Abstract
Recent experimental and computational advances in the protein folding arena have shown that the readout of the one-dimensional sequence information into three-dimensional structure begins within the first few microseconds of folding. The initiation of refolding reactions has been achieved by several means, including temperature jumps, flash photolysis, pressure jumps and rapid mixing methods. One of the most commonly used means of initiating refolding of chemically-denatured proteins is by turbulent flow mixing with refolding dilution buffer, where greater than 99% mixing efficiency has been achieved within 10’s of microseconds. Successful interfacing of turbulent flow mixers with complementary detection methods, including time-resolved Fluorescence Spectroscopy (trFL), Förster Resonance Energy Transfer (FRET), Circular Dichroism (CD), Small-Angle X-ray Scattering (SAXS), Hydrogen Exchange (HX) followed by Mass Spectrometry (MS) and Nuclear Magnetic Resonance Spectroscopy (NMR), Infrared Spectroscopy (IR), and Fourier Transform IR Spectroscopy (FTIR), has made this technique very attractive for monitoring various aspects of structure formation during folding. Although continuous-flow (CF) mixing devices interfaced with trFL detection have a dead time of only 30 µs, burst-phases have been detected in this time scale during folding of peptides and of large proteins (e.g., CheY and TIM barrels). Furthermore, a major limitation of CF mixing technique has been the requirement of large quantities of sample. In this brief communication, we will discuss the recent flurry of activity in micromachining and microfluidics, guided by computational simulations, that are likely to lead to dramatic improvements in time resolution and sample consumption for CF mixers over the next few years.
Introduction
Experiments and computer simulations show that many proteins begin folding from approximately statistical random coils to their native functional conformations in the nano- to microsecond time range. Nascent helical segments form and melt in 100’s of nanoseconds, β-hairpins and small domains do so in a few microseconds and large-scale hydrophobic collapse occurs in less than 100 µs. Of particular note are 1) chain contraction and hydrophobic collapse lead to a higher internal friction and lower intrachain diffusion constants and slow the exploration of configuration space;1 2) hydrogen bond formation and secondary structure formation occur in the nano- to microsecond timescales (coil-to-helix transitions and β-turn formation) and may compete with hydrophobic collapse to reduce the configuration space to be explored;2 3) pre-existing structure in the unfolded ensemble may initiate hydrophobic collapse and also limit the configurational search problem.
With all-atom, explicit-solvent simulations now predicting the folding trajectories of polypeptide chains of up to 100 amino acids into the micro-to-millisecond time range,3,4 it is critical to develop experimental methods that allow access to the formation of various aspects of structure in the same time range. Such techniques would allow the validation of the simulations and, especially, the refinement of algorithms for sorting the plethora of conformations into sub-ensembles that elucidate the interplay between the fundamental forces that guide the early and often determinative formation of transition state ensembles and partially-folded states.
Rapid triggering of refolding has been achieved by a variety of methods, including temperature jumps,5–9 pressure jumps,10–12 flash photolysis,13 electron transfer,14 mechanical force,15 passive diffusion out of denaturant into refolding buffer16,17 and turbulent mixing of chemically-denatured protein with refolding buffer.18–20 Although temperature jump and flash photolysis can achieve dead times on the nanosecond time scale,8,13 they have had limited application due to the requirement for cold denaturation near 0° C in the case of T-jumps and the paucity of photolyzable triggers for flash photolysis. Mechanical force measurements are limited by the response time of the piezoelectric crystal and pressure jumps by the mechanical response time, both ~50 µs. With micro-machined mixers, both laminar flow with hydrodynamic focusing and turbulent flow mixing are able to initiate folding in microseconds. Hydrodynamic focusing has achieved shorter mixing times (< 10 µs with 90% mixing efficiency)17,21 and requires significantly less sample than turbulent mixing methods (femtomoles22 vs. micromoles,18 respectively). However, the small profile of the sample illuminated (0.1 to 1 micron wide by a few microns deep) necessitates a highly focused optical beam for discrete measurements and a very high concentration of sample (~100–500 µM17 vs. 3–10 µM for turbulent flow23–25) to obtain good signal to noise ratios. CF turbulent mixers have achieved greater than 99% mixing efficiency in as little as 30 µs and can be constructed with much larger observation paths (typically 50–100 µ wide and 100–400 µ deep).20 The latter characteristic has enabled CF mixers to be interfaced with a variety of spectroscopic techniques and thus have been the most widely applied in the study of ultrafast folding reactions.
Recent advances in turbulent flow mixer technology using state of the art materials, precision laser machining and numerical computational methods, combined with more sensitive detection techniques, have set the stage for the next generation of microfluidic mixers. We can expect to achieve an order of magnitude decrease in dead time, use an order of magnitude less sample and obtain an order of magnitude higher signal to noise ratio (Figure 1). The present review focuses on turbulent flow mixers interfaced with the spectroscopic techniques widely used in the study of protein folding. We provide a brief background of the origins of this technique, what we have learned and where the field is headed. We conclude with the potential use of these mixers in answering some of the fundamental questions in the field of protein folding.
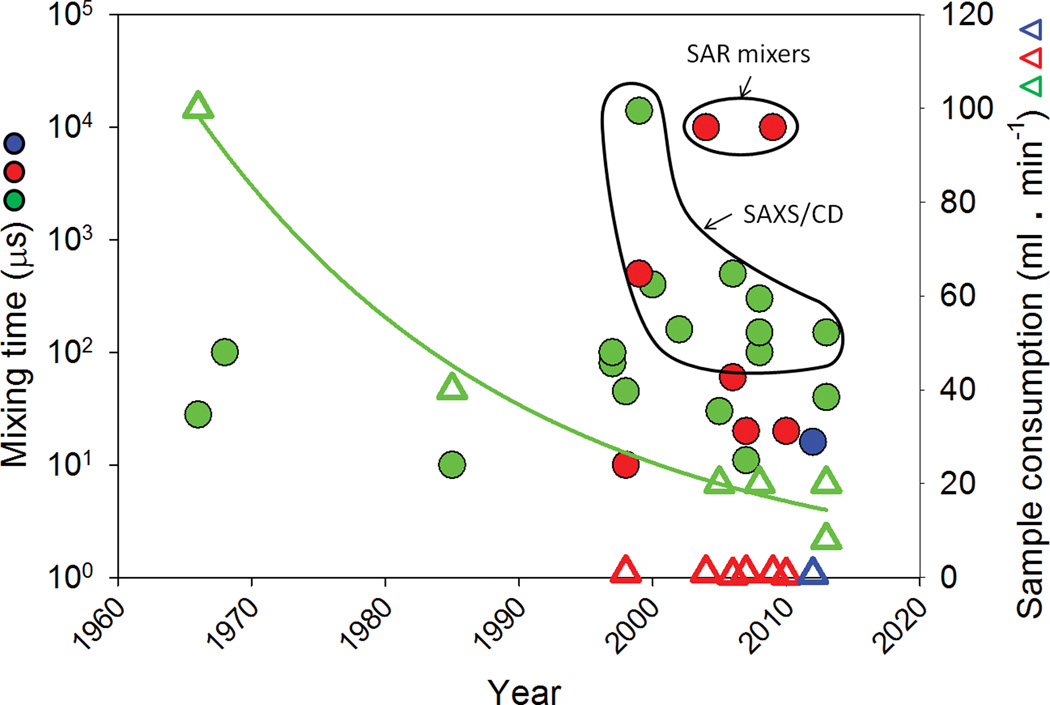
Figure 1.
Time line of mixer development – Reported mixing times of turbulent mixers (green filled circles), laminar mixers (red filled circles) and chaotic mixers (blue filled circles) are plotted as a function of the publication year. All three mixing techniques can access a mixing time of few tens of microseconds. The sample consumption rate of these mixers is also reported (open triangles) with the same color scheme. The exponential decay in the sample consumption of turbulent mixers is represented by the green line (fit of the reported flow rates). While sample consumption of laminar and chaotic mixers is an order of magnitude smaller than their modern turbulent counterparts, the sample concentrations required is correspondingly an order of magnitude higher in the laminar mixers. In the case of SAXS and CD there has been a significant improvement in the interfacing technology and a concomitant decrease in the reported mixing times. Improvements over the last 50 years have primarily been associated with a reduction in sample consumption more than a reduction in mixing times.
Historical background
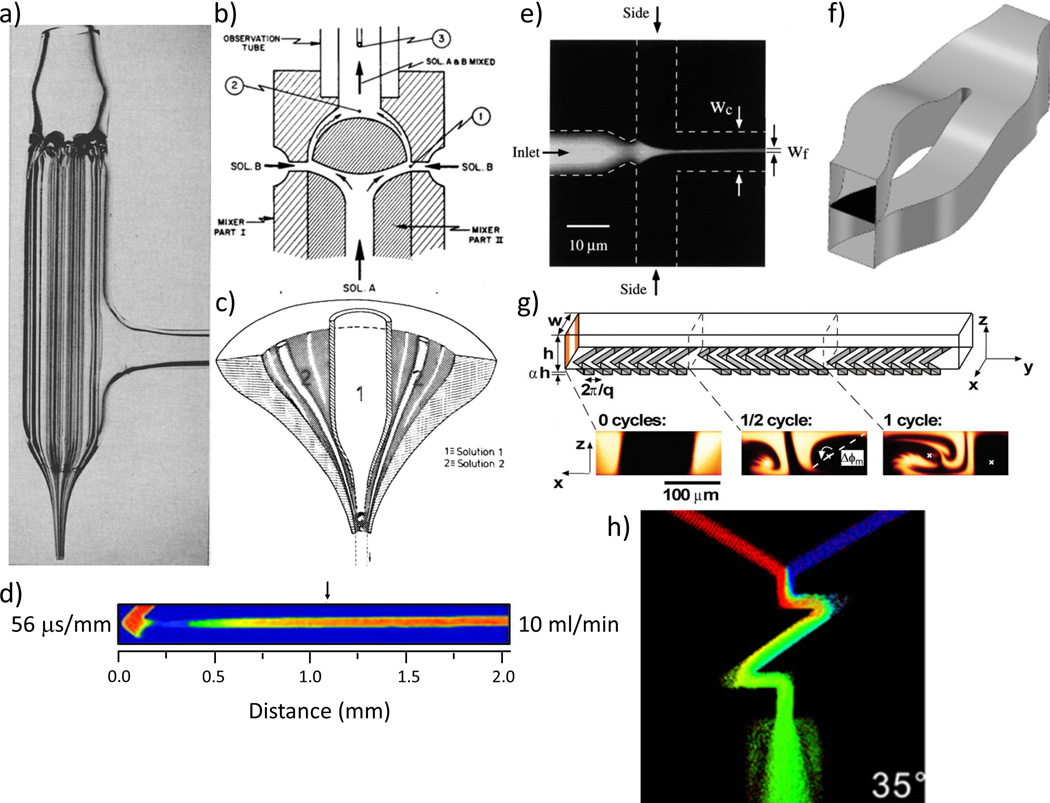
The first attempts to study sub-millisecond chemical reactions relied on the turbulent mixing of fluids that were brought into close contact with each other as thin (< 10 µm) alternating sheets,26 or as a paraxial array of alternating jets of reactants (Figure 2a).27 The emerging jet in the latter method could then be visualized by any spectroscopic technique. While these were hugely successful in achieving the goal of mixing in a few microseconds (28 µs), they required flow rates nearing 100 mL•min−1 and extremely careful manual crafting with irregular and unpredictable results. In order to overcome the problems of variability and instability of jet mixers, Berger et al.28 used the turbulence caused by obstructions placed in the flow path of mixing liquids (Figure 2b). They constructed mixers out of Kel-F and stainless steel that brought four streams of the two reactants, each of 40 µm diameter, into contact with each other just behind a hemispherical obstruction. The liquid mixture entered into an observation tube after passing around a barrier and the turbulence caused by the wake behind this ball resulted in very efficient mixing with a dead time of ~100 µs.
Figure 2.
Representative mixers. a) Multicapillary mixer,27 b) Berger ball mixer,28 c) Capillary mixer,29 d) T-mixer,20 e) hydrodynamic focusing mixer,21 (Copyright (1998) by The American Physical Society), f), split and recombine mixer,64 (Reproduced from Ref. 64 with permission from The Royal Society of Chemistry), g) herring bone mixer,72 and h) “zig-zag” mixer.77
A common feature of CF mixers is that reaction time is transduced into distance. Typically, the flow velocity and the dimensions of the flow channel or jet are known and the reaction time can be calculated by the distance from the mixing point (typically ranging from hundreds of microns to tens of millimeters). Interfacing to spectroscopic techniques is relatively straightforward although several factors need to be considered. Optical techniques (e.g., fluorescence, absorbance, CD and IR spectroscopy) have been popular owing to their sensitivity and ease of access. These also meet the requirement that the intrinsic timescale of the technique be fast enough (< 1 µs) to provide a near-instantaneous snapshot of the kinetics. The intrinsic timescale for fluorescence is typically several nanoseconds, and smearing (i.e., convolution of the detection timescale with reaction kinetics) is therefore not a concern. Absorbance, CD and x-ray scattering occur on the timescale of electronic motions in molecules and, therefore, are practically instantaneous compared to molecular vibrations and larger scale motions.
Regenfuss29 adapted the capillary mixer of Moskowitz and Bowmann27 by incorporating lessons learned from the ball mixer of Berger et al.28 for studying protein folding. Instead of the paraxial array of capillaries, Regenfuss used one pair of coaxial capillaries pulled to a fine (10–100 µm) tip, and introduced a turbulence generator at the tip of the mixer where the two reactants mix (Figure 2c). This turbulence generator was either a series of glass rods placed between the two capillaries or a small platinum ball held in place with a platinum wire running through the inner capillary. Their design used elements from both mixers and had a mixing time of 10 µs with flow rates similar to the original capillary jet mixers. These capillary mixers and their variations18,19,30 were the workhorses of turbulent mixing methods for studying sub-millisecond protein folding reactions in the 80’s and 90’s and are still proving useful,31 primarily in conjunction with fluorescence measurements. Segel et al.30 were the first to use CF mixers to monitor time-resolved SAXS of refolding proteins. The capillary mixer design was used to achieve a mixing time of 14 ms.
The Takahashi and Rousseau groups32,33 made further advances in the turbulent mixing method with the introduction of micro-machined T-mixers. The reactants in these mixers are brought together at a T-junction in 250 µm wide channels in stainless steel and the change in direction of flow at high speeds produces the turbulence required for rapid mixing. The advent of laser machining provided some additional flexibility in the manufacturing of mixers, permitting essentially arbitrary channel shapes. A simple application increased the degree of change in flow direction by using an arrowhead design in place of the T shaped junction (Figure 2d).20 These mixers are highly adaptable stemming from their stability, ease of assembly and compatibility with different window materials for the top and bottom of the observation channel. These features make them extremely suitable for interfacing with a wide range of spectroscopic techniques, e.g., Fluorescence (Tryptophan Fluorescence, FRET and trFL),19,23,24,34 CD,24,35 SAXS,24,36,37 IR spectroscopy,38,39 resonance Raman,32,40 HX-MS41 and HX-NMR. A detailed review of detection techniques used with turbulent flow mixers is provided by Roder et al.42
Major findings
In extending the observation time window down to the tens of microseconds, CF experiments have revealed several key properties of early protein folding intermediates. The major findings, illustrated with a few representative cases, are discussed below. Although temperature-jump experiments have achieved far superior time resolution8,43–46 there are distinctions between the relaxation approaches and mixing experiments. T-jump experiments, for example, initiate folding from a cold-denatured unfolded state and often monitor relaxation kinetics, where the return to equilibrium from a perturbation is monitored. In contrast, mixing experiments commence from a urea or guanidinium chloride denatured state that is the dominant species in solution (>99%). Folding, therefore, starts from a state with overall dimensions shown to be consistent with a random coil.47 The cold-denatured and pH-denatured states of most proteins are somewhat compact compared to the denaturant induced unfolded state and may contain nonrandom structure that biases the early events in folding.19
Cytochrome c as a test case
The heme-containing 104 amino acid protein, cytochrome c, has been the target of many of the initial applications of CF mixing methods to protein folding. This protein is readily available in the large quantities required by some of the experiments and has an optimal intrinsic Trp-heme FRET pair for probing early folding events. Kinetic steps (45 µs and 650 µs) in the sub-millisecond regime were observed by several groups using fluorescence19,25 and resonance Raman32 and attributed to a barrier-limited collapse.25 Elegant studies of cytochrome c have evolved from interfacing mixers (with various modifications) to SAXS,37 IR absorption,39 CD,35 and trFL.19,48 Cytochrome c is also one of the few proteins to have been studied using multiple techniques with sub-millisecond time resolution: temperature-jump,49 laminar mixing17 and turbulent mixing.19,20,25,32,35,37 The results are consistent with compaction and formation of secondary and tertiary structure occurring in a sub-100 µs kinetic step. However systematic differences in the reported time constants have been noted and consensus on the interpretation of the results has been elusive.50 Whether this originates from heterogeneity in the energy landscape of the protein or from experimental conditions (e.g., initial conditions, final conditions, protein concentration, flow conditions etc.) is not yet clear.
Rapid structure formation and local topology
Perhaps one of the most striking observations from CF studies is the extent and specificity of structure formation within the burst-phase (typically, tens of microseconds). For example, for proteins with more than 100 amino acids, FRET and SAXS studies have in many cases observed a prominent burst-phase.36,37,51 A combined FRET/SAXS study on dihydrofolate reductase showed that the adenosine binding sub-domain is ordered within the 30 µs dead-time of the experiments. Ordering of this sub-domain prior to the discontinuous loop sub-domain is believed to result from the local connectivity of a cluster of branched aliphatic side chains.23 The discontinuous domain, as the name implies, has a higher entropic penalty to overcome to stabilize the clustering of its hydrophobic side chains whose amide hydrogens were protected in H/D pulse labeling experiments with millisecond time resolution.52
Rapid formation of native-like packing in the N-terminal region of a TIM barrel protein was observed using FRET, SAXS and tryptophan rotational correlation time measurements.24 Within 30 µs, dimensions in the N-terminal half and the mobility of a single engineered tryptophan were close to those of the native state. Association with the C-terminal half of the protein and more global compaction were evident on a 100 µs timescale. The local topology of the TIM barrels (i.e., repeating βα-motif) and the predisposition of the sequence to form clusters of hydrophobic side chains are likely to lead to stabilization of this partially folded region. An earlier study also identified the N-terminal region, (βα)4, as the minimal autonomously folding fragment.53
A test of the role of topology and clusters of hydrophobic side chains in modulating the stability and frustration of the upper reaches of the energy landscape was carried out in CF SAXS studies of circular permutants of the βα flavodoxin-fold protein, CheY. By splitting a sequence-local hydrophobic cluster and stabilizing another by fusing the N- and C-termini in a circular permutant construct, the resulting microsecond intermediate showed an increased stability and a greater extent of collapse, as judged by a smaller radius of gyration, than its wild-type counterpart (Nobrega, R.P. et al., unpublished results from C.R. Matthews lab).
Heterogeneity of early events
Another common theme in early folding events has been the structural heterogeneity evident in early intermediates. Millisecond timescale studies of cytochrome c,48 CF based sub-millisecond work on TIM barrels24 and globins54 suggest that a distribution of states possibly with marginal stability may be in dynamic equilibrium. Extended and compact states were observed for cytochrome c and a TIM barrel using distance distributions from FRET. The acquisition of structure for the globins was also observed to occur gradually rather than in a cooperative manner. Additional heterogeneity in unfolded state dynamics has also been suggested from simulations and experiments, although the factors giving rise to observed reconfiguration times (< several µs) remain open questions.55 Non-random structure in the unfolded ensemble has been documented for a number of systems56–58 and the role these sub-states play in modulating the dynamics and energetics of the energy landscape is an area of active interest.
Recent trends and future directions
Laminar mixers
The most widely used laminar mixers in protein folding have employed hydrodynamic focusing (Figure 2e) to achieve mixing times of less than 30 µs.17,59,60 These mixers tend to use much smaller volumes but tend to require much higher concentrations and are challenged by low signal to noise due to small sample profiles. A major disadvantage of most mixers using laminar flow is that the flow is not uniform along the width of the channel. This is especially problematic when the focus of the beam is wide, for example in SAXS measurements.61,62 Other multi-lamination techniques, where the mixing solutions are broken up into many small interdigitating streams thereby increasing the surface area for diffusion have also been developed.63 Another mixing technique that uses flow rates in the laminar regime is the split-and-recombine (SAR) class of mixers (Figure 2f).64,65 The break in flow from repeated changes in direction results in an increased interface surface area in the SAR mixers. Among laminar mixers, hydrodynamic focusing technology is able to achieve moderate mixing efficiency (80% – 90%) in ~10 µs.66
Turbulent mixers
The capillary mixers used in the early days of turbulent mixing were probably the most efficient in achieving the fastest mixing dead times with the highest efficiency. However, the variability in the manufacturing process, the instability of jet streams and the impracticality of interfacing them with detection technologies have led to the use of more reliable, albeit less efficient T-mixers. Taking a cue from the advances in capillary mixer designs, many groups have started to improvise on the T-mixers by introducing various means of generating higher turbulence.67 Clegg and his colleagues used T-mixers that could be rapidly prototyped to generate a fully mixed jet stream.68 While these mixers are not widely used, they were successful in separating the mixing from the detection-interfacing problem. Using photolithography, Majumdar et al.69 created a series of mixers with different geometries and used obstructions in the flow path to increase turbulence by decreasing the cross-sectional area of the mixer. This modification resulted in higher velocity eddies and better mixing. These mixers are able to achieve dead times close to 10 µs at flow speeds nearing 40 ml•min−1. Takahashi and his colleagues were also able to achieve similar mixing times by introducing turbulence in the mixing solutions prior to the mixing region.33 One obvious drawback of turbulent flow mixers is the volume of sample consumed for each experiment. Although, narrowing the mixing channels to decrease the flow rates is an attractive means to reduce sample consumption, the viscous drag imposes a lower limit on the dimensions of the channels70 (typically ~ 50 µm) and results in pressures that exceed the ratings of the materials used.
Chaotic mixers
Chaotic mixers overcome the limitations of laminar flow mixing (e.g., small observation volume),71–74 while maintaining low flow rates (typically less than a couple of ml•min−1) and a close approximation to “plug flow” conditions. Chaos generators have been especially useful in mixers that employ flow rates with low (less than 2000) to transitional Reynolds numbers (up to 4000). The staggered herringbone mixers75 use a series of intersecting grooves along the fluid path as chaos generators to break up the jets of the two solutions and force a change in flow direction that exponentially increases the interaction surface (Figure 2g). Other strategies used to introduce chaos in the flow channel include serpentine channels76–78 that use a series of three-dimensional bends in flow path. A systematic study on the most efficient shapes, and flow velocities for efficient mixing using these “zig-zag” patterns77 (Figure 2h) suggests that mixing efficiencies of 90% and a dead time of 16 µs can be achieved by employing as few as two bends of 145° in the flow path at Re <300 (corresponding to a flow rate of a few µL•s−1). One significant drawback of using chaos generators in the mixing path of fluids is the significant pressure drop leads to high back pressure on the system and potential cavitation if the flow rates are increased to obtain better mixing efficiency. However, these results hold great promise and further developments in microfluidic mixer design will likely employ chaos generators under near turbulent flow regimes to reach > 95% mixing efficiency with dead times of a few microseconds. The order of magnitude lower volume requirement with nearly an order of magnitude decrease in dead-time, compared to traditional turbulent mixers, combined with the high efficiency and a “plug flow” in the flow channel, makes these mixers an ideal candidate for future development.
Computational Fluid Dynamics Simulations (CFD)
One of the challenges in designing more efficient mixers is the availability of predictive tools for validating putative design parameters such as flow regimes (e.g. turbulent, laminar or chaotic), pressure drops, mixing efficiencies at different flow rates and viscosities of the mixing solutions. Numerical simulations of the incompressible Navier-Stokes equations coupled with a transport equation for the protein concentration can be a useful tool for both the mixer design and post-experimental analysis. The two primary possibilities for the computational analysis of turbulent or chaotic mixing are: computation of the time averaged velocity field and protein concentration using Reynolds Averaged Navier-Stokes (RANS) turbulence models, or direct numerical simulation (DNS) of the fully unsteady and three-dimensional flow field and concentration field using the governing Navier-Stokes equations and no model. The Reynolds numbers are low enough for these applications that the classic third simulation alternative of large eddy simulation (LES) provides little value over DNS.
RANS calculations for these sorts of geometries take on the order of 24 hours on a high end personal computer using either commercial or open source CFD software. These simulations invoke a model about how the turbulence influences the flow and are therefore only as accurate as those model assumptions. Numerous RANS models exist in the literature and there is little consensus on which model is best.79 In contrast, DNS simulation solves the actual equations for the fluid flow, but it is considerably more expensive. A typical simulation for the mixing section of a device would require weeks on a large supercomputer. In addition, with increasing mixing (or higher Reynolds numbers), the DNS becomes increasingly more expensive to perform correctly.80 Early DNS focused on simple channel flow.81,82 More recently DNS simulations with more complex geometries and boundary conditions83 have been performed.
Microdroplet mixing
An alternative approach to rapid mixing that is very efficient in sample consumption relies on the use of microdroplets. Graceffa et al. used ink jet technology to generate two streams of microdroplets to initiate the folding reaction of cytochrome c. Microdroplets with an 80 µm diameter achieved a mixing time of ~10 ms.84 The droplets were coupled with stroboscopic detection using SAXS to record the scattering profile. Although the time resolution of the experiment and signal-to-noise are less than that achieved with more conventional mixers, the proof of principle shows great promise for future developments. An advantage of the approach, in addition to the very small sample demands, is that no optical window is necessary. Methods to generate smaller droplets using nanospray85 may provide a route to reducing the volume and mixing times by a significant margin.
Materials
One of the challenges of microsecond mixing is the design and manufacturing of the mixers. Computational approaches are sufficiently advanced to be able to simulate various mixing geometries and greatly speed the design of new mixers. The physical manufacturing process has also seen numerous advances over the past several years. One of the technologies that has greatly facilitated construction of an all quartz mixer suitable for use at relatively high pressures is the femtosecond painting method developed by Bado and colleagues.86 A femtosecond laser is used to paint an arbitrary 3-dimensional pattern in quartz via non-linear multiphoton interaction of the laser pulse with the quartz. Because the process is multiphoton, the painting is confined to an ellipsoidal volume of approximately a micron in each dimension. The quartz is etched with hydrofluoric acid after painting with the laser pulse. An advantage is of this approach is that high aspect ratios can be achieved (e.g., a deep but narrow channel) and the mixer can be a one-piece design. Construction of the channel from quartz is also advantageous because it is inert, has excellent optical properties over a wide spectral range and can also be passivated with various functional moieties (e.g., PEG-silane).87 The commercialization of this technology makes it accessible for academic laboratories that are not specialists in microfabrication. Materials that are compatible with SAXS are also being explored for construction of microfluidic devices. Dootz et al.88 have successfully manufactured a chaotic mixer out of PDMS with 53 µm thick Kapton windows. The ease of manufacture of these mixers and the adaptability of the technology to different mixer geometries makes it a promising area for innovation in the near future.
Advances in detection and interfacing
Increasing the level of detail of structural information remains a goal of CF experiments. To this end, several groups have optimized and developed tools to provide more quantitative and complementary structural probes of microsecond folding reactions.6,10,13,17–20,35,37,44–46 SAXS is attractive in this regard because of the possibility of transforming scattering profiles to three-dimensional low-resolution structural models. Signal-to-noise ratios for achieving this have so far been beyond the reach of SAXS kinetic studies. However, efforts are underway to optimize the duty cycle, sample efficiency, beam focusing and cameras to enable structural models to be obtained from scattering spectra of transient species.89 The low-resolution global picture from SAXS is complemented by studies aimed towards an amino acid level resolution of structure formation in the sub-millisecond timescale. This can be achieved with H/D pulse labeling of solvent accessible amide protons followed by mass spectrometry or NMR analysis. Mayo and colleagues introduced a multiple-mix microchannel quench flow system capable of H/D pulse labeling and quenching with a 110 µs time resolution.41 Another technique that shows great promise takes advantage of bottom-up mass spectrometry coupled with hydroxyl radical footprinting.90–92 Lapidus and colleagues have successfully interfaced this capability with a laminar-flow mixer to obtain oxidative labeling of surface exposed residues during the folding of hen egg lysozyme.93 Although the extent of labeling was not extensive, the approach provides a mechanism for probing the involvement of side chains in partially formed structures early in folding. An advantage of oxidative labeling is that, because labeling is via a covalent bond, back exchange is not a complicating factor. Therefore, structural insights can be obtained on partially folded states with marginal stability, which are likely to be present early in folding. These techniques are also likely to find applications in many enzymatic reactions to identify amino acids involved in structural rearrangements, protein-protein association, protein-RNA/DNA binding and ligand binding. Low sample consumption of laminar mixing and mass spectrometry are likely to facilitate adoption of this approach.
Conclusion
It has become abundantly clear over the past two decades that there is a significant formation of structure in most proteins within microseconds after the initiation of folding. These structural changes range from a localized chain contraction to significant hydrophobic collapse and, in some cases, correspond to direct access to specific structured intermediates or to the native state. Advances in mixer design over the past decade have resulted in significant improvement in mixing dead times and sample consumption in CF detection of protein folding events by a variety of spectroscopic methods. The field is now poised to take advantage of modern computational capabilities, robust materials and cutting-edge manufacturing techniques to develop mixers that can be interfaced to an expanding repertoire of spectroscopic methods. The insights into earliest events in folding will enable tests of various hypotheses about the driving forces and validate the results of computer simulations of protein folding reactions. Certainly, a wider range of proteins will need to be studied by a variety of spectroscopic probes in order to develop a clearer understanding of the conformational dynamics of proteins during folding in the sub-millisecond time scale. The synergy between experiments and computations will undoubtedly increase our understanding of one of the most elusive problems in biology. Ultrafast mixing technology has a very important role to play in the future of protein science.
Acknowledgements
We thank Jill A. Zitzewitz for helpful discussions. This work was supported by grants GM23303 (NIH), MCB1121942 (NSF), and in part by the National Science Foundation CBET division, grant 1032364 and by grants from the National Center for Research Resources (2P41RR008630-17) and the National Institute of General Medical Sciences (9 P41 GM103622-17) from the National Institutes of Health. Use of the Advanced Photon Source, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the U.S. DOE under Contract No. DE-AC02-06CH11357.
References
- 1.Soranno A, Buchli B, Nettels D, Cheng RR, Muller-Spath S, Pfeil SH, Hoffmann A, Lipman EA, Makarov DE, Schuler B. Proc Natl Acad Sci U S A. 2012;109:17800–17806. doi: 10.1073/pnas.1117368109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moglich A, Joder K, Kiefhaber T. Proc Natl Acad Sci U S A. 2006;103:12394–12399. doi: 10.1073/pnas.0604748103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lindorff-Larsen K, Piana S, Dror RO, Shaw DE. Science. 2011;334:517–520. doi: 10.1126/science.1208351. [DOI] [PubMed] [Google Scholar]
- 4.Lane TJ, Shukla D, Beauchamp KA, Pande VS. Curr Opin Struct Biol. 2013;23:58–65. doi: 10.1016/j.sbi.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen J, Rempel DL, Gau BC, Gross ML. J Am Chem Soc. 2012;134:18724–18731. doi: 10.1021/ja307606f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gai F, Du D, Xu Y. Methods Mol Biol. 2007;350:1–20. doi: 10.1385/1-59745-189-4:1. [DOI] [PubMed] [Google Scholar]
- 7.Jones KC, Peng CS, Tokmakoff A. Proc Natl Acad Sci U S A. 2013;110:2828–2833. doi: 10.1073/pnas.1211968110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thompson PA, Eaton WA, Hofrichter J. Biochemistry. 1997;36:9200–9210. doi: 10.1021/bi9704764. [DOI] [PubMed] [Google Scholar]
- 9.Dyer RB, Brauns EB. Methods Enzymol. 2009;469:353–372. doi: 10.1016/S0076-6879(09)69017-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prigozhin MB, Liu Y, Wirth AJ, Kapoor S, Winter R, Schulten K, Gruebele M. Proc Natl Acad Sci U S A. 2013;110:8087–8092. doi: 10.1073/pnas.1219163110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woenckhaus J, Kohling R, Thiyagarajan P, Littrell KC, Seifert S, Royer CA, Winter R. Biophys J. 2001;80:1518–1523. doi: 10.1016/S0006-3495(01)76124-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Torrent J, Font J, Herberhold H, Marchal S, Ribo M, Ruan K, Winter R, Vilanova M, Lange R. Biochim Biophys Acta. 2006;1764:489–496. doi: 10.1016/j.bbapap.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 13.Jones CM, Henry ER, Hu Y, Chan CK, Luck SD, Bhuyan A, Roder H, Hofrichter J, Eaton WA. Proc Natl Acad Sci U S A. 1993;90:11860–11864. doi: 10.1073/pnas.90.24.11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pascher T, Chesick JP, Winkler JR, Gray HB. Science. 1996;271:1558–1560. doi: 10.1126/science.271.5255.1558. [DOI] [PubMed] [Google Scholar]
- 15.Cecconi C, Shank EA, Bustamante C, Marqusee S. Science. 2005;309:2057–2060. doi: 10.1126/science.1116702. [DOI] [PubMed] [Google Scholar]
- 16.Pabit SA, Hagen SJ. Biophys J. 2002;83:2872–2878. doi: 10.1016/S0006-3495(02)75296-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lapidus LJ, Yao S, McGarrity KS, Hertzog DE, Tubman E, Bakajin O. Biophys J. 2007;93:218–224. doi: 10.1529/biophysj.106.103077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shastry MC, Luck SD, Roder H. Biophys J. 1998;74:2714–2721. doi: 10.1016/S0006-3495(98)77977-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan CK, Hu Y, Takahashi S, Rousseau DL, Eaton WA, Hofrichter J. Proc Natl Acad Sci U S A. 1997;94:1779–1784. doi: 10.1073/pnas.94.5.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bilsel O, Kayatekin C, Wallace LA, Matthews CR. Review of Scientific Instruments. 2005:76. [Google Scholar]
- 21.Knight JB, Vishwanath A, Brody JP, Austin RH. PHYSICAL REVIEW LETTERS. 1998;80:3863–- 3866. [Google Scholar]
- 22.Hertzog DE, Michalet X, Jager M, Kong X, Santiago JG, Weiss S, Bakajin O. Anal Chem. 2004;76:7169–7178. doi: 10.1021/ac048661s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arai M, Iwakura M, Matthews CR, Bilsel O. J Mol Biol. 2011;410:329–342. doi: 10.1016/j.jmb.2011.04.057. [DOI] [PubMed] [Google Scholar]
- 24.Wu Y, Kondrashkina E, Kayatekin C, Matthews CR, Bilsel O. Proc Natl Acad Sci U S A. 2008;105:13367–13372. doi: 10.1073/pnas.0802788105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shastry MC, Roder H. Nat Struct Biol. 1998;5:385–392. doi: 10.1038/nsb0598-385. [DOI] [PubMed] [Google Scholar]
- 26.Kletenik YB. J Phys Chem USSR. 1963;37:638. [Google Scholar]
- 27.Moskowitz GW, Bowmann RL. Science. 1966;153:428–429. doi: 10.1126/science.153.3734.428. [DOI] [PubMed] [Google Scholar]
- 28.Berger RL, Balko B, Chapman HF. Review of Scientific Instruments. 1968;39:493. doi: 10.1063/1.1683415. [DOI] [PubMed] [Google Scholar]
- 29.Regenfuss P, Clegg RM, Fulwyler MJ, Barrantes FJ, Jovin TM. Review of Scientific Instruments. 1985;56:283. [Google Scholar]
- 30.Segel DJ, Bachmann A, Hofrichter J, Hodgson KO, Doniach S, Kiefhaber T. J Mol Biol. 1999;288:489–499. doi: 10.1006/jmbi.1999.2703. [DOI] [PubMed] [Google Scholar]
- 31.Apetri AC, Maki K, Roder H, Surewicz WK. J Am Chem Soc. 2006;128:11673–11678. doi: 10.1021/ja063880b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahashi S, Yeh SR, Das TK, Chan CK, Gottfried DS, Rousseau DL. Nat Struct Biol. 1997;4:44–50. doi: 10.1038/nsb0197-44. [DOI] [PubMed] [Google Scholar]
- 33.Matsumoto S, Yane A, Nakashima S, Hashida M, Fujita M, Goto Y, Takahashi S. J Am Chem Soc. 2007;129:3840–3841. doi: 10.1021/ja0660958. [DOI] [PubMed] [Google Scholar]
- 34.Xu M, Beresneva O, Rosario R, Roder H. J Phys Chem B. 2012;116:7014–7025. doi: 10.1021/jp3012365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akiyama S, Takahashi S, Ishimori K, Morishima I. Nat Struct Biol. 2000;7:514–520. doi: 10.1038/75932. [DOI] [PubMed] [Google Scholar]
- 36.Arai M, Kondrashkina E, Kayatekin C, Matthews CR, Iwakura M, Bilsel O. J Mol Biol. 2007;368:219–229. doi: 10.1016/j.jmb.2007.01.085. [DOI] [PubMed] [Google Scholar]
- 37.Akiyama S, Takahashi S, Kimura T, Ishimori K, Morishima I, Nishikawa Y, Fujisawa T. Proc Natl Acad Sci U S A. 2002;99:1329–1334. doi: 10.1073/pnas.012458999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marinkovic NS, Adzic AR, Sullivan M, Kovacs K, Miller LM, Rousseau DL, Yeh S-R, Chance MR. Review of Scientific Instruments. 2000;71:4057. [Google Scholar]
- 39.Kimura T, Maeda A, Nishiguchi S, Ishimori K, Morishima I, Konno T, Goto Y, Takahashi S. Proc Natl Acad Sci U S A. 2008;105:13391–13396. doi: 10.1073/pnas.0801316105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin Y, Gerfen GJ, Rousseau DL, Yeh SR. Anal Chem. 2003;75:5381–5386. doi: 10.1021/ac0346205. [DOI] [PubMed] [Google Scholar]
- 41.Bökenkamp D, Desai A, Yang X, Tai Y-C, Marzluff EM, Mayo SL. Journal of Analytical Chemistry. 1998;70:232–236. [Google Scholar]
- 42.Roder H, Maki K, Cheng H. Chem Rev. 2006;106:1836–1861. doi: 10.1021/cr040430y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu Y, Alonso DO, Maki K, Huang CY, Lahr SJ, Daggett V, Roder H, DeGrado WF, Gai F. Proc Natl Acad Sci U S A. 2003;100:15486–15491. doi: 10.1073/pnas.2136623100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Phillips CM, Mizutani Y, Hochstrasser RM. Proc Natl Acad Sci U S A. 1995;92:7292–7296. doi: 10.1073/pnas.92.16.7292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gilmanshin R, Williams S, Callender RH, Woodruff WH, Dyer RB. Proc Natl Acad Sci U S A. 1997;94:3709–3713. doi: 10.1073/pnas.94.8.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu F, Nakaema M, Gruebele M. Journal of Chemical Physics. 2009:131. doi: 10.1063/1.3262489. [DOI] [PubMed] [Google Scholar]
- 47.Kohn JE, Millett IS, Jacob J, Zagrovic B, Dillon TM, Cingel N, Dothager RS, Seifert S, Thiyagarajan P, Sosnick TR, Hasan MZ, Pande VS, Ruczinski I, Doniach S, Plaxco KW. Proc Natl Acad Sci U S A. 2004;101:12491–12496. doi: 10.1073/pnas.0403643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kimura T, Lee JC, Gray HB, Winkler JR. Proc Natl Acad Sci U S A. 2007;104:117–122. doi: 10.1073/pnas.0609413103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hagen SJ, Eaton WA. J Mol Biol. 2000;297:781–789. doi: 10.1006/jmbi.2000.3508. [DOI] [PubMed] [Google Scholar]
- 50.Krantz BA, Mayne L, Rumbley J, Englander SW, Sosnick TR. J Mol Biol. 2002;324:359–371. doi: 10.1016/s0022-2836(02)01029-x. [DOI] [PubMed] [Google Scholar]
- 51.Kimura T, Uzawa T, Ishimori K, Morishima I, Takahashi S, Konno T, Akiyama S, Fujisawa T. Proc Natl Acad Sci U S A. 2005;102:2748–2753. doi: 10.1073/pnas.0407982102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jones BE, Matthews CR. Protein Sci. 1995;4:167–177. doi: 10.1002/pro.5560040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zitzewitz JA, Matthews CR. Biochemistry. 1999;38:10205–10214. doi: 10.1021/bi9909041. [DOI] [PubMed] [Google Scholar]
- 54.Zhu L, Kurt N, Choi J, Lapidus LJ, Cavagnero S. J Phys Chem B. 2013 doi: 10.1021/jp400174e. [DOI] [PubMed] [Google Scholar]
- 55.Voelz VA, Singh VR, Wedemeyer WJ, Lapidus LJ, Pande VS. J Am Chem Soc. 2010;132:4702–4709. doi: 10.1021/ja908369h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Klein-Seetharaman J, Oikawa M, Grimshaw SB, Wirmer J, Duchardt E, Ueda T, Imoto T, Smith LJ, Dobson CM, Schwalbe H. Science. 2002;295:1719–1722. doi: 10.1126/science.1067680. [DOI] [PubMed] [Google Scholar]
- 57.Bowler BE. Curr Opin Struct Biol. 2012;22:4–13. doi: 10.1016/j.sbi.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shan B, Eliezer D, Raleigh DP. Biochemistry. 2009;48:4707–4719. doi: 10.1021/bi802299j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gambin Y, Simonnet C, VanDelinder V, Deniz A, Groisman A. Lab on a Chip. 2010;10:598–609. doi: 10.1039/b914174j. [DOI] [PubMed] [Google Scholar]
- 60.Park HY, Qiu X, Rhoades E, Korlach J, Kwok LW, Zipfel WR, Webb WW, Pollack L. Anal Chem. 2006;78:4465–4473. doi: 10.1021/ac060572n. [DOI] [PubMed] [Google Scholar]
- 61.Kathuria SV, Guo L, Graceffa R, Barrea R, Nobrega RP, Matthews CR, Irving TC, Bilsel O. Biopolymers. 2011;95:550–558. doi: 10.1002/bip.21628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pollack L, Tate MW, Darnton NC, Knight JB, Gruner SM, Eaton WA, Austin RH. Proc Natl Acad Sci U S A. 1999;96:10115–10117. doi: 10.1073/pnas.96.18.10115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Drese KS. Chemical Engineering Journal. 2004;101:403–407. [Google Scholar]
- 64.Schonfeld F, Hessel V, Hofmann C. Lab on a Chip. 2004;4:65–69. doi: 10.1039/b310802c. [DOI] [PubMed] [Google Scholar]
- 65.Wang CT, Hu YC, Hu TY. Sensors (Basel) 2009;9:5379–5389. doi: 10.3390/s90705379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mansur EA, Ye M, Wang Y, Dai Y. Chinese Journal of Chemical Engineering. 2008;16:503–516. [Google Scholar]
- 67.Gobby D, Angeli P, Gavriilidis A. Journal of Micromechanics and Microengineering. 2001;11:126–132. [Google Scholar]
- 68.Redford GI, Majumdar ZK, Sutin JD, Clegg RM. J Chem Phys. 2005;123:224504. doi: 10.1063/1.2132283. [DOI] [PubMed] [Google Scholar]
- 69.Majumdar ZK, Sutin JDB, Clegg RM. Review of Scientific Instruments. 2005:76. [Google Scholar]
- 70.Brody JP, Yager P, Goldstein RE, Austin RH. Biophys J. 1996;71:3430–3441. doi: 10.1016/S0006-3495(96)79538-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stroock AD, Dertinger SK, Whitesides GM, Ajdari A. Anal Chem. 2002;74:5306–5312. doi: 10.1021/ac0257389. [DOI] [PubMed] [Google Scholar]
- 72.Stroock AD, Dertinger SKW, Ajdari A, Mezic I, Stone HA, Whitesides GM. Science. 2002;295:647–651. doi: 10.1126/science.1066238. [DOI] [PubMed] [Google Scholar]
- 73.Niu XZ, Lee YK. Journal of Micromechanics and Microengineering. 2003;13:454–462. [Google Scholar]
- 74.Bringer MR, Gerdts CJ, Song H, Tice JD, Ismagilov RF. Philosophical Transactions of the Royal Society of London Series a-Mathematical Physical and Engineering Sciences. 2004;362:1087–1104. doi: 10.1098/rsta.2003.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hassell DG, Zimmerman WB. Chemical Engineering Science. 2006;61:2977–2985. [Google Scholar]
- 76.Malecha K, Golonka LJ, Bałdyga J, Jasińska M, Sobieszuk P. Sensors and Actuators B: Chemical. 2009;143:400–413. [Google Scholar]
- 77.Li Y, Zhang D, Feng X, Xu Y, Liu BF. Talanta. 2012;88:175–180. doi: 10.1016/j.talanta.2011.10.028. [DOI] [PubMed] [Google Scholar]
- 78.Liu RH, Stremler MA, Sharp KV, Olsen MG, Santiago JG, Adrian RJ, Aref H, Beebe DJ. Journal of Microelectromechanical Systems. 2000;9:190–197. [Google Scholar]
- 79.Durbin PA, Pettersson-Reif BA. Statistical theory and modeling for turbulent flows. John Wiley & Sons, Ltd; 2011. [Google Scholar]
- 80.Moin P, Kim J. Scientific American. 1997;276:62–68. [Google Scholar]
- 81.Perot B, Moin P. Journal of Fluid Mechanics. 1995;295:199–227. [Google Scholar]
- 82.Moser RD, Kim J, Mansour NN. Physics of Fluids. 1999;11:943–945. [Google Scholar]
- 83.Martell MB, Rothstein JP, Perot JB. Physics of Fluids. 2010:22. [Google Scholar]
- 84.Graceffa R, Burghammer M, Davies RJ, Riekel C. Applied Physics Letters. 2012:101. [Google Scholar]
- 85.Brunelli NA, Neidholdt EL, Giapis KP, Flagan RC, Beauchamp JL. Anal Chem. 2013 doi: 10.1021/ac3032417. [DOI] [PubMed] [Google Scholar]
- 86.Bellouard Y, Colomb T, Depeursinge C, Dugan M, Said AA, Bado P. Optics Express. 2006;14:8360–8366. doi: 10.1364/oe.14.008360. [DOI] [PubMed] [Google Scholar]
- 87.Joo C, Ha T. Cold Spring Harb Protoc. 2012;2012:1104–1108. doi: 10.1101/pdb.prot071530. [DOI] [PubMed] [Google Scholar]
- 88.Dootz R, Evans H, Koster S, Pfohl T. Small. 2007;3:96–100. doi: 10.1002/smll.200600288. [DOI] [PubMed] [Google Scholar]
- 89.Graceffa R, Nobrega RP, Barrea R, Kathuria SV, Matthews CR, Guo L, Bilsel O, Irving T. J Sync Radiation. 2013 doi: 10.1107/S0909049513021833. Under review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kiselar JG, Chance MR. Journal of Mass Spectrometry. 2010;45:1373–1382. doi: 10.1002/jms.1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stocks BB, Sarkar A, Wintrode PL, Konermann L. J Mol Biol. 2012;423:789–799. doi: 10.1016/j.jmb.2012.08.019. [DOI] [PubMed] [Google Scholar]
- 92.Gau BC, Chen J, Gross ML. Biochim Biophys Acta. 2013;1834:1230–1238. doi: 10.1016/j.bbapap.2013.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wu L, Lapidus LJ. Anal Chem. 2013;85:4920–4924. doi: 10.1021/ac3033646. [DOI] [PubMed] [Google Scholar]