Abstract
The developmental stage-specific expression of the human β-like globin genes has been studied for decades, and many transcriptional factors as well as other important cis elements have been identified. However, little is known about the microRNAs that potentially regulate β-like globin gene expression directly or indirectly during erythropoiesis. In this study, we show that microRNA 23a (miR-23a) and miR-27a promote β-like globin gene expression in K562 cells and primary erythroid cells through targeting of the transcription factors KLF3 and SP1. Intriguingly, miR-23a and miR-27a further enhance the transcription of β-like globin genes through repression of KLF3 and SP1 binding to the β-like globin gene locus during erythroid differentiation. Moreover, KLF3 can bind to the promoter of the miR-23a∼27a∼24-2 cluster and suppress this microRNA cluster expression. Hence, a positive feedback loop comprised of KLF3 and miR-23a promotes the expression of β-like globin genes and the miR-23a∼27a∼24-2 cluster during erythropoiesis.
INTRODUCTION
The human β-globin locus consists of five functional globin genes (ε, Gγ, Aγ, δ, and β) within a 70-kb domain. The β-like globin genes are regulated through the locus control region (LCR), which consists of at least five DNase I hypersensitive sites (HS), HS1 to HS5, located upstream of the ε-globin gene (1). The preferential interactions between the LCR and the individual globin promoters during distinct developmental stages are pivotal for the stringent regulation of globin gene expression. These interactions are mediated by various erythroid tissue-restricted and ubiquitous transcription factors. Many transcription factors controlling β-like globin gene expression have been identified and characterized. For example, EKLF is a zinc finger transcription factor that activates the β-globin gene promoter by binding with high affinity to the CACCC element (2, 3). Whereas FKLF interacts with the CACCC box of the γ-globin gene to activate its transcription (4, 5), BCL11A functions as a silencer of the γ-globin gene and associates with HS3 and the intergenic region between Aγ- and δ-globin genes to reconfigure the β-like globin gene cluster (6, 7). Additionally, other transcription factors, such as NF-E2 (8, 9), GATA-1 (10), FOG (11), Sox6 (12), NF-E3 (13), SP1 (14, 15), KLF3/BKLF (16, 17), TR2, and TR4 (18), are involved in the control of β-like globin gene expression. Although these studies represent significant advances in the understanding of β-like globin gene regulation at the transcriptional level, only a few microRNAs (miRNAs) have been found to be regulators of the β-like globin locus (19, 20, 21).
miRNAs are endogenous, approximately 22-nucleotide (nt) RNAs that play important regulatory roles at the posttranscriptional level in animals and plants by targeting mRNAs for cleavage or translational repression (22, 23, 24). So far, miRNAs have been shown to regulate various developmental and cellular processes and are implicated in human diseases.
To understand the mechanisms of miRNAs regulating β-like globin gene expression, we analyzed miRNAs with a gene expression change correlated with the upregulation of ε- and γ-globin during hemin-induced K562 erythroid differentiation. We observed 63 miRNAs that not only gradually increased or decreased in expression level but also were in higher abundance during K562 cell erythroid differentiation. None of the miRNAs were predicted to bind to the 3′ untranslated region (UTR) of ε-, γ-, or β-globin mRNA. However, we noticed that miRNA 23a (miR-23a) and miR-27a, the levels of which increased during K562 erythroid differentiation, were potential candidates for binding to the 3′ UTR of two potential β-like globin suppressors, KLF3 and SP1, respectively. KLF3 is highly enriched in erythroid cells and is known to function as a strong transcriptional repressor (25). Furthermore, in vitro assays indicated that KLF3 could bind to the promoters of embryonic and adult β-globin genes as well as the β-globin LCR (16). The ubiquitously expressed SP1 zinc finger protein is the first described member of the Krüppel-like factors that bind to the consensus sequences of the GC and GT boxes (26). Two previous studies reported that SP1 could repress β-like globin gene transcription by binding to the LCR and globin promoter during erythroid differentiation (14, 15). These data suggest that miR-23a and miR-27a regulate β-like globin gene expression by targeting KLF3 and SP1, respectively. The implications of miR-23a and miR-27a in globin gene regulation remained to be determined, although the two miRNAs have been extensively studied in the context of cell cycle regulation, differentiation, and proliferation (27).
In this study, we show that miR-23a and miR-27a levels gradually increase during hemin-induced K562 and erythropoietin (Epo)-induced CD34+ HPCs (hematopoietic progenitor cells) erythroid differentiation. The miRNAs positively regulate β-like globin gene expression in K562 cells and primary erythroid cells by targeting the negative regulators KLF3 and SP1. Meanwhile, KLF3 interacts with the CACCC sites in the promoter of the miR-23a∼27a∼24-2 cluster (termed the miR-23a cluster), forming a positive feedback loop to upregulate the expression of β-like globin genes and the miRNA cluster during erythropoiesis.
MATERIALS AND METHODS
Bioinformatics analysis.
miRNA microarray data from K562 erythroid differentiation were obtained from our previous work, which is registered in the Gene Expression Omnibus (GEO) database (GSE30380). The miRNA microarray was carried out with Illumina microRNA expression bead chips (human V2) and was scanned by an Illumina bead array. According to the criteria, the strongest signal was >1,000, fold change was >2, the P value was <0.05, and a continuous increase or decrease in expression from 0 to 72 h was registered. We screened out 63 microRNAs that consistently and clearly increased or decreased in expression and were present in higher abundance during K562 cell erythroid differentiation (see Table S1 in the supplemental material). TargetScan software was used to predict the miRNA targets. The transcription element search system (http://www.cbil.upenn.edu/cgi-bin/tess) was used to analyze the miR-23a promoter sequence and predict the binding site of KLF3.
Cell culture and differentiation.
The human chronic myelogenous leukemia cell line K562 was maintained in RPMI 1640 supplemented with 10% fetal bovine serum (HyClone). Erythroid differentiation of K562 cells was achieved using 30 μM hemin (Sigma-Aldrich, Deisenhofen, Germany) over 24, 48, and 72 h. The degrees of differentiation were determined by benzidine staining for hemoglobin expression. Briefly, cells were collected and washed with phosphate-buffered saline (PBS) and stained with 10% freshly prepared benzidine dye (containing 2% H2O2) for 5 min, and colored cells were counted with a hemocytometer. HEK293T cells were obtained from the American Type Culture Collection (ATCC) and were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS).
Isolation and culturing of CD34+ hematopoietic progenitor cells.
The hematopoietic progenitor cell isolation system yielded approximately 90% CD34-positive cells. Human umbilical cord blood (UCB) was obtained from normal full-term deliveries after informed consent as approved by the Research Ethics Committee of the Peking Union Hospital (Beijing, China). Mononuclear cell (MNC) fractions were isolated from UCB by Percoll density (d) gradient centrifugation (d = 1.077 g/ml; Amersham Biotech, Germany). CD34+ cells were enriched from MNCs through positive immunomagnetic selection (CD34 MultiSort kit; Miltenyi Biotec, Bergisch Gladbach, Germany). The isolated CD34+ cells were cultured in Iscove's modified Dulbecco's medium (IMDM) supplemented with 30% fetal bovine serum (HyClone), 1% bovine serum albumin (BSA), 100 μM 2-mercaptoethanol (2-ME), 2 ng/ml recombinant human interleukin-3 (IL-3), 100 ng/ml recombinant human SCF (Stem Cell Technologies, Vancouver, British Columbia, Canada), 60 mg/ml penicillin, and 100 mg/ml streptomycin. For erythroid differentiation, 2 U/ml recombinant human Epo (R&D Systems, Minneapolis, MN) was added to the medium. Cells were collected and passaged every 3 to 5 days.
Oligonucleotides and transfection.
miRNA mimics, inhibitors, and negative-control molecules were obtained from Dharmacon (Austin, TX) (CN-001000-01-05 for mimic negative control, C-300494-03 for miR-23a mimic, C-300502-03 for miR-27a mimic, IN-001005-01-05 for inhibitor negative control, IH-300494-05 for miR-23a inhibitor, and IH300502-05 for miR-27a inhibitor) and were transfected with DharmaFECT1 (T-2001-03; Dharmacon) in K562 cells at a final concentration of 60 nM. Short interfering RNA (siRNA) smart pools (for KLF3 and SP1) and control siRNA pools were obtained from Dharmacon (D-001210-01-05 for nontargeting siRNA, M-006987-03 for siKLF3, and M-026959-00 for siSP1) and were transfected into K562 cells (100 nM) using DharmaFECT1. For erythroid differentiation, the transfected K562 cells were washed with PBS and plated for hemin induction the next day.
Constructs and lentivirus.
The reverse complementary sequence of miR-23a and miR-27a was inserted into the pMIR-reporter (Promega, WI) to generate a reporter system (pMIR-23a and pMIR-27a) that can detect mature miRNA expression in 293T cells. The 3′ UTRs of human KLF3 and SP1 mRNAs were PCR amplified and cloned into pMIR-reporter downstream of the firefly luciferase gene to generate the corresponding reporters. Mutations at the microRNA binding site in these mRNA sequences were created using the QuikChange site-directed mutagenesis kit (Stratagene, CA). For KLF3 and SP1 overexpression, pCMV6-KLF3 (SC101137) and pCMV6-SP1 (SC114197) were obtained from OriGene (MD). For expression of 5×His-tagged KLF3, the KLF3 open reading frame (ORF) was cloned into pcDNA6/V5-His B. The promoter of the miR-23a cluster was PCR amplified and cloned into pGL3-basic upstream of the firefly luciferase gene to generate the pGL3-23a-promoter reporter. Mutations at the KLF3 binding site in the promoter sequence were created using the QuikChange site-directed mutagenesis kit (Stratagene, CA). The primers are listed in Table S2 in the supplemental material.
The lentivirus vector expressing miR-23a (PMIRH23aPA-1) from which miR-24-2 was deleted artificially, miR-27a (PMIRH27a-onlyPA-1), and the packaging kit were purchased from System Biosciences (SBI; CA) and operated according to the manufacturer's instructions. The short hairpin RNA (shRNA) lentivirus plasmids specific to KLF3 were purchased from Santa Cruz Biotechnology (sc-44963-SH) and operated as described above. The harvested viral particles were added to K562 cells and the cultured CD34+ cells. K562 cells were then induced to erythroid differentiation by hemin after cell expansion for several days. CD34+ cells were washed with PBS the next day and plated for Epo-induced differentiation.
RNA isolation and qPCR.
Total RNA was extracted from the harvested cells using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. cDNA was synthesized via Moloney murine leukemia virus reverse transcriptase (RT; Invitrogen). Oligo(dT)18 was used as the RT primer for reverse transcription of mRNAs. 18S rRNA-specific primer was used as the RT primer for reverse transcription of 18S rRNA. Quantitative reverse transcription-PCR (qPCR) was performed in a Bio-Rad IQ5 real-time PCR system (Bio-Rad, Foster City, CA) using the SYBR premix Ex Taq kit (TaKaRa, Dalian, China) according to the manufacturer's instructions. To measure miR-23a and miR-27a expression, qPCR was performed using the following TaqMan probes according to the manufacturer's instructions (Applied Biosystems, Foster City, CA): pri-miR-23a (Hs03294931_pri), miR-27a (TM408), miR-23a (TM399), and RNU6B (TM1093). The expression of ε- and γ-globin genes was also measured by qPCR using TaqMan probes which were synthesized and labeled by Invitrogen. The comparative threshold cycle (CT) method was used to quantify the target genes relative to the endogenous control. For the mRNAs, the data were normalized with the endogenous glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and 18S rRNA control. For the miRNAs, U6 snRNA was used as the endogenous control. The oligonucleotides used for PCR are listed in Table S2 in the supplemental material. All of the PCRs were performed in triplicate.
Immunoblot analysis.
Whole-cell lysates were used for immunoblot analysis as previously described (28). The following antibodies were used for Western blot analysis: anti-γ-globin (sc-21756; Santa Cruz), anti-ε-globin (12361-1-Ap; Proteintech), anti-KLF3 (PAB6147; Abnova Corporation), anti-SP1 (17-601; Milipore), and anti-GAPDH (sc-365062; Santa Cruz). Immunoblots were quantified using ImageJ software.
Immunofluorescence and immunohistochemistry.
For the immunostaining of γ-globin in erythroid cells, cells at different induction time points were collected and smeared on glass slides. Cells were then fixed in 4% paraformaldehyde at room temperature for 10 min. After washing in PBS, cells were permeabilized in PBS containing 0.1% Triton X-100 at room temperature for 30 min and then blocked in 2.5% goat serum for 90 min. Cells were then incubated with human γ-globin antibodies (1:100; sc-21756) overnight at 4°C. As a negative control, cells were incubated with normal rabbit IgG. Detection was performed using a tetramethyl rhodamine isothiocyanate (TRITC)-conjugated goat anti-rabbit IgG (1:200; ZF-0316) at room temperature for 1 h.
Luciferase assay.
For the miRNA target analysis, 293T or K562 cells were cotransfected with 0.4 μg reporter construct, 0.02 μg pRL-TK vector, and 5 pmol miRNA mimic or scramble controls per well of a 24-well plate. To further confirm the regulation of endogenous microRNAs on the targets in K562 cells during erythroid differentiation, K562 cells were cotransfected with 0.4 μg reporter construct and 0.02 μg pRL-TK vector and then were induced to erythroid differentiation by hemin. For the functional analysis of the miR-23a promoter, 293T cells were cotransfected with 0.5 μg pGL3-basic or pGL3-23a promoter constructs, 0.02 μg pRL-TK vector and 0.5 μg pCMV6-KLF3, or empty pCMV6 vector in each well of a 24-well plate. K562 cells were also cotransfected with the reporter constructs described above, as well as 200 pmol si_KLF3 in uninduced K562 cells and 0.5 μg pCMV6-KLF3 in hemin-induced K562 cells. Cells were harvested 48 h posttransfection and assayed with a dual luciferase assay (Promega, WI) according to the manufacturer's instructions. All transfection assays were performed in triplicate.
Flow cytometry.
K562 cells and the cultured primary erythroid cells were harvested at the indicated times and were washed twice at 4°C in PBS containing 0.5% BSA. K562 cells were incubated with phycoerythrin (PE)-conjugated anti-CD71 (12-0719) and fluorescein isothiocyanate (FITC)-conjugated anti-CD235a (11-9987) antibody (eBioscience, CA). Cultured primary erythroid cells were incubated with PE-conjugated anti-CD235a (12-9987) and antigen-presenting cell (APC)-conjugated anti-CD71 (17-0719) antibody (eBioscience, CA). Flow cytometry was performed using a C6 flow cytometer instrument (BD Biosciences, Franklin Lakes, NJ).
ChIP assay.
Anti-KLF3 (PAB6147; Abnova) and anti-SP1 (17-601; Millipore) were used for chromatin immunoprecipitation (ChIP) experiments. Goat IgG (sc-34665) and rabbit IgG (sc-66931) were used as isotype antibody controls of anti-KLF3 and anti-SP1. K562 cells were induced with hemin for 0 and 48 h and then were collected and cross-linked with 1% formaldehyde for 10 min. The cells were then washed in cold PBS buffer, resuspended in lysis buffer (0.1% SDS, 0.5% Triton X-100, 20 mM Tris-HCl [pH 8.1], 150 mM NaCl, protease inhibitor), and sonicated to obtain chromatin fragments between 200 and 1,000 bp in size. The sonicated chromatin was resuspended in immunoprecipitation buffer and incubated overnight at 4°C with antibodies conjugated with magnetic beads (Santa Cruz Biotechnologies). The beads were then washed with lysis buffer, LiCl buffer (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris-HCl [pH 8.1]), and TE buffer and eluted in elution buffer (1% SDS, 0.1 M NaHCO3). The DNA was then recovered by reversing the cross-links and was purified using a Qiagen DNA purification kit. An unenriched sample of DNA was treated in a similar manner to serve as the input. An HS4 fragment containing SP1 binding sites and excluding KLF3 binding sites was used as the negative genomic region in KLF3 ChIP analysis. Similarly, fragment III of the miR-23a cluster promoter containing two KLF3 binding sites but excluding SP1 binding sites was used as a negative genomic region in SP1 ChIP analysis. The primers used for the ChIP-PCR are listed in Table S2 in the supplemental material.
EMSA.
Nuclear extracts of 293T cells transfected with pCDNA6-KLF3-His were prepared using NE-PER nuclear and cytoplasmic extraction reagents (Thermo Scientific) and stored at −80°C. The electrophoretic mobility shift assay (EMSA) was operated using a LightShift chemiluminescent EMSA kit (Thermo Scientific) according to the manufacturer's instructions. The DNA-protein binding reaction buffer was composed of 1× binding buffer supplemented with 2.5% glycerol, 5 mM MgCl2, 50 ng/ml poly(dI·dC), 0.5 μg/μl BSA, and 1 mM EDTA. Three microliters nuclear extract was used in each binding reaction in a total volume of 20 μl, and the reaction mixture was incubated at room temperature for 20 min. Wild-type and mutant probes were labeled with biotin, and 20 fmol of each probe was used per reaction. In competition experiments, a 200 M excess of unlabeled probe was added to the binding reaction 15 min before the biotin-labeled probe was added. For supershift experiments, nuclear extracts were preincubated with 2 μg of anti-His antibodies (sc-803; Santa Cruz) for 40 min at room temperature. The probes used for EMSA are listed in Table S2 in the supplemental material.
Statistics.
A Student's t test (two-tailed) was performed to analyze the data. P values of <0.05 were considered statistically significant.
RESULTS
miR-23a and miR-27a are potential regulators of β-like globin gene expression during erythroid differentiation.
We previously profiled the expression of miRNAs in an erythroid differentiation model using erythroleukemia K562 cells after hemin induction for 0, 24, 48, and 72 h. Following an analysis of differentially expressed miRNAs, we found that there were 63 miRNAs that not only gradually increased or decreased but also were in higher abundance during K562 cell erythroid differentiation (Fig. 1A; also see Table S1 in the supplemental material). Among them, none was predicted to bind to the 3′ UTR of ε-, γ-, or β-globin mRNA. However, miR-23a and miR-27a were predicted to bind to the 3′ UTR of the mRNA of two potential globin suppressors, KLF3 and SP1, respectively, suggesting that miR-23a and miR-27a regulate β-like globin gene expression through targeting KLF3 and SP1. Moreover, miR-23a and miR-27a are derived from the same miR-23a∼27a∼24 cluster, implying that they play synergistic roles in the regulation of β-like globin gene expression.
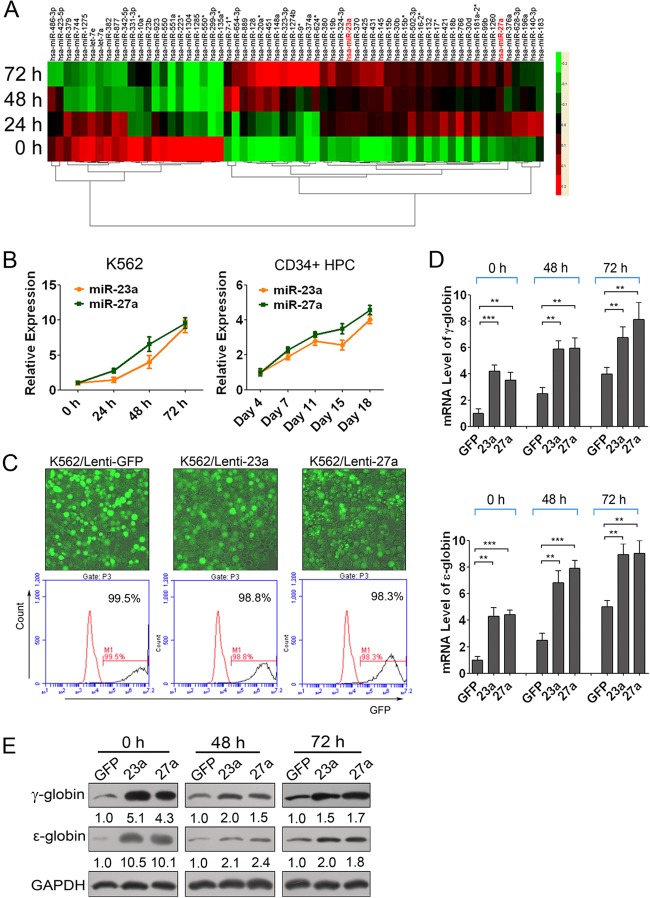
Fig 1.
miR-23a and miR-27a upregulated β-like globin genes in K562 cells. (A) Heat map representation of the expression change of 63 miRNAs that consistently increased or decreased during erythroid differentiation in the miRNA microarray analysis of hemin-induced K562 cells at 0, 24, 48, and 72 h. (B) qPCR validation of the expression of mature miR-23a and miR-27a in hemin-induced K562 cells and Epo-induced CD34+ HPCs. (C) Fluorescence image merged with phase of lentivirus-infected K562 cells and FACS analysis of GFP-positive cells. (D) qPCR analysis of ε- and γ-globin gene expression normalized with GAPDH in miR-23a- and miR-27a-overexpressing K562 cells at 0, 48, and 72 h of hemin induction. (E) Western blot analysis of ε- and γ-globin expression in miR-23a- and miR-27a-overexpressing K562 cells at 0, 48, and 72 h of hemin induction. Student's t test (two-tailed) was performed to analyze data from the experiments in triplicate. P values of <0.05 were considered significant, as indicated by the asterisks (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
To validate the expression of miR-23a and miR-27a, qPCR was used to measure the levels of mature miR-23a and miR-27a in hemin-induced K562 cells. As expected, levels of mature miR-23a and miR-27a were markedly upregulated after hemin induction (Fig. 1B). Additionally, the expression profiles of miR-23a and miR-27a in human CD34+ HPC-derived primary erythroid cultures, which resemble in vivo hematopoiesis more closely, were assessed by qPCR and showed that miR-23a and miR-27a were also upregulated during Epo-induced erythroid differentiation of human CD34+ HPCs (Fig. 1B).
Overexpression of miR-23a or miR-27a significantly promotes β-like globin gene expression in K562 cells during erythroid differentiation.
To investigate the regulation of miR-23a and miR-27a on β-like globin gene expression, we first examined the mRNA and protein levels of ε- and γ-globin, the main β-like globin products of K562 cells, in miRNA-transfected K562 cells. The introduction of both miR-23a and miR-27a increased the mRNA and protein levels of ε- and γ-globin genes and also augmented the hemoglobin-containing cell percentage compared to the negative mimic controls at the indicated time points (see Fig. S1 in the supplemental material). These results demonstrated that miR-23a and miR-27a could promote the expression of ε- and γ-globin in K562 cells. To further confirm the regulation of miR-23a and miR-27a on globin gene expression in K562 cells, we infected K562 cells with lentiviruses harboring miR-23a or miR-27a (lenti-23a and lenti-27a, respectively), which could improve the efficiency of overexpression of microRNAs. Fluorescence-activated cell sorter (FACS) analysis showed that 99% of the K562 cells infected with lenti-23a or lenti-27a were GFP positive, indicating remarkable infection efficiency of lentiviruses in K562 cells (Fig. 1C). Meanwhile, qPCR was performed to measure the levels of miR-23a or miR-27a and indicated that they were successfully overexpressed in K562 cells (see Fig. S2A in the supplemental material). Further, we examined the mRNA and protein levels of ε- and γ-globin through qPCR and immunoblotting in lentivirus-infected K562 cells. As expected, transduction of miR-23a or miR-27a significantly increased the expression levels of ε- and γ-globin genes, and the increase was far more than that in K562 cells transiently transfected with miRNA mimics (Fig. 1D and E). Thus, our data indicated the important roles of miR-23a and miR-27a on ε- and γ-globin gene expression in K562 cells. It was noteworthy that although the activity of the microRNAs on ε- and γ-globin gene expression was still obvious at both 48 and 72 h after hemin induction, it was not as significant as that in uninduced K562 cells (Fig. 1D and E). This might be due to the strong globin expression after K562 cells were induced to erythroid differentiation by hemin. All of the relative expression of globin genes detected by qPCR was normalized against both GAPDH and 18S rRNA, and the results were consistent with each other in principle. The relative expression normalized with 18S rRNA is shown in Fig. S2B in the supplemental material.
Enforced expression of miR-23a or miR-27a markedly promotes β-like globin gene expression during erythroid differentiation of CD34+ HPCs.
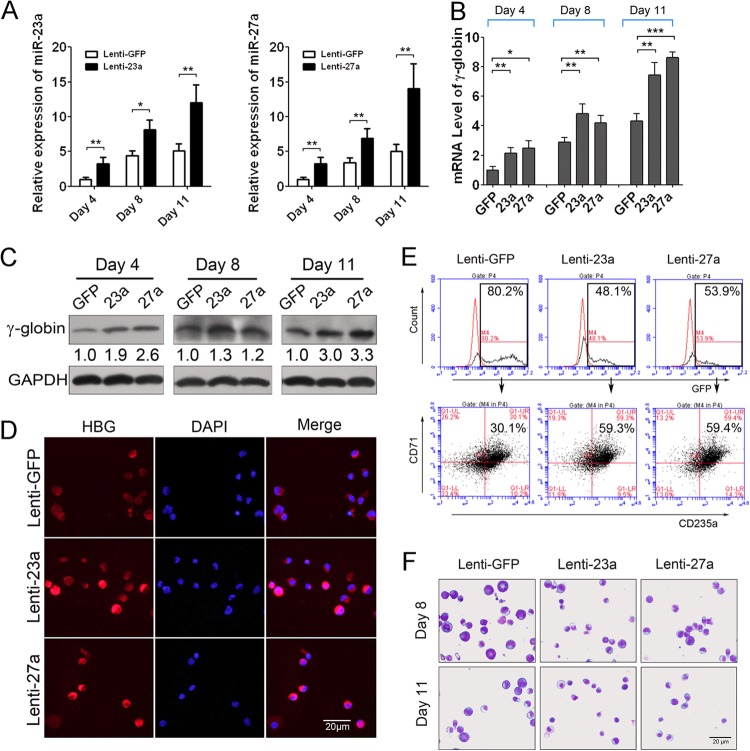
To further assess the roles of miR-23a and miR-27a in globin gene expression during normal human erythroid differentiation, we transduced CD34+ HPCs with lentivirus carrying miR-23a or miR-27a and harvested the cells after 4, 8, and 11 days of erythroid culture. qPCR was used to verify the overexpression of miR-23a and miR-27a in the CD34+ HPCs (Fig. 2A). Since γ-globin is the main β-like globin product in the UCB-derived primary erythroid cultures, qPCR and Western blotting confirmed that γ-globin gene expression was elevated in the lenti-23a- and lenti-27a-infected CD34+ cells at each time point (Fig. 2B and C; also see Fig. S3 in the supplemental material). The improvement of miR-23a and miR-27a on globin expression in early stages of induction was averaged two times, which was biologically significant because of the high expression background of γ-globin in erythroid cells. In addition, immunostaining with γ-globin antibody detected the expression of γ-globin in CD34+ HPCs at 11 days post-Epo induction and showed that γ-globin expression was obviously higher in lenti-23a- or lenti-27a-infected CD34+ cells than in control cells, consistent with the Western blotting results (Fig. 2D). Meanwhile, FACS analysis was performed to measure the percentage of CD71/CD235a double-positive cells in the GFP-positive cells at day 11 after Epo induction and showed that the percentage of double-positive cells was also augmented significantly in lenti-23a (59.3%)- and lenti-27a (59.4%)-infected cells compared to control cells (30.1%) (Fig. 2E). In addition, May-Grünwald-Giemsa staining was also employed to observe the cell and nuclear morphology of CD34+ HPCs during erythroid differentiation. As erythroid differentiation proceeds, erythroblasts display a gradual decrease in cell size and increase in nuclear condensation; indeed, the miR-23a- and miR-27a-infected erythroid cells appeared smaller with more condensed nuclei than control cells (Fig. 2F).
Fig 2.
miR-23a and miR-27a upregulated β-like globin genes in human primary erythroid cells. (A) qPCR analysis of miR-23a and miR-27a expression in lentivirus-infected human CD34+ HPCs after Epo induction for 4, 8, and 11 days. (B) qPCR analysis of γ-globin gene expression normalized with GAPDH in lentivirus-infected human CD34+ HPCs at different induction days. (C) Western blot analysis of γ-globin expression in miR-23a- and miR-27a-overexpressing CD34+ HPCs after Epo induction. (D) Immunostaining of human γ-globin in lentivirus-infected human CD34+ HPCs after Epo induction for 11 days. DAPI, 4′,6-diamidino-2-phenylindole. (E) FACS analysis of GFP-positive cells and CD71/CD235a double-positive cells among them at 11 days after Epo induction of CD34+ HPCs. (F) May-Grünwald-Giemsa staining of lentivirus-infected human CD34+ HPCs after Epo induction for 8 and 11 days. Student's t test (two-tailed) was performed to analyze the data from the experiments in triplicate. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Taken together, our data indicate that miR-23a and miR-27a promote β-like globin gene expression in K562 cells and CD34+ HPCs during erythroid differentiation.
Two potential β-like globin suppressors, KLF3 and SP1, are the direct targets of miR-23a and miR-27a, respectively.
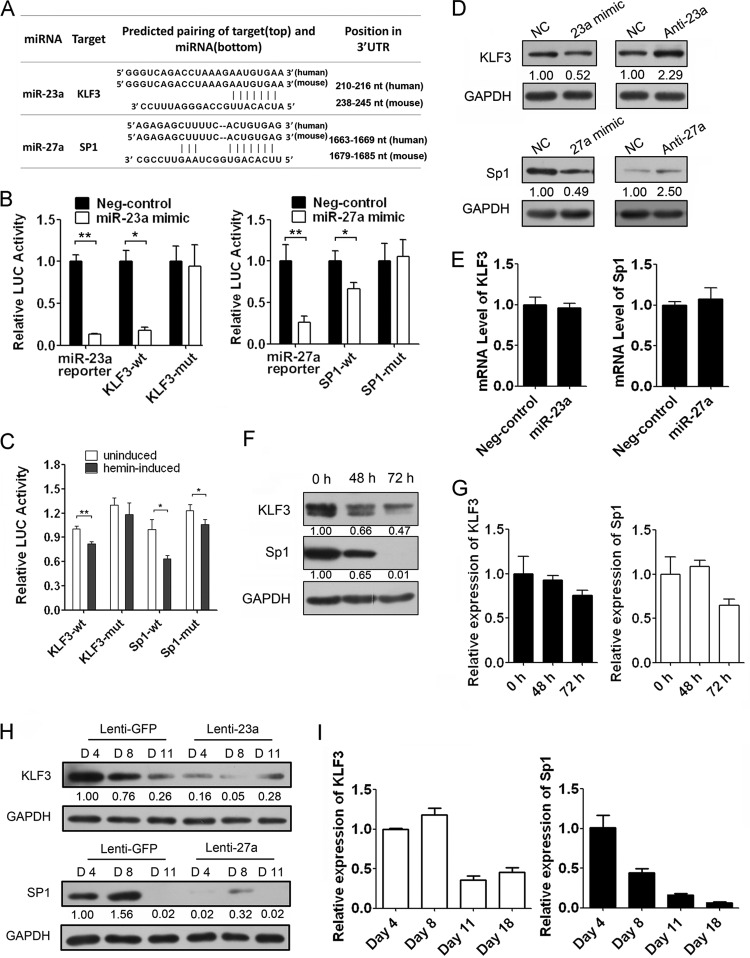
miRNAs perform the biological functions by downregulating expression of their target genes. Two algorithms, PicTar and TargetScan, were employed to predict the targets of miR-23a and miR-27a and showed they had the potential to bind to the conserved 3′ UTRs of the transcription factors KLF3 and SP1, respectively (Fig. 3A; also see Fig. S4 in the supplemental material). To validate our prediction, we cloned the 3′ UTR of KLF3 or SP1 into a luciferase reporter construct (pMIR-reporter). We also constructed it with the complete nucleotide sequence complementary to the miRNA sequence as a positive control. Reporter assays in 293T and K562 cells revealed that repression of the two 3′ UTRs was miRNA dependent, and mutation of the miRNA binding sites abrogated the reduced luciferase activity (Fig. 3B; also see Fig. S5 in the supplemental material). Furthermore, the upregulation of endogenous miR-23a and miR-27a in hemin-induced K562 cells also repressed the activity of the two 3′ UTRs, and the repression was partially dependent on the miRNA binding sites (Fig. 3C). Consistent with the reporter assays, we observed an apparent decrease in KLF3 and SP1 expression in the presence of miRNA mimics compared to that of the scramble control in K562 cells (Fig. 3D). However, there was little effect on the mRNA levels of KLF3 and SP1 in K562 cells transfected with miRNA mimics, suggesting that miR-23a and miR-27a negatively regulated the expression of their target genes through inhibiting protein translation (Fig. 3E). Conversely, KLF3 and SP1 protein levels were increased after endogenous miR-23a or miR-27a was blocked when using the corresponding miRNA inhibitors in K562 cells (Fig. 3D). Moreover, the mRNA and protein level of KLF3 and SP1 also declined along with the erythroid differentiation of K562 cells, consistent with their negative roles in globin gene expression (Fig. 3F and G). Similar to K562 cells, overexpression of miR-23a or miR-27a led to a significant decrease of KLF3 or SP1 at the indicated time points during Epo-induced CD34+ HPC differentiation, although KLF3 and SP1 levels decreased during the process (Fig. 3H and I).
Fig 3.
KLF3 and SP1 are direct targets of miR-23a and miR-27a, respectively. (A) Computer prediction of the binding of miR-23a and miR-27a on the 3′ UTR of human KLF3 and SP1, respectively. (B) The relative luciferase (LUC) activity of the reporter constructs cotransfected with scrambled control (Neg-control) or miR-23a and miR-27a mimics. Firefly luciferase activity was normalized to the activity of coexpressing renilla luciferase. (C) The relative luciferase activity of the reporter constructs in K562 cells left uninduced or induced with hemin for 48 h. (D) Western blot analysis of KLF3 and SP1 in K562 cells transfected with the scrambled control (NC) or miR-23a and miR-27a mimics and inhibitors. (E) qPCR analysis of the KLF3 and SP1 mRNA levels in K562 cells transfected with the scrambled control or miR-23a and miR-27a mimics. (F and G) The expression of KLF3 and SP1 during erythroid differentiation of hemin-induced K562 cells. (H) Western blot of KLF3 and SP1 in the lentivirus-infected CD34+ HPCs after Epo induction for 4, 8, and 11 days. (I) The mRNA level of KLF3 and SP1 during erythroid differentiation of Epo-induced CD34+ HPCs. Student's t test (two-tailed) was performed to analyze the data from the experiments in triplicate. *, P < 0.05; **, P < 0.01.
Repression of KLF3 is required for the miR-23a-mediated enhancement of globin gene expression during erythropoiesis.
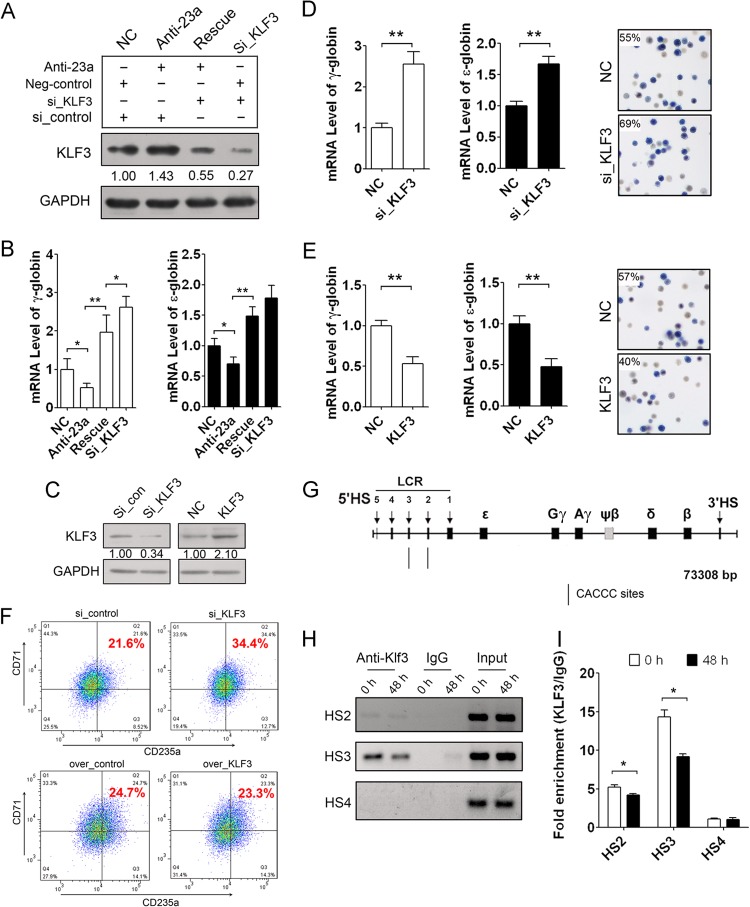
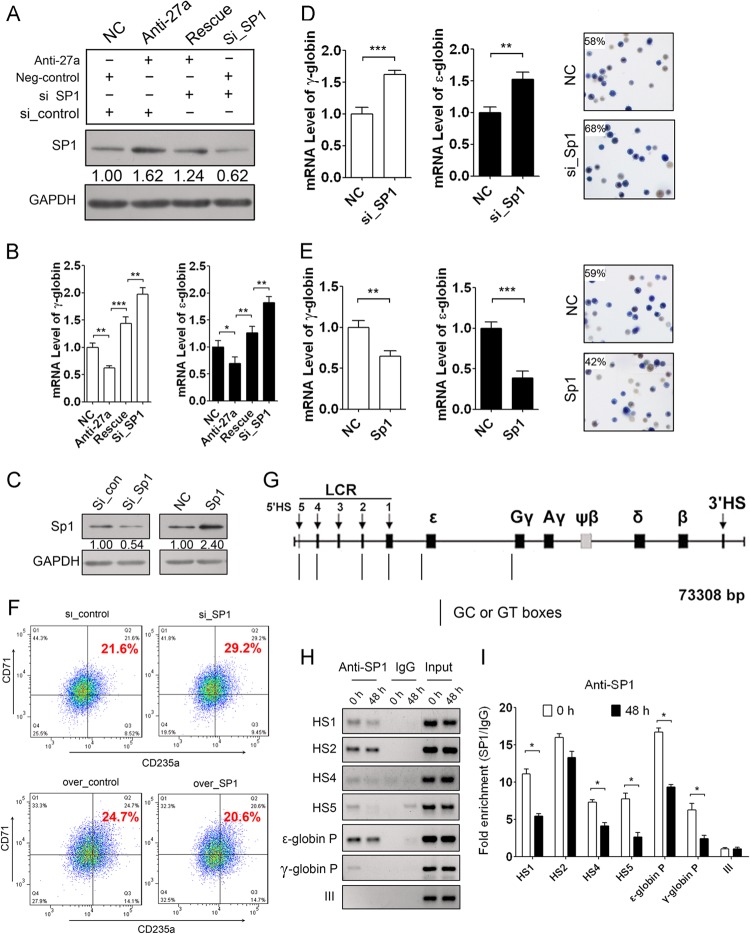
To further investigate whether miR-23a regulation of ε- and γ-globin gene expression is dependent on KLF3 targeting, we employed a rescue experiment with an miR-23a inhibitor and a KLF3 siRNA in differentiating K562 cells. A KLF3 siRNA was transfected after treatment with miR-23a inhibitor to attenuate the upregulation of KLF3, which was due to miR-23a repression. An increase in KLF3 after treatment with miR-23a inhibitor confirmed the regulatory role of miR-23a on the expression of the target. The addition of KLF3 siRNA led to further downregulation of KLF3 based on the previously described upregulation (Fig. 4A). Consistent with the restored expression of KLF3 protein, suppression of ε- and γ-globin expression by miR-23a inhibitor was rescued by the addition of KLF3 siRNA after 48 h of hemin induction (Fig. 4B). These data confirm the regulatory role of miR-23a on globin levels through targeting KLF3.
Fig 4.
Repressed KLF3 is required for miR-23a-enhanced globin gene expression. (A) Western blot analysis of KLF3 in K562 cells cotransfected with miR-23a inhibitor and/or KLF3 siRNA after 48 h of hemin induction. (B) qPCR analysis of ε- and γ-globin gene expression in the K562 cells shown in panel A. (C) Western blot analysis of KLF3 in K562 cells transfected with either KLF3 siRNA or pCMV-KLF3. (D and E) qPCR analysis of ε- and γ-globin gene expression and benzidine staining of hemoglobin-containing cells in K562 cells transfected with KLF3 siRNA (D) or pCMV-KLF3 (E) after 48 h of hemin induction. (F) FACS analysis of CD235a/CD71 double-positive cells in K562 cells transfected with KLF3 siRNA or pCMV-KLF3 after 48 h of hemin induction. (G) Schematic of the β-like globin gene locus and the KLF3 binding sites in the HS2 and HS3 regions. (H) ChIP-PCR analysis of KLF3 binding on the CACCC sites in the HS2 and HS3 regions during K562 cell erythroid differentiation. (I) ChIP-qPCR analysis of KLF3 binding on the CACCC sites in the HS2 and HS3 regions during K562 cell erythroid differentiation. Student's t test (two-tailed) was performed to analyze the data from the experiments in triplicate. *, P < 0.05; **, P < 0.01.
Furthermore, we examined the regulation of KLF3 on ε- and γ-globin gene expression in K562 cells. We knocked down or overexpressed KLF3 in K562 cells and detected ε- and γ-globin expression, the proportion of hemoglobin-containing cells, and the percentage of CD71/CD235a double-positive cells after 48 h of hemin induction. Immunoblot analysis was performed to examine the effect of KLF3 knockdown or overexpression in K562 cells (Fig. 4C). We found that knockdown of KLF3 strongly increased ε- and γ-globin expression and augmented the percentage of benzidine-positive cells (Fig. 4D). Conversely, overexpression of KLF3 decreased ε- and γ-globin expression and the percentage of benzidine-positive cells (Fig. 4E). In addition, the percentage of CD71/CD235a double-positive cells also increased when KLF3 was downregulated but decreased when KLF3 was overexpressed (Fig. 4F). KLF3 has been shown to act as a strong transcriptional repressor (25). In vitro assays have also indicated that KLF3 can bind to the CACCC sites in the HS2 and HS3 regions of the β-globin LCR (16) (Fig. 4G). Therefore, we assessed the interaction between KLF3 and the two CACCC sites during hemin-induced differentiation of K562 cells through ChIP-PCR. We found that KLF3 could bind to the CACCC sites in the HS2 and HS3 regions, and that the binding gradually decreased during K562 erythroid differentiation while exhibiting no binding with the HS4 fragment lacking KLF3 binding sites (Fig. 4H). ChIP-qPCR analysis was further employed to quantify the binding capacity and showed that the binding of KLF3 on these two CACCC sites was clearly decreased in hemin-induced K562 cells (Fig. 4I). KLF3 likely binds to the LCR to further recruit the general corepressor CtBP, which also associates with other hematopoietic transcriptional repressors, such as Evi-1 and ZEB/AREB6, to suppress ε- and γ-globin expression (25). The decrease in KLF3 is partially due to miR-23a upregulation and erythroid differentiation. The decrease in KLF3 level leads to the removal of the repressors from the globin locus and facilitates globin gene expression.
Repressed SP1 is required for miR-27a-enhanced globin gene expression during erythropoiesis.
To investigate whether miR-27a regulation of ε- and γ-globin gene expression depends on SP1 targeting, we also employed a rescue experiment as described above. An increase in SP1 after treatment with miR-27a inhibitor confirmed the negative regulation of miR-27a on target gene expression in the process. The addition of SP1 siRNA along with an miR-27a inhibitor led to further downregulation of SP1 (Fig. 5A). Consistent with the restored expression of SP1, the suppression of miR-27a inhibitor on ε- and γ-globin gene expression was rescued by SP1 siRNA after 48 h of hemin induction in K562 cells (Fig. 5B). These data confirm the specific regulatory role of miR-27a on SP1.
Fig 5.
Repressed SP1 is required for miR-27a-enhanced globin gene expression. (A) Western blot analysis of SP1 in K562 cells cotransfected with miR-27a inhibitor and SP1 siRNA. (B) qPCR analysis of ε- and γ-globin gene expression in the K562 cells shown in panel A. (C) Western blot analysis of SP1 in K562 cells transfected with SP1 siRNA or pCMV-SP1. (D and E) qPCR analysis of ε- and γ-globin gene expression and benzidine staining of hemoglobin-containing cells in K562 cells transfected with SP1 siRNA (D) or pCMV-SP1 (E) at 48 h after hemin induction. (F) FACS analysis of CD235a/CD71 double-positive cells in K562 cells transfected with SP1 siRNA or pCMV-SP1 after 48 h of hemin induction. (G) Schematic of the β-like globin gene locus and SP1 binding sites. (H) ChIP-PCR analysis of the binding of SP1 on the GC or GT boxes in HS1, HS2, HS4, HS5, and the promoter (P) of ε- and γ-globin genes during K562 cell erythroid differentiation. (I) ChIP-qPCR analysis of the binding of SP1 on these sites during K562 cell erythroid differentiation. Student's t test (two-tailed) was performed to analyze the data from the experiments in triplicate. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Furthermore, we studied the regulation of SP1 on ε- and γ-globin gene expression and the detailed molecular mechanisms in K562 cells. We knocked down and overexpressed SP1 in K562 cells and then induced the cells to undergo erythroid differentiation. Immunoblotting using an SP1 antibody showed that SP1 was successfully knocked down or overexpressed in the K562 cells (Fig. 5C). qPCR was used to detect ε- and γ-globin gene expression, benzidine staining was used for identification of hemoglobin-containing cells, and FACS was employed to identify the percentage of CD71/CD235a double-positive cells after 48 h of hemin induction. Knockdown of SP1 increased ε- and γ-globin expression and augmented the percentage of benzidine-positive cells (Fig. 5D). However, overexpression of SP1 decreased ε- and γ-globin expression and the percentage of benzidine-positive cells (Fig. 5E). Meanwhile, the downregulation of SP1 enlarged the percentage of CD71/CD235a double-positive cells, and the overexpression of SP1 decreased the percentage instead (Fig. 5F). SP1 is a ubiquitously expressed zinc finger protein that has been reported to repress β-like globin gene transcription through binding to HS1, HS2, HS3, HS4, HS5, and regions of the ε-, γ-, and β-globin gene promoter during Aγ181 cell erythroid differentiation (15) (Fig. 5G). Therefore, we further assessed the interaction between SP1 and the GC or GT box during K562 erythroid differentiation using ChIP-PCR and found that SP1 could bind to the GC or GT box in the HS1, HS2, HS4, HS5, and promoter regions of the ε- and γ-globin genes but not in the HS3 region. Moreover, these interactions were evidently decreased during erythroid differentiation of K562 cells (Fig. 5H). A negative control of the genomic region, fragment III of the miR-23a cluster promoter lacking SP1 binding sites, exhibited no binding with SP1 (Fig. 5H). The binding of SP1 with the β-globin promoter was not studied, as there was very little β-globin expression in K562 cells. ChIP-qPCR was simultaneously employed to quantify the degree of avidity and showed that the binding of SP1 on the GC or GT box in the same regions as those described in the legend to Fig. 5H was significantly downregulated after hemin induction (Fig. 5I). These results indicate that SP1 is a negative β-like globin regulator, which is in line with previous reports.
KLF3 resides in the promoter of the miR-23a cluster and represses its expression.
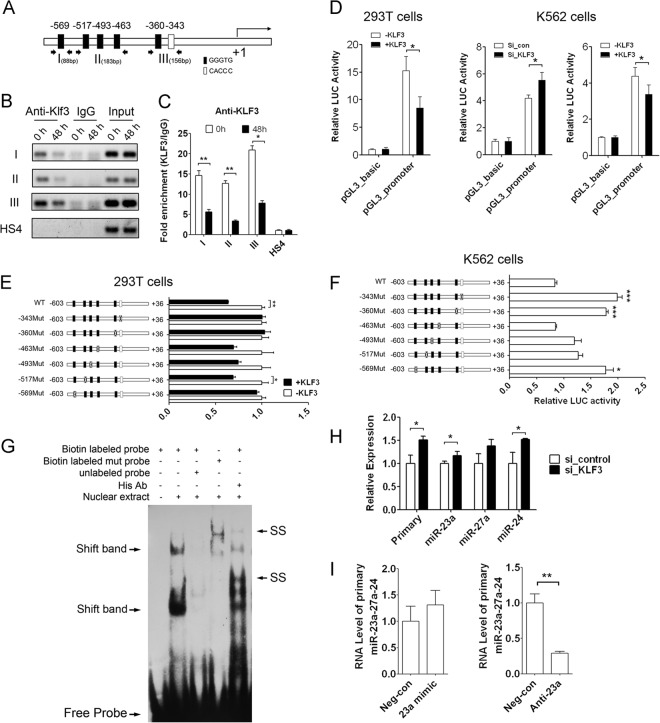
To investigate the regulation of miR-23a and miR-27a expression during erythroid differentiation, we analyzed the promoter sequences of the miR-23a cluster using a transcription element search system. The prediction revealed six putative KLF3 binding sites scattered within the promoter region of the human miR-23a cluster (Fig. 6A). ChIP-PCR was initially employed to determine whether KLF3 could bind to these sites in the miR-23a cluster promoter. Three pairs of primers were designed to detect the six putative binding sites, because several KLF3 binding sites were too close to each other to analyze (Fig. 6A). The results showed that KLF3 could bind to all three fragments, and that these interactions decreased significantly after 48 h of hemin induction, but it exhibited no binding to the HS4 region fragment lacking the KLF3 binding site (Fig. 6B). ChIP-qPCR was also performed and showed that the bindings were significantly decreased in differentiated K562 cells (Fig. 6C). To further investigate the regulation of KLF3 on the miR-23a cluster promoter, we cloned the promoter region of the miR-23a cluster into a luciferase reporter construct (pGL3-basic). A reporter assay in 293T cells showed that overexpression of KLF3 inhibited the activity of the miR-23a cluster promoter but had no effect on pGL3-basic vector activity (Fig. 6D). Furthermore, knockdown of KLF3 in K562 cells promoted the activity of the miR-23a cluster promoter, whereas overexpression of KLF3 in K562 cells inhibited the miR-23a cluster promoter activity, indicating the endogenous KLF3 repression of the activity of the miR-23a cluster promoter (Fig. 6D). To confirm the activity of each binding site, we cloned a series of luciferase reporters containing the corresponding mutated promoters. The differential cotransfection of the reporters with KLF3 in 293T cells indicated that the −569, −360, and −343 sites were important for the suppression of KLF3 on the miR-23a promoter. The inhibition was completely rescued when these three sites were mutated. However, the inhibition was moderately rescued when the −517, −493, and −463 sites were mutated, suggesting that these three sites were not as crucial as the other three sites (Fig. 6E). In the same way, repression of endogenous KLF3 on the miR-23a cluster promoter was significantly abrogated when the −569, −360, and −343 sites were mutated in K562 cells (Fig. 6F). We deduced that these sites play a coordinate role, and each of them was necessary for the repression activity of KLF3 on the promoter of the miR-23a cluster. Thus, disruption of any of the three sites (−343, −360, and −569) completely cancelled the inhibitory effect produced by KLF3. Furthermore, an EMSA using the nuclear extracts from the 293T cells overexpressing KLF3 carrying a 5×His tag and an oligonucleotide probe containing the −569 KLF3 binding site was performed and demonstrated that KLF3 could bind to the −569 GGGTG site in vitro (Fig. 6G). However, two shift bands were observed, which might be due to the interaction of KLF3 with other nuclear proteins in the 293T cells. The shift was prevented when competing with an excess of unlabeled DNA and disappeared when the GGGTG sites were mutated. In addition, supershift bands were observed behind the shift bands when the anti-His antibody was added to the binding system (Fig. 6G).
Fig 6.
KLF3 resides in the promoter of the miR-23a cluster and represses its expression. (A) The six putative KLF3 binding sites scattered within the promoter region of the human miR-23a cluster gene.(B) ChIP-PCR analysis of KLF3 binding to the six putative sites in the miR-23a cluster promoter during hemin-induced K562 erythroid differentiation. (C) ChIP-qPCR analysis of KLF3 binding to the six putative sites in the miR-23a cluster promoter during hemin-induced K562 erythroid differentiation. (D) Relative luciferase activity representing the miR-23a promoter activity in 293T cells cotransfected with KLF3 or negative control, in K562 cells cotransfected with si_KLF3/si_control, and in hemin-induced K562 cells with KLF3 overexpression. (E and F) Relative luciferase activity of a series of luciferase reporters containing the corresponding mutated promoter in 293T cells cotransfected with KLF3 (E) and also in K562 (F) cells. (G) EMSA of the binding of KLF3 with the −569 CACCC sites in vitro. Shift band stands for specific KLF3/probe complex, and SS represents the supershift band. Ab, antibody. (H) qPCR analysis of the primary transcript and the mature members of the miR-23a cluster in K562 cells transfected with KLF3 siRNA. (I) qPCR analysis of the primary miR-23a cluster in K562 cells transfected with miR-23a mimic or inhibitor. Student's t test (two-tailed) was performed to analyze the data from the experiments in triplicate. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To determine whether KLF3 could influence the expression of the miR-23a cluster in vivo, both the primary and mature levels of the miR-23a cluster were evaluated in K562 cells treated with siRNA specific to KLF3. qPCR was conducted and showed that KLF3 inhibition increased the expression of the miR-23a cluster (Fig. 6H). Therefore, we hypothesized that miR-23a could promote the expression of its own primary transcript through inhibiting KLF3. To verify the hypothesis, we detected the level of primary miR-23a cluster in K562 cells treated with miR-23a mimic or inhibitor and found that the level of the primary miR-23a cluster was upregulated upon miR-23a overexpression. Conversely, the primary miR-23a cluster was downregulated when miR-23a was inhibited in K562 cells (Fig. 6I).
KLF3 negatively regulates the expression of γ-globin and the miR-23a cluster in human primary erythroid cells.
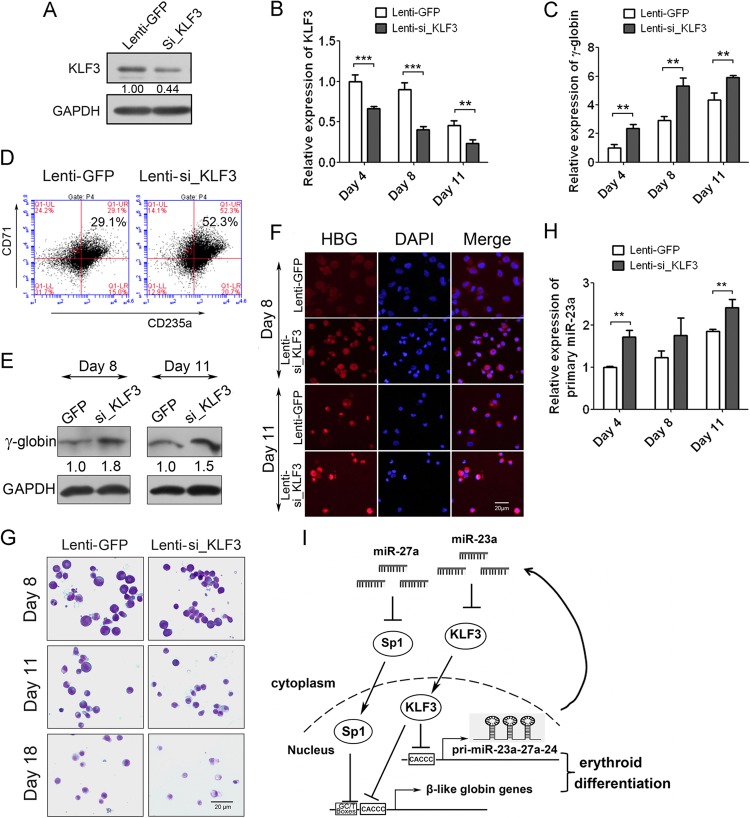
To confirm the KLF3-mediated regulation of globin genes and miR-23a cluster expression during normal erythropoiesis, we knocked down KLF3 via lentivirus-mediated shRNA in human CD34+ HPCs and validated the decreased expression of KLF3 through Western blotting and qPCR (Fig. 7A and B). Along with the decreased expression of KLF3, γ-globin was upregulated during Epo-induced erythroid differentiation of CD34+ HPCs (Fig. 7C). The relative expression of KLF3 and γ-globin detected by qPCR was normalized with both GAPDH and 18S rRNA, and the results were consistent with each other in principle. The relative expression of KLF3 and γ-globin normalized with 18S rRNA is shown in Fig. S6 in the supplemental material. FACS analysis was conducted to measure the percentage of CD71/CD235a double-positive cells and showed that knockdown of KLF3 increased the percentage of CD71/CD235a double-positive cells (52.3%) compared to 29.1% in the control cells at 11 days after Epo induction (Fig. 7D). Meanwhile, HPCs at 8 and 11 days of Epo induction were immunoblotted and stained with γ-globin antibody and clearly showed the augmentation of γ-globin expression in KLF3 shRNA-transduced CD34+ cells compared to control cells (Fig. 7E and F). May-Grünwald-Giemsa staining also indicated that the KLF3 shRNA-transduced primary erythroid cells appeared smaller with a more condensed nucleus than control cells (Fig. 7G). To further verify the negative regulation of KLF3 on miR-23a cluster expression in primary erythroid cells, we examined the expression of the primary miR-23a cluster when KLF3 was knocked down. As expected, the primary miR-23a cluster was upregulated upon KLF3 knockdown at 3 different time points (Fig. 7H).
Fig 7.
KLF3 negatively regulates the expression of γ-globin and miR-23a cluster in human primary erythroid cells. (A) Western blot analysis of KLF3 in CD34+ HPCs infected with lentivirus expressing KLF3 shRNA or negative control. (B) qPCR analysis of KLF3 mRNA normalized with GAPDH in CD34+ HPCs infected with lentivirus during Epo-induced erythroid differentiation. (C) qPCR analysis of γ-globin expression normalized with GAPDH in CD34+ HPCs shown in panel B. (D) FACS analysis of CD71/CD235a double-positive cells in CD34+ HPCs infected with lentivirus at 11 days after Epo induction. (E and F) Western blot (E) and immunostaining (F) of γ-globin expression in lentivirus-infected human CD34+ HPCs after 8 and 11 days of Epo induction. (G) May-Grünwald-Giemsa staining of the lentivirus-infected human CD34+ HPCs after 8, 11, and 18 days of Epo induction. (H) qPCR analysis of primary miR-23a cluster in lentivirus-infected human CD34+ HPCs after 4, 8, and 11 days of Epo induction. (I) Schematic of the regulation of miR-23a and miR-27a on β-like globin genes through targeting of KLF3 and SP1. Student's t test (two-tailed) was performed to analyze the data from the experiments in triplicate. **, P < 0.01; ***, P < 0.001.
Taken together, these results strongly suggest that miR-23a and miR-27a can improve β-like globin gene expression through targeting of two negative regulators of the globin gene, KLF3 and SP1. Meanwhile, KLF3 binds to the miR-23a cluster promoter and suppresses its expression, forming a positive feedback loop to improve the expression of globin genes and the microRNA cluster, which further accelerates erythroid differentiation (Fig. 7I).
DISCUSSION
The regulation of β-like globin genes has been studied for decades and is an excellent model to study eukaryotic gene regulation. However, miRNA regulation of β-like globin genes is underexplored, in part due to the short 3′ UTRs of globin genes. Very few miRNAs targeting globin have identified, except for miR-96, which was shown to directly suppress the γ-globin gene by binding to its CDS region (19). In addition, a few reports focus on miRNAs indirectly regulating globin genes through targeting of known transcription factors. For example, miR-144 was found to negatively control the embryonic α-globin by targeting klfd, an erythroid-specific Krüppel-like transcription factor (29). miR-15a and miR-16-1 also have been reported to elevate fetal hemoglobin expression in human trisomy 13 via MYB (20). In addition, the kit receptor/miR-221-222 complex may be involved in human hemoglobin switching (30). miR-210 has also been identified to be involved with the increased expression of the γ-globin genes in differentiating erythroid cells (21). In this study, we showed that miR-23a and miR-27a can increase the expression of β-like globin genes in human erythroid cells through targeting KLF3 and SP1. Our study provides new insights into miRNA-mediated globin gene regulation and extended the regulatory network of globin genes. However, miR-23a and miR-27a are not involved in the embryonic-to-fetal globin switching, because they promote the expression of both embryonic and fetal β-like globin genes. We also noticed that miR-23a and miR-27a not only upregulate β-like globin gene expression but also increase the percentage of benzidine-positive cells, indicating their roles in increasing the number of hemoglobin (tetramer)-containing cells. Since miR-23a and miR-27a are demonstrated to promote erythroid differentiation, there should be other mechanisms and targets of miR-23a and miR-27a in the regulation of erythroid differentiation or α-like globin gene expression. Of them, miR-23a was recently reported to promote erythroid differentiation through targeting SHIP2 by our laboratory (31). Although the miR-23a∼27a∼24-2 cluster was known to be involved in cell cycle regulation, proliferation, differentiation, and cardiac hypertrophy and showed deregulated expression in many diseases (27), to our knowledge, this is the first time the effects of miR-23a and miR-27a on the regulation of globin gene expression have been examined.
Globin gene expression is regulated by the LCR at two levels: the formation of an active chromatin structure and the activation of transcription. KLF3 and SP1 were previously reported to be potential globin gene suppressors through the recruitment of corepressors to block the formation of an active structure and result in the repression of globin gene transcription. KLF3 is already known to function as a strong transcriptional repressor (25). In vitro assays showed that KLF3 could bind to the β-globin locus control region (16). Reporter assays in COS-7 cells also suggested that KLF3 was able to repress the activity of the ε- and γ-globin gene promoters (17). A recent in vivo study found that KLF3 was critical during the later stages of erythroid maturation, and that Klf3−/− erythrocytes contained less mean cell hemoglobin (32). Notably, these results should be separately appreciated from the previously identified negative role for KLF3 on β-globin gene expression. We hypothesized that the difference was due to the distinct systems that were used in the studies. The study from the Crossley laboratory (32) focused on erythroid maturation but not on globin gene expression. They found that KLF3−/− erythrocytes did not cause any erythropoietic disruption until cells transitioned from CD71hi TER119hi to CD71med TER119hi in mice. The erythroid maturation mainly represents the biological process at the later stage of erythroid differentiation. However, our study on human erythropoiesis did not distinguish CD71lo CD235ahi cells from total CD71/CD235a double-positive cells, because our aim was to investigate the role of KLF3 on globin gene expression in all differentiated erythroid cells. Moreover, the effect of KLF3 knockout in the study of Crossley et al. was wholly due to the nonerythroid-specific knockout in mice. In addition, because of the species difference between humans and mice, it is possible that KLF3 exhibits a distinct expression pattern and plays different roles in human and mouse erythropoiesis. In our study, we found that KLF3 repressed human β-like globin gene expression during erythroid differentiation of hemin-induced K562 cells and Epo-induced CD34+ HPCs. We also showed the binding of KLF3 on HS2 and HS3 of the LCR in vivo. During erythroid differentiation, the decrease in the levels of KLF3 originating from the miR-23a upregulation prevented the recruitment of the CtBP corepressor and blocked KLF3 association with chromatin-modifying enzymes (25), which are required to further activate the expression of β-like globin genes.
There have been some reports elaborating the roles of SP1 in erythropoiesis. The ubiquitously expressed SP1 can bind to the consensus sequences of the GC and GT boxes (26). In A181γ cells, SP1 is found to be an inhibitor of β-like globin gene transcription through binding to the GC and GT boxes located throughout the β-locus (15). Moreover, silencing SP1 in K562 cells also resulted in the upregulation of γ-globin expression (14). Consistent with these reports, we found that SP1 levels were decreased in hemin-induced K562 cells and Epo-induced CD34+ HPCs during erythroid differentiation, and that SP1 negatively regulated ε- and γ-globin genes through binding to the GC and GT boxes in the β-locus. When SP1 was at high levels, the GC and GT boxes in the β-locus were occupied by SP1, which recruits HDAC1 to deacetylate histones and maintains the chromatin structure in a closed status (15). In our study, the downregulation of SP1 resulting from the increase in miR-27a levels led to the displacement of SP1 from the motifs. This displacement might allow other factors to recruit PCAF to acetylate histones and activate β-like globin gene transcription. The phosphorylation of SP1, which can influence its transcriptional activity and stability in regulating globin gene expression, was not addressed here and will be explored further. In conclusion, our study confirmed the negative regulation of KLF3 and SP1 on β-like globin gene expression in erythroid cells. Moreover, KLF3 and SP1 binding sites in the β-globin locus do not overlap. Thus, cointroduction of two miRNAs would additively or even synergistically increase β-like goblin gene expression, which warrants further validation.
In addition, we found that KLF3 negatively regulated not only the expression of globin genes but also that of the miR-23a cluster, forming a positive feedback loop, which might be a ubiquitous phenomenon in miRNA-mediated gene regulation. For example, in human cancers, miR-152 downregulates DNMT1 expression by targeting the 3′ UTR of its transcript. Meanwhile, depletion of DNMT1 leads to an increase in miR-152 expression via the reversion of promoter hypermethylation (33). Biological networks of similar architecture can also be found in the asymmetric differentiation of the left-right neurons in Caenorhabditis elegans (34), in granulocytic differentiation in humans (35), and in cellular transformation (36). Likewise, many negative feedback loops also occur in miRNA-mediated gene regulation to maintain a balanced state and to fine-tune gene expression, such as in the elaborate regulation of α-globin by miR-144 and KLFD (29); in the negative feedback loop between miR-200 and Sox2/E2F3 in neural progenitor cells (37); and in the case of miR-92b, which regulates drosophila muscle development (38).
In our study, miR-23a and miR-27a repress the translation of two transcription factors, KLF3 and SP1. At the initiation of erythroid differentiation, miR-23a and miR-27a are upregulated, which is partly due to the release of KLF3 from their promoter. Upregulation of miR-23a and miR-27a leads to the translational repression of KLF3 and SP1. Furthermore, the suppression of KLF3 and SP1 on β-like globin genes and the miR-23a cluster is alleviated, and the elevated expression of β-like globins and miR-23a cluster members then further accelerate erythroid differentiation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Bin Zhu from Peking Union Hospital for his assistance with the umbilical cord blood preparation.
This work was supported by grants from the National Natural Science Foundation of China (31040021 to J.Y.; 31201103 to Y.-N.M.; 31200977 to F.W.), the National Key Basic Research Program of China (2011CBA01100 to J.Y.), the IBMS, CAMS (2009RC03 to J.Y.; 2010PYB06 to J.Y.), the Beijing Municipal Science & Technology Commission (2010B071 to J.Y.), and the National Basic Research Program of China (2010CB530406 to J.-W.Z.)
Y.M., B.W., F.J., and D.W. performed the experimental design and analysis. H.L., Y.Y., H.D., and B.G. performed the luciferase reporter assays and the Western blot analysis. F.W., Y.Z., H.Y., and Z.Z. were responsible for the animal experiments. L.D. was responsible for the bioinformatics analysis. J.Y. and Y.M. designed the study and wrote the paper.
We declare no competing financial interests.
Footnotes
Published ahead of print 5 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00623-13.
REFERENCES
- 1.Crossley M, Orkin SH. 1993. Regulation of the β-globin locus. Curr. Opin. Genet. Dev. 3:232–237 [DOI] [PubMed] [Google Scholar]
- 2.Miller IJ, Bieker JJ. 1993. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Krüppel family of nuclear proteins. Mol. Cell. Biol. 13:2776–2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nuez B, Michalovich D, Bygrave A, Ploemacher R, Grosveld F. 1995. Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature 375:316–318 [DOI] [PubMed] [Google Scholar]
- 4.Asano H, Li XS, Stamatoyannopoulos G. 1999. FKLF, a novel Krüppel-like factor that activates human embryonic and fetal β-like globin genes. Mol. Cell. Biol. 19:3571–3579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asano H, Li XS, Stamatoyannopoulos G. 2000. FKLF-2: a novel Krüppellike transcriptional factor that activates globin and other erythroid lineage genes. Blood 95:3578–3584 [PubMed] [Google Scholar]
- 6.Sankaran VG, Menne TF, Xu J, Akie TE, Lettre G, Van Handel B, Mikkola HK, Hirschhorn JN, Cantor AB, Orkin SH. 2008. Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science 322:1839–1842 [DOI] [PubMed] [Google Scholar]
- 7.Xu J, Sankaran VG, Ni M, Menne TF, Puram RV, Kim W, Orkin SH. 2010. Transcriptional silencing of γ-globin by BCL11A involves long-range interactions and cooperation with SOX6. Genes Dev. 24:783–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ney PA, Sorrentino BP, McDonagh KT, Nienhuis AW. 1990. Tandem AP-1-binding sites within the human β-globin dominant control region function as an inducible enhancer in erythroid cells. Genes Dev. 4:993–1006 [DOI] [PubMed] [Google Scholar]
- 9.Forsberg EC, Downs KM, Bresnick EH. 2000. Direct interaction of NF-E2 with hypersensitive site 2 of the β-globin locus control region in living cells. Blood 96:334–339 [PubMed] [Google Scholar]
- 10.Jane SM, Cunningham JM. 1996. Molecular mechanisms of hemoglobin switching. Int. J. Biochem. Cell Biol. 28:1197–1209 [DOI] [PubMed] [Google Scholar]
- 11.Tsang AP, Visvader JE, Turner CA, Fujiwara Y, Yu C, Weiss MJ, Crossley M, Orkin SH. 1997. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell 90:109–119 [DOI] [PubMed] [Google Scholar]
- 12.Yi Z, Cohen-Barak O, Hagiwara N, Kingsley PD, Fuchs DA, Erickson DT, Epner EM, Palis J, Brilliant MH. 2006. Sox6 directly silences ε-globin expression in definitive erythropoiesis. PLoS Genet. 2:e14. 10.1371/journal.pgen.0020014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Filipe A, Li Q, Deveaux S, Godin I, Romeo PH, Stamatoyannopoulos G, Mignotte V. 1999. Regulation of embryonic/fetal globin genes by nuclear hormone receptors: a novel perspective on hemoglobin switching. EMBO J. 18:687–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu JH, Navas P, Cao H, Stamatoyannopoulos G, Song CZ. 2007. Systematic RNAi studies on the role of Sp/KLF factors in globin gene expression and erythroid differentiation. J. Mol. Biol. 366:1064–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng D, Kan YW. 2005. The binding of the ubiquitous transcription factor SP1 at the locus control region represses the expression of beta-like globin genes. Proc. Natl. Acad. Sci. U. S. A. 102:9896–9900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crossley M, Whitelaw E, Perkins A, Williams G, Fujiwara Y, Orkin SH. 1996. Isolation and characterization of the cDNA encoding BKLF/TEF-2, a major CACCC-box-binding protein in erythroid cells and selected other cells. Mol. Cell. Biol. 16:1695–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma XY, Wang MJ, Qu XH, Xing GC, Zhu YP, He FC. 2003. Transcriptional regulation of gamma- and epsilon-globin genes by basic Krüppel-like factor. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao 35:271–276 [PubMed] [Google Scholar]
- 18.Tanabe O, McPhee D, Kobayashi S, Shen Y, Brandt W, Jiang X, Campbell AD, Chen YT, Chang C, Yamamoto M, Tanimoto K, Engel JD. 2007. Embryonic and fetal β-globin gene repression by the orphan nuclear receptors, TR2 and TR4. EMBO J. 26:2295–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Azzouzi I, Moest H, Winkler J, Fauchère JC, Gerber AP, Wollscheid B, Stoffel M, Schmugge M, Speer O. 2011. MicroRNA-96 directly inhibits γ-globin expression in human erythropoiesis. PLoS One 6:e22838. 10.1371/journal.pone.0022838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sankaran VG, Menne TF, Šćepanović D, Vergilio JA, Ji P, Kim J, Thiru P, Orkin SH, Lander ES, Lodish HF. 2011. MicroRNA-15a and -16-1 act via MYB to elevate fetal hemoglobin expression in human trisomy 13. Proc. Natl. Acad. Sci. U. S. A. 108:1519–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bianchi N, Zuccato C, Lampronti I, Borgatti M, Gambari R. 2009. Expression of miR-210 during erythroid differentiation and induction of gamma-globin gene expression. BMB Rep. 42:493–499 [DOI] [PubMed] [Google Scholar]
- 22.Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297 [DOI] [PubMed] [Google Scholar]
- 23.Borel C, Deutsch S, Letourneau A, Migliavacca E, Montgomery SB, Dimas AS, Vejnar CE, Attar H, Gagnebin M, Gehrig C, Falconnet E, Dupré Y, Dermitzakis ET, Antonarakis SE. 2011. Identification of cis- and trans-regulatory variation modulating microRNA expression levels in human fibroblasts. Genome Res. 21:68–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macfarlane LA, Murphy PR. 2010. MicroRNA: biogenesis, function and role in cancer. Curr. Genomics 11:537–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turner J, Crossley M. 1999. Basic Krüppel-like factor functions within a network of interacting haematopoietic transcription factors. Int. J. Biochem. Cell Biol. 31:1169–1174 [DOI] [PubMed] [Google Scholar]
- 26.Bouwman P, Philipsen S. 2002. Regulation of the activity of SP1-related transcription factors. Mol. Cell. Endocrinol. 195:27–38 [DOI] [PubMed] [Google Scholar]
- 27.Chhabra R, Dubey R, Saini N. 2010. Cooperative and individualistic functions of the microRNAs in the miR-23a∼27a∼24-2 cluster and its implication in human diseases. Mol. Cancer 9:232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jia Y, Ryan DG, Getsios S, Oliveira-Fernandes M, Fatima A, Lavker RM. 2008. MicroRNA-184 antagonizes microRNA-205 to maintain SHIP2 levels in epithelia. Proc. Natl. Acad. Sci. U. S. A. 105:19299–19304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fu YF, Du TT, Dong M, Zhu KY, Jing CB, Zhang Y, Wang L, Fan HB, Chen Y, Jin Y, Yue GP, Chen SJ, Chen Z, Huang QH, Jing Q, Deng M, Liu TX. 2009. Mir-144 selectively regulates embryonic α-hemoglobin synthesis during primitive erythropoiesis. Blood 113:1340–1349 [DOI] [PubMed] [Google Scholar]
- 30.Gabbianelli M, Testa U, Morsilli O, Pelosi E, Saulle E, Petrucci E, Castelli G, Giovinazzi S, Mariani G. 2010. Mechanism of human Hb switching: a possible role of the kit receptor/miR 221-222 complex. Haematologica 95:1253–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu Y, Wang D, Wang F, Li T, Dong L, Liu H, Ma Y, Jiang F, Yin H, Yan W, Luo M, Tang Z, Zhang G, Wang Q, Zhang J, Zhou J, Yu J. 2013. A comprehensive analysis of GATA-1-regulated miRNAs reveals miR-23a to be a positive modulator of erythropoiesis. Nucleic Acids Res. 41:4129–4143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Funnell AP, Norton LJ, Mak KS, Burdach J, Artuz CM, Twine NA, Wilkins MR, Power CA, Hung TT, Perdomo J, Koh P, Bell-Anderson KS, Orkin SH, Fraser ST, Perkins AC, Pearson RC, Crossley M. 2012. The CACCC-binding protein KLF3/BKLF represses a subset of KLF1/EKLF target genes and is required for proper erythroid maturation in vivo. Mol. Cell. Biol. 32:3281–3292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ji W, Yang L, Yuan J, Yang L, Zhang M, Qi D, Duan X, Xuan A, Zhang W, Lu J, Zhuang Z, Zeng G. 2013. MicroRNA-152 targets DNA methyltransferase 1 in NiS-transformed cells via a feedback mechanism. Carcinogenesis 34:446–453 [DOI] [PubMed] [Google Scholar]
- 34.Johnston RJ, Jr, Chang S, Etchberger JF, Ortiz CO, Hobert O. 2005. MicroRNAs acting in a double-negative feedback loop to control a neuronal cell fate decision. Proc. Natl. Acad. Sci. U. S. A. 102:12449–12454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fazi F, Rosa A, Fatica A, Gelmetti V, De Marchis ML, Nervi C, Bozzoni I. 2005. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell 123:819–831 [DOI] [PubMed] [Google Scholar]
- 36.Iliopoulos D, Hirsch HA, Struhl K. 2009. An epigenetic switch involving NF-kappaB, Lin28, Let-7 microRNA, and IL6 links inflammation to cell transformation. Cell 139:693–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peng C, Li N, Ng YK, Zhang J, Meier F, Theis FJ, Merkenschlager M, Chen W, Wurst W, Prakash N. 2012. A unilateral negative feedback loop between miR-200 microRNAs and Sox2/E2F3 controls neural progenitor cell-cycle exit and differentiation. J. Neurosci. 32:13292–13308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Z, Liang S, Zhao Y, Han Z. 2012. miR-92b regulates Mef2 levels through a negative-feedback circuit during Drosophila muscle development. Development 139:3543–3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.