPYC | −233 (3) | N/A |
| E. coli glutaredoxin 3 | CPYC | −198 (3) | <5.5 (44) |
| Salmonella Typhimurium SrgA | CPPC | −154 (25) | 4.7 (25) |
| S. Typhimurium ScsC | CPYC | −132 [This work] | 3.4 [This work] |
| E. coli DsbG | CPYC | −129 (59) −126 (5) |
3.5 (48) |
| S. Typhimurium DsbA | CPHC | −126 (25) | 3.3 (25) |
| Homo sapiens protein disulfide isomerase | CGHCa | −110 (22) | 4.5 (35) |
| E. coli DsbA | CPHC | −124 (3) −125 [This work] |
3.5 (43) |
| S. Typhimurium DsbL | CPFC | −97 (25) | 3.8 (25) |
| E. coli DsbC | CGYC | −130 (63) | 4.3 (55) |
| Bacillus subtilis BdbD | CPSC | −75 (11) | <4.5 (11) |
Human protein disulfide isomerase has two CGHC motifs.
The current work describes the structural and biochemical characterization of the soluble, periplasmically located, S. Typhimurium ScsC (StScsC) protein, and provides evidence that the scs cluster has a role in protection against the toxic effects of redox-active copper (II) ions.
Results
Three-dimensional architecture of ScsC: ScsC is a soluble TRX-like protein
StScsC was crystallized in the monoclinic space group P21, the crystals diffracted to 2.04 Å and contained four molecules in the asymmetric unit. The structure was solved by molecular replacement using the TRX-like protein from Silicibacter pomeroyi (PDB: 3GYK) as a model. After subsequent building and refinement, the structure of StScsC corresponding to residues 9–186 of the processed peptide (178 residues) was refined to a final Rfactor of 19.9 (Rfree=25.3) (Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/ars). Pairwise comparison of the four symmetrically independent monomers showed no significant differences between molecules in the asymmetric unit; all following structural comparisons were performed with monomer A.
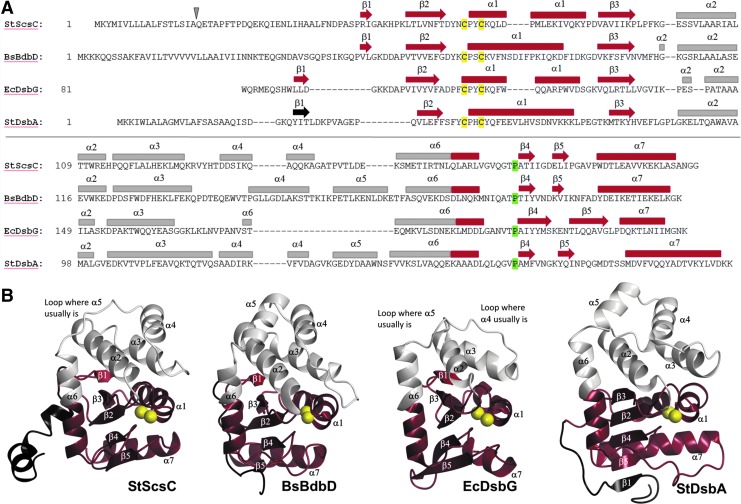
The architecture of StScsC comprises all structural hallmarks that characterize canonical thiol oxidases like StDsbA [PDB: 3L9S (25)]; it consists of a TRX domain (residues 38–75 form the βαβ motif and residues 141 to 189 form the connecting helix and the ββα motif) and a long helical insertion (residues 75–140) (Fig. 1A, B). Despite the overall structural similarity, superposition of these two proteins is poor with an r.m.s.d. value of 3.3 Å for 128 of the 189 Cα aligned. This poor match is primarily due to the different topology of StScsC, where the core β-strands (β2–β5) are structurally equivalent to those of StDsbA, but the strand β1 is hydrogen bonded to the opposite edge of the central β-sheet in the TRX domain (Figs. 1B & 2A). Another notable discrepancy between StScsC and StDsbA resides in the helical domain inserted in the TRX fold. In StDsbA, this region consists of a three helical bundle (α2–α4), plus an additional helix (α5) and an extension of the connecting helix α6, while in StScsC, a deletion in this section results in the loss of the α5 helix, which is substituted by a comparatively short loop (Fig. 1A, B). Additionally, superposition of StScsC with StDsbA (Fig. 2A) highlights a shorter loop linking β5 to α7 and a shorter α7 helix (by 1.5 turns) in StScsC compared to StDsbA. Moreover, unlike StDsbA, the StScsC β5 strand and β5-α7 loop map near the active site, a position that is stabilized by hydrogen bonds between Ile166 (O and NH) and Thr159 (NH and O) and Gly168 (NH) and cis-Pro157 (Supplementary Fig. S1). The latter modifications in StScsC alter the size and shape of the groove adjacent to the active site, which in prototypical DsbAs is defined as the hydrophobic peptide-binding groove, which binds to the redox partner DsbB (31). Other minor differences between StScsC and StDsbA include a less pronounced kink in α1 of StScsC, and longer loops linking α2, α3, and α4 in StDsbA.
FIG. 1.
Secondary structural elements of ScsC protein of Salmonella Typhimurium (StScsC) and structural homologues. (A) Structure-based sequence alignment of StScsC, BdbD from Bacillus subtilis (BsBsbD), DsbG from Escherichia coli (EcDsbG), and DsbA from S. Typhimurium (StDsbA). Cleavage site is depicted by a gray triangle, and conserved active site residues are highlighted in yellow and green. (B) Three-dimensional structures of StScsC (PDB: 4GXZ), BsBsbD [PDB: 3GHA (11)], the thioredoxin (TRX) domain of EcDsbG [PDB: 1V58 (23)], and StDsbA [PDB: 3L9S (25)]. The TRX-fold and N-terminus are shown in tan and black, respectively. Secondary structural features are labeled with the active site sulfur atoms shown as yellow spheres.
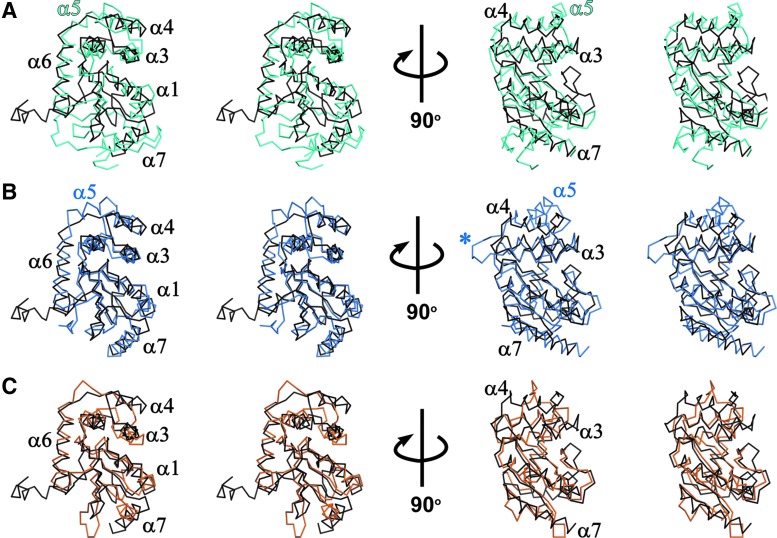
FIG. 2.
Superpositions of StScsC and structural homologues. α-carbon trace stereo views of StScsC (black) superposed with (A) StDsbA (cyan), (B) BsBdbD (blue), and (C) the TRX domain of EcDsbG (orange). α5 indicates the position of the fifth alpha helix in the inserted helical domain of DsbA-like proteins StDsbA and BsBdbD. Structures in the left panel are in the same orientation as those in Fig 1B, and in the right panel, all three structures are rotated clockwise 90° around the vertical axis, highlighting the extended loop between α3 and α4 in BsBdbD (indicated with blue asterisk).
The Dali search (29) identified 3GYK, BdbD (PDB code 3GHA), and DsbG (PDB code 1V58) as the closest structural homologues to StScsC. 3GYK and StScsC exhibit high structural similarity (r.m.s.d. value of 1.5 Å over 165 Cα aligned); this protein is a DsbA-like homologue from Silicibacter pomeroyi although no functional characterization or publication is available. Structural superposition of StScsC with Bacillus subtilis DsbA protein BsBdbD (11) (r.m.s.d. value of 2.0 Å for 148 Cα aligned) highlights features of StScsC that deviate from this close structural homologue. For example, although BsBdbD and StScsC share a similar topology (β1 interacts with β3 instead of β5), the α5 helix is present in the helical domain of BsBdbD (Fig. 1B, 2B). Rotation of these structures by 90o also shows a longer loop connecting BsBdbD α3 and α4, which protrudes into the catalytic site (Fig. 2B). StScsC also shows structural similarity with DsbG, a disulfide isomerase from E. coli (23), which has also been shown to control cysteine sulfenylation in the oxidizing periplasm (13). EcDsbG is a homodimer, where each monomer incorporates a dimerization domain and a TRX domain linked by a connecting helix. StScsC superimposes unexpectedly well with the TRX domain of EcDsbG (2.3 Å for 134 Cα aligned, Fig. 2C). Furthermore, the lack of α5 in StScsC appears to emulate the shorter insertion present in this reductase, which only contains two helices and a long loop embedded in the TRX fold (23).
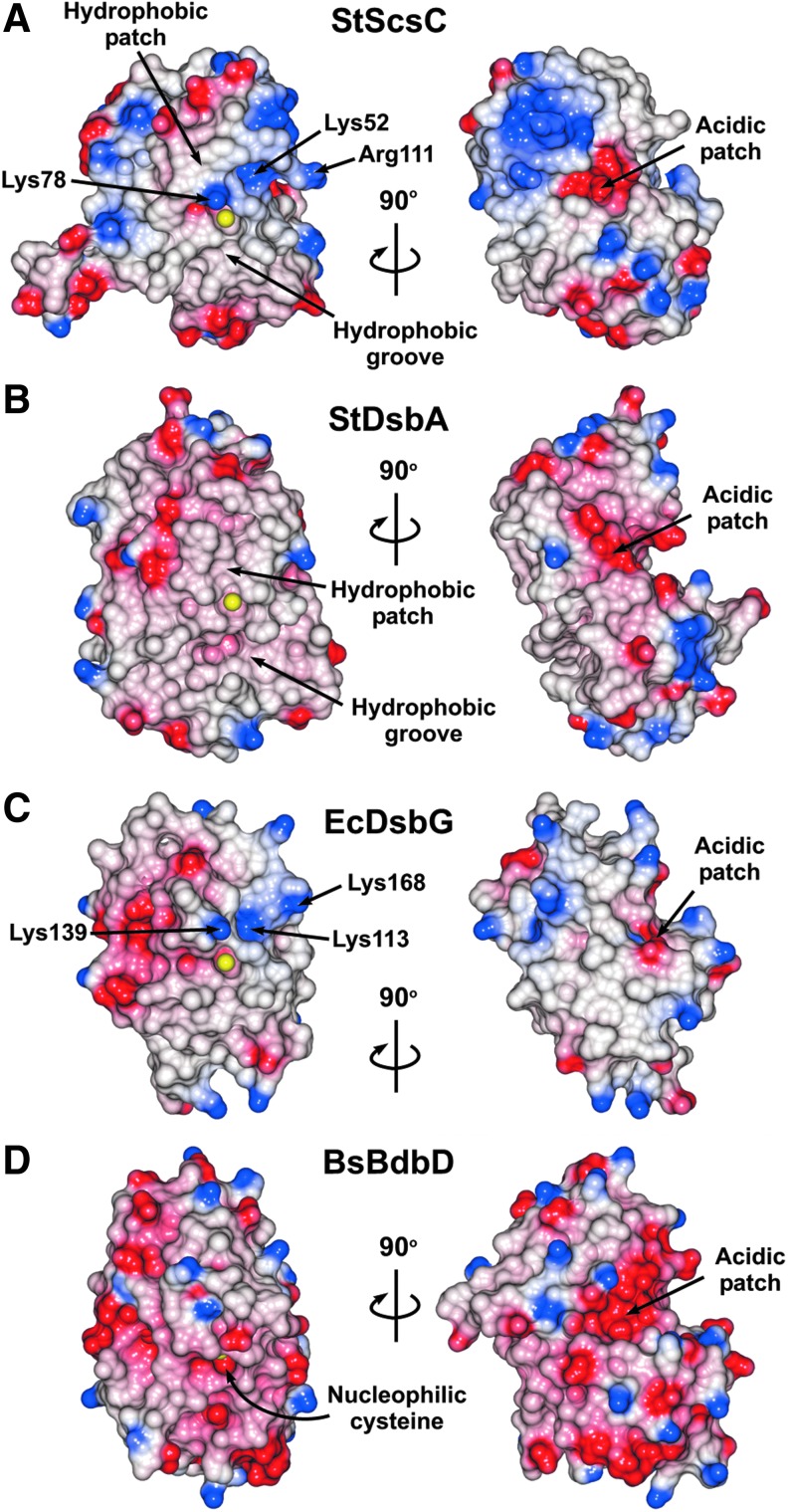
Analysis of surface electrostatics of StScsC
The electrostatic surface of StScsC partially resembles that of the thiol oxidase StDsbA, displaying a hydrophobic peptide-binding groove running along the protein surface and a hydrophobic patch, both adjacent to the CPYC catalytic motif (Fig. 3A). However, compared with StDsbA (Fig. 3B), the hydrophobic groove in StScsC is truncated. Moreover, next to the hydrophobic patch, StScsC displays a positively charged ridge above the exposed cysteine, comprising Lys52 on α1, Lys78 on the loop between β3 and α2 and Arg111 in the α3-α4 loop that overhangs the entrance to the active site cysteines (Fig. 3A). This cluster of positive residues surrounding the catalytic site is likely to stabilize the thiolate form of the nucleophilic cysteine Cys48. Interestingly, EcDsbG also has a positively charged ridge above the active site, with Lys139 on the loop between β3 and α2 directly above the exposed cysteine, although not as close as the Lys78 of StScsC. Lys168 (loop between α3 and α6) and Lys113 (α1) provide the additional positive charge to the right of Lys139 of EcDsbG (Fig. 3C).
FIG. 3.
Electrostatic surfaces of StScsC and structural homologues. Positive (blue), negative (red), and hydrophobic (white) surfaces (saturation at 20 kT/e) of (A) ScsC from S. Typhimurium, (B) DsbA from S. Typhimurium, (C) the TRX domain of DsbG form E. coli, and (D) BdbD from B. subtilis. Surface is omitted from the exposed active site cysteine residues to reveal the S atoms (shown as yellow spheres). Structures in the left panel are in the same orientation as those in Fig 1B.
Although BsBdbD is the closest structural homologue for StScsC, these two proteins are not closely related (24% sequence identity), which results in discrepancies in their surface properties. Overall, BsBdbD displays a more acidic surface, primarily in areas surrounding the catalytic CPSC motif (Fig. 3A–D). The long loop between α3 and α4 is lined with negatively charged residues that map on top of the active site (11) (Fig. 3D). Despite their different surface properties, both StScsC and BdBdbD have acidic patches in the groove between the TRX and the helical domain, which map at the opposite side of their active sites, (Fig. 3, right panels). This acidic patch is conserved in other DsbA homologues although its exact role remains unclear (40).
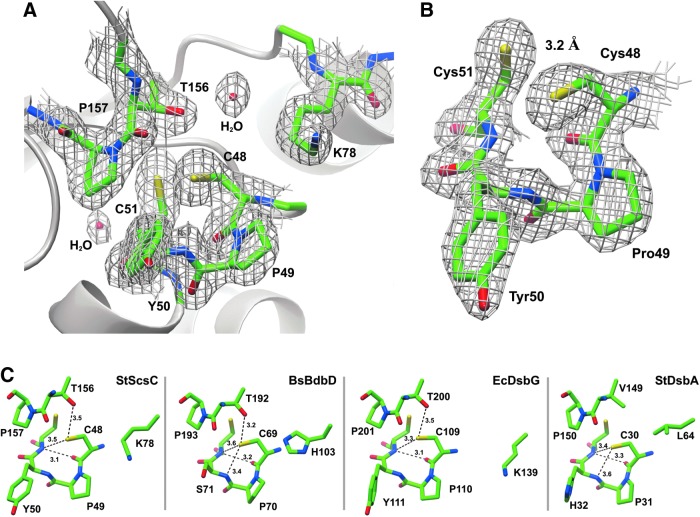
The StScsC redox active site
The catalytic active site of StScsC resembles that of the disulfide isomerase EcDsbG rather than the oxidase StDsbA, it consists of a 48CPYC51 motif located at the N-terminus of helix α1 and a cis-Pro loop (156Thr-157cisPro) in the α6-β4 connecting loop (Fig. 4A). StScsC was crystallized in its reduced form, as cleavage of the His-tag with tobacco etch virus (TEV) protease before crystallization involved incubation with 1 mM dithiothreitol (DTT). As shown in Figure 4B, the StScsC cysteine sulfur atoms are separated by a distance of 3.2 Å, which clearly indicates that these residues remained in their free thiol/thiolate forms upon crystallization. Several interactions may stabilize the reduced form of this protein; the nucleophilic N-terminal Cys48 Sγ is within hydrogen bond distance of the main chain nitrogen of the second cysteine (3.5 Å) and the side chain hydroxyl of Thr156 (3.5 Å). Furthermore, the protruding lysine residue (Lys78) forms a positively charged lip that overhangs the access route to the Cys48 residue. The close proximity between Lys78 and Cys48 could promote stabilization of the Cys48 thiolate, following a conformational rearrangement in this region; the Cδ atom of Lys78 is 3.9 Å away from the Cα atom of Cys48, but the Nζ of Lys78 is 6.6 Å from the Sγ of Cys48, so a conformational rearrangement would be required for this interaction to provide a significant stabilizing effect (Fig. 4A). The catalytic sites of BsBdbD and EcDsbG also reveal hydrogen bonds between the Sγ of the first cysteine and two acceptor groups: the second cysteine mainchain NH and the hydroxyl group of the Thr preceding the cis-Pro (Fig. 4C). The residues corresponding to StScsC Lys78 in BsBdbD and EcDsbG are His103 and Lys139, respectively. In BsBdbD, the Nδ2 of His103 is 4.8 Å away from the Sγ of Cys69, although a histidine residue is less likely to be cationic at physiological pH compared to a lysine residue, and is therefore less able to stabilize a cysteine thiolate. In EcDsbG, Lys139 generates a basic patch near the active site, which may also contribute to stabilizing the thiolate form of the cysteine (Fig. 4C).
FIG. 4.
Active site regions of StScsC and structural homologues. (A) Active sites of StScsC showing the catalytic Cys-Pro-Tyr-Cys motif, the adjacent cisPro loop (Thr156-cisPro157) and the sidechain of Lys78, which protrudes into the solvent to form a positively charged lip over the active site. The 2Fo-Fc map is also shown contoured at 1.3 σ. (B) Close-up view of the StScsC active site, refined in the reduced form (2Fo-Fc maps contoured at 1.3 σ). (C) Active site regions of StScsC, BsBdbD, EcDsbG, and StDsbA showing the hydrogen bond interactions stabilizing the side chain of the nucleophilic cysteine in each active site. Hydrogen bonds are depicted by dashed lines.
Biochemical characterization of StScsC
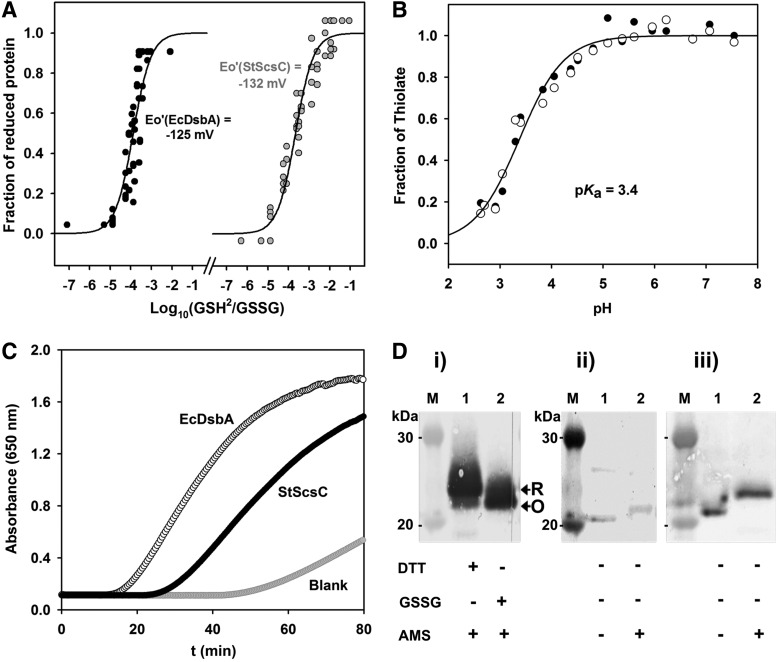
The biochemical properties of StScsC were characterized to gain further insights into its potential function. The oxidizing/reducing power of StScsC was quantified by measuring the equilibrium constant (Keq) for the disulfide exchange reaction with glutathione via monitoring changes in NMR spectra (Fig. 5A). The measured constants for StScsC and DsbA protein of E. coli (EcDsbA) were 21.7±2.6×10−5 M and 12.9±2.1×10−5 M, respectively. These Keq values convert into intrinsic redox potential values of −132 mV and −125 mV, respectively (Table 1). The redox midpoint of our EcDsbA-positive control is in close agreement with previously measured values for this protein (3). In addition, control spectra were recorded for the fully oxidized protein (using 5 mM oxidized glutathione [GSSG]) and for the fully reduced protein (using 5 mM DTT), providing confidence for the choice of peaks used to monitor the redox transition.
FIG. 5.
Biochemical characterization of StScsC. (A) Redox equilibria of EcDsbA and StScsC with glutathione. The fraction of oxidized and reduced EcDsbA and StScsC was determined using NMR spectroscopy and these data were used to calculate Keq and redox potential (Eo′) as previously described (42). Keq(StScsC)=21.7±2.6×10−5 M, Eo′(StScsC)=−132 mV, Keq(DsbA)=12.9±2.1×10−5 M, Eo′(DsbA)=−125 mV. (B) Determination of the pKa of the nucleophilic cysteine in StScsC. The pH dependence of the thiolate-specific absorbance signal (S=(A240/A280) reduced/(A240/A280) oxidized) was fitted according to the Henderson–Hasselbalch equation (the oxidized proteins were used as a reference). pKa (StScsC)=3.38±0.03. (C) Catalyzed reduction of insulin. Reduction of insulin (131 μM) was measured in a 0.1 M phosphate buffer, pH 7.0, 2 mM EDTA. The assay was performed in the absence or presence of 10 μM StScsC and 10 μM EcDsbA was used as a positive control. At time zero, DTT was added to each reaction to a final concentration of 0.35 mM, and the catalyzed reduction of insulin was measured as an increase in absorbance at 650 nm. The maximal rates have been fitted using linear regression. (D) Determination of the in vivo redox state of StScsC. (i) The fully reduced (R) and fully oxidized (O) purified StScsC is shown in lanes 1 and 2, respectively. (ii) Samples prepared from SL1344. Samples were incubated with 4-acetamino-4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS) or left untreated as indicated. The shift in the migration of StScsC following treatment with AMS demonstrates it exists in a reduced form in vivo. (iii) Samples prepared from SL1344scsC pScs. Samples were incubated with AMS or left untreated as indicated. The use of a plasmid containing the scsC gene resulted in increased expression of StScsC. As observed in the wild-type strain, shift in the migration of StScsC following treatment with AMS demonstrates it exists in a reduced form in vivo. Lane M on panels A, B, and C correspond to the 30 and 20 kDa bands of the ladder Novex Sharp protein standard (Life Technologies).
The first of the two cysteines in the CXXC motif of TRX-like proteins is a solvent-exposed, reactive nucleophilic cysteine. This reactivity is partly due to a lower pKa (∼3.5–5) compared with most cysteines (∼8.5) (43). The pKa of StScsC was measured as 3.4 (Fig. 5B), an unusually low value for a CXXC motif with such reducing power. Indeed, this is comparable to that of the oxidizing exposed thiol of EcDsbA [pKa=3.5 (43), Table 1].
Given the redox potential exhibited by StScsC is similar to that of disulfide isomerases (Table 1) and its low pKa, which is also common in highly oxidizing proteins, we investigated the in vitro oxidoreductase activity using the classic insulin reduction assay: most TRX-like oxidoreductases, including DsbC and DsbA, catalyze DTT-induced insulin reduction. StScsC catalyzed the reduction of insulin more slowly than EcDsbA, and showed an activity ∼71% of the maximal rate of EcDsbA (Fig. 5C). To further understand the function of StScsC, we also investigated its ability to catalyze Dsb isomerization using the scrambled RNaseA (scRNaseA) refolding assay (27). Under our conditions, StScsC did not show any activity in this assay (Supplementary Fig. S2). In addition, to experimentally verify that StScsC is able to perform disulfide reduction in the native host, the in vivo redox status was measured using the thiol modification reagent 4-acetamino-4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS) (Fig. 5D). Alkylation assays clearly indicate that StScsC is predominantly in the reduced state in the periplasm of S. Typhimurium.
Loss of StScsC elicits copper sensitivity in S. Typhimurium
To examine the role of StScsC in S. Typhimurium, we constructed a scsC mutant in SL1344 (referred to as SL1344scsC). Loss of StScsC expression in SL1344scsC was confirmed by Western blot analysis with a StScsC-specific antibody (data not shown). Wild-type SL1344 and SL1344scsC were assessed for copper sensitivity using disk diffusion assays. In these assays, SL1344scsC displayed enhanced sensitivity to copper, as evidenced by the larger zones of growth inhibition observed in the presence of copper (Fig. 6A). Reintroduction of the scs genes on plasmid pScs restored the wild-type phenotype, confirming a role for StScsC in resistance to copper. To confirm this role for StScsC in S. Typhimurium and to more accurately assess the concentration of copper that inhibited growth of SL1344scsC, wild-type SL1344 and SL1344scsC strains were assessed for copper sensitivity by spotting a series of 10-fold dilutions of overnight cultures onto agar plates containing a range of different copper concentrations. In these assays, wild-type SL1344 and SL1344scsC displayed significantly different sensitivity to 2.8 mM copper (Fig. 6B). Reintroduction of the scs genes on plasmid pScs restored the wild-type phenotype, confirming a role for StScsC in resistance to copper.
FIG. 6.
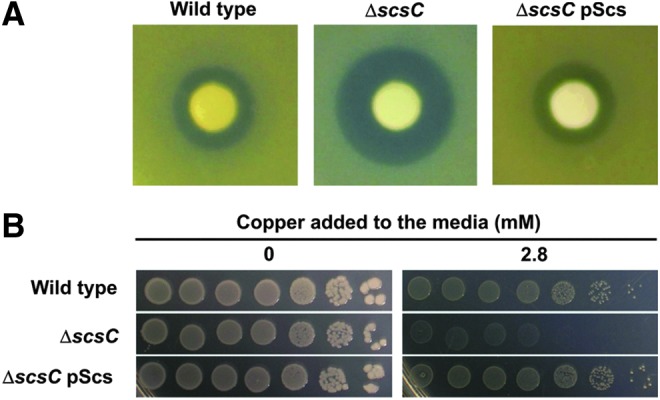
ScsC is required for S. Typhimurium copper resistance in vitro. (A) Disk diffusion assays of wild-type S. Typhimurium SL1344, SL1344scsC and SL1344scsC complemented with plasmid pScs. Sterile filter disks were placed on bacterial lawns and treated with 5 μl CuSO4 (1 M). The data presented are representative of five independent repeats. (B) Copper sensitivity and complementation of S. Typhimurium SL1344 scsC mutant was tested by spotting a series of 10-fold dilutions of cultures of the same density onto Tris agar plates containing increasing amounts of copper. Pictures were taken after a 24-h incubation at 37°C in the presence (right panel) or absence (left panel) of 2.8 mM CuSO4. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Discussion
The formation of Dsb is an essential process for the correct folding of many periplasmic and secreted proteins. Bacteria exhibit a diverse array of disulfide folding machineries (15), particularly the Gram-negative bacteria, which encode a variety of systems that have evolved to catalyze the oxidative folding of virulence proteins required for infection (18, 38). A striking example of this is S. Typhimurium, which in addition to the oxidative DsbAB system also encodes an alternative DsbLI disulfide oxidation system similar to that of UPEC strain CFT073 (18, 57), as well as a plasmid-encoded DsbA-like protein called SrgA (6). In addition to these TRX-like proteins, the scs (suppressor of copper sensitivity) locus of S. Typhimurium encodes four proteins with TRX-like CXXC motifs (19), including the soluble periplasmic ScsC protein (sequence prediction programs predict that this protein has a signal sequence and lacks membrane anchors). Scs proteins are encoded by a diverse range of bacteria, including the highly invasive UPEC strain UTI89 (7).
To date, little is known about the specific function of the Scs additional redox pathway in bacteria. A recent work on the C. crecentus Scs system showed that the membrane protein CcScsB and the soluble CcScsC form a redox pair similar to the DsbC/DsbD system. CcScsB mediates the reduction of dimeric CsScsC which, in turn, catalyzes the folding of proteins containing multiple cysteines via an isomerization mechanism (9). StScsC and CcScsC exhibit 24% sequence identity (Supplementary Fig. S3). Notable sequence differences between these two proteins include a different catalytic motif (CPYC vs. CGYC), the absence in StScsC of the long CcScsC N-terminal extension (which appears to mediate dimerization), and a deletion in CcScsC thirty residues upstream of the conserved cis-Proline present in these TRX-like proteins. To gain a better understanding of the Scs machinery in S. Typhimurium, we pursued a comprehensive structural and biochemical characterization of StScsC.
Structural characterization of StScsC revealed a canonical TRX domain with an inserted helical domain, characteristic of disulfide oxidoreductases like StDsbA or EcDsbA (25, 40). Despite this, StScsC has structural properties intermediate between DsbA-like thiol oxidases and disulfide isomerases. Indeed, a Dali search identified a DsbA homologue (BsBdbD) and an isomerase (EcDsbG) as close structural homologues for StScsC. Structural comparisons with those homologues revealed interesting peculiarities of StScsC. For example, a striking difference between StScsC and archetypal DsbA-like proteins is that the antiparallel β-strands in the TRX domain differ with respect to their topological arrangement: the β1 strand forms a β-sheet with β3 rather than β5 (Fig. 1). While this topology is uncommon in DsbA-like proteins, [only observed in few cases, including Wolbachia pipientis DsbA1 (WpDsbA1 (37)] and BsBdbD), this arrangement is present in the TRX domain of disulfide isomerases like EcDsbG and EcDsbC (23, 41). Moreover, a deletion in the helical domain of StScsC, which is not conserved in its homologue CcScsC (Fig. S2), results in StScsC lacking α5, one of the four usual alpha helices present in DsbA-like proteins like StDsbA or BsBdbD. Thus, the StScsC helical domain is somewhat reminiscent of the isomerase EcDsbG, which has a helical insertion containing two helices and a long loop substituting α4 and α5.
StScsC retains some surface features of the canonical StDsbA or EcDsbA, including a hydrophobic patch, shown to be important for substrate binding (40, 46), and a comparatively small hydrophobic groove under the active site, which in EcDsbA interacts with the partner protein EcDsbB and is key for reoxidation (31). The groove truncation in StScsC may compromise its ability to interact with StDsbB, maintaining StScsC separate from the S. Typhimurium StDsbA/StDsbB oxidative pathway. Furthermore, unlike archetypal DsbAs, the surface of StScsC also incorporates positively charged residues that generate basic protrusions flanking the 48CPYC51 active site (Fig. 3A). The latter surface properties resemble the lysines found adjacent to the also conserved 109CPYC112 active site of EcDsbG (Fig. 3C) and may contribute to stabilizing the thiolate form of the nucleophilic N-terminal cysteine.
StScsC differs from EcDsbG in that this protein is monomeric. Disulfide isomerases, EcDsbC and EcDsbG, contain extended N-terminal domains that allow dimerization, which is essential for their isomerase activity (50, 51). StScsC has an N-terminal helix that projects into the solvent (Fig. 1B, black, initial eight residues were not modeled due to weak electron density). This N-terminal helical domain is shorter than the one predicted in the C. crescentus homologue CcScsC (Supplementary Fig. S3) which is proposed to promote dimerization (9). Thus, unlike CcScsC or EcDsbG, StScsC purifies as a monomer. Furthermore, a protein interfaces, surfaces, and assemblies analysis (62) of StScsC crystal packing revealed no indication of dimer formation.
The structural analysis of ScsC revealed significant similarities to the TRX family proteins catalyzing both oxidation and isomerization/reduction. The redox properties of StScsC are similar to those of the archetypal disulfide-oxidizing DsbA and disulfide-reducing DsbG (Table 1). Furthermore, the pKa of the exposed nucleophilic cysteine of StScsC is 3.4 (Fig. 5B), a low value that is a common characteristic of Dsb-like proteins like DsbA and DsbG (Table 1). This low pKa may arise from the sulfur atom of Cys48 being hydrogen bonded to the backbone amide group of Cys51 and the hydroxyl of Thr156 in the cis-Pro loop as well as being flanked by a cluster of positively charged residues (Figs. 3 and 4) all of which contribute to the stabilization of Cys48 in the thiolate form. The low pKa value of the nucleophilic cysteine for StScsC would favor a reduced disulfide, which is consistent with the relatively oxidizing redox midpoint of −132 mV (this disulfide is relatively easy to reduce). In the case of EcDsbA, a linear relationship between the pKa of the exposed CXXC cysteine and redox midpoint of the active site thiol pair has been demonstrated, lower pKa values pairing up with higher, more oxidizing, redox midpoints (36). Hence, with a low pKa and a low redox midpoint, StScsC exhibits typical characteristics of a Dsb disulfide oxidoreductase. A common activity for TRX-like oxidoreductases is the reduction of insulin; StScsC and EcDsbA reduce insulin at a similar rate (Fig. 5C). Although some structural properties of StScsC are characteristic of E. coli disulfide isomerases, EcDsbC and EcDsbG, these features do not confer such activity to the protein. By contrast, its dimeric orthologue CcScsC catalyzes isomerization of Dsb in substrate RNase I (9). Our present work shows that StScsC is monomeric, and this probably limits the ability of the protein to reshuffle non-native disulfide linkages. Indeed, previous work has shown that modification of dimeric EcDsbC into a monomeric molecule dramatically reduces its disulfide isomerase activity (33). This suggests that there may be functional diversity within members of the ScsC family of proteins.
Copper (II) is known to drive the oxidation of thiols (53) and since StScsC is required for protection against copper stress, this suggests that StScsC is part of a system in S. Typhimurium that is dedicated to protecting periplasmic proteins against the formation of non-native Dsb induced by this transition metal. The mechanistic role that StScsC plays is still unclear; however, our results indicate that StScsC exists in the reduced state in resting cells, which may indicate that this protein is a disulfide reducing protein. The reason why S. Typhimurium would possess a system dedicated to protection against copper-catalyzed Dsb formation is not entirely clear, but it is noted that copper ions have recently been shown to play an antimicrobial role in the innate immune response and there is evidence that this ion is trafficked to the phagosome in macrophages (28, 60). In macrophage infection studies, we did not observe any difference in the bacterial load when we compared wild-type S. Typhimurium with the scsC mutant (Achard & McEwan, unpublished), but it should be noted that copper tolerance in S. Typhimurium is complex and no single locus may be essential for survival in the macrophage.
Materials and Methods
Production of purified proteins
Native ScScsC was expressed using a method similar to the one descried previously for other S. Typhimurium DsbA-like proteins (32). Briefly, the coding DNA sequence for ScScsC lacking the signal peptide (residues 19–189), was cloned into a pET21a-based LIC vector (for cloning primers, see Supplementary Table S1). The recombinant protein containing an N-terminal hexa-histidine tag was expressed using autoinduction (54), and then purified by Ni2+-affinity chromatography and gel filtration chromatography (in 50 mM HEPES pH 7.0, 150 mM NaCl). Upon removal of the tagged signal peptide by cleavage with TEV protease (in a gel filtration buffer supplemented with 1 mM DTT), uncleaved protein was removed using cobalt affinity (Talon resin) chromatography. Histidine-tagged E. coli DsbA (EcDsbA) was purified as previously described (48).
Crystallization and diffraction data collection
Purified StScsC (in 50 mM HEPES pH 7.0, 150 mM NaCl) was stored in aliquots frozen at −80°C until needed. StScsC was concentrated using a 3K Amicon centrifugal concentrator up to 26 mg·ml−1. High-throughput crystallization experiments were performed with a Mosquito robot using commercial screening kits. Initial condition screening was performed in the hanging-drop format in 96-well plates, StScsC was at 26 mg·ml−1 and the commercial screens JCSG (Molecular Dimensions), Index (Hampton), and PegRx1/PegRx2 (Hampton) were used. Drops were set using the Mosquito® robot (TTP LabTech Ltd.) by mixing 100 nL of protein with 100 nL of well condition. The plates were incubated at 20°C and monitored and scored using the RockImager and Rockmaker system (Formulatrix, Inc.). Crystals were observed in three PegRx conditions (PegRx1 #33, 0.1 M Tris pH 8.0, 28% w/v PEG 4000; PegRx2 #28, 18% v/v 2-Propanol, 0.1 M sodium citrate tribasic dihydrate pH 5.5, 20% w/v PEG 4000; PegRx2 #29, 6% v/v Tacsimate pH 6.0, 0.1 M 2-(N-morpholino)ethanesulfonic acid monohydrate pH 6.0, 25% w/v PEG 4000). Optimization using random and grid screening strategies produced crystals that diffracted to 2.04 Å. These crystals were grown in 96-well plates in gradient screens of 18%–27% (w/v) PEG 4000 and 0.1 M Tris buffer pH 8.2–9.2. A further round of optimization designed to grow larger crystals free of precipitate was performed in 24-well plates using 18%–27% (w/v) PEG 4000 with 0.1 M Tris buffer pH 8.2 with protein at 9 mg·ml−1 using microseeding. The hanging drops were prepared by adding 1 μl of protein (9 mg·ml−1) to 1 μl of well condition and were microseeded within 30 min of setup with a seed stock prepared from small crystals grown in the same condition. Diffraction data were collected on a crystal (∼300×50×50 μm) grown by this method (Supplementary Fig. S4) with a well condition of 22% (w/v) PEG 4000 in 0.1 M Tris buffer pH 8.2. The crystal was cryoprotected by the well solution supplemented with 25% ethylene glycol and flash cooled in the cryostream. Data were collected using a Rigaku FR-E Superbright X-ray generator with Osmic HiRes2 optics and a Rigaku R-Axis IV++ Image Plate detector. CrystalClear 2.0 was used to record the diffraction and indexing and scaling was performed using HKL2000 (45).
Structure determination
The crystal structure of StScsC was solved by molecular replacement using BALBES (39) with a final search model based on the crystal structure of a TRX-like oxidoreductase from Silicibacter pomeroyi (PDB: 3GYK): Model building and refinement were carried out iteratively using COOT (16) and Phenix (2). Structure validation was performed using Molprobity (8). Supplementary Table S1 provides the statistics for the X-ray data collection and final refined model. Superposition of molecules and generation of molecular figures (including electrostatic potential) were carried out using the CCP4 Molecular Graphics program (47).
Bioinformatic analyses
Pairwise sequence alignments and calculation of sequence identities were performed using Biolign (21). Structural alignments were performed using MUSTANG (34), and the secondary structure was assigned using PDBsum.
Determination of the equilibrium constants with glutathione
The redox equilibrium of StScsC with glutathione was determined alongside that of EcDsbA as a positive control, using a recently developed NMR approach (56) (using his-tagged proteins). To determine the reduction potentials, different ratios of [reduced glutathione (GSH)]/[GSSG] were used, where [GSH]+[GSSG] equalled 5 mM. All NMR data were obtained at 298 K using a 14.1 T (600 MHz 1H) Bruker Avance III NMR spectrometer equipped with a QCI-F cryoprobe. All NMR samples were 330 μL within a Shigemi NMR tube and contained 100 μM protein in a 20 mM sodium phosphate buffer at pH 6.5 containing 50 mM sodium chloride and 5% (v/v) deuterium oxide. All buffers had nitrogen bubbled through to ensure the removal of any oxygen. We found that bubbling for 30–60 min was sufficient to remove all effects of dissolved oxygen and the redox equilibrium was complete in all samples within 5 min of equilibration of buffers. All 15N-1H HSQC spectra were acquired with 2048 points in the direct F2 dimension (1H) and 256 points in the F1 dimension (15N). NMR data processing was completed using NMRpipe. NMR backbone assignments have previously been completed for EcDsbA (10) and were used to confirm analysis. Two control spectra were recorded for each protein (Supplementary Fig. S5), the fully oxidized protein (using 5 mM GSSG) and the fully reduced protein (using 5 mM DTT). To determine the reduction potential of each protein, the fraction reduced was calculated for four and six resonances for StScsC and EcDsbA, respectively. The equilibrium constant Keq and redox midpoint potential were determined from standard thermodynamic equations (42). A redox midpoint potential of −240 mV was used for the glutathione couple (49).
Determination of pKa values
The pKa of the nucleophilic cysteine in StScsC was determined by monitoring the specific absorbance of the thiolate anion at 240 nm (43). Measurements were carried out at room temperature in a buffer system consisting of 10 mM K2HPO4, 10 mM boric acid, 10 mM sodium succinate, 1 mM EDTA, and 200 mM KCl, pH 7.5. Oxidized and reduced protein samples (final concentration of 20 μM) were prepared by incubating the proteins with 20 mM GSSG or 10 mM DTT, respectively. Upon removal of the oxidizing and reducing agents using a PD10 column (GE Healthcare), the absorbance of the samples at 240 and 280 nm was recorded as the pH of the protein solution was lowered to 2.5 by the stepwise addition of aliquots of 0.2 M HCl. The pH dependence of the thiolate-specific absorbance signal (S=(A240/A280)reduced/(A240/A280)oxidized) was fitted according to the Henderson–Hasselbalch equation as previously described (42). The oxidation state of StScsC samples was assessed using the DTNB assay with an extinction coefficient of ɛ412= 13.8 mM−1 cm−1 (17).
Insulin reduction and refolding of scRNaseA assays
The disulfide oxidoreductase activity of StScsC and EcDsbA was determined by measuring the ability to catalyze insulin reduction in the presence of DTT as described previously (37). The in vitro isomerase activity of StScsC, EcDsbC, and EcDsbA was assessed using the refolding of scRNaseA assay essentially as described by Hillson et al. (27).
Bacterial strains, plasmids, and culture conditions
S. Typhimurium SL1344 strains (wild-type and deletion mutants) and E. coli strains (BL21[DE3] and DH5α) were routinely cultured at 37°C on the solid or in liquid Luria Bertani (LB) medium supplemented with the appropriate antibiotics; kanamycin (Km, 50 μg·ml−1), ampicillin (Ap, 100 μg·ml−1 or chloramphenicol (Cm, 30 μg·ml−1). Culture media were supplemented with 1 mM isopropyl β-D-thiogalactopyranoside (IPTG) to induce expression of StScsABCD from plasmid pScs. Plasmid pScs was generated by PCR amplification of the scsABCD operon (primers scsA-DF, scsA-DR) from the chromosome of SL1344 and subsequent cloning into EcoRI-XhoI digested pWSK29 (61). Expression of cloned genes was controlled by the inducible lac promoter. All primers used in this section are shown in Supplementary Table S2.
Construction of S. Typhimurium SL1344 mutants
To delete the scsC gene, a three-step PCR procedure was employed to generate an amplification product containing the Km cassette from pKD4 (12) flanked on both sides by ∼500 bp of DNA sequence homologous to the target gene to be modified. The following sets of primers were used for scsC: scsCP1F, scsCP2R, scsCP5F, and scsCP6R (Supplementary Table S2). These primers were used in combination with the Km cassette amplification primers KanP3 and KanP4 (Supplementary Table S2). The final three-step PCR product for each target gene was gel-purified and cloned into pT7Blue (Merck) according to the manufacturer's instructions, to generate a donor plasmid. SL1344 deletion mutants were constructed using the gene gorging method as previously described (26). Briefly, SL1344 cells were electrotransformed with a donor plasmid [pT7Blue::scsC::kan, and a mutagenesis plasmid (pACBSR, (26)]. Cells containing both plasmids were cultured for 9 h in the LB broth supplemented with 25 μg·ml−1 Cm and 0.2% (w/v) L-arabinose to induce the λ-Red and I-SceI genes on pACBSR. Induction of these genes leads to linearization of the donor plasmid and a double recombination event with the regions flanking scsC, resulting in disruption of the respective gene. Clones sensitive to Ap and Cm (but resistant to Km) were selected and deletion of scsC was confirmed by PCR and subsequent DNA sequencing.
Copper sensitivity assays
Copper sensitivity was assessed via growth on solid media. For disk diffusion assays, strains were grown at 37°C to mid-exponential phase, adjusted to the same cell density and plated using the agar overlay technique on LB supplemented with 1.5% agar. Sterile filter disks soaked with 5 μL of 1 M CuSO4 were placed on the center of each plate and the zones of inhibition were measured after an overnight incubation at 37°C. The data presented are representative of three independent experiments. For colony spot assays, serial 10-fold dilutions of cultures of the same density were prepared and 5 μl of each dilution was spotted on the Tris agar medium supplemented with 1.4–3.6 mM CuSO4 (at 0.2 mM increments) or left untreated. Pictures were taken after 24 h of incubation at 37°C. The data presented are representative of three independent experiments comparing growth in the presence or absence of 2.8 mM CuSO4.
Determination of the in vivo redox state of StScsC
The periplasmic protein fraction of SL1344 and SL1344scsC pScs was prepared by cold osmotic shock. Bacteria grown overnight at 37°C were washed in an ice-cold buffer containing 20% surcrose, 200 mM Tris HCl, pH8, and cells were harvested by centrifugation and resuspended in ice-cold 10 mM Tris HCl, pH8. After centrifugation, the supernatant was retained for analysis. The redox state of ScsC was measured by thiol alkylation using AMS and analysis of the molecular mass of proteins following sodium dodecyl sulfate polyacrylamide gel electrophoresis and Western blotting as previously described (30). ScsC proteins were detected using a StScsC-specific antibody (1:100) and an anti-rabbit IgG coupled with alkaline phosphatase (1:10000; Sigma) as the secondary antibody. Blots were stained with 5-bromo-4-chloro-3-indolyl-phosphate/nitro blue tetrazolium.
Supplementary Material
Abbreviations Used
- AMS
4-acetamino-4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid
- Ap
ampicillin
- BCIP/NBT
5-bromo-4-chloro-3-indolyl-phosphate/nitro blue tetrazolium
- BsBdbD
BdbD protein of B. subtilis
- CcScsC
ScsC protein of C. crescentus
- Cm
chloramphenicol
- Dsb
disulfide bond
- DTT
dithiothreitol
- EcDsbA
DsbA protein of E. coli
- GSH
reduced glutathione
- GSSG
oxidized glutathione
- IPTG
isopropyl β-D-thiogalactopyranoside
- Km
kanamycin
- MES
2-(N-morpholino)ethanesulfonic acid
- PISA
protein interfaces, surfaces, and assemblies
- SDS PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- StDsbA
DsbA protein of S. Typhimurium
- StScsC
ScsC protein of S. Typhimurium
- TRX
thioredoxin
- UPEC
uropathogenic E. coli
Acknowledgments
This work was supported by an Australian Research Council (ARC) grant DP1096395 to M.A.S., A.G.M., and B.H. M.A.S. is supported by an ARC Future Fellowship (FT100100662) and B.H. by a La Trobe Institute for Molecular Science Fellowship, (La Trobe University). The Wellcome Trust supported the reduction potential NMR experiments via Equipment Grant 091163/Z/10/Z to M.J.H. The authors acknowledge the use of the Australian Synchrotron and the UQ ROCX diffraction Facility.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Achard MES. Hamilton AJ. Dankowski T. Heras B. Schembri MA. Edwards JL. Jennings MP. McEwan AG. A periplasmic thioredoxin-like protein plays a role in defense against oxidative stress in Neisseria gonorrhoeae. Infect Immun. 2009;77:4934–4939. doi: 10.1128/IAI.00714-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Afonine PV. Mustyakimov M. Grosse-Kunstleve RW. Moriarty NW. Langan P. Adams PD. Joint X-ray and neutron refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2010;66:1153–1163. doi: 10.1107/S0907444910026582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ãslund F. Berndt KD. Holmgren A. Redox potentials of glutaredoxins and other thiol-disulfide oxidoreductases of the thioredoxin superfamily determined by direct protein-protein redox equilibria. J Biol Chem. 1997;272:30780–30786. doi: 10.1074/jbc.272.49.30780. [DOI] [PubMed] [Google Scholar]
- 4.Bader M. Muse W. Ballou DP. Gassner C. Bardwell JCA. Oxidative protein folding is driven by the electron transport system. Cell. 1999;98:217–227. doi: 10.1016/s0092-8674(00)81016-8. [DOI] [PubMed] [Google Scholar]
- 5.Bessette PH. Cotto JJ. Gilbert HF. Georgiou G. In vivo and in vitro function of the Escherichia coli periplasmic cysteine oxidoreductase DsbG. J Biol Chem. 1999;274:7784–7792. doi: 10.1074/jbc.274.12.7784. [DOI] [PubMed] [Google Scholar]
- 6.Bouwman CW. Kohli M. Killoran A. Touchie GA. Kadner RJ. Martin NL. Characterization of SrgA, a Salmonella enterica serovar typhimurium virulence plasmid-encoded paralogue of the disulfide oxidoreductase DsbA, essential for biogenesis of plasmid-encoded fimbriae. J Bacteriol. 2003;185:991–1000. doi: 10.1128/JB.185.3.991-1000.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen SL. Hung CS. Xu JA. Reigstad CS. Magrini V. Sabo A. Blasiar D. Bieri T. Meyer RR. Ozersky P. Armstrong JR. Fulton RS. Latreille JP. Spieth J. Hooton TM. Mardis ER. Hultgren SJ. Gordon JI. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: a comparative genomics approach. Proc Natl Acad Sci U S A. 2006;103:5977–5982. doi: 10.1073/pnas.0600938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen VB. Arendall WB., III Headd JJ. Keedy DA. Immormino RM. Kapral GJ. Murray LW. Richardson JS. Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho S-H. Parsonage D. Thurston C. Dutton RJ. Poole LB. Collet J-F. Beckwith J. A new family of membrane electron transporters and its substrates, including a new cell envelope peroxiredoxin, reveal a broadened reductive capacity of the oxidative bacterial cell envelope. mBio. 2012;3:e00291–11. doi: 10.1128/mBio.00291-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Couprie J. Remerowski ML. Bailleul A. Courcon M. Gilles N. Quemeneur E. Jamin N. Differences between the electronic environments of reduced and oxidized Escherichia coli DsbA inferred from heteronuclear magnetic resonance spectroscopy. Protein Sci. 1998;7:2065–2080. doi: 10.1002/pro.5560071003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crow A. Lewin A. Hecht O. Carlsson Moller M. Moore GR. Hederstedt L. Le Brun NE. Crystal structure and biophysical properties of Bacillus subtilis BdbD. J Biol Chem. 2009;284:23719–23733. doi: 10.1074/jbc.M109.005785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Datsenko KA. Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Depuydt M. Leonard SE. Vertommen D. Denoncin K. Morsomme P. Wahni K. Messens J. Carroll KS. Collet JF. A periplasmic reducing system protects single cysteine residues from oxidation. Science. 2009;326:1109–1111. doi: 10.1126/science.1179557. [DOI] [PubMed] [Google Scholar]
- 14.Depuydt M. Messens J. Collet J-F. How proteins form disulfide bonds. Antiox Redox Signal. 2011;15:49–66. doi: 10.1089/ars.2010.3575. [DOI] [PubMed] [Google Scholar]
- 15.Dutton RJ. Boyd D. Berkmen M. Beckwith J. Bacterial species exhibit diversity in their mechanisms and capacity for protein disulfide bond formation. Proc Natl Acad Sci U S A. 2008;105:11933–11938. doi: 10.1073/pnas.0804621105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emsley P. Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 17.Eyer P. Worek F. Kiderlen D. Sinko G. Stuglin A. Simeon-Rudolf V. Reiner E. Molar absorption coefficients for the reduced Ellman reagent: reassessment. Anal Biochem. 2003;312:224–227. doi: 10.1016/s0003-2697(02)00506-7. [DOI] [PubMed] [Google Scholar]
- 18.Grimshaw JPA. Stirnimann CU. Brozzo MS. Malojcic G. Gruetter MG. Capitani G. Glockshuber R. DsbL and DsbI form a specific dithiol oxidase system for periplasmic arylsulfate sulfotransferase in uropathogenic Escherichia coli. J Mol Biol. 2008;380:667–680. doi: 10.1016/j.jmb.2008.05.031. [DOI] [PubMed] [Google Scholar]
- 19.Gupta SD. Wu HC. Rick PD. A Salmonella typhimurium genetic locus which confers copper tolerance on copper-sensitive mutants of Escherichia coli. J Bacteriol. 1997;179:4977–4984. doi: 10.1128/jb.179.16.4977-4984.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hajjaji HE. Dumoulin M. Matagne A. Colau D. Roos G. Messens J. Collet J-F. The zinc center influences the redox and thermodynamic properties of Escherichia coli thioredoxin 2. J Mol Biol. 2009;386:60–71. doi: 10.1016/j.jmb.2008.11.046. [DOI] [PubMed] [Google Scholar]
- 21.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser (Oxf) 1999;41:95–98. [Google Scholar]
- 22.Hawkins HC. Denardi M. Freedman RB. Redox properties and cross-linking of the dithiol disulfide active-sites of mammalian protein disulfide-isomerase. Biochem J. 1991;275:341–348. doi: 10.1042/bj2750341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heras B. Edeling MA. Schirra HJ. Raina S. Martin JL. Crystal structures of the DsbG disulfide isomerase reveal an unstable disulfide. Proc Natl Acad Sci U S A. 2004;101:8876–8881. doi: 10.1073/pnas.0402769101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heras B. Shouldice SR. Totsika M. Scanlon MJ. Schembri MA. Martin JL. DSB proteins and bacterial pathogenicity. Nat Rev Microbiol. 2009;7:215–225. doi: 10.1038/nrmicro2087. [DOI] [PubMed] [Google Scholar]
- 25.Heras B. Totsika M. Jarrott R. Shouldice SR. Guncar G. Achard MES. Wells TJ. Argente MP. McEwan AG. Schembri MA. Structural and functional characterization of three DsbA paralogues from Salmonella enterica serovar Typhimurium. J Biol Chem. 2010;285:18423–18432. doi: 10.1074/jbc.M110.101360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herring CD. Glasner JD. Blattner FR. Gene replacement without selection: regulated suppression of amber mutations in Escherichia coli. Gene. 2003;311:153–163. doi: 10.1016/s0378-1119(03)00585-7. [DOI] [PubMed] [Google Scholar]
- 27.Hillson DA. Lambert N. Freedman RB. Formation and isomerization of disulfide bonds in proteins—protein disulfide-isomerase. Methods Enzymol. 1984;107:281–294. doi: 10.1016/0076-6879(84)07018-x. [DOI] [PubMed] [Google Scholar]
- 28.Hodgkinson V. Petris MJ. Copper homeostasis at the host-pathogen interface. J Biol Chem. 2012;287:13549–13555. doi: 10.1074/jbc.R111.316406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holm L. Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inaba K. Ito K. Paradoxical redox properties of DsbB and DsbA in the protein disulfide-introducing reaction cascade. EMBO J. 2002;21:2646–2654. doi: 10.1093/emboj/21.11.2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inaba K. Murakami S. Suzuki M. Nakagawa A. Yamashita E. Okada K. Ito K. Crystal structure of the DsbB-DsbA complex reveals a mechanism of disulfide bond generation. Cell. 2006;127:789–801. doi: 10.1016/j.cell.2006.10.034. [DOI] [PubMed] [Google Scholar]
- 32.Jarrott R. Shouldice SR. Guncar G. Totsika M. Schembri MA. Heras B. Expression and crystallization of SeDsbA, SeDsbL and SeSrgA from Salmonella enterica serovar Typhimurium. Acta Crystallogr F Struct Biol Cryst Comm. 2010;66:601–604. doi: 10.1107/S1744309110011942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ke H. Zhang S. Li J. Howlett GJ. Wang C-c. Folding of Escherichia coli DsbC: characterization of a monomeric folding intermediate. Biochemistry. 2006;45:15100–15110. doi: 10.1021/bi061511m. [DOI] [PubMed] [Google Scholar]
- 34.Konagurthu AS. Whisstock JC. Stuckey PJ. Lesk AM. MUSTANG: a multiple structural alignment algorithm. Proteins. 2006;64:559–574. doi: 10.1002/prot.20921. [DOI] [PubMed] [Google Scholar]
- 35.Kortemme T. Darby NJ. Creighton TE. Electrostatic interactions in the active site of the N-terminal thioredoxin-like domain of protein disulfide isomerase. Biochemistry. 1996;35:14503–14511. doi: 10.1021/bi9617724. [DOI] [PubMed] [Google Scholar]
- 36.Krause G. Lundstrom J. Barea JL. Delacuesta CP. Holmgren A. Mimicking the active-site of protein disulfide isomerase by substitution of proline 34 in Escherichia coli Thioredoxin. J Biol Chem. 1991;266:9494–9500. [PubMed] [Google Scholar]
- 37.Kurz M. Iturbe-Ormaetxe I. Jarrott R. Shouldice SR. Wouters MA. Frei P. Glockshuber R. O'Neill SL. Heras B. Martin JL. Structural and functional characterization of the oxidoreductase alpha-DsbA1 from Wolbachia pipientis. Antiox Redox Signal. 2009;11:1485–1500. doi: 10.1089/ars.2008.2420. [DOI] [PubMed] [Google Scholar]
- 38.Lafaye C. Iwema T. Carpentier P. Jullian-Binard C. Kroll JS. Collet J-F. Serre L. Biochemical and structural study of the homologues of the thiol-disulfide oxidoreductase DsbA in Neisseria meningitidis. J Mol Biol. 2009;392:952–966. doi: 10.1016/j.jmb.2009.07.056. [DOI] [PubMed] [Google Scholar]
- 39.Long F. Vagin AA. Young P. Murshudov GN. BALBES: a molecular-replacement pipeline. Acta Crystallogr D Biol Crystallogr. 2008;64:125–132. doi: 10.1107/S0907444907050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin JL. Bardwell JCA. Kuriyan J. Crystal structure of the DsbA protein required for disulfide bond formation in vivo. Nature. 1993;365:464–468. doi: 10.1038/365464a0. [DOI] [PubMed] [Google Scholar]
- 41.McCarthy AA. Haebel PW. Torronen A. Rybin V. Baker EN. Metcalf P. Crystal structure of the protein disulfide bond isomerase, DsbC, from Escherichia coli. Nat Struct Biol. 2000;7:196–199. doi: 10.1038/73295. [DOI] [PubMed] [Google Scholar]
- 42.Mossner E. Huber-Wunderlich M. Glockshuber R. Characterization of Escherichia coli thioredoxin variants mimicking the active-sites of other thiol/disulfide oxidoreductases. Protein Sci. 1998;7:1233–1244. doi: 10.1002/pro.5560070519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nelson JW. Creighton TE. Reactivity and ionization of the active site cysteine residues of DsbA, a protein required for disulfide bond formation in vivo. Biochemistry. 1994;33:5974–5983. doi: 10.1021/bi00185a039. [DOI] [PubMed] [Google Scholar]
- 44.Nordstrand K. Ãslund F. Meunier S. Holmgren A. Otting G. Berndt KD. Direct NMR observation of the Cys-14 thiol proton of reduced Escherichia coli glutaredoxin-3 supports the presence of an active site thiol-thiolate hydrogen bond. FEBS Lett. 1999;449:196–200. doi: 10.1016/s0014-5793(99)00401-9. [DOI] [PubMed] [Google Scholar]
- 45.Otwinowski Z. Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 46.Paxman JJ. Borg NA. Horne J. Thompson PE. Chin Y. Sharma P. Simpson JS. Wielens J. Piek S. Kahler CM. Sakellaris H. Pearce M. Bottomley SP. Rossjohn J. Scanlon MJ. The structure of the bacterial oxidoreductase enzyme DsbA in complex with a peptide reveals a basis for substrate specificity in the catalytic cycle of DsbA enzymes. J Biol Chem. 2009;284:17835–17845. doi: 10.1074/jbc.M109.011502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Potterton E. McNicholas S. Krissinel E. Cowtan K. Noble M. The CCP4 molecular-graphics project. Acta Crystallogr D Biol Crystallogr. 2002;58:1955–1957. doi: 10.1107/s0907444902015391. [DOI] [PubMed] [Google Scholar]
- 48.Ren G. Stephan D. Xu Z. Zheng Y. Tang D. Harrison RS. Kurz M. Jarrott R. Shouldice SR. Hiniker A. Martin JL. Heras B. Bardwell JCA. Properties of the Thioredoxin fold superfamily are modulated by a single amino acid residue. J Biol Chem. 2009;284:10150–10159. doi: 10.1074/jbc.M809509200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rost J. Rapoport S. Reduction-potential of glutathione. Nature. 1964;201:185. doi: 10.1038/201185a0. [DOI] [PubMed] [Google Scholar]
- 50.Segatori L. Murphy L. Arredondo S. Kadokura H. Gilbert H. Beckwith J. Georgiou G. Conserved role of the linker alpha-helix of the bacterial disulfide isomerase DsbC in the avoidance of misoxidation by DsbB. J Biol Chem. 2006;281:4911–4919. doi: 10.1074/jbc.M505453200. [DOI] [PubMed] [Google Scholar]
- 51.Segatori L. Paukstelis PJ. Gilbert HF. Georgiou G. Engineered DsbC chimeras catalyze both protein oxidation and disulfide-bond isomerization in Escherichia coli: reconciling two competing pathways. Proc Natl Acad Sci U S A. 2004;101:10018–10023. doi: 10.1073/pnas.0403003101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sinha S. Langford PR. Kroll JS. Functional diversity of three different DsbA proteins from Neisseria meningitidis. Microbiology. 2004;150:2993–3000. doi: 10.1099/mic.0.27216-0. [DOI] [PubMed] [Google Scholar]
- 53.Smith RC. Reed VD. Hill WE. Oxidation of thiols by copper(II) Phosphorus Sulfur Silicon Relat Elem. 1994;90:147–154. [Google Scholar]
- 54.Studier FW. Protein production by auto-induction in high-density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 55.Sun XX. Wang CC. The N-terminal sequence (residues 1–65) is essential for dimerization, activities, and peptide binding of Escherichia coli DsbC. J Biol Chem. 2000;275:22743–22749. doi: 10.1074/jbc.M002406200. [DOI] [PubMed] [Google Scholar]
- 56.Taylor SL. Crawley-Snowdon H. Wagstaff JL. Rowe ML. Shepherd M. Williamson RA. Howard MJ. Measuring protein reduction potentials using 15N HSQC NMR spectroscopy. Chem Commun. 2013;49:1847–1849. doi: 10.1039/c3cc38952a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Totsika M. Heras B. Wurpel DJ. Schembri MA. Characterization of two homologous disulfide bond systems involved in virulence factor biogenesis in uropathogenic Escherichia coli CFT073. J Bacteriol. 2009;191:3901–3908. doi: 10.1128/JB.00143-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Turcot I. Ponnampalam TV. Bouwman CW. Martin NL. Isolation and characterization of a chromosomally encoded disulphide oxidoreductase from Salmonella enterica serovar Typhimurium. Can J Microbiol. 2001;47:711–721. [PubMed] [Google Scholar]
- 59.van Straaten M. Missiakas D. Raina S. Darby NJ. The functional properties of DsbG, a thiol-disulfide oxidoreductase from the periplasm of Escherichia coli. FEBS Lett. 1998;428:255–258. doi: 10.1016/s0014-5793(98)00539-0. [DOI] [PubMed] [Google Scholar]
- 60.Wagner D. Maser J. Lai B. Cai ZH. Barry CE. Bentrup KHZ. Russell DG. Bermudez LE. Elemental analysis of Mycobacterium avium-, Mycobacterium tuberculosis-, and Mycobacterium smegmatis-containing phagosomes indicates pathogen-induced microenvironments within the host cell's endosomal system. J Immunol. 2005;174:1491–1500. doi: 10.4049/jimmunol.174.3.1491. [DOI] [PubMed] [Google Scholar]
- 61.Wang RF. Kushner SR. Construction of versatile low copy-number vectors for cloning, sequencing, and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 62.Xu Q. Canutescu AA. Wang G. Shapovalov M. Obradovic Z. Dunbrack RL., Jr. Statistical analysis of interface similarity in crystals of homologous proteins. J Mol Biol. 2008;381:487–507. doi: 10.1016/j.jmb.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zapun A. Missiakas D. Raina S. Creighton TE. Structural and functional characterization of DsbC, a protein involved in disulfide bond formation in Escherichia coli. Biochemistry. 1995;34:5075–5089. doi: 10.1021/bi00015a019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.