Abstract
The extent of linkage disequilibrium (LD) was studied in two small food-gathering populations—Evenki and Saami—and two larger food-producing populations—Finns and Swedes—in northern Eurasia. In total, 50 single-nucleotide polymorphisms (SNPs) from five genes were genotyped using real-time pyrophosphate DNA sequencing, whereas 14 microsatellites were genotyped in two X-chromosomal regions. In addition, hypervariable region I of the mtDNA was sequenced to shed light on the demographic history of the populations. The SNP data, as well as the microsatellite data, reveal extensive levels of LD in Evenki and Saami when compared to Finns and Swedes. mtDNA-sequence variation is compatible with constant population size over time in Evenki and Saami but indicates population expansion in Finns and Swedes. Furthermore, the similarity between Finns and Swedes in SNP allele- and haplotype-frequency distributions indicate that these two populations may share a recent common origin. These findings suggest that populations such as the Evenki and the Saami, rather than the Finns, may be particularly suited for the initial coarse mapping of common complex diseases.
Introduction
Linkage disequilibrium (LD)—the nonrandom association of alleles at different loci on the same chromosome (Lewontin and Kojima 1960)—is determined by factors, such as regional recombination rates, that are intrinsic to the genome and by the demographic history of a population (Hartl and Clark 1997). Studies of LD between microsatellites (Tishkoff et al. 1996, 1998, 2000; Kidd et al. 2000) and, more recently, between SNPs (Reich et al. 2001) have provided new insights into the origin and history of human populations. For example, LD levels much higher in non-African than in African populations (Reich et al. 2001) may reflect a population bottleneck that is associated with the origin of non-African populations (Jorde et al. 1998). LD has also been used to localize genes that are involved in monogenic disease (Jorde 2000), such as diastrophic dysplasia (Hästbacka et al. 1992). In these studies, populations believed to have been descended from a small number of founders and to have been relatively isolated over a long time have been studied. The best example of such a population is the Finnish population, in which many recessive diseases that elsewhere are rare or absent are highly enriched and in which, conversely, other diseases, such as cystic fibrosis or phenylketonuria, that are common elsewhere in Europe are rare (Peltonen et al. 1999). This unusual disease pattern suggests a past founder effect, and this seems to be corroborated by the successful mapping of >30 monogenic, generally recessive and rare diseases (Wright et al. 1999; Peltonen 2000; Peltonen et al. 2000). The successes achieved in LD mapping of monogenic diseases in Finns and other similar populations have increased hopes that these populations may be useful also for the mapping of genes underlying susceptibility to common complex diseases.
In spite of the great interest in LD mapping, few studies have compared the extent of LD among human populations, and those that have been performed have yielded somewhat bewildering results. For example, Sardinians and Finns—both considered to be isolated populations that are suitable for LD mapping—carry LD levels that are similar to other populations in Europe and the United States (Eaves et al. 2000; Jorde et al. 2000; Taillon-Miller et al. 2000). By contrast, the Saami, in northern Europe, who are not known to be characterized by any unusual spectrum of genetic diseases, have been shown to carry extensive LD between microsatellites (Laan and Pääbo 1997).
To compare LD levels among human populations that differ with regard to their demographic histories, we studied four populations (Evenki, Finns, Saami, and Swedes) that differ substantially in size and demographic history (table 1). In each population, we genotyped 50 common SNPs from five genomic loci, as well as 14 microsatellites in two X-chromosomal regions. In addition, we characterized mtDNA variation in each population.
Table 1.
Overview of Sample Sizes for the SNP, Microsatellite (STR), and mtDNA Analyses and Summary of Population Characteristics and Demographic History
|
Sample Size |
Population Characteristics |
Demographic History |
||||||
| Population | SNP | STR | mtDNA | Population Sizea | Location | Linguistic Phylumb | mtDNAGrowth? | Lifestyle |
| Evenki | 94 | 71 | 56 | ∼58,000 | Siberia | Altaic | No | Food gatherers, reindeer herders |
| Saami | 90 | 54 | 115c | ∼60,000 | Northern Europe | Uralic-Yukaghir | No | Food gatherers, reindeer herders |
| Finns | 78 | 80 | 133c | ∼5,000,000 | Northern Europe | Uralic-Yukaghir | Yes | Food producers |
| Swedes | 78 | 41 | 32c | ∼9,000,000 | Northern Europe | Indo-Hittite | Yes | Food producers |
Bertelsmann Lexikon Verlag (1996).
Ruhlen (1991). mtDNA growth was determined using Tajima's (1989) D statistic and assuming selective neutrality. The hypothesis of constant population size over time for Evenki (D=-0.88) and Saami (D=-0.95) cannot be rejected (P>.05), whereas it is rejected (P<.05) for Finns (D=-1.98) and Swedes (D=-1.87).
Subjects and Methods
Population Samples
DNA samples from the Evenki were collected in three villages located on the Amur River in northern Siberia. The Saami represent a random sample (DNA provided by Lars Beckman [Umeå University]) of unrelated individuals from the Saami community in Sweden. The Swedish DNA samples were collected by Andreas Kindmark (Uppsala University) and represent individuals from central Sweden. They were collected in the course of osteoporosis studies at the Akademiska Hospital in Uppsala. Microsatellites were genotyped in Finnish DNA samples that were kindly provided by Leena Peltonen (University of Helsinki). They were collected randomly from a panel of Finnish samples representing several different regions in Finland. The same samples were also used and described elsewhere (Varilo et al. 2000). The male Finnish samples used for SNP typing were from a diverse panel of individuals from Finland and were kindly provided by Antti Sajantila (University of Helsinki). They were collected in the course of paternity testing at the University of Helsinki and represent unrelated individuals from all over Finland. The four female samples used for SNP typing in Finns were from the collection of L. Peltonen (see above).
Whereas all 47 Evenki and 45 Saami samples used for SNP typing were from female individuals, DNA samples from Finns (35 males and 4 females) and Swedes (21 males and 18 females) were from individuals of both sexes. All Evenki (n=71), Saami (n=54), Finnish (n=80), and Swedish (n=41) samples used for the microsatellite study were from male individuals.
PCR, Genotyping, and DNA Sequencing
DNA fragments from ACE (∼7.2 kb), β-globin (∼2.8 kb), LPL (∼8.7 kb), and Xq13.3 (∼10.5 kb) were amplified by 100–200 ng of total genomic DNA with the Boehringer Expand kit and a thermal cycler (MJ Research). Buffer and enzyme concentrations, as well as cycling conditions, were as recommended by the supplier. The 300-bp fragment from the MX locus was amplified according to standard PCR procedures, by use of Taq polymerase (Perkin Elmer). One microliter of the product from these reactions was used to further amplify shorter DNA segments (i.e., for ACE, 5 fragments of 100–430 bp; for β-globin, 5 fragments of 150–650 bp; for LPL, 30 fragments of 100–630 bp; and, for Xq13.3, 6 fragments of 210–640 bp) according to standard procedures, by use of one biotinylated primer. PCRs were prepared using a Biomek 2000 robot. All primer sequences are available from the authors on request. PCR products were prepared for real-time pyrophosphate DNA sequencing (including annealing of sequencing primers) essentially as described by Alderborn et al. (2000).
The loci and primers for microsatellite typing were chosen from the Généthon genetic map (Dib et al. 1996) and multiplexed, according to PCR conditions and product sizes, such that all alleles could be separated on an ALF DNA Sequencer (Pharmacia Biotech). PCRs were performed using a hot-start procedure as described by Laan and Pääbo (1997). Allele determination was performed using Fragment Manager software (Pharmacia Biotech). The amplification sets of markers were, from Xq13-21 (preannealing at 52°C), AFM078za1/AFM311vg5/AFM285xg5, AFM116xg1/AFM207zg5/AFMa040, and AFMc020wb9/AFM234tf8 and, from Xp22 (preannealing at 55°C), AFM120xa9/AFM291wf5, AFM135xe7/AFM337wd5/AFM234yf12, and AFM164zd4. Microsatellite haplotypes were reconstructed manually. Microsatellite data from seven of the Xq13-21 loci (DXS983, DXS986, DXS8092, DXS8082, DXS1125, DXS8037, and DXS995) for the Finns, the Swedes, and the Saami were from studies described elsewhere (Laan and Pääbo 1997). PCR amplification of hypervariable region I (HVRI) of the mtDNA from Evenki was performed essentially according to standard procedures and was sequenced on ALF DNA Sequencers (Pharmacia Biotech).
Real-Time Pyrophosphate DNA Sequencing
The single-stranded PCR samples, which were annealed with their respective sequencing primers (see above), were transferred to a translucent 96-well plate for real-time pyrophosphate DNA sequencing (Ronaghi et al. 1996, 1998). DNA sequence reading was performed using a P3 PSQ 96 instrument (Pyrosequencing AB) and the PSQ 96 SNP reagent kit (Pyrosequencing AB). A unique dispensation order that was complementary to the template and included both alleles was specified for each SNP.
Selection of SNP Markers
The following markers from ACE (Rieder et al. 1999), β-globin (Harding et al. 1997), LPL (Nickerson et al. 1998), MX (Jin et al. 1999), and Xq13.3 (Kaessmann et al. 1999), which all (except for SNP 22 at Xq13.3) have minor-allele frequencies >.05, were genotyped in all populations (sequence positions as indicated in the original publications): for ACE (GenBank accession number AF118569), markers 3872, 7831, 8968, 9222, and 10514; for β-globin (GenBank accession number L26474), markers 145, 297, 1416, 1423, 2634, 2636, and 2792; for LPL (GenBank accession number AF050163), markers 736, 1216, 1220, 1286, 1571, 2131, 2500, 2996, 3022, 3723, 3843, 4343, 4346, 4418, 4576, 4872, 5441, 5554, 5687, 6250, 6595, 6678, 7315, 7360, 7754, 8089, 8292, 8393, 8644, 8755, and 8852; for MX (GenBank accession number L35674), marker 2712; and, for Xq13.3 (GenBank accession numbers AJ241023–AJ241093), SNPs 4–6, 16, 17, and 22.
LD Analyses
The 50 SNPs are in Hardy-Weinberg equilibrium in all populations (P>.01), when tested using ARLEQUIN 2.0. We used an expectation-maximization algorithm (Weir 1996) to obtain maximum-likelihood estimates of haplotype frequencies (with each haplotype consisting of a pair of SNPs). We measured LD by use of the D′ statistic (Lewontin 1964). The significance of allelic association of a pair of SNPs was calculated using Fisher's exact test. In this analysis, we excluded all monomorphic sites and singletons. A Monte Carlo approximation of Fisher's exact test was used to describe the extent of nonrandom allelic association between pairs of microsatellite loci. The tail probability (P value) of this test was computed with GenePop (Raymond and Rousset 1994) as described elsewhere (Laan and Pääbo 1997).
Haplotype Analysis
The haplotype frequencies of the different regions were estimated using the PHASE program (Stephens et al. 2001). Each run of the program was repeated 10 times with different seeds. To generate the frequency of each haplotype, the average frequency generated by these runs was calculated. Haplotypes with an average frequency <0.8 were ignored.
Statistical Analysis
To test for population differentiation, we calculated the Fst statistic (Weir 1996) with ARLEQUIN 2.0. Assuming selective neutrality, we used Tajima's (1989) D neutrality test to detect population growth in the mitochondrial HVRI sequences. Allele frequencies for each microsatellite locus were estimated by the Fstat software. Locus diversity was calculated as 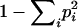 , where pi is the estimated frequency of the ith allele at the locus (Weir 1996).
, where pi is the estimated frequency of the ith allele at the locus (Weir 1996).
Results
LD
The Evenki and the Saami are comparatively small populations that traditionally practice hunting, gathering, and, for some time, reindeer herding (i.e., they are food gatherers) (table 1), whereas the Finns and the Swedes are larger populations that practice agriculture (i.e., they are food producers). In each of these populations, four loci—for which extensive comparative DNA sequence data from different populations around the world are available—were studied; these loci are ACE (located at 17q23) (Rieder et al. 1999), β-globin (located at 11p15.5) (Harding et al. 1997), LPL (located at 8p22) (Nickerson et al. 1998), and a noncoding X-chromosomal region (Xq21.1, which was formerly mapped to Xq13.3 and is so termed here) (Kaessmann et al. 1999). For each locus, SNPs (except for one SNP at Xq13.3; see “Subject and Methods”) that have a minor-allele frequency >0.05 and that together define all major haplotypes identified in previous studies were selected. In total, 5 SNPs from ACE, 7 SNPs from β-globin, 31 SNPs from LPL, and 6 SNPs from Xq13.3 were genotyped in 47 Evenki, 45 Saami, 39 Finns, and 39 Swedes; in addition, 1 SNP in the MX region (located at 21q22.3) (Jin et al. 1999) was scored.
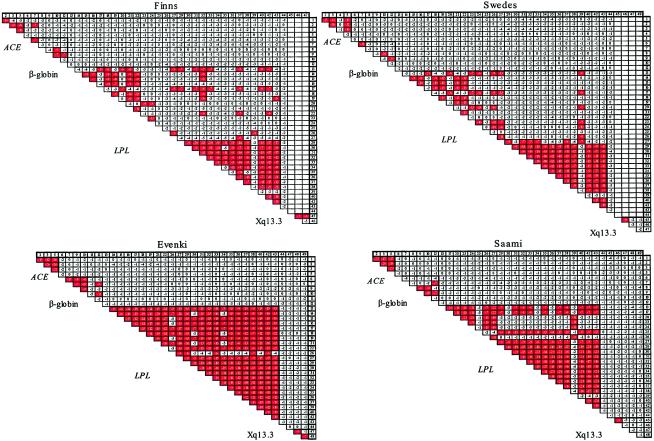
Pairwise LD was estimated for each population by use of Fisher's exact test and by excluding monomorphic sites, as well as sites where the minor allele was present in a single chromosome in a population. The fraction of linked SNP pairs that show significant LD (P<10-6) was 60% in Evenki (277/465 pairs), 48% in Saami (160/331 pairs), and 29% in both Finns (145/502 pairs) and Swedes (144/502 pairs). If only common SNPs (i.e., those for which the minor allele showed a frequency >0.1) were considered, 91% of all pairs showed LD in Evenki (273/300 pairs), 61% in Saami (161/262 pairs), 33% in Finns (141/430 pairs), and 32% in Swedes (141/437 pairs) (fig. 1). The average P values of all LD comparisons for the Evenki and Saami were four orders of magnitude smaller than those for the Finns and Swedes (i.e., P∼10-8 vs. P∼10-4, respectively). Furthermore, for all three loci where the same number of chromosomes were studied in all populations, the fraction of pairs of common SNPs showing significant LD was greater in the Evenki and Saami than in the Finns and Swedes (fig. 1).
Figure 1.
LD of SNP pairs in ACE, β-globin, LPL, and Xq13.3, for the four populations studied. For each population, only SNPs where the minor allele occurs at a frequency >0.1 are shown, since other SNPs are unlikely to show significant LD with this sample size. Orders of magnitude of significance values of pairwise associations (by Fisher's exact test) are given. SNP pairs showing significant LD (P<10-6) are shaded (red). Number 44 is the single SNP from the MX region. LD of SNP pairs in Xq13.3 is shown on the right. Interlocus LD of SNPs from Xq13.3 for Finns and Swedes was not calculated because of sample-size differences between Xq13.3 and the other loci.
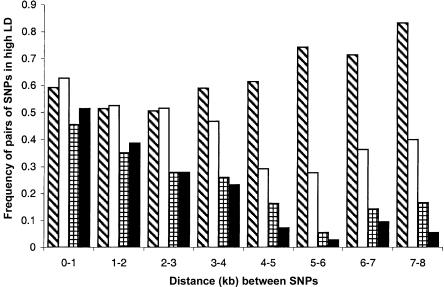
At LPL, where the most SNPs were assayed, the Evenki show significant LD for 76% (56/74) of SNP pairs separated by >5 kb, and the Saami showed significant LD for 33% (13/39) of such SNP pairs. In Finns and Swedes, 11% (8/75) and 5% (4/75), respectively, of such SNP pairs showed LD (fig. 2). Note that the significance values calculated for each population are a function of both the LD level and the sample size. However, the difference in sample size between the Evenki and Saami (47 and 45 individuals, respectively) and the Finns and Swedes (39 individuals both) is unlikely to account for the substantial differences in significance levels observed.
Figure 2.
The proportion of SNP pairs showing significant association (P<10-6) versus distance. The pattern in each bar indicates the different populations: Evenki (diagonal lines), Saami (white), Finns (horizontal and vertical lines), and Swedes (black).
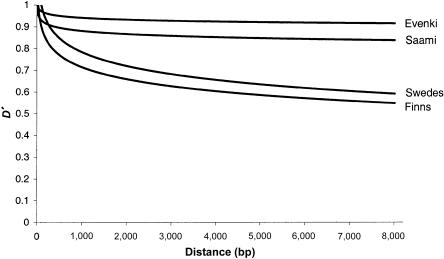
When LD was measured using the D′ statistic (Lewontin 1964), the average D′ was found to be ∼0.94 in the Evenki and ∼0.87 in the Saami, whereas it was found to be ∼0.73 in the Finns and ∼0.67 in the Swedes. When plotted as a function of the physical distance (fig. 3), D′ declines at a much lower rate in the Evenki and Saami than in the Finns and Swedes, remaining >0.8 at an intermarker distance of 8 kb in the former two populations. Indeed, not a single SNP pair in the Evenki showed D′ <0.4. Thus, substantially more LD between SNPs exists in the Evenki and Saami than in the Finns and Swedes.
Figure 3.
Relationship between mean level of LD (as measured by D′) and the distance between SNPs in ACE, β-globin, LPL, and Xq13.3 for the four populations studied.
To estimate LD over larger genetic distances, we analyzed 14 microsatellites (table 2) that are located <1–16 cM apart in two X-chromosomal regions (Xq13-21 and Xp22) (tables 2–4). Both X-chromosomal regions show elevated levels of LD in the Saami and Evenki as compared to the Finns and Swedes. In the Saami and Evenki, 20 and 12 microsatellite pairs, respectively, exhibit significant LD, whereas, in the Finns and Swedes, there is LD between a single locus pair (tables 3–5). Furthermore, there is LD between several microsatellites that are located >1 cM apart in the Evenki and Saami, whereas the single pair for which LD is seen in Finns and Swedes spans <1 cM. Thus, both short- and long-range LD is dramatically elevated in the Saami and Evenki when compared to that in Finns and Swedes.
Table 2.
Diversity and Locations of the 14 Microsatellites Analyzed[Note]
|
X-Chromosomal Location |
Diversity (No. of Alleles) for |
|||||
| Marker (Locus) | Region | Nucleotide Position | Evenki | Saami | Finns | Swedes |
| AFM078za1 (DXS983) | Xq13.1 | 66,774,153 | .536 (4) | .568 (4) | .688 (6) | .604 (6) |
| AFM285xg5 (DXS8037) | Xq13.3 | 70,811,682 | .374 (3) | .491 (5) | .712 (8) | .759 (7) |
| AFMa040 (DXS8092) | Xq13.3 | 70,812,229 | .864 (11) | .810 (8) | .842 (12) | .872 (11) |
| AFM311vg5 (DXS1225) | Xq21.1 | 77,994,863 | .698 (8) | .741 (6) | .721 (9) | .790 (8) |
| AFMc020wb9 (DXS8082) | Xq21.1 | 78,156,901 | .764 (6) | .764 (6) | .738 (9) | .741 (7) |
| AFM116xg1 (DXS986) | Xq21.1 | 79,210,352 | .870 (11) | .784 (10) | .790 (11) | .690 (9) |
| AFM234tf8 (DXS1066) | Xq21.3 | 82,569,449 | .462 (4) | .364 (3) | .378 (4) | .269 (4) |
| AFM207zg5 (DXS995) | Xq21.3 | 87,011,575 | .488 (3) | .427 (3) | .629 (6) | .551 (5) |
| AFM120xa9 (DXS987) | Xp22.2 | 13,479,069 | .794 (9) | .738 (8) | .848 (14) | .801 (11) |
| AFM164zd4 (DXS1053) | Xp22.2 | 15,106,057 | .550 (4) | .814 (8) | .658 (8) | .724 (8) |
| AFM291wf5 (DXS7163) | Xp22.1 | 18,829,368 | .555 (4) | .721 (5) | .665 (7) | .774 (7) |
| AFM234yf12 (DXS999) | Xp22.1 | 19,643,052 | .511 (4) | .616 (5) | .721 (8) | .740 (7) |
| AFM337wd5 (DXS1229) | Xp22.1 | 20,785,255 | .395 (2) | .370 (2) | .249 (4) | .460 (3) |
| AFM135xe7 (DXS989) | Xp22.1 | 22,686,265 | .848 (9) | .683 (5) | .837 (13) | .803 (10) |
Note.— Chromosomal locations were determined using the Ensembl database.
Table 3.
Significance of LD for All Pairs of Microsatellites Studied at Xq13-21
|
P Valuea for |
|||||
| Locus Pair (Markers) | Distance(cM) | Saami(n=54) | Evenki(n=71) | Swedes(n=41) | Finns(n=80) |
| DXS1066-DXS983 (234-078) | 3.9 | .1661 | .55341 | .78017 | .33848 |
| DXS995-DXS983 (207-078) | 3.2 | .01217 | .40415 | .59299 | .50813 |
| DXS1066-DXS8092 (234-040) | 2.2–2.7 | .0007 | .67842 | .15142 | .34406 |
| DXS1066-DXS8082 (234-020) | 2.2–2.7 | .00118 | .36141 | .32315 | .38662 |
| DXS1066-DXS8037 (234-285) | 2.7 | .05988 | .5895 | .9498 | .66055 |
| DXS1066-DXS1225 (234-311) | 2.2–2.7 | .25204 | .00334 | .35509 | .12499 |
| DXS1066-DXS986 (234-116) | 2.2 | .030747 | .16681 | .09235 | .23339 |
| DXS995-DXS8037 (207-285) | 2 | .12404 | .35701 | .22522 | .87376 |
| DXS995-DXS8092 (207-040) | 1.5–2 | .10293 | .15124 | .42535 | .11506 |
| DXS995-DXS8082 (207-020) | 1.5–2 | .00001 | .08884 | .49043 | .12749 |
| DXS995-DXS1225 (207-311) | 1.5–2 | .00146 | .00543 | .56353 | .15388 |
| DXS986-DXS983 (116-078) | 1.7 | .00022 | .99329 | .39951 | .82905 |
| DXS995-DXS986 (207-116) | 1.5 | 0 | .00025 | .42872 | .72858 |
| DXS1225-DXS983 (311-078) | 1.2–1.7 | 0 | .22798 | .4799 | .63015 |
| DXS8082-DXS983 (020-078) | 1.2–1.7 | 0 | .01256 | .08199 | .56539 |
| DXS8092-DXS983 (040-078) | 1.2–1.7 | .00015 | .02385 | .74607 | .31404 |
| DXS8037-DXS983 (285-078) | 1.2 | .29968 | .07627 | .92432 | .68306 |
| DXS1066-DXS995 (234-207) | .7 | .1128 | .00368 | .31729 | .42012 |
| DXS1225-DXS986 (311-116) | ⩽.5 | 0 | .00008 | .44759 | .3933 |
| DXS8037-DXS986 (285-116) | ⩽.5 | .00013 | .00001 | .25632 | .61956 |
| DXS8037-DXS8092 (285-040) | ⩽.5 | 0 | .00003 | .0283 | .18043 |
| DXS1225-DXS8092 (311-040) | ⩽.5 | 0 | .00042 | .67606 | .28304 |
| DXS8037-DXS8082 (285-020) | ⩽.5 | .01241 | .00044 | .03324 | .2377 |
| DXS1225-DXS8082 (311-020) | ⩽.5 | 0 | 0 | 0 | 0 |
| DXS8082-DXS986 (020-116) | ⩽.5 | 0 | .00039 | .61796 | .09156 |
| DXS8092-DXS986 (040-116) | ⩽.5 | 0 | 0 | .33154 | .3313 |
| DXS8082-DXS8092 (020-040) | ⩽.5 | 0 | 0 | .10228 | .07444 |
| DXS8037-DXS1225 (285-311) | ⩽.5 | .09133 | .00004 | .24217 | .83624 |
P values <.001 are shown in boldface italic.
Table 4.
Significance of LD for All Pairs of Microsatellites Studied at Xp22
|
P Valuea for |
|||||
| Locus Pair (Markers) | Distance(cM) | Saami(n=54) | Evenki(n=71) | Swedes(n=41) | Finns(n=80) |
| DXS989-987 (135-120) | 15.1 | .1092 | .57661 | .50795 | .84009 |
| DXS989-1053 (135-164) | 15 | .93402 | .04645 | .92152 | .09811 |
| DXS989-999 (135-234) | 12.5 | .061 | .29646 | .6554 | .74552 |
| DXS989-7163 (135-291) | 11.7 | .0051 | .2407 | .86224 | .88904 |
| DXS1229-987 (337-120) | 7.8 | .01604 | .00019 | .48169 | .41379 |
| DXS1229-1053 (337-164) | 7.7 | .03857 | .84375 | .45378 | .15421 |
| DXS989-1229 (135-337) | 7.3 | .3832 | .21219 | .48352 | .82206 |
| DXS989-1052 (135-163) | 7 | .14143 | .00613 | .03951 | .20042 |
| DXS1229-999 (337-234) | 5.2 | .00036 | .79923 | .13876 | .00704 |
| DXS1229-7163 (337-291) | 4.4 | .00769 | .00677 | .23943 | .01556 |
| DXS7163-987 (291-120) | 3.4 | .00916 | .00676 | .63431 | .99017 |
| DXS7163-1053 (291-164) | 3.3 | .00206 | .03497 | .96673 | .17491 |
| DXS999-987 (234-120) | 2.6 | .00026 | .00202 | .8468 | .56337 |
| DXS999-1053 (234-164) | 2.5 | .00014 | .73713 | .71497 | .5929 |
| DXS7163-999 (291-234) | .8 | 0 | .79618 | .20921 | .03075 |
| DXS1053-987 (164-120) | .1 | .00013 | .0038 | .85581 | .3116 |
P values <.001 are shown in boldface italic.
Table 5.
Overview of LD of 14 Microsatellites (Pairwise Comparisons) at Xq13-21 and Xp22[Note]
|
No. of Pairs in LD* at |
||||||||
| Xq13-21 |
Xp22 |
|||||||
| Population (Sample Size) | <1 cM | 1–2 cM | 2–4 cM | All | <2 cM | 2–5 cM | 5–16 cM | All |
| Evenki (n = 71) | 10 | 1 | 0 | 11 | 0 | 0 | 1 | 1 |
| Saami (n = 54) | 8 | 6 | 1 | 15 | 2 | 2 | 1 | 5 |
| Finns (n = 80) | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| Swedes (n = 41) | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
Note.— Significance was estimated using a Monte Carlo approximation of Fisher's exact test.
P<.001.
Allele-Frequency Distributions
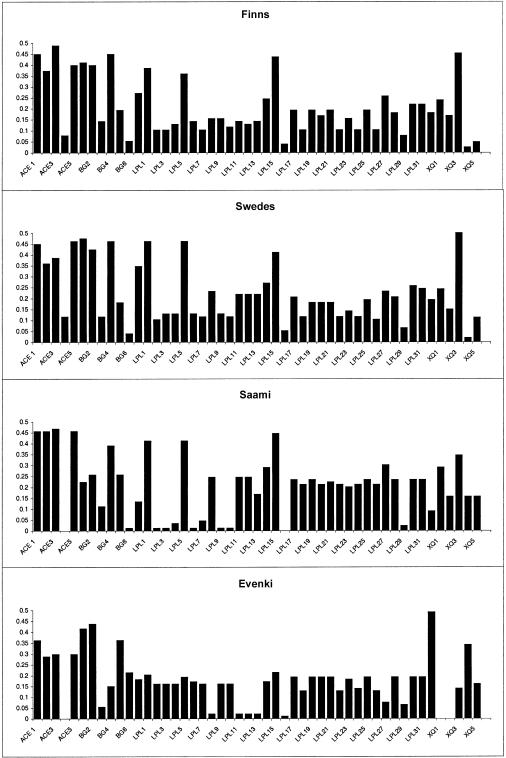
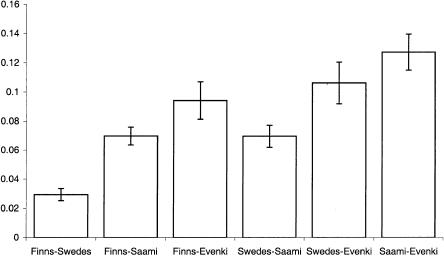
The distributions of allele frequencies in the Finns and Swedes (fig. 4) are very similar, whereas the Evenki and Saami show greater differences both with Finns and Swedes and with each other. The average differences in the allele frequencies confirm this view (fig. 5). When the F statistic (Wright 1921) is used to calculate the proportion of variation that can be attributed to between-population variation (table 6), the results similarly show that the Finns and Swedes are more alike than any of the other populations studied. In fact, all pairwise comparisons involving either the Evenki or the Saami indicate significant differentiation between populations, except for the Saami-Finns pair. Here, the hypothesis that these two groups (i.e., the Finns and the Saami) are a part of the same population cannot be rejected, although the P value is suggestive (P=.095). It is possible that the Saami and Finns have a relatively low F value because of a low degree of admixture between the populations. This is supported by the observation that, although Saami and Finns differ substantially from each other when nuclear markers are analyzed (Sajantila and Pääbo 1995; Lahermo et al. 1996), 37% of the Saami carry an mtDNA sequence motif that is absent in other European populations but is present at a very low frequency in Finns (Sajantila et al. 1995).
Figure 4.
Minor-allele frequencies of 50 SNPs from ACE, β-globin, LPL, MX, and Xq13.3 for the four populations studied
Figure 5.
Average allele-frequency differences between pairs of populations.
Table 6.
Pairwise Population Differences as Estimated by Fst Comparisons Combining SNP Data from ACE, β-Globin, LPL, and MX[Note]
| Swedes | Finns | Evenki | Saami | |
| Swedes: | ||||
| Fst | … | … | … | … |
| P | … | … | … | … |
| Finns: | ||||
| Fst | −.002 | … | … | … |
| P | .556 | … | … | … |
| Evenki: | ||||
| Fst | .058 | .055 | … | … |
| P | .000 | .000 | … | … |
| Saami: | ||||
| Fst | .022 | .012 | .085 | … |
| P | .015 | .095 | .000 | … |
Note.— Significant (P<.05) Fst values (10,100 permutations) are shown in boldface italic.
Haplotype Diversity and Distribution
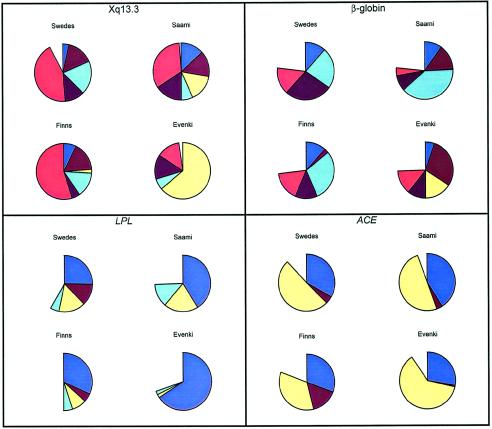
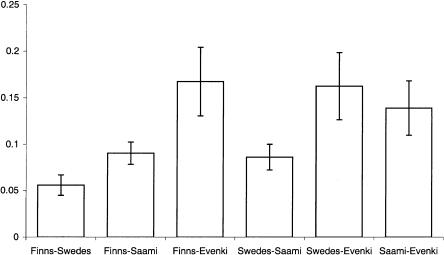
Haplotypes were estimated using the PHASE method (Stephens et al. 2001). When the haplotype distributions are compared in the four populations for the four loci (fig. 6), the distributions in the Finns and the Swedes seem to resemble each other more than those in the other populations do. This impression is confirmed when the haplotype-frequency differences between populations are calculated (fig. 7). The haplotype-frequency difference between the Finns and the Swedes is smaller than that between any other pair of populations, whereas each comparison involving the Evenki is larger than the comparisons involving the Fenno-Scandinavian populations.
Figure 6.
Overview of distributions of haplotypes that occur at a frequency >0.1 in at least one population. The missing piece of each pie chart represents haplotypes that occur in frequencies <0.1 in all populations.
Figure 7.
Average haplotype-frequency differences (for haplotypes occurring at a frequency >0.1 in at least one population) between populations studied.
mtDNA Variation
We used Tajima's (1989) D statistic to test if the variation found in mtDNA HVRI sequences is compatible with constant population size. Whereas the hypothesis of constant population size cannot be rejected in the Evenki and Saami (table 1), HVRI sequences from the Finns and Swedes show significantly negative values (P<.05), indicating population growth if selective neutrality is assumed. It is possible that the absence of growth signals in the former two populations is because low-frequency variants (Harpending et al. 1998) have been lost from the gene pool because of genetic drift (Excoffier and Schneider 1999).
Discussion
Population Differences in LD
Genetic diversity is strikingly differently distributed in the four populations studied. First, the Evenki and Saami have a much greater extent of LD over both short and long distances than do the Finns and Swedes. In the Evenki and Saami, LD extends for ⩾8 kb for SNPs and >5 cM for microsatellites (tables 3–5) . Second, although frequency distributions of SNP alleles (figs. 4 and 5 and table 6) and haplotypes (figs. 6 and 7) are similar in the Finns and Swedes, the Evenki and Saami differ both from each other and from the other populations. Third, the pattern of mtDNA-sequence variation is compatible with constant population size in the Evenki and Saami, whereas constant population size is rejected in the Finns and Swedes.
These differences can be reasonably explained by differences in the history of these populations. Whereas the Evenki (census size ∼58,000 individuals) and the Saami (census size ∼60,000 individuals) (table 1) have been of small and presumably constant size over a long period of time (see above) (Laan and Pääbo 1997), the Finns and the Swedes are larger populations that have most likely expanded. Thus, genetic drift can be expected to have been a much stronger factor in the Evenki and the Saami because of their small and constant size (Slatkin 1994; Jorde 2000; Zöllner and von Haeseler 2000). This is consistent both with the differences in mtDNA-sequence–diversity patterns in the four groups and with the greater extent of LD observed in the Evenki and the Saami and is reminiscent of the situation in subisolates that derive from larger populations (Zavattari et al. 2000; Angius et al. 2001). It is also compatible with the observation that, for LPL, the locus where the largest number of SNPs were studied, substantially more haplotypes were observed in the Finns (26) and Swedes (28) than in the Evenki (11) and Saami (15). Furthermore, the Evenki and Saami (for the three loci—ACE, β-globin, and LPL—for which sample sizes are approximately the same for all populations) have one and two monomorphic sites, respectively, as well as several singletons and doubletons, whereas there are no such sites in the Finns and Swedes (fig. 4).
One may argue that population subdivision or admixture may have contributed to the extensive LD in the Evenki and Saami. However, in a subdivided population (Hartl and Clark 1997) or after a very recent and sudden admixture event (Pritchard and Przeworski 2001), interlocus LD is expected—but not observed—in the Evenki and Saami (fig. 1). It may also be possible that low, but constant, rates of gene flow from neighboring populations with divergent allele frequencies may have increased levels of LD in the Evenki and Saami. If this were true, allele frequencies for SNPs that show extensive LD should substantially differ between the populations that contribute to the admixture. For example, if the Finns were the population that contributed divergent haplotypes to the Saami, then the allele frequency should differ between the Finns and Saami at sites that show extensive LD in the Saami. Although such a pattern is not apparent in our data (not shown), this hypothesis is difficult to test in a rigorous way. In summary, the pattern of reduction in genetic variation in the Evenki and Saami is consistent with the hypothesis that strong genetic drift has affected these populations. Although this may not be the only factor that has raised levels of LD in the Evenki and Saami, it is likely to be the major factor.
Food Production versus Food Gathering
The Evenki and Saami traditionally practice hunting and gathering food (although, since a few hundred years ago, they also practice reindeer herding), whereas the Finns and Swedes traditionally practice agriculture. Since food-gathering populations have generally been small and stationary over time, whereas food producers generally live in large populations that have recently expanded, it is possible that, in contrast to food-producing populations, a majority of food-gathering populations may show extensive levels of LD. Indeed, it is noteworthy that one hunter-gatherer population in India (Mountain et al. 1995) and three food-gathering populations in Africa (Watson et al. 1996) similarly differ from food-producing groups that live in their respective regions by showing no indication of past population growth when mtDNA sequences are analyzed. Obviously, this does not exclude the possibility that bottleneck effects (e.g., infectious diseases) may have produced high levels of LD also in some food-producing populations.
The Finnish Bottleneck
The similarity between Finns and Swedes in allele and haplotype frequencies indicates that these two populations may be descended from the same central European source population—as has been suggested by Sajantila and Pääbo (1995). Furthermore, the similar level of LD in Finns and Swedes that has been observed in this and other studies (Eaves et al. 2000; Jorde et al. 2000; Taillon-Miller et al. 2000) indicates that a strong founder effect cannot have been associated with the origin of the Finnish population. However, a “mild” bottleneck involving a founding population smaller than the Swedish founding population is compatible with the LD observed for Finns (average D′∼0.73), which was somewhat greater than that for Swedes (D′∼0.67). It is also compatible with the unusual spectrum of monogenic diseases in Finland, as well as with the reduced variation in Y-chromosomal microsatellites (Sajantila et al. 1996; Kittles et al. 1998, 1999) and mtDNA sequences (Sajantila et al. 1996) in Finns. Since they are more affected by genetic drift than autosomal and X-chromosomal loci because of their smaller effective population size, the Y chromosome and mtDNA are more likely to show the effects of a founder effect than are other parts of the genome.
Implications for Gene Mapping
Our results have important implications for the choice of populations for gene-mapping studies. We have previously suggested that LD created by genetic drift in small populations of constant size, such as the Saami, may be useful, in the identification of genetic regions involved in common disease (Laan and Pääbo 1997; Terwilliger et al. 1998). However, such populations will be unsuited for fine mapping, because markers far away from the disease locus may also show significant association with disease genes. Others have recently suggested a two-step strategy in which coarse mapping would be done with western European populations followed by fine-scale mapping with African populations (Reich et al. 2001). The extremely high levels of LD in the small and constant food-gathering populations suggest that a three-step mapping strategy may be more economical for diseases common enough to be observed in populations of an approximate size of 50,000. In such a strategy:
-
1.
A rough identification of the region of a disease locus should be performed in small and constant populations, such as the Saami or the Evenki. A map of moderate marker density is sufficient for this initial step, because markers at large distances from the disease locus will show significant LD with disease alleles in these populations. A further advantage may be that the number of disease alleles present in such populations may be smaller than that in larger populations. However, it may be advisable to perform this first step in more than one small population, since different loci may be responsible for the same disease phenotype in different populations.
-
2.
The location of a disease gene will be confirmed and further defined in non-African, food-producing populations that have expanded, such as the Finns and the Swedes. In this step, denser maps from the candidate region that was identified in the first step are required to compensate for the lower level of LD.
-
3.
African populations should be considered, since they will show significant LD only over very short distances (Reich et al. 2001). Thus, if a marker shows significant association, it is likely to be close to the disease allele so that one can hope to identify a small number of candidate genes for further study.
Obviously, the proposed mapping strategy has a number of limitations. For example, it requires that the loci underlying the disease phenotype are the same in the populations studied. It furthermore requires that the diseases studied are common enough to allow its study in a population of <100,000 individuals. However, when this is the case, we believe that populations such as the Evenki and the Saami have the potential to be extremely useful for the identification of genes underlying complex diseases, as well as for the identification of complex normal traits.
LD Landscapes
LD seems to be present in regions, or “blocks,” in the human genome (Daly et al. 2001; Rioux et al. 2001). The breakpoints between such blocks may be due to recombinational hotspots and to random events during the coalescence of human genes. The difference between small and constant populations (e.g., the Saami and the Evenki) and large, expanded populations (e.g., the Finns and the Swedes) in extent of LD is likely to be because LD blocks present in the larger populations are fused to “superblocks” in the smaller populations. For example, a clear breakpoint in LD that is seen in LPL (between sequence positions 2987 and 4872) in Finns and Swedes, as well as other populations (Templeton et al. 2000), is absent in Saami and Evenki (fig. 1).
To efficiently economize the number of SNPs necessary to perform a whole-genome screen, it will be important to characterize the LD landscape in the populations considered. That food-producing Europeans seem to share many features, including haplotype frequencies (figs. 6 and 7), makes it likely that they will all share the same or similar LD landscapes. However, for small, constant populations, it is likely that the LD landscape of each population will be different. Thus, for such populations, the LD landscapes may have to be independently characterized to economically use the LD patterns for genetic mapping. However, this will be a worthwhile undertaking if these populations are indeed useful for the mapping of genes involved in the etiology of common diseases and complex traits.
Acknowledgments
We thank L. Beckman, A. Kindmark, L. Peltonen, and A. Sajantila, for DNA samples, and K. Holmberg, J. Odeberg, and A. Åbergh, for technical support. M.L. was supported by the Alexander-von-Humboldt Stiftung. This work was supported by grants from the Bundesministerium für Bildung und Forschung and the Max-Planck-Gesellschaft.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- ARLEQUIN, http://lgb.unige.ch/arlequin/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for markers genotyped for ACE [accession number AF118569], β-globin [accession number L26474], LPL [accession number AF050163], MX [accession number L35674], and Xq13.3 [accession numbers AJ241023–AJ241093])
- Mathematical Genetics and Bioinformatics, http://www.stats.ox.ac.uk/mathgen/software.html (for PHASE program)
- Project Ensembl, http://www.ensembl.org/
References
- Alderborn A, Kristofferson A, Hammerling U (2000) Determination of single-nucleotide polymorphisms by real-time pyrophosphate DNA sequencing. Genome Res 10:1249–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angius A, Melis PM, Morelli L, Petretto E, Casu G, Maestrale GB, Fraumene C, Bebbere D, Forabosco P, Pirastu M (2001) Archival, demographic and genetic studies define a Sardinian sub-isolate as a suitable model for mapping complex traits. Hum Genet 109:198–209 [DOI] [PubMed] [Google Scholar]
- Bertelsmann Lexikon Verlag (1996) Die Völker der Erde: Bertelsmann Lexikon. Bertelsmann, Gütersloh [Google Scholar]
- Daly MJ, Rioux JD, Schaffner SF, Hudson T, Lander ES (2001) High-resolution haplotype structure in the human genome. Nat Genet 29:229–232 [DOI] [PubMed] [Google Scholar]
- Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, Lathrop M, Gyapay G, Morissette J, Weissenbach J (1996) A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380:152–154 [DOI] [PubMed] [Google Scholar]
- Eaves IA, Merriman TR, Barber RA, Nutland S, Tuomilehto-Wolf E, Tuomilehto J, Cucca F, Todd JA (2000) The genetically isolated populations of Finland and Sardinia may not be a panacea for linkage disequilibrium mapping of common disease genes. Nat Genet 25:320–323 [DOI] [PubMed] [Google Scholar]
- Excoffier L, Schneider S (1999) Why hunter-gatherer populations do not show signs of Pleistocene demographic expansions. Proc Natl Acad Sci USA 96:10597–10602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding RM, Fullerton SM, Griffiths RC, Bond J, Cox MJ, Schneider JA, Moulin DS, Clegg JB (1997) Archaic African and Asian lineages in the genetic ancestry of modern humans. Am J Hum Genet 60:772–789 [PMC free article] [PubMed] [Google Scholar]
- Harpending HC, Batzer MA, Gurven M, Jorde LB, Rogers AR, Sherry ST (1998) Genetic traces of ancient demography. Proc Natl Acad Sci USA 95:1961–1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl DL, Clark AG (1997) Principles of population genetics. Sinauer Associates, Sunderland [Google Scholar]
- Hästbacka J, de la Chapelle A, Kaitila I, Sistonen P, Weaver A, Lander E (1992) Linkage disequilibrium mapping in isolated founder populations: diastrophic dysplasia in Finland. Nat Genet 2:204–211 [DOI] [PubMed] [Google Scholar]
- Jin L, Underhill PA, Doctor V, Davis RW, Shen P, Cavalli-Sforza LL, Oefner PJ (1999) Distribution of haplotypes from a chromosome 21 region distinguishes multiple prehistoric human migrations. Proc Natl Acad Sci USA 96:3796–3800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorde LB (2000) Linkage disequilibrium and the search for complex disease genes. Genome Res 10:1435–1444 [DOI] [PubMed] [Google Scholar]
- Jorde LB, Bamshad M, Rogers AR (1998) Using mitochondrial and nuclear DNA markers to reconstruct human evolution. Bioessays 20:126–136 [DOI] [PubMed] [Google Scholar]
- Jorde LB, Watkins WS, Kere J, Nyman D, Eriksson AW (2000) Gene mapping in isolated populations: new roles for old friends? Hum Hered 50:57–65 [DOI] [PubMed] [Google Scholar]
- Kaessmann H, Heissig F, von Haeseler A, Pääbo S (1999) DNA sequence variation in a non-coding region of low recombination on the human X chromosome. Nat Genet 22:78–81 [DOI] [PubMed] [Google Scholar]
- Kidd JR, Pakstis AJ, Zhao H, Lu RB, Okonofua FE, Odunsi A, Grigorenko E, Tamir BB, Friedlaender J, Schulz LO, Parnas J, Kidd KK (2000) Haplotypes and linkage disequilibrium at the phenylalanine hydroxylase locus, PAH, in a global representation of populations. Am J Hum Genet 66:1882–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittles RA, Bergen AW, Urbanek M, Virkkunen M, Linnoila M, Goldman D, Long JC (1999) Autosomal, mitochondrial, and Y chromosomal DNA variation in Finland: evidence for a male-specific bottleneck. Am J Phys Anthropol 108:381–399 [DOI] [PubMed] [Google Scholar]
- Kittles RA, Perola M, Peltonen L, Bergen AW, Aragon RA, Virkkunen M, Linnoila M, Goldman D, Long JC (1998) Dual origins of Finns revealed by Y chromosome haplotype variation. Am J Hum Genet 62:1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laan M, Pääbo S (1997) Demographic history and linkage disequilibrium in human populations. Nat Genet 17:435–438 [DOI] [PubMed] [Google Scholar]
- Lahermo P, Sajantila A, Sistonen P, Lukka M, Aula P, Peltonen L, Savontaus ML (1996) The genetic relationship between the Finns and the Finnish Saami (Lapps): analysis of nuclear DNA and mtDNA. Am J Hum Genet 58:1309–1322 [PMC free article] [PubMed] [Google Scholar]
- Lewontin RC (1964) The interaction of selection and linkage. I. General considerations, heterotic models. Genetics 49:49–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin RC, Kojima K (1960) The evolutionary dynamics of complex polymorphisms. Evolution 14:458–472 [Google Scholar]
- Mountain JL, Hebert JM, Bhattacharyya S, Underhill PA, Ottolenghi C, Gadgil M, Cavalli-Sforza LL (1995) Demographic history of India and mtDNA-sequence diversity. Am J Hum Genet 56:979–992 [PMC free article] [PubMed] [Google Scholar]
- Nickerson DA, Taylor SL, Weiss KM, Clark AG, Hutchinson RG, Stengard J, Salomaa V, Vartiainen E, Boerwinkle E, Sing CF (1998) DNA sequence diversity in a 9.7-kb region of the human lipoprotein lipase gene. Nat Genet 19:233–240 [DOI] [PubMed] [Google Scholar]
- Peltonen L (2000) Positional cloning of disease genes: advantages of genetic isolates. Hum Hered 50:66–75 [DOI] [PubMed] [Google Scholar]
- Peltonen L, Jalanko A, Varilo T (1999) Molecular genetics of the Finnish disease heritage. Hum Mol Genet 8:1913–1923 [DOI] [PubMed] [Google Scholar]
- Peltonen L, Palotie A, Lange K (2000) Use of population isolates for mapping complex traits. Nat Rev Genet 1:182–190 [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Przeworski M (2001) Linkage disequilibrium in humans: models and data. Am J Hum Genet 69:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond M, Rousset F (1994) GenePop. Institute des Sciences de L'Evolution, Montpellier, France [Google Scholar]
- Reich DE, Cargill M, Bolk S, Ireland J, Sabeti PC, Richter D, Lavery T, Kouyoumjian R, Farhadian SF, Ward R, Lander ES (2001) Linkage disequilibrium in the human genome. Nature 411:199–204 [DOI] [PubMed] [Google Scholar]
- Rieder MJ, Taylor SL, Clark AG, Nickerson DA (1999) Sequence variation in the human angiotensin converting enzyme. Nat Genet 22:59–62 [DOI] [PubMed] [Google Scholar]
- Rioux JD, Daly MJ, Silverberg MS, Lindblad K, Steinhart H, Cohen Z, Delmonte T, et al (2001) Genetic variation in the 5q31 cytokine gene cluster confers susceptibility to Crohn's disease. Nat Genet 29:223–228 [DOI] [PubMed] [Google Scholar]
- Ronaghi M, Karamohamed S, Pettersson B, Uhlén M, Nyren P (1996) Real-time DNA sequencing using detection of pyrophosphate release. Anal Biochem 242:84–89 [DOI] [PubMed] [Google Scholar]
- Ronaghi M, Uhlen M, Nyren P (1998) A sequencing method based on real-time pyrophosphate. Science 281:363–365 [DOI] [PubMed] [Google Scholar]
- Ruhlen MA (1991) Guide to the world's languages. Edward Arnold, Kent [Google Scholar]
- Sajantila A, Lahermo P, Anttinen T, Lukka M, Sistonen P, Savontaus ML, Aula P, Beckman L, Tranebjaerg L, Gedde-Dahl T, Issel-Tarver L, DiRienzo A, Pääbo S (1995) Genes and languages in Europe: an analysis of mitochondrial lineages. Genome Res 5:42–52 [DOI] [PubMed] [Google Scholar]
- Sajantila A, Pääbo S (1995) Language replacement in Scandinavia. Nat Genet 11:359–360 [DOI] [PubMed] [Google Scholar]
- Sajantila A, Salem AH, Savolainen P, Bauer K, Gierig C, Pääbo S (1996) Paternal and maternal DNA lineages reveal a bottleneck in the founding of the Finnish population. Proc Natl Acad Sci USA 93:12035–12039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M (1994) Linkage disequilibrium in growing and stable populations. Genetics 137:331–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68:978–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taillon-Miller P, Bauer-Sardina I, Saccone NL, Putzel J, Laitinen T, Cao A, Kere J, Pilia G, Rice JP, Kwok PY (2000) Juxtaposed regions of extensive and minimal linkage disequilibrium in human Xq25 and Xq28. Nat Genet 25:324–328 [DOI] [PubMed] [Google Scholar]
- Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton AR, Clark AG, Weiss KM, Nickerson DA, Boerwinkle E, Sing CF (2000) Recombinational and mutational hotspots within the human lipoprotein lipase gene. Am J Hum Genet 66:69–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger JD, Zöllner S, Laan M, Pääbo S (1998) Mapping genes through the use of linkage disequilibrium generated by genetic drift: “drift mapping” in small populations with no demographic expansion. Hum Hered 48:138–154 [DOI] [PubMed] [Google Scholar]
- Tishkoff SA, Dietzsch E, Speed W, Pakstis AJ, Kidd JR, Cheung K, Bonne-Tamir B, Santachiara-Benerecetti AS, Moral P, Krings M (1996) Global patterns of linkage disequilibrium at the CD4 locus and modern human origins. Science 271:1380–1387 [DOI] [PubMed] [Google Scholar]
- Tishkoff SA, Goldman A, Calafell F, Speed WC, Deinard AS, Bonne-Tamir B, Kidd JR, Pakstis AJ, Jenkins T, Kidd KK (1998) A global haplotype analysis of the myotonic dystrophy locus: implications for the evolution of modern humans and for the origin of myotonic dystrophy mutations. Am J Hum Genet 62:1389–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Pakstis AJ, Stoneking M, Kidd JR, Destro-Bisol G, Sanjantila A, Lu RB, Deinard AS, Sirugo G, Jenkins T, Kidd KK, Clark AG (2000) Short tandem-repeat polymorphism/Alu haplotype variation at the PLAT locus: implications for modern human origins. Am J Hum Genet 67:901–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varilo T, Laan M, Hovatta I, Wiebe V, Terwilliger JD, Peltonen L (2000) Linkage disequilibrium in isolated populations: Finland and a young sub-population of Kuusamo. Eur J Hum Genet 8:604–612 [DOI] [PubMed] [Google Scholar]
- Watson E, Bauer K, Aman R, Weiss G, von Haeseler A, Pääbo S (1996) mtDNA sequence diversity in Africa. Am J Hum Genet 59:980–983 [PMC free article] [PubMed] [Google Scholar]
- Weir BS (1996) Genetic data analysis. Sinauer Associates, Sunderland, MA [Google Scholar]
- Wright AF, Carothers AD, Pirastu M (1999) Population choice in mapping genes for complex diseases. Nat Genet 23:397–404 [DOI] [PubMed] [Google Scholar]
- Wright S (1921) Systems of mating. Genetics 6:111–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavattari P, Deidda E, Whalen M, Lampis R, Mulargia A, Loddo M, Eaves I, Mastio G, Todd JA, Cucca F (2000) Major factors influencing linkage disequilibrium by analysis of different chromosome regions in distinct populations: demography, chromosome recombination frequency and selection. Hum Mol Genet 20:2947–2957 [DOI] [PubMed] [Google Scholar]
- Zöllner S, von Haeseler A (2000) A coalescent approach to study linkage disequilibrium between single-nucleotide polymorphisms. Am J Hum Genet 66:615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]