Abstract
Protective antigen (PA) is the primary protein antigenic component of both the currently used anthrax vaccine and related recombinant vaccines under development. An analogue of recombinant PA (2-FHis rPA) has been recently shown to block the key steps of pore formation in the process of inducing cytotoxicity in cells, and thus can potentially be used as an antitoxin or a vaccine. This rPA analogue was produced by fermentation to incorporate the unnatural amino acid 2-fluorohistidine (2-FHis). In this study, the effects of 2-FHis labeling on rPA antigen’s conformational stability and dynamic properties were investigated by various biophysical techniques. Temperature/pH stability profiles of rPA and 2-FHis rPA were analyzed by the empirical phase diagram (EPD) approach, and physical stability differences between them were identified. Results showed that rPA and 2-FHis rPA had similar stability at pH 7–8. With decreasing solution pH, however, 2-FHis rPA was found to be more stable. Dynamic sensitive measurements of the two proteins at pH 5 found that 2-FHis rPA was more dynamic and/or differentially hydrated under acidic pH conditions. The biophysical characterization and stability data provide information useful for the potential development of 2-FHis rPA as a more stable rPA vaccine candidate.
Keywords: circular dichroism, fluorescence spectroscopy, physical characterization, stability, vaccines
INTRODUCTION
Anthrax, a lethal disease caused by spores of the Gram-positive bacterium Bacillus anthracis, attacks mammalian hosts by cutaneous, inhalation and gastrointestinal routes. Mortality from anthrax disease is high in untreated cases and varies by infective routes. The inhaled form of the disease is the most severe and results from the inhalation of airborne spores. The mortality in untreated inhalational anthrax cases is almost 100%.1 There are recent serious concerns about the inhaled form due to its potential use as a biological weapon.2 Therefore, major efforts have involved the development of effective anthrax countermeasures such as vaccines.
Protective antigen (PA) is nontoxic by itself but essential for the toxic activity of Bacillus anthracis, the causative agent of anthrax. PA is an 83 kDa, four domain multifunctional protein which is recognized by anthrax toxin receptors 1 and 2 on host cells.3,4 Following receptor binding, PA is cleaved by a furin-like protease to yield a 63 kDa fragment (PA63) and an N-terminal 20 kDa fragment (PA20).5 Cleaved PA63 assembles on the surface of cells into a heptameric6 or octameric7 donut-shaped structure called a prepore. The prepore subsequently combines with edema factor (EF) and lethal factor (LF) to create a number of different, but functionally similar, complex toxins such as edema toxin (EdTx) and lethal toxin (LeTx). After receptor-mediated endocytosis, and a subsequent conformational change in PA from a prepore to a pore in acidified endosomes (pH 5~6), actively toxic proteins EF and LF can be translocated through the pore into the cytoplasm of target cells, which results in cellular toxicity causing cell killing and host death.5,8,9,10 Since PA plays an important role in the pathology of anthrax, yet is a non-toxic and a strongly immunogenic molecule, it is used as a primary antigen in anthrax vaccines.
PA is the protein antigen component in the currently used vaccine and provides a certain level of protective immunity against anthrax infection. The licensed U.S. anthrax vaccine (AVA, BioThrax) contains predominantly protective antigen adsorbed to aluminum hydroxide. It is an effective vaccine for the protection of humans anthrax5, however, there are limitations of the current AVA vaccine. The AVA vaccine has limited duration of protection, and 6 doses are required over 18 months followed by annual boosters.11 The AVA is composed of PA and an incompletely defined culture supernatant adsorbed to the adjuvant, so the vaccine purity is not easily determined. Therefore, there is a need to develop newer versions of vaccines such as those based on recombinant PA, which have more defined composition and can produce the long-lasting immunity.
A novel recombinant protein analogue of PA, designated 2-fluorohisidine recombinant protective antigen (2-FHis rPA), was recently found to have the capability to block the key step of pore formation.12,13 The preparation of the 2-FHis rPA analogue was achieved by incorporation of the unnatural amino acid 2-fluorohisidine, an isosteric analogue of histidine with a significantly reduced pKa(~1)14, during fermentation resulting in ten histidine residues in rPA being biosynthetically replaced by 2-fluorohistidine. The protein was structurally similar to the WT protein as determined by X-ray crystallography13, but this replacement was found to block low pH-induced pore formation with cells and thus protected from the lethal effects of the toxin.12,13 These results suggested that 2-FHis rPA might potentially be used as an antitoxin or a vaccine to treat anthrax infections.
We have therefore examined the effect of 2-fluorohistidine labeling of rPA on its stability and dynamics to further explore its potential use as a therapeutic entity. The conformational stability and dynamic properties of rPA and 2-FHis rPA were evaluated and compared as a function of solution pH and temperature. The physical stability of the two proteins was investigated as a function of pH (4–8) and temperature (10–85°C) using various biophysical techniques. Near and far UV circular dichroism, intrinsic and extrinsic fluorescence as well as light-scattering optical density data were collected and empirical phase diagrams were constructed to summarize the large volume of the data collected. The physical stability profiles were then compared to distinguish stability differences between the two proteins. Based on the biophysical characterization results, the dynamic properties of the two proteins at pH 5 were then studied to better understand the difference in thermal stability observed between the two proteins at pH 5. The dynamic properties of proteins were investigated by red-edge excitation shifts and high resolution ultrasonic spectroscopy.
MATERIALS AND METHODS
Materials
Recombinant protective antigen (rPA) and its 2-fluorohistidine labeled analogue (2-FHis rPA) were produced by the Laboratory of Bioorganic Chemistry at Wichita State University (Wichita, KS) as liquid solutions (~20 mg/ml) in 20 mM Tris buffer containing 150 mM NaCl at pH 8. Expression, production, and purification of rPA and 2-FHis rPA were carried out as described previously.14 All other reagents including buffers and salts were purchased from Sigma-Aldrich (St. Louis, MO).
For the physical characterization studies, high concentrations of stock protein samples (~ 20 mg/ml) were freshly prepared by dilution into 20 mM citrate phosphate buffer with an ionic strength of 0.15 (adjusted with sodium chloride) ranging from pH 4 to 8, at one pH unit intervals. The final experimental protein concentration was 0.5 mg/ml.
For dynamic studies, high concentrations of stock protein samples were freshly diluted to approximately 1.5 mg/ml using 20 mM Tris buffer containing 150 mM NaCl at pH 8. Diluted samples were then dialyzed in 20 mM citrate phosphate buffer at pH 5 at 4°C using Slide-A-Lyzer dialysis cassettes (Thermo Scientific, Rockford, IL) with a 3.5 kDa MW cutoff. The pH value of 5 for the working buffer was determined from the biophysical characterization results. Protein concentration was 0.5 mg/ml for red-edge excitation shifts experiments, and was 0.9 mg/ml for both density and high resolution ultrasonic spectroscopy measurements.
Biophysical Characterization
Far and Near UV Circular Dichroism Spectroscopy
Circular dichroism analysis was performed with a Chirascan-plus Circular Dichroism Spectrometer (Applied Photophysics Ltd, Leatherhead UK) equipped with a Peltier temperature controller and a 4-position cuvette holder. Far and near UV spectra of samples (0.5 mg/ml) were collected simultaneously in the range of 200–360 nm using a 0.2 cm path length cuvette.15 The step size of spectra scanning was 1 nm. A sampling time per point of 3 s and a bandwidth of 1 nm were used. The characteristic CD signals of proteins (pH 4–8) at 220, 262, 269, 284 and 291 nm observed in the near and far UV CD regions were monitored as a function of temperature from 10 to 85°C at 2.5°C intervals. Triplicate measurements were made, and the equilibration time at each temperature was 1 minute.
Optical Density Measurements
Ultraviolet absorbance measurements were made simultaneously during the near and far UV CD experiments using the same experimental parameters.15 The aggregation behavior of rPA and 2-FHis rPA (0.5 mg/ml) was monitored by recording the optical density at 350 nm as a function of temperature (10–85°C) and pH (4–8).
Intrinsic Fluorescence Spectroscopy
In a series of separate experiments, fluorescence emission scan data were also collected on the Chirascan-plus instrument by using an attached fluorescence detector. A 2 × 10 mm quartz cuvette was used in all experiments, and excitation of the proteins was carried out along the shorter path length (2 mm). Fluorescence emission spectra of the two proteins (0.5 mg/ml) were recorded using the same temperature program as used previously. Triplicate samples were measured using an excitation wavelength of 295 nm (> 95% tryptophan emission). Emission spectra were collected from 305 to 395 nm with a step size of 1 nm and 2 s sampling time per point. The excitation and emission slits were set at 4 and 9 nm, respectively. The position of the emission wavelength maximum was determined using a polynomial derivative fitting method executed in Origin 7.0 software.
Extrinsic (ANS) Fluorescence Spectroscopy
8-Anilino-1-naphthalene sulfonate (ANS) was used as an extrinsic probe to study conformational changes of the two proteins that result in changes in its surface polarity. The interaction of ANS with the apolar surfaces of rPA and 2-FHis rPA were monitored as a function of temperature (10–85°C) and pH (4–8) using a QuantaMaster Spectrophotometer (Photon Technology International (PTI), Inc., Birmingham, NJ). Each sample solution (0.5 mg/ml) containing 89 µM diluted ANS (a molar ratio of the ANS to the protein of 15:1) was incubated at 10°C in the dark for 20 minutes before measurement. Samples were excited at 375 nm and spectra were collected from 400 to 600 nm at 2.5°C intervals, with a 3 min equilibration time at each temperature. Duplicate measurements were made using a 2 mm path length quartz cuvette to check for inner filter effects (none were seen). Buffer baselines containing ANS were subtracted from each sample spectrum prior to analysis. Peak positions and intensities of the emission spectra were obtained using a polynomial derivative fitting method as described above (Section: Intrinsic Fluorescence Spectroscopy).
Empirical Phase Diagram (EPD)
The empirical phase diagram (EPD) was constructed as described previously16,17 to summarize and facilitate the analysis of the complex biophysical data sets obtained. The EPD is used to identify coherent parameter regions that define protein structural states as a function of solution variables (temperature and pH in this case). CD signals at 220, 262, 269, 284 and 291 nm, optical density, intrinsic Trp fluorescence peak positions, and extrinsic fluorescence intensities data were used for construction of the EPDs of rPA and 2-FHis rPA. All calculations were performed using Matlab software (MathWorks, Natick, MA).
Dynamic Properties Measurements
High Resolution Ultrasonic Spectroscopy
Ultrasonic spectroscopy measurements were performed using an HR-US 102 Spectrometer (Ultrasonic Scientific, Dublin, Ireland) with a frequency range of 2–18 MHz and a resolution of 0.2 mm/s for velocity and 0.2% for attenuation measurements. The differential velocity and attenuation between sample and reference cells were monitored at 12 MHz from 10 to 85°C and pH 5. The sample and reference solutions (0.9 mg/ml) were thoroughly degassed before each measurement. The sample and reference cells contained 1 ml of protein and buffer solution, respectively. The temperature of the cells was controlled by a Phoenix P2 water circulator (Thermo Haake, Newington, NH). Data were analyzed using HRUS v4.50.27.25 software. The coefficient of adiabatic compressibility (βs) was determined using the equations described previously.18 The adiabatic compressibility of the sample and buffer are related to the density (ρ) and ultrasonic velocity (u) by the Laplace equation, β=1/ρu2.
Density
The densities of the two protein solutions (0.9 mg/ml) and corresponding buffer solutions were measured using a DMA-5000 high precision densitometer (Anton Paar, Graz, Austria) at a precision of 1 ×10−6 g/cm3 and ±0.001°C. The densities of degassed solutions were measured from 5–22°C and 5–32°C for rPA and 2-FHis rPA in their unfolding pre-transition ranges, respectively. The increment of the temperature was 1°C. The instrument was calibrated with dry air and degassed water before analysis.
Red-Edge Excitation Shifts
Fluorescence measurements used in the determination of red-edge excitation shifts were performed with the PTI QuantaMaster Spectrophotometer. Protein concentration was 0.5 mg/ml. Excitation wavelengths of 292, 296, 300 and 304 nm were used, and the emission spectra were collected from 300 to 400 nm. The excitation and emission slit widths were set to 2 nm. The data were collected from 10 to 50°C in 2°C increments with a 3 minutes equilibration time. Triplicate measurements were made using a 2 mm path length quartz cuvette. The relative change in peak position was determined using a mean spectral center of mass (MSM) method spanning the emission spectra range of 310 to 400 nm. This range of data analysis and the lower pathlength employed for fluorescence measurements (2 mm) should minimize the contribution of Rayleigh scattering even when the data is collected at 304 nm (the highest excitation wavelength employed) as well as the lower excitation wavelengths used in these studies. The mean spectral center of mass method is a peak picking algorithm to determine the peak maximum. The maximum peak position is based on the center-of mass position of the area in a selected peak integration region (from 310- 400 nm in this case).The peak position maxima obtained by the MSM method are generally ~8–10 nm higher than the actual peak position obtained by derivative analysis but result in a better signal to noise ratio.
RESULTS
Biophysical Characterization
Near and Far UV Circular Dichroism Spectroscopy
Near and far UV CD spectra of 0.5 mg/ml rPA and 2-FHis rPA at pH 7 and 10°C are shown in Figure 1. In the far- UV region (200–250 nm), both proteins display a broad peak with a minimum at around 210 nm and a shoulder at 220 nm, suggesting that proteins contain a mixture of α-helix and β-sheet structures. In the near-UV region (250–320 nm), four characteristic peaks at 262, 269, 284 and 291 nm were observed, presumably due to the aromatic amino acid side chains.19,20
Figure 1.
Near- and far-UV CD spectra for rPA and 2-FHis rPA at pH 7 and 10°C. Near and far UV CD (A, C) and Near-UV CD (B, D) spectra are shown for (A) 0.5 mg/ml rPA at pH 7 and (B) expanded view of its near-UV region. (C) 0.5 mg/ml 2-FHis rPA at pH 7 and (D) expanded view of its near-UV region.
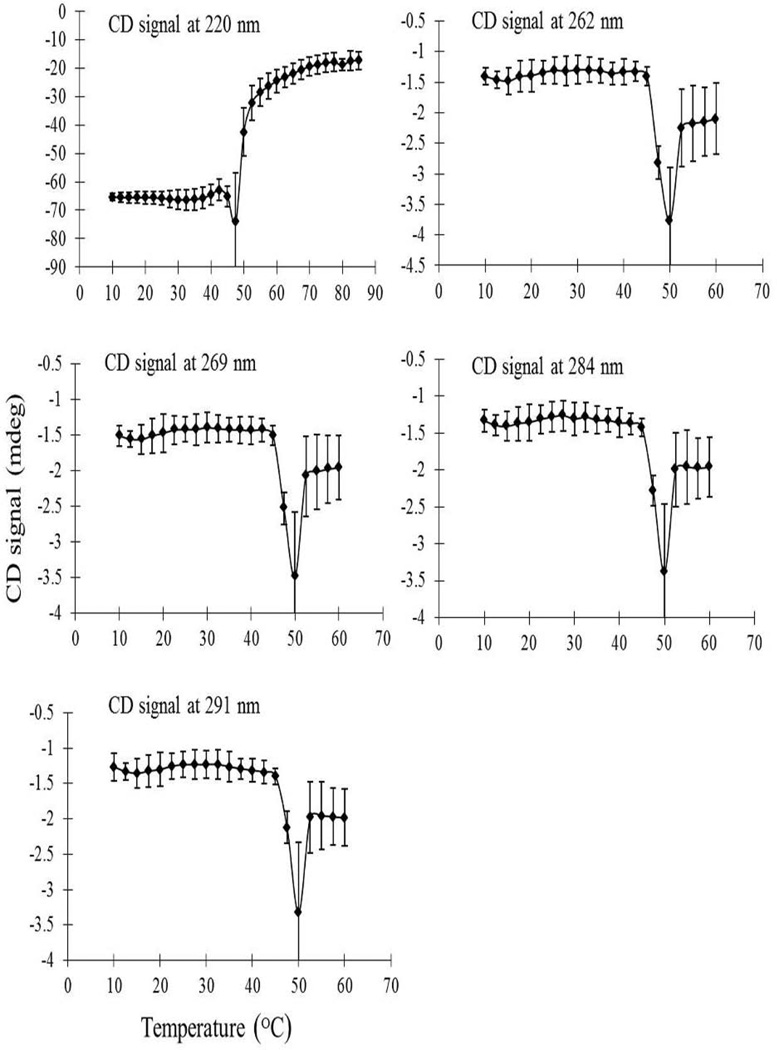
The effect of temperature on the two protein’s secondary and tertiary structures was further investigated by using thermal melt analysis. CD signals at five characteristic peaks (220, 262, 269, 284 and 291 nm) were monitored across the pH range of 4–8. Typical thermal unfolding curves of 2-FHis rPA and rPA at pH 7 are shown in Figure 2 and Supplemental Figure S1, respectively, with triplicate measurements resulting in good reproducibility. The rPA and 2-FHis rPA proteins show similar pattern in their CD thermal unfolding curves across the entire pH range examined. Although an overall loss in secondary and tertiary structures is observed as a function of increasing temperature for both proteins, the thermal transition midpoint (Tm) values vary greatly between the two proteins as the pH value decreases. For example, the two proteins show a significantly lower thermal stability at pH 5 (Supplemental Figures S2 and S3). The Tm values from CD thermal melt curves were calculated for both proteins and results are summarized in Table 1. The two proteins show similar conformational stability at pH 7 and 8. Differences in thermal stability between the two proteins are, however, observed at pH 5 and 6 (see Table 1), especially at pH 5. Interestingly, the Tm value for 2-FHis rPA at pH 5 evaluated from the Far-UV CD data was greater than that obtained from the near-UV CD data, suggesting that a molten globule like state exists in the temperature range from 35 – 41 °C. Molten globules generally possess native-like secondary structure but lack significant tertiary structure. This is seen as a gradual color change region in the EPDs. While the Tm values for both proteins decrease as a function of pH, the 2-FHis rPA exhibits a slightly higher thermal stability than rPA at pH 6, and the analogue rPA has significantly higher stability than rPA at pH 5 with an increased Tm value of approximately 10°C.
Figure 2.
Thermal stability of 2-FHis rPA (0.5 mg/ml at pH 7) as monitored by CD signals at indicated values (220, 262, 269, 284 and 291 nm) as a function of temperature. Each data point represents the mean of three independent runs, with error bars showing the standard deviation.
Table 1.
Thermal unfolding temperature values (Tm, °C) calculated from the thermal unfolding curves using different biophysical techniques. Indicated Tm values were calculated from the first derivative of the thermal unfolding curves from each technique. Values are ± 0.5°C.
| Sample | pH | Circular Dichroism | Optical Density |
Intrinsic Fluorescence |
ANS Fluorescence |
||||
|---|---|---|---|---|---|---|---|---|---|
| 220 nm |
262 nm |
269 nm |
284 nm |
291 nm |
|||||
| rPA | 8 | 49.0 | 50.0 | 50.5 | 50.5 | 50.5 | 51.0 | 46.5 | 46.5 |
| 2-FHis rPA | 8 | 47.0 | 49.0 | 49.0 | 49.0 | 49.0 | 48.5 | 45.0 | 46.5 |
| rPA | 7 | 49.0 | 48.5 | 48.5 | 48.5 | 48.5 | 48.5 | 46.5 | 48.5 |
| 2-FHis rPA | 7 | 49.0 | 47.0 | 47.0 | 48.5 | 48.5 | 46.5 | 46.5 | 50.5 |
| rPA | 6 | 43.5 | 41.0 | 41.0 | 41.0 | 41.0 | 43.5 | 39.0 | 41.0 |
| 2-FHis rPA | 6 | 44.0 | 43.5 | 43.5 | 43.5 | 43.5 | 43.5 | 43.5 | 44.0 |
| rPA | 5 | 28.5 | 26.0 | 26.0 | 26.0 | 26.0 | 28.5 | 33.5 | 29.0 |
| 2-FHis rPA | 5 | 41.0 | 34.5 | 36.5 | 36.5 | 36.5 | 39.0 | 43.0 | 41.0 |
| rPA | 4 | 36.0 | 36.0 | 36.0 | 36.0 | 36.0 | 34.0 | 29.0 | 31.0 |
| 2-FHis rPA | 4 | 36.0 | 34.0 | 34.0 | 33.5 | 34.0 | 34.0 | 34.0 | 29.5 |
Intrinsic Fluorescence Spectroscopy
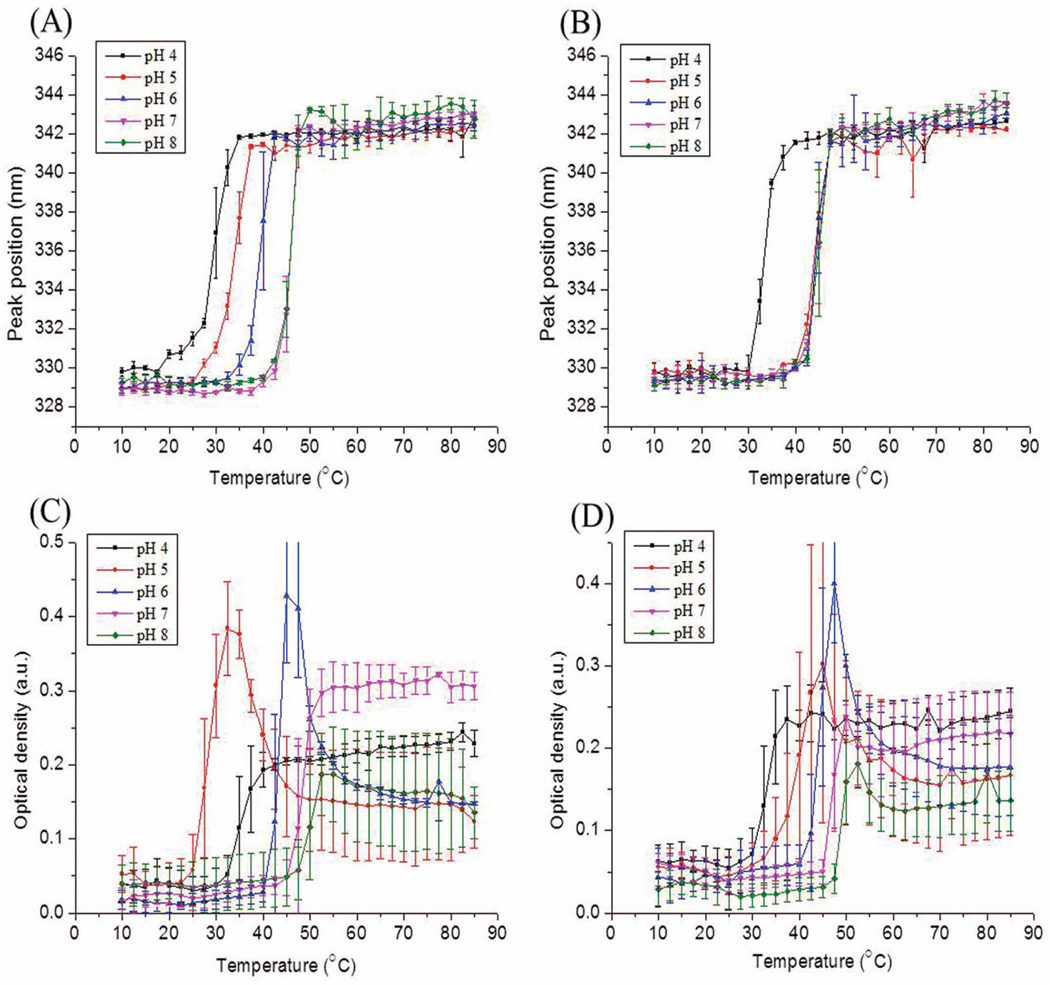
Alternations in tertiary structure as a function of temperature and solution pH of the two rPA proteins were also investigated by intrinsic fluorescence spectroscopy. Figures 3A and B show the changes in the position of the emission peak maximum and structural changes occurred in both proteins but at different temperatures depending on pH. rPA and 2-FHis rPA show their highest thermal stability at pH 7–8 and 6–8, respectively. The stability of rPA decreases much more dramatically compared to 2-FHis rPA between pH 4 and 6 (see Table 1 for the calculated Tm values). At pH 7 and 8, both proteins show comparable structural integrity with initial peak positions of about 329 nm at low temperatures. With increasing temperature, the emission peak maximum shifts to about 342 nm, indicating the proteins were conformationally altered but not completely unfolded at elevated temperatures. In summary, both proteins show their highest thermal stability at pH 7 and 8, and manifest lower thermal stability with decreasing pH.
Figure 3.
Thermal stability of rPA (Figures A and C) and 2-FHis rPA (Figures B and D) at 0.5 mg/ml as a function of solution pH: (A) and (B) Intrinsic Trp fluorescence emission maximum (peak position) was monitored; (C) and (D) Optical density at 350 nm was monitored. Each data point represents the mean of three independent samples, with error bars showing the standard deviation.
Optical Density Measurements
Optical density (OD) data at 350 nm were collected simultaneously during CD measurements. rPA and 2-FHis rPA both manifested aggregation as measured by increases in OD values with increasing temperature (Figures 3C and D). The temperature dependent OD changes (i.e., protein aggregation) are overall consistent with the observations made from the CD and intrinsic fluorescence measurements, with similar trends of pH and temperature. Again, 2-FHis rPA was significantly more stable than rPA at pH 5, where the midpoint of the aggregation values for rPA and 2-FHis rPA were 39.0°C and 28.5°C, respectively. Interestingly, the 2-FHis rPA analogue is actually more stable at pH 5 than at pH 4, in contrast to the recombinant rPA protein.
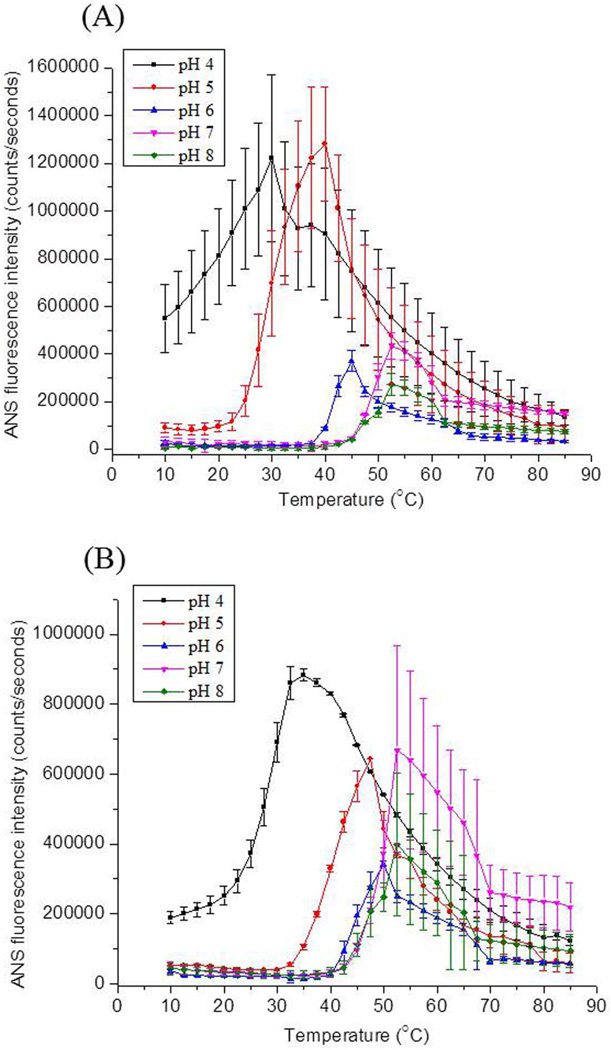
Extrinsic (ANS) Fluorescence Spectroscopy
ANS (8-Anilino-1-naphthalene sulfonate) is a fluorescent dye that binds to apolar regions in proteins, resulting in large increases in fluorescence quantum yield and a blue shift in peak position.21 This observation must always be qualified, however, by the possibility of electrostatic interactions of the negatively charged dye with a protein. Figure 4 shows the thermal melting curves of the two proteins at various pH values. These curves were obtained by monitoring the ANS emission peak intensity as a function of temperature. At pH 4, ANS data for rPA show significant fluorescence intensity even at low temperatures (Figure 4A). This increased fluorescence intensity is presumably induced by the binding of ANS to the structurally altered rPA protein under acidic conditions. Increased ANS fluorescence intensity with increases in temperature is observed over the pH range of 4 to 8. The Tm values for these transitions are summarized in Table 1. In agreement with results obtained from other biophysical techniques, both proteins show their greatest structural stability at pH 7–8. At pH 6, 2-FHis rPA is slightly more stable than rPA. Again, at pH 5, the maximum difference in the stability of two proteins is observed.
Figure 4.
ANS fluorescence intensity values in the presence (A) rPA and (B) 2-FHis rPA as a function of temperature and solution pH. Both samples were at 0.5 mg/ml. Each data point represents the mean of two independent samples, with error bars showing the standard deviation.
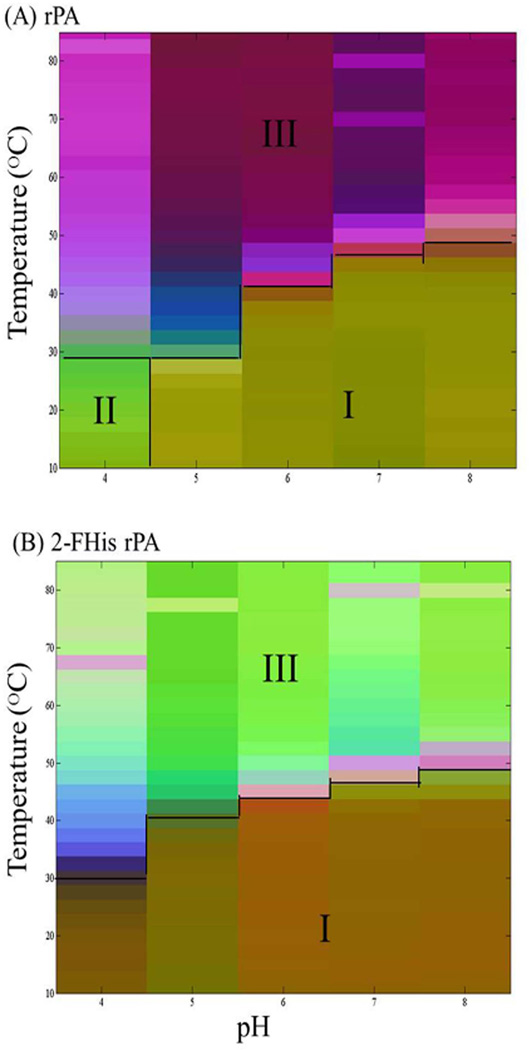
Empirical Phase Diagrams (EPDs)
Empirical phase diagrams (EPDs) are used to summarize large amounts of data to provide a more comprehensive view of the stability profiles of proteins. Such diagrams should not be confused with thermodynamic phase diagrams, however, since no equilibrium between phases is implied. The EPDs of rPA and 2-FHis rPA generated from all of the collected data are shown in Figures 5A and B, respectively. Overall, the EPD of the rPA protein (Figure 5A) shows similar defined “apparent” phases compared to those in the EPD of 2-FHis rPA (Figure 5B). There are two major phases seen for both proteins. In region 1, both proteins at pH 5–8 show a similar color below about 48°C, suggesting that they exist in similar structural states. By examining the data obtained from the specific techniques, region 1 at low temperatures from pH 5–8 is presumably the native state of protein. rPA and 2-FHis rPA show similar apparent phase boundaries across the pH range 6 to 8. In contrast, rPA shows lower stability than 2-FHis rPA at pH 6 as evidenced by a decreased Tm value of about 2.5°C (Table 1). At pH 5, there is a much more significant thermal stability difference between the two proteins. Both the EPDs and the individual experimental methods all show that 2-FHis rPA has much greater thermal stability (Tm value difference of 10–12°C). Unlike the EPD of 2-FHis rPA, the EPD of rPA displays an additional region (Region 2 in Figure 5A) based on phase color changes. In region 2, rPA samples at pH 4 show a distinct color (green) difference from region 1. This observation clearly arises from results obtained from extrinsic (ANS) fluorescence data. As discussed previously, at pH 4, rPA with ANS shows significant fluorescence intensity even at low temperature (Figure 4A), suggesting the binding of ANS to a partially unfolded protein at this acidic condition. At high temperatures, both proteins are structurally altered and aggregate as indicated by the presence of region 3. From the EPDs, we can see that the most stable region of two proteins is in the pH range of 7 to 8.
Figure 5.
Empirical phase diagrams (EPDs) of (A) rPA and (B) 2-FHis rPA. The EPD was prepared from near/far-UV CD, optical density, intrinsic fluorescence (peak position), and extrinsic (ANS) fluorescence data collected across a pH range from 4 to 8 and a temperature range from 10°C to 85°C.
Dynamic Properties
High Resolution Ultrasonic Spectroscopy
HR-US spectroscopy employs high-frequency sound waves to probe intramolecular forces in materials such as proteins.22,23 Oscillating compression (and decompression) of the ultrasonic wave causes oscillations in internal molecular rearrangement. A protein will respond by corresponding compression and expansion of its volume. In HR-US measurements, attenuation and velocity of ultrasonic waves were determined. Combined with high precision density measurements, the adiabatic compressibility of each protein sample was calculated.
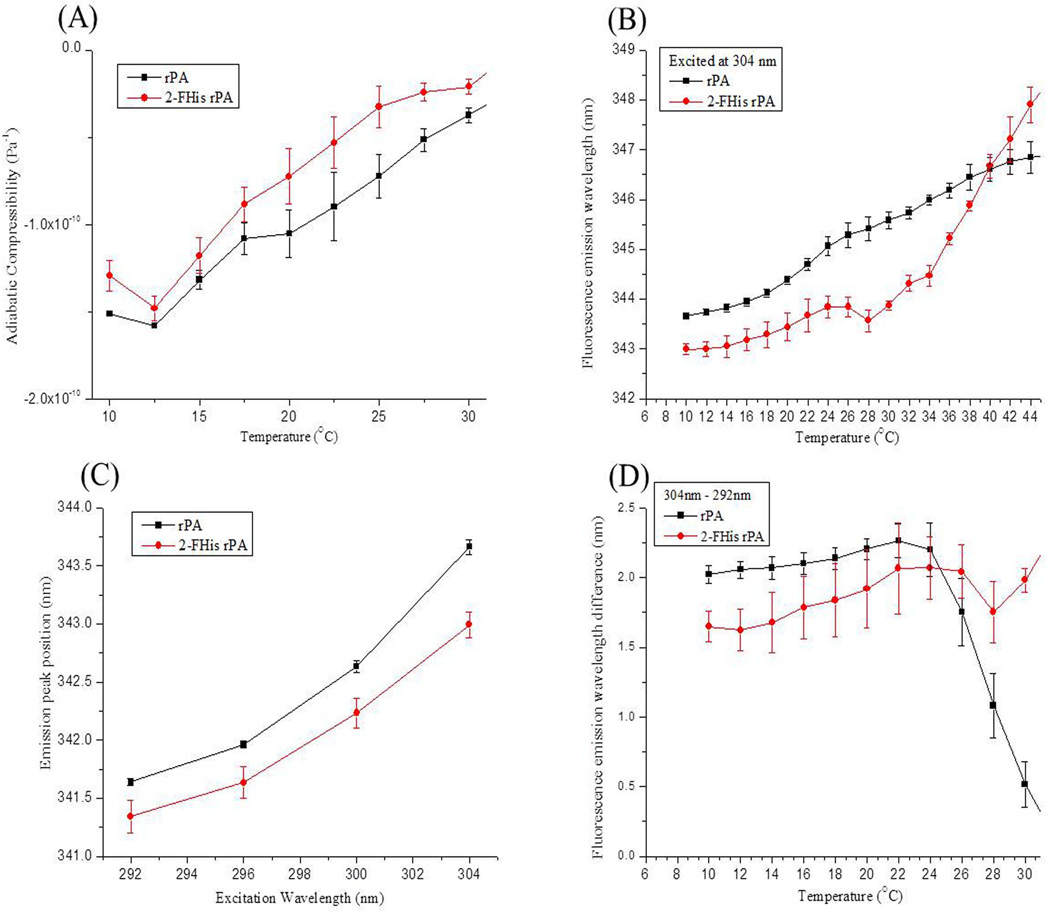
Protein compressibility is influenced by both a protein’s internal fluctuations in volume and its hydration. Because 2-FHis rPA showed greater stability than rPA at pH 5, based on the EPD characterization results discussed above, measurements of the dynamic properties of the two proteins were made under this condition. The magnitude of a protein’s apparent adiabatic compressibility has positive contributions from a protein’s intrinsic volume fluctuations and negative contributions from the hydration water.24 In Figure 6A, the apparent adiabatic compressibility values were found to increase with increases in temperature for both rPA and 2-FHis rPA. 2-FHis rPA displays higher values of adiabatic compressibility than those of rPA at low temperatures where both proteins exist in their native states. The negative compressibility, however, suggests a dominant contribution of hydration water to the experimentally determined compressibility values. The increase in compressibility with temperature for rPA and 2-FHis rPA therefore could either represent increases in their intrinsic fluctuations and/or reflect alterations in hydration dynamics around the protein’s surface due to temperature. Figure 6B shows Trp peak position shifts as a function of temperature for rPA and 2-FHis rPA when excited at 304 nm (obtained from red-edge shifts measurements). Excitation of Trp by 304 nm radiation should probe residues with greater degrees of solvent exposure which are presumably located on the surface of the protein. The Trp peak position at 10°C was red-shifted for rPA compared to 2-FHis rPA. This red-shift in peak position could be due to the differences in protein’s tertiary structure and/or hydration of the surface. The magnitude of the red-shift, however, was greater when probed with 304 nm compared to 292 nm light (data not shown), suggesting that hydration contributions to the red-shift are possible. This result along with the compressibility data (Figure 6A) suggests that the native structure of rPA and 2-FHis rPA may have differences in their hydration state. Generally, increased dynamic motions, flexibility, void volume changes and elasticity of proteins will result in higher values of adiabatic compressibility. In contrast, hydration of proteins, which reduces many of the above processes, will decrease values of compressibility.
Figure 6.
Measurements of dynamic properties of rPA and 2-FHis rPA collected at pH 5. (A) Ultrasonic measurements of the adiabatic compressibility of the two proteins. (B) Emission peak positions’ comparison between rPA and 2-FHis rPA at an excitation wavelength of 304 nm. Tryptophanyl dynamics were examined by red-edge excitation shifts of (C) rPA and 2-FHis rPA based on excitation at 292, 296, 300 and 304 nm. (D) red-edge excitation shifts compared between excitation at 292 and 304 nm for both proteins. Each data point represents the mean of three independent runs, with error bars showing the standard deviation.
Red-Edge Excitation
Red edge excitation spectroscopy is a fluorescence based technique used for studying the dynamic properties of proteins. For fluorophores under conditions where solvent relaxation is not complete (such as the viscous environment surrounding the buried protein fluorophore), emission spectra shift to longer wavelengths when excitation wavelengths move to longer wavelengths.25,26 In less viscous environments, where solvent relaxation is more complete, the magnitude of the red-edge effect decreases with shifts disappearing in fluid solvents.
The effect of red-edge excitation on tryptophan fluorescence is quantified here as the shift in the emission peak when excited at 296, 300, and 304 nm relative to 292 nm. In Figure 6C, both proteins show clear red shifts at 10°C when the excitation wavelength increases. These results show that the magnitude of the red-shifts increase as excitation is moved to longer wavelengths. Similar results were obtained throughout the pre-transition temperature range (10–20°C for rPA, and 10–30°C for 2-FHis rPA) of their thermal unfolding curves. This observation suggests that Trp residues located in restricted environments can be sampled and probed to study the differences in dynamics of such environments in both rPA and 2-FHis rPA. In Figure 6D, the red-edge excitation shifts, representing the difference in emission maxima upon red-edge excitation from 292 to 304 nm, are compared for both rPA and 2-FHis rPA. The magnitude of the shift of rPA is always larger than 2-FHis rPA at low temperatures prior to thermal structural perturbation. This observation suggests that 2-FHis rPA displays more dynamic behavior compared to rPA at pH 5. Above the unfolding temperature, rPA and 2-FHis rPA are prone to aggregate and eventually precipitate out of solution.
DISCUSSION
In the present study, the physical stability of rPA and 2-FHis rPA was investigated using spectroscopic and light scattering techniques. Temperature/pH stability profiles of the two proteins were created and compared. The proteins exhibited similar CD and fluorescence spectra at low temperatures. The estimated secondary structure contents of the two proteins are very similar to each other, although 2-FHis rPA displays an increased (about 6.5 %) α-helix content and a corresponding decrease in β-structure compared to rPA12, as estimated by the spectral deconvolution software CDNN. Similar thermal unfolding profiles of the two proteins were also obtained by different techniques at pH 6–8. We conclude that the overall structural integrity and conformational stability of the protein was not affected by incorporation of 2-FHis around neural pH values. These results are in agreement with a previous study of the equilibrium stability of rPA and 2-FHis rPA using urea in which both proteins showed similar urea denaturation profiles.12
At lower pH values, however, differences in the conformational stability of 2-FHis rPA and rPA were noted. At pH 5, and to a lesser extent at pH 6, 2-FHis rPA exhibited higher stability than rPA. This result suggests that 2-FHis incorporation into rPA inhibits pH dependent unfolding under these mildly acidic pH conditions. Thus, the 2-FHis rPA is much more thermally stable at lower pH compared to rPA (Table 1).
To better understand why the two rPA proteins demonstrate a stability difference at pH 5, the dynamic properties of the proteins were compared by red-edge excitation shifts and HR-US measurements. HR-US measurements suggest that both rPA and 2-FHis rPA may have differences in their hydration and/or intrinsic fluctuations in which rPA may be more hydrated than 2-FHis rPA. It is well known that fluorinated compounds exhibit increased hydrophobicity compared to their hydrogenated counterparts.27 Such increased apolar character in 2-FHis rPA may cause increased water mobility and less residence times near such regions.28,29 It is therefore possible that 2-FHis rPA has different hydration characteristics than rPA but this hypothesis needs further investigation. In addition, red-edge excitation shift results indicate that 2-FHis rPA has greater dynamic character than rPA at pH 5. It is often assumed that more dynamic proteins possess less stability.30,31,32 Many thermostable proteins are, however, quite dynamic, so an increase in thermostability does not necessarily correlate with a decrease in dynamic behavior.33,34,35,36,37 The relationship between protein dynamics and stability is not fully understood. The lower extent of hydration and/or higher dynamic character may increase the stability of the 2-FHis rPA at lower pH. Solution pH can potentially alter the microenvironmental pH around proteins, especially in their hydration layer. If such a phenomenon occurs at lower pH in these studies, a greater extent of hydration (with higher residence time) may explain the inverse relationship between the extent of hydration and stability for rPA and 2-FHis rPA. rPA was earlier determined to be potentially more hydrated than 2-FHis rPA using HR-US studies. The two methods used in this study both reflect the global dynamic properties of the two proteins at a specific pH condition. More detailed information about the dynamic properties of proteins can be obtained by methods such as NMR and H-D exchange mass spectrometry, which provide information about the local dynamic properties in specific regions of proteins.
This study provides important biophysical information about the effect of 2-FHis on the conformational stability of rPA in response to environmental stress factors such as pH and temperature. Our results show that the two proteins have similar stability at pH 7 to 8. The 2-FHis rPA analogue is slightly more stable at pH 6 and shows dramatically increased stability at pH 4–5. The 2-FHis rPA protein is also more dynamic than rPA at pH 5. These data provide useful information to better understand the similarities and differences in physical properties of the two proteins in solution. PA is a primary component in the currently licensed anthrax vaccine. In comparison, 2-FHis rPA possesses higher thermal stability under acidic conditions and has the capability to block key steps in its cellular cytotoxicity. These data suggest that 2-FHis rPA has the potential to for use as an alternative, more stable antigen in newer, recombinant versions of the anthrax vaccine. Given the potential need for long term storage of anthrax vaccines as a counter-terrorism measure, the increased stability of the 2-FHis analogue is an attractive property of this form of the rPA protein. The pH/temperature stability profile of 2-FHis rPA was generated through a comprehensive biophysical characterization of the secondary and tertiary structural integrity, as well aggregation behavior, of the protein. These data can be used as a starting point for further formulation development of the 2-FHis rPA as a potential anthrax vaccine or as a potential antitoxin to treat anthrax during the infection.
Supplementary Material
Supplemental Figure S1. Thermal stability of rPA (0.5 mg/ml at pH 7) as monitored by CD signals at indicated values (220, 262, 269, 284 and 291 nm) as a function of temperature. Each data point represents the mean of three independent runs, with error bars showing the standard deviation.
Supplemental Figure S2. Thermal stability of 2-FHis rPA (0.5 mg/ml at pH 5) as monitored by CD signals at indicated values (220, 262, 269, 284 and 291 nm) as a function of temperature. Each data point represents the mean of three independent runs, with error bars showing the standard deviation.
Supplemental Figure S3. Thermal stability of rPA (0.5 mg/ml at pH 5) as monitored by CD signals at indicated values (220, 262, 269, 284 and 291 nm) as a function of temperature. Each data point represents the mean of three independent runs, with error bars showing the standard deviation.
Footnotes
SUPPORTING INFORMATION
Supplemental Figures 1–3. This material is available free of charge via the Internet at www.wileyonlinelibrary.com.
REFERENCES
- 1.Donegan S, Bellamy R, Gamble CL. Vaccines for preventing anthrax. Cochrane Database Syst Rev. 2009;2:CD006403. doi: 10.1002/14651858.CD006403.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Inglesby TV, O'Toole T, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, Friedlander AM, Gerberding J, Hauer J, Hughes J. Anthrax as a biological weapon, 2002. JAMA: the journal of the American Medical Association. 2002;287(17):2236–2252. doi: 10.1001/jama.287.17.2236. [DOI] [PubMed] [Google Scholar]
- 3.Bradley KA, Mogridge J, Mourez M, Collier RJ, Young JAT. Identification of the cellular receptor for anthrax toxin. Nature. 2001;414(6860):225–229. doi: 10.1038/n35101999. [DOI] [PubMed] [Google Scholar]
- 4.Scobie HM, Rainey GJA, Bradley KA, Young JAT. Human capillary morphogenesis protein 2 functions as an anthrax toxin receptor. Proceedings of the National Academy of Sciences. 2003;100(9):5170. doi: 10.1073/pnas.0431098100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brey RN. Molecular basis for improved anthrax vaccines. Advanced drug delivery reviews. 2005;57(9):1266–1292. doi: 10.1016/j.addr.2005.01.028. [DOI] [PubMed] [Google Scholar]
- 6.Petosa C, Collier RJ, Klimpel KR, Leppla SH, Liddington RC. Crystal structure of the anthrax toxin protective antigen. Nature. 1997;85:833–838. doi: 10.1038/385833a0. [DOI] [PubMed] [Google Scholar]
- 7.Mogridge J, Cunningham K, Lacy DB, Mourez M, Collier RJ. The lethal and edema factors of anthrax toxin bind only to oligomeric forms of the protective antigen. Proceedings of the National Academy of Sciences. 2002;99(10):7045–7048. doi: 10.1073/pnas.052160199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collier RJ, Young JAT. Anthrax toxin. Annual review of cell and developmental biology. 2003;19(1):45–70. doi: 10.1146/annurev.cellbio.19.111301.140655. [DOI] [PubMed] [Google Scholar]
- 9.Bann JG. Anthrax toxin protective antigen—Insights into molecular switching from prepore to pore. Protein Science. 2012;21(1):1–12. doi: 10.1002/pro.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao-Sheridan S, Zhang S, John Collier R. Exchange characteristics of calcium ions bound to anthrax protective antigen. Biochemical and biophysical research communications. 2003;300(1):61–64. doi: 10.1016/s0006-291x(02)02771-7. [DOI] [PubMed] [Google Scholar]
- 11.Wright JG, Quinn CP, Shadomy S, Messonnier N. Use of anthrax vaccine in the United States. Morbid Mortal Wkly Rep. 2010;59(rr06):1–30. [PubMed] [Google Scholar]
- 12.Wimalasena DS, Cramer JC, Janowiak BE, Juris SJ, Melnyk RA, Anderson DE, Kirk KL, Collier RJ, Bann JG. Effect of 2-fluorohistidine labeling of the anthrax protective antigen on stability, pore formation, and translocation. Biochemistry. 2007;46(51):14928–14936. doi: 10.1021/bi701763z. [DOI] [PubMed] [Google Scholar]
- 13.Wimalasena DS, Janowiak BE, Lovell S, Miyagi M, Sun J, Zhou H, Hajduch J, Pooput C, Kirk KL, Battaile KP. Evidence that histidine protonation of receptor-bound anthrax protective antigen is a trigger for pore formation. Biochemistry. 2010;49:6973–6983. doi: 10.1021/bi100647z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eichler JF, Cramer JC, Kirk KL, Bann JG. Biosynthetic incorporation of fluorohistidine into proteins in E. coli: a new probe of macromolecular structure. ChemBioChem. 2005;6(12):2170–2173. doi: 10.1002/cbic.200500249. [DOI] [PubMed] [Google Scholar]
- 15.Hu L, Olsen CM, Maddux NR, Joshi SB, Volkin DB, Middaugh CR. Investigation of Protein Conformational Stability Employing a Multimodal Spectrometer. Analytical chemistry. 2011;83:9399–9405. doi: 10.1021/ac201995c. [DOI] [PubMed] [Google Scholar]
- 16.Kueltzo LA, Ersoy B, Ralston JP, Middaugh CR. Derivative absorbance spectroscopy and protein phase diagrams as tools for comprehensive protein characterization: A bGCSF case study. Journal of pharmaceutical sciences. 2003;92(9):1805–1820. doi: 10.1002/jps.10439. [DOI] [PubMed] [Google Scholar]
- 17.Maddux NR, Joshi SB, Volkin DB, Ralston JP, Middaugh CR. Multidimensional methods for the formulation of biopharmaceuticals and vaccines. Journal of pharmaceutical sciences. 2011;100:4171–4197. doi: 10.1002/jps.22618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramsey JD, Gill ML, Kamerzell TJ, Price ES, Joshi SB, Bishop SM, Oliver CN, Middaugh CR. Using empirical phase diagrams to understand the role of intramolecular dynamics in immunoglobulin G stability. Journal of pharmaceutical sciences. 2009;98(7):2432–2447. doi: 10.1002/jps.21619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strickland EH, Beychok S. Aromatic contributions to circular dichroism spectra of protein. Critical Reviews in Biochemistry and Molecular Biology. 1974;2(1):113–175. doi: 10.3109/10409237409105445. [DOI] [PubMed] [Google Scholar]
- 20.Kelly SM, Price NC. The use of circular dichroism in the investigation of protein structure and function. Current protein and peptide science. 2000;1(4):349–384. doi: 10.2174/1389203003381315. [DOI] [PubMed] [Google Scholar]
- 21.Hawe A, Sutter M, Jiskoot W. Extrinsic fluorescent dyes as tools for protein characterization. Pharmaceutical research. 2008;25(7):1487–1499. doi: 10.1007/s11095-007-9516-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sarvazyan AP. Ultrasonic velocimetry of biological compounds. Annual review of biophysics and biophysical chemistry. 1991;20(1):321–342. doi: 10.1146/annurev.bb.20.060191.001541. [DOI] [PubMed] [Google Scholar]
- 23.Buckin V, O'Driscoll B, Smyth C, Alting A, Visschers R. Ultrasonic spectroscopy for materials analysis: Recent advances. Spectroscopy Europe. 2003;15(1):20–25. [Google Scholar]
- 24.Gavish B, Gratton E, Hardy CJ. Adiabatic compressibility of globular proteins. Proc Natl Acad Sci U S A. 1983;80(3):750–754. doi: 10.1073/pnas.80.3.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lakowicz JR, Keating-Nakamoto S. Red-edge excitation of fluorescence and dynamic properties of proteins and membranes. Biochemistry. 1984;23(13):3013–3021. doi: 10.1021/bi00308a026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Demchenko AP. The red-edge effects: 30 years of exploration. Luminescence. 2002;17(1):19–42. doi: 10.1002/bio.671. [DOI] [PubMed] [Google Scholar]
- 27.Neil E, Marsh G. Towards the nonstick egg: designing fluorous proteins. Chem Biol. 2000;7(7):R153–R157. doi: 10.1016/s1074-5521(00)00139-3. [DOI] [PubMed] [Google Scholar]
- 28.Russo D, Hura G, Head-Gordon T. Hydration dynamics near a model protein surface. Biophys J. 2004;86(3):1852–1862. doi: 10.1016/S0006-3495(04)74252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Russo D, Murarka RK, Copley JR, Head-Gordon T. Molecular view of water dynamics near model peptides. J Phys Chem B. 2005;109(26):12966–12975. doi: 10.1021/jp051137k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vihinen M. Relationship of protein flexibility to thermostability. Protein engineering. 1987;1(6):477–480. doi: 10.1093/protein/1.6.477. [DOI] [PubMed] [Google Scholar]
- 31.Tang K, Dill KA. Native protein fluctuations: the conformational-motion temperature and the inverse correlation of protein flexibility with protein stability. Journal of biomolecular structure & dynamics. 1998;16(2):397–411. doi: 10.1080/07391102.1998.10508256. [DOI] [PubMed] [Google Scholar]
- 32.ZAvodszky PE, Kardos J, Svingor Á, Petsko GA. Adjustment of conformational flexibility is a key event in the thermal adaptation of proteins. Proceedings of the National Academy of Sciences. 1998;95(13):7406–7411. doi: 10.1073/pnas.95.13.7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hernández G, Jenney FE, Adams MWW, LeMaster DM. Millisecond time scale conformational flexibility in a hyperthermophile protein at ambient temperature. Proceedings of the National Academy of Sciences. 2000;97(7):3166–3170. doi: 10.1073/pnas.040569697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferreon JC, Volk DE, Luxon BA, Gorenstein DG, Hilser VJ. Solution structure, dynamics, and thermodynamics of the native state ensemble of the Sem-5 C-terminal SH3 domain. Biochemistry. 2003;42(19):5582–5591. doi: 10.1021/bi030005j. [DOI] [PubMed] [Google Scholar]
- 35.Durney MA, Wechselberger RW, Kalodimos CG, Kaptein R, Vorgias CE, Boelens R. An alternate conformation of the hyperthermostable HU protein fromThermotoga maritima has unexpectedly high flexibility. FEBS letters. 2004;563(1):49–54. doi: 10.1016/S0014-5793(04)00247-9. [DOI] [PubMed] [Google Scholar]
- 36.LeMaster DM, Tang J, Paredes DI, Hernández G. Enhanced thermal stability achieved without increased conformational rigidity at physiological temperatures: Spatial propagation of differential flexibility in rubredoxin hybrids. Proteins: Structure, Function, and Bioinformatics. 2005;61(3):608–616. doi: 10.1002/prot.20594. [DOI] [PubMed] [Google Scholar]
- 37.Kamerzell TJ, Middaugh CR. The complex inter-relationships between protein flexibility and stability. Journal of pharmaceutical sciences. 2008;97(9):3494–3517. doi: 10.1002/jps.21269. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1. Thermal stability of rPA (0.5 mg/ml at pH 7) as monitored by CD signals at indicated values (220, 262, 269, 284 and 291 nm) as a function of temperature. Each data point represents the mean of three independent runs, with error bars showing the standard deviation.
Supplemental Figure S2. Thermal stability of 2-FHis rPA (0.5 mg/ml at pH 5) as monitored by CD signals at indicated values (220, 262, 269, 284 and 291 nm) as a function of temperature. Each data point represents the mean of three independent runs, with error bars showing the standard deviation.
Supplemental Figure S3. Thermal stability of rPA (0.5 mg/ml at pH 5) as monitored by CD signals at indicated values (220, 262, 269, 284 and 291 nm) as a function of temperature. Each data point represents the mean of three independent runs, with error bars showing the standard deviation.